In macroautophagy, cellular components are sequestered within autophagosomes and transported to lysosomes/vacuoles for degradation. Although phosphatidylinositol 3-kinase complex I (PI3KCI) plays a pivotal role in the regulation of autophagosome biogenesis, little is known about how this complex localizes to the pre-autophagosomal structure (PAS). In Saccharomyces cerevisiae, PI3KCI is composed of PI3K Vps34 and conserved subunits Vps15, Vps30, Atg14, and Atg38. In this study, we discover that PI3KCI interacts with the vacuolar membrane anchor Vac8, the PAS scaffold Atg1 complex, and the pre-autophagosomal vesicle component Atg9 via the Atg14 C-terminal region, the Atg38 C-terminal region, and the Vps30 BARA domain, respectively. While the Atg14–Vac8 interaction is constitutive, the Atg38–Atg1 complex interaction and the Vps30–Atg9 interaction are enhanced upon macroautophagy induction depending on Atg1 kinase activity. These interactions cooperate to target PI3KCI to the PAS. These findings provide a molecular basis for PAS targeting of PI3KCI during autophagosome biogenesis.
Introduction
Macroautophagy (hereafter autophagy) is a major intracellular degradation system found in most eukaryotic cells (Nakatogawa et al., 2009; Ohsumi, 2014; Yang and Klionsky, 2010). When autophagy is induced, a membrane cisterna known as the isolation membrane or phagophore forms in the cytoplasm, and it expands, bends, and closes to form a double-membrane bound vesicle called an autophagosome. During this process, various cellular components are selectively or non-selectively sequestered within the autophagosome. The autophagosome then fuses with the lysosome or vacuole for the degradation of the contents. Autophagy-related (Atg) proteins play different roles in autophagy, e.g., the selection of degradation targets, autophagosome biogenesis, and autophagosome–lysosome/vacuole fusion (Nakatogawa, 2020). The Atg proteins responsible for autophagosome biogenesis are known as core Atg proteins and are conserved from yeasts to plants and mammals. These proteins constitute six functional units: (i) the Atg1 protein kinase complex, (ii) the Atg9 vesicle, (iii) phosphatidylinositol 3-kinase complex I (PI3KCI), (iv) the Atg16 complex, (v) the Atg8 lipidation system, and (vi) the Atg2–Atg18 complex (designations as in Saccharomyces cerevisiae). These core units localize to the site for autophagosome formation, where they organize the pre-autophagosome structure (PAS) prior to the expansion of the isolation membrane (Suzuki et al., 2001; Nakatogawa, 2020). Previous studies have revealed the mechanisms by which core Atg units are localized to the PAS during autophagy following the inactivation of Tor kinase complex 1 (TORC1) by amino acid deprivation or treatment with the specific inhibitor rapamycin. First, upon TORC1 inactivation, Atg1, Atg13, and the Atg17–Atg31–Atg29 complex are all assembled into the Atg1 complex, multiple copies of which are then phase-separated to form a liquid droplet-like structure, which serves as a scaffold for the recruitment of downstream factors (Kamada et al., 2000; Fujioka et al., 2014; Yamamoto et al., 2016; Fujioka et al., 2020). Next, Atg9 vesicles are recruited via the interaction between Atg9 and the HORMA domain of Atg13 (Suzuki et al., 2015; Jao et al., 2013). PI3KCI then localizes to the PAS in an Atg9-dependent manner (Suzuki et al., 2007), although the molecular basis of PAS recruitment remains poorly understood. Subsequently, the Atg16 complex (composed of Atg16, the ubiquitin-like protein conjugate Atg12-Atg5, and Atg21) and the Atg2–Atg18 complex localize to the PAS by binding to phosphatidylinositol 3-phosphate (PI3P), which is produced by PI3KCI, via the PI3P-binding proteins Atg21 and Atg18, respectively (Juris et al., 2015; Obara et al., 2008). In addition, interaction between Atg2 and Atg9, which is regulated by Atg1-mediated phosphorylation of Atg9, is also involved in the PAS recruitment of the Atg2–Atg18 complex (Papinski et al., 2014; Gómez-Sánchez et al., 2018), and interaction between Atg12 and Atg17 also acts to recruit the Atg16 complex to the PAS (Harada et al., 2019). Finally, Atg8 is conjugated to phosphatidylethanolamine (PE) in pre-autophagosomal membranes at the PAS via ubiquitin-like conjugation reactions, during which the Atg16 complex serves as an E3 enzyme (Ichimura et al., 2000; Hanada et al., 2007; Suzuki et al., 2007).
PI3K Vps34 forms two distinct complexes, PI3KCI and PI3KCII, which share Vps34/VPS34, Vps15/p150, and Vps30/BECN1, and contain specific factors Atg14/ATG14L and Atg38/NRBF2 for PI3KCI and Vps38/UVRAG for PI3KCII (designations as in yeasts and humans; Kihara et al., 2001; Itakura et al., 2008; Zhong et al., 2009; Araki et al., 2013; Lu et al., 2014). In S. cerevisiae, Atg14 and Vps38 target PI3KCI and PI3KCII to the PAS and endosomes, where they specifically function in autophagy and vacuolar protein sorting, respectively (Kihara et al., 2001; Obara et al., 2006). In addition, although Vps30 is a common component of both PI3K complexes, it contains a β-α repeated, autophagy-specific (BARA) domain in its C-terminal region, the deletion of which decreases the localization of PI3KCI to the PAS (Noda et al., 2012). Thus, Atg14 and the BARA domain of Vps30 are thought to be involved in the PAS targeting of PI3KCI, although the underlying mechanism remains unknown. Meanwhile, the second PI3KCI-specific component Atg38 was previously shown to stabilize PI3KCI by bridging Vps34 and Atg14 (Araki et al., 2013); however, whether Atg38 also participates in the PAS localization of PI3KCI remains unknown.
The vacuolar membrane protein Vac8 is involved in a number of events related to the vacuole, including vacuole inheritance, homotypic vacuole fusion, and the organization of nucleus-vacuole junctions (Wang et al., 1998; Veit et al., 2001; Pan et al., 2000). In addition, recent studies suggest that Vac8 also plays an important role in autophagosome formation, anchoring the Atg1 complex to the vacuolar membrane by binding to the C-terminal region of Atg13, thereby ensuring PAS organization on the vacuolar membrane (Scott et al., 2000; Hollenstein et al., 2019; Gatica et al., 2021). Moreover, more recent studies suggest that Vac8 interacts with Vps34 or the Vps34–Vps15 subcomplex to tether PI3KCI to the vacuolar membrane, facilitating its subsequent targeting to the PAS (Lei et al., 2021; Hollenstein et al., 2021). However, this interaction was shown to be independent of Atg14 and therefore does not explain how Vac8 mediates the PAS localization of PI3KCI but not PI3KCII.
In this study, we discovered three novel elements that mediate PAS targeting of PI3KCI. The C-terminal region of Atg14 is involved in PI3KCI binding to Vac8 and is important for vacuolar and PAS targeting of the complex. Meanwhile, PI3KCI also associates with the Atg1 complex and Atg9 via the C-terminal region of Atg38 and the BARA domain of Vps30, respectively, both of which are regulated by Atg1 kinase activity, and cooperate to target PI3KCI to the PAS. Thus, this study elucidated the molecular basis of PAS localization of PI3KCI.
Results
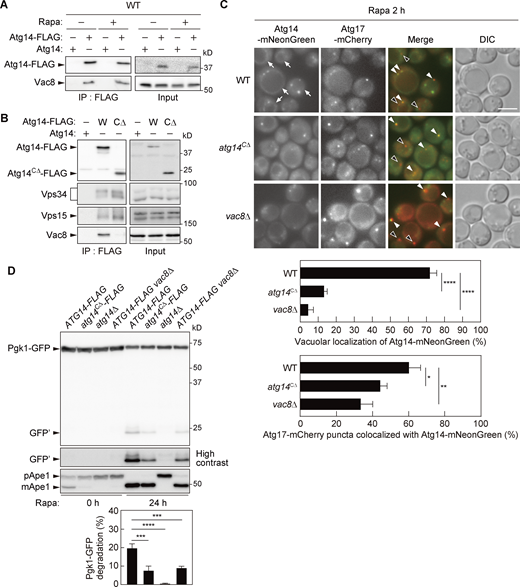
PI3KCI associates with Vac8 via the C-terminal region of Atg14 to localize to the vacuolar membrane and PAS
To determine the mechanism underlying PAS targeting of PI3KCI, we focused on proteins that interact with PI3KCI. Yeast cells expressing the PI3KCI-specific component Atg14 tagged with the 3×FLAG sequence (Atg14-FLAG) were treated with rapamycin to induce autophagosome formation, solubilized with the detergent n-dodecyl-β-D-maltoside, and subjected to immunoprecipitation using the anti-FLAG antibody. Mass spectrometry analysis of the immunoprecipitates identified the vacuolar membrane protein Vac8 (Table S1), consistent with recent reports suggesting that PI3KCI associates with Vac8 (Hollenstein et al., 2021; Lei et al., 2021). PI3KCI–Vac8 interaction was confirmed by immunoblotting of Atg14-FLAG immunoprecipitates using anti-Vac8 antibodies, which also showed that the interaction did not increase following rapamycin treatment (Fig. 1 A). While the N-terminal half of Atg14 is known to contain three coiled-coil regions that play an important role in the interactions with Vps34 and Vps30, the function of the C-terminal region (CTR) remains unknown (Obara et al., 2006). In this study, we constructed a CTR (amino acid residues 157–344)-deleted version of Atg14-FLAG (Atg14CΔ-FLAG) and confirmed that deletion of the Atg14 CTR does not affect the integrity of PI3KCI (coimmunoprecipitation of Vps34 and Vps15; Fig. 1 B; Obara et al., 2006). However, Atg14CΔ-FLAG failed to coimmunoprecipitate Vac8, suggesting that the CTR of Atg14 is important for the association between PI3KCI and Vac8. It was recently revealed that Vac8 plays an important role in targeting PI3KCI to the vacuolar membrane and PAS (Hollenstein et al., 2021; Lei et al., 2021). In this study, fluorescence microscopy revealed that, as with VAC8 knockout (vac8Δ), deletion of the Atg14 CTR abolished vacuolar localization of Atg14-mNeonGreen and decreased the colocalization of Atg14-mNeonGreen with puncta of the PAS marker Atg17-mCherry in cells treated with rapamycin (Fig. 1 C). The vacuolar localization of Atg14-mNeonGreen was also abolished by CTR deletion in cells without rapamycin treatment (Fig. S1). The ability of cells to form autophagosomes can be determined by measuring the nonselective degradation of the cytoplasmic protein Pgk1 in the cells (Welter et al., 2010). When GFP-fused Pgk1 is transported to the vacuole via autophagosomes, the fusion protein is degraded and GFP fragments relatively resistant to vacuolar proteases accumulate within the vacuole. The amount of GFP fragments that accumulated in atg14CΔ cells was significantly lower than that in wild-type cells and comparable to that in vac8Δ cells (Fig. 1 D). These results suggest that PI3KCI associates with Vac8 depending on the Atg14 CTR to localize to the vacuolar membrane and PAS for efficient autophagosome formation.
Atg14 interacts with Vac8 to target PI3KCI to the vacuolar membrane and the PAS. (A and B) Cells expressing Atg14-FLAG or Atg14CΔ-FLAG were treated with or without rapamycin (Rapa) for 2 h and the cell lysates (Input) were subjected to immunoprecipitation using anti-FLAG antibody. The immunoprecipitates (IP) were analyzed by immunoblotting using antibodies against FLAG, Vac8, Vps34, and Vps15. (C) Cells expressing Atg17-mCherry and Atg14-mNeonGreen or Atg14CΔ-mNeonGreen were treated with rapamycin for 2 h and analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. White arrowheads, Atg14-mNeonGreen-positive Atg17-mCherry puncta. Black arrowheads, Atg14-mNeonGreen-negative Atg17-mCherry puncta. Arrows, vacuolar localization of Atg14-mNeonGreen. Scale bar, 5 μm. The percentage of cells showing vacuolar localization of Atg14-mNeonGreen and the percentage of Atg17-mCherry puncta positive for Atg14-mNeonGreen are shown. Bars represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test). (D) Cells expressing Pgk1-GFP were treated with rapamycin and analyzed by immunoblotting using antibodies against GFP or Ape1. The band intensities of Pgk1-GFP and GFP fragments (GFP’) were measured, and the proportion (%) of GFP fragments to the sum of Pgk1-GFP and GFP fragments (Pgk1-GFP degradation) were determined. Bars represent means ± SD (n = 3). ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test). Source data are available for this figure: SourceData F1.
Atg14 interacts with Vac8 to target PI3KCI to the vacuolar membrane and the PAS. (A and B) Cells expressing Atg14-FLAG or Atg14CΔ-FLAG were treated with or without rapamycin (Rapa) for 2 h and the cell lysates (Input) were subjected to immunoprecipitation using anti-FLAG antibody. The immunoprecipitates (IP) were analyzed by immunoblotting using antibodies against FLAG, Vac8, Vps34, and Vps15. (C) Cells expressing Atg17-mCherry and Atg14-mNeonGreen or Atg14CΔ-mNeonGreen were treated with rapamycin for 2 h and analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. White arrowheads, Atg14-mNeonGreen-positive Atg17-mCherry puncta. Black arrowheads, Atg14-mNeonGreen-negative Atg17-mCherry puncta. Arrows, vacuolar localization of Atg14-mNeonGreen. Scale bar, 5 μm. The percentage of cells showing vacuolar localization of Atg14-mNeonGreen and the percentage of Atg17-mCherry puncta positive for Atg14-mNeonGreen are shown. Bars represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test). (D) Cells expressing Pgk1-GFP were treated with rapamycin and analyzed by immunoblotting using antibodies against GFP or Ape1. The band intensities of Pgk1-GFP and GFP fragments (GFP’) were measured, and the proportion (%) of GFP fragments to the sum of Pgk1-GFP and GFP fragments (Pgk1-GFP degradation) were determined. Bars represent means ± SD (n = 3). ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test). Source data are available for this figure: SourceData F1.
The localization of PI3KCI in cells without rapamycin treatment. Cells expressing Atg14-mNeonGreen or Atg14CΔ-mNeonGreen were analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. Scale bar, 5 μm. The percentage of cells showing vacuolar localization of Atg14-mNeonGreen is shown. Bars represent means ± SD (n = 3). ****, P < 0.0001 (Tukey’s multiple comparisons test).
The localization of PI3KCI in cells without rapamycin treatment. Cells expressing Atg14-mNeonGreen or Atg14CΔ-mNeonGreen were analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. Scale bar, 5 μm. The percentage of cells showing vacuolar localization of Atg14-mNeonGreen is shown. Bars represent means ± SD (n = 3). ****, P < 0.0001 (Tukey’s multiple comparisons test).
PI3KCI associates with multiple core Atg proteins
Immunoblotting analysis of Atg14-FLAG immunoprecipitates with antibodies against different Atg proteins showed that Atg14-FLAG also coprecipitated the core Atg proteins Atg1, Atg17, Atg9, and Atg12-Atg5 in addition to the PI3KCI components Vps34 and Vps15, but not Atg2 or Atg8 (Fig. 2 A and Fig. 3 A). Cell treatment with rapamycin increased coprecipitation of these core Atg proteins (Fig. 2 A). These results suggest that PI3KCI associates with multiple core Atg proteins under autophagosome formation-inducing conditions. When 3×FLAG-tagged Vps30, a common subunit of PI3KCI and PI3KCII, was immunoprecipitated, core Atg proteins (Atg1, Atg9, and Atg12-Atg5) and Vac8 were coprecipitated as with immunoprecipitation of Atg14-FLAG (Fig. 2 B). Coprecipitation of these proteins was abolished by the knockout of ATG14 but not by that of VPS38, which encodes the PI3KCII-specific subunit Vps38. These results suggest that these core Atg proteins and Vac8 specifically interact with PI3KCI.
PI3KCI associates with multiple core Atg proteins. (A–D) Cells expressing Atg14-FLAG (A, C, and D) or Vps30-FLAG (B) were treated with rapamycin for 2 h and subjected to immunoprecipitation using anti-FLAG antibodies, followed by immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg17, Atg9, Atg12, Atg8, Atg2, and Vac8. Source data are available for this figure: SourceData F2.
PI3KCI associates with multiple core Atg proteins. (A–D) Cells expressing Atg14-FLAG (A, C, and D) or Vps30-FLAG (B) were treated with rapamycin for 2 h and subjected to immunoprecipitation using anti-FLAG antibodies, followed by immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg17, Atg9, Atg12, Atg8, Atg2, and Vac8. Source data are available for this figure: SourceData F2.
PI3KCI associates with the Atg1 complex and Atg9 via the Atg38 C-terminal region and Vps30 BARA domain, respectively. (A, C, E, and F) Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, HA, Atg1, Atg13, Atg17, Atg9, Atg12, and Vac8. (B and D) Cells were treated with rapamycin for 2 h, and levels of Vps30BARAΔ-HA (B) and Atg38CΔ-HA (D) were examined by immunoblotting using anti-Vps30 and anti-HA antibodies, respectively. (G) PI3KCI was isolated from cells treated with rapamycin for 2 h by immunoprecipitation of Atg14-FLAG and incubated with lambda protein phosphatase with or without heat inactivation. Atg14-FLAG and Vps34 were examined by immunoblotting. Source data are available for this figure: SourceData F3.
PI3KCI associates with the Atg1 complex and Atg9 via the Atg38 C-terminal region and Vps30 BARA domain, respectively. (A, C, E, and F) Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, HA, Atg1, Atg13, Atg17, Atg9, Atg12, and Vac8. (B and D) Cells were treated with rapamycin for 2 h, and levels of Vps30BARAΔ-HA (B) and Atg38CΔ-HA (D) were examined by immunoblotting using anti-Vps30 and anti-HA antibodies, respectively. (G) PI3KCI was isolated from cells treated with rapamycin for 2 h by immunoprecipitation of Atg14-FLAG and incubated with lambda protein phosphatase with or without heat inactivation. Atg14-FLAG and Vps34 were examined by immunoblotting. Source data are available for this figure: SourceData F3.
Next, we performed coimmunoprecipitation analysis using cells lacking different core Atg proteins. The absence of Atg8 (the Atg8 lipidation system) or Atg2 (the Atg2–Atg18 complex), which was not detected in Atg14-FLAG immunoprecipitates, did not reduce coimmunoprecipitation of Atg1, Atg9, Atg12-Atg5, and Vac8 with Atg14-FLAG (coimmunoprecipitation of these Atg proteins rather increased in atg8Δ and atg2Δ cells; Fig. 2 C). Similarly, these proteins were coimmunoprecipitated with Atg14-FLAG in atg5Δ and atg9Δ cells as efficiently as they were in wild-type cells (Fig. 2, C and D). In contrast, coimmunoprecipitation of Atg9 and Atg12-Atg5 was severely impaired in cells lacking Atg1 complex components (Atg1, Atg13, or Atg17; Fig. 2 D and Fig. 3 F). These results suggest that PI3KCI associates with Atg9 and Atg12-Atg5 (the Atg16 complex) in a manner independent of each other and dependent on the Atg1 complex. Meanwhile, Atg14-FLAG failed to coimmunoprecipitate Atg1 in atg13Δ and atg17Δ cells (Fig. 2 D and Fig. 3 F). Similarly, Atg17 was not coprecipitated with Atg14-FLAG in atg1Δ cells. These findings suggest that PI3KCI associates with the Atg1 complex in a manner dependent on Atg1 complex formation and independent of other core Atg units. These experiments also revealed that the interaction between PI3KCI and Vac8 does not require any core Atg units (Fig. 2, C and D). It was also shown that Vac8 is not required for PI3KCI association with core Atg proteins (Fig. S2). It was previously revealed that Atg11 is required for most selective autophagy pathways, but not for non-selective autophagy induced by rapamycin or nitrogen starvation (Zientara-Rytter and Subramani, 2019). We showed that Atg11 is dispensable for PI3KCI association with core Atg proteins in cells treated with rapamycin (Fig. 2 D).
The interaction between PI3KCI and core Atg units does not require Vac8. Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg12, Atg9, and Vac8. Source data are available for this figure: SourceData FS2.
The interaction between PI3KCI and core Atg units does not require Vac8. Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg12, Atg9, and Vac8. Source data are available for this figure: SourceData FS2.
PI3KCI associates with the Atg1 complex and Atg9 via the Atg38 C-terminal region and Vps30 BARA domain, respectively
We further analyzed the interaction between PI3KCI and the Atg1 complex. Since PI3KCI, but not PI3KCII, interacted with the Atg1 complex, PI3KCI-specific factors were thought to be involved in this interaction. In line with this, Atg14 was found to be important for PI3KCI association with the Atg1 complex (Fig. 2 B). However, the absence of Atg14 results in the release of another PI3KCI-specific factor, Atg38, from the complex (Araki et al., 2013). We found that Atg1 and Atg17 were not coimmunoprecipitated with Atg14-FLAG in atg38Δ cells, although Atg14-FLAG coprecipitated Vps34 and Vps15 comparably with the wild-type cells (Fig. 3 A). These results suggest that Atg38 is involved in PI3KCI interaction with the Atg1 complex.
It was previously revealed that the Atg1 complex directly interacts with Atg9 via the HORMA domain of Atg13 (Suzuki et al., 2015). Therefore, we hypothesized that PI3KCI indirectly associates with Atg9 via the Atg1 complex. However, we noticed that Atg9 remained associated with PI3KCI in atg38Δ cells even though PI3KCI association with the Atg1 complex was lost (Fig. 3 A). This result suggested that PI3KCI interacts with Atg9 without mediation by the Atg1 complex. While Vps30 is contained in both PI3KCI and PI3KCII and is essential for the functions of both complexes, its C-terminal region folds into the BARA domain, which is thought to be specifically involved in the PAS localization of PI3KCI via an unknown mechanism (Noda et al., 2012). We, therefore, hypothesized that the BARA domain of Vps30 mediates the PI3KCI–Atg9 interaction, thereby contributing to PAS targeting of PI3KCI. The level of 6×HA-tagged Vps30 lacking the BARA domain (Vps30BARAΔ-HA) was comparable with that of the wild-type protein when expressed using the ADH promoter (ADHp; Fig. 3 B). Thus, we examined the PI3KCI–Atg9 interaction in atg38Δ ADHp-vps30BARAΔ-ΗΑ cells. Unfortunately, combining these mutations severely destabilized PI3KCI, and Atg14-FLAG coprecipitated Vps30BARAΔ-HA but not Vps34 and Vps15 in the mutant cells (Fig. 3 C, lane 3). This result is consistent with a previous report in which Atg38 was found to play an important role in PI3KCI stabilization by bridging Atg14 and Vps34 (Araki et al., 2013). Atg38 forms a homodimer via the CTR and binds to Atg14 and Vps34 via the N-terminal region (Araki et al., 2013). To obtain an Atg38 mutant defective in PI3KCI association with the Atg1 complex but retaining PI3KCI integrity even in vps30BARAΔ cells, we replaced the CTR of Atg38 (residues 121–226) with a dimerizing coiled-coil region of Gcn4 (Harbury et al., 1993) and engineered the chromosomal ATG38 gene to express this mutant (Atg38CΔ-Gcn4CC-HA) under the ADH promoter (Fig. 3 D). As expected, all of the PI3KCI components (Vps34, Vps15, Vps30BARAΔ-HA, and Atg38CΔ-Gcn4CC-HA) were coimmunoprecipitated with Atg14-FLAG in atg38CΔ-GCN4CCvps30BARAΔ cells (Fig. 3 C, lane 4), suggesting that the Atg38CΔ–Gcn4CC mutant was able to stabilize PI3KCI in the absence of the Vps30 BARA domain. Meanwhile, Atg9 was not coprecipitated with Atg14-FLAG in these cells, suggesting that PI3KCI associates with Atg9 in a manner dependent on the BARA domain of Vps30.
We further investigated details of the interaction between PI3KCI and the Atg1 complex using AlphaFold2 (Richard et al., 2022 Preprint), which predicted that CTRs in an Atg38 dimer interact with the C-terminal short α-helix of Atg29 (Fig. S3 A). When these regions of Atg38 (residues 210–224) or Atg29 (residues 198–213) were deleted, coimmunoprecipitation of Atg1 complex components with Atg14-FLAG decreased in the mutant cells (Fig. S3, B and C). These results suggest that PI3KCI association with the Atg1 complex is mediated by the interaction between the Atg38 CTR and the Atg29 CTR.
PI3KCI association with the Atg1 complex is mediated by the interaction between Atg38 and Atg29. (A) The structure of the Atg38–Atg29–Atg31 complex predicted by AlphaFold2 shows that residues 210–224 in Atg38 and residues 197–213 in Atg29 are involved in the interaction between these proteins. (B and C) Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg38, Atg1, Atg17, Atg9, and HA. Source data are available for this figure: SourceData FS3.
PI3KCI association with the Atg1 complex is mediated by the interaction between Atg38 and Atg29. (A) The structure of the Atg38–Atg29–Atg31 complex predicted by AlphaFold2 shows that residues 210–224 in Atg38 and residues 197–213 in Atg29 are involved in the interaction between these proteins. (B and C) Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg38, Atg1, Atg17, Atg9, and HA. Source data are available for this figure: SourceData FS3.
Regulation of PI3KCI association with the Atg1 complex and Atg9
The association of PI3KCI with the Atg1 complex was remarkably enhanced by rapamycin (Fig. 2 A). In cells expressing a kinase-defective mutant of Atg1 (atg1D211A; Matsuura et al., 1997), this intercomplex association was almost completely lost, suggesting that PI3KCI association with the Atg1 complex is strictly regulated by Atg1-mediated phosphorylation of a related factor(s) (Fig. 3 E). The interaction of PI3KCI with Atg9 also increased following rapamycin treatment (Fig. 2 A). Although PI3KCI interacts with Atg9 without mediation by the Atg1 complex, the interaction was found to be dependent on Atg1 complex components and Atg1 kinase activity (Fig. 2 D; and Fig. 3, E and F), suggesting that the PI3KCI–Atg9 interaction is also regulated by the Atg1 complex. A previous study revealed that Atg9 binds to the HORMA domain of Atg13, a subunit of the Atg1 complex, and is thereby recruited to the PAS (Suzuki et al., 2015). We found that coimmunoprecipitation of Atg9 with Atg14-FLAG decreased in atg13R213D mutant cells defective in Atg9 binding to Atg13 (Fig. 3 F), suggesting that PAS localization of Atg9 is a prerequisite for the interaction of PI3KCI with Atg9. In contrast, the association between PI3KCI and the Atg1 complex (coimmunoprecipitation of Atg1 and Atg13 with Atg14-FLAG) was unaltered in atg13R213D cells, consistent with the result showing that the association between these complexes does not involve Atg9 (Fig. 2 D).
In cells treated with rapamycin, Vps34 bands that slowly migrated during SDS-PAGE were preferentially coimmunoprecipitated with Atg14-FLAG (Fig. 2 A). These Vps34 bands were downshifted by treatment of Atg14-FLAG immunoprecipitates with lambda protein phosphatase, suggesting that these bands represent the phosphorylated forms of Vps34 (Fig. 3 G). Phosphorylated Vps34 was not found in Atg14-FLAG immunoprecipitates in cells defective in Atg1 kinase activity, such as atg1Δ, atg17Δ, and atg1D211A cells (Fig. 2 D and Fig. 3 E). In addition, the deletion of ATG14 and VPS38 specifically abolished phosphorylated and non-phosphorylated Vps34 in the Vps30-FLAG immunoprecipitates, respectively (Fig. 2 B). These results suggest that the phosphorylation of Vps34 in PI3KCI is dependent on the Atg1 kinase complex. Vps34 phosphorylation also decreased in atg38Δ and atg38CΔ-GCN4CC cells defective in PI3KCI association with the Atg1 complex (Fig. 3, A and C). Moreover, Vps34 phosphorylation in PI3KCI was defective following the deletion of ATG9 (Fig. 2 D), which impaired the localization of PI3KCI to the PAS (Suzuki et al., 2007). These results suggest that PAS localization of PI3KCI and PI3KCI association with the Atg1 complex are required for the phosphorylation of Vps34. This phosphorylation of Vps34 is thought to regulate the association between PI3KCI and other factors such as the Atg1 complex and Atg9 (see Discussion).
Interactions between the Atg1 complex and Atg9 cooperate to target PI3KCI to the PAS
While localization of the Atg1 complex and Atg9 vesicle to the PAS was found to be independent of PI3KCI, PAS localization of PI3KCI requires both (Suzuki et al., 2001; Kawamata et al., 2008; Cheong et al., 2008; Fig. 4). We, therefore, examined whether the associations of PI3KCI with the Atg1 complex and Atg9 are important for the PAS localization of PI3KCI. In atg38CΔ-GCN4CC cells, which were defective in PI3KCI association with the Atg1 complex, PAS localization of PI3KCI (colocalization of Atg14-mNeonGreen with Atg17-mCherry puncta) was also defective, as in atg1Δ cells. Deletion of the Vps30 BARA domain (vps30BARAΔ), which impaired PI3KCI-Atg9 interaction, also reduced PAS localization of PI3KCI to a level similar to that in atg9Δ cells, consistent with a previous study (Noda et al., 2012). Combining these mutations (atg38CΔ-GCN4CCvps30BARAΔ) caused more severe defects in PI3KCI localization to the PAS (Fig. 4). These results suggest that PI3KCI associations with the Atg1 complex and Atg9 cooperate during PAS targeting of PI3KCI.
Association with the Atg1 complex and Atg9 is required for PAS targeting of PI3KCI. Cells expressing Atg14-mNeonGreen and Atg17-mCherry were treated with rapamycin for 2 h and analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. White arrowheads, Atg14-mNeonGreen-positive Atg17-mCherry puncta. Black arrowheads, Atg14-mNeonGreen-negative Atg17-mCherry puncta. Scale bar, 5 μm. The percentage of Atg17-mCherry puncta positive for Atg14-mNeonGreen is shown. Bars represent means ± SD (n = 3). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test).
Association with the Atg1 complex and Atg9 is required for PAS targeting of PI3KCI. Cells expressing Atg14-mNeonGreen and Atg17-mCherry were treated with rapamycin for 2 h and analyzed by fluorescence microscopy. DIC, differential interference contrast microscopy. White arrowheads, Atg14-mNeonGreen-positive Atg17-mCherry puncta. Black arrowheads, Atg14-mNeonGreen-negative Atg17-mCherry puncta. Scale bar, 5 μm. The percentage of Atg17-mCherry puncta positive for Atg14-mNeonGreen is shown. Bars represent means ± SD (n = 3). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Tukey’s multiple comparisons test).
Finally, we examined autophagic activity in cells defective in PAS localization of PI3KCI (Fig. 5). Pgk1-GFP degradation assay showed that atg38CΔ-GCN4CC and vps30BARAΔ single mutant cells were both significantly defective in autophagy, while in atg38CΔ-GCN4CCvps30BARAΔ double mutant cells, the defect was as severe as in atg14Δ cells. These results suggest that PAS targeting of PI3KCI mediated by its association with the Atg1 complex and Atg9 is crucial for autophagosome formation.
PI3KCI association with the Atg1 complex and Atg9 is important for autophagosome formation. Cells expressing Pgk1-GFP were treated with rapamycin and analyzed by immunoblotting using anti-GFP antibodies. Pgk1-GFP degradation is shown as described in Fig. 1 D. Bars represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01 (Tukey’s multiple comparisons test). Source data are available for this figure: SourceData F5.
PI3KCI association with the Atg1 complex and Atg9 is important for autophagosome formation. Cells expressing Pgk1-GFP were treated with rapamycin and analyzed by immunoblotting using anti-GFP antibodies. Pgk1-GFP degradation is shown as described in Fig. 1 D. Bars represent means ± SD (n = 3). *, P < 0.05; **, P < 0.01 (Tukey’s multiple comparisons test). Source data are available for this figure: SourceData F5.
Discussion
Previous studies have elucidated the molecular basis of PAS localization of all core Atg units except PI3KCI. The present study revealed how PI3KCI localizes to the PAS, thus providing a full understanding of the fundamental molecular interactions underlying PAS organization. Our results showed cooperation between three distinct interactions to target PI3KCI to the PAS (Fig. 6).
A model for the PAS targeting of PI3KCI. PI3KCI localizes to the vacuolar membrane via interaction between the CTR of Atg14 and Vac8 independent of autophagy induction. When the Atg1 complex is assembled and activated upon TORC1 inactivation, PI3KCI then associates with the Atg1 complex and Atg9 via the CTR of Atg38 and the BARA domain of Vps30, respectively, dependent on Atg1 kinase activity, thereby stably accumulating at the PAS.
A model for the PAS targeting of PI3KCI. PI3KCI localizes to the vacuolar membrane via interaction between the CTR of Atg14 and Vac8 independent of autophagy induction. When the Atg1 complex is assembled and activated upon TORC1 inactivation, PI3KCI then associates with the Atg1 complex and Atg9 via the CTR of Atg38 and the BARA domain of Vps30, respectively, dependent on Atg1 kinase activity, thereby stably accumulating at the PAS.
First, the CTR of Atg14 was found to play an important role in the interaction between PI3KCI and Vac8, which is vital for targeting PI3KCI to the vacuolar membrane, and probably thereby, subsequent PAS localization (Figs. 1 and 6). This conclusion may appear contradictory to previous results suggesting an interaction between Vac8 and Vps34 or the Vps34–Vps15 subcomplex in the absence of Atg14 (Lei et al., 2021; Hollenstein et al., 2021). However, these studies examined the interactions in cells overexpressing these proteins or cells containing artificial Vac8 clusters (Lei et al., 2021; Hollenstein et al., 2021), in which Atg14 might be dispensable. Meanwhile, in our study, a small amount of Vac8 was coimmunoprecipitated even when Atg14 lacked the CTR (Fig. 1 B), consistent with the notion that PI3KCI interacts with Vac8, independent of this Atg14 region. We also confirmed that VPS34 disruption decreased the coimmunoprecipitation of Vac8 with Atg14-FLAG as well as that of PI3KCI components and other core Atg proteins (Fig. S4). In addition, Atg14-FLAG failed to coprecipitate Vac8 without association with Vps34 and Vps15 (Fig. 3 C, lane 3). Thus, the association between PI3KCI and Vac8 is thought to be mediated by multiple interactions involving the Atg14 CTR and Vps34 or the Vps34–Vps15 subcomplex. We also revealed that the Atg14 CTR-dependent interaction between PI3KCI and Vac8 occurs independent of autophagy induction (cell treatment with rapamycin). In cells not inducing autophagy, this interaction is thought to facilitate PAS targeting of PI3KCI for the cytoplasm-to-vacuole targeting (Cvt) pathway, which transports vacuolar enzymes such as the aminopeptidase Ape1 utilizing the autophagy machinery (core Atg proteins) and is active even in cells growing under nutrient-rich conditions (Lynch-Day and Klionsky, 2010). Indeed, we showed that as with Vac8 (Wang et al., 1998; Scott et al., 2000), the CTR of Atg14 is important not only for starvation-induced non-selective macroautophagy (Pgk1-GFP degradation) but also for the Cvt pathway, as assessed by the proform-to-mature form conversion of Ape1 in cells not treated with rapamycin (Fig. 1 D).
Immunoprecipitation analysis of Atg14-FLAG in VPS34 knockout cells. Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg17, Atg9, and Vac8. Source data are available for this figure: SourceData FS4.
Immunoprecipitation analysis of Atg14-FLAG in VPS34 knockout cells. Cells expressing Atg14-FLAG were treated with rapamycin for 2 h and analyzed by immunoprecipitation using anti-FLAG antibody and subsequent immunoblotting using antibodies against FLAG, Vps34, Vps15, Vps30, Atg1, Atg17, Atg9, and Vac8. Source data are available for this figure: SourceData FS4.
We also found that PI3KCI associates with the Atg1 complex and that this association involves Atg38. Since Vac8 interacts with Atg13 to recruit the Atg1 complex to the vacuolar membrane (Hollenstein et al., 2019; Gatica et al., 2021), Vac8 could serve as a hub for the association between PI3KCI and the Atg1 complex on the vacuolar membrane (Fig. 6). We further revealed that PI3KCI also interacts with Atg9 in an Atg1 complex-independent manner and that this association requires the BARA domain of Vps30 (Fig. 3). A previous study showed that Atg9 is recruited to the PAS via the interaction with the HORMA domain of Atg13 in the Atg1 complex (Suzuki et al., 2015). This interaction and PAS recruitment of Atg9 could therefore facilitate its association with PI3KCI at the PAS. Indeed, the R213D mutation in the HORMA domain of Atg13 diminished the interaction between PI3KCI and Atg9 (Fig. 3 F).
Taken together, these results allow us to propose the following model for the mechanism underlying PAS localization of PI3KCI: (i) PI3KCI is recruited to the vacuolar membrane by interacting with Vac8 in a manner dependent on the Atg14 CTR and Vps34/Vps34–Vps15. (ii) Upon TORC1 inactivation, PI3KCI, via the Atg38 CTR, binds to the Atg29 CTR in the Atg1 complex at the PAS. (iii) The BARA domain of Vps30 in PI3KCI interacts with Atg9 in the Atg9 vesicle associated with the Atg1 complex at the PAS (Fig. 6). In line with this, PAS localization of PI3KCI was modestly and severely impaired by disruption of either and both of the latter two interactions, respectively (Fig. 4), suggesting that the interactions of PI3KCI with the Atg1 complex and Atg9 collaborate to target PI3KCI to the PAS.
This study also revealed that the interactions of PI3KCI with the Atg1 complex and Atg9 are both regulated by Atg1 kinase activity. The simplest explanation for this is that the phosphorylation of related proteins by Atg1 enhances these interactions. Our results further suggested that Vps34 in PI3KCI is also phosphorylated in an Atg1 kinase activity-dependent manner. This Vps34 phosphorylation may be involved in regulating the associations of PI3KCI with the Atg1 complex and Atg9. A recent study showed that Atg1 directly phosphorylates Vps34 in vitro (Lee et al., 2023). Atg1 can also phosphorylate other PI3KCI subunits (Vps30 and Atg38), Atg1 complex subunits (Atg13, Atg29, and Atg1 itself), and Atg9 (Papinski et al., 2014; Kamber et al., 2015; Hu et al., 2019). It is therefore possible that phosphorylation of these proteins also regulates the above interactions. Future studies are required to clarify how Atg1 enhances PI3KCI associations with the Atg1 complex and Atg9; in other words, how PAS targeting of PI3KCI is upregulated upon autophagy induction.
Atg8 and the Atg2–Atg18 complex are recruited to the PAS downstream of PI3KCI (Suzuki et al., 2007). Levels of Atg1 coimmunoprecipitated with PI3KCI increased in atg8Δ and atg2Δ cells (Fig. 2 C). Atg9 was also more efficiently coimmunoprecipitated with PI3KCI in atg2Δ cells compared with wild-type cells. These results suggest that the associations of PI3KCI with the Atg1 complex and Atg9 change dynamically during the process of autophagosome formation. This regulation is also thought to involve dephosphorylation and/or additional modification of related factors. In this regard, phosphorylated Vps34 in PI3KCI increased in atg2Δ cells (Fig. 2 C), suggesting that phosphorylation/dephosphorylation of this protein is involved in the regulation. We also observed coimmunoprecipitation of the Atg16 complex (Atg12-Atg5) with PI3KCI, which was dependent on the Atg1 complex but not on Atg9 (Fig. 2 D). Since the Atg16 complex interacts with the Atg1 complex (Harada et al., 2019), it is thought that the Atg16 complex associated with the Atg1 complex is indirectly coprecipitated with PI3KCI.
In mammalian cells, a number of different mechanisms for PI3KCI regulation have been identified to date, which are more elaborate than those in yeast (Ohashi, 2021). However, little is known about how mammalian PI3KCI is targeted to the site for autophagosome formation (corresponding to the PAS in yeast). A recent study revealed that ATG101, a component of the ULK1 complex (mammalian Atg1 complex), interacts with PI3KCI and PI3KCII (Kim et al., 2018). It was also reported that ULK1 phosphorylates VPS34, BECN1 (mammalian Vps30), and ATG14L (Egan et al., 2015; Russell et al., 2013; Wold et al., 2016; Park et al., 2016). These findings support the idea that association with the ULK1 complex mediates the recruitment of PI3KCI to the autophagosome formation site in mammalian cells in a manner similar to that in yeast cells, although mechanistic details might differ. Meanwhile, the interaction between mammalian PI3KCI and ATG9 has yet to be determined, although since the BARA domain is conserved in BECN1, this domain may mediate the association between PI3KCI and ATG9 in mammalian cells.
Atg9 is an integral membrane protein localized to small vesicles known as Atg9 vesicles, which serve as a seed membrane for autophagosome formation (Yamamoto et al., 2012). In vitro studies revealed that purified S. cerevisiae PI3KCI was able to phosphorylate phosphatidylinositol in isolated Atg9 vesicles (Sawa-Makarska et al., 2020). Although whether PI3KCI produces PI3P in Atg9 vesicles in cells remains to be clarified, the interaction of PI3KCI with Atg9 depending on the BARA domain of Vps30 may also be important for PI3P production in Atg9 vesicles following PAS localization. Meanwhile, in mammalian PI3KCII, BECN1 was shown to have a membrane binding ability, requiring Phe270 and Phe274 in the BARA domain (Chang et al., 2019), both of which are conserved in S. cerevisiae Vps30. The BARA domain of Vps30/BECN1 may therefore contribute to the PAS localization of PI3KCI via binding to not only Atg9 but also the membrane of Atg9 vesicles.
In conclusion, this study established a molecular basis of PAS localization of PI3KCI during autophagosome formation in S. cerevisiae. These findings also provide a foundation for further elucidation of the mechanisms underlying autophagosome formation in other organisms including mammals. Further studies are required to determine the details of the interactions identified in this study and their regulation.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table S2. Knockout and tagging of chromosomal genes were performed based on a standard method using PCR-amplified DNA cassettes (Janke et al., 2004). Yeast strains were cultured at 30°C in YPD medium (1% yeast extract [Becton, Dickinson and Company], 2% peptone [Becton, Dickinson and Company], and 2% glucose) for immunoprecipitation analysis or in SD+CA+ATU medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate [Becton, Dickinson and Company], 0.5% ammonium sulfate, 0.5% casamino acid, 2% glucose, 3.75 mM NaOH, 0.002% adenine sulfate, 0.002% tryptophan, and 0.002% uracil) for fluorescence microscopy and the Pgk1-GFP degradation assay. Yeast cells harboring pRS316-derived plasmids were cultured in SD+CA+ATU without uracil. To induce autophagy, cells were treated with 200 ng/ml rapamycin (LCL) dissolved in 90% ethanol and 10% Tween 20.
Immunoprecipitation
Yeast cells grown to the mid-log phase were spheroplasted and incubated at 30°C in 0.5×YPD or 0.5×SD+CA+AT containing 1 M sorbitol and 200 ng/ml rapamycin. The spheroplasts were then solubilized in IP buffer (50 mM Tris-HCl [pH 7.0], 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, and 10% glycerol) containing 2×Complete protease inhibitor cocktail (Roche), 2 mM phenylmethylsulfonyl fluoride (PMSF; Sigma-Aldrich), 1×PhosSTOP phosphatase inhibitor cocktail (Roche), 500 nM microcystin (Wako), and 1% n-dodecyl-β-D-maltoside (DDM; Nacalai tesque) by rotating sample tubes at 4°C for 30 min (50 mM MES-KOH [pH 6.0] was used instead of 50 mM Tris-HCl [pH 7.0] in the experiments shown in Fig. 1 B). The lysates were clarified by centrifugation at 2,000 g for 10 min and then the supernatants were centrifuged at 15,000 g for 15 min. The resulting supernatants (input) were mixed with NHS FG-beads (Tamagawa Seiki) conjugated with anti-FLAG M2 antibody (Sigma-Aldrich) at 4°C for 2 h. The beads were washed three times with IP buffer containing 0.1% DDM and then the bound proteins were eluted by incubating the beads in SDS sample buffer (10 mM Tris-HCl [pH 7.5], 2% SDS, and 10% glycerol) at 65°C for 10 min. After the removal of the beads, 20 mM dithiothreitol (DTT; Nacalai Tesque) was added to the eluates. The input samples were then mixed with 20% trichloroacetic acid (Nacalai Tesque) on ice for 15 min and centrifuged at 15,000 g for 1 min. The pellets were then washed with ice-cold acetone (Nacalai Tesque), dried, and dissolved in SDS buffer containing 20 mM DTT by vortexing at 65°C for 10 min. The resulting samples were then analyzed by SDS-PAGE and immunoblotting with antibodies against FLAG (F1804; Sigma-Aldrich), HA (3F10; Roche), Atg1 (anti-Apg1; Matsuura et al., 1997), Atg2 (anti-Apg2; Shintani et al., 2001), Atg8 (anti-Atg8-2; Nakatogawa et al., 2012), Atg9 (anti-Atg9 N-pep; Noda et al., 2000), Atg12 (anti-Atg12 IN-3; Kuma et al., 2002), Vps15 (anti-Vps15 V15-1; Kihara et al., 2001), Vps34 (anti-Vps34 V34-1; Kihara et al., 2001), Vps30 (anti-Atg6 IN-9; Kihara et al., 2001), Vac8 (anti-Vac8p; Wang et al., 1998), and Atg38 (anti-Atg38(36-54); generated by immunizing rabbits with peptides corresponding to Atg38 residues Val36–Thr54) and HRP-conjugated secondary antibodies against Rabbit IgG (111-035-144; Jackson Immunoresearch), Rat IgG (112-035-003; Jackson Immunoresearch), and Mouse IgG (315-035-003; Jackson Immunoresearch). Chemical luminescence signals were collected using ImageQuant LAS 4000 (GE Healthcare).
Fluorescence microscopy
Yeast cells were grown to the mid-log phase and treated with rapamycin for indicated times. Cells were then concentrated by centrifugation and mounted on an agarose gel pad (SD-N containing 3.5% [wt/vol] agarose; Young et al., 2011; Graef et al., 2013). Fluorescence microscopy was performed at room temperature using an inverted fluorescence microscope (IX83; Olympus) equipped with ×150 objective lens (UAPON 150XOTIRF; Olympus). A 488-nm blue laser (50 mW; Coherent) and 588-nm yellow laser (50 mW; Coherent) were used for the excitation of mNeonGreen and mCherry, respectively. Fluorescence was filtered with a dichroic mirror reflecting 405-, 488-, and 588-nm wavelengths (Olympus), separated into two channels using the DV2 multichannel imaging system (Photometrics) equipped with a Di02-R594-25x36 dichroic mirror (Semrock), and then passed through a TRF59001-EM ET bandpass filter (Chroma) for the mNeonGreen channel and FF01-624/40-25 bandpass filter (Semrock) for the mCherry channel. Images were acquired using an electron-multiplying CCD camera (ImagEM C9100-13; Hamamatsu Photonics K.K.) and MetaMorph software (Molecular Devices) and processed using Fiji (Image J; Schneider et al., 2012).
Pgk1-GFP degradation assay
Yeast cells were cultured to the mid-log phase and treated with rapamycin. The cell pellets were then suspended in 0.2 M NaOH, incubated on ice for 5 min, and centrifuged at 3,000 g for 5 min. The pellets were suspended in urea SDS buffer (100 mM MOPS-KOH [pH 6.8], 4% SDS, 100 mM DTT, and 8 M urea) by vortexing at 65°C for 10 min. These samples were analyzed by SDS-PAGE and immunoblotting with antibodies against (11814460001; Roche) and Ape1 (anti-API-2; gifted from Dr. Yoshinori Ohsumi, Tokyo Institute of Technology, Yokohama, Japan), and HRP-conjugated secondary antibodies against Mouse IgG and Rabbit IgG. Chemical luminescence signals were collected using ImageQuant LAS 4000.
Phosphatase treatment
Cells expressing Atg14-FLAG were treated with rapamycin for 2 h. Atg14-FLAG in the cell lysates was then bound to anti-FLAG antibody-conjugated beads as described in the methods for immunoprecipitation. The beads were washed three times with NEBuffer for Protein MetalloPhosphatases (New England Biolabs, Inc.) and incubated with lambda protein phosphatase (New England Biolabs, Inc.) with or without heat inactivation (at 65°C for 1 h) at 30°C for 1 h. The beads were then washed with IP Buffer and the bound proteins were eluted by incubating the beads in SDS sample buffer at 65°C for 10 min. After removal of the beads, DTT was added to the eluates to a final concentration of 20 mM, followed by immunoblotting analysis.
AlphaFold2 prediction
The structure of S. cerevisiae Atg38 (residues 110–226) dimer complexed with Atg29 and Atg31 was predicted using AlphaFold2 v2.2.0 (downloaded on May 16, 2022, from https://github.com/deepmind/alphafold) installed on a local computer (Sun Way Technology Co., Ltd.; Jumper et al., 2021a). The predictions were run using the AlphaFold-Multimer mode (Richard et al., 2022,Preprint) with five models and a single seed per model, and default multiple sequence alignment generation using the MMSeqs2 server (Mirdita et al., 2019). The unrelaxed predicted models were subjected to an Amber relaxation procedure, and the relaxed model with the highest confidence based on predicted LDDT scores was selected as the best model and used for figure preparation (Jumper et al., 2021a). Structural figures were prepared using PyMOL (Schrödinger, L. & DeLano, W., 2020. PyMOL, Available at: http://www.pymol.org/pymol.).
Quantification and statistical analysis
Quantification of fluorescence microscopy and immunoblotting data was performed using Fiji software. Statistical tests were performed in GraphPad Prism. Data distribution was assumed to be normal, but this was not formally tested. Tukey’s multiple comparisons test was used for all statistical tests.
Online supplemental material
Fig. S1 shows the vacuolar localization of Atg14 lacking the C-terminal region in cells not treated with rapamycin. Fig. S2 shows the results of immunoprecipitation analysis of Atg14 in cells lacking Vac8. Fig. S3 shows the results related to the interaction between PI3KCI and the Atg1 complex. Fig. S4 shows the results of immunoprecipitation analysis of Atg14 in cells lacking Vps34. Table S1 lists proteins identified by mass spectrometry of Atg14 immunoprecipitates. Table S2 lists the yeast strains used in this study.
Data availability
All the data underlying this study are available in the published article and its online supplemental material.
Acknowledgments
We thank the members of our laboratory for materials, discussions, and technical and secretarial support; Dr. Lois Weisman for providing the Vac8 antibodies; and the Biomaterials Analysis Division of the Open Facility Center at the Tokyo Institute of Technology for DNA sequencing.
This work was supported in part by KAKENHI Grants-in-Aid for Scientific Research JP19H05708 (to H. Nakatogawa) and JP19H05707 (to N.N. Noda) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; AMED Grant Number JP21gm1410004 (to H. Nakatogawa); JST CREST Grant number JPMJCR20E3 (to N.N. Noda); and a STAR Grant funded by the Tokyo Tech Fund (to H. Nakatogawa).
Author contributions: K. Hitomi, T. Kotani, and H. Nakatogawa designed the project. K. Hitomi performed the experiments. Y. Kimura performed mass spectrometry. N.N. Noda predicted the structure of the Atg38–Atg29–Atg31 complex using AlphaFold2. K. Hitomi and H. Nakatogawa wrote the manuscript. All authors analyzed and discussed the results and commented on the manuscript.
References
Author notes
Disclosures: The authors declare no competing interests exist.