Regulatory T cells (Tregs) suppress the activation and subsequent effector functions of CD4 effector T cells (Teffs). However, molecular mechanisms that enforce Treg-mediated suppression in CD4 Teff are unclear. We found that Tregs suppressed activation-induced global protein synthesis in CD4 Teffs prior to cell division. We analyzed genome-wide changes in the transcriptome and translatome of activated CD4 Teffs. We show that mRNAs encoding for the protein synthesis machinery are regulated at the level of translation in activated CD4 Teffs by Tregs. Tregs suppressed global protein synthesis of CD4 Teffs by specifically inhibiting mRNAs of the translation machinery at the level of mTORC1-mediated translation control through concerted action of immunosuppressive cytokines IL-10 and TGFβ. Lastly, we found that the therapeutic targeting of protein synthesis with the RNA helicase eIF4A inhibitor rocaglamide A can alleviate inflammatory CD4 Teff activation caused by acute Treg depletion in vivo. These data show that peripheral tolerance is enforced by Tregs through mRNA translational control in CD4 Teffs.
Introduction
Most self-reactive T cells are eliminated in the thymus through the process of central tolerance. However, a small percentage of cells escape to the periphery, where they have the potential to promote autoimmunity and inflammatory responses. These cells are normally held in check by a population of CD4 T cells referred to as regulatory T cells (Tregs). Tregs are essential to maintain immune homeostasis, and the transcription factor FOXP3 has been shown to be central to the development and function of Tregs. Mutations in the FOXP3 gene in mice and human patients with IPEX (immunodysregulation polyendocrinopathy enteropathy X-linked) syndrome drive the development of a common set of autoimmune symptoms (Bennett et al., 2001; Ochs et al., 2007; Patel, 2001; Wildin et al., 2001). Mutations in the FOXP3 gene and the autoimmune phenotype are linked to a loss of Tregs or their function (Bacchetta et al., 2018; Ochs et al., 2007). Tregs have the ability to potently suppress CD4 effector T cells (Teff) either directly or through the modulation of APCs (mainly dendritic cells) to ultimately suppress activation, proliferation, and subsequent effector functions of Teffs (Josefowicz et al., 2012; Tang and Bluestone, 2008; Vignali et al., 2008). Several mechanisms have been proposed for Treg-mediated suppression, including the release of suppressive cytokines (e.g., TGFβ, IL-10, IL-35) and expression of inhibitory receptors (e.g., CTLA-4, PD-1, TIGIT; Shevach, 2009; Sojka et al., 2008). Although a block in proliferation and Teff function have been the hallmarks of Treg mediated suppression, the molecular changes in target CD4 Teffs following Treg encounter remains unclear. This is especially true for the first 24 h prior to the onset of Teff proliferation, when the biosynthetic capacity of the cell is greatly expanded (So et al., 2016; Wang et al., 2011).
Upon activation, resting T cells undergo a rapid biosynthetic and metabolic reprogramming in preparation for cell division (Manfrini et al., 2020; Ricciardi et al., 2018; Wang et al., 2011; Wolf et al., 2020). Included in this reprogramming is an increase in translational activity and capacity (Araki et al., 2017; Bjur et al., 2013). In this study, we show that Tregs suppress activation of CD4 Teffs by enforcing a global inhibition of mRNA translation. We assessed the genome-wide changes in transcriptome and translatome in activated CD4 Teffs and identified translation control of mRNAs encoding components of the protein synthesis machinery. In the first 24 h of CD4 Teff activation, a set of mRNAs encoding proteins involved in the translational machinery are shifted to polysomes, with no concomitant changes in their transcription. We found that Tregs specifically inhibit the shift of these mRNAs to polysomes by suppressing mammalian target of rapamycin complex 1 (mTORC1) signaling through two immunosuppressive cytokines IL-10 and TGFβ. In support of these findings, we provide new evidence that with direct targeting of protein synthesis using rocaglamide A (RocA), an RNA helicase eIF4A inhibitor, inflammatory CD4 Teff activation caused by in vivo Treg loss can be alleviated. In summary, we provide a novel mechanism of Treg-mediated suppression of CD4 Teff activation through the inhibition of mRNAs encoding protein synthesis machinery at the post-transcriptional level and that this biological mechanism can be therapeutically targeted using small molecule inhibitors.
Results
Tregs control the protein synthetic capacity of activated CD4 Teffs
The 24–48 h following CD4 Teff activation is critical for subsequent proliferation and expansion. This is a period during which cellular biomass is accumulated through expansion of global protein synthetic capacity in preparation for cell division. We reasoned that this period could be a target for Treg-mediated suppression in order to inhibit cell activation prior to proliferation. Overall protein synthesis rate can be quantified at the single-cell level by pulsing cells with the tRNA-analog puromycin (PMY) and intracellular staining for PMY. We co-cultured CD4 Teff cells (CD4+CD25−) and congenically marked Tregs (CD4+Foxp3+) at varying ratios with anti-CD3/CD28 coated beads and pulsed the culture with PMY. CD4 Teff cells co-cultured with Tregs exhibited marked inhibition of proliferation in a dose-dependent manner (Fig. S1 A). Interestingly, Teff cells co-cultured with Tregs clearly showed significantly less PMY incorporation in a Treg dose–dependent manner before the onset of proliferation (Fig. 1 A). The pattern of suppression was not bimodal, indicating that the protein synthesis rate of all responding conventional T cells cells was modulated by Tregs, less completely than upon cycloheximide (CHX) treatment (Fig. 1 B). Since anti-CD3/CD28 beads were used to activate both populations of cells in the co-culture system, the downregulation of protein synthesis by Tregs is independent of APC. The Treg-mediated translational inhibition was observed as early as 6 h after activation, well before any metabolic changes occur in T cells (Fig. 1 C). Suppression was primarily due to early prevention of T cell activation as we found no difference in the ability of CD4 Teffs to downregulate CD62L with or without Tregs (Fig. S1 B). Thus, the suppression of global protein synthesis in CD4 Teffs by Tregs could not be attributed to dampening or cold inhibition of general T cell activation. To test whether similar regulatory pathways were operative in human T cells, we established ex vivo Treg suppression assays using PMY incorporation as the readout. We used in vitro–expanded Tregs from a single donor, and conventional T cells (defined as CD4+CD45RA+CD127+CD25−) from five individual healthy donors. We found that 24 h of stimulation resulted in a significant increase in PMY staining, both in percentage of cells labeled and the mean fluorescence intensity (MFI) of PMY staining. In the cultures containing Tregs, PMY incorporation was significantly reduced, both in total incorporation and in the percentage of cells that incorporated PMY (Fig. 1 D). These data demonstrate that Treg-mediated inhibition of activation-induced translation is a conserved function.
Tregs do not suppress TCR-induced upregulation of expression of CD62L and TCR signaling in CD4 Teffs . (A) In vitro Treg suppression assay using different doses of purified Foxp3-YFP+ Tregs and co-culturing with CFSE-labeled YFP−CD25−CD4+CD44lowCD62L+ naive CD4 T cells as CD4 Teffs. Data are presented in two ways. Numerating the undivided CFSE peak to show percent undivided cells. Percent suppression was calculated by measuring cells that have divided at least once. (B) CD62L expression was measured in unstimulated and stimulated CD4 Teffs with or without Tregs at 24 h after stimulation. (C) Bulk CD4 T cells were purified on day 2 from control C57BL/6 mice and Foxp3DTR mice treated with DT for 2 d consecutively. Cells were labeled with CFSE and activated using anti-CD3/CD28 beads. Cell size (forward scatter) and proliferation (CFSE dilution) was measured at 72 h after stimulation. (D) CD4 Teffs stimulated in the absence and presence of Tregs were identified by congenic markers and intracellular signaling molecules were assessed by phospho-flow cytometry. (E) CD4 Teffs from Nur77-GFP mice labeled with CellTraceViolet were either activated with anti-CD3/CD28 beads alone or with titrating amounts of sorted Tregs. Some conditions received indicated concentrations of RocA for eIF4A inhibition. Proximal TCR signaling was assessed using GFP in CD4 Teffs by gating on CellTraceViolet+ population. Each datapoint represents a biological replicate (CD4 Teffs from individual Nur77-GFP mice). Data are representative of at least two independent experiments; **, P < 0.01; ***, P < 0.001.
Tregs do not suppress TCR-induced upregulation of expression of CD62L and TCR signaling in CD4 Teffs . (A) In vitro Treg suppression assay using different doses of purified Foxp3-YFP+ Tregs and co-culturing with CFSE-labeled YFP−CD25−CD4+CD44lowCD62L+ naive CD4 T cells as CD4 Teffs. Data are presented in two ways. Numerating the undivided CFSE peak to show percent undivided cells. Percent suppression was calculated by measuring cells that have divided at least once. (B) CD62L expression was measured in unstimulated and stimulated CD4 Teffs with or without Tregs at 24 h after stimulation. (C) Bulk CD4 T cells were purified on day 2 from control C57BL/6 mice and Foxp3DTR mice treated with DT for 2 d consecutively. Cells were labeled with CFSE and activated using anti-CD3/CD28 beads. Cell size (forward scatter) and proliferation (CFSE dilution) was measured at 72 h after stimulation. (D) CD4 Teffs stimulated in the absence and presence of Tregs were identified by congenic markers and intracellular signaling molecules were assessed by phospho-flow cytometry. (E) CD4 Teffs from Nur77-GFP mice labeled with CellTraceViolet were either activated with anti-CD3/CD28 beads alone or with titrating amounts of sorted Tregs. Some conditions received indicated concentrations of RocA for eIF4A inhibition. Proximal TCR signaling was assessed using GFP in CD4 Teffs by gating on CellTraceViolet+ population. Each datapoint represents a biological replicate (CD4 Teffs from individual Nur77-GFP mice). Data are representative of at least two independent experiments; **, P < 0.01; ***, P < 0.001.
Tregs suppress global protein synthesis in CD4 Teffs. (A) Purified naive CD4 T cells from Foxp3YFP-Cre mice (CD4+YFP−) were stimulated with equal number of anti-CD3/CD28 beads with or without the indicated ratio of Foxp3+Tregs (CD4+YFP+; Treg:Teff = 1:2). For cultures with CD4 Teffs only, cell number was doubled to match total cell number in the culture (2×). Cells were pulsed with PMY for 15 min before harvest and intracellular staining for incorporated PMY performed using a fluorophore-conjugated anti-PMY antibody. (B) Purified naive CD4 T cells were stimulated with anti-CD3/CD28 beads with or without FACS-sorted Foxp3-YFP+ Tregs from Foxp3YFP-Cre mice (Treg:Teff = 1:2) for 24 h, followed by analysis of PMY incorporation as in A. PMY signal and MFI were quantified. CHX was added to 24 h stimulated CD4 Teffs 5 min prior to PMY pulse to act as a negative control and set the baseline protein synthesis signal. One-way ANOVA was applied. (C) Purified naive CD4 T cells from Foxp3YFP-Cre mice as CD4 Teffs were stimulated with anti-CD3/CD28 beads with or without the indicated ratio of FACS sorted Foxp3-YFP+ Tregs from Foxp3YFP-Cre mice (Treg:Teff = 1:1) for 6, 12, and 24 h, followed by analysis of PMY incorporation as in A. (D) In vitro–expanded Tregs from a single donor, and Teff (defined as CD4+CD25−CD45RA+) from five individual healthy donors were used for the human Treg suppression assay. Cells were co-cultured and stimulated with anti-CD3/CD28 beads for 24 h, and PMY staining was performed as in A. The percent suppression of protein synthesis was calculated as described in materials and methods. (E) Foxp3DTR mice were injected with DT for 2 d consecutively, and splenocytes were harvested on day 2. Protein synthesis in CD4 T cells was measured by ex vivo PMY pulsing of splenocytes and gating on CD4 T cells. (F) CD4 Teffs stimulated in the absence and presence of Tregs were identified by congenic markers and intracellular signaling molecules were assessed by phospho-flow cytometry. (G) Purified naive CD4 T cells were treated with 5 nM rapamycin and 50 nM MLN0128 for 30 min prior to stimulation for indicated samples. The cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of iTregs (Treg:Teff = 1:1) for 24 h, subjected for analyzing PMY incorporation and phosphorylation of S6 by flow cytometry. One-way ANOVA was applied for all comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All experiments were repeated at least twice.
Tregs suppress global protein synthesis in CD4 Teffs. (A) Purified naive CD4 T cells from Foxp3YFP-Cre mice (CD4+YFP−) were stimulated with equal number of anti-CD3/CD28 beads with or without the indicated ratio of Foxp3+Tregs (CD4+YFP+; Treg:Teff = 1:2). For cultures with CD4 Teffs only, cell number was doubled to match total cell number in the culture (2×). Cells were pulsed with PMY for 15 min before harvest and intracellular staining for incorporated PMY performed using a fluorophore-conjugated anti-PMY antibody. (B) Purified naive CD4 T cells were stimulated with anti-CD3/CD28 beads with or without FACS-sorted Foxp3-YFP+ Tregs from Foxp3YFP-Cre mice (Treg:Teff = 1:2) for 24 h, followed by analysis of PMY incorporation as in A. PMY signal and MFI were quantified. CHX was added to 24 h stimulated CD4 Teffs 5 min prior to PMY pulse to act as a negative control and set the baseline protein synthesis signal. One-way ANOVA was applied. (C) Purified naive CD4 T cells from Foxp3YFP-Cre mice as CD4 Teffs were stimulated with anti-CD3/CD28 beads with or without the indicated ratio of FACS sorted Foxp3-YFP+ Tregs from Foxp3YFP-Cre mice (Treg:Teff = 1:1) for 6, 12, and 24 h, followed by analysis of PMY incorporation as in A. (D) In vitro–expanded Tregs from a single donor, and Teff (defined as CD4+CD25−CD45RA+) from five individual healthy donors were used for the human Treg suppression assay. Cells were co-cultured and stimulated with anti-CD3/CD28 beads for 24 h, and PMY staining was performed as in A. The percent suppression of protein synthesis was calculated as described in materials and methods. (E) Foxp3DTR mice were injected with DT for 2 d consecutively, and splenocytes were harvested on day 2. Protein synthesis in CD4 T cells was measured by ex vivo PMY pulsing of splenocytes and gating on CD4 T cells. (F) CD4 Teffs stimulated in the absence and presence of Tregs were identified by congenic markers and intracellular signaling molecules were assessed by phospho-flow cytometry. (G) Purified naive CD4 T cells were treated with 5 nM rapamycin and 50 nM MLN0128 for 30 min prior to stimulation for indicated samples. The cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of iTregs (Treg:Teff = 1:1) for 24 h, subjected for analyzing PMY incorporation and phosphorylation of S6 by flow cytometry. One-way ANOVA was applied for all comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. All experiments were repeated at least twice.
Next, to assess the role of Tregs in controlling protein synthesis in CD4 T cells in vivo, we acutely depleted Tregs through diphtheria toxin (DT) treatment of Foxp3DTR mice (Kim et al., 2007). Within 3 d after initial DT-induced depletion of Tregs (two consecutive DT injections on days 0 and 1), we observed rapid appearance of a CD4 T cell population with significantly elevated incorporation of PMY ex vivo compared to the PBS-treated control mice, suggesting activation of the autoreactive CD4 T cell pool in the periphery (Fig. 1 E). When CD4 T cells from spleen and lymph nodes of DT-treated Foxp3DTR mice were purified and stimulated ex vivo, they proliferated with faster kinetics and were significantly larger in size, indicating an aberrantly enhanced protein synthesis capacity in Teffs activated in vivo attributed to Treg loss (Fig. S1 C). These data suggest that Tregs are both sufficient and necessary to suppress the rapid upregulation of protein synthesis in activated CD4 T cells both in vitro and in vivo.
To uncover the underlying mechanism of Treg-mediated translational inhibition, we examined signaling pathways downstream of TCR stimulation in activated CD4 T cells. Specifically, we examined the mTOR signaling pathway as it has been shown to be critical for coordinating cell growth and proliferation in lymphocytes through the eukaryotic translation initiation factor 4E (eIF4E) in translation initiation (Wardman et al., 2016). mTOR exists in two multi-protein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2; Jhanwar-Uniyal et al., 2019; Zoncu et al., 2011). To assess mTORC1 signaling, we examined phosphorylation of ribosomal protein S6 (rpS6: S240/244) and eIF4E-binding proteins (p4EBP1/2: T37/46) at their respective mTORC1-specific phosphosites in activated CD4 T cells. mTORC2 signaling was assessed by phosphorylation of AKT at S473. As expected, both mTORC1 and mTORC2 signaling increased in activated CD4 T cells. Strikingly, Tregs significantly suppressed mTORC1 signaling (rpS6 and p4EBP1/2; Fig. 1 F). However, mTORC2 signaling (AKT S473) remained intact, as did the PDK1- and PI3K-dependent phosphosite T308, which is more proximal to TCR engagement (Fig. S1 D). Using Nur77-GFP reporter Teffs, we see no differences in CD4 Teffs alone or co-cultured with Tregs, indicating no change in proximal TCR signaling (Fig. S1 E). Furthermore, the treatment of CD4 Teffs with rapamycin or MLN0128, mTOR inhibitors, suppressed phosphorylation of rpS6 and PMY incorporation to the same extent as Treg-mediated suppression (Fig. 1 G). These data suggest that Treg-mediated translational inhibition in CD4 Teffs is associated with the reduction of mTORC1 signaling in CD4 Teffs. These findings are consistent with previous data showing that genetic or chemical inhibition of mTORC1 significantly inhibits lymphocyte proliferation in a 4EBP/eIF4E-dependent manner to control translation initiation in various cell types, including lymphocytes (So et al., 2016; Thoreen et al., 2012).
Development of a simple polysome efficient extraction and distribution (SPEED) technique to identify mRNAs that are differentially translated
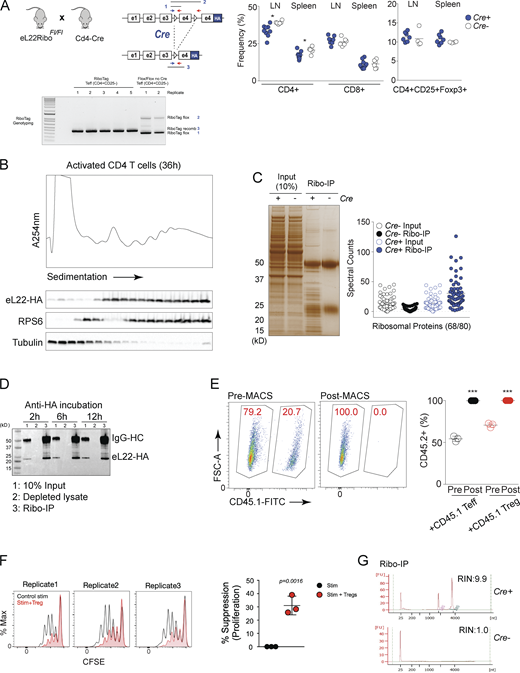
Based on our observation that Tregs suppress protein synthesis in CD4 Teffs, we investigated whether Tregs affect global mRNA translation using genome-wide approaches to interrogate changes in both the transcriptome and translatome of CD4 Teff cells. First, we used the RiboTag system to capture translatome changes in CD4 T cells. This system utilizes an HA-epitope–tagged ribosomal protein large subunit L22 (eL22) that allows for immunoprecipitation of proteins and mRNA associated with ribosomes (Ribo-IP; Sanz et al., 2009). Mature CD4 Teff cells with an HA-epitope tag on the ribosomal large subunit protein L22 (eL22; known as RiboTag) were generated using Cd4-Cre mice (eL22Ribof/fCd4Cre: T-Ribo) where exon 4 of eL22 is substituted with an identical exon containing HA epitope sequences (Fig. S2 A). No obvious defects were observed in steady-state mature T cell frequencies in the spleen and lymph nodes with only a mild reduction in CD4+ T cell frequency (Fig. S2 A). Purified CD4 T cells from T-Ribo (Cd4−Cre+) expressed eL22-HA protein that co-sedimented with polysome fractions upon activation indicating normal ribosome incorporation (Fig. S2 B). Immunoprecipitation of eL22-HA (termed Ribo-IP) followed by mass spectrometry also identified most ribosomal proteins (RPs; 68/80) suggesting eL22-HA as a bona fide RP assembled as an intact ribosome (Fig. S2 C). Ribo-IP was highly efficient with complete depletion of eL22-HA with as little as 2 h of antibody incubation (Fig. S2 D).
Preparation of the samples from RiboTag mice. (A) Genomic DNA was isolated from CD4+CD25− Teff cells from Cd4Cre-positive and -negative animals to assess RiboTag allele recombination efficiency. Three primers were used to amplify (1) RiboTag flox-small, (2) RiboTag flox-large, and (3) RiboTag recombined amplicons to assess recombination efficiency. In purified CD4+CD25− Teff cells from Cd4Cre-positive animals, we detect near 100% recombination efficiency. Non-recombined RiboTag mice (Rpl22Tag) and CMV-Cre mediated whole-body recombined RiboTag mice (Rpl22HA/+) were compared for the thymic cellularity and splenic CD4, CD8 T cells and Foxp3+CD25+ Tregs. (B) Bulk CD4 T cells from T-Ribo mice were activated and subjected for fractionation on a 15–60% sucrose gradient. Proteins were precipitated from sucrose fractions and immunoblotted using anti-HA, anti-RPS6, and anti-tubulin. (C) Samples from RPL22ΔT/+ show highly specific enrichment for RPs (9–52 kD range) as well as high molecular weight proteins that represent ribosome- associated proteins (confirmed by MS analysis). Absolute spectral counts for all RPs identified (68 out of 80). (D) Purified CD4+ T cell samples from RPL22ΔT/+ were subjected for Ribo-IP along with control T cells to show background immunoprecipitation. (E) CD45.1 congenically labeled Foxp3-YFP+ Tregs and Foxp3-YFP− Teff cells were co-cultured with CD45.2 CD4 Teff cells from T-Ribo mice. After 24 h of stimulation, CD45.1+ cells were positively selected using MACS using CD45.1-FITC microbeads. Pre- and post-MACS CD45.2+ percentage is plotted for all experiments. (F) Suppression of CD4 Teff cells by Tregs was measured using CFSE 72 h after stimulation. (G) Specificity of Ribo-IP was tested using CD4 Teff cells from T-Ribo mice and Cre− control mice. After Ribo-IP, captured total RNA was measured using Bioanalyzer (RNA pico kit). *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData FS2.
Preparation of the samples from RiboTag mice. (A) Genomic DNA was isolated from CD4+CD25− Teff cells from Cd4Cre-positive and -negative animals to assess RiboTag allele recombination efficiency. Three primers were used to amplify (1) RiboTag flox-small, (2) RiboTag flox-large, and (3) RiboTag recombined amplicons to assess recombination efficiency. In purified CD4+CD25− Teff cells from Cd4Cre-positive animals, we detect near 100% recombination efficiency. Non-recombined RiboTag mice (Rpl22Tag) and CMV-Cre mediated whole-body recombined RiboTag mice (Rpl22HA/+) were compared for the thymic cellularity and splenic CD4, CD8 T cells and Foxp3+CD25+ Tregs. (B) Bulk CD4 T cells from T-Ribo mice were activated and subjected for fractionation on a 15–60% sucrose gradient. Proteins were precipitated from sucrose fractions and immunoblotted using anti-HA, anti-RPS6, and anti-tubulin. (C) Samples from RPL22ΔT/+ show highly specific enrichment for RPs (9–52 kD range) as well as high molecular weight proteins that represent ribosome- associated proteins (confirmed by MS analysis). Absolute spectral counts for all RPs identified (68 out of 80). (D) Purified CD4+ T cell samples from RPL22ΔT/+ were subjected for Ribo-IP along with control T cells to show background immunoprecipitation. (E) CD45.1 congenically labeled Foxp3-YFP+ Tregs and Foxp3-YFP− Teff cells were co-cultured with CD45.2 CD4 Teff cells from T-Ribo mice. After 24 h of stimulation, CD45.1+ cells were positively selected using MACS using CD45.1-FITC microbeads. Pre- and post-MACS CD45.2+ percentage is plotted for all experiments. (F) Suppression of CD4 Teff cells by Tregs was measured using CFSE 72 h after stimulation. (G) Specificity of Ribo-IP was tested using CD4 Teff cells from T-Ribo mice and Cre− control mice. After Ribo-IP, captured total RNA was measured using Bioanalyzer (RNA pico kit). *, P < 0.05; ***, P < 0.001. Source data are available for this figure: SourceData FS2.
The RiboTag system allowed profiling of both the steady-state mRNA levels and the ribosome-bound mRNA levels from the same cytosolic lysate from CD4 Teff cells. CD4 Teff cells from T-Ribo mice were stimulated alone or co-cultured with isolated Tregs and stimulated with anti-CD3/CD28 coated beads for 24 h, a timepoint before the onset of proliferation. For resting conditions, we used CD4 T cells cultured in IL-7, which promotes survival but not activation, to match the timepoint for lysate preparation. Cytoplasmic lysates were prepared from pure CD4 Teffs after magnetic-activated cell sorting (MACS) to remove congenically marked CD4 Teff cells or Tregs where we obtained >99.8% purity (Fig. S2 E). A small aliquot from the lysate (10%) was taken for total input RNA extraction and Ribo-IP was performed on the remaining lysates to extract ribosome-bound mRNA. We also confirmed that Tregs faithfully suppressed CD4 Teff cell proliferation 3 d after stimulation compared to control stimulated Teff cells (Fig. S2 F). Ribosome-bound mRNA and total RNA input for each condition were subjected to RNA sequencing (RNA-seq). mRNA pulldown by Ribo-IP was highly specific as control-activated CD4 Teff cells from Cre− mice (n = 2) showed negligible traces of RNA based on Bioanalyzer analysis (Fig. S2 G). Principal component analysis separated the samples by the first component (64.1% of variance) according to their activation status (IL7 rested vs. activated) and suppressed CD4 Teff cells separated from activated CD4 Teff cells by the second component (5% of variance; Fig. S3, A and B). Importantly, the total RNA input samples were clearly separated from their corresponding Ribo-IP samples by the second component indicating discrepancies in the total RNA input and its corresponding Ribo-IP mRNA. mRNAs of the mitochondrial DNA are normally translated by mitoribosomes and not eL22 containing cytosolic ribosomes. In all conditions, we observed a significant de-enrichment of mitochondrial-derived mRNAs in the Ribo-IP samples, internally validating that the Ribo-IP faithfully captures mRNAs only bound to the cytosolic ribosome (Fig. S3 C).
Treg modulated translatome in CD4 Teff cells using the RiboTag approach. (A) Schematic of the translatome capture using the RiboTag for RNA-seq. (B) Principal component analysis of Ribo-IP RNA samples and total input RNA samples. (C) Heatmap of log2TMM value of mitochondrial genes and their enrichment in Ribo-IP samples. Mitochondrial genes are translated by mitoribosomes and are completely de-enriched in Ribo-IP, which further validates Ribo-IP capturing cytosolic ribosomes. (D) Log2 fold-change of gene expression in the input RNA is plotted against log2 fold-change of translational efficiency between samples. The colors annotate genes based on the significance of their input differential expression or translational efficiency: red if the gene changes in translational efficiency but not expression; blue if the gene changes in expression but not translational efficiency; purple if the gene changes in both; and gray if the gene changes in neither. (E) RL was calculated for each condition and mRNAs with significant changes in RL (Ribo-IP signal/total input signal) was analyzed by calculating Ribo-IP mRNA signals to its corresponding total input mRNA signal in each of the condition. Density of RLs is plotted as a histogram with low RL mRNAs (number in red bracket) and high RL mRNAs (number in blue bracket). 1.5-fold cutoff was used to highlight mRNAs with significant RLs (red open bars) in relation to non-significant mRNAs (gray filled bars). Number of mRNAs with differential RL are quantified as a histogram further supporting the idea that the RiboTag approach mainly discovers ribosome de-enriched mRNAs (negative RL) as all ribosomes are captured. (F) Venn diagram shows that mRNAs with differential RL for each condition is largely unique with minimal overlap. One-way ANOVA was applied was used for all comparisons between two groups.
Treg modulated translatome in CD4 Teff cells using the RiboTag approach. (A) Schematic of the translatome capture using the RiboTag for RNA-seq. (B) Principal component analysis of Ribo-IP RNA samples and total input RNA samples. (C) Heatmap of log2TMM value of mitochondrial genes and their enrichment in Ribo-IP samples. Mitochondrial genes are translated by mitoribosomes and are completely de-enriched in Ribo-IP, which further validates Ribo-IP capturing cytosolic ribosomes. (D) Log2 fold-change of gene expression in the input RNA is plotted against log2 fold-change of translational efficiency between samples. The colors annotate genes based on the significance of their input differential expression or translational efficiency: red if the gene changes in translational efficiency but not expression; blue if the gene changes in expression but not translational efficiency; purple if the gene changes in both; and gray if the gene changes in neither. (E) RL was calculated for each condition and mRNAs with significant changes in RL (Ribo-IP signal/total input signal) was analyzed by calculating Ribo-IP mRNA signals to its corresponding total input mRNA signal in each of the condition. Density of RLs is plotted as a histogram with low RL mRNAs (number in red bracket) and high RL mRNAs (number in blue bracket). 1.5-fold cutoff was used to highlight mRNAs with significant RLs (red open bars) in relation to non-significant mRNAs (gray filled bars). Number of mRNAs with differential RL are quantified as a histogram further supporting the idea that the RiboTag approach mainly discovers ribosome de-enriched mRNAs (negative RL) as all ribosomes are captured. (F) Venn diagram shows that mRNAs with differential RL for each condition is largely unique with minimal overlap. One-way ANOVA was applied was used for all comparisons between two groups.
As total input samples were distinct from their corresponding Ribo-IP samples, we measured “ribosome load” (RL) by calculating the ratio of Ribo-IP signal to its total RNA input signal. In all conditions, RL values were significantly biased toward the negative direction indicating the majority of mRNAs have less ribosome occupancy compared to their total input levels (Fig. S3 E and Table S1). We attribute this bias to the fact that Ribo-IP signals represent mRNAs bound to all ribosomes, even including monosomes, thereby efficiently distinguishing free mRNAs from ribosome-bound mRNAs but not necessarily resolving polysome-bound mRNAs. One question arising from this is whether mRNAs with low RLs found in all conditions have low translation efficiencies due to their intrinsic properties in T cells or whether they are dynamically regulated depending on the stimuli. We found very low overlap (<8%) between the differential RL mRNAs in all our conditions supporting the notion that the stimuli dictated the outcome of ribosome binding of a given mRNA in CD4 Teff cells (Fig. S3 F). These data show that the RiboTag did not distinguish between mRNAs with low ribosome occupancy (monosomes) from those associated with polysomes, therefore not an optimal system to measure translation efficiency (TE).
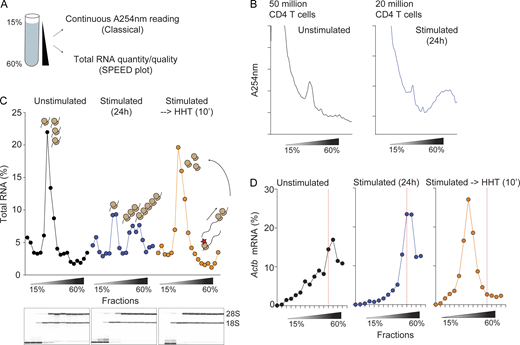
The transition of mRNAs from monosome to polysome and back is a critical aspect of translation control. To quantitatively resolve and distinguish mRNAs bound to polysomes from monosome-associated mRNAs, we optimized the classical polysome profiling approach for low input cytosolic lysates suitable for primary immune cells (Fig. 2 A). We reasoned that by assessing the quantity and quality of total RNA extracted from each fraction, we could determine the ribosome positions since total RNA is mainly composed of ribosomal RNA. Using lysates prepared from as few as 500,000 to 1 million activated CD4 Teff cells, we found that total RNA extracted from each fraction and plotted as a percent distribution plot closely resembled a classical A254 nm polysome trace obtained using >20 million activated CD4 T cells (Fig. 2 B). We have termed this a “simple polysome efficient extraction and distribution” (SPEED) plot to distinguish it from the traditional polysome traces requiring greater cellular input (Fig. 2 C). Furthermore, qualitative analysis of the extracted RNA using Bioanalyzer gave us information as to the position of the intact 80S monosome in the gradient (Fig. 2 C). The SPEED plots generated from unstimulated CD4 T cells showed enrichment of most of the total RNA in the monosome fraction (Fig. 2 C), confirming our observations that CD4 T cells prior to activation have low translational activity (Fig. 1 A). Upon stimulation, nearly 50% of the monosomes shifted towards the heavier sucrose fractions, indicating the assembly of polysomes and increased protein synthesis (Fig. 2 C). To ensure SPEED plots faithfully represent the mRNA translational status of a cell, we took advantage of the translation initiation inhibitor homoharringtonine (HHT), which only interferes with initiating ribosomes and allows elongating ribosomes to run-off (Fresno et al., 1977). Activated CD4 Teffs were treated with HHT for 10 min to allow run-off elongation of ribosomes before cytosolic lysate preparation. Remarkably, the SPEED plot from HHT-treated-activated CD4 Teffs resembled unstimulated CD4 T cells, with the majority of ribosomes enriched in the monosome fraction (Fig. 2 C). Lastly, the distribution of β-actin (Actb) mRNA was analyzed using quantitative PCR (qPCR) from each fraction. We chose Actb as it is routinely used as a housekeeping control mRNA as it has no apparent cis-regulatory sequence motif in its 5′ untranslated region (UTR) and is highly translated. Despite low polysome levels in unstimulated CD4 Teffs, Actb mRNA was abundantly enriched in the polysome fractions. Activated CD4 Teffs also translated Actb mRNA with high efficiency. As expected for a highly translated mRNA, HHT treatment led to a complete shift in Actb mRNA toward the lighter sucrose fractions, indicating successful ribosome run-off (Fig. 2 D). In summary, our SPEED technique faithfully captured the translational status of cellular lysates from low biological input, making it ideal to assess the translatome of primary immune cells, and most importantly, of Treg suppressed CD4 Teffs.
SPEED analysis as a novel polysome profiling approach. (A) Polysome profiling approach using sucrose gradients to physically stratify cellular cytosolic lysates. Classically, continuous absorbance at 254 nm (A254 nm) is used to assess monosome and polysome positions. SPEED utilizes analysis of the quantity and quality of total RNA from each fraction, making it amenable for ultra-low biological input that is below the detection limit of A254 nm reading. (B) Indicated numbers of bulk CD4 T cells were left unstimulated or stimulated with equal number of anti-CD3/CD28 beads for 24 h and subjected for classical polysome profiling using A254 nm reading to obtain polysome traces. (C) Same samples from B but equivalent to 500,000–1 million activated CD4 T cells and 2–3 million unstimulated CD4 T cells were subjected for polysome fractionation. No A254 nm traces were obtained. Total RNA was extracted from each sucrose fractions and quantified to plot total RNA percent distribution across fractions. Total RNA quality was assessed using Bioanalyzer (bottom Bioanalyzer results; only fractions #1–11 were analyzed since each RNA pico chip can analyze 11 samples at a time). (D) Equal volume of RNA from each fraction was reverse transcribed into cDNA and Actb mRNA levels were quantified using qPCR. The percent Actb mRNA across fractions was quantified and plotted. Red dashed line indicates fraction #12 in each sample. Source data are available for this figure: SourceData F2.
SPEED analysis as a novel polysome profiling approach. (A) Polysome profiling approach using sucrose gradients to physically stratify cellular cytosolic lysates. Classically, continuous absorbance at 254 nm (A254 nm) is used to assess monosome and polysome positions. SPEED utilizes analysis of the quantity and quality of total RNA from each fraction, making it amenable for ultra-low biological input that is below the detection limit of A254 nm reading. (B) Indicated numbers of bulk CD4 T cells were left unstimulated or stimulated with equal number of anti-CD3/CD28 beads for 24 h and subjected for classical polysome profiling using A254 nm reading to obtain polysome traces. (C) Same samples from B but equivalent to 500,000–1 million activated CD4 T cells and 2–3 million unstimulated CD4 T cells were subjected for polysome fractionation. No A254 nm traces were obtained. Total RNA was extracted from each sucrose fractions and quantified to plot total RNA percent distribution across fractions. Total RNA quality was assessed using Bioanalyzer (bottom Bioanalyzer results; only fractions #1–11 were analyzed since each RNA pico chip can analyze 11 samples at a time). (D) Equal volume of RNA from each fraction was reverse transcribed into cDNA and Actb mRNA levels were quantified using qPCR. The percent Actb mRNA across fractions was quantified and plotted. Red dashed line indicates fraction #12 in each sample. Source data are available for this figure: SourceData F2.
Genes controlling mRNA translation are affected in T cell activation and Treg-mediated suppression
To identify the mRNAs that shift between monosomes and polysomes following activation, we performed SPEED on CD4 Teff cells activated for 24 h with anti-CD3/CD28 beads. A portion (10%) of cellular lysate was used to isolate total RNA, while the remainder was subjected to SPEED analysis with monosome and polysome fractions collected and RNA isolated and sequenced (Fig. S4, A–C). We found ∼348 genes with differential translational efficiency (TE was calculated as polysomal/subpolysomal enrichment) when compared stimulated versus unstimulated cells, with 199 and 149 genes with lower and higher TE, respectively (Fig. 3 B). Among them, 201 genes changed in both TE and overall RNA expression in the same direction, 97 genes change in just TE and not overall input RNA and 50 gene changed in the opposite direction (Fig. 3, C and D; and Fig. S4, D and E). Gene ontology analysis of differential TE genes showed that the vast majority fell into functional categories involving ribosome biogenesis and mRNA translation, consistent with preparation for subsequent cell division following activation. It is important to note that the genes that encode these mRNAs are housekeeping genes whose expression is very high and largely unchanged by cell activation. The data presented here demonstrate that while the RNA level of transcripts encoding the transcriptional machinery is unchanged, their translation is likely increased following activation due to their shift onto polyribosomes. We next determined the fate of these mRNAs in CD4 Teffs stimulated in the presence of Tregs. We examined the mRNAs that showed increased TE in stimulated vs. resting cells and calculated their TE in CD4 Teff cells stimulated in the presence of Tregs (Stim + Treg). The set of mRNAs showing increased TE in stimulated cells were specifically reduced in their TE when the cells were stimulated in the presence of Tregs (Fig. 3 E), although the total mRNA levels of these genes remain unchanged (Fig. S4 F). These data are consistent with our hypothesis that Treg-mediated translational control targets mRNAs involved in preparing the cell for subsequent proliferation, with ribosome biogenesis and translational activity being critically important for this process. The finding that stimulation in the presence of Tregs reversed the positive TE changes in transcripts encoding proteins involved in mRNA translation is consistent with data showing that overall protein synthesis is suppressed by Tregs but rather in an active manner prior to clonal expansion (Fig. 3 F).
SPEED RNA-seq. (A) Scatterplot for each sample of quality control metrics percent alignment and median covariance of coverage, colored by fraction and treatment. Percent alignment refers to the percent of reads that can be aligned to the reference genome. Median covariance of coverage refers to the median coefficient of variation (mean/standard deviation) of gene sequence coverage for the 1,000 most highly expressed transcripts. All samples pass quality control thresholds of 75% percent alignment and 0.55 median covariance of coverage. (B) Same as A, zoomed into the region of the graph where the samples reside. (C) Bar graph of the number of millions of reads sequenced from each sample. (D) Scatterplots showing principal components 1 and 2 for each sample across the four treatment conditions and two fractions. Principal components were computed separately for each treatment condition. (E) Scatterplots showing TE (log2 fold-change of gene expression between the polysomal fraction and the subpolysomal fraction) and −log10 FDR for each stimulation condition. TE false-discovery was computed using the software package LIMMA, and the FDR refers to Benjamini-Hochberg–corrected P values for the hypotheses for each gene that the log2 fold-change between the fractions does not equal 0. Genes are colored red if their log2 translation efficiencies are less than −1 and FDRs are ≤5%. Genes are colored blue if their log2 translation efficiencies are >1 and FDRs are ≥5%. (F) Boxplots show the expression of a few translation-associated genes in the input fractions of the bead-stimulated and bead-stimulated, Treg-exposed samples. Differences in total transcript levels are not statistically significant, similar to what we observe in Fig. 4 C, our validation experiments. The FDRs reported are for differential expression between the bead-stimulated and bead-stimulated and Treg-exposed samples, multiple-test corrected across all expressed genes.
SPEED RNA-seq. (A) Scatterplot for each sample of quality control metrics percent alignment and median covariance of coverage, colored by fraction and treatment. Percent alignment refers to the percent of reads that can be aligned to the reference genome. Median covariance of coverage refers to the median coefficient of variation (mean/standard deviation) of gene sequence coverage for the 1,000 most highly expressed transcripts. All samples pass quality control thresholds of 75% percent alignment and 0.55 median covariance of coverage. (B) Same as A, zoomed into the region of the graph where the samples reside. (C) Bar graph of the number of millions of reads sequenced from each sample. (D) Scatterplots showing principal components 1 and 2 for each sample across the four treatment conditions and two fractions. Principal components were computed separately for each treatment condition. (E) Scatterplots showing TE (log2 fold-change of gene expression between the polysomal fraction and the subpolysomal fraction) and −log10 FDR for each stimulation condition. TE false-discovery was computed using the software package LIMMA, and the FDR refers to Benjamini-Hochberg–corrected P values for the hypotheses for each gene that the log2 fold-change between the fractions does not equal 0. Genes are colored red if their log2 translation efficiencies are less than −1 and FDRs are ≤5%. Genes are colored blue if their log2 translation efficiencies are >1 and FDRs are ≥5%. (F) Boxplots show the expression of a few translation-associated genes in the input fractions of the bead-stimulated and bead-stimulated, Treg-exposed samples. Differences in total transcript levels are not statistically significant, similar to what we observe in Fig. 4 C, our validation experiments. The FDRs reported are for differential expression between the bead-stimulated and bead-stimulated and Treg-exposed samples, multiple-test corrected across all expressed genes.
Tregs specifically suppress mRNAs related to the translational machinery via active translation control. (A) Scheme to capture polysome and subpolysome-associated mRNAs for next-generation sequencing. (B) TE was calculated as polysomal/subpolysomal enrichment for four conditions: (1) Unstimulated CD4 Teffs, (2) 24 h stimulated CD4 Teffs, (3) 24 h stimulated CD4 Teffs co-cultured with Tregs, and (4) 24 h stimulated CD4 Teffs treated with harringtonine for 30 min. Histograms of log2 TE for all genes were computed for the four conditions. Genes whose log2 TE were different than 0 at 5% FDR are highlighted in red. (C) Log2 fold-change of gene expression in the input RNA is plotted against log2 fold-change of translational efficiency between the stimulated and unstimulated cells. The colors annotate genes based on the significance of their input differential expression or translational efficiency: red if the gene changes in translational efficiency but not expression; blue if the gene changes in expression but not translational efficiency; purple if the gene changes in both; gray if the gene changes in neither; and orange if the gene changes in both but in opposite directions. (D) Volcano plot showing log fold-changes of TE in genes between stimulated and unstimulated CD4 Teffs and their adjusted P values. Dots in red are genes of the Reactome translation pathway. (E) Heatmap of log fold-change of TE in stimulated CD4 Teffs and stimulated CD4 Teffs co-cultured with Tregs, both against unstimulated CD4 Teffs. (F) Barcode plot ranks genes by their log fold-change of TE between Treg-exposed stimulated cells and stimulated cells (P value 0.009377618). Genes highlighted in black are genes within the Reactome Translation gene set identified to have increased TE in the stimulated vs. unstimulated cell comparison. P value are computed using the fgsea (Korotkevich et al., 2021) package that implements gene set enrichment analysis statistics.
Tregs specifically suppress mRNAs related to the translational machinery via active translation control. (A) Scheme to capture polysome and subpolysome-associated mRNAs for next-generation sequencing. (B) TE was calculated as polysomal/subpolysomal enrichment for four conditions: (1) Unstimulated CD4 Teffs, (2) 24 h stimulated CD4 Teffs, (3) 24 h stimulated CD4 Teffs co-cultured with Tregs, and (4) 24 h stimulated CD4 Teffs treated with harringtonine for 30 min. Histograms of log2 TE for all genes were computed for the four conditions. Genes whose log2 TE were different than 0 at 5% FDR are highlighted in red. (C) Log2 fold-change of gene expression in the input RNA is plotted against log2 fold-change of translational efficiency between the stimulated and unstimulated cells. The colors annotate genes based on the significance of their input differential expression or translational efficiency: red if the gene changes in translational efficiency but not expression; blue if the gene changes in expression but not translational efficiency; purple if the gene changes in both; gray if the gene changes in neither; and orange if the gene changes in both but in opposite directions. (D) Volcano plot showing log fold-changes of TE in genes between stimulated and unstimulated CD4 Teffs and their adjusted P values. Dots in red are genes of the Reactome translation pathway. (E) Heatmap of log fold-change of TE in stimulated CD4 Teffs and stimulated CD4 Teffs co-cultured with Tregs, both against unstimulated CD4 Teffs. (F) Barcode plot ranks genes by their log fold-change of TE between Treg-exposed stimulated cells and stimulated cells (P value 0.009377618). Genes highlighted in black are genes within the Reactome Translation gene set identified to have increased TE in the stimulated vs. unstimulated cell comparison. P value are computed using the fgsea (Korotkevich et al., 2021) package that implements gene set enrichment analysis statistics.
mRNAs sensitive to translational control in CD4 T cells are enriched for the terminal oligopyrimidine (TOP) motif
To identify the mechanism(s) of translation control, we first examined cis-regulatory elements shared by the 5′ UTRs with higher TEs identified by SPEED (Data S1, A–E). We searched for sequence-specific motifs within target UTRs in these Treg-sensitive genes by retrieving murine 5′ UTR sequences from the Ensembl BioMart database (Ensembl Genes78, Mus musculus) and using Multiple Em for Motif Elicitation (MEME), to uncover common cis-regulatory elements (Cunningham et al., 2014; Leppek et al., 2018; Truitt et al., 2015). Briefly, 131 genes increased in TE in stimulated cells relative to unstimulated cells at 5% false discovery rate (FDR) and also decreased in TE in Treg-exposed stimulated cells relative to the unstimulated cells (Data S1 A). Of those 131, we were able to extract 5′ untranslated mRNA sequences for 127 sequences (Data S1 B) using biomart (http://uswest.ensembl.org/biomart/martview/e728a9d413ab0dfd039a3ce666b3d3e2). We used the MEME software tool to identify a de novo motif enriched in these sequences (considering both the positive and negative strand), and the top hit closely resembles a TOP motif (Data S1 C). 109 of the 127 sequences have an instance of the motif with a P value of 0.01 or lower. 82 of the 109 show the motif in the strand orientation we expect, while 27 show the motif in the opposite orientation. To confirm that this motif is still significantly enriched in the correct orientation, the motif was tested for enrichment using the SEA tool, also in the MEME Suite package, on only the correct orientation. The motif was enriched at a P value of 0.000126 (Data S1 D), and this tool identified 67 instances of the motif among the 127 sequences (Data S1 E). We found an oligopyrimidine tract enriched in the 5′ UTR in Treg-sensitive mRNAs (Fig. 4 A). The TOP motif is a well-characterized motif which regulates key mRNAs encoding the translational machinery. The TOP motif is known to be present in the 5′ UTR of RPs and other genes required for mRNA translation (Meyuhas et al., 1987). Interestingly, TOP motif containing RP transcripts are regulated by the well-known signaling kinase mTORC1 (Schneider et al., 2013; Thoreen et al., 2012). We validated the selected genes that were differentially regulated in SPEED RNA-seq. We observed that Rps10, Rpl14, eIF3e, and Rpl8 mRNAs that contain TOP motif (Fig. S5 A), that are involved in mRNA translation, shift to polysomes upon CD4 Teff cell stimulation and back to monosomes following activation in the presence of Tregs (Fig. 4, B–D). The translational block in Treg coculture does not reflect a change in the total RNA levels of these genes (Fig. 4 C), but rather a change in the RL of these mRNAs. As predicted, the mRNA distribution shifted back to the monosome fraction when the CD4 T cells were stimulated in the presence of Tregs. As a control, ActB mRNA distribution is unchanged by Tregs. Finally, we also observed changes of TE of Rpl8 and eIF3e also reflected at the protein levels measured by flow cytometry (Fig 4 E and Fig. S5, B–E). These data confirm global changes in the translatome induced by Tregs to control CD4 T cell activation by blocking the translation of a subset of mRNAs, thereby blunting the ability of these cells to respond appropriately to stimulation.
Tregs suppress translation of TOP motif containing mRNAs. (A) Motif discovery analysis finds a TOP motif in Treg-regulated mRNAs. (B) SPEED profiling of unstimulated (unstim), stimulated (stim), and stim + iTreg cells. Total RNA of low-input samples measured at A254 nm. (C and D) mRNAs regulated by Tregs in Fig. 3 were validated using SPEED polysome qPCR. qPCR of RNA from input (C) and SPEED-fractionated (D) samples. Actb mRNA served as a control. (B and D) Green and black arrowheads highlight translation differences. (E) Purified naive CD4 T cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of iTregs for 6 h, subjected for analyzing expression of protein levels of RPL8 and eIF3e by flow cytometry. The above experiments were repeated at least twice. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Tregs suppress translation of TOP motif containing mRNAs. (A) Motif discovery analysis finds a TOP motif in Treg-regulated mRNAs. (B) SPEED profiling of unstimulated (unstim), stimulated (stim), and stim + iTreg cells. Total RNA of low-input samples measured at A254 nm. (C and D) mRNAs regulated by Tregs in Fig. 3 were validated using SPEED polysome qPCR. qPCR of RNA from input (C) and SPEED-fractionated (D) samples. Actb mRNA served as a control. (B and D) Green and black arrowheads highlight translation differences. (E) Purified naive CD4 T cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of iTregs for 6 h, subjected for analyzing expression of protein levels of RPL8 and eIF3e by flow cytometry. The above experiments were repeated at least twice. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Tregs inhibit mRNA translation through mTORC pathway and effects of RocA treatment on mice . (A) TOP motif is highlighted in red for the genes used in Fig. 4. (B–E) Purified naive CD4 T cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of iTregs for 6 h. Histogram plots shows the protein expression levels of eIF3e (B) and Rpl8 (C), analyzed by flow cytometry. Gray histograms, isotype controls; red histograms, eIF3 or RPL8. (D and E) Summary of the gMFI of the replicates. (F) Purified naive CD4 T cells as CD4 Teffs were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of Tregs for 24 h, subjected for analyzing PMY incorporation and phosphorylation of S6 and 4E-BP1, either alone or with anti–IL-2 or anti–IL-2Ra antibodies. (G) CD4 Teffs were isolated from WT mice (black circle) and IL-10Rβ–deficient mice (light blue, cyan, and deep blue circles) were cocultured with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of Tregs for 24 h and incubated with or without neutralizing antibodies for TGFβ, and analyzed for PMY incorporation, phosphorylation of S6, and 4E-BP1. (H) Mice were treated with 0.5 or 1 mg/kg of RocA or vehicle every day for 7 d, followed by isolation of bone marrow and spleen at day 8. (I) Total body weight was monitored every day after treatment. (J) Incorporation of PMY analyzed by flow cytometry after RocA treatment. (K and L) Splenocytes and bone marrow cells and were stained for neutrophils (Ly6G+), eosinophils (SiglecF+CD11c−), basophils (ckit-CD200R3+FcεRI+), monocyte (CD11b+Ly6c+), naive CD4+ T (CD4+CD3+CD44lowFoxp3−), memory CD4+ T (CD4+CD3+CD44hiFoxp3−), Treg (CD4+CD3+Foxp3+), CD8 T (CD8+CD3+), and B (CD19+) cells in indicated fractions, subjected to flow cytometry. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Tregs inhibit mRNA translation through mTORC pathway and effects of RocA treatment on mice . (A) TOP motif is highlighted in red for the genes used in Fig. 4. (B–E) Purified naive CD4 T cells were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of iTregs for 6 h. Histogram plots shows the protein expression levels of eIF3e (B) and Rpl8 (C), analyzed by flow cytometry. Gray histograms, isotype controls; red histograms, eIF3 or RPL8. (D and E) Summary of the gMFI of the replicates. (F) Purified naive CD4 T cells as CD4 Teffs were harvested with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of Tregs for 24 h, subjected for analyzing PMY incorporation and phosphorylation of S6 and 4E-BP1, either alone or with anti–IL-2 or anti–IL-2Ra antibodies. (G) CD4 Teffs were isolated from WT mice (black circle) and IL-10Rβ–deficient mice (light blue, cyan, and deep blue circles) were cocultured with or without equal number of anti-CD3/CD28 beads in the presence or absence of equal number of Tregs for 24 h and incubated with or without neutralizing antibodies for TGFβ, and analyzed for PMY incorporation, phosphorylation of S6, and 4E-BP1. (H) Mice were treated with 0.5 or 1 mg/kg of RocA or vehicle every day for 7 d, followed by isolation of bone marrow and spleen at day 8. (I) Total body weight was monitored every day after treatment. (J) Incorporation of PMY analyzed by flow cytometry after RocA treatment. (K and L) Splenocytes and bone marrow cells and were stained for neutrophils (Ly6G+), eosinophils (SiglecF+CD11c−), basophils (ckit-CD200R3+FcεRI+), monocyte (CD11b+Ly6c+), naive CD4+ T (CD4+CD3+CD44lowFoxp3−), memory CD4+ T (CD4+CD3+CD44hiFoxp3−), Treg (CD4+CD3+Foxp3+), CD8 T (CD8+CD3+), and B (CD19+) cells in indicated fractions, subjected to flow cytometry. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
IL-10 and TGFβ from Tregs block protein synthesis in Teffs
Since the activation of CD4 Teff cells in the presence of Tregs resulted in a specific inhibition of mTORC1 activation, as indicated by the lack of ribosomal protein S6 and 4E-BP1/2 phosphorylation, we hypothesized that Tregs suppress the immediate biosynthetic response to antigen-specific activation through regulation of mTORC1-mediated translational control. To define the pathways leading to the Treg-mediated inhibition of mTORC1 activation in stimulated CD4 T cells, we examined the role of soluble factors known to be produced by Tregs, using recombinant proteins, neutralizing antibodies, genetic knockouts specific for anti-inflammatory factors. It has been well known that deprivation of IL-2 is one of the mechanisms of Treg-mediated suppression of proliferation of Teffs. Consistent with this, when IL-2 signaling was blocked in stimulated T cells with αIL-2 or αCD25 neutralizing antibodies, we observed a reduction in rpS6 and PMY incorporation, while 4E-BP1 activation was not affected (Fig. S5 F).
We next examined whether IL-10 and TGFβ played a role in Treg-mediated translation inhibition. Blockade of IL-10 or TGFβ individually resulted in reduced inhibition of translation in Teff cells co-cultured with Tregs (Fig. 5 A). Importantly, neutralization of both IL-10 and TGFβ signaling significantly reduced Treg-mediated suppression of PMY incorporation compared to neutralization of each signal, respectively (Fig. 5 A). In addition, we found that blockade of IL-10 and TGFβ signaling lead to inhibition of Treg-mediated suppression of mTORC1 signaling (Fig. 5 A). Similarly, we saw a significant rescue of protein synthesis and mTORC1 pathway when TGFβ was neutralized in IL-10Rb−/− CD4 Teff cells co-cultured with Tregs (Fig. S5 G). These data suggest that Treg-mediated IL-10 and TGFβ, acting on CD4 Teff cells, result in a block in mTORC1 activation and subsequent translation inhibition. Consistent with this model, we observed that the addition of recombinant IL-10 and TGFβ to cultures of CD3+CD28-stimulated CD4 Teff cells resulted in significantly reduced PMY incorporation and mTORC1 activation (Fig. 5 B). Finally, and consistent with these data, we have also found that CD4 Teffs activated in the presence of IL-10 and TGFβ resulted in a marked reduction of the polysome fraction, similar to what was seen in Teff-Treg co-cultures (Fig. 4 B and Fig. 5 C). These data suggest that Tregs use the production of IL-10 and TGFβ in combination to disrupt mTORC1 signaling and inhibit mRNA translation in stimulated CD4 Teff cells.
IL-10 and TGFβ from Tregs block protein synthesis in T effector cells. (A) Neutralizing antibodies for IL-10 receptor and/or TGFβ was added to co-culture of CD4 Teffs and Tregs with equal number of αCD3/CD28 beads for 24 h and analyzed for PMY incorporation (left) and phosphorylation of S6 (right). (B) Recombinant IL-10 and TGFβ were added to culture of CD4 Teffs with equal number of anti-CD3/CD28 beads for 24 h and analyzed for PMY incorporation (left) and phosphorylation of S6 (right). (C) SPEED profiling of unstim, stim, and stim cells incubated with recombinant IL-10 and TGFβ. Total RNA of low-input samples measured at A254 nm. One-way ANOVA was applied for all comparisons. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. The experiments were performed at least twice.
IL-10 and TGFβ from Tregs block protein synthesis in T effector cells. (A) Neutralizing antibodies for IL-10 receptor and/or TGFβ was added to co-culture of CD4 Teffs and Tregs with equal number of αCD3/CD28 beads for 24 h and analyzed for PMY incorporation (left) and phosphorylation of S6 (right). (B) Recombinant IL-10 and TGFβ were added to culture of CD4 Teffs with equal number of anti-CD3/CD28 beads for 24 h and analyzed for PMY incorporation (left) and phosphorylation of S6 (right). (C) SPEED profiling of unstim, stim, and stim cells incubated with recombinant IL-10 and TGFβ. Total RNA of low-input samples measured at A254 nm. One-way ANOVA was applied for all comparisons. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. The experiments were performed at least twice.
mRNA translation inhibitor RocA inhibits T cell proliferation
While we show that the suppression of mRNA translation by Tregs has a direct effect on cell activation and proliferation, we next investigated whether the translation inhibition is one of the main ways Tregs exert their function downstream of mTORC1. To test this, we used a mRNA translation inhibitor RocA (RocA,1H-2,3,3a,8b-tetrahyrocyclopenta[b]benzofuran), which has been shown to reduce overall protein synthesis but to preferentially inhibit specific subset of mRNAs in cell culture models (Iwasaki et al., 2019). RocA is a secondary metabolite from the plant genus Aglaia with anti-tumor and anti-inflammatory properties (Ebada et al., 2011; Li-Weber, 2015; Bordeleau et al., 2008; Cencic et al., 2009; Li-Weber, 2015). RocA affects protein synthesis by binding to eIF4A, a DEAD-box RNA helicase, that is part of the eIF4F mRNA initiation complex (Rogers et al., 2002). RocA binds to eIF4A and clamps it on poly-purine sequences in the 5′ UTRs of mRNAs, thereby preferentially inhibiting this set of mRNAs at the level of translation control when stable RocA-eIF4A-mRNA complexes are formed to block 43S pre-initiation complex scanning (Ernst et al., 2020; Iwasaki et al., 2019). While RocA has been shown to inhibit overall protein synthesis through the translational blockade of a subset of mRNAs (Iwasaki et al., 2016), its effect on CD4 Teff cell activation and proliferation is unclear. To address this, we measured protein synthesis rate in activated CD4 Teff cells 24 h after RocA treatment using the PMY incorporation assay (Schmidt et al., 2009; Seedhom et al., 2016). RocA-treated cells showed a dose-dependent decrease in PMY incorporation, suggesting inhibition of protein synthesis. We also found that RocA treatment inhibited cell-proliferation in a dose-dependent manner at sub-nanomolar concentrations (Fig. 6, A and B). Surprisingly, early T cell activation genes such as IL-2 and CD25 (IL-2Rα) were unaffected both at the mRNA and protein level by RocA, suggesting that impacting early TCR signaling dependent on NFAT is not the mechanism by which RocA inhibits T cell proliferation. Furthermore, this supports the notion that translational blockade through RocA is not a global effect even in T cells. As expected, when cells were treated with the calcineurin inhibitor Tacrolimus (FK506), T cell proliferation was inhibited, with a corresponding downregulation of both NFAT target genes IL-2 and CD25 (IL-2Rα; Fig. 6 C). These results suggest that while both RocA and FK506 inhibit CD4 Teff cell activation and proliferation, they do so through distinct mechanisms. While our finding is different from a previous study suggesting that RocA inhibits NFAT activity in human CD4 Teff cells (Proksch et al., 2005), we propose at higher concentrations (50–100 fold) of RocA could affect transcription regulation. Nevertheless, these data demonstrated that RocA-mediated translation inhibition could suppress CD4 Teff cell proliferation ex vivo.
Acute inflammation due to Treg deficiency was reversed by an mRNA translation inhibitor. (A–C) Mouse naive CD4 T cells were isolated from the spleens of naive C57BL/6 mice and CFSE labeled. The cells were stimulated with equal number of anti-mouse CD3/CD28-conjugated beads in the presence of the indicated concentration of RocA, FK506, or vehicle for 24 h (A and C) or 72 h (B). The cells were stained for anti-CD4 (A and C) or anti-CD4 and anti-CD25 (B) for flow cytometric analysis. The data of frequency of CD25 in A was summarized in C. Murine IL-2 was assayed by ELISA from supernatants collected after 24 h after stimulation (C). (D) Incorporation of PMY analyzed by flow cytometry after RocA treatment. (E–G) Foxp3DTR mice were treated with DT every other day and 0.2 mg/kg of RocA everyday starting from 2 d before DT treatment, followed by isolation of splenocytes at day 8. The cells were stained with anti-CD4, anti-TCRβ, and anti-CD44 followed by restimulation with PMA and Ionophore for 6 h. The cells were stained by anti–IL-17A and anti-IFNγ, followed by flow cytometric analysis (E). The body weight of mice at day 8 (F). The number of Th1 cells (IFNγ+CD44hiCD4+TCR-β+) and pathogenic Th17 cells (IFNγ+IL-17A+CD44hiCD4+TCR-β+; G). (H) Human naive CD4 T cells isolated from frozen PBMC, subjected to CFSE labeling. The cells were stimulated with anti-human CD3 and anti-human CD28–conjugated beads in the presence of 10 nM of RocA or vehicle or without the beads indicated as unstim. After 3 d, cells were stained with anti-human CD25 and anti-CD4, subjected to flow cytometry to analyze the frequency of divided cells and CD25+ cells. (I) Human memory Th1 cells isolated as CD4+CD25−CD45RA−CXCR3+CCR6− cells from frozen PBMC were incubated in the presence of 10 nM of RocA or vehicle for 2 d, followed by restimulation with PMA and Ionophore for 6 h. The cells were surface stained by anti-CD4 and subjected to cytoplasmic staining for IFNγ and IL-13. The frequency of IFNγ+ cells is summarized in the dot plots. One-way ANOVA was used for C, D, and H; two-tailed t test (unpaired) was applied for F and G; two-tailed Mann–Whitney test (unpaired) was applied for I. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The above experiments were repeated at least twice.
Acute inflammation due to Treg deficiency was reversed by an mRNA translation inhibitor. (A–C) Mouse naive CD4 T cells were isolated from the spleens of naive C57BL/6 mice and CFSE labeled. The cells were stimulated with equal number of anti-mouse CD3/CD28-conjugated beads in the presence of the indicated concentration of RocA, FK506, or vehicle for 24 h (A and C) or 72 h (B). The cells were stained for anti-CD4 (A and C) or anti-CD4 and anti-CD25 (B) for flow cytometric analysis. The data of frequency of CD25 in A was summarized in C. Murine IL-2 was assayed by ELISA from supernatants collected after 24 h after stimulation (C). (D) Incorporation of PMY analyzed by flow cytometry after RocA treatment. (E–G) Foxp3DTR mice were treated with DT every other day and 0.2 mg/kg of RocA everyday starting from 2 d before DT treatment, followed by isolation of splenocytes at day 8. The cells were stained with anti-CD4, anti-TCRβ, and anti-CD44 followed by restimulation with PMA and Ionophore for 6 h. The cells were stained by anti–IL-17A and anti-IFNγ, followed by flow cytometric analysis (E). The body weight of mice at day 8 (F). The number of Th1 cells (IFNγ+CD44hiCD4+TCR-β+) and pathogenic Th17 cells (IFNγ+IL-17A+CD44hiCD4+TCR-β+; G). (H) Human naive CD4 T cells isolated from frozen PBMC, subjected to CFSE labeling. The cells were stimulated with anti-human CD3 and anti-human CD28–conjugated beads in the presence of 10 nM of RocA or vehicle or without the beads indicated as unstim. After 3 d, cells were stained with anti-human CD25 and anti-CD4, subjected to flow cytometry to analyze the frequency of divided cells and CD25+ cells. (I) Human memory Th1 cells isolated as CD4+CD25−CD45RA−CXCR3+CCR6− cells from frozen PBMC were incubated in the presence of 10 nM of RocA or vehicle for 2 d, followed by restimulation with PMA and Ionophore for 6 h. The cells were surface stained by anti-CD4 and subjected to cytoplasmic staining for IFNγ and IL-13. The frequency of IFNγ+ cells is summarized in the dot plots. One-way ANOVA was used for C, D, and H; two-tailed t test (unpaired) was applied for F and G; two-tailed Mann–Whitney test (unpaired) was applied for I. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The above experiments were repeated at least twice.
Acute inflammation due to Treg deficiency is counteracted by the mRNA translation inhibitor RocA
We next tested whether in vivo RocA administration could ameliorate inflammation-induced CD4 Teff cell activation. First, we assessed the effect of RocA on the general health of the mice and immune homeostasis by treating mice every other day for a week with RocA (0.5 and 1 mg/kg i.p.; Fig. S5 H). RocA neither impacted overall weight nor the relative contributions of various cell populations within the CD45+ compartment, including CD4 T cell subsets (Fig. S5, I–L). Thus, short term in vivo treatment with RocA-induced no deleterious effects on immune homeostasis and was well tolerated by the mice.
We next asked whether inflammatory responses were limited by RocA treatment in the absence of Tregs. For these studies, we used the Foxp3DTR mice, treated with DT to deplete Tregs as described in Fig. 1 D. To test the ability of RocA to affect acute inflammation, we dosed Foxp3DTR mice with RocA (0.2 mg/kg i.p.) or DMSO (vehicle) on days −1 and −2, and then treated with DT (100 ng i.p.) on days 0, 2, 4, and 6, followed by sacrifice and analysis on day 8 (Fig. 6 E). A hallmark feature of Treg depletion is a rapid loss of body weight, which is seen in the vehicle-treated mice on day 7, with an average weight loss of 20% of initial body weight (Fig. 6 F). We found that the RocA-treated mice showed significantly less weight loss, averaging between 85 and 90% of starting weight, suggesting that RocA treatment was blunting the inflammation caused by Treg depletion. Consistent with this, we found that numbers of splenic CD4 T cells producing IL-17A or IFNγ or both cytokines were significantly reduced in RocA-treated mice (Fig. 6 G). Especially important was the reduction in IFNγ/IL-17A co-producers, as this subset has been found to be pathogenic in inflammatory settings (Doodes et al., 2010; Duhen et al., 2013; Kaiser et al., 2016; Lee et al., 2009). These data demonstrate that RocA-mediated direct translation inhibition can ameliorate the inflammation seen in mice lacking functional Tregs, supporting our findings that one major mechanism of Treg-regulated immune tolerance is through controlling aberrant protein synthesis activity in peripheral CD4 T cells.
Lastly, we also found suppression of proliferation in primary human T cells treated with RocA and stimulated with anti-CD3/CD28 (Fig. 6 H). Similar to what was seen in mouse CD4 Teff cells, there was no difference in the surface expression of CD25 (Fig. 6 H). We also observed significant suppression of IFNγ producing cells with RocA treatment compared to the vehicle control after PMA/ionomycin stimulation (Fig. 6 I). Altogether, we show herein a novel mechanism of Treg-mediated suppression of CD4 Teff cell activation, which acts through active mRNA translation control to downregulate the protein dosage of protein synthesis machinery components in activated CD4 Teff cells. Finally, we show in a physiologically relevant setting that direct inhibition of protein synthesis using a small molecule inhibitor (RocA) can have therapeutic efficacy in alleviating unwanted inflammatory CD4 Teff cell activation.
Discussion
Regulatory T cells possess the ability to potently suppress CD4 Teff cell responses and maintain proper immune homeostasis. While Treg-mediated suppression mechanisms have been heavily studied from the Treg-centered perspective, the molecular events within CD4 Teff cells upon Treg encounter have remained unclear with some studies investigating only transcriptional changes (Akkaya and Shevach, 2020; Sojka et al., 2008; Vignali, 2012). We observed that CD4 Teffs rapidly increased their global protein synthetic rate following TCR-mediated activation, and that Tregs could suppress this global increase in translation. Interestingly, we found Tregs inhibited this process by shutting down the biosynthetic ramp-up required for subsequent cell division, as early as 6 h following T cell activation. Similarly, an acute depletion of Tregs in vivo led to the robust upregulation of protein synthesis rate in peripheral CD4 T cells (Fig. 7). These data suggest that an important aspect of Treg-mediated suppression is inhibition of the ability of CD4 Teffs to increase their protein biomass prior to cell division. The rapid increase in protein synthesis following acute loss of Tregs demonstrates a need for Tregs to hold in check the aberrant increase in protein synthesis of autoreactive T cells in the periphery. This is a highly efficient mode of regulation: inhibiting the ramp-up of biosynthesis that proceeds cell division, thereby stopping proliferation before it has begun.
A working model depicting Treg-mediated disruption of mTORC1 signaling that leads to the suppression of protein synthesis of mRNAs enriched for TOP motifs.
A working model depicting Treg-mediated disruption of mTORC1 signaling that leads to the suppression of protein synthesis of mRNAs enriched for TOP motifs.
In order to provide a broader snapshot of CD4 Teff gene expression changes following Treg encounter, we investigated both transcriptional and translational gene regulation in CD4 Teffs. We developed a novel technique called SPEED to investigate the translatome of primary CD4 Teff cells that have been suppressed by Tregs. We found that, upon activation, CD4 Teffs displayed a rapid redistribution of mRNAs from monosome to polysomes, leading to increased translation. This redistribution was independent of acute changes in the transcription of the mRNAs. Most of these mRNAs encoded for proteins important for protein biosynthesis, including RPs, RNA-binding proteins, and elongation and splicing factors, suggesting that enhanced translation following activation was under translation control itself. Importantly, we found that activation in the presence of Tregs inhibited the redistribution of these mRNAs, causing them to remain either free or associated with monosomes. As these changes in RL were independent of gene transcription and occurred within 24 h of stimulation, they would be undetectable by the transcriptional changes in gene expression or using cell division as a readout, a traditional method to study Treg-mediated suppression. The mRNAs affected at the level of TE by Tregs were enriched for 5′ UTR TOP motifs as has been shown for mRNAs involved in the core proteins of the translational machinery. Recently, it has been revealed that the TOP motif is recognized by the RNA-binding protein La-related protein 1 (LARP1), which regulates the association of these mRNAs with stress granules (Mattijssen et al., 2021; Philippe et al., 2020). The presence of this motif on Treg-affected mRNAs suggests that Tregs may regulate the trafficking of these mRNAs from ribosomes to stress granules, thus controlling their translation.
TOP motif containing mRNAs have been known to be under direct regulation by mTORC1 signaling. The mTOR pathway has been previously implicated to be critical for coordinating cell growth and proliferation in lymphocytes through the translation initiation factor eIF4E (Howie et al., 2014; So et al., 2016). Indeed, we found that activation in the presence of Tregs led to an inhibition of S6 and 4E-BP1/2 phosphorylation, both downstream targets of mTORC1. As mRNA translation is energetically demanding, an mTOR-dependent switch to aerobic glycolysis is well known (Huang et al., 2020; Makowski et al., 2020). Recent study has shown key enzymes in the pathway are translationally regulated important for metabolic reprogramming of activated cell (Ricciardi et al., 2018). These data suggest that translation remodeling occurs earlier than metabolic reprogramming in these cells. We did not detect any mRNAs linked to metabolic processes as we probed for early events of translation remodeling.
To understand how Treg encounter can impact mTORC1 signaling in CD4 Teff cells, we focused on two well-known immunosuppressive cytokines that Tregs are known to secrete. Here, we demonstrate that Treg-derived IL-10 and TGFβ were critical for the ability of Tregs to exhibit translation control in CD4 Teffs through inhibition of mTORC1 signaling output. Blockade of each individual cytokine leads to moderate restoration of translation, while blockade of both cytokines nearly recapitulated translation levels of activated CD4 Teff cells in the absence of Tregs. Similarly, addition of IL-10 or TGFβ during CD4 Teff stimulation (in the absence of Tregs) resulted in a reduction of global translation similar to that seen when CD4 Teff cells were stimulated in the presence of Tregs. Thus, IL-10 and TGFβ signaling in CD4 Teff cells is involved in Treg-mediated trans-inhibition of protein synthesis. Previous studies have shown divergent effects of IL-10 signaling on mTORC1 activity. In macrophages, IL-10 acts in a paracrine or autocrine fashion to inhibit mTORC1 activation following LPS stimulation (Baseler et al., 2016; Ip et al., 2017). The outcome was a metabolic reprogramming of the macrophages to promote oxidative phosphorylation. However, in human natural killer cells, IL-10 regulated metabolic changes that enhanced cellular function (e.g., IFNγ production), and these changes were mediated through the activation of mTORC1 (Wang et al., 2021). While these studies show opposite effects of IL-10 on cellular activity, they focus on changes in metabolism mediated through regulation of mTOR signaling. However, these studies do not address the role of IL-10 in protein synthesis. Likewise, TGFβ signaling has been shown in a variety of systems to target mTOR, with the outcome dependent on the cell type and the inflammatory context. For example, TGFβ treatment of lung fibroblasts leads to synthesis of collagen through activation of mTORC1 and 4E-BP1/2, the opposite of what we observed in Teff cells (Woodcock et al., 2019). On the other hand, TGFβ signaling was shown to inhibit mTOR activity in CD8 precursor exhausted cells (Tpex), leading to improved mitochondrial activity during chronic viral infection (Gabriel et al., 2021). Finally, work from Komai and co-workers (Komai et al., 2018) showed that IL-10 and TGFβ work synergistically to suppress both glycolysis and oxidative phosphorylation through the inhibition of mTORC1 in TLR-stimulated B cells. While it is clear from all of these studies that TGFβ and IL-10 can target the mTOR pathway for regulation, we believe our data bridge the impact of these cytokines on the mTORC1 pathway to active mRNA translation control in CD4 Teff cells. An interesting observation from our study is that the mRNA translation in Tregs itself was not affected, indicating that the Tregs have an alternative strategy to shield itself from mRNA translation suppression. Indeed, a recent study shows that Tregs utilize a non-canonical mRNA translation mechanism which uses DAP5 and eIF3d instead of canonical mTORC1-dependent eIF4E and eIF4G factors (Volta et al., 2021). These data also suggest that mRNA translation suppression is one of the major pathways through which Tregs suppress Teff function.
If CD4 Teff cells are under active translation control by Tregs, we reasoned whether autoimmune or inflammatory responses caused by Treg loss could be alleviated if we could suppress aberrant protein synthesis increase in CD4 Teff cells. We choose RocA, as it has been used in vivo in an experimental cerebral malaria mouse model with no reported toxicity (Langlais et al., 2018) and this translation inhibitor preferentially inhibits a subset of mRNAs in cell lines. Indeed, our data also showed no observable toxicity when mice were treated with repeated dosing of RocA. Consistent with our hypothesis that inhibition of mRNA translation can be an effective means of maintaining tolerance in a loss of Treg setting, treatment of these mice with RocA abrogated enhanced protein synthesis and inhibited induction of cytokine expression in CD4 Teffs. The ability of RocA to suppress inflammation through translation control in other immune cells is under active investigation.
Regardless of the variety of different mechanisms that Tregs may utilize for CD4 Teff suppression, we propose here a target cell–centered theme in that changes in intracellular signaling events in CD4 Teffs result in inhibition of proliferation that is observed later. Taken as a whole, our data show that Tregs actively control the mRNA translation process in CD4 Teff cells via inhibition of mTORC1 signaling. Furthermore, our study provides a novel mechanism for the induction and maintenance of peripheral tolerance and identifies Treg-derived IL-10 along with TGFβ as critical regulators of mRNA translation affecting proliferation and activation of CD4 Teff cells.
Materials and methods
Mice
For Treg-depletion studies, Foxp3DTR mice (B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J: Stock#016958 from Jackson Labs) were used. For Treg isolation and expansion in vivo, Foxp3YFP-Cre mice (B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J: Stock#016959 from Jackson Labs) that have been crossed to B6 Cd45.1 mice (B6.SJL-PtprcaPepcb/BoyJ: Stock#002014 from Jackson Labs) in house were used. For RiboTag studies B6J.129(Cg)-Rpl22tm1.1Psam/SjJ, B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ and B6.C-Tg(CMV-cre)1Cgn/J were used. For the studies of IL-10 in Treg-mediated suppression of protein synthesis, 7-wk-old female of IL-10Rβ−/− (B6.129S2-Il10rbtm1Agt/J: Stock#005027 from Jackson Labs) mice were used. All animal experiments were approved by the Benaroya Research Institute Institutional Animal Care and Use Committee (Protocol IACUC21-047). For all the reagent identifiers, see Table S2.
In vivo treatments
Foxp3DTR mice were intraperitoneally injected with 100 ng of DT at indicated days. For treatment of rocaglamide (RocA), mice were intraperitoneally administrated with indicated concentrations of RocA.
Murine T cell isolation, culture, and suppression assays
Bulk CD4 T cells were isolated from indicated mice and stained with antibodies against CD25, CD44, CD62L, and CD4 except for CD4 T cells from Foxp3YFP-Cre mice since Foxp3+ Tregs were identified using YFP fluorescence from these mice. Purified YFP−CD25−CD4+CD44lowCD62L+ naive CD4 T cells as CD4 Teffs were sorted using the BD FACS Aria fusion. Unless noted, all isolated T cells were cultured in T cell media (RPMI-1640, 10% fetal bovine serum, 2 mM GlutaMAX-I, 100 U/ml penicillin-streptomycin, 55 μM 2-mercaptoethanol, 1 mM sodium pyruvate, 1× non-essential amino acids, 10 mM HEPES). For Treg suppression assays, indicated numbers of tTregs (CD4+YFP+) and 5 × 105 CD4 Teffs (YFP−CD25−CD4+CD44lowCD62L+ naive CD4 T cells) were cultured in round-bottom 96-well plate for the indicated times with 5 × 105 anti-CD3/CD28–coated magnetic beads (Teff:beads = 1:1). To control for T cell density, control cultures were cultured with either the same number of total Teffs as in the Treg:Teff co-culture conditions or in some cases, YFP−CD25−CD4+CD44lowCD62L+CD45.1+ Teffs sorted from Foxp3YFP-Cre mice were used. To test proximal TCR signaling, in vitro Treg suppression assay was performed as described with modifications. CD4 Teffs were isolated from Nur77-GFP mice and Tregs were sorted from Foxp3YFP-Cre mice. Prior to co-culture, CD4 Teffs were further stained with CellTraceViolet to assess proliferative status and ensure proximal TCR signaling assessed by Nur77-GFP was prior to proliferation of these cells. Unstimulated Nur77-GFP CD4 Teffs and Tregs from wild-type C57BL/6 mice served as a negative for GFP signal.
For PMY incorporation (as a measure of global protein synthesis), PMY (10 mg/ml) was added for the last 15 min of culture, and then the cells were permeabilized and stained with Alexa647-coupled anti-PMY antibody and analyzed by flow cytometry. Controls included unstimulated Teffs also labeled with PMY and stimulated Teff given CHX at the same time as PMY (CHX results in a complete translation blockade, so this controls for non-specific uptake of PMY). For analysis of Treg-mediated suppression, we have presented the PMY incorporation data as the percent suppression of protein synthesis, relative to the PMY incorporation in the Teff alone condition (which is put at 0% inhibition). In this calculation, 100% of suppression of protein synthesis means absolute inhibition of PMY incorporation. For analysis of mTORC signaling, we have presented the geometric MFI (gMFI) data as normalized gMFI, relative to the gMFI data in the Teff alone condition (which is put at 1).
Ex vivo human Treg suppression assay
We used in vitro–expanded Tregs from a single donor and Teff (defined as CD4+CD45RA+) from five individual healthy donors for Treg suppression assay. Established protocols were used for Treg expansion (Long et al., 2017; Putnam et al., 2009) and Teff (CD4+CD45RA+) cells were purified PBMC using the naive human T cell isolation kit from Miltenyi. To determine the effect of Tregs on overall translation in Teff, we cultured Teff (104 cells) in the absence or presence of Tregs (1:4 Treg:Teff) for 24 h with anti-CD3/CD28 beads (at a ratio of 28:1 [Teff:beads]). PMY (10 μg/ml) was added for the last 15 min of culture, and then the cells were permeabilized and stained with Alexa647-coupled anti-PMY antibody and analyzed by flow cytometry. Controls included unstimulated Teffs also labeled with PMY and stimulated Teff given CHX at the same time as PMY (CHX results in a complete translation blockade; so this controls for non-specific uptake of PMY). Blood samples from healthy donors were obtained from participants in the Benaroya Research Institute Registry and Repository. The study was approved by the Benaroya Research Institute Institutional Review Board (Protocol IRB07109-431). All samples were obtained under approved research protocols with informed consent.
Ribo-IP and RNA isolation
RiboTag CD4 Teff cells (or control CD4 Teff cells: RiboTag mice with no Cd4Cre expression) were washed once with ice-cold PBS with CHX and lysed in 100 μl of polysome lysis buffer (PLB; 25 mM Tris-HCl pH 7.5 (AM9850G; Ambion), 150 mM KCl (AM9640G; Ambion), 15 mM MgCl2 (AM9530G; Ambion), 1 mM dithiothreitol (DTT; 646563; Sigma-Aldrich), 1% NP-40 (28324; Thermo Fisher Scientific), 100 μg/ml CHX, 100 U/ml SUPERaseIn RNAse Inhibitor (AM2694; Ambion), 25 U/ml TurboDNAse (AM2238; Ambion), and complete protease inhibitor EDTA-free (11836170001; Sigma-Aldrich) in nuclease-free water (10977015; Thermo Fisher Scientific). Lysates were incubated on ice for 5 min and nuclei were removed by centrifugations at 200 g, 5 min at 4°C and 13,000 g, 5 min at 4°C. A 10% aliquot (10 μl) of the lysate was harvested and kept in Trizol for total RNA extraction and the remaining lysate volume was adjusted to 400 μl with PLB for even rotation with the antibodies and beads for Ribo-IP in the subsequent steps. For each sample, 3 μl of anti-HA antibody (ab9110; Abcam) was added and samples were allowed to rotate at 4°C for 4 h in a tube rotator (88881001; Thermo Fisher Scientific) at 20 rpm speed. During rotation, 40 μl of Dynabeads Protein G (10004D; Thermo Fisher Scientific) were prepared by washing two times in ice-cold PLB. After the anti-HA binding step, Dynabeads were added to each sample for an additional 4 h rotation at 4°C (speed 20 rpm). Next, samples were magnetized on the DynaMag-2 magnet (12321D; Thermo Fisher Scientific) and washed four times with 500 μl of polysome high salt wash buffer (PHSWB; 25 mM Tris-HCl pH 7.5, 300 mM KCl, 15 mM MgCl2, 1 mM DTT, 1% NP-40, 100 μg/ml CHX, 100 U/ml SUPERaseIn RNAse Inhibitor, complete protease inhibitor EDTA-free). Usually, before the first wash, 10% of the lysate was saved to routinely check for Ribo-IP efficiency between samples by immunoblotting. For RNA extraction, Dynabeads after the final wash were resuspended in 100 μl of PHSWB and homogenized in 350 μl of RLT buffer containing 40 mM freshly added DTT (74004; Qiagen RNeasy micro kit) by vigorous vortexing. Samples were left at room temperature for 10 min before magnetizing the beads on the magnet and proceeding with the RNA extraction following the RNeasy micro kit protocol. At the last step, total RNA was eluted in 11 µl of nuclease-free water and RNA integrity and quantity was assessed using the Bioanalyzer 2100 total RNA pico kit. RNA integrity number values for all samples were >8.6 (Fig. S2) and next-generation sequencing–compatible cDNA libraries were generated according to the SMARTseq v4 Ultra-Low Input kit (634888; Clontech).
Ribo-IP and protein isolation
Ribo-IP was performed identical to the RNA isolation method (described above). After the final wash in PHSWB, Dynabeads were resuspended in 1× lithium dodecyl sulfate sample buffer and heated at 65°C for 10 min. Samples were magnetized and loaded onto 12% NuPAGE Bis-Tris gel, and proteins were separated by SDS-PAGE. Separated proteins were visualized using the Colloidal Blue Staining kit (LC6025; Thermo Fisher Scientific) and the entire lane corresponding to each sample was excised.
Low-input sucrose gradient polysome fractionation (SPEED)
Sucrose gradients (15–60%) were prepared in SW55Ti rotor-compatible Ultra-Clear ultracentrifuge tubes (344057; Beckman Coulter). Briefly, a 2 M (68.5%) sucrose solution in nuclease-free water and a 10× sucrose buffer (250 mM Tris-HCl pH 7.5, 1.5 M KCl, 150 mM MgCl2, 10 mM DTT, 1 mg/ml CHX, complete protease inhibitor EDTA-free (two tablets per 50 ml), 20 U/ml SUPERaseIn RNAse Inhibitor) was prepared. Using these two solutions, sucrose solutions with 1× sucrose buffer and different concentrations (60, 45, 30, 15%) were prepared. Each solution was added from the bottom of the tubes in the following order and quantity (60%: 750 μl, 45%: 1.5 ml, 30%: 1.5 ml, 15%: 750 μl). For each addition, tubes were kept in the −80°C for at least 15 min to freeze the sucrose solutions before adding the next sucrose solution. All tubes were sealed with parafilm and kept at −80°C forever. Tubes were allowed to thaw at 4°C for 12–16 h before the fractionation. Samples were prepared in PLB buffer similar to the Ribo-IP method and total RNA was quantified using RiboGreen in the low-range assay (1–50 ng/ml). All samples were adjusted with PLB to at least 500 ng of total RNA and layered carefully on top of thawed sucrose-gradient tubes. Tubes were ultracentrifuged at 35,000 rpm in a SW55Ti rotor using the L8-70 M ultracentrifuge (acceleration: default, deceleration: 0). Separated lysates were fractionated from top to bottom with an Auto-Densi Flow fractionator (Labconco) with continuous reading of absorbance at 254 nm (A254 nm) using a UA-6 UV/VIS detector (Teledyne Isco). A total of 16–18 fractions (250–300 μl) were fractionated with the Foxy R1 fractionator in 2 ml Eppendorf tubes and kept on ice. For digital conversion of the A254 nm signal, we attached the LabQuest Mini data-collection interface (Vernier, LQ-MINI) to the UA-6 detector. For RNA extraction, we used three volumes of Trizol LS to each sucrose fractions and vortexed vigorously. Every fraction was spiked-in with in vitro transcribed firefly Luciferase RNA (uncapped) to assess RNA extraction efficiency between fractions. Samples were either kept in −80°C at this point or proceeded with standard Trizol-mediated RNA extraction protocol or the Direct-zol 96 kit (Zymo Research, R2054) was used following the on-column DNA digestion protocol. Total RNA quantity from each fraction was measured using RiboGreen in the low-range assay to generate ribosome traces. The Bioanalyzer 2100 RNA pico kit was also used to assess the starting point of the intact 80S monosome peak (containing both 28S and 18S ribosomal RNA bands). Equal volumes of RNA from fractions corresponding to polysomes (three fractions after the 80S monosome peak) were pooled and digested with RQ1 DNase (Promega) and cleaned-up using RNA Clean&Concentrator-5 kit (Zymo Research, R1013). Finally, RNA quantity (RiboGreen) and RNA quality (Bioanalyzer) were measured again before proceeding with cDNA libraries construction using the SMARTseq v4 Ultra-Low Input kit (Clontech).
RNA-seq bioinformatics methods
Reads were aligned by STAR (Dobin et al., 2013) 2.4.2a to GRCm38.91, and genes that were not expressed in at least 20% of samples were filtered out. The reads were TPM normalized and modeled using the software LIMMA (Ritchie et al., 2015). The LIMMA model included a variable for each of the four mice and categorized samples by fractionation (input, polysomal, or sub-polysomal), whether they were stimulated, and their treatment (HHT, Treg cells, or none). Combinations of those conditions resulted in 12 categories total. TE was computed as the log fold-change in TPM between the polysomal and sub-polysomal fraction per condition, both at the individual mouse and aggregate levels. Differential TE was computed in LIMMA as differences between translation efficiencies between conditions. Gene set enrichment statistics were computed from LIMMA-computed log fold-changes and the fgsea (Korotkevich et al., 2021,Preprint) package. Motif enrichment analysis was performed on the 131 genes whose TE increased in stimulated cells relative to unstimulated cells at 5% FDR and also decreased in TE in Treg-exposed stimulated cells relative to the unstimulated cells. The 5′ UTRs for those 131 genes were queried using biomart, which returned sequences for 127 of the genes (Cunningham et al., 2022). The MEME software tool was used to identify a de novo motif enriched in these sequences (considering both the positive and negative strand; Bailey et al., 2015). The SEA software tool was used to confirm the enrichment of this motif on the strand relevant for mRNA binding (Bailey and Grant, 2021 Preprint). RNA-seq data are deposited under the accession number GSE171789.
Antibody neutralization assays
Tregs were sorted from CD45.1 Foxp3YFP-Cre mice as CD4+YFP+ cells, subjected to label with CFSE (Thermo Fisher Scientific). Indicated number of Tregs was stimulated with anti-CD3 and anti-CD28 antibody-conjugated beads for overnight prior to suppression assay. Naive CD4 T cells were isolated from WT mice or IL-10Rβ–deficient mice using Mojo sort (BioLegend), followed by staining with cell trace blue (Thermo Fisher Scientific). Indicated number of naive CD4 T cells was added into the stimulated Treg cells for suppression assay. In some experiments, those naive CD4 T cells were pre-incubated with 10 mg/ml of neutralizing antibody for IL-2 (JSE6-1A12), IL-2R (3C7; BioLegend), TGFβ (1D11; BioXcell), and/or IL-10R (1B1.3A; BioXcell) for 30 min before adding Tregs and stimulation beads.
Online supplemental material
Fig. S1 shows that Treg do not suppress TCR-induced upregulation of expression of CD62L and TCR signaling in CD4 Teffs. Fig. S2 shows preparation of the samples from RiboTag mice. Fig. S3 shows Treg-modulated translatome in CD4 Teff cells using the RiboTag approach. Fig. S4 shows SPEED analysis with monosome and polysome fractions revealed mRNA translation status of activation and suppression of T cells. Fig. S5 shows Treg inhibited mRNA translation through mTORC pathway and effects of RocA treatment on mice. Table S1 shows a list of differentially expressed genes using SPEED technique. Table S2 shows reagent identifiers, RNA-seq dataset identifiers, and computational tools links. Data S1 shows analysis and enrichment of TOP sequences.
Acknowledgments
We thank the members of the Savan and Ziegler labs for advice and input. We also thank Daniel J. Campbell for his advice, and Pamela J. Fink, Nandan Gokhale, and Stephen Anderson for their critical reading and comments.
This project was funded by National Institutes of Health grants R21AI143227 to S.F. Ziegler and R. Savan and 5T32AI106677-07 (T32) and 1F32AI145283-01A1 (F32) training grant to L. So.
Authors contributions: Conception and design: L. So, K. Obata-Ninomiya, R. Savan, and S.F. Ziegler. Development of methodology: L. So, K. Obata-Ninomiya, R. Savan, and S.F. Ziegler. Acquisition of data (provided resources and/or facilities, etc.): L. So, K. Obata-Ninomiya, A. Hu, V.S. Muir, A. Takamori, J. Song, and J.H. Buckner. Analysis and interpretation of data (statistical analysis, biostatistics, computational analysis): L. So, K. Obata-Ninomiya, A. Hu, V.S. Muir, A. Takamori, and J. Song. Writing, review, and/or revision of the manuscript: L. So, K. Obata-Ninomiya, A. Hu, V.S. Muir, A. Takamori, J. Song, J.H. Buckner, R. Savan, and S.F. Ziegler.
References
Author notes
L. So and K. Obata-Ninomiya contributed equally to this paper.
Disclosures: V.S. Muir reported “other” from Janssen R&D outside the submitted work. J.H. Buckner reported “other” from Gentibio and grants from Helmsley Charitable Trust outside the submitted work; in addition, J.H. Buckner had a patent to PCT/US2020/039445 pending and a patent to US 8,053,235 issued and is a Scientific Co-Founder and Scientific Advisory Board member of GentiBio, a company engaged in developing engineered regulatory T cell therapies. No other disclosures were reported.
L. So’s current affiliation is Kumquat Biosciences, San Diego, CA, USA.