Tumor-associated neutrophils (TANs) are heterogeneous; thus, their roles in tumor development could vary depending on the cancer type. Here, we showed that TANs affect metabolic dysfunction-associated steatohepatitis hepatocellular carcinoma (MASH-related HCC) more than viral-associated HCC. We attributed this difference to the predominance of SiglecFhi TANs in MASH-related HCC tumors. Linoleic acid and GM-CSF, which are commonly elevated in the MASH-related HCC microenvironment, fostered the development of this c-Myc–driven TAN subset. Through TGFβ secretion, SiglecFhi TANs promoted HCC stemness, proliferation, and migration. Importantly, SiglecFhi TANs supported immune evasion by directly suppressing the antigen presentation machinery of tumor cells. SiglecFhi TAN removal increased the immunogenicity of a MASH-related HCC model and sensitized it to immunotherapy. Likewise, a high SiglecFhi TAN signature was associated with poor prognosis and immunotherapy resistance in HCC patients. Overall, our study highlights the importance of understanding TAN heterogeneity in cancer to improve therapeutic development.
Introduction
Hepatocellular carcinoma (HCC) is one of the most common and lethal malignancies in the world. It arises from various etiologies, with hepatitis B virus (HBV) and hepatitis C virus (HCV) infections being primary risk factors for HCC development. However, due to the increasing prevalence of obesity globally, metabolic dysfunction-associated steatohepatitis (MASH) is emerging as the fastest-growing cause of HCC (Huang et al., 2022; Llovet et al., 2021). As a prototypical inflammation-driven cancer, the immune contexture in the tumor microenvironment (TME) plays a crucial role in HCC development (Llovet et al., 2021). Nevertheless, the influence of etiology on the immune TME remains poorly understood. Considering that the responses to immune checkpoint blockade (ICB) therapy in HCC patients are etiology-dependent, with non-viral-related patients exhibiting poorer outcomes than their viral-related counterparts (Haber et al., 2021; Pfister et al., 2021), it is vital to unravel the distinct immune dysfunctions associated with different HCC etiologies.
Neutrophils have been found to accumulate in a wide variety of solid tumors. While most studies link these tumor-associated neutrophils (TANs) to protumorigenic effects, some research also highlights their antitumorigenic properties (Engblom et al., 2017; Gungabeesoon et al., 2023; Siwicki and Pittet, 2021). Recent advancements in high-dimensional single-cell analysis have finally reconciled these differences by revealing that TANs are not homogenous as previously thought but are a heterogeneous population. In fact, they have been reprogrammed within the TME to acquire diverse transcriptomic, phenotypic, and functional properties (Hedrick and Malanchi, 2022; Ng et al., 2019; 2024; Siwicki and Pittet, 2021). TAN diversity has recently been reported in patients with HCC (Xue et al., 2022). However, numerous questions remain unanswered regarding the factors driving TAN diversity, the specific functions of TAN subsets, their prevalence in different TMEs, and their impact on therapy response.
In this study, we showed that TANs had a more deleterious effect on MASH-related HCC than on viral-related HCC. Specifically, we found that the TME of MASH-related HCC fostered the emergence of a pro-tumor TAN subpopulation defined by high sialic acid-binding immunoglobulin-like lectin F (SiglecF) expression and transforming growth factor β (TGFβ) production. In addition to their promotion of tumor cell stemness, proliferation, and migration, these SiglecFhi TANs directly suppressed MHCI presentation on tumor cells, leading to reduced tumor immunogenicity and ICB therapy resistance. This unprecedented function of SiglecFhi TANs in promoting an aggressive tumor phenotype with high proliferative capacity and enhanced immune evasion ability positions them as a crucial target for immunotherapy.
Results
Neutrophils are pathogenic in MASH-related HCC
Emerging evidence suggests that TANs play a pathogenic role in HCC (Geh et al., 2022; He et al., 2015; Leslie et al., 2022; Li et al., 2015; van der Windt et al., 2018; Xue et al., 2022; Zhou et al., 2019). Interestingly, although high expression of a TAN signature derived from a pan-cancer analysis (Zaitsev et al., 2022) significantly predicted poor prognosis in HCC patients from the Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) database, it was unable to do so in other HCC datasets, including the International Cancer Genome Consortium’s (ICGC) “Liver Cancer-RIKEN, Japan Project” (LIRI-JP), the Liver Cancer Institute (LCI) (GSE14520), and GSE76427 (Fig. 1 A). A thorough examination of their clinical details revealed an etiological bias among the HCC patients in these datasets. Of note, whilst the LIRI-JP, LCI, and GSE76427 datasets predominantly contain patients with pre-existing HBV or HCV infections, more than half of those from the TCGA cohort have a non-viral background (Fig. 1 B). These findings suggest that the effects of TANs on HCC development vary depending on the etiology. In particular, TANs may play a more pathogenic role in non-viral related HCC, of which MASH-related HCC is a major contributor (Huang et al., 2022; Llovet et al., 2021). However, one limitation of the TCGA-LIHC dataset is that the etiology of non-viral patients is insufficiently detailed. To better classify these patients, we generated a MASH score based on patients with defined etiologies in the TCGA-LIHC and metabolic dysfunction-associated fatty liver disease (MAFLD) datasets (Govaere et al., 2020). We found that the TAN signature showed significant prognostic potential only for patients with high MASH score but not for those with a low MASH score (Fig. 1 C). We further demonstrated this etiology-dependent phenotype by using two autochthonous HCC models, which were induced by hydrodynamic tail vein injections (HTVIs) of plasmid DNA in C57BL/6J mice. To mimic MASH-related HCC, we delivered transposon vectors expressing oncogenic NRAS and AKT into the liver. Consistent with our recent reports (Chen et al., 2022; Zhou et al., 2022), these NRAS/AKT tumors expressed high levels of the HCC marker α-fetoprotein (AFP) and exhibited typical MASH features such as steatosis, ballooning hepatocytes, and increased levels of liver triglycerides and diglycerides (Fig. 1 D; and Fig. S1, A and D). Given that the most common genetic alteration in viral-related HCC is the simultaneous deregulation of MYC (overexpression) and TP53 (inactivation or mutation) (Llovet et al., 2021; Péneau et al., 2022; Tornesello et al., 2016), we mimicked this etiology by delivering a MYC-expressing transposon vector, along with a CRISPR plasmid specifically targeting p53. As expected, the resulting MYC/sgp53 tumors were positive for AFP without any MASH features (Fig. 1 D; and Fig. S1, B and D). We observed that both HCC models were highly infiltrated by TANs, which accounted for around 30% of the tumor-infiltrating leukocytes (TILs) (Fig. 1 E). However, when TANs were depleted by anti-Ly6G mAb injections, following a protocol optimized for their controlled and durable elimination (Boivin et al., 2020), a significant reduction in tumor burden was only observed for NRAS/AKT tumors, but not MYC/sgp53 tumors (Fig. 1 F and Fig. S1 E), suggesting that TANs promoted tumor development in MASH-related HCCs. We further validated these findings using a dietary model of MASH-related HCC, in which C57BL/6J mice were maintained on a methionine- and choline-deficient (MCD) diet for 2 wk before being intrahepatically injected with the RIL175 HCC cell line (Brown et al., 2018) (Fig. S1, C and D). As expected, anti-Ly6G mAb injections only impaired the growth of RIL175 tumors in mice kept on an MCD diet and not in those with a chow diet (CD) (Fig. 1 G). Together, these findings from both HCC patients and murine models suggest that the pathogenic influence of TANs on HCC development is etiology-dependent and that TANs have a more deleterious effect on MASH-related HCC.
Neutrophils are pathogenic in MASH-related HCC. (A) Kaplan–Meier plots comparing HCC patients from various datasets with high (top 25%) and low (top 25%) TAN signature scores. (B) The etiologies of HCC patients in A. (C) Kaplan–Meier plots comparing different groups of TCGA-LIHC patients with high and low TAN signature scores. (D and E) Mice bearing NRAS/AKT or cMYC/sgp53 HCC tumors were sacrificed at 4 or 7 wk after tumor induction, respectively. Representative images of AFP staining (left) and neutral lipid staining (right) on liver sections. The scale bar represents 100 µm (D). Representative FACs plots and percentages of CD11b+Ly6G+ neutrophils among CD45+TILs (n = 4–5/group) (E). (F) NRAS/AKT HCC mice (top panel) and cMYC/sgp53 HCC mice (bottom panel) were treated with αLy6G or isotype control mAbs for 2 wk. Representative images of HCC livers (left); liver-to-body weight ratio (middle) and tumor nodule counts (right). Scale bar represents 1 cm. (G) Tumor volume of RIL175 orthotopic HCC mice fed with MCD diet (MCD + RIL175; top) or Chow diet (CD + RIL175; bottom), following αLy6G or isotype control mAb treatments. Each symbol represents one mouse. Data are mean ± SEM (E–G) and represent two to three independent experiments (D–G). *P < 0.05, ***P < 0.001 by Kaplan–Meier analysis (A and C) and unpaired Student’s t test (F and G). NA, not available; T, tumor; NT, non-tumor.
Neutrophils are pathogenic in MASH-related HCC. (A) Kaplan–Meier plots comparing HCC patients from various datasets with high (top 25%) and low (top 25%) TAN signature scores. (B) The etiologies of HCC patients in A. (C) Kaplan–Meier plots comparing different groups of TCGA-LIHC patients with high and low TAN signature scores. (D and E) Mice bearing NRAS/AKT or cMYC/sgp53 HCC tumors were sacrificed at 4 or 7 wk after tumor induction, respectively. Representative images of AFP staining (left) and neutral lipid staining (right) on liver sections. The scale bar represents 100 µm (D). Representative FACs plots and percentages of CD11b+Ly6G+ neutrophils among CD45+TILs (n = 4–5/group) (E). (F) NRAS/AKT HCC mice (top panel) and cMYC/sgp53 HCC mice (bottom panel) were treated with αLy6G or isotype control mAbs for 2 wk. Representative images of HCC livers (left); liver-to-body weight ratio (middle) and tumor nodule counts (right). Scale bar represents 1 cm. (G) Tumor volume of RIL175 orthotopic HCC mice fed with MCD diet (MCD + RIL175; top) or Chow diet (CD + RIL175; bottom), following αLy6G or isotype control mAb treatments. Each symbol represents one mouse. Data are mean ± SEM (E–G) and represent two to three independent experiments (D–G). *P < 0.05, ***P < 0.001 by Kaplan–Meier analysis (A and C) and unpaired Student’s t test (F and G). NA, not available; T, tumor; NT, non-tumor.
Characterization of murine models reveals MASH-HCC features in NRAS/AKT HCC and MCD + RIL175 models. (A–C) Representative H&E liver sections. The HCC tumor region is bounded by the dotted line. Scale bar represents 100 µm. (D) Untargeted lipidomic LC-MS analysis of the various types of HCC tumors and naïve liver. (E) Representative FACs plots and percentages of Ly6G+CD11b+ neutrophils (TANs) among CD45+TILs from cMYC/sgp53 tumors after 2 wk of αLy6G or isotype control mAbs treatment (top). Quantity of TANs per gram of tissue (bottom). (F and G) Gating strategies for tumor infiltrating lymphocytes (F): TANs (i), DCs (ii), CD8+T cells (iii) and their cytokines (iv), and tumor cells (G) from all the HCC models used. Each symbol represents one mouse. Data are mean ± SEM and represent two independent experiments (E). ****P < 0.0001 by unpaired Student’s t test (E). T, tumor; NT, non-tumor; TG, triglyceride; DG, diglyceride; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelins.
Characterization of murine models reveals MASH-HCC features in NRAS/AKT HCC and MCD + RIL175 models. (A–C) Representative H&E liver sections. The HCC tumor region is bounded by the dotted line. Scale bar represents 100 µm. (D) Untargeted lipidomic LC-MS analysis of the various types of HCC tumors and naïve liver. (E) Representative FACs plots and percentages of Ly6G+CD11b+ neutrophils (TANs) among CD45+TILs from cMYC/sgp53 tumors after 2 wk of αLy6G or isotype control mAbs treatment (top). Quantity of TANs per gram of tissue (bottom). (F and G) Gating strategies for tumor infiltrating lymphocytes (F): TANs (i), DCs (ii), CD8+T cells (iii) and their cytokines (iv), and tumor cells (G) from all the HCC models used. Each symbol represents one mouse. Data are mean ± SEM and represent two independent experiments (E). ****P < 0.0001 by unpaired Student’s t test (E). T, tumor; NT, non-tumor; TG, triglyceride; DG, diglyceride; PC, phosphatidylcholine; PE, phosphatidylethanolamine; SM, sphingomyelins.
TANs are heterogenous with a notable enrichment of the SiglecFhi subset in MASH-related HCC
TANs are known to be heterogeneous, consisting of functionally distinct subsets that can either support or suppress tumor growth (Hedrick and Malanchi, 2022; Siwicki and Pittet, 2021). We asked if the varying effects of TANs on different types of HCC could be attributed to differences in the TAN subset composition. To address this, we performed single-cell RNA sequencing (scRNA-seq) on TILs, splenocytes, and bone marrow (BM) cells isolated from NRAS/AKT-injected mice. A total of 14,105 neutrophils, with an average of 1,136 genes per cell, were identified using SingleR and their expression of lineage markers (Fig. S2 A). We identified six transcriptomically distinct TAN subsets and five splenic and BM neutrophil subsets (Fig. 2 A and Fig. S2 B) through unsupervised clustering. These 11 neutrophil subsets encompassed the spectrum of neutrophils observed in HCC patients (Xue et al., 2022), with the Siglecf-Hi and Siglecf-Lo subsets bearing the closest resemblance to human pro-tumor TANs (Xue et al., 2022) (Fig. 2 B). Furthermore, the gene expression profiles of these Siglecf-expressing subsets from HCC tumors were similar to those of tumor-promoting SiglecFhi TANs previously identified in lung cancers (Engblom et al., 2017; Pfirschke et al., 2020) (Fig. 2 C), suggesting the potential tumorigenic properties of these TANs. This diversity of TAN subsets in HCC was partially captured by flow cytometric analysis based on their surface expression levels of SiglecF. In particular, a group of SiglecFhi neutrophils (CD11b+Ly6G+SiglecFhi cells), corresponding to the Siglecf-Hi TAN cluster in our scRNA-seq analysis (Fig. S2 C), were highly enriched in tumors compared with blood, spleen, or BM of NRAS/AKT-injected mice (Fig. 2, D and E; and Fig. S2 D). Further morphological, phenotypical, and functional analysis revealed that SiglecFhi TANs were mature and aged neutrophils. These cells exhibited segmented nuclei and were characterized by high expression of CXCR2, CXCR4, and dcTRAIL-R1, as well as low expression of CD62L. Additionally, they displayed proficiency in phagocytosis, degranulation, and the ability to undergo NETosis (Fig. 2 F; and Fig. S2, E and F). Crucially, SiglecFhi TANs were exclusively present in MASH-related HCC tumors (NRAS/AKT; MCD + RIL175 diet) and not in their non-MASH-related counterparts (cMYC/sgp53; chow + RIL175 diet), highlighting the relevance of this population in MASH-related HCC (Fig. 2 D and Fig. S2 G). Of note, depletion of TANs through anti-Ly6G mAb injections eliminated SiglecFhi TANs while retaining some SiglecF– TANs (Fig. 2 G and Fig. S2 H), suggesting that this method could serve as a potential approach to analyze the in vivo effects of SiglecFhi TANs in subsequent investigations. Thus far, our results indicated that SiglecFhi TANs represent a TAN population that was highly enriched in murine MASH-related HCC models. To establish human relevance of these findings, we defined a gene signature representing Siglecf-Hi–like TANs (Fig. S2 I). We then applied it to the microarray data of HCC cohorts with clinically confirmed MASH and non-MASH cases (Pinyol et al., 2021; Villanueva et al., 2015). We observed that while a conventional neutrophil signature (Abbas et al., 2005) was not biased to etiology, the Siglecf-Hi–like TAN signature was significantly enriched in MASH-HCC patients (Fig. 2 H). To further investigate the impact of Siglecf-Hi–like TANs on HCC development, we assessed their prognostic potential through the TCGA-LIHC dataset, which contains patient survival information. As expected, the Siglecf-Hi–like TAN signature predicted poor prognosis in non-viral HCC patients with high MASH scores, but not in those with low MASH scores or those who were virally infected (Fig. 2 I). Together, these findings suggest that TANs are highly diverse and a pro-tumor SiglecFhi subset is predominately enriched in MASH-related HCC.
Neutrophil identification and characteristics. (A) Uniform Manifold Approximation and Projections (UMAPs) (left) and expression of neutrophil lineage markers (right) by the major immune populations identified from the scRNA-seq of the BM and spleen (top), and HCC tumors (bottom) of NRAS/AKT HCC mice. (B) Dot plot of the differently expressed genes by each neutrophil cluster in the BM and spleen (top), and HCC tumor (bottom). (C) RNA-seq analyses were performed on SiglecFhi and SiglecF– TANs purified from NRAS/AKT-HCC tumors. A gene set defining SiglecFhi TANs was generated from their top 100 differentially expressed genes. The distribution of this gene set among TAN subsets. (D) Representative FACs plots and percentage of SiglecFhi neutrophils detected in the BM, spleen, blood, and tumor of NRAS/AKT HCC mice. (E) CXCR2, CXCR4, CD62L, and dcTRAIL-R1 expression within SiglecF− and SiglecFhi TANs of NRAS/AKT HCC mice. (F) Functional characterization of BM neutrophils, SiglecF− and SiglecFhi TANs from NRAS/AKT HCC mice. (G) Representative FACs plots and percentages of SiglecFhi TANs of RIL175 orthotopic HCC mice fed with MCD diet (MCD + RIL175; left) or Chow diet (CD + RIL175; right) (n = 3/group). (H) Representative immunofluorescence images and proportion of SiglecFhi TANs (myeloperoxidase [MPO]: red; SiglecF: green; nucleus: blue) in NRAS/AKT HCC tumors after 2 wk of treatment with αLy6G or isotype control mAbs. Scale bar represents 100 µm. (I) Expression levels of genes in the Siglecf-Hi–like TAN signature across various cell types in the tumor of HCC patients (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744). Each symbol represents one mouse. Data are mean ± SEM and represent two independent experiments (D–H). **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA (F) and unpaired Student’s t test (H).
Neutrophil identification and characteristics. (A) Uniform Manifold Approximation and Projections (UMAPs) (left) and expression of neutrophil lineage markers (right) by the major immune populations identified from the scRNA-seq of the BM and spleen (top), and HCC tumors (bottom) of NRAS/AKT HCC mice. (B) Dot plot of the differently expressed genes by each neutrophil cluster in the BM and spleen (top), and HCC tumor (bottom). (C) RNA-seq analyses were performed on SiglecFhi and SiglecF– TANs purified from NRAS/AKT-HCC tumors. A gene set defining SiglecFhi TANs was generated from their top 100 differentially expressed genes. The distribution of this gene set among TAN subsets. (D) Representative FACs plots and percentage of SiglecFhi neutrophils detected in the BM, spleen, blood, and tumor of NRAS/AKT HCC mice. (E) CXCR2, CXCR4, CD62L, and dcTRAIL-R1 expression within SiglecF− and SiglecFhi TANs of NRAS/AKT HCC mice. (F) Functional characterization of BM neutrophils, SiglecF− and SiglecFhi TANs from NRAS/AKT HCC mice. (G) Representative FACs plots and percentages of SiglecFhi TANs of RIL175 orthotopic HCC mice fed with MCD diet (MCD + RIL175; left) or Chow diet (CD + RIL175; right) (n = 3/group). (H) Representative immunofluorescence images and proportion of SiglecFhi TANs (myeloperoxidase [MPO]: red; SiglecF: green; nucleus: blue) in NRAS/AKT HCC tumors after 2 wk of treatment with αLy6G or isotype control mAbs. Scale bar represents 100 µm. (I) Expression levels of genes in the Siglecf-Hi–like TAN signature across various cell types in the tumor of HCC patients (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744). Each symbol represents one mouse. Data are mean ± SEM and represent two independent experiments (D–H). **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA (F) and unpaired Student’s t test (H).
Protumorigenic SiglecF hi TANs are enriched in MASH-related HCC tumors and can be removed through αLy6G mAb injections. (A–C) scRNA-seq analysis of neutrophils from NRAS/AKT HCC mice. UMAP visualization of neutrophil subsets (A). Pearson’s correlations across neutrophil clusters in NRAS/AKT HCC mice and HCC patients (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744) (B). Distributions of SiglecFhigh neutrophil signature score derived from Engblom et al. (2017) in TAN subsets (C). (D) Representative FACs plots of SiglecFhi TANs detected in NRAS/AKT HCC (left) and cMYC/sgp53 HCC (right) tumors. (E) Representative immunofluorescence images of SiglecFhi TANs (MPO: red; SiglecF: green; nucleus: blue) in NRAS/AKT HCC tumors. Dotted oval shows the tumor region. Scale bars represent 200 and 5 μm. (F) Representative cytospin images of SiglecF− and SiglecFhi TANs. Scale bar represents 10 μm. (G) Representative FACs plots and quantities of TANs (CD11b+ Ly6Gintracellular+ (top) and TAN subsets (bottom) in NRAS/AKT HCC tumors after 2 wk of αLy6G or isotype control mAbs treatments. (H) Distributions of conventional neutrophil signature and Siglecf-Hi–like TAN signature scores in HCC patients with non-MASH (GSE63898) and MASH (GSE164760) etiologies. (I) Kaplan–Meier plots comparing different groups of TCGA-LIHC patients with high and low Siglecf-Hi–like TAN signature score. Each symbol represents one mouse. Data are mean ± SEM (D and G) and represent three independent experiments (D–G). ***P < 0.001, ****P < 0.0001 by unpaired Student’s t test (D and G-upper panel) and one sample t test (G-lower panel). Data are analyzed by Wilcoxon rank-sum test (H) and Kaplan–Meier analysis (I). hPB, human peripheral blood; hAL, human adjacent liver.
Protumorigenic SiglecF hi TANs are enriched in MASH-related HCC tumors and can be removed through αLy6G mAb injections. (A–C) scRNA-seq analysis of neutrophils from NRAS/AKT HCC mice. UMAP visualization of neutrophil subsets (A). Pearson’s correlations across neutrophil clusters in NRAS/AKT HCC mice and HCC patients (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744) (B). Distributions of SiglecFhigh neutrophil signature score derived from Engblom et al. (2017) in TAN subsets (C). (D) Representative FACs plots of SiglecFhi TANs detected in NRAS/AKT HCC (left) and cMYC/sgp53 HCC (right) tumors. (E) Representative immunofluorescence images of SiglecFhi TANs (MPO: red; SiglecF: green; nucleus: blue) in NRAS/AKT HCC tumors. Dotted oval shows the tumor region. Scale bars represent 200 and 5 μm. (F) Representative cytospin images of SiglecF− and SiglecFhi TANs. Scale bar represents 10 μm. (G) Representative FACs plots and quantities of TANs (CD11b+ Ly6Gintracellular+ (top) and TAN subsets (bottom) in NRAS/AKT HCC tumors after 2 wk of αLy6G or isotype control mAbs treatments. (H) Distributions of conventional neutrophil signature and Siglecf-Hi–like TAN signature scores in HCC patients with non-MASH (GSE63898) and MASH (GSE164760) etiologies. (I) Kaplan–Meier plots comparing different groups of TCGA-LIHC patients with high and low Siglecf-Hi–like TAN signature score. Each symbol represents one mouse. Data are mean ± SEM (D and G) and represent three independent experiments (D–G). ***P < 0.001, ****P < 0.0001 by unpaired Student’s t test (D and G-upper panel) and one sample t test (G-lower panel). Data are analyzed by Wilcoxon rank-sum test (H) and Kaplan–Meier analysis (I). hPB, human peripheral blood; hAL, human adjacent liver.
SiglecFhi TANs directly act on tumor cells and promote tumorigenesis
To gain insight into the contributions of SiglecFhi TANs in tumor development, we investigated the tumorigenic pathways that were positively correlated with the abundance of these cells in HCC patients. We observed a high correlation between angiogenesis and Siglecf-Hi–like TAN signatures, supporting prior findings that protumor TANs are one of the sources of VEGFΑ within the TME (Engblom et al., 2017; Ng et al., 2024; Ozel et al., 2022) (Fig. 3 A and Fig. S3 A). Of note, we found that pathways related to cancer stemness, cancer proliferation, and epithelial-mesenchymal transition (EMT) were also significantly correlated (Fig. 3 A). Consistently, tumor cells isolated from anti-Ly6G mAb-treated NRAS/AKT-injected mice, which lacked SiglecFhi TANs, exhibited a reduced cancer stemness signature (Fig. S3 B), a lower proportion of cancer stem cells (CSCs) (Fig. S3, C and D), and a decreased ex vivo repopulation capacity (Fig. S3 E). Together, these findings suggest that SiglecFhi TANs could promote tumorigenesis by directly affecting tumor cell behavior. To test this, we sort-purified SiglecFhi and SiglecF− TANs from NRAS/AKT tumors and co-cultured them with the HCC cell line RIL175 (Fig. 3 B). Through the limiting dilution assay (Zhou et al., 2022), we observed that SiglecFhi TANs, and not SiglecF− TANs, increased tumor-initiating cell frequency (Fig. 3 C), suggesting that SiglecFhi TANs enhanced the self-renewal potential of tumor cells. Similarly, a clonogenic assay and scratch wound assay also revealed that SiglecFhi TANs could directly promote HCC cell proliferation and migration, respectively (Fig. 3, D and E). To validate the tumor-promoting role of SiglecFhi TANs in vivo, we employed an adoptive transfer model, wherein RIL175 cells were co-injected with either SiglecFhi TANs or SiglecF− TANs into contralateral flanks of C57BL/6J mice (Fig. 3 F). In line with the ex vivo findings, co-transferring SiglecFhi TANs with RIL715 cells resulted in a significant increase in tumor proliferation and an enhanced tumor growth rate (Fig. 3, G and H). Together, these findings suggest that SiglecFhi TANs contribute to HCC development by directly enhancing tumor cell proliferation, stemness, and migration.
SiglecF hi TANs directly promote tumorigenesis. (A) Pearson’s correlation of tumorigenesis-related gene signatures with Siglecf-Hi–like TAN signature in the TCGA-LIHC dataset. (B–E) Sort-purified SiglecF– and SiglecFhi TANs were co-cultured with RIL175 cells. Schematic diagram of the sorting and co-culture strategy (B). Limiting dilution assay: representative images of spheroids formed and stem cell frequency. Scale bar represents 50 μm (C). Clonogenic assay: representative images of colonies formed and colony counts. Scale bar represents 3.5 mm (D). Scratch wound assay: representative images of the wound at indicated time points and percentage of the wound area. Scale bar represents 200 μm (E). (F–H) RIL175 cells alongside SiglecF− or SiglecFhi TANs were subcutaneously injected into C57BL/6J mice. Schematic diagram of the adoptive transfer strategy (F). Representative images and volumes of tumors at the endpoint. Scale bar represents 1 cm (G). Representative FACs plots and percentage of Ki-67+ tumor cells (H). Each symbol represents TANs isolated from two to three mice (D, E, G, and H). Data are mean ± SEM (D, E, G, and H) and represent two to three independent experiments (C, E, G, and H) or are pooled from two independent experiments (D). *P < 0.05, **P < 0.01, ***P < 0.001 by Pearson’s correlation test (A), Pearson’s chi-square test with a 95% confidence interval (C), one-way ANOVA (D and E), and unpaired Student’s t test (G and H).
SiglecF hi TANs directly promote tumorigenesis. (A) Pearson’s correlation of tumorigenesis-related gene signatures with Siglecf-Hi–like TAN signature in the TCGA-LIHC dataset. (B–E) Sort-purified SiglecF– and SiglecFhi TANs were co-cultured with RIL175 cells. Schematic diagram of the sorting and co-culture strategy (B). Limiting dilution assay: representative images of spheroids formed and stem cell frequency. Scale bar represents 50 μm (C). Clonogenic assay: representative images of colonies formed and colony counts. Scale bar represents 3.5 mm (D). Scratch wound assay: representative images of the wound at indicated time points and percentage of the wound area. Scale bar represents 200 μm (E). (F–H) RIL175 cells alongside SiglecF− or SiglecFhi TANs were subcutaneously injected into C57BL/6J mice. Schematic diagram of the adoptive transfer strategy (F). Representative images and volumes of tumors at the endpoint. Scale bar represents 1 cm (G). Representative FACs plots and percentage of Ki-67+ tumor cells (H). Each symbol represents TANs isolated from two to three mice (D, E, G, and H). Data are mean ± SEM (D, E, G, and H) and represent two to three independent experiments (C, E, G, and H) or are pooled from two independent experiments (D). *P < 0.05, **P < 0.01, ***P < 0.001 by Pearson’s correlation test (A), Pearson’s chi-square test with a 95% confidence interval (C), one-way ANOVA (D and E), and unpaired Student’s t test (G and H).
Siglecf hi TAN removal impairs tumorigenesis. (A) Visualization of Vegfa expression among TILs. (B–E) Profiling of tumor cells from NRAS/AKT HCC mice after 2 wk of αLy6G or isotype control mAbs treatments. scRNA-seq expression of mature hepatocyte and hepatic progenitor markers (B). Representative FACs plots and percentage of Hoechst-effluxing cells (polygon gate) (C). Representative FACs plots and percentage of CD13+CD133+ CSCs (polygon gate) (D). Stem cell frequency in tumor cells isolated from NRAS/AKT HCC mice 1 wk following αLy6G or isotype control mAbs treatments (E). Each symbol represents one mouse (C and D). Data are mean ± SEM and represent two independent experiments (C–E). *P < 0.05, **P < 0.01 by unpaired Student’s t test (C and D) and Pearson’s chi-square test with a 95% confidence interval (E). MQs, macrophages; DCs, dendritic cells.
Siglecf hi TAN removal impairs tumorigenesis. (A) Visualization of Vegfa expression among TILs. (B–E) Profiling of tumor cells from NRAS/AKT HCC mice after 2 wk of αLy6G or isotype control mAbs treatments. scRNA-seq expression of mature hepatocyte and hepatic progenitor markers (B). Representative FACs plots and percentage of Hoechst-effluxing cells (polygon gate) (C). Representative FACs plots and percentage of CD13+CD133+ CSCs (polygon gate) (D). Stem cell frequency in tumor cells isolated from NRAS/AKT HCC mice 1 wk following αLy6G or isotype control mAbs treatments (E). Each symbol represents one mouse (C and D). Data are mean ± SEM and represent two independent experiments (C–E). *P < 0.05, **P < 0.01 by unpaired Student’s t test (C and D) and Pearson’s chi-square test with a 95% confidence interval (E). MQs, macrophages; DCs, dendritic cells.
SiglecFhi TANs exert protumor functions via TGFβ
We next investigated the mechanism underlying the pro-tumor effects of SiglecFhi TANs. To this end, bulk RNA-seq analysis was performed on RIL175 cells cultured with and without SiglecFhi or SiglecF− TANs. We then examined the differentially expressed genes between different groups using Gene Ontology (GO) enrichment analysis. As expected, SiglecFhi TAN-treatment upregulated pathways related to cell migration, proliferation, and Wnt signaling (Fig. 4 A). Of note, genes involved in TGFβ stimulation were also significantly enriched in RIL175 cells upon SiglecFhi TANs addition (Fig. 4 A). TGFβ is a potent tumorigenic cytokine that is essential for HCC development (Chen et al., 2019). Thus, we postulated that SiglecFhi TANs promoted HCC growth through TGFβ production, which was supported by cell–cell interaction analysis of NRAS/AKT tumors. Tgfb1 was inferred as one of the top ligands affecting the tumor cell transcriptome (Fig. 4 B). Importantly, the tumor-acting TGFβ was mainly produced by TANs as its effects were significantly diminished following anti-Ly6G mAb depletion (Fig. 4 C). Upon investigating the source of TGFβ among TAN subsets, we observed a significant upregulation of genes associated with TGFβ1 production (Tgfb1, Srebf1, Plg, and Atp6ap2) among Siglecf-Hi TANs (Fig. 4 D). This finding was further supported at the protein level. Flow cytometry analysis revealed that SiglecFhi TANs significantly increased the expression of latency-associated peptides associated with TGFβ1 (LAP [TGFβ1]) compared with SiglecF− TANs (Fig. 4 E). Furthermore, freshly isolated SiglecFhi TANs, but not their SiglecF− counterparts, produced high levels of TGFβ1 even in the absence of further stimulation (Fig. 4 F).
SiglecF hi TANs secrete TGFβ. (A) GO analysis of differentially expressed genes between RIL175 cells and those co-cultured with SiglecFhi TANs. BP, biological process. (B and C) NicheNet analysis of scRNA-seq data from NRAS/AKT HCC mice treated with αLy6G or isotype control mAbs. Prioritized ligands and their target genes driving transcriptomic changes in tumor cells after αLy6G treatment (B). Log fold-change (LFC) in senders of ligands to tumor cells after αLy6G treatment (C). (D) Relative expression of indicated genes among neutrophil subsets from NRAS/AKT HCC mice. (E) Representative FACs plots (left) and percentage of LAP (TGFβ1)+ cells among TAN subsets. (F) Spontaneous TGFβ production by SiglecF– and SiglecFhi TANs. Each symbol represents one mouse (E) or TANs isolated from two to three mice (F). Data are mean ± SEM and represent two independent experiments (E and F). *P < 0.05, ****P < 0.0001 by unpaired Student’s t test (E and F).
SiglecF hi TANs secrete TGFβ. (A) GO analysis of differentially expressed genes between RIL175 cells and those co-cultured with SiglecFhi TANs. BP, biological process. (B and C) NicheNet analysis of scRNA-seq data from NRAS/AKT HCC mice treated with αLy6G or isotype control mAbs. Prioritized ligands and their target genes driving transcriptomic changes in tumor cells after αLy6G treatment (B). Log fold-change (LFC) in senders of ligands to tumor cells after αLy6G treatment (C). (D) Relative expression of indicated genes among neutrophil subsets from NRAS/AKT HCC mice. (E) Representative FACs plots (left) and percentage of LAP (TGFβ1)+ cells among TAN subsets. (F) Spontaneous TGFβ production by SiglecF– and SiglecFhi TANs. Each symbol represents one mouse (E) or TANs isolated from two to three mice (F). Data are mean ± SEM and represent two independent experiments (E and F). *P < 0.05, ****P < 0.0001 by unpaired Student’s t test (E and F).
Crucially, the protumor effects of SiglecFhi TANs on HCC cells were effectively ameliorated by blocking TGFβ signaling with Vactosertib, a TGFβ receptor I inhibitor. While Vactosertib did not influence RIL175 cells in the in vitro limiting dilution, clonogenic, and scratch wound assays, it significantly reduced the stemness properties, proliferation, and migration of RIL175 cells potentiated by the addition of SiglecFhi TANs (Fig. 5, A–C). We then extended our analysis to in vivo settings. First, to demonstrate that TGFβ is the key effector molecule of TANs, we induced NRAS/AKT tumors in Mrp8Cre.TGFβfl/fl mice, which selectively ablates TGFβ secretion in neutrophils. As expected, tumor cells from Mrp8Cre.TGFβfl/fl mice were less proliferative and contained a lower proportion of CSCs (Fig. 5, D and E). Consequently, tumor burden in Mrp8Cre.TGFβfl/fl mice was noticeably decreased compared with littermate controls (Fig. 5, F and G). Second, we confirmed that SiglecFhi TANs exerted their protumor function via TGFβ. For this, we employed the adoptive transfer model as in Fig. 3 F, but using TGFβ receptor 1 knockdown RIL175 cells (Fig. 5 H). In contrast to the tumor-promoting effects observed with WT RIL175 cells (Fig. 3, G and H), the presence of SiglecFhi TANs showed no measurable impact on the growth of RIL175_shTgfbr1 cells relative to SiglecF– TANs (Fig. 5, I and J). Collectively, these data indicate that SiglecFhi TANs exerted their protumor functions via the release of TGFβ.
SiglecF hi TANs mediate their protumorigenic functions via TGFβ secretion. (A–C) SiglecFhi TANs were co-cultured with RIL175 cells, with or without the addition of Vactosertib (10 nM). In vitro limiting dilution (A), clonogenic (B), and scratch wound (C) assays were performed. (D–G) NRAS/AKT HCC tumors were induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T). Representative FACs plots and percentage of Ki-67+ tumor cells (D). Representative FACs plots and percentage of CD13+CD133+ CSCs (E). Representative images of HCC livers and liver-to-body weight ratio. Scale bar represents 1 cm (F). Representative images of AFP+ tumors and tumor nodule counts. Scale bar represents 100 μm (G). (H–J) SiglecF– and SiglecFhi TANs were sort-purified from NRAS/AKT HCC tumors and subcutaneously injected alongside RIL175_shTgfbr1 cells into C57BL/6J mice. Schematic diagram of adoptive transfer strategy (H). Representative images and volumes of tumors at the endpoint. Scale bar represents 1 cm (I). Representative FACs plots and percentage of Ki-67+ tumor cells (J). Each symbol represents TANs isolated from two to three mice (B and C), or one mouse (D–G, I, and J). Data are mean ± SEM and represent two to three independent experiments (A, C–G, I, and J) or are pooled from two independent experiments (B). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Pearson’s chi-square test with a 95% confidence interval (A), one-way ANOVA (B and C), or unpaired Student’s t test (D–G).
SiglecF hi TANs mediate their protumorigenic functions via TGFβ secretion. (A–C) SiglecFhi TANs were co-cultured with RIL175 cells, with or without the addition of Vactosertib (10 nM). In vitro limiting dilution (A), clonogenic (B), and scratch wound (C) assays were performed. (D–G) NRAS/AKT HCC tumors were induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T). Representative FACs plots and percentage of Ki-67+ tumor cells (D). Representative FACs plots and percentage of CD13+CD133+ CSCs (E). Representative images of HCC livers and liver-to-body weight ratio. Scale bar represents 1 cm (F). Representative images of AFP+ tumors and tumor nodule counts. Scale bar represents 100 μm (G). (H–J) SiglecF– and SiglecFhi TANs were sort-purified from NRAS/AKT HCC tumors and subcutaneously injected alongside RIL175_shTgfbr1 cells into C57BL/6J mice. Schematic diagram of adoptive transfer strategy (H). Representative images and volumes of tumors at the endpoint. Scale bar represents 1 cm (I). Representative FACs plots and percentage of Ki-67+ tumor cells (J). Each symbol represents TANs isolated from two to three mice (B and C), or one mouse (D–G, I, and J). Data are mean ± SEM and represent two to three independent experiments (A, C–G, I, and J) or are pooled from two independent experiments (B). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Pearson’s chi-square test with a 95% confidence interval (A), one-way ANOVA (B and C), or unpaired Student’s t test (D–G).
We then leveraged TGFβ, the key effector molecule of SiglecFhi TANs, to further validate the clinical relevance of this TAN subset. Once again, in HCC patient datasets, TGFβ secreted by TANs was found to strongly influence gene expression in tumor cells, including those related to proliferation (e.g., CDKN1A), stemness (e.g., CD44), and metastasis (e.g., SGK1) (Fig. 6 A). We also observed a marked upregulation of TAN-derived TGFβ expression in MASH-related HCC patients compared with their non-MASH counterparts (Fig. 6 B). Importantly, we conducted co-immunofluorescence staining on tumors surgically resected from treatment-naïve HCC patients and confirmed a significantly higher percentage of TGFβ-producing TANs in MASH-related HCC tumors compared with HBV-infected HCC tumors (Fig. 6 C). Together, these data underscored the importance of SiglecFhi TANs in the tumorigenesis of MASH-related HCC patients.
TGFβ-producing TANs are enriched in MASH-related HCC patients. (A) NicheNet analysis of HCC patient dataset (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744). Prioritized ligands from neutrophils, and their target genes, driving transcriptomic differences in tumor cells between patients with high or low neutrophil infiltration. (B) Distributions of TAN-derived TGFβ production signature in HCC patients with non-MASH (GSE63898) and MASH (GSE164760) etiologies. (C) Representative immunofluorescence images and quantification of TGFβ-producing CD66b+neutrophils (CD66b: red; TGFβ: green; nucleus: blue) among all neutrophils in tumors resected from patients with HBV-related HCC or MASH-related HCC. Scale bars represent 50 and 20 μm. Each symbol represents one patient (C). Data is mean ± SEM. **P < 0.01 by Wilcoxon rank-sum test (B) and unpaired Student’s t test (C).
TGFβ-producing TANs are enriched in MASH-related HCC patients. (A) NicheNet analysis of HCC patient dataset (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744). Prioritized ligands from neutrophils, and their target genes, driving transcriptomic differences in tumor cells between patients with high or low neutrophil infiltration. (B) Distributions of TAN-derived TGFβ production signature in HCC patients with non-MASH (GSE63898) and MASH (GSE164760) etiologies. (C) Representative immunofluorescence images and quantification of TGFβ-producing CD66b+neutrophils (CD66b: red; TGFβ: green; nucleus: blue) among all neutrophils in tumors resected from patients with HBV-related HCC or MASH-related HCC. Scale bars represent 50 and 20 μm. Each symbol represents one patient (C). Data is mean ± SEM. **P < 0.01 by Wilcoxon rank-sum test (B) and unpaired Student’s t test (C).
GM-CSF and linoleic-acid-induced c-Myc transcriptional activity drives SiglecFhi TAN differentiation
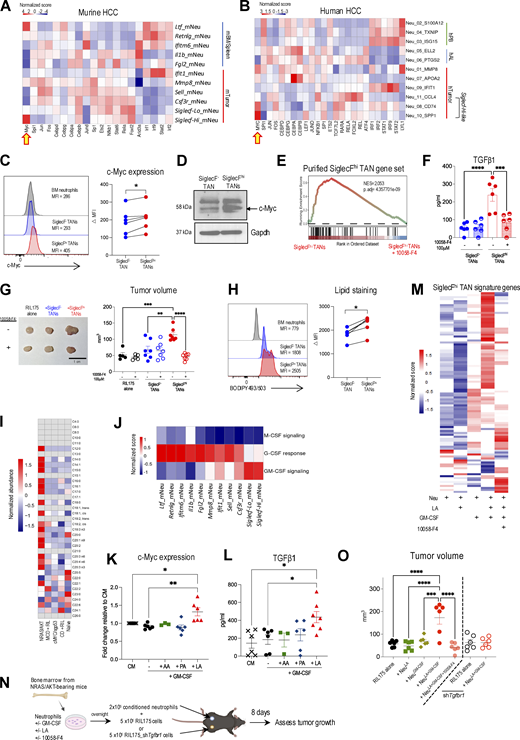
Next, we sought to identify the factors determining the emergence of SiglecFhi TANs. Tumor reprogramming is a key process enabling neutrophils to adapt to the environmental cues within the TME and consolidate their protumoral functions (Ng et al., 2024). To pinpoint the transcriptional program underscoring this dynamic in HCC tumors, we performed decoupleR analysis using the DoRothEA database (Badia-I-Mompel et al., 2022; Garcia-Alonso et al., 2019). We observed that Siglecf-Hi TANs exhibited a unique enrichment of the Myc regulon while downregulating the regulons that are associated with neutrophil commitment (Cebpa), maturation (Fos, Spi1), and activity (Jund) (Grieshaber-Bouyer et al., 2021; Gullotta et al., 2023) (Fig. 7 A). This finding was unexpected, as although c-Myc is essential for generating granulocyte/monocyte progenitors (GMPs), its repression is necessary for terminal neutrophil differentiation (Ai and Udalova, 2020), resulting in almost undetectable c-Myc levels in mature neutrophils (Poortinga et al., 2004). Similarly, the Myc regulon was also upregulated in Siglecf-Hi–like TANs present in HCC patients (Fig. 7 B), suggesting that c-Myc reactivation is a conserved phenomenon in both murine and human pro-tumor TANs. In line with the regulon analysis, we detected a significant increase in c-Myc protein levels in SiglecFhi TANs compared with SiglecF− TANs (Fig. 7, C and D). Crucially, c-Myc activation conferred the protumorigenic activity of SiglecFhi TANs as the c-Myc inhibitor 10058-F4 downregulated the expression of their signature genes (Fig. 7 E), reduced their TGFβ1 production (Fig. 7 F and Fig. S4 A), and dampened their ability to enhance tumor growth in vivo (Fig. 7 G).
GM-CSF and linoleic acid activate c-Myc regulon and drive SiglecFhiTAN development. (A and B) Relative differential transcription factor activity within each neutrophil subset in NRAS/AKT HCC mice (A) and HCC patients (B). (C) Representative histograms and c-Myc levels within SiglecF– and SiglecFhi TANs. (D) c-Myc expression assessed by western blot. Proteins were extracted from TANs subsets isolated from five mice. (E–G) SiglecF− and SiglecFhi TANs were treated with or without 10058-F4 (100 μM). GSEA analysis of genes defining SiglecFhi TANs (E). Spontaneous TGFβ production (F). RIL175 cells were s.c. injected with different TAN subsets. Tumor volume at day 8. Scale bar represents 1 cm (G). (H) Representative histograms and neutral lipid levels within SiglecF– and SiglecFhi TANs. (I) GC-MS analysis of HCC tumors and naïve liver. (J) Relative scores of M-CSF, G-CSF, and GM-CSF signaling within neutrophil subsets from NRAS/AKT HCC mice. (K and L) BM neutrophils from NRAS/AKT HCC mice were cultured overnight with or without the addition of GM-CSF (20 ng/ml) and the indicated fatty acids (50 μM). c-Myc expression (K) and TGFβ1 production (L). (M–O) BM neutrophils from NRAS/AKT HCC mice were cultured overnight with or without the addition of GM-CSF, linoleic acid, or 10058-F4. Heatmap of genes defining SiglecFhi TANs (M). Schematic diagram of adoptive transfer strategy (N). Tumor volume (O). Each symbol represents one mouse (C, H, K, L, and O) or TANs isolated from two to three mice (F and G). Data are mean ± SEM (F, G, K, L, and O) and are pooled from two (C, F–H, K, and L) or four (O) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Wilcoxon matched-pairs signed rank test (C and H) and one-way ANOVA (F, G, K, L, and O). MFI, median fluorescence intensity; CM, complete media. Source data are available for this figure: SourceData F7.
GM-CSF and linoleic acid activate c-Myc regulon and drive SiglecFhiTAN development. (A and B) Relative differential transcription factor activity within each neutrophil subset in NRAS/AKT HCC mice (A) and HCC patients (B). (C) Representative histograms and c-Myc levels within SiglecF– and SiglecFhi TANs. (D) c-Myc expression assessed by western blot. Proteins were extracted from TANs subsets isolated from five mice. (E–G) SiglecF− and SiglecFhi TANs were treated with or without 10058-F4 (100 μM). GSEA analysis of genes defining SiglecFhi TANs (E). Spontaneous TGFβ production (F). RIL175 cells were s.c. injected with different TAN subsets. Tumor volume at day 8. Scale bar represents 1 cm (G). (H) Representative histograms and neutral lipid levels within SiglecF– and SiglecFhi TANs. (I) GC-MS analysis of HCC tumors and naïve liver. (J) Relative scores of M-CSF, G-CSF, and GM-CSF signaling within neutrophil subsets from NRAS/AKT HCC mice. (K and L) BM neutrophils from NRAS/AKT HCC mice were cultured overnight with or without the addition of GM-CSF (20 ng/ml) and the indicated fatty acids (50 μM). c-Myc expression (K) and TGFβ1 production (L). (M–O) BM neutrophils from NRAS/AKT HCC mice were cultured overnight with or without the addition of GM-CSF, linoleic acid, or 10058-F4. Heatmap of genes defining SiglecFhi TANs (M). Schematic diagram of adoptive transfer strategy (N). Tumor volume (O). Each symbol represents one mouse (C, H, K, L, and O) or TANs isolated from two to three mice (F and G). Data are mean ± SEM (F, G, K, L, and O) and are pooled from two (C, F–H, K, and L) or four (O) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Wilcoxon matched-pairs signed rank test (C and H) and one-way ANOVA (F, G, K, L, and O). MFI, median fluorescence intensity; CM, complete media. Source data are available for this figure: SourceData F7.
Identification of factors contributing to the development of SiglecF hi TANs. (A) Quantities of SiglecF− and SiglecFhi TANs after overnight culture with or without 100 μM 10058-F4. (B) Representative immunofluorescence image and proximity of TAN subsets to lipid droplets (MPO: red; SiglecF: green; BODIPY 493/503: pink; nucleus: blue) in NRAS/AKT HCC tumors. Scale bar represents 10 µm. (C–E) BM neutrophils were isolated from NRAS/AKT HCC-bearing mice and cultured overnight with or without the addition of the indicated fatty acids (50 μM). Representative FACs plots of SiglecF+ neutrophils (C). Representative histograms of c-Myc expression (D). TGFβ1 production measured by ELISA following 4 h of LPS (100 ng/ml) stimulation (E). (F) Expression of Csf2 by cellular populations within NRAS/AKT HCC tumors. (G) NRAS/AKT HCC was induced in C57BL/6J or GM-CSF reporter Csf2-iCre-EGFP+/wt mice. Representative FACs plots of GM-CSF producing cells. (H) Representative FACs plots of SiglecF+ neutrophils after 20 ng/ml GM-CSF was included in C. (I and J) Neutrophils from spleen and tumor of NRAS/AKT mice were cultured overnight with or without the addition of GM-CSF (20 ng/ml) or/and linoleic acid (50 μM). c-Myc expression within splenic neutrophils (I) or SiglecF− and SiglecFhi TANs (J). (K) Pseudotime of TAN development in NRAS/AKT HCC mice, calculated with Monocle 3. (L) SiglecF− or SiglecFhi TANs isolated from CD45.1+ mice were adoptively transferred into NRAS/AKT HCC-bearing CD45.2+ recipients. After 2 days, TILs were harvested, donor TANs (CD45.1+, polygon gate) and endogenous TANs were assessed for their SiglecF expression (rectangle gate indicates SiglecFhi cells). (M and N) BM neutrophils were isolated from naïve mice and cultured with and without GM-CSF (20 ng/ml) and linoleic acid (50 μM) overnight. Representative histograms of c-Myc expression (M). TGFβ1 production measured by ELISA following 4 h of LPS (100 ng/ml) stimulation (N). Each symbol represents TANs isolated from two to three mice (A and J), or one mouse (E, I, and N), or one neutrophil from screening liver sections of eight mice (B). Data are mean ± SEM and are representative of two independent experiments (C–E, G, H, and L–N) or are pooled from two independent experiments (A, B, I, and J). *P < 0.05 by unpaired Student’s t test (B). CM, complete media.
Identification of factors contributing to the development of SiglecF hi TANs. (A) Quantities of SiglecF− and SiglecFhi TANs after overnight culture with or without 100 μM 10058-F4. (B) Representative immunofluorescence image and proximity of TAN subsets to lipid droplets (MPO: red; SiglecF: green; BODIPY 493/503: pink; nucleus: blue) in NRAS/AKT HCC tumors. Scale bar represents 10 µm. (C–E) BM neutrophils were isolated from NRAS/AKT HCC-bearing mice and cultured overnight with or without the addition of the indicated fatty acids (50 μM). Representative FACs plots of SiglecF+ neutrophils (C). Representative histograms of c-Myc expression (D). TGFβ1 production measured by ELISA following 4 h of LPS (100 ng/ml) stimulation (E). (F) Expression of Csf2 by cellular populations within NRAS/AKT HCC tumors. (G) NRAS/AKT HCC was induced in C57BL/6J or GM-CSF reporter Csf2-iCre-EGFP+/wt mice. Representative FACs plots of GM-CSF producing cells. (H) Representative FACs plots of SiglecF+ neutrophils after 20 ng/ml GM-CSF was included in C. (I and J) Neutrophils from spleen and tumor of NRAS/AKT mice were cultured overnight with or without the addition of GM-CSF (20 ng/ml) or/and linoleic acid (50 μM). c-Myc expression within splenic neutrophils (I) or SiglecF− and SiglecFhi TANs (J). (K) Pseudotime of TAN development in NRAS/AKT HCC mice, calculated with Monocle 3. (L) SiglecF− or SiglecFhi TANs isolated from CD45.1+ mice were adoptively transferred into NRAS/AKT HCC-bearing CD45.2+ recipients. After 2 days, TILs were harvested, donor TANs (CD45.1+, polygon gate) and endogenous TANs were assessed for their SiglecF expression (rectangle gate indicates SiglecFhi cells). (M and N) BM neutrophils were isolated from naïve mice and cultured with and without GM-CSF (20 ng/ml) and linoleic acid (50 μM) overnight. Representative histograms of c-Myc expression (M). TGFβ1 production measured by ELISA following 4 h of LPS (100 ng/ml) stimulation (N). Each symbol represents TANs isolated from two to three mice (A and J), or one mouse (E, I, and N), or one neutrophil from screening liver sections of eight mice (B). Data are mean ± SEM and are representative of two independent experiments (C–E, G, H, and L–N) or are pooled from two independent experiments (A, B, I, and J). *P < 0.05 by unpaired Student’s t test (B). CM, complete media.
Next, we sought to determine the tumor factors responsible for c-Myc activation in SiglecFhi TANs. Given that this TAN population only arose from MASH-related HCC models (Fig. 2), was closely positioned to lipid droplets (Fig. S4 B), and displayed a lipid-laden phenotype (Fig. 7 H), we speculated that c-Myc activation might be driven by lipids specifically accumulated within these tumors. To this end, we performed lipidomic profiling of tumors from the four HCC models used in this study (Fig. 1). As expected, several classes of fatty acids (FAs) were overexpressed in NRAS/AKT tumors (Fig. 7 I). Among these 19 FAs, we focused our analysis on: (1) palmitic acid (PA, C16:0) due to its abundant presence in MASH patients; (2) linolenic acid (LA, C18:2), which is elevated both in MASH patients and MCD+RIL175 tumors; and (3) arachidonic acid (AA, C20:4), owing to its similar enrichment in MCD+RIL175 tumors (Fig. 7 I) (Chiappini et al., 2017). BM neutrophils isolated from NRAS/AKT-injected mice were cultured with these FAs. However, no changes in neutrophil phenotype, c-Myc expression, and cytokine production were observed (Fig. S4, C–E), highlighting a requirement for additional tumor factors. For this, we delved deeper into the transcriptome of all TAN subsets and found that Siglecf-Hi TANs displayed high GM-CSF signaling signature (Fig. 7 J). GM-CSF is a neutrophil-activating cytokine that is abundant in the TME and was mainly produced by NK/NKT cells within NRAS/AKT tumors (Fig. S4, F and G). Our flow cytometry analysis revealed that GM-CSF induced SiglecF expression on BM neutrophils (Fig. S4 H). More importantly, in the presence of GM-CSF, BM neutrophils that were exposed to LA, but not PA or AA, upregulated c-Myc expression (Fig. 7 K) and increased the ability to produce TGFβ upon LPS stimulation (Fig. 7 L). RNA-seq analysis confirmed that GM-CSF and LA synergistically induced SiglecFhi TANs-associated gene signature in neutrophils through a c-Myc–mediated manner (Fig. 7 M). To further assess their functionality, we co-injected these in vitro–conditioned neutrophils with RIL175 cells into C57BL/6J mice (Fig. 7 N). As expected, GM-CSF and LA-conditioned neutrophils expedited tumor growth; however, this effect was abrogated when the Myc inhibitor 10058-F4 was added during in vitro conditioning, or when the TGFβ receptor was knocked-down in RIL175 cells (Fig. 7 O). Together, these data identified GM-CSF and LA as the key factors initiating the c-Myc–TGFβ axis in pro-tumorigenic TANs. It is noteworthy that while GM-CSF and LA induced c-Myc activation in neutrophils prior to their infiltration into tumors, they failed to do so in SiglecF− TANs (Fig. 7 K; and Fig. S4, I and J). Investigating this phenomenon, the pseudotime analysis predicted a branching in TAN development after entry into the tumor (Fig. S4 K). Furthermore, a phenotype switch between SiglecF− TANs and SiglecFhi TANs was not observed in an adoptive transfer assay (Fig. S4 L), suggesting that these two TAN subsets might originate from distinct differentiation pathways. Of note, GM-CSF and LA were unable to induce c-Myc expression and TGFβ production in BM neutrophils isolated from naïve mice (Fig. S4, M and N). Thus, a combination of GM-CSF, lipids, and additional tumor factors activates a c-Myc–dependent transcriptional program and facilitates neutrophil differentiation into protumor SiglecFhi TANs with TGFβ producing ability.
SiglecFhi TANs directly suppress HCC antigenicity via TGFβ
Surgical resection is the only curative treatment for HCC; however, it is often an unviable option as most HCC patients are diagnosed at later stages of the disease. In such cases of advanced HCC, primary treatment options typically include sorafenib, a multikinase inhibitor that suppresses tumor proliferation and angiogenesis, or anti-PD-1/PD-L1–based combination immunotherapy, which reinvigorates the body’s antitumor immune response (Gallage et al., 2021). Given the protumorigenic effects of SiglecFhi TANs, we asked whether their presence could impede the effectiveness of these therapeutic strategies. To investigate this, we reanalyzed the RNA-seq data from pretreatment tumors collected in the IMbrave150 and GO30140 clinical trials, which involved HCC patients treated with either sorafenib or combination immunotherapy with atezolizumab (anti-PD-L1) and bevacizumab (anti-VEGF) (Zhu et al., 2022). A total of 295 patients were included, with 247 receiving atezolizumab/bevacizumab and 48 treated with sorafenib, representing one of the most comprehensive transcriptional collections for prognostic analysis of HCC treatment. Interestingly, Siglecf-Hi–like TAN signature was associated with treatment failure among the immunotherapy-treated patients, but not the sorafenib-treated patients (Fig. 8 A). This led us to hypothesize that SiglecFhi TANs may foster a more aggressive HCC phenotype with heightened proliferative activity (Fig. 3), as well as an enhanced ability to evade immune detection. Indeed, SiglecFhi TANs directly suppressed MHCI expression and antigen presentation by RIL175 cells in both in vitro (Fig. 8 B) and in vivo (Fig. S5 A) settings. In line with this, MHCI expression on tumor cells was significantly upregulated following SiglecFhi TAN removal with anti-Ly6G treatment in the NRAS/AKT model (Fig. S5 B). Correspondingly, CD8+TIL effectors in these mice exhibited higher levels of genes related to TCR activation (e.g., Elf1, Zap70, and Ptpn6) compared with those in the control group (Fig. S5, C–E). In HCC patients, we also observed that the antigen processing and presentation signature in tumor cells was inversely correlated with the abundance of Siglecf-Hi–like TAN signature (Fig. 8 C) and was decreased in immunotherapy non-responders (Fig. 8 D). Thus, these results suggest that SiglecFhi TANs reduce the antigenicity of tumor cells, rendering them invisible and impervious to attack by cytotoxic T cells. To demonstrate this, we introduced ovalbumin (OVA) as a surrogate tumor antigen in the NRAS/AKT HCC model. We then evaluated tumor antigenicity by examining their surface expression of OVA/MHCI complexes and the antigen recognition by OT-1 cells (Fig. S5 F). SiglecFhi TAN removal significantly enhanced the direct presentation of OVA by tumor cells (Fig. S5 G), without affecting the cross-presentation of OVA by intratumoral dendritic cells (DCs) (Fig. S5 H). Furthermore, adoptively transferred OT-1 cells were more proliferative in mice without SiglecFhi TANs (Fig. S5 I), suggesting improved T cell recognition of tumor cells in these animals compared with their isotype-treated counterparts. To further verify that this increase in OT-1 proliferation was solely due to the difference in antigen load on tumor cells, rather than a distinct TME following SiglecFhi TAN removal, we cultured tumor cells isolated from OVA-NRAS/AKT injected animals with OT-1 cells. Once again, we found that tumor cells from mice without SiglecFhi TANs induced greater OT-1 proliferation in vitro (Fig. S5 J). Next, we set out to demonstrate that SiglecFhi TANs utilized TGFβ to modulate tumor antigenicity. We observed that blocking TGFβ signaling negated the inhibitory effects of SiglecFhi TANs on tumor MHCI expression and antigen presentation (Fig. 8 B and Fig. S5 A). Importantly, tumor cells isolated from Mrp8Cre.TGFβfl/fl mice displayed increased surface expression of OVA/MHCI complexes and were more effective in inducing OT-1 proliferation (Fig. 8, E and F). Collectively, these findings support the notion that SiglecFhi TANs are crucial in driving immunotherapy resistance as they enable immune evasion by limiting the antigenicity of tumor cells via TGFβ.
SiglecF hi TANs downregulate the antigenicity of tumor cells via TGFβ, and their removal sensitizes HCC mice to anti-PD-1 mAb treatment. (A) Distributions of Siglecf-Hi–like TAN signature score in HCC patients from the IMbrave150 and GO30140 clinical trials. (B) MHCI expression by IFNγ-treated RIL175 cells co-cultured with SiglecF− or SiglecFhi TANs, with or without Vactosertib (Vac.). (C and D) Antigen processing and presentation signature score was calculated from an imputed tumor-specific gene expression profile (tumor antigen presentation signature) in patients treated with Atezolizumab and Bevacizumab from the IMbrave150 and GO30140 clinical trials. Correlation between tumor antigen presentation signature and Siglecf-Hi–like TAN signature scores (C). Distributions of tumor antigen presentation signature score in responder (R) and non-responders (NR) (D). (E and F) OVA-expressing NRAS/AKT HCC was induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T). Representative histograms and OVA/MHCI presentation by tumor cells (E). Tumor cells were co-cultured with OT-1 cells. Representative histograms and proliferation index of OT-1 cells (F). (G and H) NRAS/AKT HCC Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T) were treated with αPD-1 or isotype control mAbs for 2 wk. Representative images of HCC livers and liver-to-body weight ratio. The scale bar represents 1 cm (G). Representative images of AFP+ tumors (brown) and tumor nodule counts. Scale bar represents 100 μm (H). (I) NRAS/AKT HCC mice were treated with isotype control mAbs or αLy6G mAbs or αPD-1 mAbs or both at the indicated time. Survival curve (n = 10/group). (J–Q) Mice were sacrificed 2 wk following treatment. Representative images of HCC livers and liver-to-body weight ratio. Scale bar represents 1 cm (J). Representative images of AFP+ tumors (brown) and tumor nodule counts. Scale bar represents 100 μm (K). MHCI expression on tumor cells (L). Number of CD8+TILs (M). Proportion of PD-1+cells among CD8+TILs (N). Proportion of terminally exhausted cells (PD-1hiCTLA4+Tox+Tcf1−) in CD8+TILs (O). Frequency of Ki-67+ CD8+PD-1+TILs (P). Percentage of CD8+PD-1+TILs co-producing IFNγ and TNFα after in vitro anti-CD3/CD28 restimulation (Q). (R–T) CD8+T cell depletion abrogated the beneficial effects of αLy6G mAb treatment in anti-PD-1 mAb administrated mice. Survival curve (n = 7–10/group) (R). Mice were sacrificed 2 wk following treatments. Liver-to-body weight ratio (S). Tumor nodule counts (T). Each symbol represents TANs isolated from two to three mice (B), one mouse (E, G, H, J–Q, S, and T), or the mean of tumor cells isolated from three mice (F). Data are mean ± SEM and are pooled from three independent experiments (B), or represent two to three independent experiments (E–T). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Wilcoxon rank-sum test (A and D), one-way ANOVA (B, G, H, J–Q, S, and T), unpaired Student’s t test (E), two-way ANOVA (F), and Kaplan–Meier analysis (I and R).
SiglecF hi TANs downregulate the antigenicity of tumor cells via TGFβ, and their removal sensitizes HCC mice to anti-PD-1 mAb treatment. (A) Distributions of Siglecf-Hi–like TAN signature score in HCC patients from the IMbrave150 and GO30140 clinical trials. (B) MHCI expression by IFNγ-treated RIL175 cells co-cultured with SiglecF− or SiglecFhi TANs, with or without Vactosertib (Vac.). (C and D) Antigen processing and presentation signature score was calculated from an imputed tumor-specific gene expression profile (tumor antigen presentation signature) in patients treated with Atezolizumab and Bevacizumab from the IMbrave150 and GO30140 clinical trials. Correlation between tumor antigen presentation signature and Siglecf-Hi–like TAN signature scores (C). Distributions of tumor antigen presentation signature score in responder (R) and non-responders (NR) (D). (E and F) OVA-expressing NRAS/AKT HCC was induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T). Representative histograms and OVA/MHCI presentation by tumor cells (E). Tumor cells were co-cultured with OT-1 cells. Representative histograms and proliferation index of OT-1 cells (F). (G and H) NRAS/AKT HCC Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T) were treated with αPD-1 or isotype control mAbs for 2 wk. Representative images of HCC livers and liver-to-body weight ratio. The scale bar represents 1 cm (G). Representative images of AFP+ tumors (brown) and tumor nodule counts. Scale bar represents 100 μm (H). (I) NRAS/AKT HCC mice were treated with isotype control mAbs or αLy6G mAbs or αPD-1 mAbs or both at the indicated time. Survival curve (n = 10/group). (J–Q) Mice were sacrificed 2 wk following treatment. Representative images of HCC livers and liver-to-body weight ratio. Scale bar represents 1 cm (J). Representative images of AFP+ tumors (brown) and tumor nodule counts. Scale bar represents 100 μm (K). MHCI expression on tumor cells (L). Number of CD8+TILs (M). Proportion of PD-1+cells among CD8+TILs (N). Proportion of terminally exhausted cells (PD-1hiCTLA4+Tox+Tcf1−) in CD8+TILs (O). Frequency of Ki-67+ CD8+PD-1+TILs (P). Percentage of CD8+PD-1+TILs co-producing IFNγ and TNFα after in vitro anti-CD3/CD28 restimulation (Q). (R–T) CD8+T cell depletion abrogated the beneficial effects of αLy6G mAb treatment in anti-PD-1 mAb administrated mice. Survival curve (n = 7–10/group) (R). Mice were sacrificed 2 wk following treatments. Liver-to-body weight ratio (S). Tumor nodule counts (T). Each symbol represents TANs isolated from two to three mice (B), one mouse (E, G, H, J–Q, S, and T), or the mean of tumor cells isolated from three mice (F). Data are mean ± SEM and are pooled from three independent experiments (B), or represent two to three independent experiments (E–T). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Wilcoxon rank-sum test (A and D), one-way ANOVA (B, G, H, J–Q, S, and T), unpaired Student’s t test (E), two-way ANOVA (F), and Kaplan–Meier analysis (I and R).
αLy6G mAb treatments downregulate the antigenicity of tumor cells and activates CD8 + TILs in HCC-bearing C57BL/6J mice, while HCC-bearing Mrp8 Cre .TGFβ fl/fl mice respond to αPD-1 mAb treatment. (A) SiglecF− and SiglecFhi TANs were adoptively transferred alongside OVA-expressing RIL175 (left) or RIL175_shTgfbr1 cells (right). OVA/MHCI presentation by tumor cells. (B) Representative histograms and MHCI expression on NRAS/AKT tumor cells following αLy6G or isotype control mAbs treatments. (C) UMAP of tumor infiltrating CD8+T cells (CD8+TILs) in NRAS/AKT-injected C57BL/6J mice treated with αLy6G or Isotype mAbs. (D) Distribution of marker genes in different CD8+TIL subsets. (E) Expression of T cell receptor signaling related genes within the Teff_CD8 subset. (F–J) OVA-expressing NRAS/AKT HCC tumors were used to assess tumor antigen presentation and T cell recognition following αLy6G or isotype control mAbs treatments. Schematic outline of the experiment (F). Representative histograms and OVA/MHCI presentation by tumor cells (G). OVA/MHCI presentation by tumor-infiltrating CD11c+MHCII+ DCs (H). Proliferation of OT-1 cells 3 days after adoptive transfer into OVA-NRAS/AKT HCC mice (I). Tumor cells from OVA-NRAS/AKT HCC mice were co-cultured with OT-1 cells at the indicated ratios for 3 days. Representative histograms and proliferation index of OT-1 cells (J). (K–P) NRAS/AKT HCC was induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T), followed by treatment with αPD-1 or isotype control mAbs. MHCI expression on tumor cells (K). Number of TCRβ+CD8+TILs per gram of HCC tissue (L). Proportion of PD-1+ cells among TCRβ+CD8+TILs (M). Frequency of proliferating TCRβ+CD8+PD-1+TILs determined by Ki-67 staining (N). Proportion of terminally exhausted cells (PD-1hiCTLA4+Tox+Tcf1−) among TCRβ+CD8+TILs (O). Percentage of TCRβ+CD8+PD-1+TILs co-producing IFNγ and TNFα after in vitro anti-CD3/CD28 restimulation (P). Each symbol represents one mouse (A, B, G–I, and K–P), or the mean of tumor cells isolated from three mice (J). Data are mean ± SEM and represent two to three independent experiments (A, B, and G–P). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired Student’s t test (A, B, and G–I), two-way ANOVA (J), and one-way ANOVA (K–P).
αLy6G mAb treatments downregulate the antigenicity of tumor cells and activates CD8 + TILs in HCC-bearing C57BL/6J mice, while HCC-bearing Mrp8 Cre .TGFβ fl/fl mice respond to αPD-1 mAb treatment. (A) SiglecF− and SiglecFhi TANs were adoptively transferred alongside OVA-expressing RIL175 (left) or RIL175_shTgfbr1 cells (right). OVA/MHCI presentation by tumor cells. (B) Representative histograms and MHCI expression on NRAS/AKT tumor cells following αLy6G or isotype control mAbs treatments. (C) UMAP of tumor infiltrating CD8+T cells (CD8+TILs) in NRAS/AKT-injected C57BL/6J mice treated with αLy6G or Isotype mAbs. (D) Distribution of marker genes in different CD8+TIL subsets. (E) Expression of T cell receptor signaling related genes within the Teff_CD8 subset. (F–J) OVA-expressing NRAS/AKT HCC tumors were used to assess tumor antigen presentation and T cell recognition following αLy6G or isotype control mAbs treatments. Schematic outline of the experiment (F). Representative histograms and OVA/MHCI presentation by tumor cells (G). OVA/MHCI presentation by tumor-infiltrating CD11c+MHCII+ DCs (H). Proliferation of OT-1 cells 3 days after adoptive transfer into OVA-NRAS/AKT HCC mice (I). Tumor cells from OVA-NRAS/AKT HCC mice were co-cultured with OT-1 cells at the indicated ratios for 3 days. Representative histograms and proliferation index of OT-1 cells (J). (K–P) NRAS/AKT HCC was induced in Mrp8Cre+ve.TGFβfl/fl mice (Mrp8Cre.TGFβfl/fl) and Mrp8Cre−ve.TGFβfl/fl littermates (W/T), followed by treatment with αPD-1 or isotype control mAbs. MHCI expression on tumor cells (K). Number of TCRβ+CD8+TILs per gram of HCC tissue (L). Proportion of PD-1+ cells among TCRβ+CD8+TILs (M). Frequency of proliferating TCRβ+CD8+PD-1+TILs determined by Ki-67 staining (N). Proportion of terminally exhausted cells (PD-1hiCTLA4+Tox+Tcf1−) among TCRβ+CD8+TILs (O). Percentage of TCRβ+CD8+PD-1+TILs co-producing IFNγ and TNFα after in vitro anti-CD3/CD28 restimulation (P). Each symbol represents one mouse (A, B, G–I, and K–P), or the mean of tumor cells isolated from three mice (J). Data are mean ± SEM and represent two to three independent experiments (A, B, and G–P). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by unpaired Student’s t test (A, B, and G–I), two-way ANOVA (J), and one-way ANOVA (K–P).
SiglecFhi TAN removal sensitizes immunotherapy-resistant HCC to anti-PD-1 mAb treatment
Finally, we asked whether the absence of SiglecFhi TANs could restore the efficacy of anti-PD-1 mAb treatment. In line with clinical observations that MASH-related HCC patients are notoriously resistant to immunotherapy (Haber et al., 2021; Pfister et al., 2021), NRAS/AKT-injected WT mice were unresponsive to anti-PD-1 mAb treatment. However, this treatment deficit was reversed in Mrp8Cre.TGFβfl/fl mice (Fig. 8, G and H; and Fig. S5, K–P), highlighting the key role of functional TANs in driving anti-PD-1 mAb therapy resistance. Similarly, SiglecFhi TAN removal with anti-Ly6G mAb also sensitized NRAS/AKT-injected WT mice to anti-PD-1 mAb therapy, resulting in enhanced tumor control and a more significant survival benefit compared with using anti-Ly6G mAb alone (Fig. 8, I–K). This enhanced anti-tumor effect was driven by the increased immune sensitivity of tumor cells, which expressed higher levels of MHCI upon receiving the anti-Ly6G/anti-PD-1 mAb combination therapy (Fig. 8 L). Correspondingly, the number of CD8+TILs increased in mice treated under this condition (Fig. 8 M). Interestingly, these CD8+TILs contained a high percentage of PD-1+ cells (Fig. 8 N); however, the proportion of exhausted cells remained the same (Fig. 8 O). This suggests that the increased presence of PD-1+cells signified elevated TCR stimulation rather than exhaustion. In support of this, CD8+PD-1+cells in mice treated with the anti-Ly6G/anti-PD-1 mAb combination therapy were more proliferative (Fig. 8 P) and secreted higher levels of effector cytokines (Fig. 8 Q). Hence, the synergy between anti-Ly6G mAb and anti-PD-1 mAb injections was predictably dependent on CD8+T cells, as their depletion with anti-CD8 mAb eliminated the treatment benefits (Fig. 8, R–T). Altogether, these results indicate that SiglecFhi TAN removal improves tumor cell recognition by rejuvenating CD8+TILs after anti-PD-1 treatment, thus enhancing the effectiveness of tumor immunotherapy.
Discussion
Several studies have explored the pathogenic role of TANs in HCC (Geh et al., 2022; He et al., 2015; Leslie et al., 2022; Li et al., 2015; van der Windt et al., 2018; Xue et al., 2022; Zhou et al., 2019), but few have examined the influence of etiology in this context, despite each HCC subtype having its own distinct pathogenesis. Our comprehensive analysis of 924 HCC patients from four public datasets revealed that the impact of TANs on disease progression is not uniform across different HCC subtypes. Notably, TANs had a more detrimental effect on MASH-related HCC than on viral-related HCC. This variation was attributed to differences in the TAN subsets emerging from these distinct TMEs. Specifically, we identified a unique subset of pro-tumor TANs that was preferentially enriched in MASH-related HCC tumors. These TANs were characterized by an elevated SiglecF expression, a c-Myc–driven transcriptional signature, and a remarkable capacity to produce TGFβ. A lipid-rich TME was crucial for the development of SiglecFhi TANs, endowing them with multiple tumor-promoting functions, such as stimulating tumor cell proliferation and promoting EMT. Beyond these well-known TAN functions, we uncovered that SiglecFhi TANs could further promote HCC aggressiveness by suppressing tumor antigen presentation via the release of TGFβ. This new insight expands our knowledge of the multifaceted roles that TANs play in tumorigenesis and holds significant implications for the advancement of cancer immunotherapy.
ICB therapy serves as the first-line treatment for advanced HCC patients; however, its efficacy is considerably limited in MASH-related HCC patients compared to those with other etiologies (Haber et al., 2021; Pfister et al., 2021). Our findings suggest that the preferential enrichment of SiglecFhi TANs in MASH-related HCC and their impact on tumor immunogenicity may account for these clinical observations. Successful ICB responses require rejuvenated CD8+T cells to recognize their targets through the presentation of tumor antigens via MHCI on tumor cells. In our study of ICB-treated HCC patients, we observed that tumor cells from non-responders displayed a lower antigen presentation signature than those from responders. Likewise, the inactivation of antigen presentation by tumor cells has been identified as a cause of ICB therapy failure in many other tumors (Jhunjhunwala et al., 2021; Montesion et al., 2021; Sade-Feldman et al., 2017). In these cases, point mutations, deletions, or loss of heterozygosity (LOH) in beta-2-microglobulin (B2M) and HLA-I are the primary contributors to antigen presentation defects. However, the prevalence of genetic alterations in B2M and HLA-I is relatively low in HCC patients (e.g., the proportion of patients with at least one incidence of HLA LOH in HCC, melanoma, and cervical cancer is 4%, 14%, and 38% respectively) (Montesion et al., 2021; Pyke et al., 2022), indicating that HCC may employ non-genetic strategies to modulate its immunogenicity. In support of this notion, we demonstrated that a high abundance of SiglecFhi TANs reduced antigen presentation by downregulating MHCI in tumor cells, thereby helping them to evade T cell recognition. Notably, SiglecFhi TANs did not completely abolish MHCI expression on tumor cells, which could otherwise result in natural killer cell-mediated tumor killing. More importantly, TAN depletion sensitized an otherwise ICB-resistant MASH-related murine HCC model to the therapy. Collectively, these findings establish SiglecFhi TAN–mediated MHCI downregulation as a key immune evasion event that confers ICB therapy resistance in MASH-related HCC and highlights the need to target this subset of TANs for immunotherapy efficacy improvements.
A key challenge that remains is the targeted depletion of SiglecFhi TANs. In line with the prevailing notion that TANs exhibit high diversity (Ng et al., 2019), our study identified six distinct TAN subsets with unique transcriptomic signatures in MASH-related HCC tumors. Leveraging the concordance of SiglecF expression at both the transcript and protein levels, we utilized this surface marker to efficiently identify and purify SiglecFhi TANs for ex vivo and adoptive transfer functional analysis. However, using an anti-SiglecF mAb for SiglecFhi TAN depletion is not ideal, as it preferentially targets eosinophils, which express high levels of SiglecF and have been shown to play a role in tumor control (Blomberg et al., 2023). Fortunately, in our study, we achieved the removal of SiglecFhi TANs through injections of anti-Ly6G mAbs (clone 1A8), possibly by disrupting TAN reprogramming within the TME. Given the accelerated turnover rate of neutrophils after anti-Ly6G mAb injections, it is widely recognized that this approach cannot completely deplete all neutrophils in the system. Nevertheless, a comprehensive study by Boivin et al. demonstrated that the remaining neutrophils are freshly derived from the BM rather than being depletion escapees (Boivin et al., 2020). Thus, the initial round of anti-Ly6G mAb treatment could effectively eliminate pre-existing SiglecFhi TANs, which are mature and aged neutrophils, in the TME, while subsequent daily injections prevented further SiglecFhi TAN formation by cutting short the in situ reprogramming of newly arrived neutrophils. This repeated interruption of TAN reprogramming resulted in the absence of SiglecFhi TANs, thus offering us the experimental opportunity to study the in vivo effects of their absence. Consistent with the ex vivo functional studies, we found that SiglecFhi TANs promoted tumor aggressiveness and dampened tumor immunogenicity in vivo. However, it is important to recognize that the depletion caused by anti-Ly6G mAb treatment is not exclusive to SiglecFhi TANs, as other TAN subsets are also affected. Hence, additional research is needed to fine-tune the selective targeting and elimination of this pro-tumorigenic population without disrupting other potentially antitumor neutrophils. Our analysis with Mrp8Cre. TGFβfl/fl mice supported the notion that TANs primarily exert their protumor effects through the release of TGFβ. Furthermore, our examination of surgically resected tumors revealed a significant enrichment of TGFβ-producing TANs in patients with MASH-related HCC. Hence, a potential strategy to reverse ICB resistance in this group of patients could involve targeting TGFβ. In fact, several clinical trials are currently underway to test the synergistic effects of ICB and TGFβ inhibitors, with promising results being reported (Derynck et al., 2021). However, due to the crucial roles of TGFβ signaling in various physiological processes (e.g., immune tolerance maintenance), systemic TGFβ suppression could lead to severe immune-related adverse events in cancer patients. Therefore, targeting TGFβ producers within the TME instead remains a superior approach.
Neutrophil plasticity is increasingly appreciated as a crucial aspect of their biology. Particularly, in cancer, tumor-driven signals initiate a rapid reprogramming process in infiltrating neutrophils, facilitating the diversification of TANs in terms of their phenotypic and functional properties (Ng et al., 2019). However, the specifics of these signals remain largely unidentified. Here, the enrichment of SiglecFhi TANs in MASH-related HCC allowed us to pinpoint their key drivers in the TME. We demonstrated that a combination of GM-CSF and LA, both elevated in the TME of MASH-related HCC (Chiappini et al., 2017; Kapanadze et al., 2013; Lewinska et al., 2021; Zaitsev et al., 2022), promoted SiglecFhi TAN development. Although the precise developmental pathway of SiglecFhi TANs remains to be elucidated, our data suggest that this process is governed by a transcriptional switch from a C/EBPα regulon to a c-Myc regulon. C/EBPα and c-Myc act as counter-regulatory factors during granulopoiesis, with c-Myc suppression in GMPs being the key event in initiating C/EBPα-mediated neutrophil commitment (Ai and Udalova, 2020; Johansen et al., 2001). More importantly, these transcriptional changes were also observed in protumor TANs identified in HCC patients, indicating that this is a conserved event during TAN reprogramming in both humans and mice. Similarly, c-Myc reactivation has also been reported in tumor-associated macrophage reprogramming (Jiang et al., 2021; Pello et al., 2012), highlighting the crucial role of c-Myc in shaping the protumorigenic immune environment. While the functions of c-Myc in granulopoiesis are well-defined, further research is needed to understand how reactivated c-Myc collaborates with other transcription factors in mature neutrophils to promote the development of SiglecFhi TANs and their ability to produce TGFβ. Nonetheless, this transcription factor shift could potentially offer additional clues for targeting SiglecFhi TANs. It is worth noting that a recent trial of MTL-CEBPA, a small activating RNA that upregulates C/EBPα, showed evidence of positive response in HCC patients with a satisfactory safety profile (Sarker et al., 2020). Although TANs are not the primary focus of this trial, it would be interesting to investigate whether this approach could counteract SiglecFhi TAN development in the TME.
In summary, our study has outlined a novel mechanism by which TANs promote immune evasion and tumorigenicity and has posited SiglecFhi TANs as a key immunotherapy target in the fight against HCC.
Materials and methods
Patient samples
Tumor samples from treatment-naïve patients with HCC who underwent primary curative resection were collected at Peking University First Hospital and Peking University People’s Hospital. This study was approved by the Research Ethics Committee of both Peking University First Hospital and Peking University People’s Hospital. Written informed consent was obtained from each patient, and their clinical information is provided in Table S1. The diagnosis of MASH-related HCC was established by two independent pathologists.
Mice
Wild type C57BL/6J mice (RRID: IMSR_JAX:000664) were purchased from the Centre for Comparative Medicine Research, Li Ka Shing Faculty of Medicine, the University of Hong Kong. OT-1 mice (C57BL/6-Tg (TcraTcrb) 1100Mjb/J, RRID: IMSR_JAX:003831), CD45.1+ mice (B6.SJL-Ptprca Pepcb/BoyJ, RRID: IMSR_JAX:002014), Mrp8Cre mice (B6.Cg-T (S100A8-cre,-EGFP)1Ilw/J, RRID: IMSR_JAX:021614), and TGFβfl/fl mice (C57BL/6J-Tgfb1em2Lutzy/Mmjax, RRID: MMRRC_065809-JAX) were purchased from Jackson Laboratory. Csf2-iCre-EGFP+/wt mice were kindly provided by B. Becher (The University of Zürich, Zürich, Switzerland). Both male and female mice were used and were 6–8 wk old at the start of experiments. All experimental procedures were performed under the guidelines and regulations of the Committee on the Use of Live Animals in Teaching and Research, the University of Hong Kong.
Cell lines and mouse tumor models
The RIL175 cell line (RRID:CVCL_B7TK) was grown in complete RPMI (RPMI 1640 medium supplemented with 10% FBS and 1% penicillin-streptomycin). RIL175_shTgfbr1 cells were generated through lentiviral transfection of pLV3-U6 plasmid containing shTgfbr1 (5′-GGACCATTGTGTTACAAGAAA-3′). For RIL175 orthotopic HCC models, mice were fed a regular CD or MCD diet (Research Diets) starting at 8 wk old. Mice were kept on diet till their endpoint. Orthotopic RIL175 tumors were established after 2 wk of feeding by injecting 5 × 104 RIL175 cells in 20 μl of a 50:50 solution of PBS and Matrigel (Corning) into the left liver lobe following an established protocol (Brown et al., 2018). Tumor volume was measured 4 wk after tumor induction and was calculated using the following formula: 1/2 (length*width2). For subcutaneous RIL175 tumors, 5 × 105 RIL175 or RIL175_shTgfbr1 cells were co-injected with 2 × 105 neutrophils in 100 μl of a 50:50 solution of PBS and Matrigel (Corning) into either flank of C57BL/6J mice. Tumor volume was measured 8 days after tumor induction using the formula above. For HTVI HCC models, plasmids overexpressing oncogenes or knocking out cancer suppressor genes were delivered into hepatocytes via HTVI (Chen et al., 2022; Molina-Sánchez et al., 2020; Zhou et al., 2022). For NRAS/AKT- and OVA-NRAS/AKT-induced HCC, 20 μg each of plasmids encoding human neuroblastoma Ras viral oncogene homolog (N-Ras G12V) and human AKT1 (myr-AKT1) or myr-AKT1-OVA were used. For cMYC/sgp53-induced HCC, 10 μg each of plasmid encoding human MYC and CRISPR/Cas9 vector targeting Trp53 were used. For both models, the plasmids were diluted in 0.9% NaCl with a 25:1 ratio of sleeping beauty transposase into a volume equivalent to 10% of the mouse’s body weight. The mixture was then filtered through a 0.22-mm filter before being injected at high pressure into the mouse’s tail vein. Tumor tissues were harvested and analyzed 4 wk after HTVI for NRAS/AKT-induced HCC or 7 wk after HTVI for cMYC/sgTP53-induced HCC. For neutrophil depletion experiments, 2 wk after HCC induction, anti-Ly6G mAb (1A8; BioXCell), or rat IgG2a isotype mAb (BioXCell) was injected intraperitoneally (i.p.). 25 μg of antibody was given daily in the first week of treatment, while 50 μg was used subsequently until the endpoint. 50 μg of anti-rat κ immunoglobulin light chain mAb was also injected i.p. on alternate days starting from the first day of depletion following an established protocol (Boivin et al., 2020). For PD-1 blockade experiments, 200 μg of anti-PD-1 mAb (BioXCell) or rat IgG2a isotype Ab (BioXCell) were injected i.p. twice a week, starting 2.5 wk after HCC induction. For treatments combining neutrophil depletion and PD-1 blockade, 50 μg of anti-Ly6G mAb or rat IgG2a isotype Ab were injected i.p. daily, whilst 200 μg of anti-PD-1 mAb were injected i.p. twice a week. For the survival experiment, each mouse was euthanized upon reaching the humane endpoint, as per our animal ethics approval. The time from tumor induction to this endpoint was recorded and used to generate the survival curve.
Tumor dissociation and cell isolation
2 g of tumor tissue were extracted from cancerous livers, cut into small pieces, and dissociated into cell suspensions using gentleMACS Dissociator (Miltenyi Biotec) in dissociation buffer containing 0.5 mg/ml of Collagenase IV (Gibco) and 100 μg/ml of DNase I (Goldbio) at 37°C. Single-cell suspensions were collected after filtering through a 70-μm cell strainer and were used directly for experiments involving tumor cells. Immune cells were enriched via a density-gradient centrifugation approach using Percoll PLUS (Cytiva) and resuspended in complete RPMI. For experiments involving purified TANs, further sorting of immune cells was performed using BD Influx (BD Biosciences), based on the lineage markers (CD11b+ Ly6G+) and subpopulation marker (SiglecF).
Neutrophil assays
Bone marrow neutrophils, SiglecF−, and SiglecFhi TANs were sort-purified from NRAS/AKT HCC mice and used in various functional assays at 1 × 105 cells/well. To assess their phagocytosis proficiency, the neutrophils were incubated with 0.1 mg/ml pHrodo Red E. coli BioParticles (Invitrogen) for 2 h and the uptake of E. coli was assessed through FACs. Their ROS production was evaluated through incubation with 5 μM CellROX Deep Red (Invitrogen) and 1 nM fMLP (Sigma-Aldrich) for 10 min followed by FACs analysis. NETosis was determined after 4 h of PMA stimulation (1 μg/ml) alongside 1 μM Helix NP Green (Biolegend). For assessment of TAN plasticity, SiglecF− and SiglecFhi TANs were sort-purified from CD45.1+ mice bearing NRAS/AKT tumors. 2 × 106 of CD45.1+ TANs were then intravenously injected into NRAS/AKT HCC-bearing CD45.2+ mice. 2 days after, tumor infiltrating leukocytes from the CD45.2+ hosts were assessed through FACs analysis.
Side population assay
Tumor cells from NRAS/AKT tumors were incubated with 5 μg/ml Hoechst 33342 (Invitrogen) for 90 min at 37°C in the absence or presence of 200 μM verapamil (Sigma-Aldrich). Through FACs analysis of live, CD45−TER-119−cells, the side population compartment was identified as the cell fraction abolished by verapamil.
In vitro and ex vivo limiting dilution assay (LDA)
For the in vitro LDA, RIL175 cells were co-cultured with purified TANs overnight, with or without the addition of 10 nM Vactosertib (MedChemExpress). Cells were then recovered and seeded at limiting dilutions (1–50 cells/well) into polyHEMA-coated plates with 100 μl of complete RPMI supplemented with 0.25% of methylcellulose (Sigma-Aldrich), B27 (1:50; Gibco), 4 μg/ml of insulin (Sigma-Aldrich), 60 ng/ml of murine epidermal growth factor (Sigma-Aldrich), 200 ng/ml of human recombinant basic fibroblast growth factor (Sigma-Aldrich). For the ex vivo LDA, tumor cells were dissociated from HCC livers as described above and were directly sorted (1,000–6,000 cells/well) with BD Influx into a polyHEMA-coated plate containing 100 μl of organoid culture media (Zhou et al., 2022). Dead cells, immune cells (CD45+), and red blood cells (TER-119+) were eliminated during the sorting process. 20 μl of culture medium was added every other day and wells with tumorsphere were counted on day 4 for the in vitro LDA or day 8 for the ex vivo LDA. The estimated stem cell frequency was calculated using ELDA software (Zhou et al., 2022).
Clonogenic assay
RIL175 cells were co-cultured with purified TANs overnight before re-seeding into a 6-well plate at a density of 500 cells/well in complete RPMI. After 5 days, colonies were visualized by fixing with methanol and staining with 0.1% of crystal violet. The plates were scanned and all colonies >0.1 mm2 were counted with Fiji.
Scratch wound assay
RIL175 cells were grown into monolayers in a 24-well plate and wounded by scraping using sterile 10 μl pipette tips. PBS was used to wash off floating cells, while those remaining were cultured in serum-reduced (1% FBS) RPMI, with the addition of purified TANs. The wound gaps were photographed at 0 and 17 h after wounding. The change in wound area was quantified using Fiji and a quantification plugin.
TGFβ1 measurement
Supernatant samples were treated with 1 N HCl for 10 min, followed by neutralization with 1.2 N NaOH/0.5 M HEPES. TGFβ1 levels in the treated samples were then measured using Simplex ProcartaPlex Kit (Invitrogen) or Mouse TGF-beta1 ELISA Kit (Proteintech) following the manufacturer’s instructions.
BM neutrophil culture
BM cells were harvested from the femurs and tibias from HCC-bearing mice. Red blood cells were removed using a Red Blood Cell Lysing Buffer (Sigma-Aldrich) and neutrophils were purified using a mouse anti-Ly-6G microbeads ultrapure kit (Miltenyi Biotec) following the manufacturer’s instructions. 1 × 105/well neutrophils were cultured in 96-well plates in complete RPMI with or without the addition of 20 ng/ml of GM-CSF (Peprotech); 50 μM of AA (Cayman); 50 μM of PA (Cayman); and/or 50 μM of LA (Cayman). Cells were collected for FACs or RNA-seq analysis or stimulated with 100 ng/ml of LPS (InvivoGen) for 4 h. Supernatants were collected for TGFβ measurement.
In vivo and ex vivo antigen presentation assays
For adoptive transfer experiment, OT-1 cells were purified from splenocytes of naïve OT-1 mice using a CD8a+T cell Isolation Kit, mouse (Miltenyi Biotech) and were labeled with eBioscience Cell Proliferation Dye eFluor 670 (Invitrogen). 5 × 106 of OT-1 cells were then injected intravenously into OVA-NRAS/AKT HCC-bearing mice 3.5 wk after tumor induction. The proliferation of OT-1 was analyzed by FACs 3 days after the adoptive transfer. For ex vivo OVA presentation assay, purified OT-1 cells were labeled with a CellTrace CFSE Cell Proliferation Kit (Invitrogen). Tumor cells from OVA-NRAS/AKT HCC livers were sort-purified as described above and were plated onto 96-well flat bottom plates at the listed densities. 100,000 CFSE-labeled OT-1 cells were then added to each well and cultured in T cell medium (RPMI 1640 medium supplemented with 10% FBS, 50 μM of β-mercaptoethanol, 1 M of HEPES, 1X non-essential amino acids, 100 mM of sodium pyruvate, and 1% penicillin-streptomycin) for 72 h before FACs analysis.
Immunohistochemistry and immunofluorescence
For histology and AFP immunostaining, murine tumor tissues were fixed in 4% paraformaldehyde at 4°C for 24 h and then dehydrated in 30% ethanol, 50% ethanol, and 70% ethanol serially at 4°C for 24 h each before embedding in paraffin wax. Patient formalin-fixed and paraffin-embedded (FFPE) biopsy tissues for co-immunofluorescence and murine FFPE tissues sectioned to 3–7 μm for histology evaluation. Paraffin sections were deparaffinized and rehydrated using xylene, xylene: 100% ethanol in 1:1 ratio, 100% ethanol, 95% ethanol, 70% ethanol, and 50% ethanol. Sectioned slides then underwent standard H&E staining or were incubated in sodium citrate buffer or EDTA buffer at 100°C for 15 min and allowed to cool to room temperature for antigen retrieval. For murine AFP staining, slides were incubated in anti-AFP polyclonal antibody (Agilent) at 4°C overnight and visualized with Metal Enhanced DAB Substrate Kit (Thermo) following the manufacturer’s procedure. Counterstaining was performed with hematoxylin. For co-immunofluorescence staining of patient samples, slides were serially incubated with anti-CD66b monoclonal antibody (GeneTex) and anti-TGFB1 polyclonal antibody (Proteintech) at 4°C overnight and visualized with AlphaTSA Multiplex IHC Kit according to manufacturer’s instructions. Slides were scanned at 10× and 20× magnification using a Vectra Polaris imaging system (Akoya), sizes of sections were measured by Fiji, and number of tumor nodules was counted blindly, while the frequency of TGFβ+CD66b+neutrophils in patient samples was quantified with Qupath.
For neutral lipid staining, snap-frozen tumor tissues were collected and sectioned. Tissue sections were incubated with an oil red O (Sigma-Aldrich) working solution for 5 min, followed by counterstaining with hematoxylin. Sections were mounted using a 50% glycerol mounting media. Images were acquired using a BX51 light microscope (Olympus) at 4× magnification.
For immunofluorescence staining, snap-frozen murine tumor tissue sections were fixed with 4% paraformaldehyde for 2 h, washed with PBS containing 0.3% Triton X-100 (Sigma-Aldrich), and then incubated for 10 min with 0.3 M glycine (Sigma-Aldrich). Blocking was performed with 10% normal goat serum (Biolegend) for 1 h. The sections were then incubated overnight at 4°C with 0.1 μg/ml BODIPY 493/503 (Invitrogen), anti-myeloperoxidase (Thermo Fisher Scientific), and anti-SiglecF (BD Biosciences) antibodies diluted at 1:100 in 1% BSA (Gibco). Following this, the sections were incubated for 2 h with secondary antibodies diluted at 1:100 in 1% BSA. Mounting media with DAPI (Vector Laboratories) was used to seal the sections for image documentation at 40× magnification using a Vectra Polaris imaging system.
For cytospin staining of TANs, SiglecFhi and SiglecF− TANs were FACs sorted from HCC tumors as described above. Cytospin was performed using a cytocentrifuge (Thermo Shandon, Inc.) with 1 × 105 cells per slide and centrifugation at 1,000 rpm for 5 min onto poly-L-lysine–coated slides. The slides were air-dried overnight before fixation with 4% paraformaldehyde and processed with standard hematoxylin staining. Images were captured with a BX51 light microscope (Olympus) at 40× magnification.
Western blotting
Sort-purified SiglecF− and SiglecFhi TANs were lysed on ice using radio immunoprecipitation assay buffer supplemented with protease inhibitors (Thermo Fisher Scientific). Protein concentration in the lysates was quantified with Pierce BCA protein assay kit (Thermo Fisher Scientific). The antibodies used were: HRP-conjugated anti-c-Myc mAb (Abcam), mouse anti-GAPDH mAb (Santa Cruz), and HRP-conjugated goat anti-mouse pAb (Invitrogen). The signals were visualized using Clarity Western ECL Substrate (Bio-Rad).
Flow cytometry analysis
To eliminate dead cells, single-cell suspensions were washed with PBS before being stained with the Zombie Aqua Fixable Viability Kit (BioLegend). Subsequent surface staining occurred in the presence of TruStain FcX anti-mouse CD16/32 (BioLegend) antibodies. For intracellular protein staining, the eBioscience Foxp3/Transcription Factor Staining Buffer Set (Invitrogen) was used. For intracellular cytokine staining, the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD) was used. After fixation, cells were stained with intracellular antibodies for 30 min at 4°C before being washed with 1X Permeabilization Buffer. Finally, the cells were resuspended in FACS buffer (1× PBS containing 0.1% BSA, 2 mM EDTA, and 0.09% sodium azide). All samples were acquired on an LSR Fortessa flow cytometer (BD Biosciences) and data were analyzed by FlowJo software version 10 (BD Life Sciences). The gating strategy is outlined in Fig. S1, F and G. During experiments involving α-Ly6G mAb injections, Ly6G was stained intracellularly. For measuring intracellular neutral lipids, cells were incubated with 0.5 μg/ml of BODIPY 493/503 (Invitrogen) at 37°C for 20 min, before live/dead and surface staining.
Total FA metabolomics analysis
Sample processing and gas chromatography–mass spectrometry (GC-MS) analysis were performed at the Centre for PanorOmic Sciences - Proteomics and Metabolomics Core, LKS Faculty of Medicine, University of Hong Kong. Briefly, around 70 mg of snap-frozen tumor sections from the HCC models used and naïve liver was submitted for profiling. A 2:1 chloroform/methanol extraction with sonication was performed, with a C19:0 FA internal standard spiked to the sample. The supernatant was cleaned by liquid–liquid extraction in 0.73% NaCl and methanol, then dried under nitrogen before transesterification with methanol and concentrated hydrochloric acid (35%, wt/wt). Fatty acid methyl esters extraction was performed with hexane and water, with the hexane phase injected for GC-MS analysis. GC-MS chromatogram was acquired in SCAN and SIM mode in an Agilent 7890B GC - Agilent 7010 Triple Quadrapole Mass Spectrometer system. The sample was separated through an Agilent DB-23 capillary column (60 m × 0.25 mm inner diameter, 0.15 µm film thickness) under constant pressure of helium at 33.4 psi. Characteristic fragment ions (m/z 55, 67, 69, 74, 79, 81, 83, 87, 91, 93, 95, 96, 97, 115, 127, 143) were monitored in SIM mode throughout the run. Mass spectra from m/z 50–350 were acquired in SCAN mode. Data analysis was performed using the Agilent MassHunter Workstation Quantitative Analysis Software. Linear calibration curves for each analyte were generated by plotting the peak area ratio of external/internal standard against standard concentration at different concentration levels. Analytes were confirmed by comparing the ratio of characteristic fragment ions in the sample and standard.
Untargeted lipidomic analysis
Sample processing and liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis were performed at the Centre for PanorOmic Sciences - Proteomics and Metabolomics Core, LKS Faculty of Medicine, University of Hong Kong. Briefly, around 70 mg of snap-frozen tumor sections from the HCC models used and naïve liver were submitted for profiling. A 2:1 chloroform/methanol extraction with sonication was performed. Each supernatant was dried under nitrogen before 1 mg was reconstituted with an isopropanol, methanol, and chloroform mixture (1:1:0.2, vol/vol) to 5 μl. Each sample was injected for LC-MS/MS analysis on a Vanquish UPLC (Thermo Fisher Scientific) and an Orbitrap Exploris 120 mass spectrometer equipped with a HESI II probe (Thermo Fisher Scientific). Mobile phases used were 10 mM ammonium formate with 0.1% formic acid in acetonitrile and water, vol/vol 6:4 (A), and 10 mM ammonium formate with 0.1% formic acid in acetonitrile and isopropanol 1:9 (B). The column was a ThermoFisher Accucore C30 (2.1 × 150 mm, 2.6 μm). An injection volume of 3 μl and a flow rate of 0.26 ml min−1 were used. The column oven temperature was set at 45°C. The gradient started at 30% B and was increased to 43% B in 2 min, then increased to 55% B in 2.1 min, 65% B in 12 min, 85% B in 18 min, and 100 % B in 20 min, then held for 5 min, and decreased linearly to 30% B for re-equilibration time at starting conditions. The MS analysis was performed in polar switching mode with source parameters set as follows: sheath gas flow rate, 60; auxiliary gas flow rate, 17; sweep gas flow rate, 1; spray voltage, +3.5/−3.0 kV; capillary temperature, 275°C; S-lens radio frequency level, 70; and heater temperature, 325°C. Data were collected at dd-MS2 mode. Data analysis was performed using Lipidsearch (Thermo Fisher Scientific/Mitsui Knowledge Industries) with the default parameters for Orbitrap MS Product Search and Alignment. After alignment, raw peak areas for all identified lipids were exported to Excel files.
RNA-seq analysis
Raw data were collected from publicly accessible datasets through their accession number. For the paired-end bulk RNA-seq of SiglecFhi and SiglecF− TANs purified from NRAS/AKT HCC tumors, of BM neutrophils reprogrammed with linoleic acid and GM-CSF, and of RIL175 after overnight co-culture with purified TANs, RNA was extracted from the cells with RNeasy Mini kit (Qiagen). Trim-galore (v0.6.10) was used to trim adaptor sequences and remove low-quality reads. STAR (v2.7.10b) aligned high-quality reads to the UCSC human genome GRCh38/hg38 or UCSC mouse genome mm10, and featureCounts (v2.0.3) quantified gene-level read counts. TPM (transcripts per million) values were calculated from count values as normalized gene expression levels. These data have been deposited at the Gene Expression Omnibus under the accession number GSE240838. The R package DESeq2 (v1.32.0) was used to perform differential gene expression analysis, and clusterProfiler (v4.0.5) (Wu et al., 2021) was used to perform GO and gene set enrichment analysis (GSEA) based on the differently expressed genes. P values and adjusted P values <0.05 were considered significant. Log (Fold change) >0 was used to identify the upregulated genes, and the downregulated genes were determined using log (fold change) <0. The R package TCGAbiolinks (v3.17) (Mounir et al., 2019) was used to retrieve clinical information and “HTSeqCounts” values from the National Cancer Institute Genomic Data Commons Data Portal for analysis of the TCGA-LIHC cohort, and patients’ viral etiology was retrieved from UCSC Xena database (https://xena.ucsc.edu/). The viral etiologies of the other datasets were retrieved from their own publications or from HCCDB (Lian et al., 2018). The pathway signature score was calculated as the average of log (TPM+0.1). The R package GSVA (v1.46.0) was used to calculate the ssGSEA enrichment score as the pathway signature score for microarray datasets. When analyzing the HCC patient datasets (IMbrave150, GO30140) (Zhu et al., 2022), to impute tumor cell-specific gene expression profiles, the transcriptomic value of each gene was multiplied by the proportion of tumor cells estimated by PUREE software in every sample (Revkov et al., 2023). Additionally, to account for variations in immune infiltration within tumor tissues, when calculating the relative SiglecFhi TAN signature score, gene expressions were normalized to that of protein tyrosine phosphatase, receptor type, C (CD45). The survminer R package (v0.4.9) was used for prognosis analysis, and the top or bottom 25% of patients with gene signature expression were categorized as “high” or “low” groups. The log-rank test was used to evaluate survival rates, and a two-tailed Pearson test was used to calculate correlations between two signature scores. The TAN signature used in Fig. 1 A was downloaded from Zaitsev et al. (2022). The gene list used to calculate MASH scores in TCGA-LIHC patients is listed in Table S2. These genes were derived from the significantly upregulated genes distinguishing MAFLD-HCC patients from HBV-HCC patients in the TCGA-LIHC dataset, overlapping with those distinguishing MAFLD patients from healthy controls (GSE135251 [Govaere et al., 2020]). This signature was able to distinguish MASH-HCC patients from non-MASH-HCC controls (GSE164760 [Pinyol et al., 2021] versus GSE63898 [Villanueva et al., 2015] P = 0.00091) and MASH patients from non-MASH controls (GSE83452 [Lefebvre et al., 2017], P = 0.043). Using this MASH signature, non-viral HCC patients in the TCGA cohort were split in half according to their MASH scores for further prognostic analysis.
scRNA-seq analysis
Sequencing data from human HCC patients was retrieved from BioProject (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744) (Xue et al., 2022). The Centre for PanorOmic Sciences (University of Hong Kong) sequenced samples from NRAS/AKT-injected mice with scRNA-seq libraries prepared using a Chromium Next GEM Single Cell 5′ Reagent Kit v2 and sequencing with Illumina NovaSeq 6000. For human and mouse samples, raw data were aligned and quantified using the CellRanger toolkit (v.6.0) against the reference genomes hg38 and mm10, respectively. The scRNAseq data generated by this study is deposited at the Gene Expression Omnibus under the accession number GSE240839. Seurat (v4.3.0), an R package, was used to analyze scRNA-seq data (Hao et al., 2021). With the help of the R package SingleR (v1.6.1) (Aran et al., 2019), major immune cell types and tumor cells were identified, with additional validation using well-known markers such as CD3D, CD4, CD8A, and CD19 for lymphoid lineages and CD14, CD68, CSF3R, and S100A8 for myeloid lineages after unsupervised clustering. To further characterize subpopulations of major immune cell types, a second round of clustering was performed. The top 10 principal components for neutrophils were chosen based on 800 highly variable genes (HVGs). The resolution of neutrophil clusters was determined based on biological features, and previously published scRNA-seq datasets of neutrophils from bone marrow, spleen, blood, and cancerous livers were also used as references (Grieshaber-Bouyer et al., 2021; Xue et al., 2022). As a result, 11 neutrophil clusters were discovered. The FindAllmarkers function was used to identify differentially expressed genes with adjusted P values <0.05. The AddModuleScore function was used to calculate the signature score in scRNA-seq analysis. To derive the Siglecf-Hi–like TAN signature, genes contributing to the similarity between the murine Siglecf-Hi TAN and the human protumor TAN it most resembled (Neu_10_SPP1) were used. FUT4 was also included to further associate the expression of these genes to TANs. The top 100 significantly upregulated genes (Table S3) from the bulk RNA-seq of sorted SiglecFhi versus SiglecF− were used to score the cells across different TAN subsets from the scRNA-seq. For tumor infiltrating CD8+ T cells from the mouse liver, the top 15 principal components were chosen according to 1500 HVGs, and six CD8+ T cell subpopulations were identified based on their differentially expressed genes. The mouse-human comparison analysis was carried out as previously described (Xue et al., 2022). In brief, sets of HVGs from human or mouse neutrophil subsets were identified, and the intersection of HVGs from human and mouse was retained. Pearson correlation analysis was performed between cell subpopulations across species using normalized average gene expression of these overlapping HVGs. To quantify transcriptional factors’ activity in mouse and human neutrophils, decoupleR (v2.5.2) (Badia-I-Mompel et al., 2022) was used to calculate the activity score of transcriptional factors defined with DoRothEA (v1.4.2). Nichenetr (v2.0.0) was used to identify the prioritized ligands from neutrophils driving the transcriptional difference between tumor cells in HCC patients with high or low neutrophil infiltration, and between NRAS/AKT HCC tumors after treatment with αLy6G or isotype control mAbs. To calculate the pseudotime trajectory of BM, spleen, and tumor neutrophils, Monocle 3 (v1.3.4) was used. M- and GM-CSF signaling genes were obtained from the molecular signatures database (MSigDB), while G-CSF response genes were derived from Pedersen et al. (2016).
Quantification and statistical analysis
Details regarding the statistical tests used, n values, and P values are described in the figure legends. Statistical tests were performed using GraphPad Prism 9 (GraphPad Software), with P < 0.05 being considered significant. Additional details regarding the quantification methods used for the various experiments are available under the method details.
Online supplemental material
Fig. S1 describes the HCC models used along with a representative FACs gating strategy. Fig. S2 complements Fig. 2, elaborating upon the identification of neutrophils from the scRNA-seq data, and describing the neutrophils’ characteristics. Fig. S3 supports the conclusions from Fig. 3 by reinforcing the consequences SiglecFhi TAN removal has on tumorigenesis. Fig. S4 pertains to Fig. 7 by further exploring the factors contributing to SiglecFhi TAN development. Fig. S5 is associated with Fig. 8 as it illustrates how αLy6G mAb treatments affect the tumor-immune landscape in HCC-bearing C57BL/6J mice, and how HCC-bearing Mrp8Cre.TGFβfl/fl mice respond to αPD-1 mAb treatment. Table S1 provides the clinical information for the patient tissue used in the co-immunofluorescence experiment of Fig. 6,.Table S2 lists the genes comprising the MASH score gene signature used in Figs. 1 and 2. Table S3 provides the top 100 differentially expressed genes defining SiglecFhi TANs. Table S4 reports the list of antibodies used, while Table S5 lists the chemicals and commercial kits used.
Data availability
The sequencing data used in this study has been deposited at the Gene Expression Omnibus under the SuperSeries accession number GSE240840. This SuperSeries includes the following SubSeries: scRNA-seq data (GSE240839) and RNA-seq data (GSE240838). All data are available in the main text or the online supplemental material.
Acknowledgments
We thank patients for their participation. We also thank Prof. Jessica Strid and Prof. Zemin Zhang for critical discussions.
This work was supported by grants from the Research Grants Council (RGC) of Hong Kong (Early Career Scheme ECS 27118520); General Research Fund (GRF 17116622) to G.S. Ling; National Natural Science Foundation of China (82322047 and 82173035), the National Key Research and Development Project of China (2022YFC3400902), the Beijing Municipal Natural Science Foundation (Z240016), the Beijing Nova Program (20230434854), and Clinical Medicine Plus X Young Scholars Project of Peking University (PKU2024LCXQ002) to R. Xue; and from RGC Research Fellow Scheme (RFS2122-7S05), RGC Research Impact Fund (R7022-20), RGC Theme-based research scheme (T12-705/24-R), and Croucher Senior Research Fellowship to S. Ma. J.M.N. Teo was supported by The Hong Kong PhD Fellowship Scheme. We also acknowledge the funding support from the Laboratory for Synthetic Chemistry and Chemical Biology and the Centre for Translational and Stem Cell Biology under the Health@InnoHK Program launched by the Innovation and Technology Commission, Hong Kong Special Administrative Region.
Author contributions: J.M.N. Teo: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing - original draft, Writing - review & editing, Z. Chen: Data curation, Formal analysis, Investigation, Validation, W. Chen: Data curation, Formal analysis, Investigation, Validation, R.J.Y. Tan: Investigation, Validation, Q. Cao: Data curation, Y. Chu: Validation, D. Ma: Resources, L. Chen: Investigation, H. Yu: Investigation, K.-H. Lam: Investigation, T.K.W. Lee: Methodology, Resources, S. Chakarov: Conceptualization, Writing - review & editing, B. Becher: Methodology, Resources, Writing - review & editing, N. Zhang: Conceptualization, Methodology, Resources, Supervision, Writing - review & editing, Z. Li: Resources, S. Ma: Methodology, Resources, Supervision, R. Xue: Data curation, Funding acquisition, Methodology, Resources, Writing - review & editing, G.S. Ling: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing - original draft, Writing - review & editing.
References
G.S. Ling is the lead contact.
Author notes
Disclosures: The authors declare no competing interests exist.






![Neutrophil identification and characteristics. (A) Uniform Manifold Approximation and Projections (UMAPs) (left) and expression of neutrophil lineage markers (right) by the major immune populations identified from the scRNA-seq of the BM and spleen (top), and HCC tumors (bottom) of NRAS/AKT HCC mice. (B) Dot plot of the differently expressed genes by each neutrophil cluster in the BM and spleen (top), and HCC tumor (bottom). (C) RNA-seq analyses were performed on SiglecFhi and SiglecF– TANs purified from NRAS/AKT-HCC tumors. A gene set defining SiglecFhi TANs was generated from their top 100 differentially expressed genes. The distribution of this gene set among TAN subsets. (D) Representative FACs plots and percentage of SiglecFhi neutrophils detected in the BM, spleen, blood, and tumor of NRAS/AKT HCC mice. (E) CXCR2, CXCR4, CD62L, and dcTRAIL-R1 expression within SiglecF− and SiglecFhi TANs of NRAS/AKT HCC mice. (F) Functional characterization of BM neutrophils, SiglecF− and SiglecFhi TANs from NRAS/AKT HCC mice. (G) Representative FACs plots and percentages of SiglecFhi TANs of RIL175 orthotopic HCC mice fed with MCD diet (MCD + RIL175; left) or Chow diet (CD + RIL175; right) (n = 3/group). (H) Representative immunofluorescence images and proportion of SiglecFhi TANs (myeloperoxidase [MPO]: red; SiglecF: green; nucleus: blue) in NRAS/AKT HCC tumors after 2 wk of treatment with αLy6G or isotype control mAbs. Scale bar represents 100 µm. (I) Expression levels of genes in the Siglecf-Hi–like TAN signature across various cell types in the tumor of HCC patients (https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA007744). Each symbol represents one mouse. Data are mean ± SEM and represent two independent experiments (D–H). **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA (F) and unpaired Student’s t test (H).](https://cdn.rupress.org/rup/content_public/journal/jem/222/1/10.1084_jem.20241442/3/m_jem_20241442_figs2.png?Expires=1773850641&Signature=OImamUzZn2a6An~apPMaKr-N~Nmcu--23M2kmmDzYvyvSx2FD~38q~El5pFZNptH6wiGGPOLTkCctBgYy3VcvaYx6BZXtvYSLcv1sdPImdv7DkyBxGhfBDeTgQvVYlTp6WnB7jee-s8WdgrS9xMh4TXaF6ivyaF9qtt82cQ0xs9V0dsxpWIrCHdRQGLLWTSF6TIYHgul39h4-iAEEJmDghgVYV~Rd~Rkox1wFNMscNPaYmSm~Zb~RFdULtAD1~Bf17CWkZSNoi1VpY19wnabt2mqEdS0hr3GZ~mLpx272NMaQJgjmrUItmBnZKZbRwiBiQVvAG8iVnni5-I6FD~wnQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)