To breach the basement membrane, cells in development and cancer use large, transient, specialized lipid-rich membrane protrusions. Using live imaging, endogenous protein tagging, and cell-specific RNAi during Caenorhabditis elegans anchor cell (AC) invasion, we demonstrate that the lipogenic SREBP transcription factor SBP-1 drives the expression of the fatty acid synthesis enzymes POD-2 and FASN-1 prior to invasion. We show that phospholipid-producing LPIN-1 and sphingomyelin synthase SMS-1, which use fatty acids as substrates, produce lysosome stores that build the AC’s invasive protrusion, and that SMS-1 also promotes protrusion localization of the lipid raft partitioning ZMP-1 matrix metalloproteinase. Finally, we discover that HMG-CoA reductase HMGR-1, which generates isoprenoids for prenylation, localizes to the ER and enriches in peroxisomes at the AC invasive front, and that the final transmembrane prenylation enzyme, ICMT-1, localizes to endoplasmic reticulum exit sites that dynamically polarize to deliver prenylated GTPases for protrusion formation. Together, these results reveal a collaboration between lipogenesis and a polarized lipid prenylation system that drives invasive protrusion formation.
Introduction
During animal development, many cells migrate to form organs and tissues (Aman and Piotrowski, 2010; Scarpa and Mayor, 2016). For example, neural crest cells undergo an epithelial-to-mesenchymal transition (EMT), detach from the neural tube, and travel throughout the embryo to form cartilage, bones, muscles, neurons, and epidermis (Szabo and Mayor, 2018). During their embryonic migrations, cells often traverse basement membrane (BM)—a dense, sheet-like, covalently crosslinked type IV collagen and laminin-rich extracellular matrix that enwraps and separates most tissues (Gros and Tabin, 2014; Jayadev and Sherwood, 2017; Kelley et al., 2014; Leonard and Taneyhill, 2020; Moser et al., 2018). Immune cells also transmigrate BM barriers to reach sites of injury and infection in adult organisms (Bahr et al., 2022). Cell invasive behavior is misregulated in many diseases, most notably cancer, where metastatic tumor cells hijack normal invasive cellular programs (Paterson and Courtneidge, 2018). To breach BM, invasive cells use small, matrix metalloproteinase (MMP)-enriched, F-actin-driven plasma membrane protrusions, termed invadosomes (Cambi and Chavrier, 2021). Because of the difficulty of visualizing rare and often stochastic cell invasion events in vivo, invadosomes have been studied most extensively in vitro on 2D surfaces that mimic the planar BM structure (Clarke et al., 2024). The events following BM breaching are less clear due to the challenge of recapitulating invasion in 3D settings. However, ex vivo invasion assays, 3D spheroid cell cultures, and tumor sections have revealed that invadosomes transition into a large, transient protrusion that clears a path through the BM to allow transmigration (Hotary et al., 2006; Leong et al., 2014; Nazari et al., 2023; Schoumacher et al., 2010). Although crucial for invasion, the mechanisms regulating this invasive structure remain poorly understood.
Invasive membrane protrusions are large and have the specialized function of BM removal, and it is unknown if lipid production or specialized lipid modification systems are required for their formation. Lipid metabolism is complex and involves lipid biosynthesis, external lipid import, lipid storage, and lipid catabolism for energy production (Snaebjornsson et al., 2020). Lipid metabolism can be regulated both at the transcriptional level, often either by the sterol regulatory element-binding protein (SREBP) family of transcription factors or by external cues (Röhrig and Schulze, 2016). Exacerbated lipid synthesis is strongly associated with metastatic cancers (Bian et al., 2021; Chen et al., 2018; Lu et al., 2022; Martin-Perez et al., 2022; Vasseur and Guillaumond, 2022). For example, SREBP transcription factors and their key lipogenic enzyme targets are overexpressed in many metastatic cancers and promote invasion in vitro through solubilized non-crosslinked BM extracts. These SREBP targets include acetyl-CoA carboxylase (ACC), which catalyzes the rate-limiting step in fatty acid synthesis, fatty acid synthase (FASN), which directs palmitate synthesis, and HMG-CoA reductase (HMGCR), which is the rate-limiting enzyme of the mevalonate pathway and necessary for the catalysis of isoprenoids for cholesterol and protein prenylation (Bao et al., 2016; Bian et al., 2021; Gao et al., 2019; Li et al., 2014; Lu et al., 2022; Wang et al., 2022; Xu et al., 2020). It is unclear, however, if these lipogenesis enzymes are required for cells to breach the dense and highly crosslinked BM found in vivo, and unknown how lipogenesis might function to promote invasion. Understanding the role of lipogenesis in cell invasion is important, as many lipid synthesis enzyme inhibitors exist that could be strategically used to target invasive behavior (Bian et al., 2021; Broadfield et al., 2021; Vasseur and Guillaumond, 2022).
The anchor cell (AC) is a specialized uterine cell that invades through the BM separating the uterine and vulval tissue in C. elegans to initiate uterine–vulval connection (Sherwood and Sternberg, 2003). AC invasion is highly stereotyped, accessible to live imaging, and allows targeted gene knockdown (Kenny-Ganzert and Sherwood, 2023). Like cancer cells, the AC uses dynamic invadosomes to breach the BM (Hagedorn et al., 2013). Following BM breaching, the netrin receptor UNC-40 (vertebrate DCC) traffics to the small hole in the BM and directs lysosome exocytosis to form a large protrusion that clears a path through the BM (Hagedorn et al., 2013; Naegeli et al., 2017). The invasive protrusion is enriched with the glycosylphosphatidylinositol (GPI)-anchored matrix metalloproteinase (MMP) ZMP-1, which helps degrade the BM (Kelley et al., 2019). The UNC-40 receptor and GPI-anchored proteins like ZMP-1 are partitioned to sphingolipid-enriched lipid rafts (Herincs et al., 2005; Sangiorgio et al., 2004). In addition, C. elegans Rac and Ras-like GTPases that promote F-actin formation for invasive protrusion outgrowth are anchored and enriched in the invasive protrusion through C-terminal lipid prenylation (Costa et al., 2023; Hagedorn et al., 2013; Lohmer et al., 2016; Wang et al., 2014b). Previous work examining energy sources that fuel invasion revealed that the AC does not contain lipid stores nor require lipid import transporters (Garde et al., 2022; Zechner et al., 2017). Whether AC invasion depends on de novo lipid synthesis and elevated GTPase lipid anchoring to form the large specialized invasive protrusion, however, is unknown. Lipid synthesis and modification enzymes are conserved in C. elegans and function similarly to mammalian orthologs (Ashrafi et al., 2003; Guzman et al., 2023, Preprint; Ranji et al., 2014; Rappleye et al., 2003; Zhang et al., 2013). Thus, examination of AC invasion offers a strong model to investigate lipid regulation during BM transmigration.
In this study, we show that the lipogenic C. elegans SREBP transcription factor SBP-1 traffics to the AC nucleus prior to invasion and drives elevated expression of genes promoting fatty acid generation for phospholipid synthesis and sphingomyelin production that promote lysosome stores and lipid rafts that build a functional invasive protrusion. We also discover that HMGR-1 (vertebrate HMGCR, mevalonate pathway) polarizes at the AC invasive front and is present in the endoplasmic reticulum (ER) and enriched in peroxisomes. HMGR-1 generates isoprenoids for prenylation, and we find that the ER-localized isoprenylcysteine carboxyl methyltransferase (ICMT-1), which finalizes protein prenylation, enriches to ER exit sites that dynamically polarize and deliver prenylated GTPases that drive protrusion formation. Collectively, these results reveal a dynamic collaboration between lipid biosynthesis and polarized lipid modification that drives invasive protrusion formation to clear BM barriers.
Results
Anchor cell (AC) invasive protrusive formation requires the SREBP ortholog SBP-1
The AC is a specialized uterine cell that invades through the juxtaposed uterine and vulval BMs during an ∼90-min period in the mid-L3 larval stage to initiate uterine–vulval attachment (Sherwood and Sternberg, 2003). AC differentiation and invasion occur in synchrony with divisions of the underlying 1° fated P6.p vulval precursor cell (VPC), which allows the staging of invasion (Fig. 1 A). The AC is specified just prior to the L2/L3 molt, and during the L3 stage expresses proinvasive actin and matrix remodeling proteins until the early-to-mid L3 when the AC initiates BM beaching with a small invadosome that penetrates the BM (P6.p 2-cell; Fig. 1 A) (Costa et al., 2023; Hagedorn et al., 2013; Kimble, 1981). The UNC-40 (DCC) receptor then localizes to the breach site and directs lysosome exocytosis to form a large protrusion that removes BM and transiently increases the surface area of the AC by as much as 40% (2–4-cell transition stage) (Morrissey et al., 2013; Naegeli et al., 2017). Following BM clearance, the protrusion retracts, and the AC nestles between the central vulval cells (4-cell stage). The AC expresses lipid-anchored proteins that enrich at the invasive plasma membrane and then concentrate in the protrusion, such as the matrix-degrading GPI-anchored MMP ZMP-1, the prenylated Rho GTPases CED-10 (Rac) and MIG-2 (Rac-like), and the Ras-like GTPase RAP-1 (Fig. 1, B–D; (Costa et al., 2023; Hagedorn et al., 2013; Kelley et al., 2019; Naegeli et al., 2017; Wang et al., 2014a). How the AC synthesizes additional lipid membranes for protrusion formation and organizes the production of lipid-anchored proteins is unknown.
The AC invasive membrane and protrusion is enriched with lipid-modified proinvasive proteins. (A) Top: A schematic diagram of AC invasion. Bottom: Merged differential interference microscopy (DIC) and fluorescence images (maximum intensity z-projections) showing the AC (cyan, mCherry::PLCδPH), the underlying BM (magenta, LAM-1:GFP), and the vulval precursor cells (VPCs, DIC). The AC is specified at the L2/L3 molt and proinvasive gene expression occurs at the P6.p 1-cell stage. AC invasion is initiated at the late P6.p 2-cell stage when an invadosome breaches the BM. During the P6.p 2–4-cell transition, a large invasive protrusion forms at the breach site that expands the BM gap and then retracts, allowing the AC to contact the central VPCs at the P6.p 4-cell stage. Timeline in hours after hatching at 20°C is shown. (B–D) In the AC of wild-type animals, the GPI-anchored matrix metalloproteinase ZMP-1 (ZMP-1::mNG), (C) lysosomes (LMP-1::mNG), (D) and the prenylated membrane reporter (GFP::CAAX), prenylated Rac GTPases (GFP::CED-10 and GFP::MIG-2), and Ras-like GTPase RAP-1 (mNG::RAP-1) are enriched at the AC basal plasma membrane prior to invasion (arrows; P6.p 2-cell stage) and within the AC invasive protrusion (arrowheads; P6.p 2–4-cell stage) (observed in n ≥ 10 animals for each). All data shown are from two or more replicates. Scale bars, 5 µm.
The AC invasive membrane and protrusion is enriched with lipid-modified proinvasive proteins. (A) Top: A schematic diagram of AC invasion. Bottom: Merged differential interference microscopy (DIC) and fluorescence images (maximum intensity z-projections) showing the AC (cyan, mCherry::PLCδPH), the underlying BM (magenta, LAM-1:GFP), and the vulval precursor cells (VPCs, DIC). The AC is specified at the L2/L3 molt and proinvasive gene expression occurs at the P6.p 1-cell stage. AC invasion is initiated at the late P6.p 2-cell stage when an invadosome breaches the BM. During the P6.p 2–4-cell transition, a large invasive protrusion forms at the breach site that expands the BM gap and then retracts, allowing the AC to contact the central VPCs at the P6.p 4-cell stage. Timeline in hours after hatching at 20°C is shown. (B–D) In the AC of wild-type animals, the GPI-anchored matrix metalloproteinase ZMP-1 (ZMP-1::mNG), (C) lysosomes (LMP-1::mNG), (D) and the prenylated membrane reporter (GFP::CAAX), prenylated Rac GTPases (GFP::CED-10 and GFP::MIG-2), and Ras-like GTPase RAP-1 (mNG::RAP-1) are enriched at the AC basal plasma membrane prior to invasion (arrows; P6.p 2-cell stage) and within the AC invasive protrusion (arrowheads; P6.p 2–4-cell stage) (observed in n ≥ 10 animals for each). All data shown are from two or more replicates. Scale bars, 5 µm.
Additional plasma membrane and lipid anchors could arise from external fatty acid import, stored lipids, or de novo synthesis (Martin-Perez et al., 2022). A previous investigation of carbon sources used to generate ATP for AC invasion found that the AC does not have lipid stores and does not depend on fatty acid or amino acid import (Garde et al., 2022). Instead, the AC imports glucose and uses mitochondrial oxidative phosphorylation to generate ATP that powers the invasive machinery (Garde et al., 2022). These observations suggest that the AC might generate de novo lipids by using citrate generated from the mitochondrial tricarboxylic acid cycle (TCA, also known as Krebs and citric acid cycle) to build new lipids (Martin-Perez et al., 2022).
Lipogenesis is a process by which cells convert carbohydrates to fatty acids via a series of highly regulated enzymatic reactions (see Fig. S1 A) (Ameer et al., 2014). Initially, citrate from the TCA cycle is converted to the two-carbon acetyl-CoA, which is then transformed into malonyl-CoA. Fatty acid synthesis enzymes then facilitate the production of lipids that serve as the precursor for glycerophospholipids (hereafter phospholipids) that form the bulk of cell membranes and specialized sphingolipids that regulate membrane stability, lipid raft formation, vesicle and protein trafficking, and signaling (Fig. S1 A) (Ameer et al., 2014; Cockcroft, 2021; Goni, 2022; Watts and Ristow, 2017). In many organisms, including C. elegans, the transcription of lipid biosynthetic enzymes is regulated by the SREBP family (sterol regulatory element-binding protein, C. elegans SBP-1) (Bertolio et al., 2019; McKay et al., 2003; Sun et al., 2020).
Fatty acid, phospholipid, and sphingolipid synthesis and the mevalonate pathway in C. elegans. (A) A schematic diagram shows the enzymes and substrates that are involved in processing acetyl-CoA (derived from glucose/mitochondrial citrate) to provide fatty acyl-CoA (red dashed box) for the phospholipid (magenta bracket) and sphingolipid (blue bracket) synthesis pathways. For reactions catalyzed by multiple enzymes with redundant functions (SMS-1, 2, 3, CGT-1, 2, 3, and PSSY-1, 2), the enzyme with expression in the AC as determined by AC-specific RNA-Seq was examined in the RNAi screen (Table S2, see Materials and methods). The enzymes whose RNAi mediated reduction led to a significant invasion defect are marked by asterisks (refer to Table S2). ACS, Acyl-CoA synthetase, represents 23 C. elegans homologs with predicted roles in Acyl-CoA synthesis; ACL, Acyl-CoA ligase, represents 14 C. elegans homologs with predicted activity as acyl-transferases; ATP, adenosine triphosphate; CTP, cytidine triphosphate; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine. (B) A schematic diagram showing the enzymes and substrates of the mevalonate pathway (green bracket) that promote Rab and CAAX geranylgeranylation, CAAX farnesylation, coenzyme Q synthesis, and protein glycosylation. The enzymes whose RNAi mediated depletion led to a significant invasion defect are marked by asterisks (refer to Table S2). COQ-1 and FDPS-1 in green dashed circle are the closest known homologs to the human geranylgeranyl diphosphate synthase. Schematics were adapted from Watts and Ristow (2017).
Fatty acid, phospholipid, and sphingolipid synthesis and the mevalonate pathway in C. elegans. (A) A schematic diagram shows the enzymes and substrates that are involved in processing acetyl-CoA (derived from glucose/mitochondrial citrate) to provide fatty acyl-CoA (red dashed box) for the phospholipid (magenta bracket) and sphingolipid (blue bracket) synthesis pathways. For reactions catalyzed by multiple enzymes with redundant functions (SMS-1, 2, 3, CGT-1, 2, 3, and PSSY-1, 2), the enzyme with expression in the AC as determined by AC-specific RNA-Seq was examined in the RNAi screen (Table S2, see Materials and methods). The enzymes whose RNAi mediated reduction led to a significant invasion defect are marked by asterisks (refer to Table S2). ACS, Acyl-CoA synthetase, represents 23 C. elegans homologs with predicted roles in Acyl-CoA synthesis; ACL, Acyl-CoA ligase, represents 14 C. elegans homologs with predicted activity as acyl-transferases; ATP, adenosine triphosphate; CTP, cytidine triphosphate; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine. (B) A schematic diagram showing the enzymes and substrates of the mevalonate pathway (green bracket) that promote Rab and CAAX geranylgeranylation, CAAX farnesylation, coenzyme Q synthesis, and protein glycosylation. The enzymes whose RNAi mediated depletion led to a significant invasion defect are marked by asterisks (refer to Table S2). COQ-1 and FDPS-1 in green dashed circle are the closest known homologs to the human geranylgeranyl diphosphate synthase. Schematics were adapted from Watts and Ristow (2017).
We hypothesized that if lipogenesis were required to build the invasive protrusion, then AC-specific loss of key lipogenesis enzymes would show defective invasion. To determine whether SBP-1 and de novo lipid biosynthesis promote AC invasive protrusion formation, we used an AC-specific RNAi strain and targeted the sbp-1 gene (see Table S1; Materials and methods) (Garde et al., 2022). Consistent with a possible role in invasive protrusion regulation, AC-specific sbp-1 RNAi severely disrupted invasion (over 65% of ACs did not remove BM by P6.p 4-cell stage compared with 10% of empty vector treated controls; Table S2 and Fig. 2 A). An sbp-1 null mutant is lethal (McKay et al., 2003); however, the slow-growing viable sbp-1(ep79) mutant (Liang et al., 2010) also showed invasion defects (Table S2). Examination of AC-specific membrane localized mCherry fluorophore (cdh-3p::mCherry::PLCδPH) in the AC-specific RNAi strain revealed that sbp-1 RNAi reduced the formation and growth rate of the invasive protrusion and also delayed and decreased BM removal, correlating with the reduced protrusion (Fig. 2 B, Fig. S2, A and B; and Video 1). These results suggest that de novo lipid synthesis regulated by SBP-1 is required for invasive protrusion formation.
SREBP (SBP-1) promotes AC invasion and invasive protrusion formation. (A) Top: Merged DIC and fluorescence images (maximum intensity z-projections) of the AC (cyan, mCherry::PLCδPH) and the underlying BM (single central confocal slice, magenta, LAM-1::GFP) at the P6.p 4-cell stage in a control (empty RNAi vector treated) and an AC-specific sbp-1 RNAi treated animal. Bottom: Fluorescence images showing BM breach (brackets) in a control animal and lack of breach after sbp-1 RNAi (arrows). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific sbp-1 RNAi treated animal (BM, orange dashed lines) with a small and delayed (see Materials and methods) breach observed corresponding to the reduced protrusion after loss of sbp-1 (middle, bracket, observed in n = 8/8 sbp-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 1). Right: Boxplot shows the maximum invasive protrusion volume in control and sbp-1 RNAi treated animals (n = 7 control and 8 sbp-1 RNAi animals, *** P ≤ 0.001, Mann–Whitney U test). For this and all subsequent boxplots, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line within the box the median value. (C) A schematic diagram illustrating SBP-1 proteolytic activation and entry into the nucleus where the SBP-1 bHLH domain binds sterol regulatory elements (SREs) to transcribe target genes. (D) Boxplot of mNG::SBP-1 mean fluorescence intensity in the AC nucleus from the L2/L3 molt stage to the P6.p 4-cell stage (n = 6 L2/L3 molt, nine 1-cell, six 2-cell, seven 2–4-cell, and six 4-cell stage animals, ** P ≤ 0.01, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (E) Top: Sum intensity z-projected fluorescence images of mNG::SBP-1 in the AC nucleus (white dashed circle) from the L2/L3 molt (time of AC specification) to the P6.p 4-cell stage. Bottom: Central plane DIC images showing dashed circles outlining the nucleus where SBP-1 fluorescence was measured. All data are from two or more replicates. Scale bars, 5 µm.
SREBP (SBP-1) promotes AC invasion and invasive protrusion formation. (A) Top: Merged DIC and fluorescence images (maximum intensity z-projections) of the AC (cyan, mCherry::PLCδPH) and the underlying BM (single central confocal slice, magenta, LAM-1::GFP) at the P6.p 4-cell stage in a control (empty RNAi vector treated) and an AC-specific sbp-1 RNAi treated animal. Bottom: Fluorescence images showing BM breach (brackets) in a control animal and lack of breach after sbp-1 RNAi (arrows). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific sbp-1 RNAi treated animal (BM, orange dashed lines) with a small and delayed (see Materials and methods) breach observed corresponding to the reduced protrusion after loss of sbp-1 (middle, bracket, observed in n = 8/8 sbp-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 1). Right: Boxplot shows the maximum invasive protrusion volume in control and sbp-1 RNAi treated animals (n = 7 control and 8 sbp-1 RNAi animals, *** P ≤ 0.001, Mann–Whitney U test). For this and all subsequent boxplots, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line within the box the median value. (C) A schematic diagram illustrating SBP-1 proteolytic activation and entry into the nucleus where the SBP-1 bHLH domain binds sterol regulatory elements (SREs) to transcribe target genes. (D) Boxplot of mNG::SBP-1 mean fluorescence intensity in the AC nucleus from the L2/L3 molt stage to the P6.p 4-cell stage (n = 6 L2/L3 molt, nine 1-cell, six 2-cell, seven 2–4-cell, and six 4-cell stage animals, ** P ≤ 0.01, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (E) Top: Sum intensity z-projected fluorescence images of mNG::SBP-1 in the AC nucleus (white dashed circle) from the L2/L3 molt (time of AC specification) to the P6.p 4-cell stage. Bottom: Central plane DIC images showing dashed circles outlining the nucleus where SBP-1 fluorescence was measured. All data are from two or more replicates. Scale bars, 5 µm.
SBP-1, LPIN-1, SMS-1, and HMGR-1 promote AC invasive protrusion growth. (A) Maximum intensity z-projected time-lapse fluorescence images showing the formation of the AC invasive protrusion (mCherry::PLCδPH, cyan) in a control and sbp-1 RNAi knockdown animal (BM, orange dashed lines). Time points indicate h::min. Isosurfaces of the AC invasive protrusions are shown below (see Video 1). The 0:45 min timepoint for control and sbp-1 RNAi is also shown in Fig. 2 A. (B–E) Left: Line graphs of invasive protrusion volume over time in control (cyan) animals and in animals after (B) sbp-1, (C) lpin-1, (D) sms-1, and (E) hmgr-1 AC-specific RNAi treatment (magenta; see Videos 1, 2, 6, and 7). Right in B–E: Boxplots of the AC invasive protrusion growth rates. In this and subsequent figures, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line inside each box the median value. AC protrusion growth rate over 60 or 64 min was calculated ([B] n = 7 control and 8 sbp-1 RNAi animals; [C] n = 7 control and 5 lpin-1 RNAi animals; [D] n = 5 control and 5 sms-1 RNAi animals; [E] n = 9 control and 7 hmgr-1 RNAi animals ** P ≤ 0.01, P = 0.1490, Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.
SBP-1, LPIN-1, SMS-1, and HMGR-1 promote AC invasive protrusion growth. (A) Maximum intensity z-projected time-lapse fluorescence images showing the formation of the AC invasive protrusion (mCherry::PLCδPH, cyan) in a control and sbp-1 RNAi knockdown animal (BM, orange dashed lines). Time points indicate h::min. Isosurfaces of the AC invasive protrusions are shown below (see Video 1). The 0:45 min timepoint for control and sbp-1 RNAi is also shown in Fig. 2 A. (B–E) Left: Line graphs of invasive protrusion volume over time in control (cyan) animals and in animals after (B) sbp-1, (C) lpin-1, (D) sms-1, and (E) hmgr-1 AC-specific RNAi treatment (magenta; see Videos 1, 2, 6, and 7). Right in B–E: Boxplots of the AC invasive protrusion growth rates. In this and subsequent figures, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line inside each box the median value. AC protrusion growth rate over 60 or 64 min was calculated ([B] n = 7 control and 8 sbp-1 RNAi animals; [C] n = 7 control and 5 lpin-1 RNAi animals; [D] n = 5 control and 5 sms-1 RNAi animals; [E] n = 9 control and 7 hmgr-1 RNAi animals ** P ≤ 0.01, P = 0.1490, Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.
SBP-1 is required for AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific sbp-1 RNAi knockdown animal. Video corresponds to Fig. 2 B. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, display rate 8 frames/sec. Scale bar, 5 µm.
SBP-1 is required for AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific sbp-1 RNAi knockdown animal. Video corresponds to Fig. 2 B. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, display rate 8 frames/sec. Scale bar, 5 µm.
SREBP proteins reside on the ER membrane in their inactive state. However, when activated, SREBP translocates to the Golgi, where two Golgi proteases cleave SREBP and release the N-terminal DNA binding basic-helix-loop-helix (bHLH) domain, which enters the nucleus and regulates lipogenic gene expression (Fig. 2 C) (DeBose-Boyd and Ye, 2018). To visualize both the expression and activity of the C. elegans SBP-1 protein in the AC, we used CRISPR-Cas9 mediated genome editing to endogenously tag the sbp-1 locus with the fluorophore mNeonGreen (mNG) at the N-terminus (mNG::SBP-1). Examination of endogenous SBP-1 in the AC revealed that SBP-1 strongly localized to the AC nucleus at the L2/L3 molt and during the early P6.p 1-cell stage, but then nuclear levels diminished rapidly after the 1-cell stage and through the time of invasion (Fig. 2, D and E). This suggests that SBP-1 is active in the AC ∼4–5 h before invasion to promote the expression of lipogenic genes required for AC invasive protrusion formation and BM breaching.
SBP-1 regulates AC expression of key lipogenic enzymes that promote invasion
The early localization of SBP-1 in the AC nucleus suggested that SBP-1 might promote the expression of lipogenic enzymes to support invasive protrusion formation. To test this, we used AC-specific RNAi against the key lipogenic transcriptional targets of SBP-1, pod-2 (vertebrate acetyl-CoA carboxylase, ACC), and fasn-1 (vertebrate FASN), which play crucial roles in the initial steps of de novo fatty acid synthesis (Fig. S1 A and Fig. 3 A) (Röhrig and Schulze, 2016; Watts and Ristow, 2017). RNAi mediated depletion of the POD-2 and FASN-1 proteins resulted in significant AC invasion defects at the P6.p 4-cell stage (∼40% and 55% respectively; Tables S1 and S2), consistent with a key role downstream of SBP-1 in invasive protrusion formation. We examined endogenously tagged GFP::POD-2 (Starich et al., 2020) and generated an endogenously tagged FASN-1::mNG. Both enzymes were highly expressed and enriched in the AC compared to neighboring non-invading uterine cells with expression increasing significantly from the P6.p 1-cell stage to the P6.p 2–4-cell stage at the time of protrusion formation (Fig. S3 A). RNAi-mediated reduction of the SBP-1 protein also significantly reduced the levels of GFP::POD-2 and FASN-1::mNG in the AC (Fig. 3 B). These observations suggest that SBP-1 might promote AC protrusion formation through transcriptional regulation of the pod-2 and fasn-1 genes.
AC expression and SBP-1 regulation of pod-2, fasn-1, elo-1, lpin-1, and sms-1. (A) A schematic diagram showing the role of the transcription factor SBP-1 in regulating phospholipid and sphingomyelin synthesis in the AC (refer to Fig. S1 A for complete pathway and Table S2 for RNAi screen results). (B) Sum intensity z-projection images of POD-2 (GFP::POD-2) and FASN-1 (FASN-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. POD-2 and FASN-1 are in the AC cytoplasm (arrows, ACs). (C) Sum intensity z-projections of ELO-1 (ELO-1::mNG, ACs outlined with white dashes) and LPIN-1 (mNG::LPIN-1, arrows, ACs) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. ELO-1 is localized to the ER (see Fig. S3 B) and LPIN-1 in the cytosol. (D) Sum intensity z-projection of SMS-1 (SMS-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage (ACs outlined with white dashes). SMS-1 localizes to the Golgi (see Fig. S3 C). (B–D) Right: Boxplots show the mean fluorescence intensity of each protein in the AC of control and sbp-1 RNAi treated animals ([POD-2] n = 10 control and 10 sbp-1 RNAi animals; [FASN-1] n = 22 control and 23 sbp-1 RNAi animals; [ELO-1] n = 15 control and 16 sbp-1 RNAi animals; [LPIN-1] n = 11 control and 10 sbp-1 RNAi animals; [SMS-1] n = 24 control and 13 sbp-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test and Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.
AC expression and SBP-1 regulation of pod-2, fasn-1, elo-1, lpin-1, and sms-1. (A) A schematic diagram showing the role of the transcription factor SBP-1 in regulating phospholipid and sphingomyelin synthesis in the AC (refer to Fig. S1 A for complete pathway and Table S2 for RNAi screen results). (B) Sum intensity z-projection images of POD-2 (GFP::POD-2) and FASN-1 (FASN-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. POD-2 and FASN-1 are in the AC cytoplasm (arrows, ACs). (C) Sum intensity z-projections of ELO-1 (ELO-1::mNG, ACs outlined with white dashes) and LPIN-1 (mNG::LPIN-1, arrows, ACs) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. ELO-1 is localized to the ER (see Fig. S3 B) and LPIN-1 in the cytosol. (D) Sum intensity z-projection of SMS-1 (SMS-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage (ACs outlined with white dashes). SMS-1 localizes to the Golgi (see Fig. S3 C). (B–D) Right: Boxplots show the mean fluorescence intensity of each protein in the AC of control and sbp-1 RNAi treated animals ([POD-2] n = 10 control and 10 sbp-1 RNAi animals; [FASN-1] n = 22 control and 23 sbp-1 RNAi animals; [ELO-1] n = 15 control and 16 sbp-1 RNAi animals; [LPIN-1] n = 11 control and 10 sbp-1 RNAi animals; [SMS-1] n = 24 control and 13 sbp-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test and Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.
POD-2, FASN-1, ELO-1, LPIN-1, and SMS-1 AC expression from P6.p 1-cell to 4-cell stage. (A) Left: Sum intensity z-projected fluorescence images of endogenously tagged GFP::POD-2, FASN-1::mNG, ELO-1::mNG, mNG::LPIN-1, and SMS-1::mNG in the AC (arrows and white dotted lines) from the P6.p 1-cell to 4-cell stages. Right: Boxplots show the mean fluorescence intensity of each protein in the AC (comparisons of P6.p 1-cell and 2–4-cell stages, [POD-2] n = 11 1-cell, 10 2-cell, 10 2–4-cell, and 11 4-cell stage animals; [FASN-1] n = 11 1-cell, 12 2-cell, 10 2–4-cell, and 10 4-cell stage animals; [ELO-1] n = 14 1-cell, 13 2-cell, 9 2–4-cell, and 13 4-cell stage animals; [LPIN-1] n = 8 1-cell, 15 2-cell, 10 2–4-cell, and 14 4-cell stage animals; [SMS-1] n = 10 1-cell, 10 2-cell, 5 2–4-cell, and 10 4-cell stage animals, * P ≤ 0.05, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test; comparisons of uterine cell [UC] to AC at P6.p 4-cell stage, [POD-2] n = 11 animals; [FASN-1] n = 10 animals; [ELO-1] n = 13 animals; [LPIN-1] n = 14 animals; [SMS-1] n = 10 animals, * P ≤ 0.05, ** P ≤ 0.001, *** P ≤ 0.0001, P = 0.10, Student’s t test). (B) Top: Single slice confocal images of mCherry::KDEL (magenta), ELO-1::mNG (cyan) and overlay in the AC show tight overlap (arrows). Bottom: Insets highlighting the overlap in the basal region (arrows; similar colocalization observed in n = 10/10 animals). (C) Top: Max intensity z-projected fluorescence images of AMAN-2::mScarlet (magenta, Golgi), SMS-1::mNG (cyan), and overlay in the AC reveal colocalization (arrows). The average percentage ± SD of colocalization of SMS-1::mNG puncta with AMAN-2::mScarlet with is indicated (n = 6 animals). Below: Insets highlight AMAN-2::mScarlet (magenta) and SMS-1::mNG (cyan) colocalization (arrows). (D) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a ppk-3 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal ZMP-1::mNG mean fluorescence signal in control and ppk-3 RNAi treated animals (n = 8 control and 9 ppk-3 RNAi animals, P ≤ 0.05, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.
POD-2, FASN-1, ELO-1, LPIN-1, and SMS-1 AC expression from P6.p 1-cell to 4-cell stage. (A) Left: Sum intensity z-projected fluorescence images of endogenously tagged GFP::POD-2, FASN-1::mNG, ELO-1::mNG, mNG::LPIN-1, and SMS-1::mNG in the AC (arrows and white dotted lines) from the P6.p 1-cell to 4-cell stages. Right: Boxplots show the mean fluorescence intensity of each protein in the AC (comparisons of P6.p 1-cell and 2–4-cell stages, [POD-2] n = 11 1-cell, 10 2-cell, 10 2–4-cell, and 11 4-cell stage animals; [FASN-1] n = 11 1-cell, 12 2-cell, 10 2–4-cell, and 10 4-cell stage animals; [ELO-1] n = 14 1-cell, 13 2-cell, 9 2–4-cell, and 13 4-cell stage animals; [LPIN-1] n = 8 1-cell, 15 2-cell, 10 2–4-cell, and 14 4-cell stage animals; [SMS-1] n = 10 1-cell, 10 2-cell, 5 2–4-cell, and 10 4-cell stage animals, * P ≤ 0.05, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test; comparisons of uterine cell [UC] to AC at P6.p 4-cell stage, [POD-2] n = 11 animals; [FASN-1] n = 10 animals; [ELO-1] n = 13 animals; [LPIN-1] n = 14 animals; [SMS-1] n = 10 animals, * P ≤ 0.05, ** P ≤ 0.001, *** P ≤ 0.0001, P = 0.10, Student’s t test). (B) Top: Single slice confocal images of mCherry::KDEL (magenta), ELO-1::mNG (cyan) and overlay in the AC show tight overlap (arrows). Bottom: Insets highlighting the overlap in the basal region (arrows; similar colocalization observed in n = 10/10 animals). (C) Top: Max intensity z-projected fluorescence images of AMAN-2::mScarlet (magenta, Golgi), SMS-1::mNG (cyan), and overlay in the AC reveal colocalization (arrows). The average percentage ± SD of colocalization of SMS-1::mNG puncta with AMAN-2::mScarlet with is indicated (n = 6 animals). Below: Insets highlight AMAN-2::mScarlet (magenta) and SMS-1::mNG (cyan) colocalization (arrows). (D) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a ppk-3 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal ZMP-1::mNG mean fluorescence signal in control and ppk-3 RNAi treated animals (n = 8 control and 9 ppk-3 RNAi animals, P ≤ 0.05, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.
De novo phospholipid and sphingolipid synthesis are required for AC invasion
Key outputs of the SBP-1 targets POD-2 and FASN-1 are palmitate, which can go on to form phospholipids and triacylglyceride for storage in lipid droplets, and 11-methyldodecanoic acid, which is a core precursor for C. elegans sphingolipids (Fig. 3 A and Fig. S1 A) (Watts and Ristow, 2017). As the AC does not form lipid droplets (Garde et al., 2022), it suggested that SBP-1 could promote invasive protrusion formation via phospholipid and sphingolipid production. To test this, we used AC-specific RNAi to target 20 key genes involved in phospholipid and sphingolipid synthesis (Table S2 and Fig. S1 A [Watts and Ristow, 2017]). RNAi against two sphingolipid biosynthetic encoding enzymes, lagr-1 and sms-1, significantly perturbed AC invasion (∼35 and 40%, respectively) and RNAi targeting a third, hyl-1, trended toward significance (Table S2 and Fig. 3 A). RNAi against the fatty acid elongases elo-1 and elo-2, as well as phosphatidic acid phosphatase lpin-1, which synthesizes diacylglycerol for phospholipid production, also resulted in significant invasion defects (Table S2 and Fig. 3 A). We examined endogenously tagged ELO-1::mNG (Costa et al., 2023) and generated endogenous mNG knock-in strains to observe mNG::LPIN-1 and SMS-1::mNG. All enzymes were present at high levels in the AC and levels peaked near the time of protrusion formation (Fig. 3, C and D; and Fig. S3 A). The enzymes also localized to expected subcellular regions: ELO-1, in perinuclear membrane in keeping with the endoplasmic reticulum (ER); LPIN-1, the cytosol; and SMS-1, in diffuse puncta consistent with dispersed Golgi stacks (Fig. 3 B) (Costa et al., 2023; D’Angelo et al., 2018; Ding et al., 2023; Naegeli et al., 2017). We confirmed the localization of ELO-1 to the ER by performing colocalization with the ER marker mCherry::KDEL and found that SMS-1 colocalized with the Golgi marker AMAN-2::mScarlet (Fig. S3, B and C) (Thomas et al., 2019). RNAi targeting sbp-1 did not affect ELO-1 or LPIN-1 levels, and only modestly reduced SMS-1 (Fig. 3 B). These results are consistent with previous studies indicating that SBP-1 regulates key upstream de novo fatty acid synthesis enzymes but not all downstream lipid synthesis genes (Nomura et al., 2010). These observations suggest that de novo–produced phospholipids and sphingolipids could promote invasive protrusion formation.
Phospholipid synthesis builds lysosome stores that construct the invasive protrusion
To investigate the role of phospholipid production in invasive protrusion formation, we targeted lpin-1 in the AC-specific RNAi strain (Table S1) and found that lpin-1 RNAi significantly reduced the formation of the invasive protrusion and decreased BM clearance (Fig. 4 A, Fig. S2 C, and Video 2). We’ve previously shown that lysosomes are exocytosed to form the invasive protrusion (Naegeli et al., 2017). Time-lapse analysis of lysosomes (AC expressed LMP-1::mNG) with images acquired at 2-min intervals, confirmed the concentration of lysosomes at the site of protrusion initiation and within the protrusion during its formation (Fig. 4 B and Video 3). Furthermore, time-lapse videos with short sampling intervals (0.74 s/frame, 91-s duration) revealed dynamic trafficking of lysosome vesicles toward the forming protrusion (Video 4, 9 of 13 vesicles moved toward protrusion, n = 3 ACs examined). Notably, lpin-1 RNAi reduced the formation of lysosome stores prior to invasive protrusion formation (Fig. 4 C and Video 5). RNAi against lpin-1, however, did not alter the prenylated Rac GTPases CED-10 and MIG-2 or the GPI-anchored ZMP-1 at the initiation of invasive protrusion formation (late P6.p 2-cell and 2–4 cell stage; Fig. 4, D and E). These results suggest that AC phospholipid production promotes invasive protrusion formation by contributing to lysosome stores that construct the protrusion but does not significantly regulate GTPase or ZMP-1 localization.
LPIN-1 promotes lysosome stores for invasive protrusion formation. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific lpin-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of lpin-1 (middle, bracket, observed in n = 5/5 lpin-1-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 2). Right: Boxplot of the maximum invasive protrusion volume in control and lpin-1 RNAi treated animals (n = 7 control and 5 lpin-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (B) Max intensity z-projected time-lapse fluorescence images (h:min, above) and isosurface images showing AC lysosomes (LMP-1::mNG) accumulating at the site of BM breach and entering the protrusion (observed in n = 3/3 time-lapses). (C) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in control and a lpin-1 RNAi treated animal at the initiation of protrusion formation (late P6.p 2-cell stage and 2–4-cell stage). Right: Boxplot of AC lysosome volume in control and lpin-1 RNAi treated animals (n = 21 control and 23 lpin-1 RNAi animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (D) Left: Sum intensity z-projected images show the polarized localization of GFP::CED-10 (Rac) and GFP::MIG-2 (Rac-like) at the AC invasive plasma membrane (arrow) in control and lpin-1 RNAi treated animals at the late P6.p 2-cell stage at the initiation of protrusion formation. Right: Boxplot showing the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 ([CED-10] n = 12 control and 13 lpin-1 RNAi animals; [MIG-2] n = 8 control and 11 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (E) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a lpin-1 RNAi treated animal at the initiation of invasive protrusion formation (late P6.p 2-cell stage and 2–4 cell stage). Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and lpin-1 RNAi treated animals (n = 10 control and 10 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.
LPIN-1 promotes lysosome stores for invasive protrusion formation. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific lpin-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of lpin-1 (middle, bracket, observed in n = 5/5 lpin-1-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 2). Right: Boxplot of the maximum invasive protrusion volume in control and lpin-1 RNAi treated animals (n = 7 control and 5 lpin-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (B) Max intensity z-projected time-lapse fluorescence images (h:min, above) and isosurface images showing AC lysosomes (LMP-1::mNG) accumulating at the site of BM breach and entering the protrusion (observed in n = 3/3 time-lapses). (C) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in control and a lpin-1 RNAi treated animal at the initiation of protrusion formation (late P6.p 2-cell stage and 2–4-cell stage). Right: Boxplot of AC lysosome volume in control and lpin-1 RNAi treated animals (n = 21 control and 23 lpin-1 RNAi animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (D) Left: Sum intensity z-projected images show the polarized localization of GFP::CED-10 (Rac) and GFP::MIG-2 (Rac-like) at the AC invasive plasma membrane (arrow) in control and lpin-1 RNAi treated animals at the late P6.p 2-cell stage at the initiation of protrusion formation. Right: Boxplot showing the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 ([CED-10] n = 12 control and 13 lpin-1 RNAi animals; [MIG-2] n = 8 control and 11 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (E) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a lpin-1 RNAi treated animal at the initiation of invasive protrusion formation (late P6.p 2-cell stage and 2–4 cell stage). Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and lpin-1 RNAi treated animals (n = 10 control and 10 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.
LPIN-1 is required for AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific lpin-1 RNAi knockdown animal. Video corresponds to Fig. 4 A. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, and display rate 8 frames/sec. Scale bar, 5 µm.
LPIN-1 is required for AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific lpin-1 RNAi knockdown animal. Video corresponds to Fig. 4 A. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, and display rate 8 frames/sec. Scale bar, 5 µm.
Time-lapse video of LMP-1::mNG lysosomes entering the invasive protrusion. Video corresponds to Fig. 4 B and shows a lateral view of LMP-1::mNG vesicles in grayscale (left) and spectral representation of fluorescence intensity (right) enriched at the invasive membrane and then entering the invasive protrusion. The total video duration was 78 min, frame interval 2 min, and display rate 6.67 frames/sec.
Time-lapse video of LMP-1::mNG lysosomes entering the invasive protrusion. Video corresponds to Fig. 4 B and shows a lateral view of LMP-1::mNG vesicles in grayscale (left) and spectral representation of fluorescence intensity (right) enriched at the invasive membrane and then entering the invasive protrusion. The total video duration was 78 min, frame interval 2 min, and display rate 6.67 frames/sec.
Time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Lateral view shows three separate high-temporal resolution videos through different lateral planes of the same AC. Arrowheads highlight destination points for vesicles traveling toward the site of invasive protrusion formation. The total video duration is 14.80 s, frame interval 0.74 s, and display rate 10.5 frames/sec.
Time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Lateral view shows three separate high-temporal resolution videos through different lateral planes of the same AC. Arrowheads highlight destination points for vesicles traveling toward the site of invasive protrusion formation. The total video duration is 14.80 s, frame interval 0.74 s, and display rate 10.5 frames/sec.
Time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Video corresponds to Fig. 4 D of control (left) and lpin-1 RNAi treated animals (right) and shows a rotating 3D projection of the raw fluorescence signal from LMP-1::mNG lysosomes and the transformation of this signal into a 3D isosurface using Imaris 9.9 software.
Time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Video corresponds to Fig. 4 D of control (left) and lpin-1 RNAi treated animals (right) and shows a rotating 3D projection of the raw fluorescence signal from LMP-1::mNG lysosomes and the transformation of this signal into a 3D isosurface using Imaris 9.9 software.
Sphingomyelin has multiple functions in building the AC’s invasive protrusion
The sphingomyelin synthase SMS-1 generates sphingomyelin and was the most downstream enzyme in sphingolipid synthesis whose loss perturbed AC invasion (Table S2 and Fig. S1 A) (Watts and Ristow, 2017). This suggested that sphingomyelin might be important for invasive protrusion formation. Sphingomyelin composes ∼5% of membrane lipids and is found in the plasma membrane and endolysosome, where it plays roles in signaling, vesicle and protein trafficking, membrane stability, and lysosome function (Duran et al., 2012; Goni, 2022; Tang et al., 2022). We knocked down the SMS-1 protein in the AC-specific RNAi strain and found sms-1 RNAi diminished the formation and growth of the invasive protrusion and decreased BM removal (Fig. 5 A, Fig. S2 D, and Video 6). Notably, sms-1 RNAi reduced lysosome stores that build the protrusion (Fig. 5 B). Rac GTPase enrichment at the initiation of protrusion formation, however, was unaltered (Fig. 5 C). Sphingomyelin is a key component of lipid rafts (Bieberich, 2018), which partition GPI-anchored proteins and the vertebrate UNC-40 receptor ortholog DCC (Herincs et al., 2005; Hernaiz-Llorens et al., 2021; Sangiorgio et al., 2004). We found that sms-1 RNAi reduced the enrichment of the GPI-anchored ZMP-1 protein at the site of invasive protrusion formation (Fig. 5 D). Further, ventral view time-lapse analysis along the BM plane revealed that sms-1 RNAi also reduced UNC-40::GFP receptor concentration at the BM breach site (Fig. 5 E), where UNC-40 directs lysosome exocytosis to form the invasive protrusion (Hagedorn et al., 2013; Morrissey et al., 2013; Naegeli et al., 2017). These results suggest that sphingomyelin plays a role in forming lysosomes that build the protrusion, is important for ZMP-1 localization for BM degradation, and promotes UNC-40 localization, which directs protrusion formation. We also hypothesized that lysosome exocytosis could transport sphingomyelin to the invasive protrusion to form lipid rafts. Consistent with this, we found that RNAi-mediated reduction of phosphatidylinositol phosphate kinase (PPK-3), which regulates the maturation and integrity of lysosomes (Nicot et al., 2006), decreased the levels of ZMP-1 at the invasive front during protrusion initiation (Fig. S3 D). Together, these studies build a network of de novo lipid synthesis genes acting downstream of SBP-1 that mediate invasive protrusion formation and function (Fig. 5 F).
SMS-1 promotes formation of lysosome stores and localization of UNC-40 and ZMP-1. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) in a control and an AC-specific sms-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of sms-1 (middle, bracket, observed in n = 5/5 sms-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 6). Right: Boxplot of the maximum invasive protrusion volume in control and sms-1 RNAi treated animals (n = 5 control and 5 sms-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in control and sms-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume in control and sms-1 RNAi treated animals (n = 24 control and 23 sms-1 RNAi animals, ** P ≤ 0.01, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projected fluorescence images of GFP::CED-10 and GFP::MIG-2. Arrows show enrichment at the AC invasive membrane in a control and sms-1 RNAi treated animals at the time of invasive protrusion initiation. Right: Boxplots show the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 fluorescence intensity in control and sms-1 RNAi treated animals ([CED-10] n = 16 control and 15 sms-1 RNAi animals; [MIG-2] n = 12 control and 11 sms-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (D) Left: Sum intensity fluorescence images of the GPI-anchored matrix metalloproteinase ZMP-1::mNG showing the AC invasive membrane (arrows) in a control and a sms-1 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and zmp-1 RNAi treated animals (n = 16 control and 13 sms-1 RNAi animals, * P ≤ 0.05, unpaired two-tailed Student’s t test). (E) Left: Ventral view of the AC-BM interface (BM visualized with LAM-1::mCherry) showing UNC-40::GFP enrichment (arrows) at the initial BM breach (orange dotted lines) in a control and a sms-1 treated RNAi animal. Right: Boxplot shows UNC-40::GFP mean fluorescence intensity at the initial BM breach (n = 6 control and 6 sms-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (F) A schematic diagram summarizing the roles of the transcription factor SBP-1, the fatty acid synthesis enzymes POD-2 and FASN-1, and phospholipid synthesizing LPIN-1 and sphingomyelin catalyzing SMS-1 in AC invasive protrusion formation. All data in the figure are from two or more replicates. Scale bars, 5 µm.
SMS-1 promotes formation of lysosome stores and localization of UNC-40 and ZMP-1. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) in a control and an AC-specific sms-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of sms-1 (middle, bracket, observed in n = 5/5 sms-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 6). Right: Boxplot of the maximum invasive protrusion volume in control and sms-1 RNAi treated animals (n = 5 control and 5 sms-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in control and sms-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume in control and sms-1 RNAi treated animals (n = 24 control and 23 sms-1 RNAi animals, ** P ≤ 0.01, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projected fluorescence images of GFP::CED-10 and GFP::MIG-2. Arrows show enrichment at the AC invasive membrane in a control and sms-1 RNAi treated animals at the time of invasive protrusion initiation. Right: Boxplots show the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 fluorescence intensity in control and sms-1 RNAi treated animals ([CED-10] n = 16 control and 15 sms-1 RNAi animals; [MIG-2] n = 12 control and 11 sms-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (D) Left: Sum intensity fluorescence images of the GPI-anchored matrix metalloproteinase ZMP-1::mNG showing the AC invasive membrane (arrows) in a control and a sms-1 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and zmp-1 RNAi treated animals (n = 16 control and 13 sms-1 RNAi animals, * P ≤ 0.05, unpaired two-tailed Student’s t test). (E) Left: Ventral view of the AC-BM interface (BM visualized with LAM-1::mCherry) showing UNC-40::GFP enrichment (arrows) at the initial BM breach (orange dotted lines) in a control and a sms-1 treated RNAi animal. Right: Boxplot shows UNC-40::GFP mean fluorescence intensity at the initial BM breach (n = 6 control and 6 sms-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (F) A schematic diagram summarizing the roles of the transcription factor SBP-1, the fatty acid synthesis enzymes POD-2 and FASN-1, and phospholipid synthesizing LPIN-1 and sphingomyelin catalyzing SMS-1 in AC invasive protrusion formation. All data in the figure are from two or more replicates. Scale bars, 5 µm.
SMS-1 is required for the AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific sms-1 RNAi knockdown animal. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 60 min, frame interval 4 min, and display rate 8 frames/sec. Scale bar, 5 µm.
SMS-1 is required for the AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific sms-1 RNAi knockdown animal. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 60 min, frame interval 4 min, and display rate 8 frames/sec. Scale bar, 5 µm.
The mevalonate pathway promotes lysosome stores and prenylated protein localization
In addition to being converted into fatty acids, acetyl-CoA can also be used to generate products of the mevalonate pathway. The mevalonate pathway converts acetyl-CoA into five-carbon branched isoprene groups that are turned into sterols and prenol lipids, such as cholesterol, coenzyme Q, dolichol, and prenylated anchors (15-carbon farnesyl and 20-carbon geranylgeranyl) for proteins such as small GTPases (Fig. 6 A and Fig. S1 B) (Rauthan and Pilon, 2011). In mammals, the mevalonate pathway produces cholesterol, and SREBP is a sensor of cholesterol levels and transcriptionally regulates the rate-limiting enzyme of the mevalonate pathway, HMG-CoA reductase (HMGCR, C. elegans hmgr-1 gene) (Rauthan and Pilon, 2011). C. elegans does not synthesize cholesterol (Vinci et al., 2008; Watts and Ristow, 2017) but instead acquires cholesterol from feeding, and it is unknown if SBP-1 regulates hmgr-1 expression (Rauthan and Pilon, 2011).
HMGR-1 promotes lysosome stores, GTPase accumulation, and invasive protrusion formation. (A) A schematic diagram showing the mevalonate pathway in the AC (see Fig. S1 B for pathway details, Table S2 for RNAi screen). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific hmgr-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of hmgr-1 (middle, bracket, observed in n = 7/7 hmgr-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 7). Right: Boxplot of the maximum invasive protrusion volume in control and hmgr-1 RNAi treated animals (n = 9 control and 7 hmgr-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (C) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in a control and an hmgr-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume of control and hmgr-1 RNAi animals (n = 17 control and 18 hmgr-1 RNAi animals, *** P ≤ 0.001, unpaired two-tailed Student’s t test). (D) Top: Sum intensity z-projected images showing invasive polarization (arrows) of GFP::CED-10, GFP::MIG-2, GFP::CAAX, and mNG::RAP-1 at the AC plasma membrane in control and hmgr-1 RNAi animals at the initiation of invasive protrusion formation. Bottom: Boxplots showing the AC basal/apical ratio of each protein’s fluorescence intensity in control and hmgr-1 RNAi-treated animals ([CED-10] n = 23 control and 23 hmgr-1 RNAi animals; [MIG-2] n = 13 control and 13 hmgr-1 RNAi animals; [CAAX] n = 16 control and 16 hmgr-1 RNAi animals; [RAP-1] n = 17 control and 18 hmgr-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (E) Top: Sum intensity projected images of ZMP-1::mNG shows ZMP-1 enrichment at the AC basal invasive plasma membrane (arrows) in a control and an hmgr-1 RNAi-treated animal at the initiation of invasive protrusion formation. Bottom: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and hmgr-1 RNAi treated animals (n = 14 control and 16 hmgr-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.
HMGR-1 promotes lysosome stores, GTPase accumulation, and invasive protrusion formation. (A) A schematic diagram showing the mevalonate pathway in the AC (see Fig. S1 B for pathway details, Table S2 for RNAi screen). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific hmgr-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of hmgr-1 (middle, bracket, observed in n = 7/7 hmgr-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 7). Right: Boxplot of the maximum invasive protrusion volume in control and hmgr-1 RNAi treated animals (n = 9 control and 7 hmgr-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (C) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in a control and an hmgr-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume of control and hmgr-1 RNAi animals (n = 17 control and 18 hmgr-1 RNAi animals, *** P ≤ 0.001, unpaired two-tailed Student’s t test). (D) Top: Sum intensity z-projected images showing invasive polarization (arrows) of GFP::CED-10, GFP::MIG-2, GFP::CAAX, and mNG::RAP-1 at the AC plasma membrane in control and hmgr-1 RNAi animals at the initiation of invasive protrusion formation. Bottom: Boxplots showing the AC basal/apical ratio of each protein’s fluorescence intensity in control and hmgr-1 RNAi-treated animals ([CED-10] n = 23 control and 23 hmgr-1 RNAi animals; [MIG-2] n = 13 control and 13 hmgr-1 RNAi animals; [CAAX] n = 16 control and 16 hmgr-1 RNAi animals; [RAP-1] n = 17 control and 18 hmgr-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (E) Top: Sum intensity projected images of ZMP-1::mNG shows ZMP-1 enrichment at the AC basal invasive plasma membrane (arrows) in a control and an hmgr-1 RNAi-treated animal at the initiation of invasive protrusion formation. Bottom: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and hmgr-1 RNAi treated animals (n = 14 control and 16 hmgr-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.
The rate limiting enzyme of the mevalonate pathway is HMG-CoA reductase (hmgr-1). HMG-CoA synthase (hmgs-1) is also a key upstream regulator (Fig. S1 B) (Watts and Ristow, 2017). Using the AC-specific RNAi strain, we found that RNAi against hmgr-1 and hmgs-1 strongly blocked AC invasion (Table S2). Time-lapse analysis of protrusion formation after hmgr-1 RNAi revealed a significant reduction in protrusion formation and growth rate and a decrease in BM removal under the AC (Fig. 6 B, Fig. S2 E, and Video 7). Examination of key regulators of protrusion formation and function after hmgr-1 RNAi further revealed that HMGR-1 is required for generating lysosome stores prior to invasion (Fig. 6 C) and is necessary for the polarized enrichment of the prenylated small GTPases CED-10, MIG-2, and RAP-1, and GFP::CAAX (Fig. 6 D), but not for the enrichment of the GPI-anchored ZMP-1 (Fig. 6 E). Interestingly, hmgr-1 RNAi also resulted in the nuclear localization of the Rac proteins CED-10 and MIG-2. Similar nuclear localization of vertebrate Rac1 occurs in the absence of prenylation, as Rho GTPases have C-terminal polybasic regions that direct nuclear localization when prenylation does not occur (Navarro-Lerida et al., 2015; Williams, 2003). Together these results implicate a key role for HMGR-1/the mevalonate pathway in targeting prenylated GTPases to the invasive protrusion, as well as the generation of lysosomes that form the protrusion.
HMGR-1 is required for the AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific hmgr-1 RNAi knockdown animal. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, display rate 8 frames/sec. Scale bar, 5 µm.
HMGR-1 is required for the AC invasive protrusion formation. Lateral view time-lapse images showing the AC invasive protrusion visualized with the membrane marker mCherry::PLCδPH (cyan) in a control and AC-specific hmgr-1 RNAi knockdown animal. BM was visualized with LAM-1:GFP (magneta). Movies were acquired with a CSU-10 spinning disc confocal microscope. Gaussian blur (1 pixel) was applied to both control and RNAi-treated animals to facilitate visualization of AC protrusion. The total video duration is 75 min, frame interval 5 min, display rate 8 frames/sec. Scale bar, 5 µm.
We were next interested in understanding what lipid derivatives that are formed from the product of HMGR-1 promote lysosome construction and GTPase localization. We thus used the AC-specific RNAi strain and knocked down 10 key enzymes acting downstream of HMGR-1 that construct diverse lipid products (Table S2 and Fig. S1 B) (Rauthan and Pilon, 2011). Notably, RNAi targeting polyprenol reductase (B0024.13), which catalyzes the conversion of polyprenol to dolichol, strongly perturbed AC invasion. Prenylation (farnesyl and geranylgeranyl) was also implicated, as RNAi against the prenyltransferases fnta-1 and fntb-1 and the isoprenylcysteine carboxylmethyltransferase icmt-1, whose gene product catalyzes the last step of prenylation, significantly disrupted AC invasion (Fig. 6 A, Fig. S1 B, and Table S2) (Wang and Casey, 2016). We thus further examined the polyprenol reductase B0024.13 and icmt-1 (the final step in prenylation) and found that B0024.13 RNAi reduced lysosome stores, while RNAi targeting of icmt-1 decreased the invasive membrane localization of the prenylated Rac CED-10 (Fig. S4, A–C). These results suggest that dolichol, which plays a key role in N-linked glycosylation, is required for forming lysosome stores that construct the protrusion, while prenylation is crucial for GTPase localization for protrusion formation.
Role and regulation of HMGR-1 and the mevalonate pathway. (A) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and in a B0024.13 (dolichol synthesis) RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot of AC lysosomal volume in control and B0024.13 RNAi treated animals (n = 20 control and 24 B0024.13 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and icmt-1 RNAi treated animal at the initiation of invasive protrusion formation. Boxplot of AC lysosome volume in control and icmt-1 RNAi treated animals (n = 23 control and 21 icmt-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projection fluorescence images of GFP::CED-10 shows enrichment at the AC basal plasma membrane (arrow) in a control, an icmt-1, and a B0024.13 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal level of GFP::CED-10 mean fluorescence intensity (n = 29 control, 22 B0024.13, and 21 icmt-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (D) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::HMGR-1 (cyan), and overlay in the AC show overlap of mNG::HMGR-1 and mCherry::KDEL signals (white arrows) and a region of enriched mNG::HMGR-1 with no overlap (yellow arrows). Below: Insets highlighting region of increased mNG::HMGR-1 fluorescence where there is no mCherry::KDEL (yellow arrows; similar colocalization observed in = 8/8 animals). (E) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::HMGR-1 (cyan), and overlay in the AC shows overlap of enriched mNG::HMGR-1 regions and mScarlet::PTS1 (white arrows). Below: Insets highlighting region of overlap (arrows; similar colocalization observed in = 3/3 animals). (F) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in two ACs with mNG::HMGR-1 enriched at the invasive basal side (arrows). Right: mNG::HMGR-1 is not concentrated at the basal side of uterine cells (UCs, arrows, n = ≥10/10). (G) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in the AC (arrows show AC and enrichment at the invasive front) in a control and a sbp-1 RNAi treated animal. Right: Boxplot of mNG::HMGR-1 fluorescence intensity in the AC of control and sbp-1 RNAi animals (n = 26 control and 27 sbp-1 RNAi animals, unpaired two-tailed Student’s t test, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (H) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 polarized (arrow) in a wild-type and mispolarized in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of mNG::HMGR-1 fluorescence intensity in wild-type and unc-6(ev400) animals (n = 48 control and 30 unc-6(ev400) animals, **** P ≤ 0.0001, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.
Role and regulation of HMGR-1 and the mevalonate pathway. (A) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and in a B0024.13 (dolichol synthesis) RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot of AC lysosomal volume in control and B0024.13 RNAi treated animals (n = 20 control and 24 B0024.13 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and icmt-1 RNAi treated animal at the initiation of invasive protrusion formation. Boxplot of AC lysosome volume in control and icmt-1 RNAi treated animals (n = 23 control and 21 icmt-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projection fluorescence images of GFP::CED-10 shows enrichment at the AC basal plasma membrane (arrow) in a control, an icmt-1, and a B0024.13 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal level of GFP::CED-10 mean fluorescence intensity (n = 29 control, 22 B0024.13, and 21 icmt-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (D) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::HMGR-1 (cyan), and overlay in the AC show overlap of mNG::HMGR-1 and mCherry::KDEL signals (white arrows) and a region of enriched mNG::HMGR-1 with no overlap (yellow arrows). Below: Insets highlighting region of increased mNG::HMGR-1 fluorescence where there is no mCherry::KDEL (yellow arrows; similar colocalization observed in = 8/8 animals). (E) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::HMGR-1 (cyan), and overlay in the AC shows overlap of enriched mNG::HMGR-1 regions and mScarlet::PTS1 (white arrows). Below: Insets highlighting region of overlap (arrows; similar colocalization observed in = 3/3 animals). (F) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in two ACs with mNG::HMGR-1 enriched at the invasive basal side (arrows). Right: mNG::HMGR-1 is not concentrated at the basal side of uterine cells (UCs, arrows, n = ≥10/10). (G) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in the AC (arrows show AC and enrichment at the invasive front) in a control and a sbp-1 RNAi treated animal. Right: Boxplot of mNG::HMGR-1 fluorescence intensity in the AC of control and sbp-1 RNAi animals (n = 26 control and 27 sbp-1 RNAi animals, unpaired two-tailed Student’s t test, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (H) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 polarized (arrow) in a wild-type and mispolarized in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of mNG::HMGR-1 fluorescence intensity in wild-type and unc-6(ev400) animals (n = 48 control and 30 unc-6(ev400) animals, **** P ≤ 0.0001, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.
HMGR-1 and protein prenylation enzymes polarize to the AC’s invasive front
Given the unique role of HMGR-1 in regulating the localization of Rac and Ras-related GTPases in the AC, we were next interested in determining the localization and regulation of HMGR-1. HMG-CoA reductases are conserved multipass transmembrane proteins that localize to the ER and peroxisomes (Fig. 7 A) (Keller et al., 1986; Wang et al., 2024). Using genome editing, we knocked-in mNG at the N-terminus of the hmgr-1 locus (mNG::HMGR-1) and found that HMGR-1 localized to the perinuclear region in many cell types and within the AC colocalized with the ER and was enriched in peroxisomes (Fig. 7 B; and Fig. S4, D and E). Notably, peroxisome-localized HMGR-1 polarized toward the AC invasive side prior to and during the invasion—a polarization not observed in neighboring uterine cells (Fig. 7 B; and Fig. S4, D and F). The mevalonate pathway in C. elegans does not contain the cholesterol synthesis branch, and it is unknown if SBP-1 regulates hmgr-1/HMG-CoA reductase expression as SREBP proteins do in organisms that produce cholesterol. RNAi-mediated reduction of the SBP-1 protein did not alter the levels of mNG::HMGR-1 in the AC (Fig. S4 G), indicating independence of HMGR-1 expression from SBP-1 regulation.
Prenylation enzymes HMGR-1, FCE-1, and ICMT-1 polarize towards BM. (A) A schematic diagram showing the mevalonate/HMGR-1 prenylation pathway (see text for detailed description). (B) Left: Sum intensity z-projected images show AC localization and invasive enrichment (arrows) of the endogenously tagged membrane-associated prenylation enzymes mNG::HMGR-1 and mNG::FCE-1, and AC expressed ICMT-1::mScarlet (lin-29p::icmt-1::mScarlet, inset shows spectral fluorescence-intensity map displaying the minimum and maximum pixel value range of the acquired data) from the P6.p 1-cell to 4-cell stages. All the subsequent spectral fluorescence-intensity maps also display the maximum (H) and minimum (L) pixel value range of the acquired data, which was independently determined for each image. Right: Boxplots show the AC basal/apical ratios of mNG::HMGR-1, mNG::FCE-1, and ICMT-1::mScarlet fluorescence intensity ([HMGR-1] n = 12 1-cell, 16 2-cell, 9 2–4-cell, and 11 4-cell stage animals; [FCE-1] n = 9 1-cell, 14 2-cell, 13 2–4-cell, and 16 4-cell stage animals; [ICMT-1] n = 10 1-cell, 15 2-cell, 10 2–4-cell, and 11 4-cell stage animals, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (C) Top: Single slice confocal images of IMCT-1::mScarlet (magenta), mNG::KDEL (cyan, ER marker) and overlay in the AC show tight overlap of localization of ICMT-1 throughout the ER (white arrows). Bottom: Insets highlight the AC basal ER region and show a puncta of intense IMCT-1 enrichment within the ER (left arrow) and a region of similar intensity to the mNG::KDEL marker (right arrow) (similar localization observed in n = 25/25 animals). All data are from two or more replicates. Scale bars, 5 µm.
Prenylation enzymes HMGR-1, FCE-1, and ICMT-1 polarize towards BM. (A) A schematic diagram showing the mevalonate/HMGR-1 prenylation pathway (see text for detailed description). (B) Left: Sum intensity z-projected images show AC localization and invasive enrichment (arrows) of the endogenously tagged membrane-associated prenylation enzymes mNG::HMGR-1 and mNG::FCE-1, and AC expressed ICMT-1::mScarlet (lin-29p::icmt-1::mScarlet, inset shows spectral fluorescence-intensity map displaying the minimum and maximum pixel value range of the acquired data) from the P6.p 1-cell to 4-cell stages. All the subsequent spectral fluorescence-intensity maps also display the maximum (H) and minimum (L) pixel value range of the acquired data, which was independently determined for each image. Right: Boxplots show the AC basal/apical ratios of mNG::HMGR-1, mNG::FCE-1, and ICMT-1::mScarlet fluorescence intensity ([HMGR-1] n = 12 1-cell, 16 2-cell, 9 2–4-cell, and 11 4-cell stage animals; [FCE-1] n = 9 1-cell, 14 2-cell, 13 2–4-cell, and 16 4-cell stage animals; [ICMT-1] n = 10 1-cell, 15 2-cell, 10 2–4-cell, and 11 4-cell stage animals, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (C) Top: Single slice confocal images of IMCT-1::mScarlet (magenta), mNG::KDEL (cyan, ER marker) and overlay in the AC show tight overlap of localization of ICMT-1 throughout the ER (white arrows). Bottom: Insets highlight the AC basal ER region and show a puncta of intense IMCT-1 enrichment within the ER (left arrow) and a region of similar intensity to the mNG::KDEL marker (right arrow) (similar localization observed in n = 25/25 animals). All data are from two or more replicates. Scale bars, 5 µm.
UNC-6 (vertebrate netrin) is secreted by the vulval P6.p cell and its descendants and polarizes Rho GTPases, other actin regulators, and metabolic components of the invasive machinery to orient the invasive protrusion toward the BM (Garde et al., 2022; Hagedorn et al., 2013; Naegeli et al., 2017; Wang et al., 2014b). We thus hypothesized that HMGR-1 might also be a component of the invasive machinery polarized by UNC-6. Consistent with this notion, HMGR-1 was no longer strongly enriched toward the BM in unc-6(ev400) null mutant animals (Fig. S4 H). These results suggest that HMGR-1 is a component of the invasive machinery for BM breaching.
After the action of the cytosolic prenyltransferases such as FNTA-1, prenylated proteins are processed by the multipass transmembrane resident RAS-converting CAAX endopeptidase 1 (RCE1, C. elegans FCE-1), which removes the -AAX residue, followed by the action of the transmembrane ICMT, which adds a methyl group onto the carboxyl terminus that increases membrane interactions of prenylated proteins (Fig. 7 A) (Wang and Casey, 2016). Previous studies have suggested that RCE1 and ICMT proteins localize to the ER (Bracha-Drori et al., 2008; Schmidt et al., 1998). Given the polarized enrichment of HMGR-1 protein toward the site of protrusion formation, we wanted to determine the localization of the cytosolic FNTA-1 and the putative ER transmembrane FCE-1 and ICMT-1 proteins. We used genome editing to tag the fnta-1 locus at the C-terminus (FNTA-1::mNG) and the fce-1 locus at the N-terminus (mNG::FCE-1) but were unable to tag icmt-1 at its endogenous locus and thus drove its expression under an AC-specific promoter (lin-29p::icmt-1::mScarlet). As expected, FNTA-1::mNG was cytosolic, but levels were elevated in the AC, and there was a consistent enrichment at the invasive front during BM breaching (Fig. S5 A). Like HMGR-1, both FCE-1 and ICMT-1 were also enriched toward the AC’s invasive side (Fig. 7 B), and like HMGR-1, ICMT-1 was polarized toward the BM by UNC-6 (Fig. S5 B). Interestingly, ICMT-1 polarization was highest at the time of protrusion formation, whereas HMGR-1 and FCE-1 showed similar polarization prior to and during invasion (Fig. 7 B). Consistent with similar regulation, FCE-1 localized to the ER and was found (but not as enriched) in peroxisomes (Fig. S5, C and D). Interestingly, ICMT strongly colocalized with the general ER marker KDEL and was uniquely found in bright ICMT puncta that localized to ER regions with lower KDEL levels, suggesting ICMT-1 concentrated in a unique subdomain of the ER (Fig. 7 C). Together, these observations indicate that the prenylation enzymes HMGR-1 and ICMT-1 are polarized to the site of invasive protrusion formation by UNC-6 (netrin), and suggest that ICMT-1, which mediates the final step of prenylation, occurs within the ER and enriches in an ER subdomain.
Regulation and localization of prenylation enzymes and GTPases during AC invasion. (A) Sum intensity z-projected images show high levels of FNTA-1::mNG in the AC at the P6.p 2-cell stage (n = 12/12 animals) and 4-cell stage (n = 12/12). FNTA-1::mNG basal enrichment was observed at the P6.p 4-cell stage (arrows, n = 12/12). Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. (B) Sum intensity z-projected images of ICMT-1::mScarlet in the AC show polarization in a wild-type animal (arrows) and lack of polarization in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of ICMT-1::mScarlet in wild-type and unc-6(ev400) animals (n = 31 control and 34 unc-6(ev400) animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (C) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::FCE-1 (cyan), and overlay in the AC show overlap of mNG::FCE-1 and mCherry::KDEL signals (white arrows) and a region of mNG::FCE-1 with no overlap with mCherry::KDEL (yellow arrows). Bottom: Insets highlighting region of mNG::FCE-1 fluorescence where there is no mCherry::KDEL (yellow arrows, similar colocalization observed in n = 13/13 animals). (D) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::FCE-1 (cyan), and overlay in the AC shows overlap of mNG::FCE-1 regions and mScarlet::PTS1 (white arrows). Bottom: Insets highlighting region of overlap (white arrows; similar colocalization observed in = 3/3 animals). (E) Top: Central AC DIC images and corresponding AC max intensity z-projected images of sfGFP::KDEL (ER marker, top) and (bottom) DIC image and AMAN-2::GFP (Golgi marker, bottom) at the time of protrusion formation show the ER and Golgi extending into the protrusion (below the BM breach indicated by dotted orange line, n = 12/12 ER and 12/14 Golgi). (F) Top: Sum intensity z-projected image of AC specific expression of sfGFP::KDEL (ER marker) at the P6.p 4-cell stage. Bottom: Line graph of AC basal/apical polarization from the P6.p 1-cell through 4-cell stages (n = 8 1-cell, 8 2-cell, 9 2–4-cell, and 6 4-cell stage animals, ns [not statistically significant], Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (G) Left: Sum intensity z-projected images showing the time course of GFP::CED-10, GFP::MIG-2, and mNG::RAP-1 polarization from the P6.p 1-cell to 4-cell stage. Right: Line graphs of AC basal/apical polarization of each protein’s fluorescence intensity ([CED-10] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 15 4-cell stage animals; [MIG-2] n = 10 1-cell, 11 2-cell, 13 2–4-cell, and 17 4-cell stage animals; [RAP-1] n = 11 1-cell, 13 2-cell, 10 2–4-cell, and 19 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). All data are from two or more replicates. Scale bars, 5 µm.
Regulation and localization of prenylation enzymes and GTPases during AC invasion. (A) Sum intensity z-projected images show high levels of FNTA-1::mNG in the AC at the P6.p 2-cell stage (n = 12/12 animals) and 4-cell stage (n = 12/12). FNTA-1::mNG basal enrichment was observed at the P6.p 4-cell stage (arrows, n = 12/12). Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. (B) Sum intensity z-projected images of ICMT-1::mScarlet in the AC show polarization in a wild-type animal (arrows) and lack of polarization in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of ICMT-1::mScarlet in wild-type and unc-6(ev400) animals (n = 31 control and 34 unc-6(ev400) animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (C) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::FCE-1 (cyan), and overlay in the AC show overlap of mNG::FCE-1 and mCherry::KDEL signals (white arrows) and a region of mNG::FCE-1 with no overlap with mCherry::KDEL (yellow arrows). Bottom: Insets highlighting region of mNG::FCE-1 fluorescence where there is no mCherry::KDEL (yellow arrows, similar colocalization observed in n = 13/13 animals). (D) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::FCE-1 (cyan), and overlay in the AC shows overlap of mNG::FCE-1 regions and mScarlet::PTS1 (white arrows). Bottom: Insets highlighting region of overlap (white arrows; similar colocalization observed in = 3/3 animals). (E) Top: Central AC DIC images and corresponding AC max intensity z-projected images of sfGFP::KDEL (ER marker, top) and (bottom) DIC image and AMAN-2::GFP (Golgi marker, bottom) at the time of protrusion formation show the ER and Golgi extending into the protrusion (below the BM breach indicated by dotted orange line, n = 12/12 ER and 12/14 Golgi). (F) Top: Sum intensity z-projected image of AC specific expression of sfGFP::KDEL (ER marker) at the P6.p 4-cell stage. Bottom: Line graph of AC basal/apical polarization from the P6.p 1-cell through 4-cell stages (n = 8 1-cell, 8 2-cell, 9 2–4-cell, and 6 4-cell stage animals, ns [not statistically significant], Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (G) Left: Sum intensity z-projected images showing the time course of GFP::CED-10, GFP::MIG-2, and mNG::RAP-1 polarization from the P6.p 1-cell to 4-cell stage. Right: Line graphs of AC basal/apical polarization of each protein’s fluorescence intensity ([CED-10] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 15 4-cell stage animals; [MIG-2] n = 10 1-cell, 11 2-cell, 13 2–4-cell, and 17 4-cell stage animals; [RAP-1] n = 11 1-cell, 13 2-cell, 10 2–4-cell, and 19 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). All data are from two or more replicates. Scale bars, 5 µm.
ICMT-1 concentrates at ER exit sites, which polarize toward the invasive protrusion
As ICMT-1 mediates the last step of prenylation and was polarized toward the BM, we were next interested in further examining the localization of ICMT-1 concentrated in puncta (Fig. 8 A). We hypothesized that these might be ER exit sites to allow the rapid trafficking of fully prenylated proteins to the invasive protrusion. To test this idea, we knocked-in mNG at the N-terminus of the sec-16a.1 locus (mNG::SEC-16A.1), whose protein marks ER exit sites (Watson et al., 2006), and found an ∼100% overlap of mScarlet::ICMT-1 with mNG::SEC-16A.1 at the time of protrusion formation (Fig. 8 A). We also observed that ICMT-1 colocalized with SEC-16A.1 at the P6.p 1-cell stage (∼3 h prior to invasion, 94.28% colocalization, n = 5 animals), suggesting ICMT-1 is an ER exit site resident. Consistent with the trafficking of prenylated proteins from ER exit sites to the cell surface, RNAi reduction of sar-1, a GTPase that mediates COPII vesicle formation at ER exit sites for trafficking of ER cargo to the Golgi and then cell surface (Van der Verren and Zanetti, 2023), led to retention of membrane vesicles harboring GFP::CED-10 near the invasive plasma membrane (Fig. 8 B).
ICMT-1 localizes to ER exit sites that dynamically polarize toward the invasive protrusion. (A) Top: Sum intensity z-projected images of ICMT-1::mScarlet (magenta), ER exit marker mNG::SEC-16A.1 (cyan), and overlay in the AC reveal colocalization at ER exit sites (arrows). The average percentage ± SD of colocalization of ER exit sites with ICMT-1 puncta is indicated (n = 17 animals). Bottom: Insets highlight ICMT-1::mScarlet and mNG::SEC-16A.1 colocalization in the ER at the AC invasive front (arrows). (B) Max intensity z-projected fluorescence images of GFP::CED-10 in a control and sar-1 RNAi treated animals shows GFP::CED-10 localized to the invasive plasma membrane (arrow) in the control animal, but predominantly localized in intracellular vesicles in sar-1 RNAi animals (brackets). Right: The relative proportion of animals with CED-10::GFP that localized predominantly to the AC basal plasma membrane or was found predominantly in the cytosol in control and sar-1 RNAi treated animals (n = 38 control and 45 sar-1 RNAi animals, ** P ≤ 0.01, Fisher’s exact test). (C) Fluorescence images showing AC ER exit sites (mNG::SEC-16A.1, Cyan; AC outlined with the plasma membrane marker mCherry::PLCδPH, red), Golgi (AMAN-2::mScarlet), and prenylated GFP::CAAX in the AC from the P6.p1-cell to 4-cell stages. Arrows indicate basal (invasive front) enrichment of each protein. Right: Quantifications of the relative distribution of the ER exit site, Golgi, and plasma membrane markers in the AC from the P6.p 1-cell to 4-cell stages. For mNG::SEC-16A.1 and AMAN-2::mScarlet, the total number of puncta in the AC apical and basal halves (green dashed line) was expressed as a basal/apical ratio. The mean fluorescence intensity of GFP::CAAX was expressed as a basal/apical ratio ([SEC-16A.1] n = 10 1-cell, 10 2-cell, 9 2–4-cell, and 10 4-cell stage animals; [AMAN-2] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 16 4-cell stage animals; [CAAX] n = 12 1-cell, 11 2-cell, 10 2–4-cell, and 11 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Mann–Whitney U test, and Brown–Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (D) A schematic diagram illustrating where GTPase prenylation occurs, with FCE-1 present in the ER and peroxisomes and ICMT-1, the final prenylation enzyme, enriched in ER exit sites (ERES) that deliver prenylated proteins (i.e., Rac and Ras-related) to the Golgi apparatus for targeting to the invasive protrusion (red arrows). Rac/Ras-related prenylated proteins might also be delivered directly from the ER to the invasive plasma membrane (magenta arrows, inset). All data in the figure are from two or more replicates. Scale bars, 5 µm.
ICMT-1 localizes to ER exit sites that dynamically polarize toward the invasive protrusion. (A) Top: Sum intensity z-projected images of ICMT-1::mScarlet (magenta), ER exit marker mNG::SEC-16A.1 (cyan), and overlay in the AC reveal colocalization at ER exit sites (arrows). The average percentage ± SD of colocalization of ER exit sites with ICMT-1 puncta is indicated (n = 17 animals). Bottom: Insets highlight ICMT-1::mScarlet and mNG::SEC-16A.1 colocalization in the ER at the AC invasive front (arrows). (B) Max intensity z-projected fluorescence images of GFP::CED-10 in a control and sar-1 RNAi treated animals shows GFP::CED-10 localized to the invasive plasma membrane (arrow) in the control animal, but predominantly localized in intracellular vesicles in sar-1 RNAi animals (brackets). Right: The relative proportion of animals with CED-10::GFP that localized predominantly to the AC basal plasma membrane or was found predominantly in the cytosol in control and sar-1 RNAi treated animals (n = 38 control and 45 sar-1 RNAi animals, ** P ≤ 0.01, Fisher’s exact test). (C) Fluorescence images showing AC ER exit sites (mNG::SEC-16A.1, Cyan; AC outlined with the plasma membrane marker mCherry::PLCδPH, red), Golgi (AMAN-2::mScarlet), and prenylated GFP::CAAX in the AC from the P6.p1-cell to 4-cell stages. Arrows indicate basal (invasive front) enrichment of each protein. Right: Quantifications of the relative distribution of the ER exit site, Golgi, and plasma membrane markers in the AC from the P6.p 1-cell to 4-cell stages. For mNG::SEC-16A.1 and AMAN-2::mScarlet, the total number of puncta in the AC apical and basal halves (green dashed line) was expressed as a basal/apical ratio. The mean fluorescence intensity of GFP::CAAX was expressed as a basal/apical ratio ([SEC-16A.1] n = 10 1-cell, 10 2-cell, 9 2–4-cell, and 10 4-cell stage animals; [AMAN-2] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 16 4-cell stage animals; [CAAX] n = 12 1-cell, 11 2-cell, 10 2–4-cell, and 11 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Mann–Whitney U test, and Brown–Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (D) A schematic diagram illustrating where GTPase prenylation occurs, with FCE-1 present in the ER and peroxisomes and ICMT-1, the final prenylation enzyme, enriched in ER exit sites (ERES) that deliver prenylated proteins (i.e., Rac and Ras-related) to the Golgi apparatus for targeting to the invasive protrusion (red arrows). Rac/Ras-related prenylated proteins might also be delivered directly from the ER to the invasive plasma membrane (magenta arrows, inset). All data in the figure are from two or more replicates. Scale bars, 5 µm.
We next wanted to determine the localization of the total ER, ER exit sites, and the Golgi (which has dispersed cisternae in C. elegans) (Sato et al., 2014), prior to and during invasion to examine the dynamics of the vesicle trafficking route during protrusion formation. While the ER was modestly enriched toward the invasive membrane prior to and during protrusion formation (Fig. S5 F), ER exit sites and the Golgi cisternae strongly polarized toward the BM, specifically during invasive protrusion formation (P6.p 2–4-cell stage) (Fig. 8 C). The ER and the Golgi also extended into the invasive protrusion (Fig. S5 E). Notably, the polarization of the vesicular apparatus coincided with the increased polarity of GTPases and GFP::CAAX (Fig. 8 C and Fig. S5 G). Taken together, these observations indicate that the prenylation machinery within the AC is regionally organized and that the final step of prenylation occurs at ER exit sites, which dynamically polarize with the Golgi to direct prenylated proteins to the invasive protrusion during BM breaching (Fig. 8 D).
Discussion
Cell invasion through BM barriers is crucial for many developmental processes and immune cell trafficking, and the cell invasion program is hijacked in cancer metastasis (Kelley et al., 2014; Paterson and Courtneidge, 2018). To transmigrate BM, cells use large membrane protrusions that harbor signaling molecules and lipid-anchored proteins to clear the dense, covalently crosslinked BM protein network (Naegeli et al., 2017; Nazari et al., 2023; Schoumacher et al., 2010). Although many lipid synthesis enzymes are strongly associated with cancer progression (Martin-Perez et al., 2022), due to the complexity of lipid metabolism and the difficulty of studying BM invasion, it has been challenging to establish the significance, regulation, and potential roles of lipid synthesis and lipid modification enzymes within invasive cells.
Using the model system of C. elegans AC invasion, endogenous gene tagging, AC-specific RNAi, genetic analysis, and live-cell imaging, we show that de novo lipid synthesis is crucial for BM invasion. We discovered that the C. elegans SREBP transcription factor ortholog SBP-1 is transiently present at high levels in the AC nucleus ∼5 h prior to invasion and is required for invasive protrusion formation. SBP-1 promotes the expression of acetyl-CoA carboxylase ACC (C. elegans POD-2), the key rate-limiting enzyme of de novo fatty acid synthesis, as well as the fatty acid synthase FASN-1 (Rohrig and Schulze, 2016). SREBP functions have primarily been studied in physiological and pathophysiological settings (Shimano and Sato, 2017). These findings add to a limited number of developmental studies indicating key cell-autonomous specific roles for SREBP proteins in morphogenetic cell behaviors, such as in the lipid-intensive process of mouse cortical neurite plasma membrane outgrowth and Drosophila sensory neuron dendrite membrane expansion (Hasegawa et al., 2012; Meltzer et al., 2017; Ziegler et al., 2017). Our studies support a model where POD-2 and FASN-1 generate fatty acids within the AC that the phosphatidic acid phosphatase lipin (LPIN-1) and sphingomyelin synthase SMS-1 use to generate phospholipids and sphingomyelin that form lysosome stores that are exocytosed to generate the invasive protrusion. Our results also suggest that lysosomes deliver SMS-1-produced sphingomyelin/lipid rafts to the invasive protrusion and that SMS-1/sphingomyelin promotes the localization of the lipid raft-associated DCC (UNC-40) receptor, which directs invasive protrusion formation, and the GPI-anchored ZMP-1 protease (Naegeli et al., 2017). In addition, our studies reveal that the HMG-CoA reductase HMGCR (C. elegans HMGR-1), the rate-limiting enzyme in the mevalonate pathway (Rauthan and Pilon, 2011), plays an important role in invasive protrusion formation by mediating the polarized prenylation and delivery of proteins, such as Rac GTPases, to the invasive protrusion. Notably, in vertebrates, the mevalonate pathway produces cholesterol, and HMGCR gene expression is under the control of sterol-responsive SREBP transcription (DeBose-Boyd and Ye, 2018). However, C. elegans lacks the cholesterol-synthetic branch of the mevalonate pathway (Rauthan and Pilon, 2011), and we found that SBP-1 does not regulate HMGR-1 expression. Although RNAi knockdown strongly depleted protein levels for each of these lipid regulators, the formation of small and less dynamic (reduced growth rate) invasive protrusions after RNAi-mediated knockdown, might have resulted from reduced but not eliminated lipid regulator activity, or from parallel pathways that promote protrusion formation. Nevertheless, these studies establish that an invasive cell uses a robust de novo lipid synthesis network that acts in concert with polarized prenylation to rapidly form a large, dynamic, and highly specialized protrusion that removes BM barriers (Fig. 9).
De novo lipid synthesis and polarized prenylation promote invasive protrusion formation. (A) A schematic diagram summarizing AC invasion. Invadosomes form and one breaches the underlying BM. The netrin receptor DCC (UNC-40) enriches at the BM breach site and directs lysosomal exocytosis, which provides membrane for protrusion growth. AC invasive protrusion growth is facilitated by the enrichment of GPI-anchored matrix degrading metalloproteinase ZMP-1 and prenylated Rac and Ras-like GTPases that direct F-actin formation. (B) A schematic diagram highlighting the roles and regulation of de novo lipid synthesis and lipid prenylation during AC invasive protrusion formation. Nuclear SBP-1 promotes expression of the fatty acid synthesis enzymes pod-2 and fasn-1 that provide fatty acid substrates for LPIN-1 and SMS-1, which synthesize phospholipids and sphingomyelin that build lysosome stores for the protrusion. Sphingomyelin, which is a key component of lipid rafts, is also required for the enrichment of the UNC-40 receptor and the GPI-anchored metalloprotease ZMP-1, which both partition to lipid rafts. Sphingomyelin and phospholipids are delivered to the protrusion via lysosomes. A specialized prenylation system, where the intermediate enzyme FCE-1 localizes to ER and peroxisomes and the final enzyme of prenylation, ICMT-1, localizes to ER exit sites (ERES), rapidly delivers Rac- and Ras-like GTPases to the AC invasive front to direct protrusion growth. Phospholipids and prenylated proteins might also be delivered directly to the invasive protrusion from the ER (magenta arrows).
De novo lipid synthesis and polarized prenylation promote invasive protrusion formation. (A) A schematic diagram summarizing AC invasion. Invadosomes form and one breaches the underlying BM. The netrin receptor DCC (UNC-40) enriches at the BM breach site and directs lysosomal exocytosis, which provides membrane for protrusion growth. AC invasive protrusion growth is facilitated by the enrichment of GPI-anchored matrix degrading metalloproteinase ZMP-1 and prenylated Rac and Ras-like GTPases that direct F-actin formation. (B) A schematic diagram highlighting the roles and regulation of de novo lipid synthesis and lipid prenylation during AC invasive protrusion formation. Nuclear SBP-1 promotes expression of the fatty acid synthesis enzymes pod-2 and fasn-1 that provide fatty acid substrates for LPIN-1 and SMS-1, which synthesize phospholipids and sphingomyelin that build lysosome stores for the protrusion. Sphingomyelin, which is a key component of lipid rafts, is also required for the enrichment of the UNC-40 receptor and the GPI-anchored metalloprotease ZMP-1, which both partition to lipid rafts. Sphingomyelin and phospholipids are delivered to the protrusion via lysosomes. A specialized prenylation system, where the intermediate enzyme FCE-1 localizes to ER and peroxisomes and the final enzyme of prenylation, ICMT-1, localizes to ER exit sites (ERES), rapidly delivers Rac- and Ras-like GTPases to the AC invasive front to direct protrusion growth. Phospholipids and prenylated proteins might also be delivered directly to the invasive protrusion from the ER (magenta arrows).
The invasive protrusion of the AC transiently increases the AC surface area by as much as ∼40% through UNC-40/t-SNARE SNAP-29–mediated lysosome exocytosis (Naegeli et al., 2017). Previous studies found that loss of the phosphatidylinositol phosphate kinase PPK-3, which regulates the maturation and integrity of lysosomes (Nicot et al., 2006), dramatically reduced the invasive protrusion, indicating that lysosomes are a crucial membrane source for protrusion formation (Naegeli et al., 2017). Notably, our time-lapse analysis confirmed that lysosome vesicles dynamically traffic into the protrusion during its formation. While a great deal is known about signaling and transcriptional control of lysosome number and size (Yang and Wang, 2021), the role of lipogenesis in lysosome construction is unclear. Our studies indicate that AC lysosome stores are reliant on de novo, cell-autonomous phospholipid synthesis, as AC LPIN-1 levels are highly upregulated and reduction of LPIN-1 dramatically reduced AC lysosomal volume prior to invasion. Given that phospholipids compose the bulk of membrane lipids (Cockcroft, 2021), this suggests that AC LPIN-1 is required to produce phospholipids used to form lysosome stores prior to invasion. Interestingly, we also found that the ER extends into the invasive protrusion, and thus it is possible that direct ER contact with the protrusion membrane could also deliver phospholipids to support protrusion growth (Fig. 9 B) (Guillen-Samander and De Camilli, 2023). Loss of the sphingomyelin synthase SMS-1 also reduced AC lysosomes prior to invasion. Although sphingomyelin composes only ∼5% of lipids in most cells, it is made in the Golgi and concentrated in the plasma membrane and within the endolysosome. Sphingomyelin is important for membrane stability (Goni, 2022; Tang et al., 2022), and thus its loss could affect lysosome stores. Sphingomyelin is also necessary for vesicle biogenesis at the Golgi (Duran et al., 2012), and sphingomyelin reduction might reduce the trafficking of phospholipids from the trans-Golgi to the lysosome (Liu et al., 2021). Finally, we discovered that the dolichol branch of the mevalonate pathway is also required for AC lysosome stores. Dolichol phosphate is critical for N-linked protein glycosylation, where dolichols function as a membrane anchor for the transfer of oligosaccharides to many newly translated proteins within the ER (Esmail and Manolson, 2021; Rauthan and Pilon, 2011). The luminal side of the lysosome membrane contains heavily glycosylated transmembrane proteins, including LAMP-1, LAMP-2, and LIMP-2 (C. elegans SCAV-3), which have been implicated in mediating lysosome integrity by forming a protective glycocalyx that prevents degradation from lysosome hydrolases (Li et al., 2016). Loss of glycosylation after perturbation of the dolichol branch of the mevalonate pathway is thus a strong candidate for the mechanism underlying reduced lysosomes within the AC. Together, these observations reveal the importance of independent lipogenesis pathways in regulating lysosome generation for invasive protrusion formation and suggest targeting lysosome stores might be a promising strategy for limiting cell invasion.
Our results also suggest that the generation of sphingomyelin within the AC is necessary for netrin receptor DCC (UNC-40) localization during invasive protrusion formation. DCC (UNC-40) localizes to the initial BM breach site, where in response to netrin (UNC-6) secreted by the underlying vulval cells, it directs lysosome exocytosis to form the invasive protrusion (Morrissey et al., 2013; Naegeli et al., 2017). Studies in vertebrate commissural neurons and immortalized cells have shown that DCC associates with lipid rafts and that lipid raft association is required for netrin-mediated activation (Herincs et al., 2005). We found that the loss of SMS-1, which generates sphingomyelin, a key component of lipid rafts (Bieberich, 2018; Goni, 2022), perturbed UNC-40 enrichment at the BM breach site. Thus, the reduction in the size and growth of the invasive protrusion after loss of SMS-1 could be in part due to perturbation in UNC-40 signaling. Like vertebrates, biochemical studies indicate that C. elegans harbor lipid rafts where GPI-anchored proteins partition (Rao et al., 2011; Sangiorgio et al., 2004). Notably, ZMP-1, a GPI-anchored protease, is concentrated in the invasive protrusion. Loss of SMS-1 disrupted ZMP-1 localization, strongly supporting the idea that the invasive cell membrane and protrusion are rich in sphingomyelin and lipid rafts that localize and regulate UNC-40, ZMP-1, and possibly other proteins that mediate invasion.
Little is known about the regulation of prenylation in cells (Wang and Casey, 2016). Endogenous tagging of the C. elegans HMG-CoA reductase (HMGR-1) protein of the mevalonate pathway that generates isoprenoids for prenylation revealed localization in the ER and enrichment of HMGR-1 in peroxisomes that polarize toward the invasive front of the AC. Similarly, the transmembrane protein FCE-1 was also found in the ER and peroxisomes. Although many studies have found that vertebrate HMG-CoA reductase (HMGCR) localizes to the ER and peroxisomes, the functional significance of its dual localization to the ER and peroxisomes is poorly understood, but has been suggested to play a role in cholesterol homeostasis (Wang et al., 2024). Given that HMGR-1 does not regulate cholesterol production in C. elegans and that HMGR-1 and the endopeptidase FCE-1, which cleaves the carboxyl-terminal three amino acids from CAAX proteins, localize to peroxisomes, our data suggests that HMG-CoA localization to peroxisomes might be important for prenylation. We further discovered that ICMT-1, which is the final enzyme of prenylation (Wang and Casey, 2016), specifically localizes to ER exit sites that dynamically orient toward the invasive protrusion during its formation. Notably, both peroxisome-localized HMGR-1 and ER-exit site localized ICMT-1 were polarized toward the invasive front by UNC-6 (netrin)—an invasive polarity cue generated by the underlying vulval cells that orients components of the invasive machinery, such as the Rac GTPases MIG-2 and CED-10, mitochondria, and the glucose transporter FGT-1 (Garde et al., 2022; Wang et al., 2014a, 2014b). These results indicate that polarized prenylation is a part of the cell invasion program. Together our results reveal a polarized prenylation and trafficking system, which directs the Rac GTPases MIG-2 and CED-10, as well as the Ras-like GTPase RAP-1 robustly to the invasive protrusion (Fig. 9). As many GTPases and signaling molecules are prenylated (Borini Etichetti et al., 2020; Rauthan and Pilon, 2011; Wang and Casey, 2016), the AC’s polarized prenylation system might be used to direct many proteins to the invasive protrusion to rapidly direct its formation and functionality.
Only ∼20% of C. elegans fatty acids are synthesized de novo, with most being acquired from the diet and imported into cells (Watts and Ristow, 2017). Further, in many organisms, fatty acid synthesis is primarily accomplished by specialized organs, such as the liver, and cells acquire fatty acids from the extracellular environment through lipid import (Cockcroft, 2021). Thus, it is notable that the invasive AC is dependent upon cell-autonomous fatty acid synthesis and uses SREBP (C. elegans SBP-1)-mediated transcription to direct high levels of expression of the fatty acid synthesis ACC (POD-2) and FASN (FASN-1) enzymes. Further, the AC requires enzymes that mediate phospholipid and sphingolipid synthesis (LPIN-1 and SMS-1) and prenylation-promoting enzymes (HMGR-1 and ICMT-1) to generate the invasive protrusion. Numerous studies have implicated the human orthologs of these enzymes in promoting metastasis (Borini Etichetti et al., 2020; Lu et al., 2022; Martin-Perez et al., 2022; Song et al., 2020; Zheng et al., 2019) and pharmacological inhibitors of these enzymes exist (Brohee et al., 2021; Companioni et al., 2021; Marchwicka et al., 2022; Röhrig and Schulze, 2016; Vasseur and Guillaumond, 2022), with HMGCR statin inhibitors showing potential promise in clinical trials targeting invasive glioblastoma in combination with chemotherapy (Afshari et al., 2021). Our results on the requirement of these lipid synthesis and modification enzymes during AC invasion offer strong in vivo support that using these inhibitors might be an effective strategy to limit the ability of tumors to breach BMs and metastasize.
Materials and methods
Strains and culture conditions
C. elegans strains were maintained on nematode growth medium (NGM) plates and fed with E. coli (OP50) at 16–20°C following standard worm growth conditions (Stiernagle, 2006). Please refer to Table S3 for strains used in this study. In this manuscript, we designate a linkage between the promoter (p) and the open reading frame with a double semicolon (::). Strains sourced from Caenorhabditis Genetics Center (CGC) are to be requested directly from CGC. Further requests for reagents should be directed to the corresponding author, D.R. Sherwood ([email protected]).
Construction of genome-edited strains
Fluorescently tagged strains were generated using CRISPR-Cas9-mediated genome editing as previously described (Dickinson et al., 2013; Keeley et al., 2020). Briefly, the mNeonGreen (mNG) coding sequence was knocked into a gene of interest via CRISPR-Cas9-triggered homologous recombination using a short guide RNA (sgRNA) and a homologous repair template that contained the fluorophore coding sequence and selection markers. Silent mutations were introduced into the homology arms using site-directed mutagenesis (refer to Table S4 for information on sgRNA sequences). All sgRNAs were cloned into the pDD122 Cas9-sgRNA expression vector (Table S5 [Dickinson et al., 2013]). The Cas9-sgRNA plasmid and homologous repair template were co-injected into the gonad of young adult N2 hermaphrodites. Recombinant animals were identified in the F3 offspring of injected animals based on the presence of selectable markers (hygromycin [Table S5] resistance and dominant-negative sqt-1 rol phenotype). Selection markers were subsequently excised from the genome through Cre-Lox recombination. Proper genome editing was confirmed by DNA sequencing of the edited genomic locus and the health of the genome-edited line was determined as previously described (Keeley et al., 2020). Please refer to Table S4 for information on genotyping primers.
CRISPR strain phenotype and viability analysis
We assessed all newly created genome-edited strains (mNG::SBP-1, FASN-1::mNG, ELO-1::mNG, mNG::LPIN-1, SMS-1::mNG, mNG::HMGR-1, mNG::FCE-1, FNTA-1::mNG and mNG::SEC-16A.1) for non-N2 (wild-type) visible plate level phenotypes (e.g., uncoordinated [Unc], lethal [Let], and dumpy [Dpy]). We also assessed AC invasion by scoring at least 20 animals of each strain and comparing invasion to wild type animals. All genome-edited strains had normal AC invasion and no visible plate-level phenotypes. Fertility and growth of edited worms were also compared with N2 at 20°C by plating five L4 animals of each strain on 200 μl of OP50 and allowing the animals to grow on the plate. One day before plate level starvation, five L4 animals were then picked to new plates for a second generation. This was then repeated for a third generation. If no differences were seen in time to starvation, the worms were considered to have N2 fertility and growth. All edited strains had N2 fertility and growth with the exception of LPIN-1::mNG, which were sterile.
Construction of fusion proteins
To express ICMT-1 in the AC, we used the cdh-3 promoter (Garde et al., 2022) and generated cdh-3p::icmt-1::mScarlet by PCR fusion. Specifically, a 1.1-kb fragment of full-length icmt-1 sequence was fused with a 1.5 kb fragment of the cdh-3 promoter and mScarlet coding region in the pAP088 vector (Table S5 [Pani and Goldstein, 2018]). The same strategy was applied using a 1.5 kb fragment of lin-29 promoter in the pAP088 vector (lin-29p::icmt-1::mScarlet). To construct the AC-specific lysosome marker cdh-3p::lmp-1::mNG using PCR fusion, a 0.9 kb fragment of the full length lmp-1 sequence was fused with a 1.5 kb fragment of the cdh3 promoter and mNG region in the pAP088 vector. To build the AC-specific membrane marker lin-29p::mKate2:: PLCδPH, a 1.5 kb fragment of lin-29 promoter was fused with mKate2 and 0.5 kbp of the PLCδPH in the pAP088 vector. To construct the AC-specific Golgi marker qyEx607[lin-29p::aman-2::mScarlet] using PCR fusion, a 0.3 kb of the full-length aman-2 sequence was fused with a 1.5 kb fragment of the lin-29 promoter and mScarlet coding region in the pAP088 vector. To build the AC-specific ER marker qyEx608[lin-29p::zmp-1sp::mCherry::KDEL], a 12-bp KDEL sequence was fused with a 1.5-kb fragment of the lin-29 promoter, a 63 bp zmp-1 signal peptide, and mCherry coding region in the PpBS vector. To make another AC-specific ER marker zmp-1p::zmp-1sp::sfGFP::KDEL, a 12-bp KDEL sequence was fused with a 2.0 kb fragment of the zmp-1 promoter, a 63-bp zmp-1 signal peptide, and sfGFP coding region in the pAP088 vector. To generate the AC-specific peroxisome marker qyEx609[lin-29p::mScarlet::PTS1], 9 bp PTS1 (peroxisomal targeting signals serine-lysine-leucine [SKL]) with 15 bp linker (Ala-Ser-Gly-Tyr-Lys) was fused with a 1.5 kb fragment of the lin-29 promoter and mScarlet coding region in the pAP088 vector. Refer to Table S4 for primer sequences. All constructs were co-injected into the gonad of young adult N2 hermaphrodites with the Cas9-sgRNA plasmid (pCFJ352; Table S5). Single-copy transgenes were knocked-in at the ttTi4348 (I) locus in chromosome I using the pAP088 vector (Frokjaer-Jensen et al., 2012). Recombinant animals were identified in the F3 offspring of P0 injected animals based on the presence of selectable markers (hygromycin resistance and dominant-negative sqt-1 rol phenotype). Selection markers were subsequently excised from the genome through Cre-Lox recombination. The AC-specific Golgi (lin-29p::aman-2::mScarlet), ER (lin-29p::zmp-1sp::mCherry::KDEL), and peroxisome (lin-29p::mScarlet::PTS1) marker constructs were expressed extrachromosomally as described (Jayadev et al., 2019).
Construction of RNAi plasmids
RNAi constructs targeting sbp-1, hmgr-1, icmt-1, B0024.13, and ppk-3 were obtained from the Ahringer laboratory and Vidal laboratory RNAi libraries (Kamath and Ahringer, 2003; Rual et al., 2004). To improve the efficiency of RNAi knockdown targeting sbp-1, hmgr-1, icmt-1, and B0024.13, new RNAi constructs were synthesized. These constructs targeted the entire mRNA transcripts of sbp-1, hmgr-1, icmt-1, and B0024.13 and were designed using the T444T RNAi vector (Sturm et al., 2018). The T444T vector was digested using SacII and HindIII restriction enzymes and ligated with the PCR-amplified coding sequences via Gibson assembly. Please refer to Table S4 for primer sequences.
RNAi experiments
Bacteria harboring the RNAi vectors were cultured in selective media (lysogeny broth with 100 mg/ml ampicillin) for 16 h at 37°C. Transcription of double-stranded RNAi was induced with 1 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG) for 1 h at 37°C. Bacteria cultures were plated onto NGM plates that were treated with 1 mM IPTG and 100 mg/ml ampicillin. Plates were dried at room temperature for 12 h before use. RNAi experiments were conducted by feeding L1 worms with HT115(DE3) bacteria expressing double-stranded RNAi for 36–42 h at 18 or 20°C. Bacteria expressing the empty RNAi vector (L4440 or T444T) served as the negative control in all experiments. Whole animal RNAi knockdown was performed in this study unless otherwise specified. AC-specific RNAi experiments were performed using the strain NK1316 (Garde et al., 2022). This strain harbors a loss of function mutation in rde-1(ne219), an argonaute protein required for RNAi. Expression of rde-1 in the AC (fos-1ap::rde-1) restores RNAi specifically in the AC. The RNAi screen was performed to identify lipid synthesis enzymes (Table S2) that are required for AC invasion (Table S2). For genes encoding enzymes with multiple family members (i.e., SMS-1, -2, -3, CGT-1, -2, -3, and PSSY-1, -2), the enzyme with expression in the AC as determined by previously published AC-specific RNA-Seq (Costa et al., 2023) was examined in the RNAi screen. To enhance RNAi efficiency, two strategies were used. RNAi was performed in the rrf-3(pk1426) background, which harbors a mutation in an RNA-directed RNA polymerase whose loss sensitizes the worms to RNAi (Costa et al., 2023; Simmer et al., 2002), or performed transgenerationally. RNAi knockdown efficiencies were determined using endogenously tagged fluorescent protein strains as described in the “Quantification and statistical analysis” section and reported in Table S1.
Microscopy and image acquisition
The AC invasion RNAi screen was performed using an Axio Imager A1 microscope (Carl Zeiss), which was equipped with a Plan-APOCHROMAT 100× (1.4 NA) differential interference contrast (DIC) oil immersion objective. Images were acquired with a Zeiss AxioCam 305 mono CMOS camera controlled by the Zen Blue version 3.2 software (Carl Zeiss). All other imaging experiments were performed using an Axio Imager A1 microscope (Carl Zeiss) equipped with either a CSU-W1 or CSU-10 confocal scanner unit (Yokogawa) and Plan-APOCHROMAT 100× (1.4 NA) and 63X (1.4 NA) oil immersion objectives. Images were acquired using either a Hamamatsu Orca-Fusion sCMOS, ORCA-Quest 2 qCMOS, or ImagEM EM-CCD camera that was controlled by µmanager software (Edelstein et al., 2010). Fluorescence images were acquired as confocal z-stacks spanning the entire cell (total z-range: 5–11 µm) using 0.37-µm optical z-slice. For low-abundance proteins that are prone to photobleaching, a shorter total z-range (5–7 µm) and larger optical z-slice (0.50 µm) were used. For single time point AC snapshots to visualize fluorescently tagged proteins in the AC, worms were anesthetized in 0.01 M sodium azide and mounted on a 5% noble agar pad with a #1.5 coverslip. For time-lapse imaging, worms were anesthetized in 5 mM of levamisole and mounted on a 5% noble agar pad with a #1.5 coverslip. VALAP was used to enclose samples to prevent evaporation. Each live imaging experiment was limited to under 2 h to prevent starvation-induced changes in physiology (Schindler et al., 2014). To track the growth of AC invasive protrusion, confocal z-stacks (total z-range: 8–11 µm; optical z-slice: 0.37 µm) of control animals were acquired from the late P6.p 2-cell stage, when the protrusion forms, until the mid 4-cell stage, when the protrusion retracts (total imaging duration ∼90 min). Due to the AC delay in BM breaching after RNAi-mediated reduction of sbp-1, lpin-1, sms-1, and hmgr-1, RNAi-treated animals were imaged beginning at the P6.p 2–4 cell stage to capture protrusion formation. The imaging frame interval for all movies was 5 min except for sms-1 RNAi-treatment and its corresponding control, which were imaged at 4-min intervals. To visualize UNC-40 recruitment to the invadosome BM breach site, worms were manually rolled to align the ventral surface along the imaging plane as previously described (Kelley et al., 2017). An imaging frame interval of 60 s was used. All image acquisition was carried out at 20°C.
Quantification and statistical analysis
RNAi knockdown efficiency and quantification of AC and UC levels of lipid regulators
The efficiency of RNAi knockdown (Table S1) and quantification of AC and UC levels of lipid regulators (Fig. 3 B and Fig. S3 A) was assessed by quantifying the fluorescence intensity of each fluorophore-tagged protein of interest (mNG::SBP-1, mNG::HMGR-1, mNG::LPIN-1, SMS-1::mNG, FNTA-1::mNG, and ICMT-1::mScarlet) in empty RNAi vector control and RNAi gene targeted knockdown animals. All z-stack images were sum intensity z-projected (total z-range: 0.74–1.48 µm) except for FNTA-1::mNG and ICMT-1::mScarlet whereby only a single z-slice that spans across the AC longitudinally was acquired. The mean fluorescence intensity of each targeted protein in the AC was quantified by drawing an AC outline with the freehand selection tool and computed via Fiji 1.53f (Schroeder et al., 2021). To account for and subtract autofluorescence signals, all fluorescence intensity comparisons were made against wild-type animals (N2). The local background mean fluorescence intensity was quantified from the adjacent uterine tissue region where autofluorescence signals were observed. The net mean fluorescence intensity was then calculated by subtracting the local background mean fluorescence intensity from the AC mean fluorescence intensity in control and RNAi knockdown animals.
Scoring of AC invasion
AC invasion was assessed via DIC microscopy as previously described (Sherwood et al., 2005). Basement membrane (BM) breach was confirmed by fluorescence microscopy using a fluorescently tagged BM laminin marker (LAM-1::GFP; NK1316). Scoring was performed at the 1° VPC P6.p 4-cell stage when the clearance of the BM is completed in wild-type animals. The invasion was scored as “full invasion” if the BM breach was at least half of the AC width. The invasion was scored as “partial invasion” if the BM breach was at most one-third of the AC width. The invasion was scored as “blocked invasion” if the BM underneath the AC was completely intact. Both partial and blocked invasions were categorized as invasion defects.
Quantification of AC invasive protrusion volume
AC invasive protrusion volume was quantified as previously described (Kelley et al., 2017; Naegeli et al., 2017). Confocal microscopy was performed to capture z-stacks (total z-range: 8–11 µm; 0.37 µm optical slice) of the AC (mCherry::PLCδPH) and the underlying BM (LAM-1::GFP). Confocal z-stacks were used to make 3D reconstructions of the AC and the BM using Imaris 9.9 (Bitplane [Naegeli et al., 2017]). Isosurface renderings of mCherry::PLCδPH were created by fluorescence intensity thresholds empirically determined in a blinded manner that outline the AC plasma membrane. To distinguish the AC invasive protrusion from the rest of the cell, the Imaris isosurface slicer tool was used to cut the AC isosurface at the AC-BM junction. AC invasive protrusion was defined by the region of the AC that breached the BM. AC protrusion volume was tracked over a 60-min period (images acquired every 5 min) in control animals and sbp-1, lpin-1, and hmgr-1 RNAi-treated animals beginning at the time when protrusion formation first occurred. In all cases, the protrusion retracted or failed to grow further over the final 15 min of analysis, which ensured that maximum protrusion volume was observed. Notably, in two control animals, protrusion formation was initiated earlier, and these were included in the analysis (see Fig. S2 B). For sms-1 RNAi-treated animals and their corresponding controls, the AC protrusion volume was tracked for 64 min (images acquired every 4 min). The AC invasive protrusion volume was quantified using the Imaris volume measurement tool in control and RNAi-treated animals. To confirm the results, each analysis was performed by two individuals.
Quantification of SBP-1 level in the AC nucleus
The level of mNG::SBP-1 in the AC nucleus over the invasion time course was assessed using Fiji 1.53f (Schroeder et al., 2021). To reduce out-of-focus fluorescence signals from neighboring tissues that were positioned above and below the AC z-plane, image sub-stacks that only span across the AC center were sum intensity z-projected (total z-range: 0.74 µm; optical z-slice: 0.37 µm). The AC nucleus was outlined with the Fiji freehand selection tool using the DIC image in the same imaging plane. The mean fluorescence intensity of mNG::SBP-1 in the outlined AC nucleus was then determined.
Quantification of AC lysosome volume
Three-dimensional reconstructions were created from confocal z-stacks of the AC expressing lin-29p::LMP-1::mNG using Imaris 9.9 (Bitplane [Naegeli et al., 2017]). Due to the variability in LMP-1::mNG expression levels within each AC, a fluorescence intensity threshold was determined empirically in a blinded manner for each AC to distinguish lysosomes from the background fluorescence signals. Isosurface renderings of LMP-1::mNG were then created and manually confirmed by assessing for overlap with lysosome fluorescence signals through 3D rotation (Video 5). The total AC lysosomal volume was quantified using the Imaris volume measurement tool in control and RNAi-treated animals. To confirm the results, each analysis was performed by two individuals.
Quantification of UNC-40 levels at the AC BM breach site
To visualize UNC-40 at the BM breach site, animals expressing UNC-40::GFP and the BM marker LAM-1::mCherry were manually rolled to align the ventral surface along the imaging plane and then imaged by confocal microscopy. Confocal z-stacks that span the BM breach and the AC invasive membrane (total z-range: 5 µm; optical z-slice: 0.37 µm) were acquired. To visualize UNC-40::GFP localization at the BM breach, a subset of z-stack images (z-range: 1.48 µm) spanning across the BM breach were sum-projected using the Z Projection Sum Slices tool in Fiji 1.53f (Schroeder et al., 2021). The mean fluorescence intensity of UNC-40::GFP at the BM breach site was quantified by using the Fiji Measure tool. The mean fluorescence intensity outside the worm was measured to account for background. The net mean fluorescence intensity of UNC-40 at the breach site was then calculated by subtracting the background mean fluorescence intensity from the AC mean fluorescence intensity in control and RNAi knockdown animals. UNC-40 localization was assessed at the P6.p 2-cell stage in control animals. Due to the AC invasion delay in sms-1 RNAi-treated animals, UNC-40 localization was assessed from the P6.p 2–4-cell stage to the 4-cell stage.
Quantification of GTPases and ZMP-1 polarization and ZMP-1 enrichment
Levels of GTPases (GFP::CED-10, GFP::MIG-2, mNG::RAP-1), GFP::CAAX, and the matrix metalloproteinase ZMP-1 (ZMP-1::mNG) at the invasive membrane were assessed using Fiji 1.53f (Schroeder et al., 2021). To reduce out-of-focus fluorescence signals from neighboring tissues that were positioned above and below the AC z-plane, image sub-stacks that only span across the AC center were sum intensity z-projected (total z-range: 1.48 µm; optical z-slice: 0.37 µm). Next, the mean fluorescence intensity of each protein at the basal and apical AC membrane was quantified with a five-pixel-wide line using the Fiji freehand line tool (Wang et al., 2014a). The local background mean fluorescence intensity outside the worm was measured and subtracted from the apical and basal membrane mean fluorescence intensities. GTPase, CAAX::GFP, and ZMP-1 polarization were then determined by calculating the ratio of the basal/apical fluorescence intensities. The mean fluorescence intensity of ZMP-1::mNG at the invasive membrane was determined using the Fiji Measure tool and taking a five-pixel wide line at the invasive membrane. The mean fluorescence intensity outside the worm was measured to account for background. The net mean fluorescence intensity at the invasive membrane was determined by subtracting the background mean fluorescence intensity from the AC mean fluorescence intensity in control and ppk-3 RNAi knockdown animals.
Analysis of the AC ER, ER exit sites, and Golgi polarization
To quantify ER polarization in the AC, sum intensity z-projections were acquired (total z-range: 2.96 µm; optical z-slice: 0.37 µm) of sfGFP::KDEL expression. Mean fluorescence intensity measurements were then made using five-pixel-wide lines at the apical and basal ER regions using the Fiji freehand line tool (Wang et al., 2014a). The local background mean fluorescence intensity outside the worm was measured and subtracted from the apical and basal membrane mean fluorescence intensities. ER polarization was then determined by calculating the ratio of the basal/apical fluorescence intensities. To quantify ER exit site polarization in the AC, three-dimensional reconstructions were created from confocal z-stacks of the AC expressing mNG::SEC-16A.1 and the AC-specific plasma membrane marker lin-29p::mKate2::PLCδPH using Imaris 9.9 (Bitplane). Isosurface renderings of mKate2::PLCδPH were created by determining fluorescence intensity thresholds that outline the AC plasma membrane. mNG::SEC-16A.1 punctae numbers in the AC basal and apical halves were counted and expressed as basal/apical ratio to assess mNG::SEC-16A.1 polarization. To quantify Golgi polarization in the AC, three-dimensional reconstructions were created from confocal z-stacks of the AC expressing the Golgi marker AMAN-2::GFP (cdh-3p::AMAN-2::GFP) using Imaris 9.9 (Bitplane). AMAN-2::GFP punctae numbers in the AC basal and apical halves were counted and expressed as basal/apical ratio to assess Golgi polarization.
ER exit site colocalization with ICMT-1 and Golgi colocalization with SMS-1
To assess colocalization between ER exit sites (mNG::SEC-16A.1) and the ER-resident prenylation enzyme (ICMT-1), three-dimensional reconstructions were first created from confocal z-stacks of endogenously tagged mNG::SEC-16A.1 and AC-expressed ICMT-1::mScarlet (lin-29p::icmt-1::mScarlet) using Imaris 9.9 software (Bitplane). ICMT-1::mScarlet punctae were then identified empirically and circled. The IMCT-1 circles were then overlayed on mNG::SEC-16A.1 punctae (ER exit sites) specifically found in the AC (identified by removing all mNG::SEC-16A.1 punctae outside of the AC using an isosurface rendering of the AC using AC expressed ICMT-1::mScarlet). The percentage of mNG::SEC-16A.1 punctae that overlapped with AC-expressed ICMT-1::mScarlet punctae in the AC was then determined. A similar analysis was performed to determine the percentage of SMS-1::mNG punctae that overlapped with AMAN-2::mScarlet marked Golgi cisternae.
Statistical analysis
All experiments were performed with two or more replicates for both control and experimental results. Statistical analysis was performed using GraphPad Prism 10 and GraphPad 2 × 2 contingency table. The normality of data distribution was evaluated with the D’Agostino-Pearson normality test. Student’s t test was performed on samples with Gaussian distribution. Mann–Whitney U test was performed on samples with non-Gaussian distribution. For comparisons of three or more datasets, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test (for samples with Gaussian distribution) or Kruskal–Wallis test followed by Dunn’s multiple comparisons tests (for samples with non-Gaussian distribution) were performed. To compare categorical data, Fisher’s exact test was performed.
Online supplemental material
Fig. S1, related to Figs. 2, 3, 4, 5, 6, 7, 8, and 9, shows the substrates and enzymes involved in de novo fatty acid synthesis, phospholipid synthesis, sphingolipid synthesis, and the mevalonate pathway in C. elegans. Fig. S2, related to Figs. 2, 4, 5, and 6, shows AC invasive protrusion formation on control empty vector L4440 RNAi and RNAi-targeting lipid synthesis regulatory genes (sbp-1, lpin-1, sms-1, and hmgr-1). Fig. S3, related to Figs. 3, 4, and 5, shows the expression of endogenously tagged phospholipid and sphingomyelin synthesis genes pod-2, fasn-1, elo-1, lpin-1, and sms-1, colocalization of ELO-1 with an ER marker, colocalization of SMS-1 with a Golgi marker, and loss of ZMP-1 at the invasive membrane after RNAi targeting ppk-3 (lysosome biogenesis). Fig. S4, related to Figs. 6, 7, and 8, shows the regulation of AC lysosomal volume and GTPase polarization by the mevalonate pathway enzymes ICMT-1 and B0024.13, colocalization of HMGR-1 with the ER and peroxisomes, independence of hmgr-1 expression from SBP-1, and polarization of HMGR-1 by UNC-6 (netrin). Fig. S5, related to Figs. 7 and 8, shows FNTA-1 polarization, ICMT-1 polarization by UNC-6 (netrin), colocalization of FCE-1 with the ER and peroxisome, polarization and extension of the ER and Golgi into the invasive protrusion, and time course of AC GTPase polarization at the invasive front. Video 1 shows that SBP-1 is required for AC invasive protrusion formation. Video 2 shows that LPIN-1 is required for AC invasive protrusion formation. Video 3 shows a time-lapse video of LMP-1::mNG lysosomes entering the invasive protrusion. Video 4 shows a time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Video 5 shows a time-lapse video of LMP-1::mNG lysosomes at high temporal resolution. Video 6 shows that SMS-1 is required for the AC invasive protrusion formation. Video 7 shows that HMGR-1 is required for the AC invasive protrusion formation. Table S1, related to Figs. 3, 4, 5, and 6, and Figs. S4 and S5, shows RNAi knockdown efficiency for respective experiments. Table S2, related to Figs. 2, 3, 4, 5, 6, 7, and 8; and Figs. S1, S2, S3, S4, and S5, shows the effect of RNAi knockdown and gene mutations on AC invasion. Table S3 outlines the strains used in this study. Table S4 shows the primer sequences used to generate genome-edited strains, transgenic constructs, and RNAi plasmids. Table S5 lists the chemical compounds and recombinant DNA reagents, and software/algorithms used in this study.
Data availability
C. elegans strains and plasmids generated in this study and original data are available upon request. All the other data are included in the manuscript and supplemental materials.
Acknowledgments
We thank members of the Sherwood laboratory for their helpful comments and discussions, P. Schedl for the helpful discussions, I. Kenny-Ganzert (Duke University, Durham, NC, USA) for making the lin-29p::mKate2::PLCδPH construct, and D. Greenstein (University of Minnesota, Minneapolis, MN, USA) for DG4324 (tn1765 (GFP::3xflag::pod-2) II).
Some strains were provided by the Cancer Genomics Centre funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). We thank the Kiehart laboratory for the use of their NIH-funded (R35GM127059) Zeiss Axiovert 200M equipped with a Yokogawa W1 spinning disk confocal head for image collection. This work was supported by the Hargitt fellowship and Jane Coffin Childs Memorial Fund for Medical Research to A.W.J. Soh and R35GM118049 and R21OD032430 to D.R. Sherwood.
Author contributions: K. Park: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing - original draft, Writing - review & editing, A. Garde: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing - original draft, Writing - review & editing, S.B. Thendral: Formal analysis, Investigation, Methodology, Validation, Visualization, A.W.J. Soh: Conceptualization, Formal analysis, Investigation, Validation, Visualization, Writing - review & editing, Q. Chi: Resources, D.R. Sherwood: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing - original draft, Writing - review & editing.
References
Author notes
Disclosures: The authors declare no competing interests exist.
Supplementary data
shows RNAi knockdown efficiency.




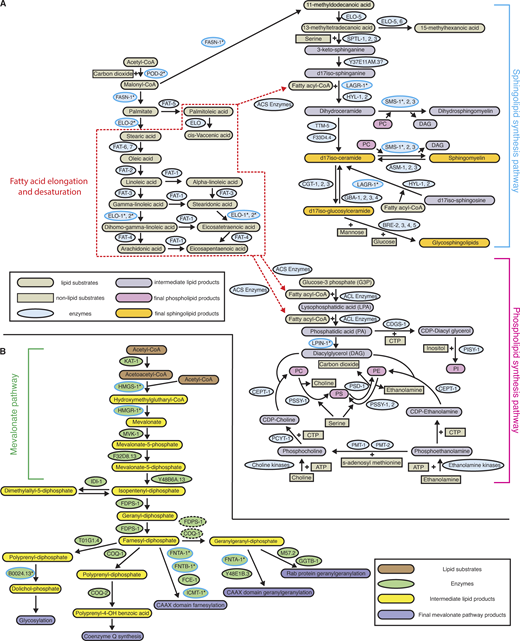
![SREBP (SBP-1) promotes AC invasion and invasive protrusion formation. (A) Top: Merged DIC and fluorescence images (maximum intensity z-projections) of the AC (cyan, mCherry::PLCδPH) and the underlying BM (single central confocal slice, magenta, LAM-1::GFP) at the P6.p 4-cell stage in a control (empty RNAi vector treated) and an AC-specific sbp-1 RNAi treated animal. Bottom: Fluorescence images showing BM breach (brackets) in a control animal and lack of breach after sbp-1 RNAi (arrows). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific sbp-1 RNAi treated animal (BM, orange dashed lines) with a small and delayed (see Materials and methods) breach observed corresponding to the reduced protrusion after loss of sbp-1 (middle, bracket, observed in n = 8/8 sbp-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 1). Right: Boxplot shows the maximum invasive protrusion volume in control and sbp-1 RNAi treated animals (n = 7 control and 8 sbp-1 RNAi animals, *** P ≤ 0.001, Mann–Whitney U test). For this and all subsequent boxplots, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line within the box the median value. (C) A schematic diagram illustrating SBP-1 proteolytic activation and entry into the nucleus where the SBP-1 bHLH domain binds sterol regulatory elements (SREs) to transcribe target genes. (D) Boxplot of mNG::SBP-1 mean fluorescence intensity in the AC nucleus from the L2/L3 molt stage to the P6.p 4-cell stage (n = 6 L2/L3 molt, nine 1-cell, six 2-cell, seven 2–4-cell, and six 4-cell stage animals, ** P ≤ 0.01, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (E) Top: Sum intensity z-projected fluorescence images of mNG::SBP-1 in the AC nucleus (white dashed circle) from the L2/L3 molt (time of AC specification) to the P6.p 4-cell stage. Bottom: Central plane DIC images showing dashed circles outlining the nucleus where SBP-1 fluorescence was measured. All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig2.png?Expires=1773919883&Signature=hnlUK0YpHbOsFZUUGewW~LqPmbHepEYgvm5OXUQNhcfoANk3c0vSKDsLGn55J~gDX2F0KjvQ-qDozKtG3Sa-xqiVlVo34I5NtTunARoQtlihk2YFWXyZS~TbeBfTOhakEtdr4BgSwi1cRqOm2qxcO4qNg8fIg60~NpNXJ1gz9npdNPaDurHg4FH5Q2LG6i16dwGeHahe8NZvpu5oXdRSum4A3wtrk1Ukbr-0pzZEUzr7G7pxqs6FUCjwBa~ALxoQPMCVoLle0U0oVclssemTCsIp9CipfIitcw~hFQNomWrx6ktvboXDmEJjWC8l2kkFxJ-SZq4SF5E5du9DNry-~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![SBP-1, LPIN-1, SMS-1, and HMGR-1 promote AC invasive protrusion growth. (A) Maximum intensity z-projected time-lapse fluorescence images showing the formation of the AC invasive protrusion (mCherry::PLCδPH, cyan) in a control and sbp-1 RNAi knockdown animal (BM, orange dashed lines). Time points indicate h::min. Isosurfaces of the AC invasive protrusions are shown below (see Video 1). The 0:45 min timepoint for control and sbp-1 RNAi is also shown in Fig. 2 A. (B–E) Left: Line graphs of invasive protrusion volume over time in control (cyan) animals and in animals after (B) sbp-1, (C) lpin-1, (D) sms-1, and (E) hmgr-1 AC-specific RNAi treatment (magenta; see Videos 1, 2, 6, and 7). Right in B–E: Boxplots of the AC invasive protrusion growth rates. In this and subsequent figures, box edges indicate the 25th and 75th percentiles, whiskers the maximum and minimum values, and the line inside each box the median value. AC protrusion growth rate over 60 or 64 min was calculated ([B] n = 7 control and 8 sbp-1 RNAi animals; [C] n = 7 control and 5 lpin-1 RNAi animals; [D] n = 5 control and 5 sms-1 RNAi animals; [E] n = 9 control and 7 hmgr-1 RNAi animals ** P ≤ 0.01, P = 0.1490, Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_figs2.png?Expires=1773919883&Signature=0P0xHL46d0Qz786J7omgl3mQ78qxU9v8apvl1fgAllLqplu2XzmoHtcELO5qEbFgQ5Cz5~a~gsVqWCWwlf9SvaTIv~zvg2sO2Vlez-cFzPJ02YtFxKiYu-iZou5e6us4hSLAZC8C5QXzsNDQu0SpJtah1Huwtw0oe5Yuv9F2w4znibHScl-nqDol5Ay1ZCzfzX0e-sYBYRsRQOIxMNuadUXbtwjTKrfkyePNIRwe1DB1GkK1hFWDrKGraSRsQ~E1fwYuJvGx5lnkC0jZm1R44zdmQtJP1oh0jxkL4EULH-VHlvP6Fbo5fhMzhLkluqJ4-LRXq299Yumgdz75ZK0XOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![AC expression and SBP-1 regulation of pod-2, fasn-1, elo-1, lpin-1, and sms-1. (A) A schematic diagram showing the role of the transcription factor SBP-1 in regulating phospholipid and sphingomyelin synthesis in the AC (refer to Fig. S1 A for complete pathway and Table S2 for RNAi screen results). (B) Sum intensity z-projection images of POD-2 (GFP::POD-2) and FASN-1 (FASN-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. POD-2 and FASN-1 are in the AC cytoplasm (arrows, ACs). (C) Sum intensity z-projections of ELO-1 (ELO-1::mNG, ACs outlined with white dashes) and LPIN-1 (mNG::LPIN-1, arrows, ACs) in control and sbp-1 RNAi animals at the P6.p 2-cell stage. ELO-1 is localized to the ER (see Fig. S3 B) and LPIN-1 in the cytosol. (D) Sum intensity z-projection of SMS-1 (SMS-1::mNG) in control and sbp-1 RNAi animals at the P6.p 2-cell stage (ACs outlined with white dashes). SMS-1 localizes to the Golgi (see Fig. S3 C). (B–D) Right: Boxplots show the mean fluorescence intensity of each protein in the AC of control and sbp-1 RNAi treated animals ([POD-2] n = 10 control and 10 sbp-1 RNAi animals; [FASN-1] n = 22 control and 23 sbp-1 RNAi animals; [ELO-1] n = 15 control and 16 sbp-1 RNAi animals; [LPIN-1] n = 11 control and 10 sbp-1 RNAi animals; [SMS-1] n = 24 control and 13 sbp-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test and Mann–Whitney U test). All data are from two or more replicates. Scale bar, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig3.png?Expires=1773919883&Signature=2s2RWlATWTRwk1QvnwSS-LlNo7fUtQJBdx86WEt6LWmRs~gicjZ6K2BezxE9z9~fjx8sso7fmvAnzekwMafxFlD9hNg3rOD8mQHh3G0CGKriNf6GLa8mkngR87iNTq57IjtwKTTrdDFPhbImX5s5l~ptsYFQa~Pmt2sqUVWDXnOWSDkRsK8n1S9AeZj5tK1zlNiZL7-9mc0xdSVCgKSOrizGvg1bqKqNZa5NhZD1uTo6SjV2MU0DcW297-bTe7qetIKC5N97zdfHe~tfEMKyemTu86SbyF~seJ8P3vtTjjjOyNAd53TaicK3crhWKlZijQUwoo2lURkK3ZIeoUTi~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![POD-2, FASN-1, ELO-1, LPIN-1, and SMS-1 AC expression from P6.p 1-cell to 4-cell stage. (A) Left: Sum intensity z-projected fluorescence images of endogenously tagged GFP::POD-2, FASN-1::mNG, ELO-1::mNG, mNG::LPIN-1, and SMS-1::mNG in the AC (arrows and white dotted lines) from the P6.p 1-cell to 4-cell stages. Right: Boxplots show the mean fluorescence intensity of each protein in the AC (comparisons of P6.p 1-cell and 2–4-cell stages, [POD-2] n = 11 1-cell, 10 2-cell, 10 2–4-cell, and 11 4-cell stage animals; [FASN-1] n = 11 1-cell, 12 2-cell, 10 2–4-cell, and 10 4-cell stage animals; [ELO-1] n = 14 1-cell, 13 2-cell, 9 2–4-cell, and 13 4-cell stage animals; [LPIN-1] n = 8 1-cell, 15 2-cell, 10 2–4-cell, and 14 4-cell stage animals; [SMS-1] n = 10 1-cell, 10 2-cell, 5 2–4-cell, and 10 4-cell stage animals, * P ≤ 0.05, *** P ≤ 0.001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test; comparisons of uterine cell [UC] to AC at P6.p 4-cell stage, [POD-2] n = 11 animals; [FASN-1] n = 10 animals; [ELO-1] n = 13 animals; [LPIN-1] n = 14 animals; [SMS-1] n = 10 animals, * P ≤ 0.05, ** P ≤ 0.001, *** P ≤ 0.0001, P = 0.10, Student’s t test). (B) Top: Single slice confocal images of mCherry::KDEL (magenta), ELO-1::mNG (cyan) and overlay in the AC show tight overlap (arrows). Bottom: Insets highlighting the overlap in the basal region (arrows; similar colocalization observed in n = 10/10 animals). (C) Top: Max intensity z-projected fluorescence images of AMAN-2::mScarlet (magenta, Golgi), SMS-1::mNG (cyan), and overlay in the AC reveal colocalization (arrows). The average percentage ± SD of colocalization of SMS-1::mNG puncta with AMAN-2::mScarlet with is indicated (n = 6 animals). Below: Insets highlight AMAN-2::mScarlet (magenta) and SMS-1::mNG (cyan) colocalization (arrows). (D) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a ppk-3 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal ZMP-1::mNG mean fluorescence signal in control and ppk-3 RNAi treated animals (n = 8 control and 9 ppk-3 RNAi animals, P ≤ 0.05, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_figs3.png?Expires=1773919883&Signature=wzfJ-MYBnwIrNxXtDZIkr4rRACwxYIpj5~4cKhzwvMUggZer5JxgX6fX6t~Q93FQpyYDV194Qz0Jyk7Z28tv88Ce2IE3wYtvp~FcMQeMOPBCLSlQIdUiB12GdPO2Z3bA1sOCDIzNnHJymJkeEHuiAMmo6hLem1W24KXksgATMhU0MyXtzHMtGCyuXqIVWsFMY9EGG7go~LzFbm7ug7~YmHi~GCZRVHzo~J67zpttuvLRTo9b7~ROcrgI1S6TT2nQ~tdAEgupVLUAflDvrIceRb9M2qHW4~JCVH-GrWsXYhr6YpPZX~eX-II1WhXzvgy83ESDVbjJw6PVUz29URl2Xg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![LPIN-1 promotes lysosome stores for invasive protrusion formation. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific lpin-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of lpin-1 (middle, bracket, observed in n = 5/5 lpin-1-1 RNAi time-lapses). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 2). Right: Boxplot of the maximum invasive protrusion volume in control and lpin-1 RNAi treated animals (n = 7 control and 5 lpin-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (B) Max intensity z-projected time-lapse fluorescence images (h:min, above) and isosurface images showing AC lysosomes (LMP-1::mNG) accumulating at the site of BM breach and entering the protrusion (observed in n = 3/3 time-lapses). (C) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in control and a lpin-1 RNAi treated animal at the initiation of protrusion formation (late P6.p 2-cell stage and 2–4-cell stage). Right: Boxplot of AC lysosome volume in control and lpin-1 RNAi treated animals (n = 21 control and 23 lpin-1 RNAi animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (D) Left: Sum intensity z-projected images show the polarized localization of GFP::CED-10 (Rac) and GFP::MIG-2 (Rac-like) at the AC invasive plasma membrane (arrow) in control and lpin-1 RNAi treated animals at the late P6.p 2-cell stage at the initiation of protrusion formation. Right: Boxplot showing the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 ([CED-10] n = 12 control and 13 lpin-1 RNAi animals; [MIG-2] n = 8 control and 11 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (E) Left: Sum intensity z-projections of ZMP-1::mNG fluorescence showing the invasive membrane enrichment (arrow) in a control and a lpin-1 RNAi treated animal at the initiation of invasive protrusion formation (late P6.p 2-cell stage and 2–4 cell stage). Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and lpin-1 RNAi treated animals (n = 10 control and 10 lpin-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig4.png?Expires=1773919883&Signature=WXblHJdDoB6Aosko-w1qYfyLW9Eme7OkijGShZ-F3XMEiEHz~Sg39KfFPc~ddw8qp~NAFmnC4GHH6vubTOaB~jxKUBjYgfIuSBZVftCcxJyxQySfM~wc11ovA7vmgVTiPh2JgNTivUpUe8-oaREQIP~~pefNySXwM0zcGEwUuzLZf83hoiGGlWslOMkf1Q7YNeKVfTNyzVxTWEQ7CzXtR9l7GmTMUFd9FoIrNJTaXDswHcWKz4zPwxLhB4auCCOH6HlUt0c0GXr3oRnOM7fOqtpxl7aWeI7Ol0DiUVyPHJewdNPtenijse4OB3pg9ZcnM3Etb2WpgeUCCgobgg~trw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![SMS-1 promotes formation of lysosome stores and localization of UNC-40 and ZMP-1. (A) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) in a control and an AC-specific sms-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of sms-1 (middle, bracket, observed in n = 5/5 sms-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 6). Right: Boxplot of the maximum invasive protrusion volume in control and sms-1 RNAi treated animals (n = 5 control and 5 sms-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in control and sms-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume in control and sms-1 RNAi treated animals (n = 24 control and 23 sms-1 RNAi animals, ** P ≤ 0.01, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projected fluorescence images of GFP::CED-10 and GFP::MIG-2. Arrows show enrichment at the AC invasive membrane in a control and sms-1 RNAi treated animals at the time of invasive protrusion initiation. Right: Boxplots show the basal/apical ratio of GFP::CED-10 and GFP::MIG-2 fluorescence intensity in control and sms-1 RNAi treated animals ([CED-10] n = 16 control and 15 sms-1 RNAi animals; [MIG-2] n = 12 control and 11 sms-1 RNAi animals, ns [not statistically significant], P > 0.05, Mann–Whitney U test). (D) Left: Sum intensity fluorescence images of the GPI-anchored matrix metalloproteinase ZMP-1::mNG showing the AC invasive membrane (arrows) in a control and a sms-1 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and zmp-1 RNAi treated animals (n = 16 control and 13 sms-1 RNAi animals, * P ≤ 0.05, unpaired two-tailed Student’s t test). (E) Left: Ventral view of the AC-BM interface (BM visualized with LAM-1::mCherry) showing UNC-40::GFP enrichment (arrows) at the initial BM breach (orange dotted lines) in a control and a sms-1 treated RNAi animal. Right: Boxplot shows UNC-40::GFP mean fluorescence intensity at the initial BM breach (n = 6 control and 6 sms-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (F) A schematic diagram summarizing the roles of the transcription factor SBP-1, the fatty acid synthesis enzymes POD-2 and FASN-1, and phospholipid synthesizing LPIN-1 and sphingomyelin catalyzing SMS-1 in AC invasive protrusion formation. All data in the figure are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig5.png?Expires=1773919883&Signature=ojcIcESkSDguBPVk-51hK4T-vkgRUEoWcCJUYLDR8CZoCqad~zSoXrReeQPrMRBh6pNfbwJUfEq03ItIPSg5cpRIfrsI6vIMQ44LnH49eu1g1FTdqc9o0~8guGScQvklRi60d0ovUkPdv4rr68CeQztBR1wEkVhEHZqkSG6IVkVhcXFuxf-r2nP-UTcZgtgsa8Wzt~5f-O~ccqHqktPwZ0XssdbPTmyKW1ZXzucXKTWPy1i8XblUn4hwq8VwmB0ryOAYg6nd14iE24OvDipmVmSbTgFLZMwYszSqvfSuIjWQP437XrBwt06-BrxI0zkQuMPSOncDwvWaSfEGGqO6VA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![HMGR-1 promotes lysosome stores, GTPase accumulation, and invasive protrusion formation. (A) A schematic diagram showing the mevalonate pathway in the AC (see Fig. S1 B for pathway details, Table S2 for RNAi screen). (B) Left: Maximum intensity z-projected fluorescence images showing the AC (cyan, mCherry::PLCδPH) protrusion (arrowheads) in a control and an AC-specific hmgr-1 RNAi treated animal (BM, orange dashed lines) with a small BM breach observed corresponding to the reduced protrusion after loss of hmgr-1 (middle, bracket, observed in n = 7/7 hmgr-1 RNAi animals). Isosurfaces of the AC invasive protrusion (below, arrowheads) at the time of maximum protrusion volume (see Video 7). Right: Boxplot of the maximum invasive protrusion volume in control and hmgr-1 RNAi treated animals (n = 9 control and 7 hmgr-1 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (C) Left: Max intensity z-projected fluorescence images and isosurfaces showing the AC lysosomes (LMP-1::mNG) in a control and an hmgr-1 RNAi-treated animal at the initiation of protrusion formation. Right: Boxplot of the AC lysosome volume of control and hmgr-1 RNAi animals (n = 17 control and 18 hmgr-1 RNAi animals, *** P ≤ 0.001, unpaired two-tailed Student’s t test). (D) Top: Sum intensity z-projected images showing invasive polarization (arrows) of GFP::CED-10, GFP::MIG-2, GFP::CAAX, and mNG::RAP-1 at the AC plasma membrane in control and hmgr-1 RNAi animals at the initiation of invasive protrusion formation. Bottom: Boxplots showing the AC basal/apical ratio of each protein’s fluorescence intensity in control and hmgr-1 RNAi-treated animals ([CED-10] n = 23 control and 23 hmgr-1 RNAi animals; [MIG-2] n = 13 control and 13 hmgr-1 RNAi animals; [CAAX] n = 16 control and 16 hmgr-1 RNAi animals; [RAP-1] n = 17 control and 18 hmgr-1 RNAi animals, ** P ≤ 0.01, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (E) Top: Sum intensity projected images of ZMP-1::mNG shows ZMP-1 enrichment at the AC basal invasive plasma membrane (arrows) in a control and an hmgr-1 RNAi-treated animal at the initiation of invasive protrusion formation. Bottom: Boxplot showing the AC basal/apical ratio of ZMP-1::mNG fluorescence intensity in control and hmgr-1 RNAi treated animals (n = 14 control and 16 hmgr-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig6.png?Expires=1773919883&Signature=eT~jp3ufW~pFH1h7jSgSjhdK9vwXSQ0Mt54Sx8DSEFtGrFZ7SibgSLw2AByD66SNJxNohtmd6b3IKyIhYK0Ww5bn6V5cHlFxnGVgbdLyCRqidMiM9aJsO16IueMQkp7iKV1-fteZmKD8~xma9chrIm56tGAohr7ZB3k7odcjUrr3KUacfUvaFRykwxj-im4n8SC9Ct1SGwy5I6rR6XnRY1NNt3TwvGJgOcI4SwtmdFmvIeO3Hy3Sl2Rlrp88iFCqc-DwrdSm3i6jlGQLuzF65UE37~lnptJfPuGV-nQfS8nMkVtt-LFaa1KcQlGAfdzOLQZFUWM38BIpWdH4xa2x4g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Role and regulation of HMGR-1 and the mevalonate pathway. (A) Left: Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and in a B0024.13 (dolichol synthesis) RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot of AC lysosomal volume in control and B0024.13 RNAi treated animals (n = 20 control and 24 B0024.13 RNAi animals, ** P ≤ 0.01, Mann–Whitney U test). (B) Max intensity z-projected fluorescence and isosurface images showing the AC lysosomes (LMP-1::mNG) in a control and icmt-1 RNAi treated animal at the initiation of invasive protrusion formation. Boxplot of AC lysosome volume in control and icmt-1 RNAi treated animals (n = 23 control and 21 icmt-1 RNAi animals, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (C) Left: Sum intensity z-projection fluorescence images of GFP::CED-10 shows enrichment at the AC basal plasma membrane (arrow) in a control, an icmt-1, and a B0024.13 RNAi treated animal at the initiation of invasive protrusion formation. Right: Boxplot showing the basal level of GFP::CED-10 mean fluorescence intensity (n = 29 control, 22 B0024.13, and 21 icmt-1 RNAi animals, * P ≤ 0.05, Mann–Whitney U test). (D) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::HMGR-1 (cyan), and overlay in the AC show overlap of mNG::HMGR-1 and mCherry::KDEL signals (white arrows) and a region of enriched mNG::HMGR-1 with no overlap (yellow arrows). Below: Insets highlighting region of increased mNG::HMGR-1 fluorescence where there is no mCherry::KDEL (yellow arrows; similar colocalization observed in = 8/8 animals). (E) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::HMGR-1 (cyan), and overlay in the AC shows overlap of enriched mNG::HMGR-1 regions and mScarlet::PTS1 (white arrows). Below: Insets highlighting region of overlap (arrows; similar colocalization observed in = 3/3 animals). (F) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in two ACs with mNG::HMGR-1 enriched at the invasive basal side (arrows). Right: mNG::HMGR-1 is not concentrated at the basal side of uterine cells (UCs, arrows, n = ≥10/10). (G) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 in the AC (arrows show AC and enrichment at the invasive front) in a control and a sbp-1 RNAi treated animal. Right: Boxplot of mNG::HMGR-1 fluorescence intensity in the AC of control and sbp-1 RNAi animals (n = 26 control and 27 sbp-1 RNAi animals, unpaired two-tailed Student’s t test, ns [not statistically significant], P > 0.05, unpaired two-tailed Student’s t test). (H) Left: Sum intensity z-projected fluorescence images of mNG::HMGR-1 polarized (arrow) in a wild-type and mispolarized in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of mNG::HMGR-1 fluorescence intensity in wild-type and unc-6(ev400) animals (n = 48 control and 30 unc-6(ev400) animals, **** P ≤ 0.0001, Mann–Whitney U test). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_figs4.png?Expires=1773919883&Signature=4Es8YLg~9KtAvvwJ9eTfaO5HFHEpqV1XoP~foyotTPX~vVqnxM6jWKDHfalW0svhwtQSzG4YRMb3oAqlO9-KReFn7LCPH2gbSgDbRrW3NpTu8nQ3KdXDmQ4FOyOm-dEkqpY7mswMO-WRGzK~WICf9oj0gr753O49cEKjI75AwxNkHiC-dkHEAcY7A3ytlBmTnK~sLHWpA7AtrDTowcjvl73rH29X2xU8UEGz6eAliUB0Vhu5AeGPzXWZj8r5fhwTsLErf3HB1PvjiHt~XFT1aKihTurF8y-ycQkFYYvKd0ma2LxUpgiuy7k6kHjibjDFaXRA8cw2rZ18P75y3ZpTJA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Prenylation enzymes HMGR-1, FCE-1, and ICMT-1 polarize towards BM. (A) A schematic diagram showing the mevalonate/HMGR-1 prenylation pathway (see text for detailed description). (B) Left: Sum intensity z-projected images show AC localization and invasive enrichment (arrows) of the endogenously tagged membrane-associated prenylation enzymes mNG::HMGR-1 and mNG::FCE-1, and AC expressed ICMT-1::mScarlet (lin-29p::icmt-1::mScarlet, inset shows spectral fluorescence-intensity map displaying the minimum and maximum pixel value range of the acquired data) from the P6.p 1-cell to 4-cell stages. All the subsequent spectral fluorescence-intensity maps also display the maximum (H) and minimum (L) pixel value range of the acquired data, which was independently determined for each image. Right: Boxplots show the AC basal/apical ratios of mNG::HMGR-1, mNG::FCE-1, and ICMT-1::mScarlet fluorescence intensity ([HMGR-1] n = 12 1-cell, 16 2-cell, 9 2–4-cell, and 11 4-cell stage animals; [FCE-1] n = 9 1-cell, 14 2-cell, 13 2–4-cell, and 16 4-cell stage animals; [ICMT-1] n = 10 1-cell, 15 2-cell, 10 2–4-cell, and 11 4-cell stage animals, **** P ≤ 0.0001, ns [not statistically significant], P > 0.05, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (C) Top: Single slice confocal images of IMCT-1::mScarlet (magenta), mNG::KDEL (cyan, ER marker) and overlay in the AC show tight overlap of localization of ICMT-1 throughout the ER (white arrows). Bottom: Insets highlight the AC basal ER region and show a puncta of intense IMCT-1 enrichment within the ER (left arrow) and a region of similar intensity to the mNG::KDEL marker (right arrow) (similar localization observed in n = 25/25 animals). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig7.png?Expires=1773919883&Signature=GuvhiMiu1ocPyczuL-m-9XXu7Zpj~oMt9IVCPnQJaovfr3tqhZ3k6Dg5JVXVCjFMDDqJxBkoqtHjBo9RTNqaZIZjxUjxL-RCLLLO5mSqIzWaUUQIB35eqdHr2N6gt2pVqTxP06He0Jr6wFgtzuvwTa1S7iEsl770KusUBDE1VMnp8HadTyvW6V-veUcew1MUbCiNrCd5Yx6QXNX0kmdcLqbJoJVv8BmN-myvrQxhVHgtx10Gsi-5FUiO1VMqnsT3abfxECJ8iwFSW9zxShe9Ouoyk~TXOAKAe4lKThAEb7shpDCoeIvGY9bEB4DBt3~FQ~4tL~JGdx1V1wKBVhs4tQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Regulation and localization of prenylation enzymes and GTPases during AC invasion. (A) Sum intensity z-projected images show high levels of FNTA-1::mNG in the AC at the P6.p 2-cell stage (n = 12/12 animals) and 4-cell stage (n = 12/12). FNTA-1::mNG basal enrichment was observed at the P6.p 4-cell stage (arrows, n = 12/12). Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. (B) Sum intensity z-projected images of ICMT-1::mScarlet in the AC show polarization in a wild-type animal (arrows) and lack of polarization in an unc-6(ev400) mutant animal at the initiation of invasive protrusion formation. Bottom panels show spectral fluorescence-intensity maps, which display the minimum (L, low) and maximum (H, high) pixel value range of the acquired data. Right: Boxplot shows AC basal/apical ratio of ICMT-1::mScarlet in wild-type and unc-6(ev400) animals (n = 31 control and 34 unc-6(ev400) animals, **** P ≤ 0.0001, unpaired two-tailed Student’s t test). (C) Top: Single slice confocal images of mCherry::KDEL (magenta, ER marker), mNG::FCE-1 (cyan), and overlay in the AC show overlap of mNG::FCE-1 and mCherry::KDEL signals (white arrows) and a region of mNG::FCE-1 with no overlap with mCherry::KDEL (yellow arrows). Bottom: Insets highlighting region of mNG::FCE-1 fluorescence where there is no mCherry::KDEL (yellow arrows, similar colocalization observed in n = 13/13 animals). (D) Top: Single slice confocal images of mScarlet::PTS1 (magenta, peroxisome marker), mNG::FCE-1 (cyan), and overlay in the AC shows overlap of mNG::FCE-1 regions and mScarlet::PTS1 (white arrows). Bottom: Insets highlighting region of overlap (white arrows; similar colocalization observed in = 3/3 animals). (E) Top: Central AC DIC images and corresponding AC max intensity z-projected images of sfGFP::KDEL (ER marker, top) and (bottom) DIC image and AMAN-2::GFP (Golgi marker, bottom) at the time of protrusion formation show the ER and Golgi extending into the protrusion (below the BM breach indicated by dotted orange line, n = 12/12 ER and 12/14 Golgi). (F) Top: Sum intensity z-projected image of AC specific expression of sfGFP::KDEL (ER marker) at the P6.p 4-cell stage. Bottom: Line graph of AC basal/apical polarization from the P6.p 1-cell through 4-cell stages (n = 8 1-cell, 8 2-cell, 9 2–4-cell, and 6 4-cell stage animals, ns [not statistically significant], Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). (G) Left: Sum intensity z-projected images showing the time course of GFP::CED-10, GFP::MIG-2, and mNG::RAP-1 polarization from the P6.p 1-cell to 4-cell stage. Right: Line graphs of AC basal/apical polarization of each protein’s fluorescence intensity ([CED-10] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 15 4-cell stage animals; [MIG-2] n = 10 1-cell, 11 2-cell, 13 2–4-cell, and 17 4-cell stage animals; [RAP-1] n = 11 1-cell, 13 2-cell, 10 2–4-cell, and 19 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Brown-Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test and Kruskal–Wallis test followed by Dunn’s multiple comparisons test). All data are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_figs5.png?Expires=1773919883&Signature=YD4wo-OcsbXRspani0TqcNfdCt3g~DtvngO-~J-Uj2UqIoX~W7mn7LAjxSc8Z~tIoONE8xVqgS7eq90InD40Dp9wElrLey5yrxXJpAEdZt1YKIpUoI7HWYuwg-k7eJ~~KRsRv8vBvvCvoj2xfVhuwtL738t28JcjDOPK297j60SI40YbGRTvJFI4K2XHZOGlYqibKp8UiZxtJnxHmgrSUb18uSV4d-3hbsLtkFFGlFBZZRHmyVt8~lZ~~zq0NcGf7TAiZeUowYTE9DDHw3D0kfBoYU0hAdrIDP1cdjZcY2Br13uPjdyN8k58QgKj8GU9pxzXOPaKUzpM2crnnmb0qg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![ICMT-1 localizes to ER exit sites that dynamically polarize toward the invasive protrusion. (A) Top: Sum intensity z-projected images of ICMT-1::mScarlet (magenta), ER exit marker mNG::SEC-16A.1 (cyan), and overlay in the AC reveal colocalization at ER exit sites (arrows). The average percentage ± SD of colocalization of ER exit sites with ICMT-1 puncta is indicated (n = 17 animals). Bottom: Insets highlight ICMT-1::mScarlet and mNG::SEC-16A.1 colocalization in the ER at the AC invasive front (arrows). (B) Max intensity z-projected fluorescence images of GFP::CED-10 in a control and sar-1 RNAi treated animals shows GFP::CED-10 localized to the invasive plasma membrane (arrow) in the control animal, but predominantly localized in intracellular vesicles in sar-1 RNAi animals (brackets). Right: The relative proportion of animals with CED-10::GFP that localized predominantly to the AC basal plasma membrane or was found predominantly in the cytosol in control and sar-1 RNAi treated animals (n = 38 control and 45 sar-1 RNAi animals, ** P ≤ 0.01, Fisher’s exact test). (C) Fluorescence images showing AC ER exit sites (mNG::SEC-16A.1, Cyan; AC outlined with the plasma membrane marker mCherry::PLCδPH, red), Golgi (AMAN-2::mScarlet), and prenylated GFP::CAAX in the AC from the P6.p1-cell to 4-cell stages. Arrows indicate basal (invasive front) enrichment of each protein. Right: Quantifications of the relative distribution of the ER exit site, Golgi, and plasma membrane markers in the AC from the P6.p 1-cell to 4-cell stages. For mNG::SEC-16A.1 and AMAN-2::mScarlet, the total number of puncta in the AC apical and basal halves (green dashed line) was expressed as a basal/apical ratio. The mean fluorescence intensity of GFP::CAAX was expressed as a basal/apical ratio ([SEC-16A.1] n = 10 1-cell, 10 2-cell, 9 2–4-cell, and 10 4-cell stage animals; [AMAN-2] n = 10 1-cell, 15 2-cell, 14 2–4-cell, and 16 4-cell stage animals; [CAAX] n = 12 1-cell, 11 2-cell, 10 2–4-cell, and 11 4-cell stage animals, * P ≤ 0.05, ** P ≤ 0.01, Mann–Whitney U test, and Brown–Forsythe and Welch ANOVA tests followed by Dunnett’s T3 multiple comparisons test). (D) A schematic diagram illustrating where GTPase prenylation occurs, with FCE-1 present in the ER and peroxisomes and ICMT-1, the final prenylation enzyme, enriched in ER exit sites (ERES) that deliver prenylated proteins (i.e., Rac and Ras-related) to the Golgi apparatus for targeting to the invasive protrusion (red arrows). Rac/Ras-related prenylated proteins might also be delivered directly from the ER to the invasive plasma membrane (magenta arrows, inset). All data in the figure are from two or more replicates. Scale bars, 5 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/10/10.1083_jcb.202402035/2/m_jcb_202402035_fig8.png?Expires=1773919883&Signature=cp4RMvh2wNTG5oTjyn5xrTNBYHEGN-DCIO8VCOMxsiAaMNAdIa2PAaJlEkZaGWZ80J0qegJUl8Korhqlphi8AaHD3xTqhz1InhYj5pv4cXVqQWc~2gh1J3MVhxc2XjNAr~mv3nQidu3V7HZTp7~lAEbxj3XKAV3J207BHiYLHHTU7hDMrRrDw7fqXbzRSRSbSq~rRUipOEnggDmCgd4rNNc~U9HeHNNG0pKLWRYxH~nE-WQWgRatjCfhxDHINB7gdpTrm-VOo8uIQ0dIEvQ71dbeKmp8OqHdxE1NbP6lCosLoiCfZbFGHMehphdJE07j6u~ECerTjrIselZiKjZ0Dg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



