The exocyst imparts spatial control during exocytic vesicle tethering through its interactions with proteins and lipids on the vesicle and the plasma membrane. One such interaction is with the vesicle tether Sro7, although the outcome of this interaction is poorly understood. Here, we describe how Sro7 binding to the Exo84 subunit results in activation of the exocyst complex which leads to an increase in avidity for the Rab GTPase Sec4 and an increase in exocyst-mediated vesicle tethering. Gain-of-function (GOF) mutations in Exo84 that mimic Sro7 activation replicate these biochemical changes and result in allosteric changes within the complex. Direct comparison of GOF mutants which mimic Sro7- and Rho/Cdc42-activation of the exocyst reveals distinct mechanisms and outcomes. We propose a model by which these two activation pathways reside within the same tethering complex but remain insulated from one another. Structural modeling suggests a related mechanism for Sro7 activation of the exocyst in yeast and Ral GTPase activation of the exocyst in animal cells.
Introduction
Cell polarity is established and maintained through the asymmetric delivery of lipids and proteins to defined places on the cell surface. The spatial and temporal control of secretory vesicle trafficking is critical for polarized growth and involves distinct regulatory events along the vesicle’s journey from the Golgi to the plasma membrane. Discrete transport, delivery, docking, and fusion events are controlled by the recruitment and activation of highly conserved protein families including Rab and Rho GTPases, tethering factors, and SNARE proteins. One of the central players in this process is the exocyst, a multi-subunit tethering complex evolutionarily conserved from yeast to mammalian cells. The exocyst is a member of the CATCHR (complexes associated with tethering containing helical rods) family of tethers and is composed of eight structurally similar subunits: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84 (Mei et al., 2018). The exocyst functions to localize and tether secretory vesicles to the proper place on the plasma membrane for subsequent SNARE assembly and exocytosis (Lepore et al., 2018; Munson and Novick, 2006).
The exocyst is thought to establish spatial specificity through its interactions with specific proteins on the vesicle and on the target plasma membrane (Wu et al., 2008). Genetic and biochemical studies in yeast suggest that the exocyst arrives at the bud tip with the secretory vesicle and localizes to a polarized site of active exocytosis on the plasma membrane (Donovan and Bretscher, 2015; Heider et al., 2016). Likewise, in mammalian cells, a majority of the exocyst subunits are preassembled into octameric complexes and associated with vesicles prior to docking on the plasma membrane (Ahmed et al., 2018; Maib and Murray, 2022). On the secretory vesicle, the exocyst directly interacts with the Rab GTPase Sec4 through a proposed binding through the exocyst Sec15 subunit (Guo et al., 1999; Jin et al., 2011), as well as the vesicle SNARE (v-SNARE) Snc2 through the exocyst Sec6 subunit (Shen et al., 2013). The requirement of the exocyst to interact with two distinct vesicle-associated proteins may be important for confirming proper vesicle identity such that the exocyst only associates with vesicles carrying both key ligands (Shen et al., 2013). To tether vesicles to the target membrane, the exocyst also binds to proteins and lipids on the plasma membrane. Specifically, the exocyst directly interacts with the phospholipid PI(4,5)P2 as well as the Rho GTPases Rho3, Cdc42, and Rho1 through the Exo70 and Sec3 subunits (He et al., 2007; Wu et al., 2010; Zhang et al., 2008). The exocyst also interacts with the target SNAREs (t-SNAREs) Sec9 and Sso1 through the Sec6 and Sec3 subunits, respectively (Dubuke et al., 2015; Yue et al., 2017). To further coordinate SNARE complex assembly, the exocyst subunit Sec6 directly binds to the SM family SNARE regulator Sec1 (Morgera et al., 2012). These interactions allow the exocyst to serve as an intermediate tether, holding the secretory vesicle in close proximity to the plasma membrane while also localizing and coordinating SNAREs to promote SNARE-mediated membrane fusion.
Unique to polarized exocytosis is another vesicle tether called Sro7 (Lehman et al., 1999; Zhang et al., 2005), which functions in concert with the exocyst. Sro7 and its paralog Sro77 are structurally related to the neuronal exocytic protein tomosyn and the Drosophila melanogaster tumor suppressor lethal giant larvae (Lgl; Lehman et al., 1999). In yeast, Sro7 is important for polarized exocytosis as high copy numbers of Sro7 suppress mutations in Rho3 and Cdc42 (Adamo et al., 2001). In addition to its role as a Sec4 effector, Sro7 binds to the t-SNARE Sec9 and loss of both SRO7 and SRO77 genes results in pronounced exocytic defects (Lehman et al., 1999). Sro7 and Sec9 are distributed in the cytosol and along the entire plasma membrane when overexpressed, but Sro7 is polarized to sites of active growth when detected at endogenous expression (Lehman et al., 1999; Rossi and Brennwald, 2011). Given Sro7’s function in homotypic clustering of post-Golgi vesicles, this localization likely represents the appearance of small clusters of vesicles which normally gather at sites of polarized growth (Preuss et al., 1992; Rossi et al., 2015). Therefore, the presence of Sro7-laden vesicle clusters may represent a polarity signal that is quite distinct from that of Cdc42/Rho3 signal on the plasma membrane.
As a vesicle tether, Sro7 interacts directly with GTP-bound Sec4 (Sec4-GTP) through a conserved cleft formed by the intersection of its two β-propeller domains (Hattendorf et al., 2007; Lehman et al., 1999). Sro7 also has a C-terminal autoinhibitory tail that regulates Sro7 activity by competitively binding to the β-propeller domains and inhibiting binding with the Qbc-SNARE domain of Sec9 (Hattendorf et al., 2007). In addition, the oligomerization of Sro7 monomers bound to Sec4-GTP likely plays a role in clustering or tethering post-Golgi vesicles at sites of polarized fusion (Rossi et al., 2018). Sro7 function likely overlaps with the function of the exocyst as part of a parallel tethering pathway. In fact, the interaction between Sro7 and Sec4 is required to rescue growth defects of the late-acting secretion mutant sec15-1. However, mutant forms of Sro7 that are unable to bind Sec4 can still serve as the sole source of sro7 and sro77, presumably because the exocyst can still tether vesicles in these yeast (Watson et al., 2015). The exocyst and Sro7 also interact directly with one another, and disruption of this interaction leads to defects in exocytosis (Zhang et al., 2005), although the specific outcome of the interaction is unknown.
We recently characterized a regulatory pathway associated with the interaction between exocyst and Rho/Cdc42 GTPases. Rho/Cdc42 GTPases are essential determinants of polarity and spatial regulators of exocytosis in yeast (Adamo et al., 2001; Adamo et al., 1999; Guo et al., 2001). Regulation of exocyst function by Rho3 and Cdc42 is independent of any effect on the localization or assembly state of the complex (Roumanie et al., 2005; Wu et al., 2010). Instead, gain-of-function mutations in the exocyst Exo70 subunit that mimic Rho/Cdc42 GTPase activation of the complex reveal a specific structural rearrangement in the exocyst which opens the “Sec6-cap” of the complex (Rossi et al., 2020). This end contains a binding site for Snc2 (Shen et al., 2013), and the rearrangement upon activation results in a significant increase in the binding of exocyst to Snc2 (Rossi et al., 2020). At the bud tip, this connection between Cdc42/Rho on the plasma membrane and Snc2 on the vesicle through their mutual interactions with the exocyst reinforces polarized exocytosis through the regulation of exocyst activity.
In the present work, we used a biochemical reconstitution assay for exocyst-mediated vesicle tethering as well as genetic and biochemical approaches to uncover a second, quite distinct allosteric mechanism for spatially restricted regulation of the exocyst. This mechanism is initiated by binding of Sro7 to the Exo84 subunit of the exocyst. This regulation results in both increased vesicle tethering activity as well as increased exocyst binding to Sec4-GTP. To identify mutants in the exocyst that might mimic Sro7 activation, we carried out a screen to identify gain-of-function mutants in Exo84 which genetically and biochemically phenocopy the effect of Sro7 engagement on the exocyst. We presented the characterization of several such alleles that demonstrate both the genetic and biochemical properties predicted for mutants acting as mimetics of this activation. Our results demonstrate the existence of a new mechanism for allosteric regulation of the exocyst that acts as a second, isolated spatial signal for activating the exocyst at the appropriate place on the plasma membrane.
Results
Sro7 interacts with Exo84 and the exocyst through the Sro7 N-terminus
To understand how the interaction of Sro7 with the exocyst contributes to vesicle tethering, we set out to determine the binding site for the exocyst within the Sro7 sequence. Previous work identified the N-terminal half of the Exo84 subunit (aa 1–326) as the site of contact within the exocyst for Sro7 (Zhang et al., 2005). To map the contact site within Sro7, we performed binding experiments using beads containing recombinant GST-Exo84 (aa 1–326) and soluble recombinant Sro7 purified from yeast. While full-length Sro7 binds readily to GST-Exo84, a truncated form of Sro7 lacking the first 50 amino acids lost most of its Exo84 binding activity (Fig. 1 A). We also examined the effect of this deletion in the N-terminus of Sro7 on the interaction with the Qbc-SNARE, Sec9, and found no detectable change in the interaction (Fig. 1 A). We then expressed and purified a peptide consisting of the first 90 residues of Sro7 (Sro7-NT) and performed pulldown experiments with GST-Exo84 immobilized on beads (Fig. 1 B). The results of this binding suggest that the Sro7-NT peptide is sufficient for interacting with GST-Exo84 beads. To identify the specific residues with the NT peptide responsible for binding, we focused our attention on a conserved region at the extreme N-terminus of Sro7, since both Sro7 and its paralog Sro77 bind directly to Exo84 and have high sequence similarity at this site (Kagami et al., 1998).
Identification of elements within the N-terminus of Sro7 required for binding to the exocyst subunit Exo84. (A) Upper panel: Purified full length Sro7 and a N-terminal deletion of Sro7(aa 50–1033) were bound to immobilized GST-Exo84(aa 1–326) or GST alone in a pull-down assay. Coomassie of the GST-Exo84 fusion protein and quantitation of binding are shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. Lower panel: Purified full-length Sro7 and an N-terminal deletion of Sro7(aa 50–1033) were bound to GST-Sec9 or GST alone in a pull-down assay. Coomassie of the GST-Sec9 fusion protein and quantitation of binding are shown to the right. Error bars represent SD; P values were obtained as above. (B) Alignment of the N-terminus of Sro7 and Sro77 shows conserved amino acids between the yeast paralogs. Amino acids 1 through 10 are numbered. His-tagged wild-type Sro7-NT(aa 1–90) peptide and mutant Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 and Sro7-NT-A(4-8) peptides were purified and then bound to immobilized GST-Exo84(aa 1–326) in a pull-down assay. Quantitation of binding is shown to the right. Error bars represent SD and P values were obtained as described above. (C) Purified His-tagged Exo84(aa 1–326) was bound to C-terminally tagged GST-Sro7-NT(aa 1–79) immobilized on beads using mutant GST-Sro7-NT-A5, A6 as a control. The additional bands in the binding sample for GST-Sro7-NT-A5,A6 were present on beads prior to binding. (D) Purified wild-type exocyst was bound to GST-Sro7-NT(aa 1–79) and mutant GST-Sro7-NT-A5,A6 immobilized on beads. Quantitation of binding for four subunits, two from each sub-complex is shown to the right. Error bars represent SD. Source data are available for this figure: SourceData F1.
Identification of elements within the N-terminus of Sro7 required for binding to the exocyst subunit Exo84. (A) Upper panel: Purified full length Sro7 and a N-terminal deletion of Sro7(aa 50–1033) were bound to immobilized GST-Exo84(aa 1–326) or GST alone in a pull-down assay. Coomassie of the GST-Exo84 fusion protein and quantitation of binding are shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. Lower panel: Purified full-length Sro7 and an N-terminal deletion of Sro7(aa 50–1033) were bound to GST-Sec9 or GST alone in a pull-down assay. Coomassie of the GST-Sec9 fusion protein and quantitation of binding are shown to the right. Error bars represent SD; P values were obtained as above. (B) Alignment of the N-terminus of Sro7 and Sro77 shows conserved amino acids between the yeast paralogs. Amino acids 1 through 10 are numbered. His-tagged wild-type Sro7-NT(aa 1–90) peptide and mutant Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 and Sro7-NT-A(4-8) peptides were purified and then bound to immobilized GST-Exo84(aa 1–326) in a pull-down assay. Quantitation of binding is shown to the right. Error bars represent SD and P values were obtained as described above. (C) Purified His-tagged Exo84(aa 1–326) was bound to C-terminally tagged GST-Sro7-NT(aa 1–79) immobilized on beads using mutant GST-Sro7-NT-A5, A6 as a control. The additional bands in the binding sample for GST-Sro7-NT-A5,A6 were present on beads prior to binding. (D) Purified wild-type exocyst was bound to GST-Sro7-NT(aa 1–79) and mutant GST-Sro7-NT-A5,A6 immobilized on beads. Quantitation of binding for four subunits, two from each sub-complex is shown to the right. Error bars represent SD. Source data are available for this figure: SourceData F1.
We created a set of charged to alanine substitutions in this region and examined the effect of these mutations on the interaction between Sro7-NT and Exo84 (Fig. 1 B). Both the Sro7-NT-A(4-8) mutant and one mutant with two conserved positive charges at residues 5 and 6 mutated to alanine (Sro7-NT-A5,A6) result in nearly complete loss of binding to Exo84 (Fig. 1 B). The interaction is disrupted to a lesser extent when residues 7 and 8 are mutated to alanine, and no disruption is observed when residues 9 and 10 are mutated to alanine. In a second assay, we examined the ability of immobilized Sro7-NT to bind both soluble, recombinant Exo84 as well as intact exocyst complex purified from yeast. To immobilize Sro7-NT, we incorporated a GST moiety at the C-terminus of the peptide (Sro7-NT-GST). As can be seen in Fig. 1 C, while wild-type Sro7-NT pulls down soluble Exo84, the Sro7-NT-A5,A6 mutant exhibits a dramatic reduction in binding. Finally, in a third assay, we examined the binding between purified exocyst complexes and either the Sro7-NT or Sro7-NT-A5,A6 GST fusion proteins. While wild-type Sro7-NT fusions pull down a readily detectable pool of intact exocyst complex, the alanine substitutions of K5 and R6 eliminate binding of the exocyst complex to the mutant GST fusion (Fig. 1 D). Taken together these data demonstrate that the primary binding site for Exo84 in the exocyst lies within a small, conserved patch of charged amino acids in the disordered N-terminus of Sro7. Moreover, the Sro7-A5,A6 mutations effectively block this interaction and therefore serve as an important control for the experiments described below.
Sro7 stimulates post-Golgi vesicle tethering activity of the exocyst
We recently established an in vitro tethering assay to examine Sro7 and exocyst function as vesicle tethers (Rossi et al., 2020). This assay measures clustering of post-Golgi vesicles isolated from yeast secretory mutant strains and labeled with GFP-Sec4 and the lipid dye FM4-64 in the presence of purified exocyst and Sro7. To analyze the clustering data presented here, we developed a protocol for automated detection and analysis of fluorescent puncta using commercially available image analysis software (see Materials and methods). Comparison of manual counting methods and automated detection method reveals a strong agreement in overall results but that the automated method identifies more puncta per field than manually counting, allowing for more efficient identification and quantitation of fluorescent puncta in a completely unbiased manner in the in vitro vesicle:vesicle clustering assay (Fig. S1). Given the well-documented interaction between Sro7 and exocyst, as well as the requirement for a low dose (0.2 μM) of Sro7 to “prime” exocyst activity in the in vitro tethering assay (Zhang et al., 2005; Rossi et al., 2020), we hypothesized that a physical interaction of Sro7 and the exocyst complex was likely to be important for exocyst-mediated tethering to occur. To test this hypothesis, we sought to disrupt this interaction using a relatively high dose (1.5 μM) of Sro7-NT peptide as a competitive inhibitor of full length Sro7 during in vitro tethering. As a control, we utilized the Sro7-NT-A5,A6 mutant peptide which should not act as a competitive inhibitor. As we observed previously, purified exocyst complexes when combined with a small amount of full-length Sro7 tether purified post-Golgi vesicles into fluorescent puncta (Rossi et al., 2020).
Establishing an automated detection and analysis of clustered vesicles. (A) Replica of Fig. 2 A showing a representative of the fields for which clusters were manually or automatically detected and quantified. Scale bar, 5 μm. (B) Manual detection and quantification of six fields of the in vitro vesicle:vesicle tethering assay shown in A. Error bars represent SD. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (C) Automated detection and quantitation of the same six fields of view quantified in B (see Materials and methods; error bars represent SD, P values obtained and reported as described for B). (D) Zoomed in image of overlayed FITC (Sec4-GFP) and TRITC (FM4-64) channels of vesicle clusters. (E) Automated detection of clusters that are outlined in yellow and filled in in blue. The fluorescence in these clusters is then reported as a percentage of the total fluorescence in the field of view.
Establishing an automated detection and analysis of clustered vesicles. (A) Replica of Fig. 2 A showing a representative of the fields for which clusters were manually or automatically detected and quantified. Scale bar, 5 μm. (B) Manual detection and quantification of six fields of the in vitro vesicle:vesicle tethering assay shown in A. Error bars represent SD. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (C) Automated detection and quantitation of the same six fields of view quantified in B (see Materials and methods; error bars represent SD, P values obtained and reported as described for B). (D) Zoomed in image of overlayed FITC (Sec4-GFP) and TRITC (FM4-64) channels of vesicle clusters. (E) Automated detection of clusters that are outlined in yellow and filled in in blue. The fluorescence in these clusters is then reported as a percentage of the total fluorescence in the field of view.
Surprisingly, when the wild-type Sro7-NT peptide was added to the standard exocyst-mediated tethering assay, we observed significant stimulation of tethering activity rather than inhibition (Fig. 2 A). This stimulatory activity appeared to require the interaction of the Sro7-NT peptide with the exocyst complex, as the Sro7-NT-A5,A6 control peptide had no effect on “primed” exocyst tethering activity in the assay (Fig. 2 A). These results suggest that the Sro7-NT peptide has the capacity to directly stimulate exocyst-mediating tethering independent of full-length Sro7. We tested this possibility by adding Sro7-NT or Sro7-NT-A5,A6 to exocyst-mediated tethering assays in the absence or presence of full-length Sro7. Addition of either full-length Sro7 protein or Sro7-NT peptide resulted in an equivalent increase in exocyst-mediated tethering although the Sro7-NT peptide was at a considerably higher concentration than full-length Sro7 (Fig. 2 B). Stimulation of tethering required Sro7-NT binding to Exo84, as the effect of Sro7-NT mutants in the assay closely mirrored the effects of these mutants in Exo84 binding. In particular, three mutants that showed a reduced interaction with Exo84 were also unable to stimulate exocyst-mediated tethering, while one mutant which demonstrated an increase in binding to Exo84 showed a significant increase in exocyst-mediated tethering (Fig. 1 B and Fig. 2 C). Additionally, the Sro7-NT has no Sec4 binding activity, and only trace levels of clustering were observed when Sro7-NT was added alone (Fig. 2 B). As Sro7-NT peptide pulled down intact exocyst complex (Fig. 1 D), these data suggest that the physical interaction of Sro7-NT with the exocyst complex creates a change in the structural state of the exocyst which stimulates its tethering activity. Taken together these results suggest that Sro7 is a direct activator of the exocyst complex, independent of its own role as a vesicle tether.
Sro7-NT peptide but not the mutant Sro7-NT-A5,A6 peptide stimulates exocyst-mediated in vitro vesicle tethering. (A) Vesicles labeled with the lipid dye FM4-64 were isolated from a sec6-4 mutant strain expressing GFP-Sec4 and used in an in vitro vesicle-vesicle tethering assay with suboptimal concentrations of purified Sro7, identical amounts of wild-type exocyst complex, and purified Sro7-NT (aa 1–90) or Sro7-NT-5A,6A peptides. Vesicle-vesicle tethered clusters were automatically detected (see Materials and methods for details) and quantified as the percentage of fluorescence seen in clusters over the total fluorescence of the image using the TRITC channel. Error bars represent SD obtained from counting images at 60× magnification. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. Scale bar, 5 μm. (B) Labeled vesicles were incubated with purified exocyst and Sro7 or exocyst and wild-type Sro7-NT peptide or mutant Sro7-NT-5A,6A peptide as described above. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. (C) Labeled vesicles were incubated with exocyst and either wild-type or mutant Sro7-NT peptides [Sro7-NT; Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 or Sro7-NT-A(4-8)]. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. Coomassie of the peptides used is shown to the right. (D) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into a sec15-1 strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. (E) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into an sro7Δ;sro77Δ strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. Source data are available for this figure: SourceData F2.
Sro7-NT peptide but not the mutant Sro7-NT-A5,A6 peptide stimulates exocyst-mediated in vitro vesicle tethering. (A) Vesicles labeled with the lipid dye FM4-64 were isolated from a sec6-4 mutant strain expressing GFP-Sec4 and used in an in vitro vesicle-vesicle tethering assay with suboptimal concentrations of purified Sro7, identical amounts of wild-type exocyst complex, and purified Sro7-NT (aa 1–90) or Sro7-NT-5A,6A peptides. Vesicle-vesicle tethered clusters were automatically detected (see Materials and methods for details) and quantified as the percentage of fluorescence seen in clusters over the total fluorescence of the image using the TRITC channel. Error bars represent SD obtained from counting images at 60× magnification. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. Scale bar, 5 μm. (B) Labeled vesicles were incubated with purified exocyst and Sro7 or exocyst and wild-type Sro7-NT peptide or mutant Sro7-NT-5A,6A peptide as described above. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. (C) Labeled vesicles were incubated with exocyst and either wild-type or mutant Sro7-NT peptides [Sro7-NT; Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 or Sro7-NT-A(4-8)]. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. Coomassie of the peptides used is shown to the right. (D) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into a sec15-1 strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. (E) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into an sro7Δ;sro77Δ strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. Source data are available for this figure: SourceData F2.
To see whether we could observe evidence of Sro7 activation of the exocyst in an in vivo genetic assay, we made use of the fact that increased dosage of SRO7 is associated with potent suppression of a number of late-acting secretory mutants including those that encode components of the exocyst (Lehman et al., 1999; Zhang et al., 2005). One of the strongest such suppression events is the ability of increased dosage of SRO7 to suppress temperature-sensitive defects in the exocyst allele sec15-1 at 37°C (Lehman et al., 1999). While we initially observed this dosage effect with high-copy plasmids, we found that even a single-copy CEN plasmid resulting in duplication of SRO7 dosage was sufficient to observe pronounced suppression of sec15-1 growth defects at 34°C (Fig. 2 D). To see whether suppression by SRO7 is mediated by the exocyst-Sro7 interaction, we examined a panel of charged-to-alanine mutants in the N-terminus of Sro7 and found excellent agreement between the binding and suppression results. In particular, we found that the expression of SRO7-A5,A6, SRO7-A7,A8, or SRO7-A(4-8) mutants, all of which are defective in binding to Exo84 in vitro, fail to rescue growth of sec15-1 at 34°C. (Fig. 2 D). The SRO7-A9,A10 mutant which had no effect on binding rescued growth similarly to wild-type SRO7 (Fig. 2 D). We also examined the ability of the Sro7 mutants to function as the sole source of Sro7/77 in the cell. As the sole source of Sro7/77, all of the mutants fully complemented the cold sensitivity of the sro7Δ; sro77Δ mutant (Fig. 2 E). This result is not surprising, as even Sec4-binding-deficient SRO7 alleles can serve as the sole source of Sro7—presumably due to redundancy with the Sec4-binding activity of the exocyst tethering complex (Watson et al., 2015). Taken together, these results demonstrate a very strong correlation between genetic suppression and the ability of Sro7 to bind to recombinant Exo84, suggesting that the interaction which we observed between these two proteins is important for both the in vitro and the in vivo functions of the exocyst.
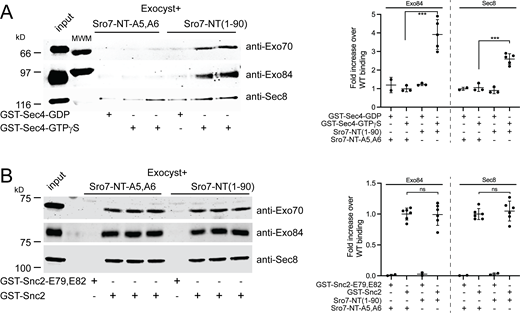
Sro7 stimulates exocyst binding to Sec4-GTP but not to Snc2
We have recently reported structural changes associated with activation of the exocyst by Rho/Cdc42 GTPases that result in increased vesicle tethering and a higher affinity of the exocyst for the v-SNARE Snc2 (Rossi et al., 2020). In addition to Snc2, the Rab GTPase Sec4 is a second post-Golgi vesicle-associated protein, which is critical for in vitro tethering (Rossi et al., 2020). Sec4-GTP on the vesicle surface is thought to bind directly to the exocyst and be involved in discriminating the appropriate vesicle:target membrane combination (Guo et al., 1999; Shen et al., 2013). To understand the effects of Sro7 activation on exocyst binding to downstream partners, we performed binding studies between Snc2 and Sec4 with purified exocyst in the presence of Sro7-NT. Previous attempts to study the interaction of the intact exocyst complex with Sec4 have been largely unsuccessful, presumably because the Sec4 binding site within the exocyst is inaccessible in the relatively compact or “closed” form of exocyst observed by CryoEM and negative stain EM approaches (Mei et al., 2018; Rossi et al., 2020). As expected, when we performed binding experiments using control 4 μM mutant peptide (Sro7-NT-A5,A6), purified exocyst, and beads containing recombinant Sec4, we were able to detect only background binding with no significant difference in binding to Sec4 exchanged with GDP or the nonhydrolyzable GTP analog, GTPγS (Fig. 3 A). However, when 4 μM Sro7-NT was added, we observed a significant and reproducible increase in exocyst binding to Sec4-GTPγS but no increase in binding to Sec4-GDP (Fig. 3 A). Increased binding was dependent on Sro7-NT interacting with the exocyst, as the addition of control peptide Sro7-NT-A5,A6 did not result in increased exocyst interaction with Sec4-GTPγS or Sec4-GDP (Fig. 3 A). We also performed exocyst binding experiments using GST beads containing the cytoplasmic domain of Snc2. We previously observed that gain-of-function mutations in the exocyst (Exo70-I114F), which mimic Rho/Cdc42 GTPase activation, result in increased affinity for Snc2 (Rossi et al., 2020). Exocyst binds readily to GST-Snc2, but not to a mutant control Snc2 (Snc2-E79,E82) containing two point mutations that block the interaction with the exocyst (Shen et al., 2013). Neither the addition of 4 μM Sro7-NT nor Sro7-NT-A5,A6 resulted in a significant increase in exocyst binding to Snc2 (Fig. 3 B). Taken together, these results demonstrate that the effect of the N-terminus of Sro7 on the exocyst and its tethering activity in vitro is likely mediated through an increase in exocyst affinity for Sec4 rather than for the vSNARE, Snc2. Furthermore, this suggests that the Sro7-mediated mechanism of activation of the exocyst through the Exo84 subunit is quite distinct from the mechanism of activation of the exocyst by Rho GTPases through the Exo70 subunit.
Sro7-NT peptide stimulates exocyst binding to Sec4-GTP but not Snc2. (A) Purified wild-type exocyst binding to immobilized GST-Sec4 beads exchanged with either GTPγS or control GDP in the presence of wild-type Sro7-NT (aa 1–90) peptide or control mutant Sro7-NT-A5,A6. Quantitation of two separate subunits one from each sub-complex is shown to the right. Error bars represent SD. P values were obtained using a two-tailed Student’s t test. *, P <0.05; **, P <0.01; *** P <0.001; ns = no significant difference. (B) Purified exocyst complex binding to immobilized GST-Snc2 or control GST-Snc2E79,E82 beads in the presence of wild-type Sro7-NT (aa 1–90) peptide or control mutant Sro7-NT-A5,A6 peptide. Quantitation of two separate subunits, one from each exocyst sub-complex is shown to the right. Error bars represent SD; P values were obtained as above. Source data are available for this figure: SourceData F3.
Sro7-NT peptide stimulates exocyst binding to Sec4-GTP but not Snc2. (A) Purified wild-type exocyst binding to immobilized GST-Sec4 beads exchanged with either GTPγS or control GDP in the presence of wild-type Sro7-NT (aa 1–90) peptide or control mutant Sro7-NT-A5,A6. Quantitation of two separate subunits one from each sub-complex is shown to the right. Error bars represent SD. P values were obtained using a two-tailed Student’s t test. *, P <0.05; **, P <0.01; *** P <0.001; ns = no significant difference. (B) Purified exocyst complex binding to immobilized GST-Snc2 or control GST-Snc2E79,E82 beads in the presence of wild-type Sro7-NT (aa 1–90) peptide or control mutant Sro7-NT-A5,A6 peptide. Quantitation of two separate subunits, one from each exocyst sub-complex is shown to the right. Error bars represent SD; P values were obtained as above. Source data are available for this figure: SourceData F3.
Isolation of dominant gain-of-function alleles of Exo84 that mimic Sro7 activation of the exocyst
We previously devised a genetic selection for gain-of-function mutants in the Exo70 subunit of the exocyst which behaved both genetically and biochemically as mimetics of Rho/Cdc42 activation (Wu et al., 2010). Like other Rho-effectors such as formins, the activation of the exocyst by Rho GTPases involves an allosteric mechanism involving “relief of autoinhibition” by blocking inhibitory interactions between Exo70 and other subunits of the complex, thus reorganizing the complex into a more active form (Rossi et al., 2020). After demonstrating biochemically distinct activities of exocyst complexes stimulated by Sro7-NT compared to exocyst complexes that mimic Rho activation, we investigated whether there were also distinct genetic consequences of activation by either the Sro7 pathway or the Rho pathway. We examined several non-exocyst post-Golgi secretory mutants for four critical properties: (1) strong suppression by multicopy (2µ) SRO7 (indicating the mutant is suppressed by increased function of Sro7); (2) lack of suppression by 2µ SRO7-A5,A6 (indicating that Sro7:Exo84 interaction is important for suppression); (3) lack of suppression by EXO70-I114F (indicating lack of sensitivity to gain-of-function through the Rho/Cdc42 activation pathway); and (4) lack of suppression by single-copy (CEN) wild-type EXO84 (critical for the selection of CEN gain-of-function alleles). Our examination of the late acting sec mutant collection revealed that the sec2-41 mutant, a temperature-sensitive allele of the guanine nucleotide exchange factor (GEF) for Sec4, satisfied all four requirements (Fig. 4 A). Importantly, because Sec2 is the sole GEF for the Sec4 GTPase (Walch-Solimena et al., 1997), we expected that use of the sec2-41 mutant in this screen would help to enrich for gain-of-function related to increased avidity of exocyst for Sec4-GTP, which would be reduced in this mutant. We chose to look for gain-of-function alleles in EXO84 for the same reasons we chose EXO70 for the Rho/Cdc42 screen; that mutations in the subunit involved in binding the activator were best placed to form gain-of-function mimetics of the effect of this binding on the complex, likely by disrupting contacts between this subunit and the rest of the complex (Rossi et al., 2020).
Isolation of dominant gain-of-function alleles of EXO84. (A) Single extra-chromosomal (CEN) copies of wild-type SRO7, wild-type EXO70, a gain-of-function allele EXO70-I114F, and wild-type EXO84 or multicopy (2μ) copies of wild-type SRO7 and SRO7-A5,A6 were transformed into the sec2-41 temperature-sensitive mutant. The growth of three independent transformants is shown for each transformation under permissive and restrictive conditions. (B) Schematic for the gap repair screen to generate and identify dominant EXO84 gain-of-function alleles in the sec2-41 mutant. (C) The N12I, L365W, and E626V gain-of-function alleles of EXO84 were transformed into the sec2-41 mutant. The growth of three independent transformants is shown for each condition. (D) Wild-type EXO84 and two gain-of-function mutations EXO84-L365W and EXO84-E626V were integrated into the EXO84 chromosomal locus marked with nourseothricin (Nat) resistance and crossed to a sec2-41 strain. Following tetrad analysis (Fig. S2), six representative segregants containing both sec2-41 and indicated genotypes of EXO84 or EXO84 gain-of-function alleles were examined for their ability to suppress the temperature-sensitive growth of sec2-41 at the indicated temperatures. (E) The structure of S. cerevisiae Exo84 (residues 169–753; PDB accession no. 5YFP, chain H) with the domains annotated and gain-of-function residues N12, L365, and E626 indicated as yellow space-filled spheres.
Isolation of dominant gain-of-function alleles of EXO84. (A) Single extra-chromosomal (CEN) copies of wild-type SRO7, wild-type EXO70, a gain-of-function allele EXO70-I114F, and wild-type EXO84 or multicopy (2μ) copies of wild-type SRO7 and SRO7-A5,A6 were transformed into the sec2-41 temperature-sensitive mutant. The growth of three independent transformants is shown for each transformation under permissive and restrictive conditions. (B) Schematic for the gap repair screen to generate and identify dominant EXO84 gain-of-function alleles in the sec2-41 mutant. (C) The N12I, L365W, and E626V gain-of-function alleles of EXO84 were transformed into the sec2-41 mutant. The growth of three independent transformants is shown for each condition. (D) Wild-type EXO84 and two gain-of-function mutations EXO84-L365W and EXO84-E626V were integrated into the EXO84 chromosomal locus marked with nourseothricin (Nat) resistance and crossed to a sec2-41 strain. Following tetrad analysis (Fig. S2), six representative segregants containing both sec2-41 and indicated genotypes of EXO84 or EXO84 gain-of-function alleles were examined for their ability to suppress the temperature-sensitive growth of sec2-41 at the indicated temperatures. (E) The structure of S. cerevisiae Exo84 (residues 169–753; PDB accession no. 5YFP, chain H) with the domains annotated and gain-of-function residues N12, L365, and E626 indicated as yellow space-filled spheres.
In the previous analysis of the Rho-Exo70 activation pathway, a key step was genetic selection and characterization of mutant EXO70 gain-of-function alleles with properties predicted of mimetics of the local Rho/Cdc42 activation of the exocyst complexes (Rossi et al., 2020; Wu et al., 2010). To carry out an analogous selection for gain-of-function alleles in an exocyst subunit that mimic Sro7 activation, we performed an extensive random mutagenesis/suppressor screen for EXO84 gain-of-function alleles which dominantly suppress sec2-41. EXO84 was randomly mutagenized by error-prone PCR amplification and introduced into the sec2-41 mutant yeast by cotransformation with a gap-repair plasmid containing the flanking sequences of the EXO84 gene (Fig. 4 B). Approximately 20,000 transformants were assayed and 22 plasmids containing dominant suppressing forms of EXO84 were ultimately isolated.
Sequence analysis of the 22 suppressors identified an average of nine coding sequence changes per allele. To identify specific amino acid substitutions likely to be linked to suppression, we scanned the mutant sequences for substitutions that appeared in two or more plasmids (Table S1). From this, we identified three single residue mutations as candidates to confer dominant suppression on Exo84. These mutations were then prepared as single residue changes and tested for dominant suppression in the sec2-41 mutant. Two of the three Exo84 substitutions, L365W and E626V, displayed clear suppression at 33.5°C, while all three single residue substitutions, including the weaker N12I substitution, demonstrated some level of suppression when tested at 31.5°C (Fig. 4 C). To determine if the suppression of sec2-41 by the EXO84 gain-of-function mutants could be observed when expressed from the EXO84 locus, we isolated strains containing chromosomal integration of the two strongest suppressors EXO84-W365 and EXO84-V626 (see Materials and methods), and crossed them along with a control EX084 integrant, to a sec2-41 strain. The results of the meiotic analysis of these crosses are shown in Fig. S2, and demonstrate that the suppression of sec2-41 following integration of these alleles is extremely similar to the suppression observed from these same alleles expressed from CEN plasmids.
Tetrad analysis of cross between sec2-41 and strains containing integrated forms of EXO84 gain-of-function alleles. Yeast strain (BY3263: α, sec2-41, his3-200) was crossed to three strains containing integrated copies of EX084 marked with nourseothricin (Nat) resistance: BY3311: a, EXO84::NatR, ura3-52, leu2-3,112; BY3312: a, EXO84-L365W::NatR, ura3-52, leu2-3,112; and BY3313: EXO84-E626V::NatR; ura3-52, leu2-3,112. Following sporulation tetrads were dissected, grown at 25C for 2 d and then replica-plated onto various media and grown at temperatures as indicated in the figure.
Tetrad analysis of cross between sec2-41 and strains containing integrated forms of EXO84 gain-of-function alleles. Yeast strain (BY3263: α, sec2-41, his3-200) was crossed to three strains containing integrated copies of EX084 marked with nourseothricin (Nat) resistance: BY3311: a, EXO84::NatR, ura3-52, leu2-3,112; BY3312: a, EXO84-L365W::NatR, ura3-52, leu2-3,112; and BY3313: EXO84-E626V::NatR; ura3-52, leu2-3,112. Following sporulation tetrads were dissected, grown at 25C for 2 d and then replica-plated onto various media and grown at temperatures as indicated in the figure.
Structural features of dominant gain-of-function Exo84 alleles
To understand our gain-of-function Exo84 mutations in the context of existing structural data, we mapped the Exo84 alleles onto the cryo-EM structure of the yeast exocyst (Mei et al. 2018). All eight exocyst subunits show a high degree of structural similarity to one another—each has a “CorEx” long coiled-coil motif, followed by 2 or 4 CATCHR domains, which are stacked alpha helices that is the main structural feature of CATCHR-family tethering complexes. However, Exo84 is the most divergent exocyst subunit from a structural standpoint. Exo84 has a disordered N-terminus (aa 1–168) before the CorEx domain, which is not seen in any structures and is predicted to be disordered in isolation. The only other subunit with a domain before its CorEx motif is Sec3, which contains a PH domain that is responsible for interaction with the plasma membrane (Zhang et al., 2008). Exo84 is also unique in that it is the only exocyst subunit that does not have 4 CATCHR motifs C-terminal to its CorEx motif. Instead, it contains a PH domain, followed by two CATCHR domains. Using the cryoEM structure of the full exocyst complex, we mapped two of the three gain-of-function mutations on Exo84 (Fig. 4 E and Fig. S3). The L365 residue resides on the surface of the PH domain at the interface between Exo84 and the third CATCHR domain of Sec10. L365 packs against a polar patch on Sec10 with the nearest interface residues being N593 and N670; it is, therefore, likely that addition of a bulky hydrophobic residue into this region of the Exo84-Sec10 interface destabilizes this interaction. Similarly, the E626 residue is surface exposed between the two CATCHR domains of Exo84 and lies at the interface with Sec5. The nearest interface residue to Exo84 E626 is Sec5 R867, although the resolution of the cryo-EM structure precludes us from saying whether these residues form a salt bridge or interact directly.
Mapping of Exo84 GOF alleles. The cryo-EM structure of yeast exocyst was used to map the location of two alleles in Exo84. E626 resides at the interface of Exo84 with Sec5, in close proximity to two basic residues, Exo84 K685 and Sec5 R867 (left). L365 is at the interface of Exo84 with Sec10, in close proximity to multiple polar residues, including Sec10 N593 and N670 (right).
Mapping of Exo84 GOF alleles. The cryo-EM structure of yeast exocyst was used to map the location of two alleles in Exo84. E626 resides at the interface of Exo84 with Sec5, in close proximity to two basic residues, Exo84 K685 and Sec5 R867 (left). L365 is at the interface of Exo84 with Sec10, in close proximity to multiple polar residues, including Sec10 N593 and N670 (right).
As the cryo-EM exocyst structure is thought to represent an auto-inhibited conformation, the L365W and E626V alleles are likely to act as activating mutations by disrupting important intra-exocyst contacts. In contrast, Exo84-N12 lies within Exo84’s disordered N-terminus and the allosteric consequences of this mutation are, therefore, more opaque than the L365 or E626 mutants. The fact that the dominant Exo84 mutations are separated by considerable distances within the exocyst suggests that there are likely to be multiple, structurally distinct mechanisms by which Exo84 gain-of-function activities can be achieved.
EXO84 dominant mutants increase exocyst vesicle tethering activity in vitro and exocyst binding to Sec4-GTP
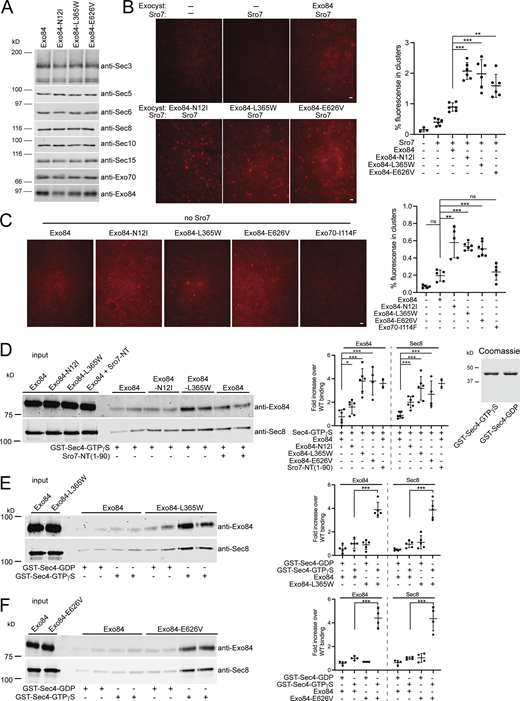
The sec2-41 screen was based on the prediction that one likely method that Exo84 gain-of-function mutations would achieve suppression is by mimicking the activation of the exocyst by Sro7. Therefore, we would expect exocyst complexes containing Exo84 gain-of-function mutants to demonstrate the properties of Sro7 activation described above, including increased tethering activity in vitro and increased binding to Sec4-GTP but no change in binding to Snc2. To test this possibility, we constructed yeast strains containing wild-type and mutant EXO84 alleles as the sole source of Exo84 in cells engineered with a C-terminal TEV/myc-tagged form of Sec8. We purified wild-type and mutant exocyst complexes from strains containing EXO84-N12I, EXO84-L365W, EXO84-E626V, or wild-type EXO84. We first assayed for the presence of the exocyst subunits by quantitative immunoblot analysis. Coimmunoprecipitation of all three of the gain-of-function alleles of EXO84 and wild-type complexes resulted in identical amounts of all eight exocyst subunits indicating that the mutants have no effect on assembly or stability of the complex (Fig. 5 A). These data also suggest that, like Rho GTPase activation, any change in the biochemical activity of the exocyst is due to an allosteric change in the packing of one or more of the subunits within the complex rather than a change in the assembly/disassembly of the complex in response to the gain-of-function mutation.
Biochemical characterization of EXO84 dominant mutants. (A) Immunoblot analysis of exocyst subunits in wild-type and EXO84 dominant mutant strains. C-terminally myc-tagged Sec8 (SEC8-myc) was introduced by transformation into strains containing wild-type or dominant mutants of EXO84 (on CEN plasmids) as the sole source of Exo84 in the cell. Cells were grown, lysed, and subjected to native immunoprecipitations with myc antibody to isolate and analyze the exocyst complex. Shown are immunoblots using antibodies against all subunits of the exocyst. (B and C) Vesicles labeled with the lipid dye FM4-64 were isolated from a sec6-4 mutant strain expressing Sec4-GFP and used in the in vitro vesicle-vesicle tethering assay with identical amounts of exocyst containing wild-type Exo84, Exo84-N12I, Exo84-L365W, Exo84-E626V, Exo70-I114F, or buffer only in the presence of Sro7 in B or in the absence of Sro7 in C. Scale bar, 5 μm. Vesicle-vesicle tethering was automatically detected and quantified as the percentage of fluorescence seen in clusters over the total fluorescence of the 60× magnification image. Error bar represents SD; P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (D) Purified wild-type exocyst and exocyst containing Exo84-N12I, Exo84-L365W, or Exo84-E626V were used in a GST pulldown assay with GST-Sec4-GTPγS immobilized on beads. Quantitation of the binding is shown to the right as well as a Coomassie stain of GST-fusion Sec4 proteins used. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (E and F) Purified wild-type exocyst, and exocyst containing Exo84-L365W or Exo84-E626V were bound to immobilized GST-Sec4-GTPγS or control GST-Sec4-GDP beads. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. Source data are available for this figure: SourceData F5.
Biochemical characterization of EXO84 dominant mutants. (A) Immunoblot analysis of exocyst subunits in wild-type and EXO84 dominant mutant strains. C-terminally myc-tagged Sec8 (SEC8-myc) was introduced by transformation into strains containing wild-type or dominant mutants of EXO84 (on CEN plasmids) as the sole source of Exo84 in the cell. Cells were grown, lysed, and subjected to native immunoprecipitations with myc antibody to isolate and analyze the exocyst complex. Shown are immunoblots using antibodies against all subunits of the exocyst. (B and C) Vesicles labeled with the lipid dye FM4-64 were isolated from a sec6-4 mutant strain expressing Sec4-GFP and used in the in vitro vesicle-vesicle tethering assay with identical amounts of exocyst containing wild-type Exo84, Exo84-N12I, Exo84-L365W, Exo84-E626V, Exo70-I114F, or buffer only in the presence of Sro7 in B or in the absence of Sro7 in C. Scale bar, 5 μm. Vesicle-vesicle tethering was automatically detected and quantified as the percentage of fluorescence seen in clusters over the total fluorescence of the 60× magnification image. Error bar represents SD; P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (D) Purified wild-type exocyst and exocyst containing Exo84-N12I, Exo84-L365W, or Exo84-E626V were used in a GST pulldown assay with GST-Sec4-GTPγS immobilized on beads. Quantitation of the binding is shown to the right as well as a Coomassie stain of GST-fusion Sec4 proteins used. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (E and F) Purified wild-type exocyst, and exocyst containing Exo84-L365W or Exo84-E626V were bound to immobilized GST-Sec4-GTPγS or control GST-Sec4-GDP beads. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. Source data are available for this figure: SourceData F5.
To determine if the tethering activity of the mutant exocyst complexes was affected by the gain-of-function Exo84 subunits, we examined wild-type Exo84 and mutant Exo84 exocyst complexes in the in vitro vesicle tethering assay under conditions described in Fig. 2. We observed a significant and substantial increase in tethering when the Exo84-N12I, Exo84-L365W, or Exo84-E626V subunits were present in the complex (Fig. 5 B). The observed similarity in increased tethering activity for all of the Exo84 gain-of-function alleles was somewhat surprising as it is in contrast to the differences that we observed in their ability to suppress sec2-41 growth defects (Fig. 4 C). Based on our prediction that the Exo84 gain-of-function mutants act as mimetics of Sro7 activation, we next tested whether Sro7 “priming” was required for the increased tethering activity we observed in Fig. 5 B. As we have shown previously, on its own wild-type exocyst has no detectable effect on vesicle clustering in the tethering assay (Rossi et al., 2020). However, when the gain-of-function exocyst complexes containing Exo84-N12I, Exo84-L365W, or Exo84-E626V were mixed with vesicles in the absence of Sro7, we detected a significant increase in fluorescent clustering relative to background seen with buffer or wild-type Exo84 containing exocyst complex (Fig. 5 C). Interestingly, no significant tethering activity was observed for gain-of-function exocyst complexes containing Exo70-I114F. Together, these results demonstrate that the Exo84 gain-of-function alleles upregulate the functional state of the exocyst both in vivo and in vitro, and now allow us to observe in vitro exocyst-mediated tethering activity in the absence of any Sro7, albeit to less of an extent than exocyst-mediated tethering in the presence of “priming” Sro7.
We next asked whether the in vivo suppression and in vitro activity of the gain-of-function mutant exocyst complexes were due to an increase in Sec4-GTP binding like wild-type exocyst stimulated with the Sro7-NT peptide (Figs. 2 and 3). We performed binding experiments using purified wild-type and Exo84 gain-of-function exocyst complexes and GST beads containing the Rab GTPase Sec4 loaded with either GDP or GTPγS. In this assay, the N-terminal peptide of Sro7 robustly stimulated exocyst binding to Sec4 in a nucleotide-dependent manner (Fig. 3 A). When the Exo84-L365W or Exo84-E626V complexes were examined, we observed a substantial increase in binding to Sec4-GTPγS relative to wild-type exocyst complex and this increase was comparable to wild-type complex stimulated with NT-Sro7 peptide (Fig. 5 D). The increase in Sec4 binding for the activated exocyst mutant complex in these binding assays is highly nucleotide-specific as only background binding was observed for wild-type, Exo84-L365W, and Exo84-E626V exocyst complexes binding to Sec4-GDP beads in side-by-side comparisons with Sec4-GTPγS beads (Fig. 5, E and F). The Exo84-N12I complexes, however, demonstrated only a small but significant increase in Sec4-GTPγS binding (Fig. 5 D). Importantly, the increased Sec4-binding capability of these Exo84 gain-of-function complexes was observed in the absence of stimulating Sro7-NT peptide, strongly supporting our hypothesis that these mutations might act as mimetics for Sro7 activation of the exocyst. Furthermore, the degree to which the three mutant alleles stimulate Sec4-binding is in strong agreement with the degree to which these mutants suppress the growth defects of the sec2-41 mutant; the strongest suppressing allele is EXO84-L365W followed by EXO84-E626V and lastly EXO84-N12I, which only weakly suppresses at 33.5°C (Fig. 4 C and Fig. 6 D). Taken together, these data support our model that Sro7-mediated activation of the exocyst results in its increased binding for GTP-bound Sec4 through specific allosteric changes that can be recapitulated by the gain-of-function alleles in the Exo84 subunit of the exocyst.
Two distinct mechanisms for activating the exocyst: Each with distinct outcomes
The suppressor screen using sec2-41 identified two strong Exo84 gain-of-function mutants (EXO84-L365W and EXO84-E626V) which appear to mimic an allosteric activation mechanism that is quite distinct from the allosteric activation mechanism we uncovered from a cdc42-6/rho3Δ suppressor screen (Wu et al., 2010; Rossi et al., 2020). To directly explore the “separation of function” of these two putative mechanistic pathways, we compared the genetic and biochemical properties of three Exo84 gain-of-function mutants with the strongest Exo70 gain-of-function mutant, Exo70-I114F, whose increased tethering activity we previously demonstrated to be linked to its increased affinity for the v-SNARE, Snc2 (Rossi et al., 2020). We first asked whether the EXO84 gain-of-function alleles resulted in measurable changes of exocyst binding to Snc2. Using the Snc2 binding assay described in Fig. 3, we measured binding of the Exo84 gain-of-function complexes to GST-Snc2 or the GST-Snc2 (E79,E82) control beads. While the L365W and E626V substitutions do not result in a change in exocyst binding for Snc2, the Exo84-N12I substitution resulted in a significant increase in exocyst binding for Snc2 (Fig. 6 A). These results suggest that the outcome of activation shown by Exo84-L365W and Exo84-E626V is distinct from Exo70-I114F or Exo84-N12I. We next directly compared the Snc2 binding affinity of exocyst complexes containing either Exo84-N12I or Exo70-I114F. As the yeast strains used to purify these complexes contain a mutant copy of the subunit gene on a CEN plasmid as the sole source, both Exo84-N12I and Exo70-I114F have their own wild type for comparison included in this binding assay. When we compared these mutant complexes side-by-side, we observed the same level of increase in binding for Snc2 (Fig. 6 B). The Exo84-N12I containing complexes, which show only a small effect on Sec4-GTP binding (Fig. 5 D), behave biochemically in a fashion that is similar to the mutants described in the cdc42-6/rho3 suppressor screen (i.e. Exo70-I114F) rather than to the other two Exo84 gain-of-function mutants (L365W and E626V). Additionally, when we measured Sec4 binding for the Exo70-I114F containing complexes, we observed only a slight increase of binding to Sec4-GTPγS relative to wild-type exocyst. In contrast, Exo84-E626V containing complexes, which show little change in Snc2 binding activity (Fig. 6 A), show a significant increase in binding to Sec4-GTPγS (and not Sec4-GDP) compared to wild-type exocyst (Fig. 6 C).
Distinct suppression and biochemical activity of exocyst gain-of-function mutants that mimic activation by Sro7 or Rho GTPases. (A) GST-Snc2 or GST-Snc2 E79,E82 were immobilized on beads and used in a pull down assay with purified wild-type exocyst or exocyst containing Exo84-N12I, Exo84-L365W, or Exo84-E626V. Quantitation of the pull-down experiments with error bars representing SD are shown to the right as well as a Coomassie stain of GST-fusion Snc2 protein used. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (B) GST-Snc2 or GST- Snc2 E79,E82 were immobilized on beads and used to bind exocyst complexes containing either wild-type subunits, Exo84-N12I, or Exo70-I114F gain-of-function alleles as performed in A. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (C) Purified wild-type exocyst and exocyst containing Exo70-I114F or Exo84-E626V were bound to beads containing immobilized GST-Sec4-GTPγS or control GST-Sec4-GDP. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (D) Wild-type EXO84, wild-type EXO70, or the gain-of-function mutants EXO84-N12I, EXO84-L365W, EXO84-E626V and EXO70-I114F, vector (CEN) and 2µSRO7 were transformed into the sec2-41 (left) and cdc42-6 (right) mutant strains. The growth of three independent transformants is shown for each mutant allele, under permissive and restrictive conditions. (E) Wild-type EXO84, EXO84-N12I, EXO84-L365W and EXO84-N12I,L365W and vector were transformed into the sec2-41(left) and cdc42-6(right) mutant strains. The growth of three independent transformants is shown at the permissive and restrictive conditions. Source data are available for this figure: SourceData F6.
Distinct suppression and biochemical activity of exocyst gain-of-function mutants that mimic activation by Sro7 or Rho GTPases. (A) GST-Snc2 or GST-Snc2 E79,E82 were immobilized on beads and used in a pull down assay with purified wild-type exocyst or exocyst containing Exo84-N12I, Exo84-L365W, or Exo84-E626V. Quantitation of the pull-down experiments with error bars representing SD are shown to the right as well as a Coomassie stain of GST-fusion Snc2 protein used. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. (B) GST-Snc2 or GST- Snc2 E79,E82 were immobilized on beads and used to bind exocyst complexes containing either wild-type subunits, Exo84-N12I, or Exo70-I114F gain-of-function alleles as performed in A. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (C) Purified wild-type exocyst and exocyst containing Exo70-I114F or Exo84-E626V were bound to beads containing immobilized GST-Sec4-GTPγS or control GST-Sec4-GDP. Quantitation of binding was conducted as described above and is shown on the right. Error bars represent SD; P values were obtained using a two-tailed Student’s t test with significance annotated as above. (D) Wild-type EXO84, wild-type EXO70, or the gain-of-function mutants EXO84-N12I, EXO84-L365W, EXO84-E626V and EXO70-I114F, vector (CEN) and 2µSRO7 were transformed into the sec2-41 (left) and cdc42-6 (right) mutant strains. The growth of three independent transformants is shown for each mutant allele, under permissive and restrictive conditions. (E) Wild-type EXO84, EXO84-N12I, EXO84-L365W and EXO84-N12I,L365W and vector were transformed into the sec2-41(left) and cdc42-6(right) mutant strains. The growth of three independent transformants is shown at the permissive and restrictive conditions. Source data are available for this figure: SourceData F6.
In order to explore these two activation pathways further in vivo, we compared the genetic properties of the EXO70 and EXO84 gain-of-function mutants side-by side. Using the sec2-41 mutant strain, we found that while the two strongest suppressors identified in the initial screen, EXO84-L365W and EXO84-E626V, clearly suppressed the temperature-sensitive growth defect of the mutant, comparatively little suppression of sec2-41 was observed for either the EXO84-N12I mutant or the EXO70-I114F mutant alleles (Fig. 6 D). We next compared these two families of exocyst gain-of-function suppressors in the cdc42-6 mutant strain. As expected, the EXO70-I114F allele clearly suppressed the growth defect as previously demonstrated (Wu et al., 2010). Surprisingly, the EXO84-N12I allele also strongly suppressed the temperature-sensitive growth defect of cdc42-6, more potently in fact than the suppression by the EXO70-I114F allele (Fig. 6 E). We noted that all of the gain-of-function mutants which demonstrate an increase in binding to Snc2 also demonstrate an ability to suppress the growth defects of a cdc42-6 mutant. Moreover, this can occur with the gain-of-function mutants in either the Exo70 or Exo84 subunits. Similarly, the gain-of-function mutants which demonstrate a strong increase in binding to Sec4-GTP also show an increase in suppression of sec2-41.
These data strongly support a model in which two distinct mechanisms activate the exocyst complex. In one mechanism, Rho/Cdc42 binding to the Exo70 subunit causes a structural rearrangement resulting in increased avidity for the v-SNARE Snc2. In the second mechanism, Sro7 binding to the Exo84 subunit leads to a different arrangement of the complex resulting in increased avidity for the Rab GTPase Sec4. For each of these mechanisms, we have identified gain-of-function mutations in exocyst subunits that mimic activation.
Since the EXO84-I12 allele strongly suppresses the cdc42-6 mutant and increases exocyst binding to Snc2, it appears to function similarly to the EXO70 gain-of-function mutants which mimic the Rho activation pathway of the complex. To ask the question of whether we could simultaneously activate both the Sro7 and Rho activation pathways, we created a double mutant, EXO84-I12,W365 and examined its ability to suppress sec2-41 and cdc42-6 temperature-sensitive growth. The results, shown in Fig. 6, F and G, demonstrate unequivocally that the EXO84-I12,W365 double mutant strongly suppresses the temperature-sensitive growth of both of these mutants. Therefore, these two regulatory pathways are distinct but are not mutually exclusive in how they operate within the complex.
Discussion
In this work, we characterized a new mechanism for allosteric regulation of the exocyst. The discovery of this regulatory pathway resulted from an unexpected biochemical effect of adding a peptide containing the N-terminus of an exocyst interacting protein Sro7 to an in vitro vesicle tethering assay to examine exocyst function (Rossi et al., 2020). Binding of this Sro7 peptide to Exo84 results in both stimulation of exocyst tethering activity and an increased avidity of the exocyst for the GTP-bound form of the Rab GTPase Sec4. We devised a genetic screen utilizing a temperature sensitive form of the Sec4 GEF Sec2, the sec2-41 mutant, and succeeded in identifying several dominant gain-of-function mutants in Exo84. Importantly, two of these Exo84 gain-of-function substitutions, L365W and E626V, appear to functionally mimic exocyst activation by Sro7-NT in their ability to stimulate both in vitro tethering activity and binding to Sec4-GTP. Further, we observed no detectable change in the assembly/disassembly state of exocyst, suggesting that this regulatory mechanism is primarily allosteric in nature.
The discovery of this new pathway allowed us to compare with our recently characterized exocyst activation pathway involving Rho GTPases and the Exo70 subunit, and we can begin to predict several important molecular features of allosteric activation of exocyst. First, prior to any spatial cue, the exocyst normally exists in a basal, inactivated state that is structurally “closed” (Fig. 7 A). At the bud tip, multiple activation signals—including Cdc42/Rho GTPases and Sro7-associated vesicle clusters—can individually initiate a specific allosteric activation mechanism. In the case of Rho/Cdc42, activation likely results from flexibility of the Exo70 subunit, which causes an opening of the “Sec6 cap” and increased availability for the Snc2-binding site (Wu et al., 2010; Rossi et al., 2020; Fig. 7 A). In the case of Sro7 activation, we suggested that Sro7 binding to Exo84 results in local structural changes where the extended “Sec15 pole” of the complex has increased flexibility or openness resulting in increased access to the Sec4-binding site at the C-terminus of Sec15 (Fig. 7 A). Previous studies suggest that the C-terminus of the Sec15 subunit is likely to be the primary binding site for the Rab GTPase Sec4 (Guo et al., 1999; Shen et al., 2013; Wu et al., 2005). The Exo84 C-terminus, which includes both sec2-41 suppressing mutations L365W and E626V, lies against the Sec15 subunit. Our gain-of-function Exo84 mutations could dissociate the Exo84 C-terminus from the rest of the exocyst complex, exposing a larger section of the Sec15 subunit and increasing exocyst affinity for Sec4 (Fig. 7 A). It is interesting to note that the Sro7 binding site is within the first half of Exo84 (aa 1–326), while the gain-of-function mutations and predicted structural changes occur in the second half of the protein, suggesting extensive allosteric crosstalk throughout the exocyst.
Models for exocyst activation mechanisms. (A) Model for dual exocyst-activation mechanism at the bud tip. During recruitment to the bud tip (red circle), the exocyst remains in a basal, autoinhibited state. While tethering a secretory vesicle to the plasma membrane (red square), the exocyst is activated by two of its binding partners. Each of the partners is color-coded to match to the subunit with which they interact. Sro7 interacts with Exo84 through its N-terminus (pink arrow) resulting in an increased binding of the exocyst for Sec4-GTP through the Sec15 subunit (red arrow). The first 168 aa of Exo84 do not appear in the CryoEM or negative stain EM electron density, likely due to flexibility/dynamics, and are depicted by a pink line. We previously reported that Rho GTPases on the plasma membrane also activate the exocyst by interacting with Exo70 (blue arrow) resulting in an increased binding affinity of the exocyst for Snc2 through the Sec6 subunit (purple arrow) and that these changes were associated with corresponding movement of Exo70 and opening up of Sec6. In the red square Sro7 is depicted as a homo-oligomer interacting with and tethering vesicles through its own interactions with Sec4-GTP. Sro7-tethered vesicle clusters primed at the site of exocytosis are shown in the yeast bud tip (shown on left) as vesicles with pink (Sro7 oligomer) lines connecting them. Putative allosteric changes associated with Sro7- or Rho-binding are modeled using the exocyst CryoEM structure (PDB accession no. 5YFP) and the Exo70 gain-of-function negative stain EM structure (PDB accession no. 6VKL). (B) Speculative Model for structurally related exocyst activation mechanisms shared between yeasts and animal cells through the Exo84 subunit resulting in increased avidity of the complex for Rab GTPase on the vesicle.
Models for exocyst activation mechanisms. (A) Model for dual exocyst-activation mechanism at the bud tip. During recruitment to the bud tip (red circle), the exocyst remains in a basal, autoinhibited state. While tethering a secretory vesicle to the plasma membrane (red square), the exocyst is activated by two of its binding partners. Each of the partners is color-coded to match to the subunit with which they interact. Sro7 interacts with Exo84 through its N-terminus (pink arrow) resulting in an increased binding of the exocyst for Sec4-GTP through the Sec15 subunit (red arrow). The first 168 aa of Exo84 do not appear in the CryoEM or negative stain EM electron density, likely due to flexibility/dynamics, and are depicted by a pink line. We previously reported that Rho GTPases on the plasma membrane also activate the exocyst by interacting with Exo70 (blue arrow) resulting in an increased binding affinity of the exocyst for Snc2 through the Sec6 subunit (purple arrow) and that these changes were associated with corresponding movement of Exo70 and opening up of Sec6. In the red square Sro7 is depicted as a homo-oligomer interacting with and tethering vesicles through its own interactions with Sec4-GTP. Sro7-tethered vesicle clusters primed at the site of exocytosis are shown in the yeast bud tip (shown on left) as vesicles with pink (Sro7 oligomer) lines connecting them. Putative allosteric changes associated with Sro7- or Rho-binding are modeled using the exocyst CryoEM structure (PDB accession no. 5YFP) and the Exo70 gain-of-function negative stain EM structure (PDB accession no. 6VKL). (B) Speculative Model for structurally related exocyst activation mechanisms shared between yeasts and animal cells through the Exo84 subunit resulting in increased avidity of the complex for Rab GTPase on the vesicle.
With these two pathways in mind, exocyst complex activation is not a simple “off/on” or “closed/open” form of binary regulation. Rather, the complexity of its eight protein subunits allows multiple discrete signals mediated by different activators to result in the opening of specific regions of the complex. Indeed, while we have described two signaling pathways using yeast, another signaling pathway mediated by Ral-family GTPases might similarly regulate exocyst in a spatially controlled manner. Ral GTPases are well-established regulators of exocytosis in animal cells (Zhang et al., 2004; Wang et al., 2004). Interestingly, RalA in humans, which does not exist in yeast (or most fungi), functions through a direct interaction with the Exo84 PH domain, which is the site of the L365W gain-of-function allele, and this residue is conserved in mammals. Docking of the rat RalA-Exo84 crystal structure (Jin et al., 2005) onto the yeast exocyst shows multiple clashes between RalA, Sec5, and Sec10 (Fig. S4). It, therefore, seems likely that Ral-family GTPases function to introduce specific conformational changes in exocyst, perhaps leading to downstream engagement of Sec15 with Rab11. Deciphering these parallel activation mechanisms (Fig. 7B) will require further structural and biochemical studies. However, the present work suggests a number of straightforward approaches to address the conservation of this regulation between yeast and animal cells.
RalA interaction with Exo84 and potential implications for exocyst activation. (A) The cryo-EM structure of yeast exocyst (PDB accession no. 5YFP) is shown colored by subunit, with an inset showing a superimposition of the X-ray structure of human RalA-Exo84 (PDB accession no. 1ZC3). (B) Model for RalA-mediated exocyst activation. RalA binding disrupts inter-subunit contacts that allosterically modulate the conformation of exocyst.
RalA interaction with Exo84 and potential implications for exocyst activation. (A) The cryo-EM structure of yeast exocyst (PDB accession no. 5YFP) is shown colored by subunit, with an inset showing a superimposition of the X-ray structure of human RalA-Exo84 (PDB accession no. 1ZC3). (B) Model for RalA-mediated exocyst activation. RalA binding disrupts inter-subunit contacts that allosterically modulate the conformation of exocyst.
CATCHR family tethering complexes are characterized by long, flexible legs composed of extended alpha helical elements (Yu and Hughson, 2010). Recent negative-stain EM of CATCHR family complexes revealed surprising differences between the exocyst and its closest evolutionary relatives. The hetero-octameric COG complex and the hetero-tetramer GARP have high levels of flexibility and legs which extend outwards from the core (Chou et al., 2016; Ha et al., 2016). By contrast, cryo-EM of exocyst revealed a compact, closed structure where the “legs” interact more extensively with one another than seen with COG or GARP (Mei et al., 2018). Early EM of the mammalian exocyst reveals striking differences in complex organization depending on whether the complexes were fixed or unfixed (Hsu et al., 1998). Unfixed complexes resembled “legs” that radiated from a central point, while fixed complexes resembled a tight core, suggesting that mammalian exocyst is capable of adopting both “closed” and “open” conformations. Given the data presented in this paper and the recent negative-stain EM of an “activated” exocyst complex (Rossi et al., 2020), it seems likely that exocyst exhibits a similar degree of conformational flexibility as other CATCHR-family tethers. Understanding the regulatory cues that drive exocyst from its basal autoinhibited state is therefore a key to deciphering the molecular function of this important tether.
Previously the extended arrangement of helical rods within the exocyst complex was proposed to play an important role in vesicle tethering but the nature of that role was unclear (Munson and Novick, 2006). However, the present work suggests a likely new purpose for the extended nature of the exocyst complex. We have shown that each of two activation pathways can function without activating the other pathway—that is that each allosteric mechanism is in some way “insulated” from activation of the other. One way of insulating these mechanisms is by physical separation within the complex. By having the vSNARE (Snc) binding site on one end of the complex (“Sec6 cap”) and the Rab GTPase (Sec4) binding site (“Sec15 pole”) at the other end of the extended complex, the exocyst allows for considerable physical separation that insulates the outputs of these two distinct regulatory pathways.
Why use one tether as an activation signal for another tethering complex? For Rho GTPase activation, the rationale for the activation and output pathway is more straightforward. A signal (Rho) on the plasma membrane stimulates the exocyst to bind with higher avidity to an output on the vesicle (Snc2), effectively tethering the vesicle to the plasma membrane. On the other hand, Sro7 is a primarily cytoplasmic protein which is enriched at the bud tip, but not solely associated with the plasma membrane (Lehman et al., 1999). Sro7 interacts with Sec4 on post-Golgi vesicles, and this interaction is required for its role as a vesicle tether in vitro (Rossi et al., 2018). As Sro7 has only a single binding site for Sec4 (Watson et al., 2015), oligomerization of Sro7 is important for the tethering process to occur (Rossi et al., 2018). It is possible that Sro7 functions primarily as a vesicle:vesicle tether; clustering together pools of exocytosis-competent vesicles at the site of polarized growth (Rossi and Brennwald, 2011). Consequently, high local concentrations of Sro7 activate the exocyst, stimulating more fusion with the plasma membrane. Interestingly, this suggests that these two discrete, insulated activation pathways for the exocyst originate in two different locations—on the plasma membrane and in the cytoplasm. Meanwhile, their outputs target the same location—increased avidity for protein signals on the post-Golgi vesicle. This dual regulation may fine tune or reinforce cell polarity. While we have shown that these two activating signals can occur simultaneously, future work will investigate whether these two signals occur sequentially or in different contexts within polarized cell growth.
Materials and methods
Plasmids and strains
Protein A-tagged Sro7 constructs BB969 (full length) and BB1991 (aa52-1033) were generated by subcloning SRO7 into BB966 (pRS426 with an ADH1 promoter and a ProteinA-TEV tag) as a BamHI-HindIII fragment as previously described (Rossi et al., 2015). These plasmids were transformed into pep4Δ background yeast to generate BY2906 (wild type) and BY3021 (aa52-1033) and used to purify Sro7.
His-tagged proteins used in this study were generated as follows. BB2321 contains Exo84-NT (aa1-326) subcloned into pET-15b (NT-6xHis tag) vector as a NdeI-BglII fragment. BB2361, BB2478, BB2479, BB2480, and BB2360 contain Sro7-NT (aa1-90); Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 and Sro7-NT-A(4-8), respectively, subcloned into pET15b (NT-6x His tag) vector as NdeI-BamHI fragments. GST-tagged proteins used in this study were generated as follows. BB2305 contains Exo84-NT (aa1-326) subcloned into the BamHI-SalI sites of a modified pGEX-6P-1 vector containing a NT-GST and a CT-6xHis tag as a BglII-SalI fragment (BglII overhangs are compatible with BamHI overhang in the vector). BB1336 contains Sec9 subcloned into the BamHI-XhoI sites of a modified pGEX-6P-1 vector containing NT-GST and a CT-6xHis tag as a BamHI-XhoI fragment. BB2485 and BB2486 contain Sro7-NT (aa1-79) and Sro7-NT-A5,A6, respectively, subcloned into the BseRI-XhoI sites of pEX-C-GST as PvuI-XhoI fragments (BseRI in this vector creates a PvuI-compatible overhang).
To study the in vivo phenotypes of the Sro7 charge-to-alanine substitutions, constructs BB2469 (SRO7-A5,A6), BB2470 (SRO7-A7,A8), BB2471 (SRO7-A9,A10), and BB2472 (SRO7-A(4-8)) expressing Sro7 on a CEN HIS3 vector were generated by site-directed mutagenesis of BB2129 (SRO7). These plasmids were transformed into a sec15-1 strain to test suppression or sro7Δ/sro77Δ plasmid shuffle strain BY3105 as the only copy of SRO7. To study high copy suppression by SRO7, construct BB2450 (wild-type SRO7) and BB2541 (SRO7-A5,A6) were generated by subcloning SRO7 as a NotI-SalI fragment into pRS425 (2μ; LEU2). The high copy SRO7 plasmids as well as BB1701 (EXO70; CEN; LEU2) and BB1622 (EXO70-I114F; CEN; LEU2) were transformed into sec2-41 and cdc42-6.
To purify exocyst complexes, a linearized plasmid (BB2563) containing SEC8-TEV-3XMYC was integrated into the yeast chromosome to generate a single functional copy of tagged SEC8 in strains containing wild-type EXO84, EXO84-N12I, EXO84-L365W, EXO84-E626V, EXO70, or EXO70-I114F as the only copy of that subunit. The resulting strains were: BY3276 (exo84Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO84), BY3284 (exo84Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO84-N12I), BY3275 (exo84Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO84-L365W), BY3285 (exo84Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO84-E626V), BY3216 (exo70Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO70), and BY3220 (exo70Δ::KANR; SEC8-TEV3XMYC::URA3; ura3-52; leu2-3,112; + pRS315-EXO70-I114F).
Media, reagents, and genetic techniques
Yeast in this study were grown in either YPD media containing 1% bacto-yeast extract, 2% bacto-peptone, and 2% glucose (Thermo Fisher Scientific) or dropout media (0.67% yeast nitrogen base without amino acids, synthetic complete amino acid supplement minus appropriate amino acid, and 2% dextrose [US Biological]). Escherichia coli in this study were grown in Terrific Broth containing 4.7% bacto-TB and 1% glycerol (Thermo Fisher Scientific). Yeast transformations were performed using the lithium acetate method (Guthrie and Fink, 1991). For genetic analyses, at least six different transformants were analyzed per experiment.
Reagents used in this study were as follows: Sigma-Aldrich: GTPγS, GDP, Triton X-100, Sepharose CL-6B, sodium azide, sodium fluoride, sorbitol, dithiothreitol (DTT), β-mercaptoethanol, pepstatin A, leupeptin, aprotinin, and antipain. US Biological: Ampicillin, 5-fluoroorotic acid (5-FOA), 4-(2-aminoethyl) benzenesulfonyl fluoride, Bovine Serum Albumin, and Zymolase (100T). Bio-Rad: Tween-20 and Precision Plus Protein Standard. GE Healthcare: Glutathione Sepharose 4B and Protein A Sepharose. Invitrogen: AlexaFluor 680 goat anti-rabbit secondary antibody for Odyssey imaging system and lipid dye FM4-64. G Biosciences: Nickel Chelating Resin. Thermo Fisher Scientific: Pierce Glutathione Agarose.
The antibodies used in this study were either commercially purchased (Anti-HIS—mouse monoclonal [Sigma-Aldrich]) or custom-designed (Rossi et al., 2020) as noted below: Anti-Sro7 (rabbit polyclonal, CUMC173; Brennwald laboratory); Anti-Sec5 (rabbit polyclonal, CUMC148; Brennwald laboratory); Anti-Sec8 (rabbit polyclonal; Munson laboratory); Anti-Sec10 (rabbit polyclonal; Munson laboratory); Anti-Exo70 (rabbit polyclonal, CUMC118; Brennwald laboratory); Anti-Exo84 (rabbit polyclonal; Munson laboratory); Anti-Sec3 (rabbit polyclonal, CUMC147; Brennwald laboratory); Anti-Sec6 (rabbit polyclonal; Munson laboratory); and Anti-Sec15 (rabbit polyclonal, CUMC140; Brennwald laboratory).
Exocyst purification
Exocyst complex purification was performed as described previously (Rossi et al., 2020). In brief, yeast strains containing a chromosomal copy of C-terminally-tagged Sec8-3xMYC and plasmids expressing wild-type Exo84 or Exo70, dominant mutations of Exo84, or the dominant Exo70-I114F mutant as the sole source of Exo84 or Exo70, respectively, were grown overnight at 30°C in synthetic media to mid-log phase (OD599 1.5). Cells were shifted to YPD (2% glucose) for 2 h to a final OD599 of 3.0. To kill the cells, sodium azide and sodium fluoride were added to 20 mM final, then cells were spun at 6,700 × gmax in a JLA10.5 rotor for 6 min and washed with cold 10:20:20 buffer (10 mM Tris, pH 7.5, 20 mM NaN3, and 20 mM NaF) before freezing on dry ice. Approximately 50 g of cells were lysed in a bead beater in lysis buffer (20 mM Pipes, pH 6.8, 120 mM NaCl, 1 mM EDTA, and 1 mM DTT) with protease inhibitors (2 μg/ml leupeptin, 2 μg/ml aprotinin, 2 μg/ml antipain, 14 μg/ml pepstatin A, 2 mM 4-[2-aminoethyl] benzene-sulfonyl fluoride, and HCl). Lysates were cleared by centrifugation at 17,418 × gmax for 12 min at 4°C in a JA25.5 rotor, and the supernatant was removed and spun again at 50,000 × gmax for 30 min in a 41Ti rotor. The final supernatant concentration was adjusted to 30 mg/ml by Bradford assay before preclearing with Sepharose beads for 1 h at 4°C to reduce nonspecific binding. The lysate was spun for 5 min to remove the Sepharose beads and incubated overnight on ice with 9E10 monoclonal anti-myc antibody. Protein A Sepharose beads were added for 2 h at 4°C. The beads were washed three times in lysis buffer and then two times in cleavage buffer (20 mM Tris, pH 7.4, 140 mM NaCl, 0.1 mM EDTA, and 1 mM DTT) before cleaving in 1 ml of cleavage buffer with TEV enzyme for 4 h at 17°C. After removal of the Protein A Sepharose beads, the cleaved exocyst complexes were collected, aliquoted, and frozen at −80°C.
Sro7 purification
Full-length Sro7 and N terminally truncated Sro7 (aa52-1033) were purified as described previously (Rossi et al., 2015). Briefly, yeast pep4Δ strains containing a high copy plasmid expressing N-terminal Protein A-tagged Sro7, or Sro7 (aa52-1033) behind a ADH1 promoter were grown overnight at 30°C in synthetic media to an OD599 of 3.0. Cells were then shifted to YPD (2% glucose) for one doubling time before harvesting and washing with ice cold 10 mM Tris, pH 7.8, 20 mM sodium azide, and 20 mM sodium fluoride. Approximately 50 g of yeast were then frozen before glass bead lysis with a bead beater and ice cold 20 mM Tris, pH 7.8, 150 mM NaCl, 0.5% Tween-20, 1 mM DTT, and protease inhibitors (as listed in Exocyst purification). Five consecutive 1 min pulses of bead beating interrupted by five 2 min intervals resulted in a yeast lysate which was centrifuged at 17,400 × gmax for 10 min at 4°C in a JA25.5 rotor and then ultracentrifuged at 140,000 × gmax for 30 min at 4°C in a type 45Ti rotor. The final protein concentration was adjusted to ∼25 mg/ml before preclearing for 30 min at 4°C with Sepharose CL-6B beads followed by binding to IgG Sepharose for 2 h at 4°C. IgG beads were then washed five times with ice cold lysis buffer, three times with lysis buffer containing 400 mM sodium chloride, and three times with cleavage buffer containing 20 mM Tris, pH 7.8, 150 mM sodium chloride, 0.1 mM EDTA and 1 mM DTT before cleavage with TEV protease for 5 h at 17°C. The supernatant containing the purified protein was then aliquoted and frozen at −80°C.
His tagged Sro7 peptide purification
6xHis-tag purification was performed by binding the bacterial lysate to a HIS-Select Nickel Affinity Gel (G-BioSciences) in 50 mM Tris, pH 7.5, 350 mM sodium chloride, 10% glycerol, 0.5% Triton X-100 and 20 mM imidazole, followed by washing in buffer with no detergent before elution of the His tagged protein with 500 mM imidazole. The 6xHistidine eluates were then pooled and dialyzed into 20 mM Tris, pH 7.5, 150 mM sodium chloride and 10% glycerol. Multi-angle light scattering coupled with size-exclusion chromatography (SEC-MALS) analysis of the purified Sro7-NT peptides gave expected results for molecular weight (Sro7-NT: predicted 11,900 Da, observed 15,570 Da; Sro7-NT-A5,A6: predicted 11,848 Da, observed 13,540 Da) and polydispersity values (Mw/Mn; Sro7-NT: 1.036; Sro7-NT-A5,A6: 1.013 when analyzed at the concentration of 300 µM.
His tagged Exo84 peptide purification
6xHis-tag purification of Exo84 (aa1-326) was performed as above for the His tagged Sro7 peptides except the protein eluted from the HIS-Select Nickel Affinity Gel was dialyzed into 20 mM Tris, pH 7.5, 200 mM sodium chloride before loading on a resource S column for FPLC. Peak fractions were dialyzed into 20 mM Tris, pH 7.5, 200 mM sodium chloride, and 5% glycerol.
Generation of GFP-Sec4- and FM4-64-labeled vesicles and the vesicle:vesicle tethering assay
To obtain an enriched vesicle fraction from the sec6-4 mutant expressing GFP-Sec4 on a CEN plasmid, yeast (BY2745) was grown overnight in selective media at the permissive temperature of 25°C to an OD599 of 0.6 then shifted into YPD at 25°C for 1 h before placing at the restrictive temperature of 36°C for 2 h to accumulate vesicles. Sodium azide and sodium fluoride were added to 20 mM final and 350 ODs were centrifuged and washed with 10 ml of 10 mM Tris, pH 7.5, 20 mM NaN3. The cells were spheroplasted in 10 ml of spheroplast buffer (100 mM Tris, pH 7.5, 1.2 M sorbitol, 10 mM sodium azide, 21 mM β-mercaptoethanol, and 0.05 mg/ml Zymolyase 100T) for 30 min at 37°C. Spheroplasts were lysed in 4 ml of ice-cold lysis buffer (10 mM triethanolamine, pH 7.2 and 0.8 M sorbitol with protease inhibitors (as listed in Exocyst purification). The lysate was centrifuged for 4 min at 450 × gmax at 4°C to remove unbroken cells, then the remaining supernatant was centrifuged for 15 min at 30,000 × gmax at 4°C to preclear larger membranes. Approximately 7 ml of the supernatant was then labeled with FM4-64 (1 μg/ml) for 10 min on ice. The labeled supernatant split and layered over a 2 ml ice-cold sorbitol cushion (20% [wt/vol] sorbitol in 10 mM triethanolamine, pH 7.2) and centrifuged at 100,000 × gmax for 1 h at 4°C. The cushion was aspirated and the vesicle-enriched fraction was resuspended in 800 μl of lysis buffer and either placed on ice or frozen immediately and then used in the vesicle:vesicle tethering assay. For the tethering assay, vesicles were pre-incubated with 3 mM MgCl2 and 1 mM GTPγS for 60 min on ice before the addition of Sro7 (0.2 μM), Sro7-NT (1.5 μM), and exocyst complex (10 nM) for 30 min at 30°C. There are no notable differences in vesicle or assay preparation relative to the prior description of this assay (Rossi et al., 2020).
Establishing an automated detection and analysis of clustered vesicles
Using the in vitro tethering assay described above, we detected and quantified vesicle clustering in the presence of tethering factors such as Sro7 and the exocyst. We defined clusters as fluorescent puncta with a diameter >1 μm. All vesicles in this study are dually labeled with FM4-64 and GFP-Sec4. Although the same clusters are observed in both the FITC and TRITC channels, the TRITC (FM4-64) channel was used for quantification.
In previous applications of this assay, we manually quantified vesicle clustering either by counting the total number of clusters per μl of reaction (Rossi et al., 2015) or by measuring the fluorescence associated with all the clusters from a single field divided by the total fluorescence in the entire field (Rossi et al., 2020). To improve vesicle detection in this assay and reduce bias in identifying puncta, we have developed a protocol for automatically counting and measuring fluorescence of puncta in a field of view using commercial software. Regardless of quantitation method, all images are acquired using Nikon E600 microscope with a Nikon Planar 60× oil-immersion lens (NA 1.4) using a Photometrics CoolSNAP Dyno charge-coupled device camera.
We first analyzed our data as performed previously by manually measuring fluorescence for the dataset with representative images shown in Fig. 2 A (Fig. S1 A). Images were visualized and manually quantitated using Nikon NIS-Elements software (v4.6). To establish threshold parameters that could automatically detect puncta, we overlayed FITC and TRITC channel images in Imaris software v9.7 (Fig. S1 C). To count only puncta that were identified in both channels, we defined clusters as surfaces using the following parameters: Surface Grain Size = 0.250 um; Manual Threshold Value = 49.9966; Manual Threshold Value B = 521.941; and filtered for surfaces ≥1 μm in any direction (Fig. S1 D). Our automated counting is as specific as manually counting clusters, but generally identified more clusters per image. This resulted in an overall higher percentage of fluorescence in clusters in our automatically quantified datasets (Fig. S1 B, partially reproduced from Fig. 2 A).
Binding assays
Bindings between wild-type Sro7 or Sro7 (aa52-1033) and GST-Exo84 were conducted using 0.15 μM GST-Exo84 immobilized on beads and 0.5 μM soluble Sro7 in a 100 μl binding assay containing 10 mM Hepes KOH, 140 mM KCl, 0.5% Triton X-100, and 2 mM MgCl2 for 1 h at 4°C. Beads were washed four times in binding buffer and then boiled in sample buffer and processed by immunoblot analysis with anti-Sro7 antibody.
Binding between wild-type Sro7 or Sro7 (aa52-1033) and GST-Sec9 was conducted using 1.0 μM GST-Sec9 immobilized on beads and 1.0 μM soluble Sro7 in a 100 μl binding assay containing 10 mM Hepes KOH, 140 mM KCl, 0.5% Triton X-100, and 2 mM MgCl2 for 1 h at 4°C. Beads were washed four times in binding buffer and then boiled in sample buffer and processed by immunoblot analysis with anti-Sro7 antibody.
Binding between GST-Exo84 and His tagged Sro7-NT, Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 and Sro7-NT-A(4-8) peptides was conducted using 0.3 μM GST-Exo84 immobilized on beads and 0.5 μM Sro7-NT peptide in a 500 μl binding assay containing 10 mM Hepes KOH, 140 mM KCl, 0.5% Triton X-100, and 2 mM MgCl2 for 1 h at 4°C. Beads were washed four times in binding buffer and then boiled in sample buffer and processed by immunoblot analysis using anti-His antibody.
Binding between C-terminally tagged wild-type GST-Sro7-NT or mutant GST-Sro7-NT-A5,A6 and His-tagged Exo84 and were conducted using 0.125 μM Sro7-NT immobilized on beads and 0.375 μM soluble His-tagged Exo84 in a 500 μl binding assay containing 10 mM Hepes KOH, 140 mM KCl, 0.5% Triton X-100, and 2 mM MgCl2 for 1 h at 4°C. Soluble Exo84 in binding buffer was pre-incubated for 10 min with Sepharose beads on ice before adding to GST-Sro7-NT beads. Beads were washed four times in binding buffer and then heated in sample buffer before analysis by SDS-PAGE and Coomassie. For binding with purified exocyst, the binding conditions were modified as follows: GST-Sro7-NT and GST-NT-A5,A6 were used at 0.625 μM and purified exocyst was used at approximately 3 nM in 100 μl binding reaction containing 10 mM Hepes KOH, 140 mM KCl, 0.5% Triton X-100, 2 mM MgCl2 and 1 mM DTT. Soluble exocyst in binding buffer was pre-incubated for 10 min with Sepharose beads on ice before adding to GST-Sro7-NT beads. Beads were washed as above but samples were then analyzed by immunoblot analysis with Sec5, Sec8, Sec10, and Exo70 antibodies.
Binding between wild-type exocyst and GST-Sec4-GTPγS or GST-Sec4-GDP in the presence of wild-type Sro7-NT or mutant Sro7-NT-A5,A6 was conducted using 3 μM GST-Sec4-GTPγS or GST-Sec4-GDP immobilized on beads and ∼12.5 nM soluble exocyst and 4 μM soluble Sro7-NT or Sro7-NT-A5,A6 in a 113 μl binding assay with buffer containing 20 mM Tris, pH 7.5, 140 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, and 1 mM DTT for 1.5 h at 4°C. Beads were washed four times and then heated in sample buffer before immunoblot analysis using both Sec8 and Exo84 antibodies. Exocyst and Sro7-NT wild-type and mutant peptides were pre-incubated with Sepharose beads for 25 min on ice before adding to the GST-Sec4 nucleotide exchanged beads. Binding between wild-type exocyst and gain-of-function Exo70 or Exo84 mutants was conducted as described above except exocyst complexes were pre-incubated with Sepharose beads in the absence of Sro7-NT peptides. GST-Sec4-GTPγS or GST-Sec4-GDP beads were prepared as follows: Glutathione agarose beads bound to GST-Sec4 in 20 mM Tris, pH 7.5, 100 mM NaCl and 5 mM MgCl2 were washed once with 20 mM Tris, pH 7.5 and 100 mM NaCl at 25°C and then incubated in exchange buffer containing 20 mM Tris, pH 7.5, 100 mM NaCl, 5 mM EDTA, 1 mM DTT, and 100 μM nucleotide for 30 min at 25°C. MgCl2 was then added to a final concentration of 25 mM for 20 min at 25°C followed by 60 mins on ice. The exchanged beads were then used for the binding assay by washing the beads into binding buffer described above.
Binding between exocyst and GST-Snc2 or GST-Snc2-E79,E82 in the presence of wild-type Sro7-NT or mutant Sro7-NT-A5,A6 was conducted as described previously (Rossi et al., 2020) with the following modifications: GST-Snc2 immobilized on beads was at 2.5 μM and the exocyst (2 nM) was pre-incubated with wild-type Sro7-NT peptide (4 μM), Sro7-NT-A5,A6 peptide (4 μM), or buffer for 25 min on ice before binding.
Dominant suppressor screen for Exo84 gain-of-function mutants
The EXO84 mutants isolated in this study were generated using a GeneMorph II random mutagenesis kit (Stratagene). 50 ng of plasmid BB1690 (EXO84, CEN, URA3 plasmid) was used as a template for PCR reaction performed under conditions that produced an average of 4 mutations/kb (low frequency of mutagenesis) according to the manufacturer’s protocol. Oligonucleotide primers were designed to generate mutagenized PCR products containing the EXO84 open reading frame for gap repair into the BamHI and XbaI digested vector (BB1670) containing 613 base pairs of 5′ and 314 base pairs of 3′ sequence surrounding EXO84.
Fifty nanograms of digested EXO84 gap repair plasmid and 3 μl of error-prone PCR reaction were transformed into the sec2-41 strain (BY31, sec2-41; ura3-52). Transformants were plated on selective media and grown directly at the restrictive temperature of 33.5°C for 4 d. Colonies isolated from these plates were grown in broth at 25°C for plasmid recovery. The plasmids were retransformed into the sec2-41 mutant strain to test if the plasmids were responsible for the gain-of-function phenotype. From 20,000 transformants, we retested 127 plasmids and identified 22 plasmids that suppressed the sec2-41 temperature sensitivity. Sequence analysis of the 22 suppressors identified an average of nine coding sequence changes per dominant suppressing allele (Table S1). The frequency of mutations under our amplifying conditions was determined to be ∼4 base pair changes/kb based on sequencing of 10 plasmids isolated from colonies plated and grown at the permissive temperature of 25°C. These yeast had no selection for EXO84 gain-of-function alleles. Single or double residue changes in Exo84 were then generated by site-directed mutagenesis of BB1690 (EXO84, CEN, URA3 plasmid) or BB2396 (EXO84, CEN, LEU2 plasmid).
Chromosomal integration of EXO84 alleles
The EXO84 gain-of-function and wild-type EXO84 alleles along with a downstream NATR marker were integrated into the genomic EXO84 locus by transformation into an EXO84 deletion strain (BY3235, MATa, exo84Δ::KANR; ura3-52; leu2-3,112; pRS316-EXO84) using the method of Li et al. (2011). Transformants resistant to Nourseothricin (clonNAT) were scored for loss of G418 resistance and ability to grow on 5-FOA plates. The strains satisfying all these criteria were subjected to genomic DNA preparation, PCR amplification, and sequencing of the EXO84 locus to confirm their identities following integration. The resulting strains were crossed to a sec2-41 strain (BY3263, MATα, sec2-41;his3-Δ200) and subjected to meiotic analysis. The phenotypes of the dissected tetrads are shown in Fig. S2 and a side-by-side comparison of the effect of the different alleles on suppression of sec2-41 is shown in Fig. 4 C.
Online supplemental material
Figs. S1, S2, S3, and S4 detail establishment of the automated vesicle cluster detection method and tetrad analysis of cross between sec2-41 and strains containing integrated forms of EXO84 gain-of-function alleles. We also included two figures with structural modeling of the Exo84-GOF alleles within the complex as well a model incorporating RalA-GTPase binding into the yeast complex. Table S1 provides more details on the sequence analysis of the suppressor mutants originally identified in the screen.
Acknowledgments
We thank Benjamin Twara, Dr. Ashutosh Tripathy, and Dr. Beverly Errede (University of North Carolina) for help with various aspects of this work. We thank Dr. Mary Munson (University of Massachusetts Chan Medical School) for helpful conversations about this manuscript and for the gift of Sec8, Sec10, Exo84, and Sec6 antibodies. We would like to thank the anonymous reviewers for insightful and helpful comments on this work.
This work was supported by National Institutes of Health award R01-GM054712 to P. Brennwald. B.K. Miller was supported by an Institutional Research and Academic Career Development Awards (IRACDA) fellowship under K12-GM000678. Dr. Ashutosh Tripathy and the UNC Macromolecular Interactions Facility are supported by National Institutes of Health award P30CA016086. The funding agency had no role in the design or execution of this study, the analysis of the data, or the decision to publish the results.
Author contributions: B. Miller, G. Rossi, S. Hudson, D. Cully, R.W. Baker, and P. Brennwald designed, performed, and analyzed experiments. B.K. Miller, G. Rossi, R.W. Baker, and P. Brennwald wrote the paper. B. Miller, G. Rossi, R.W. Baker, and P. Brennwald edited the manuscript. R.W. Baker and B. Miller performed the structural modeling experiments. B. Miller and S. Hudson performed Exo84-GOF screen. G. Rossi and B. Miller performed in vitro tethering and in vitro binding experiments, D. Cully and P. Brennwald performed the Exo84-GOF allele integration, meiotic, and suppression analysis. P. Brennwald supervised the study.
References
Author notes
B.K. Miller and G. Rossi contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.
B.K. Miller’s current affiliation is Department of Biology, Hofstra University, Hempstead, NY, USA.


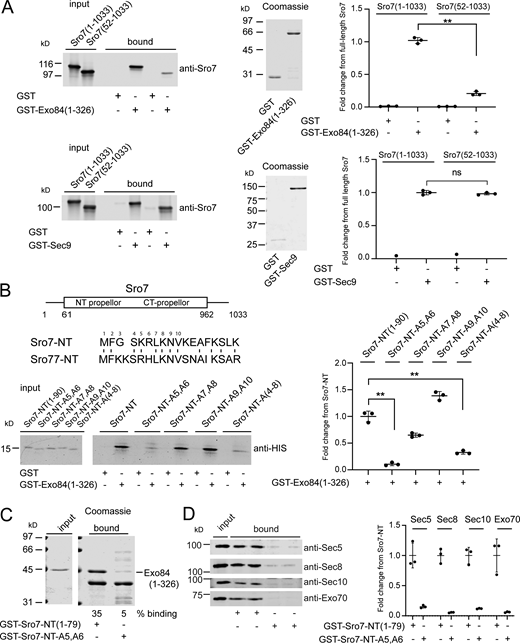

![Sro7-NT peptide but not the mutant Sro7-NT-A5,A6 peptide stimulates exocyst-mediated in vitro vesicle tethering. (A) Vesicles labeled with the lipid dye FM4-64 were isolated from a sec6-4 mutant strain expressing GFP-Sec4 and used in an in vitro vesicle-vesicle tethering assay with suboptimal concentrations of purified Sro7, identical amounts of wild-type exocyst complex, and purified Sro7-NT (aa 1–90) or Sro7-NT-5A,6A peptides. Vesicle-vesicle tethered clusters were automatically detected (see Materials and methods for details) and quantified as the percentage of fluorescence seen in clusters over the total fluorescence of the image using the TRITC channel. Error bars represent SD obtained from counting images at 60× magnification. P values were obtained using a two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; *** P < 0.001; ns = no significant difference. Scale bar, 5 μm. (B) Labeled vesicles were incubated with purified exocyst and Sro7 or exocyst and wild-type Sro7-NT peptide or mutant Sro7-NT-5A,6A peptide as described above. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. (C) Labeled vesicles were incubated with exocyst and either wild-type or mutant Sro7-NT peptides [Sro7-NT; Sro7-NT-A5,A6; Sro7-NT-A7,A8; Sro7-NT-A9,A10 or Sro7-NT-A(4-8)]. Clusters were automatically detected and quantified as a percentage of total fluorescence with significance annotated as above. Scale bar, 5 μm. Coomassie of the peptides used is shown to the right. (D) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into a sec15-1 strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. (E) Wild-type SRO7 and the Sro7 charge-to-alanine mutants SRO7-A5,A6, SRO7-A7,A8, SRO7-A9,A10, and SRO7-A(4-8), and vector only (CEN) were transformed into an sro7Δ;sro77Δ strain. The growth of three independent transformants is shown for each sro7 mutant, under permissive and restrictive conditions. Source data are available for this figure: SourceData F2.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/3/10.1083_jcb.202206108/1/m_jcb_202206108_fig2.png?Expires=1773798547&Signature=gE3IyKrAicDm20DiLt1LEh~8F7SU1qXled2AhAX6162Vkl4YpCFytJx-Vs7waphVJ2GBWbUMXJteDb6O-ozxwwe11~vKMIZHm1rRfUgx-T8Qj8a7rW9gAIHez8pePB5Abw~MThGXcdP-QkfOvPuK7z~tUEbS5rMNBnB9URJGXHL0MeoIYsZnOkOMXTNTt4lE7XF3LmnhwfbefYurhAFlKWIzWIVNidhEYt5mKbOcorsBXrHRQM8vigcjeO~CjTO0FJeAw04MgpsTmcQoNUKsbvfeuL8XW01UaYP~u6e5kq3stvdHj29dT7VJ5e7M9Bo8poxWfmmTWI7zU3ohZaRRMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)