The BRCA1-A complex contains matching lysine-63 ubiquitin (K63-Ub) binding and deubiquitylating activities. How these functionalities are coordinated to effectively respond to DNA damage remains unknown. We generated Brcc36 deubiquitylating enzyme (DUB) inactive mice to address this gap in knowledge in a physiologic system. DUB inactivation impaired BRCA1-A complex damage localization and repair activities while causing early lethality when combined with Brca2 mutation. Damage response dysfunction in DUB-inactive cells corresponded to increased K63-Ub on RAP80 and BRCC36. Chemical cross-linking coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) and cryogenic-electron microscopy (cryo-EM) analyses of isolated BRCA1-A complexes demonstrated the RAP80 ubiquitin interaction motifs are occupied by ubiquitin exclusively in the DUB-inactive complex, linking auto-inhibition by internal K63-Ub chains to loss of damage site ubiquitin recognition. These findings identify RAP80 and BRCC36 as autologous DUB substrates in the BRCA1-A complex, thus explaining the evolution of matching ubiquitin-binding and hydrolysis activities within a single macromolecular assembly.
Introduction
BRCA1- and BRCA2-dependent homologous recombination (HR) plays an essential role in preserving genome stability at DNA double-stranded breaks (DSBs), DNA interstrand crosslinks (ICLs), and stalled replication forks. Chromatin localization of BRCA1 within foci aligning DSBs or replication forks is dependent on nondegradative forms of histone ubiquitylation. The BRCA1-A complex recognizes lysine-63 polyubiquitin chains (K63-Ub) through its tandem ubiquitin interaction motifs (UIMs; Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). More recently, the BARD1 C-terminal repeats (BRCTs) were shown to provide another critical ubiquitin recognition moiety by specifically interacting with histone 2A (H2A) mono-ubiquitylated on lysine 15 (Becker et al., 2021; Dai et al., 2021; Hu et al., 2021; Krais et al., 2021; Witus et al., 2021). Ubiquitin E3 ligases RNF8 and RNF168 orchestrate damage site ubiquitylation, which enables the BRCA1-A complex and other BRCA1-BARD1 complexes to access these regions (Mirsanaye et al., 2021).
The BRCA1-A complex (also known as ARISC-RAP80) consists of five stoichiometric constituents, Abraxas1, RAP80, BRCC45, BRCC36, and MERIT40 that are collectively required to target BRCA1 to DNA damage sites and to replication forks. Structural studies on the A-complex and the related BRISC-SHMT2 complex reveal multimerization through an alpha-helical region of BRCC36 to create a dimer of pentamers (Kyrieleis et al., 2016; Rabl et al., 2019; Walden et al., 2019; Zeqiraj et al., 2015). The RAP80 tandem UIMs specifically recognize K63-Ub, while BRCC36 is a K63-Ub specific deubiquitylating enzyme (DUB). BRCC36 enzymatic activity requires interactions with other members of the A-complex (Cooper et al., 2010; Feng et al., 2010; Patterson-Fortin et al., 2010; Sobhian et al., 2007). BRCC36 is a JAMM/MPN+ domain protease that relies on Zn2+ to deliver a water molecule for the hydrolysis of the K63-Ub isopeptide bond. Biochemical and structural studies revealed the underlying basis for BRCC36 activation through its interaction with the pseudo-DUB MPN− domain protein KIAA0157/Abraxas2, a close homolog of Abraxas1 (Zeqiraj et al., 2015). This involves the positioning of BRCC36 residue E33 near the active site Zn2+ ion to coordinate the delivery of a water molecule to the scissile K63-Ub isopeptide bond to achieve hydrolysis. Rationally designed point mutations at either E33 (E33A) or H122 and H124 (H112Q, H124Q, in short QSQ; Patterson-Fortin et al., 2010; Shao et al., 2009) abolish the enzymatic activity of BRCC36. An important distinction is that the BRCC36 active site configuration is retained with E33A mutation, while the loss of Zn2+ binding in the QSQ mutation results in the distortion of the BRCC36 active site (Zeqiraj et al., 2015).
Replication stress is induced by endogenous or exogenous genotoxins that interfere with fork progression and is a major cause of genome instability. The BRCA1-A complex is present at replication forks (Jiang et al., 2015; Nakamura et al., 2021, 2019), and appears to rely on a combination of K63-Ub chain recognition by RAP80 coupled with binding of the BARD1 ankyrin and BRCT repeats to unmethylated histone 4 at lysine 20 (H4K20me0) and mono-ubiquitylated histone 2A at lysine 15 (H2AK15Ub), respectively (Becker et al., 2021; Dai et al., 2021; Hu et al., 2021; Jiang et al., 2015; Krais et al., 2021; Nakamura et al., 2019; Witus et al., 2021). The BRCA1-A complex is required for an efficient restart of stalled replication forks and for unhooking of crosslinked DNA (Jiang et al., 2015; Jones et al., 2021). Knockout of A-complex constituent, MERIT40, sensitized mice to ICL inducing agent mitomycin C (MMC) and also significantly delayed replication fork restart in response to replication stress (Jiang et al., 2015). In contrast to BRCA1 mutation, A-complex deficiency caused a paradoxical elevation in HR by increasing end-resection at DSBs and replication forks (Coleman and Greenberg, 2011; Hu et al., 2011; Jiang et al., 2015). This paradoxical increase in HR suggests a specialized role for the BRCA1-A complex in replication-associated damage responses since BRCA1 promotes HR in association with its other complexes (Greenberg et al., 2006; Nacson et al., 2020; Sy et al., 2009; Zhang et al., 2009). This function of limiting resection at forks is thought to explain how loss of the A-complex causes resistance to Topoisomerase I or PARP inhibitors (PARPi) in cells lacking ATM kinase activity (Balmus et al., 2019; Nakamura et al., 2021). As the stability of the entire A-complex is disrupted upon deletion of any of its substituents, it is unclear whether its DUB activity similarly affects these functions.
BRCC36 DUB inactivation was shown to result in IR hypersensitivity, and reductions in G2 checkpoint control and other A-complex associated damage response functions (Ng et al., 2016; Shao et al., 2009). However, the basis for DUB activity in executing damage responses remains unknown. To understand how BRCA1-A complex DUB activity contributes to the DNA damage response in a physiologic setting, we adapted CRISPR/Cas9 gene targeting technology to generate knock-in mice harboring E33A or QSQ DUB inactive BRCC36 mutations. Here, we reveal that BRCC36 DUB activity serves to relieve auto-inhibition from internal K63-Ub in the BRCA1-A complex that engages the RAP80 UIM domains in an intramolecular interaction. Autologous removal of K63-Ub by BRCC36 serves as an essential regulatory step in licensing BRCA1-A complex DNA damage response function, providing a framework to understand the association of specific ubiquitin binding and DUB activities within a single protein macromolecular assembly.
Results
BRCC36 DUB activity is important for RAP80-dependent DNA damage responses
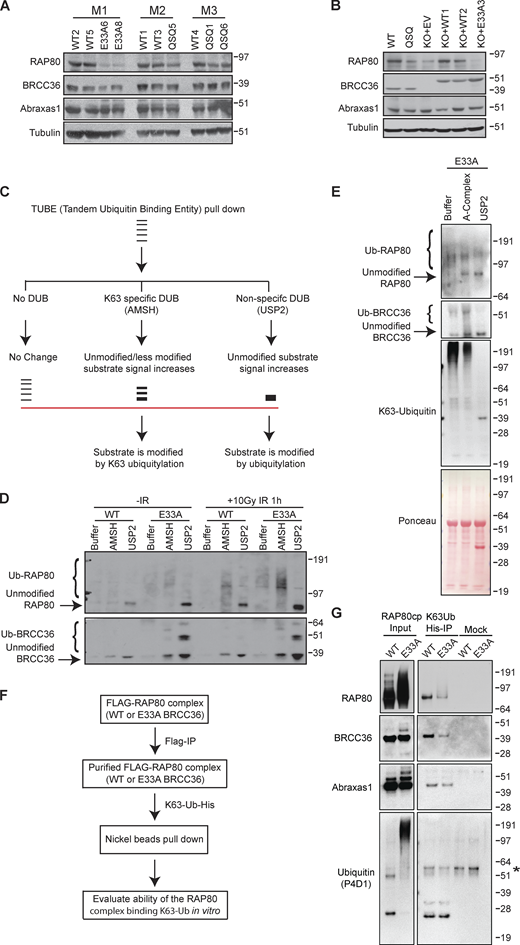
We used structural analyses to rationally design Brcc36E33A/E33A and Brcc36QSQ/QSQ knock-in mice (Fig. 1, A and B; and Fig. S1, A–C). Brcc36KO/KO mice were also produced during the targeting procedure due to the error-prone repair of Cas9 breaks. Intercrossing heterozygous mutant animals (Brcc36E33A/+, Brcc36QSQ/+ or Brcc36KO/+) yielded homozygous mutant pups Brcc36E33A/E33A (E33A), Brcc36QSQ/QSQ (QSQ), or Brcc36KO/KO (KO) at the expected Mendelian ratio. BRCC36 DUB-inactive mice were fertile and had no gross phenotypic abnormalities. Mice harboring both mutant alleles showed broadly similar protein levels to Brcc36+/+ (WT) across multiple tissues (Fig. 1 C and Fig. S1 D). Meanwhile, the cell cycle profile of primary splenocytes isolated from Brcc36 mutant or knockout mice proliferated at a similar rate as the WT cells (Fig. 1 D and Fig. S1 E), further supporting the idea that loss of BRCC36 or inactivation of DUB activity is well tolerated in mice. Mouse embryonic fibroblasts (MEFs) derived from DUB-inactive mice demonstrated reductions in BRCA1 ionizing radiation-induced foci formation (IRIF) levels that were comparable to KO cells (Fig. 1, E and F; and Fig. S1 F). While not as sensitive to MMC in clonogenic assays as KO (Fig. S1 G), E33A cells showed elevated chromosomal abnormalities following MMC treatment but not after PARP inhibition (Fig. 1 G), consistent with our published results in A-complex null cells (Jiang et al., 2015). In addition, DUB-inactive E33A mutation was found to be epistatic with RAP80 loss with respect to BRCA1 IRIF, consistent with DUB inactivation affecting BRCA1 localization through RAP80 ubiquitin recognition at DNA damage sites (Fig. 1, H and I; and Fig. S1 H).
Characterization of Brcc36 knock-in mutant mice and cells. (A) Close-up view of the superimposed BRCC36 active site from mouse (Mm; PDB accession no. 6GVW), human (Hs; PDB accession no. 6R8F), and ant (Cf; PDB accession no. 5CW3). Superimposed structures are shown as cartoons and colored wheat, pink, and light blue respectively; active site glutamate, aspartate, and histidine residues are shown as sticks. A representative Zn2+ atom and a catalytic H2O molecule obtained from the mouse and ant structures are shown as gray and red spheres; tetrahedral coordination of the Zn2+ ion by the water molecule and active site residues is indicated with dashed lines. (B) Sanger sequencing profiles of E33A and QSQ MEF cells. (C and D) Splenocytes were isolated from WT, QSQ, E33A, and KO mice and assessed for BRCC36 protein level (C) and profiled for cell cycle (D). (D) Quantification of the cell cycle data is shown. (E) Representative images of MEFs of the indicated genotypes stained with BRCA1 and γH2AX at 5 h post 6 Gy IR. Scale bar: 5 μM. (F) Quantification of foci number from E based on three independent experiments. For each experiment, more than 50 cells were quantified. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >5 BRCA1 from each replicate. *P < 0.05, **P < 0.01. Mean with SEM was used to plot the data. (G) Chromosome abnormalities were quantified in splenocytes derived from two WT and three E33A mice. Chromosomal abnormalities, including chromatid and chromosome breaks and radial chromosome structures. n > 70 metaphases per genotype were quantified. Unpaired two-sided Student’s t test was conducted based on the average chromosomal abnormalities per metaphase of each mouse. *P < 0.05. Mean with SEM was used to plot the data. (H and I) BRCA1 IRIF in control and RAP80 KO HT29 cells expressing WT or E33A BRCC36. (H) Representative images of HT29 cells of the indicated genotypes stained with BRCA1 and γH2AX at 4 h post 6 Gy IR. Scale bar: 5 μM. (I) Quantification of foci number from H based on three independent experiments. More than 100 cells were quantified in each experiment. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >10 BRCA1 foci from each experiment. **P < 0.01, ***P < 0.001. Mean with SEM was used to plot the data. Source data are available for this figure: SourceData F1.
Characterization of Brcc36 knock-in mutant mice and cells. (A) Close-up view of the superimposed BRCC36 active site from mouse (Mm; PDB accession no. 6GVW), human (Hs; PDB accession no. 6R8F), and ant (Cf; PDB accession no. 5CW3). Superimposed structures are shown as cartoons and colored wheat, pink, and light blue respectively; active site glutamate, aspartate, and histidine residues are shown as sticks. A representative Zn2+ atom and a catalytic H2O molecule obtained from the mouse and ant structures are shown as gray and red spheres; tetrahedral coordination of the Zn2+ ion by the water molecule and active site residues is indicated with dashed lines. (B) Sanger sequencing profiles of E33A and QSQ MEF cells. (C and D) Splenocytes were isolated from WT, QSQ, E33A, and KO mice and assessed for BRCC36 protein level (C) and profiled for cell cycle (D). (D) Quantification of the cell cycle data is shown. (E) Representative images of MEFs of the indicated genotypes stained with BRCA1 and γH2AX at 5 h post 6 Gy IR. Scale bar: 5 μM. (F) Quantification of foci number from E based on three independent experiments. For each experiment, more than 50 cells were quantified. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >5 BRCA1 from each replicate. *P < 0.05, **P < 0.01. Mean with SEM was used to plot the data. (G) Chromosome abnormalities were quantified in splenocytes derived from two WT and three E33A mice. Chromosomal abnormalities, including chromatid and chromosome breaks and radial chromosome structures. n > 70 metaphases per genotype were quantified. Unpaired two-sided Student’s t test was conducted based on the average chromosomal abnormalities per metaphase of each mouse. *P < 0.05. Mean with SEM was used to plot the data. (H and I) BRCA1 IRIF in control and RAP80 KO HT29 cells expressing WT or E33A BRCC36. (H) Representative images of HT29 cells of the indicated genotypes stained with BRCA1 and γH2AX at 4 h post 6 Gy IR. Scale bar: 5 μM. (I) Quantification of foci number from H based on three independent experiments. More than 100 cells were quantified in each experiment. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >10 BRCA1 foci from each experiment. **P < 0.01, ***P < 0.001. Mean with SEM was used to plot the data. Source data are available for this figure: SourceData F1.
Related toFig. 1 . (A) Schematic illustrating the strategy to knock-in BRCC36 mutations E33A, H122A, H124A at endogenous location. (B and C) Mutation of E33A abolishes BRCC36 deubiquitylase activity. (B) Schematic of an internally quenched K63-linked di-ubiquitin fluorogenic substrate (K63-diUb IQF). Cleavage of the isopeptide bond between the two ubiquitin molecules allows separation of the quencher from the fluorophore, with a subsequent increase in fluorescent signal over time. (C) Deubiquitylase activity of A-complex containing BRCC36 WT and E33A variants against a fluorogenic K63-linked di-Ub substrate. Results are the average of three independent experiments carried out in duplicates. Mean with SEM was used to plot the data. The ARISC complex contains the subunits BRCC36, Abraxas1, BRCC45 and MERIT40 that are required for maximal DUB activity (see Materials and methods for details). (D) BRCC36 protein level was examined in different tissues from one WT mouse and two E33A mice. (E) Cell cycle analysis for splenocytes with indicated genotypes. (F) Quantification of IRIF from MEFs of indicated genotypes at 5 h post 6 Gy IR based on four to eight independent experiments. More than 50 cells were quantified in each experiment. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >10 BRCA1 or MERIT40 foci from each experiment. *P < 0.05, ***P < 0.001. Mean with SEM was used to plot the data. (G) WT, E33A or KO MEFs were exposed to the indicated doses of MMC, and survival was assessed by clonogenic assay in three independent experiments. Unpaired two-sided Student’s t test was conducted based on % of cell survival at three MMC doses (4, 10, and 25 ng/ml). ***P < 0.001, ****P < 0.0001. Mean with SD was used to plot the data. (H) Immunoblots to demonstrate that specific genotypes were successfully generated in HT29 cells. Source data are available for this figure: SourceData FS1.
Related toFig. 1 . (A) Schematic illustrating the strategy to knock-in BRCC36 mutations E33A, H122A, H124A at endogenous location. (B and C) Mutation of E33A abolishes BRCC36 deubiquitylase activity. (B) Schematic of an internally quenched K63-linked di-ubiquitin fluorogenic substrate (K63-diUb IQF). Cleavage of the isopeptide bond between the two ubiquitin molecules allows separation of the quencher from the fluorophore, with a subsequent increase in fluorescent signal over time. (C) Deubiquitylase activity of A-complex containing BRCC36 WT and E33A variants against a fluorogenic K63-linked di-Ub substrate. Results are the average of three independent experiments carried out in duplicates. Mean with SEM was used to plot the data. The ARISC complex contains the subunits BRCC36, Abraxas1, BRCC45 and MERIT40 that are required for maximal DUB activity (see Materials and methods for details). (D) BRCC36 protein level was examined in different tissues from one WT mouse and two E33A mice. (E) Cell cycle analysis for splenocytes with indicated genotypes. (F) Quantification of IRIF from MEFs of indicated genotypes at 5 h post 6 Gy IR based on four to eight independent experiments. More than 50 cells were quantified in each experiment. Unpaired two-sided Student’s t test was conducted based on the average % of cells with >10 BRCA1 or MERIT40 foci from each experiment. *P < 0.05, ***P < 0.001. Mean with SEM was used to plot the data. (G) WT, E33A or KO MEFs were exposed to the indicated doses of MMC, and survival was assessed by clonogenic assay in three independent experiments. Unpaired two-sided Student’s t test was conducted based on % of cell survival at three MMC doses (4, 10, and 25 ng/ml). ***P < 0.001, ****P < 0.0001. Mean with SD was used to plot the data. (H) Immunoblots to demonstrate that specific genotypes were successfully generated in HT29 cells. Source data are available for this figure: SourceData FS1.
DUB inhibition sensitizes Brca2 mutant cells to ICL inducing agents
Loss of MERIT40 destabilizes the integrity of the A-complex and exacerbates genome instability in Brca2 mutant cells (Feng et al., 2009; Jiang et al., 2015; Shao et al., 2009; Wang et al., 2009). We tested whether DUB inactivation could produce similar effects in a mouse model harboring Brca2 exon27 deletion. These mice are viable, despite infertility in males and a shorter life span due to elevated rates of cancer (Donoho et al., 2003). Brca2Δ27/Δ27 mice lack a C-terminal RAD51 binding domain and demonstrate defective RAD51 localization to DNA damage sites, hypersensitivity to ICLs, as well as defective replication fork protection ability (Kim et al., 2014; Li et al., 2016; Navarro et al., 2006). Reduced lifespan was observed in all combinations of Brcc36 and Brca2 mutations, consistent with DUB inhibition exacerbating genome stability abnormalities in Brca2Δ27/Δ27 mice (Fig. 2 A). Plating efficiency in all three double mutant MEF clones was also significantly lower when compared to two isogenic clones of Brca2Δ27/Δ27 MEF cells (Fig. 2 B). DUB inactivity elevated levels of chromosome instability in response to ICLs, but not to PARPi, in either heterozygous or homozygous Brca2 mutant cells (Fig. 2, C–E). The effect was not related to loss of fork protection in double mutant cells (Fig. S2 A). These results suggest that BRCC36 DUB activity cooperates with BRCA2 to resolve ICLs, consistent with it contributing to BRCA1-A complex function during replication-associated DNA damage responses.
Cooperation between BRCC36 and BRCA2 in response to ICLs depends on BRCC36 DUB activity. (A) Mouse survival curve of Brca2Δ27/Δ27 mice and double mutant mice (the 21 Brca2Δ27/Δ27 Brcc36 mutant mice consist of Brca2Δ27/Δ27Brcc36KO/KO [five mice], Brca2Δ27/Δ27Brcc36QSQ/QSQ [eight mice], Brca2Δ27/Δ27Brcc36E33A/E33A [eight mice]). Mantel-Cox test was conducted to test the survival curve from Brca2Δ27/Δ27 mice and double mutant mice. (B) Plating efficiency of two Brca2Δ27/Δ27 MEF clones and three isogenic Brca2Δ27/Δ27Brcc36E33A/E33A MEF clones. Quantification of plating efficiency was conducted based on three independent experiments (duplicates were performed for each experiment). Unpaired two-sided Student’s t test was conducted based on the average plating efficiency from each experiment. **P < 0.01, ***P < 0.001. Mean with SEM was used to plot the data. (C) Representative images of metaphase spread from Brca2Δ27/Δ27 and Brcc36QSQ/QSQBrca2Δ27/Δ27 splenocytes post exposure to 60 nM MMC. Scale bar: 5 μM. (D and E) Splenocytes were treated with either 60 nM MMC (D) or 1 μM PARPi (E) for 24 h prior to metaphase harvest, n > 70 metaphases in total from three to six mice per genotype were analyzed. Unpaired two-sided Student’s t test was conducted based on the average chromosomal abnormalities per 1,000 chromosomes of each mouse. *P < 0.01, **P < 0.01, ***P < 0.001, ****P < 0.0001. Mean with SEM was used to plot the data.
Cooperation between BRCC36 and BRCA2 in response to ICLs depends on BRCC36 DUB activity. (A) Mouse survival curve of Brca2Δ27/Δ27 mice and double mutant mice (the 21 Brca2Δ27/Δ27 Brcc36 mutant mice consist of Brca2Δ27/Δ27Brcc36KO/KO [five mice], Brca2Δ27/Δ27Brcc36QSQ/QSQ [eight mice], Brca2Δ27/Δ27Brcc36E33A/E33A [eight mice]). Mantel-Cox test was conducted to test the survival curve from Brca2Δ27/Δ27 mice and double mutant mice. (B) Plating efficiency of two Brca2Δ27/Δ27 MEF clones and three isogenic Brca2Δ27/Δ27Brcc36E33A/E33A MEF clones. Quantification of plating efficiency was conducted based on three independent experiments (duplicates were performed for each experiment). Unpaired two-sided Student’s t test was conducted based on the average plating efficiency from each experiment. **P < 0.01, ***P < 0.001. Mean with SEM was used to plot the data. (C) Representative images of metaphase spread from Brca2Δ27/Δ27 and Brcc36QSQ/QSQBrca2Δ27/Δ27 splenocytes post exposure to 60 nM MMC. Scale bar: 5 μM. (D and E) Splenocytes were treated with either 60 nM MMC (D) or 1 μM PARPi (E) for 24 h prior to metaphase harvest, n > 70 metaphases in total from three to six mice per genotype were analyzed. Unpaired two-sided Student’s t test was conducted based on the average chromosomal abnormalities per 1,000 chromosomes of each mouse. *P < 0.01, **P < 0.01, ***P < 0.001, ****P < 0.0001. Mean with SEM was used to plot the data.
Related toFigs. 2,and3 . (A) Single fiber analysis of WT, QSQ, Brca2Δ27/Δ27 and Brca2Δ27/Δ27 Brcc36QSQ/QSQ splenocytes in response to HU. Cells were labeled with IdU and CldU sequentially for 30 min respectively, followed by 3 h treatment with 4 mM HU. Quantified CldU/IdU ratio was based on three mice each genotype. More than 50 fibers per mouse were quantified. Unpaired two-sided t test was conducted based on the average CldU/IdU ratio for each mouse. *P < 0.05. Mean with SEM was used to plot the data. (B) Quantified percentage of restarted replication forks for MEFs with individual genotypes were based on three independent experiments. % of restarted replication fork equals to the number of restarted forks divided by the sum of the stalled forks and restarted forks. More than 50 fibers were quantified for each replicate. Unpaired two-sided Student’s t test was conducted based on the average % of restarted replication fork from each replicate. **P < 0.01. Mean with SEM was used to plot the data. (C) HeLa S3 cells were labeled with EdU for 20 min, followed by either a chase with thymidine-containing medium for 4 h, or treatment with HU for 4 h. Cells were collected following standard iPOND protocol and then indicated proteins were examined. (D) Representative images of SIRF experiments in WT and E33A MEFs treated with no EdU, only EdU or EdU followed by HU. (E) Quantification of PCNA PLA Foci per cell in WT MEFs was based on three independent experiments. In total 30–50 cells were quantified per condition. Unpaired two-sided Student’s t test was conducted based on the average PLA foci per cell from each experiment. *P < 0.05. Mean with SEM was used to plot the data. Source data are available for this figure: SourceData FS2.
Related toFigs. 2,and3 . (A) Single fiber analysis of WT, QSQ, Brca2Δ27/Δ27 and Brca2Δ27/Δ27 Brcc36QSQ/QSQ splenocytes in response to HU. Cells were labeled with IdU and CldU sequentially for 30 min respectively, followed by 3 h treatment with 4 mM HU. Quantified CldU/IdU ratio was based on three mice each genotype. More than 50 fibers per mouse were quantified. Unpaired two-sided t test was conducted based on the average CldU/IdU ratio for each mouse. *P < 0.05. Mean with SEM was used to plot the data. (B) Quantified percentage of restarted replication forks for MEFs with individual genotypes were based on three independent experiments. % of restarted replication fork equals to the number of restarted forks divided by the sum of the stalled forks and restarted forks. More than 50 fibers were quantified for each replicate. Unpaired two-sided Student’s t test was conducted based on the average % of restarted replication fork from each replicate. **P < 0.01. Mean with SEM was used to plot the data. (C) HeLa S3 cells were labeled with EdU for 20 min, followed by either a chase with thymidine-containing medium for 4 h, or treatment with HU for 4 h. Cells were collected following standard iPOND protocol and then indicated proteins were examined. (D) Representative images of SIRF experiments in WT and E33A MEFs treated with no EdU, only EdU or EdU followed by HU. (E) Quantification of PCNA PLA Foci per cell in WT MEFs was based on three independent experiments. In total 30–50 cells were quantified per condition. Unpaired two-sided Student’s t test was conducted based on the average PLA foci per cell from each experiment. *P < 0.05. Mean with SEM was used to plot the data. Source data are available for this figure: SourceData FS2.
DUB activity promotes the function of the BRCA1-A complex during replication stress
DUB-inactive cells showed delayed recovery in response to S phase damage (Fig. 3, A and B; and Fig. S2 B). To understand the basis for this phenomenon, we utilized isolation of proteins on nascent DNA (iPOND) in HeLa S3 cells to examine BRCA1-A complex association with ongoing, previously active, and stalled replication forks. Consistent with published results (Nakamura et al., 2021), BRCC36 showed association with nascent forks by iPOND, and this increased after fork stalling in the presence of HU (Fig. S2 C). In situ analysis of protein interactions at DNA replication forks (SIRF; Roy et al., 2018) in MEFs showed similar results for increased A-complex localization to stalled forks. Cells were pulse-labeled with thymidine analog EdU for 8 min, which marks the newly synthesized DNA sites. After removing the EdU, cells were treated with HU to study protein association with stalled replication forks (Fig. 3 C). We first validated the system by examining PCNA association with EdU at ongoing forks (EdU) and stalled replication forks (EdU + HU). As expected, PCNA demonstrated strong association with nascent replication forks, and this association is reduced at stalled replication forks (Fig. S2, D and E). Next, we examined the recruitment of the A-complex to these replication forks. The A-complex (RAP80 and MERIT40) was present at replication forks under all conditions and enriched at stalled forks. E33A cells demonstrated a marked reduction in recruitment of RAP80 in comparison to WT cells after HU treatment (Fig. 3, D and E), indicating that like IR-induced foci, DUB activity also contributes to the localization of the A-complex to stalled replication forks.
Reduced recruitment of RAP80 at stalled replication forks in DUB-inactive cells. (A) Schematic of experimental design as well as representative images of replication fibers. Purple tracks: CldU; green tracks: IdU. Example of a stalled replication fork and a restarted replication fork was shown. Scale bar: 5 μM. (B) Quantified percentage of restarted replication forks for MEFs with individual genotypes were based on three independent experiments. % of restarted replication fork equals to the number of restarted forks divided by the sum of the stalled forks and restarted forks. More than 130 fibers were quantified in each replicate. Unpaired two-sided Student’s t test was conducted based on the average % of restarted replication fork from each replicate. ***P < 0.001, ****P < 0.0001. Mean with SEM was used to plot the data. (C) Schematic of SIRF experiment. MEFs were incubated with EdU to label active replication forks or stalled replication forks (cells those were also treated with HU). Click chemistry and biotin-azide allowed labeling of active or stalled forks and PLA was performed to evaluate whether certain proteins were near these forks. (D) Representative image of SIRF experiments in WT and E33A MEFs, which were treated with no EdU, only EdU or EdU followed by HU. Scale bar: 5 μM. (E) Quantification of RAP80 and MERIT40 PLA foci per cell in WT and E33A MEFs was based on three independent experiments. In total 30–50 cells were quantified per condition. Unpaired two-sided Student’s t test was conducted based on the average PLA foci per cell from each experiment. *P < 0.05, **P < 0.01. Mean with SEM was used to plot the data.
Reduced recruitment of RAP80 at stalled replication forks in DUB-inactive cells. (A) Schematic of experimental design as well as representative images of replication fibers. Purple tracks: CldU; green tracks: IdU. Example of a stalled replication fork and a restarted replication fork was shown. Scale bar: 5 μM. (B) Quantified percentage of restarted replication forks for MEFs with individual genotypes were based on three independent experiments. % of restarted replication fork equals to the number of restarted forks divided by the sum of the stalled forks and restarted forks. More than 130 fibers were quantified in each replicate. Unpaired two-sided Student’s t test was conducted based on the average % of restarted replication fork from each replicate. ***P < 0.001, ****P < 0.0001. Mean with SEM was used to plot the data. (C) Schematic of SIRF experiment. MEFs were incubated with EdU to label active replication forks or stalled replication forks (cells those were also treated with HU). Click chemistry and biotin-azide allowed labeling of active or stalled forks and PLA was performed to evaluate whether certain proteins were near these forks. (D) Representative image of SIRF experiments in WT and E33A MEFs, which were treated with no EdU, only EdU or EdU followed by HU. Scale bar: 5 μM. (E) Quantification of RAP80 and MERIT40 PLA foci per cell in WT and E33A MEFs was based on three independent experiments. In total 30–50 cells were quantified per condition. Unpaired two-sided Student’s t test was conducted based on the average PLA foci per cell from each experiment. *P < 0.05, **P < 0.01. Mean with SEM was used to plot the data.
DUB activity prevents hyper-K63-Ub of the A-complex and promotes ubiquitin binding in trans
BRCC36 DUB activity appears to be essential for BRCA1-A complex localization and function at sites of DSBs and replication stress. However, these observations are inconsistent with DUB activity exclusively being directed toward outside substrates. Since RAP80 is ubiquitylated at a steady state (Li et al., 2017; Patterson-Fortin et al., 2010), we therefore examined whether elevated K63-Ub is present on A-complex members in DUB mutant cells. Consistent with previous studies (Ng et al., 2016), protein levels of Abraxas1, MERIT40, and BRCC45 did not change in Brcc36 DUB-inactive MEFs, indicating the A-complex was not destabilized upon DUB inactivation. In contrast, three-independent isogenic MEF pairs showed reduced RAP80 intensity on immunoblots in DUB mutant cells in comparison to WT MEFs (Fig. 4 A and Fig. S3 A). To rule out the possibility that differences were due to variation in cell lines, WT and E33A BRCC36 cDNAs were introduced in the KO MEFs. Reduced RAP80 band intensity was consistently observed in cells expressing the E33A mutant (Fig. 4 B). As other A-complex members were not destabilized, it is unlikely that the RAP80 protein itself is being degraded in DUB inactive cells.
BRCC36-dependent de-ubiquitylation of RAP80 is essential for BRCA1-A complex recognition of Lys63 ubiquitylation. (A) RAP80 complex member expression was examined in three litters of paired WT and mutant MEFs. (B) RAP80 complex member protein expression level was examined in WT, QSQ MEFs as wells as in KO cells with reconstitution of either WT or E33A mutant of BRCC36. (C) Schematic of TUBE pull-down assays. Cells were incubated with TUBEs that have high affinity for ubiquitin. Post immunoprecipitation, K63 specific DUB or non-specific DUB were used to cleave off the ubiquitin. (D) MEFs were treated with either no damage or 10 Gy IR for 1 h, then harvested and processed according to the schematic of 4 C. (E) TUBE pulldown experiment in BRCC36−/− HeLa S3 cells with reconstituted E33A. TUBE pulled-down samples were treated with recombinant A-complex or USP2 followed by immunoblotting with the indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. (F) Schematic of the ubiquitylation assay. Purified ectopic RAP80 complex that containing either WT or E33A BRCC36 were incubated with non-cleavable His tagged K63 linked ubiquitylation and assessed for their ability to bind in vitro. (G) RAP80 complexes were immunoprecipitated from BRCC36−/− HeLa S3 cells reconstituted with FLAG-HA-WT BRCC36 or FLAG-HA-E33A BRCC36 by FLAG pull down and were processed according to 4F. Mock represents FLAG pulled-down RAP80 complexes incubated with nickel beads without His tagged K63-Ub-His. Star indicates a non-specific band. Source data are available for this figure: SourceData F4.
BRCC36-dependent de-ubiquitylation of RAP80 is essential for BRCA1-A complex recognition of Lys63 ubiquitylation. (A) RAP80 complex member expression was examined in three litters of paired WT and mutant MEFs. (B) RAP80 complex member protein expression level was examined in WT, QSQ MEFs as wells as in KO cells with reconstitution of either WT or E33A mutant of BRCC36. (C) Schematic of TUBE pull-down assays. Cells were incubated with TUBEs that have high affinity for ubiquitin. Post immunoprecipitation, K63 specific DUB or non-specific DUB were used to cleave off the ubiquitin. (D) MEFs were treated with either no damage or 10 Gy IR for 1 h, then harvested and processed according to the schematic of 4 C. (E) TUBE pulldown experiment in BRCC36−/− HeLa S3 cells with reconstituted E33A. TUBE pulled-down samples were treated with recombinant A-complex or USP2 followed by immunoblotting with the indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. (F) Schematic of the ubiquitylation assay. Purified ectopic RAP80 complex that containing either WT or E33A BRCC36 were incubated with non-cleavable His tagged K63 linked ubiquitylation and assessed for their ability to bind in vitro. (G) RAP80 complexes were immunoprecipitated from BRCC36−/− HeLa S3 cells reconstituted with FLAG-HA-WT BRCC36 or FLAG-HA-E33A BRCC36 by FLAG pull down and were processed according to 4F. Mock represents FLAG pulled-down RAP80 complexes incubated with nickel beads without His tagged K63-Ub-His. Star indicates a non-specific band. Source data are available for this figure: SourceData F4.
Related toFig. 4 . (A) Immunoblots showing expression level of RAP80 complex member (BRCC45 and MERIT40) in three litters of paired WT and mutant MEFs. (B and C)BRCC36−/− HeLa S3 cells were generated via CRISPR-Cas9 technology. FLAG-HA-WT, FLAG-HA-E33A BRCC36 protein were reconstituted in BRCC36−/− HeLa S3 cells. (B) Cells were subjected to TUBE pull down and further processed with USP2 followed by immunoblotting with the indicated antibodies. BRCC36 was detected using an anti-HA antibody. Ponceau S staining serves as a loading control in immunoblots. (C) Cells were treated with 500 ng/ml MMC for 24 h, then ectopic BRCC36 WT and E33A were pulled down with FLAG antibody, and probed with RAP80, BRCC36 and ubiquitin. (D) RAP80 complexes were immunoprecipitated from HeLa S3 cells expressing BRCC36 FLAG-HA-E33A by FLAG pull down. Immunoprecipitated samples were left untreated or treated with either USP2 or recombinant A-complex and subjected to Western blotting using indicated antibodies. K63-linked polyubiquitination of RAP80 complexes was determined using a K63-linkage specific polyubiquitin antibody. HeLa S3 cells harboring endogenous BRCC36 were used as mock control. Star indicates a non-specific band. (E) TUBE pulldown experiment in HeLa S3/E33A cells. TUBE pulled-down samples were treated with recombinant A-complex, USP2 or OTUB1 followed by immunoblotting with the indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. Source data are available for this figure: SourceData FS3.
Related toFig. 4 . (A) Immunoblots showing expression level of RAP80 complex member (BRCC45 and MERIT40) in three litters of paired WT and mutant MEFs. (B and C)BRCC36−/− HeLa S3 cells were generated via CRISPR-Cas9 technology. FLAG-HA-WT, FLAG-HA-E33A BRCC36 protein were reconstituted in BRCC36−/− HeLa S3 cells. (B) Cells were subjected to TUBE pull down and further processed with USP2 followed by immunoblotting with the indicated antibodies. BRCC36 was detected using an anti-HA antibody. Ponceau S staining serves as a loading control in immunoblots. (C) Cells were treated with 500 ng/ml MMC for 24 h, then ectopic BRCC36 WT and E33A were pulled down with FLAG antibody, and probed with RAP80, BRCC36 and ubiquitin. (D) RAP80 complexes were immunoprecipitated from HeLa S3 cells expressing BRCC36 FLAG-HA-E33A by FLAG pull down. Immunoprecipitated samples were left untreated or treated with either USP2 or recombinant A-complex and subjected to Western blotting using indicated antibodies. K63-linked polyubiquitination of RAP80 complexes was determined using a K63-linkage specific polyubiquitin antibody. HeLa S3 cells harboring endogenous BRCC36 were used as mock control. Star indicates a non-specific band. (E) TUBE pulldown experiment in HeLa S3/E33A cells. TUBE pulled-down samples were treated with recombinant A-complex, USP2 or OTUB1 followed by immunoblotting with the indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. Source data are available for this figure: SourceData FS3.
An alternative explanation is that the RAP80 protein is highly modified by ubiquitin upon DUB inactivation, rendering the unmodified version difficult to visualize. To test this hypothesis, tandem ubiquitin-binding entity (TUBE) immunoprecipitated proteins were treated with either AMSH, a linkage-specific DUB to remove K63-Ub chains, or a nonspecific DUB (USP2) to remove all ubiquitin. These treatments condensed RAP80 signals on the immunoblot (Fig. 4, C and D). An increased signal for a faster migrating RAP80 protein upon K63-specific DUB AMSH treatment indicated that RAP80 was modified by K63-Ub. Additionally, treatment by the non-specific DUB USP2 led to a greater increase in RAP80 protein, consistent with the removal of both K63-Ub chains and mono-ubiquitin. An increase in the RAP80 band intensity was also observed in response to DNA damage, suggesting that BRCC36-dependent deubiquitylation of RAP80 occurs both at a steady state and in response to DNA damage. Interestingly, BRCC36 was also hyper-ubiquitylated in DUB-inactive knock-in cells, suggesting that it can auto-deubiquitylate within the A-complex (Fig. 4 D). Similar results were obtained when we examined the ubiquitylation level of RAP80 in BRCC36 null HeLa S3 cell lines with different forms of BRCC36 being reconstituted to a similar level as endogenous BRCC36. In line with the results in MEFs, RAP80 and BRCC36 were hyper-ubiquitylated in E33A complexes isolated from HeLa S3 cells (Fig. S3, B and C).
To further validate K63-linked ubiquitylation of the RAP80 complex, we performed TUBE pulldown experiments in BRCC36 null HeLa S3 cell lines reconstituted with BRCC36 E33A and tested the impact of recombinant A-complex on RAP80 and BRCC36 ubiquitylation levels. Unmodified RAP80 and BRCC36 signals were found to be substantially increased upon A-complex treatment (Fig. 4 E), suggesting RAP80 modification by K63-linked polyubiquitin chains. In agreement, we detected K63-linked ubiquitylation on RAP80 complex immunoprecipitated from E33A cells using a K63-linkage specific anti-Ub antibody (Fig. S3 D). However, we did not observe any effect of K48-specific DUB OTUB1 on ubiquitylated RAP80 pulled-down from E33A cells (Fig. S3 E). Collectively, these results suggest that RAP80 is primarily modified by K63-linked polyubiquitin chains in DUB mutant cells. We hypothesized that increased A-complex ubiquitylation is inhibitory to RAP80 binding to its K63 ubiquitylated targets on chromatin. In agreement, purified E33A containing A-complex showed reduced binding to non-degradable K63-linked polyubiquitin chains compared to WT BRCC36 purified A-complex (Fig. 4, F and G). This is consistent with the premise that A-complex auto-deubiquitylation facilitates RAP80 binding in trans to ubiquitylated proteins on damaged chromatin.
Mutation of a subset of lysine residues identified by MS is inadequate to abolish K63 ubiquitylation on RAP80 and BRCC36
Next, we attempted to determine the ubiquitylated sites on RAP80 and BRCC36 by mass spectrometry (MS) using purified E33A-containing complexes from HeLa S3 cells. Gel slices that corresponded to the size of mono- or di-ubiquitylated RAP80 and BRCC36 were submitted for MS analysis (Fig. 5 A). A previous study from our lab took a similar approach and identified potential ubiquitylation sites on RAP80, including K9, K20, K31, K75, and K374 (Patterson-Fortin et al., 2010). Two replicates of such MS analyses re-identified four sites that are in common with the previous study, K20, K31, K75, and K374, as well as two novel sites on RAP80 (K19 and K382) and two novel sites on BRCC36 (K65 and K204). Most of the ubiquitylated sites were not near the UIM domains of RAP80 (except K75), suggesting ubiquitylation on these sites might lead to intramolecular binding to the RAP80 UIMs, as well as conformational changes in the protein. Sequence alignments revealed evolutionary conservation of most RAP80 sites (Fig. 5 B), suggesting these residues play an important role in modulating the functions and/or stability of this subunit. By contrast, K65 in BRCC36 localizes to a flexible loop that is uniquely conserved across mammals, while K204 localizes to a loop that is exclusively present in the human protein and situated at approximately 30 Å above the BRCC36 active site (Fig. 5, C and D).
Mutation of a subset of lysine residues identified by MS are inadequate to abolish K63 ubiquitylation on RAP80 and BRCC36. (A) FLAG and HA tandem immunoaffinity purification was performed for ectopic BRCC36 FLAG-HA-E33A from BRCC36−/− HeLa S3 nuclear extracts. The indicated gel areas (within green and red boxes) were excised and trypsin-digested prior to MS analysis. Peptide sequences obtained by MS of tryptic digests from the gel areas were highlighted. K# indicates RAP80 or BRCC36 lysine residues that are conjugated to ubiquitin. The gel picture is from the replicate II. (B and C) Multiple sequence alignments of the regions targeted by ubiquitylation in RAP80 (B) and BRCC36 (C) Sequence identity within the RAP80 and BRCC36 species is indicated with black shading and similarity with gray shading. Protein domains are shown in blue, secondary structural elements are depicted in red and ubiquitylated residues are highlighted as indicated in the legend. NLS = nuclear localization signal; SIM = SUMO (Small Ubiquitin-like Modifier)-interacting motif; UIM1 = ubiquitin-interacting motif 1; AIR = Abraxas1 interacting region; MPN+ = Mpr1/Pad1 N-terminal+, CC = coiled coil. (D) A structural model of BRCC36 is shown in cartoon representation, with secondary structural elements indicated. Close-up view highlights the position of the ubiquitylated residue K204 relative to the active site. Aspartate, glutamate and histidine residues involved in catalysis are shown as sticks; a representative Zn2+ atom and a catalytic H2O molecule (obtained using the same overlay depicted in Fig. 1 A) are shown as gray and red spheres. Tetrahedral coordination of the Zn2+ ion by the water molecule and active site residues is indicated with dashed lines. (E) GFP, GFP-RAP80, and GFP-RAP80 7KR (K9R, K19R, K20R, K31R, K75R, K374R, K382) were stably expressed in BRCC36−/− HeLa S3 cells expressing FLAG-HA BRCC36 ARA (E33A, K65R, K204A) and subjected to TUBE pulldown assay. TUBE pulled-down samples were subsequently treated with USP2 followed by immunoblotting using indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. (F) In BRCC36−/− HeLa S3 cells, FLAG tagged BRCC36 E33A and BRCC36 ARA (E33A, K65R, K204A) were expressed. FLAG immunoaffinity purification was performed on nuclear extracts from the cells listed above. Partially purified complexes were left intact or treated with USP2, blotted onto nitrocellulose membrane, and probed for FLAG-tag. Mutation of lysines, identified as targets for ubiquitylation, to arginine and alanine substantially reduces the amount of ubiquitylated species, however, does not eliminate them completely. Source data are available for this figure: SourceData F5.
Mutation of a subset of lysine residues identified by MS are inadequate to abolish K63 ubiquitylation on RAP80 and BRCC36. (A) FLAG and HA tandem immunoaffinity purification was performed for ectopic BRCC36 FLAG-HA-E33A from BRCC36−/− HeLa S3 nuclear extracts. The indicated gel areas (within green and red boxes) were excised and trypsin-digested prior to MS analysis. Peptide sequences obtained by MS of tryptic digests from the gel areas were highlighted. K# indicates RAP80 or BRCC36 lysine residues that are conjugated to ubiquitin. The gel picture is from the replicate II. (B and C) Multiple sequence alignments of the regions targeted by ubiquitylation in RAP80 (B) and BRCC36 (C) Sequence identity within the RAP80 and BRCC36 species is indicated with black shading and similarity with gray shading. Protein domains are shown in blue, secondary structural elements are depicted in red and ubiquitylated residues are highlighted as indicated in the legend. NLS = nuclear localization signal; SIM = SUMO (Small Ubiquitin-like Modifier)-interacting motif; UIM1 = ubiquitin-interacting motif 1; AIR = Abraxas1 interacting region; MPN+ = Mpr1/Pad1 N-terminal+, CC = coiled coil. (D) A structural model of BRCC36 is shown in cartoon representation, with secondary structural elements indicated. Close-up view highlights the position of the ubiquitylated residue K204 relative to the active site. Aspartate, glutamate and histidine residues involved in catalysis are shown as sticks; a representative Zn2+ atom and a catalytic H2O molecule (obtained using the same overlay depicted in Fig. 1 A) are shown as gray and red spheres. Tetrahedral coordination of the Zn2+ ion by the water molecule and active site residues is indicated with dashed lines. (E) GFP, GFP-RAP80, and GFP-RAP80 7KR (K9R, K19R, K20R, K31R, K75R, K374R, K382) were stably expressed in BRCC36−/− HeLa S3 cells expressing FLAG-HA BRCC36 ARA (E33A, K65R, K204A) and subjected to TUBE pulldown assay. TUBE pulled-down samples were subsequently treated with USP2 followed by immunoblotting using indicated antibodies. Ponceau S staining serves as a loading control in immunoblots. (F) In BRCC36−/− HeLa S3 cells, FLAG tagged BRCC36 E33A and BRCC36 ARA (E33A, K65R, K204A) were expressed. FLAG immunoaffinity purification was performed on nuclear extracts from the cells listed above. Partially purified complexes were left intact or treated with USP2, blotted onto nitrocellulose membrane, and probed for FLAG-tag. Mutation of lysines, identified as targets for ubiquitylation, to arginine and alanine substantially reduces the amount of ubiquitylated species, however, does not eliminate them completely. Source data are available for this figure: SourceData F5.
To test whether eliminating key ubiquitylation on RAP80 and BRCC36 would reduce auto-ubiquitylation in DUB-inactive cells, lysine to arginine mutations were made on seven identified sites in a GFP-RAP80 cDNA clone. GFP-RAP80 WT or mutant variant (GFP-RAP80 7KR) were stably expressed in HeLa S3 BRCC36−/− cells reconstituted with BRCC36 active site (E33A) and triple mutant (E33A + K65R + K204A, referred to as ARA). TUBE pulldown experiments in these cells revealed that GFP-RAP80 7KR maintained similar levels of ubiquitylation to WT GFP-RAP80 (Fig. 5 E). Similarly, BRCC36 ARA mutant also remained ubiquitylated as revealed by TUBE pulldowns followed by USP2 incubation (Fig. 5 F). The results demonstrate that additional ubiquitylation sites are present on both BRCC36 and RAP80 and likely contribute to autoinhibition, highlighting the considerable redundancy present in this regulatory system.
The RAP80 UIMs directly interact with ubiquitin in E33A containing A-complex
To obtain further insights on the mechanisms underlying auto-inhibition, we isolated A-complexes from WT and DUB-inactive HeLa S3 cells and performed chemical cross-linking coupled to MS analysis (Fig. 6, A and B). We employed the long-range lysine-specific cross-linker disuccinimidyl suberate (DSS) and its soluble analog bis(sulfosuccinimidyl)suberate (BS-3), both of which are characterized by a spacer arm length of 11.4 Å. Using these approaches, we identified 20 intra- and 23 intermolecular cross-links in total (Tables S1 and S2). Among the intramolecular cross-links, four were present in both complexes, six were unique for WT and 10 were unique for the E33A complex (Fig. 6, C and D; and Fig. S4 and Table S1). In addition, seven intermolecular cross-links were found in both complexes, 14 were unique for WT and two were unique to DUB-inactive complexes (Fig. 6, C–F and Fig. S5 and Table S2). These analyses suggest the structural conformation of A-complex may differ between WT and DUB-inactive complexes, most likely due to multiple ubiquitylation events targeting residues in BRCC36 and RAP80 specifically in the E33A complex.
Chemical crosslinking reveals unique ubiquitin binding of the RAP80 UIM in E33A DUB-inactive complexes. (A) Schematic of crosslinking MS experiment. WT or E33A complexes were isolated from cultured BRCC36−/− HeLa S3 cells using FLAG-tag, subjected to cross-linking, and further purified with HA-tag. Recovered material was digested with trypsin and peptides were analyzed with LC-MS/MS. Cross-link species were identified, verified, and used in structure modeling. (B) Representative image of SDS-PAGE analysis of complex isolation. Proteins bound to anti-FLAG resin were eluted with FLAG peptide in two repetitive steps (lanes 2 and 3). Resulted material was cross-linked with DSS or BS3 (lane 4) and loaded onto anti-HA media, unbound material is presented in lane 5. Complex was first eluted with low pH and 10% of it was loaded to lane 6, and then eluted with SDS solution (lane 7). Cross-linked material from first HA elution (highlighted by the red box) was used in LC-MS/MS analysis. (C and D) Chemical cross-linking and MS analysis of full-length (FL) WT (C) and DUB inactive E33A (D) A-complexes obtained from HeLa S3 cells. Intra- and intermolecular cross-links are indicated with purple arches and green lines, respectively. (E) Mass-spectrum and structure of a peptide identified as cross-link between RAP80 UIM2 and ubiquitin. Ion series derived from ubiquitin peptide are shown in green, series from RAP80 peptide—in blue. Position of linked lysines is indicated with red line. (F) Left: Cross-link between ubiquitin and RAP80 UIM is unique for E33A complex. We found 7 cross-linked peptides common for both complexes, 14 cross-linked peptides were identified only in WT complex, and 2 unique peptides in E33A complex, including RAP80 UIM2 and ubiquitin cross-link. Right: A model of the human RAP80 UIMs was overlaid on the structure of the mouse RAP80 UIMs in complex with Lys63-linked di-Ubiquitin (K63-Ub2; PDB accession no. 3A1Q), and the resulting human RAP80 UIMs:K63-Ub2 complex is shown as cartoon. The RAP80 UIMs are colored red, while the proximal (UbP) and distal (UbD) ubiquitin molecules are colored light gray. RAP80 K112, K6 in UbP and UbD, and the residues involved in isopeptide bond formation (i.e., UbP-K63 and UbD-G76) are shown as sticks. The distances between RAP80 K112 and K6 from UbD or UbP are indicated with dashed black lines and measured in Ångstrom (Å).
Chemical crosslinking reveals unique ubiquitin binding of the RAP80 UIM in E33A DUB-inactive complexes. (A) Schematic of crosslinking MS experiment. WT or E33A complexes were isolated from cultured BRCC36−/− HeLa S3 cells using FLAG-tag, subjected to cross-linking, and further purified with HA-tag. Recovered material was digested with trypsin and peptides were analyzed with LC-MS/MS. Cross-link species were identified, verified, and used in structure modeling. (B) Representative image of SDS-PAGE analysis of complex isolation. Proteins bound to anti-FLAG resin were eluted with FLAG peptide in two repetitive steps (lanes 2 and 3). Resulted material was cross-linked with DSS or BS3 (lane 4) and loaded onto anti-HA media, unbound material is presented in lane 5. Complex was first eluted with low pH and 10% of it was loaded to lane 6, and then eluted with SDS solution (lane 7). Cross-linked material from first HA elution (highlighted by the red box) was used in LC-MS/MS analysis. (C and D) Chemical cross-linking and MS analysis of full-length (FL) WT (C) and DUB inactive E33A (D) A-complexes obtained from HeLa S3 cells. Intra- and intermolecular cross-links are indicated with purple arches and green lines, respectively. (E) Mass-spectrum and structure of a peptide identified as cross-link between RAP80 UIM2 and ubiquitin. Ion series derived from ubiquitin peptide are shown in green, series from RAP80 peptide—in blue. Position of linked lysines is indicated with red line. (F) Left: Cross-link between ubiquitin and RAP80 UIM is unique for E33A complex. We found 7 cross-linked peptides common for both complexes, 14 cross-linked peptides were identified only in WT complex, and 2 unique peptides in E33A complex, including RAP80 UIM2 and ubiquitin cross-link. Right: A model of the human RAP80 UIMs was overlaid on the structure of the mouse RAP80 UIMs in complex with Lys63-linked di-Ubiquitin (K63-Ub2; PDB accession no. 3A1Q), and the resulting human RAP80 UIMs:K63-Ub2 complex is shown as cartoon. The RAP80 UIMs are colored red, while the proximal (UbP) and distal (UbD) ubiquitin molecules are colored light gray. RAP80 K112, K6 in UbP and UbD, and the residues involved in isopeptide bond formation (i.e., UbP-K63 and UbD-G76) are shown as sticks. The distances between RAP80 K112 and K6 from UbD or UbP are indicated with dashed black lines and measured in Ångstrom (Å).
Related to Fig. 7 . (A) Left: A structural model of the human A-complex (AIR) is shown as cartoon, with the BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR subunits colored blue, green, pink, wheat, and red respectively. Dashed black rectangles highlight regions in BRCC36 and Abraxas1 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites within BRCC36 (panel 1) or Abraxas1 (panels 2–4). Residues involved in cross-links are shown as sticks; the distances between cross-link sites are indicated with black dashed lines and measured in Ångstrom (Å). (B) Left: A structural model of human A-complex (AIR) is shown and depicted as described in A. Dashed black rectangles highlight regions in BRCC45 and MERIT40 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites within BRCC45 (panels 1, 2) or MERIT40 (panel 3). Residues involved in cross-links and distances between cross-link sites are indicated as in A.
Related to Fig. 7 . (A) Left: A structural model of the human A-complex (AIR) is shown as cartoon, with the BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR subunits colored blue, green, pink, wheat, and red respectively. Dashed black rectangles highlight regions in BRCC36 and Abraxas1 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites within BRCC36 (panel 1) or Abraxas1 (panels 2–4). Residues involved in cross-links are shown as sticks; the distances between cross-link sites are indicated with black dashed lines and measured in Ångstrom (Å). (B) Left: A structural model of human A-complex (AIR) is shown and depicted as described in A. Dashed black rectangles highlight regions in BRCC45 and MERIT40 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites within BRCC45 (panels 1, 2) or MERIT40 (panel 3). Residues involved in cross-links and distances between cross-link sites are indicated as in A.
Related to Fig. 7 . (A) Left: A structural model of the human A-complex (AIR) is shown as cartoon, with the BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR subunits colored blue, green, pink, wheat, and red respectively. Dashed black rectangles highlight regions in the BRCC36-Abraxas1 superdimer where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites between BRCC36 and Abraxas1. Residues involved in cross-links are shown as sticks; the distances between cross-link sites are indicated with black dashed lines and measured in Ångstrom (Å). (B) Left: A structural model of human A-complex (AIR) is depicted as described in A. Dashed black rectangles highlight regions at the interfaces between BRCC36:BRCC45, BRCC45:MERIT40, and Abraxas1:MERIT40 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites between BRCC36:BRCC45 (panel 1), BRCC45:MERIT40 (panel 2), and Abraxas1:MERIT40 (panels 3, 4). Residues involved in cross-links and distances between cross-link sites are indicated as in A. (C) Left: Structural model of the human A-complex (AIR) is shown as in A. Dashed black rectangles highlight regions at the interfaces between Abraxas1:BRCC45 and Abraxas1:MERIT40 where cross-link sites were identified. Right: Close-up views and structural modelling of the cross-linked sites between Abraxas1:BRCC45 (panel 1) and Abraxas1:MERIT40 (panel 2). BRCC45 and MERIT40 residues involved in cross-links are shown as sticks while Abraxas1 residues are indicated with green circles; black dashed lines connect cross-linked residues, and dashed green lines represent the modelled Abraxas1 C-terminus not visible in the available structures. (D) Left: Structural model of human A-complex (AIR) depicted as in A. Dashed black rectangles highlight regions where cross-link sites between RAP80 and BRCC36, Abraxas1, BRCC45, or MERIT40 were identified. Right: Close-up views and structural modelling of the cross-linked sites between RAP80:BRCC36 (panel 1), RAP80:Abraxas1 (panels 2 and 3), RAP80:BRCC45 (panel 4), and RAP80:MERIT40 (panels 5 and 6). BRCC36, Abraxas1, BRCC45, and MERIT40 residues involved in cross-links are shown as sticks; residues located at the C-terminus of Abraxas1 and within RAP80 are depicted as green and red circles, respectively. Black dashed lines connect cross-linked residues; dashed green lines indicate the modelled Abraxas1 C-terminus not visible in the available structures, whereas dashed red lines refer to modelled regions in RAP80.
Related to Fig. 7 . (A) Left: A structural model of the human A-complex (AIR) is shown as cartoon, with the BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR subunits colored blue, green, pink, wheat, and red respectively. Dashed black rectangles highlight regions in the BRCC36-Abraxas1 superdimer where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites between BRCC36 and Abraxas1. Residues involved in cross-links are shown as sticks; the distances between cross-link sites are indicated with black dashed lines and measured in Ångstrom (Å). (B) Left: A structural model of human A-complex (AIR) is depicted as described in A. Dashed black rectangles highlight regions at the interfaces between BRCC36:BRCC45, BRCC45:MERIT40, and Abraxas1:MERIT40 where cross-link sites were identified. Right: Close-up views and structural details of the cross-linked sites between BRCC36:BRCC45 (panel 1), BRCC45:MERIT40 (panel 2), and Abraxas1:MERIT40 (panels 3, 4). Residues involved in cross-links and distances between cross-link sites are indicated as in A. (C) Left: Structural model of the human A-complex (AIR) is shown as in A. Dashed black rectangles highlight regions at the interfaces between Abraxas1:BRCC45 and Abraxas1:MERIT40 where cross-link sites were identified. Right: Close-up views and structural modelling of the cross-linked sites between Abraxas1:BRCC45 (panel 1) and Abraxas1:MERIT40 (panel 2). BRCC45 and MERIT40 residues involved in cross-links are shown as sticks while Abraxas1 residues are indicated with green circles; black dashed lines connect cross-linked residues, and dashed green lines represent the modelled Abraxas1 C-terminus not visible in the available structures. (D) Left: Structural model of human A-complex (AIR) depicted as in A. Dashed black rectangles highlight regions where cross-link sites between RAP80 and BRCC36, Abraxas1, BRCC45, or MERIT40 were identified. Right: Close-up views and structural modelling of the cross-linked sites between RAP80:BRCC36 (panel 1), RAP80:Abraxas1 (panels 2 and 3), RAP80:BRCC45 (panel 4), and RAP80:MERIT40 (panels 5 and 6). BRCC36, Abraxas1, BRCC45, and MERIT40 residues involved in cross-links are shown as sticks; residues located at the C-terminus of Abraxas1 and within RAP80 are depicted as green and red circles, respectively. Black dashed lines connect cross-linked residues; dashed green lines indicate the modelled Abraxas1 C-terminus not visible in the available structures, whereas dashed red lines refer to modelled regions in RAP80.
To confirm our cross-linking analyses and gain a better view of the human A-complex (also known as ARISC-FL RAP80; see Materials and methods section for details) structure, we performed single particle cryo-EM analyses of A-complex containing full-length RAP80 (here referred to as FL), in which the phosphorylated (Brown et al., 2015) and poly-ADP ribosylated region (Guettler et al., 2011) at the N-terminus of MERIT40 (residues 1–71) was deleted to enhance protein production yields (indicated as MERIT40ΔN; refer to Materials and methods section for details). We generated cryo-EM maps ranging from 4 to 6 Å resolution (Fig. 7, A–D and Fig. S6 and Table S3). Our ensemble of structures reveals multiple RAP80 conformations (Fig. 7, A and B; and Fig. S6) and two A-complex models containing the minimal RAP80-AIR interacting region (here referred to as AIR), analogous to the previously described mouse complex (Fig. 7, C and D; Rabl et al., 2019), which can be used as a reference to identify the RAP80 cryo-EM density. Interestingly, apart from the RAP80-AIR region, RAP80 consists mostly of dynamic and intrinsically disordered regions capable of attaining multiple conformations (Fig. 7, A and B; and Fig. S6). In addition, we identified multiple structures of the A-complex, with the arm regions closer together when more RAP80 density was present (Fig. 7, A and B; and Fig. S6) and further apart when only the minimal binding RAP80-AIR region was visible (Fig. 7, C and D; and Fig. S6). This conformational heterogeneity suggests a highly mobile RAP80 subunit that may require additional interactions to stabilize its position.
Cross-linking MS and structural analyses reveal mode of A-complex interaction with ubiquitin. (A–D) Cryo-EM densities at contour levels of 0.0047 (A; 6.2 Å), 0.0054 (B; 5.1 Å), 0.0058 (C; 4.4 Å), and 0.0133 (D; 4.0 Å) are shown in transparent surface, with the regions corresponding to BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR (Abraxas1 interacting region) colored light blue, green, pink, wheat, and red respectively. The structural model of human A-complex (AIR) is depicted as cartoon and colored gray; dashed black circles indicate RAP80 density outside the AIR domain and not present in the structural model. (E) Domain architecture of RAP80 and BRCC36, with the residues found to be ubiquitylated in the DUB-inactive complex indicated. SIM = SUMO (Small Ubiquitin-like Modifier)-interacting motif; ZnF = Zinc finger; MPN+ = Mpr1/Pad1 N-terminal+; CC = coiled coil. (F) A structural model of the human A-complex (AIR) is shown as surface representation, with the different subunits colored as described in panels A–D. The residues in BRCC36, Abraxas1, BRCC45, MERIT40 and RAP80 involved in chemical cross-linking are indicated with blue, green, purple, orange, or red circles. The UIM domains of RAP80 are shown as cylinders and colored red; black asterisks indicate the position of the BRCC36 active site in the two protomers. Dashed green and red lines represent the regions in either Abraxas1 or RAP80 that are currently missing from available structures.
Cross-linking MS and structural analyses reveal mode of A-complex interaction with ubiquitin. (A–D) Cryo-EM densities at contour levels of 0.0047 (A; 6.2 Å), 0.0054 (B; 5.1 Å), 0.0058 (C; 4.4 Å), and 0.0133 (D; 4.0 Å) are shown in transparent surface, with the regions corresponding to BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR (Abraxas1 interacting region) colored light blue, green, pink, wheat, and red respectively. The structural model of human A-complex (AIR) is depicted as cartoon and colored gray; dashed black circles indicate RAP80 density outside the AIR domain and not present in the structural model. (E) Domain architecture of RAP80 and BRCC36, with the residues found to be ubiquitylated in the DUB-inactive complex indicated. SIM = SUMO (Small Ubiquitin-like Modifier)-interacting motif; ZnF = Zinc finger; MPN+ = Mpr1/Pad1 N-terminal+; CC = coiled coil. (F) A structural model of the human A-complex (AIR) is shown as surface representation, with the different subunits colored as described in panels A–D. The residues in BRCC36, Abraxas1, BRCC45, MERIT40 and RAP80 involved in chemical cross-linking are indicated with blue, green, purple, orange, or red circles. The UIM domains of RAP80 are shown as cylinders and colored red; black asterisks indicate the position of the BRCC36 active site in the two protomers. Dashed green and red lines represent the regions in either Abraxas1 or RAP80 that are currently missing from available structures.
Related to Fig. 7 . Flow-chart of data processing. Red boxes (220 pixels) in the representative micrograph indicate picked particles. Final electron microscopy maps, colored according to local resolution, are provided; scale bars are in Ångstrom (Å).
Related to Fig. 7 . Flow-chart of data processing. Red boxes (220 pixels) in the representative micrograph indicate picked particles. Final electron microscopy maps, colored according to local resolution, are provided; scale bars are in Ångstrom (Å).
We next used the structure of the mouse A-complex (AIR; Rabl et al., 2019) as a guide and our cryo-EM maps to build a model for the human A-complex (FL) and interpret the cross-linked regions. Among the intrasubunit cross-links, most of the sites identified in the A-complex subunits connect proximal residues in our structural model and lie within a distance that agrees with the spacer arm length of the cross-linker used (Fig. S4 and Table S1). Similarly, most of the identified intersubunit cross-link sites reveal proximity of complex members in good agreement with our structural model (Fig. S5, A and B; and Table S2). For instance, we found K164 of BRCC36 to be within ∼11–12 Å from residues K155 and K158 of Abraxas1 (Fig. S5 A, panel 4), and K305 of BRCC36 was close to K242 and K245 of Abraxas1 (Fig. S5 A, panel 3). Likewise, we confirmed that K382 of BRCC45 lies within 5 Å from residue K126 in MERIT40 (Fig. S5 B, panel 2). The consistency of intersubunit cross-linking and structural data further validates our approach and provides clues for positioning more mobile elements that are not visible in our cryo-EM structure (Fig. S5 A, panels 1 and 2, and Fig. S5 B, panels 1 and 3). For example, residues K51 and K65 of BRCC36 are located on a flexible loop, and our cross-linking data indicate that these residues make contacts with K245 and K256 on Abraxas1 as well as with K41 in BRCC45 (Fig. S5 A, panels 1 and 2, and Fig. S5 B, panel 1). The ability to confidently locate flexible regions in our structural model is particularly important for mapping the position of the Abraxas1 C-terminus and RAP80 relative to other members of the complex since the C-terminal region of Abraxas1 and the majority of RAP80 are not resolved on any available structures. Several identified cross-link sites suggest that the extreme C-terminus of Abraxas1 contacts residues in BRCC45 and MERIT40, while different RAP80 regions are in close proximity to the BRCC36 active site or are located near the “arm” region formed by the BRCC45-MERIT40 heterodimer (Table S2). This is supported by the extra cryo-EM density present in the A-complex (FL; Fig. 7, A and B) when compared with A-complex (AIR; Fig. 7, C and D). By combining our cross-linking data and structural modeling, we initially placed K347 of Abraxas1 in proximity to BRCC45 K270 (Fig. S5 C, panel 1). This interaction also agrees well with the position of Abraxas1 D331, the last residue visible in our structural model and located 16 amino acids before K347 (Fig. S5 C, panel 1). Taking into consideration these interactions, we subsequently positioned Abraxas1 K371 near K140 of MERIT40 (Fig. S5 C, panel 2). Overall, our data strongly support a model whereby the extreme C-terminus of Abraxas1 “wraps” around the distal end of the BRCC45-MERIT40 arm region, making contact with both subunits. This model is also consistent with the mouse A-complex (AIR) crystal structure (Rabl et al., 2019).
Having located the Abraxas1 C-terminus, we subsequently sought to determine the mode of RAP80 binding to ARISC. According to our cross-linking data, the N-terminal part of RAP80 is in proximity to the BRCC36 MPN+ domain (Fig. 7 F and Table S2). Based on the cross-linked distance restrains, we modeled K20 and K69 of RAP80 near BRCC36 K85 and K51 (Fig. 7 F and Fig. S5 D, panel 1). These interaction sites are of particular importance for the positioning of RAP80 UIM1-2 (residues 79–124; Fig. 7 E) and suggest the UIM domains of RAP80 are likely located near the active site of BRCC36 (Fig. 7 F). Importantly, BRCC36 K65 and RAP80 residues K20, K31, and K75 are ubiquitylated in the DUB-inactive complex (Fig. 5, A and B; and Fig. 7 E; and Tables S1 and S2). Together, these data suggest that positioning of the RAP80 UIMs near the active site of BRCC36 enables intramolecular interactions between the UIM domains and ubiquitin chains linked to BRCC36 and RAP80. Consistent with this hypothesis, cross-linking analyses of the E33A DUB-inactive complex define a specific interaction between K112 of RAP80 and K6 of ubiquitin (Fig. 6, D and E) that was not present in the WT BRCC36 complex. This cross-link demonstrates the direct engagement of RAP80 UIMs with ubiquitin in the DUB-inactive A-complex. Our data are consistent with a structural model (Fig. 7 F), whereby the RAP80 UIMs are engaged with K63-linked ubiquitin chains attached on RAP80 and/or BRCC36 (Fig. 8). This would prevent RAP80 UIMs from interacting with poly-Ub chains on chromatin, thus explaining the phenotypic observations with DUB-inactive alleles. Together, these data support a poly-ubiquitin dependent autoinhibition model which requires BRCC36 DUB activity as a licensing step to allow RAP80 recognition of DNA damage signals (Fig. 8).
Model for A-complex auto-inhibition mechanism and its implication in substrate recruitment. In the hyper-ubiquitylated DUB-inactive complex, the UIM domains of RAP80 are “locked” within the complex via intramolecular ubiquitin binding. This conformation inhibits A-complex interaction with ubiquitylated chromatin and recruitment of the complex to DNA damage sites. Subsequent cleavage of these ubiquitin chains by WT BRCC36 releases auto-inhibition and promotes substrate engagement by the RAP80 UIMs, allowing recruitment of A-complex to poly-ubiquitylated chromatin.
Model for A-complex auto-inhibition mechanism and its implication in substrate recruitment. In the hyper-ubiquitylated DUB-inactive complex, the UIM domains of RAP80 are “locked” within the complex via intramolecular ubiquitin binding. This conformation inhibits A-complex interaction with ubiquitylated chromatin and recruitment of the complex to DNA damage sites. Subsequent cleavage of these ubiquitin chains by WT BRCC36 releases auto-inhibition and promotes substrate engagement by the RAP80 UIMs, allowing recruitment of A-complex to poly-ubiquitylated chromatin.
Discussion
Ubiquitylation on nucleosomes flanking DNA damage sites regulates numerous downstream repair pathways (Baranes-Bachar et al., 2018; Elia et al., 2015; Mailand et al., 2007; Messick and Greenberg, 2009; Schwertman and Bekker-Jensen, 2016; Sobhian et al., 2007; Stewart et al., 2009). RNF8 and RNF168 E3 ubiquitin ligases are responsible for DNA damage response (DDR)-mediated ubiquitin chain synthesis on histones H1 and H2A (Mattiroli et al., 2012; Thorslund et al., 2015). Moreover, TRIP12 and UBR5 E3 ligases were found to restrict the spreading of polyubiquitin chains on chromatin by regulating RNF168, thus fine-tuning histone ubiquitylation near DSBs while counteracting its spread to undamaged chromatin (Gudjonsson et al., 2012). However, less is known about how DUBs mediate the DDR. The BRCA1-A complex provides an interesting example of stoichiometric pairing of K63-Ub specific binding and hydrolysis activities within a single macromolecular assembly. To examine the physiological importance of the DUB activity of BRCC36, we knocked-in several Brcc36 active site point mutations at the endogenous locus to abolish its DUB activity without disrupting the stability of the A-complex. Mutation at the E33 site generates a mutant form of BRCC36 that more closely mimics the active configuration site, as the structure is less distorted and local sites are more preserved in comparison to QSQ (Zeqiraj et al., 2015). Nonetheless, both mutants showed similar functional reductions of A-complex in response to DNA damage, thus approximating loss of function mutations in other members.
Ubiquitin binding domain containing proteins are often themselves ubiquitylated, hinting at additional layers of regulation that depend on competent ubiquitin binding. In seminal work, this was proposed to impart an intramolecular association that outcompetes interaction with ubiquitylated proteins in trans (Hoeller et al., 2006). The generality of this model and its regulation by associated DUBs, however, has remained elusive. We demonstrate that RAP80 is highly ubiquitylated and its UIMs are occupied by ubiquitin in DUB-inactive cells coinciding with diminished damage response function. This implies a need for BRCC36 to recycle the complex to an active form lacking K63-Ub. We employed an integrated structural biology approach using cross-linking of native complexes and single particle analyses by cryo-EM, which suggests a model whereby K63-ubiquitylation of multiple lysine residues in BRCC36 and RAP80 within the DUB-inactive A-complex “locks” the RAP80 UIM domains into a non-productive conformation incapable of recruiting the BRCA1-A complex to DNA damage sites (Fig. 8). This mechanism of auto-inhibition ensures that the A-complex is kept in an inactive or poised state in the absence of DNA damage. BRCC36 acts to cleave these internal ubiquitin chains from itself and RAP80, promoting the release of the RAP80 UIMs and subsequent recruitment of the complex to ubiquitylated chromatin. As BRCC36 activity is specific for K63-Ub, the resultant monoubiquitinated lysine residues appear to be insufficient to engage the UIMs and support auto-inhibition. Collectively, these data provide a rationale for how specific ubiquitin binding and hydrolysis modules function within the BRCA1-A complex. This may serve as a prototype for similar auto-inhibitory mechanisms in other complexes where ubiquitin-binding domains and DUBs co-exist (Denuc et al., 2009; Flick et al., 2006; Tyrrell et al., 2010).
Our data suggest a “tug of war” between ubiquitin conjugation by ubiquitin ligases and A-complex deubiquitylating activities that are carefully balanced during DNA damage repair. Notably, we were unsuccessful in determining the identity of an E3 ligase responsible for BRCC36 and RAP80 ubiquitylation, which could reflect the known property of UIM-containing proteins to direct their own ubiquitylation (Hoeller et al., 2006). For example, UBC13 is an E2 ubiquitin-conjugating enzyme that, in complex with UEV1A, mediates the assembly of K63-linked ubiquitin chains without the need for an E3 ligase (Deng et al., 2000; Stewart et al., 2009). We also identified lysine residues in RAP80 and BRCC36 that are selectively ubiquitylated in the DUB-inactive complex. The relatively high sequence conservation of the ubiquitylated residues in RAP80 suggests a central role in regulating A-complex activity, perhaps acting as “acceptor sites” for ubiquitin conjugation to directly modulate BRCA1-A complex recruitment to chromatin. By contrast, the ubiquitylated sites in BRCC36 appear less conserved across species, suggesting that redundant or compensatory ubiquitylation events may have evolved to regulate the abundance and activity of this subunit. We propose that the removal of these ubiquitin moieties from both RAP80 and BRCC36 is required for the correct localization and function of the complex at DNA-damaged sites. The presence of many surface lysine residues on RAP80 and BRCC36, coupled with redundancy in mediating K63-Ub engagement with RAP80 UIMs, may make it difficult to discern critical residues in this response.
In summary, we propose a mechanism whereby BRCC36 DUB activity primarily acts as a licensing step to relieve A-complex auto-inhibition and allow subsequent recognition of DNA damage signals (Fig. 8). This mechanism modulates the localization and function of the BRCA1-A complex by shielding the RAP80 UIMs with inhibitory auto-ubiquitin chains until complex accumulation in the presence of DNA damage is required. These findings reveal an additional layer of complexity in processes that govern DNA damage recognition and provide a basis to understand the importance of balanced ubiquitin binding and hydrolytic moieties within a macromolecular entity.
Materials and methods
Mouse generation
Brcc36E33A/E33A and Brcc36QSQ/QSQ knockin mice were generated via CRISPR/Cas9 technology. HR template oligos were listed as follows: Brcc36E33A/E33A: 5′-GAGTCTGACGCTTTCCTAGTTTGTCTCAACCATGCTCTGAGCACAGAAAAGGAGGCAGTGATGGGTCTGTGTATAGGCGAGGTAAGTTGGCCACCTTGGCTGAAATCTTGCTGATCAATACCCTGTGTTCCTGGAGGCGGCTGGG-3′. Brcc36QSQ/QSQ: 5′-CTTGACAAACATACAGGTTGGCTGAACTAACAGGTCGTCCCATGAGAGTTGTTGGCTGGTATCAGTCACAACCTCATATAACTGTTTGGCCTTCACATGTTGGTAAGTTTTTATATCATTTCTCCTCCATTACATATCACAGTC-3′. The guide RNA sequences for these two templates were 5′-AGTGATGGGTCTGTGTATAG-3′ and 5′-CCAAACAGTTATATGAGGGT-3′, respectively. Purified Cas9 mRNAs (100 ng/μl), gRNAs (100 ng/μl), and oligo DNAs (100 ng/μl) were injected into fertilized eggs from superovulated B6SJLF1/J females mated with B6SJLF1/J males (from JAX). The detailed procedures were done according to the standard protocol from Penn Transgenic & Chimeric Mouse Facility. For genotyping, genomic DNA was isolated by first incubating the mouse tail with alkaline lysis reagent (25 mM NaOH, 0.2 mM EDTA, pH = 12) at 95–100°C for 1 h and then neutralizing by Tris-HCl (40 mM, pH = 5). Brcc36KO/KO mice were also generated when producing Brcc36E33A/E33A mice. Specific primers were designed to only amplify WT or mutant allele at a certain temperature by PCR reaction. Reverse primer in each group can be used to amplify both WT and mutant BRCC36. Brcc36E33A/E33A mice have been backed crossed to C57BL/6J at least six generations before MEF isolation.
Brcc36E33A/E33A (anneal at 57°C): Forward E33A: 5′-GCAGTGATGGGTCTGTGTATAG-3′; Forward WT: 5′-CTGAGCACAGAAAAGGAGGA-3′; Reverse: 3′-GTTTGAGTTCTTTGCTGGGC-5′.
Brcc36QSQ/QSQ (anneal at 57°C): Forward QSQ: 5′-GGTATCAGTCACAACCTC-3′; Forward WT: 5′-TGGCTGGTATCATTCCCACC-3′; Reverse: 3′-GAGGCAGAAGCAGGCAGATCTTTC-5′.
Brcc36KO/KO (anneal at 61°C): Forward KO: 5′-GAAGTGATGGGTCTGTGTAG-3′; Forward WT: 5′-GAAGTGATGGGTCTGTGTATAGGG-3′; Reverse: 3′-GTTTGAGTTCTTTGCTGGGC-5′.
The Brca2Δ27/Δ27 mice were generously provided by Dr. Wei Tong (Philadelphia Children’s Hospital, Philadelphia, PA). Geno-typing for Brca2Δ27/Δ27 mice was conducted on genomic DNA isolated from mouse tails as suggested on the website of NCI mouse repository (https://frederick.cancer.gov/resources/repo-sitories/nci-mouse-repository/mousemodels/StrainDetails.aspx?StrainNum=01XG9&g=Brca2).
Our animal protocol number is 803170 with the title: Role of BRCA1 Interaction Network in Suppressing Cancer. The protocol has been approved by the University of Pennsylvania Office of Animal Welfare and Institutional Animal Care and Use Committee.
Isolation of mouse embryonic fibroblasts (MEFs)
Pregnant mice were sacrificed on day 13 or 14 (day post-coitum) by cervical dislocation. Uterine horns were dissected out, rinsed briefly in 70% (v/v) ethanol, and then placed into a falcon tube containing PBS without Ca2+Mg2+ (Gibco, Invitrogen). Uterine horns were placed onto a Petri dish, and each embryo was separated from its placenta and embryonic sac. Head and red organs were dissected, and the rest were washed in PBS and placed in a clean Petri dish. Tissues were minced finely using a sterile razor blade, and the after mincing, the tissues were possible to pipette 1 ml of 0.05% trypsin/EDTA (Gibco, Invitrogen) + 100 Kunitz units of DNase I (USB) per embryo. The tissues are transferred into a 50-ml falcon tube and incubated for 15 min at 37°C. After each 5 min of incubation, cells were dissociated by pipetting up and down thoroughly. One volume of freshly prepared MEF media (450 ml DMEM, 50 ml FBS, 5 ml of 200 mM L-glutamine, 5 ml of P/S) was added to inactivate the trypsin, and the cells were spun down at 300 for 5 min. Cell pellets were resuspended in warm MEF media. The plates were coated with 0.2% gelatine (gelatine from bovine skin, Type B; Sigma-Aldrich) for 2 h. The cells from each embryo were plated into one gelatine-coated plate. The cells attached to the gelatine are MEFs. Primary MEFs were immortalized by overexpression of SV40 protein.
DUB activity assay
WT and E33A A-complexes bearing FL BRCC36, FL Abraxas1, FL BRCC45, MERIT40ΔN, and FL RAP80 (50 nM for WT complex; 50 and 100 nM for the E33A variant) were assayed at 30°C in DUB reaction buffer containing 50 mM HEPES-NaOH pH 7.0, 100 mM NaCl, 0.1 mg/ml BSA, 1 mM DTT, and 0.005% Tween-20. Internally quenched fluorescent (IQF) K63-linked di-Ubiquitin (di-Ub) substrate (DU6303; Lifesensors) at a concentration of 0.5 µM was used as a reporter for DUB activity (1 part IQF di-Ub to 9 parts unlabeled K63-linked di-Ub prepared, as described by Pickart and Raasi [2005]). A total of 20 μl enzyme reactions were carried out in 384-well black flat-bottom low flange plates (35373; Corning) and cleaved di-Ub was monitored by measuring fluorescent intensity (excitation: 544 nm, emission: 575 nm) every 30 s over 1 h in a Hidex Sense microplate reader. Following baseline correction, the change in fluorescence over time was plotted using Prism 7, Version 7.0c (GraphPad Software).
Cell culture
MEFs and HeLa S3 cells were maintained in DMEM (Invitrogen) supplemented with 10% calf serum and 1% penicillin/streptomycin. Splenic B cells were maintained in RPMI (Invitrogen) supplemented with 10% FBS, 1% penicillin/streptomycin, 1% nonessential amino acids (Life Technologies), 1% sodium pyruvate (Life Technologies), 50 μM β-mercaptoethanol, 5 ng/ml murine IL-4 (Life Technologies), 0.5 μg/ml RP105 (552128; BD Pharmingen), and 25 μg/ml LPS (Sigma-Aldrich).
Cell lysis
Whole cell lysates were obtained by lysing cell pellets in cell lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol). No reducing agents were added to the lysis buffer to minimize the activity of DUBs in the lysing process. About 20 nM PR619 (cat. no. SI9619; LifeSensors), 10 mM NEM, and 10 mM 1,10-o-phenanthroline were added to the cell lysis buffer to inhibit DUB activity.
Cell cycle analysis
Cells are fixed in 70% ethanol at −20°C overnight and spun down at 300 g for 5 min at 4°C and resuspended in 200 μl PBS containing RNAse A (0.5 mg/ml) and propidium iodide (50 μg/ml). Cells were stained at room temperature (RT) for 30 min. The samples were analyzed on a caliber flow cytometry machine.
Clonogenic assay
Brcc36 WT, E33A, or KO MEFs were plated (2,000 cells) on 6-cm tissue culture dishes. MMC was added to the medium 6 h after cell seeding, and the cells were allowed to grow and form colonies in presence of MMC for 12 d. Colonies were fixed with 100% methanol and stained with a staining solution (25% methanol + 0.5% crystal violet). Percent survival was determined by the total number of colonies normalized with plating efficiency for individual genotypes.
Antibodies
The following antibodies were used for western blotting for human or mouse cells: Rabbit anti-human/mouse RAP80 (NBP1-87156; Novus Biologicals) at 1:1,000 for human cells, 1:100 for mouse cells; Rabbit anti-human RAP80 (A300-763A; Bethyl Laboratories) at 1:1,000. Rabbit anti-human MERIT40 (A302-515A; Bethyl Laboratories) at 1:1,000. Rabbit anti-human/mouse MERIT40 (D7Y5K; 12711S; Cell Signaling) at 1:1,000. Rabbit anti-human/mouse Abraxas1 (NBP1-22977; Novus Biologicals) at 1:1,000. Rabbit anti-mouse BRCC45 (EPR11858; ab177960; Abcam) at 1:1,000. Rabbit anti-human/mouse BRCC36 (ab108411; Abcam) at 1:50,000. Mouse anti-human P4D1 (sc-8017; Santa Cruz) at 1:1,000. Mouse anti-human GFP (sc-9996; Santa Cruz) at 1:500. Mouse anti-human/mouse Tubulin (3873S; Cell Signaling) at 1:10,000; Mouse anti-human Tubulin (12G10; DSHB) at 1:1,000. Mouse anti-human HA.11 (901502; BioLegend) at 1:1,000. The following antibodies were used for immunofluorescence experiment: Rabbit anti-mouse BRCA1 (homemade rabbit polyclonal antibody raised against the exon 11 region of mouse BRCA1) at 1:100; mouse anti-human BRCA1 (sc-6954; Santa Cruz) at 1:500; rabbit anti-mouse MERIT40 (polyclonal antibody raised against GST-MERIT40 as previously described [Shao et al., 2009]) at 1:200; mouse anti-mouse γH2AX (JBW301; Millipore) at 1:2,000; rabbit anti-human γH2AX (39117; Active Motif) at 1:500.
CRISPR/Cas9 knockout
sgBRCC36-1 (5′-TCTAGTTGAACGATGATACA-3′) was used to generate HeLa S3 BRCC36−/− cell lines. Two sgRNAs (sgRAP80-1: 5′-ATTGTGATATCCGATAGTGA-3′; sgRAP80-2: 5′-GTCGAATAGAGCAAAGTGTT-3′) were used to generate HT29 RAP80−/− cells.
Metaphase spread
Splenocytes were freshly isolated and cultured for 48 h and then treated with 60 nM MMC or 1 μM PARPi for 24 h. In the last 3 h before harvesting, 0.5 μM nocodazole was added. Attached as well as suspended cells were collected and swollen in 75 mM KCl at 37°C for 20 min. Cells were fixed on ice with a 3:1 volume ratio of methanol to acetic acid solution. Glass slides were washed in methanol and pre-heated in a 42°C humidity chamber. Fixed cells were dropped from heights onto slides in the humidity chamber and blown to spread. Slides were air-dried and then stained with GIEMSA staining buffer for 8 min at RT and then mounted with Permout (Thermo Fisher Scientific). Images of >70 metaphase spread from more than three individual animals were captured using 100× objective and chromosomal breaks and radial chromosomes were quantified.
Fiber analysis
Single-molecule DNA fiber analysis was performed as described previously (Chaudhuri et al., 2016; Jiang et al., 2015). Briefly, for fork restart assay, MEFs were treated with 20 μM CldU for 15 min, followed by 2 mM HU for 1 h, and after extensive washes, cells were incubated with 200 μM ldU for an additional 20 min prior to harvesting. For fork protection assay, splenocytes were cultured for 3 d after isolation for the mice. DNA was labeled by a 30 min pulse with 50 μM IdU (I7125; Sigma-Aldrich). Cells were spun down and the cell pellet was resuspended in media containing 250 μM CldU (C6891; Sigma-Aldrich) and incubated for an additional 30 min. Cells were spun down and washed three times with PBS without disturbing the cell pellet and then the cell pellet was resuspended in media containing 4 mM HU (Sigma-Aldrich) and cultured for 3 h. Cells were lysed in spreading buffer (0.5% SDS in 200 mM Tris-HCl pH 7.4, 50 mM EDTA) directly on positively charged microscope slides, incubated for 8 min, and then tipped to 30° for DNA fibers to spread. Slides were fixed with a 3:1 volume ratio of methanol to acetic acid solution overnight. DNA was denatured by 2.5 N HCl at RT for 1 h, followed by blocking (blocking buffer: 2% BSA and 0.1% Tween20 in PBS) at 4°C overnight. Cells were stained with primary antibody (α-IdU: Becton Dickinson 347580; α-CldU: ab6326) followed by secondary antibodies. DNA fibers were captured using 60× objective on a Nikon.
Microscope image acquisition
Immunofluorescence images were captured by Nikon Eclipse 80i fluorescence microscope equipped with a CoolSnap MYO camera using Plan Apo VC 60× Oil DIC N2 objective lens. The images were acquired and processed using Nikon NIS-Elements software. Bright-field images were captured by an Olympus BX41 Phase Contrast & Darkfield Microscope using 100× oil objective lens. The metaphase images were acquired using DP2-BSW software. 5 μM scale bar was included on all the images.
Statistical analysis
For Figs. 1, F and I, 2 B, 3, B and E, S1 F, and S2, B and E, at least three independent experiments were performed. An unpaired, two-sided Student’s t test was conducted based on the average value of each independent experiment. For Figs. 1 G, 2, D and E, and S2 A, at least two mice were included for each genotype. An unpaired, two-sided Student’s ttest was conducted based on the average value of each mouse. For all the figures listed above, mean with SEM was used to plot the data. For Fig. S1 G, an unpaired two-sided Student’s t test was conducted based on the percentage of cell survival at three MMC doses (4, 10, and 25 ng/ml) from three independent experiments. Mean with SD was used to plot the clonogenic survival curve. P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****) were used in the plotting of the graphs. For Fig. 2 A, Mantel-Cox test was conducted for the mouse survival curve. For all the t tests conducted in this manuscript, data distribution was assumed to be normal, but this was not formally tested.
iPOND
The experiment was performed as previously reported (Sirbu et al., 2011). Briefly, HeLa S3 cells were labeled with EdU for 20 min, followed by either a chase with thymidine-containing medium for 4 h or treatment with HU for 4 h. Cells were crosslinked with 1% formaldehyde in PBS at RT for 20 min and quenched with 0.125 M glycine in PBS for 20 min. Pellets were collected by scraping, then resuspended in permeabilization buffer (20 mM HEPES pH 7.5, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 0.5% NP40), and incubated on ice for 15 min. The pellets were spun down at 2,500 g for 10 min and then resuspended in Click Reaction Buffer (4.35 ml PBS, 50 μl 1 mM biotin azide, 500 μl 100 mM ascorbate, 100 μl 100 mM CuSO4), incubated at RT for 1 h. Pellets were then resuspended in 500 μl nuclear lysis buffer (50 mM Tris-HCl pH 8.0, 25 mM NaCl, 2 mM EDTA, 1% NP40) and incubated on ice for 15 min. The lysates were sonicated and spun at maximum speed in a tabletop centrifuge for 10 min. Lysates were diluted 1:1 with wash buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.5% NP40) and incubated with 50 μl streptavidin beads overnight at 4°C. Beads were washed 4× with wash buffer and eluted with 2× Laemmli buffer.
SIRF
Cells were seeded at ∼50% confluency the day prior to the assay. Cells were incubated with no EdU (as negative controls) or 125 μM EdU (ongoing replication forks) for 8 min before fixation. For replication stalling, EdU was washed off and 4 mM HU was added to the media for an additional 4 h. Cells were pre-extracted by PIPES pre-extraction buffer for 30 s and fixed with 3% paraformaldehyde (PFA) at RT for 10 min. Cells were then permeabilized by PIPES permeabilization buffer at 4°C for 10 min. Cells were incubated with freshly prepared click reaction cocktail (10 μM biotin-azide, 10 mM sodium ascorbate, 2 mM copper sulfate in PBS) at RT for 1 h. Cells were blocked with blocking buffer (Duolink Proximity Ligation Assay Kit; Sigma-Aldrich) at RT for 1 h, then cells were incubated with primary antibodies (conjugating protein antibody of interest with either mouse anti-biotin or rabbit anti-biotin antibody) at RT for 1 h. Cells were washed with wash buffer A (0.01 M Tris, 0.15 M NaCl, 0.05% Tween 20, pH 7.4) twice, 5 min per wash. Duolink In Situ PLA probes anti-mouse plus and anti-rabbit minus were diluted 1:5 in blocking buffer and the cells were incubated with the probe buffer at 37°C for 1 h. The wash with Wash Buffer A was repeated, 5 min per wash. Ligation stock was diluted at 1:5 in high-purity water and then ligase was diluted at 1:40 in the ligation solution. Cells were incubated with this ligation buffer at 37°C for 30 min. The wash with Wash Buffer A was repeated, 2 min per wash. Amplification stock was diluted at 1:5 in H2O, and then polymerase was diluted at 1:80 in the amplification solution. Cells were incubated with this amplification buffer at 37°C for 100 min. Cells were washed with Wash Buffer B 2× at RT, 10 min per wash. Cells were then washed with diluted Wash Buffer B for 1 min. Slides were mounted with Vectashield mounting media with DAPI (H-1200; VectaLabs) and then visualized using a Nikon Eclipse 80i fluorescent microscope.
Tandem ubiquitin binding entity (TUBE) pull-down
All the experiments were carried out following the standard manual of Ubitest (UM411; Life Sensors). AMSH and USP2 were incubated with eluted samples at RT for 2 h. In TUBE experiments using OTUB1 or purified A-complex, TUBE-beads bound samples were incubated with the respective DUB enzymes at 37°C for 4 h followed by elution using the Western blot sample buffer.
In vitro ubiquitin binding assay
RAP80 complexes were immunoprecipitated from BRCC36−/− HeLa S3 cells (∼1 × 108) containing either FLAG-HA-WT or FLAG-HA-E33A by FLAG affinity pull down. The immunoprecipitated complexes were incubated either with 40 µg of 6xHis non-cleavable K63 poly-Ub chains (2–4; J2510; UBPBio) or buffer control with slow rotation at 4°C for 4 h. Next, 30 μl Ni beads were added to the mixture and incubated in presence of 15 mM imidazole with slow rotation at 4°C for 2 h. After incubation, Ni resin beads were pelleted by centrifugation at 750 g for 2 min and washed four times with 1 ml TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl) buffer containing 30 mM imidazole. Proteins were eluted with 50 μl TBS buffer containing 300 mM imidazole.
Complex purification and cross-linking
BRCC36−/− HeLa S3 cells (∼5 × 109) stably expressing FLAG-HA tagged BRCC36 WT and BRCC36 E33A mutant constructs were pelleted, then resuspended in hypotonic buffer (10 mM Tris-HCl pH 7.4, 7.5 mM KCl, 1.5 mM MgCl2 and 1 mM PMSF) on ice for 15 min, followed by douncing to lyse the swollen cells and obtain nuclei. Nuclei was then lysed in KETNG-400 buffer (20 mM Tris-HCl pH 7.4, 400 mM KCl, 0.5 mM EDTA, 0.5% NP-40, 1.5 mM MgCl2, 10% glycerol 1 mM PMSF and protease inhibitors [Roche]) for 45 min at 4°C. The lysate was spun down at 20,000 g, and supernatant dialyzed twice against KETNG-100 (20 mM Tris-HCl pH 7.4, 100 mM KCl, 0.5 mM EDTA, 0.5% NP40, 1.5 mM MgCl2 and 10% glycerol) in 1:100 volume ratio at 4°C for 2 h. Dialyzed lysate was spun down at 20,000 g, and the supernatant was incubated with FLAG-M2 beads at 15 μl beads slurry per 1 mg protein (Sigma-Aldrich) at 4°C for 4–6 h. The beads were washed, then eluted with 0.2 mg/ml FLAG peptide (Sigma-Aldrich) at 4°C for 1 h. The eluate was either loaded on a gel for LC-MS/MS analysis or used in cross-linking experiments. FLAG-eluted complex was dialyzed twice against NEHNG-150 (20 mM HEPES pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1.5 mM MgCl2 and 10% glycerol) for 2 h at 4°C, then spun down at 20,000 g. Cleared complex was cross-linked with 0.15 mM disuccinimidyl suberate (DSS) or 0.5 mM bis(sulfosuccinimidyl)suberate (BS-3) at RT for 1 h. The reaction was quenched by adding Tris pH 7.4 to 50 mM final concentration and then cross-linked complex was incubated with 0.2 ml of settled anti-HA beads at 4°C for 4 h. The beads were washed and then protein complex was eluted with 0.1 M glycine pH 2.0 and 150 mM NaCl.
Analysis of MS data
Gel slices containing A-complex (Fig. 5 A) was submitted to the Taplin MS Facility at Harvard University for MS analysis to identify ubiquitylation sites on RAP80 and BRCC36. Cross-linked samples (Fig. 6 B) were submitted to the Quantitative Proteomics Core at the University of Pennsylvania to run an LC-MS/MS analysis. Raw data were converted to Mascot generic format files and searched against protein databases by Protein Prospector 2.0 (Trnka et al., 2014), PLINK 2.0 (Yang et al., 2012), xQuest (Rinner et al., 2008), and xiSearch (Mendes et al., 2019) software. The results obtained with 1% FDR were subjected to manual inspection for validation and exclusion of all spectra that did not show confident sequence coverage of both cross-linked peptides. Representation of MS data using 2D networks (Fig. 6, C and D) was performed using the program xiNET (Combe et al., 2015).
Cloning, protein expression, and purification of WT and E33A A-complexes
Unless otherwise indicated, the expression “A-complex” used throughout this work refers to the ARISC subunits (FL BRCC36, FL Abraxas1, FL BRCC45, and MERIT40ΔN) in complex with FL RAP80. Genes for the four subunits human ARISCΔN WT and E33A variants were cloned in the MultiBac system used for co-expression of multiprotein complexes in insect cells (Fitzgerald et al., 2006). A single 6xHis purification tag followed by a cleavable Tobacco Etch Virus (TEV) protease site was engineered at the N-terminus of BRCC45. Full-length (FL) human RAP80 was cloned into pFastBac-HTB using standard molecular biology techniques. Bacmid DNA was generated in DH10-MultiBac-Turbo cells (ATG Biosynthetics) following the manufacturer’s protocol, and virus amplification in Spodoptera frugiperda 9 (Sf9) cells (Thermo Fisher Scientific) was performed using standard procedures and as described in Zeqiraj et al. (2015). For recombinant protein expression of WT and E33A A-complexes, Trichoplusia ni (Tni) cells (Oxford Expression Technologies) were co-infected with baculoviruses encoding ARISCΔN WT or E33A and RAP80(FL) in a 1:1 ratio. Following 48 h after infection, cells were harvested by centrifugation at 500 g for 15 min and purification of WT and E33A A-complexes was carried out as previously described (Zeqiraj et al., 2015). Briefly, harvested cells were resuspended in 100 ml ice cold lysis buffer (50 mM Tris-HCl pH 7.6, 300 mM NaCl, 20 mM Imidazole, 5% glycerol, 0.075% β-mercaptoethanol, 1 mM benzamidine, and 0.8 mM phenylmethylsulfonyl fluoride) and lysed by sonication using a Sonics Vibracell instrument (1 s ON/3 s OFF, at 40% amplitude for 4 min). The cell lysates were cleared by centrifugation at 30,000 g for 30 min, and the soluble fractions were sonicated (1 s ON/3 s OFF, at 40% amplitude for 2 min) and subsequently passed through a 0.45 μm filter (Thermo Fisher Scientific). The samples were then incubated with 3 ml HisPur Ni-NTA resin (Thermo Fisher Scientific), pre-equilibrated in low salt buffer (50 mM Tris-HCl pH 7.6, 300 mM NaCl, 20 mM Imidazole, 5% glycerol, 0.075% β-mercaptoethanol, and 1 mM Benzamidine), for 1 h at 4°C in rotation. After washing the resin with 20 ml low salt buffer, 20 ml high salt buffer (low salt buffer containing 500 mM NaCl), and 20 ml low salt buffer, the complexes were eluted by subsequent washes (2 ml each) with elution buffer (low salt buffer containing 120 mM Imidazole). Fractions containing WT or E33A A-complexes were pooled and 0.3 mg of His6-TEV protease was added prior to overnight dialysis against 4 l of dialysis buffer (low salt buffer containing 150 mM NaCl). Following removal of the TEV protease and uncleaved His6-FL BRCC45 by subtraction using 1 ml HisPur Ni-NTA resin (Thermo Fisher Scientific), the cleaved complexes were concentrated and resolved on a Superose 6 10/300 Increase column (GE Healthcare) pre-equilibrated in 25 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. Eluted peaks were analyzed by SDS-PAGE, and fractions containing >95% pure WT or E33A A-complexes were combined, concentrated to 1–2 mg/ml, snap-frozen in liquid nitrogen, and stored at −80°C.
Cryo-EM grids preparation and data collection
Unless otherwise indicated, all structural biology work described in this manuscript was carried out using ARISCΔN-RAP80(FL). For consistency throughout the text, we refer to this complex as the “A-complex”. UltrAuFoil R1.2/1.3 300-mesh gold grids (Quantifoil Micro Tools GmbH) were glow-discharged using a GloQube (Quorum) glow discharge unit at 40 mA for 30 s. Purified A-complex was then diluted to 0.16 mg/ml in gel filtration buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP) supplemented with 2 mM EDTA/EGTA mixture immediately before grids preparation. Cryo-EM grids were prepared by applying 3 μl of this complex onto the glow-discharged gold grids, followed by immediate blotting and plunge-freezing in liquid ethane cooled by liquid nitrogen, using a FEI Vitrobot IV at 100% relative humidity, and with a chamber temperature set at 4°C. A dataset was collected on a FEI Titan Krios transmission electron microscope operating in counting mode at 300 keV, using a total electron dose of 43.8 e− per Å2, a magnification of 75,000×, and a final calibrated object sampling of 1.065 Å per pixel. A total of 4,392 movies were recorded using the EPU automated acquisition software on a FEI Falcon III direct electron detector (Thompson et al., 2019). Each exposure movie (one per hole) had a total exposure time of 70 s collected over 30 frames, with an electron dose of 1.46 e− per Å2 per frame and defocus values ranging from −1.7 to −3.1 μm. Detailed information on data collection is shown in Table S3.
Image processing
A schematic of the data processing pipeline is shown in Fig. S6. Image processing was carried out using a combination of cryoSPARC v3.2.0 (Punjani et al., 2017) and RELION 3.0.7 (Zivanov et al., 2018). Drift-corrected averages of each movie were created using RELION’s implementation of MotionCor2 (Zheng et al., 2017), and the contrast transfer function (CTF) of each was determined using gCTF (Zhang, 2016). Initially, ∼2,000 particles were manually picked and used to train crYOLO 1.6.1 (Zivanov et al., 2018). This trained model was used for picking on all 4,392 movies, resulting in a total number of 904,273 picked particles. Particles were extracted in RELION using a box size of 320 × 320 pixels and subsequently subjected to iterative rounds of reference-free 2D classification in cryoSPARC. Classes that contained only the intact A-complex (ARISCΔN-RAP80 FL; 386,554 particles) or a combination of the full complex and subcomplexes (638,150 particles) were used to independently generate initial 3D volumes with the ab initio reconstruction function in cryoSPARC. Based on visual inspection, quality initial models were imported into RELION for 3D classification. These comprised three models obtained in C1 symmetry—two using 2D classes that contained the intact A-complex and subcomplexes (185,287 and 118,374 particles respectively), and one using 2D classes that contained only the full complex (118,887 particles). Following 3D classification, we obtained two models that contained ARISCΔN together with RAP80(AIR) on one arm (88,975 particles) or on both (70,006 particles) while four models represented ARISCΔN with RAP80 density outside of the AIR region (92,027, 92,035, 62,516, and 56,546 particles, respectively). A-complex models constituting 62,516 and 56,546 particles were combined and subjected to a second round of 3D classification, resulting in a final model with 95,398 particles. These models were subsequently subjected to 3D refinement, per-particle CTF correction, and particle polishing in RELION (Zivanov et al., 2018). Post-processing was used to appropriately mask the models, estimate and correct for the B-factor of the maps. Final resolutions were determined using the gold-standard Fourier shell correlation criterion (FSC = 0.143). Local resolution was estimated using the local resolution feature in RELION.
Structural visualization and sequence analysis
Structural models for human BRCC36, Abraxas1, BRCC45, MERIT40, and RAP80-AIR were generated using SWISS-MODEL (Bertoni et al., 2018). All structural representations shown in Figs. 1 A, 5 D, 6, A and F, S4, and S5 were created in PyMOL (The PyMOL Molecular Graphic System, Version 1.7.2.3 Schrödinger, LLC). Structural models shown in Fig. 7, A–D and Fig. S6 were generated using UCSF Chimera version 1.12 (Pettersen et al., 2004; Pintilie et al., 2010), whereas the surface representation of the A-complex (AIR) depicted in Fig. 7 F and Fig. 8 were obtained using UCSF ChimeraX version 0.9 (Goddard et al., 2018; Pettersen et al., 2021). Superimpositions of the ant, mouse, and human BRCC36 structures shown in Fig. 1 A were performed using the program Superpose (Krissinel and Henrick, 2004), available within the CCP4 suite (Collaborative Computational Project, Number 4, 1994). Sequence alignments shown in Fig. 5, B and C were created using Aline, Version 1.0.025 (Bond and Schüttelkopf, 2009).
Online supplemental material
Fig. S1 contains characterization of Brcc36 knock-in mutant mice and cells complementary to Fig. 1. Fig. S2 contains a characterization of the Rap80 complex at stalled replication forks in DUB inactive cells complementary to Figs. 2 and 3. Fig. S3 shows how deubiquitylation of RAP80 by BRCC36 is required for BRCA1-A recognition of Lys63-linked ubiquitin chains complementary to Fig. 4. Figs. S4 and S5 are related to the details of the cross-link sites identified in this study. Fig. S6 shows the workflow employed during the processing of the cryo-EM dataset. Tables S1 and S2 are related to the intra- (Table S1) and intermolecular (Table S2) cross-linking analyses performed on WT and E33A BRCA1-A complexes, while Table S3 provides a summary of the cryo-EM data collection and image processing.
Data availability
MS data can be provided upon request. The cryo-EM maps can be accessed freely from the Electron Microscopy Data Bank under the accession codes EMD-14999 (Map 1), EMD-15000 (Map 2), EMD-15001 (Map 3), EMD-15002 (Map 4), and EMD-15003 (Map 5). All the mice strains, cell lines, and reagents generated in this study will be made available to the scientific community upon request.
Acknowledgments
This work was supported by National Institutes of Health grant no. R01 138835 and funds from the Basser Center for BRCA to R.A. Greenberg, a UKRI-MRC grant (MR/T029471/1), a Basser External Research Grant to E. Zeqiraj and M. Foglizzo, and Wellcome Trust fellowships (200523/Z/16/Z and 222531/Z/21/Z) to E. Zeqiraj. The Astbury Cryo-EM Facility is funded by a University of Leeds ABSL award and a Wellcome Trust (108466/Z/15/Z) grant.
E. Zeqiraj and R.A. Greenberg are co-founders of JAMM Therapeutics. R.A. Greenberg is also a co-founder of RADD Pharmaceuticals. No other disclosures were reported.
Author contributions: Q. Jiang: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing (original draft), writing (review and editing). M. Foglizzo: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, visualization, writing (original draft), writing (review and editing). Y.I. Morozov: data curation, formal analysis, investigation, methodology, validation, writing (review and editing). X. Yang: data curation, validation, writing (review and editing). A. Datta: data curation, formal analysis, investigation, methodology, validation, writing (review and editing). L. Tian: methodology, resources. V. Thada: methodology. W. Li: data curation. E. Zeqiraj: conceptualization, data curation, funding acquisition, investigation, supervision, writing (review and editing). R.A. Greenberg: conceptualization, data curation, funding acquisition, investigation, supervision, writing (review and editing).
References
Author notes
Q. Jiang, M. Foglizzo, Y.I. Morozov, and X. Yang contributed equally to this paper.
Q. Jiang’s present address is Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA.
Y.I. Morozov’s present address is Sanofi Pharmaceuticals, Cambridge, MA.
L. Tian’s present address is Epochal Precision Anti-Cancer Therapeutics (EPAT), Eisai Inc., Exton, PA.




![Cooperation between BRCC36 and BRCA2 in response to ICLs depends on BRCC36 DUB activity. (A) Mouse survival curve of Brca2Δ27/Δ27 mice and double mutant mice (the 21 Brca2Δ27/Δ27 Brcc36 mutant mice consist of Brca2Δ27/Δ27Brcc36KO/KO [five mice], Brca2Δ27/Δ27Brcc36QSQ/QSQ [eight mice], Brca2Δ27/Δ27Brcc36E33A/E33A [eight mice]). Mantel-Cox test was conducted to test the survival curve from Brca2Δ27/Δ27 mice and double mutant mice. (B) Plating efficiency of two Brca2Δ27/Δ27 MEF clones and three isogenic Brca2Δ27/Δ27Brcc36E33A/E33A MEF clones. Quantification of plating efficiency was conducted based on three independent experiments (duplicates were performed for each experiment). Unpaired two-sided Student’s t test was conducted based on the average plating efficiency from each experiment. **P < 0.01, ***P < 0.001. Mean with SEM was used to plot the data. (C) Representative images of metaphase spread from Brca2Δ27/Δ27 and Brcc36QSQ/QSQBrca2Δ27/Δ27 splenocytes post exposure to 60 nM MMC. Scale bar: 5 μM. (D and E) Splenocytes were treated with either 60 nM MMC (D) or 1 μM PARPi (E) for 24 h prior to metaphase harvest, n > 70 metaphases in total from three to six mice per genotype were analyzed. Unpaired two-sided Student’s t test was conducted based on the average chromosomal abnormalities per 1,000 chromosomes of each mouse. *P < 0.01, **P < 0.01, ***P < 0.001, ****P < 0.0001. Mean with SEM was used to plot the data.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/9/10.1083_jcb.202111050/2/m_jcb_202111050_fig2.png?Expires=1773815512&Signature=HXYhCbBsMI78c3ZbxsIVrlCwAFOsBdJbBbJ6IJQWoLiO2sBIM-c1uLYEos6DHoEVVSTsqhQ3ml4Q68ClCL2WgL8QJ~fcieNCqe8uK0Vxw83XMp5uJVnkSbkRrRfAzqtbTI4yarY5FoTMhSLFaCFwVCuDgPARt4G-5Vtthk8Aw1~299CPiV3zI-1MglEZxhMRWVLTrjdhoHGbbZ9K~~oimH6RYpQTBivc4~EeXG0r3AepZ6u6Kl3-~9ccHnRNS4dC5-fvdvXA0KGfiSDgyPVwUT7mNlw6EqNl087j4bFJpmHkY3-bWbqeknKDO667FeihbP2M1rUbbNj318oW2jwa7Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)