It is known that microtubule-binding proteins including the Ska1 complex and the DNA replication licensing factor, Cdt1, enable the kinetochore-localized Ndc80 complex to form robust kinetochore-microtubule attachments. However, it is not clear how the Ndc80 complex is stably coupled to dynamic spindle microtubule plus-ends. Here, we have developed a conditional auxin-inducible degron approach to reveal a function for Cdt1 in chromosome segregation and kinetochore–microtubule interactions that is separable from its role in DNA replication licensing. Further, we demonstrate that a direct interaction between Cdt1 and Ska1 is required for recruiting Cdt1 to kinetochores and spindle microtubules. Cdt1 phosphorylation by Cdk1 kinase is critical for Ska1 binding, kinetochore–microtubule attachments, and mitotic progression. Furthermore, we show that Cdt1 synergizes with Ndc80 and Ska1 for microtubule binding, including forming a diffusive, tripartite Ndc80-Cdt1-Ska1 complex that can processively track dynamic microtubule plus-ends in vitro. Taken together, our data identify the Ndc80-Cdt1-Ska1 complex as a central molecular unit that can promote processive bidirectional tip-tracking of microtubules by kinetochores.
Introduction
Bona fide DNA replication is a committed step in cell proliferation that ensures complete and precise genome duplication only once per cell cycle. Cdt1 is one of the DNA replication licensing proteins that serves a central role in this process (Pozo and Cook, 2016). Following DNA replication, cells undergo mitosis to equally partition their duplicated chromosomes between the two daughter cells. This function is achieved by the concerted function of the bipolar spindle microtubules and kinetochores (Cheeseman and Desai, 2008). Besides licensing the origins for DNA replication, a fraction of Cdt1 has been shown to localize to the kinetochores during mitosis, dependent on the Ndc80 complex (Varma et al., 2012), a key component of the core microtubule-binding site at kinetochores (Cheeseman et al., 2006; Guimaraes et al., 2008), where Cdt1 augmented the stability of kinetochore–microtubule (k-MT) attachments (Agarwal et al., 2018; Varma et al., 2012). Although our earlier work demonstrating the ability of Cdt1 to bind to microtubules directly and its regulation by Aurora B Kinase–mediated phosphorylation shed mechanistic insights into how Cdt1 promotes stable k-MT attachments (Agarwal et al., 2018), it is not clear how the function of Cdt1 and Ndc80 is coordinated in generating stable k-MT attachments. The above study also found that Cdt1 not only possessed the ability to diffuse on microtubules and bind to curved microtubule protofilaments in vitro, but also localized to the plus-ends of spindle microtubules when expressed in cells. Further, while we used an siRNA-mediated knockdown approach coupled with cell synchronization to study the mitosis-specific function of Cdt1, a more robust method that is independent of chronic long-term inhibition and chemical cell synchronization is currently lacking (Agarwal et al., 2018; Varma et al., 2012).
The spindle microtubule-binding Ska1 complex is another factor that localizes to kinetochores in an Ndc80-dependent manner and has been shown to assist Ndc80 in the formation of load-bearing k-MT attachments in metaphase (Hanisch et al., 2006; Welburn et al., 2009). Ndc80, on the other hand, was also able to strengthen the binding affinity of Ska1 to microtubules, which suggest that these proteins function synergistically and form a complex in the presence of microtubules (Schmidt et al., 2012).
In addition to their dependency on binding to the Ndc80 complex for kinetochore recruitment, it is evident from the aforementioned studies that many of the properties of Cdt1, such as its ability to bind/localize to curved microtubule protofilaments/plus-ends and diffuse on microtubules, mirror that of the Ska1 complex. Further, both the Ska1 complex and Cdt1 carry a winged-helix domain that constitutes their microtubule-binding domain (Jeyaprakash et al., 2012; Agarwal et al., 2018). Thus, we conjectured that along with binding to microtubules directly, Cdt1 might also be able to access microtubules indirectly through its interaction with the Ska1 complex. Our previous studies have demonstrated a modest interaction between Cdt1 and Ndc80 both in cells and in vitro (Agarwal et al., 2018; Varma et al., 2012); however, whether Cdt1 and the Ska1 complex could interact, remained unknown. To further understand how load-bearing k-MT attachments are generated by Ndc80, Ska1, and Cdt1, we put forth two key hypotheses: (a) The Ska1 complex and Cdt1 bind microtubules synergistically and (b) Cdt1, Ska1, and Ndc80 interact with each other to generate a trimolecular complex that is important for efficient k-MT coupling, the mechanism for which is still unknown.
In this study, we first developed an Auxin-induced Degron (AID)–mediated approach to rapidly and acutely inhibit the function of Cdt1 selectively in mitotic cells. We then used a combination of biochemical, biophysical, and cell biological approaches to investigate the above-stated hypotheses. Prior work has established Ndc80 as the major microtubule-binding complex at the kinetochore; however, individual Ndc80 complexes have demonstrated no strong preference to bind to the growing or shortening microtubule ends (Powers et al., 2009; Schmidt et al., 2012). Although Ndc80 has been shown to be able to weakly track dynamic ends in combination with Ska1, the molecular milieu of a robust tip tracker during the later stages of mitosis is still missing. Here we show that Cdt1 synergizes with Ska1 and Ndc80 to form a tripartite Ndc80-Cdt1-Ska1 complex that robustly tracks the ends of dynamic microtubules.
Results
An auxin-induced degron cellular system to assay for mitotic functions of Cdt1
We had previously demonstrated the novel mitotic function of the replication licensing protein, Cdt1, using a double-thymidine cell synchronization approach coupled with siRNA interference or by injecting a function-blocking antibody into mitotic cells. The siRNA approach took advantage of the fact that Cdt1 was degraded in the S-phase and cells entering G2/M in the presence of Cdt1 siRNA were devoid of newly accumulating Cdt1 during mitosis (Varma et al., 2012). However, it is still possible that these above-mentioned methods were not optimal in inhibiting the mitotic functions of Cdt1 and/or were also detrimental to cells. We hence developed a new AID system for Cdt1 that drives its rapid degradation upon Auxin (IAA) addition. Both the genomic Cdt1 loci were replaced with a C-terminal degron (AID)- and YFP-tagged version of Cdt1 in the resulting DLD1 stable cell line, hereafter referred to as Cdt1-AID cell line (Fig. 1 A and Fig. S1 B). Using this approach, we expected cells entering mitosis after induction with IAA not to have encountered Cdt1 inhibition in any stage of the cell cycle prior to M-phase.
Generation of an AID cellular system to assay for mitotic functions of Cdt1 (AID-Cdt1). (A) A schematic of the repair template generated to endogenously tag the Cdt1 genomic locus with YFP and AID at the C-terminal using CRISPR/CAS9-based genome editing. (B) Western-blot analysis using an anti-Cdt1 antibody showing the levels of the endogenous and tagged Cdt1 in parental and AID-Cdt1 clones, respectively, following auxin-mediated degradation. (C) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel, [−] IAA) or the Auxin, IAA, for 2 h. (middle and bottom panels, [+] IAA) followed by immunostaining using an antibody against tubulin and the chromosomes counterstained with DAPI. Scale bar, 5 µm. (D) AID-Cdt1 DLD1 cells growing at 37°C were treated with the DNA stain, Hoechst, along with either control DMSO (top panels) or IAA, for 2 h. (middle and bottom panels) followed by live-cell imaging for 2–6 h as required. Selected frames from the time series are shown for each experimental condition as indicated. Scale bar, 5 µm. (E) Quantification of live imaging data from D. n = 30 for control cells not treated with IAA and 55 for cells treated with IAA. Data presented as mean ± SD, two-tailed t test *** = P < 0.0001. (F) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel) or IAA, for 2 h (bottom panel) followed by treatment with ice-cold buffer PBS buffer for 10 min. The cells were then immunostained for antibodies against tubulin, Zwint1 (for kinetochores) with the chromosomes counterstained using DAPI. Scale bar, 5 µm. (G) Quantification of total microtubule intensities from cells in F (n = 10 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (H) Insets from the top and bottom panels of F showing samples of attached and unattached kinetochores, respectively, as indicated. Scale bar, 1 µm. (I) Quantification of the % of attached kinetochores from F under the two experimental conditions assessed (n = 10 cells). Data presented as mean ± SD. Source data are available for this figure: SourceData F1.
Generation of an AID cellular system to assay for mitotic functions of Cdt1 (AID-Cdt1). (A) A schematic of the repair template generated to endogenously tag the Cdt1 genomic locus with YFP and AID at the C-terminal using CRISPR/CAS9-based genome editing. (B) Western-blot analysis using an anti-Cdt1 antibody showing the levels of the endogenous and tagged Cdt1 in parental and AID-Cdt1 clones, respectively, following auxin-mediated degradation. (C) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel, [−] IAA) or the Auxin, IAA, for 2 h. (middle and bottom panels, [+] IAA) followed by immunostaining using an antibody against tubulin and the chromosomes counterstained with DAPI. Scale bar, 5 µm. (D) AID-Cdt1 DLD1 cells growing at 37°C were treated with the DNA stain, Hoechst, along with either control DMSO (top panels) or IAA, for 2 h. (middle and bottom panels) followed by live-cell imaging for 2–6 h as required. Selected frames from the time series are shown for each experimental condition as indicated. Scale bar, 5 µm. (E) Quantification of live imaging data from D. n = 30 for control cells not treated with IAA and 55 for cells treated with IAA. Data presented as mean ± SD, two-tailed t test *** = P < 0.0001. (F) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel) or IAA, for 2 h (bottom panel) followed by treatment with ice-cold buffer PBS buffer for 10 min. The cells were then immunostained for antibodies against tubulin, Zwint1 (for kinetochores) with the chromosomes counterstained using DAPI. Scale bar, 5 µm. (G) Quantification of total microtubule intensities from cells in F (n = 10 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (H) Insets from the top and bottom panels of F showing samples of attached and unattached kinetochores, respectively, as indicated. Scale bar, 1 µm. (I) Quantification of the % of attached kinetochores from F under the two experimental conditions assessed (n = 10 cells). Data presented as mean ± SD. Source data are available for this figure: SourceData F1.
Analyses of the AID-Cdt1 degron system to study mitotic functions of Cdt1. (A) Western blot analysis using an anti-GFP antibody showing the levels of tagged-Cdt1 in parental DLD1 cells and in the YFP-tagged AID-Cdt1 clones following auxin-mediated degradation. (B) Selection of homozygous AID-Cdt1 colonies by PCR. (C) AID-Cdt1 DLD1 cells were treated with the DNA stain, Hoechst, along with either control DMSO (top panel) or IAA (bottom panel) for 2 h and still pictures of the chromosomes and YFP were acquired. (D) AID-Cdt1 DLD1 cells were treated with ice-cold (4°C) buffer for 10 min after adding IAA for the indicated periods of time. The cells were then immunostaining for antibodies against tubulin and the chromosomes counterstained with DAPI. (E) AID-Cdt1 DLD1 cells growing at 37°C were fixed after adding IAA for the indicated periods of time. The cells were then immunostained for antibodies against tubulin and the chromosomes counterstained with DAPI. (F) Parental DLD1 cells treated with DNA damaging agent, Etoposide, for 4 h, control AID-Cdt1 DLD1 cells or those treated with IAA for 2 h were immunostained for antibodies against γ-H2AX and the chromosomes were counterstained with DAPI. (G) Control AID-Cdt1 DLD1 cells or those treated with IAA for 2 h were immunostained for antibodies against microtubules and the chromosomes were counterstained with DAPI. Scale bar, 5 µm. Source data are available for this figure: SourceData FS1.
Analyses of the AID-Cdt1 degron system to study mitotic functions of Cdt1. (A) Western blot analysis using an anti-GFP antibody showing the levels of tagged-Cdt1 in parental DLD1 cells and in the YFP-tagged AID-Cdt1 clones following auxin-mediated degradation. (B) Selection of homozygous AID-Cdt1 colonies by PCR. (C) AID-Cdt1 DLD1 cells were treated with the DNA stain, Hoechst, along with either control DMSO (top panel) or IAA (bottom panel) for 2 h and still pictures of the chromosomes and YFP were acquired. (D) AID-Cdt1 DLD1 cells were treated with ice-cold (4°C) buffer for 10 min after adding IAA for the indicated periods of time. The cells were then immunostaining for antibodies against tubulin and the chromosomes counterstained with DAPI. (E) AID-Cdt1 DLD1 cells growing at 37°C were fixed after adding IAA for the indicated periods of time. The cells were then immunostained for antibodies against tubulin and the chromosomes counterstained with DAPI. (F) Parental DLD1 cells treated with DNA damaging agent, Etoposide, for 4 h, control AID-Cdt1 DLD1 cells or those treated with IAA for 2 h were immunostained for antibodies against γ-H2AX and the chromosomes were counterstained with DAPI. (G) Control AID-Cdt1 DLD1 cells or those treated with IAA for 2 h were immunostained for antibodies against microtubules and the chromosomes were counterstained with DAPI. Scale bar, 5 µm. Source data are available for this figure: SourceData FS1.
Efficient Cdt1 degradation was observed within 1–2 h of induction with IAA (Fig. 1 B and Fig. S1 A). To validate the AID system, we sought to confirm if we could recapitulate the phenotypes which were observed using the previously employed approaches. We had demonstrated that Cdt1 depletion in mitotic cells led to substantial destabilization of k-MT attachments after cold treatment. Treating Cdt1-AID cell line with IAA, we observed a very similar phenotype. While at 30 min post-IAA exposure, there were still a considerable number of cold-stable microtubules retained in metaphase, most of these microtubules completely disappeared within 1–2 h (Fig. S1 D). Interestingly, unlike the observation with siRNA-based depletion, a substantial fraction (∼80–90%) of mitotic cells in late prometaphase or metaphase had severe defects in the structure of the mitotic spindle even at 37°C. Among these, majority of the cells (∼80%) had a disorganized metaphase spindle while in a smaller fraction (∼20%), the spindles appeared rudimentary in size. Cells in both these categories also exhibited substantial chromosome misalignment at the metaphase plate (Fig. 1 C). Further, all cells that entered anaphase exhibited at least one chromosome mis-segregation event (Fig. S1 E).
To better characterize the mitotic phenotypes of Cdt1-AID cells, we carried out live imaging of cells entering mitosis, after labeling their chromosomes with the cell permeable DNA dye, Hoechst. Live imaging was initiated at the point of nuclear envelope breakdown (NEBD) and continued for a period of 2–6 h. While the un-induced control Cdt1-AID cells treated with DMSO were YFP-positive prior to the initiation of imaging, the cells treated with IAA were YFP-negative. The control cells that were not subjected to Cdt1 degradation segregated their chromosomes in an average of 70 min after NEBD (n = 30; Video 1 and Fig. 1 D top panel, quantified in Fig. 1 E). However, the cells subjected to induced degradation of Cdt1 (n = 55) showed three broad categories of phenotypes. The first category constituted mitotic cells that exhibited mild-to-moderate (7/55) or severe (13/55) delays in mitotic progression and underwent chromosome mis-segregation at anaphase onset (Fig. 1 D, middle panel; Video 2 and Fig. 1 E, pink dots). The second category (28/55) of cells remained arrested in the mitosis for the entire period of imaging with severe chromosome misalignment after initially being able to align majority of the chromosomes (Fig. 1 D, bottom panel; Video 3 and Fig. 1 E, blue dots). The third category (7/55) of mitotic cells exhibited mild-to-moderate delays in mitosis but still segregated their chromosomes normally (Fig. 1 E, black dots). Overall, the Cdt1-AID cells treated with IAA spent extensively longer time (∼2.4 fold, 166 min) in mitosis, averaged over the entire period of imaging (Fig. 1 E).
Normal mitotic progression in AID-Cdt1 DLD1 cells treated with control DMSO. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of ∼90 min (for this example). The movie speed was up to ∼ 1,500 × and played at ∼4 frames/s. Scale bar, 5 µm.
Normal mitotic progression in AID-Cdt1 DLD1 cells treated with control DMSO. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of ∼90 min (for this example). The movie speed was up to ∼ 1,500 × and played at ∼4 frames/s. Scale bar, 5 µm.
Severe delay in mitotic progression in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of ∼350 min (for this example). The movie speed was up to ∼1,750 × and played at ∼4 frames/s. Scale bar, 5 µm.
Severe delay in mitotic progression in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of ∼350 min (for this example). The movie speed was up to ∼1,750 × and played at ∼4 frames/s. Scale bar, 5 µm.
Chromosome mis-segregation in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of 135 min (for this example). The movie speed was up to ∼1,600 × and played at ∼4 frames/s. Scale bar, 2 µm.
Chromosome mis-segregation in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Live images of chromosomes labeled with DNA dye, Hoechst, were captured every 7.5 min after, for a total period of 135 min (for this example). The movie speed was up to ∼1,600 × and played at ∼4 frames/s. Scale bar, 2 µm.
Next, we wanted to assess the stability of k-MT attachments in Cdt1-AID cells after Cdt1 degradation. We observed a 75% loss of total microtubule fluorescence in Cdt1-AID cells with IAA treatment as compared to the control untreated cells (Fig. 1, F and G). The kinetochore pairs of the treated cells were less stretched or tensed reflecting an absence of load-bearing attachments. Further, we also evaluated the frequency of physical contacts made between kinetochores and microtubules in both control and Cdt1-degraded cells. After Cdt1 was degraded, only ∼10% of the kinetochores showed proper bi-oriented attachments with spindle microtubules in contrast to the controls, where ∼90% of the kinetochores were properly attached and bi-oriented (Fig. 1, H and I). Overall, the phenotype of loss of k-MT stability seems to be stronger with the AID approach in DLD cells as compared to our previous studies that used siRNA-based approach in synchronized HeLa cells (Agarwal et al., 2018; Varma et al., 2012). Further, similar to the siRNA method, we observed no interference with relevant interphase cellular functions such as DNA replication licensing (a function of Cdt1 that induces DNA damage when perturbed; Fig. S1 F) or the organization of the microtubule cytoskeleton (Cdt1 is a microtubule-binding protein; Fig. S1 G). Based on the entire analysis of mitotic phenotypes after AID-mediated Cdt1 inhibition, our evidence points to the loss of k-MT stability as the primary cause of the spindle defects and chromosome misalignment observed at the metaphase plate.
The succeeding result sections of this work will not employ the use of AID-mediated Cdt1 inhibition as our main intention was to present this approach as an important resource for future research purposes.
Depletion of the Ska1 complex abrogates the localization of Cdt1 to kinetochores and spindle microtubules
Besides Cdt1, many other microtubule-associated proteins/protein complexes are recruited at the k-MT interface by the Ndc80 complex to facilitate stable k-MT attachments. One such critical kinetochore-localized complex is the metazoan Ska1 complex (Chan et al., 2012; Raaijmakers et al., 2009; Welburn et al., 2009; Zhang et al., 2012). Ska1 is a hetero-hexameric complex, composed of two copies each of SKA1, 2, and 3 subunits. The ability of the Ska1 complex to dock on to kinetochore-bound Ndc80 and binds to microtubules to generate robust k-MT attachments similar to Cdt1 prompted us to evaluate if Ska1 shares a functional relationship with Cdt1. How these proteins coordinate with Ndc80 either alone or in combination to modulate the strength of k-MT attachments during mitosis is yet unclear.
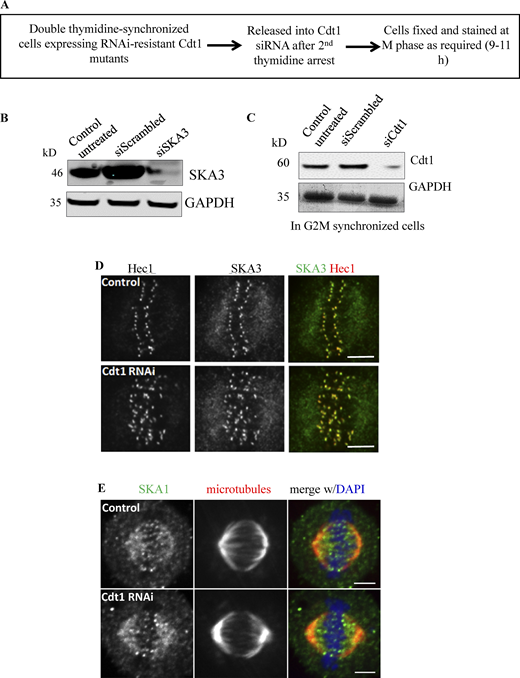
Our previous studies suggested that Ska1 levels at the kinetochores remained unaffected in the absence of Cdt1 (Varma et al., 2012); however, it is not clear if the Ska1 complex has a role in Cdt1 localization to the kinetochores. To address this question, we carried out immunostaining of endogenous Cdt1 in HeLa cells that were treated with either scrambled siRNA (control/siScrambled) or siRNA against SKA3 (siSKA3). While control cells showed distinct localization of Cdt1 to both kinetochores and the spindle poles (similar to SKA3), Cdt1 failed to localize properly to either of these structures in SKA3-depleted cells (Fig. 2 A). Quantification of Cdt1 and Ska1 levels at the kinetochores suggests a positive correlation between the loss of SKA3 and Cdt1 as the knockdown of SKA3 led to a substantial loss of Cdt1 at the kinetochores (Fig. 2 B and Fig. S2 B). However, in line with our previous findings, in the reverse experiment where mitotic cells were subjected to siRNA-mediated knockdown of Cdt1, the levels of SKA3 at the kinetochores were unaffected (Fig. S2, A, C, and D; Varma et al., 2012). Our results thus point toward a hierarchical recruitment of these proteins where Cdt1 is dependent on both the Ndc80 and the Ska1 complexes for its localization to kinetochores, while Ska1 only depended on the Ndc80 complex.
The Ska1 complex is essential for the recruitment of Cdt1 to the kinetochores and spindle microtubules. (A) HeLa cells treated with either scramble control (siControl) or siRNA against endogenous SKA3 (siSka3) were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against Ndc80/Hec1 (a kinetochore marker) in far red, SKA3 in green, and Cdt1 in red are shown. Scale bar, 5 μm. (B) A bar graph showing the quantification of kinetochore staining intensity after siRNA treatment in each case (n = 110 kinetochores, >5 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (C) HeLa cells treated with either siControl or siSKA3 were fixed using methanol. Representative images of cells immunostained with antibodies against tubulin in green and Cdt1 in red are shown; DAPI-stained chromosomes are shown in blue in the merged image. Scale bar, 5 μm. (D) A bar graph showing the quantification of Cdt1 spindle microtubule staining intensity after siSKA3 treatment as compared to siControl (n = 13 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (E–G) HeLa cells stably expressing either GFP-SKA1 full-length (E and F) or GFP-SKA1ΔMTBD (G) were treated with siSKA3 to knockdown the endogenous SKA3 and the expression of siRNA-resistant GFP-SKA1 or GFP-SKA1ΔMTBD proteins was induced by adding 2.5 µg/ml of Doxycycline (+Dox) for 48 h. Control cells were treated similarly with the siSKA3 in each case, but no doxycycline was added (−Dox). The cells were then fixed and immunostained with antibodies against GFP for SKA1-FL or SKA1 ΔMTBD (second column in E and G), Cdt1 (third column and in red in the merge images), and microtubules (fourth column and in blue in the merge images) with the chromosomes counterstained using DAPI (first column). Scale bar, 5 μm. (F) A bar graph showing the quantification of Cdt1 spindle microtubule staining intensity from E in the presence and absence of Doxycline induction (n = 22 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001.
The Ska1 complex is essential for the recruitment of Cdt1 to the kinetochores and spindle microtubules. (A) HeLa cells treated with either scramble control (siControl) or siRNA against endogenous SKA3 (siSka3) were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against Ndc80/Hec1 (a kinetochore marker) in far red, SKA3 in green, and Cdt1 in red are shown. Scale bar, 5 μm. (B) A bar graph showing the quantification of kinetochore staining intensity after siRNA treatment in each case (n = 110 kinetochores, >5 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (C) HeLa cells treated with either siControl or siSKA3 were fixed using methanol. Representative images of cells immunostained with antibodies against tubulin in green and Cdt1 in red are shown; DAPI-stained chromosomes are shown in blue in the merged image. Scale bar, 5 μm. (D) A bar graph showing the quantification of Cdt1 spindle microtubule staining intensity after siSKA3 treatment as compared to siControl (n = 13 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (E–G) HeLa cells stably expressing either GFP-SKA1 full-length (E and F) or GFP-SKA1ΔMTBD (G) were treated with siSKA3 to knockdown the endogenous SKA3 and the expression of siRNA-resistant GFP-SKA1 or GFP-SKA1ΔMTBD proteins was induced by adding 2.5 µg/ml of Doxycycline (+Dox) for 48 h. Control cells were treated similarly with the siSKA3 in each case, but no doxycycline was added (−Dox). The cells were then fixed and immunostained with antibodies against GFP for SKA1-FL or SKA1 ΔMTBD (second column in E and G), Cdt1 (third column and in red in the merge images), and microtubules (fourth column and in blue in the merge images) with the chromosomes counterstained using DAPI (first column). Scale bar, 5 μm. (F) A bar graph showing the quantification of Cdt1 spindle microtubule staining intensity from E in the presence and absence of Doxycline induction (n = 22 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001.
Depletion of Cdt1 from mitotic cells does not interfere with the normal targeting of the Ska1 complex. (A) Schematic representation of the knockdown protocol in double thymidine synchronized HeLa cells that are used to deplete Cdt1 specifically during mitosis. (B) Representative Western blots shown with indicated antibodies in each case to assess the level of Ska3 knockdown upon siRNA addition. (C) Same as in B but to assess the level of Cdt1 knockdown upon siRNA addition. (D) HeLa cells treated with either scramble control or siRNA against endogenous Cdt1 were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against Hec1 (kinetochore marker) in red and SKA3 (representing the Ska1 complex) in green are shown. Scale bar, 5 μm. (E) HeLa cells treated with either scramble control or siRNA against endogenous Cdt1 were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against SKA3 (in green) and tubulin (in red) are shown; DAPI stained the chromosomes and shown in black and white. Scale bar, 5 μm. Source data are available for this figure: SourceData FS2.
Depletion of Cdt1 from mitotic cells does not interfere with the normal targeting of the Ska1 complex. (A) Schematic representation of the knockdown protocol in double thymidine synchronized HeLa cells that are used to deplete Cdt1 specifically during mitosis. (B) Representative Western blots shown with indicated antibodies in each case to assess the level of Ska3 knockdown upon siRNA addition. (C) Same as in B but to assess the level of Cdt1 knockdown upon siRNA addition. (D) HeLa cells treated with either scramble control or siRNA against endogenous Cdt1 were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against Hec1 (kinetochore marker) in red and SKA3 (representing the Ska1 complex) in green are shown. Scale bar, 5 μm. (E) HeLa cells treated with either scramble control or siRNA against endogenous Cdt1 were fixed using paraformaldehyde. Representative images of cells immunostained with antibodies against SKA3 (in green) and tubulin (in red) are shown; DAPI stained the chromosomes and shown in black and white. Scale bar, 5 μm. Source data are available for this figure: SourceData FS2.
The next question was to test whether the binding of Cdt1 to spindle microtubules was also dependent on the Ska1 complex. To answer this question, HeLa cells were immunostained to discern the level of endogenous Cdt1 on microtubules in the absence of the Ska1 complex. Indeed, the cells treated with SKA3 siRNA demonstrated ∼55% reduction in Cdt1 localization to the spindle microtubules (Fig. 2, C and D) as compared to the controls, where Cdt1 clearly co-localized with the spindle microtubules in accordance with our previous study (Agarwal et al., 2018). However, it was plausible that the inability of Cdt1 to localize to microtubules upon SKA3 knockdown could be attributed to the disruption of spindle microtubules. To address this possibility, we performed a reverse knockdown-rescue experiment wherein, an siRNA-resistant version of the microtubule-binding SKA1 subunit of the Ska1 complex fused with GFP was stably expressed in HeLa cells in a doxycycline-inducible manner, while the endogenous SKA1 was depleted using siRNA. We then tested whether Cdt1 localization to spindles was enhanced in this scenario. The expressed SKA1-GFP localized to spindle microtubules as expected. More importantly, quantification of Cdt1 fluorescence intensity in these mitotic cells revealed a substantial enhancement of endogenous Cdt1 localization on spindle microtubules (Fig. 2, E and F), which was around fivefold higher than the uninduced control (-doxycycline) and the endogenous Cdt1 staining in normal HeLa cells (Fig. 2 C; Agarwal et al., 2018). The results demonstrate that the expressed SKA1-GFP was able to recruit substantial levels of Cdt1 to the mitotic spindle, pointing toward a possible mechanistic synergy between these two proteins for microtubule binding. Interestingly, over-expression of a SKA1-GFP fragment (SKA1ΔMTBD, aa 1–131) devoid of the microtubule-binding domain (MTBD) was not able to recruit Cdt1 to microtubules (Fig. 2 G). This could be attributed to the inability of the SKA1ΔMTBD to localize to microtubules and/or the involvement of this domain in interacting with Cdt1, thus preventing Cdt1 microtubule recruitment.
Finally, to assess if Cdt1 depletion had any effect on the ability of the Ska1 complex to interact with microtubules, we depleted Cdt1 from HeLa cells using siRNA-coupled with double thymidine synchronization and evaluated endogenous Ska1 staining on microtubules. Our results demonstrated that after Cdt1 knockdown, the localization of Ska1 to spindle microtubules remained unaffected in comparison with the control untreated cells (Fig. S2, A, C, and E). These results clearly confirm that although Cdt1 is dependent on the Ska1 complex for both its kinetochore and microtubule localization, Ska1 recruitment as such did not require Cdt1.
Cdt1 and the Ska1 complex interacts in cells and in vitro
The above data not only demonstrate the mechanism of how Cdt1 is recruited to its mitotic targets via its interaction with the Ska1 complex but also point strongly toward the scenario that these two proteins can interact with each other. Therefore, we employed several biochemical methods, including GST pull down assay, co-immunoprecipitation (coIP), in vitro Blot overlay assay and Biolayer Interferometry (BLI) to test this possibility.
For the GST pull down assay, mitotic HeLa cell lysate was incubated with equal amounts of either GST alone or GST-tagged Cdt192-546 (here after referred to as GST-Cdt1) proteins (Fig. 3 A). We had previously shown that the bacterially purified Cdt192-546 protein that lacks the N-terminal unstructured domain (aa 1–91) is competent to bind to its established mitotic interacting partners (Agarwal et al., 2018). Upon elution with reduced glutathione, the glutathione agarose beads could pull down GST-Cdt1 as expected, but SKA1 and SKA3 were also co-eluted and enriched in the bound (B) fractions. As a control, GST was not able to precipitate the Ska1 complex as is evident from the detection of SKA1 and SKA3 only in the unbound (UB) fractions (Fig. 3, A and B).
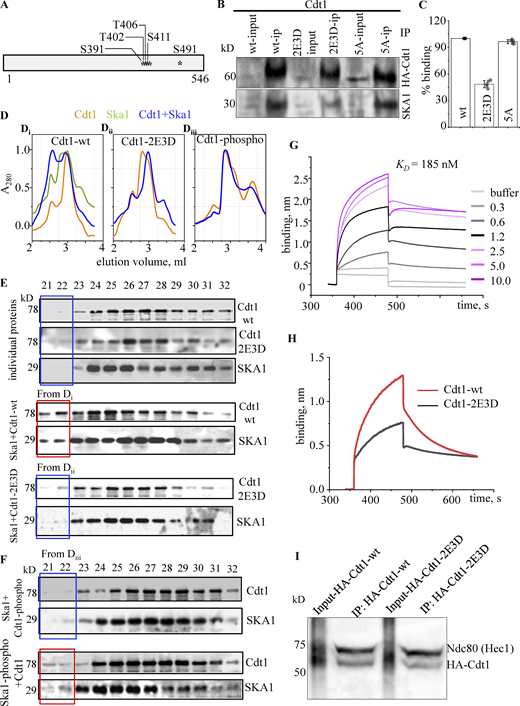
Cdt1 and the Ska1 complex physically interact with each other. (A) Diagrammatic representation of the Cdt1 (Cdt192-546) construct purified from bacteria that were used for in vitro interaction studies with the Ska1 complex. The winged-helix (WH) domains relevant for the purpose of the current study has been depicted. (B) Thymidine synchronized and nocodazole arrested mitotic HeLa cell extracts were incubated either with purified GST or GST-tagged Cdt192-546 proteins (10 μg each), followed by pull down with glutathione agarose beads. UB indicates the unbound or flow through fraction and B indicates the proteins retrieved after elution with reduced glutathione. The blots were probed with anti-GST antibody and SKA1 and SKA3 antibodies. (C) Thymidine synchronized and nocodazole arrested mitotic HeLa cell extracts were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. (D) Thymidine synchronized and nocodazole arrested mitotic HeLa cells that stably expressing GFP-SKA1 (induced by adding 2.5 µg/ml of doxycycline) were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (E) Blot overlay assay to study Cdt1-Ska1 complex interaction. 0.5 µg each of the indicated proteins were loaded as baits (top labels) on 18% SDS-PAG, transferred to nitrocellulose membrane and blocked with 5% SM-TBST. 1 µg of the indicated proteins were overlaid as prey proteins (arrows on the right-side labels) on the membrane for 12 h at 4°C. The blot was washed and probed with the indicated antibodies (bottom labels) followed by chemiluminescence. Arrows depict the proteins of interest and the required molecular mass standards are shown in kD (left labels). (F) Biolayer interferometry (BLItz) sensograms obtained using 0.2 μg/ml of GFP-tagged Cdt192-546-loaded amine-reactive biosensors and increasing concentrations (0.3–10 µM) of the Ska1 complex used as analyte to generate a series of sensograms showing the binding and dissociation phases after the baseline. Binding curves were fit globally to a 1:1 binding model to yield equilibrium dissociation constant (KD) noted above. (G) Linear diagram of the full-length and truncated Cdt1 and SKA1 proteins used to map the Cdt1-SKA1 interaction domains. (H) Table for KD values estimated from the BLI sensograms obtained for the pairwise binding of the indicated purified protein constructs. Source data are available for this figure: SourceData F3.
Cdt1 and the Ska1 complex physically interact with each other. (A) Diagrammatic representation of the Cdt1 (Cdt192-546) construct purified from bacteria that were used for in vitro interaction studies with the Ska1 complex. The winged-helix (WH) domains relevant for the purpose of the current study has been depicted. (B) Thymidine synchronized and nocodazole arrested mitotic HeLa cell extracts were incubated either with purified GST or GST-tagged Cdt192-546 proteins (10 μg each), followed by pull down with glutathione agarose beads. UB indicates the unbound or flow through fraction and B indicates the proteins retrieved after elution with reduced glutathione. The blots were probed with anti-GST antibody and SKA1 and SKA3 antibodies. (C) Thymidine synchronized and nocodazole arrested mitotic HeLa cell extracts were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. (D) Thymidine synchronized and nocodazole arrested mitotic HeLa cells that stably expressing GFP-SKA1 (induced by adding 2.5 µg/ml of doxycycline) were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (E) Blot overlay assay to study Cdt1-Ska1 complex interaction. 0.5 µg each of the indicated proteins were loaded as baits (top labels) on 18% SDS-PAG, transferred to nitrocellulose membrane and blocked with 5% SM-TBST. 1 µg of the indicated proteins were overlaid as prey proteins (arrows on the right-side labels) on the membrane for 12 h at 4°C. The blot was washed and probed with the indicated antibodies (bottom labels) followed by chemiluminescence. Arrows depict the proteins of interest and the required molecular mass standards are shown in kD (left labels). (F) Biolayer interferometry (BLItz) sensograms obtained using 0.2 μg/ml of GFP-tagged Cdt192-546-loaded amine-reactive biosensors and increasing concentrations (0.3–10 µM) of the Ska1 complex used as analyte to generate a series of sensograms showing the binding and dissociation phases after the baseline. Binding curves were fit globally to a 1:1 binding model to yield equilibrium dissociation constant (KD) noted above. (G) Linear diagram of the full-length and truncated Cdt1 and SKA1 proteins used to map the Cdt1-SKA1 interaction domains. (H) Table for KD values estimated from the BLI sensograms obtained for the pairwise binding of the indicated purified protein constructs. Source data are available for this figure: SourceData F3.
Further, to ascertain whether Cdt1 and the Ska1 complex interact in vivo, we performed coimmunoprecipitation (coIP) experiments in HeLa cells, first under endogenous conditions. Mitotic HeLa cell extracts were subjected to immunoprecipitation with non-specific IgG (as a control), or antibodies targeted against Cdt1 and SKA3. Cdt1 antibody was able to pull down endogenous Cdt1 protein as expected but simultaneously, it also immunoprecipitated both the SKA1 and the SKA3 subunits. Similarly, in a reverse pull down assay, the SKA3 antibody was also able to immunoprecipitate Cdt1 from the cell lysates along with SKA1 as expected (Fig. 3 C).
For assessing Cdt1-Ska1 interaction under conditions where one protein is over-expressed, we transfected a plasmid expressing HA/His-tagged Cdt1 into HEK293T cells followed by arresting the cells in mitosis through thymidine-synchronization and nocodazole treatment. Ni+2-NTA agarose-mediated affinity precipitation of HA/His-tagged Cdt1 led to the co-precipitation of both SKA1 and SKA3 as interacting partners (Fig. S3 A). The coIP assay was also performed in HeLa cells that were stably expressing either GFP-SKA1 (under the doxycycline inducible promoter) or GFP alone. While an anti-GFP antibody could precipitate both GFP-SKA1 and GFP as expected, Cdt1 was co-precipitated only with the former and not the latter (Fig. 3 D and Fig. S3 B) indicating that the Ska1 complex and Cdt1 indeed interacted with each other in vivo.
Analyses of binding between Cdt1, Ska1, and their fragments. (A) Pull down of the SKA1 and the SKA3 subunits of the Ska1 complex by HA/His-tagged Cdt1-WT from thymidine synchronized and nocodazole arrested mitotic HEK-293T cell extracts. The pull down was performed using Ni2+-NTA agarose beads followed by immunoblotting with either anti-HA or anti-SKA3 and SKA1 antibodies. 1% of the lysate was loaded as total protein. (B) Thymidine synchronized and nocodazole arrested mitotic HeLa cells stably expressing GFP were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (C) SDS-PAG electrophoresis of purified His-SKA1/2 dimer. (D) Linear protein diagrams of the truncated protein constructs used in the BLI-interaction assay with indicated amino acids position showing truncation sites. (E) BLI sonograms for indicated conditions and their respective Kd values. (F) SDS-PAGE gel electrophoresis of purified SKA1/2+ SKA2-GFP complex. Source data are available for this figure: SourceData FS3.
Analyses of binding between Cdt1, Ska1, and their fragments. (A) Pull down of the SKA1 and the SKA3 subunits of the Ska1 complex by HA/His-tagged Cdt1-WT from thymidine synchronized and nocodazole arrested mitotic HEK-293T cell extracts. The pull down was performed using Ni2+-NTA agarose beads followed by immunoblotting with either anti-HA or anti-SKA3 and SKA1 antibodies. 1% of the lysate was loaded as total protein. (B) Thymidine synchronized and nocodazole arrested mitotic HeLa cells stably expressing GFP were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (C) SDS-PAG electrophoresis of purified His-SKA1/2 dimer. (D) Linear protein diagrams of the truncated protein constructs used in the BLI-interaction assay with indicated amino acids position showing truncation sites. (E) BLI sonograms for indicated conditions and their respective Kd values. (F) SDS-PAGE gel electrophoresis of purified SKA1/2+ SKA2-GFP complex. Source data are available for this figure: SourceData FS3.
The results obtained so far from the pull-down experiments in cellular extracts suggested the possibility that these two proteins could certainly interact with each other but whether this interaction is direct and does not require assistance from any other extraneous protein(s) remained to be discerned. To test this, in vitro Blot overlay assay was carried out using purified recombinant proteins. The Ska1 complex was assembled by combining equimolar His-tagged SKA1/2 (Fig. S3 C) and GST-tagged SKA3. Irrespective of whether the subunits of the Ska1 complex were used as a bait and GFP-Cdt192-546 (GFP-Cdt1) was overlaid or vice versa, i.e., Cdt192-546 was used as a bait and the Ska1 complex was overlaid, the proteins showed evidence of direct binding (Fig. 3 E). GFP and GST proteins (used as negative controls) were not able to bind to the Ska1 complex or Cdt1, respectively, attesting to the specificity of the interaction.
Having demonstrated a direct interaction between Cdt1 and the Ska1 complex, we proceeded to compute the affinity of the interaction between Cdt1 and the SKA1 subunit of the Ska1 complex. To accomplish this, we took advantage of Biolayer Interferometry (BLI), which exploits changes in the interference patterns when two biomolecules interact. We chose the SKA1 subunit because our Blot overlay experiments had suggested that Cdt1 predominantly bound to the SKA1 subunit of the Ska1 complex (Fig. 3 E). GFP-Cdt1 was immobilized on the His-reactive sensor and increasing concentrations of SKA1 were used as analytes. The association and dissociation kinetics were monitored in each phase, resulting in a kinetic binding sensogram from which the association and dissociation rate constants (ka and kd) and the equilibrium-binding constant (KD) were determined. The KD for Cdt1 and SKA1 was determined to be 70 nM (Fig. 3 F). As a negative control, BSA protein used as an analyte and failed to bind to GFP-Cdt1. Further, using a combination of C- and N-terminal fragments of Cdt1 (Cdt192-546) and SKA1, we identified the domains within Cdt1 and SKA1 that were important for their interaction (Fig. 3, G and H; and Fig. S3 D). The KD of the C-terminal fragment of SKA1 binding to Cdt1 (390 nM) was considerably higher compared to that of full-length SKA1. This was, however, much lower than the N-terminal SKA1 fragment, the binding KD of which to Cdt1 was 3.1 µM (Fig. 3, G and H; and Fig. S3 E). These data suggest that the MTBD of the SKA1 subunit is primarily responsible for Cdt1-binding and for the recruitment of Cdt1 to microtubules, in agreement with our results from Fig. 2 (Fig. 2, E–G). In the context of Cdt1 binding to SKA1, both the N-terminal (aa 92–410, 220 nM) as well as the C-terminal (aa 410–546, 360 nM) regions of Cdt1 had comparable binding affinities to the full-length SKA1; albeit they were less efficient as compared to the full-length Cdt1 (Fig. 3, G and H; and Fig. S3 E). This suggests that both the central (WH-M) as well as the C-terminal (WH-C) Winged-Helix domains of Cdt1 (Fig. 3 A) were required to bind to SKA1 efficiently. It is interesting to note that both these domains were also found to be required for efficient binding of Cdt1 to microtubules in vitro (Agarwal et al., 2018).
Phosphorylation of Cdt1 by Cdk1 at G2/M transition impacts its ability to interact with the Ska1 complex
We had previously shown that the phosphorylation of Cdt1 by the Aurora B kinase impacts the ability of Cdt1 to bind microtubules. Consequently, the expression of Aurora B phosphomimetic mutants of Cdt1 (Cdt1-10D) induced severe phenotypes, including the loss of k-MT attachments and delay in mitotic progression (Agarwal et al., 2018). Based on our current results, it is possible that the phenotypes observed with these mutants could at least partly be due to their inability to interact with the Ska1 complex. To investigate this, we immunoprecipitated HA-tagged Cdt1-WT and Cdt1-10D from mitotic HeLa extracts using anti-HA antibody and assessed the interaction of both these proteins with the Ska1 complex. We find that both Cdt1-WT and Cdt1-10D were able to pull down SKA1 and SKA3 equally efficiently (Fig. S4 A), suggesting that the phosphorylation of Cdt1 by Aurora B did not affect Cdt1’s ability to interact with the Ska1 complex. Thus, unlike the direct-binding of Cdt1 to microtubules, the interaction between Cdt1 and the Ska1 complex seems to be independent of Aurora B-mediated phosphoregulation.
Characterization of WT and phosphomutant Cdt1 cell lines and proteins. (A) Thymidine synchronized and nocodazole arrested mitotic HeLa cells that were stably expressing HA-tagged Cdt1-WT or Cdt1-10D (Aurora B phosphomimetic mutant) were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (B) Western blot from the cell lysate of Hela cells to analyze the expression levels of WT-Cdt1 protein along with indicated mutant versions. (C) SDS-PAG electrophoresis of purified GST-Cdt1-2E3D protein. (D) Asynchronous (left panel) or double thymidine synchronized HeLa cells stably expressing RNAi-resistant versions of the various Cdk1 (middle panel: WT, 5A, 2E3D) or Aurora B kinase (right panel: WT, 10A, 10D) mutants of Cdt1 were depleted of endogenous Cdt1 and DNA damage was assessed in each case after immunostaining using anti-γH2AX antibody (green) with the chromosomes counterstained using DAPI (blue). (E) Same as in D but in this case the interphase microtubule network was assessed only for the Cdk1 mutants (control, WT, 5A, 2E3D) after immunostaining the cells using an anti-tubulin antibody (red) with the chromosomes counterstained using DAPI (blue). Source data are available for this figure: SourceData FS4.
Characterization of WT and phosphomutant Cdt1 cell lines and proteins. (A) Thymidine synchronized and nocodazole arrested mitotic HeLa cells that were stably expressing HA-tagged Cdt1-WT or Cdt1-10D (Aurora B phosphomimetic mutant) were immunoprecipitated (IP) and immunoblotted (IB) with the indicated antibodies; IgG was taken as a negative control. 1% of the lysate was loaded as input. (B) Western blot from the cell lysate of Hela cells to analyze the expression levels of WT-Cdt1 protein along with indicated mutant versions. (C) SDS-PAG electrophoresis of purified GST-Cdt1-2E3D protein. (D) Asynchronous (left panel) or double thymidine synchronized HeLa cells stably expressing RNAi-resistant versions of the various Cdk1 (middle panel: WT, 5A, 2E3D) or Aurora B kinase (right panel: WT, 10A, 10D) mutants of Cdt1 were depleted of endogenous Cdt1 and DNA damage was assessed in each case after immunostaining using anti-γH2AX antibody (green) with the chromosomes counterstained using DAPI (blue). (E) Same as in D but in this case the interphase microtubule network was assessed only for the Cdk1 mutants (control, WT, 5A, 2E3D) after immunostaining the cells using an anti-tubulin antibody (red) with the chromosomes counterstained using DAPI (blue). Source data are available for this figure: SourceData FS4.
Interestingly, Cdt1 is also known to acquire phosphorylation by the Cdk1 kinase at G2/M transition (Chandrasekaran et al., 2011). There are at least 5 consensus Cdk1 phosphorylation sites located toward the C-terminal region of Cdt1 (Fig. 4 A). We, therefore, generated stable HeLa cell lines expressing siRNA-resistant versions of WT, non-phosphorylatable (5A), and phosphomimetic (2E3D) Cdt1. Biochemical analysis of these cell lines indicated that the respective versions of Cdt1 were expressed in relatively equal amounts in these cell lines (Fig. S4 B). We next performed immunoprecipitation experiments using the cell lysates from these stable lines to test whether these mutants were defective in binding to either the Ska1 or the Ndc80 complexes. We observed that both the WT and the 5A mutants of Cdt1 immunoprecipitated SKA1 equally well, but remarkably, there was a 50–55% reduction in SKA1 within the immunoprecipitates of Cdt1-2E3D cell lysates (Fig. 4, B and C).
The phosphorylation of Cdt1 by the Cdk1 kinase interferes with Ska1-binding. (A) Cdk1 phosphorylation sites on Cdt1 that were mutated to obtain phophomemeitic and non-phosphorylatable versions of the same protein. (B) Western blot showing co-immunoprecipitation of HA-Cdt1 constructs and the Ska1 complex (detected by the SKA1 antibody). (C) Quantitative band intensities from B. Data presented as mean ± SD. (D) WT (Di), 2E3D mutant (Dii), or in vitro Cdk1-phosphorylated Cdt1 (Diii) were run on a Superose 6 gel filtration column after mixing with equimolar ratio of the Ska1 complex for 1 h, as described in Materials and methods. Recorded elution profiles of the above-mentioned mixture of proteins/complexes exiting the column, as indicated. The elution profiles of Cdt1 variants or the Ska1 complex run individually have not been shown. (E) Western blots of fractions from Di, Dii, or Diii above probed using primary antibodies as indicated on the right, for the proteins/protein complexes run separately or in combination as indicated on the left. The initial two fractions (21 and 22) of the elution for Cdt1-WT + Ska1 complex have been marked with a red box to highlight the observed difference with those of the individually run proteins/complexes or a mix of Cdt1-2E3D + Ska1 complex (marked in blue). (F) Same as E but with the phosphomimetic mutant variant of Cdt1 or the Ska1 complex as indicated. (G) BLI sensograms showing wavelength shifts (nm) generated by the addition of 0.5 μg/ml Cdt1-2E3D protein with increasing concentrations of SKA1 (0.3–10 µM), as indicated in the plot. (H) BLI sensograms of the indicated Cdt1 variant binding to SKA1 used at 0.6 µM concentration. (I) Nocodazole arrested mitotic HeLa cells that were stably expressing HA-tagged Cdt1-WT or Cdt1-2E3D (Cdk1 phosphomimetic mutant) were immunoprecipitated (IP) with the antibodies indicated on the right. Source data are available for this figure: SourceData F4.
The phosphorylation of Cdt1 by the Cdk1 kinase interferes with Ska1-binding. (A) Cdk1 phosphorylation sites on Cdt1 that were mutated to obtain phophomemeitic and non-phosphorylatable versions of the same protein. (B) Western blot showing co-immunoprecipitation of HA-Cdt1 constructs and the Ska1 complex (detected by the SKA1 antibody). (C) Quantitative band intensities from B. Data presented as mean ± SD. (D) WT (Di), 2E3D mutant (Dii), or in vitro Cdk1-phosphorylated Cdt1 (Diii) were run on a Superose 6 gel filtration column after mixing with equimolar ratio of the Ska1 complex for 1 h, as described in Materials and methods. Recorded elution profiles of the above-mentioned mixture of proteins/complexes exiting the column, as indicated. The elution profiles of Cdt1 variants or the Ska1 complex run individually have not been shown. (E) Western blots of fractions from Di, Dii, or Diii above probed using primary antibodies as indicated on the right, for the proteins/protein complexes run separately or in combination as indicated on the left. The initial two fractions (21 and 22) of the elution for Cdt1-WT + Ska1 complex have been marked with a red box to highlight the observed difference with those of the individually run proteins/complexes or a mix of Cdt1-2E3D + Ska1 complex (marked in blue). (F) Same as E but with the phosphomimetic mutant variant of Cdt1 or the Ska1 complex as indicated. (G) BLI sensograms showing wavelength shifts (nm) generated by the addition of 0.5 μg/ml Cdt1-2E3D protein with increasing concentrations of SKA1 (0.3–10 µM), as indicated in the plot. (H) BLI sensograms of the indicated Cdt1 variant binding to SKA1 used at 0.6 µM concentration. (I) Nocodazole arrested mitotic HeLa cells that were stably expressing HA-tagged Cdt1-WT or Cdt1-2E3D (Cdk1 phosphomimetic mutant) were immunoprecipitated (IP) with the antibodies indicated on the right. Source data are available for this figure: SourceData F4.
To further confirm the results from the immunoprecipitation experiments, we purified GFP-tagged recombinant WT and 2E3D mutants of Cdt1 in bacteria (Agarwal et al., 2018; Fig. S4 C) to perform co-fractionation experiments with the Ska1 complex using analytical size exclusion chromatography (Huis In 't Veld et al., 2019; Zhang et al., 2020). We used either the WT or 2E3D mutants of Cdt1 separately or mixed them in equimolar ratio with the Ska1 complex for 1 h, before running them on the column. Both Cdt1-WT and -2E3D as well as the Ska1 complex eluted at their normal expected molecular weights when ran individually. However, when Cdt1-WT was mixed with the Ska1 complex, we observed a different elution pattern for both these proteins. A certain fraction of the Ska1 complex appeared to coelute with Cdt1 earlier than the expected elution point of these proteins individually (Fig. 4, D and E). On the other hand, we noticed that there was considerably reduced co-fractionation of the Ska1 complex with the phosphomimetic Cdt1-2E3D (Fig. 4, D and E). Similar results were also observed when an in vitro Cdk1-phosphorylated version of Cdt1-WT (Cdt1-phospho) was used in this co-fractionation assay (Fig. 4 F, top two blots). Since Cdk1-phosphorylation of the SKA3 subunit of the Ska1 complex has been shown to be important for its association with the Ndc80 complex (Huis In 't Veld et al., 2019; Zhang et al., 2020), we also tested a Cdk1-phosphorylated version of the Ska1 complex (Ska1-phospho) in this assay. We did not observe appreciable difference in Cdt1-WT cofractionation with Ska1-phospho (Fig. 4 F, bottom two blots) as compared to the unphosphorylated Ska1 complex (Fig. 4 E). Further, binding between the SKA1 subunit and the Cdt1-2E3D protein was observed to be diminished as compared to the Cdt1-WT in a BLI experiment. The KD was reduced ∼2.5-fold for the mutant protein (70 nM for Cdt1-WT vs. 185 nM for the Cdt1-2E3D; Fig. 3 F; and Fig. 4, G and H). Finally, we carried out immunoprecipitation experiments to test whether the phosphorylation of Cdt1 by Cdk1 influenced Cdt1’s ability to bind to the Ndc80 complex, which showed that both Cdt1-WT and Cdt1-2E3D were able to immunoprecipitate the Ndc80 complex to comparably similar extents (Fig. 4 I). Taken together, our results suggest that the binding of Cdt1 and the Ska1 complex is regulated by Cdk1-mediated phosphorylation of Cdt1.
Expression of the Cdk1 phosphomutant of Cdt1 deficient in Ska1-binding exhibits defective k-MT attachments and erroneous mitotic progression
We then sought to determine whether the phosphorylation of Cdt1 by Cdk1 was physiologically relevant for mitosis. For this purpose, we utilized the stable HeLa cell lines expressing si-RNA-resistant WT, non-phosphorylable (5A), and phosphomimetic (2E3D) mutant versions of Cdt1 that we had used in our immunoprecipitation experiments of Cdt1 with the Ndc80 and Ska1 complexes. As mentioned previously, we had derived an effective protocol to perform rescue experiments in double-thymidine synchronized HeLa cell lines expressing siRNA-resistant versions of WT and mutant Cdt1, after depleting endogenous Cdt1 by siRNA-mediated knockdown (Agarwal et al., 2018; Fig. S2, A and C). The protocol exploits the fact that Cdt1 is degraded at the beginning of S phase and is freshly re-synthesized at G2/M transition. When these double thymidine-arrested stable cells expressing siRNA-resistant Cdt1 is released from thymidine into the presence of Cdt1 siRNA, these cells enter mitosis 9 h after the release from thymidine with the newly synthesized, siRNA-susceptible, endogenous Cdt1 being degraded immediately as it is being produced. We can now assess the mitotic functions of the expressed Cdt1 constructs.
The standard assay for Cdt1 mitotic function is the cold-stability assay as described previously (Fig. 1). Metaphase spindles of cells rescued by the WT or the non-phosphorylatable, Cdt1-5A was resistant to cold treatment as expected. On the other hand, spindles from the cells depleted of endogenous Cdt1 (vector) or rescued with the phosphomimetic Cdt1-2E3D were highly susceptible to cold treatment (Fig. 5 A). We then monitored mitotic progression in these cells. Normally, synchronized HeLa cells released from double thymidine arrest in S-phase will reach early mitotic prometaphase/metaphase 9 h after release from thymidine, and after 10 h, most of the mitotic cells will be in late mitotic anaphase/telophase. We observed that mitotic cells rescued by WT or the non-phosphorylable Cdt1-5A mutant entered anaphase within the expected time, but the cells depleted of endogenous Cdt1 or those rescued with the phosphomimetic Cdt1-2E3D remained in the early phases (prometaphase and metaphase) of mitosis even at the 10 h time point, suggesting that there was a considerable delay in normal mitotic progression (Fig. 5, B and C). Further, we find that carrying out rescue experiments with the WT and mutant Cdt1-expressing synchronized cell lines did not interfere with interphase cellular functions such as DNA replication licensing (Fig. S4 D) or the organization of the microtubule cytoskeleton (Fig. S4 E). Together, our results suggest that the perturbed Ska1-Cdt1 interaction in the cells rescued with the Cdk1 phosphomimetic (2E3D) mutant of Cdt1 resulted in defective k-MT attachments and delayed mitotic progression.
The phosphorylation of Cdt1 by the Cdk1 kinase impedes stable k-MT attachments and normal mitotic progression. (A) Histone H2B GFP-expressing, double thymidine synchronized, control HeLa cells (top panel) or those stably expressing RNAi-resistant WT (middle panel) or 2E3D mutant version (bottom panel) of Cdt1 were depleted of endogenous Cdt1 and the stability of K-fibers was assessed in each case by treatment with cold buffer 9 h after release from second thymidine arrest, as indicated. The cells were immunostained using anti-tubulin (green) and anti-Zwint1 (kinetochores, red) with the chromosomes counterstained using DAPI. (B) Same as in G, but the cells were fixed at both 9 and 10 h after release from second thymidine treatment followed by immunostaining using anti-tubulin (red) antibody and Histone H2B shown in green. (C) Quantification of cells in various stages of mitosis from B. Data presented as mean ± SD.
The phosphorylation of Cdt1 by the Cdk1 kinase impedes stable k-MT attachments and normal mitotic progression. (A) Histone H2B GFP-expressing, double thymidine synchronized, control HeLa cells (top panel) or those stably expressing RNAi-resistant WT (middle panel) or 2E3D mutant version (bottom panel) of Cdt1 were depleted of endogenous Cdt1 and the stability of K-fibers was assessed in each case by treatment with cold buffer 9 h after release from second thymidine arrest, as indicated. The cells were immunostained using anti-tubulin (green) and anti-Zwint1 (kinetochores, red) with the chromosomes counterstained using DAPI. (B) Same as in G, but the cells were fixed at both 9 and 10 h after release from second thymidine treatment followed by immunostaining using anti-tubulin (red) antibody and Histone H2B shown in green. (C) Quantification of cells in various stages of mitosis from B. Data presented as mean ± SD.
The Ska1 complex and Cdt1 augments each other’s ability to bind to microtubules synergistically
Having determined that Ska1-binding is critical for Cdt1 targeting during mitosis and that this interaction is physiologically relevant, we next set out to reconstitute this interaction and test its relevance in vitro. In our recently published work, we have shown that Cdt1 can independently bind to microtubules with moderate affinity in vitro (Agarwal et al., 2018). Since results from our work so far indicate a critical interaction between Cdt1 and the Ska1 complex, we postulated that the affinity of Cdt1 to microtubules could potentially be enhanced by the addition of the Ska1 complex as we had observed in the HeLa cells over-expressing SKA1-GFP (Fig. 2 E). To test this hypothesis, we employed TIRF microscopy (TIR-FM; Fig. 6 A). From our Blot overlay and BLI experiments of Cdt1-Ska1 interaction, it was evident that the SKA1 subunit of the SKA1/2 dimer, that is competent for microtubule-binding, was also key to Cdt1-binding. We hence added sub-optimal amount of GFP-Cdt1 (GFP-Cdt192-546) either alone or in combination with increasing concentrations of purified unlabeled His-SKA1/2 dimer (Fig. S3 C) on to surface-immobilized Taxol-stabilized microtubules. While GFP-Cdt1 alone showed only scarce and intermittent binding events at 1 nM concentration (Fig. 6 B), it begins to start decorating microtubule lattice as the concentration of SKA1/2 was increased. At 100 nM of SKA1/2, GFP-Cdt1 completely decorated microtubules and beyond that we observed a saturation in Cdt1 microtubule-binding (Fig. 6, B and C). Interestingly, we also noticed a similar enhancement leading to the saturation of purified SKA1/2-GFP (Fig. S3 F) binding to microtubules with increasing concentration of untagged Cdt192-546. This observation was confirmed by the quantification of intensities of Cdt1 and GFP-SKA1/2 along microtubule lattices (Fig. 6 D). Similar results were obtained for synergistic microtubule-binding with Cdt1 when we used the hexameric whole Ska1 complex, consisting of all the three subunits: SKA1, SKA2, and SKA3 (Fig. 6, E and F).
Cdt1 exhibits synergy with the Ska1 and the Ndc80 complexes in binding to microtubules in vitro. (A) Schematics representation of the single-molecule binding assay. (B) Selected images showing the binding of 1 nM GFP-tagged Cdt192-546 (indicated as GFP-Cdt1 for all in vitro assays) on the Alexa 647-biotin-labeled microtubules either alone (top) or in the presence of 100 nM untagged SKA1/2 dimer (bottom). Scale bar, 5 μm. (C) Data showing synergic-binding experiment of 1 nM GFP-Cdt1 with varying concentrations of untagged SKA1/2 (10 nM-2 µM), data are mean ± SEM. (D) Same as C but enrichment of 1 nM GFP-SKA1/2 with varying concentrations (10 nM-2 µM) of untagged Cdt192-546 (indicated as untagged Cdt1 for all in vitro experiments) was plotted in this case. N ≥ 2 experiments, n ≥ 30 microtubules for each point, data are mean ± SEM. (E) Selected images showing synergic-binding experiment of 10 nM Ska1/2/Ska3-GFP complex with varying concentrations of untagged Cdt1 (0–1 µM). (F) Quantification Ska3-GFP intensity as a function of Cdt1 concentration n ≥ 33 microtubules for each point, data are mean ± SEM. (G) Data showing synergic binding experiment of 1 nM GFP-Cdt1 with varying concentrations of HEC1/NUF2-GFP (10 nM-2 µM), data are mean ± SEM. (H) Selected images showing the binding of HEC1/NUF2-GFP (1 nM) on the Alexa 647-biotin-labeled microtubules either alone (top) or in the presence of 100 nM untagged Cdt1 (bottom). Scale bar, 5 μm. (I) Analysis of data from H showing quantification of synergistic binding between HEC1/NUF2-GFP (1 nM) and varying concentrations untagged Cdt1. N ≥ 2 experiments, n ≥ 30 microtubules for each point, data are mean ± SEM.
Cdt1 exhibits synergy with the Ska1 and the Ndc80 complexes in binding to microtubules in vitro. (A) Schematics representation of the single-molecule binding assay. (B) Selected images showing the binding of 1 nM GFP-tagged Cdt192-546 (indicated as GFP-Cdt1 for all in vitro assays) on the Alexa 647-biotin-labeled microtubules either alone (top) or in the presence of 100 nM untagged SKA1/2 dimer (bottom). Scale bar, 5 μm. (C) Data showing synergic-binding experiment of 1 nM GFP-Cdt1 with varying concentrations of untagged SKA1/2 (10 nM-2 µM), data are mean ± SEM. (D) Same as C but enrichment of 1 nM GFP-SKA1/2 with varying concentrations (10 nM-2 µM) of untagged Cdt192-546 (indicated as untagged Cdt1 for all in vitro experiments) was plotted in this case. N ≥ 2 experiments, n ≥ 30 microtubules for each point, data are mean ± SEM. (E) Selected images showing synergic-binding experiment of 10 nM Ska1/2/Ska3-GFP complex with varying concentrations of untagged Cdt1 (0–1 µM). (F) Quantification Ska3-GFP intensity as a function of Cdt1 concentration n ≥ 33 microtubules for each point, data are mean ± SEM. (G) Data showing synergic binding experiment of 1 nM GFP-Cdt1 with varying concentrations of HEC1/NUF2-GFP (10 nM-2 µM), data are mean ± SEM. (H) Selected images showing the binding of HEC1/NUF2-GFP (1 nM) on the Alexa 647-biotin-labeled microtubules either alone (top) or in the presence of 100 nM untagged Cdt1 (bottom). Scale bar, 5 μm. (I) Analysis of data from H showing quantification of synergistic binding between HEC1/NUF2-GFP (1 nM) and varying concentrations untagged Cdt1. N ≥ 2 experiments, n ≥ 30 microtubules for each point, data are mean ± SEM.
Our previous work suggests a direct interaction between Cdt1 and the Ndc80 complex (Agarwal et al., 2018; Varma et al., 2012). Having observed the synergistic behavior between Cdt1 and the Ska1 complex for microtubule-binding, we next sought to test whether we could discern a similar synergy of Cdt1 with Ndc80 for microtubule binding, employing the same TIR-FM approach. Previous work has shown that the HEC1/NDC80 subunit of the Ndc80 complex is sufficient for both microtubule- and Cdt1-binding. We hence added sub-optimal amounts of GFP-Cdt1 either alone or in combination with increasing concentrations of purified unlabeled HEC1/NUF2 dimer to microtubules. While GFP-Cdt1 at 1 nM concentration showed limited or no microtubule-binding events, we observed that it started decorating the microtubules at 100 nM HEC1/NUF2 concentration and attained saturation at even higher concentrations (Fig. 6 G). More importantly, we observed a similar enhancement leading to the saturation of HEC1/NUF2-GFP binding on microtubules with increasing concentrations of untagged Cdt192-546 (Fig. 6, H and I). These results suggest that Cdt1 exhibits synergy not only with the Ska1 complex for microtubule-binding but also with the Ndc80 complex.
A fraction of Ndc80, Ska1, and Cdt1 forms a tripartite complex
The two-way synergistic binding of Cdt1 with both the Ndc80 and Ska1 complexes prompted us to hypothesize that Cdt1 can possibly form a tripartite complex with Ndc80 and the Ska1 complexes once it is recruited to the kinetochores or microtubules and may act as a co-factor to enhance the association between these two complexes. Consistent with this possibility, we observed the both the Ndc80 and Ska1 complexes were efficiently immunoprecipitated from lysates of HeLa cells stably expressing HA-tagged WT-Cdt1 (Fig. 7 A). To analyze this further, we again carried out cofractionation experiments using analytical size-exclusion chromatography. In one case, we ran the WT Cdt1 protein, in vitro Cdk1-phosphorylated Ska1 complex (Ska1-phospho) or the Ndc80 complex separately, while in the other case, we mixed the three components in equimolar ratio for 1 h, before running all the samples on the column. Cdt1-WT and the two other complexes eluted at their normal expected molecular weights when ran individually (Fig. 7 B, top three blots as indicated). However, when Cdt1-WT was mixed with Ska1-phospho and the Ndc80 complexes, we observed a considerable forward shift in the elution profiles of all three components, indicating that they were cofractionating when mixed together (Fig. 7, B and C).
A fraction of Cdt1, Ska1, and Ndc80 form a tripartite complex that exhibit synergy in binding to microtubules in vitro. (A) HeLa cells stably expressing HA-tagged Cdt1-WT were immunoprecipitated (IP) using an anti-HA antibody and the Western blot was probed with the antibodies indicated on the right. (B) Elution profiles of indicated proteins/complexes or mixture of proteins/complexes from the co-fractionation experiments using Superose 6 gel filtration chromatography. Zoomed version from 1.5 to 2.8 ml is shown inset. (C) Western blots of fractions from B using primary antibodies as indicated on the right, for the proteins run separately or in combination as indicated on the left. The initial three elution fractions (19, 20, and 21) from a mixture of Ndc80+ Cdt1-WT + pSka1 complex have been marked with a red box to highlight the observed difference with those of the individually run proteins/complexes. (D) Selected images showing binding at single-molecule level of HEC1/NUF2-GFP, and SKA1/2Cy3 on taxol stabilized Alexa 647-biotin-labeled microtubules both in the absence (left) and in the presence (right) of 1 nM untagged Cdt1. Colocalized bindings are indicated with white arrowheads. (E) Scatter plot of the fraction of colocalized binding sites at three different concentrations of untagged Cdt1. Bar and whiskers are mean ± SEM. *** P < 0.0001, ** P < 0.001 (Mann–Whitney U test). (F) Selected images showing binding at single molecule level of GFP-Cdt1, SKA1/2Cy3, and Ndc80alexa647 proteins on taxol-stabilized biotin-labeled microtubules (not imaged). Colocalized bindings are indicated with white arrowheads. (G1and G2) Two examples showing the binding of single molecules GFP-Cdt1 (i), SKA1/2/3Cy3 (ii), and Ndc80alexa647 (iii) proteins to Taxol-stabilized biotin-labeled microtubules. Colocalized bindings are indicated with white arrowheads. Source data are available for this figure: SourceData F7.
A fraction of Cdt1, Ska1, and Ndc80 form a tripartite complex that exhibit synergy in binding to microtubules in vitro. (A) HeLa cells stably expressing HA-tagged Cdt1-WT were immunoprecipitated (IP) using an anti-HA antibody and the Western blot was probed with the antibodies indicated on the right. (B) Elution profiles of indicated proteins/complexes or mixture of proteins/complexes from the co-fractionation experiments using Superose 6 gel filtration chromatography. Zoomed version from 1.5 to 2.8 ml is shown inset. (C) Western blots of fractions from B using primary antibodies as indicated on the right, for the proteins run separately or in combination as indicated on the left. The initial three elution fractions (19, 20, and 21) from a mixture of Ndc80+ Cdt1-WT + pSka1 complex have been marked with a red box to highlight the observed difference with those of the individually run proteins/complexes. (D) Selected images showing binding at single-molecule level of HEC1/NUF2-GFP, and SKA1/2Cy3 on taxol stabilized Alexa 647-biotin-labeled microtubules both in the absence (left) and in the presence (right) of 1 nM untagged Cdt1. Colocalized bindings are indicated with white arrowheads. (E) Scatter plot of the fraction of colocalized binding sites at three different concentrations of untagged Cdt1. Bar and whiskers are mean ± SEM. *** P < 0.0001, ** P < 0.001 (Mann–Whitney U test). (F) Selected images showing binding at single molecule level of GFP-Cdt1, SKA1/2Cy3, and Ndc80alexa647 proteins on taxol-stabilized biotin-labeled microtubules (not imaged). Colocalized bindings are indicated with white arrowheads. (G1and G2) Two examples showing the binding of single molecules GFP-Cdt1 (i), SKA1/2/3Cy3 (ii), and Ndc80alexa647 (iii) proteins to Taxol-stabilized biotin-labeled microtubules. Colocalized bindings are indicated with white arrowheads. Source data are available for this figure: SourceData F7.
To conclusively demonstrate that a fraction of Cdt1, Ska1, and the Ndc80 complexes forms a tripartite complex, we resorted to single-molecule TIR-FM using sub-nanomolar concentrations of fluorescently tagged SKA1/2Cy3, HEC1/NUF2-GFP dimer, and untagged Cdt192-456 with surface-immobilized Taxol-stabilized (Alexa 647-biotin-labeled) microtubules. Indeed, we observed co-localized spots of GFP/Cy3 representing direct binding between HEC1/NUF2-GFP and SKA1/2Cy3 on the microtubules (Fig. 7 D, left panel). Interestingly, the fraction of GFP/Cy3 colocalized spots doubled in the presence of Cdt1 at concentrations as low as 1 nM (Fig. 7 D, right panel). The fraction of colocalization further increased upon increasing the concentration of Cdt1 up to 10 nM (Fig. 7 E) beyond which the microtubule lattice was completely decorated with both SKA1/2Cy3 and HEC1/NUF2-GFP and individual spots were not discernible anymore. Furthermore, we also observed a ∼30% co-localization of all the three components that were fluorescently tagged (GFP-Cdt1, SKA1/2Cy3, and full length Ndc80alexa647) after mixing them at sub-nanomolar concentrations (Fig. 7 F). Similar results were obtained when we used the SKA1/2/3 Cy3 complex with GFP-Cdt1 and Ndc80alexa647 in the same assay (Fig. 7, G1 and G2).
The tripartite Ndc80-Cdt1-Ska1 super-complex tracks the ends of dynamic microtubules
While we have made substantial progress in obtaining mechanistic details about how robust k-MT attachments are formed and maintained, we still have only very limited understanding of how kinetochores are stably coupled to dynamic microtubule plus-ends during chromosome alignment and segregation. Among the three protein components in context, Ska1 has been shown to be a weak tracker of dynamic microtubules, and although single Ndc80 molecules are not known to be a dynamic end tracker in metazoan systems, a combination of Ska1 and Ndc80 has previously been shown to be capable of tracking depolymerizing microtubule ends in vitro (Schmidt et al., 2012). Therefore, we investigated whether Cdt1 can track dynamic microtubule ends either alone or in combination with Ndc80 and/or the Ska1 complex. For these experiments, we switched to polymerizable form of GMPCPP stabilized microtubule seeds (Chakraborty et al., 2018), which can exhibit microtubule dynamics in the presence of soluble tubulin and GTP (Fig. 8 A). We also used Ska1 in both its dimeric SKA1/2 and hexameric SKA1/SKA2/SKA3 form in these assays. As expected, we observed normal binding of GFP-Cdt1 (1–100 nM) on the microtubule lattice of surface immobilized GMPCPP seeds (Fig. 8 B). However, in the presence of soluble tubulin and GTP, we did not observe any discernable tip tracking activity of GFP-Cdt1 at any of the concentrations tested. This suggests that Cdt1, by itself, might not function as a bona fide microtubule tip tracker in vitro, and possibly needs the Ska1 complex for this activity as our studies so far indicate. It is interesting, however, to note that we had previously found HA-tagged Cdt1 expressed in cells to be competent to localize to spindle microtubule plus-ends in metaphase (Agarwal et al., 2018). Consistently, when GFP-Cdt1 (1 nM) was supplemented with 1 nM untagged dimeric SKA1/2, weak tip tracking complexes were formed (4% of the total number of microtubule seeds present in the field; Fig. 8 B, boxes and arrowheads, Fig. 8 C; Video 4). These complexes appeared to form in the solution and occasionally land near the ends of dynamic microtubules. A quarter of these complexes weakly track either the depolymerizing (20%) or polymerizing (5%) phase of dynamic microtubule ends briefly. About 1% of these complexes were found to follow both the polymerizing and the depolymerizing microtubule tips. Rest of these complexes were found to diffuse near the tips (Fig. 8, B and C; and Fig. 9 A). Similar results were obtained when we used untagged Cdt1 with the hexameric Ska1 complex labeled with GFP-SKA3 (Fig. 9 B and Video 5).
The Ndc80-Cdt1-Ska1 tripartite complex exhibits potent tracking of polymerizing and depolymerizing microtubule plus ends. (A) Schematic representation of a single-molecule dynamic microtubule-binding assay. Arrow shows the direction of microtubule dynamics. (B) Representative time lapse images recorded with surface immobilized microtubules after the addition of a mixture of GFP-Cdt1 (1 nM), untagged SKA1/2 (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM), a complex of GFP-Cdt1 and SKA1/2 which lands near the plus-ends is indicated using arrowheads within the boxed areas. Time in s, and the scale bar is 5 µm. (C) A kymograph of the tip-tracking complex in B. (D) Same as in B but in the presence of a mixture of whole Ndc80 complex and Cdt1 (both untagged) with GFP-tagged hexameric SKA1/2/3 complex (1 nM each). Arrowheads indicate the dynamic plus-ends. (E) A kymograph of the tip tracking complex in D. (F) Same as in B but in the presence of a mixture of whole Ndc80 complex and dimeric SKA1/2 (both untagged) with GFP-tagged Cdt1 (1 nM each). Arrowheads indicate the dynamic plus-ends. (G) A kymograph of the tip tracking complex in F. (H) Kaplan-Meier survival curves for the total residence time (s) of tip-tracking complexes formed in the presence of indicated conditions. Calculated from n > 30 events and N > 3 independent trials in each case. (I) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of GFP-Cdt1, SKA1/2, Ndc80alexa 647 (1 nM each), and soluble tubulin (1 mg/ml), supplemented with Mg-GTP (1 mM). Arrow indicating the direction of movement of the tip-tracking complex. Time is in s, and the scale bar is 5 µm. (J) Kymographs of the tip-tracking complex in GFP-Cdt1 channel and in the Ndc80alexa 647 channel. Arrow indicating regrowth event.
The Ndc80-Cdt1-Ska1 tripartite complex exhibits potent tracking of polymerizing and depolymerizing microtubule plus ends. (A) Schematic representation of a single-molecule dynamic microtubule-binding assay. Arrow shows the direction of microtubule dynamics. (B) Representative time lapse images recorded with surface immobilized microtubules after the addition of a mixture of GFP-Cdt1 (1 nM), untagged SKA1/2 (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM), a complex of GFP-Cdt1 and SKA1/2 which lands near the plus-ends is indicated using arrowheads within the boxed areas. Time in s, and the scale bar is 5 µm. (C) A kymograph of the tip-tracking complex in B. (D) Same as in B but in the presence of a mixture of whole Ndc80 complex and Cdt1 (both untagged) with GFP-tagged hexameric SKA1/2/3 complex (1 nM each). Arrowheads indicate the dynamic plus-ends. (E) A kymograph of the tip tracking complex in D. (F) Same as in B but in the presence of a mixture of whole Ndc80 complex and dimeric SKA1/2 (both untagged) with GFP-tagged Cdt1 (1 nM each). Arrowheads indicate the dynamic plus-ends. (G) A kymograph of the tip tracking complex in F. (H) Kaplan-Meier survival curves for the total residence time (s) of tip-tracking complexes formed in the presence of indicated conditions. Calculated from n > 30 events and N > 3 independent trials in each case. (I) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of GFP-Cdt1, SKA1/2, Ndc80alexa 647 (1 nM each), and soluble tubulin (1 mg/ml), supplemented with Mg-GTP (1 mM). Arrow indicating the direction of movement of the tip-tracking complex. Time is in s, and the scale bar is 5 µm. (J) Kymographs of the tip-tracking complex in GFP-Cdt1 channel and in the Ndc80alexa 647 channel. Arrow indicating regrowth event.
GFP-Cdt1 diffusively tracks the end of dynamic microtubule in the presence of Ska1. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 visualized with 488-nm excitation laser in TIRF. A complex of GFP-Cdt1 and Ska1 (not imaged) lands near the growing end of microtubule1 at about 1 s and starts diffusively tracking the dynamic end till 12 s, then it detaches and diffuses into the solution out of field of view. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
GFP-Cdt1 diffusively tracks the end of dynamic microtubule in the presence of Ska1. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 visualized with 488-nm excitation laser in TIRF. A complex of GFP-Cdt1 and Ska1 (not imaged) lands near the growing end of microtubule1 at about 1 s and starts diffusively tracking the dynamic end till 12 s, then it detaches and diffuses into the solution out of field of view. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
Kymographs for dynamic microtubule plus-ends tracking by Ska1, Cdt1, and Ndc80 when present as indicated combinations. (A–D) Additional example kymographs for dynamic microtubule end tracking by one fluorescently tagged component when mixed with the other component(s) as indicated on top of each panel, bar 1 min, 5 μm. (E) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of the untagged whole Ndc80 complex (1 nM), GFP-tagged SKA1/2/3 hexameric complex (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM). Time is in s, and bar is 5 µm. To the right is a kymograph of the dynamic plus-ends tracking by the indicated component. (F) Additional example kymographs for dynamic microtubule end tracking by the indicated proteins on top of each panel; bars, 1 min, 5 μm. (G) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of HEC1/NUF2-GFP, untagged SKA1/2 (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM). Time is in s, and bar is 5 µm. To the right is a kympgraph of the dynamic plus-ends tracking by the indicated component. (H) More example kymographs for dynamic microtubule end tracking by GFP-Cdt1, untagged SKA1/2, and Ndc80alexa647. Panels showing kymographs for the same end-tracking events is GFP-Cdt1 channel, and in Ndc80alexa647 channel.
Kymographs for dynamic microtubule plus-ends tracking by Ska1, Cdt1, and Ndc80 when present as indicated combinations. (A–D) Additional example kymographs for dynamic microtubule end tracking by one fluorescently tagged component when mixed with the other component(s) as indicated on top of each panel, bar 1 min, 5 μm. (E) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of the untagged whole Ndc80 complex (1 nM), GFP-tagged SKA1/2/3 hexameric complex (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM). Time is in s, and bar is 5 µm. To the right is a kymograph of the dynamic plus-ends tracking by the indicated component. (F) Additional example kymographs for dynamic microtubule end tracking by the indicated proteins on top of each panel; bars, 1 min, 5 μm. (G) Representative time lapse images recorded with surface immobilized microtubules after addition of a mixture of HEC1/NUF2-GFP, untagged SKA1/2 (1 nM), soluble tubulin (1 mg/ml), and Mg-GTP (1 mM). Time is in s, and bar is 5 µm. To the right is a kympgraph of the dynamic plus-ends tracking by the indicated component. (H) More example kymographs for dynamic microtubule end tracking by GFP-Cdt1, untagged SKA1/2, and Ndc80alexa647. Panels showing kymographs for the same end-tracking events is GFP-Cdt1 channel, and in Ndc80alexa647 channel.
Cdt1 diffusively tracks the end of dynamic microtubule in the presence of SKA1/2/3-GFP complex. Video shows GMPCPP stabilized microtubules decorated with SKA1/2+ GFP-SKA3 visualized with 488-nm excitation laser in TIRF. A complex of Cdt1 (not imaged) and SKA1/2+ GFP-SKA3 diffusively tracks the dynamic end. Movie is played at 30 times faster rate than real. Scale bar, 5 µm.
Cdt1 diffusively tracks the end of dynamic microtubule in the presence of SKA1/2/3-GFP complex. Video shows GMPCPP stabilized microtubules decorated with SKA1/2+ GFP-SKA3 visualized with 488-nm excitation laser in TIRF. A complex of Cdt1 (not imaged) and SKA1/2+ GFP-SKA3 diffusively tracks the dynamic end. Movie is played at 30 times faster rate than real. Scale bar, 5 µm.
Surprisingly, when we mixed the GFP-tagged hexameric Ska1 complex together with untagged Ndc80 and Cdt1 (both at 1 nM) in this dynamic assay, we found similar landing of complexes on microtubule seeds (∼5% of the total number of microtubule seeds present in the field), but strikingly, these complexes demonstrated far better tracking (Fig. 8, D and E; and Video 6) of both polymerizing and depolymerizing microtubule ends, and for multiple polymerization/depolymerization cycles as evident form the kymographs (Fig. 8 E and Fig. 9 C). More than half (57%) of these complexes were found to be tracking either the polymerizing (21%), depolymerizing (21%), or both (15%) phases of microtubule dynamics. Most of these tip tracking complexes detached from the microtubule lattice after tracking the tip or diffusing near the tip, but some of them outlived our entire observation time of 30 min. Similar results were obtained when untagged dimeric SKA1/2 was used (instead of the GFP-tagged Ska1 complex) along with untagged Ndc80 complex and the dynamic microtubule plus-ends were marked by GFP-Cdt1 (Fig. 8, F and G; Fig. 9 D, and Video 7). To compare the residence time of these complexes and the differences between the bi/tripartite complexes, we computed survival curves, which showed that 50% of the tripartite (Ndc80-Cdt1-Ska1) complex survived for more than 8 min, whereas only 20% of the bipartite (Cdt1-Ska1) complexes survived longer than 8 min (Fig. 8 H).
SKA1/2/3-GFP mixed with untagged Cdt1 and Ndc80 show robust plus-ends tracking of dynamic microtubules. Video shows GMPCPP stabilized microtubules decorated with SKA1/2/3-GFP visualized with 488-nm excitation laser in TIRF. A complex of Cdt1, SKA1/2/3-GFP, and Ndc80 (not imaged) already bound to the grown end of a microtubule at the beginning of the movie and starts to robustly track the dynamic microtubule plus-ends for both polymerizing and depolymerizing phases and for multiple cycles. Movie is played at 20 times faster rate than real. Scale bar, 5 µm.
SKA1/2/3-GFP mixed with untagged Cdt1 and Ndc80 show robust plus-ends tracking of dynamic microtubules. Video shows GMPCPP stabilized microtubules decorated with SKA1/2/3-GFP visualized with 488-nm excitation laser in TIRF. A complex of Cdt1, SKA1/2/3-GFP, and Ndc80 (not imaged) already bound to the grown end of a microtubule at the beginning of the movie and starts to robustly track the dynamic microtubule plus-ends for both polymerizing and depolymerizing phases and for multiple cycles. Movie is played at 20 times faster rate than real. Scale bar, 5 µm.
GFP-Cdt1 robustly tracks the end of dynamic microtubule in the presence of untagged Ndc80 and SKA1/2. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 visualized with 488-nm excitation laser in TIRF. A complex of GFP-Cdt1, Ska1, and Ndc80 (not imaged) already bound to the grown end of a microtubule at the beginning of the movie and starts to robustly track the depolymerizing phase of the dynamic end at 1 s, then it remains bound on the microtubule lattice for the entire duration of the movie. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
GFP-Cdt1 robustly tracks the end of dynamic microtubule in the presence of untagged Ndc80 and SKA1/2. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 visualized with 488-nm excitation laser in TIRF. A complex of GFP-Cdt1, Ska1, and Ndc80 (not imaged) already bound to the grown end of a microtubule at the beginning of the movie and starts to robustly track the depolymerizing phase of the dynamic end at 1 s, then it remains bound on the microtubule lattice for the entire duration of the movie. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
Ndc80 has been previously shown to track the ends of dynamic microtubules together with Ska1 (Huis In 't Veld et al., 2019; Schmidt et al., 2012), and in our hand, we also observed the formation of weak tip-associated complexes with untagged Ndc80 complex and the Ska1 complex, labeled with GFP-SKA3 (Fig. 9 E, arrows and kymograph, Fig. 9 F; Video 8), or with Hec1/Nuf2-GFP and untagged SKA1/2 (Fig. 9 G arrows; Video 9) Thus, we predict that the enhanced tip tracking as observed with the tripartite complex could be ascribed to an increase of Ndc80-Ska1 binding mediated by Cdt1 and the consequent enhancement of end conversion of the tripartite complex. Using fluorescently labeled Ndc80alexa647, we were able to show that this tip-tracking tripartite complex (Video 10) actually contains both GFP-Cdt1 and the Ndc80 complexes (Fig. 8, I and J; and Fig. 9 H). Furthermore, in many instances, we have also observed tip tracking phenomenon lasting for more than one cycle of polymerization and depolymerization (Fig. 8, D and E, arrows; Fig. 8, I and J, arrows; Fig. 9 C).
SKA1/2/3-GFP mixed with untagged Ndc80 diffusively tracks the polymerizing phase of dynamic microtubules. Video shows GMPCPP stabilized microtubules decorated with SKA1/2/3-GFP visualized with 488-nm excitation laser in TIRF. A complex of SKA1/2/3-GFP and Ndc80 (not imaged) already bound to the growing end of a microtubule at the beginning of the movie and starts to diffusively tracks the polymerizing plus-ends before being bleached. Movie is played at 20 times faster rate than real. Scale bar, 2 µm.
SKA1/2/3-GFP mixed with untagged Ndc80 diffusively tracks the polymerizing phase of dynamic microtubules. Video shows GMPCPP stabilized microtubules decorated with SKA1/2/3-GFP visualized with 488-nm excitation laser in TIRF. A complex of SKA1/2/3-GFP and Ndc80 (not imaged) already bound to the growing end of a microtubule at the beginning of the movie and starts to diffusively tracks the polymerizing plus-ends before being bleached. Movie is played at 20 times faster rate than real. Scale bar, 2 µm.
Hec1/Nuf2-GFP diffusively tracks the end of dynamic microtubule in presence of untagged SKA1/2. Video shows GMPCPP stabilized microtubules decorated with Hec1/Nuf2-GFP visualized with 488-nm excitation laser in TIRF. A complex of Hec1/Nuf2-GFP and SKA1/2 (not imaged) lands near the growing end of microtubule and diffusively tracks the dynamic ends for 1–2 s then loses tracking ability for the rest of the movie. Movie is played at 20 times faster rate than real. Scale bar, 5 µm.
Hec1/Nuf2-GFP diffusively tracks the end of dynamic microtubule in presence of untagged SKA1/2. Video shows GMPCPP stabilized microtubules decorated with Hec1/Nuf2-GFP visualized with 488-nm excitation laser in TIRF. A complex of Hec1/Nuf2-GFP and SKA1/2 (not imaged) lands near the growing end of microtubule and diffusively tracks the dynamic ends for 1–2 s then loses tracking ability for the rest of the movie. Movie is played at 20 times faster rate than real. Scale bar, 5 µm.
GFP-Cdt1/Ndc80alexa647robustly tracks the end of dynamic microtubule in the presence of SKA1/2. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 and Ndc80alexa647 visualized with 488/640-nm excitation lasers in TIRF. A complex of GFP-Cdt1, Ndc80alexa647, and Ska1 (not imaged) is seen to track the dynamic end of microtubules for couple of cycles of polymerization and depolymerization. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
GFP-Cdt1/Ndc80alexa647robustly tracks the end of dynamic microtubule in the presence of SKA1/2. Video shows GMPCPP stabilized microtubules decorated with GFP-Cdt1 and Ndc80alexa647 visualized with 488/640-nm excitation lasers in TIRF. A complex of GFP-Cdt1, Ndc80alexa647, and Ska1 (not imaged) is seen to track the dynamic end of microtubules for couple of cycles of polymerization and depolymerization. Movie is played at 30 times faster rate than real. Scale bar, 3 µm.
Finally, we observed that the residence time of single-molecule Ndc80 complexes reduced ∼ twofold in the presence of the other two proteins (Ska1 and Cdt1; Fig. 10, A and B). We also note that the Ndc80 complex becomes more diffusive on microtubules (Fig. 10, A and C) when in combination with Cdt1 and Ska1 as compared to when bound to microtubules by itself (Campbell et al., 2019). Similar differences with regard to the residence time and diffusion were also observed for Cdt1 alone vs. as a part of the tripartite complex. (Fig. 10, D–F).
Processive end-tracking activity to the tripartite Ndc80-Cdt1-Ska1 complex is characterized by enhanced diffusive behavior of the complex. (A) Single-molecule diffusion kymograph for Ndc80alexa647 alone (top), and Ndc80alexa647 in the presence of GFP-Cdt1 and SKA1/2 (bottom). (B) Histogram of residence times for Ndc80alexa647 alone (left) and Ndc80alexa647 in the presence of GFP-Cdt1 and untagged-SKA1/2 (right). Optimized decay constants from the single exponential fits to the data (in red) represent the respective residence times (τres) for each condition; data from at least 189 individual Ndc80alexa647 tracks and N ≥ 2 independent trials. (C) Mean Squared Displacement (MSD) vs. time for Ndc80alexa647 alone (black plot) and Ndc80alexa647 in the presence of GFP-Cdt1 and untagged-SKA1/2 (red plot); tracks from at least 22 individual Ndc80alexa647 complexes were averaged; symbols are experimental; data lines are linear fits to the data up to 0.22 s, the slope of which is the diffusion coefficient; error bars are SEM. (D) Same as A but in this case for GFP-Cdt1; tracks from at least 250 individual GFP-Cdt1 complexes were averaged. (E) Same as D but in this case for GFP-Cdt1. (F) Same as C but in this case for GFP-Cdt1; data from at least 125 individual GFP-Cdt1 tracks and N ≥ 2 independent trials. (G) Mutual interactions between the Ndc80 complex, Cdt1, and the Ska1 complex to form a super-complex and the binding of the resulting super-complex furnishes the load bearing attachment at kinetochore. (H) Schematics (not to scale) show multi-molecular lawn of the Ndc80 complex molecules interacting with a kinetochore microtubule bundle. Propensity of Cdt1 (orange) and Ska1 (purple) proteins toward binding to curved microtubule protofilaments in conjunction with the intermolecular binding between Ndc80-Cdt1-Ska1 enables the formation of standalone microtubule tip-tracking units. Green microtubules segments showing nascent GTP-tubulin cap. (I) Zoomed in section of k-microtubule attachment shown the Ndc80-Cdt1-Ska1 trio contacting the microtubules at two point of attachments one) on the microtubule lattice primarily by the CH-domains of Ndc80 proteins and second) Ska1-Cdt1-mediated interaction with the curved protofilaments flanked by Ndc80.
Processive end-tracking activity to the tripartite Ndc80-Cdt1-Ska1 complex is characterized by enhanced diffusive behavior of the complex. (A) Single-molecule diffusion kymograph for Ndc80alexa647 alone (top), and Ndc80alexa647 in the presence of GFP-Cdt1 and SKA1/2 (bottom). (B) Histogram of residence times for Ndc80alexa647 alone (left) and Ndc80alexa647 in the presence of GFP-Cdt1 and untagged-SKA1/2 (right). Optimized decay constants from the single exponential fits to the data (in red) represent the respective residence times (τres) for each condition; data from at least 189 individual Ndc80alexa647 tracks and N ≥ 2 independent trials. (C) Mean Squared Displacement (MSD) vs. time for Ndc80alexa647 alone (black plot) and Ndc80alexa647 in the presence of GFP-Cdt1 and untagged-SKA1/2 (red plot); tracks from at least 22 individual Ndc80alexa647 complexes were averaged; symbols are experimental; data lines are linear fits to the data up to 0.22 s, the slope of which is the diffusion coefficient; error bars are SEM. (D) Same as A but in this case for GFP-Cdt1; tracks from at least 250 individual GFP-Cdt1 complexes were averaged. (E) Same as D but in this case for GFP-Cdt1. (F) Same as C but in this case for GFP-Cdt1; data from at least 125 individual GFP-Cdt1 tracks and N ≥ 2 independent trials. (G) Mutual interactions between the Ndc80 complex, Cdt1, and the Ska1 complex to form a super-complex and the binding of the resulting super-complex furnishes the load bearing attachment at kinetochore. (H) Schematics (not to scale) show multi-molecular lawn of the Ndc80 complex molecules interacting with a kinetochore microtubule bundle. Propensity of Cdt1 (orange) and Ska1 (purple) proteins toward binding to curved microtubule protofilaments in conjunction with the intermolecular binding between Ndc80-Cdt1-Ska1 enables the formation of standalone microtubule tip-tracking units. Green microtubules segments showing nascent GTP-tubulin cap. (I) Zoomed in section of k-microtubule attachment shown the Ndc80-Cdt1-Ska1 trio contacting the microtubules at two point of attachments one) on the microtubule lattice primarily by the CH-domains of Ndc80 proteins and second) Ska1-Cdt1-mediated interaction with the curved protofilaments flanked by Ndc80.
Overall, our data present a novel finding that Cdt1-Ska1 interaction facilitates the recruitment of Cdt1 to dynamic microtubule plus-ends. Cdt1 recruitment, in turn, enables the formation of a tripartite Ndc80-Cdt1-Ska1 complex that functions as a bona fide, processive k-MT coupler and is critical for accurate chromosomal segregation.
Discussion
Faithful chromosome segregation is driven by cellular mechanisms that facilitate the stabilization of k-MT attachments in metaphase and anaphase. The core microtubule-attachment machinery comprising primarily of the Ndc80 complex is established to be critical for this function. However, recent work has revealed that several accessory microtubule-binding proteins are also required for this stabilization process and more importantly for the efficient coupling of the Ndc80 complex to the dynamic plus-ends of spindle microtubules (Monda and Cheeseman, 2018; Rahi et al., 2020). The Ska1 complex has been shown to be one such factor in several metazoan systems (Daum et al., 2009; Gaitanos et al., 2009; Raaijmakers et al., 2009; Welburn et al., 2009). Our previous work had demonstrated that the DNA replication licensing protein, Cdt1, is required for stabilizing k-MT attachments by facilitating robust connections between Ndc80 and spindle microtubules in humans (Agarwal et al., 2018; Varma and Salmon, 2012). In this work, we demonstrate a viable interaction between Cdt1 and the Ska1 complex and provide evidence supporting the possibility that Cdt1 is an essential component of the k-MT plus-end coupling machinery. Further, we show that Cdt1 enables k-MT coupling by synergizing with both the Ndc80 and Ska1 complexes to form a tripartite super-complex (Fig. 7 and Fig. 10 G) that emerges as a potent tracker of dynamic microtubule plus-ends.
Both the Ska1 complex and Cdt1 have been shown to directly bind to the Ndc80 complex. Remarkably, we find that Cdt1 and the Ska1 complexes also exhibit a direct interaction that in turn seems to be important to recruit Cdt1 to its mitotic targeting sites. The targeting of these proteins to kinetochores is hierarchical with both Ska1 as well as Cdt1 being dependent on Ndc80. Cdt1 in turn is also dependent on Ska1, while Ska1 does not require Cdt1 for its localization. This relationship between Ska1 and Cdt1 also is true for spindle microtubule-binding. However, interestingly, all these three factors can bind independently to Taxol-stabilized microtubules in vitro while they also exhibit true synergy for microtubule-binding when used in pairs. The middle and C-terminal regions of Cdt1 comprising the two winged-helix domains that are required for efficient Cdt1 microtubule-binding are also required for binding to Ska1 (Rahi et al., 2020). Further, the C-terminal winged-helix domain of Ska1 that is critical for its microtubule binding is found to be important for binding and recruiting Cdt1 to spindle microtubules. Our findings suggest that Cdt1 most likely interacts primarily with the SKA1 subunit of the Ska1 complex. Finally, all three factors, Ndc80, Cdt1, and Ska1, when used in limiting concentrations show considerable colocalization on Taxol-stabilized microtubules in vitro. Taken together, these observations point toward the existence of a tri-partite Ndc80-Cdt1-Ska1 complex in mitotic cells (Fig. 7).
The k-MT end coupling-load bearing complex was long modeled upon the hill-sleeve mechanism (Hill, 1985) in higher eukaryotes or the sliding ring model (Koshland et al., 1988) for yeast, with other factors, including the Dam1 ring complex, assisting Ndc80 in this process (Monda and Cheeseman, 2018; Rahi et al., 2020). But considering that a Dam1-like complex is evolutionarily missing in metazoans and that their k-MT bundles (k-fibers) are formed of 15–20 individual microtubules, direct experimental evidence in support of either of the models has not been established. Existing studies have not revealed a model where Ndc80 and Ska1 in concert can function via either of these models. Our studies so far do not address whether Cdt1 can contribute to the formation of such a ring/sleeve around k-fibers microtubule bundles. Additional experimental approaches including cryo-electron microscopy and force measurements using optical trapping reconstitution studies are required to shed light on whether similar mechanisms exist in metazoan systems. With the existing evidence, we pose an alternate view that higher eukaryotes utilize a distinct k-MT coupling mechanism in which the three components, Ndc80, Ska1, and Cdt1, comprise single functional units of a processive end-tracking complex (Fig. 10, H and I, not to scale). This standalone unit, likely consisting of a single copy of each of the components, can independently track the ends of dynamic microtubules. Our observations suggest that this complex tracks not only the depolymerizing but also the polymerizing part of the dynamic microtubules, and importantly that the tracking persisted for multiple cycles of polymerization/depolymerization (Fig. 8, D and E, arrows; Fig. 8, I and J, arrows; Fig. 9, C and D). The fact that the observed tracking events were relatively short-lived is likely a result of a key kinetochore component missing in our assays, including the kinesin CENP-E, which is thought to be important to maintain these end-on attachments (Chakraborty et al., 2019). Interestingly, while both Ska1 and Cdt1 can bind curved protofilaments (Schmidt et al., 2012; Agarwal et al., 2018), only Ska1 seems to be able to transiently track plus-ends by itself and is required to target Cdt1 and Ndc80 to this site in vitro. However, all three components comprising the standalone units are required for long-distance processive tracking and Cdt1 seems to serve a critical function in this process.
It is quite intriguing that the tripartite complex exhibits properties such as reduced residence times and enhanced diffusivity on microtubules as compared to its individual functional units. These alterations in the properties of Ndc80 as a part of the tripartite complex may be critical to impart microtubule plus-end tracking ability to the complex as has been predicted previously by our computational simulation experiments (Campbell et al., 2019). This observation is also supported by a concomitant decrease in the residence time and increase in the diffusivity of Cdt1 as a part of the complex. These results can be explained by the fact that there are possibly multiple Ndc80-Cdt1-Ska1 (15–20 per k-MT)-mediated attachment sites at the kinetochore. Since the dynamic plus-ends of k-fiber microtubules are constantly being modified by the addition or loss of tubulin dimers, the force bearing attachment sites (mediated by Ndc80-Cdt1-Ska1) also need to be frequently modulated (change of position of protein complexes either by high diffusion or by modulating unbinding/binding rates) to sustain the attachment between the kinetochore and microtubules. As a result, the tripartite complexes need to be highly diffusive and labile with smaller residence times. The reduced residence times may result in frequent detachments, but owing to the overall stochastic nature of these attachments and considering that multiple such attachment sites are present, majority of these tripartite couplers remain bound to the dynamic MT ends at any point (Fig. 10 H), ensuring that k-MT attachments are rarely broken in metaphase and anaphase.
Chromosome segregation is a very tightly controlled process, orchestrated by the interplay of multiple kinases/phosphatases. This is especially true for the regulation of k-MT attachments where multiple kinases (Aurora B, Cdk1, etc.) and phosphatases (PP1, PP2A, etc.) have been shown to regulate the interaction of factors with each other and/or with the microtubules at the k-MT interface (Moura and Conde, 2019; Saurin, 2018). The k-MT attachment strength also needs to be spatially and temporally regulated during prometaphase and metaphase. Our studies suggest that the replication licensing protein, Cdt1, is temporally regulated by Cdk1 to modulate the strength of k-MT attachments in one such phase-dependent control mechanism during mitosis. Cdk1 phosphorylation of Cdt1 is instrumental in regulating Ska1-Cdt1 interaction and for controlling the recruitment of Cdt1 to kinetochores and spindle microtubules. Previous work has demonstrated that Cdk1 activity is highest at G2/M transition and early mitosis but reduces progressively as the cells proceed to metaphase and anaphase (Gavet and Pines, 2010). It is also known that Cdk1 phosphorylation of the SKA3 subunit is important for Ndc80-binding and kinetochore recruitment of the Ska1 complex (Huis In 't Veld et al., 2019; Zhang et al., 2020). The results from our cofractionation experiments suggest that Cdk1 phosphorylation of SKA3 is, however, not critical for Cdt1-binding of the Ska1 complex. Our results support a model where Cdk1 phosphorylation of Cdt1 reduces Cdt1 recruitment to kinetochores (Campbell et al., 2019) and the tripartite complex formation with Ska1/Ndc80 at prometaphase, thus accounting for weaker k-MT attachments earlier during mitosis. At metaphase, the Cdk1 kinase activity reduces, allowing for efficient Cdt1 recruitment via its interaction with the Ska1. Cdt1 and Ska1, in turn, strengthen k-MT attachments in synchrony with Ndc80 via the Ndc80-Cdt1-Ska1 complex formation and enabling efficient plus-ends tracking ability to this tripartite complex. Thus, while our results indicate that Cdt1 phosphorylation by Cdk1 is important for negatively regulating Ska1-binding to enable the tripartite complex formation predominantly in metaphase, Ska1 phosphorylation by the same kinase is important for positively regulating Ndc80-binding to facilitate kinetochore recruitment of a fraction of the Ska1 complex earlier in mitosis (which could be important for a yet unidentified function). The remaining fraction of Ska1 is possibly recruited to the k-MT interface in metaphase via its association with microtubule plus-ends (Radhakrishnan et al., 2023; Thomas et al., 2016).
Intriguingly, it is known that Cdt1 is not degraded at metaphase-to-anaphase transition and that the Cdk1-phosphorylated Cdt1 is not competent for origin licensing in G1 (Chandrasekaran et al., 2011; Zhou et al., 2020). This suggests that Cdt1 dephosphorylation could be critical for this cell-cycle transition and thus future work is required to identify the phosphatase that contributes to this important function. Future research is also required to understand whether other molecules including chTOG, EB1, or Astrin-SKAP function as a part of this standalone alone k-MT coupling unit or if they function with the Ndc80 complex independently of this core unit. Also remaining open is the question of how this unit tracks a bundle of parallel microtubules (Chakraborty et al., 2020) in vitro as opposed to single microtubules.
Materials and methods
Cloning, recombinant protein expression, and purification and labeling
GFP-tagged Cdt1 (aa 92–546 referred through the text as GFP-Cdt1) and its subsequent deletion fragments (aa 92–232, aa 92–410 and aa 410–546) were purified as described previously (Agarwal et al., 2018). Plasmids encoding GST-tagged SKA3, GFP-His-SKA2, and His-tagged SKA1/2 for bacterial protein purification were kind gifts from Drs. Gary Gorbsky (Oklahoma Medical Research Foundation, Oklahoma City, OK, USA), A. Arockia Jeyaprakash (University of Edinburgh, Edinburgh, UK), and P. Todd Stukenberg (University of Virginia, Charlottesville, VA, USA) laboratories, respectively. His-tagged SKA1 N-terminus (aa 1–132) and C-terminus (aa 133–255) were cloned into pET28a vector using the primer pairs 5′-GCGCGGATCCATGGCCTCGTCAGATCTGGAAC-3′, 5′-GCGCAAGCTTTCATCTTTGCTCTTTGGGAGGCTTC-3′ and 5′-GCGCGGATCCATGAGTATTAAGGAAATGCCATTTATAACTTGTG-3′, 5′-GCGCAAGCTTTCATCTTTGCTCTTTGGGAGGCTTC-3′, respectively. The proteins were purified according to published protocols and aliquots were stored in −80°C. Plasmids encoding His-SKA1/2 and GFP-His-SKA2 proteins were transfected into BL21 E. coli cells. Transformants were grown in 2xYT media at 37°C until 1.0 Optical Density is reached. Protein expression was induced by adding 0.35 mM IPTG at 18°C overnight. Cells were pelleted using centrifugation at 4,412 × g. Pellets expressing His-SKA1/2 and GFP-His-SKA2 proteins were combined, and the cells were lysed by sonication on ice in lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl, 2 mM β-ME, supplemented with 1× protease inhibitors; #AA32965; Thermo Fisher Scientific). Lysate was then clarified by centrifuge at 58,387 × g, 4°C, for 30 min. Clarified supernatant was then incubated with Ni-NTA beads (Qiagen) at 4°C for 1 h on a roller. Flow through was discarded, and the beads were then washed with 30 column volume of high salt wash buffer (20 mM Tris pH 8.0, 1,000 mM NaCl, 50 mM KCl, 10 mM MgCl2, 2 mM β-ME). The protein complex was eluted with elution buffer (20 mM Tris pH 8.0, 100 mM NaCl, 400 mM Imidazole, 2 mM β-ME). Fractions were evaluated with SDS PAGE electrophoresis, and the ones containing both His-SKA1/2+GFP-His-SKA2 subunits were pooled and further purified using size exclusion chromatography over a Superose 6 Increase 10/300 GL (GE Healthcare) column equilibrated with gel filtration buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM β-ME, 2% v/v glycerol). Fractions were analyzed again using SDS-PAGE, and the ones containing both His-SKA1/2+GFP-His-SKA2 subunits were pooled and aliquoted. Aliquots were flash-frozen and stored at −80°C. Purified HEC1/NUF2-GFP dimer of the Ndc80 complex used for TIRF imaging was a gift from Arshad Desai (University of California, San Diego, CA, USA). The His-tagged whole human Ndc80 complex and the baculovirus expression construct for purifying it (used for both Superose 6 fractionation experiments and TIRF imaging) was a gift from the Andrea Masaccio (Max Planck Institute-Dortmund, Germany) laboratory. hNdc80 complex was purified using the baculovirus expression system (Huis In 't Veld et al., 2016) and labeled as required, with Alexa 647 dye (#A20006; Thermo Fisher Scientific) using succinimide chemistry and following manufacturer’s protocol. Full-length Ska1 complex was also labeled in an analogous way with Cy3 dye (#PA13104; Amersham). The labeling reaction produced homogeneously labeled fluorescent proteins; the majority of them showed single-step photobleaching patterns, as observed with GFP-Cdt1 (Fig. S5, A–C).
Representative photobleaching steps for GFP-Cdt1, SKA1/2 Cy3 , and Ndc80 alexa647 . (A–C) Representative photobleaching steps for indicated proteins. Histogram (B) of starting fluorescence intensity of indicated single molecules and photobleaching steps (C) from n > 100 single molecule binding events and N = 3 independent trials.
Representative photobleaching steps for GFP-Cdt1, SKA1/2 Cy3 , and Ndc80 alexa647 . (A–C) Representative photobleaching steps for indicated proteins. Histogram (B) of starting fluorescence intensity of indicated single molecules and photobleaching steps (C) from n > 100 single molecule binding events and N = 3 independent trials.
Auxin-inducible degron tagging of endogenous Cdt1
The human genomic sequence of Cdt1 was downloaded from National Center for Biotechnology Information (NC_000016.10:88803778–88809258 Homo sapiens chromosome 16, GRCh38.p12) using SnapGene, and the exon/intron boundaries were located in this annotated sequence. We used the last exon sequence (+800 bp up/down stream) to design our guide RNA (gRNA) targeting the Cdt1 C-terminus, using the Broad Institute sgRNA design tool. The gRNA sequence used was 5′-GGCCCACCAGACACGTGCTG-3′. We then cloned the gRNA into PX330 (obtained from #42230; Addgene), a CAS9-EGFP sgRNA vector, using Bbs1 restriction cloning, and following established protocols (Cong et al., 2013; Li et al., 2019; Zasadzińska et al., 2018) and confirmed the insertion by DNA sequencing. For designing the Cdt1 repair template, we synthesized a DNA fragment containing the above-mentioned 5′ and-3′ 800 bp of Cdt1 homologous sequence with the sequence of the AID and YFP inserted in between these two homology arms (GeneUniversal). We then cloned this product into the vector HJURP C-terminal AID-YFP at KpnI-HindIII restriction sites (a kind gift from Dr. Daniel Foltz, Northwestern University, Evanston, IL, USA). The PAM sequence within Cdt1 homology arms were mutated and replaced with synonymous codons without altering the protein sequence. DLD-1 Tir1 parental cells (a kind gift from Andrew Holland, Johns Hopkins University, Baltimore, MD, USA) were transfected with the plasmid containing the Cdt1 gRNA/Cas9 and the Cdt1 repair template at equimolar ratio using Effectene (#301425; Qiagen) transfection agent. Following transfection, the cells were grown for 7 d, after which they were collected by centrifugation at 200 × g for 5 min. The cells were then resuspended in 500 µl of 1% FBS in PBS. A 96-well plate was prepared with 100 µl of condition media collected from a confluent plate of the parental cell line after filtering using a 0.22 µm filter. We then sorted single cells into 96-well plate using Fluorescence-Activated Cell Sorting at the Northwestern University—Flow Cytometry Core Facility. Then, single cells were allowed to grow in the 96-well plate using complete DMEM media supplemented with 20% FBS. Single colonies were then transferred into 24-well plate, followed by 12-well and then finally to 6-well plate to ensure enough propagation of the single clones. To check the transfection efficiency, we first assessed YFP expression levels for each clone using confocal microscopy, and then proceeded with making genomic DNA of each positive clone and identification of homozygous genomic insertion clones by PCR. The primers used to confirm the clones were 5′-CAAGTGCTGCCCGTGCAGGGCTG-3′, 5′-CTGGATCTGGCTTCCCTTCCCCACCAGC-3′, and 5′-GTGAGCAAGGGCGAGGAGCTGTTCACC-3′. The cloning strategy used yielded a 0.5 kb amplification for wild-type cells, a 1 kb amplification for homozygous insertions and bands of both size for heterozygous insertions (Fig. S1 B). Monoclonal lines were further screened by Western blotting and genotyping. We treated both control DLD1 cells and AID-DLD1 (clone#6) cells with 0.5 mM Indole Acetic Acid (IAA Auxin) at the following time points, 0, 1, and 2 h. Cells induced with IAA for these different time points were lysed and analyzed for the levels of YFP-Cdt1-AID with Western blotting using an anti-GFP antibody.
siRNA-mediated knockdown
For Cdt1 siRNA, HeLa cells were synchronized by treatment with 2 mM thymidine for 18 h followed by release for 9 h and then re-treatment with 2 mM thymidine for 18 h. siRNA against Cdt1 (5′-CCUACGUCAAGCUGGACAA-3′) was transfected into the cells during the second thymidine release (according to the manufacturer’s instructions) and prior to fixing or lysing the cells at designated time points as indicated in the figures. siRNA strategy used for SKA3 depletion was the ON-TARGETplus SMARTPool obtained from Dharmacon/Horizon Discovery (Cat #L-015700-00-0005). Cdt1 or Ska1 siRNA treatment in asynchronous cells was carried out for 48 h.
Immunofluorescence microscopy and live-cell imaging
For indirect immunofluorescence, the coverslips with cells were first raised three times with 1× PBS. The cells were then prefixed in ice-cold methanol or 4% PFA for 1 min. Methanol fixation was performed for 5 min at −20°C. For PFA fixation, the cells were permeabilized with Triton X-100 for 5 min followed by fixation with 4% PFA for 20 min at RT. The cells were then blocked with 0.5% BSA for 1 h. The primary antibody treatments were at 4°C overnight while the secondary antibody treatments were for 1 h at 37°C. Cells were washed with 1× PBS, three times for 10 min each after each antibody treatment. The cells were treated with 0.1 µg/ml DAPI for 10 min before being mounted on slides for fixed-cell imaging. ProLong Gold Antifade reagent (Thermo Fisher Scientific) was used for mounting the coverslips. Methanol fixation was used for spindle microtubule staining for Cdt1 while Paraformaldehyde fixation was used for kinetochore staining (Agarwal et al., 2018). The antibodies used for immunostaining include β-tubulin monoclonal (T9026, clone DM1A; Sigma-Aldrich), Zwint1 polyclonal (A300-781A; Bethyl Laboratories), Cdt1 polyclonal (H-300, sc-28262; Santa Cruz Biotech), Hec1 monoclonal (GTX70268; GeneTex), and anti-human ACA (HCT-0100; Immunovision). Guinea Pig Cdt1 antibody (Varma et al., 2012) was a gift from Jeanette Cook’s lab while SKA1 and SKA3 rabbit polyclonal antibodies were a gift from Gary Gorbsky (Oklahoma Medical Research Foundation, Oklahoma City, OK, USA).
Live-cell imaging of chromosome alignment and segregation during mitosis in DLD1 cells was carried out (Agarwal et al., 2018) on 35-mm glass-bottomed dishes (MatTek Corporation) in an incubation chamber for microscopes (Tokai Hit Co., Ltd) at 37°C and 5% CO2. However, in this case, the 35-mm glass-bottom dishes were manually coated with 10 µg/ml Fibronectin prior to using them for cell culture. The cells were incubated with IAA and 2.5 µg/ml Hoechst 33,258 dye for 1.5 h. before initiating live-cell imaging in FluoroBrite DMEM live imaging media (Life Technologies) supplemented with 2.5% FBS. Images were acquired every 7.5 min for up to 2–6 h as required based on whether the samples were controls or if Cdt1 was degraded.
Image acquisition and processing
For image acquisition, three-dimensional stacks were obtained through the cell using a Nikon Eclipse TiE inverted microscope equipped with a Yokogawa CSU-X1 spinning disc, an Andor iXon Ultra888 EMCCD camera and an ×60 or ×100 1.4 NA Plan-Apochromatic DIC oil immersion objective (Nikon). For fixed-cell experiments, images were acquired at room temperature as Z-stacks at 0.2 µm intervals controlled by NIS-elements software (Nikon). Images were processed using Fiji ImageJ or NIS-elements and represent maximum-intensity projections of the required z-stacks. Live-cell imaging of DLD1 cells was carried out in a single focal plane using identical acquisition settings and an ×60 1.4 NA Plan-Apochromatic DIC oil immersion objective. Time-lapse images were processed using the NIS-elements software (Nikon).
Analytical size exclusion chromatography
Untagged Ska1 complex and either of GFP-tagged Cdt1-WT or GFP-Cdt1-2E3D proteins were mixed at concentrations 3 mg ml−1 and 1.6 mg ml−1, respectively, and were incubated at 10°C for at least 2 h. The mixture was then clarified by centrifugation for 10 min at 16,100 × g at 4°C. The mixture was analyzed by size exclusion chromatography using Superose six Increase 5/150 GL column (GE Healthcare) mounted on an AKTApure system. The column was equilibrated with size exclusion chromatography buffer (20 mM TRIS pH 8, 200 mM NaCl, 2% v/v glycerol, 2 mM TCEP), and the column was operated at a flow rate of 0.1 ml min−1. Typical fraction volume was 0.05 ml; the fractions were collected and analyzed by SDS PAGE electrophoresis and Western blotting using anti-Cdt1 (#H-300; Santa Cruz Biotechnology, rabbit polyclonal IgG) and anti-SKA1 (#ab91550; Abcam, rabbit polyclonal) antibodies.
Kinase assays
Purified GFP-Cdt1 protein (1.6 mg ml−1) was treated with Cdk1-CyclinB Kinase (# 14–450; EMD Millipore) supplemented with 10 mM ATP in kinase buffer (20 mM Tris pH 8, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, 1× Phosphatase inhibitor [#1862495; Thermo Fisher Scientific]) at room temperature for 2 h. The phosphorylated GFP-Cdt1 protein was then incubated with the Ska1 complex for 2 h at 10°C before being used for size exclusion chromatography (as described above).
In vitro blot overlay assay
To assess interaction between the proteins of interest, 0.5 µg of one protein was loaded on to the SDS-PAGE gels as “bait” and was transferred on to the nitrocellulose membrane using standard procedure. The membrane was blocked with 5% skimmed milk in TBS buffer and 0.1% Tween20 for an hour at RT followed by the addition of the second protein (1 µg/ml in TBS) as “prey” for 18 h at 4°C with constant shaking. The blot was thoroughly washed thrice for 15 min each with TBST and antibody to detect the prey protein, which was added at indicated dilutions for 2 h with constant shaking at RT. HRP-tagged mouse/rabbit antibodies (Jackson Laboratories) were used at 1:10,000 dilution to detect the bait-prey complex, and blot was developed by Super signal west pico chemiluminescent substrate (Thermo Fisher Scientific).
Bio-layer interferometry
Bio-layer interferometry experiments were performed as in published protocols (Muller-Esparza et al., 2020) on a single-channel BLItz instrument (forteBIO). His-reactive biosensor tips (HIS1K, FORTEBIO, #185121) were hydrated in 96-well plate in BLI-buffer (20 mM HEPES pH 7.4, 1% sucrose, 150 mM NaCl, 0.5 mM TCEP, 0.02% Triton X-100) for 10 min, followed by ligand (0.2 mg/ml in BLI-buffer) immobilization for 300 s. Analyte in varying concentrations (0.3–10 μM) was placed as a drop (4 μl) onto the magnetic holder and was allowed to contact the sensor with the immobilized ligand for 120 s (association phase) followed by a dissociation phase for 180 s. Baselines for 30 s were obtained in BLI-buffer to compensate for the phase shifts. All the steps were performed at a constant shaking of 1,000 rpm. Both the immobilized and analyte proteins were subjected to buffer exchange into the BLI buffer as required and pre-cleared using high-speed ultracentrifugation to exclude the aggregates/debris prior to their use in BLI experiments. The temperature of measurement was set at 25°C unless otherwise specified. GFP and BSA proteins were taken as a negative control to rule out the possibility of non-specific interaction. Sensograms were fit globally to a 1:1 binding model by BLItz Pro version 1.1.0.28, from which the equilibrium dissociation constant (KD), association (ka), and dissociation (kd) rate constants were calculated as in Muller-Esparza et al., 2020.
Cell culture, co-immunoprecipitation, and Western blotting
HeLa, HEK293T, and GFP-expressing HeLa (kind gift from Dr. Robert Goldman, Northwestern University, Evanston, IL, USA) cells were cultured at 37°C with 5% CO2 in Dulbecco’s modified eagle’s medium (DMEM, Life Technologies) containing 10% fetal bovine serum (Seradigm, VWR Life Science), 100 U ml−1 penicillin, and 1 μg ml−1streptomycin. HeLa stable cell lines expressing GFP-SKA1 clone D1 and GFP-SKA1ΔMTBD (1-132) clone B6 were generously procured from Dr. Gary Gorbsky and were maintained similarly except for supplementing the medium with 150 µg ml−1 hygromycin.
The GFP-, SKA1-GFP, and GFP-SKA1ΔMTBD cell lines were grown till 50–60% confluence. The cells were synchronized by treatment with 2 mM thymidine for 18 h followed by release for 9 h and then addition of 0.33 µM Nocodazole for 12 h to arrest the cells in mitosis. To induce the expression of GFP-SKA1 or GFP-SKA1ΔMTBD (lacking the C-terminal residues, containing only aa 1–132) proteins, doxycycline (2.5 µg/ml) was added in the medium from the time of synchronization. The mitotic cells were collected using mitotic-shake-off and were lysed in the co-IP buffer (10 mM HEPES, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.1% Nonidet P-40 supplemented with 1 × Halt protease and phosphatase inhibitor cocktails, Thermo Fisher Scientific). One mg total cell protein was used per IP and 1% of the cell lysate volume was loaded as the input to assess the presence of desired proteins in each case. The antibodies (3 μg/IP) were used, include anti-GFP mouse monoclonal 3E6 (Thermo Fisher Scientific), anti-GFP rabbit polyclonal A-11120 (Thermo Fisher Scientific), anti-SKA3 rabbit polyclonal ab186003 (Abcam), anti-Cdt1 rabbit polyclonal H-300 (Santa Cruz Biotech), or Control IgG. Magnetic Dynabeads Protein G (Invitrogen), to isolate protein-antibody complex and were eluted by boiling the beads with 2 × Laemmli sample buffer (Bio Rad). Proteins were resolved on 4–15% gradient gel and transferred using Western blot. The interacting partners were detected by Western blotting using relevant antibodies (Cdt1 H300, SKA1, and SKA2 polyclonal antibodies received from Dr. Gary Gorbsky and GFP A-11122, all at 1:1,000 dilutions). TrueBlot-HRP antibodies against rabbit or mouse (1:1,000) were used as secondary antibody (Rockland Immunochemicals Inc.). The blots were developed by chemiluminescence.
For co-IP experiments involving WT and Cdt1 phosphomutant variants (Cdt1-5A, Cdt1-2E3D, Cdt1-10A, and Cdt-10D) and SKA1, stable HeLa cells expressing the respective proteins were grown to 70–80% confluence, followed by the addition of 0.33 µM Nocodazole for 10–12 h to arrest the cells in mitosis. Cell lysate (1 mg per input) was incubated with mouse anti-HA magnetic beads (#88836; Thermo Fisher Scientific) at a ratio of 20:1 v/v. The interacting partners were detected by Western blotting using the antibodies: anti-HA rabbit polyclonal (#H6908; Sigma-Aldrich) and anti-SKA1 rabbit polyclonal (#ab91550; Abcam) at 1:1,000 dilutions. 1% of the cell lysate volume was loaded as the input. Although the expression levels for the HA-Cdt1-WT and the phopshomutants were identical, the quantitation of the levels of WT and mutant protein bands was normalized relative to the expression levels of the WT protein. For the co-IP of Hec1, rabbit anti-HA magnetic beads (#C29F4; Cell Signaling) was used for the pull-down and anti-Hec1 9G3 (#GTX70268; GeneTex) was used for Western blotting. HRP antibodies against rabbit and mouse (1:1,000) were used as secondary antibodies (#AC2114 and #AC2115; azure biosystems, respectively) for detection, and the blots were developed by chemiluminescence.
Ni+2-NTA agarose-mediated pull down
HEK-293T cells were transfected with the plasmids (Cdt1-HASH in pQCXIP vector) for 6 h at 20–30% confluency using calcium chloride-based method. M phase synchronization was carried out by treating the cells with 2 mM thymidine for 18 h, followed by the addition of 100 ng/ml (0.33 μM) Nocodazole for 10 h. The cells were collected by mitotic-shake-off and lysed using lysis buffer (50 mM HEPES pH 8.0, 33 mM KAc, 117 mM NaCl, 20 mM Imidazole, 0.1% Triton X-100, 10% glycerol, 0.1 mM AEBSF, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM ATP, 1 mM MgCl2, 5 μg/ml phosvitin, 1 mM β-glycerol-phosphate, and 1 mM orthovanadate). The whole-cell lysate expressing the His-tagged protein was incubated with Ni+2-NTA beads for overnight with end-on rotation at 4°C. Beads were washed three times with the lysis buffer, and bound proteins were eluted by boiling for 5 min in 40 μL of 2×SDS sample buffer. For immunoblotting, proteins were electrophoresed by SDS-PAGE and transferred to the PVDF membranes. Immunoblots were developed using chemiluminescence and exposed on to the x-ray films. Cdt1 H-300 polyclonal antibody (Santa Cruz) was used at 1:1,000 dilution; SKA1 and SKA3 antibodies (1:1,000 dilution each) were generous gifts from Dr. Gary Gorbsky.
GST pull down assay
HeLa cells were synchronized at mitotic phase by adding 2 mM thymidine for 18 h followed by release for 9 h and subsequent addition of 0.33 μM Nocodazole for next 12 h. The mitotic cells were harvested by gentle shake-off and were washed twice with 1×PBS. The cell pellet was resuspended in the lysis buffer (10 mM HEPES, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.1% Nonidet P-40 supplemented with 1 × Halt protease and phosphatase inhibitor cocktails, Thermo Fisher Scientific). The lysate was incubated with 10 μg of either GST or GST-tagged Cdt192-546 proteins for 12 h at 4°C with constant shaking. The lysate was then incubated with Glutathione resin 4B FF resin for 2 h followed by elution with 30 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0. The proteins in the bound (B) and unbound (UB)/flow through were analyzed by Western blotting using anti-GST rabbit polyclonal antibody (#G7781, 1:5,000 dilution; Sigma-Aldrich).
TIR-FM
Microtubule assembly
Taxol-stabilized microtubules were prepared from mixing on ice the following: 4 µl 10 mg/ml unlabeled tubulin, 0.4 µl 5 mg/ml Alexa 647 labeled tubulin (#64705; Pursolutions), 0.4 µl of 5 mg/ml biotin labeled tubulin (#T333P-A; Cytoskeleton), 20% glycerol, and 1 mM Mg-GTP in MRB80. The mixture was then immediately placed into 37°C water bath for 30 min. After polymerization, 10 µM Taxol in MRB80 was added to the polymerized mixture. The mixture was then further incubated in 37°C water bath for additional 10 min followed by addition of 50 µl 10 µM Taxol in MRB80 (buffer B). The polymerized mix was then centrifuged at 12,000 × g for 10 min at RT. The supernatant was discarded and the resulting microtubule pellet was resuspended in 50 µl buffer B and was kept in dark for immediate use. GMPCPP-stabilized microtubule seeds were assembled in an analogous way by mixing unlabeled, labeled, and biotin-labeled tubulin 1 mM GMPCPP (# NU405S; Jena Bioscience) followed by incubation on ice for 10 min to allow nucleotide exchange and then polymerizing on the 37°C water bath for 30 min. 50 µl MRB80 was then added to the polymerization mix, and the microtubules were pelleted by centrifugation as above. The resulting microtubule seed pellet was resuspended in 50 µl MRB80 and stored as above.
Synergic-binding assay
A microscopy perfusion chamber was prepared by attaching an acid-washed coverslip (22 × 22 mm, thickness 1 1/2, Corning) over a pre-cleaned glass slide (75 × 25 mm, thickness 1 mm, Corning) using a double-sided tape (Scotch) resulting in a chamber volume of ∼10 µl. For more details, please see TIRF section in Afreen et al., 2022. Acid-washed coverslips were further cleaned by holding under flame for a couple of seconds prior to use. Solutions were then exchanged into the perfusion chamber by micropipette and filter paper. The chamber was activated by flowing in the following solution in an order in which they appear below, followed by incubation for 5 min and intermittent washing with three chamber volumes of buffer B: PLL PEG biotin (0.1 mg ml−1, Susos, AG), Streptavidin (0.625 mg ml−1, #S4762; Sigma-Aldrich), diluted solution of Taxol-stabilized microtubules (1 in 100 dilutions form the stock), and κ-Casein (1 mg ml−1). All incubations were done inside a wet chamber to prevent evaporation. The chamber was finally washed with imaging buffer:MRB80 supplemented with 10 µM Taxol, 0.6 mg ml−1 κ-casein, 4 mM DTT, 50 mM glucose (#G8270; Sigma-Aldrich), 0.2 mg ml−1, catalase (#C9322; Sigma-Aldrich), and 0.4 mg ml−1glucose oxidase (#G7141; Sigma-Aldrich). Premixed solutions of a GFP labeled protein (either Cdt1, or Ska1, or Ndc80, 1 nM) together with increased concentration untagged proteins (Cdt1, Ska1, or Ndc80, 10 nM—2 µM) in image buffer were subsequently flown into the chamber, and images in were recorded with the following microscopy settings.
Imaging was performed with a Nikon Ti-2 inverted microscope equipped with Photometrics prime 95B sCMOS camera (Teledyne photometrics); 100×, 1.49 NA objective lens. The microscope produced images of pixel size 109 nm, and with excitation laser launcher (Nikon) containing 488-, 546-, and 640-nm laser lines among other lines. NIS-elements software (Nikon) was used for data acquisition with 100 ms exposure times.
Dynamic experiment was carried out with Nikon Ti-E inverted microscope equipped with Andor iXon3 CCD camera (Cambridge Scientific), 1.49× NA, and 100× oil objective. The microscope produced a 512 × 512-pixel images with 0.16 µm per pixel resolution in both x and y directions. All experiments with dynamic microtubules were carried out at room temperature. The microscope was also equipped with differential interference contrast (DIC) imaging and microtubules were visualized using DIC mode with 100% lamp power and 300 ms exposure time.
Single-molecule co-localization assay
A microscopy perfusion chamber with immobilized Taxol-stabilized microtubules was prepared as described above. The chamber was washed with image buffer, and microtubule-binding proteins were flown into the chamber. Extremely dilute solutions of HEC1/NUF2-GFP dimer (1 nM) and Ska1Cy3 (1 nM) in image buffer was flown into chamber together with increasing concentrations (0, 1, and 10 nM) of Cdt1. For three party colocalization experiments, non-fluorescently labeled microtubule was used together with nanomolar (1 nM) concentrations of GFP-Cdt1, Ska1Cy3, and Ndc80alexa647.
Experiment with dynamic microtubules
A microscopy perfusion chamber with immobilized GMPCPP-stabilized microtubules was prepared as described above. The chamber was washed with image buffer supplemented with 1 mM GTP. A solution of 1 mg/ml tubulin was flown in image buffer (supplemented with 1 mM GTP) together with 1 nM GFP-Cdt1 and 1 nM untagged Ska1. The chamber was sealed with nail polish and a time series was recorded at room temperature in brightfield-DIC and TIRF mode (488 nm excitation, 100 ms exposure) for every 2 s time interval and for 30 min.
Single-molecule diffusion experiment
This method is same as that of the co-localization assays except that the data acquisition was done using 640-nm excitation laser, 40-ms exposure time, 999 EM gain, at 14 MHz read out speed. The data were acquired continually for 2 s. Later, kymographs were plotted using kymograph direct software (Mangeol et al., 2016), and MSD values were also obtained using the same software. Multiple MSD values were averaged and the averaged MSD vs. time plot was analyzed by linear regression to obtain the diffusion coefficient.
Statistical analysis
Origin software (version 2018) was used for all data plots and obtaining the statistical significance (Student’s t test or Mann–Whitney U test, as indicated in the respective figure legends) between data sets.
Online supplemental material
Fig. S1 shows analyses of the AID-Cdt1 degron system to study mitotic functions of Cdt1. Fig. S2 shows depletion of Cdt1 from mitotic cells does not interfere with the normal targeting of the Ska1 complex. Fig. S3 shows analyses of binding between Cdt1, Ska1, and their fragments. Fig. S4 shows characterization of WT and phosphomutant Cdt1 cell lines and proteins. Fig. S5 shows representative photobleaching steps for GFP-Cdt1, SKA1/2Cy3, and Ndc80alexa647. Video 1 shows normal mitotic progression in AID-Cdt1 DLD1 cells treated with control DMSO. Video 2 shows severe delay in mitotic progression in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Video 3 shows chromosome mis-segregation in AID-Cdt1 DLD1 cells treated with Auxin, IAA. Video 4 shows that GFP-Cdt1 diffusively tracks the end of dynamic microtubule in the presence of Ska1. Video 5 shows that Cdt1 diffusively tracks the end of dynamic microtubule in the presence of SKA1/2/3-GFP complex. Video 6 shows SKA1/2/3-GFP mixed with untagged Cdt1 and Ndc80 show robust plus-ends tracking of dynamic microtubules. Video 7 shows GFP-Cdt1 robustly tracks the end of dynamic microtubule in the presence of untagged Ndc80 and SKA1/2. Video 8 shows SKA1/2/3-GFP mixed with untagged Ndc80 diffusively tracks the polymerizing phase of dynamic microtubules. Video 9 shows Hec1/Nuf2-GFP diffusively tracks the end of dynamic microtubule in the presence of untagged SKA1/2. Video 10 shows GFP-Cdt1/Ndc80alexa647 robustly tracks the end of dynamic microtubule in the presence of SKA1/2.
Acknowledgments
We thank Dr. Gary Gorbsky (Oklahoma Medical Research Foundation, Oklahoma City, OK, USA) for GFP-SKA1 and GFP-SKA1ΔMTBD expressing stable HeLa cell lines, bacterial expression plasmid encoding GST-tagged SKA3, and for the SKA1 and SKA3 polyclonal antibodies; Dr. Andrea Musacchio (Max Plank Institute-Dortmund, Germany) for providing the purified His-human Ndc80 complex and the baculoviral expression plasmid for purifying the same; Dr. A. Arockia Jeyaprakash (University of Edinburgh, Edinburgh, UK) for bacterial expression plasmid encoding GFP-SKA2; Dr. P Todd Stukenberg (University of Virginia Charlottesville, Charlottesville, VA, USA) for the bacterial expression plasmid encoding His-SKA1/2; Dr. Arshad Desai (University of California San Diego, San Diego, CA, USA) for the HEC1/NUF2 dimeric bacterial expression plasmids; Drs. Iain Cheeseman (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA, USA) and Jeanette Cook (University of North Carolina-Chapel Hill, Chapel Hill, NC, USA), and Shashi Kumar Suman (IGBMC, University of Strasbourg, Strasbourg, France) for helpful discussions and Dr. Anita Varma (Northwestern University-Chicago, Chicago, IL, USA) for technical support. We would also like to acknowledge the Keck Biophysics and the Lurie Flow Cytometry Core Facilities at Northwestern University-Chicago for their support with our experiments.
This work was supported by National Institute of General Medical Sciences (NIGMS) grant R01GM135391 to D. Varma; and NIGMS grant R35GM124889 to R.J. McKenney.
Author contributions: D. Varma and S. Agarwal conceptualized the project. A. Rahi, M. Chakraborty, S. Agarwal, R.J. McKenney, and D. Varma performed the experiments, and also contributed to the analyses and curation of the data. A. Rahi, M. Chakraborty, K.M. Vosberg, S. Agarwal, A.Y. Wang, and D. Varma validated the data. S. Agarwal, D. Varma, M. Chakraborty, and A. Rahi wrote the original draft and also reviewed and edited the succeeding versions of the manuscript drafts. D. Varma and R.J. McKenney were instrumental in funding acquisition. D. Varma also contributed to resource procurement, project administration, and supervision.
References
Author notes
A. Rahi and M. Chakraborty contributed equally to this paper.
Disclosures: The authors declare no competing interests exist.
Shivangi Agarwal’s current affiliation is Grove Biopharma Inc., Chicago, IL, USA.
Shivani Agarwal’s current affiliation is Division of Hematology/Oncology, Robert H. Lurie Comprehensive Cancer Center, Northwestern University, Chicago, IL, USA.


![Generation of an AID cellular system to assay for mitotic functions of Cdt1 (AID-Cdt1). (A) A schematic of the repair template generated to endogenously tag the Cdt1 genomic locus with YFP and AID at the C-terminal using CRISPR/CAS9-based genome editing. (B) Western-blot analysis using an anti-Cdt1 antibody showing the levels of the endogenous and tagged Cdt1 in parental and AID-Cdt1 clones, respectively, following auxin-mediated degradation. (C) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel, [−] IAA) or the Auxin, IAA, for 2 h. (middle and bottom panels, [+] IAA) followed by immunostaining using an antibody against tubulin and the chromosomes counterstained with DAPI. Scale bar, 5 µm. (D) AID-Cdt1 DLD1 cells growing at 37°C were treated with the DNA stain, Hoechst, along with either control DMSO (top panels) or IAA, for 2 h. (middle and bottom panels) followed by live-cell imaging for 2–6 h as required. Selected frames from the time series are shown for each experimental condition as indicated. Scale bar, 5 µm. (E) Quantification of live imaging data from D. n = 30 for control cells not treated with IAA and 55 for cells treated with IAA. Data presented as mean ± SD, two-tailed t test *** = P < 0.0001. (F) AID-Cdt1 DLD1 cells growing at 37°C were treated with either control DMSO (top panel) or IAA, for 2 h (bottom panel) followed by treatment with ice-cold buffer PBS buffer for 10 min. The cells were then immunostained for antibodies against tubulin, Zwint1 (for kinetochores) with the chromosomes counterstained using DAPI. Scale bar, 5 µm. (G) Quantification of total microtubule intensities from cells in F (n = 10 cells). Data presented as mean ± SD, two-tailed t test, *** = P < 0.0001. (H) Insets from the top and bottom panels of F showing samples of attached and unattached kinetochores, respectively, as indicated. Scale bar, 1 µm. (I) Quantification of the % of attached kinetochores from F under the two experimental conditions assessed (n = 10 cells). Data presented as mean ± SD. Source data are available for this figure: SourceData F1.](https://cdn.rupress.org/rup/content_public/journal/jcb/222/8/10.1083_jcb.202208018/2/m_jcb_202208018_fig1.png?Expires=1773752024&Signature=ELoR4tQ~zSrdQI7EOp7~gHqPfCh8is~NJ6LsmqS7HNpju5d2EzhLN3lBc-WTykCaaLsTkb67RX3QIQDQ35-flJPOAAZHO3vj~NWkDmLTWETA7yaRjtH~tdLebpNPK85e9D0igDEjNG6KCU35FHrv8b03dfV-21oVcSENShttYHaNIMjVxOF8fVtBgjH9aueJhQ1deQRimYgL-NXxueUVpGNJ3eYLwG~R2~1uIFKzrBEsvpUt1P8BrpcRrYukjBpnuA9rDQdae4Mz-UT-VoBZE5unR2aRVCqI3vlp8q-IrQF5huLoZNDRTUftrO0gavQqyPBqeDOihHIa~Kj5wBKZkg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)