Our understanding of protective versus pathological immune responses to SARS-CoV-2, the virus that causes coronavirus disease 2019 (COVID-19), is limited by inadequate profiling of patients at the extremes of the disease severity spectrum. Here, we performed multi-omic single-cell immune profiling of 64 COVID-19 patients across the full range of disease severity, from outpatients with mild disease to fatal cases. Our transcriptomic, epigenomic, and proteomic analyses revealed widespread dysfunction of peripheral innate immunity in severe and fatal COVID-19, including prominent hyperactivation signatures in neutrophils and NK cells. We also identified chromatin accessibility changes at NF-κB binding sites within cytokine gene loci as a potential mechanism for the striking lack of pro-inflammatory cytokine production observed in monocytes in severe and fatal COVID-19. We further demonstrated that emergency myelopoiesis is a prominent feature of fatal COVID-19. Collectively, our results reveal disease severity–associated immune phenotypes in COVID-19 and identify pathogenesis-associated pathways that are potential targets for therapeutic intervention.
Introduction
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is an urgent public health crisis. COVID-19 has a highly variable disease course: ∼20% of infected individuals require hospitalization, ∼5% require critical care, and up to 40% of cases are asymptomatic (Huang et al., 2020; Lavezzo et al., 2020). Because the immune response to SARS-CoV-2 is a key determinant of COVID-19 severity and outcome, understanding the immunological underpinnings of COVID-19 pathogenesis is critical to predict, prevent, and treat SARS-CoV-2 infection and to prepare for the possibility of future infections caused by emerging betacoronaviruses that may be introduced from existing reservoirs (Tang et al., 2006; Cui et al., 2019; Lin et al., 2017; Banerjee et al., 2019).
Severe COVID-19 is associated with a number of distinct immunological signatures. For example, increased serum levels of pro-inflammatory cytokines such as IL-1β, IL-6, IP-10, and TNFα and the alarmins S100A8 and S100A9 are associated with worse outcomes (Silvin et al., 2020; Wilson et al., 2020; Lucas et al., 2020; Mehta et al., 2020; Arunachalam et al., 2020; Laing et al., 2020). COVID-19 also reconfigures leukocyte phenotype in a severity-specific fashion, with severe COVID-19 associated with lymphocyte exhaustion (Diao et al., 2020; Zheng et al., 2020; Wilk et al., 2020), neutrophil activation signatures (Veras et al., 2020; Wang et al., 2020; Middleton et al., 2020; Aschenbrenner et al., 2021; Meizlish et al., 2021), and hematopoietic alterations (Wilk et al., 2020; Schulte-Schrepping et al., 2020). While many of these findings have been established through transcriptomic and proteomic profiling, the gene regulatory changes underlying severe disease manifestations have not been determined.
Comparatively less is known about the features of immune responses to SARS-CoV-2 that protect against severe disease, because most cohorts profiled to date have included only hospitalized patients. Neutralizing antibodies and virus-specific T cell responses have been detected in mildly symptomatic patients, providing evidence of a successful adaptive immune response across the disease spectrum (Pepper et al., 2020,Preprint; Röltgen et al., 2020; Nielsen et al., 2020; Rydyznski Moderbacher et al., 2020; Lipsitch et al., 2020; Rodda et al., 2021). Notably, patients with mild COVID-19 have much lower levels of pro-inflammatory plasma cytokines and higher levels of tissue repair factors, suggesting that the immune response in mild disease can eradicate the virus without triggering the hyperinflammatory state observed in severe cases (Arunachalam et al., 2020; Lucas et al., 2020). Therefore, to define protective versus pathological features of the immune response, we aimed to profile both mild (World Health Organization [WHO] score 1–3, no oxygen requirement), moderate (WHO score 4–5, noninvasive oxygen requirement), and severe (WHO score 6–8, intubated) cases of COVID-19.
To map the immune response at the epigenetic, transcriptional, and proteomic levels, we performed single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq), single-cell RNA sequencing (scRNA-seq), and cytometry by time of flight (CyTOF) on the peripheral immune cells of a cohort of COVID-19 patients across the entire spectrum of disease severity. We discovered many immunological perturbations associated with disease severity, including robust signatures related to neutrophil activation along with dysfunction of monocytes, type 2 conventional dendritic cells (cDC2), and natural killer (NK) cells. In addition, we found strong evidence for emergency myelopoiesis in fatal disease. We also identified epigenetic changes correlated to these transcriptional and proteomic changes, demonstrating coordinated changes in regulatory element accessibility and transcription at key pro-inflammatory cytokine–encoding genes in monocytes. Together, this dataset reveals novel mechanistic insights into the pathological and protective mechanisms of the immune response to SARS-CoV-2.
Results
A trimodal single-cell atlas of the peripheral immune response to SARS-CoV-2
To investigate how immune responses vary between different severities of COVID-19, we profiled peripheral blood immune cells from 64 patients with COVID-19 and 12 healthy controls with three high-dimensional single-cell modalities: Seq-Well (Gierahn et al., 2017; Hughes et al., 2020)-based scRNA-seq (33 patients and 8 controls, including 7 patients previously profiled by our group; Wilk et al., 2020), scATAC-seq (18 patients, 7 controls), and CyTOF (64 patients, 12 controls; Fig. 1 A and Fig. S1). Importantly, we profiled COVID-19 patients across the full range of the disease severity spectrum, including patients with mild disease (WHO score 1–3; see Materials and methods) and hospitalized inpatients with moderate disease (WHO score 4–5), as well as critically ill patients with severe disease (WHO ordinal score 6–8). We scored patients by both peak severity (denoted by the colors representing cells/patients) and severity at the time of sample collection (separated as groups for box plots). The median age of profiled participants was 43.5 yr, and 51% were female (Fig. 1 B and Table S1). Before sampling, 14 patients received azithromycin, which has potential immunomodulatory effects (Zimmermann et al., 2018); 13 received remdesivir; and 1 received dexamethasone. No patients received tocilizumab or baricitinib before sampling (Table S1). The majority of patients were sampled during the acute phase of infection; 13 mildly and moderately ill patients were sampled in the convalescent phase (>21 d after first positive nasopharyngeal swab). Demographic information, additional clinical metadata, and the modalities applied to each sample are available in Table S1.
A trimodal single-cell atlas of the peripheral immune response to COVID-19 across a range of disease severities. (A) Pipeline for sample processing and number of patients analyzed, summarized by modality and peak disease severity score. For all display figures, scRNA-seq–derived data are boxed in blue, scATAC-seq–derived data are boxed in green, and CyTOF-derived data are boxed in orange. (B) Summary of key patient metadata, including age, peak disease severity score, and days after first positive nasopharyngeal PCR test. The vertical dotted line placed at 21 d after positive test indicates the threshold after which patient samples are considered convalescent. (C, D, and F–I) UMAP projections of complete scRNA-seq (C and D), scATAC-seq (F and G), and CyTOF (H and I) datasets colored by peak disease severity score of sample (C, F, and H) or cell type (D, G, and I). Eos, eosinophils; Prog, progenitor; Prolif Lymph, proliferating lymphocytes. (E) Cell type proportions from scRNA-seq data of PBMCs in each sample are colored by peak disease severity score. Platelets and neutrophils are excluded from the proportion calculations because their presence is related to sample processing. The x axes correspond to the disease severity score for each sample at the time of collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
A trimodal single-cell atlas of the peripheral immune response to COVID-19 across a range of disease severities. (A) Pipeline for sample processing and number of patients analyzed, summarized by modality and peak disease severity score. For all display figures, scRNA-seq–derived data are boxed in blue, scATAC-seq–derived data are boxed in green, and CyTOF-derived data are boxed in orange. (B) Summary of key patient metadata, including age, peak disease severity score, and days after first positive nasopharyngeal PCR test. The vertical dotted line placed at 21 d after positive test indicates the threshold after which patient samples are considered convalescent. (C, D, and F–I) UMAP projections of complete scRNA-seq (C and D), scATAC-seq (F and G), and CyTOF (H and I) datasets colored by peak disease severity score of sample (C, F, and H) or cell type (D, G, and I). Eos, eosinophils; Prog, progenitor; Prolif Lymph, proliferating lymphocytes. (E) Cell type proportions from scRNA-seq data of PBMCs in each sample are colored by peak disease severity score. Platelets and neutrophils are excluded from the proportion calculations because their presence is related to sample processing. The x axes correspond to the disease severity score for each sample at the time of collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Quality control of scRNA-seq and CyTOF datasets. (A and B) UMAP embeddings of complete scRNA-seq dataset, colored by cell type input (A; either PBMCs or ACK-lysed whole blood [WB]) or donor (B). (C) Upset plot depicting overlap of patient samples profiled between the three modalities, colored by peak disease severity score. (D) Top: Heatmap showing overlap in Seurat v4 annotation calls (x axis) and manual cell type annotations (y axis), colored by the percentage of a manual cell annotation within a Seurat v4 annotation (i.e., each row sums to 100%). Bottom: Bar plot showing mapping frequency of each manually assigned cell type annotation by Seurat v4. Neutrophils and developing neutrophils are the least frequently assigned cell types because they are not present in the reference dataset (Hao et al., 2021). (E) Manual gating scheme for MAIT cells in the CyTOF dataset, beginning with live singlets gated according to the scheme in L. (F) Scatter plot depicting concordance with proportions of MAITs (top) or NK cells (bottom) predicted by Seurat v4 in the scRNA-seq dataset (x axis) and proportions manually gated in the CyTOF dataset (y axis). (G) UMAP embedding of the complete PBMC CyTOF dataset, colored by cell subset. (H) Heatmap showing the z-score normalized average expression of each marker in the PBMC CyTOF panel across all cell subsets detected in that dataset. (I) UMAP embedding of our whole PBMC CyTOF dataset, colored by donor. (K and L) Gating strategies used to identify live, intact singlets (L) and live, intact, singlet NK cells (K). (J) For each marker shared between the scRNA-seq and CyTOF datasets, a linear model was used to calculate a β coefficient for the relationship between severity score at the time of sample collection and marker expression in each dataset. The scatter plots depict the correlation between these β coefficients for markers measured on monocytes (top) and NK cells (bottom). For all scatter plots, Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown.
Quality control of scRNA-seq and CyTOF datasets. (A and B) UMAP embeddings of complete scRNA-seq dataset, colored by cell type input (A; either PBMCs or ACK-lysed whole blood [WB]) or donor (B). (C) Upset plot depicting overlap of patient samples profiled between the three modalities, colored by peak disease severity score. (D) Top: Heatmap showing overlap in Seurat v4 annotation calls (x axis) and manual cell type annotations (y axis), colored by the percentage of a manual cell annotation within a Seurat v4 annotation (i.e., each row sums to 100%). Bottom: Bar plot showing mapping frequency of each manually assigned cell type annotation by Seurat v4. Neutrophils and developing neutrophils are the least frequently assigned cell types because they are not present in the reference dataset (Hao et al., 2021). (E) Manual gating scheme for MAIT cells in the CyTOF dataset, beginning with live singlets gated according to the scheme in L. (F) Scatter plot depicting concordance with proportions of MAITs (top) or NK cells (bottom) predicted by Seurat v4 in the scRNA-seq dataset (x axis) and proportions manually gated in the CyTOF dataset (y axis). (G) UMAP embedding of the complete PBMC CyTOF dataset, colored by cell subset. (H) Heatmap showing the z-score normalized average expression of each marker in the PBMC CyTOF panel across all cell subsets detected in that dataset. (I) UMAP embedding of our whole PBMC CyTOF dataset, colored by donor. (K and L) Gating strategies used to identify live, intact singlets (L) and live, intact, singlet NK cells (K). (J) For each marker shared between the scRNA-seq and CyTOF datasets, a linear model was used to calculate a β coefficient for the relationship between severity score at the time of sample collection and marker expression in each dataset. The scatter plots depict the correlation between these β coefficients for markers measured on monocytes (top) and NK cells (bottom). For all scatter plots, Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown.
Peripheral blood mononuclear cells (PBMCs) were sampled by all modalities; additionally, we processed red blood cell–lysed whole blood by scRNA-seq to profile granulocytes like neutrophils (see Fig. 8 and Fig. S4), and we processed isolated NK cells by CyTOF with a panel enabling deep interrogation of NK cell receptor expression (see Fig. 6, Fig. 7, and Fig. 1 A). In total, we analyzed ∼175,000 single transcriptomes, ∼50,000 single chromatin accessibility profiles, and >3.2 million single proteomic profiles (Fig. 1 A, Table S2, Table S3, Table S4, and Table S5). After performing modality-specific quality control procedures (see Materials and methods), we created a merged feature matrix of all profiled samples, which we subjected to dimensionality reduction, graph-based clustering, and cell type annotation (Fig. 1, C–I; Fig. S1 C; and Table S6; see Materials and methods).
We first examined how COVID-19 impacted the composition of peripheral immune cells. We saw similar trends in immune cell composition between the three modalities, including depletion of CD16 monocytes, dendritic cells (DCs), and NK cells, as well as increases in plasmablasts (PBs) in patients with severe and fatal COVID-19 (Fig. 1 E, Table S7, Table S8, and Table S9). Notably, cell subset proportions that were altered in moderate and severe disease were generally unchanged in mild cases, with the exception of plasmacytoid DCs (pDCs), which were depleted in all severity groups (Fig. 1 E, Table S7, Table S8, and Table S9). Cell type proportions where patient age was regressed as a covariate also showed similar disease severity–driven trends (Fig. S2). Further, a population of developing (or immature) neutrophils first identified in our prior study of 7 patients (Wilk et al., 2020) was confirmed in 17 additional patients (Fig. 1, C and E; and Table S7) and is similar to that observed by other groups (Schulte-Schrepping et al., 2020).
Impact of age on cell type proportions and conserved IFN signature in COVID-19 patients. (A) Scatter plots depicting correlation between each manually annotated cell type in scRNA-seq dataset and patient age. All points are colored by peak disease severity score. Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown for each cell type. (B) Proportions of manually annotated cell types in scRNA-seq dataset after regression for age. (C) Differential gene expression testing was conducted on eight major cell types from the scRNA-seq dataset, comparing each COVID-19 sample with the cells of all healthy control subjects (see Materials and methods). The plotted heatmap depicts the percentage of COVID-19 samples in which a given ISG is up-regulated in a given cell type. (D) Expression of IRF7 by pDCs in scRNA-seq dataset. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Impact of age on cell type proportions and conserved IFN signature in COVID-19 patients. (A) Scatter plots depicting correlation between each manually annotated cell type in scRNA-seq dataset and patient age. All points are colored by peak disease severity score. Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown for each cell type. (B) Proportions of manually annotated cell types in scRNA-seq dataset after regression for age. (C) Differential gene expression testing was conducted on eight major cell types from the scRNA-seq dataset, comparing each COVID-19 sample with the cells of all healthy control subjects (see Materials and methods). The plotted heatmap depicts the percentage of COVID-19 samples in which a given ISG is up-regulated in a given cell type. (D) Expression of IRF7 by pDCs in scRNA-seq dataset. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Multimodal reference mapping enables accurate annotation and analysis of cellular subtypes
Accurate identification of fine-grained immune cell subtypes is crucial to understanding how COVID-19 reconfigures the immune system; however, these cell types can be difficult to identify de novo in scRNA-seq data due to data sparsity and lack of information on canonical surface marker expression in each cell. To address this, we mapped our transcriptomic dataset to a large multimodal reference dataset introduced by Seurat v4, which incorporated extensive surface marker information to improve cell type calls (Fig. 2 A; Hao et al., 2021).
Reference-based cell subtype annotations reveal disease severity–associated perturbations in immune cell subtypes. (A) WNN projection of scRNA-seq dataset colored by cell type labels transferred from Seurat v4 (left) or by peak disease severity score (right). Eryth, erythrocyte. (B) Heatmap of cellular perturbation scores, as described by Papalexi et al. (2021), per COVID-19 sample in each Seurat v4–labeled cell type. The number of DEGs between all COVID-19 cells and healthy cells for each cell type is plotted at the left. (C) UMAP projection of all DC subsets colored by peak disease severity score (left) and Seurat v4–annotated cell type (right). (D) Dot plot depicting percentage and average expression of canonical DC genes defining the four annotated DC subsets (see Materials and methods and Table S11). (E) Box plots depicting proportions of DC subsets. (F) Box plots depicting average expression of selected DEGs (see Table S13 for complete list) by cDC2s for each sample. (G) UMAP projection of all CD8 T cells colored by peak disease severity score (left) and Seurat v4–annotated cell type (right). (H) Dot plot depicting percentage and average expression of canonical CD8 subset–defining genes (see Materials and methods). (I) Box plots depicting average expression of selected DEGs (see Table S14 for complete list) by CD8 TEM cells in each sample. (J) Box plots showing average module scores for T cell exhaustion (as reported in Miller et al., 2019) in each annotated CD8 T cell subset. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. TCM, T central memory; gdT, γδ T; dnT, double negative T cell; TEM, T effector memory cell; TCM, T central memory cell.
Reference-based cell subtype annotations reveal disease severity–associated perturbations in immune cell subtypes. (A) WNN projection of scRNA-seq dataset colored by cell type labels transferred from Seurat v4 (left) or by peak disease severity score (right). Eryth, erythrocyte. (B) Heatmap of cellular perturbation scores, as described by Papalexi et al. (2021), per COVID-19 sample in each Seurat v4–labeled cell type. The number of DEGs between all COVID-19 cells and healthy cells for each cell type is plotted at the left. (C) UMAP projection of all DC subsets colored by peak disease severity score (left) and Seurat v4–annotated cell type (right). (D) Dot plot depicting percentage and average expression of canonical DC genes defining the four annotated DC subsets (see Materials and methods and Table S11). (E) Box plots depicting proportions of DC subsets. (F) Box plots depicting average expression of selected DEGs (see Table S13 for complete list) by cDC2s for each sample. (G) UMAP projection of all CD8 T cells colored by peak disease severity score (left) and Seurat v4–annotated cell type (right). (H) Dot plot depicting percentage and average expression of canonical CD8 subset–defining genes (see Materials and methods). (I) Box plots depicting average expression of selected DEGs (see Table S14 for complete list) by CD8 TEM cells in each sample. (J) Box plots showing average module scores for T cell exhaustion (as reported in Miller et al., 2019) in each annotated CD8 T cell subset. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. TCM, T central memory; gdT, γδ T; dnT, double negative T cell; TEM, T effector memory cell; TCM, T central memory cell.
Alignment to the Seurat v4 reference dataset revealed gene expression profiles of cell subtypes matching their expected biological signatures (Fig. S1 D, Table S10, Table S11, and Table S12). For example, multiple T cell subsets, including γδ T cells, mucosal-associated invariant T (MAIT) cells, and T regulatory cells were revealed with Seurat v4 at the expected proportions (Table S10) and with the expected transcriptomic phenotype (e.g., highly specific expression of TRDC and TRGC1 by γδ T cells, and FOXP3 by T regulatory cells; Table S11 and Table S12). These annotations closely matched the manually generated labels; importantly, cell types present in our dataset but absent from the reference (i.e., neutrophils) were not successfully mapped (Fig. S1 D). To orthogonally confirm the accuracy of these annotations, we compared the abundances of two cell types that we could also identify in our CyTOF dataset that are typically difficult to distinguish in RNA space by graph-based clustering alone: NK cells and MAIT cells. This analysis revealed high concordance between modalities, supporting the accuracy of these annotations (Fig. S1, E and F).
To prioritize downstream analysis of cell subsets most affected by COVID-19, we calculated a perturbation score (Hao et al., 2021; Papalexi et al., 2021) for each cell type from each COVID-19 sample relative to healthy control subject samples (see Materials and methods). The perturbation score for each cell type is calculated by first identifying genes that display evidence of differential expression between COVID-19 samples and healthy control samples, calculating the difference of pseudobulk expression vectors of these genes between COVID-19 samples and healthy control samples, and finally projecting the whole transcriptome of each donor onto this vector. This score therefore represents the magnitude of whole-transcriptome shifts in gene expression and reveals disease severity–associated patterns in cell subtype perturbation (Fig. 2 B). This perturbation score captured phenotypic changes in major cell types such as monocytes (explored further in Figs. 3, 4, and 5) and more granular subtypes such as CD8 effector memory T (TEM) cells. We focused our downstream analysis on cell subtypes with COVID-19 severity–associated perturbation with a high number of differentially expressed genes (DEGs) relative to other cell subtypes.
Monocytes with dysfunctional and suppressive features emerge in severe and fatal COVID-19. (A) UMAP projections of monocytes from scRNA-seq dataset, colored by CD14 and FCGR3A (encoding CD16) expression (left) and colored by peak disease severity score (right). (B) Volcano plot depicting DEGs in monocytes of patients with severe and fatal COVID-19 versus healthy control subjects. (C) Box plots depicting average expression of selected DEGs by monocytes (see Table S15 for complete DEG list). (D) Box plots showing average module scores for ISG, HLA class II, bacterial sepsis (Reyes et al., 2020), and MDSC (Alshetaiwi et al., 2020) gene signatures in monocytes (see Materials and methods). (E) Box plots depicting monocyte precursor subset gene module score (see Materials and methods and Table S16; Kawamura et al., 2017), colored by peak COVID-19 severity. (F) Heatmap showing per-cell correlations between module scores plotted in E. cMoP_Mo, CD14 monocytes derived from the cMoP. (G) UMAP projection of all monocytes from CyTOF dataset, colored by peak disease severity score. (H and I) Feature plots (H) and box plots (I) depicting arcsinh-transformed expression of selected protein markers by monocytes in CyTOF dataset. (J) UMAP projections of complete scRNA-seq dataset colored by expression of stroke-predictive genes (Raman et al., 2016). (K) Box plots depicting average expression of the five stroke-predictive genes in monocytes (top) or canonical neutrophils (bottom). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. CMP, common myeloid progenitor.
Monocytes with dysfunctional and suppressive features emerge in severe and fatal COVID-19. (A) UMAP projections of monocytes from scRNA-seq dataset, colored by CD14 and FCGR3A (encoding CD16) expression (left) and colored by peak disease severity score (right). (B) Volcano plot depicting DEGs in monocytes of patients with severe and fatal COVID-19 versus healthy control subjects. (C) Box plots depicting average expression of selected DEGs by monocytes (see Table S15 for complete DEG list). (D) Box plots showing average module scores for ISG, HLA class II, bacterial sepsis (Reyes et al., 2020), and MDSC (Alshetaiwi et al., 2020) gene signatures in monocytes (see Materials and methods). (E) Box plots depicting monocyte precursor subset gene module score (see Materials and methods and Table S16; Kawamura et al., 2017), colored by peak COVID-19 severity. (F) Heatmap showing per-cell correlations between module scores plotted in E. cMoP_Mo, CD14 monocytes derived from the cMoP. (G) UMAP projection of all monocytes from CyTOF dataset, colored by peak disease severity score. (H and I) Feature plots (H) and box plots (I) depicting arcsinh-transformed expression of selected protein markers by monocytes in CyTOF dataset. (J) UMAP projections of complete scRNA-seq dataset colored by expression of stroke-predictive genes (Raman et al., 2016). (K) Box plots depicting average expression of the five stroke-predictive genes in monocytes (top) or canonical neutrophils (bottom). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. CMP, common myeloid progenitor.
Absent pro-inflammatory cytokine–encoding gene induction by monocytes in severe COVID-19. (A) Box plots depicting average expression of pro-inflammatory cytokine–encoding genes by monocytes. (B) Dot plot depicting results of iRegulon TF activity prediction analysis. Positive normalized enrichment scores (NES) indicate that the TF activity is higher in patients with severe COVID-19 relative to that in healthy control subjects. (C) Dot plot depicting average and percentage expression of NF-κB subunits. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Absent pro-inflammatory cytokine–encoding gene induction by monocytes in severe COVID-19. (A) Box plots depicting average expression of pro-inflammatory cytokine–encoding genes by monocytes. (B) Dot plot depicting results of iRegulon TF activity prediction analysis. Positive normalized enrichment scores (NES) indicate that the TF activity is higher in patients with severe COVID-19 relative to that in healthy control subjects. (C) Dot plot depicting average and percentage expression of NF-κB subunits. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Identification of putative enhancers regulating pro-inflammatory cytokine expression by monocytes in COVID-19. (A) Genome-wide footprinting of the NF-κB2 binding motif in CD14 monocytes from different severity groups shown in different colors. (B) Box plot depicting quantification of the “flanking accessibility” (Baek et al., 2017; Corces et al., 2018) for NF-κB2 motif footprints in CD14 monocytes from different samples. Each dot indicates the average “flanking accessibility” value for each sample. (C) Box plot depicting the average chromVAR z-scores of NF-κB2 binding motifs in CD14 monocytes from different samples. (D and F) The genome tracks show genomic regions near IL1B (D) and CCL2 (F) genes. The top panel indicates coverage at different peak regions for CD14 monocytes in different severity groups; the box below shows peaks called from all CD14 monocytes (dark blue) in the 100-kb region and peaks containing putative strong NF-κB2 binding sites (red); the CoAccessibility box in D shows the accessibility correlated peak pairs across all CD14 monocytes near the IL1B locus; the Genes box shows the location of IL1B (D) or CCL2 (F) together with other adjacent genes; the bottom Virtual 4C track in D shows Knight-Ruiz–normalized contact frequencies to the IL1B promoter in THP-1 monocytic cells; blue color means the gene is located on the minus strand, and red color means the gene is located on the plus strand. The arrows indicate peaks of interest whose accessibility is quantified in the corresponding box plots (E). (G and I) The genome tracks show genomic regions near the CD4 gene. The top panel indicates coverage at different peak regions for different cell subsets (G) and for CD14 monocytes in different severity groups (I); the box below shows peaks called from all PBMCs (G) or from the CD14 monocytes (I) in that region (dark blue); the bottom Genes box shows the location of CD4 and other adjacent genes; blue color means the gene is located on the minus strand, and red color means the gene is located on the plus strand. The arrows indicate monocyte-specific peaks with higher accessibility in monocytes and DCs than in CD4 T cells. (E and H) Box plots depicting the Tn5 insertions per million at the peaks marked with the corresponding arrows in CD14 monocytes. Exact P values for E: top, P = 0.0081 healthy versus severe; middle, P = 0.014 healthy versus mild; bottom, P = 0.0037. Exact P values for H: top, P = 0.0047 healthy versus severe; middle, P = 0.0047 healthy versus severe; bottom, P = 0.022 healthy versus mild. Points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni correction for multiple hypothesis testing.
Identification of putative enhancers regulating pro-inflammatory cytokine expression by monocytes in COVID-19. (A) Genome-wide footprinting of the NF-κB2 binding motif in CD14 monocytes from different severity groups shown in different colors. (B) Box plot depicting quantification of the “flanking accessibility” (Baek et al., 2017; Corces et al., 2018) for NF-κB2 motif footprints in CD14 monocytes from different samples. Each dot indicates the average “flanking accessibility” value for each sample. (C) Box plot depicting the average chromVAR z-scores of NF-κB2 binding motifs in CD14 monocytes from different samples. (D and F) The genome tracks show genomic regions near IL1B (D) and CCL2 (F) genes. The top panel indicates coverage at different peak regions for CD14 monocytes in different severity groups; the box below shows peaks called from all CD14 monocytes (dark blue) in the 100-kb region and peaks containing putative strong NF-κB2 binding sites (red); the CoAccessibility box in D shows the accessibility correlated peak pairs across all CD14 monocytes near the IL1B locus; the Genes box shows the location of IL1B (D) or CCL2 (F) together with other adjacent genes; the bottom Virtual 4C track in D shows Knight-Ruiz–normalized contact frequencies to the IL1B promoter in THP-1 monocytic cells; blue color means the gene is located on the minus strand, and red color means the gene is located on the plus strand. The arrows indicate peaks of interest whose accessibility is quantified in the corresponding box plots (E). (G and I) The genome tracks show genomic regions near the CD4 gene. The top panel indicates coverage at different peak regions for different cell subsets (G) and for CD14 monocytes in different severity groups (I); the box below shows peaks called from all PBMCs (G) or from the CD14 monocytes (I) in that region (dark blue); the bottom Genes box shows the location of CD4 and other adjacent genes; blue color means the gene is located on the minus strand, and red color means the gene is located on the plus strand. The arrows indicate monocyte-specific peaks with higher accessibility in monocytes and DCs than in CD4 T cells. (E and H) Box plots depicting the Tn5 insertions per million at the peaks marked with the corresponding arrows in CD14 monocytes. Exact P values for E: top, P = 0.0081 healthy versus severe; middle, P = 0.014 healthy versus mild; bottom, P = 0.0037. Exact P values for H: top, P = 0.0047 healthy versus severe; middle, P = 0.0047 healthy versus severe; bottom, P = 0.022 healthy versus mild. Points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni correction for multiple hypothesis testing.
This multimodal reference mapping approach enabled us to perform previously unfeasible transcriptomic analyses of fine-grained immune cell subsets. For example, we identified cDC2 cells as the principal remodeled DC subset in COVID-19; these cells are depleted in severe disease and have the greatest disease severity–associated perturbation (Fig. 2, B–E). In cDC2 cells, FCER1A, known to be involved in inflammatory DC signaling (Shin and Greer, 2015), CD83, an activation marker of mature DCs (Li et al., 2019), and CTSS, which is involved in antigen presentation (Fig. 2 F; Kim et al., 2017), were down-regulated with increased disease severity, while genes associated with tolerogenic or anti-inflammatory responses, like PKM and CD163, were up-regulated (Palsson-McDermott et al., 2017; Comi et al., 2020). Collectively, these results indicate that dysfunctional and anti-inflammatory cDC2 cells may be a feature of severe COVID-19, with important potential implications for T follicular helper cell development and mucosal immunity.
CD8 TEM cells also displayed severity-associated transcriptional perturbations (Fig. 2, B, G, and H). Notably, several markers of CD8 effector capacity, like PRF1, GZMB, and CX3CR1 (Yan et al., 2018; Gerlach et al., 2016), were down-regulated primarily in patients with mild COVID-19 (Fig. 2 I). Additionally, in severe and fatal COVID-19 patients, CD8 TEM cells retained expression of markers of effector capacity without showing features of exhaustion (Fig. 2 J). Together, these analyses provide transcriptional evidence that over-exuberant peripheral cytotoxic T cell responses may be associated with severe disease, similar to previous protein-level reports (Mathew et al., 2020).
COVID-19 acuity remodels peripheral immune phenotype in a severity-specific fashion
In light of the heterogeneity of sampling times between patients (Fig. 1 B), we examined the impact of disease time point on immune phenotype. These analyses are limited by our small sample size of acutely infected patients with mild disease in our transcriptional dataset. Nonetheless, these analyses indicate that convalescence has considerably less impact on transcriptional phenotype in patients with mild illness at peak severity (Fig. S3). These results imply that mild COVID-19 may be marked by minimal, or rapidly resolved, systemic immune responses, a finding that is orthogonally supported by our CyTOF analyses that include a greater number of subjects. Cell types most perturbed in convalescence included CD8 TEM and CD14 monocytes; in patients with moderate disease, B cells were also perturbed in convalescence, displaying down-regulation of Ig genes (Fig. S3 I). Additional longitudinal sampling across all severity groups is necessary to clarify these signatures.
Impact of disease acuity on transcriptomic phenotype of mild and moderate COVID-19 patients. (A) Upset plot depicting overlap of DEGs between acute mild, acute moderate, acute severe, and convalescent samples when each is compared with healthy control samples. DEG testing is performed on PBMCs and filtered for adjusted P < 1e-4. (B) Box plot depicting cumulative perturbation score of all cell types per patient calculated on a perturbation vector between acute and convalescent samples. Points are colored and grouped by the peak disease severity score. ***, P < 0.001 by two-sided Wilcoxon rank-sum test. (C and F) Heatmaps of cellular perturbation score, as described by Papalexi et al. (2021), per mild (C) or moderate (F) COVID-19 sample in each Seurat v4–labeled cell type. DEGs between acute and convalescent samples in each severity group are used as input for each perturbation score (see Materials and methods). (D and G) UMAP projections of all cells from mild (D) or moderate (G) COVID-19 patients colored by disease acuity (left) and Seurat v4–annotated cell type (right). (E and H) Dot plots depicting percentage and unscaled average expression for all DEGs with |log(fold-change)| > 1 in CD8 TEM cells (left) and CD14 monocytes (right) of mild (E) or moderate (H) COVID-19 patients. (I) Dot plot depicting percentage and unscaled average expression for all DEGs with |log(fold-change| > 1 in B cells of moderate COVID-19 patients.
Impact of disease acuity on transcriptomic phenotype of mild and moderate COVID-19 patients. (A) Upset plot depicting overlap of DEGs between acute mild, acute moderate, acute severe, and convalescent samples when each is compared with healthy control samples. DEG testing is performed on PBMCs and filtered for adjusted P < 1e-4. (B) Box plot depicting cumulative perturbation score of all cell types per patient calculated on a perturbation vector between acute and convalescent samples. Points are colored and grouped by the peak disease severity score. ***, P < 0.001 by two-sided Wilcoxon rank-sum test. (C and F) Heatmaps of cellular perturbation score, as described by Papalexi et al. (2021), per mild (C) or moderate (F) COVID-19 sample in each Seurat v4–labeled cell type. DEGs between acute and convalescent samples in each severity group are used as input for each perturbation score (see Materials and methods). (D and G) UMAP projections of all cells from mild (D) or moderate (G) COVID-19 patients colored by disease acuity (left) and Seurat v4–annotated cell type (right). (E and H) Dot plots depicting percentage and unscaled average expression for all DEGs with |log(fold-change)| > 1 in CD8 TEM cells (left) and CD14 monocytes (right) of mild (E) or moderate (H) COVID-19 patients. (I) Dot plot depicting percentage and unscaled average expression for all DEGs with |log(fold-change| > 1 in B cells of moderate COVID-19 patients.
Emergence of monocytes with dysfunctional features in severe COVID-19
We next analyzed the phenotypes of peripheral monocytes in COVID-19, because these cells appeared to be strongly reconfigured in nonlinear dimensionality reduction projections for all three modalities (Fig. 1, C, D, and F–I). Embedding of monocytes alone from the transcriptomic dataset recapitulated this phenotypic shift (Fig. 3 A). Similar to previous reports (Schulte-Schrepping et al., 2020; Wilk et al., 2020; Ong et al., 2020; Giamarellos-Bourboulis et al., 2020; Arunachalam et al., 2020), multiple IFN-stimulated genes (ISGs) and markers of immature and tolerogenic monocytes, such as CD163, PLAC8, and MPO (Fig. 3, B and C), were up-regulated with increasing disease severity. Notably, ARG1, encoding the myeloid-derived suppressor cell (MDSC) marker and T cell inhibitor arginase, was also up-regulated most prominently in the monocytes of fatal COVID-19 patients (Fig. 3 C). Monocytes from severe and fatal COVID-19 patients possessed additional features of an MDSC-like phenotype (Schulte-Schrepping et al., 2020), including loss of HLA class II–encoding genes (Fig. 3, C and D) and enrichment of published gene signatures from MDSCs (Alshetaiwi et al., 2020) and monocytes in the setting of bacterial sepsis (Reyes et al., 2020; Fig. 3 D). Additionally, we noted a severity-associated loss of CD4 expression (Fig. 3, B and C), which is involved in monocyte-to-macrophage differentiation and pro-inflammatory cytokine induction in CD14 monocytes (Mathy et al., 2000; Zhen et al., 2014). These results collectively suggest that suppressive and dysfunctional monocytes are a feature of severe and fatal COVID-19, in agreement with previous reports (Schulte-Schrepping et al., 2020; Arunachalam et al., 2020; Wilk et al., 2020). Importantly, mild COVID-19 generally did not lead to this shift toward suppressive and dysfunctional monocytes.
The appearance of this expanded population of monocytes with suppressor-like features led us to examine whether these cells are the result of mature circulating monocytes being exposed to the peripheral inflammatory milieu of severe COVID-19 or of immature cells that are the product of emergency myelopoiesis. To address this question, we scored monocytes in our transcriptomic dataset for gene signatures of various monocyte progenitors: common myeloid progenitor, granulocyte-monocyte progenitor, common monocyte progenitor (cMoP), premonocytes, and mature CD14 monocytes or CD14 monocytes derived from the cMoP (Fig. 3 E; Kawamura et al., 2017). This analysis reveals that the vast majority of monocytes in our dataset correspond to mature monocytes and that there is no coexpression of monocyte progenitor and MDSC gene sets (Fig. 3, E and F). This suggests that the dysfunctional and tolerogenic transcriptional signatures of monocytes in severe and fatal COVID-19 likely reflect not the immaturity of these cells but rather a phenotype acquired by mature monocytes exposed to the inflammatory milieu of COVID-19.
Proteomic profiling of COVID-19 monocytes recapitulated many of our transcriptional findings (Fig. 3, G–I). This included a loss of CD16+ monocytes as well as a distinct shift in the phenotype of CD14+ monocytes (Fig. 3, G–I; and Table S8). As observed in our transcriptional data, expression of HLA-DR and CD4 was lost in monocytes of severe COVID-19 samples. Importantly, this proteomic reconfiguration was not observed in patients with mild COVID-19, evident in nonlinear dimensionality reduction (Fig. 3 G). Patients with mild disease experienced no significant increase in expression of the stress marker CD112, nor did they up-regulate CCR2, which is involved in monocyte recruitment to the airways in the setting of severe COVID-19, although both of these markers were up-regulated in patients with severe disease (Fig. 3, G–I; Merad and Martin, 2020; Pairo-Castineira et al., 2020). Patients with mild disease also displayed a less dramatic loss of HLA-DR and CD4 expression compared with monocytes in severe cases (Fig. 3, G–I). Panel-wide analysis of COVID-19 disease severity–associated changes in monocyte phenotype between scRNA-seq and CyTOF datasets also revealed high concordance in the perturbations detected between the two modalities (Fig. S1 J). These results indicate that while monocytes are dramatically remodeled in severe COVID-19, mild COVID-19 has minimal, or rapidly resolved, impact on the monocyte proteome.
Peripheral myeloid cells up-regulate stroke risk biomarkers in severe COVID-19
We also noted that C19orf59, which encodes MCEMP1, a key biomarker for stroke risk and outcome (Wood, 2016; Raman et al., 2016), was up-regulated in the monocytes of severe and fatal COVID-19 patients (Fig. 3, J and K). Given the accumulating data that COVID-19 can drive thrombotic complications including ischemic stroke, we examined expression of other transcripts reported to predict stroke risk in the study by Raman et al. (2016). We found that each of the five most predictive transcripts for stroke risk and prognosis reported by Raman et al. (C19orf59, IRAK3, ANXA3, RBM47, and TLR5) were abundantly expressed in monocytes and neutrophils and that each of these transcripts was significantly up-regulated in severe COVID-19 in either monocytes or neutrophils (Fig. 3, J and K).
NF-κB inactivity may underlie poor pro-inflammatory cytokine production in peripheral monocytes of severe COVID-19 patients
Because cytokine production is a key antiviral function of peripheral monocytes, we next examined the expression of pro-inflammatory cytokine–encoding genes by peripheral monocytes stratified by disease severity. Interestingly, we found minimal expression of key monocyte-derived pro-inflammatory cytokine–encoding genes, particularly in severe and fatal COVID-19 patients (Fig. 4 A); in fact, IL1B and TNF were among the most significantly down-regulated genes in the monocytes of severe and fatal COVID-19 patients (Fig. 3 B and Fig. 4 A). The failure of even mild cases to significantly up-regulate many pro-inflammatory cytokine–encoding genes (Fig. 4 A) is in contrast to mild cases of similar viral infections such as influenza (Lamichhane and Samarasinghe, 2019; Vangeti et al., 2018; Hoeve et al., 2012). To explore potential regulatory mechanisms underlying this dysfunction, we performed a transcription factor (TF) activity analysis of our RNA dataset, which revealed decreased activity of NF-κB in monocytes from severe COVID-19 patients (Fig. 4 B). The NF-κB pathway is crucial for the inflammatory responses to viral infections in innate immune cells (Hetru and Hoffmann, 2009; Medzhitov and Horng, 2009; Liu et al., 2017), and its activation relies on various pro-inflammatory cytokines, including IL-1β and TNFα (Lawrence, 2009). Activated NF-κB can further induce TNF and IL1B expression (Liu et al., 2017; Hiscott et al., 1993), leading to a positive feedback loop. Our scRNA-seq data did not show significant transcriptional changes for most NF-κB family TFs, although REL and RELB are down-regulated in severe COVID-19 (Fig. 4 C). This could either reflect technical limitations of measuring lowly expressed TF transcripts, or it could indicate that our observed NF-κB activity changes are caused by post-translational modifications (Liu et al., 2017).
We next leveraged our chromatin accessibility dataset to investigate the regulatory mechanisms by which NF-κB could control expression of pro-inflammatory cytokines by monocytes in COVID-19. First, a genome-wide footprinting analysis of NF-κB motifs revealed severity-associated decreases in NF-κB binding activity (Fig. 5, A and B), consistent with our RNA-based TF activity analysis. Consistent with this hypothesis, we further observed COVID-19–associated changes in genome-wide NF-κB family TF activity. Using chromVAR analysis to quantify TF activity from the chromatin accessibility of each cell, we found increased NF-κB activity in mild cases and significantly decreased activity in severe cases (P = 0.0047, Wilcoxon test; Fig. 5 C; Schep et al., 2017).
To investigate potential gene regulatory mechanisms that could explain the down-regulation of pro-inflammatory cytokines in monocytes of severe and fatal COVID-19 patients, we examined changes in chromatin accessibility around the loci encoding IL1B and CCL2. We identified a putative enhancer downstream of IL1B, which shows linkage to the IL1B promoter via single-cell coaccessibility analysis and chromosome conformation capture Hi-C data from the THP-1 monocytic cell line (Fig. 5 D; see Materials and methods; Phanstiel et al., 2017). This putative enhancer showed significantly decreased accessibility in severe COVID-19 patients (P = 0.0081, Wilcoxon test; Fig. 5 E). Furthermore, this element contains an NF-κB binding motif, suggesting it may be regulated by NF-κB family TFs (Fig. 5 D). We also identified changes in accessibility within peaks containing NF-κB motifs at the locus for the inflammatory cytokine CCL2 (Fig. 5 F). Here, we observed an increase in the accessibilities of motifs near these loci in patients with mild disease exclusively (Fig. 5 E), similar to the pattern of NF-κB activity observed in our chromVAR analysis (Fig. 5 C); this suggests the possibility that greater accessibility of these motifs may be related to a lower burden of disease. Our results suggest that aberrant decreases in NF-κB activity in severe COVID-19 may result in loss of accessibility at putative enhancers of key cytokine genes.
We also examined the epigenetic regulation at the CD4 locus, because this gene was significantly down-regulated with increasing disease severity. Although there was no change in chromatin accessibility of the CD4 gene promoter between severity groups, we found that the accessibility of monocyte-specific CD4 gene putative regulatory regions was significantly reduced in severe samples (Fig. 5, G–I). This monocyte-specific loss of CD4 expression may provide an additional mechanism explaining the previously reported impairment of cytokine production by monocytes in COVID-19 (Arunachalam et al., 2020; Schulte-Schrepping et al., 2020), because the interaction between IL-16 and monocytic CD4 induces the expression of pro-inflammatory cytokines, including IL-1β (Mathy et al., 2000).
Peripheral NK cells are depleted in severe COVID-19 and have a highly activated phenotype
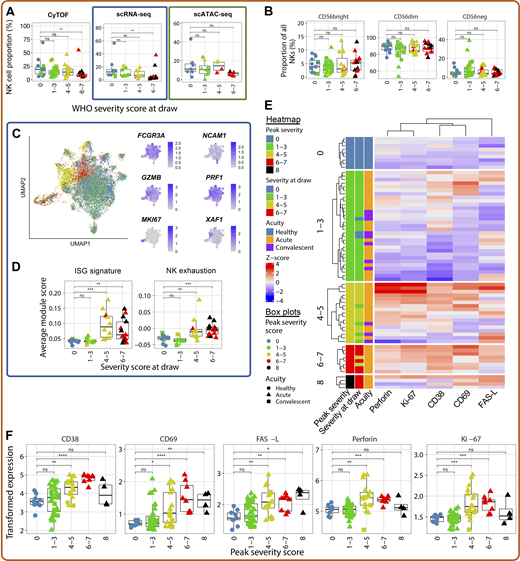
We next interrogated changes in the NK cells of COVID-19 samples. As demonstrated previously (Wilk et al., 2020; Maucourant et al., 2020), peripheral NK cells were substantially depleted across all three modalities, although the frequencies of CD56bright, CD56dim, and CD56− NK cells as a percentage of all NK cells did not change (Fig. 6, A and B). The depletion of peripheral NK cells in severe COVID-19 may reflect their trafficking to the site of infection (Liao et al., 2020). We also noted significant transcriptional reconfiguration driven by up-regulation of several canonical NK cell activation genes, including higher expression of cytotoxic effector molecule–encoding genes GZMB and PRF1, as well as proliferation marker MKI67 and ISGs like XAF1 (Fig. 6, C and D). NK cells from moderate and severe, but not mild, COVID-19 cases also displayed transcriptional evidence of exhaustion (Fig. 6 D).
NK cells of severe COVID-19 patients exhibit a unique proteomic and transcriptional profile. (A) Box plots of manually annotated NK cell proportions from CyTOF dataset (left), Seurat v4–annotated NK cell proportions from scRNA-seq dataset (center), and Seurat v4–annotated NK cell proportions from scATAC-seq dataset (right; see Materials and methods). (B) Box plots showing the frequency of CD56bright, CD56dim, and CD56− NK cells as a proportion of NK cells in the CyTOF dataset. (C) UMAP projections of NK cells from scRNA-seq dataset colored by peak disease severity score (left) and selected DEGs (right; see Table S17 for complete list). (D) Box plots of average ISG signature and NK cell exhaustion (defined as expression of LAG3, PDCD1, and HAVCR2; see Materials and methods) module scores in Seurat v4–annotated NK cells. (E) Heatmap depicting Z-score normalized protein-level expression of canonical NK cell activation and cytotoxicity markers (perforin, Ki-67, CD38, CD69, and FasL) in each sample. (F) Box plots quantifying arcsinh-transformed average expression of markers depicted in E by NK cells, grouped by peak disease severity score. For all box plots except F, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
NK cells of severe COVID-19 patients exhibit a unique proteomic and transcriptional profile. (A) Box plots of manually annotated NK cell proportions from CyTOF dataset (left), Seurat v4–annotated NK cell proportions from scRNA-seq dataset (center), and Seurat v4–annotated NK cell proportions from scATAC-seq dataset (right; see Materials and methods). (B) Box plots showing the frequency of CD56bright, CD56dim, and CD56− NK cells as a proportion of NK cells in the CyTOF dataset. (C) UMAP projections of NK cells from scRNA-seq dataset colored by peak disease severity score (left) and selected DEGs (right; see Table S17 for complete list). (D) Box plots of average ISG signature and NK cell exhaustion (defined as expression of LAG3, PDCD1, and HAVCR2; see Materials and methods) module scores in Seurat v4–annotated NK cells. (E) Heatmap depicting Z-score normalized protein-level expression of canonical NK cell activation and cytotoxicity markers (perforin, Ki-67, CD38, CD69, and FasL) in each sample. (F) Box plots quantifying arcsinh-transformed average expression of markers depicted in E by NK cells, grouped by peak disease severity score. For all box plots except F, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
We next examined this NK cell activation signature at the protein level. We corroborate previously known changes in NK cell biology, including increased expression of the activation markers CD38 and CD69 (Maucourant et al., 2020), and we also demonstrate that surface expression of the death receptor ligand FasL is increased in moderate and severe COVID-19 patients (Fig. 6, E and F). While perforin was also up-regulated in moderately and severely ill patients, surprisingly, NK cells from fatal COVID-19 patients failed to up-regulate both this cytotoxic effector and the proliferation marker Ki-67 (Fig. 6, E and F). These data, coupled with transcriptomic evidence of NK cell exhaustion in severe and fatal COVID-19 (Fig. 6 D), suggest that defects in NK cell cytotoxicity may be associated with adverse outcomes.
Dynamic changes in NK cell receptors and ligands may underlie COVID-19 severity–associated NK activation
To assess mechanisms of NK cell activation, we interrogated changes in the NK cell repertoire of surface-expressed activating and inhibitory receptors on CD38+CD69+ activated NK cells. As expected, the proportion of activated NK cells was significantly increased in moderate and severe COVID-19 (Fig. 7, A and B). Notably, surface expression of the activating receptors DNAM-1 (CD226) and NKG2D was significantly down-regulated in activated NK cells of severe COVID-19 samples compared with healthy controls (Fig. 7 C), despite no change in the expression of the genes encoding these proteins in the total NK cells within our scRNA-seq data (Fig. 7 D). Because expression of both DNAM-1 and NKG2D can be down-modulated following ligation (Carlsten et al., 2009; Molfetta et al., 2017), we investigated the abundance of a DNAM-1 ligand, Nectin-2 (CD112), and of the ULBP proteins, which are recognized by NKG2D. Both Nectin-2 and the ULBPs were significantly up-regulated on the peripheral monocytes of hospitalized COVID-19 patients compared with healthy controls (Fig. 7 E), which supports the hypothesis that SARS-CoV-2 may decrease surface expression of DNAM-1 and NKG2D through internalization following ligation of overexpressed Nectin-2 and ULBP proteins due to stress. Alternatively, activated NK cells expressing DNAM-1 or NKG2D may migrate to the tissue in the setting of severe disease, depleting these markers from the circulating population. We found no changes in the expression of either TIGIT, an inhibitory receptor that competes with DNAM-1 for binding of Nectin-2, or TACTILE (CD96), which recognizes another ligand of DNAM-1, CD155 (Fig. 7 F).
Ligation of DNAM-1 and NKG2D may drive activation of NK cells in severe COVID-19. (A) Representative flow plots showing the gating scheme used to identify activated (CD38+CD69+) NK cells in patients from each severity bin. (B) Box plot showing the proportion of CD38+CD69+ NK cells in each severity bin. (C) Box plots showing arcsinh-transformed protein-level expression of the activating receptors DNAM-1 (left) and NKG2D (right) in CD38+CD69+ NK cells. (D) Box plots showing the average expression of CD226 (which encodes DNAM-1; left) and KLRK1 (which encodes NKG2D; right) from the scRNA-seq dataset (E) Box plots depicting arcsinh-transformed protein-level expression of NK cell ligands CD112 and ULBP-1,2,5,6 in monocytes. (F) Box plots showing arcsinh-transformed expression of the inhibitory receptors TIGIT and CD96/TACTILE in CD38+CD69+ NK cells in our CyTOF dataset. (G) Box plot depicting arcsinh-transformed average protein-level expression of NK cell ligands LLT-1 in monocytes. (H) Box plots showing arcsinh-transformed protein-level expression of the inhibitory receptor CD161 on all NK cells. (I) Schematic illustrating the changes in protein-level expression of NK cell activating and inhibitory receptors as well as their ligands. Text color indicates whether a receptor/ligand is activating (green), inhibitory (red), or either, depending on the context (yellow). Arrows and dashes indicate whether abundance of a protein is increased, decreased, or unchanged in severe COVID-19 compared with healthy controls. Dashed lines indicate interactions between receptors and ligands. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Ligation of DNAM-1 and NKG2D may drive activation of NK cells in severe COVID-19. (A) Representative flow plots showing the gating scheme used to identify activated (CD38+CD69+) NK cells in patients from each severity bin. (B) Box plot showing the proportion of CD38+CD69+ NK cells in each severity bin. (C) Box plots showing arcsinh-transformed protein-level expression of the activating receptors DNAM-1 (left) and NKG2D (right) in CD38+CD69+ NK cells. (D) Box plots showing the average expression of CD226 (which encodes DNAM-1; left) and KLRK1 (which encodes NKG2D; right) from the scRNA-seq dataset (E) Box plots depicting arcsinh-transformed protein-level expression of NK cell ligands CD112 and ULBP-1,2,5,6 in monocytes. (F) Box plots showing arcsinh-transformed expression of the inhibitory receptors TIGIT and CD96/TACTILE in CD38+CD69+ NK cells in our CyTOF dataset. (G) Box plot depicting arcsinh-transformed average protein-level expression of NK cell ligands LLT-1 in monocytes. (H) Box plots showing arcsinh-transformed protein-level expression of the inhibitory receptor CD161 on all NK cells. (I) Schematic illustrating the changes in protein-level expression of NK cell activating and inhibitory receptors as well as their ligands. Text color indicates whether a receptor/ligand is activating (green), inhibitory (red), or either, depending on the context (yellow). Arrows and dashes indicate whether abundance of a protein is increased, decreased, or unchanged in severe COVID-19 compared with healthy controls. Dashed lines indicate interactions between receptors and ligands. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
We also observed a loss of LLT-1 expression on CD14+ monocytes that appears to correlate with disease severity, with a near-total loss in fatal samples (Fig. 7 G); however, we found no change in the expression of the inhibitory receptor that recognizes LLT-1, CD161, on NK cells (Fig. 7 H). The overall profile of activating and inhibitory receptors and ligands expressed in severe COVID-19 is summarized in Fig. 7 I and suggests that the activated phenotype observed in these samples may be driven by activating signals received through DNAM-1 and NKG2D as well as a lack of inhibitory signaling through CD161.
A hyperactivated neutrophil signature marks severe and fatal COVID-19
Despite evidence that neutrophils are major players in the dysregulated immune response that defines severe and fatal COVID-19 (Aschenbrenner et al., 2021; Bost et al., 2021; Meizlish et al., 2021; Barnes et al., 2020; Zuo et al., 2020; Radermecker et al., 2020; Veras et al., 2020; Middleton et al., 2020), there has been a relative dearth of deep profiling of neutrophils from COVID-19 patients, given their sensitivity to both cryopreservation and mechanical stimulation (Ekpenyong et al., 2015; Yap and Kamm, 2005). To address this, we first demonstrated that Seq-Well generated high-quality scRNA-seq data of primary human neutrophils from a healthy blood donor (Fig. S4). Although fewer genes were detected in sequenced neutrophils, the number of unique molecular identifiers (UMIs) sequenced in neutrophils was comparable to the expected recovery based on known RNA content (Xie et al., 2020; Monaco et al., 2019). We also found that neutrophils from ammonium chloride potassium (ACK)–lysed whole blood were phenotypically similar to neutrophils isolated by magnetic-activated cell sorting (Fig. S4, D and E); the former strategy was also able to uncover other granulocytic cell types, such as eosinophils (Fig. S4 C).
Seq-Well enables high-quality single-cell transcriptomic analysis of primary human neutrophils. Whole blood (WB) from a healthy donor was collected into CPT vacutainers, from which PBMCs were isolated and neutrophils were isolated from the PBMC-depleted cell pellet. Additionally, aliquots of whole blood were subjected to neutrophil isolation or red blood cell lysis with ACK buffer. These cell populations were then analyzed by Seq-Well (see Materials and methods). (A) UMAP projection colored by cell type preparation method. (B) Box plots showing comparisons of the number of UMIs sequenced (top) and the number of genes detected (bottom) in cells annotated to be PBMCs or in cells annotated as granulocytes (neutrophils and eosinophils). The median number of UMIs or genes in each group is plotted above the respective box. The difference in recovered UMIs and gene capture between PBMCs and granulocytes is comparable to that expected by RNA content (Xie et al., 2020; Monaco et al., 2019). (C) Bar plot depicting the proportions of cells from each cell sample preparation method for each annotated cell type. (D) Dot plot depicting percentage and unscaled average expression of the 15 top neutrophil-defining DEGs (see Table S24) between the three cell sample preparation methods that yielded neutrophils. (E) Dot plot depicting average and percentage expression of the top five DEGs for each cell type (see Table S24), demonstrating comparable expression patterns between PBMCs isolated through centrifugation and PBMC subsets present in ACK-lysed whole blood.
Seq-Well enables high-quality single-cell transcriptomic analysis of primary human neutrophils. Whole blood (WB) from a healthy donor was collected into CPT vacutainers, from which PBMCs were isolated and neutrophils were isolated from the PBMC-depleted cell pellet. Additionally, aliquots of whole blood were subjected to neutrophil isolation or red blood cell lysis with ACK buffer. These cell populations were then analyzed by Seq-Well (see Materials and methods). (A) UMAP projection colored by cell type preparation method. (B) Box plots showing comparisons of the number of UMIs sequenced (top) and the number of genes detected (bottom) in cells annotated to be PBMCs or in cells annotated as granulocytes (neutrophils and eosinophils). The median number of UMIs or genes in each group is plotted above the respective box. The difference in recovered UMIs and gene capture between PBMCs and granulocytes is comparable to that expected by RNA content (Xie et al., 2020; Monaco et al., 2019). (C) Bar plot depicting the proportions of cells from each cell sample preparation method for each annotated cell type. (D) Dot plot depicting percentage and unscaled average expression of the 15 top neutrophil-defining DEGs (see Table S24) between the three cell sample preparation methods that yielded neutrophils. (E) Dot plot depicting average and percentage expression of the top five DEGs for each cell type (see Table S24), demonstrating comparable expression patterns between PBMCs isolated through centrifugation and PBMC subsets present in ACK-lysed whole blood.
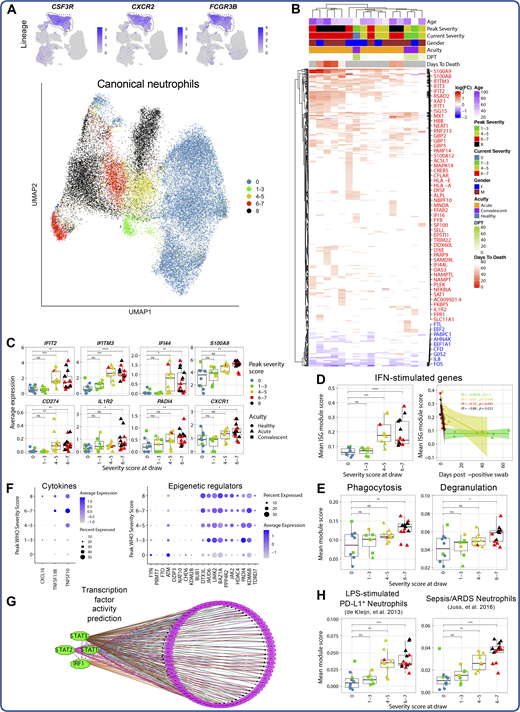
Seq-Well processing of red blood cell–lysed whole blood yielded 33,276 high-quality single transcriptomes of primary neutrophils (Fig. 8 A). These cells uniformly and specifically expressed neutrophil lineage marker–encoding genes, including CSF3R and CXCR2, indicating their identity as canonical neutrophils (Fig. 8 A and Table S6). Nonlinear dimensionality reduction revealed that neutrophil transcriptional phenotype was strongly remodeled in COVID-19 (Fig. 8 A), driven in part by up-regulation of PADI4, which is required for neutrophil extracellular trap activation and release (NETosis), the IL-8 receptor CXCR1, and multiple alarmins, including S100A8 and S100A9 (Fig. 8, B and C), which together induce neutrophil chemotaxis and adhesion (Ryckman et al., 2003). We also noted disease severity–specific induction of ISGs in moderate and severe, but not in mild, COVID-19 patients (Fig. 8 D). Although this ISG signature was detected across most cell types in moderately and severely ill patients, neutrophils up-regulated the broadest number of ISGs (Fig. S2). Importantly, the differential expression of ISGs by neutrophils between COVID-19 severity groups was not due to a difference in infection time points between patients: neutrophils from patients with mild COVID-19 did not up-regulate ISGs at any point during infection (Fig. 8 D). To examine potential sources of type I IFN, we analyzed expression of the upstream regulator of IFN in pDCs, IRF7, because type I IFN–encoding genes themselves are often undetectable at the RNA level (Kazer et al., 2020). pDCs did not display strong or consistent severity-associated up-regulation of IRF7 (Fig. S2), suggesting that the neutrophil ISG signature in moderate and severe COVID-19 is likely due to sensing of type I IFN produced at the site of infection. Additionally, gene set enrichment analysis demonstrated up-regulation of genes associated with neutrophil phagocytosis and degranulation in a disease severity–associated fashion (Fig. 8 E and Table S16).
Neutrophil activation is a hallmark of severe and fatal COVID-19. (A) UMAP projections of complete scRNA-seq dataset colored by expression of canonical neutrophil markers (top) and of canonical neutrophils alone colored by peak disease severity score (bottom). (B) Heatmap of DEGs between neutrophils of each COVID-19 sample compared with neutrophils of all healthy controls, colored by average log(fold-change). All displayed DEGs are statistically significant at the P < 0.05 confidence level by Seurat’s implementation of the Wilcoxon rank-sum test (two-sided, adjusted for multiple comparisons using Bonferroni correction). DPT, days post first positive COVID-19 test. (C) Box plots depicting average expression of selected neutrophil DEGs by severity group (see Table S18 for complete DEG list). (D) Plots depicting median ISG signature score of neutrophils in each sample grouped by disease severity score at the time of sample collection (left) and by days after first positive NP swab (right). All points are colored by peak disease severity score. For scatter plot at right, Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown for each peak disease severity score grouping. (E) Box plots depicting average module scores for genes sets of neutrophil phagocytosis and neutrophil degranulation (see Materials and methods and Table S16). (F) Dot plots depicting average and percentage expression of pro-inflammatory cytokine encoding genes (left) and epigenetic regulators (right) by canonical neutrophils. The y axis corresponds to the peak disease severity score. (G) Results of TF activity prediction analysis performed by iRegulon (Janky et al., 2014). DEGs between neutrophils from severely ill patients (peak severity 6–8) and neutrophils from healthy controls were used as input (see Materials and methods and Table S19). (H) Box plots of average module scores for PD-L1+ neutrophils in an in vitro model of endotoxemia (de Kleijn et al., 2013) and granulocytes in the setting of sepsis and ARDS (Juss et al., 2016; see Materials and methods and Table S16). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Neutrophil activation is a hallmark of severe and fatal COVID-19. (A) UMAP projections of complete scRNA-seq dataset colored by expression of canonical neutrophil markers (top) and of canonical neutrophils alone colored by peak disease severity score (bottom). (B) Heatmap of DEGs between neutrophils of each COVID-19 sample compared with neutrophils of all healthy controls, colored by average log(fold-change). All displayed DEGs are statistically significant at the P < 0.05 confidence level by Seurat’s implementation of the Wilcoxon rank-sum test (two-sided, adjusted for multiple comparisons using Bonferroni correction). DPT, days post first positive COVID-19 test. (C) Box plots depicting average expression of selected neutrophil DEGs by severity group (see Table S18 for complete DEG list). (D) Plots depicting median ISG signature score of neutrophils in each sample grouped by disease severity score at the time of sample collection (left) and by days after first positive NP swab (right). All points are colored by peak disease severity score. For scatter plot at right, Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown for each peak disease severity score grouping. (E) Box plots depicting average module scores for genes sets of neutrophil phagocytosis and neutrophil degranulation (see Materials and methods and Table S16). (F) Dot plots depicting average and percentage expression of pro-inflammatory cytokine encoding genes (left) and epigenetic regulators (right) by canonical neutrophils. The y axis corresponds to the peak disease severity score. (G) Results of TF activity prediction analysis performed by iRegulon (Janky et al., 2014). DEGs between neutrophils from severely ill patients (peak severity 6–8) and neutrophils from healthy controls were used as input (see Materials and methods and Table S19). (H) Box plots of average module scores for PD-L1+ neutrophils in an in vitro model of endotoxemia (de Kleijn et al., 2013) and granulocytes in the setting of sepsis and ARDS (Juss et al., 2016; see Materials and methods and Table S16). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
We also identified two distinct neutrophil immunophenotypes of fatal COVID-19. Neutrophils from four of six fatal COVID-19 cases had robust ISG induction and expressed CD274 (encoding PD-L1; Fig. 8, B and C), in line with previous work (Schulte-Schrepping et al., 2020). However, we also identified two fatal COVID-19 cases with less pronounced ISG induction but with up-regulation of additional neutrophil activation markers not observed in other samples, including CXCR4, CLEC12A, EGR1, and the decoy IL1β receptor IL1R2 (Fig. 8, B and C). Additional severity-associated changes in neutrophil phenotype included the up-regulation of pro-inflammatory cytokine-encoding genes, including CXCL16 and TNFSF10 (encoding TRAIL), as well as up-regulation of several epigenetic regulators involved in neutrophil activation, like PADI4, which is required for formation of neutrophil extracellular traps (Fig. 8 F; Aschenbrenner et al., 2021; Li et al., 2010; Hemmers et al., 2011).
TF activity analysis implicated STAT1, STAT2, STAT3, and IRF1 as key upstream regulators of the observed transcriptional reconfiguration, further suggesting that COVID-19 neutrophils are strongly activated by type I IFN in a disease severity–specific fashion (Fig. 8 G). To better contextualize these findings, we scored the neutrophils in our dataset against gene modules up-regulated in a model of endotoxemia (de Kleijn et al., 2013) and in acute respiratory distress syndrome (ARDS)–complicated sepsis (Juss et al., 2016). This analysis revealed that both of these signatures are up-regulated with increasing COVID-19 severity (Fig. 8 H), implying similarities in neutrophil phenotype between these clinical conditions. Collectively, profiling fresh whole blood rather than isolated PBMCs reveals a prominent neutrophil hyperactivation signature in severe and fatal COVID-19.
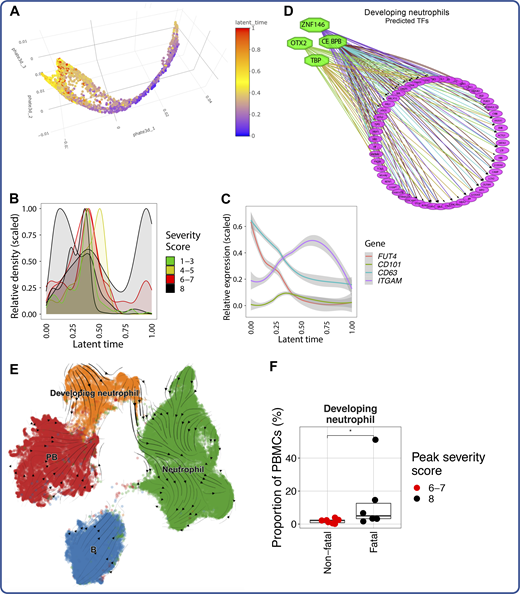
Developing neutrophils are a feature of fatal COVID-19
We next analyzed a population of developing neutrophils from the transcriptomic dataset that was enriched in patients with severe and fatal COVID-19 (Fig. 1 E and Fig. 9 A). This population has been identified in other COVID-19 datasets but is not yet well characterized (Schulte-Schrepping et al., 2020; Bost et al., 2021). These cells specifically and highly expressed genes encoding markers expressed at distinct stages in neutrophil development, including DEFA1B, LCN2, and MMP8 (Table S20), indicating that they likely represent immature neutrophils derived from emergency granulopoiesis. Because we hypothesized that these cells were not a static population but rather were dynamically differentiating, we embedded them in two and three dimensions using potential of heat diffusion for affinity-based trajectory embedding (PHATE), a dimensionality reduction method developed to visualize phenotypic continua, branches, and continual progressions (Fig. 9, B and C; and Fig. S5; Moon et al., 2019). Clustering of these cells revealed five clusters that corresponded to sequential stages in neutrophil development, beginning with cluster 0 (pro-neutrophils) expressing primary neutrophil granule protein–encoding genes, followed by clusters 1–3 (pre-neutrophils) consecutively expressing secondary and tertiary neutrophil granule protein–encoding genes, and finally cluster 4 (mature neutrophils), which expresses markers of canonical neutrophils (Fig. 9, C and D; Evrard et al., 2018). Importantly, ELANE (which encodes neutrophil elastase), was specifically expressed by pro-neutrophils, implying that these cells may be capable of NETosis (Perdomo et al., 2019; Martinod et al., 2016). An orthogonal approach ordering each cell in latent time modeled by splicing kinetics of RNA velocity (La Manno et al., 2018; Bergen et al., 2020; Fig. 9 E) also revealed a similar developmental trajectory with respect to both granule protein–encoding genes (Fig. 9 F) and TFs known to be involved in neutrophil development, such as the CCAAT-enhancer-binding protein family (CEBP; Fig. 9 G and Fig. S5). In our earlier work, we hypothesized that developing neutrophils may arise via transdifferentiation from PBs based on their phenotypic similarity in nonlinear dimensionality reduction manifold space and subsequent analysis of cellular trajectory by RNA velocity (Wilk et al., 2020). In this larger dataset, a phenotypic relationship between developing neutrophils and PBs was still evident (Fig. 1, C and D), but RNA velocity analysis of PBs, developing neutrophils, and mature neutrophils did not reveal a clear transdifferentiation bridge (Fig. S5). Orthogonal experiments are necessary to conclusively determine the developmental origins of these cells.
Emergency granulopoiesis is a feature of fatal COVID-19. (A) Box plot depicting proportions of developing neutrophils in each sample from the scRNA-seq dataset. (B and C) Two-dimensional PHATE projection of developing neutrophils colored by peak disease severity score (left) and cluster number (right). (D) Dot plot depicting percentage and average expression of DEGs between developing neutrophil clusters (see Table S20). (E) Two-dimensional PHATE projection of developing neutrophils colored by latent time calculated by scVelo (Bergen et al., 2020). (F and G) Scaled expression of selected neutrophil granule-encoding genes (F) and CCAAT-enhancer-binding protein (CEBP) TF family–encoding genes (G) by developing neutrophils across inferred latent time. (H) Bar plot representing the ranked developing neutrophil signature score (aggregated expression of DEFA1B, DEFA3, LTF, DEFA1, and S100A8; see Materials and methods) for each COVID-19 sample in a validation cohort from a publicly available bulk transcriptomic dataset (Overmyer et al., 2021), colored by the 28-d mortality. (I) ROC curve depicting sensitivity and specificity of 28-d mortality prediction of a five-gene signature of developing neutrophils (DEFA1B, DEFA3, LTF, DEFA1, and S100A8) or of Sequential Organ Failure Assessment (SOFA) score at the time of sample collection in an independent validation cohort of 103 samples where 17 cases are fatal (Overmyer et al., 2021). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. AUC, area under the curve. **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Emergency granulopoiesis is a feature of fatal COVID-19. (A) Box plot depicting proportions of developing neutrophils in each sample from the scRNA-seq dataset. (B and C) Two-dimensional PHATE projection of developing neutrophils colored by peak disease severity score (left) and cluster number (right). (D) Dot plot depicting percentage and average expression of DEGs between developing neutrophil clusters (see Table S20). (E) Two-dimensional PHATE projection of developing neutrophils colored by latent time calculated by scVelo (Bergen et al., 2020). (F and G) Scaled expression of selected neutrophil granule-encoding genes (F) and CCAAT-enhancer-binding protein (CEBP) TF family–encoding genes (G) by developing neutrophils across inferred latent time. (H) Bar plot representing the ranked developing neutrophil signature score (aggregated expression of DEFA1B, DEFA3, LTF, DEFA1, and S100A8; see Materials and methods) for each COVID-19 sample in a validation cohort from a publicly available bulk transcriptomic dataset (Overmyer et al., 2021), colored by the 28-d mortality. (I) ROC curve depicting sensitivity and specificity of 28-d mortality prediction of a five-gene signature of developing neutrophils (DEFA1B, DEFA3, LTF, DEFA1, and S100A8) or of Sequential Organ Failure Assessment (SOFA) score at the time of sample collection in an independent validation cohort of 103 samples where 17 cases are fatal (Overmyer et al., 2021). For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. AUC, area under the curve. **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing.
Additional analysis of emergency granulopoiesis in severe and fatal COVID-19. (A) Three-dimensional PHATE projection of developing neutrophils. (B) Scaled abundances of developing neutrophils present in individual COVID-19 patients across latent time. (C) Scaled expression of genes reported to define different stages of immature neutrophil development in COVID-19 (Schulte-Schrepping et al., 2020). (D) TF activity prediction analysis by iRegulon (Janky et al., 2014), using positive DEGs for developing neutrophils relative to all other cells as input (Table S19). (E) UMAP projection of developing neutrophils, canonical neutrophils, B cells, and PBs overlaid with RNA velocity stream (Bergen et al., 2020). (F) Box plot depicting proportion of developing neutrophils in patients with nonfatal severe versus fatal severe COVID-19. Points are colored and grouped by the peak disease severity score. *, P < 0.05 by two-sided Wilcoxon rank-sum test.
Additional analysis of emergency granulopoiesis in severe and fatal COVID-19. (A) Three-dimensional PHATE projection of developing neutrophils. (B) Scaled abundances of developing neutrophils present in individual COVID-19 patients across latent time. (C) Scaled expression of genes reported to define different stages of immature neutrophil development in COVID-19 (Schulte-Schrepping et al., 2020). (D) TF activity prediction analysis by iRegulon (Janky et al., 2014), using positive DEGs for developing neutrophils relative to all other cells as input (Table S19). (E) UMAP projection of developing neutrophils, canonical neutrophils, B cells, and PBs overlaid with RNA velocity stream (Bergen et al., 2020). (F) Box plot depicting proportion of developing neutrophils in patients with nonfatal severe versus fatal severe COVID-19. Points are colored and grouped by the peak disease severity score. *, P < 0.05 by two-sided Wilcoxon rank-sum test.
Because developing neutrophils were present uniformly, often at high frequencies, in patients with fatal COVID-19, and because these cells specifically expressed many genes not found in other peripheral blood cell types, we hypothesized that the developing neutrophil gene signature might be an accurate predictor of mortality in COVID-19. We therefore identified the five most positive DEGs between developing neutrophils (6,569 cells across 21 patients) and all other cells in our dataset: DEFA1B, DEFA3, LTF, DEFA1, and S100A8. We next obtained a publicly available whole blood bulk transcriptomic dataset of 103 COVID-19 patients as a validation cohort and scored each sample in this dataset by the aggregated expression of these five genes (see Materials and methods). After scoring each sample, we used the associated patient metadata to determine the 28-d mortality of each scored sample. We then constructed a receiver operating characteristic (ROC) plot using the gene score as predictor and the 28-d mortality as the response variable. We found that the developing neutrophil gene score accurately predicted 28-d mortality of the 17 patients who succumbed to infection (area under the curve, 0.81; Fig. 9, H and I). Importantly, the Sequential Organ Failure Assessment score at the time of sample collection did not strongly predict 28-d mortality (area under the curve, 0.67), indicating that the presence of developing neutrophils is a better prognostic indicator than current clinical status measured by clinically used severity scales (Fig. 9, H and I). Thus, developing neutrophils are likely enriched in the blood of fatal COVID-19 cases in other cohorts, and gene signatures from these cells have promise as a prognostic indicator.
Myeloid skewing of hematopoietic progenitors in severe and fatal COVID-19
Considering that severe and fatal COVID-19 patients displayed evidence of emergency myelopoiesis in the periphery, we hypothesized that there also may be severity-associated aberrations in a small population of hematopoietic stem and progenitor cells (HSPCs) that we identified in our transcriptomic dataset (Fig. 1, C and D; and Fig. 2, A and B). Although the frequency of these cells did not change in COVID-19 (Table S10), we found that several genes involved in HSPC maintenance and self-renewal, including CDK6, SOX4, and CHD4 (Laurenti et al., 2015; Bhullar and Sollars, 2011; Wang et al., 2019; Salvagiotto et al., 2008; Yoshida et al., 2008; Dege and Hagman, 2014; Vervoort et al., 2013), were generally up-regulated in COVID-19 patients relative to healthy controls (Fig. 10 A). These DEGs suggest that the hematopoietic progenitor compartment in COVID-19 patients has been transcriptionally reshaped to accommodate the stress of emergency hematopoiesis. To better understand the identities of these cells, we leveraged a publicly available single-cell transcriptomic dataset of hematopoiesis in healthy human blood and bone marrow (Fig. 10 B; Granja et al., 2019), into which we projected HSPCs from our COVID-19 dataset (Fig. 10 C). We found a trend toward myeloid skewing in COVID-19 circulating HSPCs with increasing disease severity, with lower frequencies of common lymphoid progenitors in severe and fatal patients, as well as granulocyte/macrophage progenitor/neutrophil-like cells appearing in severe and fatal cases (Fig. 10 D). Together, these results indicate severity-associated changes in hematopoiesis in COVID-19 with greater myeloid skewing evident in circulating HSPCs.
Myeloid skewing of circulating HSPCs and other hematopoietic abnormalities in COVID-19. (A) Box plots of average expression of selected HSPC DEGs (see Table S21). (B and C) UMAP projection of Seurat v4–annotated HSPCs from scRNA-seq dataset into a publicly available blood and bone marrow hematopoiesis dataset (Granja et al., 2019) colored by published cell type annotations (B) and with projected HSPCs colored in red (C). (D) Bar plot depicting proportions of cell type identities transferred after projection into the publicly available hematopoiesis dataset for each peak disease severity score bin. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; BMMC, bone marrow mononuclear cells; LMPP, lymphomyeloid-primed multipotent progenitor; GMP, granulocyte-monocyte progenitor; HSC, hematopoietic stem cell.
Myeloid skewing of circulating HSPCs and other hematopoietic abnormalities in COVID-19. (A) Box plots of average expression of selected HSPC DEGs (see Table S21). (B and C) UMAP projection of Seurat v4–annotated HSPCs from scRNA-seq dataset into a publicly available blood and bone marrow hematopoiesis dataset (Granja et al., 2019) colored by published cell type annotations (B) and with projected HSPCs colored in red (C). (D) Bar plot depicting proportions of cell type identities transferred after projection into the publicly available hematopoiesis dataset for each peak disease severity score bin. For all box plots, points are colored by the peak disease severity score, shaped according to disease acuity, and grouped by the disease severity score at the time of sample collection. *, P < 0.05; **, P < 0.01; ns, not significant at P = 0.05 by two-sided Wilcoxon rank-sum test with Bonferroni’s correction for multiple hypothesis testing. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; BMMC, bone marrow mononuclear cells; LMPP, lymphomyeloid-primed multipotent progenitor; GMP, granulocyte-monocyte progenitor; HSC, hematopoietic stem cell.
Discussion
In this work, we have compiled a trimodal single-cell atlas of immune cells from COVID-19 patients with a wide range of disease severities through scRNA-seq, scATAC-seq, and CyTOF. By virtue of our whole blood analyses and multimodal approach, our analysis reveals novel mechanisms of immune activation and dysregulation in the setting of severe COVID-19, in addition to providing critical validation of results from other studies. We found that neutrophils and NK cells appeared strongly activated with increasing disease severity, with heightened ISG induction and increased expression of cytotoxic machinery. Conversely, monocytes and DCs displayed dysregulated and tolerogenic features in severe and fatal COVID-19. Finally, we found dramatic changes in hematopoietic development in severe COVID-19, with the appearance of a population of developing neutrophils in the periphery and skewing of circulating hematopoietic precursors toward the myeloid lineage.
Importantly, our profiling of patients with mild COVID-19 allows us to demonstrate that many of these changes occur largely in severe and fatal COVID-19 and not in milder forms of the disease. For example, the kinetics and role of local and systemic IFN signaling in ameliorating or exacerbating SARS-CoV-2 remain controversial (Blanco-Melo et al., 2020; Giamarellos-Bourboulis et al., 2020; Broggi et al., 2020; Hadjadj et al., 2020; Lee and Shin, 2020). Here, we noted minimal induction of ISGs in mild COVID-19 cases, regardless of the time point of infection. This suggests that robust IFN responses detectable in the periphery may not be required for disease resolution. Additionally, we observed surprisingly little perturbation of monocytes and NK cells from mild COVID-19 patients, whereas mild cases of influenza are known to induce systemic activation of these cells (Andres-Terre et al., 2015; Nikitina et al., 2018). Direct comparative analyses and larger sample sizes will be necessary to identify conserved or differential features between mild COVID-19 and other mild respiratory virus infections.
Our analysis demonstrated a strong severity-associated hyperactivation phenotype in peripheral neutrophils marked by broad ISG induction, pro-inflammatory cytokine–encoding gene production, enrichment of phagocytosis and degranulation gene sets, and up-regulation of epigenetic regulators associated with inflammatory neutrophils, such as PADI4, which is required for NETosis. Neutrophils in severe COVID-19 also strongly up-regulated S100A8 and S100A9, which dimerize to form the inflammatory molecule calprotectin, which is involved in neutrophil activation and chemotaxis (Ryckman et al., 2003). Additionally, we found features associated with neutrophil exhaustion, like up-regulation of CD274, similar to other studies (Schulte-Schrepping et al., 2020). While this finding has led some groups to conclude that neutrophils become “suppressive” in severe COVID-19 (Schulte-Schrepping et al., 2020), we believe our findings, combined with accumulating evidence that NETosis contributes to tissue injury and thrombotic complications in severe COVID-19 (Barnes et al., 2020; Radermecker et al., 2020; Zuo et al., 2020; Veras et al., 2020; Middleton et al., 2020), suggest a predominantly pathogenic role for circulating neutrophils in severe COVID-19. Importantly, these data provide insight into the mechanistic pathways that drive neutrophil activation. Targeting such pathways may provide new therapeutic opportunities. For example, an agonist of a neutrophil-expressed inhibitory receptor Siglec-10 (SACCOVID) has shown promising results in suppressing hyperinflammation in severe COVID-19 and is in a phase III clinical trial (Tian et al., 2018, 2020; Chen et al., 2009). Agonists against other neutrophil-expressed inhibitory receptors, like Siglec-9, may also represent novel therapeutic candidates (Delaveris et al., 2021).
In addition to identifying features of severe COVID-19, we were also able to identify key differences between the immune responses of patients with severe COVID-19 who went on to survive the disease versus those who did not. A striking difference between fatal and nonfatal cases was the emergence of a population of developing neutrophils that was first described by our group (Wilk et al., 2020). In this earlier work, we identified these cells in 4 of 4 patients with ARDS requiring mechanical ventilation, including in one patient who died of infection. We now show the presence of these cells in 17 additional patients, including 5 of 7 patients with severe COVID-19 and 6 of 6 patients with fatal COVID-19. Our trajectory analyses demonstrate that these cells follow the stages of canonical neutrophil development, beginning with defensin-rich promyelocytes and differentiating through metamyelocytes and bands to form mature neutrophils. This process may be driven by elevated levels of circulating pro-inflammatory cytokines, such as IL-17, that may induce the formation of neutrophils (Megna et al., 2020). Although any functional or pathological role for these cells in COVID-19 pathogenesis remains unclear, their abundance in the periphery of patients with fatal COVID-19 enabled us to demonstrate that their most defining transcripts could be used to predict 28-d mortality in an independent bulk transcriptomic dataset. Although emergency myelopoiesis is known to be a feature of bacterial sepsis (Loftus et al., 2018; Scumpia et al., 2010), it is likely that emergency myelopoiesis is also an underappreciated feature of severe viral infection. For example, in an integrated multicohort analysis of viral disease severity, Zheng et al. (2021) reported a gene module that includes several markers of immature neutrophils (e.g., CEACAM8, DEFA4, LCN2) that is enriched in other viral infections, including influenza and respiratory syncytial virus. In addition, a six-mRNA classifier of viral disease severity, which includes DEFA4, trained on non–COVID-19 viral infections was found to also predict COVID-19 severity (Buturovic et al., 2021 Preprint). These results suggest that emergency myelopoiesis is a common feature of multiple severe viral infections and may be used to predict adverse outcomes.
In addition to canonical and immature neutrophils, the phenotype of NK cells in fatal COVID-19 cases was also distinct from that of severe nonfatal cases. Although circulating NK cells are known to become activated in severe COVID-19 (Maucourant et al., 2020), our work is the first to show that patients with fatal COVID-19 may fail to up-regulate the proliferation marker Ki-67 or the cytotoxic effector perforin to the same extent as patients with severe but nonfatal COVID-19. The absence of this activation could indicate a defect in the functional responses of NK cells in fatal COVID-19. This finding requires direct validation, although it has been reported that NK cells from severe and fatal COVID-19 patients appear to exhibit poor IFNγ production in response to K562 target cells (Varchetta et al., 2021). Unfortunately, we were unable to investigate potential epigenetic mechanisms behind this finding, because only one fatal case was analyzed by scATAC-seq. We did not detect significantly increased accessibility of the genes encoding perforin and Ki-67 in the NK cells of any COVID samples, possibly because factors other than promoter/enhancer accessibility may play more important roles under such pathological conditions. For example, it has been shown that IL-2 could increase not only the transcription rate of PRF1 but also the stability of the mRNAs in NK cells (Salcedo et al., 1993). We also identify NK cell receptor–ligand axes that may contribute to their activation in severe COVID-19. Our data suggest that DNAM-1–mediated recognition of Nectin-2 and NKG2D-mediated recognition of ULBP ligands could drive NK cell activation in COVID-19. These data are consistent with other viral infections in which activation of NK cells through DNAM-1 or NKG2D is important (Cifaldi et al., 2019; Kurioka et al., 2018), and they highlight pathways that should be investigated through in vivo model systems for their role in disease outcome.
Although neutrophils and NK cells are activated in COVID-19, monocytes and DCs appear to take on a tolerogenic phenotype. Perturbations in the cDC2s of patients with severe COVID-19 could inhibit the priming of T follicular helper cells and the development of mucosal immunity (Soto et al., 2020). Both cDC2s and monocytes of critically ill COVID-19 patients appeared to down-regulate or failed to up-regulate genes involved in activation and inflammation; for example, cDC2s lost expression of FCER1A and CD83, whereas CD14+ monocytes notably did not up-regulate any genes encoding pro-inflammatory cytokines such as IL-6 and CCL3, and genes encoding IL-1β and TNF were significantly down-regulated in these cells compared with those of healthy controls. This lack of pro-inflammatory cytokine expression is of note, as other viral infections such as influenza drive an increase in the production of these molecules by peripheral monocytes (Nikitina et al., 2018). We identified the inactivation of NF-κB family TFs as a possible epigenetic mechanism for this silencing of pro-inflammatory cytokine production in monocytes, because we observed a striking loss of accessibility at an NF-κB binding site within the IL1B locus. This observed inactivation is unusual, given that these TFs typically play an important role in antiviral immune responses, including through regulating the production of cytokines (Schmitz et al., 2014). Indeed, NF-κB family TFs have been implicated in driving the pro-inflammatory cytokine production in other cell types during SARS-CoV-2 infection (Hirano and Murakami, 2020). Our hypothesized linkage between IL-1B and NF-κB activity provides an avenue for future experiments.
There are several limitations to this work that should be noted. Though large for a multimodal dataset, our sample size is still limited. Moreover, we were unable to profile each patient by all three single-cell modalities, preventing us from performing cross-modality validation on a per-patient basis. Additionally, our CyTOF panels only allowed us to examine a limited number of cell types, preventing us from performing orthogonal validation of some transcriptional or epigenetic findings in proteomic space. It is also difficult to fully control for the impact of treatment on the immune profile because the standard of care varies with clinical severity, though we often collected blood before treatments were administered. This work profiled exclusively circulating immune cells; although understanding the peripheral immune system is critical to understanding aberrant and protective immune responses to SARS-CoV-2 infection, it does not capture the immune response at the site of infection. Finally, further functional experimentation is necessary to validate or refute many of the hypotheses presented here.
Collectively, our work represents the first trimodal epigenomic, proteomic, and transcriptomic cell atlas of peripheral immune responses to COVID-19 across a broad spectrum of disease severity. By identifying novel immune features associated with COVID-19 mortality, as well as the immune status of patients with mild disease, our work enhances our understanding of pathological versus protective immune responses and highlights several opportunities for therapeutic development.
Materials and methods
Subjects and specimen collection
We collected blood from 64 patients enrolled in the Stanford University COVID-19 Biobanking studies from March to June 2020 after receiving written informed consent from patients or their surrogates (Stanford Institutional Review Board approvals 28205, 55650, and 55689). Eligibility criteria included age ≥18 yr and a positive SARS-CoV-2 nasopharyngeal swab by RT-PCR. All patients who presented to the Stanford University Emergency Department were offered enrollment, regardless of admission, and patients already admitted to the wards or intensive care unit were also eligible for enrollment. Outpatients with mild COVID-19 under the care of Stanford Health Care through the care and respiratory observation of patients with novel coronavirus clinic were also eligible for enrollment. The majority of admitted patients were coenrolled in ongoing COVID-19 treatment trials at Stanford. Screening of new admissions via an electronic medical record review of all subjects was performed by the study coordinators (J. Roque, R. Mann, and H. Din) and the study principal investigators (A.J. Rogers, S. Yang, and K.C. Nadeau).
Patients were phenotyped for both peak disease severity and severity at the time of sample collection according to the WHO severity score via an electronic medical record review performed by A.J. Wilk and D. Jimenez-Morales (https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf). Briefly, the WHO severity score is an ordinal ranking score (0–8), where 0 indicates no evidence of infection. In this study, we classify patients with scores 1–3 as having “mild” disease, corresponding to no requirement for supplemental oxygen. Scores of 4–5 describe patients with “moderate” disease who are hospitalized and require noninvasive supplemental oxygen. Scores of 6–8 indicate “severe” infection requiring mechanical ventilation. Clinical data were obtained through the Stanford Research Repository, Stanford Medicine’s approved resource for working with clinical data for research purposes extracted from the Epic database management system used by the Stanford hospitals. All fatal COVID-19 cases were confirmed by the principal investigator A.J. Rogers to have been primarily the result of COVID-19 and not of any comorbidities.
To protect the identity of the COVID-19 subjects, ages are reported as ranges. For controls, blood was collected from eight asymptomatic adult donors as part of the Profiling Healthy Immunity study after written informed consent was obtained (Stanford Institutional Review Board approval 26571). All donors were asked for consent for genetic research. Blood draws from patients occurred in concert with usual care to avoid unnecessary personal protective equipment use. For both patients with COVID-19 and healthy controls, blood was collected into cell preparation tubes (CPTs) or heparin vacutainers (Becton, Dickinson and Co.; see Table S1). For samples collected in CPT tubes, PBMCs were isolated by centrifugation and washed with Ca/Mg-free PBS. For samples collected in heparin tubes, PBMCs were isolated by density gradient centrifugation using Ficoll-Paque Plus medium (GE Healthcare) and washed with Ca/Mg-free PBS. For a subset of patients, whole blood was removed from CPT vacutainers before centrifugation and treated with ACK red blood cell lysis buffer until the cell pellet appeared visually clear, and these cells were processed for single-cell transcriptomics. Blood was processed within 6 h of collection for all samples. Samples from patients with COVID-19 and from healthy controls were processed side by side to avoid variation from processing. All scRNA-seq processing was performed on samples before cryopreservation. All CyTOF and scATAC-seq processing was performed on samples cryopreserved under the vapor phase of liquid nitrogen.
scRNA-seq by Seq-Well
The Seq-Well platform for scRNA-seq was used as described previously (Gierahn et al., 2017; Hughes et al., 2019,Preprint; Wilk et al., 2020; Kazer et al., 2020). Immediately after Ficoll separation, 50,000 PBMCs were resuspended in RPMI + 10% FBS at a concentration of 75,000 cells/ml. 200 µl of this cell suspension (15,000 cells) was then loaded onto Seq-Well arrays preloaded with mRNA capture beads (ChemGenes). Following four washes with Dulbecco’s PBS to remove serum, the arrays were sealed with a polycarbonate membrane (pore size of 0.01 µm) for 30 min at 37°C and then frozen at −80°C for no less than 24 h and no more than 14 d to allow batching of samples processed at irregular hours. Next, arrays were thawed, cells lysed, transcripts hybridized to the mRNA capture beads, and beads recovered from the arrays and pooled for downstream processing. Immediately after bead recovery, mRNA transcripts were reverse transcribed using Maxima H-RT (Thermo Fisher Scientific; EPO0753) in a template-switching–based rapid amplification of cDNA ends reaction, excess unhybridized bead-conjugated oligonucleotides removed with Exonuclease I (New England Biolabs; M0293L), and second-strand synthesis performed with Klenow fragment (New England Biolabs; M0212L) to enhance transcript recovery in the event of failed template switching (Hughes et al., 2019,Preprint). Whole-transcriptome amplification was performed with KAPA HiFi PCR Master Mix (Kapa Biosystems; KK2602) using ∼6,000 beads per 50-µl reaction volume. Resulting libraries were then pooled in sets of 6 (∼36,000 beads per pool), and products were purified by Agencourt AMPure XP beads (Beckman Coulter; A63881) with a 0.6× volume wash followed by a 0.8× volume wash. Quality and concentration of whole-transcriptome amplification products was determined using an Agilent Fragment Analyzer (Stanford Functional Genomics Facility), with a mean product size >800 bp and a nonexistent primer peak indicating successful preparation. Library preparation was performed with a Nextera XT DNA library preparation kit (Illumina; FC-131-1096) with 1 ng of pooled library using dual-index primers. Tagmented and amplified libraries were again purified by Agencourt AMPure XP beads with a 0.6× volume wash followed by a 1.0× volume wash, and quality and concentration were determined by fragment analysis. Libraries between 400 and 1,000 bp with no primer peaks were considered successful and pooled for sequencing. Sequencing was performed on a NovaSeq 6000 instrument (Illumina; Chan Zuckerberg Biohub). The read structure was paired end with read 1 beginning from a custom read 1 primer (Gierahn et al., 2017) containing a 12-bp cell barcode and an 8-bp UMI, and with read 2 containing 50 bp of mRNA sequence.
Alignment and quality control of scRNA-seq data
Sequencing reads were aligned and count matrices assembled using Spliced Transcripts Alignment to a Reference (STAR; Dobin et al., 2013) and dropEst (Petukhov et al., 2018), respectively. Briefly, the mRNA reads in read 2 demultiplexed FASTQ files were tagged with the cell barcode and UMI for the corresponding read in the read 1 FASTQ file using the dropTag function of dropEst. Next, reads were aligned with STAR using the GRCh37.p13 (hg19) human reference genome from Ensembl that included the complete genome sequences for all SARS-CoV-2 strains sequenced from California before March 24, 2020 (10 SARS-CoV-2 sequences). No SARS-CoV-2 reads were aligned from these samples using this strategy, even when the outFilterMultimapNmax behavioral option of STAR was increased from 10 (default) to 20 to accommodate potential multiple-mapping SARS-CoV-2 reads. Count matrices were built from resulting BAM files using dropEst (Petukhov et al., 2018). Count matrices for intron-aligned reads were also generated in order to computationally analyze cellular trajectory. Cells that had <750 UMIs or >15,000 UMIs, as well as cells that contained >20% of reads from mitochondrial genes or rRNA genes (RNA18S5 or RNA28S5) were considered to be of low quality and removed from further analysis. To remove putative multiplets (where more than one cell may have loaded into a given well on an array), cells that expressed >75 genes per 100 UMIs were also filtered out. Genes that were expressed in <10 cells were removed from the final count matrix.
Preprocessing of scRNA-seq data
The R package Seurat (Stuart et al., 2019; Butler et al., 2018) was used for data scaling, transformation, clustering, dimensionality reduction, differential expression analysis, and most visualizations. Data were scaled and transformed and variable genes identified using the SCTransform() function, and linear regression was performed to remove unwanted variation due to cell quality (e.g., percentage mitochondrial reads, percentage rRNA reads). Principal component (PC) analysis (PCA) was performed using the 3,000 most highly variable genes, and the first 50 PCs were used to perform Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) to embed the dataset into 2 dimensions. Next, the first 50 PCs were used to construct a shared nearest neighbor graph (SNN; FindNeighbors()), and this SNN was used to cluster the dataset (FindClusters()). Although upstream quality control removed many dead or low-quality cells, some clusters in the full dataset, or in cell type subsets, were identified that were defined by few canonical cell lineage markers and enriched for genes of mitochondrial or ribosomal origin, and these clusters were removed from further analysis (Carter et al., 2018; Freytag et al., 2018). Manual annotation of cellular identity was performed by finding DEGs for each cluster using Seurat’s implementation of the Wilcoxon rank-sum test (FindMarkers()) and comparing those markers with known cell type–specific genes from previous datasets (Gutierrez-Arcelus et al., 2019; Villani et al., 2017; Kang et al., 2018; Nimmerjahn and Ravetch, 2008, 2006; Palmer et al., 2006). For per-donor DEG tests presented in Fig. 8 B, donors with <50 cells were excluded from analysis.
Automated annotation of granular cell types by Seurat v4
Seurat v4 introduced weighted nearest neighbors (WNNs) analysis as a strategy to integrate multimodal single-cell sequencing data and released a large multimodal dataset of 228 cell surface markers with simultaneous whole-transcriptome measurements (Hao et al., 2021). By using WNN to learn the relative utility of each data modality in each cell, this reference dataset contains highly robust and granular cell type annotations. The web application provided with the release of Seurat v4 (http://azimuth.satijalab.org/) was used to map our transcriptomic dataset to the annotated multimodal reference (Hao et al., 2021). Briefly, anchors between the query and reference dataset were identified using a precomputed supervised PCA on the reference dataset that maximally captures the structure of the WNN graph. Next, cell type labels from the reference dataset, as well as imputations of all measured protein markers, are transferred to each cell of the query through the previously identified anchors. Finally, the query dataset is projected onto the UMAP structure of the reference.
Calculation of transcriptomic perturbation score
To prioritize analysis of cell types of interest, we calculated a perturbation score for each cell type of each sample as previously described (Hao et al., 2021; Papalexi et al., 2021). This perturbation score is motivated by the observation that the statistical significance of per-gene differential expression tests is strongly influenced by the number of cells in each cluster or cell type. To overcome this, we first identified a set of genes for each cell type that showed evidence of differential expression (P < 0.1) between healthy controls and all COVID-19 patients. From this set of genes, we removed ISGs and Ig genes because the former are broadly up-regulated across cell types (Fig. S2) and the latter are cell type specific, not perturbation specific. Next, we defined the global perturbation vector as the average log fold change of each DEG relative to healthy control subjects normalized to length 1. Finally, we projected the transcriptome of each sample onto this vector and defined the perturbation score as the absolute value of the magnitude of this projection. This approach enables prioritization of cell type perturbations when comparing cell types of different abundances.
Gene module scoring analysis
The Seurat function AddModuleScore() was used to score single cells by expression of a list of genes of interest. This function calculates a module score by comparing the expression level of an individual query gene to other randomly selected control genes expressed at similar levels to the query genes and is therefore robust to scoring modules containing both lowly and highly expressed genes, as well as to scoring cells with different sequencing depths. Gene lists used to define each module are listed in Table S16. Methods used to select genes from published datasets varied based on the availability, format, and modality of data. For the MDSC gene set described by Alshetaiwi et al. (2020), MDSC DEGs with a log fold change >0.25 relative to monocytes were used. To estimate the expression of sepsis-related genes, all positively enriched genes in the MS1 module versus MS2 module were used (Reyes et al., 2020). For bulk transcriptomic or microarray datasets, including human monocyte precursor subsets (Kawamura et al., 2017), LPS-stimulated PD-L1+ neutrophils (de Kleijn et al., 2013) and neutrophils from ARDS-complicated sepsis (Juss et al., 2016), the top 97th percentile of DEGs relative to control samples/patients were used for scoring.
TF activity prediction analysis
The iRegulon plugin available through Cytoscape was used to predict TFs that may contribute to the observed transcriptomic changes. Gene lists supplied to iRegulon are available in Table S19. In brief, iRegulon calculates the likelihood of TF activity first by compiling for each TF motif a ranked list of genes containing that motif near the transcription start site (TSS) and then by determining the ability of each TF motif to recover the genes supplied as input. TFs with a normalized enrichment score >4 and >20 predicted targets, along with their corresponding predicted regulomes, are plotted for visualization.
Analysis of developing neutrophil trajectories
RNA velocity analysis was performed on the full dataset with the package scVelo to visualize RNA velocity field vectors and streams as well as to calculate the latent time of each developing neutrophil. Developing neutrophils were subsetted from the main object using manual cell type annotations. Upon subclustering, clusters in the subsetted object that appeared to be PBs were removed from further trajectory and DEG analysis. PHATE (Moon et al., 2019) was performed on scaled and transformed expression values for the 3,000 most highly variable genes to embed the subsetted dataset into 2 or 3 dimensions. Plots of expression of individual genes along inferred latent time are scaled at the 5th and 95th percentiles.
Projection of transcriptomic dataset onto published blood and bone marrow hematopoiesis dataset
Cells annotated as HSPCs by Seurat v4 were projected into a publicly available hematopoiesis dataset of CD34+-enriched bone marrow mononuclear cells (Granja et al., 2019) using an anchor-based integration strategy. Briefly, expression values from each dataset were normalized and variable features identified using SCTransform without covariate regression. Next, anchors were identified between the two datasets, the datasets were integrated using these anchors, and PCA and UMAP were performed as described above on the integrated gene expression matrix. To determine the identities of the projected cells, TransferData() was used to transfer cell type labels from the cells from the published dataset in the integrated object to the HSPCs from the COVID-19 dataset.
Mortality prediction using developing neutrophil DEGs
To test whether the gene signature of developing neutrophils could be used to predict COVID-19 mortality, we first developed a five-gene signature of developing neutrophils by identifying the most differentially enriched genes in developing neutrophils in our transcriptomic dataset relative to all other cells. Next, we downloaded normalized transcript counts from a publicly available whole blood bulk transcriptomic dataset published by Overmyer et al. (2021). We then scored each COVID-19 sample in this dataset by the expression of the five developing neutrophil-enriched genes (DEFA1B, DEFA3, LTF, DEFA1, and S100A8) using AddModuleScore(). Finally, we used these gene signature scores as a predictor variable and 28-d mortality as reported by metadata from Overmyer et al. as the response variable to construct an ROC curve to quantify and visualize the sensitivity and specificity of the prediction.
NK cell isolation
After thawing of PBMCs, 500,000 live PBMCs were set aside from each sample for staining with a broad lineage panel that includes NK cell ligands (ligand panel), and the remaining cells were used for NK cell isolation. NK cells were isolated from whole PBMCs via Miltenyi Biotec Human NK Cell Isolation Kits, which use negative selection, per the manufacturer’s instructions.
Mass cytometry
All antibodies not purchased directly from Fluidigm were conjugated using MaxPar X8 conjugation kits (Fluidigm). To ensure staining consistency, all antibodies except those noted were precombined into staining cocktails and either lyophilized (NK surface and intracellular cytokine staining panels) or frozen at −80°C (ligand panel) for long-term storage. Samples were barcoded with a four-choose-two scheme, using palladium isotopes (Pd102, Pd104, Pd106, Pd108) conjugated to anti-CD45 antibodies as previously described (Vendrame et al., 2020).
After isolation of NK cells, both the whole PBMCs and the isolated NK cells from each sample were stained with cisplatin for 1 min in order to allow viability determination and subsequently quenched with FBS. Samples were then stained with palladium-CD45 barcodes as previously described (Vendrame et al., 2020). After barcode staining at 4°C, samples were washed thoroughly with CyFACS buffer (PBS, 0.1% BSA, 2 mM EDTA, 0.05% sodium azide) and combined into sets of barcodes, hereafter referred to as “barcoded samples.” Barcoded samples were subsequently washed and stained with relevant surface panels (Table S22) for 30 min at 4°C. After surface staining, the barcoded samples were washed with CyFACS buffer and fixed in 2% paraformaldehyde for 20 min at room temperature. Barcoded samples were subsequently permeabilized with 1× BD Perm II (BD Biosciences), and NK cell samples were stained with the lyophilized intracellular cytokine staining panel for 45 min at 4°C (whole PBMCs were resuspended in 1× BD Perm II and left at 4°C for the same duration without any stains). All of the barcoded samples were then resuspended in iridium interchelator (DVS Sciences) in 2% paraformaldehyde until collection (within 3 d of staining). Data were collected on a Helios mass cytometer. Before collection, samples were washed with CyFACS buffer and Milli-Q water before being resuspended in 1× EQ beads (Fluidigm) for collection.
Preprocessing and data analysis of mass cytometry data
Flow cytometry standard (FCS) files were normalized and debarcoded using the Premessa package as previously described (Zunder et al., 2015; Finck et al., 2013). FlowJo version 10.7.1 was used to manually gate out EQ beads, dead cells, doublets, and debris from both whole PBMC and NK cell samples. Additional lineage markers were used to exclude contaminating non-NK cells from the samples consisting of bead-purified NK cells. Live, intact, singlet PBMCs were exported from whole PBMC samples as FCS files, whereas live, intact, singlet NK cells were exported from NK cell samples using the gating schemes described in Fig. S1, K and L. All subsequent preprocessing and downstream analysis of data were performed using the open-source software R. NK cell FCS files were normalized to account for batch effects using the CytoNorm package (Van Gassen et al., 2020); this normalization was not performed on the whole PBMC files, because no such batch effects were observed. Parameter names were altered in whole PBMC FCS files using the Premessa panel editor tool to ensure consistency across all samples. Seurat objects were created from FCS files using the Seurat and FlowCore packages. Unsupervised clustering was performed on the whole PBMC Seurat object via the phenotyping by accelerated refined community-partitioning algorithm (Stassen et al., 2020), using all proteomic parameters listed in Table S22 with the exception of HLA-Bw4 and HLA-Bw6. These parameters were excluded because their expression is mutually exclusive and can therefore drive over-clustering. Clustering was performed on whole PBMCs with a resolution of 0.19 and on subsetted monocyte clusters with a resolution of 0.21. Clustering resolutions were determined using cluster tree plots. Clustering and UMAP embeddings were performed on arcsinh-transformed data (cofactor = 5). All box plots depicting CyTOF data represent transformed per-sample mean signal.
scATAC-seq
scATAC-seq was performed by using the Chromium Next GEM Single Cell ATAC Reagent Kits version 1.1 (10x Genomics; PN-1000175) and following the demonstrated protocol (nuclei isolation for scATAC-seq) provided by 10x Genomics. Briefly, cryopreserved PBMCs were thawed, and 50,000–500,000 cells were aliquoted, washed with PBS, and then lysed with 100 µl lysis buffer for 4 min. The lysed nuclei were centrifuged after washing with 1 ml washing buffer and then resuspended in diluted nuclei buffer. The nuclei concentration was then determined with a TC20 Automated Cell Counter (Bio-Rad Laboratories; 1450102), and ∼6,000 nuclei were used for Tn5 transposition, single nuclei barcoding, and library preparation following the instructions in the kit. The final DNA libraries were sequenced with the NovaSeq 6000 system (Illumina; Chan Zuckerberg Biohub), leading to ∼200 million reads per sample.
scATAC-seq preprocessing and cell quality filtering
Demultiplexed sequencing reads were aligned to the GRCh37 (hg19) human reference genome using cellranger-atac software (10x Genomics; version 1.2) or cellranger-arc (10x Genomics; version 1.0) for the multi-omic reference dataset. Resulting fragment files were processed using ArchR (Granja et al., 2021) and filtered based on TSS enrichment and the log10 fragments per cell. Cell quality cutoff values were set on a per-sample basis to account for variable sequencing depths and sample qualities. The TSS cutoff ranged from 5 to 10, and the log10 fragment cutoff ranged from 3.2 to 4.1, such that the large number of non–cell-containing droplets were excluded from further consideration (Table S23). Cross–cell type doublets were computationally identified using ArchR’s addDoubletScores function and filtered based on a maximum doublet enrichment of 2 (i.e., regions of the manifold that are twofold enriched for simulated doublets).
scATAC-seq reference-based cell type annotation
A public multi-omic dataset from 10x Genomics on healthy PBMCs was used as an intermediate for cell type calls (https://support.10xgenomics.com/single-cell-multiome-atac-gex/datasets/1.0.0/pbmc_granulocyte_sorted_10k) because it could be readily integrated with both scRNA-seq and scATAC-seq datasets. The multi-omic dataset was realigned to hg19 using cellranger-arc version 1.0 (10x Genomics), and the resulting RNA count matrix was filtered for cells with between 1,000 and 15,000 reads and <20% mitochondrial genes. Next, cell types were annotated using the Azimuth tool from Seurat v4 in the same manner as the scRNA dataset (Hao et al., 2021). MACS2 was used to call peaks for each of the Azimuth-annotated cell types via ArchR’s addReproduciblePeakSet, a peak count matrix was calculated using addPeakMatrix, and then the peak matrix was dimensionality reduced using the addIterativeLSI function from ArchR with iterations = 3, varFeatures = 50,000, and sampleCellsPre = 50,000. This established an ATAC-based dimensionality reduction coupled to RNA-based automated cell type annotations.
Each scATAC sample from our dataset was then projected into the linear dimensionality reduction of the multi-omic dataset. Then Seurat’s internal FindAnchors function and TransferData functions (Stuart et al., 2019) were used to transfer cell type annotations from the multi-omic dataset to each scATAC dataset. Briefly, anchor pairs between the datasets are identified based on mutual nearest neighbors, then cell type calls are transferred for each cell using a distance-weighted sum of nearby anchors. The anchor filtering step was omitted (k.filter = NA) as suggested in the scATAC-seq data integration vignette from Signac (Stuart et al., 2020 Preprint). This avoids over-filtering of anchors due to the extremely sparse nature of the underlying ATAC-seq data.
scATAC batch correction and dimensionality reduction
Cells passing quality and doublet filters from each sample were combined into a linear dimensionality reduction using ArchR’s addIterativeLSI function. This dimensionality reduction was then corrected for batch effect based on processing date using the Harmony method (Korsunsky et al., 2019), via ArchR’s addHarmony function. The cells were then clustered based on the batch-corrected dimensions using ArchR’s addClusters function. Briefly, this uses a modularity-based clustering of the SNN graph as implemented in Seurat. Three doublet clusters were manually identified from these clusters, based on containing a mixture of many cell types and elevated doublet enrichment scores (although still below the doublet cutoff threshold of 2). These doublet clusters were removed from further consideration in all downstream analyses.
scATAC TF activity analysis
For TF activity analysis in CD14 monocytes, peaks were called on CD14 monocyte cells using the addReproduciblePeakSet from ArchR. Then, peaks containing NF-κB2 motifs were identified using the addPeakAnnotations function, which relies on the chromVARmotifs package’s human_pwms_v2 motif set curated from the cisBP database. Motif deviation z-scores for individual cells were calculated using ArchR’s addDeviationsMatrix function. Briefly, this method compares accessibility across all peaks containing a TF motif to accessibility across a background set of peaks matched for guanine-cytosine content and overall accessibility. This measures global changes in accessibility associated with TF activity while controlling for technical confounders.
Hi-C data
Hi-C data for monocytes was taken from Phanstiel et al. (2017), downloaded using Juicer-Tools version 1.22 (Durand et al., 2016).
Data visualization
Wrappers provided by Seurat were used to generate UMAP projections and dot plots. ComplexHeatmap was used to generate all heatmaps, and plotly was used to visualize three-dimensional PHATE projections. scVelo was used to visualize RNA velocity streams. Custom ggplot functions (see Data availability) were used to generate all other plots. For all box plots, features include minimum whisker, 25th percentile − 1.5 × interquartile range or the lowest value within; minimum box, 25th percentile; center, median; maximum box, 75th percentile; maximum whisker, 75th percentile + 1.5 × interquartile range or greatest value within.
Online supplemental material
Fig. S1 shows quality control data for the CyTOF and scRNA-seq datasets, additional information regarding the set of patient samples collected through each modality, and CyTOF cell type annotations. Fig. S2 shows the impact of age on cell type proportions and conserved features of the IFN response between patients across cell types, both in the scRNA-seq dataset. Fig. S3 shows the effect of disease acuity on the transcriptional phenotypes of mild and moderate COVID-19 patients. Fig. S4 shows quality control data demonstrating the efficacy of Seq-Well at capturing the transcriptomes of primary human neutrophils. Fig. S5 shows additional analysis of neutrophil development in fatal COVID-19. Table S1 presents patient demographics and other clinical metadata. Table S2 shows the per-donor cell counts for each manually annotated cell type in the scRNA-seq dataset. Table S3 shows the per-donor cell counts for each Seurat v4–annotated cell type in the scRNA-seq dataset. Table S4 shows the per-donor cell counts for each cell type in the CyTOF dataset. Table S5 shows the per-donor cell counts for each cell type in the scATAC-seq dataset. Table S6 shows the results of differential gene expression testing for each manually annotated cell type in the scRNA-seq dataset. Table S7 shows per-donor cell type proportions for each sample input in the scRNA-seq dataset. Table S8 shows per-donor cell type proportions for each cell type in the CyTOF dataset. Table S9 shows per-donor cell type proportions for each cell type in the scATAC-seq dataset. Table S10 shows per-donor cell type proportions for Seurat v4–annotated cell types in the scRNA-seq dataset. Table S11 shows the results of differential gene expression testing for each Seurat v4–annotated cell type in the scRNA-seq dataset. Table S12 shows the results of differential imputed protein expression testing for each Seurat v4–annotated cell type in the scRNA-seq dataset. Table S13 shows the results of differential gene expression testing for cDC2 cells in each COVID-19 severity group in the scRNA-seq dataset. Table S14 shows the results of differential gene expression testing for CD8 TEM cells in each COVID-19 severity group in the scRNA-seq dataset. Table S15 shows the results of differential gene expression testing for monocytes in each COVID-19 severity group in the scRNA-seq dataset. Table S16 lists the member genes for each module used for gene module scoring in the scRNA-seq dataset. Table S17 shows the results of differential gene expression testing for NK cells in each COVID-19 severity group in the scRNA-seq dataset. Table S18 shows the results of differential gene expression testing for neutrophils in each COVID-19 severity group in the scRNA-seq dataset. Table S19 lists the genes used as input for iRegulon analysis. Table S20 shows the results of differential gene expression testing for each cluster of developing neutrophils in the scRNA-seq dataset. Table S21 shows the DEGs for HSPCs between healthy controls and COVID-19 patients in the scRNA-seq dataset. Table S22 lists the antibodies used for CyTOF proteomic profiling. Table S23 shows quality control information for processing the scATAC-seq dataset. Table S24 shows the results of differential gene expression testing for each cell type in the healthy donor neutrophil scRNA-seq dataset presented in Fig. S4.
Data availability
FCS files (mass cytometry) with de-identified metadata supporting this publication are available at ImmPort (https://www.immport.org) under study accession no. SDY1708. Processed scRNA-seq data are hosted on the COVID-19 Cell Atlas (https://www.covid19cellatlas.org/). Data from scRNA-seq and scATAC-seq have been deposited with the Gene Expression Omnibus under accession no. GSE174072. A Github repository for all original code used for analysis and visualization is available at https://github.com/ajwilk/COVID_scMultiome.
Acknowledgments
We are grateful to all participants in this study. We thank Sopheak Sim and the Stanford Functional Genomics Facility for the use of the Fragment Analyzer, as well as Angela Detweiler and Michelle Tan at the Chan Zuckerberg Biohub for assistance with sequencing. We thank Anne-Maud Ferreira for insightful conversation on mass cytometry analysis. We thank Yuhan Hao and Rahul Satija for helpful discussions on the calculation and interpretation of the perturbation score. We thank Martin Prlic for assistance with mass cytometry gating schemes. We thank Nima Aghaeepour for his assistance in implementing the CytoNorm package. Figure illustrations were created using BioRender.com.
The Stanford ICU Biobank and A.J. Rogers are funded by National Heart, Lung, and Blood Institute grant K23 HL125663. A.J. Wilk is supported by the Stanford Medical Scientist Training Program (T32 GM007365-44) and the Stanford Bio-X Interdisciplinary Graduate Fellowship. B. Wei is supported by the Wallenberg Foundation Post-Doctoral Fellowship. B. Parks is supported by a training grant from the National Institute of Standards and Technology. C.A. Blish is supported by National Institute on Drug Abuse DP1 (DP1 DA046089), a 2019 Sentinel Pilot Project from the Bill and Melinda Gates Foundation; Bill and Melinda Gates Foundation grant OPP113682; and Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases (1016687). C.A. Blish is the Tashia and John Morgridge Faculty Scholar in Pediatric Translational Medicine from the Stanford Maternal Child Health Research Institute and an Investigator of the Chan Zuckerberg Biohub. W.J. Greenleaf acknowledges funding from the Defense Advanced Research Projects Agency, support from National Institutes of Health grants P50-HG007735 and UM1-HG009442, UM1-HG009436, and U19-AI057266, and grants from the Chan-Zuckerberg Initiative and the Rita Allen Foundation. W.J. Greenleaf is a Chan–Zuckerberg Investigator. W.J. Greenleaf acknowledges funding from Emerson Collective. This research was funded in whole or in part by the Bill and Melinda Gates Foundation. For the purpose of Open Access, the author has applied a CC-BY public copyright license to any Author Accepted Manuscript (AAM) version arising from this submission.
Author contributions: A.J. Wilk, M.J. Lee, B. Wei, B. Parks, W.J. Greenleaf, and C.A. Blish conceived the project and designed experiments. A.L. Blomkalns, R. O’Hara, E.A. Ashley, K.C. Nadeau, S. Yang, A.J. Rogers, and C.A. Blish conceived the clinical cohort, obtained clinical samples, and provided clinical input. A.J. Wilk, D. Jimenez-Morales, E.A. Ashley, and A.J. Rogers obtained metadata for clinical samples. T. Ranganath coordinated clinical sample processing. Stanford COVID-19 Biobank members enrolled, consented, and processed clinical samples. A.J. Wilk, M.J. Lee, R. Pi, and N.Q. Zhao acquired single-cell transcriptomic data. B. Wei and W. Becker acquired scATAC-seq data. M.J. Lee, R. Pi, G.J. Martínez-Colón, and T. Ranganath acquired mass cytometry data. A.J. Wilk, M.J. Lee, B. Wei, and B. Parks performed bioinformatic and statistical analyses. S. Taylor, S. Holmes, and M. Rabinovitch provided intellectual input. A.J. Wilk, M.J. Lee, B. Wei, B. Parks, W.J. Greenleaf, and C.A. Blish wrote the manuscript, which was reviewed by all authors.
References
Competing Interests
Disclosures: A.J. Wilk reported grants from Stanford University Interdisciplinary Graduate Fellowship and NIH during the conduct of the study. M.J. Lee reported grants from NIH during the conduct of the study. B. Wei reported "Stanford University." E.A. Ashley reported "other" from Personalis, Inc., DeepCell, Inc., SVEXA Inc., Astra Zeneca, Gilead, MyoKardia, Amgen, Takeda, Novartis, Genome Medical, Avive, Samsung, Apple Inc., Google, Verily, Disney Inc., Illumina Inc., PacBio, Nanopore, Foresite Capital, and Sequence Bio outside the submitted work. K.C. Nadeau reported grants from National Institute of Allergy and Infectious Diseases, National Heart, Lung, and Blood Institute, Food Allergy Research and Education, and World Allergy Organization; "other" from Cour Pharma, Before Brands, Alladapt, Latitude, IgGenix, Immune Tolerance Network, and National Institutes of Health clinical research centers outside the submitted work; in addition, K.C. Nadeau had a patent to "mixed allergen composition and methods for using the same with royalties paid (Alladapt and Before Brands), a patent to "granulocyte-based methods for detecting and monitoring immune system disorders" issued, and a patent to "methods and assays for detecting and quantifying pure subpopulations of white blood cells in immune system disorders" issued. A.J. Rogers reported personal fees from Merck outside the submitted work. W.J. Greenleaf reported personal fees from 10x Genomics during the conduct of the study, and personal fees from Guardant Health and Protillion Biosciences outside the submitted work; in addition, W.J. Greenleaf had a patent to US20160060691A1 with royalties paid (10x Genomics). C.A. Blish reported personal fees from Catamaran Bio outside the submitted work. No other disclosures were reported.
Author notes
A.J. Wilk, M.J. Lee, B. Wei, and B. Parks contributed equally to this paper.
Stanford COVID-19 Biobank members: Thanmayi Ranganath, Nancy Q. Zhao, Aaron J. Wilk, Rosemary Vergara, Julia L. McKechnie, Lauren de la Parte, Kathleen Whittle Dantzler, Maureen Ty, Nimish Kathale, Giovanny J. Martínez-Colón, Arjun Rustagi, Geoff Ivison, Ruoxi Pi, Madeline J. Lee, Rachel Brewer, Taylor Hollis, Andrea Baird, Michele Ugur, Michal Tal, Drina Bogusch, Georgie Nahass, Kazim Haider, Kim Quyen Thi Tran, Laura Simpson, Hena Din, Jonasel Roque, Rosen Mann, Iris Chang, Evan Do, Andrea Fernandes, Shu-Chen Lyu, Wenming Zhang, Monali Manohar, James Krempski, Anita Visweswaran, Elizabeth J. Zudock, Kathryn Jee, Komal Kumar, Jennifer A. Newberry, James V. Quinn, Donald Schreiber, Euan A. Ashley, Catherine A. Blish, Andra L. Blomkalns, Kari C. Nadeau, Ruth O’Hara, Angela J. Rogers, Samuel Yang.
Supplementary data
shows counts of cells in scRNA-seq dataset for each donor per manually annotated cell type.
shows counts of cells in scRNA-seq dataset for each donor per Seurat v4–annotated cell type.
shows counts of cells in CyTOF dataset for each donor per manually annotated cell type.




![Quality control of scRNA-seq and CyTOF datasets. (A and B) UMAP embeddings of complete scRNA-seq dataset, colored by cell type input (A; either PBMCs or ACK-lysed whole blood [WB]) or donor (B). (C) Upset plot depicting overlap of patient samples profiled between the three modalities, colored by peak disease severity score. (D) Top: Heatmap showing overlap in Seurat v4 annotation calls (x axis) and manual cell type annotations (y axis), colored by the percentage of a manual cell annotation within a Seurat v4 annotation (i.e., each row sums to 100%). Bottom: Bar plot showing mapping frequency of each manually assigned cell type annotation by Seurat v4. Neutrophils and developing neutrophils are the least frequently assigned cell types because they are not present in the reference dataset (Hao et al., 2021). (E) Manual gating scheme for MAIT cells in the CyTOF dataset, beginning with live singlets gated according to the scheme in L. (F) Scatter plot depicting concordance with proportions of MAITs (top) or NK cells (bottom) predicted by Seurat v4 in the scRNA-seq dataset (x axis) and proportions manually gated in the CyTOF dataset (y axis). (G) UMAP embedding of the complete PBMC CyTOF dataset, colored by cell subset. (H) Heatmap showing the z-score normalized average expression of each marker in the PBMC CyTOF panel across all cell subsets detected in that dataset. (I) UMAP embedding of our whole PBMC CyTOF dataset, colored by donor. (K and L) Gating strategies used to identify live, intact singlets (L) and live, intact, singlet NK cells (K). (J) For each marker shared between the scRNA-seq and CyTOF datasets, a linear model was used to calculate a β coefficient for the relationship between severity score at the time of sample collection and marker expression in each dataset. The scatter plots depict the correlation between these β coefficients for markers measured on monocytes (top) and NK cells (bottom). For all scatter plots, Pearson’s r, exact two-sided P values, and the 95% confidence interval are shown.](https://cdn.rupress.org/rup/content_public/journal/jem/218/8/10.1084_jem.20210582/5/m_jem_20210582_figs1.png?Expires=1773646676&Signature=Ex0hBm~XRrbTTlmn-qbtGJnWh4logHIxKLQHiCiCYWCdBGmFqVWD7oAO85zU8CEAFhAyGWdDW84cpP-I4cCq7CP4ly57G~g~EY7lsUDcU2FzEq6z1CyptW5biVIHsiRVoCrbNSp4amZJT7vHE6MFPp5eAs3EiaipkBDYEQgbF69Bmxco8Ph2rl6vmTRd8VoSDV3BVjlXNWVC1QKj0~9m0OLh61jYxGnZFA6HVvchSqR2EodCOplKh3bsysg70dpExGBCKpcgEqwFfg~XW~Lpv5jc2SE3Tk1XtwniIW3olBMnIuFtD93zHHM67l8AHYyAiQQGPS65Y90q3hLZlJGK3g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)