Ca2+ entry through CRAC channels causes fast Ca2+-dependent inactivation (CDI). Previous mutagenesis studies have implicated Orai1 residues W76 and Y80 in CDI through their role in binding calmodulin (CaM), in agreement with the crystal structure of Ca2+–CaM bound to an Orai1 N-terminal peptide. However, a subsequent Drosophila melanogaster Orai crystal structure raises concerns about this model, as the side chains of W76 and Y80 are predicted to face the pore lumen and create a steric clash between bound CaM and other Orai1 pore helices. We further tested the functional role of CaM using several dominant-negative CaM mutants, none of which affected CDI. Given this evidence against a role for pretethered CaM, we altered side-chain volume and charge at the Y80 and W76 positions to better understand their roles in CDI. Small side chain volume had different effects at the two positions: it accelerated CDI at position Y80 but reduced the extent of CDI at position W76. Positive charges at Y80 and W76 permitted partial CDI with accelerated kinetics, whereas introducing negative charge at any of five consecutive pore-lining residues (W76, Y80, R83, K87, or R91) completely eliminated CDI. Noise analysis of Orai1 Y80E and Y80K currents indicated that reductions in CDI for these mutations could not be accounted for by changes in unitary current or open probability. The sensitivity of CDI to negative charge introduced into the pore suggested a possible role for anion binding in the pore. However, although Cl− modulated the kinetics and extent of CDI, we found no evidence that CDI requires any single diffusible cytosolic anion. Together, our results argue against a CDI mechanism involving CaM binding to W76 and Y80, and instead support a model in which Orai1 residues Y80 and W76 enable conformational changes within the pore, leading to CRAC channel inactivation.
INTRODUCTION
The Ca2+ release–activated Ca2+ (CRAC) channel is the prototypical store-operated Ca2+ channel responsible for Ca2+ signals that drive activation and effector functions in many cell types, including lymphocytes, platelets, and mast cells (Parekh and Putney, 2005; Prakriya and Lewis, 2015). CRAC channels open in response to depletion of ER Ca2+ stores, and inactivate in response to local Ca2+ accumulation (Prakriya and Lewis, 2015).
Essential CRAC channel subunits include the ER Ca2+ sensor STIM1 and the pore-forming subunit Orai1. In response to depletion of ER Ca2+ stores, STIM1 undergoes oligomerization and binds directly to Orai1 to yield active CRAC channels (Liou et al., 2007; Park et al., 2009; Covington et al., 2010). At hyperpolarized potentials, Ca2+ flux through individual CRAC channels then drives local feedback regulation via Ca2+ binding to sites located nanometers from the intracellular pore mouth, and channels inactivate on a time scale of tens of milliseconds (Hoth and Penner, 1993; Zweifach and Lewis, 1995). Several regions of the CRAC channel have been reported to contribute to fast Ca2+-dependent inactivation (CDI). These include a cytosolic portion of STIM1 known as the inactivation domain of STIM (IDSTIM, aa 470–491; Derler et al., 2009; Lee et al., 2009; Mullins et al., 2009), the intracellular II-III linker of Orai1 (aa 137–173; Srikanth et al., 2010), and the calmodulin (CaM)-binding domain of Orai1 (CBDOrai1, aa 68–91; Mullins et al., 2009).
CaM is well established as the Ca2+ sensor that mediates CDI of voltage-gated Ca2+ channels (Ben-Johny and Yue, 2014). Litjens et al. (2004) initially reported that a Ca2+-insensitive CaM mutant and a CaM inhibitory peptide reduced the extent of CDI of a native CRAC-like current, suggesting that CaM might also be the Ca2+ sensor for CRAC channel CDI. After the discovery of STIM1 and Orai1 as CRAC channel subunits, we identified a putative CaM binding domain in human Orai1 (CBDOrai1, aa 68–91) and verified that it binds CaM in a Ca2+-dependent manner (Mullins et al., 2009). To test for a possible role of CaM in CDI, several Orai1 residues identified as being potentially involved in CaM binding were mutated. For a range of point mutations at the A73, W76, and Y80 positions, loss of Ca2+–CaM binding to the CBDOrai1 peptide was strongly correlated with a loss of Orai1 channel CDI, whereas mutations that did not eliminate CaM binding did not inhibit CDI. These results provided additional support for a CaM-binding model for CDI (Mullins et al., 2009). The robust biochemical interaction between Ca2+–CaM and CBDOrai1 was later confirmed by a crystal structure of the human CBDOrai1 peptide bound to the C lobe of Ca2+–CaM (CaM-C; Liu et al., 2012). The binding interface for this structure includes human Orai1 residues with strong functional effects on CDI, with the W76 side chain fitting into a hydrophobic pocket of CaM-C and a hydrogen bond between Y80 and a backbone oxygen of CaM-C providing additional stabilization. Liu et al. (2012) proposed a two-step bridging model for CDI in which binding of the C lobe of CaM to one Orai1 subunit is followed by binding of the N lobe of CaM to a neighboring Orai1 subunit.
However, several other observations raise the possibility that W76 and Y80 influence CDI in ways other than providing a binding interface for CaM. Shortly after publication of the Ca2+–CaM–CBDOrai1 structure, the crystal structure of the full Drosophila melanogaster Orai1 (dOrai) channel revealed a hexameric subunit stoichiometry and a very long pore made of TM1 helices predicted to extend >20 Å beyond the membrane into the cell cytoplasm (Hou et al., 2012). Surprisingly, the side chains of residues corresponding to human W76 and Y80 were shown to face the pore lumen rather than being freely accessible to cytosolic CaM as previously assumed. Thus, the dOrai structure could potentially impose steric constraints that interfere with CaM binding to the intact Orai1 channel, calling into question the CaM-binding model for CRAC channel CDI. In addition, the Orai1 Y80A and Y80S mutations accelerate the kinetics of CDI without affecting its extent (Mullins et al., 2009), which is not easily explained through an effect of these mutations on CaM binding, but rather raises the possibility that Orai1 pore lining residues such as Y80 may play other roles in controlling Orai1 conformational transitions leading to CDI.
In this study, we reevaluate the CaM-binding model for CRAC channel CDI by testing dominant-negative CaM mutants and an extended series of critical Orai1 mutations for their effects on CDI. Our results force a reconsideration of the CDI mechanism and of the essential roles of Orai1 residues Y80 and W76. In the simplest model compatible with our data, the importance of these two residues in CDI is not that they bind Ca2+–CaM, but instead that they participate in conformational transitions within the pore leading to inactivation.
MATERIALS AND METHODS
Cells
HEK293-H cells (Gibco) were grown in DMEM (Gemini) supplemented with 10% FBS (Gemini), 1% penicillin/streptomycin (Gemini), and 1% l-glutamine (Gemini) in a humidified, 5% CO2 incubator at 37°C. For experiments with CaM and rSK2, a previously described HEK293 cell line with an inducible mCherry-STIM1-T2A-myc-Orai1 was used (Sadaghiani et al., 2014).
Plasmid cDNA constructs
The primary constructs used in our studies were mCherry-STIM1 (Luik et al., 2006) and Orai1-GFP (Xu et al., 2006). All mutations were made by QuikChange mutagenesis (Agilent). Rat SK2 in the PCDNA6 vector was a gift from R. Aldrich (University of Texas, Austin, TX). CaM-IRES-GFP WT, CaM12-IRES-GFP, CaM34-IRES-GFP, and CAM1234-IRES-GFP were gifts from M. Tadross (Howard Hughes Medical Institute Janelia Research Campus, Ashburn, VA), and are similar in design to previously described constructs with the same aspartate-to-alanine mutations in CaM (Peterson et al., 1999; DeMaria et al., 2001). GFP-STIM1 was previously described (Wu et al., 2006). The CaV1.2 T1039Y construct was previously described (Dolmetsch et al., 2001), and was a gift from A. Rana (Stanford University, Stanford, CA). Constructs encoding the rat β1b (X61394) and α2δ1 (M86621) subunits were also gifts from A. Rana.
Transfection
HEK293-H cells were transfected with Lipofectamine 2000 (Invitrogen) 16–48 h before electrophysiology experiments. Most constructs expressed well within 24 h, but some required up to 48 h to yield currents of ideal size. STIM1- and Orai1-derived constructs were transfected in a 4:1 ratio by mass, higher than the 1:1 mass ratio used in our previous study of CDI (Mullins et al., 2009), because it is now recognized that maximal CDI requires a high STIM1/Orai1 ratio (Scrimgeour et al., 2009; Hoover and Lewis, 2011). In the standard experimental design, cells were transfected with mCh-STIM1 and Orai1-GFP, and were visualized with a filter cube that included a 470 ± 20-nm bandpass excitation filter, a 500-nm dichroic, and a 515 longpass emission filter. In this filter configuration, exciting with a Xenon lamp, both mCherry and GFP were visible, and the cells selected for recording appeared orange, consistent with a high STIM1/Orai1 ratio. Selecting cells in this manner, we saw minimal cell-to-cell variability in kinetics and extent of CDI for any given pair of STIM1- and Orai1-derived constructs, consistent with saturating levels of STIM1.
For noise analysis experiments, mCh-STIM1 and Orai1-GFP constructs were transfected at ratios as high as 12:1 to increase the yield of cells with smaller currents, as these cells tended to be more stable when held continuously at −100 mV.
In the experiments testing CaM1234 and CaM12 effects on CRAC and rSK2, inducible STIM1/Orai1 double-stable HEK293 cells were transiently transfected with CaM-IRES-GFP (or CaM1234-IRES-GFP or CaM12-IRES-GFP), rSK2, and GFP-STIM1 at a 1:1:1 ratio 2 d before recording. The additional STIM1 was transfected to ensure that cells would express a high STIM1/Orai1 ratio. mCh-STIM1 and myc-Orai1 expression were induced by addition of 1 µg/ml tetracycline 1 d before recording. Only cells with clear GFP localization in the cytosol and ER were selected for recording.
In the experiments testing CaM34 effects on CRAC and CaV1.2, HEK293H cells were transiently transfected 2 d before recording with either mCh-STIM1 and Orai1-GFP at a 4:1 ratio or CaV1.2, β1b, and α2δ1 at a 3:1:1 ratio, together with the same amount of CaM-IRES-GFP (or CaM34-IRES-GFP) as was transfected in the experiments with the STIM1/Orai1 double-stable cell line.
Recording solutions
The extracellular solution for most experiments was a 20 mM Ca2+ Ringer’s solution containing the following (in mM): 130 NaCl, 4.5 KCl, 20 CaCl2, 1 MgCl2, 10 d-glucose, and 5 HEPES (pH 7.4 with NaOH). 20 mM Ba2+ Ringer’s was made by equimolar substitution of BaCl2 for CaCl2. The 2 mM Ca2+ Ringer’s solution contained the following (in mM): 155 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose, and 5 HEPES (pH 7.4 with NaOH). For leak subtraction of CRAC currents, leak current was recorded in 2 mM Ca2+ Ringer’s solution with 100 µM LaCl3 added. The divalent-free (DVF) solution used in noise analysis experiments included the following (in mM): 150 NaCl, 10 HEDTA, 10 HEPES, 10 TEA-Cl, and 1 EDTA (pH 7.4 with NaOH). CaCl2 was added from a 1 M stock to achieve the desired free [Ca2+] as calculated by MaxChelator (Bers et al., 2010), followed by titration to pH 7.4 with NaOH.
The standard aspartate-based internal solution contained the following (in mM): 150 Cs aspartate, 8 MgCl2, 10 EGTA, and 10 HEPES (pH 7.2 with CsOH). The HEPES-based internal solution contained the following (in mM): 160 Cs HEPES, 8 MgCl2, and 10 EGTA (pH 7.2 with CsOH). The sucrose-based internal solution contained the following (in mM): 207 sucrose, 8 MgCl2, 10 HEPES, and 10 EGTA (pH 7.2 with CsOH). The chloride-based internal solution contained the following (in mM): 150 CsCl, 8 MgCl2, 10 HEPES, and 10 EGTA (pH 7.2 with CsOH). The chloride-free internal solution contained the following (in mM): 150 Cs aspartate, 8 MgATP (Sigma-Aldrich), 10 HEPES, and 10 EGTA (pH 7.2 with CsOH). For experiments with the chloride-free internal solution, the Ag/AgCl electrode in the recording pipette was ensheathed in an agar bridge containing the standard aspartate-based internal solution (with 16 mM Cl−). Given that the bridge was positioned 3 cm from the pipette tip, chloride diffusion from the bridge to the pipette tip was predicted to be negligible over the short duration of our experiments (Shao and Feldman, 2007).
For rSK2 current recordings, the extracellular solution contained (in mM): 160 KCl, 2 CaCl2, 1 MgCl2, 10 d-glucose, and 10 HEPES (pH 7.4 with KOH). To confirm rSK2 current, 100 nM apamin (Sigma-Aldrich) was added to the extracellular solution from a 250-µM stock. The internal solution contained the following (in mM): 150 K aspartate, 8 MgCl2, 10 HEPES, and 1.6 EGTA (pH 7.2 with KOH, 10−5 M free Ca2+). For CaV1.2 current recordings, the internal solution contained the following (in mM): 150 Cs aspartate, 4 MgCl2, 4 MgATP, 10 EGTA, and 10 HEPES (pH 7.2 with CsOH).
Electrophysiology
Currents were recorded with the standard whole-cell patch clamp technique at 22–25°C, as previously described (Mullins et al., 2009). We used an Axopatch 200B amplifier (Axon Instruments/Molecular Devices) interfaced to an ITC-16 I/O interface (Instrutech) and a Macintosh G3 computer. Pipettes of resistance 2–5 MΩ were fabricated from 100-µl pipettes (VWR) using a Flaming/Brown puller (model P-87; Sutter Instruments). The pipettes were fire-polished and coated with Sylgard, and cell and pipette capacitances were nulled before recording. Igor Pro (Wavemetrics) software routines developed in-house were used for stimulation, data acquisition, and analysis. Currents were filtered at 2 kHz and sampled at 5 kHz. Voltages were corrected for the junction potential of the standard aspartate-based pipette solution relative to Ringer’s solution in the bath (−10 mV). Measured junction potentials for internal solutions with other primary anions were −8 mV for HEPES, −7 mV for sucrose, and −1 mV for chloride.
For noise analysis, solutions were exchanged using a gravity-based pinch-valve perfusion system driven by the ValveLink 8.2 controller through a perfusion pencil coupled to a 19-gauge needle (AutoMate Scientific). In these experiments, Ca2+ stores were predepleted with 1 µM thapsigargin before seal formation.
For Orai1 currents, CDI was evoked by 200-ms steps to −60, −80, −100, and −120 mV delivered every 5 s from a holding potential of +30 mV. CDI was quantified as the current measured at 195 ms divided by the peak current at the beginning of the pulse. Peak current was measured 1–3 ms after the step to avoid current from residual uncompensated capacitance. For the most rapidly inactivating constructs, only those cells in which the leak-subtracted capacitance artifact decayed completely within 1–1.5 ms were used for analysis. For CaV1.2 currents in both the WT CaM and CaM34 conditions, cells were stimulated by 300-ms steps to +20, +30, +40, and +50 mV delivered every 10 s from a holding potential of −75 mV for 5 min in 20 mM Ca2+ Ringer’s before switching to 20 mM Ba2+ Ringer’s. Leak subtraction was performed with a −P/5 protocol.
Data analysis
The time course of Orai1 current inactivation was fitted with the biexponential function I = I0 + A1e−t/τ1 + A2e−t/τ2, where I is current, I0 is the steady-state current, A1 and A2 are amplitudes, and τ1 and τ2 are inactivation time constants.
For noise analysis, 200-ms sweeps collected within 1 s of DVF solution exchange were divided into 25-ms segments in which the mean current was nearly constant (<2% change). Variances were calculated for each 25-ms segment and averaged for the eight segments in each trace to give a mean value used in variance versus current plots. For some cells showing little or no depotentiation, variance was averaged for segments from multiple consecutive sweeps after each exchange. To determine the open probability in DVF solution (Po), current and variance data from single cells were corrected for leak current and background variance, and then normalized to the current and variance in DVF. Normalized data from individual cells were fitted with a modified parabolic relation (Sigworth, 1980) given by σ2 = kP(1−P), where σ2 is the normalized current variance, k is a constant, P is the open probability given by P = InormPo, and Inorm is the normalized current. The unitary Na+ current iNa was estimated after subtraction of the background variance as the extrapolated value of σ2/I at 100% block (Po = 0), where I is the macroscopic current (Prakriya and Lewis, 2006). For display purposes, data from multiple cells were combined and fitted as a group (Fig. 4, D and E).
To estimate the unitary Ca2+ current (iCa) from the unitary Na+ current (iNa), we reasoned that the ratio of macroscopic Ca2+- and Na+-CRAC currents (ICa and INa, respectively) should be proportional to the ratio of iCa and iNa:
where N is the number of activatable channels, i is the unitary current, Po is open probability and the subscripts Ca and Na refer to measurements made with extracellular solutions of 2 mM Ca2+ or immediately after a switch to DVF, respectively. We assume that N is constant immediately after a switch from 2 mM Ca2+ to DVF (though it will decline subsequently due to depotentiation); hence NCa = NNa. Po is reduced in 2 mM Ca2+ relative to DVF because of CDI, which should reverse within milliseconds (Zweifach and Lewis, 1995) after Ca2+ flux is terminated upon the switch to DVF. Thus, Po,Ca/Po,Na is given by the extent of CDI (from 0 to 1) that occurs in 2 mM Ca2+ at −100 mV. Therefore,
and
where (1-CDI) is the proportion of current remaining at the end of a voltage step to –100 mV. Measurement of the ICa/INa ratio and the CDI correction factor is illustrated in Fig. S5.
Data in all summary figures represent mean ± SEM, and statistical comparisons were made by two-sample t tests.
Structural modeling
In Fig. 1, the human CaM/CBDOrai1 structure 4EHQ (Liu et al., 2012) was aligned with the dOrai structure 4HKR (Hou et al., 2012) using the ALIGN function of MacPyMOL v1.5.0.5 and coordinates from the Protein Data Bank.
Online supplemental material
Fig. S1 shows representative data for the Cav1.2 current measurements. Fig. S2 shows representative traces for several mutations at position Y80. Fig. S3 summarizes fast and slow amplitudes from biexponential fits to CDI kinetics for mutations at the Y80 and W76 positions. Fig. S4 provides data related to the selectivity of the Y80E mutation. Fig. S5 shows representative macroscopic current data used to estimate iCa from iNa. Fig. S6 shows representative traces for several mutations at position W76. Fig. S7 summarizes fast and slow time constants for experiments in which the internal solution anions were varied.
RESULTS
Alignment of crystal structures predicts steric clash of CaM with the Orai1 pore
The relationship between the Ca2+–CaM–CBDOrai1 crystal structure (Liu et al., 2012) and the full-length hexameric dOrai crystal structure (Hou et al., 2012) is important for evaluating possible mechanisms of CRAC channel CDI. We aligned the two structures using the CBD regions of hOrai1 and dOrai to explore the plausibility of Ca2+–CaM binding to the hexameric Orai1 pore. Fig. 1 A shows a representation of the hexameric dOrai crystal structure (Hou et al., 2012) viewed from the cytosol, with an inner ring of six transmembrane TM1 domains that line the pore, surrounded by TM2, TM3, and TM4 domains; the aligned human CBDOrai derived from the Ca2+–CaM–CBDOrai1 crystal structure (Liu et al., 2012) is shown in yellow. Fig. 1 B adds a space-filling representation of the C lobe of Ca2+–CaM bound to the CBDOrai1. The alignment predicts multiple steric clashes between CaM and Orai1, the most significant of which is with an adjacent TM1 cytoplasmic region (Fig. 1, C and D). The alignment also predicts significant overlap of the N lobe of CaM with TM3 and TM4 regions, even extending into the membrane (not depicted), but as the N lobe is attached to the C lobe with a highly flexible linker (Villarroel et al., 2014), its orientation is not as firmly constrained as the C lobe bound to TM1. Therefore, we focus here on the C lobe clash, assuming the linker might be flexible enough for the N lobe to avoid overlap with surrounding regions if the C lobe were to bind.
Steric clash of CAM with the hexameric Orai pore. PYMOL-generated cartoon representations of the hexameric Drosophila Orai crystal structure are shown with and without CaM C lobe. (A) View of dOrai from the cytosolic face. The aligned human CBDOrai1 structure (with W76 side chain) is shown in yellow. Glutamate side chains near the extracellular end of the pore are shown for orientation. Helices are color coded: TM1 (red), TM2 (teal), TM3 (blue-gray), and TM4 (blue). (B) dOrai with a surface representation of the CaM C lobe (green) bound to CBDOrai1. (C) Side view of the dOrai TM1 helices with Ca2+–CaM bound to the aligned human CBDOrai1 structure (yellow) highlights the steric clash of the C lobe with the adjacent Orai pore helices. Overlap extends in the vertical direction to R83. (D) Cutaway cytosolic view of the dOrai TM1 helices in complex with Ca2+–CaM shows that the clockwise adjacent TM1 helix (arrow) overlaps completely with CaM. (E) Cutaway cytosolic view of the dOrai–CaM C lobe complex after a 15-Å radial dilation of each TM1 helix (red, compare with B), necessary to remove overlap between the adjacent TM1. Note that CaM now clashes with the TM4 cytosolic domain (blue) and overlaps completely with TM3 (blue-gray). (F) The TM1 pore regions for two opposite dOrai subunits are shown together with the pore-lining side chains (corresponding human Orai1 residues are indicated in parentheses).
Steric clash of CAM with the hexameric Orai pore. PYMOL-generated cartoon representations of the hexameric Drosophila Orai crystal structure are shown with and without CaM C lobe. (A) View of dOrai from the cytosolic face. The aligned human CBDOrai1 structure (with W76 side chain) is shown in yellow. Glutamate side chains near the extracellular end of the pore are shown for orientation. Helices are color coded: TM1 (red), TM2 (teal), TM3 (blue-gray), and TM4 (blue). (B) dOrai with a surface representation of the CaM C lobe (green) bound to CBDOrai1. (C) Side view of the dOrai TM1 helices with Ca2+–CaM bound to the aligned human CBDOrai1 structure (yellow) highlights the steric clash of the C lobe with the adjacent Orai pore helices. Overlap extends in the vertical direction to R83. (D) Cutaway cytosolic view of the dOrai TM1 helices in complex with Ca2+–CaM shows that the clockwise adjacent TM1 helix (arrow) overlaps completely with CaM. (E) Cutaway cytosolic view of the dOrai–CaM C lobe complex after a 15-Å radial dilation of each TM1 helix (red, compare with B), necessary to remove overlap between the adjacent TM1. Note that CaM now clashes with the TM4 cytosolic domain (blue) and overlaps completely with TM3 (blue-gray). (F) The TM1 pore regions for two opposite dOrai subunits are shown together with the pore-lining side chains (corresponding human Orai1 residues are indicated in parentheses).
The dOrai structure is thought to reflect the closed channel conformation, and thus may not accurately reflect the binding target for CaM if it binds to the open or inactivated state. Although these structures are not yet described, we asked whether binding of the CaM C lobe could be accommodated through conservative movements of the TM1 helices. A cutaway view from the cytosol of CaM and the six TM1 helices shows that the cytosolic domain of the clockwise-adjacent TM1 lies completely within the predicted volume of CaM (Fig. 1 D, arrow). To avoid this clash, the TM1 helices in this plane would need to move radially outward by ∼15 Å, which then creates major overlap between CaM and the outer Orai1 cytosolic domains, particularly TM3 (Fig. 1 E). Together, these considerations raised the possibility that, despite the robust biochemical and structural interactions of Ca2+–CaM with the CBDOrai1 peptide (Mullins et al., 2009; Liu et al., 2012), CaM may not bind to the full-length hexameric protein in the way it binds to the CBDOrai1 peptide, and led us to reexamine the CaM-binding model experimentally.
Mutant CaM overexpression does not affect CRAC channel inactivation
To further test the CaM-binding model for CRAC channel CDI, we measured CDI in the presence of CaM1234, a CaM mutant rendered unable to bind Ca2+ by aspartate-to-alanine mutations in all four EF hands. CaM1234 overexpression inhibits CDI of voltage-gated Ca2+ channels and activation of SK- and IK-type Ca2+-activated K+ channels by competing with WT apo–CaM for binding to the channels (Xia et al., 1998; Levitan, 1999; Erickson et al., 2001; Pitt et al., 2001; Saimi and Kung, 2002).
For this experiment, we used a double-stable HEK293 cell line with inducible expression of mCh-STIM1 and myc-Orai1 (see Materials and methods). As a positive control for knockdown of CaM function by CaM1234, we coexpressed the rat SK2 K+ channel which is known to be activated by Ca2+ binding to apo–CaM attached to the channel (Xia et al., 1998). Importantly, rSK2 and CRAC currents were measured in cells from the same transfections to ensure that CaM function was suppressed in the cells in which CRAC channel CDI was being measured. As expected, CaM1234 expression reduced rSK2 current magnitude by ∼10-fold compared with WT CaM (Fig. 2 A, top; P < 0.01 at −100, −80, and −60 mV), confirming the effective inhibition of CaM function by CaM1234. The extent of CRAC channel CDI, in contrast, was identical in cells overexpressing WT CaM or CaM1234 (Fig. 2 B; P > 0.5 at all voltages).
Mutant CaM overexpression does not affect CRAC channel inactivation. (top) A TET-inducible HEK293 cell line expressing mCh-STIM1 and myc-Orai1 (double-stable, DS) was transiently transfected with GFP-STIM1, CaM-IRES-GFP, and rSK2. (bottom) HEK293H cells were transiently transfected with Cav1.2, β1b, α2δ1, and CaM-IRES-GFP or with mCh-STIM1, Orai1-GFP, and CaM-IRES-GFP. (A) Effects on SK and CaV currents (positive controls). (top) Summary of rSK2 current elicited by 100-ms voltage ramps for transfections with WT CaM, CaM12, or CaM1234. CaM12 and CaM1234 significantly reduced rSK2 current as compared with WT CaM (P < 0.01 at −100, −80, and −60 mV). Each point represents the mean ± SEM for n = 5–9 cells. (bottom) Summary of Cav1.2 CDI in cells overexpressing WT CaM or CaM34. CaM34 significantly suppressed Cav1.2 CDI as compared with WT CaM (P < 0.01 at all voltages). Each point represents the mean ± SEM for n = 4–5 cells. Representative Cav1.2 current traces are shown in Fig. S1. (B) Effects on CRAC channel CDI. (top) Representative CRAC currents elicited by 200-ms hyperpolarizations in DS cells expressing WT CaM, CaM12, or CaM1234. Currents were recorded in 20 mM Ca2+o. Current decay is due to Ca2+-dependent inactivation (CDI). (bottom) Representative CRAC currents elicited by 200-ms hyperpolarizations for HEK293H cells expressing WT CaM or CaM34. (bottom right) Extent of CRAC channel CDI for each condition, summarized as the fraction of current remaining at the end of the pulse (see Materials and methods). Each point represents the mean ± SEM for n = 5–6 cells. There was no significant difference between CDI measured with WT CaM and any of the mutant CaMs tested (P > 0.2 compared with appropriate WT control in DS or 293H cells at all voltages).
Mutant CaM overexpression does not affect CRAC channel inactivation. (top) A TET-inducible HEK293 cell line expressing mCh-STIM1 and myc-Orai1 (double-stable, DS) was transiently transfected with GFP-STIM1, CaM-IRES-GFP, and rSK2. (bottom) HEK293H cells were transiently transfected with Cav1.2, β1b, α2δ1, and CaM-IRES-GFP or with mCh-STIM1, Orai1-GFP, and CaM-IRES-GFP. (A) Effects on SK and CaV currents (positive controls). (top) Summary of rSK2 current elicited by 100-ms voltage ramps for transfections with WT CaM, CaM12, or CaM1234. CaM12 and CaM1234 significantly reduced rSK2 current as compared with WT CaM (P < 0.01 at −100, −80, and −60 mV). Each point represents the mean ± SEM for n = 5–9 cells. (bottom) Summary of Cav1.2 CDI in cells overexpressing WT CaM or CaM34. CaM34 significantly suppressed Cav1.2 CDI as compared with WT CaM (P < 0.01 at all voltages). Each point represents the mean ± SEM for n = 4–5 cells. Representative Cav1.2 current traces are shown in Fig. S1. (B) Effects on CRAC channel CDI. (top) Representative CRAC currents elicited by 200-ms hyperpolarizations in DS cells expressing WT CaM, CaM12, or CaM1234. Currents were recorded in 20 mM Ca2+o. Current decay is due to Ca2+-dependent inactivation (CDI). (bottom) Representative CRAC currents elicited by 200-ms hyperpolarizations for HEK293H cells expressing WT CaM or CaM34. (bottom right) Extent of CRAC channel CDI for each condition, summarized as the fraction of current remaining at the end of the pulse (see Materials and methods). Each point represents the mean ± SEM for n = 5–6 cells. There was no significant difference between CDI measured with WT CaM and any of the mutant CaMs tested (P > 0.2 compared with appropriate WT control in DS or 293H cells at all voltages).
EF hand mutations in CaM can affect the stability of apo–CaM associations with some CaM targets, such as SK2 channels (Li et al., 2009), raising the possibility that the CaM1234 mutant might fail to suppress CRAC channel CDI because of an inability to efficiently replace WT CaM at the relevant apo–CaM binding site. Therefore, we tested CaM12 and CaM34 mutant proteins, in which Ca2+ binding to only the N- or C-lobe EF hands is altered, respectively. Because the CaM12 mutation is known to suppress rSK2 current in a manner similar to CaM1234 (Keen et al., 1999), we used an experimental design identical to that described in the preceding paragraph for CaM1234. Like CaM1234, CaM12 effectively suppressed rSK2 current (Fig. 2 A, top; P < 0.01 at −100, −80, and −60 mV) but did not change CRAC channel CDI (Fig. 2 B; P > 0.2 at all voltages).
The CaM34 mutant does not affect rSK2 gating (Keen et al., 1999), but it is known to suppress CDI of the voltage-gated channel CaV1.2 (Peterson et al., 1999). Thus, as a positive control for knockdown of CaM function by CaM34, we compared inactivation of CaV1.2 currents carried by Ca2+ and Ba2+ (which supports only voltage-dependent inactivation; see Materials and methods and Fig. S1). Measurements of CaV1.2 channel CDI, quantified as the change in the extent of inactivation in Ca2+ relative to Ba2+ (ΔR300), demonstrated that CaM34 suppressed CDI by >90% under our experimental conditions (Fig. 2 A, bottom; P < 0.01 at all voltages). Cells transfected with the same amount of CaM34 together with STIM1 and Orai1, displayed CRAC channel CDI that was indistinguishable from that seen for WT CaM (Fig. 2 B; P > 0.2 at all voltages).
Together, these data argue against a model in which apo–CaM prelocalized near the inner pore responds to Ca2+ accumulation by interacting with Y80 and W76 on intact Orai1 channels to cause CDI. Based on these findings, we reevaluated the functional roles of these residues in the inactivation process.
Side chain volume at hOrai1 position Y80 impacts the speed and extent of CDI
A significant complication in studies of CRAC channel CDI is that the extent of CDI increases with the STIM1/Orai1 expression ratio, presumably reflecting an effect of STIM1/Orai1 binding stoichiometry on CDI (Scrimgeour et al., 2009; Hoover and Lewis, 2011). Thus, to permit accurate comparisons of CDI among various mutants of Orai1, we transfected HEK293 cells with a 4:1 ratio of mCh-STIM1/Orai1-GFP and recorded from cells with the highest STIM1/Orai1 ratios as judged by red/green fluorescence emission (see Materials and methods). Using this protocol, WT Orai1 consistently displayed CDI kinetics and extent similar to that of native CRAC channels (Fig. 3 A; Zweifach and Lewis, 1995). At the same time, we observed phenotypes for several CDI mutants that differed somewhat from those described previously, presumably due to higher STIM1/Orai1 ratio (see Discussion).
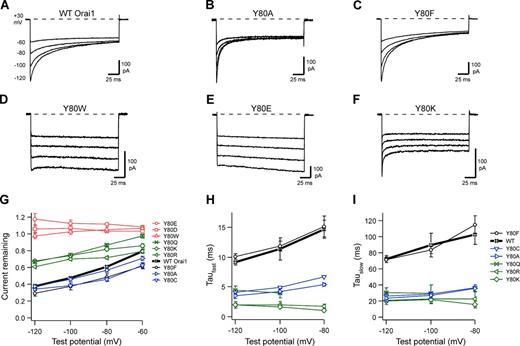
CRAC channel CDI phenotypes for mutations at Orai1 position Y80. All currents were recorded in HEK293H cells cotransfected with WT mCh-STIM1 and WT or mutant Orai1-GFP after induction of ICRAC reached a maximum (∼300 s after break-in). The representative currents shown were recorded in 20 mM Ca2+o during 200-ms hyperpolarizations to the voltages indicated for WT Orai1 (A). Substitutions included Y80A (B), Y80F (C), Y80W (D), Y80E (E), and Y80K (F), with the extent of CDI for this series summarized in G. Current traces were fit to the biexponential function I = Io + A1e-t/τ1 + A2e-t/τ2, where τ1 and τ2 represent fast and slow time constants of inactivation. Fast (H) and slow (I) time constants from biexponential fits are plotted against test potential; amplitudes of fast and slow components are shown in Fig. S3 (A and B). Each point on the summary graphs represents the mean ± SEM for n = 4–7 cells. Representative currents for additional mutations on the summary graphs are shown in Fig. S2.
CRAC channel CDI phenotypes for mutations at Orai1 position Y80. All currents were recorded in HEK293H cells cotransfected with WT mCh-STIM1 and WT or mutant Orai1-GFP after induction of ICRAC reached a maximum (∼300 s after break-in). The representative currents shown were recorded in 20 mM Ca2+o during 200-ms hyperpolarizations to the voltages indicated for WT Orai1 (A). Substitutions included Y80A (B), Y80F (C), Y80W (D), Y80E (E), and Y80K (F), with the extent of CDI for this series summarized in G. Current traces were fit to the biexponential function I = Io + A1e-t/τ1 + A2e-t/τ2, where τ1 and τ2 represent fast and slow time constants of inactivation. Fast (H) and slow (I) time constants from biexponential fits are plotted against test potential; amplitudes of fast and slow components are shown in Fig. S3 (A and B). Each point on the summary graphs represents the mean ± SEM for n = 4–7 cells. Representative currents for additional mutations on the summary graphs are shown in Fig. S2.
We first compared a series of uncharged side chains of variable size introduced at the Y80 position (Fig. 1 F) for their effects on CDI. The Y80A mutation (now measured at high STIM1/Orai1 ratio), accelerated the kinetics of CDI with minimal effects on the extent of CDI (Fig. 3 B), as we previously reported (Mullins et al., 2009). Introduction of another small side chain (Y80C) produced a similar acceleration of CDI kinetics (Fig. S2 B). In contrast, Y80F, similar in size to Y80, did not significantly affect the speed or extent of CDI (Fig. 3 C; trend toward increased CDI for Y80F; P = 0.06 at −120 mV). Finally, the Y80W mutation, which introduces an even larger side chain, nearly completely prevented CDI (Fig. 3 D; P < 10−5 at –120 mV). These results are summarized in Fig. 3 (G–I) and Fig. S3 (A and B). Overall, for uncharged substitutions, side chain volume at position Y80 is inversely related to the speed of CDI, with large aromatic side chains at the Y80 position acting to limit the speed and/or extent of inactivation.
Negative and positive charges have disparate effects at the Y80 position
Y80E channels studied at high STIM1/Orai1 ratio completely lacked CDI (Fig. 3 E), consistent with our previous results (Mullins et al., 2009). The Y80D mutation (Fig. S2 C), which also introduces a negatively charged side chain, had a noninactivating phenotype similar to that of Y80E. Introduction of positive charge at the Y80 position produced a very different effect. Both the Y80K and Y80R mutations yielded partial inactivation with accelerated kinetics (Fig. 3 F and Fig. S2 D; summary in Fig. 3, G–I and Fig. S3, A and B). Although it is uncharged, the amphipathic mutation Y80Q produced similar effects as Y80K and R (Fig. S2 E), suggesting that the effects of lysine and arginine substitutions are not caused by charge but by a property in common with glutamine, such as side chain flexibility (see Discussion).
Unitary properties of Orai1 channels with charged substitutions at position Y80
CRAC channel CDI is triggered by local Ca2+ flux through individual channels (Zweifach and Lewis, 1995). In principle, mutations that inhibit CDI could act by reducing Ca2+ accumulation near the pore or by limiting the channel’s response to a given local [Ca2+]. Particularly for mutations that introduce charge into the pore, reduced CDI could be the result of diminished unitary Ca2+ current amplitude or channel open probability. The conductance of CRAC channels is too small to allow direct measurements of single channel currents (Zweifach and Lewis, 1993). However, exposure to divalent-free solution relieves the Ca2+ block of monovalent permeation, eliciting a significantly larger current carried by Na+. Unitary properties can be estimated from ensemble variance analysis of monovalent CRAC currents in the presence of partially blocking levels of Ca2+ (Prakriya and Lewis, 2006). We applied this method to the Y80K and Y80E mutant channels to determine whether they affect channel conductance or open probability.
A representative noise analysis experiment for overexpressed WT Orai1 channels is shown in Fig. 4 A. Upon switching from 2 mM Ca2+ to divalent-free Ringer’s (DVF), current measured at the holding potential of −100 mV rapidly increased due to relief of Ca2+ block, which allows Na+ conduction. The current then declined because of depotentiation, and, after returning the cell to 2 mM Ca2+, the reduced Ca2+ current increased as channels repotentiated (Zweifach and Lewis, 1996). We subsequently perfused the cell with DVF supplemented with micromolar amounts of Ca2+ to partially block the monovalent current. Single 200-ms traces for analysis were collected immediately after changing to each DVF-based solution (Fig. 4 B). In three WT cells, Ca2+ blocked the monovalent current with an K1/2 of ∼25 µM (Fig. 4 C). For channels with a single conducting state, binomial statistics predict a parabolic relation between current amplitude and variance (see Materials and methods). A parabolic fit to a plot of normalized current variance versus mean current yielded an open probability estimate of ∼0.8 in the unblocked state (Fig. 4 D). The variance/current ratio increased linearly with degree of block, and extrapolated to a single-channel Na+ current value of −80 fA under the condition of full block (Po = 0; Fig. 4 E; see Materials and methods). These results are similar to those obtained previously for endogenous CRAC channels (Prakriya and Lewis, 2006).
Ensemble variance analysis of Orai1 channels with charged substitutions at position Y80. (A) A thapsigargin-pretreated cell expressing Ch-STIM11-469 and WT Orai1-GFP was held continuously at −100 mV in 2 mM Ca2+o, and as indicated by the bars, DVF solutions containing variable low [Ca2+]o were applied transiently to reveal and partially block the Na+-CRAC current. Colored circles indicate peak currents corresponding to the 200-ms sweeps shown in (B), which were divided into 25-ms segments for analysis (see Materials and methods). (C) Ca2+ block of DVF Na+ currents for WT Orai1, Y80K, and Y80E. Na+ current was measured immediately after applying DVF solution containing the indicated [Ca2+]o, and normalized to the value for the base DVF solution. Each point represents the mean ± SEM for n = 3–4 cells. Lines are fits of the Hill equation, block = 1/[1 + (K1/2/[Ca])nH]. (D) Normalized variance versus current plots for WT, Y80E, and Y80K were fitted by a parabolic function to determine open probability (see Materials and methods). (E) Variance/current versus block plots were fitted by a linear function to determine unitary current (see Materials and methods). Unitary channel properties are summarized in Table 1.
Ensemble variance analysis of Orai1 channels with charged substitutions at position Y80. (A) A thapsigargin-pretreated cell expressing Ch-STIM11-469 and WT Orai1-GFP was held continuously at −100 mV in 2 mM Ca2+o, and as indicated by the bars, DVF solutions containing variable low [Ca2+]o were applied transiently to reveal and partially block the Na+-CRAC current. Colored circles indicate peak currents corresponding to the 200-ms sweeps shown in (B), which were divided into 25-ms segments for analysis (see Materials and methods). (C) Ca2+ block of DVF Na+ currents for WT Orai1, Y80K, and Y80E. Na+ current was measured immediately after applying DVF solution containing the indicated [Ca2+]o, and normalized to the value for the base DVF solution. Each point represents the mean ± SEM for n = 3–4 cells. Lines are fits of the Hill equation, block = 1/[1 + (K1/2/[Ca])nH]. (D) Normalized variance versus current plots for WT, Y80E, and Y80K were fitted by a parabolic function to determine open probability (see Materials and methods). (E) Variance/current versus block plots were fitted by a linear function to determine unitary current (see Materials and methods). Unitary channel properties are summarized in Table 1.
The Orai1 mutant Y80K displayed a Ca2+ affinity, open probability, and unitary Na+ current similar to WT (Fig. 4, C–E; and Table 1; P > 0.2 for all parameters vs. WT). Whereas Y80E channels also displayed an open probability similar to that of WT (Fig. 4 D; Table 1; P = 0.27 vs. WT), the Ca2+ affinity of the selectivity filter was reduced (K1/2 ∼60 µM; Fig. 4 C; P < 0.02 vs. WT) and the unitary Na+ current increased to −148 fA (Fig. 4 E; Table 1; P < 0.01 vs. WT). Despite these changes in pore properties, the Cs+/Na+ permeability ratio for Y80E channels (as indicated by the current reversal potential in DVF) was indistinguishable from WT (Fig. S4 A), indicating that selectivity between these monovalent ions was unchanged. Y80E channels also retained high selectivity for Ca2+ over Na+, as indicated by the lack of outward current at potentials up to +100 mV (Fig. S4 B).
Unitary channel properties for WT Orai1, Y80K, and Y80E
| Orai1 channel | Po | iNa | iCa | Ca2+ affinity (K1/2) | nH | n |
| fA | fA | µM | ||||
| WT | 0.81 ± 0.02 | 79.6 ± 2.4 | 9.6 ± 0.4 | 24.2 ± 2.0 | 0.87 ± 0.10 | 3 |
| Y80K | 0.81 ± 0.02 | 91.5 ± 7.7 | 8.3 ± 1.1 | 33.5 ± 3.6 | 1.26 ± 0.10 | 4 |
| Y80E | 0.78 ± 0.02 | 147.6 ± 10.3 | 23.0 ± 4.4 | 63.5 ± 7.8 | 1.68 ± 0.20 | 4 |
| Orai1 channel | Po | iNa | iCa | Ca2+ affinity (K1/2) | nH | n |
| fA | fA | µM | ||||
| WT | 0.81 ± 0.02 | 79.6 ± 2.4 | 9.6 ± 0.4 | 24.2 ± 2.0 | 0.87 ± 0.10 | 3 |
| Y80K | 0.81 ± 0.02 | 91.5 ± 7.7 | 8.3 ± 1.1 | 33.5 ± 3.6 | 1.26 ± 0.10 | 4 |
| Y80E | 0.78 ± 0.02 | 147.6 ± 10.3 | 23.0 ± 4.4 | 63.5 ± 7.8 | 1.68 ± 0.20 | 4 |
All channel properties were measured at –100 mV.
Although our noise analysis approach yields estimates of the unitary Na+ current under DVF conditions (iNa), there is no equivalent method to determine iCa, the unitary current with Ca2+ as the current carrier. However, we can estimate iCa by multiplying iNa by the ratio of Ca2+ and Na+ macroscopic currents, with a small correction applied to remove the effect of resting inactivation on Po in 2 mM Ca2+ (see Materials and methods; Fig. S5). Although the estimated iCa for Y80K channels is slightly lower than for WT, the difference is not statistically significant (Table 1; P = 0.32). In contrast, the Y80E mutation roughly doubles the unitary current not only for Na+, but also for Ca2+ (Table 1; P = 0.01 vs. WT for both iNa and iCa). Therefore, we expect local [Ca2+] near the inner pore to be higher for Y80E channels than for WT channels. That Y80E channels fail to inactivate under these conditions implies that the Y80E mutation disrupts Ca2+ sensing or the CDI gating process (see Discussion).
Hydrophobic side chain volume at position W76 impacts the extent and kinetics of CDI
We tested a series of uncharged side chains of variable size at the W76 position for effects on CDI. W76A channels studied at high STIM1/Orai1 ratio revealed a limited extent of inactivation equivalent to ∼1/3 of the WT level, with no detectable potentiation (Fig. 5 B). All short hydrophobic side chains (W76A, W76I, W76V, and W76L; Fig. 5, B and C; and Fig. S6, B and C, respectively) produced a similar phenotype, limiting CDI to ∼1/3 of the WT extent. Larger side chains (W76F and W76Y; Fig. 5 D and Fig. S6 D) increased CDI to ∼2/3 of the WT extent, but in contrast to the pattern seen at position Y80 (Fig. 3 G), no amino acid tested at position W76 supported CDI to the level of the WT tryptophan (summary, Fig. 5 G). Inactivation kinetics were accelerated by all W76 mutations, with the smaller side chains (A, V, I, and L) producing the largest effects (Fig. 5, H and I; and Fig. S3, C and D; P < 10−3 for fast and slow time constants for A, V, I, and L vs. WT at −120 mV).
CRAC channel CDI phenotypes for mutations at Orai1 position W76. All currents were recorded and analyzed as described for Fig. 3. Representative currents are shown for WT Orai1 (A, same traces shown in Fig. 3 A) and Orai1 point mutants. W76A (B), W76I (C), W76F (D), W76E (E), and W76K (F), with the extent of CDI for this series summarized in G. Fast (H) and slow (I) time constants from biexponential fits are plotted against test potential; amplitudes of fast and slow components are shown in Fig. S3 (C and D). Each point on the summary graphs represents the mean ± SEM for n = 4–6 cells. Representative currents for additional mutations on the summary graphs are shown in Fig. S6.
CRAC channel CDI phenotypes for mutations at Orai1 position W76. All currents were recorded and analyzed as described for Fig. 3. Representative currents are shown for WT Orai1 (A, same traces shown in Fig. 3 A) and Orai1 point mutants. W76A (B), W76I (C), W76F (D), W76E (E), and W76K (F), with the extent of CDI for this series summarized in G. Fast (H) and slow (I) time constants from biexponential fits are plotted against test potential; amplitudes of fast and slow components are shown in Fig. S3 (C and D). Each point on the summary graphs represents the mean ± SEM for n = 4–6 cells. Representative currents for additional mutations on the summary graphs are shown in Fig. S6.
Disparate effects of introducing negative versus positive charge at the W76 position
W76E channels studied at high STIM1/Orai1 ratio completely lacked CDI (Fig. 5 E), consistent with our previous results (Mullins et al., 2009). The W76D mutation (Fig. S6 E), which also introduces a negatively charged side chain, had a noninactivating phenotype similar to that of W76E. In contrast, the introduction of positive charge via the W76K and W76R mutations yielded partial inactivation with accelerated kinetics (Fig. 5 F and Fig. S6 F). In terms of kinetics and extent of inactivation, the effects of introducing positive and negative charges at the W76 and Y80 positions were quite similar (compare Fig. 3, G–I; and Fig. 5, G–I).
Charge reversal or neutralization in the triple basic ring essentially eliminates CDI
A distinctive feature of the hexameric dOrai structure is a triple ring of positively charged side chains lining the central pore (R155, K159, and K163, corresponding to human Orai1 R83, K87, and R91, respectively; Hou et al., 2012). This triple ring is immediately adjacent to residues W76 and Y80. Thus, the aforementioned Y80K, Y80R, W76K, and W76R mutations (all yielding partial CDI with accelerated kinetics) introduced a fourth ring of positive charge either immediately adjacent to or near the triple basic ring. Evidence that the triple basic ring interacts with a complex anionic species in the crystal structure prompted Hou et al. (2012) to hypothesize that an interaction of this ring with an anion might stabilize the closed state.
We introduced several substitutions within the triple basic ring of human Orai1. Of the mutations that produced active Ca2+ channels (R83E, R83N, K87E, and R91E), all displayed little or no CDI (Fig. 6, B–E; see summary in F) despite retaining store-dependent activation and high Ca2+ selectivity in 20 mM Ca2+ Ringer’s. Together with the Y80 and W76 mutant data described above, these results show that introduction of negative charge at any of five consecutive pore-lining positions (Fig. 1 F) essentially eliminates CDI (summaries, Figs. 3 G, 5 G, and 6 F), as does a neutralization within the triple basic ring (Fig. 6 C, R83N).
CRAC channel CDI phenotypes for mutations of the triple basic ring. All currents were recorded as described in Fig. 3. Representative currents are shown for WT Orai1 (A, same traces shown in Fig. 3 A), and Orai1 point mutants R83E (B), R83N (C), K87E (D), and R91E (E), with the extent of CDI summarized in F. Each point represents the mean ± SEM for n = 6–10 cells.
CRAC channel CDI phenotypes for mutations of the triple basic ring. All currents were recorded as described in Fig. 3. Representative currents are shown for WT Orai1 (A, same traces shown in Fig. 3 A), and Orai1 point mutants R83E (B), R83N (C), K87E (D), and R91E (E), with the extent of CDI summarized in F. Each point represents the mean ± SEM for n = 6–10 cells.
Effects of intracellular chloride on CDI
The presence of an anion bound within the triple basic ring in the dOrai crystal structure (Hou et al., 2012), together with our observations that positive charge substitutions accelerate and negative charge substitutions eliminate CDI (Figs. 3 G, 5 G, and 6 F), raised the possibility that an anion interacting with the triple basic ring might be required for CDI to occur. CDI is stable over the course of whole-cell recordings lasting as long as 30 min; because cytosolic anions would be expected to diffuse out of the cell within this time period, endogenous diffusible anions would be reasonable candidates only if their function is maintained by entry of an anion from the pipette. The anions present in our standard pipette solution that would be expected to rapidly enter the cell are aspartate (150 mM), HEPES (10 mM), and Cl− (16 mM).
To test whether exogenous aspartate promotes CDI in our standard experimental design, we replaced aspartate in the internal solution with sucrose (Fig. 7, A and B). CDI was not impaired, arguing against a role for aspartate in CDI. Likewise, CDI was unaffected by substitution of the much larger anion HEPES for aspartate (Fig. 7 C). In both cases, the extent of CDI as compared with the standard internal solution was unchanged (P > 0.2 at −120 mV; summary, Fig. 7 D) and the kinetics trended faster, but did not reach statistical significance (0.05 < P < 0.10 for fast tau at −120 mV; Fig. S7, A and B).
CDI does not require a specific diffusible cytosolic anion. All currents were recorded from cells expressing WT mCh-STIM1 and WT Orai1-GFP as described in Fig. 3, with adjustments to the internal solution (see Materials and methods). The standard aspartate-based internal solution (A, same traces shown in Fig. 3 A) was substituted with sucrose (B) and HEPES (C), with the extent of CDI summarized in (D). Next, internal solutions were varied to include a range of chloride concentrations, from 0 (E) to 16 mM (F, same traces shown in Fig. 3 A) to 166 mM Cl− (G), with extent of CDI summarized in H. Each point on the summary graphs represents the mean ± SEM for n = 5–6 cells.
CDI does not require a specific diffusible cytosolic anion. All currents were recorded from cells expressing WT mCh-STIM1 and WT Orai1-GFP as described in Fig. 3, with adjustments to the internal solution (see Materials and methods). The standard aspartate-based internal solution (A, same traces shown in Fig. 3 A) was substituted with sucrose (B) and HEPES (C), with the extent of CDI summarized in (D). Next, internal solutions were varied to include a range of chloride concentrations, from 0 (E) to 16 mM (F, same traces shown in Fig. 3 A) to 166 mM Cl− (G), with extent of CDI summarized in H. Each point on the summary graphs represents the mean ± SEM for n = 5–6 cells.
To test whether chloride is required for CDI, we altered intracellular Cl− from 0 to 166 mM (Fig. 7, E–G). Removal of Cl− from the internal solution did not affect the extent of CDI, although it did accelerate the kinetics relative to the standard solution containing 16 mM Cl− (Fig. 7 E; and Fig. S7, C and D; P < 0.01 for fast tau at −120 mV). Thus, Cl− is apparently not required for CDI. However, we were surprised to find that increasing intracellular Cl− to 166 mM reduced the extent of CDI to ∼1/3 of the WT level (Fig. 7 G; P < 10−5). This effect occurred without a change in kinetics relative to 16 mM Cl− (Fig. S7, C and D) and without causing constitutive channel activation. Whether this is a result of a specific action on the CRAC channel pore or a general effect of high [Cl−] on protein structure through reorganization of the protein–water interface (Hofmeister, 1888; Baldwin, 1996) is unclear and was not studied further. Overall, we found no evidence that chloride, aspartate, or HEPES enabled channel inactivation (summaries, Fig. 7, D and H).
DISCUSSION
The findings of the present study prompt revisions of the current model for CRAC channel CDI. Our studies with mutant CaMs provide direct evidence that CaM does not serve as the Ca2+ sensor for CDI, and true CDI phenotypes of several Orai1 pore mutations are unmasked by our data collected at high STIM/Orai ratio. We explored in detail the effects of side chain bulk, charge, and flexibility in the inner Orai1 pore on CDI, and the results suggest roles of inner pore residues in the CDI gating process itself rather than as substrates for CaM binding. Below, we consider the ability of two possible models (conformational gating and anion binding) to explain the range of data.
The need for a high STIM/Orai ratio in studies of CDI
Measuring CDI in cells with a high STIM/Orai ratio (see Materials and methods), we observed differences in phenotypes for some Orai1 pore mutations as compared with our previous study (Mullins et al., 2009). Orai1 W76A, previously thought to eliminate CDI and support mild potentiation, is now shown to inactivate to ∼1/3 of the WT level without potentiation (Fig. 5 B). Orai1 Y80E and W76E, previously reported to produce strong potentiation in addition to eliminating CDI, are now shown to support only very limited potentiation while eliminating CDI as before (Fig. 3 E and Fig. 5 E). These differences underscore the importance of maximizing the STIM/Orai ratio (Scrimgeour et al., 2009; Hoover and Lewis, 2011) when studying the effects of mutations on CDI in heterologous expression systems. Effects of low STIM/Orai ratio may explain why several studies have reported that heterologous Orai1 channels activated by STIM1 do not inactivate at all, but instead potentiate during hyperpolarizing pulses (Zhang et al., 2008; Lee et al., 2009; Yuan et al., 2009).
A reevaluation of CaM as the Ca2+ sensor for CRAC channel CDI
The evidence in favor of the CaM-binding model for CRAC channel CDI includes the previously reported correlations between CDI of mutant Orai1 channels and the binding of Ca2+–CaM to the mutant CBDOrai1 peptides (Mullins et al., 2009), as well as the structural model for a Ca2+–CaM–CBDOrai1 peptide complex obtained by x-ray crystallography (Liu et al., 2012). The extensive steric clash revealed by alignment of the Ca2+–CaM–CBDOrai1 crystal structure (Liu et al., 2012) with the hexameric Drosophila Orai crystal structure (Fig. 1; Hou et al., 2012) calls the CaM-binding model into question.
Several possibilities might allow CaM binding without the clash. Given that the dOrai protein as crystallized lacked 131 N-terminal residues, it is possible that the cytosolic pore region in the crystal structure does not accurately reflect the native structure. Also, the inner pore structures of human Orai1 and dOrai could differ, although this seems unlikely given the high degree of sequence conservation between the two species (94% identity in the M1/pore region, corresponding to residues 76–107 of human Orai1). Further, cysteine scanning of side chain accessibility in the middle to upper pore region (R91-E106 of human Orai1; McNally et al., 2009; Zhou et al., 2010) gave results consistent with the dOrai crystal structure. The dOrai structure presumably reflects a closed channel state (Hou et al., 2012), and in theory the open or inactivated states of human Orai1 could have a more open inner pore than that seen in the dOrai structure, possibly allowing CaM binding. Our attempts to avoid the clash required large radial displacements of TM1 helices resulting in a ∼3 nm dilation of the inner pore diameter that would in turn create additional overlap between CaM and TM3. Although these results do not by themselves disprove the CaM binding model, these significant steric problems support several lines of experimental evidence that argue against it.
One line of evidence against CaM binding is that the functional effects of Orai1 point mutations are not entirely consistent with predictions of the CaM–CBDOrai1 crystal structure. In the structure, the aromatic ring of the Orai1 Y80 side chain was noted to interact with I85, A88, F141, and M145 of CaM-C via hydrophobic interactions (Liu et al., 2012). However, the Y80A and Y80C mutations, expected to have far less favorable hydrophobic interactions, support full-strength CDI with accelerated kinetics (Fig. 3 B and Fig. S2 B), contrary to expectations based on the structure. The hydroxyl group of the Orai1 Y80 side chain was noted to form a hydrogen bond with the backbone oxygen of CaM E84. If such a hydrogen bond were important to CDI, we would expect the Y80F mutation, which eliminates the hydroxyl group, to reduce the extent of CDI. Instead, speed and extent of CDI for Y80F were similar to WT (Fig. 3 C). Finally, based on the structure, positively charged substitutions such as Y80K and Y80R, and especially the W76K and W76R mutations, would be expected to disrupt hydrophobic interactions critical to CaM–CBDOrai1 binding, including the strong stabilization of W76 by five to eight side chains within a hydrophobic pocket of CaM. Instead, these mutants produce significant CDI with accelerated kinetics (Fig. 3, D and E; and Fig. 5, D and E). Together with the steric clash and the mutant CaM results (discussed below), these data argue against models in which CaM binds to CBDOrai1 in the full-length hexameric channel to drive CDI.
The most direct evidence against the CaM binding model for CDI comes from the results of overexpressing Ca2+-nonbinding mutants of CaM. To account for the rapid kinetics of CRAC channel CDI (fast time constant ∼10 ms for WT and even faster for many point mutations; Fig. 3 H and Fig. 5 H), the Ca2+ sensor must be docked at a site close to the channel before channel opening and the local increase of [Ca2+]i. Consistent with this constraint, previous work based on the differential sensitivity of CRAC channel CDI to EGTA versus BAPTA suggested that the Ca2+ sensor for CDI is located 3–4 nm from the inner pore (Zweifach and Lewis, 1995). Overexpression of Ca2+-nonbinding CaM mutants is known to displace endogenous apo–CaM from its binding sites, which allowed us to test a range of models in which apo–CaM is prebound to a site near the CRAC channel pore.
The CaM1234 mutant, in which EF hands on both the N and C lobes are nonfunctional, did not change CDI (Fig. 2 B). Because EF hand mutations in CaM can in some cases interfere with apo–CaM binding (Li et al., 2009), we also tested CaM12 and CaM34 mutants. If an unmutated N lobe is required for apo–CaM binding and the C lobe binds to the Orai1 pore to mediate CDI, then the CaM34 mutant would be expected to displace endogenous CaM, and should also prevent CDI because it lacks a functional C lobe. Liu et al. (2012) also showed evidence for N lobe binding to CBDOrai in vitro; thus, if an unmutated C lobe is required for the apoCaM binding and the N lobe binds to the Orai1 pore to mediate CDI, then the CaM12 mutation, in which N-lobe EF hands are nonfunctional, should block CDI. Counter to all predictions, none of the three mutant CaMs we tested altered CRAC channel CDI (Fig. 2), even though they effectively prevented other CaM-dependent processes including SK channel activation and CaV1.2 inactivation. These results provide the strongest evidence against the CaM binding model for CRAC channel CDI.
Our finding that heterologous CRAC channel CDI is unaffected by mutant CaMs (Fig. 2) differs from previous reports that CaM1234 suppressed CDI of CRAC current in a rat liver cell line (Litjens et al., 2004) and in RBL-1 cells (Kar et al., 2014). We note that, in both studies, CaM1234 only reduced CRAC channel CDI by ∼10%, whereas mutant CaMs nearly eliminate CDI of voltage-gated Ca2+ channels (Peterson et al., 1999; Zühlke et al., 1999). One potentially important difference in experimental designs is that the previous studies tested the effects of CaM1234 on endogenous CRAC channels, whereas we analyzed effects on heterologously overexpressed STIM1 and Orai1. Given that the STIM/Orai ratio is now recognized as a sensitive determinant of CDI (Scrimgeour et al., 2009; Hoover and Lewis, 2011), CaM1234 overexpression could have indirectly inhibited CDI if it reduced the endogenous STIM1/Orai ratio. Whether CaM is involved in regulating endogenous STIM and/or Orai expression is unknown but merits further investigation. With our experimental design, we were able to ensure that there was a high STIM1/Orai1 expression ratio (see Materials and methods) and to confirm that the mutant CaM was expressed at a high enough level to exert a maximal effect on known CaM-sensitive SK and CaV channels (Fig. 2 A).
Our current results highlight the challenges and pitfalls of building mechanistic models for processes that involve full-length proteins based on the interactions of isolated peptides. Peptide–CaM interactions merit special scrutiny, particularly when a region that may be sterically sheltered from interactions with CaM becomes accessible or changes its conformation and binding proclivity when isolated as a peptide from the parent protein (Kursula, 2014).
New functional roles for Y80 and W76 in CDI
For the reasons discussed above, we believe CaM is unlikely to bind to residues Y80 and W76 in the intact Orai1 channel. This encouraged us to assess alternate functional roles in CDI through an extensive series of mutagenesis studies. The effects of point mutations at the Y80 (Fig. 3) and W76 (Fig. 5) positions suggest distinct functions for these two residues in CDI. We discuss our main findings in the context of two models: pore blocking by an anion and conformational gating. In each case, we describe how the model would explain the results and highlight those results that are not easily explained.
Anion binding as a candidate mechanism for CDI.
A complex anion associated with the native triple basic ring was resolved in the dOrai crystal structure and was proposed to help stabilize the closed state (Hou et al., 2012). This finding led us to consider an analogous model in which CDI requires the binding of a cytosolic anion within the triple basic ring of human Orai1 (R83, K87, and R91; Fig. 1 F). An anion-binding model might explain the acceleration of CDI by small side chain substitutions at position Y80 (Fig. 3 B) and the inhibition of CDI by the bulky Y80W mutation (Fig. 3 D) as resulting from enhanced or hindered access of the blocking anion to its receptor, respectively. Similarly, small chain substitutions at W76 would accelerate CDI through speeding anion access.
The model would explain the inhibition of CDI by negative charges introduced at either Y80 (Fig. 3 E) or W76 (Fig. 5 E) as simply an electrostatic repulsion effect that prevents the anion from reaching its receptor. Conversely, positive charge introduction at these positions (Fig. 3 F and Fig. 5 F) would account for accelerated CDI by electrostatic attraction and increased local concentration of the anion near its receptor. Finally, interruption of the triple basic ring either by charge reversal or neutralization (Fig. 6) would eliminate CDI by disrupting the anion-binding site.
However, the anion-binding model has several shortcomings. First, it has difficulty explaining why the extent of CDI is reduced by the positive charge substitutions at Y80, given that an increased local concentration of anions would be expected. In addition, unlike the Y80A mutation, which preserves the extent of CDI, W76A, and other small side chain mutations reduce CDI by 2/3 (Fig. 5 B). This is difficult to explain via enhanced access of the anion to its receptor, as is possible for Y80A. Most importantly, our substitutions of diffusible intracellular anions showed no evidence for a specific anionic requirement (Fig. 7). Instead, these experiments indicated that a high concentration of chloride (166 mM) actually reduces the extent of CDI (Fig. 7 G). Together, these shortcomings lower the likelihood that anion binding within the pore drives CRAC channel CDI.
Conformational gating as a candidate mechanism for CDI.
Conformational gating refers to the general class of CDI models in which Ca2+ sensing by the channel is coupled to movements of Orai1 pore residues that are sufficient to gate the channel. The pattern of CDI phenotypes as side chain size is varied at position Y80 suggests that a motion at this position may be rate-limiting in CDI. A conformational gating model would explain the accelerated kinetics and normal extent of Y80A CDI (Fig. 3 B) as a reduction of an energy barrier for a rate-limiting step in the transition from open to inactivated. Y80F has a side chain similar in size to the WT tyrosine, and yields similar CDI kinetics and extent (Fig. 3 C), whereas the larger Y80W prevents CDI (Fig. 3 D) because it may be too bulky to permit the necessary motion. In a similar manner, the model would explain the accelerated kinetics for the range of smaller substitutions at W76 (Fig. 5) in terms of less hindered movement at that position.
What might be the nature of the conformational change at W76? The extent of CDI is graded with the size of the side chain at position W76, with small side chains permitting limited CDI, intermediate chains yielding intermediate levels of CDI, and only the WT tryptophan supporting full-strength inactivation (Fig. 5 G). This pattern is potentially compatible with conformational gating models in which native W76 side chains form a strong inactivation gate, with smaller side chains able to form only a leaky or less stable gate. The ability of tryptophan side chains to form an effective gate in the Orai pore is supported by the conduction block and structural occlusion imposed by tryptophan substitutions at R91 (hOrai1) or K163 (dOrai), respectively (Feske et al., 2006; Hou et al., 2012), and the native W76 may function similarly to occlude the pore in the presence of high local [Ca2+]. Alternatively, side chain bulk at the W76 position could be a critical determinant of closure at a gate located elsewhere in the pore, such as at V102 or the adjacent hydrophobic region (McNally et al., 2012; Gudlur et al., 2014).
Negative charge introduced at any of five consecutive pore-lining positions (W76, Y80, R83, K87, and R91; Fig. 1 F) eliminates or strongly suppresses CDI (Fig. 3 G, 5 G; and 6 F). These effects may be compatible with a conformational gating model in light of evidence that at least one of these mutations (Y80E) affects the pore conformation. The decreased Ca2+ affinity (Fig. 4 C and Table 1) and increased unitary current (Fig. 4 E and Table 1) of Orai1 Y80E suggest that in addition to abrogating CDI, this mutation also significantly alters the pore structure. The interaction of permeation and inactivation gating has been described before for the Orai1 E106D mutant channel, an outer pore mutant with markedly decreased Ca2+ affinity together with decreased CDI (Yamashita et al., 2007). Thus, an altered structure of the outer pore near the selectivity filter in Y80E may permit greater Ca2+ flux while simultaneously preventing a conformational change that underlies CDI. Further studies will be needed to test whether the other acidic mutations in the pore exhibit the same change in permeation properties.
The conformational gating model may also be able to explain the effects of the Y80K/R/Q mutations on CDI kinetics. These effects are unlikely to be a result of charge, but lysine, arginine, and glutamine all have highly flexible side chains with a particularly large number of rotamer conformations. This flexibility may allow a more rapid rearrangement of the side chains, speeding CDI kinetics. Other positively charged mutations can also be reconciled with the model. The incomplete inactivation of W76K and W76R is similar to that of W76F and W76Y (Fig. 5 G). In the framework of the conformational change model, the intermediate inactivation of W76K and W76R may be attributed to side chain size rather than charge; that is, these side chains, like W76F and W76Y, may function as partial gates. An apparent shortcoming of the conformational gating model is that it offers no simple explanation for the reduced extent of inactivation by Y80K/R/Q (Fig. 3 F; and Fig. S2, D and E).
In light of our findings, we prefer conformational gating over anion binding as a working model for CDI. It has equal if not greater explanatory power and does not invoke unknown elements outside the pore. Although this study has revealed new roles for Orai1 pore residues in the control of CDI, other regions of the CRAC channel also contribute critically to inactivation gating. In this issue (see Mullins and Lewis), we also examine the role of IDSTIM and define a functional interaction between IDSTIM and the Orai1 pore required for full-strength CDI.
Acknowledgments
The authors thank Rick Aldrich, Mike Tadross, and Anshul Rana for gifts of constructs; Luda Lokteva for technical support; members of the Lewis laboratory for their helpful comments throughout the work; Rick Aldrich for a critical reading of the manuscript; and Liang Feng for assistance with structural modeling.
This work was supported by a mentored career development award from the National Institute of General Medical Sciences (5K08GM098880 to F.M. Mullins) and by a MERIT award from NIGMS (R37GM045374 to R.S. Lewis).
The authors declare no competing financial interests.
Kenton J. Swartz served as editor.




![Figure 4. Ensemble variance analysis of Orai1 channels with charged substitutions at position Y80. (A) A thapsigargin-pretreated cell expressing Ch-STIM11-469 and WT Orai1-GFP was held continuously at −100 mV in 2 mM Ca2+o, and as indicated by the bars, DVF solutions containing variable low [Ca2+]o were applied transiently to reveal and partially block the Na+-CRAC current. Colored circles indicate peak currents corresponding to the 200-ms sweeps shown in (B), which were divided into 25-ms segments for analysis (see Materials and methods). (C) Ca2+ block of DVF Na+ currents for WT Orai1, Y80K, and Y80E. Na+ current was measured immediately after applying DVF solution containing the indicated [Ca2+]o, and normalized to the value for the base DVF solution. Each point represents the mean ± SEM for n = 3–4 cells. Lines are fits of the Hill equation, block = 1/[1 + (K1/2/[Ca])nH]. (D) Normalized variance versus current plots for WT, Y80E, and Y80K were fitted by a parabolic function to determine open probability (see Materials and methods). (E) Variance/current versus block plots were fitted by a linear function to determine unitary current (see Materials and methods). Unitary channel properties are summarized in Table 1.](https://cdn.rupress.org/rup/content_public/journal/jgp/147/2/10.1085_jgp.201511437/5/m_jgp_201511437_fig4.jpeg?Expires=1773755297&Signature=jSuozmHHKYzhIpuwDBpGouBRap-Kvs7rkB~ENFhYtxUShYbGK5Qp1R6d22werJ8U0naBj1guNMICdAEPXvoC6uroTaGuo9oiA5ZCrUqgl7HzqO5f1B4af5ypkIUkP7ujM9Nk2Gf9DS-Md~m7eIrhPwcf8a0Xb-ZqW-Iv6AiCykuWcmZmYPODCmDyDf40pFgfO5rRtfw~4r6ZnMX6aliYLlVjkPC3OPc-b0FGf0GzdPhrt6QI3Iox~bieuH-SSFd5zuaahqBeIzSIklKm0bySRyhdz3TlUyzy05giJm2Iz-EFOn~bzQO6lk52tG1x-VrB~0C8SphFt2hnGqA-lusHlw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





