The MAP kinase and motor scaffold JIP3 prevents excess lysosome accumulation in axons of vertebrates and invertebrates. How JIP3’s interaction with dynein and kinesin-1 contributes to organelle clearance is unclear. We show that human dynein light intermediate chain (DLIC) binds the N-terminal RH1 domain of JIP3, its paralog JIP4, and the lysosomal adaptor RILP. A point mutation in RH1 abrogates DLIC binding without perturbing the interaction between JIP3’s RH1 domain and kinesin heavy chain. Characterization of this separation-of-function mutation in Caenorhabditis elegans shows that JIP3–bound dynein is required for organelle clearance in the anterior process of touch receptor neurons. Unlike JIP3 null mutants, JIP3 that cannot bind DLIC causes prominent accumulation of endo-lysosomal organelles at the neurite tip, which is rescued by a disease-associated point mutation in JIP3’s leucine zipper that abrogates kinesin light chain binding. These results highlight that RH1 domains are interaction hubs for cytoskeletal motors and suggest that JIP3–bound dynein and kinesin-1 participate in bidirectional organelle transport.
Introduction
Long-range intracellular transport of vesicles and organelles by microtubule-based motors is critical for the development, survival, and function of neurons (Guedes-Dias and Holzbaur, 2019). In the axon, where most microtubules assume a plus-end-out orientation, multiple kinesins mediate anterograde transport toward synapses, and dynein is responsible for retrograde transport toward the cell body. How kinesins and dynein work together in axons to form a useful bidirectional transport system is a fundamental unanswered question.
c-Jun N-terminal kinase-interacting protein 3 (JIP3), also known as Sunday Driver or JSAP1, has emerged as an evolutionarily conserved regulator of axonal transport that associates with both kinesin-1 and dynein. JIP3 was originally identified as a scaffolding protein that binds c-Jun N-terminal kinase (JNK) and kinesin light chain (KLC; Bowman et al., 2000; Ito et al., 1999; Kelkar et al., 2000; Verhey et al., 2001), and as the product of the sunday driver (syd) gene in a D. melanogaster screen for axonal transport mutants (Bowman et al., 2000). The syd mutants accumulate synaptic vesicle cargo in segmental nerve axons, a defect that is also observed in kinesin heavy chain (khc) and klc mutants (Bowman et al., 2000). Similarly, mutants of C. elegans unc-16/JIP3, unc-116/KHC, and jnk-1/JNK mis-localize synaptic vesicles to the dorsal processes of cholinergic motor neurons (Byrd et al., 2001), and unc-16 mutants are defective in the sorting of synaptic vesicle cargo at the trans golgi (Choudhary et al., 2017). In addition to regulating composition and distribution of golgi-derived transport vesicles, the work in C. elegans established that UNC-16 acts as a negative regulator of axonal organelle abundance: mitochondria, lysosomes, early/recycling endosomes, and the golgi all accumulate in cholinergic motor neuron axons of unc-16 mutants (Edwards et al., 2013; Edwards et al., 2015), suggesting that one key function of UNC-16 is to clear organelles from axons. Studies in zebrafish sensory neurons, in mouse embryo cortical neurons, and in human iPSC-derived neurons have since shown that JIP3 also negatively regulates axonal lysosome abundance in vertebrates (Drerup and Nechiporuk, 2013; Gowrishankar et al., 2017; Gowrishankar et al., 2021), and that JIP3 shares this function with its paralog JIP4 (Gowrishankar et al., 2021). Recent work identified missense mutations in the human JIP3-encoding gene MAPK8IP3 that cause neurodevelopmental disorders and intellectual disability (Iwasawa et al., 2019; Platzer et al., 2019), and modelling in C. elegans showed that some of these mutations impair JIP3’s organelle clearance function (Platzer et al., 2019).
How JIP3’s association with microtubule-based opposite-polarity motors contributes to its role as a negative regulator of axonal organelle abundance remains poorly explored. Most studies addressing JIP3’s motor-dependent functions have focused on its interaction with kinesin-1, which is composed of a KHC dimer that binds two copies of KLC. The structure of the KLC tetratricopeptide repeat (TPR) domain bound to the mouse JIP3 leucine zipper has been determined by x-ray crystallography (Cockburn et al., 2018), and one of the recently described disease mutations in human JIP3 is predicted to interfere with KLC binding (Platzer et al., 2019). JIP3 also binds the C-terminal tail of KHC, and pull-downs of JIP3 fragments from mouse brain lysate mapped the KHC binding site to the JIP3 N-terminus (Sun et al., 2011). Studies in cultured neurons with JIP3 mutants harboring deletions in the KHC and/or KLC binding region suggest that the interaction with kinesin-1 stimulates axonal elongation (Muresan and Muresan, 2005; Sun et al., 2011; Sun et al., 2013; Watt et al., 2015). Furthermore, both the KHC and the KLC binding region of JIP3 have been implicated in the activation of kinesin-1 motility (Sato et al., 2015; Sun et al., 2011; Sun et al., 2017; Watt et al., 2015). Besides JIP3 itself (Byrd et al., 2001; Verhey et al., 2001), the only firmly established cargo of JIP3–bound kinesin-1 is the BDNF-receptor TrkB, which is transported from the cell body to synapses in the distal axon (Drerup and Nechiporuk, 2013; Huang et al., 2011; Ma et al., 2017; Sun et al., 2017). Additionally, a study in mouse hippocampal neurons reported slowed anterograde transport of mitochondria and amyloid precursor protein in a JIP3 mutant defective for kinesin-1 binding (Sato et al., 2015). There is also evidence that JIP3 and the structurally unrelated JIP1 (another MAP kinase scaffold that associates with kinesin-1 and dynein) cooperate during anterograde transport: in differentiated CAD cells, low-level overexpression of either adaptor stimulates localization of the other adaptor to the neurite tip (Hammond et al., 2008), and overexpression of either adaptor in mouse hippocampal neurons enhances TrkB localization to the axonal tip (Sun et al., 2017).
Although JIP3 has been primarily characterized as a kinesin-1 adaptor, the fact that JIP3 inhibition causes accumulation of vesicles and organelles in axons implies a role in promoting retrograde transport. The first direct evidence for such a role came from a zebrafish study, which demonstrated that retrograde motility of activated JNK and lysosomes is impaired in JIP3 mutant axons (Drerup and Nechiporuk, 2013). More recently, JIP3 inhibition was shown to impair retrograde autophagosome motility in the C. elegans AYI interneuron and in rat hippocampal neurons (Cason et al., 2021; Hill et al., 2019). Consistent with a role for JIP3 in retrograde transport, C. elegans dynein light intermediate chain (DLIC) pulls down UNC-16/JIP3 from COS-7 cell lysate, albeit only when the two proteins are coexpressed together with KLC (Arimoto et al., 2011); and JIP3’s leucine zipper region pulls down the dynein cofactor dynactin from HeLa cell lysate (Montagnac et al., 2009). The molecular details and functional significance of these interactions remain to be determined. Moreover, it is unclear whether JIP3–bound kinesin-1 and JIP3–bound dynein act on the same type of cargo, or whether JIP3 associates with the two motors in separate to carry out distinct functions, namely anterograde transport of vesicle cargo through kinesin-1 and axonal organelle clearance through dynein (Miller, 2017).
JIP3 and its vertebrate paralog JIP4 share their overall domain organization with proteins of the Rab-interacting lysosomal protein (RILP) family, which in vertebrates includes RILP, RILP-like 1 (RILPL1), and RILPL2 (Vilela et al., 2019; Wang et al., 2004). RILP recruits dynein-dynactin to late endosomes and lysosomes (Jordens et al., 2001), and RILPL2 is a cargo adaptor for myosin Va involved in ciliogenesis (Lisé et al., 2009; Schaub and Stearns, 2013). RILPL1 also plays a role in ciliogenesis (Schaub and Stearns, 2013), but an interaction between RILPL1 and cytoskeletal motors has yet to be demonstrated. The homodimeric RILP/JIP3 superfamily is characterized by an N-terminal RILP homology (RH) 1 domain of ∼100 residues, which consists of a four-helix bundle followed by a short coiled-coil segment (Wei et al., 2013); by a coiled-coil region that extends C-terminally from the RH1 domain; and by a four-helix RH2 domain located at variable distance from the RH1 domain (Fig. 1 A). RILPL2’s RH1 domain binds to myosin Va (Wei et al., 2013), JIP3’s RH1 domain includes the KHC-interacting region (Sun et al., 2011), and the RH2 domain binds small GTPases of the Rab family (Wang et al., 2004; Wu et al., 2005). Interestingly, residues 1-240 of C. elegans UNC-16 interact with DLI-1/DLIC in the yeast 2-hybrid assay (Arimoto et al., 2011), raising the question of whether the KHC binding site at the JIP3 N-terminus overlaps with that of DLIC.
Dynein adaptors of the RILP/JIP3 superfamily use their N-terminal RH1 domain to bind the C-terminal helix 1 of dynein light intermediate chain. (A) Domain organization of human dynein light intermediate chain 1 (DLIC1) and the five human cargo adaptors for cytoskeletal motors characterized by the presence of RILP homology (RH) 1 and RH2 domains. JIP3 and JIP4 also have a second coiled-coil region, here called the leucine zipper (LZ) domain. Regions implicated in direct protein-protein interactions are delineated by dashed lines. Residue numbers correspond to UniProt entries Q96NA2-1 (RILP), Q5EBL4-1 (RILPL1), Q969X0 (RILPL2), Q9UPT6-1 (JIP3), O60271-1 (JIP4), and Q9Y6G9 (DLIC1). (B and C) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs described in A except for RILPL1, which is the mouse homolog (Q9JJC6). Molecular weight is indicated in kilodaltons (kD). (D–F) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for RILP[1-106] are the same in D–F. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData F1.
Dynein adaptors of the RILP/JIP3 superfamily use their N-terminal RH1 domain to bind the C-terminal helix 1 of dynein light intermediate chain. (A) Domain organization of human dynein light intermediate chain 1 (DLIC1) and the five human cargo adaptors for cytoskeletal motors characterized by the presence of RILP homology (RH) 1 and RH2 domains. JIP3 and JIP4 also have a second coiled-coil region, here called the leucine zipper (LZ) domain. Regions implicated in direct protein-protein interactions are delineated by dashed lines. Residue numbers correspond to UniProt entries Q96NA2-1 (RILP), Q5EBL4-1 (RILPL1), Q969X0 (RILPL2), Q9UPT6-1 (JIP3), O60271-1 (JIP4), and Q9Y6G9 (DLIC1). (B and C) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs described in A except for RILPL1, which is the mouse homolog (Q9JJC6). Molecular weight is indicated in kilodaltons (kD). (D–F) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for RILP[1-106] are the same in D–F. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData F1.
Here, we show that the RH1 domains of RILP, RILPL1, and JIP3/4 are receptors for a conserved C-terminal helix in DLIC that is known to bind several functionally and structurally distinct dynein adaptors (Schroeder et al., 2014; Lee et al., 2018, 2020; Celestino et al., 2019; Renna et al., 2020). We show that the interaction occurs through a hydrophobic pocket in the RH1 domain that in RILPL2 binds myosin Va, and we validate a point mutation in the RH1 domain that specifically abrogates binding to DLIC without perturbing the interaction between JIP3 and KHC. Functional characterization of this JIP3 separation-of-function mutation in C. elegans demonstrates that UNC-16/JIP3’s interaction with DLIC promotes retrograde transport of endo-lysosomal organelles and synaptic vesicles in the anterior process of touch receptor neurons, which have an axon-like microtubule organization. Characterization of additional engineered mutants, including a disease-associated UNC-16 mutation that abrogates KLC binding, supports the idea that JIP3 uses kinesin-1 and dynein to drive bidirectional transport of endo-lysosomal organelles.
Results
Dynein light intermediate chain’s C-terminal helix 1 binds to the RH1 domain of RILP, JIP3, and JIP4
Vertebrates express five proteins with RH1/2 domains: JIP3 and its paralog JIP4, and the three members of the RILP family (Fig. 1 A). RILP and JIP3 are known to bind DLIC, yet they contain none of the three previously characterized structural elements that other cargo adaptors use to accommodate the amphipathic helix 1 in the DLIC C-terminal tail (DLIC-C). Whether JIP4 and the other two members of the RILP family, RILPL1 and RILPL2, interact with DLIC-C has not been tested. We therefore set out to examine how DLIC-C interacts with RH1/2 domain-containing proteins using purified recombinant human proteins and mouse RILPL1. Pull-downs of GST-tagged DLIC1[388-523], which corresponds to DLIC-C of isoform 1, showed that DLIC1-C interacts with the N-terminal halves of RILP and RILPL1, while no interaction was detectable with full-length RILPL2 (Fig. 1 B; note that RILPL2 is half the size of RILP/RILPL1). Pull-downs with additional RILP fragments showed that RILP[1-106], which corresponds to the RH1 domain (RILP-RH1), is sufficient to bind DLIC1-C (Fig. 1 C). Neither the subsequent coiled-coil region RILP[107-200] nor the RH2 domain RILP[240-316] bound to DLIC1-C (Fig. 1 C). We confirmed the interaction between RILP-RH1 and DLIC1-C by size exclusion chromatography (SEC; Fig. 1 D). Further SEC experiments showed that introducing two point mutations (F447A/F448A) into helix 1 of DLIC1-C abrogates binding to RILP-RH1, and that DLIC1[440-455], which corresponds to helix 1, binds RILP-RH1 (Fig. 1, E and F). Analogous experiments with the RH1 domains of JIP3 and JIP4 confirmed that they also bind DLIC1-C, and that they do so in a manner that depends on DLIC1-C helix 1 (Figs. 2 and 3; and Fig. S2).
The C-terminal amphipathic helix 1 of dynein light intermediate chain binds the RH1 domain’s hydrophobic pocket. (A) Top left: Model of the dimeric RH1 domain of human JIP3 (UniProt entry Q9UPT6-1) generated using ColabFold (Mirdita et al., 2022). Bottom left: Sequence alignment of the α3N helix for the RILP/JIP3 superfamily. Open circles denote residues participating in dimer formation in mouse RILPL2 (Wei et al., 2013). Closed circles denote residues participating in the interaction between RILPL2 and myosin Va. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and whose mutation to glutamine in RILPL2 abrogates the interaction with myosin Va. Right: Model of the JIP3 RH1 domain’s 4-helix bundle with the C-terminal helix 1 of DLIC1 (residues 440–455) docked at the hydrophobic pocket, as predicted by ColabFold. A phenylalanine and a valine side chain in DLIC1 and JIP3, respectively, which we mutate in this study, are also rendered. The models have a per-residue confidence (pLDDT) of >90% throughout and a consistently low alignment error (PAE) (Fig. S2 A; Tunyasuvunakool et al., 2021). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs. WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). (C) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. Molecular weight is indicated in kilodaltons (kD). (D and E) Sedimentation velocity AUC profiles with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved values indicate that both proteins are dimeric in solution. (F–H) Thermograms and binding isotherms of representative ITC titrations. JIP3[1-108] concentration is the concentration of the dimer. The dissociation constant (KD) and the binding stoichiometry (N) are given as mean ± SD (n = 3) and were derived from fitting to the binding isotherm (black line) with a One Set of Sites model using ORIGIN software. Source data are available for this figure: SourceData F2.
The C-terminal amphipathic helix 1 of dynein light intermediate chain binds the RH1 domain’s hydrophobic pocket. (A) Top left: Model of the dimeric RH1 domain of human JIP3 (UniProt entry Q9UPT6-1) generated using ColabFold (Mirdita et al., 2022). Bottom left: Sequence alignment of the α3N helix for the RILP/JIP3 superfamily. Open circles denote residues participating in dimer formation in mouse RILPL2 (Wei et al., 2013). Closed circles denote residues participating in the interaction between RILPL2 and myosin Va. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and whose mutation to glutamine in RILPL2 abrogates the interaction with myosin Va. Right: Model of the JIP3 RH1 domain’s 4-helix bundle with the C-terminal helix 1 of DLIC1 (residues 440–455) docked at the hydrophobic pocket, as predicted by ColabFold. A phenylalanine and a valine side chain in DLIC1 and JIP3, respectively, which we mutate in this study, are also rendered. The models have a per-residue confidence (pLDDT) of >90% throughout and a consistently low alignment error (PAE) (Fig. S2 A; Tunyasuvunakool et al., 2021). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs. WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). (C) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. Molecular weight is indicated in kilodaltons (kD). (D and E) Sedimentation velocity AUC profiles with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved values indicate that both proteins are dimeric in solution. (F–H) Thermograms and binding isotherms of representative ITC titrations. JIP3[1-108] concentration is the concentration of the dimer. The dissociation constant (KD) and the binding stoichiometry (N) are given as mean ± SD (n = 3) and were derived from fitting to the binding isotherm (black line) with a One Set of Sites model using ORIGIN software. Source data are available for this figure: SourceData F2.
Kinesin heavy chain also binds to the RH1 domain of JIP3 but does not compete for binding with dynein light intermediate chain. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs and to mouse kinesin heavy chain KIF5C (UniProt entry P28738). Molecular weight is indicated in kilodaltons (kD). (C and D) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for GST::DLIC1[388-523] are shown twice in C, and the elution profile and gel for GST::KIF5C[807-956] are shown twice in D. Molecular weight is indicated in kilodaltons (kD). WT denotes wild type. (E) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of amylose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from amylose resin after MBP pull-down. The actual amount of DLIC1[388-523] in the pull-down reaction was fivefold higher than what is shown for the input. The KIF5C fragment was used at two concentrations that differ threefold, as indicated above the 5th and 6th lanes from the left. (F) Coomassie Blue-stained SDS-PAGE gel (top) and corresponding immunoblot (bottom) of proteins eluted from amylose resin after MBP pull-down as in E. All proteins contain a 6xHis tag (see Materials and methods) that is detected on the immunoblot. Proteins are the same as in E, but amounts in the pull-down mixture were decreased relative to those in E such that DLIC1-C helix 1 peptide could be added in 150-fold molar excess over the KIF5C fragment. Source data are available for this figure: SourceData F3.
Kinesin heavy chain also binds to the RH1 domain of JIP3 but does not compete for binding with dynein light intermediate chain. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs and to mouse kinesin heavy chain KIF5C (UniProt entry P28738). Molecular weight is indicated in kilodaltons (kD). (C and D) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for GST::DLIC1[388-523] are shown twice in C, and the elution profile and gel for GST::KIF5C[807-956] are shown twice in D. Molecular weight is indicated in kilodaltons (kD). WT denotes wild type. (E) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of amylose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from amylose resin after MBP pull-down. The actual amount of DLIC1[388-523] in the pull-down reaction was fivefold higher than what is shown for the input. The KIF5C fragment was used at two concentrations that differ threefold, as indicated above the 5th and 6th lanes from the left. (F) Coomassie Blue-stained SDS-PAGE gel (top) and corresponding immunoblot (bottom) of proteins eluted from amylose resin after MBP pull-down as in E. All proteins contain a 6xHis tag (see Materials and methods) that is detected on the immunoblot. Proteins are the same as in E, but amounts in the pull-down mixture were decreased relative to those in E such that DLIC1-C helix 1 peptide could be added in 150-fold molar excess over the KIF5C fragment. Source data are available for this figure: SourceData F3.
Dynein light intermediate chain binding occurs via a hydrophobic pocket in the RH1 domain
Our pull-down experiments suggest that all RH1/2 domain-containing proteins except RILPL2 are adaptors for dynein. This is consistent with the well-established role of RILPL2 as an adaptor for myosin Va (Lisé et al., 2009; Wei et al., 2013). The globular tail domain of myosin Va interacts with the RH1 domain of RILPL2 in part by inserting a short helical segment into a hydrophobic pocket that is formed by the RH1 helical bundle, with two binding sites per RH1 dimer (Wei et al., 2013; Fig. 2 A). Since the amphipathic DLIC-C helix 1 is known to insert into hydrophobic pockets formed by the Hook domain, CC1 box, and EF-hand in different adaptors (Lee et al., 2018; Lee et al., 2020), we speculated that the hydrophobic pocket of the RH1 domain might be used to accommodate DLIC-C helix 1 in RILP, RILPL1, JIP3, and JIP4. High-confidence structure predictions, generated by the AlphaFold2-based pipeline ColabFold (Mirdita et al., 2022), suggest that this is indeed the case (Fig. 2 A and Fig. S2 A). Mutating a single residue (V59Q) in the hydrophobic pocket of mouse RILPL2-RH1 was shown to abrogate the myosin Va interaction (Wei et al., 2013). To directly test whether the other RH1 domains bind DLIC-C helix 1 through the same hydrophobic pocket, we introduced the analogous point mutation into human RILP (V59Q), JIP3 (V60Q), and JIP4 (V55Q; Fig. 2 A). GST pull-downs with purified recombinant proteins showed that full-length RILP carrying the V59Q mutation had significantly reduced affinity for GST-tagged DLIC1-C (Fig. 2 B), as did mutated RILP-RH1 in SEC experiments (Fig. S1 A). To gain a more quantitative understanding of the interaction, we sought to perform isothermal titration calorimetry (ITC). For RILP[1-106], analytical ultracentrifugation (AUC) experiments indicated that the protein was present as a mixture of dimer and trimer (data not shown). Because of this heterogeneity of RILP-RH1, we used JIP3[1-108] and JIP4[1-103] for ITC experiments, since AUC runs showed that these proteins behave as monodisperse dimers (Fig. 2, C and D; and Fig. S2, B and C). ITC measurements revealed that JIP3-RH1 and JIP4-RH1 bind to DLIC1-C with low micromolar affinity (KD of ∼1 and ∼4 µM, respectively) and with a reaction stoichiometry that indicates two DLIC1-C molecules per RH1 dimer (Fig. 2 F and Fig. S2 D). This is similar to what has been determined for the interaction between DLIC1-C and other dynein adaptors (Lee et al., 2018; Lee et al., 2020). As expected, there was no detectable signal in the thermogram when DLIC1-C helix 1 was mutated (Fig. 2 G and Fig. S2 E). Introducing the V/Q mutation into JIP3-RH1 and JIP4-RH1 had the same effect (Fig. 2 H and Fig. S2 F). Importantly, AUC confirmed that the V60Q mutation does not perturb dimerization of JIP3-RH1 (Fig. 2 E), consistent with previous analysis of this mutation in RILPL2 (Wei et al., 2013). These results suggest that RILP, JIP3, JIP4, and most likely also RILPL1, bind to DLIC1-C helix 1 via a hydrophobic pocket in their RH1 domain, which in RILPL2 accommodates helix 2 of myosin Va’s globular tail domain.
The V59Q and P56F mutations in RILP reduce the affinity for dynein light intermediate chain. (A) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. Proteins correspond to the human homologs. The elution profile and gel for GST::DLIC1[388-523] are shown twice, and the elution profile and gel for RILP[1-106] WT are the same as in Fig. 1 D. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (B) Left: Sequence alignment of the α3N helix for RILP family proteins in selected vertebrates and invertebrates. Boxed residues denote the phenylalanine in RILPL2 that is critical for myosin Va binding and the proline that typically replaces the phenylalanine in RILP and RILPL1. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and is mutated in A. Right: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input) and after elution from the resin (GST pull-down). Proteins correspond to the human homologs. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData FS1.
The V59Q and P56F mutations in RILP reduce the affinity for dynein light intermediate chain. (A) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. Proteins correspond to the human homologs. The elution profile and gel for GST::DLIC1[388-523] are shown twice, and the elution profile and gel for RILP[1-106] WT are the same as in Fig. 1 D. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (B) Left: Sequence alignment of the α3N helix for RILP family proteins in selected vertebrates and invertebrates. Boxed residues denote the phenylalanine in RILPL2 that is critical for myosin Va binding and the proline that typically replaces the phenylalanine in RILP and RILPL1. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and is mutated in A. Right: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input) and after elution from the resin (GST pull-down). Proteins correspond to the human homologs. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData FS1.
The V55Q mutation in JIP4 abrogates the binding to dynein light intermediate chain. (A) Per-residue confidence score (pLDDT; blue: high confidence, red: low confidence) of the human JIP3[1-100] dimer bound to two copies of human DLIC1[440-455] (left) and corresponding Predicted Alignment Error (PAE) plot (right). (B) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (C) Sedimentation velocity AUC profile with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved value indicates that JIP4 is dimeric in solution. (D–F) Thermograms and binding isotherms of representative ITC titrations. JIP4[1-103] concentration is the concentration of the dimer. The data in D were best described by a model corresponding to two sites with a single macroscopic dissociation constant (KD) using the SEDPHAT software package. The KD value was determined by global fitting of the two independent experiments shown in D with a 68% confidence interval of 2.8–7.4 µM. Source data are available for this figure: SourceData FS2.
The V55Q mutation in JIP4 abrogates the binding to dynein light intermediate chain. (A) Per-residue confidence score (pLDDT; blue: high confidence, red: low confidence) of the human JIP3[1-100] dimer bound to two copies of human DLIC1[440-455] (left) and corresponding Predicted Alignment Error (PAE) plot (right). (B) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (C) Sedimentation velocity AUC profile with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved value indicates that JIP4 is dimeric in solution. (D–F) Thermograms and binding isotherms of representative ITC titrations. JIP4[1-103] concentration is the concentration of the dimer. The data in D were best described by a model corresponding to two sites with a single macroscopic dissociation constant (KD) using the SEDPHAT software package. The KD value was determined by global fitting of the two independent experiments shown in D with a 68% confidence interval of 2.8–7.4 µM. Source data are available for this figure: SourceData FS2.
The high sequence homology between the RH1 domains of RILP/RILPL1 and RILPL2 raises the question of how these proteins bind specifically to DLIC and myosin Va, respectively. Mouse RILPL2 contacts myosin Va’s globular tail domain in part through the phenylalanine at position 56, which is conserved in RIPL2 proteins but is replaced by proline in RILP/RILPL1 proteins (Fig. S1 B; Wei et al., 2013). Previous work showed that mutating F56 to proline abrogates RILPL2 binding to myosin Va, while the P55F mutation enables mouse RILP to bind myosin Va with even higher affinity than RILPL2 (Wei et al., 2013). We found that the P56F mutation in human RILP reduced the binding to GST-tagged DLIC1-C in pull-down experiments, consistent with its conversion to a myosin Va binder, while the F66P mutation in human RILPL2 did not enable it to bind DLIC1-C (Fig. S1 B). Switching from myosin Va to DLIC binding therefore requires additional changes in the RH1 domain besides the phenylalanine to proline substitution.
The JIP3 V60Q mutation does not affect binding to kinesin heavy chain
JIP3 binds the C-terminal tail of KHC, and pull-downs from mouse brain lysate using JIP3 fragments previously suggested that residues 50-80 of JIP3 are required for the interaction (Sun et al., 2011). Since the JIP3 V60Q mutation falls within this region, we asked whether this mutation also affects binding to the KHC tail. Using purified recombinant proteins, we first confirmed that JIP3-RH1 binds GST-tagged mouse KIF5C[807-956] using GST pull-downs and SEC (Fig. 3, A and D). Although we could not perform ITC measurements with KIF5C[807-956] because of the poor solubility of this fragment, a side-by-side comparison of pull-downs indicated that the KHC tail binds more strongly to JIP3-RH1 than DLIC1-C (Fig. 3 A). Introducing the V60Q mutation into JIP3-RH1 abrogated binding to DLIC1-C but had no effect on the interaction with the KHC tail in GST pull-down and SEC experiments (Fig. 3, B–D). Of note, the amount of GST::KIF5C[807-956] eluting from the SEC column increased significantly when JIP3-RH1 was present, suggesting that binding to the RH1 domain improves the solubility of the KHC fragment (Fig. 3 D). The JIP3-RH1 V60Q mutant had the same stabilizing effect on GST::KIF5C[807-956], which further supports the idea that this mutation does not perturb the interaction with the KHC tail. We conclude that JIP3-RH1 binds to both DLIC and KHC, and that JIP3 V60Q is a separation-of-function mutation that specifically abrogates the interaction between JIP3 and DLIC.
We also sought to address whether the KHC tail and DLIC1-C can bind to JIP3-RH1 simultaneously. Unfortunately, at the relatively low protein concentrations, we had to use for SEC experiments with the KHC fragment (which exhibited poor solubility even when tagged with GST), JIP3-RH1 and DLIC1-C no longer coeluted robustly from the column, so we could not use SEC to determine whether JIP3-RH1 can form a tripartite complex with the KHC tail and DLIC1-C. As an alternative approach, we performed pull downs with MBP-tagged JIP3[1-108] in the presence of GST::KIF5C[807-956] alone, DLIC1[388-523] alone, or both GST::KIF5C[807-956] and DLIC1[388-523]. The KHC tail and DLIC1-C were pulled down by JIP3-RH1 in similar amounts regardless of whether or not the other fragment was present (Fig. 3 E). To further probe whether the KHC tail and DLIC1-C compete for binding to JIP3-RH1, we added an excess of DLIC-C helix 1 peptide (residues 433-458) to the pull down mixture. This displaced DLIC1-C but not the KHC tail from JIP3-RH1 (Fig. 3 F). These experiments suggest that significant overlap between the DLIC and KHC binding sites on JIP3-RH1 is unlikely.
C. elegans UNC-16 V72Q is equivalent to human JIP3 V60Q
To explore the functional significance of the interaction between DLIC and JIP3, we turned to C. elegans, whose unc-16 gene encodes the single homolog of JIP3 and JIP4. C. elegans offers the advantage that unc-16 mutants can be propagated in a homozygous state, so there is no residual maternal wild-type protein which could potentially confound phenotypic interpretation. Before proceeding to in vivo analysis, we asked whether the interactions we identified among human proteins are conserved in the nematode. Using pull-downs with purified recombinant proteins, we confirmed that UNC-16[1-120], which corresponds to the RH1 domain, binds to DLI-1[369-442], which corresponds to DLIC-C; and that the V72Q mutation in UNC-16, which corresponds to the V60Q mutation in human JIP3, abrogates binding to DLIC-C (Fig. S3 A). Furthermore, UNC-16[1-120] bound to UNC-116[675-815], which corresponds to the KHC tail, and the V72Q mutation in UNC-16 had no apparent effect on this interaction (Fig. S3 B). We conclude that human JIP3 V60Q and C. elegans UNC-16 V72Q are equivalent mutations: they abrogate RH1 domain binding to DLIC without perturbing the interaction between the RH1 domain and KHC.
Human JIP3 V60Q is equivalent to C. elegans UNC-16 V72Q. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the C. elegans homologs of JIP3 (UNC-16; UniProt entry P34609-1), kinesin heavy chain KIF5C (UNC-116; P34540), and dynein light intermediate chain (DLI-1; G5ED34). WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). MBP::UNC-16[1-120] proteins (WT and V72Q) also contain a C-terminal Strep-tag II, which is detected on the immunoblot in A along with GST::DLI-1[369-442]. Source data are available for this figure: SourceData FS3.
Human JIP3 V60Q is equivalent to C. elegans UNC-16 V72Q. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the C. elegans homologs of JIP3 (UNC-16; UniProt entry P34609-1), kinesin heavy chain KIF5C (UNC-116; P34540), and dynein light intermediate chain (DLI-1; G5ED34). WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). MBP::UNC-16[1-120] proteins (WT and V72Q) also contain a C-terminal Strep-tag II, which is detected on the immunoblot in A along with GST::DLI-1[369-442]. Source data are available for this figure: SourceData FS3.
Dynein light intermediate chain binding is essential for UNC-16 function in vivo
We next generated animals expressing UNC-16 V72Q by editing the unc-16 locus using the CRISPR/Cas9 method (Fig. 4 A). To assess UNC-16 protein levels, we generated an affinity-purified antibody, which recognizes a protein of the predicted size for full-length UNC-16 (134 kD) on immunoblots of adult worm lysate (Fig. 4 B). This protein is missing in animals carrying unc-16(ce483), a previously characterized loss-of-function mutation (Edwards et al., 2013). Immunoblotting confirmed that UNC-16 V72Q is expressed at the same levels as wild-type UNC-16 (Fig. 4 B and Fig. S4 A). To assess the effect of the unc-16(V72Q) mutation on animal behavior, we determined body bending frequency in liquid at the young adult stage. This showed that unc-16(V72Q) animals are locomotion deficient, and that this phenotype is as severe as in unc-16(ce483) animals (Fig. 4 C). We conclude that the interaction with DLI-1 is essential for UNC-16 function in vivo.
Displacing dynein light intermediate chain from C. elegans UNC-16/JIP3 through the UNC-16 V72Q mutation results in accumulation of endo-lysosomal organelles at the neurite tip of touch receptor neurons. (A) Top: Domain organization of the C. elegans UNC-16/JIP3 N-terminal region. Residue numbers correspond to isoform e (UniProt entry P34609-1). Bottom: Description of unc-16 mutants affecting the UNC-16 N-terminal region that are characterized in this study. (B) Immunoblot of adult C. elegans lysates using an affinity-purified rabbit polyclonal antibody raised against UNC-16 residues 1–506. The membrane was reprobed with an anti-α-tubulin antibody as a loading control. Molecular weight is in kilodaltons (kD). (C) Locomotion of animals at the young adult stage, assessed by determining body bending frequency (mean ± SEM) in liquid medium. n denotes the number of animals examined. Statistical significance (wild-type N2 control versus unc-16 mutants) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001. (D) Left: Location of the C. elegans anterior touch receptor neurons. ALM and AVM are the anterior lateral and anterior ventral mechanosensory neurons, respectively, which extend processes into the nose and the nerve ring. There are two ALM neurons, which are equivalent for the purpose of this study. Note that the neurite tip does not contain synapses, which are instead located in the nerve ring and were not imaged in this study. Right: Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2) or early endosomes (mKate2::RAB-5) in touch receptor neurons. Scale bar, 20 µm. (E) Number of CTNS-1::mKate2 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). n denotes the number of neurites examined (1 per animal). Statistical significance (control versus unc-16 mutants) was determined as described for C. ****P < 0.0001. (F) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing mKate2-tagged markers for endo-lysosomal organelles or synaptic vesicle precursors. n denotes the number of neurites examined (1 per animal). (G–J) Top: Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM cell body and neurite tip. Scale bars, 2 µm. Bottom: Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined (1 per animal). Statistical significance was determined as described for C. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. Source data are available for this figure: SourceData F4.
Displacing dynein light intermediate chain from C. elegans UNC-16/JIP3 through the UNC-16 V72Q mutation results in accumulation of endo-lysosomal organelles at the neurite tip of touch receptor neurons. (A) Top: Domain organization of the C. elegans UNC-16/JIP3 N-terminal region. Residue numbers correspond to isoform e (UniProt entry P34609-1). Bottom: Description of unc-16 mutants affecting the UNC-16 N-terminal region that are characterized in this study. (B) Immunoblot of adult C. elegans lysates using an affinity-purified rabbit polyclonal antibody raised against UNC-16 residues 1–506. The membrane was reprobed with an anti-α-tubulin antibody as a loading control. Molecular weight is in kilodaltons (kD). (C) Locomotion of animals at the young adult stage, assessed by determining body bending frequency (mean ± SEM) in liquid medium. n denotes the number of animals examined. Statistical significance (wild-type N2 control versus unc-16 mutants) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001. (D) Left: Location of the C. elegans anterior touch receptor neurons. ALM and AVM are the anterior lateral and anterior ventral mechanosensory neurons, respectively, which extend processes into the nose and the nerve ring. There are two ALM neurons, which are equivalent for the purpose of this study. Note that the neurite tip does not contain synapses, which are instead located in the nerve ring and were not imaged in this study. Right: Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2) or early endosomes (mKate2::RAB-5) in touch receptor neurons. Scale bar, 20 µm. (E) Number of CTNS-1::mKate2 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). n denotes the number of neurites examined (1 per animal). Statistical significance (control versus unc-16 mutants) was determined as described for C. ****P < 0.0001. (F) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing mKate2-tagged markers for endo-lysosomal organelles or synaptic vesicle precursors. n denotes the number of neurites examined (1 per animal). (G–J) Top: Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM cell body and neurite tip. Scale bars, 2 µm. Bottom: Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined (1 per animal). Statistical significance was determined as described for C. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. Source data are available for this figure: SourceData F4.
Mutant UNC-16 levels are comparable to WT; late endosomes but not synaptic vesicle precursors are distributed differently in unc-16(ce483) versus unc-16(V72Q); unc-16(ce483) is a null mutant. (A) Fluorescent immunoblot of adult C. elegans lysates with an antibody against UNC-16 (left) and corresponding quantification of the UNC-16 signal in unc-16 mutants (right). Individual measurements and the average (horizontal bar) of three immunoblots are shown. α-tubulin was used as the loading control. Molecular weight is in kilodaltons (kD). (B) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for late endosomes (mKate2::RAB-7) or synaptic vesicle precursors (SNB-1::mKate2) in touch receptor neurons. Scale bar, 20 µm. (C) Left: Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for early endosomes (mKate2::RAB-5). Right: Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined. Statistical significance was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. Source data are available for this figure: SourceData FS4.
Mutant UNC-16 levels are comparable to WT; late endosomes but not synaptic vesicle precursors are distributed differently in unc-16(ce483) versus unc-16(V72Q); unc-16(ce483) is a null mutant. (A) Fluorescent immunoblot of adult C. elegans lysates with an antibody against UNC-16 (left) and corresponding quantification of the UNC-16 signal in unc-16 mutants (right). Individual measurements and the average (horizontal bar) of three immunoblots are shown. α-tubulin was used as the loading control. Molecular weight is in kilodaltons (kD). (B) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for late endosomes (mKate2::RAB-7) or synaptic vesicle precursors (SNB-1::mKate2) in touch receptor neurons. Scale bar, 20 µm. (C) Left: Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for early endosomes (mKate2::RAB-5). Right: Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined. Statistical significance was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. Source data are available for this figure: SourceData FS4.
UNC-16(V72Q) causes prominent accumulation of endo-lysosomal organelles at the neurite tip of touch receptor neurons
To determine the consequences of the unc-16(V72Q) mutation for axonal transport, we set out to examine the distribution of organelles and vesicles in touch receptor neurons (TRNs). The six TRNs are situated underneath the cuticle and each have a long anteriorly directed neurite, in which microtubules are uniformly oriented toward the neurite tip (Arimoto et al., 2011). Therefore, kinesin-1 (along with other plus end-directed kinesins) and dynein mediate anterograde and retrograde transport in the neurite, respectively. We focused on the two ALM neurons, whose neurites extend into the head of the worm and branch off before the neurite tip to form synapses in the nerve ring (Fig. 4 D). To visualize different types of cargo, we used fluorescently tagged markers for lysosomes (CTNS-1::mKate2), late endosomes (mKate2::RAB-7), early endosomes (mKate2::RAB-5), early/recycling endosomes (mKate2::SYX-7), and synaptic vesicle precursors (SNB-1::mKate2), which were stably expressed in TRNs from the mec-7 promoter after single-copy transgene integration using the MosSCI method (Frøkjær-Jensen et al., 2012). Consistent with prior work in motor neurons (Edwards et al., 2013), the unc-16(ce483) mutation increased the amount of CTNS-1::mKate2, mKate2::RAB-7, mKate2::RAB-5, and SNB-1::mKate2 in the ALM neurite at the larval L4 stage (Fig. 4 D and Fig. S4 B), which was quantified by counting the number of CTNS-1::mKate2 puncta present in the proximal, mid-, and distal neurite (Fig. 4 E); by determining the fluorescence intensity profile of the markers along the entire neurite (Fig. 4 F); and by measuring the integrated fluorescence intensity of the markers at the neurite tip and in the cell body (Fig. 4, G–J). In all cases, an increase of signal in the neurite was accompanied by a decrease of signal in the cell body.
We considered the possibility that unc-16(ce483) may not be a true null allele, since the nonsense mutation after Q304 could, in principle, allow expression of an UNC-16 fragment that includes the RH1 domain and the first coiled-coil region. We therefore generated the knock-out allele unc-16(prt183) by removing the entire open reading frame and compared the intraneuronal distribution of mKate2::RAB-5 in unc-16(prt183) and unc-16(ce483) animals (Fig. S4 C). The two alleles had similar effects on mKate2::RAB-5 distribution, suggesting that unc-16(ce483) is a null mutant. The identical appearance of unc-16(ce483) and unc-16(prt183) on immunoblots and their identical effect on animal locomotion further supports this idea (Fig. 4, B and C). Examination of mKate2::SYX-7 localization in the unc-16(prt183) mutant showed that mKate2::SYX-7 puncta were broadly distributed throughout the ALM neurite, while mKate2::SYX-7 puncta in the control were almost exclusively found near the cell body (Fig. S5, B–D). Thus, the effect of unc-16(prt183) on mKate2::SYX-7 distribution is similar to that observed for other vesicle/organelle markers in the two unc-16 null mutants. We have not examined mitochondria in unc-16(ce483) or unc-16(prt183) animals but note that increased mitochondrial density in TRN neurites has been reported for other unc-16 mutants (Sure et al., 2018).
Recycling endosomes accumulate at the ALM neurite tip in the unc-16(V72Q) mutant and do so independently of JIP-1. (A) Genomic locus of jip-1 and deletion in the jip-1(prt187) mutant introduced by genome editing. (B) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for recycling endosomes (mKate2::SYX-7) in touch receptor neurons. Merged image on the right shows the location of the fluorescence signal relative to the differential interference contrast (DIC) image of the animal. Arrow points to the signal at the neurite tip. Note that diffuse signal in the neurite and the cell body is not readily discernable with this marker, which precludes determination of the fluorescence intensity profile along the neurite and intensity measurements in the cell body. Scale bar, 20 µm. (C) Number of mKate2::SYX-7 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). n denotes the number of neurites examined (1 per animal). Statistical significance (control versus mutants; other comparisons indicated by brackets) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. (D) Integrated fluorescence intensity (mean ± SEM, normalized to control) in the distal tip of the ALM neurite. The number of neurites examined (1 per animal) corresponds to the number n in C. Statistical significance was determined as described for C. ****P < 0.0001; **P < 0.01; ns = not significant, P > 0.05.
Recycling endosomes accumulate at the ALM neurite tip in the unc-16(V72Q) mutant and do so independently of JIP-1. (A) Genomic locus of jip-1 and deletion in the jip-1(prt187) mutant introduced by genome editing. (B) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for recycling endosomes (mKate2::SYX-7) in touch receptor neurons. Merged image on the right shows the location of the fluorescence signal relative to the differential interference contrast (DIC) image of the animal. Arrow points to the signal at the neurite tip. Note that diffuse signal in the neurite and the cell body is not readily discernable with this marker, which precludes determination of the fluorescence intensity profile along the neurite and intensity measurements in the cell body. Scale bar, 20 µm. (C) Number of mKate2::SYX-7 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). n denotes the number of neurites examined (1 per animal). Statistical significance (control versus mutants; other comparisons indicated by brackets) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. (D) Integrated fluorescence intensity (mean ± SEM, normalized to control) in the distal tip of the ALM neurite. The number of neurites examined (1 per animal) corresponds to the number n in C. Statistical significance was determined as described for C. ****P < 0.0001; **P < 0.01; ns = not significant, P > 0.05.
Similar to the two unc-16 null mutants, the unc-16(V72Q) mutant exhibited increased and decreased amounts of vesicle/organelle markers in the ALM neurite and cell body, respectively. However, with the exception of SNB-1::mKate2, the distribution profile of these markers in the neurite was distinct in the unc-16(V72Q) mutant. In contrast to unc-16 null mutants, unc-16(V72Q) resulted in prominent accumulation of CTNS-1::mKate2, mKate2::RAB-7, mKate2::RAB-5, and mKate2::SYX-7 at the neurite tip (Fig. 4, D–J; and Fig. S5, B and D). The effect was particularly striking for mKate2::RAB-5. Thus, when compared to unc-16 null mutants, unc-16(V72Q) shifts the distribution of endo-lysosomal markers in the neurite further in the anterograde direction.
The phenotype of the unc-16(V72Q) mutant is consistent with compromised retrograde transport. If this is the case, dynein mutants should result in a similar accumulation of endo-lysosomal markers at the neurite tip. We therefore directly compared unc-16(V72Q) with dli-1(F392A/F393A), which corresponds to the F447A/F448A mutation in human DLIC1 that abrogates binding to cargo adaptors (Celestino et al., 2019), including JIP3 as shown in this study (Fig. 2 G). We found that unc-16(V72Q) and dli-1(F392A/F393A) mutants have a similar distribution of CTNS-1::mKate2 and mKate2::RAB-5 along the neurite with a concomitant reduction of signal in the cell body (Fig. 5, A–C), although mKate2::RAB-5 accumulation at the neurite tip was not quite as pronounced in dli-1(F392A/F393A). We note that since dli-1(F392A/F393A) animals are sterile, we could only analyze the first generation of homozygous mutants, which are likely to retain some residual maternal wild-type DLI-1. We conclude that mutating either the DLI-1 binding site in UNC-16 or the UNC-16 binding site in DLI-1 results in a similar distribution of endo-lysosomal organelles in TRNs.
Mutations in the C-terminal helix 1 of dynein light intermediate chain and the UNC-16/JIP3 V72Q mutation result in similar transport defects in touch receptor neurons. (A and (B) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for early endosomes (mKate2::RAB-5) or lysosomes (CTNS-1::mKate2). n denotes the number of neurites examined (1 per animal). Graph in A corresponds to the data in Fig. 4 F, re-plotted with a split y-axis to highlight the difference between unc-16(ce483) and the control. (C) Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined (1 per animal). Statistical significance [control versus mutants and unc-16(V72Q) versus dli-1(F392A/F393A)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001; ns = not significant, P > 0.05. (D) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM cell body and neurite tip in L4 animals expressing transgene-encoded UNC-16::GFP [background: endogenous wild-type unc-16 with or without dli-1(F392A/F393A)] or UNC-16(V72Q)::GFP [background: endogenous unc-16(V72Q)] in touch receptor neurons. Scale bar, 2 µm. (E) Integrated fluorescence intensity in the ALM cell body and neurite tip for the GFP-tagged UNC-16 versions described in D, plotted and statistically analyzed as in C. ****P < 0.0001; ns = not significant, P > 0.05. (F) Fluorescence intensity profiles (mean ± SEM) at the ALM neurite tip for the GFP-tagged UNC-16 versions described in D. n denotes the number of neurites examined (1 per animal). (G) Fluorescence kymographs (inverted grayscale) of mKate2::RAB-5 particle motility in the ALM neurite, generated from time-lapse sequences (single z-section) recorded at the larval L2 stage. The imaged region is ∼50 µm away from the cell body, which is located to the left. Scale bar, 5 µm. (H–J) Motility parameters (mean ± SEM) of mKate2::RAB-5 particles, derived from kymograph analysis. For H, n denotes the number of neurites examined (1 per animal). For I and J, n denotes the number of track segments, framed by a pause or a reversal, from at least 15 neurites (1 per animal). Statistical significance was determined as described for C. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05.
Mutations in the C-terminal helix 1 of dynein light intermediate chain and the UNC-16/JIP3 V72Q mutation result in similar transport defects in touch receptor neurons. (A and (B) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for early endosomes (mKate2::RAB-5) or lysosomes (CTNS-1::mKate2). n denotes the number of neurites examined (1 per animal). Graph in A corresponds to the data in Fig. 4 F, re-plotted with a split y-axis to highlight the difference between unc-16(ce483) and the control. (C) Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined (1 per animal). Statistical significance [control versus mutants and unc-16(V72Q) versus dli-1(F392A/F393A)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001; ns = not significant, P > 0.05. (D) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM cell body and neurite tip in L4 animals expressing transgene-encoded UNC-16::GFP [background: endogenous wild-type unc-16 with or without dli-1(F392A/F393A)] or UNC-16(V72Q)::GFP [background: endogenous unc-16(V72Q)] in touch receptor neurons. Scale bar, 2 µm. (E) Integrated fluorescence intensity in the ALM cell body and neurite tip for the GFP-tagged UNC-16 versions described in D, plotted and statistically analyzed as in C. ****P < 0.0001; ns = not significant, P > 0.05. (F) Fluorescence intensity profiles (mean ± SEM) at the ALM neurite tip for the GFP-tagged UNC-16 versions described in D. n denotes the number of neurites examined (1 per animal). (G) Fluorescence kymographs (inverted grayscale) of mKate2::RAB-5 particle motility in the ALM neurite, generated from time-lapse sequences (single z-section) recorded at the larval L2 stage. The imaged region is ∼50 µm away from the cell body, which is located to the left. Scale bar, 5 µm. (H–J) Motility parameters (mean ± SEM) of mKate2::RAB-5 particles, derived from kymograph analysis. For H, n denotes the number of neurites examined (1 per animal). For I and J, n denotes the number of track segments, framed by a pause or a reversal, from at least 15 neurites (1 per animal). Statistical significance was determined as described for C. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05.
To examine how abrogating the interaction between DLI-1 and UNC-16 impacts the distribution of UNC-16 itself, we expressed transgenic UNC-16::GFP in TRNs from the mec-4 promoter after single-copy integration. Consistent with previous reports on UNC-16::GFP localization (Byrd et al., 2001; Choudhary et al., 2017), UNC-16::GFP was detectable in the cell body (slightly enriched at what presumably is the golgi) and at the very tip of the neurite (Fig. 5, D–F). Expression of UNC-16(V72Q)::GFP in the unc-16(V72Q) background increased neurite tip levels, and a similar effect was observed for UNC-16::GFP in the dli-1(F392A/F393A) background. This was accompanied by a decrease of signal in the cell body (Fig. 5, D–F). We conclude that abrogating the interaction between UNC-16 and DLI-1 increases the amount of UNC-16 in the neurite at the expense of cell body localization.
The unc-16(V72Q) mutant exhibits anterograde bias of mKate2::RAB-5 transport in TRN neurites
We next examined the transport kinetics of mKate2::RAB-5, the marker with the most striking difference in distribution between unc-16(V72Q) and unc-16 null mutants. Analysis of kymographs, constructed from time-lapse imaging sequences acquired in the middle segment of the ALM neurite at the larval L2 stage, showed that mKate2::RAB-5 particles exhibit bidirectional movement in control animals with ∼40% of particles remaining stationary for the duration of imaging (30 s; Fig. 5, G and H). In the unc-16(ce483) mutant, the fraction of mKate2::RAB-5 particles moving exclusively in the anterograde or retrograde direction was decreased, and there were also less particles that reversed direction during the run. Consequently, more than 75% of particles remained stationary. The unc-16(V72Q) mutant showed a similar decrease in the fraction of exclusively retrograde moving particles but not in the fraction of exclusively anterograde moving particles, and there was no significant difference in the fraction of particles that reversed direction or remained stationary (Fig. 5, G and H). We also determined run length and velocity of mKate2::RAB-5 particles, both of which were decreased for either direction in the unc-16(ce483) and unc-16(V72Q) mutant (Fig. 5, I and J). We conclude that mKate2::RAB-5 particles are more mobile in the unc-16(V72Q) mutant than in the unc-16(ce483) mutant, and that transport in the unc-16(V72Q) mutant is biased in the anterograde direction relative to the control and the unc-16(ce483) mutant. This offers an explanation for the prominent accumulation of mKate2::RAB-5 that is observed at the neurite tip in the unc-16(V72Q) mutant.
Accumulation of endo-lysosomal markers at the neurite tip in the unc-16(V72Q) mutant requires kinesin-1 activity
So far, our analysis was consistent with the idea that displacing DLI-1 from UNC-16 in the unc-16(V72Q) mutant results in enhanced anterograde transport of endo-lysosomal organelles compared to what is observed in unc-16 null mutants. To address whether kinesin-1 is implicated in this aspect of the unc-16(V72Q) phenotype, we used unc-116(e2310), a KHC reduction-of-function mutant (Patel et al., 1993). In unc-16(V72Q); unc-116(e2310) double mutants, we no longer observed accumulation of CTNS-1::mKate2, mKate2::RAB-7, or mKate2::RAB-5 at the neurite tip, and (with the exception of mKate2::RAB-7) this was accompanied by an increase of signal in the cell body relative to the unc-16(V72Q) single mutant (Fig. 6, B and C). This shows that neurite tip accumulation of endo-lysosomal markers in the unc-16(V72Q) mutant requires kinesin-1 activity. Previous characterization of unc-116(e2310) and other reduction-of-function alleles established that downregulation of kinesin-1 activity does not perturb microtubule orientation in the axon of DA9 (motor) and PHC (sensory) neurons (Yan et al., 2013), nor in the anterior neurite of ALM neurons (Arimoto et al., 2011). Thus, given that microtubule orientation likely remains correctly plus-end-out in the unc-16(V72Q); unc-116(e2310) mutant, our result suggests that neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant is the result of anterograde transport by kinesin-1.
Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires kinesin-1 activity but not MAP kinase signalling, and N-terminal UNC-16 deletions mimic the unc-16(V72Q) mutant. (A) Domain organization of the N-terminal UNC-16 region with the putative binding sites for UNC-116/KHC and JNK-1 delineated by dashed lines. (B and D) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2), late endosomes (mKate2::RAB-7), or early endosomes (mKate2::RAB-5) in touch receptor neurons. n denotes the number of neurites examined (1 per animal). (C and E) Integrated fluorescence intensity [mean ± SEM, normalized to unc-16(V72Q)] in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined in C and E corresponds to the number n in B and D, respectively. Statistical significance was determined by the Mann-Whitney test. ****P < 0.0001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. (F and G) Left: Fluorescence intensity profiles (mean ± SEM) in the ALM neurite, as described for B and D. Right: Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(V72Q) for F and to the control for G) in the cell body and at the neurite tip. The number of neurites examined corresponds to the number n in the fluorescence intensity profiles on the left. Statistical significance (control versus mutants and unc-16(V72Q) versus unc-16(V72Q); jnk-1(gk7)) was determined by the Mann-Whitney test for F and by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test for G. ****P < 0.0001; *P < 0.05; ns = not significant, P > 0.05.
Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires kinesin-1 activity but not MAP kinase signalling, and N-terminal UNC-16 deletions mimic the unc-16(V72Q) mutant. (A) Domain organization of the N-terminal UNC-16 region with the putative binding sites for UNC-116/KHC and JNK-1 delineated by dashed lines. (B and D) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2), late endosomes (mKate2::RAB-7), or early endosomes (mKate2::RAB-5) in touch receptor neurons. n denotes the number of neurites examined (1 per animal). (C and E) Integrated fluorescence intensity [mean ± SEM, normalized to unc-16(V72Q)] in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined in C and E corresponds to the number n in B and D, respectively. Statistical significance was determined by the Mann-Whitney test. ****P < 0.0001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. (F and G) Left: Fluorescence intensity profiles (mean ± SEM) in the ALM neurite, as described for B and D. Right: Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(V72Q) for F and to the control for G) in the cell body and at the neurite tip. The number of neurites examined corresponds to the number n in the fluorescence intensity profiles on the left. Statistical significance (control versus mutants and unc-16(V72Q) versus unc-16(V72Q); jnk-1(gk7)) was determined by the Mann-Whitney test for F and by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test for G. ****P < 0.0001; *P < 0.05; ns = not significant, P > 0.05.
Neurite tip accumulation of endo-lysosomal markers could be driven by kinesin-1 that is directly bound to UNC-16. Alternatively, the relevant kinesin-1 pool could be associated with another adaptor that is (co-)dependent on UNC-16. Evidence from cultured mouse neurons suggests that JIP1 can bind to and cooperate with JIP3 in anterograde axonal transport (Sun et al., 2017; Hammond et al., 2008). To ask whether C. elegans JIP-1 is the source of the kinesin-1 activity that drives neurite tip accumulation of endo-lysosomal markers in the unc-16(V72Q) mutant, we generated the putative null allele jip-1(prt187) by deleting 7.2 kb of the open reading frame and introducing two nonsense mutations followed by a frameshift after residue L410, i.e., before the binding sites for KHC and KLC (Fig. S5 A). Analysis of intraneuronal mKate2::SYX-7 distribution showed that the jip-1(prt187) single and the jip-1(prt187); unc-16(prt183) double mutant were similar to the control and the unc-16(prt183) single mutant, respectively (Fig. S5, B–D). Although the neurite tip signal of mKate2::SYX-7 in the jip-1(prt187); unc-16(V72Q) double mutant tended to be decreased relative to the neurite tip signal in the unc-16(V72Q) single mutant, the difference did not reach statistical significance (Fig. S5 D). We conclude that JIP-1–bound kinesin-1 may make a contribution but is not the main driver of endo-lysosomal organelle accumulation at the neurite tip in the unc-16(V72Q) mutant.
Combining the UNC-16 V72Q mutation with a disease-associated mutation in UNC-16 that abrogates KLC binding recapitulates the unc-16 null phenotype
Kinesin-1 interacts with JIP3/UNC-16 in two ways: through KHC, which binds to the highly conserved JIP3/UNC-16 RH1 domain (Sun et al., 2011 and this study), and KLC, which binds to the equally well conserved leucine zipper domain that precedes the RH2 domain (Fig. 1 A; Nguyen et al., 2005; Sakamoto et al., 2005). To examine the role of kinesin-1 that is bound to UNC-16, we first focused on the KHC interaction (Fig. 6 A). We reasoned that if the UNC-16 interaction with KHC is important for endo-lysosomal marker accumulation at the neurite tip in the unc-16(V72Q) mutant, this accumulation should not occur when the RH1 domain is deleted. We therefore generated animals expressing an UNC-16 mutant lacking residues 33–81, which deletes the two helices that form the four-helix bundle in the RH1 dimer (Fig. 4 A). The deletion includes most of the region previously implicated in the interaction between KHC and mouse JIP3 (Sun et al., 2011; residues 50–80 in mouse correspond to residues 62–92 in C. elegans). Immunoblotting showed that expression levels of UNC-16(∆33-81) are increased by approximately twofold relative to wild-type UNC-16 (Fig. 4 B and Fig. S4 A), and the body bending assay revealed severe locomotion deficiency in unc-16(∆33-81) animals, which was indistinguishable from that of unc-16(V72Q) animals (Fig. 4 C). Analysis of CTNS-1::mKate2, mKate2::RAB-7, and mKate2::RAB-5 distribution in the ALM neuron showed that unc-16(∆33-81) had the same effect as unc-16(V72Q) (Fig. 6, D and E). Since we could not rule out the possibility that UNC-16(∆33-81) has residual affinity for KHC, we also generated the mutant unc-16(∆33-197), which deletes the RH1 domain along with the entire first coiled-coil region of UNC-16 (Fig. 4 A). Immunoblotting showed that UNC-16(∆33-197) levels were similar to those of wild-type UNC-16 (Fig. 4 B and Fig. S4 A), and that unc-16(∆33-197) animals were as locomotion-deficient as unc-16(V72Q) animals (Fig. 4 C). Intraneuronal distribution of mKate2::RAB-5 in the unc-16(∆33-197) mutant was similar to that in the unc-16(V72Q) mutant (Fig. 6 F). This shows that simultaneous deletion of the binding sites for KHC and DLI-1 in UNC-16 mimics the specific loss of DLI-1 binding with regard to endo-lysosomal organelle distribution in TRNs.
Given that the predicted binding site for the MAP kinase JNK-1 is located just beyond the first coiled-coil region that is deleted in unc-16(∆33-197), we used the null mutant jnk-1(gk7) to ask whether JNK signaling is involved in neurite tip accumulation of endo-lysosomal markers in the unc-16(V72Q) mutant (Villanueva et al., 2001). Analysis of intraneuronal mKate2::RAB-5 distribution showed that the jnk-1(gk7) single mutant is indistinguishable from the control, while the jnk-1(gk7); unc-16(V72Q) double mutant is indistinguishable from the unc-16(V72Q) single mutant (Fig. 6 G). Thus, neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant occurs independently of JNK-1 signaling.
The results with N-terminal deletion mutants of UNC-16 implied that the interaction between UNC-16(V72Q) and KHC is not required for endo-lysosomal organelle accumulation at the neurite tip. We therefore turned our attention to the KLC binding site in UNC-16 (Fig. 7 A). KLC binds via its TPR domain to the JIP3 leucine zipper, and this interaction has been analyzed at atomic resolution for mouse JIP3 (Cockburn et al., 2018). Recent work uncovered missense mutations in the human JIP3-encoding gene MAPK8IP3 that cause neurodevelopmental disorders and intellectual disability (Iwasawa et al., 2019; Platzer et al., 2019), and one of the mutations, L444P, is predicted to perturb KLC binding to JIP3 (Platzer et al., 2019). To directly test whether this is the case, we purified recombinant human KLC1[185-501], which corresponds to the TPR domain, and mouse JIP3[186-505] with and without the L439P mutation, which is equivalent to human L444P (Fig. 7, A and B). Pull-downs with the GST-tagged KLC1 fragment demonstrated that mutating this highly conserved leucine to proline in JIP3 abrogates binding to KLC1 (Fig. 7 B).
Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires the interaction between UNC-16/JIP3 and kinesin light chain. (A) Domain organization of human JIP3 with sequence alignment of the kinesin light chain binding site in the JIP3 leucine zipper (LZ) domain. Arrowhead points to the leucine residue in JIP3 (L444 in human; L439 in mouse; L393 in C. elegans) whose mutation to proline causes neurological disease and was predicted to interfere with kinesin light chain binding (Platzer et al., 2019). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to human kinesin light chain 1 (UniProt entry Q07866-1) and mouse JIP3 (Q9ESN9-5). Molecular weight is indicated in kilodaltons (kD). (C) Immunoblot of C. elegans adult lysates using antibodies against UNC-16 and α-tubulin (loading control). Molecular weight is indicated in kilodaltons (kD). (D) Locomotion of animals at the young adult stage, assessed by determining body bending frequency (mean ± SEM) in liquid medium. n denotes the number of animals examined. Statistical significance [wild-type N2 control versus unc-16 mutants and unc-16(prt183) versus unc-16(L393P)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001. (E) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2) or early endosomes (mKate2::RAB-5) in touch receptor neurons. Scale bar, 20 µm. (F) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in the animals described in E. n denotes the number of neurites examined (1 per animal). (G) Number of CTNS-1::mKate2 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). The number of neurites examined (1 per animal) corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. (H) Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(prt183)) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined as described for G. ****P < 0.0001; ***P < 0.001; **P < 0.01; ns = not significant, P > 0.05. Source data are available for this figure: SourceData F7.
Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires the interaction between UNC-16/JIP3 and kinesin light chain. (A) Domain organization of human JIP3 with sequence alignment of the kinesin light chain binding site in the JIP3 leucine zipper (LZ) domain. Arrowhead points to the leucine residue in JIP3 (L444 in human; L439 in mouse; L393 in C. elegans) whose mutation to proline causes neurological disease and was predicted to interfere with kinesin light chain binding (Platzer et al., 2019). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to human kinesin light chain 1 (UniProt entry Q07866-1) and mouse JIP3 (Q9ESN9-5). Molecular weight is indicated in kilodaltons (kD). (C) Immunoblot of C. elegans adult lysates using antibodies against UNC-16 and α-tubulin (loading control). Molecular weight is indicated in kilodaltons (kD). (D) Locomotion of animals at the young adult stage, assessed by determining body bending frequency (mean ± SEM) in liquid medium. n denotes the number of animals examined. Statistical significance [wild-type N2 control versus unc-16 mutants and unc-16(prt183) versus unc-16(L393P)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001. (E) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2) or early endosomes (mKate2::RAB-5) in touch receptor neurons. Scale bar, 20 µm. (F) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in the animals described in E. n denotes the number of neurites examined (1 per animal). (G) Number of CTNS-1::mKate2 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). The number of neurites examined (1 per animal) corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. (H) Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(prt183)) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined as described for G. ****P < 0.0001; ***P < 0.001; **P < 0.01; ns = not significant, P > 0.05. Source data are available for this figure: SourceData F7.
In a previous effort to model the human disease, the analogous mutation (L393P) was introduced into C. elegans UNC-16, which resulted in intermediate locomotion deficiency and a moderate increase in axonal lysosome density in cholinergic motor neurons (Platzer et al., 2019). We confirmed the intermediate locomotion deficiency of unc-16(L393P) animals (Fig. 7 D) and found that unc-16(L393P) increased CTNS-1::mKate2 and mKate2::RAB-5 levels in the ALM neurite, albeit to a lesser degree than the null mutant unc-16(prt183) (Fig. 7, E–H). We conclude that the UNC-16(L393P) mutation results in a reduction-of-function phenotype, consistent with previous work (Platzer et al., 2019).
We then introduced the N-terminal mutations V72Q and ∆33-81 into the unc-16(L393P) mutant background and confirmed by immunoblot that UNC-16(L393P) and the corresponding double mutant proteins were expressed at levels comparable to those of wild-type UNC-16 (Fig. 7 C and Fig. S4 A). In contrast to the unc-16(V72Q) single mutant, the prominent neurite tip accumulation of CTNS-1::mKate2 and mKate2::RAB-5 was no longer observed in the double mutants, and the distribution in the ALM neuron now resembled that of the unc-16 null mutant (Fig. 7, E–H). These results suggest that when the interaction between UNC-16 and DLI-1 is perturbed, UNC-16 promotes neurite tip accumulation of endo-lysosomal organelles in a manner that depends on its association with KLC.
Discussion
This study identifies the RH1 domain, whose presence defines a family of cargo adaptors for cytoskeletal motors, as a binding partner of DLIC. Prior work showed that the RH1 domain of JIP3 and RILPL2 binds to KHC and myosin Va, respectively (Sun et al., 2011; Wei et al., 2013), and we demonstrate here that the RH1 domain of human JIP3, JIP4, and RILP binds to DLIC. We also show that RILPL1’s N-terminal half binds to DLIC, presumably via its RH1 domain, which identifies RILPL1 as a dynein adaptor. Based on structural work on the mouse RILPL2–myosin Va interaction (Wei et al., 2013), we mutated a conserved valine to glutamine in the RH1 domain of RILP (V59Q), JIP3 (V60Q), and JIP4 (V55Q). The point mutation abrogated the RH1–DLIC interaction, as did point mutations in helix 1 of the DLIC C-terminal tail. This suggests that the same hydrophobic pocket (one on each side of the RH1 four-helix bundle) that in RILPL2 accommodates helix 2 of the myosin Va globular tail domain is used in the dynein adaptors RILP, RILPL1, JIP3, and JIP4 to accommodate the amphipathic helix 1 of the DLIC C-terminal tail. The low micromolar affinity of the interaction is consistent with what has been determined for the three other dynein adaptor folds that bind the DLIC helix, namely the Hook domain, the CC1 box, and the EF-hand (Lee et al., 2018; Lee et al., 2020).
We show that KHC binding is not affected by the JIP3 V60Q mutation, which, taken together with binding competition assays, suggests that KHC and DLIC occupy distinct surfaces on the RH1 domain. Since the binding of DLIC and KHC to JIP3 is conserved in C. elegans, we were able to generate the separation-of-function mutant unc-16(V72Q) that specifically abrogates the interaction between UNC-16/JIP3 and DLI-1/DLIC. Characterization of unc-16(V72Q) in touch receptor neurons establishes that JIP3 binding to DLIC promotes the retrograde transport of lysosomes and endosomes and is therefore critical for JIP3’s previously described organelle clearance function (Edwards et al., 2013; Edwards et al., 2015). Additionally, our results suggest that JIP3–bound dynein promotes retrograde transport of synaptic vesicle precursors marked by SNB-1/synaptobrevin.
While both unc-16 null mutants and the unc-16(V72Q) mutant have increased amounts of endo-lysosomal organelles in the anterior neurite of touch receptor neurons, organelle distribution within the neurite is markedly distinct: abrogating the JIP3–DLIC interaction through the UNC-16(V72Q) mutation results in prominent organelle accumulation at the neurite tip, which is not observed in unc-16 null mutants. In the UNC-16(V72Q) mutant, neurite tip accumulation of organelles is rescued by the disease-associated JIP3 mutation L393P (human L444P), which we demonstrate abrogates the KLC–JIP3 interaction. This suggests that JIP3–bound kinesin-1 drives neurite tip accumulation of organelles when the interaction between JIP3 and dynein is perturbed. So far, TrkB and JIP3 itself are the only firmly established cargos for JIP3–bound kinesin-1 (incidentally, TrkB is not conserved in C. elegans). Our results argue that the cargo of JIP3–bound kinesin-1 includes endo-lysosomal organelles and that anterograde transport of organelles by JIP3–bound kinesin-1 is opposed by JIP3–bound dynein. In contrast to endo-lysosomal organelles, there was no difference in the distribution of SNB-1::mKate2 between unc-16 null mutants and the unc-16(V72Q) mutant. This implies that on SNB-1–marked vesicles JIP3–bound dynein is not opposed by JIP3–bound kinesin-1, which is consistent with the well-established fact that synaptic vesicle precursors are a cargo of kinesin-3/UNC-104 (Hall and Hedgecock, 1991). The idea that JIP3–bound dynein opposes anterograde-directed motility of UNC-104 on synaptic vesicle precursors also fits well with the observation that SNB-1::GFP mis-localization in unc-104 mutants is partially suppressed by unc-16 mutants (Byrd et al., 2001).
The JIP3-mediated bidirectional transport of endo-lysosomal organelles that we describe in this study raises the question of how the adaptor selects or switches between opposing motors. Small GTPases are likely to play a role in determining which motor is active, as previous work showed that Arf6 binding to the JIP3 leucine zipper domain promotes the interaction with the dynein cofactor dynactin and is incompatible with KLC binding (Cockburn et al., 2018; Montagnac et al., 2009). Since our results suggest that KHC and DLIC bind independently to JIP3’s RH1 domain, kinesin-1 could, in principle, remain associated with JIP3 via the RH1–KHC interaction while dynein-dynactin are engaged in transport. It is also conceivable that the RH1 domain is used for allosteric communication between the motors. JIP3 has been shown to associate with various membrane compartments by localization studies and biochemical fractionation, including lysosomes and early endosomes (Abe et al., 2009; Becker and Bonni, 2006; Cason et al., 2021; Cavalli et al., 2005; Choudhary et al., 2017; Drerup and Nechiporuk, 2013; Gowrishankar et al., 2017; Montagnac et al., 2009), but the molecular pathways through which JIP3 associates with the organelles and vesicles whose transport it regulates remain largely unknown. Defining these recruitment pathways may reveal cargo-specific regulation of JIP3–motor interactions.
We note that while combining the V72Q mutation with the L393P mutation recapitulates the endo-lysosomal organelle distribution observed in unc-16 null mutants, unc-16(L393P) on its own already causes some organelle accumulation in neurites. This is consistent with the partial loss-of-function phenotype of this mutant in motor neuron axons (Platzer et al., 2019). A plausible explanation for this effect is that kinesin-1 transports JIP3 into the neurite as cargo and that abrogating the JIP3–KLC interaction decreases the amount of JIP3 that is available in the neurite to promote retrograde transport. Interestingly, we found that deleting JIP3’s RH1 domain and the adjacent coiled-coil region, which abrogates both DLIC and KHC binding, results in the same neurite tip accumulation of mKate2::RAB-5 as UNC-16(V72Q), which specifically abrogates DLIC binding. This suggests that while the JIP3–KLC interaction is required for anterograde transport of endo-lysosomal organelles, the JIP3–KHC interaction is not. KHC binding to JIP3 has been implicated in activation of kinesin-1 motility (Sun et al., 2011; Watt et al., 2015), and it would therefore be interesting to examine how a JIP3 mutant that cannot bind KHC but still binds DLIC affects the kinetics of organelle transport. Of note, our molecular characterization of the JIP3–DLIC interaction suggests that previously characterized N-terminal deletions in mouse JIP3 (Sun et al., 2011; Sun et al., 2013; Sato et al., 2015; Watt et al., 2015; Ma et al., 2017; Sun et al., 2017), which were presumed to specifically abrogate KHC binding, also abrogate DLIC binding.
Previous characterization of JIP3 as an activator of kinesin-1 motility provides an explanation for how JIP3’s interaction with KLC could promote the anterograde transport of endo-lysosomal organelles that we describe in this study (Sun et al., 2011; Watt et al., 2015). By contrast, how the JIP3–DLIC interaction promotes retrograde transport is less obvious. Activation of processive dynein motility occurs in the context of a tripartite complex consisting of the motor, dynactin, and a cargo adaptor. In all adaptors that have been demonstrated to be bona fide activators of dynein motility, the N-terminal coiled-coil region of the adaptor runs along the entire length of the 37-nm dynactin filament. This configuration stabilizes the interaction between the dynactin filament and the N-terminal tails of the dynein heavy chain dimer, which is critical for processive motility (Urnavicius et al., 2015). Since JIP3’s N-terminal coiled-coil is only half the length of the dynactin filament (Vilela et al., 2019), it has been argued that JIP3 on its own may not be able to form motile complexes with dynein-dynactin (Reck-Peterson et al., 2018). An alternative possibility to a direct activating function is that JIP3 incorporates into the dynein-dynactin transport machine together with an activating adaptor (Chaaban and Carter, 2022,Preprint). In such a scenario, the additional binding sites for DLIC provided by the JIP3 RH1 domain may help recruit a second dynein to dynactin for increased force production and speed (Urnavicius et al., 2018). Examining how JIP3 promotes dynein motility using in vitro reconstitution, and defining the functional relationship between JIP3 and other dynein adaptors for endo-lysosomal organelles, including the structurally similar RILP, are important future research directions.
Materials and methods
C. elegans strains
Worm strains (Table S1) were maintained at 20°C on standard NGM plates seeded with OP50 Escherichia coli bacteria. The mutants unc-16(prt183), unc-16(V72Q), unc-16(∆33-81), unc-16(∆33-197), and jip-1(prt187) were generated by CRISPR/Cas9-mediated genome editing using single-stranded repair templates (Integrated DNA Technologies) with 50-bp homology regions (Paix et al., 2014). Genomic sequences targeted by guide RNAs and corresponding repair templates are listed in Table S2. For each mutation, the repair template and a ribonucleoprotein (RNP) mixture consisting of crRNA/tracrRNA (Integrated DNA Technologies) and purified recombinant Cas9 enzyme were injected into gonads of young N2 adults. To identify potentially successful edits, animals were coinjected with RNPs to generate the R92C mutation in dpy-10, which causes a dominant roller phenotype (Arribere et al., 2014). Screening was performed by PCR and edits were confirmed by sequencing. Strains were outcrossed 4–6 times against the wild-type N2 strain, and fluorescent markers or other mutant alleles were subsequently introduced by mating.
Image acquisition
Animal locomotion assay
L4 hermaphrodites were transferred to a new NGM plate with bacteria 24 h before performing the assay. For imaging, animals were transferred to a slide containing a 2-μl drop of M9. Movements were tracked at 20°C for 1 min at 40 frames per second using an SMZ 745T stereoscope (Nikon) with a QIClic CCD camera (QImaging) controlled by Micro-Manager software (Open Imaging). The wrMTrck plugin for ImageJ was used for automated counting of body bends.
Touch receptor neurons
To image mKate2-tagged organelle and vesicle markers in the ALM neuron, L4 hermaphrodites were paralyzed in M9 buffer containing 50 mM NaN3 for 5 min, transferred to a freshly prepared 2% (wt/vol) agarose pad, and covered with an 18 × 18 mm coverslip (No. 1.5H, Marienfeld). Imaging was performed at 20°C on an Axio Observer microscope (Zeiss) equipped with an Orca Flash 4.0 camera (Hamamatsu) and an HXP 200C Illuminator (Zeiss). A z-stack (step size 0.5 µm) that captured the entire neuron was acquired at 1 × 1 binning with a 40× NA 1.3 Plan-Neofluar objective (Zeiss). Image acquisition was controlled by ZEN 2.3 software (Zeiss).
For time-lapse imaging of mKate2::RAB-5 particles in the ALM neuron, L2 hermaphrodites were paralyzed with 5 mM levamisole in M9 buffer for 10 min and mounted on agarose pads as described above. Only morphologically healthy neurites were imaged. Time-lapse sequences of a region ∼50 µm away from the cell body were recorded at 20°C on a Nikon Eclipse Ti microscope coupled to an Andor Revolution XD spinning disk confocal system, composed of an iXon Ultra 897 CCD camera (Andor Technology), a solid-state laser combiner (ALC-UVP 350i, Andor Technology), and a CSU-X1 confocal scanner (Yokogawa Electric Corporation). The system was controlled by Andor IQ3 software (Andor Technology). A single image was acquired every 200 ms for a total of 30 s at 1 × 1 binning using a 100× NA 1.45 Plan-Apochromat objective (Nikon).
To image GFP-tagged UNC-16 in the ALM neuron, L4 hermaphrodites were paralyzed with NaN3 as described above and imaged on the spinning disc confocal microscope at 20°C. Z-stacks (step size 0.2 µm) that captured the neurite tip and cell body were acquired at 1 × 1 binning with a 60× NA 1.4 Plan-Apochromat objective (Nikon).
Image analysis
Image analysis was performed using Fiji software (Image J version 1.52d).
Fluorescence intensity profile along the neurite
A segmented line with a width of 10 pixels was traced on top of the ALM neurite and the fluorescence intensity profile of the entire neurite was recorded using Fiji’s “Plot Profile” function. A parallel segmented line was traced adjacent to the neurite to obtain a background intensity measurement for each position along the neurite. The final fluorescence intensity at each position was calculated by subtracting the background signal from the neurite signal. Absolute positions along the neurite were normalized to neurite length with 0% corresponding to the proximal neurite at (but not including) the cell body and 100% corresponding to the distal neurite tip. The signal was averaged over 1% intervals.
Integrated fluorescence intensity at the neurite tip and in the cell body
The integrated fluorescence intensity at the ALM neurite tip was determined for the last 20 µm of the neurite by summing up the fluorescence intensity in the profile plot described above and by subtracting the corresponding background signal. For mKate2::SYX-7, a different approach was necessary, since the diffuse signal in the neurite was too dim to accurately trace the neurite for profile plots. The integrated fluorescence intensity at the neurite tip, which was not visible in the control, was therefore measured in a 185 × 15-pixel (30 × 2.5 µm) ROI placed adjacent to the pharyngeal procorpus identified in the differential interference contrast (DIC) channel. The ROI was expanded by a few pixels on each side, and the difference in signal between the outer and inner ROI was used to define the integrated background intensity after normalization to the area of the inner ROI. The final integrated intensity was calculated by subtracting the integrated background intensity from the integrated intensity of the inner ROI. An analogous approach was used for the cell body with the inner ROI corresponding to the cell body outline.
Counting of CTNS-1::mKate2 and mKate2::SYX-7 puncta
For CTNS-1::mKate2, an empirically determined threshold (∼1.5-fold above the local diffuse background signal) was applied to the fluorescence intensity profile plot to determine the number and position of puncta, which were then grouped according to their position along the neurite: proximal neurite (≤25% length), mid-neurite (>25 to ≤75% length), and distal neurite (>75% length). Since we could not generate fluorescence intensity profile plots in neurites expressing mKate2::SYX-7, mKate2::SYX-7 puncta were counted directly in equally scaled fluorescence images, and, to define neurite length in the control, the neurite tip was estimated to lie at the transition between buccal cavity and pharyngeal procorpus identified in the DIC channel.
Motility parameters of mKate2::RAB-5 particles
Kymograph generation and analysis were performed with KymoAnalyzer (Neumann et al., 2017). Only time-lapse sequences during which there was no discernable movement of the animal and which had uniformly bright mKate2 signal along the neurite were considered. A track is defined as a single particle trajectory, and a segment corresponds to a portion within the track of a moving particle that is framed by a pause or a reversal. Values for run length and velocity reported in this study are for segments. Particles classified as “anterograde moving” or “retrograde moving” did so exclusively, i.e., their track did not contain any segments in which movement was reversed. Particles that had both anterograde and retrograde segments in their track were classified as “reversing direction.” Particles were classified as “stationary” if their track did not contain any moving segments.
Antibody against UNC-16
Polyclonal antibodies against an N-terminal fragment of UNC-16 (residues 1–506) were generated as follows: GST::UNC-16(1-506) was expressed from pGEX6P-1 in bacteria, purified as described below, and injected into rabbits (GeneCust). For affinity purification of the antibodies, the GST fusion protein was cleaved using Prescission protease, and UNC-16(1-506) was covalently coupled to a 1-ml HiTrap N-hydroxysuccinimide column (GE Healthcare).
Immunoblotting
For immunoblots of C. elegans lysate, 100 adult hermaphrodites were collected into 1 ml of M9 buffer and washed with 3 × 1 ml M9 buffer and 3 × 1 ml M9 buffer containing 0.05% Triton X-100. To 100 μl of worm suspension, 33 μl 4× SDS PAGE sample buffer (250 mM Tris-HCl pH 6.8, 30% [vol/vol] glycerol, 8% [wt/vol] SDS, 200 mM DTT, 0.04% [wt/vol] bromophenol blue), and 20 μl of glass beads were added. Samples were incubated for 3 min at 95°C and vortexed for 5 min with intermittent heating. After centrifugation at 20,000 g for 1 min at room temperature, proteins in the supernatant were resolved on a 4–20% gradient gel (Bio-Rad). Proteins were transferred to a 0.2-µm nitrocellulose membrane (GE Healthcare). The membrane was blocked with 5% (wt/vol) non-fat dry milk in TBST (20 mM Tris-Cl, 140 mM NaCl, 0.1% Tween 20, pH 7.6) and incubated overnight at 4°C in TBST/5% dry milk with rabbit polyclonal anti-UNC-16 antibody GC23 (1:1,200; made in-house), mouse monoclonal anti-α-tubulin antibody B512 (1:5,000; Sigma-Aldrich), rabbit polyclonal anti-GST antibody GC3 (1:7,500; Gama et al., 2017), mouse monoclonal anti-6xHis antibody His.H8 (1:2,500; Millipore), or anti-Strep-tag II antibody StrepMAB (1:1,000; IBA). The membrane was rinsed 5 × with TBST and incubated in TBST/5% dry milk with goat polyclonal anti-rabbit or anti-mouse IgG antibody coupled to HRP (1:10,000; 111-035-003 and 115-035-044, respectively; Jackson ImmunoResearch) for 1 h at room temperature. After three washes with TBST, proteins were visualized by chemiluminescence using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) and x-ray film (GE Healthcare). For fluorescent immunoblots, the membrane was dried for 1 h prior to the blocking step. Following incubation with anti-UNC-16 and anti-α-tubulin antibody, the membrane was washed 4 × with TBST, incubated with donkey anti-mouse IRDye 680RD (1:30,000; LI-COR) and anti-rabbit IRDye 800CW (1:20,000; LI-COR) for 1 h at room temperature, and washed again 4 × with TBST and 1 × with TBS. Fluorescent immunoblots were scanned on the Odyssey CLx Imager (LI-COR). Fluorescence intensity measurements were performed in Fiji. The integrated fluorescence intensity was determined in the ROI drawn around the UNC-16 signal (IRDye 800CW), the ROI was expanded vertically by 2 pixels in each direction, and the difference in intensity between the original and expanded ROI was used to calculate the integrated background intensity, which was subtracted from the UNC-16 intensity. The UNC-16 intensity was then normalized to the intensity of the α-tubulin signal (IRDye 680RD).
Plasmids for recombinant protein expression
cDNA encoding for the protein fragment to be expressed was inserted into a 2CT vector (N-terminal 6xHis::maltose binding protein [MBP] followed by a TEV protease cleavage site and C-terminal Strep-tag II) or into pGEX-6P-1 (N-terminal glutathione S-transferase [GST] followed by a Prescission protease cleavage site and C-terminal 6xHis). Residue numbers in text and figures correspond to the following UniProt entries: Q96NA2-1 (human RILP), Q9JJC6 (mouse RILPL1), Q969X0 (human RILPL2), Q9UPT6-1 (human JIP3), Q9ESN9-5 (mouse JIP3), O60271-1 (human JIP4), Q9Y6G9 (human DYNC1LI1), Q07866-1 (human KLC1), P28738 (mouse KIF5C), P34609-1 (C. elegans UNC-16), P34540 (C. elegans UNC-116), and G5ED34 (C. elegans DLI-1).
Recombinant protein expression and purification
Expression vectors were transformed into E. coli strains BL21, BL21-CodonPlus-RIL, or Rosetta. Expression was induced at an OD600 of 0.9 with 0.1 mM IPTG. After expression overnight at 18°C, cells were harvested by centrifugation at 4,000 g for 20 min.
For purification of proteins expressed from the 2CT vector, bacterial pellets were resuspended in lysis buffer A (50 mM HEPES, 250 mM NaCl, 10 mM imidazole, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, pH 8.0), lysed with a cell cracker, and cleared by centrifugation at 40,000 g for 45 min at 4°C. Proteins were purified by tandem affinity chromatography using HisPur Ni-NTA resin (Thermo Fisher Scientific) followed by strep-tactin sepharose resin (IBA). Ni-NTA resin was incubated in batch with the cleared lysate in the presence of 0.1% (vol/vol) Tween 20 for 1 h at 4°C and washed with wash buffer A (25 mM HEPES, 250 mM NaCl, 25 mM imidazole, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer A (50 mM HEPES, 150 mM NaCl, 250 mM imidazole, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Fractions containing the recombinant protein were pooled, incubated overnight at 4°C with TEV protease (if 6xHis::MBP needed to be cleaved off), incubated in batch with strep-tactin sepharose resin for 1 h at 4°C, and washed with wash buffer B (25 mM HEPES, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Proteins were eluted on a gravity column with elution buffer B (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 10 mM desthiobiotin, pH 8.0). Some proteins were further purified by size exclusion chromatography on a Superose 6 Increase 10/300 GL or Superdex 200 Increase 10/300 GL column (GE Healthcare) equilibrated with storage buffer (25 mM HEPES, 150 mM NaCl, pH 7.5) or ITC buffer (25 mM HEPES, 100 mM KCl, 0.5 mM TCEP, pH 7.5). Alternatively, the eluate from the gravity column was directly dialyzed against storage buffer. Glycerol and DTT were added to a final concentration of 10% (vol/vol) and 1 mM, respectively (except for ITC samples), and aliquots were flash frozen in liquid nitrogen and stored at −80°C.
For purification of proteins expressed from the pGEX-6P-1 vector, bacterial pellets were resuspended in lysis buffer B (50 mM HEPES, 250 mM NaCl, 10 mM EDTA, 10 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine-HCl, pH 8.0), lysed with a cell cracker, and cleared by centrifugation at 40,000 g for 45 min at 4°C. Proteins were purified by tandem affinity chromatography using glutathione agarose resin (Thermo Fisher Scientific) followed by Ni-NTA resin. Glutathione agarose resin was incubated in batch with the cleared lysate in the presence of 0.1% (vol/vol) Tween 20 for 1 h at 4°C, washed with wash buffer C (25 mM HEPES, 250 mM NaCl, 0.1% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0), and proteins were eluted on a gravity column with elution buffer C (50 mM HEPES, 150 mM NaCl, 10 mM reduced L-glutathione, 1 mM DTT, 2 mM benzamidine-HCl, pH 8.0). Alternatively, the resin was incubated overnight at 4°C with Prescission Protease in cleavage buffer (25 mM HEPES, 150 mM NaCl, 0.01% Tween 20, 1 mM DTT, 2 mM benzamidine-HCl, pH 7.6) to remove the GST tag. Fractions containing the (cleaved) recombinant protein were pooled, incubated in batch with Ni-NTA resin for 1 h at 4°C, washed with wash buffer A, and eluted on a gravity column with elution buffer A. Proteins were further purified by size exclusion chromatography or dialyzed before flash freezing, as described above.
Analytical size exclusion chromatography
For binding assays, proteins mixtures (20–40 µM in 25 mM HEPES, 150 mM NaCl, pH 7.5) were incubated on ice for 30 min prior to size exclusion chromatography, which was performed at room temperature on an ÄKTA Pure 25L1 system with a Superdex 200 Increase 10/300 GL column (GE Healthcare). Elution of proteins was monitored at 280 nm. 20 μl of successive 0.5-ml elution fractions were separated by SDS-PAGE and proteins were visualized by Coomassie Blue staining.
Pull-down assays
Purified recombinant proteins (300 pmol) were mixed in 45 μl pull-down buffer (50 mM HEPES, 100 mM NaCl, 5 mM DTT, pH 7.5). This initial mixture was split into two halves, one corresponding to the input and the other to the pull-down. For pull-downs, the protein mixture was incubated for 1 h at 4°C in 150 μl pull-down buffer supplemented with 15 μl glutathione agarose resin for GST pull-downs or amylose resin (New England Biolabs) for MBP pull-downs. After washing the resin with 3 × 500 μl pull-down buffer, proteins were eluted with the same buffer supplemented with 15 mM reduced L-glutathione (glutathione agarose resin) or 10 mM maltose (amylose resin). Eluted proteins were separated by SDS-PAGE gel and visualized by Coomassie Blue staining or immunoblotting.
Isothermal titration calorimetry
ITC data was recorded in a MicroCal VP-ITC calorimeter (Malvern). All proteins analyzed by ITC were subjected to a final purification step by size exclusion chromatography in ITC buffer (25 mM HEPES, 100 mM KCl, 0.5 mM TCEP, pH 7.5). Titrations were performed at 25°C and consisted of an initial 2 μl injection followed by a series of 28 injections of 10 μl each. In the cases where an interaction was detected a blank run was performed by titrating the titrant into ITC buffer. Data were processed with the ORIGIN software package (OriginLab) and in cases where an interaction was detected either the blank run was subtracted from the initial isotherm (to correct for the heat of dilution) or data were further processed for global fitting with the NITPIC, SEDPHAT, and GUSSI software packages.
Analytical ultracentrifugation (AUC)
Sedimentation velocity AUC was performed at 42,000 rpm at 20°C in an An-60 Ti rotor (Beckman Coulter) using standard double-sector centerpieces. Protein samples were diluted in AUC buffer (50 mM HEPES, 100 mM NaCl, 0.5 mM TCEP, pH 7.5). Approximately 300 radial absorbance scans at 280 nm were collected per run with a time interval of 1 min. Analysis was performed using the SEDFIT suite to obtain the continuous distribution function of sedimentation coefficients (c[S]). The software SEDNTERP was used to estimate buffer density and viscosity, as well as the protein partial specific volume. GUSSI software was used to generate the final figures.
Structure prediction
ColabFold with default parameters was used for AlphaFold2-based structure prediction. Input sequences corresponded to human JIP3(1-100) and human DLIC1(440-455). The specific settings were as follows (from settings.txt file generated by ColabFold):
sequence = MMEIQMDEGGGVVVYQDDYCSGSVMSERVSGLAGSIYREFERLIHCYDEEVVKELMPLVVNVLENLDSVLSENQ-EHEVELELLREDNEQLLTQYEREKAL:SEGVLANFFNSLLSKK
msa_method = mmseqs2; homooligomer = 2:2; pair_msa = False; max_msa = 512:1,024; subsample_msa = True; num_relax = None; use_turbo = True; use_ptm = True; rank_by = pLDDT; num_models = 5; num_samples = 1; num_ensemble = 1; max_recycles = 3; tol = 0; is_training = False; use_templates = False.
Statistical analysis
Statistical analysis was performed with Prism 8.0 software (GraphPad). Statistical significance was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test, or by a two-sided Mann-Whitney test, where ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, and ns = not significant, P > 0.05. The analytical method used is specified in the figure legends.
Online supplemental material
Fig. S1 shows that the V59Q and P56F mutations in RILP reduce the affinity for dynein light intermediate chain. Fig. S2 shows that the V55Q mutation in JIP4 abrogates the binding to dynein light intermediate chain. Fig. S3 Shows that human JIP3 V60Q is equivalent to C. elegans UNC-16 V72Q. Fig. S4 shows that mutant UNC-16 levels are comparable to WT; that late endosomes but not synaptic vesicle precursors are distributed differently in unc-16(ce483) versus unc-16(V72Q); and that unc-16(ce483) is a null mutant. Fig. S5 shows that recycling endosomes accumulate at the ALM neurite tip in the unc-16(V72Q) mutant and do so independently of JIP-1. Table S1 shows the genotype of C. elegans strains. Table S2 shows oligonucleotide sequences for CRISPR/Cas9-mediated genome editing.
Acknowledgments
The authors thank Stacey L. Edwards and Kenneth G. Miller (Oklahoma Medical Research Foundation) for C. elegans strain KG5309. Some strains were provided by the Caenorhabditis Genetics Center (CGC), which is funded by the National Institute of Health Office of Research Infrastructure Programs (P40 OD010440). The authors acknowledge the Biochemical and Biophysical Technologies Scientific Platform at i3S for support with ITC experiments.
This study was financed by the Fundação para a Ciência e a Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior through project grants PTDC/BIA-CEL/30507/2017 and PTDC/BIA-CEL/1321/2021. R. Gassmann and A.C. are supported by FCT Principal Investigator positions CEECIND/00333/2017 and CEECIND/01967/2017, respectively. R. Celestino and D.J. Barbosa are supported by FCT Junior Researcher positions DL57/2016/CP1355/CT0001 and DL57/2016/CP1355/CT0007, respectively.
The authors declare no competing financial interests.
Author contributions: Conceptualization: R. Gassmann. Formal analysis: R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa, H. Rocha, E.A. d’Amico, J.H. Morais-Cabral, R. Gassmann. Funding acquisition: R. Gassmann. Investigation: R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa, H. Rocha, E.A. d’Amico, J.H. Morais-Cabral, R. Gassmann. Project administration: R. Gassmann. Resources: R. Gassmann, A.X. Carvalho, A. Musacchio, S.L. Edwards, K.G. Miller. Supervision: R. Gassmann, A. Musacchio, A.X. Carvalho, J.H. Morais-Cabral. Validation: R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa, H. Rocha, E.A. d’Amico, J.H. Morais-Cabral, R. Gassmann. Visualization: R. Gassmann, R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa, H. Rocha, E.A. d’Amico. Writing—original draft: R. Gassmann, J.H. Morais-Cabral, A.X. Carvalho, R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa. Writing—review & editing: R. Gassmann, J.H. Morais-Cabral, A.X. Carvalho, R. Celestino, J.B. Gama, A.F. Castro-Rodrigues, D.J. Barbosa, H. Rocha, E.A. d’Amico, A. Musacchio
References
Author notes
R. Celestino, J.B. Gama, and A.F. Castro-Rodrigues contributed equally to this paper.
E.A. d’Amico’s present address is MRC Laboratory of Molecular Biology, Cambridge, UK.


![Dynein adaptors of the RILP/JIP3 superfamily use their N-terminal RH1 domain to bind the C-terminal helix 1 of dynein light intermediate chain. (A) Domain organization of human dynein light intermediate chain 1 (DLIC1) and the five human cargo adaptors for cytoskeletal motors characterized by the presence of RILP homology (RH) 1 and RH2 domains. JIP3 and JIP4 also have a second coiled-coil region, here called the leucine zipper (LZ) domain. Regions implicated in direct protein-protein interactions are delineated by dashed lines. Residue numbers correspond to UniProt entries Q96NA2-1 (RILP), Q5EBL4-1 (RILPL1), Q969X0 (RILPL2), Q9UPT6-1 (JIP3), O60271-1 (JIP4), and Q9Y6G9 (DLIC1). (B and C) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs described in A except for RILPL1, which is the mouse homolog (Q9JJC6). Molecular weight is indicated in kilodaltons (kD). (D–F) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for RILP[1-106] are the same in D–F. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData F1.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig1.png?Expires=1773840818&Signature=mty16pH9zIgIg5ypGPOkhvIh2BWwzpk3zG~~4snq2u3Cs~pAhfgQdariBDXUNryAGwgHIxJzGujnmI2Be7aPWLewGOlj1oNrpotaoyLbyzpBiO65kPn5kXhKDTaytqQGydiZMKzH7RU4FDJFwwoBgmH7LvHn0fepm8x22MakfDUKHw-ucmzI4ro1le056Y1msUjuyoXulJSP8hEfAbLL4rAsceYCP1bG7i9l9F7oxgKLxGtGwDaDnmf2T~sYEfo4wQMwoQqBXo0xDlnnD~rD5ZRBF5NklMfDTR1RcEruMZPjoGRDa~gsfkW4ITpQRGEGzjQRg82IoWwGkIYwSS4SRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The C-terminal amphipathic helix 1 of dynein light intermediate chain binds the RH1 domain’s hydrophobic pocket. (A) Top left: Model of the dimeric RH1 domain of human JIP3 (UniProt entry Q9UPT6-1) generated using ColabFold (Mirdita et al., 2022). Bottom left: Sequence alignment of the α3N helix for the RILP/JIP3 superfamily. Open circles denote residues participating in dimer formation in mouse RILPL2 (Wei et al., 2013). Closed circles denote residues participating in the interaction between RILPL2 and myosin Va. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and whose mutation to glutamine in RILPL2 abrogates the interaction with myosin Va. Right: Model of the JIP3 RH1 domain’s 4-helix bundle with the C-terminal helix 1 of DLIC1 (residues 440–455) docked at the hydrophobic pocket, as predicted by ColabFold. A phenylalanine and a valine side chain in DLIC1 and JIP3, respectively, which we mutate in this study, are also rendered. The models have a per-residue confidence (pLDDT) of >90% throughout and a consistently low alignment error (PAE) (Fig. S2 A; Tunyasuvunakool et al., 2021). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs. WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). (C) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. Molecular weight is indicated in kilodaltons (kD). (D and E) Sedimentation velocity AUC profiles with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved values indicate that both proteins are dimeric in solution. (F–H) Thermograms and binding isotherms of representative ITC titrations. JIP3[1-108] concentration is the concentration of the dimer. The dissociation constant (KD) and the binding stoichiometry (N) are given as mean ± SD (n = 3) and were derived from fitting to the binding isotherm (black line) with a One Set of Sites model using ORIGIN software. Source data are available for this figure: SourceData F2.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig2.png?Expires=1773840818&Signature=Nti6BhOJFOEU8uqPo~ywFuClwAUxXySI3jVMqMRoIdw3LgtMwtXKIynyQeQU6gmue~2Sbhye3jgwN9HOOyQhecVVYY3b2RbBB2STvq8vgMkXB7dNzdLCxUKLiPxOn-6PpI3FEE~JvzgNAR4EVsoiibh1Z0JH7Ce1hagR6zy5B3WdjVzN5pHEMH3hJN28w9zxs6nPY-t2lo7qgZOo3I0UBR4KFwtkbLcMWcqc7SURLlN5LJAkHY1yV1zjUh3Goebp6YQ0-4yOoTXlNDe-DJOU19IEIc47Q9b0ki7EWKzIeq3pvrqy6eJpIcGx86rLc4UWpGHwt3UI4~Vi2c02~kac3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Kinesin heavy chain also binds to the RH1 domain of JIP3 but does not compete for binding with dynein light intermediate chain. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the human homologs and to mouse kinesin heavy chain KIF5C (UniProt entry P28738). Molecular weight is indicated in kilodaltons (kD). (C and D) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant human proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. The elution profile and gel for GST::DLIC1[388-523] are shown twice in C, and the elution profile and gel for GST::KIF5C[807-956] are shown twice in D. Molecular weight is indicated in kilodaltons (kD). WT denotes wild type. (E) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of amylose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from amylose resin after MBP pull-down. The actual amount of DLIC1[388-523] in the pull-down reaction was fivefold higher than what is shown for the input. The KIF5C fragment was used at two concentrations that differ threefold, as indicated above the 5th and 6th lanes from the left. (F) Coomassie Blue-stained SDS-PAGE gel (top) and corresponding immunoblot (bottom) of proteins eluted from amylose resin after MBP pull-down as in E. All proteins contain a 6xHis tag (see Materials and methods) that is detected on the immunoblot. Proteins are the same as in E, but amounts in the pull-down mixture were decreased relative to those in E such that DLIC1-C helix 1 peptide could be added in 150-fold molar excess over the KIF5C fragment. Source data are available for this figure: SourceData F3.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig3.png?Expires=1773840818&Signature=bz~EVRGRmmBGnQTwr9KDUpJI1n7LiB6D9c8BWWG-O-vqsaPLgd1g5io8ULEEiQw4bYl-aVis8hIpZiMXNY6fU0xY739KTgtKWdQO~hm0drow~O~DuZVPhjsdAzSzAwQxZbDKO3Ec5FnLgiYHdaK3U0h-oEMMMTV3WHwlpX20VCcg3XZKIMR9qtPbi9YF-v-ei5EAzEkcyIHfcNHPIJ~hhugbM9G9Ce5Pn9bn8oJvTKDVwmEYlh7T8X-kk-YiB9HqoS32vy2MIRKxR9Q0JD6xWg1V3CZ-HuoRDFV1i9sDOnJ2371TR4oMyJbOpiacZXv-nsMXFmIL8bPDnzYglPuzng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The V59Q and P56F mutations in RILP reduce the affinity for dynein light intermediate chain. (A) Elution profiles (top) and Coomassie Blue-stained SDS-PAGE gels (bottom) of purified recombinant proteins after size exclusion chromatography on a Superdex 200 Increase 10/300 GL column. Proteins correspond to the human homologs. The elution profile and gel for GST::DLIC1[388-523] are shown twice, and the elution profile and gel for RILP[1-106] WT are the same as in Fig. 1 D. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (B) Left: Sequence alignment of the α3N helix for RILP family proteins in selected vertebrates and invertebrates. Boxed residues denote the phenylalanine in RILPL2 that is critical for myosin Va binding and the proline that typically replaces the phenylalanine in RILP and RILPL1. Arrowhead points to the valine residue that forms part of the RH1 domain’s hydrophobic pocket and is mutated in A. Right: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input) and after elution from the resin (GST pull-down). Proteins correspond to the human homologs. Molecular weight is indicated in kilodaltons (kD). Source data are available for this figure: SourceData FS1.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_figs1.png?Expires=1773840818&Signature=eLrnJVBkmK0vI8yOK-d9RGu2vtGLzn2hR-IzMs5vKfqfopUF6tTW72R1-DCrU6ltXH~ySuU8YD4BUPRs7SuASkSyCy9AqP9o3NG3dqsN3Cy3lnesI~WhJkbdfLSvb2Bt52~7VnRFORWcJu-7hxL9jOskmWlTYOcSo3tlag0R-AJefyqAxCGX4cAi490DJR9YkoTxeRAqvfYQshbSfo1xDu0REQP6c15JBIQhkWOOiCcCHsyqGH0Cr114TXozB4JQGB9liYkF4DQYqrMMb2nFYOMnFvRsvY14lroTjzysG-uG355kjwISy4VYONcmZn8HSagtxluNtlmJ8XIBbgtQiA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![The V55Q mutation in JIP4 abrogates the binding to dynein light intermediate chain. (A) Per-residue confidence score (pLDDT; blue: high confidence, red: low confidence) of the human JIP3[1-100] dimer bound to two copies of human DLIC1[440-455] (left) and corresponding Predicted Alignment Error (PAE) plot (right). (B) Coomassie Blue-stained SDS-PAGE gel of purified recombinant human proteins used in AUC and ITC experiments. WT denotes wild type. Molecular weight is indicated in kilodaltons (kD). (C) Sedimentation velocity AUC profile with theoretical (MWcalculated) and experimentally measured molecular mass (MWobserved). The MWobserved value indicates that JIP4 is dimeric in solution. (D–F) Thermograms and binding isotherms of representative ITC titrations. JIP4[1-103] concentration is the concentration of the dimer. The data in D were best described by a model corresponding to two sites with a single macroscopic dissociation constant (KD) using the SEDPHAT software package. The KD value was determined by global fitting of the two independent experiments shown in D with a 68% confidence interval of 2.8–7.4 µM. Source data are available for this figure: SourceData FS2.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_figs2.png?Expires=1773840818&Signature=PLfZvci9Oa~pApFQMJxZoyX~L0FTA4kBQsPIL5VsxdhYsqL7GV2F3wcv4eHSNKy~SDHwnsgVa4Sb-jGH~OWvUZeOAFkYCIuAOu83N45zaM7mhHKoGS~HMGl3goCqrp1UxafasNrgCGxw61Fcp1sxaMh6Yr0H-IzNMvpovZUzwCJ6LnBx3Am2ptYSIyxqaT1xEcBeorE7a7uJDJ58fuzijOxO3olV4Rxx-TD1hhQSDgTnrc-aaoIdSYk81pCFgJAkpiPyfxGL1D-DMKwrhrY6PFHFKay8KAMZQaVrpVkUNNhs5mPHWjv~VEeYICA1tMjvZBCRHhuUaUiKVJpP0SjHAw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Human JIP3 V60Q is equivalent to C. elegans UNC-16 V72Q. (A and B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to the C. elegans homologs of JIP3 (UNC-16; UniProt entry P34609-1), kinesin heavy chain KIF5C (UNC-116; P34540), and dynein light intermediate chain (DLI-1; G5ED34). WT denotes wild-type. Molecular weight is indicated in kilodaltons (kD). MBP::UNC-16[1-120] proteins (WT and V72Q) also contain a C-terminal Strep-tag II, which is detected on the immunoblot in A along with GST::DLI-1[369-442]. Source data are available for this figure: SourceData FS3.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_figs3.png?Expires=1773840818&Signature=ryWtR6eBOT0nCuqHQcd-si-3tMc4CoLBhxHw7FHlOdLf5gTHMqzAskuJIO7XwrPMLPG8A0KBmM65qsJUNAJxyy2oAy7KKROf42N8599DE4ZuaZnR6lLv~mH7CyG94BAooJap7sRjzUkvNvXQK7Ydi-rIAmZzTkeeFxKUbjn1J-6j9xeFDQL~qn8-Xm4kkcE2CM88mDYR~dIPHiSLMfTRFMtrp0YGWwHoCfK8rHW1NPfLjMbMb8IiXklRo0lJnNzD9bHBYMY6cv4jabiG9DPxBg1Z8ktRob54A6pc3FsY8yFNJf5kmcEAsJtffycSIoEaL1ILli17Qv7MOscxvhEQfA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

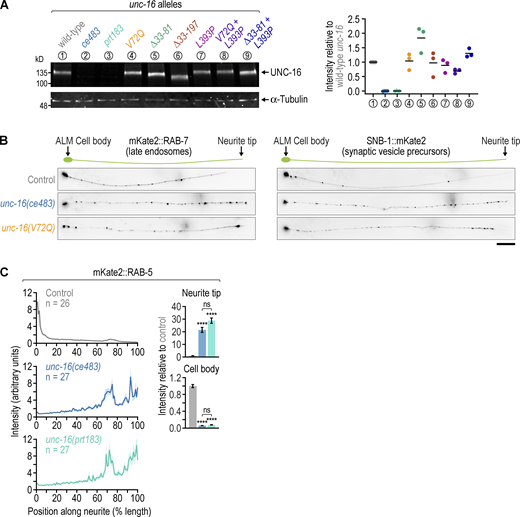

![Mutations in the C-terminal helix 1 of dynein light intermediate chain and the UNC-16/JIP3 V72Q mutation result in similar transport defects in touch receptor neurons. (A and (B) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for early endosomes (mKate2::RAB-5) or lysosomes (CTNS-1::mKate2). n denotes the number of neurites examined (1 per animal). Graph in A corresponds to the data in Fig. 4 F, re-plotted with a split y-axis to highlight the difference between unc-16(ce483) and the control. (C) Integrated fluorescence intensity (mean ± SEM, normalized to control) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). n denotes the number of neurites examined (1 per animal). Statistical significance [control versus mutants and unc-16(V72Q) versus dli-1(F392A/F393A)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001; ns = not significant, P > 0.05. (D) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM cell body and neurite tip in L4 animals expressing transgene-encoded UNC-16::GFP [background: endogenous wild-type unc-16 with or without dli-1(F392A/F393A)] or UNC-16(V72Q)::GFP [background: endogenous unc-16(V72Q)] in touch receptor neurons. Scale bar, 2 µm. (E) Integrated fluorescence intensity in the ALM cell body and neurite tip for the GFP-tagged UNC-16 versions described in D, plotted and statistically analyzed as in C. ****P < 0.0001; ns = not significant, P > 0.05. (F) Fluorescence intensity profiles (mean ± SEM) at the ALM neurite tip for the GFP-tagged UNC-16 versions described in D. n denotes the number of neurites examined (1 per animal). (G) Fluorescence kymographs (inverted grayscale) of mKate2::RAB-5 particle motility in the ALM neurite, generated from time-lapse sequences (single z-section) recorded at the larval L2 stage. The imaged region is ∼50 µm away from the cell body, which is located to the left. Scale bar, 5 µm. (H–J) Motility parameters (mean ± SEM) of mKate2::RAB-5 particles, derived from kymograph analysis. For H, n denotes the number of neurites examined (1 per animal). For I and J, n denotes the number of track segments, framed by a pause or a reversal, from at least 15 neurites (1 per animal). Statistical significance was determined as described for C. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig5.png?Expires=1773840819&Signature=flGL5QAZwVygbV4t56ElRUyEY2DfsKvTHrOP6guVm-krPVO9UrNXYnUk0xz3BclxO6GNrgv5t~fckrdRM8HZ3GntjgI~VgItlo1-VrXCvex8nP8W5DhrgLAtRP8sRXZFijXU~h6yWfml3bci0YzKpF1J5c0CCLcyy8t25J~uIegH-Hu0s-Uc8y7Jve8V-~7azmbGJvo7U0UYBtMP5d8FqO67DWN4V2~ppZcYz9zh6smdYeWv7ZKG~6XdcIPzvjrLgICQGZFGOu4cbJkAft2-ioIhaDNWfe1c~s--E07i1FSINcjNPXlhxxykRPpvmm8J-UQVHzXEbOHZqFOE8SYRHw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires kinesin-1 activity but not MAP kinase signalling, and N-terminal UNC-16 deletions mimic the unc-16(V72Q) mutant. (A) Domain organization of the N-terminal UNC-16 region with the putative binding sites for UNC-116/KHC and JNK-1 delineated by dashed lines. (B and D) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2), late endosomes (mKate2::RAB-7), or early endosomes (mKate2::RAB-5) in touch receptor neurons. n denotes the number of neurites examined (1 per animal). (C and E) Integrated fluorescence intensity [mean ± SEM, normalized to unc-16(V72Q)] in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined in C and E corresponds to the number n in B and D, respectively. Statistical significance was determined by the Mann-Whitney test. ****P < 0.0001; **P < 0.01; *P < 0.05; ns = not significant, P > 0.05. (F and G) Left: Fluorescence intensity profiles (mean ± SEM) in the ALM neurite, as described for B and D. Right: Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(V72Q) for F and to the control for G) in the cell body and at the neurite tip. The number of neurites examined corresponds to the number n in the fluorescence intensity profiles on the left. Statistical significance (control versus mutants and unc-16(V72Q) versus unc-16(V72Q); jnk-1(gk7)) was determined by the Mann-Whitney test for F and by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test for G. ****P < 0.0001; *P < 0.05; ns = not significant, P > 0.05.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig6.png?Expires=1773840819&Signature=SCEvyxGEx7RMWQICfn~sSObF6vK2XUtykjj8iFeuHHce8WcpGSwIkLGvteBzdsdZM7qzeut76FQMqDtfYumOkldeKhXwD9CnrcfH1coiGtuDyRNWmenLrQpYALJDAwnwky7L4HG6hzWGPexR9lYVvKuFn1LerD~6zBAmRiuCY39iNzcu7SE-uaGjCp9IhNS-tSyeRyhu5GaLwPfAnNpk30TgXu-hSsaSnTF~MushHk-N4z3jJm~xScNhUF3hnX5fTiEzM~75aT2JgcLvj41EXMKWk5wG1apLQqJ-PC8m4Bfg~pciRDlwMuBU5W15o7ijZhWdhpS6P3rJc3NyKH-YRw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Neurite tip accumulation of endo-lysosomal organelles in the unc-16(V72Q) mutant requires the interaction between UNC-16/JIP3 and kinesin light chain. (A) Domain organization of human JIP3 with sequence alignment of the kinesin light chain binding site in the JIP3 leucine zipper (LZ) domain. Arrowhead points to the leucine residue in JIP3 (L444 in human; L439 in mouse; L393 in C. elegans) whose mutation to proline causes neurological disease and was predicted to interfere with kinesin light chain binding (Platzer et al., 2019). (B) Left: Coomassie Blue-stained SDS-PAGE gel of purified recombinant protein mixtures prior to the addition of glutathione agarose resin (Input). Right: Coomassie Blue-stained SDS-PAGE gel of proteins eluted from glutathione agarose resin after GST pull-down. Proteins correspond to human kinesin light chain 1 (UniProt entry Q07866-1) and mouse JIP3 (Q9ESN9-5). Molecular weight is indicated in kilodaltons (kD). (C) Immunoblot of C. elegans adult lysates using antibodies against UNC-16 and α-tubulin (loading control). Molecular weight is indicated in kilodaltons (kD). (D) Locomotion of animals at the young adult stage, assessed by determining body bending frequency (mean ± SEM) in liquid medium. n denotes the number of animals examined. Statistical significance [wild-type N2 control versus unc-16 mutants and unc-16(prt183) versus unc-16(L393P)] was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ***P < 0.001. (E) Fluorescence images (maximum intensity z-stack projection, inverted grayscale) of the ALM neuron in L4 animals expressing a transgene-encoded marker for lysosomes (CTNS-1::mKate2) or early endosomes (mKate2::RAB-5) in touch receptor neurons. Scale bar, 20 µm. (F) Fluorescence intensity profiles (mean ± SEM) along the ALM neurite in the animals described in E. n denotes the number of neurites examined (1 per animal). (G) Number of CTNS-1::mKate2 puncta (mean ± SEM) in the first quarter of ALM neurite length after the cell body (proximal neurite), the middle two quarters (mid-neurite), and the last quarter (distal neurite). The number of neurites examined (1 per animal) corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined by ANOVA on ranks (Kruskal-Wallis nonparametric test) followed by Dunn’s multiple comparison test. ****P < 0.0001; ns = not significant, P > 0.05. (H) Integrated fluorescence intensity (mean ± SEM, normalized to unc-16(prt183)) in the ALM cell body and the last 20 µm of the distal neurite (neurite tip). The number of neurites examined corresponds to the number n in F. Statistical significance (control versus unc-16 mutants; other comparisons indicated by brackets) was determined as described for G. ****P < 0.0001; ***P < 0.001; **P < 0.01; ns = not significant, P > 0.05. Source data are available for this figure: SourceData F7.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/8/10.1083_jcb.202110057/2/m_jcb_202110057_fig7.png?Expires=1773840819&Signature=jJGR~KPaqAhvtEG0EpXA9mhYdL~GnsVgX8Ww73-RX9YH0gfU4~2VFVfGSSckFIJq5DPUGTAKQ7DTY002Vut1qeOlHHGbwPIe5d3qLhcK~9gze~4IpyQj-vD7u0gaAqpDjeWL7VKhqQfJ1CBZzyny1dnVjIO4Oeb2wfm~z4uLNblc0UOvI-lV-3hw6qkyNVHx4PMz5SXejGqnpZIMXAQK-6RL32zUUTymctCcu1tVibOkOGtuPyJXzqVIfgkmIL3JKykWilc4duoPfYt55wooykJY7ZNzKzU1EbBKpCCdFYMq9S-iYRC5CG7rE6lkOGAjSLmphMGxXEXajX1A7UoBww__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


