Paneth cells secrete antimicrobial peptides (AMPs) to modulate composition of gut microbiota and host defense. AMPs are typically packaged into dense core vesicles (DCVs) and secreted into the intestinal lumen. However, the mechanisms underlying DCV biogenesis and secretion are still elusive. Here we identified that ERAdP was highly expressed in Paneth cells that acted as a sensor for a bacterial second messenger c-di-AMP. ERAdP deficiency caused impaired DCV biogenesis and dysfunction of Paneth cells. Mechanistically, by sensing c-di-AMP, ERAdP interacted with NLRP6 and further recruited ANXA2 onto the DCV membrane in Paneth cells. The ERAdP–NLRP6–ANXA2 complex facilitated DCV biogenesis, which enhanced antibacterial ability of intestines. Disruption of ERAdP–NLRP6–ANXA2 axis led to loss of DCVs in Paneth cells and increased susceptibility to bacterial infection. Of note, ERAdP–NLRP6–ANXA2 proteins were lowly expressed in IBD patients, and c-di-AMP treatment enhanced antibacterial capacity in antibiotic-treated mice. Our findings reveal that c-di-AMP stimulation might provide a potential therapeutic strategy for infectious disease and gut inflammation.
Introduction
Intestinal epithelium, as the first line of defense, separates host from external environment and regulates mucosal immunity by recognizing and responding to signals from the microbiota. Intestinal homeostasis maintenance requires coordinated interactions among mucosal epithelial cells, immune cells, and gut microbiota (Lockhart et al., 2024; Caballero-Flores et al., 2023). Disruption of this balance contributes to the onset and progression of various gastrointestinal disorders, including gastrointestinal infections and inflammatory bowel disease (IBD) (Liu et al., 2025; Su et al., 2025; Kou et al., 2025; Best et al., 2025). Enteric pathogens frequently correlate with diarrheal symptoms, and acute diarrheal diseases represent a huge global health burden. Among the various cases, Salmonella infections emerge as the primary driver for hospital admissions and mortality (Cherrak et al., 2025; Yoo et al., 2024; Sun et al., 2024b; Santus et al., 2022). Intestinal epithelial cells include absorptive cells (e.g., enterocytes) and secretory cells (e.g., goblet, tuft, Paneth, and enteroendocrine cells), which are derived from intestinal stem cells (ISCs) (Beumer and Clevers, 2021). Goblet cells secrete mucus that forms a physical barrier to protect the intestinal epithelium from direct contact with microorganisms, thereby maintaining epithelial integrity (Lin et al., 2025). Tuft cells recognize succinate produced by parasitic worms to exert anti-parasitic effects (Schneider et al., 2018), while we defined that Tuft-2 cells detect bacterial metabolite N-undecanoylglycine to defend against bacterial infections (Xiong et al., 2022). Endocrine cells secrete a variety of gut hormones that regulate physiological processes, such as digestion, metabolism, and appetite, playing a critical role in maintaining metabolic balance and the stability of the body’s internal environment (Beumer et al., 2024). Paneth cells, located at the base of the crypts and interspersed with ISCs, are essential for maintaining stem cell function by providing Wnt signaling for ISC self-renewal (Sato et al., 2011). Paneth cells also secrete antimicrobial peptides (AMPs), such as lysozyme and defensins, which regulate the composition of the intestinal microbiota (Salzman et al., 2010; Yu et al., 2020) and protect against pathogens like Salmonella (Salzman et al., 2003; Vaishnava et al., 2008).
Paneth cells are distinguished by their well-developed ER and Golgi apparatus, which enable sustained high-level secretion of AMPs. Given their vital role in shaping the gut microbiota and preventing dysbiosis, precise regulation of AMP secretion is essential to the maintenance of both intestinal health and microbial balance (Pierre et al., 2023; Hu et al., 2019; Liang et al., 2019; Ehmann et al., 2019). Typically, AMPs are packaged within dense core vesicles (DCVs) and then released into the intestinal lumen. Understanding the regulatory mechanisms behind the biogenesis and secretion of DCVs in Paneth cells is critical for elucidating their roles in intestinal immunity. Dysfunction of AMP secretion in Paneth cells is closely associated with intestinal inflammation and diseases. For instance, Crohn’s disease (CD) risk allele, ATG16L1, has been shown to affect the granule exocytosis in Paneth cells (Matsuzawa-Ishimoto et al., 2022). Additionally, lack of vitamin D receptor in Paneth cells reduces lysozyme secretion and impairs the inhibition of pathogenic bacterial growth (Lu et al., 2021). The X-linked inhibitor of apoptosis protein also contributes to reduced AMP secretion and alters the gut microbiota composition (Strigli et al., 2021). NOD2 is essential for intestinal antibacterial defense by regulation of cryptdins through recognizing bacterial muramyl dipeptide in mice, whose deficiency increases susceptibility to oral infections (Kobayashi et al., 2005). In CD patients, NOD2 mutations impair Paneth cell production of α-defensins (HD-5/HD-6), weakening mucosal immunity and promoting bacterial-driven inflammation, particularly in the ileum (Wehkamp et al., 2004).
Pattern recognition receptors (PRRs), such as TLRs, nucleotide oligomerization domain (NOD)-like receptors (NLRs), and retinoic acid–inducible gene-I (RIG-I)-like receptors, play an essential role in sensing microbial components and initiating a range of immune responses. In Paneth cells, enteric bacteria directly activate TLR-MyD88 signaling, thereby initiating a multifaceted antimicrobial response (Vaishnava et al., 2008). Additionally, a report showed that TLR5 is expressed specifically in Paneth cells and its activation induces expression of various host defense genes (Price et al., 2018). However, it is still elusive how Paneth cells recognize and respond to microbial stimuli in the intestinal lumen to facilitate the generation of DCVs for the sequestration of bacteria outside the epithelial layer. NLRP6 is a kind of NLRs, which is known for its role in inflammasome activation (Shen et al., 2019; Barnett et al., 2023). In intestines, NLRP6 inflammasome mediates goblet cell mucin release to combat Salmonella Typhimurium through IL-18/IL-22 signaling (Han et al., 2024) and protects against intestinal protists by sensing bacterial sphingolipids (Winsor et al., 2025). In our previous studies, we identified that the ER membrane adaptor protein (ERAdP) acts as a direct sensor for the bacterial second messenger c-di-AMP and then initiates innate immune response against bacterial infection (Xia et al., 2018; Chen et al., 2015). Here we showed that ERAdP was highly expressed in Paneth cells of small intestines (SIs). ERAdP deficiency decreased numbers of DCVs in Paneth cells. ERAdP recruited NLRP6 and ANXA2 onto the DCV membrane to participate in the package and biogenesis of DCVs in Paneth cells. Disruption of ERAdP–NLRP6–ANXA2 axis in mice dramatically abolished antibacterial ability.
Results
c-di-AMP as an ERAdP ligand facilitates DCV generation in Paneth cells
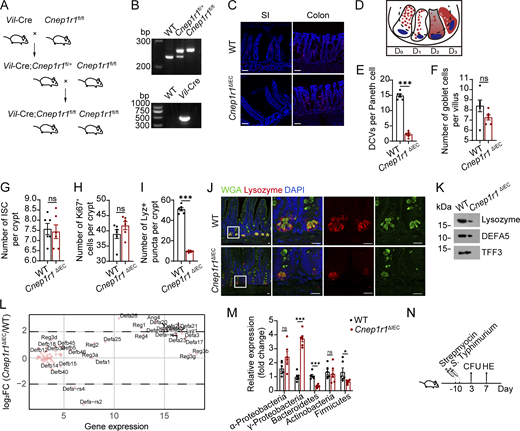
Paneth cells secrete AMPs into the intestinal lumen through DCVs to protect against intestinal microbiota. To determine whether the microbiota regulated DCV biogenesis in host Paneth cells, we compared the DCVs in Paneth cells in specific pathogen–free (SPF) and germ-free (GF) mice. We found that the numbers of DCVs in Paneth cells in GF mice were dramatically decreased (Fig. 1, A and B; and Fig. S1 A). In addition, we established an antibiotic-treated (ABX) model and assessed its efficiency in clearing commensal bacteria from intestinal contents (Fig. S1 B). Similar to GF mice, the ABX model also showed significantly reduced DCV numbers in Paneth cells (Fig. 1, C and D; and Fig. S1 C). Transmission electron microscopy revealed ultrastructural changes in Paneth cells, including reduced DCVs in both ABX and GF mice (Fig. 1 E). This observation was confirmed by 3D staining, which demonstrated decreased total DCV volume in these models (Fig. 1 F).
ERAdP promotes DCV generation in Paneth cells via c-di-AMP stimulation. (A) HE staining of intestines from SPF and GF mice. Scale bar: 20 μm. (B) AB-PAS staining of intestines from SPF and GF mice. Scale bar: 20 μm. (C) HE staining of intestines from control and ABX mice. Scale bar: 20 μm. ABX mice were treated with 2-wk antibiotic, and control mice were treated with water. (D) AB-PAS staining of intestines from control and ABX mice. Scale bar: 20 μm. ABX mice and control mice were treated as in C. (E) Left: Electron micrograph of SPF, ABX, and GF mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Numbers of DCVs per Paneth cells in SPF, ABX, and GF mice were calculated (n = 10 mice for each group). (F) Left: 3D immunofluorescence staining of ileum crypts from littermate SPF, ABX, and GF mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (G) Left: Immunofluorescence staining of intestinal organoids treated with mock, or different stimulations of PRRs: 10 μΜ c-di-AMP disodium, 10 μΜ c-di-GMP disodium, 10 μΜ 3′3′-cGAMP disodium, 10 μΜ 2′3′-cGAMP disodium, 10 μg/ml LPS, 10 μg/ml poly (I:C) sodium, and 2 μg/ml MDP for 16 h. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (H) Left: Immunofluorescence staining of ileums from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP (25 mg/kg for mouse) twice, with treatments 12 h apart. Green: UEA-1; blue: DAPI, red: EpCAM. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (I)Cnep1r1 FISH of 10 different tissues of WT mice. Red: Cnep1r1 probes; blue: DAPI. Scale bar: 35 μm. (J) ERAdP expression levels of indicated tissues from ERAdP-HA tag mice were detected by western blot using anti-HA antibody. β-Actin was used as a loading control. (K)Cnep1r1 FISH of intestine of Lgr5-EGFP mice. Red: Cnep1r1 probes, green: Lgr5EGFP, blue: DAPI, and gray: EpCAM. Scale bar: 35 μm. (L) Immunofluorescence staining showing ERAdP subcellular localization in intestines from ERAdP-HA or WT mice. Red: HA, green: WGA, blue: DAPI, and pink: lysozyme. Scale bar: 15 μm. Data in E–H are shown as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments. Source data are available for this figure: SourceData F1.
ERAdP promotes DCV generation in Paneth cells via c-di-AMP stimulation. (A) HE staining of intestines from SPF and GF mice. Scale bar: 20 μm. (B) AB-PAS staining of intestines from SPF and GF mice. Scale bar: 20 μm. (C) HE staining of intestines from control and ABX mice. Scale bar: 20 μm. ABX mice were treated with 2-wk antibiotic, and control mice were treated with water. (D) AB-PAS staining of intestines from control and ABX mice. Scale bar: 20 μm. ABX mice and control mice were treated as in C. (E) Left: Electron micrograph of SPF, ABX, and GF mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Numbers of DCVs per Paneth cells in SPF, ABX, and GF mice were calculated (n = 10 mice for each group). (F) Left: 3D immunofluorescence staining of ileum crypts from littermate SPF, ABX, and GF mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (G) Left: Immunofluorescence staining of intestinal organoids treated with mock, or different stimulations of PRRs: 10 μΜ c-di-AMP disodium, 10 μΜ c-di-GMP disodium, 10 μΜ 3′3′-cGAMP disodium, 10 μΜ 2′3′-cGAMP disodium, 10 μg/ml LPS, 10 μg/ml poly (I:C) sodium, and 2 μg/ml MDP for 16 h. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (H) Left: Immunofluorescence staining of ileums from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP (25 mg/kg for mouse) twice, with treatments 12 h apart. Green: UEA-1; blue: DAPI, red: EpCAM. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (I)Cnep1r1 FISH of 10 different tissues of WT mice. Red: Cnep1r1 probes; blue: DAPI. Scale bar: 35 μm. (J) ERAdP expression levels of indicated tissues from ERAdP-HA tag mice were detected by western blot using anti-HA antibody. β-Actin was used as a loading control. (K)Cnep1r1 FISH of intestine of Lgr5-EGFP mice. Red: Cnep1r1 probes, green: Lgr5EGFP, blue: DAPI, and gray: EpCAM. Scale bar: 35 μm. (L) Immunofluorescence staining showing ERAdP subcellular localization in intestines from ERAdP-HA or WT mice. Red: HA, green: WGA, blue: DAPI, and pink: lysozyme. Scale bar: 15 μm. Data in E–H are shown as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments. Source data are available for this figure: SourceData F1.
c-di-AMP stimulation invivo and expression of Cnep1r1. (A) Quantification of DCV numbers per Paneth cell of SPF and GF mice (n = 6 mice for each group). (B) Intestinal lumen bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums of control (Ctrl) and ABX mice for assessment of commensal bacteria clearance (n = 6 mice for each group). (C) Quantification of DCV numbers per Paneth cell of Ctrl and ABX mice (n = 6 mice for each group). (D) Small intestinal concentration of c-di-AMP, c-di-GMP, and cGAMP from Ctrl, ABX mock, and ABX with these ligands treatment mice was measured by ELISA (n = 6 mice for each group). (E) Immunofluorescence staining of ileums from ABX mice treated through intragastric gavage with c-di-AMP disodium, c-di-GMP disodium, 3′3′-cGAMP disodium, 2′3′-cGAMP disodium, LPS, poly (I:C) sodium, and MDP (25 mg/kg for mouse mouse) twice, with treatments 12 h apart. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. (F) Conservation analysis of ERAdP in indicated vertebrate animals. (G) Relative Cnep1r1 expression levels of indicated tissues were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 3 mice for each group). (H) Construction diagram of ERAdP-HA tag mouse. (I) DNA electrophoresis for ERAdP-HA tag knock-in validation. (J) DNA sequencing for ERAdP-HA tag knock-in validation. (K) Western blot for ERAdP-HA tag knock-in validation. β-Actin was used as a loading control. (L) Immunofluorescence staining of ileal crypts from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP (25 mg/kg for mouse mouse) twice, with treatments 12 h apart. Red: ERAdP-HA, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (M) Immunofluorescence staining of ileal crypts from WT mice. Green: ERAdP-HA, blue: WGA, and red: PDI. Scale bar: 5 μm. Data in A–D are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in A–E, G, and I–M are representative of at least three independent experiments. Source data are available for this figure: SourceData FS1.
c-di-AMP stimulation invivo and expression of Cnep1r1. (A) Quantification of DCV numbers per Paneth cell of SPF and GF mice (n = 6 mice for each group). (B) Intestinal lumen bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums of control (Ctrl) and ABX mice for assessment of commensal bacteria clearance (n = 6 mice for each group). (C) Quantification of DCV numbers per Paneth cell of Ctrl and ABX mice (n = 6 mice for each group). (D) Small intestinal concentration of c-di-AMP, c-di-GMP, and cGAMP from Ctrl, ABX mock, and ABX with these ligands treatment mice was measured by ELISA (n = 6 mice for each group). (E) Immunofluorescence staining of ileums from ABX mice treated through intragastric gavage with c-di-AMP disodium, c-di-GMP disodium, 3′3′-cGAMP disodium, 2′3′-cGAMP disodium, LPS, poly (I:C) sodium, and MDP (25 mg/kg for mouse mouse) twice, with treatments 12 h apart. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. (F) Conservation analysis of ERAdP in indicated vertebrate animals. (G) Relative Cnep1r1 expression levels of indicated tissues were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 3 mice for each group). (H) Construction diagram of ERAdP-HA tag mouse. (I) DNA electrophoresis for ERAdP-HA tag knock-in validation. (J) DNA sequencing for ERAdP-HA tag knock-in validation. (K) Western blot for ERAdP-HA tag knock-in validation. β-Actin was used as a loading control. (L) Immunofluorescence staining of ileal crypts from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP (25 mg/kg for mouse mouse) twice, with treatments 12 h apart. Red: ERAdP-HA, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (M) Immunofluorescence staining of ileal crypts from WT mice. Green: ERAdP-HA, blue: WGA, and red: PDI. Scale bar: 5 μm. Data in A–D are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in A–E, G, and I–M are representative of at least three independent experiments. Source data are available for this figure: SourceData FS1.
To identify candidate substrates that regulated this process, we stimulated small intestinal organoids with the ligands of common PRRs. Of note, ERAdP ligand c-di-AMP dramatically increased the number of DCVs in Paneth cells of organoids. Other molecules, such as c-di-GMP, 3′3′-cGAMP and 2′3-cGAMP (ligands of STING), LPS (ligand of TLR4), poly (I:C) (ligand of TLR3 and RIG-I), and MDP (ligand of NOD2), failed to increase the DCVs (Fig. 1 G). ABX in vivo treatment caused over 50% reduction of ligands in small intestinal crypt cells. Oral c-di-AMP supplementation could restore its concentration (Fig. S1 D), leading to recovery of DCVs in Paneth cells (Fig. 1 H). However, other ligands had no such restorative effect (Fig. S1 E).
ERAdP was highly conserved across mouse, human, and other various species (Fig. S1 F). To further investigate the function of ERAdP in Paneth cells, we assessed the expression of Cnep1r1 (gene name of ERAdP) in various tissues of mice using RNA fluorescence in situ hybridization (FISH) assays. We found that Cnep1r1 was highly expressed in SI, colon, and bone marrow (Fig. 1 I and Fig. S1 G). To test the subcellular localization of ERAdP, we generated ERAdP-HA tag mice (Fig. S1 H) and validated them (Fig. S1, I–K). In agreement with the RNA FISH results, we found ERAdP-HA was highly expressed in the SI (Fig. 1 J). We found that Cnep1r1 was highly expressed in Paneth cells, interspersed between Lgr5+ ISCs in small intestines (Fig. 1 K). Of note, ERAdP specifically localized on the DCVs of lysozyme+ Paneth cells, as evidenced by merging with wheat germ agglutinin (WGA)-positive vesicles (Fig. 1 L). Using ERAdP-HA mice, we found that ERAdP primarily localized to the DCVs rather than the ER in Paneth cells (Fig. S1, L and M). This DCV localization was reduced with ABX treatment but was restored upon oral c-di-AMP administration (Fig. S1 L). Collectively, these findings suggest that ERAdP localizes on the DCVs of Paneth cells and c-di-AMP facilitates the formation of Paneth DCVs.
ERAdP deficiency abrogates formation of DCVs in Paneth cells
To investigate the function of ERAdP in intestine, we crossed Cnep1r1fl/fl mice with Vil-Cre mice to conditionally knock-out (KO) ERAdP in epithelial cells (Cnep1r1ΔIEC) (Fig. S2, A and B). Using this mouse model, Cnep1r1 in small intestinal Paneth cells was deleted, while the signal of Cnep1r1 remained unchanged in the lamina propria, probably in macrophages (Xia et al., 2018) (Fig. S2 C). We found that proportions of normal Paneth cells in duodenum, jejunum, and ileum were all decreased in Cnep1r1ΔIEC mice compared with those of littermate WT mice, and abnormal Paneth cell ratios were increased (Fig. 2 A and Fig. S2 D). Moreover, DCVs became scarce in ERAdP-deficient Paneth cells (Fig. 2 A). However, Paneth cell numbers were not changed (Fig. 2 A). We further confirmed loss of DCVs in Paneth cells in Cnep1r1ΔIEC mice through Alcian Blue-Periodic Acid Schiff (AB-PAS) staining (Fig. 2 B and Fig. S2 E). In addition, Paneth cells DCVs were assessed through 3D imaging for visualization and quantitative analysis. Our results revealed a significant reduction of DCV volume in Cnep1r1ΔIEC mice compared with WT controls (Fig. 2 C). Furthermore, electron microscopy revealed that decreased numbers of DCVs, impaired apical polarization of DCVs, irregular DCV morphology, and weaker electron density in Cnep1r1ΔIEC mice (Fig. 2 D). In contrast, goblet cells, another secretory cell type, as well as ISCs were normal in Cnep1r1ΔIEC mice (Fig. 2, E and F; and Fig. S2, F and G). We also found that proliferation (Fig. 2 G and Fig. S2 H) and apoptosis (Fig. 2 H) of Paneth cells were not changed in Cnep1r1ΔIEC mice. Then we employed differential interference contrast (DIC) imaging (Yokoi et al., 2019) to monitor DCV generation in organoids derived from Cnep1r1ΔIEC mice and their littermate controls. At the beginning, areas of DCVs in ERAdP-deficient Paneth cells were markedly less than those of WT controls. With the stimulation of cholinergic agonist carbamylcholine (CCh), DCVs were rapidly secreted into the lumen, resulting in a reduction of DCVs in Paneth cells (Fig. 2 I). At 6 h, WT Paneth cells restored DCV area to half of unstimulated level and mostly recovered at 10 h. In contrast, ERAdP-deficient Paneth cells showed less recovery during this period (Fig. 2 I). In addition, with gene ontology (GO) analysis of RNA sequencing (RNAseq), we found that membrane-associated pathways were remarkably downregulated in Cnep1r1ΔIEC mice (Fig. 2 J). Taken together, these results reveal that ERAdP deletion abrogates the formation of DCVs in Paneth cells.
ERAdP conditional KO mice exhibits loss of DCVs and impaired antibacterial ability. (A) Cross strategy of Vil-Cre and Cnep1r1fl/fl mice. (B) DNA electrophoresis for loxP and Vil-Cre knock-in validation. (C)Cnep1r1 FISH of SI and colon of WT and Cnep1r1ΔIEC mice. Red: Cnep1r1 probes; blue: DAPI. Scale bar: 70 μm. (D) Diagram of four patterns of Paneth cells normal (D0), disordered (D1), depleted (D2), and diffuse (D3). (E–I) Quantification of DCV numbers per Paneth cell (E), goblet cells per villus (F), ISC per crypt (G), Ki67+ cells per crypt (H), and Lyz+ puncta per crypt (I) of WT and Cnep1r1ΔIEC mice (n = 6 mice for each group). (J) Immunofluorescence staining of ileums from littermate WT and Cnep1r1ΔIEC mice. Red: lysozyme, green: WGA, and blue: DAPI. Scale bar: 20 μm. (K) Western blot for lysozyme and DEFA5 of ileal intestinal mucus layer. The intestinal contents from a 5-cm ileal segment were flushed with GuHCl and scraped using glass slides, followed by supernatant collection for western blot analysis. TFF3 secreted by goblet cells was used as a loading control. (L) AMP expression analysis with RNAseq of Cnep1r1ΔIEC versus WT mice. (M) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). (N) Schematic diagram of S. Typhimurium stimulation and detection. Data in E–I and M are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in B, C, and E–M are representative of at least three independent experiments. Source data are available for this figure: SourceData FS2.
ERAdP conditional KO mice exhibits loss of DCVs and impaired antibacterial ability. (A) Cross strategy of Vil-Cre and Cnep1r1fl/fl mice. (B) DNA electrophoresis for loxP and Vil-Cre knock-in validation. (C)Cnep1r1 FISH of SI and colon of WT and Cnep1r1ΔIEC mice. Red: Cnep1r1 probes; blue: DAPI. Scale bar: 70 μm. (D) Diagram of four patterns of Paneth cells normal (D0), disordered (D1), depleted (D2), and diffuse (D3). (E–I) Quantification of DCV numbers per Paneth cell (E), goblet cells per villus (F), ISC per crypt (G), Ki67+ cells per crypt (H), and Lyz+ puncta per crypt (I) of WT and Cnep1r1ΔIEC mice (n = 6 mice for each group). (J) Immunofluorescence staining of ileums from littermate WT and Cnep1r1ΔIEC mice. Red: lysozyme, green: WGA, and blue: DAPI. Scale bar: 20 μm. (K) Western blot for lysozyme and DEFA5 of ileal intestinal mucus layer. The intestinal contents from a 5-cm ileal segment were flushed with GuHCl and scraped using glass slides, followed by supernatant collection for western blot analysis. TFF3 secreted by goblet cells was used as a loading control. (L) AMP expression analysis with RNAseq of Cnep1r1ΔIEC versus WT mice. (M) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). (N) Schematic diagram of S. Typhimurium stimulation and detection. Data in E–I and M are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in B, C, and E–M are representative of at least three independent experiments. Source data are available for this figure: SourceData FS2.
ERAdP deficiency causes loss of DCVs in Paneth cells. (A) Upper: HE staining of duodenum, jejunum, and ileum from littermate Cnep1r1fl/fl (WT) and Vil-Cre;Cnep1r1fl/fl (Cnep1r1ΔIEC) mice. Scale bar: 20 μm. Lower: Proportion of normal and abnormal Paneth cells were quantified on the basis of whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal), and numbers of Paneth cells were quantified. n = 6 mice for each group, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (B) AB-PAS staining of intestines from WT and Cnep1r1ΔIEC mice. Scale bar: 25 μm. (C) Left: 3D immunofluorescence staining of ileum crypts from littermate WT and Cnep1r1ΔIEC mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (D) Left: Electron micrograph of WT and Cnep1r1ΔIEC mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Numbers of DCVs of WT and Cnep1r1ΔIEC mice were calculated (n = 10 mice for each group). (E–G) Immunofluorescence staining of SIs from littermate WT and Cnep1r1ΔIEC mice. Green: MUC2 (E), OLFM4 (F), and Ki67 (G); red: EpCAM; blue: DAPI. Scale bar: 20 μm. (H) Cleaved caspase-3 immunohistochemistry of intestines from WT and Cnep1r1ΔIEC mice. Apoptotic epithelial cells were indicated by black asterisk. Scale bar: 50 μm. (I) Upper left: Schematic diagram of stimulation of CCh to organoids and DIC images of organoids from littermate WT and Cnep1r1ΔIEC mice. Upper right: Area of DCVs were quantified (n = 7 mice for each group). Organoids were stimulated by CCh for 10 min and then washed. Images were captured at 0, 2, 6, 10, and 24 h. Areas of DCVs were drawn with dashed red lines, and the corresponding Paneth cells were drawn with dashed black lines. Scale bar: 20 μm. (J) GO pathways enrichment analysis of downregulated genes in Cnep1r1ΔIEC mice versus littermate WT mice. Gene ratio (%) indicates proportion of genes annotated to each GO term among the total set of significantly downregulated genes in Cnep1r1ΔIEC versus WT mouse intestinal crypts. Data in A, C, D, and I are shown as means ± SEM, and significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
ERAdP deficiency causes loss of DCVs in Paneth cells. (A) Upper: HE staining of duodenum, jejunum, and ileum from littermate Cnep1r1fl/fl (WT) and Vil-Cre;Cnep1r1fl/fl (Cnep1r1ΔIEC) mice. Scale bar: 20 μm. Lower: Proportion of normal and abnormal Paneth cells were quantified on the basis of whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal), and numbers of Paneth cells were quantified. n = 6 mice for each group, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (B) AB-PAS staining of intestines from WT and Cnep1r1ΔIEC mice. Scale bar: 25 μm. (C) Left: 3D immunofluorescence staining of ileum crypts from littermate WT and Cnep1r1ΔIEC mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (D) Left: Electron micrograph of WT and Cnep1r1ΔIEC mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Numbers of DCVs of WT and Cnep1r1ΔIEC mice were calculated (n = 10 mice for each group). (E–G) Immunofluorescence staining of SIs from littermate WT and Cnep1r1ΔIEC mice. Green: MUC2 (E), OLFM4 (F), and Ki67 (G); red: EpCAM; blue: DAPI. Scale bar: 20 μm. (H) Cleaved caspase-3 immunohistochemistry of intestines from WT and Cnep1r1ΔIEC mice. Apoptotic epithelial cells were indicated by black asterisk. Scale bar: 50 μm. (I) Upper left: Schematic diagram of stimulation of CCh to organoids and DIC images of organoids from littermate WT and Cnep1r1ΔIEC mice. Upper right: Area of DCVs were quantified (n = 7 mice for each group). Organoids were stimulated by CCh for 10 min and then washed. Images were captured at 0, 2, 6, 10, and 24 h. Areas of DCVs were drawn with dashed red lines, and the corresponding Paneth cells were drawn with dashed black lines. Scale bar: 20 μm. (J) GO pathways enrichment analysis of downregulated genes in Cnep1r1ΔIEC mice versus littermate WT mice. Gene ratio (%) indicates proportion of genes annotated to each GO term among the total set of significantly downregulated genes in Cnep1r1ΔIEC versus WT mouse intestinal crypts. Data in A, C, D, and I are shown as means ± SEM, and significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
ERAdP deficiency impairs the antibacterial ability of mice
Given that AMPs were packaged into DCVs and then secreted out from Paneth cells, we examined the package and secretion of AMPs under ERAdP deficiency. We found that lysozyme could not be packaged into DCVs in Cnep1r1ΔIEC mice (Fig. 3 A and Fig. S2, I and J). Consistently, the amount of lysozyme secreted into the intestinal lumen decreased significantly (Fig. 3 B and Fig. S2 K). We analyzed other AMPs, such as defensin DEFA5, and found that the amount of secreted DEFA5 into the intestinal lumen was also declined (Fig. 3 C and Fig. S2 K), suggesting dysfunction of Paneth cells in AMP secretion. In addition, to directly measure lysozyme secretion ability of Paneth cells, we isolated intestinal crypts and stimulated Paneth cells with CCh, followed by detection of lysozyme levels and bacteriostatic effect of the secreted supernatants (Fig. 3 D). We found that a large amount of lysozyme was secreted from WT crypts, and incubation of S. Typhimurium with their secreted supernatants showed better bacteriostatic effect (Fig. 3, E and F). On the contrary, secreted lysozyme from ERAdP deficient crypts was much lower and could not inhibit S. Typhimurium expansion (Fig. 3, E and F). mRNA expression of AMPs was not reduced but even compensatively upregulated (Fig. S2 L).
ERAdP deficiency leads to abnormal lysozyme secretion. (A) Lysozyme immunohistochemistry of intestines from WT and Cnep1r1ΔIEC mice. Scale bar: 70 μm. (B and C) Whole-mount images of tissues from littermate WT and Cnep1r1ΔIEC mice taken from immediately above the ileal mucosal surface and stained. Red: lysozyme (B) and DEFA5 (C); green: WGA. Scale bar: 100 μm. (D) Schematic diagram of stimulation of small intestinal crypts by CCh and co-incubation of supernatant with S. Typhimurium. (E) Lysozyme in supernatant of stimulated crypts from littermate WT and Cnep1r1ΔIEC mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (F) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and Cnep1r1ΔIEC mice were isolated and stimulated by DMSO or CCh. The crypt-secreted supernatants were incubated with S. Typhimurium for 30 min, and CFUs were measured (n = 6 mice for each group). (G and H) Intestinal lumen (G) and tissue-associated (H) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy numbers in distal ileums (n = 6 mice for each group). (I–L) Bacterial load analysis of livers (I), spleens (J), ileal contents (K) and PPs (L) of WT and Cnep1r1ΔIEC mice. 3 days after S. Typhimurium infection, livers, spleens, ileal contents, and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (M) Body weight changes of littermate WT and Cnep1r1ΔIEC mice after infection with S. Typhimurium (n = 6 mice for each group). (N) Survival rates of littermate WT and Cnep1r1ΔIEC mice after infection with S. Typhimurium (n = 12 mice for each group). (O) Representative pathological manifestation of intestines from WT and Cnep1r1ΔIEC mice after infected by S. Typhimurium for 7 days. Scale bar: 80 μm. Data are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (E–M) and Mantel–Cox test (N) (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data in A–C and E–O are representative of at least three independent experiments.
ERAdP deficiency leads to abnormal lysozyme secretion. (A) Lysozyme immunohistochemistry of intestines from WT and Cnep1r1ΔIEC mice. Scale bar: 70 μm. (B and C) Whole-mount images of tissues from littermate WT and Cnep1r1ΔIEC mice taken from immediately above the ileal mucosal surface and stained. Red: lysozyme (B) and DEFA5 (C); green: WGA. Scale bar: 100 μm. (D) Schematic diagram of stimulation of small intestinal crypts by CCh and co-incubation of supernatant with S. Typhimurium. (E) Lysozyme in supernatant of stimulated crypts from littermate WT and Cnep1r1ΔIEC mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (F) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and Cnep1r1ΔIEC mice were isolated and stimulated by DMSO or CCh. The crypt-secreted supernatants were incubated with S. Typhimurium for 30 min, and CFUs were measured (n = 6 mice for each group). (G and H) Intestinal lumen (G) and tissue-associated (H) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy numbers in distal ileums (n = 6 mice for each group). (I–L) Bacterial load analysis of livers (I), spleens (J), ileal contents (K) and PPs (L) of WT and Cnep1r1ΔIEC mice. 3 days after S. Typhimurium infection, livers, spleens, ileal contents, and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (M) Body weight changes of littermate WT and Cnep1r1ΔIEC mice after infection with S. Typhimurium (n = 6 mice for each group). (N) Survival rates of littermate WT and Cnep1r1ΔIEC mice after infection with S. Typhimurium (n = 12 mice for each group). (O) Representative pathological manifestation of intestines from WT and Cnep1r1ΔIEC mice after infected by S. Typhimurium for 7 days. Scale bar: 80 μm. Data are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (E–M) and Mantel–Cox test (N) (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data in A–C and E–O are representative of at least three independent experiments.
In line with the reduced AMPs in the lumen, we found increased bacterial abundance in intestinal lumen and intestinal tissues in Cnep1r1ΔIEC mice through analysis of 16S copies (Fig. 3, G and H). At the phylum level, we observed significant increase in Proteobacteria of Cnep1r1ΔIEC mice, which was increased in IBD patients, while Bacteroidetes and Firmicutes were decreased in Cnep1r1ΔIEC mice, which were more abundant in healthy people (Fig. S2 M). Apart from disturbed commensal microbiota, Cnep1r1ΔIEC mice exhibited damaged anti-pathogen infection ability. After S. Typhimurium infection, much more bacterial loads were detected in livers, spleens, ileal contents, and Peyer’s patches (PPs) in Cnep1r1ΔIEC mice (Fig. 3, I–L; and Fig. S2 N). Moreover, body weight of Cnep1r1ΔIEC mice were dramatically declined, and more than half of Cnep1r1ΔIEC mice died within 10 days after infection (Fig. 3, M and N). Intestine tissues in Cnep1r1ΔIEC mice were severely damaged with S. Typhimurium infection (Fig. 3 O). These data indicate that deficiency of ERAdP impairs antibacterial ability.
ERAdP interacts with NLRP6 in DCVs
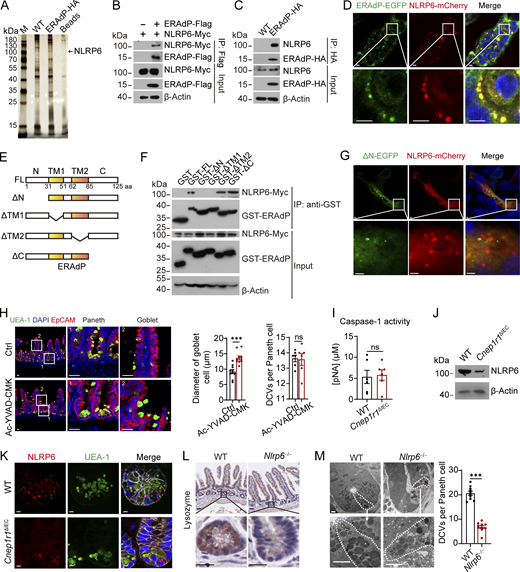
To determine how ERAdP functions in the DCVs, we identified candidate proteins interacting with ERAdP. Intestinal crypts were isolated from ERAdP-HA tag mice and WT mice, followed with cell lysis and HA-pulldown assay. We identified NLRP6 as a candidate interacting protein of ERAdP by silver staining and mass spectrometry (Fig. 4 A and Fig. S3, A and B). We next verified the interaction between ERAdP and NLRP6 by overexpressing these two proteins in HEK293T cells followed by co-immunoprecipitation (co-IP) (Fig. 4 B) and also validated their interaction in ERAdP-HA mice (Fig. 4 C). Then we overexpressed ERAdP-EGFP and NLRP6-mCherry in Caco-2 cells and found the colocalization of these two proteins (Fig. 4 D). To further determine which region of ERAdP was required for the interaction with NLRP6, we performed domain mapping assay (Fig. 4 E). We observed that the truncation of N-terminal domain (N) and transmembrane domain 1 (TM1) in ERAdP abolished the interaction between ERAdP and NLRP6 (Fig. 4 F). In addition, N-terminal domain deficiency of ERAdP disrupted the colocalization with NLRP6 (Fig. 4 G).
ERAdP interacts with NLRP6. (A) Immunoprecipitation assay was performed using small intestinal crypts from WT and ERAdP-HA mice. Crypts were lysed and incubated with anti-HA antibody and protein A/G beads or only protein A/G beads as control. Proteins precipitated on the beads were resolved by SDS-PAGE, followed by silver staining, and differential bands were cut for mass spectrometry. The band of NLRP6 was shown. (B) co-IP analysis of NLRP6-Myc and ERAdP-Flag. NLRP6-Myc and ERAdP-Flag were cotransfected into HEK293T cells for 48 h. Cell lysates were incubated with anti-Flag antibody for immunoprecipitation; proteins precipitated on the beads were analyzed by western blotting with anti-Myc and anti-Flag antibodies. β-Actin was used as a loading control. (C) Endogenous co-IP of NLRP6 and ERAdP-HA. Small intestinal crypts from WT and ERAdP-HA mice were lysed and incubated with anti-HA antibody and protein A/G beads. Proteins precipitated on the beads were analyzed with anti-NLRP6 and anti-HA antibodies. β-Actin was used as a loading control. (D) Colocalization analysis of ERAdP-EGFP and NLRP6-mCherry. ERAdP-EGFP and NLRP6-mCherry were cotransfected into Caco-2 cell line, and colocalization was analyzed with fluorescence imaging. Scale bar: 7.5 μm. (E and F) Domain mapping analysis of NLRP6-binding domains of ERAdP protein. Schematic diagram showing truncation mutants of ERAdP (E). FL, full-length; N: N-terminal; TM1, transmembrane 1, TM2, transmembrane 2; C, C-terminal. NLRP6-Myc were transfected into HEK293T cells for 48 h. Cell lysates were incubated with different truncation mutants of GST-ERAdP recombinant protein and GST beads. Proteins precipitated on the beads were analyzed with anti-Myc and anti-GST antibodies. β-Actin was used as a loading control (F). (G) Colocalization analysis of ΔN-ERAdP-EGFP and NLRP6-mCherry. Truncation mutant forms of ΔN-ERAdP-EGFP and NLRP6-mCherry were cotransfected into Caco-2 cell line, and colocalization was analyzed with fluorescence imaging. Scale bar: 3.5 μm. (H) Left: Immunofluorescence staining of SIs from mice administered with DMSO or Ac-YVAD-CMK. Representative Paneth cells and goblet cells are shown. Green: UEA-1; blue: DAPI, red: EpCAM. Scale bar: 20 μm. Right: Diameter of WGA+ cells (n = 10 mice and 20 villus-crypts were analyzed per mouse) and DCVs per Paneth cells were quantified (n = 6 mice and 20 villus-crypts were analyzed per mouse). (I) ELISA of caspase-1 activity examination of crypts from WT and Cnep1r1ΔIEC mice (n = 6 mice for each group). (J) NLRP6 expression levels of SI crypts from littermate WT and Cnep1r1ΔIEC mice were detected by western blot with anti-NLRP6 antibody. β-Actin was used as a loading control. (K) Immunofluorescence staining of ileum crypts from littermate WT and Cnep1r1ΔIEC mice. Red: NLRP6, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (L) Immunohistochemistry staining of lysozyme in intestines from WT and Nlrp6−/− mice. Scale bar: 35 μm. (M) Left: Electron micrograph of small intestinal crypts in littermate WT and Nlrp6−/− mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Number of DCVs was calculated (n = 10 mice for each group). Data in H, I, and M are expressed as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (***P < 0.00; ns, not significant). Data in A–D and F–M are representative of at least three independent experiments. Source data are available for this figure: SourceData F4.
ERAdP interacts with NLRP6. (A) Immunoprecipitation assay was performed using small intestinal crypts from WT and ERAdP-HA mice. Crypts were lysed and incubated with anti-HA antibody and protein A/G beads or only protein A/G beads as control. Proteins precipitated on the beads were resolved by SDS-PAGE, followed by silver staining, and differential bands were cut for mass spectrometry. The band of NLRP6 was shown. (B) co-IP analysis of NLRP6-Myc and ERAdP-Flag. NLRP6-Myc and ERAdP-Flag were cotransfected into HEK293T cells for 48 h. Cell lysates were incubated with anti-Flag antibody for immunoprecipitation; proteins precipitated on the beads were analyzed by western blotting with anti-Myc and anti-Flag antibodies. β-Actin was used as a loading control. (C) Endogenous co-IP of NLRP6 and ERAdP-HA. Small intestinal crypts from WT and ERAdP-HA mice were lysed and incubated with anti-HA antibody and protein A/G beads. Proteins precipitated on the beads were analyzed with anti-NLRP6 and anti-HA antibodies. β-Actin was used as a loading control. (D) Colocalization analysis of ERAdP-EGFP and NLRP6-mCherry. ERAdP-EGFP and NLRP6-mCherry were cotransfected into Caco-2 cell line, and colocalization was analyzed with fluorescence imaging. Scale bar: 7.5 μm. (E and F) Domain mapping analysis of NLRP6-binding domains of ERAdP protein. Schematic diagram showing truncation mutants of ERAdP (E). FL, full-length; N: N-terminal; TM1, transmembrane 1, TM2, transmembrane 2; C, C-terminal. NLRP6-Myc were transfected into HEK293T cells for 48 h. Cell lysates were incubated with different truncation mutants of GST-ERAdP recombinant protein and GST beads. Proteins precipitated on the beads were analyzed with anti-Myc and anti-GST antibodies. β-Actin was used as a loading control (F). (G) Colocalization analysis of ΔN-ERAdP-EGFP and NLRP6-mCherry. Truncation mutant forms of ΔN-ERAdP-EGFP and NLRP6-mCherry were cotransfected into Caco-2 cell line, and colocalization was analyzed with fluorescence imaging. Scale bar: 3.5 μm. (H) Left: Immunofluorescence staining of SIs from mice administered with DMSO or Ac-YVAD-CMK. Representative Paneth cells and goblet cells are shown. Green: UEA-1; blue: DAPI, red: EpCAM. Scale bar: 20 μm. Right: Diameter of WGA+ cells (n = 10 mice and 20 villus-crypts were analyzed per mouse) and DCVs per Paneth cells were quantified (n = 6 mice and 20 villus-crypts were analyzed per mouse). (I) ELISA of caspase-1 activity examination of crypts from WT and Cnep1r1ΔIEC mice (n = 6 mice for each group). (J) NLRP6 expression levels of SI crypts from littermate WT and Cnep1r1ΔIEC mice were detected by western blot with anti-NLRP6 antibody. β-Actin was used as a loading control. (K) Immunofluorescence staining of ileum crypts from littermate WT and Cnep1r1ΔIEC mice. Red: NLRP6, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (L) Immunohistochemistry staining of lysozyme in intestines from WT and Nlrp6−/− mice. Scale bar: 35 μm. (M) Left: Electron micrograph of small intestinal crypts in littermate WT and Nlrp6−/− mice. Paneth cells were drawn with dashed white lines, normal DCVs were indicated by yellow asterisk, and abnormal DCVs were indicated by red asterisk. Scale bar: 5 μm. Right: Number of DCVs was calculated (n = 10 mice for each group). Data in H, I, and M are expressed as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (***P < 0.00; ns, not significant). Data in A–D and F–M are representative of at least three independent experiments. Source data are available for this figure: SourceData F4.
Identification of NLRP6 and generation of NLRP6-deficient mice. (A) Western blot of input samples used in pulldown of Fig. 4 A. β-Actin was used as a loading control. (B) Peptides of NLRP6 were identified by mass spectrometry. (C) Western blot for ASC KO validation with anti-ASC antibody. β-Actin was used as a loading control. (D) Left: Immunofluorescence staining of SIs from sgCtrl and sgAsc mice. Representative Paneth cells and goblet cells are shown. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. Right: Diameter of goblet cell numbers (n = 10 mice and 20 villus-crypts were analyzed per mouse) and DCVs per Paneth cells were quantified (n = 6 mice and 20 villus-crypts were analyzed per mouse). (E) NLRP6 expression levels of small intestinal villi from littermate WT and Cnep1r1ΔIEC mice were detected by western blot with anti-NLRP6 antibody. β-Actin was used as a loading control. (F) Relative Nlrp6 expression levels of intestinal crypts from WT and Cnep1r1ΔIEC mice were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 6 mice for each group). (G) Construction diagram showing generation of Nlrp6−/− mice with CRISPR-Cas9. (H) DNA sequencing for Nlrp6 KO validation. (I) Western blot for NLRP6 KO validation with anti-NLRP6 antibody. β-Actin was used as a loading control for western blot. (J) Left: HE staining of duodenum, jejunum, and ileum from littermate WT and Nlrp6−/− mice. Scale bar: 30 μm. Middle: Proportion of normal and abnormal Paneth cells were quantified based on whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal). Right: Numbers of Paneth cells were quantified. n = 6 mice, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (K) Quantification of Lyz+ puncta per crypt of WT and Nlrp6−/− mice (n = 6 mice for each group). (L) AB-PAS staining of intestines from WT and Nlrp6−/− mice. Scale bar: 35 μm. (M) Left: 3D immunofluorescence staining of ileum crypts from littermate WT and Nlrp6−/− mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group, and 20 crypts were analyzed per mouse). (N) Immunofluorescence staining of organoids from littermate WT and Nlrp6−/− mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 20 μm. (O) Intestinal lumen (left) and tissue-associated (right) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums (n = 6 mice for each group). (P) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). (Q) Lysozyme in supernatant of stimulated crypts from littermate WT and Nlrp6−/− mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (R) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and Nlrp6−/− mice were isolated and stimulated by DMSO or CCh. The crypt secreted supernatants were incubated with S. Typhimurium for 30 min, and CFUs were measured (n = 6 mice for each group). Data in D–F, J, K, M, and O–R are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data in A–F and H–R are representative of at least three independent experiments. Source data are available for this figure: SourceData FS3.
Identification of NLRP6 and generation of NLRP6-deficient mice. (A) Western blot of input samples used in pulldown of Fig. 4 A. β-Actin was used as a loading control. (B) Peptides of NLRP6 were identified by mass spectrometry. (C) Western blot for ASC KO validation with anti-ASC antibody. β-Actin was used as a loading control. (D) Left: Immunofluorescence staining of SIs from sgCtrl and sgAsc mice. Representative Paneth cells and goblet cells are shown. Green: UEA-1, blue: DAPI, and red: EpCAM. Scale bar: 20 μm. Right: Diameter of goblet cell numbers (n = 10 mice and 20 villus-crypts were analyzed per mouse) and DCVs per Paneth cells were quantified (n = 6 mice and 20 villus-crypts were analyzed per mouse). (E) NLRP6 expression levels of small intestinal villi from littermate WT and Cnep1r1ΔIEC mice were detected by western blot with anti-NLRP6 antibody. β-Actin was used as a loading control. (F) Relative Nlrp6 expression levels of intestinal crypts from WT and Cnep1r1ΔIEC mice were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 6 mice for each group). (G) Construction diagram showing generation of Nlrp6−/− mice with CRISPR-Cas9. (H) DNA sequencing for Nlrp6 KO validation. (I) Western blot for NLRP6 KO validation with anti-NLRP6 antibody. β-Actin was used as a loading control for western blot. (J) Left: HE staining of duodenum, jejunum, and ileum from littermate WT and Nlrp6−/− mice. Scale bar: 30 μm. Middle: Proportion of normal and abnormal Paneth cells were quantified based on whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal). Right: Numbers of Paneth cells were quantified. n = 6 mice, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (K) Quantification of Lyz+ puncta per crypt of WT and Nlrp6−/− mice (n = 6 mice for each group). (L) AB-PAS staining of intestines from WT and Nlrp6−/− mice. Scale bar: 35 μm. (M) Left: 3D immunofluorescence staining of ileum crypts from littermate WT and Nlrp6−/− mice. Green: UEA-1, red: EpCAM, and yellow: DCVs’ surface fitted by Imaris software. Scale bar: 35 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group, and 20 crypts were analyzed per mouse). (N) Immunofluorescence staining of organoids from littermate WT and Nlrp6−/− mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 20 μm. (O) Intestinal lumen (left) and tissue-associated (right) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums (n = 6 mice for each group). (P) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). (Q) Lysozyme in supernatant of stimulated crypts from littermate WT and Nlrp6−/− mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (R) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and Nlrp6−/− mice were isolated and stimulated by DMSO or CCh. The crypt secreted supernatants were incubated with S. Typhimurium for 30 min, and CFUs were measured (n = 6 mice for each group). Data in D–F, J, K, M, and O–R are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data in A–F and H–R are representative of at least three independent experiments. Source data are available for this figure: SourceData FS3.
NLRP6 is a NLR that forms an inflammasome (Li and Zhu, 2020), so we wanted to know whether NLRP6 regulated DCVs through inflammasome assembling. We administrated mice with Ac-YVAD-CMK, which is a potent and irreversible inhibitor of the inflammasome downstream enzyme caspase-1. In line with a previous report (Wlodarska et al., 2014), blocking NLRP6-mediated inflammasome pathway caused accumulation of mucus in goblet cells, while DCVs in Paneth cells were not changed (Fig. 4 H). Meanwhile, we depleted ASC, an essential component of inflammasome, through adeno-associated virus (AAV) delivering sgRNA (sgAsc) (Fig. S3 C), and found mucus accumulation within goblet cells, but no change of DCVs in Paneth cells (Fig. S3 D). We measured caspase-1 activity, which reflected the activation of inflammasome. We noticed that caspase-1 activity was not significantly changed in Cnep1r1ΔIEC mice (Fig. 4 I). These results suggest that NLRP6-mediated inflammasome pathway did not contribute to abnormal DCVs of Paneth cells in ERAdP-deficient mice.
We observed NLRP6 protein level was decreased in the intestinal crypts of Cnep1r1ΔIEC mice (Fig. 4 J), while NLRP6 remained unchanged in the villi (Fig. S3 E). Notably, this reduction occurred without any relative changes in Nlrp6 mRNA levels in the crypt compartment (Fig. S3 F). In addition, NLRP6 was no longer positioned on the DCV membrane in Cnep1r1ΔIEC mice (Fig. 4 K). These data suggest that NLRP6 could regulate DCVs of Paneth cells by attachment on DCVs. Then we generated Nlrp6 KO mice (Nlrp6−/−) to investigate its function on DCV biogenesis (Fig. S3, G–I). Consistent with the phenotype of Cnep1r1ΔIEC mice, Nlrp6−/− mice displayed remarkably decreased numbers and volumes of DCVs in Paneth cells, and AMP lysozyme failed to be packaged into DCVs (Fig. 4 L and Fig. S3, J–M). Electron microscopy revealed the ultrastructure of DCVs was damaged in Nlrp6−/− mice (Fig. 4 M). In addition, organoids from Nlrp6−/− mice displayed less DCVs as well (Fig. S3 N). Nlrp6−/− mice also exhibited impaired antimicrobial capacity, characterized by increased bacterial colonization in the intestinal lumen and adhesion to the epithelium (Fig. S3 O), with a remarkable enrichment of Proteobacteria (Fig. S3 P). Lysozyme secretion was reduced in the isolated small intestinal crypts from Nlrp6−/− mice (Fig. S3 Q), and its bactericidal activity was impaired (Fig. S3 R). Taken together, these results indicate that ERAdP recruits NLRP6 to the DCV membrane to promote DCV biogenesis in Paneth cells.
The ERAdP–NLRP6 association recruits ANXA2 onto the DCV membrane for package of DCVs
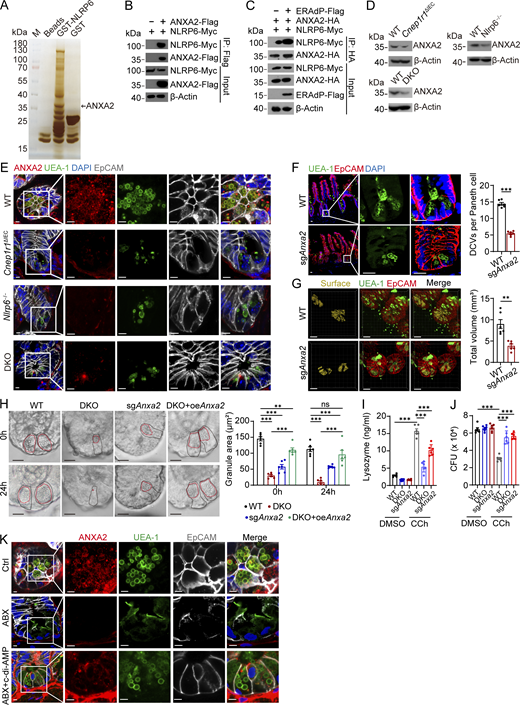
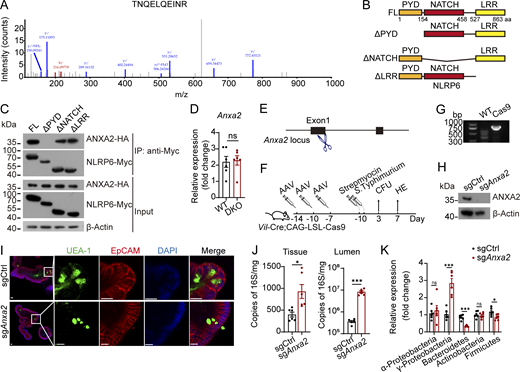
It has been reported that NLRP6 exerts antibacterial function through activation of inflammasome pathways (Wlodarska et al., 2014). However, we demonstrated that NLRP6 was located on the membrane of DCVs in Paneth cells, suggesting an alternative mechanism of antibacterial role of NLRP6. Annexin A2 (ANXA2) was identified as a candidate interacting protein of NLRP6 by GST-pulldown assay (Fig. 5 A and Fig. S4 A). ANXA2 is involved in exocytosis (Gabel et al., 2019). We further confirmed the interaction between NLRP6 and ANXA2 by cotransfection of these proteins in HEK293T cells (Fig. 5 B). To determine which region of NLRP6 was required for the interaction with ANXA2, we performed domain mapping assay (Fig. S4 B). We observed that the truncation of PYD in NLRP6 abolished the interaction between NLRP6 and ANXA2 (Fig. S4 C). Of note, ERAdP enhanced the interaction between NLRP6 and ANXA2 (Fig. 5 C). Then we tested ANXA2 protein levels and subcellular localization with ERAdP or NLRP6 deficiency. NLRP6 KO and ERAdP–NLRP6 double KO (DKO) mice showed decreased ANXA2 protein levels, and ANXA2 could not localize on the DCV membrane compared with WT mice (Fig. 5, D and E). However, absence of ERAdP and NLRP6 did not affect the mRNA level of Anxa2 (Fig. S4 D). To determine the role of ANXA2 in DCV formation, we deleted ANXA2 through AAV infection to express sgRNA-targeting ANXA2 in Cas9-expressing mice (Fig. S4, E–H). Consistent with earlier observations, ANXA2 deficiency caused decreased numbers and volumes of DCVs in Paneth cells (Fig. 5, F and G). This phenomenon was also observed in organoids, in which DCVs were scarce in ERAdP-, NLRP6-, or ANXA2-deficient Paneth cells (Fig. 5 H and Fig. S4 I). We stimulated organoids with CCh to induce DCV secretion and DCV regeneration and found that ERAdP, NLRP6, or ANXA2 deficiency disrupted replenishment of DCVs (Fig. 5 H). While ANXA2 overexpression in ERAdP- and NLRP6-deficient Paneth cells could partially restore DCV generation (Fig. 5 H). We found that bacterial burden in sgAnxa2 mice increased in the intestinal lumen and epithelium (Fig. S4 J), especially Proteobacteria (Fig. S4 K), suggesting Anxa2-deficient mice exhibited impaired antimicrobial capacity. Then we analyzed lysozyme secretion and bactericidal effect in isolated intestinal crypts. In line with ERAdP- and NLRP6-deficient mice, crypts from ANXA2-deleted mice also secreted less lysozyme with CCh stimulation, and secreted supernatants showed weaker bactericidal effect on S. Typhimurium (Fig. 5, I and J). Moreover, disruption of intestinal microbiota with ABX damaged membrane localization of ANXA2, while administration with c-di-AMP could rescue ANXA2 location on the DCV membrane (Fig. 5 K), suggesting that activation of ERAdP through c-di-AMP facilitates ANXA2 localization on the membrane of DCVs.
The ERAdP–NLRP6 association recruits ANXA2 onto the DCV membrane for generation of DCVs. (A) GST pull-down showing interaction of NLRP6 and ANXA2. Lysed small intestinal crypts from WT mice were incubated with GST-NLRP6 recombinant protein or GST recombinant protein as control. Proteins precipitated on the beads were resolved by SDS-PAGE, followed by silver staining, and differential bands were cut for mass spectrometry. Representative protein ANXA2 is shown. (B) co-IP analysis of NLRP6-Myc and ANXA2-Flag. NLRP6-Myc and ANXA2-Flag were cotransfected into HEK293T cells for 48 h. Cell lysates were incubated with anti-Flag antibody for immunoprecipitation; proteins precipitated on the beads were analyzed by western blotting with anti-Myc and anti-Flag antibodies. β-Actin was used as a loading control. (C) co-IP analysis of NLRP6-Myc and ANXA2-HA under the condition of absence or presence of ERAdP-Flag. β-Actin served as a loading control. (D) Western blot showing ANXA2 expression levels of SI crypts from littermate WT, Cnep1r1ΔIEC, Nlrp6−/−, and ERAdP–NLRP6 DKO mice with anti-ANXA2 antibody. β-Actin was used as a loading control. (E) Immunofluorescence staining showing ANXA2 localization in ileum crypts from littermate WT, Cnep1r1ΔIEC, Nlrp6−/−, and DKO mice. Red: ANXA2, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (F) Left: Immunofluorescence staining showing DCVs in Paneth cells from littermate WT and sgAnxa2 mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (G) Left: 3D immunofluorescence staining of ileum crypts from WT and sgAnxa2 mice. Green: UEA-1; red: EpCAM; yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 20 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (H) Left: DIC images of organoids from WT, Cnep1r1ΔIEC, DKO, sgAnxa2, and ANXA2-overexpressing DKO mice. Organoids were stimulated by CCh for 10 min and then washed out. Images were captured before (0 h) and after CCh stimulation (24 h). Areas of DCV were drawn with dashed red lines, and the corresponding Paneth cells were drawn with dashed black lines. Scale bar: 10 μm. Right: Area of DCVs was quantified (n = 6 mice for each group). (I) Lysozyme in supernatant of stimulated crypts from WT, DKO, and sgAnxa2 mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (J) Bacterial load analysis of S. Typhimurium in crypt supernatants. SI crypts of littermate WT, DKO and sgAnxa2 mice were isolated and stimulated by DMSO or CCh. The crypt supernatants were incubated with S. Typhimurium, and CFU was measured (n = 6 mice for each group). (K) Immunofluorescence staining of ileum crypts from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP. Green: UEA-1, blue: DAPI, red: ANXA2, and gray: EpCAM. Scale bar: 10 μm. Data in F–J are shown as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (**P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments. Source data are available for this figure: SourceData F5.
The ERAdP–NLRP6 association recruits ANXA2 onto the DCV membrane for generation of DCVs. (A) GST pull-down showing interaction of NLRP6 and ANXA2. Lysed small intestinal crypts from WT mice were incubated with GST-NLRP6 recombinant protein or GST recombinant protein as control. Proteins precipitated on the beads were resolved by SDS-PAGE, followed by silver staining, and differential bands were cut for mass spectrometry. Representative protein ANXA2 is shown. (B) co-IP analysis of NLRP6-Myc and ANXA2-Flag. NLRP6-Myc and ANXA2-Flag were cotransfected into HEK293T cells for 48 h. Cell lysates were incubated with anti-Flag antibody for immunoprecipitation; proteins precipitated on the beads were analyzed by western blotting with anti-Myc and anti-Flag antibodies. β-Actin was used as a loading control. (C) co-IP analysis of NLRP6-Myc and ANXA2-HA under the condition of absence or presence of ERAdP-Flag. β-Actin served as a loading control. (D) Western blot showing ANXA2 expression levels of SI crypts from littermate WT, Cnep1r1ΔIEC, Nlrp6−/−, and ERAdP–NLRP6 DKO mice with anti-ANXA2 antibody. β-Actin was used as a loading control. (E) Immunofluorescence staining showing ANXA2 localization in ileum crypts from littermate WT, Cnep1r1ΔIEC, Nlrp6−/−, and DKO mice. Red: ANXA2, green: UEA-1, blue: DAPI, and gray: EpCAM. Scale bar: 5 μm. (F) Left: Immunofluorescence staining showing DCVs in Paneth cells from littermate WT and sgAnxa2 mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 20 μm. Right: Number of DCVs per Paneth cells was quantified (n = 6 mice for each group). (G) Left: 3D immunofluorescence staining of ileum crypts from WT and sgAnxa2 mice. Green: UEA-1; red: EpCAM; yellow: DCVs’ surface fitted by Imaris 9 software. Scale bar: 20 μm. Right: Volume of DCVs was quantified (n = 6 mice for each group). (H) Left: DIC images of organoids from WT, Cnep1r1ΔIEC, DKO, sgAnxa2, and ANXA2-overexpressing DKO mice. Organoids were stimulated by CCh for 10 min and then washed out. Images were captured before (0 h) and after CCh stimulation (24 h). Areas of DCV were drawn with dashed red lines, and the corresponding Paneth cells were drawn with dashed black lines. Scale bar: 10 μm. Right: Area of DCVs was quantified (n = 6 mice for each group). (I) Lysozyme in supernatant of stimulated crypts from WT, DKO, and sgAnxa2 mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (J) Bacterial load analysis of S. Typhimurium in crypt supernatants. SI crypts of littermate WT, DKO and sgAnxa2 mice were isolated and stimulated by DMSO or CCh. The crypt supernatants were incubated with S. Typhimurium, and CFU was measured (n = 6 mice for each group). (K) Immunofluorescence staining of ileum crypts from mice of control, ABX mock treated, or treated through intragastric gavage with c-di-AMP. Green: UEA-1, blue: DAPI, red: ANXA2, and gray: EpCAM. Scale bar: 10 μm. Data in F–J are shown as means ± SEM, then significance was determined by unpaired two-tailed Student’s t test (**P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments. Source data are available for this figure: SourceData F5.
Identification of ANXA2 and generation of ANXA2-deficient mice. (A) Peptides of ANXA2 were identified by mass spectrometry. (B and C) Domain mapping analysis of ANXA2-binding domains of NLRP6 protein. Schematic diagram showing truncation mutants of NLRP6 (B). Different truncation mutants NLRP6-Myc and ANXA2-HA were transfected into HEK293T cells for 48 h. Cell lysates were incubated anti-Myc beads. Proteins precipitated on the beads were analyzed with anti-Myc and anti-HA antibodies. β-Actin was used as a loading control (C). (D) Relative Anxa2 expression levels of intestinal crypts from WT and DKO mice were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 6 mice for each group). (E) Construction diagram of ANXA2 KO mouse with CRISPR-Cas9. (F) Diagram of mice injected with AAV to KO ANXA2 and then infected with S. Typhimurium. (G) DNA electrophoresis for Cas9 knock-in validation. (H) Western blot for ANXA2 KO validation with ANXA2 antibody. β-Actin was used as a loading control. (I) Immunofluorescence staining of intestinal organoids from littermate sgCtr and sgAnxa2 mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 10 μm. (J) Intestinal lumen (left) and tissue-associated (right) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums of WT and sgAnxa2 mice (n = 6 mice for each group). (K) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). Data in D, J, and K are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in A, C, D, and G–K are representative of at least three independent experiments. Source data are available for this figure: SourceData FS4.
Identification of ANXA2 and generation of ANXA2-deficient mice. (A) Peptides of ANXA2 were identified by mass spectrometry. (B and C) Domain mapping analysis of ANXA2-binding domains of NLRP6 protein. Schematic diagram showing truncation mutants of NLRP6 (B). Different truncation mutants NLRP6-Myc and ANXA2-HA were transfected into HEK293T cells for 48 h. Cell lysates were incubated anti-Myc beads. Proteins precipitated on the beads were analyzed with anti-Myc and anti-HA antibodies. β-Actin was used as a loading control (C). (D) Relative Anxa2 expression levels of intestinal crypts from WT and DKO mice were detected by qRT-PCR. Fold changes were normalized to endogenous 18S (n = 6 mice for each group). (E) Construction diagram of ANXA2 KO mouse with CRISPR-Cas9. (F) Diagram of mice injected with AAV to KO ANXA2 and then infected with S. Typhimurium. (G) DNA electrophoresis for Cas9 knock-in validation. (H) Western blot for ANXA2 KO validation with ANXA2 antibody. β-Actin was used as a loading control. (I) Immunofluorescence staining of intestinal organoids from littermate sgCtr and sgAnxa2 mice. Green: UEA-1, red: EpCAM, and blue: DAPI. Scale bar: 10 μm. (J) Intestinal lumen (left) and tissue-associated (right) bacterial load analysis, quantified by qPCR of 16S rRNA gene copy number in distal ileums of WT and sgAnxa2 mice (n = 6 mice for each group). (K) qPCR detection of ileal luminal commensal bacteria classified by phylum (n = 6 mice for each group). Data in D, J, and K are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; ***P < 0.001; ns, not significant). Data in A, C, D, and G–K are representative of at least three independent experiments. Source data are available for this figure: SourceData FS4.
The ERAdP–NLRP6–ANXA2 axis is required for antibacterial function
In Paneth cells, we co-stained these three proteins and found that they were colocalized on DCVs (Fig. S5 A). To investigate the function of ERAdP–NLRP6–ANXA2 axis in Paneth cells, we generated ERAdP–NLRP6 DKO and ERAdP–NLRP6–ANXA2 triple KO (TKO) mice. DKO and TKO exhibited greater numbers of abnormal Paneth cells and loss of DCVs compared with littermate control mice (Fig. 6 A and Fig. S5 B). Consistently, lysozyme could not be packaged into DCVs in DKO and TKO mice (Fig. 6 B and Fig. S5 C), leading to reduced lysozyme secretion and diminished bactericidal activity in TKO mice (Fig. S5, D and E). However, overexpression of Anxa2 in DKO mice rescued DCV biogenesis (Fig. 6 B). We infected these mice with S. Typhimurium and found that DKO and TKO mice showed much higher bacterial loads in livers, spleens, PPs, and ileal contents; larger weight loss and mortality; and severer intestinal destruction (Fig. 6, C–G; and Fig. S5, F and G). In contrast, overexpression of Anxa2 could ameliorate these symptoms (Fig. 6, C–G). These data suggest that the ERAdP–NLRP6–ANXA2 axis plays a critical role in maintaining the normal function of DCVs and antibacterial function of Paneth cells.
DKO and TKO mice exhibit impaired antibacterial ability. (A) Immunofluorescence staining of ileal crypts from WT mice. Green: UEA-1, blue: ERAdP-HA, red: NLRP6, and pink: ANXA2. Scale bar: 5 μm. (B) Left: AB-PAS staining of intestines from ERAdP–NLRP6 DKO and ERAdP–NLRP6–ANXA2 TKO mice. Scale bar: 35 μm. Right: Quantification of DCV numbers per Paneth cell of WT, DKO, and TKO mice (n = 6 mice for each group). (C) Left: Lysozyme immunohistochemistry of intestines from WT, DKO, and TKO mice. Scale bar: 35 μm. Right: Quantification of Lyz+ puncta per crypt of WT, DKO, and TKO mice (n = 6 mice for each group). (D) Lysozyme in supernatant of stimulated crypts from littermate WT and TKO mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (E) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and TKO mice were isolated and stimulated by DMSO or CCh (n = 6 mice for each group). (F and G) Bacterial load analysis of ileal contents (F) and PPs (G) from WT, DKO, TKO, and ANXA2-overexpressing mice. 3 days after S. Typhimurium infection, ileal contents and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (H–K) Bacterial load analysis in livers (H), spleens (I), ileal contents (J), and PPs (K) of control, ABX, and ABX+c-di-AMP mice. At 3 days after S. Typhimurium infection, livers, spleens, ileal contents, and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (L) Body weight change analysis of control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium (n = 6 mice for each group). Data in B–L are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
DKO and TKO mice exhibit impaired antibacterial ability. (A) Immunofluorescence staining of ileal crypts from WT mice. Green: UEA-1, blue: ERAdP-HA, red: NLRP6, and pink: ANXA2. Scale bar: 5 μm. (B) Left: AB-PAS staining of intestines from ERAdP–NLRP6 DKO and ERAdP–NLRP6–ANXA2 TKO mice. Scale bar: 35 μm. Right: Quantification of DCV numbers per Paneth cell of WT, DKO, and TKO mice (n = 6 mice for each group). (C) Left: Lysozyme immunohistochemistry of intestines from WT, DKO, and TKO mice. Scale bar: 35 μm. Right: Quantification of Lyz+ puncta per crypt of WT, DKO, and TKO mice (n = 6 mice for each group). (D) Lysozyme in supernatant of stimulated crypts from littermate WT and TKO mice after treatment of DMSO or CCh was measured by ELISA (n = 6 mice for each group). (E) Bacterial load analysis of S. Typhimurium in secreted supernatants. SI crypts of littermate WT and TKO mice were isolated and stimulated by DMSO or CCh (n = 6 mice for each group). (F and G) Bacterial load analysis of ileal contents (F) and PPs (G) from WT, DKO, TKO, and ANXA2-overexpressing mice. 3 days after S. Typhimurium infection, ileal contents and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (H–K) Bacterial load analysis in livers (H), spleens (I), ileal contents (J), and PPs (K) of control, ABX, and ABX+c-di-AMP mice. At 3 days after S. Typhimurium infection, livers, spleens, ileal contents, and PPs were collected, and CFUs were calculated (n = 6 mice for each group). (L) Body weight change analysis of control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium (n = 6 mice for each group). Data in B–L are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
The ERAdP–NLRP6–ANXA2 axis is required for antibacterial function. (A) Upper: HE staining of duodenum, jejunum, and ileum from littermate WT, Cnep1r1ΔIEC, ERAdP–NLRP6 DKO, and ERAdP–NLRP6–ANXA2 TKO mice. Scale bar: 30 μm. Lower: Proportion of normal and abnormal Paneth cells were quantified on the basis of whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal). n = 6 mice, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (B) Immunofluorescence staining of ileums from WT, DKO, TKO, and ANXA2-overexpressing DKO mice. Red: lysozyme; green: WGA, blue: DAPI. Scale bar: 10 μm. (C and D) Bacterial load analysis of livers (C) and spleens (D) of WT, DKO, TKO, and ANXA2-overexpressing mice. 3 days after S. Typhimurium infection, livers and spleens were collected, and CFUs were calculated (n = 6 mice for each group). (E) Body weight change analysis of WT, DKO, TKO, and ANXA2-overexpressing DKO mice after infected by S. Typhimurium (n = 6 mice for each group). (F) Survival rates of WT, DKO, TKO, and ANXA2-overexpressing DKO mice after infected by S. Typhimurium (n = 12 for each group). (G) Representative pathological manifestation of intestines from WT, DKO, TKO, and DKO+oeAnxa2 mice at 7 days after S. Typhimurium infection. Scale bar: 40 μm. (H) Schematic diagram of mice treated by ABX and c-di-AMP. (I) Survival rates of control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium (n = 12 mice for each group). (J) Representative pathological manifestation of intestines from control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium for 7 days. Scale bar: 40 μm. (K) Relative expression of CNEP1R1, NLRP6, and ANXA2 of ileal CD samples detected by qRT-PCR (n = 20 clinical samples for each group). Fold changes were normalized to endogenous ACTB. Data are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (A and C–E), Mantel–Cox test (F and I), and Wilcoxon matched pairs signed rank test (K) (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
The ERAdP–NLRP6–ANXA2 axis is required for antibacterial function. (A) Upper: HE staining of duodenum, jejunum, and ileum from littermate WT, Cnep1r1ΔIEC, ERAdP–NLRP6 DKO, and ERAdP–NLRP6–ANXA2 TKO mice. Scale bar: 30 μm. Lower: Proportion of normal and abnormal Paneth cells were quantified on the basis of whether Paneth cells displayed a typical staining pattern with distinguishable granules (normal) or disordered, depleted, and/or diffuse staining (abnormal). n = 6 mice, with 20 selecting maximal crypt sections that displayed all Paneth cells within each crypt were analyzed per mouse. (B) Immunofluorescence staining of ileums from WT, DKO, TKO, and ANXA2-overexpressing DKO mice. Red: lysozyme; green: WGA, blue: DAPI. Scale bar: 10 μm. (C and D) Bacterial load analysis of livers (C) and spleens (D) of WT, DKO, TKO, and ANXA2-overexpressing mice. 3 days after S. Typhimurium infection, livers and spleens were collected, and CFUs were calculated (n = 6 mice for each group). (E) Body weight change analysis of WT, DKO, TKO, and ANXA2-overexpressing DKO mice after infected by S. Typhimurium (n = 6 mice for each group). (F) Survival rates of WT, DKO, TKO, and ANXA2-overexpressing DKO mice after infected by S. Typhimurium (n = 12 for each group). (G) Representative pathological manifestation of intestines from WT, DKO, TKO, and DKO+oeAnxa2 mice at 7 days after S. Typhimurium infection. Scale bar: 40 μm. (H) Schematic diagram of mice treated by ABX and c-di-AMP. (I) Survival rates of control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium (n = 12 mice for each group). (J) Representative pathological manifestation of intestines from control, ABX, and ABX+c-di-AMP mice after infected by S. Typhimurium for 7 days. Scale bar: 40 μm. (K) Relative expression of CNEP1R1, NLRP6, and ANXA2 of ileal CD samples detected by qRT-PCR (n = 20 clinical samples for each group). Fold changes were normalized to endogenous ACTB. Data are shown as means ± SEM; significance was determined by unpaired two-tailed Student’s t test (A and C–E), Mantel–Cox test (F and I), and Wilcoxon matched pairs signed rank test (K) (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant). Data above are representative of at least three independent experiments.
We next tested antibacterial ability of ABX mice and ABX mice treated by c-di-AMP (Fig. 6 H). We found that disruption of microbiota with ABX destroyed antibacterial ability of mice, displaying worse survival rates and tissue destruction, more bacterial loads in organs, and larger weight loss (Fig. 6, I and J; and Fig. S5, H–L). Reactivation of the ERAdP–NLRP6–ANXA2 axis with c-di-AMP partially restored antibacterial ability of mice (Fig. 6, I and J; and Fig. S5, H–L). Finally, we examined expression of these genes in IBD patients. We found that CNEP1R1, NLRP6, and ANXA2 exhibited reduced expression levels in ileal CD samples compared with normal tissues (Fig. 6 K). These data suggested that downregulation of CNEP1R1, NLRP6, and ANXA2 genes might be involved in the IBD pathogenesis. Taken together, the ERAdP–NLRP6–ANXA2 axis plays a key role in biogenesis of DCVs in Paneth cells, which participates in protection against bacterial infection.
Discussion
As the first line of defense, intestinal epithelial cells protect against pathogenic microbial infections and maintain intestinal homeostasis. One of the key mechanisms on the antimicrobial function involves Paneth cells located in the crypts, which secrete AMPs into the intestinal lumen via DCVs. In our study, we identified ERAdP as a critical nexus linking microbial sensing to effector function of Paneth cells. By sensing c-di-AMP derived from intestinal microbiota, ERAdP interacted with NLRP6 and further recruited ANXA2 onto the DCV membrane in Paneth cells. Activation of ERAdP–NLRP6–ANXA2 axis facilitated DCV biogenesis, which enhanced antibacterial ability of intestines. This work advances our understanding in intestinal immunity and provides a new avenue for targeting DCV dysfunction in treatment of infectious and inflammatory gut diseases.
Abnormalities in Paneth cell function can disrupt the balance of microbiota and host, exacerbating injury caused by pathogens. Paneth cells are regulated by multiple factors. Endogenously, lysozyme secretion is disrupted in Paneth cells in autophagy gene Atg16L1-deficient mice, which is linked to an increased risk of CD in humans (Bel et al., 2017). Additionally, the absence of TVP23B impairs the host-microbiota balance (Song et al., 2023). Regarding immune regulation, LRRK, expressed in lamina propria macrophages, impairs autophagy and contributes to Paneth cell dysfunction, which is involved in the pathogenesis of CD (Sun et al., 2024a). In another study, TNF was defined to inhibit the antimicrobial activity of Paneth cells, promoting bacterial translocation from the gut to various organs, leading to polymicrobial sepsis, organ failure, and death (Wallaeys et al., 2024). Furthermore, γδ intraepithelial lymphocytes were shown to secrete apoptosis inhibitor 5, promoting survival of Paneth cells in the absence of ATG16L (Matsuzawa-Ishimoto et al., 2022). For impact of bacterial signal on Paneth cells, some studies suggest that DCVs of Paneth cells are independent of microbiota (Jang et al., 2018). However, other studies showed the granule sizes were smaller in GF mice than conventionally raised animals (Satoh, 1988), and GF rats inoculated with bacteria from feces of SPF rats induced transient Paneth cell degranulation followed by complete restoration (Satoh et al., 1986). In addition, in the context of bacterial infections, CD74+ Paneth cells rapidly expand to exacerbate inflammatory disease progression in both mice and humans (Balasubramanian et al., 2023). Our findings show that DCVs of Paneth cells were decreased in GF and ABX mice and could be rescued by c-di-AMP derived from intestinal microbiota.
We previously defined that ERAdP was a receptor that senses c-di-AMP to combat bacteria and regulates antibacterial function of immune cells through the recognition of c-di-AMP (Xia et al., 2018; Chen et al., 2015). In this study, we observed that ERAdP was also highly expressed in Paneth cells and located on the DCV membrane of Paneth cells. ERAdP recognized c-di-AMP from the microbiota, thereby participated in assembly of DCVs that exerted antibacterial effects. Deletion of ERAdP in intestinal epithelial cells did not alter the number of Paneth cells. However, the number of DCVs was significantly reduced. Although residual DCVs were still detectable by HE staining, ultrastructural analysis via electron microscopy revealed that these remaining DCVs exhibited abnormal morphology and altered electron density in their contents. The secretion of AMPs involves multiple coordinated processes, including DCV packaging, sorting, and maturation. Thus, despite the preservation of some DCVs, their morphological abnormalities likely impair secretory function. At a molecular level, ERAdP interacted with NLRP6, which subsequently recruited ANXA2 onto the membrane of DCVs. Deficiency of NLRP6 and ANXA2 caused impaired biogenesis of DCVs. Of note, overexpression of Anxa2 in ERAdP and NLRP6 KO mice could partially restore DCV biogenesis. These data suggest that ERAdP recognized c-di-AMP derived from intestinal microbiota, subsequently activating the ERAdP–NLRP6–ANXA2 axis to promote DCV formation in Paneth cells.
NLRP6 is a NLR and a key component of the inflammasome which activates caspase-1 through ASC to promote secretion of IL-1β and IL-18 (Shen et al., 2019; Barnett et al., 2023). NLRP6 is known to influence intestinal antibacterial activity by regulating mucus secretion in goblet cells via an inflammasome pathway (Wlodarska et al., 2014) and shaping colonic commensal bacteria (Gálvez et al., 2017). A recent study showed that NLRP6 senses double-stranded RNA of virus, leading to liquid–liquid phase separation for its activation (Shen et al., 2021). Here we showed that ERAdP associated with NLRP6 to recruit ANXA2 onto DCV membrane, participating in the biogenesis of DCVs of Paneth cells. We propose that NLRP6 acted as a scaffolding protein to facilitate vesicle biogenesis. Additionally, the inability of caspase-1 inhibition to recapitulate DCV defects in ERAdP-deficient mice strongly supported an inflammasome-independent mechanism.
The annexin family members are evolutionarily conserved that regulate membrane trafficking by binding to negatively charged phospholipids in a Ca2+-dependent manner (Gabel et al., 2019). ANXA2 has been implicated in exocytosis in various cell types, including chromaffin cells, neuroendocrine cells (Gabel et al., 2019), and placental cells (Bai et al., 2022). In addition, ANXA2 plays a role in inflammatory responses, as its translocation to the cell surface promotes retinal pigment epithelial cell migration following injury (Luo et al., 2024). While ANXA2 is known to regulate membrane trafficking and mediate exocytosis. However, its role in Paneth cell function has not been defined. Here we showed that ANXA2 localization on DCV membranes takes part in the packaging and biogenesis of DCVs in Paneth cells. c-di-AMP derived from microbiota facilitated the localization of ANXA2 on the DCV membrane and promoted the DCV formation via activating ERAdP–NLRP6–ANXA2 axis. However, it needs further investigation about the precise biochemical steps by which ANXA2 modulates DCV biogenesis.
Intriguingly, we showed that expression of ERAdP, NLRP6, and ANXA2 were dramatically reduced in IBD patients. IBD is typically associated with disruption of the intestinal epithelium, leading to increased bacterial permeability. The absence of ERAdP in IBD patients could impair the antimicrobial response. In addition, we found that administration of c-di-AMP was able to promote production of AMPs, potentially enhanced antimicrobial activity, and alleviated inflammation caused by infection. These findings suggest that activating ERAdP–NLRP6–ANXA2 axis via c-di-AMP stimulation could provide a potential therapeutic strategy for infectious disease and gut inflammation.
Materials and methods
Antibodies and reagents
Anti-HA (cat# ab236632), anti-Lysozyme (cat# ab108508), anti-NLRP6 (cat# ab58705), and anti-TFF3 (cat# ab108599) were from Abcam. Alexa Fluor 647 anti-mouse CD326 (cat# 118212) was from BioLegend. Anti-PDI (3501T) was from CST. Anti-GST (cat# SC-138) was from Santa Cruz. UEA I Lectin (cat# GTX01512) and WGA Lectin (cat# GTX01500) were from GeneTex. AF594 Donkey anti-rabbit (cat# A21207) and AF647 goat anti-rabbit (cat# A21244) were from Invitrogen. Anti-Annexin A2 (cat# 11256-1-AP), anti-DEFA5 (cat# 18268-1-AP), and anti-c-Myc-tag (cat# 10828-1-AP) were from Proteintech. Anti-β-actin (cat# RM2001) was from Rayantibody. Goat anti-Mouse IgG(H+L)-HRP antibody, Goat anti-Rabbit IgG(H+L)-HRP antibody, anti-c-Myc-tag (cat# KM8003), and anti-Flag-tag (cat# KM8002) were from Sungene Biotech. Paraformaldehyde (PFA) and DAPI were from Sigma-Aldrich.
Mice
Mice we used were of the C57BL/6 background and maintained under SPF conditions. Generation of Cnep1r1fl/fl, Nlrp6−/− mice were based on the methods described before (Xia et al., 2018). In brief, vector pST1374-NLS-flaglinker-Cas9 (plasmid #44758; Addgene) expressing Cas9 and pUC57-sgRNA (plasmid #51132; Addgene) expressing sgRNAs (Table S1) for the target genes were constructed. Donor templates of loxP and HA sequences combined with homology arms were cloned into pLSODN-1. Long single-strand DNA was prepared using an LSODN Preparation Kit (Biodynamics Laboratory) according to the manufacturer’s protocol. Mixtures of Cas9 mRNA (100 ng/ml), sgRNA (50 ng/ml), and donor templates (20 ng/ml) were microinjected into the cytoplasm of C57BL/6-fertilized eggs, followed by transferring to the uterus of pseudopregnant ICR females, from which viable founder mice were obtained. Genotyping of mice were performed by indicated primers (Table S2) and verified by DNA sequencing. Cas9-KI, Vil-Cre and Lgr5-EGFP-IRES-CreERT mice were obtained from Jackson Laboratory. For generation of Anxa2 and Asc conditional KO mice, sgRNAs targeting Anxa2 or Asc were synthesized and cloned into AAV-delivering vectors. Then the AAVs were administered via tail veins of Vil-Cre;Cas9-KI mice. Targeted alleles were identified by PCR screening and DNA sequencing, followed by western blot. ERAdP-HA (Cnep1r1-C-HA) (https://www.sibcb.ac.cn/gtp/eindex.jsp, GTP ID18000056) mice were supplied by Genome Tagging Project (GTP) Center, CEMCS, Chinese Academy of Sciences. GF and its control group SPF mice were purchased from GemPharmatech Co., Ltd. To generate Cnep1r1ΔIEC mice, Cnep1r1fl/fl mice were crossed with Vil-Cre mice. To generate DKO mice, Cnep1r1ΔIEC mice were crossed with Nlrp6−/− mice. To generate TKO mice, DKO mice were crossed with Cas9-KI mice and then were administered sgAnxa2 AAVs. Mice used as control are all littermates. All mouse experiments were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committees of Institute of Biophysics, Chinese Academy of Sciences.
ABX treatment
0.05 g vancomycin, 0.1 g ampicillin, 0.1 g kanamycin, and 0.1 g metronidazole were dissolved in every 100 ml sterile water. The mice were treated for 2 wk for suppression microbiota. Control mice were treated with sterile water. For validation of microbiota suppression, we extracted bacterial DNA from the intestinal contents, and 16S levels were tested.
Cell culture
Human HEK293T and Caco-2 cell lines were purchased from National Collection of Authenticated Cell Cultures. Cells were cultured in DMEM (Macgence) supplemented with 10% (for HEK293T) or 20% (for Caco-2) fetal bovine serum (Vivacell) and Penicillin-Streptomycin Solution (VivaCell) at 37°C under a humidified atmosphere with 5% CO2. All cell lines were well established and frequently checked by monitoring morphology and functionalities. All the cell lines were authenticated by STR analysis and were routinely tested to be mycoplasma free.
co-IP assay
Cnep1r1 from murine intestinal cDNA was cloned into p3×FLAG vector, Nlrp6 from murine intestinal cDNA was cloned into pcDNA4 vector, and Anxa2 from murine intestinal cDNA was tagged by HA and cloned into pcDNA4 vector. HEK293T cells were seeded in 6-well plate one day before transfection. For transfection of each well, 2 μl transfection regent, 1 μg pcDNA4-Nlrp6, and 1 μg p3×FLAG-Cnep1r1 or 1 μg pcDNA4-Anxa2 were added. Cells were harvested 48 h after transfection and then cells were lysed with RIPA Lysis Buffer (E121-01; GenStar) for 1 h on ice. Supernatants were collected by centrifugation (15,000 g, 15 min, 4°C) and incubated with indicated antibodies for 6 h at 4°C, followed by immunoprecipitation with 20 μl Protein A/G Magnetic Beads (MCE; HY-K0202). The precipitates were completely washed with RIPA Lysis Buffer and analyzed through western blot.
Recombinant protein expression and domain mapping
Full-length ERAdP and its four truncations (ΔN, ΔTM1, ΔTM2, and ΔC) were cloned into pGEX-6P-1 vector. These plasmids were transformed into Escherichia coli strain BL21 (DE3). DE3 clones were cultured to OD600 = 0.6, followed by induction with 0.2 mM IPTG at 16°C for 18 h. Bacteria were collected and lysed by high pressure crusher, followed by successive purifications through GST-tag Purification Resin (P2253; Beyotime). Then the immunoprecipitation was administrated as described.
Colocalization analysis
NLRP6-mCherry, ERAdP-EGFP, and ΔN-EGFP were cloned into pcDNA4 vector. Caco-2 cells were seeded in 35-mm Confocal Dish (YA0572; Solarbio) 1 day before transfection. For transfection by Lipofectamine 3000 of each well, 10 μl P3000, 7.5 μl Lipo3000, 1 μg pcDNA4-Nlrp6-mCherry, and 1 μg pcDNA4-Cnep1r1-EGFP were added. Colocalization was analyzed by confocal microscopy after transfection for 48 h.
Bacterial infection
S. Typhimurium was from Institute of Microbiology, Chinese Academy of Sciences. We established bacterial infection models following standard procedures. Specifically, mice aged 8–12 wk were fasted for 4 h and subsequently administered with 20 mg of streptomycin sulfate in 200 μl of PBS via oral gavage. After 24 h, the mice were again fasted for 4 h and then received 2 × 107 CFUs of S. Typhimurium through oral gavage with a size #9 bent lavage needle. 3 days after infection, ileal contents, PPs, spleens and livers were harvested from the mice, and CFUs were quantified. Body weight changes were monitored and recorded over the 7 days following infection. Survival rates were monitored and recorded over the 10 days following infection. Intestines were collected on the seventh day after infection and subsequently paraffin embedded for further histological analysis.
Organoid culture and treatment
Intestinal organoids were cultured according to previous method (Miyoshi and Stappenbeck, 2013). After removing fat and connective tissues, a 5-cm-long section of distal ileum was opened longitudinally and rinsed with cold PBS. Villi were carefully scraped away, and the tissue was washed vigorously in 20 ml cold PBS in a 50-ml tube for 1 min and repeated for at least four times to remove intestinal contents completely. The tissues were cut into small pieces (5 mm) and incubated in 10 ml collagenase solution (DMEM/F12 medium, containing 0.1% type I collagenase (LS004197; Worthington), 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 10 mM HEPES at 37°C for 20 min. Digestion was stopped by dilution with 20 ml cold PBS when around 70% of the crypts had been released. Digests were filtered through a 70-μm cell strainer, followed by 80 g centrifugation for 5 min. Crypts were embedded in 50 μl Matrigel (BD Biosciences) and seeded on 24-well plate. After polymerization, 500 μl crypt culture medium (#6000; STEMCELL) was added and refreshed every 3 days. The organoids were passaged weekly. Following removal of the medium, organoids were suspended in 1 ml cold PBS, mechanically dissociated, and pelleted by centrifugation (900 g, 5 min, 4°C). The pelleted organoids were embedded in fresh Matrigel and seeded on plate followed by addition of culture medium.
To analyze DCV biogenesis in organoids under different ligands treatment, organoids at day 3 of culture were treated with c-di-AMP disodium (10 μΜ, MCE, HY-12326A), cGAMP disodium (10 μΜ, HY-110385; MCE), LPS (10 μg/ml, S1732; Beyotime), poly (I:C) sodium (10 μg/ml, T12516; TargetMol), and MDP (2 μg/ml, HY-127090; MCE) for 16 h. After stimulation, organoids were harvested and stained with DAPI, UEA-1, and EpCAM. Then the organoids were examined using a Nikon Eclipse A1R+ microscope.
Immunofluorescence imaging
For tissue slices, longitudinally opened ileums were coiled into a "Swiss roll" and fixed with 4% PFA (Sigma-Aldrich) at room temperature (RT) for 2 h, followed with dehydration in 30% sucrose at 4°C overnight. The fixed tissues were then embedded in OCT (# 4583; Sakura Finetek USA) and sectioned into 10-µm thick slices. Slices were permeabilized with 1% Triton X-100 (# 9002-93-1; VWR Life Science), followed by overnight incubation with primary antibodies at 4°C and a subsequent 1-h incubation with secondary antibodies at RT. Following each incubation, the slices were washed three times with PBST for 5 min each. The samples were then mounted on slices using a mounting medium that includes anti-fading agents (S2100; Solarbio) to preserve fluorescence.
For imaging organoids, after removal of the medium, organoids were suspended in 1 ml of cold PBS and pelleted by centrifugation (900 g, 5 min, 4°C), resuspended by 4% cold PFA, and transferred to Nunc Lab-Tek (155411; Thermo Fisher Scientific) for 15-minute incubation at RT. Then the organoids were permeabilized with 1% Triton X-100, followed by overnight incubation with primary antibodies at 4°C and a subsequent 1-h incubation with secondary antibodies at RT.
For tyramide signal amplification (TSA) staining, longitudinally opened ileums were prepared as Swiss rolls and then fixed in 4% PFA. The fixed tissues were embedded in paraffin and sectioned into 4-µm slices. Following deparaffinization and rehydration, the slices underwent citrate antigen retrieval at 100°C for 15 min. The TSA staining was conducted by Four-color TSA multiplex fluorescence kit (10288100020; PANOVUE). It began with deparaffinization using fresh xylene immersion for 5 min, repeated three times, followed by rehydration through graded ethanol series (100%, 95%, and 70%, 5 min each) and multiple washes in deionized water. Optional steps included refixation in 10% neutral buffered formalin for 10 min to prevent tissue detachment and endogenous peroxidase blocking with 1–3% H2O2 at RT for 5–10 min. Antigen retrieval was performed by microwaving slides in retrieval buffer at 100°C for 15 min, followed by natural cooling to RT. After blocking with appropriate buffer for 10–30 min in a humidified chamber, primary antibody incubation was conducted with optimized concentration and duration, followed by HRP-conjugated secondary antibody application for 1 h. Fluorescent tyramide staining involved 10-min incubation with working solution at RT. For multiplex experiments, microwave elution enabled iterative staining cycles through repetition of steps above. Finally, slides were incubated with WGA-biotin and then with 405-Streptavidin (YS0078S; UE). Throughout the protocol, all steps were separated by three 2-min washes in PBST unless otherwise specified, and appropriate controls were included for validation.
The slices or organoids were imaged using Nikon Eclipse A1R+ microscope, and images were analyzed with Imaris 9 software.
Stimulation of Paneth cell secretion
1000 crypts from adult mice were incubated at 37°C for 30 min in 200 μl iPIPES (#P815722; Macklin) with or without secretory stimuli of 10 μM CCh (#HY-B1208; MCE). Crypts were deposited by centrifugation (900 g, 5 min, 4°C). Bactericidal activity was assessed by incubating 1 × 105 CFU of S. Typhimurium with 100 μl of supernatant at 37°C for 30 min. The viability of bacteria was determined by plating the contents of the bacteria–supernatant incubation mixture onto nutrient agar plates and determining CFUs after growth overnight. Crypt secretion of lysozyme was determined by ELISA Kit (#KTA3020; Abbkine) according to the instruction.
Visualization of Paneth cell DCV secretion and replenishment
Visualization of Paneth cell DCV secretion and replenishment assay was performed following previous method (Yokoi et al., 2019). Organoids at day 3 of culture were harvested and transferred onto an 8-well Lab-Tek Chambered Coverglass (#155411; Thermo Fisher Scientific) at a density of 100 organoids per well. The chamber was placed on ice for 5 min to allow the organoids to settle at the bottom. After polymerization of the Matrigel, pre-warmed enteric culture medium was added. To assess Paneth cell granule secreting and refilling, organoids were stimulated with 1 μM CCh for 10 min, and the culture medium containing CCh was washed out three times with pre-warmed advanced DMEM/F12, then fresh organoid culture medium were replaced. DIC images of Paneth cells, before and after stimulation, were captured using a confocal microscope (A1, Nikon) equipped with a 0.95 NA objective lens (CFI Apo LWD 40X WI λS; Nikon).
Electron microscopic analyses of DCVs in Paneth cells
Electron microscopic analysis was conducted according to previous method (Song et al., 2023). Sections of distal ileum from euthanized mice were rinsed with cold PBS and cut into 1–2-mm pieces. The tissues were then fixed overnight at 4°C in 0.1 M sodium phosphate buffer (pH 7.2) containing 2.5% glutaraldehyde and 2% PFA. Dehydration was conducted using an ethanol gradient, followed by infiltration and embedding with the SPI-Pon 812 Epoxy Embedding Kit. Sections were sliced using Leica Ultracut Ultramicrotome (Leica EM UC6) and stained with uranyl acetate and lead citrate. Digital images were captured using a Hitachi H-7650 transmission electron microscope at 80 kV. Halo regions of DCVs were measured from a total of 500 DCVs across three animals per genotype.
Immunohistochemical staining
Longitudinally opened ileums were prepared as Swiss rolls and then fixed in 4% PFA. The fixed tissues were embedded in paraffin and sectioned into 4-µm slices. Following deparaffinization and rehydration, the slices underwent citrate antigen retrieval at 100°C for 15 min. Slices were then stained using the Anti Rabbit IgG Detection Kit (E-IR-R215; Elabscience). The slices were treated with 3% hydrogen peroxide for 10 min to block endogenous peroxidase activity, followed by a 20-min incubation with blocking serum at RT. The slices were permeabilized with 1% Triton X-100, then incubated with primary antibodies overnight at 4°C and with secondary antibodies for 1 h at RT. After each incubation, the slices were washed three times with PBST for 5 min each. Freshly prepared DAB working solution was then applied to each slide. Microscopic examination revealed positive immunoreactivity, characterized by brownish-yellow or brown staining. The reaction was terminated by rinsing the sections with distilled water. Subsequently, the sections were counterstained, dehydrated, cleared, and mounted for further analysis. For quantifications, maximal crypt sections that displayed all Paneth cells within each crypt were selected and analyzed.
AB-PAS
AB- PAS staining was performed using a commercial kit (G1285; Solarbio). Slides were dewaxed to distilled water and then rinsed in distilled water for 2 min. Subsequently, the slides were stained with Alcian Blue staining solution for 10–20 min and treated with oxidant for 5–8 min. The slides were then soaked in Schiff reagent for 10–20 min and stained with hematoxylin solution for 1–2 min. The slides were differentiated using acidic differentiation solution for 2–5 s and blued with Scott Bluing solution for 3 min. After each step, the slides were washed two–three times with distilled water. Finally, the slides were dehydrated in a series of ethanol baths, cleared with xylene, and sealed with resinene.
Whole-mount immunofluorescence staining
After removing fat and connective tissues, ileums were longitudinally opened and then fixed in 4% PFA. Subsequently, tissues were cut into small pieces and incubated with primary antibodies for 2 h at RT. Following three washes, the tissues were incubated with Alexa Fluor–conjugated secondary antibodies and WGA for 2 h at RT. Finally, tissues were mounted on glass slides and examined using a Nikon Eclipse A1R+ with a 20× objective.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues using the TriPure RNA Isolation Kit (#T6331G; Solarbio) according to the manufacturer’s protocol. The RNA was then reverse transcribed into cDNA by using All-In-One 5X RT MasterMix (#G592; abm). qRT-PCR was conducted using SuperReal PreMix Plus (SYBR Green) (# FP205; TIANGEN) and ABI 7300 Real-Time PCR System with the specified primer pairs (Table S3). Relative expression levels were calculated and normalized to the 18S rRNA expression.
FISH
Ileum samples were collected without washing and fixed in methanol-Carnoy’s fixative (60% methanol, 30% chloroform, and 10% acetic acid), embedded in paraffin, and sectioned into 4-μm slices. Subsequently, sections were dewaxed, hydrated, and incubated in hybridization buffer (750 mM NaCl, 100 mM Tris-HCl [pH 7.4], 5 mM EDTA, 0.01% BSA, and 10% dextran sulfate) supplemented with a pan-bacterial Cy3-conjugated FISH probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) at 40°C for 16 h. Sections were washed three times for 10 min each in wash buffer (50 mM NaCl, 4 mM Tris-HCl [pH 7.4], and 0.02 mM EDTA) at 45°C, followed by counterstaining with DAPI and FITC-conjugated WGA. Finally, tissues were mounted on glass slides and examined using a Nikon Eclipse A1R+ with a 20× objective.
Bacterial 16S rRNA analysis
DNA extraction for 16S rRNA analysis was performed as previously described (Birchenough et al., 2016). For isolation of luminal contents from the colon, a 4-cm section of colon was cut open longitudinally, and a fecal pellet were extracted and weighed. For analysis of tissue-associated bacteria, the same tissue samples that were used for analysis of luminal contents were washed in ice-cold PBS, and then the whole tissue was weighed. Fecal and tissue DNAs were extracted using TIANamp Stool DNA Kit (DP328; TIANGEN) following the manufacturer’s protocol.
In short, the ratio of 16S DNA to total DNA was increased by limited cycle number (LCN) PCRs amplifying the whole 16S gene. 50 μl LCN PCRs were prepared using 0.2 μM universal forward primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′), 0.2 μM universal reverse primer 1492R (5′-CGGTTACCTTGTTACGACTT-3′), and 500 ng template DNA. Thermocycling conditions were: 1 cycle of 95 ̊C for 5 min; 16 cycles of 94 ̊C for 1 min, 55 ̊C for 1 min, 72 ̊C for 1.5 min; 1 cycle of 72 ̊C for 10 min. 16S standards (quantified E. coli 16S DNA), contamination controls, and no template controls were amplified at the same time as samples. Amplified samples, standards, and controls were then analyzed by quantitative PCR (qPCR) to determine the total number of 16S copies. Briefly, 20 μl qPCRs were prepared using 2 μl of LCN PCR amplifications as template, with 0.3 μM each of universal 16S primers 926F (5′-AAACTCAAAKGAATTGACGG-3′) and 1062R (5′-CTCACRRCACGAGCTGAC-3′).
3D fluorescence imaging of DCVs and quantification
3D fluorescence imaging–based quantification of positively stained DCVs was performed as previously described (Xiong et al., 2022). Mouse intestines were harvested, rinsed in cold PBS, and fixed with 4% PFA at RT for 2 h. The intestines were then sectioned into rings and washed thrice with 1% Triton X-100 in PBS for 30 min each. The samples underwent permeabilization and blocking in a solution containing 0.1% Tween-20, 0.5% Triton X-100, 0.1% deoxycholate, 0.1% NP40, and 10% BSA in PBS at 4°C for 24 h with gentle shaking. Primary antibodies were diluted in blocking buffer (0.5% Triton X-100 and 3% BSA in PBS), and the samples were incubated for 48 h at 4°C on a shaker, followed by four washes in washing buffer (1% Triton X-100 in PBS) at intervals of 30 min, 1, 6 h, and overnight. Secondary antibodies were also diluted in blocking buffer, and the samples were incubated for 1 day on a shaker at 4°C, followed by similar washing steps. The samples were post-fixed in 4% PFA at 4°C for 30 min. For immunofluorescence staining, the samples were sequentially incubated in 20%, 40%, 60%, 80%, and 100% (wt/vol) fructose solutions for 12 h each, followed by 24 h in SeeDB buffer (80.2% wt/wt fructose with 0.5% vol/vol α-thioglycerol) at RT. Cleared samples were then imaged using a Nikon A1R+ confocal scanner. Z-stack projections and the surfaces of the DCVs were analyzed using Imaris 9 software. In Imaris 9 software, confocal results were added surface fitted by channel stained with DCV marker. The regions of interest (DCVs) were selected, and in threshold settings, we chose background subtraction, in which diameter of largest sphere is 0.2 μm.
Data reproducibility and statistical analysis
Each experiment included a minimum of three replicates, with the exact numbers specified in the figure legends. Data analysis was performed using GraphPad Prism 8, employing two-tailed unpaired Student’s t tests for mouse statistical evaluation and Wilcoxon matched pairs signed rank test for clinical statistical evaluation. P values <0.05 were considered significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001); P > 0.05, nonsignificant (ns).
Clinical samples
Human-resected ileum tissues were obtained from the Wuxi People’s Hospital with informed consent, according to the Institutional Review Board approved protocol. Genomic DNA from ileal CD samples was extracted and transcribed as described previously. qRT-PCR was performed using the indicated primer pairs (Table S3). Relative expression levels were calculated and normalized to ACTB expression.
Online supplemental material
Fig. S1 shows c-di-AMP stimulation in vivo and expression of Cnep1r1. Fig. S2 shows that ERAdP conditional KO mice exhibit loss of DCVs and impaired antibacterial ability. Fig. S3 shows identification of NLRP6 and generation of NLRP6-deficient mice. Fig. S4 shows identification of ANXA2 and generation of ANXA2-deficient mice. Fig. S5 shows that DKO and TKO mice exhibit impaired antibacterial ability. Table S1 shows sgRNAs for CRISPR/Cas9-mediated gene KO in this study. Table S2 shows mouse genotyping primers used in this study. Table S3 shows qPCR primers used in this study. Table S4 shows FISH probes used in this study.
Data availability
All reagents made for this study are available from the corresponding author upon reasonable request under a standard material transfer agreement. RNAseq data have been deposited in the Gene Expression Omnibus database under accession no. GSE294515. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD063091 (Fig. 4 A) and PXD063090 (Fig. 5 A).
Acknowledgments
We thank Yihui Xu for technical support. We thank Xiang Shi, Xing Gao, Zixin Zhao, Haocheng Liang, Ming Hao, Jiajia Hou, and Xin Wen for animal procedures. We thank Xueke Tan for technical support. We thank Jing Li (Cnkingbio Company Ltd, Beijing, China) for technical assistance.
This work was supported by National Key R&D Program of China (2020YFA0803501); National Natural Science Foundation of China (82530038, 82130088, 82271785); Natural Science Foundation of Beijing (Z231100007223013 and 5222023); the Young Elite Scientist Sponsorship Program by CAST of China (2023QNRC001); the Postdoctoral grant (2020M680712 and 2022T150685), Strategic Priority Research Programs of the Chinese Academy of Sciences (XDB0570101), and Shanghai Municipal Science and Technology Major Project; and Basic Research Program of Jiangsu (BK20231146), Program of Jiangsu Branch of the National Clinical Research Center for Digestive Diseases.
Author contributions: Cunzhen Li: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, and writing—original draft, review, and editing. Zhen Xiong: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, and writing—original draft, review, and editing. Deyuan Kong: investigation. Yuwei Xu: formal analysis and methodology. Runyuan Wu: formal analysis and software. Peikang Zhang: software and validation. Ziqi Xiao: conceptualization and data curation. Hui Guo: resources and validation. Ying Du: data curation, funding acquisition, project administration, resources, software, and supervision. JinSong Li: methodology. Yun Chen: data curation, formal analysis, and resources. Qiang Zhan: formal analysis and resources,. Zusen Fan: conceptualization, funding acquisition, resources, supervision, and writing—original draft, review, and editing.
References
Author notes
C. Li, Z. Xiong, and D. Kong contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.