Replication fork reversal protects forks from breakage after poisoning of Topoisomerase 1. We here investigated fork progression and chromosomal breakage in human cells in response to a panel of sublethal genotoxic treatments, using other topoisomerase poisons, DNA synthesis inhibitors, interstrand cross-linking inducers, and base-damaging agents. We used electron microscopy to visualize fork architecture under these conditions and analyzed the association of specific molecular features with checkpoint activation. Our data identify replication fork uncoupling and reversal as global responses to genotoxic treatments. Both events are frequent even after mild treatments that do not affect fork integrity, nor activate checkpoints. Fork reversal was found to be dependent on the central homologous recombination factor RAD51, which is consistently present at replication forks independently of their breakage, and to be antagonized by poly (ADP-ribose) polymerase/RECQ1-regulated restart. Our work establishes remodeling of uncoupled forks as a pivotal RAD51-regulated response to genotoxic stress in human cells and as a promising target to potentiate cancer chemotherapy.
Introduction
One of the most widely used approaches in cancer chemotherapy is to kill cancer cells or arrest their rapid proliferation by targeting DNA replication. As genome duplication is essential for every cell division, replication interference is inherently more toxic to rapidly proliferating cancer cells than to untransformed, mostly quiescent somatic cells. Different strategies for replication interference have been explored and are often combined in chemotherapeutic regimens. A first class of drugs target DNA topoisomerases, essential factors to release torsional stress accumulating during replication (Pommier, 2013 and references therein). Topoisomerase I (Top1) inhibitors of the class of camptothecin (CPT) are commonly used to treat ovarian, lung, and colorectal cancer and act by trapping the enzyme on the DNA after strand cleavage. The same principle of “interfacial inhibition” applies to Topoisomerase II (Top2) inhibitors, such as etoposide (ETP) and doxorubicin (DOX), both potent chemotherapeutic drugs commonly used to treat various cancers (Pommier, 2013 and references therein). ETP is the most selective Top2 inhibitor available in the clinics and, at clinically relevant doses, mostly induces single-strand breaks, by asymmetrical trapping of Top2 homodimers (Kerrigan et al., 1987). Conversely, DOX intercalates in the DNA molecule and induces “concerted” trapping of Top2 complexes, mostly leading to double-strand breaks (DSBs; Zwelling et al., 1981). A second frequent strategy for replication interference in cancer chemotherapy makes use of antimetabolites to block nucleotide biosynthesis or DNA polymerization, as for the ribonucleotide reductase inhibitor hydroxyurea (HU) or the DNA polymerase inhibitor aphidicolin (APH). HU is commonly used to treat hematological malignancies and has been extensively used in basic research to investigate the consequences of replication fork stalling (Madaan et al., 2012). Similarly, APH has been used to study chromosome fragility during replication (Arlt et al., 2012) but has also been considered to potentiate specific anticancer therapies (Michaelis et al., 2001). DNA cross-linking agents, such as mitomycin C (MMC) and cisplatin (or cis-diamminedichloroplatinum [CDDP]), are also extensively used to treat many different cancers (Deans and West, 2011). Although their cytotoxicity is commonly related to the induction of inter-strand cross-links (ICL), these drugs induce a complex combination of different adducts. ICL-inducing agents have become increasingly popular in basic research because of the isolation of numerous defects in genome stability genes sensitizing cells specifically to these agents and resulting in the cancer-prone human syndrome Fanconi anemia (FA; Deans and West, 2011). Finally, several additional treatments are known to damage the DNA bases, interfering with replication fidelity and progression (Hoeijmakers, 2009). Among the most investigated sources of base damage are UV-C irradiation, the methlyating agent methyl methanesulfonate (MMS), and oxidative DNA damage, which can be easily induced by short treatments with hydrogen peroxide (H2O2). Although this plethora of genotoxic agents share the observable ability to challenge the replication process, the mechanistic details of replication interference have been mostly studied in vitro or in model systems, and the detailed cellular responses have remained largely elusive in higher eukaryotic cells. However, mechanistic insight is required to inform the choice of specific chemotherapeutic regimens, to improve the anticancer response, and to avoid resistance or relapse of specific cancer types.
Replication fork reversal—i.e., the conversion of a replication fork into a four-way junction by reannealing of parental strands and coordinated annealing of nascent strands—was initially proposed by (Higgins et al., 1976), as a model for damage bypass during replication in human cells. Albeit conceptually attractive, the model has long remained unsubstantiated, and fork reversal has been rather associated with unscheduled transactions at unprotected replication forks in specific yeast mutants (Lopes et al., 2001, 2006; Sogo et al., 2002; Bermejo et al., 2011). More recently, however, fork reversal was reported as a strikingly frequent event upon mild Top1 poisoning in wild-type yeast cells, as well as mouse and human cells, and Xenopus laevis egg extracts (Ray Chaudhuri et al., 2012). Genetic interference with this process leads to a drastic increase in fragility of replicating chromosomes, suggesting fork reversal as a protective, evolutionarily conserved response to topological constraints in replication (Ray Chaudhuri et al., 2012). The identification of poly (ADP-ribose) polymerase (PARP) and RECQ1 as central modulators of reversed fork restart upon Top1 poisoning further implicated fork remodeling as a genetically controlled, physiological response in higher eukaryotes (Berti et al., 2013) and revived significant interest for fork reversal in genome stability and cancer (León-Ortiz et al., 2014; Zeman and Cimprich, 2014; Neelsen and Lopes, 2015). However, key biological questions remain open, such as whether reversed forks are detected upon other types of replication stress and, in that case, whether their stability and restart are controlled by a common set of cellular factors. Furthermore, although several factors were shown to induce replication fork reversal in biochemical reconstitution—including RECQ helicases, SWI/SNF (Switch/Sucrose Nonfermentable) proteins, and FANCM (Kanagaraj et al., 2006; Machwe et al., 2006; Ralf et al., 2006; Gari et al., 2008; Blastyák et al., 2010; Bugreev et al., 2011; Bétous et al., 2012, 2013; Ciccia et al., 2012; Burkovics et al., 2014)—the lack of a reliable readout for fork reversal in vivo has so far hampered the identification of fork reversing activities in the living cell.
Several homologous recombination (HR) mechanisms have been proposed to assist replication restart upon fork stalling or collapse (Petermann and Helleday, 2010). The function of HR factors in replication has been consistently related to DSB formation at stalled forks, in light of the known involvement of HR in DSB repair. However, growing evidence suggests a DSB repair-independent role for HR factors in replication stress. The central vertebrate recombinase RAD51 is detected on chromatin during unperturbed replication and is recruited to stalled forks upstream of DSB formation (Hashimoto et al., 2010; Petermann et al., 2010). Upon prolonged fork stalling, HR factors—as well as numerous FA factors—are required to prevent excessive nucleolytic degradation of nascent strands and this function can be genetically uncoupled from DSB repair (Hashimoto et al., 2010; Schlacher et al., 2011, 2012). Furthermore, HR factors reportedly involved in DSB resection (i.e., MRE11, NBS1, and CtIP) were recently involved in fork processing and ATR signaling (Shiotani et al., 2013; Murina et al., 2014; Yeo et al., 2014). Most recently, the HR cancer susceptibility gene BRCA1 was shown to promote specific recombination events at Tus/Ter-stalled mammalian forks, which can be distinguished from canonical DSB repair (Willis et al., 2014). Altogether, these recent observations suggest the mechanistic involvement of HR and possibly other FA factors in replication fork metabolism, independently from repair of chromosomal breakage.
In this work, we show that replication fork reversal is a global response to several different sources of replication stress. We suggest single-stranded DNA (ssDNA) accumulation as common precursor of fork reversal upon different types of genotoxic stress. We identify the central recombinase RAD51 as stable replisome component, independent of fork breakage, and as first cellular factor assisting in vivo the reversal process. Furthermore, we extend the role of PARP and RECQ1 to the controlled restart of reversed forks induced by different treatments.
Results
Sublethal doses of genotoxic treatments in human cells consistently induce replication fork slowing, without detectable chromosomal breakage
To investigate at the molecular level replication interference induced by different cancer chemotherapeutic drugs and other genotoxic treatments, we exposed the Rb/p53-proficient osteosarcoma cell line U-2 OS (U2OS) to a panel of clinically relevant genotoxic treatments (see Introduction), including topoisomerase inhibitors (CPT, ETP, and DOX), ICL-inducing agents (MMC and CDDP), DNA synthesis inhibitors (APH and HU), and base-damaging agents (MMS, H2O2, and UV-C irradiation, shortly UV). To allow the effective comparison of the cellular responses to these treatments, we selected for each of these genotoxic agents an appropriate dose that would induce marginal effects on cell survival and proliferation (Fig. S1 A). We next confirmed, by prolonged treatments and flow cytometric analysis, that the selected dose would permit completion of bulk genome duplication but delay transition through S phase (Fig. S1 B), indicating mild interference of these treatments with the replication process. We next used an established protocol for DNA fiber spreading analysis, after incorporation of halogenated nucleotides (Jackson and Pombo, 1998), to investigate at single molecule level the effect of these genotoxic treatments on replication fork progression (Fig. S1 C). Remarkably, despite the moderate effects on cell survival and cell cycle progression, all selected treatments quickly and markedly affected replication fork progression, spanning from 25% (H2O2) to 80% (HU) reduction in fork speed (Fig. 1 A). 1-h treatment with the selected dose of each genotoxic agent did not reveal any significant increase in the level of chromosomal breakage above background levels, as assessed by pulsed field gel electrophoresis (PFGE; Fig. 1 B). Minor DSB levels, close to the detection level of this approach (100 DSB/cell; Ray Chaudhuri et al., 2012), possibly induced by a subset of drugs are addressed by further experiments described below (see Structural determinants of ATR and ATM activation upon genotoxic treatments in human cells). Collectively, these data suggest that mild treatments with cancer chemotherapeutics and other genotoxic agents induce a marked slowdown of replication fork progression, largely uncoupled from fork breakage.
Mild genotoxic stress induces marked fork slowing in the absence of chromosomal breakage. (A) DNA fiber spreading. Statistical analysis of IdU replicated track length in U2OS cells, comparing not treated (NT) conditions with the indicated treatments. The labeling protocol and representative fibers are included in Fig. S1. At least 100 tracks were scored per sample. Horizontal lines represent the median value, and boxes and whiskers show 10–90th percentiles. Statistical analysis t test according to Mann–Whitney, results are ns, not significant; ****, P ≤ 0.0001. All experiments have been repeated at least twice, with very similar results. (B) PFGE analysis for DNA breakage detection in untreated U2OS cells and upon 1-h treatment of the indicated doses of genotoxic treatments. 1 µM camptothecin (CPT) treatment is used as a positive control for DSB formation. See also Fig. S1 for the selection of appropriate doses for each treatment. Fig. 4 and Fig. S4 include data on DDR activation possibly associated with minor levels of DSB detected in B.
Mild genotoxic stress induces marked fork slowing in the absence of chromosomal breakage. (A) DNA fiber spreading. Statistical analysis of IdU replicated track length in U2OS cells, comparing not treated (NT) conditions with the indicated treatments. The labeling protocol and representative fibers are included in Fig. S1. At least 100 tracks were scored per sample. Horizontal lines represent the median value, and boxes and whiskers show 10–90th percentiles. Statistical analysis t test according to Mann–Whitney, results are ns, not significant; ****, P ≤ 0.0001. All experiments have been repeated at least twice, with very similar results. (B) PFGE analysis for DNA breakage detection in untreated U2OS cells and upon 1-h treatment of the indicated doses of genotoxic treatments. 1 µM camptothecin (CPT) treatment is used as a positive control for DSB formation. See also Fig. S1 for the selection of appropriate doses for each treatment. Fig. 4 and Fig. S4 include data on DDR activation possibly associated with minor levels of DSB detected in B.
Fork slowing by all genotoxic treatments is associated with fork uncoupling and accumulation of postreplicative ssDNA gaps
We next used psoralen cross-linking coupled to EM (Neelsen et al., 2014) to investigate in vivo possible alterations of replication fork architecture associated with the observed fork slowing. This technique allows reliable identification of ssDNA regions on DNA molecules, based on local reduction of filament thickness (Neelsen et al., 2014 and references therein). Short (∼40 nt) ssDNA regions are expected to arise during lagging strand synthesis in eukaryotes and are promptly detected at a subset of unperturbed replication forks (untreated). However, all genotoxic treatments induced a significant accumulation of larger ssDNA stretches at replication forks, increasing their median length by 1.5–2-fold and leading to occasional ssDNA stretches up to 500-nt long (Fig. 2, A and B). Thus, whether replication stress is induced by DNA damage, topological stress, or enzymatic inhibition of DNA synthesis, replication fork uncoupling is a common structural feature associated with genotoxic treatments in human cells. It is likely that the length of these ssDNA regions reflects how strongly each treatment interferes with continuous DNA synthesis on the leading strand (Lopes et al., 2006), via modulating template availability, polymerase processivity, nucleotide abundance, and/or torsional constraints. Furthermore, careful observation of the replicated duplexes in the analyzed population of intermediates revealed that 20–30% of the replication forks exposed at least one postreplicative ssDNA gap, corresponding to a two- to threefold increase over the level observed in untreated cells (Fig. 2, C and D; and Fig. S2 A). Interestingly, the frequency of postreplicative ssDNA gaps upon different treatments generally correlated with the length of the ssDNA regions observed at the fork (Fig. 2, B and D), suggesting DNA synthesis repriming events at uncoupled replication forks, in line with previous observations in yeast (Lopes et al., 2006). However, the size of these ssDNA gaps varied significantly between different drugs (Fig. S2 B), possibly reflecting DNA synthesis restart at a different distance from the original block and/or damage-specific repair and processing events. Very similar observations on ssDNA accumulation at replication intermediates (RIs) were made on the untransformed human epithelial cell line RPE-1 treated with a subset of the genotoxic agents (Fig. S2, C and D).
Genotoxic treatments lead to extended ssDNA regions at replication forks and ssDNA gaps on replicated duplexes. (A and C) Electron micrographs of representative replication fork from U2OS cells, after 1-h treatment with 100 nM APH (A) and 50 µM MMS (C), respectively. P indicates the parental duplex, whereas D indicates daughter duplexes. The black arrow points to an ssDNA region at the fork, whereas the white arrow indicates an ssDNA gap on a replicated duplex. The relevant portions of the molecules are magnified in the insets. Bars: (main images) 0.5 kb; (insets) 0.2 kb. (B) Graphical distribution of ssDNA length at the junction (black arrow in A) in not treated (NT) U2OS cells and upon the indicated treatments (UV pulse or 1-h treatment). Only molecules with detectable ssDNA stretches are included in the analysis. The lines show the median lengths of the ssDNA regions at the fork in the specific set of analyzed molecules. Statistical analysis t test according to Mann–Whitney results are *, P ≤ 0.1; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. In brackets, the total number of analyzed molecules is given. (D) Frequency of replication forks with at least one ssDNA gap (white arrow in C) in untreated U2OS cells and upon the indicated treatments. In brackets, the total number of analyzed molecules is given. Similar results to those displayed in B and D were obtained in at least one independent experiment (see also Fig. S2 and Fig. 6 A).
Genotoxic treatments lead to extended ssDNA regions at replication forks and ssDNA gaps on replicated duplexes. (A and C) Electron micrographs of representative replication fork from U2OS cells, after 1-h treatment with 100 nM APH (A) and 50 µM MMS (C), respectively. P indicates the parental duplex, whereas D indicates daughter duplexes. The black arrow points to an ssDNA region at the fork, whereas the white arrow indicates an ssDNA gap on a replicated duplex. The relevant portions of the molecules are magnified in the insets. Bars: (main images) 0.5 kb; (insets) 0.2 kb. (B) Graphical distribution of ssDNA length at the junction (black arrow in A) in not treated (NT) U2OS cells and upon the indicated treatments (UV pulse or 1-h treatment). Only molecules with detectable ssDNA stretches are included in the analysis. The lines show the median lengths of the ssDNA regions at the fork in the specific set of analyzed molecules. Statistical analysis t test according to Mann–Whitney results are *, P ≤ 0.1; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. In brackets, the total number of analyzed molecules is given. (D) Frequency of replication forks with at least one ssDNA gap (white arrow in C) in untreated U2OS cells and upon the indicated treatments. In brackets, the total number of analyzed molecules is given. Similar results to those displayed in B and D were obtained in at least one independent experiment (see also Fig. S2 and Fig. 6 A).
Replication fork reversal is a widespread global response to replication stress in human cells
We recently reported that—upon mild, clinically relevant doses of Top1 poisons—a large fraction of forks undergo reversal (Fig. 3 A), i.e., they form a fourth regressed arm, by local reannealing of parental strands and simultaneous annealing of the newly synthesized strands (Ray Chaudhuri et al., 2012; Neelsen and Lopes, 2015). Although reversed forks were also reported upon genetic perturbations associated with early tumorigenesis (Neelsen et al., 2013a,b), a key open question was whether this DNA transaction was induced by any treatment interfering with the replication process (León-Ortiz et al., 2014). We now report high frequency of replication fork reversal (15–30%) upon all tested genotoxic treatments (Fig. 3 B). Considering the calculated number of active replication forks in a typical S phase (3,000–12,000; Ge and Blow, 2010), this corresponds to ∼500–4,000 reversed forks per cell, under different types of mild genotoxic stress compatible with cell proliferation and survival (Fig. S1 A). As previously reported for Top1 poisoning (Ray Chaudhuri et al., 2012), the observed frequency of fork reversal is already high at sublethal doses of genotoxic agents and does not significantly increase with a 10-fold higher dose (Fig. S3 A). In vivo cross-linking of RI before extraction excludes that these structures form in vitro during sample preparation (Neelsen et al., 2014). Furthermore, the relative abundance of reversed forks is not changed by omitting from the EM procedure the RI-enrichment step (Fig. S3 B; Neelsen et al., 2014). Importantly, very similar frequencies of reversed forks were induced by genotoxic treatments in RPE-1 cells (Fig. 3 C), extending our observations to noncancerous cells. Thus, replication fork reversal genuinely represents a general, widespread, physiological response to replication interference in human cells. With the exception of CPT and H2O2, which induced significantly longer regressed arms, the length of the fourth arm at reversed forks averaged around 300 bp in all conditions and only rarely exceeded 1 kb (Fig. S3 C). We also investigated the possible presence of ssDNA on the regressed arm, which may result from reversal of uncoupled forks and/or nucleolytic processing of the regressed arm. We observed that 20–50% of the regressed arms exposed ssDNA ends or gaps, whereas <15% were entirely single stranded (Fig. S3, D and E). The relative proportion of these categories shows subtle variations, but no strong bias, among the different treatments.
All tested sources of genotoxic stress lead to frequent replication fork reversal. (A) Electron micrograph of a representative reversed replication fork from U2OS cells treated for 1 h with 20 nM ETP. P indicates the parental duplex, D indicates daughter duplexes, and R indicates the regressed arm. Bar, 0.5 kb. (B and C) Frequency of reversed replication forks in U2OS (B) or RPE-1 cells (C) either not treated (NT) or upon the indicated treatments (UV pulse or 1-h treatment). In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment (see also Fig. S3 and Fig. 6 A).
All tested sources of genotoxic stress lead to frequent replication fork reversal. (A) Electron micrograph of a representative reversed replication fork from U2OS cells treated for 1 h with 20 nM ETP. P indicates the parental duplex, D indicates daughter duplexes, and R indicates the regressed arm. Bar, 0.5 kb. (B and C) Frequency of reversed replication forks in U2OS (B) or RPE-1 cells (C) either not treated (NT) or upon the indicated treatments (UV pulse or 1-h treatment). In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment (see also Fig. S3 and Fig. 6 A).
Structural determinants of ATR and ATM activation upon genotoxic treatments in human cells
Activation of the ATR-mediated replication checkpoint has been linked to excess ssDNA at RIs (Zou and Elledge, 2003). However, ATR activation requires multiple protein–protein interactions and recently revealed unexpected complexity (Nam and Cortez, 2011; Shiotani et al., 2013; Kumar et al., 2014). Furthermore, ATR activation can also be a secondary consequence of nucleolytic processing of DSB, frequently associated with prolonged replication stress, limiting our mechanistic understanding of ATR activation upon replication interference. We thus reckoned that our extensive in vivo RI visualization under mild genotoxic treatments, i.e., not associated with detectable chromosomal breakage, could provide valuable information on the structural determinants of ATR activation. We noted that, despite consistent fork slowing, frequent fork reversal, and accumulation of ssDNA upon all genotoxic treatments, several treatments (ETP, DOX, MMC, CDDP, and APH) induced no—or marginal—ATR activation, as detected by phosphorylation of its direct target CHK1 (Fig. 4 and Table 1). Furthermore, the marked CHK1 phosphorylation detected upon HU and UV treatment was not specifically associated with excessive accumulation of ssDNA regions at uncoupled forks, at postreplicative gaps, or at regressed arms (Fig. 4 A; Fig. 2, B and D; Fig. S2, A and B; Fig. S3 E; and Table 1). Differently from ssDNA visualization by EM (Figs. 2 and S2), detection of total exposed ssDNA by native BrdU staining revealed marked differences among the treatments. Although BrdU staining and γ-H2AX staining correlated at the population level, they colocalized only in a minority of the cells (Fig. 4, B and C), further uncoupling ssDNA accumulation from ATR signaling. Surprisingly, we also found no strict correlation across treatments between impairment of DNA synthesis (5′-ethynyl-2′-deoxyuridine [EdU] incorporation) and ATR activation (γ-H2AX), as APH treatment severely impairs DNA synthesis in the absence of detectable ATR activation (Fig. 4, B–D). Marked ATR activation upon HU and UV treatments is also not an indirect consequence of chromosomal breakage, as it was associated with no detectable accumulation of DSB by PFGE (Fig. 1 B) or phosphorylation of RPA32 on S4/S8 (Fig. 4; see also Fig. 6 B), a recognized DSB marker (Oakley and Patrick, 2010). Overall, ATR activation in our experimental conditions does not directly mirror the extent of replication interference, nor the amount of ssDNA detected at RIs (Table 1), and likely reflects yet-undefined signaling determinants that escape systematic cell-based and single-molecule analyses.
Differential ATR and ATM activation upon different genotoxic treatments, despite similar structural features of RIs. (A) Immunoblot for ATR (pCHK1) and ATM (pKAP1) activation and total DDR proteins (CHK1 and KAP1) in not treated (NT) U2OS cells and upon the indicated treatments (UV pulse or 1-h treatment). RPA32 (RPA) phosphorylation at S4/S8 indicates ATM/DNA-dependent protein kinase (DNA-PK) activation and is typically used as a DSB marker. Total RPA32 levels (and phosphorylation-associated mobility shift) are also displayed. 1 µM CPT treatment is used as positive control for full DDR activation. TFIIH is used as a loading control. (B) Native immunofluorescence staining for cells grown with 10 µM BrdU for 48 h and treated with the indicated drugs for 1 h. Red staining, γ-H2AX; green staining, BrdU (ssDNA); blue, DAPI. Bar, 15 µM. (C) Relative quantification of double-negative cells and cells positive for γ-H2AX, native BrdU staining (natBrdU), or both for the experiment in B. The data shown are from a single representative experiment out of three repeats, with n > 100. (D) Flow cytometry analysis of DNA synthesis (EdU), DNA content (DAPI), and DDR activation (γ-H2AX) in untreated U2OS cells and upon the indicated treatments. Dashed line indicates threshold for EdU incorporation and γ-H2AX positivity, respectively. See also Fig. S4 and Tables 1 and 2.
Differential ATR and ATM activation upon different genotoxic treatments, despite similar structural features of RIs. (A) Immunoblot for ATR (pCHK1) and ATM (pKAP1) activation and total DDR proteins (CHK1 and KAP1) in not treated (NT) U2OS cells and upon the indicated treatments (UV pulse or 1-h treatment). RPA32 (RPA) phosphorylation at S4/S8 indicates ATM/DNA-dependent protein kinase (DNA-PK) activation and is typically used as a DSB marker. Total RPA32 levels (and phosphorylation-associated mobility shift) are also displayed. 1 µM CPT treatment is used as positive control for full DDR activation. TFIIH is used as a loading control. (B) Native immunofluorescence staining for cells grown with 10 µM BrdU for 48 h and treated with the indicated drugs for 1 h. Red staining, γ-H2AX; green staining, BrdU (ssDNA); blue, DAPI. Bar, 15 µM. (C) Relative quantification of double-negative cells and cells positive for γ-H2AX, native BrdU staining (natBrdU), or both for the experiment in B. The data shown are from a single representative experiment out of three repeats, with n > 100. (D) Flow cytometry analysis of DNA synthesis (EdU), DNA content (DAPI), and DDR activation (γ-H2AX) in untreated U2OS cells and upon the indicated treatments. Dashed line indicates threshold for EdU incorporation and γ-H2AX positivity, respectively. See also Fig. S4 and Tables 1 and 2.
Relevant parameters for ATR activation upon a subset of genotoxic treatments
| Parameter | Approach | Figure | NT | MMC (200 nM) | APH (100 nM) | CPT (25 nM) | HU (0.5 mM) |
| Fork reversal | EM analysis | 3, B and C | −/+ | ++ | ++ | ++ | ++ |
| Fork slowing | DNA fiber spreading | 1 A | − | ++ | ++ | + | +++ |
| Impaired DNA synthesis | EdU incorporation (FACS) | 4 D | − | −/+ | ++ | + | +++ |
| ssDNA at forks | EM analysis | 2, A and B; S2; and S3 | − | + | + | + | + |
| Total exposed ssDNA | Native BrdU staining | 4 B | − | − | − | ++ | ++ |
| ATR signaling at forks | iPOND γ-H2AX | 6 B | − | −/+ | ND | ++ | +++ |
| ATR signaling total | WB pCHK1 | 4 A | − | − | − | + | +++ |
| ATR signaling total | IF/FACS γ-H2AX | 4, B–D | − | − | − | ++ | ++ |
| Parameter | Approach | Figure | NT | MMC (200 nM) | APH (100 nM) | CPT (25 nM) | HU (0.5 mM) |
| Fork reversal | EM analysis | 3, B and C | −/+ | ++ | ++ | ++ | ++ |
| Fork slowing | DNA fiber spreading | 1 A | − | ++ | ++ | + | +++ |
| Impaired DNA synthesis | EdU incorporation (FACS) | 4 D | − | −/+ | ++ | + | +++ |
| ssDNA at forks | EM analysis | 2, A and B; S2; and S3 | − | + | + | + | + |
| Total exposed ssDNA | Native BrdU staining | 4 B | − | − | − | ++ | ++ |
| ATR signaling at forks | iPOND γ-H2AX | 6 B | − | −/+ | ND | ++ | +++ |
| ATR signaling total | WB pCHK1 | 4 A | − | − | − | + | +++ |
| ATR signaling total | IF/FACS γ-H2AX | 4, B–D | − | − | − | ++ | ++ |
Parameters were assessed by different investigation methods, as displayed in the indicated figures. −/+, +, ++, and +++ indicate increasingly clear phenotypes. IF, immunofluorescence; NT, not treated; WB, Western blot.
We also detected phosphorylation of ATM and its target KAP1 upon mild treatments with CPT and UV, albeit not accompanied by RPA S4/S8 phosphorylation or detectable DSBs (Figs. 1 B and 4). To assess the possibility that both approaches may not be sensitive enough to reveal minor DSB levels, we increased 10-fold the dose of each genotoxic treatments and reassessed physical chromosomal breakage and cellular responses. In the case of CPT, DOX, H2O2, and UV, the higher doses did lead to detectable DSBs, expectedly associated with ATM, KAP1, and RPA32-S4/S8 phosphorylation (Fig. S4). Lack of S4/S8 phosphorylation upon DOX treatments may reflect specific effects of this drug in DSB signaling. Interestingly, at higher doses, MMS also induced ATM/KAP1 phosphorylation without detectable DSB and RPA32 S4/S8 phosphorylation (Fig. S4), as already seen for mild CPT and UV treatments (Figs. 4 A and S4), supporting the notion that under certain conditions, replication stress can activate ATM in the absence of DSB (Table 2; Ray Chaudhuri et al., 2012). However—as for the strong ATR activation upon UV and HU treatments—we could not unambiguously associate this DSB-independent ATM signaling to any specific structural feature detectable by EM analysis (Table 2). Finally, APH and ETP, despite their marked effect on DNA synthesis, fork uncoupling, and reversal already at low doses (Figs. 1 A, 2 B, 3 B, and 4 C), did not induce detectable ATR or ATM activation even at 10-fold higher doses. Altogether, these data suggest that fork slowing, fork reversal, and ssDNA accumulation are by themselves nonpredictive parameters for ATR or ATM activation upon replication stress.
Relevant parameters for ATM activation upon a subset of genotoxic treatments
| Parameter | Approach | Figure | NT | MMC (200 nM) | DOX (50 nM) | UV (5 J/m2) | CPT (25 nM) | CPT (1 µM) |
| Fork reversal | EM analysis | 3, B and C | −/+ | ++ | ++ | ++ | ++ | ++ |
| DSBs | PFGE | 1 B and S4 | − | − | −/+ | − | − | ++ |
| ATM signaling at forks | iPOND pRPA32 | 6 B | − | − | − | − | − | ++ |
| ATM signaling total | WB pRPA32 | 4 and S4 | − | − | − | − | − | ++ |
| ATM signaling total | WB pKAP1 | 4 and S4 | − | − | +/− | + | +/− | ++ |
| ATM signaling total | WB pATM | S4 | − | − | +/− | + | + | ++ |
| Parameter | Approach | Figure | NT | MMC (200 nM) | DOX (50 nM) | UV (5 J/m2) | CPT (25 nM) | CPT (1 µM) |
| Fork reversal | EM analysis | 3, B and C | −/+ | ++ | ++ | ++ | ++ | ++ |
| DSBs | PFGE | 1 B and S4 | − | − | −/+ | − | − | ++ |
| ATM signaling at forks | iPOND pRPA32 | 6 B | − | − | − | − | − | ++ |
| ATM signaling total | WB pRPA32 | 4 and S4 | − | − | − | − | − | ++ |
| ATM signaling total | WB pKAP1 | 4 and S4 | − | − | +/− | + | +/− | ++ |
| ATM signaling total | WB pATM | S4 | − | − | +/− | + | + | ++ |
Parameters were assessed by different investigation methods, as displayed in the indicated figures. −/+, +, ++, and +++ indicate increasingly clear phenotypes. NT, not treated; WB, Western blot.
RECQ1 and PARP activities regulate the restart of reversed forks induced by different types of replication stress
After identification of replication fork reversal as a frequent DNA transaction upon Top1 poisoning in human cells (Ray Chaudhuri et al., 2012), we reported reversed fork accumulation in these conditions to depend on transient PARP-mediated inhibition of the specific restart activity of the RECQ1 helicase, thus linking fork restart to DNA repair and PARP inactivation (Berti et al., 2013). We therefore decided to investigate by DNA fiber and EM analysis whether similar mechanisms would control the restart of reversed forks observed upon other genotoxic treatments. We focused this analysis on MMC and HU, as prototypes of replication stress induced by template cross-linking and DNA synthesis inhibition, respectively, thus mechanistically distinct from the replication stress induced by Top1 poisons. As previously reported for CPT (Sugimura et al., 2008; Ray Chaudhuri et al., 2012), PARP inactivation by olaparib largely abolished MMC-induced fork slowing and only partially restored replication fork progression in HU, in which nucleotide shortage cannot be overcome (HU; Fig. 5 A). Importantly, the effect of olaparib upon both treatments was dependent on the RECQ1 helicase, showing that fork slowing upon different sources of replication stress is an active process mediated by transient PARP-mediated inhibition of RECQ1 activity (Fig. 5 A). Furthermore, PARP inactivation markedly reduced reversed fork accumulation upon MMC and HU treatments. RECQ1 depletion induced by itself a threefold accumulation of reversed forks under unperturbed conditions (Fig. 5 B), associated with mild ATM/KAP1 phosphorylation and no accumulation of ssDNA gaps (Fig. S5, A and B). Importantly, RECQ1 depletion largely abolished PARP requirement for reversed fork accumulation upon both treatments and prevented the rapid decline in reversed fork frequency observed upon drug removal (Fig. 5 B). Altogether, these data strongly suggest that PARP-controlled RECQ1 activity is largely responsible to promote fork restart and progression, irrespective of whether fork reversal is induced by topological stress (CPT; Berti et al., 2013), DNA cross-linking (MMC), or DNA synthesis inhibition (HU).
RECQ1 and PARP activity control replication fork progression and accumulation/restart of reversed forks upon different types of genotoxic stress. (A) Statistical analysis of IdU track length measurements, according to the labeling protocol in Fig. S1, in U2OS cells stably transduced (shRNA) for Luciferase (shLuc) or RECQ1 (shRECQ1) depletion. 200 nM MMC and 500 µM HU were optionally added concomitantly with the second label (IdU). The PARP inhibitor olaparib (Ola; 10 µM) was optionally added 2 h before CldU labeling and maintained during labeling. At least 100 tracks were scored for each dataset. Horizontal lines represent the median value, and boxes and whiskers indicate the 10–90th percentiles. t test according to Mann–Whitney; ns, not significant; ****, P < 0.0001. Similar results were obtained in at least one independent experiment. (B) Frequency of reversed forks detected by EM in U2OS cells stably transfected (shRNA) for Luciferase or RECQ1 depletion. The cells were optionally treated for 1 h with 200 nM MMC or 500 µM HU, after an optional 2-h pretreatment with olaparib. Reversed fork restart was assessed by measuring the frequency of reversed forks 3 h after drug removal (release [Rel.]). In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment. RECQ1 levels after shRNA-mediated depletion were detected by immunoblotting. TFIIH was used as a loading control. NT, not treated.
RECQ1 and PARP activity control replication fork progression and accumulation/restart of reversed forks upon different types of genotoxic stress. (A) Statistical analysis of IdU track length measurements, according to the labeling protocol in Fig. S1, in U2OS cells stably transduced (shRNA) for Luciferase (shLuc) or RECQ1 (shRECQ1) depletion. 200 nM MMC and 500 µM HU were optionally added concomitantly with the second label (IdU). The PARP inhibitor olaparib (Ola; 10 µM) was optionally added 2 h before CldU labeling and maintained during labeling. At least 100 tracks were scored for each dataset. Horizontal lines represent the median value, and boxes and whiskers indicate the 10–90th percentiles. t test according to Mann–Whitney; ns, not significant; ****, P < 0.0001. Similar results were obtained in at least one independent experiment. (B) Frequency of reversed forks detected by EM in U2OS cells stably transfected (shRNA) for Luciferase or RECQ1 depletion. The cells were optionally treated for 1 h with 200 nM MMC or 500 µM HU, after an optional 2-h pretreatment with olaparib. Reversed fork restart was assessed by measuring the frequency of reversed forks 3 h after drug removal (release [Rel.]). In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment. RECQ1 levels after shRNA-mediated depletion were detected by immunoblotting. TFIIH was used as a loading control. NT, not treated.
The human recombinase RAD51 is recruited to replication forks in the absence of DSBs and modulates fork progression and integrity
The extensive EM analysis described in this work identifies fork uncoupling and reversal as common parallel transactions upon replication interference by various genotoxic treatments. We tested the functional correlation of these events by plotting the frequency of reversed forks versus the median size of ssDNA stretches at the forks for all EM samples analyzed (Fig. 6 A). This analysis shows that our EM measurements of fork reversal and ssDNA accumulation were highly reproducible in independent experiments for each drug. Although a general association of the two events is not unexpected, the striking correlation that we found between the two parameters (Fig. 6 A) prompted us to investigate whether they were not only correlatively associated, but rather mechanistically linked. Accumulation of ssDNA is a crucial structural feature of upstream intermediates in DSB repair by HR and is actively induced by DNA end resection for the controlled loading of the central recombinase factor RAD51, which then drives homology-directed strand invasion (Symington and Gautier, 2011). We thus tested whether, in our experimental conditions, RAD51 could be detected at replication forks, as suggested by a recent screening (Alabert et al., 2014). Using an iPOND approach with different labeling protocols (Sirbu et al., 2011; Fig. S5 C), we detected mild, but reproducible, RAD51 association with replication forks even in unperturbed conditions, which was lost—as for other replisome components, e.g., proliferating cell nuclear antigen—upon thymidine chase, and enriched by mild treatments with HU, MMC, and CPT (Fig. 6 B). Upon mild HU and CPT treatments, γ-H2AX is clearly detected at forks, confirming our results on ATR activation (Fig. 4). However, differently from acute treatments affecting fork integrity (1 µM CPT), no RPA32-S4/8 phosphorylation is present on EdU-labeled DNA upon any of the mild genotoxic treatments, confirming that the enrichment of RAD51 at forks facing replication stress is uncoupled from fork breakage (Fig. 6 B). Similarly, we observed by single-cell labeling that RAD51 is chromatin loaded in unperturbed S phase cells (EdU+) and is enriched in numerous foci upon both mild and acute genotoxic treatments (Fig. 6 C). siRNA-mediated down-regulation of RAD51 largely abolished active replication fork slowing observed upon MMC and CPT treatments, whereas had no significant effect when fork progression was physically impaired by nucleotide shortage (HU), suggesting RAD51 loading as a crucial modulator of fork progression upon genotoxic stress (Fig. 6 D). Furthermore, RAD51 depletion also impacted on replication fork integrity, leading to a significant induction of DSBs already in unperturbed conditions, with marginal further increase observable upon exogenous genotoxic stress (Fig. 6 E).
RAD51 is present at forks upon mild genotoxic stress and modulates fork progression and integrity. (A) Linear regression analysis shows strict direct correlation (P < 0.0001) between accumulation of ssDNA at the fork (median values of ssDNA regions at the junction) and frequency of fork reversal. Results from two independent experiments are displayed for untreated U2OS cells and for each genotoxic treatment. (B) HEK293T cells were EdU-labeled as indicated in Fig. S5 C and treated with sublethal doses of genotoxic drugs (0.5 mM HU, 200 nM MMC, or 25 nM CPT). Proteins and relative posttranslational modifications associated with replication forks were isolated by iPOND procedure and detected with the indicated antibodies. The thymidine (Thy; 10 µM) chase experiment is used to discriminate proteins associated with chromatin behind replicating forks. In the control (Ctrl) experiment, the click reaction is performed using DMSO instead of biotin azide. 1 µM CPT treatment is used as positive control to induce high replication stress and DSBs. (C) Immunofluorescence staining for U2OS cells grown on coverslips and treated with the indicated drugs for 1 h. Red staining, RAD51; green staining, EdU; blue, DAPI. Bar, 15 µM. (D) DNA fiber spreading. Statistical analysis of IdU replicated track length in U2OS cells, comparing not treated (NT) conditions with the indicated treatments. U2OS cells were transfected with siRNA against luciferase (siLuc) or RAD51 (siRAD51) 24 h before CldU or IdU labeling. At least 100 tracks were scored per sample. Horizontal lines represent the median value, and boxes and whiskers indicate 10–90th percentiles. Statistical analysis: one-way ANOVA; ns, not significant; ***, P ≤ 0.001. (E) PFGE analysis for DNA breakage detection in untreated U2OS cells and upon 1-h treatment with indicated doses of genotoxic treatments. U2OS cells were transfected with siRNA against luciferase or RAD51 24 h before treatments. 1 µM camptothecin (CPT) treatment is used as a positive control for DSB formation. The graph shows quantitative DSB induction from three independent experiments and includes average value and standard deviations (error bars). Statistical analysis: two-way ANOVA; ns, not significant; *, P ≤ 0.05.
RAD51 is present at forks upon mild genotoxic stress and modulates fork progression and integrity. (A) Linear regression analysis shows strict direct correlation (P < 0.0001) between accumulation of ssDNA at the fork (median values of ssDNA regions at the junction) and frequency of fork reversal. Results from two independent experiments are displayed for untreated U2OS cells and for each genotoxic treatment. (B) HEK293T cells were EdU-labeled as indicated in Fig. S5 C and treated with sublethal doses of genotoxic drugs (0.5 mM HU, 200 nM MMC, or 25 nM CPT). Proteins and relative posttranslational modifications associated with replication forks were isolated by iPOND procedure and detected with the indicated antibodies. The thymidine (Thy; 10 µM) chase experiment is used to discriminate proteins associated with chromatin behind replicating forks. In the control (Ctrl) experiment, the click reaction is performed using DMSO instead of biotin azide. 1 µM CPT treatment is used as positive control to induce high replication stress and DSBs. (C) Immunofluorescence staining for U2OS cells grown on coverslips and treated with the indicated drugs for 1 h. Red staining, RAD51; green staining, EdU; blue, DAPI. Bar, 15 µM. (D) DNA fiber spreading. Statistical analysis of IdU replicated track length in U2OS cells, comparing not treated (NT) conditions with the indicated treatments. U2OS cells were transfected with siRNA against luciferase (siLuc) or RAD51 (siRAD51) 24 h before CldU or IdU labeling. At least 100 tracks were scored per sample. Horizontal lines represent the median value, and boxes and whiskers indicate 10–90th percentiles. Statistical analysis: one-way ANOVA; ns, not significant; ***, P ≤ 0.001. (E) PFGE analysis for DNA breakage detection in untreated U2OS cells and upon 1-h treatment with indicated doses of genotoxic treatments. U2OS cells were transfected with siRNA against luciferase or RAD51 24 h before treatments. 1 µM camptothecin (CPT) treatment is used as a positive control for DSB formation. The graph shows quantitative DSB induction from three independent experiments and includes average value and standard deviations (error bars). Statistical analysis: two-way ANOVA; ns, not significant; *, P ≤ 0.05.
RAD51 is required to limit replication fork uncoupling and drive fork reversal upon different genotoxic treatments
In light of our previously reported data upon Top1 poisoning (Ray Chaudhuri et al., 2012), the observations reported here were highly suggestive of a role for RAD51 in replication fork reversal. We thus tested the hypothesis that, by analogy to HR mechanisms at DSBs, RAD51 could be loaded on extended ssDNA regions at uncoupled forks and drive fork reversal by template reannealing. EM analysis of U2OS cells treated with CPT, MMC, or HU upon siRNA-mediated RAD51 depletion revealed that effective fork reversal upon all treatments strictly requires RAD51 (Fig. 7 A). This held true using different siRAD51 oligonucleotides and extracting RI 24 h after siRNA transfection (Fig. 7 B), when protein depletion was yet incomplete and cells showed no alteration in their cell cycle and replication potential (Fig. S5, D and E). Furthermore, stable expression of exogenous, siRNA-resistant RAD51 completely restored the frequency of CPT-induced fork reversal observed in control cells (Fig. 7 C). RAD51 depletion also abolished the increased level of reversed forks observed in unperturbed RECQ1-depleted cells (Fig. S5 F), proving that RAD51 is required for replication fork reversal upon both endogenous and exogenous genotoxic stress. Importantly, upon all tested treatments, defective fork reversal was accompanied by a significant increase in RIs displaying long ssDNA regions at the fork (Fig. 7, D and E), strongly suggesting that uncoupled forks are precursors of RAD51-mediated fork reversal. Despite the effects on replication fork remodeling, RAD51 depletion had no noticeable impact on the abundance of the postreplicative ssDNA gaps induced by the genotoxic drugs (Fig. 7 F), differently from what previously shown in yeast and Xenopus egg extracts (Hashimoto et al., 2010). Altogether, these data imply RAD51-mediated recombinational mechanisms in the remodeling of uncoupled replication forks upon different types of replication stress.
RAD51 is required to convert uncoupled forks into reversed forks in response to different genotoxic treatments. (A–C) Frequency of reversed replication forks detected by EM in U2OS cells. In A, U2OS cells were transfected with Luciferase siRNA (siLuc) or RAD51siRNA (siRAD51) 72 h before DNA extraction from untreated cells or cells treated with 25 nM CPT, 200 nM MMC, or 500 nM HU for 1 h. In B, U2OS cells were transfected with Luciferase or RAD51 siRNA 24 h before treatment with 25 nM CPT for 1 h. In C, U2OS cells containing an empty vector, and U2OS cells expressing exogenous RAD51 were transfected with Luciferase or RAD51 siRNA (against 3′ UTR region of RAD51) 24 h before treatment with 25 nM CPT for 1 h. In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment. RAD51 levels after siRNA-mediated depletion were detected by immunoblotting. β-Tubulin is used as a loading control. EV, empty vector. (D) Electron micrograph of a representative replication fork with an extended ssDNA region at the junction (black arrow, magnified in the inset) upon RAD51 depletion and treatment with 25 nM CPT for 1 h. Bars: (main image) 0.5 kb; (inset) 0.2 kb. P indicates the parental duplex, and D indicates daughter duplexes. (E) Graphical distribution of ssDNA length at the junction (black arrow in C) in U2OS cells transfected with Luciferase siRNA and RAD51 siRNA and treated with 25 nM CPT, 200 nM MMC, and 500 nM HU for 1 h. The lines show the median length of the ssDNA region at the fork in the specific set of analyzed molecules. Statistical analysis t test according to Mann–Whitney, results are **, P ≤ 0.01; ***, P ≤ 0.001. Similar results were obtained in at least one independent experiment. (F) Frequency of replication forks with ssDNA gaps (Fig. 2 C and Fig. S2) in U2OS cells transfected with Luciferase or RAD51 siRNA 48 h before treatment with 25 nM CPT, 200 nM MMC, or 500 nM HU for 1 h. Similar results were obtained in at least one independent experiment. NT, not treated.
RAD51 is required to convert uncoupled forks into reversed forks in response to different genotoxic treatments. (A–C) Frequency of reversed replication forks detected by EM in U2OS cells. In A, U2OS cells were transfected with Luciferase siRNA (siLuc) or RAD51siRNA (siRAD51) 72 h before DNA extraction from untreated cells or cells treated with 25 nM CPT, 200 nM MMC, or 500 nM HU for 1 h. In B, U2OS cells were transfected with Luciferase or RAD51 siRNA 24 h before treatment with 25 nM CPT for 1 h. In C, U2OS cells containing an empty vector, and U2OS cells expressing exogenous RAD51 were transfected with Luciferase or RAD51 siRNA (against 3′ UTR region of RAD51) 24 h before treatment with 25 nM CPT for 1 h. In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment. RAD51 levels after siRNA-mediated depletion were detected by immunoblotting. β-Tubulin is used as a loading control. EV, empty vector. (D) Electron micrograph of a representative replication fork with an extended ssDNA region at the junction (black arrow, magnified in the inset) upon RAD51 depletion and treatment with 25 nM CPT for 1 h. Bars: (main image) 0.5 kb; (inset) 0.2 kb. P indicates the parental duplex, and D indicates daughter duplexes. (E) Graphical distribution of ssDNA length at the junction (black arrow in C) in U2OS cells transfected with Luciferase siRNA and RAD51 siRNA and treated with 25 nM CPT, 200 nM MMC, and 500 nM HU for 1 h. The lines show the median length of the ssDNA region at the fork in the specific set of analyzed molecules. Statistical analysis t test according to Mann–Whitney, results are **, P ≤ 0.01; ***, P ≤ 0.001. Similar results were obtained in at least one independent experiment. (F) Frequency of replication forks with ssDNA gaps (Fig. 2 C and Fig. S2) in U2OS cells transfected with Luciferase or RAD51 siRNA 48 h before treatment with 25 nM CPT, 200 nM MMC, or 500 nM HU for 1 h. Similar results were obtained in at least one independent experiment. NT, not treated.
Discussion
In this study, we have performed an unprecedented structural survey on the impact of genotoxic treatments on the replication process in human cells. The differential sensitivity of cancer and normal cells, often related to cancer-specific defects in the DNA damage response (DDR), is uncovered at relatively mild doses of the genotoxic agents used in cancer therapy (Bouwman and Jonkers, 2012 and references therein). We thus decided to investigate the effects of different genotoxic agents at doses that caused minimal lethality to the Rb/p53-proficient osteosarcoma human cell line, mostly used in this study (U2OS). These conditions enabled us to dissect the consequences of replication interference in the absence of detectably compromised chromosome integrity.
Despite diverse modes of interference with DNA synthesis (base damage, DNA intercalation and cross-linking, nucleotide depletion, polymerase inhibition, and torsional constraints), all tested genotoxic agents lead to strikingly similar mechanistic consequences on the replication process, i.e., marked fork slowing, ssDNA accumulation, and fork reversal. Importantly, these conclusions hold true in a nontransformed human cell line (RPE-1). The rapid accumulation of replication forks with extended ssDNA at the junction may represent the indirect consequence of continued helicase activity ahead of the fork, whereas DNA synthesis is asymmetrically delayed by the genotoxic treatment (Fig. 8), as reported in UV-irradiated yeast cells (Lopes et al., 2006). However, the extent of fork uncoupling may also be regulated by replisome-associated factors, specifically engaged in the replication process upon genotoxic stress, as recently suggested for the minichromosome maintenance–associated FA factor FANCD2 (Lossaint et al., 2013). Furthermore, the extent of ssDNA at replication forks challenged by genotoxic stress could be controlled by regulated nucleolytic processing of newly synthesized DNA, which becomes particularly evident after prolonged stress and pathological conditions (Schlacher et al., 2011, 2012). Interestingly, several factors previously involved in DSB resection—e.g., MRE11, NBS1, and CtIP—have been recently involved in fork metabolism and ATR activation upon genotoxic stress (Schlacher et al., 2011; Shiotani et al., 2013; Murina et al., 2014; Yeo et al., 2014), suggesting that ssDNA regions at damaged replication forks may be subjected to similar processing as DSB.
Schematic model for replication fork reversal and restart upon different types of replication stress. Template damage, DNA synthesis inhibition, or torsional stress rapidly impair symmetric elongation of nascent strands and induce replication fork uncoupling, leading to extended ssDNA regions at the fork. Controlled nascent strand resection may participate in ssDNA exposure. As characterized during DSB processing and repair, when ssDNA regions reach a critical size, the recombinase RAD51 partially replaces RPA at uncoupled forks, possibly assisted by cofactors belonging to the homologous recombination (HR) and Fanconi anemia (FA) pathways. RAD51-mediated template reannealing primes replication fork reversal, probably in concert with yet-unidentified specialized enzymatic activities, assisting template repair and limiting nucleolytic degradation of nascent strands upon prolonged stalling. PARP activation at discontinuous nascent strands and/or regressed arms stabilizes the forks in the reversed state, by transiently inhibiting the specific restart activity of RECQ1 helicase until the damage is repaired or the stress is released. RAD51 loading on regressed arms may further protect forks after reversal and promote alternative homology-mediated pathways of fork restart upon prolonged stalling.
Schematic model for replication fork reversal and restart upon different types of replication stress. Template damage, DNA synthesis inhibition, or torsional stress rapidly impair symmetric elongation of nascent strands and induce replication fork uncoupling, leading to extended ssDNA regions at the fork. Controlled nascent strand resection may participate in ssDNA exposure. As characterized during DSB processing and repair, when ssDNA regions reach a critical size, the recombinase RAD51 partially replaces RPA at uncoupled forks, possibly assisted by cofactors belonging to the homologous recombination (HR) and Fanconi anemia (FA) pathways. RAD51-mediated template reannealing primes replication fork reversal, probably in concert with yet-unidentified specialized enzymatic activities, assisting template repair and limiting nucleolytic degradation of nascent strands upon prolonged stalling. PARP activation at discontinuous nascent strands and/or regressed arms stabilizes the forks in the reversed state, by transiently inhibiting the specific restart activity of RECQ1 helicase until the damage is repaired or the stress is released. RAD51 loading on regressed arms may further protect forks after reversal and promote alternative homology-mediated pathways of fork restart upon prolonged stalling.
Indeed, one important implication of our data is that replication fork remodeling upon genotoxic stress shares an important mechanistic step with DSB repair, i.e., RAD51-mediated strand invasion, rapidly and effectively leading to replication fork reversal (Fig. 8; Neelsen and Lopes, 2015). As originally suggested by (Higgins et al., 1976), remodeling of uncoupled replication forks in human cells, besides limiting excessive ssDNA accumulation, would also allow more time for template repair and promote efficient DNA damage bypass directly at the fork, thus limiting reliance on postreplicative repair. Intriguingly, replication forks in wild-type yeast cells—which are devoid of PARPs—do not detectably undergo reversal upon most genotoxic treatments. In keeping with our model, yeast cells accumulate much longer ssDNA stretches at the junction and postreplicative ssDNA gaps, favoring fork restart by repriming (Sogo et al., 2002; Lopes et al., 2006). In line with this notion, genetic inactivation of HR-mediated repair (RAD51) upon genotoxic stress results in marked accumulation of postreplicative gaps in Saccharomyces cerevisiae (Lopes et al., 2006; Hashimoto et al., 2010) but not in human cells (Fig. 7 F).
Both aberrant RIs consistently and abundantly detected upon all tested genotoxic treatments—i.e., forks with extended ssDNA regions and reversed forks—carry intrinsic signaling potential. Excess ssDNA at replication forks has been linked to ATR activation (Zou and Elledge, 2003), whereas the formation of a new DNA end at regressed arms may potentially activate ATM in the absence of DSBs. Albeit conceptually attractive, our structural data argue against both of these models. Although ssDNA accumulation—at forks, gaps, or regressed arms—was observed at similar extents with several genotoxic treatments, some of them induced strong ATR activation (UV and HU), whereas others (APH and ETP) failed to detectably activate ATR, even at doses 10-fold higher than those required to drastically impair fork progression. Compared with EM analysis of ssDNA at RI, total ssDNA detection by native BrdU staining shows more pronounced differences among treatments and a stronger correlation with ATR activation, suggesting that ssDNA accumulation uncoupled from RI may be more relevant for ATR signaling. However, cells scoring positive in this assay still represent a minority of those showing γ-H2AX. Thus, ATR signaling in these conditions seems largely uncoupled from ssDNA/RPA accumulation, in keeping with other studies challenging this dogma (Ball et al., 2005; Recolin et al., 2012), and may reflect alternative yet-undefined mechanisms (Kumar et al., 2014). Similarly, high frequencies of reversed forks are observed with all treatments but are associated with detectable ATM activation only upon exposure to CPT and UV. Thus, besides these basic structural determinants (ssDNA and DNA ends, respectively), ATR and ATM activation at replication forks may require additional molecular features, which may be difficult to identify by EM analysis. Alternatively, checkpoint activation may entail a specific chromatin or topological context, or recruitment/removal of cellular factors at/from replication forks, which may only occur under specific conditions.
We show that the same molecular mechanism—i.e., PARP-regulated RECQ1 helicase activity—is largely responsible for the accumulation of reversed forks upon different types of genotoxic stress and to restart these forks once the stress is relieved (Figs. 5 and 8; Berti et al., 2013). An important mechanistic implication of these findings is that local PARP activation must result from a common structural determinant induced by all treatments, including genotoxic agents that do not directly cause DNA damage or breakage (e.g., HU). The unambiguous identification of this structural determinant will require further investigation. It should be noted, however, that the discontinuities present on nascent strands—as well as the DNA end at the regressed arm upon reversal—may be structurally identical to the strand breaks that are reportedly responsible for PARP activation in DNA repair (Pines et al., 2013) and that four-way junctions carry by themselves the potential to activate PARP (Lonskaya et al., 2005).
Our data clearly consolidate previous evidence that RAD51 is a stable component of replicating chromatin in metazoan, independently of fork breakage (Hashimoto et al., 2010; Petermann et al., 2010; Alabert et al., 2014). How is RAD51 recruited to uncoupled forks to promote template reannealing and thereby fork reversal (Fig. 8)? The presence of extended ssDNA regions—an ideal target for RAD51 binding—may by itself explain RAD51 loading to forks upon various genotoxic treatments. However, the competition with RPA for ssDNA binding in classical HR-dependent DSB repair implies that RAD51 loading is assisted by accessory proteins, such as the RAD51 paralogues and/or the cancer susceptibility genes BRCA1 and BRCA2 (Suwaki et al., 2011). Importantly, many of these factors have been recently reported to play a pivotal role also at stalled replication forks, promoting recombinational mechanisms that are structurally and/or genetically distinct from canonical DSB repair (Schlacher et al., 2011; Adelman et al., 2013; Willis et al., 2014). Furthermore, several additional FA factors are required for the same DSB repair-independent fork protection mechanism (Schlacher et al., 2012). Intriguingly, inactivation of the RAD51 paralogue XRCC3 in DT40 cells phenocopies PARP inactivation in suppressing fork slowing by Top1 poisons (Sugimura et al., 2008). Based on all this evidence, it will be a crucial challenge for future studies to assess in vivo the contribution of individual HR/FA factors in replication fork remodeling upon different types of genotoxic stress, by possibly mediating RAD51 loading or stabilization at fork-associated ssDNA regions (Fig. 8). In this view, these factors could contribute to genome stability by supporting DNA damage tolerance and preventing DSB formation, besides their reported role in repairing chromosomal breaks.
It will also be important to identify specific enzymatic activities required to assist RAD51 in driving replication fork reversal in vivo (Fig. 8; Neelsen and Lopes, 2015), presumably included in the list of factors showing fork remodeling activity in vitro (Kanagaraj et al., 2006; Machwe et al., 2006; Ralf et al., 2006; Gari et al., 2008; Blastyák et al., 2010; Bugreev et al., 2011; Bétous et al., 2012, 2013; Ciccia et al., 2012; Burkovics et al., 2014). Conversely, in light of our data, it will be important to extend the limited information on how the addition of RAD51 and RPA in the reactions may impact the biochemical properties of these fork remodeling proteins (Kanagaraj et al., 2006; Bugreev et al., 2011; Bétous et al., 2013; Burkovics et al., 2014).
Although we propose that the observed nascent strand degradation upon HR defects (Schlacher et al., 2011) is primarily a consequence of defective fork reversal upon prolonged fork stalling, our data do not exclude an additional role of RAD51 in stabilizing reversed forks during prolonged replication stress—as originally proposed (Schlacher et al., 2011)—by protecting the regressed arms from unscheduled nucleolytic attacks and assisting homology-directed fork restart (Fig. 8). It should be noted, however, that controlled regressed arm resection, which contributes to fork restart upon prolonged fork stalling, is genetically distinct from the extensive nascent strand degradation observed upon HR/FA defects (Schlacher et al., 2011; see Thangavel et al., in this issue).
Impairment of replication fork reversal may contribute to explain the potentiating effects of PARP inhibitors on several chemotherapeutic treatments (Rouleau et al., 2010; Ray Chaudhuri et al., 2012) and may also provide alternative mechanistic explanations for the observed synthetic lethality of PARP inhibition and HR defects (Farmer et al., 2005; Neelsen and Lopes, 2015). By analogy, the search for biochemical activities specifically required for fork reversal in vivo holds great potential to identify novel targets to potentiate cancer chemotherapy based on replication interference.
Materials and methods
Cells and cell culture
Human osteosarcoma U2OS cells, RPE-1 cells, or HEK293T cells were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin in an atmosphere containing 6% CO2 at 37°C. Cells were treated with different cancer chemotherapeutics and DNA-damaging agents as indicated, trypsinized, and processed for cell cycle analysis, Western blots, PFGE, and EM DNA extraction.
Genetic inactivation by sh/siRNA
shRNA-mediated down-regulation was achieved by cloning the sequence targeting RECQ1 (5′-GAGCTTATGTTACCAGTTA-3′) into the pLKO.1 (plasmid #10878; Addgene) lentiviral shRNA expression vector. Lentiviral particles were generated by transient cotransfection of pLKO.1 and the packaging plasmids psPAX2 (plasmid #12260; Addgene) and pM2D.G (plasmid #12259; Addgene) into HEK293T cells. Viral supernatants were filtered through a 0.45-µM filter and transduced on U2OS cells for 24 h followed by selection with 8 µg/ml puromycin for 3 d. Control transductions were performed using the pLKO.1 vector expressing a shRNA targeting Luciferase (5′-ACGCTGAGTACTTCGAAATGT-3′). For siRNA experiments, cells were transfected with the indicated siRNA using RNAiMAX (Invitrogen) according to manufacturer’s instruction. The experiments were performed 24 or 72 h after transfection. Purchased sequences were as follows: Luc siRNA (40 nM; 5′-CGUACGCGGAAUACUUCGA-3′), RAD51 #1 siRNA (40 nM; 5′-GAGCUUGACAAACUACUUC-3′), and RAD51 #2 siRNA (40 nM: 5′-GACUGCCAGGAUAAAGCUU-3′).
Drugs and reagents
CPT (Sigma-Aldrich) and cis-diammineplatinum(II)dichloride (Sigma-Aldrich) were dissolved in DMSO to yield a 20-mM (7 mg/ml) and a 15-mM (4.5 mg/ml) stock, respectively (freshly made). ETP (Sigma-Aldrich) and DOX (Sigma-Aldrich) were dissolved in DMSO to a stock concentration of 10 mM (6 mg/ml) and 5 mM, respectively, with aliquots stored at 4°C, protected from light. APH (Sigma-Aldrich) was dissolved in DMSO to yield a 3-mM stock, and aliquots were stored at −20°C. HU (Sigma-Aldrich) and MMC (Sigma-Aldrich) were prepared in double-distilled H2O to obtain a 100-mM (7.6 mg/ml) and a 3-mM (1 mg/ml) stock (freshly made), respectively. MMS purchased as a 10-M solution was stored at 4°C. Hydrogen peroxide solution (H2O2) 35% by weight in H2O (Sigma-Aldrich) was dissolved in double-distilled H2O. UV irradiation was administered using a UV 254-nm lamp.
Cell cycle analysis
Asynchronous subconfluent cultures of U2OS cells were treated with the indicated dose of the genotoxic agents for 8, 24, and 48 h. Time point 0 h represents cell cycle distribution of nontreated cells. The cells were then trypsinized, collected, and spun down at 400 g for 5 min (using 12 × 75–mm falcon tubes). The cells were washed with 1–2 ml PBS and spun down at 400 g for 5 min. After discarding PBS, control and treated cells were fixed with 70% ethanol at 4°C (for ≥30 min), washed, digested with 100 µg/ml RNase A, stained with 25 µg/ml propidium iodide, subjected to flow microfluorimetry on a flow cytometer (FACScan; BD), and analyzed by the FlowJo software (Tree Star). Flow cytometric analysis for γ-H2AX/EdU/DAPI, cells were labeled for 30 min with 10 µM EdU, harvested, and fixed for 10 min with 4% formaldehyde/PBS. Cells were washed with 1% BSA/PBS, pH 7.4, permeabilized with 0.5% saponin/1% BSA/PBS, and stained with mouse anti–γ-H2AX antibody (05-636; EMD Millipore) for 2 h followed by incubation with a suitable secondary antibody for 30 min. Incorporated EdU was labeled according to the manufacturer’s instructions (Invitrogen). DNA was stained with 1 µg/ml DAPI. Samples were measured on a flow cytometer (CyAn ADP; Beckman Coulter) and analyzed with Summit software v4.3 (Beckman Coulter).
Immunofluorescence microscopy
Cells were grown on coverslips in 10 µM BrdU for 48 h before the treatment with drugs. Cells were then treated with the aforementioned drugs for 1 h. After treatment, cells were washed with PBS and preextracted (25 mM Hepes, pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose, and 0.5% Triton X-100) on ice. Cells were then fixed using 4% formaldehyde for 15 min at RT. Fixed cells were then incubated with primary antibodies against BrdU mouse (347580; BD) and γ-H2AX rabbit (9718; Cell Signaling Technology) in a moist chamber for 1 h. Cells were incubated with secondary antibodies (anti–mouse 488 [A11029; Invitrogen] and anti–rabbit 594 [A11037; Invitrogen]) in a moist chamber for 1 h. For RAD51 and EdU, coverslips were preextracted in preextraction buffer (80 mM NaCl and 3 mM MgCl2) and fixed with 4% paraformaldehyde in PBS followed by permeabilization with 0.5% Triton X-100 in PBS. Subsequently, Click-iT reaction was performed using the manufacturer’s protocol (Invitrogen). Next, cells were blocked in 1% BSA and incubated with primary antibody against RAD51 rabbit (gift from F. Esashi, Sir William Dunn School of Pathology, University of Oxford, Oxford, England, UK) in a moist chamber. Coverslips were then incubated with secondary antibody (anti–rabbit 594; A11037). Coverslips were then washed with PBS, mounted with 4 µl Vectashield/DAPI, and sealed with nail polish. Cells were washed between all steps.
Microscopy was performed with a fluorescence microscope (DMRB; Leica; objective lenses: HCX Plan Apochromat 63×/1.40-0.60 NA oil) and acquired with a camera (DFC 360FX; Leica). The images were processed with Leica Application Suite Version 3.3.0.
Cell proliferation and viability
CellTiter blue reagent was used to estimate the number of viable cells present in multiwell plates after treating 2,000 cells/well with the doses of the genotoxic agents indicated in Fig. S1. 20 µl of CellTiter blue reagent was added to 100 µl of medium in the 96-well plate followed by incubation for 3 h at 37°C. The fluorescent signal was measured by recording fluorescence (560 nm(20)Ex/590 nm(10)Em) using a plate reader (Fluoroskan Ascent; Labsystems).
Human fork progression by DNA fiber analysis
The procedure was essentially performed according to Jackson and Pombo (1998), with previously described modifications (Ray Chaudhuri et al., 2012). In brief, asynchronously growing U2OS cells were labeled with 30 µM chlorodeoxyuridine (CldU; Sigma-Aldrich), a thymidine analogue, for 30 min, washed twice with PBS, treated with appropriate dosage with any of the genotoxic agents (or nontreated as control), and exposed to 250 µM 5-iodo-2′-deoxyuridine (IdU). The cells were quickly trypsinized and resuspended in PBS at 2.5 × 105 cells/ml. The labeled cells were diluted 1:8 with unlabeled cells, and 2.5 µl of cells were mixed with 7.5 µl of lysis buffer (200 mM Tris-HCl, pH 7.5, 50 mM EDTA, and 0.5% [wt/vol] SDS) on a glass slide. After 9 min, the slides were tilted at 15–45°, and the resulting DNA spreads were air dried, fixed in 3:1 methanol/acetic acid, and refrigerated overnight. The DNA fibers were denatured with 2.5 M HCl for 1 h, washed with PBS, and blocked with 2% BSA in PBST (PBS and Tween 20) for 40 min. The newly replicated CldU and IdU tracks were labeled (for 2.5 h in the dark, at RT) with anti-BrdU antibodies recognizing CldU (rat; Abcam) and IdU mouse (BD), respectively. After washing for 5 × 3 min in PBST (0.2%), the following secondary antibodies were used (incubated for 1 h in the dark, at RT): anti–mouse Alexa Fluor 488 (Molecular Probes) and anti–rat Cy3 (Jackson ImmunoResearch Laboratories, Inc.). After washing for 5 × 3 min in PBST (0.2%), the slides were air dried completely, mounted with 20 µl/slide Antifade gold (Invitrogen), and sealed to a coverslip by transparent nail polish. Microscopy was performed with a fluorescence microscope (IX81; Olympus; objective lenses: LC Plan Fluor 60×, 1.42 NA oil) and acquired with a charge-coupled device camera (Orca AG; Hamamatsu Photonics). The images were processed with CellR software (version2.6; Olympus). Statistical analysis of track length was performed using Prism (GraphPad Software). The significance of the difference between the means was determined by t test or by one-way analysis of variance (ANOVA).
DSB detection by PFGE
The procedure was essentially performed as previously described (Ray Chaudhuri et al., 2012). Asynchronous subconfluent cultures of U2OS cells were treated with defined doses of the genotoxic agents for 1 h. Cells were harvested by trypsinization, and agarose plugs of 2.5 × 105 cells were prepared in a disposable plug mold (Bio-Rad Laboratories). Plugs were then incubated in lysis buffer (100 mM EDTA, 1% [wt/vol] sodium lauroyl sarcosinate, 0.2% [wt/vol] sodium deoxycholate, and 1 mg/ml proteinase K) at 37°C for 72 h. Plugs were then washed four times in 20 mM Tris-HCl, pH 8.0, and 50 mM EDTA before loading onto an agarose gel. Electrophoresis was performed for 21 h at 14°C in 0.9% (wt/vol) Pulse Field Certified Agarose (Bio-Rad Laboratories) containing Tris-borate/EDTA buffer in a PFGE apparatus (CHEF DR III; Bio-Rad Laboratories), according to the following protocol (block I: 9 h, 120° included angle, 5.5 V/cm, 30 to 18-s switch; block II: 6 h, 117° included angle, 4.5 V/cm, 18 to 9-s switch; block III: 6 h, 112° included angle, 4.0 V/cm, 9 to 5-s switch). The gel was then stained with ethidium bromide and analyzed by the AlphaImager system (ProteinSimple). Relative DSB levels were assessed by comparing DSB signals for each treatment to the background levels observed in untreated conditions. Statistical analysis was performed using Prism. The significance was determined by using two-way ANOVA.
Protein extraction and Western blotting
Levels of intracellular pATM, pCHK1, pKAP1, and pRPA proteins were determined by Western blot analysis of cell extracts. Mammalian cell extracts were prepared in Laemmli sample buffer (4% SDS, 20% glycerol, and 120 mM Tris-HCl, pH 6.8). 50 µg total protein from cell isolates was loaded onto 9% polyacrylamide gel. Proteins were separated electrophoretically at 12 mA (for one gel; two gels at 24 mA) for 15–30 min and then at 18 mA until the end (for one gel; two gels at 36 mA) followed by transferring the proteins to Immobilon-P membrane (Thermo Fisher Scientific) for 2 h at 100 V (4°C) in a transfer buffer (25 mM Tris and 192 mM glycine) containing 15% methanol. Before addition of primary antibodies, membranes were blocked for 1 h in TBS containing 0.1% Tween 20 and 2% ECL blocking solution (GE Healthcare). Membranes were probed for pATM, total ATM, pChk1, total Chk1, pKAP1, total KAP1, RPA32 (S4/S8), total RPA, Rad51, RecQ1, β-Tubulin (loading control), and TFIIH (loading control). ATM p1981 rabbit (2152-1; Epitomics), ATM (2C1) mouse (GTX70103; GeneTex), CHK1 pS345 rabbit (2348; Cell Signaling Technology), CHK1 mouse (sc-8408; Santa Cruz Biotechnology, Inc.), KAP1 pS824 rabbit (A300-767A; Bethyl Laboratories, Inc.), KAP1 rabbit (A300-274A; Bethyl Laboratories, Inc.), phospho-RPA32 (S4/S8) rabbit (A300-245A; Bethyl Laboratories, Inc.), RPA32 rabbit (A300-244A; Bethyl Laboratories, Inc.), RAD51 (H-92) rabbit (sc-8349; Santa Cruz Biotechnology, Inc.), RECQ1 rabbit (NB100-618; Novus Biologicals), β-Tubulin (H-235) rabbit (sc-9104; Santa Cruz Biotechnology, Inc.), and TFIIH p89 rabbit (S-19; sc-293; Santa Cruz Biotechnology, Inc.). Secondary antibodies were ECL anti–mouse/rabbit IgG and horseradish peroxidase–linked whole antibody from sheep (GE Healthcare). The membrane was then exposed to an ECL system (detection reagent final volume equivalent to 0.125-ml/cm2 membrane; GE Healthcare), and a charge-coupled device image analyzer was used to visualize immunoreactive bands.
EM analysis of DNA RIs in human cells
The procedure was essentially performed as previously described (Neelsen et al., 2014). Asynchronous subconfluent cultures of U2OS cells were treated with defined doses of the genotoxic agents for 1 h. In vivo psoralen cross-linking of the DNA was achieved by a repetitive exposure of living cells to 4,5′,8-trimethylpsoralen (10 µg/ml final concentration) followed by irradiation pulses with UV 365-nm monochromatic light (UV Stratalinker 1800; Agilent Technologies). The cells were then lysed with cell lysis buffer (buffer C1: 1.28 M sucrose, 40 mM Tris-Cl, pH 7.5, 20 mM MgCl2, and 4% Triton X-100; QIAGEN) and then digested by digestion buffer (QIAGEN buffer G2: 800 mM guanidine–HCl, 30 mM Tris–HCl, pH 8.0, 30 mM EDTA, pH 8.0, 5% Tween 20, and 0.5% Triton X-100) and 1 mg/ml proteinase K at 50°C for 2 h. Chloroform/Isoamyl alcohol (24:1) was used to collect DNA via phase separation (centrifugation at 8,000 rpm for 20 min) followed by DNA precipitation by adding 0.7× volume of isopropanol. The DNA was then washed with 70% ethanol, air dried, and resuspended in 200 µl TE (Tris-EDTA) buffer. 100 U restriction enzyme PvuII high-fidelity was used for 12 µg mammalian genomic DNA digestion (4–5-h incubation). Poly-Prep chromatography columns were used for RI enrichment. Benzoylated naphthoylated DEAE–cellulose granules were resuspended in 10 mM Tris-HCl, pH 8.0, and 300 mM NaCl to a final concentration of 0.1 g/ml. The columns were washed and equilibrated with 10 mM Tris-HCl, pH 8.0, and 1 M NaCl and 10 mM Tris-HCl, pH 8.0, and 300 mM NaCl, respectively. The sample DNA was then loaded and incubated for 0.5 h. After washing the columns (10 mM Tris-HCl, pH 8.0, and 1 M NaCl), the DNA was eluted in caffeine solution (10 mM Tris-HCl, pH 8.0, 1 M NaCl, and 1.8% [wt/vol] caffeine) for 10 min followed by sample collection. DNA is then purified and concentrated, using an Amicon size-exclusion column and resuspended in TE. With DNA spreading by the “BAC method,” the DNA was loaded on carbon-coated 400-mesh copper grids. The DNA was then coated with platinum by platinum-carbon rotary shadowing (High Vacuum Evaporator MED 020; Bal-Tec). Microscopy was performed with a transmission electron microscope (Tecnai G2 Spirit; FEI; LaB6 filament; high tension ≤120 kV) and acquired with a side mount charge-coupled device camera (2,600 × 4,000 pixels; Orius 1000; Gatan, Inc.). The images were processed with DigitalMicrograph Version 1.83.842 (Gatan, Inc.) and analyzed with ImageJ (National Institutes of Health).
iPOND
iPOND was essentially performed as originally described (Sirbu et al., 2011, 2012) with minor modifications. HEK293T cells were labeled with 10 µM EdU (Life Technologies) and treated with the different drugs as indicated. For the pulse-chase experiments with thymidine, cells were washed with cell culture medium and incubated for 45 min in medium supplemented with 10 µM thymidine (Sigma-Aldrich). Then, the cells were cross-linked with 1% formaldehyde for 15 min at RT, quenched with 0.125 M glycine for 5 min, and washed three times with PBS. For the conjugation of EdU with biotin azide, cells were permeabilized with 0.25% Triton X-100/PBS, washed twice with PBS, and incubated in click reaction buffer (10 mM sodium-l-ascorbate, 20 µM biotin azide [Life Technologies], and 2 mM CuSO4) for 1 h at RT. DMSO was used instead of biotin azide for the “no click” control. Cells were washed twice with PBS, resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, and 1% SDS) supplemented with protease inhibitors (Sigma-Aldrich), and chromatin was solubilized by sonication in a Bioruptor (Diagenode) at 4°C at the highest setting for 10 min (30 s on and 45 s off cycles). After centrifugation for 30 min at 14,000 rpm, supernatants were diluted with 1:1 PBS (vol/vol) containing protease inhibitors and incubated overnight with streptavidin-agarose beads (EMD Millipore). Beads were washed once with lysis buffer, once with 1 M NaCl, twice with lysis buffer, and once with PBS, and captured proteins were eluted by boiling beads in 2× NuPAGE LDS Sample Buffer (Life Technologies) containing 100 mM DTT for 30 min at 95°C. Proteins were resolved by electrophoresis using NuPAGE Novex 4–12% Bis-Tris gels and detected by Western blotting with the indicated antibodies: RAD51 rabbit polyclonal (1:1,000; H92; Sigma-Aldrich), proliferating cell nuclear antigen mouse monoclonal (F2; 1:2,000; Sigma-Aldrich), RPA32 mouse monoclonal (NA19L; 1:1,000; EMD Millipore), RPA32-S4/S8 rabbit polyclonal (A300-245A; 1:1,000; Bethyl Laboratories), γ-H2AX–S139 rabbit monoclonal (1:1,000; 20E3; Cell Signaling Technology), and H3 rabbit polyclonal (1:1,000; ab1791; Abcam).
Online supplemental material
Fig. S1 shows cell proliferation (CellTiter blue) and cell cycle analyses (FACS) performed to identify mild doses of different genotoxic treatments interfering with DNA replication but not significantly affecting cell survival. Fig. S2 shows frequency and size of ssDNA gaps arising on replicated duplexes of U2OS or RPE-1 cells upon different genotoxic treatments. Fig. S3 shows control experiments for the frequency of reversed forks, the size of the regressed arms, and their possible exposure of ssDNA at forks reversed by different genotoxic stresses, providing examples of different categories of reversed forks. Fig. S4 shows Western blot and PFGE analyses upon genotoxic treatments at different doses and complement similar observations published in Figs. 1 and 5. Fig. S5 includes checkpoint activation and ssDNA gap accumulation in RECQ1-depleted cells, the labeling scheme for the iPOND experiments, cell cycle profiles upon progressive RAD51 depletion, and the dependency on RAD51 of reversed forks accumulating in RECQ1-depleted cells.
Acknowledgments
We thank the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance with EM. We are grateful to F. Esashi for sharing multiple reagents, to R. Santoro for indirect support, to K. Neelsen for critical reading of the manuscript, and to all current and past members of the Lopes group for useful discussions.
This work was supported by the Swiss National Science Foundation grant 31003A_146924 to M. Lopes and by the National Institutes of Health grant R01GM108648 to A. Vindigni.
The authors declare no competing financial interests.
References
- ANOVA
analysis of variance
- APH
aphidicolin
- CDDP
cis-diamminedichloroplatinum
- CldU
chlorodeoxyuridine
- CPT
camptothecin
- DDR
DNA damage response
- DNA-PK
DNA-dependent protein kinase
- DOX
doxorubicin
- DSB
double-strand break
- EdU
5′-ethynyl-2′-deoxyuridine
- ETP
etoposide
- FA
Fanconi anemia
- HR
homologous recombination
- HU
hydroxyurea
- ICL
interstrand cross-link
- IdU
5-iodo-2′-deoxyuridine
- MMC
mitomycin C
- MMS
methyl methanesulfonate
- PARP
poly (ADP-ribose) polymerase
- PFGE
pulsed field gel electrophoresis
- RI
replication intermediate
- ssDNA
single-stranded DNA
- Top1
Topoisomerase I
- Top2
Topoisomerase II
Author notes
R. Zellweger and D. Dalcher contributed equally to this paper.


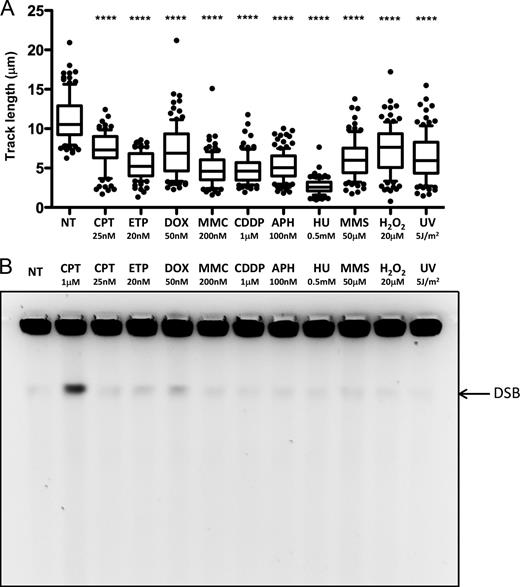

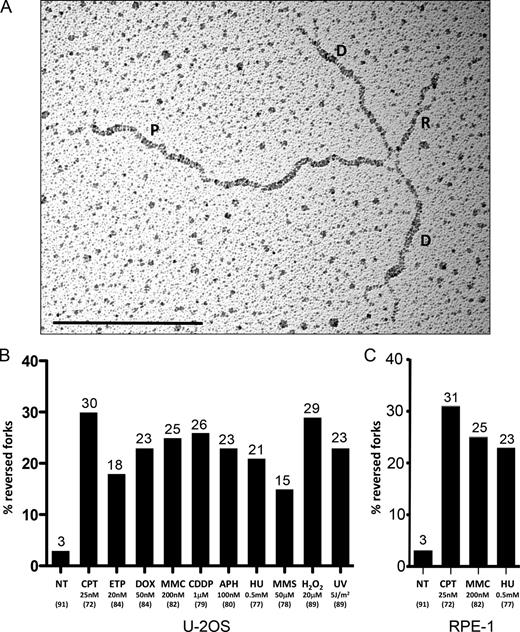

![Figure 5. RECQ1 and PARP activity control replication fork progression and accumulation/restart of reversed forks upon different types of genotoxic stress. (A) Statistical analysis of IdU track length measurements, according to the labeling protocol in Fig. S1, in U2OS cells stably transduced (shRNA) for Luciferase (shLuc) or RECQ1 (shRECQ1) depletion. 200 nM MMC and 500 µM HU were optionally added concomitantly with the second label (IdU). The PARP inhibitor olaparib (Ola; 10 µM) was optionally added 2 h before CldU labeling and maintained during labeling. At least 100 tracks were scored for each dataset. Horizontal lines represent the median value, and boxes and whiskers indicate the 10–90th percentiles. t test according to Mann–Whitney; ns, not significant; ****, P < 0.0001. Similar results were obtained in at least one independent experiment. (B) Frequency of reversed forks detected by EM in U2OS cells stably transfected (shRNA) for Luciferase or RECQ1 depletion. The cells were optionally treated for 1 h with 200 nM MMC or 500 µM HU, after an optional 2-h pretreatment with olaparib. Reversed fork restart was assessed by measuring the frequency of reversed forks 3 h after drug removal (release [Rel.]). In brackets, the total number of analyzed molecules is given. Above each column, the percentage of reversed forks is indicated. Similar results were obtained in at least one independent experiment. RECQ1 levels after shRNA-mediated depletion were detected by immunoblotting. TFIIH was used as a loading control. NT, not treated.](https://cdn.rupress.org/rup/content_public/journal/jcb/208/5/10.1083_jcb.201406099/6/m_jcb_201406099_fig5.jpeg?Expires=1772621055&Signature=VV0~hK65UjKUrkkZM4BTXe5qx0p-fCbtLw5WLW0FA0P4FkBOt1mW~9L-CajUE3~mOI0c5Q5NSqnfA-CgvWpdlah5Nwv4rlG4Ar5aT2cS9giVEZQJTzx9JspwjI22DGgnsQyvbchh2wdwAFGZx3pCcDolK~X4TiDz8JRJE1bpuHigszMon82BrTbtnxM-yo4tcrAaIWj9e2qSEVIFJjbBwyZYORVpaVChuY61ahn72BctdZ~lA7CkVuImL1AL4IxdrP~203o3j5AOQo7BZIckySKj39xGIiiqX1IGS~rEdMX-p-yobh0gGGUqu8WsMmV~XWMosVmhRjjZ3lZpmTyc-w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





