The cell nucleus contains distinct biomolecular condensates that form at specific genetic loci, organize chromosomes in 3D space, and regulate RNA processing. Among these, Cajal bodies (CBs) require key “scaffolding” proteins for their assembly, which is not fully understood. Here, we employ proximity biotinylation, mass spectrometry, and functional screening to comprehensively identify and test the functions of CB components. We document 144 protein interactors of coilin, of which 70 were newly detected, and establish 25 players needed for CB assembly and/or maintenance. Surprisingly, the depletion of nine coilin interactors—mostly constituents of the 60S ribosome (RPLs)—increased CB number and caused subdomains defined by coilin and the survival motor neuron protein (SMN) to merge. These phenotypes were traceable to altered nuclear levels of dimethylarginine. Our data implicate RPL24 and other players in the regulation of CBs by modulating posttranslational modifications. Moreover, the prevalence of transcription factors among the identified components highlights roles for gene activity in CB assembly and nuclear positioning.
Introduction
The cell nucleus is compartmentalized by distinct nuclear bodies, which are biomolecular condensates (BMCs) that concentrate specific proteins and nucleic acids. The Cajal body (CB) is an RNA-rich nuclear body discovered in vertebrate neurons in 1903 by Ramon y Cajal and present in most metabolically active metazoan cells (Gall, 2000). CBs concentrate regulatory non-coding RNAs—namely, small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs)—and promote maturation of their corresponding ribonucleoprotein complexes (snRNPs and snoRNPs), including RNA modifications of snRNAs and even some tRNAs (Bizarro et al., 2021; Darzacq et al., 2002; Jády et al., 2003; Machyna et al., 2014; Matera and Ward, 1993; Stanĕk and Neugebauer, 2004; Vitali and Kiss, 2019). CBs form at the sites of transcription and/or processing of snRNAs, snoRNAs, and histone mRNAs (Sawyer et al., 2017), and their presence has been implicated in the productive transcription of snRNAs (Wang et al., 2016). In addition, CBs appear to be responsible for the clustering of discrete regions of chromosomes; this is due to the association of CBs with transcriptionally active snRNA and histone genes scattered throughout the genome on different chromosomes (Frey et al., 1999; Gao et al., 1997; Wang et al., 2016). Morphological alterations in CBs in the context of several human diseases highlight the significance of these cellular roles (Sawyer et al., 2017). To fully understand the biological functions of CBs, a comprehensive understanding of their composition, structure, and assembly dynamics is necessary.
The recent discovery that CBs harbor prominent substructure, with different molecular components localizing to two distinct compartments (Courchaine et al., 2021; Novotny et al., 2015), highlights the need to understand how CB composition relates to substructure. It is well known that CB composition overlaps with several nuclear BMCs: nucleoli, histone locus bodies (HLBs), and gems (Machyna et al., 2013). It was previously thought that CBs containing coilin, Nopp140, snRNPs, and scaRNAs could be distinct from or completely overlapping with gems, depending on coilin methylation related to the level of arginine methyltransferases expressed in the cell type or line (Hebert et al., 2002). The recent work using super-resolution or STED microscopy in tissue culture cells has shown that the coilin-containing compartment wraps around the gem, without the contents mixing (Courchaine et al., 2021). This indicates that HeLa cell CBs defined by coilin exclude the well-defined contents of gems, including Survival Motor Neuron protein (SMN), U1-70K protein, and gemins (Šimčíková et al., 2023; Stejskalová and Staněk, 2014). Gems and CBs can be forced to intermix when asymmetric dimethylarginine, one class of ligands for SMN, is inhibited (Courchaine et al., 2021). Under stress conditions, CB components disperse and can instead accumulate at the nuclear periphery (Barentine et al., 2023). Thus, these findings reveal substructure and dynamics that are important for function related to human disease that we currently do not fully understand.
Like most BMCs, the discovery of CB components (e.g., snRNP proteins, SMN, Nopp140) has been based primarily on colocalization of individual new players with previously identified CB proteins, like coilin, often in different types of cells (Machyna et al., 2013). CBs are presumed to be BMCs because their known constituents exchange on a timescale of seconds to minutes between the CB and nucleoplasm as well as within amphibian oocyte CBs (Deryusheva and Gall, 2004; Dundr et al., 2004). Accordingly, biochemical purification of CBs has not been possible to date because their integrity is compromised by the mildest detergent conditions (Lam et al., 2002; Stanĕk et al., 2003). Our lab previously immunopurified human coilin and conducted an interactome study by mass spectrometry, but only a few coilin interactors were identified (Machyna et al., 2013, 2015). This may be due to the low affinity of some CB components for coilin and/or to the association of coilin and its interactors with chromatin, making their extraction from cells more difficult. Although coilin has provided a molecular handle for marking and perturbing CBs, identifying CB components by colocalization depends on having good antibodies for a protein of interest, as well as a comprehensive list of candidates. A previous study identified four novel CB proteins de novo by performing a global-scale, microscopy-based screen of HA-Flag tagged proteins and analyzing their localization in nuclear bodies (Fong et al., 2013). This screen relied on tagging the candidates and excluded the identification of proteins that did not brightly or exclusively colocalize with coilin. Because CB components often overlap with other BMCs (nucleoli, HLBs, and gems) and can sometimes be less concentrated in CBs compared to other structures, detection by fluorescence imaging may not be entirely comprehensive.
Here, we use proximity biotinylation followed by mass spectrometry to comprehensively identify coilin interactors, which we consider to be candidate CB components, some of which may be transiently associated. In addition, proximity biotinylation has the potential to identify nucleoplasmic interactors, as suggested in recent reports of coilin nucleoplasmic activities from a variety of cell types (Bártová et al., 2014; Kanno et al., 2016; McLaurin et al., 2023). APEX2 is an engineered peroxidase that uses hydrogen peroxide to catalyze the formation of biotin-phenoxyl radicals from biotin-phenol (Lam et al., 2015; Qin et al., 2021). These biotin-phenoxyl radicals then “tag” nearby endogenous proteins at electron-rich amino acids, covalently biotinylating proteins regardless of their binding affinity for coilin (see Fig. 1 A). APEX2 biotinylation occurs within 1 min of peroxide addition and within 20 nm of the APEX2 fusion protein. Due to its rapid reaction, we reasoned that APEX2 would allow for comprehensive identification of transient interactors in highly dynamic CBs in situ given the ∼1-min residence time of coilin and other CB components in the CB where coilin and snRNPs are ∼20-fold enriched (Dundr et al., 2004; Klingauf et al., 2006). As a secondary screen for the functionality of identified components, each of the 144 known and newly identified CB proteins was depleted individually in an unbiased siRNA screen and quantitatively analyzed for changes in CB number and coilin localization. This work refines our understanding of the roles of known CB components and establishes those of newly detected by parameterizing our image data for sensitive, side-by-side comparison.
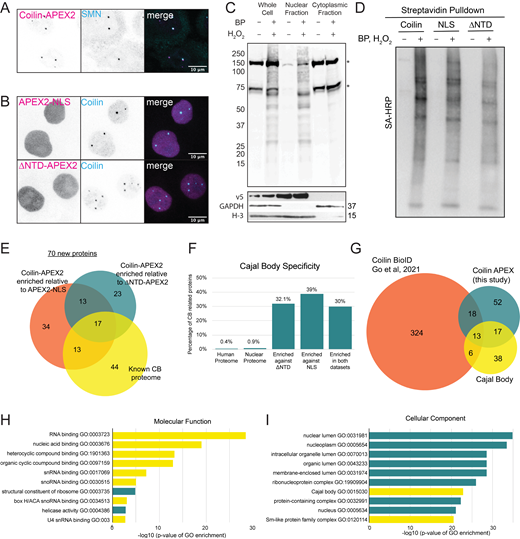
Enrichment of CB proteins using proximity biotinylation identifies 70 new CB proteins. (A) Schematic of APEX2 biotinylation and experimental pipeline for APEX2 MS experiments used in this study. (B) Immunofluorescent staining of coilin-APEX2 HeLa cell line with construct labeled by α-V5 antibody (magenta) and α-coilin, α-Sm (Y12), or α-Nopp140 antibody (cyan). Scale bars = 10 µm. (C) Immunofluorescent staining of coilin-APEX2, ΔNTD-APEX2, and APEX2-NLS after the addition of biotin phenol (BP) (30 min) and hydrogen peroxide (1 min), cells were stained with α-V5 antibody (magenta) and fluorescent streptavidin to label for biotin (cyan). Scale bars = 10 µm. (D) Normalized biotin intensity measured in CBs, nucleolus and nucleoplasm of Coilin-APEX2 cell line. The biotin intensities are normalized to mean signal in the nucleoplasm. Median of normalized biotin intensity is shown by the cyan line. P values obtained by unpaired t test: ****, P < 0.0001; ns, not significant. (E and F) Label-free quantification (LFQ) of CB proteome identification using coilin-APEX2 relative to APEX2-NLS (E) or relative to ΔNTD-APEX2 (F). Number of replicates: coilin-APEX2 = 4; APEX2-NLS = 4; ΔNTD-APEX2 = 3. Known CB proteins are labeled and colored orange, new significant hits are colored cyan.
Enrichment of CB proteins using proximity biotinylation identifies 70 new CB proteins. (A) Schematic of APEX2 biotinylation and experimental pipeline for APEX2 MS experiments used in this study. (B) Immunofluorescent staining of coilin-APEX2 HeLa cell line with construct labeled by α-V5 antibody (magenta) and α-coilin, α-Sm (Y12), or α-Nopp140 antibody (cyan). Scale bars = 10 µm. (C) Immunofluorescent staining of coilin-APEX2, ΔNTD-APEX2, and APEX2-NLS after the addition of biotin phenol (BP) (30 min) and hydrogen peroxide (1 min), cells were stained with α-V5 antibody (magenta) and fluorescent streptavidin to label for biotin (cyan). Scale bars = 10 µm. (D) Normalized biotin intensity measured in CBs, nucleolus and nucleoplasm of Coilin-APEX2 cell line. The biotin intensities are normalized to mean signal in the nucleoplasm. Median of normalized biotin intensity is shown by the cyan line. P values obtained by unpaired t test: ****, P < 0.0001; ns, not significant. (E and F) Label-free quantification (LFQ) of CB proteome identification using coilin-APEX2 relative to APEX2-NLS (E) or relative to ΔNTD-APEX2 (F). Number of replicates: coilin-APEX2 = 4; APEX2-NLS = 4; ΔNTD-APEX2 = 3. Known CB proteins are labeled and colored orange, new significant hits are colored cyan.
Results
Enrichment of Cajal body-associated proteins using APEX2
To determine the CB proteome, we created a stable HeLa cell line expressing moderate levels of an APEX2-tagged coilin protein (coilin-APEX2). As described above, APEX2 is a peroxidase that uses biotin phenol and hydrogen peroxide to covalently biotinylate nearby proteins in situ within 1 min (Fig. 1 A). HeLa cells were chosen because they contain the previously described substructure that we are interested in functionally understanding (Courchaine et al., 2021; Novotny et al., 2015). To validate the behavior of this fusion protein, we tested the localization of coilin-APEX2, making use of an included V5 epitope tag. As expected, the coilin-APEX2 fusion protein was concentrated in endogenous CBs containing snRNPs, SMN, and Nopp140 (Fig. 1 B and Fig. S1 A). We created two control cell lines with APEX2-tagged constructs to aid in downstream analysis. The first, an APEX2-NLS construct, enables us to determine the proteins enriched in CBs relative to the nuclear background. The second control cell line expresses a coilin lacking the N-terminal domain (NTD), which is required for CB localization; this ΔNTD-APEX2 protein distinguishes CB proteins from other proteins that may interact with nucleoplasmic coilin. Both APEX2-NLS and ΔNTD-APEX2 fusion proteins localized to the nucleoplasm and did not form or associate with nuclear bodies, as expected (Fig. S1 B). Importantly, we validated the biotinylation of CB components by using biotin phenol and hydrogen peroxide on all APEX2-expressing cell lines; the fixed cells were then stained with streptavidin to characterize the sites of biotinylation. Coilin-APEX2 cells showed robustly biotinylated CBs, while APEX2-NLS and ΔNTD-APEX2 biotinylated proteins throughout the entire nucleus without enrichment in any nuclear body (Fig. 1 C). To independently assess the localization of tagged coilin in CBs, nucleoplasm, and nucleoli, biotin intensities were normalized to the mean signal in the nucleoplasm and showed to be 20-fold enriched in CBs with no significant enrichment in nucleoli (Fig. 1 D).
Enrichment of CB proteins using proximity biotinylation identifies seventy new CB proteins. (A) Immunofluorescent staining of coilin-AEPX2 labeled by α-V5 antibody (magenta) and α-SMN antibody (cyan). (B) Immunofluorescent staining of APEX2-NLS and ΔNTD-APEX2 labeled by α-V5 antibody (magenta) and α-coilin (cyan). (C) Streptavidin Blot of fractionated coilin-APEX2 cells ± BP and H2O2 showing endogenously biotinylated proteins (*) and biotinylated proteins after treatment (top). Primary antibodies showing localization of proteins in fractions; α-V5 antibody is a marker for coilin-APEX2, α-GAPDH antibody is a marker for cytoplasm, α-H3 antibody is a marker for nucleus. (D) Streptavidin Blot of biotinylated proteins pulled down from nuclear lysates ± BP and H2O2 treatment. (E) Venn diagram of statistically enriched CB proteins relative to ΔNTD-APEX2 (teal) and APEX2-NLS (orange) compared to known CB proteome (yellow). (F) CB proteins that are enriched in the 100 hits. The percent of proteins in the human and nuclear proteome and of the enriched hits that localize to CBs. (G) Venn diagram of statistically enriched CB proteins against both controls (teal) compared to coilin-BioID (orange) (Go et al., 2021) and known CB proteome (yellow). (H and I) GO-term analysis of proteins enriched in coilin-APEX2. The 10 most significant hits along with the −log10 of their P values are displayed, CB-associated GO terms are highlighted in yellow. Source data are available for this figure: SourceData FS1.
Enrichment of CB proteins using proximity biotinylation identifies seventy new CB proteins. (A) Immunofluorescent staining of coilin-AEPX2 labeled by α-V5 antibody (magenta) and α-SMN antibody (cyan). (B) Immunofluorescent staining of APEX2-NLS and ΔNTD-APEX2 labeled by α-V5 antibody (magenta) and α-coilin (cyan). (C) Streptavidin Blot of fractionated coilin-APEX2 cells ± BP and H2O2 showing endogenously biotinylated proteins (*) and biotinylated proteins after treatment (top). Primary antibodies showing localization of proteins in fractions; α-V5 antibody is a marker for coilin-APEX2, α-GAPDH antibody is a marker for cytoplasm, α-H3 antibody is a marker for nucleus. (D) Streptavidin Blot of biotinylated proteins pulled down from nuclear lysates ± BP and H2O2 treatment. (E) Venn diagram of statistically enriched CB proteins relative to ΔNTD-APEX2 (teal) and APEX2-NLS (orange) compared to known CB proteome (yellow). (F) CB proteins that are enriched in the 100 hits. The percent of proteins in the human and nuclear proteome and of the enriched hits that localize to CBs. (G) Venn diagram of statistically enriched CB proteins against both controls (teal) compared to coilin-BioID (orange) (Go et al., 2021) and known CB proteome (yellow). (H and I) GO-term analysis of proteins enriched in coilin-APEX2. The 10 most significant hits along with the −log10 of their P values are displayed, CB-associated GO terms are highlighted in yellow. Source data are available for this figure: SourceData FS1.
All three cell lines showed faint levels of biotinylation in the cytoplasm due to endogenously biotinylated proteins, as well as endogenous peroxidases that increase background signal regardless of whether cells have been exposed to biotin phenol and hydrogen peroxide (Fig. S1 C). To exclude this background signal from our analysis, biotinylated cells were fractionated into nuclear and cytoplasmic compartments and only the nuclear fractions were analyzed. Successful depletion of biotinylated cytoplasmic proteins was confirmed by blotting with streptavidin (Fig. S1 C). Robust biotinylation of other proteins was only seen in the nuclear fraction, corresponding to the nuclear localization of the APEX2-tagged proteins. After biotinylation and fractionation of cells, we enriched biotinylated proteins from nuclear fractions using streptavidin magnetic beads and confirmed successful enrichment (Fig. S1 D).
Identification of Cajal body–associated proteins by mass spectrometry
Enriched proteins from nuclear fractions were identified using liquid chromatography followed by tandem mass spectrometry (LC MS-MS). In total, 100 unique proteins were significantly enriched in Coilin-APEX2 compared with ΔNTD-APEX2 (53 hits) and APEX2-NLS (77 hits) conditions (Fig. 1, E and F; and Tables 1 and S1). Among the proteins identified, 30 of those enriched were shared between the two datasets (Fig. S1 E). As expected, there were fewer proteins enriched when compared with the ΔNTD-APEX2 control since the coilin C-terminus alone has the potential to interact with CB proteins in the nucleoplasm. Of the 100 total enriched proteins, 30 were previously identified CB proteins (Fig. S1, E and F; and Table 1). One published study using BioID identified 342 coilin-interacting proteins when coilin was highly overexpressed (Go et al., 2021); 31 of these proteins were also enriched in our Coilin-APEX2 dataset (Fig. S1 G). Gene ontology (GO) analysis of the 100 proteins enriched in our dataset revealed a strong association with functions and complexes known to be in CBs such as snRNA binding, snoRNA binding, and Sm-like protein family complex (Fig. S1, H and I). Interestingly, the gem protein SMN was not identified in our current analysis, suggesting at least partial specificity for the CB itself and/or that some known coilin interactors may not label well.
List of 100 CB Proteins identified by Coilin-APEX2 enrichment
| Function . | Proteins enriched in Coilin-APEX2 . |
|---|---|
| Cell cycle progression | ANLN, ANXA11, BUB3, KIF4A, NCAPD3, NUMA1, ZNF207 |
| DNA replication and repair | ATAD2, POLD1, RFC5, RECQL, RIF1, USP7 |
| Transcription | CDC73, ERCC3, FUBP1, GTF2F2, GTF3C5, HTATSF1, IRF2BP1, MED1, MED14, MED17, POLR2C, SMARCA4, SMARCC1, SSRP1, SUPT5H, SUPT6H, UBR5, ELL, ICE1, ICE2, POLR2A, POLR2B |
| Pre-mRNA splicing and cleavage | CLP1, CPSF6, DHX38, IK, KHSRP, NUDT21, PPAN, RBM10, SON, SRRM2, EFTUD2, NHP2L1, PRPF3, PRPF4, SART1, SF3A1, SF3B2, SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, TARDBP |
| snRNA maturation and snRNP assembly | COIL, DDX20, SART3, TGS1, TOE1 |
| rRNA processing and ribosome biogenesis | GAR1, GNL3L, GTPBP4, NSA2, TEX10, DKC1, NOLC1, NOP56, NOP58 |
| Histone mRNA processing | LSM11, NPAT |
| miRNA processing | SRRT |
| RNA helicase | DDX18 |
| RNA exosome | EXOSC2 |
| Ribosomal protein | RPL13, RPL14, RPL15, RPL24, RPL3, RPL34, RPL7A, RPL8, RPS2, RPS24 |
| Other | AP2A2, ATP6V1A, CSNK2A1, CSNK2B, CYB5R1, HIST1H2AJ, NUP54, OXA1L, PPIL4, SFXN3, SLC25A13, SPCS3, PSME3 |
| Function . | Proteins enriched in Coilin-APEX2 . |
|---|---|
| Cell cycle progression | ANLN, ANXA11, BUB3, KIF4A, NCAPD3, NUMA1, ZNF207 |
| DNA replication and repair | ATAD2, POLD1, RFC5, RECQL, RIF1, USP7 |
| Transcription | CDC73, ERCC3, FUBP1, GTF2F2, GTF3C5, HTATSF1, IRF2BP1, MED1, MED14, MED17, POLR2C, SMARCA4, SMARCC1, SSRP1, SUPT5H, SUPT6H, UBR5, ELL, ICE1, ICE2, POLR2A, POLR2B |
| Pre-mRNA splicing and cleavage | CLP1, CPSF6, DHX38, IK, KHSRP, NUDT21, PPAN, RBM10, SON, SRRM2, EFTUD2, NHP2L1, PRPF3, PRPF4, SART1, SF3A1, SF3B2, SNRPB, SNRPD1, SNRPD2, SNRPD3, SNRPE, TARDBP |
| snRNA maturation and snRNP assembly | COIL, DDX20, SART3, TGS1, TOE1 |
| rRNA processing and ribosome biogenesis | GAR1, GNL3L, GTPBP4, NSA2, TEX10, DKC1, NOLC1, NOP56, NOP58 |
| Histone mRNA processing | LSM11, NPAT |
| miRNA processing | SRRT |
| RNA helicase | DDX18 |
| RNA exosome | EXOSC2 |
| Ribosomal protein | RPL13, RPL14, RPL15, RPL24, RPL3, RPL34, RPL7A, RPL8, RPS2, RPS24 |
| Other | AP2A2, ATP6V1A, CSNK2A1, CSNK2B, CYB5R1, HIST1H2AJ, NUP54, OXA1L, PPIL4, SFXN3, SLC25A13, SPCS3, PSME3 |
Statistically enriched proteins identified in the Coilin-APEX2 dataset relative to ΔNTD-APEX2 and relative to APEX2-NLS. Proteins are identified by their Gene ID, and Gene IDs in bold represent previously known CB components.
Although some coilin interactors may be CB components or regulators without concentrating on CBs, one expectation for potential new CB components is that immunofluorescence analysis might reveal colocalization. Thus, we sought to test new “hits” enriched in coilin-APEX2 by immunostaining fixed cells with antibodies specific to the candidate and coilin. One limitation is the lack of suitable commercially available antibodies for many of our hits. Nevertheless, highly enriched significant hits, such as TOE1 and newly detected IRF2BP1 (interferon regulatory factor binding protein 1), were strongly colocalized with coilin in CBs by immunofluorescence (Fig. 2, A and B). Other less highly enriched hits, such as EFTUD2 and the new hit SRRT, showed diffuse nuclear staining but were not excluded from CBs. To better determine the localization of our hits in CBs, we created an image analysis tool to analyze the degree to which the target protein signal overlapped with the coilin signal. Briefly, CBs within the nucleus were automatically selected from the coilin channel. Sub-images were then generated by cropping symmetrically around the CB center and averaged. Colocalization was measured by calculating the Pearson’s correlation coefficient (PCC) between the averaged images from different channels. As a validation of our algorithm, cells transiently transfected with coilin-EGFP were immunostained with antibodies specific for the trimethylguanosine (TMG) cap on snRNAs (Fig. S2 A). As expected, snRNAs colocalized with coilin in CBs and had a PCC value of 0.92. Applying this method to our hits, IRF2BP1 was also significantly enriched in CBs (Fig. 2 C). In contrast, EFTUD2 and SRRT were less concentrated in CBs owing to their presence at higher levels in the nucleoplasm. Thus, among these four hits, CB localization correlated with enrichment levels detected by mass spectrometry (Fig. 2, A–C).
IRF2BP1 and SRRT, identified using Coilin-APEX2, are enriched in CBs. (A) Volcano plot showing the location of TOE1 (known component) and IRF2BP1 (new component) in teal and EFTUD2 (known component) and SRRT (new component) in orange. (B) Immunofluorescent staining of previously known and new CB components using α-coilin (magenta) and antibodies specific to either high enrichment, high significance components (TOE1 and IRF2BP1, top) or low enrichment, low significance components (EFTUD and SRRT, bottom) (cyan). Scale bars = 10 µm. (C) Colocalization analysis of coilin with IRF2BP1, EFTUD2, and SRRT. CBs were identified from the coilin channel and cropped symmetrically around their center of mass. Sub-images at the same position in the other channel were also cropped. Averaged images were generated for each channel, and Pearson’s correlation coefficient (PCC) was calculated from the averaged images (see Materials and methods). Scale bars = 0.5 µm. (D) Immunofluorescent staining of SMN or coilin (magenta) and IRF2BP1 (cyan), showing IRF2BP1 is not in gems. Representative single nucleus with accompanying line profile plots showing intensities through a single CB. (E) Immunofluorescent staining of coilin (magenta), IRF2BP1 (cyan) or snRNPs (cyan) after siRNA knockdown of coilin or IRF2BP1, as indicated. Scale bars = 10 µm.
IRF2BP1 and SRRT, identified using Coilin-APEX2, are enriched in CBs. (A) Volcano plot showing the location of TOE1 (known component) and IRF2BP1 (new component) in teal and EFTUD2 (known component) and SRRT (new component) in orange. (B) Immunofluorescent staining of previously known and new CB components using α-coilin (magenta) and antibodies specific to either high enrichment, high significance components (TOE1 and IRF2BP1, top) or low enrichment, low significance components (EFTUD and SRRT, bottom) (cyan). Scale bars = 10 µm. (C) Colocalization analysis of coilin with IRF2BP1, EFTUD2, and SRRT. CBs were identified from the coilin channel and cropped symmetrically around their center of mass. Sub-images at the same position in the other channel were also cropped. Averaged images were generated for each channel, and Pearson’s correlation coefficient (PCC) was calculated from the averaged images (see Materials and methods). Scale bars = 0.5 µm. (D) Immunofluorescent staining of SMN or coilin (magenta) and IRF2BP1 (cyan), showing IRF2BP1 is not in gems. Representative single nucleus with accompanying line profile plots showing intensities through a single CB. (E) Immunofluorescent staining of coilin (magenta), IRF2BP1 (cyan) or snRNPs (cyan) after siRNA knockdown of coilin or IRF2BP1, as indicated. Scale bars = 10 µm.
Details of the high throughput functional screen. (A) Validation of the colocalization analysis. MEF coilin knockout cell lines transiently expressing fluorescently tagged coilin (magenta) were immunostained with TMG (tri-methyguanosine cap) antibody (cyan). Colocalization analysis result is shown on the right. The total number of CBs analyzed (n) and the Pearson’s correlation coefficient (PCC) are listed. (B) Frequency histogram of Pearson’s correlation coefficient between nucleophosmin and coilin fluorescent signal per siRNA in screen. Values of siRNAs that relocalize coilin to nucleoli are highlighted in purple (Pearson’s > 0.75). (C) Representative images of hits showing relocalization of coilin to nucleoli (siMED14 and siUSPL1). Immunofluorescent images show α-coilin antibody in magenta and α-nucleophosmin antibody in cyan. Histograms for each hit quantifying the number of CBs per nucleus from their respective well and plate in the screen (12 fields of view). (D) Analysis of cell ploidy and CB integrity after knockdown of proteins that increase CB number. Representative histograms of Hoechst log2 integrated intensity in samples treated with siNT or siRNAs targeting hits that increase CB numbers upon depletion normalized to siNT control. G0/G1 phase (0.75–1.25) highlighted in orange, S phase (1.25–1.75) in white/black font, G2/M phase (1.75–2.25) highlighted in purple, >4N (>2.5) highlighted in teal. On the right are cell cycle values showing % of total in each phase, quantified from values from the left.
Details of the high throughput functional screen. (A) Validation of the colocalization analysis. MEF coilin knockout cell lines transiently expressing fluorescently tagged coilin (magenta) were immunostained with TMG (tri-methyguanosine cap) antibody (cyan). Colocalization analysis result is shown on the right. The total number of CBs analyzed (n) and the Pearson’s correlation coefficient (PCC) are listed. (B) Frequency histogram of Pearson’s correlation coefficient between nucleophosmin and coilin fluorescent signal per siRNA in screen. Values of siRNAs that relocalize coilin to nucleoli are highlighted in purple (Pearson’s > 0.75). (C) Representative images of hits showing relocalization of coilin to nucleoli (siMED14 and siUSPL1). Immunofluorescent images show α-coilin antibody in magenta and α-nucleophosmin antibody in cyan. Histograms for each hit quantifying the number of CBs per nucleus from their respective well and plate in the screen (12 fields of view). (D) Analysis of cell ploidy and CB integrity after knockdown of proteins that increase CB number. Representative histograms of Hoechst log2 integrated intensity in samples treated with siNT or siRNAs targeting hits that increase CB numbers upon depletion normalized to siNT control. G0/G1 phase (0.75–1.25) highlighted in orange, S phase (1.25–1.75) in white/black font, G2/M phase (1.75–2.25) highlighted in purple, >4N (>2.5) highlighted in teal. On the right are cell cycle values showing % of total in each phase, quantified from values from the left.
Although IRF2BP1 showed strong colocalization with coilin, it did not colocalize with SMN-containing gems (Fig. 2 D), as emphasized by line graphs drawn through CBs in cells stained for IRF2BP1 and coilin versus IRF2BP1 and SMN. This highlights the specific spatial organization of the coilin-containing CBs relative to SMN-containing gems, which are not merged in these Hela cells (Courchaine et al., 2021). Knockdown of coilin leads to diffuse IRF2BP1 signal, demonstrating that the presence of IRF2BP1 in CBs is dependent on coilin expression (Fig. 2 E). Conversely, the knockdown of IRF2BP1 does not affect CB number or recruitment of snRNPs to CBs, indicating that IRF2BP1 is not required for CB assembly.
An siRNA screen of CB-associated proteins reveals regulators of Cajal body number
To analyze the importance of each CB protein in the assembly and maintenance of CBs, we established an unbiased, imaging-based siRNA screen. We compiled a list of 144 total CB proteins—including the 70 new proteins identified by mass spectrometry—and generated a CB-specific siRNA library. We then transfected HeLa cells to deplete individual CB proteins and assayed for changes in coilin localization (Fig. 3 A). Coilin has previously been shown to interact with nucleoli under stress and in disease states (Trinkle-Mulcahy and Sleeman, 2017). Anticipating this relationship between CBs and nucleoli, we counterstained for nucleoli with anti-nucleophosmin and calculated the intensity of coilin staining in the nucleolus. Coilin relocalization to nucleoli was measured by determining the PCC between the anti-nucleophosmin and anti-coilin intensities. Thereby, we identified 11 siRNAs that caused coilin to relocalize to nucleoli, precluding quantification of nucleoplasmic CBs (Fig. S2, B and C; and Table 2).
siRNA screen of CB components identifies 34 proteins that change CB number in HeLa cells. (A) Schematic of microscopy-based CB-specific siRNA screen. (B) Frequency histogram of percent effect per siRNA in screen. Values of siRNAs that decreased the number of CBs per nucleus are highlighted in yellow (PE > 60). Values of siRNAs that increased the number of CBs per nucleus are highlighted in orange (PE < −60). (C and D) Representative images of wells from siRNA screen. Immunofluorescent staining shows coilin (magenta) and nucleophosmin (cyan). Histograms for each siRNA condition quantifying the number of CBs per nucleus from their respective well and plate in the screen (12 fields of view/well). (C) Representative image of control conditions (siNT and siCoilin). (D) Representative image of hits showing a decrease in CBs per nucleus (siSPT6 and siTRIM22). Representative hit shown in teal with siNT (non-targeting siRNA) in light gray. Scale bars = 10 µm.
siRNA screen of CB components identifies 34 proteins that change CB number in HeLa cells. (A) Schematic of microscopy-based CB-specific siRNA screen. (B) Frequency histogram of percent effect per siRNA in screen. Values of siRNAs that decreased the number of CBs per nucleus are highlighted in yellow (PE > 60). Values of siRNAs that increased the number of CBs per nucleus are highlighted in orange (PE < −60). (C and D) Representative images of wells from siRNA screen. Immunofluorescent staining shows coilin (magenta) and nucleophosmin (cyan). Histograms for each siRNA condition quantifying the number of CBs per nucleus from their respective well and plate in the screen (12 fields of view/well). (C) Representative image of control conditions (siNT and siCoilin). (D) Representative image of hits showing a decrease in CBs per nucleus (siSPT6 and siTRIM22). Representative hit shown in teal with siNT (non-targeting siRNA) in light gray. Scale bars = 10 µm.
List of proteins altering CB number and coilin localization
| Change in CB number . | Change in coilin localization . | ||||
|---|---|---|---|---|---|
| Decrease in CBs . | Percent effect . | Increase in CBs . | Percent effect . | Nucleolar coilin . | Pearson’s R . |
| NPAT | 120 | ANLN | −179 | MED14 | 0.87 |
| SLBP | 115 | RPL14 | −95 | SNRPG | 0.84 |
| CASP8AP2 | 104 | RPL24 | −94 | POLR2B | 0.84 |
| POLR2A | 101 | NUMA1 | −90 | POLR2A | 0.84 |
| COIL | 100 | RPL13 | −80 | POLR2C | 0.83 |
| SNRPD3 | 99 | RPL8 | −77 | ICE1 | 0.83 |
| SNU13 | 98 | NSA2 | −76 | USPL1 | 0.82 |
| SNRPB | 94 | EFTUD2 | −75 | CYB5R1 | 0.81 |
| SMN1 | 94 | RPL7A | −75 | UBR5 | 0.78 |
| IK | 93 | MED17 | 0.77 | ||
| TRIM22 | 91 | SNRPD1 | 0.76 | ||
| SNRPD2 | 90 | ||||
| NOLC1 | 90 | ||||
| SUPT6H | 86 | ||||
| NOP58 | 83 | ||||
| CDC73 | 77 | ||||
| CSNK2B | 77 | ||||
| SNRPF | 74 | ||||
| PRPF4 | 68 | ||||
| TGS1 | 66 | ||||
| EAF1 | 66 | ||||
| SNRPG | 65 | ||||
| SON | 65 | ||||
| Change in CB number . | Change in coilin localization . | ||||
|---|---|---|---|---|---|
| Decrease in CBs . | Percent effect . | Increase in CBs . | Percent effect . | Nucleolar coilin . | Pearson’s R . |
| NPAT | 120 | ANLN | −179 | MED14 | 0.87 |
| SLBP | 115 | RPL14 | −95 | SNRPG | 0.84 |
| CASP8AP2 | 104 | RPL24 | −94 | POLR2B | 0.84 |
| POLR2A | 101 | NUMA1 | −90 | POLR2A | 0.84 |
| COIL | 100 | RPL13 | −80 | POLR2C | 0.83 |
| SNRPD3 | 99 | RPL8 | −77 | ICE1 | 0.83 |
| SNU13 | 98 | NSA2 | −76 | USPL1 | 0.82 |
| SNRPB | 94 | EFTUD2 | −75 | CYB5R1 | 0.81 |
| SMN1 | 94 | RPL7A | −75 | UBR5 | 0.78 |
| IK | 93 | MED17 | 0.77 | ||
| TRIM22 | 91 | SNRPD1 | 0.76 | ||
| SNRPD2 | 90 | ||||
| NOLC1 | 90 | ||||
| SUPT6H | 86 | ||||
| NOP58 | 83 | ||||
| CDC73 | 77 | ||||
| CSNK2B | 77 | ||||
| SNRPF | 74 | ||||
| PRPF4 | 68 | ||||
| TGS1 | 66 | ||||
| EAF1 | 66 | ||||
| SNRPG | 65 | ||||
| SON | 65 | ||||
We initially addressed CB morphology by staining and quantifying the number of CBs per nucleus (Fig. 3 A). These data enabled a ranking of the component-specific siRNAs according to the relative change in CB number (Fig. 3 B). For each sample, the average number of CBs per nucleus was calculated. Non-targeting siRNA (siNT) and coilin siRNA (siCOIL) were used as negative and positive controls respectively for the inhibition of CB assembly (Fig. 3 C). The mean number of CBs per nucleus was 2.04 ± 0.25 after siNT transfection and 0.36 ± 0.15 after siCOIL. In total, 46 proteins (32% of CB proteins) affected CB formation detectably (Tables 2 and S2). To assess changes in the average number of CBs in the cell, we calculated the percent effect (PE) of CB knockdown by setting the average CB/nucleus of siNT to zero (0) PE and siCOIL to 100. Specifically, 25 of these hits decreased the number of CBs per nucleus (PE > 60), consistent with their role in the assembly and/or maintenance of CBs (Fig. 3 B). These hits included SMN, Nopp140 (NOLC1), and Sm proteins, as expected (Courchaine et al., 2022; Girard et al., 2006; Lemm et al., 2006; Renvoisé et al., 2009; Shpargel et al., 2009; Strzelecka et al., 2010). The strongest decreases in CB number were observed upon knockdown of coilin itself, POLR2A in line with the transcription-dependence of CBs (Carmo-Fonseca et al., 1992), and two histone locus body proteins (SLBP and NPAT), reflecting the overlap of HLBs with CBs in HeLa cells (Machyna et al., 2013, 2014). Of the newly identified CB proteins, SPT6 and TRIM22 had the strongest effect in reducing CB number (Fig. 3 D). Surprisingly, we detected 10 hits that showed a negative percent effect (PE < −60), indicating an increase in CB number (Fig. 3 B) and suggesting a possible regulatory role. Taken together, 35 siRNAs in our screen regulate the number of CBs per nucleus (Fig. 3 B; and Tables 2 and S2).
Evidence that six 60S ribosomal proteins regulate CB number and structure
Focusing on the depletions that increased CB numbers, all of them new hits from the APEX2 dataset except EFTUD2, (Fig. 4 A and Table 2), we were surprised that this group included five ribosomal proteins found in the large 60S subunit and one protein involved in biogenesis of the 60S subunit (Fig. 4 B). Interestingly, all of the RPL subunits that increased CB numbers are either present on the external surface of the human 60S ribosome or alternatively on the internal surface, facing the small subunit during translation (Fig. 4 C). This indicates that these RPLs could in principle interact with coilin in the nucleus in the context of pre-60S subunits. Images showing the increases in CB number are shown in Fig. 4 D. These effects were specific for the 60S subunit because the two RPS protein components (RPS2 & RPS24; see Table 1) had no effect on CB number when depleted (Table S2). Moreover, depletion of the RPL15 subunit, which does not localize to the 60S surface but was also biotinylated by coilin-APEX2, did not lead to this phenotype (Table S2).
siRNA screen of CB components identifies large ribosomal subunit proteins that increase CB number. (A) Table of siRNA hits listed by Gene ID that increase CB number and the PE (percent effect) value. Large ribosomal subunit and ribosome biogenesis proteins are highlighted in teal. (B) Nucleolar and ribosomal proteins identified in LFQ comparing coilin-APEX2 relative to APEX2-NLS. Known CB proteins are labeled and colored orange, known dual CB and nucleolar proteins are colored cyan, new significant nucleolar protein hits are colored purple, and new significant ribosomal protein hits are colored pink. (C) Structure of human 60S subunit in complex with Tetracenomycin X (PDB ID: 6Y6X). Selected ribosomal proteins are shown as ribbons, other ribosomal proteins and rRNAs are shown as a white surface. Proteins found to affect CB number (RPL13, 14, 24, 7A, and 8) are colored orange. The protein that did not show an effect (RPL15) on CB number upon knockdown is colored marine and mostly buried. Two additional surface ribosomal proteins (RPL6 and RPL17) not enriched in proximity biotinylation are colored cyan. (D) Representative images of the top three hits showing an increase in CBs per nucleus (siANLN, siRPL14, and siRPL24). Immunofluorescent staining shows α-coilin antibody in magenta and α-nucleophosmin antibody in cyan. Scale bars = 10 µm. Histograms for each hit quantifying the number of CBs per nucleus from their respective well and plate in the screen shown in teal with siNT control in light grey (12 fields of view/well). (E) DNA content analysis. Representative histograms of Hoechst log2 integrated intensity in siNT, siANLN, and siRPL14 normalized to siNT control. G0/G1 phase (0.75–1.25) highlighted and quantified in orange, S phase (1.25–1.75) in white/black font, G2/M phase (1.75–2.25) highlighted and quantified in purple, >4N (>2.5) highlighted and quantified in teal.
siRNA screen of CB components identifies large ribosomal subunit proteins that increase CB number. (A) Table of siRNA hits listed by Gene ID that increase CB number and the PE (percent effect) value. Large ribosomal subunit and ribosome biogenesis proteins are highlighted in teal. (B) Nucleolar and ribosomal proteins identified in LFQ comparing coilin-APEX2 relative to APEX2-NLS. Known CB proteins are labeled and colored orange, known dual CB and nucleolar proteins are colored cyan, new significant nucleolar protein hits are colored purple, and new significant ribosomal protein hits are colored pink. (C) Structure of human 60S subunit in complex with Tetracenomycin X (PDB ID: 6Y6X). Selected ribosomal proteins are shown as ribbons, other ribosomal proteins and rRNAs are shown as a white surface. Proteins found to affect CB number (RPL13, 14, 24, 7A, and 8) are colored orange. The protein that did not show an effect (RPL15) on CB number upon knockdown is colored marine and mostly buried. Two additional surface ribosomal proteins (RPL6 and RPL17) not enriched in proximity biotinylation are colored cyan. (D) Representative images of the top three hits showing an increase in CBs per nucleus (siANLN, siRPL14, and siRPL24). Immunofluorescent staining shows α-coilin antibody in magenta and α-nucleophosmin antibody in cyan. Scale bars = 10 µm. Histograms for each hit quantifying the number of CBs per nucleus from their respective well and plate in the screen shown in teal with siNT control in light grey (12 fields of view/well). (E) DNA content analysis. Representative histograms of Hoechst log2 integrated intensity in siNT, siANLN, and siRPL14 normalized to siNT control. G0/G1 phase (0.75–1.25) highlighted and quantified in orange, S phase (1.25–1.75) in white/black font, G2/M phase (1.75–2.25) highlighted and quantified in purple, >4N (>2.5) highlighted and quantified in teal.
Anillin (ANLN) and nuclear mitotic apparatus protein 1 (NUMA1) depletion cause increases in ploidy, which was previously shown to increase the nucleolar number (Farley-Barnes et al., 2018). We validated the increase in ploidy through ANLN depletion by measuring cellular DNA content with Hoechst dye in terms of fluorescence intensity per nucleus (Fig. 4 E). As expected, ANLN depletion led to an accumulation of cells with >4N genomes. Since CBs and nucleoli form on active genes, it is not surprising that increased ploidy would lead to increased numbers of both BMCs. Depletion of specific RPL proteins can lead to defects in cell cycle progression due to extraribosomal functions, for example, human RPL24 binds and inhibits cyclin-dependent kinases (Bhavsar et al., 2010; Warner and McIntosh, 2009). In contrast, NUMA1 increased the number of cells in G2 without inducing aneuploidy. However, none of the ribosomal protein depletions analyzed altered either cell ploidy or the percent of cells in different cell cycle stages (Fig. 4 E and Fig. S2 D).
To address the possibility that the RPL proteins are bound to coilin and/or localized to CBs, we attempted to image the RPL proteins using immunocytochemistry. However, commercially available antibodies specific for RPL proteins had poor signal-to-noise and were not suitable for immunolocalization (data not shown). Therefore, we created stable Hela cell lines expressing low levels of GFP-RPL13 and GFP-RPL14, using methods previously established for RPS2-YFP (Zemp et al., 2009), and coimmunostained with coilin (Fig. 5 A). We then applied our colocalization analysis approach to the stable HeLa cell lines expressing fluorescently tagged ribosomal proteins (see Fig. 2 C). However, we did not observe colocalization between coilin and the ribosomal proteins tested, and PCC values ranged from 0.19 to 0.5 (Fig. 5, A and B). Likewise, immunostaining for RPL24 did not reveal colocalization with coilin (Fig. S3 A). To test for potential direct or indirect interactions between coilin and ribosomal proteins, we performed immunoprecipitation experiments using the fluorescent protein “handles” on each of the tested ribosomal proteins to assess coilin binding (Fig. S3, B and C); interactions between coilin and these ribosomal proteins were not detectable.
Ribosomal subunit proteins do not localize to CBs. (A) Immunofluorescent images of stable HeLa cell lines expressing fluorescently tagged ribosomal proteins. Ribosomal protein expression is induced by 0.5 µg/ml doxycycline for 16 h before fixing the cell for immunostaining with coilin (magenta). GFP or YFP fluorescence is shown in cyan. RPL13 and RPL14 are hits found to increase CB number, RPS2 is enriched in proximity biotinylation but was found to not affect CB number. The nucleus outline is shown in blue (white background) or white (dark background) lines. Scale bars = 10 µm. (B) Colocalization analysis of coilin with the ribosomal proteins. The total number of CBs analyzed (n) and the Pearson’s correlation coefficient (PCC) are listed on the right. Scale bars = 0.5 µm.
Ribosomal subunit proteins do not localize to CBs. (A) Immunofluorescent images of stable HeLa cell lines expressing fluorescently tagged ribosomal proteins. Ribosomal protein expression is induced by 0.5 µg/ml doxycycline for 16 h before fixing the cell for immunostaining with coilin (magenta). GFP or YFP fluorescence is shown in cyan. RPL13 and RPL14 are hits found to increase CB number, RPS2 is enriched in proximity biotinylation but was found to not affect CB number. The nucleus outline is shown in blue (white background) or white (dark background) lines. Scale bars = 10 µm. (B) Colocalization analysis of coilin with the ribosomal proteins. The total number of CBs analyzed (n) and the Pearson’s correlation coefficient (PCC) are listed on the right. Scale bars = 0.5 µm.
Ribosomal subunit proteins do not localize to CBs or interact with coilin. (A) Left: Immunofluorescent staining of wildtype HeLa cell using RPL24 (cyan) and coilin (magenta) antibody. Scale bar = 10 μm. Right: colocalization analysis of RPL24 and coilin. Scale bars = 1 μm. (B) Coimmunoprecipitation using GFP antibody with RPL13, RPL14, or RPS2 cell line. Stable HeLa cell lines expressing fluorescently tagged ribosomal proteins were induced by 0.5 μg/ml doxycycline for 20 h before harvest. Cell lysate was incubated with IgG or GFP antibody overnight. GAPDH was used as negative control. 2.2% of sample were loaded as input. (*) indicates IgG heavy chain, which is below fluorescently tagged RPL13 and RPS2 and above RPL14. (C) Coimmunoprecipitation using RPL24 or Lsm8 antibody in wildtype HeLa cells. HeLa cell lysate was incubated with IgG, RPL24, or Lsm8 antibody overnight. GAPDH was used as negative control. 2% of sample were loaded as input. Two replicates are shown for each IP. Source data are available for this figure: SourceData FS3.
Ribosomal subunit proteins do not localize to CBs or interact with coilin. (A) Left: Immunofluorescent staining of wildtype HeLa cell using RPL24 (cyan) and coilin (magenta) antibody. Scale bar = 10 μm. Right: colocalization analysis of RPL24 and coilin. Scale bars = 1 μm. (B) Coimmunoprecipitation using GFP antibody with RPL13, RPL14, or RPS2 cell line. Stable HeLa cell lines expressing fluorescently tagged ribosomal proteins were induced by 0.5 μg/ml doxycycline for 20 h before harvest. Cell lysate was incubated with IgG or GFP antibody overnight. GAPDH was used as negative control. 2.2% of sample were loaded as input. (*) indicates IgG heavy chain, which is below fluorescently tagged RPL13 and RPS2 and above RPL14. (C) Coimmunoprecipitation using RPL24 or Lsm8 antibody in wildtype HeLa cells. HeLa cell lysate was incubated with IgG, RPL24, or Lsm8 antibody overnight. GAPDH was used as negative control. 2% of sample were loaded as input. Two replicates are shown for each IP. Source data are available for this figure: SourceData FS3.
The lack of evidence for direct interactions between coilin and RPL proteins led us to consider that the effect of ribosomal protein depletion on CB number could be indirect. Searching for evidence that ribosomal proteins could have other functions in eukaryotic cells, we discovered that an interaction between RPL24 and the U6 protein Lsm8 had previously been discovered in a yeast two-hybrid assay (Lehner and Sanderson, 2004). We found that coilin and RPL24 were detectable in an anti-Lsm8 pulldown (Fig. S3 C), consistent with this finding. Intriguingly, U6 snRNP is targeted to the CB by its constituent protein, SART3, which promotes U6 annealing to U4 snRNA together with the Lsm ring that assembles specifically on U6 (Staněk, 2017). We could not find evidence that any of the other RPL hits had a relationship with the snRNP assembly. This caused us to scrutinize the differences between the RPL hits. As new specificity controls, we included RPL6 and RPL17, large subunit proteins on the surface of the ribosome (Fig. 4 C), which were not enriched in the coilin-APEX2 proximity biotinylation and therefore not tested in our image-based screen. RPL24 depletion led to the greatest increase in CBs visually and upon quantification (Fig. 6 A and Fig. S4 A), with 3.73 ± 1.87 CBs compared with 2.58 ± 1.43 in the non-targeting siRNA control. RPL6 depletion also significantly increased the number of CBs showing that RPL proteins that were not labeled by coilin-APEX2 have the capacity to influence CB number.
RPL24 has the greatest effect on CB number and the least effect on ribosome integrity. (A) Quantification of CB numbers per nucleus in HeLa cells after siRNA knockdown of control (NT) or ribosomal proteins for 72 h. RPL14 and RPL24 are positive hits identified in the siRNA screen to increase the CB number. RPL6 and RPL17 were not enriched in proximity biotinylation. Number of replicates: siNT = 7; siRPL14 = 6; siRPL24, siRPL6 and siRPL17 = 5. P values are obtained by Kruskal–Wallis test followed by the Dunn test with Bonferonni correction. **, P < 0.01, ****, P < 0.0001. Whiskers are plotted by the Tukey method. (B) 28S/18S rRNA ratio of RNA extracted from HeLa cells after respective siRNA knockdown. Number of replicates = 3. P values are obtained by unpaired t tests with siNT. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bar, SD. (C) Western blot analysis of ribosomal protein expression in HeLa cells after respective siRNA knockdown. Tubulin is used as a loading control. Number 1–3 stands for independent replicates. Source data are available for this figure: SourceData F6.
RPL24 has the greatest effect on CB number and the least effect on ribosome integrity. (A) Quantification of CB numbers per nucleus in HeLa cells after siRNA knockdown of control (NT) or ribosomal proteins for 72 h. RPL14 and RPL24 are positive hits identified in the siRNA screen to increase the CB number. RPL6 and RPL17 were not enriched in proximity biotinylation. Number of replicates: siNT = 7; siRPL14 = 6; siRPL24, siRPL6 and siRPL17 = 5. P values are obtained by Kruskal–Wallis test followed by the Dunn test with Bonferonni correction. **, P < 0.01, ****, P < 0.0001. Whiskers are plotted by the Tukey method. (B) 28S/18S rRNA ratio of RNA extracted from HeLa cells after respective siRNA knockdown. Number of replicates = 3. P values are obtained by unpaired t tests with siNT. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bar, SD. (C) Western blot analysis of ribosomal protein expression in HeLa cells after respective siRNA knockdown. Tubulin is used as a loading control. Number 1–3 stands for independent replicates. Source data are available for this figure: SourceData F6.
Large ribosomal protein depletion leads to increased CB number, impaired ribosomal biogenesis and translation inhibition. (A) Immunofluorescent images showing coilin signal in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The nucleus outline is shown in blue. (B) Electropherograms of total RNA isolated from HeLa cells after respective siRNA knockdown. 28S/18S rRNA intensity ratio is listed below each lane, and a representative electropherogram in arbitrary intensity unit is shown for each siRNA. (*) marks sample reruns. (C) WB of HeLa cell lysate harvested after respective siRNA knockdown. Tubulin was used as the loading control. Numbers 1–3 stand for independent replicates. (D) WB of puromycin incorporation assay. HeLa cell was treated with the siRNA of control or ribosomal proteins for 72 h, followed by incubation with or without 1 μM puromycin for 1 h before harvest. “−” indicates no puromycin control, “+” indicates puromycin addition. Ponceau stain was shown as the loading control. Two replicates are shown for each RPL knockdown condition. Source data are available for this figure: SourceData FS4.
Large ribosomal protein depletion leads to increased CB number, impaired ribosomal biogenesis and translation inhibition. (A) Immunofluorescent images showing coilin signal in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The nucleus outline is shown in blue. (B) Electropherograms of total RNA isolated from HeLa cells after respective siRNA knockdown. 28S/18S rRNA intensity ratio is listed below each lane, and a representative electropherogram in arbitrary intensity unit is shown for each siRNA. (*) marks sample reruns. (C) WB of HeLa cell lysate harvested after respective siRNA knockdown. Tubulin was used as the loading control. Numbers 1–3 stand for independent replicates. (D) WB of puromycin incorporation assay. HeLa cell was treated with the siRNA of control or ribosomal proteins for 72 h, followed by incubation with or without 1 μM puromycin for 1 h before harvest. “−” indicates no puromycin control, “+” indicates puromycin addition. Ponceau stain was shown as the loading control. Two replicates are shown for each RPL knockdown condition. Source data are available for this figure: SourceData FS4.
Taken together, depletion of a long and likely incomplete list of protein constituents of the large ribosomal subunit (RPLs 5, 6, 7A, 13, 14, 24 as well as the 60S biogenesis protein NSA2) detectably increases CB numbers in cells. Previous reports have suggested that depletion of any of the large subunit proteins leads to loss of 60S subunits and a repression of translation (Luan et al., 2022; Robledo et al., 2008). We address these possibilities with the following three experiments. First, the ratio of 28S/18S rRNA was measured in control and depletion conditions, showing that the abundance of 60S subunits is reduced to ∼30% in knockdowns of RPL6, 14, and 17 and ∼60% in RPL24 knockdown (Fig. 6 B and Fig. S4 B). Second, western blot analysis of RPL proteins from extracts of cells under depletion conditions revealed the loss of RPL proteins not directly targeted by siRNA (Fig. 6 C and Fig. S4 C). For example, RPL14 depletion leads to loss of RPLs 6, 14, 17 and 24, in agreement with the strong loss of 60S subunits. On the other hand, RPLs 3, 6, 14, and 17 remained detectable upon RPL24 depletion, consistent with a higher level of remaining large subunits. Third, the depletion of each of the RPL proteins investigated here decreased the amount of translation measured by the puromycin assay, with the exception that an intermediate level of protein synthesis remained after RPL24 depletion (Fig. S4 D). We conclude that depletion of most of the RPL proteins that increase CB number decreases the levels of functional ribosomes and therefore translation.
We then wanted to consider the composition of the increased CBs in these depletion conditions to ensure they were not empty coilin foci and to determine if RPL proteins are detectable in CBs. We stained for common CB components (snRNPs, Nopp140, and dyskerin) upon depletion of the top two ribosomal protein hits (RPL14 and RPL24). We found that these CBs in RPL depletions contain prominent, function-related members Nopp140, dyskerin, and Sm proteins (Fig. S5, A–C), as expected for bona fide CBs. Therefore, we concluded that the “extra” CBs present when RPL proteins are depleted contain key signature proteins of CBs.
Large ribosomal protein depletion does not affect CB composition but leads to an altered dimethylarginine profile. (A and B) Immunofluorescent images showing the presence of (A) nopp140 and (B) dyskerin (cyan) in CBs (α-coilin antibody in magenta) in control or ribosomal protein knockdown cells. (C) Immunofluorescent images showing the presence of snRNPs (α-Sm, Y12 antibody in cyan) in CBs (α-coilin antibody in magenta). Scale bars = 10 µm. (D) Representative single nucleus immunofluorescent staining of coilin (magenta) and SMN (cyan) in siNT and siRPL14 with accompanying line profile plots showing intensities through a single CB. (E) Immunofluorescent images showing asymmetric dimethylarginine (aDMA) signal in HeLa cell after respective siRNA knockdown. CB outline is shown in magenta. Scale bars = 5 μm. (F and G) Quantification of aDMA intensity in the nucleus or CB in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The relative DMA intensity is calculated by dividing mean DMA intensity in each nucleus (F) or CB (G) to the respective mean coilin intensity. The median of the relative DMA intensity is shown by the magenta (F) or cyan (G) line. Number of replicates = 4. P values are obtained by Kruskal–Wallis test followed by Dunn test with Bonferroni correction. **, P < 0.01; ****, P < 0.0001.
Large ribosomal protein depletion does not affect CB composition but leads to an altered dimethylarginine profile. (A and B) Immunofluorescent images showing the presence of (A) nopp140 and (B) dyskerin (cyan) in CBs (α-coilin antibody in magenta) in control or ribosomal protein knockdown cells. (C) Immunofluorescent images showing the presence of snRNPs (α-Sm, Y12 antibody in cyan) in CBs (α-coilin antibody in magenta). Scale bars = 10 µm. (D) Representative single nucleus immunofluorescent staining of coilin (magenta) and SMN (cyan) in siNT and siRPL14 with accompanying line profile plots showing intensities through a single CB. (E) Immunofluorescent images showing asymmetric dimethylarginine (aDMA) signal in HeLa cell after respective siRNA knockdown. CB outline is shown in magenta. Scale bars = 5 μm. (F and G) Quantification of aDMA intensity in the nucleus or CB in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The relative DMA intensity is calculated by dividing mean DMA intensity in each nucleus (F) or CB (G) to the respective mean coilin intensity. The median of the relative DMA intensity is shown by the magenta (F) or cyan (G) line. Number of replicates = 4. P values are obtained by Kruskal–Wallis test followed by Dunn test with Bonferroni correction. **, P < 0.01; ****, P < 0.0001.
Ribosomal proteins link nuclear dimethylarginine levels to CB number and structure
It is striking that depletion of RPL24 has the strongest effect on CB number and the weakest effect on ribosome and translation levels. RPL24 is also strongly codepleted when the other RPLs are knocked down. Together with the suggestion that RPL24 may be involved with U6 snRNP trafficking (see above), we hypothesized that RPL24 may be the true effector of CB number and morphology. A striking bipartite substructure of HeLa cell CBs was recently discovered (Courchaine et al., 2021; Novotny et al., 2015), showing that the coilin-containing CB is not a sphere in HeLa cells but rather forms a pocket around an SMN-containing gem with which its contents do not mix. We noticed that the bipartite morphology was affected by RPL14 and RPL24 protein depletion by STED microscopy (Fig. 7 A). It is possible to observe the shift in the position of CBs and gems with confocal imaging (Fig. S5 D), enabling quantification and statistical evaluation of measured distances between the center of CBs and gems under normal or RPL protein knockdown conditions (Fig. 7 B). Depletion of all RPL proteins tested led to a significant decrease in the distance between CB and gems, suggesting a mixing of the two compartments. Previous work showed that the bipartite structure is dependent on protein modification by dimethylarginine (DMA); symmetric DMA (sDMA) is a prominent posttranslational modification on the Sm protein components in snRNPs, which localizes to the coilin-rich portion of CBs (Courchaine et al., 2021; Šimčíková et al., 2023). The Y12 antibody used to detect the Sm proteins on snRNPs is specific for sDMA and revealed elevated levels of antigen in the whole nucleus (Fig. S5 C). We therefore undertook a detailed analysis of sDMA and asymmetric DMA (aDMA) levels upon RPL depletion (Fig. 7 C and Fig. S5 E). To compensate for uneven staining errors, the DMA signals were normalized to coilin signals during quantification. Levels of sDMA in the nucleus and specifically in CBs increased upon RPL depletion, with RPL24 consistently trending to have the strongest effects (Fig. 7, D and E). Interestingly, aDMA was also significantly increased in both nuclei and CBs (Fig. S5, E–G), indicating overall misregulation of nuclear DMA levels. Given that the effects of RPL depletion—most notably RPL24—mimic the effects of inhibiting the protein methyltransferases (PRMTs) that install DMA (Courchaine et al., 2021; Hebert et al., 2002), we suggest that RPL depletion and/or downstream effects of reduced translation increases nuclear levels of DMA and accounts for the increased number of CBs and changes in their morphology.
Large ribosomal protein depletion impacts CB substructure and nuclear dimethylarginine profile. (A) Representative single CB immunofluorescent staining of coilin (magenta) and SMN (cyan) using STED microscopy in siNT, siRPL14, and siRPL24 cells. Two CBs per condition are shown. (B) Quantification of offset distance between CB and gems in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The offset distance between CB and gems is calculated from coilin and SMN fluorescent intensity-weighted center of mass. The cyan line stands for the median offset distance in each sample. Number of CB-gem pairs measured: siNT = 356; siRPL14 = 460; siRPL24 = 229; siRPL6 = 152; siRPL17 = 178. P values are obtained by Kruskal–Wallis test followed by the Dunn test with Bonferroni correction. **, P < 0.01; ****, P < 0.0001. (C) Immunofluorescent images showing symmetric dimethylarginine (sDMA) signal in HeLa cell after respective siRNA knockdown. CB outline is shown as the magenta line. Scale bars = 5 μm. (D and E) Quantification of sDMA intensity in HeLa cell nucleus or CB after respective siRNA knockdown. The relative sDMA intensity is calculated by dividing mean DMA intensity in each nucleus (D) or CB (E) to the respective mean coilin intensity. Median of the relative DMA intensity is shown by the magenta (D) or cyan (E) line. Number of replicates = 4. P values are obtained in the same way as in B.
Large ribosomal protein depletion impacts CB substructure and nuclear dimethylarginine profile. (A) Representative single CB immunofluorescent staining of coilin (magenta) and SMN (cyan) using STED microscopy in siNT, siRPL14, and siRPL24 cells. Two CBs per condition are shown. (B) Quantification of offset distance between CB and gems in HeLa cells after siRNA knockdown of control or ribosomal proteins for 72 h. The offset distance between CB and gems is calculated from coilin and SMN fluorescent intensity-weighted center of mass. The cyan line stands for the median offset distance in each sample. Number of CB-gem pairs measured: siNT = 356; siRPL14 = 460; siRPL24 = 229; siRPL6 = 152; siRPL17 = 178. P values are obtained by Kruskal–Wallis test followed by the Dunn test with Bonferroni correction. **, P < 0.01; ****, P < 0.0001. (C) Immunofluorescent images showing symmetric dimethylarginine (sDMA) signal in HeLa cell after respective siRNA knockdown. CB outline is shown as the magenta line. Scale bars = 5 μm. (D and E) Quantification of sDMA intensity in HeLa cell nucleus or CB after respective siRNA knockdown. The relative sDMA intensity is calculated by dividing mean DMA intensity in each nucleus (D) or CB (E) to the respective mean coilin intensity. Median of the relative DMA intensity is shown by the magenta (D) or cyan (E) line. Number of replicates = 4. P values are obtained in the same way as in B.
Discussion
Here, we have undertaken proteomic and functional screens to comprehensively determine the protein composition of Cajal bodies, leveraging the scaffolding protein coilin for proximity labeling with APEX2. We gain insight into the range of activities these interactors have in CB assembly and maintenance, expanding the number of proteins that may concentrate in or otherwise modulate CBs. This “CB proteome” comprises 144 proteins, including 70 newly detected proteins. We chose APEX2 biotinylation over other robust proximity biotinylation techniques, such as BioID or TurboID, due to its short labeling time. Indeed, we were able to achieve robust biotinylation of CBs with 1-min labeling, similar to the residence time of coilin in CBs (Dundr et al., 2004). For comparison, a recent large-scale study by the Gingras lab included coilin as one of the bait proteins for a BioID-based map of the human cell to identify proteins that concentrate in nuclear bodies (Go et al., 2021). While their study identified more labeled prey proteins (385), the dataset is less specific for CB components because only 19 known CB components (5%) are among this large cohort (see Fig. S1 G). This difference likely reflects higher levels of biotinylation by BioID throughout the nucleus and is not restricted to CBs (Go et al., 2021). In contrast, 30% of the CB proteins in our dataset were previously known, and the new components fell into functional groups that are linked to known CB functions. The most enriched proteins identified in our dataset have previously been shown to have longer residence times in the CB than other components (Dundr et al., 2004). For example, coilin and TGS1 both have long residence times in CBs and are among the top hits, while snRNP proteins have shorter residence times in CBs and show less enrichment in our data. This demonstrates that our approach is more specific for CB-localized proteins, possibly also due to the modest levels of coilin-APEX2 expressed in our stable cell lines.
The range of CB components identified by our approach changes how we think about the overall composition and function of this BMC. Previously, the “parts list” emphasized snRNP and snoRNP components (Machyna et al., 2013). Our updated list of 144 components reveals 18 additional proteins involved in transcription, bringing the total number of components to 23, equivalent to the number involved in pre-mRNA splicing and cleavage (see Table 1). One new hit, SRRT, was somewhat enriched in CBs relative to nucleoplasm (see Fig. 2), consistent with the known distribution of many CB components in both compartments (e.g., snRNPs). Another new hit, IRF2BP1 showed strong enrichment against both APEX2-NLS and ΔNTD-APEX2 and was highly concentrated in CBs relative to nucleoplasm. IRF2BP1 is an understudied protein originally identified as an interactor of Interferon Regulatory Factor 2, IRF2 (Childs and Goodbourn, 2003). IRF2BP1 has a zinc finger and a C3HC4 RING domain at the N- and C-terminus, respectively, and has been shown to inhibit transcription of various genes (Barysch et al., 2021; Faresse et al., 2008; Kimura, 2008; Yeung et al., 2011). Due to its role as a transcriptional regulator, we speculate that it may bind DNA at or near CB-associated genes.
Among the CB components discovered here, our functional screen identified proteins that are necessary for proper CB formation using an imaging-based screen after siRNA depletion. Depleting each of all 144 CB components and conducting quantitative image analyses, 25 proteins were identified as positive regulators of CB assembly because CB numbers decreased when they were depleted (see Fig. 3 B and Table S2). While CB components required for CB formation and maintenance, like coilin, have been known for a long time (Sawyer et al., 2017), these were well detected and expanded, owing to the sensitivity of our imaging-based assay and our ability to quantify even partial effects. Additionally, the depletion of 11 components caused coilin to relocalize in nucleoli. Among the CB components in the transcription category, 3/23 decreased CB number and 7/23 caused coilin to relocalize. These data underscore the importance of transcription as a crucial determinant of CB assembly, maintenance, or positioning relative to the genome.
Previously, Pols I, II, and III were found to colocalize in Xenopus laevis oocyte CBs, which are numerous and largely extrachromosomal (Gall, 2000), suggesting that CBs may have an intrinsic affinity for RNA polymerases independent of transcription. This could explain why CBs are localized to active snRNA and histone genes in somatic cells (Frey et al., 1999; Wang et al., 2016), the significance of which can be due to two mechanisms that are not mutually exclusive. First, the association of CBs with snRNA genes may enhance their transcription (Wang et al., 2016). More recently, CBs have been implicated in Pol II elongation at transcription end sites of replication-dependent histone genes, which require SLBP and NPAT for their expression (Suzuki et al., 2022). In our screen, SLBP and NPAT were detected as CB components that reduced CB number upon depletion. Second, coilin binding to nascent RNA at active snRNA and histone genes may help nucleate CBs (Arias Escayola and Neugebauer, 2018; Frey et al., 1999; Machyna et al., 2014). Although CBs can associate with target genes after their formation elsewhere in the nucleus (Dundr et al., 2007), the initial nucleation sites of CBs are still unknown. The only known transcriptionally inactive cells with CBs are those of early zebrafish, where maternally loaded extrachromosomal CBs are numerous even though the zygote’s genome is completely silent (Arias Escayola and Neugebauer, 2018; Heyn et al., 2017; Strzelecka et al., 2010). Developing oocytes and embryos are also known to have even more numerous extrachromosomal nucleoli, suggesting that germ cells are a special case. As discussed above, the detection of transcription factors by coilin-APEX2 proximity labeling and the effects of their depletion on CB number or coilin localization suggest important relationships between CBs and active genomic loci.
Unexpectedly, we discovered a limited group of components including five 60S ribosomal subunit proteins (RPLs) and one 60S biogenesis factor (NSA2) that appears to restrict the number of CBs per nucleus, a regulatory activity that was previously unknown. Three proteins (EFTUD2, ANLN, and NUMA1) do not fit into this group. EFTUD2 (hSnu114) is a component of the U4•U6/U5 spliceosomal tri-snRNP. CB numbers have previously been shown to increase when protein components of the tri-snRNP are depleted (Novotny et al., 2015), suggesting that EFTUD2 depletion directly leads to the accumulation of immature snRNPs in CBs rather than influencing CB assembly per se. Because RPL proteins are not present in spliceosomes or required for snRNP assembly, we presume this is not the explanation for the identification of RPL proteins in our screen. ANLN and NUMA1 had previously been shown to alter ploidy and cell cycle (Farley-Barnes et al., 2018).
Remarkably, the number of CBs per interphase nucleus approximately doubled when each of the RPL factors was depleted without changes in ploidy or cell cycle. In addition to this increase in CB abundance, the bipartite substructure of CBs collapsed into one compartment, in which coilin was mixed with the gem component SMN. Non-ribosomal nucleolar proteins frequently associate with CBs and can even be integral to their assembly and function (Machyna et al., 2013), as in the case of Nopp140 (Bizarro et al., 2019, 2021; Courchaine et al., 2021). In contrast, a role for ribosomal proteins had not been investigated largely due to the lack of optimal reagents for visualization in cells. Interestingly, some of the same RPL proteins were detected in the Gingras BioID study discussed above (Go et al., 2021). However, we were unable to detect GFP-RPL13 or GFP-RPL14 in CBs using rigorously established stable cell lines, making it unlikely that RPL proteins exert a direct influence on CB assembly, structure, or maintenance.
A series of experiments leads us to offer the following explanation for why RPL depletion causes CB numbers to increase and changes their substructure. In all cases, RPL depletion increases the amount of aDMA and sDMA posttranslational modifications relative to coilin in both the nucleus and the CB (see Fig. 7 and Fig S5). CB substructure in HeLa cells is determined by SMN binding to nuclear ligands that are dimethylated on arginine, and the balance between aDMA and sDMA levels determines if gems are separate or merged with coilin-positive CBs (Courchaine et al., 2021; Hebert et al., 2002). Disruption of the balance of aDMA and sDMA modifications installed, respectively, by Type I and Type II protein methyl transferases (PRMTs) can lead to the phenomenon of “PRMT scavenging,” in which a protein that is primarily modified by aDMA could shift to sDMA or vice versa (Maron et al., 2021; Šimčíková et al., 2023). Because many proteins are modified by DMA, it is therefore difficult to track down which nuclear DMA ligands are changing in response to RPL knockdown. This understanding nevertheless highlights the possibility that depletion of RPL proteins may alter the constellation of aDMA- and sDMA-modified ligands in the nucleus and CBs such that their number and substructure are altered.
Protein components of the large and small subunits of the ribosome are largely disordered and associate either with the solvent-exposed surface or at the interface between the two subunits (Yusupov et al., 2001). The RPL proteins we identified in our screen are located on the surface of the 60S subunit, meaning they could interact with coilin/CBs in the context of assembled large subunits, and we validated that other RPLs (RPL6 and RPL17) with similar properties also lead to higher DMA levels and greater numbers of CBs upon depletion. A recent study showed that SMN binds ribosomes and preferentially associates with the 60S subunit in the context of translation in the cytoplasm (Lauria et al., 2020), suggesting the possibility that SMN association with RPLs in the nucleolus, nucleoplasm, or cytoplasm could indirectly modulate CB assembly. Because (1) RPL24 has the greatest effect on aDMA and sDMA levels and (2) each of the other RPLs tested depletes cells of 60S ribosomes as well as RPL24, we propose that the increase in DMA occurs through an as-yet-unknown mechanism linked most closely to RPL24. In support of this working model, we show that depletion of RPL24 leads to the greatest increase in CBs while having the least effect on the expression of other RPL proteins, the levels of the 60S ribosomes, and overall protein synthesis (see Fig. 6 and Fig. S4). Knocking down of 60S ribosomal proteins suppresses the expression of ribosomal proteins of both subunits and broadly impacts cellular functions (Luan et al., 2022). In contrast, RPL24 assembles onto the 60S subunit in the cytoplasm, while RPL6, 14, and 17 join in the nucleolus (de la Cruz et al., 2015), and its depletion does not lead to nucleolar stress (Nicolas et al., 2016). Moreover, the evidence that RPL24 interacts with the U6 Lsm ring, which is instrumental for U4/U6 assembly within the CB, suggests an additional or alternative possibility for the observed dependency of CB number and substructure on RPL levels. These points highlight the possibility that RPL24 and other ribosomal proteins may “moonlight” in other cellular metabolic roles. Since the biological function of CBs is the assembly and recycling of spliceosomal snRNPs, the coordination of mRNA biosynthesis with translation by ribosomes can be viewed in the context of growth control. Our study therefore underscores this potential by suggesting that mRNA metabolism and translation may be coordinated through CBs.
Materials and methods
Cell culture and cell line generation
HeLa KYOTO cells were obtained from ATTC and grown in DMEM GlutaMAX medium (Gibco) supplemented with 10% heat-inactivated FBS (Gibco) and 1% penicillin-streptomycin (Gibco). Cells were incubated in humidified 5% CO2 at 37°C. Coilin-APEX2, APEX2-NLS, and ΔNTD-NLS constructs were generated with the InFusion HD kit (Takara) by inserting sequences after the EF-1α promoter into plasmid backbone pWPI (generated by Didier Trono, #12254; Addgene). Lentiviruses from these constructs were prepared by transfecting confluent HEK293FT cells with pWPI containing the desired insert along with pMD2.G and pCMV R8.74 (generated by Dider Trono, #12250 and #22036, respectively; Addgene) using Fugene HD reagent (Promega). After 72 h, the viral supernatant was harvested, filtered through a 0.45-μm filter, and used to transduce HeLa cells. After three passages, GFP-positive cells were sorted using FACSAria II. Coilin-APEX2 expressing cells were sorted into single cells and validated by staining for V5 and coilin to ensure that CB formation was not altered. APEX2-NLS and ΔNTD-NLS were sorted into low-GFP expressing pools and validated by staining for V5 to confirm the nuclear expression of the construct. Stable HeLa cell lines expressing GFP-RPL13 and GFP-RPL14 were constructed using the same strategy used to establish the cell line expressing RPS2-YFP (Zemp et al., 2009). The cells were selected with 300 µg/ml hygromycin once per week, and ribosomal protein expression was induced by 0.5 µg/ml doxycycline for 16 h before immunofluorescent staining or 20 h before GFP co-IP (see below).
Antibodies used
Mouse monoclonal α-coilin (#ab87913 and # ab210785; Abcam), α-TMG (#NA02; Calbiochem), mouse monoclonal a-SMN/Gemin 1 (2B1) (#ab5831; Abcam), rabbit monoclonal recombinant a-NOLC1 (EPR14896) (#ab184550; Abcam); mouse monoclonal a-V5 tag antibody (R960-25; Invitrogen); rabbit polyclonal a-DKC1 (HPA000447; Sigma-Aldrich); rabbit polyclonal a-IRF2BP1 (HPA049480; Sigma-Aldrich); rabbit polyclonal a-SRRT (HPA042858; Sigma-Aldrich); rabbit polyclonal a-EFTUD2 (HPA022021; Sigma-Aldrich); Y12 mouse monoclonal a-Sm antibody (generous gift from J. Steitz). α-RPL14 (ab181200; Abcam), α-RPL24 (SAB2700500; Sigma-Aldrich), α-RPL3 (11005-1-AP; Proteintech), α-RPL6 (15387-1-AP; Proteintech), α-RPL17 (14121-1-AP; Proteintech), α-tubulin (ab6161; Abcam), α-puromycin (Kf-Ab02366-1.1; Kerafast), α-Lsm8 (10134-1-AP; Proteintech), α-aDMA (07-414; Sigma-Aldrich), α-sDMA (07-412; Sigma-Aldrich), α-GFP (ab290; Abcam), α-GAPDH (#ab9485; Abcam), mouse monoclonal α-histone H3 (1G1) (sc-517576; Santa Cruz). Secondary antibodies: α-mouse IgG-HRP (A9917; Sigma-Aldrich), α-rabbit IgG-HRP (A9169; Sigma-Aldrich), α-rat IgG-HRP (ab97057; Abcam), α-rabbit IgG-Alexa594 (Jackson Immunoresearch, 711-585-152), α-mouse IgG-Alexa594 (715-585-150; Jackson Immunoresearch), α-rabbit IgG-Alexa488 (711-545-152; Jackson Immunoresearch), α-mouse IgG-Alexa488 (715-545-150; Jackson Immunoresearch).
APEX2 labeling
APEX2 labeling was performed as described (Hung et al., 2016). Briefly, confluent HeLa cells were incubated with 500 μM biotinyl tyramide (Chemodex) for 30 min at 37°C with 5% CO2. H2O2 was then added to a final concentration of 1 mM for 1 min. The reaction was immediately quenched by washing three times with a “quencher solution” (10 mM sodium ascorbate, 10 mM sodium azide, and 5 mM Trolox in 1XPBS). Cells were then either fixed for imaging or pelleted for nuclei isolation and enrichment with streptavidin.
Nuclear isolation and streptavidin enrichment
Cells from five 15-cm plates (∼1 × 108 cells) were labeled as previously described and scraped from plates using a quencher solution. Cells were centrifuged at 3,000 g for 10 min at 4°C. Cells were then washed with cold PBS, transferred to a fresh tube, and spun down at 250 g for 5 min at 4°C. Cells were then gently resuspended in 5 ml of “Buffer A” (10 mM Hepes pH 7.4, 10 mM KCl, 340 mM sucrose, 10% glycerol, 4 mM MgCl2, 1 mM DTT, 1X protease inhibitors [Roche], 10 mM beta-GP, and 0.1% RNAse OUT [Invitrogen]). 5 ml of “Buffer B” (Buffer A + 0.2% Triton X-100) was added and samples were incubated on ice for 10 min and centrifuged at 1,200 g for 5 min at 4°C. A fraction of the supernatant (cytoplasmic fraction) was saved for downstream analysis while the rest was discarded. Cells were resuspended in 5 ml Nuclear Resuspension Buffer (10 mM Hepes pH 7.4, 50% glycerol, 75 mM NaCl, 1 mM DTT) and centrifuged at 1,000 g for 5 min at 4°C 2X. The supernatant was discarded and the remaining pellet (nuclei) lysed in 1 ml RIPA lysis buffer (50 mM Tris pH 7.5, 150 mM NaCL, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with 1X protease inhibitors (Roche) and 1 mM PMSF. Nuclei were sonicated at 30% AMP, 10sON 20sOFF 30X. Lysates were clarified by centrifuging at 15,000 g for 10 min at 4°C. Protein concentration was quantified using Pierce 660 nm Protein Assay Reagent (Thermo Fisher Scientific). Streptavidin enrichment was performed as previously described (Hung et al., 2016); 4 mg of protein was incubated with 50 μl of magnetic streptavidin beads (Pierce) for 3 h at 4°C. Beads were pelleted and the “flow-through” was set apart for downstream analysis. Then, beads were sequentially washed 2X with RIPA lysis buffer, 1X with 1 M KCl, 1X with 0.1 M Na2CO3, 1 × 2 M urea in 10 mM Tris-HCl pH8.0, and 2X with RIPA lysis buffer. Biotinylated proteins were eluted from beads by boiling for 10 min in 4X NuPAGE LDS Sample Buffer (Invitrogen) supplemented with 2 mM biotin and 20 mM DTT. For western blotting, proteins were resolved on 4–12% Bis-Tris polyacrylamide gels (Invitrogen) and transferred to a 0.45-μm nitrocellulose membrane (BioRad). After blocking, membranes were probed with the following antibodies: Streptavidin-HRP (Jackson Immunoresearch), GAPDH (Santa Cruz), H3 (Santa Cruz), and V5 (Invitrogen).
Mass spectrometry
Samples were separated on a 4–12% NOVEX NuPage gradient SDS gel (Thermo Fisher Scientific) for 10 min at 180 V in 1X MES buffer (Thermo Fisher Scientific). Proteins were fixed and stained with Coomassie G250 brilliant blue (Carl Roth). The gel lanes were cut, and each lane was minced into ∼1 × 1 mm pieces. Gel pieces were destained with a 50% ethanol/50 mM ammonium bicarbonate (ABC) solution. Proteins were reduced in 10 mM DTT (Sigma-Aldrich) for 1 h at 56°C and then alkylated with 50 mM iodoacetamide (Sigma-Aldrich) for 45 min at room temperature. Proteins were digested with mass spec grade trypsin (Sigma-Aldrich) overnight at 37°C. Peptides were extracted from the gel by two incubations with 30% ABC/acetonitrile and three subsequent incubations with pure acetonitrile. The acetonitrile was subsequently evaporated in a concentrator (Eppendorf) and loaded on StageTips (Rappsilber et al., 2007) for desalting and storage.
For mass spectrometric analysis, peptides were separated on a 20-cm self-packed column with a 75-µm inner diameter filled with ReproSil-Pur 120 C18-AQ (Dr. Maisch GmbH) mounted to an EASY HPLC 1000 (Thermo Fisher Scientific) and sprayed online into a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific). We used a 94-min gradient from 2% to 40% acetonitrile in 0.1% formic acid at a flow of 225 nl/min. The mass spectrometer was operated with a top 10 MS/MS data-dependent acquisition scheme per MS full scan. Mass spectrometry raw data were searched using the Andromeda search (Cox et al., 2011) integrated into MaxQuant suite 1.5.2.8 engine (Cox and Mann, 2008) using the Uniprot Homo sapiens database (January 2020; 42,338 entries). In all analyses, carbamidomethylation at cysteine was set as fixed modification while methionine oxidation and protein N-acetylation were considered as variable modifications. Match between run option was activated. Prior to bioinformatics analysis, reverse hits, proteins only identified by site, protein groups based on one unique peptide, and known contaminants were removed. For further bioinformatics analysis, the LFQ values were log2-transformed and the median across the replicates was calculated. This enrichment was plotted against the – log 10 transformed P value (Welch t test) using the ggplot2 package in the R environment.
High throughput screen by siRNA knockdown
The high throughput screen was conducted at the Yale Center for Molecular Discovery, where target genes were knocked down by reverse transfection using ON-Targetplus siRNA in SMARTpool format (Horizon). A CB-specific siRNA library was generated, including all previously annotated CB proteins as well as the proteins identified in this study by proximity labeling and mass spectrometry (see Table S3 for oligo information). Additionally, this library contained 18 wells of a negative control siRNA (non-targeting SMARTpool) and 18 wells of coilin siRNA as a positive control. 100 nM siRNAs in 1X siRNA buffer were incubated for 30 min at room temperature with 1:100 Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) in Opti-MEM (Gibco). HeLa cells were reverse-transfected with a transfection mix (final siRNA concentration 20 nM) and incubated for 72 h before experimental assays. After the knockdown of target genes, cells were fixed and stained for coilin to mark CBs and nucleophosmin to mark nucleoli, as described above. Cells were imaged on In Cell Analyzer 2200 Imaging System (GE). 12 fields of view (40×; 332.8 × 332.8 μm) were acquired per well. Image analysis was performed using Fiji (Schindelin et al., 2012). Nuclei and nucleoli were segmented from the Hoechst and nucleophosmin staining respectively. The nucleoli mask was then subtracted from the coilin channel to avoid counting nucleolar coilin as CBs. CBs per nuclei were counted by finding the local maxima as single points in the coilin staining. Percent effect (PE) of each siRNA was calculated by setting the average CBs/nucleus siNT wells to 0% and the average CBs/nucleus in the positive siCOIL control to 100%. Cutoffs to define “hits” of decreased (>60 PE) or increased (less than −70 PE) CB/nucleus were determined by eye. Nucleolar coilin levels were measured by calculating a Pearson’s correlation coefficient between the nucleophosmin and coilin immunostainings. Cutoffs for nucleolar coilin “hits” (>0.75 Pearson’s correlation coefficient) were determined by eye.
Cell cycle analysis
Cell cycle analysis of siRNA hits that increased CBs per nucleus was done as previously described (Chan et al., 2013). Briefly, the integrated intensity of the Hoechst DNA stain per nucleus was used to determine cell cycle distribution. The log2 integrated intensities of the siNT controls were plotted as a histogram and the value of the G1 peak set to 1 and G2 peak set to 2. Integrated intensities for all other siRNAs analyzed were then normalized to siNT.
Confocal and STED imaging
Cells were grown in No 1.5 coverslips (Zeiss) in either 6- or 24-well plates, fixed in 4% paraformaldehyde, and permeabilized in 0.5% Triton X-100 (American Bioanalytical) in 1XPBS. Cells were then blocked in 3% Bovine Serum Albumin (Sigma-Aldrich) and probed with the following antibodies in blocking buffer Coilin (Abcam), SMN (Abcam), V5 (Invitrogen), Streptavidin-Cy5 (Jackson Immunoresearch), Y12 (gift from J. Steitz), Nopp140 (abcam), Dyskerin (Sigma-Aldrich), IRF2BP1 (Sigma-Aldrich), SRRT (Sigma-Aldrich), and EFTUD2 (Sigma-Aldrich). Cell nuclei were stained with 0.25 μg/ml Hoechst 34580 (Thermo Fisher Scientific) in 1XPBS and mounted using Prolong Diamond Antifade Mountant (Thermo Fisher Scientific). Imaging was done using a Leica SP8 Laser Scanning Confocal microscope, using a 63× or 100× objective lens (1.4NA, oil immersion).
STED imaging was carried out on the same confocal microscope enabled with a 775 nm depletion laser as described previously (Courchaine et al., 2021). Atto 594 and Atto 647N secondary antibodies were used to facilitate two-color STED. A mounting medium (Prolong Diamond; Thermo Fisher Scientific) was used to supply anti-fade protection and to reduce the refractive index difference between the sample and the objective’s immersion silicon oil. Analysis was done using FIJI software (Schindelin et al., 2012). STED and confocal image stacks of Coilin and SMN were segmented using a linear-bin Otsu threshold applied to the entire volume of each channel after smoothing by a Gaussian filter (of 40 nm in x and y, 43.87 nm in z; Sigma-Aldrich). The STED mask in both colors was taken to be the intersection of the smoothed and thresholded STED channels and their paired confocal channel masks. The original STED images were then analyzed using these masks. The intensity-weighted and masked center of mass was calculated in 3D for both coilin and SMN, and the Euclidean distance separation was calculated as the offset. To calculate the fractional intensity overlap, the intersection of coilin and SMN masks was taken, and the intensity in this region was summed over both channels and divided by the combined sum of the intensity in each of the individual channels within their own mask.
Image analysis: Colocalization and offset distance calculation
The Hoechst signal in confocal images was used to segment nuclei with the Cellpose neural network (Stringer et al., 2021). Next, coilin foci were automatically detected in the maximum intensity projection within the nuclear area using a spot-finding algorithm (https://github.com/ejh516/pystachio-smt) that finds their center coordinates using a 2D Gaussian model. From each field of view, several sub-images were then generated by cropping symmetrically around the center of each focus, excluding cropped regions that contain multiple foci. Sub-images originating from several fields of view were then averaged and Pearson’s correlation of pixel values were calculated between the indicated fluorophore channels, yielding correlation coefficients (PCC). To calculate the offset between different nuclear bodies, we algorithmically detected foci as 3D Gaussians as described above, but this time in both fluorescent channels. We then found colocalizing foci by selecting pairs where the Gaussian fits overlap within one standard deviation each. Foci that overlap with multiple other foci were excluded. We then calculated the Euclidean distance between the centers of both overlapping foci and reported that value as the offset.
Image analysis: CB number quantification
Z-stack images for each sample were collected on Leica SP8 Laser Scanning Confocal. Maximum intensity Z projection was done on the central four to eight slices by Fiji. The projected images were then processed by CellProfiler 4.2.6. CBs were segmented from nuclei masks after enhancing for speckles. Nucleus with >10 CBs counted were removed as outliers. CB number in different samples were analyzed by the Kruskal–Wallis test followed by the Dunn test with Bonferroni correction.
Image analysis: Relative aDMA/sDMA signal quantification
Single z-slice confocal images for each sample were collected on Leica SP8 Laser Scanning Confocal. CellProfiler was used to segment nuclei and CBs as in CB number quantification, and then the mean, maximum and minimum intensity of aDMA/sDMA, and coilin in segmented nuclei and CB were measured. Saturated nuclei (maximum intensity of DMA or coilin = 1) or CBs were removed, and nuclei with >10 CBs were removed from further analysis. The mean DMA intensity in each CB or nuclei was divided over the respective coilin intensity to calculate the relative DMA signal. Relative DMA signals in different samples were compared by the Kruskal–Wallis test followed by the Dunn test with Bonferroni correction.
Image analysis: Normalized biotin signal quantification
Single z-slice confocal images for each sample were collected on Leica SP8 Laser Scanning Confocal. CellProfiler was used to segment nuclei. CBs were then identified from the nuclei mask in the coilin channel. To ensure that CBs were not measured in nucleoli or nucleoplasm, they were each expanded by three pixels (CBexp). Nucleoli were segmented as dark holes in the DNA stain, and then CBexp was removed from the identified region. Nucleoplasm was marked as the nucleus region outside CBexp and nucleoli. Mean biotin intensity was measured for each CB, nucleoli, and nucleoplasm separately. Then the mean nucleoli intensity per nucleus was calculated. Saturated nuclei (maximum intensity of coilin = 1) or CBs were removed. Normalized biotin signal was calculated by dividing the biotin intensity in any segmented region to the mean intensity of nucleoplasm. Statistical significance was calculated by unpaired t test between CB or nucleoli and nucleoplasm.
GFP co-IP
Cells were grown to confluency on a 15-cm plate. Harvested cells were resuspended in 1 ml cold NET-2 buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 0.05% NP-40, 1x cOmplete, and EDTA-free protease inhibitor [Roche]). Samples were sonicated at 20% amplitude for three pulses of 10 s “on,” 20 s “off,” on W-450D sonifier (Branson) and cleared by centrifugation (15,000 g, 10 min, 4°C). 2.2% of lysate was kept as input and the remaining amount was equally divided between samples. The lysates are incubated with either 1 μl (5 µg) GFP antibody (#ab290; Abcam) or 1 μl (1 µg) rabbit IgG (#CST-2729; Cell Signaling Technology) overnight on a rotating wheel at 4°C. 40-μl SureBeads Protein G Magnetic Beads (Bio-Rad) were used per IP. The beads were washed twice with NET-2 buffer, incubated with 0.5% BSA (wt/vol) in NET-2 buffer on a rotating wheel for 10 min at room temperature, washed once with NET-2 buffer, incubated twice with NET-2 buffer on a rotating wheel for 5 min at room temperature, and washed once with NET-2 buffer before being resuspended in 40 μl NET-2. The beads were then incubated with the sample for 2 h on a rotating wheel at 4°C. The beads were washed five times with cold NET-2 buffer, resuspended in 20 μl NET-2 buffer and 20 μl 4x Laemmli buffer, and incubated for 15 min at 95°C to release bound proteins. The samples were then analyzed by western blot and probed with α-coilin, α-GFP, and α-GAPDH antibody.
Puromycin incorporation assay
Levels of ongoing translation were measured as described (Farley-Barnes et al., 2018). Briefly, HeLa cells were transfected with different siRNAs for 72 h before changing the media to contain 1 μM puromycin (A1113803; Thermo Fisher Scientific). Cells were incubated for 1 h at 37°C, then aspirated off the media, washed twice with PBS before being harvested for western blot analysis.
Bioanalyzer
HeLa cells were transfected with different siRNAs for 72 h before extracting total RNA using Monarch Total RNA Miniprep Kit (#T2010S; NEB). 1 μg total RNA for each sample was sent to Yale Center for Genome Analysis (YCGA) to analyze on BioAnalyzer (5200 Fragment Analyzer; Agilent Technologies).
Online supplemental material
Fig. S1 (linked to Fig. 1) shows the identification of known and new CB proteins by proximity biotinylation. Fig. S2 (linked to Figs. 2, 3, and 4) shows the effect of siRNA knockdown on coilin relocalization and cell cycle. Fig. S3 (linked to Fig. 5) shows the ribosomal proteins tested do not colocalize or interact with coilin. Fig. S4 (linked to Fig. 6) shows the effect of large ribosomal protein depletion on CB number and ribosome function. Fig. S5 (linked to Fig. 7) shows that large ribosomal protein depletion leads to intact CBs with altered substructure and changed dimethylarginine modification profile. Table S1 shows all comparisons of enrichment between coilin-APEX2, ∆NTD-APEX2, and APEX2-NLS. Table S2 shows the values for the percent effect and the Pearson’s correlation coefficient between nucleophosmin and coilin for all siRNAs screened. Table S3 provides all the siRNA sequences used in the high throughput screen.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD042406.
Acknowledgments
We thank S. Baserga, C. Bryant, L. Ogawa, L. Fuentes, R. Wu, and Ivo Zemp for helpful discussion and technical assistance. We thank the Yale Center for Molecular Discovery for their assistance with the high-throughput screen and microscopy.
The Core is supported in part by a National Cancer Institute Cancer Center Support Grant # National Institutes of Health (NIH) P30 CA016359. Equipment and libraries were supported in part by the Program in Innovative Therapeutics for Connecticut’s Health. We thank the Yale Center for Advanced Light Microscopy Facility for their assistance with confocal and STED microscopy. The microscopes used were funded by shared instrument grant # NIH S10 OD023598. We are grateful for funding from NIH awards U01CA200147 TCPA-2017-Neugebauer and OPAS (to K.M. Neugebauer) and R01NS128358-01 (to K.M. Neugebauer). This work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author contributions: D. Arias Escayola: Conceptualization, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, C. Zhang: Conceptualization, Formal analysis, Investigation, Validation, Visualization, Writing - review & editing, E. Nischwitz: Formal analysis, Investigation, Visualization, L. Schärfen: Formal analysis, Software, Visualization, K. Dörner: Resources, K. Straube: Investigation, Methodology, Validation, U. Kutay: Resources, F. Butter: Data curation, Supervision, Writing - review & editing, K.M. Neugebauer: Conceptualization, Data curation, Funding acquisition, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing.
References
Author notes
D. Arias Escayola and C. Zhang contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.