Septins are cytoskeletal proteins conserved from algae and protists to mammals. A unique feature of septins is their presence as heteromeric complexes that polymerize into filaments in solution and on lipid membranes. Although animal septins associate extensively with actin-based structures in cells, whether septins organize as filaments in cells and if septin organization impacts septin function is not known. Customizing a tripartite split-GFP complementation assay, we show that all septins decorating actin stress fibers are octamer-containing filaments. Depleting octamers or preventing septins from polymerizing leads to a loss of stress fibers and reduced cell stiffness. Super-resolution microscopy revealed septin fibers with widths compatible with their organization as paired septin filaments. Nanometer-resolved distance measurements and single-protein tracking further showed that septin filaments are membrane bound and largely immobilized. Finally, reconstitution assays showed that septin filaments mediate actin–membrane anchoring. We propose that septin organization as octamer-based filaments is essential for septin function in anchoring and stabilizing actin filaments at the plasma membrane.
Introduction
Septins comprise a family of cytoskeletal proteins conserved from algae and protists to mammals (Cao et al., 2007; Momany et al., 2008; Nishihama et al., 2011; Pan et al., 2007). Septins were discovered in budding yeast as mutants that result in cytokinesis defects (Hartwell, 1971; Hartwell et al., 1970) and were later shown to be also required for animal cell division (Echard et al., 2004; Estey et al., 2010; Founounou et al., 2013; Kechad et al., 2012; Kinoshita et al., 1997; Neufeld and Rubin, 1994; Surka et al., 2002). However, septins are expressed in practically all human tissues, including non-dividing neurons (Karlsson et al., 2021). Septins play roles in a wide range of biological processes in non-dividing cells and tissues, including cell motility, sperm integrity, neuron development, tissue morphogenesis, and host–pathogen interactions (Fares et al., 1995; Finger et al., 2003; Gilden et al., 2012; Ihara et al., 2005; Kim et al., 2010; Kissel et al., 2005; Kuo et al., 2012; Mostowy et al., 2010; Mostowy et al., 2011; Nguyen et al., 2000; Shindo and Wallingford, 2014; Steels et al., 2007; Tada et al., 2007; Tooley et al., 2009; Xie et al., 2007). The embryonic lethality of mouse and Drosophila septin knock-outs (Adam et al., 2000; Fuchtbauer et al., 2011; Menon et al., 2014; Roseler et al., 2011) emphasizes their essential contribution to animal physiology, yet the precise molecular basis of septin function remains elusive.
Biochemical isolation of native septins revealed that septins exist as stable heteromeric complexes that can polymerize into filaments (Field et al., 1996; Frazier et al., 1998; Hsu et al., 1998; Kim et al., 2011; Kinoshita et al., 2002; Sellin et al., 2011). The isolation of recombinant septin complexes established that septin complexes are palindromes, with each septin in two copies and in a specific position within the complex. Each monomer interacts with its neighbors by alternating interfaces, named NC (from the N- and C-terminal domains) and G (from the GTP-binding domain; Bertin et al., 2008; DeRose et al., 2020; Farkasovsky et al., 2005; Garcia et al., 2011; Huijbregts et al., 2009; Iv et al., 2021; John et al., 2007; Kinoshita et al., 2002; Kumagai et al., 2019; Mavrakis et al., 2014; Mendonca et al., 2019; Rosa et al., 2020; Sala et al., 2016; Sirajuddin et al., 2007; Soroor et al., 2021; Versele and Thorner, 2004). Human septins are classified in four homology groups, namely, the SEPT2 group (SEPT1, 2, 4, and 5), SEPT6 group (SEPT6, 8, 10, 11, and 14), SEPT7 group (SEPT7), and SEPT3 group (SEPT3, 9, and 12; Kinoshita, 2003). Cell-isolated human septins exist as stable hexamers and octamers (Kim et al., 2011; Sellin et al., 2011; Sellin et al., 2014), with hexamers composed of septins from the SEPT2, SEPT6, SEPT7 groups, and octamers containing additional septins from the SEPT3 group (Fig. 1 A).
The most convincing evidence that septins form filaments in vivo and thereby contribute to cell viability comes from electron microscopy and functional data in budding yeast (Bertin et al., 2012; Byers and Goetsch, 1976; McMurray et al., 2011; Ong et al., 2014; Rodal et al., 2005). The conservation of septins and the ability of mammalian septin hexamers and octamers (hereafter referred to as protomers) to form filaments in solution and on lipid membranes (DeRose et al., 2020; Iv et al., 2021; Leonardo et al., 2021; Soroor et al., 2021; Szuba et al., 2021) has led to the assumption that human septins also organize as filaments in cells, but formal evidence for this is scarce. Immunogold electron microscopy has shown septins closely apposed to cortical actin filaments and to the plasma membrane organizing in linear arrays (Hagiwara et al., 2011; Kinoshita et al., 1997). Septin protomers along actin filaments or the membrane would, however, result in a similar pattern. It is reasonable to assume that septin rings and fiber-looking segments that form in the cytoplasm of mammalian cells upon actin depolymerization correspond to septin filaments or bundles thereof (Joo et al., 2007; Kim et al., 2011; Kinoshita et al., 2002; Schmidt and Nichols, 2004). However, it is unknown if these fibers originate from direct end-to-end septin polymerization. Whether all septins in cells function as filaments, and how hexamers and octamers contribute to septin filament formation and function is not known.
Actin-binding domains on septins have not yet been identified. It is hence unclear if actin-septin binding involves direct interactions or if it occurs through myosin-II (Joo et al., 2007; Mostowy et al., 2010) or Borg proteins (Calvo et al., 2015; Farrugia et al., 2020; Joberty et al., 2001; Liu et al., 2014; Salameh et al., 2021). Similarly, although mammalian septins bind lipid membranes (Bridges et al., 2016; Dolat and Spiliotis, 2016; Szuba et al., 2021; Tanaka-Takiguchi et al., 2009; Yamada et al., 2016), whether there is direct septin-membrane binding in cells has not been shown; the identification of the membrane-binding site of septins is a matter of debate (Cavini et al., 2021). It is thus not known if septin-decorated actin fibers and membranes in cells reflect membrane-bound septins.
To elucidate the interplay between human septin organization and function in non-dividing cells, we used actin stress fibers in U2OS cells as a model system. Septins in mammalian cells have been reported to decorate stress fibers in multiple studies (Calvo et al., 2015; Connolly et al., 2011; Dolat et al., 2014; Joo et al., 2007; Kim et al., 2011; Kinoshita et al., 2002; Kinoshita et al., 1997; Liu et al., 2014; Salameh et al., 2021; Schmidt and Nichols, 2004; Surka et al., 2002; Verdier-Pinard et al., 2017; Xie et al., 1999; Zhang et al., 1999). Subsets of stress fibers are lost upon septin disruption or septin relocalization to microtubules (Calvo et al., 2015; Kinoshita et al., 2002; Kuzmic et al., 2022; Salameh et al., 2021; Schmidt and Nichols, 2004; Targa et al., 2019) suggesting an essential, yet still unclear, role of septins in stress fiber integrity. To test how septins organize in cells, we combined a tripartite split-GFP complementation assay with mutants disrupting specific septin–septin interfaces to selectively perturb hexamers or octamers, or abolish polymerization altogether. Atomic force microscopy nanoindentation was used to assess the specific contribution of hexamers vs. octamers to cell stiffness. We employed super-resolution structured illumination microscopy to decipher the higher-order assembly of septin filaments. Moreover, to determine whether septin filaments are membrane-bound and if they can bridge membrane-actin interactions, we combined nanometer-resolved distance measurements and single protein tracking in cells with cell-free reconstitution assays using supported lipid bilayers. Our findings demonstrate that all actin-associated septins in cells organize as paired membrane-bound filaments whose integrity and function depend on octamers.
Results
Septins associate with contractile stress fibers
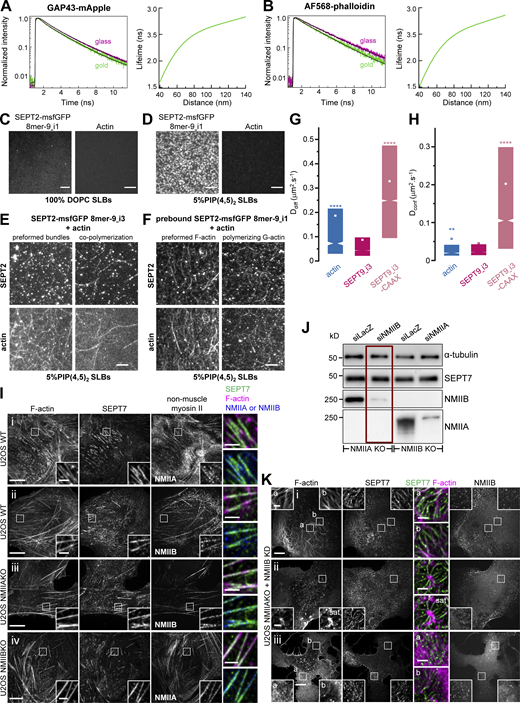
Whether septins associate preferentially with specific types of SFs and if septin organization differs among SFs is not known. To answer these questions, we examined how septins distribute in U2OS cells, with respect to peripheral, dorsal, and ventral SFs, transverse arcs and the perinuclear actin cap (Fig. 1 A). Given that SFs are classified based on their subcellular localization and their anchoring at one or both ends by focal adhesions (FAs; Tojkander et al., 2012), we co-stained for septins, actin filaments and the FA protein, paxillin. We examined the distribution of SEPT2 and SEPT7, which are common to both hexamers and octamers, and SEPT9, which is specific to octamers (Fig. 1, A and B; and Fig. S1). U2OS cells express two SEPT9 isoforms, SEPT9_i1 and SEPT9_i3 (Kuzmic et al., 2022), both of which are detected by our SEPT9 antibodies. Both septin immunostainings and live imaging of septin-GFP fusions showed identical distributions of all three septins with respect to SFs. They all decorated myosin-II containing contractile SFs (Fig. 1 B i–iii; Fig. S1 A i, ii, and iv; and Fig. S1 B i–v), but not the non-contractile dorsal ones (Fig. 1, B iii and b; Fig. S1, A iii; and Fig. S1, B iv and a). Although septins decorated contractile SFs throughout their length, they were systematically excluded from FAs (Fig. 1, B i and a; Fig. S1, A i and c; and Fig. S1, B i and c). Septins localized to peripheral and ventral SFs, transverse arcs and perinuclear actin caps, and also associated with geodesic actin nodes on the ventral plasma membrane and with actin nodes in transverse arcs. Actin nodes were enriched in F-actin and α-actinin, while actin filaments interconnecting nodes were decorated by septins and myosin-II in an aster-like pattern (Fig. S1, C and D).
A tripartite split-GFP complementation assay detects SEPT2-SEPT2 interactions on septin-decorated actin stress fibers in U2OS cells. (A) The scheme on the left depicts septin-decorated stress fibers (SFs) in a mammalian cell. (B) Septins (green) decorate different types of actin SFs (red), notably peripheral and ventral SFs (i), perinuclear actin caps (ii), and transverse arcs (iii), shown in the respective panels i–iii in B. The schematics on the right show human septin octamers and hexamers associating with SFs either as single protomers (top), as octamer and hexamer-driven filaments (middle), or as hybrid filaments from octamer and hexamer co-polymerization (bottom). Single protomers (top) could co-exist with filaments (middle and bottom). Septins can associate exclusively with SFs or also with the plasma membrane. (B) Representative confocal micrographs of SEPT7 immunostainings showing examples of SEPT7 localizing (i) to ventral (a and b) and peripheral (c) SFs and excluded from focal adhesions (FA; a), (ii) to perinuclear actin caps (a and b), and (iii) to transverse arcs (a) and excluded from dorsal SFs (b). Cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. (C) Schematic representation of the tripartite split-GFP complementation assay for probing SEPT2-SEPT2 interactions. The transparency of the SEPT7 subunits is used to suggest that the polymerizing protomers can be hexamers or/and octamers. (D and E) Representative confocal micrographs of SEPT2-SEPT2 reconstituted GFP (rGFP) distribution in fixed cells (D) co-stained for F-actin (phalloidin) and in live cells (E). Examples in fixed cells show rGFP localizing (i) to ventral SFs (a and b) and (ii) to actin caps (a and b). Examples in live cells show rGFP localizing (i) to peripheral (a) and ventral (b) SFs, (ii) to transverse arcs (a and b), and (iii) to actin caps (a and b). (F) Representative confocal micrograph of SEPT2-SEPT2 rGFP in fixed cells using recombinant purified GFP1-9. The example shows rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm.
A tripartite split-GFP complementation assay detects SEPT2-SEPT2 interactions on septin-decorated actin stress fibers in U2OS cells. (A) The scheme on the left depicts septin-decorated stress fibers (SFs) in a mammalian cell. (B) Septins (green) decorate different types of actin SFs (red), notably peripheral and ventral SFs (i), perinuclear actin caps (ii), and transverse arcs (iii), shown in the respective panels i–iii in B. The schematics on the right show human septin octamers and hexamers associating with SFs either as single protomers (top), as octamer and hexamer-driven filaments (middle), or as hybrid filaments from octamer and hexamer co-polymerization (bottom). Single protomers (top) could co-exist with filaments (middle and bottom). Septins can associate exclusively with SFs or also with the plasma membrane. (B) Representative confocal micrographs of SEPT7 immunostainings showing examples of SEPT7 localizing (i) to ventral (a and b) and peripheral (c) SFs and excluded from focal adhesions (FA; a), (ii) to perinuclear actin caps (a and b), and (iii) to transverse arcs (a) and excluded from dorsal SFs (b). Cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. (C) Schematic representation of the tripartite split-GFP complementation assay for probing SEPT2-SEPT2 interactions. The transparency of the SEPT7 subunits is used to suggest that the polymerizing protomers can be hexamers or/and octamers. (D and E) Representative confocal micrographs of SEPT2-SEPT2 reconstituted GFP (rGFP) distribution in fixed cells (D) co-stained for F-actin (phalloidin) and in live cells (E). Examples in fixed cells show rGFP localizing (i) to ventral SFs (a and b) and (ii) to actin caps (a and b). Examples in live cells show rGFP localizing (i) to peripheral (a) and ventral (b) SFs, (ii) to transverse arcs (a and b), and (iii) to actin caps (a and b). (F) Representative confocal micrograph of SEPT2-SEPT2 rGFP in fixed cells using recombinant purified GFP1-9. The example shows rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm.
SEPT2, SEPT7 and SEPT9 distribution on different types of stress fibers in U2OS cells. (A) Representative confocal micrographs of SEPT2 immunostained cells (i–iii) and cells expressing SEPT2-msfGFP (iv and v). SEPT2 immunostained cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. Examples show SEPT2 localizing (i) to peripheral (a) and ventral (b and c) SFs and excluded from focal adhesions (FAc) , (ii) to peripheral (a) and perinuclear actin caps (b), (iii) to transverse arcs (b) and excluded from dorsal SFs (a and b), (iv) to transverse arcs (a and b) and excluded from dorsal SFs (a), and (v) showing a diffuse cytosolic phenotype. (B) Representative confocal micrographs of SEPT9 immunostained cells (i–iii) and cells expressing SEPT9_i3-msfGFP (iv and v). SEPT9 immunostained cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. Examples show SEPT9 localizing (i) to peripheral (a) and ventral (b and c) SFs and excluded from focal adhesions (FA; c), (ii) to perinuclear actin caps (a and b), (iii) to transverse arcs (a) and ventral SFs (b), (iv) to transverse arcs (a) and excluded from dorsal SFs (a) and to ventral SFs (b), and (v) to peripheral (a) and perinuclear actin caps (b). (C and D) Representative confocal micrographs of SEPT7 immunostainings showing SEPT7 localizing to ventral actin nodes. Cells are co-stained for F-actin (phalloidin) and non-muscle myosin heavy chain IIA (NMIIA; C) or α-actinin (D). Yellow arrowheads point to two actin nodes in each example. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm (A and B) and 5 μm (C and D). Related to Fig. 1 B.
SEPT2, SEPT7 and SEPT9 distribution on different types of stress fibers in U2OS cells. (A) Representative confocal micrographs of SEPT2 immunostained cells (i–iii) and cells expressing SEPT2-msfGFP (iv and v). SEPT2 immunostained cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. Examples show SEPT2 localizing (i) to peripheral (a) and ventral (b and c) SFs and excluded from focal adhesions (FAc) , (ii) to peripheral (a) and perinuclear actin caps (b), (iii) to transverse arcs (b) and excluded from dorsal SFs (a and b), (iv) to transverse arcs (a and b) and excluded from dorsal SFs (a), and (v) showing a diffuse cytosolic phenotype. (B) Representative confocal micrographs of SEPT9 immunostained cells (i–iii) and cells expressing SEPT9_i3-msfGFP (iv and v). SEPT9 immunostained cells are co-stained for F-actin (phalloidin) and the FA protein paxillin. Examples show SEPT9 localizing (i) to peripheral (a) and ventral (b and c) SFs and excluded from focal adhesions (FA; c), (ii) to perinuclear actin caps (a and b), (iii) to transverse arcs (a) and ventral SFs (b), (iv) to transverse arcs (a) and excluded from dorsal SFs (a) and to ventral SFs (b), and (v) to peripheral (a) and perinuclear actin caps (b). (C and D) Representative confocal micrographs of SEPT7 immunostainings showing SEPT7 localizing to ventral actin nodes. Cells are co-stained for F-actin (phalloidin) and non-muscle myosin heavy chain IIA (NMIIA; C) or α-actinin (D). Yellow arrowheads point to two actin nodes in each example. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm (A and B) and 5 μm (C and D). Related to Fig. 1 B.
The presence of SEPT9 on SFs does not exclude that septin hexamers are also present. Furthermore, the diffraction-limited optical resolution of our setup cannot distinguish septin protomers from filaments. Septin decoration of SFs may therefore reflect the presence of protomers (hexamers and/or octamers) or/and of filaments (with hexamers and octamers forming separate or hybrid filaments; Fig. 1 A).
Septins organize as filaments on contractile SFs
Both protomers have an exposed SEPT2 NC interface at their termini (Fig. 1 A; Iv et al., 2021; Mendonca et al., 2019; Soroor et al., 2021). To determine whether septins are present as filaments, we therefore designed a tripartite split-GFP complementation assay for probing SEPT2–SEPT2 interactions as a molecular readout of end-to-end septin polymerization in situ in living cells (Fig. 1 C). This protein–protein interaction assay involves the fusion of the proteins of interest to the two last beta-strands of GFP, β10 and β11: in the presence of specific protein–protein interactions in cells expressing GFP1-9 (GFP strands β1-β9), the GFP barrel is reconstituted leading to fluorescence (Cabantous et al., 2013; see Materials and methods and Fig. S2, A–C for the assay design). We generated β10- and β11-strand fusions with SEPT2 that we co-expressed using an inducible bidirectional vector in U2OS cells constitutively expressing GFP1-9 (Fig. 1 C and Fig. S2 D). To minimize the risk of not detecting SEPT2–SEPT2 interactions due to endogenous untagged SEPT2 and given that the expression levels of SEPT2-β10/β11 fusions were kept low to minimize overexpression artifacts (Fig. S2 E), we consistently knocked down endogenous SEPT2 in all subsequent experiments (Fig. S2 F).
Design of the tripartite split-GFP complementation assay for probing septin organization. (A) Sequences of the β10- and β11-tags used for all split assays and of the linker sequences tested in screening experiments (B and C); 14-residue linkers were used throughout this study. (B) Schematic of N- and C-terminal β10- and β11-tag septin fusions tested in screening experiments (C) using short or long linkers (A). (C) Schematic of β10- and β11-septin fusion combinations for screening tripartite split GFP complementation. Combinations with the same pattern were considered to be equivalent (for example, SEPT2-14-β10/β11-14-SEPT2 and SEPT2-14-β11/β10-14-SEPT2). The combinations in green are the ones tested experimentally. (D) Schematic of the pTRIP TRE Bi vector bearing a bidirectional tetracycline response element (TRE) promoter for the doxycycline-inducible co-expression of β10- and β11-tagged septins. An IRES-TagBFP cassette was used for monitoring septin expression. Restriction sites used for subcloning are indicated (see methods for details). (E) Left: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT7, anti-β10 and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT7 (siSEPT7), and targeting SEPT7 while co-expressing wild-type β10- and β11-SEPT7 (WT), β10- and β11-SEPT7Gmut1 (Gmut1), and β10- and β11-SEPT7Gmut2 (Gmut2). Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT7 and β10 blots are also shown saturated on purpose for displaying weaker bands. Molecular weight markers are shown on the left. Middle: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT2, anti-β10, anti-β11, and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT2 (siSEPT2), and targeting SEPT2 while co-expressing wild-type SEPT2-β10 and -β11 (WT) or SEPT2NCmut-β10 and -β11 (NCmut). Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT2 blot is also shown saturated on purpose for displaying weaker bands. Right: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT9, anti-β10, anti-β11, and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT9 (siSEPT9), and targeting SEPT9 while co-expressing wild-type SEPT9-β10 and -β11 (WT) or SEPT9NCmut-β10 and -β11 (NCmut) for both SEPT9_i1 and SEPT9_i3. Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT9 blot is also shown saturated on purpose for displaying weaker bands. Asterisks point to SEPT9 degradation products. (F) Western blot of U2OS cell lysates probed with anti-SEPT2 and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl) or SEPT2 (siSEPT2). Molecular weight markers are shown on the left. Bottom, respective quantification of SEPT2 protein levels (mean + SD). Mean values (normalized to 1 for siCtrl) are from three independent siCtrl and nine independent siSEPT2 treatments. SEPT2 was knocked down on average by 92%. (G) Same as F for SEPT9. The asterisk points to a SEPT9 degradation product. Mean values (normalized to 1 for siCtrl) are from three independent siCtrl and three independent siSEPT9 treatments. SEPT9 was knocked down on average by 92%. (H) Same as F for SEPT7. Mean values (normalized to 1 for siCtrl) are from 3 independent siCtrl and 12 independent siSEPT7 treatments. SEPT7 was knocked down on average by 86%. Source data are available for this figure: SourceData FS2.
Design of the tripartite split-GFP complementation assay for probing septin organization. (A) Sequences of the β10- and β11-tags used for all split assays and of the linker sequences tested in screening experiments (B and C); 14-residue linkers were used throughout this study. (B) Schematic of N- and C-terminal β10- and β11-tag septin fusions tested in screening experiments (C) using short or long linkers (A). (C) Schematic of β10- and β11-septin fusion combinations for screening tripartite split GFP complementation. Combinations with the same pattern were considered to be equivalent (for example, SEPT2-14-β10/β11-14-SEPT2 and SEPT2-14-β11/β10-14-SEPT2). The combinations in green are the ones tested experimentally. (D) Schematic of the pTRIP TRE Bi vector bearing a bidirectional tetracycline response element (TRE) promoter for the doxycycline-inducible co-expression of β10- and β11-tagged septins. An IRES-TagBFP cassette was used for monitoring septin expression. Restriction sites used for subcloning are indicated (see methods for details). (E) Left: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT7, anti-β10 and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT7 (siSEPT7), and targeting SEPT7 while co-expressing wild-type β10- and β11-SEPT7 (WT), β10- and β11-SEPT7Gmut1 (Gmut1), and β10- and β11-SEPT7Gmut2 (Gmut2). Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT7 and β10 blots are also shown saturated on purpose for displaying weaker bands. Molecular weight markers are shown on the left. Middle: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT2, anti-β10, anti-β11, and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT2 (siSEPT2), and targeting SEPT2 while co-expressing wild-type SEPT2-β10 and -β11 (WT) or SEPT2NCmut-β10 and -β11 (NCmut). Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT2 blot is also shown saturated on purpose for displaying weaker bands. Right: Western blots of U2OS-Tet-On-GFP1-9 cell line lysates probed with anti-SEPT9, anti-β10, anti-β11, and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT9 (siSEPT9), and targeting SEPT9 while co-expressing wild-type SEPT9-β10 and -β11 (WT) or SEPT9NCmut-β10 and -β11 (NCmut) for both SEPT9_i1 and SEPT9_i3. Yellow and orange arrowheads point to bands correspond to β10- and β11-fusions. The SEPT9 blot is also shown saturated on purpose for displaying weaker bands. Asterisks point to SEPT9 degradation products. (F) Western blot of U2OS cell lysates probed with anti-SEPT2 and anti-α-tubulin antibodies upon treatment with siRNAs targeting LacZ (siCtrl) or SEPT2 (siSEPT2). Molecular weight markers are shown on the left. Bottom, respective quantification of SEPT2 protein levels (mean + SD). Mean values (normalized to 1 for siCtrl) are from three independent siCtrl and nine independent siSEPT2 treatments. SEPT2 was knocked down on average by 92%. (G) Same as F for SEPT9. The asterisk points to a SEPT9 degradation product. Mean values (normalized to 1 for siCtrl) are from three independent siCtrl and three independent siSEPT9 treatments. SEPT9 was knocked down on average by 92%. (H) Same as F for SEPT7. Mean values (normalized to 1 for siCtrl) are from 3 independent siCtrl and 12 independent siSEPT7 treatments. SEPT7 was knocked down on average by 86%. Source data are available for this figure: SourceData FS2.
Confocal imaging revealed the presence of the reconstituted GFP (rGFP) on peripheral and ventral SFs, transverse arcs and perinuclear actin caps (Fig. 1, D and E), with the rGFP distribution closely resembling endogenous SEPT2 immunostainings and SEPT2–GFP distribution (Fig. S1 A). The presence of SEPT2-SEPT2 rGFP on SFs was further detected in fixed wild-type U2OS cells co-expressing SEPT2-β10 and SEPT2-β11 upon incubation with recombinant purified GFP1-9 (Fig. 1 F), confirming the presence of SEPT2–SEPT2 interactions in situ on SFs. Given that recombinant animal septins form both single and paired filaments (Szuba et al., 2021), we generated structure models to examine GFP complementation both from direct SEPT2–SEPT2 interactions within a filament (Fig. 2 A), and from SEPT2 facing another SEPT2 in apposed protomers of a paired filament (Fig. 2 B). Examination of the distances and the flexibility of the SEPT2 C-termini and the linkers showed that GFP reconstitution could occur either way.
All septins on SFs organize as filaments. (A and B) Structure models of rGFP through direct SEPT2-SEPT2 interactions of two polymerizing septin protomers within a filament (A) or from SEPT2 in two apposed protomers (B). Only the end-to-end interacting halves of the protomers (hexamers or/and octamers) are shown in A for simplicity. SEPT6 and SEPT7 coiled-coils are not shown in B for simplicity. The transparency of the terminal SEPT2 subunits in B is used to suggest that the paired protomers could be found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark gray. The colors of septin subunits in the structure models correspond to the ones in the color-coded sphere representation of hexamers and octamers. The second half of the octamer is not shown in the rotated filament pair in B for the sake of simplicity. (C) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of octamers-9_i3 in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (left panel) or SEPT2NCmut (right panel). Images use an inverted grayscale. (D) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin octamers in solution. Protomers contained either wild-type SEPT2 (left panel) or SEPT2NCmut (right panel). Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin (top row) and septin (bottom row) signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (E) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT7 (left) and anti-SEPT2 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT2 (siSEPT2), and targeting SEPT2 while expressing wild-type SEPT2-msfGFP (WT) or SEPT2NCmut-msfGFP (NCmut). Molecular weight markers are shown on the left. The overexpression of the msfGFP fusions leads to SEPT2 monomers and dimers in addition to hexamers and octamers (arrowheads). (F) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT2-β10 and -β11 or SEPT2NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT2 distributed among the two phenotypes. (G) Representative example of a GFP1-9 cell co-expressing SEPT2NCmut-β10 and -β11 and co-stained for F-actin (phalloidin) showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (H) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT2-msfGFP or SEPT2NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT2 distributed among the two phenotypes. (I) Representative example of a cell expressing SEPT2NCmut-msfGFP and co-stained for F-actin (phalloidin) showing a diffuse cytosolic phenotype. Scale bar, 10 μm. Source data are available for this figure: SourceData F2.
All septins on SFs organize as filaments. (A and B) Structure models of rGFP through direct SEPT2-SEPT2 interactions of two polymerizing septin protomers within a filament (A) or from SEPT2 in two apposed protomers (B). Only the end-to-end interacting halves of the protomers (hexamers or/and octamers) are shown in A for simplicity. SEPT6 and SEPT7 coiled-coils are not shown in B for simplicity. The transparency of the terminal SEPT2 subunits in B is used to suggest that the paired protomers could be found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark gray. The colors of septin subunits in the structure models correspond to the ones in the color-coded sphere representation of hexamers and octamers. The second half of the octamer is not shown in the rotated filament pair in B for the sake of simplicity. (C) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of octamers-9_i3 in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (left panel) or SEPT2NCmut (right panel). Images use an inverted grayscale. (D) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin octamers in solution. Protomers contained either wild-type SEPT2 (left panel) or SEPT2NCmut (right panel). Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin (top row) and septin (bottom row) signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (E) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT7 (left) and anti-SEPT2 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT2 (siSEPT2), and targeting SEPT2 while expressing wild-type SEPT2-msfGFP (WT) or SEPT2NCmut-msfGFP (NCmut). Molecular weight markers are shown on the left. The overexpression of the msfGFP fusions leads to SEPT2 monomers and dimers in addition to hexamers and octamers (arrowheads). (F) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT2-β10 and -β11 or SEPT2NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT2 distributed among the two phenotypes. (G) Representative example of a GFP1-9 cell co-expressing SEPT2NCmut-β10 and -β11 and co-stained for F-actin (phalloidin) showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (H) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT2-msfGFP or SEPT2NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT2 distributed among the two phenotypes. (I) Representative example of a cell expressing SEPT2NCmut-msfGFP and co-stained for F-actin (phalloidin) showing a diffuse cytosolic phenotype. Scale bar, 10 μm. Source data are available for this figure: SourceData F2.
The structural models highlighted that paired protomers would lead to GFP reconstitution whether they polymerize or not. Hence to test if SEPT2-SEPT2 rGFP originates from direct SEPT2–SEPT2 interactions, we designed a double point SEPT2 NC interface mutant (SEPT2 F20D, V27D, hereafter SEPT2NCmut) to prevent end-to-end association and thereby abolish polymerization (Fig. S3 A; Kuzmic et al., 2022; Sirajuddin et al., 2007). Reconstitution assays using purified recombinant protomers bearing these mutations confirmed that this mutant abolishes polymerization, although it can still bind actin filaments in vitro (Fig. 2, C and D; and Fig. S3, B and C). Native PAGE in cell lysates expressing SEPT2NCmut confirmed that protomers were intact: the expression of either wild-type SEPT2 or SEPT2NCmut in SEPT2 knockdown cells rescued equally well the hexamer and octamer distribution in control cells (Fig. 2 E). Strikingly, using this mutant in the context of the split SEPT2-SEPT2 assay completely abolished SF localization, as indicated by purely diffuse cytosolic fluorescence (Fig. 2, F and G; hereafter referred to as “diffuse cytosolic”). Given that wild-type SEPT2-SEPT2 rGFP was occasionally found as diffuse cytosolic, we quantified the distribution of diffuse cytosolic and non-diffuse phenotypes in cells expressing wild-type SEPT2- vs. SEPT2NCmut-β10/β11 fusions. While 95% of wild-type SEPT2-SEPT2 rGFP localized to SFs and only 5% was diffuse cytosolic, 100% of SEPT2NCmut-SEPT2NCmut rGFP was diffuse cytosolic (Fig. 2, F and G). Thus direct end-to-end septin polymerization is required for septin localization to SFs. We attributed the fact that the split-GFP assay with the NC mutant still produced fluorescence to the plasticity of septins, which are able to use both NC and G interfaces when either one is compromised (Kim et al., 2012). Our native PAGE shows the presence of SEPT2NC homodimers (Fig. 2 E), which we speculate are G-homodimers, thus enabling GFP complementation.
Septin interface mutants used in this study and cell-free reconstitution of septin assembly. (A) Left, top: Conserved residues in the SEPT2-SEPT2 NC interface are shown in the crystal structure of human SEPT2 homodimers (PDB accession no. 2QA5; Sirajuddin et al., 2007). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues F20 from the hook-loop of one SEPT2 subunit (orange) interact with the hydrophobic cleft formed by V86, I88, V93, and I281 of the adjacent SEPT2 subunit (blue). The importance of this phenylalanine in anchoring the α0 helix at the NC interface was emphasized only recently (Cavini et al., 2021). The blue subunit's surface representation highlights the complementary of shape between the two SEPT2 subunits in this interface. The interaction between the α0 helices of each subunit is also stabilized through a hydrophobic interaction between their respective V27. Left, bottom: Sequence alignment of the regions including the residues shown in the NC interface structure for SEPT2 and SEPT9_i3. The structural elements (α0, β2, α5) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties; period, conservation between residues with weakly similar physicochemical properties). We note that the residues described above are strictly or physicochemically conserved (except for V86), highlighting their importance in stabilizing the SEPT2-SEPT2 NC interface. The NC interface mutants used in this study are listed on the right of the crystal structure. A mutation of F20D/I263D is expected to destabilize the hydrophobic pocket depicted above, whereas a V27D/M270D is expected to destabilize the α0 helices interface. Importantly, a strictly conserved aspartate (SEPT2 E90, corresponding to SEPT6 E90 and SEPT7 E102 which are well defined in the cryo-EM structure of the SEPT6-SEPT7 NC interface [Mendonca et al., 2019]) in the loop connecting β2 and β3 is pointing to the hydrophobic cleft where the phenylalanine resides. The F20D mutation is thus expected to result in a repulsion between the aspartate and glutamate and contribute further to the destabilization of the NC interface. Right, top: Conserved residues in the SEPT7-SEPT7 G interface are shown in the crystal structure of human SEPT7 homodimers (PDB accession no. 6N0B; Brognara et al., 2019). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues W269 of one SEPT7 subunit (yellow) interact with residues W269, A272 and H279 in the adjacent SEPT7 subunit (green; Sirajuddin et al., 2007; Zent et al., 2011). W269 from adjacent subunits interact through a water molecule bridge through hydrogen bonds. In addition, each W269 is engaged in π-π interactions with H279 and CH-π interactions with A272 of the opposite subunit. Right, bottom: Sequence alignment of the regions including the residues shown in the G interface structure for SEPT7 and SEPT9_i3. The structural elements (β7, β8) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties). Notice that W269 and H279 are both strictly conserved, showing their importance in stabilizing this interface. The G interface mutants that were used in this study are listed on the right of the crystal structure. The presence of both mutations W269A and H279D in SEPT7 and SEPT9 is expected to destabilize the SEPT7-SEPT7 and SEPT7-SEPT9 G-interfaces. The loss of the aromatic cycle properties in the mutant W269A does not allow the abovementioned critical interactions mediated by the wild-type Trp. W269A is expected to destabilize H279 and potentially change its orientation. In addition, the much smaller size of the alanine will poorly mimic the hydrophobic interaction between W269 and H279, weakening the G-interface. Note that W269 is in the vicinity of Y267 of the same subunit. This tyrosine interacts with the nucleotide buried within the G-interface. Consequently, any mutations destabilizing W269 could dramatically destabilize the overall G-interface because of a domino effect. Similarly, H279D is expected to preclude hydrophobic interactions with W269 and thus destabilize the latter. The single mutation H279D in SEPT7 is expected to destabilize the SEPT7-SEPT7 G-interface when present in both SEPT7 subunits, but not the SEPT7-SEPT9 interface with wild-type SEPT9. (B) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of hexamers in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (top panel) or SEPT2NCmut (bottom panel). Images use an inverted grayscale. Related to Fig. 2 C. (C) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin hexamers in solution. Protomers contained either wild-type SEPT2 or SEPT2NCmut. Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. The image for actin in the presence of wild-type hexamers is contrast-enhanced on purpose in order to saturate the actin bundles so that weaker-intensity single actin filaments are also visible. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin and septin signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (D) Representative spinning disk fluorescence images of septin filament assembly upon co-polymerization of hexamers containing SEPT2-msfGFP and octamers-9_i3 containing SEPT2-sfCherry2 at the indicated final protomer concentration. Images use an inverted grayscale. Scale bars in all large fields of views, 10 μm.
Septin interface mutants used in this study and cell-free reconstitution of septin assembly. (A) Left, top: Conserved residues in the SEPT2-SEPT2 NC interface are shown in the crystal structure of human SEPT2 homodimers (PDB accession no. 2QA5; Sirajuddin et al., 2007). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues F20 from the hook-loop of one SEPT2 subunit (orange) interact with the hydrophobic cleft formed by V86, I88, V93, and I281 of the adjacent SEPT2 subunit (blue). The importance of this phenylalanine in anchoring the α0 helix at the NC interface was emphasized only recently (Cavini et al., 2021). The blue subunit's surface representation highlights the complementary of shape between the two SEPT2 subunits in this interface. The interaction between the α0 helices of each subunit is also stabilized through a hydrophobic interaction between their respective V27. Left, bottom: Sequence alignment of the regions including the residues shown in the NC interface structure for SEPT2 and SEPT9_i3. The structural elements (α0, β2, α5) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties; period, conservation between residues with weakly similar physicochemical properties). We note that the residues described above are strictly or physicochemically conserved (except for V86), highlighting their importance in stabilizing the SEPT2-SEPT2 NC interface. The NC interface mutants used in this study are listed on the right of the crystal structure. A mutation of F20D/I263D is expected to destabilize the hydrophobic pocket depicted above, whereas a V27D/M270D is expected to destabilize the α0 helices interface. Importantly, a strictly conserved aspartate (SEPT2 E90, corresponding to SEPT6 E90 and SEPT7 E102 which are well defined in the cryo-EM structure of the SEPT6-SEPT7 NC interface [Mendonca et al., 2019]) in the loop connecting β2 and β3 is pointing to the hydrophobic cleft where the phenylalanine resides. The F20D mutation is thus expected to result in a repulsion between the aspartate and glutamate and contribute further to the destabilization of the NC interface. Right, top: Conserved residues in the SEPT7-SEPT7 G interface are shown in the crystal structure of human SEPT7 homodimers (PDB accession no. 6N0B; Brognara et al., 2019). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues W269 of one SEPT7 subunit (yellow) interact with residues W269, A272 and H279 in the adjacent SEPT7 subunit (green; Sirajuddin et al., 2007; Zent et al., 2011). W269 from adjacent subunits interact through a water molecule bridge through hydrogen bonds. In addition, each W269 is engaged in π-π interactions with H279 and CH-π interactions with A272 of the opposite subunit. Right, bottom: Sequence alignment of the regions including the residues shown in the G interface structure for SEPT7 and SEPT9_i3. The structural elements (β7, β8) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties). Notice that W269 and H279 are both strictly conserved, showing their importance in stabilizing this interface. The G interface mutants that were used in this study are listed on the right of the crystal structure. The presence of both mutations W269A and H279D in SEPT7 and SEPT9 is expected to destabilize the SEPT7-SEPT7 and SEPT7-SEPT9 G-interfaces. The loss of the aromatic cycle properties in the mutant W269A does not allow the abovementioned critical interactions mediated by the wild-type Trp. W269A is expected to destabilize H279 and potentially change its orientation. In addition, the much smaller size of the alanine will poorly mimic the hydrophobic interaction between W269 and H279, weakening the G-interface. Note that W269 is in the vicinity of Y267 of the same subunit. This tyrosine interacts with the nucleotide buried within the G-interface. Consequently, any mutations destabilizing W269 could dramatically destabilize the overall G-interface because of a domino effect. Similarly, H279D is expected to preclude hydrophobic interactions with W269 and thus destabilize the latter. The single mutation H279D in SEPT7 is expected to destabilize the SEPT7-SEPT7 G-interface when present in both SEPT7 subunits, but not the SEPT7-SEPT9 interface with wild-type SEPT9. (B) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of hexamers in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (top panel) or SEPT2NCmut (bottom panel). Images use an inverted grayscale. Related to Fig. 2 C. (C) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin hexamers in solution. Protomers contained either wild-type SEPT2 or SEPT2NCmut. Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. The image for actin in the presence of wild-type hexamers is contrast-enhanced on purpose in order to saturate the actin bundles so that weaker-intensity single actin filaments are also visible. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin and septin signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (D) Representative spinning disk fluorescence images of septin filament assembly upon co-polymerization of hexamers containing SEPT2-msfGFP and octamers-9_i3 containing SEPT2-sfCherry2 at the indicated final protomer concentration. Images use an inverted grayscale. Scale bars in all large fields of views, 10 μm.
Septin protomers do not associate with SFs
The presence of septin filaments does not exclude that septin protomers are also present on SFs. To test whether septin protomers associate with SFs, we examined the cellular distribution of SEPT2NCmut fused to full-length GFP. Cells expressing this mutant exhibited a diffuse cytosolic localization, demonstrating that this mutant does not bind SFs (Fig. 2, H and I). Wild-type SEPT2-GFP fusions also showed diffuse cytosolic localization in addition to SF localization (Fig. S1 A v). However, while SEPT2-GFP was diffuse cytosolic in only ∼50% of cells, 100% of the cells expressing SEPT2NCmut showed this phenotype (Fig. 2, H and I). This result showed that septin protomers in cells do not associate with SFs, meaning that all septins decorating SFs are filamentous.
SF-associated septin filaments contain predominantly octamers
As SEPT2 is common to both protomers (Fig. 1 A), the results described above did not inform us on the composition of septin filaments. Recombinant hexamers and SEPT9-containing octamers have the capacity to co-polymerize in vitro (Fig. S3 D and Soroor et al., 2021). To explicitly visualize the presence of octamers on SFs, we probed SEPT7-SEPT9 and SEPT9-SEPT9 interactions as molecular signatures of octamers (Fig. 3, A and B). Expression levels of all β10/β11 fusions were kept low to minimize overexpression artifacts (Fig. S2 E), and endogenous SEPT7 and SEPT9 were consistently knocked down in all subsequent experiments (Fig. S2, G and H).
SF-associated septin filaments contain octamers. (A and B) Schematic (top) and respective structure model (bottom) of rGFP through SEPT9–SEPT9 interactions (A) and SEPT7–SEPT9 interactions (B) within an octamer. The transparency of the terminal SEPT2 subunits is used to suggest that the protomers are found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark gray. The colors of septin subunits in the structure models match the ones in the color-coded sphere representation of octamers. (C) Representative confocal micrographs of SEPT9_i3-SEPT9_i3 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Examples of rGFP in fixed cells localizing (i) to ventral (a) and peripheral (b) SFs and (ii) to transverse arcs (b) and excluded from dorsal SFs (a). Examples in live cells show rGFP localizing (iii) to ventral SFs (a and b) and (iv) to actin caps (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Representative confocal micrographs of SEPT7-SEPT9_i3 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Example of rGFP in fixed cells localizing (i) to ventral SFs (a and b). Example in live cells showing rGFP localizing (ii) to transverse arcs (a and b). The arrowhead points to a ring. Such cytoplasmic rings were ∼0.5–1.6 μm in diameter (0.9 μm on average from 19 measured rings). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (E) Additional examples of rings (arrowheads) in GFP1-9 cells co-expressing β11-SEPT7 and SEPT9_i3-β10. Scale bar, 2 μm. (F) Representative confocal micrographs of SEPT7-SEPT9_i1 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Examples in fixed and live cells show rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm.
SF-associated septin filaments contain octamers. (A and B) Schematic (top) and respective structure model (bottom) of rGFP through SEPT9–SEPT9 interactions (A) and SEPT7–SEPT9 interactions (B) within an octamer. The transparency of the terminal SEPT2 subunits is used to suggest that the protomers are found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark gray. The colors of septin subunits in the structure models match the ones in the color-coded sphere representation of octamers. (C) Representative confocal micrographs of SEPT9_i3-SEPT9_i3 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Examples of rGFP in fixed cells localizing (i) to ventral (a) and peripheral (b) SFs and (ii) to transverse arcs (b) and excluded from dorsal SFs (a). Examples in live cells show rGFP localizing (iii) to ventral SFs (a and b) and (iv) to actin caps (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Representative confocal micrographs of SEPT7-SEPT9_i3 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Example of rGFP in fixed cells localizing (i) to ventral SFs (a and b). Example in live cells showing rGFP localizing (ii) to transverse arcs (a and b). The arrowhead points to a ring. Such cytoplasmic rings were ∼0.5–1.6 μm in diameter (0.9 μm on average from 19 measured rings). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (E) Additional examples of rings (arrowheads) in GFP1-9 cells co-expressing β11-SEPT7 and SEPT9_i3-β10. Scale bar, 2 μm. (F) Representative confocal micrographs of SEPT7-SEPT9_i1 rGFP distribution in fixed cells (left and middle columns) co-stained for F-actin (phalloidin) and in live cells (right column). Examples in fixed and live cells show rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm.
As expected, rGFP from SEPT9_i3-SEPT9_i3 localized to contractile SFs (Fig. 3 C), similarly to SEPT9 immunostainings (Fig. S1 B), confirming that septin filaments contain SEPT9_i3-octamers. Split-GFP assays probing SEPT7-SEPT9_i3 interactions confirmed these findings (Fig. 3 D), with rGFP additionally labeling cytoplasmic rings of ∼0.9 μm in diameter (Fig. 3, D ii and E). To confirm that SF-localized rGFP from SEPT7-SEPT9_i3 and SEPT9_i3-SEPT9_i3 reflect direct interactions, we designed a double point SEPT9_i3 NC interface mutant (SEPT9_i3 M263D, I270D, hereafter SEPT9_i3NCmut), a double point SEPT9_i3 G interface mutant (SEPT9_i3 W502A, H512D, hereafter SEPT9_i3Gmut) and a double point SEPT7 G interface mutant (SEPT7 W269A, H279D, hereafter SEPT7Gmut1; Fig. S3 A)(Kuzmic et al., 2022; Sirajuddin et al., 2007; Zent et al., 2011). Native PAGE in cells expressing these mutants confirmed that SEPT9_i3NCmut completely disrupted octamers (Fig. 4 A), whereas SEPT7Gmut1 completely disrupted octamers and hexamers (Fig. 4 B). Split-GFP assays using these mutants completely abolished SF localization (Fig. 4, C–H), confirming that SF localization requires intact SEPT7-SEPT9_i3 and SEPT9_i3-SEPT9_i3 interfaces. All above assays gave identical results for SEPT9_i1, confirming the presence of both SEPT9_i1- and SEPT9_i3-containing octamers in SF-associated septin filaments (Fig. 3 F, Fig. 4, A and H; and Fig. S4, A and B).
Intact SEPT9 NC and G interfaces are required for septin localization to SFs. (A) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT9 (left and middle) and anti-SEPT7 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT9 (siSEPT9), and targeting SEPT9 while expressing wild-type SEPT9-msfGFP (WT) or SEPT9NCmut-msfGFP (NCmut) for both SEPT9_i1 and SEPT9_i3. The SEPT9 blot is also shown saturated on purpose for displaying weaker bands. Arrowheads point to the sizes of the indicated complexes. The asterisks point to SEPT9 degradation. Molecular weight markers are shown on the left. (B) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT9 (left) and anti-SEPT7 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT7 (siSEPT7), and targeting SEPT7 while expressing wild-type msfGFP-SEPT7 (WT), msfGFP-SEPT7Gmut1 (Gmut1), or msfGFP-SEPT7Gmut2 (Gmut2). Arrowheads point to the sizes of the indicated complexes. Molecular weight markers are shown on the left. (C) Representative example of a GFP1-9 cell co-expressing SEPT9_i3NCmut-β10 and -β11, co-stained for F-actin (phalloidin), showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (D) Representative examples of a fixed (i) and a live (ii) GFP1-9 cell co-expressing β11-SEPT7Gmut1 and SEPT9_i3Gmut-β10 showing a diffuse cytosolic phenotype. The fixed cell is co-stained for F-actin (phalloidin). Scale bar, 10 μm. (E) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT9_i3-msfGFP or SEPT9_i3NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (F) Representative example of a cell expressing SEPT9_i3NCmut-msfGFP, co-stained for F-actin (phalloidin), showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (G) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT9_i3-β10 and -β11 or SEPT9_i3NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (H) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing the indicated combinations of β11-SEPT7 and SEPT9-β10 fusions. Data points are from a total of 40 cells each for each combination, distributed among the two phenotypes. Source data are available for this figure: SourceData F4.
Intact SEPT9 NC and G interfaces are required for septin localization to SFs. (A) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT9 (left and middle) and anti-SEPT7 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT9 (siSEPT9), and targeting SEPT9 while expressing wild-type SEPT9-msfGFP (WT) or SEPT9NCmut-msfGFP (NCmut) for both SEPT9_i1 and SEPT9_i3. The SEPT9 blot is also shown saturated on purpose for displaying weaker bands. Arrowheads point to the sizes of the indicated complexes. The asterisks point to SEPT9 degradation. Molecular weight markers are shown on the left. (B) Western blot following native PAGE of U2OS cell lysates probed with anti-SEPT9 (left) and anti-SEPT7 (right) antibodies upon treatment with siRNAs targeting LacZ (siCtrl), SEPT7 (siSEPT7), and targeting SEPT7 while expressing wild-type msfGFP-SEPT7 (WT), msfGFP-SEPT7Gmut1 (Gmut1), or msfGFP-SEPT7Gmut2 (Gmut2). Arrowheads point to the sizes of the indicated complexes. Molecular weight markers are shown on the left. (C) Representative example of a GFP1-9 cell co-expressing SEPT9_i3NCmut-β10 and -β11, co-stained for F-actin (phalloidin), showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (D) Representative examples of a fixed (i) and a live (ii) GFP1-9 cell co-expressing β11-SEPT7Gmut1 and SEPT9_i3Gmut-β10 showing a diffuse cytosolic phenotype. The fixed cell is co-stained for F-actin (phalloidin). Scale bar, 10 μm. (E) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT9_i3-msfGFP or SEPT9_i3NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (F) Representative example of a cell expressing SEPT9_i3NCmut-msfGFP, co-stained for F-actin (phalloidin), showing a diffuse cytosolic phenotype. Scale bar, 10 μm. (G) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT9_i3-β10 and -β11 or SEPT9_i3NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (H) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing the indicated combinations of β11-SEPT7 and SEPT9-β10 fusions. Data points are from a total of 40 cells each for each combination, distributed among the two phenotypes. Source data are available for this figure: SourceData F4.
All septins on microtubules organize as octamer-based filaments. (A) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT9_i1-msfGFP or SEPT9_i1NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (B) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT9_i1-β10 and -β11 or SEPT9_i1NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (C) Representative confocal micrograph of SEPT9_i1-SEPT9_i1 rGFP distribution in fixed cells co-stained for microtubules (α-tubulin). (D) Representative example of a GFP1-9 cell co-expressing β11-SEPT7 and SEPT9_i1-β10 and co-stained for α-tubulin, with rGFP localizing to microtubules. (E) Representative example of GFP1-9 cell co-expressing β11-SEPT7Gmut1 and SEPT9_i1Gmut-β10 co-stained for α-tubulin showing a diffuse cytosolic phenotype. (F) Representative confocal micrograph of SEPT2-SEPT2 rGFP distribution in a live cell co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (G) Representative example of a cell (bottom left) co-expressing msfGFP-SEPT7 and mCherry-SEPT9_i1 and labeled for microtubules (SiR-tubulin) showing msfGFP-SEPT7 localizing to microtubules (b). A cell expressing only msfGFP-SEPT7 (top right) shows msfGFP-SEPT7 localizing to ectopic bundles not co-localizing with microtubules (a). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. Related to Fig. 5 E. (H) Representative confocal micrograph of SEPT7-SEPT7 rGFP distribution in a live cell co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (I) Representative examples of GFP1-9 cells co-expressing β11-SEPT7Gmut2 and SEPT9_i1-β10 with rGFP localizing to microtubules. (J) Representative example of a GFP1-9 cell co-expressing β10- and β11-SEPT7Gmut2 co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (C–J) Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (K) Representative SIM micrographs of cells (i, ii) expressing mCherry-SEPT9_i1 (not shown) co-stained for SEPT7 and α-tubulin. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (L) Box plots depict the distributions of measured widths, as the full width at half maximum (FWHM), of microtubules (MT) and MT-associated septins (SEPT7). The data points are plotted on top of the respective box plots; data points correspond to width measurements at multiple positions along MT and septin fibers and in multiple MT and septin fibers per cell in a total of 8 cells. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements is n = 128 and 112 for MTs and septins, respectively. The respective median values are 128 and 119 nm for MTs and septins, respectively.
All septins on microtubules organize as octamer-based filaments. (A) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type SEPT9_i1-msfGFP or SEPT9_i1NCmut-msfGFP. Data points are from a total of 90 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (B) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type SEPT9_i1-β10 and -β11 or SEPT9_i1NCmut-β10 and -β11. Data points are from a total of 40 cells each for wild-type and mutant SEPT9 distributed among the two phenotypes. (C) Representative confocal micrograph of SEPT9_i1-SEPT9_i1 rGFP distribution in fixed cells co-stained for microtubules (α-tubulin). (D) Representative example of a GFP1-9 cell co-expressing β11-SEPT7 and SEPT9_i1-β10 and co-stained for α-tubulin, with rGFP localizing to microtubules. (E) Representative example of GFP1-9 cell co-expressing β11-SEPT7Gmut1 and SEPT9_i1Gmut-β10 co-stained for α-tubulin showing a diffuse cytosolic phenotype. (F) Representative confocal micrograph of SEPT2-SEPT2 rGFP distribution in a live cell co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (G) Representative example of a cell (bottom left) co-expressing msfGFP-SEPT7 and mCherry-SEPT9_i1 and labeled for microtubules (SiR-tubulin) showing msfGFP-SEPT7 localizing to microtubules (b). A cell expressing only msfGFP-SEPT7 (top right) shows msfGFP-SEPT7 localizing to ectopic bundles not co-localizing with microtubules (a). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. Related to Fig. 5 E. (H) Representative confocal micrograph of SEPT7-SEPT7 rGFP distribution in a live cell co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (I) Representative examples of GFP1-9 cells co-expressing β11-SEPT7Gmut2 and SEPT9_i1-β10 with rGFP localizing to microtubules. (J) Representative example of a GFP1-9 cell co-expressing β10- and β11-SEPT7Gmut2 co-expressing mCherry-SEPT9_i1 (not shown) and labeled for microtubules (SiR-tubulin). (C–J) Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (K) Representative SIM micrographs of cells (i, ii) expressing mCherry-SEPT9_i1 (not shown) co-stained for SEPT7 and α-tubulin. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (L) Box plots depict the distributions of measured widths, as the full width at half maximum (FWHM), of microtubules (MT) and MT-associated septins (SEPT7). The data points are plotted on top of the respective box plots; data points correspond to width measurements at multiple positions along MT and septin fibers and in multiple MT and septin fibers per cell in a total of 8 cells. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements is n = 128 and 112 for MTs and septins, respectively. The respective median values are 128 and 119 nm for MTs and septins, respectively.
To test whether hexamers are also present on SFs, we probed SEPT7–SEPT7 interactions. Strikingly, rGFP from SEPT7–SEPT7 interactions was unexpectedly difficult to detect on SFs: although it localized to SFs (Fig. 5, A i, and B i), the majority was found on ectopic short, needle-like bundles (Fig. 5, A ii, and B ii), similar to the localization of full-length GFP-SEPT7 fusions (Fig. 5, C and D). These ectopic bundles did not localize to SFs (Fig. 5, A ii, and D) and contained SEPT2 but not SEPT9 (Fig. 5 C). These bundles thus most likely consist of hexamers, in line with the capacity of recombinant hexamers to form septin filament bundles in vitro (DeRose et al., 2020; Iv et al., 2021; Kinoshita et al., 2002; Leonardo et al., 2021). The presence of rGFP on the ectopic bundles thus showed that the split SEPT7-SEPT7 assay readily detects SEPT7–SEPT7 interactions originating from hexamers.
Exogenous SEPT7 and SEPT9 expression affect SEPT7 distribution. (A) Representative confocal micrographs of SEPT7-SEPT7 rGFP distribution in fixed cells co-stained for F-actin (phalloidin) localizing (i) to ventral (a and b) SFs and (ii) to ectopic bundles devoid of phalloidin staining (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (B) Representative confocal micrographs of SEPT7-SEPT7 rGFP distribution in live cells localizing (i) to transverse arcs (a and b) and (ii) to ectopic bundles (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (C) Examples of cells expressing msfGFP-SEPT7 and co-stained for SEPT2 (i) or for SEPT9 (ii). msfGFP-SEPT7 localizing to ectopic bundles contained SEPT2 (i; a and b) but not SEPT9 (ii; a). A non-transfected cell in (ii) shows SEPT9-stained SFs (b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Representative confocal micrograph of a cell expressing msfGFP-SEPT7 co-stained for F-actin (phalloidin) localizing to ventral SFs (a) and to ectopic bundles devoid of phalloidin staining (b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (E) Representative example of a cell (top right) co-expressing msfGFP-SEPT7 and SEPT9_i3-mApple and labeled for F-actin (SiR-actin). Example shows msfGFP-SEPT7 localizing to ventral SFs (b). A cell expressing only msfGFP-SEPT7 (bottom left) in (ii) shows msfGFP-SEPT7 localizing to ectopic bundles that are devoid of F-actin (a). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (F) Representative examples of GFP1-9 cells co-expressing β10- and β11-SEPT7, SEPT9_i3-mApple and labeled for F-actin (SiR-actin). Example in (i) shows rGFP localization to ventral SFs. Example in (ii) shows rGFP localization to peripheral (a), ventral SFs (b) and transverse arcs (c). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (G and H) Schematic (top) and respective structure model (bottom) of rGFP through SEPT7–SEPT7 interactions within a hexamer (G) or from SEPT7 in two apposed octamers within a paired filament (H). The transparency of the terminal SEPT2 subunits is used to suggest that the protomers are found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark grey. The colors of septin subunits correspond to the ones in the color-coded sphere representation of hexamers and octamers. The second half of the octamer is not shown in the rotated filament pair in H for the sake of simplicity. Only SEPT7 subunits are shown in the zoom-in of the reconstituted GFP barrel in H for the sake of simplicity.
Exogenous SEPT7 and SEPT9 expression affect SEPT7 distribution. (A) Representative confocal micrographs of SEPT7-SEPT7 rGFP distribution in fixed cells co-stained for F-actin (phalloidin) localizing (i) to ventral (a and b) SFs and (ii) to ectopic bundles devoid of phalloidin staining (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (B) Representative confocal micrographs of SEPT7-SEPT7 rGFP distribution in live cells localizing (i) to transverse arcs (a and b) and (ii) to ectopic bundles (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (C) Examples of cells expressing msfGFP-SEPT7 and co-stained for SEPT2 (i) or for SEPT9 (ii). msfGFP-SEPT7 localizing to ectopic bundles contained SEPT2 (i; a and b) but not SEPT9 (ii; a). A non-transfected cell in (ii) shows SEPT9-stained SFs (b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Representative confocal micrograph of a cell expressing msfGFP-SEPT7 co-stained for F-actin (phalloidin) localizing to ventral SFs (a) and to ectopic bundles devoid of phalloidin staining (b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (E) Representative example of a cell (top right) co-expressing msfGFP-SEPT7 and SEPT9_i3-mApple and labeled for F-actin (SiR-actin). Example shows msfGFP-SEPT7 localizing to ventral SFs (b). A cell expressing only msfGFP-SEPT7 (bottom left) in (ii) shows msfGFP-SEPT7 localizing to ectopic bundles that are devoid of F-actin (a). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (F) Representative examples of GFP1-9 cells co-expressing β10- and β11-SEPT7, SEPT9_i3-mApple and labeled for F-actin (SiR-actin). Example in (i) shows rGFP localization to ventral SFs. Example in (ii) shows rGFP localization to peripheral (a), ventral SFs (b) and transverse arcs (c). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (G and H) Schematic (top) and respective structure model (bottom) of rGFP through SEPT7–SEPT7 interactions within a hexamer (G) or from SEPT7 in two apposed octamers within a paired filament (H). The transparency of the terminal SEPT2 subunits is used to suggest that the protomers are found within a filament. β10 and β11 strands are shown in yellow and orange, respectively. Linker sequences between septins and the β-strands, delimited by arrowheads, are shown in dark grey. The colors of septin subunits correspond to the ones in the color-coded sphere representation of hexamers and octamers. The second half of the octamer is not shown in the rotated filament pair in H for the sake of simplicity. Only SEPT7 subunits are shown in the zoom-in of the reconstituted GFP barrel in H for the sake of simplicity.
An observation that could explain the difficulty to detect SEPT7-SEPT7 on SFs was the dependence of SEPT7 localization on SEPT9 expression levels. We consistently detected ectopic bundles when we exogenously expressed only SEPT7, either GFP-SEPT7 or split SEPT7-SEPT7 (Fig. 5, A–D), but not when we co-expressed SEPT9 (Fig. 5, E and F). We reasoned that in the absence of exogenous SEPT9, the slightest excess of SEPT7 leads to ectopic hexamer-based bundles, also reducing the availability of SEPT7 for forming octamers to bind SFs. Exogenous co-expression of SEPT9, on the other hand, would cause incorporation of the exogenous SEPT7 into octamers, thus preventing the formation of ectopic hexamer bundles. Consistent with this hypothesis, SEPT7-SEPT7 rGFP was readily detectable on SFs under conditions of exogenous SEPT9 co-expression (Fig. 5 F). Furthermore, it was difficult to find SEPT9-decorated SFs in cells also displaying ectopic hexamer-based bundles (Fig. 5 C). These observations raised the possibility that septin filaments on SFs contain mostly, if not exclusively, octamers.
To explore the origin of SEPT7-SEPT7 rGFP on SFs, we generated structure models of septin protomers in order to examine GFP complementation from SEPT7–SEPT7 interactions within one hexamer (Fig. 5 G), as well as from SEPT7 facing another SEPT7 in apposed hexamers or octamers in a paired filament (Fig. 5 H). Examination of the distances and the flexibility of the SEPT7 N-termini and the linkers showed that GFP reconstitution can occur either way.
To identify the sources of the SEPT7-SEPT7 rGFP signal on SFs, we aimed at perturbing hexamers while preserving octamers. To this end, we generated a single point SEPT7 G interface mutant (SEPT7 H279D, hereafter SEPT7Gmut2; Fig. S3 A) that should destabilize the SEPT7-SEPT7 G-interface when present in both SEPT7 subunits, but preserve the SEPT7-SEPT9 G-interface if SEPT7 is mutated but SEPT9 is wild-type. In line with these predictions, native PAGE showed that octamers are not affected by the expression of SEPT7Gmut2 (Fig. 4 B), and rGFP from SEPT7Gmut2-SEPT9_i3 recapitulated normal septin localization on SFs (Fig. 6 A). Importantly, rGFP from SEPT7Gmut2-SEPT7Gmut2 localized to SFs but did not show any ectopic bundles (Fig. 6 B) indicating that SEPT7Gmut2 completely abolished SEPT7–SEPT7 interactions within hexamers in the bundles. Thus our findings show that the SF-localized rGFP from SEPT7Gmut2-SEPT7Gmut2 originates from paired octamers (Fig. 5 H).
Intact SEPT7 G interfaces are required for septin localization to SFs. (A) Representative examples of fixed (i and ii) and live (iii–vi) GFP1-9 cells co-expressing β11-SEPT7Gmut2 and SEPT9_i3-β10. Fixed cells are co-stained for F-actin (phalloidin). Examples shows rGFP localizing (i and ii) to ventral SFs (a and b), (iii) to perinuclear actin caps (a and b), (iv) to ventral SFs (a and b), and (v and vi) rings (arrowheads). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (B) Representative examples of GFP1-9 cells (i–iv) co-expressing β10- and β11-SEPT7Gmut2. The fixed cell is co-stained for F-actin (phalloidin). Examples show diffuse cytosolic phenotypes (i and iii) of the rGFP and rGFP localizing to SFs (ii and iv). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (C) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type msfGFP-SEPT7 or msfGFP-SEPT7NCmut. Data points are from a total of 71 cells for wild-type, 68 cells for SEPT7Gmut1 and 90 cells for SEPT7Gmut2 distributed among the two phenotypes. (D) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type β10- and β11-SEPT7, β10- and β11-SEPT7Gmut1, or β10- and β11-SEPT7Gmut2. Data points are from a total of 40 cells for wild-type, 33 cells for β10- and β11-SEPT7Gmut1 and 29 cells for β10- and β11-SEPT7Gmut2 distributed among the two phenotypes. (E) Representative examples of GFP1-9 cells (i and ii) co-expressing β10- and β11-SEPT7Gmut1 showing a diffuse cytosolic phenotype. The fixed cell is co-stained for F-actin (phalloidin). Scale bar, 10 μm.
Intact SEPT7 G interfaces are required for septin localization to SFs. (A) Representative examples of fixed (i and ii) and live (iii–vi) GFP1-9 cells co-expressing β11-SEPT7Gmut2 and SEPT9_i3-β10. Fixed cells are co-stained for F-actin (phalloidin). Examples shows rGFP localizing (i and ii) to ventral SFs (a and b), (iii) to perinuclear actin caps (a and b), (iv) to ventral SFs (a and b), and (v and vi) rings (arrowheads). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (B) Representative examples of GFP1-9 cells (i–iv) co-expressing β10- and β11-SEPT7Gmut2. The fixed cell is co-stained for F-actin (phalloidin). Examples show diffuse cytosolic phenotypes (i and iii) of the rGFP and rGFP localizing to SFs (ii and iv). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (C) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the msfGFP signal in cells expressing wild-type msfGFP-SEPT7 or msfGFP-SEPT7NCmut. Data points are from a total of 71 cells for wild-type, 68 cells for SEPT7Gmut1 and 90 cells for SEPT7Gmut2 distributed among the two phenotypes. (D) Violin plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes as a function of the intensity of the rGFP signal in GFP1-9 cells co-expressing wild-type β10- and β11-SEPT7, β10- and β11-SEPT7Gmut1, or β10- and β11-SEPT7Gmut2. Data points are from a total of 40 cells for wild-type, 33 cells for β10- and β11-SEPT7Gmut1 and 29 cells for β10- and β11-SEPT7Gmut2 distributed among the two phenotypes. (E) Representative examples of GFP1-9 cells (i and ii) co-expressing β10- and β11-SEPT7Gmut1 showing a diffuse cytosolic phenotype. The fixed cell is co-stained for F-actin (phalloidin). Scale bar, 10 μm.
These observations altogether strongly suggest that the detected rGFP from SEPT7-SEPT7 on SFs originates from paired octamers. Split-GFP assays using the SEPT7Gmut1 mutant resulted in diffuse cytosolic distributions (Fig. 4, D and H; and Fig. 6, C–E), confirming that SF localization requires intact SEPT7 G interfaces. We concluded that septins on SFs organize as paired filaments containing mostly, or even exclusively, octamers.
Polymerization-competent septin octamers, but not hexamers, are essential for the integrity and function of SF-associated septin filaments
To further test the contribution of hexamers vs. octamers to septin filament formation, we examined septin filaments under three conditions: (a) the presence of hexamers and octamers (control condition), (b) the absence of octamers, by knocking down SEPT9, and (c) the presence of octamers only, by expressing SEPT7Gmut2. As a readout of septin filaments, we examined rGFP from SEPT2-SEPT2 in live cells while imaging stress fibers (Fig. 7, A–C). To assess the effects of the perturbations, we quantified the distribution of non-diffuse vs. diffuse cytosolic phenotypes and calculated Pearson and Manders correlation coefficients for actin-septin co-localization (Fig. 7, D and E). Strikingly, removing octamers by knocking down SEPT9 entirely removed the SEPT2-SEPT2 rGFP signal from all SFs, leaving behind a punctate pattern not localizing to SFs, suggesting that filamentous septin integrity depends entirely on octamers. On the other hand, preserving octamers in the absence of hexamers preserved septin filaments on SFs, showing that the absence of hexamers does not compromise septin filament integrity.
Septin octamers are essential for the integrity of SF-associated septin filaments. (A–C) Representative confocal micrographs of SEPT2-SEPT2 rGFP distribution in live cells co-labeled for F-actin (SiR-actin). Cells were treated with siRNA targeting SEPT2 (A), with siRNAs targeting both SEPT2 and SEPT9 (B), or with siRNA targeting both SEPT2 and SEPT7 and co-transfected with mApple-SEPT7Gmut2 (C). Examples in A and C show rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Scatter dot plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes in cells under the same conditions as in A–C, also shown as pie graphs. Data points are from a total of 59 cells for wild-type and 60 cells for each perturbation condition, distributed among the two phenotypes. (E) Scatter dot plots (mean ± SD) depicting the distributions of calculated Pearson (left) and Manders (right) correlation coefficients for actin-septin colocalization in cells under the same conditions as in A–C. Data points for each plot, from left to right, are from a total of 30, 37, and 46 cells, respectively. Kruskal–Wallis test; *** P < 0.001, **** P < 0.0001.
Septin octamers are essential for the integrity of SF-associated septin filaments. (A–C) Representative confocal micrographs of SEPT2-SEPT2 rGFP distribution in live cells co-labeled for F-actin (SiR-actin). Cells were treated with siRNA targeting SEPT2 (A), with siRNAs targeting both SEPT2 and SEPT9 (B), or with siRNA targeting both SEPT2 and SEPT7 and co-transfected with mApple-SEPT7Gmut2 (C). Examples in A and C show rGFP localizing to ventral SFs (a and b). Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. (D) Scatter dot plots depicting the distribution of diffuse cytosolic (red datapoints) vs. non-diffuse (green datapoints) phenotypes in cells under the same conditions as in A–C, also shown as pie graphs. Data points are from a total of 59 cells for wild-type and 60 cells for each perturbation condition, distributed among the two phenotypes. (E) Scatter dot plots (mean ± SD) depicting the distributions of calculated Pearson (left) and Manders (right) correlation coefficients for actin-septin colocalization in cells under the same conditions as in A–C. Data points for each plot, from left to right, are from a total of 30, 37, and 46 cells, respectively. Kruskal–Wallis test; *** P < 0.001, **** P < 0.0001.
To test the functional contribution of octamers and hexamers to SFs, we quantified the number of cells presenting ventral SFs in cells containing hexamers and octamers, hexamers only, octamers only, and containing hexamers and octamers that cannot polymerize. Knocking down SEPT9 removed both SEPT9 and SEPT7 from all ventral SFs (compare Fig. 8, A and B). Importantly, whereas 82% of control cells, and 81% of cells with octamers only, contained ventral SFs, only 44% of cells in the absence of octamers, and 46% of cells with hexamers and octamers that could not polymerize, presented ventral SFs (Fig. 8, B and C). Total actin levels did not change among these conditions (Fig. 8 D). Thus, polymerization-competent septin octamers are essential for the integrity of SFs.
Ventral SFs are significantly reduced in the absence of octamers or upon impairing septin polymerization. (A) Representative confocal micrograph of cells co-stained for SEPT7, SEPT9, and F-actin (phalloidin). Examples show septins localizing to ventral SFs (a and b). (B) Representative confocal micrographs of wild type U2OS cells treated with siRNA targeting SEPT9 and co-stained for SEPT7, SEPT9, and F-actin (phalloidin). Example (i) depicts cells with no ventral SFs, whereas example (ii) shows cells that have ventral SFs that are devoid of septins (a and b; see Materials and methods for classification). (C) Box plots showing the frequency of ventral SF presence in wild-type cells treated with siRNA targeting LacZ (6mer+8mer), siRNA targeting SEPT9 (6mer), siRNA targeting SEPT7 and co-transfected with msfGFP-SEPT7Gmut2 (8mer), and siRNA targeting SEPT2 and co-transfected with SEPT2NCmut-msfGFP (no filaments). The data points are plotted on top of the respective box plots; each data point corresponds to one round of experiments. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of cells analyzed per experiment in each box plot, from left to right, is n = 46; 87; 104; 152; 18; 31; 26; 12 (6mer+8mer), n = 51; 83; 117; 165 (6mer), n = 66; 79; 84; 82 (8mer), and n = 99; 60; 67; 84; 51 (no filaments). The respective median frequencies are 82% (6mer+8mer), 44% (6mer), 81% (8mer), and 46% (no filaments). One-way ANOVA; ns = not significant; *** P < 0.001, **** P < 0.0001. (D) Top: Western blots of cell lysates probed with anti-actin and anti-α-tubulin antibodies under the same conditions as in C. Molecular weight markers are shown on the left. Bottom: Respective quantification of actin protein levels (mean + SD). Mean values (normalized to 1 for 6mer+8mer) are from three to five independent experiments. One-way ANOVA; ns = not significant. (E) Atomic force microscopy nanoindentation on cells under the conditions “6mer+8mer,” “6mer,” and “8mer” as described in C. Left: Example of an experimental force-indentation curve. Right and left arrows correspond to the approach and retraction curves, respectively. The solid red lines represent the fits to the viscolelastic model (see Materials and methods). The inset depicts the indentation of the cell, also showing ventral and dorsal SFs in red. The image on the right shows the cantilever tip indenting the dorsal membrane of a micropatterned wild-type cell. Right: Box plots showing the distributions of cell stiffness (E0) and cell fluidity (β). E0 values are plotted on a log scale. The data points are plotted on top of the respective box plots; each data point corresponds to one cell. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 31, 29, 23. The respective median cell stiffness values are 656, 479, and 719 Pa, and the respective median cell fluidity values are 0.21, 0.23, and 0.19. One-way ANOVA for log(E0) and for β; ns = not significant; ** P < 0.01. Source data are available for this figure: SourceData F8.
Ventral SFs are significantly reduced in the absence of octamers or upon impairing septin polymerization. (A) Representative confocal micrograph of cells co-stained for SEPT7, SEPT9, and F-actin (phalloidin). Examples show septins localizing to ventral SFs (a and b). (B) Representative confocal micrographs of wild type U2OS cells treated with siRNA targeting SEPT9 and co-stained for SEPT7, SEPT9, and F-actin (phalloidin). Example (i) depicts cells with no ventral SFs, whereas example (ii) shows cells that have ventral SFs that are devoid of septins (a and b; see Materials and methods for classification). (C) Box plots showing the frequency of ventral SF presence in wild-type cells treated with siRNA targeting LacZ (6mer+8mer), siRNA targeting SEPT9 (6mer), siRNA targeting SEPT7 and co-transfected with msfGFP-SEPT7Gmut2 (8mer), and siRNA targeting SEPT2 and co-transfected with SEPT2NCmut-msfGFP (no filaments). The data points are plotted on top of the respective box plots; each data point corresponds to one round of experiments. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of cells analyzed per experiment in each box plot, from left to right, is n = 46; 87; 104; 152; 18; 31; 26; 12 (6mer+8mer), n = 51; 83; 117; 165 (6mer), n = 66; 79; 84; 82 (8mer), and n = 99; 60; 67; 84; 51 (no filaments). The respective median frequencies are 82% (6mer+8mer), 44% (6mer), 81% (8mer), and 46% (no filaments). One-way ANOVA; ns = not significant; *** P < 0.001, **** P < 0.0001. (D) Top: Western blots of cell lysates probed with anti-actin and anti-α-tubulin antibodies under the same conditions as in C. Molecular weight markers are shown on the left. Bottom: Respective quantification of actin protein levels (mean + SD). Mean values (normalized to 1 for 6mer+8mer) are from three to five independent experiments. One-way ANOVA; ns = not significant. (E) Atomic force microscopy nanoindentation on cells under the conditions “6mer+8mer,” “6mer,” and “8mer” as described in C. Left: Example of an experimental force-indentation curve. Right and left arrows correspond to the approach and retraction curves, respectively. The solid red lines represent the fits to the viscolelastic model (see Materials and methods). The inset depicts the indentation of the cell, also showing ventral and dorsal SFs in red. The image on the right shows the cantilever tip indenting the dorsal membrane of a micropatterned wild-type cell. Right: Box plots showing the distributions of cell stiffness (E0) and cell fluidity (β). E0 values are plotted on a log scale. The data points are plotted on top of the respective box plots; each data point corresponds to one cell. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 31, 29, 23. The respective median cell stiffness values are 656, 479, and 719 Pa, and the respective median cell fluidity values are 0.21, 0.23, and 0.19. One-way ANOVA for log(E0) and for β; ns = not significant; ** P < 0.01. Source data are available for this figure: SourceData F8.
To further question the functional contribution of protomers, we turned to atomic force microscopy (AFM) nanoindentation for measuring cell stiffness. Septin depletion has been shown to reduce cell stiffness, using AFM, in cultured mammalian cells (Mostowy et al., 2011), but the specific contribution of hexamers vs. octamers was not explored. To address this question, we indented cells containing hexamers and octamers, hexamers only and octamers only with an AFM cantilever tip and determined the elastic modulus (E0) and the fluidity (β) of the cells by fitting the experimental force-indentation curves to a viscoelastic model (Fig. 8 E, see Materials and methods). While removing hexamers did not have any effect, the depletion of octamers resulted in a statistically significant decrease in cell stiffness and a corresponding increase in cell fluidity (Fig. 8 E). Cells with significantly reduced ventral SFs in the absence of octamers are predicted to generate less prestress and thus lower stiffness (Chowdhury et al., 2021; Wang et al., 2002). We concluded that polymerization-competent octamers are essential for their function in cell mechanics through the generation or/and maintenance of SFs.
Super-resolution microscopy reveals septin fibers running longitudinally along and around SFs and interconnecting SFs
Having shown that all septins associated with SFs are filamentous, we aimed at visualizing how septin filaments organize on the different types of SFs. We employed super-resolution structured illumination (SIM) microscopy in cells co-stained for SEPT7 (as a pan-septin filament marker), actin filaments, and α-actinin or non-muscle myosin heavy chain IIA (NMIIA). We examined septin filament organization on perinuclear actin caps, transverse arcs, including at arc nodes, on ventral SFs and at ventral actin nodes (Fig. 9, A–E). Regardless of the type of SF, we noticed that septin filament morphology was very different from that of the corresponding actin filament bundles. While actin filament bundles typically appeared as straight, rigid fibers, septin fibers consistently appeared less straight and with lower orientational persistence (Note: we choose to use septin “fibers” instead of “filaments” in this section to avoid confusion with single or paired septin filaments or bundles thereof; we discuss the composition of septin fibers below). Unlike core SF components like myosin and α-actinin, which displayed a sarcomere-like punctate distribution (Fig. 9, E i), septin fibers were distinctly separate from SFs, organizing in three manners: (a) septin fibers running longitudinally along SFs, either on the side of SFs with their signal segregated from the F-actin signal, or overlapping with SFs with the septin and F-actin signals merging (Fig. 9, A a, and E i and iii, a, b), (b) septin fibers running longitudinally along SFs and diagonally across their width, as if wrapping around the SFs (Fig. 9, A b, and E ii a,b), and (c) septin fibers running longitudinally along segments of SFs while interconnecting different SFs and connecting to other septin fibers (Fig. 9, C and D, and E iv b). Interconnecting septin fiber segments in between SFs frequently colocalized with F-actin signal, but in many instances there was no detectable F-actin signal along these segments.
Super-resolution structured illumination (SIM) microscopy of septin filaments on SFs. (A–E) Representative SIM micrographs of SEPT7 immunostained cells co-stained for F-actin (phalloidin; A–E), and additionally for α-actinin (C and D) or non-muscle myosin heavy chain IIA (NMIIA; E). Examples show septin filament localization to perinuclear actin caps (A), arcs (B) and arc nodes (C), ventral nodes (D), and ventral SFs (E, cells i–iv). The insets adjacent to the full field-of-views depict regions of interest (solid outlined boxes) shown at high magnification. Dashed outlined boxes and their respective insets show specific features at higher magnification. Scale bars in all large fields of views, 10 μm. Scale bars in insets, 1 or 2 μm as indicated. (F–I) Fiber width measurements and real size estimations from SIM images. Box plots in F depict the distributions of measured widths, as the full width at half maximum (FWHM), of microtubules (MT;inset shows an example SIM image of MTs) and septins associated with peripheral SFs (“plasma membrane”), perinuclear actin caps (“cap”), arc and ventral actin nodes (“aster”), arcs and ventral SFs; widths from thin and thick ventral septin fibers were plotted separately. The data points are plotted on top of the respective box plots; data points correspond to width measurements at multiple positions along MT and septin fibers and in multiple MT and septin fibers per cell in a total of 10 cells for MT and 10 cells for septin fiber measurements. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 180, 123, 175, 88, 184, 330, 114. The respective median values are 115, 123, 137, 131, 133, 134, and 231 nm. Kruskal–Wallis test; ns = not significant; **** P < 0.0001. (G) Numerical simulations of the expected FWHM in SIM images (“image diameter”) as a function of the real fiber diameter. The curve was generated from the convolution of a Gaussian point spread function (PSF) of 115 nm with an increasing fiber size. Fiber sizes above ∼200 nm scale linearly with the image sizes. These simulations were used together with FWHM measurements in SIM images (F) to estimate an upper width limit for septin fibers associated with the different types of SFs (H). These estimations were then compared to the real width ranges one expects from IgG antibody-decorated septins organizing as single or double filaments (I). Primary and fluorophore (cyan asterisk)-conjugated secondary antibodies are depicted in green and magenta, respectively. The primary SEPT7 antibody used in our immunostainings binds the very C-terminus of SEPT7. The narrow and wide spacings of paired filaments, the presence of homodimeric coiled coils for SEPT2, SEPT6 and SEPT7, and of heterodimeric coiled coils for SEPT6 and SEPT7 are based on experimental evidence from (de Almeida Marques et al., 2012; Leonardo et al., 2021; Low and Macara, 2006; Sala et al., 2016). (J) Scatter dot plots of length distributions for septin fibers on the indicated types of SFs. Bars depict median values. The number of measurements in each plot, from left to right, is n = 151, 97, 227, 249. The respective median values are 2.8, 1.3, 2.0, and 3.8 μm.
Super-resolution structured illumination (SIM) microscopy of septin filaments on SFs. (A–E) Representative SIM micrographs of SEPT7 immunostained cells co-stained for F-actin (phalloidin; A–E), and additionally for α-actinin (C and D) or non-muscle myosin heavy chain IIA (NMIIA; E). Examples show septin filament localization to perinuclear actin caps (A), arcs (B) and arc nodes (C), ventral nodes (D), and ventral SFs (E, cells i–iv). The insets adjacent to the full field-of-views depict regions of interest (solid outlined boxes) shown at high magnification. Dashed outlined boxes and their respective insets show specific features at higher magnification. Scale bars in all large fields of views, 10 μm. Scale bars in insets, 1 or 2 μm as indicated. (F–I) Fiber width measurements and real size estimations from SIM images. Box plots in F depict the distributions of measured widths, as the full width at half maximum (FWHM), of microtubules (MT;inset shows an example SIM image of MTs) and septins associated with peripheral SFs (“plasma membrane”), perinuclear actin caps (“cap”), arc and ventral actin nodes (“aster”), arcs and ventral SFs; widths from thin and thick ventral septin fibers were plotted separately. The data points are plotted on top of the respective box plots; data points correspond to width measurements at multiple positions along MT and septin fibers and in multiple MT and septin fibers per cell in a total of 10 cells for MT and 10 cells for septin fiber measurements. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 180, 123, 175, 88, 184, 330, 114. The respective median values are 115, 123, 137, 131, 133, 134, and 231 nm. Kruskal–Wallis test; ns = not significant; **** P < 0.0001. (G) Numerical simulations of the expected FWHM in SIM images (“image diameter”) as a function of the real fiber diameter. The curve was generated from the convolution of a Gaussian point spread function (PSF) of 115 nm with an increasing fiber size. Fiber sizes above ∼200 nm scale linearly with the image sizes. These simulations were used together with FWHM measurements in SIM images (F) to estimate an upper width limit for septin fibers associated with the different types of SFs (H). These estimations were then compared to the real width ranges one expects from IgG antibody-decorated septins organizing as single or double filaments (I). Primary and fluorophore (cyan asterisk)-conjugated secondary antibodies are depicted in green and magenta, respectively. The primary SEPT7 antibody used in our immunostainings binds the very C-terminus of SEPT7. The narrow and wide spacings of paired filaments, the presence of homodimeric coiled coils for SEPT2, SEPT6 and SEPT7, and of heterodimeric coiled coils for SEPT6 and SEPT7 are based on experimental evidence from (de Almeida Marques et al., 2012; Leonardo et al., 2021; Low and Macara, 2006; Sala et al., 2016). (J) Scatter dot plots of length distributions for septin fibers on the indicated types of SFs. Bars depict median values. The number of measurements in each plot, from left to right, is n = 151, 97, 227, 249. The respective median values are 2.8, 1.3, 2.0, and 3.8 μm.
Regardless of the type of SFs, the majority of septin fibers appeared thinner than their associated SFs. Septin fibers were often thicker on the SF segments adjacent to FAs (Fig. 9, E iii b and E iv a), but thicker septin fibers were also found on arcs, caps, and ventral SFs. Thicker septin fibers did not exceed the width of the associated SF, and appeared either as single thick fibers, or what looked like two closely apposed thin fibers (dashed rectangle in the SEPT7 channel of Fig. 9, E iii). To compare septin fiber thicknesses across the different SF types we measured the width of septin fibers for each SF type. The full width at half maximum (FWHM) was calculated from fluorescence intensity line profiles (Fig. 9 F). Widths of the thick septin fibers found on ventral SFs were plotted separately. All thin septin fiber populations had median FWHM values in a narrow range of 123–137 nm, whereas the thick ones showed an almost twofold higher median FWHM value of 231 nm (Fig. 9, F and H). There was no statistically significant difference between thin septin fiber widths on caps, asters, arcs, and ventral SFs (Fig. 9 F).
To determine whether septin fibers are single or paired filaments (“double septin filaments”), or bundles thereof, we checked the FWHM values of microtubules (MTs) in the same cells. MTs are 25-nm wide tubes and are routinely used as the gold standard for assessing the performance of super-resolution microscopy techniques. MTs were stained using whole primary and fluorophore-coupled secondary IgG antibodies, just like for septin stainings, leading to an estimated real MT width of ∼60 nm (Fig. 9 I; Weber et al., 1978). We found an average FWHM value of 115 nm for the MT width in our cells, in line with reported values (Hamel et al., 2014; Wegel et al., 2016), given that the lateral resolution of SIM is roughly half of the diffraction limit, i.e., ∼110 nm. Given that the observed size in our images is the convolution of the real object size with the point spread function (PSF) of the SIM microscope, we simulated the predicted image size as a function of the real fiber size (Fig. 9 G). The comparison of the estimated real widths of primary and secondary IgG-decorated septins, assumed to organize as single or as paired filaments with either narrow (∼5 nm) or wide (∼20 nm) spacing (Leonardo et al., 2021; Fig. 9 I), with the widths predicted from our FWHM measurements of immunostained septins (Fig. 9 H), suggests that the thin septin fiber widths are compatible with single or paired septin filaments, whereas the thick septin fibers could correspond to two single or two double septin filaments. We note that these estimations assume that septin filaments in cells are present as single filaments with flexible coiled-coils, or paired filaments mediated by coiled-coil pairing (Fig. 9 I) in line with published literature (Cavini et al., 2021). The flexibility of coiled-coils combined with the presence of primary and secondary antibodies suggests that septin fibers cannot correspond to more than a few filaments even if septins interact directly through their GTP-binding domains, a condition previously observed only in the absence of coiled-coils (Bertin et al., 2010; Szuba et al., 2021).
We also wondered about the length of the SF-associated septin fibers. Short septin fibers associated with and interconnecting actin nodes had lengths on the order of 0.5–3.5 μm, whereas septin fibers on arcs, actin caps and ventral SFs were as short as 0.5–1 μm and as long as 10–15 μm (Fig. 9 J). We note that these values provide an upper bound since, given the lateral resolution limit of SIM, we cannot be sure that what appears as continuous fiber signal originates from a single fiber or from adjacent fibers overlapping at their ends.
All microtubule-associated septins organize as filaments containing predominantly octamers
To examine if our results on SF-associated septin filaments also hold for MT-associated septins, which contain specifically SEPT9_i1 (Kuzmic et al., 2022), we probed SEPT9_i1-SEPT9_i1, SEPT7-SEPT9_i1, and SEPT2–SEPT2 interactions using the split-GFP assay. Our results confirmed that MT localization requires intact SEPT7-SEPT9_i1 and SEPT9_i1-SEPT9_i1 interfaces and direct SEPT2–SEPT2 interactions (Fig. S4, A–F and Fig. 4 H), allowing us also to visualize septin filaments on MTs in situ in cells (Fig. S4 F). We further tested the importance of hexamers for septin-MT association. In line with our results on exogenously expressed SEPT7, it was difficult to find SEPT9-decorated MTs in cells also displaying ectopic hexamer-based bundles (Fig. S4 G). rGFP from SEPT7-SEPT7 was, however, readily detected on MTs upon exogenous SEPT9_i1 co-expression (Fig. S4 H), suggesting that the rGFP signal may originate from paired filaments (Fig. 5 H). rGFP from SEPT7Gmut2-SEPT7Gmut2, in the presence of exogenous SEPT9_i1, and SEPT7Gmut2-SEPT9_i1 were readily detected on MTs (Fig. S4, I and J), reflecting SEPT7-SEPT7 rGFP from paired octamers. Finally, SIM imaging revealed thin septin fibers running along MTs over several micrometers (Fig. S4 K). Different from the presence of both thin and thick actin-associated septin fibers, all MT-associated septin fibers appeared homogeneous in their width. FWHM measurements (Fig. 9 G and Fig. S4 L) predicted MT and septin fiber widths in the ranges of 65–135 and 85–130 nm, respectively, compatible with MT-associated septins organizing as paired septin filaments (Fig. 9 I). Our findings reveal that all MT- associated septins are exclusively in the form of octamers and filamentous, consistent with septins on actin SFs.
SF-associated septin filaments are closely apposed to the plasma membrane
Having shown that all SF-associated septins are filamentous, we wondered how septin function relates to septins being filamentous. Recombinant human septins can bind and cross-link actin filaments, but can also bind lipid membranes, raising the hypothesis that septin filaments in cells anchor SFs to the plasma membrane. A first indication that septins might be membrane-bound came from live cell extraction experiments. While extracting the plasma membrane after fixation entirely preserved septin localization to SFs, live-cell extraction removed septins from all SFs while preserving myosin on SFs (Fig. 10 A). Septins thus did not behave like core components of SFs, and their sensitivity to the detergent suggested they might be bound to the membrane.
Septin filaments are closely apposed to the plasma membrane, are largely immobilized on actin stress fibers, and can mediate actin-membrane anchoring. (A) Representative confocal micrographs of SEPT7 immunostained cells co-stained for F-actin and non-muscle myosin heavy chain IIA (NMIIA). Cells were either extracted after fixation (left panel) or were live-extracted right before fixation (right panel). Scale bars, 10 μm. (B) Schematic of the metal-induced energy transfer (MIET) assay in cells for probing fluorophore (mApple or AlexaFluor 568) distances from a gold-coated coverslip using fluorescence lifetime measurements. (C–E) C depicts representative examples of lifetime decay traces for SEPT9_i3-mApple on glass and in the presence of gold (left) and for SEPT9_i3-mApple and SEPT9_i3-mApple-CAAX in the presence of gold (right). The solid lines represent the numerical fits, showing the lifetime reduction due to the MIET process. The calculated lifetime-distance dependence for SEPT9_i3-mApple (D, see Materials and methods) was used to calculate the distance of SEPT9_i3-fused mApple, with or without the CAAX lipid anchor, from the coverslip (E). Lifetime decay traces and lifetime-distance dependence curves for GAP43-mApple (plasma membrane) and AF568-phalloidin (F-actin) are shown in Fig. S5, A and B. Box plots in E depict the distributions of calculated distances for SEPT9_i3-mApple-CAAX, SEPT9_i3, GAP43-mApple (plasma membrane, PM) and AF568-phalloidin (F-actin). The data points are plotted on top of the respective box plots; each data point corresponds to one cell. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 13, 11, 8, 8. The respective median values are 90, 88, 86, and 115 nm. One-way ANOVA; ns = not significant; *** P < 0.001. (F) TIRF images of SEPT2-msfGFP 8mer-9_i1 (top row) and F-actin (bottom row), either co-polymerized on top of a supported lipid bilayer (SLB), or co-polymerized in solution to form preformed bundles that were then flushed onto the supported lipid bilayer. The supported lipid bilayer was composed either of 5% of PI(4,5)P2, a septin-interacting lipid, and 95% DOPC (left panels), or 100% DOPC (right panels). Due to the shallow penetration depth (∼100 nm) of TIRF together with the absence of crowding agents, only truly membrane-associated structures are visible. Scale bar, 5 μm. (G–K) Septins are primarily immobilized and confined on actin stress fibers but also undergo very slow lateral free diffusion in the vicinity of the plasma membrane. (G–I) Left: Super-resolution PALM intensity images of mEos2-Actin (G), SEPT9_i3-mEos3.2 (H) and SEPT9_i3-mEos3.2-CAAX (I) in mouse embryonic fibroblasts obtained from a sptPALM sequence (50 Hz, >80 s). Insets: low resolution images of GFP-actin (G) or EYFP-paxillin (H and I), which were co-expressed for FA labeling. Scale bars, 3 µm. Right: color-coded trajectories overlaid on FAs labeled by EYFP-paxillin or on FAs and SFs labeled by GFP-actin (grayscale) show the diffusion modes: free diffusion (yellow), confined diffusion (green) and immobilization (red). (J) Distributions of the diffusion coefficient D computed from the trajectories of mEos2-actin (blue), SEPT9_i3-mEos3.2 (magenta), and SEPT9_i3-CAAX-mEos3.2 (light magenta) obtained outside FAs, are shown in a logarithmic scale. The gray area including D values inferior to 0.011 µm2.s−1 corresponds to immobilized proteins. Values represent the average of the distributions obtained from different cells. (K) Fraction of mEos2-actin (blue), SEPT9_i3-mEos3.2 (magenta) and SEPT9_i3-mEos3.2-CAAX (light magenta) undergoing free diffusion, confined diffusion or immobilization outside FAs. Values represent the average of the fractions obtained from different cells (error bars: SEM). Results for SEPT9_i3-mEos3.2 (14 cells) correspond to pooled data from two independent experiments with n, the number of trajectories analyzed: SEPT9_i3-mEos3.2 nSEPT9_i3 = 72,720. Results for mEos2-actin (9 cells) and SEPT9_i3-mEos3.2-CAAX (5 cells) correspond each to data from one experiment with n, the number of trajectories analyzed: mEos2-actin nactin = 34,715; SEPT9_i3-mEos3.2-CAAX nSEPT9_i3-CAAX = 37,339. Statistical significance in K was obtained using two-tailed, non-parametric Mann–Whitney rank sum test. The different conditions were compared to the SEPT9_i3-mEos3.2 condition. The resulting P values are indicated as follows: *** P < 0.001; **** P < 0.0001. (L) Working model supported by the results of this study. Septins in cells organize as paired, octamer-based filaments mediating actin-membrane anchoring.
Septin filaments are closely apposed to the plasma membrane, are largely immobilized on actin stress fibers, and can mediate actin-membrane anchoring. (A) Representative confocal micrographs of SEPT7 immunostained cells co-stained for F-actin and non-muscle myosin heavy chain IIA (NMIIA). Cells were either extracted after fixation (left panel) or were live-extracted right before fixation (right panel). Scale bars, 10 μm. (B) Schematic of the metal-induced energy transfer (MIET) assay in cells for probing fluorophore (mApple or AlexaFluor 568) distances from a gold-coated coverslip using fluorescence lifetime measurements. (C–E) C depicts representative examples of lifetime decay traces for SEPT9_i3-mApple on glass and in the presence of gold (left) and for SEPT9_i3-mApple and SEPT9_i3-mApple-CAAX in the presence of gold (right). The solid lines represent the numerical fits, showing the lifetime reduction due to the MIET process. The calculated lifetime-distance dependence for SEPT9_i3-mApple (D, see Materials and methods) was used to calculate the distance of SEPT9_i3-fused mApple, with or without the CAAX lipid anchor, from the coverslip (E). Lifetime decay traces and lifetime-distance dependence curves for GAP43-mApple (plasma membrane) and AF568-phalloidin (F-actin) are shown in Fig. S5, A and B. Box plots in E depict the distributions of calculated distances for SEPT9_i3-mApple-CAAX, SEPT9_i3, GAP43-mApple (plasma membrane, PM) and AF568-phalloidin (F-actin). The data points are plotted on top of the respective box plots; each data point corresponds to one cell. On each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements in each box plot, from left to right, is n = 13, 11, 8, 8. The respective median values are 90, 88, 86, and 115 nm. One-way ANOVA; ns = not significant; *** P < 0.001. (F) TIRF images of SEPT2-msfGFP 8mer-9_i1 (top row) and F-actin (bottom row), either co-polymerized on top of a supported lipid bilayer (SLB), or co-polymerized in solution to form preformed bundles that were then flushed onto the supported lipid bilayer. The supported lipid bilayer was composed either of 5% of PI(4,5)P2, a septin-interacting lipid, and 95% DOPC (left panels), or 100% DOPC (right panels). Due to the shallow penetration depth (∼100 nm) of TIRF together with the absence of crowding agents, only truly membrane-associated structures are visible. Scale bar, 5 μm. (G–K) Septins are primarily immobilized and confined on actin stress fibers but also undergo very slow lateral free diffusion in the vicinity of the plasma membrane. (G–I) Left: Super-resolution PALM intensity images of mEos2-Actin (G), SEPT9_i3-mEos3.2 (H) and SEPT9_i3-mEos3.2-CAAX (I) in mouse embryonic fibroblasts obtained from a sptPALM sequence (50 Hz, >80 s). Insets: low resolution images of GFP-actin (G) or EYFP-paxillin (H and I), which were co-expressed for FA labeling. Scale bars, 3 µm. Right: color-coded trajectories overlaid on FAs labeled by EYFP-paxillin or on FAs and SFs labeled by GFP-actin (grayscale) show the diffusion modes: free diffusion (yellow), confined diffusion (green) and immobilization (red). (J) Distributions of the diffusion coefficient D computed from the trajectories of mEos2-actin (blue), SEPT9_i3-mEos3.2 (magenta), and SEPT9_i3-CAAX-mEos3.2 (light magenta) obtained outside FAs, are shown in a logarithmic scale. The gray area including D values inferior to 0.011 µm2.s−1 corresponds to immobilized proteins. Values represent the average of the distributions obtained from different cells. (K) Fraction of mEos2-actin (blue), SEPT9_i3-mEos3.2 (magenta) and SEPT9_i3-mEos3.2-CAAX (light magenta) undergoing free diffusion, confined diffusion or immobilization outside FAs. Values represent the average of the fractions obtained from different cells (error bars: SEM). Results for SEPT9_i3-mEos3.2 (14 cells) correspond to pooled data from two independent experiments with n, the number of trajectories analyzed: SEPT9_i3-mEos3.2 nSEPT9_i3 = 72,720. Results for mEos2-actin (9 cells) and SEPT9_i3-mEos3.2-CAAX (5 cells) correspond each to data from one experiment with n, the number of trajectories analyzed: mEos2-actin nactin = 34,715; SEPT9_i3-mEos3.2-CAAX nSEPT9_i3-CAAX = 37,339. Statistical significance in K was obtained using two-tailed, non-parametric Mann–Whitney rank sum test. The different conditions were compared to the SEPT9_i3-mEos3.2 condition. The resulting P values are indicated as follows: *** P < 0.001; **** P < 0.0001. (L) Working model supported by the results of this study. Septins in cells organize as paired, octamer-based filaments mediating actin-membrane anchoring.
To test if septins on SFs are close to the plasma membrane of the U2OS cells, we employed a metal-induced energy transfer (MIET) assay (Chizhik et al., 2014). In MIET, the fluorescence lifetime is dependent on the distance of fluorophores from a metal layer, allowing us to deduce the axial distance of fluorophores from a gold-coated coverslip surface with an axial resolution of a few nanometers (Fig. 10 B). We hypothesized that septins could either associate with the plasma membrane while interacting with SFs, or that septins interact with SFs in the absence of any septin-membrane association. To distinguish these scenarios, we compared distances of the fluorescent protein, mApple, in three conditions: (a) mApple N-terminally fused to the 20 N-terminal residues of neuromodulin/GAP43 that contains palmitoylated cysteines (GAP43-mApple); as a reference for fluorophores localizing directly at the plasma membrane, (b) SEPT9_i3-mApple as a reference for ventral SF-associated septin octamers, and (c) SEPT9_i3-mApple-CAAX as a reference for septins targeted to the plasma membrane through the H-Ras CAAX motif which functions as a membrane targeting signal. Representative lifetime decay traces are shown in Fig. 10 C; and Fig. S5, A and B. Strikingly, the distance of mApple from the metal surface, derived from the lifetime-distance dependence curve (Fig. 10 D), was the same for SF-associated septins, membrane-bound mApple, and membrane-bound septins, meaning that septins are closely apposed to the plasma membrane (Fig. 10 E). Lifetime measurements of AF568-phalloidin bound to ventral SFs under the same conditions placed SFs significantly further away, by ∼25 nm, from the plasma membrane (Fig. 10 E). MIET assays being limited to probing interactions within 200 nm from the metal surface, it was not feasible to probe septin populations on transverse arcs and perinuclear actin caps, which are localized further away.
Septin filaments are closely apposed to the plasma membrane, are immobilized on actin stress fibers, and can mediate actin-membrane anchoring in the absence of myosin-II. (A and B) depict representative examples of lifetime decay traces for GAP43-mApple (A, left) and AF568-phalloidin (F-actin; B, left) on glass and in the presence of gold. The solid lines represent the numerical fits, showing the lifetime reduction due to the MIET process. The respective calculated lifetime-distance dependences, used to calculate the distances of the fluorophores from the coverslip (Fig. 10 E) are shown in the respective right panels. Related to Fig. 10, B–E. (C and D) TIRF images of SEPT2-msfGFP 8mer-9_i1 alone (left) or F-actin alone (right) on top of a supported lipid bilayer (SLB) composed of 100% DOPC (C) or 5% PI(4,5)P2 and 95% DOPC (D). Scale bar, 5 μm. Related to Fig. 10 F. (E) TIRF images of SEPT2-msfGFP 8mer-9_i3 and F-actin, either co-polymerized on top of an SLB, or co-polymerized in solution to form preformed bundles that were then flushed onto the supported lipid bilayer. The supported lipid bilayer was composed of 5% of PI(4,5)P2, a septin-interacting lipid, and 95% DOPC. Scale bar, 5 μm. Related to Fig. 10 F. (F) TIRF images of SEPT2-msfGFP 8mer-9_i1 prepolymerized on top of a supported lipid bilayer (SLB), with actin added in a second step to the membrane-bound septin filaments, either as prepolymerized actin filaments or as G-actin. The supported lipid bilayer was composed of 5% PI(4,5)P2 and 95% DOPC. Scale bar, 5 μm. Related to Fig. 10 F. (G and H) Box plots displaying the median (notch) and mean (square) ± percentile (25–75%) of diffusion coefficients corresponding to the free diffusion (Ddiff) (G) and confined diffusion (Dconf) (H) trajectories outside FAs from sptPALM. Related to Fig. 10, G–K. Statistical significance was obtained using two-tailed, non-parametric Mann–Whitney rank sum test. The different conditions were compared to the SEPT9_i3-mEos3.2 condition. The resulting P values are indicated as follows: ** P < 0.01; **** P < 0.0001. (I) Representative confocal micrographs of U2OS cells co-stained for F-actin (phalloidin), SEPT7 and non-muscle myosin heavy chain isoforms IIA (NMIIA) or IIB (NMIIB). Wild-type U2OS cells were co-stained for either NMIIA (i) or NMIIB (ii). NMIIA KO U2OS cells were co-stained for NMIIB (iii). NMIIB KO U2OS cells were co-stained for NMIIA (iv). (J) Western blot of NMIIA KO and NMIIB KO U2OS cell lysates probed with anti-α-tubulin, anti-SEPT7, anti-NMIIA and anti-NMIIB antibodies. NMIIA KO cells were treated either with LacZ or NMIIB siRNA. NMIIB KO cells were treated either with LacZ or NMIIA siRNA. Molecular weight markers are shown on the left. The condition shown with the red rectangle corresponds to the images shown in K. (K) Representative confocal micrographs of NMIIA KO U2OS cells (i–iii) treated with NMIIB siRNA and co-stained for F-actin (phalloidin), SEPT7 and NMIIB. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. Source data are available for this figure: SourceData FS5.
Septin filaments are closely apposed to the plasma membrane, are immobilized on actin stress fibers, and can mediate actin-membrane anchoring in the absence of myosin-II. (A and B) depict representative examples of lifetime decay traces for GAP43-mApple (A, left) and AF568-phalloidin (F-actin; B, left) on glass and in the presence of gold. The solid lines represent the numerical fits, showing the lifetime reduction due to the MIET process. The respective calculated lifetime-distance dependences, used to calculate the distances of the fluorophores from the coverslip (Fig. 10 E) are shown in the respective right panels. Related to Fig. 10, B–E. (C and D) TIRF images of SEPT2-msfGFP 8mer-9_i1 alone (left) or F-actin alone (right) on top of a supported lipid bilayer (SLB) composed of 100% DOPC (C) or 5% PI(4,5)P2 and 95% DOPC (D). Scale bar, 5 μm. Related to Fig. 10 F. (E) TIRF images of SEPT2-msfGFP 8mer-9_i3 and F-actin, either co-polymerized on top of an SLB, or co-polymerized in solution to form preformed bundles that were then flushed onto the supported lipid bilayer. The supported lipid bilayer was composed of 5% of PI(4,5)P2, a septin-interacting lipid, and 95% DOPC. Scale bar, 5 μm. Related to Fig. 10 F. (F) TIRF images of SEPT2-msfGFP 8mer-9_i1 prepolymerized on top of a supported lipid bilayer (SLB), with actin added in a second step to the membrane-bound septin filaments, either as prepolymerized actin filaments or as G-actin. The supported lipid bilayer was composed of 5% PI(4,5)P2 and 95% DOPC. Scale bar, 5 μm. Related to Fig. 10 F. (G and H) Box plots displaying the median (notch) and mean (square) ± percentile (25–75%) of diffusion coefficients corresponding to the free diffusion (Ddiff) (G) and confined diffusion (Dconf) (H) trajectories outside FAs from sptPALM. Related to Fig. 10, G–K. Statistical significance was obtained using two-tailed, non-parametric Mann–Whitney rank sum test. The different conditions were compared to the SEPT9_i3-mEos3.2 condition. The resulting P values are indicated as follows: ** P < 0.01; **** P < 0.0001. (I) Representative confocal micrographs of U2OS cells co-stained for F-actin (phalloidin), SEPT7 and non-muscle myosin heavy chain isoforms IIA (NMIIA) or IIB (NMIIB). Wild-type U2OS cells were co-stained for either NMIIA (i) or NMIIB (ii). NMIIA KO U2OS cells were co-stained for NMIIB (iii). NMIIB KO U2OS cells were co-stained for NMIIA (iv). (J) Western blot of NMIIA KO and NMIIB KO U2OS cell lysates probed with anti-α-tubulin, anti-SEPT7, anti-NMIIA and anti-NMIIB antibodies. NMIIA KO cells were treated either with LacZ or NMIIB siRNA. NMIIB KO cells were treated either with LacZ or NMIIA siRNA. Molecular weight markers are shown on the left. The condition shown with the red rectangle corresponds to the images shown in K. (K) Representative confocal micrographs of NMIIA KO U2OS cells (i–iii) treated with NMIIB siRNA and co-stained for F-actin (phalloidin), SEPT7 and NMIIB. Scale bars in large fields of views, 10 μm. Scale bars in insets, 2 μm. Source data are available for this figure: SourceData FS5.
Septin filaments anchor actin filaments to lipid membranes
Since SF-associated septins are closely apposed to the membrane, we wondered if septin filaments could function to anchor stress fibers to the plasma membrane. In the absence of available septin membrane-binding and actin-binding mutants, we turned to reconstitution assays on supported lipid bilayers (SLBs), comparing only phosphatidylcholine- (PC) vs. phosphatidylinositol(4,5)-bisphosphate (PI(4,5)P2)-containing membranes (Fig. 10 F), PI(4,5)P2 being a septin-interacting lipid (Szuba et al., 2021). To image only truly membrane-associated structures, we used total internal reflection fluorescence (TIRF) microscopy in the absence of crowding agents. Actin filaments alone did not bind lipid membranes, whereas septin octamers alone specifically bound PI(4,5)P2-containing membranes (Fig. S5, C and D), in line with previous reports for mammalian septin hexamers (Szuba et al., 2021). To test if septins can anchor actin to membranes, we either preformed actin–septin bundles in solution and then added them to SLBs, co-polymerized septins and actin on SLBs, or added actin to preassembled septin filaments on SLBs. In all cases, and specifically on PI(4,5)P2-containing membranes but not on membranes composed only of PC, actin filaments and actin filament bundles were anchored to the lipid bilayers (Fig. 10 F; and Fig. S5, E and F), showing that septin filaments can indeed at the same time bind membranes and actin and thus mediate membrane-actin anchoring.
Single protein tracking reveals that septins are immobilized on actin stress fibers
To determine whether the molecular dynamics of septins at the plasma membrane are consistent with a SF-anchoring function, we combined photoactivated localization microscopy (PALM) with live-cell single protein tracking of SEPT9_i3 and actin fused to photoswitchable mEos fluorescent proteins using sptPALM (Manley et al., 2008; Rossier et al., 2012). Cells were co-transfected with mEos-fused proteins and EYFP-paxillin as a FA reporter, or GFP-actin as a SF and FA reporter. Using TIRF microscopy, we detected and tracked sparse photo-activated proteins within 200 nm above the coverslip surface at high-frequency (50 Hz acquisition), allowing us to reconstruct thousands of protein trajectories (Fig. 10, G–I). For trajectories lasting at least 260 ms, we computed the mean square displacement (MSD). We then sorted trajectories according to their diffusion modes (immobile, confined, free-diffusive), and extracted diffusion coefficients (Ddiff, Dconf; Fig. 10, J and K; and Fig. S5, G and H; see Materials and methods). We first looked at the dynamic behavior of mEos2-actin in SFs labeled with GFP-actin (Fig. 10 G). mEos2-actin was found inside FAs and also linearly organized along SFs between FAs, as expected. Actin mostly displayed immobilized and confined behaviors, as illustrated by the large fractions of immobilization and confined diffusion (Fig. 10, G and K; immobile: 88.5 ± 0.5%, confined: 6.8 ± 0.3%, mean ± SEM) and a distribution of diffusion coefficients centered around 1.5–2.5.10−3 µm2.s−1 (Fig. 10 J). In line with septin immunostainings, single SEPT9_i3-mEos3.2 molecules were rarely found inside FAs, but were linearly organized between FAs decorating SFs (Fig. 10 H). Like actin, also SEPT9_i3-mEos3.2 was found to be primarily immobilized and confined (Fig. 10, H and K; immobile: 70.0 ± 1.9%, confined: 13.9 ± 0.8%, mean ± SEM). Contrary to actin, however, SEPT9_i3-mEos3.2 also displayed a significant freely diffusing population (Fig. 10, H and K; diffusive: 16.1 ± 1.3%, mean ± SEM). However, septin free-diffusion was very slow (Fig. 10 J and Fig. S5 G) with a diffusion constant Ddiff = 0.087 ± 0.001 µm2.s−1 (mean ± SEM) that is comparable to that of free diffusing transmembrane proteins (integrins: Rossier et al., 2012) or of a lipid-anchored protein bound to the plasma membrane by its PH domain (kindlin: Orre et al., 2021). Confined SEPT9_i3-mEos3.2 also diffused very slowly (Fig. S5 H) with a diffusion constant Dconf = 0.044 ± 0.001 µm2.s−1, comparable to that of mEos2-actin (0.057 ± 0.003 µm2.s−1; means ± SEM). Overall these results suggest that septins, when immobilized and confined, could indeed be anchoring actin SFs to the plasma membrane, while the free-diffusing septins display a diffusivity that is consistent with them being membrane-anchored.
Similarly to the MIET experiments, we used SEPT9_i3-mEos3.2-CAAX as a reference for septins targeted to the plasma membrane. SEPT9_i3-mEos3.2-CAAX did not localize specifically to SFs but decorated the whole plasma membrane (Fig. 10 I). In comparison with the behavior of SEPT9_i3-mEos3.2, SEPT9_i3-mEos3.2-CAAX displayed a smaller immobilized fraction (Fig. 10, I–K; immobile: 32.6 ± 0.5%) but increased free diffusion and confined diffusion fractions with an increased diffusion constant (Ddiff: 0.328 ± 0.002 µm2.s−1 ; Dconf: 0.202 ± 0.002 µm2.s−1, mean ± SEM; Fig. S5, G and H). Interestingly, being stably anchored to the plasma membrane allowed SEPT9_i3-mEos3.2-CAAX to diffuse inside FAs (Fig. 10 I). The much lower diffusion coefficient of SEPT9_i3-mEos3.2 compared to that of SEPT9_i3-mEos3.2-CAAX is in line with freely diffusing SEPT9_i3 being fully incorporated into septin filaments and could reflect hop diffusion of septin filaments alternating between SFs and the plasma membrane.
Septins can associate with actin filaments in cells in a myosin-II independent manner
Although septins decorate myosin-II containing SFs in cells, reconstituted septin filaments can mediate membrane-actin anchoring in the absence of myosin-II. To address if myosin-II is required for binding septins to SFs, we examined the distribution of F-actin, septins, and the two most abundant non-muscle myosin heavy chain isoforms IIA (NMIIA) and IIB (NMIIB) in wild-type U2OS cells, in CRISPR-mediated NMIIA and NMIIB knock-out (KO) U2OS lines (Kage et al., 2022), as well as in NMIIA KO cells treated with NMIIB siRNA to remove both isoforms. Both NMIIA and NMIIB were present on septin-decorated SFs in wild-type cells (Fig. S5, I i and ii). In NMIIA KO cells, NMIIB still associated with septin-decorated SFs (Fig. S5, I iii), and NMIIA still associated with septin-decorated SFs in NMIIB KO cells (Fig. S5, I iv). Thus septin association with SFs does not depend specifically on NMIIA or on NMIIB. However, there is 78% identity among NMIIA and NMIIB, including within the coiled-coil region previously implicated in septin binding (Joo et al., 2007), raising the possibility that a shared septin-binding sequence could bind septins to SFs. To assess septin-SF binding in the absence of myosin-II, we turned to NMIIA KO cells treated with NMIIB siRNA (Fig. S5, J and K). NMIIA- and NMIIB-depleted cells showed a loss of SFs with dramatic cell shape changes, including “C” and dendritic shapes previously reported upon NMII inhibition or silencing (Cai et al., 2010). Interestingly, F-actin appeared as a dense mesh of loosely cross-linked filaments as well as arrays of asters, with septin filaments associating extensively with actin filaments in the meshes and asters (Fig. S5 K). Septins are thus able to associate with actin filaments in cells in a myosin-II independent manner.
Discussion
Employing a tripartite split-GFP complementation assay to probe SEPT2–SEPT2 interactions as a molecular readout for end-to-end septin polymerization, we showed that all septins on SFs organize as filaments. Mutants that disrupt specifically the SEPT7–SEPT7 interface allowed us to distinguish the contributions of hexamers vs. octamers. Our results showed that septin filaments on SFs contain exclusively octamers, which are essential for the integrity of septin filaments and the integrity of SFs. Septin fiber widths measured by SIM microscopy are compatible with their organization as paired septin filaments. Nanometer-resolved distance measurements and single-protein tracking showed that septin filaments are closely apposed to the plasma membrane and largely immobilized on SFs. Finally, reconstitution assays showed that septin filaments mediate actin-membrane anchoring. We propose that septin filaments anchor and stabilize actin fibers at the plasma membrane (working model in Fig. 10 L).
Whether septins organize as filaments in a specific cellular context can now be tested using the tools we have developed in this study, including in genetic animal model systems. Our mutants and split-GFP assays are easily adapted for other septins within the SEPT2 group when required by cell- and tissue-specific expression (Karlsson et al., 2021; Uhlen et al., 2015). Split-GFP assays can also be used as readouts for filament formation in the context of studies aiming to identify regulators or inhibitors of septin polymerization or septin mutants related to disease.
An unexpected result of our study is that human septin filament integrity in cells depends entirely on octamers, questioning the functional importance of hexamers. We speculate that it is the SEPT3 group septin, which is absent from hexamers, that dictates septin function. The observation that SEPT7 assembly is most sensitive to SEPT7 and SEPT9 expression levels, and the fact that SEPT7–SEPT7 interactions are stronger than SEPT7-SEPT9 ones (Rosa et al., 2020) suggest that SEPT9, and possibly the other SEPT3 group septins, may help prevent SEPT7 from forming ectopic bundles. It is intriguing that Drosophila does not have any SEPT3 group septins and thus contains only hexamers (Field et al., 1996). Interestingly, Drosophila septins occasionally form cytoplasmic bundles devoid of Anillin (Hickson and O'Farrell, 2008). Such bundles also form in the absence of Anillin’s septin-binding domain that recruits septins to the plasma membrane (Kechad et al., 2012). We speculate that the formation of these bundles is analogous to the ectopic hexamer bundles we observed, forming at limiting amounts of a physiological partner.
Our findings suggest that actin-associated septin filaments in mammalian cells organize as paired septin filaments, with thicker septin fibers consisting of two to three double septin filaments. One single-molecule localization microscopy study of SF-associated septins has reported septin bundles with 25–150 filaments (Vissa et al., 2019), but this study assumed septin GTP-binding domains associating laterally without considering the presence of coiled-coils and the spacing occupied by coiled-coil pairing (Leonardo et al., 2021), so the number of filaments could be overestimated. We thus hypothesize that human septins in cells, like budding yeast septins, organize in a rather narrow range of assembly geometries. We speculate that the wide range of assembly geometries found for reconstituted septins in solution reflects their plasticity, but that the presence of a physiological partner leads to their native assembly into paired filaments (Bertin et al., 2010; Ong et al., 2014; Szuba et al., 2021).
Mammalian septins are distinct from other membrane-bound actin-binding and -crosslinking proteins in that they form filaments. Their capacity to polymerize, catalyzed by membrane binding (Szuba et al., 2021) and coupled with their ability to bind and cross-link actin filaments (Iv et al., 2021; Mavrakis et al., 2014), provides them with the unique potential to stabilize actin filament bundles and meshes at the plasma membrane over considerable distances. We propose that septins function by stabilizing ventral SFs, transverse arcs and perinuclear actin caps at the respective ventral and dorsal plasma membrane, participating in their generation or/and maintenance. It will be interesting to explore if cortical actin meshworks are also membrane attached through septins (Vadnjal et al., 2022). Septin enrichment adjacent to FAs lets us propose that they also contribute a stabilization function at the connection between FAs and SFs and thus impact FA maturation indirectly (Calvo et al., 2015; Dolat et al., 2014; Kang et al., 2021) by affecting the accumulation of mechanical tension on SFs.
The findings of this study lead to several remaining open questions. Whether septins in cells bind membranes and actin filaments directly or indirectly remains to be shown. Our data show that septins can associate with actin fibers in cells in the absence of myosin-II, supporting the possibility of direct septin-actin interactions in the context of SFs. The fact that septins are found only on contractile SFs suggests that myosin-II related signaling might be involved in their recruitment to SFs. Also what regulates septin polymerization in cells is still unknown. Cell-free reconstitution approaches and animal model systems promise to provide important further insights into the link between animal septin organization and function.
Materials and methods
Design of septin fusions for the tripartite split-GFP complementation assay
For the tripartite complementation assay to report SEPT-SEPT interactions with stringency, the amino acid linker length between SEPT and the β10- and β11-strands should not be too short in order to allow for the necessary proximity and flexibility for the β10- and β11-strands to orient in an antiparallel fashion for complementing GFP1-9, but it should be short enough to minimize reporting longer-range interactions. We used fluorescence imaging to test the dependence of split-GFP complementation on the linker length and on the position of the β10- and β11-tags by screening different homo- and hetero-septin combinations as shown in Fig. S2, A–C. All the combinations we tested resulted in fluorescence, reflecting the inherent flexibility of the N- and C-termini of SEPT2, 7 and 9. To allow for the most stringent complementation, we chose to use C-terminal fusions with 14-residue linkers for SEPT2-β10- and -β11 tags and for SEPT9-β10- and -β11 tags, and N-terminal fusions with 14-residue linkers for β10- and β11-SEPT7 tags. This short linker is comparable in length to the 10-residue-long β10- and β11-strands and thus long enough to allow the antiparallel arrangement of the latter. Protein structure models of human septin hexamers and octamers bearing full-length β10- and β11-tagged septins (see Materials and methods section “Modeling of human septin complexes”) confirmed the efficiency of GFP complementation for the final chosen linker length and β10/β11-tag positioning (Fig. 2, A and B; Fig. 3, A and B; and Fig. 5, G and H).
Plasmids and cloning
Septin and msfGFP cDNAs were as described in (Iv et al., 2021). mApple and sfCherry2 cDNAs were PCR-amplified from Addgene plasmids #54862 and #83031, respectively. Three types of mammalian expression plasmids were used in this study. A pCMV backbone (Clontech) was used for the expression of full-length fluorescent protein (msfGFP/mApple) fusions. A pcDNA3.1 backbone (Thermo Fisher Scientific), also with a CMV promoter, was used for the expression of β10- or β11-tagged septins. Finally, a pTRIP TRE Bi vector, modified from pTRE-Tight-BI (Takara-Bio; Koraichi et al., 2018), bearing a bidirectional tetracycline response element (TRE) promoter and an IRES-TagBFP cassette downstream β10-tagged septins, was used for the doxycycline-inducible co-expression of β10- and β11-tagged septins (Fig. S2, D and E). pCMV and pTRIP TRE Bi plasmids were used for all results presented in the figures. The pcDNA3.1 plasmids were used only for the initial screening (Fig. S2, A–C).
All pCMV plasmids, SEPT2 constructs in pTRIP TRE Bi plasmids and all interface mutants in pTRIP TRE Bi plasmids were cloned using seamless cloning (In-Fusion HD Cloning Plus Kit from Takara Bio, 638910). All pcDNAs and all wild-type SEPT7 and SEPT9-containing constructs in pTRIP TRE Bi plasmids were generated with classical cloning. In this latter case, DNA fragments were amplified by PCR using the PCR Master Mix from Thermo Fisher Scientific (K0171), TaqFast DNA polymerase (G277; Applied Biological Materials) or Phusion High-Fidelity DNA Polymerase (M0530S; New England Biolabs) and ligated into double digested plasmids with the Rapid DNA Ligation Kit from Thermo Fisher Scientific (K1422). pCMV constructs were cloned into a NheI/BamHI linearized vector. pcDNA constructs were cloned into a NheI/XbaI linearized vector. pTRIP TRE Bi constructs were cloned in two steps: first the β10-tagged septins were cloned into a SacII/NheI digested vector, then the β11-tagged septins were cloned into a NdeI/XbaI digested vector carrying the β10-tagged septin. The starting methionine of septin sequences is included in the N-terminal β10- and β11-tagged versions.
Bacterial expression plasmids for generating wild-type SEPT2-msfGFP hexamers and octamers-9_i3 were described in Iv et al. (2021) and are available through Addgene (#174492, 174498, 174499, 174501). pnEA-vH plasmids for the bacterial expression of SEPT2NCmut-msfGFP and SEPT2-sfCherry2 were generated using seamless cloning following the same strategy described in Iv et al. (2021). All primers for seamless cloning were Cloning Oligo (<60 bp) or EXTREmer (>60 bp) synthesis and purification quality from Eurofins Genomics and are listed in Table S1. All restriction enzymes were FastDigest enzymes from Thermo Fisher Scientific or from New England Biolabs. All plasmids were verified by sequencing (Eurofins Genomics) after each cloning step. We have deposited all plasmids with the nonprofit repository Addgene. Note that the SEPT9_i1NCmut in this study is the same as the SEPT9_i1NCmut2 in Kuzmic et al. (2022). Plasmid mCherry-SEPT9_i1 was from Addgene (#71622).
Cell lines, cell culture, and transfection
U2OS osteosarcoma cells for the expression of full-length fluorescent protein septin fusions were from ATCC (HBT-96). For the inducible co-expression of β10- and β11-tagged septins in the context of the tripartite split-GFP complementation system, we generated an inducible U2OS-Tet-On-GFP1-9 cell line which expresses constitutively a GFP1-9 fragment and an anti-GFP VHH intrabody that enhances split-GFP fluorescence. To generate this cell line, U2OS cells were successively transduced with lentiviruses encoding rtTA, GFP1-9 (#130271; Addgene) and anti-GFP VHH G4 (#182236; Addgene) and tested for complementation efficiency using transient expression of a GFP10-zipper-GFP11 domain (Koraïchi et al., 2018). One additional round of transduction with GFP1-9 lentivirus led to an optimized U2OS-Tet-On-GFP1-9 cell line that showed 80% GFP positive cells upon expression of the GFP10-zipper-GFP11 domain. An IRES-TagBFP cassette downstream β10-tagged septins was used for monitoring septin expression. Cells were maintained in McCoy’s medium (16600082; Gibco) supplemented with 10% fetal bovine serum (S181H; Dominique Dutscher), 100 U/ml penicillin and 100 μg/ml streptomycin antibiotics (P4333; Sigma-Aldrich) in a humidified atmosphere at 37°C containing 5% CO2.
Transfections with pcDNAs, for the screening of β10- and β11-tag combinations (Fig. S2, A–C), were performed 16 h prior to immunostainings using jetPRIME (101000015; PolyPlus). To obtain single cells for imaging, 50 × 103 U2OS-Tet-On-GFP1-9 cells were typically grown on 18 mm coverslips (Knittel Glass MS0010), previously cleaned by sonication in 70% ethanol, and placed into a 12-well plate a day prior to the day of transfection, for allowing an optimal number of cells to attach and spread. A total of 0.4 μg of DNA and a 4:1 ratio of jetPRIME (μl) : DNA (μg) were used per reaction. To minimize septin overexpression artifacts, the total amount of DNA was composed by 30 ng of β10-septin, 30 ng of β11-septin and 340 ng of empty vector.
Transfections with either pCMV or pTRIP TRE Bi plasmids and siRNAs were performed through electroporation using the Neon Transfection System (MPK5000; Thermo Fisher Scientific). For pCMVs, a single 100-μl reaction using 1.8 × 106 U2OS cells, 300 pmol of each siRNA and 6 μg of each DNA was electroporated within the dedicated tip (MPK10096; Thermo Fisher Scientific). Electroporation parameters consisted in four pulses of 10 ms width and a voltage of 1,230 V. The electroporated cells were then inoculated in 5 ml of culture medium without antibiotics, and immediately divided for native-PAGE, SDS-PAGE/Western blots and immunostaining as follows: 3 ml in a 6-cm dish containing 2 ml of medium without antibiotics, 2 ml in a 6-cm dish containing 3 ml of medium without antibiotics, and 100 μl in the well of a 12-well plate containing a 18-mm coverslip in 900 μl of medium without antibiotics, respectively. A satisfactory septin knockdown efficiency was achieved within 48–96 h after electroporation. Typically, immunostaining and protein extraction were performed 72 h post electroporation.
For pTRIP TRE Bi plasmids, a single 100-μl reaction using 1.8 × 106 U2OS-Tet-On-GFP1-9 cells, 300 pmol of each siRNA and 6 μg of each DNA was electroporated within the dedicated tip using the same electroporation parameters described previously. The electroporated cells were then inoculated in 5.5 ml of culture medium without antibiotics and immediately divided for SDS-PAGE/Western blots, immunostaining and live cell imaging as follows: 5 ml in a 6-cm dish, 400 μl in the well of a 12-well plate containing a 18-mm coverslip in 600 μl of medium without antibiotics, and 200 μl in the well of a 24-well glass bottom plate (Cellvis, P24-1.5H-N) containing 800 μl of medium without antibiotics, respectively. After either 48 h, for samples intended for live cell imaging or immunostainings, or 72 h, for samples for biochemical analysis, protein expression was induced using 1 μg/ml of doxycycline (D9891; Sigma-Aldrich) for 16 h.
Septin mutant phenotype classification
The diffuse cytosolic vs. non-diffuse phenotype classification analysis for mutant characterization with pCMV plasmids (Fig. 2 H, Fig. 4 E, Fig. 6 C, and Fig. S4 A) was done from three independent experiments. Transfected cells were fixed and co-stained for actin and α-tubulin. Each round of experiments was composed of the 30 first fluorescent cells found randomly in the sample, with the exception of one round containing 11 cells for msfGFP-SEPT7 and 8 cells for msfGFP-SEPT7Gmut1. Acquired images were classified as “diffuse cytosolic” in the presence of purely diffuse cytosolic signal or as “non-diffuse” in the presence of structure-like signal; no differentiation was applied for SF-, microtubule-, membrane-like or punctate signals in the latter case. The violin graphs representing the phenotype distributions show the mean intensity distribution calculated on the whole field of view from maximum intensity projections of all z-planes. The phenotype classification for reconstituted split-GFP fluorescence distribution (Fig. 2 F, Fig. 4, G and H; Fig. 6 D; and Fig. S4 B) was identical, in terms of the used criteria and graph display, but the data was generated from two independent experiments, with each experimental round composed of the 20 first fluorescent cells, in live cell imaging, with the exception of one round containing 13 cells for SEPT7Gmut1-SEPT7Gmut1 and 9 cells for SEPT7Gmut2-SEPT7Gmut2. For the septin-actin colocalization analysis, the diffuse cytosolic vs non-diffuse phenotype sorting is displayed both as scatter dot plots and as a pie graph to highlight the diffuse cytosolic vs. non-diffuse proportion from each condition. Bars in scatter dot plots depict means, and error bars represent SD. Violin plots, scatter dot plots, and pie graphs were prepared using GraphPad Prism. The number of cells used to assess the phenotypes for each condition is indicated in the respective legends.
RNA interference
Control synthetic small interfering RNA (siRNA) targeting the coding region of LacZ (5′-GCGGCUGCCGGAAUUUACC-3′) and siRNA targeting the 3′UTR region of all SEPT9 mRNA variants (5′-GGAUCUGAUUGAGGAUAAA-3′) were previously validated (Verdier-Pinard et al., 2017). The siRNA sequences targeting the 3′UTR regions of SEPT2 and SEPT7 were 5′-ACACUUUCCUGGAUAAAAA-3′ and 5′-GCAUUUAGCUGUAUUCAUA-3′, respectively. All siRNAs were designed to hybridize with 19-bp sequences in the 3′UTR regions of septin genes, thus knocking down endogenous septins while allowing the expression of the transfected plasmids. 21mer siRNAs, 20 nmol each, were synthesized with dTdT overhangs by Eurofins, and delivered as annealed and ready-to-use siRNA duplexes. siRNAs targeting the coding regions of non-muscle myosin heavy chain IIA (NMIIA; 5′-GCCACGCCCAGAAGAACGAGAAUGC-3′) and non-muscle myosin heavy chain IIB (NMIIB; 5′-UCAAUAAAGCUCUGGAUAGGACCAA-3′) were previously validated (Kage et al., 2022). 20 nmol of these 25mer siRNAs without overhangs were synthesized by Eurofins, and delivered as annealed and ready-to-use siRNA duplexes.
SDS-PAGE and Western blotting of cell lysates
The dish containing the cells was placed on ice and the cells were washed twice with PBS, without Ca2+ and Mg2+, before being detached with 40 μl of ice-cold lysis buffer (10 mM Tris-HCl, pH 7.5, 148.5 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1× PhosSTOP Roche, 5× cOmplete protease inhibitor cocktail Roche, 1 mM DTT) using a cell scraper (TPP 99003). The lysate was collected in a 1.5 ml tube and incubated on ice for 30 min. The lysates were then centrifuged at 20,000 g for 20 min at 4°C for removing cell debris. An aliquot of 6 μl was collected for protein quantification using the BCA Protein Assay (23227; Thermo Fisher Scientific) and the remaining clarified lysates were kept at −20°C until SDS-PAGE analysis.
The lysates were analyzed by 4–20% SDS-PAGE using Mini-PROTEAN TGX Precast Protein Gels (4561095; BioRad). Molecular mass markers were Precision Plus Protein All Blue Standards (1610373; BioRad) or Amersham ECL Rainbow Marker (RPN800E; Cytiva). For the Western blot, the gel, the PVDF Immobilon-PSQ membrane (ISEQ85R; MERCK), filter pads, and filter papers were all incubated in transfer buffer (25 mM Tris, 192 mM glycine and 20% of methanol) for 15 min before assembly in the Mini Trans-Blot transfer cell (1703935; BioRad). The transfer was done at 4°C for 16 h at 110 mA constant current. The transfer efficiency was checked by Ponceau S staining (P7170; Sigma-Aldrich). The membrane was then blocked in a 3% wt/vol dry milk TBS-T solution (20 mM Tris-HCl pH7.5, 200 mM NaCl and 0.1% vol/vol Tween20) for 90 min under constant agitation. Primary and secondary antibodies were diluted in the same blocking solution and incubated over the membrane for 90 and 60 min, respectively. In between antibody incubations, membranes were washed three times for 10 min with TBS-T, and the very last wash right before ECL detection was done only with TBS.
The loaded amount of extracted protein in the gels was adapted depending on the expression promoter and the analyzed septin. For pCMV plasmids used to assess the knockdown efficiency (Fig. S2, F–H), a total of 4 μg of extracted protein was used for detecting endogenous SEPT2, 8 μg for endogenous SEPT7, and 4 μg for endogenous SEPT9. For pTRIP TRE Bi plasmids, a total of 8 μg of extracted protein was used for all analysis. To detect specific septins, we used rabbit anti-SEPT2 (1:2,500, HPA018481; Sigma-Aldrich), rabbit anti-SEPT7 (1:200, sc-20620; Santa Cruz Biotechnology) and rabbit anti-SEPT9 (1:4,000, 10769-1-AP; Proteintech). For detecting β10- and β11-tag expression, we used rabbit anti-β10 (1:5,000) and rabbit anti-β11 (1:5,000; Koraichi et al., 2018). For detecting tubulin as a loading control, we used mouse anti-α-tubulin (1:2,500, T9026; Sigma-Aldrich). For total actin level quantification (Fig. 8 D), we used mouse anti-actin (1:1,000, AC-40, ab11003; Abcam). Secondary HRP-conjugated antibodies were either anti-rabbit-IgG (1:10,000, GENA934; Cytiva) or anti-mouse-IgG (1:10,000, GENA931; Cytiva). Chemiluminescent detection was performed with an Amersham ImageQuant 800 imager (29399481; Cytiva) using Amersham ECL Select Western Blotting Detection Reagent (RPN2235; Cytiva) diluted five times in Milli-Q water. The membrane was incubated with the diluted reagent for 30 s, and washed for 10 s in TBS right before image acquisition. Images were collected in time series mode every 10 s, for a total of 50 images, and processed with ImageQuantTL software for quantification of the band intensities to measure expression levels. Expression quantification graphs for assessing septin knockdown efficiency were prepared using GraphPad Prism and are shown as mean values (normalized to 1 for siCtrl) with the error bar representing SD. Data are from at least three independent siRNA treatments. Data distribution for total actin level quantification (Fig. 8 D) was assumed to be normal, but this was not formally tested.
Native PAGE and Western blotting of cell lysates
The dish containing the cells was placed on ice and the cells were washed twice with PBS, without Ca2+ and Mg2+, before being detached with 40 μl of ice-cold native lysis buffer (80 mM PIPES, pH 6.9, 2 mM MgCl2, 4 mM EGTA, 0.2% saponin, 5× cOmplete protease inhibitor cocktail Roche). The lysate was collected in 1.5 ml tube and incubated on ice for 10 min. The lysates were then centrifuged at 14,000 g for 10 min at 4°C for removing cell debris. To prevent septin polymerization, clarified lysates were supplemented with NaCl, adding 10 μl of NaCl 5 M for each 100 μl of lysate. After 15 min of incubation on ice, the lysates were clarified in a second centrifugation step of 10 min, 14,000 g at 4°C. An aliquot of 12 μl was collected for protein quantification using the BCA Protein Assay (23227; Thermo Fisher Scientific), and the remaining clarified lysates were kept at −20°C until Native PAGE analysis.
The lysates were analyzed by 4–16% Native PAGE using precast Bis-Tris Mini Protein Protein Gels (BN1003BOX; Invitrogen) following the manufacturer’s instructions. The molecular mass marker was NativeMark Unstained Protein Standard (LC0725; Invitrogen). For the Western blot, the gel, the PVDF Immobilon-PSQ membrane, filter pads, and filter papers were all incubated in NuPAGE transfer buffer for 15 min before assembly. The transfer was done at 4°C for 16 h at 20 V constant voltage. The transfer efficiency was checked by destaining the membrane with an aqueous solution containing 25% of methanol and 10% of acetic acid. The protein marker was identified and the membrane completely destained with pure methanol for 3 min. The membrane was then blocked and stained with the respective antibodies as described for SDS-PAGE Western blots.
The loaded amount of extracted protein in the gels was again adapted depending on the analyzed septin. A total of 10 μg of extracted protein was used for detecting endogenous or exogenously expressed SEPT2 and SEPT7, and 10 or 4 μg for endogenous or exogenously expressed SEPT9, respectively. To detect specific septins, we used mouse anti-SEPT2 (1:7,500, 60075-1; Proteintech), rabbit anti-SEPT7 (1:200, sc-20620; Santa Cruz Biotechnology) and rabbit anti-SEPT9 (1:2,000, 10769-1-AP; Proteintech). Secondary HRP-conjugated antibodies were either anti-rabbit IgG (1:10,000, GENA934; Cytiva) or anti-mouse IgG (1:10,000, GENA931; Cytiva). Chemiluminescent detection was done with an Amersham ImageQuant 800 imager (29399481; Cytiva) using Amersham ECL Select Western Blotting Detection Reagent (RPN2235;Cytiva) as described previously for SDS-PAGE Western blot.
Immunofluorescence
Cells were fixed for 15 min with 4% paraformaldehyde (15714; Electron Microscopy Sciences) in 37°C-prewarmed cytoskeleton buffer (10 mM MES pH 6.1, 150 mM NaCl, 5 mM EGTA, 5 mM MgCl2, 5 mM glucose), followed by 2 × 5 min wash steps in phosphate-buffered saline (PBS) solution, and a subsequent permeabilization and blocking step with PBS containing 0.1% saponin and 1% IgG-free/protease free bovine serum albumin (BSA; 001-000-161; Jackson ImmunoResearch) for 1 h at RT. Cells were incubated successively with primary antibodies for 16 h at 4°C in a humidified chamber, followed by secondary Alexa Fluor-conjugated IgG antibodies combined with 0.165 μM Alexa Fluor 647-phalloidin (A22287; Thermo Fisher Scientific) for 2 h at RT. Antibody solutions were prepared in PBS containing 0.1% saponin and 1% BSA, and 3 × 10 min wash steps in the same buffer were performed in between antibody incubations. Coverslips with stained cells were washed 2 × 5 min in PBS and then mounted with 15 μl Fluoromount (F4680; Sigma-Aldrich) for image acquisition. Primary antibodies were rabbit anti-SEPT2 (1:500, HPA018481; Sigma-Aldrich), rabbit anti-SEPT7 (1:500, 18991; IBL), rabbit anti-SEPT9 (1:200, 10769-1-AP; Proteintech), mouse anti-α-tubulin (1:10,000, T9026; Sigma-Aldrich), mouse anti-paxillin (1:500, 05–417; Merck Millipore). Secondary antibodies were donkey AlexaFluor488-conjugated anti-rabbit IgGs (1:500, A10037; Thermo Fisher Scientific) and donkey AlexaFluor568-conjugated anti-mouse IgGs (1:500, A21206; Thermo Fisher Scientific). For F-actin, SEPT9, and SEPT7 co-stainings, performed for the quantification of the number of ventral SFs in cells (see Materials and methods section “Quantification of the frequency of ventral SF presence upon septin perturbation”), rabbit anti-SEPT9 (1:200, 10769-1-AP; Proteintech) was combined with rat anti-SEPT7 (1:150, clone 10A7, described in Kuzmic et al. [2022]). Secondary antibodies in this case were donkey AlexaFluor488-conjugated anti-rabbit IgGs (1:500, A10037; Thermo Fisher Scientific) and goat AlexaFluor647-conjugated anti-rat IgGs (1:500, A21247; Thermo Fisher Scientific), and were incubated together with 0.165 μM Atto590-phalloidin (ATTO-TEC AD 590-81).
Immunostaining after live-cell extraction vs. after extraction post-fixation
To live-extract cells (Fig. 10 A), we incubated cells in 37°C-prewarmed cytoskeleton buffer containing 0.1% v/v Triton X-100 for 1 min, then replaced immediately with 37°C-prewarmed cytoskeleton buffer containing 4% paraformaldehyde (43368; Alfa Aesar) and fixed cells for 15 min. Cells were rinsed with PBS and incubated in a permeabilization/blocking solution of PBS containing 0.1% saponin and 5% goat serum (16210064; Thermo Fisher Scientific) overnight at 4°C. Cells were incubated successively with primary antibodies for 2 h at RT in a humidified chamber, followed by secondary Alexa Fluor-conjugated IgG antibodies combined with 0.165 μM Alexa Fluor 546-phalloidin (A22283; Thermo Fisher Scientific) for 1 h at RT. Antibody solutions were prepared in PBS containing 0.1% saponin and 5% goat serum, and 3 × 10 min wash steps in the same buffer were performed in between antibody incubations. Coverslips with stained cells were washed 2 × 5 min in PBS and then mounted with Fluoromount-G (0100-01; Southern Biotech) for image acquisition. Primary antibodies were rabbit anti-SEPT7 (1:400, 18991; IBL) and mouse anti-non-muscle myosin heavy chain IIA (NMIIA; 1:200, ab55456; Abcam). Secondary antibodies were goat AlexaFluor488-conjugated anti-mouse IgGs (1:400, A11001; Thermo Fisher Scientific) and goat AlexaFluor633-conjugated anti-rabbit IgGs (1:400, A21070; Thermo Fisher Scientific). The respective control experiment, i.e., extracting cells post-fixation (Fig. 10 A), involved fixing cells in 37°C-prewarmed cytoskeleton buffer containing 4% paraformaldehyde for 15 min, then extracting cells with PBS containing 0.5% Triton X-100 and 5% goat serum for 10 min before overnight permeabilization/blocking and antibody incubations as described above. This last protocol was also used for the immunostainings shown in Fig. 9; and Fig. S1, C and D using the additional primary antibody mouse anti-α-actinin-1 (1:200, clone BM 75.2; Thermo Fisher Scientific). Cells shown in Fig. 9, Fig. 10 A; and Fig. S1, C and D were plated on fibronectin-coated coverslips and left to attach and spread for 6 h before immunostainings. Human plasma fibronectin was from Millipore (FC010) and was used at 20 μg/ml in 100 mM bicarbonate buffer, pH 8.5, for coating coverslips overnight at 4°C.
Split-GFP complementation in cells using recombinant purified GFP1-9
Recombinant GFP1-9 expression was achieved from a pET-28a (+) vector coding for GFP1–9 OPT (Cabantous et al., 2013) with an in-frame 6xHis coding sequence (#182240; Addgene). For protein expression, the plasmid was transformed into Escherichia coli BL21(DE3; C2527; New England Biolabs). A 3 ml overnight culture was inoculated into 50 ml of Luria-Bertani (LB) medium containing 35 μg/ml kanamycin and was grown at 37°C to A600nm ∼0.6. The temperature was reduced to 25°C prior to induction with 0.1 mM isopropyl β-D-thiogalactoside (IPTG) for 24 h. Bacterial cultures were collected by centrifugation at 4,200 g for 10 min. Cell pellets were resuspended in 20 ml lysis buffer (150 mM NaCl, 150 mM Tris-HCl, pH 7.2, 5% vol/vol glycerol, 0.2 mM TCEP, 10 mM imidazole) and lysed by sonication on ice with a 0.5-inch diameter probe with 30 s ON, 30 s OFF pulses for 4 min total. The lysate was centrifuged at 30,000 g for 20 min. The supernatant was loaded on a column with 1 ml of TALON Metal Affinity Resin (635503; Takara Bio) that was equilibrated with the lysis buffer. After two washes with 10 volumes of lysis buffer, recombinant GFP1–9 was eluted with the elution buffer (200 mM imidazole in the same buffer). Recombinant GFP1-9 fractions were pooled and dialyzed in TNG buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.2, 5% vol/vol glycerol). Protein concentration was determined with the Bio-Rad Protein Assay (5000002; Bio-Rad).
For split-GFP complementation in cells, wild-type U2OS cells were treated with SEPT2 siRNA and also co-transfected with SEPT2-β10- and SEPT2-β11-encoding pCMV plasmids, and plated on 18-mm coverslips as described above for immunostainings. 48 h post-electroporation, cells were fixed and permeabilized as described for immunofluorescence, and incubated with the recombinant purified GFP1-9 solution (0.2 mg/ml in TNG buffer) diluted twofold in the permeabilization buffer for 4 h at RT followed by overnight incubation at 4°C. After 2 × 10 min wash steps, cells were stained with 0.165 μM Alexa Fluor 647-phalloidin for 1 h at RT. Fluorescence images of SEPT2-SEPT2 reconstituted GFP in the phalloidin-stained cells were acquired as described in the confocal fluorescence microscopy methods section.
Myosin-II-dependence of septin recruitment to stress fibers
For septin costainings with non-muscle myosin heavy chain IIA (NMIIA) and non-muscle myosin heavy chain IIB (NMIIB), wild-type U2OS cells were treated with siLacZ siRNA and prepared for immunofluorescence as described in the respective sections. Primary antibodies were rabbit anti-NMIIA (1:400, 909801; BioLegend), rabbit anti-NMIIB (1:200, 3404; Cell Signaling), and rat anti-SEPT7 (1:150, clone 10A7, described in Kuzmic et al. [2022]), and were incubated for 4 h at RT. Secondary antibodies were donkey AlexaFluor488-conjugated anti-rabbit IgGs (1:400, A10037; Thermo Fisher Scientific) and goat AlexaFluor568-conjugated anti-rat IgGs (1:400, A11077; Thermo Fisher Scientific), and were incubated together with 0.165 μM Alexa Fluor 647-phalloidin (A22287; Thermo Fisher Scientific) for 1 h at RT.
To examine septins in the absence of a given NMII isoform, CRISPR-mediated NMIIA and NMIIB KO U2OS lines (Kage et al., 2022), a kind gift from Frieda Kage and Henry Higgs (Dartmouth College, Hanover, NH, USA), were treated with siLacZ siRNA and immunostained for SEPT7, F-actin and the myosin isoform that is not knocked-out, i.e., for NMIIB in the NMIIA KO line and for NMIIA in the NMIIB KO line, using the same antibodies described in this section. NMIIA KO and NMIIB KO U2OS lines were cultured as wild-type U2OS cells.
Given that NMIIA is by far the most abundant NMII in U2OS cells (Kage et al., 2022) and that the efficiency of knocking down NMIIB (∼ 96–98%) was much higher than knocking down NMIIA in our hands (∼ 60–76%), we chose to treat NMIIA KO cells with NMIIB siRNA to examine septins under conditions of minimal presence of NMII in U2OS cells. Treatment with NMIIB siRNA was performed as described in (Kage et al., 2022) but with electroporation. SDS-PAGE and Western blotting in the NMIIA KO and NMIIB KO cell lysates was performed as described in the respective Materials and methods section using the same rabbit NMII antibodies described in this section at 1:1,000 each, rabbit anti-SEPT7 (1:200, sc-20620; Santa Cruz Biotechnology), and mouse anti-α-tubulin (1:2,500, T9026; Sigma-Aldrich).
Quantification of the frequency of ventral SF presence upon septin perturbation
The frequency of ventral SF presence in wild-type U2OS cells treated with siLacZ siRNA (6mer+8mer), with SEPT9 siRNA (6mer), with SEPT7 siRNA and also transfected with msfGFP-SEPT7Gmut2 (8mer), and in U2OS cells treated with SEPT2 siRNA and also transfected with SEPT2NCmut-msfGFP (no filaments), was quantified from at least four independent experiments (Fig. 8 C). Electroporated cells were either incubated with SiR-actin for live cell imaging (two independent experiments for 6mer+8mer, 6mer and 8mer conditions) or fixed and co-stained for F-actin, SEPT9 and SEPT7 (from two to six independent experiments for 6mer+8mer, 6mer, 8mer and “no filaments” conditions). Each round of experiments was composed of at least 20 and 10 fields of view for live and fixed cell imaging, respectively. Only cells with most of their surface in the acquired field of view were considered for the quantification, and were classified regarding the presence, in the case of at least one detected ventral SF, or absence of ventral SFs based on the phalloidin or SiR-actin signal; no differentiation was applied for the density/number of ventral SFs, or the presence of other SF subtypes, as no clear effect could be detected in the latter. Each dot in the scatter plots represents the percentage of cells presenting ventral SFs in a given round of experiments. Bars in scatter dot plots depict means, and error bars represent SD. Scatter dot plots were prepared using GraphPad Prism. The number of cells used to assess the phenotypes for each condition is indicated in the respective legends. Data distribution was assumed to be normal but this was not formally tested. For the quantification of the percentage of SEPT9 siRNA-treated cells containing SEPT7 vs. SEPT9-decorated SFs, two rounds of experiments using fixed cells were used. Only 2–3% of SEPT9 siRNA-treated cells showed SEPT7-decorated SFs, and these same SFs contained also SEPT9, corresponding to the small percentage of cells where SEPT9 was not knocked down.
Confocal fluorescence microscopy of cells and image processing
For live cell imaging, right before microscopy and due to the absence of CO2 control on our microscope setup, the culture medium was exchanged by Leibovitz medium (21083027; Gibco) supplemented with 10% fetal bovine serum and antibiotics. Cells were kept at 37°C in a heating chamber (H301-T-UNIT-BL; OkoLab). Fluorescence images of live or fixed cells were acquired using a spinning disk unit (CSU-X1-M1 from Yokogawa) connected to the side-port of an inverted microscope (Eclipse Ti2-E from Nikon Instruments) using a Nikon Plan Apo ×100/1.45 NA oil immersion objective lens, 488- 561- and 641-nm laser lines (Coherent) and an iXon Ultra 888 EMCCD camera (1,024 × 1,024 pixels, 13 × 13 μm pixel size, Andor, Oxford Instruments) resulting in an image pixel size of 65 nm. Z-stacks were acquired with a Δz interval of 0.4 μm. Exposure times were in the range of 0.5–3.0 s depending on the exact condition. For the non-diffuse vs. diffuse cytosolic phenotype classification for septin mutant characterization, acquisition parameters were kept the same among imaging sessions. For actin or microtubule co-labeling in live cells, cells were incubated for 30 min with 0.5 μM of SiR-actin or 60 min with 0.5 μM of SiR-tubulin and 10 μM of verapamil in culture medium (SiR Cytoskeleton Kit, Spirochrome SC006).
Images were processed with the open-source image processing software ImageJ/Fiji. All shown images, except for the ones used for the septin-actin co-localization analysis that were acquired as single z-planes, are maximum intensity projections of two consecutive z-planes contrasted manually in order to optimize the image display. For septin-actin co-localization measurements, acquired channels of single z-planes, for septin and actin, were individually processed as follows: images were subjected to automatic contrast enhancement, allowing 0.1% of saturated pixels, then to a blurring with a Gaussian filter of radius 1.0 and a subsequent background subtraction using a rolling ball radius of seven pixels. A manual intensity threshold was used when calculating Pearson and Manders co-localization coefficients, using the JACoP plugin for ImageJ (Bolte and Cordelières, 2006).
Images shown in Fig. 10 A and Fig. S1, C and D were acquired on a Zeiss LSM 710 laser scanning confocal microscope using a PlanApochromat 100×/1.4 NA oil immersion objective lens, 488- 543- and 633-nm laser lines for excitation, with all channels at 1AU for the pinholes. Z-stacks were acquired with a Δz interval of 0.48 μm. All shown images are single z-planes and were processed with ImageJ/Fiji.
Super-resolution structured illumination microscopy
Sample preparation and image acquisition
Cells for super-resolution structured illumination (SIM) microscopy were plated on high precision (170 ± 5 µm thick) 18 × 18 mm glass coverslips from Zeiss (474030-9000-000) and prepared for immunostainings as detailed in the section “Immunostaining after live-cell extraction vs. after extraction post-fixation”; all images shown in Fig. 9 and Fig. S4 K employ extraction post-fixation. For microtubule stainings used for microtubule width measurements in Fig. 9, F and H, cells were fixed with −20°C-prechilled methanol for 2 min at −20°C and rinsed with PBS before overnight permeabilization/blocking and antibody incubations as described in the above section. Primary antibodies were mouse tubulin (1:1,000, T9026; Sigma-Aldrich) and rabbit anti-SEPT7 (1:500, 18991; IBL). Secondary antibodies were goat AlexaFluor488-conjugated anti-mouse IgGs (1:400, A11001; Thermo Fisher Scientific) and goat AlexaFluor633-conjugated anti-rabbit IgGs (1:400, A21070; Thermo Fisher Scientific). Images in Fig. 9 were acquired on a Zeiss Elyra PS.1 super-resolution microscope using an alpha PlanApochromat 100×/1.46 NA DIC M27 Elyra oil immersion objective lens, 488-, 561-, and 642-nm laser lines for excitation and respective BP495-550, BP570-620 and LP655 emission filters. Z-stacks were acquired with a Δz interval of 0.101 μm. Images were processed and channel-aligned with the Zeiss ZEN Black software. Images in Fig. S4 K were acquired on a DeltaVision OMX SR (Leica Microsystems/Cytiva) super-resolution microscope using an Olympus PlanApo N 60×/1.42 NA oil immersion objective lens, 488- and 640-nm laser lines for excitation and respective 528/48 and 683/40 emission filters. Z-stacks were acquired with a Δz interval of 0.125 μm. Images were processed and channel-aligned with the DeltaVision softWoRx 7.0.0 software. All shown images are single z-planes and were prepared with ImageJ/Fiji.
Septin fiber diameter and length measurements
Full width at half maximum (FWHM) measurements for measuring the diameter of microtubules (MT) and septin fibers in SIM images (Fig. 9, F and H) were made with a custom-generated Matlab code (FilamentAnalysis.mlx), the source code of which is available at https://github.com/cchandre/Polarimetry. A line was drawn perpendicular to the axis of the MT or to the long axis of the septin fiber, and the FWHM was extracted from the intensity profile using the findpeaks Matlab function. We measured the width of MTs and septin fibers at multiple positions along their length and in multiple microtubules and multiple septin fibers for each SF type per cell. Box plots depicting the distribution of FWHM measurements (Fig. 9 F) were prepared using GraphPad Prism (one data point corresponds to one width measurement). The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of measurements per condition (MT or SF type) is indicated in the respective legend. A Kruskal–Wallis test followed by a multiple comparison test was used for comparing the distributions.
Length measurements were implemented in the same custom-generated code. A line was drawn parallel to the long axis of the septin fiber and the length extracted with the curveLength Matlab function. We measured the length of multiple septin fibers for each SF type per cell. Scatter dot plots depicting the distribution of length measurements (Fig. 9 J) were prepared using GraphPad Prism (one data point corresponds to one length measurement). The number of measurements per SF type is indicated in the respective legend. Bars depict median values.
Numerical simulations for fiber size estimation
Numerical simulations of the expected FWHM in SIM images (“image diameter” in Fig. 9 G) as a function of the real fiber diameter (“fiber diameter” in Fig. 9 G) were made with a custom-generated Matlab code (Convolution_1D.m), the source code of which is available at https://github.com/cchandre/Polarimetry/tree/master/AdditionalFiles. A Gaussian point spread function (PSF) was used, and the curve was generated from the convolution of this PSF with an increasing fiber diameter size, using the conv Matlab function. Assuming a real antibody-decorated MT diameter size of ∼60 nm (Weber et al., 1978), the convolution curve permits to deduce the PSF size from the measured FWHM in isolated microtubule fibers (the median value is used). This PSF size being linearly dependent on the emission wavelength, it is then rescaled to account for the wavelength difference used in MT vs. septin imaging: MTs and septins were imaged at 488 and 642 nm, respectively, for SIM in Fig. 9, whereas MTs and septins for SIM in Fig. S4 K were imaged at 640 and 488 nm, respectively. To predict the real width of the respective septin fiber diameters (Fig. 9 H), the convolution curve was finally used for the estimated PSF, using as input the measured septin FWHM.
Production and purification of recombinant human septin complexes
Wild-type nonfluorescent and SEPT2-msfGFP hexamers and octamers-9_i3, SEPT2NCmut-msfGFP hexamers and octamers-9_i3, and SEPT2-sfCherry2 octamers-9_i3 were produced and purified as follows. Plasmids expressing SEPT2, SEPT2-msfGFP or SEPT2NCmut-msfGFP, and plasmids co-expressing SEPT2, SEPT2-msfGFP or SEPT2NCmut-msfGFP and SEPT6, were co-transformed with plasmids co-expressing SEPT6 and SEPT7 (#174499; Addgene), or SEPT7 and SEPT9_i3 (#174501; Addgene), for generating recombinant nonfluorescent, SEPT2-msfGFP, or SEPT2NCmut-msfGFP hexamers and octamers-9_i3 (Iv et al., 2021). Plasmids co-expressing SEPT2-sfCherry2 and SEPT6 were co-transformed with plasmids co-expressing SEPT7 and SEPT9_i3 (#174501; Addgene) to generate recombinant SEPT2-sfCherry2 octamers-9_i3. The N-terminus of SEPT2 is tagged with a His6-tag, and the C-terminus of SEPT7 (for isolation of hexamers), or the C-terminus of SEPT9 (for isolation of octamers), is tagged with a Strep-tag. A purification scheme comprising a Strep-Tactin affinity column to capture Strep-tagged complexes, followed by a nickel affinity column to retain the Strep-tagged complexes that also bear His6-tagged septins isolates hexamers and octamers (Iv et al., 2021).
Co-transformed E. coli BL21(DE3) were selected on LB agar plates with carbenicillin and spectinomycin each at 100 µg/ml. A single colony was selected to prepare an overnight LB medium preculture at 37°C with antibiotics at 100 µg/ml. Terrific Broth with antibiotics at 50 µg/ml, typically 3.5–5 L, was inoculated with the pre-culture and incubated at 37°C. Bacteria were left to grow to A600nm ∼ 0.6–0.8 before inducing expression with 0.5 mM IPTG for overnight expression at 17°C. The culture was stopped by centrifuging at 3,400 g for 15 min and 4°C, and the supernatants were pooled and further centrifuged at 5,000 g for 10 min and 4°C. Bacteria pellets were stored at −20°C until protein purification. Bacteria expressing msfGFP- and sfCherry2-tagged septins yield yellow-greenish and pink-reddish pellets, respectively.
On the day of purification, the pellet was resuspended in ice-cold lysis buffer (50 mM Tris-HCl, pH 8, 300 mM KCl, 5 mM MgCl2, 0.25 mg/ml lysozyme, 1 mM PMSF, cOmplete protease inhibitor cocktail [1 tablet per 50 ml], 10 mg/l DNase I, 20 mM MgSO4) and lysed on ice using a tip sonicator with 5 cycles of 30 s “ON”, 15 s “OFF”. The lysate was clarified by centrifugation for 30 min at 20,000 g and 4°C, and the supernatant loaded on a StrepTrap HP column. Strep-tag-II-containing septin complexes were eluted with 50 mM Tris-HCl, pH 8, 300 mM KCl, 5 mM MgCl2, and 2.5 mM desthiobiotin. The pooled fractions were then loaded to a HisTrap HP column, and His6-tag-containing complexes eluted with 50 mM Tris-HCl at pH 8, 300 mM KCl, 5 mM MgCl2, and 250 mM imidazole. Only the highest-concentration peak fractions were collected. Both affinity steps were performed on an ÄKTA pure protein purification system at 4°C (Cytiva). To remove imidazole, we either performed overnight dialysis or used a PD-10 column, also including DTT in this last step. The final elution buffer, in which septins are stored, was 50 mM Tris-HCl, pH 8, 300 mM KCl, 5 mM MgCl2, and 1 mM DTT. Protein concentration was assessed with absorbance measurements at 280 nm from the calculated extinction coefficients using ExPASy, and protein aliquots were flash-frozen in liquid nitrogen and stored at −80°C until further use.
Chemicals used for recombinant septin complex production and purification are as follows. E. coli BL21(DE3) from Agilent (200131). Carbenicillin (C3416), spectinomycin (S4014), LB broth medium (L3022), LB agar (L2897), SOC medium (S1797) from Sigma-Aldrich. Terrific Broth from MP Biomedicals (091012017). IPTG (EU0008-C) and lysozyme (5933) from Euromedex. Imidazole from Thermo Fisher Scientific (Fisher Chemical I/0010/53). PMSF (78830), cOmplete Protease Inhibitor Cocktail Tablets (11836145001; Roche), DNase I (10104159001; Roche), d-Desthiobiotin (D1411), and DTT (D0632) from Sigma-Aldrich. HisTrap HP 1 ml columns (17524701) and StrepTrap HP 1 ml columns from Cytiva (28907546). 20 K MWCO Slide-A-Lyzer cassettes from Thermo Fisher Scientific (87735). PD-10 desalting columns from Cytiva (17085101).
Sample preparation for fluorescence microscopy of in vitro reconstituted actin and septins
To prepare flow cells, glass slides and coverslips were cleaned for 15 min in base-piranha solution (Milli-Q water, 30% ammonium hydroxide, 35% hydrogen peroxide at a 5:1:1 volume ratio), rinsed with Milli-Q water and stored in 0.1 M KOH up to 1 mo. Right before assembling flow cells, slides and coverslips were rinsed with Milli-Q water and dried with synthetic air. Flow cells with ∼10 μl channels were assembled by sandwiching ∼2-mm-wide and ∼2.5-cm-long strips of Parafilm between a cleaned glass slide and coverslip and melting on a hot plate at 120°C. The resulting chambers were passivated by incubating for 45 min with 1 M KOH, rinsing with actin polymerization buffer (5 mM Tris-HCl, pH 8, 50 mM KCl, 1 mM MgCl2, 0.2 mM Na2ATP, 1 mM DTT), incubating for another 45 min with 0.2 mg/ml PLL-PEG, and rinsing with actin polymerization buffer. Flow cells were placed in a Petri-dish along with tissue paper soaked in water to prevent flow channels from drying during the incubation steps and until use.
Lyophilized rabbit skeletal muscle G-actin was resuspended to 5 mg/ml (119 μM) in G-buffer (5 mM Tris-HCl pH 8, 0.2 mM Na2ATP, 0.1 mM CaCl2, 1 mM DTT), aliquots snap-frozen in liquid nitrogen and stored at −80°C. Frozen aliquots were thawed and centrifuged for 30 min at 120,000 g in a benchtop Beckman air-driven ultracentrifuge (Beckman Coulter Airfuge, 340401) to clear the solution from aggregates. Clarified G-actin was kept at 4°C and used within 3–4 wk.
For actin-septin reconstitution experiments, thawed septin aliquots were cleared for 15 min at 120,000 g in a Beckman airfuge right before use. To polymerize G-actin in the presence of septins, we mixed G-actin, previously diluted with G-buffer to 5 μM, with septins, either nonfluorescent ones or msfGFP-labeled septins (at 20% msfGFP molar ratio for wild-type septins, and 100% GFP for SEPT2NC septins) to a final actin concentration of 1 μM and a final septin concentration of 0.3 μM, right before polymerization in actin polymerization buffer, additionally containing 1 mM Trolox, 2 mM protocatechuic acid (PCA), 0.1 μM protocatechuate 3,4-dioxygenase (PCD) and 0.1% wt/vol methylcellulose. To fluorescently label actin filaments, we polymerized G-actin in the presence of 1 μM Alexa Fluor 568-conjugated phalloidin.
Actin-septin samples were prepared with a final volume of 10 μl, were loaded immediately into passivated flow channels upon mixing of the components to start polymerization, and flow channels were sealed with VALAP (1:1:1 vasoline:lanoline:paraffin). The contributions of KCl and MgCl2 from the septin elution buffer were taken into account to yield the same final composition of actin polymerization buffer. Actin-septin samples were incubated overnight at RT in the dark before observation. To polymerize septins in the absence of actin, we followed the same procedure as above, but replaced the G-actin solution with G-buffer. Septins were used at 20% msfGFP and 20% sfCherry2 molar ratio for wild-type septins and at 100% GFP for SEPT2NC septins.
The sources and identifiers for proteins, materials and chemicals are as follows. Glass slides (26 × 76 mm; AA00000102E01FST20) and glass coverslips (24 × 60 mm; BB02400600A113FST0) from Thermo Scientific. Ammonium hydroxide solution (221228) and hydrogen peroxide solution (95299) from SIGMA. PLL-PEG from SuSoS AG (PLL(20)-g[3.5]-PEG(2)). Rabbit skeletal muscle G-actin from Cytoskeleton, Inc. (AKL99). Alexa Fluor 568-phalloidin from Thermo Fisher Scientific (A12380). Methylcellulose (M0512), Trolox (238813), protocatechuic acid (03930590), protocatechuate 3,4-dioxygenase (P8279) from Sigma-Aldrich.
Confocal fluorescence microscopy of reconstituted actin-septins and image processing
Reconstituted actin-septin assemblies were imaged on the same spinning disk microscope setup described for imaging cells using the same objective lens and camera. Images were acquired with an exposure time of 0.1 s. Actin-septin bundles were imaged close to the surface. Septin filament bundles were also found at the surface, but the clusters of interconnected filament bundles were observed floating in the bulk of the flow channels. To capture such clusters, z-stacks were acquired over 10–50 μm using a Δz interval of 0.5 μm. Images were processed with ImageJ/Fiji. Images of actin-septin bundles are from single planes. Images of septin filament bundles are from maximum-intensity z projections. The contrast of all images shown was adjusted post-acquisition so that both dim and bright structures are visible without saturation. All images use an inverted grayscale, with bright signals appearing black in a white background.
Metal-induced energy transfer assays
U2OS cells were transfected with SEPT9_i3-mApple, SEPT9_i3-mApple-CAAX, or GAP43-mApple with FuGeneHD (E2311; Promega). 16 h post-transfection, cells were plated on glass coverslips (for obtaining reference lifetime measurements, see below) and on gold-coated glass coverslips, previously cleaned with 70% ethanol. Cells were left to attach and spread for 24 h, then fixed for 15 min using 4% paraformaldehyde (15714; Electron Microscopy Sciences) in cytoskeleton buffer (10 mM MES – pH 6.1 with NaOH, 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM MgCl2). The excess of cytosolic protein content was washed out with a permeabilization/blocking step (0.1% saponin, 1% BSA in PBS) for 1 h at room temperature. Labeling of F-actin in U2OS cells was achieved with AlexaFluor568-phalloidin (A12380; Invitrogen) at 165 nM in permeabilization/blocking solution for 1 h. The samples were maintained in PBS until and throughout the measurements.
Metal-induced energy transfer (MIET) was performed following the concept introduced by Enderlein and coworkers (Chizhik et al., 2014). Briefly, we measured the fluorescence lifetime of emitters in the vicinity of a 18 nm-thick gold film. From the calibration of the fluorescence lifetime dependence with the distance to the gold film (Chizhik et al., 2014), the distance between the fluorophore and the metal is recovered. The MIET calibration curve was computed using the MIET-GUI Matlab code developed by the Enderlein group (https://www.uni-goettingen.de/de/software/532664.html). For mApple, we used a peak emission wavelength at 610 nm and a quantum yield of 49%. For Alexa Fluor 568, the emission peak was 603 nm and the quantum yield 69%. Our calculation used the fluorescence lifetime of the dyes measured on a glass coverslip, in the absence of the metal layer, to account for the slight 0.1 ns lifetime change induced by the functionalization of the dye to septin, GAP43 or phalloidin. An isotropic orientation of the fluorophores is assumed (Chizhik et al., 2014).
We used a gold film of 18 nm thickness deposited by electron-beam assisted evaporation of gold on a borosilicate glass coverslip (Bühler Syrus Pro 710). A 2 nm-thick chromium layer is used to promote the adhesion of gold on the glass coverslip. For the MIET calibration, the refractive indexes of the gold and chromium layers were taken from Rosenblatt et al. (2020) and Johnson and Christy (1974), respectively, while the refractive index of 1.52 for the borosilicate glass coverslip was provided by the supplier (D 263 M glass by Schott AG).
The fluorescence lifetime measurements were performed with a home built confocal microscope with a 557 nm iChrome-TVIS laser (Toptica GmbH, pulse duration 3 ps, 40 MHz repetition rate) and a Zeiss C-Apochromat 63×, 1.2 NA water immersion objective. The excitation power remained below 2 μW on the sample to avoid photobleaching during the measurement. The fluorescence light was collected by the same microscope objective and filtered using a dichroic mirror (ZT 405/488/561/640rpc; Chroma), long-pass filter (ET570LP; Chroma) and bandpass filter (ET595/50 m; Chroma). The confocal pinhole diameter was 50 μm. The photon counting detection used an avalanche photodiode (MPD-5CTC; Picoquant) connected to a time correlated counting module (HydraHarp400; PicoQuant). The temporal resolution (full width at half maximum of the instrument response function) was measured to be 38 ps. The fluorescence lifetime histograms were fitted using SymPhoTime 64 software (PicoQuant GmbH) with a reconvolution taking into account the measured instrument response function. All the histograms were fitted using a biexponential function which provided a better fit to the intensity decay than a single exponential decay. About 20% of the total detected intensity corresponded to the short lifetime component (below 0.5 ns), which was not considered further for the analysis. The MIET distance measurements were taken on the long lifetime component which represented more than 80% of the total detected photons. The distribution of calculated distances from lifetime measurements for each condition is represented in box plots using GraphPad Prism (one data point per cell for each condition). The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of cells per condition is indicated in the respective legend. One-way ANOVA followed by a multiple comparison test was used for comparing the distributions. Data distribution was assumed to be normal, but this was not formally tested.
Supported lipid bilayer assays
Small unilamellar vesicle formation
We used three types of lipids, 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate; ammonium salt; PI(4,5)P2; 850155P; Sigma-Aldrich), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC; 850375C; Sigma-Aldrich), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(Cyanine 5; DOPE-Cy5; 810335C; Sigma-Aldrich), all from Avanti Polar Lipids. The lipids were mixed in chloroform, or, in case PI(4,5)P2 was present, in a 20:9:1 chloroform/methanol/water mixture in a glass vial. The organic solvent was then evaporated completely using a stream of N2 followed by overnight incubation in a dessicator. The dried lipid film was resuspended in buffer to give a total lipid concentration of 0.25 mM. We used a sodium citrate buffer of pH 4.8 (50 mM citrate, made of equal molarity trisodium citrate and citric acid mixed in a 2:3 volume ratio, 50 mM KCl, 0.1 mM ethylenediaminetetraacetic acid) in case PI(4,5)P2 was present, and otherwise F-buffer of pH 7.4 (20 mM Tris-HCl, 2 mM MgCl2, 50 mM KCl, 1 mM DTT). The lipids were dissolved by four cycles of 1 min vortexing and 5 min incubation. Finally, small unilamellar vesicles (SUVs) were obtained by sonicating the lipid solution using an Ultrasonic homogeniser series HD 2000.2 sonicator equipped with a BR30 cup resonator (Bandelin) at 10% amplitude for 30 min with pulses of 5 s on and 5 s off to avoid excessive heating.
Protein preparation
Unlabeled septin octamers and SEPT2-msfGFP octamers were purified in house as previously reported (Iv et al., 2021). The protein was stored in aliquots in septin storage buffer (20 mM Tris HCl, pH 7.4, 2 mM MgCl2, 300 mM KCl, 1 mM DTT), at −80°C. Before each experiment, unlabeled and labeled septin octamers were mixed in a 9:1 molar ratio in septin storage buffer at a total concentration of 1,800 nM. Lyophilized monomeric actin (G-actin) from rabbit skeletal muscle (8101-03; Hypermol) was resuspended following the manufacturer’s instructions and dialyzed against G-buffer (5 mM Tris-HCl, pH 7.8, 0.1 mM CaCl2, 0.2 mM ATP, and 1 mM DTT) to remove residual disaccharides from the freeze-drying process. Protein aggregates were removed by centrifugation at 148,000 × g for 1 h and the supernatant was snap-frozen and stored in aliquots at −80°C. Fluorescently tagged G-actin was prepared by covalent modification with Alexa Fluor 594 Carboxylic Acid (15461054; Thermo Fisher Scientific; Alvarado and Koenderink, 2015). Before experiments, G-actin aliquots were thawed, and any aggregates were removed by leaving the protein on ice for at least 2 h and subsequently centrifuging at 148,000 × g for 20 min. Unlabeled and fluorescent actin were mixed in a 9:1 molar ratio in G-buffer at a total G-actin concentration of 5 μM.
Sample preparation
Supported lipid bilayers (SLB) were formed in custom-made flow channels made of nr. 1 Menzel coverslips (11961988; Thermo Fisher Scientific) and glass slides (11879022; Thermo Fisher Scientific). The coverslips and glass slides were first cleaned in base piranha solution (5% hydrogen peroxide, 5% ammonium hydroxide) at 70°C for 10 min, extensively washed with Milli-Q water, and stored in Milli-Q water for a maximum of 5 d. Just before use, a coverslip and a slide were dried with a stream of N2 gas. Flow channels were prepared by sandwiching 2 × 20 mm parafilm strips separated by ∼3 mm between the glass slide and the coverslip. The parafilm was then melted by placing the chambers on a hot plate at 120°C and gently pressing on top with clean tweezers. After cooling down, an SUV solution (7–12 μl, depending on the distance between the parafilm strips) was pipetted into the channels and incubated in a humid chamber for at least 20 min to promote SUV rupture and SLB formation. Residual SUVs were removed by washing with four channel volumes of F-buffer for DOPC SLBs or with two channel volumes of sodium citrate buffer followed by 2 channel volumes of F-buffer for 5% PI(4,5)P2 SLBs. DOPC SLBs contained 99.7% DOPC and 0.3% DOPE-Cy5; 5%PIP2 SLBs also contained 94.7% DOPC and 0.3% DOPE-Cy5.
Septin octamers and actin were co-polymerized at room temperature in polymerization buffer (20 mM Tris-HCl, pH 7.4, 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.5 mM ATP, 1 mM GTP) supplemented with 1 mM Trolox to suppress blinking, and an oxygen scavenging system composed of 1 mM protocatechuic acid and 0.05 μM of procatechuate 3,4-dioxygenase to minimize photobleaching. We first prepared a 5× master buffer (100 mM Tris HCl, pH 7.4, 10 mM MgCl2, 5 mM DTT, 2.5 mM ATP, 5 mM GTP, 5 mM Trolox and 5 mM protocatechuic acid). To prepare the sample, we mixed the master buffer (fivefold dilution), 0.05 μM of procatechuate 3,4-dioxygenase, the G-actin mix (fivefold dilution to give a final concentration of 1 μM), and the septin mix (sixfold dilution, to give a final concentration of 300 nM), in that order. The mixture was either immediately added to the flow channels containing the SLBs and incubated for 1 h in a humid environment, or first incubated in the tube for 1 h to promote septin-actin bundle formation and then added to the flow channels using a cut pipette tip to minimize bundle disruption. For the sequential addition of septin octamers and actin, first septin octamers were diluted into the polymerization buffer, immediately added to the flow channels, and incubated for ∼30 min in a humid environment. Afterwards, the channels were washed with 1× channel volumes of F-buffer to remove unbound septin octamers; only 1× channel volume is used in this case to minimize disruption of the membrane-bound septin filament mesh. Finally, either pre-polymerized F-actin in polymerization buffer, or G-actin freshly added to polymerization buffer, was flushed into the flow channels with a cut tip to minimize actin filament disruption and incubated for 1 h. The channels were then sealed with Dow Corning high-vacuum silicone grease (Z273554; Sigma-Aldrich) to avoid drying while imaging.
Image acquisition
The samples were immediately imaged using a Nikon Ti2-E microscope complemented with a Gataca iLAS2 azimuthal TIRF illumination system. The sample was illuminated with 488- and 561-nm lasers (Gataca laser combiner iLAS2) to visualize the septin and the actin signals, respectively. The fluorescence signal was split with a Cairn Research Optosplit II ByPass containing a Chroma ZT 543 rdc dichroic mirror and filtered with either a 525/50 or a 600/50 chroma bandpass filter. The images were collected with a Nikon Apo TIRF 100× oil, NA 1.49 objective and recorded with an Andor iXon Ultra 897 EM-CCD camera using an exposure time of 50 ms. To check that the SLBs were uniform and free of defects, we examined DOPE-Cy5 distribution by illuminating with a 642-nm laser filtered with a 708/75 chroma bandpass filter and recorded using an exposure time of 20 ms. We checked SLB fluidity by fluorescence recovery after photobleaching of DOPE-Cy5.
Atomic force microscopy
Sample preparation
Measurements were made on cells plated on Y shape-micropatterned substrates to minimize variability due to size and shape differences among cells (Rigato et al., 2015). 12-mm glass coverslips were coated with 0.1 mg/ml PLL-PEG (PLL[20]-g[3.5]-PEG[2], Susos) before being illuminated with a deep-UV lamp through a quartz-chrome photomask bearing the micropattern features (Front Range Photomask) designed using AutoCAD (Autodesk). We used Y-shaped micropatterns with a spread area of ∼1,500 mm2. Micropatterned coverslips were then incubated with 25 µg/ml fibronectin and 5 µg/ml fibrinogen-GFP, the latter for visualizing micropatterns. Wild type U2OS cells treated with siLacZ siRNA (6mer + 8mer), with SEPT9 siRNA (6mer), and U2OS cells treated with SEPT7 siRNA and also transfected with msfGFP-SEPT7Gmut2 (8mer) were seeded on fibronectin-coated micropatterns 48 h post-electroporation. Cells were incubated for 5–7 h to attach and spread adopting a triangular shape. The expression of msfGFP-SEPT7Gmut2 for the 8mer condition was confirmed through the detection of fluorescence in each measured cell.
Force spectroscopy experiments and data analysis
The values of log10(E0) and β extracted from each force measurement were pooled by cell and then averaged. The data was reproduced in three independent experiments and their distribution represented in box plots using GraphPad Prism (one data point per cell for each condition). The central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the minimum and maximum values. The number of cells per condition is indicated in the respective legend. E0 values (in Pa) were plotted on a log scale. One-way ANOVA followed by a multiple comparison test was used for comparing the distributions of E0 values using the log10(E0) values, given the log-normal distribution of E0.
Single particle tracking Photo-activated localization microscopy (sptPALM)
Cell culture
Mouse embryonic fibroblasts were cultured in DMEM (10313-021; Gibco) with 10% fetal calf serum (FCS, CVFSVF00-01; Eurobio scientific). Transient transfections of plasmids were performed 2 d before experiments using the Amaxa nucleofector (VPD-1004; Lonza). The cells were detached with trypsin/EDTA, the trypsin was inactivated using DMEM with 10% FCS, and the cells were washed and suspended in serum-free Ringer solution (150 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES-Na pH 7.4, 2 g/liter glucose), then incubated for 30 min in Ringer solution before plating on fibronectin-coated glass coverslips (human plasma fibronectin at 10 µg/ml, 10838039001; Roche).
Plasmids
SEPT9_i3-mEos3.2 and SEPT9_i3-mEos3.2-CAAX were cloned in a pCMV plasmid backbone with seamless cloning into a NheI/BamHI linearized vector (primers in Table S1). EGFP-human β-actin was provided by A. Matus (Friedrich Miescher Institute for Biomedical Research, Switzerland). The mEos2-actin construct was generated from EGFP-actin as described in Rossier et al. (2012). EYFP-human paxillin (isoform alpha) was used as described in Rossier et al. (2012).
Optical setup and image acquisition
sptPALM acquisitions were steered by MetaMorph software (Molecular Devices) with an inverted motorized microscope (Nikon Ti) equipped with a temperature control system (The Cube, The Box, Life Imaging Services), a Nikon CFI Apo TIRF 100× oil, NA 1.49 objective and a Perfect Focus System, allowing long acquisition in TIRF illumination mode.
Imaging was performed at least 3 h after seeding the cells on fibronectin-coated coverslips mounted in a Ludin chamber (Life Imaging Services). For photoactivation localization microscopy, cells expressing mEos2 and mEos3.2 tagged constructs were photoactivated using a 405 nm laser (Omicron) and the resulting photoconverted single molecule fluorescence was excited with a 561 nm laser (Cobolt Jive). Both lasers illuminated the sample simultaneously. Their respective power was adjusted to keep the number of the stochastically activated molecules constant and well separated during the acquisition. Fluorescence was collected by the combination of a dichroic and emission filters (D101-R561 and F39-617 respectively, Chroma) and a sensitive EMCCD (electron-multiplying charge-coupled device, Evolve, Photometric). The acquisition was performed in streaming mode at 50 Hz. Either EYFP-paxillin or GFP-actin were imaged using a conventional GFP filter cube (ET470/40, T495LPXR, ET525/50; Chroma). Using this filter cube does not allow spectral separation of the unconverted pool of mEos from the GFP fluorescent signal. However, with all of the constructs used, whether the mEos signal was highly or poorly enriched in FAs, we were still able to detect FAs.
Single molecule segmentation and tracking
A typical sptPALM experiment leads to a set of at least 4,000 images per cell, analyzed in order to extract molecule localization and dynamics. Single molecule fluorescent spots were localized and tracked over time using a combination of wavelet segmentation and simulated annealing algorithms (Izeddin et al., 2012; Racine et al., 2006; Racine et al., 2007). Under the experimental conditions described above, the resolution of the system was quantified to 59 nm (Full Width at Half Maximum, FWHM). This spatial resolution depends on the image signal to noise ratio and the segmentation algorithm (Cheezum et al., 2001) and was determined using fixed mEos2 samples. We analyzed 130 2D distributions of single molecule positions belonging to long trajectories (>50 frames) by bi-dimensional Gaussian fitting, the resolution being determined as 2.3 sxy, where sxy is the pointing accuracy.
Modeling of human septin complexes
Models of full-length human septin complexes were built for analyzing and interpreting split-GFP experiments. The septin GTP-binding domains (GBDs) used as templates for the SEPT2, 6 and 7 models using SWISS-MODEL homology modeling software (Waterhouse et al., 2018) were from PDB accession no. 7M6J (Leonardo et al., 2021), the most complete human septin hexamer structure to date, which includes α0 helices for SEPT6 and 7. As solved in its integrity, the SEPT6 GBD remained unchanged and was used as is. The SEPT7 GBD structure was completed using SWISS-MODEL. As the use of the SEPT2 GBD from 7 M6J for modeling SEPT2 led to clashes in the modeled SEPT2-SEPT2 NC interface, the SEPT2 GBD subunit was modeled using the SEPT7 GBD structure from 7 M6J as a template. The lack of structural information for the short N-terminal extensions of SEPT2, 6, and 7 prompted us to model them as disordered segments using Phyre2 (Kelley et al., 2015). The C-terminal domains of SEPT2, 6, and 7 were modeled with CCFold (Guzenko and Strelkov, 2018) for the coiled–coil (CC) parts and Phyre2 for the flexible parts, as detailed in Iv et al. (2021). The homodimeric parallel SEPT2CC was used unaltered with respect to Iv et al. (2021). The previously modeled SEPT6 and 7 helices in the SEPT6-SEPT7 parallel coiled–coil in Iv et al. (2021) were repositioned slightly after comparison with the only parallel septin CC structure to date (PDB accession no. 6WCU; Leonardo et al., 2021). GBDs, N- and C-terminal extensions were then combined with PyMOL open-source software. When necessary, the disordered segments were manually modified to avoid steric clashes and to adjust distances. The SEPT9_i3 model used was the one built for (Iv et al., 2021) and included already N- and C-terminal extensions. Hexameric SEPT2-SEPT6-SEPT7-SEPT7-SEPT6-SEPT2 and octameric SEPT2-SEPT6-SEPT7-SEPT9-SEPT9-SEPT7-SEPT6-SEPT2 complexes were built by fitting the modeled structures to the hexamer from the PDB accession no. 7M6J.
To analyze and interpret the split-GFP experiments, the entire constructs used in the assays, including β10- and β11-tagged septins and the reconstituted GFP, were modeled. To this aim, the split GFP structure (PDB accession no. 4KF5) was added to the modeled septin complexes. The flexible linkers linking the reconstituted GFP to the septin of interest were built manually using PyMOL; their straight-ish appearance in the models is due to the polypeptide chains being built as linear structures. To mimic paired septin filaments with narrow spacing (Leonardo et al., 2021), mediated by homodimeric SEPT2 antiparallel CCs (Fig. 5 H), septin complexes were duplicated and placed parallel to each other with a gap of ∼5 nm. The bent conformation of the septin was built by rotating the CC domain manually by 90° relative to the GBD. The helices within the homodimeric SEPT2 antiparallel CC were positioned using the antiparallel SEPT4CC structure from PDB accession no. 6WB3 as a reference (Leonardo et al., 2021). All manual interventions were realized using PyMOL.
Statistics and reproducibility
The distributions of measurements, or of phenotypes in the case of septin mutant characterization, are represented with GraphPad Prism using box plots, violin plots, and scatter dot plots as indicated in the respective methods sections and legends. Bars, error bars (SD or SEM), and box plot features are as indicated in the respective figure legends. The number of measurements in each plot and the numbers of experiments are indicated in the respective figure legend or methods. Statistical significance tests were performed with GraphPad Prism. The tests applied and the obtained P values are mentioned in the respective figure legend. Experiments were repeated at least three times independently to ensure reproducibility. Experiments from Fig. 1 F, Fig. 2, C and D; Fig. 10, C–F, Fig. 10 H; Fig. S3, B and C; and Fig. S5, I–K were performed twice. Experiments from Fig. 10, G and I were performed once. No data were excluded from the analyses.
Online supplemental material
Fig. S1 shows examples of SEPT2, SEPT7, and SEPT9 distribution in cells from immunostainings and live imaging of GFP fusions. Fig. S2 contains details on the design of the tripartite split-GFP complementation assay for probing septin organization. Fig. S3 depicts the mutants used in this study, as well as examples of cell-free reconstitution of actin-septin assembly. Fig. S4 contains data that support that all septins on microtubules organize as octamer-based filaments. Fig. S5 contains data that support that septin filaments can mediate actin-membrane anchoring in the absence of myosin-II. Table S1 shows oligonucleotide primer sequences used for the generation of the plasmids used in this study.
Data availability
All data supporting the findings of this study are available within the article and its supporting information files. The source datasets generated and analyzed during the current study are available from the corresponding author on reasonable request. The source codes for the custom-generated Matlab codes for measurements of fiber diameter (FWHM) and length, and for numerical simulations of expected fiber diameter (FWHM) from SIM images has been deposited to Github. The respective links are mentioned in the relevant Materials and methods sections.
Acknowledgments
We thank Josette Perrier and Cendrine Nicoletti (iSm2) for hosting protein purification. The authors thank Artemis Kosta, Hugo Le Guenno and the Microscopy Core Facility of IMM for SIM microscopy. We thank R. Sterling for technical assistance and the IINS Cell culture facility, especially E. Verdier and N. Retailleau for technical help (IINS Cell Biology Facility, grant no. ANR-10-LABX-43). We also thank J.B. Sibarita (IINS) for support with sptPALM analysis.
This research received funding from the Agence Nationale de la Recherche (ANR grants ANR-17-CE13-0014 SEPTIMORF to M. Mavrakis; ANR-17-CE09-0026-01 AntennaFRET to J. Wenger; ANR-20-CE42-0003 3DPolariSR to V. Milanović and O. Rossier), the Fondation ARC pour la recherche sur le cancer (grant ARCDOC42020010001242 to C.S. Martins), and from the Cancéropôle PACA, Institut National du Cancer and Conseil Régional PACA (Bourse mobilité to M. Mavrakis). We further acknowledge financial support from the French Ministry of Research, CNRS and the Conseil Régional Nouvelle-Aquitaine (grant MechanoStem to O. Rossier and V. Milanović). We acknowledged the France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreements No. 723241 to J. Wenger and No. 772257 to F. Rico). This project has received funding from the European Union’s Horizon 2020 research and Innovation program under the H2020-MSCA-ITN-2018 Grant Agreement n. 812772. This work was supported in part by the National Institutes of Health (R01GM122375 to S. Kumar). Confocal microscopy with the Zeiss LSM710 microscope and SIM with the Zeiss Elyra PS.1 microscope was performed at the UC Berkeley Biological Imaging Facility, which was supported in part by the National Institutes of Health S10 program under award numbers 1S10RR026866-01 and 1S10OD018136-01. G. Castro-Linares and G.H. Koenderink gratefully acknowledge financial support by the Netherlands Organization for Scientific Research (NWO/OCW) through the “BaSyC—Building a Synthetic Cell” Gravitation grant (024.003.019).
Author contributions: C.S. Martins: investigation, conceptualization, methodology, funding acquisition, writing—original draft, writing—review & editing; C. Taveneau, G. Castro-Linares: investigation, writing—original draft, writing—review & editing; M. Baibakov, N. Buzhinsky, M. Eroles, V. Milanović, S. Omi, J.-D. Pedelacq, F. Iv, L. Bouillard, A. Llewellyn, M. Gomes, M. Belhabib, M. Kuzmić, S. Lee: investigation; P. Verdier-Pinard, A. Badache: investigation, writing—review & editing; S. Kumar: supervision, funding acquisition, resources; C. Chandre: software; S. Brasselet: software, writing—original draft; F. Rico: supervision, funding acquisition, writing—original draft, writing—review & editing; O. Rossier, J. Wenger: investigation, supervision, funding acquisition, writing—original draft; G.H. Koenderink: supervision, funding acquisition, writing—review & editing; S. Cabantous, M. Mavrakis: investigation, conceptualization, methodology, funding acquisition, project administration, supervision, writing—original draft, writing—review & editing.
References
Author notes
Disclosures: The authors declare no competing interests exist.
P. Verdier-Pinard's and A. Badache’s current affiliation is Aix Marseille Univ, INSERM, MMG, U1251, Marseille, France.





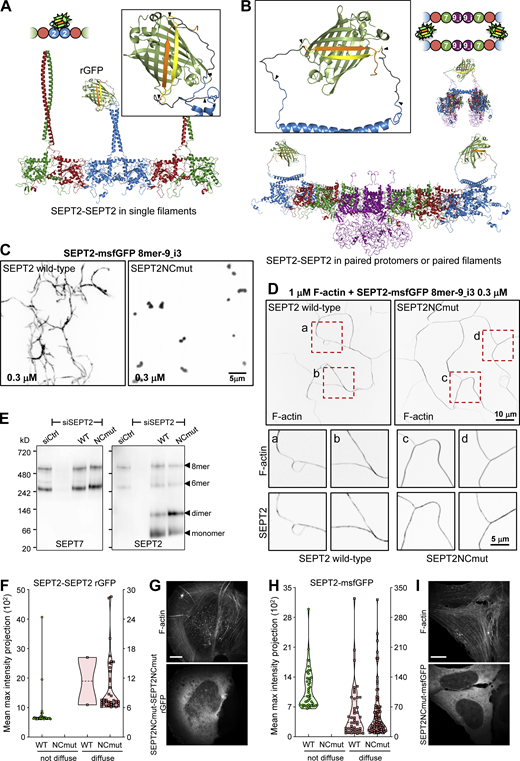
![Septin interface mutants used in this study and cell-free reconstitution of septin assembly. (A) Left, top: Conserved residues in the SEPT2-SEPT2 NC interface are shown in the crystal structure of human SEPT2 homodimers (PDB accession no. 2QA5; Sirajuddin et al., 2007). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues F20 from the hook-loop of one SEPT2 subunit (orange) interact with the hydrophobic cleft formed by V86, I88, V93, and I281 of the adjacent SEPT2 subunit (blue). The importance of this phenylalanine in anchoring the α0 helix at the NC interface was emphasized only recently (Cavini et al., 2021). The blue subunit's surface representation highlights the complementary of shape between the two SEPT2 subunits in this interface. The interaction between the α0 helices of each subunit is also stabilized through a hydrophobic interaction between their respective V27. Left, bottom: Sequence alignment of the regions including the residues shown in the NC interface structure for SEPT2 and SEPT9_i3. The structural elements (α0, β2, α5) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties; period, conservation between residues with weakly similar physicochemical properties). We note that the residues described above are strictly or physicochemically conserved (except for V86), highlighting their importance in stabilizing the SEPT2-SEPT2 NC interface. The NC interface mutants used in this study are listed on the right of the crystal structure. A mutation of F20D/I263D is expected to destabilize the hydrophobic pocket depicted above, whereas a V27D/M270D is expected to destabilize the α0 helices interface. Importantly, a strictly conserved aspartate (SEPT2 E90, corresponding to SEPT6 E90 and SEPT7 E102 which are well defined in the cryo-EM structure of the SEPT6-SEPT7 NC interface [Mendonca et al., 2019]) in the loop connecting β2 and β3 is pointing to the hydrophobic cleft where the phenylalanine resides. The F20D mutation is thus expected to result in a repulsion between the aspartate and glutamate and contribute further to the destabilization of the NC interface. Right, top: Conserved residues in the SEPT7-SEPT7 G interface are shown in the crystal structure of human SEPT7 homodimers (PDB accession no. 6N0B; Brognara et al., 2019). The backbone structure is displayed as a cartoon representation in PyMOL, with critical residues represented as sticks (deep blue and red for nitrogen and oxygen atoms, respectively). Residues W269 of one SEPT7 subunit (yellow) interact with residues W269, A272 and H279 in the adjacent SEPT7 subunit (green; Sirajuddin et al., 2007; Zent et al., 2011). W269 from adjacent subunits interact through a water molecule bridge through hydrogen bonds. In addition, each W269 is engaged in π-π interactions with H279 and CH-π interactions with A272 of the opposite subunit. Right, bottom: Sequence alignment of the regions including the residues shown in the G interface structure for SEPT7 and SEPT9_i3. The structural elements (β7, β8) related to these residues are underlined and shown above the sequences. The consensus symbols are from ClustalW alignments of all human septins (*, fully conserved residue; colon, conservation between residues with strongly similar physicochemical properties). Notice that W269 and H279 are both strictly conserved, showing their importance in stabilizing this interface. The G interface mutants that were used in this study are listed on the right of the crystal structure. The presence of both mutations W269A and H279D in SEPT7 and SEPT9 is expected to destabilize the SEPT7-SEPT7 and SEPT7-SEPT9 G-interfaces. The loss of the aromatic cycle properties in the mutant W269A does not allow the abovementioned critical interactions mediated by the wild-type Trp. W269A is expected to destabilize H279 and potentially change its orientation. In addition, the much smaller size of the alanine will poorly mimic the hydrophobic interaction between W269 and H279, weakening the G-interface. Note that W269 is in the vicinity of Y267 of the same subunit. This tyrosine interacts with the nucleotide buried within the G-interface. Consequently, any mutations destabilizing W269 could dramatically destabilize the overall G-interface because of a domino effect. Similarly, H279D is expected to preclude hydrophobic interactions with W269 and thus destabilize the latter. The single mutation H279D in SEPT7 is expected to destabilize the SEPT7-SEPT7 G-interface when present in both SEPT7 subunits, but not the SEPT7-SEPT9 interface with wild-type SEPT9. (B) Representative spinning disk fluorescence images of septin filament assembly upon polymerization of hexamers in solution at the indicated final protomer concentration. Protomers contained either wild-type SEPT2 (top panel) or SEPT2NCmut (bottom panel). Images use an inverted grayscale. Related to Fig. 2 C. (C) Representative spinning disk fluorescence images of reconstituted actin filaments, polymerizing in the presence of septin hexamers in solution. Protomers contained either wild-type SEPT2 or SEPT2NCmut. Actin filaments are visualized with AlexaFluor568-conjugated phalloidin, and septins with SEPT2-msfGFP. One example of large fields of view are shown for each condition, depicting cross-linking of actin filaments; only actin labeling is shown. The image for actin in the presence of wild-type hexamers is contrast-enhanced on purpose in order to saturate the actin bundles so that weaker-intensity single actin filaments are also visible. Insets on the bottom show higher magnifications of selected regions of interest on the top (dashed squares in red). Two regions of interest (a and b for wild-type SEPT2 and c and d for SEPT2NCmut) are shown in each case, depicting both the actin and septin signals. Scale bars in all large fields of views, 10 μm. Scale bars in all insets, 5 μm. (D) Representative spinning disk fluorescence images of septin filament assembly upon co-polymerization of hexamers containing SEPT2-msfGFP and octamers-9_i3 containing SEPT2-sfCherry2 at the indicated final protomer concentration. Images use an inverted grayscale. Scale bars in all large fields of views, 10 μm.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/3/10.1083_jcb.202203016/1/m_jcb_202203016_figs3.png?Expires=1773834935&Signature=L-85UqnN8VnUKh6uzzZsHez5Dohv6NxCFnNljcprSFN~Y2n~c1wRgy86JRhOd6yeY50HCFlfPb380F9~n~6gtBR2gc86qsaIYYYpjnprH4fot~6xE1fPqbWvh~vLVd4TtxJAMfy5CFS74XYISCjZeaeaYEwpmY8pPxFDsqVCwfc8TawtaDhZJ9ryMD0qU~zBbXuloGmOYLJjT8gaQjezHePoSrxYhcR3wrkgehUSCTZgHq7WJUGlCCDJPyYCejACFzLrsqWkm2X-XhpzZaxXBzlQag9zz~YleYaxkjZSKINZbY25JFVvkX7hNbHLE51hEnHd8vNiljzjUGVOBQBnQg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)