N6, 2′-O-dimethyladenosine (m6Am) is a widespread RNA modification catalyzed by the methyltransferase PCIF1 (phosphorylated CTD interacting factor 1). Despite its prevalence, the biological functions of m6Am in RNA remain largely elusive. Here, we report a critical role of PCIF1-dependent m6Am RNA modification in ciliogenesis in RPE-1 cells. Our findings demonstrate that PCIF1 acts as a negative regulator of ciliation through its m6Am methyltransferase activity. A quantitative proteomic analysis identifies BICD2 as a downstream target of PCIF1, with PCIF1 depletion resulting in a significant increase in BICD2 levels. BICD2 depletion leads to a significant reduction in ciliation. Crucially, the ciliary phenotype in PCIF1-depleted cells is reversed upon BICD2 knockdown. Further investigations reveal that PCIF1 regulates BICD2 protein levels through its m6Am catalytic activity, which reduces the stability and translation efficiency of BICD2 mRNA. Single-base resolution LC-MS analysis identifies the m6Am site on BICD2 mRNA modified by PCIF1. These findings establish the essential involvement of PCIF1-dependent m6Am modification in ciliogenesis.
Introduction
N6, 2′-O-dimethyladenosine (m6Am) is an evolutionarily conserved mRNA modification located at the transcription start site of capped mRNAs and adjacent to the cap structure (Keith et al., 1978; Wei et al., 1975; Cowling, 2019). The methyltransferase PCIF1 (phosphorylated carboxyl-terminal domain interacting factor 1) has been identified as the specific methyltransferase responsible for the terminal m6Am of mRNAs, influencing mRNA stability and cap-dependent translation (Akichika et al., 2019; Boulias et al., 2019; Sendinc et al., 2019; Sun et al., 2019). While complete loss of Pcif1 in mice does not impact viability or fertility, Pcif1 knockout mice exhibited a reduction in body weight (Pandey et al., 2020). Our previous work and other groups found that m6Am and PCIF1 were involved in regulating gastric and colorectal tumorigenesis (Zhuo et al., 2022; Wang et al., 2023). Additionally, PCIF1 is involved in the modulation of vesicular stomatitis virus infection, HIV infection, and SARS-CoV-2 susceptibility (Tartell et al., 2021; Zhang et al., 2021a, 2021b; Wang et al., 2023). However, the physiological significance of m6Am RNA modification remains largely unclear.
Primary cilia are microtubule-based organelles extending from the basal body in most mammalian cell types (Anvarian et al., 2019). They function as sensors of chemical and physical cues in the extracellular environment, regulating a variety of cellular and developmental processes (Nachury and Mick, 2019). Dysfunction of cilia due to genetic mutations or environmental factors leads to a range of human diseases known as ciliopathies, including primary ciliary dyskinesia and polycystic kidney disease (Reiter and Leroux, 2017). The m6A (N6-methyladenosine) demethylase FTO (fat mass and obesity-associated protein) has been implicated in regulating motile ciliogenesis by demethylating FOXJ1 (forkhead box protein J1) mRNA (Kim et al., 2021). Our previous work demonstrated that the m1A demethylase ALKBH3 significantly inhibits non-motile ciliogenesis through the removal of m1A sites on Aurora A mRNA (Kuang et al., 2022). However, the role of m6Am modification in the formation of primary cilia remains unknown.
The BICD2 protein, also known as Bicaudal D homolog 2, plays a pivotal role in intracellular transport processes within eukaryotic cells (Suter et al., 1989; Hoogenraad et al., 2001, 2003; Huynh and Vale, 2017; Schlager et al., 2014). As a key adaptor protein, it connects the dynein motor complex to various cellular cargoes, facilitating their movement along microtubules toward the minus ends. BICD2’s interaction with the small GTPase Rab6 enables it to participate in the recruitment of the dynein motor to Rab6-positive membranes in the Golgi apparatus and cytoplasmic vesicles (Matanis et al., 2002; Short et al., 2002; Reck-Peterson et al., 2018).
In this study, we reveal that PCIF1 negatively modulates ciliogenesis in mammalian cells through its catalytic activity. PCIF1 modifies the m6Am site on BICD2 mRNA, promoting the degradation and inhibition of translation of BICD2 mRNA, thereby attenuating ciliation. Our data highlight the significance of the PCIF1-BICD2 axis in ciliogenesis through an m6Am RNA modification-dependent mechanism.
Results
Depletion of pcif1 promotes ciliation in mammalian cells
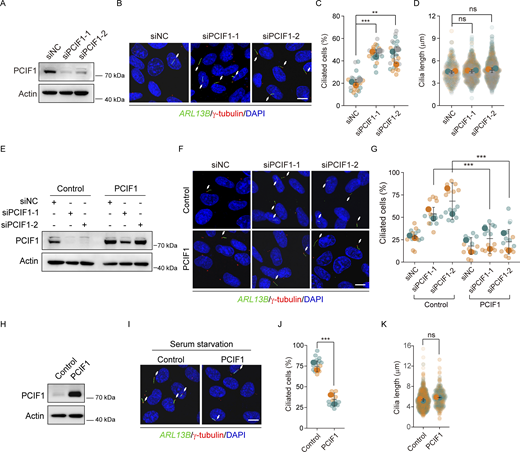
To elucidate the biological role of m6Am in ciliogenesis, we employed two distinct small interfering RNAs (siRNAs) targeting the m6Am methyltransferase PCIF1 in human retinal pigmented epithelial 1 (RPE-1) cells. Immunofluorescence experiments revealed a marked increase in the percentage of ciliated cells following PCIF1 knockdown (Fig. 1, A–D), a phenomenon significantly rescued by the ectopic expression of PCIF1 (Fig. 1, E–G). Furthermore, serum starvation, a condition known to induce ciliation in cultured cells, provided additional validation of PCIF1’s inhibitory role in ciliogenesis, as the exogenous expression of PCIF1 significantly impeded ciliation under serum-starvation conditions (Fig. 1, H–K). These findings collectively signify that PCIF1 functions as a negative regulator of cilia formation in RPE-1 cells.
PCIF1 suppresses ciliogenesis in an m6Am-dependent manner
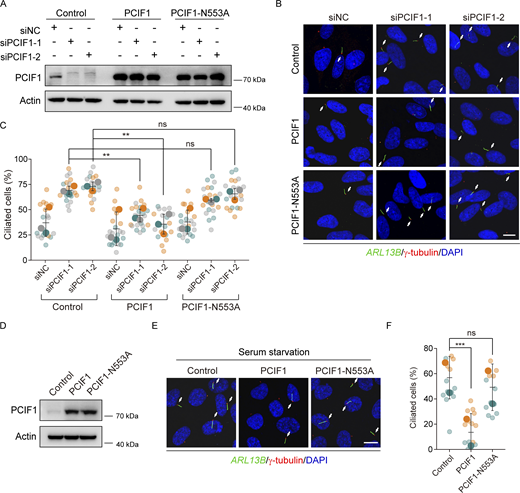
To interrogate whether the m6Am catalytic activity of PCIF1 underscores its role in ciliogenesis, we utilized a catalytically inactive mutant of PCIF1, incorporating an Asn553→Ala substitution in the NPPF motif, rendering it incapable of m6Am modification (Akichika et al., 2019). Subsequently, we conducted rescue experiments by reintroducing either the wild-type or PCIF1-N553A variant into PCIF1-depleted RPE-1 cells. The rescue experiments unequivocally demonstrated that wild-type PCIF1, but not the PCIF1-N553A mutant, significantly rectified the ciliary defects in PCIF1-depleted cells (Fig. 2, A–C). Furthermore, the overexpression of wild-type PCIF1 impeded ciliogenesis, whereas overexpression of the PCIF1-N553A mutant showed no discernible effect under serum starvation conditions (Fig. 2, D–F). These findings collectively signify that the m6Am methyltransferase PCIF1 inhibits ciliogenesis in a manner contingent on its m6Am-dependent catalytic activity in mammalian cells.
PCIF1 inhibits ciliation through suppression of BICD2 expression
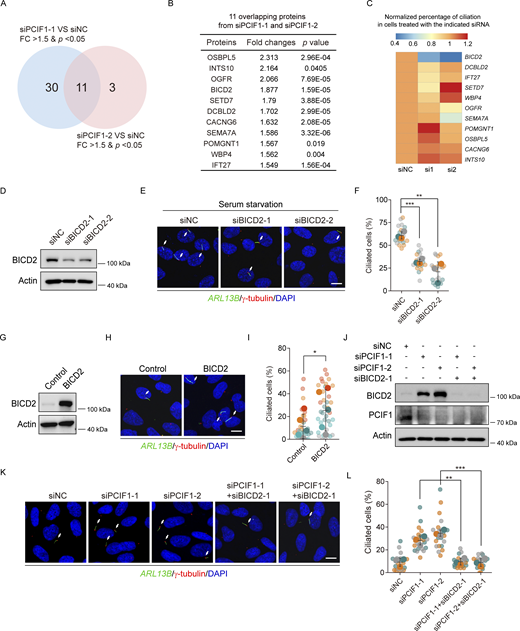
In light of our observation that PCIF1 negatively regulates the expression of the target protein (Zhuo et al., 2022), we conducted a thorough investigation into the underlying mechanism by employing tandem mass tag (TMT)-based quantitative proteomics in RPE-1 cells to elucidate how PCIF1 contributes to ciliation attenuation. The single knockdown experiment on cells was replicated in three independent experiments for TMT proteomics analysis. We comprehensively analyzed the proteomics data (Table S1) and identified 41 significantly upregulated hits (Fold Change >1.5, P value <0.05) between siPCIF1-1 and its control (Table S2), along with 14 significant upregulated hits (Fold Change >1.5, P value <0.05) between siPCIF1-2 and its control (Table S3). As a result, we identified 11 upregulated proteins in RPE-1 cells treated with either siPCIF-1 or siPCIF1-2 (Fig. 3, A and B).
Given their upregulation in PCIF1-depleted cells and the negative regulatory role of PCIF1 in ciliation, we hypothesized that downstream targets of PCIF1 should have the potential to promote ciliogenesis. As a logical extension of our investigation, we initiated immunofluorescence experiments to delineate the role of these eleven upregulated proteins in ciliogenesis. Among them, only BICD2 exhibited a significant effect on ciliation (Fig. 3, C–F; and Figs. S1 and S2). To further investigate the role of BICD2 in ciliogenesis, we conducted immunostaining experiments in cells overexpressing BICD2, revealing a significant promotion of ciliogenesis (Fig. 3, G–I). Importantly, rescue experiments showed that depletion of BICD2 significantly reversed the enhanced ciliation induced by PCIF1 knockdown (Fig. 3, J–L), implying that PCIF1 inhibits ciliation through the inhibition of BICD2 expression.
Depletion of PCIF1 increases the stability and translation of BICD2 mRNA
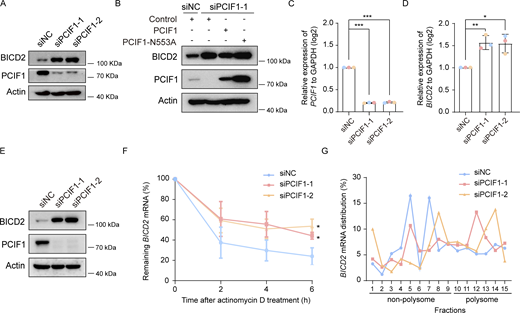
Subsequently, we investigated whether PCIF1 modulates the protein level of BICD2 through its m6Am catalytic activity. Western blot analysis revealed a noticeable elevation of BICD2 in PCIF1-depleted cells (Fig. 4 A), a phenomenon rescued by the ectopic expression of wild-type PCIF1 but not by PCIF1-N553A mutants (Fig. 4 B). These findings suggest that PCIF1 inhibits BICD2 expression through its m6Am catalytic activity. We further explored the impact of PCIF1 depletion on the expression of BICD2 mRNA. Quantitative real-time PCR (qRT-PCR) analysis demonstrated a significant increase in BICD2 mRNA levels upon silencing of PCIF1 in RPE-1 cells (Fig. 4, C and D).
Given the known involvement of m6Am in mRNA stability and translation regulation, we hypothesized that PCIF1 might regulate the expression of BICD2 by affecting the stability or translation of BICD2 mRNA. mRNA decay assay revealed that the decay rate of BICD2 mRNA was significantly reduced in cells depleted of PCIF1 when the transcription was halted with actinomycin D in RPE-1 cells (Fig. 4, E and F). Furthermore, polysome profiling analysis demonstrated that PCIF1 knockdown resulted in increased BICD2 mRNA abundance in high-molecular-weight polysome fractions, indicative of high translation efficiency (Fig. 4 G). Collectively, these data indicate that PCIF1 plays a negative regulatory role in both the stability and translation of BICD2 mRNA.
PCIF1 modifies BICD2 mRNA with m6Am sites
In light of PCIF1’s role in suppressing ciliation through the downregulation of BICD2 expression, and recognizing that PCIF1’s impact on ciliogenesis hinges, in part, on its m6Am methyltransferase activity, we postulated the presence of m6Am sites on BICD2 mRNA. To precisely identify the m6Am site on BICD2 mRNA, we employed a single-base resolution method for targeted m6A and m6Am detection based on liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Fig. 5 A). The results revealed distinct molecular weight differences of 14 or 28 Da in the fragments containing m6A or m6Am, respectively, indicative of the specific modification at this site (Fig. 5, B–E). Our LC-MS/MS analysis achieved the identification of the m6Am site on BICD2 mRNA with single-base resolution. Notably, we observed a noteworthy decrease in the signal abundance of RNA fragments containing m6Am, as depicted in the total ion flow (TIC) diagram following PCIF1 knockdown (Fig. 5 F), while the signal abundance of RNA fragments containing m6A remained unaltered (Fig. 5 G), demonstrating the sensitivity of detection of m6Am modification at this site.
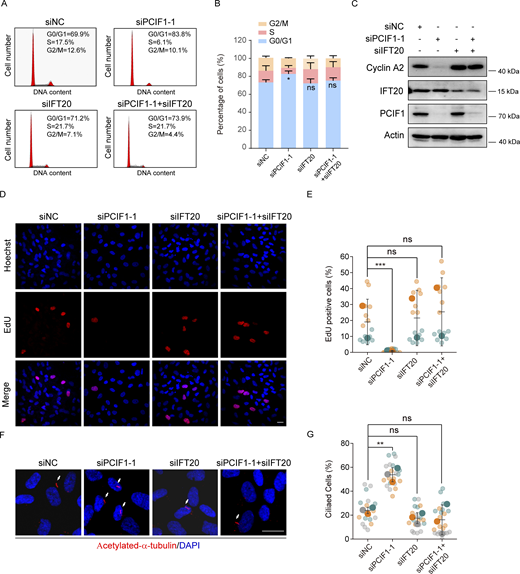
Knockdown of PCIF1 induces cell cycle arrest by enhancing ciliation
Primary cilia represent dynamic organelles intricately linked to cell cycle progression, with their assembly and disassembly tightly regulated (Kasahara and Inagaki, 2021). To investigate whether PCIF1’s impact on ciliation is associated with cell cycle regulation, we initially examined the consequences of PCIF1 deficiency on cell cycle progression in RPE-1 cells. Flow cytometry (FACS) analysis revealed that PCIF1 depletion increased the percentage of cells in the G0/G1 phase, indicative of cell cycle arrest (Fig. 6, A and B). Immunoblotting analyses unveiled reduced levels of cyclin A in PCIF1-depleted cells, suggesting a decrease in cells at the S or G2/M phase (Fig. 6 C). EdU-labeling experiments further demonstrated a decrease in the number of proliferating cells upon PCIF1 depletion (Fig. 6, D and E). These findings collectively indicate that PCIF1 depletion induces cell cycle arrest specifically at the G0/G1 phase.
Considering that IFT20, a crucial intraflagellar transport protein, is essential for cilia formation but has no substantial effect on cell cycle progression (Inaba et al., 2016; Zhang et al., 2021a, 2021b), we depleted IFT20 to impede cilia growth in PCIF1-depleted, aiming to discern whether the observed cell cycle arrest induced by PCIF1 depletion is mediated by enhanced ciliation. The data demonstrated a significant reduction in the enhanced ciliation in PCIF1-depleted cells upon IFT20 depletion (Fig. 6, F and G). Moreover, FACS profiles, immunoblotting, and EdU incorporation experiments revealed that the elimination of cilia induced by IFT20 depletion effectively reversed the G0/G1 arrest in PCIF1-depleted cells (Fig. 6), suggesting that PCIF1 depletion causes cell cycle arrest at G0/G1 in a cilia-dependent manner.
Discussion
Our study shows the biological significance of PCIF1-mediated regulation of BICD2 mRNA in mammalian cell ciliogenesis. PCIF1 adds m6Am to BICD2 mRNA, leading to reduced mRNA stability and translation, which suppresses BICD2 protein expression to inhibit ciliogenesis in mammalian cells (Fig. 7). These findings contribute valuable insights into the impact of RNA epigenetics on ciliogenesis, advancing our understanding of this complex biological process.
Previous studies suggest that PCIF1 regulates the accumulation of the homeodomain transcription factor PDX1, influencing the expression of insulin and other genes crucial for maintaining β cell mass and function in adult mice (Claiborn et al., 2010). Additionally, Pandey et al. (2020) demonstrated that Pcif1 mutant mice exhibited reduced body weight without affecting viability and fertility (Pandey et al., 2020). Here, we found that PCIF1 negatively regulates ciliogenesis, and given the reported roles of primary cilia in controlling mammalian energy homeostasis and body weight (Vaisse et al., 2017), it is intriguing to explore whether PCIF1’s contribution to ciliation plays a role in these physiological events.
The roles of m6Am modifications in regulating mRNA transcription or translation remain unclear and context dependent. Suzuki’s group reported that m6Am promotes the translation of capped mRNAs in an eIF4E-independent manner without affecting transcriptome alteration (Akichika et al., 2019). Conversely, Sendinc et al. found that m6Am negatively impacted cap-dependent translation of methylated mRNAs in PCIF1 knockout MEL624 cells (Sendinc et al., 2019). Other studies propose that m6Am might modulate mRNA stability. Hirose et al. reported that PCIF1 negatively regulates transcription, although the mechanism remains unclear (Hirose et al., 2008). Additionally, it is found that m6Am modifications at the beginning of mRNAs increase stability, with an average increase in the half-life of around 2.5 h compared with non-methylated bases (Mauer et al., 2017). Boulias et al. also demonstrated that depleting PCIF1 minimally affects mRNA translation but leads to reduced stability of a subset of m6Am-annotated mRNAs in HEK293T cells (Boulias et al., 2019). Moreover, the loss of cap-specific m6Am RNA methylation in mice resulted in transcript destabilization (Pandey et al., 2020). These contrasting findings necessitate further investigations to unravel the context-dependent roles of m6Am and PCIF1 in mRNA regulation.
Dynein-2, also known as IFT dynein, is a motor protein complex essential for the structure and function of cilia (Vuolo et al., 2018). Since BICD2 is not a component in the dynein-2 complex (Asante et al., 2014), it may not directly enter cilia or regulate the retrograde IFT. However, BICD2 is an adaptor protein that facilitates the interaction between cytoplasmic dynein-1 and its cargo (Hoogenraad et al., 2001). The altered levels of BICD2 due to PCIF1 dysfunction will impact various cellular processes mediated by dynein-1, including membrane traffic, organelle dynamics, and centrosome and nuclear positioning (Grigoriev et al., 2007; Matanis et al., 2002; Short et al., 2002; Splinter et al., 2012). Though dynein-1 is not directly involved in the movement of cilia, it influences ciliogenesis by facilitating the intracellular transport of key components required for cilia assembly and maintenance (Palmer et al., 2011; Roberts et al., 2013). Consequently, the aberrance production of BICD2 and resultant dynein-1 dysfunction may affect ciliogenesis. Additionally, BICD2 has been identified as a component of centriolar satellite proteins (Quarantotti et al., 2019), which play a crucial role in basal body maturation and function (Tollenaere et al., 2015). This suggests that the localization of BICD2 in centriolar satellites may also contribute to the regulation of ciliation. Future investigations are clearly needed to decipher the molecular mechanisms governing the regulation of ciliogenesis orchestrated by BICD2.
Materials and methods
Cell culture and transfection
Human telomerase reverse transcriptase–immortalized retinal pigment epithelial cells (hTERT RPE-1) were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM/F12 medium (Corning) supplemented with 10% fetal bovine serum (FBS; ExCell Bio) at 37°C in a 5% CO2 incubator. Serum starvation involved culturing cells in DMEM/F12 without FBS at 37°C in a 5% CO2 incubator. For transfections, Lipofectamine iMAX (Invitrogen) or Lipofectamine 3000 (Invitrogen) was used as per the manufacturer’s instructions. Transfection efficiencies were assessed via Western blot or qRT-PCR analysis. The siRNA sequences are provided in Table S4.
Plasmids
The full-length human PCIF1 gene was amplified from cDNAs and subsequently cloned into the pCS2+ vector. The PCIF1-N553A mutation was generated by PCR amplified from wild-type PCIF1 plasmid.
RNA extraction and quantitative real-time PCR
Total RNA was extracted using the TRIzol reagent (Ambion), and cDNA was synthesized using the HiScript II first strand cDNA Synthesis kit (Vazyme). Quantitative real-time PCR (qRT-PCR) was performed on a Roche 480Ⅱ-384 System using the HiScript Q RT SuperMix (Vazyme). Primer sequences are provided in Table S5.
Western blot
For Western blot assays, cells were lysed with TBSN buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5% NP-40, 5 mM EGTA, 1.5 mM EDTA, 0.5 mM Na3VO4, and 20 mM p-nitrophenyl phosphate) and supplemented with protease inhibitor cocktails (04693132001; Roche). The cell lysates were then subjected to SDS-PAGE, and the proteins were transferred onto polyvinylidene fluoride membranes (Millipore). The membranes were blocked with 5% non-fat milk in PBST (PBS containing 0.1% Tween-20) for 1 h at room temperature and then incubated overnight at 4°C with the respective primary antibodies. After washing three times with PBST for 5 min each, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, followed by three additional washes with PBST for 5 min each. Protein bands were visualized using the ChemiDoc Touch Imaging System (Bio-Rad).
Immunofluorescence staining
Immunofluorescence assays were carried out as described previously (Liu et al., 2021). Cells cultured on coverslips were fixed with 100% methanol at −20°C for 10 min and then blocked with 5% BSA in PBSTX (PBS containing 0.1% Triton X-100) for 1 h at room temperature. Subsequently, the cells were incubated overnight with the respective primary antibodies diluted in the blocking buffer at 4°C. After three washes with PBSTX for 5 min each, the cells were incubated with species-specific Alexa 488/555 fluorochrome-conjugated secondary antibodies diluted in a blocking buffer for 1 h at room temperature in a dark room. Following three additional washes with PBSTX for 5 min each, the nuclei were stained with DAPI (564907; BD Biosciences). Confocal images were randomly acquired using an OLYMPUS FV3000 OSR laser scanning confocal microscope at room temperature using a 60×/1.42 oil objective (Fig. 6, D and F; Fig. S1, B, E, H, K, and N; and Fig. S2, B, E, H, K, and N) or a 63×/1.40 oil objective on Zeiss LSM 880 microscope (Fig. 1, B, F, and I; and Fig. 2, B and E; Fig. 3, D, G, and J). The acquisition settings were consistent for both the experimental and control groups within the same experiment. For the quantification process, we consistently selected seven to nine random fields within each experimental group to count both the number of cells (based on DAPI staining) and the cilia signal. Subsequently, we utilized Photoshop software to quantify the number of cells and cilia. Specifically, cilia length was measured from the base to the tip of the cilia.
Tandem mass tag quantitative MS and data analysis
RPE-1 cells were transfected with control or PCIF1 siRNAs in a 10% serum-containing DMEM/F12 medium for 48 h. Following transfection, cells were collected and underwent TMT quantitative proteomic analysis by Jingjie PTM BIO. Peptides were processed using NSI and analyzed by MS/MS on a Q ExactiveTM Plus instrument coupled with UPLC. Maxquant search engine (v.1.5.2.8) was used for data analysis, with spectra interrogated against the human Uniprot database and a reverse decoy database. Quantitative analysis employed TMT 10-plex, with both protein and PSM identification filtered rigorously at a 1% FDR threshold.
5,856 proteins were identified in total. To facilitate data sharing and accessibility, the raw proteomics data and search results were deposited in the PRIDE archive, accessible through the ProteomeXchange accession number PXD044338.
The relative quantitative value of proteins was used for differential expressed proteins analysis. Student’s t test was performed to calculate the statistical significance of difference between groups. Proteins with P value <0.05 and Fold Change >1.5 was regarded as significantly upregulated proteins, while the protein with P value <0.05 and Fold Change <0.667 were regarded as significantly downregulated proteins.
Single-base resolution m6Am site detection
The m6A RNA immunoprecipitation (MeRIP) was conducted using the Magna MeRIP m6A kit (17-10499; Millipore) following the manufacturer’s instructions. Subsequently, the resulting complementary DNA fragments were utilized as probes to enrich targeted mRNAs containing m6Am sites. The length of the DNA probe was 25 nt and it had biotin at the terminal. The DNA probe had a length of 25 nucleotides with biotin at the terminal, and the specific sequences for targeting BICD2 were 5′-AGGGCGGCGGCGGCGGCGGCAGCGG-3′ and 5′-TCGGCCACCATGTCGGCGCCGTCGG-3′. The hybridization process lasted for 3 h, maintaining a probe-to-mRNA ratio of 1.25:1 (mol/ml). To obtain ∼25 nucleotide fragments, the targeted digestion of mRNA was performed using the RNase T1/A mix kit (EN0551; Thermo Fisher Scientific), following the instructions provided with the kit. The digestion method was according to the instructions of the RNase mix kit. Finally, the digested mRNA fragments were eluted with 50% formamide and analyzed using Nano-liquid chromatography-high resolution mass spectrometry (LC-HRMS) on a Thermo QE HF instrument. The MS/MS data was processed using Thermo Xcalibur 2.7 software, and RNA fragments with a signal-to-noise ratio (SNR) >10 were considered valid peaks. The sequencing analysis of these RNA fragments was performed using an online software tool (https://Ariadne.riken.jp/html/search.html).
mRNA decay assay
The mRNA decay assay was conducted following the protocol described in Kuang et al. (2022). After transfection with control or PCIF1 siRNAs for 48 h, RPE-1 cells were treated with 5 μg/ml actinomycin D (HY-17559; MedChemExpress) to inhibit global mRNA transcription. Subsequently, cells were harvested at the designated time points and total RNAs were extracted for reverse transcription. The expression levels of BICD2 mRNA, following transcription inhibition, were analyzed using qRT-PCR.
Polysome fractionation
Polysome fractionation was carried out following the protocol described in Gandin et al. (2014). Briefly, control or PCIF1-depleted RPE-1 cells (three 150-mm culture dishes) were treated with cycloheximide (HY-12320; MedChemExpress) at a final concentration of 100 µg/ml in growth media for 5 min at 37°C and 5% CO2. The cells were then washed twice with 10 ml of ice-cold 1× PBS containing 100 µg/ml cycloheximide. After that, the cells were collected and resuspended in 425 μl of hypotonic buffer (5 mM Tris-HCl [pH 7.5], 2.5 mM MgCl2, 1.5 mM KCl, and 1× protease inhibitor cocktail [EDTA-free]). To the cell suspension, 5 μl of 10 mg/ml cycloheximide, 1 μl of 1 M DTT, and 100 U of RNAse inhibitor were added. The mixture was vortexed for 5 s, followed by the addition of 25 μl of 10% Triton X-100 (final concentration 0.5%) and 25 μl of 10% sodium deoxycholate (final concentration 0.5%). The mixture was vortexed again for 5 s. After centrifugation of the lysates at 16,000 g for 7 min at 4°C, the supernatant was transferred, and the absorbance was measured at 260 nm to determine the RNA concentration. Subsequently, the supernatant was loaded onto a 5–50% sucrose gradient and centrifuged at 36,000 rpm for 2 h at 4°C using an SW41Ti rotor (Beckman). The gradients were fractionated and absorbance at 254 nm was monitored using a fraction collector (Biocomp). Polysome-associated and cytosolic RNAs from each fraction were extracted using TRIzol protocol according to the manufacturer’s instructions, and their levels were monitored by qRT-PCR.
Flow cytometry analysis
For flow cytometry analysis, RPE-1 cells were harvested and washed once with ice-cold 1× PBS. Subsequently, the cells were resuspended in ice-cold 1× PBS and fixed by incubating in ice-cold 95% ethanol at −20°C overnight. To stain the DNA, the cells were treated with 100 μg/ml propidium iodide and 50 μg/ml RNase A for 30 min at 37°C. Finally, the samples were analyzed using an FC 500 MCL Flow Cytometer (Beckman Coulter). We conducted three biological replicates for the flow cytometry analysis, involving the transfection of cells with siRNAs and subsequent flow cytometry analysis on separate occasions. Each occasion utilized distinct batches of cell samples.
EdU assay
Before and during Edu experiments, the cells were cultured in normal conditions (DMEM/F12 medium with 10% serum). RPE-1 cells were incubated with 50 μM EdU (5-Ethynyl-2′-deoxyuridine) diluted in DMEM/F12 medium with 10% serum for 2 h. After incubation, the cells were fixed in 4% PFA for 30 min at room temperature. EdU staining was performed using the Cell-Light EdU Apollo567 In Vitro Kit (C10310-1; Ribobio) following the manufacturer’s instructions.
Antibodies
Antibodies used in this study for Western blot included rabbit anti-PCIF1 (1:1,000, ab300487; Abcam), rabbit anti- BICD2 (1:1,000, NBP1-81488; Novus Biologicals), rabbit anti- cyclin A2 (1:1,000, 18202-1-AP; Proteintech), rabbit anti- IFT20 (1:500, 13615-1-AP; Proteintech), mouse anti-ACTIN (1:2,000, 66009-1-Ig; Proteintech), HRP Goat anti-Mouse IgG (1:5,000, AS003; Abclonal), and HRP Goat anti-Rabbit IgG (1:5,000, AS014; Abclonal). Antibodies used in this study for immunofluorescence included rabbit anti-ARL13B (1:200, 17711-1-AP; Proteintech), mouse anti-γ-tubulin (1:1,000, T6557; Sigma-Aldrich), mouse anti-acetylated-α-tubulin (1:1,000, T7451; Sigma-Aldrich), Alexa Fluor 488-conjugated anti-rabbit IgG (1:200, A21206; Invitrogen), and Alexa Fluor 555-conjugated anti-mouse IgG (1:200, A31570; Invitrogene).
Statistical analysis
The means and standard deviations (SD) were calculated and are shown in the graphs. Student’s t test was applied to assess statistical significance. SuperPlots (Lord et al., 2020) and heatmaps were produced in R programming language (version 4.2.3).
Language editing
ChatGPT3.5 was employed to refine the language of the manuscript using the command “Below is an academic paper. Polish the writing to meet the academic style, improve spelling, grammar, clarity, and overall readability.”
Online supplemental material
Fig. S1 shows the effect of OSBPL5, NTS10, OGFR, SETD7, and DCBLD2 knockdown on ciliation. Fig. S2 shows the effect of CACNG6, SEMA7A, POMGNT1, WBP4, and IFT27 genes knockdown on ciliation. Table S1 shows analysis of TMT proteomics. Table S2 shows significant hit proteins for siPCIF1-1 versus its control. Table S3 shows significant hit proteins for siPCIF1-2 versus its control. Table S4 shows sequences of siRNAs. Table S5 shows sequences of primers.
Data availability
Data is accessible in the main text or supplementary materials. For additional details, resource inquiries, and reagent requests, please contact the lead, Tianhua Zhou ([email protected]).
Acknowledgments
We are grateful to Shuangshuang Liu, Guifeng Xiao, Junli Xuan, and Wei Yin for their help in the confocal laser scanning microscope. We thank Wanling Zheng (Zhejiang University) for helping with the polysome profiling assay.
This work was supported by the National Natural Science Foundation of China (U21A20197 and 32270723) and the National Key Research and Development Program of China (2019YFA0802202).
Author contributions: S. Xie and T. Zhou conceived the work. S. Xie and W. Kuang designed and performed most of the experiments. M. Guo conducted the experiments on m6Am site analysis. F. Yang performed the bioinformatics and statistical analysis. H. Jin and X. Chen performed part of the experiments about BICD2. C. Huo and Z. Xu carried out the polysome profiling assay. X. Chen and L. Yi performed cell cycle analysis. S. Xie and T. Zhou wrote the manuscript. Q. Shu, J. Mao, W. Liu, and A. Lin edited the manuscript.
References
Author notes
S. Xie, W. Kuang, M. Guo, and F. Yang contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.
Supplementary data
shows analysis of TMT proteomics.
shows sequences of siRNAs.
shows sequences of primers.