Migrasomes, organelles crucial for cell communication, undergo distinct stages of nucleation, maturation, and expansion. The regulatory mechanisms of migrasome formation, particularly through biological cues, remain largely unexplored. This study reveals that calcium is essential for migrasome formation. Furthermore, we identify that Synaptotagmin-1 (Syt1), a well-known calcium sensor, is not only enriched in migrasomes but also indispensable for their formation. The calcium-binding ability of Syt1 is key to initiating migrasome formation. The recruitment of Syt1 to migrasome formation sites (MFS) triggers the swelling of MFS into unstable precursors, which are subsequently stabilized through the sequential recruitment of tetraspanins. Our findings reveal how calcium regulates migrasome formation and propose a sequential interaction model involving Syt1 and Tetraspanins in the formation and stabilization of migrasomes.
Introduction
The migrasome, an organelle recently identified in migrating cells, originates from their retraction fibers (RFs) (Ma et al., 2015; Fan et al., 2022). These organelles are pivotal in facilitating cell–cell communication, playing an essential role in transferring messenger RNA and proteins between adjacent cells and delivering signaling molecules to specific spatial locations (Jiang et al., 2019; Zhu et al., 2021). Additionally, migrasomes are capable of evicting damaged mitochondria from cells, thereby maintaining mitochondrial homeostasis (Jiao et al., 2021). Physiologically, migrasomes are crucial in a variety of functions, especially in organ morphogenesis (Jiang et al., 2019) and in promoting angiogenesis during embryonic development (Zhang et al., 2022). They also play a significant role in maintaining mitochondrial homeostasis and survival in neutrophils after their entry into circulation (Jiao et al., 2021).
Migrasome formation, a complex cellular process, is orchestrated by molecular mechanisms and influenced by physical forces. Migrasome biogenesis involves three phases: nucleation, maturation, and expansion. Nucleation is driven by sphingomyelin (SM) synthesis, with sphingomyelin synthase 2 (SMS2) playing a crucial role in converting ceramide to SM at the plasma membrane, a process integral to migrasome structural integrity (Liang et al., 2023). Maturation involves the de novo synthesis of PI(4,5)P2 at the migrasome formation site (MFS). PI(4,5)P2 targets and concentrates Rab35, a PI(4,5)P2-binding protein, on the MFS. Through Rab35, integrins are recruited and concentrated on MFS, thereby firmly adhering MFS to the extracellular matrix (ECM). Expansion is marked by an influx of tetraspanins to MFS, leading to the coalescence of tetraspanin-enriched microdomains (TEMs) into the migrasomes (Huang et al., 2019; Wu et al., 2017; Ding et al., 2023). Mechanistically, the coalescence of TEMs enhances the membrane binding rigidity of the migrasome membrane, thereby increasing its stability (Huang et al., 2019).
Recent studies using live cell imaging and a biomimetic system designed for migrasomes and RFs have shown that migrasome expansion is a sequential, two-stage process. In the initial stage, mediated by membrane tension in the biomimetic system, localized swellings largely devoid of the Tetraspanin 4 (TSPAN4) protein emerge on the RFs. Subsequently, in the second stage, TSPAN4 molecules migrate toward these initial swellings, leading to their enlargement, potentially reaching several microns in size, and culminating in their transformation into fully formed migrasomes. TSPAN4 stabilizes membrane swellings and facilitates their maturation into migrasomes (Huang et al., 2019; Dharan et al., 2023). The recruitment of TSPAN4 to these swellings is identified as a critical factor for the continued growth and eventual stabilization of the migrasomes. It is currently unknown whether additional steps are needed to facilitate the transition from initial swelling to tetraspanins-mediated expansion and stabilization.
Calcium is vital for cell migration, orchestrating cytoskeletal dynamics, adhesion, and directional movement. It functions as a signal transducer (Streb et al., 1983), influencing cell protrusions and adhesions, and it plays a key role in coordinating cellular movement through interactions with various signaling pathways (Sechi and Wehland, 2000). Calcium’s actions are detected by numerous sensors, such as Synaptotagmin-1 (Syt1), which is pivotal in neurotransmitter release (Geppert et al., 1994). Syt1, responding to calcium influx, triggers synaptic vesicle fusion, thereby facilitating efficient synaptic transmission. Beyond this, Syt1 is involved in other cellular activities, including endocytosis and plasma membrane repair (Chapman, 2002; Schapire et al., 2008). However, the specific roles of calcium and Syt1 in migrasome formation remain unexplored.
Results
Ca2+-binding Syt1 is required for migrasome formation
We discovered that calcium is required for migrasome formation. Treating L929 cells with increasing doses of BAPTA, a calcium chelator, blocks migrasome formation. Despite the widespread use of 10–20 μM BAPTA as a calcium chelator in the literature (Zhao et al., 2019; Xu et al., 2004), our findings reveal that a lower concentration of 5 μM BAPTA is sufficient to significantly impact migrasome formation in our system (Fig. 1, A and B). Furthermore, this concentration exerts a relatively minor effect on RF formation (Fig. 1 A). This data suggests that calcium may play a direct role in migrasome biogenesis.
Ca 2+ -binding Syt1 is required for migrasome formation. (A) Confocal image of migrasomes of L929 cells treated with 0, 2.5, 5, and 10 μM BAPTA and cell migrate on chamber overnight for 12 h. L929 are labeled with wheat germ agglutinin (WGA) (green). Scale bar, 10 μm. (B) The number of migrasomes per 100 μm RF length per cell from images like those shown in A was quantified. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (C) Confocal image of migrasomes from NRK and NRK Syt1-GFP. Cells are labeled with WGA (magenta). Scale bar, 10 µm or 2 µm (inset). (D) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK and NRK Syt1-GFP cells shown in C. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (E) Quantification of the Max Feret’s diameter of migrasomes per cell in NRK and NRK Syt1-GFP cells shown in C. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (F) Confocal image of migrasomes from NRK Syt1-GFP treated with/without 1 mM Ca2+ for 12 h. Scale bar, 10 µm. (G) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK Syt1-GFP cells shown in F. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 for NRK Syt1-GFP; n = 50 for NRK Syt1-GFP treated with Ca2+. The experiments were performed three times. (H) Confocal image of migrasomes from NRK, NRK Syt1-GFP, and NRK Syt1-GFP treated with 5 μM BAPTA for 12 h. NRK are labeled with WGA (magenta). Scale bar, 10 μm. (I) The number of migrasomes per 100 μm RF length per cell from H was quantified. Data shown represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (J) Schematic representation of Syt1 showing transmembrane domain, C2A domain, C2B domain, the phospholipid binding region, the calcium-binding sites, and the sites on the tips of membrane-binding loops. (K) Confocal image of migrasomes from NRK, NRK Syt1-mCherry, NRK Syt1(△C2A)-mCherry, NRK Syt1(△C2B)-mCherry, and NRK Syt1(△C2AB)-mCherry. The △C2A represents the truncation of C2A, the △C2B represents the truncation of C2B. The △C2AB represents the truncation of the phospholipid binding region. Cells are labeled with WGA (green). Scale bar, 10 µm. (L) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK, NRK Syt1-mCherry, NRK Syt1(△C2A)-mCherry, NRK Syt1(△C2B)-mCherry, and NRK Syt1(△C2AB)-mCherry cells shown in K. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 51 cells per group. The experiments were performed three times. (M) Confocal image of migrasomes from NRK, NRK Syt1-GFP, NRK Syt1(C2A*)-GFP, NRK Syt1(C2B*)-GFP, and NRK Syt1(C2A*B*)-GFP. The C2A* represents the calcium—binding site mutant (D178A, D230A, and D232A) on C2A, the C2B* represents the calcium—binding site mutant (D309A, D363A, and D365A) on C2B, the C2A*B* represents the calcium—binding site mutant (D178A, D230A, D232A, D309A, D363A, and D365A) on C2AB. Cells are labeled with WGA (magenta). Scale bar, 10 µm. (N) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK, NRK Syt1-GFP, NRK Syt1(C2A*)-GFP, NRK Syt1(C2B*)-GFP, and NRK Syt1(C2A*B*)-GFP cells shown in M. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (O) Confocal image of migrasomes from NRK Syt1-GFP and NRK Syt1(4A)-GFP. The Syt1(4A) represent the mutant on the tips of the Syt1 membrane-binding loops (M173A, F234A, V304A, and I367A). Scale bar, 10 µm. (P) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK Syt1-GFP and NRK Syt1 (4A)-GFP shown in O. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 51 cells per group. The experiments were performed three times.
Ca 2+ -binding Syt1 is required for migrasome formation. (A) Confocal image of migrasomes of L929 cells treated with 0, 2.5, 5, and 10 μM BAPTA and cell migrate on chamber overnight for 12 h. L929 are labeled with wheat germ agglutinin (WGA) (green). Scale bar, 10 μm. (B) The number of migrasomes per 100 μm RF length per cell from images like those shown in A was quantified. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (C) Confocal image of migrasomes from NRK and NRK Syt1-GFP. Cells are labeled with WGA (magenta). Scale bar, 10 µm or 2 µm (inset). (D) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK and NRK Syt1-GFP cells shown in C. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (E) Quantification of the Max Feret’s diameter of migrasomes per cell in NRK and NRK Syt1-GFP cells shown in C. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (F) Confocal image of migrasomes from NRK Syt1-GFP treated with/without 1 mM Ca2+ for 12 h. Scale bar, 10 µm. (G) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK Syt1-GFP cells shown in F. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 for NRK Syt1-GFP; n = 50 for NRK Syt1-GFP treated with Ca2+. The experiments were performed three times. (H) Confocal image of migrasomes from NRK, NRK Syt1-GFP, and NRK Syt1-GFP treated with 5 μM BAPTA for 12 h. NRK are labeled with WGA (magenta). Scale bar, 10 μm. (I) The number of migrasomes per 100 μm RF length per cell from H was quantified. Data shown represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (J) Schematic representation of Syt1 showing transmembrane domain, C2A domain, C2B domain, the phospholipid binding region, the calcium-binding sites, and the sites on the tips of membrane-binding loops. (K) Confocal image of migrasomes from NRK, NRK Syt1-mCherry, NRK Syt1(△C2A)-mCherry, NRK Syt1(△C2B)-mCherry, and NRK Syt1(△C2AB)-mCherry. The △C2A represents the truncation of C2A, the △C2B represents the truncation of C2B. The △C2AB represents the truncation of the phospholipid binding region. Cells are labeled with WGA (green). Scale bar, 10 µm. (L) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK, NRK Syt1-mCherry, NRK Syt1(△C2A)-mCherry, NRK Syt1(△C2B)-mCherry, and NRK Syt1(△C2AB)-mCherry cells shown in K. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 51 cells per group. The experiments were performed three times. (M) Confocal image of migrasomes from NRK, NRK Syt1-GFP, NRK Syt1(C2A*)-GFP, NRK Syt1(C2B*)-GFP, and NRK Syt1(C2A*B*)-GFP. The C2A* represents the calcium—binding site mutant (D178A, D230A, and D232A) on C2A, the C2B* represents the calcium—binding site mutant (D309A, D363A, and D365A) on C2B, the C2A*B* represents the calcium—binding site mutant (D178A, D230A, D232A, D309A, D363A, and D365A) on C2AB. Cells are labeled with WGA (magenta). Scale bar, 10 µm. (N) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK, NRK Syt1-GFP, NRK Syt1(C2A*)-GFP, NRK Syt1(C2B*)-GFP, and NRK Syt1(C2A*B*)-GFP cells shown in M. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (O) Confocal image of migrasomes from NRK Syt1-GFP and NRK Syt1(4A)-GFP. The Syt1(4A) represent the mutant on the tips of the Syt1 membrane-binding loops (M173A, F234A, V304A, and I367A). Scale bar, 10 µm. (P) Quantification of the number of migrasomes per 100 μm RF length per cell in NRK Syt1-GFP and NRK Syt1 (4A)-GFP shown in O. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 51 cells per group. The experiments were performed three times.
We hypothesize that for calcium to have a direct role in promoting migrasome formation, calcium sensors likely localize on the migrasome. Syt1, a well-characterized calcium sensor, plays an essential role in neurotransmitter release. We found that Syt1 is enriched on migrasome, and overexpression of Syt1 markedly enhances migrasome (Fig. 1, C and D). It is important to note that migrasome numbers are elevated even when the length of RFs is normalized. This suggests that Syt-1 plays a direct role in facilitating migrasome formation. Additionally, overexpression of Syt1 is associated with an increase in migrasome size (Fig. 1 E). Moreover, elevating the calcium concentration in the culture medium can further boost Syt1-induced migrasome formation (Fig. 1, F and G). In contrast, overexpression of Syt1 showed a reduced capacity to promote migrasome formation in the presence of BAPTA, suggesting that calcium is required for Syt1-promoted migrasome formation (Fig. 1, H and I).
Syt1 is composed of a single-pass transmembrane domain linked to two C2 domains (C2A and C2B) that bind calcium (Chapman, 2002). In addition, the C2A and C2B domains are the Ca2+-dependent membrane interaction regions (Fig. 1 J). We found that both the C2A and C2B domains are required for promoting migrasome formation, suggesting that Syt1 needs calcium to facilitate this process (Fig. 1, K and L). To further confirm this hypothesis, we mutated three calcium-binding sites on C2A and C2B, individually and in combination. We found that the mutant proteins showed a reduced capacity to promote migrasome formation, further supporting our hypothesis that Syt1 needs to bind calcium to promote migrasome formation (Fig. 1, M and N). Calcium binding causes the insertion of four loops from the C2 domains into the lipid bilayer (Hui et al., 2006; Herrick et al., 2006), with M173, F234, V304, and I367 located on the tips of the membrane-binding loops mediating the membrane insertion (Fig. 1 J). To test whether membrane insertion is required for migrasome formation, we generated an M173A-F234A-V304A-I367A mutant (4A mutant). We found that the 4A mutant showed a reduced capacity to promote migrasome formation (Fig. 1, O and P), suggesting that membrane insertion of Syt-1 is required for migrasome formation.
Syt1 promotes the formation of migrasome-like structures in vitro
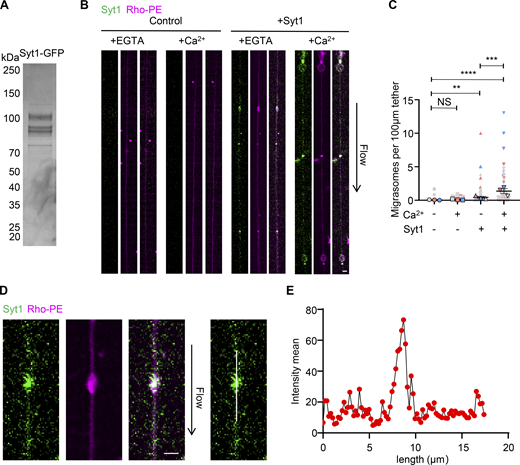
To test whether Syt1 alone is sufficient to drive the swelling of migrasomes, we used the in vitro reconstitution system we previously established (Huang et al., 2019). In this system, the giant unilamellar vesicles (GUVs) or Syt1-GFP GUVs transform into membrane tethers by flow in a flow chamber. As we previously showed, control GUVs rarely form migrasomes (Huang et al., 2019). In contrast, in Syt1-embedded GUVs, in the presence of Ca2+, migrasome-like structures are induced, while in the presence of EGTA, the formation of migrasome-like structures is significantly reduced (Fig. 2, A–C). Moreover, compared with the membrane tether, Syt1 is concentrated on migrasome-like structures, similar to what we observed in Syt1-GFP expressing cells (Fig. 2, D and E). Together, these data suggest that in the presence of Ca2+, Syt1 is sufficient to cause swelling of the membrane tether to form migrasome-like structures.
Syt1 promotes the formation of migrasome-like structures in vitro. (A) Recombinant Syt1-GFP was analyzed on an SDS-PAGE gel and stained with Coomassie Brilliant Blue. (B) Control GUVs and GUVs embedded with Syt1-GFP treated with or without Ca2+ were subjected to the in vitro reconstitution assay. Green signal, Syt1-GFP. Magenta signal, Rhodamine-PE. Scale bar, 2 µm. (C) Quantification of the number of migrasome-like structures per 100 µm of the tether from images in B. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 102 (no Syt1, no Ca2+), 108 (no Syt1, with Ca2+), 172 (with Syt1, no Ca2+), and 213 (with Syt1, with Ca2+) tethers. Quantifications are pooled from three independent experiments. Each biological replicate is color-coded: the statistical data from one experimental run is red, another independent experiment is represented by gray, and a third experiment is shown as blue. The dots, squares, triangles, and inverted triangles represent the four groups respectively. The bordered shapes represent those three means. (D) Image of Syt1-containing GUVs in vitro reconstitution assay. The mean intensity was generated using ImageJ. Scale bar, 2 µm. (E) Mean fluorescence intensity of Syt1 along the white lines in D. Source data are available for this figure: SourceData F2.
Syt1 promotes the formation of migrasome-like structures in vitro. (A) Recombinant Syt1-GFP was analyzed on an SDS-PAGE gel and stained with Coomassie Brilliant Blue. (B) Control GUVs and GUVs embedded with Syt1-GFP treated with or without Ca2+ were subjected to the in vitro reconstitution assay. Green signal, Syt1-GFP. Magenta signal, Rhodamine-PE. Scale bar, 2 µm. (C) Quantification of the number of migrasome-like structures per 100 µm of the tether from images in B. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 102 (no Syt1, no Ca2+), 108 (no Syt1, with Ca2+), 172 (with Syt1, no Ca2+), and 213 (with Syt1, with Ca2+) tethers. Quantifications are pooled from three independent experiments. Each biological replicate is color-coded: the statistical data from one experimental run is red, another independent experiment is represented by gray, and a third experiment is shown as blue. The dots, squares, triangles, and inverted triangles represent the four groups respectively. The bordered shapes represent those three means. (D) Image of Syt1-containing GUVs in vitro reconstitution assay. The mean intensity was generated using ImageJ. Scale bar, 2 µm. (E) Mean fluorescence intensity of Syt1 along the white lines in D. Source data are available for this figure: SourceData F2.
The recruitment of Syt1 induces an unstable swelling of migrasomes prior to the recruitment of TSPAN4
Previously, we showed that migrasome formation is driven by the assembly of a tetraspanin-enriched microdomain (Huang et al., 2019), which stabilizes the bulging of RFs caused by changes in tension. To study the molecular mechanism of Syt1-mediated migrasome formation, we first checked the recruitment of Syt1 compared with the recruitment of TSPAN4-GFP, which is considered the final step of migrasome formation. We found that Syt1 is recruited to migrasomes prior to the recruitment of TSPAN4, indicating that Syt1 works in a step before TSPAN4-mediated stabilization of the migrasome (Fig. 3, A and B; and Video 1).
The recruitment of Syt1 induces an unstable swelling of migrasomes prior to the recruitment of TSPAN4. (A) Time-lapse imaging of NRK cells stably expressing Syt1-mCherry and TSPAN4-GFP. Confocal microscopy images were captured every 2 min 54 s. Scale bar, 2 μm. (B) Statistical analysis of normalized fluorescence intensity of Syt1 and TSPAN4 at migrasome formation sites during migrasome formation shown in A. Data are presented as mean ± SEM; n = 30. (C) Time-lapse imaging of NRK Syt1-GFP migrasome formation. Green signal, Syt1-GFP. The time-lapse imaging from top to bottom represent three independent migrasomes. Scale bar, 2 µm. (D) Time-lapse imaging of migrasomes from NRK Syt1-mCherry, NRK TSPAN4-GFP, and NRK Syt1-mCherry TSPAN4-GFP. Scale bar, 2 µm. (E) Statistical analysis of the shrinking back migrasomes to all migrasomes ratio shown in D. Data are presented as mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50. The experiments were performed three times. (F) Quantification of the life time of migrasome shown in D. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (G) Time-lapse imaging of migrasomes from NRK Syt1-GFP treated with/without 1 mM Ca2+ for 12 h. Scale bar, 2 µm. (H) Quantification of the life time of migrasome shown in G. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 100 cells per group. The experiments were performed three times.
The recruitment of Syt1 induces an unstable swelling of migrasomes prior to the recruitment of TSPAN4. (A) Time-lapse imaging of NRK cells stably expressing Syt1-mCherry and TSPAN4-GFP. Confocal microscopy images were captured every 2 min 54 s. Scale bar, 2 μm. (B) Statistical analysis of normalized fluorescence intensity of Syt1 and TSPAN4 at migrasome formation sites during migrasome formation shown in A. Data are presented as mean ± SEM; n = 30. (C) Time-lapse imaging of NRK Syt1-GFP migrasome formation. Green signal, Syt1-GFP. The time-lapse imaging from top to bottom represent three independent migrasomes. Scale bar, 2 µm. (D) Time-lapse imaging of migrasomes from NRK Syt1-mCherry, NRK TSPAN4-GFP, and NRK Syt1-mCherry TSPAN4-GFP. Scale bar, 2 µm. (E) Statistical analysis of the shrinking back migrasomes to all migrasomes ratio shown in D. Data are presented as mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50. The experiments were performed three times. (F) Quantification of the life time of migrasome shown in D. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (G) Time-lapse imaging of migrasomes from NRK Syt1-GFP treated with/without 1 mM Ca2+ for 12 h. Scale bar, 2 µm. (H) Quantification of the life time of migrasome shown in G. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 100 cells per group. The experiments were performed three times.
Movie of an NRK cell expressing Syt1-mCherry and TSPAN4-GFP. Time-lapse images were taken every 2 min 54 s for 1 h 33 min 48 s. Images were captured by FV3000. Scale bars, 2 μm. Playback speed is 5 frames per second.
Movie of an NRK cell expressing Syt1-mCherry and TSPAN4-GFP. Time-lapse images were taken every 2 min 54 s for 1 h 33 min 48 s. Images were captured by FV3000. Scale bars, 2 μm. Playback speed is 5 frames per second.
NRK cells only generate a very small amount of migrasome and thus have been extensively used to study the effect of overexpression of migrasome genes. When compared with the overexpression of TSPAN4-GFP, which promotes the formation of stable, spherical migrasomes, we found that the shape of Syt1-expressing induced migrasomes is highly unstable (Fig. 3 C). The shape of these migrasomes keeps changing, transforming between spherical and cylindrical shapes, which is uncommon in TSPAN4-GFP-induced migrasomes. Moreover, the Syt1-induced migrasomes often shrink back after swelling, a behavior relatively uncommon in TSPAN4-induced migrasomes (Fig. 3, D and E). Unsurprisingly, Syt1-induced migrasomes have a significantly shorter lifespan compared with TSPAN4-GFP–induced migrasomes, and co-expression of Tspan4-GFP with Syt-1 prolongs the lifespan of these migrasomes (Fig. 3, D and F). Moreover, adding 1 mM Ca2+ can further shorten the lifespan of Syt1-induced migrasomes (Fig. 3, G and H). Together, these data suggest that Syt1 and TSPAN4 work sequentially: the recruitment of Syt1 induces an unstable swelling of migrasomes, followed by a stabilization stage driven by TSPAN4.
Syt1 is necessary for migrasome biogenesis
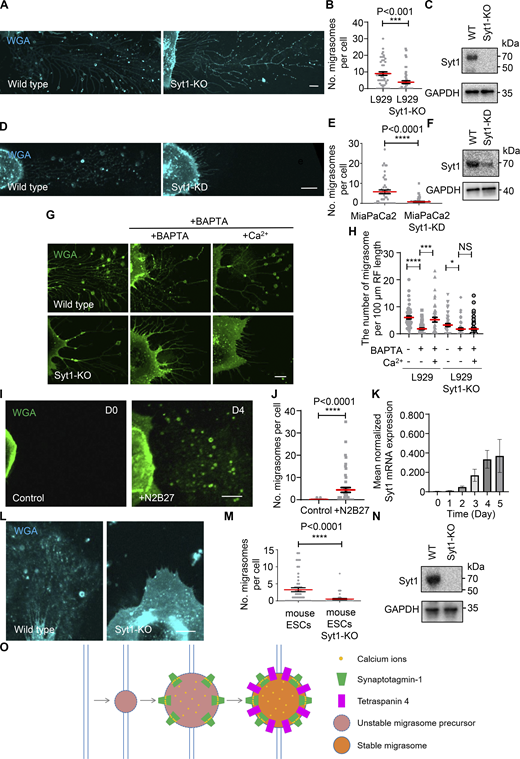
Next, we tested whether Syt1 is necessary for migrasome biogenesis. While Syt1 is known for its role in neurotransmitter release, we found that the expression of Syt1 is not restricted to neurons. In L929 cells, which have been extensively used to study migrasome biogenesis (Zhu et al., 2021; Jiao et al., 2021; Liang et al., 2023), Syt1 is expressed. We found that knocking out Syt1 significantly reduces migrasome formation in L929 cells (Fig. 4, A–C). Similarly, Syt1 is expressed in MiaPaCa2 cells and is required for migrasome formation, suggesting that Syt1 is necessary for this process (Fig. 4, D–F). We found that in BAPTA-pretreated wild-type cells, replacing BAPTA with a calcium-containing medium can trigger migrasome formation. In contrast, Syt1 knockout cells are not able to respond to calcium-triggered migrasome formation, suggesting that calcium promotes migrasome formation via Syt1 (Fig. 4, G and H).
Syt1 is necessary for migrasome biogenesis. (A) Confocal images of migrasomes of cells derived from WT and Syt1 knockout L929. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (B) Quantification of the number of migrasomes in WT and Syt1 knockout L929 shown in A. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (C) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockout L929. (D) Confocal images of migrasomes of cells derived from WT and Syt1 knockdown MiaPaCa2. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (E) Quantification of the number of migrasomes in WT and Syt1 knockdown MiaPaCa2 shown in D. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (F) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockdown MiaPaCa2. (G) Confocal images of migrasomes of cells from WT and Syt1 knockout L929 with 0.5 μM BAPTA pretreatment and following with/without 1 mM Ca2+ rescue. Left panels: Cells cultured overnight without BAPTA pretreatment. Middle panels: Cells underwent BAPTA pretreatment overnight without Ca2+ rescue after 6 h. Right panels: Cells experienced BAPTA pretreatment, 6 h after BAPTA pretreatment, Ca2+ was added into culture medium to substitute for the medium. Cells were labeled with WGA (green). Scale bar represents 10 µm. (H) Quantification of the number of migrasomes per 100 μm RF length per cell in WT and Syt1 knockout L929 shown in G. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (I) Confocal image of migrasomes of mouse embryonic stem cells treated with N2B27 at D0 and D4. Cells are labeled with WGA (green). Scale bar, 10 µm. (J) The number of migrasomes per cell from images like those shown in I was quantified. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (K) mRNA levels of Syt1, determined by quantitative PCR analysis, of D0-D5 in mouse embryonic stem cells treated with N2B27. Quantitative PCR data were normalized to Gapdh mRNA level and data are reported as the mean ± SEM. n = 3 independent experiments. (L) Confocal images of migrasomes of cells derived from WT and Syt1 knockout mouse embryonic stem cells treated with N2B27 at D4. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (M) Quantification of the number of migrasomes in WT and Syt1 knockout group shown in L. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (N) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockout mouse embryonic stem cells treated with N2B27 at D4. (O) Model of the role of Syt1 in migrasome biogenesis. Source data are available for this figure: SourceData F4.
Syt1 is necessary for migrasome biogenesis. (A) Confocal images of migrasomes of cells derived from WT and Syt1 knockout L929. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (B) Quantification of the number of migrasomes in WT and Syt1 knockout L929 shown in A. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (C) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockout L929. (D) Confocal images of migrasomes of cells derived from WT and Syt1 knockdown MiaPaCa2. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (E) Quantification of the number of migrasomes in WT and Syt1 knockdown MiaPaCa2 shown in D. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (F) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockdown MiaPaCa2. (G) Confocal images of migrasomes of cells from WT and Syt1 knockout L929 with 0.5 μM BAPTA pretreatment and following with/without 1 mM Ca2+ rescue. Left panels: Cells cultured overnight without BAPTA pretreatment. Middle panels: Cells underwent BAPTA pretreatment overnight without Ca2+ rescue after 6 h. Right panels: Cells experienced BAPTA pretreatment, 6 h after BAPTA pretreatment, Ca2+ was added into culture medium to substitute for the medium. Cells were labeled with WGA (green). Scale bar represents 10 µm. (H) Quantification of the number of migrasomes per 100 μm RF length per cell in WT and Syt1 knockout L929 shown in G. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (I) Confocal image of migrasomes of mouse embryonic stem cells treated with N2B27 at D0 and D4. Cells are labeled with WGA (green). Scale bar, 10 µm. (J) The number of migrasomes per cell from images like those shown in I was quantified. Data shown represent the mean ± SEM; P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (K) mRNA levels of Syt1, determined by quantitative PCR analysis, of D0-D5 in mouse embryonic stem cells treated with N2B27. Quantitative PCR data were normalized to Gapdh mRNA level and data are reported as the mean ± SEM. n = 3 independent experiments. (L) Confocal images of migrasomes of cells derived from WT and Syt1 knockout mouse embryonic stem cells treated with N2B27 at D4. Cells are labeled with WGA (cyan). Scale bar, 10 µm. (M) Quantification of the number of migrasomes in WT and Syt1 knockout group shown in L. Data represent the mean ± SEM. P values were calculated using a two-tailed, unpaired t test. n = 50 cells per group. The experiments were performed three times. (N) Western blot analysis of Syt1 and GAPDH in WT and Syt1 knockout mouse embryonic stem cells treated with N2B27 at D4. (O) Model of the role of Syt1 in migrasome biogenesis. Source data are available for this figure: SourceData F4.
Elevated Syt-1 expression levels promote migrasome formation during the differentiation of mouse embryonic stem cells into neuronal cells
We found that when we treat mouse embryonic stem cells (mESCs) with N2B27, which induces ES cells to differentiate into neuronal cells, migrasome formation is induced 4 days after N2B27 treatment when ES cells have differentiated into neuroectodermal precursors (Fig. 4, I and J). Interestingly, when we checked the expression level of Syt1, we found that ES cells barely express Syt1. However, 3 days after N2B27 treatment, the cells start to express Syt1 and continue increasing its expression level, suggesting the possibility that the formation of migrasomes during ES to neuronal cell differentiation may be driven by the expression of Syt1 (Fig. 4 K). To test this hypothesis, we knocked out Syt1 in ES cells and then treated the cells with N2B27. We found that in Syt1 knockout cells, N2B27 treatment cannot induce migrasome formation anymore (Fig. 4, L–N). Put together, these data suggest that Syt1 is necessary for migrasome formation.
Discussion
In this study, we have elucidated the role of Ca2+ in facilitating migrasome formation. Our findings demonstrate that Syt1, a Ca2+ sensor, is essential for this process. The capacity of Syt1 to bind Ca2+ is critical for its function in promoting migrasome formation. Live cell imaging analyses revealed that Syt1 is recruited to sites of migrasome formation before the recruitment of TSPAN4. This suggests that Syt1 functions upstream of the assembly of the TEM. The recruitment of Syt1 leads to swelling of the MFS, resulting in the creation of an unstable migrasome precursor. This precursor can be further stabilized by the subsequent recruitment of the TEM (Fig. 4 O). Our research uncovers a pivotal step that facilitates the transition from tension-induced swelling to the establishment of a stable migrasome.
Ca2+ is widely recognized for its significant role in various aspects of cell migration, including cytoskeletal reorganization, adhesion, and the dynamics of focal adhesions. This study extends our understanding of Ca2+ functions by highlighting its involvement in another migration-related process—migrasome formation. The enrichment of Syt1 in migrasomes and the necessity of its Ca2+ binding capacity for migrasome formation imply that Ca2+ is likely sensed locally at the migrasome by Syt1 in cells expressing this protein. We hypothesize that other calcium sensors may play similar roles in cells lacking Syt1 expression.
The mechanism by which Syt1 promotes the formation of unstable migrasome precursors remains to be fully elucidated. However, the fact that Syt1 overexpression can promote the formation of unstable, Syt1-positive migrasome precursors, along with the observation that the presence of Syt1 on GUVs induces the formation of migrasome-like structures in our in vitro reconstitution system, suggests that Syt1 may facilitate tension-induced swelling, thereby promoting migrasome formation. Future research is required to explore the exact mechanisms through which Syt1 facilitates initial swelling.
Materials and methods
Cell culture, transfection, and lentivirus infection
NRK (Number: CRL-6509; ATCC), L929 (Number: CCL-1; ATCC), MiaPaCa2 (Number: CRL-1420; ATCC), 293T (Number: CRL-3216; ATCC), and their derivatives were cultured at 37°C and 5% CO2 in DMEM supplemented with 10% serum, 1% Glutamax, and 1% penicillin–streptomycin. Suspensions of 293F (gift from Laboratory of Hongwei Wang, School of Life Sciences, Tsinghua University, Beijing, China) cells were cultured at 37°C, 8% CO2, and 125 rpm in SMM293-TI supplemented with 1% penicillin–streptomycin.
Mouse mESCs (gift from Laboratory of Xiaohua Shen, School of Medicine, Tsinghua University, Beijing, China) were adapted to and maintained on 0.1% gelatin-coated tissue plates, and cells were maintained in complete ESC culture medium (DMEM; Dulbecco’s Modified Eagle’s Medium) supplemented with 15% heat-inactivated fetal calf serum, 0.1 mM 2-mercaptoethanol, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1% nucleoside mix (100X stock; Sigma-Aldrich), 1,000 U/ml recombinant leukemia inhibitory factor (LIF2010; Merck Millipore), and 50 U/ml penicillin–streptomycin. Differentiation of ES cells was induced by LIF withdrawal. Cells were dissociated and plated onto fibronectin-coated tissue culture plastic dishes at a density of 0.5–1.5 × 10^4/cm^2 in N2B27 medium. The medium was renewed every 2 days. N2B27 is a 1:1 mixture of DMEM/F12 (11320033; Gibco) supplemented with modified N2 (CA3018012; 100X stock) and neurobasal medium supplemented with B27 (CA3015008; 50X stock), and the medium was supplemented with 1% Glutamax and 1% penicillin–streptomycin. ES cells were differentiated for 4–5 days in the N2B27 medium.
For NRK transfection, one-third of cells from a full 6-cm dish were transfected with 3–5 μg plasmid via Amaxa nucleofection using program NRK and electroporation solution. For transfection of L929 cells, 80–90% confluent cultured cells from a 3.5-cm dish were transfected with 5 μg DNA via Vigofect (T001; Vigorous). For purification of Syt1-GFP, 1 liter of 293F cells was transfected by adding 2 mg DNA with 3 mg polyethylenimine (1 mg/ml).
For lentivirus production, 293T cells were seeded in 12-well plates and cultured to 80% confluency; each well of cells was then transfected with 4 µg DNA (PSPAX2:pMDZ.G:shRNA-plasmid, 3:2:2) and 1.6 µl Vigofect in 150 µl NaCl (150 mM). The transfection mix was replaced with fresh medium after 12 h. The 293T cells were cultured for another 24 h. For virus infection, the 12-well plate was pre-seeded with MiaPaCa2 cells. A 1:1 mixture of 500 µl of the 293T cells supernatant medium and 500 µl fresh medium was prepared and transferred to a 12-well plate on which MiaPaCa2 cells were cultured for 24 h. MiaPaCa2 cells were treated with polybrene (8 µg/ml). After 24 h, the medium was replaced with fresh medium, and the cells were cultured for another 24 h. Medium containing 3 µg/ml puromycin was then added to select resistant colonies. The resistant colonies were seeded in plates pre-coated with fibronectin (10 µg/ml) in full medium for 12 h.
Stable cell generation
For transposon-mediated overexpression to establish stable cell lines, overexpression vectors were cotransfected with pBASE transposase into cells (the primer sequences mentioned in Table S1). Cells that stably expressed genes through transposon-mediated random insertion were selected with hygromycin. The resistant colonies were maintained for subsequent experiments.
Generation of Syt1 knockout cell lines
To generate KO cell lines, the Syt1 gene in L929 cells and ESCs was deleted using a modified PX458 plasmid (provided by W. Guo, International Campus Zhejiang University, Jiaxing City, Zhejiang Province, China) that contains two guide RNAs coupled with Cas9 nuclease. Compared with mutations generated by single guide RNAs (sgRNAs), mutations generated by targeting with dual sgRNAs are more likely to yield visibly shorter PCR fragments when amplified from genomic DNA. The sgRNA sequences used for CRISPR–Cas9 were 5′-GGTTCAAGCGGAACTGTGAT-3′ (sgRNA-1) and 5′-GAGCACGAAGTCTAACCCGT-3′ (sgRNA-2) for L929 cells and mESCs (the primer sequences mentioned in Table S1). After 48 h of transfection, cells were seeded into 96-well plates using fluorescence-activated cell sorting (FACS) for enhanced GFP signal. Syt1 KO clones were identified by western blotting, and the correct clones were selected.
Generation of plasmids
The shRNAs originated from the Custom Glycerol-mouse shRNA library at the Center of Biomedical Analysis, Tsinghua University, initially purchased from Sigma-Aldrich. Information about the human Syt1 shRNA includes: TRC_ID is TRCN0000053856; PLATE Name, Row, and Column as AAH19, E, and 6, respectively. The Olig_Seq is 5′-CCGGGCGACTGTTCTGCCAAGCAATCTCGAGATTGCTTGGCAGAACAGTCGCTTTTTG-3′. Vector Control is the Empty Vector Control SHC001 (pLKO.1-puro). For stable cell generation, the Syt1 gene was cloned from mouse cDNA and inserted into pB-GAG-BGH. Syt1 point mutations and truncation mutants were derived from the original plasmid. For protein purification, Syt1-GFP were cloned into pCAG-strep. The Syt1 genes were originally cloned from mouse cDNAs. The primer sequences are mentioned in Table S1.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (15596026; Invitrogen). Complementary DNA was synthesized using a ReverTra Ace qPCR RT kit (FSQ-101; TOYOBO). Quantitative real-time PCR was performed using 2×T5 fast qPCR mix (TSINGKE Biological Technology, TSE202; SYBR Green I). The primer sequences are mentioned in Table S1.
Protein purification
pCAG-strep-Syt1-GFP was transfected into 293F cells using a polyethylenimine-based transfection protocol. For the transfection of 1 liter of 293F cells, 2 mg of DNA was used. After transfection, the 293F cells were collected after 3 days and lysed using a Dounce tissue grinder (100 strokes) in a buffer containing 25 mM HEPES (pH 7.4), 400 mM NaCl, protease inhibitor cocktail (Roche), and 1 mM DTT. After centrifugation at 30,000 g for 1 h at 4°C, the pellet was lysed in a buffer containing 25 mM HEPES (pH 7.4), 400 mM NaCl, protease inhibitor cocktail (Roche), 1 mM DTT, and 1% β-OG for 1 h at 4°C. After another centrifugation at 30,000 g for 1 h, the supernatant was loaded onto a column containing Strep-Tactin resin (IBA) and incubated for 1 h at 4°C. The resin was then washed four times, and the protein was eluted with 10 mM desthiobiotin (Sigma-Aldrich).
Liposome preparation and protein insertion
First, we prepared small unilamellar vesicles using POPC (1-palmitoyl-2-oleoyl-glycero-3-phosphocholine), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), POPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine), DOPE-rhodamine, and PE-biotin, all purchased from Avanti Polar Lipids, with cholesterol from Sigma-Aldrich. The lipids were mixed in a ratio of POPC:DOPE:POPS:cholesterol (4:2.5:2.5:1) with 0.1% DOPE-rhodamine and 1% PE-biotin, dried under a nitrogen stream, and further dried for 1 h at 37°C. The lipid film was then hydrated with a buffer containing 25 mM HEPES (pH 7.4) and 400 mM NaCl and subjected to 13 cycles of freezing in liquid nitrogen and thawing in a 42°C water bath. Subsequently, the liposomes were extruded 21 times through a polycarbonate film with a 1,000-nm pore size to produce small unilamellar vesicles (SUVs). For protein insertion, 1% β-OG was added to loosen the SUV membranes at 4°C for 30 min, followed by the addition of proteins with 35 rpm rotation at 4°C for 1 h. The β-OG was removed by adding 8–15 mg BIO-beads (Bio-Rad) per 100 µl system, repeated four times. The supernatant was then stored.
Preparation of GUVs with embedded proteins
Giant unilamellar vesicles (GUVs) were prepared following the protocols (Garten et al., 2015). We prepared a 1% agarose solution by dissolving 100 mg of agarose in 10 ml of pure water and heated it until boiling to ensure complete dissolution. A cover slide was plasma-cleaned (air plasma) for 1 min to enhance the spreading of the agarose solution. Quickly using the cover slides, within 10 min, we added 200 µl of warm agarose solution to a 22 × 22 mm^2 slide, ensuring the solution covered the entire surface to leave a thin, smooth layer of agarose. The slide was then placed in an oven at 60°C and left to dry further for 30 min. After cooling to room temperature, the slides were placed in a 3.5-cm dish and used immediately. The SUV solution was applied in very small drops onto the agarose surface and further dried under a gentle stream of nitrogen. Once the SUVs had dried, a buffer (5 mM KCl, 1 mM HEPES [pH 7.4]) was added to cover the slide surface, allowing the swelling to proceed for 30 min. The GUVs were then collected and stored at 4°C to be used within 3 days.
Migrasome reconstitution assay via flow channel
The GUVs were incubated with either 1 mM CaCl2 or 1 mM EGTA for 5 h. The flow channels were prepared as per the previously published motility assay protocols using hydrophilic coverslips (Wang et al., 2015; Du W et al., 2016; Su et al., 2016). The channels were blocked with 20 µg/ml streptavidin at room temperature for 6 min. Then, GUVs (20 µl) were incubated in the channels for 6 min at room temperature. Subsequently, 60 µl of buffer (5 mM KCl, 1 mM HEPES [pH 7.4]) was flowed through the channel. Images were acquired using an Olympus FV-1200 confocal microscope.
Cell imaging and image analysis
Confocal imaging and analysis were conducted using 3.5-cm dishes pre-coated with fibronectin (10 µg/ml) at 37°C for 30 min. Cells were cultured in these fibronectin-precoated dishes for 10–12 h. For mESCs differentiation from day 0 to day 5, cells were cultured in fibronectin-precoated plastic confocal dishes. After differentiation, neuroectodermal precursors were collected by Accutase digestion and then cultured in fibronectin-precoated dishes for 10–12 h.
Confocal snapshot images and long-term time-lapse images of living cells were acquired using an Olympus FV-3000 (60×/1.30 silicone oil objective) or NIKON A1 confocal microscope (60×/1.40 oil objective). Images were collected at 1,024 × 1,024 pixels resolution. The cells were maintained at 37°C in an atmosphere containing 5% CO2. Images were processed with ImageJ (RRID:SCR_003070), and statistical analyses were conducted using GraphPad Prism 8.0.1 (RRID: SCR_002798).
Western blot
Cells or migrasomes were lysed with SDS lysis buffer (2.5% in PBS) and then boiled for 20 min at 95°C. Proteins were separated on SDS-PAGE gels and transferred electrophoretically onto nitrocellulose membranes. The membranes were blocked with 5% non-fat milk in TBST buffer and incubated with the primary antibody in solution 1 (TOYOBO) overnight at 4°C. Nitrocellulose membranes were then incubated with HRP-conjugated secondary antibody for 1 h at room temperature. Signals were detected using a WESTAR NOVA2.0 kit (CYANAGEN). The primary antibodies used were Rabbit anti-Syt1 (1:1,000; ab302627; abcam), Rabbit anti-Syt1 (1:1,000; 14558S; CST), and Rabbit anti-GAPDH (1:5,000; ab313650; abcam).
Statistics and reproducibility
All experiments were performed independently at least three times. The quantification of migrasome numbers per 100 µm RF length, the migrasome numbers, and the Max Feret’s diameter of migrasomes were carried out three times. To calculate migrasome numbers per 100 µm RF length, the total number of migrasomes was counted, and the total length of RFs was measured in at least 50 randomly selected cells. For statistical analysis of migrasomes number, at least 50 random cells were chosen and the numbers of migrasomes were counted. For statistical analysis of the Max Feret’s diameter of migrasomes, at least 50 migrasomes were chosen. For statistical analysis of the time course of the normalized mean fluorescence intensity of TSPAN4–GFP and Syt1-mCherry during migrasome biogenesis, at least 30 random migrasomes were chosen. To calculate the number of migrasome-like structures per 100 µm tether length in vitro, the length of each tether was measured and the total number of migrasome-like structures on the tether was counted for more than 102 randomly selected tethers. Statistical analyses were conducted using the unpaired two-tailed t test in GraphPad Prism 8.0.1. Error bars represent the mean ± SEM Significance levels are denoted as *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS = not significant.
Online supplemental material
Data availability
The data are presented in the published article, and additional data files are available upon reasonable request.
Acknowledgments
We thank the State Key Laboratory of Membrane Biology for confocal microscopy imaging and facility support. We thank the Xiaohua Shen lab (School of Medicine, Tsinghua University, Beijing, China) for sharing the mESCs with us. We thank the Hongwei Wang Lab (School of Life Sciences, Tsinghua University, Beijing, China) for sharing the 293F with us.
This research was supported by Tsinghua-Toyota Joint Research Fund (grant no. 20233930058 to L. Yu), the National Natural Science Foundation of China (grant no. 92354306, 32330025, and 32030023 to L. Yu), and Beijing Municipal Science and Technology Commission, Administrative Commission of Zhongguancun Science Park (grant no. Z221100003422012 to L. Yu).
Author contributions: Funding acquisition, resources, and supervision: L. Yu; Data curation, formal analysis, investigation, methodology, project administration, validation, visualization: Y. Han; Conceptualization, Writing–original draft and writing–review and editing: L. Yu and Y. Han.
References
Author notes
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Y. Han reported “L. Yu is the scientific founder of Migrasome Therapeutics Inc.” L. Yu reported “L. Yu is the scientific founder of Migrasome Therapeutics Inc.”