Deubiquitinating enzymes (DUBs) negatively regulate protein ubiquitination and play an important role in diverse physiological processes, including mitotic division. The BRCC36 isopeptidase complex (BRISC) is a DUB that is specific for lysine 63–linked ubiquitin hydrolysis; however, its biological function remains largely undefined. Here, we identify a critical role for BRISC in the control of mitotic spindle assembly in cultured mammalian cells. BRISC is a microtubule (MT)-associated protein complex that predominantly localizes to the minus ends of K-fibers and spindle poles and directly binds to MTs; importantly, BRISC promotes the assembly of functional bipolar spindle by deubiquitinating the essential spindle assembly factor nuclear mitotic apparatus (NuMA). The deubiquitination of NuMA regulates its interaction with dynein and importin-β, which are required for its function in spindle assembly. Collectively, these results uncover BRISC as an important regulator of the mitotic spindle assembly and cell division, and have important implications for the development of anticancer drugs targeting BRISC.
Introduction
The mitotic spindle is a bipolar array of microtubules (MTs) required for the symmetrical distribution of chromosomes to each daughter cell (Merdes et al., 2000; Silk et al., 2009). The process of bipolar spindle formation is controlled by both the centrosome- and chromatin-mediated pathways. Whereas the minus ends of spindle MTs cluster together at the spindle poles, their plus ends grow toward the cell equator and capture the kinetochores (Gadde and Heald, 2004; Wong et al., 2006; Radulescu and Cleveland, 2010). Ubiquitination is a widespread modification that ensures fidelity of mitotic progression (Fournane et al., 2012). Ubiquitination is highly dynamic and reversible, and is determined by ubiquitin ligases and deubiquitinating enzymes (DUBs) (Komander et al., 2009; Komander and Rape, 2012). Despite recent advances in our understanding of the E3 ubiquitin ligases, the precise roles and substrate specificity of DUBs in the regulation of mitosis are only beginning to be understood (Fournane et al., 2012).
BRCC36 was identified as a component of the BRCA1–BRCA2-containing complex (BRCC) (Dong et al., 2003). It is a JAMM/MPN+-containing DUB that preferentially cleaves K63-linked polyubiquitin chains (K63Ubs) (Cooper et al., 2009) and exists in at least two distinct complexes, the Rap80 complex (also called the BRCA1-A complex) and the BRCC36 isopeptidase complex (BRISC) (Feng et al., 2010; Hu et al., 2011). The Rap80 complex consists of five proteins (Rap80, BRCC36, MERIT40/NBA1, BRE/BRCC45, and Abraxas) and has been shown to disassemble K63Ub upon targeting to DNA double-strand breaks (Sobhian et al., 2007; Feng et al., 2009; Shao et al., 2009b; Wang et al., 2009). The BRISC complex contains four stoichiometric subunits: ABRO1/KIAA0157, BRCC36, MERIT40/NBA1, and BRCC45/BRE (Cooper et al., 2009; Feng et al., 2010; Hu et al., 2011). BRCC36 and ABRO1 are the two most important components, as they control BRISC DUB activity and cytoplasmic localization, whereas the other two contribute to the integrity and stability of the complex (Cooper et al., 2010; Feng et al., 2010; Hu et al., 2011). The biochemical activity of BRISC has been well characterized, and it has been shown to function as a DUB that specifically cleaves K63Ubs (Cooper et al., 2009, 2010). BRISC was recently shown to deubiquitinate IFNAR1 and thereby regulate interferon response (Zheng et al., 2013); however, its biological function during cell division is largely undefined.
Here, we report that BRISC ensures the fidelity of mitosis by regulating mitotic spindle assembly. We provide evidence that BRISC is a MT-associated protein (MAP) with a unique localization during mitosis and that the DUB activity of BRISC is essential for the spindle assembly, by specifically removing K63Ubs from nuclear mitotic apparatus (NuMA), one of the most important spindle assembly factors (SAFs), thus regulating the interaction of NuMA with its partners, dynein and importin-β, thereby promoting proper bipolar spindle assembly.
Results
BRISC is important for normal mitosis in mammalian cells
To investigate the function of BRISC, we inhibited its expression by using two individual siRNAs specific for each of the BRISC components, including ABRO1, BRCC36, and MERIT40, respectively. The RNA interference efficiency was confirmed by Western blotting and immunofluorescence, using an antibody against the C terminal of ABRO1 peptide (261–415 aa) or antibodies against BRCC36/MERIT40 generated using a method described previously (Sobhian et al., 2007; Shao et al., 2009b) (Fig. S1 A and Fig. 1, A–C). Each of these siRNAs efficiently silenced the corresponding protein expression in HeLa cells and were both used in the experiments with consistent results (Fig. S1 A).
BRISC is important for normal mitosis in mammalian cells. (A–C) Mitotic defects in ABRO1 siRNA-transfected HeLa cells. Cells transfected with control or ABRO1 siRNA were fixed in cold methanol and immunostained for α-tubulin (green) and ABRO1 (red); DNA was stained with DAPI. ABRO1 was efficiently silenced. Spindle structures were categorized based on the stage of mitosis. Bars, 5 µm. (A) Representative images of multipolar defects in ABRO1-depleted metaphase cells. (B) Representative examples of lagging chromosomes and aberrant cytokinesis in ABRO1-depleted anaphase and telophase cells, respectively. (C) Representative images of multinuclei defects in ABRO1-depleted postmitotic interphase cells. (D–F) Quantitative analysis of the mitotic spindle structures that are shown in A–C. Error bars show mean ± SD of three independent experiments. **, P < 0.01; ***, P < 0.001; Student’s t test. (G) Representative images of the rescue efficiency in CFP-ABRO1res–transfected metaphase cells. ABRO1 siRNA-transfected HeLa cells were cotransfected with plasmid CFP-ABRO1 WT, which is sensitive to ABRO1 siRNA, or CFP-ABRO1res, which is resistant to ABRO1 siRNA. The mitotic defects shown in A–C were observed. Bars, 5 µm. (H) Quantification of the mitotic spindle structures shown in G. Data from three independent experiments. ***, P < 0.001; Student’s t test. (I) Protein samples from cells treated as shown in H were analyzed by Western blotting. Actin was used as a loading control. Error bars show mean ± SD.
BRISC is important for normal mitosis in mammalian cells. (A–C) Mitotic defects in ABRO1 siRNA-transfected HeLa cells. Cells transfected with control or ABRO1 siRNA were fixed in cold methanol and immunostained for α-tubulin (green) and ABRO1 (red); DNA was stained with DAPI. ABRO1 was efficiently silenced. Spindle structures were categorized based on the stage of mitosis. Bars, 5 µm. (A) Representative images of multipolar defects in ABRO1-depleted metaphase cells. (B) Representative examples of lagging chromosomes and aberrant cytokinesis in ABRO1-depleted anaphase and telophase cells, respectively. (C) Representative images of multinuclei defects in ABRO1-depleted postmitotic interphase cells. (D–F) Quantitative analysis of the mitotic spindle structures that are shown in A–C. Error bars show mean ± SD of three independent experiments. **, P < 0.01; ***, P < 0.001; Student’s t test. (G) Representative images of the rescue efficiency in CFP-ABRO1res–transfected metaphase cells. ABRO1 siRNA-transfected HeLa cells were cotransfected with plasmid CFP-ABRO1 WT, which is sensitive to ABRO1 siRNA, or CFP-ABRO1res, which is resistant to ABRO1 siRNA. The mitotic defects shown in A–C were observed. Bars, 5 µm. (H) Quantification of the mitotic spindle structures shown in G. Data from three independent experiments. ***, P < 0.001; Student’s t test. (I) Protein samples from cells treated as shown in H were analyzed by Western blotting. Actin was used as a loading control. Error bars show mean ± SD.
The effect of BRISC depletion on cell cycle progression was first examined in HeLa cells. Depletion of ABRO1 induced a significant decrease in the percentage of cells in the G1 phase and an increase of cells in the G2/M phase, as compared with the control cells (Fig. S1 B). The protein level of cyclin B and cyclin D1 was increased, whereas the protein level of p27 was decreased in ABRO1-silenced cells, helping to promote the cell cycle into the G2/M phase, which is consistent with the previous report (Zhang et al., 2014) (Fig. S1 C). The percentage of mitotic cells determined by immunofluorescence staining of phospho-histone H3 (pH3) was also higher in ABRO1-silenced cells than that in control. Similar results were observed when other BRISC components including BRCC36 and MERIT40 were knocked down with siRNAs (Fig. S1, B–D). Collectively, these data suggest that BRISC plays an important role in mitosis.
The mitotic role of BRISC was further confirmed by indirect immunofluorescence microscopy examination. The majority of ABRO1-depleted cells delayed in mitosis displayed multiple defects, which were categorized and calculated based on the stage of mitosis and the extent to which they were defective (Fig. 1, A–F). Depletion of ABRO1 reduced the abundance of normal metaphase cells with bipolar spindles and properly aligned chromosomes, whereas it induced a higher proportion of mitotic cells with multipolar spindles (30 ± 1.4% vs. 6.5 ± 1.1%) (Fig. 1, A and D). In addition, ABRO1 siRNA-treated cells exhibited severe defects in chromosome segregation during anaphase and a dramatic increase in the number of lagging chromosomes (Fig. 1, B, the left two panels, and E). At telophase, some lagging chromosomes became enclosed in nuclear envelopes, giving rise to multinuclei (Fig. 1, B, the right two panels, and E). Furthermore, knockdown of ABRO1 led to an increase of binucleated and multinucleated postmitotic cells (Fig. 1, C and F), which may be a consequence of spindle pole defects (Fant et al., 2004) or cytokinesis failure (Fededa and Gerlich, 2012). Similar results were observed when other BRISC components including BRCC36 and MERIT40 were knocked down with siRNA or shRNA (Fig. S1, E and F). To further confirm that the mitotic defects in siABRO1-treated cells were specifically caused by ABRO1 depletion, we generated a siRNA-resistant ABRO1 plasmid (CFP-ABRO1res) for rescue experiments. Reintroduction of CFP-ABRO1res remarkably rescued the mitotic defects caused by siABRO-1 treatment, which depleted endogenous but not the ectopic expressed CFP-ABRO1 (Fig. 1, G–I). These results demonstrate an important role for ABRO1 in mitosis.
Multipolar spindle could be a result of the amplification of centrosomes. To exclude this possibility, we compared the number of centrosomes in control and ABRO1-siRNA–depleted cells, and no significance was found (Fig. S1 G).
BRISC is required for proper spindle assembly
To explore the effects of BRISC depletion on mitosis, we followed spindle assembly by using live-cell imaging in H2B-GFP/tubulin-mRFP HeLa cells, 48 h after transfection with control or ABRO1 siRNA. Control cells (n = 37) built a spindle, visualized with tubulin-mRFP, and divided in less than 1 h (Fig. 2, A and B; and Video 1). In contrast, ABRO1-silenced cells (n = 96) took a longer time to assemble the spindles (Fig. 2, A and B). 22.9% (22/96) of the cells examined had a prolonged metaphase-like arrest, and the spindles assembled in these cells were highly unstable, collapsed, and reassembled into multipolar spindles. Of those cells with multipolar spindles, 31.8% (7/22) entered cytokinesis and completed mitosis, resulting in cells with aberrant nuclei (Fig. 2,AandC; and Video 2), whereas the remaining 68.2% (15/22) underwent apoptosis or mitosis catastrophe (Fig. 2,AandC; and Video 3). Meanwhile, the chromosomes visualized with GFP-H2B in control cells properly congressed at the metaphase plate. In contrast, 60.4% (58/96) of the ABRO1-silenced cells exhibited misaligned chromosomes that had not been congressed at the metaphase plate. Of those cells with misaligned chromosomes, 41.3% (24/58) entered cytokinesis and completed mitosis (Fig. 2 D), whereas the remaining 58.6% (34/58) underwent apoptosis or mitosis catastrophe (Fig. 2 D). These mitotic defects were carefully categorized, calculated, and summarized in Fig. 2 (B–D). Importantly, these mitotic defects observed were partially rescued in cells cotransfected with RNAi-resistant CFP-ABRO1res plasmid (Fig. 2,A–D; and Video 4), indicating that ABRO1 is required for proper spindle assembly.
Time-lapse imaging of dividing control siRNA–treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with control siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing control siRNA–treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with control siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-siRNA–treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-siRNA–treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-siRNA treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-siRNA treated cells, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-depleted cells rescued by CFP-ABRO1res, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with ABRO1-siRNA and CFP-ABRO1res that is resistant to ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing ABRO1-depleted cells rescued by CFP-ABRO1res, based on Fig. 2. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with ABRO1-siRNA and CFP-ABRO1res that is resistant to ABRO1-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
BRISC is required for proper spindle assembly. (A) Representative live-cell still images of dividing H2B-GFP/α-tubulin-mRFP HeLa cells, transfected with control siRNA (see Video 1), ABRO1-siRNA (see Videos 2 and 3), or cotransfected with ABRO1-siRNA and CFP-ABRO1res (see Video 4), respectively. Bars, 5 µm. NEBD indicates the first frame after NEBD, based on the chromatin marker H2B-GFP. Times are shown in hours:minutes. (B) Box-and-whisker plots showing the duration cells spent in mitosis from NEBD to anaphase onset or cell death in the cells shown in A. Boxes show the upper and lower quartiles (25–75%) with a line at the median. Whiskers extend from the 10th to the 90th percentile, and dots correspond to outliers. *, P < 0.05; ***, P < 0.001; Student’s t test. (C) Quantification of the cells with bipolar or multipolar spindles shown in A, whereas cells with multipolar spindles were further classified, based on with or without complete mitosis, into multipolar with cytokinesis and multipolar with cell death. (D) Quantification of the cells with aligned or misaligned chromosomes shown in A, whereas cells with misaligned chromosomes were further classified into misaligned with cytokinesis and misaligned with cell death based on with or without complete mitosis. Error bars show mean ± SD.
BRISC is required for proper spindle assembly. (A) Representative live-cell still images of dividing H2B-GFP/α-tubulin-mRFP HeLa cells, transfected with control siRNA (see Video 1), ABRO1-siRNA (see Videos 2 and 3), or cotransfected with ABRO1-siRNA and CFP-ABRO1res (see Video 4), respectively. Bars, 5 µm. NEBD indicates the first frame after NEBD, based on the chromatin marker H2B-GFP. Times are shown in hours:minutes. (B) Box-and-whisker plots showing the duration cells spent in mitosis from NEBD to anaphase onset or cell death in the cells shown in A. Boxes show the upper and lower quartiles (25–75%) with a line at the median. Whiskers extend from the 10th to the 90th percentile, and dots correspond to outliers. *, P < 0.05; ***, P < 0.001; Student’s t test. (C) Quantification of the cells with bipolar or multipolar spindles shown in A, whereas cells with multipolar spindles were further classified, based on with or without complete mitosis, into multipolar with cytokinesis and multipolar with cell death. (D) Quantification of the cells with aligned or misaligned chromosomes shown in A, whereas cells with misaligned chromosomes were further classified into misaligned with cytokinesis and misaligned with cell death based on with or without complete mitosis. Error bars show mean ± SD.
BRISC is a MAP with cell cycle–dependent localization
To gain mechanistic insight into the function of BRISC, we analyzed the distribution of ABRO1 during cell cycle. In addition to cytoplasmic localization, as reported previously (Feng et al., 2010; Hu et al., 2011; Zhang et al., 2014), ABRO1 was shown to be located at the centrosomes in interphase (Figs. 3 A and S2 A), associated with spindle poles from prophase to telophase, and accumulated at midbody in telophase (Fig. S2 B). Similar localization pattern of MERIT40, a component of the BRISC complex that is essential for the integrity of the complex, was also observed in eYFP-MERIT40 stably expressing HeLa cells (Fig. S2 C). Consistently, live-cell images also detected the centrosome and spindle pole localization of eYFP-ABRO1 (Fig. S2 D and Video 9) and CFP-BRCC36 wild type (WT; Fig. 8 G). Biochemistry study showed that the total amount of ABRO1 was stable during cell cycle. Interestingly, a shift band of ABRO1 was found in the G2/M phase of the cells, suggesting that ABRO1 may be modified in mitosis (Fig. S2 E).
The localization of eYFP-ABRO1 during mitosis, based on Fig. S2 D. α-tubulin–mRFP HeLa cells were transiently transfected with eYFP–ABRO1 for 48 h. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon). Frames were taken every 5 min for 24 h at 37°C.
The localization of eYFP-ABRO1 during mitosis, based on Fig. S2 D. α-tubulin–mRFP HeLa cells were transiently transfected with eYFP–ABRO1 for 48 h. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon). Frames were taken every 5 min for 24 h at 37°C.
BRISC is a MAP with cell cycle–dependent localization. (A) The localization of ABRO1 in cell cycle. Exponential HeLa cells were fixed in cold methanol and immunostained for γ-tubulin (red) and ABRO1 (green); DNA was stained with DAPI. BRISC localizes at centrosomes, spindle poles, and midbody during mitosis. (B) K-fiber minus ends localization of ABRO1. Control (top), cold-treated (middle), and NUF2 siRNA–transfected (bottom) HeLa cells were fixed and stained with anti–α-tubulin (red) and anti-ABRO1 (green) antibodies. DNA was stained with DAPI. Bars, 5 µm. (C) MT sedimentation assay with purified recombinant FH–ABRO1. Purified FH–ABRO1 was mixed with in vitro–produced MTs (+MTs) or with buffer (−MTs) as a control. Subsequently, these samples were spun through a glycerol cushion, and the supernatant (S) and pellet (P) fractions were then analyzed for the presence of FH–ABRO1 by Western blotting. MTs produced in vitro with pure TAMRA rhodamine–labeled tubulin were spotted onto slides for visualization (right). FH–ABRO1 binds taxol-stabilized MTs in vitro. In parallel, the negative control experiment was performed with BSA. (D) MT sedimentation assay with HeLa mitotic lysate. Concentrated mitotic lysate was mixed with pure TAMRA rhodamine–labeled tubulin (+MT) or buffer (−MT) as a control, and the reactions were performed as described in Materials and methods. Subsequently, the samples were separated as described in C, and the sedimented MTs were analyzed for the presence of BRISC and NuMA by Western blotting. MTs produced in the mitotic lysate were spotted onto slides for visualization (bottom). BRISC and NuMA cosedimentation with MTs formed in the mitotic lysate.
BRISC is a MAP with cell cycle–dependent localization. (A) The localization of ABRO1 in cell cycle. Exponential HeLa cells were fixed in cold methanol and immunostained for γ-tubulin (red) and ABRO1 (green); DNA was stained with DAPI. BRISC localizes at centrosomes, spindle poles, and midbody during mitosis. (B) K-fiber minus ends localization of ABRO1. Control (top), cold-treated (middle), and NUF2 siRNA–transfected (bottom) HeLa cells were fixed and stained with anti–α-tubulin (red) and anti-ABRO1 (green) antibodies. DNA was stained with DAPI. Bars, 5 µm. (C) MT sedimentation assay with purified recombinant FH–ABRO1. Purified FH–ABRO1 was mixed with in vitro–produced MTs (+MTs) or with buffer (−MTs) as a control. Subsequently, these samples were spun through a glycerol cushion, and the supernatant (S) and pellet (P) fractions were then analyzed for the presence of FH–ABRO1 by Western blotting. MTs produced in vitro with pure TAMRA rhodamine–labeled tubulin were spotted onto slides for visualization (right). FH–ABRO1 binds taxol-stabilized MTs in vitro. In parallel, the negative control experiment was performed with BSA. (D) MT sedimentation assay with HeLa mitotic lysate. Concentrated mitotic lysate was mixed with pure TAMRA rhodamine–labeled tubulin (+MT) or buffer (−MT) as a control, and the reactions were performed as described in Materials and methods. Subsequently, the samples were separated as described in C, and the sedimented MTs were analyzed for the presence of BRISC and NuMA by Western blotting. MTs produced in the mitotic lysate were spotted onto slides for visualization (bottom). BRISC and NuMA cosedimentation with MTs formed in the mitotic lysate.
The MT distribution of ABRO1 was further examined in cold-treated cells, in which only the MT bundles of K-fibers were preserved (Rieder, 1981). Strikingly, ABRO1 decorated a small area at the distal part of the K-fibers, indicating that it specifically accumulates at their minus ends (Fig. 3 B white box). Moreover, ABRO1 localization was K-fiber specific, because the spindle pole localization of ABRO1 was nearly absent in NUF2-silenced metaphase cells (Fig. 3 B), in which MT–kinetochore attachment was abolished (DeLuca et al., 2002), whereas the localization of TPX2 was not affected (Fig. S2 F), as demonstrated previously (Manning and Compton, 2007). Instead, ABRO1 was found in numerous widespread foci.
Given the specific localization of BRISC with K-fiber minus ends, direct binding of BRISC to MTs was also tested. To this end, FLAG- and HA-tagged ABRO1 (FH–ABRO1) was expressed in HeLa cells. The FH–ABRO1 complex was purified, incubated with or without taxol-stabilized MTs generated in vitro, and centrifuged through a glycerol cushion. In the presence of MTs, most FH–ABRO1 was in the pellet fraction, whereas it remained soluble in the absence of MTs (Fig. 3 C). Furthermore, we performed a MT cosedimentation assay in a mitotic lysate system from HeLa cells, in which MTs assemble in response to physiologically relevant cues, rather than taxol (Chang et al., 2009). To facilitate observation, we added TAMRA rhodamine–labeled tubulin (Cytoskeleton, Inc.) to the concentrated lysate. The lysate was incubated to allow assembly of MTs, centrifuged through a glycerol cushion, and spotted onto slides for visualization. BRISC also cosedimented with tubulin assembled in this mitotic lysate system (Fig. 3 D). It is noteworthy that ABRO1 contains a coiled-coil domain (aa 218–254) that is also present in many centrosomal proteins (Cep135) and MAPs (NuMA), which suggests that ABRO1 may use its coiled-coil domain to mediate protein–protein interactions and perform its MT-dependent function. Together with its unique localization to MTs and K-fibers in cultured mammalian cells, we identify BRISC as a MAP that is essential for the assembly of a functional bipolar spindle.
BRISC is required for chromosomal MT assembly
Mitotic spindle assembly in centrosome-containing cells relies on two MT nucleation mechanisms: one centrosome dependent and one chromosome dependent (Rieder, 2005). Based on the observation that ABRO1 accumulates around/at kinetochores from prophase to prometaphase and rapidly decreases at metaphase (Fig. S3 A), we investigated the effect of BRISC depletion on spindle assembly by using a MT regrowth assay, in which MT formation at centrosomes and chromosomes can be examined separately (Tulu et al., 2006; Meunier and Vernos, 2011). In brief, HeLa cells were treated with nocodazole (NOC) to completely disassemble MTs, the drug was carefully removed, and then the initial stages of spindle reassembly were followed over time. In control cells, MT asters formed immediately after NOC release, and their numbers increased over the first 10 min, during which time the MT asters progressively coalesced and were eventually organized into a bipolar spindle (Fig. S3 B). In contrast, although two initial MT asters were formed in BRISC-silenced cells, either no additional asters or only a few weak asters appeared over time (Fig. S3 B).
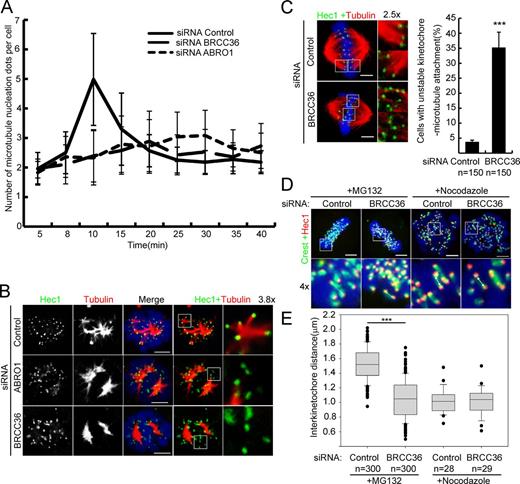
To quantify the above phenotypes, we counted the number of MT asters, indicated by γ-tubulin staining, in each cell at each time point after NOC release. At the early time points, all cells had two MT asters. At later time points, control cells contained a mean of five asters per cell, whereas ABRO1-silenced cells only had a mean of two asters per cell. At the final stage (25–40 min), the number of asters in control cells was decreased to two, reflecting the formation of a bipolar spindle, whereas the number of asters in ABRO1-silenced cells increased, suggesting organization of a multipolar spindle (Fig. 4 A). As expected, the similar effects were observed in BRCC36-silenced cells (Fig. 4 A).
BRISC is required for chromosomal MT assembly. (A) Quantification of the number of MT asters in each mitotic cell fixed at the indicated times after NOC washout, as described in Fig. S3 B (n = 120 for each time point; means ± SD; three independent experiments). (B) Representative confocal images of K-fiber formation in control ABRO1/BRCC36–silenced cells. HeLa cells released from NOC for 6 min, as described in Fig. S3 B, were fixed and stained for Hec1 (green) and α-tubulin (red). K-fiber formation is significantly reduced in ABRO1/BRCC36-silenced cells compared with that in controls. Insets show K-fibers. (C) Representative confocal images of the kinetochore–MT attachment in control and BRCC36-silenced cells. Cells were treated with 10 µM MG132 for 4 h, fixed, and stained for Hec1 (green) and α-tubulin (red). End-on amphitelic attachment of MTs to kinetochores is observed in control cells. However, kinetochore–MT attachment was deficient or impaired in BRCC36-silenced cells, and many kinetochores were laterally bound to MT bundles. (D) Representative confocal images of the interkinetochore distance in control and BRCC36-silenced cells. Control or BRCC36-silenced HeLa cells were treated with 10 µM MG132 or 30 ng/ml NOC for 4 h, and then fixed and stained for the outer kinetochore marker Hec1 (red) and CREST (green). CREST sera are human autoantibodies against centromeric proteins. The images correspond to confocal microscopy maximum projections for NOC-treated cells and single confocal microscopy sections for 10 µM of MG132-treated cells. The white arrows show paired sister kinetochores. (E) Box-and-whiskers plots showing the quantification of the interkinetochore distance. Boxes show the upper and lower quartiles (25–75%) with a line at the median. Whiskers extend from the 10th to the 90th percentile, and dots correspond to outliers. Results are means ± SD. The mean interkinetochore distance is significantly lower in BRCC36-silenced cells than in the controls (1.10 vs. 1.51 µm). This effect is dependent on MT forces, as the interkinetochore distance in control and BRCC36-silenced cells is similar after NOC treatment (1.01 vs. 1.02 µm). ***, P < 0.001; Student’s t test. Bars, 5 µm. Error bars show mean ± SD.
BRISC is required for chromosomal MT assembly. (A) Quantification of the number of MT asters in each mitotic cell fixed at the indicated times after NOC washout, as described in Fig. S3 B (n = 120 for each time point; means ± SD; three independent experiments). (B) Representative confocal images of K-fiber formation in control ABRO1/BRCC36–silenced cells. HeLa cells released from NOC for 6 min, as described in Fig. S3 B, were fixed and stained for Hec1 (green) and α-tubulin (red). K-fiber formation is significantly reduced in ABRO1/BRCC36-silenced cells compared with that in controls. Insets show K-fibers. (C) Representative confocal images of the kinetochore–MT attachment in control and BRCC36-silenced cells. Cells were treated with 10 µM MG132 for 4 h, fixed, and stained for Hec1 (green) and α-tubulin (red). End-on amphitelic attachment of MTs to kinetochores is observed in control cells. However, kinetochore–MT attachment was deficient or impaired in BRCC36-silenced cells, and many kinetochores were laterally bound to MT bundles. (D) Representative confocal images of the interkinetochore distance in control and BRCC36-silenced cells. Control or BRCC36-silenced HeLa cells were treated with 10 µM MG132 or 30 ng/ml NOC for 4 h, and then fixed and stained for the outer kinetochore marker Hec1 (red) and CREST (green). CREST sera are human autoantibodies against centromeric proteins. The images correspond to confocal microscopy maximum projections for NOC-treated cells and single confocal microscopy sections for 10 µM of MG132-treated cells. The white arrows show paired sister kinetochores. (E) Box-and-whiskers plots showing the quantification of the interkinetochore distance. Boxes show the upper and lower quartiles (25–75%) with a line at the median. Whiskers extend from the 10th to the 90th percentile, and dots correspond to outliers. Results are means ± SD. The mean interkinetochore distance is significantly lower in BRCC36-silenced cells than in the controls (1.10 vs. 1.51 µm). This effect is dependent on MT forces, as the interkinetochore distance in control and BRCC36-silenced cells is similar after NOC treatment (1.01 vs. 1.02 µm). ***, P < 0.001; Student’s t test. Bars, 5 µm. Error bars show mean ± SD.
To further confirm the function of BRISC in chromosomal MT assembly, we examined MT formation around the kinetochores by observing anti-Hec1 and α-tubulin antibody-labeled control and ABRO1- or BRCC36-siRNA–treated cells that were recovered from NOC and incubated at 37°C for 6 min by confocal microscopy. As observed above, short α-tubulin fibers coming from kinetochores were evident in control cells, whereas a clear defect was seen in ABRO1- or BRCC36-silenced cells (Fig. 4 B). These results demonstrate that BRISC is not required for centrosomal aster formation but is essential for chromosomal MT assembly.
BRISC participates in MT kinetochore attachment
Next, we investigated whether BRISC is involved in kinetochore–MT attachment. Kinetochore–MT interactions were stable in control cells without NOC treatment, whereas these interactions were severely impaired in BRCC36-silenced cells (Fig. 4 C). The number of MTs that attached to kinetochores was reduced, and some lateral contacts (instead of typical end-on attachments) between MTs and kinetochores were observed in BRCC36-silenced cells (Fig. 4 C). The lack of attachment to MTs may abolish tension across sister kinetochores. To test this hypothesis, we measured the interkinetochore distance between pairs of Hec1/CREST foci as a measurement of tension. In control cells, metaphase chromosomes under full tension had relatively long interkinetochore distances (1.51 ± 0.21 µm), whereas in BRCC36-silenced cells, the interkinetochore distance was significantly shortened (1.10 ± 0.16 µm; Fig. 4,DandE), indicating that the chromosomes were misaligned and not under tension, which mimics the chromosome congression observed in the absence of K-fibers (Cai et al., 2009).
Collectively, these results demonstrate that BRISC is critical for chromosome-associated MT assembly, suggesting that in the absence of BRISC, cells fail to establish stable kinetochore–MT interactions, leading to defects in kinetochore tension, chromosome alignment/congression and segregation, and the formation of multipolar spindles.
DUB activity of BRISC contributes to the mitotic spindle assembly
BRISC is a DUB that specifically deubiquitinates K63Ubs. Whether the DUB activity of BRISC is required for its function in spindle assembly was further explored. We used time-lapse imaging in H2B-GFP/α-tubulin-mRFP HeLa cells transfected with control, BRCC36 siRNA, or BRCC36 siRNA plus WT, RNAi-resistant CFP-BRCC36 plasmid, or DUB-deficient mutant QSQ (Shao et al., 2009b), and the mitotic progression was followed.
Similar to that of ABRO1 as shown in Fig. 2 (B–D), depletion of BRCC36 by siRNA resulted in prolonged mitotic progression and multiple abnormal mitotic phenotypes (Fig. 5). In brief, depletion of BRCC36 resulted in a significantly higher portion of cells with multipolar spindles (20.4%, 20/98). Of these multipolar spindle cells examined, 60% (12/20) entered cytokinesis and completed mitosis (Fig. 5, A and B), whereas the remaining 40% (8/20) underwent apoptosis or mitosis catastrophe. Meanwhile, 59.2% (58/98) of the BRCC36-silenced cells exhibited misaligned chromosomes. Of these misaligned cells examined, 58.6% (34/58) entered cytokinesis and completed mitosis (Fig. 5, A and C), whereas the remaining 41.4% (24/58) underwent apoptosis or mitosis catastrophe. Importantly, these mitotic defects raised by BRCC36 depletion were partially rescued by the coexpression of CFP-BRCC36res WT, but not that of the DUB-inactive CFP-BRCC36res QSQ (Fig. 5, A–C). In addition, BRCC36 depletion significantly prolonged the mitotic progression (273 min in siBRCC36 vs. 60 min in control); both the duration from nuclear envelope breakdown (NEBD) to metaphase and metaphase to anaphase were increased in BRCC36-depleted cells. These defects were partially rescued by the reexpression of WT, but not the DUB-inactive mutant (QSQ) of CFP-BRCC36res (Fig. 5 D, Fig. S4 B, and Videos 5–8). Collectively, these results demonstrate that the DUB activity of BRCC36 is critical for BRISC’s function in spindle assembly, and further suggest an important role for the protein degradation–independent signaling of K63 ubiquitination in mitosis.
Time-lapse imaging of dividing control siRNA–treated cells, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with control siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing control siRNA–treated cells, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with control siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-siRNA–treated cells, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with BRCC36-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-siRNA–treated cells, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were transfected with BRCC36-siRNA. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-silenced cells rescued by DUB-active CFP-BRCC36res WT, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with BRCC36-siRNA and CFP-BRCC36res WT plasmids. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-silenced cells rescued by DUB-active CFP-BRCC36res WT, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with BRCC36-siRNA and CFP-BRCC36res WT plasmids. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-silenced cells, which were rescued by DUB-inactive mutant CFP-BRCC36res QSQ, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with BRCC36-siRNA and CFP-BRCC36res QSQ plasmids. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
Time-lapse imaging of dividing BRCC36-silenced cells, which were rescued by DUB-inactive mutant CFP-BRCC36res QSQ, based on Fig. 5. H2B-GFP/α-tubulin–mRFP HeLa cells were cotransfected with BRCC36-siRNA and CFP-BRCC36res QSQ plasmids. Images were analyzed by time-lapse confocal microscopy using a laser-scanning confocal microscope (40× oil objective, Plan-Apochromat, NA 1.4; Eclipse Ti-E; Nikon) 36 h after transfection. Frames were taken every 5 min for 24 h at 37°C.
DUB activity of BRISC contributes to the proper mitotic spindle assembly. (A) Representative live-cell still images of H2B-GFP/α-tubulin-mRFP HeLa cells transfected with control (see Video 5) or BRCC36 siRNA (see Video 6), or cotransfected with BRCC36 siRNA and CFP-BRCC36res WT (see Video 7) or BRCC36 siRNA and CFP-BRCC36res QSQ, without DUB activity (see Video 8). NEBD indicates the first frame after NEBD, based on the chromatin marker H2B-GFP. Times are shown in hours:minutes. (B) Quantification of the cells with bipolar or multipolar spindles as shown in A, calculated as described in Fig. 2 C. (C) Quantification of the cells with aligned or misaligned chromosomes as shown in A, calculated as described in Fig. 2 D. (D) Box-and-whisker plots showing the duration of the cells, shown in Fig. 5 A, spent in mitosis from NEBD to metaphase and metaphase to anaphase onset or cell death. *, P < 0.05; ***, P < 0.001; Student’s t test. ns, no significance. Error bars show mean ± SD.
DUB activity of BRISC contributes to the proper mitotic spindle assembly. (A) Representative live-cell still images of H2B-GFP/α-tubulin-mRFP HeLa cells transfected with control (see Video 5) or BRCC36 siRNA (see Video 6), or cotransfected with BRCC36 siRNA and CFP-BRCC36res WT (see Video 7) or BRCC36 siRNA and CFP-BRCC36res QSQ, without DUB activity (see Video 8). NEBD indicates the first frame after NEBD, based on the chromatin marker H2B-GFP. Times are shown in hours:minutes. (B) Quantification of the cells with bipolar or multipolar spindles as shown in A, calculated as described in Fig. 2 C. (C) Quantification of the cells with aligned or misaligned chromosomes as shown in A, calculated as described in Fig. 2 D. (D) Box-and-whisker plots showing the duration of the cells, shown in Fig. 5 A, spent in mitosis from NEBD to metaphase and metaphase to anaphase onset or cell death. *, P < 0.05; ***, P < 0.001; Student’s t test. ns, no significance. Error bars show mean ± SD.
BRISC associates with the SAF NuMA
Functional bipolar spindle assembly requires the activation of several SAFs such as TPX2 (Wittmann et al., 2000; Gruss and Vernos, 2004), NuMA (Wiese et al., 2001), and others. NuMA is a coiled-coil structural protein that localizes to the interphase nucleus and the mitotic spindle poles. Concomitant with NEBD, NuMA is liberated from importin-β, and then mediates centrosome-independent MT polymerization and spindle assembly (Compton and Cleveland, 1993; Merdes et al., 2000; Silk et al., 2009; Radulescu and Cleveland, 2010). During mitotic progression, NuMA is transported to spindle poles (Merdes et al., 1996, 2000), where it is involved in the maintenance and establishment of spindle poles (Khodjakov et al., 2003; Silk et al., 2009). Aberrant expression of NuMA has been linked to formation of multipolar spindles (Wong et al., 2006) and less efficient formation of kinetochore fibers (Haren et al., 2009). The similar localization pattern and function between BRISC and NuMA, as well as the result from mass spectrum analysis, in which NuMA was identified as a FH–ABRO1-interacting protein (Fig. S5 A), prompted us to investigate whether NuMA, one of the most important SAFs, is associated with BRISC.
To this end, we performed immunoprecipitation on mitotic HeLa cells stably expressing FH–ABRO1. As expected, NuMA was detected in the FH–ABRO1 immunoprecipitates (Fig. 6 A). Consistent with the ectopic FH–ABRO1, NuMA could also coimmunoprecipitate with endogenous ABRO1 (Fig. 6 B). In addition, NuMA was directly pulled down by GST–ABRO1/BRCC36 recombinant complex (Fig. 6 C). In agreement with these biochemical data, EYFP-NuMA was also shown to colocalize with ABRO1 at the spindle poles (Fig. 6 D). Collectively, these results indicate that BRISC is associated with the key SAF, NuMA.
BRISC associates with the SAF NuMA during mitosis. (A) The ectopic FH–ABRO1 complex interacts with endogenous NuMA. Immunoprecipitate assays were performed on mock or FH–ABRO1 stably expressing HeLa cells with anti–FLAG M2 agarose gel, and analyzed by immunoblotting with the indicated antibodies. (B) The endogenous ABRO1 interacts with endogenous NuMA during mitosis. Immunoprecipitate assays were performed with anti-ABRO1 antibody in HeLa cells synchronized by double thymidine block or a sequential thymidine/NOC block release protocol. Immunoprecipitates were detected by immunoblotting with the indicated antibodies. For ABRO1 detection, a secondary antibody (A25022; Abbkine) specific against the light chain of IgG was used to exclude the interference of IgG heavy chain. Cyclin B1 was used as a G2/M index. (C) NuMA directly binds to the BRISC complex in vitro. GST–MERIT40 and GST–ABRO1/BRCC36 fusion proteins were purified from bacterial or insect cells. GST pull-down assays were performed using the purified GST fusion proteins and HeLa mitotic cell lysate. The bound protein complexes were analyzed by SDS-PAGE and visualized by Ponceau staining (bottom) and immunoblotting with an anti-NuMA antibody. (D) ABRO1 colocalizes with eYFP-NuMA at spindle poles. eYFP-NuMA stably expressing HeLa cell lines were fixed by cold methanol and stained with anti-ABRO1 antibody (red). DNA was stained with DAPI. Bar, 5 µm.
BRISC associates with the SAF NuMA during mitosis. (A) The ectopic FH–ABRO1 complex interacts with endogenous NuMA. Immunoprecipitate assays were performed on mock or FH–ABRO1 stably expressing HeLa cells with anti–FLAG M2 agarose gel, and analyzed by immunoblotting with the indicated antibodies. (B) The endogenous ABRO1 interacts with endogenous NuMA during mitosis. Immunoprecipitate assays were performed with anti-ABRO1 antibody in HeLa cells synchronized by double thymidine block or a sequential thymidine/NOC block release protocol. Immunoprecipitates were detected by immunoblotting with the indicated antibodies. For ABRO1 detection, a secondary antibody (A25022; Abbkine) specific against the light chain of IgG was used to exclude the interference of IgG heavy chain. Cyclin B1 was used as a G2/M index. (C) NuMA directly binds to the BRISC complex in vitro. GST–MERIT40 and GST–ABRO1/BRCC36 fusion proteins were purified from bacterial or insect cells. GST pull-down assays were performed using the purified GST fusion proteins and HeLa mitotic cell lysate. The bound protein complexes were analyzed by SDS-PAGE and visualized by Ponceau staining (bottom) and immunoblotting with an anti-NuMA antibody. (D) ABRO1 colocalizes with eYFP-NuMA at spindle poles. eYFP-NuMA stably expressing HeLa cell lines were fixed by cold methanol and stained with anti-ABRO1 antibody (red). DNA was stained with DAPI. Bar, 5 µm.
BRISC regulates the ubiquitination of NuMA
NuMA is an essential mitotic component with distinct functions in the establishment and maintenance of focused spindle poles (Merdes et al., 2000; Silk et al., 2009; Radulescu and Cleveland, 2010). The function of NuMA is tightly regulated by posttranscriptional modifications, including phosphorylation and PARsylation (Saredi et al., 1997; Fant et al., 2004; Chang et al., 2005a,,b). Despite the wealth of information known about the function of NuMA, little is known about the role of its ubiquitination in mitosis. The association of BRISC with NuMA raises the possibility that NuMA might be modified by K63Ubs and regulated by BRISC. To test this hypothesis, we first characterized NuMA ubiquitination in HEK293T cells cotransfected with His-tagged NuMA, hemagglutinin-tagged ubiquitin, or a mutant (HA-K63Ub) that only mediates K63 linkage. His-NuMA was immunoprecipitated, and the precipitates were analyzed. As shown, NuMA was ubiquitinated through a K63 linkage (Fig. S5 B). Next, we investigated whether BRISC regulates the ubiquitination of NuMA. 293T cells were cotransfected with His-NuMA, and HA-K63Ub, BRCC36, or ABRO1 was simultaneously depleted by treatment with specific siRNAs. His-tagged NuMA was then immunoprecipitated under denaturing conditions and analyzed by immunoblotting. Knockdown of either BRCC36 or ABRO1 noticeably increased K63-linked ubiquitination of NuMA (Figs. 7 A and S5 C). Furthermore, the increased K63Ub modification of NuMA was also detected within endogenous NuMA immunoprecipitated from ABRO1-depleted mitotic HeLa lysates using an anti-NuMA antibody (Fig. 7 B). Collectively, these results suggest that NuMA is a substrate of BRISC. To confirm this, an in vitro deubiquitination assay was performed. First, endogenous NuMA was immunoprecipitated from mitotic HeLa cells by using anti-NuMA antibody (Fig. 7 C), and WT or DUB-inactive mutant (QSQ) of GST–BRCC36 complex was purified from Sf9 insect cells (Fig. 7 D). Next, endogenous NuMA immunoprecipitions was used as substrate and incubated with either the purified GST–BRCC36–WT or GST–BRCC36–QSQ complex at 37°C for 2 h. As shown in Fig. 7 E, the abundance of K63Ub decreased significantly in these NuMAs incubated with GST–BRCC36–WT, but not the DUB-inactive GST–BRCC36–QSQ complex. These data suggest that NuMA might be modified with K63Ubs and could be deubiquitinated by BRISC.
NuMA is a substrate for BRISC DUB activity. (A) Ubiquitination of NuMA increased upon inhibition of BRISC. 293T cells cotransfected with His-tagged NuMA and HA-K63Ub were simultaneously treated with control, ABRO1, or BRCC36 siRNAs. Cell lysates were subjected to Ni-NTA pull-down assays under denaturing conditions. His-NuMA pull-downs were then analyzed by immunoblotting with the indicated antibodies. (B) Endogenous K63-linked ubiquitination of NuMA was increased in ABRO1-silenced HeLa cells. HeLa cells were transfected with control or ABRO1 siRNAs and synchronized at G2/M phase. Cell lysates were subjected to immunoprecipitation by using anti-NuMA antibody and probed with the indicated antibodies. (C) Immunoprecipitations from mitotic HeLa cells by using anti-NuMA antibody were separated by SDS-PAGE and silver stained. (D) Purified GST, GST–ABRO1/BRCC36 WT or GST–ABRO1/BRCC36 QSQ proteins from Sf9 insect cells were separated by SDS-PAGE and stained using Coomassie brilliant blue. Lane 1, GST; lane 2, GST–ABRO1/BRCC36 WT; lane 3, GST–ABRO1/BRCC36 QSQ. (E) In vitro DUB assay. Endogenous NuMA immunoprecipitations, as described in C, were incubated with the purified GST–BRCC36 WT or GST–BRCC36 QSQ complex, as described in D, in DUB buffer. The abundances of K63Ub decreased significantly with the incubation in the presence of GST–BRCC36 WT, but not the inactive GST–BRCC36 QSQ complex.
NuMA is a substrate for BRISC DUB activity. (A) Ubiquitination of NuMA increased upon inhibition of BRISC. 293T cells cotransfected with His-tagged NuMA and HA-K63Ub were simultaneously treated with control, ABRO1, or BRCC36 siRNAs. Cell lysates were subjected to Ni-NTA pull-down assays under denaturing conditions. His-NuMA pull-downs were then analyzed by immunoblotting with the indicated antibodies. (B) Endogenous K63-linked ubiquitination of NuMA was increased in ABRO1-silenced HeLa cells. HeLa cells were transfected with control or ABRO1 siRNAs and synchronized at G2/M phase. Cell lysates were subjected to immunoprecipitation by using anti-NuMA antibody and probed with the indicated antibodies. (C) Immunoprecipitations from mitotic HeLa cells by using anti-NuMA antibody were separated by SDS-PAGE and silver stained. (D) Purified GST, GST–ABRO1/BRCC36 WT or GST–ABRO1/BRCC36 QSQ proteins from Sf9 insect cells were separated by SDS-PAGE and stained using Coomassie brilliant blue. Lane 1, GST; lane 2, GST–ABRO1/BRCC36 WT; lane 3, GST–ABRO1/BRCC36 QSQ. (E) In vitro DUB assay. Endogenous NuMA immunoprecipitations, as described in C, were incubated with the purified GST–BRCC36 WT or GST–BRCC36 QSQ complex, as described in D, in DUB buffer. The abundances of K63Ub decreased significantly with the incubation in the presence of GST–BRCC36 WT, but not the inactive GST–BRCC36 QSQ complex.
BRISC modulates the function of NuMA in spindle pole assembly
Concomitant with NEBD, NuMA is released from the inhibition of importin-α/β and relocalizes to spindle poles by associating with the minus end–directed motor protein dynein during mitosis. NuMA, in complex with dynein and dynactin, functions as a tether that links the bulk MTs of the spindle to centrosomes (Khodjakov et al., 2003; Silk et al., 2009), providing an essential stabilizing structure at spindle pole. To gain insight into the functional significance of BRISC-mediated regulation of NuMA ubiquitination, we examined whether BRISC modulates the interaction between NuMA and dynein. Immunoprecipitation revealed that depletion of ABRO1 or BRCC36 increased the interaction of NuMA with dynein, as well as with importin-β in HeLa cells (Fig. 8 A), indicating that BRISCs negatively regulate the association of NuMA with dynein and importin-β.
BRISC modulates the function of NuMA in spindle pole assembly. (A) Depletion of BRISC regulates the interaction of NuMA with dynein and importin-β. HeLa cells transfected with control, ABRO1, or BRCC36 siRNA for 72 h were collected, and protein samples were analyzed by Western blotting with the indicated antibodies. (B) Depletion of BRISC decreased the incorporation of NuMA into the spindle poles during mitosis. HeLa cells transfected with control or BRCC36 siRNA were treated as described in MT regrowth assay in Materials and methods, and immunostained for α-tubulin (red) and NuMA (green). (C) Quantitative analysis of cells in which NuMA did not completely incorporate into the spindle poles within 25 min after NOC release. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (D) Representative images of the K-fiber minus ends distribution of NuMA. Control, ABRO1, or BRCC36-silenced HeLa cells were arrested by treatment with STLC and immunostained for NuMA (green) and α-tubulin (red). DNA was stained with DAPI. In control cells, NuMA accumulated and was orderly arranged as a ring-like structure surrounding the spindle pole body, whereas in ABRO1/BRCC36-depleted cells, this kind of structure was significantly disrupted. (E) Quantification of cells with normal (ring-like) NuMA distribution at monopolar poles, as shown in D. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (F) Quantitative analysis of the diameter in the z axis for NuMA accumulated at monopolar spindles in 3D reconstructions. Z stacks comprising 8–17 0.5-µm sections were acquired, and deconvolution of 3D image stacks were performed using LAS-AF software (Leica). Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (G) BRCC36 siRNA-induced irregular distribution of NuMA at spindle poles was rescued by CFP-BRCC36res WT. HeLa cells were cotransfected with BRCC36 siRNA and CFP-BRCC36res WT or CFP-B36res QSQ, treated with STLC, and then examined as described in D. (F) Graph showing the quantification of cells with normal NuMA distribution at monopolar poles, as shown in G. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. Bars, 5 µm. Error bars show mean ± SD.
BRISC modulates the function of NuMA in spindle pole assembly. (A) Depletion of BRISC regulates the interaction of NuMA with dynein and importin-β. HeLa cells transfected with control, ABRO1, or BRCC36 siRNA for 72 h were collected, and protein samples were analyzed by Western blotting with the indicated antibodies. (B) Depletion of BRISC decreased the incorporation of NuMA into the spindle poles during mitosis. HeLa cells transfected with control or BRCC36 siRNA were treated as described in MT regrowth assay in Materials and methods, and immunostained for α-tubulin (red) and NuMA (green). (C) Quantitative analysis of cells in which NuMA did not completely incorporate into the spindle poles within 25 min after NOC release. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (D) Representative images of the K-fiber minus ends distribution of NuMA. Control, ABRO1, or BRCC36-silenced HeLa cells were arrested by treatment with STLC and immunostained for NuMA (green) and α-tubulin (red). DNA was stained with DAPI. In control cells, NuMA accumulated and was orderly arranged as a ring-like structure surrounding the spindle pole body, whereas in ABRO1/BRCC36-depleted cells, this kind of structure was significantly disrupted. (E) Quantification of cells with normal (ring-like) NuMA distribution at monopolar poles, as shown in D. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (F) Quantitative analysis of the diameter in the z axis for NuMA accumulated at monopolar spindles in 3D reconstructions. Z stacks comprising 8–17 0.5-µm sections were acquired, and deconvolution of 3D image stacks were performed using LAS-AF software (Leica). Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. (G) BRCC36 siRNA-induced irregular distribution of NuMA at spindle poles was rescued by CFP-BRCC36res WT. HeLa cells were cotransfected with BRCC36 siRNA and CFP-BRCC36res WT or CFP-B36res QSQ, treated with STLC, and then examined as described in D. (F) Graph showing the quantification of cells with normal NuMA distribution at monopolar poles, as shown in G. Mean ± SD of three independent experiments. ***, P < 0.001; Student’s t test. Bars, 5 µm. Error bars show mean ± SD.
Next, the effect of BRISC depletion on NuMA function in spindle pole assembly was further analyzed. Cells were first treated with NOC to completely disassemble MTs, and then the drug was removed followed by the dynamic incorporation of NuMA into spindle poles. As reported previously (Merdes et al., 2000; Khodjakov et al., 2003; Kisurina-Evgenieva et al., 2004; Rousselet, 2009), small NuMA-containing particles were transported from the periphery of the cell toward the spindle pole after NOC release. In control cells, small aggregates of NuMA formed in the first several min (0–8 min), coalescing toward the spindle poles in 10–25 min. However, in BRCC36-silenced cells, a large proportion of NuMA aggregates was retained in the peripheral areas distant from the spindle poles (Fig. 8, B and C; 25–30 min), and supernumerary poles were formed, indicating that depletion of BRCC36 decreases the incorporation of NuMA into spindle poles during mitosis, presumably by aberrant dynamic interaction between NuMA and its partner dynein.
Furthermore, the K-fiber minus ends distribution of NuMA, which is required for the capture and incorporation of the K-fibers into the mitotic spindle (Khodjakov et al., 2003), was examined in HeLa cells arrested in mitosis, using the Eg5 inhibitor S-trityl-l-cysteine (STLC; Skoufias et al., 2006). STLC-treated cells produced a high frequency of monopolar spindles that appeared almost exclusively as chromosome rosettes surrounding a central pole. Although NuMA bound efficiently to MTs and was concentrated at spindle poles in both the control and ABRO1/BRCC36-silenced cells, there was a profound difference in their architecture. In control siRNA-treated cells, NuMA accumulated and was orderly arranged as a ring-like structure surrounding the spindle pole (∼85%; Fig. 8, D and E). In contrast, in ABRO1/BRCC36-depleted cells, NuMA accumulated disorderly at the spindle poles in either a compact or aggregated manner (∼70–80%; Fig. 8, D and E). Inspection of NuMA images acquired in z-axis sections from monopolar spindles assisted by 3D reconstructions revealed that NuMA was recruited to the spindle poles and arranged into an olive-like structure in control cells (diameter in z axis, 6.66 ± 0.71 µm; Fig. 8 F), aggregated into an olive-like structure with small holes in ABRO1-silenced cells (diameter in z axis, 5.47 ± 0.51 µm; Fig. 8 F), or compacted into a gengon-like structure in BRCC36-silenced cells (diameter in z axis, 4.95 ± 0.58 µm; Fig. 8 F).
In addition, BRCC36 siRNA-induced irregular distribution of NuMA at spindle poles was rescued by the reexpression of WT but not the DUB-inactive QSQ mutant of CFP-BRCC36 (Fig. 8, G and H). These results demonstrate that the DUB activity of BRISC is essential for the proper spindle pole organization, most likely by regulating the K63Ub-mediated NuMA–dynein interactions. During NEBD, K63Ub-modified NuMA associates with BRISC and dynein. Dynein carries the K63Ub–NuMA in a minus end–directed fashion and deposits it at the K-fiber minus ends with the removal of K63Ubs from NuMA, which is catalyzed by active BRISC. Dimeric or oligomeric NuMA complexes and NuMA–dynein complexes in the “spindle pole matrix” are responsible for tethering MT minus ends to the poles (Merdes et al., 2000; Silk et al., 2009; Radulescu and Cleveland, 2010). In CFP-BRCC36 QSQ cells, the lack of DUB activity may increase the interaction of NuMA with dynein (Fig. 8 A), which in turn could affect the timely unload of NuMA from dynein and deposition to “spindle pole matrix,” as well as the dynamics and focusing activity of NuMA complexes and NuMA–dynein complexes at K-fiber minus ends, leading to the formation of defective spindle poles (Fig. S5, D and E).
Discussion
Faithful division of eukaryotic cells requires temporal and spatial coordination of morphological transitions, which are finely modulated by E3 ubiquitin ligases and DUBs. Here, we describe the identification and initial characterization of BRISC, a K63Ub-specific DUB (Cooper et al., 2009), as a novel essential mitotic regulator that coordinates mitotic spindle assembly.
Our data, especially those obtained by rescue experiments with CFP-ABRO1res, CFP-B36WT, and CFP-B36 QSQ mutant, establish a crucial role for BRISC in the control of mitotic spindle assembly in cultured mammalian cells via its dual functions as a MAP protein and DUB. BRISC distributes along and directly binds to MTs during mitosis and associates with NuMA, another MAP with cross-linking properties, to form a functional complex to participate in spindle assembly. The DUB activity of BRISC can catalyze the removal of the K63Ubs from its substrates and therefore is essential to control their ubiquitination status, which in turn may affect the interaction between these substrates and their partners. Consistent with this interpretation, in this study we found that the K63Ub modification of NuMA and the association of NuMA with dynein and importin-β are regulated by BRISC, suggesting an important role for K63Ub modification in proper functional spindle assembly.
A recent study demonstrated that CYLD, another K63Ub-specific DUB, is also involved in regulating cortical NuMA function via a mechanism mediated by K63Ubs (Yang et al., 2014). NuMA was also reported to be complexed with tumor suppressor BRCA1, whose E3 ligase activity is K63- and K6-linkage specific and is required for Ran-dependent spindle assembly (Wu-Baer et al., 2003; Joukov et al., 2006). These studies together with our current findings suggest that the K63Ub status of NuMA is crucial for its function, and is most likely to be regulated by both E3 ligases and DUBs. Further studies addressing the functional link between BRISC, E3 ligase, and the ubiquitination of NuMA will be essential for our understanding the mechanisms of mitotic spindle assembly.
The diverse localization of BRISC and the pleiotropic effects arising from depletion of BRISC suggest that BRISC may form various functional complexes with distinct substrates to regulate mitosis progression, which supports the notion of “substrate-inducible localization” (Zheng et al., 2013). Although our data establish NuMA as a substrate of BRISC in the control of spindle assembly, it would not be surprising if other mitotic spindle factors could be identified as substrate for BRISC in the future. The identification of these potential BRISC substrates that are involved in spindle assembly will improve our understanding of the mechanism by which BRISC regulates mitosis.
The dynamic balance between ubiquitination and deubiquitination is controlled by both E3s and DUBs, and has been shown to contribute to the generation of the switch-like transition controlling mitotic progression (Stegmeier et al., 2007a; Fournane et al., 2012). Therefore, inactivation/down-regulation of these enzymes may cause mitotic defects, leading to the production of aneuploid progeny. Thus, many DUBs and E3s act as tumor suppressors or promoters and have been implicated in tumorigenesis (Stegmeier et al., 2007b; Holland and Cleveland, 2012). Indeed, BRCC36, a core BRISC subunit, is found to be overexpressed in many types of cancers, further underscoring its importance in the pathogenesis of human cancer (Dong et al., 2003).
Materials and methods
Antibodies
The following antibodies were used: rabbit anti-NuMA (ab36999; Abcam; NB100-74636; Novus Biologicals); mouse anti-TPX2 (ab32795; Abcam); rabbit anti–α tubulin (DM1A; Cell Signaling Technology); rabbit anti–importin-β (no. 8673S; Cell Signaling Technology); rabbit anti–Dynein HC (sc-9115; Santa Cruz Biotechnology, Inc.); rabbit anti-BRE (GTX105364; Genetex); mouse anti-HA (MMS-101P; Covance); rabbit anti-K63Ub (Apu3; EMD Millipore); mouse anti-pH3 (Ser10; 05-806; EMD Millipore); human anti–centromere positive serum (no. 15-235-0001; Antibodies Inc.); mouse Hec1 (ab3613; Abcam); mouse anti–γ-tubulin (T6557; Sigma-Aldrich); rabbit anti–γ-tubulin (ab84355; Abcam); rabbit anti-p27 (sc-528; Santa Cruz Biotechnology, Inc.); rabbit anti-p16 (sc-759; Santa Cruz Biotechnology, Inc.); rabbit anti–cyclin D1 (ab16663; Abcam); and rabbit normal IgG (sc-2027; Santa Cruz Biotechnology, Inc.). ABRO1 rabbit polyclonal antibody was raised against C termini of ABRO1 (261–415 aa) fused with GST–BRCC36 and subsequently affinity purified; the rabbit polyclonal anti-MERIT40 and anti-BRCC36 were generated as described previously (Sobhian et al., 2007; Shao et al., 2009b). In brief, anti-BRCC36 was generated using the C-terminal 20 amino acids of BRCC36 conjugated with KLH as immunogen. The antibody was affinity purified using a Sepharose column coupled with the C-terminal 20 amino acids of BRCC36 peptide. Anti-MERIT40 was generated against a GST–MERIT40 fusion protein.
Plasmid construction
Full-length cDNA encoding NuMA or ABRO1 was amplified by PCR with Phusion DNA polymerase (New England Biolabs, Inc.) and subcloned into the plasmids pEYFP-C1 (CMV promoter; Takara Bio Inc.) or pRK5-His (CMV promoter; provided by J. Tang, China Agricultural University, Beijing, China) to generate pEYFP-NuMA, pRK5-His6-NuMA, and pEYFP-ABRO1. For construction of pcDNA3.1-Flag-HA-ABRO1, pcDNA3.1-Flag-HA/Hygro plasmid was first constructed by introducing a Flag-HA double tag into pcDNA3.1(−)/Hygro (CMV promoter). ABRO1 ORF was then amplified by PCR and subcloned into pcDNA3.1-Flag-HA/Hygro. pLKO.1-shRNA-MERIT40 and pLKO.1-shRNA-Luc (used as control) plasmids were generated using pLKO.1-GFP (hU6 promoter) and oligo DNA sequences that target either MERIT40 (5′-AAGGGTCAACTGTCCAGAGAA-3′) or luciferase (GUGCGCUGCUGGUGCCAAC). RNAi-resistant plasmids pcDNA3.1-CFP-BRCC36res WT or QSQ were constructed by subcloning RNAi-resistant DNA encoding BRCC36 WT or QSQ mutant into pcDNA3.1-CFP plasmid (CMV promoter) using pEYFP-C1-BRCC36res WT/QSQ plasmids as templates for PCR. pGEX-6P-3-MERIT40 and pGEX-6P-3-ABRO1 (261–415 aa) were constructed by subcloning cDNA encoding the full-length MERIT40 or the C-terminal ABRO1 (261–415 aa) into pGEX-6P-3 plasmid (tac promoter; GE Healthcare). The pcDNA3.1-CFP-ABRO1res plasmid was designed to contain two silent point mutations (C244T and T250A), which rendered them resistant to ABRO1-1 siRNA, and generated by site-directed mutagenesis using the following oligonucleotides and site-directed mutagenesis kit: 5′-AAGTGAATGAGGAGAGTTTGGATAGGATACTTAAAGATCGGAGAAAGAAAGTCA-3′, 5′-CCAATGACTTTCTTTCTCCGATCCTTAAGAATTCTGTCCAAACTCTCCTCATTCAC-3′. pEYFP-C1-BRCC36res WT and QSQ pLKO.1-shRNA-GFP were provided by R. Greenberg (University of Pennsylvania, Philadelphia, PA). pmRFP-a-Tubulin-IRES_puro2b plasmid and GFP-H2B/RFP-a-Tubulin HeLa cell line were provided by D.W. Gerlich (Institute of Biochemistry, Swiss Federal Institute of Technology Zurich [ETHZ], Switzerland.).
Cell culture and synchronization
Human cervical carcinoma HeLa and human kidney 293T cell lines were from ATCC and kept in our laboratory. The cell lines were cultured in DMEM (Gibco) supplemented with 10% FBS (Hyclone) and 100 µg/ml penicillin/streptomycin at 37°C with 5% CO2 in a humid atmosphere.
To generate cells at a different stage of cell cycle, exponential cell lines were synchronized by a standard double thymidine block or by a sequential thymidine block/NOC release protocol, in which cells were double blocked by thymidine and released into fresh medium containing 30 ng/ml NOC (Sigma-Aldrich) for 12 h (Chang et al., 2005b). Cells were harvested for analysis at intervals from 0 to 12 h during the NOC incubation. After 12 h in NOC, cells were collected by mitotic shake-off, replated in fresh medium, and harvested for analysis at intervals from 0 to 4 h.
siRNA and transfection
siRNA (Table S1) was synthesized from Invitrogen. Transfections were performed in 6-well plate according to the manufacturer’s instructions using Lipofectamine RNAiMax (Invitrogen).
Immunoprecipitation
As described previously (Wang et al., 2009), cells were harvested and lysed in NETN-400 buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40) with protease inhibitors and protein phosphatase inhibitors, for 20 min on ice. The samples were centrifuged at 13,000 rpm for 15 min, and the supernatants were diluted with the same buffer without NaCl (NETN-0) to obtain a final concentration of NaCl at 150 mM (NETN-150; 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40). The samples were then cleared by centrifugation and incubated with the appropriate antibodies at 4°C with rocking for 2 h. Protein G agarose (Roche) was then added, and the incubation was continued for an additional 2 h. Beads were then washed three times using the NETN-150 buffer. The bound proteins were eluted with 100 mM glycine, pH 2.5, and then neutralized by adding 1/10 vol of 1 M Tris-Cl, pH 8.0. Eluted proteins were separated on 4–12% SDS-PAGE and blotted with the corresponding antibodies as indicated.
Purification of BRISC complex
HeLa cells stably expressing FLAG-HA–tagged ABRO1 were used for purification of BRISC as described previously (Shao et al., 2009a,,b). In brief, cell pellets were lysed with (10 vol of pellet) NETNG400 (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 10% glycerol, and 1 mM PMSF) and proteinase inhibitor cocktail (Roche), and centrifuged at 25,000 rpm for 30 min at 4°C. The supernatants were then diluted with the same buffer without NaCl (NETNG-0) to obtain a final concentration of NaCl at 150 mM (NETNG-150). The diluted samples were subjected to affinity purification using anti–FLAG M2 antibody–coupled beads (Sigma-Aldrich) and eluted with NETENG-150 containing FLAG peptides (Sigma-Aldrich). The FLAG-peptide elutes were further purified using HA antibody–coupled beads (Sigma-Aldrich). The bound beads were extensively washed and eluted in NETENG-150 with HA peptides (Sigma-Aldrich). The final purified protein complexes were used for in vitro DUB assays.
Purification of GST fusion proteins and GST pull-down assay
GST, GST–ABRO1 (261–415 aa), and GST–MERIT40 fusion proteins were expressed and purified according to standard protocols as described previously (Shao et al., 2009a,,b). In brief, Escherichia coli BL21 (DE3) cells (New England Biolabs, Inc.) transformed with pGEX-6P-3, pGEX-6P-3-ABRO1, and pGEX-6P-3-MERIT40 were cultured at 37°C until an optical density at 600 nm of ∼0.6–1.0 was obtained, and induced by the addition of 0.1 mM IPTG for 6 h at 28°C. The cells were harvested, and the pellets were resuspended in PBS containing protease inhibitors, sonicated, and centrifuged. After centrifugation, recombinant proteins in the supernatant were purified by glutathione–Sepharose 4B according to the manufacturer’s instructions (GE Healthcare). To express GST–ABRO1 and/or GST–BRCC36 proteins in Sf9 cells, pFastBac1-GST–ABRO1 and pFastBac1-GST-BRCC36 plasmids were constructed, and the Bac-to-Bac Baculovirus Expression System (Invitrogen) was used according to the manufacturer’s instructions. Sf9 cells were infected with GST–ABRO1 and GST–BRCC36, respectively, or coinfected with GST–ABRO1 and GST–BRCC36 baculovirus stocks and harvested 48 h later (Feng et al., 2010; Hu et al., 2011). Cells were lysed in NETN150 buffer containing protease inhibitors and purified with glutathione–Sepharose 4B according to the manufacturer’s instructions (GE Healthcare). For in vitro GST pull-down, GST, GST–MERIT40, and GST–ABRO1/GST–BRCC36 immobilized on glutathione-agarose beads were incubated with mitotic HeLa cell lysate at 4°C for 2 h, respectively. The beads were washed extensively, and the proteins were eluted by adding 1× SDS loading buffer, boiled, and analyzed by Western blotting.
Immunofluorescence microscopy
For Hec1 staining, cells were permeabilized in PHEM buffer supplemented with 0.1% Triton X-100 and protease inhibitors for 1 min on ice, fixed with 3.7% paraformaldehyde in PHEM buffer for 5 min at room temperature, washed three times with PBS, and immunostained as appropriate (Hua et al., 2011). For all other fluorescence microscopy applications, cells grown on coverslips were fixed in −20°C methanol for 10 min, blocked with 10% goat serum at 37°C for 30 min, incubated at 37°C with the primary antibodies for 2 h, washed extensively and probed with the FITC- and Rhodamine red–conjugated goat anti–rabbit or anti–mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) at 37°C for 30 min. Coverslips were mounted in VECTASHIELD Mounting Medium with DAPI (Vector Laboratories). Fluorescence images of fixed cells shown in Figs. 1 (A–C and G), 2 (A and B), 6 D, 8 B, S1 (D–G), S2 (A–C and F), and S3 (A and B) were acquired in a DM Digital Microscope (DM5000; AF6000 E acquisition software; Leica) at room temperature, using a 63× HC Plan-Apochromat, NA 1.40 oil-immersion objective. Images shown in Figs. 4 (B–D) and 8 (D and G) were acquired in a laser confocal microscope (TCS SP8; LAS-AF acquisition software; Leica) at room temperature, using a 63× HC Plan-Apochromat, NA 1.40 oil-immersion objective. Images were processed using Photoshop CS5 (version 12.0; Adobe).
Live-cell imaging
Transfected H2B-GFP/α-tubulin-mRFP HeLa cells were seeded onto Petri dishes with a 15-mm glass base (NEST) and grown in DMEM media supplemented with 10% FBS without any antibiotics. Fluorescence time-lapse images were taken every 5 min for 24 h at 37°C in 5% CO2 by using a laser-scanning confocal microscope (Eclipse Ti-E; Nikon), with a 40× oil objective or 20× objective (HC Plan-Apochromat, NA 1.4; Nikon), a camera (iXon3 DU-897 EM CCD; Andor), and acquisition software (V4.00; NIS-Elements). GFP, RFP, and CFP fluorescence were examined using the conventional laser excitation and filter sets. Photoshop CS5 (version 12.0; Adobe) was used for image processing.
MT regrowth assay
The MT regrowth experiment was based on a method described previously (Yang et al., 2007; Meunier and Vernos, 2011). In brief, cells were plated 2 d before imaging. 30 ng/ml NOC was added to the medium for 4 h and washed out four times with PBS and once with medium at 0°C for 10 min to synchronize MT aster regrowth, and then fixed after recovery for the indicated times at 37°C and immunostained as appropriate. Images were taken using a laser-scanning microscope (TCS SP8; Leica) and a 63× oil objective (HC PL APO, NA 1.40; Leica), analyzed, and quantified by counting the number of MT asters in at least 100 cells to obtain the mean per cell.
In vitro MT cosedimentation assay
The in vitro MT cosedimentation assay was performed as described previously (Silljé et al., 2006) with a modification. In brief, 100 mM TAMRA rhodamine–labeled porcine tubulin (Cytoskeleton, Inc.) was incubated with 2 mM GTP (Amresco) in BRB80 buffer (80 mM K-pipes, 1 mM EGTA, and 1 mM MgCl2, pH 6.8) containing 50% glycerol for 40 min at 37°C, and then 20 µM (final concentration) taxol (Sigma-Aldrich) was added to stabilize the formed MTs, and incubated for an additional 40 min. Then the formed rhodamine-labeled MTs were mixed with and without purified BRISC in BRB80 for 15 min at 25°C. In parallel, the same was done with BSA as a negative control. Samples were then centrifuged through a 40% glycero-BRB80 cushion at 120,000 g for 30 min at 25°C (Optima MAX Ultracentrifuge; Beckman Coulter). Proteins in the pellet and supernatant fractions were separated by SDS-PAGE, visualized by Ponceau staining, and analyzed by Western blot.
MT sedimentation assay with mitotic lysates
Concentrated mitotic lysate were prepared as described previously (Chang et al., 2009). In brief, HeLa cells were treated with 10 µM STLC for 11 h and then 1 µg/ml latrunculin A for 1 h at 37°C. Cells were pelleted at 400 g for 5 min and washed three times in ice-cold HBS. Pellets were flash-frozen in liquid nitrogen and thawed twice, and then centrifuged at 5,000 g at 4°C for 30 min. Supernatants were obtained and stored on ice, and 1 µg/ml cytochalasin B was added. Extracts were prepared fresh for each experiment. To facilitate observation, concentrated mitotic lysate was supplemented with 1 mg/ml TAMRA rhodamine–labeled tubulin (Cytoskeleton, Inc.). 1 mM GTP and 20% glycerol were added, and the reaction was incubated at 37°C for 30 min to allow assembly of MTs, and then centrifuged through a glycerol cushion as described above or spotted onto slides for visualization.
In vitro DUB assay
NuMA immunoprecipitated from mitotic HeLa cells by using anti-NuMA antibody was used as the substrate and incubated with purified GST–BRCC36 WT or GST–BRCC36 QSQ complex in DUB buffer at 37°C for 2 h. The reactions were stopped by adding SDS loading buffer, and samples were separated by 4–12% SDS PAGE and analyzed by Western blotting with anti–K63 ubiquitination antibody and anti-NuMA antibody.
Statistics
Data are presented as means ± SD of three independent experiments. Statistical significance was determined using Student’s t test, or χ2 test (Fig. 4 D) with the SPSS16.0 software.
Online supplemental material
Fig. S1 shows the RNAi depletion efficiencies of ABRO1, MERIT40, and BRCC36, as well as the mitotic defects upon depletion of BRISC. Fig. S2 shows the localization of endogenous ABRO1, eYFP-ABRO1 and eYFP-MERIT40. Fig. S3 shows the ratio of the ABRO1 intensity localized at kinetochores/centrosomes during spindle reassembly. Fig. S4 shows Western blotting analysis of H2B-GFP/α-tubulin-mRFP HeLa cells transfected with control or BRCC36 siRNA, or cotransfected with BRCC36 siRNA and CFP-BRCC36res WT or with BRCC36 siRNA and CFP-BRCC36res QSQ. Fig. S5 shows mass spectrometry analysis of ABRO1-associated proteins, and the proposed model for BRISC’s function in spindle assembly. Video 1 (based on Fig. 2) shows time-lapse imaging of dividing control siRNA–treated H2B-GFP/α-tubulin-mRFP HeLa cells. Videos 2 and 3 (based on Fig. 2) show time-lapse imaging of dividing ABRO1-silenced H2B-GFP/α-tubulin-mRFP HeLa cells. Video 4 (based on Fig. 2) shows time-lapse imaging of dividing ABRO1-silenced H2B-GFP/α-tubulin-mRFP HeLa cells, which was rescued by CFP-ABRO1 res. Video 5 (based on Fig. 5) shows time-lapse imaging of dividing control siRNA–treated H2B-GFP/α-tubulin-mRFP HeLa cells. Video 6 (based on Fig. 5) shows time-lapse imaging of dividing BRCC36-silenced H2B-GFP/α-tubulin-mRFP HeLa cells. Video 7 (based on Fig. 5) shows time-lapse imaging of dividing BRCC36-silenced H2B-GFP/α-tubulin–mRFP HeLa cells, which was rescued by DUB-active CFP-BRCC36res WT. Video 8 (based on Fig. 5) shows time-lapse imaging of dividing BRCC36-silenced H2B-GFP/α-tubulin-mRFP HeLa cells, which was rescued by DUB-inactive CFP-BRCC36res QSQ. Video 9 (based on Fig. S2 D) shows localization of eYFP-ABRO1 in α-tubulin-mRFP HeLa cells.
Acknowledgments
We thank Dr. Roger Greenberg for his support. Many plasmids in this study are either provided by him or derived from those plasmids he provided. We also thank Dr. Zhong Zhang (Beijing Key Laboratory of Tumor Systems Biology, Peking University) for microscope support.
This study was supported by grants from the National Natural Science Foundation of China (3117192, 81071655, and 81430056); the Doctoral Program of Higher Education (20110001110013); Beijing Natural Science Foundation (5112016); The Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (JWSL44-8); The Project for Extramural Scientists of State Key Laboratory of Agrobiotechnology (2015SKLAB6-27); and 985 Program, Ministry of Education of China.
The authors declare no competing financial interests.
References
Author notes
K. Yan and L. Li contributed equally to this paper.