Inhibition of αvβ3 integrin or expression of oncogenic mutants of p53 promote invasive cell migration by enhancing endosomal recycling of α5β1 integrin under control of the Rab11 effector Rab-coupling protein (RCP). In this paper, we show that diacylglycerol kinase α (DGK-α), which phosphorylates diacylglycerol to phosphatidic acid (PA), was required for RCP to be mobilized to and tethered at the tips of invasive pseudopods and to allow RCP-dependent α5β1 recycling and the resulting invasiveness of tumor cells. Expression of a constitutive-active mutant of DGK-α drove RCP-dependent invasion in the absence of mutant p53 expression or αvβ3 inhibition, and conversely, an RCP mutant lacking the PA-binding C2 domain was not capable of being tethered at pseudopod tips. These data demonstrate that generation of PA downstream of DGK-α is essential to connect expression of mutant p53s or inhibition of αvβ3 to RCP and for this Rab11 effector to drive the trafficking of α5β1 that is required for tumor cell invasion through three-dimensional matrices.
Introduction
The poor clinical outcome of many cancers is caused by dissemination of metastatic tumor cells and the outgrowth of secondary tumors at distant sites. To metastasize, cancer cells must cross ECM barriers, such as basement membranes and the stromal tissue that surrounds tumors, as well as acquire the ability to extravasate and insinuate themselves into their metastatic target organs (Sahai, 2005; Rowe and Weiss, 2009). Many of these steps to metastasis require cancer cells to acquire particular migratory characteristics, and this issue is now becoming a major focus for researchers keen to understand how cancer progresses.
A cell’s migratory properties depend on the way in which it interacts with and responds to the surrounding ECM, and much of this is dictated by the integrin family of ECM receptors. Integrins are heterodimeric transmembrane receptors that not only physically link the intracellular actin cytoskeleton to the ECM but are also signaling molecules that transduce signals bidirectionally across the plasma membrane (Hynes, 2002). There are several ways in which a cell can control integrin behavior. For instance, FERM domain–containing proteins, such as talin and kindlin, can be recruited to integrin cytotails to activate their ECM ligand-binding capacity (Moser et al., 2009). Moreover, surface integrins are continuously endocytosed and then returned (or recycled) back to the plasma membrane, and it is now apparent that the way in which integrins are trafficked through the endosomal pathway is key to how they function (Caswell and Norman, 2006, 2008; Pellinen and Ivaska, 2006; Ramsay et al., 2007; Caswell et al., 2009). There is a reciprocal relationship between the trafficking of αvβ3 and α5β1 integrins such that when αvβ3 cycling or ligand engagement is compromised, recycling of α5β1 integrin is strongly promoted (White et al., 2007; Caswell et al., 2008). Furthermore, it is now clear that oncogenic mutant forms of p53 strongly promote α5β1 recycling and that this is achieved via mutant p53’s ability to inhibit p63 function (Muller et al., 2009). Importantly, whether achieved by αvβ3 inhibition or by expression of mutant p53s, the migratory consequences of increased α5β1 recycling depend on the ECM environment. Thus, when cells are plated onto 2D surfaces, increased α5β1 recycling causes cells to switch from directional to random migration (White et al., 2007; Caswell et al., 2008). On the other hand, when tumor cells are in 3D microenvironments, activated α5β1 recycling promotes the extension of invasive pseudopodial structures, leading to increased invasiveness of the type associated with metastatic cancers (Caswell et al., 2008; Muller et al., 2009).
Small GTPases of the Rab11 family, including Rab11a and Rab25, are known to regulate α5β1 recycling (Roberts et al., 2001; Caswell et al., 2007). The Rab11 family–binding proteins, known as the Rab11–family-interacting proteins (FIPs), are key to Rab11 function (Prekeris, 2003; Horgan and McCaffrey, 2009), and recently, we have established that one of these, Rab-coupling protein (RCP), is required to link expression of mutant p53 (and inhibition of αvβ3) to increased α5β1 recycling (Caswell et al., 2008; Muller et al., 2009). Furthermore, the migratory consequences of increased α5β1 recycling, such as increased invasiveness and the acquisition of random migration, depend on RCP and its ability to recruit α5β1. Consistent with this, RCP has recently been identified to be located within a genomic region (8p11-12) that is frequently amplified in breast cancer and to contribute to the progression of certain forms of this disease (Zhang et al., 2009). The class I Rab11-FIPs (Rip11, RCP, and Rab11-FIP2) contain a C2 domain at the N-terminal end of the protein, and this has been shown to bind to the acidic phospholipids, phosphatidic acid (PA) and phosphatidylinositol 3,4,5-trisphosphate (Lindsay and McCaffrey, 2004). Moreover, there are indications that PA synthesis may be required for translocation of RCP from perinuclear endosomes to a subplasmalemmal location after treatment of cells with phorbol esters (Lindsay and McCaffrey, 2004).
PA serves as an important second messenger that can be found at various locations within the cell, including the plasma membrane, Golgi, and endosomes, and PA can be produced in cells by two different enzyme families, diacylglycerol kinases (DGKs) and PLDs (Jenkins and Frohman, 2005; Mérida et al., 2008). DGKs phosphorylate DAG to yield PA, thereby regulating the levels of both these lipid second messengers in a reciprocal manner. Thus, DGKs act both as terminators of DAG-mediated signals as well as activators of PA-mediated ones. DGKs comprise a family of 10 distinct enzymes, grouped in five classes each featuring a highly conserved catalytic domain preceded by two cysteine-rich C1-like domains and distinct regulatory domains (Mérida et al., 2008).
PA generated by activation of either DGKs or PLDs mediates cell migration by regulating the function of small GTPases through a variety of mechanisms (Nishikimi et al., 2009; Chianale et al., 2010), and Src-dependent activation of DGK-α mediates hepatocyte growth factor (HGF)– and VEGF-induced cell migration by regulating atypical PKCs and Rac (Cutrupi et al., 2000; Chianale et al., 2007, 2010; Baldanzi et al., 2008). Several lines of evidence indicate that DGKs are involved in membrane traffic. A large-scale kinome-wide siRNA screen has identified three DGKs (β, δ, and γ) as potential regulators of endocytosis or recycling (Pelkmans et al., 2005). In T cells, DGK-α may be required for multivesicular body polarization and exosome release (Alonso et al., 2011), and knockdown of DGK-ζ has been shown to promote transferrin recycling (Rincón et al., 2007). In addition, DGK-δ suppresses endoplasmic reticulum to Golgi traffic in epithelial cells (Nagaya et al., 2002). Conversely, DGKs have been largely ruled out as the source of PA required for Golgi coatomer recruitment (Yang et al., 2008).
Here, we show that DGK-α is required to support RCP-dependent α5β1 integrin recycling. Moreover, the migratory consequences of this RCP-regulated trafficking event (such as the acquisition of random migration on 2D surfaces and invasion into fibronectin-containing 3D matrices) are DGK-α dependent. Consistently, RCP must be capable of binding to PA to be mobilized to and tethered at the tips of invasive pseudopods, and expression of a constitutively active mutant of DGK-α is sufficient to drive RCP-dependent invasiveness. Our data suggest that by providing a source of PA to maintain RCP function, DGK-α forms an essential component of the pathway by which mutant p53s drive tumor cell invasion.
Results
DGK-α is required for RCP-dependent α5β1 recycling
We have previously shown that inhibition of αvβ3 integrin or expression of oncogenic mutants of p53 (such as p53175H or p53273H) increases the rate at which internalized α5β1 integrin is returned to the plasma membrane; an event that requires recruitment of RCP to the integrin (Caswell et al., 2008; Muller et al., 2009). Given the presence of a PA-binding C2 domain within the N-terminal region of RCP, we investigated whether DGK-α was required for RCP-dependent integrin trafficking. We treated A2780 ovarian carcinoma cells with either cRDGfNMeV or cRGDfV (cyclic peptides known to bind to and inhibit αv integrins; Dechantsreiter et al., 1999) and measured the rate at which internalized α5β1 returned to the plasma membrane. Consistent with our previous observations, addition of cRGDfNMeV (Fig. 1, A and B) or cRGDfV (Fig. 1 C) enhanced the rate at which α5β1 was recycled to the plasma membrane. This enhancement of α5β1 recycling was completely opposed by siRNA of DGK-α (Figs. 1 A and S1) or treatment of cells with low concentration of the class I DGK inhibitor, R59949 (Fig. 1 B; Jiang et al., 2000), indicating that enzymatic activity of DGK-α was required. Moreover, the ability of DGK-α knockdown to oppose α5β1 recycling was negated by expression of an siRNA-resistant rescue mutant of DGK-α (Figs. 1 C and S1 A). Accordingly, we found that addition of R59949 significantly inhibited α5β1 recycling in H1299 lung carcinoma cells expressing the 273H mutant of p53 but did not diminish integrin recycling in p53-null H1299 cells (Fig. 1 D). DGK-α knockdown did not inhibit 125I-transferrin recycling—an RCP-independent event in A2780 and H1299 cells (Caswell et al., 2008; Muller et al., 2009)—although the return of internalized 125I-transferrin to the plasma membrane was opposed by addition of receptor recycling inhibitors, such as chloroquine or primaquine (Fig. 1 E). Furthermore, internalization of both α5β1 (Fig. S1 D) and the transferrin receptor (TfnR; Fig. S1 E) was unaffected by DGK-α knockdown.
DGK-α is required for α5β1 recycling in response to αvβ3 inhibition or expression of mutant p53. (A–E) The recycling of internalized α5β1 (A–D) or 125I-transferrin (TFN; E) was determined in A2780 (A–C and E) or empty vector and mutant p53-expressing H1299 (D) cells. In A and E, A2780 cells were transfected with a nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα). In C, A2780 cells were transfected with an siRNA targeting DGK-α or siRNA targeting DGK-α in combination with an siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) or empty vector control (pMT2). (B and D) The DGK inhibitor, 1 µM R59949, or DMSO control was included during the internalization and recycling periods as indicated. (E) 120 µM chloroquine (CQ) was included during the internalization and recycling periods, whereas 0.3 mM primaquine (PQ) was only in the recycling period. (A–C) 36 nM cRGDfNMeV (A and B) or 2.5 µM cRGDfV (C) was included as indicated during the recycling period. Values are means ± SEM (A–D) and means ± SD (E) of at least nine replicates. †††, P < 0.0001 for si-con+cRGDfNMeV versus si-DGK-α+cRGDfNMeV; Mann–Whitney test. ***, P < 0.0001 for si-DGK-α+cRGDfNV versus si-DGK-α/DGK-α(resc)+cRGDfV; Mann–Whitney test. §§, P < 0.0001 for DMSO+cRGDfNMeV versus R59949+cRGDfNMeV; Mann–Whitney test. ‡‡‡, P < 0.0001 for p53273H DMSO versus p53273H R59949; Mann–Whitney test. $$$, P = 0.0002 for vehicle versus primaquine/chloroquine; Mann–Whitney test.
DGK-α is required for α5β1 recycling in response to αvβ3 inhibition or expression of mutant p53. (A–E) The recycling of internalized α5β1 (A–D) or 125I-transferrin (TFN; E) was determined in A2780 (A–C and E) or empty vector and mutant p53-expressing H1299 (D) cells. In A and E, A2780 cells were transfected with a nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα). In C, A2780 cells were transfected with an siRNA targeting DGK-α or siRNA targeting DGK-α in combination with an siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) or empty vector control (pMT2). (B and D) The DGK inhibitor, 1 µM R59949, or DMSO control was included during the internalization and recycling periods as indicated. (E) 120 µM chloroquine (CQ) was included during the internalization and recycling periods, whereas 0.3 mM primaquine (PQ) was only in the recycling period. (A–C) 36 nM cRGDfNMeV (A and B) or 2.5 µM cRGDfV (C) was included as indicated during the recycling period. Values are means ± SEM (A–D) and means ± SD (E) of at least nine replicates. †††, P < 0.0001 for si-con+cRGDfNMeV versus si-DGK-α+cRGDfNMeV; Mann–Whitney test. ***, P < 0.0001 for si-DGK-α+cRGDfNV versus si-DGK-α/DGK-α(resc)+cRGDfV; Mann–Whitney test. §§, P < 0.0001 for DMSO+cRGDfNMeV versus R59949+cRGDfNMeV; Mann–Whitney test. ‡‡‡, P < 0.0001 for p53273H DMSO versus p53273H R59949; Mann–Whitney test. $$$, P = 0.0002 for vehicle versus primaquine/chloroquine; Mann–Whitney test.
Given the requirement for DGK-α in RCP-dependent recycling, we used an in vitro kinase assay to determine whether inhibition of αvβ3 or expression of mutant p53 led to increased DGK-α activity. However, although addition of EGF strongly activated both endogenous (Fig. S1 F) and overexpressed myc–DGK-α (Fig. S1 G), neither addition of cRGDfV (or cRGDfNMeV; not depicted) nor expression of mutant p53 affected DGK-α activity (Fig. S1, F and G). We also used immunofluorescence to look at the distribution of endogenous DGK-α and found that the enzyme was distributed diffusely in the cytoplasm in both the presence and absence of cRGDfV (Fig. S1 H), consistent with the observation that inhibition of αvβ3 does not, per se, lead to activation of the kinase (Fig. S1 F).
Inhibition of αvβ3 (Fig. S2 A; Caswell et al., 2008) and expression of mutant p53 (Muller et al., 2009) lead to recruitment of RCP to α5β1 integrin, and this is reflected in an increased ability to coimmunoprecipitate these two proteins. To determine whether DGK-α was required for association of RCP with α5β1, we expressed GFP-RCP (or GFP control) and immunoprecipitated with an antibody recognizing GFP. However, inhibition of DGK-α did not reduce the quantity of α5β1 that coimmunoprecipitated with GFP-RCP after inhibition of αvβ3 (Fig. S2 A) or expression of mutant p53 (Fig. S2 B). We also performed the immunoprecipitation the other way around in mutant p53-expressing H1299 cells and found that addition of the DGK inhibitor did not affect the quantity of endogenous RCP that coimmunoprecipitated with α5 integrin (Fig. S2 C).
Collectively, these data indicate that although DGK-α has no role in the internalization of α5β1, its activity is permissive for RCP-dependent events that enhance the integrin’s return to the cell surface. Moreover, as addition of DGK-α inhibitors did not influence the ability of RCP to coimmunoprecipitate with α5β1 after addition of αvβ3 inhibitors or expression of mutant p53 (Fig. S2, A–C), we propose that DGK-α acts at a step downstream of integrin-RCP association to promote the delivery of RCP-α5β1 recycling vesicles to the plasma membrane.
DGK-α is required for α5β1-RCP–driven random migration
We have previously shown that increased RCP-dependent α5β1 recycling (whether evoked by inhibition of αvβ3 or expression of mutant p53s) is associated with a switch from persistent to random migration when cells are plated onto 2D surfaces (White et al., 2007; Caswell et al., 2008; Muller et al., 2009). We used fluorescence time-lapse microscopy to look at the organization of the actin cytoskeleton and persistence of cell movement during migration into scratch wounds. Consistent with our previous observations, A2780 (Fig. 2 A) and H1299 (not depicted) cells adopted a fanlike morphology as they migrated persistently into wounds (Fig. 2, B–E), and GFP-actin imaging indicated the presence of a stable actin-rich lamellipodium at the cell front (Fig. 2 A and Video 1). However, after inhibition of αvβ3 (Fig. 2, A–D; and Video 2) or expression of mutant p53 (Fig. 2, E–G; and not depicted), the ability of cells to maintain a stable lamellipodium and to migrate persistently was disrupted; rather, the cells moved randomly while extending highly protrusive actin-rich structures in all directions. Indeed, the acquisition of random migration was reflected in a marked reduction in migratory persistence (Fig. S3, A and B) and a lowered forward migration index, a quantitative measure of persistent migration perpendicular to the wound’s edge (Fig. 2, C, D, F, and G). Interestingly, the ability of cRGDfNMeV and cRGDfV (Figs. 2, A–C; and S3 A; and Video 3) or mutant p53 (Figs. 2, E–G; and S3 B; and not depicted) to disrupt the lamellipodium and induce random migration (i.e., to reduce the forward migration index and migratory persistence) was opposed by siRNA or pharmacological inhibition of DGK-α. Moreover, random migration was restored in DGK-α knockdown cells by reexpression of an siRNA-resistant mutant of DGK-α (Fig. 2, D and F). However, it must be noted that overexpression of DGK-α did lead to a small (but significant) decrease in forward migration index, even in the absence of αvβ3 inhibition (Fig. 2 D). Importantly, knockdown of DGK-α had no detectable effect on the migratory characteristics of p53-null H1299 (H1299 vector) or A2780 cells that were not treated with αvβ3 inhibitors (Fig. 2).
GFP-actin localization in A2780 cells transfected with a nontargeting siRNA. A2780 cells were transfected with GFP-actin in combination with a nontargeting siRNA, and a time-lapse video was recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
GFP-actin localization in A2780 cells transfected with a nontargeting siRNA. A2780 cells were transfected with GFP-actin in combination with a nontargeting siRNA, and a time-lapse video was recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
GFP-actin localization in A2780 cells treated with cRGDfNMeV in the presence of a nontargeting siRNA. A2780 cells were transfected with GFP-actin in combination with a nontargeting siRNA and treated with 160 nM cRGDfNMeV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
GFP-actin localization in A2780 cells treated with cRGDfNMeV in the presence of a nontargeting siRNA. A2780 cells were transfected with GFP-actin in combination with a nontargeting siRNA and treated with 160 nM cRGDfNMeV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
GFP-actin localization in A2780 cells treated with cRGDfNMeV in the presence an siRNA against DGK-α. A2780 cells were transfected with GFP-actin in combination with an siRNA targeting DGK-α and treated with cRGDfNMeV, and a time-lapse video was recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-actin localization in A2780 cells treated with cRGDfNMeV in the presence an siRNA against DGK-α. A2780 cells were transfected with GFP-actin in combination with an siRNA targeting DGK-α and treated with cRGDfNMeV, and a time-lapse video was recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
DGK-α is required for cRGDfNMeV- and mutant p53-driven random migration on 2D surfaces. (A) A2780 cells were transfected with GFP-actin in combination with either nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and allowed to grow to confluence on glass surfaces. Confluent monolayers were wounded with a plastic pipette tip, and the cells were allowed to migrate into the wound in the absence (−) or presence of 36 nM cRGDfNMeV. Time-lapse videos were recorded (Videos 1, 2, and 3), and representative stills are displayed. Bar, 20 µm. (B) A2780 cells were transfected with nontargeting siRNA or an siRNA targeting DGK-α, grown to confluence, and wounded as for A. The cells were observed by time-lapse video microscopy, the movement of individual cells was followed using cell-tracking software, and representative trackplots of trajectories described by cells during the first 8 h of migration into the wound are presented. (C and D) A2780 cells were transfected with nontargeting siRNA, an siRNA targeting DGK-α, or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)), and their migration into scratch wounds in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV both with or without 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots. (E–G) H1299 cells expressing p53273H or a control vector were transfected as for C and D, and their migration into scratch wounds in the presence and absence of 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots as for C. (C, D, F, and G) Data are represented as box and whisker plots (black lines shows medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
DGK-α is required for cRGDfNMeV- and mutant p53-driven random migration on 2D surfaces. (A) A2780 cells were transfected with GFP-actin in combination with either nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and allowed to grow to confluence on glass surfaces. Confluent monolayers were wounded with a plastic pipette tip, and the cells were allowed to migrate into the wound in the absence (−) or presence of 36 nM cRGDfNMeV. Time-lapse videos were recorded (Videos 1, 2, and 3), and representative stills are displayed. Bar, 20 µm. (B) A2780 cells were transfected with nontargeting siRNA or an siRNA targeting DGK-α, grown to confluence, and wounded as for A. The cells were observed by time-lapse video microscopy, the movement of individual cells was followed using cell-tracking software, and representative trackplots of trajectories described by cells during the first 8 h of migration into the wound are presented. (C and D) A2780 cells were transfected with nontargeting siRNA, an siRNA targeting DGK-α, or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)), and their migration into scratch wounds in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV both with or without 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots. (E–G) H1299 cells expressing p53273H or a control vector were transfected as for C and D, and their migration into scratch wounds in the presence and absence of 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots as for C. (C, D, F, and G) Data are represented as box and whisker plots (black lines shows medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
DGK-α is required to tether RCP vesicles at pseudopod tips
When A2780 or H1299 cells are plated into the cell-derived matrix (CDM), a planar 3D matrix that recapitulates many of the characteristics of native fibrillar fibronectin/collagen-rich ECM (Cukierman et al., 2001), they migrate in a sluglike fashion with a relatively broad cell front, and RCP is localized primarily to the cytosol and to vesicles in the cell body (Figs. 3 and 4). However, inhibition of αvβ3 (Fig. 3, A and B) or expression of mutant p53 (Fig. 3, C and D) promoted extension of long invasive pseudopods in the direction of cell migration, and fluorescence live-cell imaging clearly reveals a population of RCP vesicles that are recruited to the tips of these structures (Fig. 4 and Videos 4 and 6). Furthermore, by coexpressing GFP-RCP with an untagged mCherry protein, we confirmed that pseudopodial enrichment of RCP vesicles did not simply reflect increased cytoplasmic volume within this cellular extremity (Fig. S4 A).
DGK-α is required for pseudopod elongation evoked by inhibition of αvβ3 or expression of mutant p53. (A–D) A2780 cells (A and B) or vector and p53273H-expressing H1299 cells (C and D) were transfected with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα) or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) and plated onto CDM. Time-lapse videos were recorded in the absence (−) or presence of 36 nM cRGDvNMeV or 2.5 µM cRGDfV in combination with 10 µM R59022 or its vehicle (DMSO) and 0.5% butan-1-ol (But-1-ol; or secondary alcohol control [0.5% butan-2-ol]). Representative stills are displayed. (A and C) The arrows indicate elongated pseudopods. Bars, 20 µm. In B and D, the distance between the center of the nucleus and the cell front (with respect to the direction of migration) was measured using ImageJ. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
DGK-α is required for pseudopod elongation evoked by inhibition of αvβ3 or expression of mutant p53. (A–D) A2780 cells (A and B) or vector and p53273H-expressing H1299 cells (C and D) were transfected with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα) or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) and plated onto CDM. Time-lapse videos were recorded in the absence (−) or presence of 36 nM cRGDvNMeV or 2.5 µM cRGDfV in combination with 10 µM R59022 or its vehicle (DMSO) and 0.5% butan-1-ol (But-1-ol; or secondary alcohol control [0.5% butan-2-ol]). Representative stills are displayed. (A and C) The arrows indicate elongated pseudopods. Bars, 20 µm. In B and D, the distance between the center of the nucleus and the cell front (with respect to the direction of migration) was measured using ImageJ. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
DGK-α is necessary for RCP recruitment to pseudopod tips during migration on CDM. (A and B) A2780 cells (A) or vector and p53273H-expressing H1299 cells (B) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV. Images were captured at one frame/second over a period of 60 s, and videos were generated from these (Videos 4, 5, 6, and 7). Single-section confocal image stills corresponding to individual frames from these videos are presented. The arrows indicate the direction of migration, and the portion of the cell within the white square is presented in the bottom images. Bars, 10 µm.
DGK-α is necessary for RCP recruitment to pseudopod tips during migration on CDM. (A and B) A2780 cells (A) or vector and p53273H-expressing H1299 cells (B) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV. Images were captured at one frame/second over a period of 60 s, and videos were generated from these (Videos 4, 5, 6, and 7). Single-section confocal image stills corresponding to individual frames from these videos are presented. The arrows indicate the direction of migration, and the portion of the cell within the white square is presented in the bottom images. Bars, 10 µm.
The RCP-positive structures at pseudopod tips appeared to be relatively immobile, as if a population of GFP-RCP molecules were tethered at the cell front (Videos 4 and 6). To test this, we photobleached GFP-RCP at the front of cells and looked at the recovery of fluorescence into the photobleached area (termed FRAP). When A2780 or H1299 cells were migrating on the CDM, GFP-RCP fluorescence recovered quickly and completely at the front of photobleached pseudopod tips, indicating that RCP was relatively mobile under these circumstances. In contrast, inhibition of αvβ3 (Fig. 5, A and B; and Video 8) or expression of mutant p53 (Fig. 5 C) increased the proportion of GFP-RCP that was immobile at the pseudopod tip and that did not recover after photobleaching. This indicates that the RCP-positive structures at the tips of pseudopods are tightly tethered within this cellular locale and that the population of RCP molecules involved in this process do not exchange rapidly with those in the cytosol. Moreover, GFP that was not conjugated to RCP recovered completely after photobleaching, irrespective of whether or not cells had been treated with cRGDfV, indicating that inhibition of αvβ3 did not nonspecifically affect the mobility of cytosolic proteins within the pseudopod tip (Fig. S4 B).
GFP-RCP localization in A2780 cells treated with cRGDfV and transfected with a nontargeting siRNA. A2780 cells were transfected with GFP-RCP in combination with a nontargeting siRNA, plated onto CDM, and treated with 2.5 µM cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-RCP localization in A2780 cells treated with cRGDfV and transfected with a nontargeting siRNA. A2780 cells were transfected with GFP-RCP in combination with a nontargeting siRNA, plated onto CDM, and treated with 2.5 µM cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-RCP localization in p53273H-expressing H1299 cells transfected with a nontargeting siRNA. p53273H-expressing H1299 cells were transfected with GFP-RCP in combination with a nontargeting siRNA and plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
GFP-RCP localization in p53273H-expressing H1299 cells transfected with a nontargeting siRNA. p53273H-expressing H1299 cells were transfected with GFP-RCP in combination with a nontargeting siRNA and plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min. sicon, control siRNA.
The recovery from photobleaching of GFP-RCP in A2780 cells transfected with a nontargeting siRNA or an siRNA against DGK-α. A2780 cells were transfected with GFP-RCP and a nontargeting si-RNA (left) or an siRNA targeting DGK-α (right), plated onto CDM, and treated with 2.5 µM cRGDfV. The areas indicated by the yellow circles were subjected to photobleaching as indicated (frame 4), and the recovery from photobleaching followed by time-lapse fluorescence microscopy was performed using an inverted confocal microscope (FV1000). Frames were captured every second. sicon, control siRNA.
The recovery from photobleaching of GFP-RCP in A2780 cells transfected with a nontargeting siRNA or an siRNA against DGK-α. A2780 cells were transfected with GFP-RCP and a nontargeting si-RNA (left) or an siRNA targeting DGK-α (right), plated onto CDM, and treated with 2.5 µM cRGDfV. The areas indicated by the yellow circles were subjected to photobleaching as indicated (frame 4), and the recovery from photobleaching followed by time-lapse fluorescence microscopy was performed using an inverted confocal microscope (FV1000). Frames were captured every second. sicon, control siRNA.
DGK-α is required for the tethering of RCP vesicles at pseudopod tips. (A–C) A2780 (A and B) or vector and p53273H-expressing H1299 cells (C) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 2.5 µM cRGDfV. Photobleaching was performed as described in the Materials and methods section. Stills from a video (Video 8) at the indicated time points are shown. (A) The arrows indicate the direction of cell migration, whereas the circles highlight the bleaching region. Recovery curves were generated using the FV10-ASW software (Olympus), and the immobile fraction (percentile) was extracted from these. Values are means ± SEM; ***, P < 0.0001. Bar, 10 µm.
DGK-α is required for the tethering of RCP vesicles at pseudopod tips. (A–C) A2780 (A and B) or vector and p53273H-expressing H1299 cells (C) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 2.5 µM cRGDfV. Photobleaching was performed as described in the Materials and methods section. Stills from a video (Video 8) at the indicated time points are shown. (A) The arrows indicate the direction of cell migration, whereas the circles highlight the bleaching region. Recovery curves were generated using the FV10-ASW software (Olympus), and the immobile fraction (percentile) was extracted from these. Values are means ± SEM; ***, P < 0.0001. Bar, 10 µm.
Given that extension of invasive pseudopods depends on both α5β1 and RCP (Caswell et al., 2008), we wished to determine the requirement for DGK-α in this process. Indeed, siRNA of DGK-α or inhibition of its catalytic activity impaired pseudopodial extension after inhibition of αvβ3 (Fig. 3, A and B) or expression of mutant p53 (Fig. 3, C and D). Moreover, DGK-α knockdown markedly reduced the number of RCP-positive structures (Fig. 4) and the immobile fraction of GFP-RCP (Fig. 5) at pseudopod tips in a way that could be rescued by reexpression of siRNA-resistant DGK-α. It is interesting to note that DGK-α knockdown increased the population of RCP vesicles in the perinuclear region (Fig. 4) without apparently restricting the overall mobility of RCP endosomes within the pseudopod shaft (Videos 5 and 7). Importantly, inhibition of DGK-α by siRNA or pharmacological means did not affect the morphology (Figs. 3 and 4) or migration speed (not depicted) of p53-null H1299 (H1299 vector) or A2780 cells that were not treated with αvβ3 inhibitors.
Influence of DGK-α on DAG and PA lipid species
Mass spectrometry was used to determine the cellular levels of DAG and PA. Table 1 and Table 2 show that suppressing the level of DGK-α by siRNA reduced the total concentration of PA and increased that of DAG; though in each case, the effects were relatively small. The DGK inhibitor surprisingly reduced cellular DAG and had a minimal effect upon PA. Determination of relatively small total cellular lipid concentrations does not give a true reflection of changes in lipid-mediated responses and their effects upon membranes because individual lipid species within a lipid class function in a distinct manner. Indeed, many DAG and PA species are metabolic intermediates rather than signals (Hodgkin et al., 1998). It has been shown that 36:1 PA is the major species produced in cells by PLD activity (Pettitt et al., 2001). In contrast, 38:4 is the major DAG and PA species generated by phosphatidylinositol-4,5 bisphosphate hydrolysis and, as such, is predominantly a plasma membrane lipid. Similarly, 38:3 and 38:5 DAG and PA are generated through this route, though to a lesser extent. Examination of the individual DAG and PA species demonstrates that inhibition of DGK activity with R59022 and siRNA of DGK-α had minimal effects upon 36:1 lipids, demonstrating that PLD activity was unaffected. The most significant effects of the DGK inhibitor and siRNA against DGK-α were upon the C38 lipids, in particular 38:3, 38:4, and 38:5, pointing to selectivity of the kinase for substrate lipids and to its effects being predominantly at the plasma membrane. Collectively, these data point to the generation of plasma membrane 38:4 PA, in particular, as being an important step in α5β1 recycling. It is perhaps important in this regard that 38:4 PA will induce greater membrane fluidity and curvature than 38:1 PA or 38:4 DAG.
Influence of DGK-α on total DAG and PA lipid content
| Sample | Total DAG | Total PA |
| nmol | nmol | |
| si-con | 253.4 ± 24.7 | 1,382.5 ± 42.5 |
| si-DGK-α | 282.5 ± 5.3 | 1,308.5 ± 82.6 |
| R59022 | 197.4 ± 16.5 | 1,365.4 ± 126.9 |
| Sample | Total DAG | Total PA |
| nmol | nmol | |
| si-con | 253.4 ± 24.7 | 1,382.5 ± 42.5 |
| si-DGK-α | 282.5 ± 5.3 | 1,308.5 ± 82.6 |
| R59022 | 197.4 ± 16.5 | 1,365.4 ± 126.9 |
The lipids were separated and analyzed by HPLC mass spectrometry, and the total amount of each DAG and PA in each sample was determined. si-con, siRNA control; si-DGK-α, siRNA targeting DGK-α.
Influence of DGK-α on particular DAG and PA lipid species
| Lipid | Lipid species in cells treated with si-con | Lipid species in cells treated with si-DGK-α | Lipid species in cells treated with R59022 |
| % | % | % | |
| DAG 36:1 | 6.09 ± 0.91 | 8.16 ± 0.42 | 5.71 ± 0.37 |
| DAG 38:1 | 0.09 ± 0.01 | 0.14 ± 0.01b | 0.08 ± 0.14 |
| DAG 38:2 | 0.20 ± 0.07 | 0.50 ± 0.09b | 0.21 ± 0.06 |
| DAG 38:3 | 1.03 ± 0.64 | 1.29 ± 0.27 | 0.71 ± 0.37 |
| DAG 38:4 | 4.08 ± 0.87 | 4.40 ± 0.57 | 5.86 ± 0.45a |
| DAG 38:5 | 1.32 ± 0.63 | 1.01 ± 0.19 | 0.63 ± 0.24 |
| DAG 38:6 | 0.68 ± 0.96 | 0.62 ± 0.16 | 0.43 ± 0.25 |
| PA 36:1 | 13.88 ± 0.14 | 14.65 ± 0.50 | 13.89 ± 0.51 |
| PA 38:1 | 0.15 ± 0.01 | 0.11 ± 0.01b | 0.12 ± 0.01b |
| PA 38:2 | 1.10 ± 0.21 | 0.95 ± 0.05 | 0.99 ± 0.06 |
| PA 38:3 | 4.82 ± 0.36 | 3.88 ± 0.35a | 3.39 ± 0.24b |
| PA 38:4 | 25.28 ± 0.34 | 21.41 ± 1.12b | 15.85 ± 0.97b |
| PA 38:5 | 2.59 ± 0.09 | 2.41 ± 0.14 | 2.22 ± 0.05b |
| PA 38:6 | 1.35 ± 0.03 | 1.47 ± 0.05 | 1.32 ± 0.08 |
| Lipid | Lipid species in cells treated with si-con | Lipid species in cells treated with si-DGK-α | Lipid species in cells treated with R59022 |
| % | % | % | |
| DAG 36:1 | 6.09 ± 0.91 | 8.16 ± 0.42 | 5.71 ± 0.37 |
| DAG 38:1 | 0.09 ± 0.01 | 0.14 ± 0.01b | 0.08 ± 0.14 |
| DAG 38:2 | 0.20 ± 0.07 | 0.50 ± 0.09b | 0.21 ± 0.06 |
| DAG 38:3 | 1.03 ± 0.64 | 1.29 ± 0.27 | 0.71 ± 0.37 |
| DAG 38:4 | 4.08 ± 0.87 | 4.40 ± 0.57 | 5.86 ± 0.45a |
| DAG 38:5 | 1.32 ± 0.63 | 1.01 ± 0.19 | 0.63 ± 0.24 |
| DAG 38:6 | 0.68 ± 0.96 | 0.62 ± 0.16 | 0.43 ± 0.25 |
| PA 36:1 | 13.88 ± 0.14 | 14.65 ± 0.50 | 13.89 ± 0.51 |
| PA 38:1 | 0.15 ± 0.01 | 0.11 ± 0.01b | 0.12 ± 0.01b |
| PA 38:2 | 1.10 ± 0.21 | 0.95 ± 0.05 | 0.99 ± 0.06 |
| PA 38:3 | 4.82 ± 0.36 | 3.88 ± 0.35a | 3.39 ± 0.24b |
| PA 38:4 | 25.28 ± 0.34 | 21.41 ± 1.12b | 15.85 ± 0.97b |
| PA 38:5 | 2.59 ± 0.09 | 2.41 ± 0.14 | 2.22 ± 0.05b |
| PA 38:6 | 1.35 ± 0.03 | 1.47 ± 0.05 | 1.32 ± 0.08 |
A2780 cells were transfected with a non-targeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGK-α) or treated with 10 µM R59022 for 1 h. Cell pellets were collected, and lipids were extracted in the presence of appropriate standards to control for extraction efficiency and to identify the lipids on mass spectrometry. Analysis of each individual molecular species allowed determination of the percentage of each within the total lipid class demonstrating which lipids changed. In each sample, 21 distinct DAG species and 32 distinct PA species were identified and quantified; the species not shown in the table demonstrated minimal changes.
P < 0.05; unpaired t test.
P < 0.01; unpaired t test.
PA production downstream of DGK-α and RCP’s C2 domain are required to tether recycling vesicles at pseudopod tips
To directly determine whether PA-binding by RCP was required for α5β1 recycling and the extension of invasive pseudopods, we deployed a mutant of RCP (RCP200–649), which lacks the PA-binding C2 domain (Lindsay and McCaffrey, 2004). Although expression of an siRNA-resistant form of full-length RCP (RCPWT) effectively restored α5β1 recycling in RCP knockdown cells, RCP200–649 was only partially able to do this, indicating that RCP needs to bind PA to properly drive integrin trafficking (Figs. 6 A; and S4 C). Consistent with this, expression of full-length RCP rescued cRGDfV-driven pseudopod extension in RCP knockdown cells, but RCP200–649 was ineffective in this regard (Fig. 6 B). We also investigated the recruitment and tethering of GFP-RCP200–649 at the tips of pseudopods in cells in which RCP had not been knocked down. Indeed, although αvβ3 inhibition (Fig. 6 C) and mutant p53 expression (Fig. 6 E) induced pseudopod elongation in cells expressing GFP-RCP200–649 (indicating that RCP200–649 does not exert dominant-negative inhibition over endogenous RCP function), the truncated RCP mutant was not concentrated at the tips of these structures (Fig. 6, C and E; and Videos 9 and 10) despite its ability to coimmunoprecipitate with α5β1 (Fig. S2 D). Moreover, FRAP analyses indicated that GFP-RCP200–649 was not tethered at the cell front in cRGDfV-treated or p53273H-expressing cells (Fig. 6, D and F).
RCP’s C2 domain controls recruitment to pseudopod tips. (A and B) A2780 cells were transfected with an siRNA targeting RCP (si-RCP) in combination with either GFP, siRNA-resistant GFP-RCPWT (GFP-RCPWT(resc)), or siRNA-resistant GFP-RCP200–649 (GFP-RCP200–649(resc)). In A, 2.5 µM cRGDfV was included during the recycling period. Values are means ± SEM of ≥12 replicates. ***, P < 0.0005 GFP versus GFP-RCPWT; Mann–Whitney test. ##, P < 0.0012 GFP versus GFP-RCP200–649. In B, cells were plated onto the CDM, and videos were collected in the absence (−) or presence of 2.5 µM cRGDfV as for Fig. 3. Pseudopod length was determined as for Fig. 3. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test. (C–F) A2780 cells (C and D) or vector and p53273H-expressing H1299 cells (E and F) were transfected with GFP-RCP200–649. Cells were plated onto the CDM, and videos were collected in the absence (−) or presence of 2.5 µM cRGDfV as for Fig. 4 (Videos 9 and 10). Single-section confocal image stills corresponding to individual frames from these videos are presented. The yellow arrows indicate the direction of migration, and the portion of the cell within the squares is presented in the bottom images. Bars, 10 µm. (D and F) Photobleaching was performed as described in the Material and methods section. Recovery curves were generated using the FV10-ASW software and the immobile fraction (percentage) was extracted from these. In D and F, the recovery curves and immobile fraction of GFP-RCP200–649, which was photoactivated in either vesicular structures (vesic.) or diffusely cytoplasmic regions (cyto.), were analyzed either in combination (all) or separately (vesicular structures and cytoplasmic regions)—see Fig. S5 B for examples of stills from these videos. The number of cells represented by each analysis is depicted in parentheses. Values are means ± SEM.
RCP’s C2 domain controls recruitment to pseudopod tips. (A and B) A2780 cells were transfected with an siRNA targeting RCP (si-RCP) in combination with either GFP, siRNA-resistant GFP-RCPWT (GFP-RCPWT(resc)), or siRNA-resistant GFP-RCP200–649 (GFP-RCP200–649(resc)). In A, 2.5 µM cRGDfV was included during the recycling period. Values are means ± SEM of ≥12 replicates. ***, P < 0.0005 GFP versus GFP-RCPWT; Mann–Whitney test. ##, P < 0.0012 GFP versus GFP-RCP200–649. In B, cells were plated onto the CDM, and videos were collected in the absence (−) or presence of 2.5 µM cRGDfV as for Fig. 3. Pseudopod length was determined as for Fig. 3. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test. (C–F) A2780 cells (C and D) or vector and p53273H-expressing H1299 cells (E and F) were transfected with GFP-RCP200–649. Cells were plated onto the CDM, and videos were collected in the absence (−) or presence of 2.5 µM cRGDfV as for Fig. 4 (Videos 9 and 10). Single-section confocal image stills corresponding to individual frames from these videos are presented. The yellow arrows indicate the direction of migration, and the portion of the cell within the squares is presented in the bottom images. Bars, 10 µm. (D and F) Photobleaching was performed as described in the Material and methods section. Recovery curves were generated using the FV10-ASW software and the immobile fraction (percentage) was extracted from these. In D and F, the recovery curves and immobile fraction of GFP-RCP200–649, which was photoactivated in either vesicular structures (vesic.) or diffusely cytoplasmic regions (cyto.), were analyzed either in combination (all) or separately (vesicular structures and cytoplasmic regions)—see Fig. S5 B for examples of stills from these videos. The number of cells represented by each analysis is depicted in parentheses. Values are means ± SEM.
The PA/DGK-α–dependent slowing of fluorescence recovery may be owing not only to tethering of RCP vesicles at pseudopod tips but also to its recruitment from the cytosol to vesicular membranes. GFP-RCP200–649, although unable to bind to PA and tether vesicles at the extremities of pseudopods, can still associate with α5β1 and be recruited to vesicular structures that are located within the cell body and the pseudopod shaft (Fig. 6, C and E). We photobleached GFP-RCP200–649 located at vesicular structures in the pseudopod shaft near to the tip and GFP-RCP200–649, which exhibited a diffuse cytoplasmic distribution at the cell front, and compared the recovery times of these two pools of mutant RCP (Fig. S5 B). The recovery time of vesicular GFP-RCP200–649 was slightly slower (and the immobile fraction marginally larger) than GFP-RCP200–649, which was diffusely cytoplasmic (Figs. 6, D and F; and S5 B). Nevertheless, the immobile fraction of vesicular GFP-RCP200–649 (immobile fraction ≈ 27–34%) was much less than that of GFP-RCPWT at pseudopod tips (immobile fraction ≈ 63–65%; compare Fig. 5 [B and C] with Fig. 6 [D and F]). Thus, recruitment of RCP from the cytoplasm to vesicles leads to only a small decrease in its mobility—indicating that GFP-RCP200–649 can exchange quite rapidly between the cytosol and the vesicle membrane. On the other hand, GFP-RCPWT is tightly tethered at pseudopod tips in a PA/DGK-α–dependent fashion and cannot exchange rapidly with a cytoplasmic pool of RCP. Collectively, these data indicate that PA generated downstream of DGK-α acts via a mechanism involving RCP’s C2 domain to properly recycle α5β1 integrin and to recruit and tether RCP vesicles to the front of long invasive protrusions as cells migrate on CDMs.
At this stage, we assessed whether PA that is generated downstream of DGK isoforms other than DGK-α or that was produced via PLDs had a role in RCP-dependent events. Quantitative PCR indicated that A2780 cells expressed (in addition to DGK-α) the δ, ε, η, θ, and ι isoforms of DGK and that all of these could be knocked down using the appropriate siRNA oligonucleotides (Fig. S3 E). However, of these DGKs, only DGK-ι had any significant effect on pseudopod extension (Fig. S3 E). Interestingly, we recently showed that DGK-ι is required for anchorage-independent growth of Kaposi sarcoma cells (Baldanzi et al., 2011). To look at the contribution made by PLDs, we used a primary alcohol (butan-1-ol) to inhibit the lipases or a control secondary alcohol (butan-2-ol). We found that pseudopod extension (Fig. 3 B) and the recruitment of GFP-RCP to the tips of these structures (Fig. S5 A) were unaffected by addition of butan-1-ol. These data indicate that although DGK-α (and possibly DGK-ι) plays a role in RCP function, the majority of DGK isoforms and the PLDs are not required for RCP-dependent invasive processes in these cell types.
DGK-α is necessary for α5β1-dependent tumor cell invasion
Up-regulation of α5β1 recycling through the RCP pathway (whether by inhibition of αvβ3 or by expression of mutant p53s) is associated with increased tumor cell invasiveness into plugs of collagen I or Matrigel (Caswell et al., 2008; Muller et al., 2009). Moreover, RCP-dependent invasion is only apparent after addition of fibronectin to the ECM plug, thus highlighting the requirement for α5β1 in this type of cell migration. Consistent with our previous observations, we found that inhibition of αvβ3 or expression of mutant p53 increased invasion into collagen I plugs and that this was particularly apparent when fibronectin was added to the matrix (Fig. 7, A and B). Interestingly, invasion that was driven by inhibition of αvβ3 or by expression of mutant p53 (and that was fibronectin dependent) was strongly opposed by siRNA of DGK-α, whereas the basal invasiveness of A2780 and H1299 cells was not significantly affected by knockdown of the kinase (Fig. 7, A and B). Furthermore, invasion of MDA-MB-231 breast carcinoma cells (that endogenously express the DNA-binding R280K mutant of p53) into fibronectin-containing collagen I plugs was inhibited by siRNA of DGK-α (Figs. 7 C; and S1 C) to a similar extent as by knockdown of mutant p53 (not depicted; Muller et al., 2009) in this cell type.
DGK-α is required for invasion into fibronectin-containing collagen I hydrogels evoked by inhibition of αvβ3 or expression of mutant p53. (A–C) A2780 (A), vector, and p53273H-expressing H1299 (B) or MDA-MB-231 (C) cells were transfected with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and plated onto the bottom of collagen I plugs. In A, the collagen plugs were formed in the presence and absence of 25 µg/ml fibronectin (FN) as indicated, and in B and C, 25 µg/ml fibronectin was included for all conditions. The cells were allowed to invade into the plugs for 48 h toward a gradient of 30 ng/ml EGF and 10% FCS. 2.5 µM cRGDfV was added to both the upper and lower chambers of the inverted collagen plug as indicated in A. Invading cells were stained with calcein acetoxymethyl ester and visualized by confocal microscopy. Serial optical sections were captured at 15-µm intervals and are presented as a sequence in which the individual optical sections are placed alongside one another with increasing depth from left to right as indicated. Migration was quantitated by measuring the fluorescence intensity of cells penetrating the collagen/fibronectin to depths of 60 µm (A) or 30 µm (B and C) and greater and expressing this as a percentage of the total fluorescence intensity of all cells within the plug. Data are represented as box and whiskers plot (black lines show medians; whiskers show minimum to maximum values); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
DGK-α is required for invasion into fibronectin-containing collagen I hydrogels evoked by inhibition of αvβ3 or expression of mutant p53. (A–C) A2780 (A), vector, and p53273H-expressing H1299 (B) or MDA-MB-231 (C) cells were transfected with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and plated onto the bottom of collagen I plugs. In A, the collagen plugs were formed in the presence and absence of 25 µg/ml fibronectin (FN) as indicated, and in B and C, 25 µg/ml fibronectin was included for all conditions. The cells were allowed to invade into the plugs for 48 h toward a gradient of 30 ng/ml EGF and 10% FCS. 2.5 µM cRGDfV was added to both the upper and lower chambers of the inverted collagen plug as indicated in A. Invading cells were stained with calcein acetoxymethyl ester and visualized by confocal microscopy. Serial optical sections were captured at 15-µm intervals and are presented as a sequence in which the individual optical sections are placed alongside one another with increasing depth from left to right as indicated. Migration was quantitated by measuring the fluorescence intensity of cells penetrating the collagen/fibronectin to depths of 60 µm (A) or 30 µm (B and C) and greater and expressing this as a percentage of the total fluorescence intensity of all cells within the plug. Data are represented as box and whiskers plot (black lines show medians; whiskers show minimum to maximum values); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.
A membrane-anchored DGK-α promotes recruitment of RCP to invasive pseudopods and drives RCP-dependent invasion
Although DGK-α itself was not activated by inhibition of αvβ3 or by expression of mutant p53s (Fig. S1, F and G), it was clear that addition of exogenous PA (but not DAG or lyso-PA) was able to drive pseudopod extension (Fig. 8 A). Moreover, this was not opposed by pharmacological inhibition of DGK, indicating that PA is acting downstream of the kinase (Fig. 8 A). A mutant of DGK-α containing the Src myristoylation sequence at its C terminus (myr–DGK-α) has previously been shown to be constitutively membrane-associated, catalytically active, and able to induce membrane ruffles and migration of epithelial cells in the absence of chemotactic factors (Baldanzi et al., 2008; Chianale et al., 2010). Indeed, expression of myr-DGK-α increased α5β1 recycling even in the absence of αvβ3 inhibition (Fig. 8 B). Strikingly, expression of myr–DGK-α promoted the extension of very long pseudopods when cells were migrating on the CDM (Fig. 8, C and D), and large numbers of RCP vesicles were concentrated toward the tips of these structures (Fig. 8 D). Moreover, myr–DGK-α–driven pseudopod extension was opposed by knockdown of RCP, and this was rescued by expression of an siRNA-resistant form of full-length RCP (RCPWT) but not by the RCP200–649 mutant that lacks the PA-binding C2 domain (Fig. 8 C). Consistently, myr–DGK-α increased invasiveness of A2780 cells through collagen I plugs, in a way that required addition of fibronectin to the ECM (Fig. 8 F), and this was completely opposed by siRNA of RCP (Fig. 8, E and G). Collectively, these data show that PA production downstream of DGK-α drives the invasive behavior of tumor cells and that this is achieved through the control of RCP function and its regulation of α5β1 integrin recycling.
myr–DGK-α promotes RCP-dependent integrin recycling, invasion, and recruitment of RCP to pseudopod tips. (A) A2780 cells were plated onto the CDM, time-lapse videos were collected in the absence (–) or presence of lyso-PA (LPA), DAG, PA, and R59022 (all at 10 µM), and pseudopod length was determined as for Fig. 3. Blue boxes indicate addition of PA. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test. (B) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector), and the recycling of α5β1 was determined as for Fig. 1. **, P = 0.004; ***, P = 0.0008; Mann–Whitney test. (C) A2780 cells were transfected with myr–DGK-α or pMT2 together with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting RCP (si-RCP) in combination with GFP, siRNA-resistant GFP-RCPWT (GFP-RCPWT(resc)), or siRNA-resistant GFP-RCP200–649 (GFP-RCP200–649(resc)). Cells were plated onto the CDM, and pseudopod length was determined as for Fig. 3. ***, P = 0.0008; Mann–Whitney test. (D) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector) in combination with GFP-RCP. Transfected cells were plated onto CDM, and videos were captured as for Fig. 4. Representative stills are displayed. The yellow arrows indicate the direction of migration and the portion of the cell within the white square, which is presented at the bottom. Bar, 20 µm. (E–G) A2780 cells were transfected with myr–DGK-α or pMT2 in combination with either nontargeting siRNA or an siRNA targeting RCP and plated into inverted invasion assays in the presence (E and G) or absence (F) of 25 µg/ml fibronectin (FN) as indicated. Invasion was determined as for Fig. 7. Data are represented as box and whiskers plots (black lines show medians; whiskers: minimum to maximum). ***, P < 0.0001; Mann–Whitney test. Values are means ± SEM.
myr–DGK-α promotes RCP-dependent integrin recycling, invasion, and recruitment of RCP to pseudopod tips. (A) A2780 cells were plated onto the CDM, time-lapse videos were collected in the absence (–) or presence of lyso-PA (LPA), DAG, PA, and R59022 (all at 10 µM), and pseudopod length was determined as for Fig. 3. Blue boxes indicate addition of PA. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test. (B) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector), and the recycling of α5β1 was determined as for Fig. 1. **, P = 0.004; ***, P = 0.0008; Mann–Whitney test. (C) A2780 cells were transfected with myr–DGK-α or pMT2 together with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting RCP (si-RCP) in combination with GFP, siRNA-resistant GFP-RCPWT (GFP-RCPWT(resc)), or siRNA-resistant GFP-RCP200–649 (GFP-RCP200–649(resc)). Cells were plated onto the CDM, and pseudopod length was determined as for Fig. 3. ***, P = 0.0008; Mann–Whitney test. (D) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector) in combination with GFP-RCP. Transfected cells were plated onto CDM, and videos were captured as for Fig. 4. Representative stills are displayed. The yellow arrows indicate the direction of migration and the portion of the cell within the white square, which is presented at the bottom. Bar, 20 µm. (E–G) A2780 cells were transfected with myr–DGK-α or pMT2 in combination with either nontargeting siRNA or an siRNA targeting RCP and plated into inverted invasion assays in the presence (E and G) or absence (F) of 25 µg/ml fibronectin (FN) as indicated. Invasion was determined as for Fig. 7. Data are represented as box and whiskers plots (black lines show medians; whiskers: minimum to maximum). ***, P < 0.0001; Mann–Whitney test. Values are means ± SEM.
Discussion
Tumor cells expressing gain-of-function p53 mutants have altered cell migration and increased invasiveness (Adorno et al., 2009; Muller et al., 2009), and similar changes in tumor cell motility may be evoked by inhibition of αvβ3 integrin (Caswell et al., 2008). These alterations to cell migration depend on the ECM environment in which cell movement is studied. For instance, on plastic surfaces, mutant p53 expression and inhibition of αvβ3 both decrease the persistence of migration. However, on CDMs, migratory persistence is not affected, but expression of mutant p53 or inhibition of αvβ3 drives extension of long pseudopods in the direction of migration. Importantly, in 3D microenvironments, mutant p53 and inhibition of αvβ3 both drive increased invasiveness in a way that is dependent on α5β1 and its ligand fibronectin. However, it is clear that, irrespective of the microenvironmental context, increased RCP-dependent recycling of α5β1 integrin underlies this altered migratory behavior. This study describes a role played by DGK-α in RCP function, and—importantly—knockdown or inhibition of DGK-α prevents RCP-dependent receptor recycling and cell migration but has no effect on related events that are RCP independent. Critical to RCP’s ability to localize to the tips of invasive structures is the PA-binding C2 domain that lies within its N-terminal region. Moreover, expression of a constitutively active plasma membrane–anchored DGK-α mutant is a powerful driver of tumor cell invasion in a way that is dependent on RCP and the ability of RCP to bind to PA. These data show that the role of DGK-α during the invasion of mutant p53-expressing cells is to generate PA to enable RCP function, and without DGK-α, the migratory behavior of mutant p53-expressing tumor cells is indistinguishable from their p53-null counterparts.
PA can be generated by the activity of either PLDs or DGKs, and both these enzyme families have been linked to membrane trafficking events (Jenkins and Frohman, 2005; Mérida et al., 2008). PA generation has a well-established role in the budding of transport vesicles from several cellular membranes, including the plasma membrane and Golgi complex. Much of PA’s ability to promote vesicle budding is thought to be caused by the curvature that the lipid imparts when generated in restricted membrane regions, and a recent study has shown that this is mediated via PA’s collaboration with membrane-binding proteins, such as BARS, to bend membranes to promote vesicle fission (Yang et al., 2008). Indeed, PA directly binds proteins, such as Sos, and this interaction mediates the potentiation of Ras signaling by PLD (Zhao et al., 2007). Furthermore, several proteins known to control membrane traffic, such as Arfs1 and 6, PIP4K, and NSF, have been reported to bind to and/or be activated by PA (Ktistakis et al., 2003). The C2 domains of the class I FIPs—RCP, FIP2, and Rip11—bind to PA (Lindsay and McCaffrey, 2004), so potentially, these Rab11 effectors may be controlled by PLDs and/or DGKs. However, of the class I FIPs, only RCP is capable of binding to α5β1 (Caswell et al., 2008), thus highlighting a potentially selective connection between PA signaling and integrin trafficking. So how do PA and RCP collaborate in integrin recycling? DGK-α activity and the C2 domain are not required for RCP to bind to α5β1 (Fig. S3 D) or Rab11 nor for its recruitment to endosomal membranes (Lindsay and McCaffrey, 2004). However, deletion of the C2 domain or inhibition of DGK-α signaling reduces the number of RCP-positive structures that are tethered at pseudopod tips and correspondingly increases the vesicle population in the perinuclear region without apparently restricting the overall mobility of RCP endosomes and their ability to move up and down the pseudopod shaft. An immunofluorescence study indicates that active DGK-α is localized to the plasma membrane (Baldanzi et al., 2008), and consistently, we cannot detect DGK-α localized to RCP endosomes (Fig. S1 H). Moreover, myr–DGK-α, which so potently drives RCP-dependent pseudopodial extension and invasion, is localized exclusively to the plasma membrane (unpublished data; Baldanzi et al., 2008; Chianale et al., 2010), indicating that it is here that PA acts to influence RCP function. Established vesicle tethers commonly possess a Rab GTPase/cargo-binding portion, which is linked by an extended coiled-coil region to a domain that senses membrane curvature (Brown and Pfeffer, 2010). Collectively, these observations suggest that RCP may function as a vesicle tether that interacts C terminally with Rab11 and the integrin cargo vesicle and N terminally with a highly concave PA-rich region of the plasma membrane at the pseudopod tip. Indeed, the selective generation of longer chain, more unsaturated PA species (38:3, 38:4, and 38:5), such as we have detected in our mass spectrometry analyses, would be expected to promote localized membrane curvature. Further work is required to determine precisely where in the cell DGK-α acts to produce PA during migration and the role played by RCP in recruiting and tethering integrin transport vesicles at the tips of invading pseudopods.
Several groups have shown that PA production downstream of PLDs contributes to endosomal transport and also to actin remodelling that is associated with cell migration. However, we show that siRNA knockdown or pharmacological inhibition of DGK-α consistently opposes recruitment of RCP to pseudopod tips, whereas treatment of cells with butan-1-ol to inhibit PLDs has no effect on these processes, indicating that DGK-α is more important as a source of PA to enable RCP function than are PLDs; a conclusion supported by there being no change in 38:1 PA in our mass spectrometry analyses. Moreover, the way in which DGK-α influences tumor cell migration is not implemented via an overarching requirement for the kinase in cell motility—indeed DGK-α knockdown A2780 and H1299 cells migrate persistently on plastic surfaces, indicating that the cell’s migratory machinery is working properly—but by controlling RCP/α5β1-dependent events that dictate the particular mode of migration adopted by the cell. α5β1 integrin signaling through RhoA stimulates rapid random migration on 2D surfaces (Danen et al., 2005), and we have shown that up-regulation of α5β1 recycling evokes Rho-dependent motile responses, indicating that RCP recycling enhances α5β1’s communication with RhoA (White et al., 2007). Consistently, we find that the ability of cRGDfV and mutant p53 to increase random cell migration (a DGK-α–dependent process) is opposed by inhibition of Rho kinase (Fig. S3, C and D). DGK-α has previously been shown to link HGF receptor activation to membrane targeting and the protrusive activity of Rac in MDCK epithelial cells (Chianale et al., 2007). This occurs via a mechanism whereby PA production downstream of DGK-α acts on atypical PKC, which, in turn, allows Rac release from Rho GDP dissociation inhibitor and delivery to the plasma membrane (Chianale et al., 2010). However, the migration of nontransformed epithelial cells and that of tumor cells is controlled differently, and further work will be needed to determine the relationship between HGF-induced Rac activation in epithelial cells and DGK-α/RCP/α5β1-dependent RhoA-associated random migration in mutant p53-expressing tumor cells.
DGK-α has been linked to both positive and negative roles in cancer. On the one hand, a study in lung cancer has identified a correlation between elevated DGK-α expression and good prognosis (Berrar et al., 2005). This indicates a potential tumor suppressor role for DGK-α and may be related to its ability to oppose DAG signaling. On the other hand, many studies indicate that DGK-α contributes to cancer progression. For instance, DGK-α is up-regulated in lymph node metastases with respect to primary breast tumors, indicating that it may be a prometastatic gene (Hao et al., 2004). Gain-of-function mutants of p53 are clearly prometastatic, and interestingly, we find that DGK-α is required for these p53 mutants to influence tumor cell migration and invasion. Mutant p53s act by inhibiting p63 to drive association of RCP with internalized α5β1 integrin and EGF receptor (EGFR), and these processes do not require DGK-α. However, DGK-α is required for subsequent RCP-dependent delivery of α5β1 and EGFR (Fig. S4 D) to the plasma membrane. This indicates that, although DGK-α is neither activated nor up-regulated by mutant p53, the kinase forms an essential part of the mutant p53–RCP–integrin axis, as it is required for RCP-recycling vesicles to be recruited to the appropriate plasma membrane regions during invasive cell migration (Fig. 9).
Schematic to summarize DGK-α’s role in RCP-dependent events. Inhibition of αvβ3 or expression of mutant p53273H promotes recruitment of RCP to endosomal α5β1. Association of α5β1 with RCP is not a DGK-dependent event and does not require RCP’s PA-binding C2 domain. RCP/integrin recycling vesicles can then move up and down the pseudopod shaft, and the role of DGK-α is to generate PA species that enable the tethering of RCP at pseudopod tips, an event that requires RCP’s C2 domain. Of the lipid species detected in the mass spectrometric analysis, inhibition or silencing of DGK-α most significantly affects the interconversion of 38:4 DAG to 38:4 PA. The 38:4 species of PA is therefore depicted as the most likely to be involved in tethering RCP at pseudopod tips.
Schematic to summarize DGK-α’s role in RCP-dependent events. Inhibition of αvβ3 or expression of mutant p53273H promotes recruitment of RCP to endosomal α5β1. Association of α5β1 with RCP is not a DGK-dependent event and does not require RCP’s PA-binding C2 domain. RCP/integrin recycling vesicles can then move up and down the pseudopod shaft, and the role of DGK-α is to generate PA species that enable the tethering of RCP at pseudopod tips, an event that requires RCP’s C2 domain. Of the lipid species detected in the mass spectrometric analysis, inhibition or silencing of DGK-α most significantly affects the interconversion of 38:4 DAG to 38:4 PA. The 38:4 species of PA is therefore depicted as the most likely to be involved in tethering RCP at pseudopod tips.
These findings not only offer mechanistic insight into the control of tumor cell invasion and motility but also provide evidence for an essential role for DGK-α and specific molecular species of PA in delivery of recycling vesicles to the plasma membrane at the tips of invasive pseudopodial structures. Whether DGK-α–dependent integrin recycling impacts tumor invasion in vivo remains to be determined, but the contribution made by this kinase to integrin trafficking and signaling needs to be considered in the context of cancer metastasis.
Materials and methods
Cell culture and transfection
Stable clones of H1299 vector– and p53273H-expressing cells were generated by transfecting an empty vector (PCB6+) or construct expressing p53273H (in PCB6+) with transfection reagent (Effectene; QIAGEN). Cells were selected by medium containing 600 mg/ml G418. The p53273H mutant was created using site-directed mutagenesis as described previously (Muller et al., 2009). A2780 cells were grown in RPMI 1640 containing 10% serum, and H1299 and MDA-MB-231 cells were grown in Dulbecco’s minimal essential medium containing 10% serum at 37°C and 10% CO2. All constructs and siRNA oligonucleotides were transfected into cells using the transfection system (Nucleofector Solution T, program A-23, for A2780 cells and Nucleofector Solution V, program X-01, for H1299 cells; Lonza) according to the manufacturer’s instruction. DGK inhibitors I (R59022) and II (R59949), both obtained from Sigma-Aldrich, were used in the presence and absence of serum, respectively. R59949, although more specific for DGKs, cannot be used in the presence of serum owing to its tendency to be sequestered onto serum lipids. The recycling assays were performed on serum-starved cells, whereas all other experiments were conducted in the presence of serum. C6-PA, C6–lyso-PA, and C8-DAG were purchased from Avanti Polar Lipids, Inc. Lipids were dissolved in chloroform, dried under nitrogen, and then dissolved in PBS by sonication before dilution in serum-containing medium to the appropriate concentration (10 µM). siRNA targeting DGK-δ, -ε, -η, -θ, and -ι and primers used for RT-PCR were previously described in Baldanzi et al. (2011).
Plasmid constructs and siRNA sequences
The constructs were previously described as follows: pEGFP-C3 RCP was generated by subcloning the open reading frame of RCP from pGADGH RCP into pEGFP-C3 (Damiani et al., 2004), GFP-RCP200–649 was constructed by ligating the SalI–BamHI fragment from pEGFP-C3 RCP into the SalI–BamHI site of pEGFP-C1 (Lindsay and McCaffrey, 2004), and myc–DGK-α was generating by cloning DGK-α cDNA into pMT2 expression vector (Cutrupi et al., 2000); to obtain myr-myc–DGK-α, the N terminus of myc–DGK-α was modified by adding the Src myristoylation sequence (Baldanzi et al., 2008). The siRNA sequence targeting DGK-α was 5′-GGUCAGUGAUGUCCUAAAG-3′, and the silent mutations introduced into the DGK-α PMT2 rescue construct were C345A, T351C, and A357C. The siRNA targeting RCP was 5′-AGUGAGAACUUGAACAAUG-3′, and the silent mutations introduced into the GFP-RCPWT and GFP-RCP200–649 rescue constructs were T1757C, C1763T, and C1769T.
Immunoprecipitation and Western blotting
Cells were lysed in lysis buffer (200 mM NaCl, 75 mM Tris-HCl, pH 7, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 0.15% [vol/vol] Tween 20, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride). Lysates were passed three times through a 27-gauge needle and clarified by centrifugation at 10,000 g for 10 min at 4°C. Magnetic beads conjugated to sheep anti–mouse IgG (Invitrogen) were bound to the anti-GFP (Abcam) antibody. Antibody-coated beads were incubated with lysates for 2 h at 4°C with constant rotation. Unbound proteins were removed by extensive washing in lysis buffer, and specifically associated proteins were eluted from the beads by boiling for 10 min in Laemmli sample buffer. Proteins were resolved by SDS-PAGE and analyzed by Western blotting as described previously (Roberts et al., 2001). Antibodies used for Western blotting were as follows: anti–α5 integrin (BD), anti-RCP (in-house rabbit antibody raised against RCP379–649), anti-GFP (Abcam), and anti–β-actin (Sigma-Aldrich). Monoclonal mouse anti–DGK-α was provided by W.J. van Blitterswijk (Netherlands Cancer Institute, Amsterdam, Netherlands).
DGK assay
DGK-α was assayed as previously described (Cutrupi et al., 2000). In brief, anti-Myc (Santa Crutz Biotechnology, Inc.) or anti–DGK-α (Abcam) immunoprecipitates were incubated with the immunocomplexes at 30°C with 3 mg/ml sn-1,2-dioleiyl-glycerol (Sigma-Aldrich), 5 mM ATP, 10 µCi/ml γ-[32P]ATP (PerkinElmer), 10 mM MgCl2, 1 mM ZnCl2, and 1.2 mM EGTA in 7.5 mM Hepes, pH 8. After 5 min, the reaction was stopped with 1 N HCl, and lipids were extracted with chloroform/methanol at 1:1 and separated by TLC on sheets coated with 1.3% potassium oxalate and 5 mM EDTA/methanol (3:2) in chloroform/methanol/water/32% ammonium hydroxide (60:47:10:3). γ-[32P]PA was identified by co-migration with nonradioactive PA standards stained by incubation in an iodine chamber. Radioactive signals were detected and quantified with a molecular imager (GS-250; Bio-Rad Laboratories) and Phosphor Analyst software (Bio-Rad Laboratories).
Mass spectrometric determination of DAG and PA species
Cell pellets were spiked with 12:0/12:0-DAG and 12:0/12:0-PA (100 ng each) as internal standards and extracted with 4 ml chloroform/2 ml methanol/2 ml of 0.88% NaCl followed by a second extraction of upper phase with 3 ml synthetic lower phase of chloroform/methanol/0.88% NaCl at 2:1:1. The combined lower phase of lipid extract was dried at room temperature under a vacuum, dissolved in 50 µl chloroform/methanol at 1:1, and subjected to liquid chromatography–mass spectrometry analysis using a liquid chromatography–tandem mass spectrometry system (IT-TOF; Shimadzu). Accurate mass (with mass accuracy of ∼5 ppm) and tandem mass spectrometry were used for molecular species identification and quantification. All solvents used for lipid extraction and liquid chromatography–tandem mass spectrometry analysis were liquid chromatography–mass spectrometry grade obtained from Thermo Fisher Scientific.
Generation of CDM
The CDM was generated as described previously (Cukierman et al., 2001). In brief, gelatin-coated tissue cultureware was cross-linked with glutaraldehyde, quenched, and equilibrated in DME containing 10% FCS. Primary cultured human dermal fibroblasts were seeded at near confluence (∼2 × 104 cells/cm2) and grown for 10 d in DME containing 10% FCS and 50 µg/ml ascorbic acid. Matrices were denuded of living cells by incubation with PBS containing 20 mM NH4OH and 0.5% Triton X-100, and DNA residue was removed by incubation with DNase I. Matrices were blocked with 0.1% heat-denatured BSA before seeding of cells.
Microscopy
For time-lapse imaging, cells were plated onto 6-well plates coated with CDM and incubated at 37°C until cells adhered and began migrating (4 h). Cells were imaged using a 10× Plan Fluor objective, NA 0.3, and an inverted microscope (TE200; Nikon) with a camera (CoolSNAP HQ; Photometrics) and Metamorph software (Molecular Devices) in an atmosphere of 5% CO2 at 37°C for 16 h. Videos were generated using ImageJ (National Institutes of Health). The movement of individual cells was followed using ImageJ software, and the forward migration index was calculated by the Chemotaxis and Migration Tool from ImageJ (version 1.01). For fluorescence time-lapse imaging, cells were plated onto glass-bottomed 3-cm plates coated with CDM and incubated at 37°C for 24 h. Cells were imaged using a U Plan S Apochromat 60× objective, NA 1.35, and an inverted confocal microscope (FluoView FV1000; Olympus) with FluoView software (Olympus) in an atmosphere of 5% CO2 at 37°C. Brightness and contrast of the videos and stills from the videos were adjusted using ImageJ. No nonlinear adjustments were used.
FRAP
0.75 × 105 cells were seeded onto CDM and treated as indicated. The experiments were performed using a confocal microscope (FV1000). Effective photobleaching was achieved with 18% 405-nm laser power with one frame bleach time. Images were captured every second for 60 s. Up to 30 cells per condition were measured. Analysis of unbleached regions showed that total photobleaching was <6% over the time course of recovery.
Receptor internalization and recycling assays
Internalization.
Cells were serum starved for 1–2 h, transferred to ice, washed twice in cold PBS, and surface labeled at 4°C with 0.133 mg/ml NHS-SS-Biotin (Thermo Fisher Scientific) in PBS for 1 h. Labeled cells were washed in cold PBS and transferred to serum-free medium at 37°C in the presence of 0.6 mM primaquine to allow internalization. After internalization, the medium was aspirated, and the dishes were rapidly transferred to ice and washed twice with ice-cold PBS. Biotin was removed from proteins remaining at the cell surface by incubation with a solution containing 20 mM MesNa (sodium 2-mercaptoethanesulphonate) in 50 mM Tris, pH 8.6, and 100 mM NaCl for 15 min at 4°C. MesNa was quenched by addition of 20 mM iodoacetamide for 10 min, and the cells were lysed in 200 mM NaCl, 75 mM Tris, 15 mM NaF, 1.5 mM Na3VO4, 7.5 mM EDTA, 7.5 mM EGTA, 1.5% Triton X-100, 0.75% IGEPAL CA-630, 50 µg/ml leupeptin, 50 µg/ml aprotinin, and 1 mM 4-(2-aminoethyl)benzynesulphonyl fluoride. Lysates were passed through a 25-guage needle and clarified by centrifugation at 10,000 g for 10 min. Levels of biotinylated integrin were determined by capture ELISA as described previously (Roberts et al., 2001). In brief, 96-well plates (MaxiSorp; Life Technologies) were coated overnight with 5 µg/ml anti–α5 integrin antibody (no. 555651) in 0.05 M Na2CO3, pH 9.6, at 4°C and blocked in PBS containing 0.05% Tween 20 (PBS-T) with 5% BSA for 1 h at room temperature. Integrins were captured by overnight incubation of 50 µl cell lysate at 4°C. Unbound material was removed by extensive washing with PBS-T, and wells were incubated with streptavidin-conjugated horseradish peroxidase (GE Healthcare) in PBS-T containing 1% BSA for 1 h at 4°C. After further washing, biotinylated integrins were detected by chromogenic reaction with orthophenylenediamine.
Recycling.
After surface labeling, cells were transferred to serum-free medium for 30 min at 37°C to allow internalization of tracer. Cells were returned to ice and washed twice with ice-cold PBS, and biotin was removed from proteins remaining at the cell surface by reduction with MesNa. The internalized fraction was then chased from the cells by returning them to 37°C in serum-free medium. After recycling, the cells were returned to ice, and biotin was removed from recycled proteins by a second reduction with MesNa. Biotinylated integrins were then determined by capture ELISA as described previously (Roberts et al., 2001). The antibodies used for capture ELISA detection of α5β1 integrin (clone VC5) and EGFR (clone EGFR.1) were obtained from BD. cRGDfNMeV and cRGDfV were added to the cells only during the recycling period.
Transferrin recycling.
To measure recycling of internalized transferrin, serum-starved cells were incubated with 125I-transferrin (0.1 µCi/well) for 1 h at 4°C in PBS with 1% BSA. The tracer was allowed to internalize for 30 min at 37°C. Tracer remaining at the cell surface was removed by incubation with an excess of 10 µg/ml of unlabeled transferrin at 4°C for 1 h, and the tracer was allowed to recycle at 37°C in serum-free medium supplemented with 10 µg/ml of unlabeled transferrin. To provide a positive control for inhibition of recycling, 0.3 mM primaquine was included in the recycling period only, and 120 µM chloroquine was added during both the internalization and recycling periods. The quantity of 125I recycled into the medium is expressed as a percentage of the number of counts incorporated during the internalization period.
Inverted invasion assays
Inverted collagen I invasion assays were performed as described previously (Hennigan et al., 1994; Caswell et al., 2008). In brief, collagen supplemented with 25 µg/ml fibronectin as indicated was allowed to polymerize in transwell inserts (Corning) for 1 h at 37°C, cells were seeded on the opposite side of the filter, and 10% FBS + 30 ng/ml EGF was placed on top of the matrix. 2.5 µM cRGDfV was added to the collagen before plug polymerization and also in the medium. 48 h after seeding, migrating cells were stained with calcein acetoxymethyl ester and visualized by confocal microscopy with serial optical sections being captured at 15-µm intervals with a microscope (20× HC Plan Fluotar objective, NA 0.5; SP2; Leica) using LCS software (Leica).
Online supplemental material
Fig. S1 shows that the expression of DGK-α is reduced upon transfection of an siRNA against DGK-α in A2780, H1299, and MDA-MB-231 cells, DGK-α is not required for the internalization of α5β1 or TfnR, and inhibition of αvβ3 or expression of mutant p53 did not affect the enzymatic activity or the localization of DGK-α. Fig. S2 shows that DGK activity and the C2 domain of RCP are not required for the interaction of RCP with α5β1. Fig. S3 shows the effects of DGK-α knockdown on speed and persistence of cell migration, the effects of Rho kinase inhibition on random migration induced by inhibition of αvβ3 or expression of mutant p53, and the role of other DGK isoforms in pseudopod elongation. Fig. S4 shows that the accumulation of RCP at the tip of pseudopods does not reflect increased cytoplasmic volume at these cellular extremities. Fig. S5 shows that PLD activity is not required for RCP recruitment to pseudopod tips. Videos 1–3 show GFP-actin localization in A2780 cells transfected with a nontargeting siRNA (Video 1), treated with cRGDfNMeV in presence of a nontargeting siRNA (Video 2), or an siRNA against DGK-α (Video 3). Videos 4 and 5 show GFP-RCP localization in A2780 cells treated with cRGDfV and transfected with a nontargeting siRNA (Video 4) or an siRNA against DGK-α (Video 5). Videos 6 and 7 show GFP-RCP localization in p53273H-expressing H1299 cells transfected with a nontargeting siRNA (Video 6) or an siRNA against DGK-α (Video 7). Video 8 shows the recovery from photobleaching of GFP-RCP in A2780 cells transfected with a nontargeting siRNA or an siRNA against DGK-α. Videos 9 and 10 show the localization of GFP-RCP200–649 in A2780 (Video 9) and p53273H-expressing H1299 cells (Video 10).
GFP-RCP localization in A2780 cells treated with cRGDfV and transfected with an siRNA against DGK-α. A2780 cells were transfected with GFP-RCP in combination with an siRNA targeting DGK-α and plated onto CDM. Cells were treated with 2.5 µM cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-RCP localization in A2780 cells treated with cRGDfV and transfected with an siRNA against DGK-α. A2780 cells were transfected with GFP-RCP in combination with an siRNA targeting DGK-α and plated onto CDM. Cells were treated with 2.5 µM cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-RCP localization in p53273H-expressing H1299 cells transfected with an siRNA against DGK-α. p53273H-expressing H1299 cells were transfected with GFP-RCP in combination with an siRNA targeting DGK-α. Cells were plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
GFP-RCP localization in p53273H-expressing H1299 cells transfected with an siRNA against DGK-α. p53273H-expressing H1299 cells were transfected with GFP-RCP in combination with an siRNA targeting DGK-α. Cells were plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
The localization of GFP-RCP200–649 in A2780. A2780 cells were transfected with GFP-RCP200–649, plated onto CDM, and treated with cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
The localization of GFP-RCP200–649 in A2780. A2780 cells were transfected with GFP-RCP200–649, plated onto CDM, and treated with cRGDfV, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
The localization of GFP-RCP200–649 in p53273H-expressing H1299 cells. p53273H-expressing H1299 cells were transfected with GFP-RCP200–649 and plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
The localization of GFP-RCP200–649 in p53273H-expressing H1299 cells. p53273H-expressing H1299 cells were transfected with GFP-RCP200–649 and plated onto CDM, and time-lapse videos were recorded using an inverted confocal microscope (FV1000). Frames were taken every second for 1 min.
Acknowledgments
The work at the Beatson Institute in the J.C. Norman and K.H. Vousden laboratories is funded by Cancer Research UK. A. Graziani is funded by a grant from the Association for International Cancer Research and from Fondazione Cariplo. M.W. McCaffrey is supported by a Science Foundation Ireland Programme grant (reference 09/IN.1/B2629). A European Molecular Biology Organization short-term fellowship (ASTF 391.00-2008) to E. Rainero enabled this work to be initiated. E. Rainero is currently funded by the West of Scotland Women’s Bowling Association. Work at the Babraham Institute is funded by the Biotechnology and Biosciences Research Committee.
References
Author notes
P.T. Caswell’s present address is Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester M13 9PT, England, UK.


![Figure 1. DGK-α is required for α5β1 recycling in response to αvβ3 inhibition or expression of mutant p53. (A–E) The recycling of internalized α5β1 (A–D) or 125I-transferrin (TFN; E) was determined in A2780 (A–C and E) or empty vector and mutant p53-expressing H1299 (D) cells. In A and E, A2780 cells were transfected with a nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα). In C, A2780 cells were transfected with an siRNA targeting DGK-α or siRNA targeting DGK-α in combination with an siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) or empty vector control (pMT2). (B and D) The DGK inhibitor, 1 µM R59949, or DMSO control was included during the internalization and recycling periods as indicated. (E) 120 µM chloroquine (CQ) was included during the internalization and recycling periods, whereas 0.3 mM primaquine (PQ) was only in the recycling period. (A–C) 36 nM cRGDfNMeV (A and B) or 2.5 µM cRGDfV (C) was included as indicated during the recycling period. Values are means ± SEM (A–D) and means ± SD (E) of at least nine replicates. †††, P < 0.0001 for si-con+cRGDfNMeV versus si-DGK-α+cRGDfNMeV; Mann–Whitney test. ***, P < 0.0001 for si-DGK-α+cRGDfNV versus si-DGK-α/DGK-α(resc)+cRGDfV; Mann–Whitney test. §§, P < 0.0001 for DMSO+cRGDfNMeV versus R59949+cRGDfNMeV; Mann–Whitney test. ‡‡‡, P < 0.0001 for p53273H DMSO versus p53273H R59949; Mann–Whitney test. $$$, P = 0.0002 for vehicle versus primaquine/chloroquine; Mann–Whitney test.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112_fig1.jpeg?Expires=1771330469&Signature=lCFJ0Zok6-eLfsJkQ0rSid56V7FFQd10m5DpoIHxVm0NfD6rtIGgfS~ycy5J0OhMWI6qQvpCh6sgsHluq06XLZtSsHE1lISFLMjCevZo9id01mcA0L8Fo2oAxQlbUUjftV2Dk7c2J3HT6e~egS7LdluXoLLxrQRlNxWAtWghcD9iJm4h8UlErsx7e8YD35R7fCBvfcQKUTvTLqeqWLrLVB8MCCFYmtPoECveGqweZr48nMIrMZRsXEXeRZTxfDVQMY-MXViewr-67H1~Cbz8dcN7oj3-B9fpjDs186xbuXbmGhJGEGhxKHUTVRs49tMATItWOM15r8DU7V4kdgkf~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. DGK-α is required for cRGDfNMeV- and mutant p53-driven random migration on 2D surfaces. (A) A2780 cells were transfected with GFP-actin in combination with either nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and allowed to grow to confluence on glass surfaces. Confluent monolayers were wounded with a plastic pipette tip, and the cells were allowed to migrate into the wound in the absence (−) or presence of 36 nM cRGDfNMeV. Time-lapse videos were recorded (Videos 1, 2, and 3), and representative stills are displayed. Bar, 20 µm. (B) A2780 cells were transfected with nontargeting siRNA or an siRNA targeting DGK-α, grown to confluence, and wounded as for A. The cells were observed by time-lapse video microscopy, the movement of individual cells was followed using cell-tracking software, and representative trackplots of trajectories described by cells during the first 8 h of migration into the wound are presented. (C and D) A2780 cells were transfected with nontargeting siRNA, an siRNA targeting DGK-α, or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)), and their migration into scratch wounds in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV both with or without 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots. (E–G) H1299 cells expressing p53273H or a control vector were transfected as for C and D, and their migration into scratch wounds in the presence and absence of 10 µM R59022 or vehicle control (DMSO) was recorded as for B. The forward migration index was extracted from the trackplots as for C. (C, D, F, and G) Data are represented as box and whisker plots (black lines shows medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112r_fig2.jpeg?Expires=1771330469&Signature=OFM4XyUK-BMeNKwFQECYf5tvLzKagYc2SK11Dr7SXF3W-Y4LWEMyDQeKxpsORWukD3MQG-8-3l5C7Tp6LjnmiQjjCh8wtfhCyEvGC2ZSjYwRwyhzO03jrzxPEIEpuGouNrJKjKWEY1Dx6Sb~~xb-VWQe6gxWGOry6DbXoTaSAZDzlf-AXw0jnosPEie1Rm4oC1zTlLycB3Qv~~O13qKGNiC1IY~Y1yYWqsN0QcztYgtBqGpxhXsC1Xz0MxYEqEvBVH65qdXVR9lYmHeOyJEsQY4LjnG~5sdG1xTuurVub544qbjjCcuc~70EcsOIvla8o8Lg-uAoUVScBlxGuerMtQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. DGK-α is required for pseudopod elongation evoked by inhibition of αvβ3 or expression of mutant p53. (A–D) A2780 cells (A and B) or vector and p53273H-expressing H1299 cells (C and D) were transfected with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα) or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)) and plated onto CDM. Time-lapse videos were recorded in the absence (−) or presence of 36 nM cRGDvNMeV or 2.5 µM cRGDfV in combination with 10 µM R59022 or its vehicle (DMSO) and 0.5% butan-1-ol (But-1-ol; or secondary alcohol control [0.5% butan-2-ol]). Representative stills are displayed. (A and C) The arrows indicate elongated pseudopods. Bars, 20 µm. In B and D, the distance between the center of the nucleus and the cell front (with respect to the direction of migration) was measured using ImageJ. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112r_fig3.jpeg?Expires=1771330469&Signature=5K6pL~SN~xuLigwm3~EqvRLrOjQfUi4rBtLOlarIoxbAJa-eRbabBDHd7fH9-OxY6HQ2OpMxBPHgJxaWpfoylOUsdh-4SPvfvJwJq3ENAf3Jforyqk5zPskMNVG588LZE-uAbfv6JC-fEiQuOtaLJdfzhAA~2FBX-ZIPSklJV2B2NHCuzul6QPk7PTYdPet6OUKUXyFlcDPZGyK-BqUqfQt6P8Qici5xy6drQvw8Dt52IZcu746Hudw7Qk~WIlrzXyqvYZmYcicMCNPnbjQdDGTjcySoue-ZdwoTpr2q3JUEkS5-N0saBO8~5KLXHSLjrDQDOBa7rm4Rk2O1HR4weA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. DGK-α is necessary for RCP recruitment to pseudopod tips during migration on CDM. (A and B) A2780 cells (A) or vector and p53273H-expressing H1299 cells (B) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 36 nM cRGDfNMeV or 2.5 µM cRGDfV. Images were captured at one frame/second over a period of 60 s, and videos were generated from these (Videos 4, 5, 6, and 7). Single-section confocal image stills corresponding to individual frames from these videos are presented. The arrows indicate the direction of migration, and the portion of the cell within the white square is presented in the bottom images. Bars, 10 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112_fig4.jpeg?Expires=1771330469&Signature=Hhov3DfXbaKAqO6N3rQdNBiyGpq1o1c8oGqjoQvA5X0wyBuinXPTUrsy-1p4gX1LLpfUBrAl1wEdb0X6y2dS8EFevErZX6KokQNuXW6t-brdVz6EbXf4eIfEQhLa17EKj9Ve~ZXE1nnHDG5MpUWjT~GycVKalk-Lc3oSLQ3D8rGIfJs8vbNO1xdMq-VUX4Xh8e5savoZOPNQLogGDEwmS8efAiqXwJzUFb5n3a~gO--6E8ZJhnYvLV8oXD3Jyyzp~AbZne37amK2AXcVpvjw~YMQb2TPOH0b~9HrFRzRjgvv-cPOKpkEiMw7Gjelzv4CvFRvP46InlbYY-q5GMU1TA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. DGK-α is required for the tethering of RCP vesicles at pseudopod tips. (A–C) A2780 (A and B) or vector and p53273H-expressing H1299 cells (C) were transfected with GFP-RCP in combination with nontargeting siRNA (siRNA control [si-con]), an siRNA targeting DGK-α (si-DGKα), or siRNA targeting DGK-α in combination with siRNA-resistant DGK-α (si-DGKα/DGKα(resc)). Cells were plated onto CDM, mounted onto the stage of a confocal microscope (FV1000), and incubated in the absence (−) or presence of 2.5 µM cRGDfV. Photobleaching was performed as described in the Materials and methods section. Stills from a video (Video 8) at the indicated time points are shown. (A) The arrows indicate the direction of cell migration, whereas the circles highlight the bleaching region. Recovery curves were generated using the FV10-ASW software (Olympus), and the immobile fraction (percentile) was extracted from these. Values are means ± SEM; ***, P < 0.0001. Bar, 10 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112_fig5.jpeg?Expires=1771330469&Signature=f-QTR4n4hn9Q1tAwNRdfFsP4gbcgEw~QZQp-luzXlb-bV1g3vu5ar1y5ulID~29lrDSe3tODRkxt6fLCw4ryOyjVLV283bZ~hzwwaNOhE5nItlgzMh~obftjuEbDKRgF-CT2Nufea5DfWK1OJniHTIvm2DCy6scVDsCgdJl3MB52Y-R2GACsb8uS~qxBJ~8WM7qZ5mC0-XwjcFqsef6K3bxEq7tSLPoRVPPGFwRkfPgG5wv1A~SoJKmzEAxCyZ--wvRY1E6U~j~uxUbwW~TVGDDaV6lSVy7iQVt0YTKGPmmn~QwKNPiAzYOAe6mh3ehs~ywPy1f5QjWH~p11rHZTLw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
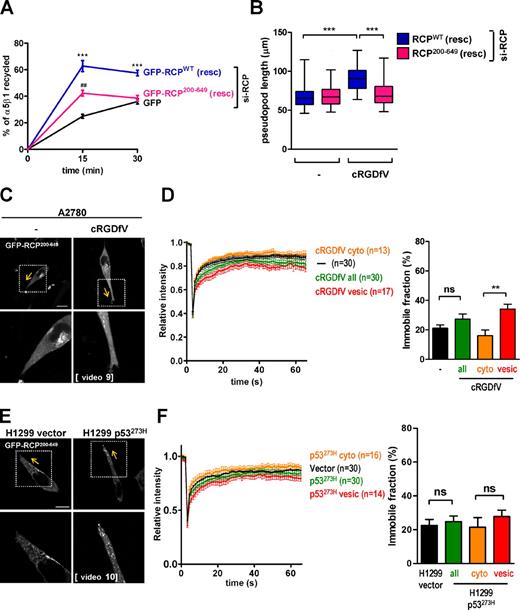
![Figure 7. DGK-α is required for invasion into fibronectin-containing collagen I hydrogels evoked by inhibition of αvβ3 or expression of mutant p53. (A–C) A2780 (A), vector, and p53273H-expressing H1299 (B) or MDA-MB-231 (C) cells were transfected with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting DGK-α (si-DGKα) and plated onto the bottom of collagen I plugs. In A, the collagen plugs were formed in the presence and absence of 25 µg/ml fibronectin (FN) as indicated, and in B and C, 25 µg/ml fibronectin was included for all conditions. The cells were allowed to invade into the plugs for 48 h toward a gradient of 30 ng/ml EGF and 10% FCS. 2.5 µM cRGDfV was added to both the upper and lower chambers of the inverted collagen plug as indicated in A. Invading cells were stained with calcein acetoxymethyl ester and visualized by confocal microscopy. Serial optical sections were captured at 15-µm intervals and are presented as a sequence in which the individual optical sections are placed alongside one another with increasing depth from left to right as indicated. Migration was quantitated by measuring the fluorescence intensity of cells penetrating the collagen/fibronectin to depths of 60 µm (A) or 30 µm (B and C) and greater and expressing this as a percentage of the total fluorescence intensity of all cells within the plug. Data are represented as box and whiskers plot (black lines show medians; whiskers show minimum to maximum values); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112_fig7.jpeg?Expires=1771330469&Signature=NhvL7vLYebcTj7KksVfnDg9tbw5Y4jeeVyKxTE5R-5AtfCFHbcmlupZopECM43hZ0EWU8Mq8OM6ZQM1NFnt4lWJ5MeMxxP~Ja~DYPlWew4V-7JiITm8uebGzcyJUU1nARJSKDrMoQL1lHmg4~trIA727KygIe8LdjODwX9pAu-pDj99A2x6GkVshJKMKpuYxzm77YC36zMSEu24B5FDbAytL1DqkKLkaw4juRTmqLVAehPVqTkKk9hdoEUeJ7SPlkUCdnAUdXv61BXD~jcYyN9dA9-twuxOTAImMdv0LsTueMGX6VMd7RdBfPOCwPAG793yyX2lBxdYbdYs92H94Lw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 8. myr–DGK-α promotes RCP-dependent integrin recycling, invasion, and recruitment of RCP to pseudopod tips. (A) A2780 cells were plated onto the CDM, time-lapse videos were collected in the absence (–) or presence of lyso-PA (LPA), DAG, PA, and R59022 (all at 10 µM), and pseudopod length was determined as for Fig. 3. Blue boxes indicate addition of PA. Data are represented as box and whiskers plots (black lines show medians; whiskers: 5–95 percentile); n = 3 independent experiments. ***, P < 0.0001; Mann–Whitney test. (B) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector), and the recycling of α5β1 was determined as for Fig. 1. **, P = 0.004; ***, P = 0.0008; Mann–Whitney test. (C) A2780 cells were transfected with myr–DGK-α or pMT2 together with nontargeting siRNA (siRNA control [si-con]) or an siRNA targeting RCP (si-RCP) in combination with GFP, siRNA-resistant GFP-RCPWT (GFP-RCPWT(resc)), or siRNA-resistant GFP-RCP200–649 (GFP-RCP200–649(resc)). Cells were plated onto the CDM, and pseudopod length was determined as for Fig. 3. ***, P = 0.0008; Mann–Whitney test. (D) A2780 cells were transfected with myr–DGK-α or pMT2 (empty vector) in combination with GFP-RCP. Transfected cells were plated onto CDM, and videos were captured as for Fig. 4. Representative stills are displayed. The yellow arrows indicate the direction of migration and the portion of the cell within the white square, which is presented at the bottom. Bar, 20 µm. (E–G) A2780 cells were transfected with myr–DGK-α or pMT2 in combination with either nontargeting siRNA or an siRNA targeting RCP and plated into inverted invasion assays in the presence (E and G) or absence (F) of 25 µg/ml fibronectin (FN) as indicated. Invasion was determined as for Fig. 7. Data are represented as box and whiskers plots (black lines show medians; whiskers: minimum to maximum). ***, P < 0.0001; Mann–Whitney test. Values are means ± SEM.](https://cdn.rupress.org/rup/content_public/journal/jcb/196/2/10.1083_jcb.201109112/6/m_jcb_201109112_fig8.jpeg?Expires=1771330469&Signature=btINB2O2TVocLs8E1hMntUSwCimJY4-Jc~whncB1oUUlOU900iinh~l2x9XLaEnHyCjXQsKL3ZZ~g-7kjwLCNzOQOyltvXIaVDA4O8L7h17iWHsNBnPLdKDg2R9up4ywyvXpAQAyKmHJ-FjHnWpcBi4h1tZOXSy8-67HnHcq3AEQM0i1ZsigCb4d7~LbkjIZMIy6YyyVvT~YCfvk6pxYCWF6tNTMT2Ckm8QR5sWfAQDkof880NlDbuB72c29HIfLE-w6zMf6gH3jq8GVMijhdmdi6NEJH6Ri1589ySYC~-V3t9bhfNjS9g73UJlD4D9hGiTHJsfFSsavDzr30ZfeQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



