Dendritic cells (DCs) contribute a small fraction of the tumor microenvironment but are emerging as an essential antitumor component based on their ability to foster T cell immunity and immunotherapy responses. Here, we discuss our expanding view of DC heterogeneity in human tumors, as revealed with meta-analysis of single-cell transcriptome profiling studies. We further examine tumor-infiltrating DC states that are conserved across patients, cancer types, and species and consider the fundamental and clinical relevance of these findings. Finally, we provide an outlook on research opportunities to further explore mechanisms governing tumor-infiltrating DC behavior and functions.
Introduction
Dendritic cells (DCs) were first described by Steinman, Cohn, and colleagues in a series of reports in this journal almost half a century ago (Steinman and Cohn, 1973, 1974; Steinman et al., 1974, 1975). The work that followed recognized these cells as key orchestrators of antigen-specific immunity and tolerance, despite their limited abundance in the body (Steinman, 2012). DCs were initially identified in mouse secondary lymphoid organs (SLOs), namely spleen and lymph nodes (Steinman and Cohn, 1973), which is significant considering that this localization enables two events that are necessary for antigen-specific immunity: (1) the capture and processing of antigens that originate from peripheral tissues, and (2) physical encounters with (including antigen cross-presentation to and activation of) rare clones of specific T cells that travel through lymphoid tissues and accumulate in so-called T cell areas where DCs are found.
The last several decades of work have revealed many additional features of DCs, including their origins, phenotypes, and functions (Steinman et al., 2003; Colonna et al., 2004; Geissmann et al., 2010; Merad et al., 2013; Guilliams et al., 2014; Murphy et al., 2016; Eisenbarth, 2019). These findings have also served to divide DCs operationally into several states, including two classical DC (cDC) states (cDC1 and cDC2), a plasmacytoid DC (pDC) state, and a monocyte-like inflammatory DC (MoDC) state. Here, cell states are broadly defined as multidimensional vectors (McKinley et al., 2020) and can include various measurable dimensions such as protein and mRNA expression or bodily location.
cDC1s are commonly thought to specialize in antigen cross-presentation and CD8+ T cell activation and are often described as XCR1+ CLEC9A+ CD141+ in humans. cDC2s can activate different types of CD4+ T helper cells, but also CD8+ T cells, and are often defined as CD11b+ SIRPα+ CD1c+. The pDC state may be less capable than the cDC states at priming T cells but can produce type I IFNs that foster antitumor immunity and are often described as CD123+ CLEC4C+. Finally, MoDCs are defined based on their similarity to monocytes, are typically produced in response to inflammation, and may promote various effector T cell responses, but can be challenging to discriminate from cDC2s (Eisenbarth, 2019). Of note, MoDCs found in vivo differ from monocyte-derived DCs generated in vitro with GM-CSF and IL-4 (Alcántara-Hernández et al., 2017). Other DC states may exist depending on bodily location or exposure to exogenous stimuli.
In the context of cancer, human DCs have long been considered cells that can cross-present tumor cell–derived antigens and stimulate tumor-specific T cell responses (Albert et al., 1998; Nouri-Shirazi et al., 2000). More than 10 yr ago, mechanistic studies in mice uncovered Batf3 as a critical transcription factor for the production of DCs that cross-present antigens and participate in the elimination of immunogenic tumors (Hildner et al., 2008). Additional molecules are likely to be required for cross-presentation of tumor-associated antigens, including the protein WDFY4 expressed by cDC1s in mice (Theisen et al., 2018).
Besides their presence in SLOs, DCs can be found within solid tumors (Broz et al., 2014). Some of these cells migrate through afferent lymph vessels while carrying tumor material and reach tumor-draining lymph nodes, where they can present tumor antigens, thereby activating tumor-specific T cells. Transfer of tumor material from migratory DCs to lymph node–resident DCs can also lead to T cell activation (Ruhland et al., 2020). These processes enhance the cancer immunity cycle (Chen and Mellman, 2013). Tumor-infiltrating DCs can also act locally by promoting antitumor T cell immunity and licensing immunotherapy responses (Engblom et al., 2016; Wculek et al., 2020). These findings suggest that important DC functions exist beyond SLOs, and consequently interest has been sparked in studying tumor-infiltrating DCs in more detail. The development of single-cell RNA sequencing (scRNA-seq) provides the opportunity to map tumor-infiltrating DC states through measurements of single-cell transcriptomes without having to rely on prior assumptions of a limited set of markers that define them. Besides expanding our view of tumor-infiltrating DC heterogeneity across cancer types in both patients and experimental models, measurements of single-cell transcriptomes encode information about multiple dimensions of tumor-infiltrating DC states, such as their cell state–specific gene signature, the cells they might interact with, and their putative ontogeny.
Conserved tumor-infiltrating DC states across solid human cancers
Tumor-infiltrating DC states have started to be defined by scRNA-seq in various human tumors, including non–small cell lung cancer (NSCLC; Zilionis et al., 2019; Kim et al., 2020; Maier et al., 2020; Qian et al., 2020), head and neck squamous cell carcinoma (Cillo et al., 2020), hepatocellular carcinoma (Zhang et al., 2019), melanoma (Nirschl et al., 2017; Brown et al., 2019), cutaneous squamous cell carcinoma (Ji et al., 2020), colorectal cancer (Qian et al., 2020; Zhang et al., 2020), ovarian cancer (Qian et al., 2020), and breast cancer (Qian et al., 2020). When considering NSCLC, scRNA-seq analysis of a given tumor typically reveals transcriptionally distinct DC states, which are resolved by clustering of single-cell transcriptomes based on their similarity. Repeating this analysis with tumors from additional NSCLC patients essentially rediscovers the same tumor-infiltrating DC states and finds no others (Fig. 1, A and B). These findings suggest the existence of a conserved structure of tumor-infiltrating DCs across NSCLC patients and support the view that DCs should be consistently classified using a discrete set of marker genes. Also, the markers that are commonly used to characterize the cDC1 state (e.g., CLEC9A, XCR1, and CADM1), cDC2 state (e.g., FCER1A), and pDC state (e.g., TCF4, LILRA4, CLEC4C, and IRF7) are differentially expressed by distinct tumor-infiltrating DC states, further indicating the possibility to relate scRNA-seq data with previous cytometric studies that used cDC markers for subpopulation gating.
scRNA-seq–based detection of tumor-infiltrating DC states in human NSCLC. (A) Uniform manifold approximation and projection of tumor-infiltrating DC states identified in NSCLC by scRNA-seq. (B) Cumulative plots show the total number of identifiable DC states (left) or tumor cell states (right), with every patient added. The order of patients is determined by accumulating number of cell states detected. All panels are adapted from Zilionis et al. (2019).
scRNA-seq–based detection of tumor-infiltrating DC states in human NSCLC. (A) Uniform manifold approximation and projection of tumor-infiltrating DC states identified in NSCLC by scRNA-seq. (B) Cumulative plots show the total number of identifiable DC states (left) or tumor cell states (right), with every patient added. The order of patients is determined by accumulating number of cell states detected. All panels are adapted from Zilionis et al. (2019).
Similar scRNA-seq analyses of head and neck squamous cell carcinoma, hepatocellular carcinoma, colorectal cancer, ovarian cancer, and breast cancer also show the existence of tumor-infiltrating DC states. These states may be either related to the tumor tissue in which they reside (and thus differ across tissues) or conserved across tissues and cancer types. Addressing this question is complicated by the fact that scRNA-seq studies of human tumors were performed by different investigators who may have used varying tissue dissociation protocols and bioinformatic approaches, and who may have given distinctive names to newly observed DC states. Furthermore, cell state detection can depend on the scRNA-seq platform used (Zhang et al., 2019). Here we performed a meta-analysis of currently available scRNA-seq datasets with the goal of generating a map of tumor-infiltrating DC states across human cancers (Fig. 2 A). This meta-analysis allows direct comparison of all DC states to each other and resolution of possible measurement bias of individual studies and differences in tumor-infiltrating DC state annotation.
Five tumor-infiltrating DC states are conserved across solid human cancer types.(A) Cartoon illustrating the human tumor tissues and scRNA-seq studies that were used to compare the transcriptome of tumor-infiltrating DC states. Specific references are as follows: zi, Zilionis et al. (2019); ma, Maier et al. (2020); qi, Qian et al. (2020); zh_l, Zhang et al. (2019); zh_c, Zhang et al. (2020). (B) Heatmap showing a reciprocal similarity score r for each tumor-infiltrating DC state comparison pair, as defined in the supplemental text. The score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. Five conserved states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). (State 8 is heterogeneous: its reclustering reveals two new states that are similar to other cDC1 and DC3 states based on marker gene comparison; Zhang et al., 2020.)
Five tumor-infiltrating DC states are conserved across solid human cancer types.(A) Cartoon illustrating the human tumor tissues and scRNA-seq studies that were used to compare the transcriptome of tumor-infiltrating DC states. Specific references are as follows: zi, Zilionis et al. (2019); ma, Maier et al. (2020); qi, Qian et al. (2020); zh_l, Zhang et al. (2019); zh_c, Zhang et al. (2020). (B) Heatmap showing a reciprocal similarity score r for each tumor-infiltrating DC state comparison pair, as defined in the supplemental text. The score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. Five conserved states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). (State 8 is heterogeneous: its reclustering reveals two new states that are similar to other cDC1 and DC3 states based on marker gene comparison; Zhang et al., 2020.)
To compare tumor-infiltrating DC states across studies, we defined a reciprocal similarity score for each DC state comparison pair using machine-learning classifier models fitted to each scRNA-seq dataset (for further details, see supplemental text at the end of this article). This analysis is analogous to “reciprocal best hit” approaches used to identify orthologous genes between species (Tatusov et al., 1997); here we instead look for orthologous cell states and use gene expression rather than sequence. The results reveal the existence of five tumor-infiltrating DC states across patients and cancer types, indicating DC state conservation regardless of the tissue in which these cells reside, the genetic makeup of tumor cells, or the composition of the tumor microenvironment (Fig. 2 B). Three of these states are particularly well conserved across cancer types: cDC1 (color-coded in red), pDC (color-coded violet), and DC3 (color-coded dark red). The latter was initially identified by scRNA-seq in all seven NSCLC patients tested (Zilionis et al., 2019); the meta-analysis presented here confirms the existence of this state in additional NSCLC patients and in all other cancer types tested. Other names used to refer to the tumor-infiltrating DC3 state include mature DCs enriched in immunoregulatory molecules (mregDCs; Maier et al., 2020), LAMP3+ DCs (Zhang et al., 2019), CCR7+ DCs (Qian et al., 2020), and BATF3+ DCs (Zhang et al., 2020). (In the latter study, so-called BATF3+ DCs contain both cDC1 and DC3 states.) The remaining two DC states are less conserved: one of them identifies as cDC2 (color-coded orange) and was found in at least two studies (Zilionis et al., 2019; Qian et al., 2020); the other is referred to here as cDC2/MoDC (color-coded orange-green striped), as it contains the cDC2 state identified in various cancer types (Zhang et al., 2019, 2020; Maier et al., 2020; Qian et al., 2020) and the MoDC state identified in NSCLC (Zilionis et al., 2019).
All five tumor-infiltrating DC states express distinctive gene expression signatures based on an analysis comprising five cancer types and eight different scRNA-seq datasets (Fig. 3 A). The top enriched genes include many that have been previously used to define DC states, but also new ones that warrant investigation (Fig. 3 B). The tumor-infiltrating cDC1, cDC2, DC3, and pDC states are distinct from their monocyte and macrophage counterparts, whereas the cDC2/MoDC state shows similarities with these cells (Fig. S1). Evidence supports the existence of a cDC2-to-monocyte continuum instead of discrete subsets (Alcántara-Hernández et al., 2017; Villani et al., 2017; Dutertre et al., 2019; Bourdely et al., 2020; Leader et al., 2020). The two ends of this continuum include CD1C+ CD5+ CD14– CD88– CD163– cDC2s and CD88+ CD14+ CD163+ CD1C– CD5– monocytes, respectively; the cells found in between, which we refer to as cDC2/MoDCs, may variably express these and other markers (Dutertre et al., 2019; Leader et al., 2020; Bourdely et al., 2020). This complexity, as well as interindividual heterogeneity in the abundance of cells across this continuum, may explain some of the differences observed for cDC2 and cDC2/MoDC states between studies. Nevertheless, the overall conservation of tumor-infiltrating DC states across cancers should facilitate their investigation, and findings made in the context of a given cancer may be relatively generalizable. Although conserved, the abundance of specific DC states can vary substantially across individual patients; this variation is potentially important, because it may affect clinical outcome. Also, it allows for correlative analyses between DC state abundances and patient prognosis or response to different therapeutic modalities.
Human tumor-infiltrating DC states show distinctive gene expression profiles across solid cancer types.(A) Enriched genes in human tumor-infiltrating DC states. The identification of these genes is detailed in the supplemental text. The DC populations numbered 1 to 36 refer to previously published states (see Fig. 2 B for definitions) and are detailed in Table S1. The five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). (B) Highlight of the 10 most differentially expressed genes for each human tumor-infiltrating DC state across cancer types. Additional differentially expressed genes of interest are also shown. (C) List of the enriched genes from Fig. 3, A and B, that are also conserved between mouse tumor-infiltrating DC states, based on data obtained in a murine lung adenocarcinoma model driven by KrasG12D and loss of Tp53 (Zilionis et al., 2019).
Human tumor-infiltrating DC states show distinctive gene expression profiles across solid cancer types.(A) Enriched genes in human tumor-infiltrating DC states. The identification of these genes is detailed in the supplemental text. The DC populations numbered 1 to 36 refer to previously published states (see Fig. 2 B for definitions) and are detailed in Table S1. The five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). (B) Highlight of the 10 most differentially expressed genes for each human tumor-infiltrating DC state across cancer types. Additional differentially expressed genes of interest are also shown. (C) List of the enriched genes from Fig. 3, A and B, that are also conserved between mouse tumor-infiltrating DC states, based on data obtained in a murine lung adenocarcinoma model driven by KrasG12D and loss of Tp53 (Zilionis et al., 2019).
Tumor-infiltrating cDC1, cDC2, DC3, and pDC states are distinct from monocyte and macrophage states, whereas the tumor-infiltrating cDC2/MoDC state is not. Heatmap showing a reciprocal similarity score r for each tumor-infiltrating DC, monocyte, and macrophage state. This score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. The monocyte (Mono) and macrophage (Mø) populations lettered a to dd also refer to previously published states, also referenced in Table S1. Five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). Monocyte and macrophage states are highlighted in green.
Tumor-infiltrating cDC1, cDC2, DC3, and pDC states are distinct from monocyte and macrophage states, whereas the tumor-infiltrating cDC2/MoDC state is not. Heatmap showing a reciprocal similarity score r for each tumor-infiltrating DC, monocyte, and macrophage state. This score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. The monocyte (Mono) and macrophage (Mø) populations lettered a to dd also refer to previously published states, also referenced in Table S1. Five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet). Monocyte and macrophage states are highlighted in green.
It remains to be defined whether the entire tumor-infiltrating DC landscape has been captured. It is likely that the frame of the spectrum has been set, although it cannot formally be excluded that additional tumor-infiltrating DC states can be found. These states may have been missed in previous studies because of their very low abundance or because they exist in locations or under conditions that have not yet been studied. For example, the current body of evidence mainly stems from scRNA-seq analyses of treatment-naive tumors, and additional DC states may emerge in the context of therapeutic intervention. The number of patients considered in any study is also small, allowing for potential DC subsets unique to rare tumor subtypes. More heterogeneity may also be revealed as scRNA-seq improves in sensitivity and when additional variables are considered (i.e., beyond the transcriptome, as discussed further below).
Conserved tumor-infiltrating DC states between humans and mice
To study tumor-infiltrating DC states in vivo in a way that yields translatable findings, there should be similar cellular diversity among tumor-infiltrating DC states in human and laboratory organism models. As mice remain the primary model used to investigate immunological processes underlying human disease, it is important to compare tumor-infiltrating DC states between the tumor microenvironment of human cancer and murine tumor models. Direct side-by-side analysis has been performed in NSCLC (Zilionis et al., 2019; Maier et al., 2020) and colorectal cancer (Zhang et al., 2020). Based on these reports, conservation between mouse and human can be appreciated for tumor-infiltrating DC states. For example, unsupervised hierarchical clustering relates each tumor-infiltrating DC state in human NSCLC one-to-one to those found in their mouse counterparts (at least those tumor mouse models driven by Kras activation and loss of Tp53; Zilionis et al., 2019; Maier et al., 2020). Furthermore, many of the genes that are enriched in human tumor-infiltrating DC states are conserved in their mouse counterparts (Fig. 3 C). These findings are remarkable, considering that the two species diverged ∼100 million years ago (Nei et al., 2001), and underline the value of murine tumor models in studying DC functions.
Increasing evidence indicates that the variable abundance of tumor-infiltrating DC states observed in patients may be reproduced, and thus studied, in mice. For example, mouse models of melanoma show that expression of active β-catenin in tumor cells limits cDC1 recruitment due to decreased production of the cDC1 chemoattractant CCL4. This results in T cell exclusion from the tumor, which recapitulates clinical observations of melanomas with active β-catenin signaling (Spranger et al., 2015; Nsengimana et al., 2018). Similarly, both human pancreatic ductal adenocarcinomas and spontaneous mouse models of this disease typically resist therapy, with resistance being linked to a scarcity of pancreatic ductal adenocarcinoma–infiltrating cDC1s and cDC2s (Hegde et al., 2020; Lin et al., 2020). These studies support the relevance of using murine tumor models to study DCs, particularly when DC abundance within murine tumors reflects that of their human counterparts.
Tumor-infiltrating DC states: Origins
The conservation of tumor-infiltrating DC states suggests that the ontogeny of these cells may also be conserved. The origins of cDC1s, cDC2s, MoDCs, and pDCs has been discussed in detail elsewhere (Sichien et al., 2017; Reizis, 2019). Interestingly, meta-analysis of scRNA-seq data shows that tumor-infiltrating cDC1 and pDC states have their counterparts in peripheral blood (Fig. S2). This similarity suggests that the cDC1 and pDC states are defined phenotypically before the cells enter the tumor, although it cannot be formally excluded that cells that recirculate after residing in tissue contribute to the peripheral blood states. These findings also align with the notion that these tumor-infiltrating DC states have distinct developmental pathways. Conversely, both tumor-infiltrating cDC2 and cDC2/MoDC states resemble a circulating cDC2 state (Fig. S2; Villani et al., 2017; Dutertre et al., 2019; Bourdely et al., 2020), and the tumor-infiltrating cDC2/MoDC state additionally resembles a circulating cDC2/MoDC state (Fig. S2; Villani et al., 2017; Dutertre et al., 2019; Bourdely et al., 2020). Considering not only this overlap, but also that cDC2s and cDC2/MoDCs belong to a continuum of states, these states are likely more closely connected ontogenically (Yáñez et al., 2017; Bourdely et al., 2020; Weinreb et al., 2020). An additional blood DC state, referred to as DC5 (Villani et al., 2017), has not been detected in the tumor stroma, suggesting that it defines precursor cells that are never recruited to tumors or that acquire a distinctive phenotype upon tissue entry. Favoring the latter possibility, the DC5 state expresses markers that are associated with preDCs (Dutertre et al., 2019).
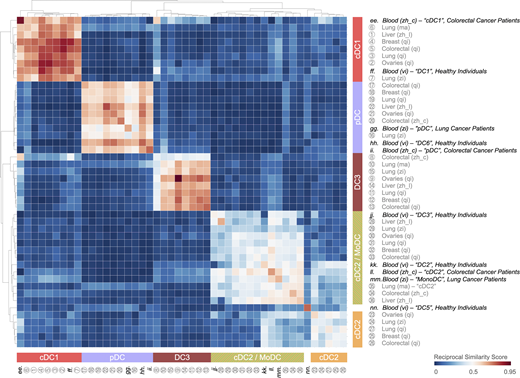
Tumor-infiltrating cDC1, cDC2, cDC2/MoDC, and pDC states resemble circulating DC states in peripheral blood of cancer patients and healthy individuals, whereas the tumor-infiltrating DC3 state does not. Heatmap showing a reciprocal similarity score r for each tumor-infiltrating and blood DC state. This score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. The blood DC states lettered ee to nn also refer to previously published states, which are also referenced in Table S1. Five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet).
Tumor-infiltrating cDC1, cDC2, cDC2/MoDC, and pDC states resemble circulating DC states in peripheral blood of cancer patients and healthy individuals, whereas the tumor-infiltrating DC3 state does not. Heatmap showing a reciprocal similarity score r for each tumor-infiltrating and blood DC state. This score was calculated using the probability estimates returned by the Linear Support Vector Machine classifier on log-transformed data. The DC populations numbered 1 to 36 refer to previously published states, which are referenced in Table S1. The blood DC states lettered ee to nn also refer to previously published states, which are also referenced in Table S1. Five conserved DC states are identified as follows: cDC1 (red), cDC2 (orange), cDC2/MoDC (orange-green striped), DC3 (dark red), and pDC (violet).
The tumor-infiltrating DC3 state is found exclusively in tissue, hence without a similar counterpart in the blood (Fig. S2). Computational analyses based on RNA velocity suggest that the DC3 state may derive from either the cDC1 or cDC2/MoDC state (Zhang et al., 2019) and possibly the cDC2 state, the contribution of which may vary among tumor types or locations. Accordingly, tumor-infiltrating DC3s identified by scRNA-seq can express protein markers that are normally associated with cDC1s (e.g., XCR1 and CD103) or cDC2s (e.g., CD11b; Maier et al., 2020). These findings suggest that maturation into DC3s supersedes ontogeny differences for cDC1 and cDC2 differentiation. Consequently, in previous studies the use of, for example, XCR1, CD103, and CD11b markers to delineate cDC1s and cDC2s may have included cells in the DC3 state. In fact, only a fraction of CD103+ DCs (often referred to as cDC1s) may express CCR7 intratumorally (Roberts et al., 2016); this division of CCR7 expression among CD103+ DCs may define cDC1 and DC3 states, which lack and express CCR7, respectively (Zhang et al., 2019, 2020; Zilionis et al., 2019; Maier et al., 2020; Qian et al., 2020).
The concept that tissue DCs can share a common gene signature, regardless of tissue location or lineage, has been suggested before (Miller et al., 2012; Dalod et al., 2014; Nirschl et al., 2017); this phenomenon may apply to the DC3 state. Moreover, the DC3 state is not unique to tumors. For example, DC3-like cells can be found in Crohn’s disease lesions (Martin et al., 2019): these cells have a reciprocity similarity score of ∼0.6 compared with all other DC3-like states identified in tumors and a reciprocal similarity score ∼0.05 compared with all other non-DC3 states. Thus, DC3 amplification in tissue may be a common feature of inflammation. Preferential localization of DC3s close to blood vessels, at least in the context of anti–programmed cell death protein 1 (anti–PD-1) therapy (Garris et al., 2018), further suggests that DC3 maturation in tissue is spatially compartmentalized. Additional studies, for example using intravital imaging as discussed below, may help address DC3s’ origins and fate.
Tumor-infiltrating DC states: Regulation and functions
Tumor-infiltrating DCs can regulate key aspects of tumor-associated immunity (Fig. 4, A and B). Experimental evidence suggests that tumor-infiltrating cDC1s can foster tumor control in different ways, including production of the chemokines CXCL9 (Chow et al., 2019) and CXCL10 (Spranger et al., 2017) that recruit CD8+ T cells to tumors and enable local (re)activation of these cells through antigen cross-presentation (Spranger et al., 2017; Chow et al., 2019), and through production of various factors such as IFN-λ (Hubert et al., 2020). cDC1s are also required for successful immunotherapy in various murine tumor models (Moynihan et al., 2016; Salmon et al., 2016; Sánchez-Paulete et al., 2016; Mao et al., 2020; Morrison et al., 2020). Accordingly, cDC1 gene expression signatures in human tumors often correlate with improved patient survival and clinical response to immunotherapy (Böttcher et al., 2018; Barry et al., 2018). Inhibitors of cDC1 responses include prostaglandin E2, IL-10, XBP1-mediated endoplasmic reticulum stress, and lipid peroxidation (Ruffell et al., 2014; Cubillos-Ruiz et al., 2015; Zelenay et al., 2015; Veglia et al., 2017; Böttcher et al., 2018), whereas enhancers of cDC1 responses include type I IFNs (Fuertes et al., 2011), the chemokines CCL4, CCL5, and XCL1 (Spranger et al., 2015; Böttcher et al., 2018; Williford et al., 2019) and the growth factor Flt3L (Broz et al., 2014; Salmon et al., 2016; Barry et al., 2018), some of which are produced by natural killer (NK) cells.
Relevance, regulation, and function of tumor-infiltrating DC states. (A) Overall clinical and experimental observations related to tumor-infiltrating DCs, T cell infiltration, and outcome. (B) Each DC state can be positively or negatively regulated by tumor microenvironment–derived factors that influence their antitumor capacity, as identified in experimental studies. Thin dashed lines/arrows represent cell migration or differentiation into another state. Solid arrows and inhibitory signs identify factors that regulate target cell function either positively or negatively. The text boxes describe key functions of the respective DC states. The cDC2 and cDC2/MoDC names represent the states identified in the meta-analysis, and their functions might differ from those of classically gated cDC2s and MoDCs. For instance, classically gated cDC2s from prior studies in mice may include cells from both the cDC2 and cDC2/MoDC states, and classically gated MoDCs from prior studies may include cells from the cDC2/MoDC state.
Relevance, regulation, and function of tumor-infiltrating DC states. (A) Overall clinical and experimental observations related to tumor-infiltrating DCs, T cell infiltration, and outcome. (B) Each DC state can be positively or negatively regulated by tumor microenvironment–derived factors that influence their antitumor capacity, as identified in experimental studies. Thin dashed lines/arrows represent cell migration or differentiation into another state. Solid arrows and inhibitory signs identify factors that regulate target cell function either positively or negatively. The text boxes describe key functions of the respective DC states. The cDC2 and cDC2/MoDC names represent the states identified in the meta-analysis, and their functions might differ from those of classically gated cDC2s and MoDCs. For instance, classically gated cDC2s from prior studies in mice may include cells from both the cDC2 and cDC2/MoDC states, and classically gated MoDCs from prior studies may include cells from the cDC2/MoDC state.
Classically gated tumor-infiltrating cDC2s, which may include cells that fall within the tumor-infiltrating cDC2 or cDC2/MoDC states identified in the meta-analysis, have been less studied, perhaps due to a lack of cDC2-specific depletion strategies in mice and the difficulty of distinguishing these cells from MoDCs. Experimental evidence indicates that migration of tumor-infiltrating cDC2s to tumor-draining lymph nodes enables presentation of tumor-derived antigens predominantly to CD4+ conventional T cells; however, the presence of regulatory T cells (T reg cells) limits CD4+ T cell differentiation into proinflammatory antitumor cells (Laoui et al., 2016; Binnewies et al., 2019). Furthermore, depletion of intratumoral T reg cells leads to increased migration to tumor-draining lymph nodes of only one of two cDC2 states identified in murine melanoma (Binnewies et al., 2019), suggesting that cDC2 heterogeneity may have functional implications. Corresponding to this finding, studies in healthy murine spleen, where two cDC2 states were also identified, show that one cDC2 state is more anti-inflammatory than the other (Brown et al., 2019), in which differentially expressed gene RUNX3 may play a role (Fainaru et al., 2004). In patients, the increased presence of tumor-infiltrating BDCA-1+ cDC2s in head and neck cancer has been associated with improved progression-free survival, and this gain was even more prominent when tumor-infiltrating T reg cell counts were low (Binnewies et al., 2019). The increased expression of cDC2 marker genes in tumors has also been associated with improved patient survival in NSCLC (Zilionis et al., 2019).
Emerging from recent scRNA-seq studies, tumor-infiltrating DC3s contain a transcriptional module lacking key cDC1 and cDC2 markers and resemble “activated” Ccr7+ cDCs in mice (Ardouin et al., 2016). Because the chemokine CCR7 guides tumor-infiltrating cDC trafficking to lymph nodes (Roberts et al., 2016), it is likely that a proportion of DC3s eventually migrate to tumor-draining lymph nodes, where they may activate tumor antigen-specific T cells (Zhang et al., 2019). Yet, in the context of successful immunotherapy, some tumor-infiltrating DC3s may persist locally for at least several days (Garris et al., 2018), suggesting they can acquire aberrant trafficking behavior or are retained due to intratumoral interactions, such as via attraction to stromal cells or tumor cells expressing CCR7 ligands (Novak et al., 2007; Cheng et al., 2018; Whyte et al., 2020). Within tumors, DC3s can produce high amounts of IL-12 upon sensing IFN-γ released by neighboring T or NK cells (Garris et al., 2018; Maier et al., 2020); in turn, IL-12 stimulates antitumor CD8+ T cells (Garris et al., 2018) or NK cells (Mittal et al., 2017), indicating that DC3s can license full-fledged antitumor immunity through a molecular cross-talk involving IFN-γ and IL-12. DC3s may also activate tumor-specific CD4+ T cell responses (Maier et al., 2020). The increased expression of DC3 marker genes in tumors has been associated with improved patient survival in NSCLC and colorectal cancer (Zilionis et al., 2019; Zhang et al., 2020). Furthermore, this DC state was found to be selectively enriched in microsatellite instability–high colorectal cancer (Zhang et al., 2020), which responds better to immune checkpoint blockade therapy than its microsatellite instability–low counterpart (Le et al., 2015).
The tumor-infiltrating DC3 state can be amplified and activated through noncanonical NF-κB signaling, for example with CD40 agonists (Garris et al., 2018). Sensing of IFN-γ may play a role, since DCs in IFN-γ receptor–deficient mice show decreased expression of noncanonical NF-κB pathway genes (Nirschl et al., 2017). Correspondingly, defects in the noncanonical NF-κB pathway impair IL-12 production (Katakam et al., 2015). Tumor-infiltrating DC3s (also referred to as mregDCs; Maier et al., 2020) also express an immunoregulatory program that is characterized by programmed death ligand 1 (PD-L1), PD-L2, and IL-4i1 expression. This program is independent of CCR7 signaling, and PD-L1 expression is driven by the phagocytic receptor tyrosine kinase AXL. IL-4 signaling suppresses IL-12 production by DC3s, and IL-4 blockade can induce a therapeutic response against lung tumors in mice that otherwise resist immune checkpoint blockade treatment (Maier et al., 2020). These findings suggest that inhibitory molecules such as PD-L1 and IL-4i1 represent DC analogues to lymphocytes’ coinhibitory checkpoints and may serve as their own self-limiting programs. In line with this notion, selective deletion of PD-L1 in Clec9a-dependent DCs enhances antitumor immunity (Oh et al., 2020).
At steady-state, tissue-derived migratory DCs also share a unique regulatory transcriptional program across humans and mice, regardless of tissue or cell of origin. This intrinsic regulatory module may help maintain tolerance under normal conditions (Miller et al., 2012; Anandasabapathy et al., 2014) and depends at least in part on NF-κB–dependent activity, because mice develop autoimmunity in its absence (Dalod et al., 2014; Baratin et al., 2015). It is possible that such DC tissue programming is coopted in tumor and other chronic inflammatory settings, such as with the DC3 state.
Classically gated tumor-infiltrating MoDCs, which may include cells that fall within the cDC2/MoDC state identified in the meta-analysis, and pDC states also require investigation. At present, there is evidence that MoDCs can be recruited to inflamed sites such as tumors upon sensing the chemokine MCP-1, also referred to as CCL2 (Ma et al., 2014; Laoui et al., 2016), or immunogenic tumor cell death–derived factors (Ma et al., 2013). MoDCs may help protect the host against cancer by acquiring the ability to cross-present tumor antigens to CD8+ T cells (Ma et al., 2013) or by licensing CD8+ T cells independently of MHC class I (Santana-Magal et al., 2020); however, several tumor microenvironmental factors, including lactic acid, IL-6, and prostaglandin E2 (Chomarat et al., 2000; Sombroek et al., 2002; Gottfried et al., 2006), may functionally suppress MoDCs. Tumor-infiltrating pDCs can be activated through pattern recognition receptors comprising TLR7 and TLR9 (Stary et al., 2007; Gungor et al., 2014), display antitumor functions such as secretion of type I IFNs that can promote tumor cell lysis (Stary et al., 2007; Le Mercier et al., 2013), and may recruit NK cells and enhance their cytotoxic activity in tumors (Liu et al., 2008). However, tumor-infiltrating pDCs may be suppressed by the cytokines TNF-α and TGF-β (Labidi-Galy et al., 2011; Sisirak et al., 2013) and express the inhibitory receptor TIM-3 (Chiba et al., 2012); they may also actively promote cancer by inducing and activating T reg cells (Ito et al., 2007; Sisirak et al., 2012) or fostering neoangiogenesis (Curiel et al., 2004). Accordingly, tumor-infiltrating pDCs have been identified as a negative prognostic marker in melanoma (Aspord et al., 2013) and ovarian (Labidi-Galy et al., 2012) cancers; although it is possible that therapeutically targeting these cells may trigger antitumor functions.
Integration of scRNA-seq landscapes with functional studies
We need to continue using scRNA-seq approaches to assess tumor-infiltrating DC states in more cancer types and to expand these studies to (pre)clinical trials to better understand how therapeutic interventions impact the tumor microenvironment at the cellular and molecular levels. The use of single-cell resolution genomic approaches in clinical trials for the development of next-generation immunotherapy is reviewed elsewhere (Yofe et al., 2020). It is also important that information produced with scRNA-seq approaches is used to generate models for assessing the functional relevance of newly defined cell states, which we discuss below (Fig. 5).
Increasing the number of measured dimensions of tumor-infiltrating DC states. Computational tools can predict multiple dimensions of tumor-infiltrating DC states from single-cell transcriptomes such as fate potential (trajectory inference and RNA velocity), cell–cell interactions, and downstream targets of these ligand–receptor cell–cell interactions (ligand–target modeling), active transcription factors and signaling pathways (transcription factor–target and pathway–target modeling), and conservation of DC states (meta-analyses as performed in this review). These computational prediction tools are instrumental to generate testable hypotheses about tumor-infiltrating DC states; however, multimodal approaches that measure both single-cell transcriptomes and other dimensions are emerging. Available and emerging tools permit the evaluation of other intrinsic features of cells, their location, origins, fate, and functions. *, tools applicable to exploratory models only; GEMM, genetically engineered mouse model.
Increasing the number of measured dimensions of tumor-infiltrating DC states. Computational tools can predict multiple dimensions of tumor-infiltrating DC states from single-cell transcriptomes such as fate potential (trajectory inference and RNA velocity), cell–cell interactions, and downstream targets of these ligand–receptor cell–cell interactions (ligand–target modeling), active transcription factors and signaling pathways (transcription factor–target and pathway–target modeling), and conservation of DC states (meta-analyses as performed in this review). These computational prediction tools are instrumental to generate testable hypotheses about tumor-infiltrating DC states; however, multimodal approaches that measure both single-cell transcriptomes and other dimensions are emerging. Available and emerging tools permit the evaluation of other intrinsic features of cells, their location, origins, fate, and functions. *, tools applicable to exploratory models only; GEMM, genetically engineered mouse model.
First, the information encoded in single-cell transcriptome data should be exhausted to generate hypotheses about the function of tumor-infiltrating DC states and to guide the design of relevant functional experiments that can test these hypotheses. For example, differentiation hierarchies in the tumor-infiltrating DC landscape discussed above can be predicted using single-cell trajectory inference methods (Saelens et al., 2019) or with measurements of nascent mRNA abundance (La Manno et al., 2018; Zhang et al., 2019). Interactions of tumor-infiltrating DCs with other cells and downstream intracellular targets of these receptor–ligand interactions may be predicted with ligand–target modeling (Browaeys et al., 2020). The transcription factors and signaling pathways that are active in tumor-infiltrating DCs may also be predicted with transcription factor–target modeling and pathway–target modeling, respectively (Schubert et al., 2018; Garcia-Alonso et al., 2019; Holland et al., 2020), and further tested functionally.
Second, multimodal approaches integrating single-cell transcriptomes with other dimensions of cell states allow for the mapping of these dimensions onto the high-resolution transcriptionally defined tumor-infiltrating DC landscape. Such multimodal approaches may help design functional experiments. For example, conventional scRNA-seq data, such as those discussed in the previous sections, lack spatial context; yet defining the location of a cell state in a given tissue may identify its neighbors and help understand how these cells functionally regulate each other. Combined measurements of single-cell transcriptomes with protein cell surface expression (Stoeckius et al., 2017; Baron et al., 2019; Dutertre et al., 2019) enables the identification of cell-state protein markers, which may then be used to locate cells of interest in tissue by histology, multiplexed single-cell mass cytometry imaging (Jackson et al., 2020), or multiplexed immunofluorescence (Gut et al., 2018; Lin et al., 2018). The identification of surface markers may additionally assist in developing reagents that facilitate labeling or depletion of cell states of interest in experimental models for in vivo functional studies.
Spatial context may further be assessed in combination with time, such as with intravital imaging, to track cell behavior in complex tissue environments (Pittet et al., 2018), or with cell state morphology, such as with correlative light electron microscopy (Begemann and Galic, 2016), to interrogate whether cell–cell interactions imprint cell identities (Bonnardel et al., 2019). In addition to the approaches mentioned above, spatial identification of tumor-infiltrating DC states may be measured with spatial transcriptomics (Chen et al., 2015). Multimodal approaches may also integrate single-cell transcriptomes with cell surface and lineage markers to help map DC state ontogeny (Wagner and Klein, 2020; Weinreb et al., 2020), with chromatin accessibility to uncover accessible transcription factor motifs in cell states of interest (Cusanovich et al., 2018), or with cell sorting of physically interacting cells to discover preferential interactions within organs (Boisset et al., 2018; Giladi et al., 2020). Using these combined approaches should increase our understanding of the unfolding of tumor-associated DC responses, as well as the intracellular molecular wiring and cellular crosstalk with neighboring cells that dictate DC functions. For example, these approaches may help better understand the relationships between DC3s with cDC1s and/or cDC2s, and DC3 programming in tissue. Altogether, the experimental aims of producing multidimensional information are to generate informed, testable hypotheses that encompass fundamental and translational aspects of DC function and to develop animal models that closely model human biology and disease to allow for biologically and clinically relevant investigation.
The DC states discussed here likely contribute differentially to antitumor immunity but remain underinvestigated. cDC1s have attracted increased attention, based on their ability to induce tumor-specific CD8+ T cell responses; yet, all other states may contribute nonoverlapping functions, which require study. Additionally, there is increasing evidence that tumor immune evasion involves crippling normal DC functions, which can be expected when considering the central role these cells can play in fostering antitumor immunity. Studying mechanisms leading to dysfunctional or tolerogenic DC states warrant further investigation, especially with information suggesting that at least some DC states can be suppressed or negatively regulated. Ultimately, the goal of studying DCs in cancer and related therapeutic interventions is to discover aspects of their cellular identity that can be targeted to favor a functional antitumor environment and boost treatment response. This knowledge may also facilitate the design of DC vaccines and the identification of predictive biomarkers of clinical outcome and treatment response. The incorporation of multimodal single-cell approaches into translational research projects, especially as part of clinical trials, will certainly shed more light on dimensional aspects of tumor-infiltrating DC identity (and that of other cell types), better enabling the accomplishment of these goals.
Online supplemental material
Fig. S1 shows that tumor-infiltrating cDC1, cDC2, DC3, and pDC states are distinct from monocyte and macrophage states, whereas the tumor-infiltrating cDC2/MoDC state is not. Fig. S2 shows that tumor-infiltrating cDC1, cDC2, cDC2/MoDC, and pDC states resemble circulating DC states in peripheral blood of cancer patients and healthy individuals, whereas the tumor-infiltrating DC3 state does not. Table S1 lists DC, monocyte, and macrophage states that were identified in previous publications.
Acknowledgments
We thank Nicolas A. Gort-Freitas for his feedback on the reciprocal similarity meta-analysis.
This work was supported in part by the Massachusetts General Hospital Samana Cay MGH Research Scholar Fund, the ISREC Foundation, and National Institutes of Health grants R01-AI084880, R01-CA206890, NIH-U01-CA224348, P01-CA240239 (to M.J. Pittet), and R01-CA218579 (to A.M. Klein and M.J. Pittet). R. Bill was funded by a Postdoctoral Mobility Fellowship of the Swiss National Science Foundation (P400PM_183852).
Author contributions: G.M. Gerhard, R. Bill, M. Messemaker, and M.J. Pittet wrote the manuscript. M. Messemaker performed meta-analyses. M. Messemaker and A.M. Klein designed the meta-analyses. M.J. Pittet directed the study.
References
Competing Interests
Disclosures: R. Bill reported that is wife is an employee and stockholder of CSL Behring. His salary is funded by a postdoc fellowship of the Swiss National Science Foundation (SNSF; P400PM_183852). M.J. Pittet reported personal fees from Aileron Therapeutics, AstraZeneca, Cygnal Therapeutics, Elstar Therapeutics, KSQ Therapeutics, Merck, and Siamab Therapeutics outside the submitted work. No other disclosures were reported.
Author notes
G.M. Gerhard, R. Bill, and M. Messemaker contributed equally to this paper.