Glutathionylation is a posttranslational modification involved in various molecular and cellular processes. However, it remains unknown whether and how glutathionylation regulates nervous system development. To identify critical regulators of synapse growth and development, we performed an RNAi screen and found that postsynaptic knockdown of glutathione transferase omega 1 (GstO1) caused significantly more synaptic boutons at the Drosophila neuromuscular junctions. Genetic and biochemical analysis revealed an increased level of glass boat bottom (Gbb), the Drosophila homolog of mammalian bone morphogenetic protein (BMP), in GstO1 mutants. Further experiments showed that GstO1 is a critical regulator of Gbb glutathionylation at cysteines 354 and 420, which promoted its degradation via the proteasome pathway. Moreover, the E3 ligase Ctrip negatively regulated the Gbb protein level by preferentially binding to glutathionylated Gbb. These results unveil a novel regulatory mechanism in which glutathionylation of Gbb facilitates its ubiquitin-mediated degradation. Taken together, our findings shed new light on the crosstalk between glutathionylation and ubiquitination of Gbb in synapse development.
Introduction
Various posttranslational modifications (PTMs), including glutathionylation, phosphorylation, ubiquitination, methylation, and acetylation, affect various aspects of development, physiology, and pathogenesis (Ali et al., 2018; Cohen, 2002; Dalle-Donne et al., 2009; DiAntonio and Hicke, 2004; Rashdan et al., 2020; Walsh, 2006). S-glutathionylation is a specific PTM of protein cysteine residues involving the reversible addition of the tripeptide glutathione (GSH), the most abundant and important low-molecular-mass thiol (Dalle-Donne et al., 2009; Xiong et al., 2011). Recent development of tools and strategies to analyze protein glutathionylation, including proteomics and advanced mass spectrometry, has improved the ability to identify targets and molecular pathways regulated by glutathionylation (Anashkina et al., 2020; Dalle-Donne et al., 2008; Gould et al., 2015; Hill et al., 2010; Mullen et al., 2015). S-glutathionylation protects protein cysteines from irreversible oxidation and, at the same time, regulates various molecular and cellular pathways by affecting the activity of target proteins under physiological and pathological conditions (Adachi et al., 2004; Ghezzi, 2013; Shelton and Mieyal, 2008). For example, glutathionylation of titin inhibits its folding and enhances the elasticity of human cardiomyocytes (Alegre-Cebollada et al., 2014). In hypertensive vessels, glutathionylation of endothelial nitric oxide synthase (eNOS) is increased, leading to decreased activity and impaired endothelium-dependent vasodilation (Chen et al., 2010). Glutathionylated proteins appear uniformly distributed in the cerebral and cerebellar cortex of the human brain (Sparaco et al., 2006). However, how glutathionylation regulates neural development remains unclear.
Cytosolic glutathione S-transferases (GSTs), consisting of multiple classes including Alpha and Omega based on sequence similarity, are best known for their ability to catalyze the conjugation of the reduced form of glutathione (GSH) to xenobiotic substrates for the purpose of detoxification (Hayes et al., 2005; Whitbread et al., 2005). There are two members of the GST Omega class GSTO1 and GSTO2 in mammals, one of which is involved in the glutathionylation cycle (Menon and Board, 2013). Polymorphisms in GSTO1 have been amply reported to affect the onset and progress of several neurological diseases (Menon and Board, 2013). However, how GSTOs affect the activity and function of target proteins in neural development remains largely unknown.
Bone morphogenetic proteins (BMPs) play a crucial role in the development of both the central and peripheral nervous systems in vertebrates (Liu and Niswander, 2005). For example, BMP7 deletion results in reduced cortical thickening and impaired neurogenesis (Segklia et al., 2012). Dysregulation of BMP signaling has also been characterized in neurodevelopmental diseases such as fragile X syndrome, the most common heritable form of intellectual disability and autism spectrum disorder (Kashima et al., 2016). The Drosophila neuromuscular junction (NMJ) has been extensively employed to investigate how BMP signaling is regulated at various steps (Fuentes-Medel et al., 2012; Harris and Littleton, 2015; Kim et al., 2019; Li et al., 2016; McCabe et al., 2003; Wang et al., 2007). For example, the BMP receptor Thickveins (Tkv) is ubiquitinated by Ube3a (Li et al., 2016). Glass bottom boat (Gbb), the Drosophila homolog of mammalian BMPs, acts as a muscle-derived retrograde signal that promotes NMJ synaptic growth (McCabe et al., 2003). However, little is known about how Gbb is regulated at the protein level.
In the present study, we conducted an unbiased RNA interference (RNAi) screen to identify genes in postsynaptic muscles that regulate synapse growth and identified GstO1 as a positive candidate. To determine how GstO1 regulates NMJ synapse development, we performed multiple independent assays and demonstrated negative regulation of Gbb by GstO1 in vivo and in cultured cells. Our results revealed a new mechanism of crosstalk between glutathionylation and ubiquitination, i.e., glutathionylation of Gbb enhanced by GstO1 facilitates its binding to the E3 ligase Ctrip and subsequent ubiquitin-mediated degradation.
Results
Genetic screen to identify new regulators of synapse development
To identify genes in postsynaptic muscles that regulate synapse development, we performed an unbiased screen for the first time by knocking down genes via C57-Gal4-driven expression of RNAi lines obtained from the Tsinghua Fly Center (https://thfc.zzbd.org/en/dl.html). Drosophila third instar larvae were dissected and stained with an antibody-recognizing cysteine string protein (CSP), a synaptic vesicle-associated protein, and anti-horseradish peroxidase (HRP), which labels neuronal membranes (Fig. 1). We manually screened 1,152 randomly selected transgenic RNAi lines for 1,022 genes using NMJ morphology as a readout and identified 21 genes (2% of the total genes screened) associated with apparently either more boutons including satellite boutons or fewer boutons when knocked down by RNAi compared with genetic controls (Fig. 1; and Tables S1 and S2). Specifically, RNAi knockdown of 19 out of the 21 positive genes produced overgrown NMJs, while RNAi knockdown of two uncharacterized genes CG17078 and CG42594 led to the opposite (Fig. 1 and Table S2). The initial results of positive RNAi lines were verified by independent RNAi lines obtained from Bloomington Stock Center whenever possible (Table S2). Several components of BMP (Gbb and Dad) and Wnt (Wnt5, Arr, Dsh, sgg, Arm, and pan; listed in Table S1) pathways that regulate NMJ growth were screened but not identified as positive genes, probably because some, i.e., Dad, Wnt5, and sgg act presynaptically or the RNAi phenotypes were too weak to be detected. For validation of the screen, we used Wsp, which encodes a protein-promoting F-actin formation, as a positive control; knockdown of Wsp in postsynaptic muscles by C57-Gal4-driven RNAi resulted in more boutons as well as more satellite boutons as reported previously (Fig. 1; Nahm et al., 2010a). Among the 21 positive candidate genes, we focused on GstO1 encoding GST omega 1 because its mutant phenotype at NMJs was apparent and robust, and the function of GstO1 and its mammalian homolog GSTO1 is poorly understood.
Altered NMJ terminals caused by postsynaptic knockdown of specific genes. A diagram of an NMJ bouton marked by presynaptic CSP, HRP, and postsynaptic muscle-specific C57-Gal4 is shown at the upper left corner. C57-Gal4-mediated knockdown of specific genes of various functions led to altered NMJ terminals double-labeled with anti-HRP (green) and anti-CSP (magenta). Scale bar, 5 µm.
Altered NMJ terminals caused by postsynaptic knockdown of specific genes. A diagram of an NMJ bouton marked by presynaptic CSP, HRP, and postsynaptic muscle-specific C57-Gal4 is shown at the upper left corner. C57-Gal4-mediated knockdown of specific genes of various functions led to altered NMJ terminals double-labeled with anti-HRP (green) and anti-CSP (magenta). Scale bar, 5 µm.
GstO1 null mutants are viable and fertile
GstO1 is highly conserved from Caenorhabditis elegans to humans. Drosophila GstO1 (CG6662) is 37.3% identical and 62.2% similar to human GSTO1. CG6673 was originally named DmGSTO1 (Kim et al., 2012), but later renamed as GstO2 (Kim and Yim, 2013). To understand the role of GstO1 in NMJ synapse development, we generated GstO16-11, a small deletion generated by CRISPR/Cas9 in which a 121-bp DNA fragment starting from the second base of the start codon ATG is deleted (Fig. 2 A). The GstO1PBac mutant carries a piggyBac transposon insertion in the second exon (Lee et al., 2015). There was no detectable GstO1 protein in hemizygous GstO16-11/Df larval muscles by Western blotting with a self-made rabbit antibody against the C-terminal 228−242 amino acid residues of GstO1 (Fig. 2 B), indicating antibody specificity. There was a faint band (a leaky expression or a non-specific band, as the PBac insertion is expected to lead to a truncation upstream of the antigen) in GstO1PBac mutants but an intense band when UAS-GstO1 was driven by C57-Gal4 (Fig. 2 B). Immunostaining showed that GstO1 was substantially expressed in WT larval muscle cells with a gradual increase in GstO1 abundance from first instar to third instar larvae, but no expression of GstO1 was detected in GstO16-11 while a residual GstO1 expression was detected in GstO1PBac mutants (Fig. 2 C and Fig. S1). Hemizygous GstO16-11/Df mutants were fully viable with no obvious developmental abnormalities, and both males and females were fertile, indicating that GstO1 is not required for viability or fertility.
GstO1 inhibits NMJ synapse development postsynaptically. (A) Intron–exon organization and different mutations of GstO1. Exon and intron are indicated by boxes and gaps, respectively. The antigen for antibody production is indicated by a gray bar. P-element insertion GstO1PBac was indicated by a triangle. (B) Western analysis of different genotypes using an anti-GstO1 antibody. No GstO1 expression was detected in hemizygous GstO16-11/Def mutants. GstO1 OE denotes UAS-GstO1/+; C57-Gal4/+. Flag denotes a Flag tag knocked in at the C-terminus of the endogenous GstO1. Tubulin was used as the loading control. The loading of GstO1 OE was one-seventh of the other. (C) Images of coimmunostaining with anti-GstO1 (green) and nuclear marker To-Pro-I3 (blue) of muscles of different genotypes. The genotypes are WT, GstO1PBac/GstO1PBac, GstO16-11/Def, and GstO1 OE (UAS-GstO1/+; C57-Gal4/+). Scale bar, 5 μm. (D) All images were projections of confocal z-stacks of synapses double-stained with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, GstO1PBac, GstO16-11/Def, Mus RNAi: C57-Gal4/GstO1 RNAi, Neu RNAi: OK6-Gal4/+; GstO1 RNAi, Mus res: UAS-GstO1/+; C57-Gal4 GstO16-11/Def, and Neu res: UAS-GstO1/OK6-Gal4; GstO16-11/Def. Scale bar, 10 μm. (E and F) Statistical results of total bouton number (E) and satellite bouton number (F) in different genotypes; n ≥11 for each genotype; arrows indicate satellite boutons; ***P < 0.001 by one-way ANOVA with Tukey post hoc test; ns means no significance; error bars denote SEM. Source data are available for this figure: SourceData F2.
GstO1 inhibits NMJ synapse development postsynaptically. (A) Intron–exon organization and different mutations of GstO1. Exon and intron are indicated by boxes and gaps, respectively. The antigen for antibody production is indicated by a gray bar. P-element insertion GstO1PBac was indicated by a triangle. (B) Western analysis of different genotypes using an anti-GstO1 antibody. No GstO1 expression was detected in hemizygous GstO16-11/Def mutants. GstO1 OE denotes UAS-GstO1/+; C57-Gal4/+. Flag denotes a Flag tag knocked in at the C-terminus of the endogenous GstO1. Tubulin was used as the loading control. The loading of GstO1 OE was one-seventh of the other. (C) Images of coimmunostaining with anti-GstO1 (green) and nuclear marker To-Pro-I3 (blue) of muscles of different genotypes. The genotypes are WT, GstO1PBac/GstO1PBac, GstO16-11/Def, and GstO1 OE (UAS-GstO1/+; C57-Gal4/+). Scale bar, 5 μm. (D) All images were projections of confocal z-stacks of synapses double-stained with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, GstO1PBac, GstO16-11/Def, Mus RNAi: C57-Gal4/GstO1 RNAi, Neu RNAi: OK6-Gal4/+; GstO1 RNAi, Mus res: UAS-GstO1/+; C57-Gal4 GstO16-11/Def, and Neu res: UAS-GstO1/OK6-Gal4; GstO16-11/Def. Scale bar, 10 μm. (E and F) Statistical results of total bouton number (E) and satellite bouton number (F) in different genotypes; n ≥11 for each genotype; arrows indicate satellite boutons; ***P < 0.001 by one-way ANOVA with Tukey post hoc test; ns means no significance; error bars denote SEM. Source data are available for this figure: SourceData F2.
Increased GstO1 expression during larval development. (A–C) Representative confocal images of WT muscle cells at different larval stages (L1, L2, and L3) triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue, circled by white dash). Scale bar, 5 µm. (D) Quantification of GstO1 and Gbb levels at different larval stages in arbitrary units (a.u.). n = 10 muscle cells from 10 larvae, one cell per larva. *P < 0.05, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
Increased GstO1 expression during larval development. (A–C) Representative confocal images of WT muscle cells at different larval stages (L1, L2, and L3) triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue, circled by white dash). Scale bar, 5 µm. (D) Quantification of GstO1 and Gbb levels at different larval stages in arbitrary units (a.u.). n = 10 muscle cells from 10 larvae, one cell per larva. *P < 0.05, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
GstO1 inhibits NMJ development postsynaptically
To explore the role of GstO1 at synapses, we examined NMJ morphology in GstO1 mutants. WT muscle 4 NMJ (NMJ4) showed normal morphology when co-stained with anti-CSP and anti-HRP. NMJ synapses in hemizygous GstO16-11/Df and homozygous GstO1PBac mutants were obviously overgrown with more boutons, including satellite boutons (Fig. 2, D–F). The mean total number of synaptic boutons per NMJ4 was 40.81 ± 0.55 in hemizygous GstO16-11/Df mutants, significantly higher than 24.87 ± 0.56 in WT (P < 0.001; Fig. 2, D–F). Homozygous GstO1PBac mutants also showed more total boutons than WT control (Fig. 2, D–F). The mean number of satellite boutons in both GstO16-11/Df and GstO1PBac mutants was more than eightfold higher than WT (P < 0.001; Fig. 2, D–F).
To determine whether GstO1 acts on the pre- or postsynaptic side, we conducted tissue-specific knockdown of GstO1 expression and examined its effect on NMJ development. Specific knockdown of GstO1 in post-synaptic muscles by C57-Gal4-driven RNAi resulted in more boutons as well as more satellite boutons compared with WT (24.87 ± 0.56 boutons for WT and 36.07 ± 0.37 boutons for C57-Gal4/GstO1 RNAi, P < 0.001; 1.78 ± 0.22 satellite boutons for WT and 14.27 ± 0.30 for C57-Gal4/GstO1 RNAi, P < 0.001; Fig. 2, D–F). By contrast, RNAi knockdown in presynaptic neurons by OK6-Gal4 revealed no significant effect on overall bouton number (24.87 ± 0.56 boutons for WT and 24.73 ± 0.44 boutons for OK6-Gal4/+; GstO1 RNAi/+, P < 0.001; Fig. 2, D–F). These tissue-specific knockdown results suggest that GstO1 postsynaptically regulates NMJ synapse development. To further verify this possibility, we carried out tissue-specific rescue experiments. Postsynaptic overexpression of GstO1 driven by the muscle-specific C57-Gal4 in GstO16-11 mutant background restored the overgrown NMJs to the WT level (24.87 ± 0.56 total boutons for WT and 25.20 ± 0.32 total boutons for UAS-GstO1/+; C57-Gal4 GstO16-11/Df; Fig. 2, D–F), while overexpression of GstO1 in presynaptic neurons driven by OK6-Gal4 showed no rescue of the mutant phenotype (Fig. 2, D–F). These results demonstrate that GstO1 inhibits NMJ growth postsynaptically.
Increased BMP signaling leads to NMJ overgrowth in GstO1 mutants
The role of multiple signaling pathways such as BMP, Wingless, and c-Jun N-terminal kinase (JNK) in regulating synaptic growth has been well characterized at the Drosophila NMJs (Harris and Littleton, 2015; Huang et al., 2018; Korkut and Budnik, 2009; Mathew et al., 2005; Milton et al., 2011). For example, mutations in retrograde BMP signaling components lead to a decrease in total boutons including satellite boutons (McCabe et al., 2003), while increased BMP signaling leads to NMJ overgrowth with more synaptic boutons (Kim et al., 2019; Li et al., 2016), similar to what we observed in GstO1 mutants. Therefore, we wondered whether GstO1 participates in the regulation of BMP signaling at NMJ synapses. BMP activation leads to phosphorylation of the transcriptional activator Mad (pMad), which then translocates to the nucleus to activate target gene transcription. Since pMad accumulation at NMJ termini is an indicator of retrograde BMP signaling at the NMJ (Li et al., 2016; Zhao et al., 2015), we tested whether pMad intensity was affected in GstO1 mutants. In homozygous GstO1PBac and hemizygous GstO16-11/Df mutant NMJs, the intensity of pMad-positive punctae was apparently increased compared with WT controls (Fig. 3, A–C, and F). Consistent with the increased pMad intensity in mutants, muscle-specific RNAi knockdown of GstO1 also caused increased pMad intensity (Fig. 3, A, D, and F). Statistically, pMad signal intensity (a.u.) at NMJ synapses was significantly increased by 6.54 folds in GstO1PBac mutants, 6.7 folds in GstO16-11/Df mutants, and 5.59 folds following RNAi knockdown compared with WT controls (P < 0.001; Fig. 3 F). Postsynaptic re-expression of GstO1 by the muscle-specific C57-Gal4 in GstO16-11 mutant background restored pMad to WT levels (1.136 ± 0.056 a.u. for WT and 1.129 ± 0.045 a.u. for UAS-GstO1/+; C57-Gal4 GstO16-11/Df; Fig. 3, A, C, E, and F). These results show that GstO1 inhibits pMad levels at NMJ synapses.
Increased BMP signaling leads to overgrown NMJs in GstO1 mutants. (A–E) Confocal images of NMJ4 co-labeled with anti-pMad (green) and anti-HRP (magenta). pMad level was obviously upregulated in homozygous GstO1PBac, hemizygous GstO16-11/Def, and GstO1 RNAi knockdown by C57-Gal4 compared with WT. (F) Quantification of the relative fluorescence intensities of pMad at NMJ terminals of different genotypes (n ≥10 NMJs). Scale bar, 10 µm. The genotypes are WT, GstO1PBac, GstO16-11/Def, C57-Gal4/GstO1 RNAi, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. (G–N) Confocal images of NMJ4 double-labeled with anti-HRP (green) and anti-CSP (magenta). Heterozygous mutations of gbb4, tkv8, and mad12 rescued synaptic overgrowth of GstO1 mutants to WT level. (O) Quantification of synaptic bouton numbers of different genotypes including WT, GstO16-11/Def, gbb4/+, tkv8/+, mad12/+, gbb4/+; GstO16-11/Def, tkv8/+; GstO16-11/Def, and mad12/+; GstO16-11/Def. Scale bar, 10 µm; Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM.
Increased BMP signaling leads to overgrown NMJs in GstO1 mutants. (A–E) Confocal images of NMJ4 co-labeled with anti-pMad (green) and anti-HRP (magenta). pMad level was obviously upregulated in homozygous GstO1PBac, hemizygous GstO16-11/Def, and GstO1 RNAi knockdown by C57-Gal4 compared with WT. (F) Quantification of the relative fluorescence intensities of pMad at NMJ terminals of different genotypes (n ≥10 NMJs). Scale bar, 10 µm. The genotypes are WT, GstO1PBac, GstO16-11/Def, C57-Gal4/GstO1 RNAi, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. (G–N) Confocal images of NMJ4 double-labeled with anti-HRP (green) and anti-CSP (magenta). Heterozygous mutations of gbb4, tkv8, and mad12 rescued synaptic overgrowth of GstO1 mutants to WT level. (O) Quantification of synaptic bouton numbers of different genotypes including WT, GstO16-11/Def, gbb4/+, tkv8/+, mad12/+, gbb4/+; GstO16-11/Def, tkv8/+; GstO16-11/Def, and mad12/+; GstO16-11/Def. Scale bar, 10 µm; Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM.
Increased BMP signaling may mediate the overgrowth of synaptic boutons in GstO1 mutants. To test this possibility, we examined genetic interactions between GstO1 and BMP signaling components such as the BMP homolog Gbb, the receptor Tkv, and the downstream effector Mad. Mutating one copy of gbb (heterozygous gbb4 mutation) had no effect on NMJ growth but significantly reduced the number of excess boutons in GstO1 mutants (Fig. 3, G, I, L, and O). Similarly, loss of one copy of tkv and mad showed no effect on bouton number, but reduced the number of boutons to WT levels in GstO1 mutants (Fig. 3, G, J, K, and M–O). Together, these results demonstrate that synaptic overgrowth in GstO1 mutants results from increased BMP signaling.
GstO1 downregulates Gbb protein levels in vivo and in S2 cells
As GstO1 acts in postsynaptic muscles in which Gbb is produced and secreted as a retrograde signal, we hypothesized that GstO1 may negatively regulate Gbb expression to inhibit BMP signaling. Indeed, we found that Gbb levels were upregulated in GstO16-11/Df mutants compared with WT (8.358 ± 0.187 a.u. versus 2.068 ± 0.174 a.u. for Gbb intensity; Fig. 4, A, B and E). Conversely, overexpression of GstO1 in the WT background reduced Gbb intensity (2.068 ± 0.174 a.u. for WT and 0.485 ± 0.095 a.u. for UAS-GstO1/+; C57-Gal4/+; Fig. 4, A, C, and E). Moreover, postsynaptic re-expression of GstO1 driven by the muscle-specific C57-Gal4 in GstO16-11 mutant background restored Gbb protein to WT levels (2.068 ± 0.174 a.u. for WT and 2.25 ± 0.17 a.u. for UAS-GstO1/+; C57-Gal4 GstO16-11/Df; Fig. 4, A–E).
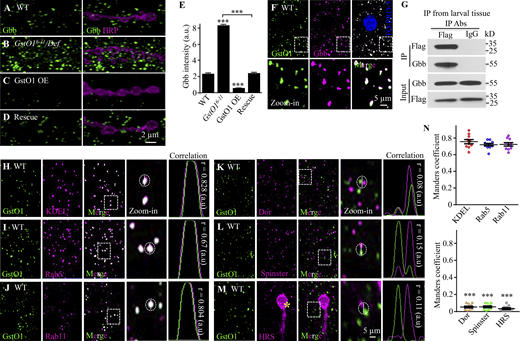
GstO1 negatively regulates Gbb protein levels and co-localizes with Gbb. (A–D) Confocal images of NMJ 4 colabeled with anti-Gbb (green) and anti-HRP (magenta). The genotypes are WT, GstO16-11/Def, UAS-GstO1/+; C57-Gal4/+, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. Scale bar, 2 µm. (E) Quantification of cytoplasmic Gbb level of different genotypes in arbitrary units (a.u.). n = 12 NMJs, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (F) Representative confocal images of WT muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-I3 (blue). The zoom-in views of the boxed area are shown in the bottom row. Scale bar, 5 µm. (G) Gbb interacts with Flag-tagged endogenous GstO1 in larval muscles as detected by co-IP. Gbb was coimmunoprecipitated by anti-Flag from larvae expressing endogenous GstO1 tagged with Flag. (H–J) GstO1 is colocalized with the ER marker KDEL (H), the early endosomal marker Rab5 (I), and the recycling endosomal marker Rab11 (J) in muscle cells. (K–M) GstO1 is not colocalized with the late endosomal markers Dor (K), Spinster (L), and Hrs (M) in muscle cells. The yellow asterisk in M denotes NMJ boutons. Scale bar, 5 µm. (N) Manders coefficient as a measure of colocalization of GstO1 with different markers. n = 10 muscle cells from 10 larvae. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F4.
GstO1 negatively regulates Gbb protein levels and co-localizes with Gbb. (A–D) Confocal images of NMJ 4 colabeled with anti-Gbb (green) and anti-HRP (magenta). The genotypes are WT, GstO16-11/Def, UAS-GstO1/+; C57-Gal4/+, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. Scale bar, 2 µm. (E) Quantification of cytoplasmic Gbb level of different genotypes in arbitrary units (a.u.). n = 12 NMJs, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (F) Representative confocal images of WT muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-I3 (blue). The zoom-in views of the boxed area are shown in the bottom row. Scale bar, 5 µm. (G) Gbb interacts with Flag-tagged endogenous GstO1 in larval muscles as detected by co-IP. Gbb was coimmunoprecipitated by anti-Flag from larvae expressing endogenous GstO1 tagged with Flag. (H–J) GstO1 is colocalized with the ER marker KDEL (H), the early endosomal marker Rab5 (I), and the recycling endosomal marker Rab11 (J) in muscle cells. (K–M) GstO1 is not colocalized with the late endosomal markers Dor (K), Spinster (L), and Hrs (M) in muscle cells. The yellow asterisk in M denotes NMJ boutons. Scale bar, 5 µm. (N) Manders coefficient as a measure of colocalization of GstO1 with different markers. n = 10 muscle cells from 10 larvae. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F4.
In addition to the negative regulation of cytoplasmic Gbb by GstO1 (Fig. 4, A–E), we determined the level of secreted extracellular Gbb upon altered expressions of GstO1 by immunostaining in a detergent-free system following published protocols (James et al., 2014). Extracellular Gbb was robustly expressed at the postsynaptic NMJ termini (Fig. S2), in agreement with a previous report (James et al., 2014). We found that extracellular Gbb levels at NMJs were also upregulated in GstO16-11/Df mutants (8.145 ± 0.158 a.u.) compared with WT (2.708 ± 0.124 a.u.; Fig. S2, A, B, and E). Overexpression of GstO1 driven by C57-Gal4 reduced extracellular Gbb levels compared with WT (2.708 ± 0.124 a.u. for WT and 1.25 ± 0.197 a.u. for UAS-GstO1/+; C57-Gal4/+; Fig. S2, A, C, and E). As expected, postsynaptic re-expression of GstO1 driven by C57-Gal4 in the GstO16-11 mutant background restored extracellular Gbb to WT levels (2.708 ± 0.124 a.u. for WT and 2.603 ± 0.124 a.u. for UAS-GstO1/+; C57-Gal4 GstO16-11/Df; Fig. S2, A–E). The negative regulation of Gbb by GstO1 was at the post-transcriptional level, as the mRNA level of Gbb remained unchanged when GstO1 expression was altered (Fig. S2 F).
GstO1 negatively regulates Gbb levels in vivo and in S2 cells. (A–D) Confocal images of NMJs co-stained with anti-Gbb (green) and anti-HRP (magenta) in detergent-free conditions. Gbb was upregulated in GstO1 mutants. Scale bar, 10 µm. (E) Quantification of extracellular Gbb intensity of different genotypes including WT, GstO16-11/Def, UAS-GstO1/+; C57-Gal4/+, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. Scale bar, 10 µm. n = 10 NMJs, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (F) The gbb mRNA level normalized to the RpL28 mRNA level in larval muscles of WT, GstO16-11/Df mutants, and GstO1 OE (UAS-GstO1/+; C57-Gal4/+) animals. No significant difference in gbb mRNA levels between the genotypes by one-way ANOVA with Tukey’s post hoc test. n = 4, error bars indicate SEM. (G) Gbb proteins in cell lysates and culture medium of S2 cells with altered expression of GstO1 were detected by Western analysis using anti-Gbb. Both precursor Gbb and processed Gbb protein levels were increased in GstO1 RNAi knockdown cells. On the contrary, reduced precursor Gbb and processed Gbb protein levels were observed in GstO1-overexpressing cells. Tubulin was used for loading control. (H) Quantitative analysis of the level of processed Gbb. n = 3, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData FS2.
GstO1 negatively regulates Gbb levels in vivo and in S2 cells. (A–D) Confocal images of NMJs co-stained with anti-Gbb (green) and anti-HRP (magenta) in detergent-free conditions. Gbb was upregulated in GstO1 mutants. Scale bar, 10 µm. (E) Quantification of extracellular Gbb intensity of different genotypes including WT, GstO16-11/Def, UAS-GstO1/+; C57-Gal4/+, and UAS-GstO1/+; C57-Gal4 GstO16-11/Def. Scale bar, 10 µm. n = 10 NMJs, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (F) The gbb mRNA level normalized to the RpL28 mRNA level in larval muscles of WT, GstO16-11/Df mutants, and GstO1 OE (UAS-GstO1/+; C57-Gal4/+) animals. No significant difference in gbb mRNA levels between the genotypes by one-way ANOVA with Tukey’s post hoc test. n = 4, error bars indicate SEM. (G) Gbb proteins in cell lysates and culture medium of S2 cells with altered expression of GstO1 were detected by Western analysis using anti-Gbb. Both precursor Gbb and processed Gbb protein levels were increased in GstO1 RNAi knockdown cells. On the contrary, reduced precursor Gbb and processed Gbb protein levels were observed in GstO1-overexpressing cells. Tubulin was used for loading control. (H) Quantitative analysis of the level of processed Gbb. n = 3, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData FS2.
To further verify the in vivo negative regulation of GstO1 on Gbb, we knocked down GstO1 in Drosophila S2 cells and assessed the level of intracellular and extracellular Gbb by Western analysis. Unprocessed full-length cytoplasmic Gbb in cell lysates was increased when GstO1 was knocked down by double-stranded RNA (dsRNA)-mediated RNAi compared with controls (Fig. S2 G). Knockdown of GstO1 also increased the level of processed, secreted mature Gbb in the culture medium (Fig. S2, G and H), consistent with the in vivo staining results (Fig. S2, A, B, and E). These results from in vivo and S2 cell studies together demonstrate that GstO1 negatively regulates the Gbb protein level.
Did the negative regulation of Gbb by GstO1 occur only in the neuromusculature or in other tissues such as the wing disc as well? Immunostaining showed that both GstO1 and Gbb were evenly expressed in the wing disc. When GstO1 was knocked down by the ubiquitous act-Gal4, we observed an increased level of Gbb in the wing disc compared with WT (Fig. S3), demonstrating a negative regulation of Gbb by GstO1 as in muscle cells and S2 cells.
Negative regulation of Gbb by GstO1 in the wing disc. (A and B) Representative confocal images of WT and act-Gal4/GstO1 RNAi wing disc cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue). Scale bar, 50 µm. (C) Quantification of GstO1 and Gbb levels in wing disc in arbitrary units (a.u.). n = 10 pairs of wing discs from 10 larvae, one pair per larva. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
Negative regulation of Gbb by GstO1 in the wing disc. (A and B) Representative confocal images of WT and act-Gal4/GstO1 RNAi wing disc cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue). Scale bar, 50 µm. (C) Quantification of GstO1 and Gbb levels in wing disc in arbitrary units (a.u.). n = 10 pairs of wing discs from 10 larvae, one pair per larva. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
GstO1 interacts and colocalizes with Gbb
Given the negative regulation of Gbb by GstO1 in vivo and in S2 cells (Fig. 4, A–E and Fig. S2), we hypothesized that Gbb may be a target for GstO1-mediated glutathionylation. This hypothesis predicts a physical interaction between the two. Immunostaining showed perfect colocalization of endogenous GstO1 and Gbb in muscle cells (Fig. 4 F). Furthermore, we found a physical interaction between GstO1 and Gbb by coimmunoprecipitation (co-IP). In third-instar muscles, Gbb was co-IPed with Flag-tagged endogenous GstO1 by anti-Flag but was absent in mouse IgG IPed controls (Fig. 4 G). Together, the colocalization and co-IP results support a physical interaction between GstO1 and Gbb.
GstO1-positive punctae were suggestive of a membrane organelle distribution. To test this possibility, we analyzed the colocalization of GstO1 with markers of various organelles such as ER and endosomes. We observed a near-perfect colocalization of GstO1 with the ER marker KDEL, and substantial overlap with the early endosomal marker Rab5 and the recycling endosomal marker Rab11 in third-instar larval muscle cells (Fig. 4, H–J, and N). As a control, we did not observe any colocalization or juxtaposition of GstO1 with the late endosomal markers Dor, Spinster, and HRS (Fig. 4, K−N). These results together show that GstO1 is localized at ER and early/recycling endosomes, but not at late endosomes.
The negative regulation of Gbb by GstO1 occurs under hypoxia and in hyperexcitable mutants
Did the negative regulation of Gbb by GstO1 occur upon intrinsic and extrinsic stimuli? To address this possibility, we examined GstO1 and Gbb expression levels in third instar larval muscles under hypoxic conditions (2.5% O2) for 16 h and normal conditions (21% O2). The larvae were sluggish and grew slower under hypoxia, but the locomotion recovered when put back to a normal oxygen concentration. We found that GstO1 expression was reduced while Gbb increased under hypoxic conditions (Fig. 5, A–C). Consistent with an increased level of Gbb, the NMJ terminals were overgrown with more boutons and satellite boutons under hypoxia (Fig. 5, D and E).
Reduced GstO1 expression induced by hypoxia and increased neural activity. (A and B) Representative confocal images of WT muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue; circled by white dash) in normal oxygen (21% O2; A) and hypoxia (2.5% O2; B). Scale bar, 5 µm. (C) Quantification of GstO1 and Gbb levels in arbitrary units (a.u.) under normal oxygen and hypoxia. n = 10 muscle cells from 10 larvae, one muscle per larva. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (D) Confocal images of WT NMJ4 co-labeled with anti-CSP (magenta) and anti-HRP (green) in normal oxygen and hypoxia. Scale bar, 10 µm. The zoom-in views of the boxed area are shown in the right panels. (E) Statistical results of total bouton number and satellite bouton number in normal oxygen and hypoxia; n ≥10 larvae for each genotype; ***P < 0.001 by one-way ANOVA with Tukey post hoc test; error bars denote SEM. (F and G) Representative confocal images of WT and eag1Sh120 mutant muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue, circled by white dash). Scale bar, 5 µm. (H) Quantification of GstO1 and Gbb levels in arbitrary units (a.u.). n = 10 muscle cells from 10 larvae, one cell per larva. ***P <0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
Reduced GstO1 expression induced by hypoxia and increased neural activity. (A and B) Representative confocal images of WT muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue; circled by white dash) in normal oxygen (21% O2; A) and hypoxia (2.5% O2; B). Scale bar, 5 µm. (C) Quantification of GstO1 and Gbb levels in arbitrary units (a.u.) under normal oxygen and hypoxia. n = 10 muscle cells from 10 larvae, one muscle per larva. ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. (D) Confocal images of WT NMJ4 co-labeled with anti-CSP (magenta) and anti-HRP (green) in normal oxygen and hypoxia. Scale bar, 10 µm. The zoom-in views of the boxed area are shown in the right panels. (E) Statistical results of total bouton number and satellite bouton number in normal oxygen and hypoxia; n ≥10 larvae for each genotype; ***P < 0.001 by one-way ANOVA with Tukey post hoc test; error bars denote SEM. (F and G) Representative confocal images of WT and eag1Sh120 mutant muscle cells triple-stained with anti-GstO1 (green), anti-Gbb (magenta), and nuclear marker anti-To-Pro-3 Iodide (blue, circled by white dash). Scale bar, 5 µm. (H) Quantification of GstO1 and Gbb levels in arbitrary units (a.u.). n = 10 muscle cells from 10 larvae, one cell per larva. ***P <0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM.
We also examined the effect of neural activity on GstO1 and Gbb expression. Immunostaining of the hyperexcitable eag1Sh120 mutants in which neuronal activity is increased (Budnik et al., 1990; Zhong and Wu, 2004) showed that GstO1 expression was reduced concomitant with increased Gbb in eag1Sh120 mutants compared with WT (Fig. 5, F–H). The upregulated Gbb was consistent with the increased NMJ terminals observed in eag1Sh120 mutants (Budnik et al., 1990). The reason for the reduced GstO1 expression in hyperexcitable mutants is currently unknown.
GstO1 is required and sufficient for Gbb glutathionylation in vivo and in S2 cells
S-glutathionylation involves mixed disulfide bond formation between cysteine residues and GSH, a tripeptide consisting of glycine, cysteine, and glutamate (Dalle-Donne et al., 2009). To determine if Gbb S-glutathionylation was affected by GstO1, we examined Gbb glutathionylation levels when GstO1 expression was altered (Fig. 6 A). Endogenous Gbb was IPed from larval muscles using an anti-Gbb antibody followed by Western blotting with anti-GSH, which detects glutathione–protein complexes under non-reducing conditions. We found that Gbb was glutathionylated in WT (Fig. 6 A), but no glutathionylation of Gbb was detected in the absence of GstO1; conversely, Gbb glutathionylation was elevated when GstO1 was overexpressed in postsynaptic muscles compared with WT (Fig. 6 A).
Glutathionylation of Gbb at Cys354 and Cys420 mediated by GstO1. (A) GstO1 positively regulates glutathionylation of Gbb. Gbb was immunoprecipitated by anti-Gbb, electrophoresed by non-reducing SDS-PAGE, and probed with anti-GSH and anti-Gbb antibodies. (B) Detection of glutathionylation at Cys420 and Cys354 of Gbb by mass spectrometry. Cys420 and Cys354 in the peptides of aa 352–363 and aa 414–424 of Gbb, respectively, are glutathionylated and illustrated along the peptide sequence shown at the top. The b- and y-type product ions are marked on the spectrum. (C) Verification of Gbb glutathionylation sites by mutational analysis. S2 cells were co-transfected with plasmids encoding Flag-tagged GstO1 and Myc tagged WT or mutant Gbb. Anti-GSH was used to detect glutathionylated Gbb. Tubulin was used as a loading control. (D) Quantitative analysis of the relative Gbb-SSG/Gbb ratio and Gbb/tubulin ratio. n = 3, ns, no significance, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F6.
Glutathionylation of Gbb at Cys354 and Cys420 mediated by GstO1. (A) GstO1 positively regulates glutathionylation of Gbb. Gbb was immunoprecipitated by anti-Gbb, electrophoresed by non-reducing SDS-PAGE, and probed with anti-GSH and anti-Gbb antibodies. (B) Detection of glutathionylation at Cys420 and Cys354 of Gbb by mass spectrometry. Cys420 and Cys354 in the peptides of aa 352–363 and aa 414–424 of Gbb, respectively, are glutathionylated and illustrated along the peptide sequence shown at the top. The b- and y-type product ions are marked on the spectrum. (C) Verification of Gbb glutathionylation sites by mutational analysis. S2 cells were co-transfected with plasmids encoding Flag-tagged GstO1 and Myc tagged WT or mutant Gbb. Anti-GSH was used to detect glutathionylated Gbb. Tubulin was used as a loading control. (D) Quantitative analysis of the relative Gbb-SSG/Gbb ratio and Gbb/tubulin ratio. n = 3, ns, no significance, ***P < 0.001 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F6.
To determine if GstO1-mediated glutathionylation affected the trafficking of Gbb, we analyzed colocalization of Gbb with various membrane organelles when GstO1 was knocked down (KD). The percentages of Gbb colocalization with different membrane organelles were 62.22% for ER, 85.71% for Rab5, and 74.41% for Rab11 in WT controls (Fig. S4, A, C, and E). In GstO1 KD cells, the percentages of Gbb colocalization with different membrane organelles were 37.11% for ER, 25.21% for Rab5, and 35.92% for Rab11 (Fig. S4, B, D and F), apparently lower than that in WT controls, probably due to more Gbb positive puncta in GstO1 KD animals. As with GstO1 (Fig. 4, K−N), no co-localization of Gbb with late endosomes labeled by anti-HRS and anti-Spinster (Fig. S4, G and H) was observed. As a substantial subset of Gbb puncta was not co-localized with any membrane organelles in both WT and GstO1 KD animals, i.e., these Gbb puncta were not on the trafficking route, we were unable to conclude definitively if there was a defect in Gbb trafficking when GstO1-mediated glutathionylation was disrupted.
Colocalization of Gbb with ER and endosomes. (A–F) Representative confocal images of WT and GstO1 knockdown muscle cells double-stained with anti-Gbb (green) and different organelle markers (magenta). Gbb is colocalized with a subset of ER (KDEL positive; A and B), early endosome (Rab5 positive; C and D), and recycling endosome (Rab11 positive; E and F) in muscle cells of WT controls (A, C, and E) and GstO1 KD (B, D, and F) animals. Scale bar, 5 µm. (G and H) Gbb does not co-localize with late endosomes labeled by anti-HRS (G) and anti-Spinster (H). The yellow asterisk indicates NMJ boutons. (I) Increased ratio of GSH/GSSG in GstO1 mutants (GstO16-11/Def) and rescued by postsynaptic overexpression of GstO1. OE denotes overexpression of GstO1 driven by C57-Gal4.
Colocalization of Gbb with ER and endosomes. (A–F) Representative confocal images of WT and GstO1 knockdown muscle cells double-stained with anti-Gbb (green) and different organelle markers (magenta). Gbb is colocalized with a subset of ER (KDEL positive; A and B), early endosome (Rab5 positive; C and D), and recycling endosome (Rab11 positive; E and F) in muscle cells of WT controls (A, C, and E) and GstO1 KD (B, D, and F) animals. Scale bar, 5 µm. (G and H) Gbb does not co-localize with late endosomes labeled by anti-HRS (G) and anti-Spinster (H). The yellow asterisk indicates NMJ boutons. (I) Increased ratio of GSH/GSSG in GstO1 mutants (GstO16-11/Def) and rescued by postsynaptic overexpression of GstO1. OE denotes overexpression of GstO1 driven by C57-Gal4.
Similar to other forms of PTMs, glutathionylation is site-specific, occurring only at certain cysteine residues of a target protein. There are seven cysteine residues in Gbb protein. To identify the potential glutathionylated cysteines, we purified His-tagged, C-terminal Gbb fragment of amino acids 336–455 containing all seven cysteines from Escherichia coli; the Gbb fragment was then glutathionylated in vitro with diamide and GSH, and subjected to proteolytic digestion and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Peptides with a mass difference of 305 Da, representing one glutathione moiety, were detected by LC-MS. We identified two glutathionylated cysteine residues in His-Gbb, namely Cys354 (C354) and Cys420 (C420; Fig. 6 B). To verify the specific sites of Gbb glutathionylated by GstO1, we mutated cysteine to alanine to generate C354A and C420A single mutants, as well as C419A as a control. Glutathionylation analysis by anti-GSH showed that C419A but not C354A, C420A, or C354A C420A double mutant Gbb was as sensitive as WT for glutathionylation in S2 cells (Fig. 6 C). Concomitant with increased glutathionylation after GstO1 overexpression, there was an appreciable reduction in Myc-tagged WT and C419A Gbb protein levels, while Gbb proteins with C354A, C420A, or C354A C420A double mutations were stable (Fig. 6 C). Statistically, the ratio of glutathionylated WT or C419A Gbb to the total Gbb was significantly higher when GstO1 was overexpressed than that of the GFP overexpression control (Fig. 6 D). We noticed the effect of C354A and C420A single mutation on glutathionylation level and Gbb stability was similar to that of double mutations (Fig. 6, C and D). The reason for this is currently unknown, but possible explanations are that disruption of glutathionylation at one site might affect the glutathionylation of the other, the changes in glutathionylation levels between single and double mutant Gbb were subtle to be detected by Western analysis, or both.
To further determine the in vivo functions of the S-glutathionylation sites in Gbb, we generated mutant lines carrying C354A, C419A, and C420A single mutations. C420A mutation led to significantly more total boutons (35.50 ± 0.65) compared with WT (26.17 ± 0.72; Fig. 7, A, C, and E). As with the C420A mutation, the C354A mutation also resulted in synapse overgrowth whereas C419A did not (Fig. 7, B–E). Consistent with the overgrown NMJs, we found that Gbb intensities in both C420A and C354A mutants were significantly increased (5.57 ± 0.37 a.u. for gbbC420A/+ and 5.17 ± 0.27 a.u. for gbbC354A/+), whereas the Gbb intensity in C419A mutants remained normal (1.90 ± 0.17 a.u. for gbbC419A/+ versus 2.38 ± 0.31 a.u for WT; Fig. 7, F−J). These results from in vivo and cell culture studies together demonstrate that Gbb is selectively glutathionylated at C354 and C420 but not at C419.
Mutation of Gbb glutathionylation sites leads to overgrown NMJ synapses and an increased Gbb protein level. (A–D) Confocal images of NMJs double-stained with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, gbbC419A/+, gbbC420A/+, gbbC354A/+. Both gbbC420A/+ and gbbC354A/+ showed synaptic overgrowth with more total boutons including satellite boutons. (E) Quantification of the total bouton numbers of different genotypes. Scale bar, 10 µm. n ≥12 NMJs for each genotype, ***P < 0.001; error bars denote SEM. (F–I) Confocal images of NMJs from different genotypes co-stained with anti-Gbb (green) and anti-HRP (magenta). Intracellular Gbb intensity was upregulated when one of the glutathionylation sites was mutated. (J) Quantification of Gbb intensity of different genotypes in arbitrary units (a.u.). Scale bar, 2 µm. n ≥10 NMJs for each genotype. ***P < 0.001; error bars denote SEM.
Mutation of Gbb glutathionylation sites leads to overgrown NMJ synapses and an increased Gbb protein level. (A–D) Confocal images of NMJs double-stained with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, gbbC419A/+, gbbC420A/+, gbbC354A/+. Both gbbC420A/+ and gbbC354A/+ showed synaptic overgrowth with more total boutons including satellite boutons. (E) Quantification of the total bouton numbers of different genotypes. Scale bar, 10 µm. n ≥12 NMJs for each genotype, ***P < 0.001; error bars denote SEM. (F–I) Confocal images of NMJs from different genotypes co-stained with anti-Gbb (green) and anti-HRP (magenta). Intracellular Gbb intensity was upregulated when one of the glutathionylation sites was mutated. (J) Quantification of Gbb intensity of different genotypes in arbitrary units (a.u.). Scale bar, 2 µm. n ≥10 NMJs for each genotype. ***P < 0.001; error bars denote SEM.
S-glutathionylation is a reversible process. Glutaredoxins (Grxs) are generally involved in the reduction of oxidative modifications using glutathione and represent the major enzymes responsible for deglutathionylation (Stroher and Millar, 2012). We sought to identify the gene that acts to deglutathionylate Gbb by a genetic approach. In the Drosophila genome, there are seven putative genes encoding glutaredoxins: Grx1, glutaredoxin 1 testis-specific (Grx1t), CG14407, CG9147, CG6523, CG31559, and CG12206. We hypothesized that overexpression of a specific Grx might recapitulate the overgrown NMJ phenotype and increase Gbb in GstO1 mutants. Indeed, overexpression of Grx1 but no other glutaredoxins produced more total boutons compared with WT (24.80 ± 0.84 boutons for WT and 40.60 ± 2.60 boutons for C57-Gal4/UAS-Grx1; Fig. 8, A, B, and E). The increased bouton number was rescued by co-overexpression of GstO1 and Grx1 in postsynaptic muscles (UAS-GstO1/+; UAS-Grx1/C57-Gal4; Fig. 8, A, D, and E). Consistently, we observed significantly upregulated cytoplasmic Gbb in C57-Gal4/UAS-Grx1 muscles (6.70 ± 0.28 a.u.) compared with WT (2.45 ± 0.21 a.u.; Fig. 8, F, G, and J). Postsynaptic co-overexpression of GstO1 and Grx1 by C57-Gal4 restored cytoplasmic Gbb to WT levels (Fig. 8, F−J). Thus, Grx1 appears to be the main, if not the only, deglutathionylating enzyme for Gbb and acts antagonistically with GstO1 in regulating Gbb level and NMJ growth in Drosophila.
Co-overexpression of GstO1 and Grx1 reciprocally rescues the Gbb level to WT level. (A–D) Confocal images of NMJs double-labeled with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, Grx1 OE: UAS-Grx1/C57-Gal4, Grx1 OE: UAS-GstO1/+; C57-Gal4/+, and Grx1+GstO1 OE: UAS-GstO1/+; UAS-Grx1/C57-Gal4. Overexpression of Grx1 led to synaptic overgrowth which was rescued by co-overexpression of GstO1 to the WT level. (E) Quantification of the total bouton numbers of different genotypes. Scale bar, 10 µm. n = 11 NMJs for each genotype, Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (F–I) Confocal images of NMJs co-stained with anti-Gbb (green) and HRP (magenta). Intracellular Gbb was upregulated when Grx1 was overexpressed but rescued when GstO1 was co-overexpressed. (J) Quantification of intracellular Gbb intensity in different genotypes in arbitrary units (a.u.). Scale bar, 2 µm. n = 10 NMJs for each genotype. Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM.
Co-overexpression of GstO1 and Grx1 reciprocally rescues the Gbb level to WT level. (A–D) Confocal images of NMJs double-labeled with anti-HRP (green) and anti-CSP (magenta). The genotypes are WT, Grx1 OE: UAS-Grx1/C57-Gal4, Grx1 OE: UAS-GstO1/+; C57-Gal4/+, and Grx1+GstO1 OE: UAS-GstO1/+; UAS-Grx1/C57-Gal4. Overexpression of Grx1 led to synaptic overgrowth which was rescued by co-overexpression of GstO1 to the WT level. (E) Quantification of the total bouton numbers of different genotypes. Scale bar, 10 µm. n = 11 NMJs for each genotype, Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (F–I) Confocal images of NMJs co-stained with anti-Gbb (green) and HRP (magenta). Intracellular Gbb was upregulated when Grx1 was overexpressed but rescued when GstO1 was co-overexpressed. (J) Quantification of intracellular Gbb intensity in different genotypes in arbitrary units (a.u.). Scale bar, 2 µm. n = 10 NMJs for each genotype. Statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM.
Gbb is degraded by the proteasomal degradation pathway
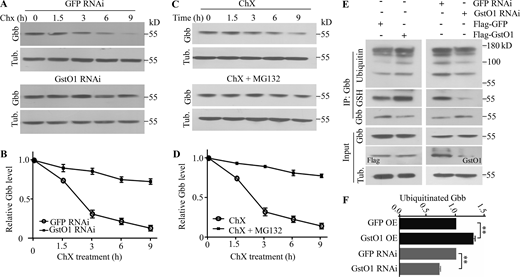
Since Gbb protein levels were upregulated in GstO1 mutants but reduced when GstO1 was overexpressed (Fig. 4, A−E), we explored the mechanism mediating the negative regulation of Gbb by GstO1 by examining the stability of Gbb protein in S2 cells at various time points after treatment with the protein synthesis inhibitor cycloheximide (ChX). In control cells transfected with dsRNAs against green fluorescent protein (GFP), the Gbb level decreased markedly from 3 to 9 h upon ChX treatment (Fig. 9, A and B). However, in GstO1 knockdown cells, a relatively stable level of Gbb was observed when treated with ChX for 9 h (Fig. 9, A and B). These results support a role for GstO1 in the downregulation of Gbb stability.
Gbb is degraded by the proteasomal degradation pathway. (A) Gbb protein levels in S2 cells treated with dsRNAs targeting GstO1 or GFP control at various time points after blocking protein synthesis by cycloheximide (ChX). (B) A slowed decline in Gbb protein levels in S2 cells after the knockdown of GstO1 by dsRNA. Values are shown as the ratio of Gbb intensity to tubulin control, normalized to the untreated cells at time zero. n = 3. Data are presented as means ± SEM. (C) Treatment of S2 cell with ChX and with or without proteasome inhibitor MG132. (D) A slowed decline in Gbb protein levels in S2 cells treated with CHX and MG132. Values are shown as the ratio of Gbb intensity to tubulin control, normalized to the cells only treated with CHX at time zero. n = 3. Data are presented as means ± SEM. (E) GstO1 facilitates Gbb glutathionylation and ubiquitination. Gbb ubiquitination together with glutathionylation upon RNAi knockdown or overexpression of GstO1 in S2 cells. Anti-ubiquitin was used to detect ubiquitinated Gbb. Glutathionylated Gbb was detected by anti-GSH after immunoprecipitation with anti-Gbb. Tubulin was used as a loading control. (F) Quantitative analysis of ubiquitinated Gbb. n = 3, **P < 0.01 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F9.
Gbb is degraded by the proteasomal degradation pathway. (A) Gbb protein levels in S2 cells treated with dsRNAs targeting GstO1 or GFP control at various time points after blocking protein synthesis by cycloheximide (ChX). (B) A slowed decline in Gbb protein levels in S2 cells after the knockdown of GstO1 by dsRNA. Values are shown as the ratio of Gbb intensity to tubulin control, normalized to the untreated cells at time zero. n = 3. Data are presented as means ± SEM. (C) Treatment of S2 cell with ChX and with or without proteasome inhibitor MG132. (D) A slowed decline in Gbb protein levels in S2 cells treated with CHX and MG132. Values are shown as the ratio of Gbb intensity to tubulin control, normalized to the cells only treated with CHX at time zero. n = 3. Data are presented as means ± SEM. (E) GstO1 facilitates Gbb glutathionylation and ubiquitination. Gbb ubiquitination together with glutathionylation upon RNAi knockdown or overexpression of GstO1 in S2 cells. Anti-ubiquitin was used to detect ubiquitinated Gbb. Glutathionylated Gbb was detected by anti-GSH after immunoprecipitation with anti-Gbb. Tubulin was used as a loading control. (F) Quantitative analysis of ubiquitinated Gbb. n = 3, **P < 0.01 by one-way ANOVA with Tukey’s post hoc test. Data are presented as means ± SEM. Source data are available for this figure: SourceData F9.
Given the increased Gbb level in GstO1 mutants and in S2 cells expressing a reduced level of GstO1 through RNAi knockdown (Fig. 4, A−E; and Fig. S2, G and H), we suspected that Gbb stability might be regulated by the proteasomal degradation pathway, and this was indeed the case. Gbb was relatively stable when S2 cells were cotreated with ChX and the proteasome inhibitor MG132, while the Gbb level in control cells treated with ChX alone was decreased markedly from 3 to 9 h after treatment (Fig. 9, C and D), suggesting that Gbb is degraded primarily by the proteasomal pathway, at least in S2 cells. Furthermore, we determined total and ubiquitinated Gbb levels in S2 cells and found that GstO1 overexpression resulted in more ubiquitination of Gbb (1.37 ± 0.04 a.u.), while GstO1 knockdown led to the opposite (0.65 ± 0.03 a.u.) compared with GFP control manipulations (1.0 a.u.) by western analysis with an antibody against ubiquitin (Fig. 9, E and F). These results indicate that GstO1-mediated Gbb glutathionylation promotes its degradation through the proteasomal pathway.
The E3 ligase Ctrip mediates Gbb ubiquitination
Given that GstO1 promoted Gbb glutathionylation and subsequent degradation by the proteasome pathway (Fig. 6 A and Fig. 9, C−E), we sought to identify the specific E3 ligase that ubiquitinates Gbb using a genetic approach. Ubiquitin E3 ligases catalyze the last step of ubiquitin conjugation reactions by transferring ubiquitin from ubiquitin-conjugating E2 enzymes to substrates. In the Drosophila genome, there are 17 distinct E3 ligases expressed in the carcass of third-instar larvae, based on bioinformatics analysis of publically available genomics data at FlyBase (https://flybase.org/). The 10 E3s with RNAi lines publically available were Ctrip, Cnot4, mi-2, rocla, CG9934, tn, CG11984, th, MG53 (Trim72), and Nedd4 (Table S3). Knockdown of ctrip but not the other genes encoding the nine E3 ligases exhibited more synaptic boutons, mimicking the phenotype of GstO1 mutants (Fig. 10, A–D). Consistent with more synaptic boutons, there was also an increased level of intracellular Gbb when ctrip was knocked down (Fig. 10, E–H).
E3 ligase Ctrip preferentially interacts with glutathionylated Gbb. (A–C) Confocal images of NMJs double-labeled with anti-HRP (green) and anti-CSP (magenta). The genotypes are: WT, ctrip RNAi1 (THU0789)/C57-Gal4, ctrip RNAi2 (TH01600.N)/C57-Gal4. Scale bar, 10 µm. (D) Quantification of total bouton numbers of different genotypes. n = 10 NMJs for each genotype, statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (E–G) Confocal images of NMJs colabeled with anti-Gbb (green) and anti-HRP (magenta). Knockdown of ctrip by C57-Gal4 resulted in increased intracellular Gbb level compared with WT. (H) Quantification of cytoplasmic Gbb level of different genotypes in arbitrary units (a.u.). n = 12 larvae; statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (I) Gbb preferentially interacts with Ctrip2. Lysates of S2 cells co-expressing Myc-Gbb and individual Flag-tagged Ctrip fragments were subjected to immunoprecipitation with anti-Flag antibody. Gbb was detected with anti-Myc antibody. Tubulin was used as a loading control. (J) Ctrip2 binds weakly to non-glutathionylated C354A and C420A double mutation Gbb. S2 cell lysates co-overexpressing Flag-Ctrip2 and WT or C354A and C420A double mutated Gbb. Myc-Gbb was subjected to co-immunoprecipitation with anti-Flag antibody and detected with anti-Myc antibody. Tubulin was used as a loading control. (K) A working model. GstO1 promotes glutathionylation and subsequent proteasome-mediated degradation of Gbb, while Grx1 does the opposite. GstO1 and Ctrip negatively regulate NMJ synapse development by down-regulating Gbb protein level. Source data are available for this figure: SourceData F10.
E3 ligase Ctrip preferentially interacts with glutathionylated Gbb. (A–C) Confocal images of NMJs double-labeled with anti-HRP (green) and anti-CSP (magenta). The genotypes are: WT, ctrip RNAi1 (THU0789)/C57-Gal4, ctrip RNAi2 (TH01600.N)/C57-Gal4. Scale bar, 10 µm. (D) Quantification of total bouton numbers of different genotypes. n = 10 NMJs for each genotype, statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (E–G) Confocal images of NMJs colabeled with anti-Gbb (green) and anti-HRP (magenta). Knockdown of ctrip by C57-Gal4 resulted in increased intracellular Gbb level compared with WT. (H) Quantification of cytoplasmic Gbb level of different genotypes in arbitrary units (a.u.). n = 12 larvae; statistical significance was calculated using one-way ANOVA; ***P < 0.001; error bars denote SEM. (I) Gbb preferentially interacts with Ctrip2. Lysates of S2 cells co-expressing Myc-Gbb and individual Flag-tagged Ctrip fragments were subjected to immunoprecipitation with anti-Flag antibody. Gbb was detected with anti-Myc antibody. Tubulin was used as a loading control. (J) Ctrip2 binds weakly to non-glutathionylated C354A and C420A double mutation Gbb. S2 cell lysates co-overexpressing Flag-Ctrip2 and WT or C354A and C420A double mutated Gbb. Myc-Gbb was subjected to co-immunoprecipitation with anti-Flag antibody and detected with anti-Myc antibody. Tubulin was used as a loading control. (K) A working model. GstO1 promotes glutathionylation and subsequent proteasome-mediated degradation of Gbb, while Grx1 does the opposite. GstO1 and Ctrip negatively regulate NMJ synapse development by down-regulating Gbb protein level. Source data are available for this figure: SourceData F10.
Ctrip E3 ligase contains a HECT (homologous to E6AP C-terminus) domain. Haploinsufficiency of TRIP12, the human homolog of Drosophila Ctrip, causes intellectual disability with or without autism spectrum disorders, speech delay, and dysmorphic features (Zhang et al., 2017). Given the involvement of the proteasome in Gbb degradation (Fig. 9, C and D) and the increased Gbb in ctrip knockdown flies (Fig. 10, E–H), we hypothesized that Gbb may be a direct target for Ctrip-mediated ubiquitination resulting in proteasome-mediated degradation. This model predicts a physical interaction between Ctrip and Gbb. Because the coding region of ctrip is long with 8,127 bp, we expressed the full-length Ctrip in three tandem fragments Ctrip1, Ctrip2, and Ctrip3. We examined the physical interaction of Ctrip fragments with Gbb in S2 cells by co-IP and found that Ctrip2 interacted with Gbb, but Ctrip1 and Ctrip3 did not (Fig. 10 I). We further investigated whether the interaction between Ctrip2 and Gbb was influenced by Gbb glutathionylation and found that Ctrip2 bound weakly to C354A and C420A double mutated Gbb compared with WT controls (Fig. 10, I and J), suggesting that Gbb glutathionylation may promote Ctrip and Gbb interaction. Together, these results of the genetic and biochemical analysis indicate that Ctrip may be the E3 ligase mediating the ubiquitination of Gbb.
Discussion
In the present study, we uncovered from a genetic screen that Drosophila GstO1, a previously uncharacterized member of the GST family, inhibits NMJ growth by suppressing BMP signaling. We further demonstrated that GstO1 suppresses BMP signaling by promoting glutathionylation and subsequent proteasome-mediated degradation of Gbb (Fig. 10 K), the Drosophila homolog of mammalian BMPs. Our results reveal the in vivo function of GstO1 and shed new light on the mechanism of an intersection between glutathionylation and ubiquitination of Gbb in synapse development.
GstO1 inhibits NMJ synapse development by glutathionylation of Gbb
The postsynaptic muscle-derived Gbb promotes NMJ growth in a retrograde manner. Regulation of Gbb signaling occurs at a number of steps, including transcriptional and post-translational trafficking. For example, Dawdle (Daw), a Drosophila homolog of mammalian activin, promotes Gbb transcription in muscles (Ellis et al., 2010). Meanwhile, dRich, a conserved Cdc42-selective guanosine triphosphatase-activating protein, regulates synaptic growth and function by promoting Gbb release from postsynaptic muscles (Nahm et al., 2010b). In addition, dCIP4, short for Drosophila Cdc42-interacting protein 4, inhibits Gbb secretion by restraining Cdc42-Wsp-Arp2/3-induced actin polymerization on the postsynaptic side (Nahm et al., 2010a). Our genetic data reveal that postsynaptic GstO1 restrains synaptic growth by attenuating retrograde BMP signaling (Figs. 3 and 4). Specifically, presynaptic pMad levels at NMJs were significantly increased upon GstO1 mutation and postsynaptic RNAi-mediated knockdown of GstO1 (Fig. 3, A–D and F). Moreover, the Gbb level at NMJ synapses was negatively regulated by GstO1 (Fig. 4, A–E). Thus, we favor a model in which BMP signaling is upregulated leading to overgrown NMJs in GstO1 mutants.
While other types of PTM such as ubiquitination are closely involved in synapse development and function (DiAntonio and Hicke, 2004; Harris and Littleton, 2015; Li et al., 2016), we showed for the first time in the present study that glutathionylation regulates synapse development by promoting proteasome-mediated degradation of Gbb. Specifically, GstO1 mutants showed decreased glutathionylation of Gbb, while overexpression of GstO1 increased glutathionylation of Gbb (Fig. 6 A). Further, mutational and biochemical analyses demonstrated that glutathionylation of Gbb occurs specifically at C354 and C420, but not C419 (Fig. 6, B–D).
The thiol group of cysteine permits a number of PTMs including S-glutathionylation, which is favored within the oxidizing environment of ER (Yoboue et al., 2018). While the cellular localization of GstO1 has not been reported, GST Pi (GSTP) localizes to the ER where it forms a protein complex with resident proteins such as BiP and calnexin, and catalyzes their S-glutathionylation (Ye et al., 2017). Other proteins such as Grx and thioredoxin that participate in the S-glutathionylation cycle are also localized to the ER (Ye et al., 2017). In the present study, we showed that GstO1 is localized at the ER and early/recycling endosomes, but not late endosomes (Fig. 4, H–N). Based on these findings, we propose that within the ER and possibly early/recycling endosomes, GstO1 may maintain the environment that facilitates Gbb glutathionylation. The cellular conditions and molecular signals that affect Gbb glutathionylation are largely unknown. Our results show that GstO1 expression was reduced while Gbb increased under hypoxia and in hyperexcitable mutants (Fig. 5). Because glutathionylation may act as a sensor of redox state, nitric oxide, or reactive oxygen species during normal cellular processes (Dalle-Donne et al., 2009; Xiong et al., 2011; Zhang et al., 2016), it is possible that the redox state in the ER may affect the level of Gbb glutathionylation. Indeed, as expected from the biochemical functions of GST family members, we found a higher ratio of GSH/GSSG in GstO1 mutants (Fig. S4 I).
Glutathionylation facilitates proteasome-mediated degradation of Gbb
Different types of PTM may coordinate to regulate the functions of target proteins. For example, sequential phosphorylation of the transcription co-activator TAZ at Ser311 and Ser314 by the Hippo pathway components of large tumor suppressor kinase LATS and casein kinase 1 (CK1-ε), respectively, is essential for its E3 ligase SCFβ-TrCP-mediated ubiquitination and degradation (Liu et al., 2010). TAZ protein stability is controlled by a C-terminal phosphodegron recognized by the F-box protein β-TrCP and ubiquitinated by the SCFβ-TrCP E3 ligase, leading to proteasome-mediated degradation (Liu et al., 2010).
The effects of glutathionylation on proteins vary dramatically. S-glutathionylation on hypoxia-inducible factor (HIF)-1α inhibits protein interaction with an E3 ligase called von Hippel-Lindau tumor suppressor resulting in HIF-1α stabilization (Watanabe et al., 2016). S-glutathionylation of the adipogenic transcription factor CCAAT enhancer-binding protein β (C/EBPβ) also suppresses its attachment to the small ubiquitin-like modifier (SUMO) E3 ligase PIAS1, thus protecting it from degradation via the proteasome pathway (Watanabe et al., 2020). However, there are also reports documenting that glutathionylation leads to proteolytic degradation of target proteins such as mitochondrial thymidine kinase 2 by unknown proteases (Sun et al., 2012). In the present study, we show that WT Gbb exhibited a higher binding affinity with the E3 ligase Ctrip, while glutathionylation site-mutated Gbb displayed reduced binding with Ctrip (Fig. 10 J), resulting in higher stability of Gbb. Based on these findings, together with the negative regulation of Gbb protein abundance by GstO1, we propose that glutathionylation of Gbb promotes its proteasome-mediated degradation (Fig. 10 K). The exact molecular and structural mechanism by which glutathionylation promotes ubiquitination remains to be elucidated.
We demonstrate that GstO1 is required and sufficient for Gbb glutathionylation. It is currently unknown how GstO1 mediates the process. There are two possibilities, not mutually exclusive. One is that GstO1 may directly glutathionylate Gbb; the other is that GstO1 may maintain the redox status which facilitates Gbb glutathionylation. In addition to glutathionylation, cysteines are also targets for other modifications such as disulfide bond formation. It is not yet clear how different modifications on cysteines of Gbb are coordinated in vivo. Furthermore, how the E3 ligase Ctrip recognizes and preferentially binds glutathionylated Gbb needs to be explored in the future. As GstO1 negatively regulates the Gbb level in both muscle cells and wing discs, it is interesting to know if this regulation is conserved in mammals.
Materials and methods
Drosophila strains and genetics
All fly strains were maintained on standard cornmeal food at 25°C. w1118 was used as the WT control unless otherwise indicated. We generated a 121-bp DNA deletion named GstO16-11 by the CRISPR/Cas9 method for germ-line specific targeting with two sgRNAs 5′-GTGCTGAGTATTGCTCATATTGG-3′ and 5′-TCATCCGGAAATACGGGCTTTGG-3′ (Ren et al., 2013) at Qidong Fungene Biotechnology Co., Ltd (www.fungene.tech). To generate a GstO1-Flag stock, we used two overlapping sgRNAs 5′-GTCAAATTGGGGTAGTCTACGGG-3′ and 5′-GGGGTAGTCTACGGGATGATGG-3′, and a 1.9-kb donor DNA including 3×Flag sequence (…5′-GGAGGTGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAG-3′…). The Flag tag was knocked in at the C-terminus of the endogenous GstO1 on the w1118 background by combining ends-out gene targeting along with DNA integration using phage integrase φC31 (Huang et al., 2009). Drosophila lines carrying single mutations of Cys354A, Cys419A, and Cys420A in Gbb were generated by homology-directed repair procedures using optimized CRISPR/Cas tools (Port et al., 2014) at Qidong Fungene Biotechnology Co., Ltd (www.fungene.tech). Transgenic flies carrying UAS-GstO1 were generated as follows. The full-length cDNA of GstO1 was amplified by PCR from a cDNA prepared from the adult w1118 strain and cloned into a pUAST-attB vector. The following primers were used (forward: 5′-GCAACTACTGAAATCTGCCA-3′, reverse: 5′-AGTTCCATAGGTTGGAATCT-3′). The plasmid was injected into flies carrying an attP40 docking site. There are seven predicted glutaredoxins glutaredoxin 1 (Grx1), glutaredoxin 1 testis-specific (Grx1t), CG14407, CG9147, CG6523, CG31559, and CG12206 in Drosophila. We generated UAS lines for all seven predicted glutaredoxins using flySAM by a single vector encoding both sgRNA and UAS:Cas9-activator following a previously established protocol (Jia et al., 2018) at the Tsinghua Fly Center (https://thfc.zzbd.org/en/dl.html). Tissue-specific Gal4 drivers C57-Gal4 (muscle-specific) and OK6-Gal4 (motor-neuron-specific) were used for tissue-specific expression experiments as previously reported (Li et al., 2016; Huang et al., 2018). GstO1PBacLL04542 (GstO1PBac thereafter) flies were a gift from Dr. Kiyoung Kim (Soonchunhyang University, Asan, Republic of Korea; Lee et al., 2015). eag1Sh120 double mutants were obtained from Y. Zhang (Tsinghua University, Beijing, China; Budnik et al., 1990). GstO1 RNAi (BL#34727), Df(3L)BSC219 (abbreviated as Df; BL#9696), gbb4 (BL#63053), tkv8 (BL#34509), mad12 (BL#51286), and Df(2R)BSC600 for gbb (BL# 26512) were obtained from the Bloomington Stock Center. RNAi lines for the genetic screen (Table S1) and ten E3 ligases including Ctrip (Table S3) were obtained mostly from Tsinghua Fly Center.
Hypoxia condition: Third instar larval development in 2.5% O2 for 16 h was achieved in a hypoxia chamber (Maworde, GC-CT) following a previous report (Zhou et al., 2008).
Immunohistochemistry
Immunostaining of larval muscles was done as described previously (Li et al., 2016). Especially, wandering third instar larvae were dissected in calcium-free HL-3 saline (110 mM NaCl, 5 mM KCl, 10 mM NaHCO3, 5 mM Hepes, 30 mM sucrose, 5 mM trehalose, and 10 mM MgCl2) on Petri dishes. Tissues were fixed either in Bouin’s solution (HT10132; Sigma-Aldrich) for 5 min (for anti-Rab11 and extracellular Gbb staining) or 4% paraformaldehyde for 30–60 min (for all other antibodies), followed by washing with 0.2% Triton X-100 in phosphate-buffered saline. Anti-HRP staining was used as an internal control for the quantification of Gbb and pMad intensity presented as arbitrary units (a.u.; Zhao et al., 2015). For the detection of secreted Gbb, we followed a previous protocol (James et al., 2014) in which detergent-free buffers were used. The following primary antibodies were used: mouse anti-CSP (1:100; 6D6, Developmental Studies Hybridoma Bank [DSHB]), rabbit polyclonal anti-GstO1 (1:100, this study), mouse anti-Gbb (1:50; 3D6-24; DSHB), rabbit anti-pMad (PS1, 1:500, Leiden University Medical Center, Netherlands; Persson et al., 1998), mouse anti-Rab11 (1:50; 610656; BD Biosciences), mouse anti-Rab5 (1:50, AR038; Beyotime Biotechnology), guinea pig anti-Spinster (1:200, G. Davis, University of California, San Francisco; Sweeney and Davis, 2002), guinea pig anti-Dor (1:500, UT Southwestern Medical Center, Dallas, USA; Pulipparacharuvil et al., 2005), guinea pig anti-Hrs (1:1,000, GP 30, Baylor College of Medicine, Houston, USA; Lloyd et al., 2002), rat anti-KDEL (1:100, ab50601; Abcam), and Alexa 649-conjugated anti-horseradish peroxidase (anti-HRP, 1:100; Jackson Immuno Research). Alexa 488-conjugated goat anti-mouse (RRID: AB_2633275), anti-rabbit (RRID: AB_2576217), anti-rat (RRID: AB_2534074) IgG (Molecular Probes), Alexa 568-conjugated goat anti-mouse (RRID: AB_141371), anti-rabbit (RRID: AB_143011), and anti-rat (RRID: AB_141874) IgG (Molecular Probes) were used at 1:1,000. To-PRO-3 Iodide (T3605; Molecular Probes) was used at 1:1,000 for labeling the nucleus.
Generation of rabbit anti-GstO1 antibody
A polyclonal rabbit antibody against GstO1 was raised by immunizing rabbits with synthetic peptides containing C-terminal 228-242 amino acid residues of GstO1 (accession number AY071499). The antibody was used at 1:100 for immunostaining and 1:750 for Western blotting.
DNA constructs and mutagenesis
GstO1, ctrip1, ctrip2, and ctrip3 (coding sequence of 1–3,147 bp, 3,127–5,655 bp, and 5,629–8,127 bp of the registered accession NM_001300234.1 for ctrip) were amplified by RT-PCR using Superscript III reverse transcriptase (18080044; Invitrogen) from total RNA extracted from Drosophila larval muscles. gbb was PCR-amplified and cloned from UAS-gbb flies (Khalsa et al., 1998). For cell culture expression in S2 cells, ctrip fragments and gbb full-length cDNA were subcloned into pAC5.1 with N-terminal 3x Flag and pMT with N-terminal 6x Myc tag, respectively. Mutations at specific codons for cysteines in Gbb were introduced with a seamless cloning kit (C5891-25; Clone Smarter). The integrity of all constructs was verified by DNA sequencing.
Quantitative PCR of gbb transcripts
Total RNA was extracted from the muscles of 30 third instar larvae following the standard Trizol reagent protocol (15596026; Invitrogen). First-strand cDNA was synthesized from 1 μg total RNA using Superscript III reverse transcriptase (18080044; Invitrogen). Power SYBR Green PCR Master Mix (A25742; Applied Biosystems) was used for quantitative real-time PCR. RpL28 cDNA was amplified as the internal control with primers 5′-GCAACCTCTTCGCACCTCAAT-3′ and 5′-CACGACTCCCAGGGTCTTCT-3′, whereas gbb cDNA was amplified using primers 5′-ATCAGGATGAGGACGACGAC-3′ and 5′- GTCCAGGTCGGTGATGAAGT-3′.
Western blotting and immunoprecipitation (IP)
Western analysis was performed as described previously (Li et al., 2016; Metwally et al., 2021). Specifically, third instar larvae muscles were homogenized in ice-cold RIPA lysis buffer (50 mM Tris HCl at pH 7.4, 150 mM NaCl, 0.1% SDS, 1% NP-40) with a protease inhibitor cocktail (Set 1, Roche). Western blotting was carried out with the following primary antibodies: mouse anti-Gbb (1:300; 3D6-24-S; DSHB), mouse anti-ubiquitin (1:1,000; P4D1 from Cell Signaling Technologies), mouse anti-glutathione antibody (anti-GSH; 1:500; AB19534; Abcam), mouse anti-Flag (1:1,000; F3165; Sigma-Aldrich), and mouse anti-α-tubulin (1:50,000; CP06; Millipore). The secondary antibody was HRP-conjugated goat anti-mouse IgG (A4416-5; Sigma-Aldrich) or goat anti-rabbit IgG (A0545; Sigma-Aldrich) used at 1:50,000. Protein bands were visualized on x-ray films by chemiluminescence. To quantify the levels of target proteins, positive signals on Western blots from multiple independent repeats were calculated using ImageJ and normalized to the loading control. For IP of in vivo glutathionylation experiments, larval muscle lysates were incubated with appropriate antibodies (4 µg antibodies of anti-Gbb, anti-IgG, anti-Flag, and anti-Myc) followed by precipitating with Dynabeads (10003D; Thermo Fisher Scientific). For detection of Gbb glutathionylation, larval muscle or S2 cell lysates in non-reducing lysis buffer (20 mM HEPES, pH 7.6; 100 mM KCl; 5 mM MgCl2; 0.5% NP-40) were immune-precipitated, followed by Western analysis with anti-GSH.
Cell culture and RNA interference
Drosophila S2 cells (RRID:CVCL_Z232) were cultivated at 25°C in Schneider’s medium (S0146; Sigma-Aldrich). The cultured cells were resuspended in a fresh medium with the final concentration at 1×106 cells/ml medium and plated at 1 ml/well in six-well culture plates. Cells were transfected with dsRNAs using Cellfectin II reagent (T-IVGN-10362100; Invitrogen). After 48 h incubation, transfected cells were lysed in RIPA buffer (EA20-188; Millipore) containing 50 mM Tris, pH 7.5, 150 mM NaCl, and 1% Triton X-100 for 1 h on ice. Double-stranded RNAs (dsRNAs) against GstO1 and GFP were generated by in vitro transcription of PCR-amplified templates containing T7 promoter sequences at both ends using a Megascript T7 kit (AM1333; Invitrogen) and purified using a RNeasy kit (74104; Qiagen). The primers used for the production of dsRNAs were: 5′-TAATACGACTCACTATAGGGCGCCAAAGCCCGTATTTCC-3′, and 5′-TAATACGACTCACTATAGGGGCCACATCATGTAGTCAAGCAT-3′ for GstO1 RNAi 1, 5′-TAATACGACTCACTATAGGGCTC GCCAAAGCCCGTATTT-CC-3′ and 5′-TAATACGACTCACTATAGGGCGTCGCTTCAGTTCCTCCTCATA-3′ for GstO1 RNAi 2, and 5′-TAATACGACTCACTATAGGGATGGTGAGCAAGGGCGAGG-3′ and 5′-TAATACGACTCACTATAGGGGTGCCCCAGGATGTTGCCGTC-3′ for control GFP RNAi. The protein in the culture medium was condensed by centrifugal filters (Centriprep 3K device; Millipore) followed by Western analysis using a mouse anti-Gbb.
LC-MS/MS analysis to identify glutathionylation sites of Gbb
For identification of glutathionylation sites of Gbb proteins, we produced and purified a fusion protein containing the C-terminal amino acids 336–455 of the Drosophila Gbb (Swiss-Prot: P27091.1) with an N-terminal 6×His tag on pET-28a vector from E. coli. Preparation of glutathionylated protein and mass spectrometry analysis were performed as described previously (Zhang et al., 2016). Specifically, the truncated Gbb peptide of 336–455 residues containing all cysteine residues in Gbb affinity purified from E. coli was treated with 1 mM diamide and 2 mM GSH to allow the glutathionylation reaction to proceed. For immunodetection of Gbb-glutathione, non-reducing SDS-PAGE loading buffer (50 mM Tris-HCl, pH 6.8, 10% glycerol, 20 g/l SDS, 0.02% bromophenol blue) was added to the reaction. The samples were separated by non-reducing SDS-PAGE followed by Coomassie blue staining. The protein band of 17 kD was excised and digested with trypsin. The solution was then transferred to a sample vial for LC-MS/MS analysis. All nano LC-MS/MS experiments were performed on a Q Exactive (Thermo Fisher Scientific) equipped with an Easy n-LC 1000 HPLC system (Thermo Fisher Scientific). The MS data were acquired at a high resolution 70,000 (m/z 200) across the mass range of 300–1,600 m/z, with the data-dependent acquisition mode. The raw data from Q Exactive were analyzed with Proteome Discovery version 1.4 using Sequest HT search engine for protein identification and Percolator for FDR (false discovery rate) analysis against a Uniprot Drosophila melanogaster protein database (https://www.uniprot.org/proteomes/UP000000803; updated on February 2017).
Measurement of GSH/GSSG ratio
For the measurement of the GSH/GSH disulfide (GSSG) ratio of third instar larval carcasses, we used a GSH and GSSG assay kit (Cat #S0053, Beyotime Biotechnology, China) following the product instructions.
Image collection and data analysis
Images were collected according to previous reports (Li et al., 2016; Metwally et al., 2021). Specifically, all staining images were collected using an Olympus Fluoview FV1000 confocal microscope with a 40×/3NA or 60×/3NA oil objective and FV10-ASW software (https://www.olympus-lifescience.com/en/, RRID: SCR_014215) or with a Leica confocal microscope using a 40×/3NA oil objective and LAS AF software (http://www.leica-microsystems.com/, RRID:SCR_013673). All images of muscle 4 type Ib NMJs of abdominal segments A2 to A4 for a specific experiment were captured using identical settings for statistical comparison among different genotypes. Immunostaining intensities were quantified using ImageJ (National Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/ij/, RRID: SCR_003070), with anti-HRP staining as an internal control for quantification. The intensity of synaptic Gbb signal within the region of interest (ROI) defined by HRP staining was normalized to the average HRP intensity. An arbitrary threshold was set for each channel and used for all relevant images. For quantification analysis of NMJ growth, individual boutons were defined according to the discrete staining signal of anti-CSP. Satellite boutons were defined as extensions of two or more small boutons emanating from the main branch of the NMJ terminals.
GstO1 localization at intracellular organelles such as ER, early endosome, late endosome, and recycling endosome was quantified by calculating the percentage of co-localization punctae in 20 µm2 area using ImageJ. The extent of colocalization was analyzed by Manders coefficients and Pearson correlation coefficient (r; https://imagej.nih.gov/ij///plugins/colocalization.html; Metwally et al., 2021).
Statistical analysis
Statistical comparisons were performed using GraphPad Prism 9. All the data are expressed as mean ± SEM. The differences between multiple group means were evaluated by one-way ANOVA with Tukey’s post hoc tests for pair-wise comparisons. All comparisons were between a specific genotype and the control unless otherwise indicated. P < 0.05 were considered statistically significant.
Online supplemental material
Fig. S1, related to Fig. 2, shows increased GstO1 expression during larval development.Fig. S2, related to Fig. 4, shows increased Gbb levels in vivo and in S2 cells when GstO1 is mutated and knocked down by RNAi, respectively, but normal gbb mRNA when GstO1 expression is altered. Fig. S3, also related to Figs. 4 and 5, shows representative microscopy images of GstO1 and Gbb staining in wing discs of WT and GstO1 knockdown larvae. Fig. S4, related to Fig. 4, shows colocalization of Gbb with various membrane organelles including ER and endosomes in WT, GstO1 knockdown, and gbb354/Df(2R)BSC600 mutants, as well as increased GSH/GSSG ratio in GstO1 mutants. Table S1, related to Fig. 1, shows the list of 1,022 genes (1,152 RNAi lines) screened. Table S2, related to Fig. 1, shows the list of positive genes regulating NMJ synapse growth. Table S3, related to Fig. 10, shows the list of 10 E3 ligases screened.
Acknowledgments
We thank H. Yuan and Prof. Y. Zhang for the database analysis of expression patterns of E3 ligases in larvae. We thank Dr. Kiyoung Kim for GstO1PBac flies. We thank Prof. Seungbok Lee, Yuhang Chen, and Sean T. Sweeney for the discussion. We thank Bloomington and Tsinghua stock centers for providing fly stocks, and Qidong Fungene Biotechnology Co., Ltd (www.fungene.tech) for generating gene-edited stocks. We are also grateful to the members of the Yong Q. Zhang lab for their discussion and suggestions.
This work was supported in part by the Ministry of Science and Technology of China (2019YFA0707100, 2017YFA0504000, and 2021ZD0203900), the National Natural Science Foundation of China (31830036, 31921002, 91849203, 31700885, and 31900893), the Chinese Academy of Sciences Strategic Priority Research Program B grants (XDBS1020100 and XDB39000000), and Spring City Plan: The High-level Talent Promotion and Training Project of Kunming (2022SCP001).
Author contributions: M.S. Hossain designed research and performed most experiments. A. Yao designed research, performed experiments, and supervised the projects. X. Qiao and T. Xie performed LC-MS analysis and GSH/GSSG ratio measurement experiments. W. Shi helped with the GstO1 antibody and indel mutant generation. Y.Q. Zhang, C. Chen, and A. Yao supervised research. M.S. Hossain and A. Yao wrote the whole manuscript, and Y.Q. Zhang and C. Chen finalized the paper.
References
Author notes
M.S. Hossain, A. Yao, and X. Qiao contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.
M.S. Hossain’s current affiliation is Department of Pharmacy, Manarat International University, Ashulia, Dhaka, Bangladesh.