IRF8 and IRF1 are transcriptional regulators that play critical roles in the development and function of myeloid cells, including activation of macrophages by proinflammatory signals such as interferon-γ (IFN-γ). Loss of IRF8 or IRF1 function causes severe susceptibility to infections in mice and in humans. We used chromatin immunoprecipitation sequencing and RNA sequencing in wild type and in IRF8 and IRF1 mutant primary macrophages to systematically catalog all of the genes bound by (cistromes) and transcriptionally activated by (regulomes) IRF8, IRF1, PU.1, and STAT1, including modulation of epigenetic histone marks. Of the seven binding combinations identified, two (cluster 1 [IRF8/IRF1/STAT1/PU.1] and cluster 5 [IRF1/STAT1/PU.1]) were found to have a major role in controlling macrophage transcriptional programs both at the basal level and after IFN-γ activation. They direct the expression of a set of genes, the IRF8/IRF1 regulome, that play critical roles in host inflammatory and antimicrobial defenses in mouse models of neuroinflammation and of pulmonary tuberculosis, respectively. In addition, this IRF8/IRF1 regulome is enriched for genes mutated in human primary immunodeficiencies and with loci associated with several inflammatory diseases in humans.
IRF8 and IRF1 play an important dual role in the development of the myeloid lineage and in the activation of mature myeloid cells by microbial products and cytokines. Mouse strains carrying complete (Irf8−/−) or severe (Irf8m/m; BXH2) loss-of-function alleles at Irf8 either lack all DC subsets (Irf8−/−) or lack CD8α+ conventional DCs (BXH2; Aliberti et al., 2003; Turcotte et al., 2005). Moreover, Irf8 mutant mice develop a chronic myelogenous leukemia–like syndrome characterized by the expansion of immature granulocytes (Holtschke et al., 1996; Turcotte et al., 2004). IRF8 is also required for the differentiation of other myeloid lineages, including osteoclasts (Zhao et al., 2009), microglia (Kierdorf et al., 2013), basophils, and mast cells (Sasaki et al., 2015). In humans, we reported that autosomal recessive IRF8 deficiency is a life-threatening pediatric immunodeficiency (IRF8K108E) featuring a complete absence of CD14+ and CD16+ monocytes and of all circulating CD11c+ DCs, as well as concomitant granulocytic hyperplasia (Hambleton et al., 2011). Studies of the IRF8K108E patient’s peripheral blood mononuclear cells additionally reveals that the primary myeloid defect is associated with an absence of mature antigen-experienced T cells in this patient (Salem et al., 2014b). Autosomal dominant IRF8 deficiency (IRF8T80A) is a milder immunodeficiency featuring reduced numbers of CD11c+CD1+ IL-12–producing DCs that presents as recurrent mycobacterial infections (Hambleton et al., 2011). Hence, IRF8 promotes the differentiation of myeloid progenitors toward the mononuclear phagocyte lineages (monocytes, macrophages, and DCs) by acting as an antagonist of the polymorphonuclear granulocyte pathway (Tamura et al., 2000; Kurotaki et al., 2014).
IRF1 is another member of the IFN regulatory factor (IRF) family that plays an important role in myeloid cell development. Irf1−/− mice harbor myeloid defects, such as immature macrophages and DCs, constitutive granulocytic hyperplasia (Abdollahi et al., 1991; Testa et al., 2004), and altered function of osteoclasts (Salem et al., 2014a). IRF1 is also important for the maturation of the lymphoid lineage: Irf1−/− mice show reduced numbers and altered activity of NK cells (Duncan et al., 1996; Nozawa et al., 1999), along with defective intrathymic maturation and reduced numbers of circulating CD8+ T cells (Matsuyama et al., 1993; Penninger et al., 1997). Importantly, the combined impact of myeloid and lymphoid perturbations associated with the loss of IRF8 or IRF1 function leads to impaired production of IL-12 by DCs and macrophages, to defective IFN-γ production by lymphoid and NK cells, and to defective Th1 polarization of the immune response that leads to hypersusceptibility to viral, bacterial, and parasitic infections in vivo (Tamura et al., 2008).
IRF1 and IRF8 also play an important role in the amplification of myeloid cell response to IFN-γ. Binding of IFN-γ to its receptor causes activation of JAK1/JAK2 kinases, which leads to phosphorylation, nuclear translocation, and binding of STAT1 homodimers to GAS elements. Engagement of the IFN-γ receptor also activates expression, nuclear translocation, and transcriptional activity of IRF1 and IRF8, which are essential to activate the full microbicidal potential of macrophages (Hu and Ivashkiv, 2009). Indeed, Irf1 or Irf8 mutant macrophages are susceptible to infection with intracellular pathogens (Fehr et al., 1997; Cooper et al., 2000; Marquis et al., 2009b). Transactivation experiments with target genes have shown that IRF8 and IRF1 functionally interact for IFN-γ–induced activation of certain genes involved in macrophage antimicrobial defenses and in production of inflammatory cytokines that activate early immune responses (Dror et al., 2007). At the molecular level, IRF8 is known to be corecruited to ternary complexes with other transcription factors (TFs) such as (a) IRF1 and IRF2 that bind to IFN-stimulated response elements (ISREs; GAAAnnGAAA; Bovolenta et al., 1994), (b) AP-1 family members that bind to AP1-IRF composite elements (TGAnnnGAAA or GAAATGA; Glasmacher et al., 2012; Li et al., 2012), or (c) Ets family members such as PU.1 that are required for the development of lymphoid and myeloid lineages (Scott et al., 1994; McKercher et al., 1996) and that bind to Ets-IRF composite elements (EICEs; GGAAnnGAAA).
In addition to a shared role in host defenses against infections, IRF1, IRF8, and their partner STAT1 are important regulators of pathological inflammation in humans, including rheumatoid arthritis, multiple sclerosis, primary biliary cirrhosis, systemic sclerosis, systemic lupus erythematosus, and inflammatory bowel disease (Cunninghame Graham et al., 2011; Jostins et al., 2012; Beecham et al., 2013; Okada et al., 2014). Likewise, mouse mutants bearing loss-of-function alleles at Irf1, Irf8, or Stat1 are resistant to neuroinflammation in the cerebral malaria model induced by infection with Plasmodium berghei (Berghout et al., 2013) as well as in the experimental allergic encephalitis model (Buch et al., 2003). Hence, a better understanding of the transcriptional programs activated by IRF8, IRF1, and STAT1 in response to IFN-γ may provide novel insight into molecular pathways of pathological inflammation, including possible novel targets for therapeutic intervention.
We investigated the genome-wide distribution of IRF1, IRF8, STAT1, and PU.1 in chromatin from resting and from IFN-γ–activated macrophages and characterized the associated gene expression programs in WT and in Irf1 and Irf8 mutants. Our results show that IRF8 is mostly bound constitutively to DNA with PU.1 and that IFN-γ treatment has only a modest effect on its DNA binding. In contrast, IFN-γ strongly induces recruitment of IRF1 and STAT1 to cis-regulatory elements prebound by PU.1. We have identified two binding combinations (IRF8/IRF1/STAT1/PU.1 and IRF1/STAT1/PU.1) that have a major role in controlling macrophage transcriptional programs both at the basal level and in response to IFN-γ activation. Loss of IRF8 and IRF1 activity leads to reduced basal and IFN-γ–induced expression of different subsets of genes that play critical roles in macrophage activity and function, and that we tentatively designate the IRF8/IRF1 regulome. Importantly, these are genes that (a) are activated in response to infections, with several being mutated in patients with primary immunodeficiencies (PIDs), and (b) are activated during pathological states of inflammation, mapping within risk loci for common human inflammatory diseases.
RESULTS
Considering the critical role of IRF8 and IRF1 in antimicrobial defenses of macrophages, we set out to identify all of the genes in which expression is regulated by these two factors both at steady state and in response to IFN-γ. We then investigated the molecular basis of this regulation and determined whether these groups of genes are associated with disease in mouse models of infection and in available human genetic datasets.
Different engagement of IRF8 and IRF1 in response to IFN-γ in macrophages
We first established the genome-wide binding profiles of IRF8 and IRF1 by chromatin immunoprecipitation sequencing (ChIP seq) in WT (C57BL/6J) BMDMs before or after exposure to IFN-γ. We also generated binding profiles for PU.1 and incorporated published STAT1 datasets into our analysis (Ng et al., 2011). In agreement with published data (Ghisletti et al., 2010; Heinz et al., 2010), PU.1 was bound constitutively to thousands of sites in BMDM chromatin, and this binding was unaffected by IFN-γ (binding peak heights of 0 vs. 3 h for IFN-γ; Fig. 1 A); however, STAT1 recruitment was enhanced by IFN-γ treatment (Ng et al., 2011). We observed that IRF8 binding was mostly constitutive in macrophages with <15% of sites showing a more than twofold increase in response to IFN-γ (Fig. 1 A). In contrast, IRF1 recruitment was the most responsive to IFN-γ (Fig. 1 A) with ∼80% of the binding sites at 3 h being either novel or showing a more than twofold increase over untreated cells. IFN-γ–dependent recruitment of IRF1 to chromatin was relatively slow (Fig. S2), likely reflecting prerequisite activation of Irf1 transcription (see Fig. 3 B) and protein synthesis (see Fig. 3 C). Genome wide, binding of IRF1 and IRF8 is highly enriched in the vicinity of gene transcription start sites (Fig. 1 B). De novo motif analysis in our datasets identified the core sequence GGAA as being enriched at PU.1 peaks (Fig. 1 C), in agreement with recently published data (Pham et al., 2013; Barozzi et al., 2014). In addition, the EICE motif (GGAAnnGAAA) was strongly enriched at IRF8 peaks; this motif was previously shown by x-ray crystallography (Escalante et al., 2002) to recruit PU.1 on the first half site (GGAA) and an IRF family member (IRF4 or IRF8) on the second half site (GAAA), with the guanine at position 2 being specific for PU.1 binding. Under IRF1 peaks, the ISRE motif (GAAAnnGAAA) containing two IRF, binding half sites is predominant (Fig. 1 C). In addition, the ISRE was identified as the top motif in the subset of IFN-γ–activated IRF8 binding sites (Fig. 1 C), strongly suggesting corecruitment of IRF8 and IRF1. Together, these results suggest different binding characteristics for IRF1 and IRF8 in BMDMs: (a) IRF8 binds DNA constitutively with PU.1 as a partner on EICE motifs, and (b) IRF1 recruitment is strongly induced in response to IFN-γ and binds DNA either as a homodimer (Spink and Evans, 1997; Escalante et al., 1998; Kirchhoff et al., 1998) or as a heterodimer with IRF8 (Bovolenta et al., 1994) or possibly with STAT1 (Chatterjee-Kishore et al., 2000).
Differential effect of IFN-γ on recruitment of IRF8 and IRF1 to chromatin. (A) Peak heights (number per 107 reads) for IRF8, IRF1, STAT1, and PU.1 binding sites determined by ChIP seq before and after IFN-γ stimulation of BMDMs. (B) Distribution of the individual TF binding sites (after IFN-γ) relative to the closest annotated gene. (C) De novo motif analysis was performed for the IRF8, IRF1, STAT1, and PU.1 binding peaks and after treatment with IFN-γ. Shown are the top motifs identified for each TF, reference to their published names, and the fraction of peaks containing these motifs within a 100-bp region of the binding peak. (D) Volcano plot showing pairwise analysis of differential gene expression (RNA seq data) in BMDMs after 3 h of IFN-γ treatment (fold change ≥|2|; adjusted p-value ≤10−5). RNA seq data were validated by RT-qPCR for a subset of 25 transcripts on independent biological samples (Fig. S1 A). (E) Mean number of peaks per gene (per 10-kb intervals) plotted for all expressed genes and for up- or down-regulated genes after IFN-γ treatment, and for control random (Rdm) gene sets. TSS, transcription start site.
Differential effect of IFN-γ on recruitment of IRF8 and IRF1 to chromatin. (A) Peak heights (number per 107 reads) for IRF8, IRF1, STAT1, and PU.1 binding sites determined by ChIP seq before and after IFN-γ stimulation of BMDMs. (B) Distribution of the individual TF binding sites (after IFN-γ) relative to the closest annotated gene. (C) De novo motif analysis was performed for the IRF8, IRF1, STAT1, and PU.1 binding peaks and after treatment with IFN-γ. Shown are the top motifs identified for each TF, reference to their published names, and the fraction of peaks containing these motifs within a 100-bp region of the binding peak. (D) Volcano plot showing pairwise analysis of differential gene expression (RNA seq data) in BMDMs after 3 h of IFN-γ treatment (fold change ≥|2|; adjusted p-value ≤10−5). RNA seq data were validated by RT-qPCR for a subset of 25 transcripts on independent biological samples (Fig. S1 A). (E) Mean number of peaks per gene (per 10-kb intervals) plotted for all expressed genes and for up- or down-regulated genes after IFN-γ treatment, and for control random (Rdm) gene sets. TSS, transcription start site.
To assess the impact of recruitment of IRF8, IRF1, and STAT1 on the regulation of gene expression in BMDMs in response to IFN-γ, we performed RNA sequencing (RNA seq) at 0 and at 3 h after stimulation. This analysis revealed that the presence and numbers of binding sites for these factors are correlated with increased gene expression (Fig. S1 B). Differential gene expression analysis revealed that treatment of macrophages with IFN-γ caused the up-regulation of 611 genes (more than or equal to twofold; adjusted P ≤ 10−5) and the repression of 308 genes (Fig. 1 D). Strikingly, IFN-γ–activated genes harbor IRF1, IRF8, and/or STAT1 binding in their vicinity, whereas down-regulated genes do not show this association (Fig. 1 E). As previously shown (Ghisletti et al., 2010), PU.1 establishes the macrophage transcriptional program and is not particularly associated with differentially expressed genes, but rather with macrophage-expressed genes by themselves (Fig. 1 E). Overall, these results show that the genes up-regulated by IFN-γ are enriched for IRF8, IRF1, and STAT1 binding.
IRF8 and IRF1 genomic binding schemes and associated chromatin status in macrophages
To evaluate the functional interplay between IRF8, IRF1, STAT1, and PU.1 in regulating gene expression in macrophages, we examined their corecruitment by consolidating colocalized binding sites defined as having peak maxima within 100 bp of the other factors (Fig. 2 A). This analysis readily suggested preferential association of these TFs with each other. Indeed, IRF8 is never found bound to DNA alone, in agreement with poor intrinsic DNA binding capacity (Bovolenta et al., 1994), whereas IRF8 and IRF1 are corecruited with STAT1 and PU.1 on 3,584 genomic regions (Fig. 2 A). To explore possible binding combinations of these TFs, we extracted sequence read densities around each genomic position bound by IRF8, IRF1, STAT1, or PU.1 and assembled them into different binding combinations (Fig. 2 B). This clustering analysis identified nine different binding schemes (Fig. S2), seven of which involved IRF8 and/or IRF1 (Fig. 2 B). Cluster 1 (combining IRF8/IRF1/STAT1/PU1) and cluster 5 (combining IRF1/STAT1/PU.1) showed the highest sequence conservation across mammals, possibly suggesting functional relevance (an extended version of the heatmap containing additional TFs and mammalian sequence conservation is shown in Fig. S2, and a list of cluster binding peaks is in Table S1). Another key feature of clusters 1 and 5 is the presence of IFN-γ–induced recruitment of IRF1 (with additional recruitment of STAT1; Fig. 2 B). Thus, IRF8 and IRF1 are recruited to macrophage cis-regulatory regions with PU.1 and STAT1 in different combinations.
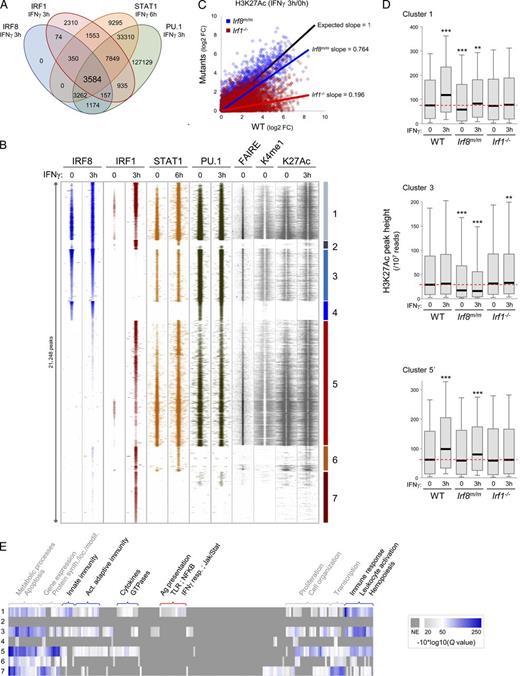
Different IRF8/IRF1 binding combinations in macrophages and association with epigenetic profiles. (A) Venn diagram depicting overlaps between IRF8, IRF1, STAT1, and PU.1 binding sites genome wide (sites located <100 bp from each other). (B) Clustering analysis of the 21,248 unique IRF1- and/or IRF8-containing regulatory regions (including STAT1 and PU.1) before or after IFN-γ treatment. Each horizontal line presents the read density in a ±1-kb region around a unique position; DNA accessibility (FAIRE; Ostuni et al., 2013) and H3K4me1 and H3K27Ac epigenetic datasets are shown for a ±2-kb region surrounding the cluster peaks. Different binding combinations before or after IFN-γ treatment are shown (clusters numbered 1–7). TF enrichment at various cluster peaks was validated by qPCR (Fig. S1 C). (C) Global changes in H3K27Ac levels at IRF8- or IRF1-bound sites in response to IFN-γ in Irf1−/− and in Irf8m/m mutant macrophages (log2 fold changes of H3K27Ac peak heights). Linear regressions are shown for Irf8m/m and Irf1−/− cells and are compared with an expected regression (black) if Irf mutations had no effect. (D) Box plots of H3K27Ac ChIP seq read density for TF binding clusters 1 and 3 and for the subset IFN-γ–activated sites of cluster 5 (5′); the dotted red lines identify median K27Ac levels in untreated WT BMDMs; p-values (Wilcoxon rank sum test; **, P ≤ 0.01; ***, P < 0.001) were calculated for each group compared with untreated WT controls. (E) GO category enrichment analysis for genes in the different TF binding clusters; the gray/white/blue color gradient indicates the significance of category enrichment (−10*log10 of the FDR q-value using a minimal threshold of 0.01; NE, nonenriched).
Different IRF8/IRF1 binding combinations in macrophages and association with epigenetic profiles. (A) Venn diagram depicting overlaps between IRF8, IRF1, STAT1, and PU.1 binding sites genome wide (sites located <100 bp from each other). (B) Clustering analysis of the 21,248 unique IRF1- and/or IRF8-containing regulatory regions (including STAT1 and PU.1) before or after IFN-γ treatment. Each horizontal line presents the read density in a ±1-kb region around a unique position; DNA accessibility (FAIRE; Ostuni et al., 2013) and H3K4me1 and H3K27Ac epigenetic datasets are shown for a ±2-kb region surrounding the cluster peaks. Different binding combinations before or after IFN-γ treatment are shown (clusters numbered 1–7). TF enrichment at various cluster peaks was validated by qPCR (Fig. S1 C). (C) Global changes in H3K27Ac levels at IRF8- or IRF1-bound sites in response to IFN-γ in Irf1−/− and in Irf8m/m mutant macrophages (log2 fold changes of H3K27Ac peak heights). Linear regressions are shown for Irf8m/m and Irf1−/− cells and are compared with an expected regression (black) if Irf mutations had no effect. (D) Box plots of H3K27Ac ChIP seq read density for TF binding clusters 1 and 3 and for the subset IFN-γ–activated sites of cluster 5 (5′); the dotted red lines identify median K27Ac levels in untreated WT BMDMs; p-values (Wilcoxon rank sum test; **, P ≤ 0.01; ***, P < 0.001) were calculated for each group compared with untreated WT controls. (E) GO category enrichment analysis for genes in the different TF binding clusters; the gray/white/blue color gradient indicates the significance of category enrichment (−10*log10 of the FDR q-value using a minimal threshold of 0.01; NE, nonenriched).
The epigenetic profile of a cis-regulatory region is an important indicator of its current or potential transcriptional activity. The chromatin status of the seven IRF-containing binding schemes was examined for K4me1 (a mark of enhancer regions) and H3K27Ac (transcriptional activity) and for open chromatin conformation as determined by formaldehyde-assisted isolation of regulatory elements (FAIREs; Figs. 2 B and S2; Ostuni et al., 2013). Distinctively, only clusters 1, 3, and 5 show characteristics of active regulatory regions before IFN-γ treatment with open chromatin and monomethylated H3K4, with an increase of H3K27 acetylation at clusters 1 and 5 sites 3 h after IFN-γ stimulation (Fig. 2 B). These results strongly suggest that IRF-bound clusters 1, 3, and 5 are major regulatory hotspots of the macrophage transcriptional program. Moreover, clusters lacking IRF1 do not display increased K27Ac in response to IFN-γ, suggesting that IRF1 plays a critical role in transcriptional activation in response to IFN-γ in macrophages.
To assess the importance of chromatin-bound IRF8 and IRF1 on the transcriptional activity of these regulatory regions, we measured the level of H3K27Ac at the different clusters in chromatin from WT, Irf1−/−, and Irf8 mutant (Irf8m/m; R294C) macrophages; we used Irf8m/m because Irf8−/−-null mice produce few F4/80+ mature macrophages, whereas these cells are present in mice bearing the hypomorphic IRF8R294C allele (see IRF8 ChIP seq in Irf8m/m in Fig. S2). Flow cytometry analysis confirmed that F4/80+ BMDMs can be obtained in similar numbers from WT, Irf8m/m, and Irf1−/− mice (Fig. 3 A). Also, abrogation of IRF8 function had no effect on levels of IRF1 and STAT1 mRNA and proteins, and likewise, elimination of IRF1 did not alter STAT1 and IRF8 expression (Fig. 3, B and C). Importantly, the loss of IRF8 function results in a decreased K27Ac steady-state level at IRF8-containing clusters 1 and 3 (Figs. 2 D and S2) without affecting K27Ac deposition in response to IFN-γ (Fig. 2 C), in agreement with a functional role of IRF8 constitutively bound at these sites. Conversely, the loss of IRF1 has little effect on constitutive K27Ac levels, but it eliminates K27Ac deposition at clusters 1 and 5 sites in response to IFN-γ (5.1-fold decrease compared with WT; Fig. 2, C and D; and Fig. S2). Collectively, these results strongly suggest that in macrophages, IRF8 binding is important to maintain basal K27Ac levels (clusters 1 and 3), whereas IRF1 is required for activation of regulatory regions in response to IFN-γ (clusters 1 and 5). In agreement with this proposal, chromatin from Irf8m/m BMDMs shows low but IFN-γ–inducible K27Ac deposition at cluster 1 sites, whereas chromatin from Irf1−/− BMDMs shows normal basal levels but lacks IFN-γ–inducible K27Ac deposition at cluster 1 sites (Figs. 2 D and S2).
Characteristics of Irf8m/m and Irf1−/− mutant BMDMs. (A) The differentiation of BM cells from WT B6 and Irf8 and Irf1 mutants into macrophages (BMDMs) was assessed by flow cytometry. The fraction of F4/80 positive cells (±SD) for each genotype (representative of at least of five independent experiments). (B) Basal and IFN-γ–stimulated RNA expression levels of the four studied TFs in BMDMs from each genotype; data are presented relative to Hprt mRNA (±SD of biological replicates) used as an internal control (representative of three independent experiments). (C) Western blot analysis of IRF8, IRF1, STAT1, PU.1, inducible nitric oxide synthase, Cxcl10, and Cxcl9 protein expression in WT and Irf8m/m and Irf1−/− mutants before and after treatment with IFN-γ. (D) Volcano plot of differential gene expression (RNA seq data) of Irf8m/m and Irf1−/− BMDMs in response to 3 h of IFN-γ treatment; responsive genes were identified using a fold change ≥|2| and an adjusted p-value ≤10−5.
Characteristics of Irf8m/m and Irf1−/− mutant BMDMs. (A) The differentiation of BM cells from WT B6 and Irf8 and Irf1 mutants into macrophages (BMDMs) was assessed by flow cytometry. The fraction of F4/80 positive cells (±SD) for each genotype (representative of at least of five independent experiments). (B) Basal and IFN-γ–stimulated RNA expression levels of the four studied TFs in BMDMs from each genotype; data are presented relative to Hprt mRNA (±SD of biological replicates) used as an internal control (representative of three independent experiments). (C) Western blot analysis of IRF8, IRF1, STAT1, PU.1, inducible nitric oxide synthase, Cxcl10, and Cxcl9 protein expression in WT and Irf8m/m and Irf1−/− mutants before and after treatment with IFN-γ. (D) Volcano plot of differential gene expression (RNA seq data) of Irf8m/m and Irf1−/− BMDMs in response to 3 h of IFN-γ treatment; responsive genes were identified using a fold change ≥|2| and an adjusted p-value ≤10−5.
Gene ontology (GO) enrichment analysis (Fig. 2 E) showed that clusters 1, 3, and 5 are enriched for genes associated with immune response, innate immunity, activation of adaptive immunity, cytokine production, and small GTPases. Strikingly, cluster 1–associated genes defined by corecruitment of IRF8/IRF1/STAT1/PU.1 and by strong epigenetic activation in response to IFN-γ (Fig. 2, B and D) are uniquely enriched for markers and functional features of myeloid cells, including TLR, NF-κB, Jak/Stat, and IFN-γ response pathways, and all aspects of antigen presentation.
In summary, IRF8 and IRF1 were found to be recruited to chromatin regions with STAT1 and PU.1 in different binding combinations. Of them, three clusters (1, 3, and 5) show characteristics of active enhancers, demonstrating greater evolutionary sequence conservation and harboring accessible chromatin domains, including H3K4me1 and H3K27Ac histone modifications (Heintzman et al., 2007). These clusters, bound by PU.1, STAT1, and IRF8 and/or IRF1, also recruit several other TFs at steady state or in response to stimuli (i.e., AP-1, NF-κB, C/EBP-α and -β, LXR-β, and STAT2 [unpublished data]) and behave as so-called “regulatory hotspots,” previously described in macrophages and other cell types as having the strongest regulatory potential among the enhancer repertoire (Lawrence and Natoli, 2011; Garber et al., 2012). Despite recruiting all of these TFs, IRF8-bound clusters exhibited defective K27Ac status in resting Irf8 mutant macrophages. Moreover, only clusters 1 and 5 show increased K27 acetylation after 3 h of IFN-γ activation, which is strongly indicative of coactivator recruitment and of enhancer activity, and this activation is dependent on the presence of IRF1. At specific gene promoters, it was shown that IRF1 binding to DNA is a key step to bring histone acetylases (such as p300/CBP and pCAF) to chromatin (Masumi et al., 1999) and is a key step in IFN-dependent transcriptional activation (Ramsauer et al., 2007). Herein, we show that this critical role of IRF1 in cis-regulatory region activation is widespread and important for genome-wide IFN-γ response.
Regulation of basal expression of target genes by IRF8 and IRF1 in macrophages
To further characterize the role of IRF1 or IRF8 in macrophage transcriptional programs, we performed RNA seq in BMDMs from WT controls and from Irf1−/− and Irf8m/m mice (Fig. 3 D). Principal component analysis (Fig. 4 A) clearly illustrates a major effect of Irf1 and Irf8 genotypes (PC2) and of IFN-γ treatment (PC1) on macrophage gene expression programs. In addition, the prominent effect of the loss of IRF8 and IRF1 function (PC2) observed before treatment (basal gene expression) is transposed after activation by IFN-γ (Fig. 4 A).
IRF8 and IRF1 are required for macrophage basal transcriptional programs. (A) Principal component analysis of RNA seq data from WT, IRF8-deficient (Irf8m/m), and IRF1-deficient (Irf1−/−) BMDMs before and after 3 h of stimulation with IFN-γ. (B) Fraction of genes showing decreased (down) or increased (up) expression in Irf8m/m and Irf1−/− BMDMs and that harbor IRF8 or IRF1 binding sites within 20 kb of their transcription start site (p-values are Fisher’s exact test relative to random [Rdm] expectations). ns, not significant. (C) Expression level changes for genes significantly dysregulated at the basal level in mutant Irf8m/m and Irf1−/− BMDMs (fold change ≥|2|; adjusted p-value ≤10−5). Genes were grouped (a–f) for the effect of IRF8 and IRF1 loss of function (enriched GO categories are shown). (D) Bubble histogram showing the association of TF binding clusters with genes showing reduced (a–c) or increased (d–f) expression. The bubble size reflects the strength of the –log10 Fisher’s exact test p-value for the association of dysregulated genes (compared with random gene sets). The color gradient reflects the ratio in the number of peaks associated with dysregulated genes versus control sets of randomly selected genes.
IRF8 and IRF1 are required for macrophage basal transcriptional programs. (A) Principal component analysis of RNA seq data from WT, IRF8-deficient (Irf8m/m), and IRF1-deficient (Irf1−/−) BMDMs before and after 3 h of stimulation with IFN-γ. (B) Fraction of genes showing decreased (down) or increased (up) expression in Irf8m/m and Irf1−/− BMDMs and that harbor IRF8 or IRF1 binding sites within 20 kb of their transcription start site (p-values are Fisher’s exact test relative to random [Rdm] expectations). ns, not significant. (C) Expression level changes for genes significantly dysregulated at the basal level in mutant Irf8m/m and Irf1−/− BMDMs (fold change ≥|2|; adjusted p-value ≤10−5). Genes were grouped (a–f) for the effect of IRF8 and IRF1 loss of function (enriched GO categories are shown). (D) Bubble histogram showing the association of TF binding clusters with genes showing reduced (a–c) or increased (d–f) expression. The bubble size reflects the strength of the –log10 Fisher’s exact test p-value for the association of dysregulated genes (compared with random gene sets). The color gradient reflects the ratio in the number of peaks associated with dysregulated genes versus control sets of randomly selected genes.
At steady state, genes displaying reduced basal expression in Irf1−/− and Irf8m/m mutant BMDMs are substantially associated with direct binding of the respective TF (Fig. 4 B). This is not seen for genes for which expression is increased in either of the two mutants (Fig. 4 B), in agreement with IRF8 and IRF1 functioning primarily as transcriptional activators in macrophages. A clustering analysis of transcripts dysregulated in either Irf1−/− or Irf8m/m mutant BMDMs at steady-state identifies groups of genes for which expression (a) requires both IRF1 and IRF8 (groups a and d; common genes) or (b) is only affected by specific loss (specific genes) of either IRF8 (groups b and e) or IRF1 (groups c and f; Fig. 4 C and Table S2). In agreement with the significant constitutive binding of IRF8 (which is minimal for IRF1) at target regulatory regions (Fig. 2, B and D), loss of IRF8 affects basal expression of a greater number of genes than the loss of IRF1 (Figs. 4 C and 5 C). GO analysis associates group a genes (IRF8/IRF1; n = 210) with immune response functions and cytokine production and group b genes (IRF8; n = 265) with basal macrophage functions, antigen presentation, cell adhesion, and lymphocyte activation molecules, whereas group c genes (IRF1; n = 104) fall into immune response, cytokine production, and cell proliferation annotations.
IRF1 and IRF8 are required for transcriptional activation by IFN-γ in macrophages. (A) Heatmap identifying three groups of genes (labeled I–III) in which activation by IFN-γ is altered by more than or equal to twofold in BMDMs mutated for Irf8 (II), Irf1 (III), or both (I). (B) Bubble histogram showing the association of TF binding clusters with groups of genes dysregulated in Irf8m/m and Irf1−/− mutant BMDMs in response to IFN-γ (groups I–III), other IFN-γ activated (Act), or repressed (Rep) genes; see Fig. 4 D. (C) Proportions of dysregulated genes in Irf8m/m and Irf1−/− mutant macrophages at steady state and in response to IFN-γ (p-values calculated using the Fisher’s exact test for TF-specific dysregulated genes vs. total number of dysregulated genes). (D) Schematic models for IRF8- and IRF1-dependent regulation of basal and IFN-γ–induced transcriptional programs (see Results for details).
IRF1 and IRF8 are required for transcriptional activation by IFN-γ in macrophages. (A) Heatmap identifying three groups of genes (labeled I–III) in which activation by IFN-γ is altered by more than or equal to twofold in BMDMs mutated for Irf8 (II), Irf1 (III), or both (I). (B) Bubble histogram showing the association of TF binding clusters with groups of genes dysregulated in Irf8m/m and Irf1−/− mutant BMDMs in response to IFN-γ (groups I–III), other IFN-γ activated (Act), or repressed (Rep) genes; see Fig. 4 D. (C) Proportions of dysregulated genes in Irf8m/m and Irf1−/− mutant macrophages at steady state and in response to IFN-γ (p-values calculated using the Fisher’s exact test for TF-specific dysregulated genes vs. total number of dysregulated genes). (D) Schematic models for IRF8- and IRF1-dependent regulation of basal and IFN-γ–induced transcriptional programs (see Results for details).
We investigated a possible correlation between the effect of Irf genotypes on gene expression and the specific IRF binding clusters identified by ChIP seq in Fig. 2 B. We observed (bubble histogram; Fig. 4 D) a highly significant association between genes showing reduced expression in both Irf1−/− and Irf8m/m mutants (group a) and the cluster 1 binding scheme (cobinding IRF8/IRF1/STAT1/PU.1) and to a lesser degree to clusters 3 and 5, which recruit IRF8 or IRF1 uniquely in conjunction with STAT1 and PU.1 (Fig. 4 D). However, group c transcripts (reduced in Irf1−/−) showed enrichment for clusters 1 and 5, whereas group b (reduced in Irf8m/m) was enriched in clusters 1 and 3 (see Fig. S3 [A–C] for the C1q locus example). Noticeably, no IRF clusters were found closely associated to genes with increased expression in either Irf1−/− or Irf8m/m mutants. Moreover, the other IRF-bound clusters (2, 4, 6, and 7) were not associated with dysregulated genes. Together, these results highlight the critical role of IRF1- and IRF8-containing clusters 1, 3, and 5 in regulating basal expression of a subset of ∼600 immune genes in macrophages.
Regulation of IFN-γ–induced expression of target genes by IRF8 and IRF1 in macrophages
The contribution of IRF8 and IRF1 to IFN-γ–induced transcriptional responses in macrophages was investigated next (Fig. 5). As predicted from the preferential association of IRF8 and IRF1 binding sites with IFN-γ–activated genes (Fig. 1 E), macrophages from either Irf1−/− or Irf8m/m mutants are defective in transcriptional activation in response to IFN-γ. Genes showing significantly reduced IFN-γ–dependent activation (more than or equal to twofold; n = 204) in either Irf1−/− or Irf8m/m mutants could be organized into three groups (Fig. 5 A). Group I genes are less activated in the absence of either IRF8 or IRF1, whereas activation of group II and group III genes is dampened by the absence of IRF8 and IRF1, respectively (see complete gene lists in Table S3). In agreement with the significant IFN-γ–induced binding of IRF1 at target regulatory regions (Fig. 2, B and D), the loss of IRF1 affects the expression of a greater number of genes compared with the loss of IRF8 (Fig. 5, A and C). Furthermore, transcripts with defective activation (I, II, and III) in mutants are found to be globally enriched for cluster 1 binding schemes and to a lesser extent for the cluster 5 binding scheme (Fig. 5 B). None of the IRF binding clusters are associated with IFN-γ–repressed genes (Fig. 5 B). Of note, there is significant overlap between group I–III transcripts and genes for which basal expression is affected by elimination of either IRF1 or IRF8 (Fig. 4 C, group a).
The relative role of IRF8 and IRF1 in macrophage transcriptional regulation is recapitulated in a proposed model shown in Fig. 5 D. At steady state (basal), IRF8 is constitutively bound with PU.1 to EICE motifs in cluster 1 and 3 sites and maintains basal H3K27 acetylation level and target gene expression; some basal STAT1 binding is also observed. In response to IFN-γ, macrophages increase STAT1 genomic binding without affecting the constitutive PU.1 deployment. In turn, macrophages strongly activate, in a STAT1-dependent manner (Li et al., 1996), the transcription of the Irf1 gene, and the consequent increase in IRF1 protein results in the massive recruitment of IRF1 at cluster 1 and 5 sites on ISRE motifs. Based on our data and on previous biochemical studies, we suggest that IRF1 is mainly recruited as a homodimer on the cluster 5 ISRE motif, but also as a heterodimer with IRF8 on the cluster 1 ISRE motif; this is supported by the absence of IFN-γ–induced IRF8 binding to cluster 1 sites in Irf1−/− macrophages (unpublished data). Importantly, IRF1 binding is required to increase K27Ac deposition and target gene expression in response to IFN-γ stimulation.
The three major IRF binding combinations (1, 3, and 5; Fig. 2) do not function in isolation and are often found clustered at the promoter and enhancer regions of individual genes, providing further complexity and flexibility in fine-tuning the expression of individual genes at basal level or in response to stimuli such as IFN-γ. Representative examples of group I–III genes that illustrate transcriptional regulation by clusters (1, 3, and 5) in response to IFN-γ are reviewed in Figs. 6, 7, and 8. Group I contains several genes that are critical for the innate immune functions of macrophages, including Tnf, Ctsc, Nos2, and Tlr9 (Table S3). The Tlr9 gene is a good example of the functional dichotomy between IRF8 and IRF1. First, the locus contains the three major TF binding schemes (1, 3, and 5) linked to transcriptional regulation. Second, IRF8 binds constitutively to sites 1 and 3, which is essential to establish basal Tlr9 gene expression and locus activity (K27Ac) that is permissive to IFN-γ–dependent transcriptional activation. Third, IRF1 is significantly recruited to sites 1 and 5 only after IFN-γ treatment, and Irf1 mutation has little effect on basal Tlr9 expression while reducing IFN-γ–dependent activation (Fig. 6, A, B, and D). The Nos2 gene (coding for inducible nitric oxide synthase) is another example of a group I gene, where the loss of IRF8 and in particular IRF1 (Kamijo et al., 1994) causes a severe reduction of Nos2 locus activation and transcription (Fig. 6, C, E, and F) and of inducible nitric oxide synthase protein expression (Fig. 3 C). This results in severely impaired nitric oxide production by mutant BMDMs (Fig. 6 G) and mutant splenocytes challenged ex vivo for 48 h with IFN-γ (Fig. 6 H).
Tlr9 and Nos2 are transcriptionally coregulated by IRF8 and IRF1 in macrophages. (A) Genomic snapshot of the Tlr9 locus (group I gene). Density of ChIP seq reads (for IRF8, IRF1, STAT1, PU.1, and H3K27Ac) and RNA seq reads in BMDMs (WT, and Irf1−/− and Irf8m/m mutants) are shown, with arrows pointing to cluster 1, 3, and 5 binding schemes. (B) RT-qPCR validation of Tlr9 gene expression in response to IFN-γ in WT and mutant BMDMs (mean ± SD of independent biological replicates; Student’s t test relative to WT). HPRT, hypoxanthine-guanine phosophoribosyltransferase. (C) Genomic snapshot (as in A) of the Nos2 locus (group I gene). (D) ChIP-qPCR validation of H3K27Ac levels (relative to histone H3) at Tlr9 cluster 3 and 1 peaks (results are representative of three independent experiments). (E) RT-qPCR validation of Nos2 expression (as in B). (F) H3K27Ac levels (ChIP-qPCR relative to histone H3) at three cluster 5 peaks at the Nos2 locus (designated A, B, and C) showing increased K27Ac deposition in WT BMDMs after IFN-γ stimulation, which is diminished in Irf8m/m and abolished in Irf1−/− BMDMs (results are representative of three independent experiments). (G) Nitric oxide production by WT, Irf8m/m, and Irf1−/− mutant BMDMs in vitro in response to IFN-γ (mean of biological replicates ± SD; p-values were calculated using Student’s t test). (H) Nitric oxide (NO) production by splenocytes from Irf8m/m and Irf1−/− Plasmodium berghei ANKA-infected mice (compared with WT) treated with 400 U/ml IFN-γ for 48 h and 1.5 µg/ml TLR9 ligand CpG oligonucleotides. Box plots show the mean nitric oxide production from four to five mice per strain (whiskers extending to 5–95 percentiles; p-values were calculated using Student’s t test). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Tlr9 and Nos2 are transcriptionally coregulated by IRF8 and IRF1 in macrophages. (A) Genomic snapshot of the Tlr9 locus (group I gene). Density of ChIP seq reads (for IRF8, IRF1, STAT1, PU.1, and H3K27Ac) and RNA seq reads in BMDMs (WT, and Irf1−/− and Irf8m/m mutants) are shown, with arrows pointing to cluster 1, 3, and 5 binding schemes. (B) RT-qPCR validation of Tlr9 gene expression in response to IFN-γ in WT and mutant BMDMs (mean ± SD of independent biological replicates; Student’s t test relative to WT). HPRT, hypoxanthine-guanine phosophoribosyltransferase. (C) Genomic snapshot (as in A) of the Nos2 locus (group I gene). (D) ChIP-qPCR validation of H3K27Ac levels (relative to histone H3) at Tlr9 cluster 3 and 1 peaks (results are representative of three independent experiments). (E) RT-qPCR validation of Nos2 expression (as in B). (F) H3K27Ac levels (ChIP-qPCR relative to histone H3) at three cluster 5 peaks at the Nos2 locus (designated A, B, and C) showing increased K27Ac deposition in WT BMDMs after IFN-γ stimulation, which is diminished in Irf8m/m and abolished in Irf1−/− BMDMs (results are representative of three independent experiments). (G) Nitric oxide production by WT, Irf8m/m, and Irf1−/− mutant BMDMs in vitro in response to IFN-γ (mean of biological replicates ± SD; p-values were calculated using Student’s t test). (H) Nitric oxide (NO) production by splenocytes from Irf8m/m and Irf1−/− Plasmodium berghei ANKA-infected mice (compared with WT) treated with 400 U/ml IFN-γ for 48 h and 1.5 µg/ml TLR9 ligand CpG oligonucleotides. Box plots show the mean nitric oxide production from four to five mice per strain (whiskers extending to 5–95 percentiles; p-values were calculated using Student’s t test). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
T cell costimulatory and chemoattractive genes are regulated in an IRF8-dependent fashion in macrophages. Results are shown as in the legend to Fig. 6. (A) ChIP seq and RNA seq profiles at the Cxcl9-10 locus show IRF8-dependent regulation (group II gene). (B) RT-qPCR validation of Cxcl9 expression in WT and Irf mutant BMDMs (mean ± SD; p-values were calculated using Student’s t test). HPRT, hypoxanthine-guanine phosophoribosyltransferase. (C) Quantification of CXCL9 secretion by macrophages 44 h after IFN-γ treatment. (D) ChIP seq and RNA seq read density profiles at the Cd40 locus showing IRF8-dependent regulation (group II gene). (E) qPCR validation of Cd40 expression. (F) Irf8m/m BMDMs show altered H3K27Ac deposition at both IRF8-bound cluster 1 and 3 peaks (identified by the arrows in D; results are representative of three independent experiments). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, from independent biological replicates.
T cell costimulatory and chemoattractive genes are regulated in an IRF8-dependent fashion in macrophages. Results are shown as in the legend to Fig. 6. (A) ChIP seq and RNA seq profiles at the Cxcl9-10 locus show IRF8-dependent regulation (group II gene). (B) RT-qPCR validation of Cxcl9 expression in WT and Irf mutant BMDMs (mean ± SD; p-values were calculated using Student’s t test). HPRT, hypoxanthine-guanine phosophoribosyltransferase. (C) Quantification of CXCL9 secretion by macrophages 44 h after IFN-γ treatment. (D) ChIP seq and RNA seq read density profiles at the Cd40 locus showing IRF8-dependent regulation (group II gene). (E) qPCR validation of Cd40 expression. (F) Irf8m/m BMDMs show altered H3K27Ac deposition at both IRF8-bound cluster 1 and 3 peaks (identified by the arrows in D; results are representative of three independent experiments). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, from independent biological replicates.
Gbp4 and Clic5 are transcriptionally regulated in an IRF1-dependent fashion in macrophages. Results are shown as in the legend to Fig. 6. (A–C) Gbp4 (A) and Clic5 (C) are regulated in an IRF1-specific fashion (group III genes). The inset at the right side of A is a blowup of the key Gbp4 regulatory sites. (B) qPCR validation of Gbp4 gene expression under different conditions in WT and mutant Irf BMDMs (mean ± SD of independent biological replicates). (C) Clic5 as an IRF1-regulated gene (group III gene), including a cluster 1 peak 0.9 kb upstream of the transcription start site. (D and E) Clic5 mRNA expression (RT-qPCR; D) and H3K27Ac deposition (E) in response to IFN-γ in WT and Irf mutant BMDMs (ChIP-qPCR results are representative of three independent experiments). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; Student’s t test relative to WT.
Gbp4 and Clic5 are transcriptionally regulated in an IRF1-dependent fashion in macrophages. Results are shown as in the legend to Fig. 6. (A–C) Gbp4 (A) and Clic5 (C) are regulated in an IRF1-specific fashion (group III genes). The inset at the right side of A is a blowup of the key Gbp4 regulatory sites. (B) qPCR validation of Gbp4 gene expression under different conditions in WT and mutant Irf BMDMs (mean ± SD of independent biological replicates). (C) Clic5 as an IRF1-regulated gene (group III gene), including a cluster 1 peak 0.9 kb upstream of the transcription start site. (D and E) Clic5 mRNA expression (RT-qPCR; D) and H3K27Ac deposition (E) in response to IFN-γ in WT and Irf mutant BMDMs (ChIP-qPCR results are representative of three independent experiments). *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; Student’s t test relative to WT.
Group II contains genes dysregulated in an IRF8-specific fashion and that are important for activation of antigen-presenting cells and T lymphocytes (Cd86 and Cd40) for lymphoid and myeloid chemotaxis (Cxcl9, Cxcl10, Ccl4, Ccl8, and Ccl12), as well as other signaling molecules (Epha1, Tlr1, Hbegf, and P2y12; Table S3). A representative example of this group is the Cxcl9 locus that contains three cluster 1 binding sites, one proximal and two further upstream of the Cxcl10 adjacent gene (Fig. 7 A). Even though these cluster 1 sites corecruit IRF8 and IRF1, the expression of Cxcl9 in response to IFN-γ is strongly diminished only in Irf8 mutant cells, whereas Irf1 mutants have normal K27Ac on these sites and produce near-WT levels of Cxcl9 transcripts and secrete Cxcl9 (Fig. 7, A–C), suggesting that the absence of IRF1 binding in response to IFN-γ might be compensated by other TF such as STAT1. Cd40 is shown (Fig. 7, D–F) as an additional illustrative example, where a locus recruiting type 1, 3, and 5 TF combinations is only affected by the loss of IRF8 function both at the basal level and in response to IFN-γ.
Finally, the list of genes regulated in an IRF1-dependent manner (group III) contains several important proteins for myeloid cell antimicrobial activity, such as members of the family of IFN-γ–inducible intracellular GTPases (guanylate-binding proteins [GBPs]; Kim et al., 2012) implicated in phagocytosis and in phagosome maturation. The p65 subgroup of GBPs contains 11 members that map to two gene clusters on chr3 (Gbp1, 2, 3, 5, 6, and 13) and chr5 (Gbp4, 8, 9, 10, 11, and 12). Gbp mRNAs are induced by IFN in macrophages in vitro and in the spleen, liver, and lungs of mice infected with intracellular pathogens. The chr5 locus is seeded with strongly IFN-γ–induced IRF1 and STAT1 peaks from cluster 1 and 5 configurations (Fig. 8 A). Interestingly, although the low basal expression is reduced in both Irf8m/m and Irf1−/− macrophages (Table S2), the loss of IFN-γ transcriptional response is seen only in Irf1−/− macrophages (Fig. 8, A and B) and not in Irf8m/m mutant cells. The same is true for the chr3 Gbp gene cluster (Table S3). In addition, group III contains the antiviral GTPases from the Mx and p47 families and several well-known innate immune signaling and apoptosis-related molecules that are regulated in an IRF1-dependent fashion (Table S3). Clic5, Tnfsf10 (Trail), Casp1, and Casp12 are shown as additional examples of IRF1-dependent IFN-γ transcriptional activation (Fig. 8, C–E; and Fig. S3, D–I).
DISCUSSION
The objective of the present study was to understand the role of IRF8 and IRF1 in regulation of global gene expression during activation of macrophages by IFN-γ.
The analysis of the IRF8 and IRF1 cistromes confirmed a major role of these IRF family members in macrophage function, including response of these cells to infectious and inflammatory stimuli (Kubosaki et al., 2010; Berghout et al., 2013; Mancino et al., 2015). The loss of IRF8 binding leads to defective expression of genes critical for antigen presentation pathway and instructive molecules for T cell activation, but also response to stimuli such as cytokine receptors, pathogen-associated molecular pattern receptors, complement factors, and others. However, the loss of IRF1 function inactivates IFN-γ–dependent transcriptional activation of several key macrophage functions, including expression of costimulatory molecules, cytokines and chemokines, TLRs and nucleotide-binding oligomerization domain–like receptor signaling pathways, antigen processing and presentation machinery, and most small antiviral GTPases. These results define the macrophage pathways that are activated by IRF1 and IRF8 either intrinsically or in response to IFN-γ, hereby designated the IRF8/IRF1 regulome.
In RNA expression studies, we have previously observed significant overlap between genes activated in situ in infected lungs during pulmonary tuberculosis and in the brain during lethal encephalitis associated with cerebral malaria (Marquis et al., 2009a; Berghout et al., 2013). In these animal models, activation of this common transcriptome is required for protective immune response against tuberculosis, but its engagement is associated with pathological acute neuroinflammation. In agreement with the observed tuberculosis hypersusceptibility and cerebral malaria resistance phenotypes of Irf1, Irf8, and Stat1 mouse mutants in these experimental models (Fehr et al., 1997; Marquis et al., 2009b; Berghout et al., 2013), we note a strong association of genes activated during these infections with clusters 1 (9–12-fold; P < 10−19) and 5 (five- to sevenfold; P < 10−11) TF binding combinations (Fig. 9 A). Furthermore, the transcripts of the IRF8/IRF1 regulome are found significantly enriched in situ in pulmonary tuberculosis and in cerebral malaria–activated genes (Fig. 9 B), highlighting the critical role of cluster 1 and 5 cistromes and associated transcripts in host response to infectious and inflammatory stimuli. We further note considerable overlap between the IRF8/IRF1 regulome and genes in which mutations cause PIDs and that are associated with susceptibility to infections in humans (Fig. 9 D). Accordingly, the PIDs caused by IRF8K108E mutation (Salem et al., 2014b) is associated with reduced expression of genes of the IRF8/IRF1 regulome in the patient’s peripheral blood mononuclear cells (Fig. 9 C). Recent advances in whole exome and whole genome sequencing have identified >260 genes in which mutations cause different types of PIDs (Al-Herz et al., 2014). The IRF8/IRF1 regulome is overrepresented among the PID-causing genes (n = 32; 13%; P = 2.5 × 10−4; Fig. 9, D and E); the enrichment is significant and limited to genes that are positively regulated by IRF1/IRF8 and activated by IFN-γ. The PID genes involved (see complete list in Table S4) are from most major types of PIDs (Fig. 9 E) and include TFs (CIITA, IRF7, and STAT2), cytokine signaling (CXCR4, IL-21R, IL-7R, IL-17RA, and IL-12RB1), components of the complement systems (C1q, C3, C4, and CFB), an inflammasome platform (CARD11) and other proteins involved in microbial product sensing (TLR3), antigen presentation (CD40, TAP2, and TAPBP), and early innate responses of myeloid cells. Hence, it is tempting to speculate that mutations in additional poorly annotated members of the IRF8/IRF1 regulome may also cause novel immunodeficiencies where myeloid cells may be involved in causation. The IRF8/IRF1 regulome gene list may be particularly useful to prioritize candidate genes and associated protein variants found in complex datasets stemming from whole exome and whole genome sequencing of sporadic cases of PIDs and that lack family history and/or consanguinity to facilitate the search for morbid gene and associated mutation.
The IRF8/1 regulome in response to infections and in inflammatory diseases. (A) Bubble histograms showing a strong enrichment of TF binding clusters 1, 3, and 5 at genes activated in mouse lungs infected with Mycobacterium tuberculosis (TB) and in mouse brains during cerebral malaria–associated encephalitis (data are presented as in Fig. 4 D). (B) Enrichment of IRF8/IRF1 regulome transcripts among genes activated during pulmonary tuberculosis and during cerebral malaria. Histograms contrast enrichment of activated genes (Act) versus repressed genes (Rep) compared with sets of randomly selected genes (Rdm); p-values were calculated using Fisher’s exact test relative to random expectations (***, P < 0.001). (C) Same as B, but for genes with decreased expression in peripheral blood mononuclear cells from an immunodeficient patient homozygote for an IRF8K108E mutation. (D and E) Enrichment of the IRF8/1 regulome (D) among genes mutated in all eight categories (E) of human PIDs. (F) Enrichment of genes from the IRF8/IRF1 regulome within loci detected in GWASs of human chronic inflammatory diseases (red) compared with other noninflammatory diseases and other phenotypes (gray; Wilcoxon rank sum test; P = 1.3 × 10−6).
The IRF8/1 regulome in response to infections and in inflammatory diseases. (A) Bubble histograms showing a strong enrichment of TF binding clusters 1, 3, and 5 at genes activated in mouse lungs infected with Mycobacterium tuberculosis (TB) and in mouse brains during cerebral malaria–associated encephalitis (data are presented as in Fig. 4 D). (B) Enrichment of IRF8/IRF1 regulome transcripts among genes activated during pulmonary tuberculosis and during cerebral malaria. Histograms contrast enrichment of activated genes (Act) versus repressed genes (Rep) compared with sets of randomly selected genes (Rdm); p-values were calculated using Fisher’s exact test relative to random expectations (***, P < 0.001). (C) Same as B, but for genes with decreased expression in peripheral blood mononuclear cells from an immunodeficient patient homozygote for an IRF8K108E mutation. (D and E) Enrichment of the IRF8/1 regulome (D) among genes mutated in all eight categories (E) of human PIDs. (F) Enrichment of genes from the IRF8/IRF1 regulome within loci detected in GWASs of human chronic inflammatory diseases (red) compared with other noninflammatory diseases and other phenotypes (gray; Wilcoxon rank sum test; P = 1.3 × 10−6).
The established role of IRF8 and IRF1 in the ontogeny, response to microbial products, and activation of myeloid cells has suggested a parallel critical role of IRF8/IRF1-dependent transcriptional programs in proinflammatory responses and, as noted in the previous paragraph, in pathological inflammation. Indeed, different association studies have identified IRF8- and/or IRF1-associated variants as genetic risk factors for inflammatory bowel disease, multiple sclerosis, rheumatoid arthritis, and other chronic inflammatory diseases in humans (Jostins et al., 2012; Beecham et al., 2013; Okada et al., 2014). Of interest is a single nucleotide polymorphism near IRF8 (SNP accession number rs17445836) that regulates IRF8 mRNA expression (eSNP) in peripheral blood mononuclear cells and that is also the lead risk factor single nucleotide polymorphism associated with multiple sclerosis (Fairfax et al., 2014). Likewise, a similar regulatory single nucleotide polymorphism located near IRF1 behaves as a risk factor for Crohn’s disease (Fairfax et al., 2014). These suggest that modest changes in the expression of IRF8/IRF1 regulome transcripts may also be associated with inflammatory disease in humans. We investigated this possibility by looking for concordance between genes included in the IRF8/IRF1 regulome and genetic loci contributing to susceptibility to inflammatory diseases and mapped by genome-wide association studies (GWASs; Fig. 9 F). In this analysis, we have included all GWASs (from the National Human Genome Research Institute GWAS catalog) that had at least 15 significant loci (P < 10−8); these included 15 inflammatory and 37 noninflammatory traits. We noted a strong and significant three- to fourfold enrichment for loci that contain IRF1/IRF8-regulated genes among inflammatory disease risk loci (association of inflammatory vs. noninflammatory traits has a P = 1.3 × 10−6), with the most striking effects being seen for celiac disease, type I diabetes, multiple sclerosis, asthma, rheumatoid arthritis, and inflammatory bowel disease. Such enrichment was not seen in other GWASs that monitor physical or physiological traits unrelated to inflammation. Because the IRF8/1 regulome identified herein was from macrophages, its particular enrichment in a subset of inflammatory diseases may be indicative of a central role for this cell type in the establishment or progression of these pathologies.
Together, these results confirm the importance of IRF1/IRF8 in regulating the intensity of inflammatory responses, including contribution of pathological inflammation. Moreover, it suggests that the IRF8/1 regulome contains genes of unknown functions that could have critical implication in the defense response against infections or in the etiology of chronic inflammatory diseases. Indeed, this regulome contains several genes and associated biochemical and immune pathways that play critical cell-specific functions in myeloid cells, such as antigen recognition (Fc receptors, TLRs, and nucleotide-binding oligomerization domains), antigen processing and presentation (class I and class II pathways), and phagosome maturation (IRGM1 and GBPs; Tables S2 and S3). Conversely, we anticipate that other members of the IRF8/IRF1 regulome (n > 600), including poorly annotated genes (GM-annotated genes and Rik transcripts), proteins defined by a single motif (PHF11), or genes with nonimmune annotations (PLIN4), may encode protein building blocks of established immune pathways or may define novel pathways that play important and possibly unsuspected roles in intrinsic or IFN-γ–stimulated macrophage function, including defenses against infections. Many such examples can be found in the complete listings provided in Tables S4 and S5.
Collectively, we have demonstrated that IRF8 and IRF1 are important to control macrophage transcriptional programs. IRF8 plays a prominent role in the maintenance of the steady-state epigenetic and transcriptional level of critical macrophage pathways, whereas IRF1 is necessary for the induction of cis-regulatory region activity and target gene transcriptional activation in response to IFN-γ. Genes dependent on IRF8 and IRF1 for proper transcriptional regulation, the IRF8/IRF1 regulome, are significantly implicated in response to infections and in inflammatory diseases, and mutation in some of these genes causes a variety of PIDs in humans. Hence, the characterization of the IRF8/IRF1 regulome provided herein (Tables S2 and S3) should help prioritize the search for morbid genes in PIDs and in inflammatory conditions.
MATERIALS AND METHODS
Ethics statement
All mice were kept under specific pathogen-free conditions and handled according to the guidelines and regulations of the Canadian Council on Animal Care. Experimental protocols were approved by the McGill University Institutional Animal Care Committee (protocol number 3659).
Primary BMDMs
BMDMs were differentiated from BM isolated from femurs and tibias of 8–16-wk-old C57BL/6 (B6), BXH2 (Ifr8m/m), or Irf1−/− female mice (The Jackson Laboratory). In brief, BM cells were cultured in DMEM (Wisent) containing 10% heat-inactivated FBS, antibiotics, and 20% L cell–conditioned medium (LCCM) as a source of macrophage CSF. The cells were supplemented with 10% LCCM 4 d later, and at day 6, cells were harvested by gentle washing of the monolayer with PBS-citrate. Cells were plated in tissue culture–grade dishes in DMEM containing 10% FBS, 20% LCCM, and antibiotics and used the next day.
Flow cytometry analysis
To monitor in vitro differentiation of BMDMS, cells were analyzed by flow cytometry (FACSCalibur; BD) for expression of CD11b (M1/70; eBioscience), F4/80 (BM8; eBioscience), and Ly6G (1A8, BioLegend). More than 98% of cells were positive for F4/80 (Fig. 3 A) and CD11b (not depicted), and they were negative for the neutrophil marker Ly6G (not depicted), confirming their differentiation into macrophages.
ChIP seq and quantitative PCR (qPCR)
20 million BMDMs (B6, Irf8m/m, and Irf1−/−) were plated in 15-cm dishes and the next day were treated or not treated with 400 U/ml IFN-γ for 30 min or 3 h. ChIPs were performed as previously described (Langlais et al., 2011) with few modifications. In brief, BMDMs were cross-linked for 10 min at room temperature with 1% formaldehyde added in a culture medium, and cross-link was stopped with ice-cold PBS containing 0.125-M glycine for 5 min. Nuclei were prepared by sequential incubation on ice for 5 min in buffer A (10-mM Tris-HCl, pH 8, 10-mM EDTA, and 0.25% Triton X-100) and for 30 min in buffer B (10-mM Tris-HCl, pH 8, 1-mM EDTA, and 200-mM NaCl; all buffers included protease inhibitors). Nuclei were resuspended in a sonication buffer (10-mM Tris-HCl, pH 8, 1-mM EDTA, 0.5% SDS, 0.5% Triton X-100, 0.05% NaDOC, and 140-mM NaCl) and sonicated with a digital sonifier (Branson Ultrasonics) to a mean size of 250 bp. Sonicated chromatin was incubated overnight on a rotating platform at 4°C with a mixture of 20 µl protein A and 20 µl protein G Dynabeads (Invitrogen) prebound with 6 µg of control IgG (sc-2028) or IRF8 (sc-6058), IRF1 (sc-640), and PU.1 (sc-352) antibodies from Santa Cruz Biotechnology or 3 µg H3 (ab1791), H3K4me1 (ab8895), and H3K27Ac (ab4729) from Abcam. Immune complexes were washed sequentially for 2 min at room temperature with 1 ml of the following buffers: wash B (1% Triton X-100, 0.1% SDS, 150-mM NaCl, 2-mM EDTA, and 20-mM Tris-HCl, pH 8), wash C (1% Triton X-100, 0.1% SDS, 500-mM NaCl, 2-mM EDTA, and 20-mM Tris-HCl, pH 8), wash D (1% NP-40, 250-mM LiCl, 1-mM EDTA, and 10-mM Tris-HCl, pH 8), and TEN buffer (50-mM NaCl, 10-mM Tris-HCl, pH 8, and 1-mM EDTA). After de–cross-linking by overnight incubation at 65°C, the DNA was purified with QIAquick PCR purification columns (QIAGEN) according to the manufacturer’s instructions. ChIP seq libraries were prepared using the Illumina TruSeq kit and sequenced on a HiSeq 2000 in a paired-end 50-bp configuration according to the manufacturer’s recommendations. TF ChIP efficiency relative to the IgG control and H3K27Ac enrichment relative to H3 levels were assessed by qPCR using the Perfecta SYBR green PCR kit (Quanta Bioscience) for known TF binding sites using oligonucleotide primers listed in Table S5.
ChIP seq peak finding analysis
The following ChIP seq datasets were analyzed: IRF8 (WT and Irf1−/− BMDM), IRF1, PU.1 (WT and Irf8m/m BMDM), H3K4me1 (WT), and H3K27Ac (WT, Irf8m/m, and Irf1−/− BMDM); published STAT1 datasets were also included in this analysis (Ng et al., 2011). The sequence reads were mapped to the mouse mm9 reference genome assembly with Bowtie 1.0.0 using the following settings: –best–trim5 2 mm9 (Langmead et al., 2009). To identify significant TF binding events, we processed the mapped sequence reads with MACS 1.4.1 using a matching number of IgG control reads to TF samples (Zhang et al., 2008). The resulting TF peak lists were filtered to remove regions longer than 3 kb because they were artifacts generated from high background sequences, and contiguous peaks were separated with PeakSplitter (Salmon-Divon et al., 2010). The files containing mapped reads were converted from SAM to BAM format using samtools 0.1.18 (Li et al., 2009) and then converted into Tag directories for further analysis using the Homer tool kit (Heinz et al., 2010). Sequence read density profiles (bigwig) were generated using the Homer tool and normalized to counts per 107 reads. The Integrative Genomics Viewer was used to visualize sequence read density profiles and to extract genome browser snapshots for figures (Thorvaldsdóttir et al., 2013). To compare the genomic binding profiles of IRF8, IRF1, and STAT1, we overlapped the list of TF binding peak maxima in each condition using the Homer mergePeaks function and then generated a list of unique chromosomal positions by removing closely overlapping peaks (±100 bp). From this list of 63,853 unique chromosomal positions, we retrieved the total number of sequence reads (normalized per 107 reads) within 200 bp in each ChIP seq dataset (not filtered for duplicate reads). Thereafter, the binding regions were clustered by flagging for the presence or absence of binding of each TF and by ordering them relative to this flag and peak heights. The different binding schemes were attributed a cluster number from 1 to 9 (Figs. 2 B and S2). The Homer tool kit was used to extract the genomic characteristics of the binding cluster (i.e., position relative to the closest transcription start site, mammalian sequence conservation, and guanine–cytosine content) and the sequence read densities around each binding site; these were then used to generate heatmaps (Figs. 2 B and S2) using TreeView (Java; Saldanha, 2004). The raw sequence files of supplemental ChIP seq datasets shown in Fig. S2 were downloaded from the Gene Expression Omnibus database and reanalyzed using the parameters described for the other datasets above; these include K3K4me1 (0, 4, or 24 h of IFN-γ) and FAIRE at steady state (Ostuni et al., 2013) and H4Ac (Chen et al., 2012).
De novo motif analyses
To define TF binding preferences and putative novel binding motifs, a de novo motif analysis was performed on 100 bp of DNA sequence surrounding the binding peak maxima of each IRF8, IRF1, STAT1, and PU.1 dataset using the Homer findMotifGenome tool (Heinz et al., 2010). The graphical representations of the position weight matrices obtained from these analyses were generated with WebLogo (Crooks et al., 2004). The top motifs identified for each TF are shown in Fig. 1 C.
GO analyses
To determine whether specific biological functions were enriched among genes associated with ChIP seq TF binding clusters, the genomic coordinates of each cluster peak were submitted to GREAT 2.0.2 tool (McLean et al., 2010). We used 20 kb as the proximal interval and 200 kb as the distal interval to identify genes potentially regulated by these binding sites. Representative GO (biological processes) categories were selected to remove redundancy, and the resulting Hyper FDR q-values were –Log10 transformed (q-values greater than 10−2 were considered not significant and set to 0) and clustered in MeV (Saeed et al., 2003) with the Manhattan distance algorithm (Fig. 2 E). GO enrichment analysis for gene expression profiling results was performed using the DAVID website (Huang et al., 2009).
RNA seq and qPCR
Independent duplicates of 5 × 105 WT, Irf8m/m, and Irf1−/− BMDMs were treated or not treated with 400 U/ml IFN-γ for 3 h, and RNA was extracted using RNeasy columns (QIAGEN). RNA integrity was assessed on a Bioanalyzer RNA pico chip, followed by poly A mRNA enrichment and library preparation using the TruSeq RNA sample preparation kit (Illumina). The RNA seq libraries were sequenced on a HiSeq 2000 sequencer (Illumina) in a paired-end 50-bp configuration. BMDM response to IFN-γ treatment was validated by qPCR using a Perfecta SYBR green PCR kit (Quanta Biosciences), and the gene expression was normalized to Hprt.
RNA seq analyses
The quality of sequence reads obtained for each sequence read was confirmed using the FastQC tool (Babraham Bioinformatics). Reads were mapped to the mouse UCSC mm9 reference assembly using TopHat v2.0.9 in conjunction with the Bowtie 1.0.0 algorithm (Langmead et al., 2009; Trapnell et al., 2009; Kim et al., 2013), and gene expression was quantified by counting the number of uniquely mapped reads with featureCounts using default parameters (Liao et al., 2014). Normalization and differential expression analysis was conducted using the edgeR Bioconductor package (Robinson and Oshlack, 2010). We retained genes that had an expression level of a minimum of five counts per million reads in at least 2 of the 12 samples, and pairwise differential gene expression analysis was performed by comparing IFN-γ–treated samples versus untreated samples. Genes with changes in expression ≥|2| and adjusted p-values (<10−5) were considered significant. For RNA seq data visualization in genome snapshots, duplicate data were combined, and bigwig scaled per million reads mapped were generated using genomeCoverageBed and wigToBigWig tools. Relative gene expression change (log2 fold change; Figs. 4 C and 5 A) heatmaps were created using MeV (Saeed et al., 2003).
BMDM function
BMDMs were prepared from two independent B6, Irf8m/m, and Irf1−/− mice. The derived macrophages were plated in 24-well dishes and were stimulated or not stimulated the next day in duplicates with 400 U/ml IFN-γ. Culture mediums were recovered 44 h later and cleared by centrifugation. CXCL9 secretion was assessed using a commercially available ELISA (R&D Systems). Nitric oxide production was assessed by a Greiss assay on the culture supernatants (Karpuzoglu et al., 2006).
Data access
ChIP seq and RNA seq data are available in the National Center for Biotechnology Information GEO database under the accession number GSE77886.
Online supplemental material
Fig. S1 shows the validation of RNA seq and ChIP seq data and the association of gene expression with the presence of TF binding sites. Fig. S2 includes a heatmap showing genomic binding profiles of selected TFs and epigenetic characteristics of the corresponding binding clusters. Fig. S3 shows examples of IRF8-dependent basal regulation and of IRF1-dependent IFN-γ–induced transcription in BMDMs. Table S1 is available as an Excel file and lists the genomic positions for the selected TF binding clusters. Table S2 is available as an Excel file and lists the genes that showed altered expression at steady state in Irf8m/m and Irf1−/− mutant macrophages. Table S3 is available as an Excel file and lists the genes regulated in response to IFN-γ treatment in Irf8m/m and Irf1−/− mutant macrophages. Table S4 is available as an Excel file and lists the genes mutated in human PIDs and the forming part of the IRF8/IRF1 regulome. Table S5 is available as an Excel file and lists oligonucleotide primers used for ChIP-qPCR and RT-qPCR in this study.
ACKNOWLEDGMENTS
We acknowledge Susan Gauthier for technical assistance. We are grateful to Jessica Brinkworth for her help in RNA seq library preparation.
This work was supported by a research grant to P. Gros from the National Institutes of Health (National Institute of Allergy and Infectious Disease; R01AI035237). P. Gros is supported by a James McGill Professorship salary award. D. Langlais is supported by fellowships from the Fonds de Recherche du Québec Santé, the Canadian Institutes of Health Research Neuroinflammation Training Program, and the McGill Integrated Cancer Research Training Program. L.B. Barreiro is supported by grants from the Canadian Institutes of Health Research (301538 and 232519), the Human Frontiers Science Program (CDA-00025/2012), and the Canada Research Chairs Program (950-228993).
The authors declare no competing financial interests.
References
- ChIP
chromatin immunoprecipitation
- EICE
Ets-IRF composite element
- FAIRE
formaldehyde-assisted isolation of regulatory element
- GBP
guanylate-binding protein
- GO
gene ontology
- GWAS
genome-wide association study
- IRF
IFN regulatory factor
- ISRE
IFN-stimulated response element
- LCCM
L cell–conditioned medium
- PID
primary immunodeficiency
- qPCR
quantitative PCR
- TF
transcription factor




![Figure 4. IRF8 and IRF1 are required for macrophage basal transcriptional programs. (A) Principal component analysis of RNA seq data from WT, IRF8-deficient (Irf8m/m), and IRF1-deficient (Irf1−/−) BMDMs before and after 3 h of stimulation with IFN-γ. (B) Fraction of genes showing decreased (down) or increased (up) expression in Irf8m/m and Irf1−/− BMDMs and that harbor IRF8 or IRF1 binding sites within 20 kb of their transcription start site (p-values are Fisher’s exact test relative to random [Rdm] expectations). ns, not significant. (C) Expression level changes for genes significantly dysregulated at the basal level in mutant Irf8m/m and Irf1−/− BMDMs (fold change ≥|2|; adjusted p-value ≤10−5). Genes were grouped (a–f) for the effect of IRF8 and IRF1 loss of function (enriched GO categories are shown). (D) Bubble histogram showing the association of TF binding clusters with genes showing reduced (a–c) or increased (d–f) expression. The bubble size reflects the strength of the –log10 Fisher’s exact test p-value for the association of dysregulated genes (compared with random gene sets). The color gradient reflects the ratio in the number of peaks associated with dysregulated genes versus control sets of randomly selected genes.](https://cdn.rupress.org/rup/content_public/journal/jem/213/4/10.1084_jem.20151764/5/m_jem_20151764_fig4.jpeg?Expires=1773749806&Signature=4VOEzXuV7pMH8jT1GnLZoETVwBjvtX4WeYcr~LhAvyH8kpEB84~w4fkIoraeGxoALUDJpm-dU~PdI9YFbUHRzYID0Nab6U-8qifjWxqKiBeYrdFAsvWTCCAzZqzvqM7o0NehfNPd-TGv9PBSNEQOjNr1~ohOA~Eca1EYhRAc8vbmeEYJ-jDy514EQTFSada9~ePhLxI5joAu7qMNCGDHGpCqhJwre1~qusjJdx6N1zCB~h9dL~roSsjq9j0jkQA7ve1rlQRiJZzwVDHaVk9gvhIHd54ddrXh0rlR72HETbgJ~NOyZjFxWxmtU~vNkogNIP5cagVg~GkYmX4nRxA13A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)







