Membrane contact sites between organelles are organized by protein bridges. Among the components of these contacts, the VAP family comprises ER–anchored proteins, such as MOSPD2, that function as major ER–organelle tethers. MOSPD2 distinguishes itself from the other members of the VAP family by the presence of a CRAL-TRIO domain. In this study, we show that MOSPD2 forms ER–lipid droplet (LD) contacts, thanks to its CRAL-TRIO domain. MOSPD2 ensures the attachment of the ER to LDs through a direct protein–membrane interaction. The attachment mechanism involves an amphipathic helix that has an affinity for lipid packing defects present at the surface of LDs. Remarkably, the absence of MOSPD2 markedly disturbs the assembly of lipid droplets. These data show that MOSPD2, in addition to being a general ER receptor for inter-organelle contacts, possesses an additional tethering activity and is specifically implicated in the biology of LDs via its CRAL-TRIO domain.
Introduction
The ER, a major membrane-bound organelle of eukaryotic cells, ensures diverse functions such as lipid synthesis, protein synthesis and folding, calcium storage, etc. The ER has a network architecture spreading throughout the cytosol, and is in physical contact with other organelles such as mitochondria, endosomes/lysosomes, autophagic structures, peroxisomes, lipid droplets, and the plasma membrane (Wu et al., 2018). These contacts, which do not lead to organelle fusion, are named membrane contact sites. They are scaffolded by protein bridges connecting the two membranes and involving protein–membrane and protein–protein interactions (Gatta and Levine, 2017).
Many molecular players involved in the formation of contact sites have been identified in recent years (Di Mattia et al., 2020b; Prinz et al., 2019; Wu et al., 2018). A few of them act as general receptors that recruit a variety of tethering partners and hold a central role in the biology of inter-organelle contacts. The ER-resident VAMP-associated protein (VAP) protein family plays a major role in the formation of contacts between the ER and the other organelles, as well as to the plasma membrane. This family comprises six proteins divided into two subfamilies. The first sub-family comprises VAP-A, VAP-B, and motile sperm domain-containing protein 2 (MOSPD2) that are anchored in the ER membrane by a transmembrane helix (Murphy and Levine, 2016; Di Mattia et al., 2018). VAP-A, VAP-B, and MOSPD2 act as ER receptors that bind multiple protein ligands, either cytosolic or localized on the surface of other organelles, to connect them with the ER (Di Mattia et al., 2018; Murphy and Levine, 2016). VAP-A, VAP-B, and MOSPD2 have a major sperm protein (MSP) domain exposed to the cytosol; this domain hooks proteins that possess a small linear motif named FFAT (two phenylalanines in an acidic tract; Mikitova and Levine, 2012; Loewen et al., 2003; Di Mattia et al., 2018). Recently, a novel subfamily comprising MOSPD1 and MOSPD3 was characterized; these proteins have an MSP domain that recognizes FFNT (two phenylalanines in a neutral tract) motifs (Cabukusta et al., 2020). Lastly, the sixth family member named CFAP65 (Cilia- and flagella-associated protein 65) is involved in ciliogenesis (Zhao et al., 2022).
The functions of VAP-A and VAP-B are well studied; they are central proteins for the formation of inter-organelle contacts and function in lipid transport, ion homeostasis, and autophagy (Murphy and Levine, 2016; Wilhelm et al., 2017; Zhao et al., 2018; Mesmin et al., 2013; Kirmiz et al., 2018; Johnson et al., 2018). In contrast, the function of MOSPD2 is still elusive. MOSPD2 is a genuine member of the VAP family: It shares with VAP-A and VAP-B a large number of tethering partners and promotes the formation of contacts between the ER and many organelles (Di Mattia et al., 2018, 2020a). Unlike VAP-A, VAP-B, MOSPD1, and MOSPD3, which only possess an MSP domain in addition to their transmembrane (TM) domain, MOSPD2 possesses an additional cytosolic domain named CRAL-TRIO (cellular retinaldehyde-binding protein [CRALBP] and triple functional domain protein [TRIO]) at its amino terminus. The CRAL-TRIO domain (also called the Sec14 domain) is present in 28 proteins in human and contains a hydrophobic pocket that, in the case of Sec14p and α-tocopherol transfer protein, can bind/transport small lipophilic molecules such as phospholipids or tocopherols (Chiapparino et al., 2016). Knowing that MOSPD2 contains a CRAL-TRIO domain, we hypothesized that it may have a broader function than the VAP proteins. Here, we show that in addition to serving as a VAP-like tether that establishes ER–organelle contacts through protein–protein interactions, MOSPD2 also tethers the ER to lipid droplets (LDs) by protein–membrane interactions. LDs are ubiquitous organelles that serve as universal lipid stores in cells; they consist of a neutral lipid oil core surrounded by a monolayer of phospholipids associated with peripheral proteins (Olzmann and Carvalho, 2019; Thiam and Beller, 2017; Thiam et al., 2013). In this report, we found that the absence of MOSPD2 compromises LD assembly, showing that MOSPD2 is involved in the biology of LDs.
Results
MOSPD2 is involved in the biology of LDs
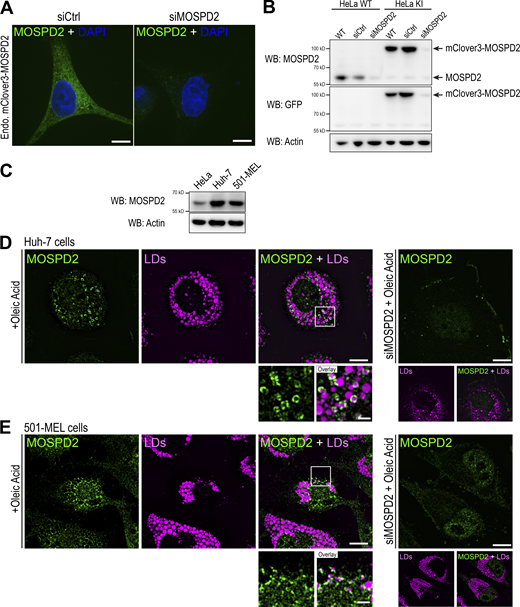
MOSPD2 is an ER-resident protein (Di Mattia et al., 2018). When expressed in HeLa cells, GFP-labeled MOSPD2 exhibited a distinctive reticular localization pattern throughout the cytoplasm and co-localized with the ER marker Calnexin (Fig. 1 A, a). Remarkably, in about half of the cells, MOSPD2 was additionally found in ring- and comma-shaped structures that were also positive for the ER marker Calnexin (Fig. 1 A, b and c). This shows that in some cells, MOSPD2 can be enriched in sub-regions of the ER. Importantly, VAP-A and VAP-B were never observed in similar ring-like structures, suggesting that only MOSPD2 can specifically populate these ER subdomains (Fig. 1 B). We then checked whether these MOSPD2-enriched areas corresponded to contact sites between the ER and a particular organelle by performing co-localization experiments using markers of LDs (Nile Red; Fig. S1 A), early endosomes (EEA1), late endosomes/lysosomes (Lamp1), mitochondria (OPA1), and Golgi (GM130; Fig. S2, A–D). MOSPD2-positive rings did not overlap with endosomes, Golgi, and mitochondria. In contrast, ring- and comma-shaped MOSPD2-positive structures were found to be around LDs labeled with Nile Red. To confirm this observation, LD biogenesis in HeLa cells was stimulated by oleic acid (OA) treatment (Listenberger and Brown, 2007). After this treatment, most HeLa cells had numerous and large LDs massively surrounded by ring- and comma-shaped structures positive for MOSPD2 (Fig. 1 C). This accumulation of MOSPD2-positive ER around LDs was also found in other tested cell lines, including hepatocytes (Huh7), melanoma (501-mel), and mammary epithelial (MCF7) cells (Fig. S1, B–D).
MOSPD2 is an ER-resident protein found enriched around LDs. (A) HeLa cells expressing GFP-MOSPD2 (green) were labeled with an anti-Calnexin antibody (magenta). GFP-MOSPD2 exhibited a reticular pattern (a), with additional ring- and comma-shaped structures (b). c: Percentage of cells with GFP-positive ring- or comma-shaped structures in the absence of treatment (NT) or after OA treatment. Mean ± SD; n = 3 independent experiments (NT: 300 cells, OA: 136 cells). (B) Confocal images of HeLa cells expressing GFP-VAP-A (green; a) and GFP-VAP-B (green; b), labeled as in A. c: Quantification as in panel A, c of VAP-A and VAP-B expressing cells. Mean ± SD; n = 3 independent experiments (GFP-VAP-A: NT: 119 cells, OA: 126 cells; GFP-VAP-B: NT: 140 cells, OA: 141 cells). (C) a: HeLa cells expressing GFP-MOSPD2 were treated with OA (400 µM, overnight) and LDs were labeled with Nile Red. b: 3D reconstruction from confocal images of MOSPD2-positive ER (green) around LDs (magenta). Images generated with the surface representation tool of the Chimera software (Pettersen et al., 2004). Scale bar: 500 nm. (D) a: Live imaging of CRISPR/Cas9-edited HeLa cells expressing mClover3-MOSPD2 (green) at the endogenous level, treated with OA, and labeled with LipidTOX (magenta). b: 3D reconstruction from confocal images of MOSPD2-positive ER (green) around LDs (magenta) using Imaris (white dashed rectangle from overlay panel). Scale bar: 500 nm. (A, B, and D) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bars: 10 μm (insets, 2 μm). (C) Confocal microscope (Leica SP5, 63× NA 1.4) images, scale bars: 10 μm (insets, 2 μm).
MOSPD2 is an ER-resident protein found enriched around LDs. (A) HeLa cells expressing GFP-MOSPD2 (green) were labeled with an anti-Calnexin antibody (magenta). GFP-MOSPD2 exhibited a reticular pattern (a), with additional ring- and comma-shaped structures (b). c: Percentage of cells with GFP-positive ring- or comma-shaped structures in the absence of treatment (NT) or after OA treatment. Mean ± SD; n = 3 independent experiments (NT: 300 cells, OA: 136 cells). (B) Confocal images of HeLa cells expressing GFP-VAP-A (green; a) and GFP-VAP-B (green; b), labeled as in A. c: Quantification as in panel A, c of VAP-A and VAP-B expressing cells. Mean ± SD; n = 3 independent experiments (GFP-VAP-A: NT: 119 cells, OA: 126 cells; GFP-VAP-B: NT: 140 cells, OA: 141 cells). (C) a: HeLa cells expressing GFP-MOSPD2 were treated with OA (400 µM, overnight) and LDs were labeled with Nile Red. b: 3D reconstruction from confocal images of MOSPD2-positive ER (green) around LDs (magenta). Images generated with the surface representation tool of the Chimera software (Pettersen et al., 2004). Scale bar: 500 nm. (D) a: Live imaging of CRISPR/Cas9-edited HeLa cells expressing mClover3-MOSPD2 (green) at the endogenous level, treated with OA, and labeled with LipidTOX (magenta). b: 3D reconstruction from confocal images of MOSPD2-positive ER (green) around LDs (magenta) using Imaris (white dashed rectangle from overlay panel). Scale bar: 500 nm. (A, B, and D) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bars: 10 μm (insets, 2 μm). (C) Confocal microscope (Leica SP5, 63× NA 1.4) images, scale bars: 10 μm (insets, 2 μm).
MOSPD2 is enriched around LDs in different cell lines. (A) HeLa cells expressing GFP-MOSPD2 (green) and not treated with OA were labeled with Nile Red to stain LDs (magenta). (B–D) Localization of GFP-MOSPD2 (green) in Huh-7 (B), 501-MEL (C), and MCF7 (D) cells. Cells were either treated with OA (right) or not treated (left). LDs were stained using Nile Red (magenta). Subpanels on the right are higher magnification images of the outlined areas. The overlay panel shows merged channels. The coloc panel displays a colocalization mask in which pixels of the green and magenta channels that co-localize are shown in white. Linescan shows fluorescence intensities of the green and magenta channels along the white arrow from the overlay subpanel. Black rectangles indicate the position of LDs. (A–D) Scale bars: 10 µm (insets, 2 µm). Confocal microscope (Leica SP5, 63× NA 1.4) images.
MOSPD2 is enriched around LDs in different cell lines. (A) HeLa cells expressing GFP-MOSPD2 (green) and not treated with OA were labeled with Nile Red to stain LDs (magenta). (B–D) Localization of GFP-MOSPD2 (green) in Huh-7 (B), 501-MEL (C), and MCF7 (D) cells. Cells were either treated with OA (right) or not treated (left). LDs were stained using Nile Red (magenta). Subpanels on the right are higher magnification images of the outlined areas. The overlay panel shows merged channels. The coloc panel displays a colocalization mask in which pixels of the green and magenta channels that co-localize are shown in white. Linescan shows fluorescence intensities of the green and magenta channels along the white arrow from the overlay subpanel. Black rectangles indicate the position of LDs. (A–D) Scale bars: 10 µm (insets, 2 µm). Confocal microscope (Leica SP5, 63× NA 1.4) images.
Colocalization of MOSPD2 with different organelles. (A–D) Colocalization in HeLa cells transfected with GFP-MOSPD2 (green) of MOSPD2 and endogenous EEA1 (A, magenta), Lamp1 (B, magenta), OPA-1 (C, magenta), and GM130 (D, magenta). Subpanels on the right are higher magnification images of the outlined areas. The overlay panel shows merged channels. The coloc panel displays a colocalization mask in which pixels of the green and magenta channels that co-localize are shown in white. Linescan shows fluorescence intensities of the green and magenta channels along the white arrow from the overlay subpanel. Black rectangles indicate the position of early endosomes (EE; A), late endosomes (LE; B), mitochondria (Mito.; C), and the Golgi apparatus (Golgi; D). Scale bars: 10 µm (insets, 2 µm). Images were acquired on a confocal microscope (Leica SP5, 63× NA 1.4).
Colocalization of MOSPD2 with different organelles. (A–D) Colocalization in HeLa cells transfected with GFP-MOSPD2 (green) of MOSPD2 and endogenous EEA1 (A, magenta), Lamp1 (B, magenta), OPA-1 (C, magenta), and GM130 (D, magenta). Subpanels on the right are higher magnification images of the outlined areas. The overlay panel shows merged channels. The coloc panel displays a colocalization mask in which pixels of the green and magenta channels that co-localize are shown in white. Linescan shows fluorescence intensities of the green and magenta channels along the white arrow from the overlay subpanel. Black rectangles indicate the position of early endosomes (EE; A), late endosomes (LE; B), mitochondria (Mito.; C), and the Golgi apparatus (Golgi; D). Scale bars: 10 µm (insets, 2 µm). Images were acquired on a confocal microscope (Leica SP5, 63× NA 1.4).
To examine whether endogenous MOSPD2 can be seen in association with LDs, we generated a reporter cell line. The endogenous MOSPD2 gene was modified in HeLa cells using the CRISPR/Cas9 method to fuse the coding sequence of the fluorescent protein mClover3 in frame with that of MOSPD2 (Fig. S3, A and B). As observed by expressing GFP-MOSPD2, endogenous mClover3-MOSPD2 was present in structures in contact with LDs (Fig. 1 D). Next, in Huh-7 and 501-MEL cells, which express higher levels of endogenous MOSPD2 than HeLa cells (Fig. S3 C), we could analyze this protein using a specific antibody (Fig S3, D and E). We found that the endogenous MOSPD2 did accumulate in ring- and comma-shaped structures around some LDs in Huh7 and 501-MEL cells treated with OA (Fig. S3, D and E). These data indicate that endogenous MOSPD2 is localized around a subset of LDs.
Characterization of CRISPR/Cas9 knock-in HeLa cells and endogenous localization of MOSPD2. (A) Live confocal images of HeLa cells expressing mClover3-MOSPD2 (green) at the endogenous level and transfected with control siRNAs (siCtrl) and siRNAs targeting MOSPD2 (siMOSPD2) to confirm the specificity of mClover3 signal. Scale bars: 10 µm. (B) Western blot (WB) analysis of MOSPD2 expression in WT (HeLa WT) and mClover3-MOSPD2 knock-in (HeLa KI) HeLa cells transfected with control siRNAs (siCtrl) and siRNAs targeting MOSPD2 (siMOSPD2). mClover3 was detected using anti-GFP antibodies. (C) Western blot analysis of MOSPD2 expression in HeLa, Huh-7, and 501-MEL cells. (D and E) Colocalization of endogenous MOSPD2 (labeled with anti-MOSPD2, in green) and LDs (labeled with LipidTOX, in magenta) in Huh-7 cells (D) or 501-MEL (E) after OA treatment. Panels on the right show the background signal of anti-MOSPD2 antibodies in cells transfected with siRNAs targeting MOSPD2 (siMOSPD2). Subpanels show the LD staining. Left: Subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels. Scale bars: 10 µm (insets, 2 µm). (A, D, and E) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Source data are available for this figure: SourceData FS3.
Characterization of CRISPR/Cas9 knock-in HeLa cells and endogenous localization of MOSPD2. (A) Live confocal images of HeLa cells expressing mClover3-MOSPD2 (green) at the endogenous level and transfected with control siRNAs (siCtrl) and siRNAs targeting MOSPD2 (siMOSPD2) to confirm the specificity of mClover3 signal. Scale bars: 10 µm. (B) Western blot (WB) analysis of MOSPD2 expression in WT (HeLa WT) and mClover3-MOSPD2 knock-in (HeLa KI) HeLa cells transfected with control siRNAs (siCtrl) and siRNAs targeting MOSPD2 (siMOSPD2). mClover3 was detected using anti-GFP antibodies. (C) Western blot analysis of MOSPD2 expression in HeLa, Huh-7, and 501-MEL cells. (D and E) Colocalization of endogenous MOSPD2 (labeled with anti-MOSPD2, in green) and LDs (labeled with LipidTOX, in magenta) in Huh-7 cells (D) or 501-MEL (E) after OA treatment. Panels on the right show the background signal of anti-MOSPD2 antibodies in cells transfected with siRNAs targeting MOSPD2 (siMOSPD2). Subpanels show the LD staining. Left: Subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels. Scale bars: 10 µm (insets, 2 µm). (A, D, and E) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Source data are available for this figure: SourceData FS3.
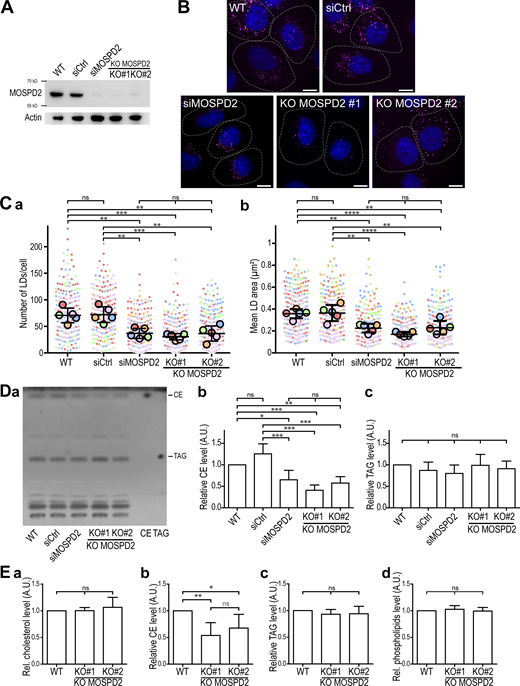
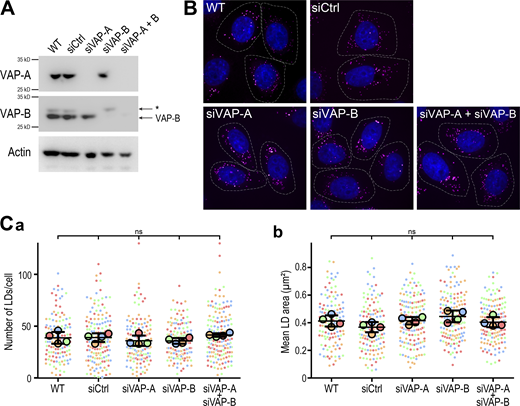
These observations prompted us to investigate whether MOSPD2 has a role in LD biology. We first established several cell models of MOSPD2 deficiency. MOSPD2 was either knocked-down using a pool of siRNAs or knocked-out using a CRISPR/Cas9 approach in HeLa cells (Fig. 2 A); two independent knock-out clones (KO#1 and KO#2) were analyzed. The number and size of LDs labeled with BODIPY 493/503 were then quantified in MOSPD2 knocked-down, knocked-out, and control cells (Fig. 2, B and C; and S4 A). Strikingly, compared with control cells, MOSPD2-deficient cells contained fewer (∼2-fold) and smaller (∼40% decrease) LDs. To compare the effect of MOSPD2 deficiency with that of VAP-A and VAP-B, we silenced VAP-A and VAP-B either individually or together using pools of siRNAs (Fig. S5 A). The LDs were then labeled (Fig. S5 B), and their number and size quantified (Fig. S5 C). The silencing of VAP-A and/or VAP-B had no effect on the number and size of LDs, thus showing that among the FFAT-binding proteins of the VAP family, only MOSPD2 has a specific role in LD biology.
MOSPD2 is involved in LD homeostasis. (A) Western blot analysis of MOSPD2 protein level in control HeLa cells (WT), HeLa cells transfected with control siRNAs (siCtrl), siRNAs targeting MOSPD2 (siMOSPD2), and in two MOSPD2-deficient HeLa cell clones (KO#1 and KO#2) established by CRISPR/Cas9 gene editing. (B) Representative confocal images of parental HeLa cells (WT), of HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting MOSPD2 (siMOSPD2), and of MOSPD2-deficient HeLa cell clones (KO#1 and KO#2) labeled with BODIPY 493/503 (LDs, magenta) and Hoechst 33258 (nuclei, blue). Scale bars: 10 µm. Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). The contour of each cell is delimited by a white dotted line. (C) Number (a) and area (b) of LDs in cells shown in B. Data are displayed as Superplots (Lord et al., 2020) showing the mean number and area of LDs per cell (small dots) or per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 398 (WT), 323 (siCtrl), 280 (siMOSPD2), 333 (KO#1), and 413 (KO#2) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n = 5 independent experiments). (D) a: TLC analysis of lipids extracted from cells shown in A. Neutral lipids were separated with hexane/diethylether/AcOH (80:20:2 vol/vol) and revealed with primuline. CE and TAG were used as standards. b and c: Relative levels of CE (b) and TAG (c) detected by TLC. Means and error bars (SD) are shown. One-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 4 independent experiments). (E) Enzymatic quantification of cholesterol (a), cholesterol ester (b), triacylglycerol, (c) and phospholipid (d) in control HeLa cells (WT) and MOSPD2-deficient HeLa cell clones (KO#1 and KO#2). Means and error bars (SD) are shown. One-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05; **, P < 0.01; n = 6 independent experiments). A.U., arbitrary unit. Source data are available for this figure: SourceData F2.
MOSPD2 is involved in LD homeostasis. (A) Western blot analysis of MOSPD2 protein level in control HeLa cells (WT), HeLa cells transfected with control siRNAs (siCtrl), siRNAs targeting MOSPD2 (siMOSPD2), and in two MOSPD2-deficient HeLa cell clones (KO#1 and KO#2) established by CRISPR/Cas9 gene editing. (B) Representative confocal images of parental HeLa cells (WT), of HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting MOSPD2 (siMOSPD2), and of MOSPD2-deficient HeLa cell clones (KO#1 and KO#2) labeled with BODIPY 493/503 (LDs, magenta) and Hoechst 33258 (nuclei, blue). Scale bars: 10 µm. Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). The contour of each cell is delimited by a white dotted line. (C) Number (a) and area (b) of LDs in cells shown in B. Data are displayed as Superplots (Lord et al., 2020) showing the mean number and area of LDs per cell (small dots) or per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 398 (WT), 323 (siCtrl), 280 (siMOSPD2), 333 (KO#1), and 413 (KO#2) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n = 5 independent experiments). (D) a: TLC analysis of lipids extracted from cells shown in A. Neutral lipids were separated with hexane/diethylether/AcOH (80:20:2 vol/vol) and revealed with primuline. CE and TAG were used as standards. b and c: Relative levels of CE (b) and TAG (c) detected by TLC. Means and error bars (SD) are shown. One-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 4 independent experiments). (E) Enzymatic quantification of cholesterol (a), cholesterol ester (b), triacylglycerol, (c) and phospholipid (d) in control HeLa cells (WT) and MOSPD2-deficient HeLa cell clones (KO#1 and KO#2). Means and error bars (SD) are shown. One-way ANOVA with Tukey’s multiple comparisons test (*, P < 0.05; **, P < 0.01; n = 6 independent experiments). A.U., arbitrary unit. Source data are available for this figure: SourceData F2.
Image analysis workflows for LD and ER quantifications. (A) Cells stained with BODIPY 493/503 (LDs) and Hoechst (nuclei) were imaged on multiple z slices using a confocal microscope. Z-stack projection (max intensity) images were generated using Fiji and processed using CellProfiler. Cells were manually segmented and LDs identified as objects ≥2 pixels of diameter. Multi-parametric object measurements were performed on the identified LDs. (B) Cells were treated with OA at 50 µM for 6 h before imaging. Three channels were acquired: LDs (stained with HCS DeepRed LipidTox), the ER (stained with the ER marker mScarlet-ER), and MOSPD2/VAP-A (tagged with GFP). Cells were manually segmented and masks of the cytoplasm (i.e., without the nuclei) were generated with CellProfiler. LDs were identified as objects ≥4 pixels of diameter. Multiple areas (2-pixel wide) from 0 to 20 pixels around each LD were added. Multi-parametric measurements were performed for each area around LDs in the red (mScarlet-ER) and green (GFP-MOSPD2/VAP) channels.
Image analysis workflows for LD and ER quantifications. (A) Cells stained with BODIPY 493/503 (LDs) and Hoechst (nuclei) were imaged on multiple z slices using a confocal microscope. Z-stack projection (max intensity) images were generated using Fiji and processed using CellProfiler. Cells were manually segmented and LDs identified as objects ≥2 pixels of diameter. Multi-parametric object measurements were performed on the identified LDs. (B) Cells were treated with OA at 50 µM for 6 h before imaging. Three channels were acquired: LDs (stained with HCS DeepRed LipidTox), the ER (stained with the ER marker mScarlet-ER), and MOSPD2/VAP-A (tagged with GFP). Cells were manually segmented and masks of the cytoplasm (i.e., without the nuclei) were generated with CellProfiler. LDs were identified as objects ≥4 pixels of diameter. Multiple areas (2-pixel wide) from 0 to 20 pixels around each LD were added. Multi-parametric measurements were performed for each area around LDs in the red (mScarlet-ER) and green (GFP-MOSPD2/VAP) channels.
VAP-A and VAP-B are not involved in LD homeostasis. (A) Western blot analysis of VAP proteins level in control HeLa cells (WT), HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting VAP-A (siVAP-A), VAP-B (siVAP-B), and both (siVAP-A + B). The band labeled with an * on VAP-B blot corresponds to cross-reactivity with VAP-A. (B) Representative confocal images of parental HeLa cells (WT) and HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting VAP-A (siVAP-A), VAP-B (siVAP-B), or both (siVAP-A + B). Cells were labeled with BODIPY 493/503 (LD, magenta) and Hoechst 33258 (nuclei, blue). The cell contour is delimited by a white dotted line. Scale bars: 10 µm. Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). (C) Number (a) and area (b) of LDs in cells shown in B. Data are displayed as Superplots showing the mean number and area of LDs per cell (small dots), and the mean number and area of LDs per independent experiment (large dots). Independent experiments (n = 4) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 98 (WT), 118 (siCtrl), 134 (siVAP-A), 129 (siVAP-B), and 135 (siVAP-A + VAP-B) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; n = 4 independent experiments). Source data are available for this figure: SourceData FS5.
VAP-A and VAP-B are not involved in LD homeostasis. (A) Western blot analysis of VAP proteins level in control HeLa cells (WT), HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting VAP-A (siVAP-A), VAP-B (siVAP-B), and both (siVAP-A + B). The band labeled with an * on VAP-B blot corresponds to cross-reactivity with VAP-A. (B) Representative confocal images of parental HeLa cells (WT) and HeLa cells transfected with control siRNAs (siCtrl), and with siRNAs targeting VAP-A (siVAP-A), VAP-B (siVAP-B), or both (siVAP-A + B). Cells were labeled with BODIPY 493/503 (LD, magenta) and Hoechst 33258 (nuclei, blue). The cell contour is delimited by a white dotted line. Scale bars: 10 µm. Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). (C) Number (a) and area (b) of LDs in cells shown in B. Data are displayed as Superplots showing the mean number and area of LDs per cell (small dots), and the mean number and area of LDs per independent experiment (large dots). Independent experiments (n = 4) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 98 (WT), 118 (siCtrl), 134 (siVAP-A), 129 (siVAP-B), and 135 (siVAP-A + VAP-B) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; n = 4 independent experiments). Source data are available for this figure: SourceData FS5.
We next determined the level of neutral lipids in MOSPD2-deficient cells by quantifying total cellular triacylglycerols (TAG) and cholesterol esters (CE) using thin-layer chromatography (TLC; Fig. 2 D). In MOSPD2-deficient cells, TAG levels were unchanged while CE were reduced by ∼40%. To further substantiate this phenotype, cholesterol, phospholipids, CE, and TAG were quantified by enzymatic methods in wild type and MOSPD2 knock-out cells (Fig. 2 E). While cholesterol, phospholipids, and TAG remained unchanged in MOSPD2-deficient cells, CE levels were reduced by ∼40%. These data show that in the absence of MOSPD2, the level of neutral lipids, especially CE, is decreased.
Together, these data suggest that MOSPD2 is present in ER sub-domains in contact with LDs and show that MOSPD2 is involved in the biology of LDs.
MOSPD2 is dynamically recruited to ER–LD contacts
MOSPD2 is present in comma-shaped structures that are also positive for the ER marker Calnexin. This suggests both that MOSPD2 is in ER sheets that do not completely encircle LDs, and that it does not translocate at the surface of LDs, meaning that it remains in ER–LD contact sites. To examine this, we performed correlative light and electron microscopy (CLEM). GFP-MOSPD2–expressing cells were processed for electron microscopy and embedded in a fluorescence-preserving resin. Thick sections were imaged by spinning-disk confocal microscopy and then by transmission electron microscopy (TEM; Fig. 3 A). GFP-MOSPD2 fluorescence correlated with the presence of ER sheets in contact with LDs. Noteworthy, we confirmed that comma-shaped fluorescent structures corresponded to areas of ER in contact with LDs. Fluorescence was absent from the surface of LDs making no contact with the ER. These data indicate that MOSPD2 accumulates in ER regions in contact with LDs.
MOSPD2 is dynamically recruited to ER–LD contact sites. (A) CLEM of HeLa/GFP-MOSPD2 cells. a: fluorescence microscopy image; b: EM image; c: correlation of GFP-MOSPD2 and EM images (scale bar: 2 µm); d: composite showing higher magnification images of the area outlined in black in c (scale bar: 500 nm); bottom right: interpretation scheme showing contacts between organelles; ER and lipid droplets are in cyan and pink, respectively. Mitochondria and endosomes/lysosomes are in light yellow and gray, respectively. (B–D) FRAP analysis of MOSPD2 and PLIN2 mobility. GFP-MOSPD2 (B) and GFP-PLIN2 (C) expressing cells were treated with OA and labeled with LipidTOX. GFP fluorescence was photobleached in the area outlined in white, and images acquired every second over a 1-min period. Scale bars: 2 µm. (D) Lineplot showing the relative fluorescence intensity in the photobleached region of GFP-MOSPD2 (green) and GFP-PLIN2 (purple) expressing cells. The grey curve shows the relative fluorescence intensity of GFP-positive LDs that were not bleached. Means and error bars (SD) of relative fluorescence intensities of 56 (GFP-MOSPD2), 57 (GFP-PLIN2), and 72 (unbleached control) regions of interest from 20, 13, and 26 cells, respectively. Data from three independent experiments. (E and F) HeLa cells expressing GFP-MOSPD2 (green) were treated with OA and labeled with anti-PLIN3 antibodies (magenta). Images were acquired by confocal microscopy (Leica SP8, 63× NA 1.4; E), or by STED super-resolution microscopy (F). MOSPD2 and PLIN3 were heterogeneously distributed around LDs. Scale bar: 10 µm (insets, 2 µm) in E and 5 µm (insets, 1 µm) in F. Subpanels on the right are higher magnification images of the area outlined. The overlay panel shows merged channels. In E, linescan shows fluorescence intensities of the green and magenta channels along the white circular arrow of the overlay subpanel (i.e., at the surface of LDs). (G) HeLa cells expressing GFP-MOSPD2 were imaged live during LDs induction (stained with LipidTOX) by OA addition. The white arrow shows an enrichment of MOSPD2 signal before the appearance of LipidTOX staining. The yellow arrow shows the growth of a LD positive for MOSPD2 before the start of the induction. Images were acquired every 90 s (t0-900) on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bar: 2 µm.
MOSPD2 is dynamically recruited to ER–LD contact sites. (A) CLEM of HeLa/GFP-MOSPD2 cells. a: fluorescence microscopy image; b: EM image; c: correlation of GFP-MOSPD2 and EM images (scale bar: 2 µm); d: composite showing higher magnification images of the area outlined in black in c (scale bar: 500 nm); bottom right: interpretation scheme showing contacts between organelles; ER and lipid droplets are in cyan and pink, respectively. Mitochondria and endosomes/lysosomes are in light yellow and gray, respectively. (B–D) FRAP analysis of MOSPD2 and PLIN2 mobility. GFP-MOSPD2 (B) and GFP-PLIN2 (C) expressing cells were treated with OA and labeled with LipidTOX. GFP fluorescence was photobleached in the area outlined in white, and images acquired every second over a 1-min period. Scale bars: 2 µm. (D) Lineplot showing the relative fluorescence intensity in the photobleached region of GFP-MOSPD2 (green) and GFP-PLIN2 (purple) expressing cells. The grey curve shows the relative fluorescence intensity of GFP-positive LDs that were not bleached. Means and error bars (SD) of relative fluorescence intensities of 56 (GFP-MOSPD2), 57 (GFP-PLIN2), and 72 (unbleached control) regions of interest from 20, 13, and 26 cells, respectively. Data from three independent experiments. (E and F) HeLa cells expressing GFP-MOSPD2 (green) were treated with OA and labeled with anti-PLIN3 antibodies (magenta). Images were acquired by confocal microscopy (Leica SP8, 63× NA 1.4; E), or by STED super-resolution microscopy (F). MOSPD2 and PLIN3 were heterogeneously distributed around LDs. Scale bar: 10 µm (insets, 2 µm) in E and 5 µm (insets, 1 µm) in F. Subpanels on the right are higher magnification images of the area outlined. The overlay panel shows merged channels. In E, linescan shows fluorescence intensities of the green and magenta channels along the white circular arrow of the overlay subpanel (i.e., at the surface of LDs). (G) HeLa cells expressing GFP-MOSPD2 were imaged live during LDs induction (stained with LipidTOX) by OA addition. The white arrow shows an enrichment of MOSPD2 signal before the appearance of LipidTOX staining. The yellow arrow shows the growth of a LD positive for MOSPD2 before the start of the induction. Images were acquired every 90 s (t0-900) on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bar: 2 µm.
The fact that MOSPD2 is both distributed throughout the entire ER membrane and enriched in subdomains of the ER surrounding LDs suggests that it is in equilibrium between ER–LD contact sites and the remainder of the ER. To analyze the dynamics of MOSPD2 movement between these two regions, we performed fluorescence recovery after photobleaching (FRAP) experiments. GFP-MOSPD2 was expressed in HeLa cells, and individual LDs were bleached (Fig. 3, B and D). GFP-MOSPD2 fluorescence rapidly recovered (t1/2 of ∼8 s) and reached a plateau equivalent to ∼50% of the initial fluorescence (Fig. 3, B and D). In comparison, the LD protein Perilipin-2 (PLIN2) fused with GFP remained permanently associated with LDs with no recovery observed for the duration of the experiment (Fig. 3, C and D). The localization of MOSPD2 is balanced between distinct areas of the ER, some of which are in contact with the LDs, and half of the protein pool can rapidly move in and out these two regions.
We noted that the MOSPD2 signal was not uniform around LDs, suggesting that the surface of LDs might have sub-domains. To examine this, we performed co-localization experiments between GFP-MOSPD2 and endogenous PLIN3, a coat protein of LDs. Of interest, while both proteins were present around LDs, they exhibited a different distribution, the two signals being mostly mutually exclusive, as observed by confocal and super-resolution microscopy (Fig. 3, E and F). This supports the notion that, in terms of protein composition, the surface of LDs has distinct territories, either sticking to the ER (MOSPD2 territory) or being free (PLIN3 territory), and that contacts define them.
MOSPD2 is recruited around mature LDs. Next, to determine whether MOSPD2 is already recruited early in the life of LDs, we visualized the location of GFP-MOSPD2 following the induction of LD biogenesis by OA using live cell imaging (Fig. 3 G). As shown in Fig. 3 G, GFP-MOSPD2 was recruited early during LD biogenesis: GFP-MOSPD2 was already present in areas that were not yet detected by the LD marker LipidTox. Thus, MOSPD2 is associated with LDs at different stages of their life.
Overall, these results demonstrate that MOSPD2 dynamically distributes between specific subdomains of the ER: ER membranes in contact with LDs and the remainder of the ER.
MOSPD2 tethers the ER to LDs
Since associations between the ER and LDs are frequent events, we wondered whether MOSPD2 merely populates existing ER–LD contacts or, instead, contributes to making these contacts. To address this question, we determined by TEM whether the overexpression of MOSPD2 generates more ER–LD contacts. In control cells, LDs made few focal contacts with the ER (Fig. 4 A, a). In contrast, in cells expressing MOSPD2, LDs were frequently associated with long segments of ER encircling them (Fig. 4 A, b).
MOSPD2 regulates ER–LD contact sites. (A) TEM images of control HeLa (a) and HeLa/GFP-MOSPD2 cells (b) with their interpretation scheme; the ER and LDs are in green and magenta, respectively. Mitochondria and endosomes are in light yellow and gray, respectively. Scale bars: 500 nm. (B) HeLa cells stably expressing the mScarlet-ER marker (green) were either not transfected (NT, top), transfected with GFP-MOSPD2 (gray, middle), or with GFP-VAP-A (gray, bottom). Cells were treated with OA (50 µM for 6 h) and LDs stained with LipidTOX (magenta). Images were acquired on a confocal microscope (Leica SP5; 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm). (C) Schematic representation of the method used for fluorescence quantification around LDs: two pixels-wide areas were segmented around LDs (here represented for a 1-µm-wide LD), and the mean mScarlet fluorescence intensity was measured in each area. (D) Fluorescence intensity of the ER marker mScarlet-ER around LDs in untransfected (NT, red), GFP-MOSPD2 (green), and GFP-VAP-A (purple) transfected cells. Means ± SD (NT: 39 cells; GFP-MOSPD2: 42 cells; GFP-VAP-A: 46 cells; from four independent experiments). The relative mScarlet fluorescence intensity corresponds to the mean fluorescence intensity of mScarlet in each area, divided by the mean fluorescence intensity in the cytoplasm away from LDs (10–20 pixels distance from LDs). (E) Relative enrichment of GFP-MOSPD2 and GFP-VAP-A around LDs. The Peri-LD enrichment ratio is the ratio of the mean GFP fluorescence intensity (GFP-MOSPD2 or GFP-VAP-A) in the vicinity of LDs (0–4 pixels distance from LDs; see C), to the mean fluorescence intensity of GFP at a distance from LDs (10–20 pixels distance from LDs). MOSPD2 fluorescence is twice as high around LDs as in the remainder of the cytoplasm, whereas VAP-A fluorescence is at the same level next to LDs and in the rest of the cytoplasm. Means ± SD (GFP-MOSPD2: 42 cells; GFP-VAP-A: 46 cells; data from four independent experiments).
MOSPD2 regulates ER–LD contact sites. (A) TEM images of control HeLa (a) and HeLa/GFP-MOSPD2 cells (b) with their interpretation scheme; the ER and LDs are in green and magenta, respectively. Mitochondria and endosomes are in light yellow and gray, respectively. Scale bars: 500 nm. (B) HeLa cells stably expressing the mScarlet-ER marker (green) were either not transfected (NT, top), transfected with GFP-MOSPD2 (gray, middle), or with GFP-VAP-A (gray, bottom). Cells were treated with OA (50 µM for 6 h) and LDs stained with LipidTOX (magenta). Images were acquired on a confocal microscope (Leica SP5; 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm). (C) Schematic representation of the method used for fluorescence quantification around LDs: two pixels-wide areas were segmented around LDs (here represented for a 1-µm-wide LD), and the mean mScarlet fluorescence intensity was measured in each area. (D) Fluorescence intensity of the ER marker mScarlet-ER around LDs in untransfected (NT, red), GFP-MOSPD2 (green), and GFP-VAP-A (purple) transfected cells. Means ± SD (NT: 39 cells; GFP-MOSPD2: 42 cells; GFP-VAP-A: 46 cells; from four independent experiments). The relative mScarlet fluorescence intensity corresponds to the mean fluorescence intensity of mScarlet in each area, divided by the mean fluorescence intensity in the cytoplasm away from LDs (10–20 pixels distance from LDs). (E) Relative enrichment of GFP-MOSPD2 and GFP-VAP-A around LDs. The Peri-LD enrichment ratio is the ratio of the mean GFP fluorescence intensity (GFP-MOSPD2 or GFP-VAP-A) in the vicinity of LDs (0–4 pixels distance from LDs; see C), to the mean fluorescence intensity of GFP at a distance from LDs (10–20 pixels distance from LDs). MOSPD2 fluorescence is twice as high around LDs as in the remainder of the cytoplasm, whereas VAP-A fluorescence is at the same level next to LDs and in the rest of the cytoplasm. Means ± SD (GFP-MOSPD2: 42 cells; GFP-VAP-A: 46 cells; data from four independent experiments).
To evaluate the capacity of MOSPD2 to drive ER–LD contact formation more quantitatively, we examined by light microscopy the recruitment of the ER around LDs in cells where MOSDP2 was overexpressed. The ER surface was labeled with a red fluorescent marker (mScarlet-ER) and the radial distribution of the fluorescence signal was measured around LDs (Fig. 4, B and C; and Fig. S4 B). In cells expressing the mScarlet-ER marker alone, the fluorescent signal was evenly distributed in the cytosol, with no enrichment around LDs (Fig. 4, B and D). In contrast, in cells expressing MOSPD2, the ER marker accumulated in the periphery of LDs, thus showing that MOSPD2 expression promotes the formation of ER–LD contacts. This was specific to MOSPD2 since VAP-A expression did not result in ER accumulation around the LDs (Fig. 4, B and D). Furthermore, consistent with data from Fig. 1, the relative fluorescence of MOSPD2 was 2-fold higher at the periphery of LDs than in the remaining part of the cytoplasm, whereas VAP-A fluorescence was homogeneously distributed; this confirmed that MOSPD2 accumulates around LDs (Fig. 4 E).
We next addressed whether MOSPD2 association with LDs relies on Seipin. Seipin, a protein encoded by the BSCL2 gene, which is mutated in lipodystrophy, is a major tether localized in ER–LD junctions tightly controlling LD assembly (Szymanski et al., 2007; Salo et al., 2016; Gao et al., 2019). Since we found MOSPD2 at ER–LD contacts, especially during the early stages of LD formation (Fig. 3 G), we asked whether this localization relies on Seipin. We found that in cells knocked-out or silenced for Seipin, MOSPD2 still localized to ER–LD contacts (Fig. 5, A and B), indicating that Seipin was not required for MOSPD2-mediated ER–LD contact formation.
Seipin is dispensable for MOSPD2-mediated ER–LD contact formation and the GFP-MOSPD2 RD/LD mutant is localized in ER–LD contacts. (A) Representative confocal images of the GFP-MOSPD2 WT (green) localization in cells transfected with control siRNAs (left) and siRNAs targeting Seipin (right) and left untreated (a) or treated with OA (b). LDs were stained with Nile Red (magenta). (B) Representative confocal images of the GFP-MOSPD2 WT (green) localization in WT (left) and Seipin knock-out (right) cells treated with OA. LDs were stained with LipidTox (magenta). Note that Seipin silencing or knock-out results in heterogeneous lipid droplet size. In absence of Seipin, MOSPD2 still mediates ER–LD contact formation. (C) a: Representative confocal images of GFP-MOSPD2 WT and RD/LD mutant expressing cells. Cells were not treated with OA. LDs were stained with Nile Red (magenta). b: percentage of cells with GFP-positive ring- or comma-shaped structures. Mean ± SD; n = 4 independent experiments (WT: 156 cells; RD/LD: 162 cells). (D) CLEM of a GFP-MOSPD2 RD/LD expressing cell. a: GFP-MOSPD2 RD/LD fluorescence microscopy image; b: EM image; c: correlation of GFP-MOSPD2 RD/LD fluorescence and EM images (scale bar: 2 µm); d: higher magnification images of the area outlined in black (scale bar: 500 nm); bottom right: interpretation scheme showing contacts between organelles; ER and lipid droplets are in cyan and pink, respectively. Mitochondria, endosomes/lysosomes and nucleus are in light yellow, gray and light blue, respectively. (Aand C) Confocal microscope (Leica SP8, 63× NA 1.4) images. (B) Zeiss LSM800 Airyscan images. Scale bars: 10 μm (insets, 2 μm).
Seipin is dispensable for MOSPD2-mediated ER–LD contact formation and the GFP-MOSPD2 RD/LD mutant is localized in ER–LD contacts. (A) Representative confocal images of the GFP-MOSPD2 WT (green) localization in cells transfected with control siRNAs (left) and siRNAs targeting Seipin (right) and left untreated (a) or treated with OA (b). LDs were stained with Nile Red (magenta). (B) Representative confocal images of the GFP-MOSPD2 WT (green) localization in WT (left) and Seipin knock-out (right) cells treated with OA. LDs were stained with LipidTox (magenta). Note that Seipin silencing or knock-out results in heterogeneous lipid droplet size. In absence of Seipin, MOSPD2 still mediates ER–LD contact formation. (C) a: Representative confocal images of GFP-MOSPD2 WT and RD/LD mutant expressing cells. Cells were not treated with OA. LDs were stained with Nile Red (magenta). b: percentage of cells with GFP-positive ring- or comma-shaped structures. Mean ± SD; n = 4 independent experiments (WT: 156 cells; RD/LD: 162 cells). (D) CLEM of a GFP-MOSPD2 RD/LD expressing cell. a: GFP-MOSPD2 RD/LD fluorescence microscopy image; b: EM image; c: correlation of GFP-MOSPD2 RD/LD fluorescence and EM images (scale bar: 2 µm); d: higher magnification images of the area outlined in black (scale bar: 500 nm); bottom right: interpretation scheme showing contacts between organelles; ER and lipid droplets are in cyan and pink, respectively. Mitochondria, endosomes/lysosomes and nucleus are in light yellow, gray and light blue, respectively. (Aand C) Confocal microscope (Leica SP8, 63× NA 1.4) images. (B) Zeiss LSM800 Airyscan images. Scale bars: 10 μm (insets, 2 μm).
Collectively, these data show that MOSPD2 favors the tethering of the ER with LDs.
The MSP domain of MOSPD2 is dispensable for the formation of ER–LD contacts
MOSPD2 contains an MSP domain involved in protein–protein interactions and a CRAL-TRIO domain, which is potentially a lipid transfer domain (Di Mattia et al., 2018; Chiapparino et al., 2016). We first reasoned that the MSP domain could mediate the formation of ER–LD contacts to position the CRAL-TRIO domain at the ER/LD interface. To test this hypothesis, we constructed a deletion mutant lacking the MSP domain (Fig. 6 A). This mutant was transfected in HeLa cells in which LD biogenesis was induced by OA, and we analyzed its ability to form ring- and comma-shaped structures around LDs, i.e., to form ER–LD contacts. Unexpectedly, we observed that the MOSPD2 ΔMSP mutant localized in ring- and comma-shaped structure around LDs like the WT protein; in fact, we noted that the recruitment around LDs was even more frequent than that of the WT protein (97 vs. 77%, Fig. 6, B and C). Likewise, the mutation of two key residues in the MSP domain (R404D/L406D referred to here as RD/LD mutant), precluding the recognition of FFAT motifs (Di Mattia et al., 2018), resulted in a massive recruitment of MOSPD2 at the periphery of LDs in most cells, both in the presence (Fig. 6, A–C) and in the absence of OA (Fig. 5 C). Consistent with this observation, CLEM experiments performed with cells expressing GFP-MOSPD2 RD/LD mutant (Fig. 5 D) showed that the protein accumulated in ER strands in tight contact with LDs.
ER–LD contact sites mediated by MOSPD2 depends on its CRAL-TRIO and TM domains. (A) Schematic representation of the different WT and mutant proteins used in the study. Two kinds of mutants were utilized: deletions of specific domains (ΔCRAL-TRIO, ΔMSP, ΔTM) and point mutation (RD/LD) impairing the MSP domain function. (B) Representative confocal images of the GFP-MOSPD2 WT and mutants (green) localization. Cells were treated with OA and LDs stained with Nile Red (magenta). (C) Quantification of cells presenting ring- and comma-shaped staining. Mean ± SD; n = 3 independent experiments (WT: 67 cells; ΔMSP: 138 cells; RD/LD: 152 cells; ΔCRAL-TRIO: 140 cells; ΔTM: 64 cells). (D) EM images of HeLa/GFP-MOSPD2, HeLa/GFP-MOSPD2 ΔMSP, HeLa/GFP-MOSPD2 RD/LD, and HeLa/GFP-MOSPD2 ΔCRAL-TRIO cells (top) and their interpretation scheme (bottom); the ER and LDs are in green and magenta, respectively. Mitochondria and endosomes are in light yellow and gray, respectively. Scale bars: 500 nm. (E) Left: schematic representation of the different chimeric constructs in which the MOSPD2 TM domain is replaced by the TM of SAC1 (purple). Right: localization of these chimeric proteins (green) and LDs stained with Nile Red (magenta) in HeLa cells treated with OA. In B and E, subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels. (B and E) Images were acquired on a confocal microscope (Leica SP8, 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm).
ER–LD contact sites mediated by MOSPD2 depends on its CRAL-TRIO and TM domains. (A) Schematic representation of the different WT and mutant proteins used in the study. Two kinds of mutants were utilized: deletions of specific domains (ΔCRAL-TRIO, ΔMSP, ΔTM) and point mutation (RD/LD) impairing the MSP domain function. (B) Representative confocal images of the GFP-MOSPD2 WT and mutants (green) localization. Cells were treated with OA and LDs stained with Nile Red (magenta). (C) Quantification of cells presenting ring- and comma-shaped staining. Mean ± SD; n = 3 independent experiments (WT: 67 cells; ΔMSP: 138 cells; RD/LD: 152 cells; ΔCRAL-TRIO: 140 cells; ΔTM: 64 cells). (D) EM images of HeLa/GFP-MOSPD2, HeLa/GFP-MOSPD2 ΔMSP, HeLa/GFP-MOSPD2 RD/LD, and HeLa/GFP-MOSPD2 ΔCRAL-TRIO cells (top) and their interpretation scheme (bottom); the ER and LDs are in green and magenta, respectively. Mitochondria and endosomes are in light yellow and gray, respectively. Scale bars: 500 nm. (E) Left: schematic representation of the different chimeric constructs in which the MOSPD2 TM domain is replaced by the TM of SAC1 (purple). Right: localization of these chimeric proteins (green) and LDs stained with Nile Red (magenta) in HeLa cells treated with OA. In B and E, subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels. (B and E) Images were acquired on a confocal microscope (Leica SP8, 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm).
To further ascertain that the MSP domain of MOSPD2 is dispensable for the formation of ER–LD contacts, cells expressing GFP-tagged MOSPD2 MSP deletion mutant (GFP-MOSPD2 ΔMSP) and the RD/LD mutant (GFP-MOSPD2 RD/LD) were processed for TEM (Fig. 6 D). In cells expressing MOSPD2 mutants lacking the MSP domain (GFP-MOSPD2 ΔMSP) or having a defective MSP domain (GFP-MOSPD2 RD/LD), the ER remained extensively attached to LDs (Fig. 6 D).
Thus, the MSP domain is not involved in the formation of ER–LD contacts, and may even limit the recruitment of MOSPD2 at ER–LD contacts.
The CRAL-TRIO and TM domains of MOSPD2 mediate the formation of ER–LD contacts
Since the MSP domain of MOSPD2 is not necessary for the formation of ER–LD contacts, we alternatively examined the contribution of the TM and CRAL-TRIO domains by testing deletion mutants (GFP-MOSPD2 ΔCRAL-TRIO and GFP-MOSPD2 ΔTM; Fig. 6 A). In contrast to WT MOSPD2 that accumulated in ring- and comma-shaped structures, MOSPD2 devoid of the CRAL-TRIO domain had a reticular-only localization, thus showing that the CRAL-TRIO domain is necessary for the recruitment of MOSPD2 to LDs (Fig. 6, B and C).
Cells expressing the GFP-MOSPD2 ΔCRAL-TRIO deletion mutant were further analyzed by TEM. Compared with cells expressing WT MOSPD2 in which extended and frequent ER–LD contacts were observed, cells expressing this mutant only harbored focal ER–LD contacts (Fig. 6 D). Jointly, these results point to a crucial role of the CRAL-TRIO domain for the ability of MOSPD2 to create ER–LD contacts.
In addition, we observed that the MOSPD2 mutant devoid of TM remained cytosolic and did not accumulate on the LD surface (Fig. 6, B and C). To better understand this, the TM domain of MOSPD2 was substituted with the TM of the ER-anchored phosphatase SAC1 that comprises two transmembrane helices (named TM(SAC1); Fig. 6 E). While the fusion protein GFP-TM(SAC1) was evenly localized in the ER, the chimeric protein composed of the MOSPD2 CRAL-TRIO and MSP domains fused with the TM(SAC1) domain was present in the ER and accumulated around LDs. Similarly, the fusion protein comprising only the CRAL-TRIO domain of MOSPD2 and the TM(SAC1) domain was also present in the ER and enriched around LDs (Fig. 6 E). Thus, MOSPD2 needs an ER-anchor to be localized in contact with LDs.
Together, these data show that the TM and CRAL-TRIO domains are necessary for MOSPD2 binding to LDs.
An amphipathic helix in the CRAL-TRIO domain of MOSPD2 is required for binding to LDs
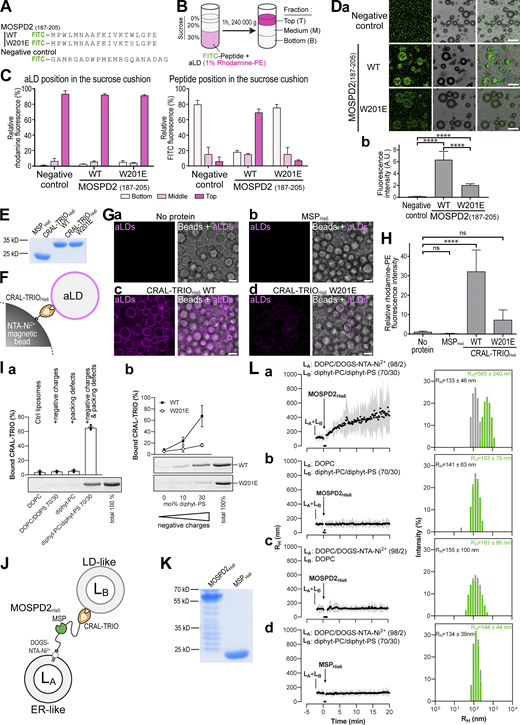
Proteins that associate with LDs do so via at least two known modalities: Class I proteins are embedded in the ER bilayer and can diffuse laterally to the LD monolayer, while Class II proteins translocate from the cytosol to the surface of LDs (Olzmann and Carvalho, 2019; Kory et al., 2016). Most Class II proteins associate with LDs through an amphipathic α-helix (AH), in which hydrophobic and polar residues are segregated to form two distinct faces along the helix axis. This topology allows AH to efficiently bind membranes because hydrophobic residues can insert between lipid acyl chains whereas polar residues can make polar contacts with lipid headgroups. The LD surface has more packing defects than a bilayer, i.e., gaps in the phospholipid layer, which are favorable for the insertion of hydrophobic residues and thus the association of Class II proteins (Giménez-Andrés et al., 2018; Chorlay and Thiam, 2020; Chorlay et al., 2021). Because MOSPD2 is anchored to the ER and does not diffuse to the LD monolayer, we hypothesized that the CRAL-TRIO domain might behave like a Class II protein and thus possess an AH. As no experimental structure of the CRAL-TRIO domain of MOSPD2 was available, we built structural models of the protein using SWISS-MODEL and AlphaFold (Waterhouse et al., 2018; Jumper et al., 2021), and identified AHs in the models. We determined their hydrophobicity and hydrophobic moment using HeliQuest (Gautier et al., 2008) and identified an AH at the end of the CRAL-TRIO domain (Fig. 7, A–D), that is exposed at the surface of the protein and thus potentially able to insert into a membrane. Sequence analyses showed that the helix is highly conserved from Cnidaria to Human (Fig. 7 C). To determine whether this AH is responsible for MOSPD2 binding to LDs, we replaced the bulky hydrophobic residue tryptophan 201 in the middle of the nonpolar face by the negatively charged residue glutamate (mutant W201E) which would perturb the membrane partitioning of this helix. Compared with WT MOSPD2, which is found both in the ER and in ER–LD contacts when expressed in cells, the MOSPD2 W201E mutant was evenly distributed in the ER (Fig. 7, F and G). Moreover, replacing the CRAL-TRIO domain by the AH only (GFP-AH-MOSPD2 ΔCRAL-TRIO fusion protein) was sufficient to recruit MOSPD2 on LDs (Fig. 7, F and G).
An amphipathic helix in the CRAL-TRIO domain of MOSPD2 mediates its localization at ER–LD contacts. (A) Schematic representation of MOSPD2 showing the position of the amphipathic helix (red) and its sequence. The arrowhead shows the position of residue W201. (B) Helical wheel representation of the WT (left) and W201E mutant (right) AH (aa 189-203) generated with HeliQuest (http://heliquest.ipmc.cnrs.fr/; left). The W201E mutation alters the amphipathic character of the helix by reducing its hydrophobic moment (µH) from 0.436 to 0.254. (C) WebLogo generated from an alignment of MOSPD2 AH sequence from 44 species. The AH is highlighted in light orange and 10 flanking residues from either side are shown. (D) Ribbon diagram of the structure model of the CRAL-TRIO domain of human MOSPD2 (Uniprot Q8NHP6; residues 1-241) obtained with AlphaFold (Jumper et al., 2021). The domain is in light grey except for the amphipathic helix depicted in stick model with residues colored as in B. (E) Far-UV circular dichroism spectrum of the MOSPD2 CRAL-TRIO domain and its W201E variant (6.7 μM) in 20 mM Tris, pH 7.4, 120 mM NaF buffer. MRE, mean residue ellipticity. The percentage of α-helix, β-sheet and turn, deriving from the analysis of the spectrum (WT) are given as well as the values deriving from the structure model (AlphaFold) using the Define Secondary Structure of Protein algorithm. (F) Left: Schematic representation of GFP-MOSPD2 constructs either WT (GFP-MOSPD2), bearing a mutation in the AH (GFP-MOSPD2 W201E), or containing a deletion of the CRAL-TRIO domain together with an insertion of the AH (AH-MOSPD2-ΔCRAL-TRIO). Right: Localization of these constructs in HeLa cells treated with OA; LDs were stained with Nile Red (magenta). Images were acquired on a confocal microscope (Leica SP5, 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm). (G) Quantification of cells showing ring- or comma-shaped staining for these constructs. Mean ± SD; n = 3 independent experiments (MOSPD2 WT:117 cells; MOSPD2 W201E: 156 cells; AH-MOSPD2-ΔCRAL-TRIO: 113 cells). (H) Left: Schematic representation of WT GFP-VAP-A and GFP-AH-VAP-A chimera in which the AH of MOSPD2 was fused at the N-terminus of VAP-A. Right: localization of the different constructs. LDs were stained with Nile Red in HeLa cells treated with OA. Confocal microscope (Leica SP5, 63× NA 1.4) images. Scale bars: 10 µm (insets, 2 µm). (I) Quantification of cells showing ring- or comma-shaped staining for GFP-VAP-A and GFP-AH-VAP-A chimera. Mean ± SD; n = 3 independent experiments (VAP-A WT: 109 cells; AH-VAP-A: 102 cells). In F and H, composite subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels.
An amphipathic helix in the CRAL-TRIO domain of MOSPD2 mediates its localization at ER–LD contacts. (A) Schematic representation of MOSPD2 showing the position of the amphipathic helix (red) and its sequence. The arrowhead shows the position of residue W201. (B) Helical wheel representation of the WT (left) and W201E mutant (right) AH (aa 189-203) generated with HeliQuest (http://heliquest.ipmc.cnrs.fr/; left). The W201E mutation alters the amphipathic character of the helix by reducing its hydrophobic moment (µH) from 0.436 to 0.254. (C) WebLogo generated from an alignment of MOSPD2 AH sequence from 44 species. The AH is highlighted in light orange and 10 flanking residues from either side are shown. (D) Ribbon diagram of the structure model of the CRAL-TRIO domain of human MOSPD2 (Uniprot Q8NHP6; residues 1-241) obtained with AlphaFold (Jumper et al., 2021). The domain is in light grey except for the amphipathic helix depicted in stick model with residues colored as in B. (E) Far-UV circular dichroism spectrum of the MOSPD2 CRAL-TRIO domain and its W201E variant (6.7 μM) in 20 mM Tris, pH 7.4, 120 mM NaF buffer. MRE, mean residue ellipticity. The percentage of α-helix, β-sheet and turn, deriving from the analysis of the spectrum (WT) are given as well as the values deriving from the structure model (AlphaFold) using the Define Secondary Structure of Protein algorithm. (F) Left: Schematic representation of GFP-MOSPD2 constructs either WT (GFP-MOSPD2), bearing a mutation in the AH (GFP-MOSPD2 W201E), or containing a deletion of the CRAL-TRIO domain together with an insertion of the AH (AH-MOSPD2-ΔCRAL-TRIO). Right: Localization of these constructs in HeLa cells treated with OA; LDs were stained with Nile Red (magenta). Images were acquired on a confocal microscope (Leica SP5, 63× NA 1.4). Scale bars: 10 µm (insets, 2 µm). (G) Quantification of cells showing ring- or comma-shaped staining for these constructs. Mean ± SD; n = 3 independent experiments (MOSPD2 WT:117 cells; MOSPD2 W201E: 156 cells; AH-MOSPD2-ΔCRAL-TRIO: 113 cells). (H) Left: Schematic representation of WT GFP-VAP-A and GFP-AH-VAP-A chimera in which the AH of MOSPD2 was fused at the N-terminus of VAP-A. Right: localization of the different constructs. LDs were stained with Nile Red in HeLa cells treated with OA. Confocal microscope (Leica SP5, 63× NA 1.4) images. Scale bars: 10 µm (insets, 2 µm). (I) Quantification of cells showing ring- or comma-shaped staining for GFP-VAP-A and GFP-AH-VAP-A chimera. Mean ± SD; n = 3 independent experiments (VAP-A WT: 109 cells; AH-VAP-A: 102 cells). In F and H, composite subpanels on the bottom are higher magnification images of the area outlined. The overlay panel shows merged channels.
As mentioned before, VAP-A is not recruited in ER–LD contacts (Figs. 1 B and 4 E). To know whether the AH of MOSPD2 could allow the recruitment of VAP-A on LDs, we created a chimeric protein composed of the AH of MOSPD2 fused to VAP-A. Unlike VAP-A, which is distributed evenly in the ER, the fusion protein AH-VAP-A accumulated in ER subdomains around LDs (Fig. 7, H and I).
Combined together, these data show that the AH of MOSPD2 is necessary and sufficient for this ER-bound protein to mediate the formation of ER–LD contacts.
The amphipathic helix of MOSPD2 directly interacts with the surface of LDs
To directly test whether the AH of MOSPD2 could bind LDs, we carried out flotation assays using artificial LDs (aLDs) and fluorescein isothiocyanate (FITC)–labeled synthetic peptides encompassing the AH of MOSPD2, either wild type or with the W201E mutation (Fig. 8 A). A peptide with a random sequence was used as negative control (Fig. 8 A). aLDs composed of a mix of triolein and surrounded by a monolayer of phosphatidylcholine with di-oleyl (DOPC) and phosphatidylethanolamine with di-oleyl (DOPE) labeled with a fluorescent lipid (Rhodamine-PE) were prepared. The peptides were incubated with these aLDs, mixed with sucrose, and allowed to float over this sucrose cushion (Fig. 8 B). Three fractions corresponding to the top, middle, and bottom position of the cushion were collected, and the fluorescence signal of aLDs and of the peptides were measured. After ultracentrifugation, Rhodamine-labeled aLDs were in the top fraction (Fig. 8 C). The control peptide remained in the bottom fraction, while the peptide corresponding to the AH of MOSPD2 was in the top fraction with aLDs. Unlike the WT AH of MOSPD2, the W201E mutant behaved like the negative control peptide and remained in the bottom fraction. To further characterize the association of the AH of MOSPD2 to aLDs, we performed aLD–peptide interaction assays (Fig. 8 D). aLDs composed of triolein were mixed with the fluorescent peptides. aLDs were imaged and fluorescence on the surface of aLDs were quantified. In agreement with the flotation assays, the control peptide did not bind to the aLDs, while the peptide corresponding to the AH of MOSPD2 was found attached to the aLDs. The W201E mutant was only minimally retained by the aLDs. Together, these experiments show that the AH of MOSPD2 directly binds to LDs.
The CRAL-TRIO domain of MOSPD2 directly interacts with aLDs. (A) Peptides used for aLDs flotation assays. Peptides corresponding to the WT or W201E mutant AH of MOSPD2 (residues 187-205), and negative control composed of a random sequence, were coupled with FITC at their amino-terminal end. (B) Principle of aLDs flotation assays. Fluorescent peptides were incubated with aLDs containing 1 mol% Rhodamine-PE, then ultracentrifuged to allow aLDs to float on the sucrose cushion. Top, middle and bottom fractions were collected and FITC and rhodamine fluorescence quantified. (C) aLDs flotation assays. Left: Relative rhodamine fluorescence (i.e., aLDs); right: Relative FITC fluorescence (i.e., peptides), in the bottom (light pink), middle (pink) and top (dark pink) fractions. Means (± SD) from n = 5 independent experiments. (D) aLDs peptide interaction assay. a: Representative images of aLDs incubated with peptides shown in A. Scale bars: 10 μm. b: quantification of peptide fluorescence on aLDs. Means (± SD); n = 2 independent experiments (negative control: 1,397; WT AH : 231; W201E AH: 198 aLDs) . Student’s t test (****, P < 0.0001). (E) Coomassie blue staining of the recombinant MSPHis6, WT CRAL-TRIOHis6, and mutant CRAL-TRIOHis6 W201E proteins after SDS-PAGE. (F) Principle of aLDs pull-down assay. Proteins were immobilized on magnetic NTA-Ni2+ beads, owing to their His6 tag, and incubated with fluorescent aLDs. (G) Representative confocal images of NTA-Ni2+ beads either not coated with recombinant proteins (a, no protein) or coated with recombinant domains of MOSPD2 (b, MSPHis6; c, WT CRAL-TRIOHis6; and d, mutant CRAL-TRIOHis6 W201E) and incubated with fluorescent aLDs (magenta). Left: Confocal section of aLD fluorescence; right: Superposition with brightfield images showing the beads. Spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4) images. Scale bars: 10 µm. (H) Quantification of aLDs recruitment on NTA-Ni2+ beads. Rhodamine fluorescence was measured using a fluorimeter. Means ± SD. Kruskal–Wallis with Tukey’s multiple comparisons test (ns, not significant; ****, P < 0.0001; n = 6 independent experiments). (I) a: Flotation assays. CRAL-TRIOHis6 (0.75 µM) was mixed with liposomes (0.75 mM lipids) only made of DOPC or diphyt-PC or composed of DOPC/DOPS (7/3 mol/mol) or diphyt-PC/diphyt-PS (7/3 mol/mol) in HK buffer at 25°C for 10 min. After centrifugation, the liposomes were recovered at the top of a sucrose cushion and analyzed by SDS-PAGE. The amount of protein recovered in the top fraction (lane 1–4) was quantified and the fraction of liposome-bound CRAL-TRIOHis6 was determined using the content of lane 5 (total 100%) as a reference. Data are represented as mean ± SEM (error bars; n = 4). b: Flotation assays. WT (closed circle) and W201E mutant (open circle) MOSPD2 CRAL-TRIOHis6 proteins (0.75 µM) were mixed for 10 min with liposomes (0.75 mM lipids) only made of diphyt-PC or additionally containing 10 or 30 mol% diphyt-PS. Data are represented as mean ± SEM (error bars; n = 3–5). (J) Principle of the membrane tethering assay. (K) Coomassie blue staining of the recombinant MOSPD2His6 and MSPHis6 proteins after SDS-PAGE. (L) Membrane tethering assays. LA liposomes (50 µM total lipids) composed of DOPC/DOGS-NTA-Ni2+ (98/2 mol/mol; a, b, and d) or DOPC (b) were mixed with LB liposomes (50 µM), composed of diphyt-PC/diphyt-PS (70/30 mol/mol) (a, b, and d) or DOPC (c) in HK buffer at 25°C. After 2 min, MOSPD2His6 (a–c) or MSPHis6 (d; 0.4 µM) was added and the size of liposomes was measured for 23 min. Left: Mean radius (dots) and polydispersity (shaded area) over time. Right: Hydrodynamic radius (RH) distribution before (gray bars) and after the reaction (green bars). These experiments are representative of several independent experiments (n = 3–5). Source data are available for this figure: SourceData F8.
The CRAL-TRIO domain of MOSPD2 directly interacts with aLDs. (A) Peptides used for aLDs flotation assays. Peptides corresponding to the WT or W201E mutant AH of MOSPD2 (residues 187-205), and negative control composed of a random sequence, were coupled with FITC at their amino-terminal end. (B) Principle of aLDs flotation assays. Fluorescent peptides were incubated with aLDs containing 1 mol% Rhodamine-PE, then ultracentrifuged to allow aLDs to float on the sucrose cushion. Top, middle and bottom fractions were collected and FITC and rhodamine fluorescence quantified. (C) aLDs flotation assays. Left: Relative rhodamine fluorescence (i.e., aLDs); right: Relative FITC fluorescence (i.e., peptides), in the bottom (light pink), middle (pink) and top (dark pink) fractions. Means (± SD) from n = 5 independent experiments. (D) aLDs peptide interaction assay. a: Representative images of aLDs incubated with peptides shown in A. Scale bars: 10 μm. b: quantification of peptide fluorescence on aLDs. Means (± SD); n = 2 independent experiments (negative control: 1,397; WT AH : 231; W201E AH: 198 aLDs) . Student’s t test (****, P < 0.0001). (E) Coomassie blue staining of the recombinant MSPHis6, WT CRAL-TRIOHis6, and mutant CRAL-TRIOHis6 W201E proteins after SDS-PAGE. (F) Principle of aLDs pull-down assay. Proteins were immobilized on magnetic NTA-Ni2+ beads, owing to their His6 tag, and incubated with fluorescent aLDs. (G) Representative confocal images of NTA-Ni2+ beads either not coated with recombinant proteins (a, no protein) or coated with recombinant domains of MOSPD2 (b, MSPHis6; c, WT CRAL-TRIOHis6; and d, mutant CRAL-TRIOHis6 W201E) and incubated with fluorescent aLDs (magenta). Left: Confocal section of aLD fluorescence; right: Superposition with brightfield images showing the beads. Spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4) images. Scale bars: 10 µm. (H) Quantification of aLDs recruitment on NTA-Ni2+ beads. Rhodamine fluorescence was measured using a fluorimeter. Means ± SD. Kruskal–Wallis with Tukey’s multiple comparisons test (ns, not significant; ****, P < 0.0001; n = 6 independent experiments). (I) a: Flotation assays. CRAL-TRIOHis6 (0.75 µM) was mixed with liposomes (0.75 mM lipids) only made of DOPC or diphyt-PC or composed of DOPC/DOPS (7/3 mol/mol) or diphyt-PC/diphyt-PS (7/3 mol/mol) in HK buffer at 25°C for 10 min. After centrifugation, the liposomes were recovered at the top of a sucrose cushion and analyzed by SDS-PAGE. The amount of protein recovered in the top fraction (lane 1–4) was quantified and the fraction of liposome-bound CRAL-TRIOHis6 was determined using the content of lane 5 (total 100%) as a reference. Data are represented as mean ± SEM (error bars; n = 4). b: Flotation assays. WT (closed circle) and W201E mutant (open circle) MOSPD2 CRAL-TRIOHis6 proteins (0.75 µM) were mixed for 10 min with liposomes (0.75 mM lipids) only made of diphyt-PC or additionally containing 10 or 30 mol% diphyt-PS. Data are represented as mean ± SEM (error bars; n = 3–5). (J) Principle of the membrane tethering assay. (K) Coomassie blue staining of the recombinant MOSPD2His6 and MSPHis6 proteins after SDS-PAGE. (L) Membrane tethering assays. LA liposomes (50 µM total lipids) composed of DOPC/DOGS-NTA-Ni2+ (98/2 mol/mol; a, b, and d) or DOPC (b) were mixed with LB liposomes (50 µM), composed of diphyt-PC/diphyt-PS (70/30 mol/mol) (a, b, and d) or DOPC (c) in HK buffer at 25°C. After 2 min, MOSPD2His6 (a–c) or MSPHis6 (d; 0.4 µM) was added and the size of liposomes was measured for 23 min. Left: Mean radius (dots) and polydispersity (shaded area) over time. Right: Hydrodynamic radius (RH) distribution before (gray bars) and after the reaction (green bars). These experiments are representative of several independent experiments (n = 3–5). Source data are available for this figure: SourceData F8.
We then tested whether the CRAL-TRIO domain binds to LDs via its AH. We produced in Escherichia coli and purified the WT and W201E mutant CRAL-TRIO domains of MOSPD2 fused with a His6 tag (Fig. 8 E). We also purified the MSP domain of MOSPD2 fused with a His6 tag as a control (Fig. 8 E). By circular dichroism spectroscopy, we established that secondary structure content of the W201E mutant was identical to that of the WT CRAL-TRIO domain, indicating that the mutation did not impair the folding of the domain (Fig. 7 E). To assess the ability of these three recombinant proteins to bind aLDs, we performed aLD pull-down assays. Each protein was immobilized on magnetic NTA-Ni2+ beads, owing to its His6 tag, and incubated with fluorescent aLDs (Fig. 8 F). After several washes to remove unbound aLDs, the beads were imaged (Fig. 8 G) and fluorescence-quantified using a fluorimeter (Fig. 8 H). In the absence of protein or in the presence of the MSPHis6, no fluorescence was measured, meaning that aLDs were not retained on the beads (Fig. 8 G, a and b; and 8 H). In contrast, when the wild type CRAL-TRIOHis6 was attached to the beads, a high fluorescence was detected showing that aLDs were retained by the protein (Fig. 8 G, c and 8 H). In comparison, a much lower aLD retention was observed with the W201E mutant (Fig. 8 G, d and 8 H). These data indicate that the AH of the CRAL-TRIO domain is instrumental for the protein to bind LDs.
To better define which membrane determinants facilitate MOSPD2 binding to LDs, we performed flotation assays with membranes that differ in terms of lipid packing defect and electrostatics (Fig. 8 I). The recombinant CRAL-TRIO domain of the protein was tested with different types of liposomes made of phospholipids and with or without negative charges and/or packing defects. Control liposomes with few packing defects and no charge were composed of DOPC. Negative charges were provided by replacing 30% of phosphatidylcholine by the anionic lipid phosphatidylserine (PS). Finally, packing defects were generated by using phospholipids containing diphytanoyl (diphyt-PC) acyl chains; diphytanoyl is a 16:0 acyl-chain with branched methyl groups that forms large packing defects. The CRAL-TRIOHis6 protein was poorly bound by control liposomes (no charges, no packing defects). It did not associate either with liposomes having negative charges only (DOPC/DOPS 70/30), or having packing defects only (diphyt-PC; <6% membrane-bound protein; Fig. 8 I, a). In contrast, more than 60% of the protein was associated with liposomes containing both negative charges and packing defects (diphyt-PC/diphyt-PS 70/30; Fig. 8 I, a). Flotation assays performed with liposomes bearing packing defects and increasing concentration of negatively charged phospholipids showed that the binding of the CRAL-TRIOHis6 protein was proportional to the amount of charges (Fig. 8 I, b). Thus, the binding of the CRAL-TRIO domain to packing defects bearing liposomes is tuned by electrostatics. Noteworthy, almost no binding was seen with MOSPD2 CRAL-TRIO W201E even in the presence of more packing defects and negative charges (Fig. 8 I, b). Collectively, these data show that in vitro the association of the CRAL-TRIO domain of MOSDP2 with a membrane is facilitated by the presence of very large packing defects and negatively charged lipids, both of which are characteristics of the LD surface.
Finally, we examined in vitro whether MOSPD2 was able to directly connect the ER with LDs by performing membrane tethering assays. These were done using two populations of liposomes, LA and LB, mimicking the ER and LDs, respectively. The association of liposomes into large particles as a result of membrane tethering was measured by dynamic light scattering (DLS). LA liposomes made of DOPC and doped with DOGS-NTA-Ni2+ were mixed with LB liposomes composed of diphyt-PC/diphyt-PS (70/30 mol/mol; Fig. 8 J). Then, MOSPD2His6, corresponding to the cytosolic part of MOSPD2 tagged with a C-terminal His6 tag (Fig. 8 K), was added so that LA liposomes were covered by the protein and constituted ER-like liposomes. A rapid increase in the initial mean radius of liposomes was observed suggesting that MOSPD2, once attached to LA liposomes, connected them with LB liposomes (Fig. 8 L, a). In contrast, no aggregation occurred when LA liposomes were devoid of attached MOSPD2His6 (Fig. 8 L, b), or when they were covered by the MSP domain of MOSPD2 (MSPHis6; Fig. 8 L, d). Moreover, no aggregation was observed when LB liposomes were replaced with liposomes that did not mimic LDs (i.e., without packing defects and negative charges; Fig. 8 L, c).
These data showed that MOSPD2, anchored to the ER by its C-terminus, directly connects this compartment with a second one delimited by a membrane with large packing defects and anionic lipids, such as LDs, owing to its CRAL-TRIO domain.
The formation of ER–LD contacts mediated by the CRAL-TRIO domain is essential for the function of MOSPD2 in the biology of LDs, while the MSP domain is dispensable
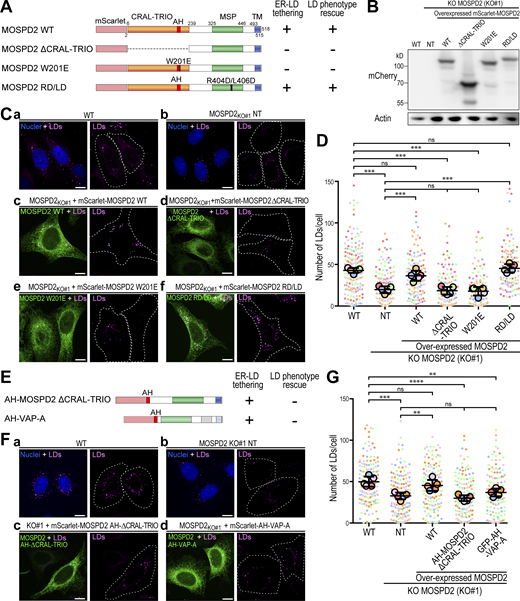
In the absence of MOSPD2, lipid droplets are fewer (Fig. 2 C). Having identified the molecular mechanism of ER–LD contact formation mediated by MOSPD2, we asked whether its ability to form ER–LD contacts was required for its role in LD biology. To answer this question, we performed rescue experiments by restoring MOSPD2 expression in knock-out cells (KO#1) using mScarlet-tagged WT or mutant MOSPD2 (Fig. 9, A and B). LDs were then labeled and their number quantified (Fig. 9, C and D). Consistent with data from Fig. 2, B and C, MOSPD2 knock-out cells had two times fewer LDs than wild-type cells (Fig. 9 C, a and b; and 9 D). When mScarlet-MOSPD2 was re-expressed in knock-out cells, the number of LDs was similar to that of WT cells (Fig. 9 C, c and 9 D). Thus, the ectopic expression of MOSPD2 rescues the LD phenotype of MOSPD2 knock-out cells.
The capacity of MOSPD2 to form ER–LD contact sites is necessary but not sufficient to regulate LDs. (A) Schematic representation of mScarlet-MOSPD2 constructs either WT (mScarlet-MOSPD2) or containing a deletion of the MSP domain (ΔMSP) or the CRAL-TRIO domain (ΔCRAL-TRIO), a mutation in the MSP domain (RD/LD) or in the CRAL-TRIO domain (W201E). For each construct, the LD tethering activity and the rescue (see panels below) are summarized as + or −. (B) Western Blot analysis of WT and MOSPD2 knock-out (KO#1) HeLa cells. MOSPD2 expression was rescued in MOSPD2 knock-out cells using mScarlet-MOSPD2 expression constructs either WT or mutant (mScarlet-MOSPD2 ΔCRAL-TRIO, W201E, and RD/LD). NT, non-transfected. (C) Representative confocal images of WT and MOSPD2 knock-out (KO#1) HeLa cells in which MOSPD2 expression was restored using mScarlet-MOSPD2 constructs (c, d, e, and f; green) depicted in panel A. As control, untransfected WT (a) and MOSPD2 knock-out (b) cells were imaged. LDs were stained with BODIPY 493/503 (magenta) and nuclei with Hoechst (blue). (D) Quantification of the number of LDs in cells shown in B. Data are displayed as Superplots showing the mean number of LDs per cell (small dots) and the mean number of LDs per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 213 (WT), 200 (KO#1), 150 (KO + mScarlet-MOSPD2 WT), 238 (KO + mScarlet-MOSPD2 ΔCRAL-TRIO), 126 (KO + mScarlet-MOSPD2 W201E), and 118 (KO + mScarlet-MOSPD2 RD/LD) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; ***, P < 0.001; n = 5 independent experiments). (E) Schematic representation of mScarlet constructs containing the deletion of the CRAL-TRIO domain together with an insertion of the AH (AH-MOSPD2-ΔCRAL-TRIO), and of the GFP-AH-VAP-A chimera in which the AH of MOSPD2 was fused at the N-terminus of VAP-A. For both constructs, the LD tethering activity and the rescue (see panels below) are summarized as + or −. (F) Representative confocal images of WT and MOSPD2 knock-out (KO#1) of HeLa cells in which constructs (green) from panel E were expressed (c and d). As control, untransfected WT (a) and MOSPD2 knock-out (b) cells were imaged. LDs were stained with BODIPY 493/503 (magenta) and nuclei with Hoechst (blue). (G) Quantification of the number of LDs in cells shown in F. Data are displayed as Superplots showing the mean number of LDs per cell (small dots) and the mean number of LDs per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 202 (WT), 192 (KO#1), 156 (KO + mScarlet-MOSPD2 WT), 147 (KO + mScarlet-AH-MOSPD2-ΔCRAL-TRIO), and 155 (KO + mScarlet-AH-VAP-A). One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; **, P < 0.01; ***, P < 0.001; n = 5 independent experiments). (C and F) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bars: 10 µm. The cell contour is shown with a white dotted line. Source data are available for this figure: SourceData F9.
The capacity of MOSPD2 to form ER–LD contact sites is necessary but not sufficient to regulate LDs. (A) Schematic representation of mScarlet-MOSPD2 constructs either WT (mScarlet-MOSPD2) or containing a deletion of the MSP domain (ΔMSP) or the CRAL-TRIO domain (ΔCRAL-TRIO), a mutation in the MSP domain (RD/LD) or in the CRAL-TRIO domain (W201E). For each construct, the LD tethering activity and the rescue (see panels below) are summarized as + or −. (B) Western Blot analysis of WT and MOSPD2 knock-out (KO#1) HeLa cells. MOSPD2 expression was rescued in MOSPD2 knock-out cells using mScarlet-MOSPD2 expression constructs either WT or mutant (mScarlet-MOSPD2 ΔCRAL-TRIO, W201E, and RD/LD). NT, non-transfected. (C) Representative confocal images of WT and MOSPD2 knock-out (KO#1) HeLa cells in which MOSPD2 expression was restored using mScarlet-MOSPD2 constructs (c, d, e, and f; green) depicted in panel A. As control, untransfected WT (a) and MOSPD2 knock-out (b) cells were imaged. LDs were stained with BODIPY 493/503 (magenta) and nuclei with Hoechst (blue). (D) Quantification of the number of LDs in cells shown in B. Data are displayed as Superplots showing the mean number of LDs per cell (small dots) and the mean number of LDs per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 213 (WT), 200 (KO#1), 150 (KO + mScarlet-MOSPD2 WT), 238 (KO + mScarlet-MOSPD2 ΔCRAL-TRIO), 126 (KO + mScarlet-MOSPD2 W201E), and 118 (KO + mScarlet-MOSPD2 RD/LD) cells. One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; ***, P < 0.001; n = 5 independent experiments). (E) Schematic representation of mScarlet constructs containing the deletion of the CRAL-TRIO domain together with an insertion of the AH (AH-MOSPD2-ΔCRAL-TRIO), and of the GFP-AH-VAP-A chimera in which the AH of MOSPD2 was fused at the N-terminus of VAP-A. For both constructs, the LD tethering activity and the rescue (see panels below) are summarized as + or −. (F) Representative confocal images of WT and MOSPD2 knock-out (KO#1) of HeLa cells in which constructs (green) from panel E were expressed (c and d). As control, untransfected WT (a) and MOSPD2 knock-out (b) cells were imaged. LDs were stained with BODIPY 493/503 (magenta) and nuclei with Hoechst (blue). (G) Quantification of the number of LDs in cells shown in F. Data are displayed as Superplots showing the mean number of LDs per cell (small dots) and the mean number of LDs per independent experiment (large dots). Independent experiments (n = 5) are color-coded. Means and error bars (SD) are shown as black bars. Data were collected from 202 (WT), 192 (KO#1), 156 (KO + mScarlet-MOSPD2 WT), 147 (KO + mScarlet-AH-MOSPD2-ΔCRAL-TRIO), and 155 (KO + mScarlet-AH-VAP-A). One-way ANOVA with Tukey’s multiple comparisons test (ns, not significant; **, P < 0.01; ***, P < 0.001; n = 5 independent experiments). (C and F) Images were acquired on a spinning-disk confocal microscope (Nikon CSU-X1, 100× NA 1.4). Scale bars: 10 µm. The cell contour is shown with a white dotted line. Source data are available for this figure: SourceData F9.
Next, we repeated the rescue experiment by expressing two MOSPD2 mutants unable to form ER–LD contacts, a deletion mutant devoid of the CRAL-TRIO domain (mScarlet-MOSPD2 ΔCRAL-TRIO) and a point mutant with a defective AH (mScarlet-MOSPD2 W201E), in MOSPD2 knock-out cells (Fig. 9, A and B). These two mutants failed to rescue the absence of MOSPD2 (Fig. 9 C, d and e; and 9 D).
In contrast, the expression of a mutant MOSPD2 having an MSP domain unable to bind FFAT motifs (mScarlet-MOSPD2 RD/LD) restored the number of LDs to a level similar to that of WT cells (Fig. 9 C, f; and 9 D). Thus, the ability of MOSPD2 to bind FFAT motifs is dispensable for its function in LDs. These experiments show that the ability of MOSPD2 to form ER–LD contacts by its CRAL-TRIO domain contributes to LD biology.
Finally, we tested whether promoting ER–LD tethering was sufficient to recapitulate the function of MOSPD2. We expressed two constructs lacking a CRAL-TRIO domain but capable of promoting ER–LD contact formation (Fig. 7, F–I): a MOSPD2 mutant in which the CRAL-TRIO domain was replaced by the AH only (AH-MOSPD2 ΔCRAL-TRIO fusion protein), and the chimeric protein composed of the AH of MOSPD2 fused to VAP-A (Fig. 9 E). While these two proteins promoted ER–LD contact formation (Fig. 7, F–I), they did not rescue the number of LDs in MOSPD2-deficient cells (Fig. 9, F and G). These data show that MOSPD2 must connect the ER with the LDs to exert its activity, but that the membrane tethering ability by itself is not sufficient to recapitulate the activity of MOSPD2.
Together, these data show that the ability of MOSPD2 to create ER–LD contacts is crucial for the activity of the protein in the biology of LDs. Moreover, it shows that the CRAL-TRIO domain is instrumental for MOSPD2 function in LDs, while the MSP domain is dispensable.
Discussion
Organelles are no longer considered as isolated compartments but as active units able to constantly communicate and function with each other. The ER plays a key role in the interactions between organelles, as it is a meshwork of membrane tubes and sheets that extend throughout the cytosol and make extensive contacts with other organelles (Wu et al., 2018). LDs, which are involved in cellular energy storage, have a very unique relationship with the ER. They are generated from the ER and maintain regular physical contacts with it throughout their life cycle (Olzmann and Carvalho, 2019; Walther et al., 2017). LD biogenesis starts with the synthesis of neutral lipids in the ER membrane. These newly made lipids nucleate into oil lenses in the ER bilayer that ultimately bud toward the cytosol and grow further. At the beginning of their life, LDs are attached to the ER as their monolayer is continuous with the cytosolic leaflet of the ER membrane (Hugenroth and Bohnert, 2020; Salo and Ikonen, 2019). Eventually, LDs bud toward the cytosol and detach from the ER but continue to establish contacts with the ER by other mechanisms. This physical connectivity might ensure the functional interplay between the ER and LDs. In this study, we reveal that MOSPD2 contributes to this process by forming contacts between the ER and LDs, and participates in the biology of these organelles (Fig. 10).
Schematic representation of ER–LD contact sites mediated by MOSPD2. MOSPD2 tethers the ER to LDs thanks to its TM and CRAL-TRIO domains. The amphipathic helix located in the CRAL-TRIO domain directly interacts with the surface of LDs. The CRAL-TRIO domain of MOSPD2 might also be involved in lipid binding and/or transport between the ER and LDs.
Schematic representation of ER–LD contact sites mediated by MOSPD2. MOSPD2 tethers the ER to LDs thanks to its TM and CRAL-TRIO domains. The amphipathic helix located in the CRAL-TRIO domain directly interacts with the surface of LDs. The CRAL-TRIO domain of MOSPD2 might also be involved in lipid binding and/or transport between the ER and LDs.
We previously showed that MOSDP2 mediates the formation of contacts between the ER and endosomes, the Golgi, and mitochondria (Di Mattia et al., 2018) by a mechanism relying on its MSP domain. By binding to FFAT motifs present in proteins on the surface of these organelles, the MSP domain of MOSPD2 attaches the ER to the other organelle, as do the VAP-A and VAP-B proteins (Di Mattia et al., 2018, 2020a; Murphy and Levine, 2016). Here, we identified that MOSPD2 has a second tethering activity to specifically create ER–LD contacts by a mechanism that does not rely on its MSP domain, but surprisingly on its CRAL-TRIO domain.
The association of this domain with LDs is mediated by an amphipathic helix that is conserved in other members of the CRAL-TRIO family (Bankaitis et al., 2010), and notably its archetypical member, Sec14p (Sha et al., 1998). In this protein, this helix acts as a gate regulating access to the hydrophobic cavity of the protein (Ryan et al., 2007; Bankaitis et al., 1990; Sha et al., 1998). AHs are found in a variety of proteins interacting with membranes; they do not have a consensus sequence, and they were shown to interact with diverse polar/non-polar interfaces (Giménez-Andrés et al., 2018). Depending on the type of hydrophobic residues on their apolar side, the nature and charge of residues on their polar side, and their length, they can interact with membranes having distinct properties in terms of lipid composition, charge, curvature, etc. (Giménez-Andrés et al., 2018). AHs binding to LDs have affinity for packing defects owing to the presence of large hydrophobic residues on their apolar face (Prévost et al., 2018; Chorlay and Thiam, 2020; Čopič et al., 2018). In agreement with this notion, mutation of W201 in the AH of MOSPD2 abolishes its binding to LDs. Besides, the CRAL-TRIO domain of MOSDP2 has no or little affinity for other cellular membranes, such as the ER membrane, which is expected to be more packed than the LD surface and contain no more than 15 mol% negatively charged lipids (van Meer et al., 2008; Chorlay and Thiam, 2020; Chorlay et al., 2021).
The main feature of the CRAL-TRIO domain is that it possesses a cavity to specifically host a lipophilic molecule. CRAL-TRIO domain-containing proteins belong to the large category of lipid transfer proteins (Chiapparino et al., 2016). Sec14p can exchange glycerophospholipids (PI and PC) between membranes, whereas retinaldehyde-binding protein 1 and α-tocopherol transfer protein use a CRAL/TRIO domain to convey retinaldehyde and α-tocopherol (vitamin E), respectively. Thus, an appealing hypothesis is that MOSPD2 could transport fatty acids to fuel LD enzymes that build neutral lipids or transports phospholipids to the monolayer of LDs to allow their proper growth. The reduced level of sterol esters in MOSPD2-deficient cells points to a link between MOSPD2 and the metabolism of cholesterol and its derivatives. Therefore, one possibility is that MOSPD2 could transport sterols toward LDs. Interestingly, other lipid transfer proteins are present in ER–LD contacts such as ORP5 and ORP8 (Guyard et al., 2021; Du et al., 2020), thus raising the possibility that ER–LD contacts are a major platform for the non-vesicular exchange of lipids. If MOSPD2 is a lipid transporter, we can speculate that the AH of the CRAL-TRIO domain of MOSPD2 has a dual function: interacting with the surface of LDs to mediate the formation of ER–LD contacts and being a gate to access the hydrophobic cavity of the CRAL-TRIO domain of the protein. However, a debate still exists on whether Sec14p exchanges lipids between organelles in yeast. As proposed for Sec14p, rather than transporting lipids, MOSPD2 could act by presenting its lipid substrate to an enzyme and thus increase its activity (Lete et al., 2020). Accordingly, we cannot exclude that MOSPD2 acts directly on sterol or sterol-ester metabolism. Further work will be needed to determine whether MOSPD2 mediates lipid transport at the ER/LD interface.
It is intriguing that a single protein possesses two tethering mechanisms targeting distinct organelles: on the one hand, the MSP domain which allows the formation of contacts with many organelles via protein-protein interactions and on the other hand the CRAL-TRIO domain, which contacts only one organelle, the LD, via a protein–membrane interaction. The ability of MOSPD2 to tether the ER to other organelles using two alternate molecular mechanisms is a new illustration of the plasticity of inter-organelle contacts. Indeed, other tether proteins alternate between distinct contacts. For instance, the mitochondria-bound protein MIGA2 is present in mitochondria–ER contacts when bound to VAP proteins, thanks to its Phospho-FFAT motif, and alternatively in mitochondria–LD contacts by directly binding to the surface of LDs, thanks to an amphipathic helix. These two localizations of MIGA2 most probably reflect alternate functions of the protein related to cellular metabolism (Klemm, 2021; Freyre et al., 2019). Similarly, some members of the VPS13 family are present in different inter-organelle contacts, in contact with LDs using an AH (Yeshaw et al., 2019; Wang et al., 2021; Kumar et al., 2018; Ramseyer et al., 2018), and with the ER using FFAT and Phospho-FFAT (Guillén-Samander et al., 2021; Wang et al., 2021). In the case of MOSPD2, either deleting the MSP domain or preventing its binding to FFAT motifs, unexpectedly promoted the formation of ER–LD contacts. This observation suggests that MOSPD2 is balanced between two kinds of membrane contact sites: MSP domain-dependent contacts that involve FFAT-containing partners of MOSPD2 and CRAL-TRIO domain-dependent contacts that involve the direct recognition of the surface of LDs by MOSPD2. It is plausible that regulation mechanisms exist to control the repartition of MOSPD2 between the two types of contact sites in which the protein might then play distinct roles. In line with this, we have recently shown that phosphorylation of FFAT-like motifs, which we named Phospho-FFAT, allows a regulatable MSP-dependent formation of contact sites (Di Mattia et al., 2020a). Thus, the Phospho-FFAT phosphorylation status of MOSPD2 partners can probably indirectly control pools of MOSPD2 in and out of ER–LD contacts. We also observed that in cells producing large amounts of LDs, MOSPD2 accumulated in ER–LD contacts, suggesting that ER–LD contacts are privileged and that the metabolic state of the cell probably dictates the localization and function of MOSPD2. Moreover, the binding of MOSPD2 to LDs might also be tuned by the lipid composition of LDs; in agreement with our in vitro data, changes in the phospholipid monolayer or in the neutral core of LDs most probably regulate the affinity of binding of MOSPD2. Accordingly, endogenous MOSPD2 is not enriched on the surface of all LDs, raising the possibility that it preferentially attaches to a subpopulation of LDs (e.g., sterol-rich LDs), as previously shown for some members of the Perilipin family (Hsieh et al., 2012). It is not yet known whether the absence of MOSPD2 affects the amount of ER–LD contacts. Moreover, it is unclear whether the association of MOSPD2 with LDs is at the expense of other specific contacts or simply reduces the amount of MOSPD2 that can be recruited by FFAT-containing partners. In either case, this would represent an additional level of control of contacts between the ER and other organelles.
To summarize, we report that MOSPD2 builds ER–LD contacts and thereby affects LD homeostasis. MOSPD2 shares many similarities with VAP-A and VAP-B: these three proteins recognize FFAT and Phospho-FFAT motifs, they have many partners in common, and by this molecular mechanism are involved in the formation of contacts between the ER and many organelles. The finding that MOSPD2 makes additional contacts through a mechanism distinct to that of VAP-A and VAP-B provides specificity and expands the repertoire of contacts that this protein can make. The formation of contacts between the ER and other organelles is a plastic phenomenon involving complex networks of interactions, and future work should provide evidence on how the recruitment of MOSPD2 in a given inter-organelle contact is orchestrated.
Materials and methods
Cloning and constructs
The GFP-MOSPD2 (WT and RD/LD mutant), GFP-VAP-A, and GFP-TM(SAC1) expression vectors were previously described (Alpy et al., 2013; Di Mattia et al., 2018).
The GFP-MOSPD2 ∆MSP, GFP-MOSPD2 ∆TM, GFP-MOSPD2 ∆CRAL-TRIO, GFP-MOSPD2 W201E expression vectors were constructed by overlap extension PCR using GFP-MOSPD2 as a template and the following central primers: GFP-MOSPD2 ∆MSP: 5′-AGTGTATTTAAAGGCCCCGAAAGCAGTAAACCAAAC-3′ and 5′-GTTTGGTTTACTGCTTTCGGGGCCTTTAAATACACT-3′; GFP-MOSPD2 ∆TM: 5′-CAGCGTTGTATCTGAATTCCAGCAGCTGCTGCTTTCC-3′ and 5′-CAGCTGCTGGAATTCAGATACAACGCTGAACTTGGTC-3′; GFP-MOSPD2 ∆CRAL-TRIO: 5′-ATCATCTACTAGTGGTGGATAGCTAGAATTCGAAGCTTGAGCTCGAGA-3′ and 5′-TCTCGAGCTCAAGCTTCGAATTCTAGCTATCCACCACTAGTAGATGAT-3′; GFP-MOSPD2 W201E: 5′-ATTGTGAAAACCGAACTTGGTCCAGAAGCAGTGAGC-3′ and 5′-TTCTGGACCAAGTTCGGTTTTCACAATTTTGAAAGC-3′; and the peripheral primers 5′-GAGACGGCCGATGGTGAGCAAGGGCGAGGAGCTG-3′, and 5′-GAGAGGATCCTTAACTGTACAATAAATAGAAG-3′. PCR fragments were cloned by ligation into the BamHI and EagI-linearized pQCXIP vector.
The GFP-AH-MOSPD2 ∆CRAL-TRIO expression vector was obtained by PCR using GFP-MOSPD2 as template and the following central primers 5′-TGGACCAAGCCAGGTTTTCACAATTTTGAAAGCAGCATTCATTAACCAAGGCATAGAATTCGAAGCTTGAGCTCGAGA-3′ and 5′-TGGACCAAGCCAGGTTTTCACAATTTTGAAAGCAGCATTCATTAACCAAGGCATAGAATTCGAAGCTTGAGCTCGAGA-3′ and the peripheral primers 5′-GGAATTGATCCGCGGCCGCCGATGGTGAGCAAGGGCGAGGAGCTGT-3′ and 5′-GGGCGGAATTCCGGATCTTAACTGTACAATAAATAGAAGAAAGAGGTGACAAAAGCAAG-3′. PCR fragments were cloned using the SLiCE (Seamless Ligation Cloning Extract) method (Okegawa and Motohashi, 2015) into the NotI and BamHI-linearized pQCXIP vector.
GFP-AH-VAP-A construct was obtained by PCR using the following primers: 5′-GGAATTGATCCGCGGCCGCCGATGGTGAGCAAGGGCGAGGAGCTGT-3′, 5′-TGGACCAAGCCAGGTTTTCACAATTTTGAAAGCAGCATTCATTAACCAAGGCATAGAATTCGAAGCTTGAGCTCGAGA-3′, and 5′-TGCCTTGGTTAATGAATGCTGCTTTCAAAATTGTGAAAACCTGGCTTGGTCCAGCGTCCGCCTCAGGGGCCATG-3′, 5′-GGGCGGAATTCCGGATCCTACAAGATGAATTTCCCTAG-3′, and GFP-VAP-A as a template. PCR fragments were cloned by SLiCE into the NotI and BamHI-linearized pQCXIP vector.
The GFP-MOSPD2-TM(SAC1) construct was obtained by PCR using the following primers: 5′-GGAATTGATCCGCGGCCGCCGATGGTGAGCAAGGGCGAGGAGCTGT-3′, 5′-AACAACCATGATAATAGGCAAAGCCAGGAAGATACAACGCTGAACTTGGTCTTCAAGCTT-3′, and 5′-TTCCTGGCTTTGCCTATTATCATGGTTGTT-3′, 5′-GGGCGGAATTCCGGATCTCAGTCTATCTTTTCTTTCTGGACCAGTCT-3′, and GFP-MOSPD2 and GFP-TM(SAC1) as a template, respectively. PCR fragments were cloned by SLiCE into the NotI and BamHI-linearized pQCXIP vector.
GFP-CRALTRIO-TM(SAC1) construct was obtained by PCR using the following primers: 5′-GGAATTGATCCGCGGCCGCCGATGGTGAGCAAGGGCGAGGAGCTGT-3′, 5′-AACAACCATGATAATAGGCAAAGCCAGGAAGGGGCCTTTAAATACACTCAATGG-3′, and 5′-TTCCTGGCTTTGCCTATTATCATGGTTGTT-3′, 5′-GGGCGGAATTCCGGATCTCAGTCTATCTTTTCTTTCTGGACCAGTCT-3′, and GFP-MOSPD2 and GFP-TM(SAC1) as a template, respectively. PCR fragments were cloned using the SLiCE method into the NotI and BamHI-linearized pQCXIP vector.
mScarlet-ER construct was obtained by PCR using the following primers: 5′-GGAATTGATCCGCGGCCGCCACCATGGTGAGCAAGGGC-3′, 5′-CCGGACTTGTACAGCTCGTCCATGCCGCCGGTGGAGTGGCGGCCCTCGGAGC-3′, and 5′-GCTGTACAAGTCCGGATTCCTGGCTTTGCCTATTATCATGGTTGTTGCCTTT-3′, 5′-GGGGGGGGCGGAATTCTCAGTCTATCTTTTCTTTCTGGACCAGTCTGGGAGC-3′ and pmScarlet-C1 (gift from Dorus Gadella, University of Amsterdam, Amsterdam, Netherlands; Addgene plasmid # 85042; http://n2t.net/addgene:85042; RRID:Addgene_85042) and GFP-TM(SAC1) as a template, respectively. PCR fragments were cloned using the SLiCE method into the NotI and BamHI-linearized pQCXIP vector.
mScarlet-MOSPD2 WT, mScarlet-MOSPD2 W201E, mScarlet-MOSPD2 RD/LD, mScarlet-MOSPD2 AH-∆CRAL-TRIO, and mScarlet-AH-VAP-A were obtained from GFP-MOSPD2 WT, GFP-MOSPD2 W201E, GFP-MOSPD2 RD/LD, GFP-MOSPD2 AH-∆CRAL-TRIO, and GFP-AH-VAP-A in which the GFP cassette was excised by SbfI and XhoI digestion and replaced using SLiCE by the coding sequence of mScarlet amplified by PCR using the primers: 5′-TGCATTGGAACGGACCTGCAGCCACCATGGTGAGCAAGGGCGAGGCAGTGATCAA-3′ and 5′-GCAGAATTCGAAGCTTGAGCTCGAGATCTGAGTCCGGACTTGTACAGCTCGTCCAT-3′ and pmScarlet-C1 as a template.
mScarlet-MOSPD2 ∆CRAL-TRIO was obtained from mScarlet-MOSPD2 WT in which the coding sequence of MOSPD2 was excised by XhoI and BamHI digestion and replaced using SLiCE by MOSPD2 ∆CRAL-TRIO coding sequence amplified by PCR using the primers: 5′-TCCGGACTCAGATCTCGAAGCTATCCACCACTAGTAGATGATGACTTCCAGACCCCACTGTGTGAG-3′ and 5′-GGGCGGAATTCCGGATCTTAACTGTACAATAAATAGAAGAAAGAGGTGACAAAAGCAAG-3′.
All constructs were verified by DNA sequencing (Eurofins Genomics).
Cell culture, transfection, and infection
HeLa cells (American Type Culture Collection [ATCC] CCL-2, RRID:CVCL_0030) were maintained in DMEM (4.5 g/liter glucose) with 5% FCS and 40 µg/ml gentamycin. 293T cells (ATCC CRL-3216) were maintained in DMEM (4.5 g/liter glucose) with 10% FCS, 100 UI/ml penicillin, and 100 µg/ml streptomycin. Huh-7 cells (JCRB0403, RRID:CVCL_0336) were maintained in DMEM (4.5 g/liter glucose) with 10% FCS, 0.1 mM Non Essential Amino Acids, 1 mM sodium pyruvate, and 40 µg/ml gentamycin. 501-MEL cells (RRID:CVCL_4633; obtained from Dr. Colin Goding, University of Oxford, Oxford, UK) were maintained in RPMI without Hepes with 10% FCS and 40 µg/ml gentamycin. MCF-7 cells (ATCC HTB-22) were maintained in DMEM (1 g/liter glucose) with 10% FCS, 0.6 µg/ml insulin, and 40 µg/ml gentamycin.
HeLa WT and Seipin-KO (kind gift from Hongyuan Robert Yang, University of New South Wales, Sydney, Australia) were maintained in DMEM High Glucose (Dutscher) with 10% FBS and 1% penicillin–streptomycin. They were transfected using jetPEI transfection reagent (PolyPlus #101-10N).
Cells were transfected using X-tremeGENE 9 DNA transfection reagent (Roche). To generate retroviral particles, pQCXIP vectors were co-transfected with pCL-Ampho vector (Imgenex) into 293T retroviral packaging cell line. Retroviral infections were used to generate HeLa/Ctrl, HeLa/GFP-MOSPD2, HeLa/GFP-MOSPD2 RD/LD, HeLa/GFP-MOSPD2 ∆MSP, HeLa/GFP-MOSPD2 ∆TM, HeLa/GFP-MOSPD2 ∆CRAL-TRIO, and HeLa/mScarlet-ER cell lines. The HeLa/Ctrl cell line was obtained using the empty pQCXIP plasmid.
siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Control siRNA (D-001810-10) and MOSPD2-targeting siRNAs (J-017039-09) were SMARTpool ON-TARGETplus obtained from Horizon Discovery.
Oleic acid was complexed with fatty acid-free BSA, as described in Listenberger and Brown (2007), and diluted in cell culture medium. Unless otherwise stated, cells were treated overnight with 400 µM OA.
CRISPR/Cas9-mediated genome editing
To generate MOSPD2 KO clones, HeLa cells were plated in 100-mm dishes and transfected with pX751 mCherry-Cas9 HF plasmid (gRNA deleting MOSPD2 exon 5: 5′-CAAGTGCAACAGTTTCTCATT-3′/5′-TGTTTGACTACACTCACACT-3′) using X-tremeGENE 9 DNA Transfection Reagent (Roche). 48 h after transfection, clones were sorted and isolated in 96-well plates using FACS (Fusion). Clones were then screened by PCR (5′-CATCTTAGCTACCACCACCTGAACAGTTTAC-3′/5′-GCCTCGACATGCTACCTCTCC-3′ and 5′-CATCTTAGCTACCACCACCTGAACAGTTTAC-3′/5′-AATTGCTGCTGAAGGGTTTGTAGGTATC-3′) and further analyzed by Western Blot (anti-MOSPD2; HPA003334; Sigma-Aldrich, RRID:AB_2146004) and Sanger sequencing (Eurofins Genomics).
To generate endogenous mClover3-MOSPD2 knock-in cells, HeLa cells were plated in 100-mm dishes and transfected with the pX852 plasmid (encoding mCherry-Cas9 and two gRNAs: 5′-AACCGCAATCACATCCACGA-3′/5′-CACCTCTGCCATGATCACCG-3′) and the repair template (synthesized by ProteoGenix). The repair template was composed of two homology arms of 1,000 bp flanking a puromycin resistance gene and mClover3 coding sequence separated by a P2A cleavage site. The insertion was made in the first exon of MOSPD2 genomic sequence to allow expression of a fusion protein with mClover3 at the N-terminus of MOSPD2. 3 d after transfection, cells were selected with medium containing 0.5 µg/μl of puromycin. After 5 d of selection, cells were sorted in 96-well plates. Clones were screened by PCR (5′-GTGAATTTTCATGTACACTGGAGGATGTTTGGCAGC-3′/5′-GCGAGGCGCACCGTGGGCTTGTACTCGGTC-3′ and 5′-ACACATGGCATGGACGAGCTGTACAAGTCC-3′/5′-GCTTAACTCCTTTCACAGTAACCAAAATGAC-3′) and analyzed by Western Blot (anti-GFP and anti-MOSPD2) and Sanger sequencing (Eurofins Genomics).
Immunofluorescence
Cells were grown on glass coverslips, fixed in 4% paraformaldehyde in PBS for 15 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking with 1% bovine serum albumin in PBS (PBS-BSA), cells were incubated overnight at 4°C with the primary antibody in PBS-BSA. Primary antibodies were: rabbit anti-MOSPD2 (1:250; HPA003334; Sigma-Aldrich, RRID:AB_2146004), rabbit anti-GFP (1:1,000; TP401; Torrey Pine Biolabs, RRID:AB_10013661), mouse anti-GFP (1:1,000; 2A3; IGBMC), rabbit anti-Calnexin (1:1,000; 10427-2-AP; Proteintech, RRID:AB_2069033), anti-PLIN3 (1:1,000; GP36; Progen), mouse anti-EEA1 (1:1,000; 610457; BD Biosciences, RRID:AB_397830), rabbit anti-ORP1 (1:200; EPR8646; Abcam), rabbit anti-GM130 (1:500; 11308-1-AP; ProteinTech, RRID:AB_2115327), mouse anti-Lamp1 H4A3 (1:50; DSHB, RRID:AB_2296838) and mouse anti-OPA-1 (1:1,000; 1A8; IGBMC). Cells were washed twice in PBS and incubated for 30 min with the secondary antibody (AlexaFluor 488 [RRID: AB_2535792 and AB_141607], AlexaFluor 555 [RRID: AB_2762848 and AB_162543], AlexaFluor 647 [RRID: AB_2536183 and AB_162542] from Thermo Fisher Scientific and Abberior STAR 580 [RRID: AB_2620153] from Abberior). After two washes with PBS, the slides were mounted in ProLong Gold (Invitrogen). Observations were made with a Leica TCS SP5 inverted confocal microscope (63×, NA 1.4), a Leica SP8 UV inverted confocal microscope (63×, NA 1.4), and a spinning-disk confocal microscope (CSU-X1; Nikon, 100×, NA 1.4). 2D-STimulated Emission Depletion (STED) imaging was performed with a Leica SP8 STED 3× microscope in a thermostated chamber at 21°C and equipped with a STED motorized oil immersion objective (HC PL Apo 100×/NA 1.40 CS2). Excitation was performed with white-light laser and depletion with a 775 nm pulsed laser (STED 775). Excitation and depletion lasers were calibrated with the STED auto beam alignment tool during imaging sessions. HeLa WT and KO Seipin cells were observed on a Carl ZEISS LSM 800 Airyscan microscope.
To stain LDs, cells were incubated after permeabilization with either BODIPY 493/503 (0.5 µg/ml in 150 mM NaCl; Thermo Fisher Scientific), Nile Red (1:8,000 in 150 mM NaCl; Thermo Fisher Scientific), or HCS DeepRed LipidTOX (1:1,000 in PBS; Invitrogen) for 20 min at room temperature.
Quantification of ring-like/coma-shaped structures
HeLa WT cells were plated in 24-well plates (30,000 cells/well) and transfected the same day. 2 d after transfection, cells were fixed using 4% PFA in PBS, washed two times with PBS, and mounted on glass slides in ProLong Gold (Invitrogen).
Images were acquired on a Leica SP5 inverted confocal (63× oil objective, NA 1.4). The presence of enrichments (coma- and ring-shaped structures) was confirmed by eyes from two individuals.
Quantification of LD number and size
Two million cells were plated in T75 flasks and allowed to grow for 48 h. 30,000 cells were then plated in 24-well plates on glass coverslips. For the rescue experiment, HeLa MOSPD2 KO#1 cells were transfected the same day with different plasmids as previously described. After 48 h, cells were fixed in 4% PFA PBS and permeabilized as described above. LDs were stained using BODIPY 493/503 and nuclei with Hoechst for 20 min. Cells were mounted on glass slides in ProLong Gold (Invitrogen). Images were acquired on a spinning-disk CSU-X1 (Nikon, 100× NA 1.4) using the same setup every time (laser power, number of z-slices, exposition length). Cells were selected based on the nuclei channel to avoid any bias.
For image processing (illustrated in Fig. S4 A), a z-stack projection (max intensity) was performed on ImageJ Fiji (Schindelin et al., 2012). These images were then processed using CellProfiler (McQuin et al., 2018). First, cells were manually segmented to create cell masks. Then, LDs were identified as objects with a diameter ≥2 pixels (i.e., 220 nm) within cell masks using the global threshold strategy and the minimum cross-entropy method. Multiple parameters (object intensity, object neighbors, and object size/shape) were analyzed on identified LDs and these data were treated using Spyder 4.1 (Python 3.7) and GraphPad Prism. See https://github.com/mzouiouich/Quantifications_MOSPD2.
Quantification of ER fluorescence signal around LDs
Stable mScarlet-ER HeLa cells were plated in 24-well plates (30,000 cells/well) and transfected the same day with either GFP-VAP-A or GFP-MOSPD2. 36 h later, cells were treated with 50 µM of OA for 6 h. Cells were fixed in PFA 4% PBS, permeabilized with Triton X-100 0.1%, and LDs were stained using HCS DeepRed LipidTOX as described above. Images were acquired on a Leica SP5 inverted confocal (63× oil objective, NA 1.4) and cells were selected based on the GFP signal.
Image processing (illustrated in Fig. S4 B) was performed on CellProfiler. In brief, cells were manually selected and the nucleus was excluded (cell mask). The LDs were identified as objects superior or equal to 4 pixels of diameter using the global threshold strategy and the minimum cross-entropy method. Then, 2 pixels-wide areas were added from 0 to 20 pixels around LDs. These areas are mutually exclusive, meaning that the same pixel can be measured only once (i.e., for one LD). Multi-parametric measurements were performed for each area around LDs in the GFP channel (MOSPD2 and VAP-A) and mScarlet channel (ER marker). Data were analyzed on Excel and GraphPad. See https://github.com/mzouiouich/Quantifications_MOSPD2.
CLEM
EM was performed as previously described (Di Mattia et al., 2018; Wilhelm et al., 2017; Alpy et al., 2013). Cells grown on carbon-coated sapphire disks were cryoprotected with DMEM containing 10% FCS and frozen at high pressure (HPM 10 Abra Fluid AG). Samples were then freeze-substituted and embedded in lowicryl HM20. Thick sections (∼250 nm) were collected on carbon-coated copper grids (200 Mesh; AGS160; Agar Scientific).
EM grids were placed on a MatTek glass bottom dish in a drop of water and imaged with a spinning-disk confocal microscope (CSU-X1, Nikon, 100× oil objective, NA 1.4). The position of the imaged cells was determined using the asymmetrical center mark of the grid. Then, samples were imaged with a transmission electron microscope (Philips CM12) coupled to an Orius 1000 CCD camera (Gatan). Images were processed and merged with the open-source software Icy (de Chaumont et al., 2012) using the eC-CLEM plugin (Paul-Gilloteaux et al., 2017).
FRAP
Cells were plated on 35-mm glass bottom dishes (MatTek), transfected with plasmids encoding GFP-MOSPD2 or GFP-PLIN2 (gift from Elina Ikonen, University of Helsinki, Helsinki, Finland; Addgene plasmid # 87161; http://n2t.net/addgene:87161; RRID:Addgene_87161), and allowed to grow for 24 h. Cells were then treated with 400 µM OA overnight. HCS LipidTOX Deep Red Neutral Lipid Stain (H34477; Invitrogen) at a 1:2,000 dilution was added to the medium without phenol red 10 min prior imaging. Experiments were performed using the spinning-disk CSU-X1 (Nikon; 100× oil objective, NA 1.4). A region of interest was photobleached with the 405-nm laser line at 15% laser power and 5 repetitions. Recovery of fluorescence was monitored every second for 1 min immediately after photobleaching.
Protein expression and purification
The recombinant WT, the mutants CRAL-TRIOHis6 (MOSPD2 2-246) and MSPHis6 (MOSPD2 315-445) proteins, and the recombinant MOSPD2His6 (MOSPD2 1-490) protein corresponding to the full-length protein with its carboxyl-terminal transmembrane region deleted were expressed in E. coli BL21 (DE3) at 20°C for 16 h upon induction with 1 mM IPTG (at an OD600nm = 0.5). Cells were suspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 30 mM imidazole, 1 mM dithiothreitol, protease inhibitors tablets cOmplete; Roche). Cells were lysed by a Cell Disruptor TS SERIES (Constant Systems Ltd.) and the lysate was first centrifuged at 3,500 g for 15 min, then at 50,000 g for 45 min, and filtered through a 0.22-µm membrane. Purification was performed on an ÄKTA Start chromatography system (GE Healthcare Life Sciences) using HisTrap HP 1 ml columns. Proteins were eluted with Elution buffer (50 mM Tris-HCl, pH 7.5, 300 mM imidazole, 1 mM dithiothreitol) and further purified by gel filtration (HiLoad 16/60 Superdex 200; GE) in GF Buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM dithiothreitol). Proteins were concentrated with an Amicon Ultra-4 10 kD centrifugal filter unit (Merck). Protein concentration was determined by UV-spectroscopy.
Peptide synthesis
Peptides were synthesized on an Applied biosystem 433A peptide synthesizer using standard Fmoc chemistry and purified by reverse phase HPLC using a preparative scale column (Phenomenex: Kinetex EVO C18, 100 Å, 5 µM, 250 × 21.2 mm). Molecular weight and purity of the peptides were confirmed by mass spectrometry.
SDS-PAGE, Western blot, and Coomassie blue staining
SDS-PAGE and Western blot analysis were performed as previously described (Alpy et al., 2005) using the following antibodies: rabbit anti-GFP (1:2,000; TP401; Torrey Pine Biolabs), rabbit anti-MOSPD2 (1:250; HPA003334; Sigma-Aldrich), rabbit anti-mCherry (1:1,000; ab167453; Abcam), and mouse anti-actin (1:5,000; ACT-2D7; IGBMC).
Protein gels were stained with Coomassie blue (PageBlue Protein Staining Solution; Thermo Fisher Scientific).
Lipids
DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), diphytanoyl-PC (1,2-diphytanoyl-sn-glycero-3-phosphocholine), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine), diphytanoyl-PS (1,2-diphytanoyl-sn-glycero-3-phospho-L-serine), NBD-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl), 18:1 Liss Rhod PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[lissamine rhodamine B sulfonyl]), and DOGS-NTA-Ni2+ (1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl) iminodiacetic acid) succinyl] [nickel salt]) were purchased from Avanti Polar Lipids. Glyceryl trioleate was purchased from Sigma-Aldrich.
TLC
Two million cells were plated in T75 flask and allowed to grow for 48 h. Then, 500,000 cells were plated in 6-well plates. After 24 h, cells were lysed with 0.1% SDS. Proteins were quantified, and the lysate equivalent to 450 µg of proteins was transferred into a new tube and the volume adjusted to 800 μl. Lipids were extracted using Bligh & Dyer protocol (water/chloroform/methanol with a ratio of 1.8:2:2 vol/vol) in glass tubes (Bligh and Dyer, 1959). After 5 min of centrifugation at 400 g, the lower-phase containing the lipids was transferred into a disposable glass tube and dried under a nitrogen stream. Lipids were solubilized in three drops of a chloroform/methanol mix (9:1 vol/vol) and applied to an HPTLC plate by capillarity. CE and TAG standards were also applied to the plate. The plate was then developed in a neutral lipid solvent (hexane/diethylether/AcOH with a ratio of 80:20:2 vol/vol) and primuline solution was used for revelation. Images were acquired on an ImageQuant LAS 4000 mini (GE Healthcare Life Sciences) using EtBr as fluorescence and the 605DF40 filter. The intensity of CE and TAG bands were quantified using Fiji.
Enzymatic quantification of lipids
Two million cells were plated in T75 flask and allowed to grow for 48 h. Then, 500,000 cells were plated in 6-well plates. After 24 h, cells from two wells were scraped in PBS and lipids extracted using the same protocol as for TLC (Bligh & Dyer protocol). After drying under nitrogen, lipids were solubilized in 200 μl of ethanol 95%. Quantifications were then performed according to the manufacturers’ instructions (Cholesterol Amplex Red from Thermo Fisher Scientific [A12216], High Sensitivity Triglyceride Fluorometric Assay Kit from Sigma-Aldrich [MAK264-1KT,] and Phospholipid quantification Assay Kit from Sigma-Aldrich [CS0001-1KT]).
aLDs preparation
aLDs were prepared as described in Prévost et al. (2018) with slight modifications. Phospholipids (DOPC, DOPE, and Rhodamine-DOPE with a ratio of 73:25:2 mol/mol, respectively) were added to triolein at a 0.5% molar ratio in a glass tube. The solvent was evaporated under a stream of nitrogen and 1 ml of HK buffer (50 mM Hepes, pH 7.2, 120 mM K-acetate) was added. The solution was vortexed for 3 min. Then, aLDs were extruded 11 times through polycarbonate filters (Nuclepore Track-Etch Membrane; Whatman) with a pore diameter of 100 nm using a mini-extruder (Avanti Polar Lipids). The size was verified by dynamic light scattering measurements on a DynaPro (Protein Solutions). The preparation was used in a couple of hours after extrusion.
aLDs flotation assay
Each peptide (1 µM) was mixed with aLDs (1 mM), vortexed, and incubated at room temperature for 15 min. The solution (volume of 390 μl) was adjusted to 30% (wt/wt) sucrose by mixing with 260 μl of a 75% sucrose HK buffer. Two layers (520 μl of 25% sucrose and 130 μl of sucrose-free HK buffer) were gently added on top. Samples were centrifuged at 240,000 g for 1 h in a swing rotor (SWTi 60) at 20°C and decelerated without brake. The bottom (520 μl), middle (520 μl), and top (260 μl) fractions were collected and the fluorescence was measured using a fluorimeter (Pherastar FSX; BMG LABTECH).
aLDs and peptide interaction assay
aLDs were prepared following the protocol developed in Chorlay and Thiam (2020). Briefly, 5 μl of triolein was mixed with 70 μl of HKM buffer (50 mM Hepes, 120 mM K-acetate, and 1 mM MgCl2 at pH 7.4), vortexed for 5 s, and sonicated in a bath sonicator for 10 s. Then each peptide was separately mixed and incubated with aLDs at a concentration of 1 µM. The resulting emulsions were then introduced in a glass chamber for visualization. Fluorescence data were acquired 30 min after incubation with a laser scanning microscope (LSM 800; Carl Zeiss). Fluorescence intensity was measured after segmentation of individual aLDs.
aLDs pull-down assay
aLDs pull-down assays were performed as described in Kassas et al. (2017). NTA-Ni2+ beads (PureProteome Nickel Magnetic Beads, LSKMAGH10; Millipore) were first washed with HK buffer. Then, 1 µM of recombinant proteins (MSPHis6, WT, and W201E CRAL-TRIOHis6) was added to the beads and incubated 20 min under agitation at 4°C. To remove the excess of proteins, beads were washed twice with HK buffer. Afterward, aLDs (1 mM) were added to the beads and incubated 20 min again under agitation at 4°C. Beads were then washed three times with HK buffer and resuspended in a final volume of 30 μl of HK buffer. For imaging, 10 μl of the suspension were dropped on a glass bottom dish (MatTek) and imaged on a spinning-disk CSU-X1 (Nikon; 100× NA 1.4). Fluorescence was measured with a fluorimeter (Pherastar FSX; BMG LABTECH) using 20 μl of suspension.
Liposome preparation
Lipids stored in stock solutions in CHCl3 were mixed at the desired molar ratio in glass tubes. The solvent was removed in a dryer-block at 33°C under a nitrogen flow. If the mixture contained DOGS-NTA-Ni2+, it was pre-warmed at 33°C for 5 min prior to drying. The lipid film was hydrated in HK buffer to obtain a suspension of multi-lamellar vesicles. The multi-lamellar vesicles suspensions were frozen and thawed five times and then extruded through polycarbonate filters of 0.2 µm pore size using a mini-extruder (Avanti Polar Lipids). Liposomes were stored at 4°C and in the dark when containing fluorescent lipids and used within 2 d.
Flotation experiment
The CRAL-TRIOHis6 protein (0.75 µM) was incubated with liposomes (0.75 mM total lipids) with a given lipid composition doped with 0.1 mol% NBD-PE in 150 μl of HK buffer at 25°C for 10 min under agitation at 800 rpm. The suspension was adjusted to 28% (wt/wt) sucrose by mixing 100 μl of a 60% (wt/wt) sucrose solution in HK buffer and overlaid with 200 μl of HK buffer containing 24% (wt/wt) sucrose and 50 μl of sucrose-free HK buffer. The sample was centrifuged at 240,000 g in a swing rotor (TLS-55 Beckmann) for 1 h. The bottom (250 μl), middle (150 μl), and top (100 μl) fractions were collected. The bottom and top fractions were analyzed by SDS-PAGE by direct fluorescence and after staining with SYPRO Orange, using a FUSION FX fluorescence imaging system.
Circular dichroism
The experiments were performed on a Jasco J-815 spectrometer at room temperature with a quartz cell of 0.05 cm path length (Starna). The CRAL-TRIOHis6 protein (WT or W201E mutant) was dialysed against a 20 mM Tris, pH 7.4, 120 mM NaF buffer to remove glycerol. Each spectrum is the average of 10 scans recorded from 185–260 nm with a bandwidth of 1 nm, a step size of 0.5 nm, and a scan speed of 50 nm min−1. The protein concentration was 6.7 µM. A control spectrum of buffer was subtracted from each protein spectrum. The spectra were analyzed in the 185–240 nm range using the BeStSel method provided online (Micsonai et al., 2015).
Dynamic light scattering measurements of liposome aggregation
The experiments were performed at 25°C in a Dynamo apparatus (Protein Solutions). LA liposomes (50 µM total lipids) made only of DOPC or composed of DOPC/DOGS-NTA-Ni2+ (98/2 mol/mol) were mixed with LB liposomes (50 µM, made of DOPC or composed of diphytanoyl-PC and diphytanoyl-PS [70/30 mol/mol]) in 20 μl of a freshly degassed HK buffer in a quartz cell. A first set of 12 autocorrelations curves was acquired to determine the size distribution of the initial liposome suspensions. Then, MOSPD2His6 or MSPHis6 (0.4 µM final concentration) was added manually and mixed thoroughly. The kinetics of aggregation was measured during 23 min by acquiring one autocorrelation curve every 10 s. At the end of the experiment, a set of 12 autocorrelation functions was acquired. The data were analyzed using two different algorithms provided by the Dynamics v6.1 software (Protein Solutions). During the kinetics, the autocorrelation functions were fitted assuming that the size distribution is a simple Gaussian function. This mode, referred to as the monomodal or cumulant algorithm, gives a mean hydrodynamic radius and the width (or polydispersity). Before and after the aggregation process, the autocorrelation functions were fitted using a more refined algorithm, referred to as a regularization algorithm. This algorithm is able to resolve several populations of different sizes, such as free liposomes and liposome aggregates.
Statistical analyses
Statistical analyses were performed using the Mann–Whitney or the Kruskal–Wallis non-parametric test and with the One-way ANOVA or Student’s t test parametric tests (Prism; GraphPad). Conditions were compared with the Dunn’s and Tukey’s multiple comparisons tests, respectively. P-values <0.05, <0.01, <0.001, and <0.0001 are identified with one, two, three, and four asterisks, respectively. ns: P ≥ 0.05.
Online supplemental material
Fig. S1 shows data that support the findings presented in Fig. 1 on the localization of MOSPD2 around LDs in different cell lines. Fig. S2 shows the co-labeling of MOSPD2 and markers of early/late endosomes, mitochondria, and Golgi. Fig. S3 shows data that support the findings shown in Fig. 1 on the localization of endogenous MOSPD2. Fig. S4 shows the image analysis workflows used for LD and ER quantifications. Fig. S5 shows that the silencing of VAP-A and VAP-B does not affect LDs.
Acknowledgments
We thank Alastair McEwen for his critical reading of the manuscript. We thank the members of the Molecular and Cellular Biology of Breast Cancer team (IGBMC) for helpful advice and discussions. We are grateful to the members of the IGBMC Imaging Center, especially Elvire Guiot for her help with FRAP analysis and Bertrand Vernay for his help with CellProfiler. We thank Paolo Ronchi from the Electron Microscopy Core Facility at the European Molecular Biology Laboratory Heidelberg and Coralie Speigelhalter and Danièle Spehner from the IGBMC for their help with electron microscopy. We thank Marko Lampe from the Advanced Light Microscopy Facility at the European Molecular Biology Laboratory Heidelberg for his help with STED microscopy. We thank the IGBMC cell culture facility (Betty Heller), the peptide synthesis facility (Pascal Eberling), the Flow Cytometry facility (Claudine Ebel and Muriel Philipps), Molecular Biology and Virus Service (Paola Rossolillo and Nicole Jung), the Integrated Structural Biology platform (Catherine Birck and Pierre Poussin-Courmontagne), and the polyclonal and monoclonal antibody facility (Mustapha Oulad-Abdelghani) for their excellent technical assistance.
M. Zouiouich received a fellowship from ITMO Cancer AVIESAN (Alliance Nationale pour les Sciences de la Vie et de la Santé/National Alliance for Life Sciences and Health) within the framework of the Cancer Plan (https://itcancer.aviesan.fr/). T. Di Mattia received a fellowship from the Fondation pour la Recherche Médicale (https://www.frm.org/). A. Martinet received a fellowship from the Fondation ARC pour la recherche sur le cancer (https://www.fondation-arc.org/). J. Eichler received a fellowship from EUR Integrative Molecular & Cellular Biology (IMCBio; ANR-17-EURE-0023; https://imcbio.unistra.fr/). This work was supported by grants from the Agence Nationale de la Recherche (grant ANR-21-CE13-0014-01 and ANR-19-CE44-0003; https://anr.fr/), and from the Ligue Contre le Cancer (Conférence de Coordination Interrégionale du Grand Est; https://www.ligue-cancer.net). This work of the Interdisciplinary Thematic Institute IMCBio, as part of the ITI 2021-2028 program of the University of Strasbourg (http://www.unistra.fr), Centre national de la recherche scientifique (http://www.cnrs.fr/), and Inserm (http://www.inserm.fr/), was supported by IdEx Unistra (ANR-10-IDEX-0002) and by SFRI-STRAT’US project (ANR 20-SFRI-0012) and EUR IMCBio (ANR-17-EURE-0023) under the framework of the French Investments for the Future Program.
The authors declare no competing financial interests.
Author contributions: M. Zouiouich, T. Di Mattia, C. Tomasetto, and F. Alpy conceptualized the study. Investigations were performed by M. Zouiouich (immunofluorescence, image analysis and quantifications, CLEM, lipid biochemistry, FRAP, aLD pull-down assay, generation of knock-in cell lines and molecular biology), T. Di Mattia (immunofluorescence, FRAP, CLEM, and molecular biology), A. Martinet (immunofluorescence, lipid biochemistry, and aLD pull-down assay), J. Eichler (aLD flotation experiment and aLD pull-down assay), C. Wendling (generation of knock-out cell lines and molecular biology), N. Tomishige (TLC experiment), E. Grandgirard (FRAP and CLEM), N. Fuggetta (liposome flotation assays and DLS), C. Ramain (molecular biology), G. Mizzon (CLEM), C. Dumesnil (aLD peptide interaction assay), M. Carpentier (immunofluorescence of Seipin KO cells), A.R. Thiam (aLD peptide interaction assay and immunofluorescence of Seipin KO cells), G. Drin (circular dichroism), and F. Alpy (CLEM, electron microscopy, aLD pull-down assay, and molecular biology). Validation of experiments was done by M. Zouiouich, T. Di Mattia, A. Martinet, J. Eichler, C. Wendling, N. Fuggetta, C. Dumesnil, M. Carpentier, A.R. Thiam, G. Drin, and F. Alpy. Formal analysis was done by M. Zouiouich, T. Di Mattia, J. Eichler, C. Dumesnil, G. Drin, and F. Alpy. Methodology was developed by M. Zouiouich, T. Di Mattia, J. Eichler, C. Wendling, N. Tomishige (lipid biochemistry), B. Reina-San-Martin (CRISPR-Cas9 genome editing), Y. Schwab (CLEM), A.R. Thiam (aLDs and peptide interaction assay), T. Kobayashi (lipid biochemistry), G. Drin (liposome flotation assay and DLS experiment), C. Tomasetto, and F. Alpy. Funding acquisition obtained by C. Mathelin, A.R. Thiam, G. Drin, C. Tomasetto, and F. Alpy. Supervision by A.R. Thiam, T. Koyabashi, G. Drin, C. Tomasetto, and F. Alpy. The original draft was written by M. Zouiouich, T. Di Mattia, G. Drin, C. Tomasetto, and F. Alpy. Review and editing of manuscript by M. Zouiouich, T. Di Mattia, J. Eichler, N. Tomishige, C. Mathelin, A.R. Thiam, T. Kobayashi, G. Drin, C. Tomasetto, and F. Alpy. Project administration managed by F. Alpy.