An exome sequencing strategy employed to identify pathogenic variants in patients with pediatric-onset systemic lupus or Evans syndrome resulted in the discovery of six novel monoallelic mutations in PTPN2. PTPN2 is a phosphatase that acts as an essential negative regulator of the JAK/STAT pathways. All mutations led to a loss of PTPN2 regulatory function as evidenced by in vitro assays and by hyperproliferation of patients’ T cells. Furthermore, patients exhibited high serum levels of inflammatory cytokines, mimicking the profile observed in individuals with gain-of-function mutations in STAT factors. Flow cytometry analysis of patients’ blood cells revealed typical alterations associated with autoimmunity and all patients presented with autoantibodies. These findings further supported the notion that a loss of function in negative regulators of cytokine pathways can lead to a broad spectrum of autoimmune manifestations and that PTPN2 along with SOCS1 haploinsufficiency constitute a new group of monogenic autoimmune diseases that can benefit from targeted therapy.
Introduction
The challenge facing the immune system is to deploy effective defenses against infection by pathogenic microorganisms, while at the same time maintaining tolerance to components of the self and limiting collateral damage. This delicate balance is ensured by a set of mechanisms that negatively regulate immune cell activation pathways. Dysfunctions in these mechanisms can result in inappropriate responses of innate or adaptive immune cells driving tissue damage and inflammation. In particular, the onset of autoimmune or inflammatory diseases in childhood is often linked to inborn errors of immunity (IEI), caused by damaging germline variants in single genes (Omarjee et al., 2019; Di Donato et al., 2021). The identification of the genetic cause underlying these diseases has largely contributed to pinpointing key actors in immune tolerance and non-redundant regulators of human immunity. This is notably the case of the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathways, which orchestrate immune response downstream of cytokine activation. Upon engagement of cytokine receptors, the JAK kinases (including JAK1, 2, 3 and tyrosine kinase 2) constitutively bound to their cytoplasmic tail, are transactivated, and phosphorylate the cytokine receptors, allowing the recruitment of the STAT transcription factors. Once phosphorylated by the JAKs, the STATs homo or heterodimerize and translocate to the nucleus where they induce the transcription of multiple genes whose promoters contain STAT-responsive elements (Philips et al., 2022). In recent years, an increasing number of IEI have been linked to dysregulated JAK/STAT pathways, including gain-of-function (GOF) mutations in STAT1, STAT2, STAT3, STAT4, and STAT6 (Ott et al., 2023). GOF mutations in STAT2 have been linked to interferonopathies associated with brain calcifications (Gruber et al., 2020; Duncan et al., 2019; Duncan and Hambleton, 2021). Heterozygous GOF mutations in STAT1 and STAT3 transcription factors (Zhang et al., 2021; van de Veerdonk et al., 2011; Fabre et al., 2019; Flanagan et al., 2014; Faletti et al., 2021) and in JAK1 and JAK3 kinases (Del Bel et al., 2017; Lesmana et al., 2020) result in a broad spectrum of autoimmune manifestations, demonstrating that hyperactivation of the JAK/STAT pathways invariably leads to autoimmunity.
The JAK/STAT pathways are tightly regulated by multiple negative feedback loops, including the suppressor of cytokine signaling (SOCS) family of proteins, which are rapidly induced upon activation of the pathway and promote signal termination (Ilangumaran and Rottapel, 2003). In particular, SOCS1 modulates JAK/STAT activity through various mechanisms, including the direct inhibition of JAK kinase activity, the limitation of STAT recruitment, and the ubiquitination of STATs, which results in their degradation by the proteasome. Accordingly, we recently reported germline heterozygous loss-of-function (LOF) mutations in SOCS1, leading to hyperactivation of the signaling cascade, in 10 patients with early onset autoimmune manifestations and immune-mediated cytopenia as the most common feature (Hadjadj et al., 2020). To date, 18 cases of SOCS1 haploinsufficiency have been described to be associated with early-onset autoimmunity (Hadjadj et al., 2020; Lee et al., 2020; Thaventhiran et al., 2020; Rodari et al., 2023; Körholz et al., 2021; Michniacki et al., 2022).
In addition, negative regulation of the JAK/STAT pathways is also achieved by members of the protein tyrosine phosphatases (PTPs) family via direct dephosphorylation of JAKs and STATs, which prevents their sustained activation. PTP non-receptor 2 (PTPN2) is ubiquitously expressed in humans, especially in immune cells (Song et al., 2022a), and exists in two isoforms, which share the same catalytic domain but differ in their C-terminal tail. They appear to have different substrate specificities due to their different localizations, although their respective roles remain poorly understood. The 48-kDa isoform (called T cell 48 [TC48]) is composed of 415 amino acids and is preferentially localized in the endoplasmic reticulum, although a nuclear localization has already been observed (Kamatkar et al., 1996). The 45-kDa isoform (TC45) is composed of 387 amino acids and shuttles between nucleus and cytoplasm. The C-terminal domain of PTPN2 appears to play an essential role in regulating subcellular localization of PTPN2 and tyrosine phosphatase activity (Hao et al., 1997; Singh et al., 2022). The 34 hydrophobic amino acids present in the TC48 isoform mask the nuclear localization signal (NLS), directing the protein in the endoplasmic reticulum. Also, in cells, PTPN2 exists in an inactive basal state due to the folding of its C-terminal domain over the catalytic site. Activation of the enzyme could be achieved by the cellular environment, notably through competitive interaction with other cellular proteins, such as integrin alpha-1, that would displace the C-terminal tail and allow accessibility of the catalytic site (Singh et al., 2022; Tun et al., 2022). The important and non-redundant role of PTPN2 in the regulation of inflammation has been first shown in mouse models. Ptpn2−/− mice die shortly after birth from systemic inflammation due to lymphocyte hyperactivation and increased IFN-γ secretion (Heinonen et al., 2004; You-Ten et al., 1997). In humans, genome-wide association studies have shown that single nucleotide polymorphisms in the PTPN2 gene are associated with inflammatory manifestations such as inflammatory bowel disease, type I diabetes, and rheumatoid arthritis (Wellcome Trust Case Control Consortium, 2007; Todd et al., 2007). Recently, both heterozygous and homozygous mutations resulting in PTPN2 LOF have been identified in patients with autoimmune enteropathy (Parlato et al., 2020; Awwad et al., 2023) and with primary immune deficiency and autoimmunity with variable manifestation (Thaventhiran et al., 2020).
In this study, we report the identification of six novel PTPN2 monoallelic variants in six unrelated families, associated with various autoimmune manifestations (juvenile lupus or Evans syndrome). All six variants led to functional deficiency of PTPN2, either by loss of expression or by changes in its phosphatase activity resulting in hyperactivation of the JAK/STAT pathways.
Results
Clinical features of patients
We assessed six unrelated patients exhibiting features of systemic autoimmunity. Main clinical characteristics are summarized in Table 1. All patients presented with symptoms at pediatric age. Patient A1 developed a juvenile systemic lupus erythematosus (SLE) beginning at the age of 5, with skin rash, lupus nephritis, hematological system, and liver involvements with chronic hepatitis and cholangitis (Metavir score A3F3) (Fig. 1 A). First-degree relatives remained asymptomatic. Patients B1, C1, D1, E1, and F1 had association of autoimmune thrombocytopenia and hemolytic anemia (Evans syndrome). Two patients (B1 and D1) had recurrent infections (otitis and pneumonitis) and two (D1 and E1) had clinical lymphoproliferation characterized by lymphoid hyperplasia or splenomegaly. Laboratory exams revealed variable anomalies. Serum levels of Ig classes IgG and IgA were normal before supplementation in three patients, and five had detectable platelet autoantibodies (A1, B1, C1, E1, and F1). We also monitored other autoantibodies in sera from patients when available. Patient A1 had positive antinuclear antibodies (ANA), anti-β2 glycoprotein-1 (β2GP1), and anti-C1q. B1 and F1 had positive ANA, C1 had positive ANA and antineutrophil cytoplasmic antibodies (ANCA), and E1 had positive anti-HLA I. Three patients required intermittent immunoglobulins perfusions and three received an immunosuppressive drug (azathioprine and sirolimus). The clinical presentations are more detailed in Data S1.
Clinical phenotype of patients with PTPN2 mutations
| . | Family A . | Family B . | Family C . | Family D . | Family E . | Family F . | |
|---|---|---|---|---|---|---|---|
| Patient | A1 (Proband) | A2 (Father) | B1 (Proband) | C1 (Proband) | D1 (Proband) | E1 (Proband) | F1 (Proband) |
| Gender | F | M | F | F | F | F | M |
| Genotype | c.1209delT (p.Phe403Leufs*25) | c.1209delT (p.Phe403Leufs*25) | c.293G>A (p.Trp98*) | c.866G>A (p.Trp289Ter) | c.70-2A>T (p.Glu24Metfs*20) | c.376T>A (p.Tyr126Asn) | c.647G>C (p.Cys216Ser) |
| Age at onset (years) | 5 | 9 | 6 | Infancy | Infancy | 8 | |
| Clinical features | |||||||
| Autoimmune manifestations | Lupus (renal, cutaneous, hematological, hepatitis) | Asymptomatic | Evans syndrome | Evans syndrome | ITP Common variable immune deficiency | Evans syndrome, autoimmune neutropenia Hashimoto thyroiditis Interstitial pneumonitis Mild cognitive impairment | Evans syndrome (leuco-neutropenia and thrombopenia) Recurrent mouth aphthae Pediatric psychiatric disorders |
| Clinical lymphoproliferation | No | No | No | No | Yes (lymphoid hyperplasia) | Yes (splenomegaly) | No |
| Metabolic disorders | No | No | No | No | No | No | No |
| Growth | Normal | na | na | na | na | na | No |
| Infections | No | No | Yes (recurrent otitis) | No | Yes (recurrent pneumonitis) | No | Rare |
| Therapy | CS, HC, AZA | No | Intermittent Ig | CS, intermittent Ig, VIN, COL, AZA | Intermittent Ig | SIR, relayed by AZA | Antibiotic prophylaxis, AZA |
| Laboratory values | |||||||
| Antibodies (UA/ml) | ANA+ Anti double-stranded DNA and ENA-Anti B2Gp1+ Anti C1q+ Coombs: + | na Coombs: + | ANA+ Coombs: + | ANA+ ANCA+ Coombs: + | na Coombs: + | Anti-HLA I Coombs: + | ANA+ Coombs: + |
| Complement (g/L; normal range) | Lowered | Normal | Normal | na | na | na | Normal |
| Platelet antibodies | Yes | na | Yes | Yes | na | Yes | Yes |
| Type 1 IFN | Positive (7.3) | Negative | na | na | na | na | na |
| Ig | Normal Type III cryoglobulinemia | Inc. IgG Normal IgA and IgM | Normal | Normal IgG and IgA Dec. IgM (0.39) | Dec. IgG, IgA, IgM | Normal | Normal IgG, IgM Dec. IgA, IgG2 partial deficiency |
| Immunophenotyping | Inc. Treg | Normal | Normal Treg Dec. TCD4+; Th1, Th2, Th17; NK Inc. Tfh, auto-reactive B cells | Global lymphopenia, predominant on TCD4+ and NK | TCD4+ moderate lymphocytosis Dec. memory B cells Inc. auto-reactive B cells | Inc. naive B cells Inc. B cells CD21 low | Mild CD4 T and NK lymphopenia, deficiency in switched memory B cells; significant excess of double-negative T cells (TCRab CD8− CD4−) |
| . | Family A . | Family B . | Family C . | Family D . | Family E . | Family F . | |
|---|---|---|---|---|---|---|---|
| Patient | A1 (Proband) | A2 (Father) | B1 (Proband) | C1 (Proband) | D1 (Proband) | E1 (Proband) | F1 (Proband) |
| Gender | F | M | F | F | F | F | M |
| Genotype | c.1209delT (p.Phe403Leufs*25) | c.1209delT (p.Phe403Leufs*25) | c.293G>A (p.Trp98*) | c.866G>A (p.Trp289Ter) | c.70-2A>T (p.Glu24Metfs*20) | c.376T>A (p.Tyr126Asn) | c.647G>C (p.Cys216Ser) |
| Age at onset (years) | 5 | 9 | 6 | Infancy | Infancy | 8 | |
| Clinical features | |||||||
| Autoimmune manifestations | Lupus (renal, cutaneous, hematological, hepatitis) | Asymptomatic | Evans syndrome | Evans syndrome | ITP Common variable immune deficiency | Evans syndrome, autoimmune neutropenia Hashimoto thyroiditis Interstitial pneumonitis Mild cognitive impairment | Evans syndrome (leuco-neutropenia and thrombopenia) Recurrent mouth aphthae Pediatric psychiatric disorders |
| Clinical lymphoproliferation | No | No | No | No | Yes (lymphoid hyperplasia) | Yes (splenomegaly) | No |
| Metabolic disorders | No | No | No | No | No | No | No |
| Growth | Normal | na | na | na | na | na | No |
| Infections | No | No | Yes (recurrent otitis) | No | Yes (recurrent pneumonitis) | No | Rare |
| Therapy | CS, HC, AZA | No | Intermittent Ig | CS, intermittent Ig, VIN, COL, AZA | Intermittent Ig | SIR, relayed by AZA | Antibiotic prophylaxis, AZA |
| Laboratory values | |||||||
| Antibodies (UA/ml) | ANA+ Anti double-stranded DNA and ENA-Anti B2Gp1+ Anti C1q+ Coombs: + | na Coombs: + | ANA+ Coombs: + | ANA+ ANCA+ Coombs: + | na Coombs: + | Anti-HLA I Coombs: + | ANA+ Coombs: + |
| Complement (g/L; normal range) | Lowered | Normal | Normal | na | na | na | Normal |
| Platelet antibodies | Yes | na | Yes | Yes | na | Yes | Yes |
| Type 1 IFN | Positive (7.3) | Negative | na | na | na | na | na |
| Ig | Normal Type III cryoglobulinemia | Inc. IgG Normal IgA and IgM | Normal | Normal IgG and IgA Dec. IgM (0.39) | Dec. IgG, IgA, IgM | Normal | Normal IgG, IgM Dec. IgA, IgG2 partial deficiency |
| Immunophenotyping | Inc. Treg | Normal | Normal Treg Dec. TCD4+; Th1, Th2, Th17; NK Inc. Tfh, auto-reactive B cells | Global lymphopenia, predominant on TCD4+ and NK | TCD4+ moderate lymphocytosis Dec. memory B cells Inc. auto-reactive B cells | Inc. naive B cells Inc. B cells CD21 low | Mild CD4 T and NK lymphopenia, deficiency in switched memory B cells; significant excess of double-negative T cells (TCRab CD8− CD4−) |
M: male; F: female; ITP: idiopathic thrombocytopenic purpura; AIHA: autoimmune hemolytic anemia; ENA: extractable nuclear antigen antibodies; GP: glycoprotein; LB: B lymphocytes; AZA: azathioprine; CS: corticosteroids; HC: hydroxychloroquine; VIN: vinblastine; COL: colchicine; SIR: sirolimus; na: nonavailable; Inc.: increased; Dec.: decreased.
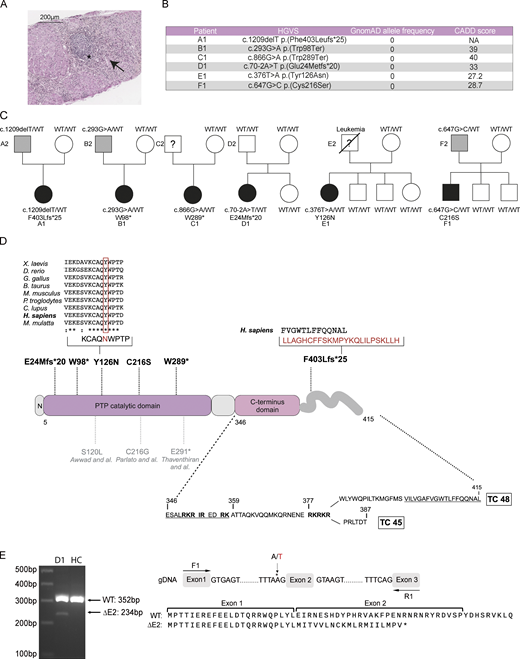
Pedigrees and genetics of families carrying PTPN2 mutations. (A) Anatomopathological section of liver biopsy showing lesions of chronic hepatitis and cholangitis (Metavir A3F3) with an inflammatory infiltrate compatible with a lupus-type autoimmune etiology. Hematoxylin-eosin saffron staining: portal tract inflammatory infiltrate with lymphoid follicles (*) and piecemeal necrosis (arrows). HGVS, Human Genome Variation Society. (B) cDNA mutations, GnomAD allele frequency (v2.1), and CADD score. (C) Family pedigrees with probands indicated by A1, B1, C1, D1, E1, F1. Squares: males; circles: females; black: affected mutation carriers; grey: unaffected mutation carriers. WT: WT PTPN2 allele. Under each patient is the genotype at the specified locus. (D) Linear protein model showing the location of the described PTPN2 variants (represented in bold) together with previously published variants (in grey). Multiple alignments of PTPN2 orthologs from different species using the Clustal Omega software. The affected amino acid for Y126N is boxed in red. Residues corresponding to bipartite NLS are underlined. TC, T cell. (E) cDNA from EBV-transformed B cells of patient D1 and one HC were amplified around exon 2. Migration and sequencing of PCR products are represented along with the expected result of RNA translation with exon 2 skipping. Source data are available for this figure: SourceData F1.
Pedigrees and genetics of families carrying PTPN2 mutations. (A) Anatomopathological section of liver biopsy showing lesions of chronic hepatitis and cholangitis (Metavir A3F3) with an inflammatory infiltrate compatible with a lupus-type autoimmune etiology. Hematoxylin-eosin saffron staining: portal tract inflammatory infiltrate with lymphoid follicles (*) and piecemeal necrosis (arrows). HGVS, Human Genome Variation Society. (B) cDNA mutations, GnomAD allele frequency (v2.1), and CADD score. (C) Family pedigrees with probands indicated by A1, B1, C1, D1, E1, F1. Squares: males; circles: females; black: affected mutation carriers; grey: unaffected mutation carriers. WT: WT PTPN2 allele. Under each patient is the genotype at the specified locus. (D) Linear protein model showing the location of the described PTPN2 variants (represented in bold) together with previously published variants (in grey). Multiple alignments of PTPN2 orthologs from different species using the Clustal Omega software. The affected amino acid for Y126N is boxed in red. Residues corresponding to bipartite NLS are underlined. TC, T cell. (E) cDNA from EBV-transformed B cells of patient D1 and one HC were amplified around exon 2. Migration and sequencing of PCR products are represented along with the expected result of RNA translation with exon 2 skipping. Source data are available for this figure: SourceData F1.
Identification of PTPN2 heterozygous mutations in patients using next-generation sequencing
Exome sequencing analysis in these patients led to the identification of six different heterozygous germline PTPN2 mutations, including three truncating variants (one frameshift and two non-sense), two missense, and one affecting a canonical splice site. All variants were absent from the Genome Aggregation Database, gnomAD (Fig. 1 B). For patients A1 (F403Lfs*25), B1 (W98*), and F1 (C216S), the variant was inherited from the asymptomatic father, suggesting autosomal dominant inheritance with incomplete clinical penetrance (Fig. 1 C). Variant from patient D1 (E24Mfs*20) was likely a de novo mutation, and genetic material from C1 and E1’s father was not available.
The A1 (F403Lfs*25) variant was a single nucleotide deletion, leading to a frameshift and the replacement of the last 13 C-terminus amino acids of the long isoform by a neo sequence of 24 amino acids (Fig. 1 D). Patients B1 (W98*) and C1 (W289*) carried non-sense mutations predicted to encode truncated proteins of 98 and 289 amino acids, respectively. The E1 (Y126N) mutation was a missense variant, with a tyrosine replaced by asparagine in a highly conserved region across species (Fig. 1 D). This substitution was predicted to be deleterious by in silico tools such as the combined annotation-dependent depletion (CADD) score (Fig. 1 B). The D1 (E24Mfs*20) mutation affected the canonical exon 2 splice acceptor site, thus resulting in exon 2 skipping. Using primers on flanking exon 1 and 3, we observed a shorter RT-PCR product confirming the exon 2 skipping at the cDNA level. Sanger sequencing confirmed the predicted frameshift resulting in a 43–amino acids truncated protein (Fig. 1 E). The F1 (C216S) mutation was a missense variant. A mutation at this position was already described, but with the substitution of the cysteine by a glycine (Parlato et al., 2020). This mutation was associated with the development of Crohn’s disease. Except for patient A1 (F403Lfs*25), all mutations were located in the catalytic domain of the phosphatase (Fig. 1 D).
PTPN2 isoforms expression and impact of mutations on protein expression
PTPN2 is expressed as two isoforms: the TC48 isoform referred to as the canonical sequence and located preferentially in the endoplasmic reticulum, and the TC45 isoform located preferentially in the nucleus and the cytoplasm. The expression pattern and the function of both isoforms have not been extensively studied so far. We therefore evaluated PTPN2 protein expression in different immune cells and cell lines. We sorted T, B, natural killer (NK), and monocytes from the blood of healthy donors and evaluated PTPN2 protein expression using automated quantitative western blot (Fig. 2 A, upper panel). We could only detect the TC48 isoform in these cells, as well as in control EBV-transformed B cell lines. In the Jurkat–T cell line, the TC48 isoform was the predominant form, while the TC45 isoform was present at a low level (Fig. 2 A, lower panel).
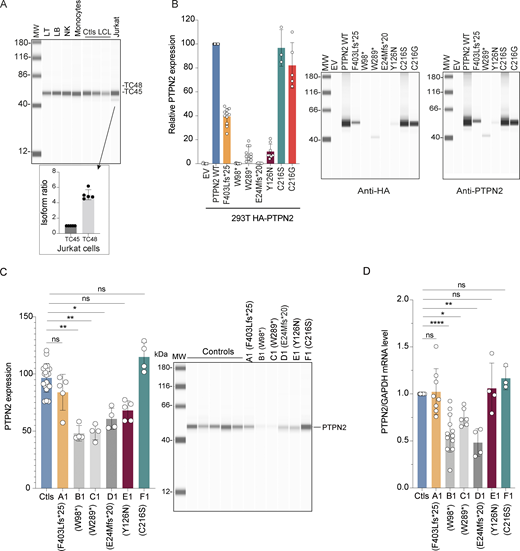
PTPN2 regulation and expression. (A) PTPN2 protein expression in human primary T (LT) and B lymphocytes (LB), NK cells, monocytes, Jurkat–T cell line, and EBV-derived control B cell lines (LCL). PTPN2 expression was analyzed using automated quantitative western blot. Relative expression of TC48 and TC45 isoforms in Jurkat–T cells is indicated in the lower panel (mean and SDs from five different experiments are represented). (B) PTPN2 expression in HEK293T cell transfected with different constructs. Protein expression was revealed with anti-HA (left panel) or anti-PTPN2 (right panel) antibodies and analyzed using automated quantitative western blot. Protein quantification was calculated on anti-HA expression normalized on total protein expression. Mean and SDs are shown and representative of at least three different experiments. (C) Endogenous PTPN2 protein expression in control and patient in vitro–activated T cells analyzed using automated quantitative western blot. Quantification was calculated on anti-PTPN2 expression normalized on total protein expression. Mean and SDs are shown and representative of three different experiments (ns, non-significant, *P < 0.05, **P < 0.01, one-way analysis of variance, Kruskal–Wallis test). (D) Real-time RT-QPCR assays of PTPN2 in control and patient in vitro activated T cells. Results represent the PTPN2 expression normalized to GAPDH expression, related to the expression of controls. Mean and SDs are shown and representative of at least three different experiments (ns, nonsignificant, *P < 0.05, **P < 0.01, ****P < 0.0001, one-way analysis of variance, Kruskal–Wallis test). All MW are in kDa. Source data are available for this figure: SourceData F2.
PTPN2 regulation and expression. (A) PTPN2 protein expression in human primary T (LT) and B lymphocytes (LB), NK cells, monocytes, Jurkat–T cell line, and EBV-derived control B cell lines (LCL). PTPN2 expression was analyzed using automated quantitative western blot. Relative expression of TC48 and TC45 isoforms in Jurkat–T cells is indicated in the lower panel (mean and SDs from five different experiments are represented). (B) PTPN2 expression in HEK293T cell transfected with different constructs. Protein expression was revealed with anti-HA (left panel) or anti-PTPN2 (right panel) antibodies and analyzed using automated quantitative western blot. Protein quantification was calculated on anti-HA expression normalized on total protein expression. Mean and SDs are shown and representative of at least three different experiments. (C) Endogenous PTPN2 protein expression in control and patient in vitro–activated T cells analyzed using automated quantitative western blot. Quantification was calculated on anti-PTPN2 expression normalized on total protein expression. Mean and SDs are shown and representative of three different experiments (ns, non-significant, *P < 0.05, **P < 0.01, one-way analysis of variance, Kruskal–Wallis test). (D) Real-time RT-QPCR assays of PTPN2 in control and patient in vitro activated T cells. Results represent the PTPN2 expression normalized to GAPDH expression, related to the expression of controls. Mean and SDs are shown and representative of at least three different experiments (ns, nonsignificant, *P < 0.05, **P < 0.01, ****P < 0.0001, one-way analysis of variance, Kruskal–Wallis test). All MW are in kDa. Source data are available for this figure: SourceData F2.
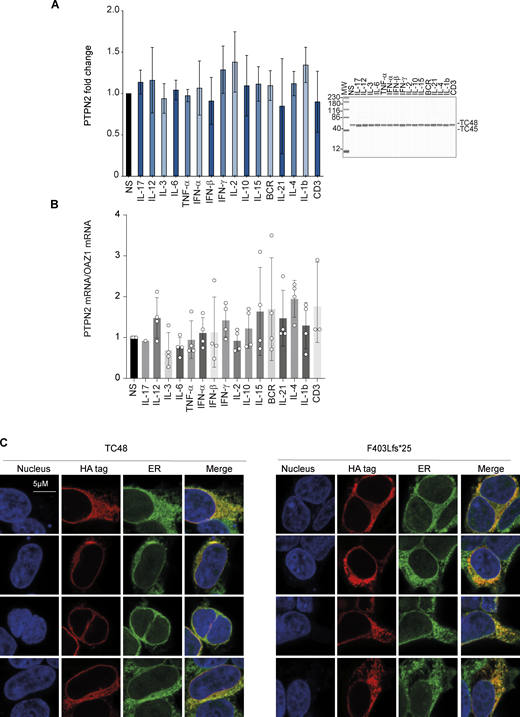
To investigate the transcriptional regulation of PTPN2, we stimulated peripheral blood mononuclear cells (PBMCs) from healthy donors with different cytokines or with anti-CD3 or anti-BCR for 6 h and assessed PTPN2 expression at the protein or mRNA levels. Results showed that PTPN2 was expressed at steady state at the protein (Fig. S1 A) and mRNA (Fig. S1 B) levels, and its expression was not increased following cytokine or BCR/TCR stimulation, at least at the time point analyzed.
PTPN2 expression following PBMC stimulation and intracellular localization. (A) PTPN2 protein in PBMCs stimulated 6 h with indicated cytokines or ligands was quantified using automated quantitative western blot. Protein quantification was calculated on anti-PTPN2 expression normalized on total protein expression. Mean and SDs are shown and representative of three different experiments. All MW are in kDa. (B) PTPN2 mRNA was measured in control PBMCs stimulated 6 h with indicated cytokines or ligands. Mean and SDs are shown and representative of three different experiments. (C) Confocal microscopy analysis of PTPN2 subcellular localization. Constructs were overexpressed in HEK293T cells and detected with anti-HA antibody together with endoplasmic reticulum and nucleus staining. Source data are available for this figure: SourceData FS1.
PTPN2 expression following PBMC stimulation and intracellular localization. (A) PTPN2 protein in PBMCs stimulated 6 h with indicated cytokines or ligands was quantified using automated quantitative western blot. Protein quantification was calculated on anti-PTPN2 expression normalized on total protein expression. Mean and SDs are shown and representative of three different experiments. All MW are in kDa. (B) PTPN2 mRNA was measured in control PBMCs stimulated 6 h with indicated cytokines or ligands. Mean and SDs are shown and representative of three different experiments. (C) Confocal microscopy analysis of PTPN2 subcellular localization. Constructs were overexpressed in HEK293T cells and detected with anti-HA antibody together with endoplasmic reticulum and nucleus staining. Source data are available for this figure: SourceData FS1.
We then assessed the impact of PTPN2 mutations identified in patients on protein expression. To do so, corresponding cDNA sequences were cloned in a mammalian expression vector with the addition of an HA-Tag sequence in the N-terminal part of the protein. Since TC48 seems to be the main isoform expressed in primary immune cells, we focused on this isoform (hereafter referred to as PTPN2 WT). As a control, we also analyzed the expression of the PTPN2 loss of function variant (C216G) previously reported to be involved in Crohn’s disease (Parlato et al., 2020). Expression plasmids were transfected into the HEK293T cell line and PTPN2 expression was quantified using automated quantitative western blot using anti-HA or anti-PTPN2 specific antibodies (Fig. 2 B). Mutations resulting in truncated proteins of 98 and 43 amino acids (found in patients B1 and D1, respectively) did not yield any detectable expression of the proteins. W289* or Y126N variants (patients C1 and E1, respectively) were weakly expressed at the protein level (10% of the PTPN2 WT). The frameshift mutation (patient A1) encoded a detectable protein expressed approximately at half the level of the PTPN2 WT. The C216S (patient F1) and control C216G mutations found in Crohn’s patient had no effect on PTPN2 expression as compared with PTPN2 WT. For mutant proteins that were still expressed, we checked that they were also detected by an anti-PTPN2 antibody (Fig. 2 B, right panel). In particular, a truncated band at 40 kDa was detected for W289* variant with anti-PTPN2 antibody. We then quantified PTPN2 expression at protein and mRNA levels in in vitro–activated T cells from patients or healthy donors (Fig. 2, C and D, respectively). PTPN2 protein expression was 50% lower in patients B1, C1, and D1, suggesting haploinsufficiency, and 15% and 35% lower in patients A1 and E1, respectively. No decreased PTPN2 expression was observed in patient F1. Importantly, we did not detect the truncated form of PTPN2 in patient C1’s cells, contrary to what was observed when overexpressing W289* cDNA in HEK293T cells (Fig. 2 C). These results were consistent with those obtained at the mRNA level.
Impact of mutations on PTPN2 regulatory functions
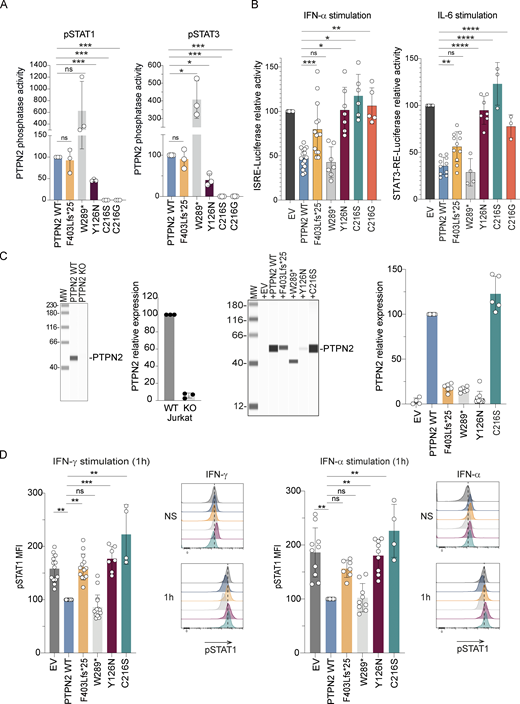
We then assessed the intrinsic tyrosine phosphatase activity of PTPN2 variants that exhibited residual or normal expression (F403Lfs*25, W289*, Y126N, and C216S) as compared with PTPN2 WT and the previously described Crohn variant (C216G). PTPN2 variants were overexpressed in HEK293T cells and immunoprecipitated using an anti-HA antibody. Intrinsic tyrosine phosphatase activity of these immunoprecipitated proteins was then quantified using phospho-STAT1 or STAT3 peptides as substrates (Fig. 3 A). As previously described, the C216G variant had a complete loss of tyrosine phosphatase activity compared with PTPN2 WT, and we obtained the same result with our C216S variant. We also observed a decrease in intrinsic tyrosine phosphatase activity for the Y126N variant (to 50% of WT levels). The phosphatase activity of the F403Lfs*25 variant was equivalent to that of PTPN2 WT. On the contrary, the W289* variant was five times more efficient than the PTPN2 WT in dephosphorylating STAT1 and STAT3 peptides. This is likely due to the loss of the autoinhibitory C-terminal domain of the protein in this variant since a previous study reported that the removal of the last 20 C-terminal amino acids of PTPN2 was sufficient to fully activate the enzyme (Hao et al., 1997).
Effects of mutations on PTPN2 phosphatase activity and JAK-STAT signaling. (A) Tyrosine PTPN2 phosphatase activity of immunoprecipitated WT or mutant PTPN2 toward fluorescent tyrosine-phosphorylated STAT1 or STAT3 peptide by RP-UFLC. Mean and SDs are shown and represent three different experiments (ns, nonsignificant, *P < 0.05, ***P < 0.001, Paired t test). (B) STAT1 transcriptional activity following 24-h activation with IFN-α of HEK293T cells expressing IFN-sensitive responsive element (ISRE) and transfected with the indicated constructs (left panel). Mean and SDs are shown and representative of at least five different experiments in triplicate STAT3 transcriptional activity following 24-h activation with IL-6 of HEK293T cells expressing STAT3 responsive element and transfected with the indicated constructs (right panel). Results were normalized on Renilla. Mean and SDs are shown and representative of at least three different experiments in triplicate (ns, nonsignificant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ordinary one-way ANOVA, followed by Dunnett’s multiple comparisons test). (C) PTPN2 protein expression in Jurkat–T cells and PTPN2 Crispr KO Jurkat–T cells. PTPN2 expression was quantified using anti-PTPN2 antibody and automated quantitative western blot (left panels). Mean and SDs are shown and representative of three different experiments. PTPN2 constructs were re-expressed in Jurkat–T cells following lentiviral transduction and protein expression was verified using anti-HA antibody and automated quantitative western blot (right panels). Mean and SDs are shown and representative of five different experiments. (D) Modified Jurkat–T cells were stimulated with IFN-α or IFN-γ as indicated and phosphorylation of STAT1 was measured by intracellular staining and analyzed by spectral cytometry. pSTAT1 phosphorylation in Jurkat transduced with PTPN2 WT was used as reference. Histograms representative of one experiment out of four are illustrated on the right of each quantification (ns, nonsignificant, *P < 0.05, **P < 0.01, one-way ANOVA, Kruskal–Wallis test). All MW are in kDa. Source data are available for this figure: SourceData F3.
Effects of mutations on PTPN2 phosphatase activity and JAK-STAT signaling. (A) Tyrosine PTPN2 phosphatase activity of immunoprecipitated WT or mutant PTPN2 toward fluorescent tyrosine-phosphorylated STAT1 or STAT3 peptide by RP-UFLC. Mean and SDs are shown and represent three different experiments (ns, nonsignificant, *P < 0.05, ***P < 0.001, Paired t test). (B) STAT1 transcriptional activity following 24-h activation with IFN-α of HEK293T cells expressing IFN-sensitive responsive element (ISRE) and transfected with the indicated constructs (left panel). Mean and SDs are shown and representative of at least five different experiments in triplicate STAT3 transcriptional activity following 24-h activation with IL-6 of HEK293T cells expressing STAT3 responsive element and transfected with the indicated constructs (right panel). Results were normalized on Renilla. Mean and SDs are shown and representative of at least three different experiments in triplicate (ns, nonsignificant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ordinary one-way ANOVA, followed by Dunnett’s multiple comparisons test). (C) PTPN2 protein expression in Jurkat–T cells and PTPN2 Crispr KO Jurkat–T cells. PTPN2 expression was quantified using anti-PTPN2 antibody and automated quantitative western blot (left panels). Mean and SDs are shown and representative of three different experiments. PTPN2 constructs were re-expressed in Jurkat–T cells following lentiviral transduction and protein expression was verified using anti-HA antibody and automated quantitative western blot (right panels). Mean and SDs are shown and representative of five different experiments. (D) Modified Jurkat–T cells were stimulated with IFN-α or IFN-γ as indicated and phosphorylation of STAT1 was measured by intracellular staining and analyzed by spectral cytometry. pSTAT1 phosphorylation in Jurkat transduced with PTPN2 WT was used as reference. Histograms representative of one experiment out of four are illustrated on the right of each quantification (ns, nonsignificant, *P < 0.05, **P < 0.01, one-way ANOVA, Kruskal–Wallis test). All MW are in kDa. Source data are available for this figure: SourceData F3.
As PTPN2 acts as a negative regulator of the JAK/STAT pathways, we evaluated the ability of the different variants to regulate the cellular response to different cytokines. We first used a luciferase reporter plasmid carrying the luciferase gene under the control of an IFN-stimulated response element (ISRE)–dependent promoter or a STAT3 response element–dependent promoter that was transfected in HEK293T together with PTPN2 variants. While in the control condition (empty vector), stimulation with IFN-α or IL-6 strongly induced luciferase expression (Fig. 3 B), the PTPN2 WT overexpression led to a significant reduction in the luciferase signal, thus illustrating the strong inhibitory function of PTPN2 on the JAK/STAT pathways. However, this inhibitory activity was partially or totally impaired for the F403Lfs*25, Y126N, and C216S variants following IFN-α or IL-6 stimulation (Fig. 3 B). The W289* PTPN2 variant showed similar activity as the PTPN2 WT protein, which is likely due to the loss of its autoinhibitory C-tail and subsequent increased catalytic activity thus compensating for its reduced expression (Fig. 3 A) (Hao et al., 1997).
To confirm these results, we used another cellular model based on the Jurkat–T cell line. We knocked out endogenous PTPN2 (both isoforms) using Crispr-Cas9–based genome editing and verified loss of PTPN2 protein expression (Fig. 3 C, left panel). We then stably complemented this PTPN2-deficient cell line with the PTPN2 WT or the different PTPN2 variants using lentivectors (Fig. 3 C, right panel). The transduced cells were stimulated with IFN-α or IFN-γ, and pSTAT1 was measured by flow cytometry. As shown in Fig. 3 D, STAT1 phosphorylation was strongly induced by both stimulations in PTPN2 KO Jurkat–T cells. Moreover, STAT1 phosphorylation was unchanged when F403Lfs*25, Y126N, or C216S were expressed, whereas it was decreased following expression of PTPN2 WT or W289* mutants, thus confirming the results obtained with the luciferase reporter assay.
Since we failed to identify a decreased phosphatase activity for the F403Lfs*25 variant in vitro, while its activity was reduced in the luciferase assay and in the Jurkat model, we reasoned that this mutation could impair the PTPN2 subcellular localization. To test this possibility, we expressed the WT TC48 PTPN2 or the F403Lfs*25 variant in HEK293T cells. We then analyzed their subcellular localization by fluorescence microscopy. As expected, the WT TC48 isoform was mainly localized in the endoplasmic reticulum. Yet, the F403Lfs*25 mutant was also located in the endoplasmic reticulum, suggesting that the C-terminal modification of this variant did not impair its intracellular localization (Fig. S1 C).
Enhanced JAK/STAT signaling in patients’ PBMCs in response to cytokines
Experiments in cell lines suggested a severe impact of the different mutations on PTPN2 expression or function on the JAK/STAT pathways downstream of cytokine receptors. To confirm these results, we compared the cytokine response of blood cells from patients to that of healthy donors. We first analyzed STAT5 phosphorylation of CD4+ and CD8+ T cells after ex vivo stimulation of whole blood with IL-2. We observed a higher phospho-STAT5 staining after 15 min of IL-2 stimulation of T cells from patients compared with controls (Fig. 4 A). We also observed a higher phospho-STAT1 staining on patients’ monocytes after 15 min of IFN-γ stimulation (Fig. 4 A).
Study of JAK/STAT signaling pathway response in in vitro–activated T cell from patients. (A) Phospho-STAT5 analysis at steady state (NS) in CD4+ T cells and CD8+ T cells from HCs (blue bars) or patients (grey bars) after ex vivo stimulation of whole blood with IL-2 (104 U/ml) for 15 min. Phospho-STAT1 analysis at steady state (NS) in CD14+ monocytes from HC (blue bars) or patients (grey bars) after ex vivo stimulation of whole blood with IFN-γ (104 U/ml) for 15 min. Mean and SDs are shown and representative of four different experiments, with a total height of HCs. P values were obtained with two-way ANOVA: <0.0001 (****), 0.0002 (***), 0.0021 (**), 0.1234 (ns). (B) Phosphorylation of STAT5 was assessed by intracellular staining in in vitro–activated T cells from HC and patients without stimulation (left histograms), upon 15 min IL-2 stimulation (250 U/ml) (middle histograms) or 2 h following IL-2 removal after IL-2 stimulation (250 U/ml) (right histograms). Representative flow cytometry histograms are shown on the left. Normalized pSTAT5 mean fluorescence intensity (MFI) to the mean of controls from the day of the experiment for each condition, n = 7 patients, n = 13 controls. The results were obtained from five separate experiments. P values were obtained with two-way ANOVA: <0.0332 (*), 0.1234 (ns). NS, nonstimulated. (C) Proliferation of in vitro–activated T cells stimulated or not with different concentrations of IL-2 for 4 days. T cell proliferation was determined from the level of CellTrace Violet (CTV) dye dilution. Representative histograms (left panel) show cell divisions of activated CD8 T cells from HCs or PTPN2 patients as indicated. Percentage of CD8+ dividing cells from HCs and patients (right panel). Dividing cells represent cells having undergone at least one division (data pooled from n = 5 independent experiments including a total of 18 HCs and 7 patients). P values were determined in a two-way ANOVA: <0.0002 (***), 0.0021(**), and 0.1234 (ns). (D) Proliferation of patients’ IL2-stimulated activated T cells with or without incubation with JAK1/JAK3 inhibitor tofacitinib for 4 days (data pooled from n = 2 independent experiments including a total of seven HCs and seven patients). P values were determined in a two-way ANOVA: <0.0332 (*).
Study of JAK/STAT signaling pathway response in in vitro–activated T cell from patients. (A) Phospho-STAT5 analysis at steady state (NS) in CD4+ T cells and CD8+ T cells from HCs (blue bars) or patients (grey bars) after ex vivo stimulation of whole blood with IL-2 (104 U/ml) for 15 min. Phospho-STAT1 analysis at steady state (NS) in CD14+ monocytes from HC (blue bars) or patients (grey bars) after ex vivo stimulation of whole blood with IFN-γ (104 U/ml) for 15 min. Mean and SDs are shown and representative of four different experiments, with a total height of HCs. P values were obtained with two-way ANOVA: <0.0001 (****), 0.0002 (***), 0.0021 (**), 0.1234 (ns). (B) Phosphorylation of STAT5 was assessed by intracellular staining in in vitro–activated T cells from HC and patients without stimulation (left histograms), upon 15 min IL-2 stimulation (250 U/ml) (middle histograms) or 2 h following IL-2 removal after IL-2 stimulation (250 U/ml) (right histograms). Representative flow cytometry histograms are shown on the left. Normalized pSTAT5 mean fluorescence intensity (MFI) to the mean of controls from the day of the experiment for each condition, n = 7 patients, n = 13 controls. The results were obtained from five separate experiments. P values were obtained with two-way ANOVA: <0.0332 (*), 0.1234 (ns). NS, nonstimulated. (C) Proliferation of in vitro–activated T cells stimulated or not with different concentrations of IL-2 for 4 days. T cell proliferation was determined from the level of CellTrace Violet (CTV) dye dilution. Representative histograms (left panel) show cell divisions of activated CD8 T cells from HCs or PTPN2 patients as indicated. Percentage of CD8+ dividing cells from HCs and patients (right panel). Dividing cells represent cells having undergone at least one division (data pooled from n = 5 independent experiments including a total of 18 HCs and 7 patients). P values were determined in a two-way ANOVA: <0.0002 (***), 0.0021(**), and 0.1234 (ns). (D) Proliferation of patients’ IL2-stimulated activated T cells with or without incubation with JAK1/JAK3 inhibitor tofacitinib for 4 days (data pooled from n = 2 independent experiments including a total of seven HCs and seven patients). P values were determined in a two-way ANOVA: <0.0332 (*).
Next, we analyzed STAT5 phosphorylation upon IL-2 stimulation of in vitro preactivated T cells from patients. These analyses were performed following 15 min stimulation and also 2 h after IL-2 removal to assess STAT5 dephosphorylation kinetic. After 15 min of stimulation, we observed an increase in STAT5 phosphorylation level similar in patients compared with controls (Fig. 4 B). However, 2 h after IL-2 washout, we observed a significantly delayed STAT5 dephosphorylation in T cells from patients compared with T cells from healthy controls (HCs). This delay in STAT5 dephosphorylation was also observed in patient C1 carrying the W289* PTPN2 mutation, further confirming haploinsufficiency for this variant.
Increased proliferation of patients’ T cells upon IL-2 stimulation
To evaluate the functional impact of PTPN2 mutations in patients’ lymphocytes, we analyzed the proliferation of in vitro–preactivated T cells upon IL-2 stimulation. We observed an increased proliferation of patients’ T cells compared with controls after IL-2 stimulation at different concentrations (Fig. 4 C). Patients’ T cell proliferations were, however, comparable with that of controls following TCR stimulation using anti-CD3 coated beads (OKT3) (Fig. S2), suggesting that PTPN2 regulates JAK/STAT but not TCR signaling, consistent with previous published data (Parlato et al., 2020).
Proliferation of patients’ T cells following TCR activation. Proliferation of activated T cells stimulated or not with different concentrations of anti-CD3–coated beads (OKT3) for 4 days. T cell proliferation was determined from the level of CellTrace Violet dye dilution. Percentage of CD8+ dividing cells from HCs and patients. Dividing cells represent cells having undergone at least one division (data pooled from n = 5 independent experiments including a total of 13 HCs and 7 patients). P values were determined in a two-way ANOVA (ns: not significant). NS, nonstimulated.
Proliferation of patients’ T cells following TCR activation. Proliferation of activated T cells stimulated or not with different concentrations of anti-CD3–coated beads (OKT3) for 4 days. T cell proliferation was determined from the level of CellTrace Violet dye dilution. Percentage of CD8+ dividing cells from HCs and patients. Dividing cells represent cells having undergone at least one division (data pooled from n = 5 independent experiments including a total of 13 HCs and 7 patients). P values were determined in a two-way ANOVA (ns: not significant). NS, nonstimulated.
JAK inhibitors were recently introduced in the clinic for different conditions including type I interferonopathies. We evaluated the impact of tofacitinib (JAK1/JAK3 inhibitor) on T cell proliferation induced by IL-2 using the same conditions as described above. Results show that tofacitinib treatment normalized the proliferation of PTPN2 mutant T cells, reducing it to HC levels when used at 25 nM concentration (Fig. 4 D). These data confirmed that PTPN2 inhibited T cell proliferation by targeting the JAK/STAT pathways and suggested that JAK inhibitors could be interesting treatments for PTPN2-deficient patients.
Augmentation of transcriptional responses to cytokines in patients’ T cells
To get a broader and unbiased view of the impact of PTPN2 mutations on the response to cytokines, we performed bulk RNA sequencing (RNA-seq) analysis on in vitro–preactivated T cells from patients or HCs. Cells were either left unstimulated or stimulated 3 h with IL-2 or IFN-α. We then used the Gene Set Enrichment Analysis (GSEA) method to functionally annotate the differentially expressed genes between patients and healthy donors. The cytokine/cytokine receptor interaction pathway from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database was predicted to be more activated in patients compared with HCs for the unstimulated condition and the IL-2 and IFN-α–stimulated conditions (Fig. 5 A). The IL-2/STAT5 and the JAK/STAT signaling pathways were enriched only in patients’ cells at basal state and after IL-2 stimulation but not after IFN-α stimulation (Fig. 5 A). These results suggested that, in this experimental setting, PTPN2 was an important regulator of the IL-2 signaling pathway. Then regarding the gene significantly overexpressed in patients’ group compared with healthy donors, we found that signatures of cytokine receptor signaling pathways, including IL-27, IL-2, GM-CSF, and IL6, were strongly enriched in patients’ T cells after IL-2 stimulation (Fig. 5 B). Interestingly, the functional term “signaling events mediated by PTPN2,” a gene set including JAK1, JAK3, and STAT1, was also enriched in PTPN2 mutated patients’ cells, in particular in A2, B1, C1, and D1 cells (Fig. 5, B and C). All data from RNA-seq are available online (http://flowrepository.org/id/FR-FCM-Z7P5).
Global cytokine responses in in vitro–activated T cells from patients. (A) RNA-seq analysis of in vitro–activated T cells from patients (n = 5) compared with HCs (n = 3) in unstimulated condition or upon IL-2 or IFN-α stimulation for 3 h. Significant pathways enrichment in patients (A1, A2, B1, C1, and D1) on a list of 36 pathways from MSigDB database are represented. NS, nonstimulated. (B) Significantly upregulated pathways from NCI Nature database in in vitro–activated T cells from patients (n = 5) (A1, A2, B1, C1, and D1) as compared with HCs (n = 3) after IL-2 stimulation (104 U/ml for 3 h). *P < 0.05, **P < 0.01. (C) Heatmap of the normalized gene expressions found upregulated in the signaling events mediated by PTPN2 for patients (A1, A2, B1, C1, and D1) and HC cells after IL-2 stimulation. (D) Heatmap of cytokine dosage in supernatant of resting or activated T cells stimulated by IFN-α or IL-2 (104 U/ml) for 24 h. Z-score of log10 cytokine levels normalized to HC means in indicated stimulation conditions. Data from n = 2 independent experiments including a total of 13 HCs and 6 patients.
Global cytokine responses in in vitro–activated T cells from patients. (A) RNA-seq analysis of in vitro–activated T cells from patients (n = 5) compared with HCs (n = 3) in unstimulated condition or upon IL-2 or IFN-α stimulation for 3 h. Significant pathways enrichment in patients (A1, A2, B1, C1, and D1) on a list of 36 pathways from MSigDB database are represented. NS, nonstimulated. (B) Significantly upregulated pathways from NCI Nature database in in vitro–activated T cells from patients (n = 5) (A1, A2, B1, C1, and D1) as compared with HCs (n = 3) after IL-2 stimulation (104 U/ml for 3 h). *P < 0.05, **P < 0.01. (C) Heatmap of the normalized gene expressions found upregulated in the signaling events mediated by PTPN2 for patients (A1, A2, B1, C1, and D1) and HC cells after IL-2 stimulation. (D) Heatmap of cytokine dosage in supernatant of resting or activated T cells stimulated by IFN-α or IL-2 (104 U/ml) for 24 h. Z-score of log10 cytokine levels normalized to HC means in indicated stimulation conditions. Data from n = 2 independent experiments including a total of 13 HCs and 6 patients.
We also monitored cytokine levels in supernatants of T cells from patients and controls at basal level or upon type I IFN or IL-2 stimulation using the Legendplex technology. In the absence of any stimulation, most of the patients exhibited an increased expression of inflammatory cytokines compared with healthy donors (Fig. 5 D, upper heatmap). This was even more pronounced after IFN-α or IL-2 (except for C1) stimulations. Upon IL-2 stimulation, we observed a notably robust increase in several cytokines, including IP-10, TNF-α, MCP1, IFN-γ, IL-8, and IL-10 (Fig. 5 D). These results were consistent with the RNA-seq analysis (Fig. 5 A).
These analyses confirmed that cytokine signaling pathways, in particular those involving the JAK/STAT modules, are hyperactivated in cytokine-stimulated patients’ T cells, which led to increased cytokine production by these cells. This exaggerated response could potentially set the stage for autoimmune reactions.
Immunophenotyping of patients’ PBMC and serum cytokine levels
Since the JAK/STAT pathways are involved in the signal transduction of numerous cytokines, shaping immune cell development and response, we conducted a detailed mass cytometry immunophenotyping on available whole blood samples (patients B1, C1, D1, and F1) in comparison with both adult and pediatric (for B1 patient) controls. Our findings revealed a decrease in NK cells’ percentage and an elevated proportion of total monocytes primarily driven by an increase in classical monocytes (Fig. 6, A and B).
Immunophenotyping of circulating patients’ cells and cytokine signatures. (A) Representative UMAP plots were generated from immunophenotyping CyTOF data of whole blood from adult controls (n = 4), pediatric controls (n = 5), and patients’ cells (B1, C1, D1 and F1). At the day of prelevement, the B1 patient is considered as pediatric (9 years old) while patients C1, D1, and F1 were adults (22, 26, and 20 years old, respectively). pDC: plasmacytoid dendritic cell; mDC: myeloid dendritic cell; CM: central memory; EM: effector memory; TEM: terminal effector memory; TEMRA: terminally differentiated effector memory; MAIT: mucosal-associated invariant T cell; GD: gamma delta. (B) Quantification of immune cell populations in adult (n = 4) or pediatric (n = 5) control or patient (B1, C1, D1 and F1) groups. Patient’s age group is indicated on the graph’s legend (A: adult; P: pediatric). P values were obtained using Kristal–Wallis test: <0.0332 (*), 0.1234 (ns). (C) Heatmap of cytokine dosage in plasma of HCs (n = 7) and patients with STAT1 GOF (n = 3), STAT3 GOF (n = 2), SOCS1 LOF (n = 3), and PTPN2 mutation (n = 5). Z-score of log10 cytokine levels normalized to HC mean. D1 received corticoids and B1 Imurel before sampling.
Immunophenotyping of circulating patients’ cells and cytokine signatures. (A) Representative UMAP plots were generated from immunophenotyping CyTOF data of whole blood from adult controls (n = 4), pediatric controls (n = 5), and patients’ cells (B1, C1, D1 and F1). At the day of prelevement, the B1 patient is considered as pediatric (9 years old) while patients C1, D1, and F1 were adults (22, 26, and 20 years old, respectively). pDC: plasmacytoid dendritic cell; mDC: myeloid dendritic cell; CM: central memory; EM: effector memory; TEM: terminal effector memory; TEMRA: terminally differentiated effector memory; MAIT: mucosal-associated invariant T cell; GD: gamma delta. (B) Quantification of immune cell populations in adult (n = 4) or pediatric (n = 5) control or patient (B1, C1, D1 and F1) groups. Patient’s age group is indicated on the graph’s legend (A: adult; P: pediatric). P values were obtained using Kristal–Wallis test: <0.0332 (*), 0.1234 (ns). (C) Heatmap of cytokine dosage in plasma of HCs (n = 7) and patients with STAT1 GOF (n = 3), STAT3 GOF (n = 2), SOCS1 LOF (n = 3), and PTPN2 mutation (n = 5). Z-score of log10 cytokine levels normalized to HC mean. D1 received corticoids and B1 Imurel before sampling.
Additionally, our investigation revealed a bias in B cell profiles, with more CD11c+ B cells for all patients. Further analysis of this population by subclustering with CD38, CD27, and IgD showed a marked increase in the proportion of CD27− IgD+ activated naive B cells.
Furthermore, we subclustered memory CD4+ T cells (excluding regulatory T cells [Tregs]) to delineate T helper (Th) cell subsets based on the expression patterns of CXCR5, CCR6, CXCR3, and CCR4. Our analysis showed a significant increase in the proportion of T follicular helper-like (Tfh) CD4+ CXCR5+ cells, a feature commonly observed in lupus patients (Walker, 2022). Th1 subset was also increased in patients. Conversely, Th2 and Th17 subsets were decreased in comparison with the control group (Fig. 6 B).
All data from immunophenotyping analyses are detailed in Fig. S3.
Quantification of immune subsets defined following CyTOF analysis. Barplot showing the proportion of each identified immune subset across all groups (x-axis) with dots shaped and colored by patient identification and bar colored by groups. Indicated proportion is the frequency among all singlet viable PBMCs. A, adult; P, pediatric. P values were obtained using Kruskal–Wallis test: <0.0332(*), and 0.1234 (ns). DN: double negative; cDC: conventional dendritic cell; CM: central memory; EM: effector memory; TEM: terminal effector memory; TEMRA: terminally differentiated effector memory; pDC: plasmacytoid dendritic cell; GD: gamma delta.
Quantification of immune subsets defined following CyTOF analysis. Barplot showing the proportion of each identified immune subset across all groups (x-axis) with dots shaped and colored by patient identification and bar colored by groups. Indicated proportion is the frequency among all singlet viable PBMCs. A, adult; P, pediatric. P values were obtained using Kruskal–Wallis test: <0.0332(*), and 0.1234 (ns). DN: double negative; cDC: conventional dendritic cell; CM: central memory; EM: effector memory; TEM: terminal effector memory; TEMRA: terminally differentiated effector memory; pDC: plasmacytoid dendritic cell; GD: gamma delta.
Subsequently, we measured the levels of various cytokines in patients’ serum, comparing them with healthy individuals and patients with GOF mutations in STAT factors. Strikingly, we observed an increase in various cytokines like IL-10, IP-10, or IFN-γ in the serum of patients compared with HCs (Fig. 6 C). Interestingly, this profile was similar to that of patients with STAT1 GOF mutations or SOCS1 LOF mutations, indicating an overlap in the pathogenesis of these IEI impacting the JAK/STAT pathway.
Altogether, the analysis of PBMCs revealed typical alterations associated with autoimmunity, and the analysis of serum cytokines showed a remarkable similarity in the profile observed among patients mutated in PTPN2, SOCS1, or STAT1.
Discussion
In this study, we identified private LOF heterozygous PTPN2 mutations in six patients from six unrelated families. They were variably associated with the development of SLE in one family and Evans syndrome in five families, thus extending the spectrum of clinical manifestations associated with PTPN2 deficiency. Previously published cases included an individual with common variable immunodeficiency (Thaventhiran et al., 2020) and two others with inflammatory bowel disease (Parlato et al., 2020; Awwad et al., 2023). While PTPN2 was reported to be critical for maintaining intestinal barrier polarization and enterocyte adhesion (Marchelletta et al., 2021), none of the six affected individuals in our study exhibited intestinal features, despite the defective regulatory function of the identified mutants. This suggested that PTPN2 deficiency alone was not sufficient to drive intestinal inflammation and that additional factors such as gut dysbiosis could be important as previously suggested (Denoth et al., 2021).
A2, B2, and F2 fathers are asymptomatic carriers of the mutation, indicating autosomal dominant inheritance with incomplete clinical penetrance. We have also collected the genetic data from the siblings of all patients and none of them carried the proband’s PTPN2 mutation and were all healthy. Incomplete clinical penetrance is relatively common in immunological diseases, particularly when haploinsufficiency mechanisms are involved (e.g., CTLA4, SOCS1) (Hadjadj et al., 2020; Rieux-Laucat and Casanova, 2014). In other diseases, like COPA syndrome, the penetrance is around 60%. Here, we detected PTPN2 mutations in nine individuals, with six exhibiting symptoms, resulting in an estimated penetrance of 67%. Of note, we did not identify any other predicted pathogenic variants in PTPN2 or other related genes in our cohort of six symptomatic patients. In addition, it is also very important to emphasize the rarity, if not absence of these variants in control population databases, highlighting the fact that these variants are private to the families. However, it is worth mentioning that homozygous mutations in PTPN2 have been recently identified in one patient (Awwad et al., 2023). Among the JAK/STAT regulating molecules, while PTPN2 biallelic defect seems possible, such homozygous mutations have not been reported for SOCS1, another master regulator of this pathway, associated with Evans syndrome and lupus. This observation may suggest compensatory mechanisms for PTPN2 complete LOF, which is not redundant for SOCS1.
Two variants described in this study (W98* and E24Mfs*20) resulted in a truncated protein with no detectable expression in all cellular models tested. The Y126N variant also led to a profound decrease in protein expression and phosphatase activity, whereas the C216S variant led to normal expression associated with a complete loss of phosphatase activity, thus showing that haploinsufficiency in PTPN2 predisposes to autoimmunity.
In contrast, the F403Lfs*25 variant (affecting the PTPN2 TC48 isoform only) showed residual expression. The mutant protein displayed normal intrinsic phosphatase activity in in vitro assays despite reduced phosphatase activity in cellular assays. Thanks to its C-terminal tail, the TC48 isoform was described to specifically localize in the endoplasmic reticulum (Lorenzen et al., 1995; Kamatkar et al., 1996). This localization was not modified by the F403Lfs*25 mutation. Since the C-terminal tail of the protein was also described as an autoinhibitory domain of PTPN2 phosphatase activity, we speculated that the functional defect of the F403Lfs*25 variant could be due to a conformational anomaly in the 3D structure of PTPN2. Cellular context and the interaction of PTPN2 with mediators of signal transduction have been shown to impact the folding of the C-terminal tail and its auto-inhibitory function (Tun et al., 2022), and this may be dysregulated in the context of F403Lfs*25 variant. Along the same line, the overexpressed W289* variant showed an intrinsic tyrosine phosphatase activity fivefold higher than the PTPN2 WT when using the in vitro phosphatase assay. These results were consistent with a previous work showing that C-terminus truncated forms of PTPN2 exhibited tyrosine phosphatase activities 100 times more elevated than the WT protein (Hao et al., 1997). The W289* variant expression was very low in transfected cells (around 10% of the WT), but most likely absent in patients’ cells since total PTPN2 expression was around 50% of the control cells, which is evocative of haploinsufficiency, and overall no truncated band could be detected in in vitro–activated T cells derived from the C1 patient. These results suggest that this variant is not expressed under physiological conditions and mRNA is probably eliminated by nonsense mediated decay mechanism due to the presence of a premature stop codon. Overall, our data supported the critical regulatory role of the C-terminal domain of PTPN2.
We demonstrated that the PTPN2 mutations described here led to JAK/STAT pathways hyperactivation, as indicated by increased STATs phosphorylation in response to various cytokines, notably IL-2 and type I and II IFN. Patients’ T cells also exhibited increased proliferation under these conditions and produced more proinflammatory cytokines (including IP-10, TNF-α, MCP1, IFN-γ, IL-8, and IL-10). This was reminiscent of the hypersensitivity to IL-6, IL-15, and IL-21 previously reported in PTPN2-deficient patients (Parlato et al., 2020). Of note, TCR stimulation did not lead to an increased proliferation of patients’ T cells. This was consistent with the previous description of the C216G PTPN2 variant, which was not associated with hyperphosphorylation of lymphocyte cell–specific protein-tyrosine kinase or ERK mediators upon TCR stimulation (Parlato et al., 2020). The elevated production of inflammatory cytokines (in particular IP-10, TNF-α, MCP1, IFN-γ, IL-8, and IL-10) by T cells in vitro was correlated with the elevated concentration of these cytokines in patients’ sera. These results confirmed the role of the PTPN2 phosphatase as a crucial modulator of inflammation. This was in agreement with the systemic inflammation and increased expression of TNF-α, IFN-γ, and IL-12 described in Ptpn2 KO mice (Heinonen et al., 2004; You-Ten et al., 1997). Last, the dysregulated response to cytokines was corroborated by the RNA-seq analysis showing an enhanced activation of JAK/STAT pathways in patients’ T cells in response to IL-2 or type I IFN.
To assess the potential impact of these molecular and cellular dysregulations, we performed in-depth immunophenotyping on blood cells from patients B1, C1, D1, and F1 using mass cytometry. These analyses revealed an increased proportion of CD11c+ B cells, also known as “age-associated B cells” in mice (Rubtsov et al., 2011). These cells were already reported to be associated with several infectious and autoimmune diseases (Stepensky et al., 2015; Wang et al., 2018; Sanz et al., 2019). Upon stimulation, these overactivated cells were prone to differentiate into antibody-secreting cells, especially plasmocytes, and can be responsible for the production of autoantibodies (Golinski et al., 2020; Wang et al., 2018). Detailed analysis of this CD19+ CD11c+ subpopulation revealed an increased proportion of CD27–IgD+ activated naive B cells, suggesting a chronic activation of the extrafollicular pathway previously described in SLE (Sanz et al., 2019; Jenks et al., 2019; Stepensky et al., 2015). Interestingly, an elevated proportion of CD11c+ B cells was also described in patients with SOCS1 LOF, STAT1 GOF, and STAT3 GOF, thus highlighting that the JAK/STAT pathways are key in the differentiation process of this B cell subset (Keller et al., 2021). We also observed an increased proportion of circulating Tfh cells in PTPN2-deficient patients, a finding consistent with previous studies in mice showing that Ptpn2 KO mice expand Tfh cells and develop autoimmune manifestations (Wiede et al., 2017, 2019; Walker, 2022). This population could be involved in the excess of CD11c+ B cells in PTPN2 patients, thereby contributing to the onset of autoimmunity in patients (Song et al., 2022b). An increase in classical monocytes was also observed. Even if the role of monocytes in the development of autoimmune diseases was not fully elucidated, there was a consensus suggesting that their abnormal activation could play a key role, possibly through uncontrolled secretion of proinflammatory cytokines (Ożańska et al., 2020). Immunophenotyping of patients also revealed a reduction in NK cell population, consistent with previous results describing a decrease in frequency and cytotoxic activity of NK cells from SLE patients (Erkeller-Yuksel et al., 1997; Yabuhara et al., 1996).
The molecular and functional analyses of PTPN2 variants demonstrated that defects in negative regulation downstream of cytokines promote autoimmune manifestations. We previously described variants in the SOCS1 gene in patients presenting with similar autoimmune diseases (Hadjadj et al., 2020). SOCS1 was described as the main negative regulator of the JAK/STAT pathways. It primarily acts by ubiquitinating effectors of these pathways, targeting them for degradation by the proteasome, or by directly inhibiting JAK kinases through interaction with their killer Ig receptor domain (Ilangumaran and Rottapel, 2003). Similar to what we described for PTPN2, haploinsufficiency of SOCS1 led to early onset autoimmune manifestations in patients, as a consequence of hyperactivation of the JAK/STAT pathways. The expression profile of proinflammatory cytokines in PTPN2 and SOCS1 mutated patients was similar to that observed in patients carrying GOF mutations in the STAT1 and STAT3 transcription factors.
The central role of PTPN2 in the regulation of inflammation is confirmed by its potential as an antitumor target (Tiganis, 2016; Wiede and Tiganis, 2017). Indeed, in our study, E2 (E1’s father) is deceased from leukemia. As we now know that E1’s mother is PTPN2 WT/WT, one can hypothesize that the father’s leukemia was associated with PTPN2. Studies have shown that PTPN2 KO tumors are more responsive to immunotherapies due to their hypersensitivity to IFN-γ (Manguso et al., 2017). Additionally, PTPN2 KO T cells are hyperactivated following IL-2 stimulation and are therefore all the more effective in their antitumor action. Recently, a compound capable of blocking the phosphatase activity of PTPN2 and PTPN1 has been identified and clinical trials are underway to evaluate its antitumor activity (Baumgartner et al., 2023).
Overall, we showed that PTPN2 plays a pivotal role as a regulator of immune system activation preventing not only inflammatory bowel diseases but also systemic autoimmunity. Detailed analysis of signaling pathways regulated by PTPN2 will enable personalized therapeutical strategies, relying in particular on JAK inhibition, which will be promising for patients bearing PTPN2 mutations.
Materials and methods
Study participants
The probands were included in clinical studies of early-onset autoimmune diseases by different institutions (Lyon Hospital, Imagine Institute, Paris, France). Written informed consent was obtained from all patients from Lyon Hospital included in this research involving human blood samples. Ethical approval was obtained from the Ethic Committee CPP-SUD-Est III on February 5, 2013 (ID number 2012.769) and approval of French competent authority on May 2, 2013 (ref 130588-B). This study was registered under EudraCT number (2012-A01449-34) and NCT01992666 (Genetic & Immunologic Abnomalities in SLE). The study was conducted in accordance with the French legislation, the Good Clinical Practice, and the Declaration of Helsinki.
Biological samples from Imagine Institute (Paris, France) are from INSERM UMR1163/Imagine collection, declared to Research Ministry number CODECOH no. DC-2020-3994. Written informed consent was obtained from all patients from Imagine Institute (Paris, France).
The information and consent form of patients from Imagine Institute (Paris, France) has received approval from the ethics Committee Assistance Publique Hôpitaux de Paris Centre (Institutional Review Board registration: #00011928).
Clinical study
Exome sequencing
DNA was extracted from whole blood samples using standard methods. Exome sequencing was performed on the genomic DNA of patients. Whole exome sequencing was performed using an AmpliSeq kit, with libraries analyzed on a Life Technologies Proton instrument. P3 was sequenced using SureSelect Human All Exon kit (Agilent Technologies) for targeted enrichment and an Illumina HiSeq 2000. Variants were assessed using the in silico programs CADD (https://cadd.gs.washington.edu), SIFT (http://sift.jcvi.org), and Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/); summarized in Varcards (https://www.varcard.io/); and population allele frequencies obtained from the ExAC (https://exac.broadinstitute.org) and gnomAD (https://gnomad.broadinstitute.org) databases. Variants were annotated on the PTPN2 NM_002828 transcript. Variants were confirmed using Sanger sequencing and studied in parents when DNA was available.
Cell culture and reagents
Jurkat–T cell line and lymphoblastic B cell line derived from patients were cultured in RPMI 1640 GlutaMax (Thermo Fisher Scientific) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS) (Dutscher), 10 mM HEPES pH 7.5 (Thermo Fisher Scientific), and 0.04 mg/ml gentamycin (Thermo Fisher Scientific), and incubated at 37°C in 5% CO2. HEK293T cell line was cultured in DMEM High Glucose GlutaMax supplement (Thermo Fisher Scientific) supplemented with 10% (vol/vol) heat-inactivated FCS (Dutscher), 10 mM HEPES pH 7.5 (Thermo Fisher Scientific), and 0.04 mg/ml gentamycin (Thermo Fisher Scientific), and incubated at 37°C in 5% CO2.
HEK293T cells were transiently transfected with indicated constructs using JetPEI reagent (PolyPlus transfection) according to the manufacturer’s instructions. Cells were plated at 1 × 106 cells/well in 2 ml culture medium (6-well plate).
Blood was obtained from healthy donors from the Etablissement Français du Sang Auvergne Rhône Alpes, France under the convention EFS 16–2066. PBMCs were isolated from 450 ml of freshly drawn venous human peripheral blood by Ficoll (Eurobio, Courtaboeuf, France) density gradient centrifugation. Cells were washed three times in sterile PBS (Thermo Fisher Scientific), and 100 × 106 cells were stained for cell sorting. The pellet was resuspended in staining buffer (PBS, 2% FCS, 2 mM EDTA [Thermo Fisher Scientific]) and incubated with anti-CD3 V500 (BD), anti-CD56 APC (Beckman), anti-CD19 PE Texas red (BD), anti-CD14 FITC (Miltenyi Biotech), and near infrared immunofluorescence (Thermo Fisher Scientific) for viability. B and T lymphocytes, monocytes, and NK cells were then sorted on an ARIA II cytometer and lysed in radioimmunoprecipitation assay (RIPA) buffer for automated western blot analysis.
Plasmid and lentiviral particle generation
Plasmid containing PTPN2 full sequence (isoform TC48, NM_002828) was obtained from OriGene and subcloned in lentiviral vector p197 pRRLSIN-MND-IRES2-ZsGreen-WPRE from Vect’UB (vectorology platform, INSERM US 005-CNRS UAR 3427-TBM-Core, Université de Bordeaux, Bordeaux, France). HA tag was added in the N-terminal part of the cDNA sequence. All PTPN2 variants were also cloned in this vector with N-terminal HA tag sequence by directed mutagenesis and all cDNA sequences were confirmed by Sanger sequencing (Genewiz). Lentiviral particles for cell transduction were obtained from ANIRA vectorology platform at the SFR Biosciences in Lyon.
PTPN2 immunoprecipitation and reverse phase-ultra fast liquid chromatography (RP-UFLC) activity assay
Immunoprecipitation of transfected PTPN2 and measurement of tyrosine phosphatase activity were carried out as previously described (Nian et al., 2022; Duval et al., 2015).
Briefly, HEK293T cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 1 mM L-glutamine at 37°C under 5% CO2. For transfection, HEK293T cells were seeded at 30,000 cells/cm2 in a 100-cm2 Petri dish and directly transfected with 5 µg of plasmid and 15 μl of Metafectene (Biontex). 48 h after transfection, transfected cells were harvested and lysed using cell lysis buffer (PBS 150 mM NaCl pH 7.5, 1% Triton X-100, 1× protease inhibitors) for 30 min at 4°C under agitation. The lysate was sonicated for 2 s (10% power) and then centrifuged for 15 min at 15,600 g at 4°C.
For PTPN2 immunoprecipitation, 800 μl of whole-cell extracts (1,400 µg of total protein extract) were incubated overnight at 4°C under agitation with 2 µg of HA-Tag (C29F4; Cell Signaling) monoclonal rabbit antibody (Cell Signaling) and protease inhibitor cocktail (Roche). Samples were then rocked for 2 h at 4°C in the presence of 40 μl of protein A-Agarose (sc-2001; Santa Cruz). The immunobeads were harvested by centrifugation and washed three times with cell lysis buffer and two times with phosphatase buffer (100 mM sodium acetate, pH 6). One-fifth of the beads were centrifuged (4,000 rpm, 5 min) and mixed with 20 μl Laemmli sample buffer containing 400 mM β-mercaptoethanol for subsequent use in immunoblot analysis with HA-Tag (C29F4; Cell Signaling) monoclonal rabbit and α-tubulin (T5168; Sigma-Aldrich) monoclonal mouse as primary antibodies. The rest of the beads was spun down by centrifugation (700 g, 3 min, 4°C) and then incubated with 100 μl of phosphatase buffer containing 75 µM fluorescent substrate peptide pSTAT3, 1 mM DTT, and protease inhibitors at 37°C. Every 5 min, 20 μl of supernatant was taken and mixed with 50 μl of HClO4 (vol/vol) and 20 μl of the mixture was analyzed by UFLC (Shimadzu) on a reverse-phase column (Shim-pack XR ODS 100 × 2 mm; 2.2 µm, Shimadzu). The mobile phase composition was water with 0.12% trifluoracetic acid (TFA) for mobile phase A and acetonitrile with 0.12% TFA for mobile phase B. Compounds were separated by an isocratic flow (80% A/20% B) rate of 0.6 ml min−1. The peptides were monitored by fluorimetry (excitation at 485 nm, emission at 530 nm) and quantified with LabSolution software by integration of the peak absorbance area. RP-HPLC activity assay were performed using the Bioprofiler platform of the Unit “Biologie Fonctionnelle et Adaptative,” Université Paris Cité, BFA, UMR 8251 CNRS, F-75205 Paris, France.
Immunofluorescence assay
HEK293T cells were transfected with HA-tagged PTPN2 constructs. 24 h after transfection, cells (2 × 105 per well) were dispensed on coverslip tissue culture (Ibidi), pretreated for 1 h with poly-L-lysine hydrobromide (Sigma-Aldrich), and left to adhere for 16 h. Then medium was discarded and cells were fixed with 4% PFA (Sigma-Aldrich) for 1 h at 37°C, permeabilized in 0.05% Triton X-100 for 7 min at room temperature (RT), and blocked for 30 min in blocking buffer (PBS 3% BSA, 5% FCS). Then cells were stained 16 h at 4°C with primary antibodies (anti-HA [Cell Signaling Technologies], anti-Calreticulin [Thermo Fisher Scientific]), and then with secondary antibodies (anti-rabbit AF568 and anti-mouse AF647 [Thermo Fisher Scientific]) and DAPI (Thermo Fisher Scientific) for nucleus staining. Images were acquired on an inverted confocal microscope (LSM800; ZEISS).
Luciferase reporter assays
HEK293T cells were dispensed into a 6-well cell-culture plate (1 × 106 cells/well), left to adhere for 6 h, and transiently transfected using JET-PEI (Kit JetPEI Polyplus Transfection) according to the manufacturer’s instructions. Cells were then transfected with PTPN2 constructs expressed in p197 lentiviral vector together with firefly luciferase reporter plasmid under the control of the ISRE promoter or STAT3 induced element promoter as indicated) and Renilla luciferase normalization vector (prL-SV40; Promega). 24 h after transfection, cells were detached, dispensed in a 96-well plate (50,000 cells/well), and left to adhere for 6 h. Transfected cells were then stimulated or not with IFN-α (103 international unit (UI)/ml; Miltenyi biotech), IFN-β (103 UI/ml Rebif), IFN-γ (103 UI/ml; Miltenyi biotech), or IL-6 (250 UI/ml; Miltenyi biotech) for 24 h. Then luciferase activity was measured using Dual-Glo assay (Promega) and firefly luciferase activity was normalized against Renilla luciferase activity. All transfection experiments were performed in triplicate. Data are presented as percent of luciferase activity obtained in empty vector (EV) condition. Luminescence was measured on a TRISTAR 3 multimode Berthold microplate reader (Berthold Technologies GmbH & Co. KG). Data are expressed as fold-induction, relative to unstimulated cells.
Quantitative RT-PCR (RT-QPCR)
For RT-QPCR on PBMCs
RNA was extracted from whole blood with Direct-zol RNA MicroPrep (Ozyme), and cDNA was generated using the high-capacity RNA-to-cDNA kit (Applied Biosystems). PCR was carried out with a FastStart Universal SYBR Green Master (Roche) on a StepOne plus instrument (Applied Biosystems). Primers were designed using Primerblast web tool. The following primers were used for RT-QPCR: PTPN2: Forward: 5′-AGCGGGAGTTCGAAGAGTTG-3′; Reverse 5′-ACGACTGTGATCATATGGGCT-3′; OAZ1: Forward: 5′-GGATAAACCCAGCGCCAC-3′; Reverse: 5′-TACAGCAGTGGAGGGAGACC-3′. The expression PTPN2 mRNA was normalized to the level of OAZ1.
For RT-QPCR on in vitro–activated T cells
RNA was extracted from in vitro–activated T cells with RNeasy Mini Kit (#74106; Qiagen) and cDNA was generated using the QuantiTect Reverse Transcription Kit (#205313; Qiagen). QPCR was carried out with SYBR Green PCR Master Mix (#4472918; Thermo Fisher Scientific) on ViiA 7 Real-Time PCR System (Applied Biosystems). The following primers were used for RT-QPCR: PTPN2: Forward: 5′-ACATCTTAACACAGGGTCCAC-3′; Reverse 5′-TGCTGCCAAACCATAAGCCA-3’. The expression PTPN2 mRNA was normalized to the level of GAPDH by the following calculation: 2(Ct GAPDH−Ct PTPN2)*100.
CRISPR/Cas9 genome editing
Jurkat–T cells were knocked out for PTPN2 gene using CRISPR/Cas9 genome editing protocol. CRISPR RNAs (crRNAs) were designed using either Crispor.tefor internet tool or IDT design tool. We selected three crRNA targeting PTPN2 exon 1 (3′-CTCTTCGAACTCCCGCTCGA-5′), exon 2 (3′-CCACTCTATGAGGATAGTCA-5'), and exon 6 (3′-GCACTACAGTGGATCACCGC-5′). Equivalent volumes of crRNA (100 µM) and TracrRNA (100 µM) (IDT) were mixed and annealed for 5 min at 95°C and cooled to RT (15–25°C) on the bench top to obtain three different single guide RNA (sgRNA) at 50 µM final concentration. RNP complex was then prepared by mixing 1 μl of each sgRNA with 2 µM of 62 µM Cas9 enzyme (IDT) and incubating at RT for 20 min.
Jurkat-T cells were prepared for electroporation according to SE-Cell Line 4D-Nucleofector Kit recommendation. Briefly, 1 million cells per condition were washed once in PBS buffer and pellet was resuspended in appropriate buffer. 5 μl of RNP complex and 1.5 μl of electroporation enhancer solution (IDT) were added to the cells. The mixture was transferred to a nucleofector cuvette for electroporation using 4D-Nucleofector Core Unit (Lonza).
75 μl of warm medium was added to the cuvette and cells were transferred in 96-well plates and amplified for 4–5 days. The PTPN2 KO efficiency was determined by quantitative automated western blot (ProteinSimple) and confirmed by Sanger sequencing (Genewiz).
Jurkat PTPN2 KO cells were thereafter transduced with lentiviral particles enabling the expression of PTPN2 WT or variants.
Immunohistochemical analyses
Formalin-fixed paraffin-embedded liver tissue sections from patients were stained with hematoxylin and eosin.
Quantitative automated capillary western immunoblotting
Cells were lysed in RIPA lysis buffer (25 mM Tris, HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS containing protease and phosphatase Inhibitors [Sigma-Aldrich]) for 30 min at 4°C. The supernatant was collected following 10 min centrifugation at 16,000 g, 4°C, and protein content was quantified using the μBCA quantification kit (Thermo Fisher Scientific). Protein expression was detected using the automated Jess Simple Western system (ProteinSimple). Briefly, cell lysate was diluted to 0.5 µg/μl in 0.1× sample buffer and Fluorescent 5× Master mix containing 400 mM DTT (ProteinSimple). Samples were denatured at 95°C for 5 min and proteins were separated in capillaries using 12–230-kDa Jess separation module and following the manufacturer’s standard method (ProteinSimple).
Anti-PTPN2 antibody diluted 1:40 (Bio-techne), anti-HA tag antibody (1:40), anti-phospho STAT1 (Y701) (1:40) (Bio-techne), and anti-phospho STAT3 (Y705) (1:50) (Ozyme) were used for protein detection and detected using secondary antibodies coupled to HRP (ProteinSimple). Digital image of chemiluminescence of the capillary was analyzed using Compass Simple Western software (version 4.1.0, Protein Simple) that calculated automatically heights (chemiluminescence intensity), area, and signal/noise ratio. Results could be visualized as electropherograms representing the peak of chemiluminescence intensity and as lane view from signal of chemiluminescence detected in the capillary.
Protein of interest levels were normalized on total protein expression using the JESS Replex total protein assay and according to the manufacturer’s instructions.
STAT1 phosphorylation assay
Cells were fixed with Lyse/Fix buffer (5×; BD Biosciences phosflow) and permeabilized with Perm Buffer III (BD Biosciences Pharmingen). Cells were stained for pSTAT1 pY701 with mouse anti-pSTAT1-PE (BD Biosciences) for 30 min in ice, then resuspended with decomplemented FCS and washed with FACS buffer four times. Flow cytometry was carried out on a Cytek Aurora, allowing spectral flow cytometry (Cytek Bioscience). Data were analyzed using FlowJo (Treestar).
Statistical analysis
All statistical analyses were performed using GraphPad Prism (version 9.5.1). For all analyses, the threshold for statistical significance was set to P < 0.05. One-way ANOVA was used when more than two experimental groups were compared, and the statistical significance was determined using Tukey’s or Dunnett’s multiple comparisons post hoc test.
Activation and expansion of primary human T cells
Fresh or thawed PBMCs were stimulated in complete medium containing Panserin 401 (#P04-710401; PanBiotech), supplemented with 5% human male AB serum (#S4190-50; BioWest), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (Thermo Fischer Scientific) for 72 h with CD3/CD28 beads (at the ratio of one bead for two cells (#11161D; Thermo Fischer Scientific). After 3 days, dead cells were removed by Ficoll density gradient, and activated T cells were expanded in complete Panserin supplemented with IL-2 (100 U/ml). Cells were split every 2–3 days with complete medium containing IL-2 (100 U/ml) to maintain a cell concentration of 1 × 106/ml.
Proliferation assay
Day 10 activated T cells were starved of IL-2 for 72 h, washed, incubated with CellTrace Violet reagent (#c34557; Invitrogen) for 8 min at 37°C in the dark and washed twice more. A total of 2 × 105 cells were seeded into a 96-well round plate and subjected to different stimuli (plate-bound with anti-CD3 antibody [Invitrogen], IL-2 at the concentration indicated in the figures) in the presence or absence of tofacitinib CP-690550 (100 μM) (#S2789; SelleckChem). The cells were cultured for 4 days, washed with PBS, and stained with anti-CD3-PeCy7, CD4-PerCP5.5, CD8-APC, CD25-BV650, and CD69-PE antibodies prior to flow cytometry measurement (BD LSRFortessa X-20 SORP Cell Analyzer cytometer). Analysis was performed on FlowJo V.10.9.0 software.
STAT5 dephosphorylation assay
Activated T cells were starved of IL-2 for 24 h, washed, and stained for extracellular labeling with anti-CD3-PeCy7, CD4-BV510, and CD8-APC during 15 min at RT and washed again. Cells were then incubated with IL-2 (250 U/ml) in Panserin medium only at a cell concentration of 5 × 106 cells/ml at 37°C. After 15 min of stimulation, cells were washed and resuspended in fresh medium always at the same cell concentration. Cells for 15-min time points are taken whereas others are incubated during 2 h at 37°C. Directly after these time points, cells were fixed, permeabilized using PerFix EXPOSE kit (#B26976; Beckman Coulter), and stained with anti-pSTAT5-BV421 (pY694, #562984; BD) or control isotype antibodies. The level of pSTAT5 was determined by flow cytometry (Novocyte Agilent) and analyzed on FlowJo V.10.9.0 software.
RNA-seq
RNA preparation
The expanded T cells were starved of IL-2 for 24 h, plated at cell concentration of 1 × 106 cells/ml in Panserin medium only, and then were left unstimulated or stimulated with IFN-α (104 U/ml, #IF007; Millipore) or IL-2 (104 U/ml) for 3 h at 37°C. RNA was extracted using RNeasy Mini Kit (Qiagen) and quality was assessed using Fragment Analyzer (Agilent Technologies) with RNA Integrity Number >8 and 28S/18S ratio around 2. mRNA was finally converted to complementary DNA by capture of poly-A+ RNA, amplified, and sequenced to a depth of at least 50 million reads using the Illumina technology (Novaseq 6000 sequencer) at the Genomic Platform of the Imagine Institute.
Data analysis
R v3.6.1 was used for analysis, FASTQ files were mapped to the HG38reference using HISAT2 and counted by featureCounts from the Subread R package. Read count normalization and group comparisons were performed by three independent and complementary statistical methods: DESeq2 (v1.24.0), edgeR (v3.26.8), and Limma Voom (v3.40.6). Flags were computed from counts normalized to the mean coverage. All normalized counts <20 were considered as background (flag 0) and ≥20 as signal (flag = 1). The results of the three methods were filtered at P value ≤0.05, and lists of genes statistically overexpressed with a fold change of 1.2 comparing patient and control groups were used for pathway enrichments. Enrichment for Gene Ontology (GO) Cellular Component 2021, GO Molecular Function 2021, GO Biological Process 2021, KEGG 2021 Human, BioCarta 2015, Reactome 2016, WikiPathway 2021 Human, and NCI Nature 2015 gene sets libraries were performed using EnrichR (Chen et al., 2013). Selected groups of genes corresponding to the “Signaling events mediated by TCPTP” gene set from the NCI Nature 2015 gene set library (Table S1) were chosen for plotting heatmap after performing normalization of each gene expression with scale function.
GSEA using GSEA package (Subramanian et al., 2005) was performed on the gene list of differential genes expressed with 1.2-fold change between patient and control groups for each stimulation condition. All types of gene sets were exported from Human MSigDB (Table S2). Collections used for this analysis are reported in Table S2 and gene sets enriched with adjusted P value <0.05 were reported in Table S3.
Phosphoflow on whole blood
Fresh whole blood of patients and controls were either left unstimulated or stimulated with IL-2, IFNg, or IFNa (104 U/ml) for 15 min or 1 h at 37°C. Extracellular staining was performed 15 min before fixation of cells using antibodies anti-CD3-PeCy7; CD4-BV510; CD8-BV711; CD19-APC; and CD14-PE (BD). Cells were then fixed and permeabilized with the kit PerFix EXPOSE (#B26976; Beckman Coulter) according to the manufacturer's instructions. Intracellular staining was performed 2 h at RT using antibodies anti pSTAT1-BV421 (pY701, #562985; BD) or pSTAT5-BV421 (pY694, #562984; BD). Cells were analyzed on the BD LSRFortessa X-20 SORP Cell Analyzer cytometer. CD4+ and CD8+ cells were gated on CD3+ cells. CD14+ cells were gated on CD3− and CD19− cells. The results were analyzed with FlowJo V.10.9.0 software.
Mass cytometry
Samples staining
Immune phenotyping on whole blood was carried out using the Maxpar Direct Immune Profiling kit (Cat#201325; Fluidigm) with an antibody panel of 30 markers for CyTOF (cytometry by time of flight) analysis. Eight additional antibodies were added to detect FAS and certain immune checkpoints (TIM3, TIGIT, ICOS, GITR, and PD-1). 300 μl of whole blood was used per labeling. The cells were incubated for 20 min at RT with 3 μl of heparin (Cat#H3149-10KU; Sigma-Aldrich) at 10,000 U/ml and 5 μl of Human TruStain FcX (Cat#422302; Biolegend) and then incubated for 30 min at RT with the antibody cocktail for extracellular labeling. Blood lysis was performed using Cell Cal-lyse buffer (GAS-010S100; Thermo Fischer Scientific) according to the manufacturer’s instructions. Finally, cells were incubated in the Fix&Perm buffer (Cat#201325; Fluidigm) with the Iridium intercalator at 1:1,000 dilution (Cat#201325; Fluidigm) overnight at 4°C. Cell solutions were frozen at −80°C prior to acquisition.
Acquisition
Cells were washed and resuspended at a concentration of 1 × 106/ml in Maxpar Cell Acquisition Solution, a high ion concentration solution, and mixed with 10% EQ beads (allowing for calibration automatic device) immediately before the acquisition. The acquisition of the events was carried out on the Helios mass cytometer 8 (Fluidigm) coupled with the CyTOF software version 6.7.1014 (Fluidigm) at the Pitié-792 Salpetrière Cytometry Platform. The acquired data were normalized using the Fluidigm normalization algorithm. Cells were selected by cell selection (Ir191+Ir193+), cell doublets were removed (time/offset, time/width, time/center, and time/residual), and dead cells were removed (Ir193+Rh103+). This selection is done automatically with the Pathsetter software.
Data analysis
FCS files containing viable singlet cells were uploaded in OMIQ software, https://app.omiq.ai/.
After control (number of cells per sample, expression pattern of all markers across samples), all cells were submitted to gating using the Maxpar Pathsetter analysis pipeline (Fluidigm).
To go further in the detail of memory CD4 T cells, they were submitted to subclustering using CXCR3, CXCR5, CCR6, and CCR4 as type markers defining Th1-like, Th2-like, Th17-like, and Tfh-like cells. The number for each subpopulation was divided per the number of memory non-Treg CD4 T cells.
To go further in the detail of B cells, they were subclustered using CD38, CD27, IgD, and CD11c to gate activated naive B cells (CD19+, CD38−, CD27−, IgD+, and CD11c+) or double-negative B cells (CD19+, CD38−, CD27−, IgD−, CD11c+).
Representation of immune cell populations was done on Prism 10 software; Kruskal–Wallis test was performed to assess statistical differences between control groups and PTPN2 group.
Visualization on Uniform Manifold Approximation and Projections (UMAPs) was performed using OMIQ software to see immune cell repartition in different groups (adult controls, pediatric controls, and PTPN2). Data were deposited in the FlowRepository public website (accession number: FR-FCM-Z7P5).
Cytokine assays
Plasma of controls or patients were collected by centrifugation of heparin blood sample. Mutations corresponding to STAT1, STAT3 GOF, or SOCS1 LOF patients are listed in Table S4.
For cytokine dosage in activated T cell supernatant, overnight-starved T cells from PTPN2 patients or controls were either unstimulated or stimulated by IL-2 or IFNα 104 U/ml during 24 h before supernatant collection.
Cytokine concentrations have been measured with the Legendplex Human Essential Immune Response Panel (#740930; Biolegend) according to the manufacturer’s instructions. The results have been analyzed on https://legendplex.qognit.com/ and then the decimal logarithms of the concentrations were normalized as follows: median concentrations obtained in the nine HCs for plasma dosage or control activated T cells for supernatant dosage were defined as 0, and the X-fold standard deviation or Z-score above this median (0 to +3.5, coded in red) or below (0 to −3.5, coded in blue) was calculated for each individual cytokine.
Online supplemental material
Fig. S1 characterizes PTPN2 induction at protein and mRNA levels following different stimuli on human PBMCs as well as phosphatase subcellular localization. Fig. S2 measures in vitro–activated T cell proliferation following TCR stimulation. All data from mass spectrometry analysis are available in Fig. S3 and all data from RNA-seq are available online (http://flowrepository.org/id/FR-FCM-Z7P5). Table S1 lists all pathways analyzed in NCI Nature 2015 gene set with corresponding genes found upregulated in patient cells compared with controls. Table S2 lists gene sets from https://www.gsea-msigdb.org/gsea/msigdb/ with corresponding gene set libraries they come from. Table S3 shows significantly enriched gene sets from GSEA analysis. Table S4 shows the genotype of patients from Fig. 6 C. Complete clinical data descriptions are available in Data S1.
Data availability
RNA-seq data that support the findings of this study have been deposited in the GEO repository public website (accession number: GSE270302). Mass spectrometry data were deposited in the FlowRepository public website (accession number: FR-FCM-Z7P5). All other data are available in the main text or the supplementary materials.
Acknowledgments
The authors thank the staff of Vect’UB, the vectorology platform (INSERM US 005 – CNRS UAR 3427- TBM-Core, Université de Bordeaux, France) for technical assistance. They thank the technical platform BIOPROFILER for providing UFLC facilities. We acknowledge the contribution of Structure Fédérative de Recherche Biosciences (UAR3444/CNRS, US8/Inserm, Ecole Normale Supérieure de Lyon, Université Claude Bernard) facilities: LYMIC-Platim-Microscopy/Ani-Rhone-Alpes (ANIRA)-Cytometry/ANIRA Vectorology and Protein Science Facility. We acknowledge the contributions of the Creation, Breeding, Phenotyping, Distribution, and Archiving of Model Organisms Infrastructure (https://www.celphedia.eu/), especially the center AniRA in Lyon.
This work was supported by INSERM and by government grants managed by the Agence National de la Recherche as part of the “Investment for the Future” program (Institut Hospitalo-Universitaire Imagine, grant ANR-10-IAHU-01, Recherche Hospitalo-Universitaire, grant ANR-18-RHUS-0010), the Centre de Référence Déficits Immunitaires Héréditaires, the Agence National de la Recherche (ANR-14-CE14-0026-01 “Lumugene”; ANR-18-CE17-0001 “Action”; ANR-22-CE15-0047-02 “BREAK-ITP”), the Fondation ARC pour la recherche sur le Cancer, the Fondation pour la recherche Médicale (EQU202103012670), by Agence National de la Recherche (ANR-21-CE17-0064 [SOCSIMMUNITY]), ANR-21-RHUS-08 [COVIFERON]) from the Agence National de la Recherche–Recherche Hospitalo Universitaire Program; by the Horizon Europe (01057100 [UNDINE]) from the HORIZON-HLTH-2021-DISEASE-04; le Centre de référence des rhumatismes inflammatoires, des interféronopathies et des maladies autoimmunes, the CNRS, the Université Paris Cité, and the Ligue Nationale Contre le Cancer (Ile-de-France committee).
Author contributions: M. Jeanpierre: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft, Writing—review & editing, Q. Riller: Data curation, Formal analysis, Software, Visualization, Writing—review & editing, L.-C. Bui: Investigation, Methodology, Writing—original draft, Writing—review & editing, J. Berthelet: Formal analysis, Investigation, Validation, Writing—original draft, A. Laurent: Writing—review & editing, E. Crickx: Investigation, Resources, Writing—original draft, Writing—review & editing, M. Parlato: Writing—review & editing, M.-C. Stolzenberg: Data curation, Investigation, F. Suarez: Data curation, G. Leverger: Resources, N. Aladjidi: Data curation, Resources, Writing—review & editing, S. Collardeau-Frachon: Investigation, Resources, Visualization, C. Pietrement: Resources, M. Malphettes: Resources, A. Froissart: Resources, N. Cagnard: Data curation, Formal analysis, T. Walzer: Conceptualization, Funding acquisition, Writing—review & editing, F. Rieux-Laucat: Conceptualization, Funding acquisition, Resources, Supervision, Validation, Writing—original draft, Writing—review & editing, A. Belot: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing—original draft, Writing—review & editing, A.-L. Mathieu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing, J. Cognard: Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing—original draft, Writing—review & editing, M. Tusseau: Formal analysis, Supervision, C. Bole-Feysot: Methodology, F. Rodrigues Lima: Conceptualization, Methodology, Writing—review & editing.
References
Author notes
M. Jeanpiere and J. Cognard contributed equally to this paper and share first authorship.
F. Rieux-Laucat, A. Belot, and A.-L. Mathieu share last authorship.
Disclosures: E. Crickx reported personal fees from Novartis, Amgen, UCB, and Sanofi outside the submitted work. A. Belot reported grants from Boehringer Ingelheim and personal fees from Abbvie, Kabi, and GlaxoSmithKline during the conduct of the study. No other disclosures were reported.