Communication usually applies feedback loop–based filters and amplifiers to ensure undistorted delivery of messages. Such an amplifier acts during Drosophila melanogaster midoogenesis, when oskar messenger ribonucleic acid (mRNA) anchoring depends on its own locally translated protein product. We find that the motor regulator Klar β mediates a gain-control process that prevents saturation-based distortions in this positive feedback loop. We demonstrate that, like oskar mRNA, Klar β localizes to the posterior pole of oocytes in a kinesin-1–dependent manner. By live imaging and semiquantitative fluorescent in situ hybridization, we show that Klar β restrains oskar ribonucleoprotein motility and decreases the posterior-ward translocation of oskar mRNA, thereby adapting the rate of oskar delivery to the output of the anchoring machinery. This negative regulatory effect of Klar is particularly important for overriding temperature-induced changes in motility. We conclude that by preventing defects in oskar anchoring, this mechanism contributes to the developmental robustness of a poikilothermic organism living in a variable temperature environment.
Introduction
Complex systems must function predictably even in the face of external and internal perturbations (Kitano, 2004). Coping with varying ambient temperature is one of the greatest challenges, as it directly impacts on physicochemical properties of any machinery. For example, when embryos of various fruit fly species are exposed to temperatures ranging from 17.5 to 27.5°C, they develop at dramatically different rates; yet, they successfully complete embryogenesis at all these temperatures, and the relative timing of major developmental events remains invariant (Kuntz and Eisen, 2014). As the underlying chemical reactions all scale uniquely according to temperature, dedicated regulatory mechanisms must operate to establish such thermal robustness.
In both engineering applications and biological systems, robustness is often achieved by combinations of negative and positive feedback loops: negative feedback returns a system to its original state, whereas positive feedback loops act as amplifiers to support switchlike decision making or to ensure reliable information transfer in communication, such as in cell-to-cell signaling (Guyton, 1991). However, amplifiers generate output predictably only within a certain range of input. Thus, to avoid risks of instability, the gain of these processes must be carefully controlled.
During Drosophila melanogaster oogenesis, positive feedback loops play a critical role in germ plasm assembly in midstage oocytes (Zimyanin et al., 2007). The germ (pole) plasm is essential for determining the germline and the anteroposterior (AP) axis of the future embryo; its assembly is initiated by the protein products of oskar mRNA (Ephrussi et al., 1991; Kim-Ha et al., 1991; St Johnston et al., 1991; Breitwieser et al., 1996; Mahowald, 2001). Over many hours, oskar mRNA and its associated proteins (mRNPs) continue to arrive at the posterior pole (Sinsimer et al., 2011) where the mRNA is selectively translated, as a result of localized translational derepression (Kim-Ha et al., 1995; Yoshida et al., 2004), and anchored to the cortex, in part via the long Oskar isoform (Markussen et al., 1995; Vanzo and Ephrussi, 2002). This sets up a self-reinforcing process whereby more anchored oskar mRNA leads to more Oskar protein, which—in turn—anchors additional oskar mRNA. This positive feedback loop is likely instrumental for robust assembly of the germ plasm. Presumably, there are mechanisms that prevent uncontrolled output of this loop, but they are unknown.
The gain of this feedback loop depends on the delivery of oskar mRNA to the posterior pole. In the oocyte, oskar mRNPs actively move along microtubules, undergoing long, directed movements in a seemingly random fashion with a slight posterior-ward bias, probably as a result of polarization of the microtubule network (Zimyanin et al., 2008; Parton et al., 2011; Ghosh et al., 2012). Posterior-ward transport of oskar depends on kinesin-1, a molecular motor that moves cargo toward the plus ends of microtubules (Vale et al., 1985; Palacios and St Johnston, 2002; Zimyanin et al., 2008; Loiseau et al., 2010). Although previous studies have identified numerous components essential for oskar mRNA localization, the regulation of kinesin-1 in oskar transport and thus the timing of mRNA accumulation remain poorly understood.
Kinesin-1 also powers the motion of lipid droplets in Drosophila embryos (Welte et al., 1998; Shubeita et al., 2008). Correct temporal regulation depends on the Klar (Klarsicht) protein, in particular the Klar β isoform (Guo et al., 2005). In the present work, we demonstrate that Klar β—which forms a complex with kinesin-1—modulates the motility of oskar mRNPs and the distribution of oskar mRNA in the oocyte. This modulation adapts the rate of RNP arrival to the output of the anchoring machinery, ensuring proper coupling between elements of a developmentally critical positive feedback loop. We find that without Klar, this feedback loop is sensitive to thermal perturbations: even a mild reduction in growth temperature causes a dramatic disruption of the germ plasm assembly process and of embryogenesis, resulting in substantial lethality. We conclude that Klar overrides temperature-dictated changes in mRNP motility and thus contributes to overall robustness of development.
Results
Klar β accumulates at the oocyte posterior pole during midoogenesis
Klar plays important regulatory roles during development, such as in lipid droplet motion in early embryos (Welte et al., 1998), nuclear positioning in the optical neuroepithelium and in myofibers (Fischer-Vize and Mosley, 1994; Elhanany-Tamir et al., 2012), and apical membrane growth in embryonic salivary glands (Myat and Andrew, 2002). Klar is also prominently expressed during oogenesis (Guo et al., 2005), enriched around the nuclei of nurse cells, oocytes, and follicle cells (Technau and Roth, 2008; Xie and Fischer, 2008). When labeling egg chambers with anti-Klar antibodies (Kracklauer et al., 2007), we detected—in addition to the previously reported perinuclear signal—transient accumulation of Klar puncta in a tight crescent at the posterior pole of stage 9–10 oocytes (Fig. 1, A and B; and Fig. S1, M and R). During stages 6–8, a strong Klar signal was present throughout the oocyte cytoplasm (Fig. 1 A and Fig. S1 C), and from stage 10B onward, posterior enrichment of Klar could no longer be detected (Fig. S1 W).
The Klar β isoform accumulates at the posterior pole of midstage oocytes. (A and B) Klar accumulates in oocytes during stages 7 and 8 and prominently localizes at the posterior pole starting from early stage 9, as revealed by Klar-MK antibodies. Higher magnification (B) reveals distinct Klar puncta. (C) Schematic representation of the klar locus, showing the five Klar isoforms, promoters (blue arrows), noncoding exons (gray), and coding exons (red). klar alleles are indicated in green: deletions (klarYG3; klarSC2), chromosomal breaks (klarmBX18; klarmBX13), and nonsense mutations (klarB; klarmBX14; klar1). Epitopes recognized by antibodies Klar-MK, Klar-M, and Klar-C are shown in magenta, and the two characterized domains of Klar (lipid droplet–targeting domain [LD] and KASH) are shown in blue. See Kim et al. (2013) for details. Ruler indicates distance (in kilobases) to the telomeric tip of the left arm of the 3rd chromosome. (D–F) Posteriorly accumulated Klar is detected by Klar-MK (D) and Klar-M (E) but not by the Klar-C antibody (F). (G) In oocytes, GFP–Klar β accumulates at the posterior pole. In oocytes with higher expression levels, GFP–Klar β accumulates both at the posterior and ectopically (not depicted). (H) No posterior Klar is detected in klarYG3, in which the promoter for both Klar α and Klar β is deleted. Posterior Klar is also absent in klarmBX18 and klarmBX14 (not depicted). (I) In klarmBX13, Klar accumulation at the posterior is normal. (J) In midway mutants, in which lipid droplets are essentially absent from oocytes (Buszczak et al., 2002), posterior Klar localization is normal. (K) Klar localizes to the posterior pole in klar1 mutant oocytes that express a truncated molecule lacking LD and KASH domains (Guo et al., 2005). (L) At high magnification, posterior Klar is present in distinct puncta that show minimal overlap with GFP-LD, a marker for lipid droplets (Yu et al., 2011). (M) By immunoblotting, Klar-M antibody recognizes two bands >250 kD in the ovarian lysate, presumably Klar isoforms α and β, as γ, δ, and ε have apparent molecular masses <200 kD (Guo et al., 2005; Kim et al., 2013). Because only the top band is recognized by antibody Klar-C, we propose that it represents Klar α, and the bottom band represents Klar β. (N) Klar-M Western blot of ovarian samples. The klarYG3 allele abolished both α and β isoforms. For klarmBX13, the top Klar band is shifted downward, consistent with a truncated Klar α protein encoded by this allele. Df indicates Df(3L)emcE12. Khc was used as a loading control (L and M, bottom bands). wt, wild type. Bars: (A, B, and D–K) 20 µm; (L) 5 µm.
The Klar β isoform accumulates at the posterior pole of midstage oocytes. (A and B) Klar accumulates in oocytes during stages 7 and 8 and prominently localizes at the posterior pole starting from early stage 9, as revealed by Klar-MK antibodies. Higher magnification (B) reveals distinct Klar puncta. (C) Schematic representation of the klar locus, showing the five Klar isoforms, promoters (blue arrows), noncoding exons (gray), and coding exons (red). klar alleles are indicated in green: deletions (klarYG3; klarSC2), chromosomal breaks (klarmBX18; klarmBX13), and nonsense mutations (klarB; klarmBX14; klar1). Epitopes recognized by antibodies Klar-MK, Klar-M, and Klar-C are shown in magenta, and the two characterized domains of Klar (lipid droplet–targeting domain [LD] and KASH) are shown in blue. See Kim et al. (2013) for details. Ruler indicates distance (in kilobases) to the telomeric tip of the left arm of the 3rd chromosome. (D–F) Posteriorly accumulated Klar is detected by Klar-MK (D) and Klar-M (E) but not by the Klar-C antibody (F). (G) In oocytes, GFP–Klar β accumulates at the posterior pole. In oocytes with higher expression levels, GFP–Klar β accumulates both at the posterior and ectopically (not depicted). (H) No posterior Klar is detected in klarYG3, in which the promoter for both Klar α and Klar β is deleted. Posterior Klar is also absent in klarmBX18 and klarmBX14 (not depicted). (I) In klarmBX13, Klar accumulation at the posterior is normal. (J) In midway mutants, in which lipid droplets are essentially absent from oocytes (Buszczak et al., 2002), posterior Klar localization is normal. (K) Klar localizes to the posterior pole in klar1 mutant oocytes that express a truncated molecule lacking LD and KASH domains (Guo et al., 2005). (L) At high magnification, posterior Klar is present in distinct puncta that show minimal overlap with GFP-LD, a marker for lipid droplets (Yu et al., 2011). (M) By immunoblotting, Klar-M antibody recognizes two bands >250 kD in the ovarian lysate, presumably Klar isoforms α and β, as γ, δ, and ε have apparent molecular masses <200 kD (Guo et al., 2005; Kim et al., 2013). Because only the top band is recognized by antibody Klar-C, we propose that it represents Klar α, and the bottom band represents Klar β. (N) Klar-M Western blot of ovarian samples. The klarYG3 allele abolished both α and β isoforms. For klarmBX13, the top Klar band is shifted downward, consistent with a truncated Klar α protein encoded by this allele. Df indicates Df(3L)emcE12. Khc was used as a loading control (L and M, bottom bands). wt, wild type. Bars: (A, B, and D–K) 20 µm; (L) 5 µm.
The klar locus encodes multiple isoforms (Fig. 1 C): Klar α, β, γ, δ, and ε (Guo et al., 2005; Kim et al., 2013). To identify the isoform enriched at the oocyte posterior, we compared the labeling pattern produced by three Klar-specific antibodies that recognize different subsets of isoforms (Fig. 1 C). Klar-M and Klar-MK, but not Klar-C, detected Klar at the posterior pole (Fig. 1, D and E), even though Klar-C labeling was prominent around nuclei (Fig. 1 F; see also Guo et al., 2005; Technau and Roth, 2008). Deletion of the α/β promoter (klarYG3) abolished both perinuclear and posterior Klar signal (Fig. 1 H), whereas alleles that truncate Klar α, γ, and δ (klarmBX13) or delete the promoter for δ and ε (klarSC2) still exhibited Klar accumulation at the posterior pole (Fig. 1 I and not depicted). These findings indicate that it is the Klar β isoform that accumulates at the posterior pole. Consistent with this conclusion, Western analysis showed that Klar β is indeed expressed in ovaries (Fig. 1, M and N), and when expressed in the female germline, GFP–Klar β accumulated in puncta at the posterior pole of stage 9 and 10 oocytes, even in the absence of endogenous Klar (Fig. 1 G).
The only previously described function of Klar β is in the motion of embryonic lipid droplets. Although in early embryos this protein is droplet associated (Guo et al., 2005), in the oocyte, we failed to detect colocalization of Klar and droplets at the posterior pole (Fig. 1 L). In addition, Klar distribution was unaltered in oocytes lacking lipid droplets (Fig. 1 J) or in a Klar mutant lacking the lipid droplet–targeting domain (Fig. 1 K; Yu et al., 2011). We conclude that in oocytes, posterior-bound Klar β is not associated with lipid droplets, suggesting a novel cargo and a novel function for this Klar isoform.
Klar β accumulation at the posterior requires kinesin-1
Kinesin-1 is responsible for the microtubule-dependent transport of most cargoes localized at the oocyte posterior pole. In midstage oocytes, the pattern of Klar localization resembled the distribution of kinesin-1 reported previously (Clark et al., 1994; Brendza et al., 2002): kinesin-1 accumulated uniformly throughout stage 6–8 oocytes (Fig. S1, A and F) and, by stage 9, concentrated in a posterior crescent (Fig. 2 A and Fig. S1 K). Kinesin-1 and Klar signals largely overlapped at the posterior pole during stages 9 and 10 (Fig. 2 A).
Klar is in a complex with kinesin-1, and its posterior accumulation depends on kinesin-1. (A) Klar and kinesin-1 colocalize at the posterior pole of an early stage 10 oocyte. (B) In the wild-type ovarian lysate, Klar (detected by Klar-M antibody) coprecipitated with kinesin-1 but was not precipitated by generic IgG. No Klar coprecipitation with Khc was observed in two klar alleles (klarB and klarYG3) lacking α and β isoforms. (C) Anti-GFP antibody coprecipitated Klar (Klar-M) from GFP-Khc–expressing lysates but not from wild-type lysates. All blots are stainings from the same gel. (D) Posterior localization of Klar was abolished in Khc27 germline clones. Instead, Klar was mislocalized to the anterior (arrows). Ab, antibody; IP, immunoprecipitation; wt, wild type. Bars, 20 µm.
Klar is in a complex with kinesin-1, and its posterior accumulation depends on kinesin-1. (A) Klar and kinesin-1 colocalize at the posterior pole of an early stage 10 oocyte. (B) In the wild-type ovarian lysate, Klar (detected by Klar-M antibody) coprecipitated with kinesin-1 but was not precipitated by generic IgG. No Klar coprecipitation with Khc was observed in two klar alleles (klarB and klarYG3) lacking α and β isoforms. (C) Anti-GFP antibody coprecipitated Klar (Klar-M) from GFP-Khc–expressing lysates but not from wild-type lysates. All blots are stainings from the same gel. (D) Posterior localization of Klar was abolished in Khc27 germline clones. Instead, Klar was mislocalized to the anterior (arrows). Ab, antibody; IP, immunoprecipitation; wt, wild type. Bars, 20 µm.
To determine whether kinesin-1 and Klar are part of the same complex, we immunoprecipitated kinesin-1 from ovarian lysates using an anti–kinesin heavy chain (Khc) antibody. In the case of wild-type lysates, a Klar band was present in the pellet fraction after precipitation with anti-Khc but not with a generic immunoglobulin control (Fig. 2 B). This band co-migrated with Klar β and was absent from ovarian lysates of klar mutants that express no Klar α and Klar β. We also used fly lines expressing GFP-tagged Khc (Sung et al., 2008). After precipitation with anti-GFP, we detected Klar β in the pellet (Fig. 2 C).
To test whether Klar posterior localization depends on the kinesin-1–based transport machinery, we generated oocytes lacking Khc. In Khc27 homozygous germline clones, Klar was enriched near the anterior margin of the oocyte and failed to accumulate at the posterior (Fig. 2 D). In summary, our results reveal that Klar and kinesin-1 are in a common complex and that posterior accumulation of Klar depends on kinesin-1 activity.
Absence of Klar β causes temperature-dependent mislocalization of oskar mRNA
oskar mRNA is the most prominent cargo to localize at the posterior pole of midstage oocytes in a kinesin-1–dependent manner (Brendza et al., 2002; Palacios and St Johnston, 2002). Klar and Staufen, a double-strand RNA binding protein and obligatory partner of oskar mRNA during oogenesis (St Johnston et al., 1991; Palacios and St Johnston, 2002), colocalized at the posterior during stage 9 (Fig. 3 B and Fig. S1, K–N) as well as during stage 10A (Fig. S1, P–S), when oskar RNPs display long, unidirectional runs in that area (Fig. S1, O and T; Zimyanin et al., 2008). Klar and Staufen also coprecipitated from ovarian lysates (Fig. 3, D and E), and oskar mRNA was specifically enriched when GFP–Klar β was precipitated from ovarian extract (Fig. 3 F). This interaction is apparently not critical for normal Klar localization: Klar accumulated robustly at the posterior of oocytes lacking oskar RNA (Fig. 3 A). These observations indicate that oskar RNPs are among the posterior-bound cargoes with which Klar β associates.
Klar is in complex with Staufen, but its posterior accumulation does not depend on Oskar or Staufen. (A) oskar RNA–null oocytes in which the oskar 3′UTR was overexpressed (UASoskar3′UTR; oskar-Gal4; oskA87/Df(3R)pXT103) to overcome the early arrest of oogenesis in these mutants (Jenny et al., 2006). The oskar 3′UTR on its own fails to localize at the posterior pole, as reflected by a fairly homogenous distribution of Staufen (Stau). Klar still robustly accumulates at the posterior. (B) Staufen and Klar colocalize at the posterior pole in the presence of oskar mRNA. (A and B) Colocalizing pixels are shown in white. (C) In klarYG3 homozygous oocytes, both Staufen and Khc localize to the posterior pole. The arrowhead indicates ectopic Staufen patch. (D) In the wild-type ovarian lysate, Klar (detected by the Klar-M antibody) coprecipitated with Staufen but was not precipitated by generic IgG. (E) Anti-GFP antibody coprecipitated Staufen from ovarian samples expressing GFP–Klar β (detected by GFP antibody) in the female germline but not from the wild type. (F) qRT-PCR results of RNA immunoprecipitation experiments amplifying oskar and rp49 mRNAs from anti-GFP and control IgG precipitates of EGFP-, GFP–Klar β-, and Staufen-GFP–containing ovarian lysates. Bars represent the mean difference of take-off cycles (Ct) relative to the EGFP immunoprecipitation (first lane) normalized to the Ct values measured in the input (ΔΔCt method). Error bars indicate SDs. ***, P < 0.001 indicates significant difference of Ct values compared with EGFP control (two-sample Student’s t test). Ab, antibody; IP, immunoprecipitation; wt, wild type. Bars, 20 µm.
Klar is in complex with Staufen, but its posterior accumulation does not depend on Oskar or Staufen. (A) oskar RNA–null oocytes in which the oskar 3′UTR was overexpressed (UASoskar3′UTR; oskar-Gal4; oskA87/Df(3R)pXT103) to overcome the early arrest of oogenesis in these mutants (Jenny et al., 2006). The oskar 3′UTR on its own fails to localize at the posterior pole, as reflected by a fairly homogenous distribution of Staufen (Stau). Klar still robustly accumulates at the posterior. (B) Staufen and Klar colocalize at the posterior pole in the presence of oskar mRNA. (A and B) Colocalizing pixels are shown in white. (C) In klarYG3 homozygous oocytes, both Staufen and Khc localize to the posterior pole. The arrowhead indicates ectopic Staufen patch. (D) In the wild-type ovarian lysate, Klar (detected by the Klar-M antibody) coprecipitated with Staufen but was not precipitated by generic IgG. (E) Anti-GFP antibody coprecipitated Staufen from ovarian samples expressing GFP–Klar β (detected by GFP antibody) in the female germline but not from the wild type. (F) qRT-PCR results of RNA immunoprecipitation experiments amplifying oskar and rp49 mRNAs from anti-GFP and control IgG precipitates of EGFP-, GFP–Klar β-, and Staufen-GFP–containing ovarian lysates. Bars represent the mean difference of take-off cycles (Ct) relative to the EGFP immunoprecipitation (first lane) normalized to the Ct values measured in the input (ΔΔCt method). Error bars indicate SDs. ***, P < 0.001 indicates significant difference of Ct values compared with EGFP control (two-sample Student’s t test). Ab, antibody; IP, immunoprecipitation; wt, wild type. Bars, 20 µm.
In the absence of Klar, oskar mRNA and Staufen still accumulated at the posterior (Fig. 3 C, Fig. 4 B, and Fig. S3 A) but not perfectly normally. When the flies were kept at 25°C, we occasionally observed ectopic patches of oskar RNA (3/42; Fig. 4 B and see Fig. 6 C) and Staufen protein (8%; Fig. 3 C) away from the cortex. These defects were minor; qualitatively, oskar accumulation appeared almost identical between wild-type and mutant oocytes (Fig. 4, A and B). However, when the flies were kept at 18°C, ectopic oskar patches were much more frequent (31/79; Fig. 4, C and D; and see Fig. 6 N), and oskar mislocalization was severe. These observations suggest that (a) oskar localization is an inherently temperature-sensitive process and that (b) Klar is an important component that provides thermal robustness to this process. As very little is known about the mechanisms that allow Drosophila to develop consistently in its variable temperature environment, understanding Klar’s role in oskar localization provides a unique opportunity to unravel strategies that support robust development.
oskar mRNA and Oskar protein localization. (A–D) oskar mRNA (osk) and Oskar protein (Osk) localization in stage 10 wild-type (A and C) and klarYG3/klarYG3 (B and D) oocytes at 25°C (A and B) and 18°C (C and D). wt, wild type. Bars, 20 µm.
oskar mRNA and Oskar protein localization. (A–D) oskar mRNA (osk) and Oskar protein (Osk) localization in stage 10 wild-type (A and C) and klarYG3/klarYG3 (B and D) oocytes at 25°C (A and B) and 18°C (C and D). wt, wild type. Bars, 20 µm.
Klar β restrains oskar mRNP motility in a temperature-dependent manner
Given that Klar is a regulator of microtubule-based transport (Welte et al., 1998), we investigated whether the observed mislocalization of oskar mRNA might result from altered oskar RNP trafficking. We used the oskMS2:MS2 coat protein–GFP system that allows quantification of the parameters of RNP motility (Zimyanin et al., 2008).
At 25°C, oskar transport was grossly normal in the absence of Klar: the majority of runs occurred near the posterior pole, as in wild-type stage 9 oocytes (Fig. 5, A and B); the fraction and the net velocity vector (Zimyanin et al., 2008; Ghosh et al., 2012) of motile oskar RNP particles also did not differ significantly from wild type (Table 1). However, the mean speed of moving RNPs was slightly, but significantly, reduced in the klar mutant background (Table 1). Strikingly, RNP travel distance was significantly increased (Table 1 and Fig. 5 C). At 18°C, we observed a reduction in motility for both genotypes compared with 25°C (Table 1); however, the displacements in the mutant oocytes were still significantly longer than in wild type (Fig. 5 F and Table 1). In addition, within a given imaging period, we observed twice as many motile particles in the absence of Klar as in the control (Fig. 5, D, E, and G; and Table 1). Identical changes in oskar RNP motility were observed in klarYG3/klar1 trans-heterozygous egg chambers (Table 1).
Motility of oskar RNPs in stage 9 oocytes. (A, B, D, and E) oskar RNP runs collected from several oocytes (number of oocytes is shown in top right corners) projected relative to the posterior pole (right) in wild-type (A and D) and klarYG3/klarYG3 (B and E) oocytes at 25°C (A and B) and 18°C (D and E). Colors represent mean velocity (see key in E); arrows represent length and directionality of runs. Vertical lines indicate distance from the posterior pole, in 10-µm increments. Bar, 10 µm. Length scales for the x and y axis are identical. Gray curves represent the outline of imaginary oocytes drawn arbitrarily to aid visual orientation. (C and F) Histogram of the length distribution of linear displacements of oskar RNP runs (thin steps) overlaid with continuous distribution (thick curves) at 25°C (C) and 18°C (F). The last bin of the histogram indicates the fraction of runs longer than 5 and 4 µm, respectively. Open arrowheads indicate the single mode of run length distribution of oskar RNPs acquired in wild-type oocytes. Closed arrows point to the “shoulders” caused by the disappearance of the single major mode in absence of Klar β. (see also Table 1). (G) Fraction of motile oskar mRNPs during time-lapse acquisition at 18°C. The fraction of motile RNPs decreases by ∼25% in the presence of Khc17 when Klar is lacking. This is expected to be the fraction of Khc17 homodimeric kinesin-1 within these oocytes, which might drive runs short and slow enough to be missed during analysis at 18°C. *, P < 0.05 and ***, P < 0.001 indicate significant difference from wild type; #, P < 0.05 compared with klarYG3 homozygotes using one-way analysis of variance with post hoc Dunnett’s test. Error bars represent SEMs. wt, wild type.
Motility of oskar RNPs in stage 9 oocytes. (A, B, D, and E) oskar RNP runs collected from several oocytes (number of oocytes is shown in top right corners) projected relative to the posterior pole (right) in wild-type (A and D) and klarYG3/klarYG3 (B and E) oocytes at 25°C (A and B) and 18°C (D and E). Colors represent mean velocity (see key in E); arrows represent length and directionality of runs. Vertical lines indicate distance from the posterior pole, in 10-µm increments. Bar, 10 µm. Length scales for the x and y axis are identical. Gray curves represent the outline of imaginary oocytes drawn arbitrarily to aid visual orientation. (C and F) Histogram of the length distribution of linear displacements of oskar RNP runs (thin steps) overlaid with continuous distribution (thick curves) at 25°C (C) and 18°C (F). The last bin of the histogram indicates the fraction of runs longer than 5 and 4 µm, respectively. Open arrowheads indicate the single mode of run length distribution of oskar RNPs acquired in wild-type oocytes. Closed arrows point to the “shoulders” caused by the disappearance of the single major mode in absence of Klar β. (see also Table 1). (G) Fraction of motile oskar mRNPs during time-lapse acquisition at 18°C. The fraction of motile RNPs decreases by ∼25% in the presence of Khc17 when Klar is lacking. This is expected to be the fraction of Khc17 homodimeric kinesin-1 within these oocytes, which might drive runs short and slow enough to be missed during analysis at 18°C. *, P < 0.05 and ***, P < 0.001 indicate significant difference from wild type; #, P < 0.05 compared with klarYG3 homozygotes using one-way analysis of variance with post hoc Dunnett’s test. Error bars represent SEMs. wt, wild type.
Motility statistics of oskar RNPs in the presence and absence of Klar
| Genotype | Oocytes | Runs | Fraction of motile RNPs | Displacement | Duration | Velocity | Net velocity vector |
| n | n | % | µm | s | µm/s | nm/s | |
| 25°C | |||||||
| Wild type | 13 | 681 | 31.95 ± 2.90 | 1.054 ± 0.682 (1.0) | 2.179 ± 1.558 (1.0) | 0.55 ± 0.01 (1.0) | 51.2 ± 107.8 |
| klarYG3/klarYG3 | 15 | 811 | 28.97 ± 3.53 | 1.368 ± 0.963 (<0.001) | 3.058 ± 2.098 (<0.001) | 0.49 ± 0.01 (<0.001) | 0.0 ± 62.70 |
| Khc17/+; klarYG3/klarYG3 | 12 | 802 | 37.82 ± 4.51 | 1.235 ± 0.887 (<0.001)c | 2.421 ± 1.817 (0.068)c | 0.56 ± 0.01 (1.0)c | 4.65 ± 73.77 |
| Khc17/+ | 7 | 273 | 31.45 ± 2.45 | 1.058 ± 0.788 (0.240) | 2.157 ± 1.702 (0.417) | 0.54 ± 0.02 (0.945) | −28.05 ± 75.3 |
| pat1r/pat1r klarYG3/klarYG3 | 15 | 660 | 33.49 ± 4.56 | 1.116 ± 0.937 (0.319) | 2.363 ± 2.071 (0.915) | 0.52 ± 0.01 (0.070) | 55.05 ± 23.27e |
| pat1r/pat1r | 6 | 499 | 41.56 ± 4.84 | 1.093 ± 0.817 (0.947)c | 2.453 ± 1.830 (<0.001)c | 0.49 ± 0.01 (<0.001) | 42.70 ± 21.20 |
| klarYG3/klar1 | 10 | 851 | 37.6 ± 2.24 | 1.458 ± 1.255 (1.0)d | 3.240 ± 2.805 (0.817)d | 0.49 ± 0.01 (1.0)d | 13.21 ± 109.64 |
| 18°C | |||||||
| Wild type | 15 | 175 | 10.2 ± 1.3 | 0.728 ± 0.543 (1.0) | 2.992 ± 2.861 (1.0) | 0.34 ± 0.02 (1.0) | 4.6 ± 116.8 |
| klarYG3/klarYG3 | 16 | 552 | 22.7 ± 2.8a | 0.902 ± 0.611 (<0.001) | 3.271 ± 2.635 (0.029) | 0.36 ± 0.01 (0.717) | 39.0 ± 90.3 |
| Khc17/+; klarYG3/klarYG3 | 15 | 319 | 15.0 ± 1.7a,b | 0.945 ± 0.763 (0.004) | 2.683 ± 2.276 (0.667)c | 0.41 ± 0.01 (0.012)c | −42.7 ± 101.3 |
| klarYG3/klar1 | 11 | 220 | 19.1 ± 2.4 | 1.109 ± 0.850 (0.005)d | 3.146 ± 2.698 (0.30)d | 0.43 ± 0.02 (<0.001)d | 15.8 ± 126.4 |
| Genotype | Oocytes | Runs | Fraction of motile RNPs | Displacement | Duration | Velocity | Net velocity vector |
| n | n | % | µm | s | µm/s | nm/s | |
| 25°C | |||||||
| Wild type | 13 | 681 | 31.95 ± 2.90 | 1.054 ± 0.682 (1.0) | 2.179 ± 1.558 (1.0) | 0.55 ± 0.01 (1.0) | 51.2 ± 107.8 |
| klarYG3/klarYG3 | 15 | 811 | 28.97 ± 3.53 | 1.368 ± 0.963 (<0.001) | 3.058 ± 2.098 (<0.001) | 0.49 ± 0.01 (<0.001) | 0.0 ± 62.70 |
| Khc17/+; klarYG3/klarYG3 | 12 | 802 | 37.82 ± 4.51 | 1.235 ± 0.887 (<0.001)c | 2.421 ± 1.817 (0.068)c | 0.56 ± 0.01 (1.0)c | 4.65 ± 73.77 |
| Khc17/+ | 7 | 273 | 31.45 ± 2.45 | 1.058 ± 0.788 (0.240) | 2.157 ± 1.702 (0.417) | 0.54 ± 0.02 (0.945) | −28.05 ± 75.3 |
| pat1r/pat1r klarYG3/klarYG3 | 15 | 660 | 33.49 ± 4.56 | 1.116 ± 0.937 (0.319) | 2.363 ± 2.071 (0.915) | 0.52 ± 0.01 (0.070) | 55.05 ± 23.27e |
| pat1r/pat1r | 6 | 499 | 41.56 ± 4.84 | 1.093 ± 0.817 (0.947)c | 2.453 ± 1.830 (<0.001)c | 0.49 ± 0.01 (<0.001) | 42.70 ± 21.20 |
| klarYG3/klar1 | 10 | 851 | 37.6 ± 2.24 | 1.458 ± 1.255 (1.0)d | 3.240 ± 2.805 (0.817)d | 0.49 ± 0.01 (1.0)d | 13.21 ± 109.64 |
| 18°C | |||||||
| Wild type | 15 | 175 | 10.2 ± 1.3 | 0.728 ± 0.543 (1.0) | 2.992 ± 2.861 (1.0) | 0.34 ± 0.02 (1.0) | 4.6 ± 116.8 |
| klarYG3/klarYG3 | 16 | 552 | 22.7 ± 2.8a | 0.902 ± 0.611 (<0.001) | 3.271 ± 2.635 (0.029) | 0.36 ± 0.01 (0.717) | 39.0 ± 90.3 |
| Khc17/+; klarYG3/klarYG3 | 15 | 319 | 15.0 ± 1.7a,b | 0.945 ± 0.763 (0.004) | 2.683 ± 2.276 (0.667)c | 0.41 ± 0.01 (0.012)c | −42.7 ± 101.3 |
| klarYG3/klar1 | 11 | 220 | 19.1 ± 2.4 | 1.109 ± 0.850 (0.005)d | 3.146 ± 2.698 (0.30)d | 0.43 ± 0.02 (<0.001)d | 15.8 ± 126.4 |
Fraction of motile RNPs are means ± SEM; the fraction of motile particles is statistically not different among the different genotypes at 25°C (Kruskal–Wallis test: P = 0.238; α = 0.05). Displacement and duration are means ± SD; SD is used to represent variance, as the data distribution is not greatly different from normal (Fig. 4 G); the numbers in parentheses indicate the p-value of a pairwise Mann–Whitney U test (α = 0.005) against the wild-type control after the equivalence of the different genotypes was rejected in a Kruskal–Wallis test (P < 0.001 for both displacement and duration). Velocity measurements are in means ± SEM; the numbers in parentheses indicate the p-value of a post hoc Dunnett’s T3 test (α = 0.005, equal variances are not assumed; Levene’s test: P < 0.001) against the wild type. The last column represents the resulting net velocity vector after vectorially averaging the velocities of oskar RNP runs in individual oocytes of a given genotype. None of the net velocity vectors are significantly different from 0 (two-tailed Wilcoxon sum ranks test: P > 0.05; α = 0.05) with the exception of pat1r/pat1r;;klarYG3/klarYG3.
Significant difference of motile fraction to wild-type control at 18°C (Mann–Whitney U test: α = 0.05; P < 0.05).
Significant difference of the motile fraction to klarYG3 at 18°C (Mann–Whitney U test: α = 0.05; P < 0.05).
Significant difference between statistics of oskar RNP motility in the presence and absence of mutations impairing kinesin activity (Khc17 and pat1r) in klarYG3 homozygous oocytes; P < 0.005 and α = 0.005 for all statistics using the appropriate statistical probe.
Pairwise comparison of klarYG3/klar1 with klarYG3/klarYG3 mutant oocytes using the appropriate tests.
Statistically different value from 0.
To determine whether these motility differences arise from altered cytoskeletal tracks, we analyzed polarity and persistence lengths of microtubules using EB1 tracking. The spatial organization of the microtubule network (i.e., the polarity of the network) was statistically indistinguishable between the klar mutant and wild-type oocytes (Fig. S2, A–C and G). For both genotypes, the microtubules were generally long enough within the imaging window to support start-to-end tracking of the particles. Intriguingly, microtubule persistence length was reduced in the klar mutants, suggesting a role of Klar in microtubule dynamics (as proposed by Long et al., 2013). Our analysis, however, demonstrates that the observed increase of run lengths in the absence of Klar is not simply because tracks are longer. We conclude that Klar restrains oskar RNP motility in a temperature-sensitive manner by limiting both transporter persistence and initiation of new runs.
Klar β limits the net posterior-ward transport and ensures tight localization of oskar mRNA
To understand the functional consequence of altered RNP motility, we measured the effect of lack of Klar β on overall oskar mRNA distribution. We performed semiquantitative FISH analysis in fixed oocytes (Ghosh et al., 2012). We determined the center of mass of the mRNA (Fig. 6 A and Fig. S3, A and B) and the dimensions of the posterior domain, which we defined as the area where the relative enrichment of oskar uninterruptedly exceeds the double of the expected concentration (Fig. S3, A and B).
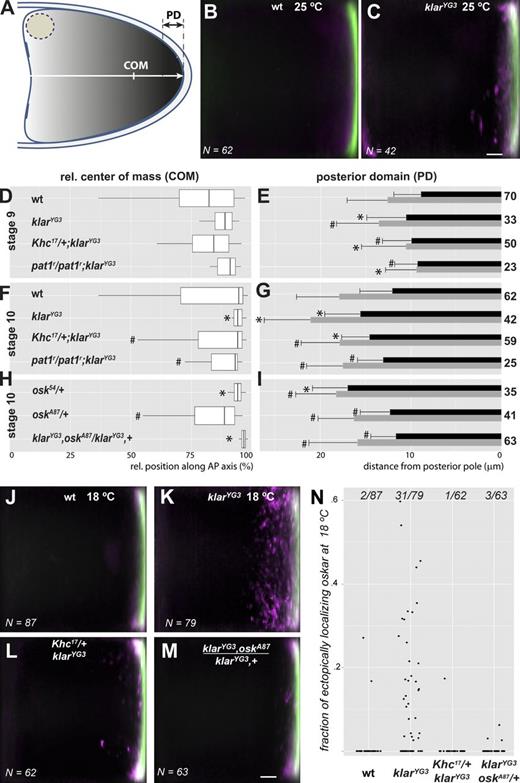
Semiquantitative analysis of oskar mRNA localization. (A) Schematic representation of the center of mass (COM) of the oskar signal and the posterior domain (PD), where oskar mRNA accumulates (see also Fig. S3). (B and C) Relative oskar mRNA distribution in stage 10 wild-type (B) and klarYG3/klarYG3 (C) oocytes. oskar mRNA distribution within similarly developed oocytes of the same genotype was averaged (see details in Fig. S3), mean oskar signal is shown in green, and SD of the signal is in magenta to indicate variability (e.g., ectopically localizing mRNA). N (bottom left corner) indicates the number of oocytes analyzed and plotted. The images in B and C are shown again alongside additional genotypes in Fig. S3 (F and G), respectively. Bar indicates 10% of relative oocyte length. (D, F, and H) Boxplots of the distribution of the center of mass of bulk oskar mRNA in stage 9 (D) and stage 10 (F and H) oocytes along the AP axis in percentages relative (rel.) to the length of the AP axis at 25°C. The boxplots indicate the 10th, 25th, 50th, 75th, and the 90th percentiles of datasets, from left to right. (E, G, and I) The size of the posterior domain of oskar mRNA (black horizontal bars) and Oskar protein (gray horizontal bars) in stage 9 (E) and stages 10–11 (G and I) oocytes at 25°C. Bars indicate the mean; error bars represent SDs of the size of the posterior domain. Genotypes are indicated left of the boxplots, and number of oocytes analyzed are to the right of the bar plots. Asterisk and hash signs indicate significant difference relative to wild-type or klarYG3 homozygous oocytes, respectively (α = 0.05; see also Table 2). In stage 10 osk54/+ oocytes, the center of mass of oskar mRNA is closer to the posterior pole than in wild type, similar to klarYG3. This shift might be a result of the reduced stability of the nonlocalized mutant oskar mRNA (oskar levels are 72 ± 5% of wild type in osk54/+ ovaries as determined by qRT-PCR; mean ± SD). (see also Fig. S3, Fig. S4, Fig. S5, and Table 2). (J–M) Relative oskar mRNA distribution in stage 10 wild-type (J), klarYG3/klarYG3 (K), Khc17/+; klarYG3/klarYG3 (L), and klarYG3,oskA87/klarYG3,osk+ (M) oocytes. (M) Bar indicates 10% of relative oocyte length. (K) Note the “ragged” posterior domain in the absence of Klar β. (N) The relative amount of oskar found ectopically localizing outside the posterior domain. Ectopic localization is defined in Fig. S3. Dots represent individual oocytes; fractions on the top represent the number of oocytes with ectopically localizing oskar. wt, wild type.
Semiquantitative analysis of oskar mRNA localization. (A) Schematic representation of the center of mass (COM) of the oskar signal and the posterior domain (PD), where oskar mRNA accumulates (see also Fig. S3). (B and C) Relative oskar mRNA distribution in stage 10 wild-type (B) and klarYG3/klarYG3 (C) oocytes. oskar mRNA distribution within similarly developed oocytes of the same genotype was averaged (see details in Fig. S3), mean oskar signal is shown in green, and SD of the signal is in magenta to indicate variability (e.g., ectopically localizing mRNA). N (bottom left corner) indicates the number of oocytes analyzed and plotted. The images in B and C are shown again alongside additional genotypes in Fig. S3 (F and G), respectively. Bar indicates 10% of relative oocyte length. (D, F, and H) Boxplots of the distribution of the center of mass of bulk oskar mRNA in stage 9 (D) and stage 10 (F and H) oocytes along the AP axis in percentages relative (rel.) to the length of the AP axis at 25°C. The boxplots indicate the 10th, 25th, 50th, 75th, and the 90th percentiles of datasets, from left to right. (E, G, and I) The size of the posterior domain of oskar mRNA (black horizontal bars) and Oskar protein (gray horizontal bars) in stage 9 (E) and stages 10–11 (G and I) oocytes at 25°C. Bars indicate the mean; error bars represent SDs of the size of the posterior domain. Genotypes are indicated left of the boxplots, and number of oocytes analyzed are to the right of the bar plots. Asterisk and hash signs indicate significant difference relative to wild-type or klarYG3 homozygous oocytes, respectively (α = 0.05; see also Table 2). In stage 10 osk54/+ oocytes, the center of mass of oskar mRNA is closer to the posterior pole than in wild type, similar to klarYG3. This shift might be a result of the reduced stability of the nonlocalized mutant oskar mRNA (oskar levels are 72 ± 5% of wild type in osk54/+ ovaries as determined by qRT-PCR; mean ± SD). (see also Fig. S3, Fig. S4, Fig. S5, and Table 2). (J–M) Relative oskar mRNA distribution in stage 10 wild-type (J), klarYG3/klarYG3 (K), Khc17/+; klarYG3/klarYG3 (L), and klarYG3,oskA87/klarYG3,osk+ (M) oocytes. (M) Bar indicates 10% of relative oocyte length. (K) Note the “ragged” posterior domain in the absence of Klar β. (N) The relative amount of oskar found ectopically localizing outside the posterior domain. Ectopic localization is defined in Fig. S3. Dots represent individual oocytes; fractions on the top represent the number of oocytes with ectopically localizing oskar. wt, wild type.
At 25°C, in klarYG3/klarYG3 and klarYG3/klar1 stage 10 oocytes, when the effects of altered motility of oskar mRNPs are expected to be manifest, the measured center of mass of oskar was significantly closer to the posterior pole than in wild type (Fig. 6, D and F; and Table 2), and a significantly larger portion of oskar mRNA was localized within the posterior domain (Table 2). Because we found no difference in the amount of oskar mRNA in klarYG3 homozygous ovaries relative to wild type (97 ± 25% of wild-type, mean ± SD, n = 4; measured by quantitative RT-PCR [qRT-PCR]), we conclude that the amount of oskar arriving in the posterior domain within a given period is increased when Klar is absent.
Semiquantitative analysis of oskar mRNA and Oskar protein distribution in fixed oocytes raised at 25°C
| Genotype | Oocytes | oskar mRNA | Oskar protein | ||
| Off center | Width of PD | Localized fraction of mRNA | Width of PD | ||
| n | µm | µm | % | µm | |
| Stage 9 | |||||
| Wild type | 70 | 26.41 ± 1.80 (1.0) | 8.89 ± 0.36 (1.0) | 51.2 ± 2.2 (1.0) | 12.21 ± 0.58 (1.0) |
| klarYG3/klarYG3 | 33 | 31.73 ± 2.97 (0.13) | 10.51 ± 0.76 (0.037) | 62.3 ± 3.3 (0.007) | 13.54 ± 0.83 (0.359) |
| Khc17/+; klarYG3/klarYG3 | 50 | 27.78 ± 2.14 (0.718)a | 9.98 ± 0.45 (0.05)a | 53.8 ± 2.8 (0.536)a | 10.34 ± 0.72 (0.012)a |
| pat1r/pat1r; klarYG3/klarYG3 | 23 | 30.20 ± 2.33 (0.235) | 9.26 ± 0.53 (0.401)b | 64.3 ± 2.9 (0.002)b | 9.36 ± 0.76 (0.004)a,b |
| pat1r/pat1r | 19 | 22.00 ± 4.513 (0.195) | 18.93 ± 1.77 (<0.001) | 47.1 ± 4.6 (0.614) | 18.40 ± 2.86c (0.015) |
| osk54/+ | 33 | 32.83 ± 2.76 (0.06) | 11.95 ± 0.64 (<0.001) | 62.2 ± 2.9 (0.005) | 12.53 ± 0.98 (0.343) |
| oskA87/+ | 39 | 25.60 ± 1.79 (0.937) | 9.50 ± 0.43 (0.061) | 56.2 ± 2.5 (0.047) | 9.669 ± 0.607 (0.002) |
| klarYG3, oskA87/klarYG3, + | 88 | 37.77 ± 1.31a (<0.001) | 9.70 ± 0.26 (0.032) | 74.7 ± 0.8 (<0.001)a | 11.125 ± 0.441 (0.040)a |
| klarYG3/klar1 | 32 | 31.30 ± 1.90 (0.870)d | 8.44 ± 0.55 (0.013)d | 77.5 ± 1.2 (<0.001)d | ND |
| Stage 10 | |||||
| Wild type | 62 | 56.05 ± 4.31 (1.0) | 12.05 ± 0.45 (1.0) | 51.7 ± 2.9 (1.0) | 17.87 ± 0.61 (1.0) |
| klarYG3/klarYG3 | 42 | 68.57 ± 5.49 (0.026) | 15.54 ± 0.61 (<0.001) | 67.5 ± 3.1 (<0.001) | 21.03 ± 0.80 (<0.001) |
| Khc17/+; klarYG3/klarYG3 | 59 | 64.24 ± 3.08 (0.440)a | 14.59 ± 0.40 (<0.001) | 57.6 ± 2.7 (0.116)a | 17.85 ± 0.56 (0.809)a |
| pat1r/pat1r; klarYG3/klarYG3 | 25 | 61.37 ± 2.91 (0.551)a | 13.03 ± 0.57 (0.214)a,b | 49.1 ± 3.9 (0.649)a | 17.45 ± 0.83 (0.855)a,b |
| pat1r/pat1r | 41 | 62.03 ± 4.915 (0.461) | 16.64 ± 0.81 (<0.001) | 46.9 ± 3.1 (0.306) | 22.62 ± 0.96 (<0.001) |
| osk54/+ | 35 | 73.38 ± 1.98 (0.033) | 16.95 ± 0.66 (<0.001) | 67.0 ± 2.5 (0.001) | 18.25 ± 0.76 (0.496) |
| oskA87/+ | 41 | 56.33 ± 3.38 (0.164) | 12.30 ± 0.51 (0.654) | 39.1 ± 2.3 (0.007) | 16.26 ± 0.63 (0.166) |
| klarYG3, oskA87/klarYG3, + | 63 | 76.74 ± 2.89 (<0.001) | 11.64 ± 0.35a (0.647) | 58.8 ± 2.3 (0.077)a | 15.86 ± 0.67 (0.112)a |
| klarYG3/klar1 | 28 | 76.44 ± 4.16 (0.390)d | 13.85 ± 0.64 (0.094)d | 67.3 ± 3.1 (0.686)d | ND |
| Genotype | Oocytes | oskar mRNA | Oskar protein | ||
| Off center | Width of PD | Localized fraction of mRNA | Width of PD | ||
| n | µm | µm | % | µm | |
| Stage 9 | |||||
| Wild type | 70 | 26.41 ± 1.80 (1.0) | 8.89 ± 0.36 (1.0) | 51.2 ± 2.2 (1.0) | 12.21 ± 0.58 (1.0) |
| klarYG3/klarYG3 | 33 | 31.73 ± 2.97 (0.13) | 10.51 ± 0.76 (0.037) | 62.3 ± 3.3 (0.007) | 13.54 ± 0.83 (0.359) |
| Khc17/+; klarYG3/klarYG3 | 50 | 27.78 ± 2.14 (0.718)a | 9.98 ± 0.45 (0.05)a | 53.8 ± 2.8 (0.536)a | 10.34 ± 0.72 (0.012)a |
| pat1r/pat1r; klarYG3/klarYG3 | 23 | 30.20 ± 2.33 (0.235) | 9.26 ± 0.53 (0.401)b | 64.3 ± 2.9 (0.002)b | 9.36 ± 0.76 (0.004)a,b |
| pat1r/pat1r | 19 | 22.00 ± 4.513 (0.195) | 18.93 ± 1.77 (<0.001) | 47.1 ± 4.6 (0.614) | 18.40 ± 2.86c (0.015) |
| osk54/+ | 33 | 32.83 ± 2.76 (0.06) | 11.95 ± 0.64 (<0.001) | 62.2 ± 2.9 (0.005) | 12.53 ± 0.98 (0.343) |
| oskA87/+ | 39 | 25.60 ± 1.79 (0.937) | 9.50 ± 0.43 (0.061) | 56.2 ± 2.5 (0.047) | 9.669 ± 0.607 (0.002) |
| klarYG3, oskA87/klarYG3, + | 88 | 37.77 ± 1.31a (<0.001) | 9.70 ± 0.26 (0.032) | 74.7 ± 0.8 (<0.001)a | 11.125 ± 0.441 (0.040)a |
| klarYG3/klar1 | 32 | 31.30 ± 1.90 (0.870)d | 8.44 ± 0.55 (0.013)d | 77.5 ± 1.2 (<0.001)d | ND |
| Stage 10 | |||||
| Wild type | 62 | 56.05 ± 4.31 (1.0) | 12.05 ± 0.45 (1.0) | 51.7 ± 2.9 (1.0) | 17.87 ± 0.61 (1.0) |
| klarYG3/klarYG3 | 42 | 68.57 ± 5.49 (0.026) | 15.54 ± 0.61 (<0.001) | 67.5 ± 3.1 (<0.001) | 21.03 ± 0.80 (<0.001) |
| Khc17/+; klarYG3/klarYG3 | 59 | 64.24 ± 3.08 (0.440)a | 14.59 ± 0.40 (<0.001) | 57.6 ± 2.7 (0.116)a | 17.85 ± 0.56 (0.809)a |
| pat1r/pat1r; klarYG3/klarYG3 | 25 | 61.37 ± 2.91 (0.551)a | 13.03 ± 0.57 (0.214)a,b | 49.1 ± 3.9 (0.649)a | 17.45 ± 0.83 (0.855)a,b |
| pat1r/pat1r | 41 | 62.03 ± 4.915 (0.461) | 16.64 ± 0.81 (<0.001) | 46.9 ± 3.1 (0.306) | 22.62 ± 0.96 (<0.001) |
| osk54/+ | 35 | 73.38 ± 1.98 (0.033) | 16.95 ± 0.66 (<0.001) | 67.0 ± 2.5 (0.001) | 18.25 ± 0.76 (0.496) |
| oskA87/+ | 41 | 56.33 ± 3.38 (0.164) | 12.30 ± 0.51 (0.654) | 39.1 ± 2.3 (0.007) | 16.26 ± 0.63 (0.166) |
| klarYG3, oskA87/klarYG3, + | 63 | 76.74 ± 2.89 (<0.001) | 11.64 ± 0.35a (0.647) | 58.8 ± 2.3 (0.077)a | 15.86 ± 0.67 (0.112)a |
| klarYG3/klar1 | 28 | 76.44 ± 4.16 (0.390)d | 13.85 ± 0.64 (0.094)d | 67.3 ± 3.1 (0.686)d | ND |
Means ± SEM; the numbers in parentheses indicate the p-value of a pairwise Mann–Whitney U test against the wild-type control of a given developmental stage performed after a Kruskal–Wallis test rejecting similarity of the data distributions (α = 0.05). Off center represents the difference between the observed and the calculated center of mass along the AP axis. Width of PD is the onset of oskar mRNA or Oskar protein enrichment, when the observed signal uninterruptedly exceeds the double of the calculated or expected signal. Distance is measured from the posterior pole. Localized fraction of mRNA is the fraction of the oskar mRNA signal found in the posterior domain.
Pairwise Mann–Whitney U test against the klarYG3/klarYG3 control with P < 0.05.
Pairwise Mann–Whitney U test against the pat1r/pat1r control with P < 0.05.
Only 9 out of 19 oocytes had detectable Oskar protein.
Pairwise comparison of klarYG3/klar1 with klarYG3/klarYG3 mutant oocytes using the appropriate tests.
However, although the overall oskar distribution was shifted toward the posterior in the absence of Klar, at the posterior pole itself, oskar failed to accumulate as tightly as in the wild type. The posterior domain itself was enlarged in stage 10 oocytes, with its anterior margin further from the posterior pole than in the wild type (Fig. 6 G and Table 2). This expansion was observed for both oskar mRNA and Oskar protein. Additionally, the rate at which the posterior domain expands between stages 9 and 10 was significantly increased in klarYG3 (Fig. S4 B), and—unlike in the wild type—expansion continued during stage 10 (Fig. S4 D).
When klarYG3 females were kept at 18°C, oskar mRNA mislocalization increased dramatically: in ∼40% of the observed stage 10 klarYG3 oocytes, oskar was found in large ectopic patches close to but outside the posterior domain (Fig. 6, K and N). Similar to what was observed at 25°C, the posterior domain did not cease to expand during stage 10 and became significantly larger than in wild type (Fig. S5, D and H). Similar defects were apparent for the Oskar protein.
To test whether the altered distribution of oskar mRNA in klar mutants is caused by the increased transport efficiency of oskar RNPs, we impaired the activity of kinesin-1 in vivo by either expressing the Khc17-encoded slow ATPase mutant of Khc (Brendza et al., 1999; Shubeita et al., 2008; Zimyanin et al., 2008) or by removing the kinesin auxiliary factor, Pat1 (Loiseau et al., 2010). Oocytes heterozygous for Khc17/+ or lacking Pat1 (pat1r homozygotes) showed no major difference in oskar RNP motility relative to the wild type (Table 1). However, when Klar was also lacking, the mean run length of RNPs was intermediate between that observed in wild-type and klarYG3 homozygous oocytes (Table 1), and the run length distribution reverted to wild type (Fig. 5, C and F; and Table 1). Also, the bulk distribution of oskar mRNA was found to be intermediate between klarYG3 homozygous and wild-type oocytes, and the posterior-ward shift of the center of mass was slightly, but significantly, reduced (Fig. 6 F and Table 2), both in the presence of Khc17 and in the absence of Pat1. The presence of a Khc17 allele did not significantly reduce the size of the posterior domain in klarYG3 mutant oocytes at 25°C (Fig. 6 G), presumably a result of the still excessive oskar mRNP motility (Table 1); however, the occurrence of ectopically localized oskar mRNA greatly diminished at both 25 and 18°C (Fig. 6, L and N; and Fig. S3 H). At 25°C, the posterior domain of pat1r, klarYG3 double mutant egg chambers returned to wild-type dimensions (Fig. 6 G), likely as the result of complete restoration of wild type–like motility (Table 1).
In summary, absence of Klar and the consequent excessive delivery of oskar RNPs causes improper formation of the oskar posterior domain, resulting in an increase in its size and, probably due to detachment, in ectopic oskar accumulation. These localization defects, in turn, cause a similar mislocalization of Oskar protein (Fig. 4 D).
Proper anchoring may depend on the oskar mRNA/Oskar protein ratio
Intuitively, the increased transport efficiency of oskar RNPs in the absence of Klar should result in better than normal localization. However, we observed that enhanced delivery of oskar RNPs negatively affects their proper localization. One possibility is that the mechanisms ensuring oskar mRNA anchoring at the posterior pole cannot cope with the increased rate of delivery of mRNPs in klarYG3 mutant oocytes and get saturated, resulting in a larger posterior domain. It is well established that cortical actin is required for long-term anchoring of oskar mRNA and the polar granules at the posterior cortex at stage 10, when ooplasmic streaming commences (Markussen et al., 1995; Vanzo and Ephrussi, 2002; Babu et al., 2004; Vanzo et al., 2007; Tanaka et al., 2011). However, phalloidin staining of klarYG3 egg chambers revealed no obvious defects in the actin network (Fig. S2, I and J).
We therefore focused on the role of Oskar protein production in this process. On the one hand, the act of translation is expected to at least transiently restrain oskar mRNA, with engaged ribosomes acting as short-term RNP-capturing platforms (Katz et al., 2012), especially on the surface of the rough ER. On the other hand, Oskar protein is required for long-term anchoring of oskar mRNA: for example, in oocytes homozygous for osk54, a nonsense allele that produces mRNA but no protein, the initially arriving oskar mRNA localizes correctly but detaches from the posterior cortex at stage 10, when ooplasmic streaming commences (Markussen et al., 1995; Vanzo and Ephrussi, 2002; Babu et al., 2004; Vanzo et al., 2007; Tanaka et al., 2011). Thus, if Oskar translation cannot keep up with all the newly arriving mRNA, the excess RNA might not be retained at the cortex.
To test whether a relative excess of oskar mRNA over Oskar protein can indeed cause posterior domain expansion, we analyzed the distribution of the mRNA in ovaries heterozygous for osk54. Such oocytes displayed a significantly enlarged posterior domain of oskar mRNA at both 25 and 18°C (Fig. 6 I and Tables 2 and 3), suggesting that the amount of Oskar protein produced, and presumably the rate of ribosome recycling, is limiting for proper oskar mRNA anchoring. Consistent with the hypothesis of a temporally limited anchor, removal of one dose of endogenous oskar mRNA in klarYG3 oocytes (klarYG3,+/klarYG3,oskA87) restored the size of the posterior domain to wild-type dimensions and greatly reduced the frequency of ectopic oskar patches both at 25°C (Fig. 6 I and Table 2) and at 18°C (Fig. 6, M and N). Interestingly, in osk54/+ oocytes, no ectopic patches of oskar mRNA were observed at either 25 or 18°C (Fig. S3, K and L), most likely because the unanchored—mostly osk54 mutant—mRNA molecules cannot produce sufficient Oskar protein to create ectopic platforms of germ plasm assembly. These observations indicate that the anchoring mechanisms can handle the half-dose of rapidly arriving mRNA and highlights the existence of a critical threshold for anchoring. oskar RNPs arriving in excess above this threshold appear to progressively enlarge the domain of final localization and—most probably through the same positive feedback loop mechanism (Zimyanin et al., 2007)—create discontinuous patches of oskar mRNA and Oskar protein.
Semiquantitative analysis of oskar mRNA and Oskar protein distribution in fixed oocytes raised at 18°C
| Genotype | Oocytes | oskar mRNA | Oskar protein | ||
| Off center | Width of PD | Localized fraction of mRNA | Width of PD | ||
| n | µm | µm | % | µm | |
| Stage 9 | |||||
| Wild type | 95 | 31.20 ± 1.72 | 9.49 ± 0.36 (1.0) | 56.9 ± 2.4 (1.0) | 10.64 ± 0.65 (1.0) |
| klarYG3/klarYG3 | 78 | 34.09 ± 1.60a | 9.11 ± 0.38 (0.650) | 67.2 ± 2.2 (0.002) | 12.02 ± 0.54a |
| Khc17/+; klarYG3/klarYG3 | 50; 25 for Oskar protein | 32.68 ± 2.25a | 10.57 ± 0.50 (0.026)b | 72.9 ± 2.1 (<0.001) | 11.73 ± 0.68a |
| klarYG3, oskA87/klarYG3, + | 67 | 32.28 ± 1.34a,b | 8.80 ± 0.32 (0.271)a | 71.6 ± 0.01 (<0.001) | 10.81 ± 0.0.51a |
| klarYG3/klar1 | 46 | 37.06 ± 1.91a,c | 9.64 ± 0.45 (0.193)c | 86.1 ± 4.0 (<0.001)c | 11.58 ± 0.56a,c |
| osk54/+ | 20 | 32.63 ± 2.59a | 9.94 ± 0.63 (0.228) | 73.0 ± 1.1 (0.002) | 12.44 ± 1.06a |
| Stage 10 | |||||
| Wild type | 87 | 65.57 ± 3.39 (1.0) | 12.43 ± 0.30 (1.0) | 60.7 ± 2.4 (1.0) | 18.58 ± 0.75 (1.0) |
| klarYG3/klarYG3 | 79 | 73.75 ± 2.19 (0.426) | 17.06 ± 0.69 (<0.001) | 66.0 ± 2.3 (0.115) | 20.24 ± 0.81 (0.146) |
| Khc17/+; klarYG3/klarYG3 | 62; 29 for Oskar protein | 80.68 ± 3.36 (<0.001)b | 15.21 ± 0.51 (<0.001) | 73.8 ± 1.5 (<0.001)b | 19.59 ± 1.04 (0.759) |
| klarYG3, oskA87/klarYG3, + | 63 | 73.49 ± 2.49 (0.137) | 13.07 ± 0.38b (0.162b) | 62.3 ± 1.8 (0.761) | 16.57 ± 0.64 (0.012)b |
| klarYG3/klar1 | 25 | 71.37 ± 2.46 (0.428)c | 22.52 ± 1.38 (<0.001)c | 79.5 ± 2.5 (0.001)c | 23.39 ± 1.54 (0.033)c |
| osk54/+ | 30 | 79.64 ± 2.39 (0.017) | 14.78 ± 0.65 (0.002) | 68.4 ± 1.9 (0.324) | 20.2 ± 0.94 (0.191) |
| Genotype | Oocytes | oskar mRNA | Oskar protein | ||
| Off center | Width of PD | Localized fraction of mRNA | Width of PD | ||
| n | µm | µm | % | µm | |
| Stage 9 | |||||
| Wild type | 95 | 31.20 ± 1.72 | 9.49 ± 0.36 (1.0) | 56.9 ± 2.4 (1.0) | 10.64 ± 0.65 (1.0) |
| klarYG3/klarYG3 | 78 | 34.09 ± 1.60a | 9.11 ± 0.38 (0.650) | 67.2 ± 2.2 (0.002) | 12.02 ± 0.54a |
| Khc17/+; klarYG3/klarYG3 | 50; 25 for Oskar protein | 32.68 ± 2.25a | 10.57 ± 0.50 (0.026)b | 72.9 ± 2.1 (<0.001) | 11.73 ± 0.68a |
| klarYG3, oskA87/klarYG3, + | 67 | 32.28 ± 1.34a,b | 8.80 ± 0.32 (0.271)a | 71.6 ± 0.01 (<0.001) | 10.81 ± 0.0.51a |
| klarYG3/klar1 | 46 | 37.06 ± 1.91a,c | 9.64 ± 0.45 (0.193)c | 86.1 ± 4.0 (<0.001)c | 11.58 ± 0.56a,c |
| osk54/+ | 20 | 32.63 ± 2.59a | 9.94 ± 0.63 (0.228) | 73.0 ± 1.1 (0.002) | 12.44 ± 1.06a |
| Stage 10 | |||||
| Wild type | 87 | 65.57 ± 3.39 (1.0) | 12.43 ± 0.30 (1.0) | 60.7 ± 2.4 (1.0) | 18.58 ± 0.75 (1.0) |
| klarYG3/klarYG3 | 79 | 73.75 ± 2.19 (0.426) | 17.06 ± 0.69 (<0.001) | 66.0 ± 2.3 (0.115) | 20.24 ± 0.81 (0.146) |
| Khc17/+; klarYG3/klarYG3 | 62; 29 for Oskar protein | 80.68 ± 3.36 (<0.001)b | 15.21 ± 0.51 (<0.001) | 73.8 ± 1.5 (<0.001)b | 19.59 ± 1.04 (0.759) |
| klarYG3, oskA87/klarYG3, + | 63 | 73.49 ± 2.49 (0.137) | 13.07 ± 0.38b (0.162b) | 62.3 ± 1.8 (0.761) | 16.57 ± 0.64 (0.012)b |
| klarYG3/klar1 | 25 | 71.37 ± 2.46 (0.428)c | 22.52 ± 1.38 (<0.001)c | 79.5 ± 2.5 (0.001)c | 23.39 ± 1.54 (0.033)c |
| osk54/+ | 30 | 79.64 ± 2.39 (0.017) | 14.78 ± 0.65 (0.002) | 68.4 ± 1.9 (0.324) | 20.2 ± 0.94 (0.191) |
Means ± SEM; numbers in parentheses indicate the p-value of a pairwise Mann–Whitney U test against the wild-type control of a given developmental stage performed after a Kruskal–Wallis test rejecting similarity of the data distributions (α = 0.05). Off center is the difference between the observed and the calculated center of mass along the AP axis. Width of PD is the onset of oskar mRNA or Oskar protein enrichment, when the observed signal uninterruptedly exceeds the double of the calculated or expected signal. Distance from the posterior pole. Localized fraction of mRNA shows the fraction of the oskar mRNA signal found in the posterior domain.
P-value of a Kruskal–Wallis test failing to reject similarity of the data distributions (α = 0.05).
Pairwise Mann–Whitney U test against the klarYG3/klarYG3 control with P < 0.05.
Pairwise comparison of klarYG3/klar1 with klarYG3/klarYG3 mutant oocytes using the appropriate tests.
Klar-mediated oskar anchoring is necessary for proper embryogenesis
The severely disrupted oskar localization in klarYG3 mutant oocytes was associated with defects in embryogenesis and female fecundity; embryogenesis is also disrupted when Oskar protein is not confined to the posterior pole as a result of oskar mRNA overexpression (Zimyanin et al., 2007). Although hatching rates of embryos from wild-type mothers were high at both 18 and 25°C (>90%), they were significantly reduced for embryos produced by klarYG3 females: to ∼78% at 25°C and ∼50% at 18°C (Fig. 7 F). In these experiments, the genotype of the embryos was identical (klarYG3/+); thus, decreased hatching must be caused by the maternal genotype.
Effects of Klar on embryonic development. (A–C″) Oskar (Osk) and Vasa (Vas) staining in embryos of wild-type (A) and klarYG3 (B and C) mothers freshly laid at 18°C. The embryos are preblastoderm stage; thus, no nuclei are present close to the embryonic cortex (A″ and B″). Dashed box in B indicates magnified area shown in C. In the absence of Klar, Oskar puncta are frequently present throughout the cytoplasm and appear to be associated with large vesicles, possibly yolk granules. These Oskar-positive structures do not enrich Vasa (C′ and C″). Dashed white lines in A–B″ indicate where the images were stitched (see Materials and methods). (D–D‴) Representative images of early embryonic phenotypes observed in embryos of klarYG3 mothers, such as early arrest/unfertilized embryos (D), posterior developmental defect reflected by the alteration of the expression pattern of a pair-rule gene, Even-skipped (Eve; D′), a lack of cells at the anterior tip of the embryo (anterior hole; D″), and completely normal embryos (laid by a wild-type mother; D‴). (E) Distribution of early embryonic phenotypes described in D. Black and white bars represent the fraction of early arrest (D) and normal-looking (D‴) embryos, respectively. Red and blue bars indicate the frequency of posterior (D′) and anterior defects (D″), respectively. Numbers indicate embryo counts. (F) Hatching rate of embryos laid by mothers of the indicated genotypes. Black bars represent the fraction of dead embryos; numbers indicate counts of embryos. (E and F) ***, P < 0.001 indicates significant difference compared with wild type; #, P < 0.05 and ###, P < 0.001 indicate significant difference compared with klarYG3 using χ2 test, omitting categories with <10 counts (E). (G and H) Cuticle preparations of unhatched klarYG3/+ embryos laid by wild-type (G) and klarYG3 females (H) at 18°C. Note that only 10% of klarYG3-derived embryos form a cuticle yet fail to hatch, and among those, the majority show patterning defects. wt, wild type. Bars: (A″, B″, D–D‴, G, and H) 50 µm; (C″) 5 µm.
Effects of Klar on embryonic development. (A–C″) Oskar (Osk) and Vasa (Vas) staining in embryos of wild-type (A) and klarYG3 (B and C) mothers freshly laid at 18°C. The embryos are preblastoderm stage; thus, no nuclei are present close to the embryonic cortex (A″ and B″). Dashed box in B indicates magnified area shown in C. In the absence of Klar, Oskar puncta are frequently present throughout the cytoplasm and appear to be associated with large vesicles, possibly yolk granules. These Oskar-positive structures do not enrich Vasa (C′ and C″). Dashed white lines in A–B″ indicate where the images were stitched (see Materials and methods). (D–D‴) Representative images of early embryonic phenotypes observed in embryos of klarYG3 mothers, such as early arrest/unfertilized embryos (D), posterior developmental defect reflected by the alteration of the expression pattern of a pair-rule gene, Even-skipped (Eve; D′), a lack of cells at the anterior tip of the embryo (anterior hole; D″), and completely normal embryos (laid by a wild-type mother; D‴). (E) Distribution of early embryonic phenotypes described in D. Black and white bars represent the fraction of early arrest (D) and normal-looking (D‴) embryos, respectively. Red and blue bars indicate the frequency of posterior (D′) and anterior defects (D″), respectively. Numbers indicate embryo counts. (F) Hatching rate of embryos laid by mothers of the indicated genotypes. Black bars represent the fraction of dead embryos; numbers indicate counts of embryos. (E and F) ***, P < 0.001 indicates significant difference compared with wild type; #, P < 0.05 and ###, P < 0.001 indicate significant difference compared with klarYG3 using χ2 test, omitting categories with <10 counts (E). (G and H) Cuticle preparations of unhatched klarYG3/+ embryos laid by wild-type (G) and klarYG3 females (H) at 18°C. Note that only 10% of klarYG3-derived embryos form a cuticle yet fail to hatch, and among those, the majority show patterning defects. wt, wild type. Bars: (A″, B″, D–D‴, G, and H) 50 µm; (C″) 5 µm.
Embryos laid by klarYG3 females displayed defects already very early in development. Many appeared arrested before the syncytial blastoderm stage (Fig. 7, D and E) or displayed abnormal nuclear distribution around cellularization (Fig. 7 D″, arrow). Consistent with early defects, the majority of unhatched embryos (∼40% of all embryos at 18°C; Fig. 7 F) failed to form a cuticle, and those that did displayed defects in overall patterning (Fig. 7 H).
The molecular basis of the observed defects is currently unknown, but the available evidence suggests that they are—at least partly—caused by aberrant Oskar activity or localization: halving the dose of oskar mRNA (klarYG3,+/klarYG3,oskA87) reduced the frequency of early developmental defects and improved the hatching rate (Fig. 7, E and F). Finally, in preblastoderm stage klarYG3 embryos, we frequently observed Oskar-positive puncta scattered throughout the entire cytoplasm (Fig. 7, B and C), whereas in wild-type embryos, Oskar is restricted to the posterior pole.
Discussion
The dynamic and accurate distribution of intracellular cargoes requires precise control of the cellular transport machinery. Previously described forms of regulation are on/off switches or regulators that enhance motor activity, such as Ensconsin, necessary for efficient kinesin-1 landing on microtubules (Sung et al., 2008), or Pat1, which increases stepping rate and processivity of kinesin-1 (Loiseau et al., 2010). The absence of such regulators impairs motor activity. In contrast, absence of Klar unleashes the motors underlying oskar mRNA transport in the Drosophila oocyte. Our analysis thus reveals a novel mode of regulation, which is to restrain motor activity to ensure an optimal ratio of two components in a positive feedback loop (Fig. 8).
Proposed model of restraining activity of Klar β on transport of oskar mRNPs. (A) In wild-type oocytes, Klar β present on oskar RNPs limits the length of active runs and thus ensures that the arriving oskar mRNPs do not saturate the translation-dependent anchoring machinery (indicated by the ribosome) either at 25 or 18°C. Translating ribosomes act as short-term RNP-capturing platforms that hand over oskar mRNA to the Oskar protein–based long-term anchoring (curved blue arrows) and get recycled. Orange-tinted background indicates motility at 25°C, and orange and cyan background areas together indicate motility at 18°C. (B) In the absence of Klar β, the elongated—and at 18°C, more frequent—runs result in a greater posterior-ward displacement of oskar mRNPs than in wild type, and oskar mRNA levels at the posterior pole rise above saturation (orange and cyan arrows). These “excessive” mRNPs get translationally derepressed and, as they are not anchored, may move away from the posterior either by active transport or passive diffusion (indicated by wavy arrows), increasing the domain occupied by oskar mRNA and Oskar protein. In extreme cases, which happen frequently at 18°C, this increase results in interruptions in the continuity of the posterior domain, creating ectopically localized patches of oskar and the germ plasm (see also Fig. 4 D).
Proposed model of restraining activity of Klar β on transport of oskar mRNPs. (A) In wild-type oocytes, Klar β present on oskar RNPs limits the length of active runs and thus ensures that the arriving oskar mRNPs do not saturate the translation-dependent anchoring machinery (indicated by the ribosome) either at 25 or 18°C. Translating ribosomes act as short-term RNP-capturing platforms that hand over oskar mRNA to the Oskar protein–based long-term anchoring (curved blue arrows) and get recycled. Orange-tinted background indicates motility at 25°C, and orange and cyan background areas together indicate motility at 18°C. (B) In the absence of Klar β, the elongated—and at 18°C, more frequent—runs result in a greater posterior-ward displacement of oskar mRNPs than in wild type, and oskar mRNA levels at the posterior pole rise above saturation (orange and cyan arrows). These “excessive” mRNPs get translationally derepressed and, as they are not anchored, may move away from the posterior either by active transport or passive diffusion (indicated by wavy arrows), increasing the domain occupied by oskar mRNA and Oskar protein. In extreme cases, which happen frequently at 18°C, this increase results in interruptions in the continuity of the posterior domain, creating ectopically localized patches of oskar and the germ plasm (see also Fig. 4 D).
In the absence of Klar, we observe an increase in the mean distance traveled by oskar RNPs and enhanced posterior accumulation of bulk oskar mRNA. Although Klar stabilizes microtubules in cultured cells (Long et al., 2013), we find no striking differences in microtubule organization close to the posterior pole in the absence of Klar. Rather, we propose that Klar affects oskar RNPs directly because Klar immunoprecipitates contain oskar mRNA and Staufen, and Klar and Staufen display developmentally restricted, oskar mRNA–dependent colocalization at the posterior pole, coinciding with the peak in motility of oskar RNPs during stages 9–10 (this study; Zimyanin et al., 2008; Ghosh et al., 2012). Because Klar accumulates at the posterior pole even in the absence of oskar mRNA, Klar likely also modulates the transport of other, as yet unknown, cargoes.
Our data show that in ovaries, Klar is in complex with kinesin-1, the dominant motor involved in oskar RNP transport (Palacios and St Johnston, 2002; Zimyanin et al., 2008). The increase in both number and travel distance of motile oskar mRNPs indicates that in the absence of Klar, the motors initiate runs more frequently and gain processivity. Although the mechanisms behind this dual regulation remain to be defined, Klar appears to limit the activity of individual mechanoenzymes. Consistent with this notion, the presence of Khc17-encoded slow ATPase Khc molecules resulted in reduction of the mean travel distance of oskar RNPs only when the restraining activity of Klar was simultaneously lacking (Khc17/+; klarYG3/klarYG3 oocytes), indicating that the Klar-mediated regulation can mask subtle malfunctions of individual motors.
Intriguingly, although lack of Klar β increases travel distances for oskar transport (this study), for lipid droplets, travel distances are decreased (Welte et al., 1998). Lipid droplets, unlike oskar RNP particles, constantly alternate between runs powered by kinesin-1 and by cytoplasmic dynein (Gross et al., 2000; Shubeita et al., 2008). It has been proposed that for lipid droplets, Klar acts as a motor coordinator, turning kinesin-1 off while cytoplasmic dynein is active and vice versa. The unrestrained tug-of-war events in the absence of Klar reduce the efficient run length of lipid droplets and might mask a second, travel-restraining activity of Klar β.
It was previously shown that reductions in oskar RNP motility coincide with a reduced rate of posterior-ward transport of the bulk of oskar mRNA, leading to a failure in oskar mRNA localization (Ghosh et al., 2012). Our analysis of klar mutant oocytes indicates that an excess rate of RNP transport also interferes with proper oskar localization. Because the majority of motile oskar mRNPs are found in the vicinity of the posterior domain (<20 µm) during stage 9, the longer and more frequent runs in the absence of Klar will increase the likelihood of encountering the anchors at the posterior cortex, even in the absence of a net directionality of RNP motility. We hypothesize that the availability of the anchors, which involve cortical actin (Babu et al., 2004; Vanzo et al., 2007; Tanaka et al., 2011), Oskar protein (Markussen et al., 1995; Rongo et al., 1995; Vanzo and Ephrussi, 2002), and for short-term anchoring, the translational machinery, is temporally limited and cannot cope with increased delivery of oskar mRNPs. This idea is supported by the observation that similar oskar localization defects result from an increase in the oskar mRNA/Oskar protein ratio (osk54/+). Moreover, in the absence of Klar, reducing oskar mRNA levels restores the size of the posterior domain to wild-type dimensions. These observations indicate that the source of the observed oskar mislocalization is indeed the excess of delivered RNPs, which presumably saturates one of the oskar anchoring mechanisms, the Oskar translation machinery.
The effect of Klar on oskar mRNP motility is fairly subtle, even more than it is for lipid droplet transport (Welte et al., 1998). This apparent “weakness” of modulation might suggest that Klar’s role as a regulator is rather unimportant. On the contrary, for both oskar RNP and lipid droplet transport, these seemingly minor quantitative differences in individual transport parameters result in dramatic net outcomes of intracellular transport. In fact, the apparent weakness of the modulation is a key feature of the type of regulation we have uncovered: according to our data, Klar limits motility of oskar RNPs up to the point at which excessive delivery of mRNPs is abolished but without overshooting and compromising normal delivery.
Only through this “subtle” but adaptable modulation can the system achieve a very precise amount of oskar RNP delivery under a range of different temperatures. This precision is apparently crucial for the fitness of the animals—more than 40% of the mutant eggs will not hatch at low temperature as a result of randomly mislocalized oskar mRNA and Oskar protein within the embryonic cytoplasm. Our findings suggest that the temperature-dictated changes in the relative speed of oskar RNP arrival and of the generation of new anchors result in asynchrony, with deleterious consequences if left uncorrected. To ensure robust development across the variable temperature environments encountered by fruit flies, Klar β protects the pole plasm from distortions by its temperature-sensitive motility-modulating activity.
In a recent study, Kuntz and Eisen (2014) have described that different Drosophila species are not only capable of developing normally under a variety of different temperatures but also maintain the relative timing of key morphological hallmarks of embryogenesis. However, the mechanisms by which synchrony of the multitude of temperature-sensitive biological processes is maintained remain elusive. Considering Klar’s critical role in several transport processes during development (Fischer-Vize and Mosley, 1994; Myat and Andrew, 2002; Elhanany-Tamir et al., 2012; discussed in Kim et al., 2013), our discovery that Klar β improves developmental robustness by limiting the activity of microtubule motors may provide a paradigm for understanding how transport and other cellular processes can be kept in synchrony even in the face of thermal perturbations.
Materials and methods
Fly stocks
All flies were raised and maintained at 25°C unless indicated otherwise. To study localization and function of Klar α, β, γ, δ, and ε (FBtr0072565, FBtr0112808, FBtr0112807, FBtr0333851, and FBtr0343526, respectively; Guo et al., 2005; Kim et al., 2013), the following alleles were used in this study: klarYG3 (FBal0194848), klarB (FBal0194849), klarSC2 (FBal0285001), klar1 (FBal0005769), klarmBX13 (FBal0039628), klarmBX14 (FBal0039629), klarmBX18 (FBal0039633), klarmBX3 (FBal0039638), and the deficiency Df(3L)emcE12 (FBab0002367) to alter Klar expression (some of these strains were gifts from J. Fischer, The University of Texas at Austin, Austin, TX; Fischer-Vize and Mosley, 1994; Welte et al., 1998; Guo et al., 2005; Yu et al., 2011; Kim et al., 2013). We used mdyqx25 (FBal0032113) and mdyrf48 (FBal0032114; gift of T. Schupbach, Princeton University, Princeton, NJ) to remove lipid droplets (Buszczak et al., 2002); Khc27 (FBal0101625; gift of W. Saxton, University of California, Santa Cruz, Santa Cruz, CA) to generate Khc-null germline clones (Brendza et al., 1999); Khc>Khc-GFP (FBal0230204; gift of P. Rørth, Institute of Molecular and Cell Biology, Singapore; Sung et al., 2008) to test interaction with Klar; Khc17 (FBal0101630; Brendza et al., 1999); pat1robin (pat1r; FBal0246618), a protein-null allele of pat1 (Loiseau et al., 2010), to impair kinesin-1 activity; osk54 (FBal0013303) to reduce Oskar protein levels; and oskA87 (FBal0141009) to reduce oskar mRNA levels and in combination with Df(3R)pXT103 (FBab0002842) to generate oskar mRNA–null oocytes (Jenny et al., 2006). Oregon R (FBst0000005) and w1118 (FBst0003605) were used as the wild type. oskar RNP motility was analyzed using the oskar>oskarMS2.10 (FBtp0070262) and hsp83>MS2 coat protein–GFP (FBtp0017709) system (Zimyanin et al., 2008). Lipid droplets were labeled with UASp>GFP-lipid droplet–targeting domain (LD), with the last 114 amino acids of Klar β—encoded by exon 15ext—C-terminally fused to EGFP (Yu et al., 2011). As controls for RNA immunoprecipitations, we used flies carrying a matαtub>Staufen-GFP (FBtp0009650; Schuldt et al., 1998) transgene or UASp>EGFP constructs (gift of L. Cooley, Yale University, New Haven, CT) in combination with the matαtub>Gal4-VP16 driver (FBtp0009293; Wakefield et al., 2000).
To generate the GFP–Klar β-expressing flies, GFP was amplified from pEGP-C1 (Takara Bio Inc.) and cloned into pUASp (Rørth, 1998). A tobacco etch virus protease site (5′-GAGAATTTGTATTTTCAGGGT-3′) was generated by oligonucleotide (oligo) synthesis and cloned 3′ to the GFP gene. A Klar β cDNA was used to clone the entire Klar β open reading frame—encoded by exon 2 through exon 15ext—in frame 3′ to the tobacco etch virus site. The resulting plasmid was injected into Drosophila embryos by Genetic Services, Inc. Transgenic flies were selected by eye color. To express the fusion protein, transgenic animals were crossed with lines carrying matαtub>Gal4-VP16 drivers.
The fertility of females mated to w1118 males was assayed by counting the number of unhatched embryos 2 d after overnight egg laying, both at 18 and 25°C. w1118 females crossed to klarYG3 males were used as a control.
Immunostaining
Ovaries of flies fed for 24 h were dissected into PBST (PBS + 0.1% Triton X-100) and were heat fixed in boiling PBST for 60 s (Guo et al., 2005). Ovarioles were separated using fine tungsten needles, and the specimen was blocked overnight at 4°C in PBS, 1 mg/ml BSA, and 0.5% Triton X-100.
Embryos were collected on apple juice plates and were fixed with heptane-saturated methanol (30 s) or heptane-saturated 4% PFA (15 min) in BRB80 (80 mM Pipes, pH 6.9, 1 mM EGTA, and 2 mM MgCl2) for immunostaining using anti-Nanos/anti-Oskar or –Eve (Even-skipped) antibodies, respectively. Fixed specimens were blocked with 10% goat serum diluted in PBS + 0.1% Tween 20.
Primary antibodies were applied overnight at 4°C in blocking solution at the following dilutions: mouse monoclonal Klar-M (Guo et al., 2005) at 1:50, mouse monoclonal Klar-C at 1:5 (Guo et al., 2005), guinea pig polyclonal anti-Klar (gift of J. Fischer and M. Kracklauer, The University of Texas at Austin, Austin, TX; referred to here as Klar-MK) at 1:1,000 (Kracklauer et al., 2007), rabbit anti-Khc at 1:1,000 (Cytoskeleton, Inc.), mouse anti-GFP at 1:200 (Roche), goat anti-Staufen at 1:200 (sc-15823; Santa Cruz Biotechnology, Inc.), rabbit anti-Vasa at 1:500 (gift of R. Pflanz, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany), and mouse anti-Eve at 1:200 (Developmental Studies Hybridoma Bank). Actin labeling was performed as described in Leibfried et al. (2013) using rhodamine-phalloidin.
To develop the signal, Alexa Fluor 488–conjugated secondary antibodies were used in single, Alexa Fluor 488 and Alexa Fluor 594 in double, and Alexa Fluor 488, Alexa Fluor 555, and Cy5 in triple labelings. Secondary antibodies diluted 1:1,000 in blocking solution were applied for 2 h at room temperature. Images were acquired at room temperature on laser-scanning microscopes (TCS SP5 [Leica] and LSM 780 [Carl Zeiss]), using 63×, 1.4 NA and 40×, 1.2 NA oil immersion objectives, respectively, using the standard image acquisition software provided by the manufacturer (LAS AF [Leica] and ZEN [Carl Zeiss]). High magnification images of Klar puncta were acquired using a 100×, 1.4 NA oil immersion objective on TCS SP5. Embryos were imaged on TCS SP5 using 40×, 1.1 NA water immersion objective in 2 × 1 or 2 × 2 tile scans. Images were created by using built-in tile-stitching algorithm of LAS AF. Darkfield images of cuticles were acquired by a zoom stereomicroscope (SZX16; Olympus) equipped with SDF Plan Apochromat 1× parfocal objective and a camera (14.2 Color Mosaic; Visitron Systems). Contrast and brightness of all images presented were linearly adjusted using ImageJ (National Institutes of Health).
Immunoprecipitation and immunoblotting
Adult ovaries were dissected and homogenized in homogenizing buffer containing 125 mM Tris-HCl, pH 7.4, 375 mM NaCl, 0.5% Triton X-100, 0.5 mM EDTA, 12% glycerol, 10 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and protease inhibitor cocktail (Sigma-Aldrich). After homogenizing, samples were centrifuged at 13,000 rpm at 4°C for 10 min. The protein concentration of the supernatant was determined with the bicinchoninic acid protein assay (Thermo Fisher Scientific). For immunoprecipitation, ∼4 mg of protein sample was incubated with 1–2 µg antibody overnight at 4°C. Then, 20 µl G protein–coupled Dynabeads (Invitrogen) was added to each sample and incubated for 4 h. Afterward, the beads were washed five times in homogenizing buffer, boiled in 2× SDS sample buffer for 10 min, and subjected to immunoblotting. Primary antibodies used were mouse monoclonal Klar-M at 1:50, mouse monoclonal Klar-C at 1:5, rabbit anti-Khc at 1:100,000, mouse anti-GFP at 1:1,000, and rabbit anti-Staufen at 1:10,000 (gift of D. St Johnston, Gurdon Institute, Cambridge, England, UK). For Klar-M and Klar-C, we sometimes used both secondary (rat anti–mouse at 1:1,000) and tertiary (goat anti–rat-HRP at 1:1,000) antibodies (applied sequentially, with a 2-h incubation time each) to amplify the signal, as previously described (Guo et al., 2005).
Live-cell imaging
Ovaries were dissected in BRB80 and were transferred into a drop of S2 cell culture medium supplemented with 10% FCS and 200 µg/ml insulin (Invitrogen). Ovarioles containing stage 9 egg chambers were separated, and the preparation was covered with Voltalef 10 S oil (VWR International) to prevent drying out of the specimen (Ghosh et al., 2012). Images of 44.95 µm × 22.48 µm × 0.9 µm × 65 s were recorded at 5.2 frames/s with a laser-scanning confocal microscope (LSM 780) and ZEN acquisition software using a 63×, 1.2 NA water immersion objective after 15 min of preincubation of the specimen at 18 or 25°C. Motile oskar mRNPs under all conditions were tracked using a custom ImageJ plugin that assists tracking by segmenting images, determining the centroid of particles based on difference of Gaussian filters with 21 ± 13 nm (mean ± SD) precision, and interconnecting manually determined nodes to generate trajectories. These trajectories were subsequently fitted with a minimal number of lines that described the entire trajectory with a goodness of fit (R2) of 0.85 for each individual fitted line. Multiple lines fitted to one trajectory were considered as different tracks. Each line was then split into runs at pauses and reversals: pauses are defined as events when the absolute value of instantaneous velocity dropped <50 nm/s for at least two consecutive frames; reversals are events when the negative instantaneous velocity was >50 nm/s for at least two consecutive frames. Leading and trailing low motile components (instantaneous velocity < 50 nm/s) were trimmed off the runs before determining the linear, Euclidian displacement, duration (minimum three frames long), and the mean travel velocity of the RNP run. The durations and run lengths in this study are considerably shorter than reported in Ghosh et al. (2012) and Zimyanin et al. (2008) because track changes, pauses, and reversals break the trajectory and because Euclidian start-to-end distance was measured instead of summing frame-to-frame displacements. However, as not the entire duration of the trajectory can be attributed to runs (as pauses, time spent on track changes, and the leading and trailing low motility components are not considered as active motion), velocities reported here are consequently higher than in the previous two studies. Rare events, such as fusion of mRNPs, were not scored. To calculate the motile fraction, the quotient of motile and all present oskar mRNPs was calculated. The number of all oskar mRNPs present in the images was estimated by manually counting the traces of mRNPs on maximal intensity projections. On average, we observed 147 ± 12 oskar particles in wild type and 149 ± 14 in klarYG3 homozygotes per recording (mean ± SEM). Runs originating from the same oocyte were placed into the oocyte coordinate system, the x axis of which was defined as the AP axis of the oocyte, to extract the net velocity vector by vectorially averaging the velocities of the runs.
To visualize microtubule growth, EB1-mCherry (gift of D. Brunner, University of Zürich, Zürich, Switzerland) was overexpressed in the female germline using oskar-Gal4 (Telley et al., 2012). Dissected oocytes were imaged as described in the previous paragraph, recording 64 µm × 64 µm × 0.9 µm × 65 s images at ∼1 frame/s. EB1 comets were manually tracked in ImageJ using the MTrackJ plugin.
Semiquantitative FISH
Ovaries were dissected from females kept at 25 or 18°C for 1 or 2 d. Standard FISH protocol was performed (Ghosh et al., 2012) on whole mount egg chambers fixed overnight in 4% EM-grade PFA using a 400-nucleotide-long digoxigenin-labeled antisense oskar probe. The signal was developed by HRP-tyramide-Cy5 signal amplification kit (PerkinElmer). Oskar protein was stained with anti-Oskar primary antibody (Anix; 1:4,000; Suyama et al., 2009) detected by Alexa Fluor 488 (Invitrogen). 3D stacks of the specimen were taken on a confocal laser-scanning microscope (LSM 780 and SP5 TCS) using 20×, 0.8 NA dry and 20×, 0.8 NA water immersion objectives and ZEN and LAS AF acquisition softwares, respectively, avoiding detector saturation. 8–14 optical sections (representing 7–13-µm thickness) representing the maximal sagittal section of the oocyte were summed. Parameters describing the spatial distribution of the signal were extracted using a custom ImageJ plugin (Ghosh et al., 2012) with some modifications. The oocyte boundaries and the AP axis were manually determined. The recorded, background-corrected signal was then redistributed in 100 × 100% images, with one dimension being the distance from the anterior membrane along the AP axis and the other representing the distance from the left lateral membrane normal to the AP axis. These relative signal distribution images were then categorized based on the sagittal area of the selected oocyte as stage 9 (1,500–7,000 µm2) or stage 10 (>7,000 µm2). These cluster boundaries were selected to best match the morphological characteristics of the appropriate stages. After classification, images representing the mean and SD of each category were calculated. Simultaneously, parameters describing localization, such as the center of mass, the off center (the difference of the observed and the expected center of mass), the anterior margin of the posterior domain (the point along the AP axis where the observed signal density first uninterruptedly exceeds the double of the expected density), the width, and the area of the posterior domain were determined.
qRT-PCR
RNA was isolated from dissected ovaries using TRI Reagent according to the manufacturer’s recommendation (Molecular Research Center, Inc.), and cDNA was synthesized by first-strand synthesis kit (SuperScript III; Invitrogen) using random hexamer priming. oskar cDNA was amplified by using two oligos flanking the first splice junction (5′-GCAACTATATATCCGTGCGCG-3′ and 5′-CCCGTCAGTTTTCGATATTCA-3′), and the resulting signal was normalized to rp49, encoding for a ribosomal protein (oligos: 5′-GCTAAGCTGTCGCACAAA-3′ and 5′-TCCGGTGGGCAGCATGTG-3′). qRT-PCR was performed on a quantitative PCR machine (7500; Applied Biosystems). Take-off cycle (Ct) values were determined by using the automated threshold function of StepOne software (Applied Biosystems).
RNA immunoprecipitation
Adult ovaries were dissected and homogenized in lysis buffer containing 10 mM Hepes, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.25% Triton X-100, 1 mM DTT, 10 mM NaF, 2 mM Na3VO4, protease inhibitor cocktail (Sigma-Aldrich), and 100 U/ml RNase inhibitor (Thermo Fisher Scientific). After homogenizing, samples were centrifuged at 13,000 rpm at 4°C for 10 min. Supernatant was treated with DNase I and precleared with 5 ml G protein–coupled Dynabeads (Invitrogen). The same amount, ∼4 mg of total protein sample, was incubated with 1–2 mg antibody for 3 h at 4°C. Then, 25 ml G protein–coupled Dynabeads was added to each sample and incubated for 1 h. The beads were washed five times in lysis buffer. Subsequently, RNA was extracted using TRIZOL (Invitrogen) reagent, purified by RNA miniprep kit (Direct-zol; Zymo Research), and reverse-transcribed using first-strand cDNA synthesis kit (Maxima H Minus; Thermo Fisher Scientific). Finally, the cDNA was subjected to qRT-PCR (see previous section), and results were analyzed using the comparative threshold cycle method.
Statistical analyses
All statistics of FISH and motility analysis, except speed, were compared using the nonparametric Kruskal–Wallis test and subsequently pairwise Mann–Whitney U tests as indicated in the table legends. Speed of the runs was compared using one-way analysis of variance followed by a post hoc Dunnett’s T3 test. Slopes of the regression lines were compared by using Student’s t test. α values are indicated in table legends. All statistical tests were performed in R 2.15.2 (R Development Core Team, 2012). Graphs were created using ggplot2 in R.
Online supplemental material
Fig. S1 shows the time course of Klar and Khc localization during midoogenesis, with respect to oskar mRNA (Staufen) and oskar RNP motility. Fig. S2 provides information about the microtubule and actin cytoskeletons in wild-type and klar mutant oocytes, as they are critical for oskar mRNP motility and anchoring, respectively. Fig. S3 gives a detailed description of our semiquantitative FISH approach and shows the remainder of the mean oskar mRNA localization images not depicted in Fig. 6. Fig. S4 shows kinetics of oskar mRNA distribution in oocytes. Fig. S5 provides additional evidence on the detrimental effect of excessive oskar RNP motility on the size of the posterior domain at 25 and 18°C. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201310010.dv.
Acknowledgments
We thank the Bloomington Drosophila Stock Center, Janice Fischer, Pernille Rørth, Damian Brunner, Lynn Cooley, William Saxton, and Trudi Schupbach for fly stocks and Martin Kracklauer, Janice Fischer, Daniel St Johnston, Ralf Pflanz, and the Developmental Studies Hybridoma Bank at the University of Iowa for antibodies. We are grateful to Florence Besse, Ivo Telley, Bernhard Hampoelz, and David Lambert for critical comments on the manuscript and to Trudi Schupbach, Elizabeth Gavis, Amanda Norvell, and Dipak Manna for helpful discussions. We also thank the European Molecular Biology Laboratory (EMBL) Advanced Light Microscopy Facility and Carl Zeiss for use of their microscopes and the EMBL Gene Core Facility for assistance with qRT-PCR.
This work was supported by start-up support from the University of Rochester and National Institute of General Medical Sciences grant GM64687 to M.A. Welte and by the EMBL (A. Ephrussi). Y.V. Yu was in part supported through National Institute of General Medical Sciences grant GM064624 to Steven P. Gross (University of California, Irvine, Irvine, CA). I. Gaspar was supported by an EMBL Interdisciplinary Postdoctoral fellowship and a European Molecular Biology Organization Long-Term Fellowship.
The authors declare no competing financial interests.
Author contributions: Y.V. Yu performed the immunostainings of Klar, Vasa, and Khc, Western blotting, immunoprecipitation experiments, and the generation of GFP–Klar β-expressing transgenic flies. I. Gaspar performed the Klar, Staufen, and Khc triple stainings, the time-lapse imaging of live oocytes, the semiquantitative FISH, analysis of embryos, and the statistical analyses. S.L. Cotton performed anti-Klar immunostaining for multiple klar genotypes. D.-H. Kim cloned Klar β and expressed Klar β fusions in oogenesis. Y.V. Yu, I. Gaspar, A. Ephrussi, and M.A. Welte conceived the experiments, analyzed the data, and wrote the manuscript.
References
Author notes
I. Gaspar and Y.V. Yu contributed equally to this paper.
A. Ephrussi and M.A. Welte contributed equally to this paper.
Y.V. Yu’s present address is Dept. of Biology, Brandeis University, Waltham, MA 02454.
S.L. Cotton’s present address is The Forsyth Institute, Boston, MA 02142.
D.-H. Kim’s present address is Dept. of Neurology, Seoul National University Hospital, Seoul 110-744, South Korea.
M.A. Welte’s present address is University of Rochester, Rochester, NY 14627.


![Figure 1. The Klar β isoform accumulates at the posterior pole of midstage oocytes. (A and B) Klar accumulates in oocytes during stages 7 and 8 and prominently localizes at the posterior pole starting from early stage 9, as revealed by Klar-MK antibodies. Higher magnification (B) reveals distinct Klar puncta. (C) Schematic representation of the klar locus, showing the five Klar isoforms, promoters (blue arrows), noncoding exons (gray), and coding exons (red). klar alleles are indicated in green: deletions (klarYG3; klarSC2), chromosomal breaks (klarmBX18; klarmBX13), and nonsense mutations (klarB; klarmBX14; klar1). Epitopes recognized by antibodies Klar-MK, Klar-M, and Klar-C are shown in magenta, and the two characterized domains of Klar (lipid droplet–targeting domain [LD] and KASH) are shown in blue. See Kim et al. (2013) for details. Ruler indicates distance (in kilobases) to the telomeric tip of the left arm of the 3rd chromosome. (D–F) Posteriorly accumulated Klar is detected by Klar-MK (D) and Klar-M (E) but not by the Klar-C antibody (F). (G) In oocytes, GFP–Klar β accumulates at the posterior pole. In oocytes with higher expression levels, GFP–Klar β accumulates both at the posterior and ectopically (not depicted). (H) No posterior Klar is detected in klarYG3, in which the promoter for both Klar α and Klar β is deleted. Posterior Klar is also absent in klarmBX18 and klarmBX14 (not depicted). (I) In klarmBX13, Klar accumulation at the posterior is normal. (J) In midway mutants, in which lipid droplets are essentially absent from oocytes (Buszczak et al., 2002), posterior Klar localization is normal. (K) Klar localizes to the posterior pole in klar1 mutant oocytes that express a truncated molecule lacking LD and KASH domains (Guo et al., 2005). (L) At high magnification, posterior Klar is present in distinct puncta that show minimal overlap with GFP-LD, a marker for lipid droplets (Yu et al., 2011). (M) By immunoblotting, Klar-M antibody recognizes two bands >250 kD in the ovarian lysate, presumably Klar isoforms α and β, as γ, δ, and ε have apparent molecular masses <200 kD (Guo et al., 2005; Kim et al., 2013). Because only the top band is recognized by antibody Klar-C, we propose that it represents Klar α, and the bottom band represents Klar β. (N) Klar-M Western blot of ovarian samples. The klarYG3 allele abolished both α and β isoforms. For klarmBX13, the top Klar band is shifted downward, consistent with a truncated Klar α protein encoded by this allele. Df indicates Df(3L)emcE12. Khc was used as a loading control (L and M, bottom bands). wt, wild type. Bars: (A, B, and D–K) 20 µm; (L) 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/206/2/10.1083_jcb.201310010/5/m_jcb_201310010_fig1.jpeg?Expires=1773781714&Signature=qIatiFbm7nnc0lxOFSva0Vjhi6uPMSZacOnWZ~Axbm6ZfzVFhnSgP5yXDre9IHpa8U9y6-Ws99pccgB1u0ydfmK1TV169st8CNBqQtIeiKCJOkZfSffUxgUtTPUU40NI4Q28~JJOKatGgPEbP-XvmcJ3LQmlDan07-LT422h~6Yf2a57JfqN8AS7GMyGuBpBCYvq~7IzizZwXfpga1KcIRhClWG5s5ZiK3Kw1-jb-mrdq5wXuJZT9POa1zJJ-QQ89e9UQO4XR3qzzEsvn1i2qqhN~H7mgdYU1eS45~nue-UFzoEl1kCQ9t9bbIrDZpSCc6ICjAcv9LMTgneLvGO8yw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)