In autophagy, autophagosomes deliver the lumenal contents to lysosomes for degradation via autophagosome–lysosome fusion. In contrast, autophagosome outer membrane components were recycled via autophagosomal components recycling (ACR), which is mediated by the recycler complex. The recycler complex, composed of SNX4, SNX5, and SNX17, cooperate with the dynein–dynactin complex to mediate ACR. However, how ACR is regulated remains unknown. Here, we found that Rab32 family proteins localize to autolysosomes and are required for ACR, rather than other autophagosomal or lysosomal Rab proteins. The GTPase activity of Rab32 family proteins, governed by their guanine nucleotide exchange factor and GTPase-activating protein, plays a key role in regulating ACR. This regulation occurs through the control of recycler complex formation, as well as the connection between the recycler-cargo and dynactin complex. Together, our study reveals an unidentified Rab32 family-dependent regulatory mechanism for ACR.
Introduction
Autophagy is a highly conserved intracellular process that scavenges and selectively degrades cytoplasmic components. Defects in autophagy have been implicated in numerous diseases, including neurodegenerative and infection-related diseases (Fleming et al., 2022; Mizushima and Levine, 2020). During autophagy, isolation membranes seal and close to form complete autophagosomes, which then deliver the lumenal contents to lysosomes for degradation (Bas et al., 2018; Gao et al., 2018; Itakura et al., 2012; Klionsky et al., 2021; Matsui et al., 2018; Takáts et al., 2013, 2018; Yamamoto et al., 2023). Although internal cargoes are degraded in lysosomes, we recently found that outer membrane components of autophagosomes are recycled from autolysosomes via autophagosomal components recycling (ACR) by the recycler complex, but not by the retromer and retriever complex, which are required for recycling of membrane proteins from endosomes to Golgi apparatus or cell surface (Zhou et al., 2022). However, the destination for these recycled cargoes remained largely unknown. A prior study demonstrated that ATG9A on lysosomes is retrieved and sent to endosomes during starvation (Ravussin et al., 2021).
Recycler comprises the SNX4, SNX5, and SNX17 subunits, among which SNX4 forms a heterodimer with SNX5 that induces curvature in the autolysosomal membrane via their BAR domains to facilitate ACR. These BAR domains are also responsible for the recycler’s recognition of autophagosomal outer membrane components, such as STX17 and ATG9A. SNX17 connects the dynein–dynactin complex with SNX4–SNX5–cargo to mediate the minus-end movement of retrieved budding vesicles that shuttles cargoes along microtubules. Disabled ACR leads to the accumulation of autophagosomal outer membrane proteins as well as SNX4 and SNX5 on autolysosomes (Zhou et al., 2022). Although the recycler complex responsible for ACR has been characterized, it remains unknown how ACR is regulated.
Rab proteins are well-known for their participation in various intracellular membrane trafficking pathways, such as vesicle budding, vesicle tethering, vesicle fusion, and vesicle trafficking, by acting as molecular switches that cycle between inactive GDP-bound and active GTP-bound states (Borchers et al., 2021). Among Rab proteins, Rab32 family proteins, thus far including only Rab32 and Rab38, are conserved between vertebrate and invertebrate animals (Ohbayashi et al., 2017). Both Rab32 and Rab38 have been shown to localize to lysosome-related organelles in melanocytes under physiological conditions (Bultema and Di Pietro, 2013). They share ∼75% amino acid sequence similarity and show function redundancy in many physiological/pathological processes (Bultema and Di Pietro, 2013; Ohbayashi et al., 2017). RUTBC1 functions as a GTPase-activating protein (GAP) to inactivate Rab32 and Rab38 by stimulating their GTPase activity (Marubashi et al., 2016; Nottingham et al., 2011). The biogenesis of lysosome-related organelles complex 3 (BLOC-3), comprising HPS1 and HPS4, functions as the guanine nucleotide exchange factor (GEF) to activate Rab32 and Rab38 (Gerondopoulos et al., 2012). Mutations of BLOC-3 have been linked to Hermansky–Pudlak syndrome (Chiang et al., 2003; Martina et al., 2003; Nazarian et al., 2003). The large majority of studies examining the physiological function of Rab32 and Rab38 have focused on melanosomes (Sitaram and Marks, 2012); however, it still remains unknown whether Rab32 and Rab38 function on autolysosomes in non-melanocyte cells.
Based on our previous work, which showed that some Rab proteins were biotinylated by APEX2-STX17-TM under starvation conditions (Zhou et al., 2022); in the current study, we screened 32 Rab proteins that potentially interact with STX17. Rab32 and Rab38 were identified to localize on autolysosomes. Further, we found that Rab32 and Rab38 deficiency resulted in the accumulation of STX17 and ATG9A on autolysosomes, inhibited the recycler complex formation, disrupted the connection between recycler and dynactin complex, and ultimately blocked ACR in a Rab-GTPase activity-dependent manner.
Results
Both Rab32 and Rab38 are required for STX17 retrieval from autolysosomes
Given the essential roles that Rab proteins (hereafter Rabs) play in various membrane and protein trafficking pathways, we investigated whether Rabs were also involved in ACR by first searching our previous mass spectral results generated by the APEX2-STX17-TM proximal biotinylation assay (Zhou et al., 2022). In total, 32 Rab proteins were identified in this dataset (Table S1). Immunofluorescent costaining assays with these 32 GFP-fusion Rabs revealed that Rab7A, Rab9A, Rab9B, and Rab32 colocalized with LC3 and LAMP1, suggesting that these Rabs localize to autolysosomes (i.e., LC3+/LAMP1+ vesicles) (Fig. S1, A and B; and Fig. 1 A).
Examination the localization of Rab proteins. (A) HeLa cells were transfected with GFP-tagged Rab proteins. 24 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against GFP, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 with mKO-Rab32 or mKO-Rab38 were starved with EBSS for 2 h and stained with antibodies against Flag and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (C and D) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, cells were transfected with HA-STX17 with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData FS1.
Examination the localization of Rab proteins. (A) HeLa cells were transfected with GFP-tagged Rab proteins. 24 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against GFP, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 with mKO-Rab32 or mKO-Rab38 were starved with EBSS for 2 h and stained with antibodies against Flag and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (C and D) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, cells were transfected with HA-STX17 with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData FS1.
Rab32 and Rab38 are required for ACR. (A) U2OS cells stably expressing Flag-STX17 with mKO-Rab32 were starved with EBSS for 2 h, and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNAs against Rab32, Rab38, or Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (C) Quantification of STX17-positive autolysosome number. Images in B and (S2C) were analyzed. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001, ***P < 0.001. Two-way ANOVA. (D) U2OS cells stably expressing Flag-STX17 with mKO-Rab38 were starved with EBSS for 2 h, and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) HEK293T cells were treated with EBSS for 2 h. Then Lyso-IP was performed. The resultant samples were subjected to immunoblot analysis with the indicated antibodies. * indicates a non-specific band. (F) The samples from Lyso-IP were split into three aliquots and subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. Then immunoblot analysis was performed with the indicated antibodies. * indicates a non-specific band. (G) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (H) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (I) Quantification of SNX4 or SNX5 localization to autolysosomes in G and H. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. Source data are available for this figure: SourceData F1.
Rab32 and Rab38 are required for ACR. (A) U2OS cells stably expressing Flag-STX17 with mKO-Rab32 were starved with EBSS for 2 h, and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNAs against Rab32, Rab38, or Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (C) Quantification of STX17-positive autolysosome number. Images in B and (S2C) were analyzed. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001, ***P < 0.001. Two-way ANOVA. (D) U2OS cells stably expressing Flag-STX17 with mKO-Rab38 were starved with EBSS for 2 h, and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) HEK293T cells were treated with EBSS for 2 h. Then Lyso-IP was performed. The resultant samples were subjected to immunoblot analysis with the indicated antibodies. * indicates a non-specific band. (F) The samples from Lyso-IP were split into three aliquots and subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. Then immunoblot analysis was performed with the indicated antibodies. * indicates a non-specific band. (G) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (H) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (I) Quantification of SNX4 or SNX5 localization to autolysosomes in G and H. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. Source data are available for this figure: SourceData F1.
As shown in our previous work, STX17 is translocated from autophagosomes to autolysosomes via autophagosome–lysosome fusion and retrieved from autolysosomes by the recycler complex (Zhou et al., 2022). We next investigated whether these four Rabs were involved in this recycling process by knocking down (KD) these four genes in cells stably expressing Flag-STX17. In Rab7A KD cells, colocalization between STX17 and LAMP1 was rare (Fig. S2, A and B), indicating severe blockage of autophagosome–lysosome fusion, as reported by previous studies (Yu et al., 2018; Zhao and Zhang, 2019). Focusing on the other three Rabs, we observed no accumulation of STX17 on autolysosomes in either Rab9A KD or Rab9B KD cells (Fig. S2, A and B). Nonetheless, STX17 clearly accumulated on autolysosomes of Rab32 KD cells with autophagosomes forming and fusing with lysosomes at 2 h after starvation (Fig. 1, B and C; and Fig. S2 C). Additional siRNA against Rab32 also significantly inhibited STX17 retrieval, and the retrieval defect was rescued by Rab32 WT (Fig. S3, A–E). These data suggest that, among these candidate Rab proteins, Rab32 is involved in STX17 retrieval from autolysosomes.
The effects of Rab7A, Rab9A or Rab9B on STX17 location to autolysosomes. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC) or siRNA against Rab7A, Rab9A, or Rab9B. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. Two-way ANOVA. (C) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against Rab32, Rab38 or Rab32, and Rab38. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm.
The effects of Rab7A, Rab9A or Rab9B on STX17 location to autolysosomes. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC) or siRNA against Rab7A, Rab9A, or Rab9B. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. Two-way ANOVA. (C) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against Rab32, Rab38 or Rab32, and Rab38. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm.
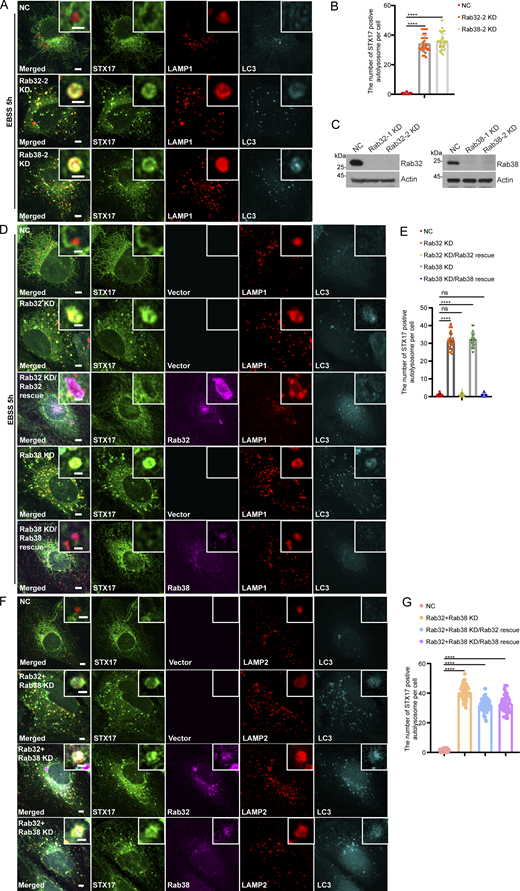
The STX17 retrieval defect caused by Rab32/Rab38 knockdown is rescued by the expression of wild-type Rab32/Rab38. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (C) U2OS cells stably expressing Flag-STX17 were transfected with the indicated siRNAs. 48 h after transfection, cells were subjected to immunoblot with antibodies against Rab32, Rab38, and Actin. (D) U2OS cells stably expressing Flag-STX17 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of STX17 positive autolysosomes in D. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. One-way ANOVA. (F) U2OS cells stably expressing Flag-STX17 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of STX17 positive autolysosomes in F. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. Source data are available for this figure: SourceData FS3.
The STX17 retrieval defect caused by Rab32/Rab38 knockdown is rescued by the expression of wild-type Rab32/Rab38. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (C) U2OS cells stably expressing Flag-STX17 were transfected with the indicated siRNAs. 48 h after transfection, cells were subjected to immunoblot with antibodies against Rab32, Rab38, and Actin. (D) U2OS cells stably expressing Flag-STX17 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 or Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of STX17 positive autolysosomes in D. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. One-way ANOVA. (F) U2OS cells stably expressing Flag-STX17 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of STX17 positive autolysosomes in F. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. Source data are available for this figure: SourceData FS3.
Since Rab32 and Rab38 in the Rab32 family are known to have redundant functions in some physiological processes (Bultema and Di Pietro, 2013; Ohbayashi et al., 2017), we therefore investigated whether Rab38 also participates in STX17 retrieval. Colocalization assays showed that Rab38 also localized to STX17+ autolysosomes (Fig. 1 D and Fig. S1 B). Further, STX17 retrieval was significantly inhibited under Rab38 KD, and autophagosomes formed and fused with lysosomes at 2 h after starvation (Fig. 1, B and C; and Fig. S2 C). Knockdown of Rab38 with additional siRNA also significantly inhibited STX17 retrieval, and the retrieval defect was rescued by Rab38 WT (Fig. S3, A–E). The autolysosomes extracted via Lyso-IP contain Rab32 and Rab38 and they were digested by proteinase K, suggesting that Rab32 and Rab38 are present on the cytoplasmic side of autolysosomal membranes (Fig. 1, E and F). All these results suggest the direct involvement of Rab32 and Rab38 in STX17 retrieval from autolysosomes. Further, double knockdown (DKD) of Rab32 and Rab38 further increased STX17 accumulation on autolysosomes albeit to a limited extent (Fig. 1, B and C). Interestingly, re-expression of Rab32 or Rab38 in Rab32/Rab38 double knockdown cells failed to rescue the retrieval defect of STX17, respectively (Fig. S3, F and G). Taken together, these results strongly suggest that Rab32 and Rab38 are both required for STX17 retrieval from autolysosomes. They do not function redundantly, but probably in a cascade of some form.
Rab32 and Rab38 deficiency results in accumulation of SNX4 and SNX5 on autolysosomes
Since SNX4 and SNX5 accumulate on autolysosomes when ACR is disabled (Zhou et al., 2022), we also examined the localizations of SNX4 and SNX5 in cells with Rab32 or Rab38 knockdown. Taking into account that the presence of SNX4 or SNX5 in autolysosomal buds is more distinctly observable in live cells and that GFP-STX17 inhibits autophagosome–lysosome fusion, whereas GFP-STX17-TM has no impact on this fusion process and can be successfully delivered to autolysosomes and efficiently recycled (Uematsu et al., 2017; Zhou et al., 2022), we assessed the localizations of SNX4 and SNX5 in live cells with stable expression of GFP-STX17-TM along with mKATE2-SNX4 or mKATE2-SNX5. Consistent with the deficient STX17 retrieval in Rab32 or Rab38 KD cells, both SNX4 and SNX5 accumulate to significantly higher levels on STX17-positive autolysosomes under Rab32 or Rab38 KD compared with that in RNAi scramble control cells (Fig. 1, G–I). These results suggest that Rab32 and Rab38 are dispensable for the recruitment of SNX4 and SNX5, but instead supported that the deficiency of Rab32 and Rab38 led to failed retrieval of SNX4 and SNX5 from autolysosomes. Noticeably, interactions between STX17 and SNX4 or SNX5 markedly increased in Rab32 or Rab38 KD cells (Fig. S1, C and D), likely due to STX17 co-accumulation with SNX4 and SNX5 on autolysosomes in Rab32 or Rab38 KD cells.
BLOC-3 activation of Rab32 and Rab38 is required for STX17 retrieval from autolysosomes
Since the BLOC-3 complex, which consists of HPS1 and HPS4, serves as the guanine nucleotide-exchange factor (GEF) that activates Rab32 and Rab38 (Martina et al., 2003), we next investigated whether BLOC-3 activation of Rab32 and Rab38 functions in STX17 retrieval from autolysosomes in cells with HPS1 or HPS4 KD. Similar to the above results, autophagosome formation and autophagosome–lysosome fusion occur after 2 h of starvation in HPS1 or HPS4 KD cells, while STX17 significantly accumulated on autolysosomes (Fig. 2, A and B; and Fig. S4 A). Consistently, we also observed greater accumulation of SNX4 and SNX5 on autolysosomes in HPS1 or HPS4 KD cells compared with RNAi scramble controls (Fig. 2, C–E). Additionally, interactions between STX17 and SNX4, SNX5, and SNX17 also increased in HPS1 or HPS4 KD cells (Fig. 2, F and G), indicating that Recycler subunits could still interact with STX17. These increased interactions were likely due to the accumulation of both STX17 and Recycler subunits on autolysosomes. Together, these cumulative results suggest that BLOC-3 activation of Rab32 and Rab38 is required for STX17 retrieval from autolysosomes and the disassociation of SNX4 and SNX5 from autolysosomes but is dispensable for the recruitment of SNX4 and SNX5 to autolysosomes.
BLOC3 activation of Rab32 and Rab38 is required for STX17 retrieval from autolysosomes. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A and (S4A). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. Two-way ANOVA. (C) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (D) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (E) Quantification of SNX4 or SNX5 localization to autolysosomes in C and D. Data are means ± SD for (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (F and G) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with HA-STX17 with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData F2.
BLOC3 activation of Rab32 and Rab38 is required for STX17 retrieval from autolysosomes. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of STX17 positive autolysosomes in A and (S4A). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. Two-way ANOVA. (C) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (D) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h. Scale bar, 5 μm. (E) Quantification of SNX4 or SNX5 localization to autolysosomes in C and D. Data are means ± SD for (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (F and G) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with HA-STX17 with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData F2.
Inactivation of Rab32 and Rab38 has no effect on the first round of autophagosome–lysosome fusion. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm.
Inactivation of Rab32 and Rab38 has no effect on the first round of autophagosome–lysosome fusion. (A) U2OS cells stably expressing Flag-STX17 were transfected with non-targeting siRNA (NC), siRNA against HPS1 or HPS4. 48 h after transfection, cells were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells stably expressing Flag-STX17 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for the indicated hours and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm.
RUTBC1 inhibits STX17 retrieval from autolysosomes depending on its GAP activity
RUTBC1 is a known GTPase-activating protein (GAP) for Rab32 and Rab38 (Marubashi et al., 2016; Nottingham et al., 2011). To investigate whether its GAP activity also regulates STX17 retrieval, we first examined the localization of RUTBC1 inside cells since its localization in non-melanocytes has not been determined. We found that exogenously expressed RUTBC1 diffusely distributed throughout the cytosol, with some degree of localization to lysosomes, and maintained its localization to autolysosomes after starvation (Fig. 3 A and Fig. S4 B). The GAP activity deficient mutant, RUTBC1-R803A, showed similar localization to WT RUTBC1 (Fig. 3 A and Fig. S4 B), suggesting that its GAP activity is not required for its localization to lysosomes or autolysosomes. Furthermore, both the C-terminal and N-terminal regions of RUTBC1 were found to localize to lysosomes and autolysosomes (Fig. S4 B), suggesting these two regions contribute interdependently to its localization. Moreover, the RUTBC1-N variant was observed to be localized to lysosomes and autolysosomes, whereas the RUTBC1-RUN variant exhibited greatly reduced localization to these organelles (Fig. S4 B), indicating that the region between RUTBC1-RUN and RUTBC1-C (amino acids that span from 185 to 533) is essential for the localization of RUTBC1-N. Together, these data strongly support that RUTBC1 localizes to lysosomes and autolysosomes.
RUTBC1 inhibits the recycling of STX17 from autolysosomes. (A) U2OS cells stably expressing Flag-STX17 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Schematic diagram of RUTBC1 truncated variants. (C) Quantification of STX17 positive autolysosomes in (A) and (S4B). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. Two-way ANOVA. (D) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 with vector or V5-RUTBC1 were starved with EBSS for 2 h. Scale bar, 5 μm. (E) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 with vector or V5-RUTBC1 were starved with EBSS for 2 h. Scale bar, 5 μm. (F) Quantification of SNX4 or SNX5 localization to autolysosomes in D and E. Data are means ± SD for (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. Unpaired two-tailed t test. (G) HEK293T cells were transfected with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17 with or without V5-RUTBC1. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData F3.
RUTBC1 inhibits the recycling of STX17 from autolysosomes. (A) U2OS cells stably expressing Flag-STX17 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for 5 h and stained with antibodies against Flag, LC3, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Schematic diagram of RUTBC1 truncated variants. (C) Quantification of STX17 positive autolysosomes in (A) and (S4B). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. Two-way ANOVA. (D) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX4 with vector or V5-RUTBC1 were starved with EBSS for 2 h. Scale bar, 5 μm. (E) U2OS cells stably expressing GFP-STX17-TM, LAMP1-CFP, and mKATE2-SNX5 with vector or V5-RUTBC1 were starved with EBSS for 2 h. Scale bar, 5 μm. (F) Quantification of SNX4 or SNX5 localization to autolysosomes in D and E. Data are means ± SD for (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. Unpaired two-tailed t test. (G) HEK293T cells were transfected with vector, Flag-SNX4, Flag-SNX5, or Flag-SNX17 with or without V5-RUTBC1. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were blotted with the indicated antibodies. Source data are available for this figure: SourceData F3.
To investigate whether the GAP activity of RUTBC1 is involved in STX17 retrieval, we expressed RUTBC1 to inactivate Rab32 and Rab38 and then examined its effects on STX17 retrieval. Consistent with the effects of BLOC-3 knockdown on STX17 retrieval through the inactivation of Rab32 and Rab38, we observed that this retrieval process was significantly blocked by RUTBC1 overexpression (Fig. 3, A and C). Expression of exogenous RUTBC1 also led to greater accumulation of SNX4 and SNX5 on autolysosomes (Fig. 3, D–F). Moreover, the interaction of STX17 with Recycler subunits also markedly increased upon expression of exogenous RUTBC1 (Fig. 3 G). These results suggest the negative regulation of RUTBC1 on STX17 retrieval from autolysosomes.
The TBC domain of RUTBC1 is responsible for its GAP activity. A construct containing only the TBC domain (RUTBC1-C) significantly inhibited STX17 retrieval (Fig. 3, A and C). This suggests that the inhibition of STX17 retrieval is dependent on its GAP activity. Consistently, the mutant RUTBC1-R803A, which lacks GAP activity, and the mutants RUTBC1-RUN and RUTBC1-N, which lack the TBC domain, failed to inhibit STX17 retrieval from autolysosomes (Fig. 3, A and C). Collectively, these results suggest that the GAP activity of RUTBC1 negatively regulates STX17 retrieval from autolysosomes.
Rab32/Rab38 activation is required for recycler complex formation
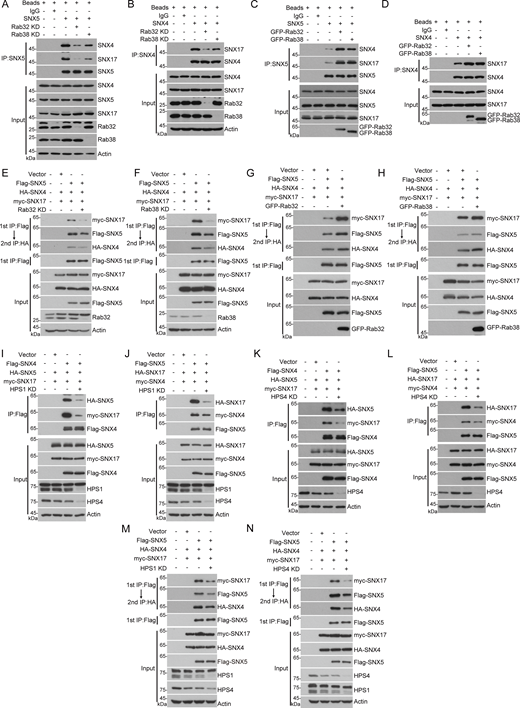
Since Recycler, composed of SNX4, SNX5, and SNX17, is responsible for STX17 retrieval from autolysosomes (Zhou et al., 2022), we sought to determine whether Rab32 and Rab38 regulate this process through the Recycler complex. First, we found that both exogenous and endogenous Rab32 and Rab38 could interact with exogenous and endogenous recycler subunits SNX4, SNX5, and SNX17, respectively (Fig. S5, A–F), supporting the likelihood that Rab32 and Rab38 might regulate the Recycler complex. Consistent with this hypothesis, immunoprecipitation assays showed that SNX5 interactions with SNX4 and SNX17, as well as SNX4 interactions with SNX17, were dramatically reduced in Rab32 KD or Rab38 KD cells (Fig. 4, A and B; and Fig. S5, G–J). As Rab32 KD or Rab38 KD led to the inhibition of STX17 retrieval, we inferred that Rab32 KD or Rab38 KD inhibits the recycler complex formation. Tandem affinity purification (TAP) results further confirmed that the Recycler complex formation was dramatically inhibited in Rab32 KD or Rab38 KD cells (Fig. 4, E and F). Conversely, overexpression of either Rab32 or Rab38 promoted the interactions among the Recycler complex subunits (Fig. 4, C and D) and Recycler complex formation as shown in the TAP results (Fig. 4, G and H). These results suggest that both Rab32 and Rab38 are required for the Recycler complex formation.
Rab32 and Rab38 interact with the subunits of the recycler complex. (A) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (B–D) HEK293T cells were lysed for immunoprecipitation using SNX4, SNX5, or SNX17 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (E) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (F) HEK293T cells were lysed for immunoprecipitation using Rab38 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. HE and LE indicate high exposure and low exposure, respectively. (G–J) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (K–P) Glutathione Sepharose beads, bound with GST-Rab32 or GST-Rab38, were incubated with His-SNX4, His-SNX5, or His-SNX17 in the presence of 100 μM GDP or 100 μM GTP-γ-S, followed by elution for immunoblotting analysis. Source data are available for this figure: SourceData FS5.
Rab32 and Rab38 interact with the subunits of the recycler complex. (A) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (B–D) HEK293T cells were lysed for immunoprecipitation using SNX4, SNX5, or SNX17 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (E) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (F) HEK293T cells were lysed for immunoprecipitation using Rab38 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. HE and LE indicate high exposure and low exposure, respectively. (G–J) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (K–P) Glutathione Sepharose beads, bound with GST-Rab32 or GST-Rab38, were incubated with His-SNX4, His-SNX5, or His-SNX17 in the presence of 100 μM GDP or 100 μM GTP-γ-S, followed by elution for immunoblotting analysis. Source data are available for this figure: SourceData FS5.
Rab32/Rab38 and its GTPase activity are required for the formation of recycler complex. (A and B) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 48 h after transfection, cells were lysed for immunoprecipitation using SNX5 or SNX4 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (C and D) HEK293T cells were transfected with GFP-Rab32 or GFP-Rab38. 24 h after transfection, cells were lysed for immunoprecipitation assay using SNX5 or SNX4 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (E and F) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, HEK293T cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. (G and H) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. (I–L) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (M and N) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, HEK293T cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F4.
Rab32/Rab38 and its GTPase activity are required for the formation of recycler complex. (A and B) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 48 h after transfection, cells were lysed for immunoprecipitation using SNX5 or SNX4 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (C and D) HEK293T cells were transfected with GFP-Rab32 or GFP-Rab38. 24 h after transfection, cells were lysed for immunoprecipitation assay using SNX5 or SNX4 as a bait protein and IgG as the negative control. Immunoblotting was performed with the indicated antibodies. (E and F) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38. 24 h after transfection, HEK293T cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. (G and H) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. (I–L) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (M and N) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, HEK293T cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The elution from the first immunoprecipitation was subjected to a second round of immunoprecipitation with anti-HA antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F4.
Subsequent Co-IP assays showed a notable reduction in the interactions between SNX4 and SNX5 or SNX17, as well as between SNX5 and SNX4 or SNX17, upon HPS1 or HPS4 knockdown (Fig. 4, I–L). These findings suggest that HPS1 or HPS4 depletion hinders the formation of the Recycler complex. The TAP results further corroborated that the formation of the Recycler complex was remarkably inhibited by HPS1 or HPS4 KD (Fig. 4, M and N). Taken together, these results suggest that the activation of both Rab32 and Rab38 is essential for the formation of the Recycler complex.
DCTN1-recycler connection is regulated by Rab32 and Rab38 GTPase activity
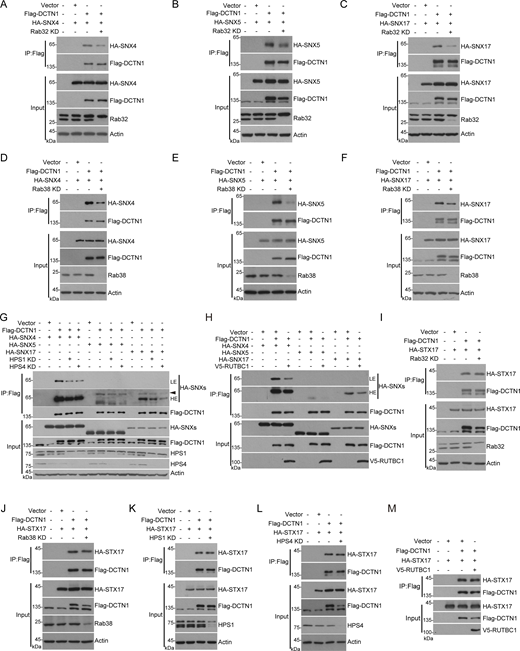
As an adaptor subunit of the dynactin complex, DCTN1 plays a crucial role in linking the recycler complex to the dynein–dynactin complex (Zhou et al., 2022). To investigate the impact of Rab32 and Rab38 on DCTN1 interactions with Recycler, we examined the binding between DCTN1 and SNX4, SNX5, or SNX17 in Rab32 or Rab38 KD cells. Our data showed that the interactions between DCTN1 and these subunits were significantly reduced in Rab32 or Rab38 KD cells compared with their scramble controls (Fig. 5, A–F). This suggests that both Rab32 and Rab38 positively regulate the binding between DCTN1 and the recycler complex subunits. Furthermore, our results also revealed that the knockdown of HPS1 or HPS4 or expression of exogenous RUTBC1 resulted in a remarkable decrease in DCTN1 binding with SNX4, SNX5, and SNX17 (Fig. 5, G and H). However, the interaction between DCTN1 and STX17 was found to be unaffected in cells with Rab32 KD, Rab38 KD, HPS1 KD, HPS4 KD, or with the expression of exogenous RUTBC1 (Fig. 5, I–M). These findings suggest that the activation of Rab32 and Rab38 does not participate in the regulation of the DCTN1–STX17 interaction. Therefore, these results suggest that the interaction between DCTN1 and Recycler subunits requires the activation of Rab32 and Rab38.
Rab32 and Rab38 positively regulate the interaction between DCTN1 and the recycler complex. (A–C) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (D–F) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab38. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (G) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. LE, low exposure. HE, high exposure. The arrow indicates the non-specific bands for SNX5 and SNX17. (H) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. LE, low exposure. HE, high exposure. (I–L) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (M) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F5.
Rab32 and Rab38 positively regulate the interaction between DCTN1 and the recycler complex. (A–C) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (D–F) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab38. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (G) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against HPS1 or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. LE, low exposure. HE, high exposure. The arrow indicates the non-specific bands for SNX5 and SNX17. (H) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. LE, low exposure. HE, high exposure. (I–L) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (M) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F5.
Rab32 and Rab38 regulate ATG9A retrieval from autolysosomes
As the other known cargo of ACR, we then investigated whether ATG9A retrieval from autolysosomes requires Rab32 and Rab38 (Zhou et al., 2022). Similar to the findings on STX17, we observed a significant accumulation of ATG9A puncta on autolysosomes in Rab32 or Rab38 KD cells compared with the RNAi scramble controls (Fig. 6, A and B). Notably, double knockdown of Rab32 and Rab38 further intensified the accumulation of ATG9 on autolysosomes compared with the single Rab32 or Rab38 knockdown cells, although the extent of this increase was limited (Fig. 6, A and B). These observations suggest that Rab32 and Rab38 were also required for ATG9A retrieval from autolysosomes. Moreover, HPS1 KD, HPS4 KD, or RUTBC1 expression leads to a significant elevation of ATG9A puncta on autolysosomes compared with control cells (Fig. 6, A and B; and Fig. 7, A and B), suggesting that the activation of Rab32/Rab38 is required for efficient retrieval of ATG9A from autolysosomes. Similar to STX17, re-expression of Rab32 or Rab38 in Rab32/Rab38 double knockdown cells failed to rescue the retrieval defect of ATG9A, respectively, suggesting that Rab32 and Rab38 participate in a cascade of functions (Fig. 6, C and D).
Rab32 and Rab38 are required for the retrieval of ATG9A from autolysosomes. (A) U2OS cells stably expressing GFP-LC3 were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, Rab32 and Rab38, HPS1, or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against GFP, ATG9A, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of ATG9A puncta on autolysosomes in (A). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (C) U2OS cells stably expressing GFP-LC3 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against ATG9A and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (D) Quantification of ATG9A puncta on autolysosomes in C. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (E–H) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (I) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F6.
Rab32 and Rab38 are required for the retrieval of ATG9A from autolysosomes. (A) U2OS cells stably expressing GFP-LC3 were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, Rab32 and Rab38, HPS1, or HPS4. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against GFP, ATG9A, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of ATG9A puncta on autolysosomes in (A). Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (C) U2OS cells stably expressing GFP-LC3 with vector or siRNA resistant mKO-Rab32/Rab38 were transfected with non-targeting siRNA (NC), siRNA against Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against ATG9A and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (D) Quantification of ATG9A puncta on autolysosomes in C. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001. One-way ANOVA. (E–H) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (I) HEK293T cells were transfected with non-targeting siRNA (NC) or siRNA against Rab32, Rab38, HPS1, or HPS4. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. Source data are available for this figure: SourceData F6.
Both RUTBC1 and AP4 regulate the retrieval of ATG9A from autolysosomes. (A) U2OS cells stably expressing GFP-LC3 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for 2 h and stained with antibodies against GFP, ATG9A, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of ATG9A puncta on autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. One-way ANOVA. (C and D) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (E) U2OS cells stably expressing GFP-LC3 were transfected with indicated siRNAs. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against ATG9A and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (F) Quantification of ATG9A positive autolysosomes in E. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). *P < 0.05, ****P < 0.0001. ns, no significance. One-way ANOVA. Source data are available for this figure: SourceData F7.
Both RUTBC1 and AP4 regulate the retrieval of ATG9A from autolysosomes. (A) U2OS cells stably expressing GFP-LC3 with wild-type mKO-RUTBC1 or its truncated variants were starved with EBSS for 2 h and stained with antibodies against GFP, ATG9A, and LAMP1. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of ATG9A puncta on autolysosomes in A. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). ns, no significance. ****P < 0.0001. One-way ANOVA. (C and D) HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. The precipitates were analyzed with the indicated antibodies. (E) U2OS cells stably expressing GFP-LC3 were transfected with indicated siRNAs. 48 h after transfection, cells were starved with EBSS for 2 h and stained with antibodies against ATG9A and LAMP2. Scale bar, 5 μm. Inset scale bar, 1 μm. (F) Quantification of ATG9A positive autolysosomes in E. Data are means ± SD (n = 3, 60 cells from three independent experiments were quantified). *P < 0.05, ****P < 0.0001. ns, no significance. One-way ANOVA. Source data are available for this figure: SourceData F7.
As observed with STX17, knockdown of Rab32, Rab38, HPS1, HPS4, or RUTBC1 expression resulted in an increase in ATG9A interactions with Recycler subunits (Fig. 6, E–H; and Fig. 7 C). This may be due to the accumulation of ATG9A and unassembled Recycler subunits on autolysosomes. Similar to the effects of Rab32 or Rab38 on STX17-DCTN1 interaction, no obvious impacts on ATG9A–DCTN1 interactions were detected in cells undergoing knockdown of Rab32, Rab38, HPS1, HPS4, or overexpressing RUTBC1 (Fig. 6 I and Fig. 7 D). These results suggest the activation of Rab32 and Rab38 does not regulate ATG9A–DCTN1 interaction.
AP4 regulates the location of ATG9A on autolysosomes upstream of Rab32
AP5 has been demonstrated to play a role in recycling from late endosomal compartments, while AP4, but not AP5, is specifically necessary for the export of ATG9A from the trans-Golgi network (TGN) to peripheral compartments (Hirst et al., 2011, 2013; Mattera et al., 2017). Consistent with previous studies, we also observed that ATG9A became more concentrated in the perinuclear region, with a dramatic reduction in dispersed cytoplasmic ATG9A puncta in AP4E1 KD and AP4M1 KD cells. This phenomenon was not observed in AP5Z1 KD cells (Fig. 7 E). Furthermore, the localization of ATG9A on autolysosomes was significantly decreased in AP4E1/Rab32 double knockdown and AP4M1/Rab32 double knockdown cells, but not in AP5Z1/Rab32 double knockdown cells (Fig. 7, E and F). These observations suggest that the role of AP4 in regulating the location of ATG9A to autolysosomes occurs upstream of Rab32.
Rab32 and Rab38 positively regulate autophagy
To investigate the roles of Rab32 and Rab38 in autophagy, we assessed autophagic flux by examining the levels of LC3 and p62, as well as the acidification of RFP-GFP-LC3 or RFP-GFP-p62 in both image-based and flow cytometry-based assays. Western blot results revealed significant inhibition in the turnover of LC3 and p62 in Rab32 KD, Rab38 KD, or Rab32/Rab38 DKD cells after 2 h of starvation, with more pronounced inhibition observed after 5 h of starvation (Fig. 8, A–F). In line with this, the acidification of RFP-GFP-LC3 was significantly inhibited in Rab32 KD, Rab38 KD, or Rab32/Rab38 DKD cells (Fig. 8, G and H). Results from the flow cytometry assay also indicated that the acidification of RFP-GFP-LC3 or RFP-GFP-p62 was inhibited in Rab32 KD, Rab38 KD, or Rab32/Rab38 DKD cells, and these inhibitions could be rescued by the re-expression of Rab32 or Rab38 in Rab32 KD or Rab38 KD cells, respectively (Fig. 8, I and J). Therefore, all these findings suggest that the deficiency of Rab32 or Rab38 inhibits autophagic flux.
Rab32 and Rab38 positively regulate autophagic flux. (A and B) Wild-type 293T cells, as well as 293T cells with Rab32 or Rab38 knockdown, were subjected to starvation with EBSS and treated with or without 100 nM Bafilomycin A1 for 2 or 5 h. Subsequently, immunoblots were conducted using the specified antibodies. * indicates a non-specific band. (C–F) Quantification of LC3-II and p62 band intensity in A and B. The intensity of LC3-II and p62 bands were normalized to actin. Data are means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA. (G) U2OS cells stably expressing RFP-GFP-LC3 were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38 or Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 4 h. Scale bar, 5 μm. (H) Quantification of the ratio of GFP+RFP+-LC3/total LC3 puncta in G. Data are mean ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. (I and J) Quantification of the GFP/RFP intensity ratio was performed using flow cytometry. Data are mean ± SD of three independent experiments. ns, no significance, ****P < 0.0001. Two-way ANOVA. Source data are available for this figure: SourceData F8.
Rab32 and Rab38 positively regulate autophagic flux. (A and B) Wild-type 293T cells, as well as 293T cells with Rab32 or Rab38 knockdown, were subjected to starvation with EBSS and treated with or without 100 nM Bafilomycin A1 for 2 or 5 h. Subsequently, immunoblots were conducted using the specified antibodies. * indicates a non-specific band. (C–F) Quantification of LC3-II and p62 band intensity in A and B. The intensity of LC3-II and p62 bands were normalized to actin. Data are means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA. (G) U2OS cells stably expressing RFP-GFP-LC3 were transfected with non-targeting siRNA (NC) or siRNA against Rab32 or Rab38 or Rab32 and Rab38. 48 h after transfection, cells were starved with EBSS for 4 h. Scale bar, 5 μm. (H) Quantification of the ratio of GFP+RFP+-LC3/total LC3 puncta in G. Data are mean ± SD (n = 3, 60 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. (I and J) Quantification of the GFP/RFP intensity ratio was performed using flow cytometry. Data are mean ± SD of three independent experiments. ns, no significance, ****P < 0.0001. Two-way ANOVA. Source data are available for this figure: SourceData F8.
Discussion
ACR was recently discovered as an autophagy-related process in which components of autophagosome outer membrane are recycled from autolysosomes after autophagosome–lysosome fusion (Zhou et al., 2022). However, the regulatory mechanism responsible for coordinating the players in ACR remains largely unknown. Here, we identified Rab32 and Rab38, which localize to autolysosomes, as essential regulators for the activity of ACR, the recycler complex formation, and dynactin–Recycler interaction, rather than other autophagosomal or lysosomal Rabs. More specifically, knockdown of their GEF activator, BLOC-3, or overexpression of their GAP, RUTBC1, supports the requirement for Rab32/Rab38 activation in ACR. In addition, Rab32, Rab38, inactivation of Rab32 and Rab38 by knockdown of BLOC3, or RUTBC1 expression does not participate in cargo-DCTN1 interaction. Collectively, our study revealed that Rab32 and Rab38 regulate ACR in an activity-dependent manner from different aspects.
Previous studies have shown that Rab32 and Rab38 are redundant in the biogenesis of lysosome-related organelles in various animal models (Aguilar et al., 2019; Loftus et al., 2002). Rab32/Rab38 double knockout in melanocytes also results in more severe hypopigmentation due to blocked trafficking of melanogenic enzymes to the melanosome (Bultema et al., 2012; Wasmeier et al., 2006). In addition, Rab32 and Rab38 are both considered regulators of lysosomal protease, cathepsin D, trafficking to phagosomes containing Mycobacterium tuberculosis (Seto et al., 2011). Unlike the redundancy of Rab32 and Rab38 in melanosome biogenesis and cathepsin D trafficking, Rab32 and Rab38 do not function redundantly in ACR but are likely involved in a cascade of some form. Furthermore, the pull-down assay revealed no significant difference in the interactions of Rab32 or Rab38 with SNX4, SNX5, or SNX17 in the presence of GTP-γ-S or GDP (Fig. S5, K–P). This suggests the potential existence of other, yet-to-be-identified, effectors for Rab32 and Rab38. The identification of these effectors in the future will aid in discerning potential functional differences between Rab32 and Rab38. Additionally, Rab32 and Rab38 may have additional regulatory functions independent of their GTPase activity.
Although the recycler complex is disassembled in cells with knockdown of Rab32, Rab38, HPS1, HPS4, or overexpression of RUTBC1, the interaction between Recycler subunits and STX17/ATG9A was found to markedly increase. This suggests that despite the recycler complex being disassembled, unassembled Recycler subunits were still able to interact with STX17/ATG9A. The increased interactions were likely due to co-accumulation of both STX17/ATG9A and unassembled Recycler subunits on autolysosomes.
In the recycler complex, the interaction of STX17 with DCTN1 depends on SNX17 because the interaction between STX17 and DCTN1 decreased in the absence of SNX17 (Zhou et al., 2022). However, either Rab32 or Rab38 knockdown results in an increased interaction between SNX17 and STX17 and a decreased interaction between SNX17 and DCTN1. These opposing changes may counterbalance each other and lead to the indiscernible interaction between STX17 and DCTN1. The same applies to ATG9A. It is still possible that a direct interaction between cargo (unassociated with the recycler complex) and DCTN1 occurs when the recycler complex is disassembled, potentially offsetting the reduced interaction between DCTN1 and the cargo. Further investigations are required to resolve this issue.
Rab21 is known to be regulated by the GEF activity of VARP, which is a downstream effector of Rab32 and Rab38. We found that Rab21 does not localize to autolysosomes, suggesting that Rab32 and Rab38 do not function via VARP and Rab21 in ACR. Another study reported that Rab9 regulates melanogenesis via BLOC3 activation of Rab32/Rab38 in melanocytes (Marubashi et al., 2016). In our study, Rab9 localizes to autolysosomes, but it is not required for ACR, while BLOC3 activation of Rab32 and Rab38 is required for ACR. This suggests that the upstream regulation of Rab9 on the BLOC3-Rab32/Rab38 pathway may not exist in ACR, similar to our speculation that Rab9 is also dispensable for Rab32/BLOC-3-dependent Salmonella killing (Balci et al., 2020) and that Rab9 also can regulate melanogenesis independently of BLOC3 activation of Rab32/Rab38 (Ohishi et al., 2019).
Given the possible role of Rab32 family proteins in pathological conditions such as hypopigmentation, platelet defects, pulmonary homeostasis, Parkinson’s disease, brain inflammation, or pathogen replication, the findings in this study not only provide a framework for understanding the Rab32/Rab38 activity-dependent regulatory mechanisms in ACR but also may provide a potential link between impaired ACR and Rab32 family protein–related diseases.
Materials and methods
Reagents and antibodies
Rabbit polyclonal anti-Rab32 (Cat#HPA025731, Lot#A104621, RRID: AB_1856001), Anti-Flag M2 Affinity Gel (Cat#A2220, Lot#SLCL1176, RRID: AB_10063035), Anti-Flag M2 magnetic beads (Cat#M8823, Lot#SLCH4826, RRID: AB_2637089), Mouse monoclonal anti-Flag M2 (Cat#F3165, Lot#SLCF4933, RRID: AB_259529), Rabbit polyclonal anti-HA (Cat#H6908, Source #0000086963, RRID: AB_260070), Rabbit polyclonal anti-LC3 (Cat#L7543, Lot#084M4798V, RRID: AB_796155), and Rabbit polyclonal anti-LAMP1 (Cat#L1418, Lot#128M4803V, RRID: AB_477157) were purchased from Sigma-Aldrich. Rabbit monoclonal anti- ATG9A (Cat#ab108338, Lot#GR3216593-13, RRID: AB_10863880), Chicken polyclonal anti-GFP (Cat#ab13970, Lot#GR3361051-7, RRID: AB_300798), and Goat Anti-Chicken IgY H&L (FITC) (Cat#ab46969, Lot#GR3179274-12, RRID: AB_2338589) were purchased from Abcam. Mouse monoclonal anti-LAMP2 (Cat#sc-18822, Lot#L3216, RRID: AB_626858), Rabbit monoclonal anti-myc (Cat#sc-789, Lot#K1615, RRID: AB_631274), Mouse monoclonal anti-Rab38 (Cat#sc-390176, Lot#E3116, RRID: AB_2798463), and protein A/G PLUS-Agarose (Cat#sc-2003, Lot#C2822, RRID: AB_10201400) were purchased from Santa Cruz Biotechnology. Rabbit Polyclonal SEC61β (Cat#A15788, Lot#0161690201, RRID: AB_2763208), Mouse monoclonal anti-GST (Cat#AE001, Lot#4000057011, RRID: AB_2770403), Rabbit Control IgG (Cat#AC005, Lot#3500000108, RRID: AB_2771930), and Mouse Control IgG (Cat#AC011, Lot#9100011106, RRID: AB_2770414) were purchased from Abclonal. Mouse monoclonal anti- SQSTM1/p62 (Cat#88588S, Lot#1, RRID: AB_2800125) was purchased from CST. Glutathione Sepharose 4B glutathione-sepharose Resin (Cat#17-0756-04) and Ni-NTA agarose resin (Cat#17-5318-06) were purchased from GE Healthcare. DMEM (Cat#SH30022.01B) was purchased from Hyclone. Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor647 (Cat#A21235, Lot#1915807, RRID: AB_2535804), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 405 (Cat#A31556, Lot#2273716, RRID: AB_221605), and Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody and Alexa Fluor 647 (Cat#A21245, Lot#1445259, RRID: AB_2535813) were purchased from Invitrogen. Cy3 AffiniPure Goat Anti-Mouse IgG (H+L) (Cat#115-165-003, Lot#117093, RRID: AB_2338680) and Cy3 AffiniPure Goat Anti-Rabbit IgG (H+L) (Cat#111-165-003, Lot#128284, RRID: AB_2338000) were purchased from Jackson. Rabbit polyclonal anti-LC3 (Cat#PM036, Lot#036, RRID: AB_2274121) and Mouse monoclonal anti-LC3 (Cat#M152-3, Lot#056, RRID: AB_1279144) were purchased from MBL. Bafilomycin A1(BFA) (Cat#HY-100558) was purchased from MCE. Rabbit polyclonal anti-HPS1 (Cat#15077-1-AP, Lot#00025232, RRID: AB_10694669), Rabbit polyclonal anti-HPS4 (Cat#14627-1-AP, Lot#00020715, RRID: AB_2878071), TOM20 Polyclonal antibody (Cat#11802-1-AP, Lot#00068680, RRID: AB_2207530), Rabbit polyclonal anti-HSP60 (Cat#15282-1-AP, Lot#0001350201, RRID: AB_2121440), and Rabbit polyclonal anti-TGN46 (Cat#13573-1-AP, Lot#00044796, RRID: AB_10597396) were purchased from Proteintech. Rabbit polyclonal anti-GFP (Cat#D110008, Lot#l531AA0031) and Rabbit polyclonal anti-Flag (Cat#D110005, Lot#G928AA0012-0200) were purchased from Sangon Biotech. Rabbit polyclonal anti-Actin (Cat#GB11001, Lot#Ac2111012A, RRID: AB_2801259) was purchased from Service Bio. Earle’s balanced salt solution (EBSS) (Cat#H2020) was purchased from Solarbio. Goat Anti-Mouse IgG(H+L)-HRP (Cat#1036-05, Lot#D1912-SL71D, RRID: AB_2794348) and Goat Anti-Rabbit IgG-HRP (Cat#4010-05, Lot#A0422-PL42, RRID: AB_2687483) were purchased from SouthernBiotech. Mouse monoclonal anti-6×His (Cat#A2050, Lot#ATUSE1801) was purchased from Abbkine. 3×Flag peptide was synthesized by ChinaPeptides Co., Ltd.
Plasmids
The RUTBC1-related plasmids were kindly provided by Suzanne R. Pfeffer (Stanford University, Stanford, CA, USA). cDNAs encoding human STX17, STX17TMD, Rab32, Rab38, and LAMP1 were amplified by PCR and subcloned into the pLV-EF1a-IRES (Addgene #85132). The human SNX4-, SNX5-, and SNX17-related plasmids were described previously (Zhou et al., 2022).
Cell culture and transfection
HEK293T, U2OS, and HeLa cells (Zhou et al., 2022) were cultured in DMEM (Hyclone) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin solution (Beyotime) at 37°C with 5% CO2. Transient transfection of plasmids in HEK293T cells was performed using PEI according to the manufacturer’s protocol, while HeLa cells were transiently transfected using Lipofectamine 3000 according to the manufacturer’s instructions. Cells were analyzed 24 h after transfection. To perform RNA interference, siRNA duplexes were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. After 48 h of transfection, cells were starved for analysis. To induce starvation, cells were washed three times with PBS (Hyclone) and then incubated with EBSS for a specific period of time, as indicated in the experiment.
siRNA sequences
For siRNA-mediated gene knockdown, siRNA duplexes were purchased from GenePharma. Cells were analyzed 48 h after transfection of siRNA. The targeting sequences were NC: 5′-UUCUUCGAACGUGUCACGUTT-3′; Rab32-1: 5′-CCAAAGCTTTCCTAATGAA-3′; Rab32-2: 5′-CAUUUGAGGCAGUCUUAAAAU-3′; Rab38-1: 5′-CAATGGCCTCAAGATGGAC-3′; Rab38-2: 5′-GCAAAUGAGUGUGACCUAA-3′; Rab38-3: 5′-UAUCGCAGGUCAAGAAAGA-3′; HPS1-1: 5′-AAUGGCAACUUCCUGUAUG-3′; HPS1-2: 5′-GCGUCUUGGUGGCCACUGA-3′; HPS1-3: 5′-UGAAGUUCGGGCAGUCAGA-3′; HPS1-4: 5′-CUGGACAGAUCAGGAGUUU-3′; HPS4: 5′-AAGCGGTTTCTGGATCAGCTA-3′; Rab7A: 5′-CACGTAGGCCTTCAACACAAT-3′; Rab9A: 5′-CCAGCTCTTCCATACAATA-3′; Rab9B: 5′-GCTTATGAACCGTTACGTA-3′; AP4E1-1: 5′-GCUGCCUCCUUGCAUAUAUUU-3′; AP4E1-2: 5′-GGCAUAUGAAGAUGAUUAUUU-3′; AP4E1-3: 5′-GGCGCACUCUUCUAAUACAUU-3′; AP4M1-1: 5′-GGCUCCAGGUUUAUCUAAAUU-3′; AP4M1-2: 5′-GCCGUCAUUUCAUUCACAUUU-3′; AP4M1-3: 5′-GCUGGACUAUGGCUAUGUAUU-3′; AP5Z1-1: 5′-GCCUCAUUGAGCAAAGUAAUU-3′; AP5Z1-2: 5′-GCCUCUUUAUUGCUGUCAAUU-3′; AP5Z1-3: 5′-GUUCCCGGAUCUGUAAACUUU-3′.
Immunostaining assay
The cells grown on coverslips were washed with PBS and fixed in 4% paraformaldehyde in PBS for 15 min at room temperature. After washing three times with PBS, the cells were permeabilized with 0.1% saponin in PBS for 10 min and then blocked with 10% FBS in PBS for 1 h. The cells were then incubated with primary antibodies for 1 h or overnight at 4°C. After washing three times with PBS, the cells were incubated with the appropriate secondary antibodies at room temperature for 1 h and washed three times with PBS. Finally, the coverslips were mounted, and images of the stained cells were acquired under a laser scanning confocal microscope (FV3000; Olympus), Plan Apochromat N 100×/1.49 oil at room temperature. Images were processed and analyzed using OlyVIA and ImageJ 1.52a.
Live cell imaging
Cells were placed on a four-chambered cover glass (syD35-20-1-N; In Vitro Scientific) and knocked down with siRNAs for 2 d before observation. During live imaging, the culture dish was mounted on an inverted microscope (FV3000; Olympus) to maintain incubation conditions at 37 °C and 5% CO2 using a Plan Apochromat N 100×/1.49 oil. Images were recorded using a confocal laser microscope system and then further processed and analyzed using ImageJ 1.52a.
Western blotting
The cells were lysed using an SDS sample buffer and heated at 110°C for 10 min. The resulting samples were loaded onto an SDS-PAGE gel for electrophoresis. After electrophoresis, the proteins were transferred onto a PVDF membrane (162-0177; BIO-RAD), which was then blocked with 5% skimmed milk at room temperature for 1 h. The membrane was then incubated with primary antibodies at 37°C for 1 h or overnight at 4°C. After washing the membrane three times, it was incubated with secondary antibodies conjugated with horseradish peroxidase at a dilution of 1/10,000 for 1 h at room temperature. Finally, visualization was carried out using enhanced chemiluminescence (P0018M-2; Beyotime) according to the manufacturer’s protocol, and the resulting images were developed using an OPTIMAX x-ray film processor (Protec GmbH and Co.). Quantitative analysis of the protein bands was carried out using ImageJ 1.52a.
Immunoprecipitation assay
HEK293T cells were washed with ice-cold PBS and lysed in lysis buffer for 10 min at 4°C. The lysates were then centrifuged at 12,000 g for 10 min at 4°C, and the resulting supernatant was subjected to immunoprecipitation using anti-Flag M2 Affinity Gel for 12 h at 4°C to isolate the protein of interest. The precipitated immunocomplexes were washed three times in lysis buffer and eluted with the 3×Flag peptide for 2 h at 4°C before being boiled in 2× sample buffer. The samples were then subjected to SDS-PAGE and analyzed by immunoblotting to study the protein of interest. For coimmunoprecipitation, antibodies against SNX4, SNX5, SNX17, or Rab38 were used to isolate the interacting proteins. The lysates were subjected to immunoprecipitation using these antibodies for 12 h at 4°C, with IgG as the negative control. The precipitated immunocomplexes were then incubated with protein A/G PLUS-agarose for 12 h at 4°C before being washed three times in lysis buffer and boiled in 2× sample buffer for immunoblot analysis.
Immunopurification of lysosomes (LysoIP) and proteinase K protection assay
HEK293T cells stably expressed LAMP1-Flag from one 15-cm dish were starved for 2 h with EBSS. Then the cells were harvested and washed twice with ice-cold PBS. The cell pellets were collected after centrifugation at 1,000 g for 1 min and resuspended in 1 ml ice-cold homogenization buffer (250 mM sucrose, 20 mM HEPES-KOH, pH 7.4, 1 mM EDTA, and complete EDTA-free protease inhibitor). Cells were homogenized in a 2 ml Dounce homogenizer. The cell homogenate was then centrifuged at 1,000 × g for 10 min at 4°C, and 50 μl of samples was saved as input. The rest of the supernatant was incubated with 150 μl of anti-Flag M2 magnetic beads on a gentle rotator shaker overnight. Immunoprecipitates were then washed three times and eluted in the SDS loading buffer. Western blotting for proteins indicated in the figures was done as described in the above section.
One 15-cm dish of HEK293T cells expressing LAMP1-Flag were starved for 2 h with EBSS and then homogenized as described above. The supernatant was mixed with 45 μl of 5 M NaCl and 70 μl Anti-FLAG M2 magnetic beads and rotated overnight at 4°C. The magnetic beads were washed three times with 250 mM NaCl and resuspended in ice-cold PS200 (20 mM K-PIPES, pH 6.8, 250 mM sucrose, 5 mM MgCl2). The resultant product was split into three aliquots and treated as follows: (A) PS200 alone (no treatment control), (B) 0.1–10 µg/ml proteinase K, and (C) 0.1–10 µg/ml proteinase K with 0.2% Triton X-100. The samples were incubated on ice for 5 min. The final products were dissolved in a sample buffer for immunoblotting.
In vitro binding assay
Genes were cloned into pGEX-4T-1 or pET-28a vector for expression in E. coli T7. The recombinant proteins were purified by glutathione sepharose resin or Ni-affinity resin. Purified bacterial GST-Rab32, GST-Rab38 proteins, or GST was incubated in Buffer A (20 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM DTT, 5 mM EDTA) at 4°C overnight for applying to GST resin. The beads were washed three times in Buffer B (20 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM DTT) and 100 μM GTP-γ-S or GDP was added and incubated for 30 min at room temperature. Then the beads were incubated with His-SNX4, His-SNX5, or His-SNX17, respectively, in the presence of 20 mM Mg2+ for 2 h at room temperature. After washing three times with Buffer C (20 mM HEPES [pH 7.5], 250 mM NaCl, 1 mM DTT, 10 μM GTP-γ-S or GDP, 5 mM MgCl2), proteins were eluted with Buffer D (20 mM HEPES [pH 7.5], 1.5 M NaCl, 20 mM EDTA, 1 mM DTT, 5 mM GDP) for 2 h at room temperature and dissolved in sample buffer for SDS-PAGE and then immunoblotted.
Statistical analysis
The data from independent experiments were subjected to statistical analysis using Prism software from GraphPad. The standard deviation (SD) was used to indicate the variability or spread of the data, and error bars were included in the figures to represent this measure. All the images were captured randomly and all imaged cells were included in the analysis. The analysis was conducted in a single-blind manner using Image J, with a sample size of n = 3, encompassing a total of 60 cells from three independent experiments. To determine the statistical significance of the results, unpaired two-tailed t test, one-way ANOVA, and two-way ANOVA were performed. Significance levels were determined as the indicated P values. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows the localization of Rab proteins. Fig. S2 shows the effects of Rab7A, Rab9A, or Rab9B on STX17 location to autolysosomes. Fig. S3 shows that the STX17 retrieval defect caused by Rab32/Rab38 knockdown is rescued by the expression of wild-type Rab32/Rab38. Fig. S4 shows that inactivation of Rab32 and Rab38 has no effect on the first round autophagosome–lysosome fusion. Fig. S5 shows that Rab32 and Rab38 interact with the subunits of the recycler complex.
Data availability
The authors declare that all relevant data supporting the findings of this study are available within the paper and its supplemental files. The proteomics data have been deposited in the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org/) via the PRIDE partner repository with the dataset identifier PXD031183.
Acknowledgments
We are deeply grateful to Liang Ge (Tsinghua University), Qiming Sun (Zhejiang University), Rui Chen (Wuhan University), and Meisheng Ma (Huazhong University of Science and Technology) for helpful suggestions on this study. We thank Suzanne R. Pfeffer (Stanford University) (Nottingham et al., 2011) for gifting us the RUTBC1 related plasmids.
The work was supported by grants from the National Natural Science Foundation of China 92254302, 91854116, 31771529, and 32170685 (to Y. Rong) and the Fundamental Research Funds for the Central Universities 5003510089 and 2023BR028 (to Y. Rong).
Author contributions: Y. Rong and Z. Wu designed the experiments and conducted the experiments. Z. Wu, H. Que, and C. Li performed the biological and biochemical experiments. Z. Wu, L. Yan, S. Wang, and Y. Rong analyzed the data and wrote the manuscript with the help of all authors.
References
Author notes
Disclosures: The authors declare no competing interests exist.