The spatiotemporal regulation of E-cadherin expression is important during body plan development and carcinogenesis. We found that Tara (Trio-associated repeat on actin) is enriched in cadherin-based adherens junctions (AJs), and its knockdown in MDCK cells (Tara-KD cells) significantly decreases the expression of E-cadherin. Tara-KD activates Rac1 through the Trio RhoGEF, which binds to E-cadherin and subsequently increases the phosphorylation of p38 and Tbx3, a transcriptional E-cadherin repressor. Accordingly, the decrease in E-cadherin expression is abrogated by ITX3 and SB203580 (specific inhibitors of Trio RhoGEF and p38MAPK, respectively), and by dephosphomimetic Tbx3. Despite the decreased E-cadherin expression, the Tara-KD cells do not undergo an epithelial–mesenchymal transition and remain as an epithelial cell sheet, presumably due to the concomitant up-regulation of cadherin-6. Tara-KD reduces the actin-belt density in the circumferential ring, and the cells form flattened cysts, suggesting that Tara functions to modulate epithelial cell sheet formation and integrity by up-regulating E-cadherin transcription.
Introduction
E-cadherin functions as a Ca2+-dependent adhesion molecule through its extracellular region (Takeichi, 1995); its cytoplasmic portion is a cortical organizer that binds catenins and various cytoskeletal and signaling molecules (Braga, 2002; Perez-Moreno et al., 2003; Mège et al., 2006). Overall, E-cadherin is a system developer that integrates cell–cell adhesion and intracellular events to support the complex functional organization of epithelial cell sheets, which are essential in multicellular organisms (Perez-Moreno et al., 2003; Lecuit, 2005; Brembeck et al., 2006). Accumulating evidence indicates that the EMT (epithelial–mesenchymal transition)-dependent or -independent regulation of E-cadherin expression is critical for essential biological events, including development, differentiation, and regeneration, and for carcinogenesis (Gumbiner, 2005; Halbleib and Nelson, 2006; Jeanes et al., 2008). However, in contrast to E-cadherin’s EMT-dependent transcriptional regulation, the EMT-independent mechanism is not well understood, although it generally occurs when epithelial sheets are modulated without their cell–cell adhesions being disrupted.
E-cadherin and other types of classical cadherins are highly enriched in the adherens junctions (AJs) and integrated into various signaling cascades, which allows epithelial cell sheets to change their configurations as required for various biological events (Wheelock et al., 2008; Nishimura and Takeichi, 2009). The mixed expression of classical cadherins changes cell–cell interactions both by homophilic interactions through the cadherins’ extracellular domains and by altering the properties of the signal platform through their cytoplasmic domains. In this respect, the regulation of E-cadherin expression and that of other classical cadherins is critical for determining the dynamic properties of epithelial cell sheets (Perez-Moreno et al., 2003; Nishimura and Takeichi, 2009). Although the integrity of epithelial sheets is not maintained during EMT, the EMT-independent regulation of cadherin expression has recently attracted considerable attention because of its role in remodeling epithelial cell sheets without destroying the integrity of the cell sheet configuration (Wheelock et al., 2008). However, our knowledge about the EMT-independent regulation of cadherin expression is still fragmentary.
There is accumulating evidence that the ras-like GTP-binding protein (Rho) family members Rac, Rho, and Cdc42 spatiotemporally regulate the dynamic molecular organization of AJ components (Braga, 2002; Etienne-Manneville and Hall, 2002; Heasman and Ridley, 2008). A number of guanine nucleotide exchange factors (GEFs) are associated with AJs, and are thought to specify the spatiotemporally restricted actions of Rho family members (Schmidt and Hall, 2002; Otani et al., 2006). In addition, GEF-binding proteins may help specify the activities of their corresponding GEFs. However, the detailed contribution of Rho-related proteins to E-cadherin expression has not been reported.
Tbx3 is a member of the T-box family, a family of transcription factors with at least 22 members, each of which is involved in regulating particular developmental stages and in cancer-related processes (Papaioannou and Silver, 1998; Rodriguez et al., 2008). Although the T-box proteins are thought to be regulated downstream of WNT and/or BMP signaling pathways (Renard et al., 2007), the signaling cascades that regulate the transcriptional activity or other events downstream of the T-box proteins are not fully understood. A recent report showed that, in melanoma cells, E-cadherin expression is directly repressed by Tbx3 and not by EMT-related transcription factors, such as Snail and Slug (Rodriguez et al., 2008).
One prominent feature of E-cadherin is its integration into the apical belt-like AJ, with which the actin circumferential ring (CR) is associated (Yonemura et al., 1995). The AJ/CR complex is considered to play a critical role in belt-like arrangement of AJ in epithelial cells, which is spatiotemporally regulated in order to integrate various extracellular and intracellular components (Itoh et al., 1997; Perez-Moreno et al., 2003; Lecuit, 2005; Pokutta and Weis, 2007; Abe and Takeichi, 2008). Because E-cadherin– and/or other classical cadherin-based catenin/actin frameworks are distributed in cell–cell contacts, it is plausible that a critical mechanism for integrating these frameworks into the AJ/CR system exists at AJs.
Here we show that the Trio-associated repeat on actin (Tara) protein is enriched at AJs through its association with Trio RhoGEF, a binding partner of E-cadherin. In addition, Tara down-regulates the activity of Tbx3, a transcriptional suppressor of E-cadherin, by negatively regulating Trio’s RhoGEF activity EMT independently, through a novel signaling cascade mediated by Tara/Trio/Rac1/p38/Tbx3. This finding provides a conceptually new paradigm for the regulation of E-cadherin, in which a specific signaling cascade initiated at the AJ controls the activity of a transcriptional suppressor of E-cadherin.
We also found that Tara-KD reduced the packing density of actin in the AJ-associated CR by decreasing the level of E-cadherin expression and concomitantly increasing the protein level of cadherin-6. We propose that Tara is a critical regulator of E-cadherin expression and that Tara modulates the properties of the cadherin-based CR and of epithelial cell sheets through its regulation.
Results
Tara is an AJ component that affects the levels of AJ-associated E-cadherin
In an ongoing proteomic screen of junctional fractions of the liver membrane (Tsukita and Tsukita, 1989; Yamazaki et al., 2008a), to find junctional components whose exogenous expression and/or knockdown alter the junctional levels of E-cadherin and/or claudins in cultured epithelial cells, we identified a molecule that was enriched in junctional fractions and whose knockdown decreased E-cadherin; its sequence matched that of Tara (Fig. 1 A). We then made stable knockdown cell lines of Tara by transfecting MDCK cells with a Tara-specific RNAi (Tara-KD cells). To confirm that the effects of Tara-KD were specific, we examined two independent constructs for Tara-KD, and obtained the same results. As a control for RNAi-KD, stable cell lines expressing the empty RNAi expression vector (mock transfection) were used; cell lines expressing scrambled constructions of the nucleotides for RNAi-KD were also established, and showed the same results as the mock-transfected cells (Fig. 1 D; Fig. S1 A). Immunoblotting and immunofluorescence analyses, using an anti-Tara antibody that we produced, showed that Tara colocalized with E-cadherin (Fig. 1 B; Fig. S1, B and C).
To compare the intensity of E-cadherin signals precisely between mock and Tara-KD cells, equal numbers of control and Tara-KD cells were mixed and seeded on a cover glass. In confluent sheets of Tara-KD cells, the E-cadherin expression was lower than in mock-transfected control cell sheets (Fig. 1, C and D), and the sheet-like configuration of the cells was maintained. In addition, the protein level of cadherin-6, the second major cadherin in MDCK cells (Stewart et al., 2000; Capaldo and Macara, 2007), was up-regulated by Tara-KD (Fig. 1, C and D), suggesting that the down-regulation of Tara at least partly induced cadherin switching in these cells. In contrast, the N-cadherin signal, which was very weak, was unchanged in the Tara-KD cells (unpublished data). Neither the protein levels nor the intensities of the immunofluorescent signals for cell–cell AJ-associated proteins, such as the α-, β-, and p120-catenins, occludin, and ZO-1, were altered in the Tara-KD cells (Fig. 1, C and E).
Consistent with these findings, thin-section electron microscopy showed that the ultrastructure of the AJ was unaffected in the Tara-KD cells (Fig. S1 D). Furthermore, the cell proliferation rate, in vitro wound healing, and cell–cell adhesion were all unaffected in these cells (Fig. S2, A–C). Collectively, our results indicate that knocking down Tara decreases the expression of E-cadherin, which is accompanied by the up-regulation of cadherin-6, without affecting the AJ-based epithelial cell sheet organization.
Molecular basis for the association of Tara with cell–cell AJs
Given the colocalization of Tara with E-cadherin, we further examined their mode of interaction. We transiently expressed tagged forms of Tara, E-cadherin, and Trio, which is a reported binding partner of Tara, in HEK293 cells and examined the binding affinities between them by coprecipitation assay. We found that Tara did not bind directly to E-cadherin, but it did bind to Trio, which bound to E-cadherin (Fig. 2 A, a). The linkage between Trio and E-cadherin was confirmed by binding experiments between Trio and a GST fusion protein of the cytoplasmic domain of E-cadherin (Fig. 2 A, b), and was consistent with the biochemical enrichment of Trio RhoGEF into the junctional fractions and AJ localization of the HA-tagged construct of Trio (Fig. 2 A, c; Fig. S2 D).
These results were in accordance with previous reports that Trio RhoGEF is associated with cadherin-11 (Kashef et al., 2009) and M-cadherin (Charrasse et al., 2007) and that Tara binds to the RhoG/Rac1-GEF domain (Trio GEF-D1) of Trio RhoGEF (Seipel et al., 2001). To address whether Trio plays a role in the junctional recruitment of Tara, we knocked down Trio in MDCK cells. We observed that the staining for Tara at AJs was lost in the Trio knockdown MDCK cells, suggesting that the junctional localization of Tara was dependent on the E-cadherin/Trio linkage (Fig. S2 E).
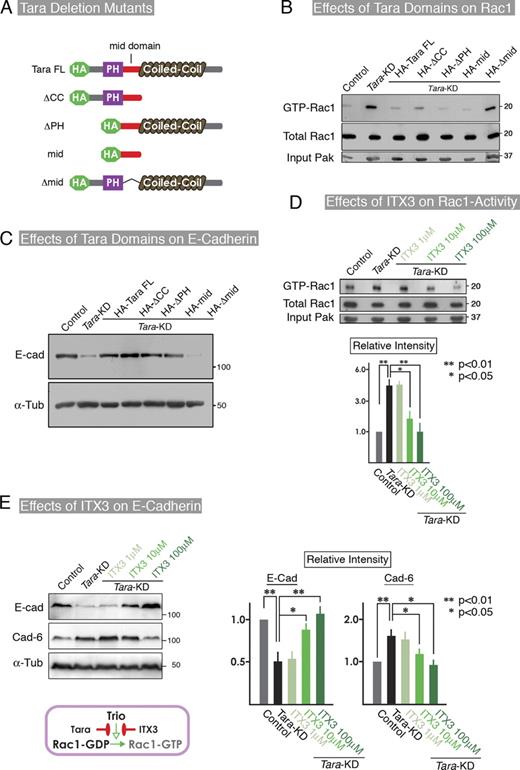
In the present study, we used mouse Tara sequence as RNAi-resistant sequence and found by immunofluorescence and pull-down analyses that the “mid-domain,” between Tara’s PH and coiled-coil domains, was responsible for its AJ localization and Trio RhoGEF binding (Fig. 2, B–D). In an immunofluorescence experiment using a series of Tara deletion constructs, all the constructs except the one that lacked the mid-domain increased the E-cadherin expression levels in Tara-KD cells, indicating that the mid-domain of Tara was required to offset the Tara-KD–induced down-regulation of E-cadherin (Fig. 2, B and C).
Tara-KD decreases E-cadherin expression by activating Rac1
To examine Tara-KD’s effect on epithelial cell sheets near confluence, we treated MDCK cells with EDTA/trypsin, replated them, and cultured them for 24–48 h to obtain 80–100% confluency. By immunofluorescence, E-cadherin and Tara were consistently localized to cell–cell AJs at every time point (Fig. S3 A). Moreover, when the cells were separated into insoluble and soluble fractions, both E-cadherin and Tara were predominantly recovered in the insoluble fraction, supporting the immunofluorescence results (Fig. S3 B). Immunofluorescence and immunoblotting showed that the E-cadherin levels were clearly lower in the Tara-KD than in the control cells at 24 and 48 h after replating (Fig. 3 A; Fig. S3 A). The E-cadherin mRNA was also decreased in the Tara-KD cells at 24 and 48 h after replating, suggesting that E-cadherin was regulated at the transcription level. In contrast, the mRNA level of cadherin-6 did not change in the Tara-KD cells (Fig. 3 B). We performed a dual luciferase assay using the minimal reporter of E-cadherin and found a ∼40% decrease in reporter expression in the Tara-KD cells 48 h after replating (Fig. 3 C). Moreover, cycloheximide, but not MG132 (a proteolysis inhibitor), suppressed the Tara-KD–induced up-regulation of cadherin-6 (Fig. S3 C). These results indicate that E-cadherin is transcriptionally down-regulated by the knockdown of Tara, and its down-regulation is accompanied by the biosynthesis-dependent post-transcriptional increase in cadherin-6 protein. We next asked whether Tara-KD induced EMT. We found that the mRNA levels of EMT-related genes like Snail, Slug, Twist, Zeb-1, Zeb-2, N-cadherin, α-smooth muscle actin, and vimentin were not elevated in the Tara-KD cells (unpublished data), and even if Snail was exogenously expressed, no changes in expression of Tara was detected. Collectively, the decreased expression of E-cadherin in the Tara-KD cells was probably not caused by a typical EMT.
Because Tara binds the RhoG/Rac1-GEF domain (Trio GEF-D1) of Trio RhoGEF (Seipel et al., 2001), we monitored the levels of active Rac1 and RhoG in control and Tara-KD MDCK cells after EDTA/trypsin treatment and replating. By a conventional pull-down assay for the active form of Rac1, we found that the level of Rac1 activation was much higher in the Tara-KD than the control cells at 24 and 48 h after replating (Fig. 3 D), although we could not detect the activated RhoG signal in either the control or the Tara-KD cells (Fig. S3 D), consistent with a previous report (Backer et al., 2007). In parallel, fluorescence resonance energy transfer (FRET) analyses of Rac1 were performed to observe the spatiotemporal activation patterns in cells, using the Raichu-Rac1 probe (Fig. 3 E; Videos 1–6). Activation of Rac1 was observed at the cell–cell contacts of the control and Tara-KD cells, 12 and 24 h after replating. However, although active Rac1 was still easily observed at cell–cell contacts in the Tara-KD cells 48 h after replating, it was scarce in the control cells at this time point (Fig. 3 E; Videos 1–6). To confirm that the up-regulation of Rac1 activity in the Tara-KD cells was specific, we transfected these cells with full-length and deletion constructs of Tara. All of the constructs except the one lacking the mid-domain decreased the level of active Rac1 (Fig. 4, A and B) and restored the E-cadherin levels (Fig. 3 C). We also introduced a dominant-negative Rac1 into the Tara-KD cells, and found that it suppressed the Tara-KD–induced decrease in E-cadherin expression (Fig. S4 A), confirming that Tara’s regulation of Rac1’s activity specifically affected the E-cadherin expression.
Tara-KD increases Rac1 activity via Trio RhoGEF, which leads to the phosphorylation of p38
To examine whether the Rac1 activity was specifically enhanced by Trio RhoGEF in Tara-KD cells, we used ITX3, a specific inhibitor of the Rac1-GEF activity of Trio RhoGEF (Bouquier et al., 2009). ITX3 repressed the Rac1 activity and dose dependently up-regulated the E-cadherin protein level in the Tara-KD cells (Fig. 4 D), thereby reversing the effects of Tara-KD (Fig. 4 E). The quantification of GTP-Rac1, E-cadherin, and cadherin-6 by densitometry (as shown in Fig. S4 B) in three independent experiments showed that 100 µM ITX3 significantly increased the E-cadherin expression (by 80%) in the Tara-KD cells compared with untreated Tara-KD cells (Fig. 4 E). To complement the Trio inhibitor data with an independent maneuver that targets Trio, we knocked down Rac1 or Trio in the Tara-KD cells and found that the E-cadherin expression was recovered (Fig. S4, C and D). These results indicate that the Trio RhoGEF-mediated Rac1 activation specifically decreased the E-cadherin expression. Together, our findings support the idea that AJ-associated Tara/Trio RhoGEF spatiotemporally regulates Rac1 activity to alter the expression level of E-cadherin.
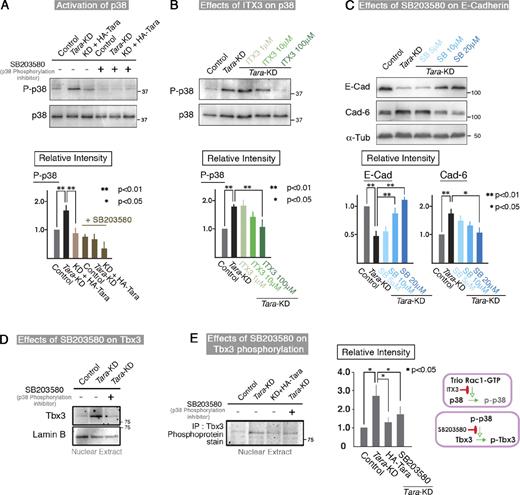
Because the Trio RhoGEF-dependent Rac1 activation was specifically up-regulated in Tara-KD cells, we sought to identify the signaling cascade that linked the Rac1 activation to the decreased E-cadherin expression. In this respect, Rac1 activation regulates MAP kinases such as ERK, JNK, and p38 (Rygiel et al., 2008; Zhang et al., 2009). We therefore examined the phosphorylation levels of these MAP kinases by immunoblotting, using antibodies specific for their phosphorylated form. We found that the phosphorylation level of p38 was up-regulated in the Tara-KD cells compared with controls (Fig. 5 A). When the Trio RhoGEF-mediated Rac1 activity was repressed by ITX3 in the Tara-KD cells, the phospho-p38 signal decreased (Fig. 5 B). Therefore, the Trio RhoGEF-mediated activation of Rac1 apparently increased the phosphorylation of p38. Consistent with this scenario, SB203580, an inhibitor of p38 phosphorylation, suppressed the Tara-KD–induced down-regulation of E-cadherin in a dose-dependent manner (Fig. 5 C). Quantification by densitometry (Fig. 5 C; Fig. S4 B) showed that the E-cadherin signal was significantly higher in the SB203580-treated cells (∼100% increase with 10 and 20 µM SB203580). Furthermore, when SB202190 was used to inhibit the activity of p38 (Fig. 5 C; Fig. S4 E), almost the same results were obtained as those by SB203580. Thus, it is likely that Trio RhoGEF-mediated Rac1 activation increases the phosphorylation of p38, leading to a decreased level of E-cadherin in Tara-KD cells.
Tara-KD–based phosphorylation of p38 phosphorylates Tbx3, a transcriptional repressor of E-cadherin that functions downstream of Tara/Trio/Rac1/p38
We next investigated how the phosphorylation of p38 decreases the expression of E-cadherin. After the EMT-related transcription factors had been excluded, the T-box protein family seemed the most likely candidates (Rodriguez et al., 2008). Among them, Tbx2 is reported to be phosphorylated by p38 (Abrahams et al., 2008), which suggested that Tbx2 could be a downstream effector of Tara/Trio/Rac1/p38. However, we did not detect Tbx2 signals in MDCK cells by immunoblotting, although we did detect Tbx3, which is the most closely related T-box member to Tbx2 (Papaioannou and Silver, 1998). Furthermore, the protein level of Tbx3 was increased in the nuclei as well as in whole Tara-KD cells compared with control cells, and this effect was counteracted by SB203580, the p38 phosphorylation inhibitor (Fig. 5 D). When we immunoprecipitated Tbx3 from nuclear extracts and analyzed it using a phosphoprotein detection kit, we found that its phosphorylation was elevated in the Tara-KD cells, but SB203580 reduced this effect (Fig. 5 E). These results indicated that Tbx3 is a downstream target of Tara/Trio/Rac1/p38 and that the phosphorylation and dephosphorylation of Tbx3 are critical for the transcriptional control of E-cadherin.
The phosphorylation of Tbx3 decreases the transcription of E-cadherin downstream of Tara/Trio/p38, and cadherin-6 is up-regulated
To examine the involvement of Tbx3 phosphorylation in the transcriptional control of E-cadherin, we analyzed the effects of a dephosphomimetic Tbx3 on the Tara-KD–induced decrease in E-cadherin. The likely phosphorylation sites on Tbx3 were deduced by comparing its amino acid sequence with that of Tbx2 (Fig. 6 A), for which three phosphorylation sites have been reported: S336, S623, and S675 (Abrahams et al., 2008). There were two corresponding serine residues in Tbx3 (S334 and S692 with no candidate for S623) that might be phosphorylated. Of these, Tbx3 (S692A) had a dominant-negative effect on Tbx3’s transcriptional repression of E-cadherin expression in Tara-KD cells, and the dephosphomimetic mutation of this serine in Tbx3 increased the signals for E-cadherin in the Tara-KD cells as shown in immunofluorescence and immunoblotting, respectively (Fig. 6, A and B). These findings led us to conclude that Tbx3 is activated by phosphorylation downstream of p38 to transcriptionally repress E-cadherin.
In the Tara-KD cells, although E-cadherin was decreased, the levels of the β- and p120-catenins were unchanged. Because the β- and p120-catenins, respectively, control the ER exit and plasma membrane association of classical cadherins (Chen et al., 1999; Ireton et al., 2002), these findings led us to hypothesize that the translation of cadherin-6, another classical cadherin that is expressed in MDCK cells, was controlled downstream of the E-cadherin transcription. We therefore knocked down E-cadherin in MDCK cells, and found that the level of cadherin-6 increased (Fig. 6 C). Furthermore, the protein levels of the β- and p120-catenins did not change in the E-cadherin-KD MDCK cells, suggesting there were no changes in the association of the classical cadherins with the ER exit or plasma membrane. Hence, if the protein expression of one classical cadherin decreases, the other cadherins have more p120 available to them, and their levels rise, which is referred to as “p120 sharing.” In the present case, the cadherin switching was probably driven by the transcriptional control of E-cadherin expression. That is, when E-cadherin transcription decreased, the unchanged levels of p120- and β-catenins drove the concomitant up-regulation of cadherin-6, which does not conform to the more canonical EMT-like mechanisms.
Tara-KD affects the organization of the CR
We next investigated the role of the decrease in E-cadherin and increase in cadherin-6 at the AJs in MDCK cells. Because AJs are highly organized into belt-like arrangement by the peripheral actin structure in epithelial cells, which forms the CR, we carefully examined the actin intensities at E-cadherin–rich AJs in control and Tara-KD MDCK cell sheets by confocal fluorescence microscopy (Fig. 7, A–C). To compare the actin intensities precisely between them, equal numbers of control and Tara-KD cells were mixed and seeded on a cover glass. Actin staining with rhodamine-phalloidin was less intense in the Tara-KD MDCK cells than in control cells. The actin intensity was restored to normal when exogenous Tara or E-cadherin was expressed in the Tara-KD MDCK cell sheets (Fig. 7, B and C). Furthermore, because the actin CRs form in association with the AJs, we examined the CRs stereoscopically by scanning electron microscopy (Fig. 7 D). At the AJs of control MDCK cell sheets, the actin filaments were closely packed into CRs. In contrast, the packing density of the actin filaments in the CRs was decreased in the Tara-KD MDCK cells. These findings suggested that the E-cadherin–based junctional actin filaments of CRs, which cooperate to organize the total AJ, are more densely packed at AJs than are the cadherin-6–based ones.
To further explore the biological relevance of Tara we used various morphogenetic approaches because E-cadherin is generally involved in morphogenesis. Among our trials, we observed Tara-KD–dependent morphogenetic phenotypes in three-dimensional (3D) cultures of control and Tara-KD MDCK cells (Fig. 7 E; Fig. S5). Control MDCK cells formed spherical cysts in collagen gels with Y/X ratios (length/width radii) of nearly 1.0 and Z/X ratios (vertical/horizontal radii) of 0.92 ± 0.07. In contrast, the Tara-KD MDCK cells formed 3D cysts that were more elliptical, with Z/X ratios of 0.62 ± 0.08. Immunofluorescence microscopy revealed that, although the intensity of the apical actin remained unchanged in the Tara-KD cells, the intensity of actin and E-cadherin at the AJs and lateral membranes of the Tara-KD MDCK cell cysts decreased relative to the intensities in control cysts. Collectively, these findings indicate that Tara probably plays a role in fine-tuning the actin-based tension through the AJs.
Discussion
E-cadherin is acknowledged to play critical roles in numerous biological events (Perez-Moreno et al., 2003; Lecuit, 2005; Brembeck et al., 2006), which has spurred considerable interest in researching E-cadherin–related signaling pathways with a particular emphasis on their diversity and specificity. Here, we identified a novel, specific signaling pathway, Tara/Trio/Rac1/p38/Tbx3/E-cadherin. In this pathway, Tara functions as an AJ component and a binding partner of RhoG/Rac1GEF-Trio, which up-regulates the transcription of E-cadherin (Fig. 7 F). In this signaling system, the Tara-KD–induced down-regulation of E-cadherin transcription is independent of EMT in the MDCK cells, which express Tbx3. Because the Tbx3-dependent regulation of E-cadherin transcription apparently occurs in many important biological events, future studies should examine whether the Tara/Trio/Rac1/p38/Tbx3 signaling cascade is involved in these cases. An important aspect of the current findings is that the initiation of an E-cadherin regulatory signal at the AJ is conceptually new.
The current work reveals a mechanism by which Rac can reduce E-cadherin activity. The source of the Rac activation signal is often not clear (Lozano et al., 2008; Hage et al., 2009), and Rac1 signaling as it pertains to cadherins is still largely unknown. In this context, many GEFs are known to be directly or indirectly associated with cadherins. In particular, Trio RhoGEF is reported to bind M-cadherin and cadherin-11 (Charrasse et al., 2007; Kashef et al., 2009), which play distinct roles in development through Rac1-related signaling. However, the details of the signaling pathways that use RhoG/Rac1 GEF-Trio are still fragmentary.
In the present study, we showed for the first time that the Trio RhoGEF-dependent Rac1 signaling that down-regulates E-cadherin transcription was dampened by Tara through an interaction between the mid-domain of Tara and the RhoG/Rac1-GEF domain of Trio. In this respect, Trio bound to the cytoplasmic domain of E-cadherin, and Trio knockdown decreased the level of Tara at AJs. It is possible that Rac1 activation is at least partly mediated by Tara-free E-cadherin–associated Trio RhoGEF. Tara functions to maintain E-cadherin transcription by spatiotemporally inhibiting Rac activation in cell–cell adhesion-based events through the Tara/Trio RhoGEF/Rac1/p38/Tbx3 signaling cascade. The relationship of Rac1 to E-cadherin activity has been poorly understood, and this work adds new insight to the story.
Interestingly, Trio RhoGEF is reportedly anchored to LAR (Debant et al., 1996), a transmembrane protein with a phosphatase domain in its cytoplasmic region that binds β- and γ-catenin. This finding suggests that LAR-related signals may modulate the link between Tara and Trio RhoGEF, to activate its Rac1-GEF activity. It is also possible that cell–cell adhesion-based signals modulate the link between Trio RhoGEF and E-cadherin, which will be a subject of future study.
Although the general idea that Rac1 activation alters the localization and/or the total protein level of E-cadherin in cells is being established (Lozano et al., 2008; Hage et al., 2009), this is the first example of Rac1 regulating the transcription of E-cadherin, as indicated by our finding that Trio RhoGEF-based Rac1 activity up-regulates the phosphorylation level of p38 independent of EMT, to down-regulate E-cadherin transcription. This function of the Rac1/p38 pathway may have conceptual implications for understanding Rac1/p38 pathways in general because only a few cases involve a signaling route from p38 to cadherins, and in these cases, the phosphorylation of p38 was reported to affect the post-transcriptional down-regulation of cadherins (Zohn et al., 2006; Khanna et al., 2010). Recently, p38 was reported to phosphorylate Tbx2 (Abrahams et al., 2008), and Tbx2/3 were reported to down-regulate the transcription of E-cadherin (Rodriguez et al., 2008). However, these distinct events have not been linked, as they have been in this study, where we show that Rac1/p38 directly regulates the transcription of E-cadherin.
Despite the decreased expression of E-cadherin caused by the Tara knockdown, the Tara-KD cells did not induce an EMT, and retained their configuration as an epithelial cell sheet, presumably because of the concomitant up-regulation of cadherin-6. These findings were reminiscent of “cadherin switching,” the phenomenon in which two or more cadherins show a seesaw pattern of protein expression, although cadherin switching under non-EMT circumstances is relatively undocumented (Wheelock et al., 2008). In this respect, transfection of Snail did not show any influence on the expression of Tara (Ikenouchi et al., 2003; unpublished data). Our present data show a detailed example of molecular signaling mechanisms leading to cadherin switching in a non-EMT situation.
A final issue relates to the cadherin-based junctional CR. Classical cadherins, with which β-catenin is associated, are linked to the CR through α-catenin/eplin/actin (Abe and Takeichi, 2008). Our present findings suggest that each type of classical cadherin has a distinct CR-organizing activity. These findings provide a new perspective on the higher-order structures of AJs. Because the AJ-based CR was first observed in various types of epithelia in vivo, CRs of epithelial cell sheets have been thought to contain E-cadherin and/or classical cadherins in a ring-like arrangement (Mooseker, 1976; Owaribe et al., 1981). The density of the actin belt in CRs varies with the cell/tissue type, but the mechanism for regulating actin packing in CRs is not well understood. We propose here that the density of the actin belt in CRs might be determined by the balance between the classical cadherins expressed in the AJs of a given cell type. If this is the case, Tara might contribute to the morphogenesis of various parts of the body by spatiotemporally modulating the balance of the classical cadherins.
In conclusion, we found that Tara/Trio RhoGEF/Rac1/p38/Tbx3 is a novel signaling cascade involved in the remodeling of epithelial cell sheets by regulating the transcription of E-cadherin. These findings reveal for the first time that the AJ-associated E-cadherin complex regulates the transcription of E-cadherin. These discoveries will be critical to fully understand E-cadherin’s diverse and specific functions, and to suggest new research directions on the roles of E-cadherin–related signaling cascades. Further analyses of this cascade in vivo and of other signals that regulate E-cadherin transcription should elucidate ways to manipulate development-, regeneration-, and/or tumor-related processes for novel strategies in medical therapies. Studies along these lines are being conducted in our laboratory.
Materials and methods
Antibodies and reagents
To generate an anti-Tara pAb, rabbits were immunized with a GST-Tara (111–190 aa) fusion protein. Mouse anti–cadherin-6 pAb was a special gift from Dr. M. Takeichi (Center for Developmental Biology, Kobe, Japan) and was generated using cadherin-6 knockout mice. Rat anti-occludin (MOC37) and anti-ZO1 mAbs were generated and characterized in our laboratory (Saitou et al., 1998; Kitajiri et al., 2004). Rat anti–E-cadherin mAb was kindly provided by Dr. M. Takeichi. Mouse anti-HA mAb (Covance), goat anti–ZO-2 pAb, rabbit anti-Trio pAb, rabbit anti-Tbx3 pAb, anti–lamin B pAb (Santa Cruz Biotechnology, Inc.), anti-p38, anti–phospho p38 (Cell Signaling Technology), rabbit anti–claudin-3 pAb (Invitrogen), mouse anti-Rac1 mAb (Millipore), mouse anti-desmoplakin mAb (Progen), mouse anti-Trio pAb (Abnova), and mouse and rabbit anti-Tbx3 pAbs (Abcam) were purchased. Rabbit anti–α-catenin pAb, rabbit anti–β-catenin pAb, mouse anti-vinculin mAb, mouse anti-Flag mAb, and mouse anti–α-tubulin mAb were from Sigma-Aldrich. SB203580 was from EMD. ITX3 was described previously (Bouquier et al., 2009). The Phosphoprotein Colorimetric Assay kit, Pro-Q, was from Invitrogen.
Plasmids and primers
The cDNA clones encoding Tara and Tbx3 were isolated from a mouse liver and kidney cDNA library. The Tara cDNA was cloned into the pCAGGS-NHA vector, and Tbx3 and its mutant were cloned into the pAcGFP-C1 vector (Takara Bio Inc.). The pGEX4T1-CRIB-Pak used for the pull-down assays of Rac1 was described previously (Matsuo et al., 2002; Ikenouchi et al., 2007). The primers for E-cadherin and GAPDH used for quantitative RT-PCR and the E-cadherin promoter used for luciferase assays were described previously (Behrens et al., 1991; Youn et al., 2005).
Knockdown constructs
To suppress the expression of Tara in MDCK cells, DNA oligonucleotides of target sequences encoding two distinct portions of the respective ORFs were cloned into an H1 promoter-driven RNAi vector (Brummelkamp et al., 2002). The vectors were transfected into MDCK cells, and suppressed the expression of Tara. Four clones obtained from the two constructs were selected. The probe sequences were: Tara (112–130), 5′-GGAGAGTGGAAGAAGCATT-3′; Tara (117–135), 5′-GTGGAAAGAAGCATTGGTTT-3′; Tara Scramble, 5′-TTACGAAGAAGGTGAGAGG-3′; Trio (305–323), 5′-GCATTCATGTTGCACTGAT-3′; Rac1 (552–570), 5′-GAGGAAGAGAAAATGCCTG-3′. The probe sequences for E-cadherin and cadherin-6 were reported previously (Capaldo and Macara, 2007).
Cell culture and transfections
MDCK-II and HEK293 cells were grown in DME supplemented with 10% fetal calf serum. Transfection was performed using the Lipofectamine Plus Reagent (Invitrogen), according to the manufacturer’s instructions. Most results shown were from transfected MDCK-II cells.
Immunofluorescence microscopy
MDCK cells were fixed in 1% formalin for 5 min at room temperature. The cells were then treated with 0.2% Triton X-100 in PBS for 5 min and processed for immunofluorescence microscopy as described previously (Yamazaki et al., 2008b). Alexa Fluor 488– or 568–conjugated secondary antibodies were used. For actin staining, Alexa Fluor 568 phalloidin (Invitrogen) was added to the secondary antibody. Specimens were observed at RT with a photomicroscope (BX51 and BX70; Olympus) equipped with a 100x, 1.4 NA oil immersion lens; 60x, 1.42 NA oil immersion lens; and 20x, 0.5 NA lens, and with a confocal microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a Plan-Apochromat (63x/1.40 NA oil immersion objective) with appropriate binning of pixels and exposure time. Photographs were recorded with a cooled CCD camera (ORCA-ER, Hamamatsu Photonics; or CoolSnap HQ, Photometrics). The images were analyzed with IPLab version 3.9.5 (BD), MetaMorph (Molecular Devices), LSM510 Meta version 3.0 (Carl Zeiss, Inc.), and Deltavision software (DeltaVision Core; Applied Precision). Images shown represent maximal intensity projections. Equivalent exposure conditions and scaling was used between controls and Tara-KD cells, and adjusted with only contrast and brightness adjustment.
GST pull-down and immunoprecipitation
HEK293 cells were transfected with expression vectors. The lysates from these cells were mixed with GST 4b beads (GE Healthcare) preabsorbed with GST fusion proteins. The nuclei were isolated from MDCK cells as reported previously (Sadowski and Gilman, 1993). In brief, cells were rinsed with ice-cold PBS contaning 1 mM Na3VO4 and 5 mM sodium fluoride. Ice-cold hypotonic buffer with 0.2% Nonidet P-40 was used as cell lysis buffer. After centrifugation, the nuclei pellets were treated with high salt buffer to remove DNA. The nuclear extracts were prepared and incubated with antibody-conjugated beads for 5 h. Each sample was analyzed by immunoblotting or with a phosphoprotein detection kit.
Rac1 activity assay
GTP-loaded Rac1 was measured using a commercially available kit (Thermo Fisher Scientific), as described previously (Nojima et al., 2008).
Immunoblotting
To prepare total cell lysates for immunoblotting, MDCK or HEK293 cells were lysed with SDS-PAGE sample buffer, sonicated, and boiled. The protein samples were separated by one-dimensional SDS-PAGE, transferred onto a nitrocellulose or PVDF membrane, and blotted with the appropriate antibodies. For quantification of signals in immunoblotting, the densitometric quantification of immunoblot bands with loading control in the same immunoblotting membranes was performed using Photoshop 7.0 software (Adobe). The immunoblot signals were normalized to the corresponding α-tubulin signal from the same filter, basically by being immunoblotted on the different portions of the same membranes or by being reprobed one by one. In some cases, they were normalized by being immunoblotted at the same time by mixure of antibodies, as specially mentioned in the legends.
FRET analyses
Raichu FRET probes for Rac1 were generous gifts from M. Matsuda (Kyoto University, Kyoto, Japan). Raichu probes were transiently transfected into MDCK cells, or Tara-KD cells by using Lipofectamine (Invitrogen) following the manufacturer’s recommended procedure. 24 h later, the cells were replated on the glass-bottom dishes. 12, 24, and 48 hours after replating, cells were imaged every 5 min for 2 h with a microscope (IX-81; Olympus) equipped with a 75-W xenon arc lamp, two filter changers, a temperature-controlled chamber, and a cooled charge-coupled device camera (CoolSNAP HQ; Photometrics), controlled by MetaMorph software (Invitrogen). The ratio image of YFP/CFP was created with MetaMorph software and used to represent FRET efficiency.
In the IMD mode images (Ratio), eight colors from red to blue are used to represent the YFP/CFP ratio, with the intensity of each color indicating the mean intensity of YFP and CFP. The upper and lower limits of the ratio range are shown on the right.
3D culture in collagen gel
3D cultures were performed in collagen gels (Nitta Gelatin) according to the manufacturer’s instructions. In brief, cells were added to a collagen I mixture, gently mixed, and plated onto 12-well transwell insert plates at 5 × 103 cells/well. 10-d-old cysts were examined for the general cyst architecture as described previously (Pollack et al., 1998).
Scanning and transmission electron microscopy
Cells were cultured in Transwell inserts or collagen gels. For scanning electron microscopy, the cells were permeabilized with RIPA buffer after 1% formalin treatment. The images were obtained with a microscope (S-800; Hitachi). The transmission electron microscopy was performed as described previously (Umeda et al., 2006).
Statistical analysis
Data are presented as mean ± SD. Whenever necessary, statistical significance of the data was analyzed by performing one-sample t tests. The specific types of tests and the P values, when applicable, are indicated in the figure legends.
Online supplemental material
Fig. S1 shows additional data on the AJ localization of Tara and additional characterization of Tara-KD cells. Fig. S2 shows additional characterization of Tara-KD cells and of Trio as a binding partner of Tara. Fig. S3 shows additional characterization of the mechanism of Tara-KD–induced down-regulation of E-cadherin. Fig. S4 shows additional analysis of Tara-KD–mediated signals. Fig. S5 shows characterization of cyst formation of Tara-KD cells. Video 1 shows FRET analysis for Raichu-Rac1 in the mock control cells between 11 and 13 h after being seeded. Video 2 shows FRET analysis for Raichu-Rac1 in the mock control cells between 23 and 25 h after being seeded. Video 3 shows FRET analysis for Raichu-Rac1 in the mock control cells between 47 and 49 h after being seeded. Video 4 shows FRET analysis for Raichu-Rac1 in the Tara-KD cells between 11 and 13 h after being seeded. Video 5 shows FRET analysis for Raichu-Rac1 in the mock control cells between 23 and 25 h after being seeded. Video 6 shows FRET analysis for Raichu-Rac1 in the mock control cells between 47 and 49 h after being seeded.
Acknowledgments
We are grateful to Dr. Masatoshi Takeichi for the generous gift of the mouse anti–cadherin-6 pAb and for valuable discussions during this study; Drs. Kohei Miyazono and Akira Nagafuchi for helpful discussions; and Dr. Hisashi Nojima for providing the Trio expression plasmids. We also thank Chiyomi Inoue and Orie Koga for their technical assistance and Drs. Grace Gray and Leslie Miglietta for proofreading the manuscript. We further thank all the members of our laboratory for helpful discussions.
This study was supported in part by a Grant-in-Aid for Creative Scientific Research and a Grant-in-Aid for Cancer Research from the Ministry of Education, Science and Culture of Japan to Sachiko Tsukita.
References
Author notes
Dr. Shoichiro Tsukita died on 11 December 2005.
T. Yano and Y. Yamazaki contributed equally to this work.