Autophagy is a conserved and tightly regulated intracellular quality control pathway. ULK is a key kinase in autophagy initiation, but whether ULK kinase activity also participates in the late stages of autophagy remains unknown. Here, we found that the autophagosomal SNARE protein, STX17, is phosphorylated by ULK at residue S289, beyond which it localizes specifically to autophagosomes. Inhibition of STX17 phosphorylation prevents such autophagosome localization. FLNA was then identified as a linker between ATG8 family proteins (ATG8s) and STX17 with essential involvement in STX17 recruitment to autophagosomes. Phosphorylation of STX17 S289 promotes its interaction with FLNA, activating its recruitment to autophagosomes and facilitating autophagosome–lysosome fusion. Disease-causative mutations around the ATG8s- and STX17-binding regions of FLNA disrupt its interactions with ATG8s and STX17, inhibiting STX17 recruitment and autophagosome–lysosome fusion. Cumulatively, our study reveals an unexpected role of ULK in autophagosome maturation, uncovers its regulatory mechanism in STX17 recruitment, and highlights a potential association between autophagy and FLNA.
Introduction
Autophagy is a conserved pathway in eukaryotic cells responsible for the scavenging and removal of unwanted materials, damaged organelles, and protein aggregates. Dysfunction in autophagy has been linked to many human diseases (Klionsky et al., 2021b; Matsui et al., 2018; Menzies et al., 2015; Mizushima and Levine, 2020). ULK, the mammalian homolog of Atg1 in yeast, is a serine/threonine protein kinase essential for autophagy initiation. It includes two homologs, ULK1 and ULK2 (Mizushima et al., 2011), that function in the most upstream step of autophagy (Itakura and Mizushima, 2010). The ULK complex, composed of ULK, ATG13, FIP200, and ATG101 in mammals, is critical for the initiation of autophagy and functions primarily through the phosphorylation of its substrates (Mizushima et al., 2011). Moreover, ULK is well known to phosphorylate many autophagy initiation-related proteins including AMBRA1 (Di Bartolomeo et al., 2010), ATG14 (Park et al., 2016; Wold et al., 2016), BECN1 (Russell et al., 2013), ATG13 (Chan et al., 2009; Hosokawa et al., 2009; Jung et al., 2009), FIP200 (Jung et al., 2009), BNIP3 (Poole et al., 2021), FUNDC1 (Wu et al., 2014), ZIPK/Sqa (Tang et al., 2011), VPS34 (Egan et al., 2015), and VPS15 (Mercer et al., 2021). The kinase activity of the ULK complex is regulated by upstream signals, such as mTOR, AMPK, PKA, DAPK3, and Rab2 (Ding et al., 2019; Ganley et al., 2009; Hosokawa et al., 2009; Jung et al., 2009; Li et al., 2021). In addition, RAPTOR and all three AMPK subunits serve as ULK1 substrates in positive- and negative-feedback regulation, respectively (Dunlop et al., 2011; Loffler et al., 2011). However, whether ULK kinase activity is involved in the late stages of autophagy remains unknown.
After phagophore closure, autophagosomes mature and fuse with lysosomes to degrade substrates. SNARE component recruitment is a critical event in the autophagosome maturation process (Yim and Mizushima, 2020). STX17 is an essential component of a SNARE complex responsible for autophagosome–lysosome fusion that is not conserved in yeast; in this complex, STX17 is recruited to sealed autophagosomes where it forms a ternary SNARE complex with SNAP29 and VAMP7/8 to mediate autophagosome–lysosome fusion (Itakura et al., 2012; Takats et al., 2013; Tsuboyama et al., 2016). Recently, we found that WT STX17 interacts with STING at ER/ERGIC. Upon autophagy induction, this interaction is interrupted and STX17 is released, after which STX17 recruitment to autophagosomes increases (Rong et al., 2022); STX17 is subsequently recycled by the recycler complex after autophagosome–lysosome fusion (Zhou et al., 2022). However, the mechanism by which STX17 is recruited to autophagosomes remains largely unknown.
Filamins, including filamin A (FLNA), filamin B (FLNB), and filamin C (FLNC), are actin-binding proteins that are conserved across vertebrate species (Chakarova et al., 2000). Mutations in filamins have been linked to many hereditary diseases (Robertson et al., 2003), including otopalatodigital syndrome types 1 and 2, frontometaphyseal dysplasia, Melnick–Needles syndrome, periventricular nodular heterotopias, and osteochondrodysplasias (Giuliano et al., 2005; Moro et al., 2002; Robertson et al., 2003; Rosa et al., 2019; Zenker et al., 2004). Filamins carry an actin-binding domain (ABD) and 24 repeat immunoglobulin-like domains (Ig), which are interrupted by flexible hinge region 1 (H1), between Ig15 and Ig16, and flexible hinge region 2 (H2), between Ig23 and Ig24 (Iwamoto et al., 2018; Seo et al., 2009). In addition to their function as potent F-actin crosslinking proteins, filamins also participate in intracellular signal transduction (Ohta et al., 2006), cell migration and transport (Ehrlicher et al., 2011; Li et al., 2010; van Vliet et al., 2017; Whitmarsh, 2013), and drug resistance (Cheng and Tong, 2021; Mantovani et al., 2019) via binding with a variety of structural and signaling proteins (Chantaravisoot et al., 2015; Segura et al., 2016).
While the roles of ULK kinase activity in the initiation of autophagy have been extensively studied, it remains unknown whether ULK kinase activity is involved in the late stages of autophagy. Any investigation of this question would clearly require the identification of ULK phosphorylation substrates during the later stages of autophagy. Here, we unexpectedly found that ULK also functions in autophagosome maturation by phosphorylating the autophagosomal SNARE STX17 at S289. This phosphorylation increases upon autophagy induction and is required for STX17 recruitment to autophagosomes and subsequent autophagosome maturation. Further investigations identified FLNA as a linker between STX17 and ATG8 family proteins (collectively referred to as ATG8s) that mediates STX17 recruitment to autophagosomes. Phosphorylation of the STX17 residue S289 by ULK promotes its interaction with FLNA and therefore promotes its recruitment to autophagosomes. Examination of disease-causative mutations occurring in the ATG8s- or STX17-binding regions of FLNA revealed that mutations of these sites result in disrupted FLNA interactions with ATG8s or STX17, respectively, and consequently limit STX17 recruitment to autophagosomes and autophagosome maturation. In addition to revealing this unexpected role of ULK kinase activity in autophagosome maturation, this study also defines a ULK-kinase-activity-dependent and FLNA-dependent mechanism responsible for STX17 recruitment in starvation-induced autophagy.
Results
STX17 is phosphorylated by ULK
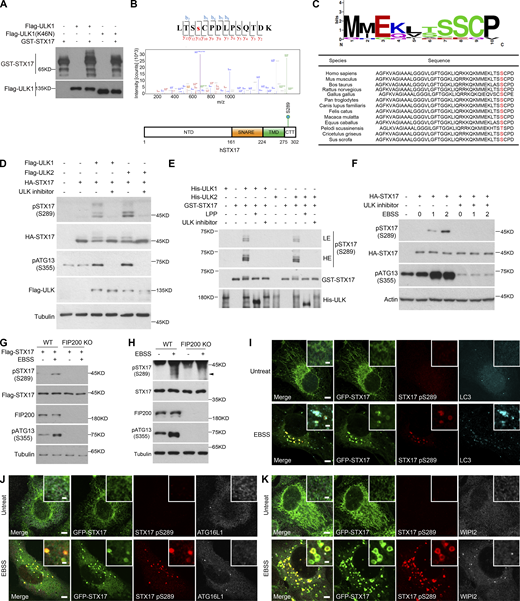
To investigate whether ULK kinase activity is involved in autophagosome–lysosome fusion, we first examined whether ULK phosphorylates any of the key autophagosome–lysosome fusion machinery. Since ULK1 is predicted to be the primary mammalian homolog of Atg1 kinase in mammals (Chan et al., 2007; Young et al., 2006), we coexpressed ULK1 with STX17, SNAP29, or VAMP8 and found that only STX17 showed a dramatic shift in its Western blot band (Fig. 1 A). The STX17 band also shifted following the expression of ULK2 (Fig. 1 B). In both cases, the STX17 band shift could be abolished by either ULK inhibitor MRT68921 or the dominant negative form of ULK1 (ULK1 K46N; Fig. 1 B). The band shift caused by ULK1 or ULK2 was observed for both exogenously and endogenously expressed STX17 (Fig. 1, C and D). These cumulative results suggest that ULK kinase activity affects STX17 electrophoretic mobility. Furthermore, these band shifts were lost following exposure to lambda protein phosphatase (Fig. 1, C and D), supporting that the band shift reflects the phosphorylation of STX17 caused by ULK. Subsequent in vitro kinase assays further confirmed that ULK1 and ULK2 directly phosphorylated STX17 (Fig. 1 E).
ULK directly phosphorylates STX17. (A) ULK1 causes STX17 band shift. HEK293T cells were transfected with Flag-STX17, Flag-SNAP29, or Flag-VAMP8, with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (B) ULK1 and ULK2 lead to band shift of STX17. HEK293T cells were transfected with HA-STX17 with Flag-ULK1, Flag-ULK2, or Flag-ULK1 (K46N). 24 h after transfection, cells were treated with or without the ULK inhibitor (MRT68921) for 2 h. Cells were then subjected to immunoblot with the indicated antibodies. (C and D) ULK leads to the phosphorylation of STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, HEK293T cells were lysed, and the supernatant was treated with or without lambda protein phosphatase (LPP) for 2 h at 37°C and then subjected to immunoblot with the indicated antibodies. (E) ULK directly phosphorylates STX17. An in vitro kinase assay was performed with purified GST-STX17 and His-ULK1 or His-ULK2 in the presence or absence of LPP or the ULK inhibitor. Samples were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. (F) ULK1 localizes to the autophagosome. HeLa cells stably expressing GFP-STX17 were transfected with Flag-ULK1. Cells were then starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of the percentage of ULK1+/STX17−/LC3+ and ULK1+/STX17+/LC3+ puncta in total LC3 puncta upon EBSS treatment in F. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). (H) ULK2 localizes to the autophagosome. HeLa cells stably expressing GFP-STX17 were transfected with Flag-ULK2. Cells were then starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Quantification of the percentage of ULK2+/STX17−/LC3+ and ULK2+/STX17+/LC3+ puncta in total LC3 puncta upon EBSS treatment in H. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). (J and K) ULK1 or ULK2 interacts with exogenous STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (L and M) ULK1 or ULK2 interacts with endogenous STX17. HEK293T cells were lysed and immunoprecipitated with anti-ULK1 or anti-STX17 antibody. IgG was used as the negative control. (N and O) Direct binding of STX17 with ULK1 or ULK2. Ni-NTA agarose resin bound with His-ULK1 or His-ULK2 was incubated with purified GST-STX17 for 16 h and then eluted for immunoblot.
ULK directly phosphorylates STX17. (A) ULK1 causes STX17 band shift. HEK293T cells were transfected with Flag-STX17, Flag-SNAP29, or Flag-VAMP8, with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (B) ULK1 and ULK2 lead to band shift of STX17. HEK293T cells were transfected with HA-STX17 with Flag-ULK1, Flag-ULK2, or Flag-ULK1 (K46N). 24 h after transfection, cells were treated with or without the ULK inhibitor (MRT68921) for 2 h. Cells were then subjected to immunoblot with the indicated antibodies. (C and D) ULK leads to the phosphorylation of STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, HEK293T cells were lysed, and the supernatant was treated with or without lambda protein phosphatase (LPP) for 2 h at 37°C and then subjected to immunoblot with the indicated antibodies. (E) ULK directly phosphorylates STX17. An in vitro kinase assay was performed with purified GST-STX17 and His-ULK1 or His-ULK2 in the presence or absence of LPP or the ULK inhibitor. Samples were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. (F) ULK1 localizes to the autophagosome. HeLa cells stably expressing GFP-STX17 were transfected with Flag-ULK1. Cells were then starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of the percentage of ULK1+/STX17−/LC3+ and ULK1+/STX17+/LC3+ puncta in total LC3 puncta upon EBSS treatment in F. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). (H) ULK2 localizes to the autophagosome. HeLa cells stably expressing GFP-STX17 were transfected with Flag-ULK2. Cells were then starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Quantification of the percentage of ULK2+/STX17−/LC3+ and ULK2+/STX17+/LC3+ puncta in total LC3 puncta upon EBSS treatment in H. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). (J and K) ULK1 or ULK2 interacts with exogenous STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (L and M) ULK1 or ULK2 interacts with endogenous STX17. HEK293T cells were lysed and immunoprecipitated with anti-ULK1 or anti-STX17 antibody. IgG was used as the negative control. (N and O) Direct binding of STX17 with ULK1 or ULK2. Ni-NTA agarose resin bound with His-ULK1 or His-ULK2 was incubated with purified GST-STX17 for 16 h and then eluted for immunoblot.
ULK1 and ULK2 are established phagophore markers and, as such, colocalized with most LC3+/STX17− puncta. However, we still observed that 13.6 and 8.9% of LC3 vesicles were positive for both STX17 and either ULK1 or ULK2, respectively (Fig. 1, F–I), suggesting ULK localization on autophagosomes. In addition, both exogenous and endogenous STX17 interacted with exogenous or endogenous ULK1 or ULK2, respectively (Fig. 1, J–M), and these interactions were shown to be direct (Fig. 1, N and O). Taken together, these results suggest that ULK phosphorylates an autophagosome–lysosome fusion machinery, STX17.
ULK phosphorylates STX17 at Ser289
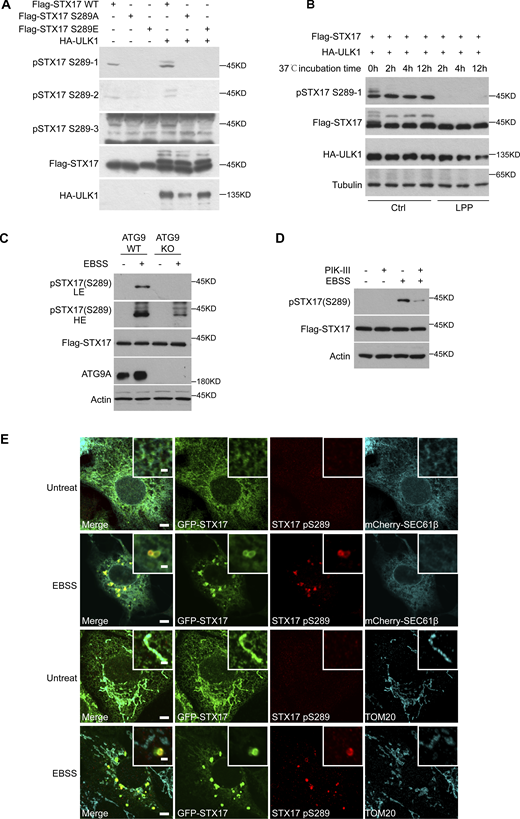
To identify the specific site phosphorylated by ULK, in vitro kinase assays were performed with ULK1 immunoprecipitated from 293T cells and STX17 purified from Escherichia coli (Fig. 2 A). Mass spectrometric analysis revealed multiple candidate phosphorylation sites (Table S1). We then examined the potential role(s) of each candidate STX17 phospho-site in its recruitment to autophagosomes by converting serine or threonine residues at each site to alanines. Immunofluorescent staining for LC3 and Flag indicated that all of these phospho-site mutants colocalized with LC3, except STX17 S289A (Fig. S1 A). Mass spectrometric analysis was used to further confirm the phosphorylation of Serine 289 in the C-terminal tail of STX17 (CTT; Fig. 2 B), while sequence alignments indicated that this Serine 289 was highly conserved across species (Fig. 2 C). Interestingly, the S289A mutation had no obvious effect on the observed band shift in STX17 (Fig. S1 B).
ULK directly phosphorylates STX17 at Ser289. (A) In vitro kinase assay to detect STX17 phosphorylation by ULK1. Flag-ULK1 or Flag-ULK1 (K46N) proteins immunoprecipitated from transfected HEK293T cells were incubated with GST-STX17 purified from E. coli at 30°C for 30 min. Samples were subjected to immunoblot with the indicated antibodies. (B) Identification of STX17 phosphorylation at Ser289. Phosphorylated proteins of GST-STX17 in A were analyzed by mass spectrometry. Results show GST-STX17 phosphorylation by Flag-ULK1 at Ser289 residue. The schematic diagram of the STX17 phosphorylation site at Ser289 is shown. (C) Sequence alignment shows that S289 in different species is highly conserved (analyzed by WebLogo). (D) ULK leads to the phosphorylation of STX17 at Ser289. HEK293T cells were transfected with HA-STX17 and either Flag-ULK1 or Flag-ULK2. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (E) ULK directly phosphorylates STX17 at Ser289. An in vitro kinase assay was performed with purified GST-STX17 and either His-ULK1 or His-ULK2 in the presence or absence of LPP or the ULK inhibitor. Samples were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. (F) EBSS starvation promotes the phosphorylation of STX17 at Ser289. HEK293T cells were transfected with HA-STX17. 24 h after transfection, HEK293T cells were starved with or without EBSS for the indicated hours in the presence or absence of ULK inhibitor. Cells were subjected to immunoblot with the indicated antibodies. (G) Starvation-induced phosphorylation of exogenous STX17 at Ser289 is abolished in FIP200 KO cells. WT and FIP200 KO MEF cells stably expressing Flag-STX17 were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (H) Starvation-induced phosphorylation of endogenous STX17 at Ser289 is abolished in FIP200 KO cells. WT and FIP200 KO MEF cells were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. The black arrowhead indicates the phosphorylation band of STX17. (I) U2OS cells stably expressing GFP-STX17 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) U2OS cells stably expressing GFP-STX17 and Flag-ATG16L1 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (K) U2OS cells stably expressing GFP-STX17 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. Source data are available for this figure: SourceData F2.
ULK directly phosphorylates STX17 at Ser289. (A) In vitro kinase assay to detect STX17 phosphorylation by ULK1. Flag-ULK1 or Flag-ULK1 (K46N) proteins immunoprecipitated from transfected HEK293T cells were incubated with GST-STX17 purified from E. coli at 30°C for 30 min. Samples were subjected to immunoblot with the indicated antibodies. (B) Identification of STX17 phosphorylation at Ser289. Phosphorylated proteins of GST-STX17 in A were analyzed by mass spectrometry. Results show GST-STX17 phosphorylation by Flag-ULK1 at Ser289 residue. The schematic diagram of the STX17 phosphorylation site at Ser289 is shown. (C) Sequence alignment shows that S289 in different species is highly conserved (analyzed by WebLogo). (D) ULK leads to the phosphorylation of STX17 at Ser289. HEK293T cells were transfected with HA-STX17 and either Flag-ULK1 or Flag-ULK2. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (E) ULK directly phosphorylates STX17 at Ser289. An in vitro kinase assay was performed with purified GST-STX17 and either His-ULK1 or His-ULK2 in the presence or absence of LPP or the ULK inhibitor. Samples were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. (F) EBSS starvation promotes the phosphorylation of STX17 at Ser289. HEK293T cells were transfected with HA-STX17. 24 h after transfection, HEK293T cells were starved with or without EBSS for the indicated hours in the presence or absence of ULK inhibitor. Cells were subjected to immunoblot with the indicated antibodies. (G) Starvation-induced phosphorylation of exogenous STX17 at Ser289 is abolished in FIP200 KO cells. WT and FIP200 KO MEF cells stably expressing Flag-STX17 were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (H) Starvation-induced phosphorylation of endogenous STX17 at Ser289 is abolished in FIP200 KO cells. WT and FIP200 KO MEF cells were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. The black arrowhead indicates the phosphorylation band of STX17. (I) U2OS cells stably expressing GFP-STX17 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) U2OS cells stably expressing GFP-STX17 and Flag-ATG16L1 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (K) U2OS cells stably expressing GFP-STX17 were treated with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. Source data are available for this figure: SourceData F2.
The localization to autophagosomes and band shift of STX17 dephosphorylation mimic variants. (A) Examination of co-localization between LC3 and STX17 dephosphorylation mimic variants. AScore of STX17 phosphorylation sites above 10 were selected for localization examination. HeLa cells were transfected with the indicated Flag-STX17 mutants. 24 h after transfection, cells were starved with or without EBSS for 2 h. Then cells were stained with antibodies against LC3 and Flag. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) The effect of STX17 dephosphorylation mimic variants on STX17 band shift. HEK293T cells were transfected with the indicated Flag-STX17 mutants with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. Source data are available for this figure: SourceData FS1.
The localization to autophagosomes and band shift of STX17 dephosphorylation mimic variants. (A) Examination of co-localization between LC3 and STX17 dephosphorylation mimic variants. AScore of STX17 phosphorylation sites above 10 were selected for localization examination. HeLa cells were transfected with the indicated Flag-STX17 mutants. 24 h after transfection, cells were starved with or without EBSS for 2 h. Then cells were stained with antibodies against LC3 and Flag. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) The effect of STX17 dephosphorylation mimic variants on STX17 band shift. HEK293T cells were transfected with the indicated Flag-STX17 mutants with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. LE, low exposure; HE, high exposure. Source data are available for this figure: SourceData FS1.
We next generated and screened three antibodies to detect phosphate modifications at S289 of STX17. Western blot analysis of STX17 in cell lysates identified one antibody (pSTX17 S289-1) that presented the strongest signal for distinguishing between phosphorylation or dephosphorylation at STX17 residue S289 (Fig. S2, A and B). It could also be used to reveal subsequent changes in phosphorylation status as mediated by ULK1. Using this antibody, we found that STX17 phosphorylation at S289 was dramatically increased by ULK1 or ULK2 overexpression in vivo and in vitro, whereas the addition of a ULK inhibitor abolished such phosphorylation (Fig. 2, D and E). EBSS starvation also promoted this phosphorylation, while it was also abolished by a ULK inhibitor (Fig. 2 F). STX17 S289 phosphorylation was also lost in FIP200 KO cells which lack both ULK1 and ULK2 kinase activity (Fig. 2, G and H), suggesting that ULK phosphorylates STX17 at S289. This phosphorylation was also detected in ATG9 KO cells or in the presence of PIK-III (a VPS34 inhibitor), both of which inhibit autophagosome formation, although the level of STX17 pS289 was dramatically lower (Fig. S2, C and D). These results supported that STX17 phosphorylation at S289 by ULK occurs independently of autophagosome formation, but autophagosome formation promotes this phosphorylation.
pSTX17 S289 does not localize to ER and mitochondria. (A) Screening of three antibodies to detect STX17 pS289. HEK293T cells were transfected with Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with antibodies against Flag, HA, and pSTX17 S289(1-3), respectively. (B) Distinguishing the phosphorylation or dephosphorylation at the residue S289 of STX17 by pSTX17 S289-1. HEK293T cells were transfected with Flag-STX17 WT and HA-ULK1. 24 h after transfection, cells were lysed and the supernatant was treated with or without lambda protein phosphatase for the indicated hours at 37°C and then subjected to immunoblot with the indicated antibodies. (C) WT and ATG9 KO HeLa cells stably expressing Flag-STX17 were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (D) HeLa cells stably expressing Flag-STX17 were treated with PIK-III for 30 min, or with EBSS for 2 h, or pretreated with PIK-III for 30 min and then treated with EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (E) STX17 pS289 does not localize to the ER or mitochondria. U2OS cells stably expressing GFP-STX17 were transfected with or without mCherry-Sec61β. 24 h after transfection, cells were starved with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Pseudo colors were used for mCherry-Sec61β and STX17 pS289 in the upper panel. Scale bar, 5 μm. Inset scale bar, 1 μm. Source data are available for this figure: SourceData FS2.
pSTX17 S289 does not localize to ER and mitochondria. (A) Screening of three antibodies to detect STX17 pS289. HEK293T cells were transfected with Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E with or without HA-ULK1. 24 h after transfection, cells were subjected to immunoblot with antibodies against Flag, HA, and pSTX17 S289(1-3), respectively. (B) Distinguishing the phosphorylation or dephosphorylation at the residue S289 of STX17 by pSTX17 S289-1. HEK293T cells were transfected with Flag-STX17 WT and HA-ULK1. 24 h after transfection, cells were lysed and the supernatant was treated with or without lambda protein phosphatase for the indicated hours at 37°C and then subjected to immunoblot with the indicated antibodies. (C) WT and ATG9 KO HeLa cells stably expressing Flag-STX17 were treated with or without EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (D) HeLa cells stably expressing Flag-STX17 were treated with PIK-III for 30 min, or with EBSS for 2 h, or pretreated with PIK-III for 30 min and then treated with EBSS for 2 h. Cells were subjected to immunoblot with the indicated antibodies. (E) STX17 pS289 does not localize to the ER or mitochondria. U2OS cells stably expressing GFP-STX17 were transfected with or without mCherry-Sec61β. 24 h after transfection, cells were starved with or without EBSS for 2 h. Cells were stained with the indicated antibodies. Pseudo colors were used for mCherry-Sec61β and STX17 pS289 in the upper panel. Scale bar, 5 μm. Inset scale bar, 1 μm. Source data are available for this figure: SourceData FS2.
In addition, we found that STX17 phosphorylated at S289 (i.e., STX17 pS289) did not localize to the endoplasmic reticulum or mitochondria, but instead specifically localized to STX17+/LC3+ vesicles (Fig. S2 E and Fig. 2 I). Notably, STX17 pS289 showed no colocalization with WIPI2 or ATG16L1 (Fig. 2, J and K), suggesting that STX17 pS289 localizes to autophagosomes but not phagophores. Collectively, these results suggest that ULK phosphorylates STX17 at S289 and that STX17 pS289 specifically localizes to autophagosomes.
Dephosphorylation of STX17 at S289 inhibits its localization to autophagosomes
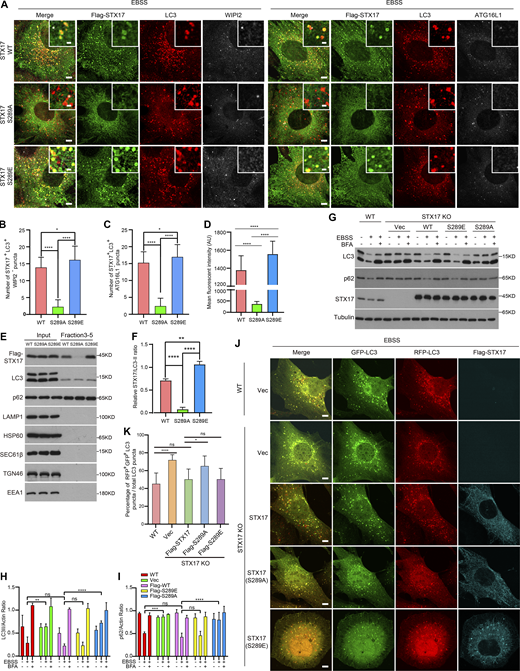
We subsequently tested whether STX17 S289 phosphorylation affected STX17 localization to autophagosomes. Immunostaining assays showed that the STX17 S289A conversion variant exhibited almost completely abolished its localization to sealed autophagosomes, whereas the STX17 S289E variant showed greater localization to autophagosomes than STX17 WT (Fig. 3, A–D). Gradient fractionation, to isolate autophagosomes from cells stably expressing STX17 WT, STX17 S289A, or STX17 S289E, revealed that STX17 S289A level dramatically decreased in the autophagosome fractions relative to that of STX17 WT, while such levels for STX17 S289E was markedly greater than that of WT (Fig. 3, E and F). These results suggest that phosphorylation of STX17 at S289 is not only required for STX17 recruitment to autophagosomes but also promotes its autophagosomal recruitment.
The dephosphorylation of STX17 at Ser289 inhibits its localization to autophagosomes. (A) The dephosphorylation of STX17 at Ser289 fails to translocate to the autophagosome. STX17 KO MEF cells stably expressing Flag-STX17 (WT), Flag-STX17 (S289A), or Flag-STX17 (S289E) were treated with EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of the STX17+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, one-way ANOVA. (C) Quantification of the STX17+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, one-way ANOVA. (D) Quantification of the mean fluorescent intensity of STX17 on autophagosomes in A. Data are mean ± SD (n = 3; 300 autophagosomes from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. (E) Gradient fractionation analysis of STX17 in autophagosome enriched fractions. HEK293T cells stably expressing Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved for 2 h with EBSS, and then separated using the OptiPrep gradient. The fractions from 3 to 5, in a total of 14 fractions, were collected and combined for immunoblot. (F) Quantification of the intensity ratio of STX17/LC3-II in E. The intensity of STX17 mutant bands was normalized to LC3-II. Data are mean ± SEM of three independent experiments. ****P < 0.0001, **P < 0.01, one-way ANOVA. (G) The effect of STX17 S289 phosphorylation on autophagic flux. WT and STX17 KO MEF cells complemented with or without Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved with EBSS in the presence or absence of 100 nM BFA for 2 h. Immunoblot was then performed with the indicated antibodies. (H and I) Quantification of LC3-II and p62 band intensity in G. The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance, two-way ANOVA followed by multiple comparison tests. (J) The effect of STX17 S289 phosphorylation on RFP-GFP-LC3 acidification. WT and STX17 KO MEF cells complemented with or without Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved with EBSS for 2 h. Cells were then stained with the antibody against Flag and images were taken. Scale bar, 5 μm. (K) Quantification of the percentage of GFP+/RFP+ autophagosomes in J. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, ns, no significance, one-way ANOVA. Source data are available for this figure: SourceData F3.
The dephosphorylation of STX17 at Ser289 inhibits its localization to autophagosomes. (A) The dephosphorylation of STX17 at Ser289 fails to translocate to the autophagosome. STX17 KO MEF cells stably expressing Flag-STX17 (WT), Flag-STX17 (S289A), or Flag-STX17 (S289E) were treated with EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of the STX17+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, one-way ANOVA. (C) Quantification of the STX17+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, one-way ANOVA. (D) Quantification of the mean fluorescent intensity of STX17 on autophagosomes in A. Data are mean ± SD (n = 3; 300 autophagosomes from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. (E) Gradient fractionation analysis of STX17 in autophagosome enriched fractions. HEK293T cells stably expressing Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved for 2 h with EBSS, and then separated using the OptiPrep gradient. The fractions from 3 to 5, in a total of 14 fractions, were collected and combined for immunoblot. (F) Quantification of the intensity ratio of STX17/LC3-II in E. The intensity of STX17 mutant bands was normalized to LC3-II. Data are mean ± SEM of three independent experiments. ****P < 0.0001, **P < 0.01, one-way ANOVA. (G) The effect of STX17 S289 phosphorylation on autophagic flux. WT and STX17 KO MEF cells complemented with or without Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved with EBSS in the presence or absence of 100 nM BFA for 2 h. Immunoblot was then performed with the indicated antibodies. (H and I) Quantification of LC3-II and p62 band intensity in G. The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001, ns, no significance, two-way ANOVA followed by multiple comparison tests. (J) The effect of STX17 S289 phosphorylation on RFP-GFP-LC3 acidification. WT and STX17 KO MEF cells complemented with or without Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved with EBSS for 2 h. Cells were then stained with the antibody against Flag and images were taken. Scale bar, 5 μm. (K) Quantification of the percentage of GFP+/RFP+ autophagosomes in J. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, *P < 0.05, ns, no significance, one-way ANOVA. Source data are available for this figure: SourceData F3.
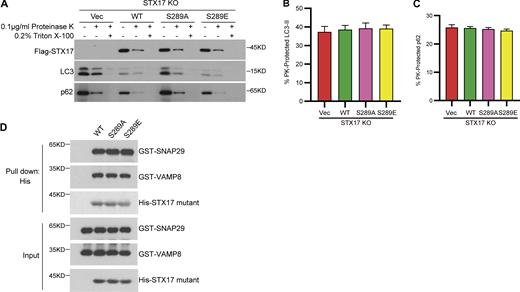
To further examine the effects of STX17 pS289 on autophagy, autophagic flux was determined by examining LC3 and p62 degradation and the acidification of RFP-GFP-LC3. Consistent with previous reports, which showed inhibition of LC3, p62 degradation, and decreased acidification of GFP in RFP-GFP-LC3 in STX17 knockout (KO) cells, these inhibitory effects were rescued in STX17 WT and STX17 S289E expressing cells, but not in those expressing STX17 S289A (Fig. 3, G–K). Proteinase protection assays showed no difference in the cleavage of LC3 and p62 among STX17 KO cells expressing empty vector, STX17 WT, STX17 S289A, or STX17 S289E mutants (Fig. S3, A–C). These results suggest that dephosphorylation of STX17 at S289 inhibits autophagosome maturation rather than autophagosome formation. In addition, in vitro protein binding assays showed that neither STX17 S289A nor STX17 S289E had any noticeable effect on STX17-SNAP29-VAMP8 SNARE complex assembly (Fig. S3 D), possibly due to the location of pS289 being in the C-terminal tail rather than the SNARE domain. These results together suggest that the dephosphorylation of STX17 at S289 confers inhibitory effects on its localization to autophagosomes and consequently inhibits autophagosome maturation.
Dephosphorylation of STX17 at S289 does not affect autophagosome formation and SNARE assembly.(A) STX17 KO MEF cells complemented with empty vector, Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were treated with EBSS for 2 h. Cells were homogenized, split into three aliquots, and subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. The samples were then subjected to immunoblot analysis with the indicated antibodies. (B and C) Quantification of the autophagosome-protected LC3-II and p62 in A. Data are mean ± SEM of three independent experiments. (D) STX17 S289 phosphorylation does not affect STX17-SNAP29-VAMP8 SNARE complex assembly. Ni-NTA agarose resin bound with His-STX17 WT, His-STX17 S289A, or His-STX17 S289E were incubated with purified GST-SNAP29 and GST-VAMP8 for 16 h and then eluted for immunoblot. Source data are available for this figure: SourceData FS3.
Dephosphorylation of STX17 at S289 does not affect autophagosome formation and SNARE assembly.(A) STX17 KO MEF cells complemented with empty vector, Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were treated with EBSS for 2 h. Cells were homogenized, split into three aliquots, and subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. The samples were then subjected to immunoblot analysis with the indicated antibodies. (B and C) Quantification of the autophagosome-protected LC3-II and p62 in A. Data are mean ± SEM of three independent experiments. (D) STX17 S289 phosphorylation does not affect STX17-SNAP29-VAMP8 SNARE complex assembly. Ni-NTA agarose resin bound with His-STX17 WT, His-STX17 S289A, or His-STX17 S289E were incubated with purified GST-SNAP29 and GST-VAMP8 for 16 h and then eluted for immunoblot. Source data are available for this figure: SourceData FS3.
FLNA interacts with STX17 and localizes to autophagosomes
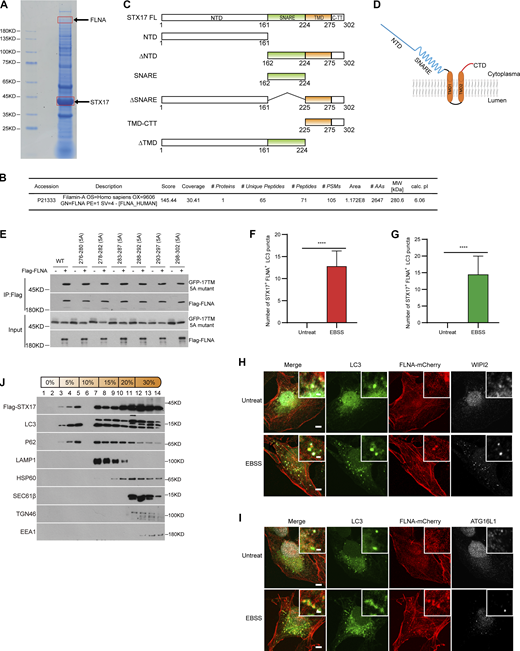
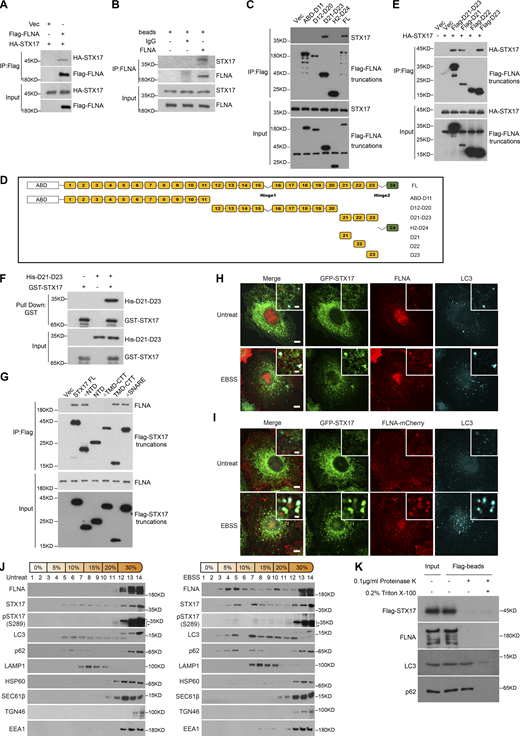
To investigate the detailed mechanism by which the ULK phosphorylation of STX17 S289 regulates its localization to autophagosomes, we next sought to identify STX17-interacting proteins through immunopurification of STX17. Interestingly, an actin-binding protein, FLNA, was identified (Fig. S4, A and B), and STX17-FLNA interactions were confirmed in both exogenous and endogenous proteins (Fig. 4, A and B). Mapping analysis with truncation variants showed that the region between D21 and D23 of FLNA was responsible for this interaction (Fig. 4, C and D). Further analysis revealed that both D21 and D23 independently bound STX17 (Fig. 4 E), and this interaction was direct (Fig. 4 F). Truncation analysis revealed that NTD (N-terminal domain) and SNARE domain were both dispensable for this interaction, while the TMD–CTT (transmembrane domain + C-terminal tail) of STX17 was required for its interaction with FLNA (Fig. 4 G; and Fig. S4, C and D). Alanine scanning analysis of the CTT indicated that it was not required for FLNA–STX17 interaction (Fig. S4 E). These results suggest that TMD was necessary for FLNA–STX17 interaction.
FLNA interacts with STX17 and does not localize to isolation membranes. (A) Coomassie blue staining of the STX17 immunoprecipitation sample. HEK293T cells stably expressing Flag-STX17 were lysed and immunoprecipitated with anti-Flag antibody and eluted for Coomassie blue staining. (B) Mass spectrometric analysis of FLNA in Flag-STX17 immuno-precipitates. (C) Schematic diagram of STX17 truncated variants. (D) Schematic diagram of STX17 topology structure. (E) Alanine scanning analysis of STX17 C-terminal domain. HEK293T cells were transfected with GFP-STX17 TMD-CTT variants with or without Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag. Immunoblot was then performed with the indicated antibodies. (F and G) Quantification of the number of STX17+LC3+FLNA− puncta during EBSS starvation in Fig. 4 H and 4 I. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (H and I) U2OS cells stably expressing FLNA-mCherry were starved with or without EBSS for 2 h and then permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) HEK293T cells stably expressing Flag-STX17 were starved for 2 h with EBSS and subjected to OptiPrep fractionation. 14 fractions were collected for immunoblot. Source data are available for this figure: SourceData FS4.
FLNA interacts with STX17 and does not localize to isolation membranes. (A) Coomassie blue staining of the STX17 immunoprecipitation sample. HEK293T cells stably expressing Flag-STX17 were lysed and immunoprecipitated with anti-Flag antibody and eluted for Coomassie blue staining. (B) Mass spectrometric analysis of FLNA in Flag-STX17 immuno-precipitates. (C) Schematic diagram of STX17 truncated variants. (D) Schematic diagram of STX17 topology structure. (E) Alanine scanning analysis of STX17 C-terminal domain. HEK293T cells were transfected with GFP-STX17 TMD-CTT variants with or without Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag. Immunoblot was then performed with the indicated antibodies. (F and G) Quantification of the number of STX17+LC3+FLNA− puncta during EBSS starvation in Fig. 4 H and 4 I. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (H and I) U2OS cells stably expressing FLNA-mCherry were starved with or without EBSS for 2 h and then permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) HEK293T cells stably expressing Flag-STX17 were starved for 2 h with EBSS and subjected to OptiPrep fractionation. 14 fractions were collected for immunoblot. Source data are available for this figure: SourceData FS4.
FLNA interacts with STX17 and localizes to autophagosomes. (A) The interaction between exogenous STX17 and FLNA. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (B) The interaction between endogenous STX17 and FLNA. HEK293T cells were lysed and immunoprecipitated with anti-FLNA antibody and IgG was used as the negative control. Immunoblotting was performed with the indicated antibodies. (C) The D21-D23 domain is required for the interaction of FLNA with STX17. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (D) Schematic diagram of full-length and truncated variants of FLNA. (E) STX17 interacts with the D21 and D23 domains of FLNA. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (F) FLNA binds STX17 directly. Glutathione Sepharose beads bound with GST-STX17 were incubated with purified His-D21-D23 for 16 h and then eluted for immunoblot. (G) FLNA interacts with the TMD-CTT domain of STX17. HEK293T cells were transfected with empty vector or truncated variants of Flag-STX17. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblot was then performed with the indicated antibodies. (H) Endogenous FLNA localizes to autophagosomes. U2OS cells stably expressing GFP-STX17 were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Exogenous FLNA localizes to autophagosomes. U2OS cells stably expressing GFP-STX17 and FLNA-mCherry were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) FLNA co-fractionates with autophagosome components. MEF cells were treated with or without EBSS for 2 h, and then separated using the OptiPrep gradient. Fourteen fractions were collected for immunoblot. * indicates non-specific bands. (K) FLNA is presented on the autophagosome outer membrane. HEK293T cells stably expressing Flag-STX17 were starved for 2 h with EBSS and subjected to OptiPrep fractionation. The autophagosome-containing fraction 3 was incubated with anti-Flag M2 magnetic beads, split into three aliquots, then subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. The samples were then subjected to immunoblot analysis with the indicated antibodies. Source data are available for this figure: SourceData F4.
FLNA interacts with STX17 and localizes to autophagosomes. (A) The interaction between exogenous STX17 and FLNA. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (B) The interaction between endogenous STX17 and FLNA. HEK293T cells were lysed and immunoprecipitated with anti-FLNA antibody and IgG was used as the negative control. Immunoblotting was performed with the indicated antibodies. (C) The D21-D23 domain is required for the interaction of FLNA with STX17. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (D) Schematic diagram of full-length and truncated variants of FLNA. (E) STX17 interacts with the D21 and D23 domains of FLNA. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (F) FLNA binds STX17 directly. Glutathione Sepharose beads bound with GST-STX17 were incubated with purified His-D21-D23 for 16 h and then eluted for immunoblot. (G) FLNA interacts with the TMD-CTT domain of STX17. HEK293T cells were transfected with empty vector or truncated variants of Flag-STX17. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblot was then performed with the indicated antibodies. (H) Endogenous FLNA localizes to autophagosomes. U2OS cells stably expressing GFP-STX17 were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Exogenous FLNA localizes to autophagosomes. U2OS cells stably expressing GFP-STX17 and FLNA-mCherry were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) FLNA co-fractionates with autophagosome components. MEF cells were treated with or without EBSS for 2 h, and then separated using the OptiPrep gradient. Fourteen fractions were collected for immunoblot. * indicates non-specific bands. (K) FLNA is presented on the autophagosome outer membrane. HEK293T cells stably expressing Flag-STX17 were starved for 2 h with EBSS and subjected to OptiPrep fractionation. The autophagosome-containing fraction 3 was incubated with anti-Flag M2 magnetic beads, split into three aliquots, then subjected to different conditions: No treatment, 0.1 μg/ml proteinase K (PK), or 0.1 μg/ml proteinase K in the presence of 0.2% Triton X-100. The samples were then subjected to immunoblot analysis with the indicated antibodies. Source data are available for this figure: SourceData F4.
We also observed colocalization of endogenous or exogenous FLNA with STX17 and LC3 (Fig. 4, H and I; and Fig. S4, F and G), while FLNA did not co-localize with phagophore markers WIPI2 or ATG16L1 (Fig. S4, H and I). In line with these results, gradient fractionation analysis further revealed the cofractionation of FLNA with STX17, LC3, and STX17 pS289 (Fig. 4 J). In addition, a dramatically greater proportion of FLNA in fraction 3 (the autophagosome-enriched fraction) was cleaved by proteinase K in the absence of Triton X-100, while p62 remained intact (Fig. 4 K and Fig. S4 J). All such data together suggest that FLNA localizes to the autophagosome outer membrane and interacts with STX17.
FLNA is required for STX17 recruitment to autophagosomes
To further investigate the effects of FLNA on STX17 localization to autophagosomes, we next examined its localization patterns in cells with FLNA knockdown (KD). In FLNA KD cells, upon EBSS starvation, STX17’s colocalization with LC3 was almost completely abolished, and LC3 puncta showed no obvious colocalization with phagophore makers (WIPI2 and ATG16L1; Fig. 5, A–C). In addition, the abolished localization of STX17 on autophagosomes was rescued by the re-expression of FLNA (Fig. 5, D and E). Interestingly, colocalization between STX17 and LC3 was unaffected by the knockdown of the FLNA homologs FLNB and FLNC (Fig. S5, A–D), suggesting that FLNA is specifically required for STX17 recruitment to autophagosomes. Moreover, FLNA colocalization with LC3 was unaffected in STX17 KO cells (Fig. 5, F and G), suggesting that the localization of FLNA to LC3 puncta was independent of STX17. It is noteworthy that treatment with Latrunculin B, an F-actin depolarizing agent, had no effect on STX17 localization to autophagosomes (Fig. 5, H and I), suggesting the regulatory role of FLNA toward STX17 to be independent of its F-actin related function.
FLNA is required for STX17 recruitment to autophagosomes. (A) FLNA knockdown abolishes the localization of STX17 to autophagosomes. U2OS cells stably expressing GFP-STX17 were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of the STX17+/LC3+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (C) Quantification of the STX17+/LC3+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (D) Wild-type FLNA rescues the deficiency of STX17 recruitment to autophagosomes in FLNA knockdown cells. U2OS cells stably expressing GFP-STX17 and vector or siRNA resistant FLNA-mCherry WT were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h, permeabilized before fixation, and then stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of the number of STX17 positive autophagosomes. Images in D were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (F) Localization of FLNA to autophagosomes is independent of STX17. Wild-type and STX17 KO U2OS cells were starved with or without EBSS for 2 h and permeabilized before fixation. Then cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of the FLNA positive autophagosome number. Images in F were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ns, no significance, unpaired two-tailed t test. (H) STX17 recruitment to autophagosomes is actin-independent. U2OS cells stably expressing GFP-STX17 were starved with EBSS and were treated with or without Latrunculin B for 2 h. Then cells were stained with phalloidin and the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Quantification of the STX17 positive autophagosome number. Images in H were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ns, no significance, unpaired two-tailed t test. (J) Autophagic flux is inhibited in FLNA knockdown cells. WT or FLNA knockdown U2OS cells were starved with EBSS and were treated with or without 100 nM BFA for 2 h and immunoblots were performed with the indicated antibodies. (K and L) Quantification of LC3-II and p62 band intensity in (J). The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA followed by multiple comparison tests. (M) RFP-GFP-LC3 acidification is inhibited in FLNA-deficient cells. U2OS cells stably expressing RFP-GFP-LC3 were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h. Scale bar, 5 μm. Inset scale bar, 1 μm. (N) Quantification of the percentage of GFP+/RFP+ autophagosomes in M. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. Source data are available for this figure: SourceData F5.
FLNA is required for STX17 recruitment to autophagosomes. (A) FLNA knockdown abolishes the localization of STX17 to autophagosomes. U2OS cells stably expressing GFP-STX17 were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) Quantification of the STX17+/LC3+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (C) Quantification of the STX17+/LC3+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. (D) Wild-type FLNA rescues the deficiency of STX17 recruitment to autophagosomes in FLNA knockdown cells. U2OS cells stably expressing GFP-STX17 and vector or siRNA resistant FLNA-mCherry WT were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h, permeabilized before fixation, and then stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of the number of STX17 positive autophagosomes. Images in D were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (F) Localization of FLNA to autophagosomes is independent of STX17. Wild-type and STX17 KO U2OS cells were starved with or without EBSS for 2 h and permeabilized before fixation. Then cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (G) Quantification of the FLNA positive autophagosome number. Images in F were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ns, no significance, unpaired two-tailed t test. (H) STX17 recruitment to autophagosomes is actin-independent. U2OS cells stably expressing GFP-STX17 were starved with EBSS and were treated with or without Latrunculin B for 2 h. Then cells were stained with phalloidin and the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (I) Quantification of the STX17 positive autophagosome number. Images in H were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ns, no significance, unpaired two-tailed t test. (J) Autophagic flux is inhibited in FLNA knockdown cells. WT or FLNA knockdown U2OS cells were starved with EBSS and were treated with or without 100 nM BFA for 2 h and immunoblots were performed with the indicated antibodies. (K and L) Quantification of LC3-II and p62 band intensity in (J). The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA followed by multiple comparison tests. (M) RFP-GFP-LC3 acidification is inhibited in FLNA-deficient cells. U2OS cells stably expressing RFP-GFP-LC3 were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h. Scale bar, 5 μm. Inset scale bar, 1 μm. (N) Quantification of the percentage of GFP+/RFP+ autophagosomes in M. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, unpaired two-tailed t test. Source data are available for this figure: SourceData F5.
Depletion of FLNB and FLNC does not affect the recruitment of STX17 to autophagosomes. (A) Knockdown of FLNA, but not FLNB and FLNC, abolishes the STX17 recruitment to autophagosomes. U2OS cells stably expressing GFP-STX17 were transfected with non-targeting siRNA (NC) or siRNA against FLNA, FLNB, or FLNC, respectively. 48 h after transfection, cells were starved with EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells were transfected with the indicated siRNAs. 48 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (C) Quantification of the STX17+/LC3+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (D) Quantification of the STX17+/LC3+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (E) Knockdown of FLNA does not alter cathepsin D processing. U2OS cells were transfected with no-targeting siRNA (NC) or siRNAs against FLNA. 48 h after transfection, cells were starved with EBSS or treated with 100 nM BFA for 24 h. Then cells were collected for immunoblot with the indicated antibodies. Source data are available for this figure: SourceData FS5.
Depletion of FLNB and FLNC does not affect the recruitment of STX17 to autophagosomes. (A) Knockdown of FLNA, but not FLNB and FLNC, abolishes the STX17 recruitment to autophagosomes. U2OS cells stably expressing GFP-STX17 were transfected with non-targeting siRNA (NC) or siRNA against FLNA, FLNB, or FLNC, respectively. 48 h after transfection, cells were starved with EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (B) U2OS cells were transfected with the indicated siRNAs. 48 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (C) Quantification of the STX17+/LC3+/WIPI2− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (D) Quantification of the STX17+/LC3+/ATG16L1− autophagosome number. Images in A were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (E) Knockdown of FLNA does not alter cathepsin D processing. U2OS cells were transfected with no-targeting siRNA (NC) or siRNAs against FLNA. 48 h after transfection, cells were starved with EBSS or treated with 100 nM BFA for 24 h. Then cells were collected for immunoblot with the indicated antibodies. Source data are available for this figure: SourceData FS5.
Consistent with the disappearance of STX17 localization on autophagosomes in FLNA KD cells, LC3 and p62 degradation, and the acidification of GFP in RFP-GFP-LC3 were both inhibited in the absence of FLNA (Fig. 5, J–N). Conversely, FLNA knockdown had no effect on cathepsin D processing (Fig. S5 E), suggesting that decreased degradation of autophagic substrates and RFP-GFP-LC3 acidification was unrelated to lysosomal activity. Together, these results suggest that FLNA is required for STX17 localization to autophagosomes and subsequent autophagosome maturation.
Phosphorylation at STX17 S289 increases STX17 interactions with FLNA
Given the similar effects of FLNA knockdown and STX17 S289A, we hypothesized that interplay might occur between the regulation of FLNA and STX17 phosphorylation. Western blot analysis showed that the phosphorylation of STX17 S289 remained unaffected by knockdown of FLNA (Fig. 6 A), which suggests that FLNA is not involved in STX17 phosphorylation of S289. Interestingly, interactions between FLNA and STX17 not only increased upon the induction of autophagy or under ULK1 overexpression but were also inhibited upon the addition of ULK1 inhibitor or by expression of the dominant negative ULK1 K46N variant (Fig. 6 B). These findings raised the possibility that STX17 phosphorylation by ULK1 may regulate STX17 interactions with FLNA. To test this possibility, we examined the binding between FLNA and the phosphorylation mimic STX17 S289E or the dephosphorylation mimic STX17 S289A. Immunoprecipitation assays showed that FLNA interactions were markedly increased with the STX17 S289E variant compared with STX17 WT, while there were fewer interactions with STX17 S289A than with STX17 WT (Fig. 6 C). Consistent with these results, immunostaining assays confirmed that STX17 S289E showed increased colocalization with FLNA and LC3, while the STX17 S289A variant exhibited decreased colocalization (Fig. 6, D and E). These results suggest that ULK phosphorylation of STX17 residue S289 promotes interaction between STX17 and FLNA.
STX17 pS289 increases its interaction with FLNA. (A) FLNA knockdown does not affect the phosphorylation of STX17 at S289. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with indicated plasmids. Another 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (B) ULK activation increases the interaction of FLNA with STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were treated with or without Torin2 or ULK inhibitor for 2 h. Cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (C) The effect of STX17 Ser289 phosphorylation on its interaction with FLNA. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (D) STX17 S289A and STX17 S289E decrease and increase their colocalization with FLNA compared to STX17 WT, respectively. U2OS cells stably expressing FLNA-mCherry with GFP-STX17 WT, GFP-STX17 S289A, or GFP-STX17 S289E were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of the FLNA+/STX17+ autophagosome number. Images in D were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. Source data are available for this figure: SourceData F6.
STX17 pS289 increases its interaction with FLNA. (A) FLNA knockdown does not affect the phosphorylation of STX17 at S289. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with indicated plasmids. Another 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (B) ULK activation increases the interaction of FLNA with STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were treated with or without Torin2 or ULK inhibitor for 2 h. Cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (C) The effect of STX17 Ser289 phosphorylation on its interaction with FLNA. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblotting was performed with the indicated antibodies. (D) STX17 S289A and STX17 S289E decrease and increase their colocalization with FLNA compared to STX17 WT, respectively. U2OS cells stably expressing FLNA-mCherry with GFP-STX17 WT, GFP-STX17 S289A, or GFP-STX17 S289E were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (E) Quantification of the FLNA+/STX17+ autophagosome number. Images in D were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. Source data are available for this figure: SourceData F6.
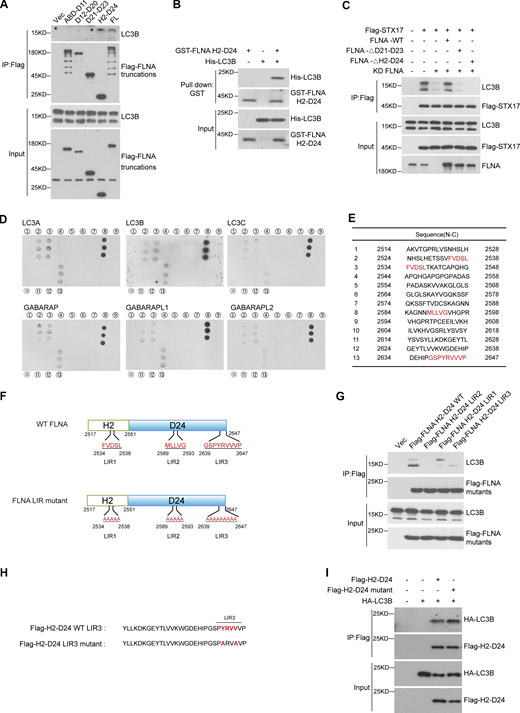
FLNA links STX17 to ATG8s
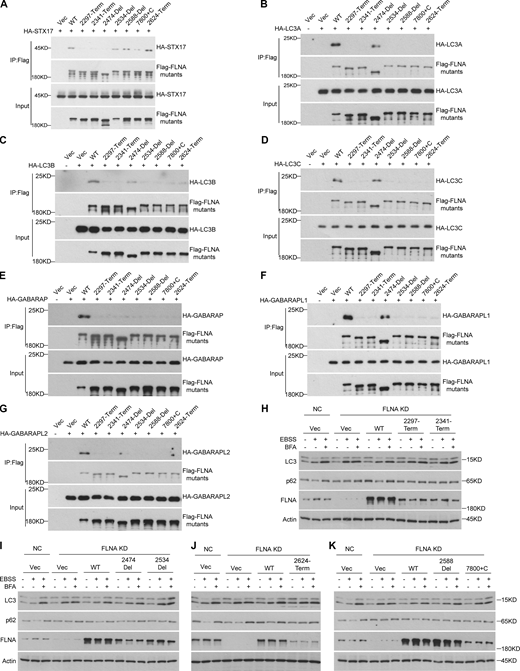
We next investigated how FLNA recruits STX17 to autophagosomes. STX17 and ATG8 family proteins are present on autophagic vacuoles (Yim and Mizushima, 2020; Zhao and Zhang, 2019). Since FLNA localization to autophagosomes occurred independently of STX17 (Fig. 5, F and G), we hypothesized that ATG8 family proteins could be responsible for FLNA localization to autophagosomes. To test this possibility, we first examined interactions between FLNA and each ATG8 homolog. Co-immunoprecipitation assays showed that all ATG8 homologs interacted with FLNA (Fig. 7 A). Consistently, Flag-FLNA interacted with the endogenous ATG8 homolog, LC3B (Fig. S6 A). Mapping analysis also showed that the H2-D24 region of FLNA interacted with LC3B, and this was a direct interaction (Fig. S6, A and B). Deletion of the H2-D24 in FLNA resulted in disruption of STX17-LC3B interactions (Fig. S6 C). To pinpoint the site(s) within the FLNA H2-D24 region necessary for interaction with ATG8s, we performed an in vitro peptide screening and found that all ATG8 homologs bound to peptides 2, 3, 8, and 13 of the H2-D24 region (Fig. S6, D and E). Thus, three potential binding sites (2- or 3-FVDSL, 8-MLLVG, and 13-GSPYRVVVP hereafter referred to as LIR1, LIR2, and LIR3, respectively) were identified in this region (Fig. S6 F). Mutation of any one of these sites disrupted interactions between the FLNA H2-D24 region and ATG8 family proteins (Fig. 7, B–G and Fig. S6 F). In addition, the LIR1, LIR2, and LIR3 FLNA mutants also showed decreased interactions with endogenous LC3B (Fig. S6 G). These cumulative findings suggest that ATG8 family proteins directly interact with FLNA via LIR motifs in its H2-D24 region.
FLNA mediates the interaction between STX17 and ATG8s. (A) FLNA interacts with ATG8s. ATG8s KO HeLa cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (B–G) FLNA LIR mutants disrupt their interactions with ATG8s. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H) The interactions between STX17 and ATG8s are inhibited by FLNA knockdown. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (I) FLNA mutants fail to rescue the localization of STX17 to autophagosomes. U2OS cells stably expressing GFP-STX17 with vector, siRNA-resistant FLNA-mCherry WT or FLNA-mCherry mutants were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) Quantification of the number of STX17 positive autophagosomes. Images in I were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (K) Quantification of the number of STX17+/FLNA+ positive autophagosomes. Images in I were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. Source data are available for this figure: SourceData F7.
FLNA mediates the interaction between STX17 and ATG8s. (A) FLNA interacts with ATG8s. ATG8s KO HeLa cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (B–G) FLNA LIR mutants disrupt their interactions with ATG8s. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H) The interactions between STX17 and ATG8s are inhibited by FLNA knockdown. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (I) FLNA mutants fail to rescue the localization of STX17 to autophagosomes. U2OS cells stably expressing GFP-STX17 with vector, siRNA-resistant FLNA-mCherry WT or FLNA-mCherry mutants were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and permeabilized before fixation. Cells were stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (J) Quantification of the number of STX17 positive autophagosomes. Images in I were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (K) Quantification of the number of STX17+/FLNA+ positive autophagosomes. Images in I were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, one-way ANOVA. Source data are available for this figure: SourceData F7.
Mapping analysis of the interaction region of FLNA with ATG8s. (A) LC3B interacts with the H2-D24 domain of FLNA. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (B) LC3B binds FLNA H2-D24 directly. Glutathione Sepharose beads bound with GST-FLNA H2-D24 were incubated with purified His-LC3B for 16 h and then eluted for immunoblot. (C) FLNA deficiency leads to disrupted interaction of STX17 with LC3B. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (D) Mapping the interaction region of FLNA with ATG8s. The peptides with the overlapping sequence in the FLNA H2-D24 domain were synthesized, fixed on the nitrocellulose filter membrane, and then incubated with different His-ATG8s. A dot blot was performed with the antibody against His. (E) The peptide sequences of FLNA H2-D24 used in D. (F) Schematic diagram of hypothetical LIR motifs in FLNA. (G) FLNA H2-D24 LIR mutants show disrupted interaction with endogenous LC3B. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H) Schematic diagram of the FLNA LIR3 motif. (I) The interaction between the FLNA H2-D24 (LIR3-2A) mutant and LC3B remains unaffected. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. Source data are available for this figure: SourceData FS6.
Mapping analysis of the interaction region of FLNA with ATG8s. (A) LC3B interacts with the H2-D24 domain of FLNA. HEK293T cells were transfected with empty vector or truncated variants of Flag-FLNA. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (B) LC3B binds FLNA H2-D24 directly. Glutathione Sepharose beads bound with GST-FLNA H2-D24 were incubated with purified His-LC3B for 16 h and then eluted for immunoblot. (C) FLNA deficiency leads to disrupted interaction of STX17 with LC3B. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with the indicated plasmids. Another 24 h after transfection, cells were subjected to immunoblot with the indicated antibodies. (D) Mapping the interaction region of FLNA with ATG8s. The peptides with the overlapping sequence in the FLNA H2-D24 domain were synthesized, fixed on the nitrocellulose filter membrane, and then incubated with different His-ATG8s. A dot blot was performed with the antibody against His. (E) The peptide sequences of FLNA H2-D24 used in D. (F) Schematic diagram of hypothetical LIR motifs in FLNA. (G) FLNA H2-D24 LIR mutants show disrupted interaction with endogenous LC3B. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H) Schematic diagram of the FLNA LIR3 motif. (I) The interaction between the FLNA H2-D24 (LIR3-2A) mutant and LC3B remains unaffected. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. Source data are available for this figure: SourceData FS6.
Furthermore, the finding that STX17 interacts with all ATG8s was consistent with a previous report (Kumar et al., 2018), and STX17-ATG8 interactions decreased under FLNA KD (Fig. 7 H). Given that FLNA interacts with both STX17 and ATG8 family proteins and deletion in FLNA of either the H2-D24 region (required for ATG8-FLNA interaction) or the D21-D23 region (required for STX17-FLNA interaction) of FLNA disrupted interactions between STX17 and LC3B (Fig. S6 C), it was reasonable to hypothesize that FLNA likely functions as a linker between ATG8s and STX17. Supporting this conclusion, the deletion of the H2-D24, D21-D23 regions, or the LIR2 mutation in FLNA each resulted in abolishing LC3 colocalization with STX17 and FLNA compared with cells harboring FLNA WT (Fig. 7, I–K). Collectively, all these results suggest FLNA functions as a linker between ATG8s and STX17 to recruit STX17 to autophagosomes.
FLNA deficiency-related disease mutations inhibit autophagy
Interestingly, among the disease-associated mutations distributed throughout FLNA, some mutations present in or near regions responsible for its interactions with ATG8 or STX17 (e.g., 2297 Tyr-Term, 2341 Gln-Term, 2474-Del, 2534-Del, 2588-Del, 7800+C, and 2624 Trp-Term) have been linked to diseases such as periventricular nodular heterotopia and frontometaphyseal dysplasia (Fig. 9 A; Jefferies et al., 2010; Moro et al., 2002; Parrini et al., 2006; Robertson et al., 2006; Sheen et al., 2001; Sole et al., 2009). Since abnormalities in autophagy have been implicated in neurological diseases (Frake et al., 2015; Menzies et al., 2015), we further tested whether these disease-causative mutations in FLNA also affected autophagy. Coimmunoprecipitation assays revealed that these mutations (2297-Term, 2341-Term, and 2474-Del) in or around STX17 binding regions could disrupt FLNA interaction with STX17, while mutations in the regions responsible for interactions with ATG8s (2534-Del, 2588-Del, 7800+C, and 2624-Term) had no obvious effect on FLNA binding with STX17 (Fig. 8 A). Conversely, mutations in or around the STX17- or ATG8-binding regions disrupted interactions between FLNA and ATG8s, although the FLNA 2474-Del mutant displayed only marginally decreased interactions (Fig. 8, B–G). In line with these results, colocalization between STX17 and LC3 decreased in FLNA KD cells complemented with disease-causative FLNA variants (Fig. 9, B and C). Moreover, autophagic flux was also decreased in cells expressing these FLNA mutants compared with that in WT cells (Fig. 8, H–K and Fig. S7). Collectively, these results support the likelihood that disease-causing mutations in the FLNA binding regions with STX17 or ATG8s inhibit autophagy.
FLNA deficiency disease causative mutations inhibit autophagy. (A) FLNA deficiency disease causative mutations in or around D21-D23 regions disrupt the interaction of FLNA with STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were then performed with the indicated antibodies. (B–G) FLNA deficiency disease causative mutations in or around H2-D24 regions disrupt its interaction with ATG8s. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H–K) FLNA deficiency disease causative mutations fail to rescue the autophagic flux in FLNA deficient cells. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with different FLNA mutant plasmids. Cells were starved with EBSS with or without 100 nM BFA for 2 h and immunoblots were performed with the indicated antibodies. Source data are available for this figure: SourceData F8.
FLNA deficiency disease causative mutations inhibit autophagy. (A) FLNA deficiency disease causative mutations in or around D21-D23 regions disrupt the interaction of FLNA with STX17. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were then performed with the indicated antibodies. (B–G) FLNA deficiency disease causative mutations in or around H2-D24 regions disrupt its interaction with ATG8s. HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed and immunoprecipitated with anti-Flag antibody. Immunoblots were performed with the indicated antibodies. (H–K) FLNA deficiency disease causative mutations fail to rescue the autophagic flux in FLNA deficient cells. HEK293T cells were transfected with non-targeting siRNA (NC) or siFLNA#2. 24 h after transfection, cells were transfected with different FLNA mutant plasmids. Cells were starved with EBSS with or without 100 nM BFA for 2 h and immunoblots were performed with the indicated antibodies. Source data are available for this figure: SourceData F8.
The disease causative mutants of FLNA fail to rescue the STX17 localization to autophagosomes. (A) Schematic diagram of FLNA deficiency disease causative mutants. (B) The FLNA deficiency disease causative mutants fail to rescue STX17 localization to autophagosomes. U2OS cells stably expressing GFP-STX17 with vector or FLNA mutants were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (C) Quantification of the number of STX17 positive autophagosomes. Images in B were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (D) Sequence alignment among FLNA, FLNB, and FLNC. The locations of FLNA deficiency disease causative mutations are marked with red arrows. Conserved percentage of FLNA, FLNB, and FLNC: red 100%; green 80%; yellow 60%.
The disease causative mutants of FLNA fail to rescue the STX17 localization to autophagosomes. (A) Schematic diagram of FLNA deficiency disease causative mutants. (B) The FLNA deficiency disease causative mutants fail to rescue STX17 localization to autophagosomes. U2OS cells stably expressing GFP-STX17 with vector or FLNA mutants were transfected with non-targeting siRNA (NC) or siFLNA#2. 48 h after transfection, cells were starved with or without EBSS for 2 h and stained with the indicated antibodies. Scale bar, 5 μm. Inset scale bar, 1 μm. (C) Quantification of the number of STX17 positive autophagosomes. Images in B were analyzed. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, one-way ANOVA. (D) Sequence alignment among FLNA, FLNB, and FLNC. The locations of FLNA deficiency disease causative mutations are marked with red arrows. Conserved percentage of FLNA, FLNB, and FLNC: red 100%; green 80%; yellow 60%.
Quantification of autophagic flux in Fig. 8,. (A–D) Quantification of LC3-II and p62 band intensity in Fig. 8, H–K. The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. ns, no significance, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA followed by multiple comparison tests.
Quantification of autophagic flux in Fig. 8,. (A–D) Quantification of LC3-II and p62 band intensity in Fig. 8, H–K. The intensity of LC3-II and p62 bands were normalized to actin. Data are mean ± SEM of three independent experiments. ns, no significance, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA followed by multiple comparison tests.
Discussion
As a key kinase in autophagy, ULK is well-known to play an essential role in initiating autophagy by phosphorylating autophagy-related machinery. However, the role of kinase activity in the late stages of autophagy has remained completely unknown due to the lack of identifiable substrates. Here, we demonstrated that ULK1 regulates autophagosome maturation by phosphorylating STX17 to promote STX17 recruitment to autophagosomes. FLNA, which was identified as a STX17-interacting protein in the current study, functions as a linker molecule to bridge STX17 and ATG8 proteins and is therefore required for STX17 recruitment to autophagosomes. The phosphorylation of STX17 by ULK increases its interactions with FLNA, which then facilitates its recruitment to autophagosomes and autophagosome maturation. Disease-causative mutations in FLNA in or around the regions necessary for binding STX17 or ATG8 proteins decrease FLNA interactions with STX17 and ATG8s, respectively, thus inhibiting STX17 recruitment and autophagosome maturation.
The Atg1 complex functions in a kinase-dependent or -independent manner to regulate autophagy in yeast (Cheong et al., 2008; Kabeya et al., 2005; Kamada et al., 2000). Recently, ULK1 was shown to recruit STX17 to autophagosomes and facilitate its interaction with another autophagosomal SNARE protein, SNAP29, independent of its kinase activity in mammalian cells (Wang et al., 2018). These findings suggest that ULK also functions in a kinase-dependent or -independent manner in mammals similarly. In mammals, the effects of ULK are primarily mediated by its kinase activity in autophagy (Mizushima et al., 2011). The identification of physiologically relevant substrate(s) of Atg1/ULK will be essential for understanding the precise role of this complex. Here, we report the first known target of ULK phosphorylation activity in the autophagosome maturation stage and describe its kinase activity-mediated regulation of autophagosome maturation. Ongoing exploration of other potential ULK functions in the late stages of autophagy depends on the identification of other possible substrates.
STX17 is a component of the STX17-SNAP29-VAMP7/8 SNARE complex which is conserved in mammals, Drosophila, and C. elegans, but are not found in yeast. The yeast autophagosomal R-SNARE protein, Ykt6, mediates autophagosome–vacuole fusion in conjunction with the vacuolar Q-SNAREs Vam3, Vam7, and Vti1 (Bas et al., 2018; Gao et al., 2018). The Atg1 complex phosphorylates the Ykt6 SNARE domain to preserve its inactive state and negatively regulate SNARE bundling, thereby inhibiting autophagosome–lysosome fusion (Barz et al., 2020; Gao et al., 2020). However, in the current work, we found that the mammalian SNARE STX17 is phosphorylated by ULK at residue S289 in its C-terminal tail. While phosphorylation of STX17 S289 has no effect on SNARE complex assembly, STX17 pS289 promotes its recruitment to autophagosomes and positively regulates autophagosome–lysosome fusion. These results suggest that Atg1/ULK regulates SNARE function through distinctly different mechanisms in yeast and mammals.
STX17 has been previously shown to participate in autophagy initiation (Arasaki et al., 2018; Arasaki et al., 2015; Hamasaki et al., 2013). Although our proteinase protection assays showed that the RFP-GFP-LC3 puncta which accumulate in STX17 knockout cells are sealed autophagosomes, not phagophores, we cannot exclude the possibility that a few unsealed autophagosomes are also present in STX17 knockout cells. These unsealed autophagosomes, if present, may be undetectable by protein protection assay due to a combination of insufficient sensitivity and the weak effect of STX17 knockout on autophagy initiation.
In addition, TBK1 phosphorylates STX17 at Ser202 and STX17 pS202 and then abolishes its interaction with STING, and this pool of STX17 provides a substantial contribution to autophagosome formation (Kumar et al., 2019; Rong et al., 2022). Here, we found that STX17 is phosphorylated by ULK at S289 in mammalian cells and that STX17 pS289 specifically localizes to autophagosomes, but not the mPAS (mammalian pre-autophagosomal structure), the putative mammalian equivalent of the PAS (Kumar et al., 2019; Mizushima et al., 2011). STX17 pS289 does not contribute to autophagosome formation but is involved in STX17 recruitment to the autophagosome; it therefore plays a most significant role in autophagosome maturation. This functional bifurcation suggests the presence of at least two pools of STX17 in cells and illustrates how different kinases are responsible for maintaining a corresponding pool of STX17 required at different subcellular destinations and for different functions. The autophagosomal STX17 pool can thus be distinguished by specifically labeling STX17 pS289. Further, ULK-mediated phosphorylation of STX17 residue S289 still occurs in cells deficient for autophagosome formation, although with dramatically decreased STX17 pS289 phosphorylation levels. These findings thus suggest that STX17 phosphorylation by ULK can occur independently of autophagosome formation, although autophagosome formation appears to promote STX17 phosphorylation by ULK.
It was noteworthy that FLNA interaction with the STX17 288-292 (5A) variant is the same as its interaction with STX17 WT, seemingly contradicting findings that FLNA exhibits decreased interaction with the STX17 S289A variant. We are inclined to speculate that differences in peptide sequence between STX17 S289A and STX17 288-292 (5A) may have resulted in these varying results. It is also possible that STX17 phosphorylation at S289 may regulate STX17-FLNA interaction indirectly (e.g., phosphorylation of STX17 at S289 may cooperate with an unidentified binding partner to regulate STX17-FLNA interaction).
In our study, STX17 recruitment to autophagosomes requires ATG8 family proteins in starvation-induced autophagy, which is consistent with a previous report (Kumar et al., 2018). However, in another previous study, GFP-STX17 localized around mitochondria in a manner comparable between WT and ATG8s KO HeLa cell lines during mitophagy (Nguyen et al., 2016). These findings together suggest that the regulatory mechanisms responsible for STX17 recruitment to autophagosomes may differ between mitophagy and starvation-induced autophagy. In addition, ATG8 lipidation is dispensable for STX17 recruitment to autophagosomes in starvation-induced autophagy (Tsuboyama et al., 2016). In our current study, we found that FLNA functions as a linker between STX17 and ATG8s, recruiting STX17 to autophagosomes. However, we observed phenotypic discrepancies in STX17 recruitment that were cell-type-specific. Specifically, FLNA knockdown only inhibited STX17 recruitment in U2OS cells but not in MEF or HeLa cells (Fig. S8), while Tsuboyama et al. (2016) reported their findings using MEF cells. Collectively, these findings suggest that autophagosomes could employ different recruitment mechanisms for STX17 in various types of cells. These mechanisms may be dependent on ATG8, independent of ATG8, dependent on FLNA, or independent of FLNA.
FLNA-dependent STX17 recruitment is a cell-specific mechanism. (A, D, and G) U2OS, HeLa, and MEF cells stably expressing GFP-STX17 were transfected with siRNA against FLNA. 48 h after transfection, cells were starved with EBSS for 2 h and then stained with LC3B antibody. Scale bar, 5 μm. Inset scale bar, 1 μm. (B, E, and H) Quantification of the number of STX17/LC3 positive puncta in U2OS, HeLa, and MEF cells stably expressing GFP-STX17. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, unpaired two-tailed t test. (C, F, and I) FLNA knockdown efficiency in U2OS, HeLa and MEF cells stably expressing GFP-STX17. Cells were transfected with the siFLNA. 48 h after transfection, cells were subjected to immunoblot with the indicated antibodies. Source data are available for this figure: SourceData FS8.
FLNA-dependent STX17 recruitment is a cell-specific mechanism. (A, D, and G) U2OS, HeLa, and MEF cells stably expressing GFP-STX17 were transfected with siRNA against FLNA. 48 h after transfection, cells were starved with EBSS for 2 h and then stained with LC3B antibody. Scale bar, 5 μm. Inset scale bar, 1 μm. (B, E, and H) Quantification of the number of STX17/LC3 positive puncta in U2OS, HeLa, and MEF cells stably expressing GFP-STX17. Data are mean ± SD (n = 3; 100 cells from three independent experiments were quantified). ****P < 0.0001, ns, no significance, unpaired two-tailed t test. (C, F, and I) FLNA knockdown efficiency in U2OS, HeLa and MEF cells stably expressing GFP-STX17. Cells were transfected with the siFLNA. 48 h after transfection, cells were subjected to immunoblot with the indicated antibodies. Source data are available for this figure: SourceData FS8.
ATG8 family proteins commonly interact with their target proteins via canonical LIR/AIM motifs [W/F/Y]xx[L/I/V] (Birgisdottir et al., 2013). Two other recent studies have identified two recognition motifs (UIM and VLIR) that are distinct from the canonical LIR/AIM motifs (Ma et al., 2022; Marshall et al., 2019). Here, we found that ATG8 family proteins interact with FLNA via three ATG8 binding regions, which we designated LIR1, LIR2, and LIR3. Among them, LIR1 (FVDSL) does not conform to any known LIR motifs. LIR2 (MLLVG) shows similarity to the recently identified VLIR (VLL or VLI) motif, and LIR3 (GSPYRVVVP) matches canonical LIR/AIM motifs. Although LIR3 contains the classical LIR motif (YRVV), mutation of LIR3 (YRVV→ARVA) was not sufficient to disrupt interactions between FLNA and LC3B (Fig. S6, H and I), suggesting that the additional amino acids around the YRVV recognition site remain essential for this interaction. In addition, all LIR mutants of FLNA show a marked decrease in interactions with ATG8s, potentially due to the synergism of ATG8s’ binding activity by these three motifs. Disruption of any single motif could therefore negatively impact the interactions of the other two LIR’s. These results collectively portray a highly sophisticated mode for interactions between ATG8 family proteins and their target proteins. Further structural biology analyses will be required to identify any mechanistic differences in their interaction modes. In addition, immunity-related GTPase M (IRGM) can directly interact with STX17 and ATG8 family proteins and is involved in the recruitment of STX17 to autophagosomes (Kumar et al., 2018). However, the relationship between IRGM-related and FLNA-related mechanisms requires further investigation.
Filamin has three homologs including FLNA, FLNB, and FLNC (Chakarova et al., 2000). However, we found that only FLNA, but not FLNB or FLNC, functions in STX17 recruitment, suggesting that FLNA performs a specialized function in autophagy. STX17 interacts with the FLNA Ig D21-D23 domain, while ATG8s interact with the FLNA H2-Ig D24 domain. The sequence identity in these regions is low (8.67% for A vs. B, 6.09% for A vs. C) and mutations in FLNA described in this study have not been found in either FLNB or FLNC (Fig. 9 D). It is, therefore, possible that sequence divergence among the three filamin homologs in these two regions may be responsible for their observed functional divergence. Further structural investigations of the FLNA-STX17-ATG8 complex may provide more insight into their homolog-specific activities. In addition, one major function of FLNA is to crosslink F-actin (Nakamura et al., 2007). However, we found that disruption of F-actin by latrunculin had no effect on STX17 recruitment to autophagosomes, suggesting that the FLNA functions in autophagy in a manner independent of its role in F-actin. Our discoveries in this study thus significantly expand the scope of known FLNA functions.
To address some possible technical effects of overexpressing STX17 variants, a phenotypic analysis should also be performed in STX17 mutant knock-in cell lines in future works. In addition, differences in basal levels of LC3-II observed among STX17 knockout MEF, FLNA knockdown HEK 293T, and U2OS cells, which all exhibited a marked increase in LC3-II in starvation-induced autophagy compared with control cells, are possibly caused by cell-type-specific expression or differences in treatment or culture conditions (Klionsky et al., 2021a).
Our study thus reveals a previously unknown function of ULK during the late stages of autophagy in mammals and expands our understanding of the actin-independent functions of FLNA. In particular, this study illustrates how FLNA participates as a likely structural component in ATG8 recruitment of STX17 to autophagosomes and suggests that future investigation is warranted to explore a possible role of autophagy in FLNA-related diseases.
Materials and methods
Antibodies and reagents
Mouse monoclonal anti-WIPI2 (Cat#ab237956, Lot#GR3225460.2), Chicken polyclonal anti-GFP (Cat#ab13970, Lot#GR3361051-7,RRID: AB_300798), Rabbit monoclonal anti-FLNA (Cat#ab76289, Lot#GR212664-10, RRID: AB_1523618), and Goat Anti-Chicken IgY H&L (FITC; Cat#ab46969, Lot#GR3179274-12, RRID: AB_2338589) were purchased from Abcam. Rabbit polyclonal anti-pSTX17 SerS289 (1-3), Rabbit polyclonal anti-ULK2 (Cat#A15244, Lot#35166103, RRID: AB_2762141), Rabbit polyclonal anti-FLNB (Cat#A2481, Lot#0203610101, RRID: AB_2764376), Rabbit polyclonal anti-FLNC (Cat#A13018, Lot#0095980201, RRID: AB_2759865), Rabbit Polyclonal SEC61β (Cat#A15788, Lot#0161690201, RRID:AB_2763208), Mouse monoclonal anti-GST (Cat#AE001, Lot#4000057011, RRID: AB_2770403), Rabbit Control IgG (Cat#AC005, Lot#3500000108, RRID: AB_2771930), and Mouse Control IgG (Cat#AC011, Lot#9100011106, RRID: AB_2770414) were purchased from Abclonal. Rabbit monoclonal anti-ULK1 (Cat#8054S, Lot#7), Rabbit monoclonal anti-pAtg13 (Ser355; Cat#46329, Lot#3561064212), Rabbit monoclonal anti-Atg16L1 (Cat#8089, Lot#2), Mouse monoclonal anti- SQSTM1/p62 (Cat# 88588S, Lot#1, RRID: AB_2800125), and Rabbit anti-Cathepsin D (Cat#2284, Lot#2, RRID: AB_10694258) were purchased from CST. Glutathione Sepharose 4B glutathione-sepharose Resin (Cat#17-0756-04), and Ni-NTA agarose resin (Cat#17-5318-06) were purchased from GE healthcare. DMEM (Cat#SH30022.01B) was purchased from Hyclone. Goat anti-Mouse IgG(H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 405 (Cat#A-31553, Lot#2231671, RRID: AB_221604), Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor647 (Cat#A21235, Lot#1915807, RRID:AB_2535804), Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 405 (Cat#A31556, Lot#2273716, RRID: AB_221605) and Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647 (Cat#A21245, Lot#1445259, RRID: AB_2535813) were purchased from Invitrogen. Fluorescein (FITC) AffiniPure Goat Anti-Mouse IgG (H+L; Cat#115-095-003, Lot#136596, RRID: AB_2338589), Fluorescein (FITC) AffiniPure Goat Anti-Rabbit IgG (H+L; Cat#111-095-003, Lot#133027, RRID:AB_2337972), Cy3 AffiniPure Goat Anti-Mouse IgG (H+L; Cat#115-165-003, Lot#117093, RRID: AB_2338680), and Cy3 AffiniPure Goat Anti-Rabbit IgG (H+L; Cat#111-165-003, Lot#128284, RRID: AB_2338000) were purchased from Jackson. Rabbit polyclonal anti-LC3 (Cat#PM036, Lot#036, RRID: AB_2274121), and Mouse monoclonal anti-LC3 (Cat#M152-3, Lot#056, RRID: AB_1279144) were purchased from MBL. Bafilomycin A1 (BFA; Cat#HY-100558) and MRT68921 dihydrochloride (Cat#HY-100006A) were purchased from MCE. Latrunculin B (Cat#MZ5810, Lot#2606Z210844) was purchased from MKBIO. Lambda PP (Cat#P0753L), and ATP (Cat#P0756s, Lot#10153222) were purchased from NEB. Rabbit polyclonal anti-STX17 (Cat#17815-1-AP, Lot#00097453, RRID: AB_2255542), TOM20 Polyclonal antibody (Cat#11802-1-AP, Lot#00068680, RRID: AB_2207530), Rabbit polyclonal anti-HSP60 (Cat#15282-1-AP, Lot#0001350201, RRID: AB_2121440), Rabbit polyclonal anti-TGN46 (Cat#13573-1-AP, Lot#00044796, RRID: AB_10597396), and Mouse monoclonal anti-α-Tubulin (Cat#66031-1-Ig, Lot#10004185, RRID: AB_11042766) were purchased from Proteintech. Rabbit polyclonal anti-GFP (Cat#D110008, Lot#l531AA0031), and Rabbit polyclonal anti-Flag (Cat#D110005, Lot#G928AA0012-0200) were purchased from Sangon Biotech. Protein A/G PLUS-Agarose (Cat#sc-2003, Lot#C2822, RRID: AB_10201400) was purchased from Santa Cruz Biotechnology. PIK-III (Cat#S7683) was purchased from Selleck. Rabbit polyclonal anti-KIAA0652(ATG13; Cat#GB11591, Lot#1), and Rabbit polyclonal anti-Actin (Cat#GB11001, Lot#Ac2111012A, RRID: AB_2801259) were purchased from Service Bio. Anti-Flag M2 Affinity Gel (Cat#A2220, Lot#SLCL1176, RRID: AB_10063035), Anti-Flag M2 magnetic beads (Cat#M8823, Lot#SLCH4826, RRID: AB_2637089), Rabbit monoclonal anti-STX17 (Cat#HPA001204, Lot#000007890, RRID: AB_1080118), Mouse monoclonal anti-Flag M2 (Cat#F3165, Lot#SLCF4933, RRID: AB_259529), Rabbit polyclonal anti-HA (Cat#H6908, Lot#0000086963, RRID: AB_260070), Rabbit polyclonal anti-LC3 (Cat#L7543, Lot#084M4798V, RRID: AB_796155), and Rabbit polyclonal anti-LAMP1 (Cat#L1418, Lot#128M4803V, RRID: AB_477157) were purchased from Sigma-Aldrich. Earle’s balanced salt solution (EBSS; Cat#H2020) was purchased from Solarbio. Goat Anti-Mouse IgG(H+L)-HRP (Cat#1036-05, Lot#D1912-SL71D, RRID: AB_2794348), and Goat Anti-Rabbit IgG-HRP (Cat#4010-05, Lot#A0422-PL42, RRID: AB_2687483) were purchased from SouthernBiotech. Phalloidin-iFluor 647 Conjugate (Cat#23127, Lot#268003) was purchased from AAT Bioquest. Mouse monoclonal anti-6×His (Cat#A2050, Lot#ATUSE1801) was purchased from Abbkine.
Cell culture and transfection
HEK293T, U2OS, MEF, HeLa cells (Zhou et al., 2022), and HeLa-ATG8 KO cells (Nguyen et al., 2016) were grown in DMEM (Hyclone) containing 10% FBS (Gibco) and 1% penicillin-streptomycin solution (Beyotime) at a temperature of 37°C and a CO2 level of 5%. Transient transfection of plasmids in HEK293T and HeLa cells was performed using PEI following the manufacturer’s instructions, and the cells were analyzed after 24 h. Lipofectamine RNAiMAX (Invitrogen) was used to introduce siRNA duplexes into cells, and the cells were harvested 48 h after transfection. For starvation treatment, cells were washed with PBS (Hyclone) three times and then incubated with EBSS for 2 h or treated with Torin2 (100 nM) for 2 h. Cells were incubated with bafilomycin A1 for 2 h to block autophagy, MRT68921 dihydrochloride (1 μM) was used to inhibit ULK kinase activity, and latrunculin B (10 μM) was used to inhibit actin polymerization.
siRNA sequences
Individual siRNAs (GenePharma) were transfected by using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. The targeting sequences were: NC: 5′-UUCUUCGAACGUGUCACGUTT-3′; hFLNA#1: 5′-GUGACCGCCAAUAACGACA-3′; hFLNA#2: 5′-CCCACCCACTTCACAGTAAAT-3′; hFLNB#1: 5′-AAGCUCCCUUAAAGAUAUUUG-3′; hFLNB#2: 5′-AAGGUCCUUCCCACAUAUGAU-3′; hFLNB#3: 5′-AACACCUGAAGGGUACAAAGU-3′; hFLNC#1: 5′-GAGCAUUCUGUUAUGACCUACCUGU-3′; hFLNC#2: 5′-UCAAAGCCACCAUUCGGCCUGUGUU-3′.
Immunoprecipitation assays
HEK293T cells were rinsed in ice-cold PBS and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 1 μg/ml Aprotinin, 1 μg/ml Pepstatin A, 1 μg/ml Leupeptin, 200 mM NaF, and 200 mM Na3VO4) for 10 min at 4°C. The supernatants were then subjected to immunoprecipitation using anti-Flag M2 Affinity Gel for 12 h at 4°C after centrifugation at 12,000 g for 10 min at 4°C. The precipitated immunocomplexes were washed three times in lysis buffer and eluted with the 3× Flag peptide for 2 h at 4°C, followed by boiling in 2× sample buffer. The samples were then subjected to SDS-PAGE and analyzed by immunoblotting. For co-immunoprecipitation, HEK293T cells were washed with ice-cold PBS, lysed in lysis buffer for 10 min at 4°C, and centrifugated at 12,000 g for 10 min at 4°C. The supernatants were immunoprecipitated using antibodies against ULK1, STX17, or FLNA for 12 h at 4°C, with IgG as the negative control. The precipitated immunocomplexes were incubated with protein A/G PLUS-agarose for 12 h at 4°C, and then the samples were washed three times in lysis buffer and boiled in 2× sample buffer for immunoblotting. The samples were then subjected to SDS-PAGE and analyzed by immunoblotting.
Western Blotting and dot blot
After being heated to 110°C for 10 min, the SDS sample buffer was used to lyse the cells. The resulting samples were subjected to SDS-PAGE electrophoresis and transferred to a polyvinylidene fluoride (PVDF) membrane (162-0177; Bio-Rad). The membrane was then blocked with 5% (m/v) skimmed milk at room temperature for 1 h and then incubated with primary antibodies at 37°C for 1 h or 4°C overnight. After washing three times, membranes were incubated with secondary antibodies that were conjugated with horseradish peroxidase (1/10,000) for 1 h at room temperature. Visualization was performed using enhanced chemiluminescence (P0018M-2; Beyotime), and the resulting images were developed using an OPTIMAX x-ray film processor (Protec GmbH & Co.). Quantitative analysis of the protein bands was carried out using ImageJ software.
Synthesized peptides with an overlapping sequence in the FLNA H2-D24 domain were immobilized on the nitrocellulose membrane (HATF00010; Sigma-Aldrich) and allowed to dry at 37°C for 30 min. The membrane was blocked with 5% (m/v) skimmed milk and incubated with His-tagged LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2 at a concentration of 10 μg/ml for 24 h at 4°C. The membrane was then washed three times and visualized by immunoblotting using an anti-His antibody.
Immunofluorescence assays
Coverslip-grown cells were rinsed with PBS and fixated for 15 min at room temperature in 4% paraformaldehyde. After washing three times with PBS, the cells were incubated with primary antibodies overnight at 4°C or for 1 h at room temperature. The primary antibodies were diluted in a solution containing 10% FBS and 0.1% saponin. After washing three times with PBS, the cells were incubated with Alexa Fluor 405/488/Cy3/647–conjugated goat anti-mouse, anti-chicken, or anti-rabbit IgG secondary antibodies for 1 h at room temperature. Prior to fixation, the cells were treated with 50 μg/ml digitonin (D141; Sigma-Aldrich) in KHM buffer (125 mM potassium acetate, 25 mM Hepes, pH 7.2, and 2.5 mM magnesium acetate) for 1 min for U2OS and 3 min for HeLa cells. Following two washes with KHM buffer, the cells were fixed. The slides were mounted, and images were acquired using a laser scanning confocal microscope (FV3000, Olympus), Plan Apochromat N 100×/1.49 oil at room temperature. Using a confocal laser microscope system, images were captured, processed, and analyzed using OlyVIA and ImageJ 1.52a.
In vitro dephosphorylation assays
In this experiment, HEK293T cells were used to investigate the effect of ULK1 and ULK2 on the phosphorylation of HA-STX17 and endogenous STX17 proteins. The cells were either transfected with ULK1 or ULK2 and HA-STX17 or left without transfection. After 24 h, the cells were lysed and the supernatants containing phosphorylated HA-STX17 or endogenous STX17 proteins were incubated with Lambda Protein Phosphatase (10 U/μl) to dephosphorylate the proteins at 37°C for 2 h. The samples were then boiled and subjected to immunoblotting analysis.
Protein purification and in vitro binding assay
E. coli T7 was modified to express the various genes (STX17, SNAP29, VAMP8, ULK1, ULK2, LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2) from bacterial expression constructs (pGEX-6P-1 or pET-28a). Cells were induced to protein overexpression under 0.4 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) at 16°C overnight. After that, the cells were then resuspended in either GST lysis buffer (350 mmol NaCl, 20 mmol Tris-HCl, pH 7.5, 1 mmol EDTA, and 1% Triton X-100) or His lysis buffer (350 mmol NaCl, 20 mmol Tris HCl, pH 7.5, 20 mmol imidazole, and 1% Triton X-100) and ultrasonically processed. Using Glutathione-Sepharose resin or Ni-Affinity resin and following the manufacturer's instructions, the proteins were purified in a single step. In pull-down assays, His or GST-tagged proteins were applied to Ni-Affinity resin or GST resin, respectively, and then incubated with 1 μg of the corresponding proteins in binding buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40) and supplemented with protease inhibitor cocktail (Roche) for 2 h at 4°C. After being washed three times, proteins were eluted and dissolved in sample buffer for SDS-PAGE and immunoblotted.
In vitro kinase assay
Purified bacterial GST-STX17 proteins (5 μg) were used for kinase assay as substrates to determine phosphorylation sites. For the kinase assay, ULK1 and ULK1 (K46N) were prepared from HEK293 cells, and His-ULK1 and His-ULK2 were prepared from E. coli. The purified ULK proteins were incubated in kinase assay buffer: 50 mM HEPES at pH 7.4, 2 mM EDTA, 10 mM MgCl2, 2 mM DTT (dithiothreitol), and 20 µM ATP. The kinase reaction was performed at 30°C for 30 min and the reaction was terminated by adding SDS loading buffer and subjected to immunoblot with antibodies against Flag, His, and GST.
Mass spectrometry
All samples were redissolved in 0.1% formic acid and ∼100 ng of each sample was injected into a nanoElute high-performance liquid chromatography (HPLC) system (Bruker) connected to a Bruker timsTOF Pro mass spectrometer (Bruker). The peptide sample was loaded directly onto the analytical column (Aurora, 25 cm × 75 μm, 1.6 μm; IonOpticks). The elution gradient was set to 2–28% buffer B for 100 min, 28%–45% for 10 min at 300 nL/min (mobile phases A: 0.1% formic acid in water; mobile phases B: 0.1% formic acid in acetonitrile). Tryptic peptides were ionized at a spray voltage of 1.5 kV and subsequently injected into the mass spectrometer (MS) system. Mass spectra were obtained through data-dependent acquisition (DDA) in PASEF (Parallel Accumulation Serial Fragmentation) model. All raw data were analyzed using the PEAKS Online toolkit (Bioinformatics Solutions). Methylation modification of Cys residues was set as the fixed modification, while methionine oxidation and phosphorylation of serine, threonine, or tyrosine residues were considered variable modifications. The false discovery rate for proteins and peptides was set to 0.01, and mass tolerance thresholds were set to 10 ppm for both MS ions and MS/MS fragment ions.
Cell fractionation and proteinase K protection assay
Cell fractionation was carried out as previously described (Matsui et al., 2018; Uematsu et al., 2017). Briefly, MEF cells from 10 15-cm dishes, or HEK293T cells from 3 15-cm dishes stably expressing Flag-STX17 WT, Flag-STX17 S289A, or Flag-STX17 S289E were starved for 2 h with or without EBSS. The harvested cells were then rinsed twice with ice-cold PBS. After centrifugation at 500 g for 5 min, the cell pellets were collected and dispersed in 1 ml ice-cold homogenization buffer (250 mM sucrose, 20 mM Hepes-KOH, pH 7.4, 1 mM EDTA, and complete EDTA-free protease inhibitor). 2 ml Dounce homogenizer was used to homogenize cells. The cell homogenate was then centrifuged for 10 min at 1,000 g to remove cell debris and intact cells. The resultant supernatant was centrifuged at 10,000 g for 10 min, followed by centrifugation at 20,000 g for 20 min to collect the pellet. Subsequently, the pellet was then diluted with 1 ml 30% OptiPrep. In a centrifuge tube (326819; Beckman Coulter) for ultracentrifuge rotors (MLS 50; Beckman Coulter), discontinuous OptiPrep gradients were generated by overlaying the following OptiPrep solutions in homogenization buffer: 982 μl of the supernatant in 30% OptiPrep (sample), 738 μl 20% OptiPrep, 820 μl 15% OptiPrep, 820 μl 10% OptiPrep, 820 μl 5% OptiPrep, and 820 μl 0% OptiPrep. The gradient was centrifuged at 150,000 g using ultracentrifuge for 5 h and ultimately 14 fractions (350 μl each) were collected from the top.
Three 15-cm dishes of HEK293T cells expressing Flag-STX17 WT were starved for 2 h with EBSS and then subjected to cell fractionation as described above. Fractions from the three (0.9 ml) were mixed with 45 μl of 5 M NaCl and 70 μl anti-Flag M2 magnetic beads and rotated overnight at 4°C. The magnetic beads were washed three times with 250 mM NaCl and resuspended in ice-cold PS200 buffer (20 mM K-PIPES, pH 6.8, 250 mM sucrose, 5 mM MgCl2). The resultant product was divided into three aliquots and treated as follows: (A) PS200 alone (no treatment control); (B) 0.1–10 μg/ml proteinase K; (C) 0.1–10 μg/ml proteinase K with 0.2% Triton X-100. Samples were then incubated for 5 min on ice. The final products were dissolved in the sample buffer for immunoblotting.
Statistical analysis
The statistical analysis was performed using Prism software (GraphPad) and the error bars in the figures represented the SD or SEM. Unpaired t tests, one-way ANOVA, and two-way ANOVA were performed for statistical analysis. The experiments were repeated at least three times. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Table S1 shows the mass spectrometric analysis of STX17 phosphorylation by ULK1. Fig. S1 shows the localization to autophagosomes and band shift of STX17 dephosphorylation mimic variants. Fig. S2 shows the absence of colocalization between STX17 pS289 and ER or mitochondria. Fig. S3 shows that dephosphorylation of STX17 at S289 does not affect autophagosome formation and SNARE assembly. Fig. S4 shows FLNA interacts with STX17 and does not localize to isolation membranes. Fig. S5 shows that the depletion of FLNB and FLNC does not affect the recruitment of STX17 to autophagosomes. Fig. S6 shows the mapping analysis of the interaction region of FLNA with ATG8s. Fig. S7 shows the quantification of autophagic flux in Figure 8. Fig. S8 shows that FLNA-dependent STX17 recruitment is a cell-specific mechanism.
Data availability
The authors declare that all relevant data supporting the findings of this study are available within the paper and its supplemental files. The proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifiers PXD041897 and PXD043082.
Acknowledgments
We are deeply grateful to Li Yu (Tsinghua University), Wei Liu (Zhejiang University), Qing Zhong (Shanghai Jiao Tong University), Qiming Sun (Zhejiang University), Liang Ge (Tsinghua University), and Yixian Cui (Wuhan University) for helpful suggestions on this study. We thank Peter Carmeliet (KU Leuven; Vesalius Research Center; Leuven, Belgium; [Segura et al., 2016]) and E. Peverelli (Milan University, Milan, Italy [Peverelli et al., 2018]) for gifting us the FLNA-related plasmids. We thank Liang Ge (Tsinghua University, Beijing, China [Ma et al., 2022]) for gifting us the mATG8s plasmids.
The work was supported by grants from the National Natural Science Foundation of China (92254302, 91854116, 31771529, and 32170685 to Y. Rong) and Fundamental Research Funds for the Central Universities (2023BR028 to Y. Rong).
Author contributions: Y. Rong and Y. Wang designed the experiments and conducted the experiments. Y. Wang, C. Li, Z. Wu, H. Que, F. Jian, and Y. Zhao performed the biological and biochemical experiments. S. Gao, C.L. Wong., C. Zhao, and H. Tang conducted mass spectrometry analyses. Y. Wang and Y. Rong analyzed the data and wrote the manuscript with the help of all authors.
References
Author notes
Disclosures: The authors declare no competing interests exist.