Cell biologists typically focus on conserved regions of a protein, overlooking innovations that can shape its function over evolutionary time. Computational analyses can reveal potential innovations by detecting statistical signatures of positive selection that lead to rapid accumulation of beneficial mutations. However, these approaches are not easily accessible to non-specialists, limiting their use in cell biology. Here, we present an automated computational pipeline FREEDA that provides a simple graphical user interface requiring only a gene name; integrates widely used molecular evolution tools to detect positive selection in rodents, primates, carnivores, birds, and flies; and maps results onto protein structures predicted by AlphaFold. Applying FREEDA to >100 centromere proteins, we find statistical evidence of positive selection within loops and turns of ancient domains, suggesting innovation of essential functions. As a proof-of-principle experiment, we show innovation in centromere binding of mouse CENP-O. Overall, we provide an accessible computational tool to guide cell biology research and apply it to experimentally demonstrate functional innovation.
Introduction
Purifying selection eliminates deleterious non-synonymous mutations, leading to conservation of amino acid sequence. In contrast, positive selection results in the accumulation of non-synonymous mutations that lead to functional innovation and adaptation (reviewed in Nielsen et al., 2005). Compelling examples of how positive selection has regulated protein function come from studying host–pathogen genetic conflicts. In these evolutionary arms races, positive selection leads to rapid accumulation of mutations in both viral proteins that help infect the host and host proteins that help evade the infection (reviewed in Daugherty and Malik, 2012; Sironi et al., 2015). To experimentally test functional innovation, evolutionary biologists swap protein regions (or entire alleles) from closely related species that are suspected to have diverged due to positive selection. This approach generates an “evolutionary mismatch” between the divergent protein and the cellular environment, revealing which protein function might have evolved adaptively (reviewed in Brand and Levine, 2021). For example, swapping a region of the TRIM5 protein between human and rhesus monkey suggested that positive selection shaped its role in fighting species-specific retroviral infections (Sawyer et al., 2005; Stremlau et al., 2005; Yap et al., 2005). Remarkably, variation at even single residues under positive selection can lead to functional changes, as in the human MAVS (Mitochondrial Antiviral Signaling) protein that has evolved to evade infection with hepaciviruses (Patel et al., 2012). These examples illustrate that innovation-guided functional analyses can complement more traditional conservation-guided approaches in revealing regulation of protein function.
Genetic conflicts, like those between host and pathogen, can result in recurrently changing selection pressure and recurrent adaptation of proteins regulating essential cellular processes. For example, pressure to maintain genome integrity at fertilization is thought to fuel a sexual conflict between paternal proteins that adapt to maximize the chance of fertilizing the egg and maternal proteins that adapt to prevent entry of more than one sperm (reviewed in Carlisle and Swanson, 2020). Similarly, selfish genetic elements such as transposons constantly disrupt genome integrity, leading to intragenomic conflicts and recurrent adaptation of DNA packaging proteins (reviewed in Brand and Levine, 2021). Centromere DNA sequences have also been proposed to act as selfish elements, raising the possibility of intragenomic conflict with centromere-associated proteins. Centromeres are repetitive DNA regions that direct chromosome segregation in mitosis and meiosis by assembling kinetochores, multiprotein structures that connect to spindle microtubules. Despite their essential function, centromeric DNA and proteins evolve rapidly across taxa, suggesting an evolutionary pressure to recurrently innovate. The centromere drive hypothesis proposes that selfish centromeric DNA sequences achieve non-Mendelian segregation during asymmetric female meiosis, increasing their transmission to the egg. Fitness costs imposed by this selfish behavior would lead to recurrent adaptation of centromeric proteins to suppress the costs (Henikoff et al., 2001). While there is experimental evidence for selfish centromeric DNA, the impact of positive selection on centromeric protein function remains largely untested (reviewed in Dudka and Lampson, 2022).
The scarcity of experimental studies of adaptive evolution in centromeric proteins, in contrast to our increasingly detailed understanding of their conserved functions (Kixmoeller et al., 2020; McKinley and Cheeseman, 2016; Mellone and Fachinetti, 2021), reflects the general focus of cell biology research on protein conservation rather than innovation. This discrepancy is due in part to challenges in designing experiments to infer functional consequences of positive selection, but also to the complexity of methods needed to distinguish positive selection from neutral evolution of protein-coding sequences (reviewed in Anisimova and Liberles, 2012). A widely used method calculates the rate ratio of non-synonymous (dN) to synonymous (dS) substitutions per codon (dN/dS ratio; Goldman and Yang, 1994; Kimura, 1977; Muse and Gaut, 1994) using multiple sequence alignment of closely related orthologs, which are homologous genes that arise when speciation occurs. This approach assumes that synonymous mutations are neutral, while deleterious non-synonymous mutations are purged by purifying selection. An enrichment of non-synonymous relative to synonymous substitutions within the alignment suggests recurrent adaptation to a constantly changing selection pressure (types of recurrent evolution are discussed in Maeso et al., 2012). Well-established computational suites such as PAML (Phylogenetic Analysis by Maximum Likelihood; Yang, 2007) and HyPhy (Hypothesis Testing using Phylogenies; Pond et al., 2005) offer a number of tools that can reliably detect statistical signatures of positive selection but are seldom used by cell biologists because expertise in computational biology and molecular evolution is required to generate the input data, and the output is rarely provided in an intuitive visual format.
Automated molecular evolution pipelines that incorporate the abovementioned tools have been developed (see Fig. S1 for a non-exhaustive list), but their complexity and the need for user-provided input still render them inaccessible to experimental cell biologists with limited computational skills. Increasing this access requires a “one-click” application that (1) offers a simple graphical user interface, (2) fully automates input preparation, (3) finds orthologs despite the lack of genomic annotations, (4) reduces parameterization, and (5) provides intuitive visual representation of the output. Here, we present FREEDA (Finder of Rapidly Evolving Exons in Diverse Assemblies), a fully automated, end-to-end pipeline designed for cell biologists seeking to apply an evolutionary lens by testing for statistical evidence of positive selection in their favorite proteins. FREEDA provides the key functionalities listed above, including a unique ability to map positively selected residues onto any predicted protein structure. As a proof-of-principle, we first use FREEDA to map positive selection across centromeric proteins in rodents, as mice are currently the only experimentally tractable cell biological model system for centromere drive (reviewed in Dudka and Lampson, 2022). Guided by these computational analyses, we use the evolutionary mismatch approach to provide experimental evidence of functional innovation in the centromeric protein CENP-O.
Features of published automated pipelines used to detect signatures of positive selection. Only automated pipelines are represented: Selecton (Stern et al., 2007), IDEA (Egan et al., 2008), JCoDA (Steinway et al., 2010), PhyleasProg (Busset et al., 2011), POTION (Hongo et al., 2015), PosiGene (Sahm et al., 2017), PoSeiDon (Hölzer and Marz, 2021), MEGA (Tamura et al., 2021), and DGINN (Picard et al., 2020).
Features of published automated pipelines used to detect signatures of positive selection. Only automated pipelines are represented: Selecton (Stern et al., 2007), IDEA (Egan et al., 2008), JCoDA (Steinway et al., 2010), PhyleasProg (Busset et al., 2011), POTION (Hongo et al., 2015), PosiGene (Sahm et al., 2017), PoSeiDon (Hölzer and Marz, 2021), MEGA (Tamura et al., 2021), and DGINN (Picard et al., 2020).
Results
Overview of the FREEDA pipeline
FREEDA is a stand-alone application with an intuitive graphical user interface (GUI) operating on UNIX systems (MacOS and Linux; Windows users, please see documentation). An overview and documentation of the pipeline are provided at https://ddudka9.github.io/freeda/ with a more detailed walkthrough in Materials and methods. FREEDA first downloads the reference genome of the user-selected species and prepares input data for the gene of interest by connecting to genomic, protein, and protein structure databases (Fig. 1; blue). Next, FREEDA downloads a preselected set of non-annotated genome assemblies related to the reference species, performs a BLAST (Basic Local Alignment Search Tool) search for orthologs of the gene of interest, and uses the reference species data to find orthologous sequences (Fig. 1; orange). Combining several well-established molecular evolution tools, FREEDA aligns all coding sequences, builds phylogenetic trees, determines the likelihood that positive selection has shaped the evolution of the gene, and estimates the probability that given residues have evolved under positive selection (Fig. 1; brown). Key results are displayed within the GUI (Fig. 2 A) and all results are saved into the “Results-current-date” folder generated in a location selected by the user (“Set directory”; Fig. 2 A). These files include the BLAST output, nucleotide alignment, phylogenetic tree, protein alignment, residue mapping onto reference coding sequence, and residue mapping onto protein structure. The raw data and intermediate alignment files are saved in the “Raw_data” folder. Since FREEDA finds orthologs by downloading entire genomic assemblies, the user is advised to select an external data storage device (e.g., a hard drive) when setting the directory. A stable internet connection is also required to allow communication with various databases (Fig. 1).
Overview of the FREEDA pipeline. The schematic shows the main steps, with more details in Materials and methods. Launching the FREEDA application opens a graphical user interface (gray), which prompts selection of a reference species and a gene name. First, operating on the selected reference species (blue path), FREEDA downloads the genome and uses it to curate input for the gene of interest (protein sequence, exon sequences, coding sequence, and gene sequence). Second, operating on closely related species (orange path), FREEDA downloads non-annotated genomes, searches for putative orthologous loci, retrieves these loci, finds syntenic (homologous to the reference locus) exons, and assembles coding sequences of orthologous genes based on the intron–exon boundaries known for the reference gene. Third, operating on the multiple coding sequences (brown path), FREEDA makes and curates a multiple sequence alignment, generates a phylogenetic gene tree, and detects statistical signatures of positive selection using established models measuring the rate ratio of non-synonymous to synonymous substitutions. Fourth, FREEDA maps sites with the highest probability of positive selection onto both the reference coding sequence and the structure prediction of the reference protein.
Overview of the FREEDA pipeline. The schematic shows the main steps, with more details in Materials and methods. Launching the FREEDA application opens a graphical user interface (gray), which prompts selection of a reference species and a gene name. First, operating on the selected reference species (blue path), FREEDA downloads the genome and uses it to curate input for the gene of interest (protein sequence, exon sequences, coding sequence, and gene sequence). Second, operating on closely related species (orange path), FREEDA downloads non-annotated genomes, searches for putative orthologous loci, retrieves these loci, finds syntenic (homologous to the reference locus) exons, and assembles coding sequences of orthologous genes based on the intron–exon boundaries known for the reference gene. Third, operating on the multiple coding sequences (brown path), FREEDA makes and curates a multiple sequence alignment, generates a phylogenetic gene tree, and detects statistical signatures of positive selection using established models measuring the rate ratio of non-synonymous to synonymous substitutions. Fourth, FREEDA maps sites with the highest probability of positive selection onto both the reference coding sequence and the structure prediction of the reference protein.
Example analysis of the primate MX1 gene. (A) FREEDA’s graphical user interface is divided into an input window (left half) and an output window (right half). The input window is used to provide a gene name (a), select the reference species (b), indicate where to save the data (c), and start the analysis (d). Optionally, the user can select advanced features (“Duplication expected,” “Tandem duplication expected,” “Long introns expected (>50 kb),” and “Common domains expected”; see online documentation: https://ddudka9.github.io/freeda/; e), label up to three regions of choice on the protein structure (f), select an additional codon frequency model (g), exclude species from the analysis (h), select a subgroup (i), and abort the analysis (j). The output window shows current tasks (“Events window,” top) and key results for each gene (“Results window,” bottom). The bottom part of the output window (green font) displays interactive messages guiding the user on how to provide input. (B) Putative adaptive sites are mapped onto the reference coding sequence. Graphs show recurrently changing residues (top, black bars), residues that are likely to have evolved under positive selection (middle, blue bars, probability ≥ 0.7), and most likely targets of positive selection (bottom, magenta bars, probability ≥ 0.9). Gray bars in all graphs show residues removed from the analysis. (C) Residues with the highest probability of positive selection (magenta) are mapped on the structural prediction model of the MxA protein (encoded by MX1) from AlphaFold. The N- and C-termini and known domains are automatically annotated, in addition to the user-specified region (“My motif of interest”). Regions removed from the analysis (arrowhead) are colored dark gray. To clearly show residues under positive selection, labels were modified manually in PyMOL (raw output is shown in Fig. S2 E).
Example analysis of the primate MX1 gene. (A) FREEDA’s graphical user interface is divided into an input window (left half) and an output window (right half). The input window is used to provide a gene name (a), select the reference species (b), indicate where to save the data (c), and start the analysis (d). Optionally, the user can select advanced features (“Duplication expected,” “Tandem duplication expected,” “Long introns expected (>50 kb),” and “Common domains expected”; see online documentation: https://ddudka9.github.io/freeda/; e), label up to three regions of choice on the protein structure (f), select an additional codon frequency model (g), exclude species from the analysis (h), select a subgroup (i), and abort the analysis (j). The output window shows current tasks (“Events window,” top) and key results for each gene (“Results window,” bottom). The bottom part of the output window (green font) displays interactive messages guiding the user on how to provide input. (B) Putative adaptive sites are mapped onto the reference coding sequence. Graphs show recurrently changing residues (top, black bars), residues that are likely to have evolved under positive selection (middle, blue bars, probability ≥ 0.7), and most likely targets of positive selection (bottom, magenta bars, probability ≥ 0.9). Gray bars in all graphs show residues removed from the analysis. (C) Residues with the highest probability of positive selection (magenta) are mapped on the structural prediction model of the MxA protein (encoded by MX1) from AlphaFold. The N- and C-termini and known domains are automatically annotated, in addition to the user-specified region (“My motif of interest”). Regions removed from the analysis (arrowhead) are colored dark gray. To clearly show residues under positive selection, labels were modified manually in PyMOL (raw output is shown in Fig. S2 E).
Advantages over existing automated pipelines
Several features distinguish FREEDA from currently available automated pipelines (Fig. S1). First, FREEDA is fully automated, requiring only a gene name, and distributed as a self-contained application that does not require installation or compilation of any additional programs, except for a straightforward installation of the widely used protein structure viewer PyMOL (The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC) for MacOS users. Second, FREEDA uses a defined set of non-annotated genomic assemblies that ensure high statistical power of the analysis while absolving the users from manually curating their input. As new genomic assemblies become available, they will be incorporated into new FREEDA releases. Third, FREEDA automatically maps residues with the highest probabilities of having evolved under positive selection onto protein structure models by querying the AlphaFold database containing structure predictions for nearly all known proteins (Jumper et al., 2021). Finally, by providing a simple GUI, FREEDA minimizes complexity compared to currently available pipelines, while offering a restrained number of advanced options (Fig. 2 A). Therefore, the user may consider FREEDA as an entry point to performing the first molecular evolution analyses of their proteins of interest.
FREEDA validation: Finding orthologous genes
To demonstrate that FREEDA’s simplicity does not compromise its functionality, we first tested its ability to find orthologous sequences in non-annotated assemblies—a notoriously challenging task in the field. To remain unbiased, we randomly selected five genes from reference assemblies of rodents (Murinae; mouse), primates (Simiiformes; human), carnivores (Carnivora; dog), and birds (Phasianidae; chicken) and compared the FREEDA-identified orthologs to their annotations in 26 species available in the highly curated Ensembl database (Cunningham et al., 2022). While only 26/74 species used by FREEDA for these clades have Ensembl-annotated assemblies, we managed to analyze >120 orthologs. We confirmed the identities of all the FREEDA-identified orthologs by showing that they share (1) genomic location with Ensembl-annotated flanking genes and (2) an average of 99.9% (without insertions/deletions [indels]) or 90.3% (with indels) nucleotide sequence identity with Ensembl-annotated orthologs. The use of alternative exons and start codons explains the vast majority of sequence differences when indels are not excluded (see Table S1). These unbiased analyses validate FREEDA’s ability to reliably detect orthologous genes.
To ensure rigor in detecting orthologs, we tested if FREEDA can distinguish them from paralogs, which form by a duplication event and may evolve under different selective pressures. FREEDA demonstrated the ability to resolve ancient duplications by correctly distinguishing HERC5 orthologs from HERC6 paralogs present within a dataset of previously curated human immune genes used to validate the DGINN pipeline (Detect Genetic INNovations; Picard et al., 2020; see raw data of the analyzed dataset in additional online supplemental material: https://doi.org/10.5281/zenodo.7997737). Additionally, we tested if FREEDA could resolve tandem duplications (duplicated genes located side by side) and retro-duplications (intron-less mRNA that was reverse-transcribed and inserted back into the genome). Using the “Tandem duplication expected” option (see GUI; Fig. 2 A), FREEDA successfully distinguished primate genes H4C1 and H4C2, both encoding histone H4 and located merely 5 kb apart with 85% nucleotide sequence identity (additional supplementary materials). Visual examination of the nucleotide alignment revealed that one H4C2 ortholog (in Plecturocebus donacophilus) lost the ancestral start codon and likely pseudogenized. In such cases, the user may choose to rerun the analysis using the “Exclude species” option (see GUI; Fig. 2 A). We further used the “Duplication expected” option (see GUI; Fig. 2 A) to show that FREEDA could correctly distinguish KIF4A, encoding a kinesin motor, from its retroduplicate KIF4B. These genes are an example of a recent duplication that occurred in a common ancestor of primates, retaining 96% nucleotide sequence identity (Florio et al., 2018; additional supplementary materials). We also found that KIF4B, and not KIF4A, has likely evolved under positive selection, suggesting that a duplication event spurred adaptive evolution of this kinesin. While human KIF4A regulates chromosome segregation (Mazumdar et al., 2004), cellular transport (Peretti et al., 2000), and anti-viral response (Gad et al., 2022), KIF4B remains poorly studied. Together, these analyses demonstrate that FREEDA reliably finds orthologous sequences even when a gene has undergone duplication.
FREEDA validation: Detecting statistical signatures of positive selection
We then tested if FREEDA could accurately detect statistical signatures of positive selection in genes with known evolutionary histories. To do so, we used a dataset of 23 primate (Simiiformes) genes whose statistical signatures of positive selection (or lack thereof) have been previously defined. The dataset included 19 genes curated to validate the DGINN pipeline (Picard et al., 2020). Analyzing a set of 19 primate species, FREEDA found 18 orthologs with 98% coding sequence coverage (median values; Table S2). Consistent with the literature, FREEDA found statistical signatures of positive selection in TRIM5, MAVS, SAMHD1, IFI16, ZC3HAV1, RSAD2, GBP5, MX1, APOBEC3F, and NBN (Table S2). Although previous studies also reported that positive selection has likely shaped the evolution of BST2 (using nine primate species; Gupta et al., 2009; van der Lee et al., 2017), FREEDA only found a weak statistical signature of positive selection in that gene (P = 0.0864; Table S2), which likely stems from the lineage of New World monkeys (Liu et al., 2010). Of six genes whose evolutionary history is less clear, with results dependent on the method used (Picard et al., 2020), FREEDA found statistical evidence of positive selection in only one (SERINC3; Table S2), highlighting the stringency of the analysis. Of six genes whose adaptive evolution has been previously deemed unlikely, FREEDA detected a signature of positive selection in one, TREX1, a nuclease that guards genome integrity (Picard et al., 2020; Table S2). One of the residues with the highest probability of positive selection (serine at position 166 in human; probability = 0.97; Table S2) is proximal to a primate-specific DNA-binding site (arginine at position 164 in humans; Zhou et al., 2022), suggesting that adaptive evolution has shaped DNA recognition. Consistent with our finding, divergent DNA binding sites in TREX1 regulate DNA recognition (Zhou et al., 2022). We suspect that differences in regions removed from the analysis are responsible for the disparity between published results (Picard et al., 2020) and ours. Altogether, using previously curated datasets allowed us to objectively validate our pipeline and provided additional insight into the evolutionary history of these genes.
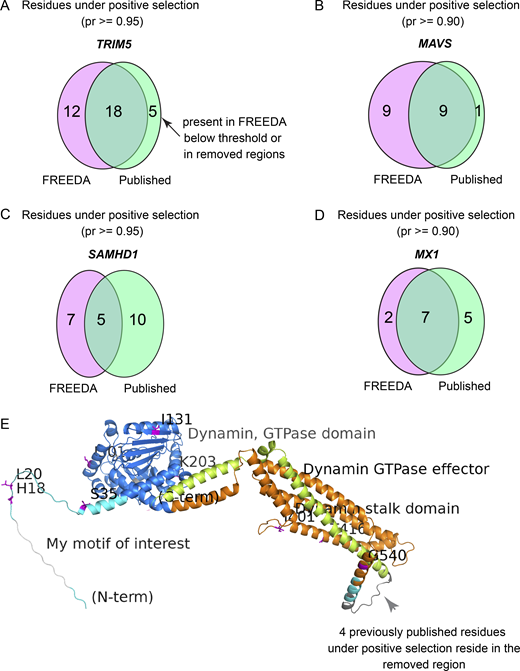
To further validate accuracy of the pipeline at the level of single residues, we compared specific sites that have likely evolved under positive selection found by FREEDA to those previously mapped in MAVS, MX1, SAMHD1, and TRIM5 (Laguette et al., 2012; Lim et al., 2012; Mitchell et al., 2012; Patel et al., 2012; Sawyer et al., 2005; van der Lee et al., 2017). Exact matching of probabilities for each residue was not expected due to differences in algorithms for aligning orthologous sequences (see Materials and methods for details). Nevertheless, FREEDA found statistical signatures of positive selection in all published sites, except for those located in regions removed from the alignment to ensure its high quality (five residues in each SAMHD1 and MX1; two residues in TRIM5; Fig. S2, A–E; and Table S3). Using MX1 as an example (Fig. 2 A), FREEDA maps detected sites onto the reference coding sequence (Fig. 2 B) and onto structural prediction models generated by AlphaFold (Fig. 2 C; Jumper et al., 2021). Overall, these analyses demonstrate that FREEDA can retrieve expected sites with previously reported signatures of positive selection and showcase FREEDA’s key results visualization features.
Pipeline validation at the level of single residues. (A)TRIM5 analysis of 16 species with 96% coding sequence (CDS) coverage as compared with Sawyer et al. (2005) and van der Lee et al. (2017). (B)MAVS analysis of 19 species with 97% CDS coverage as compared with Patel et al. (2012). (C)SAMHD1 analysis of 18 species with 92% CDS coverage as compared with Laguette et al. (2012), Lim et al. (2012), and van der Lee et al. (2017). (D)MX1 analysis of 18 species with 97% CDS coverage as compared with Mitchell et al. (2012). See Table S3 for detailed analyses. Only the number of residues from M8 vs. M7 analysis are reported (see Materials and methods for details). Comparisons are made using similar phylogenetic branches of Simiiformes (hominoids, Old World monkeys, and New World monkeys) in most cases. Probability thresholds are set to match those used by the referenced studies. Number of species used varies due to the elimination of incomplete sequences from FREEDA analyses. Magenta ellipses, number of sites matching the threshold found by FREEDA; green ellipses, number of published sites. (E) Raw FREEDA-annotated image of the MxA protein. Note the region removed from the analysis due to alignment uncertainty (dark grey arrowhead) where four residues under positive selection have previously been identified (Mitchell et al., 2012).
Pipeline validation at the level of single residues. (A)TRIM5 analysis of 16 species with 96% coding sequence (CDS) coverage as compared with Sawyer et al. (2005) and van der Lee et al. (2017). (B)MAVS analysis of 19 species with 97% CDS coverage as compared with Patel et al. (2012). (C)SAMHD1 analysis of 18 species with 92% CDS coverage as compared with Laguette et al. (2012), Lim et al. (2012), and van der Lee et al. (2017). (D)MX1 analysis of 18 species with 97% CDS coverage as compared with Mitchell et al. (2012). See Table S3 for detailed analyses. Only the number of residues from M8 vs. M7 analysis are reported (see Materials and methods for details). Comparisons are made using similar phylogenetic branches of Simiiformes (hominoids, Old World monkeys, and New World monkeys) in most cases. Probability thresholds are set to match those used by the referenced studies. Number of species used varies due to the elimination of incomplete sequences from FREEDA analyses. Magenta ellipses, number of sites matching the threshold found by FREEDA; green ellipses, number of published sites. (E) Raw FREEDA-annotated image of the MxA protein. Note the region removed from the analysis due to alignment uncertainty (dark grey arrowhead) where four residues under positive selection have previously been identified (Mitchell et al., 2012).
Finally, we tested if FREEDA can reliably detect statistical signatures of positive selection in rodent genomes (Murinae). As a test dataset, we selected 104 centromeric genes, 42 of which have been previously analyzed using a smaller number of species (up to 11; Kumon et al., 2021). Analyzing a set of 19 Murinae species, FREEDA found 16 orthologs with 94% coding sequence coverage (median values; Table S4). Consistent with our previous findings of pervasive evolutionary innovation across the rodent centromere (Kumon et al., 2021), FREEDA found that 36/104 genes have likely evolved under positive selection (Fig. 3 and Table S4). Corroborating our previous results, FREEDA detected statistical signatures of positive selection in genes encoding CENP-C, CENP-I, CENP-T, HJURP, INCENP, MIS18BP1, KNL1, and SGO2. In contrast, DSN1 and HEC1 did not show statistical signatures of positive selection. This discrepancy is likely due to a difference in coding sequence coverage between the analyses (higher in FREEDA) or it reflects a higher statistical power due to more orthologs (up to 19 used by FREEDA), which facilitates distinguishing between positive selection and relaxation of purifying selection. This high statistical power revealed several previously unknown targets of positive selection, including components of the fibrous corona, which helps capture microtubules (CENP-F, SPINDLY, ZWILCH, ROD, NUP85, NUP98, and ELYS; reviewed in Kops and Gassmann, 2020), microtubule motors (CENP-E, KIF2B, and KIF18A), and protein kinases (AURKC and HASPIN). To further validate our findings, we repeated the analyses with rat as reference species. Since the quality of the available rat genome annotation is lower than that of mouse, FREEDA was able to collect reliable input data for only 89/104 genes. As expected, we found statistical evidence (or lack thereof) of positive selection in almost exactly the same genes as when using mouse as reference (85/89 genes; see Discussion; Table S4). Overall, these tests show that despite its simplicity for the user, FREEDA is a fully functional and dependable tool to detect statistical signatures of positive selection.
Positive selection across the rodent centromere. Centromere-associated proteins are grouped to reflect their approximate positions between chromatin and microtubules. Relative protein placement is informed by binding partners, but not all known interactions are depicted. Proteins forming complexes are grouped together. Magenta: proteins that have likely evolved under positive selection.
Positive selection across the rodent centromere. Centromere-associated proteins are grouped to reflect their approximate positions between chromatin and microtubules. Relative protein placement is informed by binding partners, but not all known interactions are depicted. Proteins forming complexes are grouped together. Magenta: proteins that have likely evolved under positive selection.
Using FREEDA to derive evolution-guided hypotheses
To test if FREEDA can help derive evolution-guided hypotheses, we leveraged its ability to map residues that have likely evolved under positive selection onto protein structures. We found statistical evidence of positive selection within ancient (retained across long evolutionary timescales) protein domains of centromeric proteins, suggesting that adaptive evolution shaped essential protein functions (Fig. 4, A–F). For instance, we detected residues with high probability of having evolved under positive selection in the ancient Yippee domain (Roxström-Lindquist and Faye, 2001) of MIS18β (encoded by Oip5 in mouse), which participates in centromere chromatin assembly (reviewed in Zasadzińska and Foltz, 2017). In addition to its divergent N- and C-termini, one of the most likely adaptive residues (arginine at position 76 in mouse; probability = 0.98) is located within the loop-forming CXXC motif of the Yippee domain (Fig. 4, A and B), which is required for MIS18 complex assembly at centromeres (Fujita et al., 2007; Stellfox et al., 2016; Subramanian et al., 2016).
Positive selection in loops and turns within ancient protein domains. (A, C, and E) Ribbon diagrams show annotated structural prediction models of mouse proteins, generated automatically by FREEDA and visualized in PyMOL without manual modifications. Residues with the highest probability of having evolved under positive selection are colored magenta, and a subset of these is shown in snippets of the multiple sequence alignments in Murinae. (B, D, and F) Dark gray: highly conserved residues; gray: less conserved residues; white: non-synonymous substitutions. (A and B) MIS18β (MmMIS18β) shows the Yippee domain (orange) and two CXXC motifs (labeled by the user within the GUI, gray and yellow). The label for CXXC motif 1 is not visible to accommodate labeling of the R76 residue. The two CXXC motifs are enlarged with the R76 residue (magenta) within motif 1. (C and D) AURKC (MmAURKC) shows the protein kinase domain (orange) and activation loop (labeled by the user within the GUI, gray). A loop within the protein kinase domain is enlarged, with N150 shown in the multiple sequence alignment. (E and F) CENP-O (MmCENP-O) shows the C-terminal RWD domain (labeled by the user within the GUI, gray). The most likely adaptive residues of C-terminal RWD domain are shown in the multiple sequence alignment.
Positive selection in loops and turns within ancient protein domains. (A, C, and E) Ribbon diagrams show annotated structural prediction models of mouse proteins, generated automatically by FREEDA and visualized in PyMOL without manual modifications. Residues with the highest probability of having evolved under positive selection are colored magenta, and a subset of these is shown in snippets of the multiple sequence alignments in Murinae. (B, D, and F) Dark gray: highly conserved residues; gray: less conserved residues; white: non-synonymous substitutions. (A and B) MIS18β (MmMIS18β) shows the Yippee domain (orange) and two CXXC motifs (labeled by the user within the GUI, gray and yellow). The label for CXXC motif 1 is not visible to accommodate labeling of the R76 residue. The two CXXC motifs are enlarged with the R76 residue (magenta) within motif 1. (C and D) AURKC (MmAURKC) shows the protein kinase domain (orange) and activation loop (labeled by the user within the GUI, gray). A loop within the protein kinase domain is enlarged, with N150 shown in the multiple sequence alignment. (E and F) CENP-O (MmCENP-O) shows the C-terminal RWD domain (labeled by the user within the GUI, gray). The most likely adaptive residues of C-terminal RWD domain are shown in the multiple sequence alignment.
Similarly, we found strong statistical evidence of positive selection in one of the loops of an ancient protein kinase domain (reviewed in Taylor and Kornev, 2011) in the meiosis-specific Aurora kinase C (AURKC, asparagine at position 150 in mouse, probability = 0.98; Fig. 4, C and D), which helps correct erroneous kinetochore-microtubule attachments (Balboula and Schindler, 2014). In contrast, we found no recurrent changes in the related AURKA and AURKB kinases (Fig. S3, A–C). These data suggest that positive selection has uniquely tuned the kinase activity of the specialized meiotic Aurora kinase, consistent with previous reports of adaptive evolution of reproduction genes (Jagadeeshan and Singh, 2005; Nielsen et al., 2005; Swanson and Vacquier, 2002).
Aurora kinases differ in the number of recurrently changing residues. (A–C) Recurrently changing residues, mapped onto reference coding sequences, are not detected in AURKA (A) and AURKB (B), as compared with AURKC (C). Orange frames: protein kinase domain. Structural models highlight high structural conservation of Aurora protein kinase domains (orange). Parts of the AURKA N-terminus were cropped to allow better visualization and comparison of kinase domains.
Aurora kinases differ in the number of recurrently changing residues. (A–C) Recurrently changing residues, mapped onto reference coding sequences, are not detected in AURKA (A) and AURKB (B), as compared with AURKC (C). Orange frames: protein kinase domain. Structural models highlight high structural conservation of Aurora protein kinase domains (orange). Parts of the AURKA N-terminus were cropped to allow better visualization and comparison of kinase domains.
Finally, we found statistical evidence that positive selection shaped evolution of the ancient double RWD domain (RING-WD-DEAD; Tromer et al., 2019) of CENP-O, which regulates kinetochore–microtubule attachments by forming the CENP-OPQUR complex (Amaro et al., 2010; Bancroft et al., 2015; Chen et al., 2021; Hori et al., 2008; Singh et al., 2021; Fig. 4, E and F). RWD domains are prevalent structural modules that facilitate protein–protein interactions across the centromere (Schmitzberger and Harrison, 2012; Tromer et al., 2019). CENP-O shares a high structural similarity with its binding partner CENP-P, which also shows statistical signatures of positive selection within its double RWD domain (Fig. S4, A and B). Furthermore, some of the residues that have evolved under positive selection with the highest probability are located in or near loops and turns flanking highly structured α-helices and β-sheets in CENP-O and -P C-terminal RWD domains (Fig. 4 E and Fig. S4 A). Based on these results, we propose that positive selection has regulated essential functions of centromeric proteins by acting on loops and turns of ancient domains, consistent with previous reports of frequent innovation of flexible regions in other proteins (Afanasyeva et al., 2018; Nilsson et al., 2011; Ridout et al., 2010). Altogether, we demonstrate that FREEDA can help derive evolution-guided hypotheses by highlighting protein domains whose function has likely been shaped by adaptive evolution.
Signatures of positive selection in the CENP-P C-terminal RWD domain. (A) Annotated structural prediction model of MmCENP-P, generated automatically by FREEDA and visualized in PyMOL without manual modifications. The C-terminal RWD domain (labeled by the user within the GUI) is in gray. (B) Three residues (magenta) with the highest probability of having evolved under positive selection (≥0.9) are shown in snippets of the multiple sequence alignment in Murinae. Dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
Signatures of positive selection in the CENP-P C-terminal RWD domain. (A) Annotated structural prediction model of MmCENP-P, generated automatically by FREEDA and visualized in PyMOL without manual modifications. The C-terminal RWD domain (labeled by the user within the GUI) is in gray. (B) Three residues (magenta) with the highest probability of having evolved under positive selection (≥0.9) are shown in snippets of the multiple sequence alignment in Murinae. Dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
Using FREEDA to infer molecular mechanisms regulated by positive selection
Each of the proteins discussed above (MIS18β, AURKC, and CENP-OP) functions as part of a complex. To infer mechanisms regulated by positive selection in this context, we aligned FREEDA-annotated protein structure predictions of mouse proteins (Fig. 4) to experimentally solved structures of their orthologs in complex with binding partners (see Materials and methods for details). Two loops formed by CXXC motifs within the Yippee domain of MIS18β together give rise to a tetrahedral module whose four conserved cysteines bind a zinc ion (Subramanian et al., 2016), likely stabilizing protein conformation (Nguyen et al., 2020). Aligning mouse MIS18β to the crystal structure of the fission yeast MIS18 Yippee-like domain (Subramanian et al., 2016; Fig. 5, A and B) shows that the side chain of arginine at the positively selected position 76 in mouse likely faces the opening of the tetrahedral module. This finding is consistent with XX residues regulating the function of CXXC motifs in other proteins (Quan et al., 2007). Alternatively, R76 could mediate MIS18α and MIS18β heterodimerization (Subramanian et al., 2016). Therefore, we hypothesize that positive selection favored amino acid changes within the CXXC motif to modulate MIS18 complex stability. Consistent with functional innovation of CXXC motifs, we also found recurrently changing residues within the second CXXC motif of MIS18β (glycine at position 135 in mouse; Fig. 5, A and B) and in the first CXXC motif of its binding partner MIS18α (serine at position 57 in mouse; Fig. S5, A–C), albeit the probability that they have evolved under positive selection was lower (probabilities = 0.88 and 0.77, respectively). These data suggest that positive selection in the loops of the ancient Yippee domains regulated centromere assembly by modulating stability of the MIS18 complex.
Putative molecular interactions of likely adaptive residues. (A) Yippee domain (orange) of MmMIS18β aligned to the Yippee-like domain of fission yeast MIS18 (PDB 5JH0; Subramanian et al., 2016). FREEDA automatically annotates residues with the highest probability of having evolved under positive selection (probability ≥ 0.9, magenta). Manual annotations show residues with lower probability (probability ≥ 0.7, blue) and conserved cysteins (green). (B) Enlarged CXXC motifs forming a tetrahedral module holding a zinc ion. (C) MmAURKC protein kinase domain (orange, with activation loop in gray) and AURK-binding domain of mouse INCENP (MmINCENP; yellow), both aligned to the human AURKC-INCENP complex with the BRD-7880 inhibitor bound to the ATP-binding site (black; PDB 6GR8; Abdul Azeez et al., 2019). Annotations show likely adaptive residues (magenta, probability ≥ 0.9; blue, probability ≥ 0.7) and conserved residue Y827 of MmINCENP (yellow). (D) Enlarged ATP-binding site. Dashed lines show the closest distance from side chains of residues that likely evolved under positive selection (S156 or N150) to the BRD-7880 inhibitor or to Y827 of MmINCENP. (E) MmCENP-O (blue) and MmCENP-P (yellow) aligned to human CENP-O and -P (HsCENP-O and -P) from the human CENP-OPQUR complex (PDB 7PB8; Yatskevich et al., 2022). Gray: C-terminal RWD domains; magenta: the most likely adaptive residues (probability ≥ 0.9). Magenta arcs highlight most likely adaptive residues within the C-terminal RWD domains facing opposite sides of the heterodimer, which likely interface with other centromeric proteins.
Putative molecular interactions of likely adaptive residues. (A) Yippee domain (orange) of MmMIS18β aligned to the Yippee-like domain of fission yeast MIS18 (PDB 5JH0; Subramanian et al., 2016). FREEDA automatically annotates residues with the highest probability of having evolved under positive selection (probability ≥ 0.9, magenta). Manual annotations show residues with lower probability (probability ≥ 0.7, blue) and conserved cysteins (green). (B) Enlarged CXXC motifs forming a tetrahedral module holding a zinc ion. (C) MmAURKC protein kinase domain (orange, with activation loop in gray) and AURK-binding domain of mouse INCENP (MmINCENP; yellow), both aligned to the human AURKC-INCENP complex with the BRD-7880 inhibitor bound to the ATP-binding site (black; PDB 6GR8; Abdul Azeez et al., 2019). Annotations show likely adaptive residues (magenta, probability ≥ 0.9; blue, probability ≥ 0.7) and conserved residue Y827 of MmINCENP (yellow). (D) Enlarged ATP-binding site. Dashed lines show the closest distance from side chains of residues that likely evolved under positive selection (S156 or N150) to the BRD-7880 inhibitor or to Y827 of MmINCENP. (E) MmCENP-O (blue) and MmCENP-P (yellow) aligned to human CENP-O and -P (HsCENP-O and -P) from the human CENP-OPQUR complex (PDB 7PB8; Yatskevich et al., 2022). Gray: C-terminal RWD domains; magenta: the most likely adaptive residues (probability ≥ 0.9). Magenta arcs highlight most likely adaptive residues within the C-terminal RWD domains facing opposite sides of the heterodimer, which likely interface with other centromeric proteins.
Signatures of positive selection in the CXXC motif of the Yippee domain of MIS18α. (A) Annotated structural prediction model of mouse MIS18α (MmMIS18α), generated automatically by FREEDA and visualized in PyMOL without any manual modifications. Orange: Yippee domain, magenta: highly likely adaptive residues (probability ≥ 0.9). (B) Enlarged CXXC motifs with the S57 residue (blue) within motif 1. Green: conserved cysteine residues. (C) Snippet of the multiple sequence alignment of CXXC motif 1 in Murinae. Dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
Signatures of positive selection in the CXXC motif of the Yippee domain of MIS18α. (A) Annotated structural prediction model of mouse MIS18α (MmMIS18α), generated automatically by FREEDA and visualized in PyMOL without any manual modifications. Orange: Yippee domain, magenta: highly likely adaptive residues (probability ≥ 0.9). (B) Enlarged CXXC motifs with the S57 residue (blue) within motif 1. Green: conserved cysteine residues. (C) Snippet of the multiple sequence alignment of CXXC motif 1 in Murinae. Dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
AURKB and AURKC kinase activity requires binding to a conserved domain of INCENP (INner-CENtromere Protein; reviewed in Krenn and Musacchio, 2015). Aligning the Mus musculus (Mm) AURKC protein kinase domain and MmINCENP AURK-binding domain to the crystal structure of the orthologous human domains (Abdul Azeez et al., 2019) shows the side chain of positively selected asparagine at position 150 in mouse in close proximity to tyrosine at conserved position 827 in MmINCENP. This finding suggests modulation of INCENP binding and, therefore, kinase activity by positive selection (Fig. 5, C and D). The rodent AURKC activation loop also contains a recurrently changing, albeit less likely adaptive residue (serine at position 156 in mouse; probability = 0.77; Fig. 5, C and D) whose side chain reaches toward the AURKC ATP-binding site (marked by the inhibitor BRD-7880; Abdul Azeez et al., 2019; Fig. 5, C and D). These data suggest that positive selection in the loop of the ancient protein kinase domain of AURKC regulated meiotic functions by modulating kinase activity.
Double RWD domains mediate the formation of CENP-OP heterodimers, allowing recruitment of the CENP-OPQUR complex to centromeres (Pesenti et al., 2018; Schmitzberger and Harrison, 2012). Aligning the FREEDA-annotated CENP-O and -P C-terminal RWD domains to the experimentally solved human CENP-OPQUR complex (Yatskevich et al., 2022) suggests that positive selection shaped opposite sides of the CENP-OP heterodimer (Fig. 5 E) and therefore is unlikely to have impacted heterodimerization. In yeast, C-terminal RWD domains of CENP-O and -P orthologs bind to CENP-Q and -U orthologs to form the COMA complex (Hinshaw and Harrison, 2019; Schmitzberger and Harrison, 2012). We were unable to reliably align mouse CENP-Q and -U to the human CENP-OPQUR complex, likely due to long unstructured regions in CENP-Q and -U, but the striking pattern of likely adaptive residues in C-terminal RWD domains facing the outside of the heterodimer suggests that positive selection regulated binding to nearby centromeric components (Fig. 5 E). We find statistical signatures of positive selection in CENP-Q and -U in rodents (Fig. 3), suggesting that positive selection regulated interactions between CENP-OPQUR complex components. Altogether, these analyses of multiple centromere proteins demonstrate how FREEDA-annotated structures can be used to generate hypotheses for how positive selection might have regulated essential protein functions.
Experimental testing of functional protein innovation
To test our hypothesis that loops and turns in ancient protein domains regulate their essential functions, we chose to focus on CENP-O because FREEDA suggests that positive selection operated on residues flanking α-helices and β-sheets of both rodent and primate C-terminal RWD domains (Fig. S6, A–D), and centromere binding provides a straightforward functional assay. We used mouse oocytes for these experiments because they are an established model system for centromere drive, the most likely selective pressure sculpting evolution of centromeric proteins, and thus a natural context to probe for functional protein innovation. To create an evolutionary mismatch (see Introduction), we introduced GFP-tagged full-length mouse (control) or rat (divergent) CENP-O at similar expression levels (Fig. S7 A). Mouse CENP-O localized to centromeres as expected, but rat CENP-O was nearly undetectable at mouse centromeres (Fig. 6, A and B), indicating functional innovation in centromere binding. To test if the C-terminal RWD domain is responsible for that innovation, we compared three chimeric rat CENP-O constructs with different regions of mouse CENP-O: N-terminal (N-terminal tail and N-terminal helix), central (N-terminal RWD domain and central helix), or C-terminal (C-terminal RWD domain; Fig. 6, A and B; and Fig. S7 A). Only the mouse C-terminal RWD domain could rescue, albeit not fully, the localization of rat CENP-O to mouse centromeres. In an inverse experiment, a chimera of mouse CENP-O with the rat C-terminal RWD domain failed to localize to mouse centromeres (Fig. 6, C and D; and Fig. S7 B). Together, these results demonstrate that mouse-specific innovation in the C-terminal RWD domain is required for CENP-O binding to mouse centromeres. Within this domain, 10 out of 13 residues that differ between mouse and rat are putatively adaptive (probability ≥ 0.5; Fig. S8). Almost all (9/10) of these residues flank highly structured α-helices or β-sheets (±1 amino acid), consistent with our hypothesis that positive selection drives functional innovation of ancient domains in centromeric proteins by acting on their loops and turns. Swapping five of the most likely adaptive residues in the mouse C-terminal RWD domain to those found in rat did not, however, reduce centromere localization of mouse CENP-O (Fig. S9, A–C). Similarly, swapping equivalent rat residues within the C-terminal RWD domain of rat CENP-O to mouse-specific ones (in addition to six mutations in other parts of the protein) did not restore its centromere localization (Fig. S10, A–C). These analyses highlight the difficulty in attributing innovation to specific residues given the number of possible combinations as well as the potential for epistasis (Starr and Thornton, 2016). Altogether, we show that our fully automated molecular evolution pipeline can guide experimental testing of functional protein innovation.
Signatures of positive selection in loops and turns of rodent and primate CENP-O C-terminal RWD domains. (A and B) Most likely adaptive residues in rodent (A) or primate (B) CENP-O mapped to the mouse or human CENP-O coding sequence, respectively, by FREEDA. Blue frames were manually added to outline C-terminal RWD domains. (C and D) FREEDA-annotated C-terminal RWD domains of mouse (C) and human (D) CENP-O. Magenta: highly likely adaptive residues (probability ≥ 0.9).
Signatures of positive selection in loops and turns of rodent and primate CENP-O C-terminal RWD domains. (A and B) Most likely adaptive residues in rodent (A) or primate (B) CENP-O mapped to the mouse or human CENP-O coding sequence, respectively, by FREEDA. Blue frames were manually added to outline C-terminal RWD domains. (C and D) FREEDA-annotated C-terminal RWD domains of mouse (C) and human (D) CENP-O. Magenta: highly likely adaptive residues (probability ≥ 0.9).
Expression levels of microinjected CENP-O–GFP are equal between constructs. (A and B) Quantification of CENP-O–GFP levels in cytoplasm for constructs analyzed in Fig. 6, A and B (A) and Fig. 6, C and D (B). Each spot represents one cell. For each construct, 38 cytoplasmic ROIs per cell were analyzed (see Materials and methods for details) from ≥19 cells from three (A) or two (B) independent experiments. Bars: mean intensity with standard deviation. *P < 0.05; ns: not significant; one-way ANOVA with Tukey’s multiple comparison test (A) or two-tailed Student's T test (B).
Expression levels of microinjected CENP-O–GFP are equal between constructs. (A and B) Quantification of CENP-O–GFP levels in cytoplasm for constructs analyzed in Fig. 6, A and B (A) and Fig. 6, C and D (B). Each spot represents one cell. For each construct, 38 cytoplasmic ROIs per cell were analyzed (see Materials and methods for details) from ≥19 cells from three (A) or two (B) independent experiments. Bars: mean intensity with standard deviation. *P < 0.05; ns: not significant; one-way ANOVA with Tukey’s multiple comparison test (A) or two-tailed Student's T test (B).
Experimental evidence of functional innovation in the CENP-O C-terminal RWD domain. Mouse oocytes expressing mouse, rat, or chimeric CENP-O–GFP were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). (A and C) Images show maximum intensity projections; scale bars, 5 μm. (B and D) Graphs show CENP-O–GFP intensity at centromeres; for each construct, n ≥ 722 centromeres from ≥19 cells from three (B) or two (D) independent experiments. Each spot represents one cell; bars: mean intensities with standard deviation; ****P < 0.0001, ns: not significant, one-way ANOVA with Tukey’s multiple comparison test (B) or two-tailed Student's T test (D).
Experimental evidence of functional innovation in the CENP-O C-terminal RWD domain. Mouse oocytes expressing mouse, rat, or chimeric CENP-O–GFP were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). (A and C) Images show maximum intensity projections; scale bars, 5 μm. (B and D) Graphs show CENP-O–GFP intensity at centromeres; for each construct, n ≥ 722 centromeres from ≥19 cells from three (B) or two (D) independent experiments. Each spot represents one cell; bars: mean intensities with standard deviation; ****P < 0.0001, ns: not significant, one-way ANOVA with Tukey’s multiple comparison test (B) or two-tailed Student's T test (D).
Multiple sequence alignment of the rodent CENP-O C-terminal domain. Schematic shows α-helices and β-sheets (gray boxes) and loops and turns (gray lines). Asterisks indicate highly likely adaptive residues (magenta, probability ≥ 0.9), less likely adaptive residues (blue, probability ≥ 0.5), and residues that differ between mouse and rat but do not evolve under positive selection (black). Magenta arrows indicate residues swapped in the experiment. Alignment: dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
Multiple sequence alignment of the rodent CENP-O C-terminal domain. Schematic shows α-helices and β-sheets (gray boxes) and loops and turns (gray lines). Asterisks indicate highly likely adaptive residues (magenta, probability ≥ 0.9), less likely adaptive residues (blue, probability ≥ 0.5), and residues that differ between mouse and rat but do not evolve under positive selection (black). Magenta arrows indicate residues swapped in the experiment. Alignment: dark gray: highly conserved residues, gray: less conserved residues, white: non-synonymous substitutions.
Swapping five most likely adaptive residues in the CENP-O C-terminal RWD domain is insufficient to reduce centromere binding. Mouse oocytes expressing the indicated CENP-O–GFP constructs were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). One construct is rat CENP-O with the mouse C-terminal RWD domain, and the other is identical except for five mutations in the RWD domain swapping mouse-specific residues with the highest probability of having evolved under positive selection (probability ≥ 0.9; R225G, C226T, T228A, N247G, P261H) to the corresponding rat-specific residues. (A–C) Images (A) show maximum projections; scale bars, 5 μm. Graphs show CENP-O-GFP intensity at centromeres (B) or in the cytoplasm (C); for each construct, n ≥ 950 centromeres from ≥25 cells from three independent experiments. Each spot represents one cell; bars, mean intensities with standard deviation; ns: not significant, two-tailed Student's T test.
Swapping five most likely adaptive residues in the CENP-O C-terminal RWD domain is insufficient to reduce centromere binding. Mouse oocytes expressing the indicated CENP-O–GFP constructs were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). One construct is rat CENP-O with the mouse C-terminal RWD domain, and the other is identical except for five mutations in the RWD domain swapping mouse-specific residues with the highest probability of having evolved under positive selection (probability ≥ 0.9; R225G, C226T, T228A, N247G, P261H) to the corresponding rat-specific residues. (A–C) Images (A) show maximum projections; scale bars, 5 μm. Graphs show CENP-O-GFP intensity at centromeres (B) or in the cytoplasm (C); for each construct, n ≥ 950 centromeres from ≥25 cells from three independent experiments. Each spot represents one cell; bars, mean intensities with standard deviation; ns: not significant, two-tailed Student's T test.
Swapping 11 most likely adaptive residues in rat CENP-O is insufficient to rescue centromere binding. Mouse oocytes expressing the indicated CENP-O–GFP constructs were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). The constructs are: mouse CENP-O, rat CENP-O, and rat CENP-O with 11 mouse-specific residues with the highest probability of having evolved under positive selection (probability ≥ 0.9; L99V, Y123C, A141V, S177R, S179W, V186A, G225R, T226C, A228T, G247N, and H261P). (A–C) Images (A) show maximum projections; scale bars, 5 μm. Graphs show CENP-O–GFP intensity at centromeres (B) or in the cytoplasm (C); for each construct, n ≥ 684 centromeres from ≥18 cells from two independent experiments. Each spot represents one cell; bars, mean intensities with standard deviation; ****P < 0.0001, ns: not significant, one-way ANOVA with Tukey’s multiple comparison test.
Swapping 11 most likely adaptive residues in rat CENP-O is insufficient to rescue centromere binding. Mouse oocytes expressing the indicated CENP-O–GFP constructs were fixed in meiosis I and stained for centromeres (CREST) and DNA (DAPI). The constructs are: mouse CENP-O, rat CENP-O, and rat CENP-O with 11 mouse-specific residues with the highest probability of having evolved under positive selection (probability ≥ 0.9; L99V, Y123C, A141V, S177R, S179W, V186A, G225R, T226C, A228T, G247N, and H261P). (A–C) Images (A) show maximum projections; scale bars, 5 μm. Graphs show CENP-O–GFP intensity at centromeres (B) or in the cytoplasm (C); for each construct, n ≥ 684 centromeres from ≥18 cells from two independent experiments. Each spot represents one cell; bars, mean intensities with standard deviation; ****P < 0.0001, ns: not significant, one-way ANOVA with Tukey’s multiple comparison test.
Discussion
The motivation to develop FREEDA was to catalyze participation of the cell biology community in testing functional consequences of protein innovation. We demonstrate that detecting statistical signatures of positive selection, which implicates functional innovation, can be fully automated by compiling widely used bioinformatic and molecular evolution tools into a single pipeline (Fig. 1). FREEDA’s simple and user-friendly GUI makes it a suitable entry point for experimentalists who may have limited programming skills (Fig. 2). Moreover, by leveraging the ever-growing pool of newly sequenced but not yet annotated genomic assemblies, FREEDA bypasses the requirement for obtaining tissue samples and cloning the genes of interest to have sufficient numbers of orthologs from closely related species to detect signatures of positive selection. Nevertheless, as with any fully automated tool, FREEDA has limitations. First, by inferring orthologs based on the annotated reference sequence, rather than experimentally validated transcripts, FREEDA does not account for tissue-specific splicing, shifts in intron–exon boundaries, or the use of alternative exons (see Table S1). Despite this caveat, using independently annotated rat sequences as reference led to the same result as using mouse annotations in 85/89 centromeric genes (note that relatively poorer rat genome annotation quality prevented reliable input generation for 15 genes; Table S4). Nevertheless, isoforms that substantially differ from the reference coding sequence might interfere with the accurate detection of positive selection. As an example, annotated variants of the rat NUP37 nucleoporin substantially differ at their C-termini from the reference mouse NUP37 sequence, suggesting the use of alternative exons (additional supplementary materials), which likely led to inconsistent signals of positive selection (P = 0.510 with mouse as reference vs. P = 0.0499 with rat as reference; Table S4). Second, to prioritize computational speed and reduce output complexity, FREEDA does not test for possible recombination events known to increase the probability of false positives when recombination rates are high (Anisimova et al., 2003). While estimated recombination frequencies are lower in vertebrates and insects as compared to yeast and protozoa, we cannot exclude the influence of recombination events (Stapley et al., 2017; Wilfert et al., 2007). Third, while FREEDA can robustly resolve gene duplications present in the ancestor of a selected taxon (e.g., primates), caution is advised when analyzing lineage-specific genes. For example, primate MICA (MHC class I chain-related gene A) is known to have duplicated from MICB in the common ancestor of hominoids and Old World monkeys (Florio et al., 2018). Therefore, searching for MICA orthologs across the entire primate taxon yields MICB coding sequences in New World monkeys (additional supplementary materials). In case lineage-specificity is suspected, we suggest using the “Subgroup” option (currently supporting: hominoidea, catarrhini, caniformia, and melanogaster subgroup) and/or the “Exclude species” option (Fig. 2 A). Fourth, FREEDA is designed to test for signatures of recurring (pervasive) positive selection acting on the entire taxon (e.g., primates) rather than episodic selection that may have led to adaptation in a specific branch (e.g., hominoids). Nevertheless, using the aforementioned “Subgroup” or “Exclude species” options allows narrowing of the phylogenetic window if needed. Finally, mapping FREEDA’s results onto protein structures is not yet fully supported for carnivores (Carnivora) and birds (Phasianidae).
Our analyses of genes with known evolutionary histories demonstrate that FREEDA is reliable. Building on this validation, we provide the most detailed characterization of signatures of positive selection at rodent centromeres to date. Consistent with previous analyses (Kumon et al., 2021), we infer pervasive evolutionary innovation in domains of centromeric proteins that do not directly touch DNA, such as RWD (Fig. 3). Therefore, our data support the idea that fitness costs of centromere drive are suppressed by innovation in protein–protein interactions (Kumon et al., 2021; Rosin and Mellone, 2016) as well as protein–DNA interactions (Henikoff et al., 2001; Malik et al., 2002; Vermaak et al., 2002). Furthermore, by mapping regions that are likely under positive selection onto protein structures, we derive a hypothesis that positive selection acting on loops and turns of ancient domains impacts essential protein functions. For example, recurrent amino acid changes within the loops formed by CXXC motifs could regulate MIS18 complex formation. Similarly, recurrent changes in loops of the ancient AURKC protein kinase domain could modulate kinetochore–microtubule attachment dynamics, specifically in meiosis I. Both centromere assembly and microtubule detachment (in meiosis I) represent mechanisms potentially hijacked by selfish centromeres (Akera et al., 2019; Henikoff et al., 2001; Rosin and Mellone, 2016; Wu et al., 2018). These analyses provide a starting point for future experiments probing the functional impacts of innovation, including testing whether positive selection reduces fitness costs associated with centromere drive (reviewed in Dudka and Lampson, 2022).
Previous experiments in fruit flies, using evolutionarily mismatches of the L1 loop within the ancient histone fold domain of CidCENP-A, suggested functional innovation in a DNA-binding domain of a centromere protein (Rosin and Mellone, 2016; Vermaak et al., 2002). Here, we propose that positive selection in CENP-O may have regulated centromere binding via recurrent changes in loops and turns within an ancient C-terminal RWD domain (Fig. 6), which does not interact with DNA (Pesenti et al., 2022; Yatskevich et al., 2022). While we are unable to pinpoint the exact combination of residues responsible for functional innovation, the observation that most recurrently changing residues are within regions that flank the highly structured α-helices or β-sheets of that domain supports our hypothesis. CENP-O is expected to dock the CENP-OPQUR complex to centromeres (Eskat et al., 2012; Pesenti et al., 2018), promoting kinetochore–microtubule attachment stability (Amaro et al., 2010; Bancroft et al., 2015; Chen et al., 2021; Hori et al., 2008; Singh et al., 2021). Therefore, we propose that innovation in the CENP-O C-terminal RWD domain modulated interactions with other centromeric components (possibly CENP-Q and -U) necessary to form a stable CENP-OPQUR complex at centromeres (Foltz et al., 2006; Hori et al., 2008; Kagawa et al., 2014; Minoshima et al., 2005; Okada et al., 2006; Pesenti et al., 2018), potentially stabilizing kinetochore–microtubules to counteract destabilizing activities associated with driving centromeres (Akera et al., 2019; Wu et al., 2018). Future work using centromere drive models (reviewed in Dudka and Lampson, 2022) will be needed to experimentally test this idea. Overall, we show how FREEDA can help derive evolutionary hypotheses and guide experimental testing of functional innovation, starting from just a gene name, making it a powerful tool for incorporating evolutionary analyses into cell biology research and generating new insights into essential cellular processes.
Materials and methods
Resources and datasets
FREEDA was written in Python and compiled into a stand-alone application using pyinstaller (https://pyinstaller.org/en/stable/). Core packages used for the compilation (Table S5) were installed using standard package managers: pip (https://pypi.org/project/pip/) and conda (https://docs.conda.io/en/latest/). All selected genomic assemblies and Ensembl releases used to generate datasets are listed in Table S5. All data were collected using desktop computers: iMac (late 2015) with MacOS Monterey 12.6 (8 GB RAM; 4 CPU cores; 2.8 GHz Quad-Core Intel Core i5) and iMac (2017) with MacOS Ventura 13.2 (16 GB RAM, 2 CPU cores; 2.3 GHz Intel Core i5), or a laptop MacBook Pro (mid-2014) with MacOS Mojave 10.14.6 (16 GB RAM, 2 CPU cores; 3 GHz Intel Core i7).
Input extraction and identification of potential orthologous exons
FREEDA can be downloaded from an open-source repository: https://github.com/DDudka9/freeda/releases. When running the app for the first time, MacOS users will be prompted to download PyMOL, which renders a 3D result of the FREEDA analysis. To run the pipeline, the user needs to provide at least one gene name, select a reference species, and select a location where all the data will be stored. At least 100 GB of storage space is needed to analyze a single vertebrate taxon (e.g., primates), 20 GB is sufficient to analyze only flies, and 500 GB is recommended to analyze all taxons. Optionally, the user can (1) specify the coordinates of residues or domains of interest that will be labeled on the protein structure prediction, (2) customize the BLAST search and ortholog finding (see below), (3) exclude selected species from the analysis, and (4) narrow the analysis to a specific subgroup (e.g., catarrhini). Advanced users can also specify the codon frequency model used (F3X4 or F3X4 and F61; see PAML manual for details: http://abacus.gene.ucl.ac.uk/software/pamlDOC.pdf). The pipeline starts with downloading the reference genome (using NCBI Datasets [Sayers et al., 2021]) and then retrieving all possible UniProt IDs (UniProt Consortium, 2021) for a protein encoded by the gene of interest and matching them to AlphaFold database entries (Jumper et al., 2021). Next, FREEDA extracts protein sequence and coding sequence (using pyensembl package: https://github.com/openvax/pyensembl) from the Ensembl database (Cunningham et al., 2022) and exon sequences and gene sequence (using pybedtools [Dale et al., 2011]) from the downloaded reference genome. Visualization of residues that have likely evolved under positive selection requires that the protein sequence of the structural prediction and the protein sequence from the Ensembl database are identical (tested using the Biopython package; Cock et al., 2009). If the proteins are not identical, FREEDA performs the analysis without mapping the residues onto structure predictions. The first run triggers downloading of the selected reference genome (e.g., human), followed by the genomes of closely related species (e.g., Simiiformes), and then building of local BLAST databases (using BLAST + applications; Camacho et al., 2009). FREEDA queries these databases to find genomic coordinates of putative orthologous regions using tblastn algorithms (default identity threshold is set at 60% [or 30% for Drosophila] but can be increased to 80% [or 60% for Drosophila] by selecting the advanced option “Common domains expected”) and retrieves corresponding nucleotide sequences (using pybedtools) from downloaded related genomes. Overall, these features allow fully automated generation of the input data needed to find orthologous coding sequences in non-annotated genomes.
Finding orthologous exons
FREEDA performs a multiple sequence alignment of each region found during the BLAST search to the reference coding sequence, BLAST sequences stitched together, the genomic locus these sequences reside in (contig), and the reference gene sequence using MAFFT (Multiple Alignment using Fast Fourier Transform; Katoh and Standley, 2013). Regions aligning to both the coding sequence and the reference gene sequence are considered putative exons. To determine if the putative exon is syntenic (resides in a homologous locus), the flanking sequence is compared with the introns of the reference gene separately at 5′ and 3′ ends. An exon is considered syntenic if at least one of the flanking regions is at minimum 75% (60% for Drosophila) identical to the reference intron over a stretch of at least 50 bp (30 bp for Drosophila). The identity is calculated as the Hamming distance (the number of different bases in a pair-wise comparison of two aligned sequences; Hamming, 1950; divided by the sequence length). The putative exon is called as not syntenic and discarded from the analysis if none of the flanking regions reaches the identity threshold and the exon itself is <80% (70% for Drosophila) identical to the reference exon over a stretch of the first 30 bp. Since introns are generally less conserved than exons, when the identity of a flanking region is uncertain (66–75%; 50–60% for Drosophila), a longer sequence is compared. Lowered values for detecting synteny in Drosophila genomes are due to an observed high divergence of intragenic regions and high rates of indels within orthologous loci.
To increase stringency in detecting synteny and reduce uncertainty resolving segmental gene duplications, the user can additionally select an advanced option “Duplication expected” that penalizes any exon that is syntenic only at one end (e.g., recent segmental duplication whose introns have not yet diverged significantly). Segmental duplications that lead to duplicated genes residing next to each other (tandem duplications) will not only have similar flanking regions but might also be difficult to align if residing on the same contig. To ensure robust analysis of tandem duplications, the “Tandem duplication expected” option limits the flanking region of each blast hit (leading to smaller contigs), decreasing the chance of tandemly duplicated genes residing on the same contig. In addition, to avoid retroduplications (mRNA-derived gene duplications; Kaessmann et al., 2009), FREEDA always discards exons that are at least 80% (70% for Drosophila) identical to the reference exon but lack the flanking regions (intron-less).
To preserve intron–exon boundaries, each putative exon is given a number and directly aligned to a reference exon of the same number. Therefore, exons do not need to reside on the same contig to form a complete coding sequence, which is helpful when querying genomic assemblies with short contigs. Very small exons (microexons; reviewed in Ustianenko et al., 2017) shorter than 18 bp cannot be reliably aligned and are discarded from the analysis. If the same putative exon is found on different contigs (e.g., due to a duplication), the contig containing fewer putative exons is discarded. If both contigs carry the same number of putative exons (likely due to heterozygosity of the orthologous locus or a very recent duplication), these are compared to corresponding reference exons, and the contig with a higher overall identity of exons is considered orthologous. Rare cases of mistakes in aligning intron–exon boundaries may lead to indels, which are resolved at later stages (e.g., the entire codon is removed in case of a 1-nt indel). To allow manual review, all the above-mentioned steps are logged and all the intermediate alignments are saved as raw data (“Raw_data” folder).
Manual verification of detected orthologs
Genomic location and nucleotide sequence identity of >120 FREEDA-identified orthologs representing 20 randomly selected genes (five per clade: Murinae, Simiiformes, Carnivora, Phasianidae) were compared with their annotations found in the Ensembl database. Genomic location (contig number) logged in the “FREEDA-current-date.log” file was compared to that of the expected flanking genes that were also analyzed by FREEDA. Nucleotide sequence identity with Ensembl-annotated orthologous coding sequences was measured using pairwise alignment and MAFFT protocol designed to limit over-aligning errors (Katoh and Standley, 2016). This approach generates large indels in the alignment and facilitates detection of alternative exons and start codons. Each alignment was visually inspected without manual curation. See detailed analysis and commentary in Table S1.
Manual verification of known recent gene duplications
The ability to distinguish tandem duplication (H4C1 from H4C2) and recent retro-duplication (KIF4A from KIF4B) was tested by manual BLAST (blastn) of the nucleotide sequence of each ortholog identified by FREEDA (“GENE_raw_nucleotide_alignment.fasta” file) against the primate NCBI gene database (Simiiformes; taxid: 314294). For each gene, an orthologous sequence was always the highest scoring hit (by similarity) as opposed to a paralogous sequence. Exceptions were sister species Aotus nancymaae and Callithrix jacchus, whose identified KIF4B coding sequences were more similar to KIF4A than KIF4B. However, all the KIF4B orthologs detected by FREEDA were intron-less, consistent with KIF4B being a primate-specific retro-duplication of KIF4A (Florio et al., 2018), which FREEDA called correctly. Therefore, we are confident that FREEDA identified KIF4B orthologs for all species and not KIF4A paralogs (additional supplementary materials).
Building the multiple sequence alignment and phylogenetic gene tree
Detection of recurring amino acid substitutions requires a gapless, in-frame multiple sequence alignment. To avoid large gaps (suggesting incomplete coding sequences), FREEDA first removes entire coding sequences that are shorter than 90% compared with the reference sequence. To ensure high-quality alignment of the remaining sequences, we tested the commonly used aligners: MUSCLE (Edgar, 2004), PRANK (Löytynoja and Goldman, 2005), MACSE (Ranwez et al., 2011), and MAFFT (Katoh et al., 2002). We decided to use a modified MAFFT protocol designed to limit over-aligning errors (Katoh and Standley, 2016) as we found it both fast and accurate. To curate the alignment, FREEDA removes insertions that are defined as regions missing in the reference coding sequence and deletes stop codons (including premature ones). At this point, coding sequences that are <69% (60% for Drosophila) identical to the reference sequence are discarded as likely too divergent to produce a reliable alignment (based on Jeffares et al., 2015; Sievers et al., 2011) or misaligned. Additionally, remaining small gaps (deletions) and ambiguously aligned codons are removed from the alignment (using Gblocks; Castresana, 2000; Talavera and Castresana, 2007). Note that MAFFT is not codon-aware, which allows aligning incomplete sequences (e.g., missing some of the exons or containing indels) that cannot be expected to have a length of multiplication of 3. Therefore, to ensure that the aligning process and alignment curation did not alter the open reading frame of the aligned sequences, FREEDA compares the identity of the translated reference sequence within the curated alignment to the original reference protein sequence from the Ensembl database. Once 100% identity is confirmed, FREEDA translates the orthologous sequences and checks if >10 contiguous non-synonymous substitutions compared with the reference sequence are present. FREEDA considers such rare cases as likely frameshift events and removes the entire sequences from the alignment. Final in-frame multiple nucleotide sequence alignment is then used to build a phylogenetic gene tree (using RAxML; Stamatakis, 2014), which guides the widely used CODEML program from the PAML suite (Yang, 2007) to infer the enrichment of recurrent non-synonymous substitutions suggestive of positive selection. We strongly urge the user to manually verify the final protein alignment (“Results-Current-Date/Results/Protein_alignments/GENE_protein_alignment.fasta” file), ensuring that there are no obvious misaligned regions before considering the results of the PAML analysis (e.g., using the free software Unipro UGENE; Okonechnikov et al., 2012). In case of apparent misalignments, we suggest simply rerunning the analysis using the “Exclude species” option (Fig. 2 A). An example of a misaligned sequence can be found in the documentation.
Detection of positive selection
To detect statistical signatures of positive selection, FREEDA relies on the rate ratio of non-synonymous (dN) to synonymous (dS) substitutions (dN/dS > 1 suggests positive selection). However, most genes contain conserved regions that evolve under purifying selection (dN/dS < 1), which usually decreases gene-wide dN/dS below 1. Therefore, to find specific regions that likely evolved under positive selection, FREEDA uses “site models” of the CODEML program that allow for varying dN/dS between different codons. Each model describes a set of parameters (including dN/dS per codon; for details, see the official PAML guide, http://abacus.gene.ucl.ac.uk/software/pamlDOC.pdf, or a beginners guide; Jeffares et al., 2015) and either allows for sites (codons) with a dN/dS ratio of >1 (signature of positive selection; M8 and M2a models) or not (null hypothesis; M7 and M1a models). Using a maximum likelihood approach, CODEML then fits the parameters estimated from the data to each model. Significantly more likely fit (based on the likelihood ratio test) to the model that allows for codons with dN/dS > 1 indicates the presence of sites that have likely evolved under positive selection. Bayesian statistics (Bayes Empirical Bayes) are then used to estimate probabilities of positive selection acting on specific codons. FREEDA outputs the key results of the CODEML analysis within the GUI’s “Results window” (likelihood ratio test value for M7 vs. M8 comparison, P value, and number of codons with the highest probability to have evolved under positive selection). Additionally, the results of the M1a vs. M2a comparison and the identity of specific codons under positive selection are saved in an Excel sheet (“Results-Current-Date/Results/Results_sheet” folder).
Visualization of residues under positive selection
FREEDA maps the protein sequence of the reference species from the multiple sequence alignment to the expected reference protein sequence and, if appropriate, introduces gaps that represent residues excluded from the analysis (“GENE_protein_alignment.fasta” file). Based on that mapping, FREEDA provides both 2D and 3D visual representations of residues that likely evolved under positive selection. 2D bar graphs are provided in the “Results-Current-Date/Results/Graphs” folder. These graphs display the positions of recurrently changing residues (“Posterior mean omega,” top) and the probability of positive selection acting on each codon (“Prob. positive selection,” middle, probability 0.7–1.0; “High prob. positive selection,” bottom, probability 0.9–1.0). Codons excluded from the analysis are marked in gray. 3D representation of the most likely adaptive residues is found in the “Results-Current-Date/Results/Structures” folder, provided that the prediction model from the AlphaFold database matches the protein sequence extracted from the Ensembl database. For clarity, only residues with the highest probability (≥0.9) of having evolved under positive selection are mapped and their side chains are shown. The residues excluded from the analysis are colored in gray, and the N-terminal and C-terminal ends are labeled. Additionally, any domain annotation available in the Interpro database (Blum et al., 2021) is automatically marked with a distinct color and labeled allowing quick visual identification.
Manual alignment of structural prediction models
FREEDA-annotated protein structure prediction models from AlphaFold designated by their UniProt entries (MmMIS18β—A2AQ14; MmAURKC—O88445; MmINCENP—Q9WU62; MmCENP-O—Q8K015; MmCENP-P—Q9CZ92) were aligned to PDB entries (SpMIS18—5HJ0; HsAURKC—6GR8; HsINCENP—6GR8; HsCENP-OP—7PB8) using an aligner module in PyMOL. Briefly, a FREEDA-annotated structure (e.g., “Aurkc_Mm.pse” found in “Results-Current-Date/Results/Structures”) was opened in PyMOL, the selected PDB entry was downloaded (e.g., “fetch 6GR8”), and the two structures were aligned (e.g., “align Aurkc_Mm, 6GR8”). The align module first aligns protein sequences and then superimposes their structures, returning RMSD (Root-Mean-Square-Deviation). Lower RMSD values indicate better alignment. All alignments presented here returned RMSD below 2 Å.
Generation of CENP-O constructs
All CENP-O coding sequences were cloned from testis or liver samples. The use of rat CENP-O to represent a divergent ortholog was motivated by the availability of a rat (Rattus norvegicus) tissue sample for cloning. Chimeric CENP-O constructs were designed based on their 3D structure (AlphaFold database). Tissue was mechanically homogenized and total mRNA was isolated using TRIzol reagent (15596026; Invitrogen); cDNA was prepared using reverse transcription (18080051; SuperScript III First-Strand Synthesis System), amplified using construct-specific PCR primers (KK2502; KAPA HiFi Hot Start plus dNTPs; Roche), and inserted into the pGEMHE plasmid backbone (638948; In-fusion kit; Takara). Primers were designed using SnapGene (Dotmatics) software and are listed in Table S6. Each CENP-O construct was tagged with GFP at the C-terminus, separated by a linker of five glycines. Site-directed mutagenesis was performed using the Quik-Change Multisite Directed Mutagenesis kit (200515; Agilent) to introduce R225G, C226T, T228A, N247G, and P261H mutations. Rat CENP-O with 11 mouse point mutations was synthesized (Genewiz). The identity of all constructs was confirmed using Sanger sequencing of the entire coding sequence, including the reporter gene.
Oocyte isolation, microinjection, and in vitro maturation
Mus musculus mice (CF-1 strain) were purchased from Envigo NSA stock # 033. Females were primed with 5 U of pregnant mare somatic gonadotropin (367222; Calbiochem) injected into the intraperitoneal cavity 44–48 h prior to oocyte collections to induce superovulation. The ovaries were isolated using M2 medium (M7167; Sigma-Aldrich) supplemented with 2.5 mM of the maturation-blocking phosphodiesterase 3 inhibitor milrinone (M4659; 2.5 mM; Sigma-Aldrich Milipore). Germinal-vesicle oocytes were collected, denuded mechanically from cumulus cells, and incubated for at least 1 h prior to microinjection on a hot plate (38°C) under mineral oil (9305; FUJIFILM Irvine Scientific). Oocytes were then microinjected with ∼5 pl of mRNAs in M2 medium with 2.5 mM milrinone and 3 mg/ml BSA at RT with a micromanipulator TransferMan NK 2 (Eppendorf) and a picoinjector (Medical Systems Corp). Oocytes were then incubated in 30–50 μl drops of Chatot-Ziomek-Bavister medium (MR019D; Thermo Fisher Scientific) under mineral oil (M5310; Sigma-Aldrich Milipore) at 37.8°C and 5% CO2 (Airgas) for 16 h to allow protein expression. The concentration of mRNA (15–30 ng/μl) used was selected to ensure similar cytoplasmic expression. mRNAs were synthesized using the T7 mScriptTM Standard mRNA Production System (C-MSC100625; CELLSCRIPT).
Immunofluorescence imaging
Oocyte maturation was induced in vitro by washing out milrinone 7.5 h before fixation. MI oocytes were fixed in freshly prepared 2% paraformaldehyde in PBS (pH 7.4) with 0.1% Triton X-100 for 20 min at RT, permeabilized in PBS with 0.1% Triton X-100 for 15 min at RT, placed in blocking solution (PBS with 0.3% BSA and 0.01% Tween-20) overnight at 4°C, incubated 1 h with primary antibody in blocking solution, washed three times for 15 min each, incubated 1 h with secondary antibody, washed three times for 15 min each, and mounted in Vectashield with DAPI (H-1200; Vector) to visualize chromosomes. Centromeres were labeled with CREST (human anti-human anti-centromere antibody, 1:200, HCT-0100; Immunovision) and an Alexa Fluor 594–conjugated goat anti-human secondary antibody (A-110014; Thermo Fisher Scientific). Confocal images were collected as 31 z-stacks at 0.5-μm intervals to visualize the entire meiotic spindle, using a confocal microscope (DMI4000B; Leica) equipped with a 63× 1.3 NA glycerol-immersion objective lens, an xy piezo Z stage (Applied Scientific Instrumentation), a spinning disk (Yokogawa Corporation of America), and an electron multiplier charge-coupled device camera (ImageEM C9100-13; Hamamatsu Photonics), controlled by MetaMorph software (Molecular Devices). Excitation was done with a Vortran Stradus VersaLase 4 laser module with 405-, 488-, 561-, and 639-nm lasers (Vortran Laser Technology). Panels of microscopic images were prepared using ImageJ (National Institutes of Health) free software.
Automated image analysis
Confocal images were analyzed using a custom-built Python-based automated program “Centrocalc” available here: https://github.com/DDudka9/Centrocalc. First, the program performs whole chromosome segmentation using the chromosome channel as a mask by grayscale dilation at a width of 16 pixels to ensure the centromeres are included. Then, a threshold is calculated using the Iterative Self-Organizing Data Analysis Technique method. If no chromosome channel is given, the entire cell is considered. Second, the program identifies centromeres using an approach based on (Vermolen et al., 2008). A difference of Gaussians algorithm is used to isolate spots of 150 nm. Third, a local maxima algorithm is used to identify centromeres. Up to 38 spots are chosen (the mouse 2n genome contains 40 chromosomes), separated by a minimum of two pixels by Chebyshev distance. The spots must be away from the edges of the image (20 pixels in x and y; 2 pixels in z). Fourth, 3D ellipsoid regions of interest (4 × 4 × 3 pixels) are drawn using the local maxima. Background regions of interest (ROIs) are drawn as volumes around the centromere ROIs (expanding the 3D ellipsoids by 1 pixel in all dimensions). Overlapping centromere ROIs are resolved by distance, where each pixel is assigned to the closest maxima. Fifth, centromere and background intensities are calculated as average grayscale pixel values and saved. ImageJ ROI files are also created to reference back to the original images. Modifying the Centrocalc source code can customize most of the described parameters.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.3.1 (GraphPad Software). Statistical significance was assessed using a two-tailed T test (data distribution was normal under Kolmogorov–Smirnov and Shapiro–Wilk tests) or one-way ANOVA with Tukey’s multiple comparison test (data distribution was assumed normal but was not formally tested). Graphs display means with standard deviations. P values indicated on graphs are P ≥ 0.05, not significant (ns); P < 0.05, *; and P < 0.0001, ****.
Online supplemental material
Comparison of FREEDA with other pipelines is outlined in Fig. S1. Results of FREEDA validation are listed in Table S1 (validation of ortholog detection based on Ensembl database), Table S2 (accuracy in detecting positive selection using published datasets), Fig. S2, and Table S3 (accuracy in detecting sites under positive selection based on published data). Results of positive selection detection in rodent centromere and kinetochore proteins are shown in Table S4. The list of all genomic assemblies used by the FREEDA pipeline and the core packages comprising the pipeline are listed in Table S5. Table S6 lists all primers used to generate CENP-O constructs. Figs. S3, S4, S5, and S6 show structural analyses of selected centromeric proteins. Experimental analyses of rodent CENP-O are shown in Figs. S7, S9, and S10. Fig. S8 indicates sites that differ between mouse and rat CENP-O.
Data availability
FREEDA pipeline can be downloaded from the public GitHub repository https://github.com/DDudka9/freeda/releases. FREEDA documentation is available at https://ddudka9.github.io/freeda/. Supplementary materials including FREEDA validation results for all proteins analyzed in the manuscript are available on the open-access Zenodo repository at https://doi.org/10.5281/zenodo.7997737. These also include ortholog detection validation, analysis of rodent centromere proteins, and additional analyses of KIF4A, KIF4B, histone H4, MICA, MICB, NUP73, and HERC5 as mentioned in the text. Manually aligned structural prediction models for MIS18A, MIS18B, AURKC, CENP-O, and CENP-P are also included. Centrocalc is available here: https://github.com/DDudka9/Centrocalc.
Acknowledgments
We would like to thank all members of Michael Lampson’s and Mia Levine’s labs (University of Pennsylvania) for scientific discussions, especially Mia Levine and Cara Brand who provided critical feedback on the manuscript and FREEDA development. We extend special thanks to Piero Lamelza, Arunika Das, Hyuk-Joon Jeon, Cara Brand, Xueqi Sun, Sung-Ya Lin, Geng-Yuan Chen, and Katelyn Boese for testing the pipeline. Finally, we would like to acknowledge the vibrant community of the StackOverflow server for providing feedback on the code underlying our pipeline.
This work was supported by the Swiss National Science Foundation Early Postdoc Mobility grant P2GEP3_187772 awarded to D. Dudka, and National Institutes of Health grant R35GM122475 awarded to M.A. Lampson.
Author contributions: Conceptualization: D. Dudka and M.A. Lampson, Data curation: D. Dudka and R.B. Akins, Formal analysis: D. Dudka, Funding Acquisition: D. Dudka and M.A. Lampson, Investigation: D. Dudka and R.B. Akins, Project administration: D. Dudka and M.A. Lampson, Resources: M.A. Lampson, Software: D. Dudka and R.B. Akins, Supervision: D. Dudka and M.A. Lampson, Validation: D. Dudka and R.B. Akins, Visualization: D. Dudka and R.B. Akins, Methodology: D. Dudka and R.B. Akins, Writing–Original draft: D. Dudka and M.A. Lampson, Writing–Review and editing: D. Dudka, R.B. Akins and M.A. Lampson.
References
Author notes
Disclosures: The authors declare no competing interests exist.