Autonomous circadian clocks exist in nearly every mammalian cell type. These cellular clocks are subjected to a multilayered regulation sensitive to the mechanochemical cell microenvironment. Whereas the biochemical signaling that controls the cellular circadian clock is increasingly well understood, mechanisms underlying regulation by mechanical cues are largely unknown. Here we show that the fibroblast circadian clock is mechanically regulated through YAP/TAZ nuclear levels. We use high-throughput analysis of single-cell circadian rhythms and apply controlled mechanical, biochemical, and genetic perturbations to study the expression of the clock gene Rev-erbα. We observe that Rev-erbα circadian oscillations are disrupted with YAP/TAZ nuclear translocation. By targeted mutations and overexpression of YAP/TAZ, we show that this mechanobiological regulation, which also impacts core components of the clock such as Bmal1 and Cry1, depends on the binding of YAP/TAZ to the transcriptional effector TEAD. This mechanism could explain the impairment of circadian rhythms observed when YAP/TAZ activity is upregulated, as in cancer and aging.
Introduction
The vast majority of organisms display daily physiological and behavioral changes in adaptation to the cyclic environment that arises from the Earth’s rotation (Bass and Lazar, 2016). These rhythms, termed circadian, are endogenous and self-sustained at the cell level by a clock directed by a set of proteins whose expression is regulated via a transcription–translation feedback loop with a period close to 24 h (Cox and Takahashi, 2019). In mammals, up to 25% of the proteome is estimated to be influenced, in a cell-type-specific manner, by the circadian clock, which deeply conditions dynamics and function of tissues and organs throughout the day (Storch et al., 2002; Rijo-Ferreira and Takahashi, 2019; Beytebiere et al., 2019).
The central element of the mammalian circadian clock is the transcription factor BMAL1:CLOCK, which activates the transcription of, among many others, Cry and Per. In turn, the sustained expression of CRY and PER generates a transcriptional loop by binding to BMAL1:CLOCK and blocking its activity (Shearman et al., 2000). A second stabilizing layer of regulation of the clock is provided by REV-ERB, which, on one hand, is transcriptionally promoted by BMAL1:CLOCK and, on the other hand, acts as a transcriptional inhibitor of Bmal1 (Preitner et al., 2002).
For every cell residing in a peripheral tissue, the robustness of the circadian clock and its synchrony with the environment are subjected to multilayered regulation (Finger et al., 2020; Albrecht, 2012). Decades of work have unveiled the biochemical side of this regulatory network which, mainly through neurocrine and paracrine signaling, drives important changes in the gene expression landscape of the different cell types (Dibner et al., 2010; Reinke and Asher, 2019; Koronowski et al., 2019). Besides biochemical regulation, recent evidence suggests a reciprocal communication between the circadian clock and mechanobiological factors such as actin dynamics, cell–cell adhesions, and the stiffness of the extracellular matrix (Yang et al., 2017; Hoyle et al., 2017; Streuli and Meng, 2019; Gerber et al., 2013). However, how and to what extent mechanotransduction impacts on the transcriptional regulation of the circadian network at the single-cell level is still largely unknown.
The main mechanotransduction pathway proposed to have an impact on the circadian clock is the one mediated by MAL (also known as MRTF) and the serum response factor. MAL, whose intracellular localization is regulated by actin dynamics, has been shown to modulate the transcription of the circadian genes Per2, Rev-erbα, and Nfil3 (Gerber et al., 2013; Xiong et al., 2022), which has been suggested as a mechanism to entrain the clock. By contrast, the potential role of the central components of the Hippo pathway YAP/TAZ as regulators of the circadian clock has been so far overlooked. YAP/TAZ translocate in a mechanoresponsive and phosphorylation-dependent manner between the cytosol and the nucleus (Dupont et al., 2011; Aragona et al., 2013; Benham-Pyle et al., 2015; Mason et al., 2019), where they bind, among others, the transcriptional factor TEAD. The nuclear localization of YAP/TAZ has a profound impact in cell physiology and disease, since it causes reprograming of cell metabolism and enhanced cell proliferation among other effects (Totaro et al., 2018).
Here, by combining confocal microscopy, microfabrication, and a customized computational toolkit, we show that high nuclear levels of the transcriptional regulators YAP/TAZ cause a dramatic change in the expression of Rev-erbα and the subsequent malfunction of the fibroblast circadian clock. This novel molecular mechanism, which we show to be TEAD-dependent, could explain the highly impaired circadian clock in circumstances typically associated with high nuclear YAP/TAZ translocation such as cancer or aging (Stearns-Reider et al., 2017; Zanconato et al., 2016; Mattis and Sehgal, 2016; Blakeman et al., 2016).
Results
Rev-erbα basal expression and circadian oscillations depend on cell density
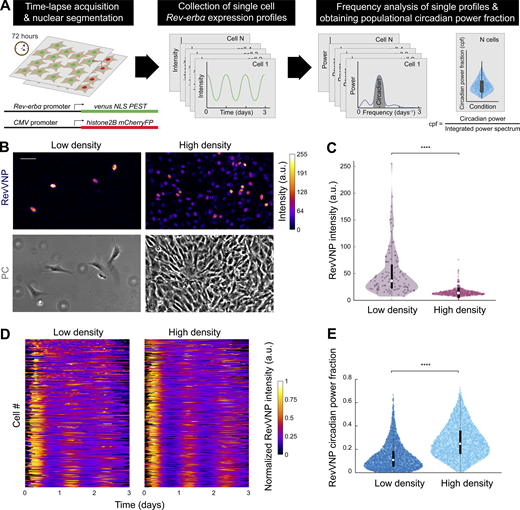
We conducted our work using NIH3T3 fibroblasts, a cellular model extensively reported to express a robust self-sustained circadian molecular clock (Nagoshi et al., 2004; Bieler et al., 2014). We generated a stable clonal cell line carrying both a reporter of Rev-erbα transcription (RevVNP; Nagoshi et al., 2004) and a chimeric histone2B-mCherry that we used as a constitutive nuclear marker (Fig. 1 A, left). To avoid the effect of specific signaling pathways derived from hormonal shocks (Balsalobre et al., 1998) and reveal underlying mechanobiological signals, cells were neither reset nor entrained exogenously during our experiments. As a consequence of this, instead of performing the common analysis of populational oscillations, we systematically tracked and measured the single-cell expression of RevVNP, which was obtained over the course of three-day time-lapses performed by confocal microscopy. We then developed a method to quantify and compare to what extent the unsynchronized cells display a 24-h oscillatory behavior. Performing Fourier analysis of the fluorescence emission of each cell we obtained individual power spectral densities. From these, we quantified the circadian power fraction, defined as the fraction of power found in a window between 0.7 and 1.3 d−1 of the cell’s RevVNP signal (Fig. 1 A and Fig. S1, A–F; Materials and methods). This analysis allows us to quantitatively distinguish between conditions that result in robust circadian oscillations (circadian power fraction tending to 1) from conditions where the RevVNP signal is affected by non-circadian modes, including non-rhythmic contributions (circadian power fraction tending to 0). Another method commonly used to analyze circadian oscillations, PyBOAT (Mönke et al., 2020 Preprint), provides similar results (Fig. S1).
Rev-erbα basal expression and circadian oscillations depend on cell density. (A) Schematics of the systematic computational analysis pipeline used to calculate the circadian power fraction (cpf) of a population of cells expressing RevVNP and H2B-mCherry and imaged during 72 h via time-lapse confocal microscopy. (B) Confocal microscopy (top) and phase contrast (PC; bottom) images of RevVNP-expressing cells grown at low (left) and high (right) density. Scale bar, 50 µm. (C) Violin plots representing the distribution of the single-cell RevVNP intensities of low- and high-density populations of a typical experiment of 14; n = 112 cells and n = 622 for low density and high density, respectively; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value < 0.0001. Full P values are reported in Table S1. (D) Raw data of the RevVNP intensities over time, represented in kymograph style, of 325 cells grown and tracked under low- or high-density conditions, from a representative experiment of 14. The single tracks are ordered from lower (top) to higher (bottom) circadian power fraction and aligned along the time axis according to maximum cross-correlation with the median track. (E) Violin plots representing the distribution of the single-cell RevVNP circadian power fraction of low- and high-density populations; n = 2,726 and 2,389 cells, respectively, from four experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value <0.0001. Full P values are reported in Table S1.
Rev-erbα basal expression and circadian oscillations depend on cell density. (A) Schematics of the systematic computational analysis pipeline used to calculate the circadian power fraction (cpf) of a population of cells expressing RevVNP and H2B-mCherry and imaged during 72 h via time-lapse confocal microscopy. (B) Confocal microscopy (top) and phase contrast (PC; bottom) images of RevVNP-expressing cells grown at low (left) and high (right) density. Scale bar, 50 µm. (C) Violin plots representing the distribution of the single-cell RevVNP intensities of low- and high-density populations of a typical experiment of 14; n = 112 cells and n = 622 for low density and high density, respectively; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value < 0.0001. Full P values are reported in Table S1. (D) Raw data of the RevVNP intensities over time, represented in kymograph style, of 325 cells grown and tracked under low- or high-density conditions, from a representative experiment of 14. The single tracks are ordered from lower (top) to higher (bottom) circadian power fraction and aligned along the time axis according to maximum cross-correlation with the median track. (E) Violin plots representing the distribution of the single-cell RevVNP circadian power fraction of low- and high-density populations; n = 2,726 and 2,389 cells, respectively, from four experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value <0.0001. Full P values are reported in Table S1.
Calculation of circadian power fraction from single-cell RevVNP signals. (A) Representative RevVNP fluorescence traces obtained from single-cell tracking of cells cultured at high density, showing raw data (yellow) and low-pass filtered data (black). (B) Representative RevVNP fluorescence traces obtained from single-cell tracking of cells cultured at low density, showing raw data (orange) and low-pass filtered data (black). (C–F) Schematic representation of the data analysis pipeline. A dataset of single-cell tracks (C) is pre-processed by low-pass filtering, mean subtraction and division by SD (D), using Fast Fourier Transform average PSD of the dataset is calculated (E) and the peak circadian frequency fc is identified. This frequency is then used as the center of the integration window (dark gray) on the single-cell PSDs to calculate the circadian power fraction (F). (G–J) Analysis of single-cell tracks at high (G and I) and low (H and J) density, using the continuous wavelet analysis software pyBOAT, with signal pre-processing by mean subtraction and normalization by standard deviation. (G and H) Wavelet spectra. (I and J) Ridge detected from wavelet spectra. (K–N) Analysis of single-cell tracks at high (K and M) and low (L and N) density, using signal pre-processing by pyBOAT’s signal detrending. (K and L) Wavelet spectra. (M and N) Ridge detected from wavelet spectra. Wavelet spectra show a decrease in power after ∼30 h due to the prevalence of high RevVNP signals at the beginning of single-cell traces.
Calculation of circadian power fraction from single-cell RevVNP signals. (A) Representative RevVNP fluorescence traces obtained from single-cell tracking of cells cultured at high density, showing raw data (yellow) and low-pass filtered data (black). (B) Representative RevVNP fluorescence traces obtained from single-cell tracking of cells cultured at low density, showing raw data (orange) and low-pass filtered data (black). (C–F) Schematic representation of the data analysis pipeline. A dataset of single-cell tracks (C) is pre-processed by low-pass filtering, mean subtraction and division by SD (D), using Fast Fourier Transform average PSD of the dataset is calculated (E) and the peak circadian frequency fc is identified. This frequency is then used as the center of the integration window (dark gray) on the single-cell PSDs to calculate the circadian power fraction (F). (G–J) Analysis of single-cell tracks at high (G and I) and low (H and J) density, using the continuous wavelet analysis software pyBOAT, with signal pre-processing by mean subtraction and normalization by standard deviation. (G and H) Wavelet spectra. (I and J) Ridge detected from wavelet spectra. (K–N) Analysis of single-cell tracks at high (K and M) and low (L and N) density, using signal pre-processing by pyBOAT’s signal detrending. (K and L) Wavelet spectra. (M and N) Ridge detected from wavelet spectra. Wavelet spectra show a decrease in power after ∼30 h due to the prevalence of high RevVNP signals at the beginning of single-cell traces.
We first addressed the effect of cell density on RevVNP expression by culturing cells on hydrogels (30 kPa in stiffness) at two distinct densities (∼30 and 1,000 cells/mm2, hereafter referred to as low and high density, respectively), which we kept approximately constant over time by arresting the cell cycle with the addition of thymidine, a DNA synthesis inhibitor (Jackman and O’ Connor, 1998). We observed a dramatic cell density–dependent difference in Rev-erbα expression, with cells cultured at low density displaying much higher RevVNP nuclear intensity than confluent cells (Fig. 1, B and C). These differences were ratified at the protein level via immunostainings against REV-ERBα (Fig. S2, A and B). The single-cell frequency analysis of the RevVNP signal revealed that the population of cells grown at high density mostly showed stable oscillations and had a higher circadian power fraction with an average period of 24.7 ± 1.9 h (Fig. 1, D and E; and Fig. S2 C). By contrast, cells at low density displayed low RevVNP circadian power fraction, with many cells exhibiting poor circadian oscillations, missing one or several peaks or even experiencing non-periodic fluctuations rather than oscillatory behavior (Fig. 1, D and E; and Fig. S2 C).
REV-ERBα levels decrease with cell density in a cell division–independent manner. (A) Confocal microscopy images of low- and high-density cell populations immunostained against REV-ERBα. All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (B) Violin plots representing the single-cell levels of nuclear REV-ERBα. n = 104 for low density and 142 for high density, from one representative of five independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001. (C) Violin plots representing the distribution of the single-cell RevVNP circadian power fraction of low- and high-density populations for three alternative clonal RevVNP cell lines; from left to right: n = 73, 197, 2,342, 3,857, 1,424, and 4,297, from three independent experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value <0.001. (D) Kymograph of Fig. 2 B with a modified color map dynamic range so the oscillations of the bulk population after the gap closure are more conspicuous. (E–G) RevVNP-expressing cells were cultured in two adjacent compartments separated by a barrier until they reached high density. Next, thymidine 2 mM was added, the barrier was released, and time-lapse confocal microscopy was performed during and after gap closure every 15 min for 4 d. A selection of time-lapse images showing the closure of the gap is depicted in E while F is a kymograph representing the average RevVNP intensity of all the cells along the x axis per time point. The average RevVNP intensity over time of the cells at the edge in comparison to those residing in the confluent zone is shown in G. The prior location of the barrier is depicted in the left image as a white dashed line. Notice that inhibition of proliferation causes a delay in gap closure and a decrease of overall cell density, which has a potential effect on RevVNP expression and rhythmicity. Scale bar, 100 µm.
REV-ERBα levels decrease with cell density in a cell division–independent manner. (A) Confocal microscopy images of low- and high-density cell populations immunostained against REV-ERBα. All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (B) Violin plots representing the single-cell levels of nuclear REV-ERBα. n = 104 for low density and 142 for high density, from one representative of five independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001. (C) Violin plots representing the distribution of the single-cell RevVNP circadian power fraction of low- and high-density populations for three alternative clonal RevVNP cell lines; from left to right: n = 73, 197, 2,342, 3,857, 1,424, and 4,297, from three independent experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; **** indicates a P value <0.001. (D) Kymograph of Fig. 2 B with a modified color map dynamic range so the oscillations of the bulk population after the gap closure are more conspicuous. (E–G) RevVNP-expressing cells were cultured in two adjacent compartments separated by a barrier until they reached high density. Next, thymidine 2 mM was added, the barrier was released, and time-lapse confocal microscopy was performed during and after gap closure every 15 min for 4 d. A selection of time-lapse images showing the closure of the gap is depicted in E while F is a kymograph representing the average RevVNP intensity of all the cells along the x axis per time point. The average RevVNP intensity over time of the cells at the edge in comparison to those residing in the confluent zone is shown in G. The prior location of the barrier is depicted in the left image as a white dashed line. Notice that inhibition of proliferation causes a delay in gap closure and a decrease of overall cell density, which has a potential effect on RevVNP expression and rhythmicity. Scale bar, 100 µm.
Rev-erbα circadian oscillations do not depend on cell–cell adhesions or paracrine signaling
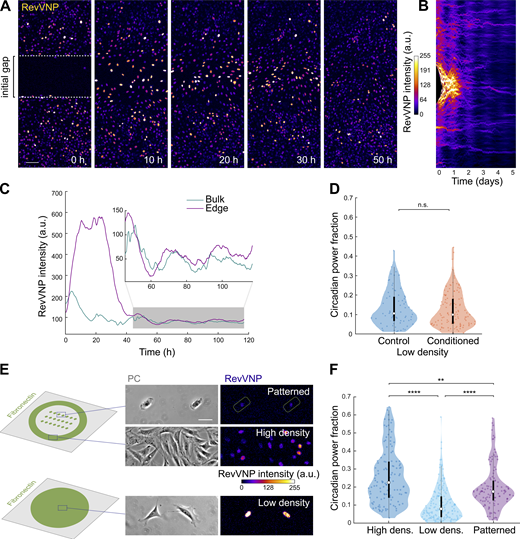
We next asked whether the detected differences in circadian power fraction are caused by paracrine signals, as previously reported for intercellular circadian coupling and synchronization (Noguchi et al., 2013; Finger et al., 2021). To address this question, we exposed closely packed cells to an abrupt drop in cell density. To do so, two cell monolayers were grown to confluence separated by a polydimethylsiloxane (PDMS) barrier. Upon lifting the barrier (Fig. 2 A, left), cells at the monolayer edges begun to spread and migrate toward the freely available gap. These cells experienced a sudden increase in RevVNP intensity that transiently disrupted their circadian oscillations until the gap was closed (Fig. 2, A–C; and Video 1). This disruption contrasted with the behavior of cells far from the edge, which oscillated robustly throughout the entire experiment as shown in the kymograph depicted in Fig. 2 B and Fig. S2 D. We obtained similar results in presence or absence of thymidine (Fig. S2, E–G), which confirms that the increase seen in Rev-erbα transcription is not a side effect of a cell cycle re-entry at the monolayer edge upon a loss of cell–cell contact. These experiments further demonstrate that cell density regulates the circadian clock. They suggest as well that this phenomenon does not depend on paracrine signaling, since high- and low-density cells shared the same extracellular chemical environment (i.e., potential cyclic paracrine signals secreted by the rhythmic bulk cells do not impose rhythmicity on the cells at the gap edge). We confirmed that absence of paracrine factors in the medium is not the cause of RevVNP circadian impairment by growing cells at low density with conditioned medium obtained from a high-density culture. We observed no increase in circadian power fraction compared to the low-density cells grown in fresh medium (Fig. 2 D). These results suggest that in our system cell density does not affect the circadian expression of Rev-erbα through paracrine signals, which led us to hypothesize that the observed behavior had a mechanochemical origin.
Rev-erbα circadian oscillations do not depend on cell–cell adhesions or paracrine signaling. (A) RevVNP-expressing cells were cultured in two adjacent compartments separated by a barrier until they reached high density. Next, the barrier was removed, and time-lapse confocal microscopy was performed during and after gap closure every 15 min for 5 d. A sequence of time-lapse images corresponding to the closure of the gap is shown. The prior location of the barrier is depicted in the left image as a white dashed line. Scale bar, 100 µm. (B) Kymograph representing the average RevVNP intensity per time point of all the cells from A along the x axis. The same kymograph with a dynamic range that enhances the fluorescence of the bulk after the gap closure is shown in Fig. S2 D. (C) Average RevVNP intensity over time of the cells at the edge in comparison to those residing in the confluent zone. The inset shows a magnification of the area shaded in gray. This figure shows an example of n = 6 independent experiments. (D) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of low-density cells grown in fresh medium and conditioned medium; n = 72 and 116 cells, respectively, from three experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively; two-sided Wilcoxon rank sum test; P value = 0.4168. (E) Schematics of the strategy followed to isolate single cells via micropatterning of fibronectin on glass. On the right, phase contrast (PC) and confocal microscopy images of RevVNP cells in stadium-shaped patterns (top), confluent cells cultured in the same well (middle), and non-confined cells cultured on a homogenous fibronectin-coated surface in a density as low as that of the micropatterned cells (bottom). Scale bar, 50 µm. (F) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of the conditions depicted in E; n = 121, 174, and 115 cells for the high-density, low-density, and micropatterned cells, respectively, from three experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; ** indicates P value <0.01; **** indicates P values <0.0001. Full P values are reported in Table S1.
Rev-erbα circadian oscillations do not depend on cell–cell adhesions or paracrine signaling. (A) RevVNP-expressing cells were cultured in two adjacent compartments separated by a barrier until they reached high density. Next, the barrier was removed, and time-lapse confocal microscopy was performed during and after gap closure every 15 min for 5 d. A sequence of time-lapse images corresponding to the closure of the gap is shown. The prior location of the barrier is depicted in the left image as a white dashed line. Scale bar, 100 µm. (B) Kymograph representing the average RevVNP intensity per time point of all the cells from A along the x axis. The same kymograph with a dynamic range that enhances the fluorescence of the bulk after the gap closure is shown in Fig. S2 D. (C) Average RevVNP intensity over time of the cells at the edge in comparison to those residing in the confluent zone. The inset shows a magnification of the area shaded in gray. This figure shows an example of n = 6 independent experiments. (D) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of low-density cells grown in fresh medium and conditioned medium; n = 72 and 116 cells, respectively, from three experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively; two-sided Wilcoxon rank sum test; P value = 0.4168. (E) Schematics of the strategy followed to isolate single cells via micropatterning of fibronectin on glass. On the right, phase contrast (PC) and confocal microscopy images of RevVNP cells in stadium-shaped patterns (top), confluent cells cultured in the same well (middle), and non-confined cells cultured on a homogenous fibronectin-coated surface in a density as low as that of the micropatterned cells (bottom). Scale bar, 50 µm. (F) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of the conditions depicted in E; n = 121, 174, and 115 cells for the high-density, low-density, and micropatterned cells, respectively, from three experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; ** indicates P value <0.01; **** indicates P values <0.0001. Full P values are reported in Table S1.
The origin of the Rev-erbα circadian impairment at low density is mechanical rather than paracrine. Time-lapse confocal imaging of RevVNP-expressing cells during and after gap closure. Cells were cultured on hydrogels at high density in two adjacent compartments separated by a PDMS barrier, which was removed just before imaging for 5 d every 15 min. Cells were not entrained by serum or hormonal shocks, although some synchronization is observed, possibly arising from the unavoidable medium washes after seeding and after barrier removal. Scale bar, 100 μm. Frame rate, 50 frames per second. hh:mm:ss, h:min:s.
The origin of the Rev-erbα circadian impairment at low density is mechanical rather than paracrine. Time-lapse confocal imaging of RevVNP-expressing cells during and after gap closure. Cells were cultured on hydrogels at high density in two adjacent compartments separated by a PDMS barrier, which was removed just before imaging for 5 d every 15 min. Cells were not entrained by serum or hormonal shocks, although some synchronization is observed, possibly arising from the unavoidable medium washes after seeding and after barrier removal. Scale bar, 100 μm. Frame rate, 50 frames per second. hh:mm:ss, h:min:s.
To test this hypothesis, we first asked whether the differences in RevVNP circadian oscillations between high and low density depend on cell–cell adhesion. To address this question, we confined individual cells in fibronectin micropatterns of 1,200 µm2 (a size comparable to that of fibroblasts within a monolayer; Fig. 2 E). The confined individual cells showed a significant reduction in RevVNP intensity together with an increase in circadian power fraction when compared to free cells grown at equivalent low density, a behavior resembling that of high-density cells (Fig. 2, E and F; and Video 2). This result implies that contact-based mechanisms in general, and intercellular adhesion in particular, do not explain the dependence of the Rev-erbα circadian oscillations on cell density.
Cell confinement in microprinted fibronectin areas rescues the Rev-erbα circadian impairment observed at low density. Phase contrast and confocal microscopy time-lapse movies of cells confined in stadium-shaped fibronectin-coated patterns. Images were taken every 15 min for 3 d. Scale bar, 100 µm. Frame rate, 50 frames per second. hh:mm, h:min.
Cell confinement in microprinted fibronectin areas rescues the Rev-erbα circadian impairment observed at low density. Phase contrast and confocal microscopy time-lapse movies of cells confined in stadium-shaped fibronectin-coated patterns. Images were taken every 15 min for 3 d. Scale bar, 100 µm. Frame rate, 50 frames per second. hh:mm, h:min.
Rev-erbα circadian power fraction anticorrelates with nuclear YAP
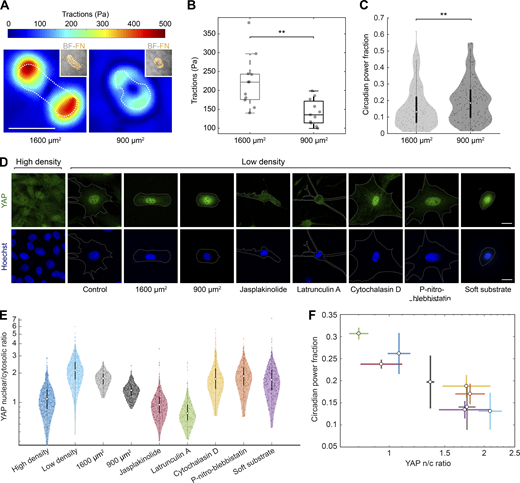
To explore whether circadian oscillations were regulated mechanically, we seeded single cells on stadium-shaped fibronectin micropatterns of two different areas while keeping all other variables constant. In agreement with previous studies (Oakes et al., 2014; Alisafaei et al., 2019), we found that cells spread on the larger patterns of 1,600 µm2 exerted more traction forces than cells constrained on the smaller 900 µm2 patterns (Fig. 3, A and B). This increase in traction forces was paralleled by a decrease in RevVNP circadian power fraction, supporting our hypothesis of a mechanical regulation of the circadian clock (Fig. 3 C).
Rev-erbα circadian power fraction anticorrelates with nuclear YAP. (A) Representative maps of traction force modulus on micropatterns of 1,600 and 900 µm2. The dashed line indicates the contour of the cell. Inset: brightfield (BF) image of corresponding cell and fibronectin (FN) micropattern (orange). Scale bar is 50 µm. (B) Box plot representing the mean traction force modulus of cells on micropatterns of 1,600 and 900 µm2. Two sample t test; ** indicates P value <0.01. (C) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of cells cultured in fibronectin stadium-shaped micropatterns of 1,600 and 900 µm2; n = 154 and 161, respectively, from four experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; ** indicates P value <0.01; full P values are reported in Table S1. (D) Confocal microscopy images of cells under different conditions (high-density control, low-density control, micropatterned cells of 1,600 and 900 µm2, low-density treated for 24 h with jasplakinolide 1 µM, latrunculin A 200 nM, cytochalasin D 1 µM, para-nitro-blebbistatin 10 µM, and low-density cells grown on polyacrylamide gels with a stiffness of 300 Pa). The cells were stained with an anti-YAP antibody (green, top) and Hoechst (blue, bottom); the cell perimeter is represented with a dashed white line. Scale bar, 20 µm. (E) Violin plots representing the distribution of the single-cell YAP nuclear to cytosolic ratios for the conditions depicted in C; n = 554, 589, 208, 331, 203, 171, 238, 153, and 315 cells, respectively, from three to seven experiments depending on the condition. Medians and interquartile ranges are depicted as white circles and black bars, respectively. The P values of the comparisons of every condition against the low-density, obtained with a two-sided Wilcoxon rank sum test, are represented in Table S1. (F) Correlation between circadian power fraction and YAP nuclear to cytosolic (n/c) ratio of all the aforementioned conditions. The values represented are the means of the medians of each independent experiment for every condition. The error bars refer to the corresponding SD. Pearson’s correlation coefficient is r = −0.93 with a P value of 0.0003.
Rev-erbα circadian power fraction anticorrelates with nuclear YAP. (A) Representative maps of traction force modulus on micropatterns of 1,600 and 900 µm2. The dashed line indicates the contour of the cell. Inset: brightfield (BF) image of corresponding cell and fibronectin (FN) micropattern (orange). Scale bar is 50 µm. (B) Box plot representing the mean traction force modulus of cells on micropatterns of 1,600 and 900 µm2. Two sample t test; ** indicates P value <0.01. (C) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of cells cultured in fibronectin stadium-shaped micropatterns of 1,600 and 900 µm2; n = 154 and 161, respectively, from four experiments; medians and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; ** indicates P value <0.01; full P values are reported in Table S1. (D) Confocal microscopy images of cells under different conditions (high-density control, low-density control, micropatterned cells of 1,600 and 900 µm2, low-density treated for 24 h with jasplakinolide 1 µM, latrunculin A 200 nM, cytochalasin D 1 µM, para-nitro-blebbistatin 10 µM, and low-density cells grown on polyacrylamide gels with a stiffness of 300 Pa). The cells were stained with an anti-YAP antibody (green, top) and Hoechst (blue, bottom); the cell perimeter is represented with a dashed white line. Scale bar, 20 µm. (E) Violin plots representing the distribution of the single-cell YAP nuclear to cytosolic ratios for the conditions depicted in C; n = 554, 589, 208, 331, 203, 171, 238, 153, and 315 cells, respectively, from three to seven experiments depending on the condition. Medians and interquartile ranges are depicted as white circles and black bars, respectively. The P values of the comparisons of every condition against the low-density, obtained with a two-sided Wilcoxon rank sum test, are represented in Table S1. (F) Correlation between circadian power fraction and YAP nuclear to cytosolic (n/c) ratio of all the aforementioned conditions. The values represented are the means of the medians of each independent experiment for every condition. The error bars refer to the corresponding SD. Pearson’s correlation coefficient is r = −0.93 with a P value of 0.0003.
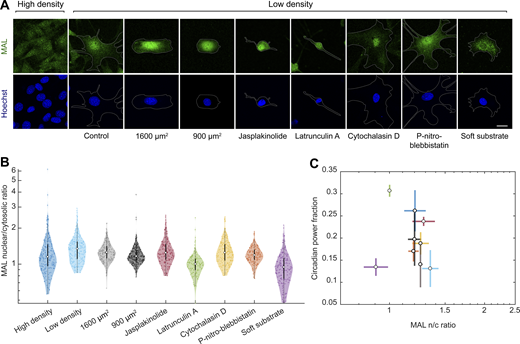
To identify the mechanotransduction pathway that regulates Rev-erbα expression, we focused on two key mechanosensitive transcriptional regulators: MAL and YAP. We measured both the circadian power fraction and the subcellular localization of MAL and YAP in a series of conditions known to impact their nucleocytoplasmic shuttling. These conditions include: the already mentioned high and low cell density and single-cell micropatterning on different areas; treatment with drugs that alter the actomyosin cytoskeleton such as jasplakinolide, latrunculin A, para-nitro-blebbistatin, and cytochalasin D; and the growth of cells on very soft hydrogels (300 Pa; Wada et al., 2011; Gegenfurtner et al., 2018; Elosegui-Artola et al., 2017; Sun et al., 2021). As expected, these conditions had a variety of effects on cell morphology (Fig. 3 D and Fig. S3). We found no correlation between the nuclear to cytoplasmic ratio of MAL and the circadian power fraction (Fig. S3). By contrast, the circadian power fraction showed a progressive decrease with increasing YAP nuclear to cytoplasmic ratio (Fig. 3). This striking anticorrelation implies a potentially causal link between the robustness of the Rev-erbα circadian expression and the mechanosensitive regulation and nucleocytoplasmic transport of YAP.
Rev-erbα circadian power fraction does not correlate with nuclear MAL. (A) Confocal microscopy images of cells under different conditions (high-density control, low-density control, micropatterned cells of 1,600 and 900 µm2, low-density treated for 24 h with jasplakinolide 1 µM, latrunculin A 200 nM, cytochalasin D 1 µM, para-nitro-blebbistatin 10 µM, and low-density cells grown on polyacrylamide gels with a stiffness of 300 Pa). The cells were stained with an anti-MAL antibody (green, top) and Hoechst (blue, bottom); the cell perimeter is represented with a dashed white line. Scale bar, 20 µm. (B) Violin plots representing the distribution of the single-cell MAL nuclear to cytosolic ratios of the conditions depicted in A; n = 215, 254, 208, 332, 163, 140, 165, 157, and 162, respectively, from three to four experiments depending on the condition. Medians and interquartile ranges are depicted as white circles and black bars, respectively. The P values of the comparisons of every condition against the low-density, obtained with a two-sided Wilcoxon rank sum test, are represented in Table S1. (C) Dependence of circadian power fraction with MAL nuclear to cytosolic ratio of all the aforementioned conditions. The values represented are the means of the medians of each independent experiment for every condition. The error bars refer to the corresponding SD. Pearson’s correlation coefficient is r = −0.19 with a P value of 0.63.
Rev-erbα circadian power fraction does not correlate with nuclear MAL. (A) Confocal microscopy images of cells under different conditions (high-density control, low-density control, micropatterned cells of 1,600 and 900 µm2, low-density treated for 24 h with jasplakinolide 1 µM, latrunculin A 200 nM, cytochalasin D 1 µM, para-nitro-blebbistatin 10 µM, and low-density cells grown on polyacrylamide gels with a stiffness of 300 Pa). The cells were stained with an anti-MAL antibody (green, top) and Hoechst (blue, bottom); the cell perimeter is represented with a dashed white line. Scale bar, 20 µm. (B) Violin plots representing the distribution of the single-cell MAL nuclear to cytosolic ratios of the conditions depicted in A; n = 215, 254, 208, 332, 163, 140, 165, 157, and 162, respectively, from three to four experiments depending on the condition. Medians and interquartile ranges are depicted as white circles and black bars, respectively. The P values of the comparisons of every condition against the low-density, obtained with a two-sided Wilcoxon rank sum test, are represented in Table S1. (C) Dependence of circadian power fraction with MAL nuclear to cytosolic ratio of all the aforementioned conditions. The values represented are the means of the medians of each independent experiment for every condition. The error bars refer to the corresponding SD. Pearson’s correlation coefficient is r = −0.19 with a P value of 0.63.
YAP/TAZ perturbs the circadian clock via TEAD
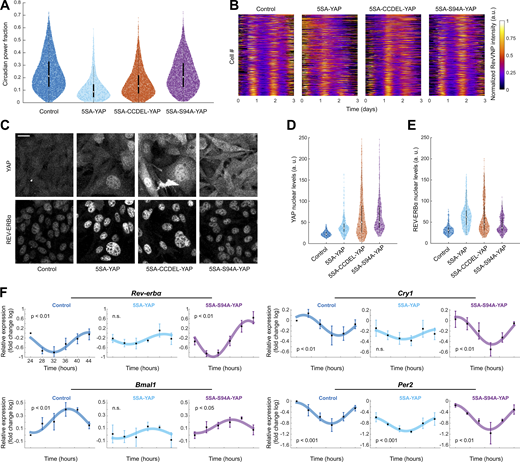
To test whether this dependence is indeed causal, we induced in high-density cells a sustained overexpression of 5SA-YAP, a mutated version of YAP whose five main phosphorylation sites have been removed resulting in an abnormally high nuclear retention (Zhao et al., 2007). The high levels of 5SA-YAP led to a severe reduction of RevVNP circadian power fraction (Fig. 4, A and B) and an increase in REV-ERBα basal levels when compared to high-density control cells that expressed only low basal nuclear levels of endogenous YAP (Fig. 4, C–E). A similar result was obtained when overexpressing the TAZ mutant 4SA-TAZ, which is also retained in the nucleus due to the mutation of its four main phosphorylation sites (Park et al., 2015; Lei et al., 2008; Fig. S4). Together, these results establish that the fibroblast circadian clock is controlled through both YAP and TAZ.
YAP perturbs the circadian clock via TEAD. (A) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of YAP-overexpressing cells carrying different mutations and the control; n = 6,466, 440, 5,320, and 4,571 for control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP, respectively, from five independent experiments; medians (0.21, 0.09, 0.13, and 0.19) and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; all comparisons show statistically significant differences; full P values are reported in Table S1. (B) Raw data of the RevVNP intensities over time, represented in kymograph style, of 440 cells grown and tracked under low- or high-density conditions, from a representative experiment of three. The single tracks are ordered from less (top) to more (bottom) circadian power fraction and aligned along the time axis according to maximum cross-correlation with the median track. (C) Confocal microscopy images of control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP cells immunostained for YAP (left) and a REV-ERBα (right). All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (D and E) Violin plots representing the single-cell levels of nuclear YAP (D) and nuclear REV-ERBα (E) for all the conditions depicted in C. n = 294, 400, 383, and 393 for control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP, respectively, from three independent experiments. Medians (21.99, 36.35, 50.13, and 50.77 for YAP; 30.75, 61.89, 45.58, and 36.51 for REV-ERBα) and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; all comparisons show statistically significant differences; full P values are reported in Table S1. (F) mRNA expression of Rev-erbα, Bmal1, Cry1, and Per2 in high-density cells subjected to constitutive overexpression of an empty vector (control), 5SA-YAP, and 5SA-S94A-YAP, measured by RT-qPCR. The values refer to the logarithms of the fold change relative to the first time point in the control condition. Cells were synchronized with a 2-h horse serum shock. The first time point corresponds to cells collected 24 h after the shock. The graphs represent the mean of n = 3 biological repeats, and the error bars represent the SD. The depicted sinusoidal fits and the statistical analysis were performed using single cosinor analysis. The reported P values (P) refer to an F test against a null hypothesis of zero amplitude. Exact values are listed in Table S1.
YAP perturbs the circadian clock via TEAD. (A) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of YAP-overexpressing cells carrying different mutations and the control; n = 6,466, 440, 5,320, and 4,571 for control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP, respectively, from five independent experiments; medians (0.21, 0.09, 0.13, and 0.19) and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; all comparisons show statistically significant differences; full P values are reported in Table S1. (B) Raw data of the RevVNP intensities over time, represented in kymograph style, of 440 cells grown and tracked under low- or high-density conditions, from a representative experiment of three. The single tracks are ordered from less (top) to more (bottom) circadian power fraction and aligned along the time axis according to maximum cross-correlation with the median track. (C) Confocal microscopy images of control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP cells immunostained for YAP (left) and a REV-ERBα (right). All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (D and E) Violin plots representing the single-cell levels of nuclear YAP (D) and nuclear REV-ERBα (E) for all the conditions depicted in C. n = 294, 400, 383, and 393 for control, 5SA-YAP, 5SA-CCDEL-YAP, and 5SA-S94A-YAP, respectively, from three independent experiments. Medians (21.99, 36.35, 50.13, and 50.77 for YAP; 30.75, 61.89, 45.58, and 36.51 for REV-ERBα) and interquartile ranges are depicted as white circles and black bars, respectively. Two-sided Wilcoxon rank sum test; all comparisons show statistically significant differences; full P values are reported in Table S1. (F) mRNA expression of Rev-erbα, Bmal1, Cry1, and Per2 in high-density cells subjected to constitutive overexpression of an empty vector (control), 5SA-YAP, and 5SA-S94A-YAP, measured by RT-qPCR. The values refer to the logarithms of the fold change relative to the first time point in the control condition. Cells were synchronized with a 2-h horse serum shock. The first time point corresponds to cells collected 24 h after the shock. The graphs represent the mean of n = 3 biological repeats, and the error bars represent the SD. The depicted sinusoidal fits and the statistical analysis were performed using single cosinor analysis. The reported P values (P) refer to an F test against a null hypothesis of zero amplitude. Exact values are listed in Table S1.
Overexpression of a constitutively active TAZ mutant affects REV-ERBα expression. (A) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of control and TAZ-overexpressing cells carrying a 4SA dominant positive mutation; n = 3,097 and 2,359, respectively, from five independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001. (B) Confocal microscopy images of control and 4SA-TAZ cells immunostained against REV-ERBα. All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (C) Violin plots representing the single-cell levels of nuclear REV-ERBα. n = 134 for the control and 123 for 4SA-TAZ, from one representative of three independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001.
Overexpression of a constitutively active TAZ mutant affects REV-ERBα expression. (A) Violin plots representing the distribution of the single-cell RevVNP circadian power fractions of control and TAZ-overexpressing cells carrying a 4SA dominant positive mutation; n = 3,097 and 2,359, respectively, from five independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001. (B) Confocal microscopy images of control and 4SA-TAZ cells immunostained against REV-ERBα. All images are displayed under the same brightness and contrast settings. Scale bar, 20 µm. (C) Violin plots representing the single-cell levels of nuclear REV-ERBα. n = 134 for the control and 123 for 4SA-TAZ, from one representative of three independent experiments. Two-sided Wilcoxon rank sum test; **** indicates P value <0.0001.
The activity of YAP/TAZ as gene expression coactivators requires the interaction with other transcription factors. Since YAP interacts with REV-ERBα through its coiled-coil (CC) domain (Zhang et al., 2017), we tested if this interaction gives rise to the deleterious effect of nuclear YAP on the circadian clock. For that, we overexpressed 5SA-YAP but with its CC domain deleted (5SA-CCDEL-YAP), a mutation that had been previously shown to disable the molecular interaction between YAP and REV-ERBα (Zhang et al., 2017). We found that the CCDEL mutation only rescued slightly the impaired RevVNP circadian oscillations (Fig. 4 A) and the REV-ERBα basal levels (Fig. 4, B–D) observed upon 5SA-YAP overexpression. This result shows that the direct molecular binding through the YAP CC domain is not the main regulatory pathway connecting YAP and the circadian clock.
Alternatively, we studied whether this regulation was mediated by the well-known interaction of YAP/TAZ with the TEAD family (Vassilev et al., 2001). We confirmed by real-time quantitative PCR (RT-qPCR) that the transcriptional levels of Cyr61 and CTGF, two well-known TEAD target genes, were abnormally high upon the overexpression of 5SA-YAP or 5SA-CCDEL-YAP (Fig. S5), which suggests that hyperactive TEAD-driven transcription is behind the RevVNP circadian deregulation. To corroborate this, we overexpressed 5SA-S94A-YAP, a mutant version of YAP which, besides being non-phosphorylatable, is unable to interact with TEAD (Zhao et al., 2008). We observed that 5SA-S94A-YAP cells, which showed normal levels of TEAD activity (Fig. S5), recovered to a large extent both the RevVNP circadian power fraction and the REV-ERBα basal levels displayed by the wild-type high-density population (Fig. 4). This result establishes that the Rev-erbα circadian deregulation in nuclear YAP-enriched cells is controlled by the YAP-TEAD transcriptional cascade.
TEAD-associated signaling is activated under 5SA-YAP and 5SA-CCDEL-YAP but not 5SA-S94A-YAP overexpression. mRNA expression of the TEAD targets Cyr61 and CTGF in high-density cells subjected to constitutive overexpression of an empty vector (control), 5SA-YAP, 5SA-CCDDEL-YAP, and 5SA-S94A-YAP, measured by RT-qPCR. The values refer to the fold change compared to the control condition. GAPDH was used as the housekeeping gene. The graphs represent the mean of n = 5–6 biological repeats (black dots), and the error bars represent the SD. Ordinary one-way ANOVA test; **** indicates P value <0.0001; ** indicates P value <0.01, * indicates P value <0.1; ns indicates P value >0.5.
TEAD-associated signaling is activated under 5SA-YAP and 5SA-CCDEL-YAP but not 5SA-S94A-YAP overexpression. mRNA expression of the TEAD targets Cyr61 and CTGF in high-density cells subjected to constitutive overexpression of an empty vector (control), 5SA-YAP, 5SA-CCDDEL-YAP, and 5SA-S94A-YAP, measured by RT-qPCR. The values refer to the fold change compared to the control condition. GAPDH was used as the housekeeping gene. The graphs represent the mean of n = 5–6 biological repeats (black dots), and the error bars represent the SD. Ordinary one-way ANOVA test; **** indicates P value <0.0001; ** indicates P value <0.01, * indicates P value <0.1; ns indicates P value >0.5.
We finally investigated if the observed YAP-TEAD regulation has implications on the cellular circadian landscape beyond Rev-erbα. Analysis of published chromatin immunoprecipitation sequencing and microarray databases listed Bmal1 and Cry1 as YAP and TEAD targets (Zhao et al., 2008; Lee et al., 2016; Zanconato et al., 2016; Rajbhandari et al., 2018), providing a potential direct mechanism for YAP/TAZ to control the circadian core clock, which could concomitantly impact the expression of Rev-erbα. We tracked, via RT-qPCR, the transcription of Rev-erbα and the circadian core clock components Bmal1, Per2, and Cry1 in conditions where YAP-TEAD signaling is deregulated. Specifically, we synchronized, through a serum shock, high-density cells overexpressing either 5SA-YAP, 5SA-S94A-YAP, or a control empty vector, and compared their circadian transcriptional profiles over the course of 24 h. The RT-qPCR results were statistically analyzed using single cosinor rhythmometry (Fig. 4 F; Cornelissen, 2014) and JTK-Cycle (Table S1; Hughes et al., 2010; Zielinski et al., 2014). This analysis showed that high expression of 5SA-YAP disrupted the circadian oscillation of Rev-erbα in a TEAD-dependent manner (Fig. 4 F), which agrees with the results shown in Fig. 4, A and B regarding the RevVNP circadian power fraction of non-shocked individual cells. Importantly, high levels of 5SA-YAP, but not of 5SA-S94A-YAP, also significantly disrupted the rhythmicity of the master clock regulator Bmal1 and its repressor Cry1 (Fig. 4 F). Intriguingly, Per2 expression was still cyclic under YAP overactivation, although the amplitude of the oscillation was lower than in the other conditions (Fig. 4 F), which is in line with a general perturbation of the core clock regulation by YAP. Taken together, these results demonstrate that robust circadian homeostasis in fibroblasts requires low levels of nuclear YAP/TAZ and its subsequent TEAD signaling.
Discussion
The blooming of mechanobiology in this century has established mechanics as an essential player in the regulation of key biological processes in physiology and disease (Hayward et al., 2021). However, it was not until very recently that the influence of mechanical cues on the circadian clock has begun to be addressed, partly due to large differences in experimental approaches and timescales between the fields of chronobiology and cell mechanics. Traditionally, circadian experiments have mainly been carried out at the populational level and, when performed in vitro, they often relied on cell synchronization by the application of zeitgebers (Balsalobre et al., 1998; Balsalobre et al., 2000; Gerber et al., 2013). These treatments are biochemically invasive and could mask the effect of non-canonical signaling cascades like those triggered by mechanical factors. For example, typically used serum shocks have been described to strongly activate the Hippo pathway (Hergovich, 2019), while glucocorticoids are known to have a profound impact on, among many other intracellular processes, the actin cytoskeleton dynamics, cell shape, and cell fate (Ren et al., 2012). For the most part of our work, we studied the mechanical regulation of the circadian clock at the single-cell level and without need for synchronization strategies. This approach allowed us, for example, to study oscillations in single cells confined in desired shapes and sizes and to closely link cell density with the oscillation robustness. We expect the present work to open new avenues of research to address how mechanosensing signaling cascades impact the clock. This approach can be fostered by (a) the development of techniques allowing a precise mechanical manipulation of the cells over prolonged periods of time; (b) the increasing availability of new fluorescent circadian reporters through genome-editing techniques like CRISPR knock-in (Gabriel et al., 2021; Smyllie et al., 2022); and (c) the continuous development of new algorithms to analyze noisy oscillatory signals in single cells (Manella et al., 2021).
An intricate relationship between cellular mechanobiology and the circadian clock has recently begun to emerge. For example, two recent studies have shown that actin dynamics and the secretion of extracellular matrix are subjected to circadian regulation (Chang et al., 2020; Hoyle et al., 2017). The groups of C.H. Streuli and Q.J. Meng have studied the complementary regulation, showing that the robustness of the circadian clock is influenced by the stiffness of the extracellular environment in a cell-type-dependent manner, with a mechanistic focus on integrins and Rho/ROCK (Yang et al., 2017; Williams et al., 2018). With our finding that nuclear YAP/TAZ disrupts the fibroblast circadian clock via TEAD, we now provide a transcriptional explanation of how mechanical signals, independently of their nature and how they are triggered, can be transduced into alterations of the circadian clock. Given the widespread role of YAP/TAZ as core mechanosensors, the potential implications of this new mechanism are multiple. We believe this study will contribute to explain the loss of circadian rhythms in aging and cancer, two processes associated with an impaired clock, with an alteration in the mechanical properties of the tissues and with an abnormal activation of YAP/TAZ (Stokes et al., 2021; Blakeman et al., 2016; Broadberry et al., 2018; Dupont et al., 2011; Kondratov et al., 2006; Rivera-Reyes et al., 2018; Salzer et al., 2018; Stearns-Reider et al., 2017). Since the expression and localization of YAP/TAZ vary highly among and within tissues (The Human Protein Atlas, https://www.proteinatlas.org/ENSG00000137693-YAP1/tissue; van Soldt et al., 2019), future research should address how that heterogeneity impacts the different cellular clocks and to what extent they are sensitive to mechanical deregulation through this pathway. Such experiments should involve not only overexpression of YAP and TAZ and the modulation of their nuclear levels, as performed here, but also their selective knockout.
Our result that YAP/TAZ and TEAD dramatically affect the basal expression of REV-ERBα in non-entrained individual cells has potential implications beyond the field of chronobiology. REV-ERB proteins are nuclear receptors that, acting as strong transcriptional repressors, play a key role in a large variety of physiological processes and have been extensively used as drug targets (Everett and Lazar, 2014; Uriz-Huarte et al., 2020). For example, high levels of REV-ERB have been correlated with increased insulin secretion and to altered lipid metabolism, which has opened new strategies for the treatment of diabetes (Uriz-Huarte et al., 2020). REV-ERB proteins are also involved in inflammation regulation, muscle function through the control of mitochondrial activity, bone loss, and Alzheimer’s disease (Uriz-Huarte et al., 2020). In light of this, we expect this study to serve as a meeting point between the largely disconnected fields of chronobiology, mechanobiology, and metabolism.
Remarkably, our work shows that YAP/TAZ and TEAD signaling has a rather transversal impact on the core clock, since the mRNA circadian expression of not only Rev-erbα but also Bmal1, Cry1, and—at to lower extent—Per2, is altered under high levels of 5SA-YAP in a S94-dependent manner. Noticeably, Cry1 and Bmal1 have been detected as targets of YAP/TAZ and/or TEAD in several studies (Zhao et al., 2008; Lee et al., 2016; Zanconato et al., 2016; Rajbhandari et al., 2018), which hints at the potential molecular sites where the negative feedback loop that governs the circadian clock is altered upon YAP-TEAD activation. Further experiments and theoretical work should be conducted to decipher the precise kinetics that underlie the transcriptional regulation of the circadian clock by the TEAD family.
Materials and methods
Cell culture
We cultured NIH3T3 mice fibroblasts in DMEM with pyruvate (41966-029; Thermo Fisher Scientific) supplemented with 10% or 2% FBS (for cell lines maintenance or for experimental purposes, respectively; 10270-106; Thermo Fisher Scientific), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, and 292 μg ml−1 of L-glutamine (10378016; Thermo Fisher Scientific). For all the experiments except the gap closure one, thymidine 2 mM (T9250-1G; Sigma-Aldrich) was added 24 h before fixation or image acquisition. The experiment with conditioned medium was carried out with medium composed by 50% collected medium from a high-density population of cells after 2 d of culture and 50% fresh medium. To prevent undesired entrainments caused by medium changes like described in Guenthner et al. (2014), we maintained the same medium for the cell culture on the plates and the entire image acquisition. However, the conveyance of a certain level of phase synchrony in RevVNP expression was unavoidable in some cases, mainly in the gap closure experiments, which required intensive cell culture manipulation.
Cell lines and constructs
For the circadian analyses, we generated a stable cell line expressing Venus fluorescent protein with a nuclear localization signal (NLS) and a destabilization PEST domain under the promoter of Rev-erbα. We used specifically the plasmid rev12-ex2-Venus-NLS-PEST1, courtesy of Prof. Ueli Schibler (Nagoshi et al., 2004), and random genomic integration via, first, lipofectamine-based transfection (Lipofectamine 3000; Thermo Fisher Scientific) and second, several rounds of fluorescence-based flow cytometry sorting, until stable cassette expression was observed. Lentiviral particles carrying plenti6-H2B-mCherry (Addgene plasmid #89766 [Pemble et al., 2017]) were generated by transient transfection of Hek293T cells together with the corresponding envelope and packaging vectors. Several clonal lines were obtained by transduction of the RevVNP cell line with the mentioned particles and flow cytometry sorting (Beckman Coulter). After testing that all of them responded similarly to cell density (Fig. S2 C), one was selected and used for all the subsequent experiments.
The 5SA-YAP, 5SA-CCDEL-YAP, 5SA-S94A-YAP, 4SA-TAZ, and control cell lines were obtained by transduction with retroviral particles generated in HEK293T cells and sustained selection with puromycin 2 μg ml−1 (A1113803; Thermo Fisher Scientific). Specifically, the retroviral FLAG-5SA-YAP and FLAG-5SA-S94A-YAP plasmids were generated via restriction-based cloning from pQCXIH-Myc-YAP-5SA (Addgene plasmid #33093 [Zhao et al., 2007]) and pCMV-Flag-YAP-5SA/S94A (Addgene plasmid #33103 [Zhao et al., 2008]), respectively, and pBABE-PURO (Addgene plasmid #1764 [Morgenstern and Land, 1990]). The CCDEL deletion was introduced in the latter via overlapping PCR with the primers YAP-DEL-CC-HA-F and YAP-DEL-CC-HA-R as described in Zhang et al. (2017). The FLAG-5SA-S94A-YAP plasmid corresponds to the Addgene plasmid The FLAG-4SA-TAZ plasmid was a kind gift of Dr. Hyun Woo Park (Yonsei University, Seoul, Korea [Park et al., 2015]). All the cell lines were checked by PCR, DNA sequencing, and/or Western blot.
The cytoskeletal drugs used throughout the study were: jasplakinolide 1 µM (J4580-100UG; Sigma-Aldrich), cytochalasin D 1 µM (C8273-1MG; Sigma-Aldrich), latrunculin A 200 nM (L5163-100UG; Sigma-Aldrich), and para-nitro-blebbistatin 10 µM (DR-N-111; OptoPharma). The control conditions were performed with similar concentrations of DMSO (D8418-50 Ml; Sigma-Aldrich). All of them were added 24 h before the initiation of the live imaging or fixation.
Time-lapse imaging of circadian oscillations of NIH3T3 fibroblasts
Time-lapse acquisitions were performed on a scanning confocal microscope (Nikon Ti Eclipse) equipped with thermal, CO2, and humidity regulation. 12-well or 6-well glass-bottomed plates were fixed on the stage and imaging was performed using a 10× objective (NA = 0.30, air) with focusing maintained by the Perfect Focus System (Nikon). The software NIS Elements was used to image every 15 min during experiments lasting for 3–5 d. Multiple wells, and multiple positions within the different wells, were imaged by phase contrast and in laser scanning mode using a 488 and a 561 nm excitation. For the 488 nm acquisition, two different sets of settings were used, to better capture the large variations in fluorescence signal seen in the RevVNPchannel: a low-intensity channel with low excitation power, and a high-intensity channel with five times higher excitation power. All confocal images were acquired by scanning 1,024 × 1,024 pixels on the objective’s field of view, yielding a pixel-size of 1.28 µm. Pixel dwell times were adjusted independently in each experiment.
Single-cell tracking and frequency analysis of single-cell circadian oscillations
Tiff stacks of time-lapse confocal microscopy images were analyzed using Fiji (Schindelin et al., 2012). Images were not treated prior to tracking. Fluorescent nuclei visible in the high-intensity channel were tracked in space and time using the Fiji plugin TrackMate (Tinevez et al., 2017). Within TrackMate, firstly nuclei were localized using the LoG detector, after which the LAP tracker was used, with a maximum linking distance of 15 pixels and gap closing set to two frames. This yielded individual tracks of the average intensity of each nucleus present in the time-lapse. The tracks were then filtered by length, keeping only those that lasted at least 60 h. In the rare cases of cell division, only one daughter cell was tracked. The spatial trajectories were then used to additionally obtain tracks for the low-intensity channel. Finally, the position and average nuclear intensity data for both channels were exported as .csv files.
All analyses were performed with custom software created using MATLAB R2017a (Fig. 1 and Fig. S1, A–F). We relied on Fourier analysis to assess how the circadian oscillations of RevVNP were affected by different experimental conditions. We analyzed datasets of single-cell time series according to the following procedure, implemented in MATLAB 2017a. Individual time series Ii(t) were analyzed to find any missing time-points. These occurred rarely and were replaced by nearest-neighbor linear interpolations using the MATLAB function interp1. Then, the tracks were analyzed individually to remove any dying cells: these were defined as intensity traces whose standard deviation in the final 24 h is <5% of the SD during the rest of the experiment. Following this, the individual time series were denoised by a low-pass filter, using the MATLAB zero-phase filter filtfilt, with parameters a = 1 and b = (0.2, 0.2, 0.2, 0.2, 0.2). The time series were then rescaled by subtracting the mean and normalized by dividing by their SD. The power spectrum density PSDi(f) of each time series was obtained using MATLAB’s fft function and then the average power spectrum of the whole dataset was computed. The ensemble circadian frequency fc was identified as the coordinate of the peak in the interval [0.7, 1.3] d−1 in . Then, we went back to the single-cell PSDs and individually analyzed them as follows: the integral of the PSD in the frequency interval W = [fc – 0.2, fc + 0.2] d−1 was divided by the full integral of that individual track (i.e., the total power), giving what we termed the circadian power fraction of a single-cell oscillation. Note that to set the width of W we used pooled experiments with cells at high-density, identified the ensemble circadian frequency fc and measured the full width at half maximum of the corresponding peak.
As a means of control, we compared our custom frequency analysis with a recently published software for time-frequency analysis of biological oscillations called pyBOAT (Mönke et al., 2020 Preprint). It uses continuous wavelet analysis to extract time dependent frequency information from oscillatory signals. We used pyBOAT to analyze the circadian behavior of cells cultured at high density and low density, using the same data reported in Fig. 1, D and E. We used a set of 330 single-cell oscillations per condition, and pre-processed them in two different ways: in one case, we applied the same normalization we used before running our custom frequency analysis, in the other case we used the unnormalized traces and applied a detrending step in pyBOAT. In all cases, analysis with pyBOAT confirmed the trends observed by our custom analysis method (Fig. S1, G–N).
Kymography
The denoised and rescaled single-cell tracks were further analyzed by pairwise cross-correlation, and the phases of the signals were obtained as the lag values giving maximum cross-correlation. By virtue of the periodicity of the signals, these lags were restricted to the interval [−1,1] d. Tracks were ordered by decreasing circadian power fraction and the phases (taken with respect to the median track) were used to shift (in time) the other tracks and thus artificially synchronize the dataset. These data were plotted as a kymograph of fluorescence intensity by time and circadian power fraction rank, with the highest circadian power fraction tracks at the bottom.
For the gap closure experiments, the nuclei in time-lapse fluorescence images were segmented using a combination of TrackMate’s spot detection algorithm and intensity thresholding. The kymographs were generated by averaging the intensity of the segmented nuclei in each frame along the direction parallel to the wound edge. These lines were then assembled into a kymograph representing fluorescence intensity as a function of time and distance from the center of the wound.
Polyacrylamide (PAA) gels
PAA gels with Young modulus E = 30 kPa—which matches approximately the stiffness of the majority of abdominal organs and the skin (Guimarães et al., 2020)—were produced according to a previously published protocol (Bazellières et al., 2015). A PBS solution containing 12% acrylamide (161-0140; Bio-Rad), 0.15% bis-acrylamide (161-0142; Bio-Rad), plus 0.05% ammonium persulfate (A3678; Sigma-Aldrich) and 0.05% tetramethyl ethylenediamine (T9281-25 Ml; Sigma-Aldrich) was prepared and allowed to polymerize between a coverslip and a glass-bottomed dish (Mattek). Alternatively, 3% acrylamide and 0.03% bis-acrylamide were used to make 300 Pa gels. The PAA gel surface was then incubated with a solution of 2 mg ml−1 Sulfo-SANPAH (4822589; Thermo Fisher Scientific) under ultraviolet light for 5 min (wavelength of 365 nm at 5 cm distance). After that, the excess Sulfo-SANPAH was removed by three consecutive 3-min washes with PBS. A solution of fibronectin 0.1 mg ml−1 (F0895; Sigma-Aldrich) was added on top of the gels and left overnight at 4°C.
Gap closure
The gap closure experiments were performed following the protocol described in Rodríguez-Franco et al. (2017). Magnetic PDMS stencils consisting in two hollow regions separated by a barrier of 600–900 µm were obtained using a customized 3D-printed mold. The cured, autoclaved, passivated, and dried magnetic PDMS stencils were then deposited on a fibronectin-coated PAA gel polymerized in a glass-bottomed dish. After this, the dish was placed on top of a custom-made magnetic holder to keep the gasket in place and avoid medium leakage. The hollow regions were then filled with 40,000 cells contained in 120 µl of medium. After 1 h, excess of cells was removed by two consecutive washes with medium. The attached cells were left overnight at 37°C before the removal of the magnetic PDMS gasket and the filling of the whole glass-bottomed dish with medium.
Immunostainings
Cells were fixed using paraformaldehyde 4% for 15 min and washed with PBS three times. Then, they were permeabilized with 0.1% Triton X-100 for 45 min, incubated with the primary antibodies for 90 min at room temperature, washed and incubated with the secondary antibodies for another 90 min at room temperature. After this step, Hoechst 33342 (H3570; Thermo Fisher Scientific) was added at a concentration of 1 μg ml−1 for 10 min. Finally, cells were mounted in Mowiol reagent (81381; Merck). The buffers used during the whole procedure had fish gelatine 1.6% (vol/vol) as a blocking agent (G7765; Merck). The primary antibodies used were rabbit anti-NR1D1 (ab174309; Abcam), rabbit anti-MKL1 (ab49311; Abcam) and mouse anti-YAP 63.7 (sc-101199; Santa Cruz). The secondary antibodies used were Alexa Fluor-488 goat anti-mouse (A-11029; Thermo Fisher Scientific), Alexa Fluor-488 donkey anti-rabbit (A-21206; Thermo Fisher Scientific), Alexa Fluor-555 goat anti-mouse (A-21424; Thermo Fisher Scientific), and Alexa Fluor-555 goat anti-rabbit (A-21429; Thermo Fisher Scientific). All the antibodies were incubated in a 1:200 dilution. The acquisition of z-stacks (with a z-axis step of 0.7 µl) was done using a 40× water immersion LD LCI Plan Apo objective (NA = 1.2) in a scanning confocal microscope (Zeiss LSM880) or a 60× oil immersion objective (NA = 1.40) in an inverted microscope (Nikon Eclipse Ti). Maximal projections of the stacks were obtained with Fiji before quantification of nuclear and cytosolic intensity levels or, for the case of YAP and MAL nuclear to cytosolic ratios, they were calculated after measuring the mean intensity of two adjacent regions of identical size, inside and outside the nucleus as in Elosegui-Artola et al. (2017), using the Hoechst image as a reference for the nuclear position.
Fabrication and coating of soft PDMS gels
Soft elastomeric silicone gels (PDMS) gels with Young modulus E = 12 kPa were used in the traction force microscopy (TFM) experiments described below. Gels were prepared using a protocol based on previous publications (Macminn et al., 2014; Latorre et al., 2018). Briefly, they were synthesized by mixing a 9:10 weight ratio of CY52-276A and CY52-276B polydimethylsiloxane (Dow Corning Toray). After degassing for 30–60 min on ice, the gel was spin-coated on glass-bottom dishes (35 mm, no. 0 coverslip thickness, Mattek) for 90 s at 400 rpm. The samples were then cured at 65°C overnight and maintained in a clean, dust-free, and dry environment. They were always used within 4 wk from fabrication.
The coating with fluorescent beads required for TFM was performed as follows. The soft PDMS was treated with 5% (3-Aminopropyl)triethoxysilane (cat. no. A3648; Sigma-Aldrich), diluted in absolute ethanol for 3 min and rinsed three times with absolute ethanol and three times with water. Samples were incubated for 5 min with a solution of 200-nm-diameter red fluorescent Carboxylate-Modified Microspheres (FluoSpheres, #F8810; Invitrogen) in sodium tetraborate (3.8 mg/ml; Sigma-Aldrich), boric acid (5 mg/ml; Sigma-Aldrich), and 1-ethyl-3-(3- dimethylaminopropyl)carbodiimide (0.1 mg/ml; Sigma-Aldrich). Next, based on the indications of Strale et al. (2016), gels were incubated with 1% Poly-L-lysine (#P2636; Sigma-Aldrich) for 1 h. Gels were washed 3× with 10 mM Hepes buffer, pH = 8.24, (#H3375; Sigma-Aldrich). Finally, the surface of soft PDMS gels was passivated by incubating with 50 mg/ml solution of PEG coupled with succinimidyl valerate (#MPEG-SVA-5000; Laysan Bio) diluted in 10 mM Hepes buffer, pH = 8.24, for 1 h. Gels were washed 3× with water and preserved at 4°C.
Alternatively, when the purpose of the micropatterning was not TFM, glass-bottom dishes without PDMS gels were directly passivated as explained above.
Micropatterning
The micropatterning protocol was an adaptation of Strale et al. (2016). After the passivation, a ring of PDMS with an inner diameter of 7 mm was attached to the glass surface or the PDMS gel to delimitate the micropatterning area, and that area was immediately filled with 50 µl of poly(L-lysine)-grafted-poly(ethylene glycol) (PLL-g-PEG) solution (PLL (20 Kda)-g[3,5]-PEG(2), SuSoS) at a concentration of 0.1 mg ml−1 in PBS, which was incubated for 1 h at room temperature. Then, the solution was carefully removed, the surface was washed three times with PBS, and 30 µl of photoinitiator (4-benzoylbenzyl-trimethylammonium chloride; Alvéole) at a concentration of 14.5 mg ml−1 were added. After that, micropatterning was performed in an inverted microscope (Nikon Ti Eclipse), using the PRIMO system controlled by the Leonardo software (Alvéole) and a 20× objective (CFI S Plan Fluor ELWD ADM, Nikon), directing a UV power of 1,050 mJ mm−2 following the patterns predesigned in ImageJ. When the patterning was completed, filtered PBS (concentration 1×) was used to clean three times and the PDMS ring was removed. The whole glass surface was then treated for 10 min with a solution of fibronectin (F0895; Sigma-Aldrich) 100 µg ml−1 and Alexa Fluor 647 fibrinogen (F35200; Thermo Fisher Scientific) 5 µg ml−1, which caused attachment of the coating proteins at both the patterns and the area where the PDMS ring had previously been placed. After three washes, the glass-bottom dishes were stored, filled with PBS, at 4°C for a maximum of 48 h before the experiments. 2 × 105 cells were seeded diluted in DMEM with 0.2% FBS to avoid cell attachment in unwanted areas. After 1 h, excess of cells was removed by three intensive washes with the same medium. 24 h before imaging or fixation the medium was replaced with 2% FBS-containing DMEM.
TFM
TFM measurements were performed on a Zeiss LSM880 confocal microscope running the software Zeiss ZEN2.3 SP1 FP3 (black, version 14.0.24.201) and using a Plan Apochromat 20 × 0.8 NA objective. Fourier transform traction microscopy was used to measure traction forces, using previously established algorithms (Trepat et al., 2009). All computations and analyses were performed with custom-written MATLAB scripts. The displacement fields of the fluorescence microspheres were obtained using a homemade particle imaging velocimetry algorithm using square interrogation windows of side 40 pixels with an overlap of 0.8. The pixel size was 0.2075 µm.
RT-qPCR
To assess the expression of circadian and YAP-TEAD target genes, RT-qPCR experiments were carried out. Cells were seeded on glass-bottom dishes at high density and cultured during 2 d in DMEM with 10% FBS. Once they were fully confluent, the medium was replaced by DMEM with 2% FBS. 24 h later, the cells were subjected to a shock with 1:1 horse serum and DMEM during 2 h, after which 2% FBS-containing DMEM was added again. Cells were cultured in this medium for 24 h prior to the initiation of the sequential cell lysis, which was done every 4 h. RNA extraction was performed using the Qiagen RNeasy Micro Kit, in accordance with the manufacturer’s guidelines. Nanodrop ND-1000 Spectrophotometer was utilized to measure the concentration of the obtained total mRNA, and equal quantities were utilized for reverse transcription, which was done with the iScript Reverse Transcription Supermix Kit (#1708841; Bio-Rad). SYBR Green RT-qPCR assays (#4385612; Applied Biosystems) were carried out in technical triplicates, and in at least three biological replicates, using a StepOnePlus System (Applied Biosystems) with standard parameters. The 2−ΔΔCt method was used to compute relative gene expression, with normalization of all ΔΔCt values to the housekeeping gene GAPDH and, in the case of the circadian experiments depicted in Fig. 4 F, to the first time point of the control condition. The sequences of the primers used are listed in Table S2.
Statistical analysis
All violin plots were generated using violinplot, an open-source MATLAB script (Bechtold, 2016). Two-sided Wilcoxon rank sum test was used when testing for statistical significance, and P values smaller than 0.05 were considered as significant, unless specified otherwise.
The results of the RT-qPCR were analyzed using the single cosinor approach presented in Cornelissen (2014). The analysis was performed on the mean values of the repetitions of each time point, using an open-source MATLAB script (cheart 2023, Cosinor Analysis, https://www.mathworks.com/matlabcentral/fileexchange/20329-cosinor-analysis, MATLAB Central File Exchange. Retrieved February 20, 2023). P values are relative to an F test taking as null hypothesis a zero-amplitude signal. The same data were also analyzed using JKT-Cycle, through the online platform BioDare (https://biodare2.ed.ac.uk; Hughes et al., 2010; Zielinski et al., 2014), using the BD2 eJTK implementation.
Online supplemental material
Fig. S1 shows calculation of Circadian Power fraction from single-cell RevVNP signals. Fig. S2 shows REV-ERBα levels decrease with cell density in a cell division–independent manner. Fig. S3 shows Rev-erbα circadian power fraction does not correlate with nuclear MAL. Fig. S4 shows overexpression of a constitutively active TAZ mutant affects REV-ERBα expression. Fig. S5 shows TEAD-associated signaling is activated under 5SA-YAP and 5SA-CCDEL-YAP but not 5SA-S94A-YAP overexpression. Table S1 shows exact P values of the statistical tests performed for the data shown in Figs. 1, 2, 3, 4, and S3. Table S2 shows primers used for quantitative real-time PCR. Video 1 shows the origin of the Rev-erbα circadian impairment at low density is mechanical rather than paracrine. Video 2 shows cell confinement in microprinted fibronectin areas rescues the Rev-erbα circadian impairment observed at low density.
Data availability
The data that support the findings of this study and the MATLAB analysis procedures are available from the corresponding authors on reasonable request.
Acknowledgments
We thank S. Aznar-Benitah, G. Solanas, F. Peixoto, M. Uroz, A. Elósegui-Artola, C. Pérez-González, D. Zalvidea, S. Conti, J. Oliver-De la Cruz, I.C. Fortunato, B. Bano-Otalora, M. Belle, and all the members of the Roca-Cusachs and Trepat laboratories for their discussions and technical support. We thank A. Menéndez, S. Usieto, and N. Castro for daily technical assistance. We also thank N. Montserrat (Institute for Bioengineering of Catalonia, Barcelona, Spain), E. Martínez (Institute for Bioengineering of Catalonia, Barcelona, Spain), U. Schibler (University of Geneva, Geneva, Switzerland), H.W. Park (Yonsei University, Seoul, Korea), and Y.S. Choi (University of Western Australia, Perth, Australia) for sharing some of the cell lines and plasmids used in this work.
This paper was funded by The Generalitat de Catalunya (AGAUR SGR-2017-01602 to X. Trepat, AGAUR Beatriu de Pinós 2014 BP-B 00105 to J.F. Abenza, the CERCA Programme, and “ICREA Academia” awards to P. Roca-Cusachs and J. García-Ojalvo); Spanish Ministry for Science and Innovation MICINN/FEDER (PGC2018-099645-B-I00 to X. Trepat, BFU2016-79916-P and PID2019-110298GB-I00 to P. Roca-Cusachs, and PID2021-127311NB-I00 to J. García-Ojalvo); European Research Council (Adv-883739 to X. Trepat); Fundació la Marató de TV3 (project 201903-30-31-32 to X. Trepat); European Commission (H2020-FETPROACT-01-2016-731957 to P. Roca-Cusachs and X. Trepat, H2020 Marie Sklodowska-Curie Actions MECHADIAN - IF/750557 to J.F. Abenza); La Caixa Foundation (LCF/PR/HR20/52400004 to P. Roca-Cusachs and X. Trepat); Institute for Bioengineering of Catalonia is recipient of a Severo Ochoa Award of Excellence from the Ministry of Economy and Competitiveness. DCEXS-UPF is recipient of a Maria de Maeztu Award of Excellence from the Ministry of Economy and Competitiveness.
Author contributions: J.F. Abenza and X. Trepat conceived the project. J.F. Abenza, M. Mouelhi, L. Rossetti, and I. Andreu performed experiments and analyzed data. L. Rossetti and J. Burgués developed software and analyzed data. K. Kennedy, P. Roca-Cusachs, S. Marco, and J. García-Ojalvo contributed technical expertise, materials, and discussion. J.F. Abenza, L. Rossetti, and X. Trepat wrote the manuscript. J.F. Abenza and X. Trepat supervised the project. All authors revised the completed manuscript.
References
Author notes
J.F. Abenza and L. Rossetti contributed equally to this paper.
Disclosures: K. Kennedy reported grants from Agencia de Gestión de Ayudas Universitarias y de Investigación outside the submitted work. J. García-Ojalvo reported grants from Ministerio de Ciencia e Innovación, Spain, and grants from Institució Catalana de Recerca i Estudis Avançats outside the submitted work. No other disclosures were reported.