Kinetochore protein phosphorylation promotes the correction of erroneous microtubule attachments to ensure faithful chromosome segregation during cell division. Determining how phosphorylation executes error correction requires an understanding of whether kinetochore substrates are completely (i.e., all-or-none) or only fractionally phosphorylated. Using quantitative mass spectrometry (MS), we measured phospho-occupancy on the conserved kinetochore protein Hec1 (NDC80) that directly binds microtubules. None of the positions measured exceeded ∼50% phospho-occupancy, and the cumulative phospho-occupancy changed by only ∼20% in response to changes in microtubule attachment status. The narrow dynamic range of phospho-occupancy is maintained, in part, by the ongoing phosphatase activity. Further, both Cdk1–Cyclin B1 and Aurora kinases phosphorylate Hec1 to enhance error correction in response to different types of microtubule attachment errors. The low inherent phospho-occupancy promotes microtubule attachment to kinetochores while the high sensitivity of kinetochore–microtubule attachments to small changes in phospho-occupancy drives error correction and ensures high mitotic fidelity.
Introduction
The kinetochore is a complex protein structure that provides the link between chromosomes and microtubules during mitosis (Cheeseman et al., 2006). Attachments between kinetochores and microtubules are highly regulated to correct attachment errors that frequently occur in early mitosis to prevent aneuploidy (Cimini 2003; Godek et al., 2015; Funabiki and Wynne, 2013; DeLuca et al., 2011; Zaytsev et al., 2015). The Knl1–Mis12–NDC80 (KMN) complex has emerged as a critical target for regulation at the kinetochore as this complex plays an essential role in microtubule attachment (Welburn et al., 2010). The Aurora A and B kinases have been shown to promote error correction through phosphorylation of these proteins, while the PP1 and PP2A-B56 phosphatases have been shown to oppose these changes. Phosphorylation of the KMN network by these kinases reduces their affinity for microtubules, enabling kinetochore–microtubule (k-MT) detachment to promote error correction (DeLuca et al., 2011; Welburn et al., 2010; DeLuca et al., 2018; Cheeseman et al., 2002; Foley et al., 2011; Suijkerbuijk et al., 2012). Therefore, it has been suggested that this network “tunes” the phosphorylation state on kinetochores to precisely regulate the stability of k-MT attachments (Godek et al., 2015; Bakhoum and Compton, 2012). However, there is currently no systems-level understanding of how these kinases and phosphatases coordinate to phosphorylate kinetochore substrates to provide this tuning of k-MT stability. Current models suggest that phosphorylation acts as an all-or-none mechanism and it remains unknown how the net (sum of kinase and phosphatase activities) phosphorylation changes in specific sites in response to k-MT attachment errors (Funabiki and Wynne, 2013; Lampson and Cheeseman, 2011). Although plausible, it seems improbable that kinetochore phosphorylation operates through an all-or-none mechanism because saturation of the phosphorylation sites on kinetochore substrates would be anticipated to severely impair k-MT attachment, and conversely, a complete lack of phosphate would be anticipated to create hyper-stable k-MT attachments that impede error correction.
Hec1 has an N-terminal tail comprising the first 70 amino acids that provides a primary microtubule attachment site in kinetochores. Up to nine sites among this short sequence are phosphorylated in human cells, yet mutational analysis has revealed that wild-type function can be provided by Hec1 possessing only one to two phosphorylation mimicking sites (DeLuca et al., 2011; Zaytsev et al., 2015; Welburn et al., 2010; DeLuca et al., 2006; Nousiainen et al., 2006; Ciferri et al., 2008). Therefore, it is unclear how to reconcile the functional sufficiency provided by only two phosphorylation sites with the evidence that nine sites serve as phosphoacceptors, underscoring our lack of understanding of the actual phosphorylation occupancy of these sites. Conceptually, the determination of post-translational modification site occupancy is necessary to explain the mechanisms through which these modifications regulate biological processes and determine how kinase and phosphatase networks create the net phospho-occupancy on specific sites to “tune” the stability of k-MT attachments. Despite the potential for mechanistic insight, occupancy analysis is not yet routine and instead, differences in phosphorylation are routinely reported as a relative fold-change (Prus et al., 2019). However, a twofold change could arise from a change in absolute phospho-occupancy of either 1–2% or 50–100%, and these two scenarios have different implications for how sensitive the system is in regulating k-MT attachment stability. This issue cannot be resolved using amino acid substitution because mutant proteins exist in one extreme (alanine mimics 0% phosphorylation occupancy) or the other (aspartic acid or glutamic acid mimic 100% phosphorylation occupancy) relative to mimicking phosphorylation status.
To investigate the phosphorylation-dependent mechanisms that govern mitotic fidelity, we selected the protein Hec1 as a candidate to measure phospho-occupancy because it is crucial for microtubule binding at the k-MT interface and because previous models posit that phosphorylation at multiple sites facilitates k-MT detachment to correct attachment errors (DeLuca et al., 2011; Zaytsev et al., 2015; DeLuca et al., 2006; Ciferri et al., 2008). We determined that Hec1 is only partially phosphorylated, due in part to constant phosphatase activity, at all sites queried under a variety of microtubule attachment conditions, and that only small changes in phospho-occupancy are associated with changes in k-MT attachment status including erroneous attachments. We also discovered that Cdk1–Cyclin B1 plays a direct role in k-MT error correction through the phosphorylation of the Hec1 tail, which is critical for accurate chromosome segregation. Thus, the system of kinases and phosphatases operates to limit the extent of phosphorylation to both allow for initial k-MT attachments and ensure efficient error correction.
Results
Hec1 is phosphorylated at only partial occupancy during mitosis
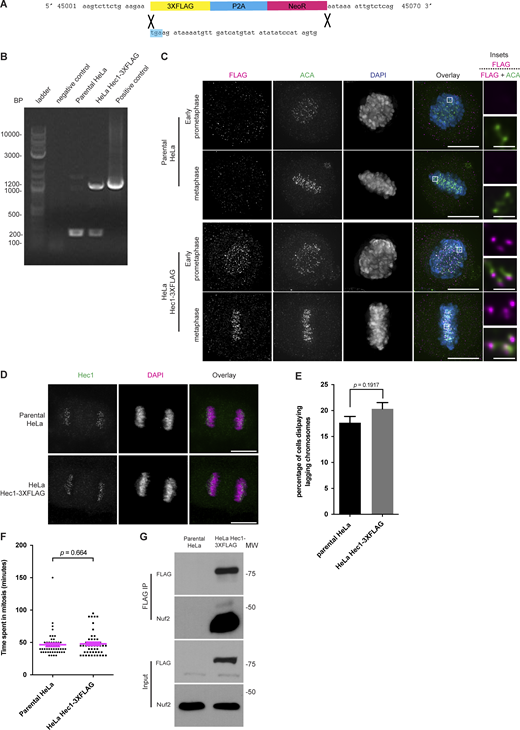
To purify Hec1, we created cells expressing Hec1 fused to a 3XFLAG tag using CRISPR/Cas9 to insert the tag into the endogenous locus at the 3′ end (Fig. S1 A). Genotyping analysis revealed heterozygous modification of Hec1 alleles (Fig. S1 B). The tag did not perturb Hec1 kinetochore localization, the lagging chromosome rate in anaphase, mitotic duration, or binding to the known Hec1 binding partner Nuf2 (Fig. S1, C–G).
Addition of a 3XFLAG tag to Hec1 does not perturb mitosis. (A) Schematic showing the addition of a 3XFLAG-P2A-Neo cassette to the 3′ of the endogenous Hec1 gene. The blue highlight indicates the original stop codon. (B) Genotyping of HeLa-3XFLAG cells. PCR was performed on genomic DNA from the indicated cell lines. The negative control is the water used for the reaction. The positive control is 10 ng of the Donor plasmid construct used to create the cell line. A 240 base-pair amplicon is expected for unmodified alleles, and 1,240 base-pair amplicon is expected for modified alleles. PCR reactions were run on 1% agarose gel stained with ethidium bromide and imaged. The genotyping was performed once. (C) Immunofluorescence images of HeLa Hec1-3XFLAG and parental HeLa cells. The cells were fixed and stained for FLAG, ACA, and DAPI. The FLAG and ACA channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images from two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. (D) Immunofluorescence images of HeLa Hec1-3XFLAG and parental HeLa cells. The cells were fixed and stained for Hec1 and DAPI. The Hec1 and DAPI channels were adjusted evenly for brightness and contrast for presentation. Representative images from three independent biological experiments are shown. The scale bars are 10 μm. (E) Quantification of the percentage of lagging chromosomes in cells undergoing anaphase from an asynchronous population of HeLa Hec1-3XFLAG and parental HeLa cells. 100 cells per cell line were scored for the presence of lagging chromosomes for each of three independent biological experiments. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (F) Quantification of the time spent in mitosis from the onset of cell rounding to anaphase from an asynchronous population of HeLa Hec1-3XFLAG and parental HeLa cells. 50 cells for each population were quantified from a single experiment. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG and parental HeLa cells. The cells were synchronized by thymidine-NOC and then lysed in NETN buffer. Hec1-3XFLAG was then immunoprecipitated, and the purified proteins then separated by SDS-PAGE, transferred to nitrocellulose membrane and blotted for Nuf2 and FLAG. A single experiment was performed. Source data are available for this figure: SourceData FS1.
Addition of a 3XFLAG tag to Hec1 does not perturb mitosis. (A) Schematic showing the addition of a 3XFLAG-P2A-Neo cassette to the 3′ of the endogenous Hec1 gene. The blue highlight indicates the original stop codon. (B) Genotyping of HeLa-3XFLAG cells. PCR was performed on genomic DNA from the indicated cell lines. The negative control is the water used for the reaction. The positive control is 10 ng of the Donor plasmid construct used to create the cell line. A 240 base-pair amplicon is expected for unmodified alleles, and 1,240 base-pair amplicon is expected for modified alleles. PCR reactions were run on 1% agarose gel stained with ethidium bromide and imaged. The genotyping was performed once. (C) Immunofluorescence images of HeLa Hec1-3XFLAG and parental HeLa cells. The cells were fixed and stained for FLAG, ACA, and DAPI. The FLAG and ACA channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images from two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. (D) Immunofluorescence images of HeLa Hec1-3XFLAG and parental HeLa cells. The cells were fixed and stained for Hec1 and DAPI. The Hec1 and DAPI channels were adjusted evenly for brightness and contrast for presentation. Representative images from three independent biological experiments are shown. The scale bars are 10 μm. (E) Quantification of the percentage of lagging chromosomes in cells undergoing anaphase from an asynchronous population of HeLa Hec1-3XFLAG and parental HeLa cells. 100 cells per cell line were scored for the presence of lagging chromosomes for each of three independent biological experiments. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (F) Quantification of the time spent in mitosis from the onset of cell rounding to anaphase from an asynchronous population of HeLa Hec1-3XFLAG and parental HeLa cells. 50 cells for each population were quantified from a single experiment. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG and parental HeLa cells. The cells were synchronized by thymidine-NOC and then lysed in NETN buffer. Hec1-3XFLAG was then immunoprecipitated, and the purified proteins then separated by SDS-PAGE, transferred to nitrocellulose membrane and blotted for Nuf2 and FLAG. A single experiment was performed. Source data are available for this figure: SourceData FS1.
We purified Hec1-3XFLAG via immunoprecipitation (Fig. S2 A) and analyzed it by mass spectrometry (MS) to determine phospho-occupancy. Since it cannot be assumed that a phospho-peptide and cognate non–phospho-peptide will be detected equally, we monitored the change of non–phospho-peptide cognates with and without calf intestinal phosphatase (CIP) treatment to dephosphorylate half of each sample by differential, isotopically encoded reductive dimethyl labeling. The two fractions are then remixed and analyzed by MS (Fig. 1 A and Fig. S2 B). MS analysis confirmed that CIP treatment completely dephosphorylates Hec1 peptides (Fig. S2 D). Notably, this approach cannot distinguish site-specific occupancies for peptides containing more than one phosphorylated residue, but instead integrates them to a single value. To explore if phosphorylation occupancy changes under different conditions of k-MT attachment, we purified total Hec1-3XFLAG from mitotic cells lacking microtubules (500 ng/ml nocodazole [NOC]), with k-MT attachment errors and no tension (100 ng/ml NOC or 25 µM S-Trityl-L-cysteine [STLC]; Skoufias et al., 2006), or with predominantly correct bi-oriented k-MT attachments under tension (25 µM proTAME; Zeng et al., 2010; Fig. S2, E and F).
Both soluble and chromatin localized Hec1 is only partially phosphorylated; Hec1 is purified from cells treated to generate different types of k-MT attachment. (A) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The Hela-3XFLAG cells were synchronized by thymidine-NOC and then prepared as a total cell lysate or fractionated into soluble and chromatin fractions. Control parental HeLa cells remained asynchronous and were prepared as a total cell lysate. Hec1-3XFLAG was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and finally blotted as indicated. The upper band of the Hec1 (endogenous epitope) input blot represents Hec1-3XFLAG, while the lower band represents unmodified Hec1. * indicates a non-specific band. The ponceau stained panel was adjusted for brightness and contrast. A single experiment was performed. (B) Cartoon illustration depicting the calculation of absolute phosphorylation occupancy. The ratio of the unphosphorylated peptide (colored magenta) between the heavy and light channels is used to calculate phosphorylation occupancy. (C) Graphs showing the absolute phosphorylation occupancy of various Hec1 peptides purified from cells fractionated into soluble and chromatin fractions as determined by MS. Individual and average values from three independent biological experiments are shown. The subcellular fractions are derived from the same cells as used for the total protein analysis presented in Fig. 1 B. Error bars indicate mean ± SEM. *, P < 0.05; **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) MS peaks for the Hec1 peptide LSINKPTSER purified from the chromatin fraction of cells synchronized by thymidine then 100 ng/ml NOC. A representative trace from one of the three replicate experiments presented in Fig. S2 C is shown. The magenta trace indicates the CIP treated, heavy labeled peptides and the blue trace indicates the sham treated light labeled peptides. The absence of peaks in the magenta trace indicates complete removal of phosphates. (E) Immunofluorescence images of HeLa Hec1-3XFLAG cells synchronized by thymidine and then NOC, STLC, or proTAME. The cells were fixed and stained for ACA, tubulin, and DAPI. The tubulin and ACA channels were adjusted evenly for brightness and contrast for presentation. DAPI channel was adjusted independently for presentation. The scale bars are 10 μm. Representative images from a single experiment are shown. (F) Quantification of inter-centromere distances of cells prepared as in E. 100 pairs of centromeres were measured across 10 cells from a single experiment. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS2.
Both soluble and chromatin localized Hec1 is only partially phosphorylated; Hec1 is purified from cells treated to generate different types of k-MT attachment. (A) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The Hela-3XFLAG cells were synchronized by thymidine-NOC and then prepared as a total cell lysate or fractionated into soluble and chromatin fractions. Control parental HeLa cells remained asynchronous and were prepared as a total cell lysate. Hec1-3XFLAG was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and finally blotted as indicated. The upper band of the Hec1 (endogenous epitope) input blot represents Hec1-3XFLAG, while the lower band represents unmodified Hec1. * indicates a non-specific band. The ponceau stained panel was adjusted for brightness and contrast. A single experiment was performed. (B) Cartoon illustration depicting the calculation of absolute phosphorylation occupancy. The ratio of the unphosphorylated peptide (colored magenta) between the heavy and light channels is used to calculate phosphorylation occupancy. (C) Graphs showing the absolute phosphorylation occupancy of various Hec1 peptides purified from cells fractionated into soluble and chromatin fractions as determined by MS. Individual and average values from three independent biological experiments are shown. The subcellular fractions are derived from the same cells as used for the total protein analysis presented in Fig. 1 B. Error bars indicate mean ± SEM. *, P < 0.05; **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) MS peaks for the Hec1 peptide LSINKPTSER purified from the chromatin fraction of cells synchronized by thymidine then 100 ng/ml NOC. A representative trace from one of the three replicate experiments presented in Fig. S2 C is shown. The magenta trace indicates the CIP treated, heavy labeled peptides and the blue trace indicates the sham treated light labeled peptides. The absence of peaks in the magenta trace indicates complete removal of phosphates. (E) Immunofluorescence images of HeLa Hec1-3XFLAG cells synchronized by thymidine and then NOC, STLC, or proTAME. The cells were fixed and stained for ACA, tubulin, and DAPI. The tubulin and ACA channels were adjusted evenly for brightness and contrast for presentation. DAPI channel was adjusted independently for presentation. The scale bars are 10 μm. Representative images from a single experiment are shown. (F) Quantification of inter-centromere distances of cells prepared as in E. 100 pairs of centromeres were measured across 10 cells from a single experiment. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS2.
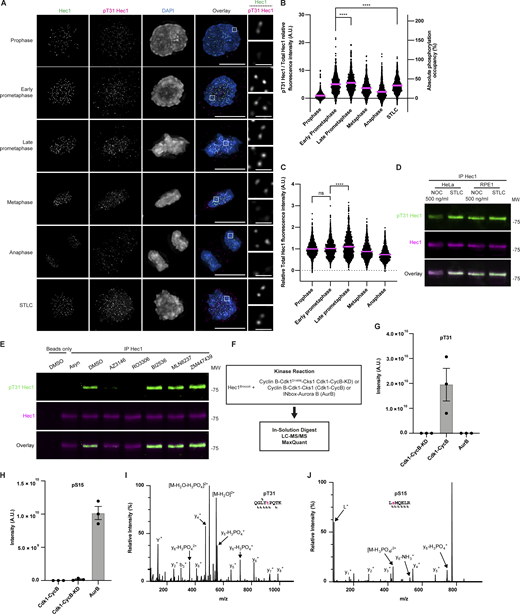
Hec1 is phosphorylated at only partial occupancy during mitosis. (A) Cartoon illustration depicting the experimental scheme used to determine protein phosphorylation occupancy. Mitotic cells are lysed. Hec1 is then immunoprecipitated from the lysate and purified by SDS-PAGE. The band of Hec1 is then excised, and the protein is subjected to tryptic digest. The sample is then divided in two, and half is treated with CIP and labeled with a heavy isotope. The sample is then recombined and analyzed by MS. (B) Histograms showing the absolute phosphorylation occupancy of various Hec1 peptides purified from total cell lysate as determined by MS. The individual and average values from three independent biological experiments are shown. Error bars indicate mean ± SEM. *, P < 0.05; **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Amino acid sequence alignment of Hec1 orthologs from residues 1–76 (relative to the human sequence). Residues highlighted in yellow indicate Aurora kinase consensus sites. Residues highlighted in blue indicate Cdk1 consensus sites.
Hec1 is phosphorylated at only partial occupancy during mitosis. (A) Cartoon illustration depicting the experimental scheme used to determine protein phosphorylation occupancy. Mitotic cells are lysed. Hec1 is then immunoprecipitated from the lysate and purified by SDS-PAGE. The band of Hec1 is then excised, and the protein is subjected to tryptic digest. The sample is then divided in two, and half is treated with CIP and labeled with a heavy isotope. The sample is then recombined and analyzed by MS. (B) Histograms showing the absolute phosphorylation occupancy of various Hec1 peptides purified from total cell lysate as determined by MS. The individual and average values from three independent biological experiments are shown. Error bars indicate mean ± SEM. *, P < 0.05; **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Amino acid sequence alignment of Hec1 orthologs from residues 1–76 (relative to the human sequence). Residues highlighted in yellow indicate Aurora kinase consensus sites. Residues highlighted in blue indicate Cdk1 consensus sites.
We obtained sequence coverage and determined the phospho-occupancy of five known sites within the N-terminal tail of Hec1: S15, S44, T49/S50, S55, and S69, as well as the previously identified, but uncharacterized sites, T31, S75, and S76 (Kettenbach et al., 2011; Fig. 1 B). None of these were detected in previous “omic-scale” analyses of phospho-occupancy in human cultured cells or fission yeast, except for a peptide containing pS69 and/or pS72, which was reported at 63% phospho-occupancy in NOC-treated human cells (Olsen et al., 2010; Rao and Møller, 2012; Carpy et al., 2012). We were unable to determine the occupancy of the sites S4, S5, S8, and S62 because their peptides are very hydrophilic and appeared only sporadically in the data. The Aurora B phosphorylation site S15 and the Aurora A/B phosphorylation site S55 were confidently localized and quantified. Multiple peptides spanning the Aurora B phosphoacceptors S44, T49, and S50 were identified as singly phosphorylated at S44 and separately at either T49 or S50, which could not be confidently localized. It is likely that either site could be phosphorylated, as determined previously (Kettenbach et al., 2011; Malik et al., 2009; Santamaria et al., 2011). We did not observe any peptides doubly phosphorylated on S44 and S49/T50. Singly phosphorylated peptides containing the Aurora A and/or B phosphoacceptors S69, S75, and S76 were observed, in which S69 phosphorylation was unambiguously assigned on some peptides, as well as on either S75 or S76, which has been previously observed (Kettenbach et al., 2011). S75/S76 is decreased under Aurora B inhibition (Kettenbach et al., 2011), although the adjacent amino acid sequence is not characteristic of mitotic kinases (Alexander et al., 2011), suggesting that it may be an atypical Aurora B consensus motif. Finally, we determined phospho-occupancy on a single peptide containing T31, a site that is conserved and conforms to a Cdk1 consensus site (Alexander et al., 2011; Fig. 1 C).
The experiment revealed three categories of response (Fig. 1 B). One category includes S44/T49/S50 and S69/S75/S76, which displayed maximum occupancy in cells without microtubule attachments (500 ng/ml NOC) and with approximately twofold decrease of phospho-occupancy in each of the other conditions. Another category is defined by the Aurora B site S15 and the Cdk1 site T31. These sites display maximum occupancy under conditions of erroneous k-MT attachments (STLC and 100 ng/ml NOC), and exceptionally low occupancy in cells with bioriented k-MT attachments (proTAME). A third category is defined by the peptide containing S55, which displayed switch-like behavior with maximum phosphorylation in the absence of microtubules (500 ng/ml NOC) and almost no phosphorylation in the other conditions. These findings are broadly consistent with previous experiments showing that Hec1 phosphorylation is lowest in cells with bioriented k-MT attachments (DeLuca et al., 2011), but demonstrate that these sites respond independently to different types of k-MT attachments. Furthermore, the phospho-occupancy of individual sites does not uniformly correlate with the inter-kinetochore distance that has previously been proposed to influence the extent of Hec1 phosphorylation (compare S15 and S44/T49/S50 at 500 versus 100 ng/ml NOC; Fig. 1 B and Fig. S2, C and F).
Mutational and domain swapping experiments suggest that the Hec1 N-terminal tail phosphorylation sites participate interchangeably in causing k-MT detachment (DeLuca et al., 2018). Therefore, the functional detachment of microtubules is likely sensitive to the cumulative phospho-occupancy of these sites, and our data can express the phospho-occupancies within this domain as a cumulative score. The theoretical maximum cumulative occupancy for the Hec1 peptides as measured by MS is 5.00 (five peptides, each with 100% occupancy). Surprisingly, the cumulative phosphorylation of all peptides did not approach this maximum, and none of the individual peptides that we sampled exceeded 50%. The condition displaying the highest total occupancy was cells treated with 500 ng/ml of NOC, which equaled 1.46, or ∼29.2% of theoretical maximum occupancy. Cells treated with STLC and 100 ng/ml NOC that have erroneous microtubule attachments, displayed a cumulative occupancy of 1.37 (27.4% of theoretical maximum) and 1.08 (21.6% of theoretical maximum), respectively. Cells under proTAME arrest displayed a cumulative occupancy of 0.46 (9.2% of theoretical maximum). Similar trends were observed on Hec1 purified from soluble or chromatin-associated cell fractions (S2C), indicating that phospho-occupancy is not dependent on specific protein subpopulations. Thus, the cumulative occupancy of these Hec1 peptides changes by only 20% (from ∼29 to 9%) between mitotic cells with no k-MT attachments and those with bioriented k-MT attachments.
Hec1 phosphorylation by Cdk1 on T31 during mitosis is temporally regulated
To investigate how Hec1 phospho-occupancy is set by the competing activities of protein kinases and phosphatases, we generated an antibody against phosphorylated T31 (pT31) because T31 is the only predicted Cdk1 site in the N-terminal tail of Hec1 and it is uncharacterized. We validated its specificity toward pT31 in mitotic cells (Fig. S3, A and B). We also used immunoblotting to show that the antibody binds only to phosphorylated Hec1 (Fig. S3 C), that both soluble and chromatin-bound Hec1 is phosphorylated equally (Fig. S3 D), and that 3XFLAG-tagged and unmodified Hec1 are equally recognized by the anti-pT31 antibody (Fig. S3 E).
The antibody raised against pT31 binds only to pT31; Hec1 is phosphorylated on T31 in prometaphase RPE1 cells; pT31 is maximally phosphorylated in cells arrested in STLC when analyzed by IF. (A) Cartoon illustration of the Hec1-GFP constructs used in Fig. S3 showing the mutations made and the remaining potential phosphorylation sites. (B) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells in early prometaphase transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained for pT31 Hec1 and DAPI. The GFP and pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of two independent biological experiments are shown. (C) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate. Hec1-3XFLAG was then immunoprecipitated. Following washes, the samples were mock treated or treated with Lambda phosphatase for 20 min at 30°C. The purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (D) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG and parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate or fractionated into soluble and chromatin fractions. Hec1-3XFLAG was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (E) Western blots showing either anti-FLAG or total Hec1 immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (F) Immunofluorescence images of RPE1 cells in various stages of mitosis taken from an asynchronous population. 48 h after plating the cells were fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of two independent biological experiments are shown. (G) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in F. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Quantification of kinetochore total Hec1 levels from the conditions from F and G. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC, STLC, or proTAME. The cells were then fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (J) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in I. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (K) Quantification of kinetochore total Hec1 levels from the conditions from I and J. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS3.
The antibody raised against pT31 binds only to pT31; Hec1 is phosphorylated on T31 in prometaphase RPE1 cells; pT31 is maximally phosphorylated in cells arrested in STLC when analyzed by IF. (A) Cartoon illustration of the Hec1-GFP constructs used in Fig. S3 showing the mutations made and the remaining potential phosphorylation sites. (B) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells in early prometaphase transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained for pT31 Hec1 and DAPI. The GFP and pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of two independent biological experiments are shown. (C) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate. Hec1-3XFLAG was then immunoprecipitated. Following washes, the samples were mock treated or treated with Lambda phosphatase for 20 min at 30°C. The purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (D) Western blots showing an anti-FLAG immunoprecipitation from HeLa Hec1-3XFLAG and parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate or fractionated into soluble and chromatin fractions. Hec1-3XFLAG was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (E) Western blots showing either anti-FLAG or total Hec1 immunoprecipitation from HeLa Hec1-3XFLAG or parental HeLa cells. The cells were synchronized by thymidine-NOC and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and finally blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (F) Immunofluorescence images of RPE1 cells in various stages of mitosis taken from an asynchronous population. 48 h after plating the cells were fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of two independent biological experiments are shown. (G) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in F. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Quantification of kinetochore total Hec1 levels from the conditions from F and G. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC, STLC, or proTAME. The cells were then fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (J) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in I. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (K) Quantification of kinetochore total Hec1 levels from the conditions from I and J. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS3.
To explore the functional role of T31 phosphorylation, we measured the pT31 intensity at kinetochores in human cells as they underwent unperturbed mitosis. We found that in both HeLa and non-transformed RPE1 cells, maximal phosphorylation occurs in prometaphase (Fig. 2, A and B; and Fig. S3, F and G). We also observed fluctuation in the quantity of Hec1 localized to kinetochores during different mitotic phases in HeLa (Fig. 2 C) and RPE1 (Fig. S3 H) cells as previously reported (Magidson et al., 2015), and therefore we normalized the phospho-signal intensity to total Hec1 in all experiments (Fig. 2 B; Fig. 5, B, G, and J; Fig. 8, B and D; Fig. 9, B and D; Fig. 10, C and D; Fig. S3, G and J; Fig. S4, F and G; and Fig. S5, B and D). To compare the extent of phosphorylation of Hec1 between HeLa and RPE1 cells, we immunoprecipitated equal quantities of Hec1 from each cell line treated with either 500 ng/ml NOC or STLC and blotted for pT31 Hec1 (Fig. 2 D). In HeLa cells, the signal intensity when blotted for pT31 was slightly higher in STLC compared to NOC-treated cells, which mirrors the phospho-occupancy of this site when measured by MS (Fig. 1 B). In RPE1 cells, the signal intensity when blotted for pT31 was equivalent to that of HeLa cells (Fig. 2 D). Furthermore, the intensity of pT31 relative to total Hec1 as measured by immunofluorescence staining in these two cell lines was comparable (compare Fig. 2, A and B to Fig. S3, F and G). Thus, the phospho-occupancy of this site on Hec1 is similar between HeLa (transformed and aneuploid) and RPE1 (immortalized, non-transformed, and near-diploid) cells. We also measured the pT31 levels by IF in cells arrested under the same conditions that we used for MS analysis. Importantly, the relative pT31 intensity at kinetochores in imaged cells broadly paralleled the occupancy values determined using MS in that T31 is robustly phosphorylated in cells arrested in 100 ng/ml NOC or STLC, but poorly phosphorylated in cells arrested in 500 ng/ml NOC or proTAME (Fig. 1 B; Fig. S2 C; and Fig. S3, I and J). That pT31 levels appear higher when measured in individual mitotic cells by IF compared to values obtained by MS likely reflects that the population of proTAME-synchronized cells contained some contaminating non-mitotic cells. Since the cells were treated identically for both IF and MS, the phospho-occupancy values determined by MS can be used to calibrate the fluorescence intensities at kinetochores in cells progressing through mitosis. Specifically, the fluorescence intensity at kinetochores in cells arrested in STLC would reflect ∼33% T31 phospho-occupancy. Using this calibration, the pT31 levels range from <10% (prophase) to ∼40% (late prometaphase; Fig. 2, A and B).
Hec1 phosphorylation on T31 by Cdk1 is temporally regulated during mitosis. (A) Immunofluorescence images from an asynchronous population of HeLa cells in various stages of mitosis or arrested with STLC stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel of each condition was adjusted independently. Representative images from three independent biological experiments are shown. The scale bars of main images are 10 μm, and insets are 1 μm. (B) Quantification of the relative kinetochore pT31 Hec1/Hec1 intensities from the conditions from A. The condition with the lowest level of pT31 Hec1/Hec1 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. The source data and images for the STLC condition originate from the experiment shown in Fig. S3, I and J. (C) Quantification of kinetochore total Hec1 levels from the conditions from A. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Western blots showing an anti-Hec1 immunoprecipitation from HeLa or RPE1 cells. The cells were synchronized by thymidine (HeLa) or Palbociclib (RPE1)-NOC or STLC and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (E) Western blots showing an anti-Hec1 immunoprecipitation from HeLa cells. The cells were synchronized by thymidine-NOC, treated as indicated and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (F) Schematic of kinase complexes tested for their ability to phosphorylate Hec1Broccoli in an in vitro kinase assay. (G) Intensities for peptides containing phosphorylated T31 (n = 3). The repeats were independent kinase reactions performed with separate aliquots of Hec1 and kinase derived from the same batch of protein purifications. Error bars indicate the mean ± SEM. (H) Same as for E except for phosphorylated S15. (I) Annotated MS/MS spectra for monophosphorylated T31. (J) Annotated MS/MS spectra for monophosphorylated S15. Source data are available for this figure: SourceData F2.
Hec1 phosphorylation on T31 by Cdk1 is temporally regulated during mitosis. (A) Immunofluorescence images from an asynchronous population of HeLa cells in various stages of mitosis or arrested with STLC stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel of each condition was adjusted independently. Representative images from three independent biological experiments are shown. The scale bars of main images are 10 μm, and insets are 1 μm. (B) Quantification of the relative kinetochore pT31 Hec1/Hec1 intensities from the conditions from A. The condition with the lowest level of pT31 Hec1/Hec1 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. The source data and images for the STLC condition originate from the experiment shown in Fig. S3, I and J. (C) Quantification of kinetochore total Hec1 levels from the conditions from A. The levels of Hec1 shown from the prophase condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Western blots showing an anti-Hec1 immunoprecipitation from HeLa or RPE1 cells. The cells were synchronized by thymidine (HeLa) or Palbociclib (RPE1)-NOC or STLC and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane and blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (E) Western blots showing an anti-Hec1 immunoprecipitation from HeLa cells. The cells were synchronized by thymidine-NOC, treated as indicated and then prepared as a total cell lysate. Hec1 was then immunoprecipitated and the purified proteins were then separated by SDS-PAGE, transferred to nitrocellulose membrane, and blotted as indicated. The panels were adjusted for brightness and contrast for presentation. An experiment representative of two independent biological repeats is shown. (F) Schematic of kinase complexes tested for their ability to phosphorylate Hec1Broccoli in an in vitro kinase assay. (G) Intensities for peptides containing phosphorylated T31 (n = 3). The repeats were independent kinase reactions performed with separate aliquots of Hec1 and kinase derived from the same batch of protein purifications. Error bars indicate the mean ± SEM. (H) Same as for E except for phosphorylated S15. (I) Annotated MS/MS spectra for monophosphorylated T31. (J) Annotated MS/MS spectra for monophosphorylated S15. Source data are available for this figure: SourceData F2.
Cyclin A2 knockdown does not obviously affect Cyclin B1 function; Hec1 phosphorylation is repressed by protein phosphatases. (A) Immunofluorescence images of HeLa cells in mid-prometaphase from an asynchronous population transfected with non-silencing control or siCyclin A2 and then fixed and stained for lamin A/C pS22, Cyclin A2, and DAPI. The lamin A/C pS22 and Cyclin A2 panels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. Representative images from two independent biological experiments are shown. (B) Cells were prepared as for A, except that they were transfected with non-silencing or siCyclin B1 and stained for lamin A/C pS22, Cyclin B1, and DAPI. Representative images from two independent biological experiments are shown. (C) Quantification of the images from A. 83 control cells and 88 siCyclin A2 cells over two independent biological experiments were analyzed for total cellular content of lamin A/C pS22 and Cyclin A2 levels. 93 control cells and 100 siCyclin B1 cells over two independent biological experiments were analyzed for lamin A/C pS22 and Cyclin B1 levels. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC as indicated. The cells were then treated for 2 h with CalA or DMSO control and finally fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of two independent biological experiments are shown. (E) Immunofluorescence images of cells prepared as in D, but stained for pS44 instead of pT31. (F) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in D. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) Quantification of the relative pS44 Hec1/total Hec1 kinetochore intensities from the conditions in E. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Immunofluorescence images of HeLa cells synchronized by thymidine and then 100 ng/ml NOC as indicated. The cells were then treated for 2 h with CalA or DMSO control and finally fixed and stained for ACA and α tubulin. Images representative of two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets.
Cyclin A2 knockdown does not obviously affect Cyclin B1 function; Hec1 phosphorylation is repressed by protein phosphatases. (A) Immunofluorescence images of HeLa cells in mid-prometaphase from an asynchronous population transfected with non-silencing control or siCyclin A2 and then fixed and stained for lamin A/C pS22, Cyclin A2, and DAPI. The lamin A/C pS22 and Cyclin A2 panels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. Representative images from two independent biological experiments are shown. (B) Cells were prepared as for A, except that they were transfected with non-silencing or siCyclin B1 and stained for lamin A/C pS22, Cyclin B1, and DAPI. Representative images from two independent biological experiments are shown. (C) Quantification of the images from A. 83 control cells and 88 siCyclin A2 cells over two independent biological experiments were analyzed for total cellular content of lamin A/C pS22 and Cyclin A2 levels. 93 control cells and 100 siCyclin B1 cells over two independent biological experiments were analyzed for lamin A/C pS22 and Cyclin B1 levels. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC as indicated. The cells were then treated for 2 h with CalA or DMSO control and finally fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of two independent biological experiments are shown. (E) Immunofluorescence images of cells prepared as in D, but stained for pS44 instead of pT31. (F) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in D. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of two independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) Quantification of the relative pS44 Hec1/total Hec1 kinetochore intensities from the conditions in E. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Immunofluorescence images of HeLa cells synchronized by thymidine and then 100 ng/ml NOC as indicated. The cells were then treated for 2 h with CalA or DMSO control and finally fixed and stained for ACA and α tubulin. Images representative of two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets.
Depletion of the BubR1 pool of PP2A-B56 phosphatase increases Hec1 phosphorylation and decreases k-MT stability; description and effect of Hec1 mutant constructs. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against BubR1 and then stained for Hec1 pS44, Hec1, and DAPI. The Hec1 and Hec1 pS44 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pS44 Hec1/total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Immunofluorescence images of cells prepared as in A, except stained for pT31 instead of pS44. Representative images of three independent biological experiments are shown. (D) Quantification of the relative pT31 Hec1/total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against BubR1. The media was then changed for media at 4°C, and the cells placed at 4°C for 15 min prior to fixation and staining with antibodies against ACA and α tubulin. Representative images of three independent biological experiments are shown. (F) Quantification of the relative intensity of α tubulin from the conditions shown in E. The levels of the control condition were set to 1 and the other condition shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) HeLa Cyclin B1-GFP cells were transfected with control or siRNA against BubR1. The cells were then fixed and stained for ACA, tubulin, and DAPI. The channels were adjusted independently for presentation. Representative images of three independent biological experiments are shown. (H) Quantification of the percentage of cells displaying Cyclin B1-GFP localization at kinetochores from G. At least 20 cells were imaged per experimental condition for each of three independent biological experiments. All cells were scored for having Cyclin B1 at kinetochores or not. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) HeLa cells transfected with control or siRNA against BubR1. They were then harvested, lysed, and run on SDS-PAGE and transferred onto membrane. The membranes were then blotted as indicated. A single experiment was performed. (J) Cartoon illustration of the Hec1-GFP constructs used in Figs. 5 and 10 showing the mutations made and the remaining potential phosphorylation sites. (K) Quantification of inter-kinetochore distances of cells shown in Fig. 10, A and B. 660, 650, 640, 660, 660, and 670 total pairs of kinetochores for the Wt, 8A, 8D (early prometaphase) and Wt, 8A, 8D (late prometaphase), respectively, were measured across 20 cells in each of three independent biological experiments. Error bars indicate the mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS5.
Depletion of the BubR1 pool of PP2A-B56 phosphatase increases Hec1 phosphorylation and decreases k-MT stability; description and effect of Hec1 mutant constructs. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against BubR1 and then stained for Hec1 pS44, Hec1, and DAPI. The Hec1 and Hec1 pS44 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pS44 Hec1/total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Immunofluorescence images of cells prepared as in A, except stained for pT31 instead of pS44. Representative images of three independent biological experiments are shown. (D) Quantification of the relative pT31 Hec1/total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against BubR1. The media was then changed for media at 4°C, and the cells placed at 4°C for 15 min prior to fixation and staining with antibodies against ACA and α tubulin. Representative images of three independent biological experiments are shown. (F) Quantification of the relative intensity of α tubulin from the conditions shown in E. The levels of the control condition were set to 1 and the other condition shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) HeLa Cyclin B1-GFP cells were transfected with control or siRNA against BubR1. The cells were then fixed and stained for ACA, tubulin, and DAPI. The channels were adjusted independently for presentation. Representative images of three independent biological experiments are shown. (H) Quantification of the percentage of cells displaying Cyclin B1-GFP localization at kinetochores from G. At least 20 cells were imaged per experimental condition for each of three independent biological experiments. All cells were scored for having Cyclin B1 at kinetochores or not. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) HeLa cells transfected with control or siRNA against BubR1. They were then harvested, lysed, and run on SDS-PAGE and transferred onto membrane. The membranes were then blotted as indicated. A single experiment was performed. (J) Cartoon illustration of the Hec1-GFP constructs used in Figs. 5 and 10 showing the mutations made and the remaining potential phosphorylation sites. (K) Quantification of inter-kinetochore distances of cells shown in Fig. 10, A and B. 660, 650, 640, 660, 660, and 670 total pairs of kinetochores for the Wt, 8A, 8D (early prometaphase) and Wt, 8A, 8D (late prometaphase), respectively, were measured across 20 cells in each of three independent biological experiments. Error bars indicate the mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. Source data are available for this figure: SourceData FS5.
We next sought to identify which kinase is responsible for the phosphorylation of Hec1 on T31 in mitosis. We synchronized cells in mitosis using thymidine-NOC and treated them with inhibitors of the following protein kinases: Mps1 (AZ3146), Cdk1 (RO3306), Plk1 (BI2536), Aurora A (MLN8237), or Aurora B (ZM447439) for 2 h before harvest (except RO3306; 30 min). We then immunoprecipitated Hec1 and analyzed pT31 levels by Western blot. This experiment revealed robust Hec1 T31 phosphorylation in control cells and a profound decrease in the presence of the Mps1 inhibitor or Cdk1 inhibitor. The Plk1 and Aurora kinase inhibitors did not affect pT31 levels (Fig. 2 E). Consistent with the consensus sequence at this site, Cdk1 complexes likely phosphorylate Hec1 on T31. The reduction in pT31 levels caused by Mps1 inhibition is likely to be indirect since Mps1 is required for the recruitment of Cdk1–Cyclin B1 complexes to kinetochores (Alfonso-Pérez et al., 2019; Allan et al., 2020).
To confirm that Cdk1 complexes phosphorylate Hec1 on T31, we performed an in vitro phosphorylation experiment using either a kinase-dead Cdk1–Cyclin B1 complex, wild-type Cdk1–Cyclin B1 complex, or Aurora B to phosphorylate Hec1Broccoli (Fig. 2 F). We then analyzed Hec1Broccoli phosphorylation status by MS and observed robust phosphorylation of Hec1Broccoli on T31 only in the presence of wild-type Cdk1–Cyclin B1 complex. For comparison, we observed robust phosphorylation of the S15 site only in the presence of Aurora B (Fig. 2, G–J). Therefore, we conclude that Cdk1 phosphorylates Hec1 on T31 in vivo and in vitro.
Human cells enter mitosis with Cdk1–Cyclin B1 and Cdk1–Cyclin A2 complexes, which phosphorylate residues at the same motif (Alexander et al., 2011). We therefore asked which of these complexes phosphorylates Hec1 on T31 by siRNA silencing. Strikingly, T31 phosphorylation was abolished at kinetochores of cells lacking either Cyclin A2 or Cyclin B1 (Fig. 3, A and B). However, we also observed the failure of Cyclin B1-GFP to localize to kinetochores in cells lacking Cyclin A2 (Fig. 3 C). To confirm that Cyclin A2 knockdown does not affect Cdk1–Cyclin B1 activity, we assessed the levels of lamin A/C pS22, a well-described Cdk1–Cyclin B1 target (Liu and Ikegami, 2020) in cells depleted of Cyclin A2. We observed that Cyclin A2 depletion did not reduce lamin A/C pS22 levels, whereas Cyclin B1 depletion did significantly reduce lamin A/C pS22 levels. Therefore, Cdk1–Cyclin B1 complexes remain active in cells despite the loss of Cyclin A2 (Fig. S4, A–C). Lastly, we examined the dependence of Cyclin B1 kinetochore localization on Cdk1 and Mps1 kinase activity. Consistent with previous results (Fig. 2 E and Alfonso-Pérez et al. [2019]), either Mps1 or Cdk1 inhibition abolished Cyclin B1 kinetochore localization. Notably, we also observed that Cdk1 activity is required for full kinetochore localization of Hec1 (Fig. 3 D). These results indicate that Cyclin A2 and Mps1 are upstream factors required for Cyclin B1 kinetochore localization and that Cdk1–Cyclin B1 is likely to be directly responsible for Hec1 T31 phosphorylation.
Cyclin B1 is required for Hec1 T31 phosphorylation downstream of Cyclin A2. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against Cyclin A2 and then stained for Cyclin A2, pT31 Hec1, and DAPI. The Cyclin A2 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (B) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against Cyclin B1 and then stained for Cyclin B1, pT31 Hec1, and DAPI. The Cyclin B1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (C) Immunofluorescence images from an asynchronous population of HeLa Cyclin B1-GFP cells transfected with control or siRNA against Cyclin A2 and then stained for Cyclin A2, ACA, and DAPI. The GFP, Cyclin A2, and ACA channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (D) Immunofluorescence images from of HeLa Cyclin B1-GFP cells arrested with 100 ng/ml NOC and treated with DMSO control, RO3306, or AZD3146. The Hec1 and Cyclin B1-GFP channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown.
Cyclin B1 is required for Hec1 T31 phosphorylation downstream of Cyclin A2. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against Cyclin A2 and then stained for Cyclin A2, pT31 Hec1, and DAPI. The Cyclin A2 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (B) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or siRNA against Cyclin B1 and then stained for Cyclin B1, pT31 Hec1, and DAPI. The Cyclin B1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. (C) Immunofluorescence images from an asynchronous population of HeLa Cyclin B1-GFP cells transfected with control or siRNA against Cyclin A2 and then stained for Cyclin A2, ACA, and DAPI. The GFP, Cyclin A2, and ACA channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (D) Immunofluorescence images from of HeLa Cyclin B1-GFP cells arrested with 100 ng/ml NOC and treated with DMSO control, RO3306, or AZD3146. The Hec1 and Cyclin B1-GFP channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown.
Next, we assessed the localization of Cyclins A2 and B1 relative to Hec1 and pT31. Cyclin A2-GFP is cytosolic and does not obviously localize to kinetochores (Fig. 4, A and B), while Cyclin B1-GFP localized to kinetochores, microtubules, and spindle poles from early prometaphase until metaphase as seen previously (Kabeche and Compton, 2013; Alfonso-Pérez et al., 2019; Bentley et al., 2007; Chen et al., 2008). Kinetochore-localized Cyclin B1-GFP largely co-localized with Hec1 and pT31, although it occupies a larger volume than Hec1 (Fig. 4, C–E).
Cyclin B1 but not Cyclin A2 co-localizes with Hec1 during prometaphase. (A) Immunofluorescence images from an asynchronous population of HeLa Cyclin A2-GFP cells stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (B) Line scan through the indicated pair of kinetochores from the cells in A. (C) Immunofluorescence images from an asynchronous population of HeLa Cyclin B1-GFP cells stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (D) Line scan through the indicated pair of kinetochores from the cells in C). (E) Orthogonal line scan through the upper kinetochore from the cells in C.
Cyclin B1 but not Cyclin A2 co-localizes with Hec1 during prometaphase. (A) Immunofluorescence images from an asynchronous population of HeLa Cyclin A2-GFP cells stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (B) Line scan through the indicated pair of kinetochores from the cells in A. (C) Immunofluorescence images from an asynchronous population of HeLa Cyclin B1-GFP cells stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (D) Line scan through the indicated pair of kinetochores from the cells in C). (E) Orthogonal line scan through the upper kinetochore from the cells in C.
Cdk1-dependent phosphorylation of Hec1 promotes error correction
The T31 location within the amino acid sequence of Hec1 and the temporal profile of pT31 suggest a role in k-MT attachment error correction. To explore this idea, we first measured the pT31 intensity at laterally attached compared to end-on attached kinetochores (Fig. 5, A and B) which revealed an approximately twofold increase in pT31. Since some phosphorylation sites implicated in error correction show increased phosphorylation at pole-proximal kinetochores (Chmátal et al., 2015; Ye et al., 2015), we tested the relationship between distance from the pole and level of pT31 in cells arrested by STLC. No relationship was observed (Fig. 5, D and E). We then induced k-MT attachment errors using the CENP-E inhibitor GSK-923295, which induces clustering of chromosomes at the spindle poles and k-MT attachment errors through persistent monotelic attachments (Wood et al., 2010; McEwen et al., 2001). This treatment induced an approximately twofold increase in pT31 at pole-arrested compared to aligned kinetochores (Fig. 5, F and G). We also utilized a mutant of Hec1 (Hec1 8A; Zaytsev et al., 2015; Guimaraes et al., 2008; Fig. S5 J) which prevents complete chromosome alignment through hyperstable k-MT attachments that cannot be corrected. In cells expressing Hec1 8A, unaligned chromosomes are frequently present (Fig. 5 I). Inter-kinetochore distance measurements confirm hyperstabilization of k-MT attachments in mitotic cells expressing Hec1 8A (Fig. S5 K; DeLuca et al., 2011; DeLuca et al., 2006; Zaytsev et al., 2014; Wimbish et al., 2020). In these cells, pT31 levels were also elevated in pole-arrested compared to aligned kinetochores (Fig. 5, I and J). Taken together, these data show increased phospho-occupancy at T31 on improper lateral and monotelic attachment configurations compared to end-on k-MT attachments.
Hec1 pT31 is increased on erroneously attached kinetochores. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with wild-type Hec1-GFP in an early stage of prometaphase stained for pT31 Hec1, tubulin, and DAPI. The images were adjusted for brightness and contrast for presentation. Representative images from two independent biological experiments are shown. The scale bars of main images are 10 μm, and in the insets are 1 μm. (B) Quantification of the relative pT31 Hec1/Total Hec1kinetochore intensities of end-on attached kinetochores compared to laterally attached kinetochores as shown in A. The levels of the end-on attached kinetochores were set to 1 and the levels of the laterally attached kinetochores shown as a fold-change. 827 (laterally attached) and 505 (end-on attached) total kinetochores from 10 cells for each of two independent biological experiments were measured. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Quantification of kinetochore total Hec1 levels from the conditions from A. The levels of Hec1 shown from the lateral condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa cells synchronized by thymidine-STLC and stained for Hec1 and pT31 Hec1. The images were adjusted for brightness and contrast for presentation. The cells presented in this experiment are from the same experiment presented in Fig. S3 I but re-analyzed to determine the relationship of pT31 to spindle pole distance. Representative images from three independent biological experiments are shown. The scale bars of main images are 10 μm, and in the insets are 1 μm. (E) Quantification of the relative pT31 Hec1/Hec1 kinetochore intensities of kinetochores versus distance from the spindle poles. The levels of pT31 Hec1 and Hec1were measured together with the distance from the spindle poles. 674 total kinetochores from 20 cells for each of three independent biological experiments were measured. The equation of the regression line and R2 values are shown. The cells presented in this experiment are from the same experiment presented in Fig. S3 I but re-analyzed to determine the relationship of pT31 to spindle pole distance. (F) Immunofluorescence images of HeLa cells in late prometaphase from an asynchronous population treated with GSK-923295 for 2 h and then fixed and stained for Hec1, pT31 Hec1, and DAPI. The images were adjusted for brightness and contrast for presentation. The top insets show aligned kinetochores, and the lower insets show unaligned kinetochores. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of three independent biological experiments are shown. (G) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in F. The levels of the aligned kinetochores were set to 1 and the intensity of the unaligned kinetochores shown as a fold-change. 1,509 (aligned) and 1,355 (unaligned) total kinetochores were quantified from 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Quantification of kinetochore total Hec1 levels from the conditions from F and G. The levels of Hec1 shown from the aligned condition were set to 1 and the other condition shown as a fold-change. 1,509 (aligned) and 1,355 (unaligned) total kinetochores were quantified from 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the Hec1-8A construct. The cells are in late prometaphase taken from an asynchronous population. 72 h after transfection the cells were fixed and stained for pT31 Hec1 and DAPI. The GFP and pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The top inset shows aligned kinetochores and the bottom inset shows unaligned kinetochores. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. The cells presented in this experiment are from the same experiment presented in Fig. 10 B but reanalyzed to compare the levels of pT31 in unaligned versus aligned kinetochores. (J) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in I. The levels of the aligned kinetochores were set to 1 and the intensity of the unaligned kinetochores shown as a fold-change. 957 (aligned) and 625 (unaligned) total kinetochores were quantified from at least 10 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. The cells analyzed in this experiment are the same cells presented in Fig. 10 B but reanalyzed to compare the levels of pT31 in unaligned versus aligned kinetochores. (K) Quantification of kinetochore total Hec1 levels from the conditions in I. The levels of Hec1 shown from the aligned condition were set to 1 and the other condition shown as a fold-change. 957 (aligned) and 625 (unaligned) total kinetochores were quantified from at least 10 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (L) HeLa Cyclin B1-GFP cells were treated as in F, but then stained for ACA, tubulin, and DAPI. The images were adjusted for brightness and contrast for presentation. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (M) Line scans through the kinetochores shown in the insets in L.
Hec1 pT31 is increased on erroneously attached kinetochores. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with wild-type Hec1-GFP in an early stage of prometaphase stained for pT31 Hec1, tubulin, and DAPI. The images were adjusted for brightness and contrast for presentation. Representative images from two independent biological experiments are shown. The scale bars of main images are 10 μm, and in the insets are 1 μm. (B) Quantification of the relative pT31 Hec1/Total Hec1kinetochore intensities of end-on attached kinetochores compared to laterally attached kinetochores as shown in A. The levels of the end-on attached kinetochores were set to 1 and the levels of the laterally attached kinetochores shown as a fold-change. 827 (laterally attached) and 505 (end-on attached) total kinetochores from 10 cells for each of two independent biological experiments were measured. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Quantification of kinetochore total Hec1 levels from the conditions from A. The levels of Hec1 shown from the lateral condition were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa cells synchronized by thymidine-STLC and stained for Hec1 and pT31 Hec1. The images were adjusted for brightness and contrast for presentation. The cells presented in this experiment are from the same experiment presented in Fig. S3 I but re-analyzed to determine the relationship of pT31 to spindle pole distance. Representative images from three independent biological experiments are shown. The scale bars of main images are 10 μm, and in the insets are 1 μm. (E) Quantification of the relative pT31 Hec1/Hec1 kinetochore intensities of kinetochores versus distance from the spindle poles. The levels of pT31 Hec1 and Hec1were measured together with the distance from the spindle poles. 674 total kinetochores from 20 cells for each of three independent biological experiments were measured. The equation of the regression line and R2 values are shown. The cells presented in this experiment are from the same experiment presented in Fig. S3 I but re-analyzed to determine the relationship of pT31 to spindle pole distance. (F) Immunofluorescence images of HeLa cells in late prometaphase from an asynchronous population treated with GSK-923295 for 2 h and then fixed and stained for Hec1, pT31 Hec1, and DAPI. The images were adjusted for brightness and contrast for presentation. The top insets show aligned kinetochores, and the lower insets show unaligned kinetochores. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of three independent biological experiments are shown. (G) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in F. The levels of the aligned kinetochores were set to 1 and the intensity of the unaligned kinetochores shown as a fold-change. 1,509 (aligned) and 1,355 (unaligned) total kinetochores were quantified from 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (H) Quantification of kinetochore total Hec1 levels from the conditions from F and G. The levels of Hec1 shown from the aligned condition were set to 1 and the other condition shown as a fold-change. 1,509 (aligned) and 1,355 (unaligned) total kinetochores were quantified from 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the Hec1-8A construct. The cells are in late prometaphase taken from an asynchronous population. 72 h after transfection the cells were fixed and stained for pT31 Hec1 and DAPI. The GFP and pT31 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The top inset shows aligned kinetochores and the bottom inset shows unaligned kinetochores. The scale bars are 10 μm. Representative images of three independent biological experiments are shown. The cells presented in this experiment are from the same experiment presented in Fig. 10 B but reanalyzed to compare the levels of pT31 in unaligned versus aligned kinetochores. (J) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in I. The levels of the aligned kinetochores were set to 1 and the intensity of the unaligned kinetochores shown as a fold-change. 957 (aligned) and 625 (unaligned) total kinetochores were quantified from at least 10 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. The cells analyzed in this experiment are the same cells presented in Fig. 10 B but reanalyzed to compare the levels of pT31 in unaligned versus aligned kinetochores. (K) Quantification of kinetochore total Hec1 levels from the conditions in I. The levels of Hec1 shown from the aligned condition were set to 1 and the other condition shown as a fold-change. 957 (aligned) and 625 (unaligned) total kinetochores were quantified from at least 10 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (L) HeLa Cyclin B1-GFP cells were treated as in F, but then stained for ACA, tubulin, and DAPI. The images were adjusted for brightness and contrast for presentation. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (M) Line scans through the kinetochores shown in the insets in L.
To test if Cyclin B1 specifically localizes to kinetochores with improper k-MT attachments, we treated cells with GSK-923295 to generate k-MT attachment errors. We frequently observed pole-arrested kinetochores with lateral syntelic attachments with Cyclin B1 present on both sister kinetochores. We did not observe any Cyclin B1 at kinetochores with end-on amphitelic attachments (Fig. 5, L and M). Furthermore, the intensity of Cyclin B1-GFP at kinetochores was high in mitotic cells arrested with 100 ng/ml NOC or STLC, which have k-MT attachment errors, and low in cells arrested with 500 ng/ml NOC that do not have k-MT attachments or proTAME that have bioriented attachments (Fig. 6, A and B), a pattern similar to pT31 levels in cells arrested under those conditions (Fig. S3, I and J). Thus, Cyclin B1 localization is enriched on improper k-MT attachments coinciding with increased Hec1 pT31. Previous results suggest that Cyclin B1 localizes specifically to unattached kinetochores (Alfonso-Pérez et al., 2019; Bentley et al., 2007; Chen et al., 2008). Our results suggest that it also localizes to kinetochores with improper attachments.
Cyclin B1 only localizes strongly to kinetochores that have improper k-MT attachments. (A) Immunofluorescence images from HeLa Cyclin B1-GFP cells synchronized with thymidine and then NOC, STLC, or proTAME. The cells were then fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (B) Line scans through the indicated pairs of kinetochores from the cells shown in A.
Cyclin B1 only localizes strongly to kinetochores that have improper k-MT attachments. (A) Immunofluorescence images from HeLa Cyclin B1-GFP cells synchronized with thymidine and then NOC, STLC, or proTAME. The cells were then fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and Hec1 pT31 channels were adjusted evenly for brightness and contrast. The GFP and DAPI channels were adjusted independently. The scale bars for the main images are 10 and 1 μm for the insets. Representative images of two independent biological experiments are shown. (B) Line scans through the indicated pairs of kinetochores from the cells shown in A.
Persistent errors in k-MT attachment manifest as chromosomes lagging in anaphase and increase chromosome mis-segregation (Thompson and Compton, 2011). To test if Hec1 T31 phosphorylation participates in error correction, we expressed Hec1-GFP mutants in cells with doxycycline-inducible Hec1 knockout. We used Hec1-GFP in which T31 was mutated to either alanine (T31A) or aspartic acid (T31D). For comparison, we also generated S15A and S15D mutants because S15 is phosphorylated by a different kinase (Aurora B; Fig. 2 H), and it demonstrated a similar phospho-occupancy profile as T31 (Fig. 1 B and Fig. S2 C). Hec1 T31A expression resulted in elevated lagging chromosome rates (Fig. 7, A and B). Hec1 T31D expression showed no change relative to wild-type Hec1, suggesting that this substitution may not have a functional effect. Next, we increased k-MT attachment errors via the treatment of cells via STLC washout and measured the lagging chromosome frequency. As expected, the overall lagging chromosome rate was elevated. Hec1 T31A expression resulted in very high lagging chromosome rates in cells recovering from STLC, suggesting that Hec1 T31 phosphorylation is required for efficient k-MT attachment error correction (Fig. 7 C). Interestingly, Hec1 S15A or S15D expression had no significant effect on the rate of lagging chromosomes in either asynchronous cells or cells recovering from STLC treatment (Fig. 7, A–C), suggesting perhaps that Aurora kinases can compensate for the absence of one phosphorylation site by increasing phosphorylation at other sites.
Hec1 T31 phosphorylation is critical for k-MT attachment error correction. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained with DAPI. The GFP and DAPI channels were adjusted evenly for brightness and contrast for presentation. A lagging chromosome is indicated with a yellow arrow. Representative images from three independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. (B) Quantification of the percentage of lagging chromosomes in cells from an asynchronous population undergoing anaphase and expressing the indicated Hec1 constructs. At least 319 cells per construct were scored for the presence of lagging chromosomes over three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Quantification of the percentage of lagging chromosomes in anaphase for cells expressing the indicated Hec1 constructs following an STLC washout. 120 cells per construct were scored for the presence of lagging chromosomes for each of three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 48 h post-transfection the cells were fixed and stained for Ska3 and DAPI. The GFP, Ska3, and DAPI channels were adjusted evenly for brightness and contrast for presentation. Representative images from three independent biological experiments are shown. The scale bars are 10 and 1 μm for insets. (E) Quantification of the relative Ska3 kinetochore intensities from the conditions in D. The Ska3 levels of Wt expressing cells were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (F) Immunofluorescence images from an asynchronous population of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 48 h after transfection the media was then changed for media at 4°C, and the cells were placed at 4°C for 15 min prior to fixation and staining with antibodies against α tubulin. Representative images of three independent biological experiments are shown. The images were adjusted evenly for brightness and contrast for presentation. (G) Quantification of the relative intensity of α tubulin from the conditions shown in F. The levels of the Hec1 Wt expressing cells were set to 1 and the other conditions shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests.
Hec1 T31 phosphorylation is critical for k-MT attachment error correction. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained with DAPI. The GFP and DAPI channels were adjusted evenly for brightness and contrast for presentation. A lagging chromosome is indicated with a yellow arrow. Representative images from three independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. (B) Quantification of the percentage of lagging chromosomes in cells from an asynchronous population undergoing anaphase and expressing the indicated Hec1 constructs. At least 319 cells per construct were scored for the presence of lagging chromosomes over three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Quantification of the percentage of lagging chromosomes in anaphase for cells expressing the indicated Hec1 constructs following an STLC washout. 120 cells per construct were scored for the presence of lagging chromosomes for each of three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 48 h post-transfection the cells were fixed and stained for Ska3 and DAPI. The GFP, Ska3, and DAPI channels were adjusted evenly for brightness and contrast for presentation. Representative images from three independent biological experiments are shown. The scale bars are 10 and 1 μm for insets. (E) Quantification of the relative Ska3 kinetochore intensities from the conditions in D. The Ska3 levels of Wt expressing cells were set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (F) Immunofluorescence images from an asynchronous population of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 48 h after transfection the media was then changed for media at 4°C, and the cells were placed at 4°C for 15 min prior to fixation and staining with antibodies against α tubulin. Representative images of three independent biological experiments are shown. The images were adjusted evenly for brightness and contrast for presentation. (G) Quantification of the relative intensity of α tubulin from the conditions shown in F. The levels of the Hec1 Wt expressing cells were set to 1 and the other conditions shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests.
To determine how Hec1 T31 phosphorylation promotes error correction, we first tested if T31 phosphorylation influenced recruitment of the Ska complex to kinetochores by staining cells in metaphase with a Ska3-specific antibody following expression of Hec1-GFP wild-type, T31A, or T31D. There was no detectable difference in the intensity of Ska3 at kinetochores in the presence of these mutant versions of Hec1 relative to wild-type Hec1 (Fig. 7, D and E). Next, we performed a cold MT stability assay to determine if the Hec1 T31A mutant contributes to elevated rates of lagging chromosomes by altering k-MT stability (Fig. 7, F and G). In this experiment we included the Hec1 9A mutant because it has been shown previously to stabilize k-MT attachments (Zaytsev et al., 2014). We observed a significant increase in the quantity of cold-stable MTs in cells expressing either Hec1 T31A or 9A mutants relative to wild-type Hec1. Thus, Cdk1–Cyclin B1-dependent phosphorylation of Hec1 on T31 destabilizes k-MT attachments to promote error correction required for faithful chromosome segregation.
Kinetochore Hec1 phosphorylation levels are influenced by phosphatase activity
To determine the phosphatase contribution to the net phospho-occupancy of the Cdk1–Cyclin B1 site T31, we arrested cells with 100 or 500 ng/ml NOC and then treated them with the phosphatase inhibitors okadaic acid (OA; 200 nM) or Calyculin A (CalA; 5 nM) for 2 h before fixation. In both conditions, the pT31 intensity at kinetochores increased in the presence of OA or CalA (Fig. 8, A and B; and Fig. S4, D and F) despite poor localization of Cyclin B1 to kinetochores lacking microtubule attachments (500 ng/ml NOC; Fig. 6, A and B) suggesting that elevated Hec1 phospho-occupancy occurs under these conditions because partial localization of Cyclin B1 is sufficient and/or Hec1 can be phosphorylated by soluble Cyclin–B1 complexes. We also tested the contribution of phosphatases to pS44 levels. Interestingly, the extent of phosphorylation was markedly increased in cells treated with the high dose of NOC, but only to a lesser extent in cells treated arrested in the lower dose (Fig. 8, C and D; and Fig. S4, E and G). These data show that the phospho-occupancy of these Aurora and Cdk1 phosphoacceptor sites is differentially regulated. Notably, the phosphorylation levels at these two sites did not reach the theoretical maximum following phosphatase inhibition, possibly due to charge repulsion among closely spaced phosphoacceptor sites as previously described (Rao and Møller, 2012).
Hec1 phosphorylation is repressed by phosphatase activity to bias kinetochores toward a state of attachment. (A) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC as indicated. The cells were then treated for 2 h with OA or DMSO control and finally fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm for main images and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pT31 Hec1/total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pT31 was set to 1 and the other conditions are shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three biological independent repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t-tests. (C) Immunofluorescence images of cells prepared as in A but stained for pS44 instead of pT31. (D) Quantification of the relative pS44 Hec1/Total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images of HeLa cells synchronized by thymidine and then 100 ng/ml NOC as indicated. The cells were then treated for 2 h with OA or DMSO control and finally fixed and stained for ACA and α tubulin. Images representative of two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. The ACA and tubulin channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently.
Hec1 phosphorylation is repressed by phosphatase activity to bias kinetochores toward a state of attachment. (A) Immunofluorescence images of HeLa cells synchronized by thymidine and then NOC as indicated. The cells were then treated for 2 h with OA or DMSO control and finally fixed and stained for Hec1, pT31 Hec1, and DAPI. The Hec1 and pT31 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars are 10 μm for main images and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pT31 Hec1/total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pT31 was set to 1 and the other conditions are shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three biological independent repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t-tests. (C) Immunofluorescence images of cells prepared as in A but stained for pS44 instead of pT31. (D) Quantification of the relative pS44 Hec1/Total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images of HeLa cells synchronized by thymidine and then 100 ng/ml NOC as indicated. The cells were then treated for 2 h with OA or DMSO control and finally fixed and stained for ACA and α tubulin. Images representative of two independent biological experiments are shown. The scale bars for main images are 10 and 1 μm for insets. The ACA and tubulin channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently.
We also noticed in these experiments that in cells treated with 100 ng/ml NOC, the kinetochores were clustered around and attached to short microtubule stub-spindles (Fig. 3 D; Fig. 6 A; Fig. 8, A, C, and E; Fig. S2 E; Fig. S3 I; and Fig. S4, D and E). However, following treatment with OA, the kinetochores were distributed throughout the cytoplasm, suggesting that the kinetochores had detached from microtubules. To determine if this was the case, we stained cells treated with or without phosphatase inhibitors for α tubulin and examined the extent of kinetochore attachment. This experiment confirmed that in cells treated with NOC, all chromosomes were attached to the stub spindles by at least one kinetochore. However, in cells treated with both NOC and OA almost all kinetochores were detached from the spindles (Fig. 8 E), suggesting that Hec1 phosphorylation levels are repressed by phosphatases to prevent k-MT detachment despite ongoing kinase activity. Given that only a modest 1.3-fold increase in phosphorylation was observed at S44 in cells treated with OA, k-MT attachments are likely very sensitive to phosphorylation. Interestingly, cells treated with CalA (limited by toxicity to 5 nM) did not show k-MT detachment (Fig. S4 H). This may be because CalA treatment did not cause as high an increase in phospho-Hec1 levels compared to OA (Fig. 8 and Fig. S4, D–G).
K-MT attachments have previously been shown to be regulated by the PP2A-B56 family of phosphatases (Foley et al., 2011; Suijkerbuijk et al., 2012; Kruse et al., 2013). To determine if Hec1 tail phosphorylation is regulated by PP2A-B56, we took advantage of the fact that kinetochore isoforms of B56 are recruited to the kinetochore by the protein BubR1 (Suijkerbuijk et al., 2012). Therefore, the knockdown of BubR1 removes them from the kinetochore. We silenced the expression of BubR1 using siRNA (Fig. S5 I) and examined Hec1 phosphorylation. Interestingly, we observed an ∼1.2-fold increase in phosphorylation of both S44 and T31 positions on Hec1 (Fig. S5, A–D), which suggests that Hec1 phosphorylation by both Aurora and Cdk1 kinases are repressed by the PP2A-B56 group of phosphatases, consistent with previous findings (Foley et al., 2011; Suijkerbuijk et al., 2012). In these experiments, we also observed a large proportion of the BubR1 knockdown cells that displayed chromosome alignment defects (Fig. S5, A, C, E, and G) as reported previously (Suijkerbuijk et al., 2012). Since it had been previously determined that these defects are due to hyper–k-MT attachment as a result of decreased KMN network protein phosphorylation (Foley et al., 2011; Suijkerbuijk et al., 2012), we also examined the stability of MTs using a cold MT stability assay under these conditions. Despite only a 1.2-fold increase in phosphorylation at S44 and T31 on Hec1, this experiment revealed a 0.4-fold loss of MTs (Fig. S5, E and F), suggesting that the k-MT attachment state is hypersensitive to KMN network protein phosphorylation.
To confirm that Hec1 phosphorylation is repressed by PP2A-B56, we silenced the expression of the B56 subunit isoforms using a pool of siRNAs (Foley et al., 2011; Fig. 9 I) and examined the phosphorylation state of Hec1 at S44 and T31. This experiment showed a 1.6-fold increase in pS44 levels (Fig. 9, A and B). Interestingly, under the same conditions, we observed a slight decrease in pT31 levels (Fig. 9, C and D), although we continued to observe increased phosphorylation at pole-arrested kinetochores (Fig. 9 C). This might indicate additional OA-sensitive phosphatases acting to regulate the phospho-occupancy of the T31 site. Consistent with previous findings (Foley et al., 2011), we also observed that cells with B56 knockdown display moderate (5–10 misaligned chromosomes) to severe (>10 misaligned chromosomes) chromosome alignment defects (Fig. 9, A, C, E, and G). To test if the defects in chromosome alignment are due to a loss of K-MT attachment stability, we again performed a cold MT stability assay in cells with B56 knockdown compared to controls. This experiment revealed that cells with B56 knockdown have only 35% of the cold-stable microtubule fluorescence compared to control cells, likely explaining the defects in chromosome alignment (Fig. 9, E and F). As we and others have noted previously, the presence of (improper) k-MT attachments is required both for Cyclin B1 localization to kinetochores and for robust phosphorylation of T31 (Fig. 6; Bentley et al., 2007; Chen et al., 2008). We, therefore, examined if kinetochores have such weak attachments under conditions of total phosphatase loss that Cdk1–Cyclin B1 is unable to efficiently localize to and phosphorylate Hec1 under conditions of phosphatase loss at kinetochores. We compared the localization pattern of Cyclin B1-GFP during prometaphase under conditions of BubR1 knockdown or B56 knockdown. Interestingly, most cells with BubR1 knockdown maintained Cyclin B1 at kinetochores (Fig. S5, G and H), as did cells with B56 knockdown and moderate chromosome alignment defects. However, most cells with B56 knockdown and severe alignment defects did not have Cyclin B1 at kinetochores (Fig. 9, G and H). This suggests that Cyclin B1 might only localize to kinetochores that are strongly bound to microtubules to destabilize them, but it would not further destabilize attachments that are already weak.
Hec1 phosphorylation is repressed by PP2A-B56 with differential effects on pT31 and pS44. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or a pool of siRNA against B56 α-ε and then stained for Hec1 pS44, Hec1, and DAPI. The Hec1 and pS44 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pS44 Hec1/Total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Immunofluorescence images of cells prepared as in A except stained for pT31 instead of pS44. (D) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or a pool of siRNA against B56 α–ε. The media was then changed for media at 4°C and the cells were placed at 4°C for 15 min before fixation and staining with antibodies against ACA and α tubulin. The ACA and tubulin channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images of three independent biological experiments are shown. (F) Quantification of the relative intensity of cold-stable α tubulin from the conditions shown in E. The levels of the control condition were set to 1 and the other condition shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) HeLa Cyclin B1-GFP cells were transfected with control or a pool of siRNA against the five B56 isoforms. The cells were then fixed and stained for ACA, tubulin, and DAPI. The channels were adjusted independently for presentation. Representative images of three independent biological experiments are shown. (H) Quantification of the percentage of cells displaying Cyclin B1-GFP localization at kinetochores from G. At least 20 cells were imaged per experimental condition for each of three independent biological experiments. The cells with B56 knockdown were identified as having either moderate or severe alignment defects. All cells were scored for having Cyclin B1 at kinetochores or not. Error bars indicate the mean ± SEM. ***, P < 0.001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) HeLa cells transfected with control or a pool of siRNA against B56 α–ε. They were then harvested, lysed and separated by SDS-PAGE and transferred onto membranes. The membranes were then blotted as indicated. A single experiment was performed. Source data are available for this figure: SourceData F9.
Hec1 phosphorylation is repressed by PP2A-B56 with differential effects on pT31 and pS44. (A) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or a pool of siRNA against B56 α-ε and then stained for Hec1 pS44, Hec1, and DAPI. The Hec1 and pS44 Hec1 channels were adjusted evenly for brightness and contrast. The DAPI channel was adjusted independently. The scale bars for main images are 10 and 1 μm for insets. Representative images of three independent biological experiments are shown. (B) Quantification of the relative pS44 Hec1/Total Hec1 kinetochore intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (C) Immunofluorescence images of cells prepared as in A except stained for pT31 instead of pS44. (D) Quantification of the relative pT31 Hec1/Total Hec1 kinetochore intensities from the conditions in C. The condition with the lowest level of pT31 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (E) Immunofluorescence images from an asynchronous population of HeLa cells transfected with control or a pool of siRNA against B56 α–ε. The media was then changed for media at 4°C and the cells were placed at 4°C for 15 min before fixation and staining with antibodies against ACA and α tubulin. The ACA and tubulin channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images of three independent biological experiments are shown. (F) Quantification of the relative intensity of cold-stable α tubulin from the conditions shown in E. The levels of the control condition were set to 1 and the other condition shown as a fold-change. At least 50 cells per condition from each of three independent biological experiments were quantified. Error bars indicate the mean ± SEM. ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (G) HeLa Cyclin B1-GFP cells were transfected with control or a pool of siRNA against the five B56 isoforms. The cells were then fixed and stained for ACA, tubulin, and DAPI. The channels were adjusted independently for presentation. Representative images of three independent biological experiments are shown. (H) Quantification of the percentage of cells displaying Cyclin B1-GFP localization at kinetochores from G. At least 20 cells were imaged per experimental condition for each of three independent biological experiments. The cells with B56 knockdown were identified as having either moderate or severe alignment defects. All cells were scored for having Cyclin B1 at kinetochores or not. Error bars indicate the mean ± SEM. ***, P < 0.001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (I) HeLa cells transfected with control or a pool of siRNA against B56 α–ε. They were then harvested, lysed and separated by SDS-PAGE and transferred onto membranes. The membranes were then blotted as indicated. A single experiment was performed. Source data are available for this figure: SourceData F9.
The aurora and Cdk1 phosphorylation networks respond differentially to stimuli
We next sought to compare how the Aurora B and CDK1 signaling networks respond to alterations in k-MT stability. We used the Hec1 8A and 8D mutants (Fig. S5 J) because they preserve an Aurora B site (S44) and the Cdk1 site (T31), allowing us to monitor the phosphorylation status of these two specific sites in cells with k-MT attachment errors. We observed a significant increase in pS44 levels at kinetochores in early and late prometaphase cells expressing Hec1 8A compared to wild-type Hec1. We also observed a slight increase in pS44 levels in late prometaphase cells expressing the 8D mutant (Fig. 10, A and C). In contrast, there was a significant decrease in pT31 levels in early prometaphase cells expressing Hec1 8A (Fig. 10, B and D). Since this Hec1 mutant promotes the formation of precocious end-on k-MT attachments (Wimbish et al., 2020), this is consistent with our results showing decreased pT31 at end-on attached kinetochores (Fig. 5, A and B). There was no overall significant change in pT31 levels in late prometaphase Hec1 8A expressing cells (although we observe increased phosphorylation at pole-arrested kinetochores), suggesting a temporal change in Cdk1 or phosphatase activity for T31 under these conditions. Notably, we observed a slight decrease in pT31 levels in cells expressing the 8D mutant in both early and late prometaphase cells (Fig. 10, B and D), which is consistent with our data showing that phosphatase inhibition reduces pT31 levels due to a loss of k-MT attachment and Cyclin B1 localization at kinetochores (Fig. 9 and Fig. S5, A–H). Thus, the Aurora B and Cdk1 signaling networks respond independently to changes in k-MT attachments, which is consistent with our phospho-occupancy measurements determined by MS (Fig. 1 B).
Hec1 pT31 and pS44 sites are differentially regulated. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained for pS44 Hec1 and DAPI. The GFP and pS44 Hec1 channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images from three independent biological experiments are shown. The scale bars are 10 μm. (B) As in A, except that the cells were stained for pT31 instead of pS44. (C) Quantification of the relative kinetochore pS44 Hec1/Total Hec1 intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01; ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) As in C, but for pT31 instead of pS44. (E) Cartoon illustration showing the kinase and phosphatase network regulating Hec1 phosphorylation. In early prometaphase, cells without k-MT attachments have elevated levels of fractional phosphorylation on several Hec1 sites that are phosphorylated by Aurora kinases (green circles) or Cdk1–Cyclin B1 kinase (magenta circles). When lateral k-MT attachments occur, the levels of phosphorylation generally decrease. Finally, when end-on k-MT attachments occur, levels of phosphorylation decrease further. Phosphorylation levels at Hec1 S4,5,8 and S62 are speculative or based on partial data coverage.
Hec1 pT31 and pS44 sites are differentially regulated. (A) Immunofluorescence images of HeLa doxycycline inducible Hec1 knockout cells transfected with the indicated Hec1-GFP constructs. 72 h after transfection the cells were fixed and stained for pS44 Hec1 and DAPI. The GFP and pS44 Hec1 channels were adjusted evenly for brightness and contrast for presentation. The DAPI channel was adjusted independently. Representative images from three independent biological experiments are shown. The scale bars are 10 μm. (B) As in A, except that the cells were stained for pT31 instead of pS44. (C) Quantification of the relative kinetochore pS44 Hec1/Total Hec1 intensities from the conditions in A. The condition with the lowest level of pS44 was set to 1 and the other conditions shown as fold-changes. 25 kinetochores were quantified from each of 20 cells for each of three independent biological repeats. Error bars indicate the mean ± SEM. **, P < 0.01; ****, P < 0.0001. Statistical significance was calculated between the indicated conditions using two-tailed t tests. (D) As in C, but for pT31 instead of pS44. (E) Cartoon illustration showing the kinase and phosphatase network regulating Hec1 phosphorylation. In early prometaphase, cells without k-MT attachments have elevated levels of fractional phosphorylation on several Hec1 sites that are phosphorylated by Aurora kinases (green circles) or Cdk1–Cyclin B1 kinase (magenta circles). When lateral k-MT attachments occur, the levels of phosphorylation generally decrease. Finally, when end-on k-MT attachments occur, levels of phosphorylation decrease further. Phosphorylation levels at Hec1 S4,5,8 and S62 are speculative or based on partial data coverage.
Discussion
Our data demonstrate that phosphorylation of the population of Hec1 molecules does not follow an all-or-none mechanism even though phosphorylation of individual amino acids on individual molecules is binary. Importantly, the phospho-occupancy that we measured on multiple sites on Hec1 was similar between the soluble and insoluble (chromatin bound) populations of Hec1 in mitotic cells. Moreover, we observed partial phospho-occupancy on multiple sites of Hec1 in cells treated with high doses of NOC, a condition where all kinetochores lack microtubule attachments and are in an equivalent state (the idea of equivalency is supported by a smaller standard deviation amongst kinetochores; Fig. S3 J). Thus, fractional phospho-occupancy reflects an equal distribution of the phosphorylated Hec1 amongst the total population of molecules as opposed to reflecting different sub-populations of molecules where phospho-occupancy depends on the nature of the sub-population. In other words, in a case where a specific site demonstrates 25% phospho-occupancy, our data indicate that 25% of all Hec1 molecules are phosphorylated rather than specific sub-populations of Hec1 molecules, where one sub-population is comprised of 25% of Hec1 molecules that are 100% phospho-occupied at that site and another sub-population that is comprised of 75% of Hec1 molecules that are 0% phospho-occupied.
We envisage two mechanisms acting together to limit the extent of phosphorylation of Hec1. One is direct dephosphorylation catalyzed by ongoing protein phosphatase activity, and our data illustrate the essential role of the ongoing phosphatase activity in regulating k-MT attachments. The second is steric charge repulsion, which acts to hinder phosphorylation of very closely spaced phosphoacceptor sites and thereby favor a lower overall phospho-occupancy state (Rao and Møller, 2012).
These findings carry numerous implications for how phosphorylation regulates k-MT attachments to both favor efficient microtubule capture by kinetochores and support robust error correction to ensure high mitotic fidelity (Fig. 10 E). Some models predict that phospho-occupancy of proteins like Hec1 at the kinetochore–microtubule interface would be nearly saturated under conditions where kinetochores are not under tension (Funabiki and Wynne, 2013; Lampson and Cheeseman, 2011; Liu et al., 2009). However, we demonstrate that in mitotic cells treated with low dose NOC, which induces the formation of stub-spindles with erroneous k-MT attachments lacking tension, that chromosomes detach from the stub-spindles upon phosphatase inhibition (Fig. 8 and Fig. S4, D–H). Thus, in testing one of the two mechanisms that serve to limit the extent of Hec1 phosphorylation, we demonstrate the sensitivity of this system to subtle changes by showing that ongoing phosphatase activity is essential to bias kinetochores to remain attached to the stub spindles. Moreover, when kinetochores completely lack microtubule attachments induced by treatment with high dose NOC, the combination of ongoing phosphatase activity and steric hindrance maintains a relatively low level of Hec1 phospho-occupancy (Fig. 1 B; Fig. 8, A–D; Fig. S2 C; and Fig. S4, D–G). In support of this idea, it has been shown that mitotic cells in which phosphatases have been inactivated are unable to properly congress chromosomes to the metaphase plate (Foley et al., 2011; Suijkerbuijk et al., 2012).
These observations resolve a previously described paradox (Lampson and Cheeseman, 2011; Liu et al., 2009; Biggins et al., 1999; Tanaka et al., 2002) that predicted high Hec1 phospho-occupancy in the absence of microtubule attachments. Resolving this paradox is particularly important to illuminate how conditions favor initial k-MT attachments at the prophase to prometaphase transition. At the time of nuclear envelope breakdown, kinetochores lack microtubule attachments and experience no tension. If kinase activity were unconstrained on unattached kinetochores, then complete phosphorylation of the sites at the prophase to prometaphase transition would substantially disfavor the formation of initial microtubule attachments. Our data provides a mechanistic explanation for how Hec1 (and possibly other proteins) phosphorylation is dampened to favor initial attachments.
We also provide the first direct evidence that Cdk1–Cyclin B1 plays a role in promoting k-MT attachment error correction, and that Cdk1-mediated error correction is independent from the established role of Aurora kinases. Our data show that these two enzymes recognize different types of k-MT attachment errors with Cdk1 focused on the conversion of sidewall to end-on attachments and Aurora focused on k-MT attachments with improper tension. Consistently, sites phosphorylated by these enzymes show opposing changes in response to the same stimulus (Fig. 10, A–D). This supports the view that different types of k-MT attachment errors rely on different mechanisms of detection and/or response for correction. Perhaps it is surprising that these two kinases, which respond to different types of k-MT attachment defects, phosphorylate closely located sites on the same peptide of Hec1. However, the output of these kinases is not expected to be limited to changes in phosphorylation of only Hec1 at kinetochores.
This new role of Cdk1–Cyclin B1 in error correction fits within a feed-forward mechanism of mitotic control whereby Cdk1–Cyclin B1 targeting kinetochores through Mad1 (Alfonso-Pérez et al., 2019; Allan et al., 2020; Jackman et al., 2020) regulates cell cycle timing through the satisfaction of the spindle assembly checkpoint and also ensures efficient correction of erroneous k-MT attachments. Notably, the targeting of Cdk1–Cyclin B1 to kinetochores with attachment errors in early mitosis depends on Cdk1–Cyclin A2. Previously, we demonstrated that Cdk1–Cyclin A2 activity destabilizes k-MT attachment stability coordinately on all kinetochores to promote error correction in prometaphase (Kabeche and Compton, 2013), which reinforces this feed-forward mechanism involving cell cycle timing with k-MT attachment error correction. The data presented here provide further insights into how a distributive system that regulates all kinetochores coordinately through Cdk1–Cyclin A2 forms direct mechanistic links to regulatory molecules that operate the chromosome autonomously (Cyclin B1 & MYPT1/Plk1; Dumitru et al., 2017).
Our data fit well with previous studies that determined phospho-occupancies in mitosis. Interestingly, the average phospho-occupancy of the fission yeast proteome during mitosis is only 44%, whereas in human cells it is over 70%. These studies revealed that while many sites are phosphorylated at high occupancy (e.g., Lamin A pS22; 99%), many are not (e.g., PLK1 pT210; 47.6%). High occupancy is likely required in cases where a strong, unidirectional effect is desirable, whereas moderate occupancy likely reflects participation in signaling feedback loops and the need for up- or down-regulation. Fittingly, whereas Cdk sites show a mean occupancy of 88%, Aurora kinase sites show modest occupancy in human mitotic cells (mean occupancy = 58%; Olsen et al., 2010; Carpy et al., 2012). Since no site on Hec1 that we measured exceeded 50% phospho-occupancy under any condition that we tested and the cumulative phospho-occupancy among the sites measured changed by only 20% in response to the full spectrum of k-MT attachment status, Hec1 fits into the modest occupancy and highly regulated category. These data indicate that the correction of k-MT attachment errors responds to very small changes in phosphorylation, at least with respect to the phospho-occupancy of Hec1. As a mechanism of control, this indicates that phosphorylation displays properties of high sensitivity at the kinetochore–microtubule binding interface because small changes in phosphorylation are associated with substantial changes in microtubule attachment status (i.e., MT detachment). The system appears strongly biased toward maintaining low phospho-occupancy, which is important to both promote initial attachments and re-attachment following a detachment event related to error correction. Our data provide direct biochemical evidence for results based on amino acid substitution indicating that one to two phosphates per Hec1 molecule are sufficient for normal kinetochore/chromosome behavior, and that kinetochore behavior is most sensitive between zero and four phosphates (Zaytsev et al., 2015). Importantly, we demonstrate that Hec1 molecules are phosphorylated at partial occupancy over several sites, rather than an all-or-none at one to two sites. Tangibly, stoichiometric assessments show that single microtubule ends are surrounded by eight Hec1 molecules at kinetochores (Joglekar et al., 2006), and our data indicate that phosphorylation of as few as three of the eight molecules of Hec1 surrounding a given microtubule (37.5% phospho-occupancy) would be sufficient to favor k-MT detachment.
Materials and methods
Cell culture
HeLa cells (CCL-2) and RPE1 (CRL-4000) were obtained from the ATCC. All cells were grown at 37°C in a humidified environment with 5% CO2. The unmodified cell lines were grown in DMEM (#15-017-CM; Corning) containing 10% FCS (#82013-586; Hyclone), 250 µg/l Amphotericin B (#82026-728; VWR), 50 U/ml penicillin, and 50 µg/ml streptomycin (#15140122; Thermo Fisher Scientific). Doxycycline-inducible Hec1 knockout HeLa cells (gift of Iain Cheeseman, Massachusetts Institute of Technology, Cambridge, MA) were grown in media containing 10% tetracycline-free FBS (#1005807; Denville Scientific), the same antibiotics as for unmodified cells, 500 µg/ml G418 (#ant-gn-5; Invivogen), and 5 µg/ml puromycin (#A1113803; Thermo Fisher Scientific). HeLa Cyclin B1-GFP cells were grown in media containing 4 μg/ml blasticidin (# ant-bl-05; Invivogen). HeLa Cyclin A2-GFP cells (Alfonso-Pérez et al., 2019) were grown in media containing 9 μg/ml blasticidin (gifts of Francis Barr, University of Oxford, Oxford, UK). Cells were verified to be free of mycoplasma by frequent staining of plated cells with DAPI. No contamination was observed.
Inhibitors and reagents
The CENP-E inhibitor GSK-923295 (# HY-10299; MedChemExpress LLC) was used at 200 nM. NOC (#80058-500; VWR) was used at 100–500 ng/ml. STLC (#2191; Tocris) was used at 25 μM. ProTAME (Concept Life Sciences custom synthesis) was used at 25 μM. Okadaic acid (O-5857; LC Labs) was used at 200 nM. Doxycycline hyclate (#24390-14-5; Sigma-Aldrich) was used at 2 μg/ml. RO-3306 (#S7747; Selleck) was used at 10 μΜ. ΑΖ3146 (#3994/10; R&D Systems) was used at 2 μM. ΒΙ2536 (synthesized in-house) was used at 100 nM. MLN8237 (#S1133; Selleck) was used at 50 nM. ZM447439 (#2458/10; Tocris Bioscience) was used at 2 μM. Calyculin A (#9902S; Cell Signaling Technology) was used at 5 nm. Palbociclib (#PD-0332991; Selleck) was used at 500 nM.
Plasmids and cloning
pFETCh_Donor (EMM0021) was a gift from Eric Mendenhall and Richard M. Myers (plasmid # 63934; Addgene; HudsonAlpha Institute for Biotechnology, Huntsville, AL). pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang (plasmid # 48138; Addgene; Broad Institute, Massachusetts Intstitute of Technology). The Hec1-GFP Wt and 9A mutant plasmids were gifts from Jennifer DeLuca (Colorado State University, Fort Collins, CO). Additional mutations were generated via site-directed mutagenesis using the following primers:
S44 Wt/8A FD: 5′-GAGAAACCAACCTTTGGAAAGTTGAGTATAAACAAACCGGCCTCTGAAAGA-3′
RV: 5′-TCTTTCAGAGGCCGGTTTGTTTATACTCAACTTTCCAAAGGTTGGTTTCTC-3′
T31A FD: 5′-AAACAAGGCCTCTATGCCCCTCAAACCAAAGAGAAACC-3′
RV: 5′-GGTTTCTCTTTGGTTTGAGGGGCATAGAGGCCTTGTTT-3′
T31D FD: 5′-AAACAAGGCCTCTATGACCCTCAAACCAAAGAGAAACC-3′
RV: 5′-GGTTTCTCTTTGGTTTGAGGGTCATAGAGGCCTTGTTT-3′
S15A FD: 5′-GGTGCTGGCCGCCTCGCCATGCAGGAGTTAAGATCC-3′
RV: 5′-GGATCTTAACTCCTGCATGGCGAGGCGGCCAGCACC-3′
S15D FD: 5′-GGTGCTGGCCGCCTCGACATGCAGGAGTTAAGATCC-3′
RV: 5′-GGATCTTAACTCCTGCATGTCGAGGCGGCCAGCACC-3′
The DNA constructs were then verified by Sanger sequencing and amplified by maxiprep (#12162; Qiagen).
Transfections
100,000 HeLa-inducible Hec1 knockout cells were plated on glass coverslips in 12-well plates. After 24 h of plating, they were transfected with 1 μg plasmid DNA and 2.5 μl Lipofectamine 2000 (#11668027; Thermo Fisher Scientific) in a total of 1 ml Opti-MEM media (#31985070; Thermo Fisher Scientific). 4 h following transfection, the media was changed for DMEM containing doxycycline, puromycin, and G418. The cells were then assayed 48–72 h following transfection.
Mitotic error rate
100,000 HeLa inducible Hec1 knockout cells were plated on glass coverslips in 12-well plates. For determination of the error rate in asynchronous cells, the cells were fixed 72 h after transfection, stained with DAPI, and then analyzed. For determination of the error rate from STLC washout, the cells were transfected as described above, allowed to recover for 24 h, and then synchronized by thymidine addition for 24 h. The cells were then released into media containing STLC for 14 h. The cells were then released from STLC by two PBS washes and then fresh medium for 2.5 h before the cells were fixed.
CRISPR/Cas9 genomic editing and genotyping
We employed a strategy described previously (Savic et al., 2015) to tag Hec1 at the endogenous locus (NCBI accession no. NC_000018.10). HeLa cell genomic DNA was purified using a kit (#69504; Qiagen). The homology arms were then amplified by PCR using NEB Phusion polymerase (#M0530S) using the following primers:
Hom1 FD: 5′-TCCCCGACCTGCAGCCCAGCTGATAAATACGAAGGTGAAAACCAGG-3′
Hom1 RV: 5′-CCGGAACCTCCTCCGCTCCCTTCTTCAGAAGACTTAATTAGAG-3′
Hom2 FD: 5′-AGTTCTTCTGATTCGAACATCGAATAAAATTGTCTCAGTAAAGTG-3′
Hom2 RV: 5′-TGGAGAGGACTTTCCAAGGGATTTGGCTCACATGATGTAGG-3′
A donor plasmid was then created via Gibson assembly according to the manufacturer protocol (#E2611S; New England Biolabs). Separately, a vector expressing the gRNA and Cas9 was created as previously described (Ran et al., 2013) using the following DNA fragments:
5′-CACCGTGAGACAATTTTATTCACTA-3′
5′-AAACTAGTGAATAAAATTGTCTCAC-3′
The DNA constructs were then verified by sequencing and amplified by maxiprep. 200,000 HeLa cells were then plated on a 6-well plate. 24 h later they were transfected with 2 μg donor plasmid, 1 μg gRNA/Cas9 plasmid, and 7.5 μl lipofectamine 2000 in 2 ml opti-MEM media. 4 h following transfection, the media was changed for DMEM, and 24 h later, G418 was added to select for successfully edited cells. Once the cells reached colonies of 50–100 cells, they were trypsinized and plated on 96-well plates at a density of 0.5 cells per well. Single-cell clones were then amplified and tested for expression of the edited gene via IF and Western blot. Genotyping of Hec1-3XFLAG cells was performed using the following primers:
FD: 5′-AAGAATGCATGTCAGAAGATCTCTCG-3′
RV: 5′-CAACCTGTATTTTGCCGACTTCTGG-3′
Genotyping was performed using 200 ng of genomic DNA purified as above and NEB Phusion polymerase in a total of 25 μl using the supplied buffers. The PCR reaction conditions were 98°C for 3′, 98°C for 30″, 55°C for 30″, 72°C for 3′, 40 cycles, 72°C for 5’. PCR reactions were then run on 1% agarose gel stained with ethidium bromide and imaged.
Purification of Hec1-3XFLAG
HeLa Hec1–3XFLAG cells were expanded to 16–15 cm dishes. When they reached 70–80% confluency, Thymidine (#T1895-5G; Sigma-Aldrich) was added to a concentration of 2.5 mM. After 24 h, the cells were washed twice with PBS and released into NOC or STLC. The cells were then harvested 14 h after release. For proTAME synchronization, the cells were released from thymidine as above, but the drug was added 8 h later for a further 4 h before harvesting. Mitotic cells were then harvested by washing the detached and semi-detached cells off the plate by pipetting. The cells were then washed with PBS and then divided by 60–40% for subcellular fractionation and total protein purification, respectively. Cells used for total protein purification were lysed with 2 ml of a buffer containing 1% SDS; #436143; Sigma-Aldrich), 50 mM Tris pH 7.5, and 1 mM EDTA. The cells were then crudely homogenized using a 5-ml syringe and 18-gauge needle. The lysate was then immediately boiled for 5 min. After cooling, the DNA was sheared until the viscosity was acceptably reduced in a 15-ml tube using a 5-ml syringe and 18-gauge needle. The SDS was sequestered by the addition of 13 ml extraction buffer (3% Triton X-100 [#T8787; Sigma-Aldrich], 50 mM Tris, pH 7.5, 250 mM NaCl, and 1 mM EDTA), and cooled on ice. Concurrently, subcellular fractionation was performed on the remaining cells by suspending them in 10 ml cold hypotonic buffer (10 mM Tris, pH 7.5, 10 mM KCl, 1 mM MgCl2, 10 mM glycerol 2-phosphate [#G9422; Sigma-Aldrich], and 1 mM EDTA) on ice, followed by douncing every 5 min. After 20 min, the lysate was aliquoted into 1.5 ml tubes and centrifuged at 4°C at maximum speed for 20 min. The supernatant containing soluble protein was then removed, SDS added to a concentration of 0.2% and boiled for 5 min. After cooling to room temperature, 5 ml of 3× extraction buffer was added to the soluble fraction, which was set aside on ice. The chromatin-containing pellets were crudely homogenized via a syringe in 2 ml SDS lysis buffer and boiled for 5 min. The DNA was then sheared via a syringe in a 15-ml tube. The SDS was then sequestered by adding 13 ml extraction buffer and cooled on ice. All three fractions were then centrifuged at maximum speed in a benchtop centrifuge for 10 min to remove any remaining cell debris from the lysates. Then, 30 μl of packed M2 anti-FLAG affinity gel (#A2220-5ML; Sigma-Aldrich) was added to the lysates, which were then incubated overnight on a rotating platform at 4°C. The affinity gel was then transferred to a 1.5-ml tube and washed twice with extraction buffer. The volume of extraction buffer was then reduced to 30 μl, and 70 μl water and 1 mM MnCl2 were added together with 1 μl DNaseI (#M0303S; NEB) for 10 min at room temperature to degrade any DNA that precipitated overnight. Any separable debris was aspirated using a 30-gauge needle. The affinity gel was then washed an additional three times with cold extraction buffer, incubating for 10 min between each wash. The gel was then dried using a 30-gauge needle connected to a vacuum line. To elute the purified protein, the affinity gel was suspended in elution buffer (2% SDS, 50 mM Tris, pH 7.5, 50 mM NaCl, 20% glycerol, and 2 mM dithiothreitol (#D0632-5G; Sigma-Aldrich; fresh) and incubated at 65°C for 20 min. The samples were then cooled to room temperature. Iodoacetamide (#I6125-5G; Sigma-Aldrich) was added at a concentration of 6 mM, vortexed, and the samples incubated in the dark for 1 h. Then, an additional 2 mM dithiothreitol was added and incubated for 15 min. The samples were then run on 8% SDS-PAGE gel and stained with Coomassie blue. The gels were imaged with a Gel Doc Ez (#1708270; Bio-Rad).
SDS-PAGE and Western blot
Gels and blots were performed as described previously (Kucharski et al., 2017). Prior to loading the samples on the gel, 4× Laemmli buffer (200 mM Tris, pH 6.8, 4% SDS, 40% glycerol, 4% 2-mercaptoethanol, and 0.12 mg/ml bromophenol blue) was added to a final dilution of 1×, and the samples were boiled for 5 min. Proteins were separated on SDS-PAGE gel using stacking gel (4% 29:1 acrylamide: Bis-acrylamide, 125 mM Tris, pH 6.8, 0.1% SDS, 0.1% ammonium persulfate, and 0.1% TEMED and 8–15% 29:1 acrylamide: Bis-acrylamide, 400 mM Tris pH 8.8, 0.1% SDS, 0.1% ammonium persulfate, and 0.1% TEMED) at 120 V until the bromophenol blue has run off or longer, as needed. Transfer onto nitrocellulose membrane was performed for at least 24 h at 30 V under wet conditions in 1× transfer buffer (14.4 g/l glycine, 3.0 g/l Tris, and 20% methanol). Conditions for Western blots include the use of 5% nonfat dry milk in TBS-T 0.5% (50 mM Tris, pH 7.2, 150 mM NaCl, and 0.5% Tween 20) for blocking and TBS-T 0.5% for washing. Blocking and primary antibody incubation were performed in 5% BSA for the anti-pT31 Hec1 antibody. Four washes for 10 min each were performed after primary and secondary antibody incubation periods. The bands were visualized by enhanced chemiluminescence using Clarity (#1705060; Bio-Rad) and detected on film or using fluorescent secondary antibodies in the LI-COR Odyssey-CLX system.
MS for phospho-occupancy
Gel bands were excised, destained to clarity, digested using trypsin (#V5113; Promega), and the resulting peptides were extracted as described previously (Lundby and Olsen, 2011). Each sample was desalted by STAGE tip (Rappsilber et al., 2007) and then dried by vacuum centrifugation. Samples were resuspended in 220 μl of buffer (100 mM TEAB [#17902; Sigma-Aldrich], 10 mM MgCl2, and 100 mM NaCl) by vortexing and split into pairs of 100 μl each. One of each pair was treated with 0.5 μl of CIP (#M0290; New England Biolabs) before the pair was incubated at 37°C for 1 h. Since the Aurora kinases phosphorylate proteins adjacent to lysine and arginine residues, which also represent trypsin cleavage sites, both CIP-treated and untreated samples were then re-digested with an additional 0.1 μg trypsin overnight at 37°C to prevent missed cleavages due to protein phosphorylation. The CIP-treated and untreated samples were then labeled by heavy and light reductive dimethyl labeling, respectively, as previously described (Boersema et al., 2009; Kettenbach et al., 2015; #252549, #156159, #190090, Sigma-Aldrich; #CDLM-4599-1, Cambridge Isotope Laboratories). After labeling, the heavy and light pairs were mixed, desalted together by STAGE tip, and dried by vacuum centrifugation. Dried samples were resuspended in 5% methanol (#34860; Sigma-Aldrich) /1.5% formic acid (# 64-18-6; Honeywell), and analyzed by online microcapillary liquid chromatography–tandem MS using either a Thermo Fisher Scientific QE +, Orbitrap Fusion, or an Orbitrap Lumos nLC-MS/MS platform as previously described (Grassetti et al., 2017). The mass spectra were searched using the COMET search algorithm (Eng et al., 2013) to generate peptide spectral matches as previously described for dimethyl labeled samples (Kettenbach et al., 2015). Peptide spectral matches were filtered to a ∼1% FDR using the target-decoy strategy (Elias and Gygi, 2007) and reported. Peptide masses and retention times were then used to manually quantify the extracted ion chromatograms of unphosphorylated heavy and light labeled pairs in all runs using Xcalibur Qual Browser software (Thermo Fisher Scientific). Phosphorylation occupancy was calculated for candidate sites using these areas as described previously (Wu et al., 2011).
RNA interference
Gene knockdown was performed as described previously (Kucharski et al., 2017) with the following siRNAs purchased from Sigma-Aldrich:
Cyclin B1: 5′-GACCAUGUACAUGACUGUCUC[dT][dT]-3′
Cyclin A2: 5′-UAUACCCUGGAAAGUCU[dT][dT]-3′
BubR1: 5′-AAAGAUCCUGGCUAACUGUUC[dT][dT]-3′
B56 Alpha: 5′-UGAAUGAACUGGUUGAGUA[dT][dT]-3′
B56 Beta: 5′-GAACAAUGAGUAUAUCCUA[dT][dT]-3′
B56 Gamma: 5′-GGAAGAUGAACCAACGUUA[dT][dT]-3′
B56 Delta: 5′-UGACUGAGCCGGUAAUUGU[dT][dT]-3′
B56 Epsilon: 5′-GCACAGCUGGCAUAUUGUA[dT][dT]-3′
NSC: 5′-AAUUCUCCGAACGUGUCACGU[dT][dT]-3′
Immunofluorescence
Following treatments, cells were fixed in methanol at −20°C except for experiments using GFP constructs, cold stable MT assays, and those involving Cyclins A2 or B1, which were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were pre-extracted in PHEM buffer (60 mM Pipes, 25 mM Hepes, pH 6.9, 10 mM EGTA, 4 mM MgSO4, 1% Triton X-100, and 10 mM glycerol 2-phosphate) for 10 min at 4°C. The remaining steps were performed at room temperature. Experiments involving Cyclin A2 or Cyclin B1 were not pre-extracted and instead were permeabilized post-fixation with PHEM buffer for 10 min. The cells were then re-hydrated in PBS for 10 min before blocking with TrueBlack IF blocking buffer (#23012B; Biotium) for 10 min. Primary antibodies were then diluted in Trueblack and used at the indicated dilution. Secondary antibodies were diluted in 10% FCS, 0.1% Triton X-100 in PBS. After incubation with antibodies, the cells were washed three times with PBS. Finally, the cells were counterstained with DAPI and mounted on slides using ProLong Gold antifade reagent (#P36934; Thermo Fisher Scientific).
Microscopy
Images were acquired with a Nikon Eclipse Ti microscope equipped with a cooled charge-coupled device Clara camera (Andor Technology) controlled by Nikon NIS-Elements software version 4.30.02. Images were acquired in 0.15–0.4 μm sections using a plan apo 1.4 numerical aperture 100× (kinetochore analysis) or 60× (lagging chromosome analysis) oil-immersion objective using 1 × 1 binning. Samples were illuminated using an X-cite light source (Excelitas Technologies Corp). All image analysis, adjustment, and cropping were performed using Fiji software (Schindelin et al., 2012). Image deconvolution was performed using Nikon Elements batch deconvolution software version 5.21.00 on automatic mode. All kinetochore intensity analysis was performed on the raw images, except for Figs. 5, A–C; Fig. 7, F and G; Fig. 9, E and F; and Fig. S5, E and F for which deconvolution was performed before analysis. Line scans were performed on the deconvoluted images in Fiji and the data was then exported to Graphpad Prism. All images selected for presentation are deconvolved maximum-intensity Z-stack projections. Where shown, insets are from single optical Z-slices. Images were selected to represent the mean quantified data. Quantification of kinetochore staining intensity was performed by first determining the brightest plane for which a given kinetochore appears. Then, an outline was drawn using the ellipse tool. The integrated intensity was determined by multiplying the average intensity by the area of the kinetochore. Background levels of staining were determined by saturating the brightness and contrast settings to find the darkest spot within the chromatin-containing area and were subtracted from the intensity of the kinetochore. Lamin A/C pS22 was quantified by first creating a sum projection of the image. Then, an outline was drawn around the whole cell, and the fluorescence intensity was measured. The background-subtracted, normalized integrated intensities for all cells were then plotted. To quantify levels of cold-stable microtubules, images were first deconvolved. Then, A sum-intensity projection was created in Fiji. Viewing the ACA channel, a rectangle was drawn that was 2 μm wide in the pole–pole axis, and as long as the furthest centromeres in the metaphase plate. Then, switching to the tubulin channel, the average tubulin intensity was measured in the rectangle. From the average intensity value, the minimum intensity value was used as the background level and subtracted. The background-subtracted intensity values were then averaged across all of the cells that were analyzed for each condition and experimental repeat.
Antibodies
The following antibodies were used for immunofluorescence (IF) and/or immunoblotting (IB): human anti-ACA (Geisel School of Medicine; IF at 1:2,000), mouse anti-Hec1 (C-11; IF,IB at 1:1,000; Santa Cruz), rabbit anti-PP2A B56 Alpha (https://www.MyBioSource.com; MBS8524809; IB at 1:500), mouse anti-PP2A B56 Beta (E-6; IB at 1:500; Santa Cruz), mouse anti-PP2A B56 Gamma (A-11; IB at 1:500; Santa Cruz), mouse anti-PP2A B56 Delta (H-11; IB at 1:500; Santa Cruz), rabbit anti-PP2A B56 Epsilon (RP56694_P050; IB at 1:500; Aviva Systems Biology), mouse anti-Nuf2 (E-6; IB at 1:1,000; Santa Cruz), rabbit anti-Hec1 pS44 (gift of Jennifer DeLuca; IF at 1:500), mouse anti-GAPDH (G-9; IB at 1:1,000; Santa Cruz), rabbit anti-Histone H3 pS10 (#9701; IB at 1:2,000; Cell Signaling Technology), mouse anti-FLAG M2 (IF and IB at 1:1,000; Sigma-Aldrich), mouse anti-Cyclin A2 (GT2547; IF at 1:1,000; Genetex), mouse anti-Cyclin B1 (GNS1; IF at 1:500; Santa Cruz), mouse anti-α-Tubulin DM1α (IF at 1:4,000-1:10,000; Sigma-Aldrich), rabbit anti-Lamin A/C pS22 (#2026S; IF at 1:250; Cell Signaling Technology), rabbit anti-Ska3 (#91559; IF at 1:1,000; Abcam). The rabbit anti-Hec1 pT31 antibody was generated by Pacific Immunology where two rabbits were immunized with the peptide corresponding to amino acids 25–37: Cys-NKQGLY-pT-PQTKEK, followed by affinity purification against the non-phospho and phospho peptides (IF at 1:100,000, WB at 1:1,000). Secondary antibodies used were highly cross-adsorbed Alexa Fluor 488, 594, and 647 raised in donkey or goat (IF at 1:1,000; Invitrogen), horseradish peroxidase (IB at 1:10,000-10,0000; Bio-Rad), Mouse TrueBlot ULTRA: rat Anti-Mouse IgG (18-8817-33; IB at 1:1,000; Rockland), LI-COR IRDye 800CW Goat anti-Mouse IgG (H + L), LI-COR IRDye 800CW Goat anti-Rabbit IgG (H + L), LI-COR IRDye 680RD Goat anti-Mouse IgG (H + L), LI-COR IRDye 680RD Goat anti-Rabbit IgG (H + L; 1:10,000–1:40,000).
In vitro kinase assay
Recombinant cyclin B1-CDK1-Cks1 (Cdk1-CycB) was expressed and purified as described previously (Brown et al., 2015). To purify the Aurora B/Inbox complex, BL21 Rosetta (DE) cells were co-transformed with two plasmids (kindly gifted by the Arockia Jeyaprakash lab) carrying either the sequence of 6× Histidine-SUMO N-terminally tagged Aurora B55-344 or the C-terminus of INCENP (INCENP835-903). Cultures were grown at 37°C up to a density of ∼0.5 OD, after which, the temperature was decreased to 18°C, and the expression was induced with 350 µM IPTG for 18 h. Cell pellets were then lysed using a bio-disruptor in a lysis buffer (25 mM Hepes, 500 mM NaCl, 25 mM Imidazole, 2 mM β-mercaptoethanol, 1 × cOMPLETE protease inhibitor cocktail [#11836170001; Sigma-Aldrich], and 50 U Benzonase [#70664-10KUN; EMD Millipore]; pH 7.5) and spun at 22,500 rpm for 50 min at 4°C. To pull down the Aurora B/Inbox complex, the supernatant was incubated with Ni-NTA agarose beads (#25214; ThermoFisher Scientific) for 2 h at 4°C. This was followed by washes with the lysis buffer, a chaperone buffer (25 mM Hepes, 1,000 mM NaCl, 30 mM Imidazole, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, and 2 mM β-mercaptoethanol; pH 7.5) to remove non-specifically bound proteins and then a low salt buffer (25 mM Hepes, 200 mM NaCl, 25 mM Imidazole, and 2 mM β-mercaptoethanol; pH 7.5). Aurora B/Inbox was then eluted from the beads by an incubation with Imidazole elution buffer (25 mM Hepes, 200 mM NaCl, 500 mM imidazole, and 2 mM β -mercaptoethanol; pH 7.5) for 2 h at 4°C that was followed by four elutions more. Representative samples from the elution fractions were evaluated by SDS-PAGE, and those containing the two subunits of the complex were pooled together and desalted in a HiPrep 26/10 Sephadex G-25 fine resin-packed desalting column (Cytiva) fitted on ÄKTA liquid chromatography instrument and eluted in dialysis buffer (25 mM Hepes, 200 mM NaCl, and 4 mM DTT; pH 7.5). To cleave the 6× Histidine-SUMO tag, the desalted Aurora B/Inbox complex was incubated overnight at 4°C with SENP2 protease (kindly gifted by the Arockia Jeyaprakash lab). For the final purification step, the cleaved Aurora B/Inbox was concentrated into 300 µl, loaded into a Superdex 75 10/300 column (Cytiva) pre-equilibrated with 25 mM Hepes, 200 mM NaCl, 4 mM dithiothreitol (DTT) and 5% glycerol; pH 7.5. Elution fractions were identified by SDS-PAGE then snap frozen in liquid nitrogen and stored at −80°C until the complex was used in the in vitro kinase experiments. The pST39-Hec1 “Broccoli” vector containing Hec11-506 and Nuf21-348 (gift from I. Cheeseman, Whitehead Institute, Cambridge, MA; Schmidt et al., 2012) was expressed in BL21 Rosetta (DE) cells using 2XTY media. Cultures were grown at 37°C until OD 0.8 and induced overnight with 0.35 mM IPTG at 18°C. Cells were lysed in lysis buffer (50 mM potassium phosphate, pH 8, 300 mM NaCl, 35 mM imidazole, and 2 mM β-mercaptoethanol) and purified using 5 ml of Ni-NTA agarose beads (Qiagen). The protein-bound beads were washed with 50 column volumes (cv) of lysis buffer and 20 cv of high salt buffer (20 mM Tris-HCl, pH 8, 1 M NaCl, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, 35 mM imidazole, and 2 mM β-mercaptoethanol), followed by a 5 cv wash with lysis buffer. Hec1 and Nuf2 proteins were eluted with elution buffer (50 mM potassium phosphate, pH 8, 300 mM NaCl, 400 mM imidazole, and 2 mM β-mercaptoethanol) and dialyzed overnight against 20 mM Hepes, pH 8, 150 mM NaCl, 1 mM DTT, and 5% glycerol. The protein complex was further purified by anion exchange chromatography (HiTrap Q HP; Cytiva) and collected in the flow-through. The complex was then concentrated using a Vivaspin centrifugal concentrator with a 10 kD molecular weight cut-off (Sartorius) and loaded onto a Superdex 200 increase 10/300 column (Cytiva) preequilibrated with 20 mM Hepes, pH 7.5, 250 mM NaCl, 1 mM DTT, and 5% glycerol. Peak fractions were pooled, concentrated using the Vivaspin centrifugal concentrator, and snap-frozen at a concentration of 2 mg/ml. Cyclin B1-CDK1(D146N)-Cks1 (kinase dead CDK mutant, Cdk1-CycB-KD) was purified and assembled in the same manner as Cdk1-CycB, but without CAK activation. Hec1Broccoli was incubated with ATP and each of the recombinant kinases: Cdk1-CycB, Cdk1-CycB-KD, and AurB. Reactions (n = 3) were incubated for 30 min at 21 and 30°C for the CDK complexes and AurB, respectively. Reactions were then stopped by the addition of 2% SDS. Reaction products were precipitated by acetone–ethanol, double digested in solution with trypsin (2 × 1:50) in 0.1 M triethylammonium bicarbonate (TEAB), and desalted using home-made C18 StageTips (Rappsilber et al. 2007). Peptides were analyzed by LC-MS/MS using an Ultimate 3000 RSLCnano HPLC (Dionex) coupled to an Orbitrap Eclipse (Thermo Fisher Scientific) instrument using an EasySpray source. Peptides were separated using a 50 cm C18 EasySpray column on a 20 min, two-step linear gradient, starting from 2 to 8% over 6 min, and then to 60% over 14 min (A: 0.1% formic acid, B: 80% ACN +0.1% formic acid). Peptides were analyzed using a data-dependent acquisition method. The duty cycle consisted of a MS1 Orbitrap scan at 120k resolution, followed by isolation, HCD fragmentation on the top 20 peptide-like precursors having charge states between +2 and +7, and mass analysis using the linear ion trap in rapid scan mode. Peptide identification and quantitation was performed using MaxQuant v2.0.3.0 (Cox and Mann, 2008) using a FASTA database containing human Hec1(NDC80), Nuf2, and common contaminant proteins, at 1% FDR. Raw summed label-free intensities from three repeats are shown.
Statistical analysis
All statistical tests were performed using Graphpad Prism version 8.4.2. Except where indicated, all experiments were performed as three independent biological repeats. In most IF experiments, 20 cells per repeat were analyzed, and 25 kinetochores for each cell were measured. Experiments with other numbers are noted in the figure legends. For anaphase error rates, 120 anaphases per condition were assessed for each of the three independent biological experiments. Statistical significance was calculated between indicated conditions using two-tailed t tests. For these tests, the data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 (in support of Fig. 1) shows that the addition of a 3XFLAG tag to the c-terminus of Hec1 does not perturb mitotic progression in HeLa cells. Fig. S2 (in support of Fig. 1) shows that Hec1 exists in soluble and insoluble forms in mitotic cells and that both fractions are similarly phosphorylated. Fig. S2 also shows effective synchronization of cells under the conditions used for experimentation. Fig. S3 (in support of Fig. 2) shows that the antibody raised against pT31 Hec1 is specific for Hec1 phosphorylation at that site. Fig. S3 also shows that Hec1 is phosphorylated on T31 in RPE1 cells similarly to HeLa cells and that Hec1 is maximally phosphorylated under the STLC synchronization condition. Fig. S4 (in support of Fig. 3 and Fig. 8) shows that knockdown of Cyclin A2 does not significantly affect Cyclin B1 function. Fig. S4 also shows that treatment of cells with Calyculin A increases levels of Hec1 phosphorylation. Fig. S5 (in support of Figs. 4, 9, and 10) shows that knockdown of BubR1 reduces phosphatase activity against Hec1. It also shows cartoons of the Hec1 constructs used in Figs. 5 and 10. Table S1 summarizes MS data in support of Fig. 1 and Fig. S2.
Data availability
Acknowledgments
We thank Jennifer DeLuca, Francis Barr, and Iain Cheeseman for providing reagents. We also thank the members of the Compton laboratory for helpful discussions and feedback. We thank Ann Lavanway, Zdenek Svindrych, and the Dartmouth College Life Sciences Light Microscopy Facility for assistance with microscopy and image deconvolution.
Recombinant cyclin B-CDK1 was a gift from Jane Endicott and Svitlana Korolchuk (University of Newcastle, Tyne, UK), supported by the MRC MR/N009738/1. T.J. Kucharski was supported by fellowships from the Fonds de Recherche du Québec—Santé and the Canadian Institute of Health Research. This work was also supported by grants from the National Institutes of Health # P20-GM113132 to the BioMT facility of Dartmouth College, #GM122846 to S.A. Gerber, #1R01HD101436 to K.M. Godek and #GM051542 to D.A. Compton, Wellcome and the Royal Society grants 206211/A/17/Z to T. Ly, Wellcome 202811 to A.A. Jeyaprakash, the Darwin Trust of Edinburgh to A. Al-Rawi, Wellcome PhD programme funding (218520/Z/19/Z) to E. Kaye. The authors acknowledge the following Shared Resources facilities (LI-COR Odyssey CLX) at the Norris Cotton Cancer Center at Dartmouth with NCI Cancer Center Support Grant 5P30 CA023108-40.
The authors declare no competing financial interests.
Author contributions: Conceptualization: D.A. Compton and T.J. Kucharski. Investigation: T.J. Kucharski, R. Hards, S.E. Vandal, A. al-Rawi . Formal analysis: T.J. Kucharski, R. Hards, A. al-Rawi , T. Ly. Methodology: T.J. Kucharski, K.M. Godek, R. Hards, M.A. Abad, S.A. Gerber, T. Ly, D.A. Compton. Funding acquisition: T.J. Kucharski, T. Ly, A.A. Jeyaprakash, E. Kaye, K.M. Godek, S.A. Gerber, D.A. Compton. Resources: M.A. Abad, A.A. Jeyaprakash, E. Kaye. Supervision: S.A. Gerber, T. Ly, D.A. Compton. Writing- original draft: T.J. Kucharski. Writing- review & editing: T.J. Jucharski, K.M. Godek, D.A. Compton. Project administration: D.A. Compton.