β-Coronaviruses remodel host endomembranes to form double-membrane vesicles (DMVs) as replication organelles (ROs) that provide a shielded microenvironment for viral RNA synthesis in infected cells. DMVs are clustered, but the molecular underpinnings and pathophysiological functions remain unknown. Here, we reveal that host fragile X–related (FXR) family proteins (FXR1/FXR2/FMR1) are required for DMV clustering induced by expression of viral non-structural proteins (Nsps) Nsp3 and Nsp4. Depleting FXRs results in DMV dispersion in the cytoplasm. FXR1/2 and FMR1 are recruited to DMV sites via specific interaction with Nsp3. FXRs form condensates driven by liquid–liquid phase separation, which is required for DMV clustering. FXR1 liquid droplets concentrate Nsp3 and Nsp3-decorated liposomes in vitro. FXR droplets facilitate recruitment of translation machinery for efficient translation surrounding DMVs. In cells depleted of FXRs, SARS-CoV-2 replication is significantly attenuated. Thus, SARS-CoV-2 exploits host FXR proteins to cluster viral DMVs via phase separation for efficient viral replication.
Introduction
β-Coronaviruses are a group of positive single-strand RNA viruses that include the pathogenic strains severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), SARS-CoV, murine hepatitis virus, the Middle East respiratory syndrome coronavirus (MERS-CoV), etc. Upon entering host cells, the viruses transcribe and replicate their RNA using a common strategy characterized by the remodeling of host endomembranes into organelle-like structures, termed replication organelles (ROs). Among the viral nonstructural proteins (Nsps) that are cleaved from polyprotein pp1a and pp1ab by two viral proteases (Thiel et al., 2003; Prentice et al., 2004), the two membrane-integral proteins Nsp3 and Nsp4 are essential for RO biogenesis. The most prominent ROs are double-membrane vesicles (DMVs) that can be induced by ectopic expression of Nsp3 and Nsp4 in various types of cells (Knoops et al., 2008; Snijder et al., 2020; Cortese et al., 2020; Mohan and Wollert, 2021). The interaction of Nsp3 and Nsp4 via their luminal domains brings together endoplasmic reticulum (ER) membranes and the zippered ER subsequently curves into closed vesicles (Ji et al., 2022; Oudshoorn et al., 2017; Twu et al., 2021; Wolff et al., 2020). Previous studies showed that most DMVs are tightly packed into clusters both during virus infection (Cortese et al., 2020; Knoops et al., 2008; Snijder et al., 2020) and in cells coexpressing Nsp3/4 (Ji et al., 2022; Ricciardi et al., 2022). The molecular mechanism underlying the spatial confinement of DMVs remains unknown.
Liquid–liquid phase separation (LLPS)–mediated formation of protein condensates has been recently shown to drive the spatial organization of membrane-bound organelles. LLPS, driven by multivalent interactions among constituents, concentrates proteins and RNA molecules into liquid droplets (Banani et al., 2017; Wang and Zhang, 2019; Wu et al., 2020). Phase-separated condensates can interact with vesicles to regulate their release, trafficking, and storage (Zhao and Zhang, 2020). For example, assembly of synaptic vesicles by phase separation of synapsin 1 ensures their spatial confinement at the presynaptic membranes in neurons, allowing their rapid release for fusion with the plasma membrane in response to stimuli (Milovanovic et al., 2018; Zhao and Zhang, 2020). mRNAs and organelles, including mitochondria, ER, and Golgi, are stored in membraneless compartments called Balbiani bodies during oocyte dormancy (Kloc et al., 2014).
The fragile X–related (FXR) protein family, consisting of FXR1, FXR2, and FMR1/FMRP, plays an important role in RNA metabolism (Kao et al., 2010), neural homeostasis (Zhou et al., 2017), muscle development (Mientjes et al., 2004), and tumorigenesis (Majumder et al., 2016; Qian et al., 2015). The FXR family proteins are highly homologous RNA-binding proteins that undergo LLPS, possibly mediated by the K-homology 2 (KH2)) domain and the intrinsically disordered regions (IDRs) at the C terminus (Kang et al., 2022; Kim et al., 2019; Kirkpatrick et al., 2001). In addition to their role in translational repression, FXRs also positively regulate adenylate-uridylate–rich element-mediated translation (Vasudevan and Steitz, 2007) and recruit translation initiation complexes for activation of stored mRNAs during spermiogenesis (Kang et al., 2022). FXRs were previously identified as host factors specifically important for the replication of New World alphaviruses, such as Venezuelan equine encephalitis virus (VEEV), but the underlying mechanism is unknown (Götte et al., 2019; Kim et al., 2016).
Here, we found that LLPS of FXRs plays an important role in clustering viral ROs for efficient viral replication. Through interaction with the DMV outer membrane protein Nsp3, FXRs are recruited to the DMV sites and form condensates to capture and cluster DMVs. In cells depleted of FXRs, scattered DMVs are distributed throughout the cytoplasm. In vitro reconstitution demonstrates that phase separation of FXR1 is sufficient to concentrate Nsp3 and Nsp3-labeled liposomes. FXR condensates enrich translation machinery to promote synthesis of viral proteins. SARS-CoV-2 replication is greatly attenuated after depleting FXRs. Our findings reveal that β-coronaviruses hijack host FXR proteins for DMV clustering through phase separation to facilitate viral replication.
Results
FXR family proteins interact with Nsp3
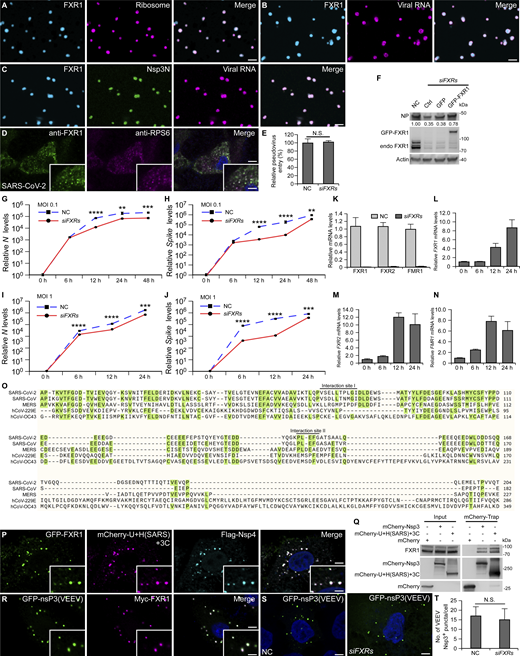
To identify host factors involved in DMV formation, we performed interactome analysis of the two viral DMV inducers, Nsp3 and Nsp4. GFP-Trap was conducted with lysates from HeLa cells coexpressing mCherry-Nsp3 and Nsp4-GFP, in which DMVs are formed, while lysates from cells expressing Nsp4-GFP alone, which fail to generate DMVs, served as a control (Fig. 1 A). After mass spectrometry (MS) analysis, two fragile X family proteins, FXR1 and FMR1, appeared as top hits of the interactor list (Fig. 1 B). Coimmunoprecipitation (co-IP) results showed that when expressed alone, Nsp3, but not Nsp4, bound to endogenous FXR1, FXR2, and FMR1 (Fig. 1, C and D). These results suggest that in the interactome analysis of cells expressing mCherry-Nsp3 and Nsp4-GFP, Nsp4 pulled down FXRs indirectly via its interaction with Nsp3 (Fig. 1 E). We previously showed that Nsp3 and Nsp4 localize on the outer and inner membranes of DMVs, respectively (Ji et al., 2022). Since FXRs are cytosolic proteins, the direct interaction of FXRs with Nsp3, but not Nsp4, is consistent with the location of Nsp3 on the outer DMV membrane. FXR2 did not appear in the MS results, probably due to its lower expression levels (Fig. S1 A). Furthermore, both endogenous and GFP-tagged FXR1, FXR2, and FMR1 showed near-complete colocalization with Nsp3/4+ foci in Nsp3/4-coexpressing cells (Fig. 1 F and Fig. S1 B).
FXRs are recruited to DMV sites through binding to Nsp3. (A) The schematic diagram shows the experimental design for identifying DMV-related factors. Lysates from cells expressing Nsp4-GFP and mCherry-Nsp3 or Nsp4-GFP alone (serving as control) are subjected to GFP-Trap, followed by MS analysis. N, nucleus. (B) The volcano plot from the MS data demonstrates quantitative changes in proteins pulled down by Nsp4-GFP in Nsp3/4-expressing cells compared with cells expressing Nsp4-GFP alone. The horizontal dashed line shows where P value is 0.05 (−log 10 [0.05] = 1.3), and the vertical dashed lines show where the fold change is 2 (log 2 [2] = 1) or 0.5 (log 2 [0.5] = −1). These parameters were used as the threshold cutoff. Upregulated proteins are shown in red and downregulated proteins are shown in blue. FXR1 and FMR1 are labeled. (C) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are immunoprecipitated by mCherry-Nsp3. (D) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are not immunoprecipitated by Nsp4-mCherry. (E) The schematic shows the specific localization of Nsp3 and Nsp4 on DMVs and the Nsp3-mediated recruitment of FXRs to DMVs. (F) Endogenous FXR1, FXR2, and FMR1 colocalize well with Nsp3/4+ foci in mCherry-Nsp3/Flag-Nsp4-coexpressing HeLa cells. FXR1, FXR2, and FMR1 show diffuse cytoplasmic localization under control conditions. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (G) Endogenous FXR1 forms puncta that colocalize well with Nsp3 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (H) Endogenous FXR2 and FMR1 form puncta that colocalize well with FXR1 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F1.
FXRs are recruited to DMV sites through binding to Nsp3. (A) The schematic diagram shows the experimental design for identifying DMV-related factors. Lysates from cells expressing Nsp4-GFP and mCherry-Nsp3 or Nsp4-GFP alone (serving as control) are subjected to GFP-Trap, followed by MS analysis. N, nucleus. (B) The volcano plot from the MS data demonstrates quantitative changes in proteins pulled down by Nsp4-GFP in Nsp3/4-expressing cells compared with cells expressing Nsp4-GFP alone. The horizontal dashed line shows where P value is 0.05 (−log 10 [0.05] = 1.3), and the vertical dashed lines show where the fold change is 2 (log 2 [2] = 1) or 0.5 (log 2 [0.5] = −1). These parameters were used as the threshold cutoff. Upregulated proteins are shown in red and downregulated proteins are shown in blue. FXR1 and FMR1 are labeled. (C) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are immunoprecipitated by mCherry-Nsp3. (D) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are not immunoprecipitated by Nsp4-mCherry. (E) The schematic shows the specific localization of Nsp3 and Nsp4 on DMVs and the Nsp3-mediated recruitment of FXRs to DMVs. (F) Endogenous FXR1, FXR2, and FMR1 colocalize well with Nsp3/4+ foci in mCherry-Nsp3/Flag-Nsp4-coexpressing HeLa cells. FXR1, FXR2, and FMR1 show diffuse cytoplasmic localization under control conditions. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (G) Endogenous FXR1 forms puncta that colocalize well with Nsp3 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (H) Endogenous FXR2 and FMR1 form puncta that colocalize well with FXR1 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F1.
FXRs, but not SGs, are involved in DMV biogenesis, related to Fig. 1 . (A) Transcriptional levels of FXR2 are much lower than FXR1 and FMR1 in HeLa cells. Quantitative data are presented as mean ± SD (n = 3). (B) GFP-FXR1, FXR2-GFP, and GFP-FMR1 concentrate at Nsp3/4+ foci in Nsp3/4-overexpressing cells. GFP-FXR1, FXR2-GFP, and GFP-FMR1 show diffuse cytoplasmic localization under control conditions. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (C) The schematic shows the domain organization of FXR1, FXR2, and FMR1. (D) Immunostaining with anti-G3BP shows that G3BP does not colocalize with Nsp3/4+ puncta in cells treated without or with arsenite. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm.
FXRs, but not SGs, are involved in DMV biogenesis, related to Fig. 1 . (A) Transcriptional levels of FXR2 are much lower than FXR1 and FMR1 in HeLa cells. Quantitative data are presented as mean ± SD (n = 3). (B) GFP-FXR1, FXR2-GFP, and GFP-FMR1 concentrate at Nsp3/4+ foci in Nsp3/4-overexpressing cells. GFP-FXR1, FXR2-GFP, and GFP-FMR1 show diffuse cytoplasmic localization under control conditions. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (C) The schematic shows the domain organization of FXR1, FXR2, and FMR1. (D) Immunostaining with anti-G3BP shows that G3BP does not colocalize with Nsp3/4+ puncta in cells treated without or with arsenite. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm.
To investigate if FXR1, FXR2, and FMR1 are recruited to DMV sites during viral infection, HeLa cells stably expressing the SARS-CoV-2 receptor protein human ACE2 (ACE2-HeLa) were infected with SARS-CoV-2, and Nsp3 and double stranded RNA (dsRNA) antibodies were used as DMV markers (Knoops et al., 2008; Snijder et al., 2020; Ulasli et al., 2010). FXR1 formed puncta that colocalized well with Nsp3 and dsRNA in SARS-CoV-2–infected cells (Fig. 1 G). FXR2 and FMR1 were also targeted to DMVs (Fig. 1 H). These data demonstrate that FXRs are recruited to DMV sites induced by both ectopic Nsp3/4 expression and virus infection. Since FXR1, FXR2, and FMR1 share high structural and functional similarity (Fig. S1 C) (Zhang et al., 1995), we mainly used FXR1 in the following study.
As FXR proteins were previously identified as stress granule (SG) components (Mazroui et al., 2002) and SGs have been reported to be involved in viral infections (Reineke and Lloyd, 2013), we examined if SGs act in DMV formation. The SG marker G3BP1 did not colocalize with Nsp3/4+ puncta in cells treated with or without the SG inducer arsenite, and the formation of Nsp3/4+ foci was not affected in arsenite-treated cells (Fig. S1 D). These data indicate that FXRs, but not SGs, may specifically be involved in DMV biogenesis.
FXR family proteins are required for DMV clustering
To interrogate the potential role of FXRs in DMV formation, we treated cells with non-targeting (negative control, NC) small interfering RNA (siRNA) or siRNAs targeting FXR1, FXR2, and FMR1, and simultaneously cotransfected the cells with Nsp3 and Nsp4. FXR1, FXR2, and FMR1 were efficiently depleted by RNAi (Fig. 2 A). In control cells, Nsp3/4+ formed a few big puncta, while depleting all FXR mRNAs in cells resulted in numerous small Nsp3/4+ dots (Fig. 2, B and C). Single or double knockdown (KD) of FXRs did not cause obvious changes in DMV formation, except that KD of FXR1 and FMR1 together showed partial defects (Fig. S2, A–I). These observations indicate functional redundancy of the three genes.
FXRs are essential for forming and maintaining gathered DMVs. (A) Western blotting shows the KD efficiency of siFXRs. (B and C) Fluorescence images reveal numerous small Nsp3/4+ puncta in cells after KD of all FXR genes, while a few big Nsp3/4+ puncta are formed in control cells (B). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 30; siFXRs, n = 30) in C. ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (D–F) TEM images show that dispersed DMVs are observed in siFXRs cells, while clustered DMVs are present in NC-treated cells (D). Percentages of cells with clustered DMVs (at least five DMVs gathering together) are quantified in E, and numbers and sizes of DMVs are shown as mean ± SD in F (NC, n = 85; siFXRs, n = 99). ****, P < 0.0001; N.S., not significant. Bars: 500 nm. (G and H) Re-expressing RNAi-resistant GFP-FXR1, but not GFP, induces big Nsp3/4+ puncta in siFXRs cells (G). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 30; GFP, n = 25; GFP-FXR1, n = 27) (H). ****, P < 0.0001; N.S., not significant. Bars: 5 μm; insets, 2 μm. (I–K) Big Nsp3/4+ puncta are formed in cells coexpresssing mCherry-Nsp3 and Nsp4-GFP for 2 days (I). When the cells are treated with NC or siFXRs, big Nsp3/4+ puncta remain present in NC cells while a large number of Nsp3/4+ dots are induced after KD of FXRs (J). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 34; siFXRs, n = 41) (K). ****, P < 0.0001. Bar: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F2.
FXRs are essential for forming and maintaining gathered DMVs. (A) Western blotting shows the KD efficiency of siFXRs. (B and C) Fluorescence images reveal numerous small Nsp3/4+ puncta in cells after KD of all FXR genes, while a few big Nsp3/4+ puncta are formed in control cells (B). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 30; siFXRs, n = 30) in C. ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (D–F) TEM images show that dispersed DMVs are observed in siFXRs cells, while clustered DMVs are present in NC-treated cells (D). Percentages of cells with clustered DMVs (at least five DMVs gathering together) are quantified in E, and numbers and sizes of DMVs are shown as mean ± SD in F (NC, n = 85; siFXRs, n = 99). ****, P < 0.0001; N.S., not significant. Bars: 500 nm. (G and H) Re-expressing RNAi-resistant GFP-FXR1, but not GFP, induces big Nsp3/4+ puncta in siFXRs cells (G). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 30; GFP, n = 25; GFP-FXR1, n = 27) (H). ****, P < 0.0001; N.S., not significant. Bars: 5 μm; insets, 2 μm. (I–K) Big Nsp3/4+ puncta are formed in cells coexpresssing mCherry-Nsp3 and Nsp4-GFP for 2 days (I). When the cells are treated with NC or siFXRs, big Nsp3/4+ puncta remain present in NC cells while a large number of Nsp3/4+ dots are induced after KD of FXRs (J). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 34; siFXRs, n = 41) (K). ****, P < 0.0001. Bar: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F2.
FXRs are essential for DMV clustering, related toFigs. 2 and 3 . (A–H) Compared with NC-treated cells (A), the number of Nsp3/4+ puncta is slightly increased in siFXR1, siFXR2, siFMR1, siFXR1/2, or siFXR2/FMR1 cells (B–F). Cells with double KD of FXR1 and FMR1 show an obvious increase in the number of Nsp3/4+ puncta (G). Depleting all FXR genes causes dramatic DMV dispersion (H). Bars: 10 μm. (I) Numbers of DMVs are shown as mean ± SD (NC, n = 25; siFXR1, n = 35; siFXR2, n = 34; siFMR1, n = 35; siFXR1/2, n = 32; siFXR2/FMR1, n = 41; siFXR1/FMR1, n = 38; siFXRs, n = 39). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (J) Fluorescence images reveal numerous small Nsp3/4+ puncta in cells coexpressing GFP-Nsp6, mCherry-Nsp3, and Nsp4-BFP after KD of all FXR genes, while a few big Nsp3/4+ puncta are formed in control cells. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (K) TEM images show that dispersed DMVs are observed in siFXRs cells coexpressing GFP-Nsp6, mCherry-Nsp3, and Nsp4-BFP, while clustered DMVs are present in NC-treated cells. Bars: 500 nm. (L) In a GFP-Trap assay, FXR1, FXR2, and FMR1 are immunoprecipitated by GFP-Nsp3, but not Nsp6. (M and N) FRAP results show that after photobleaching part of a punctum (arrows), most of the GFP-FXR1 fluorescent signal recovers, while the mCherry-Nsp3 signal fails to recover (M). Quantitative data are presented as mean ± SEM (n = 12) in N. Bars: 2 μm. (O) GFP-FXR1 with the L351P mutation, which disrupts the phase separation ability of FXR1, fails to accumulate at Nsp3/4+ sites. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (P) Most of the GFP-FXR1 signal is diffuse in cells treated with 1,6-hexanediol. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (Q) Nsp3N does not form liquid droplets in vitro. Bar: 5 μm. (R) In a GFP-Trap assay, mCherry-Nsp3 is precipitated by GFP-FXR1, but not GFP-FXR1(L351P). Source data are available for this figure: SourceData FS2.
FXRs are essential for DMV clustering, related toFigs. 2 and 3 . (A–H) Compared with NC-treated cells (A), the number of Nsp3/4+ puncta is slightly increased in siFXR1, siFXR2, siFMR1, siFXR1/2, or siFXR2/FMR1 cells (B–F). Cells with double KD of FXR1 and FMR1 show an obvious increase in the number of Nsp3/4+ puncta (G). Depleting all FXR genes causes dramatic DMV dispersion (H). Bars: 10 μm. (I) Numbers of DMVs are shown as mean ± SD (NC, n = 25; siFXR1, n = 35; siFXR2, n = 34; siFMR1, n = 35; siFXR1/2, n = 32; siFXR2/FMR1, n = 41; siFXR1/FMR1, n = 38; siFXRs, n = 39). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (J) Fluorescence images reveal numerous small Nsp3/4+ puncta in cells coexpressing GFP-Nsp6, mCherry-Nsp3, and Nsp4-BFP after KD of all FXR genes, while a few big Nsp3/4+ puncta are formed in control cells. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (K) TEM images show that dispersed DMVs are observed in siFXRs cells coexpressing GFP-Nsp6, mCherry-Nsp3, and Nsp4-BFP, while clustered DMVs are present in NC-treated cells. Bars: 500 nm. (L) In a GFP-Trap assay, FXR1, FXR2, and FMR1 are immunoprecipitated by GFP-Nsp3, but not Nsp6. (M and N) FRAP results show that after photobleaching part of a punctum (arrows), most of the GFP-FXR1 fluorescent signal recovers, while the mCherry-Nsp3 signal fails to recover (M). Quantitative data are presented as mean ± SEM (n = 12) in N. Bars: 2 μm. (O) GFP-FXR1 with the L351P mutation, which disrupts the phase separation ability of FXR1, fails to accumulate at Nsp3/4+ sites. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (P) Most of the GFP-FXR1 signal is diffuse in cells treated with 1,6-hexanediol. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (Q) Nsp3N does not form liquid droplets in vitro. Bar: 5 μm. (R) In a GFP-Trap assay, mCherry-Nsp3 is precipitated by GFP-FXR1, but not GFP-FXR1(L351P). Source data are available for this figure: SourceData FS2.
Transmission electron microscopy (TEM) was performed to examine the ultrastructures of DMVs in cells treated with NC siRNA and siRNAs against all three FXRs (siFXRs). In control cells, most of the DMVs were organized into clusters as previously reported (Ji et al., 2022; Ricciardi et al., 2022), whereas DMVs with comparable sizes were homogeneously distributed in cells without FXRs (Fig. 2, D and E). The dispersion of DMVs in siFXRs cells was not because of defective DMV formation, as the total numbers of DMVs were comparable with those in control cells (Fig. 2 F). Furthermore, the dispersed Nsp3/4+ puncta in siFXRs cells was rescued by re-expressing RNAi-resistant FXR1 (Fig. 2, G and H). These data suggest that FXRs are critical for concentrating DMVs.
We then asked whether FXRs are also essential for maintaining clustered DMVs. To test this, cells were allowed to coexpress Nsp3/4 for 2 days to induce big punctate structures (Fig. 2 I) and then subjected to KD of FXRs. The results demonstrated that depleting FXRs caused the appearance of numerous small Nsp3/4+ puncta, similar to those formed when FXRs are depleted at the beginning (Fig. 2, B, C, J, and K). These results demonstrated the functional importance of FXRs in both forming and maintaining DMV clusters.
Nsp6 was reported to organize DMVs in both infected and Nsp3/4-transfected cells (Ricciardi et al., 2022). We found that Nsp6 formed punctate structures associated with Nsp3/4+ foci in Nsp3/4/6-coexpressing control cells, as in the previous study (Ricciardi et al., 2022). When FXRs were knocked down, Nsp3/4+ structures still became numerous dispersed small dots, and their association with Nsp6 was unaffected (Fig. S2 J). Consistent results were observed in TEM images (Fig. S2 K). We also investigated the interaction between Nsp6 and FXR proteins, and no binding was detected (Fig. S2 L). These results suggest that FXRs and Nsp6 may have distinct functions in assembling DMVs: FXRs may be the main drivers for clustering DMVs, while Nsp6 may be critical for connecting DMVs with the ER.
FXRs undergo phase separation to capture DMVs
FXRs were reported to form condensates through phase separation (Kang et al., 2022; Kim et al., 2019). To assess whether the foci formed by FXRs at Nsp3/4+ sites were liquid droplets, we analyzed their dynamics using fluorescence photobleaching recovery (FRAP) assays. After photobleaching, most of the GFP-FXR1 fluorescence signals recovered within ∼120 s, while mCherry-Nsp3 signals did not recover, no matter whether partial or whole puncta were bleached (Fig. 3, A and B; and Fig. S2, M and N). Within cells, GFP-FXR1 became largely diffuse after its phase separation ability was disrupted by L351P mutation (Kang et al., 2022) or 1,6-hexanediol treatment (Fig. S2, O and P). Liquid droplets such as SGs often exhibit electron-dense structures under TEM (Souquere et al., 2009). TEM analysis also revealed that electron-dense patches were frequently detected around clustered DMVs (Fig. 2 D). To verify whether these patches were formed by FXR1, we transfected cells with GFP-FXR1 with GFP-binding protein (GBP)–linked APEX2, which is a peroxidase that catalyzes DAB polymerization to generate highly electron-dense signals in the presence of OsO4 during TEM analysis. The dark APEX2-FXR1 signal was enriched around DMV clusters (Fig. 3 C). Hence, instead of forming stable complexes with Nsp3/4, FXRs concentrate at the DMV sites as liquid droplets.
LLPS of FXR1 drives DMV clustering. (A and B) FRAP results in cells show that after photobleaching the whole punctum (arrows), most of the fluorescent signal of GFP-FXR1 recovers, while the mCherry-Nsp3 signal fails to recover (A). Quantification data are presented as mean ± SEM (n = 12) in B. Bars: 2 μm. (C) APEX2-FXR1 signal is detected surrounding clustered DMVs in cells. Bars: 500 nm. (D) FXR1 undergoes LLPS in vitro. Bar: 5 μm. (E) The schematic shows the domain organization of Nsp3. Nsp3N is indicated by the dashed box. (F) Nsp3N partitions into FXR1 liquid droplets in vitro. Bar: 5 μm. (G) Top row: In the absence of Nsp3N, FXR1 droplets (cyan) are associated with a few liposomes (green) in vitro. Bottom row: FXR1 droplets capture high levels of Nsp3N-decorated liposomes. Bars: 5 μm. (H) In a GFP-Trap assay, FKBP-GFP-FXR1(L351P) immunoprecipitates comparable levels of FRB-mCherry-Nsp3 as WT proteins treated with 1.5 μM rapamycin. Quantifications of levels of Nsp3 (normalized by FXR1 levels) are shown. (I and J) Re-expressing RNAi-resistant FKBP-GFP-FXR1, but not FKBP-GFP-FXR1(L351P), induces big Nsp3/4+ puncta in siFXRs cells (I). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 18; FKBP-GFP-FXR1, n = 18; FKBP-GFP-FXR1(L351P), n = 12) (J). ****, P <0.0001; N.S., not significant. Bars: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F3.
LLPS of FXR1 drives DMV clustering. (A and B) FRAP results in cells show that after photobleaching the whole punctum (arrows), most of the fluorescent signal of GFP-FXR1 recovers, while the mCherry-Nsp3 signal fails to recover (A). Quantification data are presented as mean ± SEM (n = 12) in B. Bars: 2 μm. (C) APEX2-FXR1 signal is detected surrounding clustered DMVs in cells. Bars: 500 nm. (D) FXR1 undergoes LLPS in vitro. Bar: 5 μm. (E) The schematic shows the domain organization of Nsp3. Nsp3N is indicated by the dashed box. (F) Nsp3N partitions into FXR1 liquid droplets in vitro. Bar: 5 μm. (G) Top row: In the absence of Nsp3N, FXR1 droplets (cyan) are associated with a few liposomes (green) in vitro. Bottom row: FXR1 droplets capture high levels of Nsp3N-decorated liposomes. Bars: 5 μm. (H) In a GFP-Trap assay, FKBP-GFP-FXR1(L351P) immunoprecipitates comparable levels of FRB-mCherry-Nsp3 as WT proteins treated with 1.5 μM rapamycin. Quantifications of levels of Nsp3 (normalized by FXR1 levels) are shown. (I and J) Re-expressing RNAi-resistant FKBP-GFP-FXR1, but not FKBP-GFP-FXR1(L351P), induces big Nsp3/4+ puncta in siFXRs cells (I). Numbers of large and all Nsp3/4+ puncta are shown as mean ± SD (NC, n = 18; FKBP-GFP-FXR1, n = 18; FKBP-GFP-FXR1(L351P), n = 12) (J). ****, P <0.0001; N.S., not significant. Bars: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F3.
We next determined if Nsp3 partitions into the FXR liquid droplets in vitro. Purified FXR1 underwent LLPS (Fig. 3 D), as previously reported (Kang et al., 2022). Nsp3 possesses 10 N-terminal cytoplasmic domains and two C-terminal transmembrane (TM) regions (Lei et al., 2018) (Fig. 3 E). The purified N-terminal cytosolic stretch of Nsp3 (Nsp3N, Fig. 3 E), labeled with iFluor 488, did not form condensates (Fig. S2 Q). However, when mixed with Cy3-labeled FXR1, Nsp3N partitioned into FXR1 liquid droplets (Fig. 3 F).
If the phase separation feature of FXR1 is involved in DMV clustering, FXR1 condensates should be capable of capturing vesicles through Nsp3. We thus generated Nsp3N-decorated liposomes with a lipid composition mimicking the ER where DMVs were generated. In the absence of Nsp3N, small amounts of empty liposomes stuck to the surface of FXR1 droplets, while Nsp3N-decorated liposomes entered into FXR1 droplets at a much higher level (Fig. 3 G). We further examined whether the L351P mutant FXR1 was able to rescue the DMV defects in siFXRs cells. However, L351P disrupted the interaction of FXR1 with Nsp3 (Fig. S2 R). To solve this problem, we generated FKBP–rapamycin binding (FRB)-mCherry-Nsp3 and FKBP-GFP-FXR1(L351P), which interacted with each other at a level comparable with WT proteins upon rapamycin treatment (Fig. 3 H). In the presence of rapamycin, FKBP-GFP-FXR1(L351P) failed to reverse the dispersed DMVs in siFXRs cell coexpressing FRB-mCherry-Nsp3 and Nsp4 (Fig. 3, I and J). Taken together, the above findings suggest that FXR1 undergoes LLPS to drive clustering of DMVs.
The UBL1 and hypervariable region (HVR) domains in Nsp3 are essential for recruiting FXR1
To dissect the domain(s) of Nsp3 responsible for its binding with FXRs, we generated a series of Nsp3 deletions lacking one or two of the 10 N-terminal domains (Fig. 4 A). Interestingly, both fluorescence and TEM images showed that dispersed DMVs were induced in cells expressing Nsp3(ΔUBL1) or Nsp3(ΔHVR) or Nsp3(ΔUBL1ΔHVR) with Nsp4 (Fig. 4, B–D), which is similar to the phenotype in siFXRs cells (Fig. 2, C and D). Deletion of any one of the other eight domains of Nsp3 resulted in no obvious defects (Fig. S3).
DMV clustering depends on the UBL1 and HVR domains in Nsp3. (A) The schematic shows the domain organization of Nsp3 deletion mutants. (B and C) Cells with coexpression of mCherry-Nsp3(ΔUBL1), Nsp3(ΔHVR), or Nsp3(ΔUBL1ΔHVR) and Nsp4-GFP contain a large number of small Nsp3/4+ puncta that fail to colocalize with endogenous FXR1, while full-length mCherry-Nsp3 forms big puncta with Nsp4, which colocalize well with FXR1 foci (C). Numbers of DMVs are shown as mean ± SD (mCherry-Nsp3, n = 33; mCherry-Nsp3(ΔUBL1), n = 22; mCherry-Nsp3(ΔHVR), n = 30; mCherry-Nsp3(ΔUBL1ΔHVR), n = 41) (B). ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (D) TEM images show that coexpressing Nsp3(ΔUBL1), Nsp3(ΔHVR), or Nsp3(ΔUBL1ΔHVR) with Nsp4 induces DMV dispersion throughout the cytoplasm, while clustered DMVs are present in cells expressing full-length (FL) Nsp3 and Nsp4. Bars: 500 nm; insets, 200 nm. (E) In a GFP-Trap assay, GFP-FXR1 immunoprecipitates full-length mCherry-Nsp3, but not mCherry-Nsp3(ΔUBL1), mCherry-Nsp3(ΔHVR), or mCherry-Nsp3(ΔUBL1ΔHVR). (F) Deleting either the UBL1 or HVR domain of Nsp3N slightly reduces the entry of Nsp3 into FXR1 condensates, while deleting both domains completely eliminates Nsp3 from FXR1 droplets. Bars: 5 μm. Source data are available for this figure: SourceData F4.
DMV clustering depends on the UBL1 and HVR domains in Nsp3. (A) The schematic shows the domain organization of Nsp3 deletion mutants. (B and C) Cells with coexpression of mCherry-Nsp3(ΔUBL1), Nsp3(ΔHVR), or Nsp3(ΔUBL1ΔHVR) and Nsp4-GFP contain a large number of small Nsp3/4+ puncta that fail to colocalize with endogenous FXR1, while full-length mCherry-Nsp3 forms big puncta with Nsp4, which colocalize well with FXR1 foci (C). Numbers of DMVs are shown as mean ± SD (mCherry-Nsp3, n = 33; mCherry-Nsp3(ΔUBL1), n = 22; mCherry-Nsp3(ΔHVR), n = 30; mCherry-Nsp3(ΔUBL1ΔHVR), n = 41) (B). ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (D) TEM images show that coexpressing Nsp3(ΔUBL1), Nsp3(ΔHVR), or Nsp3(ΔUBL1ΔHVR) with Nsp4 induces DMV dispersion throughout the cytoplasm, while clustered DMVs are present in cells expressing full-length (FL) Nsp3 and Nsp4. Bars: 500 nm; insets, 200 nm. (E) In a GFP-Trap assay, GFP-FXR1 immunoprecipitates full-length mCherry-Nsp3, but not mCherry-Nsp3(ΔUBL1), mCherry-Nsp3(ΔHVR), or mCherry-Nsp3(ΔUBL1ΔHVR). (F) Deleting either the UBL1 or HVR domain of Nsp3N slightly reduces the entry of Nsp3 into FXR1 condensates, while deleting both domains completely eliminates Nsp3 from FXR1 droplets. Bars: 5 μm. Source data are available for this figure: SourceData F4.
The UBL1 and HVR domains of Nsp3 are required for DMV formation, related to Fig. 4 . (A–I) In cells expressing Nsp4, puncta formed by coexpression of Nsp3 with a deletion of the Mac1 (B), Mac2 (C), Mac3 (D), DPUP (E), UBL2 (F), PL2 (G), NAB (H), or βSM (I) domain show no obvious difference compared with full-length Nsp3 (A). Bars: 5 μm; insets, 2 μm. (J) TEM images of cells coexpressing Nsp3(ΔMac1), Nsp3(ΔMac2), Nsp3(ΔMac3), Nsp3(ΔDPUP), Nsp3(ΔUBL2), Nsp3(ΔPL2), Nsp3(ΔNAB), or Nsp3(ΔβSM) with Nsp4. Bars: 500 nm; insets, 200 nm.
The UBL1 and HVR domains of Nsp3 are required for DMV formation, related to Fig. 4 . (A–I) In cells expressing Nsp4, puncta formed by coexpression of Nsp3 with a deletion of the Mac1 (B), Mac2 (C), Mac3 (D), DPUP (E), UBL2 (F), PL2 (G), NAB (H), or βSM (I) domain show no obvious difference compared with full-length Nsp3 (A). Bars: 5 μm; insets, 2 μm. (J) TEM images of cells coexpressing Nsp3(ΔMac1), Nsp3(ΔMac2), Nsp3(ΔMac3), Nsp3(ΔDPUP), Nsp3(ΔUBL2), Nsp3(ΔPL2), Nsp3(ΔNAB), or Nsp3(ΔβSM) with Nsp4. Bars: 500 nm; insets, 200 nm.
The above data suggest that the UBL1 and HVR domains of Nsp3 may be responsible for the interaction of Nsp3 with FXRs. GFP-Trap assays showed that deleting either the UBL1 or HVR domain or both domains greatly suppressed the binding of FXR1 to Nsp3 (Fig. 4 E). Nsp3 containing only the UBL1 (mCherry-Nsp3[UBL1+C]) or HVR (mCherry-Nsp3[HVR+C]) domain followed by the C-terminal TM domains were marginally precipitated by Myc-FXR1, while Nsp3(UBL1+HVR+C) showed strong interaction with FXR1 (Fig. S4, A and B). Moreover, FXR1 showed a diffuse pattern and failed to colocalize with the tiny puncta induced by Nsp3(ΔUBL1), Nsp3(ΔHVR), Nsp3(ΔUBL1ΔHVR), Nsp3(UBL1+C), or Nsp3(HVR+C) (Fig. 4 C and Fig. S4 C). Expressing mCherry-Nsp3(UBL1+HVR+C)/Nsp4-GFP induced big Nsp3/4+ puncta positive for FXR1 (Fig. S4 D). Thus, both UBL1 and HVR domains are required and sufficient for recruitment of FXR1.
Both the UBL1 and HVR domains of Nsp3 are required for recruitment of FXRs, related toFigs. 5 and 6 . (A) The schematic shows the domain organization of the truncated forms of Nsp3. (B) In a Myc-Trap assay, mCherry-Nsp3(UBL1+C) or mCherry-Nsp3(HVR+C) is marginally precipitated by Myc-FXR1. Myc-FXR1 pulls down similar levels of mCherry-Nsp3(UBL1+HVR+C) as WT mCherry-Nsp3. (C and D) Cells coexpressing mCherry-Nsp3(UBL1+C) or mCherry-Nsp3(HVR+C) with Nsp4-GFP contain numerous tiny Nsp3/4+ puncta that do not colocalize with endogenous FXR1 (C). FXR1+ puncta colocalize well with mCherry-Nsp3(UBL1+HVR+C)/Nsp4-GFP double-positive dots (D). Bars: 5 μm; insets, 2 μm. (E) The UBL1 or HVR domain of Nsp3 enters into FXR1 droplets at a lower level than Nsp3N. A fragment containing both HVR and UBL1 domains strongly segregates into FXR1 phases. Bars: 5 μm. (F) The schematic shows full-length FXR1 and mutants with deletions of different domains. (G–M) Full-length FXR1 (G), GFP-FXR1(ΔAL1ΔAL2) (H), GFP-FXR1(KH1-C) (I), GFP-FXR1(KH2-C) (J), and GFP-FXR1 (ΔKH2) (L) all show similar colocalization with Nsp3/4+ puncta, while GFP-FXR1(IDR1-C) (K) or GFP-FXR1 (ΔNIR) (M) fails to be recruited to Nsp3/4+ foci. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (N) Full-length FXR1, GFP-FXR1(ΔAL1ΔAL2), GFP-FXR1(KH1-C), GFP-FXR1(KH2-C), GFP-FXR1(IDR1-C), GFP-FXR1 (ΔKH2), and GFP-FXR1(ΔNIR) express well in cells. Source data are available for this figure: SourceData FS4.
Both the UBL1 and HVR domains of Nsp3 are required for recruitment of FXRs, related toFigs. 5 and 6 . (A) The schematic shows the domain organization of the truncated forms of Nsp3. (B) In a Myc-Trap assay, mCherry-Nsp3(UBL1+C) or mCherry-Nsp3(HVR+C) is marginally precipitated by Myc-FXR1. Myc-FXR1 pulls down similar levels of mCherry-Nsp3(UBL1+HVR+C) as WT mCherry-Nsp3. (C and D) Cells coexpressing mCherry-Nsp3(UBL1+C) or mCherry-Nsp3(HVR+C) with Nsp4-GFP contain numerous tiny Nsp3/4+ puncta that do not colocalize with endogenous FXR1 (C). FXR1+ puncta colocalize well with mCherry-Nsp3(UBL1+HVR+C)/Nsp4-GFP double-positive dots (D). Bars: 5 μm; insets, 2 μm. (E) The UBL1 or HVR domain of Nsp3 enters into FXR1 droplets at a lower level than Nsp3N. A fragment containing both HVR and UBL1 domains strongly segregates into FXR1 phases. Bars: 5 μm. (F) The schematic shows full-length FXR1 and mutants with deletions of different domains. (G–M) Full-length FXR1 (G), GFP-FXR1(ΔAL1ΔAL2) (H), GFP-FXR1(KH1-C) (I), GFP-FXR1(KH2-C) (J), and GFP-FXR1 (ΔKH2) (L) all show similar colocalization with Nsp3/4+ puncta, while GFP-FXR1(IDR1-C) (K) or GFP-FXR1 (ΔNIR) (M) fails to be recruited to Nsp3/4+ foci. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (N) Full-length FXR1, GFP-FXR1(ΔAL1ΔAL2), GFP-FXR1(KH1-C), GFP-FXR1(KH2-C), GFP-FXR1(IDR1-C), GFP-FXR1 (ΔKH2), and GFP-FXR1(ΔNIR) express well in cells. Source data are available for this figure: SourceData FS4.
Deleting either the HVR or UBL1 domain slightly decreased the entry of Nsp3 into FXR1 condensates, while deleting both domains completely abolished the partitioning of Nsp3 into FXR1 droplets (Fig. 4 F). The UBL1 or HVR domain of Nsp3 alone weakly colocalized with FXR1 liquid droplets, while fragments containing both domains segregated into FXR1 phases even more efficiently than Nsp3N (Fig. S4 E). Therefore, the recruitment of Nsp3 into FXR1 droplets depends on the UBL1 and HVR domains.
The region between the KH2 and IDR domains in FXR1 is critical for interaction with Nsp3
FXR family proteins contain two tandem agenet-like (AL) domains and two KH domains followed by two IDR domains (Fig. 5 A). A series of FXR1 truncations was constructed to determine which region mediates recruitment of FXR1 by Nsp3 (Fig. S4 F). The results demonstrate that the fragment between the KH2 and IDR1 domains is required for Nsp3 binding (Fig. S4, G–N). This fragment was named as Nsp3-interacting region (NIR).
The region between the KH2 and IDR domains in FXR1 is essential for FXR1-Nsp3 interaction. (A) The schematic shows the domain organization of FXR1. The Nsp3-interacting region (NIR) is located between the KH2 and IDR1 domains. (B and C) Alphafold predictions show that Nsp3 (green) interacts with FXR1 (cyan) via an α-helix (aa V68–I77) in the UBL1 domain (B) and a β-strand (aa 142–146) in the HVR domain (C). Key residues are labeled in the images. (D and E) Cells cotransfected with mCherry-Nsp3(E70A/L71A) or mCherry-Nsp3(Δ142–146) and Nsp4 show a huge abundance of small puncta that fail to colocalize with endogenous FXR1, while WT mCherry-Nsp3 and Nsp4 form big dots and recruit FXR1 (D). Numbers of DMVs are shown as mean ± SD [mCherry-Nsp3, n = 33; mCherry-Nsp3(E70A/L71A), n = 37; mCherry-Nsp3(Δ142–146), n = 30] (E). ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (F) In a GFP-Trap assay, GFP-FXR1 immunoprecipitates dramatically reduced levels of mCherry-Nsp3(E70A/L71A) compared to WT mCherry-Nsp3. (G) In a GFP-Trap assay, GFP-FXR1(I355A/L358A/K359A) immunoprecipitates dramatically reduced levels of mCherry-Nsp3 compared to WT GFP-FXR1. (H) In a GFP-Trap assay, the interaction between Nsp3 and FXR1 is inhibited by expression of mCherry-Nsp3(Δ142–146) or FXR1(R315D/R317D). Source data are available for this figure: SourceData F5.
The region between the KH2 and IDR domains in FXR1 is essential for FXR1-Nsp3 interaction. (A) The schematic shows the domain organization of FXR1. The Nsp3-interacting region (NIR) is located between the KH2 and IDR1 domains. (B and C) Alphafold predictions show that Nsp3 (green) interacts with FXR1 (cyan) via an α-helix (aa V68–I77) in the UBL1 domain (B) and a β-strand (aa 142–146) in the HVR domain (C). Key residues are labeled in the images. (D and E) Cells cotransfected with mCherry-Nsp3(E70A/L71A) or mCherry-Nsp3(Δ142–146) and Nsp4 show a huge abundance of small puncta that fail to colocalize with endogenous FXR1, while WT mCherry-Nsp3 and Nsp4 form big dots and recruit FXR1 (D). Numbers of DMVs are shown as mean ± SD [mCherry-Nsp3, n = 33; mCherry-Nsp3(E70A/L71A), n = 37; mCherry-Nsp3(Δ142–146), n = 30] (E). ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (F) In a GFP-Trap assay, GFP-FXR1 immunoprecipitates dramatically reduced levels of mCherry-Nsp3(E70A/L71A) compared to WT mCherry-Nsp3. (G) In a GFP-Trap assay, GFP-FXR1(I355A/L358A/K359A) immunoprecipitates dramatically reduced levels of mCherry-Nsp3 compared to WT GFP-FXR1. (H) In a GFP-Trap assay, the interaction between Nsp3 and FXR1 is inhibited by expression of mCherry-Nsp3(Δ142–146) or FXR1(R315D/R317D). Source data are available for this figure: SourceData F5.
We then performed structural prediction with the UBL1 and HVR domains of Nsp3 and the NIR of FXR1 using Alphafold 2.0. The result indicated two potential interaction sites: an α-helix (aa V68–I77) in the UBL1 domain (Fig. 5 B) and a β-strand (aa 142–146) in the HVR domain, which is induced by a β-sheet of FXR1 (Fig. 5 C).
To test the role of these sites, we disrupted the predicted interaction interfaces by mutating Glu70 and Leu71 in the UBL1 domain into Ala or deleting aa 142–146 in the HVR domain. Cells cotransfected with mCherry-Nsp3(E70A/L71A) or mCherry-Nsp3(Δ142–146) and Nsp4 contained large numbers of small Nsp3/4+ puncta (Fig. 5, D and E), resembling the phenotype of FXRs-depleted cells or cells expressing Nsp3 without the UBL1 or HVR domain (Fig. 2, B and C; and Fig. 4, B and C). The interaction between Nsp3 and FXR1 was also disrupted by mutations of Nsp3 or FXR1 within these interfaces (Fig. 5, F–H). All the above data suggest that the FXR1-Nsp3 binding occurs through multivalent interactions between the UBL1 and HVR domains of Nsp3 with the NIR of FXR1.
FXRs facilitate recruitment of translation machinery
Given that FXRs play an important role in activating RNA translation by recruiting ribosomes (Siomi et al., 1996), we speculated that FXR condensates at the DMV sites may benefit the virus by concentrating translation machinery in the proximity of viral replication. To determine the ability of FXR1 droplets to assemble ribosomes and viral RNA in vitro, we mixed Cy5-labeled FXR1 protein with iFluor 488–labeled 80 ribosome subunits and Cy3-labeled 1-kb viral RNA. FXR1 granules were capable of enriching ribosomes and RNAs individually and simultaneously (Fig. 6 A; and Fig. S5, A and B). Furthermore, ribosomes and viral RNA were able to enter FXR1 droplets together with Nsp3N (Fig. 6 B and Fig. S5 C).
FXR1 facilitates recruitment of translation machinery. (A) Ribosomes (green) and viral RNA (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (B) Ribosomes (green) and Nsp3N (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (C) The schematic diagram shows the TRICK assay to assess translation of viral RNA. The viral RNA is engineered to contain PP7 and MS2 sequences for two RNA-binding reporter proteins: GFP-NLS-PCP (PP7 coat protein) in the translated region and RFP-NLS-MCP (MS2 coat protein) in the 3′ untranslated region. If the viral RNA is actively translated, ribosomes will displace GFP-NLS-PCP but not RFP-NLS-MCP (top). If translation is inhibited (for example, by puromycin), both reporter proteins will be present on the RNA (bottom). (D) RFP signal, but not GFP, is detected at foci positive for FXR1 and Nsp3/4. Stalled translation, induced by puromycin treatment, causes both GFP and RFP signals to localize in FXR1 condensates. GFP-NLS-PCP, PCP fused to an NLS and GFP; RFP-NLS-MCP, MCP fused to an NLS and RFP. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (E) In siFXRs cells expressing RNAi-resistant BFP-FXR1, Nsp3/4 forms big puncta positive for FXR1 and RFP-NLS-PCP, but not GFP-NLS-PCP. Small dispersed Nsp3/4+ puncta are negative for FXR1, RFP-NLS-MCP, or GFP-NLS-PCP RFP in cells expressing RNAi-resistant BFP-FXR1(I355A/L358A/K359A) or BFP-FXR1(R315D/R317D). Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm.
FXR1 facilitates recruitment of translation machinery. (A) Ribosomes (green) and viral RNA (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (B) Ribosomes (green) and Nsp3N (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (C) The schematic diagram shows the TRICK assay to assess translation of viral RNA. The viral RNA is engineered to contain PP7 and MS2 sequences for two RNA-binding reporter proteins: GFP-NLS-PCP (PP7 coat protein) in the translated region and RFP-NLS-MCP (MS2 coat protein) in the 3′ untranslated region. If the viral RNA is actively translated, ribosomes will displace GFP-NLS-PCP but not RFP-NLS-MCP (top). If translation is inhibited (for example, by puromycin), both reporter proteins will be present on the RNA (bottom). (D) RFP signal, but not GFP, is detected at foci positive for FXR1 and Nsp3/4. Stalled translation, induced by puromycin treatment, causes both GFP and RFP signals to localize in FXR1 condensates. GFP-NLS-PCP, PCP fused to an NLS and GFP; RFP-NLS-MCP, MCP fused to an NLS and RFP. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (E) In siFXRs cells expressing RNAi-resistant BFP-FXR1, Nsp3/4 forms big puncta positive for FXR1 and RFP-NLS-PCP, but not GFP-NLS-PCP. Small dispersed Nsp3/4+ puncta are negative for FXR1, RFP-NLS-MCP, or GFP-NLS-PCP RFP in cells expressing RNAi-resistant BFP-FXR1(I355A/L358A/K359A) or BFP-FXR1(R315D/R317D). Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm.
FXR1 droplets recruit translation factors and are important for viral proliferation, related toFigs. 6 and 7 . (A) Ribosomes (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (B) Viral RNA (magenta) partitions into FXR1 liquid droplets (cyan). Bar: 5 μm. (C) Nsp3N (green) and viral RNA (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (D) Immunostaining with anti-RPS6 shows that ribosome component RPS6 is detected at FXR1 and dsRNA double-positive punctate structures 24 h after SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (E) Control cells (NC) and siFXRs cells were infected with SARS-CoV-2-Spike–packaged pseudoviruses expressing luciferase. The pseudovirus entry efficiency is presented as luciferase activity normalized relative to control levels. N.S., not significant. (F) Compared with FXRs KD cells, reintroducing RNAi-resistant FXR1 significantly increases NP levels after SARS-CoV-2 infection with MOI = 0.5 for 24 h. Quantifications of levels of NP (normalized by Actin levels) are shown. (G–J) Compared with control cells (NC), viral N (G and I) and Spike (H and J) RNA levels are significantly decreased in FXRs KD cells after SARS-CoV-2 infection with MOI = 0.1 (G and H) or MOI = 1 (I and J). Quantitative data are presented as mean ± SD (n = 3). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (K)FXR1, FXR2, and FMR1 mRNA levels are efficiently depleted by siFXRs. Quantitative data are presented as mean ± SD (n = 3). (L–N) Transcription levels of FXR1 (L), FXR2 (M), and FMR1 (N) are dramatically increased upon SARS-CoV-2 infection with MOI = 1. (O) Amino acid alignment of the UBL1 and HVR domains of Nsp3 from SARS-CoV-2, SARS-CoV, MERS, hCoV-229E, and hCoV-OC43. The alignment was generated using SnapGene. Identical residues are indicated with green boxes. The black lines above the SARS-CoV-2 Nsp3 sequence show the two FXR1 interaction regions. (P) GFP-FXR1 colocalizes well with Nsp3/4+ puncta in cells coexpressing mCherry-U+H(SARS)+3C with Flag-Nsp4 (cyan). Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (Q) In an mCherry-Trap assay, levels of endogenous FXR1 immunoprecipitated by mCherry-U+H(SARS)+3C are comparable with those precipitated by mCherry-Nsp3. (R) Myc-FXR1 (magenta) puncta colocalize well with GFP-nsP3(VEEV) foci. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (S and T) Formation of GFP-nsP3(VEEV) foci is comparable in NC and siFXRs-treated cells (S). Images are representative of at least 10 cells. Quantitative data are presented as mean ± SD (NC, n = 32; siFXRs, n = 43) (T). Bars: 5 μm. Source data are available for this figure: SourceData FS5.
FXR1 droplets recruit translation factors and are important for viral proliferation, related toFigs. 6 and 7 . (A) Ribosomes (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (B) Viral RNA (magenta) partitions into FXR1 liquid droplets (cyan). Bar: 5 μm. (C) Nsp3N (green) and viral RNA (magenta) partition into FXR1 liquid droplets (cyan). Bar: 5 μm. (D) Immunostaining with anti-RPS6 shows that ribosome component RPS6 is detected at FXR1 and dsRNA double-positive punctate structures 24 h after SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (E) Control cells (NC) and siFXRs cells were infected with SARS-CoV-2-Spike–packaged pseudoviruses expressing luciferase. The pseudovirus entry efficiency is presented as luciferase activity normalized relative to control levels. N.S., not significant. (F) Compared with FXRs KD cells, reintroducing RNAi-resistant FXR1 significantly increases NP levels after SARS-CoV-2 infection with MOI = 0.5 for 24 h. Quantifications of levels of NP (normalized by Actin levels) are shown. (G–J) Compared with control cells (NC), viral N (G and I) and Spike (H and J) RNA levels are significantly decreased in FXRs KD cells after SARS-CoV-2 infection with MOI = 0.1 (G and H) or MOI = 1 (I and J). Quantitative data are presented as mean ± SD (n = 3). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (K)FXR1, FXR2, and FMR1 mRNA levels are efficiently depleted by siFXRs. Quantitative data are presented as mean ± SD (n = 3). (L–N) Transcription levels of FXR1 (L), FXR2 (M), and FMR1 (N) are dramatically increased upon SARS-CoV-2 infection with MOI = 1. (O) Amino acid alignment of the UBL1 and HVR domains of Nsp3 from SARS-CoV-2, SARS-CoV, MERS, hCoV-229E, and hCoV-OC43. The alignment was generated using SnapGene. Identical residues are indicated with green boxes. The black lines above the SARS-CoV-2 Nsp3 sequence show the two FXR1 interaction regions. (P) GFP-FXR1 colocalizes well with Nsp3/4+ puncta in cells coexpressing mCherry-U+H(SARS)+3C with Flag-Nsp4 (cyan). Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (Q) In an mCherry-Trap assay, levels of endogenous FXR1 immunoprecipitated by mCherry-U+H(SARS)+3C are comparable with those precipitated by mCherry-Nsp3. (R) Myc-FXR1 (magenta) puncta colocalize well with GFP-nsP3(VEEV) foci. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (S and T) Formation of GFP-nsP3(VEEV) foci is comparable in NC and siFXRs-treated cells (S). Images are representative of at least 10 cells. Quantitative data are presented as mean ± SD (NC, n = 32; siFXRs, n = 43) (T). Bars: 5 μm. Source data are available for this figure: SourceData FS5.
We then utilized a TRICK (translating RNA imaging by coat protein knock-off) reporter system as previously described (Halstead et al., 2015) to examine if FXR1 condensates promote translation at DMV sites (Fig. 6 C). For this assay, we inserted two bacteriophage PP7 and MS2 sequences into the translated region and the 3′ untranslated region, respectively, of a 1-kb viral RNA. The PP7 and MS2 sequences could be detected by the RNA-binding proteins PP7 coat protein (PCP) and MS2 coat protein (MCP), respectively. We then cotransfected cells with viral RNA, GFP-NLS-PCP (PCP fused to a nuclear localization sequence [NLS] and GFP), RFP-NLS-MCP (MCP fused to an NLS and RFP), Flag-Nsp3/4, and BFP-FXR1. If the viral RNA is actively translated, ribosomes will displace GFP-NLS-PCP but not RFP-NLS-MCP. Indeed, only RFP signal was detected at Nsp3/4/FXR1+ foci, indicating translation of viral mRNA at DMV sites recruited by FXR1 granules. When translation was stalled by puromycin treatment, both GFP and RFP signals were detected at FXR1 condensates (Fig. 6 D). Moreover, in siFXRs cells expressing FXR1 mutants that disrupt FXR1-Nsp3 binding (FXR1[I355A/L358A/K359A] or FXR1[R315D/R317D]), the small dispersed Nsp3/4+ puncta were negative for FXR1, RFP-NLS-MCP, or GFP-NLS-PCP (Fig. 6 E). This suggests that FXRs may not only activate translation but also concentrate RNA at the DMV sites. Thus, FXRs may act as a bridge between DMVs and translation machineries to promote local translation activity in the close vicinity of viral ROs.
FXRs promote SARS-CoV-2 replication
During SARS-CoV-2 infection, signals from the ribosome components RPS6 and RPL7a and the translation initiation factor EIF4G3 were detected at the FXR1-positive punctate structures (Fig. 7, A and B; and Fig. S5 D). To further verify active local translation at FXR1 condensates, we exploited the SUnSET (surface sensing of translation) method to allow detection of newly synthesized polypeptides by a puromycin-specific antibody based on incorporation of puromycin into nascent chains (Schmidt et al., 2009). After pulse labeling with puromycin for 30 min, strong puromycin signals were evident at FXR1 puncta upon viral infection (Fig. 7 C), which suggests that FXRs may facilitate translation at the viral replication sites.
FXRs promote viral replication. (A and B) Signals from the ribosome component RPL7a (A) and translation initiation factor EIF4G3 (B) are detected at FXR1 punctate structures after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (C) After 24 h SARS-CoV-2 infection with MOI=1, ACE2-HeLa cells were treated with 10 μg/ml puromycin for 30 min. Puromycin signals colocalize with FXR1 foci. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (D and E) Levels of viral subgenomic E RNA are significantly decreased in cells depleted of FXRs compared with control cells after SARS-CoV-2 infection with MOI = 0.1 (D) or MOI = 1 (E). Quantification data are presented as mean ± SD (n = 3). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (F) Puromycin incorporation assays with ACE2-HeLa cells show that signals from anti-puromycin antibody are dramatically suppressed upon SARS-CoV-2 infection for 12 h. After 24 h infection, slightly recovered anti-puromycin signals are detected in NC-treated but not siFXRs cells. Immunoblotting shows that viral NP levels are suppressed by siFXRs after 12 and 24 h SARS-CoV-2 infection with MOI = 0.1 or 0.5. (G) TEM images show clustered DMVs in control cells (NC), while DMVs (arrow) are only occasionally observed in siFXRs cells. Bars, 500 nm. (H) Immunoblotting shows that FXR1, FXR2, and FMR1 levels are dramatically increased after 8 h SARS-CoV-2 infection with MOI = 1. (I) The schematic model shows that LLPS of FXRs drives clustering of DMVs via interaction with the UBL1 and HVR domains of Nsp3, which further facilitates recruitment of translation machinery for viral replication. Source data are available for this figure: SourceData F7.
FXRs promote viral replication. (A and B) Signals from the ribosome component RPL7a (A) and translation initiation factor EIF4G3 (B) are detected at FXR1 punctate structures after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (C) After 24 h SARS-CoV-2 infection with MOI=1, ACE2-HeLa cells were treated with 10 μg/ml puromycin for 30 min. Puromycin signals colocalize with FXR1 foci. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (D and E) Levels of viral subgenomic E RNA are significantly decreased in cells depleted of FXRs compared with control cells after SARS-CoV-2 infection with MOI = 0.1 (D) or MOI = 1 (E). Quantification data are presented as mean ± SD (n = 3). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (F) Puromycin incorporation assays with ACE2-HeLa cells show that signals from anti-puromycin antibody are dramatically suppressed upon SARS-CoV-2 infection for 12 h. After 24 h infection, slightly recovered anti-puromycin signals are detected in NC-treated but not siFXRs cells. Immunoblotting shows that viral NP levels are suppressed by siFXRs after 12 and 24 h SARS-CoV-2 infection with MOI = 0.1 or 0.5. (G) TEM images show clustered DMVs in control cells (NC), while DMVs (arrow) are only occasionally observed in siFXRs cells. Bars, 500 nm. (H) Immunoblotting shows that FXR1, FXR2, and FMR1 levels are dramatically increased after 8 h SARS-CoV-2 infection with MOI = 1. (I) The schematic model shows that LLPS of FXRs drives clustering of DMVs via interaction with the UBL1 and HVR domains of Nsp3, which further facilitates recruitment of translation machinery for viral replication. Source data are available for this figure: SourceData F7.
Given the above findings, we further investigated the role of FXRs in the viral life cycle. A pseudovirus entry assay demonstrated that depletion of FXRs does not affect virus entry (Fig. S5 E). However, after infection with SARS-CoV-2, both Nucleocapsid (N) and Spike gene levels were significantly decreased after depleting FXRs in ACE2-HeLa compared with controls (Fig. S5, G–K). Furthermore, transcription of subgenomic E RNA, which is a marker for monitoring active viral replication (Dagotto et al., 2021; Kim et al., 2020), was more dramatically reduced in siFXRs cells at all time points compared with NC cells (Fig. 7, D and E). Accordingly, the levels of N protein (NP) were also obviously lower than in control cells (Fig. 7 F), which could be rescued by reintroducing FXR1 (Fig. S5 F). To directly examine the function of FXRs in translation during viral infection, we utilized the puromycin incorporation assay, which labels newly synthesized polypeptide chains. The results showed that 12 h after virus infection, translation was almost completely blocked, as indicated by immunoblotting with anti-puromycin antibody. This is probably due to the inhibitory effect of viral protein Nsp1 on host ribosome function (Jaafar and Kieft, 2019). 24 h after virus infection, translation was detected in control cells but not in siFXRs cells (Fig. 7 F), which suggests that FXRs are crucial for viral translation. TEM analysis was performed to investigate the DMV morphology in virus-infected cells. Although clustered DMVs were formed in control cells, very few DMVs were observed in siFXRs cells (Fig. 7 G), which probably results from greatly suppressed viral protein synthesis in cells depleted of FXRs. Interestingly, both transcription and translation of FXRs were greatly increased upon virus infection (Fig. 7 H and Fig. S5, L–N). These results suggest that FXRs are recruited to DMV sites during viral infection and facilitate SARS-CoV-2 replication possibly via concentration of the translation machinery.
In a recent study comparing the interactomes of different β-coronavirus strains, FXRs were identified as Nsp3-interacting proteins specifically in SARS-CoV-2 and SARS-CoV (Almasy et al., 2021; Garvanska et al., 2024). The amino acid sequences of the FXR1-binding sites in Nsp3 identified in our study are conserved in SARS-CoV, but not in MERS, hCoV-229E, or hCoV-OC43 (Fig. S5 O). The SARS-CoV UBL1 and HVR domains at the N-terminus of Nsp3C are sufficient for recruiting FXR1 to Nsp3/4 puncta and for interacting with FXR1 (Fig. S5, P and Q). DMV clustering is generally observed in cells infected by β-coronaviruses (Orenstein et al., 2008; Oudshoorn et al., 2017; Snijder et al., 2020; Wolff et al., 2020), which indicates that distinct β-coronaviruses may employ different host proteins to organize ROs. FXRs were previously identified as host factors important for the replication of New World alphaviruses, and FXRs colocalize with the foci formed by VEEV nsP3 (Fig. S5 R) (Kim et al., 2016). However, KD of FXRs does not cause dispersion of these puncta (Fig. S5, S and T). Therefore, during alphavirus infection, FXRs may play different roles or may only act in recruiting translation complexes.
Discussion
Compartments formed by membrane-bound and membraneless organelles ensure that specific biochemical processes occur in a spatially restricted site separated from the complex intracellular environment. Viruses also compartmentalize host cells upon infection. The efficient proliferation of β-coronaviruses depends on the formation of viral ROs, mainly DMVs, via remodeling of the host ER network, which provides a safe environment for viral RNA synthesis (Cortese et al., 2020; Knoops et al., 2008; Mohan and Wollert, 2021; Snijder et al., 2020). Moreover, in virus-infected cells, DMVs always exist as grape-like clusters with no restraining boundary (Cortese et al., 2020; Ji et al., 2022; Ricciardi et al., 2022; Snijder et al., 2020), which indicates that there is another layer of organization with unknown mechanism and function. Emerging evidence shows that liquid droplets formed by LLPS concentrate proteins and RNAs related to a specific function into compartments devoid of delimiting membranes. Biocondensates also modulate the functions of membrane-bound organelles, such as storage, transportation, and release.
Here, our study uncovers that DMV clustering is driven by phase separation of host FXR proteins both in vivo and in vitro (Fig. 7 I). FXRs are recruited by the DMV outer membrane Nsp3 through its N-terminal UBL1 and HVR domains. This is consistent with a recently published study which reported that a 20-aa stretch in the Nsp3 HVR domain is required for binding to FXRs (Garvanska et al., 2024). In the absence of FXRs, DMVs appear as numerous individual vesicles dispersed throughout the cytoplasm. Re-expression of the L351P mutant FXR1, which fails to undergo LLPS, did not rescue the scattered DMVs in FXRs-depleted cells, which confirms that the phase-separation capability of FXRs is the main driver for capturing DMVs. LLPS is facilitated by increasing the concentration of constituents (Wang and Zhang, 2019; Wu et al., 2020; Zhao and Zhang, 2020). The transcription and translation of FXRs are dramatically increased upon virus infection. Interestingly, a previous study showed that SARS-CoV-2 Nsp1 blocked the ribosomal accessibility of host mRNA, except for transcripts from genes with 5′ terminal oligopyrimidine tracts, including FXR1 (Rao et al., 2021). This further supports the importance of FXRs during virus proliferation. Nsp3 was found to suppress the incorporation of FXRs into SGs by blocking their binding with SG protein UBSP2L (Garvanska et al., 2024). Although the authors could not confirm the effect of Nsp3-FXRs binding on SG function, this may explain how FXRs concentrate at DMV sites during viral infection.
As viruses lack biosynthetic capability, they have developed elaborate strategies to utilize host translational apparatus to ensure efficient viral protein synthesis (Eriani and Martin, 2022; Jaafar and Kieft, 2019). Here, we describe a novel mechanism whereby SARS-CoV-2 utilizes host FXRs to concentrate translation machinery at the sites of viral RNA replication. FXRs have been shown to drive active translation of C-Myc and spermiogenesis-related genes by recruiting translation initiation factors and ribosomal complexes in cancer cells and late spermatids, respectively (George et al., 2021; Kang et al., 2022). We found that FXR1 droplets are capable of capturing ribosomes, viral RNA, and Nsp3 in vitro and triggering active local translation of viral RNA at DMV sites in vivo. During viral infection, translation initiation and ribosomal proteins, as well as active translation, are detected at the FXR1 condensates in the close vicinity of DMVs. Moreover, cryo-electron tomography reveals abundant ribosomes surrounding DMVs (Wolff et al., 2020). Accordingly, depletion of FXRs significantly suppresses viral RNA replication and protein synthesis during SARS-CoV-2 infection. Compartmentalization of translation around the replication sites would provide several benefits for viruses. First, it concentrates translation of viral RNA in close proximity to the ROs where RNAs are synthesized and thereby ensures efficient gene expression via physically linking the two processes. Second, the overlap of viral RNA and protein production sites, which are largely associated with the ER, could be convenient for their subsequent delivery to the nearby virus assembly site, the ER–Golgi intermediate compartment (V’kovski et al., 2021). Third, immediate translation at the replication sites may be a protective mechanism that shields viral RNA from being recognized by host factors to trigger mRNA decay mechanisms or innate immune responses.
Taking all the data together, our mechanistic study reveals a novel model in which LLPS of FXRs mediates DMV compartmentalization for assembly of viral replication centers and activation of viral RNA translation. This advance uncovers a potential therapeutic target for treating infectious diseases caused by SARS-CoV-2 or other viruses using similar mechanisms.
Materials and methods
Plasmids
GFP-Nsp3, mCherry-Nsp3, Nsp4-GFP, and Flag-Nsp4 were generated as previously described (Ji et al., 2022). GFP-Nsp3 was constructed by inserting SARS-CoV-2 Nsp3 into the pEGFP-C1 vector. mCherry-Nsp3 was generated by replacing the GFP fragment of GFP-Nsp3 with mCherry. Nsp4-GFP was constructed by inserting SARS-CoV-2 Nsp4 into pEGFP-N1 vector. Nsp4-mCherry was generated by replacing the GFP fragment of Nsp4-GFP with mCherry. mCherry-Nsp3(ΔUBL1, Δaa 1–111), mCherry-Nsp3(ΔHVR, Δaa 112–198), mCherry-Nsp3(ΔUBL1ΔHVR, Δaa 1–198), mCherry-Nsp3(ΔMac1, Δaa 199–387), mCherry-Nsp3(ΔMac2, Δaa 413–548), mCherry-Nsp3(ΔMac3, Δaa 549–676), mCherry-Nsp3(ΔDPUP, Δaa 677–743), mCherry-Nsp3(ΔUBL2, Δaa 746–805), mCherry-Nsp3(ΔPL2, Δaa 806–1059), mCherry-Nsp3(ΔNAB [nucleic-acid binding], Δaa 1089–1203), mCherry-Nsp3(ΔβSM [β-coronavirus-specific marker], Δaa 1226–1341), mCherry-Nsp3(E70A/L71A), and mCherry-Nsp3(Δ142–146) were generated by PCR-based mutagenesis from mCherry-Nsp3. mCherry-Nsp3C was generated by inserting the C-terminal fragment of Nsp3 (aa 1356–1946) into mCherry-C1. mCherry-Nsp3(UBL1+HVR+C), mCherry-Nsp3(UBL1+C), and mCherry-Nsp3(HVR+C) were generated by inserting the corresponding domain between mCherry and Nsp3C of mCherry-Nsp3C. Nsp4-BFP was generated by replacing GFP with BFP. GFP-FXR1 and GFP-FMR1 were constructed by inserting human FXR1 and FMR1, respectively, into the pEGFP-C1 vector. FXR2-GFP was constructed by inserting human FXR2 into the pEGFP-N1 vector. GFP-FXR1(L351P), GFP-FXR1(Δ314–318), GFP-FXR1(ΔAL1ΔAL2, Δaa 1–156), GFP-FXR1(KH1-C, aa 217–621), GFP-FXR1(KH2-C, aa 280–621), GFP-FXR1(IDR1-C, aa 375–621) GFP-FXR1(ΔKH2, Δaa 285–314), GFP-FXR1 (I355A/L358A/K359A), and GFP-FXR1(R315D/R317D) were generated by PCR-based mutagenesis from GFP-FXR1. BFP-FXR1 was generated by replacing GFP with BFP. RNAi-resistant GFP-FXR1 and GFP-FXR1(L351P) were generated by PCR-based mutagenesis from GFP-FXR1 and GFP-FXR1(L351P), respectively, using the primer (5′-GCAGCGTGCGATGCGACG-3′). FRB-mCherry-Nsp3 was generated by inserting FKBP–rapamycin binding (FRB) into the carboxyl-terminus of Nsp3. RNAi-resistant FKBP-GFP-FXR and FKBP-GFP-FXR(L351P) were generated by inserting FKBP at the carboxyl-terminus of RNAi-resistant GFP-FXR and GFP-FXR(L351P), respectively (Choi et al., 1996). GFP-NLS-PCP, RFP-NLS-MCP, and the Gpd2 TRICK reporter were kind gifts from Dr. Mofang Liu (Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, Shanghai, China) (Kang et al., 2022). The viral RNA TRICK reporter was generated by replacing the Gpd2 sequence with the Nsp3 sequence (nt 5505–6518 of SARS-CoV-2 ORF1ab, accession no. BS007403). mCherry-U+H(SARS)-3C was generated by inserting the UBL1 and HVR domains (aa 1–178) between mCherry and Nsp3C of mCherry-Nsp3C. APEX2-GBP was generated by inserting APEX2 and GBP into pEGFP-C1 with the GFP sequence deleted. GFP-Nsp6 was generated by inserting code-optimized Nsp6 into pEGFP-C1 vector. Nsp3N (aa 1–1341), Nsp3N(ΔUBL1, Δaa 1–111), Nsp3N(ΔHVR, Δaa 112–198), Nsp3(UBL1, aa 1–111), Nsp3(HVR, aa 112–198), and Nsp3(UBL1+HVR, aa 1–198) were generated by inserting the corresponding fragments of SARS-CoV-2 Nsp3 into a modified pET-32a vector with an N-terminal Trx-His8 tag and an HRV-3C protease cutting site. MBP-FXR1 was constructed by inserting FXR1 into a modified pET-32a vector with an N-terminal MBP-His6 tag and an HRV-3C protease cutting site.
Antibodies
The following primary antibodies were used in this study: rabbit anti-FXR1 (ab129089; Abcam), goat anti-FXR1 (ab51970; Abcam), rabbit anti-FXR2 (7098; Cell Signaling Technology), rabbit anti-FMR1 (ab17722; Abcam), mouse anti-dsRNA (10010200; SCICONS), rabbit anti-G3BP1 (13057-2-AP; Proteintech), rabbit anti-SARS-CoV-2 Nsp3 (GTX135589; Genetex), rabbit anti-SARS-CoV-2 N protein (40143-T62; SinoBiological), rabbit anti-EIF4G3 (NBP2-16309; NOVUS), rabbit anti-RPL7a (15340-1-AP; Proteintech), rabbit anti-RPS6 (ab40820; Abcam), rabbit anti-Myc (HX1821; Huaxingbio), ABfluo 488 rabbit anti-puromycin (A23130; Abclonal), mouse anti-GFP (11814460001; Roche), mouse anti-Flag (F1804; Sigma-Aldrich), rabbit anti-mCherry (GTX59788; Genetex), and mouse anti-Actin (60008-1-Ig; Proteintech).
The following secondary antibodies were used: rabbit anti-goat IgG-HRP (BE0103; EASYBIO), goat anti-mouse-HRP (BE0102; EASYBIO), goat anti-rabbit IgG-HRP (BE0101; EASYBIO), goat anti-mouse-Rhodamine (TRITC) (115-025-003; Jackson), goat anti-rabbit- TRITC (111-025-003; Jackson), goat anti-rabbit-Fluorescein (FITC) (111-095-003; Jackson), goat anti-mouse-FITC (115-095-003; Jackson), rabbit anti-goat-hyperFluor 488 (K1214; Apexbio), goat anti-rabbit-Alexa Fluor 647 (A-21245; Invitrogen), and goat anti-mouse-Alexa Fluor 647 (A-21236; Invitrogen).
Cell lines
HeLa cells used in this study were obtained from ATCC. HeLa cells stably expressing human ACE2 (ACE2-HeLa) were generated previously (Xu et al., 2022). Cells were cultured in DMEM (C11965500BT; Thermo Fisher Scientific), supplemented with 10% FBS (10099-141C; Thermo Fisher Scientific) and penicillin-streptomycin at 37°C with 5% CO2. For arsenite treatment, cells were incubated with 500 μM arsenite (S7400; Sigma-Aldrich) for 1 h. To inhibit ribosomal translation, cells were treated with 10 μg/ml puromycin (ST551; Beyotime) for 30 min. To induce FRB-FKBP interaction, cells were incubated with 1.5 μM rapamycin (V900930; Sigma-Aldrich) for 30 min.
SARS-CoV-2 virus amplification and infection
All experiments involving the infection of cells with SARS-CoV-2 were conducted within the biosafety level-3 laboratory located at Shenzhen Third People’s Hospital following standard operating procedures. The SARS-CoV-2 WT strain was isolated from nasopharyngeal aspirate specimens sourced from COVID-19 patients at Shenzhen Third People’s Hospital. The virus was expanded in Vero-E6 cells and maintained at −80°C. For the infection of ACE2-HeLa cells (MOI as indicated in the figure legends), the virus was added directly to the cells and incubated for the indicated time (specified in the figure legends) at 37°C with 5% CO2 for viral adsorption. Following this, the virus-containing medium was removed and the cells were incubated in 2% FBS medium at 37°C with 5% CO2 before being harvested for analysis at the indicated time points.
Generation of SARS-CoV-2 pseudoviruses
The Spike gene of WT SARS-CoV-2 was synthesized by GenScript and then inserted into the pVAX1 vector. SARS-CoV-2 pseudovirus was generated by co-transfection of the Spike plasmid and env-deficient HIV-1 backbone vector (pNL4-3.Luc.R-E-) into HEK-293T cells. All culture supernatants were collected 2 days later and centrifuged to remove the precipitation. The infectious titer was determined by measuring the luciferase activity in the ACE2-HeLa cells using Bio-Lite Luciferase Assay System (DD1201; Vazyme).
Quantitative RT-PCR
RNA was isolated using TRIzol (15596018; Thermo Fisher Scientific), then cDNA was generated by reverse transcription using a SuperRT cDNA Synthesis Kit (CW0741M; CWBIO). Quantitative RT-PCR was carried out using 2*TSINGKE Master qPCR Mix (TSE201; TSINGKE) on an Applied Biosystems 7500 Real-Time PCR System. The primers used in the experiment were as follows:
F-FXR1, 5′-AGGTCTTATAGCGGAAGAGGC-3′;
R-FXR1, 5′-AACTGATGGAGGATTTGCCACC-3′;
F-FXR2, 5′-TAGAGCAGCTTCGCTTGGAG-3′;
R-FXR2, 5′-TCCTCTCTCTTCTCTGACTCAGTC-3′;
F-FMR1, 5′-AGCTTGCCTCGAGATTTCATG-3′;
R-FMR1, 5′-TCCTTGGAACTTGTATTACATCTTCAGC-3′;
F-Subgenomic E, 5′-CGATCTCTTGTAGATCTGTTCTC-3′;
R-Subgenomic E, 5′-ATATTGCAGCAGTACGCACACA-3′;
F-Spike, 5′-GTGACACTTGCAGATGCTGGCT-3′;
R-Spike, 5′-GCACCTGCACCAAAGGTCCAA-3′;
F-N, 5′-TCTACGCAGAAGGGAGCAGAGG-3′;
R-N, 5′- AAGAGCAGCATCACCGCCATTG-3′;
F-ACTB, 5′-GCACTCTTCCAGCCTTCCTTCC-3′;
R-ACTB, 5′- AGACAGCACTGTGTTGGCGTAC-3′.
Transfection and RNAi
To perform transient transfection, cells were transfected with the desired plasmids using Lipofectamine 2000 (12566014; Thermo Fisher Scientific). For RNA interference experiments, cells were transfected with either NC or specific siRNA oligos obtained from GenePharma using Lipofectamine RNAiMAX (13778150; Thermo Fisher Scientific). The cells were collected 72 h after transfection for further analysis. The siRNA sequences used in the experiment were:
NC, 5′-UUCUCCGAACGUGUCACGUTT-3′;
Human FXR1, 5′-GCUGCUUGUGACGCUACUUTT-3′;
Human FXR2, 5′-GCAGAGUGAGAGACAAAUUTT-3′;
Human FMR1, 5′-GCUAGAAGCUUUCUCGAAUTT-3′.
Immunostaining
Cells cultured on coverslips (C015001; Matsunami) were fixed with 4% paraformaldehyde (PFA) for 30 min and then permeabilized using 0.1% Triton for 20 min at room temperature. Subsequently, the cells were blocked with 1% BSA for 1 h and incubated overnight at 4°C with the specified primary antibodies, which were diluted in 1% BSA. After washing thrice with PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), cells were exposed to fluorescently tagged secondary antibodies for 1 h at room temperature. Finally, DAPI in 50% glycerol was used to mount the coverslips, and the cells were imaged using a confocal microscope (LSM 980 Meta plus Zeiss Axiovert zoom, Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromat, Zeiss) and a point detector (GaAsP PMT, Zeiss) using Zeiss ZEN software (blue edition) at room temperature. The fluorochromes utilized in the imaging were FITC, TRITC, Alexa Fluor 647, DAPI, EGFP, RFP, and mCherry.
Co-IP and immunoblotting
To perform co-IP analysis, cells were transfected with the appropriate plasmids for 24–48 h and subsequently harvested with lysis buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100 supplemented with a protease inhibitor cocktail (11836170001; Roche). The lysates were incubated on ice for 30 min and then centrifuged at 13,000 rpm for 10 min at 4°C. The resulting supernatants were collected and incubated with GFP-Trap (KTSM1334; Shenzhen KT Life Technology), mCherry-Trap (KTSM1334; Shenzhen KT Life Technology), or Myc-Trap (KTSM1334; Shenzhen KT Life Technology) agarose beads at 4°C for 1.5 h. The proteins that bound to the beads were eluted with SDS sample buffer and subsequently analyzed via immunoblotting.
For immunoblotting, cells were lysed with lysis buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, and a protease inhibitor cocktail, followed by a 30-min incubation on ice. The homogenates were then centrifuged at 13,000 rpm for 10 min at 4°C, and the supernatants were mixed with SDS sample buffer and subjected to SDS-PAGE electrophoresis. Hydrophobic polyvinylidene fluoride transfer membranes with 0.45 µm pore size (IPVH00010; Millipore) were used. The protein signals were detected using specific primary and secondary antibodies, and the resulting bands were visualized on an imaging system (5200SF; Tanon). The images were subsequently analyzed using ImageJ software.
MS analysis
Protein bands on the SDS-PAGE gel were shrunk with destaining solution (50 mM ammonium bicarbonate/acetonitrile [1:1, vol/vol]) at room temperature for 15 min. The shrunk gel fragments were swollen with 10 mM DTT solution (dissolved in 50 mM ammonium bicarbonate) at 56°C for 15 min and cooled to room temperature. After a brief centrifugation, the liquid was discarded and 100 mM iodoacetamide solution (dissolved in 50 mM ammonium bicarbonate) was added to the gel pieces. The mixture was allowed to stand for 15 min in a dark place, and then the gel pieces were shrunk with destaining solution again and dried in a speed vac. The destained gel fragments were digested with sequencing-grade trypsin (10 ng/μl trypsin, 50 mM ammonium bicarbonate) overnight at 37°C. Peptides were extracted with 25 mM ammonium bicarbonate and 5% formic acid/50% acetonitrile sequentially and then the collected samples were dried in a speed vac. Before analysis, the samples were dissolved in 0.5% formic acid and desalinated in 50% acetonitrile and 100% acetonitrile in sequence with ZipTip micro chromatographic columns (ZTC18S096; Merck). After another speed vac drying step, the final samples were dissolved in 1% formic acid.
The extracted peptides were separated on an analytical capillary column (100 μm inner diameter × 20 cm) packed with 1.9 μm and 120 Å ReproSil-Pur C18 resins (Dr. Maisch GmbH). An EASY-nLC 1000 system (Thermo Fisher Scientific) was used to generate the following HPLC gradient: 3–7% B for 2 min, 7–22% B for 50 min, and 22–35% B for 10 min (A = 0.1% formic acid in water, B = 0.1% formic acid in acetonitrile). The eluted peptides were sprayed into an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with a nano-ESI ion source. The mass spectrometer was operated in data-dependent mode in top speed mode with higher-energy collision dissociation (target 5e4 ions, max ion injection time 40 ms, isolation window 1.6 m/z, normalized collision energy 30%). The dynamic exclusion time was set to 30 s. Database searches were performed on MaxQuant (version 1.5.8.3) against the human Uniprot fasta database with default parameters. Two missed cleavage sites were allowed for trypsin digestion. Cysteine carbamidomethylation was set as a fixed modification, while methionine oxidation, and asparagine and glutamine deamidation were set as variable modifications. False discovery rate was set to 1% for protein and peptide spectrum matches.
TEM and APEX2 labeling
Cells were harvested and fixed in 2.5% glutaraldehyde in PBS at 4°C overnight. Following two washes with PBS and one wash with distilled water, cells were post-fixed with 1% OsO4 and 1.5% K3Fe(CN)6 for 90 min at room temperature. Cells were then washed with water and incubated in chilled 2% aqueous uranyl acetate for 1 h at room temperature. After washing with distilled water, the cells were gradually dehydrated with increasing concentrations of ethanol and embedded in epoxy EMBED-812 resin. Images were obtained using a 120-kV electron microscope (Tecnai Spirit, FEI) and a CCD camera (MoradaG3; EMSIS) with RADIUS software at room temperature.
For DAB staining, cells were transfected with GFP-APEX2, mCherry-Nsp3, Nsp4-BFP, APEX2-GBP. Cells were fixed with 2.5% glutaraldehyde in PBS overnight at 4°C and rinsed 3 × 10 min in PBS. Then cells were incubated with a freshly diluted solution of 0.5 mg/ml (1.4 mM) DAB tetrahydrochloride for 8–15 min. After rinsing three times with chilled buffer, cells were post-fixed in 1% OsO4 for 8 min on ice. Cells were rinsed 3 × 10 min in chilled distilled water, and then placed in chilled 2% aqueous uranyl acetate for 30 min. Subsequent steps (washing, dehydration, embedding, and imaging) were the same as above.
Protein expression and purification
Proteins were expressed in Escherichia coli BL21 (Codon Plus) cells in lysogeny broth medium at 16°C for 18 h via induction by isopropyl-β-D-thiogalactoside at a final concentration of 0.2 mM. The cells were harvested by centrifugation at 4°C and then lysed with a buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, and 1 mM phenylmethanesulfonyl fluoride) for Trx-tagged proteins (or 50 mM Tris-HCl, pH 8.0, 1.5 M NaCl, 10% glycerol, and 1 mM phenylmethanesulfonyl fluoride for MBP-tagged proteins). Recombinant proteins were purified using a Ni2+-NTA affinity column followed by Superdex 200 26/60 size exclusion chromatography with the column buffer of 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM DTT, 1 mM EDTA for Trx-tagged proteins (or 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM TCEP, 10% glycerol for MBP-tagged proteins).
Fluorophore labeling of proteins
Purified proteins were subjected to buffer exchange using a HiTrap desalting column, with the exchange buffer comprising 50 mM NaHCO3 (pH 8.3), 300 mM NaCl, 1 mM EDTA, and 1 mM DTT for Trx-tagged proteins (or 50 mM NaHCO3 [pH 8.3], 500 mM NaCl, 1 mM TCEP, 10% glycerol for MBP-tagged proteins). The fluorophores, including iFluor 488 maleimide (1062; AAT Bioquest), GE Cy3 (271; AAT Bioquest), and GE Cy5 NHS ester (280; AAT Bioquest), were dissolved in DMSO, then added to the protein solution in a 1:1 M ratio, and the mixture was incubated at room temperature for 1 h. The labeled protein was buffer-exchanged into 50 mM Tris (pH 8.0), 300 mM NaCl, 1 mM EDTA, and 1 mM DTT(or 50 mM Tris [pH 8.0], 500 mM NaCl, 1 mM TCEP, 10% glycerol for MBP-tagged proteins) using a HiTrap desalting column. The labeling efficiency was determined by measuring the fluorescence intensity using a Nanodrop 2000 instrument (ND-2000; Thermo Fisher Scientific).
LLPS assays
For LLPS, the indicated purified proteins were directly mixed to reach specified concentrations. HRV-3C proteinase was added at a final concentration of 0.045 mg/ml to remove the MBP tag for FXR1 fragments and the Trx tag for Nsp3 fragments. For LLPS assay mixtures, the final buffer was 50 mM Tris, pH 8.0, 150 mM NaCl. For the microscope-based LLPS assay, each mixture was loaded into a custom-made chamber comprising a coverslip and a glass slide assembled with a layer of double-sided tape. Fluorescence images were acquired at room temperature using a confocal microscope (LSM 980 Meta plus Zeiss Axiovert zoom, Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromat, Zeiss) and a point detector (GaAsP PMT, Zeiss) at room temperature.
Liposome preparation
All the lipids (from Avanti Polar Lipids) were dissolved in chloroform and stored at −20°C. Liposomes consisted of 58.5:16.7:13.5:10:1.4 mole percent of POPC:DOPE:DOPS:DGS-NTA(Ni):488-PE. Lipids were mixed with a glass pipette in a 2-ml glass vial and dried under a stream of nitrogen gas. The samples were then vacuum-treated for at least 1 h to ensure removal of residual chloroform. The lipid mixture was resuspended in reaction buffer (1× TBS buffer) supplemented with 1% sodium cholate (#27029; Sigma-Aldrich). Lipids completely dissolved in a 1% sodium cholate environment should be transparent. The dissolved lipid mixture was loaded onto a HiTrap desalting column (GE Healthcare) with the reaction buffer. Small unilamellar vesicles (SUVs) were generated during the detergent removal process. The concentration of SUVs was estimated by measuring the fluorophore concentration using a Nanodrop 2000 spectrophotometer (ND-2000; Thermo Fisher Scientific).
Purification of ribosomes
Sucrose solutions (10% and 50%) were prepared in buffer containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 600 mM KCl, and 2 mM DTT in RNase-free conditions. 293F cells were pelleted and resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM KCl, and 2 mM DTT). The cells were disrupted with a dounce homogenizer and then centrifuged for 10 min at 14,000 rpm at 4°C. Clear supernatants from lysed cells were pelleted over sucrose cushions (1 M sucrose, 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM KCl, and 2 mM DTT) in a Beckman TLA120.2 rotor for 1 h at 100,000 rpm, 4°C and resuspended in lysis buffer. Then samples were loaded onto a 10–50% sucrose gradient and centrifuged at 38,000 rpm, 4°C for 2 h 40 min using a Beckman SW40 Ti rotor. The ribosome peaks were pelleted and resuspended in buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl), labeled by iFluor 488 maleimide, and stored at −80°C.
Viral RNA synthesis
A 1,000-bp viral DNA template was generated by PCR from a cDNA library prepared from SARS-CoV-2–infected cells. The following primers were used:
F-Viral RNA-1k: 5′-TAATACGACTCACTATAGGGACGTGGTGTGTAAAACTTGTGGA-3′;
R-Viral RNA-1k: 5′-TCTGTAATTTTTAAACTATTATTTGCTGGTTTAAGTATAATGTCTCC-3′.
Viral RNA was transcribed from the DNA template by T7 RNA polymerase (catalog #P1300; Promega). Cy3-labeled UTP (catalog #B8330; APExBio) was added into the reaction system for incorporation into the RNA. The synthesized RNA was then purified with the MEGAclear kit (catalog #AM1908; Thermo Fisher Scientific). The concentration of RNA was estimated using the Nanodrop 2000 spectrophotometer (ND-2000; Thermo Fisher Scientific).
FRAP assays
Cells for FRAP were cultured on 3.5-mm glass-bottom dishes (801001; NEST) and transfected with specific plasmids. FRAP was performed in no phenol red DMEM (21063029; Thermo Fisher Scientific) supplemented with 10% FBS using a confocal microscope (LSM 980 Meta plus Zeiss Axiovert zoom, Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromatlan, Zeiss) and a point detector (GaAsP PMT, Zeiss) using Zeiss ZEN software (blue edition) at room temperature. The region of interest (ROI) for photobleaching was selected using the Zeiss Zen software and the laser intensity was set at 100%. The bleach duration was set to 200 ms. Photobleaching of mCherry- or GFP-labeled proteins was achieved using 561-nm or 488-nm laser beams, respectively, at 100% power. The recovery of fluorescence in the bleached region was monitored over time using the same microscope and imaging settings. A total of 20 images were taken at 2-s intervals immediately after photobleaching, followed by an additional 80 images at 10-s intervals. The images were analyzed using ImageJ software to quantify the fluorescence intensity within the ROI over time.
SUnSET assay
Cells were cultured in 24-well plates with coverslips. Upon infection with SARS-CoV-2 for 24 h, puromycin was added to a final concentration of 10 µg/ml, and cells were incubated for 30 min at 37°C. After three washes with PBS, samples were fixed with 4% PFA and permeabilized with 0.1% Triton, followed by blocking with 1% BSA. Cells were then immunostained with anti-puromycin and anti-FXR1 antibodies, and subsequently fluorescent secondary antibodies. Slides were imaged using a confocal microscope (LSM 980 Meta plus Zeiss Axiovert zoom, Zeiss) with a 63×/1.40 oil-immersion objective lens (Plan-Apochromat, Zeiss) and a point detector (GaAsP PMT, Zeiss) using Zeiss ZEN software (blue edition) at room temperature.
Statistical analysis
Co-IP and immunoblotting results were obtained from no fewer than three independent experiments. FRAP data analysis involved normalizing the intensity at the prebleach point to 100% and setting the intensity right after the bleaching to 0% using ImageJ. The signal intensities of protein bands of interest from immunoblotting were analyzed using ImageJ. The sample size was determined based on preliminary experiments. The statistical parameters, such as n, SEM, and SD, are provided in the figure legends. Data were assumed to follow a normal distribution without formal testing. One-way ANOVA analysis was employed to assess statistical significance, with a P value of <0.05 considered statistically significant. The Chi-square test was used to compare percentages between control and KD cells and to determine significance.
Online supplemental material
Fig. S1 shows that GFP-tagged FXR family proteins are localized to DMV sites, where the SG marker G3BP is absent. Fig. S2 shows that concentration of DMVs requires FXRs and depends on their phase separation ability. Fig. S3 shows that all eight domains of Nsp3 following the HVR domain are dispensable for DMV clustering. Fig. S4 shows that the UBL1 and HVR domains in Nsp3 and the NIR domain in FXR1 are responsible for NSP3/FXR1 interaction. Fig. S5 shows that FXR family proteins are critical for efficient SARS-CoV-2 replication.
Data availability
All data are available in the main text or the supplemental materials. Further reasonable requests should be directed to and will be fulfilled by the lead contact, Yan G. Zhao ([email protected]).
Acknowledgments
We are grateful to Dr. Isabel Hanson for editing work.
This work was supported by the National Natural Science Foundation of China (32222021, 32170753 to Y.G. Zhao, and 82025022 to Z. Zhang), the National Key Research and Development Program (2021YFA1300800 to Y.G. Zhao), the Guangdong Innovative and Entrepreneurial Research Team Program (2021ZT09Y104 to Y.G. Zhao), the Shenzhen Talent Program (KQTD20210811090115021 to Y.G. Zhao), the Shenzhen–Hong Kong Institute of Brain Science–Shenzhen Fundamental Research Institutions (2023SHIBS0002 to Y.G. Zhao), the Shenzhen Science and Technology Program (ZDSYS20220402111000001 to Y.G. Zhao, and JCYJ20220818103017036 to Z. Zhang), R&D Program of Guangzhou Laboratory (GZNL2024A01008 to Y.G. Zhao), the Guangdong Program (2019CX01Y222 to Y.G. Zhao), and Guangdong Science and Technology Plan Project, Construction of high-level biosafety laboratories (2021B1212030010 to Z. Zhang).
Author contributions: Y.G. Zhao and Z. Zhang designed the project. M. Li, Y. Hou, Y. Zhou, and Z. Yang performed the experiments. H. Zhao carried out the TEM experiments. Q. Yu and F. Zeng carried out the ribosome purification experiments. T. Jian and X. Liu performed protein purification. All authors analyzed and discussed the results. Y.G. Zhao wrote the manuscript.
References
Author notes
M. Li, Y. Hou, Y. Zhou, and Z. Yang contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.


![FXRs are recruited to DMV sites through binding to Nsp3. (A) The schematic diagram shows the experimental design for identifying DMV-related factors. Lysates from cells expressing Nsp4-GFP and mCherry-Nsp3 or Nsp4-GFP alone (serving as control) are subjected to GFP-Trap, followed by MS analysis. N, nucleus. (B) The volcano plot from the MS data demonstrates quantitative changes in proteins pulled down by Nsp4-GFP in Nsp3/4-expressing cells compared with cells expressing Nsp4-GFP alone. The horizontal dashed line shows where P value is 0.05 (−log 10 [0.05] = 1.3), and the vertical dashed lines show where the fold change is 2 (log 2 [2] = 1) or 0.5 (log 2 [0.5] = −1). These parameters were used as the threshold cutoff. Upregulated proteins are shown in red and downregulated proteins are shown in blue. FXR1 and FMR1 are labeled. (C) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are immunoprecipitated by mCherry-Nsp3. (D) In an mCherry-Trap assay, FXR1, FXR2, and FMR1 are not immunoprecipitated by Nsp4-mCherry. (E) The schematic shows the specific localization of Nsp3 and Nsp4 on DMVs and the Nsp3-mediated recruitment of FXRs to DMVs. (F) Endogenous FXR1, FXR2, and FMR1 colocalize well with Nsp3/4+ foci in mCherry-Nsp3/Flag-Nsp4-coexpressing HeLa cells. FXR1, FXR2, and FMR1 show diffuse cytoplasmic localization under control conditions. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. (G) Endogenous FXR1 forms puncta that colocalize well with Nsp3 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bar: 5 μm; inset, 2 μm. (H) Endogenous FXR2 and FMR1 form puncta that colocalize well with FXR1 and dsRNA foci after 24 h SARS-CoV-2 infection with MOI = 1. Images are representative of at least 10 cells. Bars: 5 μm; insets, 2 μm. Source data are available for this figure: SourceData F1.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/6/10.1083_jcb.202309140/1/m_jcb_202309140_fig1.png?Expires=1773846788&Signature=OvGLmhBJqFdF4x7fhrayygpVdQzm4RyXRI1BA2MYnLvX70rBYAFF5sK3Z4WO1sMEFZzpNF59ECm44JEfqVBbE5ru-BfPYEE~OJEYPy1RjTuwXaVmrrZbJgu4r6xHmMZXHzB3Tm5hus5xszyDSHj46Oblmq3uLUIyMQIjOglSuOBFhd~8ezWb-jrnoENhXYXDBeCTzyJhr0HoPL6MlGZFuQxrTh3K13kmUAswPqlhdFjpouticHN1p9AuLz2kOnX7mZH4sog7MtkEev5-EfYBc9HbYqaQ~53XcnOKVko-SWufQVw7Y2cJwW4p9AOCPMyw2Tn9eVPRPn2uLuWEcDd66Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

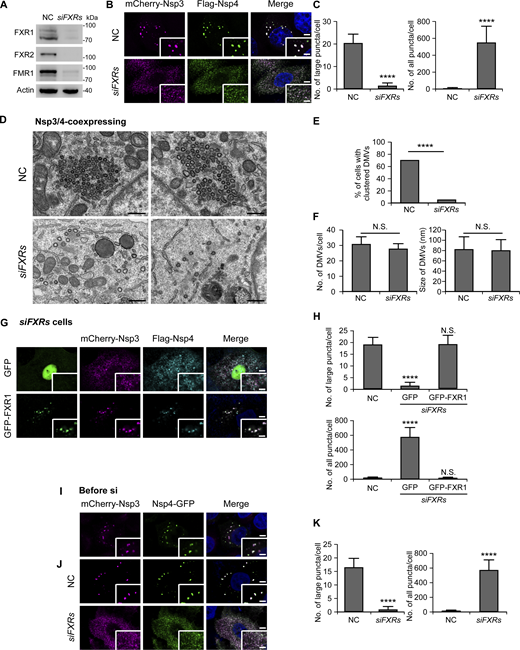





![The region between the KH2 and IDR domains in FXR1 is essential for FXR1-Nsp3 interaction. (A) The schematic shows the domain organization of FXR1. The Nsp3-interacting region (NIR) is located between the KH2 and IDR1 domains. (B and C) Alphafold predictions show that Nsp3 (green) interacts with FXR1 (cyan) via an α-helix (aa V68–I77) in the UBL1 domain (B) and a β-strand (aa 142–146) in the HVR domain (C). Key residues are labeled in the images. (D and E) Cells cotransfected with mCherry-Nsp3(E70A/L71A) or mCherry-Nsp3(Δ142–146) and Nsp4 show a huge abundance of small puncta that fail to colocalize with endogenous FXR1, while WT mCherry-Nsp3 and Nsp4 form big dots and recruit FXR1 (D). Numbers of DMVs are shown as mean ± SD [mCherry-Nsp3, n = 33; mCherry-Nsp3(E70A/L71A), n = 37; mCherry-Nsp3(Δ142–146), n = 30] (E). ****, P < 0.0001. Bars: 5 μm; insets, 2 μm. (F) In a GFP-Trap assay, GFP-FXR1 immunoprecipitates dramatically reduced levels of mCherry-Nsp3(E70A/L71A) compared to WT mCherry-Nsp3. (G) In a GFP-Trap assay, GFP-FXR1(I355A/L358A/K359A) immunoprecipitates dramatically reduced levels of mCherry-Nsp3 compared to WT GFP-FXR1. (H) In a GFP-Trap assay, the interaction between Nsp3 and FXR1 is inhibited by expression of mCherry-Nsp3(Δ142–146) or FXR1(R315D/R317D). Source data are available for this figure: SourceData F5.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/6/10.1083_jcb.202309140/1/m_jcb_202309140_fig5.png?Expires=1773846788&Signature=nSwEkRTI~voqAtVeM7tx9wXXx6gZM0MZL57NDmDFGdAp-E3MgY2R5Hj~i~ZPjFBWAd49Dhlu17iTvwGHYKxIZQTX1azHxQBG8moakLYUgAdtst6rCI-hU9a~mpB8fSnCEu6UJdpk5e2VpgnOjATgHGVFGKo6dE449g6J72AZdM75a2GAeMdNzoDgZLcTZiIg7UJBLthXhxhk0CLkER81oy2beQN3sxdXCfg0Q7gbsys9En-EtsS2JvQLIaH1d534jSLikg0opmSWoNmU6E78HkfRy~qq-5kJmpg05R6U4nYciDuQ4db2BzkWttt7737YuOO6Fm0vDSwJguh3pqQFFg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)