Transforming growth factor β (TGF-β) and HER2 signaling collaborate to promote breast cancer progression. However, their molecular interplay is largely unclear. TGF-β can activate mitogen-activated protein kinase (MAPK) and AKT, but the underlying mechanism is not fully understood. In this study, we report that TGF-β enhances HER2 activation, leading to the activation of MAPK and AKT. This process depends on the TGF-β type I receptor TβRI kinase activity. TβRI phosphorylates HER2 at Ser779, promoting Y1248 phosphorylation and HER2 activation. Mice with HER2 S779A mutation display impaired mammary morphogenesis, reduced ductal elongation, and branching. Furthermore, wild-type HER2, but not S779A mutant, promotes TGF-β-induced epithelial–mesenchymal transition, cell migration, and lung metastasis of breast cells. Increased HER2 S779 phosphorylation is observed in human breast cancers and positively correlated with the activation of HER2, MAPK, and AKT. Our findings demonstrate the crucial role of TGF-β-induced S779 phosphorylation in HER2 activation, mammary gland development, and the pro-oncogenic function of TGF-β in breast cancer progression.
Introduction
TGF-β plays a critical role in regulating cellular processes such as proliferation, differentiation, survival, and organ development. However, its dysregulation can lead to various diseases, including cancer (Massagué, 2008, 2012). The biological effects of TGF-β are mediated through Smad-dependent transcription regulation or non-Smad pathways (Derynck and Zhang, 2003; Moustakas and Heldin, 2005; Yu and Feng, 2019; Zhang, 2009). Smad2 and Smad3, when phosphorylated and activated by the TGF-β type I receptor (TβRI), collaborate with Smad4 and other factors to regulate gene expression in the nucleus. Additionally, TGF-β receptors can also activate other signaling pathways, including mitogen-activated protein kinases (MAPKs: ERKs, p38, and JNK), AKT, and small GTPases, in a cell type– and context-dependent manner. These pathways are crucial for TGF-β-induced processes such as epithelial–mesenchymal transition (EMT) and cell migration. However, the precise underlying mechanisms of these processes remain to be elucidated.
HER2 (ERBB2, Neu), a member of the epidermal growth factor receptor (EGFR) family, can promote cell proliferation, migration, and cancer progression (Arteaga and Engelman, 2014), and plays a significant role in various physiological processes, including mammary morphogenesis (Andrechek et al., 2005; Jackson-Fisher et al., 2004). Pathological mutations or overexpression of HER2 have been identified in several malignancies, including lung, breast, colorectal, esophageal carcinomas, and hepatomas (Arteaga and Engelman, 2014; Roskoski, 2014; Slamon et al., 1989). HER2 can form homodimers or heterodimers to activate AKT and MAPKs (Arteaga and Engelman, 2014). Its kinase activity is tightly regulated. The αC-β4 loop (amino acids 776–783) within the kinase domain of HER2 is responsible for its autoinhibition, and its loss leads to oncogenic activation of HER2 (Arteaga and Engelman, 2014; Bose and Zhang, 2009; Ding et al., 2019; Fan et al., 2008; Mitani et al., 2022; Stephens et al., 2004; Wen et al., 2015).
TGF-β and HER2 signaling pathways can modulate each other at various levels (Chow et al., 2011; Shi and Chen, 2017), and their cooperation accelerates tumorigenesis and tumor metastasis (Chow et al., 2011; Huang et al., 2018; Muraoka-Cook et al., 2006; Seton-Rogers et al., 2004; Shi and Chen, 2017; Wang, 2011). TGF-β signaling can promote HER2-induced lung metastasis of breast cancer in transgenic mouse models (Muraoka et al., 2003; Muraoka-Cook et al., 2006; Siegel et al., 2003). Furthermore, HER2 is necessary for the promigratory effect of TGF-β on HER2-overexpressing mammary epithelial cells (Wang et al., 2008). However, it remains unclear whether TGF-β can directly regulate HER2/EGFR activity. In this study, we present evidence showing that TGF-β enhances HER2 activity via the direct phosphorylation of HER2 at Ser779 by TβRI. The Ser779 site of HER2 was found to be essential for mammary gland development, specifically regarding ductal elongation and epithelium branching. TGF-β-regulated HER2 activity was found to promote EMT, cell migration, and lung metastasis of breast cancer. Furthermore, we observed that inhibition of HER2 suppressed the TGF-β-induced activation of AKT and MAPKs. These findings reveal the novel interplay between TGF-β and HER2 signaling in mammary gland development and breast cancer progression.
Results
TGF-β-induced activation of AKT and ERK requires HER2 activity
In addition to activating Smad proteins, TGF-β can also signal via non-Smad pathways, such as PI3K/AKT and MAPK (Derynck and Zhang, 2003; Yu and Feng, 2019; Zhang, 2009). To investigate the role of HER2 in TGF-β-induced activation of AKT and MAPKs, we examined the effects of lapatinib, a dual HER2 and EGFR kinase inhibitor, on NMuMG cells (normal mouse mammary epithelial cells), SKBR3 cells (HER2-positive human breast cancer cells), MCF10A cells (normal human mammary epithelial cells), MDA-MB-231 cells (triple-negative breast cancer cells), and BT474 cells (luminal B-like). Our results demonstrated that lapatinib effectively blocked the TGF-β-stimulated phosphorylation of AKT and ERK while having no effect on Smad2 phosphorylation (Fig. 1, A–C; and Fig. S1, A and B). As a control, lapatinib inhibited HER2 phosphorylation at tyrosine 1248 (Y1248), an indicator of HER2 activation (Hazan et al., 1990). Similar findings were obtained when using the HER2 inhibitor CP724714 on SKBR3 cells and NMuMG cells (Fig. S1 C). Furthermore, HER2 knockdown resulted in a significant attenuation of TGF-β-induced AKT and ERK signaling (Fig. 1 D). Interestingly, we also observed an augmentation of HER2 activation following TGF-β treatment, as evidenced by increased phosphorylation at Y1248 in both mammary epithelial cells and breast cancer cells (Fig. 1 and Fig. S1, A–D). The increased phosphorylation of PLC-γ1 at the Y783 site by TGF-β-induced enhancement of the HER2 kinase activity can also be detected (Fig. S1 E). These results suggest that the enhanced activation of HER2 induced by TGF-β contributes to the activation of AKT and ERK.
TGF-β-induced activation of AKT and ERK requires HER2 activity. (A–C) NMuMG, MCF10A, and SKBR3 cells were treated with 2 μM (A and B) or 0.1 μM (C) Lapatinib (Lap) for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) Control and HER2 knockdown NMuMG cells were treated with 100 pM TGF-β1 for 1 h and then harvested for immunoblotting. (E) MCF10A cells were treated with 2 μM Lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h, followed by anti-HER2 Y1248 immunofluorescence (red) and anti-HER2 immunofluorescence (green). The nuclei were counterstained by DAPI (blue). Bar, 20 μm. Source data are available for this figure: SourceData F1.
TGF-β-induced activation of AKT and ERK requires HER2 activity. (A–C) NMuMG, MCF10A, and SKBR3 cells were treated with 2 μM (A and B) or 0.1 μM (C) Lapatinib (Lap) for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) Control and HER2 knockdown NMuMG cells were treated with 100 pM TGF-β1 for 1 h and then harvested for immunoblotting. (E) MCF10A cells were treated with 2 μM Lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h, followed by anti-HER2 Y1248 immunofluorescence (red) and anti-HER2 immunofluorescence (green). The nuclei were counterstained by DAPI (blue). Bar, 20 μm. Source data are available for this figure: SourceData F1.
Inhibition of HER2 impairs TGF-β-induced activation of AKT and ERK. (A) The relative p-Y1248, p-AKT, p-ERK, and p-Smad2 levels of three repeated experiments in Fig. 1 A were quantified. (B and C) Cells were treated as indicated (MDA-MB-231 cells: 5 μM lapatinib; BT474 cells: 0.01 μM lapatinib; SKBR3 cells: 1 μM CP-724714 [CP]; NMuMG cells: 10 μM CP-724714) for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) NMuMG (Left) and SKBR3 (Right) cells were treated with 2 or 0.01 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h, followed by anti-HER2 Y1248 immunofluorescence (red) and anti-HER2 immunofluorescence (green). The nuclei were counterstained by DAPI (blue). Bar, 20 μm. (E) NMuMG cells were treated with 2 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (F) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-p-Y1248 immunoblotting. (G) MCF10A or SKBR3 cells were treated with 5 μM SB-431542 (SB) for 2 h, 100 pM TGF-β1 was added in the last 1 h before being harvested for immunoblotting. (H and I) HEK293T cells were transfected with indicated plasmids before being harvested for anti-HA pulldown and then for immunoblotting. (J) NMuMG cells were treated with 100 pM TGF-β1 for the indicated time before being harvested for anti-TβRI pulldown and then for immunoblotting. (K) MCF10A cells were treated with 100 pM TGF-β1 for 1 h before being harvested for anti-TβRI pulldown and then for immunoblotting. (L) The purified MBP-tagged proteins (1, WT TβRI ICD; 2, GGD-ICD; 3, HER2 ICD) are shown with asterisks. M: Protein markers. The band intensity of phosphorylated proteins was normalized to total proteins. The band intensity of pull-down HER2 was normalized to pull-down TβRI. Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. Source data are available for this figure: SourceData FS1.
Inhibition of HER2 impairs TGF-β-induced activation of AKT and ERK. (A) The relative p-Y1248, p-AKT, p-ERK, and p-Smad2 levels of three repeated experiments in Fig. 1 A were quantified. (B and C) Cells were treated as indicated (MDA-MB-231 cells: 5 μM lapatinib; BT474 cells: 0.01 μM lapatinib; SKBR3 cells: 1 μM CP-724714 [CP]; NMuMG cells: 10 μM CP-724714) for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) NMuMG (Left) and SKBR3 (Right) cells were treated with 2 or 0.01 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h, followed by anti-HER2 Y1248 immunofluorescence (red) and anti-HER2 immunofluorescence (green). The nuclei were counterstained by DAPI (blue). Bar, 20 μm. (E) NMuMG cells were treated with 2 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (F) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-p-Y1248 immunoblotting. (G) MCF10A or SKBR3 cells were treated with 5 μM SB-431542 (SB) for 2 h, 100 pM TGF-β1 was added in the last 1 h before being harvested for immunoblotting. (H and I) HEK293T cells were transfected with indicated plasmids before being harvested for anti-HA pulldown and then for immunoblotting. (J) NMuMG cells were treated with 100 pM TGF-β1 for the indicated time before being harvested for anti-TβRI pulldown and then for immunoblotting. (K) MCF10A cells were treated with 100 pM TGF-β1 for 1 h before being harvested for anti-TβRI pulldown and then for immunoblotting. (L) The purified MBP-tagged proteins (1, WT TβRI ICD; 2, GGD-ICD; 3, HER2 ICD) are shown with asterisks. M: Protein markers. The band intensity of phosphorylated proteins was normalized to total proteins. The band intensity of pull-down HER2 was normalized to pull-down TβRI. Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. Source data are available for this figure: SourceData FS1.
TGF-β-enhanced HER2 activity depends on TβRI kinase activity
As previously reported, TβRI kinase activity was required for AKT and MAPK activation (Huang and Chen, 2012; Rodríguez-García et al., 2017; Zhang, 2009). We then examined whether TGF-β-enhanced HER2 activation was dependent on the kinase activity of TβRI. In NMuMG cells, TGF-β stimulation resulted in HER2 tyrosine phosphorylation, which was blocked by lapatinib and the TβRI inhibitor SB-431542 (Fig. 2 A). Furthermore, HER2 tyrosine phosphorylation, particularly at the Y1248 site, was significantly enhanced by constitutively activated TβRI (GGD), but not wild-type (WT) TβRI or kinase-deficient TβRI (KR) (Fig. 2 B and Fig. S1 F). Similarly, in NMuMG cells, inhibition of TβRI effectively blocked TGF-β-induced enhancement of HER2 Y1248 phosphorylation and AKT or ERK activation (Fig. 2 C and Fig. S1 G). Moreover, re-expression of TβRI in TβRI-deficient mink lung epithelial R1B/L17 cells promoted TGF-β-induced HER2 activation (Fig. 2 D). These findings confirm that TGF-β-augmented HER2 activation is dependent on the kinase activity of TβRI.
TGF-β-enhanced HER2 activity depends on TβRI kinase activity. (A) NMuMG cells were treated with 2 μM Lapatinib or 5 μM SB431542 (SB) for 2 h, and 100 pM TGF-β1 was added in the last 1 h before being harvested for anti-HER2 pulldown and then for anti-phospho-tyrosine immunoblotting. (B) HEK293T cells were transfected with indicated plasmids before being harvested for anti-Myc pulldown and then for anti-phospho-Tyrosine immunoblotting. (C) NMuMG cells were treated with 5 μM SB431542 for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) R1B/L17 cells overexpressed with empty vector or TβRI were treated with 100 pM TGF-β1 for 1 h before being harvested for immunoblotting. (E) SPR analysis of TβRI WT ICD—HER2 ICD and GGD ICD—HER2 ICD interactions. HER2 ICD protein was immobilized on Series S sensor chips CM5, and the binding experiments were carried out using a Biacore S200. The estimated Kd values were derived by fitting the association and dissociation signals with a 1:1 (Langmuir) model using the Biacore S200 Evaluation Software. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F2.
TGF-β-enhanced HER2 activity depends on TβRI kinase activity. (A) NMuMG cells were treated with 2 μM Lapatinib or 5 μM SB431542 (SB) for 2 h, and 100 pM TGF-β1 was added in the last 1 h before being harvested for anti-HER2 pulldown and then for anti-phospho-tyrosine immunoblotting. (B) HEK293T cells were transfected with indicated plasmids before being harvested for anti-Myc pulldown and then for anti-phospho-Tyrosine immunoblotting. (C) NMuMG cells were treated with 5 μM SB431542 for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) R1B/L17 cells overexpressed with empty vector or TβRI were treated with 100 pM TGF-β1 for 1 h before being harvested for immunoblotting. (E) SPR analysis of TβRI WT ICD—HER2 ICD and GGD ICD—HER2 ICD interactions. HER2 ICD protein was immobilized on Series S sensor chips CM5, and the binding experiments were carried out using a Biacore S200. The estimated Kd values were derived by fitting the association and dissociation signals with a 1:1 (Langmuir) model using the Biacore S200 Evaluation Software. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F2.
Given that both TβRI and HER2 are transmembrane proteins, we proceeded to examine their interaction. Compared with WT TβRI or kinase-deficient TβRI, constitutively activated TβRI (GGD) exhibited a stronger interaction with HER2 (Fig. S1 H). Furthermore, we found that both the intracellular and extracellular domains of HER2 interacted with TβRI (Fig. S1 I). Consistently, TGF-β significantly enhanced the endogenous interaction between HER2 and TβRI (Fig. S1, J and K). To confirm whether the two receptors directly interacted, we performed surface plasmon resonance (SPR) analysis. MBP-tag-fused intracellular domain (ICD) of TβRI (WT, GGD) and HER2 were purified (Fig. S1 L). HER2 ICD protein was immobilized on Series S sensor chips CM5, and the solution containing TβRI ICD was passed over the sensor chip. The Kd values were calculated from the equilibrium binding isotherms using the 1:1 Langmuir binding model. The affinity constant of WT TβRI to HER2 was 4.24 nM, and GGD to HER2 was 2.68 nM (Fig. 2 E). The data indicate the direct interaction between intracellular domains of TβRI and HER2, and the lower Kd value of GGD-HER2 also confirmed the TGF-β-enhanced TβRI-HER2 interactions.
TGF-β induces HER2 phosphorylation at the S779 site
As TβRI physically interacted with HER2, we then examined if TβRI directly phosphorylated HER2. Immunoprecipitation analysis demonstrated that the active TβRI(GGD) enhanced HER2 serine phosphorylation, concomitant with an increase in HER2 autophosphorylation at the Y1248 site (Fig. 3 A).
TGF-β induces HER2 phosphorylation at Ser779. (A–C) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-phospho-serine and anti-p-Y1248 (A and B), anti-p-S779, and anti-p-Y1248 (C) immunoblotting. (D) Flag-tagged activated intracellular domain of TβRI (caTβRI, 148 aa–503 aa) protein (200 ng) was incubated with 2 μg WT or S779A mutant HER2 protein containing intracellular domain (C-term, 712 aa–1255 aa) in an in vitro kinase assay, and the phosphorylation of HER2 at S779 and Y1248 sites were detected with anti-p-S779 and anti-p-Y1248 immunoblotting. (E) NMuMG cells were treated with 100 pM TGF-β1 for indicated times before being harvested for immunoblotting. p-HER2 (p-S779 and p-Y1248) levels were quantified. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F3.
TGF-β induces HER2 phosphorylation at Ser779. (A–C) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-phospho-serine and anti-p-Y1248 (A and B), anti-p-S779, and anti-p-Y1248 (C) immunoblotting. (D) Flag-tagged activated intracellular domain of TβRI (caTβRI, 148 aa–503 aa) protein (200 ng) was incubated with 2 μg WT or S779A mutant HER2 protein containing intracellular domain (C-term, 712 aa–1255 aa) in an in vitro kinase assay, and the phosphorylation of HER2 at S779 and Y1248 sites were detected with anti-p-S779 and anti-p-Y1248 immunoblotting. (E) NMuMG cells were treated with 100 pM TGF-β1 for indicated times before being harvested for immunoblotting. p-HER2 (p-S779 and p-Y1248) levels were quantified. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F3.
Mass spectrometry analysis showed that the residue Ser779, located within the αC-β4 loop of the HER2 kinase domain, was the phosphorylation target of activated TβRI (Fig. S2 A). Subsequent mutation of Ser779 to alanine (S779A) in HEK 293T cells resulted in a pronounced inhibition of HER2 serine and tyrosine phosphorylation, as well as a reduction in AKT and ERK activation (Fig. 3, B and C). Conversely, the phosphorylation-mimic mutant (S779D) retained phosphorylation capabilities similar to WT HER2 (Fig. 3 B). Moreover, an in vitro kinase assay using purified caTβRI intracellular domain protein confirmed its direct serine phosphorylation within the WT HER2 kinase domain (KD), which was abolished by the S779A mutation (Fig. S2 B). Notably, both the S779A and S779D mutations did not affect the interaction between HER2 and activated TβRI (Fig. S2 C), ruling out the possibility of HER2 conformational damage due to Ser779 mutations. Furthermore, we observed that TGF-β enhanced HER2 Y1248 phosphorylation, as well as AKT and ERK activation in HER2-knockdown NMuMG and MDA-MB-231 cells that overexpressed WT HER2 but not the HER2 (S779A) mutant (Fig. S2, D and E).
TGF-β-enhanced HER2 S779 phosphorylation is important for the activation of HER2 AKT and ERK. (A) HEK293T cells transfected with HER2-myc and GGD-HA were harvested for anti-myc immunoprecipitation. After digestion, the HER2 peptides were analyzed with LC-MS/MS to identify the phosphorylation sites. The residue S779 was marked with a red asterisk on the top-right corner. (B) Flag-tagged intracellular domain of activated TβRI protein (caTβRI, 148 aa–503 aa) (200 ng) was incubated with 2 μg WT or S779A mutant HER2 protein containing the kinase domain (KD, 712 aa–990 aa) in an in vitro kinase assay, and the total serine phosphorylation of HER2 (marked by black asterisk) and HER2 phosphorylation at Y877 site were detected with immunoblotting. (C) HEK293T cells were transfected with indicated plasmids before being harvested for anti-HA pulldown and then for anti-myc immunoblotting. (D) HER2-knockdown NMuMG cells expressed with GFP, WT HER2, S779A mutant (SA), or S779D mutant (SD) were treated with 100 pM TGF-β for 1 h before harvesting for immunoblotting. (E) MDA-MB-231 cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 1 h before harvested for immunoblotting. (F) SKBR3 cells were treated with 100 pM TGF-β1 for 1 h before being harvested for immunoblotting. (G) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-p-S779 and anti-p-Y1248 immunoblotting. The band intensity of phosphorylated proteins was normalized to pull-down HER2 proteins. Source data are available for this figure: SourceData FS2.
TGF-β-enhanced HER2 S779 phosphorylation is important for the activation of HER2 AKT and ERK. (A) HEK293T cells transfected with HER2-myc and GGD-HA were harvested for anti-myc immunoprecipitation. After digestion, the HER2 peptides were analyzed with LC-MS/MS to identify the phosphorylation sites. The residue S779 was marked with a red asterisk on the top-right corner. (B) Flag-tagged intracellular domain of activated TβRI protein (caTβRI, 148 aa–503 aa) (200 ng) was incubated with 2 μg WT or S779A mutant HER2 protein containing the kinase domain (KD, 712 aa–990 aa) in an in vitro kinase assay, and the total serine phosphorylation of HER2 (marked by black asterisk) and HER2 phosphorylation at Y877 site were detected with immunoblotting. (C) HEK293T cells were transfected with indicated plasmids before being harvested for anti-HA pulldown and then for anti-myc immunoblotting. (D) HER2-knockdown NMuMG cells expressed with GFP, WT HER2, S779A mutant (SA), or S779D mutant (SD) were treated with 100 pM TGF-β for 1 h before harvesting for immunoblotting. (E) MDA-MB-231 cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 1 h before harvested for immunoblotting. (F) SKBR3 cells were treated with 100 pM TGF-β1 for 1 h before being harvested for immunoblotting. (G) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-p-S779 and anti-p-Y1248 immunoblotting. The band intensity of phosphorylated proteins was normalized to pull-down HER2 proteins. Source data are available for this figure: SourceData FS2.
To specifically detect S779 phosphorylation, we generated polyclonal antibodies, and this antibody specifically detected S779 phosphorylation of HER2 as there was no signal on the S779A mutant (Fig. 3 C). Additionally, the in vitro kinase assay demonstrated that caTβRI protein directly enhanced S779 and Y1248 phosphorylation of HER2 intracellular domain (712 aa–1255 aa), while this effect was not observed in HER2 (S779A) mutant (Fig. 3 D). In NMuMG cells, TGF-β stimulation induced rapid phosphorylation of HER2 at S779, as early as 2.5 min, a robust increase of p-S779 level at 15 min, whereas TGF-β-induced phosphorylation at Y1248 appeared at 5 min, reaching the highest level at 60 min (Fig. 3 E). These data indicate that TGF-β induces a quick and direct phosphorylation of HER2 at S779, which precedes Y1248 phosphorylation. Furthermore, TGF-β-induced HER2 S779 phosphorylation was also observed in other breast cell lines, including SKBR3 cells (Fig. S2 F).
Previous studies have indicated that the introduction of two glycine residue mutations (G776S and G778D) within the αC-β4 loop of HER2 significantly enhances its kinase activity and eliminates autoinhibition (Fan et al., 2008). Considering this, we investigated whether S779 is involved in the regulation of HER2 kinase activity. Indeed, we observed that the presence of G776S, G778D, or both mutations notably increased HER2 phosphorylation at S779 and Y1248 (Fig. S2 G). Interestingly, Y1248 phosphorylation was abolished upon the introduction of the S779A mutation, further confirming the critical role of S779 in HER2 activation.
We consistently observed increased HER2 phosphorylation at both the S779 and Y1248 sites in response to TGF-β stimulation in various cell types expressing HER2, including epithelial cells (HaCaT and HeLa), mouse embryonic fibroblast cells (NIH 3T3), non-small cell lung cancer cells (A549 and H1299), hepatoma carcinoma cells (HepG2), esophageal carcinoma cells (KYSE410), and breast cells (EpH4-Ras) (Fig. S3). Importantly, this effect was effectively blocked by the inhibitors SB431542 and lapatinib. It is noteworthy that TGF-β-induced AKT activation was observed in these cell lines and ERK activation in most of them, except HeLa cells and H1299 cells (Ras mutant) (Fig. S3). Overall, these findings highlight the universality of TGF-β-induced HER2 activation via S779 phosphorylation in multiple HER2-positive epithelial, fibroblast, and tumor cells.
Enhancement of HER2 activity by TGF-β is observed in multiple epithelial cells and cancer cells. HaCaT, HeLa, NIH 3T3, EpH4, HepG2, KYSE410, A549, and H1299 cells were treated with 5 μM SB-431542, 2 μM Lapatinib, 10 μM CP-724714, or 8 μM Nystatin for 2 h, and 100 pM TGF-β1 was added in the last 1 h before harvested for immunoblotting. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData FS3.
Enhancement of HER2 activity by TGF-β is observed in multiple epithelial cells and cancer cells. HaCaT, HeLa, NIH 3T3, EpH4, HepG2, KYSE410, A549, and H1299 cells were treated with 5 μM SB-431542, 2 μM Lapatinib, 10 μM CP-724714, or 8 μM Nystatin for 2 h, and 100 pM TGF-β1 was added in the last 1 h before harvested for immunoblotting. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData FS3.
HER2 S779 site is important for mammary epithelial morphogenesis
HER2 plays a crucial role in mammary gland development (Andrechek et al., 2005; Jackson-Fisher et al., 2004; Schroeder and Lee, 1998). To further investigate the physiological function of HER2 S779 (corresponding to mouse ERBB2 S780) in mammary gland development, we generated an inducible ERBB2CKI-S780A knock-in mice (Fig. S4 A). These mice were then crossed with mice carrying the MMTV-Cre allele to generate ERBB2S780A/+;MMTV-Cre (SA) mice (Fig. S4 B).
The residue S779 of HER2 is crucial for maintaining mammary epithelium branching and for promoting EMT and cell migration. (A) Schematic diagram of knock-in donor vector to generate inducible ERBB2CKI-S780A KI mice. The red asterisk marks the location of the S780A mutation corresponding to the human S779A mutation. ERBB2CKI-S780A knock-in allele can be generated after Cre-mediated excision of the floxed wide-type ERBB2 cDNA coding for exon 20–27. (B) Schematic summary of three primer sets and DNA gel patterns of the different genotypes ([1] ERBB2+/+;MMTV-Cre with positive band of ; [2] ERBB2S780A/+ with positive band of ; [3] ERBB2S780A/+;MMTV-Cre with positive band of ; [4] Negative control with no positive band). M: DNA marker. The thickened dark line indicates the sequencing region. (C) Representative images showing reduced branching and distribution of K14+ basal epithelial cells in organoids from the SA group. Bar, 100 μm. (D) Representative images of organoids derived from sorted basal epithelial cells (CD24medCD29hi) (5,000 cells per well in a 48-well plate) at day 3 after culture. The average number of buds per organoid (#budding) and organoid size were quantified. Bar, 500 μm. (E) The expression levels of WT HER2 and SA mutant in MCF10A cells (Fig. 5 A). (F) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with DMSO or 100 pM TGF-β1 for 48 h and imaged before subjected to immunoblotting with indicated antibodies. (G) mRNA levels of Fibronectin and PAI-1 were examined by quantitative real-time RT-PCR. (H) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 48 h, followed by immunofluorescence with anti-E-cadherin antibody (green) and rhodamine-phalloidin to stain F-actin (red). The nuclei were counterstained by DAPI (blue). The relative E-cadherin signals were quantified and normalized to the GFP group without TGF-β1 treatment. Bar, 20 μm. (I) The expression levels of WT HER2 and SA mutant in MDA-MB-231 cells (Fig. 5 B). (J) Subconfluent MDA-MB-231 cells stably expressed with GFP, WT HER2, S779A (SA), or S779D mutant (SD) were wounded by a 1,000-μl pipette tip, and then treated with 100 pM TGF-β1 for 48 h and imaged (Left). Right is the expression levels of WT HER2, SA and SD mutants in MDA-MB-231 cells (Fig. 5 C). Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. The band intensity of each protein was normalized to the first group. Source data are available for this figure: SourceData FS4.
The residue S779 of HER2 is crucial for maintaining mammary epithelium branching and for promoting EMT and cell migration. (A) Schematic diagram of knock-in donor vector to generate inducible ERBB2CKI-S780A KI mice. The red asterisk marks the location of the S780A mutation corresponding to the human S779A mutation. ERBB2CKI-S780A knock-in allele can be generated after Cre-mediated excision of the floxed wide-type ERBB2 cDNA coding for exon 20–27. (B) Schematic summary of three primer sets and DNA gel patterns of the different genotypes ([1] ERBB2+/+;MMTV-Cre with positive band of ; [2] ERBB2S780A/+ with positive band of ; [3] ERBB2S780A/+;MMTV-Cre with positive band of ; [4] Negative control with no positive band). M: DNA marker. The thickened dark line indicates the sequencing region. (C) Representative images showing reduced branching and distribution of K14+ basal epithelial cells in organoids from the SA group. Bar, 100 μm. (D) Representative images of organoids derived from sorted basal epithelial cells (CD24medCD29hi) (5,000 cells per well in a 48-well plate) at day 3 after culture. The average number of buds per organoid (#budding) and organoid size were quantified. Bar, 500 μm. (E) The expression levels of WT HER2 and SA mutant in MCF10A cells (Fig. 5 A). (F) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with DMSO or 100 pM TGF-β1 for 48 h and imaged before subjected to immunoblotting with indicated antibodies. (G) mRNA levels of Fibronectin and PAI-1 were examined by quantitative real-time RT-PCR. (H) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 48 h, followed by immunofluorescence with anti-E-cadherin antibody (green) and rhodamine-phalloidin to stain F-actin (red). The nuclei were counterstained by DAPI (blue). The relative E-cadherin signals were quantified and normalized to the GFP group without TGF-β1 treatment. Bar, 20 μm. (I) The expression levels of WT HER2 and SA mutant in MDA-MB-231 cells (Fig. 5 B). (J) Subconfluent MDA-MB-231 cells stably expressed with GFP, WT HER2, S779A (SA), or S779D mutant (SD) were wounded by a 1,000-μl pipette tip, and then treated with 100 pM TGF-β1 for 48 h and imaged (Left). Right is the expression levels of WT HER2, SA and SD mutants in MDA-MB-231 cells (Fig. 5 C). Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. The band intensity of each protein was normalized to the first group. Source data are available for this figure: SourceData FS4.
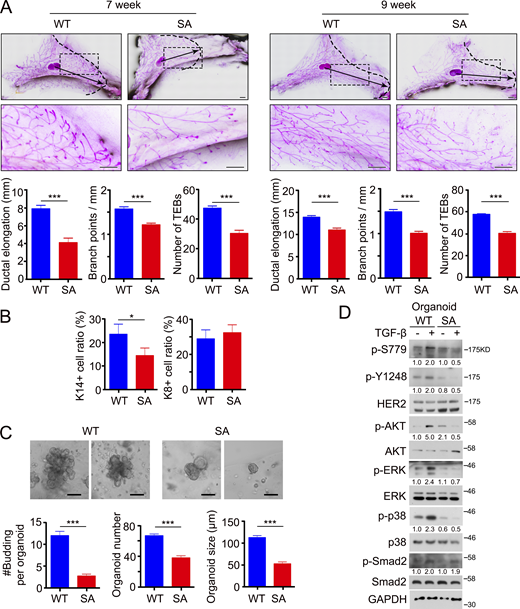
Histological analysis of mammary gland sections did not reveal significant differences between the mammary glands of ERBB2+/+;MMTV-Cre (WT) and ERBB2S780A/+;MMTV-Cre (SA) mice. However, at 7 and 9 wk of age, SA mice exhibited delayed mammary morphogenesis (Fig. 4 A). In WT mice, the epithelium extended into the fat pad, and the mammary glands displayed increased numbers of terminal end buds and branching. In contrast, mammary glands in SA mice exhibited reduced duct extension and branching. As the proportions of basal and luminal epithelial cell populations are linked to branching (Macias et al., 2011), we performed flow cytometry analysis of enzyme-digested mammary glands to assess whether the ratio was altered in the ductal network of SA mice. The relative fraction of CD24+CD29high basal (K14+) mammary epithelial cells was significantly lower in SA mice (13.56%) compared with WT mice (28.91%), while no significant differences were observed in the CD24+CD29low luminal (K8+) mammary epithelial cell populations (Fig. 4 B). The decreased basal/luminal ratio in mammary glands from SA mice correlated with the suppressed epithelial branching (Fig. 4 A) (Macias et al., 2011), underscoring the importance of the HER2 S779 site in mammary epithelial morphogenesis.
The residue S779 in HER2 is important for mammary epithelial morphogenesis. (A) Branching trees of the #4 mammary glands from control (WT: ERBB2+/+;MMTV-Cre) mice and mammary-specific HER2 S779A mutant (SA: ERBB2S780A/+;MMTV-Cre) mice at 7- and 9-wk old. Arrows indicate the extent of ductal penetration in the fat pad. The dotted black line illustrates the epithelial invasion front (n ≥ 3/genotype). Below: Quantitation of ductal elongation, branching and number of TEBs (terminal end buds) between WT and SA glands. Bar, 1 mm. (B) Representative flow cytometry plots and quantification of K14+ basal (CD24+ CD29high) and K8+ luminal (CD24+ CD29low) mammary epithelial cells in WT and SA glands, respectively. (C) Representative images of organoids at day 4 after removing FGF2. Average number of buds (#Budding) per organoid, organoid size, and organoid number per well (24-well plate) were quantified. Mammary fragments from WT and SA mice at 9-wk-old were isolated and cultured for 4 days before removing FGF2. Bar, 100 μm. (D) Immunoblotting of HER2 phosphorylation at S779 and Y1248 sites, AKT, ERK, and p38 phosphorylation in response to 1 h-TGF-β stimulation in the organoids from WT and SA glands. Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F4.
The residue S779 in HER2 is important for mammary epithelial morphogenesis. (A) Branching trees of the #4 mammary glands from control (WT: ERBB2+/+;MMTV-Cre) mice and mammary-specific HER2 S779A mutant (SA: ERBB2S780A/+;MMTV-Cre) mice at 7- and 9-wk old. Arrows indicate the extent of ductal penetration in the fat pad. The dotted black line illustrates the epithelial invasion front (n ≥ 3/genotype). Below: Quantitation of ductal elongation, branching and number of TEBs (terminal end buds) between WT and SA glands. Bar, 1 mm. (B) Representative flow cytometry plots and quantification of K14+ basal (CD24+ CD29high) and K8+ luminal (CD24+ CD29low) mammary epithelial cells in WT and SA glands, respectively. (C) Representative images of organoids at day 4 after removing FGF2. Average number of buds (#Budding) per organoid, organoid size, and organoid number per well (24-well plate) were quantified. Mammary fragments from WT and SA mice at 9-wk-old were isolated and cultured for 4 days before removing FGF2. Bar, 100 μm. (D) Immunoblotting of HER2 phosphorylation at S779 and Y1248 sites, AKT, ERK, and p38 phosphorylation in response to 1 h-TGF-β stimulation in the organoids from WT and SA glands. Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. The band intensity of phosphorylated proteins was normalized to total proteins. Source data are available for this figure: SourceData F4.
Next, we generated organoids derived from mammary gland fragments to investigate the role of the HER2 S779 site in epithelial branching in vitro. Mammary organoids from 9-wk-old WT mice displayed evident branching within 4 d after FGF2 removal, as indicated by the number of buds per organoid, organoid size, and number, while SA organoids had no branching (Fig. 4 C), further supporting the critical function of the HER2 S779 site in mammary epithelial branching. Immunostaining results also revealed the absence of K14+ basal epithelial cells in organoids from SA mice (Fig. S4 C). In addition, we generated WT and SA organoids from sorted mammary basal epithelial cells. WT organoids were larger and had more buds at day 3 after culture, while SA organoids almost had no buds. The results verified the importance of the HER2 S779 site on the proliferation and organoid-forming capacity (Fig. S4 D). Furthermore, TGF-β-induced HER2 phosphorylation at the S779 and Y1248 sites was observed in organoids from WT mice, accompanied by increased activation of AKT, ERK, and p38, but absent in SA organoids (Fig. 4 D) (Donnelly et al., 2014).
TGF-β-enhanced HER2 activation promotes EMT, migration, and lung metastasis of breast cancer cells
To investigate the role of TGF-β-regulated HER2 activity in breast cancer progression, we first examined the effects of TGF-β on epithelial-mesenchymal transition (EMT) and cell migration in mammary epithelial cells and breast cancer cells overexpressing either HER2 WT or S779A mutant. Expectedly, TGF-β-induced EMT was further enhanced in MCF10A and NMuMG cells overexpressing WT HER2, but not S779A mutant, as evidenced by changes in cell morphology, loss of E-cadherin, and upregulation of mesenchymal markers (N-cadherin, Fibronectin, Snail, and PAI-1) (Fig. 5 A and Fig. S4, E–H). Moreover, overexpression of WT HER2, but not S779A mutant, facilitated TGF-β-stimulated migration of MDA-MB-231 cells (Fig. 5, B and C; and Fig. S4, I and J).
TGF-β-enhanced HER2 activation promotes EMT, cell migration, and lung metastasis. (A) MCF10A cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 48 h, followed by immunofluorescence with anti-E-cadherin antibody (green) and rhodamine-phalloidin to stain F-actin (red). The nuclei were counterstained by DAPI (blue). The relative E-cadherin signals were quantified and normalized to the GFP group without TGF-β1 treatment. Bar, 20 μm. (B) MDA-MB-231 cells stably expressed with GFP, WT HER2, or S779A mutant (SA) in Transwell plates were treated with 100 pM TGF-β1 for 36 h. The invasion cells were stained with crystal violet and counted. The number was normalized to the GFP group without TGF-β1 treatment. (C) Subconfluent MDA-MB-231 cells stably expressed with GFP, WT HER2, S779A mutant (SA), or S779D mutant (SD) were wounded by a 1,000-μl pipette tip and then treated with 100 pM TGF-β1 for 48 h, and gap width at 48 h post-wounding was normalized to the one at the start time. (D and E) MDA-MB-231 cells (2 × 105) stably expressed with GFP, WT HER2, or S779A mutant (SA) pretreated with DMSO or 500 pM TGF-β1 for 24 h were injected into tail vein to assess lung metastasis. The color scale depicts the photon flux (photons per second) emitted from the mice (D). n = 8 mice per group. Bioluminescent signal and surface metastases in the lungs were quantified (E). Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments.
TGF-β-enhanced HER2 activation promotes EMT, cell migration, and lung metastasis. (A) MCF10A cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 48 h, followed by immunofluorescence with anti-E-cadherin antibody (green) and rhodamine-phalloidin to stain F-actin (red). The nuclei were counterstained by DAPI (blue). The relative E-cadherin signals were quantified and normalized to the GFP group without TGF-β1 treatment. Bar, 20 μm. (B) MDA-MB-231 cells stably expressed with GFP, WT HER2, or S779A mutant (SA) in Transwell plates were treated with 100 pM TGF-β1 for 36 h. The invasion cells were stained with crystal violet and counted. The number was normalized to the GFP group without TGF-β1 treatment. (C) Subconfluent MDA-MB-231 cells stably expressed with GFP, WT HER2, S779A mutant (SA), or S779D mutant (SD) were wounded by a 1,000-μl pipette tip and then treated with 100 pM TGF-β1 for 48 h, and gap width at 48 h post-wounding was normalized to the one at the start time. (D and E) MDA-MB-231 cells (2 × 105) stably expressed with GFP, WT HER2, or S779A mutant (SA) pretreated with DMSO or 500 pM TGF-β1 for 24 h were injected into tail vein to assess lung metastasis. The color scale depicts the photon flux (photons per second) emitted from the mice (D). n = 8 mice per group. Bioluminescent signal and surface metastases in the lungs were quantified (E). Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments.
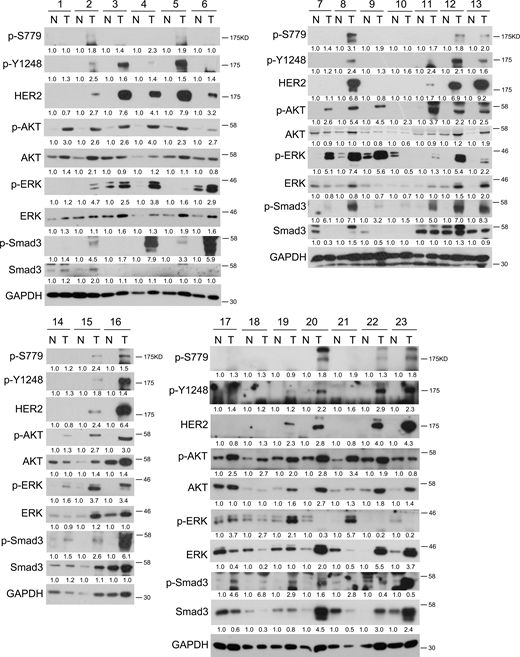
Intravenous injection of TGF-β-treated MDA-MB-231 cells resulted in lung metastasis, and the overexpression of WT HER2, but not HER2 (S779A) mutant, further promoted metastasis (Fig. 5, D and E). These findings underscore the important function of the HER2 S779 site in TGF-β-induced EMT, cell migration, and lung metastasis of breast cancer cells. We further examined the levels of S779 and Y1248 phosphorylation of HER2 in human breast cancer samples. Compared with adjacent normal tissues, cancer tissues exhibited significantly higher levels of HER2 and its phosphorylation (p-S779 and p-Y1248), along with increased levels of C-terminally phosphorylated Smad3, p-AKT, and p-ERK (Fig. 6 A and Fig. S5). This positive correlation between activated TGF-β signaling and HER2 S779 phosphorylation suggests a promoting role of the TGF-β-HER2-AKT/MAPK axis in breast cancer progression.
The TGF-β-HER2-AKT/MAPK axis promotes breast cancer progression. (A) The correlation among HER2 p-S779, HER2 p-Y1248, p-AKT, p-ERK, or p-Smad3 levels in breast cancer patient samples and their adjacent normal tissues was analyzed with GraphPad Prism 5.0 (the Pearson test) after band intensity quantification. (B) A model depicting how the TGF-β-HER2-AKT/MAPK axis promotes EMT, cell migration, and lung metastasis of breast cells. Activated TβRI directly phosphorylates HER2 at the S779 site, which enhances the autophosphorylation of HER2 at Y1248 and thus HER2 activation, leading to activation of PI3K/AKT and ERK and promotion of EMT, cell migration, and metastasis of breast cells.
The TGF-β-HER2-AKT/MAPK axis promotes breast cancer progression. (A) The correlation among HER2 p-S779, HER2 p-Y1248, p-AKT, p-ERK, or p-Smad3 levels in breast cancer patient samples and their adjacent normal tissues was analyzed with GraphPad Prism 5.0 (the Pearson test) after band intensity quantification. (B) A model depicting how the TGF-β-HER2-AKT/MAPK axis promotes EMT, cell migration, and lung metastasis of breast cells. Activated TβRI directly phosphorylates HER2 at the S779 site, which enhances the autophosphorylation of HER2 at Y1248 and thus HER2 activation, leading to activation of PI3K/AKT and ERK and promotion of EMT, cell migration, and metastasis of breast cells.
High levels of HER2 S779 phosphorylation are detected in breast cancer samples. 23 pairs of breast cancer samples (T) and their adjacent normal tissues (N) were harvested for immunoblotting. Sample 1 and 9 are triple-negative breast cancer; Sample 10 and 18 are Luminal A subtype of breast cancer; Sample 2, 4, 7, 11, 12, 15, 17, 20, 21, and 22 are Luminal B subtype of breast cancer; Sample 3, 5, 6, 8, 13, 14, 16, 19, and 23 are HER2-positive breast cancer. The band intensity of phosphorylated proteins was normalized to total proteins. The band intensity of total proteins was normalized to GAPDH. Source data are available for this figure: SourceData FS5.
High levels of HER2 S779 phosphorylation are detected in breast cancer samples. 23 pairs of breast cancer samples (T) and their adjacent normal tissues (N) were harvested for immunoblotting. Sample 1 and 9 are triple-negative breast cancer; Sample 10 and 18 are Luminal A subtype of breast cancer; Sample 2, 4, 7, 11, 12, 15, 17, 20, 21, and 22 are Luminal B subtype of breast cancer; Sample 3, 5, 6, 8, 13, 14, 16, 19, and 23 are HER2-positive breast cancer. The band intensity of phosphorylated proteins was normalized to total proteins. The band intensity of total proteins was normalized to GAPDH. Source data are available for this figure: SourceData FS5.
Discussion
In addition to the well-established TGF-β-Smad pathway, non-Smad effectors also play a crucial role in TGF-β-mediated pathophysiological events (Moustakas and Heldin, 2005). The adaptor proteins ShcA and TRAF6 have been shown to interact with TβRI to transmit TGF-β signals and activate downstream ERK/MAPK and AKT kinases (Hong et al., 2011; Northey et al., 2008; Yamashita et al., 2008; Yu et al., 2002). As HER2/EGFR signaling also activates AKT and MAPKs, the interplay between TGF-β and HER2/EGFR signals in activating these kinases is not fully understood. HER2/EGFR signaling and TGF-β signaling are known to interact and modulate each other at multiple levels (Chow et al., 2011; Shi and Chen, 2017). TGF-β signaling can influence the expression levels of ligands and receptors involved in HER2/EGFR signaling and enhance its oncogenic functions, such as promoting cell survival, migration, and drug resistance. On the other hand, HER2/EGFR signaling can regulate the expression of TGF-β ligands, modulate Smad activity, and affect Smad-dependent transcription. Our study reveals that TGF-β enhances HER2 activity to activate AKT and MAPKs, which requires TβRI. These findings provide an insight into the crosstalk between TGF-β and HER2/EGFR pathways in cellular responses. The TGF-β-HER2-AKT/ERK axis and the HER2/EGFR-AKT-Smad3 axis (Huang et al., 2018) upregulates EMT, migration, and lung metastasis to promote breast cancer malignancy, emphasizing the synergy and interplay of TGF-β-SMAD and non-SMAD pathways during this process (Ali et al., 2023; Hao et al., 2019; Tsubakihara and Moustakas, 2018).
We found that activated TβRI directly phosphorylates HER2 at the S779 site in the αC-β4 loop of its kinase domain. This phosphorylation event enhances the autophosphorylation of HER2 at Y1248, leading to its kinase activation. Mutations in the αC-β4 loop of HER2 have been shown to induce conformational changes in both the αC-helix and activation loop, which are key regulatory elements for kinase activity (Bose and Zhang, 2009; Fan et al., 2008; Huse and Kuriyan, 2002). Additionally, the formation of asymmetrical dimers has been proposed as a contributing factor to HER2 activation (Zhang et al., 2006). It is unclear whether S779 phosphorylation influences the conformational changes of the αC-helix and activation loop, or the potential role of HER2 dimer formation, which needs further investigation.
Activating mutations in ERBB2 have been detected in multiple cancer types (Connell and Doherty, 2017; Zheng et al., 2023). ERBB2 exon 20 mutations, including the S779 location, are frequently found in the small intestine, lung, and breast cancers (Robichaux et al., 2019). These mutations often confer resistance to many tyrosine kinase inhibitors (TKIs) (Nagano et al., 2018; Robichaux et al., 2018). Unfortunately, there are currently no approved targeted therapies specifically tailored to tumors harboring ERBB2 mutations. HER2 (G776S) mutation was reported in gastric and colorectal cancer (Mitani et al., 2022; Stephens et al., 2004). Additionally, HER2 S779F mutation was identified in breast carcinoma (Ding et al., 2019). Consistently, we observed elevated levels of HER2 p-S779 in breast cancer tissues, which positively correlated with HER2 p-Y1248, p-Smad3, p-AKT, and p-ERK. These data indicate that S779 could serve as a novel therapeutic target in HER2-expressing cancer cells.
Materials and methods
Generation of ERBB2CKI-S780A KI mice and ERBB2S780A/+; MMTV-Cre mice
The sequences of the two sgRNAs targeting the exon 20 of ERBB2 were 5′-CCCCCTTTAACCTCCTCCTT-3′ and 5′-TCCATATGTGTCCCGCCTCC-3′. ERBB2CKI-S780A KI mice that can conditionally express the ERBB2S780A allele were commercially generated by CRISPR/Cas9 in GemPharmatech Co., Ltd. In the genetically engineered mice, we replaced the ERBB2 exon 20 with a targeting vector composed of a LoxP-flanked cDNA cassette coding for exon 20–27 and a transcriptional stop cassette. The modified wild-type exon 20 was followed by a copy of ERBB2 exon 20 encoding the S780A mutation (TCT to GCC). Expression of the mutant allele was induced by Cre recombinase. Cas9, sgRNAs, and donor vector were simultaneously injected into the fertilized eggs. Mammary tissue-specific ERBB2S780A knock-in was achieved by intercrossing mice carrying ERBB2S780A and MMTV-Cre alleles. Knock-in was confirmed by genomic DNA sequencing. For the ERBB2S780A KI allele, the PCR product showed a 937, 1,267, and 750 bp band respectively produced by primer set1 (F1: 5′-ACAGTGCAGATTAACAAGGTGGGTT-3′; R1:5′-GTGATCTTCACAATCCCAATAACTTCG-3′), primer set2 (F2: 5′-CGGAACCATAACTTCGTATAGCATAC-3′; R2: 5′-TTCTCCAGCAAATCAGGGATCT-3′), and primer set3 (F3: 5′-ACGGCTACTCCTCAGTGTCTTAATG-3′; R3: 5′-CAAAGCACATACCTTGGCAATCTG-3′), while for the endogenous ERBB2 allele, the PCR product showed a single 750 bp band produced by primer3. The primer sets Cre-F (5′-ATTTGCCTGCATTACCGGTC-3′) and Cre-R (5′-ATCAACGTTTTCTTTTCGG-3′) were used to genotype the MMTV-Cre allele.
Plasmids, antibodies, and reagents
The human HER2 was subcloned into pcDNA-4TO-myc and pEGFP-N1 plasmids. Human TβRI cDNA was subcloned into pCMV5-HA plasmid. The constitutively active TβRI (GGD) harbors three point mutations (L193G, P194G, and T204D) (Chen et al., 1997). CaTβRI were generated from the intracellular domain of GGD (148 aa–503 aa). The validity of all constructs was confirmed by DNA sequencing.
Antibodies were used in this study were as follows: mouse anti-HER2 (Cat# MA5-13675; Thermo Fisher Scientific); rabbit anti-p-HER2 Y1248 (Cat# BS4090; Bioworld); rabbit anti-p-HER2 Y877 (Cat# BS4091; Bioworld); rabbit anti-p-serine (Cat# 61-8100; ZYMED); mouse anti-Fibronectin (Cat# 610077), mouse anti-E-Cadherin (Cat# 610181), and mouse anti-N-Cadherin (Cat# 610920) (BD Biosciences); rabbit anti-p-Smad2 (Cat# 566415; Millipore); rabbit anti-p-Smad3 (Cat# 9520s), rabbit anti-Smad3 (Cat# 9523s), rabbit anti-Smad2 (Cat# 5339), rabbit anti-AKT (Cat# 9272s), rabbit anti-p-AKT S473 (Cat# 4060s), rabbit anti-p-p38 (Cat# 4511), rabbit anti-p38 (Cat# 8690), rabbit anti-p-PLC-γ1 Y783 (D6M9S) (Cat# 14008), rabbit anti-PLC-γ1 (D9H10) (Cat# 5690), rabbit p-EGFR Y1173 (Cat# 4407s), mouse anti-myc (Cat# 2276), rabbit anti-HA (Cat# 3724T), and rabbit anti-Snail (Cat# 3879s) (Cell Signaling Technology); anti-p-tyrosine (PY20) HRP (Cat# sc-508), mouse anti-p-ERK (Cat# sc-7383), rabbit anti-ERK (Cat# sc-93), rabbit anti-TβRI (V22) (Cat# sc-398), and rabbit anti-PAI-1 (Cat# sc-8979) (Santa Cruz Technology); anti-Flag (Cat# bsm-33346M; Bioss Antibodies); rat anti-K8 (TROMA-1, DSHB); rabbit anti-K14 (Cat# ab181595; Abcam); and APC anti-mouse/rat CD29 (Cat# 102215; BioLegend), PE/Cyanine7 anti-mouse CD24 (Cat# 101821; BioLegend).
To generate the rabbit polyclonal antibody against phosphoSer779 on HER2, phosphorylated and nonphosphorylated peptides (CAGVG{pSer}PYVSR and CAGVGSPYVSR) (20 mg, ≥90% purity) were designed and synthesized by MBL BEIJING BIOTECH CO., LTD, followed by coupling with carrier protein. The rabbits were immunized with the phosphorylated peptide. The polyclonal antibody against phosphor-Ser779 was purified from the serum against the nonphosphorylated peptide.
Lapatinib (S2111), CP-724714 (S1167), SB-431542 (S1067), and Y27632 (S1049) were from Selleck. Recombinant human TGF-β1 was purchased from R&D Systems, EGF was from Invitrogen, and FGF2 was from Thermo Fisher Scientific.
Cells and transfection
HEK293T was from ATCC. Mouse mammary epithelial NMuMG cells were a gift from Dr. Ying E. Zhang (National Cancer Institute, NIH, Bethesda, MD, USA), and MCF-10A cells were a gift from Dr. Xiao-Fan Wang (Duke University School of Medicine, Durham, NC, USA) and Dr. Yongfeng Shang (Peking University, Beijing, China). Other cell lines were from China Infrastructure of Cell Line Resource (Beijing). All the cell lines had been confirmed without mycoplasma contamination. MDA-MB-231 and H1299 were cultured in RMPI1640 (Gibco) with 10% fetal bovine serum (FBS) (Hyclone); SKBR3, HEK293T, NMuMG, EpH4, BT474, HaCaT, Hela, NIH3T3, A549, and KYSE410 cells in DMEM (Gibco) with 10% FBS; HepG2 cells in MEM (Gibco) with 10% FBS; MCF-10A cells in DMEM/F12 (Gibco) supplemented with 5% horse serum (Hyclone), 20 ng/ml EGF, 0.5 μg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), and 10 μg/ml insulin.
Transfection of DNA plasmids into HEK293T, NMuMG, MCF10A, and MDA-MB-231 cells was performed with Lipofectamine 2000 (Invitrogen) or Vigofect (Vigorous Biotechnology). ShRNAs of HER2 were expressed in breast cells with a lentivrial system (Sigma-Aldrich), and the shRNA sequences were 5′-CCGGTGTCAGTATCCAGGCTTTGTACTCGAGTACAAAGCCTGGATACTGACATTTTTG-3′ and 5′-CCGGCAGTGCCAATATCCAGGAGTTCTCGAGAACTCCTGGATATTGGCACTGTTTTTG-3′, and the stable cell lines were selected with 0.3 μg/ml puromycin (Sigma-Aldrich). Wild-type HER2 or its mutants were cloned into pENTR1A plasmid under the control of CMV promoter in the p2k7bsd lentiviral backbone (a gift from Dr. Kehkooi Kee at Tsinghua University, Beijing, China). The viral supernatants were used to infect breast cell lines. Blasticidin (3 μg/ml, Sigma-Aldrich) was added to the culture medium for 5 days to select cells with stable viral integration. Genes expressing the intracellular domain of activated TβRI (148 aa–503 aa, caTβRI), the kinase domain of HER2 (712 aa–990 aa, KD), or the intracellular domain of HER2 (712aa-1255aa, C-term) were PCR-amplified and cloned into pCAG-C-TEV-Strep-Flag vector (a gift from Dr. Li Yu at Tsinghua University, Beijing, China) to produce Strep-tag- and Flag-tag-fused recombinant proteins. Point mutations were introduced by the site-directed mutagenesis approach.
Immunoblotting, immunoprecipitation, immunofluorescence
These experiments were described as previously reported (Zuo and Chen, 2009). Cells for immunoprecipitation (IP) were lysed on ice with lysis solution (50 mM Tris˙HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 10 mM NaF, 1 mM sodium orthovanadate dodecahydrate, and protease inhibitors) and rotated for >30 min at 4°C. The cell lysates were cleared by centrifugation at 16,000 g. After an aliquot was taken for protein expression analyses, the left cell lysates containing equivalent amounts of total proteins were precleared with protein A-Sepharose (GE Healthcare) at 4°C. Immmunoprecipitation was carried out by the addition of appropriate antibodies and protein A-Sepharose followed by incubation at 4°C overnight with gentle rotation. Then the immune complex was isolated by centrifugation and repeated washes with lysis buffer, analyzed by SDS-PAGE and immunoblotting, and detected with the enhanced chemiluminescent substrate (Pierce) according to the manufacturer’s instructions.
For immunofluorescence, cells were grown on coverslips, fixed with 4% paraformaldehyde (PFA) for 20 min, permeabilized for 5 min with 0.1% Triton X-100 in phosphate-buffered saline (PBS, pH 7.0), and blocked with 5% bovine serum albumin in PBS (pH 7.4) for 30 min at room temperature. The cells were then incubated with primary antibodies overnight, followed by fluorescein isothiocyanate (FITC)- or Tetramethylrhodamine (TRITC)-conjugated secondary antibodies for 1 h at room temperature. The nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). The following primary antibodies were used: anti-p-Y1248 (1:100), anti-HER2 (1:300), and anti-E-cadherin (1:300). One drop of Antifade Mounting Medium (Cat# P0126; Beyotime Biotechnology) was added to the slide and the coverslip containing cells was carefully covered. Images were obtained at room temperature using the NIKON A1 HD25 microscope equipped with a 100×/1.45 NA oil objective (WD 0.13 mm) at 1,024 × 1,024 pixels. Images were analyzed with NIS-Elements AR Analysis and processed with Adobe Photoshop CC 2017 software.
Protein expression and purification and SPR
Genes expressing the intracellular domain (ICD) of WT or activated (GGD) TβRI (148 aa–503 aa) and HER2 ICD (712 aa–1255 aa) were PCR-amplified and cloned into pETMBP.3C vector to produce MBP-tag-fused recombinant proteins. All recombinant proteins were expressed in E. coli BL21-CodonPlus (DE3) with induction by 1 M IPTG for 16 h at 18°C. Following induction, E. coli cells were resuspended in binding buffer (50 mM Tris-Cl, pH 7.9, 1 M NaCl, and 10 mM imidazole), lysed with a high-pressure homogenizer, and sedimented at 18,000 rpm for 30 min. The supernatant lysates were purified on Amylose Resin (NEB). After extensive washing with binding buffer, proteins were eluted with MBP elution buffer (50 mM Tris-Cl, pH 7.9, 1 M NaCl, and 10 mM imidazole for MBP-tagged proteins), then purified with a HiPrep 26/60 Sephacryl S-200 HR column (17-1195-01; GE Healthcare) on an AKTA purifier (GE Healthcare), and eluted with PBS buffer for SPR experiment.
HER2 ICD was immobilized on Series S sensor chips CM5 (GE Healthcare Bio-Sciences), and the binding experiments were carried out using a Biacore S200. TβRI WT or GGD ICD solutions were diluted in PBS to the final concentrations of 31.25, 15.63, 7.81, 3.91, 1.95, and 0.98 nM. Diluted samples were injected in duplicate over each immobilized protein for 60 s at a flow rate of 30 μl/min. The running buffer was then flushed for 60 s at a flow rate of 30 μl/min. Control samples were used to monitor the sensor chip surface stability. The estimated Kd values were derived by fitting the association and dissociation signals with a 1:1 (Langmuir) model using the Biacore S200 Evaluation Software.
Wound-healing and transwell assays
For wound healing assay, the confluent cell monolayer in a six-well plate was wounded by manually scraping the cells with a 1,000-μl pipette tip and then treated with 100 pM TGF-β1 for the indicated time. Cell migration into the wound surface was monitored in the culture medium at room temperature by the 10 × 10 microscopy (Roper Scientific) at various times. Quantitation was done by measuring the distance of the wound edge of the migrating cells from the start point to the migrated point from three independent experiments. For transwell assay, cells were plated in a medium with 1% FBS in the upper chamber of a transwell plate (Corning), while the media containing 10% FBS ± TGF-β1 was placed in the lower well. After incubation for the indicated time, fixed with methanol for 20 min, stained with crystal violet dye for 30 min, and photographed in PBS at room temperature under the 10 × 10 microscope (Roper Scientific). The images were then processed with Adobe Photoshop CC 2017 software. Experiments were carried out at least three times.
Quantitative real-time RT-PCR
Total cell RNA was isolated with TRIzol (Invitrogen), and cDNA was synthesized with Revertra Ace (Toyobo). Real-time PCR was performed with a Roche480 system (Roche). Expression values were normalized to GAPDH expression. The PCR oligo sequences of Fibronectin are 5′-CCATCGCAAACCGCTGCCAT-3′ and 5′-AACACTTCTCAGCTATGGGCTT-3′. The PCR oligo sequences of PAI-1 are 5′-ACCGCAACGTGGTTTTCTCA-3′ and 5′-TTGAATCCCATAGCTGCTTGAA-3′.
In vitro phosphorylation assay
Flag-tagged caTβRI protein (100 ng) was incubated with 2 μg wild-type Flag-tagged HER2 (KD) or its S779A mutant in a 50-μl reaction buffer containing 25 mM Tris.HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 5 mM ATP for 1 h at room temperature. 100 ng Flag-tagged CaTβRI protein was incubated with 2 μg wild-type Flag-tagged HER2 (C-term) or its S779A mutant in 50 μl-reaction buffer for 1 h at room temperature. The reaction was stopped by adding the SDS loading buffer. HER2 phosphorylation was detected by immunoblotting.
Tumor metastasis assays
For lung metastasis formation, 2 × 105 MDA-MB-231 cells stably expressing firefly luciferase expressed with GFP, wild-type HER2 or HER2 (S779A) mutant pretreated with DMSO or 500 pM TGF-β for 24 h were injected into the lateral tail vein of nude mice (n = 8 per group) in a volume of 0.1 ml. Bioluminescence images were collected after 30 days by using the Lumina II imaging system (PerkinElmer). The color scale depicts the photon flux (photons per second) emitted from xenografted mice. After scarification, the lungs were excised for metastatic foci analysis, and the number of metastatic nodules of each group was analyzed.
Whole mount carmine staining of mammary glands
Mammary glands were harvested and fixed with 4% PFA overnight, washed with PBS for 5 min, rehydrated with 70%, 35%, and 17% ethanol for 15 min respectively, and immersed in flowing water for 10 min followed by carmine staining buffer overnight. The following day, mammary glands were decolored using 1% HCl in 70% ethanol for 10 min, dehydrated with 70%, 90%, and 100% ethanol for 1 h respectively, degreased with xylene until the mammary fat pad was transparent, and finally submerged in methyl salicylate for storage. Images were captured at room temperature using the NIKON Ti2E microscope with NIKON DS-Ri2 equipped with a 4×/0.2 NA objective (WD 20 mm), stitched together to show the entire tissue section, analyzed with NIS-Elements AR Analysis and processed with Adobe Photoshop CC 2017 software.
Organoid generation from mammary gland
Mouse mammary glands were finely chopped and the mince was digested in collagenase buffer (RPMI 1640 with 2 mg/ml collagenase I [Invitrogen] and 2,000 U/μl DNase I [Kaien Biotechnology]) for 30 min at 37°C. Next, DMEM/F12 (Corning) with 5% FBS (Hyclone) was added to stop the reaction, gently mixed, and centrifuge at 450 g for 5 min to obtain a pellet containing mammary epithelial cells and fibroblasts. After 5 min of treatment with red blood cell lysing buffer (Sigma-Aldrich) at room temperature, pellets were washed with DMEM/F12 and subjected to a short-pulse centrifugation. The supernatant was collected and pellets were resuspended in DMEM/F12 for another round of differential centrifugation. After five rounds of differential centrifugation, pellets containing mammary organoids were resuspended with 30 μl 100% growth factor-reduced Matrigel (Corning) and placed in a 24-well plate and allowed to solidify for 15 min at 37°C in a 5% CO2 incubator before top up with 1 ml organoid culture medium (DMEM/F12 supplemented with 1% penicillin-streptomycin [Thermo Fisher Scientific], 1% ITS [Thermo Fisher Scientific], 50 ng/ml EGF, 2.5 ng/ml FGF2 and 10 μM Y27632). FGF2 was only included in the culture medium for the first 4 days. The images of organoids (in the culture medium) were captured at 37°C supplemented with 5% CO2 with a NIKON A1 HD25 microscope equipped with a 10× objective/0.45 NA objective (WD 4 mm), analyzed with NIS-Elements AR Analysis, and processed with Adobe Photoshop CC 2017 software.
Immunostaining of cultured organoids
Matrigel-containing organoids were gently lifted loose from the bottom of the well by blunted pipette tip and then fixed with 4% PFA for 15 min, permeabilized for 1 h with 1% Triton X-100 and 3% BSA in PBS, blocked, and incubated with primary antibodies (anti-K14: 1:50; anti-K8: 1:50) in PBS containing 0.01% Triton X-100 and 3% BSA overnight at 4°C, followed by FITC- or TRITC-conjugated secondary antibodies overnight at 4°C. The nuclei were counterstained with DAPI (1 mg/ml, 1:500). Stained organoids were transferred to the slide using a cut pipette tip and mounted with an antifade mounting medium. The stained organoid images were captured at room temperature with a NIKON A1 HD25 microscope equipped with a 10× objective/0.45 NA objective (WD 4 mm), analyzed with NIS-Elements AR Analysis, and processed with Adobe Photoshop CC 2017 software.
Fluorescence-activated cell sorting and flow cytometric analysis
Mouse mammary glands were finely chopped and the mince was digested in collagenase buffer (RPMI 1640 with 2 mg/ml collagenase I [Invitrogen] and 2,000 U/μl DNase I [Kaien Biotechnology]) for 30 min at 37°C. Next, DMEM/F12 (Corning) with 5% FBS (Hyclone) was added to stop the reaction, gently mixed, and centrifuged at 450 g for 3 min to obtain a pellet containing mammary epithelial cells and fibroblasts. The supernatant was removed and washed with PBS, gently mixed, and centrifuged at 450 g for 3 min to get the pellet. The pellets were digested with tryple (Beyotime Biotechnology) at 37°C for 15 min (in a 60-mm dish). DMEM/F12 with 5% FBS was added to stop the reaction, gently mixed, and centrifuged at 450 g for 3 min. The digested cells were washed with PBS, followed by filtering with a 40-µm strainer. Single-cell suspensions were stained for CD24 (1:100) and CD29 (1:100) in PBS for 30 min at 4°C, followed by staining with DAPI (1 mg/ml, 1:500) in PBS for Live/Dead cell staining.
Organoids generation from sorted basal epithelial cells
Freshly isolated mammary tissues were trypsinized to obtain single-cell suspension, stained for CD24 (1:100) and CD29 (1:100) in PBS for 30 min at 4°C, followed by staining with DAPI (1 mg/ml, 1:500) in PBS for Live/Dead cell staining. Basal epithelial cells (CD24medCD29hi) were sorted and resuspended with 20 μl 100% growth factor-reduced Matrigel (Corning) and placed in a 48-well plate (5,000 cells per well) and allowed to solidify for 15 min at 37°C in a 5% CO2 incubator before top up with 500 μl organoid culture medium. The images of organoids (in the culture medium) were captured at room temperature under the 10 × 10 microscope (Roper Scientific) and then processed with Adobe Photoshop CC 2017 software.
Human breast cancer samples
A total of 23 pairs of breast cancer specimens (2 Luminal A: ER+PR+HER2−, 10 Luminal B: ER+PR+HER2+, 9 HER2+: ER−PR−HER2+, and 2 triple-negative breast cancer [TNBC]: ER−PR-HER2−), mostly from invasive breast carcinomas, were analyzed by immunoblotting. All tumors were primary and untreated before surgery. All subjects received radical mastectomy or modified radical mastectomy. The use of clinical samples was approved by the Institutional Review Board at the First Affiliated Hospital of Nanchang University. Informed consent was obtained from all subjects or their relatives.
Statistical analysis
Unpaired, two-sided Student’s t test was used. *P < 0.05 were considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001. For the bar charts, data were plotted as mean ± SD of at least three independent experiments. For immunoblot gel quantification, the band intensity of phosphorylated proteins normalized to the total proteins, and the band intensity of total proteins was normalized to GAPDH after the gels were scanned and band intensity was qualified with ImageJ.
Online supplemental material
Fig. S1 shows that inhibition of HER2 impairs TGF-β-induced activation of AKT and ERK. Fig. S2 shows that TGF-β-enhanced HER2 S779 phosphorylation is important for the activation of HER2 AKT and ERK. Fig. S3 shows that enhancement of HER2 activity by TGF-β is observed in multiple epithelial cells and cancer cells. Fig. S4 shows that the residue S779 of HER2 is crucial for maintaining mammary epithelium branching and for promoting EMT and cell migration. Fig. S5 shows that high levels of HER2 S779 phosphorylation are detected in breast cancer samples.
Data availability
All data used in this study are available upon request.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31988101 to Y.-G. Chen, 32100566 to Q. Shi) and the Natural Science Foundation of Jiangxi Province (20224ACB209001 to Y.-G. Chen).
Author contributions: Conceptualization: Q. Shi, F. Huang, and Y.-G. Chen. Data curation: Q. Shi and Y.-G. Chen. Formal analysis: Q. Shi and Y.-G. Chen. Funding acquisition: Q. Shi and Y.-G. Chen. Investigation: Q. Shi, F. Huang, and Y.-G. Chen. Methodology: Q. Shi, H.T. Deng, and Y.-G. Chen. Project administration: Q. Shi and Y.-G. Chen. Supervision: Y.-G. Chen. Writing–original draft: Q. Shi and Y.-G. Chen. Writing–review & editing: Q. Shi, H. Liu, Y. Wang, and Y.-G. Chen.
References
Author notes
Disclosures: The authors declare no competing interests exist.




![Inhibition of HER2 impairs TGF-β-induced activation of AKT and ERK. (A) The relative p-Y1248, p-AKT, p-ERK, and p-Smad2 levels of three repeated experiments in Fig. 1 A were quantified. (B and C) Cells were treated as indicated (MDA-MB-231 cells: 5 μM lapatinib; BT474 cells: 0.01 μM lapatinib; SKBR3 cells: 1 μM CP-724714 [CP]; NMuMG cells: 10 μM CP-724714) for 2 h, and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (D) NMuMG (Left) and SKBR3 (Right) cells were treated with 2 or 0.01 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h, followed by anti-HER2 Y1248 immunofluorescence (red) and anti-HER2 immunofluorescence (green). The nuclei were counterstained by DAPI (blue). Bar, 20 μm. (E) NMuMG cells were treated with 2 μM lapatinib for 2 h and 100 pM TGF-β1 was added in the last 1 h. Cells were then harvested for immunoblotting. (F) HEK293T cells were transfected with indicated plasmids before being harvested for anti-myc pulldown and then for anti-p-Y1248 immunoblotting. (G) MCF10A or SKBR3 cells were treated with 5 μM SB-431542 (SB) for 2 h, 100 pM TGF-β1 was added in the last 1 h before being harvested for immunoblotting. (H and I) HEK293T cells were transfected with indicated plasmids before being harvested for anti-HA pulldown and then for immunoblotting. (J) NMuMG cells were treated with 100 pM TGF-β1 for the indicated time before being harvested for anti-TβRI pulldown and then for immunoblotting. (K) MCF10A cells were treated with 100 pM TGF-β1 for 1 h before being harvested for anti-TβRI pulldown and then for immunoblotting. (L) The purified MBP-tagged proteins (1, WT TβRI ICD; 2, GGD-ICD; 3, HER2 ICD) are shown with asterisks. M: Protein markers. The band intensity of phosphorylated proteins was normalized to total proteins. The band intensity of pull-down HER2 was normalized to pull-down TβRI. Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. Source data are available for this figure: SourceData FS1.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/4/10.1083_jcb.202307138/1/m_jcb_202307138_figs1.png?Expires=1773744616&Signature=Jgw5NrtBqVEsuvW9npJKuh1Etg3B810LF74bX6hKtVm-8FQx-g1xLs7wNoshjHhAV-I5ozDlq5b~6yJKGAQ6dgSD6t9pxfAnbGVkKFWua5DWoB3AYrMyCE3P-N7LqRuUmppQIllwCpqhbrr6qf0nz-wHrbEa4xUtZkkCSRL-PyaQPNUWUQhtn1itXXkKoCRNHuBzUzOX9HBPocUwCV7K7bQzWjsM0NWK3OSDb-~-nAgES5C7dA2D-ebimzqfRocnfJl6Ufh70oKar8x-HlF2j9TBt4YLrCxB7f9Ny5kfW-38D6jXkOXyKfHV1DOeXZBxN5xSiMe-EY-ExNV-873ddg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![The residue S779 of HER2 is crucial for maintaining mammary epithelium branching and for promoting EMT and cell migration. (A) Schematic diagram of knock-in donor vector to generate inducible ERBB2CKI-S780A KI mice. The red asterisk marks the location of the S780A mutation corresponding to the human S779A mutation. ERBB2CKI-S780A knock-in allele can be generated after Cre-mediated excision of the floxed wide-type ERBB2 cDNA coding for exon 20–27. (B) Schematic summary of three primer sets and DNA gel patterns of the different genotypes ([1] ERBB2+/+;MMTV-Cre with positive band of ③and④; [2] ERBB2S780A/+ with positive band of ①,②,and③; [3] ERBB2S780A/+;MMTV-Cre with positive band of ①,②,③,and④; [4] Negative control with no positive band). M: DNA marker. The thickened dark line indicates the sequencing region. (C) Representative images showing reduced branching and distribution of K14+ basal epithelial cells in organoids from the SA group. Bar, 100 μm. (D) Representative images of organoids derived from sorted basal epithelial cells (CD24medCD29hi) (5,000 cells per well in a 48-well plate) at day 3 after culture. The average number of buds per organoid (#budding) and organoid size were quantified. Bar, 500 μm. (E) The expression levels of WT HER2 and SA mutant in MCF10A cells (Fig. 5 A). (F) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with DMSO or 100 pM TGF-β1 for 48 h and imaged before subjected to immunoblotting with indicated antibodies. (G) mRNA levels of Fibronectin and PAI-1 were examined by quantitative real-time RT-PCR. (H) NMuMG cells overexpressed with GFP, WT HER2, or S779A mutant (SA) were treated with 100 pM TGF-β1 for 48 h, followed by immunofluorescence with anti-E-cadherin antibody (green) and rhodamine-phalloidin to stain F-actin (red). The nuclei were counterstained by DAPI (blue). The relative E-cadherin signals were quantified and normalized to the GFP group without TGF-β1 treatment. Bar, 20 μm. (I) The expression levels of WT HER2 and SA mutant in MDA-MB-231 cells (Fig. 5 B). (J) Subconfluent MDA-MB-231 cells stably expressed with GFP, WT HER2, S779A (SA), or S779D mutant (SD) were wounded by a 1,000-μl pipette tip, and then treated with 100 pM TGF-β1 for 48 h and imaged (Left). Right is the expression levels of WT HER2, SA and SD mutants in MDA-MB-231 cells (Fig. 5 C). Statistical analyses were performed with Student’s t test (two-sided, *P < 0.05, **P < 0.01, ***P < 0.001). For the bar charts, data were plotted as mean ± SD of at least three independent experiments. The band intensity of each protein was normalized to the first group. Source data are available for this figure: SourceData FS4.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/4/10.1083_jcb.202307138/1/m_jcb_202307138_figs4.png?Expires=1773744616&Signature=gNMl68KzWxjxqibMyurYpKCPaA2VgPLXsIuScQOAW6jvENukpAqZtga1aUsgFxQwOWlpgYp4elpTuO1mPfK0epaYrrdeneKwj19CB14g-etkgWp0fsCAqlJbToTeC~Ry~No4e9dOqRgWmSqLjlhUfEpE30j-q3VesH9GNiIuewVYBEgDXyEZZYMoTB7ddW7WTm7sK-QEerOhvPoy338a5cE7wJEIt3VqMAPIGpo63qU8A7s7DYbTa0uxCp33Ki57mwW5d272ve21TNSkpuolPcJRaFnrVoq8OnWDdZdQy~T9z~z5MKyXSiEETASz9PXV-JW86DPoaepRNY4alwFNMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)