We used a genome-wide screen in mutagenized mice to identify genes which inactivation protects against lethal neuroinflammation during experimental cerebral malaria (ECM). We identified an ECM-protective mutation in coiled-coil domain containing protein 88b (Ccdc88b), a poorly annotated gene that is found expressed specifically in spleen, bone marrow, lymph nodes, and thymus. The CCDC88B protein is abundantly expressed in immune cells, including both CD4+ and CD8+ T lymphocytes, and in myeloid cells, and loss of CCDC88B protein expression has pleiotropic effects on T lymphocyte functions, including impaired maturation in vivo, significantly reduced activation, reduced cell division as well as impaired cytokine production (IFN-γ and TNF) in response to T cell receptor engagement, or to nonspecific stimuli in vitro, and during the course of P. berghei infection in vivo. This identifies CCDC88B as a novel and important regulator of T cell function. The human CCDC88B gene maps to the 11q13 locus that is associated with susceptibility to several inflammatory and auto-immune disorders. Our findings strongly suggest that CCDC88B is the morbid gene underlying the pleiotropic effect of the 11q13 locus on inflammation.
Inflammation is a normal physiological response to tissue injury caused by infections, burns, trauma, and other insults. Tight regulation of this response is important for initial recognition of danger signals, elimination of the causative lesion, and restoration of tissue homeostasis (Serhan et al., 2010). This involves a complex cascade of events including recruitment of neutrophils, basophils, monocytes, macrophages, and CD4+ and CD8+ T lymphocytes to the site of injury. These infiltrates release soluble mediators (histamine, leukotrienes, and nitric oxide), cytokines (TNF, IFN-γ, and IL-1), chemokines (IL-8, MCP1, and KC), and enzymes (lysosomal proteases) that together establish and amplify the inflammatory response. Timely production of antiinflammatory molecules (PGE2, IL-10, TGF-β, and IL-1Rα) dampens and terminates this response (Lawrence et al., 2002). In the presence of persistent tissue injury or of an unusual infectious/environmental insult, overexpression of proinflammatory mediators or insufficient production of antiinflammatory signals results in an acute or chronic state of inflammation (Serhan et al., 2010). Acute inflammatory conditions, such as septic shock and encephalitis, are difficult to manage clinically and have high mortality rates. Chronic inflammatory diseases such as rheumatoid arthritis (RA; Majithia and Geraci, 2007), inflammatory bowel disease (IBD; Loftus, 2004), systemic lupus erythematosus (SLE; Rahman and Isenberg, 2008), psoriasis (PS; Gelfand et al., 2005), multiple sclerosis (MS; Ramagopalan et al., 2010), type 1 diabetes (T1D; Green et al., 2000), and celiac disease (Trynka et al., 2011) are common and debilitating conditions.
The etiology of acute or chronic inflammatory diseases involves the interaction between intrinsic genetic risk factors of the host, and environmental triggers (Koch et al., 2013; Wang et al., 2014). Environmental triggers are complex, heterogeneous and poorly understood, and may include microbial products such as commensal flora or opportunistic pathogens and/or certain enticing self-antigens which underlie the autoimmune aspect associated with certain chronic inflammatory diseases (Koch et al., 2013; Wang et al., 2014). Linkage and genome-wide association studies (GWAS) have identified a strong but complex genetic component to inflammatory diseases with >400 loci mapped to date for IBD, MS, RA, SLE, PA and others (Cooper et al., 2008; Raychaudhuri et al., 2008; Strange et al., 2010; Jostins et al., 2012; Beecham et al., 2013; Martin et al., 2013). Interestingly, nearly a quarter of the mapped loci are shared in common between 2 or more of these diseases. This shared core of genetic risk factors points to common aspects of pathophysiology among these diseases. Characterization of the corresponding proteins and pathways may provide a better understanding of the mechanisms underlying pathological inflammation in multiple such conditions.
Cerebral malaria (CM) is the most severe complication of Plasmodium falciparum infection in humans; it is an acute and rapidly fatal form of encephalitis with a predominant neuroinflammatory component. CM is characterized by high fever, progressing rapidly to severe cerebral symptoms, including impaired consciousness, seizures, and coma, ultimately leading to lethality in ∼20% of all cases (Newton et al., 2000; Mishra and Newton, 2009). During CM, parasitized erythrocytes (pRBCs) become trapped in the brain microvasculature, triggering a strong inflammatory response featuring recruitment of immune cells and activated platelets, and leading to loss of integrity of the blood brain barrier (Brown et al., 1999; Miller et al., 2002). In mice, experimental CM (ECM) can be induced by infection with Plasmodium berghei ANKA (P. berghei). P. berghei infection in mice mimics several aspects of P. falciparum–induced CM in humans, (de Souza and Riley, 2002; Lamb et al., 2006) including development of acute neuroinflammation and encephalitis within 6–7 d characterized by ataxia, paraplegia, seizures, and coma leading to uniform lethality by day 8–10 after infection.
Recent studies have identified substantial protection against ECM in mouse mutants bearing loss-of-function alleles for key proinflammatory cytokines and chemokines (including IFN-γ and LTα) and transcription factors that activate their expression (STAT1, IRF1, or IRF8). Likewise, elimination of immune cells (NK cells, CD4+, and, in particular, CD8+ T cells) producing these cytokines causes ECM protection (Nitcheu et al., 2003; Rénia et al., 2006; Longley et al., 2011; Hansen, 2012). In addition, transcript profiling studies in brain tissues from P. berghei–infected mice and ChIP sequencing data in primary cells from normal and from ECM-resistant Irf8 mutant mice have identified a core transcriptome activated during ECM (Berghout et al., 2013). Several members of the identified network are bound and regulated by IRF1, IRF8, and STAT1 and their targeted ablation causes ECM resistance. This network also contains genes recently identified as risk factors in acute and chronic human inflammatory conditions (Berghout et al., 2013). These results suggest that genetic studies in the ECM model may identify critical regulatory or rate-limiting steps that underlie common etiology and pathogenesis of inflammatory diseases.
We have used an unbiased genome-wide screen in chemically mutagenized mice (N-ethyl N-nitrosourea; ENU mutagenesis) to identify genes that become resistant to P. berghei–induced ECM after inactivation. In the breeding scheme we have used, ENU-induced mutations are bred to homozygosity in informative pedigrees. High-throughput screening of such pedigrees uncovers informative mutations that manifest themselves as rare pheno-deviant pedigrees with significant, heritable ECM resistance (Richer et al., 2008). Positional cloning of the mutated gene responsible for these effects is facilitated by the de novo nature of the mutation, which can be readily identified by exome sequencing of multiple ECM-resistant animals from the same pedigree (Richer et al., 2010; Bongfen et al., 2012). Using this approach, we report the identification and characterization of a novel, otherwise poorly annotated protein, CCDC88B (coiled-coil domain [CCD] containing protein 88b) that is expressed at high level in immune cells of the lymphoid lineage, and that is required for T cell maturation and activation.
RESULTS
A mutation in Ccdc88b protects against experimental cerebral malaria
We screened ENU-mutagenized mice, looking for the appearance of ECM-resistant pheno-deviant pedigrees on the otherwise ECM-susceptible genetic background of C57BL/6J (B6) and C57BL/10J (B10). In brief, mutagenized B6 males (G0) were crossed to B10 (to facilitate subsequent genetic mapping), and the resulting G1 males were backcrossed to B10. The resulting G2 females were backcrossed to their G1 father to produce G3 pedigrees where mutations are fixed to homozygosity in 25% of the animals (Fig. 1 A). G3 pedigrees were infected with P. berghei, and were monitored for presence of pheno-deviant progeny that fail to develop cerebral symptoms. Consecutive G3 offspring derived from G1 male Deric and G2 female Lilyan were found to segregate ECM-resistant progeny (9 resistant, 18 susceptible; Fig. 1 B). These G3 mice were genotyped for 196 polymorphic markers informative for the B6 versus B10 parental strains, and genome-wide linkage analysis (R/qtl; Broman et al., 2003) detected a single linkage peak on the proximal portion of Chr.19 with a LOD score of 2.98 (peak marker rs31112038; Fig. 1 C). Homozygosity for B6 alleles (derived from mutated G0 male) at this locus was associated with ECM-resistance, whereas these genotypes were under-represented in ECM-susceptible siblings. Conversely, homozygosity for nonmutated B10-derived alleles at this locus was only found in ECM-susceptible mice, whereas B6/B10 heterozygotes were found in both phenotypic groups (Fig. 1 D).
An ENU-induced mutation in the Ccdc88b gene protects mice from P. berghei ANKA-induced cerebral malaria. (A) Breeding scheme used to identify recessive, chemically (ENU)-induced mutations that protect mice against lethal experimental cerebral malaria (ECM). (B) Survival plot of P. berghei–infected G3 animals generated by independent mating of the Deric G1 male to G2 females Renée and Lilyan (10 ECM-resistant in 35 G3 mice tested). Renee, n = 8; Lilyan, n = 27; B6, n = 21. (C) Genome-wide linkage mapping in 27 G3 mice (9R, 18S) from G2 female Lilyan detects linkage of the ECM-resistance trait to Chr.19. (LOD = 2.9; P = 0.144). (D) Haplotype analysis for proximal Chr.19 markers (A, mutant B6 homozygote; B, WT B10 homozygote; H, mutant B6/B10 heterozygote). (E) Schematic representation of CCDC88B protein showing the N-terminal microtubule-binding domain (N), two central CCDs and the C-terminal organelle binding domain (top). Exon-intron structure of the Ccdc88b gene, highlighting exons 21 and 22 where an ENU-induced mutation is located (middle). Sequence alignment of the genomic DNA segment overlapping exons 21/22 (boxed) and intron 21 donor splice site (lower case) from WT controls (B6) and from Deric-derived G3 mice (Ccdc88b Hmz Mut); the mutated nucleotide is identified (bold, star), and the position of the alternate donor splice site used to produce the Ccdc88bm1PGrs mRNA transcript is shaded. The sequence of Ccdc88b mRNA (from RT-PCR sequencing) produced by the WT and G1-derived mutant alleles are shown, along with the predicted amino acid sequence of the encoded polypeptides, and associated termination codons at positions 1483 (for WT), and 1286 (Ccdc88bm1PGrs; bottom). (F) Effect of G1-male–derived B6 Chr. 19 haplotypes on survival of F2S mice bred by intercrossing to 129S1 mice illustrates genetic background independence of the ECM resistance trait. Homozygotes, n = 15; heterozygotes, n = 15; 129S1, n = 8; B6, n = 16.
An ENU-induced mutation in the Ccdc88b gene protects mice from P. berghei ANKA-induced cerebral malaria. (A) Breeding scheme used to identify recessive, chemically (ENU)-induced mutations that protect mice against lethal experimental cerebral malaria (ECM). (B) Survival plot of P. berghei–infected G3 animals generated by independent mating of the Deric G1 male to G2 females Renée and Lilyan (10 ECM-resistant in 35 G3 mice tested). Renee, n = 8; Lilyan, n = 27; B6, n = 21. (C) Genome-wide linkage mapping in 27 G3 mice (9R, 18S) from G2 female Lilyan detects linkage of the ECM-resistance trait to Chr.19. (LOD = 2.9; P = 0.144). (D) Haplotype analysis for proximal Chr.19 markers (A, mutant B6 homozygote; B, WT B10 homozygote; H, mutant B6/B10 heterozygote). (E) Schematic representation of CCDC88B protein showing the N-terminal microtubule-binding domain (N), two central CCDs and the C-terminal organelle binding domain (top). Exon-intron structure of the Ccdc88b gene, highlighting exons 21 and 22 where an ENU-induced mutation is located (middle). Sequence alignment of the genomic DNA segment overlapping exons 21/22 (boxed) and intron 21 donor splice site (lower case) from WT controls (B6) and from Deric-derived G3 mice (Ccdc88b Hmz Mut); the mutated nucleotide is identified (bold, star), and the position of the alternate donor splice site used to produce the Ccdc88bm1PGrs mRNA transcript is shaded. The sequence of Ccdc88b mRNA (from RT-PCR sequencing) produced by the WT and G1-derived mutant alleles are shown, along with the predicted amino acid sequence of the encoded polypeptides, and associated termination codons at positions 1483 (for WT), and 1286 (Ccdc88bm1PGrs; bottom). (F) Effect of G1-male–derived B6 Chr. 19 haplotypes on survival of F2S mice bred by intercrossing to 129S1 mice illustrates genetic background independence of the ECM resistance trait. Homozygotes, n = 15; heterozygotes, n = 15; 129S1, n = 8; B6, n = 16.
Whole exome sequencing of 2 ECM-resistant G3 Deric mice revealed 2 ENU-specific homozygous mutations common to both animals. The first mapped in the Nbea1 gene on Chr.1 (A:G 60,274,052 mm9 assembly), while the second was found in a gene annotated as Ccdc88b (Fig. 1 E) on proximal Chr.19 (T:C, position 6,922,670) located under the mapped linkage peak. The mutation affects the second nucleotide (GT) in the donor splice site of intron 21; RT-PCR analysis of thymic cDNA overlapping the junction between exons 21 and 22 from B6 controls and Ccdc88bm1PGrs homozygote mutants (Fig. 1 E) showed complete abrogation of normal splicing of exon 21 in the Ccdc88bm1PGrs mutant. Instead, the mutant Ccdc88bm1PGrs allele produces an mRNA that makes use of a cryptic splice site located 5 nucleotides upstream of the natural exon 21/intron 21 boundary, causing a 5-nt deletion in the transcript, and a frame-shift resulting in a predicted premature termination codon at aa 1286 (Fig. 1 E). Annotation of the CCDC88B protein indicates presence of an N-terminal microtubule-binding domain, two large coiled-coil domains (CCDs), and a C-terminal organelle-binding domain (Le-Niculescu et al., 2005). The mutant allele is predicted to produce a protein that lacks a portion of the C-terminal CCD and the organelle-binding domain, causing a probable loss of function (Fig. 1 E). The Deric G1 male was outcrossed to ECM-susceptible 129S1/SvImJ (129S1) to produce [Deric X 129S1] F2 mice, which were phenotyped for susceptibility to P. berghei. We confirmed that homozygosity for G1-derived B6 alleles at proximal Chr.19 confers ECM-resistance in this cross, independently of the genetic background of the mating stock (B10 and 129S1; Fig. 1 F).
The Ccdc88b gene is expressed in hematopoietic tissues
To gain insight into the role of CCDC88B in pathological inflammation, the organ, tissue, and cell-specific expression of Ccdc88b mRNA was investigated by whole-mount in situ hybridization (Fig. 2, A–F) using whole body cryosections of juvenile (P10) and adult mice (Ad). Specificity of the Ccdc88b signal was ascertained by parallel hybridization to the corresponding sense cRNA probe (unpublished data). Ccdc88b is found expressed in a tissue-specific fashion and only in hematopoietic organs. At P10, Ccdc88b is detected in the thymus, spleen, and bone marrow (hip/femur; Fig. 2 A). In adult mice, it is expressed in the spleen, LNs, and bone marrow associated with skeletal structures including ribs, hip bones, and femurs (Fig. 2, B and C). Additional analysis at higher magnification and increased exposure times suggests that Ccdc88b is expressed in germinal centers and lymphatic nodules of the spleen (Fig. 2 D), and in the medullar areas of LNs (Fig. 2 E), whereas both the medullar and cortical regions of the thymus are positive for expression (Fig. 2 F). These results indicate that Ccdc88b is expressed in a tissue- and cell-specific fashion in myeloid cells and T lymphocytes, two cell populations that are key mediators of inflammation, whereas it is not expressed in B lymphocytes (negative staining detected in lymphoid follicles of LNs).
Organ and cell-specific expression of Ccdc88b mRNA and CCDC88B protein. Cryosections prepared from newborn (post-natal day 10, p10) (A) and adult male (B) and female (C) mice were used for in situ hybridization. Following hybridization, sections were exposed to photographic emulsion and developed. Parallel sections were stained with cresyl violet (CV), and are shown as references (with scale bar). Ccdc88b mRNA expression is detected in thymus (Th), spleen (Spl), LNs, and BM. Higher magnification images of hybridization signals (longer exposures) detected in the spleen (D), LNs (E), and thymus (F). In the spleen, Ccdc88b mRNA is expressed in the lymphatic nodules (LN) and germinal centers (GC), whereas in thymus it is detected in the medulla (Me) and the cortex (Cx). In LNs, Ccdc88b is expressed in the medulla (Me). Cryosections from spleen, LNs, and thymus were stained and analyzed by immunofluorescence for organ (G) and cell-specific (H) expression of CCDC88B (CCDC; red) and T lymphocyte–specific marker CD3 (CD3; green), showing that CD3+ cells in these organs express CCDC88B. (I) Parallel analysis of the same organs stained with the B lymphocyte marker B220 (green) show a lack of localization of CCDC88B to B220+ B lymphocytes zones (last column). Bars: (A–C) 1 cm; (D–F), 1 mm; (G) 100 µm; (H) 25 µm; (I) 100 µm. Abbreviations used: Adr, adrenal gland; Ar, artery; B, bone; Br, brain; Cb, cerebellum; H, heart; In, incisor; KB, knee bone; Ki, kidney; Li, liver; LI, large intestine; Lu, lung; Ov, ovary; Ovi, oviducts; PB, pelvis bone; R, ribs; Sk, skin; SI, small intestine; SM, skeletal muscle; Spl, spleen; St, stomach; Th, thymus; U, uterus; Ve, vertebrae.
Organ and cell-specific expression of Ccdc88b mRNA and CCDC88B protein. Cryosections prepared from newborn (post-natal day 10, p10) (A) and adult male (B) and female (C) mice were used for in situ hybridization. Following hybridization, sections were exposed to photographic emulsion and developed. Parallel sections were stained with cresyl violet (CV), and are shown as references (with scale bar). Ccdc88b mRNA expression is detected in thymus (Th), spleen (Spl), LNs, and BM. Higher magnification images of hybridization signals (longer exposures) detected in the spleen (D), LNs (E), and thymus (F). In the spleen, Ccdc88b mRNA is expressed in the lymphatic nodules (LN) and germinal centers (GC), whereas in thymus it is detected in the medulla (Me) and the cortex (Cx). In LNs, Ccdc88b is expressed in the medulla (Me). Cryosections from spleen, LNs, and thymus were stained and analyzed by immunofluorescence for organ (G) and cell-specific (H) expression of CCDC88B (CCDC; red) and T lymphocyte–specific marker CD3 (CD3; green), showing that CD3+ cells in these organs express CCDC88B. (I) Parallel analysis of the same organs stained with the B lymphocyte marker B220 (green) show a lack of localization of CCDC88B to B220+ B lymphocytes zones (last column). Bars: (A–C) 1 cm; (D–F), 1 mm; (G) 100 µm; (H) 25 µm; (I) 100 µm. Abbreviations used: Adr, adrenal gland; Ar, artery; B, bone; Br, brain; Cb, cerebellum; H, heart; In, incisor; KB, knee bone; Ki, kidney; Li, liver; LI, large intestine; Lu, lung; Ov, ovary; Ovi, oviducts; PB, pelvis bone; R, ribs; Sk, skin; SI, small intestine; SM, skeletal muscle; Spl, spleen; St, stomach; Th, thymus; U, uterus; Ve, vertebrae.
The CCDC88B protein is expressed in T lymphocytes
To identify the cell populations that express the CCDC88B protein, we produced a polyclonal anti-CCDC88B antibody. For this, an 88-aa segment from the first CCD of CCDC88B, which shows low sequence similarity to other CCDC88 proteins and to CCDC88B relatives in other species, was fused in-frame to glutathione-S-transferase and used as an immunogen (Fig. S1 A). The anti-CCDC88B antibody was affinity purified, and its specificity was tested by immunoblotting against cell extracts from HEK293T cells expressing full-length CCDC88B protein modified by addition of a hemagglutinin (HA) epitope at its C terminus (Fig. S1B). The anti-CCDC88B antibody detects a major ∼170 kD immunoreactive band, which is also recognized by the anti-HA monoclonal antibody (Fig. S1B). Discrete degradation products detected by both, anti-CCDC88B and anti-HA antibodies, suggest a protease-sensitive site located in the intervening region separating the two CCDs of CCDC88B. Immunofluorescence studies of the same cells stained with either anti-HA and anti-CCDC88B antibodies confirmed the specificity of the antibody, and bright intracellular staining which is absent from control nontransfected cells (Fig. S1 C).
The identity of the cells expressing the CCDC88B protein was investigated in spleen, LNs, and thymus (Fig. 2, G–I). We used double immunofluorescence on frozen tissue sections stained with a combination of antibodies against CCDC88B, CD3 (for T cells), B220 (for B lymphocytes), and CD11b (for myeloid cells). Most obviously, there was strong CCDC88B protein expression in both the LNs and the thymus. In these organs, the majority of the staining was associated with CD3+ T lymphocytes (Fig. 2, G and H), with strong co-localization of CCDC88B with the membrane-bound CD3 protein, which forms part of the T cell receptor complex. In the spleen, CCDC88B staining was noted in the red pulp (Fig. 2 G), whereas germinal centers rich in B220+ B lymphocytes were largely negative for CCDC88B (Fig. 2 I). In the spleen, we also detected CCDC88B protein expression in some, but not all, CD11b+ myeloid cells, as well as in other cells, which were negative for CD11b, CD3, B220, and for the erythroid marker Ter119 (unpublished data). These experiments establish that CCDC88B is strongly expressed in CD3+ lymphocytes, with additional presence in myeloid cells.
Resistance to experimental cerebral malaria is associated with absence of CCDC88B protein in hematopoietic cells
The effect of the Ccdc88bm1PGrs mutation on protein expression was investigated by immunoblotting, and by immunofluorescence using primary cells and on frozen tissue sections. Immunoblotting of total protein extracts from spleen, thymus, and LNs detected the ∼170 kD CCDC88B WT protein, whereas no mature CCDC88B protein was detected in tissue extracts from the mutant; kidney extracts were used as negative control for CCDC88B expression (Fig. 3 A). Parallel immunostaining of frozen sections of the same tissues confirmed the immunoblotting results and indicated that CCDC88B protein expression is lost in the mutant (Fig. 3 B). These results suggest that the ECM-protective Ccdc88bm1PGrs mutation results in either absence of translated protein or production of a CCDC88B variant which is unstable, and possibly targeted for degradation. In turn, this also suggests that the Ccdc88bm1PGrs mutant allele behaves as a complete loss of function.
The CCDC88B protein is expressed in CD8+ and CD4+ T lymphocytes and in NK cells of WT mice and is lost in mutant mice. (A) Immunoblotting analysis of total organ extracts from spleen, thymus, LNs, and kidney (used as a negative control) prepared from WT B6 controls (WT) and from Ccdc88bm1PGrs mice (Mutant), and probed with anti-CCDC88B antibody shows absence of CCDC88B protein in mutant animals. Similar loading was verified by probing the same immunoblot with an anti-actin antibody. (B) Immunofluorescence analysis of primary tissues from WT and mutant animals with the anti-CCDC88B antiserum fails to detect expression of CCDC88B protein in tissues from mutants, compared with WT controls. Nuclei were stained with DAPI to localize the cells. Bars, 100 µm. (C) CCDC88B protein expression in primary cells determined by FACS analysis of spleen cell suspensions from WT B6 controls and Ccdc88bm1PGrs mutant mice. Single-cell suspensions were incubated with a cocktail of antibodies directed against T lymphocytes (CD3, CD4, and CD8), B lymphocytes (CD19), and NK cells (NKp46), and then intracellularly stained with the anti-CCDC88B affinity purified antiserum. Numbers in each quadrant indicate the percentage of the total cell population. (D) Survival of bone marrow chimeric mice after infection with PbA (mutant to B6, n = 14; B6 to B6, n = 14; B6 to mutant, n = 4; Mutant to mutant, n = 4). (E) Whole splenocyte and total T cell transfer into Jak3−/− mice (Spl., splenocytes; C57BL6J WT, n = 5; B6 T cells to Jak3−/−, n = 5; Mutant T cells to Jak3−/−, n = 5; Mutant Spl. in Jak3−/−, n = 5; B6 Spl. in Jak3−/−, n = 3; Jak3−/−, n = 5).
The CCDC88B protein is expressed in CD8+ and CD4+ T lymphocytes and in NK cells of WT mice and is lost in mutant mice. (A) Immunoblotting analysis of total organ extracts from spleen, thymus, LNs, and kidney (used as a negative control) prepared from WT B6 controls (WT) and from Ccdc88bm1PGrs mice (Mutant), and probed with anti-CCDC88B antibody shows absence of CCDC88B protein in mutant animals. Similar loading was verified by probing the same immunoblot with an anti-actin antibody. (B) Immunofluorescence analysis of primary tissues from WT and mutant animals with the anti-CCDC88B antiserum fails to detect expression of CCDC88B protein in tissues from mutants, compared with WT controls. Nuclei were stained with DAPI to localize the cells. Bars, 100 µm. (C) CCDC88B protein expression in primary cells determined by FACS analysis of spleen cell suspensions from WT B6 controls and Ccdc88bm1PGrs mutant mice. Single-cell suspensions were incubated with a cocktail of antibodies directed against T lymphocytes (CD3, CD4, and CD8), B lymphocytes (CD19), and NK cells (NKp46), and then intracellularly stained with the anti-CCDC88B affinity purified antiserum. Numbers in each quadrant indicate the percentage of the total cell population. (D) Survival of bone marrow chimeric mice after infection with PbA (mutant to B6, n = 14; B6 to B6, n = 14; B6 to mutant, n = 4; Mutant to mutant, n = 4). (E) Whole splenocyte and total T cell transfer into Jak3−/− mice (Spl., splenocytes; C57BL6J WT, n = 5; B6 T cells to Jak3−/−, n = 5; Mutant T cells to Jak3−/−, n = 5; Mutant Spl. in Jak3−/−, n = 5; B6 Spl. in Jak3−/−, n = 3; Jak3−/−, n = 5).
The immune cell populations expressing the CCDC88B protein and that may underlie ECM resistance in the Ccdc88bm1PGrs mutant, were further investigated by flow cytometry. For this, splenocytes from WT and from Ccdc88bm1PGrs mutants were labeled with antibodies, which included CCDC88B, and markers for T cells (CD3, CD8, and CD4), B cells (CD19), and NK cells (NKp46). CCDC88B was strongly expressed in T cells (CD3+) in both the CD4+ and CD8+ populations, and was also expressed in NK cells (NKp46+; Fig. 3 C). The specificity of CCDC88B staining was further validated by the loss of staining in the same cell populations isolated from the Ccdc88bm1PGrs mutant animals (Fig. 3 C). In agreement with results from Fig. 2, B lymphocytes (CD19+, B220+) do not express CCDC88B. These results demonstrate that CCDC88B is expressed primarily in T cells, and that the Ccdc88bm1PGrs mutation causes a loss-of-protein expression in these cells. These results suggested that CCDC88B might play a role in the proinflammatory function of T cells during neuroinflammation. In agreement with a hematopoietic origin for the ECM-protective effect of the Ccdc88bm1PGrs mutation, studies in bone marrow chimeras demonstrated that ECM resistance could be transferred from the Ccdc88bm1PGrs mutant into lethally irradiated WT B6 mice. Similarly, ECM susceptibility could be transferred from B6 into irradiated Ccdc88bm1PGrs mutant animals (Fig. 3 D). These results established that the loss of CCDC88B expression in the hematopoietic cell compartment is required and sufficient to cause ECM resistance. The role of T cells in the ECM resistance phenotype of Ccdc88bm1PGrs mutant mice was investigated by further adoptive transfer studies and using either total splenocytes or purified T cells. In these experiments, we used as recipient an immunodeficient Jak3−/− mutant stock that we have previously shown to be ECM resistant, a phenotype that can be corrected to susceptibility by T cell transfer (Bongfen et al., 2012). We observed that although ECM-susceptibility could be conferred to Jak3−/− mice by total splenocytes or purified T cells, from normal B6 mice, Ccdc88bm1PGrs mutant splenocytes or purified T cells, could not do so in the same experimental setting (Fig. 3 E). These results establish that the cell population responsible for the ECM resistance phenotype of in the Ccdc88bm1PGrs mutant is the T cells compartment.
CCDC88B-deficient T lymphocytes are functionally impaired
We investigated the effect of the loss of CCDC88B expression on the number and activity of different T cells subsets. First, we did not detect significant changes in the frequencies and number of splenic CD4+ and CD8+ T cells present in Ccdc88bm1PGrs mutants compared with WT mice (unpublished data). Hence, the proportion and number of the antigen-experienced T cells (CD44hi) was analyzed. Spleens of Ccdc88bm1PGrs mutant mice displayed a major decrease in the total number and proportion (ninefold) of CD44hi CD8+ T cells compared with WT mice (Fig. 4 A). These numbers suggest a role for CCDC88B protein in training T cells for antigen recognition in the context of Class I MHC. On the other hand, a similar percentage and number of CD44hi CD4+ T cells was noted in mutant animals and WT controls (Fig. 4 B). We then investigated whether the decreased CD44hi CD8+ T cells subpopulation in Ccdc88bm1PGrs mutant mice is associated with a reciprocal increase of naive T cells in the mutant (CD62L+CD44low). We noted a significant increase of CD62L+CD44low proportion and numbers in CD8+ T cells from Ccdc88bm1PGrs mutant mice compared with WT mice (Fig. 4 C). In the CD4+ compartment, no statistical differences between WT and mutant mice were found in CD62L+CD44low cell numbers (Fig. 4 D), although frequency of CD44low CD4+ T cells were greater in mutant than WT mice (Fig. 4 B). Thus, splenic peripheral circulating CD8+ T cells display a reduced number and fraction of antigen-experienced T cells in Ccdc88bm1PGrs mutant mice. This is concomitant with an increased proportion of naive T cells, and that is again most striking in the CD8+ compartment.
CCDC88B is required for maturation of T lymphocytes. Spleen cell suspensions from WT and from Ccdc88bm1PGrs mutant mice were incubated with a cocktail of antibodies including T cell–specific anti-CD3, anti-CD4, and anti-CD8, as well as markers of T cell maturation CD44 and CD62L. (A) Expression of CD44 within TCRβ+ CD8+ T cells population (left), total numbers (right), and proportion of CD44hi CD8+ T cells (center) in the gated areas of histograms is shown for WT (B6) and Ccdc88bm1PGrs (Mutant) animals. Similarly, the effect of the Ccdc88b mutation on the expression of CD44 on TCRβ+ CD4+ T cells is shown in B. (C and D) Spleen cells were stained as in A and B for the expression of CD62L and CD44. Dot plots (left) identify naive T cells as CD62L+CD44low (bottom right quadrant). Total numbers (right) and proportion of CD62L+CD44low cells (center) in the gated areas of the dot plot is shown for WT (B6) and Ccdc88bm1PGrs (Mutant) animals for CD4+ and CD8+ T cells. n = 6 for all experiments. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Students t test: *, P < 0.05; ***, P < 0. 001).
CCDC88B is required for maturation of T lymphocytes. Spleen cell suspensions from WT and from Ccdc88bm1PGrs mutant mice were incubated with a cocktail of antibodies including T cell–specific anti-CD3, anti-CD4, and anti-CD8, as well as markers of T cell maturation CD44 and CD62L. (A) Expression of CD44 within TCRβ+ CD8+ T cells population (left), total numbers (right), and proportion of CD44hi CD8+ T cells (center) in the gated areas of histograms is shown for WT (B6) and Ccdc88bm1PGrs (Mutant) animals. Similarly, the effect of the Ccdc88b mutation on the expression of CD44 on TCRβ+ CD4+ T cells is shown in B. (C and D) Spleen cells were stained as in A and B for the expression of CD62L and CD44. Dot plots (left) identify naive T cells as CD62L+CD44low (bottom right quadrant). Total numbers (right) and proportion of CD62L+CD44low cells (center) in the gated areas of the dot plot is shown for WT (B6) and Ccdc88bm1PGrs (Mutant) animals for CD4+ and CD8+ T cells. n = 6 for all experiments. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Students t test: *, P < 0.05; ***, P < 0. 001).
Next, splenocytes were subjected to TCR-mediated activation by incubation with anti-CD3 and anti-CD28 (CD3/CD28) antibodies or by activation with PMA and ionomycin (PMA/Iono), and the effect of the mutation on expression of activation marker CD69 was analyzed. CD69 expression revealed a much lower T cell activation in both CD4+ and CD8+ T cells from Ccdc88bm1PGrs mutant mice than controls (Fig. 5, A and B). This was particularly obvious for CD8+ cells and after stimulation via the T cell receptor (CD3/CD28). To address the question whether the decreased activation status seen in Ccdc88bm1PGrs mutants affects T cell proliferation, the proliferative capabilities of CD8+ and CD4+ T cells in response to CD3/CD28 stimulation or PMA/Iono activation were assessed by the analysis of the CFSE dye dilution after 48 h of splenocyte culture. Ccdc88bm1PGrs mutant CD8+ and CD4+ T cells activated with PMA/Iono show no difference in terms of proliferation compared with WT controls (Fig. 5, C and D). In contrast, when T cells were stimulated by engagement of the T cell receptor (anti-CD3 and anti-CD28), both CD8+ and CD4+ T cells showed lower proliferative capacity than WT animals (Fig. 5, C and D). To ensure that the difference in cell proliferation between WT and Ccdc88bm1PGrs mutant mice is not the consequence of increased cell death of the Ccdc88bm1PGrs mutant T cells, cell viability was analyzed. The percentage of dead and viable cells was not different between WT and CCDC88B T cells (Fig. 5, E and F).
Reduced activity of Ccdc88bm1PGrs mutant T cells in response to nonspecific stimuli or to engagement of T cell receptor. (A and B) Splenocytes from WT (B6) and Ccdc88bm1PGrs mutants were incubated with either PMA/Iono or with anti-CD3 and anti-CD28 antibodies for 6 h. Spleen cells were examined for expression of the activation marker CD69, before (gray line), and after (black line) stimulation CD8+ T cells (A), and CD4+ (B) T cells (n = 4). (C and D) Cells were incubated with CFSE and cell proliferation in response to indicated stimuli was analyzed by FACS for CD8+ (C) and CD4+ (D) T cells after PMA/Iono treatment or TCR engagement (red traces compared with blue traces; n = 3); Quantification is provided using the same color code. (E and F) Cell death was analyzed using a fixable viability, live/dead stain, and gated on live cells. Each peak representing viable (left) and nonviable (right) cells (n = 3). CD8+ (G) and CD4+ (H) T cells from spleen of WT (B6) and Ccdc88bm1PGrs mutants were incubated with either PMA/Iono or with anti-CD3 and anti-CD28 antibodies for 6 h, followed by intracellular staining of TNF and IFN-γ. Results are quantified and expressed as percentage of CD8+ (G) or CD4+ (H) T cells that are positive for TNF, IFN-γ (graphs at the right), or both (from scatter plot) for indicated stimuli. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; *** P < 0.001).
Reduced activity of Ccdc88bm1PGrs mutant T cells in response to nonspecific stimuli or to engagement of T cell receptor. (A and B) Splenocytes from WT (B6) and Ccdc88bm1PGrs mutants were incubated with either PMA/Iono or with anti-CD3 and anti-CD28 antibodies for 6 h. Spleen cells were examined for expression of the activation marker CD69, before (gray line), and after (black line) stimulation CD8+ T cells (A), and CD4+ (B) T cells (n = 4). (C and D) Cells were incubated with CFSE and cell proliferation in response to indicated stimuli was analyzed by FACS for CD8+ (C) and CD4+ (D) T cells after PMA/Iono treatment or TCR engagement (red traces compared with blue traces; n = 3); Quantification is provided using the same color code. (E and F) Cell death was analyzed using a fixable viability, live/dead stain, and gated on live cells. Each peak representing viable (left) and nonviable (right) cells (n = 3). CD8+ (G) and CD4+ (H) T cells from spleen of WT (B6) and Ccdc88bm1PGrs mutants were incubated with either PMA/Iono or with anti-CD3 and anti-CD28 antibodies for 6 h, followed by intracellular staining of TNF and IFN-γ. Results are quantified and expressed as percentage of CD8+ (G) or CD4+ (H) T cells that are positive for TNF, IFN-γ (graphs at the right), or both (from scatter plot) for indicated stimuli. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; *** P < 0.001).
To investigate whether the T cell phenotype in Ccdc88bm1PGrs mutant mice is associated with decreased effector function, the production of inflammatory cytokines was assessed after activation with anti-CD3 and anti-CD28 or treatment with PMA/Iono, followed by intracellular staining of TNF and IFN-γ. We observed a reduction in the numbers of TNF+ T cells in the Ccdc88bm1PGrs mutant compared with WT, and this for both the CD8+ and the CD4+ T cell compartments (Fig. 5, G and H). Ccdc88b mutants also showed strong reduction of IFN-γ production by CD8+ T cells in response to either treatment compared with their WT counterpart (Fig. 5 G). Interestingly, the reduction of TNF and IFN-γ was observed in both CD44hi and CD44low mutant T cells of the CD8+ and CD4+ compartments (unpublished data).
Finally, we investigated the effect of CCDC88B deficiency on T cell function during P. berghei infection. At day 5 after infection, splenocytes were subjected to TCR-mediated activation or activation by PMA/Iono, and the level of CD69 expression and production of TNF and IFN-γ were analyzed (Fig. 6). CD69 expression was significantly reduced in both CD8+ (Fig. 6 A) and CD4+ (Fig. 6 B) T cells of mutant infected mice, either in absence of in vitro treatment or after stimulation with CD3/CD28 or with PMA/Iono (Fig. 6, A and B). In addition, TNF and IFN-γ production by CD8+ (Fig. 6 C) and by CD4+ (Fig. 6 D) T cells from PbA-infected CCDC88B-deficient animals was reduced compared to controls.
Reduced T cell activation in Ccdc88bm1PGrs mutant mice during P. berghei–induced ECM. Mice were infected with P. berghei and 5 d later, splenocytes were harvested and incubated with indicated stimuli (anti-CD3/anti-CD28; PMA/Iono) and T cell activation (CD69 expression) was assessed in CD8+ (A) and CD4+ (B) T cells, as described Fig. 5. Intracellular production of TNF, and IFN-γ in CD8+ (C) and CD4+ (D) was also measured in the same cells. All results are quantified and expressed as percentage of CD8+ (A and C) or CD4+ (B and D) T cells that are positive for CD69, TNF, and/or IFN-γ (graphs at the right) for the indicated stimuli. n = 4 in all experiments. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Reduced T cell activation in Ccdc88bm1PGrs mutant mice during P. berghei–induced ECM. Mice were infected with P. berghei and 5 d later, splenocytes were harvested and incubated with indicated stimuli (anti-CD3/anti-CD28; PMA/Iono) and T cell activation (CD69 expression) was assessed in CD8+ (A) and CD4+ (B) T cells, as described Fig. 5. Intracellular production of TNF, and IFN-γ in CD8+ (C) and CD4+ (D) was also measured in the same cells. All results are quantified and expressed as percentage of CD8+ (A and C) or CD4+ (B and D) T cells that are positive for CD69, TNF, and/or IFN-γ (graphs at the right) for the indicated stimuli. n = 4 in all experiments. Data are representative of at least two independent experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT B6 mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Collectively, these data indicate that CCDC88B plays an important role in T cell maturation, production of proinflammatory cytokines, and proliferation, in response to specific (T cell receptor dependent) or nonspecific stimulation, and during PbA-induced ECM.
CCDC88B expression in myeloid cells
CCDC88B expression was also noted in myeloid cells, and the effect of Ccdc88b mutation on the function of these cells was investigated both at steady state and after infection with P. berghei. Comparative FACS analysis in control and mutant mice (Fig. 7, A–C) detected CCDC88B expression in CD11b+ cells, particularly in Ly6G+ neutrophils and in Ly6Chi inflammatory monocytes at steady state, with additional expression in F4/80+ macrophages. At steady-state, there was no effect of Ccdc88b mutation on the number of CD11b+ myeloid cells (Ly6G+, Ly6C+, F4/80+) or on their activation status monitored by expression of CD40 and CD80 (FACS analysis). After P. berghei infection, we noted an increase in the number of CD11b+ cells in the spleen of Ccdc88b mutant mice, with reduced expression of CD40/CD80 in the expanded populations of CD11b+Ly6G+, CD11b+F4/80+, CD11b+Ly6C+ cells. These results demonstrate CCDC88B expression in myeloid cells, suggesting a possible functional role in these cells. However, the limited effect of Ccdc88bm1PGrs mutation on myeloid cells function, together with the obvious effect of Ccdc88bm1PGrs mutation on T cell function and results from T cells transfer studies, indicate that the protective effect of Ccdc88bm1PGrs mutation on P. berghei–induced neuroinflammation is most likely through altered T cell proinflammatory function.
Expression of CCDC88B in myeloid cells and effect of Ccdc88bm1PGrs deficiency. (A) Immunophenotyping of subsets of myeloid cell populations in WT and Ccdc88bm1PGrs mutant mice was as described in Fig. 3 C. Spleen cells from WT and Ccdc88bm1PGrs mutants were stained with a cocktail of anti-CCDC88B, anti-CD11b, anti-Ly6G, anti-F4/80, and anti-Ly6C, and were analyzed by FACS. (B and C) Splenocytes from control (NI) and from P. berghei–infected mice (PbA) were stained with a cocktail of antibodies against myeloid cells (CD11b, Ly6G, Ly6C, and F4/80) markers (A), and the number of each cell type (B) and the proportion of these cells expressing CD40 and CD80 (C) was analyzed. Data are representative of two independent experiments. n = 4 in all experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; ***, P < 0.001).
Expression of CCDC88B in myeloid cells and effect of Ccdc88bm1PGrs deficiency. (A) Immunophenotyping of subsets of myeloid cell populations in WT and Ccdc88bm1PGrs mutant mice was as described in Fig. 3 C. Spleen cells from WT and Ccdc88bm1PGrs mutants were stained with a cocktail of anti-CCDC88B, anti-CD11b, anti-Ly6G, anti-F4/80, and anti-Ly6C, and were analyzed by FACS. (B and C) Splenocytes from control (NI) and from P. berghei–infected mice (PbA) were stained with a cocktail of antibodies against myeloid cells (CD11b, Ly6G, Ly6C, and F4/80) markers (A), and the number of each cell type (B) and the proportion of these cells expressing CD40 and CD80 (C) was analyzed. Data are representative of two independent experiments. n = 4 in all experiments. Data are represented as mean ± SD. Results are considered significant at P < 0.05 when comparing WT mice to the Ccdc88bm1PGrs mutants (two-tailed Student’s t test: *, P < 0.05; ***, P < 0.001).
DISCUSSION
In this study, we have shown that an ENU-induced mutation in the mouse Ccdc88b gene causes protection against P. berghei–induced lethal ECM. We establish that the CCDC88B protein is expressed almost exclusively in the hematopoietic system, primarily in the lymphoid and myeloid lineages. Ccdc88b mutant mice develop normally, have no gross anatomical defects, and present with seemingly normal hematopoietic organs with respect to number of myeloid and lymphoid cells. However, we demonstrate that loss of CCDC88B protein in the hematopoietic compartment is sufficient to induce resistance to CM. Moreover, the Ccdc88bm1PGrs mutation causes defects in T cell functions, which are concomitant to reduced production of proinflammatory cytokines upon specific or nonspecific stimulation of these cells. These results suggest that CCDC88B acts as a positive regulator of T cell maturation and T cell function.
CD4+ and CD8+ T cells play a critical role in early inflammatory and immune responses of the host (Miyakoda et al., 2008; Villegas-Mendez et al., 2012), and partial or complete elimination of these cells or of their key components or of their secreted products causes ECM resistance (Yañez et al., 1996; Nitcheu et al., 2003; Rénia et al., 2006; Hansen, 2012). The mechanism by which CCDC88B affects T cell maturation and function remains to be fully elucidated. However, it does not appear to be linked to the thymic production of T cells per se, as mutant and WT animals show similar numbers of both CD4+ and CD8+ T cells in spleen, LNs, and thymus (unpublished data). However, Ccdc88bm1PGrs mutant T cells show several striking differences. First, Ccdc88b-deficient T cells show a maturation defect, which is characterized by a reduced number of antigen-experienced CD44hi T cells (DeGrendele et al., 1997), a phenotype that is most striking in the CD8+ compartment. CD44hi T cells represent a heterogeneous population that includes central memory cells (Tcm) and effector memory cells (Tem; Sallusto et al., 2004). We found that in CD8+ T cells, Tcm (CD62Lhi CD44hi) and Tem (CD66Llo CD44hi) populations are both dramatically decreased in the absence of the CCDC88B protein. Previous data has shown that CD8+ memory T cells (Tcm and Tem) are programmed to rapidly proliferate under antigen stimulation, and the efficiency of cytokine production by T cells correlates with the CD44hi cell population (Cho et al., 1999; Veiga-Fernandes et al., 2000; Krieg et al., 2007). Second, our study shows that CCDC88B is required for several functions of T cells, in both the CD4+ and the CD8+ compartments. This includes expression of cell surface markers of activation, proliferation, and cytokine production in response to TCR cross-linking or to nonspecific stimuli (PMA/Iono), and during infection with P. berghei. Accumulating evidence indicates that inflammatory responses mediated by cytokines such as TNF, IFN-γ, and effector cells including CD4+ and CD8+ T cells contribute to neuroinflammation (Nitcheu et al., 2003; Rénia et al., 2006; Hansen, 2012). Third, we also detected CCDC88B protein expression in CD11b+Ly6G+ (neutrophils), CD11b+Ly6Ghi (inflammatory monocytes) and CD11b+F4/80+ myeloid cells, as well as in NKp46+ NK cells. Although these cells represent a small proportion of CCDC88B+ total hematopoietic cells, CD11b+ dendritic cells (Berghout et al., 2013), Ly6G+ neutrophils (Porcherie et al., 2011), and NK cells (Hansen et al., 2003, 2007) have all been shown to contribute to pathological neuroinflammation in ECM. In addition, we noted expansion of the Ly6G+, Ly6Ghi, and F4/80+ myeloid cells (CD11b+) in the spleen of P. berghei–infected Ccdc88bm1PGrs mutant mice compared with B6 controls, although the state of activation of these cells (frequency of CD40+, CD80+) was reduced compared with the same cells present in infected wild-type mice. These results suggest that CCDC88B may play a role in myeloid cells, and that a defect in this compartment may additionally contribute to a primary T cell defect causing ECM resistance in Ccdc88bm1PGrs mutants.
Little is known about the structure and biochemical function of CCDC88B. It belongs to the so-called hook related protein family that includes CCDC88B (Gipie), CCDC88A (GIV, Girdin, and HkRP1), and CCDC88C (DAPLE and HkRP2). This family is defined by shared secondary structure motifs, including an N-terminal hook-related microtubule-binding domain (MBD), a central CCD, and a C-terminal domain thought to be involved in binding different subcellular organelles (Le-Niculescu et al., 2005). Whereas the MBD and CCD domains show sequence conservation, the divergent C-terminal domain is thought to confer protein-specific functions (Le-Niculescu et al., 2005; Simpson et al., 2005). The overall primary sequence similarity is low among members of this family, but includes recognizable leucine zipper and peroxisomal targeting signal sequences (PTS2) in the CCD (Le-Niculescu et al., 2005). CCDC88B and CCDC88A appear to be more closely related, sharing 48% sequence identity and 80% similarity in a portion of the CCD that contains a Gα-protein–binding motif (Le-Niculescu et al., 2005) and 29% identity and 52% similarity in the hooklike MBD, previously shown in other proteins to mediate interactions with microtubules such as in the case of the C. elegans relative ZYG-12 (Walenta et al., 2001; Malone et al., 2003; Le-Niculescu et al., 2005).
CCDC88A has been detected on ER-Golgi vesicles and helps support trafficking of these vesicles (Le-Niculescu et al., 2005), whereas CCDC88C binds actin and interacts with the scaffolding protein Disheveled (Dvl) through its PDZ binding motif, acting in the Wnt signaling pathway (Oshita et al., 2003). In an endothelial cell line, Gipie (CCDC88B) was detected in ER-Golgi vesicles where, upon ER stress (unfolded protein response), it can bind GRP78 and IRE1 to help suppress ER stress–induced apoptosis (Matsushita et al., 2011). The relationship of these results, if any, to findings in the current study is unclear. We did not detect CCDC88B mRNA expression in vascular structures and associated endothelial cells, and cellular and subcellular localization studies of CCDC88B by co-immunofluorescence in T lymphocytes are not consistent with ER-Golgi localization but rather suggest overlapping staining with the membrane protein CD3. Likewise, we did not detect major differences between WT and mutant T cells in cell death after stimulations with either anti-CD3 and anti-CD28 or with PMA and ionomycin. Therefore, the molecular mechanism of action of CCDC88B in regulating T cell function remains to be elucidated, but does not appear linked to regulation of ER stress in these cells.
The human CCDC88B gene maps to distal chromosome 11 (11q13; position 64.1 Mb). Recent genome-wide association studies, meta-analyses and immunochip experiments have detected ubiquitous association of a 11q13 locus with susceptibility to several inflammatory diseases, including sarcoidosis (SC; Fischer et al., 2012), IBD (Jostins et al., 2012), PS (Tsoi et al., 2012), Alopecia areata (Petukhova et al., 2010), MS (Beecham et al., 2013), and primary biliary cirrhosis (PBC; Juran et al., 2012). The morbid genes underlying the 11q13 effect in these studies has remained uncertain, as the top linked marker (top SNP) is in linkage disequilibrium with several candidate genes (23–36 genes) in a >500-kb gene-dense chromosomal segment. In these studies, the causative candidate gene has been alternately suggested to be PRDX5 (PBC, PS, and MS), RPS6KA4 (PS), and CCDC88B (MS, IBD, PBC, and SC).
The PRDX5 gene (64.08 Mb) encodes a member of the peroxiredoxin family of antioxidant enzymes, which use cytoplasmic and mitochondrial thioredoxins to reduce hydrogen peroxide and alkyl hydroperoxides. The encoded protein may play an antioxidant-protective role in different tissues under normal conditions and during inflammatory processes (Sun et al., 2010; Knoops et al., 2011). RPS6KA4 (64.12 Mb) encodes a member of the ribosomal S6 kinase family of serine/threonine kinases (also known as p90S6K; MSK2, RSK-B), which is required for phosphorylation of numerous intracellular substrates in response to mitogens and stress, and is an important downstream effector of mitogen-activated protein kinase (MAPK) for regulation of transcription factors such as CREB1 and ATF1, and RELA (member of the NF-κB family) in response to TNF (Leighton et al., 1995). Therefore, both PRDX5 and RPSKA4 represent good positional candidates, although their role in pathological inflammation has not yet been validated in humans or mice. However, our results in mice establish that the neighboring gene, Ccdc88b, is required in vivo for T cell–dependent and IFN-γ/TNF–driven inflammatory response during neuroinflammation.
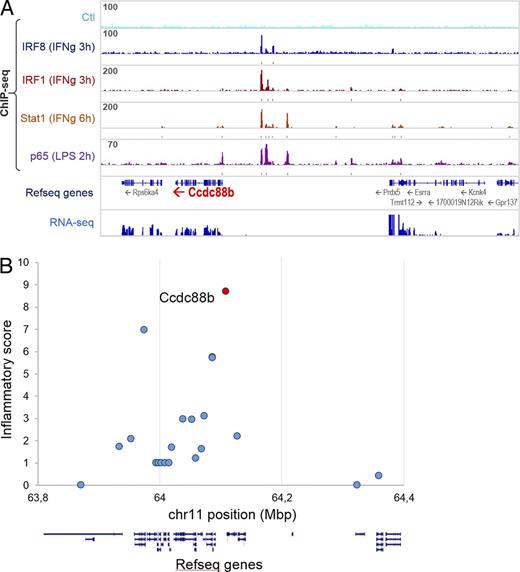
Inactivation of proinflammatory transcription factors Irf1, Irf8, and Stat1 blunts neuroinflammation and causes protection against P. berghei–induced ECM (Longley et al., 2011). Using ChIP-seq, we identified and quantified the intensity of Irf1, Irf8, Stat1, and NF-κB (p65) binding peaks in the vicinity of the 22 mouse orthologues (on mouse Chr. 19) that correspond to the 23 human genes in linkage disequilibrium (LD) at the 11q13 locus associated with inflammatory diseases (IBD) in humans. The number of peaks, the peak height, and the proximity of the peaks to the transcription start site were used to give each gene a peak score (Table S1 and Fig. 8 A). In addition, RNA-seq was used to establish baseline and IFN-γ responsive mRNA expression in myeloid cells, and used to produce an “expression score” of each gene (Table S1). The peak score and expression scores were tabulated to produce an “Inflammation Score” (IS) for the 22 genes. This analysis showed that CCDC88B has the highest IS and ranks number 1 among the 23 genes at 11q13 (Fig. 8 B). Finally, careful examination of cis effects from the top diseases-associated markers (eSNP) from 11q13 shows a strong correlation with expression of CCDC88B (eQTL) in lymphoblastoid cell lines and primary tissues (Dimas et al., 2009; Nica et al., 2011; Grundberg et al., 2012), with the risk allele for Crohn’s, Alopecia areata, and PBC being associated with increased expression. Therefore, the combined observations that (a) inactivation of Ccdc88b protects against acute neuroinflammation in lethal ECM, (b) the protein is expressed in, and is essential for, proinflammatory function of T cells, and (c) the Ccdc88b gene expression is regulated by binding of transcription factors whose inactivation causes ECM resistance, together strongly suggest that CCDC88B is the morbid gene at 11q13 locus.
Candidate gene analysis of the human 11q13 locus associated with susceptibility to inflammatory diseases. (A) Chromatin immunoprecipitation sequencing (ChIP-seq) identifies IFN-γ–induced binding peaks for Irf1, Irf8, and Stat1, and LPS-induced binding peaks for p65 (NF-κB) predominantly in the 5′ region of the Ccdc88b gene. (B) An Inflammatory score was calculated for each of the 22 human genes with mouse orthologues in the 600Mb 11q13 locus. This score is based in part on the presence and on the intensity of binding peaks for proinflammatory transcription factors Irf1, Irf8, Stat1, and p65 (NF-κB) as determined by ChIP-seq, as well as expression in inflammatory cells (RNA-seq) and as described in Table S1. The position of each of the genes in the locus listed in Table S1 is shown as exon/intron arrangement and the inflammatory score is indicated at position of their transcriptional start site.
Candidate gene analysis of the human 11q13 locus associated with susceptibility to inflammatory diseases. (A) Chromatin immunoprecipitation sequencing (ChIP-seq) identifies IFN-γ–induced binding peaks for Irf1, Irf8, and Stat1, and LPS-induced binding peaks for p65 (NF-κB) predominantly in the 5′ region of the Ccdc88b gene. (B) An Inflammatory score was calculated for each of the 22 human genes with mouse orthologues in the 600Mb 11q13 locus. This score is based in part on the presence and on the intensity of binding peaks for proinflammatory transcription factors Irf1, Irf8, Stat1, and p65 (NF-κB) as determined by ChIP-seq, as well as expression in inflammatory cells (RNA-seq) and as described in Table S1. The position of each of the genes in the locus listed in Table S1 is shown as exon/intron arrangement and the inflammatory score is indicated at position of their transcriptional start site.
Overall, our studies in mice identify Ccdc88b as a novel and important regulator of T cell maturation and inflammatory function. Structural or regulatory alterations in human CCDC88B may be a key determinant to the onset, progression, and severity of common inflammatory diseases, including IBD and MS.
MATERIALS AND METHODS
ENU mutagenesis and mouse breeding.
N-ethyl-N-nitrosourea (ENU) mutagenesis and breeding of informative pedigrees was performed as previously described (Bongfen et al., 2012) and as summarized in Fig. 1 A. C57BL/6J, (B6) C57BL10/J (B10), and 129S1/SvImJ (129S1) mice were purchased from The Jackson Laboratory. All mice were maintained under pathogen-free conditions at the animal care facility of the Goodman Cancer Research Centre (McGill University, Montreal, Canada) under the guidelines and regulations of the Canadian Council of Animal Care.
Parasites and infections.
Mice were infected with 106 P. berghei parasitized erythrcoytes (i.v). Animals showing severe symptoms of ECM-associated encephalitis were euthanized. Mice displaying such clinical symptoms within 5–8 d after infection were deemed susceptible, whereas mice surviving beyond 13 d after infection were considered to be resistant to ECM (de Souza and Riley, 2002).
Genetic mapping and exome sequencing.
27 mice from the Deric pedigree were genotyped with a panel of 196 polymorphic markers informative for the B6/B10 parental combination (Illumina Mouse Low Density Linkage panel), and genotypes were analyzed using the R/QTL software package using a binary phenotypic model. This analysis revealed linkage of ECM resistance to proximal Chr.19. Genomic DNA from 2 ECM-resistant Deric mice were subjected to whole-exome sequencing. Exome capture was performed using a SureSelect Mouse All Exon kit (Agilent Technologies) and parallel sequencing on an Illumina HiSeq 2000 (100-bp paired-end reads). Reads were aligned to mouse genome assembly July 2007 (NCBI37/mm9) with Burrows-Wheeler Alignment (BWA) tool (Li and Durbin, 2009) and coverage was assessed with BEDTools (Quinlan and Hall, 2010). Single nucleotide variants were called using samtools pileup and varFilter (Li and Durbin, 2009), and were then quality filtered to require at least 20% of reads supporting the variant call. Variants were annotated using Annovar (Wang et al., 2010) and custom scripts to identify whether they had previously been seen in mouse dbSNP128 or in any of 2 mouse exomes sequenced in parallel.
RNA expression studies.
The expression of the Ccdc88b gene was monitored by whole mount in situ hybridization. PCR amplification of the designed Ccdc88b probe was performed using primers: 5′-GCGCTATAATACGACTCACTATAGGGAGATCCGAATCTTTGGACCTGCCTTCT-3′ and 3′-GCATTAATTTAGGTGACACTATAGAAGCGAAGCTAGCCGTATCCACTGCTTCA-5′, which contain embedded T7 and SP6 polymerases promoter sites (underlined). 35S-UTP labeled cRNA was synthesized in the presence of SP6 (antisense) and T7 (sense) RNA polymerases, and using 35S-UTP (>1,000 Ci/mmol; PerkinElmer). Tissues were fixed in 4% formaldehyde and hybridized with 35S-labeled cRNA antisense and sense probes. The final slides were dehydrated, and then dipped in Kodak NTB nuclear track emulsion, and exposed for 18 d. The slides were lightly counterstained with cresyl violet and analyzed under both light and dark field optics.
For reverse transcription PCR experiments, total spleen RNA was reverse transcribed into cDNA using oligo-d(T) primers and M-MuLV reverse transcription (Invitrogen). PCR amplification of exons 21–22 of Ccdc88b was performed using primers 5′-CCTGCAGGCTGAAAAGTCA-3′ and 5′-GCTCTCGTCGCTCTCATGGA-3′, and Taq DNA polymerase (Invitrogen), for 28 cycles at 30 s/94°C, 30 s/58°C and 30 s/72°C).
Production and affinity purification of anti-CCDC88B antibodies.
The preparation of rabbit polyclonal anti-CCDC88B antiserum was performed as we have previously described (Canonne-Hergaux et al., 1999). A recombinant protein consisting of glutathione-S-transferase (GST) fused in frame to a CCDC88B segment corresponding to mouse amino acid positions 654 to 774, was expressed in E. coli BL21, followed by affinity purification with glutathione-agarose beads (GE Healthcare). Antisera were raised in New Zealand white rabbits using purified protein (0.5 mg per rabbit per injection) emulsified in Freund’s incomplete adjuvant. Affinity purification of the anti-CCDC88B antibody (Canonne-Hergaux et al., 1999) was using a recombinant CCDC88B protein (positions 654 to 774) comprising poly-histidine tails (His)6 fused in-frame at its N terminus, and purified by chromatography onto Ni-NTA agarose (QIAGEN). The immobilized protein was used to capture the anti-CCDC88B fraction of the hyperimmune serum, which was then released by washing in imidazole (Canonne-Hergaux et al., 1999).
Immunofluorescence analysis.
LNs, kidney, spleen, and thymus were collected from WT and Ccdc88bm1PGrs mutant mice, fixed in 4% paraformaldehyde (6 h, 4°C), cryo-protected in 20% sucrose (16 h, 4°C), embedded, and frozen in OCT. Primary antibodies used for immunofluorescence were as follows: affinity purified rabbit anti-CCDC88B antiserum (rabbit, 1:400), anti-CD3 (17A2, rat, 1:50), anti-CD11b (M1/70, rat, 1:50), and anti-CD45R/B220 (RA3-6B2, rat, 1:200). Anti–mouse and anti–rabbit secondary antibodies conjugated with Cy3 or DyLight 488 were used at a 1:3,000 dilution. The B220 primary antibody was conjugated to eFluor 660. Immunostained sections were counterstained with DAPI (1:1,000) to visualize nuclei. All images were acquired on a Zeiss LSM710 Meta Laser Scanning Confocal microscope. All antibodies are from eBioscience unless otherwise stated.
Cell transfer.
For the bone marrow transfer, mice were lethally irradiated twice with 450 rads in a 3-h interval on a X-Ray RS-2000 Biological irradiator. After 3 h, mice were injected i.v. with 107 red blood cell–depleted bone marrow from indicated sex-matched donors. The engraftment was verified after 6 wk by FACS staining with anti-CD45 and anti-B220 antibodies. At 7 wk after engraftment, mice were infected with P. berghei ANKA as described above. For whole splenocyte and T cells transfers, 8–10-wk-old WT or mutant mice were injected i.v. with 106 P. berghei ANKA and 5 d later, spleens were collected and single-cell suspensions were prepared as previously described (Bongfen et al., 2012). Total T cells were enriched by magnetic cell sorting (MACS; Miltenyi Biotec), and 100 million total splenocytes and 10 million enriched T cells were transferred i.v. into Jak3−/− animals. 2 h later, control and reconstituted mice were infected with 106 P. berghei ANKA and were monitored for survival.
Cellular immunophenotyping.
Spleen cell suspensions were prepared, and incubated with the following antibody cocktails: APC eFluor 780 anti-CD3 (I7A2), APC anti-NKp46 (29A1.4; BioLegend), BV421 anti-CD19 (6D5, BioLegend), PE anti-CD8α (53–6.7), PerCP Cy 5.5 anti-CD4 (Rm4-5) for lymphoid cells or PE anti-CD11b (M1/70), PerCP Cy 5.5 anti-Ly6C (HK1.4), eFluor 660 anti-F4/80 (BM8), BV421 anti-Ly6G (1A8; BioLegend), PE anti-F4/80 (BM8), APC anti-CD40 (1C10), APC anti-CD80 (16-10A1) for myeloid cells. Cells were fixed then permeabilized and stained with anti-CCDC88B antibody (30 min at 4°C), followed by incubation with Alexa Fluor 488 coupled goat anti–rabbit IgG (30 min at 4°C). For analysis of T cell activation status, cells were incubated with the following antibody cocktails: PerCP Cy 5.5 anti-CD4 (RM4-5), APC eFluor 780 anti-CD8α (53–6.7), APC anti-TCRβ (H57-597), PE anti-CD44 (IM7; BioLegend), FITC anti-CD69 (H1.2F3), and eFluor 450 anti-CD62L (MeL-1.4). Cells were analyzed with an eight-color FACSCanto II using FACS Diva software (BD); the data were analyzed using FlowJo software (Tree Star). 50,000 cells were acquired, and aggregates were removed based on forward scatter height (FSC-H) versus area (FSC-A). Live cells were determined by side scatter area (SSC-A) versus FSC-A followed by live/dead gating and leukocytes were isolated as CD45+ cells. All antibodies are from eBioscience unless otherwise stated. For cytokine production, spleen cell suspensions from spleen were incubated at 37°C for 6 h with either precoated anti-CD3 (5 µg/ml; eBioscience) and soluble anti-CD28 (2 µg/ml; eBioscience) antibodies, or were stimulated with PMA (50 ng/ml) and ionomycin (500 ng/ml). Cells were then incubated with the antibody cocktail: PE anti-CD4 (GK1.5), BV421 anti-CD8α (53–6.7; BioLegend), and FITC anti-TCRβ (H57-597). Cells were fixed and permeabilized and stained intracellularly (30 min at 4°C) with the following antibody cocktail: PerCP Cy 5.5 anti-TNF (MPG-XT22; BioLegend), APC anti–IFN-γ (XMG1.2). For T cell proliferation assays, cells were suspended in 2.5 µM cytosolic CFSE dye (10 min at 37°C) and stimulated (48 h at 37°C) with either coated anti-CD3 (5 µg/ml) and soluble anti-CD28 (2 µg/ml) or with PMA (50 ng/ml) and ionomycin (500 ng/ml). Viable cells were surface stained with the following antibody cocktail: APC Cy7 anti-CD4 (GK1.5, BioLegend), BV421 anti-CD8α (53–6.7, BioLegend), and APC anti-TCRβ (H57-597, eBioscience). Cells were analyzed by FACS.
Inflammatory score.
Chromatin immunoprecipitation sequencing (ChIP-seq) and RNA sequencing (RNA-seq) from myeloid cells before and after IFN-γ stimulation were performed to identify and quantify binding of pro-inflammatory transcription factors IRF1, IRF8, STAT1 (Ng et al., 2011) and NF-κB (p65; Barish et al., 2010) in the context of IFN-γ stimulation. This analysis was conducted using mouse cells for the 22 orthologous genes (on mouse Chr19) that correspond to the 23 human genes in linkage disequilibrium with the top SNP at the 11q13 locus associated with susceptibility to IBD in humans (Jostins et al., 2012). We computed a “ChIP-seq score” for each gene and that summarizes association of core inflammatory transcription factors IRF8, IRF1, STAT1 and NF-κB (p65). This score was weighed with 2/3 coming from peak proximity to gene transcriptional start site (TSS) using a 20kb window, and 1/3 from peak height relative to the median of peak heights for each transcription factor. ChIP-seq score:
where Pi = hi/2H (if Pi >1, then Pi = 1); tf = transcription factor; g = gene; w = genomic window size (20 kb); d = distance of peak from TSS; n = number of peaks in the window; h = peak height; H = median of peak height. Genes showing detectable mRNA expression in myeloid mouse cells (from RNA-seq data; FPKM ≥ 20) were attributed an “Expression score” of 1. The ChIP-seq score and RNA-seq scores were added to attribute each gene an “inflammation score.” This inflammation score was used as an indicator of a possible participation of the gene in inflammatory processes.
Supplemental material.
Table S1 shows the inflammatory score for candidate genes at the human 11q13 locus associated with susceptibility to multiple inflammatory diseases. Fig. S1 shows the production and characterization of the anti-CCDC88B antibody used to carry out experiments.
Acknowledgments
This work was supported by a team grant to SV and an operating grant PG from the Canadian Institutes of Health Research. The authors are indebted to Maya Saleh and Jorg Fritz for critical comments and suggestions for this manuscript, and to Patricia D’Arcy, Genevieve Perreault, Vanessa Guay, Leigh Piercey-Brunet, Cynthia Villeda-Herrera, Irena Radovanovic, Susan Gauthier, and Normand Groulx for expert technical assistance.
The authors declare no conflict of interest.
References
- CCD
coiled-coil domain
- CCDC88B
CCD containing protein 88B
- ChIP
chromatin immunoprecipitation
- CM
cerebral malaria
- ECM
experimental CM
- ENU
N-ethyl-N-nitrosourea
- GWAS
genome-wide association studies
- HA
hemagglutinin
- IBD
inflammatory bowel disease Iono, Ionomycin
- IRF1/8
interferon regulatory factor-1/8
- KC
keratinocyte chemoattractant
- LD
linkage disequilibrium
- LOD
logarithm of odds
- LTα
lymphotoxin α
- MBD
microtubule binding domain
- MCP-1
monocyte chemoattractant protein-1
- MS
multiple sclerosis
- P.bA
Plasmodium berghei ANKA
- PBC
primary biliary cirrhosis
- PGE2
prostaglandin E2
- PMA
phorbol 12-myristate 13-acetate
- PS
psoriasis
- PTS
peroxisomal targeting sequence
- RA
rheumatoid arthritis
- SC
sarcoidosis
- SLE
systemic lupus erythematosus
- STAT1
signal transducer and activator of transcription 1
- T1D
type 1 diabetes
Author notes
J.M. Kennedy and N. Fodil contributed equally to this paper.