Late endosomes/lysosomes (LELs) are crucial for numerous physiological processes and their dysfunction is linked to many diseases. Proteomic analyses have identified hundreds of LEL proteins; however, whether these proteins are uniformly present on each LEL, or if there are cell-type-dependent LEL subpopulations with unique protein compositions is unclear. We employed quantitative, multiplexed DNA-PAINT super-resolution imaging to examine the distribution of seven key LEL proteins (LAMP1, LAMP2, CD63, Cathepsin D, TMEM192, NPC1, and LAMTOR4). While LAMP1, LAMP2, and Cathepsin D were abundant across LELs, marking a common population, most analyzed proteins were associated with specific LEL subpopulations. Our multiplexed imaging approach identified up to eight different LEL subpopulations based on their unique membrane protein composition. Additionally, our analysis of the spatial relationships between these subpopulations and mitochondria revealed a cell-type-specific tendency for NPC1-positive LELs to be closely positioned to mitochondria. Our approach will be broadly applicable to determining organelle heterogeneity with single organelle resolution in many biological contexts.
Introduction
The endosomal–lysosomal system is a dynamic network of membrane-bound compartments that plays a critical role in maintaining cellular homeostasis (Klumperman and Raposo, 2014; van Meel and Klumperman, 2008). Lysosomes, notable for their acidic lumen containing an array of degradative enzymes (De Duve et al., 1955), mature from late endosomes and represent the final stage in the endosomal–lysosomal pathway (Ballabio and Bonifacino, 2020; Bonifacino and Traub, 2003; Yang and Wang, 2021). Although historically regarded as the cell’s waste disposal system, lysosomes are now recognized as functionally diverse organelles (Bussi and Gutierrez, 2024) that regulate nutrient sensing, metabolic signaling, membrane repair, and several other cellular processes (Lawrence and Zoncu, 2019; Reddy et al., 2001; Settembre et al., 2013; Trivedi et al., 2020). Additionally, lysosomes are increasingly implicated in numerous neurodegenerative diseases and the aging process (Chen et al., 2019; Malik et al., 2019; Settembre et al., 2013; Tan and Finkel, 2023; Udayar et al., 2022). Given their emerging importance beyond the canonical degradative function, a more in-depth knowledge of lysosomal homeostasis will enable a better understanding of their vital roles in both health and disease.
Early electron microscopy studies, including those utilizing immunoelectron microscopy, have suggested the presence of significant heterogeneity among late endocytic organelles, including lysosomes (Geuze et al., 1984; Griffiths et al., 1989). However, despite its high spatial resolution, EM is low throughput and not well-suited to revealing the protein composition and dynamic behavior of these organelles. Consequently, light microscopy is indispensable for studying the dynamic interconversion of endosomal/lysosomal compartments, their motility, subcellular positioning, and function. These studies often use fluorescent markers like Lysotracker to track endosomal/lysosomal compartments, but pH-dependent dyes fail to distinguish between late endosomes and lysosomes (LELs) (Barral et al., 2022). Surface markers, which reflect stage-specific molecular machinery, provide a more accurate method (Klumperman and Raposo, 2014; van Meel and Klumperman, 2008). For example, EEA1 and Rab5 mark early endosomes, while Rab7 indicates late endosomes (Lakadamyali et al., 2006; Nielsen et al., 1999; Rink et al., 2005; Vanlandingham and Ceresa, 2009; Wilson et al., 2000).
The most abundant lysosomal membrane proteins are the lysosome-associated membrane proteins 1 (Lippincott-Schwartz and Fambrough, 1986) and 2 (LAMP1 and LAMP2), lysosomal integral membrane protein 2 (LIMP2), and CD63 (LIMP1/LAMP3) (Lübke et al., 2009; Schröder et al., 2010; Schwake et al., 2013; Winchester, 2001). LAMP1/2 plays roles in lysosome biogenesis (Schwake et al., 2013) and in regulating lysosomal pH (Zhang et al., 2023). CD63, a member of the tetraspanin superfamily, is upregulated in many cancers (Pols and Klumperman, 2009) and may play roles in extracellular vesicle production and endosomal cargo sorting (Hurwitz et al., 2018; van Niel et al., 2011). Due to their high abundance, LAMP1 and LAMP2 are commonly overexpressed to visualize lysosomes in light microscopy studies, though this may alter lysosomal dynamics, distribution, pH, and functionality.
Beyond LAMP and LIMP proteins, proteomic studies have identified over 100 different lysosomal membrane proteins, including ion channels, transporters, and exchangers (Akter et al., 2023; Bagshaw et al., 2005; Lübke et al., 2009; Muthukottiappan and Winter, 2021; Schröder et al., 2010; Yu et al., 2024). The lysosomal membrane also serves as a hub for various proteins that dynamically and transiently assemble on it including components of the nutrient-sensing mTOR (mechanistic target of rapamycin) pathway such as mTORC1, Ragulator, and Raptor (Perera and Zoncu, 2016; Rogala et al., 2019; Sancak et al., 2008, 2010; Settembre et al., 2013; Zoncu et al., 2011). Additionally, various degradative enzymes like Cathepsin D are present within the lysosomal lumen (Trivedi et al., 2020).
A key question remains: Are all these proteins equally abundant within every lysosome? Addressing this key question requires a method that can visualize and quantify many lysosome-associated proteins in a multiplexed fashion with high molecular specificity, sensitivity, and spatial resolution to resolve small, densely packed lysosomes within cells. Previous work used correlative light and electron microscopy to reveal differences in the molecular composition of early and late endosomes (van der Beek et al., 2022). However, this approach is low-throughput and technically challenging. In addition, the reliance on low-resolution light microscopy limits it to evaluating subcellular compartments that are spatially well-separated within the cell.
Super-resolution light microscopy enables visualization of the inner architecture of cells with nanoscale spatial resolution (Bond et al., 2022). Among various super-resolution methods, DNA Point Accumulation in Nanoscale Topography (DNA-PAINT) stands out for its ability to multiplex (Jungmann et al., 2014). Multiplexed DNA-PAINT (Jungmann et al., 2014) employs DNA-barcoded antibodies to detect and image multiple proteins. DNA-PAINT’s single-molecule detection efficiency also makes it highly sensitive to even lowly abundant proteins. Importantly, the well-defined binding kinetics of the imager oligonucleotides ensure that the number of detected localizations is directly and linearly proportional to the abundance of the target protein (Jungmann et al., 2016). This quantitative aspect of DNA-PAINT makes it ideally suited for accurate analysis of protein levels across different lysosomal compartments.
Here, we developed a quantitative pipeline using multiplexed DNA-PAINT imaging to analyze the abundance and heterogeneity of LEL proteins at the endogenous level. Our findings reveal that the canonical lysosomal proteins LAMP1, LAMP2, and Cathepsin D mark the same population of organelles. Therefore, we used LAMP1 as a reference to determine the abundance of other lysosomal proteins in these LAMP1-positive compartments, which we refer to as LELs. Our results revealed substantial heterogeneity in LEL subpopulations containing unique combinations of proteins including Niemann Pick Disease Type C1 protein (NPC1), which plays a role in cholesterol trafficking on the lysosomal membrane (Infante et al., 2008; Pfeffer, 2019) and LAMTOR4, a subunit of the Ragulator complex involved in mTOR activation (Sancak et al., 2008, 2010; Zoncu et al., 2011). Overexpression of LAMP1 as well as treatment with drugs such as EN6 and Bafilomycin A1 (BafA1) impacted NPC1 and LAMTOR4-positive LEL subpopulations. Spatial analysis also provided insights into the subcellular localization of these distinct subsets in relation to the nucleus, mitochondria, and the trans-Golgi network (TGN).
Overall, our study offers quantitative tools and a novel framework for characterizing the protein composition of individual organelles within cells with high sensitivity and spatial resolution, revealing the heterogeneity of LELs characterized by both unique combinations of resident proteins and variability in their abundance. This method can be widely applied to investigate organelle heterogeneity in various cellular contexts.
Results
Quantitative DNA-PAINT pipeline for characterizing LEL proteins
We first developed a comprehensive quantitative pipeline designed for broad application in quantifying the abundance of various organelle-associated proteins and applied it to characterize the abundance of seven LEL proteins: LAMP1, LAMP2, CD63, Cathepsin D, TMEM192, NPC1, and LAMTOR4 (Fig. 1). Since we aimed to characterize LEL proteins at endogenous levels, we used immunofluorescence (IF) labeling with commercially available antibodies previously validated in IF studies (Cason et al., 2022; Eapen et al., 2021; Gallagher and Holzbaur, 2023; Hiragi et al., 2022; Ishii et al., 2019; Keren-Kaplan et al., 2022; Rebsamen et al., 2015; Wang et al., 2020b; Weng et al., 2022). To further validate antibody specificity, we overexpressed target proteins fused to a tag (when available) and compared the antibody staining to that of the tag (Fig. S1, A–F). In all cases, we observed a high degree of colocalization between the tagged protein and the antibody stain on vesicular compartments (Fig. S1, A–F), validating the specificity of the used antibodies. For proteins where a tagged construct was unavailable as well as for low-abundance proteins, we additionally validated antibodies with knockdown and knockout (KO) cell lines.
Quantitative DNA-PAINT pipeline for characterizing LEL membrane proteins. Schematic shows LELs with target proteins imaged in this study. Target proteins (e.g., LAMP1 and LAMP2) are labeled with primary and secondary antibodies for DNA-PAINT imaging. Step 1 shows the data acquisition step in which a dual-color DNA-PAINT image is acquired (in this case, LAMP1 and LAMP2). Subsequently, in Step 2, LAMP1 is segmented to be used as the reference channel, and the LAMP2 raw localizations (target channel) are overlaid onto the segmented LELs. During colocalization (Step 3), the target localization density is measured in the region of the segmented LELs (colocalization region), as well as a local background region surrounding the LEL. If the density within the colocalization region is significantly (>3 standard deviations) enriched over the background density that LEL is determined to have both target and reference proteins. Several quantitative outputs are obtained as a result of this pipeline. Cell scale bars = 10 µm. Inset scale bars = 500 nm.
Quantitative DNA-PAINT pipeline for characterizing LEL membrane proteins. Schematic shows LELs with target proteins imaged in this study. Target proteins (e.g., LAMP1 and LAMP2) are labeled with primary and secondary antibodies for DNA-PAINT imaging. Step 1 shows the data acquisition step in which a dual-color DNA-PAINT image is acquired (in this case, LAMP1 and LAMP2). Subsequently, in Step 2, LAMP1 is segmented to be used as the reference channel, and the LAMP2 raw localizations (target channel) are overlaid onto the segmented LELs. During colocalization (Step 3), the target localization density is measured in the region of the segmented LELs (colocalization region), as well as a local background region surrounding the LEL. If the density within the colocalization region is significantly (>3 standard deviations) enriched over the background density that LEL is determined to have both target and reference proteins. Several quantitative outputs are obtained as a result of this pipeline. Cell scale bars = 10 µm. Inset scale bars = 500 nm.
Overexpression of target proteins confirms antibody (AB) specificity. (A–F) Overexpression of tagged constructs of (A) LAMP1-Flag, (B) LAMP2-BFP, (C) CD63-HA, (D) TMEM192-3xHA, (E) NPC1-Flag, and (F) LAMTOR4-Flag were used to verify antibody specificity in HeLa cells. Widefield images showing the co-staining with antibodies to the target protein (magenta) and the overexpression tag (yellow) colocalized for all proteins listed. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Overexpression of target proteins confirms antibody (AB) specificity. (A–F) Overexpression of tagged constructs of (A) LAMP1-Flag, (B) LAMP2-BFP, (C) CD63-HA, (D) TMEM192-3xHA, (E) NPC1-Flag, and (F) LAMTOR4-Flag were used to verify antibody specificity in HeLa cells. Widefield images showing the co-staining with antibodies to the target protein (magenta) and the overexpression tag (yellow) colocalized for all proteins listed. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Given the high abundance of LAMP1 and LAMP2 on LEL membranes, we validated our quantitative pipeline using DNA-PAINT imaging of these two proteins in two cell types: HeLa (Fig. 1 and Fig. 2 A) and ARPE-19 (Fig. 3 A). As expected, both proteins predominantly localized to vesicular compartments resembling LELs. We first optimized fixation and permeabilization by comparing two different methods aiming to minimize disruption to the LEL localization of LAMP proteins. Previous studies showed that fixation with −20°C methanol disrupts membranes and is not suitable for visualizing organelles (Whelan and Bell, 2015). We instead evaluated aldehyde-based fixation using 4% PFA against glyoxal fixation (Fig. S2 A), the latter suggested to be a quicker fixative (Richter et al., 2018). In addition, we compared the standard Triton-X100 permeabilization with the gentler saponin (Jamur and Oliver, 2010; Lacaille-Dubois and Wagner, 1996) (Fig. 2, B–D) (see Materials and methods). Our results indicated that a combination of 4% PFA with saponin most effectively maintained the vesicular enrichment of LAMP1 and LAMP2 (Fig. S2, A–D). Consequently, we adopted this combination for all subsequent experiments.
Dual-color DNA-PAINT in HeLa cells identifies heterogeneity in the enrichment of canonical LEL proteins. (A–F) Representative DNA-PAINT images of LAMP1 or LAMP2 reference channel (magenta) and target protein channel (yellow) in HeLa cells for (A) LAMP2, (B) CD63, (C) Cathepsin D, (D) TMEM192, (E) NPC1, and (F) LAMTOR4. Arrows indicate LELs positive for both LAMP1/LAMP2 reference and target protein, arrowheads indicate LELs positive for LAMP1/LAMP2 reference and negative for the target protein, and the dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Dual-color DNA-PAINT in HeLa cells identifies heterogeneity in the enrichment of canonical LEL proteins. (A–F) Representative DNA-PAINT images of LAMP1 or LAMP2 reference channel (magenta) and target protein channel (yellow) in HeLa cells for (A) LAMP2, (B) CD63, (C) Cathepsin D, (D) TMEM192, (E) NPC1, and (F) LAMTOR4. Arrows indicate LELs positive for both LAMP1/LAMP2 reference and target protein, arrowheads indicate LELs positive for LAMP1/LAMP2 reference and negative for the target protein, and the dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Dual-color DNA-PAINT in ARPE-19 cells identifies heterogeneity in the enrichment of canonical LEL proteins. (A–F) Representative DNA-PAINT images of LAMP1 or LAMP2 reference channel (magenta) and target protein channel (yellow) in ARPE-19 cells for (A) LAMP2, (B) CD63, (C) Cathepsin D, (D) TMEM192, (E) NPC1, and (F) LAMTOR4. Arrows indicate LELs positive for both LAMP1/LAMP2 reference and target protein, arrowheads indicate LELs positive for LAMP1/LAMP2 reference and negative for target protein, and the dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Dual-color DNA-PAINT in ARPE-19 cells identifies heterogeneity in the enrichment of canonical LEL proteins. (A–F) Representative DNA-PAINT images of LAMP1 or LAMP2 reference channel (magenta) and target protein channel (yellow) in ARPE-19 cells for (A) LAMP2, (B) CD63, (C) Cathepsin D, (D) TMEM192, (E) NPC1, and (F) LAMTOR4. Arrows indicate LELs positive for both LAMP1/LAMP2 reference and target protein, arrowheads indicate LELs positive for LAMP1/LAMP2 reference and negative for target protein, and the dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Controls demonstrate optimal fixation and permeabilization conditions as well as the robustness of DNA-PAINT imaging. (A) Representative DNA-PAINT image of glyoxal-fixed HeLa cells stained for LAMP1 and LAMP2 shows significant disruption of LAMP2 staining. (B and C) Representative DNA-PAINT image of HeLa cells fixed with warm 4% PFA and permeabilized with (B) 0.2% Triton X-100 or (C) 0.1% saponin. (D) Quantitative analysis of protein density shows a significant reduction with 0.2% Triton X-100. Mann Whitney U-Test was performed to compare LAMP2 protein density in saponin-treated cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with Triton X-100 treated cells (N = 1 biological replicate, n = 6 cells) (*** P = 0.0002). (E and F) Plots show the percent area of an LEL calculated from the reference image (LAMP2 for Cathepsin D, LAMP1 for all other targets) covered by the localizations from the target protein in HeLa and ARPE-19 cells. (E) shows LAMP1 calculated as the reference channel while (F) shows all other targets calculated as the target channel on a LAMP1/LAMP2 reference channel. Colored lines show traces from 500 randomly selected LELs per target. Solid black line indicates average over all traces for that target. As LAMP1 was used as the reference channel, these traces always reached 100% coverage. Other targets cover a variable percent of LEL area, and importantly individual traces approach a plateau by 25,000 frames for all targets, indicating majority of target protein localizations have been captured by this imaging time. (G and H) (G) Representative dual-color DNA-PAINT images in HeLa cells and (H) quantification in HeLa and ARPE-19 cells of LAMP1 and EEA1 shows distinct organelle populations labeled by each marker, indicating that LAMP1 does not label EEA1-positive early endosomes. N = 3 biological replicates per cell type, n = 14 HeLa cells, n = 14 ARPE-19 cells. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (I and J) LAMP1 and LAMP2 levels on individual LELs in (I) HeLa and (J) ARPE-19 cells show minimal correlation. Pearson’s correlation coefficients (R2) were 0.138 and 0.263 for HeLa and ARPE-19 cells, respectively. (K) LAMP1 localization density is consistent across multiple distinct dual-color experiments in HeLa cells. Kruskal–Wallis test was performed on median LAMP1 density per cell across experiments where LAMP1 was used as the reference channel, P = 0.5342, not significant.
Controls demonstrate optimal fixation and permeabilization conditions as well as the robustness of DNA-PAINT imaging. (A) Representative DNA-PAINT image of glyoxal-fixed HeLa cells stained for LAMP1 and LAMP2 shows significant disruption of LAMP2 staining. (B and C) Representative DNA-PAINT image of HeLa cells fixed with warm 4% PFA and permeabilized with (B) 0.2% Triton X-100 or (C) 0.1% saponin. (D) Quantitative analysis of protein density shows a significant reduction with 0.2% Triton X-100. Mann Whitney U-Test was performed to compare LAMP2 protein density in saponin-treated cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with Triton X-100 treated cells (N = 1 biological replicate, n = 6 cells) (*** P = 0.0002). (E and F) Plots show the percent area of an LEL calculated from the reference image (LAMP2 for Cathepsin D, LAMP1 for all other targets) covered by the localizations from the target protein in HeLa and ARPE-19 cells. (E) shows LAMP1 calculated as the reference channel while (F) shows all other targets calculated as the target channel on a LAMP1/LAMP2 reference channel. Colored lines show traces from 500 randomly selected LELs per target. Solid black line indicates average over all traces for that target. As LAMP1 was used as the reference channel, these traces always reached 100% coverage. Other targets cover a variable percent of LEL area, and importantly individual traces approach a plateau by 25,000 frames for all targets, indicating majority of target protein localizations have been captured by this imaging time. (G and H) (G) Representative dual-color DNA-PAINT images in HeLa cells and (H) quantification in HeLa and ARPE-19 cells of LAMP1 and EEA1 shows distinct organelle populations labeled by each marker, indicating that LAMP1 does not label EEA1-positive early endosomes. N = 3 biological replicates per cell type, n = 14 HeLa cells, n = 14 ARPE-19 cells. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (I and J) LAMP1 and LAMP2 levels on individual LELs in (I) HeLa and (J) ARPE-19 cells show minimal correlation. Pearson’s correlation coefficients (R2) were 0.138 and 0.263 for HeLa and ARPE-19 cells, respectively. (K) LAMP1 localization density is consistent across multiple distinct dual-color experiments in HeLa cells. Kruskal–Wallis test was performed on median LAMP1 density per cell across experiments where LAMP1 was used as the reference channel, P = 0.5342, not significant.
We next developed a robust colocalization analysis to determine the extent of colocalization between LAMP1 and LAMP2-positive LELs in dual-color DNA-PAINT images. While several colocalization algorithms for super-resolution microscopy exist, they predominantly rely on the cross-correlation of point localizations (Hugelier et al., 2023; Malkusch et al., 2012; McCall, 2024; Stone and Veatch, 2015). While these approaches provide an average colocalization index, they do not provide information on specific characteristics of individual objects (in this case, LELs) such as the protein density on each LEL, the size, or the spatial location within the cell of LELs containing specific proteins. To address this, we developed an object-based colocalization algorithm (see Materials and methods for details). We first segmented individual LELs in a reference channel (e.g., LAMP1, ChREF) using Voronoi-based clustering and segmentation with a minimum size filter of 250 nm (Fig. 1). The segmentation results were verified by visual inspection, which confirmed that individual LELs were well-segmented using the chosen parameters. Next, we applied these segmented compartments as a mask to compute the density of localizations from the second, target channel (ChTARGET) within the masked area (Fig. 1). Furthermore, we calculated a background density, representative of the local background density proximal to the mask (see Materials and methods) (Fig. 1). When the localization density inside the reference mask exceeded three standard deviations above the background density, we interpreted this as a significant signal surpassing background levels, indicative of positive colocalization.
This analysis enabled us to determine the percentage of LAMP1-positive LELs that exhibited colocalization with LAMP2-positive LELs and the density of LAMP2 protein within each LAMP1-positive LEL (Fig. 4, A–D). Localization density correlates with imaging duration in DNA-PAINT. Hence, we standardized this duration across all proteins imaged to guarantee reproducibility and consistent comparison. Additionally, if the imaging time is not long enough, only a subset of the target protein’s localizations will be captured, leading to incomplete images of the target protein. To ensure imaging time was long enough to avoid undersampling, we computed the percentage of the LEL area covered by the target protein localizations within the reference mask over the imaging duration (see Materials and methods: Estimating imaging completeness and Fig. S2, E and F). While the coverage percentage varied between individual LELs depending on the abundance of the target protein, we observed levels approaching saturation for both LAMP1 and LAMP2 for each LEL (Fig. S2, E and F), confirming imaging duration was sufficient for capturing the majority of relevant localizations.
Dual-color DNA-PAINT identifies LEL subpopulations with variable protein makeup that persist across epithelial cell types. ( A and B) Box and whisker plots showing the percent colocalization of target proteins with (A) LAMP1 used as reference or (B) LAMP2 used as a reference in HeLa (magenta), HeLa LAMP1-GFP overexpressing (green), or ARPE-19 cells (cyan). All targets were imaged in three independent biological replicates. (A) For LAMP2 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells, n = 11 ARPE-19 cells), CD63 (n = 14 HeLa cells, n = 13 ARPE-19 cells), TMEM192 (n = 13 HeLa cells, n = 14 ARPE-19 cells), NPC1 (n = 16 HeLa cells, n = 15 HeLa LAMP1-GFP overexpressing cells, n = 14 ARPE-19 cells), and LAMTOR4 (n = 16 HeLa cells, n = 14 ARPE-19 cells). CBC refers to the target protein colocalization as analyzed using a coordinate-based colocalization analysis. (B) For Cathepsin D (n = 15 HeLa cells, n = 13 ARPE-19 cells) and LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells, n = 11 ARPE-19 cells). Plot line color indicates cell type, black circles indicate individual cells. Mann–Whitney U test was performed to compare percent colocalization of LAMP2 with LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (**** P < 0.0001), NPC1 with LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (P = 0.3527, no significance), and LAMP1 with LAMP2 (n = 16 HeLa cells, n = 12 HeLa LAMP1-GFP overexpressing cells) (**** P < 0.0001) in HeLa versus HeLa LAMP1-GFP overexpressing cells. Vertical dashed lines separate different target proteins. (C and D) Violin plots of target protein density for (C) high-density and (D) low-density targets. Plot line color indicates cell type, black line indicates median target density on LELs positive for a given target. Black circles, squares, and diamonds represent individual cells from three different biological replicates. Mann–Whitney U test was performed to compare median target densities in HeLa versus HeLa LAMP1-GFP overexpressing cells for LAMP2 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (* P = 0.0172) and NPC1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (P = 0.0683). Vertical dashed lines separate different target proteins. (E–G) Pattern analysis of target proteins on individual LEL membranes. (E) Representative zoomed-in DNA-PAINT image of an individual LEL as defined by LAMP1 or LAMP1-GFP localizations (magenta, first panel), raw localizations of target protein on the LEL (yellow, second panel), nanoclusters of the target channel segmented using DBSCAN and pseudocolored to indicate distinct nanoclusters (third panel), and these nanoclusters overlaid on LEL reference channel alphashape bounding area calculated from the LAMP1 or LAMP1-GFP localizations (fourth panel). Scale bar, 100 nm. (F) Violin plots show nanocluster density (number of localizations of the target protein per unit area of the nanocluster). Plot line color indicates cell type, black line indicates median nanocluster density on LELs positive for a given target. Black circles, squares, and diamonds represent individual cells from three different biological replicates. Vertical dashed lines separate different target proteins. Mann-Whitney U test was performed to compare median nanocluster densities in HeLa cells between NPC1 (n = 16 cells) and LAMTOR4 (n = 16 cells) (**** P < 0.0001) and for NPC1 in HeLa (n = 16 cells) versus HeLa LAMP1-GFP overexpressing cells (n = 13 cells) (**** P < 0.0001). (G) Violin plots show nanocluster diameter. Vertical dashed lines separate different target proteins. Mann–Whitney U test was performed to compare median nanocluster diameters in HeLa cells between NPC1 (n = 16 cells) and LAMTOR4 (n = 16 cells) (**** P < 0.0001) and for NPC1 in HeLa (n = 16 cells) versus HeLa LAMP1-GFP overexpressing cells (n = 13 cells) (* P < 0.0251). (H) FRC analysis (left y-axis) and localization precision measurements (right y-axis) for NPC1 and LAMTOR4 target protein images. Circles indicate individual fields of view. Solid line indicates data plotted on the left versus right y-axis.
Dual-color DNA-PAINT identifies LEL subpopulations with variable protein makeup that persist across epithelial cell types. ( A and B) Box and whisker plots showing the percent colocalization of target proteins with (A) LAMP1 used as reference or (B) LAMP2 used as a reference in HeLa (magenta), HeLa LAMP1-GFP overexpressing (green), or ARPE-19 cells (cyan). All targets were imaged in three independent biological replicates. (A) For LAMP2 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells, n = 11 ARPE-19 cells), CD63 (n = 14 HeLa cells, n = 13 ARPE-19 cells), TMEM192 (n = 13 HeLa cells, n = 14 ARPE-19 cells), NPC1 (n = 16 HeLa cells, n = 15 HeLa LAMP1-GFP overexpressing cells, n = 14 ARPE-19 cells), and LAMTOR4 (n = 16 HeLa cells, n = 14 ARPE-19 cells). CBC refers to the target protein colocalization as analyzed using a coordinate-based colocalization analysis. (B) For Cathepsin D (n = 15 HeLa cells, n = 13 ARPE-19 cells) and LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells, n = 11 ARPE-19 cells). Plot line color indicates cell type, black circles indicate individual cells. Mann–Whitney U test was performed to compare percent colocalization of LAMP2 with LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (**** P < 0.0001), NPC1 with LAMP1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (P = 0.3527, no significance), and LAMP1 with LAMP2 (n = 16 HeLa cells, n = 12 HeLa LAMP1-GFP overexpressing cells) (**** P < 0.0001) in HeLa versus HeLa LAMP1-GFP overexpressing cells. Vertical dashed lines separate different target proteins. (C and D) Violin plots of target protein density for (C) high-density and (D) low-density targets. Plot line color indicates cell type, black line indicates median target density on LELs positive for a given target. Black circles, squares, and diamonds represent individual cells from three different biological replicates. Mann–Whitney U test was performed to compare median target densities in HeLa versus HeLa LAMP1-GFP overexpressing cells for LAMP2 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (* P = 0.0172) and NPC1 (n = 16 HeLa cells, n = 13 HeLa LAMP1-GFP overexpressing cells) (P = 0.0683). Vertical dashed lines separate different target proteins. (E–G) Pattern analysis of target proteins on individual LEL membranes. (E) Representative zoomed-in DNA-PAINT image of an individual LEL as defined by LAMP1 or LAMP1-GFP localizations (magenta, first panel), raw localizations of target protein on the LEL (yellow, second panel), nanoclusters of the target channel segmented using DBSCAN and pseudocolored to indicate distinct nanoclusters (third panel), and these nanoclusters overlaid on LEL reference channel alphashape bounding area calculated from the LAMP1 or LAMP1-GFP localizations (fourth panel). Scale bar, 100 nm. (F) Violin plots show nanocluster density (number of localizations of the target protein per unit area of the nanocluster). Plot line color indicates cell type, black line indicates median nanocluster density on LELs positive for a given target. Black circles, squares, and diamonds represent individual cells from three different biological replicates. Vertical dashed lines separate different target proteins. Mann-Whitney U test was performed to compare median nanocluster densities in HeLa cells between NPC1 (n = 16 cells) and LAMTOR4 (n = 16 cells) (**** P < 0.0001) and for NPC1 in HeLa (n = 16 cells) versus HeLa LAMP1-GFP overexpressing cells (n = 13 cells) (**** P < 0.0001). (G) Violin plots show nanocluster diameter. Vertical dashed lines separate different target proteins. Mann–Whitney U test was performed to compare median nanocluster diameters in HeLa cells between NPC1 (n = 16 cells) and LAMTOR4 (n = 16 cells) (**** P < 0.0001) and for NPC1 in HeLa (n = 16 cells) versus HeLa LAMP1-GFP overexpressing cells (n = 13 cells) (* P < 0.0251). (H) FRC analysis (left y-axis) and localization precision measurements (right y-axis) for NPC1 and LAMTOR4 target protein images. Circles indicate individual fields of view. Solid line indicates data plotted on the left versus right y-axis.
The colocalization analysis revealed 93.5 ± 7.8% and 95.0 ± 4.0% overlap between LAMP1-positive LELs and LAMP2-positive ones both in HeLa and ARPE-19 cells (Fig. 4 A), respectively, as expected. This result held true when LAMP2 was used as a reference mask instead of LAMP1 (97.7 ± 2.7% overlap for HeLa, 94.9 ± 6.0% for ARPE-19 cells) (Fig. 4 B), underscoring the robustness of our analysis pipeline. As a negative control, the early endosomal marker EEA1 displayed minimal colocalization with LAMP1 (14.1 ± 6.1% overlap for HeLa, 10.7 ± 5.1% for ARPE-19 cells) (Fig. S2, G and H), further validating the colocalization analysis pipeline.
To ensure our IF labeling was not impeded by steric effects, we determined the localization density of LAMP1 and LAMP2 on each LEL using our pipeline. If steric effects were present, an inverse correlation would be expected, where the high density of one protein would coincide with the low density of the other. However, we observed a low correlation between the densities of LAMP1 and LAMP2 proteins on LELs (Fig. S2, I and J), ruling out concerns of steric interference.
Overall, we have successfully developed a robust and quantitative DNA-PAINT imaging pipeline, enabling precise determination of protein levels and their colocalization within LELs.
Dual-color DNA-PAINT identifies LEL subpopulations with variable protein makeup
We applied our quantitative pipeline to determine whether five additional LEL proteins, crucial for different aspects of LEL biology, are present on all LAMP1-positive compartments or if there exist subpopulations of LELs with distinct protein compositions (Fig. 2, B–F, Fig. 3, B–F, and Fig. 4, A–D). We once again verified that the imaging time was long enough to not lead to undersampling (see Materials and methods: Estimating imaging completeness and Fig. S2 F). The dual-color imaging additionally gave us the opportunity to test the quantitative robustness of DNA-PAINT imaging as LAMP1 was independently imaged as a reference channel while concurrently evaluating LAMP2, CD63, TMEM192, NPC1, and LAMTOR4. When we compared the average protein density of LAMP1 across these five distinct biological replicate experiments in HeLa cells, we found no statistically significant differences in its abundance (Fig. S2 K). This consistency highlights the quantitative robustness inherent in our imaging and analysis pipeline.
We found that CD63, another prevalent LEL protein (Schwake et al., 2013), was present on 87.0 ± 6.8% of LAMP1-positive LELs in HeLa cells (Fig. 2 B and Fig. 4 A). Interestingly, in ARPE-19 cells, CD63 displayed more variation in its colocalization with LAMP1, ranging from as low as 40% in some cells to almost complete colocalization in others with an average colocalization of 70.7 ± 20.2% (Fig. 3 B and Fig. 4 A). This result may reflect differences in the maturity or function of LELs in the two different cell types.
When visualizing Cathepsin D, a luminal degradative enzyme, we found that this enzyme is present within 86.4 ± 15.4% of LAMP2-positive LELs in HeLa and 84.7 ± 11.4% in ARPE-19 cells (Fig. 2 C, Fig. 3 C, and Fig. 4 B), suggesting that the majority of visualized LELs contain degradative enzymes. However, a limitation in interpreting these results is that the antibody used, while confirmed to be specific to Cathepsin D using knockdown analysis (Fig. S3, A, F, I, and J), could not differentiate between the uncleaved procathepsin D (pCD) and the cleaved, active form of the enzyme within degradative LELs (Fig. S3 F) (Di et al., 2021).
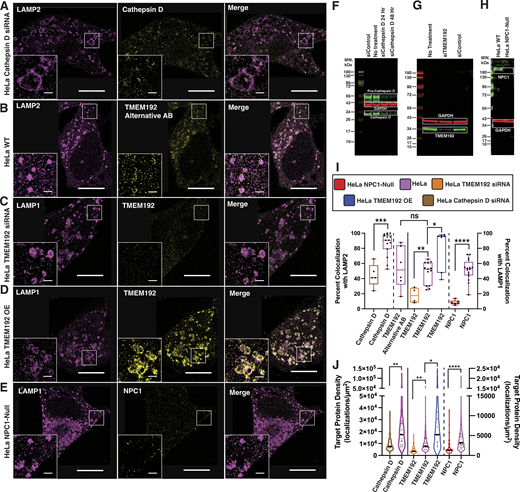
Cathepsin D, TMEM192, and NPC1 antibodies are robust and specific. (A) Representative DNA-PAINT image of LAMP2 and Cathepsin D when the Cathepsin D protein is knocked down via siRNA in HeLa cells shows a significant reduction in Cathepsin D signal on LAMP2 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (B) Representative DNA-PAINT image of LAMP2 and TMEM192 in HeLa cells imaged using an alternative TMEM192 antibody shows a staining pattern consistent with results from the main antibody used. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (C) Representative DNA-PAINT image of LAMP1 and TMEM192 when the TMEM192 protein is knocked down via siRNA in HeLa cells shows a significant reduction in TMEM192 signal on LAMP1 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (D) Representative DNA-PAINT image of LAMP1 and TMEM192 in TMEM192-3xHA overexpressing (OE) HeLa cells shows near-complete overlap with LAMP1. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (E) Representative DNA-PAINT image of LAMP1 and NPC1 in NPC1-null HeLa cells shows minimal overall signal and overlap with LAMP1 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (F–H) WB analyses of (F) Cathepsin D, (G) TMEM192, and (H) NPC1 levels in HeLa cells. (F) Cathepsin D antibody recognizes both uncleaved pro-Cathepsin D and cleaved Cathepsin D protein products, as expected. Cathepsin D siRNA knockdown in HeLa cells shows a reduction in Cathepsin D signal. (G) TMEM192 siRNA knockdown in HeLa cells shows a reduction in TMEM192 signal. (H) NPC1-null HeLa cells show loss of NPC1 signal. (I) Quantification of colocalization of Cathepsin D, TMEM192, and NPC1 in HeLa cells. Plot line color indicates cell type, black circles indicate individual cells, solid black line indicates data plotted on left Y-axis (LAMP2 reference) versus right y-axis (LAMP1 reference). Vertical dashed lines separate different target proteins. Mann–Whitney U-tests were performed to compare Cathepsin D siRNa knockdown HeLa cells (N = 1 biological replicate, n = 5 cells) with HeLa cells (N = 3 biological replicates, n = 15 cells, replotted from Fig. 4) (*** P = 0.0005), HeLa cells imaged with TMEM192 main antibody (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4), or alternative antibody (N = 1 biological replicate, n = 6 cells) (P = 0.6388, no significance), TMEM192 siRNA knockdown HeLa cells (N = 1 biological replicate, n = 4 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (** P = 0.0034), TMEM192-3xHA overexpressing cells (N = 1 biological replicate, n = 7 cells) with wildtype HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (* P = 0.0297), and NPC1-null HeLa cells (N = 1 biological replicate, n = 6 cells) with HeLa cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) (**** P < 0.0001). (J) Protein density of Cathepsin D, TMEM192, and NPC1 in HeLa cells. Plot line color indicates cell type, black circles indicate individual cell medians, solid black line indicates data plotted on the left y-axis (LAMP2 reference) versus the right y-axis (LAMP1 reference). Vertical dashed lines separate different target proteins. Mann–Whitney U-tests were performed to compare: Cathepsin D siRNa knockdown HeLa cells (N = 1 biological replicate, n = 5 cells) with HeLa cells (N = 3 biological replicates, n = 15 cells, replotted from Fig. 4) (** P = 0.0015), TMEM192 siRNA knockdown HeLa cells (N = 1 biological replicate, n = 4 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (** P = 0.0034), and TMEM192-3xHA overexpressing cells (N = 1 biological replicate, n = 7 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (* P = 0.0236), and NPC1-null HeLa cells (N = 1 biological replicate, n = 6 cells) with HeLa cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) (**** P < 0.0001). Source data are available for this figure: SourceData FS3.
Cathepsin D, TMEM192, and NPC1 antibodies are robust and specific. (A) Representative DNA-PAINT image of LAMP2 and Cathepsin D when the Cathepsin D protein is knocked down via siRNA in HeLa cells shows a significant reduction in Cathepsin D signal on LAMP2 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (B) Representative DNA-PAINT image of LAMP2 and TMEM192 in HeLa cells imaged using an alternative TMEM192 antibody shows a staining pattern consistent with results from the main antibody used. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (C) Representative DNA-PAINT image of LAMP1 and TMEM192 when the TMEM192 protein is knocked down via siRNA in HeLa cells shows a significant reduction in TMEM192 signal on LAMP1 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (D) Representative DNA-PAINT image of LAMP1 and TMEM192 in TMEM192-3xHA overexpressing (OE) HeLa cells shows near-complete overlap with LAMP1. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (E) Representative DNA-PAINT image of LAMP1 and NPC1 in NPC1-null HeLa cells shows minimal overall signal and overlap with LAMP1 with respect to wildtype conditions. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (F–H) WB analyses of (F) Cathepsin D, (G) TMEM192, and (H) NPC1 levels in HeLa cells. (F) Cathepsin D antibody recognizes both uncleaved pro-Cathepsin D and cleaved Cathepsin D protein products, as expected. Cathepsin D siRNA knockdown in HeLa cells shows a reduction in Cathepsin D signal. (G) TMEM192 siRNA knockdown in HeLa cells shows a reduction in TMEM192 signal. (H) NPC1-null HeLa cells show loss of NPC1 signal. (I) Quantification of colocalization of Cathepsin D, TMEM192, and NPC1 in HeLa cells. Plot line color indicates cell type, black circles indicate individual cells, solid black line indicates data plotted on left Y-axis (LAMP2 reference) versus right y-axis (LAMP1 reference). Vertical dashed lines separate different target proteins. Mann–Whitney U-tests were performed to compare Cathepsin D siRNa knockdown HeLa cells (N = 1 biological replicate, n = 5 cells) with HeLa cells (N = 3 biological replicates, n = 15 cells, replotted from Fig. 4) (*** P = 0.0005), HeLa cells imaged with TMEM192 main antibody (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4), or alternative antibody (N = 1 biological replicate, n = 6 cells) (P = 0.6388, no significance), TMEM192 siRNA knockdown HeLa cells (N = 1 biological replicate, n = 4 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (** P = 0.0034), TMEM192-3xHA overexpressing cells (N = 1 biological replicate, n = 7 cells) with wildtype HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (* P = 0.0297), and NPC1-null HeLa cells (N = 1 biological replicate, n = 6 cells) with HeLa cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) (**** P < 0.0001). (J) Protein density of Cathepsin D, TMEM192, and NPC1 in HeLa cells. Plot line color indicates cell type, black circles indicate individual cell medians, solid black line indicates data plotted on the left y-axis (LAMP2 reference) versus the right y-axis (LAMP1 reference). Vertical dashed lines separate different target proteins. Mann–Whitney U-tests were performed to compare: Cathepsin D siRNa knockdown HeLa cells (N = 1 biological replicate, n = 5 cells) with HeLa cells (N = 3 biological replicates, n = 15 cells, replotted from Fig. 4) (** P = 0.0015), TMEM192 siRNA knockdown HeLa cells (N = 1 biological replicate, n = 4 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (** P = 0.0034), and TMEM192-3xHA overexpressing cells (N = 1 biological replicate, n = 7 cells) with HeLa cells (N = 3 biological replicates, n = 13 cells, replotted from Fig. 4) (* P = 0.0236), and NPC1-null HeLa cells (N = 1 biological replicate, n = 6 cells) with HeLa cells (N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) (**** P < 0.0001). Source data are available for this figure: SourceData FS3.
The transmembrane protein 192 (TMEM192) is a lesser-known LEL protein, initially identified through organellar proteomics (Chapel et al., 2013; Nguyen et al., 2017; Schröder et al., 2007, 2010). Although the function of TMEM192 is unclear (Nguyen et al., 2017), it is widely used in Lyso-IP (immunoprecipitation) studies to immunoprecipitate lysosomes as it preserves lysosomal association when overexpressed (Abu-Remaileh et al., 2017). We aimed to determine the LEL localization of this protein under native conditions. We found that TMEM192, while lowly abundant (Fig. 4 D), was consistently present above background levels on 47.9 ± 14.6% of LAMP1-positive LELs in HeLa cells and 59.4 ± 13.1% in ARPE-19 cells (Fig. 2 D, Fig. 3 D, and Fig. 4 A). This result was corroborated using a second, alternative TMEM192 antibody (Fig. S3, B and I). When TMEM192 was knocked down, there was a marked reduction in both the colocalization percentage and protein density on LELs (Fig. S3, C, G, I, and J). Conversely, overexpression of TMEM192 led to a significant increase in protein density and colocalization (81.1 ± 26.0%), consistent with expectations for LEL localization upon overexpression (Fig. S3, D, I, and J). These results demonstrate the capability of our pipeline to accurately identify and quantify even lowly abundant proteins on LELs.
NPC1 is an important protein essential for cholesterol export from LELs, with genetic mutations in NPC1 leading to Niemann-Pick type C disease (Infante et al., 2008; Pfeffer, 2019). Analysis showed that NPC1 is also lowly abundant on the membrane of 51.6 ± 14.0% of LELs in HeLa cells and 46.5 ± 15.8% in ARPE-19 cells (Fig. 2 E, Fig. 3 E; and Fig. 4, A and D). DNA-PAINT imaging in NPC1 KO HeLa cells once again confirmed the specificity of the antibody (Fig. S3, E and H–J). Notably, NPC1 localizations formed tightly packed nanoscale domains on the LEL membrane in DNA-PAINT images (Fig. 4, E–G). We employed density-based spatial clustering of applications with noise (DBSCAN) clustering to segment and quantitatively analyze these nanoclusters (Fig. 4 E), finding an average of five NPC1 nanoclusters per LEL with a median diameter of 55 nm (Fig. 4 G), which was above the spatial resolution limit of our imaging as computed using Fourier ring correlation (FRC) (Nieuwenhuizen et al., 2013) or using localization precision (Fig. 4 H). These results suggest that NPC1 organizes into multiple nanoscale platforms on LEL membranes, potentially facilitating cholesterol export through clustering.
Finally, LAMTOR4, a component of the pentameric Ragulator complex, plays a pivotal role as a scaffold for Rag GTPases crucial in the recruitment and activation of mTORC1 on LEL membranes (Sancak et al., 2008, 2010; Zoncu et al., 2011). Like TMEM192 and NPC1, LAMTOR4 was found in low abundance on LELs (Fig. 2 F, Fig. 3 F; and Fig. 4, A and D). However, in contrast to these proteins, LAMTOR4 was detected on a much higher percentage of LELs (79.9 ± 8.1% in HeLa, 83.0 ± 5.3% in ARPE-19 cells) (Fig. 4 A). DBSCAN clustering analysis revealed distinct characteristics of LAMTOR4 distribution; compared with NPC1, LAMTOR4 formed larger (83 nm and larger than the spatial resolution limit computed by FRC, Fig. 4 H), less dense nanoplatforms on the LEL membrane, indicating a different organizational pattern for this protein (Fig. 4, E–G).
Given that TMEM192 and NPC1 were enriched only on LEL subsets, we wanted to ensure that the low colocalization was not due to the limitations of the object-based colocalization algorithm. We thus compared the colocalization results to the coordinate-based colocalization (CBC) method (Malkusch et al., 2012) (Fig. 4 A, TMEM192 CBC and NPC1 CBC). The colocalization index was 33.2 ± 7.0% and 33.3 ± 7.1% for TMEM192 and NPC1, respectively. These numbers were slightly lower but close to those obtained using the object-based colocalization method, suggesting that the low colocalization is not due to the limits of the object-based colocalization analysis.
TMEM192 and NPC1 were also less abundant than LAMP1, LAMP2, and CD63; hence, we further wanted to ensure that the low colocalization was not due to undersampling of these proteins. Indeed, deliberately undersampling a protein of interest by using low antibody concentration can lead to low colocalization, as evidenced by the colocalization between LAMP1 and LAMP2 dropping to 48.3 ± 7.0% when the LAMP2 antibody concentration was reduced 20-fold (Fig. S4, A and B). Therefore, to ensure low colocalization is not a result of undersampling, we doubled the concentration of TMEM192 and NPC1 antibodies. This did not affect the density or colocalization percentage of these proteins with LAMP1, indicating that the antibody concentrations were already saturating (Fig. S4, A and B). Additionally, there was no correlation between the median protein density of LAMP2, NPC1, or TMEM192 in a given cell and their colocalization percentage with LAMP1 (Fig. S4, C and D). Some cells exhibited similar protein densities for NPC1 compared with LAMP2; however, even in these cells, LAMP2 consistently showed a much higher colocalization percentage with LAMP1 than NPC1 (Fig. S4 C, red rectangles). Similarly, when TMEM192 was overexpressed, there was a wide range of protein densities per cell, some overlapping with the protein density in wildtype cells (Fig. S4 D, red rectangles). Even for these cells where the protein densities were similar, TMEM192 overexpressing cells exhibited a higher colocalization percentage of TMEM192 compared with wildtype cells (Fig. S4 D, red rectangles). This is likely due to a spillover effect, where upon overexpression, TMEM192 becomes localized to LAMP1-positive compartments that normally lack TMEM192. Additionally, when we lowered the threshold for colocalization from three standard deviations to two standard deviations above the background density, there was no change in the percent colocalization between TMEM192, NPC1, and LAMP1 (Fig. S4 E).
Controls show the robustness of object-based colocalization analysis for low-density targets, and LAMP1-GFP overexpression (OE) in HeLa cells induces changes in LELs. (A) Box and whisker plots of percent colocalization of target proteins on LELs in HeLa cells imaged with varying primary antibody (AB) concentrations. The solid line indicates data plotted on the left y-axis (LAMP1 reference) versus the right y-axis (LAMP2 reference). Mann–Whitney U tests were performed to compare LAMP2 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with LAMP2 1:2,000 (20-fold lower concentration, N = 1 biological replicate, n = 7 cells) (**** P < 0.0001), NPC1 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with NPC1 1:50 (twofold higher concentration, N = 1 biological replicate, n = 7 cells) (P = 0.5245, no significance), TMEM192 1:25 (standard dilution, N = 3 biological replicates, n = 13 cells) with TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) (P = 0.21, no significance), and TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) with TMEM192 imaged with an alternative antibody (N = 1 biological replicate, n = 6 cells, replotted from Fig. S3) (P = 0.4848, no significance). (B) Violin plots of target protein density on LELs in HeLa cells imaged with varying primary antibody concentrations. Mann–Whitney U tests were performed to compare LAMP2 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with LAMP2 1:2,000 (20-fold lower concentration, N = 1 biological replicate, n = 7 cells) (**** P < 0.0001), NPC1 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with NPC1 1:50 (twofold higher concentration, N = 1 biological replicate, n = 7 cells) (P = 0.249, no significance), and TMEM192 1:25 (standard dilution, N = 3 biological replicates, n = 13 cells) with TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) (P = 0.5789, no significance). (C) Colocalization percentage versus median protein density per cell plots for NPC1 and LAMP2 imaged on LAMP1 reference LELs in HeLa cells. Black circles represent individual cells. Red boxes indicate protein density values between 3,500 and 6,500 localizations/area where both NPC1 and LAMP2 targets have similar median densities but LAMP2 shows much higher percent colocalization with LAMP1. (D) Colocalization percentage versus median density per cell plots for TMEM192 in HeLa cells and TMEM192 in TMEM192-3xHA overexpressing HeLa cells imaged on LAMP1 reference LELs. Black circles represent individual cells. Red boxes indicate protein density values between 1,000–4,000 localizations/area where both WT and TMEM192 overexpressing cells have similar median protein densities but TMEM192 shows much higher percent colocalization with LAMP1 in overexpressing cells. (E) Box and whisker plots of colocalization percentage of target proteins with LAMP1 analyzed with two different colocalization thresholds. NPC1 and TMEM192 both show minimal differences in colocalization when analyzed using the standard cutoff threshold for colocalization of three standard deviations above background density when compared to the less-stringent two standard deviation above background density threshold. Mann-Whitney U tests were performed to compare percent colocalization with LAMP1 for NPC1 using three standard deviation versus two standard deviation filter (N = 3 biological replicates, n = 16 cells) (P = 0.0849, no significance) and TMEM192 using three standard deviation versus two standard deviation filter (N = 3 biological replicates, n = 13 cells) (P = 0.2642, no significance). (F and G) Representative DNA-PAINT images of (F) LAMP2 and (G) NPC1 with LAMP1 in HeLa cells overexpressing LAMP1-GFP. Arrows indicate LELs positive for both LAMP1-GFP and target protein, arrowheads indicate LELs positive for LAMP1 and negative for target protein, dotted line indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (H) Comparison of LEL area in wild type versus LAMP1-GFP overexpressing HeLa cells. Black circles indicate individual cell medians. Mann-Whitney U-test was performed to compare cell medians of HeLa cells (n = 75 cells) with LAMP1-GFP overexpressing HeLa cells (n = 26 cells) (* P = 0.0197).
Controls show the robustness of object-based colocalization analysis for low-density targets, and LAMP1-GFP overexpression (OE) in HeLa cells induces changes in LELs. (A) Box and whisker plots of percent colocalization of target proteins on LELs in HeLa cells imaged with varying primary antibody (AB) concentrations. The solid line indicates data plotted on the left y-axis (LAMP1 reference) versus the right y-axis (LAMP2 reference). Mann–Whitney U tests were performed to compare LAMP2 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with LAMP2 1:2,000 (20-fold lower concentration, N = 1 biological replicate, n = 7 cells) (**** P < 0.0001), NPC1 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with NPC1 1:50 (twofold higher concentration, N = 1 biological replicate, n = 7 cells) (P = 0.5245, no significance), TMEM192 1:25 (standard dilution, N = 3 biological replicates, n = 13 cells) with TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) (P = 0.21, no significance), and TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) with TMEM192 imaged with an alternative antibody (N = 1 biological replicate, n = 6 cells, replotted from Fig. S3) (P = 0.4848, no significance). (B) Violin plots of target protein density on LELs in HeLa cells imaged with varying primary antibody concentrations. Mann–Whitney U tests were performed to compare LAMP2 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with LAMP2 1:2,000 (20-fold lower concentration, N = 1 biological replicate, n = 7 cells) (**** P < 0.0001), NPC1 1:100 (standard dilution, N = 3 biological replicates, n = 16 cells, replotted from Fig. 4) with NPC1 1:50 (twofold higher concentration, N = 1 biological replicate, n = 7 cells) (P = 0.249, no significance), and TMEM192 1:25 (standard dilution, N = 3 biological replicates, n = 13 cells) with TMEM192 1:12.5 (twofold higher concentration, N = 1 biological replicate, n = 6 cells) (P = 0.5789, no significance). (C) Colocalization percentage versus median protein density per cell plots for NPC1 and LAMP2 imaged on LAMP1 reference LELs in HeLa cells. Black circles represent individual cells. Red boxes indicate protein density values between 3,500 and 6,500 localizations/area where both NPC1 and LAMP2 targets have similar median densities but LAMP2 shows much higher percent colocalization with LAMP1. (D) Colocalization percentage versus median density per cell plots for TMEM192 in HeLa cells and TMEM192 in TMEM192-3xHA overexpressing HeLa cells imaged on LAMP1 reference LELs. Black circles represent individual cells. Red boxes indicate protein density values between 1,000–4,000 localizations/area where both WT and TMEM192 overexpressing cells have similar median protein densities but TMEM192 shows much higher percent colocalization with LAMP1 in overexpressing cells. (E) Box and whisker plots of colocalization percentage of target proteins with LAMP1 analyzed with two different colocalization thresholds. NPC1 and TMEM192 both show minimal differences in colocalization when analyzed using the standard cutoff threshold for colocalization of three standard deviations above background density when compared to the less-stringent two standard deviation above background density threshold. Mann-Whitney U tests were performed to compare percent colocalization with LAMP1 for NPC1 using three standard deviation versus two standard deviation filter (N = 3 biological replicates, n = 16 cells) (P = 0.0849, no significance) and TMEM192 using three standard deviation versus two standard deviation filter (N = 3 biological replicates, n = 13 cells) (P = 0.2642, no significance). (F and G) Representative DNA-PAINT images of (F) LAMP2 and (G) NPC1 with LAMP1 in HeLa cells overexpressing LAMP1-GFP. Arrows indicate LELs positive for both LAMP1-GFP and target protein, arrowheads indicate LELs positive for LAMP1 and negative for target protein, dotted line indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (H) Comparison of LEL area in wild type versus LAMP1-GFP overexpressing HeLa cells. Black circles indicate individual cell medians. Mann-Whitney U-test was performed to compare cell medians of HeLa cells (n = 75 cells) with LAMP1-GFP overexpressing HeLa cells (n = 26 cells) (* P = 0.0197).
Even highly abundant proteins like CD63 could be present in LEL subpopulations, as evidenced by CD63 being found on average on 70.7 ± 20.2% of LELs (and as low as 40% in some cells) in ARPE-19 cells (Fig. 4, A and C). Conversely, lowly abundant proteins could be on a high percentage of LELs as shown by the lowly abundant LAMTOR4 being found on 79.9 ± 8.1% of LELs in HeLa and 83.0 ± 5.3% of LELs in ARPE-19 cells (Fig. 4, A and D).
Taken together, these results strongly argue against undersampling as the cause of low colocalization. However, we cannot entirely rule out the possibility that some LELs have protein densities below our detection limit, which may result in them being categorized as negative for a given protein due to the sensitivity limitations of our method.
Protein overexpression and drug-based perturbations impact LEL subpopulations
After establishing the baseline abundance and heterogeneity of LEL proteins, we explored how specific perturbations impact these subpopulations. We first asked how the overexpression of LAMP1, a common practice in cell biology studies, might influence these baseline populations. We overexpressed LAMP1 fused to GFP and used a GFP nanobody to simultaneously visualize LAMP1 and either LAMP2 or NPC1 in dual-color DNA-PAINT images (Fig. S4, F and G). We picked LAMP2 and NPC1 for our analysis as they provide examples of both high- and low-abundance LEL proteins. Quantitative analysis showed that overexpression differentially affected these two proteins. Notably, the levels of both LAMP2 and NPC1 slightly decreased on LEL membranes following LAMP1 overexpression (Fig. 4, C and D). This decrease was a result of an increase in the size of LELs rather than a decrease in the absolute amount of LAMP1 or NPC1 (Fig. S4 H). Interestingly, while the overlap percentage of LAMP1-positive LELs with NPC1 remained unchanged, there was a significant reduction in the colocalization of LAMP1-positive LELs with LAMP2—decreasing from 93.5 ± 7.8% to 76.8 ± 16.2% (Fig. 4 A). Similar trends were observed when LAMP2 was used as the reference, showing a decrease in the overlap with LAMP1 (Fig. 4 B). Additionally, the overexpression of LAMP1 significantly influenced NPC1 nanoclusters, resulting in increased size and reduced packing density on the LEL membrane (Fig. 4, E–G). These results highlight that the overexpression of LEL proteins can affect not only the levels of the overexpressed protein itself, but also the size of the organelle, the density, and the organization of other membrane proteins. Moreover, overexpression can lead to the emergence of LEL subpopulations absent under native conditions such as those that are LAMP1-positive but LAMP2-negative.
We next explored the impact of two drug-based perturbations, EN6 and Bafilomycin A1 (BafA1) (Fig. 5). EN6 treatment increases lysosomal acidity by enhancing vacuolar ATPase (vATPase) activity, which activates autophagy and blocks mTORC1 activation (Chung et al., 2019). In contrast, BafA1 reduces vATPase activity and prevents autophagosome–lysosome fusion, leading to enlarged LELs with reduced acidity (Mauvezin and Neufeld, 2015). We analyzed the impact of these treatments on NPC1 and LAMTOR4-positive LEL subpopulations, given the relevance of these proteins to cholesterol homeostasis (NPC1) and mTORC1 signaling (LAMTOR4).
Drug-based perturbations impact LEL subpopulations. (A–D) Representative DNA-PAINT images of LAMP1 reference channel (magenta) and target protein channel (yellow) in (A and B) HeLa cells treated for 16 h with either 15 μM EN6 or (C and D) 100 nm BafA1. Arrows indicate LELs positive for both LAMP1 and target protein, arrowheads indicate LELs positive for LAMP1 and negative for target protein, dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (E) Box and whisker plots showing the percent colocalization of target proteins with LAMP1 reference in HeLa (re-plotted from Fig. 4), EN6-treated HeLa, or BafA1-treated HeLa cells. All targets were imaged in three or four independent biological replicates. Plot line color indicates cell type, black circles indicate individual cells. Cells were clustered using a standard minimum area filter unless otherwise specified. Mann–Whitney U test was performed to compare the percent colocalization of NPC1 in untreated HeLa with NPC1 in EN6-treated HeLa (n = 16 HeLa cells, n = 14 EN6-treated HeLa cells) (**** P < 0.0001), NPC1 in untreated HeLa with NPC1 in BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (*** P = 0.001), NPC1 in BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.058), LAMTOR4 in untreated HeLa with LAMTOR4 in EN6-treated HeLa (n = 16 HeLa cells, n = 13 EN6-treated HeLa cells) (*** P = 0.0005), LAMTOR4 in untreated HeLa with LAMTOR4 in BafA1-treated HeLa (n = 16 HeLa cells, n = 12 BafA1-treated HeLa cells) (**** P < 0.0001), and LAMTOR4 in BafA1-treated HeLa cells with untreated HeLa using 350-nm-diameter filter (n = 12 BafA1-treated HeLa cells, n = 16 HeLa cells) (**** P < 0.0001). (F) Violin plots of target protein density on reference LELs in HeLa (re-plotted from Fig. 4), EN6-treated HeLa, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using 350-nm-diameter filter. Plot line color indicates cell type, black line indicates median target density on LELs positive for a given target. Black circles represent individual cells from three or four independent biological replicates. Mann-Whitney U test was performed to compare median target densities in untreated HeLa with NPC1 in EN6-treated HeLa (n = 16 HeLa cells, n = 14 EN6-treated HeLa cells) (P = 0.061), NPC1 in untreated HeLa with NPC1 in BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (P = 0.8204, no significance), NPC1 in BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.8678, no significance), LAMTOR4 in untreated HeLa with LAMTOR4 in EN6-treated HeLa (n = 16 HeLa cells, n = 13 EN6-treated HeLa cells) (**** P < 0.0001), LAMTOR4 in untreated HeLa with LAMTOR4 in BafA1-treated HeLa (n = 16 HeLa cells, n = 12 BafA1-treated HeLa cells) (*** P = 0.0004), and LAMTOR4 in BafA1-treated HeLa cells with untreated HeLa using 350-nm-diameter filter (n = 12 BafA1-treated HeLa cells, n = 16 HeLa cells) (**** P < 0.0001). (G) Violin plots of median LEL area in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using a 350-nm-diameter filter. All cells imaged with LAMP1 and NPC1 as well LAMP1 and LAMTOR4 were combined for each condition. Mann–Whitney U test was performed to compare median LEL areas per cell in untreated HeLa with BafA1-treated HeLa (n = 32 HeLa cells, n = 33 BafA1-treated HeLa cells) (**** P < 0.0001), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 32 HeLa cells, n = 33 BafA1-treated HeLa cells) (**** P < 0.0001). (H) Box and whisker plots showing the total NPC1-positive LEL area per cell in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using a 350-nm-diameter filter. Mann–Whitney U test was performed to compare the total NPC1-positive LEL area per cell in untreated HeLa with BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (* P = 0.0346), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (** P = 0.0064). (I) Box and whisker plots showing the total NPC1-negative LEL area per cell in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using 350-nm-diameter filter. Mann-Whitney U test was performed to compare NPC1-negative LEL area per cell in untreated HeLa with BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (* P = 0.0162), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.089).
Drug-based perturbations impact LEL subpopulations. (A–D) Representative DNA-PAINT images of LAMP1 reference channel (magenta) and target protein channel (yellow) in (A and B) HeLa cells treated for 16 h with either 15 μM EN6 or (C and D) 100 nm BafA1. Arrows indicate LELs positive for both LAMP1 and target protein, arrowheads indicate LELs positive for LAMP1 and negative for target protein, dotted circle indicates which LEL arrow or arrowhead refers to. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (E) Box and whisker plots showing the percent colocalization of target proteins with LAMP1 reference in HeLa (re-plotted from Fig. 4), EN6-treated HeLa, or BafA1-treated HeLa cells. All targets were imaged in three or four independent biological replicates. Plot line color indicates cell type, black circles indicate individual cells. Cells were clustered using a standard minimum area filter unless otherwise specified. Mann–Whitney U test was performed to compare the percent colocalization of NPC1 in untreated HeLa with NPC1 in EN6-treated HeLa (n = 16 HeLa cells, n = 14 EN6-treated HeLa cells) (**** P < 0.0001), NPC1 in untreated HeLa with NPC1 in BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (*** P = 0.001), NPC1 in BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.058), LAMTOR4 in untreated HeLa with LAMTOR4 in EN6-treated HeLa (n = 16 HeLa cells, n = 13 EN6-treated HeLa cells) (*** P = 0.0005), LAMTOR4 in untreated HeLa with LAMTOR4 in BafA1-treated HeLa (n = 16 HeLa cells, n = 12 BafA1-treated HeLa cells) (**** P < 0.0001), and LAMTOR4 in BafA1-treated HeLa cells with untreated HeLa using 350-nm-diameter filter (n = 12 BafA1-treated HeLa cells, n = 16 HeLa cells) (**** P < 0.0001). (F) Violin plots of target protein density on reference LELs in HeLa (re-plotted from Fig. 4), EN6-treated HeLa, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using 350-nm-diameter filter. Plot line color indicates cell type, black line indicates median target density on LELs positive for a given target. Black circles represent individual cells from three or four independent biological replicates. Mann-Whitney U test was performed to compare median target densities in untreated HeLa with NPC1 in EN6-treated HeLa (n = 16 HeLa cells, n = 14 EN6-treated HeLa cells) (P = 0.061), NPC1 in untreated HeLa with NPC1 in BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (P = 0.8204, no significance), NPC1 in BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.8678, no significance), LAMTOR4 in untreated HeLa with LAMTOR4 in EN6-treated HeLa (n = 16 HeLa cells, n = 13 EN6-treated HeLa cells) (**** P < 0.0001), LAMTOR4 in untreated HeLa with LAMTOR4 in BafA1-treated HeLa (n = 16 HeLa cells, n = 12 BafA1-treated HeLa cells) (*** P = 0.0004), and LAMTOR4 in BafA1-treated HeLa cells with untreated HeLa using 350-nm-diameter filter (n = 12 BafA1-treated HeLa cells, n = 16 HeLa cells) (**** P < 0.0001). (G) Violin plots of median LEL area in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using a 350-nm-diameter filter. All cells imaged with LAMP1 and NPC1 as well LAMP1 and LAMTOR4 were combined for each condition. Mann–Whitney U test was performed to compare median LEL areas per cell in untreated HeLa with BafA1-treated HeLa (n = 32 HeLa cells, n = 33 BafA1-treated HeLa cells) (**** P < 0.0001), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 32 HeLa cells, n = 33 BafA1-treated HeLa cells) (**** P < 0.0001). (H) Box and whisker plots showing the total NPC1-positive LEL area per cell in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using a 350-nm-diameter filter. Mann–Whitney U test was performed to compare the total NPC1-positive LEL area per cell in untreated HeLa with BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (* P = 0.0346), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (** P = 0.0064). (I) Box and whisker plots showing the total NPC1-negative LEL area per cell in untreated HeLa cells, BafA1-treated HeLa cells, or HeLa cells with LELs segmented using 350-nm-diameter filter. Mann-Whitney U test was performed to compare NPC1-negative LEL area per cell in untreated HeLa with BafA1-treated HeLa (n = 16 HeLa cells, n = 21 BafA1-treated HeLa cells) (* P = 0.0162), and BafA1-treated HeLa with untreated HeLa using 350-nm-diameter filter (n = 21 BafA1-treated HeLa cells, n = 16 HeLa cells) (P = 0.089).
Interestingly, EN6 treatment led to a small but significant increase in the density of both NPC1 and LAMTOR4 on LELs (Fig. 5 A, B, E, and F). This increase in protein abundance was consistent with the results of western blot (WB) analysis of these proteins (Fig. S5, A and B). The percentage of NPC1-positive LELs increased with EN6 treatment (from 51.6 ± 14.0% to 85.9 ± 4.7), along with a small but significant increase in the percentage of LAMTOR4-positive LELs (from 79.9 ± 8.1 to 90.0 ± 6.0) (Fig. 5 E).
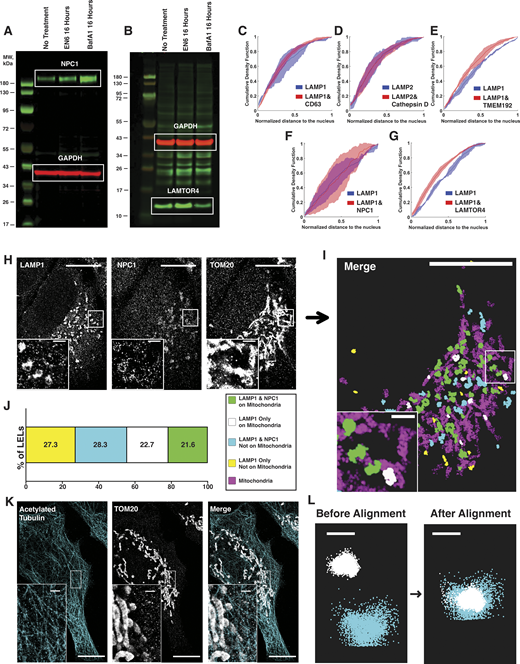
Bulk analysis of drug-treated HeLa cells, examining LEL subpopulations with additional cellular context in ARPE-19 cells, and multiplexed imaging controls. (A and B) Representative WBs of (A) NPC1 and (B) LAMTOR4 in untreated, EN6-treated, or BafA1-treated conditions in HeLa cells. EN6 treatment leads to an increase in both NPC1 and LAMTOR4. BafA1 treatment leads to an increase in NPC1 but a decrease in LAMTOR4. (C–G) Cumulative density function plots of LEL distance from the nucleus from dual-color DNA-PAINT imaging experiments in ARPE-19 cells for subpopulations containing LAMP1 or LAMP2 only or LAMP1/LAMP2 and target protein: (C) CD63, (D) Cathepsin D, (E) TMEM192, (F) NPC1, and (G) LAMTOR4. Distance to the nucleus was normalized per cell to the maximum distance from the nucleus of an LEL in that cell, with values closest to zero indicating greatest proximity to the nucleus. Line indicates median with standard deviation between biological replicates. Kolmogorov–Smirnov tests were performed on the mean distributions from three independent biological replicates: for CD63 n = 13 cells, P = 0.193, no significance; for Cathepsin D n = 13 cells, P = 0.794, no significance; for TMEM192 n = 14 cells, P = 0.261, no significance; for NPC1 n = 14 cells, P = 0.140, no significance; for LAMTOR4 n = 14 cells, P = 0.193, no significance. (H–J) Representative three-color DNA-PAINT image of LAMP1, NPC1, and mitochondria (TOM20) in ARPE-19 cells. (H and I) (H) Raw images and (I) post-processed image showing a spatial map of LELs with or without NPC1 in relation to mitochondria. (J) Quantification shows combined subpopulations of seven cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (K) Acetylated tubulin and mitochondria (TOM20) in HeLa cells imaged using two rabbit primary antibodies pre-labeled with secondary rabbit nanobodies, showing minimal cross-talk between the two targets. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (L) Localizations corresponding to Tetraspeck beads imaged in a multicolor DNA-PAINT acquisition were used for image registration. Localizations are shown before and after post-processing alignment. Scale bar, 100 nm, Source data are available for this figure: SourceData FS5.
Bulk analysis of drug-treated HeLa cells, examining LEL subpopulations with additional cellular context in ARPE-19 cells, and multiplexed imaging controls. (A and B) Representative WBs of (A) NPC1 and (B) LAMTOR4 in untreated, EN6-treated, or BafA1-treated conditions in HeLa cells. EN6 treatment leads to an increase in both NPC1 and LAMTOR4. BafA1 treatment leads to an increase in NPC1 but a decrease in LAMTOR4. (C–G) Cumulative density function plots of LEL distance from the nucleus from dual-color DNA-PAINT imaging experiments in ARPE-19 cells for subpopulations containing LAMP1 or LAMP2 only or LAMP1/LAMP2 and target protein: (C) CD63, (D) Cathepsin D, (E) TMEM192, (F) NPC1, and (G) LAMTOR4. Distance to the nucleus was normalized per cell to the maximum distance from the nucleus of an LEL in that cell, with values closest to zero indicating greatest proximity to the nucleus. Line indicates median with standard deviation between biological replicates. Kolmogorov–Smirnov tests were performed on the mean distributions from three independent biological replicates: for CD63 n = 13 cells, P = 0.193, no significance; for Cathepsin D n = 13 cells, P = 0.794, no significance; for TMEM192 n = 14 cells, P = 0.261, no significance; for NPC1 n = 14 cells, P = 0.140, no significance; for LAMTOR4 n = 14 cells, P = 0.193, no significance. (H–J) Representative three-color DNA-PAINT image of LAMP1, NPC1, and mitochondria (TOM20) in ARPE-19 cells. (H and I) (H) Raw images and (I) post-processed image showing a spatial map of LELs with or without NPC1 in relation to mitochondria. (J) Quantification shows combined subpopulations of seven cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (K) Acetylated tubulin and mitochondria (TOM20) in HeLa cells imaged using two rabbit primary antibodies pre-labeled with secondary rabbit nanobodies, showing minimal cross-talk between the two targets. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (L) Localizations corresponding to Tetraspeck beads imaged in a multicolor DNA-PAINT acquisition were used for image registration. Localizations are shown before and after post-processing alignment. Scale bar, 100 nm, Source data are available for this figure: SourceData FS5.
BafA1 treatment led to a visually noticeable enlargement in LEL size and the appearance of partially damaged LELs (Fig. 5, C and D). Due to the enlarged LELs, a larger minimum size filter was necessary to properly segment intact LELs in BafA1-treated cells (350 nm instead of 250 nm). Consequently, we compared our quantitative metrics after BafA1 treatment with untreated cells, where LELs were segmented using either the original size filter (250 nm) or the larger size filter (350 nm) for consistency. This comparison corroborated the visual impression that the median LEL size was indeed larger in BafA1-treated cells (Fig. 5 G). NPC1 density per LEL did not change after BafA1 treatment, though there was a slight increase in the percentage of NPC1-positive LELs (67.8 ± 11.8%) compared with untreated cells (51.6 ± 14.0%) (Fig. 5, E and F). WB analysis showed an upregulation of NPC1 protein levels after BafA1 treatment (Fig. S5 A), consistent with previous findings (Mauvezin and Neufeld, 2015). Since NPC1 protein density per LEL was unchanged after BafA1 treatment, to reconcile the imaging and WB results, we determined the total area of NPC1-positive and NPC1-negative LELs (Fig. 5, H and I). This analysis revealed an increase in the total area of NPC1-positive LELs after BafA1 treatment (Fig. 5 H). Overall, these data suggest that while the protein density on LELs remains unchanged, the total amount of NPC1 is higher after BafA1 treatment due to an increase in both the area and percentage of NPC1-positive LELs, consistent with the WB analysis. On the other hand, there was a significant downregulation of LAMTOR4, as evidenced by decreased LAMTOR4 density and a reduction in the percentage of LAMTOR4-positive LELs (Fig. 5, E and F), consistent with WB results for this protein (Fig. S5 B).
Overall, these results show that the observed LEL subpopulations are sensitive to various perturbations.
Spatial analysis reveals the relationship of LEL subsets to other organelles
The spatial positioning of lysosomes within a cell is crucial for their function (Pu et al., 2016) and has been shown to be linked to anabolic and catabolic responses and nutrient availability (Jia and Bonifacino, 2019; Korolchuk et al., 2011; Pu et al., 2017; Walton et al., 2018). We thus investigated whether specific LEL subpopulations occupy distinct spatial locations relative to the cell nucleus or other organelles. To explore this, we developed a quantitative method to measure the normalized distance of each LEL from the cell nucleus (Fig. 6, A–E and Fig. S5, C–G), identifiable in our super-resolution images as the empty, dark spaces within the cell (Materials and methods). There were no significant differences in the distance of LEL subpopulations from the nucleus (Fig. S5, E and G), suggesting that, under homeostatic conditions, different subpopulations of LELs are distributed across a wide range of spatial locations.
Spatial analysis reveals the location of LEL subsets with respect to the nucleus, mitochondria, and the TGN . (A–E) Cumulative density function plots of LEL distance from the nucleus calculated from dual-color DNA-PAINT images in HeLa cells for subpopulations containing LAMP1 or LAMP2 only or LAMP1/LAMP2 and target protein: (A) CD63, (B) Cathepsin D, (C) TMEM192, (D) NPC1, and (E) LAMTOR4. Distance to the nucleus was normalized per cell to the maximum distance from the nucleus of an LEL in that cell, with values closest to zero indicating the greatest proximity to the nucleus. The line indicates median with standard deviation between biological replicates. Kolmogorov–Smirnov tests were performed on the mean distributions from three independent biological replicates: for CD63, n = 14 cells, P = 0.261, no significance; for Cathepsin D, n = 15 cells, P = 0.193, no significance; for TMEM192, n = 13 cells, P = 0.794, no significance; for NPC1, n = 16 cells, P = 0.261, no significance; for LAMTOR4, n = 16 cells, P = 0.069, no significance. (F–H) Three-color DNA-PAINT image of LAMP1, NPC1, and mitochondria (TOM20) in HeLa cells. (F and G) (F) Raw images and (G) post-processed image showing a spatial map of LELs with or without NPC1 in relation to mitochondria. (H) Quantification shows combined subpopulations of five cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (I–L) Three-color DNA-PAINT image of LAMP1, NPC1, and TGN (P230) in HeLa cells. (I and J) (I) Raw images and (J) post-processed images showing a spatial map of LELs with or without NPC1 in relation to TGN. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (K and L) Quantification of five cells from three independent biological replicates shows (K) percent of NPC1-positive LELs colocalized with the TGN compared to LELs independent from the TGN and (L) distance of each LEL subtype to the TGN normalized to the maximum distance in each cell. Mann-Whitney U test was used to compare percent colocalization with NPC1 for LELs on TGN with LELs independent from TGN (n = 5 HeLa cells) (P = 0.4206, no significance).
Spatial analysis reveals the location of LEL subsets with respect to the nucleus, mitochondria, and the TGN . (A–E) Cumulative density function plots of LEL distance from the nucleus calculated from dual-color DNA-PAINT images in HeLa cells for subpopulations containing LAMP1 or LAMP2 only or LAMP1/LAMP2 and target protein: (A) CD63, (B) Cathepsin D, (C) TMEM192, (D) NPC1, and (E) LAMTOR4. Distance to the nucleus was normalized per cell to the maximum distance from the nucleus of an LEL in that cell, with values closest to zero indicating the greatest proximity to the nucleus. The line indicates median with standard deviation between biological replicates. Kolmogorov–Smirnov tests were performed on the mean distributions from three independent biological replicates: for CD63, n = 14 cells, P = 0.261, no significance; for Cathepsin D, n = 15 cells, P = 0.193, no significance; for TMEM192, n = 13 cells, P = 0.794, no significance; for NPC1, n = 16 cells, P = 0.261, no significance; for LAMTOR4, n = 16 cells, P = 0.069, no significance. (F–H) Three-color DNA-PAINT image of LAMP1, NPC1, and mitochondria (TOM20) in HeLa cells. (F and G) (F) Raw images and (G) post-processed image showing a spatial map of LELs with or without NPC1 in relation to mitochondria. (H) Quantification shows combined subpopulations of five cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (I–L) Three-color DNA-PAINT image of LAMP1, NPC1, and TGN (P230) in HeLa cells. (I and J) (I) Raw images and (J) post-processed images showing a spatial map of LELs with or without NPC1 in relation to TGN. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (K and L) Quantification of five cells from three independent biological replicates shows (K) percent of NPC1-positive LELs colocalized with the TGN compared to LELs independent from the TGN and (L) distance of each LEL subtype to the TGN normalized to the maximum distance in each cell. Mann-Whitney U test was used to compare percent colocalization with NPC1 for LELs on TGN with LELs independent from TGN (n = 5 HeLa cells) (P = 0.4206, no significance).
Considering NPC1’s crucial role in cholesterol export and the established proximity of LELs to other cellular organelles, we conducted three-color multiplexed DNA-PAINT imaging to investigate the likelihood of NPC1-positive LELs being in close proximity to mitochondria (Fig. 6, F–H and Fig. S5, H–J) or the TGN (Fig. 6, I–L). We then adapted our colocalization analysis to differentiate between LELs with partial mitochondrial or TGN overlap and those showing no mitochondrial or TGN overlap (see Materials and methods and Fig. 6, H and K and Fig. S5 J). As a substantial number of LELs overlapped with mitochondria but only a small fraction overlapped with the TGN, we additionally measured the distance of LEL subpopulations from the TGN to determine if there was a spatial bias in their distance to the TGN (Fig. 6 L). We note that partial overlap in our images does not imply the presence of LEL-mitochondria or LEL-TGN membrane contact, as the spatial resolution of our DNA-PAINT imaging is not sufficient to infer membrane contact sites. We spatially mapped and visualized the subpopulations within each cell, providing a depiction of both their unique protein composition and spatial relationship to other organelles (Fig. 6, G and J and Fig. S5 I). The analysis revealed that in HeLa cells ∼50% of NPC1-positive LELs overlapped with mitochondria, in contrast to only 30% of NPC1-negative LELs (Fig. 6 H). These findings suggest a higher propensity for NPC1-positive LELs to be in close spatial proximity to mitochondria in HeLa cells. Interestingly, this trend was not true in ARPE-19 cells (Fig. S5 J), showing cell-type-specific subcellular positioning of distinct LEL subpopulations with respect to other organelles. Finally, NPC1-positive LELs did not show preferential overlap with TGN (Fig. 6 K), and NPC1-positive as well as NPC1-negative LELs had a similar distance distribution from the TGN (Fig. 6 L).
These results further highlight the power of our quantitative pipeline in uncovering inter-relationships among various molecularly distinct organelle subpopulations.
Higher-order multiplexing reveals molecularly distinct LEL subsets
Given that only specific subsets of LAMP1-positive LELs carry certain LEL proteins, we employed higher-order multiplexing with these markers to further uncover the diversity within LEL subpopulations. However, one major hurdle in multiplexing beyond two or three targets is the limited availability of high-quality antibodies from unique species. To overcome this, we leveraged a newly developed workflow where DNA-PAINT labeled secondary nanobodies, each tagged with unique oligo barcodes, were preincubated with their respective primary antibodies to create a stable antibody–nanobody complex (Sograte-Idrissi et al., 2020). We first validated that this protocol did not lead to target crosstalk by targeting two distinct structures—mitochondria and microtubules (Fig. S5 K).
Another challenge associated with multiplexed imaging is the fact that only two spectrally distinct fluorophores are routinely used for DNA-PAINT, necessitating the removal and exchange of imager oligos between imaging rounds. To maintain precise alignment between sequentially imaged targets, we employed fluorescent beads as fiducial markers and conducted image alignment (Fig. S5 L).
We next applied this approach to examine protein targets in HeLa cells that exhibited heterogeneous colocalization with LAMP1 (i.e., NPC1 and LAMTOR4) (Fig. 7, A–C). A similar approach was applied to ARPE-19 cells with three proteins that showed heterogeneous colocalization with LAMP1 (i.e., CD63, NPC1, and LAMTOR4) (Fig. 7, D–F). Our colocalization analysis again enabled us to spatially map the unique LEL subpopulations within each individual cell (Fig. 7, B and E). In HeLa cells, the predominant LEL subpopulation comprised all three proteins (∼40% contained LAMP1, NPC1, and LAMTOR4) (Fig. 7 C). Given the 93.5 ± 7.8% colocalization of LAMP2 with LAMP1 and 87.0 ± 6.8% colocalization of CD63 with LAMP1 in HeLa cells, this subpopulation most likely also contains LAMP2 and CD63. However, we identified a significant (∼27%) subpopulation of LELs that were solely positive for LAMP1 (and presumably LAMP2/CD63) but lacked both NPC1 and LAMTOR4. These findings imply that LEL proteins identified in proteomic studies are not uniformly present in every LEL subpopulation, revealing significant heterogeneity in the protein composition of canonical LAMP1/2-positive LELs.
Higher-order multiplexing reveals molecularly distinct LEL subsets. (A–C) Three-color DNA-PAINT image of LAMP1, LAMTOR4, and NPC1 in HeLa cells reveals four distinct subpopulations. (A and B) (A) Raw images and (B) post-processed image showing a spatial map of these four subsets. (C) Quantification shows combined subpopulations of four cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (D–F) Four-color DNA-PAINT image of LAMP1, CD63, LAMTOR4, and NPC1 in ARPE-19 cells reveals eight distinct subpopulations. (D and E) (D) Raw images and (E) post-processed images showing a spatial map of these eight subsets. (F) Quantification shows combined subpopulations of four cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
Higher-order multiplexing reveals molecularly distinct LEL subsets. (A–C) Three-color DNA-PAINT image of LAMP1, LAMTOR4, and NPC1 in HeLa cells reveals four distinct subpopulations. (A and B) (A) Raw images and (B) post-processed image showing a spatial map of these four subsets. (C) Quantification shows combined subpopulations of four cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm. (D–F) Four-color DNA-PAINT image of LAMP1, CD63, LAMTOR4, and NPC1 in ARPE-19 cells reveals eight distinct subpopulations. (D and E) (D) Raw images and (E) post-processed images showing a spatial map of these eight subsets. (F) Quantification shows combined subpopulations of four cells from three biological replicates. Cell scale bars, 10 µm. Inset scale bars, 1 µm.
This diversity was similarly observed in ARPE-19 cells, where cells displayed up to eight distinct LEL subpopulations based on their protein makeup (Fig. 7, E and F). The most common subpopulation again included all four proteins (40% of LELs had LAMP1, CD63, LAMTOR, and NPC1) (Fig. 7 F). However, there were also subpopulations missing one to three of the examined proteins. Furthermore, we observed variability among individual cells in terms of these subpopulations, with certain cells devoid of specific minority subpopulations, potentially indicating these subpopulations may lack significant functional importance.
Discussion
Here, we introduce a novel application of multiplexed and quantitative DNA-PAINT imaging as a robust and effective method for determining the heterogeneity of individual organelles under native conditions. While multiplexed DNA-PAINT has very recently been applied to visualize synaptic protein heterogeneity (Unterauer et al., 2024) as well as the distribution of Golgi protein complexes (Schueder et al., 2024), to our knowledge, this is the first application of this approach to visualize the heterogeneity of native organelle subpopulations in the cellular context. Our work provides several key resources including (1) thoroughly validated antibodies that can be used for super-resolution visualization of LEL membrane proteins, (2) a robust, object-based colocalization analysis, and (3) comprehensive datasets that quantitatively profile seven LEL proteins across two different cell types.
Using this approach, we found that abundant and canonical LEL proteins (LAMP1 and LAMP2) exhibit a high degree of colocalization in the same compartments. Thus, these proteins act as general markers of the LEL population. We also identified that LELs segregate into distinct subpopulations based on the presence or absence of other membrane proteins like TMEM192, NPC1, and LAMTOR4 in both HeLa and ARPE-19 cells. Notably, the largest subpopulations were those containing all the visualized proteins (LAMP1/NPC1/LAMTOR4 in HeLa cells and LAMP1/CD63/NPC1/LAMTOR4 in ARPE-19 cells), which may represent a functionally distinct subset of LELs. Heterogeneity in LEL characteristics is indeed increasingly recognized as reviewed recently (Bussi and Gutierrez, 2024). For example, previous studies in neurons showed that not all LAMP1-positive LELs contain degradative enzymes like Cathepsin D (Cheng et al., 2018), although this was not the case in HeLa and ARPE-19 cells examined here. Additionally, recent work uncovered two distinct LEL populations characterized by their distinct subcellular positioning, morphology, lipid composition, and mTORC1 activity (Ebner et al., 2023). These studies support the notion of functionally distinct LEL subsets. Recent metabolomic profiling of single lysosomes revealed five distinct subpopulations of lysosomes based on their metabolomes (Zhu et al., 2021). In the future, it would be exciting to link these subpopulations based on metabolomic characterization to the proteomic-based subpopulations we have identified using DNA-PAINT imaging.
Proteins like NPC1 and LAMTOR4 exhibited distinct nanoscale organization patterns on the LEL membranes. Understanding the mechanisms that govern the organization of these proteins into these specific nanoscale platforms is crucial for determining whether their spatial organization influences their function. For example, NPC1, which plays a key role in cholesterol homeostasis, might segregate into cholesterol-enriched lipid domains (Wang et al., 2020a) on the LEL membrane. This spatial arrangement could be linked to lysosomal contact sites with other organelles including the ER, peroxisomes, Golgi, and mitochondria to facilitate cholesterol delivery to these organelles (Radulovic et al., 2022). Additionally, LAMTOR4 is important for mTORC1 recruitment to the LEL membrane (Sancak et al., 2010) and the LAMTOR4 nanodomains may serve as platforms for efficient mTORC1 recruitment.
Proteins like TMEM192 and NPC1 were present only on a limited subset of LELs, potentially indicating unique properties and functions for these subpopulations. TMEM192 overexpression is used in Lyso-IP studies to biochemically isolate lysosomes for proteomic and metabolomic analysis. We showed that when overexpressed, this protein associates with all LAMP1-positive LELs, as previously reported (Abu-Remaileh et al., 2017). These results highlight a limitation of current Lyso-IP approaches, namely the inability to isolate and analyze distinct LEL subpopulations. Consequently, proteomic and metabolomic analyses fail to differentiate between molecularly diverse LEL subpopulations. In addition, a recent comparative proteomic analysis showed that TMEM192 overexpression has pronounced effects on the expression of lysosomal membrane proteins (Bonini and Winter, 2024). To address these limitations, new strategies are required, such as using CRISPR tagging (Chen et al., 2018) to label and isolate specific LEL subpopulations.
Our results also revealed that overexpression of LEL proteins as well as perturbation with drugs that impact LEL acidity and function can significantly alter various characteristics of LELs, changes that might be challenging to detect with conventional, diffraction-limited microscopy. Overexpressing LAMP1 resulted in a small but significant enlargement of LELs, reducing the surface density of LAMP2 and NPC1 proteins on their membranes and influencing NPC1 nanodomains. These observations suggest the need for caution in interpreting data from overexpression experiments, as they can subtly but significantly affect LEL biology. Treatment with EN6, which increases lysosomal acidity via increased vATPase activity, led to an increase in LAMTOR4-positive LEL subpopulations and an increase in LAMTOR4 density on LEL membranes. The regulation of mTORC1 on LEL membranes is complex and the mechanisms coupling vATPase to mTORC1 recruitment are not fully elucidated. We speculate that the increase in LAMTOR4 may be a compensatory upregulation of the components needed to dock mTORC1 to the lysosomal membrane. Contrary to EN6 treatment, BafA1 treatment, which reduces vATPase activity and prevents autophagosome–lysosome fusion, decreased the percentage of LAMTOR4-positive LELs and LAMTOR4 protein density on LEL membranes. We hypothesize that lysosomal damage induced by BafA1 treatment (Mauvezin and Neufeld, 2015) may result in a disruption of LAMTOR4 since it is not an integral transmembrane protein embedded within the lysosomal membrane. Both treatments led to an increase in the amount of NPC1 and the percentage of NPC1-positive LELs. The mechanisms and the biological outcomes of this increase are unclear and should be further elucidated in future studies.
One limitation of our approach is that it was performed in 2D rather than 3D. Given the 3D nature of LELs, it is possible that LELs overlapping in 3D may be mis-segmented or misclassified. In addition, for LELs that are not fully captured within the 2D plane, protein density may be under or overestimated. While we tried to avoid such issues by focusing on LELs within an appropriate size range (>250 nm) that are fully imaged within the focal plane, in the future extending this approach to 3D imaging (Bond et al., 2022; Hugelier et al., 2024) will further improve the precision of the analysis.
Here, we profiled seven LEL proteins, though proteomic studies have identified hundreds more (Bagshaw et al., 2005; Chapel et al., 2013; Lübke et al., 2009; Schröder et al., 2010). The main limitation in expanding our multiplexing approach to a larger set of proteins is the scarcity of high-quality antibodies for endogenous-level labeling. We evaluated a wide range of commercially available antibodies against numerous LEL proteins, many of which did not meet our stringent validation criteria due to lack of specificity. To broaden our investigation to include more targets, there is a critical need for the development of new, high-quality labeling reagents suitable for DNA-PAINT. Emerging advancements in the creation of synthetic nanobodies (or sybodies) (Misson Mindrebo et al., 2023; Zimmermann et al., 2020) are particularly promising in this context. With the availability of such advanced reagents, our pipeline will be broadly applicable to profiling organelle heterogeneity at an unprecedented level of detail in the future.
Materials and methods
Cell culture
Wildtype HeLa (ATCC CCL-2, RRID:CVCL_0030) and ARPE-19 (ATCC CRL-2302, RRID:CVCL_0145) cell lines were obtained from the American Type Culture Collection (ATCC). The HeLa NPC1-null cell line was a kind gift from Prof. Neale Ridgway, PhD (Dalhousie University, Halifax, Canada) (Zhao and Ridgway, 2017). NPC1-null cells were verified using WB analysis. Other cell lines were not further authenticated. HeLa cells were derived from a female and ARPE-19 cells from a male. Sex as a biological variable was not considered in the manuscript. HeLa cells were propagated in DMEM, and ARPE-19 cells in DMEM:F12 media. All culture media (GIBCO Laboratories) were supplemented with 10% (vol/vol) fetal bovine serum and antibiotics. Cells were maintained at 37°C in the presence of 5% CO2. To overexpress or knockdown a protein of interest, cells were transiently transfected at 50–60% confluency with plasmid-expressing protein of interest (Table 1) and/or target-specific siRNA using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s protocol. Cells were subjected to experimental treatments 24 h after transfection.
Plasmids and siRNAs
| Plasmid name . | Identifier . | Insert . | Species . |
|---|---|---|---|
| LAMP1-mGFP | RRID:Addgene_34831 | LAMP1 | Homo sapiens (human) |
| pRK5-LAMP1-FLAG | RRID:Addgene_71868 | LAMP1 | H. sapiens (human) |
| LAMP2-BFP | Gift from Holzbaur lab (University of Pennsylvania, Philadelphia, PA, USA) (Gallagher and Holzbaur, 2023) | LAMP2 | H. sapiens (human) |
| pMEH371 CD63 HA | RRID:Addgene_168220 | CD63 | H. sapiens (human) |
| pLJC5-Tmem192-3xHA | RRID:Addgene_102930 | TMEM192 | H. sapiens (human) |
| pcDNA 3.1(+) – hNPC1(WT)-FLAG | Custom generated by Genscript, re-cloned from RRID:Addgene_164972 | NPC1 | H. sapiens (human) |
| pRK5-FLAG-C7orf59 | RRID:Addgene_42332 | LAMTOR4 | H. sapiens (human) |
| TMEM192 siRNA | Santa Cruz Biotech #sc-89327 | siRNA to TMEM192 | H. sapiens (human) |
| Cathepsin D siRNA | Santa Cruz Biotech #sc-29239 | siRNA to Cathepsin D | H. sapiens (human) |
| Control siRNA | Santa Cruz Biotech #sc-37007 | Control siRNA | H. sapiens (human) |
| Plasmid name . | Identifier . | Insert . | Species . |
|---|---|---|---|
| LAMP1-mGFP | RRID:Addgene_34831 | LAMP1 | Homo sapiens (human) |
| pRK5-LAMP1-FLAG | RRID:Addgene_71868 | LAMP1 | H. sapiens (human) |
| LAMP2-BFP | Gift from Holzbaur lab (University of Pennsylvania, Philadelphia, PA, USA) (Gallagher and Holzbaur, 2023) | LAMP2 | H. sapiens (human) |
| pMEH371 CD63 HA | RRID:Addgene_168220 | CD63 | H. sapiens (human) |
| pLJC5-Tmem192-3xHA | RRID:Addgene_102930 | TMEM192 | H. sapiens (human) |
| pcDNA 3.1(+) – hNPC1(WT)-FLAG | Custom generated by Genscript, re-cloned from RRID:Addgene_164972 | NPC1 | H. sapiens (human) |
| pRK5-FLAG-C7orf59 | RRID:Addgene_42332 | LAMTOR4 | H. sapiens (human) |
| TMEM192 siRNA | Santa Cruz Biotech #sc-89327 | siRNA to TMEM192 | H. sapiens (human) |
| Cathepsin D siRNA | Santa Cruz Biotech #sc-29239 | siRNA to Cathepsin D | H. sapiens (human) |
| Control siRNA | Santa Cruz Biotech #sc-37007 | Control siRNA | H. sapiens (human) |
Molecular cloning
A flag-tagged human NPC1 construct, pcDNA 3.1 (+) – hNPC1(WT)-FLAG, was custom-ordered from Genscript. The NPC1-insert sequence was recloned from the pLVX-NPC1(WT)-FLAG construct (#164972; Addgene) into the EcoRI and NotI restriction site of the pcDNA 3.1 (+) mammalian expression vector (Genscript). Kozak sequence (GCCACC) was included before the start codon. Correct insertion was verified by whole plasmid sequencing (Eurofins Genomics).
WB analysis
WB analysis was performed using the two-color Odyssey LI-COR technique according to the manufacturer’s protocol. All antibodies and corresponding dilutions used can be found in Table 2.
Antibodies, DNA imager oligomers, and drug treatments
| Antibody/oligo . | Identifier . | Experiments used . | Dilutions . |
|---|---|---|---|
| Sheep Anti-LAMP1 | RRID:AB_1026176 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Mouse Anti-LAMP2 | RRID:AB_626858 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Mouse Anti-CD63 | RRID:AB_2884028 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Goat anti-Cathepsin D | RRID:AB_2087218 | DNA-PAINT; WB | 1:100; 1:1,000 |
| Rabbit Anti-TMEM192 | RRID:AB_3095683 | DNA-PAINT; widefield imaging; WB | 1:25; 1:500; 1:1,000 |
| Rabbit Anti-NPC1 | RRID:AB_10001101 | DNA-PAINT | 1:100 |
| Rabbit Anti-LAMTOR4 | RRID:AB_2798129 | DNA-PAINT; widefield imaging; WB | 1:100; 1:500 |
| Mouse Anti-TOM20 | RRID:AB_628381 | DNA-PAINT | 1:100 |
| Mouse Anti-P230 (TGN) | RRID:AB_398808 | DNA-PAINT | 1:25 |
| Mouse Anti-flag | RRID:AB_262044 | Widefield imaging | 1:500 |
| Rabbit Anti-HA | RRID:AB_307019 | Widefield imaging | 1:500 |
| Rabbit Anti-EEA1 | RRID:AB_2096811 | DNA-PAINT | 1:100 |
| Rabbit Anti-TMEM192 (alternative antibody) | RRID:AB_2902909 | DNA-PAINT | 1:100 |
| Rabbit Anti-NPC1 | RRID:AB_2734695 | WB | 1:1,000 |
| Mouse Anti-GAPDH | RRID:AB_2107436 | WB | 1:1,000 |
| Rabbit Anti-TOM20 | RRID:AB_2207530 | DNA-PAINT | 1:100 |
| Rabbit anti-tubulin, acetylated | RRID:AB_477585 | DNA-PAINT | 1:100 |
| Anti-Mouse IgG+ Docking site 1 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 1:100 |
| Anti-Rabbit IgG+ Docking site 2 | Massive Photonics Massive-AB 2-Plex | DNA-PAINT | 1:100 |
| Donkey anti-sheep IgG+ Docking site E2 | RRID:AB_2340704 | DNA-PAINT | 1:100 |
| Donkey anti-Goat IgG+ Docking site E2 | RRID:AB_2340384 | DNA-PAINT | 1:100 |
| Tag-Q Anti-GFP single-Domain Antibody+ Docking site 3 | Massive Photonics, Massive-TagQ Anti-GFP | DNA-PAINT | 1:100 |
| Anti-Rabbit single-Domain Antibody+ Docking site F1 | Massive Photonics, custom order | DNA-PAINT | 1:25 |
| Anti-Rabbit single-Domain Antibody+ Docking site F2 | Massive Photonics, Massive SDAB fast one-Plex | DNA-PAINT | 1:25 |
| Donkey anti-Mouse IgG+ Alexa fluor 647 | RRID:AB_162542 | Widefield imaging | 1:500 |
| Goat anti-Rabbit IgG+ Cy3 | RRID:AB_10563288 | Widefield imaging | 1:500 |
| IRDye800CW Donkey anti-Rabbit | RRID:AB_621848 | WB | 1:10,000 |
| IRDye680RD Donkey anti-Mouse | RRID:AB_10953628 | WB | 1:10,000 |
| IRDye800CW Donkey anti-Goat | RRID:AB_2687553 | WB | 1:10,000 |
| Imager oligo for Docking site 1+ Cy3B or ATTO655 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 500 pM |
| Imager oligo for Docking site 2+ Cy3B or ATTO655 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 500 pM |
| Imager oligo for Docking site 3+ Cy3B or ATTO655 | Massive Photonics, Massive-TagQ Anti-GFP | DNA-PAINT | 100–300 pM |
| Imager oligo for Docking site E2+ Cy3 | IDT, custom order | DNA-PAINT | 500 pM |
| Imager oligo for Docking site F1+ Cy3B or ATTO655 | Massive Photonics, custom order | DNA-PAINT | 1.5 nM |
| Imager oligo for Docking site F2+ Cy3B or ATTO655 | Massive Photonics, Massive SDAB fast one-Plex | DNA-PAINT | 1.5 nM |
| EN6 | Tocris Biosciences #7474 | DNA-PAINT; WB | 15 µM, 16 h |
| BafA1 | Sigma-Aldrich #SML1661 | DNA-PAINT; WB | 100 nM, 16 h |
| Antibody/oligo . | Identifier . | Experiments used . | Dilutions . |
|---|---|---|---|
| Sheep Anti-LAMP1 | RRID:AB_1026176 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Mouse Anti-LAMP2 | RRID:AB_626858 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Mouse Anti-CD63 | RRID:AB_2884028 | DNA-PAINT; widefield imaging | 1:100; 1:500 |
| Goat anti-Cathepsin D | RRID:AB_2087218 | DNA-PAINT; WB | 1:100; 1:1,000 |
| Rabbit Anti-TMEM192 | RRID:AB_3095683 | DNA-PAINT; widefield imaging; WB | 1:25; 1:500; 1:1,000 |
| Rabbit Anti-NPC1 | RRID:AB_10001101 | DNA-PAINT | 1:100 |
| Rabbit Anti-LAMTOR4 | RRID:AB_2798129 | DNA-PAINT; widefield imaging; WB | 1:100; 1:500 |
| Mouse Anti-TOM20 | RRID:AB_628381 | DNA-PAINT | 1:100 |
| Mouse Anti-P230 (TGN) | RRID:AB_398808 | DNA-PAINT | 1:25 |
| Mouse Anti-flag | RRID:AB_262044 | Widefield imaging | 1:500 |
| Rabbit Anti-HA | RRID:AB_307019 | Widefield imaging | 1:500 |
| Rabbit Anti-EEA1 | RRID:AB_2096811 | DNA-PAINT | 1:100 |
| Rabbit Anti-TMEM192 (alternative antibody) | RRID:AB_2902909 | DNA-PAINT | 1:100 |
| Rabbit Anti-NPC1 | RRID:AB_2734695 | WB | 1:1,000 |
| Mouse Anti-GAPDH | RRID:AB_2107436 | WB | 1:1,000 |
| Rabbit Anti-TOM20 | RRID:AB_2207530 | DNA-PAINT | 1:100 |
| Rabbit anti-tubulin, acetylated | RRID:AB_477585 | DNA-PAINT | 1:100 |
| Anti-Mouse IgG+ Docking site 1 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 1:100 |
| Anti-Rabbit IgG+ Docking site 2 | Massive Photonics Massive-AB 2-Plex | DNA-PAINT | 1:100 |
| Donkey anti-sheep IgG+ Docking site E2 | RRID:AB_2340704 | DNA-PAINT | 1:100 |
| Donkey anti-Goat IgG+ Docking site E2 | RRID:AB_2340384 | DNA-PAINT | 1:100 |
| Tag-Q Anti-GFP single-Domain Antibody+ Docking site 3 | Massive Photonics, Massive-TagQ Anti-GFP | DNA-PAINT | 1:100 |
| Anti-Rabbit single-Domain Antibody+ Docking site F1 | Massive Photonics, custom order | DNA-PAINT | 1:25 |
| Anti-Rabbit single-Domain Antibody+ Docking site F2 | Massive Photonics, Massive SDAB fast one-Plex | DNA-PAINT | 1:25 |
| Donkey anti-Mouse IgG+ Alexa fluor 647 | RRID:AB_162542 | Widefield imaging | 1:500 |
| Goat anti-Rabbit IgG+ Cy3 | RRID:AB_10563288 | Widefield imaging | 1:500 |
| IRDye800CW Donkey anti-Rabbit | RRID:AB_621848 | WB | 1:10,000 |
| IRDye680RD Donkey anti-Mouse | RRID:AB_10953628 | WB | 1:10,000 |
| IRDye800CW Donkey anti-Goat | RRID:AB_2687553 | WB | 1:10,000 |
| Imager oligo for Docking site 1+ Cy3B or ATTO655 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 500 pM |
| Imager oligo for Docking site 2+ Cy3B or ATTO655 | Massive Photonics, Massive-AB 2-Plex | DNA-PAINT | 500 pM |
| Imager oligo for Docking site 3+ Cy3B or ATTO655 | Massive Photonics, Massive-TagQ Anti-GFP | DNA-PAINT | 100–300 pM |
| Imager oligo for Docking site E2+ Cy3 | IDT, custom order | DNA-PAINT | 500 pM |
| Imager oligo for Docking site F1+ Cy3B or ATTO655 | Massive Photonics, custom order | DNA-PAINT | 1.5 nM |
| Imager oligo for Docking site F2+ Cy3B or ATTO655 | Massive Photonics, Massive SDAB fast one-Plex | DNA-PAINT | 1.5 nM |
| EN6 | Tocris Biosciences #7474 | DNA-PAINT; WB | 15 µM, 16 h |
| BafA1 | Sigma-Aldrich #SML1661 | DNA-PAINT; WB | 100 nM, 16 h |
Sample preparation
Generation of anti-sheep and anti-goat DNA-PAINT secondary antibodies
Affinipure donkey anti-sheep secondary antibody or rabbit anti-goat secondary antibody (Jackson Immunoresearch Labs) was conjugated to 5′-TTATCTACATA-3′ for DNA-PAINT. This docking site is referred to as docking site E2 and imaged with corresponding imager strand E2 (custom ordered from IDT). DNA was conjugated to the antibody via DBCO-sulfo-NHS ester chemistry according to the protocol described previously (Schnitzbauer et al., 2017). Briefly, the antibody was incubated with a 10-fold excess of bifunctional DBCO-sulfo-NHS ester (Cat.# CLK-A124-10; Jena Bioscience) for 2 h at +40°C. The unbound linker was removed using Zeba Spin Desalting columns (0.5 ml, 7K molecular weight cutoff; 89882; Thermo Fisher Scientific). Azide-modified DNA was added to the DBCO-antibody in a 15 M excess and incubated for 1 h at room temperature, protected from light. At the end of incubation, the buffer was exchanged for PBS using Amicon centrifugal filters (100,000 molecular weight cutoff). Antibody labeling was confirmed using the NanoDrop spectrophotometer by the shift of the peak signal from 280 nm toward 260 nm.
Standard DNA-PAINT immunostaining
Cells were seeded onto LabTek-II imaging chambers (Nalge Nunc; Thermo Fisher Scientific) before fixation. Glyoxal was tested as a fixative for 10 min at room temperature (Richter et al., 2018) but reduced staining quality when compared with 4% PFA in PBS prewarmed to 37°C for 20 min at room temperature, thus PFA was used for all experiments. Cells were then washed 3× with PBS and permeabilized for 10 min in 0.1% saponin in PBS. 0.2% Triton X-100 was tested as an alternative for permeabilization but reduced retention of target proteins on LEL membranes; thus, saponin was used for all experiments. Blocking was done in a primary blocking buffer (10% donkey serum, 0.1% saponin, 0.05 mg/ml sonicated salmon sperm DNA [Stratagene]) in PBS for 1 h at room temperature. Following blocking, cells were incubated for 1 h at room temperature in primary antibodies described in Table 2 diluted in primary blocking buffer. After primary antibody staining, cells were washed briefly 1× with PBS and 3× for 5 min with 1× wash buffer (Massive Photonics). Secondary antibodies were diluted in antibody incubation buffer (Massive Photonics) as described in Table 2 and incubated for 1 h at room temperature. Subsequently, cells were washed briefly 1× with PBS, 3× for 7 min with 1× wash buffer (Massive Photonics), and 1× for 5 min with PBS and stored at 4° in fresh PBS until imaging. For LAMP1 and LAMTOR4, staining quality was improved by using this protocol sequentially with a 15-min post-fixation using 4% PFA between staining for each target. For LAMP1-mGFP overexpression samples, anti-GFP single-domain antibody was used to detect this population and was added during the secondary antibody incubation.
EN6 and BafA1 treatments
For experiments using drug-based perturbations, HeLa cells were treated with either 15 μM EN6 or 100 nm BafA1 for 16 h before fixation and staining.
Immunostaining for widefield imaging
Immunostaining for widefield imaging was done as above but with lower antibody concentrations (see Table 2) and slightly altered blocking buffers. It was not necessary to add sonicated salmon sperm DNA to the blocking buffer to block off-target DNA-PAINT imager oligo binding in the nucleus, and thus primary and secondary antibody incubation was performed with antibodies diluted in Widefield Imaging Buffer (10% donkey serum, 0.1% saponin in PBS).
DNA-PAINT immunostaining with nanobody premixing
Multiplexed imaging of LAMTOR4 and NPC1 required an alternative staining protocol as the antibodies to both these targets were raised in rabbits. We utilized a previously described protocol (Sograte-Idrissi et al., 2020) for premixing primary antibodies with secondary nanobodies to circumvent this issue. Excess DNA conjugated nanobodies at a fourfold higher concentration than saturating levels of secondary antibody were premixed with each primary antibody for 30 min at room temperature in PBS before they were subsequently mixed and incubated with the cells during the secondary antibody step. To minimize the possibility of crosstalk as much as possible, these samples were imaged immediately following staining completion. As an additional validation, this protocol was tested on acetylated tubulin and TOM20 using rabbit primary antibodies for each (Table 2).
Widefield imaging
Widefield images were acquired using a Nanoimager (ONI) equipped with 405-, 488-, 561- and 640-nm lasers, 498–551- and 576–620-nm band-pass filters in channel 1, 666–705-nm band-pass filters in channel 2, a 100× 1.45 NA oil immersion objective (Olympus), and a Hamamatsu Flash 4 V3 sCMOS camera. Widefield images were captured at 100–200-ms exposure using epifluorescence illumination and exported from the NimOS software (ONI) for visualization in FIJI (Schindelin et al., 2012). For visualization purposes only, a rolling ball background subtraction with a radius of 5–10 pixels was performed on these images.
DNA-PAINT imaging
DNA-PAINT images were acquired on the same Nanoimager system as was used for widefield imaging, and all experiments were conducted at 30°C using HiLo illumination. The laser power used for DNA-PAINT imaging was 10 mW for the 647-nm laser and 2.5 mW for the 560-nm laser. In DNA-PAINT, the final image could only be rendered upon the completion of data acquisition and no information on the final image is known a priori. Thus, images were acquired randomly.
Dual-color DNA-PAINT of LEL targets
Before imaging, appropriate imager oligo strands (see Table 2) for both labeled targets were diluted to a final concentration of 500 pM in an imaging buffer (Massive Photonics) and added to imaging chambers. For imaging of LAMP1-GFP with GFP nanobody, 100–300-pM imager was used, as 500 pM yielded overlapping fluorescent signal and was deemed too high. For dual-color experiments, one imager with conjugated Cy3 or Cy3B and one imager with conjugated ATTO655 were used. The sample was imaged at 100-ms exposure using a laser program alternating between 561-nm excitation and 640-nm excitation every 100 frames for a total of 50,000 frames and 25,000 frames for each target.
Three-color DNA-PAINT of LAMP1, NPC1, and TOM20
Before imaging, samples were incubated with 0.1 µm Tetraspeck microspheres (Invitrogen; Thermo Fisher Scientific) in PBS to use as a fiducial marker. Appropriate imager oligo strands for LAMP1 and NPC1 were diluted to 500 pM in an imaging buffer and added to imaging chambers. The sample was imaged at 100 ms exposure using a laser program designed for sequential imaging of targets, beginning with 25,000 frames with 561-nm excitation and followed by 25,000 frames with 640-nm excitation. The third step in the laser program was 2,500–5,000 frames with no laser excitation to allow for imager oligo exchange for imaging of TOM20. The imager oligo solution on the sample was removed and the sample was washed 3× with PBS and replaced with 500 pM of the appropriate imager oligo strand for TOM20 conjugated to either Cy3B or ATTO655. The fourth step in the laser program turned on the appropriate laser corresponding to the fluorophore used (561 nm for Cy3B or 640 nm for ATTO655) for 10,000 frames.
Three-color DNA-PAINT of LAMP1, NPC1, and TGN
Three-color imaging with the TGN was done as described with TOM20 with minor modifications. For these experiments, the TGN (P230) was imaged first for 25,000 frames with 500-pM imager oligo, followed by LAMP1 for 25,000 frames with 500-pM imager oligo. Next, 3× PBS washes were completed and 500 pM of imager oligo for NPC1 was introduced, and NPC1 was imaged for 25,000 frames.
Higher-order multiplexed DNA-PAINT using secondary nanobodies
Before imaging, samples were incubated with 0.1 µm Tetraspeck microspheres in PBS to be used as a fiducial marker. For both three-color imaging of LAMP1, NPC1, and LAMTOR4 in HeLa cells and four-color imaging of LAMP1, CD63, NPC1, and LAMTOR4 in ARPE-19 cells, appropriate imager oligos for LAMTOR4 and NPC1 were first diluted to 500 pM in Imaging Buffer and added to the imaging chambers. The sample was imaged at 50-ms exposure, per manufacturer recommendation for the nanobodies, with a laser program designed for sequential imaging of targets. First, LAMTOR4 was imaged for 25,000 frames. Next, NPC1 was imaged for 50,000 frames. The third step in the laser program was 5,000 frames with no lasers enabled to allow for imager oligo exchange to image remaining targets. The imager oligo sample on the stage was removed and the sample was washed 3× with PBS and replaced with 500 pM of appropriate imager oligo for LAMP1 in HeLa cells, or LAMP1 and CD63 in ARPE-19 cells. The fourth step in the laser program imaged LAMP1 for 25,000 frames, after which three-color imaging in HeLa cells was completed. For four-color imaging in ARPE-19 cells, a fifth step imaged CD63 for 25,000 frames.
DNA-PAINT image analysis
For all experiments, data were exported from NimOS software (ONI) with the following initial filtering parameters applied: photon counts above 300, an X/Y localization precision of 30 nm, and a Sigma X/Y between 0 and 250 nm. These parameters were identical for all targets in all experiments. All following analyses were performed in MATLAB R2022b (The MathWorks) using custom-made code (Github: https://github.com/LakGroup/Data_Analysis_Browser_Bond_etal).
Segmentation and clustering
Individual cells were cropped and reference channel localizations were Voronoi-segmented and clustered with a minimum of 25 localizations and a maximum Voronoi area threshold, based on which target protein was used as the reference channel and visual inspection of the segmentation quality. For LAMP1, thresholds ranged from 68 to 548 nm2. For LAMP2, thresholds ranged from 178 to 342 nm2. For LAMP1-GFP imaged with GFP-nanobody, thresholds ranged from 178 to 1,369 nm2. To remove the background antibody signal, clusters were then filtered above a minimum clustered area of 49,912 nm2, which corresponds to a circle with a diameter of 250 nm, as previous super-resolution imaging showed lysosome size ranges between 200 and 800 nm (Verdeny-Vilanova et al., 2017). For experiments using BafA1 treatment, LEL size was increased and segmentation was more accurate using a larger area filter of 96,211 nm2, corresponding to a circle with a radius of 175 nm. For dual-color experiments with wildtype HeLa and ARPE-19 cells using all standard imaging parameters (100-ms exposure, 25,000 frames, and 500-pM imager oligo) and LAMP1 as the reference, clusters were additionally filtered for a minimum of 1,500 localizations.
For experiments examining colocalization of LELs with mitochondria, TOM20 was used as a mitochondrial reference channel and Voronoi-segmented and clustered with a threshold of 3,422 nm2 and post-processed for a minimum of 500 localizations and minimum clustered area of 49,912 nm2. For experiments examining colocalization of LELs with the TGN, P230 was used as a TGN reference channel and Voronoi-segmented and clustered with a threshold of 1,232–2,053 nm2 and post-processed for a minimum of 500–1,000 localizations and minimum clustered area of 8,775–40,950 nm2. The higher Voronoi thresholds for mitochondria and the TGN compared with LAMP1/2 ensured that these organelles were clustered as the large networks they represent in cells rather than separate segments.
Estimating imaging completeness and resolution
The imaging of an organelle is considered complete if the area of the localizations that describe the organelle no longer changes with an increasing number of frames. This area evolution is considered in a pair-wise way, for each cluster of the reference channel (ChREF) and the corresponding target channel (ChTARGET) cluster.
In practice, the localizations in ChREF (LAMP2 for Cathepsin D experiments, LAMP1 for all others) were Voronoi-segmented and clustered using the most broadly applicable parameters used in the colocalization analysis (LAMP1 or LAMP2 clustering threshold of 178 nm2). The localizations in ChTARGET were not Voronoi-segmented and clustered, as segmentation of protein targets that show low density and sparse localization is challenging. Instead, the localizations in this channel that were spatially located within a cluster of ChREF were considered as a single “cluster.” The evolution of the area of each individual cluster in ChREF (and its associated cluster in ChTARGET) was followed with respect to the number of frames (by considering the area of their alphashape object), beginning at the frame that had the first detected localization. Each curve was normalized to its final area and visualized in percentage to remove size dependencies. For the localizations of the ChTARGET channel, the area evolution was visualized in terms of the final area of the associated organelle of ChREF.
Image resolution was calculated based on FRC analysis using the FRCRes Plugin in ImageJ (Nieuwenhuizen et al., 2013).
Colocalization of LEL targets
To perform the colocalization analysis, the data in the reference channel (LAMP1 unless otherwise indicated; ChREF) were considered Voronoi-segmented and clustered data, whereas the data from the target channel (ChTARGET) were considered as localizations, for the same reasons as described above.
First, the localizations in ChTARGET that are spatially located within the boundaries of ChREF clusters were grouped together and considered as a cluster. Then, the local background density distribution was calculated by considering the area that was not directly adjacent to the ChREF clusters, but proximal to them (i.e., between 234 and 585 nm around the border). The area adjacent to ChREF clusters (i.e., between 0 and 234 nm around the border) was not considered as some targets have a locally increased density in this region (i.e., LAMTOR4, discussed below). ChTARGET clusters were then considered to be statistically significant clusters and colocalized if they had a density that was >3 standard deviations higher than the mean of the local background distribution.
Additional output of this analysis is given as properties of the clusters. For the individual clusters in ChREF, the area, the number of localizations, its density, and its distance to the nucleus (see next section) were calculated. For the colocalized clusters of ChTARGET, the number of localizations and their densities were calculated.
For all experiments using LAMTOR4 as ChTARGET, a small alteration was implemented, as LAMTOR4 is part of a protein complex that sits on the lysosomal membrane rather than an integral membrane protein, and thus the localization pattern from this target showed a clear bias to extend beyond the edges of the LAMP1 ChREF cluster. To accommodate for this, the clusters of ChREF were slightly expanded (with 20 nm), but the rest of the analysis described above remained the same.
To showcase the usefulness of our colocalization analysis code, we compared with a CBC analysis method (Malkusch et al., 2012), which is based on calculating the Spearman correlation between the local densities of the two channels. We refer to the reference for more information.
Distance from nucleus analysis
The location of the nuclei could easily be discerned visually from the DNA-PAINT images and therefore manually annotated. Their coordinates were then extracted and used to determine the distance of the clusters in ChREF to this nucleus (nearest border-to-border distance). In rare occurrences where an LEL was inside the nucleus area, its distance was considered as 0.
To accommodate for different cell sizes, the distances of the individual lysosomes to the nucleus were normalized between 0 and 1 by dividing by the maximum distance in each cell. This was appropriate as all cells showed a distribution of LEL spread throughout the entire cell spanning both the perinuclear and peripheral regions. Then, the data were split up into two colocalized categories (see previous section; colocalized versus non-colocalized clusters), and a histogram of distances for each category was constructed using 10% bins by pooling data from all cells.
Colocalization of LELs with mitochondria/TGN
For analysis of LEL overlap with mitochondria, it is possible to segment and cluster the data from both second organelle (TOM20 for mitochondria, P230 for TGN) and LEL reference (LAMP1) channels, and a different strategy than previously described can be used for the colocalization analysis. Here, all channels have clearly clustered localizations, and thus a polyshape/alphashape can be constructed for the clusters in both the reference channel (TOM20 or P230, ChREF) and the target channel (LAMP1, ChTARGET). These objects can then be compared to each other to determine the degree of overlap between them and are considered as colocalized when there is at least 10% overlap between them. The percentage of overlap is defined as the percentage of localizations of the ChTARGET cluster that is spatially located within a cluster of the ChREF channel with respect to its total number of localizations. In the case that a ChTARGET cluster overlaps with multiple ChREF clusters, it is associated to the ChREF cluster that it overlaps the most with. For colocalization with the TGN, we also calculated the distance of each ChTARGET LEL cluster to ChREF TGN. This distance was normalized to the maximum distance of an LEL from the TGN per cell for consistent comparison.
Drift correction for higher-order multiplexing
For three- and four-color DNA-PAINT acquisitions, drift correction was performed after export from the NimOS software to ensure each channel was drift-corrected separately and any changes in localization not due to drift (i.e., small changes in position during manual imager swaps) were not improperly corrected (see below section on alignment). To correct for drift throughout the measurement, the Drift at Minimum Entropy method was used (Cnossen et al., 2021). In brief, drift will increase the reconstruction uncertainty in localization microscopy, and therefore the entropy will increase as well. This property is associated with the fact that the localization microscopy reconstruction is an approximation of the true distribution of the molecules (according to a probability distribution). Thus, to remove the drift from the localized molecules, the entropy of this reconstruction can be minimized, which can be estimated by minimizing an upper bound of the statistical entropy of the probability distribution of all the localizations. For more details about practical implementations, we referred to the original work (Cnossen et al., 2021). One small change was made to the original algorithm which was to calculate the drift with respect to the first frame for each channel rather than with respect to the mean drift.
Channel alignment for higher-order multiplexing
For three- and four-color measurements, a change of imager had to be made in the middle of the measurement. Due to the manual intervention, a slight misalignment of the sample (before versus after) can occur that could not be corrected using the drift correction method described above as the change was abrupt and not gradual. To correct for this, Tetraspeck microspheres were included during sample preparation that emit fluorescent signals in each channel that was measured for the entire acquisition duration. The localizations of this signal could then be used to align the channels correctly and remove the disruption introduced by the manual intervention. To align the channels, beads were manually selected and the distance between the centroids of the beads (in a pairwise way) was then calculated. These distances were then averaged over the selected beads and the channels were corrected accordingly.
Pattern analysis of NPC1 and LAMTOR4 on LEL membranes
To analyze the clustering of proteins on LEL membranes, we used DBSCAN clustering on the colocalized localizations of ChTARGET (Ester et al., 1996), given a search radius and a minimum number of points. The appropriate search radius was determined for each target using visual inspection as follows: NPC1 in wildtype HeLa cells 17.6 nm, NPC1 in LAMP1 overexpressing HeLa cells 23.4 nm, and LAMTOR4 in wildtype HeLa cells 23.4 nm, and a minimum number of points of 5. Clusters were post-filtered for a minimum of 10 localizations per cluster for all targets.
Once the localizations were clustered, we additionally determined the number of localizations per cluster, cluster area, cluster density, and cluster diameter.
Statistical analysis
Statistical analysis was performed in Prism 10. Mann–Whitney U tests were performed to compare cell medians for colocalization and protein-density measurements. Kruskal–Wallis test was used for multiple comparisons of LAMP1 density across different experiments. Kolmogorov–Smirnov tests were used to compare cumulative density functions of LEL subset distance from the nucleus. The precise number of biological and technical replicates, as well as test performed and P-value obtained are found in figure legends for each experiment.
Online supplemental material
Fig. S1 shows overexpression validation of target antibodies. Fig. S2 shows permeabilization and fixation controls, imaging completeness analysis, minimal colocalization of LAMP1 with EEA1, low correlation between imaging targets, and consistency of LAMP1 density measurements across experiments. Fig. S3 shows knockdown, overexpression, and alternative antibody validations for Cathepsin D, TMEM192, and NPC1. Fig. S4 shows analyses validating the object-based colocalization analysis and LAMP1-GFP overexpression imaging and quantification. Fig. S5 shows WB analysis of drug treatments, distance from nucleus analysis in ARPE-19 cells, imaging and analysis of LEL subsets with respect to mitochondria in ARPE-19 cells, and multiplexed imaging controls. All custom analysis codes written in Matlab are available online on Github (https://github.com/LakGroup/Data_Analysis_Browser_Bond_etal).
Data availability
The raw imaging data is available upon request as it is too large to deposit in a data repository. All other data used in making the plots in the main and supplementary figures have been deposited to Figshare (https://doi.org/10.6084/m9.figshare.25429189.v1).
Acknowledgments
We would like to thank Qing Tang, Patricia Colosi, Elizabeth Gallagher, Vera Moiseenkova-Bell, and Claire H. Mitchell at the University of Pennsylvania for critical reading and detailed comments on the manuscript.
This work is funded by National Institutes of Health grants R01 GM133842 (to M. Lakadamyali), RM1 GM136511 and R35 GM152111 (to M. Lakadamyali), and National Science Foundation grant CMMI-1548571 (to M. Lakadamyali).
Author contributions: C. Bond: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing - original draft, Writing - review & editing, S. Hugelier: Data curation, Formal analysis, Methodology, Software, Writing - review & editing, J. Xing: Data curation, Formal analysis, Investigation, Validation, E.M. Sorokina: Investigation, Resources, Validation, Visualization, Writing - review & editing, M. Lakadamyali: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing - original draft, Writing - review & editing.
References
Author notes
Disclosures: The authors declare no competing interests exist.