Axonal mitochondria are recruited to synaptic terminals in response to neuronal activity, but the mechanisms underlying activity-dependent regulation of mitochondrial transport are largely unknown. In this paper, using genetic mouse model combined with live imaging, we demonstrate that syntaphilin (SNPH) mediates the activity-dependent immobilization of axonal mitochondria through binding to KIF5. In vitro analysis showed that the KIF5–SNPH coupling inhibited the motor adenosine triphosphatase. Neuronal activity further recruited SNPH to axonal mitochondria. This motor-docking interplay was induced by Ca2+ and synaptic activity and was necessary to establish an appropriate balance between motile and stationary axonal mitochondria. Deleting snph abolished the activity-dependent immobilization of axonal mitochondria. We propose an “Engine-Switch and Brake” model, in which SNPH acts both as an engine off switch by sensing mitochondrial Rho guanosine triphosphatase-Ca2+ and as a brake by anchoring mitochondria to the microtubule track. Altogether, our study provides new mechanistic insight into the molecular interplay between motor and docking proteins, which arrests axonal mitochondrial transport in response to changes in neuronal activity.
Introduction
Neurons require specialized mechanisms to transport mitochondria to axons and to maintain their retention near synaptic terminals, where energy production and calcium homeostatic capacity are in high demand. The loss of mitochondria from axon terminals results in impaired synaptic transmission (Guo et al., 2005; Verstreken et al., 2005; Ma et al., 2009). Axonal mitochondria display complex motility patterns characterized by frequent pauses, changes in direction, and stationary docking (Hollenbeck and Saxton, 2005), suggesting that mitochondria are coupled to molecular motors kinesin-1 for anterograde transport and dyneins for retrograde movement together with docking machinery (MacAskill and Kittler, 2010; Sheng and Cai, 2012). Kinesin-1 is a tetramer consisting of a homodimer of one of three kinesin-1 heavy chains (KHCs; KIF5A, -B, and -C) and two kinesin light chains (KLCs; KLC1 and KLC2; Hirokawa et al., 2010). KIF5 motors use adaptors for cargo recognition and binding. In Drosophila melanogaster, mitochondrial Rho GTPase (Miro; dMiro) connects mitochondria to kinesin motors through Milton (Stowers et al., 2002; Guo et al., 2005). In mammals, two isoforms of Miro (Miro1 and 2) act similarly (Fransson et al., 2006). Trak1 and 2 (trafficking kinesin-binding protein 1 and 2), mammalian Milton orthologues, serve as an adaptor linking mitochondria to KIF5 through Miro1 (Smith et al., 2006; MacAskill et al., 2009a). Motile mitochondria are recruited to synapses in response to elevated Ca2+ and synaptic activity (Rintoul et al., 2003; Yi et al., 2004; MacAskill et al., 2009b; Wang and Schwarz, 2009). Synaptic structure and function are highly plastic and undergo activity-dependent remodeling, thereby dynamically altering mitochondrial motility and distribution.
How are motile mitochondria recruited to the stationary pool in response to synaptic activity? Recent work characterized Miro’s role as a Ca2+ sensor to regulate mitochondrial motility (Saotome et al., 2008; MacAskill et al., 2009b; Cai and Sheng, 2009; Wang and Schwarz, 2009). Miro has two EF-hand Ca2+ binding domains (Fransson et al., 2006). Activity-triggered Ca2+ influx recruits motile mitochondria to the stationary phase adjacent to activated synapses through a Miro-Ca2+ sensor that inactivates KIF5-driven transport machineries. Although this model is attractive, the mechanistic insight into how Miro-Ca2+ sensing inactivates KIF5 motor is unclear. Recently, we identified syntaphilin (SNPH) as a “static anchor” specific for axonal mitochondria (Kang et al., 2008; Chen et al., 2009). SNPH targets to axonal mitochondria and mediates their docking by anchoring them to the microtubule (MT)-based cytoskeleton. Deleting snph in mice robustly increases axonal mitochondrial motility. Thus, identifying SNPH as a docking protein provides a molecular target to investigate how motile axonal mitochondria are recruited to the stationary pool in response to changes in neuronal activity.
Using snph mouse models and time-lapse imaging analysis in live neurons, we demonstrate that SNPH mediates the activity-dependent immobilization of axonal mitochondria by physical displacement of KIF5 from the Miro–Track (trafficking kinesin-binding protein) complex. Such a KIF5–SNPH coupling inhibits KIF5 ATPase and is controlled by a Miro-Ca2+ sensing switch in response to neuronal activity. We propose the “Engine-Switch and Brake” model, which nicely reconciles the current dispute in explaining how Miro-Ca2+ sensing arrests mitochondrial transport. Our study elucidates a new molecular mechanism underlying the complex regulation of axonal mitochondrial transport, thereby advancing our knowledge that may be essential for maintaining axonal and synaptic homeostasis.
Results
SNPH is required for activity-dependent regulation of mitochondrial transport
To determine whether SNPH is involved in Ca2+-dependent immobilization of axonal mitochondria, we conducted time-lapse imaging in live hippocampal neurons to record the transport of both mitochondria and late endosomes along the same axons of snph+/+ and snph−/− neurons expressing DsRed–mitochondria targeting sequence (Mito) and YFP-Rab7. To elevate cytosolic Ca2+, the neurons were treated briefly (2 min) with 50 mM KCl and 10 µM FPL 64176, an L-type Ca2+ channel agonist. This combined application was shown to raise synaptic Ca2+ to levels similar to those produced by 30 µM glutamate and result in an inhibitory effect on mitochondrial motility without inducing neuronal toxicity (MacAskill et al., 2009b). As we previously reported (Kang et al., 2008; Chen et al., 2009; Cai et al., 2010), axons were distinguished from dendrites based on known morphological characteristics: greater length, thin and uniform diameter, and sparse branching (Banker and Cowan, 1979). Kymographs were used to quantify stationary and motile organelles (Miller and Sheetz, 2004). The KCl treatment in snph+/+ neurons decreased axonal mitochondria motility (35 ± 2% before and 15 ± 2% after treatment; mean ± SEM, P < 0.001; Fig. 1, A and C). In contrast, the same treatment of snph−/− neurons failed to affect axonal mitochondria motility (81 ± 5% before and 76 ± 4% after treatment, respectively; P = 0.22; Fig. 1, B and C). As a control, the KCl treatment had no significant effect on the motility of late endosomes labeled with YFP-Rab7 (Fig. 1, A, B, and D; and Videos 1 and 2) or early endosomes labeled with GFP-Rab5 (Fig. S1, A and B) along the same axons of snph+/+ and snph−/− neurons. Furthermore, overexpressing SNPH in wild-type neurons immobilized axonal mitochondria but had no effect on late endosomes (Fig. S1 C and Video 3). In dendrites, however, mitochondrial motility in both snph+/+ and snph−/− neurons decreased significantly after the same treatments (P < 0.001; Fig. 1 E), consistent with the role of SNPH as a docking receptor specific for axonal, but not dendritic, mitochondria (Kang et al., 2008).
Overexpressing SNPH selectively immobilizes axonal mitochondria, but not late endosome, along the same axon. Representative time-lapse images demonstrating that overexpressing SNPH in wild-type hippocampal neurons immobilizes almost all axonal mitochondria (red) but has no observable role in the motility of late endosomes (green) labeled with GFP-Rab7 along the same axons. Hippocampal neurons were cotransfected with DsRed-Mito, SNPH, and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. S1 C.
Overexpressing SNPH selectively immobilizes axonal mitochondria, but not late endosome, along the same axon. Representative time-lapse images demonstrating that overexpressing SNPH in wild-type hippocampal neurons immobilizes almost all axonal mitochondria (red) but has no observable role in the motility of late endosomes (green) labeled with GFP-Rab7 along the same axons. Hippocampal neurons were cotransfected with DsRed-Mito, SNPH, and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. S1 C.
SNPH is required for activity-dependent immobilization of axonal mitochondria. (A–D) Kymographs (A and B) and quantitative analysis (C and D) showing the motility of axonal mitochondria and late endosomes along the same axons of snph+/+ and snph−/− neurons before and after treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Hippocampal neurons were cotransfected with DsRed-Mito and YFP-Rab7 at DIV9 followed by imaging at DIV12. Vertical lines in kymographs represent stationary mitochondria; slanted lines or curves to the right (negative slope) represent retrograde movement; curves to the left (positive slope) indicate anterograde movement (also see Fig. S1 and Videos 1, 2, and 3). (E) The relative motility of dendritic mitochondria in snph+/+ and snph−/− hippocampal neurons before and after treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Data were quantified from numbers of neurons indicated in parentheses (C and D) or within bars (E) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
SNPH is required for activity-dependent immobilization of axonal mitochondria. (A–D) Kymographs (A and B) and quantitative analysis (C and D) showing the motility of axonal mitochondria and late endosomes along the same axons of snph+/+ and snph−/− neurons before and after treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Hippocampal neurons were cotransfected with DsRed-Mito and YFP-Rab7 at DIV9 followed by imaging at DIV12. Vertical lines in kymographs represent stationary mitochondria; slanted lines or curves to the right (negative slope) represent retrograde movement; curves to the left (positive slope) indicate anterograde movement (also see Fig. S1 and Videos 1, 2, and 3). (E) The relative motility of dendritic mitochondria in snph+/+ and snph−/− hippocampal neurons before and after treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Data were quantified from numbers of neurons indicated in parentheses (C and D) or within bars (E) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
We applied electrical field stimulation (100 Hz for 2 s) to increase firing rates and excitatory synaptic currents (MacAskill et al., 2009b). Enhanced synaptic activity in snph+/+ neurons dramatically decreased axonal mitochondrial motility (37 ± 2% before and 15 ± 3% after stimulation; P < 0.001). However, the same treatment failed to change mitochondrial motility in snph−/− neurons (77 ± 3% before and 73 ± 4% after stimulation; P = 0.10; Fig. 2 and Videos 4 and 5), suggesting that deleting snph abolishes activity-dependent regulation of axonal mitochondrial transport. Removing extracellular Ca2 or blocking the action potential using 1 µM tetrodotoxin (TTX), a sodium channel blocker, abolished mitochondrial immobilization in response to field stimulation in wild-type neurons (Fig. 2). Thus, SNPH is required to arrest axonal mitochondrial movement in response to synaptic activity.
SNPH is required to arrest axonal mitochondrial movement in response to synaptic activity. (A and B) Kymographs (A) and quantitative analysis (B) showing the motility of axonal mitochondria in snph+/+ and snph−/− hippocampal neurons before and after 100-Hz field stimulation in the presence or absence of 1.2 mM CaCl2 or 1 µm TTX. Neurons were transfected with DsRed-Mito at DIV9 followed by time-lapse imaging at DIV12. Vertical lines in kymographs represent stationary mitochondria; slanted lines or curves to the right (negative slope) represent retrograde movement (also see Videos 4 and 5). Data were quantified from numbers of neurons indicated within bars in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
SNPH is required to arrest axonal mitochondrial movement in response to synaptic activity. (A and B) Kymographs (A) and quantitative analysis (B) showing the motility of axonal mitochondria in snph+/+ and snph−/− hippocampal neurons before and after 100-Hz field stimulation in the presence or absence of 1.2 mM CaCl2 or 1 µm TTX. Neurons were transfected with DsRed-Mito at DIV9 followed by time-lapse imaging at DIV12. Vertical lines in kymographs represent stationary mitochondria; slanted lines or curves to the right (negative slope) represent retrograde movement (also see Videos 4 and 5). Data were quantified from numbers of neurons indicated within bars in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
KIF5–SNPH interaction mediates activity-induced immobilization of axonal mitochondria
Trak2 links mitochondrial receptor Miro1 to KIF5 motors to assemble mitochondrial transport complexes (MacAskill et al., 2009a). Elevating Miro1 expression increases recruitment of Trak2 and KIF5 to mitochondria, thus enhancing mitochondrial transport (MacAskill et al., 2009b). Both Miro1 and Trak2 target axonal mitochondria (Fig. S2). Knocking down Trak2 or expressing its dominant-negative mutants reduces mitochondrial motility in axons (Brickley and Stephenson, 2011). To determine whether Miro1/Trak2 and SNPH play opposite roles in the regulation of axonal mitochondrial motility, we conducted six panels of experiments in neurons. Expressing SNPH alone immobilized almost all axonal mitochondria (motility: 3 ± 1%), whereas expressing Miro1 or Trak2 alone robustly increased axonal mitochondrial motility (77 ± 3% [P < 0.001] and 81 ± 4% [P < 0.001], respectively) relative to control neurons (33 ± 2%). Interestingly, coexpressing SNPH and Miro1 or SNPH and Trak2 dominantly immobilized axonal mitochondria (16 ± 2% [P < 0.001] and 14 ± 2% [P < 0.001], respectively; Fig. 3 and Videos 6 and 7).
SNPH dominantly immobilizes axonal mitochondria. (A and B) Kymographs (A) and quantitative analysis (B) showing axonal mitochondrial motility in snph+/+ neurons expressing SNPH, Miro1, or Trak2 alone or coexpressing Miro1 + SNPH or Trak2 + SNPH. Hippocampal neurons were transfected at DIV9 and imaged at DIV12. Data were quantified from numbers of neurons (indicated in parentheses) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test. Also see Fig. S2 and Videos 6 and 7.
SNPH dominantly immobilizes axonal mitochondria. (A and B) Kymographs (A) and quantitative analysis (B) showing axonal mitochondrial motility in snph+/+ neurons expressing SNPH, Miro1, or Trak2 alone or coexpressing Miro1 + SNPH or Trak2 + SNPH. Hippocampal neurons were transfected at DIV9 and imaged at DIV12. Data were quantified from numbers of neurons (indicated in parentheses) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test. Also see Fig. S2 and Videos 6 and 7.
The opposite effects of Miro1/Trak2 and SNPH in shifting the balance between motile and stationary axonal mitochondria raise two questions: whether (1) there is molecular interplay between motors and docking receptors and (2) Miro-Ca2+ sensing immobilizes axonal mitochondria by switching the KIF5 interaction from motor adaptors to SNPH. To address these questions, we performed six lines of biochemical analysis. First, we performed a pull-down assay using a truncated GST-SNPH (203–469 aa), lacking the MT binding domain and Mito (Fig. 4 E). GST-SNPH (203–469 aa) pulled down the kinesin-1 heavy chain (KHC), but not its light chain (KLC), and the dynein intermediate chain (IC74) from mouse brain homogenates (Fig. 4 A). To confirm a native SNPH–KIF5 interaction in neurons, we applied an anti-SNPH antibody to pull down endogenous KHC, but not KLC and IC74, from adult mouse brain homogenates (Fig. 4 B). Conversely, SNPH was specifically pulled down by the anti-KHC antibody from wild type, but not from snph knockout, mouse brains (Fig. 4 C), providing convincing evidence for a native SNPH–KIF5 complex. Our pull-down and coimmunoprecipitation assays suggest that SNPH interacts with KIF5 independent of KLC. In contrast, a native SNPH–dynein heavy chain (DHC) complex was not detected by either an anti-SNPH or an anti-DHC antibody from mouse brains under the same conditions (Fig. S3).
SNPH interacts with KIF5. (A) GST-SNPH (203–469 aa) pulls down the kinesin-1 heavy chain (KHC or KIF5) from mouse brain homogenates (BH). (B) An anti-SNPH antibody, but not preimmune serum (PIS), immunoprecipitates KHC from brain homogenates. KLC and the dynein intermediate chain IC74 are not detected in the same assays. (C) Coimmunoprecipitation showing a native SNPH–KIF5 complex in mouse brains. SHPN was pulled down by an anti-KHC antibody from wild-type (WT) mouse brain homogenates followed by detection with an anti-SNPH antibody. As a control, the same antibody failed to detect the SHPN–KIF5 complex from snph knockout (KO) mouse brains. (D) GST pull-down showing SNPH binding to the C-terminal tail (814–963 aa) of KIF5B. GST-SNPH (203–469 aa) or GST was incubated with His-tagged KIF5B truncated mutants. The bound protein complexes were blotted with an anti–H7-His antibody. Coomassie blue staining shows the relative purity of the recombinant proteins. (E) Schematic diagram of truncated SNPH mutants. MTB, MT binding domain; KBD, KHC binding domain; Mito, mitochondrial targeting sequence. (F) Coimmunoprecipitation showing KIF5 binding to the SNPH sequence (381–466). GFP-KIF5C and GFP-SNPH truncated mutants were coexpressed in HEK293 cells. The cell lysates were immunoprecipitated (IP) with an anti-KHC antibody and immunoblotted (IB) with an anti-GFP antibody. Note that the 86-residue segment (381–466 aa) of SNPH is sufficient to bind to KIF5C and is thus referred to as the KBD. (G) Deleting KBD abolishes SNPH binding to KIF5C. GFP-tagged KIF5C, SNPH, and its KBD deletion mutant, or KBD alone, were coexpressed in HEK293 cells. The cell lysates were immunoprecipitated with an anti-KHC antibody and immunoblotted with an anti-GFP antibody. Red boxes indicate SNPH, its KBD deletion mutant, or KBD alone. Asterisks show degraded bands.
SNPH interacts with KIF5. (A) GST-SNPH (203–469 aa) pulls down the kinesin-1 heavy chain (KHC or KIF5) from mouse brain homogenates (BH). (B) An anti-SNPH antibody, but not preimmune serum (PIS), immunoprecipitates KHC from brain homogenates. KLC and the dynein intermediate chain IC74 are not detected in the same assays. (C) Coimmunoprecipitation showing a native SNPH–KIF5 complex in mouse brains. SHPN was pulled down by an anti-KHC antibody from wild-type (WT) mouse brain homogenates followed by detection with an anti-SNPH antibody. As a control, the same antibody failed to detect the SHPN–KIF5 complex from snph knockout (KO) mouse brains. (D) GST pull-down showing SNPH binding to the C-terminal tail (814–963 aa) of KIF5B. GST-SNPH (203–469 aa) or GST was incubated with His-tagged KIF5B truncated mutants. The bound protein complexes were blotted with an anti–H7-His antibody. Coomassie blue staining shows the relative purity of the recombinant proteins. (E) Schematic diagram of truncated SNPH mutants. MTB, MT binding domain; KBD, KHC binding domain; Mito, mitochondrial targeting sequence. (F) Coimmunoprecipitation showing KIF5 binding to the SNPH sequence (381–466). GFP-KIF5C and GFP-SNPH truncated mutants were coexpressed in HEK293 cells. The cell lysates were immunoprecipitated (IP) with an anti-KHC antibody and immunoblotted (IB) with an anti-GFP antibody. Note that the 86-residue segment (381–466 aa) of SNPH is sufficient to bind to KIF5C and is thus referred to as the KBD. (G) Deleting KBD abolishes SNPH binding to KIF5C. GFP-tagged KIF5C, SNPH, and its KBD deletion mutant, or KBD alone, were coexpressed in HEK293 cells. The cell lysates were immunoprecipitated with an anti-KHC antibody and immunoblotted with an anti-GFP antibody. Red boxes indicate SNPH, its KBD deletion mutant, or KBD alone. Asterisks show degraded bands.
Next, we examined the direct interaction between SNPH and KHC. Mammals have three KHCs (KIF5A, -B, and -C), which share a highly conserved sequence homology, particularly in their C-terminal cargo binding domains (Hirokawa et al., 2010). SNPH specifically binds His-KIF5B (814–963 aa), a C-terminal tail containing the cargo binding domain (Fig. 4 D and Fig. S4). In contrast, no SNPH binding was observed for His-KIF5B (1–554 aa), the N-terminal motor domain (N-MD), or His-KIF5B (555–813 aa), the middle stalk coils. Furthermore, we located a KHC binding sequence in SNPH by performing coimmunoprecipitation in HEK293 cells coexpressing GFP-tagged KIF5C, a neuron-specific Kinesin-1 isoform, and various truncated SNPH mutants. The 86-residue segment (381–466 aa) of SNPH is sufficient to bind to KIF5C and is hereafter referred to as KHC binding domain (KBD; Fig. 4, E and F). In addition, we generated an SNPH mutant lacking KBD (SNPH-ΔKBD). Although KBD alone is sufficient to interact with KIF5C, no binding was detected for SNPH-ΔKBD to KIF5C (Fig. 4 G), suggesting direct interaction between SNPH and KIF5 via KBD.
Next, we tested whether KIF5–SNPH interaction is required to immobilize axonal mitochondria. Although GFP-SNPH arrested almost all axonal mitochondria (Fig. 5, A and B), a significant portion (∼24%) of mitochondria labeled with SNPH-ΔKBD moved dynamically. Given the fact that SNPH-ΔKBD contains the MT binding domain (103–203 aa) and the Mito (466–532 aa; Fig. 4 E), overexpressing this mutant on mitochondria would enhance the static link between mitochondria and MTs. We alternatively performed immunoprecipitation using cell lysates from transfected HEK cells. Although the KIF5C–SNPH complex was readily detectable in the absence of expression of KBD, coexpressing KBD reduced the KIF5C–SNPH complex (Fig. 5 C), suggesting that KBD interferes with the KIF5–SNPH interaction. To validate the biochemical observations, we performed live-neuron imaging. Field stimulation (100 Hz for 2 s) of neurons expressing KBD failed to arrest axonal mitochondrial transport (40 ± 2% before and 37 ± 4% after stimulation; P = 0.30; Fig. 5, D and E; and Video 8). However, the same stimulation in neurons expressing GFP control results in a rapid reduction in mitochondrial movement (34 ± 2% before and 14 ± 2% after stimulation; P < 0.001). In contrast, expressing KBD did not impair activity-dependent immobilization of dendritic mitochondria (32 ± 9% before and 6 ± 5% after stimulation; P < 0.001). These results suggest that KIF5–SNPH coupling is crucial for activity-dependent immobilization of axonal but not dendritic mitochondria.
Disrupting KIF5–SNPH interaction reduces activity-dependent immobilization of axonal mitochondria. Representative time-lapse images showing axonal mitochondrial transport in hippocampal neurons expressing GFP-KBD during ∼16 min of observation. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied during the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. Note that expressing KBD impairs the immobilization of axonal mitochondria in response to neuronal activity relative to the control neurons (Video 4). The kymograph of this video is presented in Fig. 5 D.
Disrupting KIF5–SNPH interaction reduces activity-dependent immobilization of axonal mitochondria. Representative time-lapse images showing axonal mitochondrial transport in hippocampal neurons expressing GFP-KBD during ∼16 min of observation. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied during the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. Note that expressing KBD impairs the immobilization of axonal mitochondria in response to neuronal activity relative to the control neurons (Video 4). The kymograph of this video is presented in Fig. 5 D.
Disrupting KIF5–SNPH interaction abolishes activity-dependent immobilization of axonal mitochondria. (A and B) Kymographs (A) and quantitative analysis (B) showing the motility of axonal mitochondria in neurons expressing GFP-SNPH or GFP-SNPH-ΔKBD. (C) An inhibitory role of KBD in the KIF5C–SNPH interaction. HEK293 cells were transfected with GFP-KIF5C alone or cotransfected with GFP-SNPH or GFP-SNPH + GFP-KBD. The KIF5C–SNPH complex was immunoprecipitated (IP) from the lysates using an anti-KHC antibody followed by immunoblotting (IB) with an anti-GFP antibody. Note that KBD interferes with the interaction of SNPH with KIF5C (top red box) by competitively binding to KIF5C (bottom red box). (D and E) The motility of axonal and dendritic mitochondria in wild-type neurons before and after 100-Hz (2 s) field stimulation. Expressing GFP-KBD abolishes the activity-dependent immobilization of axonal but not dendritic mitochondria (also see Video 8). Data were quantified from numbers of neurons (indicated in parentheses in B and within bars in E) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
Disrupting KIF5–SNPH interaction abolishes activity-dependent immobilization of axonal mitochondria. (A and B) Kymographs (A) and quantitative analysis (B) showing the motility of axonal mitochondria in neurons expressing GFP-SNPH or GFP-SNPH-ΔKBD. (C) An inhibitory role of KBD in the KIF5C–SNPH interaction. HEK293 cells were transfected with GFP-KIF5C alone or cotransfected with GFP-SNPH or GFP-SNPH + GFP-KBD. The KIF5C–SNPH complex was immunoprecipitated (IP) from the lysates using an anti-KHC antibody followed by immunoblotting (IB) with an anti-GFP antibody. Note that KBD interferes with the interaction of SNPH with KIF5C (top red box) by competitively binding to KIF5C (bottom red box). (D and E) The motility of axonal and dendritic mitochondria in wild-type neurons before and after 100-Hz (2 s) field stimulation. Expressing GFP-KBD abolishes the activity-dependent immobilization of axonal but not dendritic mitochondria (also see Video 8). Data were quantified from numbers of neurons (indicated in parentheses in B and within bars in E) in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test.
Miro-Ca2+ sensing facilitates KIF5–SNPH interaction
We then examined whether the KIF5–SNPH interaction has any impact on the complex KIF5–Trak2–Miro1. First, we coexpressed GFP-KIF5C and Flag-Trak2 in the absence or presence of SNPH or SNPH-ΔKBD in HEK293 cells followed by immunoprecipitation with an anti-Flag antibody. Although KIF5C–Trak2 complexes were readily detectable in the absence of SNPH, coexpressing SNPH, but not SNPH-ΔKBD, reduced KIF5C–Trak2 complexes (Fig. 6, A and B). As the KIF5C C-terminal tail binds both Trak2 (Smith et al., 2006) and SNPH, it is likely that SNPH and Trak2 competitively bind KIF5C. Second, expressing Trak2 alone in neurons significantly increased axonal mitochondrial motility (81 ± 4%) relative to wild-type control (P < 0.001). When coexpressing Trak2 with SNPH, only a small portion (14 ± 2%) of axonal mitochondria was motile. In contrast, when coexpressing Trak2 with SNPH-ΔKBD, a larger portion (50 ± 3%; P < 0.001) of axonal mitochondria was motile (Fig. 6, C and D). Based on the in vitro biochemical analyses in soluble conditions and live-neuron imaging, we hypothesize that SNPH-mediated regulation of axonal mitochondrial transport is likely by competing with Trak2 for binding KIF5.
SNPH competes with Trak2 to bind KIF5. (A and B) Representative blots (A) and quantitative analysis (B) showing competition of SNPH and Trak2 for binding KIF5. HEK293 cells were cotransfected as indicated. The KIF5C–Trak2 complex was coimmunoprecipitated (IP) from cell lysates with an anti-Flag antibody and immunoblotted (IB) with an anti-GFP antibody (bottom). Note that SNPH, but not SNPH-ΔKBD, inhibits the formation of the KIF5C–Trak2 complex (red box). Data were quantified from three independent experiments. The asterisk denotes degraded GFP-SNPH. (C and D) SNPH, but not SNPH-ΔKBD, competes with Trak2 to immobilize axonal mitochondria. (E and F) Immunoprecipitation (E) and quantitative analysis (F) showing activity-induced and Ca2+-dependent formation of the native SNPH–KHC complex in mature cortical neurons. The cultured neurons (DIV14) were treated for 10 min with 50 mM KCl and 10 µM FPL 64176 in modified Tyrode’s solution with or without 1.2 mM Ca2+. The native SNPH–KHC complexes were coimmunoprecipitated from neuronal lysates with an anti-SNPH antibody and subsequently immunoblotted with an anti-KHC and LC8 antibody. Coimmunoprecipitated levels of KIF5 and LC8 after stimulation were normalized against their levels before stimulation, respectively. Note that removing [Ca2+]ext from the medium abolished the activity-induced SNPH–KIF5 interaction. (G and H) Kymographs (G) and quantitative analysis (H) showing enhanced motility of axonal mitochondria after Ca2+ depletion. Mitochondrial motility in hippocampal neurons were recorded before and 20 min after removing 1.2 mM [Ca2+]ext from the medium on the same axons (also see Video 9). Bars, 10 µm. Data in D and H were quantified from numbers of axons (indicated in bars). Student’s t test in D and H and Mann–Whitney U test in B and F. Error bars show SEM.
SNPH competes with Trak2 to bind KIF5. (A and B) Representative blots (A) and quantitative analysis (B) showing competition of SNPH and Trak2 for binding KIF5. HEK293 cells were cotransfected as indicated. The KIF5C–Trak2 complex was coimmunoprecipitated (IP) from cell lysates with an anti-Flag antibody and immunoblotted (IB) with an anti-GFP antibody (bottom). Note that SNPH, but not SNPH-ΔKBD, inhibits the formation of the KIF5C–Trak2 complex (red box). Data were quantified from three independent experiments. The asterisk denotes degraded GFP-SNPH. (C and D) SNPH, but not SNPH-ΔKBD, competes with Trak2 to immobilize axonal mitochondria. (E and F) Immunoprecipitation (E) and quantitative analysis (F) showing activity-induced and Ca2+-dependent formation of the native SNPH–KHC complex in mature cortical neurons. The cultured neurons (DIV14) were treated for 10 min with 50 mM KCl and 10 µM FPL 64176 in modified Tyrode’s solution with or without 1.2 mM Ca2+. The native SNPH–KHC complexes were coimmunoprecipitated from neuronal lysates with an anti-SNPH antibody and subsequently immunoblotted with an anti-KHC and LC8 antibody. Coimmunoprecipitated levels of KIF5 and LC8 after stimulation were normalized against their levels before stimulation, respectively. Note that removing [Ca2+]ext from the medium abolished the activity-induced SNPH–KIF5 interaction. (G and H) Kymographs (G) and quantitative analysis (H) showing enhanced motility of axonal mitochondria after Ca2+ depletion. Mitochondrial motility in hippocampal neurons were recorded before and 20 min after removing 1.2 mM [Ca2+]ext from the medium on the same axons (also see Video 9). Bars, 10 µm. Data in D and H were quantified from numbers of axons (indicated in bars). Student’s t test in D and H and Mann–Whitney U test in B and F. Error bars show SEM.
Next, we asked whether elevated Ca2+ levels facilitate formation of the native SNPH–KIF5 complex in mature cortical neurons (day in vitro [DIV] 14) treated for 10 min with 50 mM KCl and 10 µM FPL 64176 in the presence or absence of 1.2 mM Ca2+ in modified Tyrode’s solution. Removing [Ca2+]ext from the medium abolished the activity-enhanced SNPH–KIF5 interaction (Fig. 6, E and F). As an internal control, neuronal activity has no influence on the SNPH-LC8 binding. We further examined the relative motility of axonal mitochondria under the resting conditions in the absence of [Ca2+]ext. Mitochondrial motility was recorded before and 20 min after removing [Ca2+]ext from medium on the same axons. In the resting state, one third (33 ± 3%) of axonal mitochondria were motile in the presence of 1.2 mM [Ca2+]ext. Removing [Ca2+]ext from the medium significantly increased the motility (56 ± 3%; P < 0.001; Fig. 6, G and H; and Video 9), supporting the notion that the SNPH–KIF5 interaction is induced by Ca2+. Depleting Ca2+ facilitates the dissociation of SNPH from KIF5 motors, thus relieving the inhibitory role of SHPN on the motor activity.
Ca2+ depletion enhances the axonal mitochondrial motility. Representative time-lapse images showing enhanced motility of axonal mitochondria under the resting conditions after Ca2+ depletion. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Mitochondrial motility in hippocampal neurons were recorded before and 20 min after removing 1.2 mM [Ca2+]ext from modified Tyrode’s solution on the same axons. The kymograph of this video is presented in Fig. 6 G.
Ca2+ depletion enhances the axonal mitochondrial motility. Representative time-lapse images showing enhanced motility of axonal mitochondria under the resting conditions after Ca2+ depletion. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Mitochondrial motility in hippocampal neurons were recorded before and 20 min after removing 1.2 mM [Ca2+]ext from modified Tyrode’s solution on the same axons. The kymograph of this video is presented in Fig. 6 G.
SNPH–KIF5 coupling inhibits motor activity
Although two thirds of axonal mitochondria are in the stationary phase, deleting snph in mice results in >70% of axonal mitochondria in the motile state (Kang et al., 2008). To address whether this robustly enhanced motility is caused by increased recruitment of molecular motors to mitochondria, we isolated axonal mitochondria from sciatic nerves of snph+/+ and snph−/− mice as previously described (Frezza et al., 2007). Immunoblot analysis showed no significant change in the normalized intensity of axonal mitochondria-associated KIF5 (P = 0.28) and dynein intermediate chain IC74 (P = 0.88) between snph+/+ and snph−/− mice (Fig. 7, A and B), suggesting that KIF5 and dynein motors attach to both motile and stationary mitochondria. This study raises a question as to whether motile mitochondria are recruited to the stationary pool by inactivating motors on mitochondria.
SNPH inhibits KIF5 motor ATPase activity. (A and B) Deleting snph has no effect on the motor association with axonal mitochondria. Axonal mitochondria were isolated from sciatic nerves of snph+/+ and snph−/− mice. 15 µg of total proteins from supernatant and mitochondria-enriched fractions was loaded in SDS-PAGE and sequentially immunoblotted on the same membrane after stripping between each antibody application. The intensities of KIF5 and dynein IC74 were normalized to mitochondrial marker Tom20. Relative purity of the mitochondrial fraction was assessed by sequential immunoblotting mitochondrial marker Tom20, SNPH, and other markers including EEA1 (endosomes), Grp78 (ER), and actin. Data were collected from four paired littermates. Error bars show SEM; Mann–Whitney U test. (C) SNPH and Trak2 play opposite roles in regulating KIF5C ATPase. ATPase activity of full-length KIF5C or its motor domain (KIF5C–N-MD) was measured using Kinesin ATPase Endpoint Biochem kit and expressed as hydrolyzed ATP. MT-stimulated ATPase activity of both KIF5C (blue) and KIF5C–N-MD (green) was used as a reference value for subsequent endpoint assays by adding SNPH (or its mutant) or Trak2 under the same conditions. Incubating SNPH (1–469 aa) significantly reduces the motor ATPase activity (P = 0.029), whereas Trak2 increases the activity (P = 0.028). Deleting the KIF5 binding domain (ΔKBD) abolishes its inhibitory role. Error bars show SEM. Mann–Whitney U test, n = 4. (D) Representative time-lapse imaging showing the movement of EGFP-KIF5C along MTs labeled with mCherry–α-tubulin in a live COS cell. (E) Mean velocity of EGFP-KIF5C puncta along MTs in living cells with or without expression of SNPH, its ΔKBD mutant, or Trak2. Note that coexpressing SNPH immobilizes KIF5C puncta. Data were quantified from a total number of EGFP-KIF5C puncta (indicated in parentheses above the bars). Error bars show SEM. Student’s t test. WT, wild type; KO, knockout.
SNPH inhibits KIF5 motor ATPase activity. (A and B) Deleting snph has no effect on the motor association with axonal mitochondria. Axonal mitochondria were isolated from sciatic nerves of snph+/+ and snph−/− mice. 15 µg of total proteins from supernatant and mitochondria-enriched fractions was loaded in SDS-PAGE and sequentially immunoblotted on the same membrane after stripping between each antibody application. The intensities of KIF5 and dynein IC74 were normalized to mitochondrial marker Tom20. Relative purity of the mitochondrial fraction was assessed by sequential immunoblotting mitochondrial marker Tom20, SNPH, and other markers including EEA1 (endosomes), Grp78 (ER), and actin. Data were collected from four paired littermates. Error bars show SEM; Mann–Whitney U test. (C) SNPH and Trak2 play opposite roles in regulating KIF5C ATPase. ATPase activity of full-length KIF5C or its motor domain (KIF5C–N-MD) was measured using Kinesin ATPase Endpoint Biochem kit and expressed as hydrolyzed ATP. MT-stimulated ATPase activity of both KIF5C (blue) and KIF5C–N-MD (green) was used as a reference value for subsequent endpoint assays by adding SNPH (or its mutant) or Trak2 under the same conditions. Incubating SNPH (1–469 aa) significantly reduces the motor ATPase activity (P = 0.029), whereas Trak2 increases the activity (P = 0.028). Deleting the KIF5 binding domain (ΔKBD) abolishes its inhibitory role. Error bars show SEM. Mann–Whitney U test, n = 4. (D) Representative time-lapse imaging showing the movement of EGFP-KIF5C along MTs labeled with mCherry–α-tubulin in a live COS cell. (E) Mean velocity of EGFP-KIF5C puncta along MTs in living cells with or without expression of SNPH, its ΔKBD mutant, or Trak2. Note that coexpressing SNPH immobilizes KIF5C puncta. Data were quantified from a total number of EGFP-KIF5C puncta (indicated in parentheses above the bars). Error bars show SEM. Student’s t test. WT, wild type; KO, knockout.
KIF5 motors undergo a mechanochemical cycle by hydrolyzing ATP into ADP and Pi; the energy released from the phosphate drives motor translocation along the MT filament. The N-terminal domain of KIF5 is comprised of a motor domain with ATPase and MT-binding activity (Vale, 2003; Hirokawa et al., 2010). We next examined whether the binding of SNPH or Trak2 to the KIF5 tail could have any impact on its motor ATPase activity. First, we performed endpoint ATPase activity assays as previously described (Cho et al., 2009). Flag-tagged full-length KIF5C or its N-MD (KIF5–N-MD) was purified with anti-Flag magnetic beads and incubated with SNPH or its mutants or Trak2 in the presence or absence of MTs (Fig. 7 C). MT-stimulated ATPase activity was used as a reference value. Coincubating SNPH (1–469 aa), which contains the KBD, significantly reduces KIF5C ATPase activity (P = 0.029), whereas Trak2 plays an opposite role by increasing the motor ATPase activity (P = 0.028) under the same conditions. Deleting the KBD (ΔKBD) abolishes its inhibitory role (P = 0.147). In contrast, coincubating SNPH or Trak2 has no significant effect on ATPase activity of KIF5–N-MD, which lacks the binding domains for SNPH and Trak2. Thus, by binding to the C-terminal domain of KIF5, the docking adaptor SNPH inhibits motor activity while Trak2 stimulates the motor ATPase.
To evaluate the functional role of SNPH in inactivating KIF5 motors, we examined the velocity of KIF5 along MT tracks in live COS cells expressing EGFP-KIF5C alone or coexpressing with SNPH. A previous study characterized movements of vesicular EGFP-KIF5C structures along MTs in COS cells with a velocity similar to that measured in vivo (Dunn et al., 2008). Our live-imaging analysis showed motile EGFP-KIF5C vesicular structures along MTs labeled with mCherry–α-tubulin (Fig. 7 D). The mean velocity of the motor puncta in cells expressing KIF5C alone is 0.47 ± 0.05 µm/s. However, coexpressing SNPH, but not SNPH-ΔKBD, immobilized KIF5C puncta (0.03 ± 0.01 µm/s; P < 0.001), further supporting our hypothesis that SNPH inactivates KIF5 motor function.
SNPH is recruited to axonal mitochondria in response to neuronal activity
We then examined whether SNPH-labeled mitochondria are recruited to presynaptic boutons in response to synaptic activity. Colocalization of SNPH-labeled mitochondria with synaptic marker synaptophysin was increased after the KCl treatment (P < 0.001; Fig. 8, A and B), supporting the hypothesis that action potential firing recruits axonal mitochondria to synapses. SNPH was recruited to axonal mitochondria after a 100-Hz field stimulation. Before stimulation, 24 ± 2% of axonal mitochondria display low levels of SNPH staining (defined as SNPH/cytochrome c intensity ratio <0.5). These low levels of SNPH were only visible when the image contrast was enhanced. This staining pattern is consistent with our previous study that under regular imaging contrast settings, one third of axonal mitochondria had no detectable SNPH staining (Kang et al., 2008). Sustained neuronal activity increased SNPH intensity on axonal mitochondria (Fig. 8, C and D). These data suggest that (a) SNPH enrichment on axonal mitochondria is heterogeneous, and (b) SNPH undergoes dynamic recruitment to axonal mitochondria in response to neuronal activity although the underlying mechanism directing SNPH translocation is unclear.
SNPH is recruited to axonal mitochondria in response to neuronal activity. (A and B) Representative images (A) and quantitative analysis (B) showing the relative colocalization of SNPH-labeled axonal mitochondria with synapses. Neurons before and after the KCl treatment were fixed and coimmunostained with antibodies against SNPH and the synaptic marker synaptophysin (synapto). Note that action potential firing recruits SNPH-labeled mitochondria to synapses. Data were quantified from a total number of neurons (indicated within the bars) in three independent experiments. (C and D) Representative images (C) and quantitative analysis (D) showing increased SNPH enrichment into axonal mitochondria after 100-Hz field stimulation. Cultured hippocampal neurons were coimmunostained with anti-SNPH and anti–cytochrome c (cyto c). Arrows point to axonal mitochondria with low or no SNPH staining. Mean SNPH intensity on each mitochondrion was normalized against cytochrome c intensity. Relative SNPH enrichment on mitochondria was expressed as an intensity ratio (SNPH/cytochrome c; D, top) and the percentage of low SNPH-labeled axonal mitochondria (defined by the SNPH/cytochrome c ratio <0.5; D, bottom). Note that those tiny SNPH puncta not colabeled with mitochondria may represent shuttle cargoes delivering SNPH from axoplasm to mitochondria. Data were quantified from several axonal mitochondria (M) from the total number of neurons (N) examined in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test. (E) Proposed “Engine-Switch and Brake” model. SNPH plays a critical role in docking mitochondria via directly binding KIF5 and inhibiting motor ATPase activity. KIF5–SNPH molecular interplay is triggered by a Miro-Ca2+ sensing that releases KIF5 to bind to SNPH. Thus, SNPH turns off the “engine” (KIF5 motor) by sensing a “stop sign” (elevated Ca2+) and arrests mitochondria moving along the MT track. When in their stationary phase, mitochondria remain bound to the MT tracks, whereas KIF5 is in an inactive state.
SNPH is recruited to axonal mitochondria in response to neuronal activity. (A and B) Representative images (A) and quantitative analysis (B) showing the relative colocalization of SNPH-labeled axonal mitochondria with synapses. Neurons before and after the KCl treatment were fixed and coimmunostained with antibodies against SNPH and the synaptic marker synaptophysin (synapto). Note that action potential firing recruits SNPH-labeled mitochondria to synapses. Data were quantified from a total number of neurons (indicated within the bars) in three independent experiments. (C and D) Representative images (C) and quantitative analysis (D) showing increased SNPH enrichment into axonal mitochondria after 100-Hz field stimulation. Cultured hippocampal neurons were coimmunostained with anti-SNPH and anti–cytochrome c (cyto c). Arrows point to axonal mitochondria with low or no SNPH staining. Mean SNPH intensity on each mitochondrion was normalized against cytochrome c intensity. Relative SNPH enrichment on mitochondria was expressed as an intensity ratio (SNPH/cytochrome c; D, top) and the percentage of low SNPH-labeled axonal mitochondria (defined by the SNPH/cytochrome c ratio <0.5; D, bottom). Note that those tiny SNPH puncta not colabeled with mitochondria may represent shuttle cargoes delivering SNPH from axoplasm to mitochondria. Data were quantified from several axonal mitochondria (M) from the total number of neurons (N) examined in three independent experiments. Bars, 10 µm. Error bars show SEM. Student’s t test. (E) Proposed “Engine-Switch and Brake” model. SNPH plays a critical role in docking mitochondria via directly binding KIF5 and inhibiting motor ATPase activity. KIF5–SNPH molecular interplay is triggered by a Miro-Ca2+ sensing that releases KIF5 to bind to SNPH. Thus, SNPH turns off the “engine” (KIF5 motor) by sensing a “stop sign” (elevated Ca2+) and arrests mitochondria moving along the MT track. When in their stationary phase, mitochondria remain bound to the MT tracks, whereas KIF5 is in an inactive state.
Discussion
KIF5–SNPH coupling turns off “engine switch” to immobilize mitochondria
Recent studies characterized Miro’s role as a Ca2+ sensor for activity-dependent regulation of mitochondrial transport (Saotome et al., 2008; MacAskill et al., 2009b; Wang and Schwarz, 2009; Sheng and Cai, 2012). In the current study, we demonstrate that SNPH mediates the activity-dependent immobilization of mitochondria in axons. The SNPH–KIF5 coupling is triggered by a Miro-Ca2+ sensor, which releases the C-terminal tail of KIF5 to bind SNPH, resulting in inhibition of the motor ATPase. Thus, SNPH arrests axonal mitochondrial movement in response to synaptic activity by (a) interfering with the motor transport complex and (b) inactivating the motor ATPase. Deleting the snph gene abolishes the activity-induced immobilization of mitochondria in axons but not in dendrites. Here, we propose a new “Engine-Switch and Brake” model (Fig. 8 E), in which SNPH acts both as an engine off switch by sensing stop sign-Ca2+ and as a brake by anchoring mitochondria to the MT track. When in their stationary phase, both the KIF5 motor and mitochondria remain bound to the MT track.
Our model nicely reconciles the standing dispute regarding how Miro-Ca2+ sensing arrests mitochondrial transport. MacAskill et al. (2009b) suggested that Ca2+ binding to Miro releases KIF5 motors from mitochondria. Glutamate application led to a 45% decrease in the amount of KIF5 motors bound to mitochondria. Because glutamate acts predominantly on dendrites where SNPH is absent, glutamate evokes immobilization of dendritic mitochondria likely via a Ca2+-dependent dissociation of KIF5 from mitochondria. Alternatively, Wang and Schwarz (2009) found that in axons, KIF5 remained associated with stationary and motile mitochondria at both resting and elevated Ca2+ conditions. These discrepant findings may account for their selective observations of mitochondrial motility in axons versus dendrites. SNPH is an axonal mitochondrial outer membrane protein (Kang et al., 2008). Thus, upon Miro-Ca2+ sensing, KIF5 motors remain on axonal but not dendritic mitochondria, likely through its coupling to SNPH. Our findings suggest that motor–cargo association is insufficient for the motor-driven transport, consistent with a recent in vivo study on prion protein vesicles, which showed that both KIF5 and dynein motors remain bound to organelles regardless of whether they were moving or stationary (Encalada et al., 2011).
One third of axonal mitochondria displayed low levels or undetectable SNPH (Kang et al., 2008). Sustained neuronal activity may recruit more SNPH to axonal mitochondria. Synaptic localization of mitochondria in snph−/− hippocampal neurons was significantly lower (P < 0.001). Thus, during prolonged stimulation in the snph mutant neurons, short-term facilitation is enhanced as a result of impaired [Ca2+] buffering at presynaptic terminals. This phenotype is fully rescued by reintroducing the snph gene into the mutant neurons (Kang et al., 2008). One of remaining questions is how stationary mitochondria move again. The sequence features of SNPH, 12% residues as serine or threonine and numerous phosphorylation sites, suggest that its role as a brake could be turned off through signal transduction pathways. Given that SNPH is a cellular target of the E3 ubiquitin ligase Cullin 1 (Yen and Elledge, 2008), mitochondria-targeted SNPH may also be terminated by ubiquitin-mediated degradation pathways.
SNPH arrests bidirectional transport of axonal mitochondria
The Miro-Ca2+ sensing pathway equally affects both anterograde and retrograde mitochondrial transport (Cai and Sheng, 2009; MacAskill et al., 2009b; Wang and Schwarz, 2009), leaving a question as how the KIF5 adaptor complex affects dynein-driven retrograde transport. Dynein has been observed colocalizing with mitochondria moving in both the anterograde and retrograde directions (Hirokawa et al., 1990). Inhibiting kinesin-1 in Drosophila also reduces retrograde mitochondrial movement (Pilling et al., 2006). Thus, the kinesin and dynein motors may coordinate bidirectional transport rather than simply competing against one another (Ligon et al., 2004). Any change in this coordination may alter the motor processivity, time spent in stationary phases, run lengths, or pause frequencies (Gross et al., 2007; Encalada et al., 2011). Miro is one candidate for regulating bidirectional transport. dMiro in Drosophila regulates both anterograde and retrograde mitochondrial transport (Russo et al., 2009). Mitochondrial fusion proteins mitofusins (Mfn1 and Mfn2) interact with Miro, and this interaction is necessary to regulate axonal mitochondrial transport. Deleting Mfn2 or expressing Mfn2 mutants results in axonal mitochondria spending more time paused and undergoing slower anterograde and retrograde transport (Misko et al., 2010).
Identifying SNPH as a motor “brake” provides new mechanistic insights into how Miro-Ca2+ sensing arrests mitochondrial bidirectional movement. Miro-Trak2 and SNPH can displace one another when binding to the KIF5 motor, and SNPH–KIF5 coupling inhibits the motor ATPase. Deleting snph impairs the motor brake, thus mitochondria are unable to be immobilized upon the Miro-Ca2+ sensing. Given the fact that no native SNPH–dynein complex is detected in mouse brains, SNPH may affect retrograde transport through inhibiting KIF5 motor. Our model is similar to a general “Molecular Brakes” model (Gross et al., 2007), in which the cargo-MT linkage via specific “coupling proteins” could act as molecular brakes to stop bidirectional movement.
SNPH-mediated mitochondrial immobilization and synaptic transmission
Mitochondria maintain synaptic transmission by producing ATP and maintaining Ca2+ homeostasis at synapses (Tang and Zucker, 1997; Kang et al., 2008). SNPH was originally isolated through a yeast two-hybrid screening using the coiled-coil domain of syntaxin-1A as bait (Lao et al., 2000). SNPH coiled-coil domain (130–203 aa) shares ∼80% structural similarity to the syntaxin coiled-coil domain. Earlier studies showed that SNPH plays a role in reducing synaptic transmission after acute overexpression (6–12 h) of recombinant SNPH in autaptic hippocampal neurons through Semliki Forest virus infection or after microinjection of highly concentrated coiled-coil domain (50 mg) into superior cervical ganglion neurons (Lao et al., 2000). It is likely that there is a competitive interference between the SNPH coiled-coil domain and the SNARE machinery. We further characterized the snph knockout mice combined with gene rescue experiments and found that deleting snph had no significant impact on mini-AMPA events and basal synaptic transmission. However, deleting snph results in increased presynaptic [Ca2+] levels during prolonged high-frequency stimulation (Kang et al., 2008). This phenotype is attributed to fewer mitochondria being tethered at terminals, thus reducing the Ca2+ buffering capacity. In the same snph−/− neurons, dendritic mitochondria are immobilized in response to neuronal activity as efficiently as in the wild-type neurons. Therefore, the defects in activity-dependent immobilization of axonal mitochondria are less likely a result of adaptation of the mutant neurons to Ca2+ influx, lack of appropriate receptors, or downstream signaling molecules.
Defective mitochondrial trafficking was implicated in the pathological processes of major neurodegenerative diseases (Sheng and Cai, 2012). Selective mitochondrial degradation through mitophagy plays an important role in the quality control of mitochondria (Chen and Chan, 2009; Cai et al., 2012). Whether and how altered mitochondrial motility impacts the turnover of dysfunctional mitochondria in axons and at terminals are unclear. Our current findings provide molecular targets for future studies investigating into how disease-related mutations impair mitochondrial transport and docking machineries. These studies may advance our understanding of human neurodegenerative disorders.
Materials and methods
Mouse line
The snph−/− mouse line was generated by targeted gene replacement in embryonic stem (ES) cells as previously described (Kang et al., 2008). All four coding exons of the snph gene were replaced by the selection cassette phosphoglycerate kinase (PGK) neo (neomycin resistance gene under the PGK promoter). The herpes simplex virus-1 thymidine kinase gene under the PGK promoter (thymidine kinase cassette) was placed at the end of the long arm of the construct. The vector was linearized and electroporated into ES cell line TC1 (derived from a 129/SvEvTacfBR mouse) and then selected with G418 and fialuridine. Homologous recombination events were identified by Southern blot screening using a PCR-generated probe. ES cells heterozygous for the targeted mutation were independently microinjected into C57BL/6J blastocysts, generating chimeric mice that transmitted the mutation through the germline (germline transmission). Heterozygous and knockout mice were generated and identified by Southern blotting and PCR amplification. Both homozygote and heterozygote mutant animals were viable, fertile, and morphologically normal. Animal care and use were performed in accordance with National Institutes of Health guidelines and were approved by the National Institutes of Health National Institute of Neurological Disorders and Stroke/National Institute on Deafness and Other Communication Disorders Animal Care and Use Committee.
DNA constructs
The following DNA constructs were gifts: pRK5Myc-Miro1 obtained from P. Aspenström (Karolinska Institutet, Stockholm, Sweden); pEGFP-KIF5C obtained from M. Peckham (University of Leeds, Leeds, England, UK); pCMVFlag-Trak2 obtained from F.A. Stephenson (University College London, London, England, UK); and pET-28a-KHC truncated mutants obtained from R.J. Diefenbach (The University of Sydney and Westmead Hospital, Westmead, Australia). Various truncated SNPH mutants were generated by PCR and subcloned into pEGFP-C and pGEX-4T vectors (GE Healthcare). An SNPH (381–466 aa) deletion was introduced by mutagenesis (QuikChange; Agilent Technologies). The full-length coding sequences of Miro1 and Trak2 were cloned by PCR and inserted into the pEGFP-C vector. pEYFP-tubulin containing full-length human α-tubulin was purchased from Takara Bio Inc. and subcloned into pmCherry-C1 vectors.
Antibodies and reagents
The purified rabbit polyclonal antibody against SNPH residues 225–428 was described previously (Kang et al., 2008). Sources of other antibodies or reagents are as follows: monoclonal anti-KIF5 (heavy chain) and anti–dynein IC74 antibodies (EMD Millipore); polyclonal anti-Flag, monoclonal anti-actin, and anti–β-tubulin (Sigma-Aldrich); monoclonal anti–cytochrome c (BD); polyclonal anti-EEA1, anti-TOM20, anti-KLC, anti-DHC, and monoclonal anti-synaptophysin (Santa Cruz Biotechnology, Inc.); monoclonal anti-GFP (JL-8; Takara Bio Inc.); ECL-HRP–linked secondary antibodies (GE Healthcare); Alexa fluor 546– or 633–conjugated secondary antibodies (Invitrogen); Pepstatin A (EMD Millipore); and leupeptin (Sigma-Aldrich).
Hippocampal neuronal culture and transfection
Hippocampi were dissected from postnatal day 1 mouse or rat pups and cultured as previously described (Ilangovan et al., 2005). In brief, after dissociation with papain (Worthington), hippocampal cells recovered by centrifugation were plated onto 12-mm coverslips in 2 ml of plating medium in 35-mm dishes (for 10 ml of plating medium: 9.5 ml Neurobasal medium [Invitrogen], 0.5 mM glutamine, 0.25 mg insulin, 5% fetal bovine serum, 2% B-27 medium supplement, and G-5 supplement). Beginning on the second day in culture, half of the medium was replaced with feeding medium twice a week (10 ml of feeding medium: 9.8 ml Neurobasal medium and 2% B-27 medium supplement). To label mitochondria and early and late endosomes, DsRed-Mito and YFP-Rab7 or GFP-Rab5 constructs were cotransfected into neurons at DIV9 using the calcium phosphate method (Jiang and Chen, 2006). In brief, the DNA-Ca2+ and phosphate solutions were gently mixed and incubated at room temperature for 20 min. After incubation with cultured neurons on the coverslip for 45 min, the coverslip was washed with prewarmed DMEM and transferred to a new dish containing original culture medium followed by time-lapse imaging in live neurons 3–4 d after transfection.
COS and HEK293 cell cultures and transfection
COS and HEK293 cells were cultured in high-glucose DMEM containing sodium pyruvate and l-glutamine supplemented with 10% FBS and penicillin–streptomycin (Invitrogen) until the cells became confluent. Transient transfection was performed using Lipofectamine 2000 (Invitrogen). After transfection, cells were cultured for an additional 1–2 d before immunocytochemistry or harvesting for biochemical analysis.
Immunocytochemistry
Cultured cells were fixed with 4% paraformaldehyde and 4% sucrose at RT for 15 min, washed three times with PBS for 5 min each, permeabilized in 0.1% Triton X-100 for 15 min, and then incubated in 5% normal goat serum in PBS for 1 h. Fixed cells were incubated with primary antibodies in PBS with 1% BSA at 4°C overnight or at RT for 4 h. Cells were washed four times with PBS at RT for 5 min each, incubated with secondary fluorescent antibodies in PBS with 1% BSA for 30 min, rewashed with PBS, and mounted with antifade mounting medium (Fluoro-Gel; Electron Microscopy Sciences) for imaging.
Recombinant protein preparation
GST-tagged proteins were expressed in Escherichia coli BL21 (DE3). The BL21 cells were grown to an OD600 of 0.6; expressed protein was induced with IPTG to a final concentration of 0.5 mM at 30°C for 4–6 h. The cell pellet was resuspended in 20 mM phosphate buffer, pH 7.6, containing 150 mM NaCl, 0.5 mM DTT, and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, and 3 µg/ml each of leupeptin and pepstatin). After sonication and centrifugation, the crude extract was purified using GST–Sepharose beads (GE Healthcare). Expression of His-KIF5C and its truncated fragments were purified with Ni–nitrilotriacetic acid beads (QIAGEN) according to the manufacturer’s instructions.
GST pull-down assay
Rat brains were homogenized in buffer A (25 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.5% Triton X-100, and protease inhibitors including 1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, and 3 µg/ml each of leupeptin and pepstatin). The crude homogenate was centrifuged at 15,000 g for 40 min; the supernatant was carefully collected. GST beads coated with 4 µg GST, GST-SNPH, or its truncated mutants were mixed with the brain homogenates and incubated for 3 h with gentle rotation. The beads were then extensively washed with buffer B (25 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.1% Nonidet P-40, and protease inhibitors). The beads were dissolved in SDS-PAGE sample buffer and heated at 95°C for 10 min. Proteins were resolved by SDS-PAGE and processed for Western blot analysis. For multiple detections with different antibodies on the same membranes, blots were stripped in a solution of 62.5 mM Tris-HCl, pH 7.5, 20 mM DTT, and 1% SDS for 20 min at 50°C with agitation and washed twice with TBS/0.1% Tween 20 for 15 min each time.
In vitro binding assay and coimmunoprecipitation
10 µl glutathione–Sepharose resin were added to 4 µg of GST-SNPH or its truncated and deleted mutants; the mixture was incubated on ice for 30 min before adding 8 µg purified His-KIF5C. The mixture was incubated on ice for another 1–3 h. The resin was then washed four times. Bound protein was eluted from the resin with 30 µl of 2× SDS sample buffer and heated at 95°C for 10 min. After brief centrifugation, the supernatant was subjected to SDS-PAGE followed by Western blot analysis.
Mouse brains or HEK293 cells or cultured cortical neurons were homogenized in TBS with 1% Triton X-100 and protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 0.1 mM benzamidine, and 3 µg/ml each of leupeptin and pepstatin). The crude lysates were centrifuged at 13,000 g for 30 min, and the supernatant was collected. The lysates were incubated with 3 µg antibody or preimmune serum in 0.5 ml TBS with 0.1% Triton X-100 and protease inhibitors and incubated on a rotator at 4°C for 2 h. Protein A–Sepharose CL-4B resin (GE Healthcare) was added to each sample; incubation continued for an additional 3 h followed by three washes with TBS/0.1% Triton X-100. The beads were dissolved in SDS-PAGE sample buffer and heated at 95°C for 10 min. The proteins were resolved by SDS-PAGE and processed for Western blot analysis. For semiquantitative analysis, protein bands detected by ECL were scanned into Photoshop CS3 (Adobe) and analyzed using protein imaging software (Gel-Pro 4.5; Media Cybernetics). Care was taken during exposure of the ECL film to ensure that intensity readouts were in a linear range of standard curve blot detected by the same antibody.
Analysis of mitochondrial motility and imaging data acquisition
Both axonal and dendritic processes were selected for mitochondrial motility analysis. As we previously reported (Kang et al., 2008; Cai et al., 2010), axons in live images were distinguished from dendrites based on known morphological characteristics: greater length, thin and uniform diameter, and sparse branching (Banker and Cowan, 1979). In addition, axonal mitochondria are more sparsely distributed along axons and display distinct morphology patterns as small, vesicular, or short vesicular–tubular structures. In contrast, dendritic mitochondria are elongated, tubular, or filamentous structures occupying a large fraction of dendritic processes (Popov et al., 2005; Chang et al., 2006). We selected the proximal region of axons and dendrites for time-lapse imaging analysis. Only those that appeared to be single axons and separate from other processes in the field were chosen for recording axonal mitochondrial transport. Regions where crossing or fasciculation occurred were excluded from analysis. Axonal processes were further confirmed by retrospective immunostaining with an anti-MAP2 antibody.
Time-lapse imaging was performed at 37°C using a confocal microscope (LSM 510 META; Carl Zeiss) with a C-Apochromat 40×/1.2 NA water Corr objective (Carl Zeiss) and a perfusion system (0.4 ml/min) with modified Tyrode’s solution (10 mM Hepes, 10 mM glucose, 3 mM KCl, 145 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, and 1 µM glycine, pH 7.4). Time-lapse images were collected at a 512 × 512–pixel resolution (8 bit). A relatively long interval (10 s) for a total of 100 images was used to minimize laser-induced cellular damage. Kymographs were made with extra plug-ins for ImageJ (National Institutes of Health) as previously described (Kang et al., 2008). In brief, we used plug-ins “LSM_Reader” to read Carl Zeiss LSM images, “Straighten” to straighten curved axons, “Grouped_ZProjector” to z-axially project resliced time-lapse images, “Time_Stamper” to make time-stamped images/frames, and “QT_Movie_Writer” to make the video files as MOV format. The height of the kymographs represents recording time, and the width of the kymographs represents length (micrometers) of the axon imaged. A mitochondrion was considered stationary if it remained immotile for the entire recording period; a motile one was counted only if the displacement was ≥5 µm.
Electric field stimulation
During time-lapse imaging, cultured hippocampal neurons transfected with DsRed-Mito were treated with 100-Hz field stimulation, which was generated with a stimulator (Master 8; A.M.P.I.) and amplified with stimulus isolation (A385; World Precision Instruments). 200 biphasic pulses were delivered through platinum electrodes positioned on opposite sides of the field stimulation chamber (Warner Instruments) at 1 ms of stimulus duration and 50 mA of constant current. 1.2 mM CaCl2 was removed from Tyrode’s solution for control experiments in the absence of extracellular calcium. 1 µm TTX was added to Tyrode’s solution for the action potential blocking experiment.
Preparation of axonal mitochondria-enriched membrane fractions
Axonal mitochondria were prepared from sciatic nerves of 9-month-old snph+/+ and snph−/− mice as previously described (Frezza et al., 2007). Mouse sciatic nerves were homogenized in mitochondria isolation buffer (10 mM Tris-MOPS, 1 mM EGTA, and 20 mM sucrose plus protease inhibitors, pH 7.4) centrifuged at 600 g for 10 min at 4°C. The supernatants were collected and further centrifuged at 7,000 g for 10 min at 4°C to pellet mitochondria. The pellet was washed in isolation buffer. Protein concentration was determined by the bicinchoninic acid protein assay (Thermo Fisher Scientific). Samples were subjected to SDS-PAGE followed by immunoblot analysis. The intensities of KIF5 and dynein IC74 were normalized to the mitochondrial marker Tom20. Relative purity of the mitochondrial fraction was assessed by sequentially immunoblotting the same membrane with mitochondrial marker TOM20 and other organelle markers, including EEA1 (endosomes), Grp78 (ER), and actin.
Kinesin ATPase activity assay and motor velocity measurement
Flag-KIF5C, Flag-SNPH and its truncated mutants, and Flag-Trak2 were expressed in HEK293 cells and then purified using anti-Flag M2 magnetic beads (Sigma-Aldrich). The ATPase activity of KIF5C motor was measured by the Kinesin ATPase Endpoint Assay Biochem kit (Cytoskeleton). In brief, 0.2 µg of purified Flag-KIF5C was incubated in the presence or absence of 0.5 µg Flag-SNPH or Flag-Trak2 in 30 µl of reaction buffer (15 mM Pipes, 5 mM MgCl2, 20 µM Taxol, and 20 µM acetyl–coenzyme A) for 30 min at room temperature. After adding 2 µg MTs and 0.3 mM ATP to initiate the reaction, the reaction continued for 5 min before termination by adding 70 µl CytoPhos for 10 min. The OD650 was measured by absorbance microplate reader (SpectraMax 190; Molecular Devices). For KIF5 velocity measurement, COS cells were transfected with EGFP-KIF5C and mCherry–α-tubulin or coexpressing with SNPH or Trak2 for 24 h. Time-lapse imaging was performed using a confocal microscope (LSM 510 META) with a C-Apochromat 63×, 1.4 NA oil immersion lens in modified Tyrode’s solution (10 mM Hepes, 10 mM glucose, 3 mM KCl, 145 mM NaCl, 1.2 mM CaCl2, 1.2 mM MgCl2, and 1 µM glycine, pH 7.4). Time-lapse images were collected at 256 × 256–pixel resolution (8 bit) with a 2.5-s interval. The software RETRAC (Carter and Cross, 2001) was used to measure the velocity of EGFP-KIF5C by tracking the EGFP-KIF5C puncta along MTs labeled with mCherry–α-tubulin.
Statistical analysis
Images are representative of >10 cells as indicated from each figure legend from at least three repeats. All immunoblots are representative of at least three experiments. For significance evaluation, two-tailed Student’s t test was performed for normal distributed datasets. If the dataset did not follow normal distribution, nonparametric Mann–Whitney U test was performed for two groups by Prism 5 (GraphPad Software). Measurements are presented as means ± SEM.
Online supplemental material
Fig. S1 shows that SNPH selectively immobilizes axonal mitochondria. Fig. S2 shows axonal mitochondria targeting of Miro1/Trak2. Fig. S3 shows that there is no binding between SNPH and DHC. Fig. S4 shows a characterization of the SNPH–KIF5 interaction. Video 1 shows activity-dependent immobilization of axonal mitochondria, but not late endosomes, in snph+/+ neurons. Video 2 shows that deleting snph abolishes activity-dependent immobilization of axonal mitochondria. Video 3 shows that overexpressing SNPH selectively immobilizes axonal mitochondria, but not late endosome, along the same axon. Video 4 shows immobilization of axonal mitochondria by applying field stimulation to wild-type neurons. Video 5 shows that deleting snph abolishes field stimulation–induced immobilization of axonal mitochondria. Video 6 shows that expressing Trak2 increases axonal mitochondrial transport. Video 7 shows that SNPH competes with Trak2 and abolishes axonal mitochondrial motility. Video 8 shows that disrupting the KIF5–SNPH interaction reduces activity-dependent immobilization of axonal mitochondria. Video 9 shows that Ca2+ depletion enhances the axonal mitochondrial motility.
Activity-dependent immobilization of axonal mitochondria, but not late endosomes, in snph+/+ neurons. Representative time-lapse images showing the relative motility of both mitochondria (red) and late endosomes (green) along the same axons of snph+/+ neurons before and after brief treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Hippocampal neurons were cotransfected with DsRed-Mito and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12 (8 min each before and after treatment). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. Note that the brief treatment effectively reduces both anterograde and retrograde transport of axonal mitochondria but not of late endosomes. The kymograph of this video is presented in Fig. 1 A.
Activity-dependent immobilization of axonal mitochondria, but not late endosomes, in snph+/+ neurons. Representative time-lapse images showing the relative motility of both mitochondria (red) and late endosomes (green) along the same axons of snph+/+ neurons before and after brief treatment (2 min) with 50 mM KCl and 10 µM FPL 64176. Hippocampal neurons were cotransfected with DsRed-Mito and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12 (8 min each before and after treatment). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. Note that the brief treatment effectively reduces both anterograde and retrograde transport of axonal mitochondria but not of late endosomes. The kymograph of this video is presented in Fig. 1 A.
Deleting snph abolishes activity-dependent immobilization of axonal mitochondria. Representative time-lapse images showing the relative motility of both mitochondria (red) and late endosomes (green) along the same axons of snph−/− neurons before and after brief treatment with 50 mM KCl and 10 µM FPL 64176. Hippocampal snph−/− neurons were cotransfected with DsRed-Mito and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12 (8 min each before and after treatment). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 1 B.
Deleting snph abolishes activity-dependent immobilization of axonal mitochondria. Representative time-lapse images showing the relative motility of both mitochondria (red) and late endosomes (green) along the same axons of snph−/− neurons before and after brief treatment with 50 mM KCl and 10 µM FPL 64176. Hippocampal snph−/− neurons were cotransfected with DsRed-Mito and GFP-Rab7 at DIV9 followed by ∼16-min time-lapse imaging at DIV12 (8 min each before and after treatment). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 1 B.
Immobilization of axonal mitochondria by applying field stimulation to wild-type neurons. Representative time-lapse images showing axonal mitochondrial transport in snph+/+ hippocampal neurons during ∼16 min of observations (8 min each before and after stimulation). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied during the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 2 A.
Immobilization of axonal mitochondria by applying field stimulation to wild-type neurons. Representative time-lapse images showing axonal mitochondrial transport in snph+/+ hippocampal neurons during ∼16 min of observations (8 min each before and after stimulation). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied during the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 2 A.
Deleting snph abolishes field stimulation–induced immobilization of axonal mitochondria. Representative time-lapse images showing axonal mitochondria transport in snph−/− hippocampal neurons during ∼16 min of observations (8 min each before and after stimulation). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied while imaging the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 2 A.
Deleting snph abolishes field stimulation–induced immobilization of axonal mitochondria. Representative time-lapse images showing axonal mitochondria transport in snph−/− hippocampal neurons during ∼16 min of observations (8 min each before and after stimulation). Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). The 100-Hz (2 s) field stimulation was applied while imaging the 50th frame, as indicated by the flash. Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 2 A.
Expressing Trak2 increases axonal mitochondrial transport. Representative time-lapse images showing increased axonal mitochondria transport in hippocampal neurons expressing Flag-Trak2 during ∼16 min of observations. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 3 A.
Expressing Trak2 increases axonal mitochondrial transport. Representative time-lapse images showing increased axonal mitochondria transport in hippocampal neurons expressing Flag-Trak2 during ∼16 min of observations. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 3 A.
SNPH competes with Trak2 and abolishes axonal mitochondrial motility. Representative time-lapse images showing axonal mitochondrial transport in hippocampal neurons coexpressing Flag-Trak2 and SNPH during ∼16 min of observations. Note that coexpressing SNPH dominantly immobilizes axonal mitochondria. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 3 A.
SNPH competes with Trak2 and abolishes axonal mitochondrial motility. Representative time-lapse images showing axonal mitochondrial transport in hippocampal neurons coexpressing Flag-Trak2 and SNPH during ∼16 min of observations. Note that coexpressing SNPH dominantly immobilizes axonal mitochondria. Time-lapse sequences of 100 frames were collected at 10-s intervals using a laser-scanning confocal microscope (LSM 510 META). Movement toward the bottom indicates retrograde transport. The kymograph of this video is presented in Fig. 3 A.
Acknowledgments
We thank Q. Cai and other members of the Sheng laboratory for helpful discussions; M. Davis for editing; T.L. Schwarz, J.T. Kittler, P. Aspenström, M. Peckham, F.A. Stephenson, R.J. Youle, and R.J. Diefenbach for DNA constructs and reagents; and the National Institute of Neurological Disorders and Stroke (NINDS) Facility for DNA Sequencing and Protein Sequencing.
This work was supported by the Intramural Research Program of NINDS, National Institutes of Health (Z.-H. Sheng) and an NINDS Competitive Fellowship (Y. Chen).


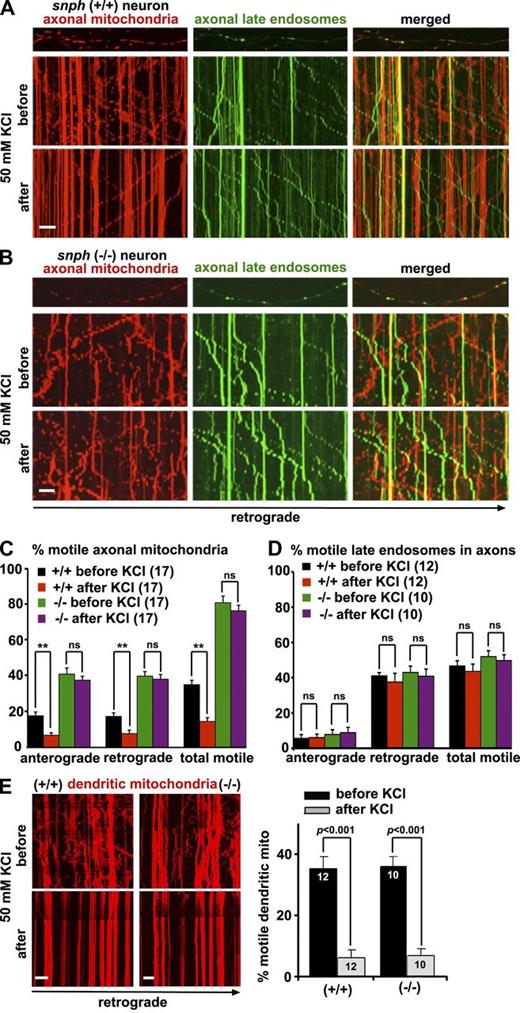
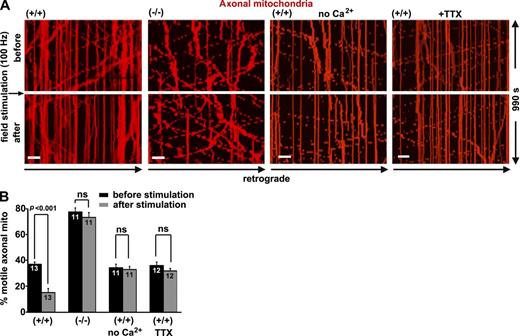



![Figure 6. SNPH competes with Trak2 to bind KIF5. (A and B) Representative blots (A) and quantitative analysis (B) showing competition of SNPH and Trak2 for binding KIF5. HEK293 cells were cotransfected as indicated. The KIF5C–Trak2 complex was coimmunoprecipitated (IP) from cell lysates with an anti-Flag antibody and immunoblotted (IB) with an anti-GFP antibody (bottom). Note that SNPH, but not SNPH-ΔKBD, inhibits the formation of the KIF5C–Trak2 complex (red box). Data were quantified from three independent experiments. The asterisk denotes degraded GFP-SNPH. (C and D) SNPH, but not SNPH-ΔKBD, competes with Trak2 to immobilize axonal mitochondria. (E and F) Immunoprecipitation (E) and quantitative analysis (F) showing activity-induced and Ca2+-dependent formation of the native SNPH–KHC complex in mature cortical neurons. The cultured neurons (DIV14) were treated for 10 min with 50 mM KCl and 10 µM FPL 64176 in modified Tyrode’s solution with or without 1.2 mM Ca2+. The native SNPH–KHC complexes were coimmunoprecipitated from neuronal lysates with an anti-SNPH antibody and subsequently immunoblotted with an anti-KHC and LC8 antibody. Coimmunoprecipitated levels of KIF5 and LC8 after stimulation were normalized against their levels before stimulation, respectively. Note that removing [Ca2+]ext from the medium abolished the activity-induced SNPH–KIF5 interaction. (G and H) Kymographs (G) and quantitative analysis (H) showing enhanced motility of axonal mitochondria after Ca2+ depletion. Mitochondrial motility in hippocampal neurons were recorded before and 20 min after removing 1.2 mM [Ca2+]ext from the medium on the same axons (also see Video 9). Bars, 10 µm. Data in D and H were quantified from numbers of axons (indicated in bars). Student’s t test in D and H and Mann–Whitney U test in B and F. Error bars show SEM.](https://cdn.rupress.org/rup/content_public/journal/jcb/202/2/10.1083_jcb.201302040/6/m_jcb_201302040_fig6.jpeg?Expires=1773382444&Signature=jMRUPcPmh51XEoDkEoiQOfaLP4~k8Jr6F58XHcPurud~1i1eMIu3QNoeLSMmTHTByynaI6Y81KUeTaJZOGSKiu-in671H4YU2ipZRfoXMUz~TuI-Ljl~v0lnjASpe50fQEw8hNXopCkEy3K5eVL9Q3zhXbPiy3lQZCfjmM3Kn5qWBXc5OlCAUUv6M1Y9ipYPiKsPygkta~HsaXqjW5CtScnF~itYxDbaSEizxKgrezuQUHU7qSimcGPEcaE-IFGsM4a6Z70eqgK~W8F~av9TMxdIYLtv3Ujmb53oGLpv2qD~e6QaR8ZAX0ETYCG7wZFalfToAvRY3HqKzaefKkjoRw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




