Cell growth is required for cell cycle progression. The amount of growth required for cell cycle progression is reduced in poor nutrients, which leads to a reduction in cell size. In budding yeast, nutrients can influence cell size by modulating the extent of bud growth, which occurs predominantly in mitosis. However, the mechanisms are unknown. Here, we used mass spectrometry to identify proteins that modulate bud growth in response to nutrient availability. This led to the discovery that nutrients regulate numerous components of the mitotic exit network (MEN), which controls exit from mitosis. A key component of the MEN undergoes gradual multisite phosphorylation during bud growth that is dependent upon bud growth and correlated with the extent of growth. Furthermore, activation of the MEN is sufficient to override a growth requirement for mitotic exit. The data suggest a model in which the MEN ensures that mitotic exit occurs only when an appropriate amount of bud growth has occurred.
Introduction
In all orders of life, cell cycle progression is dependent upon cell growth, which ensures that dividing cells attain sufficient volume and mass so that cell division produces viable cells of an appropriate size (Jorgensen and Tyers, 2004; Turner et al., 2012; Ginzberg et al., 2015). The threshold amount of growth required for cell cycle progression is reduced when cells are growing under poor nutrient conditions, which leads to a large reduction in cell size (Kellogg and Levin, 2022). The reduced growth threshold in poor nutrients likely confers a competitive advantage, as it allows cells to proliferate more rapidly when nutrients are limited. The mechanisms that set and modulate the amount of growth required for cell cycle progression are largely unknown.
In budding yeast, most cell growth takes place during bud growth and most bud growth takes place during mitosis (Leitao and Kellogg, 2017). Bud growth is required for progression through mitosis. Moreover, the rate, duration, and extent of bud growth during mitosis are strongly influenced by nutrient availability (Leitao and Kellogg, 2017). Thus, the rate of bud growth is reduced in poor nutrients and the duration of bud growth is extended, presumably to allow more time for bud growth. Furthermore, the threshold amount of bud growth required for the completion of mitosis is reduced, leading to the birth of very small daughter cells. These observations suggest the existence of nutrient and growth-sensing mechanisms that modulate the duration and extent of bud growth to ensure that daughter cells are born at an appropriate size.
As the bud grows during mitosis, approximately equal volumes are added during metaphase and anaphase (Leitao and Kellogg, 2017). Nutrient availability modulates the duration and extent of bud growth in both intervals, and recent work suggests that the two intervals are controlled by different mechanisms (Leitao et al., 2019; Jasani et al., 2020). The metaphase interval is influenced by a pair of related kinases called Gin4 and Hsl1. Both kinases are required for normal control of bud growth and undergo gradual hyperphosphorylation and activation that appear to be dependent upon bud growth and proportional to the extent of bud growth, which suggests that they play roles in measuring growth (Jasani et al., 2020). Once they have been fully activated, Gin4 and Hsl1 promote mitotic progression by inhibiting Swe1, the budding yeast homolog of the conserved Wee1 kinase that directly phosphorylates and inhibits mitotic Cdk1/cyclin complexes (Ma et al., 1996; Longtine et al., 2000). In both mammalian cells and budding yeast, it is thought that Wee1 keeps mitotic Cdk1 activity low during metaphase and that inhibition of Wee1 leads to full activation of Cdk1 that drives the metaphase to anaphase transition (Deibler and Kirschner, 2010; Harvey et al., 2011). Although Gin4 and Hsl1 strongly influence bud growth in metaphase they do not influence the duration and extent of growth during anaphase (Leitao et al., 2019; Jasani et al., 2020). These data suggest that growth in anaphase is regulated by unknown mechanisms that are distinct from those that work in metaphase.
Here, we used proteome-wide mass spectrometry to search for proteins that influence the duration and extent of bud growth in late mitosis. The results point to the mitotic exit network (MEN), an essential signaling network that includes highly conserved proteins, as a likely target of signals that link mitotic exit to bud growth. Previous studies suggested that the MEN links mitotic exit to mitotic spindle orientation and elongation (Bardin et al., 2000; Pereira et al., 2000; Campbell et al., 2020). Our studies establish for the first time that key components of the MEN also respond to signals associated with bud growth and nutrient availability.
Results
Nutrient-dependent signals modulate the phosphorylation of multiple proteins in the MEN
Changes in nutrient availability immediately influence cell cycle progression and the threshold amount of growth required for cell cycle progression (Fantes and Nurse, 1977; Leitao and Kellogg, 2017). Therefore, to identify signals that could play a role in modulating the duration and extent of bud growth in mitosis, we used proteome-wide mass spectrometry to search for proteins that undergo large changes in phosphorylation in response to a rapid shift from rich to poor carbon during late mitosis. We reasoned that these could include proteins that play roles in setting the threshold amount of bud growth required for cell cycle progression.
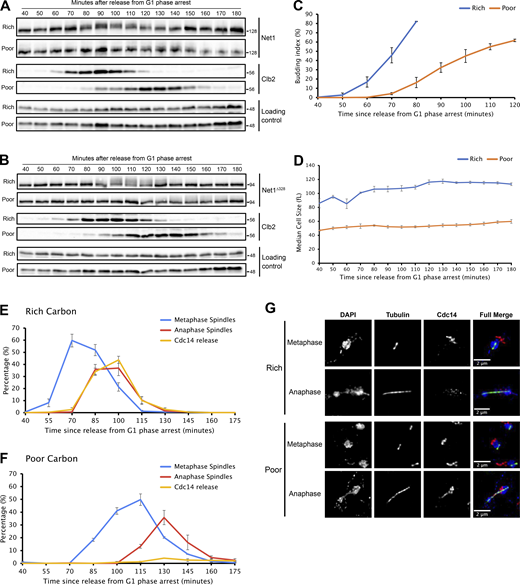
Cells growing in a medium containing a rich carbon source (YP+2% dextrose) were released from a G1 phase arrest and rapidly shifted to a poor carbon medium (YP+2% glycerol/2% ethanol) at 90 min, when the mitotic cyclin Clb2 reached peak levels (Fig. 1). Proteins were isolated from the shifted and unshifted control cells 10 min after the shift, and proteolytic peptides were analyzed by quantitative proteome-wide mass spectrometry to search for changes in phosphorylation. The ratio of phosphorylated to unphosphorylated peptides in shifted versus unshifted control cells was log2 transformed. Thus, a negative log2 ratio indicates a loss of phosphorylation in response to a shift to poor carbon, whereas a positive log2 ratio indicates a gain of phosphorylation. A total of three biological replicates were analyzed, which allowed the calculation of average log2 ratios and standard deviation. The complete data set is shown in Table S1.
Analysis of the effects of a shift from rich to poor carbon during metaphase. Cells were grown overnight in YPD medium and arrested in G1 phase with alpha factor. The cells were then released from the arrest into fresh YPD at 25°C. At the point indicated by the arrow, half of the culture was shifted to poor carbon (bottom panel). Samples were collected at the indicated time points to analyze Clb2 levels by western blot. Samples for mass spectrometry analysis were collected 10 min after the shift to poor carbon. Source data are available for this figure: SourceData F1.
Analysis of the effects of a shift from rich to poor carbon during metaphase. Cells were grown overnight in YPD medium and arrested in G1 phase with alpha factor. The cells were then released from the arrest into fresh YPD at 25°C. At the point indicated by the arrow, half of the culture was shifted to poor carbon (bottom panel). Samples were collected at the indicated time points to analyze Clb2 levels by western blot. Samples for mass spectrometry analysis were collected 10 min after the shift to poor carbon. Source data are available for this figure: SourceData F1.
We searched the mass spectrometry data for proteins involved in mitotic progression and cell growth, which are summarized in Table 1. A log2 ratio of ±1.32, corresponding to a 2.5-fold change in phosphorylation occupancy, was used as an arbitrary cutoff to define substantial changes in phosphorylation. Numerous components of a signaling network that regulates exit from mitosis, referred to as the mitotic exit network (MEN), were amongst the proteins that showed the largest loss of phosphorylation after a shift from rich to poor nutrients in mitosis. These proteins include Net1, Cdc14, Lte1, Cdc15, Bfa1, Nud1, and Zds1. Previous work suggested that the MEN triggers mitotic exit by initiating the destruction of the mitotic cyclin Clb2 (Visintin et al., 1998, 1999; Jaspersen et al., 1999; Shou et al., 1999; Stegmeier et al., 2002). It is thought that the MEN initiates mitotic exit in response to signals generated by proper orientation of the mitotic spindle in the daughter bud (Bardin et al., 2000; Pereira et al., 2000). Here, the discovery that numerous components of the MEN show large changes in phosphorylation in response to a shift to poor nutrients indicates that the MEN is strongly regulated by nutrient-dependent signals. Since the MEN can influence the duration of late mitosis, the data further suggested that the MEN could be the target of nutrient-dependent signals that influence the duration and extent of bud growth in late mitosis.
A summary of key peptides that undergo substantial changes in phosphorylation in response to a shift to poor carbon during mitosis
| Cellular Process . | Site Position . | Sequence . | A-score . | # of peptides . | Log2(Poor/Rich) . |
|---|---|---|---|---|---|
| MEN | |||||
| NET1 | |||||
| 212 | VST#PLAR | 0 | 3 | −4.497 | |
| 259 | ISS#GIDAGK | 24.43 | 6 | −4.409 | |
| 280 | S#ATVDPDK | 30.83 | 2 | −4.343 | |
| 362 | ITS#GM*LK | 24.43 | 3 | −4.259 | |
| 388 | EGPSSPAS#ILPAK | 5.08 | 3 | −3.419 | |
| 270 | SS#IVEEDIVSR | 9.34 | 1 | −3.180 | |
| 447 | S#QSSIADNNGS#PVK | 100.02 | 3 | −3.059 | |
| 301;304 | LLSGT#PIM*STMT#PNR | 4.08 | 3 | −2.740 | |
| 166 | LNNGS#PQSVQPQQQIPSSSGVLR | 15.01 | 2 | −1.640 | |
| 1164 | S#ASAALGK | 30.83 | 2 | −1.331 | |
| 48 | TNM*AQSAGDAS#LQYANLR | 33.66 | 2 | −1.315 | |
| CDC14 | |||||
| 537 | RTTS#AAGGIR | 6.59 | 1 | −2.903 | |
| 494;497 | KNDISS#ASS#SR | 17.01;9.34 | 1 | −1.590 | |
| 429 | SSAVPQTS#PGQPR | 30.83 | 5 | −1.533 | |
| LTE1 | |||||
| 789;793 | VQS#IAIT#PTK | 110.63 | 3 | −2.413 | |
| 718 | ST#IDGLEK | 24.44 | 1 | −2.220 | |
| 779 | QAVVRPAS#GR | 1000 | 1 | −1.857 | |
| 799 | ELS#IVDPEQNK | 1000 | 2 | −1.481 | |
| CDC15 | |||||
| 940 | VSSVT#AAIGSS#PTK | 2.81 | 1 | −1.668 | |
| BFA1 | |||||
| 317 | HCS#NQNVQLNGPAK | 1000 | 1 | −1.614 | |
| NUD1 | |||||
| 352 | APS#ILDK | 1000 | 1 | −1.469 | |
| TORC2 Signaling | |||||
| AVO2 | |||||
| 305 | VNS#INVK | 1000 | 1 | −3.872 | |
| 273 | SS#ITNPVFNPR | 17.01 | 2 | −2.293 | |
| 330;333 | HSAT#PTS#PHNNIALINR | 4.08;6.59 | 1 | −2.061 | |
| 310 | T#PVGVS#PK | 1000 | 1 | −1.432 | |
| 310;315 | T#PVGVS#PK | 1000;1000 | 2 | −1.387 | |
| TAP42 | |||||
| 251;260 | ESNDDDS#TGFT#DK | 85.26;13.38 | 1 | −3.872 | |
| GIN4 | |||||
| 471 | RAS#VINVEK | 1000 | 3 | −3.100 | |
| 382;389 | RQS#ISSVS#VSPSK | 20.17 | 1 | −2.569 | |
| 805 | HFS#ESNK | 30.83 | 2 | −2.338 | |
| 639 | SIS#APM*ENEEK | 0 | 1 | −1.648 | |
| 384 | RQS#IS#SVSVSPSK | 13.21 | 1 | −1.594 | |
| YPK1 | |||||
| 57 | GT#INPSNSSVVPVR | 59.30 | 5 | −2.807 | |
| 57;64 | GT#INPS#NSSVVPVR | 59.30;17.01 | 4 | −2.017 | |
| HSL1 | |||||
| 899 | NIS#QPVNSK | 9.30 | 1 | −2.790 | |
| 1210 | ETT#EEILSK | 9.34 | 2 | −2.082 | |
| 641 | AIHAS#PSTK | 33.66 | 1 | −1.893 | |
| 629;631;632 | SPS#RY#S#LSR | 30.83;24.24;17.01 | 1 | −1.848 | |
| BIT61 | |||||
| 164 | AS#GFFNR | 1000 | 1 | −2.721 | |
| TSC11 | |||||
| 19 | S#VTNTTPLLTPR | 48.87 | 1 | −2.372 | |
| 24;28 | SVTNT#TPLLT#PR | 9.34;65.44 | 1 | −2.176 | |
| 50;52 | NITSSS#PS#TITNESSK | 24.43;9.34 | 1 | −1.936 | |
| 251 | NSQQNLNRNS#TVNSR | 6.59 | 1 | −1.742 | |
| 19;28;32 | S#VTNTTPLLT#PRHS#R | 48.87;65.44;63.08 | 1 | −1.586 | |
| AVO1 | |||||
| 537 | DTVIS#GK | 33.66 | 1 | −2.332 | |
| 543 | EPTS#LTSSNR | 9.34 | 2 | −1.872 | |
| 588 | VSDS#VLHR | 6.59 | 3 | −1.704 | |
| 326 | SHFPTS#QK | 0 | 1 | −1.632 | |
| MSS4 | |||||
| 341 | RSES#ATAEIK | 20.17 | 3 | −2.039 | |
| 238 | HS#QILPM*DDSDVIK | 88.82 | 2 | −2.025 | |
| PKH2 | |||||
| 1005 | THSQS#PSIS#K | 11.01 | 4 | −1.585 | |
| PRK1 | |||||
| 488 | LS#PTIT#SK | 30.83 | 1 | −1.506 | |
| SLM2 | |||||
| 7;11 | NS#ARAS#LDLR | 1000;1000 | 1 | −1.466 | |
| Secretory Pathway | |||||
| SEC16 | |||||
| 1961 | AS#TNQYR | 24.44 | 2 | −3.849 | |
| 759 | GVS#NAS#VGSSASFGAR | 62.66 | 3 | −3.163 | |
| 2144 | TSPSPTGPNPNNSPSPS#S#PISR | 13.38 | 3 | −2.022 | |
| 759;762 | GVS#NAS#VGSSASFGAR | 62.66; 87.26 | 5 | −1.992 | |
| 806 | YAPVS#PTVQQK | 6.59 | 5 | −1.882 | |
| 766;768 | GVSNASVGSS#AS#FGAR | 4.08; 17.01 | 1 | −1.407 | |
| SEC31 | |||||
| 980 | APSS#VSM*VS#PPPLHK | 71.34 | 1 | −1.611 | |
| 988;992 | NS#RVPS#LVATSESPR | 113.18; 99.28 | 6 | −1.514 | |
| 988 | NS#RVPS#LVAT#SESPR | 113.18 | 3 | −1.475 | |
| Ribosome Biogenesis | |||||
| DOT6 | |||||
| 322;326;331 | RSS#FNVS#SNNT#SR | 24.44; 0; 9.34 | 1 | −4.104 | |
| 322 | RSS#FNVSSNNTSR | 24.44 | 2 | −3.133 | |
| 247 | SNS#HSFTNSLNQDPIVR | 11.01 | 2 | −1.687 | |
| 322;331 | RSS#FNVS#SNNTSR | 24.44; 9.34 | 4 | −1.424 | |
| TOD6 | |||||
| 300;304 | NSHS#VISS#R | 37.54; 9.34 | 1 | −3.938 | |
| 298;304 | NS#HSVIS#SR | 13.38; 9.34 | 1 | −3.484 | |
| 318;329 | RSS#FNSHAPTEPIS#R | 0; 12.43 | 1 | −2.103 | |
| 298 | NS#HSVIS#SR | 13.38 | 1 | −1.956 | |
| 298;300 | NS#HSVISS#R | 13.40; 37.54 | 1 | −1.382 | |
| STB3 | |||||
| 337;341 | LTATS#EPTS#R | 13.38; 17.25 | 4 | −3.382 | |
| 341 | LTATS#EPTS#R | 17.25 | 2 | −3.144 | |
| 336;341 | LTAT#SEPT#SRR | 24.44; 17.25 | 6 | −2.754 | |
| 336 | LTAT#SEPT#SRR | 24.44 | 1 | −1.767 | |
| IFH1 | |||||
| 1041 | RQS#MVEAAAENLR | 1000 | −2.340 | ||
| ENP1 | |||||
| 5;6 | M*ARAS#S#TK | 4.08; 6.59 | 1 | −1.504 | |
| SCH9 | |||||
| 288 | S#S#SQLDQLNSCSSVTDPSK | 5.75 | 4 | −1.397 |
| Cellular Process . | Site Position . | Sequence . | A-score . | # of peptides . | Log2(Poor/Rich) . |
|---|---|---|---|---|---|
| MEN | |||||
| NET1 | |||||
| 212 | VST#PLAR | 0 | 3 | −4.497 | |
| 259 | ISS#GIDAGK | 24.43 | 6 | −4.409 | |
| 280 | S#ATVDPDK | 30.83 | 2 | −4.343 | |
| 362 | ITS#GM*LK | 24.43 | 3 | −4.259 | |
| 388 | EGPSSPAS#ILPAK | 5.08 | 3 | −3.419 | |
| 270 | SS#IVEEDIVSR | 9.34 | 1 | −3.180 | |
| 447 | S#QSSIADNNGS#PVK | 100.02 | 3 | −3.059 | |
| 301;304 | LLSGT#PIM*STMT#PNR | 4.08 | 3 | −2.740 | |
| 166 | LNNGS#PQSVQPQQQIPSSSGVLR | 15.01 | 2 | −1.640 | |
| 1164 | S#ASAALGK | 30.83 | 2 | −1.331 | |
| 48 | TNM*AQSAGDAS#LQYANLR | 33.66 | 2 | −1.315 | |
| CDC14 | |||||
| 537 | RTTS#AAGGIR | 6.59 | 1 | −2.903 | |
| 494;497 | KNDISS#ASS#SR | 17.01;9.34 | 1 | −1.590 | |
| 429 | SSAVPQTS#PGQPR | 30.83 | 5 | −1.533 | |
| LTE1 | |||||
| 789;793 | VQS#IAIT#PTK | 110.63 | 3 | −2.413 | |
| 718 | ST#IDGLEK | 24.44 | 1 | −2.220 | |
| 779 | QAVVRPAS#GR | 1000 | 1 | −1.857 | |
| 799 | ELS#IVDPEQNK | 1000 | 2 | −1.481 | |
| CDC15 | |||||
| 940 | VSSVT#AAIGSS#PTK | 2.81 | 1 | −1.668 | |
| BFA1 | |||||
| 317 | HCS#NQNVQLNGPAK | 1000 | 1 | −1.614 | |
| NUD1 | |||||
| 352 | APS#ILDK | 1000 | 1 | −1.469 | |
| TORC2 Signaling | |||||
| AVO2 | |||||
| 305 | VNS#INVK | 1000 | 1 | −3.872 | |
| 273 | SS#ITNPVFNPR | 17.01 | 2 | −2.293 | |
| 330;333 | HSAT#PTS#PHNNIALINR | 4.08;6.59 | 1 | −2.061 | |
| 310 | T#PVGVS#PK | 1000 | 1 | −1.432 | |
| 310;315 | T#PVGVS#PK | 1000;1000 | 2 | −1.387 | |
| TAP42 | |||||
| 251;260 | ESNDDDS#TGFT#DK | 85.26;13.38 | 1 | −3.872 | |
| GIN4 | |||||
| 471 | RAS#VINVEK | 1000 | 3 | −3.100 | |
| 382;389 | RQS#ISSVS#VSPSK | 20.17 | 1 | −2.569 | |
| 805 | HFS#ESNK | 30.83 | 2 | −2.338 | |
| 639 | SIS#APM*ENEEK | 0 | 1 | −1.648 | |
| 384 | RQS#IS#SVSVSPSK | 13.21 | 1 | −1.594 | |
| YPK1 | |||||
| 57 | GT#INPSNSSVVPVR | 59.30 | 5 | −2.807 | |
| 57;64 | GT#INPS#NSSVVPVR | 59.30;17.01 | 4 | −2.017 | |
| HSL1 | |||||
| 899 | NIS#QPVNSK | 9.30 | 1 | −2.790 | |
| 1210 | ETT#EEILSK | 9.34 | 2 | −2.082 | |
| 641 | AIHAS#PSTK | 33.66 | 1 | −1.893 | |
| 629;631;632 | SPS#RY#S#LSR | 30.83;24.24;17.01 | 1 | −1.848 | |
| BIT61 | |||||
| 164 | AS#GFFNR | 1000 | 1 | −2.721 | |
| TSC11 | |||||
| 19 | S#VTNTTPLLTPR | 48.87 | 1 | −2.372 | |
| 24;28 | SVTNT#TPLLT#PR | 9.34;65.44 | 1 | −2.176 | |
| 50;52 | NITSSS#PS#TITNESSK | 24.43;9.34 | 1 | −1.936 | |
| 251 | NSQQNLNRNS#TVNSR | 6.59 | 1 | −1.742 | |
| 19;28;32 | S#VTNTTPLLT#PRHS#R | 48.87;65.44;63.08 | 1 | −1.586 | |
| AVO1 | |||||
| 537 | DTVIS#GK | 33.66 | 1 | −2.332 | |
| 543 | EPTS#LTSSNR | 9.34 | 2 | −1.872 | |
| 588 | VSDS#VLHR | 6.59 | 3 | −1.704 | |
| 326 | SHFPTS#QK | 0 | 1 | −1.632 | |
| MSS4 | |||||
| 341 | RSES#ATAEIK | 20.17 | 3 | −2.039 | |
| 238 | HS#QILPM*DDSDVIK | 88.82 | 2 | −2.025 | |
| PKH2 | |||||
| 1005 | THSQS#PSIS#K | 11.01 | 4 | −1.585 | |
| PRK1 | |||||
| 488 | LS#PTIT#SK | 30.83 | 1 | −1.506 | |
| SLM2 | |||||
| 7;11 | NS#ARAS#LDLR | 1000;1000 | 1 | −1.466 | |
| Secretory Pathway | |||||
| SEC16 | |||||
| 1961 | AS#TNQYR | 24.44 | 2 | −3.849 | |
| 759 | GVS#NAS#VGSSASFGAR | 62.66 | 3 | −3.163 | |
| 2144 | TSPSPTGPNPNNSPSPS#S#PISR | 13.38 | 3 | −2.022 | |
| 759;762 | GVS#NAS#VGSSASFGAR | 62.66; 87.26 | 5 | −1.992 | |
| 806 | YAPVS#PTVQQK | 6.59 | 5 | −1.882 | |
| 766;768 | GVSNASVGSS#AS#FGAR | 4.08; 17.01 | 1 | −1.407 | |
| SEC31 | |||||
| 980 | APSS#VSM*VS#PPPLHK | 71.34 | 1 | −1.611 | |
| 988;992 | NS#RVPS#LVATSESPR | 113.18; 99.28 | 6 | −1.514 | |
| 988 | NS#RVPS#LVAT#SESPR | 113.18 | 3 | −1.475 | |
| Ribosome Biogenesis | |||||
| DOT6 | |||||
| 322;326;331 | RSS#FNVS#SNNT#SR | 24.44; 0; 9.34 | 1 | −4.104 | |
| 322 | RSS#FNVSSNNTSR | 24.44 | 2 | −3.133 | |
| 247 | SNS#HSFTNSLNQDPIVR | 11.01 | 2 | −1.687 | |
| 322;331 | RSS#FNVS#SNNTSR | 24.44; 9.34 | 4 | −1.424 | |
| TOD6 | |||||
| 300;304 | NSHS#VISS#R | 37.54; 9.34 | 1 | −3.938 | |
| 298;304 | NS#HSVIS#SR | 13.38; 9.34 | 1 | −3.484 | |
| 318;329 | RSS#FNSHAPTEPIS#R | 0; 12.43 | 1 | −2.103 | |
| 298 | NS#HSVIS#SR | 13.38 | 1 | −1.956 | |
| 298;300 | NS#HSVISS#R | 13.40; 37.54 | 1 | −1.382 | |
| STB3 | |||||
| 337;341 | LTATS#EPTS#R | 13.38; 17.25 | 4 | −3.382 | |
| 341 | LTATS#EPTS#R | 17.25 | 2 | −3.144 | |
| 336;341 | LTAT#SEPT#SRR | 24.44; 17.25 | 6 | −2.754 | |
| 336 | LTAT#SEPT#SRR | 24.44 | 1 | −1.767 | |
| IFH1 | |||||
| 1041 | RQS#MVEAAAENLR | 1000 | −2.340 | ||
| ENP1 | |||||
| 5;6 | M*ARAS#S#TK | 4.08; 6.59 | 1 | −1.504 | |
| SCH9 | |||||
| 288 | S#S#SQLDQLNSCSSVTDPSK | 5.75 | 4 | −1.397 |
The A-score indicates the likelihood a phosphorylation site is correctly localized within a peptide. A given phosphorylation site is correctly assigned with >95% confidence when the A-score is >13. An A-score of 0 indicates that the phosphorylated residue within the peptide sequence cannot be precisely determined. An asterisk (*) represents oxidation of the preceding methionine, an artifact of the sample preparation. A hashtag indicates that the preceding amino acid is likely the relevant phosphorylation site.
The mass spectrometry analysis also identified multiple components of TOR Complex 2 (TORC2) and its surrounding signaling network (Table 1). These proteins include Avo2, Tsc11, Ypk1, Mss4, Pkh2, Gin4, Hsl1, Rts1, and Prk1. The identification of components of the TORC2 network is consistent with previous studies, which found that TORC2 signaling is required for nutrient modulation of cell size and growth rate (Lucena et al., 2018). As an example, the loss of Rts1, a regulatory subunit for PP2A, causes misregulation of TORC2 signaling, as well as a complete loss of the proportional relationship between cell size and growth rate during bud growth (Lucena et al., 2018; Leitao et al., 2019).
Finally, the analysis identified several core components of the COPII complex that are required for ER to Golgi transport (Sec16 and Sec31) as well as a key regulator of ribosome biogenesis (Sch9) (Table 1). Since a shift to poor nutrients leads to a rapid reduction in growth rate, these findings suggest that poor nutrients trigger signals that rapidly reduce the rates of both ribosome biogenesis and membrane growth to match reduced biosynthetic rates in poor nutrients.
Net1 hyperphosphorylation occurs before mitotic exit
The discovery that multiple components of the MEN are regulated in response to changes in carbon source suggested that the MEN could play a role in linking mitotic exit to cell growth and nutrient availability. In this context, the discovery that Net1 phosphorylation is strongly modulated by carbon source was particularly interesting. Net1 showed large decreases in phosphorylation at 11 sites. Net1 was also identified as a top hit in a mass spectrometry screen for proteins that underwent large changes in phosphorylation in response to an arrest of plasma membrane growth during early stages of bud growth (Clarke et al., 2017). Net1 plays a critical role in the initiation of mitotic exit and also controls ribosome biogenesis, which is a major facet of cell growth (Shou et al., 1999, 2001; Straight et al., 1999; Visintin et al., 1999; Shou and Deshaies, 2002; Hannig et al., 2019). Net1 is localized in the nucleolus early in the cell cycle, where it binds and inhibits Cdc14 (Shou et al., 1999; Visintin et al., 1999; Traverso et al., 2001). Hyperphosphorylation of Net1 is thought to be a critical step that helps release Cdc14 from Net1 so that it can leave the nucleolus to initiate mitotic exit (Shou et al., 1999; Yoshida and Toh-e, 2002; Visintin et al., 2003).
In previous studies, we found evidence that the events of bud growth generate signals that are dependent upon growth and proportional to growth (Anastasia et al., 2012; Jasani et al., 2020). We, therefore, considered a working hypothesis in which the events of bud growth generate a signal that contributes to Net1 phosphorylation and helps trigger activation of the MEN, while nutrient-dependent signals modulate the strength of the growth-dependent signal needed to initiate mitotic exit.
Since hyperphosphorylation of Net1 is thought to be a key early step in the initiation of mitotic exit, we carried out new experiments to determine whether Net1 phosphorylation is influenced by nutrients or growth, which would begin to test the hypothesis that the MEN links mitotic exit to cell growth. Previous studies reported that hyperphosphorylation of Net1 can be detected via electrophoretic mobility shifts; however, we found that the large size of Net1 made reproducible detection of these shifts difficult. We therefore developed an additional tool for the detection of Net1 phosphorylation. We reasoned that the expression of a truncated version of Net1 could provide a readout of phosphorylation while also allowing easier detection due to the reduced size of the truncated Net1. We analyzed proteome-wide mass spectrometry data from multiple studies that have been collated on BioGrid and the Saccharomyces Genome Database, which indicated that most phosphorylation of Net1 occurs in the first two-thirds of the protein. We therefore constructed a plasmid that expresses a version of Net1 that lacks the last 328 amino acids and is tagged with 3xHA (Net1∆328-3xHA). The truncated Net1 is expressed from the endogenous promoter and can be integrated at URA3 so that cells express both full-length Net1 and the truncated reporter. The reporter includes an N-terminal Cdc14 binding site that is thought to undergo phosphorylation that helps release Cdc14 (Traverso et al., 2001). Hyperphosphorylation of Net1 that can be detected via electrophoretic mobility shifts could reflect the aggregate effects of multiple kinases.
Current models suggest that phosphorylation of Net1 is a critical event that helps initiate mitotic exit, which would suggest that Net1 phosphorylation is initiated during mitotic exit. However, in a previous study, it appears that Net1 phosphorylation is initiated early in mitosis as mitotic cyclin levels are rising, which could suggest that Net1 phosphorylation is initiated before mitotic exit (Visintin et al., 2003). To further investigate, we tested whether arresting cells at metaphase, before mitotic exit, blocked hyperphosphorylation of Net1. To do this, we released cells from a G1 phase arrest into YPD medium containing benomyl, which depolymerizes microtubules and activates a mitotic spindle checkpoint that arrests cells before metaphase. Depolymerization of microtubules also eliminated any signals generated by the proper orientation of the mitotic spindle, which is thought to be a critical trigger for mitotic exit. As a control, we also released cells into a medium that does not contain benomyl. Phosphorylation of both full-length Net1 and Net1∆328 was analyzed by western blot to detect electrophoretic mobility shifts, and levels of the mitotic cyclin Clb2 were analyzed as a marker for mitotic progression. In the control cells without benomyl, both Net1 and Net1∆328 underwent hyperphosphorylation as cells progressed through mitosis, with peak phosphorylation appearing to occur shortly after peak Clb2 levels, which likely corresponds to anaphase/telophase (Fig. 2, A and B). Both Net1 and Net1∆328 underwent extensive hyperphosphorylation when cells were arrested before mitotic exit. The extent of Net1 hyperphosphorylation was greater in benomyl-arrested cells compared with the control cells, which will be addressed further below.
Net1 hyperphosphorylation occurs before mitotic exit. (A)NET1-6xHA cells growing in YPD were released from a G1 arrest at 25°C into YPD or YPD containing benomyl. Samples were collected at the indicated time points to assay Net1 phosphorylation and Clb2 protein levels by western blot. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. (B)NET1∆328-3xHA cells growing in YPD were released from a G1 arrest at 20°C into YPD or YPD containing benomyl. Samples were collected at the indicated time points to assay Net1 phosphorylation and Clb2 protein levels by western blot. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. (C)NET1∆328-3xHA GAL1-CDC20 cells were grown overnight to log phase in YPGal medium at room temperature and were released from a G1 phase arrest into YPGal or YPD at 25°C. Samples were collected at the indicated time intervals to assay for NET1∆328-3xHAand Clb2 by western blot. In all panels, the Net1 blots and the Clb2 blots are from the same samples, so the Net1 blots serve as a loading control for the Clb2 blots. Source data are available for this figure: SourceData F2.
Net1 hyperphosphorylation occurs before mitotic exit. (A)NET1-6xHA cells growing in YPD were released from a G1 arrest at 25°C into YPD or YPD containing benomyl. Samples were collected at the indicated time points to assay Net1 phosphorylation and Clb2 protein levels by western blot. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. (B)NET1∆328-3xHA cells growing in YPD were released from a G1 arrest at 20°C into YPD or YPD containing benomyl. Samples were collected at the indicated time points to assay Net1 phosphorylation and Clb2 protein levels by western blot. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. (C)NET1∆328-3xHA GAL1-CDC20 cells were grown overnight to log phase in YPGal medium at room temperature and were released from a G1 phase arrest into YPGal or YPD at 25°C. Samples were collected at the indicated time intervals to assay for NET1∆328-3xHAand Clb2 by western blot. In all panels, the Net1 blots and the Clb2 blots are from the same samples, so the Net1 blots serve as a loading control for the Clb2 blots. Source data are available for this figure: SourceData F2.
To further test whether Net1 phosphorylation occurs before mitotic exit, we used depletion of Cdc20 as an alternative means of arresting cells in metaphase (Fig. 2 C). Again, Net1 underwent full hyperphosphorylation in metaphase-arrested cells.
These results show that hyperphosphorylation of Net1 does not depend on mitotic exit or any functions of the mitotic spindle.
Net1 phosphorylation and release of Cdc14 from the nucleolus are reduced when cells are grown in poor carbon
We next analyzed whether Net1 phosphorylation is influenced by the carbon source. In previous studies, we showed that the durations of both metaphase and anaphase are increased in poor carbon, while the extent of bud growth during both intervals is reduced (Leitao and Kellogg, 2017; Jasani et al., 2020). Cells growing in rich or poor carbon were released from a G1 phase arrest and Net1 phosphorylation was assayed by western blot (Fig. 3, A and B). Levels of Clb2, median cell size, and the percentage of cells that had undergone bud emergence were also assayed (Fig. 3, A–D). In rich carbon, both full-length Net1 and Net1∆328 underwent hyperphosphorylation during mitosis. In poor carbon, the rate of growth was reduced and there was a reduction in the extent of Net1 phosphorylation as well as an increase in the duration of Net1 phosphorylation. Since the rate and extent of bud growth were strongly reduced in poor carbon (Fig. 3 D and Leitao and Kellogg, 2017), these observations suggest that the extent of Net1 phosphorylation is correlated with the rate and extent of bud growth. Previous work found that Gin4 and Hsl1, which are thought to relay growth-dependent signals that measure bud growth before mitotic exit, also undergo hyperphosphorylation that is correlated with the rate and extent of bud growth in early mitosis (Jasani et al., 2020).
Net1 hyperphosphorylation is reduced in poor carbon. (A and B)NET1-6xHA (A) or NET1∆328-3xHA (B) cells grown overnight in YPD or YPG/E were released from a G1 phase arrest into a fresh medium at 25°C. Samples were collected at the indicated time points to assay for Net1 and Clb2 by western blot. An anti-Nap1 antibody was used for a loading control. Alpha factor was added back to the cultures 60 min after release to prevent a second cell cycle. (C and D) Samples from multiple biological replicates of the experiment shown in A were analyzed to determine the percentage of cells undergoing bud emergence (C) and to measure median cell size with a Coulter Counter (D). (E and F) Samples from multiple biological replicates of the experiment shown in A were analyzed by immunofluorescence to determine the percentage of cells with short metaphase spindles or long anaphase/telophase spindles, and the percentage of cells that showed release of Cdc14 from the nucleolus. (G) Examples of Cdc14 localization during metaphase and anaphase for cells growing in rich or poor carbon. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F3.
Net1 hyperphosphorylation is reduced in poor carbon. (A and B)NET1-6xHA (A) or NET1∆328-3xHA (B) cells grown overnight in YPD or YPG/E were released from a G1 phase arrest into a fresh medium at 25°C. Samples were collected at the indicated time points to assay for Net1 and Clb2 by western blot. An anti-Nap1 antibody was used for a loading control. Alpha factor was added back to the cultures 60 min after release to prevent a second cell cycle. (C and D) Samples from multiple biological replicates of the experiment shown in A were analyzed to determine the percentage of cells undergoing bud emergence (C) and to measure median cell size with a Coulter Counter (D). (E and F) Samples from multiple biological replicates of the experiment shown in A were analyzed by immunofluorescence to determine the percentage of cells with short metaphase spindles or long anaphase/telophase spindles, and the percentage of cells that showed release of Cdc14 from the nucleolus. (G) Examples of Cdc14 localization during metaphase and anaphase for cells growing in rich or poor carbon. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F3.
Since hyperphosphorylation of Net1 is thought to drive the release of Cdc14 from the nucleolus, we next analyzed Cdc14 release from the nucleolus in rich versus poor carbon. In rich carbon, the release of Cdc14 occurred during anaphase spindle elongation, as previously described (Fig. 3 E) (Visintin et al., 1999). In poor carbon, very few cells showed the release of Cdc14 from the nucleolus (Fig. 3 F). In these cells, the localization of Cdc14 during anaphase was similar to the previously reported localization of Net1 during anaphase (Fig. 3 G) (de Los Santos-Velázquez et al., 2017). The data do not rule out the possibility that a low undetectable amount of Cdc14 is released during mitotic exit in cells growing in poor carbon.
These observations indicate that the extent of Net1 phosphorylation required for mitotic exit is reduced in poor carbon. There also appears to be a reduced requirement for the release of Cdc14 from the nucleolus. Previous studies found that poor carbon medium also appears to cause a substantial reduction in growth-dependent signals required for passage through G1 phase and early mitosis, which suggests that nutrient availability influences global signals that modulate the extent of growth required for cell cycle progression at multiple stages of the cell cycle (Schneider et al., 2004; Jasani et al., 2020; Sommer et al., 2021).
Hyperphosphorylation of Net1 is correlated with the rate and extent of bud growth
To further investigate the relationship between Net1 phosphorylation and cell growth, we took advantage of the fact that bud growth continues during a prolonged mitotic arrest, which leads to the growth of unusually large buds during an unusually long interval of bud growth (Gihana et al., 2021). We also took advantage of the fact that the rate and extent of bud growth can be modulated by the quality of the carbon source in the culture medium (Leitao and Kellogg, 2017). Thus, we tested whether the extent of Net1 hyperphosphorylation was correlated with growth when the duration and extent of bud growth were increased by a benomyl arrest and when the rate of growth during the arrest was modulated by carbon source.
Cells were grown in carbon sources of varying quality and released from a G1 phase arrest into a medium containing benomyl to arrest the cell cycle before metaphase. The cells were then maintained at the arrest point for a prolonged interval of time. We utilized dextrose as a rich carbon source, galactose as an intermediate-quality carbon source, and glycerol/ethanol as a poor carbon source. Samples were taken at regular intervals and phosphorylation of the Net1∆328 reporter was measured by western blot (Fig. 4 A). In addition, median cell size was plotted as a function of time to provide a measure of the rate and extent of bud growth (Fig. 4 B).
Hyperphosphorylation of Net1 is correlated with the rate and extent of bud growth. (A)NET1∆328-3xHA cells grown overnight to log phase in YPD, YPGal, or YPG/E were released from a G1 phase arrest at 25°C into fresh media containing benomyl. Samples were taken at the indicated times to assay for Net1∆328-3xHA and Clb2. The Net1 blots and the Clb2 blots are from the same samples, so the Net1 blots serve as a loading control for the Clb2 blots. (B) A Coulter Counter was used to analyze cell size as a function of time for the experiments shown in A. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F4.
Hyperphosphorylation of Net1 is correlated with the rate and extent of bud growth. (A)NET1∆328-3xHA cells grown overnight to log phase in YPD, YPGal, or YPG/E were released from a G1 phase arrest at 25°C into fresh media containing benomyl. Samples were taken at the indicated times to assay for Net1∆328-3xHA and Clb2. The Net1 blots and the Clb2 blots are from the same samples, so the Net1 blots serve as a loading control for the Clb2 blots. (B) A Coulter Counter was used to analyze cell size as a function of time for the experiments shown in A. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F4.
In rich carbon, buds underwent rapid growth. Net1∆328 phosphorylation appeared to go to completion during the prolonged arrest, and the maximal extent of Net1∆328 phosphorylation achieved during the prolonged benomyl arrest was greater than in cells going through a normal cell cycle (compare top panels in Fig. 4 A and Fig. 2 B). Since daughter buds grow to be much larger in benomyl-arrested cells, this observation suggests that the extent of Net1 phosphorylation is correlated with the extent of bud growth. In galactose, the rate of bud growth was reduced, and it took longer for Net1∆328 phosphorylation to go to completion. In glycerol/ethanol, the rate and extent of bud growth were strongly reduced, and Net1 failed to reach full hyperphosphorylation. Together, these results provide further evidence that Net1 phosphorylation is correlated with the rate and extent of bud growth.
Hyperphosphorylation of Net1 is dependent upon membrane trafficking events that drive bud growth
To further investigate the relationship between Net1 phosphorylation and bud growth, we tested whether hyperphosphorylation of Net1 is dependent upon bud growth. In previous studies, we tested for the dependency of signaling events on bud growth by blocking plasma membrane growth in the daughter bud, which can be achieved by inactivating Sec6, a component of the exocyst complex that is required for the fusion of vesicles with the plasma membrane (Anastasia et al., 2012). A temperature-sensitive allele of SEC6 (sec6-4) provides rapid conditional inactivation of Sec6.
Inactivation of Sec6 before bud emergence causes cells to arrest at metaphase, due most likely to a failure in bud growth during early mitosis (Anastasia et al., 2012). Swe1, the budding yeast homolog of Wee1, is required for the arrest and is thought to respond to signals that measure bud growth before metaphase. Loss of Swe1 allows cells that lack Sec6 function to proceed through metaphase to late anaphase/telophase, and the cells show normal mitotic spindle assembly and chromosome segregation within the mother cell (Anastasia et al., 2012). Therefore, we analyzed Net1 phosphorylation in wild-type and sec6-4 swe1∆ cells, which ensured that any effects on Net1 phosphorylation were not due to a metaphase arrest. Cells were shifted to the restrictive temperature 20 min after release from a G1 phase arrest, before bud emergence. Hyperphosphorylation of full-length Net1 was assayed by western blot, and levels of Clb2 were assayed as a marker for mitotic progression. Hyperphosphorylation of Net1 was substantially reduced when sec6-4 was inactivated (Fig. 5). Controls showed that sec6-4 alone also caused a failure in Net1 phosphorylation, while swe1∆ alone had no effect on Net1 phosphorylation (not shown). Thus, hyperphosphorylation of Net1 is independent of mitotic exit and mitotic spindle function but is completely dependent upon membrane trafficking events that are required for bud growth. Furthermore, previous work found that Clb2 accumulates to high levels in sec6-4 swe1∆ cells and is capable of driving mitotic spindle assembly and segregation of chromosomes in anaphase (Anastasia et al., 2012), which indicates that Clb2/Cdk1 is active in this context and not sufficient to drive Net1 phosphorylation. This finding is consistent with a previous study, which found that Clb2/Cdk1 activity likely plays a minor role in the regulation of Net1, and only after MEN signaling has been initiated (Azzam et al., 2004).
Net1 hyperphosphorylation is dependent upon membrane trafficking events that are required for bud growth. (A)NET1∆328-3xHA and NET1∆328-3xHA sec6-4 swe1∆ cells grown overnight to log phase in YPD were released from a G1 phase arrest at room temperature and were then shifted to the restrictive temperature (34°C) 20 min after release. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. Samples were taken at the indicated time intervals to assay for Net1∆328-3xHA and Clb2 by western blot. An anti-Nap1 antibody was used for a loading control. (B and C) Samples from multiple biological replicates of the experiment shown in A were analyzed by immunofluorescence to determine the percentage of cells with short metaphase spindles or long anaphase/telophase spindles and the percentage of cells that show localization of Cdc14 to the nucleolus. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F5.
Net1 hyperphosphorylation is dependent upon membrane trafficking events that are required for bud growth. (A)NET1∆328-3xHA and NET1∆328-3xHA sec6-4 swe1∆ cells grown overnight to log phase in YPD were released from a G1 phase arrest at room temperature and were then shifted to the restrictive temperature (34°C) 20 min after release. Alpha factor was added back to the cultures at 60 min after release to prevent a second cell cycle. Samples were taken at the indicated time intervals to assay for Net1∆328-3xHA and Clb2 by western blot. An anti-Nap1 antibody was used for a loading control. (B and C) Samples from multiple biological replicates of the experiment shown in A were analyzed by immunofluorescence to determine the percentage of cells with short metaphase spindles or long anaphase/telophase spindles and the percentage of cells that show localization of Cdc14 to the nucleolus. Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F5.
We next tested whether the localization of Cdc14 is disrupted in sec6-4 cells. We again released sec6-4 and wild-type control cells from a G1 phase arrest and shifted to the restrictive temperature at 20 min. We then used immunofluorescence to analyze the localization of Cdc14. This showed that the inactivation of sec6-4 caused rapid loss of Cdc14 from the nucleolus well before mitosis (Fig. 5 B). Previous studies found that disrupting membrane trafficking events that are required for plasma membrane growth causes an arrest of ribosome biogenesis, as well as disruption of nucleolar structure, which could explain the loss of Cdc14 localization (Mizuta and Warner, 1994; Nanduri and Tartakoff, 2001). The dependence of ribosome biogenesis on membrane traffic could help ensure that the rate of ribosome biogenesis is matched to the rate of plasma membrane growth.
An allele of CDC14 that is not inhibited by Net1 bypasses the late mitotic arrest caused by a failure in bud growth
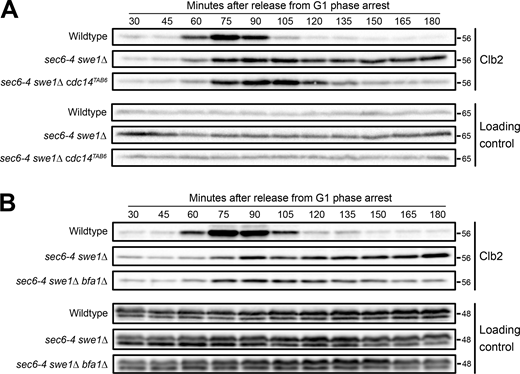
The data thus far suggest that hyperphosphorylation of Net1 is dependent upon bud growth and correlated with the extent of bud growth. To explain the data, we hypothesized that hyperphosphorylation of Net1 is driven by growth-dependent signals. We further hypothesized that growth-dependent hyperphosphorylation of Net1 is required for activation of the MEN, which would ensure that mitotic exit occurs only when sufficient growth has occurred. This model suggests that the late anaphase arrest in sec6-4 swe1∆ cells is caused by a failure in the release of Cdc14 from Net1. To test this model, we determined whether an allele of Cdc14 that shows reduced binding to Net1 (cdc14TAB6) is sufficient to bypass the late mitotic arrest in sec6-4 swe1∆ cells. The cdc14TAB6 allele bypasses the telophase arrest caused by loss of Cdc15 function, which indicates that the allele can bypass a failure to activate the MEN (Shou et al., 2001; Shou and Deshaies, 2002). We released sec6-4 swe1∆ cdc14TAB6 cells and control cells from a G1 phase arrest at the permissive temperature and then shifted to the restrictive temperature 20 min later and assayed Clb2 protein levels by western blot to assess mitotic progression (Fig. 6 A). The cdc14TAB6 allele fully abrogated the late anaphase arrest caused by sec6-4.
An allele of CDC14 that is not inhibited by Net1 bypasses the anaphase arrest caused by a failure in bud growth. (A)NET1-6xHA, NET1-6xHA sec6-4 swe1∆, and NET1-6xHA sec6-4 swe1∆ cdc14TAB6 cells grown overnight in YPD to log phase were released from a G1 phase arrest into fresh media at 25°C. 20 min after release the cultures were shifted to 34°C to inactivate sec6-4 and samples were collected at the indicated time points to assay levels of Clb2. A background band on the Clb2 blots was used as a loading control. (B)NET1-6xHA, NET1-6xHA sec6-4 swe1∆, and NET1-6xHA sec6-4 swe1∆ bfa1∆ cells grown overnight in YPD to log phase were released from a G1 phase arrest into fresh media at 25°C. 20 min after release the cultures were shifted to 34°C to inactivate sec6-4 and samples were collected at the indicated time points to assay levels of Clb2. An anti-Nap1 antibody was used as a loading control. Source data are available for this figure: SourceData F6.
An allele of CDC14 that is not inhibited by Net1 bypasses the anaphase arrest caused by a failure in bud growth. (A)NET1-6xHA, NET1-6xHA sec6-4 swe1∆, and NET1-6xHA sec6-4 swe1∆ cdc14TAB6 cells grown overnight in YPD to log phase were released from a G1 phase arrest into fresh media at 25°C. 20 min after release the cultures were shifted to 34°C to inactivate sec6-4 and samples were collected at the indicated time points to assay levels of Clb2. A background band on the Clb2 blots was used as a loading control. (B)NET1-6xHA, NET1-6xHA sec6-4 swe1∆, and NET1-6xHA sec6-4 swe1∆ bfa1∆ cells grown overnight in YPD to log phase were released from a G1 phase arrest into fresh media at 25°C. 20 min after release the cultures were shifted to 34°C to inactivate sec6-4 and samples were collected at the indicated time points to assay levels of Clb2. An anti-Nap1 antibody was used as a loading control. Source data are available for this figure: SourceData F6.
To further test the model, we analyzed the effects of deleting the BFA1 gene, which encodes a GAP thought to inhibit the MEN via inhibition of Tem1 (Pereira et al., 2000; Wang et al., 2000; Ro et al., 2002). Loss of BFA1 partially abrogated the late mitotic arrest in sec6-4 swe1∆ cells (Fig. 6 B).
These results suggest that the late mitotic arrest caused by sec6-4 is due to a failure to activate the MEN, consistent with a model in which the events of bud growth generate signals that are required for Net1 phosphorylation and activation of the MEN.
Components of the TORC2 signaling network are required for normal hyperphosphorylation of Net1
A key question concerns the nature of the signals that drive Net1 hyperphosphorylation. However, these signals are poorly understood. Over 90 phosphorylation sites have been detected on Net1, and multiple kinases have been implicated (Shou and Deshaies, 2002; Azzam et al., 2004; Mah et al., 2005; Stark et al., 2010; Zhou et al., 2021). Mitotic Cdk1 activity is thought to contribute to Net1 phosphorylation; however, only 10 of the many sites that have been detected on Net1 correspond to the minimal Cdk1 consensus site (S/TP). Furthermore, a previous study showed that mitotic spindle assembly, chromosome segregation, and mitotic spindle elongation occur normally in sec6-4 swe1∆ cells, despite the failure in bud growth (Anastasia et al., 2012). Since all these events require mitotic Cdk1 activity, the fact that they occur normally in sec6-4 swe1∆ cells that show a complete failure in Net1 hyperphosphorylation suggests that mitotic Cdk1 activity does not drive hyperphosphorylation of Net1. Another kinase thought to phosphorylate Net1 is the Cdc5/Polo kinase; however, mutation of sites thought to be phosphorylated by Cdc5 has no effect on release of Cdc14 or mitotic progression in vivo (Shou and Deshaies, 2002; Zhou et al., 2021). No kinase has been shown to be required for the extensive hyperphosphorylation of Net1 seen in vivo. Although kinases have been identified that are capable of disrupting the Net1-Cdc14 complex in vitro, it remains unclear whether they work directly to disrupt the complex in vivo (Shou and Deshaies, 2002; Azzam et al., 2004).
To investigate further, we first tested whether kinases previously proposed to phosphorylate Net1 are required for Net1 phosphorylation in vivo. Cells harboring an analog-sensitive allele of CDC5 (cdc5-as1) were released from a G1 phase arrest into medium containing benomyl and analog inhibitor was added 30 min after release. These cells arrested with hyperphosphorylated Net1∆328, which indicates that Cdc5 is not required for Net1 hyperphosphorylation (Fig. S1 A). Similarly, inhibition of an analog-sensitive allele of the CDC15 kinase (cdc15-as1), a key component of the MEN signaling cascade, had no effect on Net1∆328 phosphorylation in cells released from a G1 phase arrest into a medium containing benomyl (Fig. S1 B). Since Cdc15 is required for the activation of Dbf2, these results indicate that none of the key kinases in the MEN are required for hyperphosphorylation of Net1, which suggests that the hyperphosphorylation of Net1 that can be detected via electrophoretic mobility shifts is not due to MEN activity. This is consistent with previous studies, which found that the MEN is not active in metaphase-arrested cells (Fesquet et al., 1999; Jaspersen and Morgan, 2000; Hu et al., 2001; Visintin and Amon, 2001; Hu and Elledge, 2002). The results do not rule out the possibility that MEN kinases phosphorylate a subset of sites that do not influence the electrophoretic mobility of Net1.
Analysis of candidate Net1 kinases in vivo. For all experiments, cells were grown in YPD medium and released from a G1 phase arrest into fresh YPD media containing benomyl. Inhibitor was added at the times indicated by the arrow. Samples were then taken at the indicated time points to assay for NET1∆328 and Clb2 by western blot. (A)NET1∆328-3xHA and NET1∆328-3xHA cdc5-as1 cells were released from G1 phase at 30°C. 10 μM CMK was added 30 min after release. (B)NET1∆328-3xHA and NET1∆328-3xHA cdc15-as1 cells were released from G1 phase arrest at 25°C. 10 μM 1-NA-PP1 was added 40 min after release. (C)NET1∆328-3xHA cells were released from G1 phase arrest at 25°C. The culture was split into two aliquots and 0.22 μM rapamycin was added to one aliquot 90 min after release. (D)NET1∆328-3xHA and NET1∆328-3xHA sch9-as1 cells were released from G1 phase at 25°C. 5 μM of 1-NM-PP1 was added 30 min after release. Source data are available for this figure: SourceData FS1.
Analysis of candidate Net1 kinases in vivo. For all experiments, cells were grown in YPD medium and released from a G1 phase arrest into fresh YPD media containing benomyl. Inhibitor was added at the times indicated by the arrow. Samples were then taken at the indicated time points to assay for NET1∆328 and Clb2 by western blot. (A)NET1∆328-3xHA and NET1∆328-3xHA cdc5-as1 cells were released from G1 phase at 30°C. 10 μM CMK was added 30 min after release. (B)NET1∆328-3xHA and NET1∆328-3xHA cdc15-as1 cells were released from G1 phase arrest at 25°C. 10 μM 1-NA-PP1 was added 40 min after release. (C)NET1∆328-3xHA cells were released from G1 phase arrest at 25°C. The culture was split into two aliquots and 0.22 μM rapamycin was added to one aliquot 90 min after release. (D)NET1∆328-3xHA and NET1∆328-3xHA sch9-as1 cells were released from G1 phase at 25°C. 5 μM of 1-NM-PP1 was added 30 min after release. Source data are available for this figure: SourceData FS1.
The links between Net1 phosphorylation and bud growth suggest that signals associated with cell growth could play a role in the hyperphosphorylation of Net1. We therefore tested whether signals known to be associated with cell growth are required for hyperphosphorylation of Net1. Previous work has shown that TOR kinases play important roles in the control of cell growth. The TOR kinases are assembled into two distinct multiprotein complexes, referred to as Target of Rapamycin Complexes 1 and 2 (TORC1 and TORC2) (Loewith and Hall, 2011). A key target of TORC1 is the conserved Sch9 kinase, which controls the transcription of ribosome biogenesis genes. To test whether TORC1 is required for the hyperphosphorylation of Net1, we synchronized cells and added rapamycin shortly after bud emergence. Rapamycin had no effect on the hyperphosphorylation of Net1∆328 in cells released from a G1 phase arrest into medium containing benomyl (Fig. S1 C). Similarly, inhibition of an analog-sensitive version of Sch9 (sch9-as) had no effect on hyperphosphorylation of Net1∆328 (Fig. S1 D).
Recent studies have shown that TORC2 activity is correlated with growth rate and the extent of growth, and our mass spectrometry data indicated that multiple components of TORC2 and its surrounding signaling network underwent substantial changes in phosphorylation when cells in late mitosis were shifted from rich to poor carbon. Moreover, TORC2 signaling is required for normal control of cell size and nutrient modulation of cell size (Alcaide-Gavilán et al., 2018; Lucena et al., 2018; Leitao et al., 2019). A key downstream target of TORC2 that is required for normal control of cell growth and size is a pair of redundant kinase paralogs called Ypk1 and Ypk2, which are the yeast homologs of mammalian SGK kinases (Niles et al., 2012). Loss of Ypk1 causes a large reduction in cell size, while loss of Ypk2 has no apparent phenotype. Loss of both is lethal. To test whether Ypk1/2 influence Net1 phosphorylation, we released cells that carry an analog-sensitive allele of YPK1 in a ypk2∆ background (ypk1-as ypk2∆) from a G1 phase arrest into media that contains benomyl. We then assayed hyperphosphorylation of Net1∆328 as well as bud emergence and cell size as a function of time (Fig. 7). Inhibition of ypk1-as caused a substantial reduction in Net1 phosphorylation. Bud emergence occurred with normal timing, consistent with a previous study (Clarke et al., 2017), but the rate of growth after bud emergence was reduced. Previous studies have shown that the mother cell stops growing at bud emergence (Jasani et al., 2020; Ferrezuelo et al. 2012), which indicates that the reduced growth rate observed after inhibition of ypk1-as is due to a reduced rate of bud growth. Together, these observations provide further evidence that hyperphosphorylation of Net1 is strongly associated with bud growth. They also show for the first time that normal bud growth is dependent upon key targets of TORC2 signaling. Our discovery that multiple components of the TORC2 signaling network are rapidly regulated in response to a shift from rich to poor carbon during mitosis (Table 1) provides further evidence that TORC2 signaling plays important roles in controlling bud growth.
Ypk1/2 is required for normal hyperphosphorylation of Net1 in vivo. (A)NET1∆328-3xHA and NET1∆328-3xHA ypk2∆ ypk1-as1 cells grown overnight in YPD were released from a G1 phase arrest at 25°C into YPD medium containing benomyl. 3MOB-PP1 was added to both cultures to a final concentration of 10 uM at 40 min after release. Samples were taken at the indicated time intervals to assay for Net1∆328-3xHA and Clb2 by western blot. An anti-Nap1 antibody was used as a loading control. (B and C) Samples from multiple biological replicates of the experiment shown in A were analyzed to determine the percentage of cells undergoing bud emergence (B) and to measure median cell size with a Coulter Counter (C). Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F7.
Ypk1/2 is required for normal hyperphosphorylation of Net1 in vivo. (A)NET1∆328-3xHA and NET1∆328-3xHA ypk2∆ ypk1-as1 cells grown overnight in YPD were released from a G1 phase arrest at 25°C into YPD medium containing benomyl. 3MOB-PP1 was added to both cultures to a final concentration of 10 uM at 40 min after release. Samples were taken at the indicated time intervals to assay for Net1∆328-3xHA and Clb2 by western blot. An anti-Nap1 antibody was used as a loading control. (B and C) Samples from multiple biological replicates of the experiment shown in A were analyzed to determine the percentage of cells undergoing bud emergence (B) and to measure median cell size with a Coulter Counter (C). Error bars indicate SEM for three biological replicates. Source data are available for this figure: SourceData F7.
Discussion
Numerous components of the MEN are controlled by nutrient-dependent signals
Previous studies suggested that bud growth is required for mitotic progression and that the threshold amount of growth required for mitotic progression is reduced in poor nutrients, which would explain why daughter cells are born at a very small size in poor nutrients (Anastasia et al., 2012; Jasani et al., 2020; Leitao and Kellogg, 2017). Here, we searched for molecular mechanisms that link the completion of mitosis to bud growth. To do this, we took advantage of the fact that changes in nutrient availability rapidly reset the threshold amount of growth required for cell cycle progression (Fantes and Nurse, 1977; Leitao and Kellogg, 2017). We reasoned that proteins involved in measuring bud growth and setting the growth threshold should show rapid changes in phosphorylation in response to a shift to poor nutrients. Thus, we used proteome-wide mass spectrometry to identify proteins that undergo rapid changes in phosphorylation when cells in mitosis are shifted from rich to poor carbon. This revealed that numerous components of the MEN are regulated by nutrient availability. Since activation of the MEN initiates exit from mitosis, the MEN is well-positioned to receive physiological signals related to nutrients and cell growth to ensure that mitotic exit is initiated only when an appropriate amount of growth has occurred. A role for the MEN in controlling mitotic exit in response to nutrient and growth cues would be compatible with previously proposed models in which the MEN responds to the migration of the daughter nucleus into the daughter bud. For example, signals associated with cell growth and nutrient availability could influence the initiation of mitotic exit, while migration of the nucleus into the daughter bud could initiate positive feedback that fully activates the MEN to complete mitotic exit.
An alternative hypothesis could be that a shift to poor carbon reduces the availability of ATP, leading to a general slowdown in cellular events and a decrease in the rate of phosphorylation of proteins. Several observations argue against this interpretation. First, a shift to poor carbon late in mitosis does not cause a delay in mitotic exit, which suggests that the shift does not cause a non-specific slowdown of mitotic events (Leitao and Kellogg, 2017). Rather, a shift to poor carbon in late mitosis may not cause a delay because sufficient bud growth has already occurred, especially as the shift should immediately lower the threshold amount of growth required for cell cycle progression (Fantes and Nurse, 1977). A further argument against a general reduction in ATP is that the top proteins identified by mass spectrometry are enriched in proteins involved in the MEN, TORC2 signaling, ribosome biogenesis, and membrane trafficking, all of which are involved in growth or cell cycle progression and would be expected to change when growth rate is slowed by poor nutrients. In contrast, few proteins involved in mitotic spindle dynamics, which are known to undergo phosphorylation during mitosis, were identified.
Hyperphosphorylation of Net1 occurs before mitotic exit
Among the MEN components identified by mass spectrometry, Net1 was amongst the most prominent with 11 phosphorylation sites that showed substantial loss of phosphorylation. It is thought that hyperphosphorylation of Net1 promotes dissociation of Cdc14 to help initiate mitotic exit, although there is evidence that hyperphosphorylation of Net1 may not be sufficient to release Cdc14 (Traverso et al., 2001; Visintin et al., 2003; Azzam et al., 2004; Zhou et al., 2021). Here, we found that Net1 undergoes full hyperphosphorylation in metaphase-arrested cells, which indicates that Net1 hyperphosphorylation is not strictly correlated with mitotic exit and does not depend upon the events of mitotic exit. A previous study found that Cdc14 is not released in metaphase-arrested cells (Stegmeier et al., 2002). Together these findings are consistent with previously proposed models in which hyperphosphorylation of Net1 can contribute to mitotic exit but is not sufficient for mitotic exit.
Phosphorylation of Net1 is correlated with the rate and extent of bud growth
What are the physiological signals that drive hyperphosphorylation of Net1 before mitotic exit? Several observations suggest that signals associated with cell growth influence Net1 phosphorylation. First, hyperphosphorylation of Net1 occurs gradually during bud growth, which indicates that Net1 phosphorylation is correlated with bud growth. Furthermore, we found that metaphase-arrested cells undergo unusually extensive bud growth, which leads to abnormally large daughter buds as well as unusually extensive hyperphosphorylation of Net1. Similarly, we used growth in poor carbon sources to show that reducing the rate and extent of bud growth leads to a corresponding reduction in the rate at which hyperphosphorylated forms of Net1 accumulate as well as a reduction in the maximal extent of Net1 phosphorylation. Thus, hyperphosphorylation of Net1 is strongly correlated with the rate and extent of bud growth, which suggests that the events of growth generate signals that help drive hyperphosphorylation of Net1.
The amount of Cdc14 released from the nucleolus is strongly reduced in slow-growing cells
In cells growing slowly in poor carbon, the extent of Net1 phosphorylation and Cdc14 release from the nucleolus are strongly reduced. This discovery strengthens the correlation between Net1 phosphorylation and the release of Cdc14. In addition, the data suggest that the threshold amount of Cdc14 that needs to be released from the nucleolus to promote mitotic exit is reduced in poor carbon. In previous studies, we found that growth-dependent signals required for cell cycle progression at other stages of the cell cycle are also strongly reduced (Sommer et al., 2021; Jasani et al., 2020). Together, these observations suggest that there may be a global nutrient-modulated signal that sets the threshold amount of growth required for cell cycle progression. Previous studies suggest that the TORC2 signaling network could be responsible for generating such a signal (Lucena et al., 2018).
Hyperphosphorylation of Net1 is dependent upon membrane trafficking events that drive bud growth
Further evidence for a link between Net1 phosphorylation and cell growth came from our finding that hyperphosphorylation of Net1 is dependent upon membrane trafficking events that drive bud growth. In sec6-4 swe1∆ cells, the initial steps of mitotic exit occur normally as the spindle elongates during anaphase (Anastasia et al., 2012), yet hyperphosphorylation of Net1 fails to occur. These observations provide further evidence that hyperphosphorylation of Net1 is closely associated with bud growth, rather than with early steps of anaphase and mitotic exit. Previous studies have shown that an arrest of membrane trafficking events that are required for plasma membrane growth leads to inhibition of ribosome biogenesis and a disruption of nucleolar structure (Mizuta and Warner, 1994; Li et al., 2000). Furthermore, we found that inactivation of Sec6 causes loss of Cdc14 from the nucleolus long before mitotic exit, consistent with a rapid disruption of nucleolar function. The failure in Net1 phosphorylation in sec6-4 swe1∆ cells could be a consequence of a restructuring of the nucleolus that accompanies an arrest of ribosome biogenesis, although it is unclear from previous studies whether arresting membrane growth causes an arrest of ribosome biogenesis on a sufficiently rapid time scale to account for the observed effects on Net1 phosphorylation. It is also possible that the inhibition of ribosome biogenesis caused by blocking membrane trafficking events is due to a failure in Net1 phosphorylation, as previous studies have shown that Net1 is required for normal transcription of rRNA genes (Shou et al., 2001; Hannig et al., 2019).
The fact that Net1 is intimately connected to ribosome biogenesis further suggests a potential connection between Net1 hyperphosphorylation and cell growth (Straight et al., 1999; Shou et al., 2001; Hannig et al., 2019). An interesting possibility is that gradual phosphorylation of Net1 during bud growth ensures that the rate of rDNA gene transcription increases during growth to meet the increased protein synthesis requirements of larger cells. Consistent with this, a previous study found that phosphorylation of Net1 promotes the binding and activation of RNA polymerase PolI (Hannig et al., 2019). This would suggest a model in which the events of growth generate feedback signals that promote further growth and also provide a measure of the extent of growth.
Bypassing the MEN relieves a late mitotic arrest caused by a failure in bud growth
We previously found that sec6-4 swe1∆ cells arrest in late anaphase with long mitotic spindles, separated nuclei, and high levels of mitotic cyclin, which indicates that they fail to complete mitotic exit (Anastasia et al., 2012). The cause of this late mitotic arrest was unknown. Here, we found that the late anaphase arrest in sec6-4 swe1∆ cells can be bypassed by preventing Net1-dependent inhibition of Cdc14. Together, these observations suggest that the late mitotic arrest caused by blocking bud growth is due to a failure to activate the MEN, consistent with the hypothesis that activation of the MEN is dependent upon bud growth.
Net1 undergoes extensive hyperphosphorylation that is not dependent upon key components of the MEN
Net1 is one of the most highly phosphorylated proteins in budding yeast. Various mass spectrometry analyses have identified 91 phosphosites. The functions of Net1 phosphorylation remain poorly understood. Many of the 91 sites were identified in non-quantitative proteome-wide analyses, which makes it unclear when they are phosphorylated in a cell cycle-dependent manner or whether they are phosphorylated at significant stoichiometries that are likely to be biologically relevant. A NET1 mutant in which all 91 sites were mutated (net1-91A) is viable and causes only a slight delay in mitotic exit (Zhou et al., 2021). However, whether the net1-91A mutant eliminates phosphorylation-induced electrophoretic mobility shifts in vivo was not tested, so it is unclear whether relevant sites were mutated. Furthermore, mutating 91 phosphorylation sites almost certainly eliminates regulation by multiple kinases, which makes interpretation of the results difficult. For example, if Net1 plays both positive and negative roles in controlling ribosome biogenesis and mitotic exit, as suggested by previous studies, the elimination of 91 phosphorylation sites could cause both gain- and loss-of-function effects that cancel each other out (Straight et al., 1999; Shou et al., 2001; Hannig et al., 2019). The fact that mutating subsets of sites appears to cause stronger effects is consistent with this possibility (Zhou et al., 2021).
Previous studies suggested that Cdc5 and Dbf2, two key components of the MEN, could phosphorylate Net1 (Shou and Deshaies, 2002; Yoshida and Toh-e, 2002; Visintin et al., 2003; Mah et al., 2005; Zhou et al., 2021). Previous studies mapped phosphorylation sites on Net1 that are dependent upon Cdc5 or Cdc15. In some cases, mutation of these sites causes mitotic delays, especially in sensitized genetic backgrounds (Loughrey Chen et al., 2002; Shou and Deshaies, 2002; Shou et al., 2002; Zhou et al., 2021). However, the phosphorylation site mutants do not cause a failure in mitotic exit or a loss of viability, and they appear to have only mild effects in an otherwise wild-type background, which indicates that they are not essential for mitotic exit.
No previous studies have tested whether Cdc5, Cdc15 or Dbf2 are required for hyperphosphorylation of Net1 that can be detected via electrophoretic mobility shifts. Here, we found that Cdc5 and Cdc15 are not required for hyperphosphorylation of Net1 in vivo. Since Cdc15 is required for activation of Dbf2, these findings suggest that none of the protein kinases in the MEN are required for hyperphosphorylation of Net1. Previous studies found that key components of the MEN are inactive during a metaphase arrest, which provides further evidence that hyperphosphorylation of Net1 in metaphase-arrested cells is not due to MEN signaling (Fesquet et al., 1999; Jaspersen and Morgan, 2000; Hu et al., 2001; Visintin and Amon, 2001; Hu and Elledge, 2002). Furthermore, the fact that full hyperphosphorylation of Net1 occurs even when mitotic spindle assembly is blocked shows that hyperphosphorylation of Net1 is not dependent upon any events associated with the function or orientation of the mitotic spindle. A potential explanation for these observations is that hyperphosphorylation of Net1 is the consequence of signals that are necessary but not sufficient for initiating the full activation of the MEN signaling network.
TORC2 signaling is required for growth-dependent hyperphosphorylation of Net1
Since hyperphosphorylation of Net1 is correlated with growth and dependent upon growth, we tested whether it is influenced by the activity of TORC1 or TORC2. Inactivation of TORC1 or Sch9, a key target of TORC1 that controls ribosome biogenesis, had no effect on the extent of Net1 phosphorylation. In contrast, the inactivation of Ypk1 and Ypk2, key targets of TORC2, caused a large reduction in both Net1 hyperphosphorylation and growth rate, which provides additional evidence that Net1 phosphorylation is associated with cell growth.
We found that multiple components of the TORC2 network are regulated in response to a shift from rich to poor carbon during mitosis. Previous studies found that TORC2 signaling, as measured via phosphorylation of Ypk1/2, is modulated by nutrient availability and increases throughout the interval of bud growth (Alcaide-Gavilán et al., 2018; Lucena et al., 2018). Furthermore, multiple components of the TORC2 signaling network are required for normal control of cell size, nutrient modulation of cell size, and the proportional relationship between cell size and growth rate (Lucena et al., 2018; Leitao et al., 2019). Together, these observations suggest the possibility that gradually rising TORC2 signaling during bud growth could drive growth-dependent phosphorylation of Net1 and that nutrient modulation of TORC2 signaling could explain the effects of nutrient availability on Net1 phosphorylation and cell size at the completion of mitosis. However, it remains unclear how directly TORC2 signaling influences Net1 phosphorylation.
What are the signals that drive mitotic exit?
A puzzling aspect of the MEN has been that Cdc14, which plays perhaps the most critical role in the initiation of mitotic exit, is conserved across eukaryotic cells, yet the role of Cdc14 in promoting mitotic exit is not conserved, as budding yeast is the only known organism in which Cdc14 functions in mitotic exit. A potential explanation could come from the fact that budding yeast is the only well-understood organism in which most cell growth occurs during mitosis. Thus, it seems possible that Cdc14 has been co-opted in budding yeast to function in an unusual pathway that links mitotic exit to bud growth. Our results support this model, as they suggest that the MEN responds to growth-dependent signals that define the duration and extent of bud growth in late mitosis. The data do not rule out previously suggested models in which the MEN ensures that mitotic exit does not occur until the daughter nucleus has entered the bud. Indeed, the existence of a mechanism to ensure that nuclear segregation occurs only when the bud has undergone sufficient growth to accommodate the daughter nucleus would appear to be essential for yeast cells that rely on a highly unusual growth mode (i.e., bud growth in mitosis) to reproduce.
Materials and methods
Strain construction, media, and reagents
All strains are in the W303 background (MATa leu2-3,112 ura3-1 can1-100 ade2-1 his3-11,15 trp1-1 GAL+ ssd1-d2). Additional genetic features are listed in Table 2. Gene deletions and epitope tagging were performed by standard PCR-based methods (Longtine et al., 2000; Janke et al., 2004).
Strains used in this study
| Strain . | Genotype . | Figure . |
|---|---|---|
| DK2017 | CLN3-6XHA::His3MX6, bar1 | Fig. 1 |
| DK3822 | NET1-6XHA::kanMX4, bar1 | Figs. 2, 3, and 6 |
| DK4616 | net1∆328-3XHA::URA, bar1 | Figs. 2, 4, 7, and S1 |
| DK4661 | net1∆328-3XHA::URA, pGAL-CDC20::kanMX5, bar1 | Fig. 2 |
| DK4563 | NET1-6XHA::kanMX4, net1∆328-3XHA::URA, bar1 | Fig. 3 |
| DK4066 | NET1-6XHA:: hphNT1 | Figs. 5 and 6 |
| DK4180 | NET1-6XHA:: hphNT1, sec6-4::kanMX6, swe1∆::His5 | Figs. 5 and 6 |
| DK4005 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, cdc14TAB6-1 | Fig. 6 |
| DK3838 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, bar1 | Fig. 6 |
| DK4442 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, bfa1::caURA, bar1 | Fig. 6 |
| DK4708 | net1∆328-3XHA::URA, ypk2::His3, ypk1-as1, bar1 | Fig. 7 |
| DK4624 | net1∆328-3XHA::URA, cdc5-as1, bar1 | Fig. S1 |
| DK4706 | net1∆328-3XHA::URA, cdc15-as1, bar1 | Fig. S1 |
| DK4654 | net1∆328-3XHA::URA, sch9-as1 | Fig. S1 |
| Strain . | Genotype . | Figure . |
|---|---|---|
| DK2017 | CLN3-6XHA::His3MX6, bar1 | Fig. 1 |
| DK3822 | NET1-6XHA::kanMX4, bar1 | Figs. 2, 3, and 6 |
| DK4616 | net1∆328-3XHA::URA, bar1 | Figs. 2, 4, 7, and S1 |
| DK4661 | net1∆328-3XHA::URA, pGAL-CDC20::kanMX5, bar1 | Fig. 2 |
| DK4563 | NET1-6XHA::kanMX4, net1∆328-3XHA::URA, bar1 | Fig. 3 |
| DK4066 | NET1-6XHA:: hphNT1 | Figs. 5 and 6 |
| DK4180 | NET1-6XHA:: hphNT1, sec6-4::kanMX6, swe1∆::His5 | Figs. 5 and 6 |
| DK4005 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, cdc14TAB6-1 | Fig. 6 |
| DK3838 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, bar1 | Fig. 6 |
| DK4442 | NET1-6XHA::hphNT1, sec6-4::kanMX6, swe1∆::His5, bfa1::caURA, bar1 | Fig. 6 |
| DK4708 | net1∆328-3XHA::URA, ypk2::His3, ypk1-as1, bar1 | Fig. 7 |
| DK4624 | net1∆328-3XHA::URA, cdc5-as1, bar1 | Fig. S1 |
| DK4706 | net1∆328-3XHA::URA, cdc15-as1, bar1 | Fig. S1 |
| DK4654 | net1∆328-3XHA::URA, sch9-as1 | Fig. S1 |
Strains were grown in YEP medium (1% yeast extract, 2% peptone, and 40 mg/ml adenine) containing 2% dextrose (YPD), 2% galactose (YPGal), or 2% glycerol/2% ethanol (YPG/E), as indicated in the text. In experiments involving analog-sensitive alleles of kinases, cells were grown in media that was not supplemented with adenine.
The Net1∆328 phosphorylation reporter was created by amplifying a region of the NET1 gene that includes the promoter and the first 806 amino acids (oligos: 5′-ATACC-CGGGCGTAGGGAGCGATATGTGCATTATG-3′ and 5′-ATAGGTACCCTTCTTGTTTGAA-TTCGGATAAAAGCTTTTCGCC-3′). The resulting PCR fragment was cloned into the KpnI and XmaI sites of pDK51, which expresses proteins with a C-terminal 3XHA tag and carries the URA3 marker, to create pDK131. The plasmid is cut with StuI to target integration at the URA3 gene. The addition of the Net1∆328-3xHA reporter to cells did not cause detectable differences in cell cycle progression, as assayed by the timing of mitotic cyclin expression in synchronized cells (not shown).
Benomyl was solubilized in 100% DMSO to prepare a 20 mg/ml stock solution. Media containing benomyl was prepared by first heating the media to 100°C before adding the drug to a final concentration of 30 μg/ml. The media was then allowed to cool at room temperature while stirring.
Cell cycle time courses and western blotting
For cell cycle time courses, cultures were grown overnight at room temperature to log phase (OD600 = 0.4–0.7). The cells were then arrested in G1 phase by the addition of 0.5 μg/ml or 15 μg/ml alpha factor for bar1 or BAR1 strains, respectively. Cultures were arrested for 3–3.5 h and then released from the arrest by three washes in fresh media not containing mating pheromone. All time courses were performed at 25°C, except for experiments involving sec6-4 strains, which were carried out at 34°C. Alpha factor was added back to cultures 60 min after release to prevent the initiation of a second cell cycle. At each time point, 1.6 ml of the sample was collected in screw cap tubes and centrifuged at 15,000 rpm for 15 s. The supernatant was then removed and 200 μl of acid-washed glass beads were added before freezing samples in liquid nitrogen. Cell pellets were lysed in 140 μl 1x SDS-PAGE sample buffer (65 mM Tris-HCl pH 6.8, 3% SDS, 10% glycerol, 100 mM β-glycerophosphate, 50 mM NaF, 5% β-mercaptoethanol, 3 mM PMSF, and bromophenol blue) by bead beating in a Biospec Mini-Beadbeater-16 at 4°C for 2 min. Lysed samples were spun down in a microcentrifuge for 15 s at 15,000 rpm before incubating in a 100°C water bath for 6 min followed by centrifugation at 15,000 rpm for 10 min.
SDS-PAGE was carried out as previously described (Harvey et al., 2011). 10% polyacrylamide gels with 0.13% bis-acrylamide were used for the analysis of Net1, Clb2, and Nap1 (loading control). Blots were probed with the primary antibody at 1–2 μg/ml at 4°C overnight in PBST (phosphate-buffered saline, 250 mM NaCl, and 0.1% Tween-20) containing 4% nonfat dry milk. Primary antibodies used to detect Clb2 and Nap1 were rabbit polyclonal antibodies generated as described previously (Kellogg and Murray, 1995; Sreenivasan and Kellogg, 1999; Mortensen et al., 2002). Net1-6xHA and Net1∆328-3xHA were detected by a mouse monoclonal antibody (12CA5). Primary antibodies were detected by an HRP-conjugated donkey anti-rabbit secondary antibody (#NA934V; GE Healthcare) or HRP-conjugated donkey anti-mouse secondary antibody (#NXA931; GE Healthcare) incubated in PBST containing 4% nonfat dry milk for 1 h at room temperature. Blots were rinsed in PBS before detection via chemiluminescence using ECL reagents (#K-12045-D50; Advansta) with a Bio-Rad ChemiDoc imaging system.
Cell size analysis and immunofluorescence
For Fig. 4 B, cell cultures were grown overnight at room temperature in YPD, YPGal, or YPG/E to an OD600 between 0.4 and 0.6. During the time course, 450 μl of culture was collected at each time point and fixed by the addition of 50 μl of 37% formaldehyde to the culture medium followed by incubation at room temperature for 1 h. Cells were then pelleted and resuspended in 0.25 ml PBS containing 0.02% sodium azide and 0.1% Tween-20. Cell size was measured on the same day using a Coulter Counter (Coulter Counter Z2; Beckman).
Cdc14 localization was assayed via immunofluorescence using a rabbit polyclonal antibody raised against full-length Cdc14 protein. The Cdc14 protein was expressed in bacteria as a GST fusion protein (Jaspersen et al., 1999). The antibody was purified from serum by first depleting anti-GST antibodies with an anti-GST affinity column. Anti-Cdc14 antibody was then purified using a GST-Cdc14 affinity column. Tubulin was detected with a fluorescein-conjugated secondary antibody, and Cdc14 was detected with a rhodamine-conjugated secondary antibody.
All images were acquired using a DeltaVision Personal DV system (Applied Precision) equipped with a 100X N.A. 1.40 oil-immersion objective (Olympus), resulting in an effective XY pixel spacing of 0.064 or 0.040 μm. Three-dimensional image stacks were collected at 0.2-μm Z-spacing and processed by constrained, iterative deconvolution. ImageJ was used to generate additive projection images.
Tandem mass spectrometry analysis
Cells were grown overnight to log phase in YPD medium and were then arrested in the G1 phase with alpha factor. The cells were released from the arrest into fresh YPD medium, and at 90 min after release, one-half of the culture was shifted to poor carbon (YPG/E) by pelleting the cells and resuspending in YPG/E three times. At 10 min after the first wash into poor carbon, the cells from 50 ml of both cultures (O.D. 600 of 0.5) were pelleted in a 50 ml conical tube, resuspended in 1 ml of media, transferred to a new 1.6 ml screw top, and then pelleted again for 15 s in a microfuge. After removing the supernatant, 200 μl of acid-washed beads were added and the samples were frozen on liquid nitrogen. Parallel samples were taken for western blotting to ensure that the shift to poor carbon occurred at peak Clb2 levels. A total of two biological replicates were carried out and analyzed by mass spectrometry. A strain carrying Cln3-6XHA (DK2017) was used for these experiments because we wanted to test whether the Cln3 protein present in mitosis responds rapidly to a shift to poor carbon. Western blotting showed that Cln3 rapidly disappeared when the cells were shifted to poor carbon within 5 min, as observed previously for Cln3 in the G1 phase. Since the shifted cells were pelleted and resuspended but the control cells were not, we cannot rule out the possibility that some of the changes in phosphorylation were due to the physical manipulations of washing the cells.
Cell pellets were lysed in a Biospec Mini-Beadbeater-16 at 4°C in 500 μl of Lysis Buffer (8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.0, 50 mM B-glycerolphosphate, 1 mM NaVO3, 10 mM sodium pyrophosphate, and 1 mM PMSF) by two 1-min rounds of bead-beating at 4°C, with 1 min of chilling in an ice water bath in between. The lysate was then centrifuged at 14,000 rpm at 4°C for 10 min, and the supernatant was transferred to a new tube and frozen on liquid nitrogen. The samples were prepared for quantitative proteome and phosphoproteome profiling following the SL-TMT protocol that includes the “mini-phos” phosphopeptide enrichment strategy (Navarrete-Perea et al., 2018).
Online supplementary material
Data availability
The data for all figures and mass spectrometry analysis are available in the figures, supplementary material, and the source data.
Acknowledgments
We thank the Shokat lab for providing 3MOB-PP1 and Bhumil Patel and Needhi Bhalla for assistance with microscopy and image analysis. We also thank members of the Kellogg lab for advice and critical reading of the manuscript.
This work was supported by the National Institutes of Health grant R35 GM131826.
Author contributions: B.E. Prichard: Investigation. R.A. Sommer: Investigation. R.M. Leitao: Investigation. C.J. Sarabia: Investigation. S. Hazir: Investigation. J.A. Paulo: Investigation and Methodology. S.P. Gygi: Investigation and Methodology. D.R. Kellogg: Conceptualization, Data Curation, Funding acquisition, Investigation, Project administration, Supervision, Methodology, Writing—original draft, and Writing—review and editing.
References
Author notes
Disclosures: The authors declare no competing interests exist.