Although mutations in the SCRIB gene lead to multiple morphological organ defects in vertebrates, the molecular pathway linking SCRIB to organ shape anomalies remains elusive. Here, we study the impact of SCRIB-targeted gene mutations during the formation of the gut epithelium in an organ-on-chip model. We show that SCRIB KO gut-like epithelia are flatter with reduced exposed surface area. Cell differentiation on filters further shows that SCRIB plays a critical role in the control of apical cell shape, as well as in the basoapical polarization of myosin light chain localization and activity. Finally, we show that SCRIB serves as a molecular scaffold for SHROOM2/4 and ROCK1 and identify an evolutionary conserved SHROOM binding site in the SCRIB carboxy-terminal that is required for SCRIB function in the control of apical cell shape. Our results demonstrate that SCRIB plays a key role in epithelial morphogenesis by controlling the epithelial apical contractility during cell differentiation.
Introduction
Tissue folding drives embryo remodeling and assembly of internal organs during animal development. The planar cell polarity pathway (PCP) participates in tissue folding at the cellular level by promoting the planar polarized distribution of contractile actomyosin structures that leads to apical constriction, polarized cell intercalation, and neighbor exchanges (Zallen, 2007). Disruption of PCP signaling genes in mice impacts numerous organs with a particularly strong and specific effect on neural tube development (Curtin et al., 2003; Kibar et al., 2001; Murdoch et al., 2003; Paudyal et al., 2010; Wang et al., 2006a, 2006b; Wansleeben et al., 2010). In PCP mutant mice, the neural tube fails to bend and fuse dorsally, causing most of the brain and the spinal cord to stay open, a phenotype known clinically as craniorachischisis (Davey and Moens, 2017). The inability of the neural tube to fold is partly due to the failure of the neuroepithelium to correctly perform apical planar polarized actomyosin contraction (Nikolopoulou et al., 2017). Inactivation of the SCRIB gene in mice causes craniorachischisis, thus positioning SCRIB into the PCP gene family in vertebrates (Murdoch et al., 2003). SCRIB KO mice embryos have additional organ deficiencies, such as shorter intestines, a defect observed in other animal models with PCP pathway deficiencies (Carnaghan et al., 2013; Dush and Nascone-Yoder, 2019). In humans, mutations in the gene SCRIB are associated with craniorachischisis and Spina Bifida (Robinson et al., 2012; Lei et al., 2013). However, the molecular mechanism by which SCRIB controls organ morphology is still not completely clear.
SCRIB was first identified in Drosophila, where it plays an important role in apicobasal polarity (Bilder and Perrimon, 2000; Bilder et al., 2000). In vertebrates, SCRIB partakes in a very large number of cellular functions including cell migration, polarity proliferation, differentiation, apoptosis, stem cell maintenance, and vesicle trafficking (Humbert et al., 2008). In cells, SCRIB is frequently observed at cell–cell junctions (Navarro et al., 2005; Qin et al., 2005). The SCRIB protein has a tripartite domain organization that consists of an N-terminal region composed of leucine-rich repeats (LRR), four PDZ domains, and a C-terminal region with no identified protein domain. While the LRR region is crucial for membrane targeting, the PDZ domains are implicated in protein–protein interactions with >50 direct protein partners identified so far (Stephens et al., 2018). The C-terminal region of SCRIB directly binds to the spectrin/actin cortical cytoskeleton through three short linear motifs (Boëda and Etienne-Manneville, 2015). SCRIB regulates front-to-rear polarity during cell migration through its interaction with the Rho-GEF β-Pix and the local activation of small GTPase at the leading edge of migrating cells (Osmani et al., 2006; Nola et al., 2008). SCRIB physically and genetically interacts with the PCP gene Vangl, but how this interaction influences PCP signaling remains unclear (Montcouquiol et al., 2003). The PCP pathway, downstream of the noncanonical Wnt signaling, is known to promote myosin light chain phosphorylation and actomyosin contractility through the local accumulation and activation of the RhoA/ROCK pathway at the apical side of epithelia (Schlessinger et al., 2009). An interaction between SCRIB and the protein SHROOM2, a direct regulator of ROCK and actomyosin contractility expressed at the level of tight junctions (Etournay et al., 2007), has been recently characterized (Amano et al., 2015), suggesting a potential role of SCRIB in the control of actomyosin contractility.
To clarify the role played by SCRIB on planar epithelium morphogenesis, we generated SCRIB KO Caco-2 cells and took advantage of intestine-on-chip (IOC) and filter culture systems. In the IOC system, Caco-2 cells differentiate into a 3D intestine-like structure composed of villus and crypts (Kim and Ingber, 2013; Kim et al., 2016). When grown on filters for 21 d, Caco-2 cells spontaneously differentiate into a planar epithelium composed of polarized columnar cells sharing many properties with enterocytes (Lea, 2015). Using a combination of in vitro culture models and biochemical experiments, we further analyzed the role of SCRIB and its interaction with SHROOM2 in the control of the morphological differentiation of epithelial monolayers.
Results and discussion
Effects of SCRIB gene inactivation on an IOC system
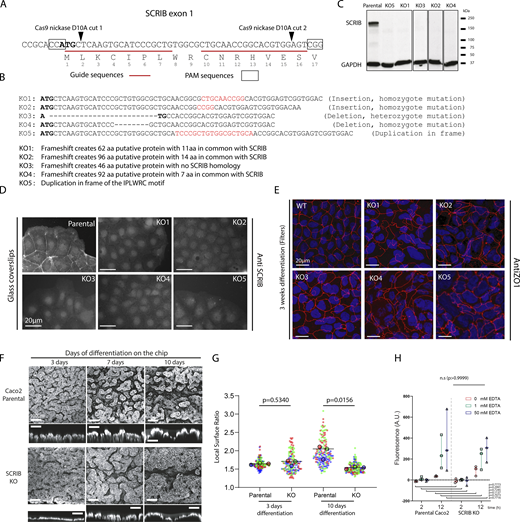
To characterize the SCRIB function in epithelial cells, we used the CRISPR/Cas9 D10A double-nicking system to target the SCRIB gene exon 1 in the human intestinal epithelial Caco-2 cell line (Fig. S1 A). CRISPR/Cas9 D10A system mediates highly specific genome editing and has been shown to reduce off-target activity up to 1,000 fold in cell lines when compared with the CRISPR/Cas9 WT system (Ran et al., 2013). We selected multiple clones (KO-1 to KO-5) possessing short genomic frameshifts in SCRIB coding sequence and in which SCRIB expression could not be detected by Western blotting and immunofluorescence assays (Fig. S1, B and C). To analyze the impact of SCRIB loss at the tissue level, we grew SCRIB KO Caco-2 cells in an IOC system. IOC is made of a flexible silicone polymer (polydimethylsiloxane [PDMS]) chip in which cells are grown in the upper channel. There, continuous medium perfusion at a physiological rate emulates the shear stress on the apical cell surface that is observed in the human intestinal lumen (Fig. 1 A). After 7 d of culture in these conditions, the parental Caco-2 cells organized into numerous short finger-shaped villi and crypt-like structures covered with microvilli (Fig. 1, B and C; left). SCRIB KO cells grown under similar conditions differentiated into fewer but larger villi and crypt-like structures (Fig. 1, B and C; right). Large strips of epithelia were recovered from the IOC system, stained with phalloidin and DAPI, and analyzed by confocal microscopy (Fig. 1, D–F). Local epithelial height and surface area were calculated from square patches (0.4 mm2) along the epithelia and a depth map was extracted (Fig. 1 G). While no statistical difference could be observed after 3 d of growth, comparison showed that intestine folds obtained from the SCRIB KO cells after 7 d of growth were flatter and had significantly reduced exposed surface area compared with villi structures derived from parental Caco-2 cells (Fig. 1 H; and Fig. S1, F and G). KO SCRIB villi formed after 10 d still displayed reduced surface area when compared with the control (Fig. S1 G), suggesting that the differences observed after 7 d were not due to delayed morphological changes of the SCRIB KO epithelium. To determine whether the altered morphology of the epithelium had any impact on the epithelial barrier function, we performed a dextran permeability assay in the IOC system. Both parental and SCRIB KO epithelia villi-like structures formed functional and tight barriers (Fig. S1 H). These results show that, in the IOC system, inactivation of the SCRIB gene in human intestinal epithelial Caco-2 cells reduced the cell ability to form invaginations and villi-like structures without affecting the intestinal barrier function. Interestingly, the intestines of SCRIB KO mice embryos display a significant reduction in length and weight compared to WT mice, but villus and crypt morphology remain to be investigated (Carnaghan et al., 2013). At the cellular level, ZO-1 staining confirmed epithelial cohesion but suggested that cell–cell contact morphology was perturbed in SCRIB KO cells compared with the parental cells (Fig. 1 I), possibly indicative of a role of SCRIB in the control of epithelial cell shape.
CRISPR D10A SCRIB exon 1 gene targeting and IOC quantifications. (A) CRISPR D10A targeted region in SCRIB exon 1 is represented with the sgRNA sequences 1 and 2 underlined in red and the PAM (Protospacer Adjacent Motif) boxed. (B) Genomic SCRIB exon 1 region was amplified by PCR and cloned by TA cloning. For each five clones, a total of 20 cloned PCR products were sequenced and the resulting genomic modifications are described. (C) Western blot on parental and SCRIB KO Caco-2 cell extracts stained with anti-SCRIB and anti-GAPDH antibodies. (D) Immunofluorescence on parental and SCRIB KO Caco-2 cells grown on glass for 3 d and stained with anti-SCRIB antibody. (E) Immunofluorescence on parental and SCRIB KO Caco-2 cells differentiated on filters for 21 d and stained with DAPI and anti-ZO-1 antibody. (F) Intestine-like tissues derived from parental (top) or SCRIB KO Caco-2 cells (bottom) grown on IOC system for 3, 7, and 10 d, dissected from the PDMS chips, stained with DAPI, and observed with confocal microscopy (XY and Z sections). (G) Quantification of the local surface ratio (surface of the folded gut divided by the corresponding plane surface) of intestine-like tissues derived from parental and SCRIB KO clones grown for 3 and 10 d on the IOC system. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments. (Mean WT 3 d: 1.640 versus mean KO 3 d: 1.671; mean WT 10 d: 1.970 versus mean KO 10 d:1.532). (H) Quantification of fluorescent dextran permeability assays at 2 and 12 h after EDTA treatment (0, 1, and 50 mM EDTA) on parental and SCRIB KO cells. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments. Source data are available for this figure: SourceData FS1.
CRISPR D10A SCRIB exon 1 gene targeting and IOC quantifications. (A) CRISPR D10A targeted region in SCRIB exon 1 is represented with the sgRNA sequences 1 and 2 underlined in red and the PAM (Protospacer Adjacent Motif) boxed. (B) Genomic SCRIB exon 1 region was amplified by PCR and cloned by TA cloning. For each five clones, a total of 20 cloned PCR products were sequenced and the resulting genomic modifications are described. (C) Western blot on parental and SCRIB KO Caco-2 cell extracts stained with anti-SCRIB and anti-GAPDH antibodies. (D) Immunofluorescence on parental and SCRIB KO Caco-2 cells grown on glass for 3 d and stained with anti-SCRIB antibody. (E) Immunofluorescence on parental and SCRIB KO Caco-2 cells differentiated on filters for 21 d and stained with DAPI and anti-ZO-1 antibody. (F) Intestine-like tissues derived from parental (top) or SCRIB KO Caco-2 cells (bottom) grown on IOC system for 3, 7, and 10 d, dissected from the PDMS chips, stained with DAPI, and observed with confocal microscopy (XY and Z sections). (G) Quantification of the local surface ratio (surface of the folded gut divided by the corresponding plane surface) of intestine-like tissues derived from parental and SCRIB KO clones grown for 3 and 10 d on the IOC system. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments. (Mean WT 3 d: 1.640 versus mean KO 3 d: 1.671; mean WT 10 d: 1.970 versus mean KO 10 d:1.532). (H) Quantification of fluorescent dextran permeability assays at 2 and 12 h after EDTA treatment (0, 1, and 50 mM EDTA) on parental and SCRIB KO cells. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments. Source data are available for this figure: SourceData FS1.
Parental and SCRIB KO Caco-2 cells growth on an IOC system. (A) IOC is made of a flexible silicone polymer (polydimethylsiloxane [PDMS]; left panel) in which cells are grown in the upper central channel under continuous perfusion (right panel). (B) Scanning electronic microscopy image of parental (left) and SCRIB KO (right) Caco-2 cells grown 7 d on the IOC system under continuous perfusion. (C) Higher magnifications of SEM images of parental (left) and mutant (right) intestine-like villi. (D) Intestine-like tissues were grown on IOC for 7 d, dissected from the PDMS chips, stained with DAPI, and observed with confocal microscopy. Quantification of folds was performed on central portions of the intestine strips as indicated by the white square. (E) Maximum intensity projection of confocal acquisition of parental (left) and mutant (right) intestine-like villi grown 7 d on the IOC system and stained with phalloidin and DAPI used for depth map quantification. (F) Z-section confocal images corresponding to the double arrow dotted lines shown in E. (G) Depth map of images presented in E. (H) Quantification of the local surface ratio (surface of the folded gut divided by the corresponding plane surface) intestine-like tissues derived from parental and SCRIB KO Caco-2 cells. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (means WT: 2.364, 2.043, and 2.297 versus means KO: 1.814, 1.839, and 1.859). (I) Maximum intensity projection of confocal acquisition of parental (top) and mutant (bottom) intestine-like villi stained with phalloidin/ZO1. Zooms indicated by the white squares (left) are shown on the right (ZO1 only).
Parental and SCRIB KO Caco-2 cells growth on an IOC system. (A) IOC is made of a flexible silicone polymer (polydimethylsiloxane [PDMS]; left panel) in which cells are grown in the upper central channel under continuous perfusion (right panel). (B) Scanning electronic microscopy image of parental (left) and SCRIB KO (right) Caco-2 cells grown 7 d on the IOC system under continuous perfusion. (C) Higher magnifications of SEM images of parental (left) and mutant (right) intestine-like villi. (D) Intestine-like tissues were grown on IOC for 7 d, dissected from the PDMS chips, stained with DAPI, and observed with confocal microscopy. Quantification of folds was performed on central portions of the intestine strips as indicated by the white square. (E) Maximum intensity projection of confocal acquisition of parental (left) and mutant (right) intestine-like villi grown 7 d on the IOC system and stained with phalloidin and DAPI used for depth map quantification. (F) Z-section confocal images corresponding to the double arrow dotted lines shown in E. (G) Depth map of images presented in E. (H) Quantification of the local surface ratio (surface of the folded gut divided by the corresponding plane surface) intestine-like tissues derived from parental and SCRIB KO Caco-2 cells. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (means WT: 2.364, 2.043, and 2.297 versus means KO: 1.814, 1.839, and 1.859). (I) Maximum intensity projection of confocal acquisition of parental (top) and mutant (bottom) intestine-like villi stained with phalloidin/ZO1. Zooms indicated by the white squares (left) are shown on the right (ZO1 only).
SCRIB loss alters cell morphology during epithelial differentiation in Caco-2 cells
To further examine the role of SCRIB in epithelial cell shape, the parental and SCRIB KO Caco-2 cell lines were grown to confluence on glass coverslips for 3 d (Fig. 2, A and B; left) or on polycarbonate transwell filters for 21 d to promote epithelial differentiation (Fig. 2, A and B; right). On glass, both cell types were indistinguishable by eye and automated cell segmentation, and cell morphology analysis (Etournay et al., 2016) revealed that the cell area of parental and SCRIB KO Caco-2 cells were in the same order of magnitude (285.9 ± 2.4 versus 274.7 ± 3.1 µm2 respectively, Fig. 2 C) as were their cell shape index (perimeter/√area; 4.5 versus 4.7 respectively, Fig. 2 D; Wang et al., 2020). During the 21 d of differentiation, parental Caco-2 cells progressively transitioned from a flat heterogeneous epithelium to a homogenous columnar epithelium (Fig. 2 E). Analysis of the apical cell area and the shape of parental cells using automated cell segmentation on large patches of the epithelial cell surface area revealed a strong decrease over the 3 wk of culture (261 ± 4 to 152 ± 1 µm2), suggesting an apical constriction associated with epithelial differentiation. In contrast, when cultivated in these conditions, SCRIB KO cells failed to undergo apical constriction and showed an extended apical area compared with glass-cultivated cells (265 ± 5 versus 363 ± 8 µm2; Fig. 2, B, C, and E; Fig. S1 E; and Fig. S2, B and C). They also displayed striking contour anomalies quantified by shape index measurement (Fig. 2 D and Fig. S2) reminiscent of but more pronounced than the abnormal cell contours observed in IOC (Fig. 1 I). Images acquired at different stages of epithelial growth showed that SCRIB KO cells developed defects early on with delayed nuclear condensation (day 5 WT versus day 9 KO) and E-cadherin recruitment (day 12 WT versus day 18 KO; Fig. S3 A). Trans-epithelial electrical resistance (TEER) measurement showed that both parental and KO cells displayed typical values (comprised between 600 and 1,000 Ω/cm2 [Hellinger et al., 2012]) after 21 d of differentiation, indicative of a functional epithelial tight junction barrier (Fig. 2 F). Phalloidin staining and scanning electron microscopy revealed the presence of an apical brush border with a high density of microvilli in both parental and SCRIB KO cells, showing that enterocytic differentiation was not impaired in SCRIB KO cells (Fig. 2 G). SCRIB has been previously implicated in the establishment of the basolateral identity of vertebrate epithelial cells, in collaboration with Lano and Erbin, each playing partially redundant roles (Choi et al., 2019). Optical sectioning along the Z-axis of differentiated cells stained with both tight (ZO-1) and E-cadherin adherens junctions showed that the SCRIB KO cells exhibit a reduction in the size of the lateral domain and an increase in the cross-sectional area that extends into the basal part of the cells (Fig. 2 H and Fig. S2 A). Analysis of the shape index along the Z-axis reveals a higher susceptibility to contour anomalies in the apical region of the cell (Fig. 2 I). To confirm the role of SCRIB in the control of the apical morphology during epithelial differentiation, we infected SCRIB KO cells with lentivirus expressing untagged SCRIB (Fig. 2 J). In contrast to neighboring SCRIB KO cells, clonal patches of SCRIB-positive cells, revealed by anti-SCRIB staining, displayed a shape index similar to WT cells (Fig. 2, K and L). Together, our results indicate that SCRIB loss of function strongly affected cell height, cell area, and apical cell contour during Caco-2 cell differentiation.
SCRIB KO results in cell shape and size defects in filter-grown Caco-2 cells. (A) Immunofluorescence images of Caco-2 cells parental (top panels) and SCRIB KO (bottom panels) cultivated 3 d on glass and stained with ZO-1. (B) Immunofluorescence images of Caco-2 cells parental (top panels) and SCRIB KO (bottom panels) cultivated 21 d on polycarbonate membranes filters and stained with ZO-1. (C) Scatter plot representation of the apical areas of Caco-2 cells cultivated in the indicated conditions. Single data points are represented in black or gray. Blue dots represent the mean of independent experiments. Red line marks the cumulated mean of three repeats (parental Caco-2 on glass: total mean = 285.9 µm2, total nb of cells = 4,431; parental on filters: mean = 156.8 µm2, n = 7,930; SCRIB KO on glass: mean = 274.7 µm2, n = 4,468, and SCRIB KO on filters: mean = 345.7 µm2, n = 2,985. For easier readability, areas above 1,000 and below 10 µm2 were excluded from the graphical representation (< 1% of total data). The P value from unpaired two-tailed t test statistics calculated from the mean of three repeats are indicated. (D) Scatter plot representation of the shape index areas of Caco-2 cells cultivated in the indicated conditions. Single data points are represented in black or gray. Blue dots represent the mean of independent experiments. The red line marks the cumulated mean of three repeats (parental Caco-2 on glass: total mean = 4.571, total nb of cells n = 4,508, parental on filters: mean = 4,556, n = 8,024, SCRIB KO on glass: mean = 4,784, n = 3,240, and SCRIB KO on filters: mean = 5,424 µm2, n = 3,151). For practicality, shape indexes above 10 were excluded from the graphical presentation (< 1%). The P value from unpaired two-tailed t test statistics calculated from the mean of three repeats are indicated. (E) Frequency distribution (in %) of the apical cell area of parental and SCRIB KO cells grown in different indicated conditions. (F) Transepithelial electrical resistance measurements were performed on 21-d filter-differentiated parental and SCRIB KO Caco-2 cells. The dashed red line indicates the upper and down limit between which Caco-2 cells TEER is known to oscillate (Hellinger et al., 2012). (G) Left panels, immunofluorescence of WT (top) and SCRIB KO (bottom) filter differentiated Caco-2 cells stained with phalloidin (magenta) and a ZO-1 (green). Right panels, the apical surface of WT (top) and SCRIB KO (bottom) filter differentiated Caco-2 epithelia detected by scanning electronic microscopy. (H) Confocal 2 µm optical sections from the apical to the basal sides of parental (top panels) and SCRIB KO Caco-2 cells (bottom panels) differentiated 21 d on filters, fixed, and stained with antibodies against ZO1 (green) and E-cadherin (red). (I) Left: Schematic representation of parental (top) and SCRIB KO (bottom) Caco-2 cells. Right: Graphical representation of the cell shape index as a function of the distance from the cell apex for both parental and SCRIB KO filter-differentiated Caco-2 cells. The cell shape index was calculated either from ZO-1 staining (green) or cadherin staining (red) acquired from successive 2 µm confocal optical sections. Each mean is statistically different (ANOVA, P < 0.0001, 2,210 WT cells, 675 SCRIB KO cells). Data distribution was assumed to be normal, but this was not formally tested. (J and K) SCRIB KO Caco-2 cells infected with WT SCRIB expressing lentiviruses and differentiated 21 d on filters. Anti-SCRIB immunostaining was used to detect SCRIB rescued cellular clones (red). ZO1 immunostaining (green) was used to detect apical cell shape. The right column shows ZO1 staining of cells KO outside (top and bottom) or inside (middle) the SCRIB rescued cell clonal patch. (L) Shape index quantification and comparison of neighbor SCRIB KO cells (left) bordering rescued WT SCRIB clonal cells patches (right). The P value from a paired two-tailed t test statistic calculated from nine points over three repeats (red, green, and blue) is indicated. Paired experiments are connected with dashed lines.
SCRIB KO results in cell shape and size defects in filter-grown Caco-2 cells. (A) Immunofluorescence images of Caco-2 cells parental (top panels) and SCRIB KO (bottom panels) cultivated 3 d on glass and stained with ZO-1. (B) Immunofluorescence images of Caco-2 cells parental (top panels) and SCRIB KO (bottom panels) cultivated 21 d on polycarbonate membranes filters and stained with ZO-1. (C) Scatter plot representation of the apical areas of Caco-2 cells cultivated in the indicated conditions. Single data points are represented in black or gray. Blue dots represent the mean of independent experiments. Red line marks the cumulated mean of three repeats (parental Caco-2 on glass: total mean = 285.9 µm2, total nb of cells = 4,431; parental on filters: mean = 156.8 µm2, n = 7,930; SCRIB KO on glass: mean = 274.7 µm2, n = 4,468, and SCRIB KO on filters: mean = 345.7 µm2, n = 2,985. For easier readability, areas above 1,000 and below 10 µm2 were excluded from the graphical representation (< 1% of total data). The P value from unpaired two-tailed t test statistics calculated from the mean of three repeats are indicated. (D) Scatter plot representation of the shape index areas of Caco-2 cells cultivated in the indicated conditions. Single data points are represented in black or gray. Blue dots represent the mean of independent experiments. The red line marks the cumulated mean of three repeats (parental Caco-2 on glass: total mean = 4.571, total nb of cells n = 4,508, parental on filters: mean = 4,556, n = 8,024, SCRIB KO on glass: mean = 4,784, n = 3,240, and SCRIB KO on filters: mean = 5,424 µm2, n = 3,151). For practicality, shape indexes above 10 were excluded from the graphical presentation (< 1%). The P value from unpaired two-tailed t test statistics calculated from the mean of three repeats are indicated. (E) Frequency distribution (in %) of the apical cell area of parental and SCRIB KO cells grown in different indicated conditions. (F) Transepithelial electrical resistance measurements were performed on 21-d filter-differentiated parental and SCRIB KO Caco-2 cells. The dashed red line indicates the upper and down limit between which Caco-2 cells TEER is known to oscillate (Hellinger et al., 2012). (G) Left panels, immunofluorescence of WT (top) and SCRIB KO (bottom) filter differentiated Caco-2 cells stained with phalloidin (magenta) and a ZO-1 (green). Right panels, the apical surface of WT (top) and SCRIB KO (bottom) filter differentiated Caco-2 epithelia detected by scanning electronic microscopy. (H) Confocal 2 µm optical sections from the apical to the basal sides of parental (top panels) and SCRIB KO Caco-2 cells (bottom panels) differentiated 21 d on filters, fixed, and stained with antibodies against ZO1 (green) and E-cadherin (red). (I) Left: Schematic representation of parental (top) and SCRIB KO (bottom) Caco-2 cells. Right: Graphical representation of the cell shape index as a function of the distance from the cell apex for both parental and SCRIB KO filter-differentiated Caco-2 cells. The cell shape index was calculated either from ZO-1 staining (green) or cadherin staining (red) acquired from successive 2 µm confocal optical sections. Each mean is statistically different (ANOVA, P < 0.0001, 2,210 WT cells, 675 SCRIB KO cells). Data distribution was assumed to be normal, but this was not formally tested. (J and K) SCRIB KO Caco-2 cells infected with WT SCRIB expressing lentiviruses and differentiated 21 d on filters. Anti-SCRIB immunostaining was used to detect SCRIB rescued cellular clones (red). ZO1 immunostaining (green) was used to detect apical cell shape. The right column shows ZO1 staining of cells KO outside (top and bottom) or inside (middle) the SCRIB rescued cell clonal patch. (L) Shape index quantification and comparison of neighbor SCRIB KO cells (left) bordering rescued WT SCRIB clonal cells patches (right). The P value from a paired two-tailed t test statistic calculated from nine points over three repeats (red, green, and blue) is indicated. Paired experiments are connected with dashed lines.
Z sections and workflow of data acquisition and quantification. (A) Top: Confocal acquisition of parental, SCRIB KO1, and 2 Caco-2 cells grown of filters for 21 d and stained with anti-ZO1 (green), E-cadherin (red), phalloidin (magenta), and DAPI. Bottom: Z projections follow the dashed line. (B) Parental and SCRIB KO1 and two Caco-2 cells grown on filters for 21 d and stained with anti-ZO1 antibody. Confocal acquisition of 25 images was taken with a 40× objective and assembled into a single square panel using 20% overlap. Cell perimeters and area were extracted based on cell segmentation using the TissueMiner program (Etournay et al., 2016). (C) Scatter plot representation of the apical areas of parental and SCRIB KO Caco-2 cell lines 1 and 2 (parental Caco-2: mean = 156.9 µm2, n = 7,930; SCRIB KO1: mean = 345.7 µm2, n = 2,985; SCRIB KO2: mean = 368.1 µm2, n = 1,542). The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (blue dots). (D) Histograms of shape index of parental and SCRIB KO 1 and 2 Caco-2 cells. Parental Caco-2: mean = 4.601, n = 10,223; SCRIB KO1: mean = 5.310, n = 3,704; SCRIB KO2: mean = 5.764, n = 2,484). The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (blue dots).
Z sections and workflow of data acquisition and quantification. (A) Top: Confocal acquisition of parental, SCRIB KO1, and 2 Caco-2 cells grown of filters for 21 d and stained with anti-ZO1 (green), E-cadherin (red), phalloidin (magenta), and DAPI. Bottom: Z projections follow the dashed line. (B) Parental and SCRIB KO1 and two Caco-2 cells grown on filters for 21 d and stained with anti-ZO1 antibody. Confocal acquisition of 25 images was taken with a 40× objective and assembled into a single square panel using 20% overlap. Cell perimeters and area were extracted based on cell segmentation using the TissueMiner program (Etournay et al., 2016). (C) Scatter plot representation of the apical areas of parental and SCRIB KO Caco-2 cell lines 1 and 2 (parental Caco-2: mean = 156.9 µm2, n = 7,930; SCRIB KO1: mean = 345.7 µm2, n = 2,985; SCRIB KO2: mean = 368.1 µm2, n = 1,542). The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (blue dots). (D) Histograms of shape index of parental and SCRIB KO 1 and 2 Caco-2 cells. Parental Caco-2: mean = 4.601, n = 10,223; SCRIB KO1: mean = 5.310, n = 3,704; SCRIB KO2: mean = 5.764, n = 2,484). The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (blue dots).
Overtime growth of parental and SCRIB KO cells and schematic representation of real-time biacore experiments. (A) Immunofluorescence analysis of parental and SCRIB KO cells using anti-ZO1 (lanes 1 and 4), anti-E-cadherin antibodies (lanes 2 and 5), and DAPI staining (lanes 3 and 6) at 2, 5, 9, 12, 15, 18, and 21 d of differentiation on filters. (B) SCRIB on SHROOM. The GFP-tagged SHROOM ASD2 domain was captured on a Biacore sensorchip surface using anti-GFP nanobodies (steps 1 and 2). After PBS wash, 50 nM of the SCRIB C-terminal fragment (1,478–1,630) was injected over the functionalized surface in a constant buffer flow (steps 3 and 4). (C) SCRIB on SHROOM/ROCK. The GFP-tagged SHROOM ASD2 domain was captured on a Biacore sensorchip surface using anti-GFP nanobodies (steps 1 and 2). After PBS wash, 5 µM of the ROCK SBD domain (834–913) was injected over the functionalized surface in a constant buffer flow (steps 3 and 4). After PBS wash, 50 nM of SCRIB C-terminal fragment (1,478–1,630) was injected over the SHROOM/ROCK surface in a constant buffer flow (steps 5 and 6).
Overtime growth of parental and SCRIB KO cells and schematic representation of real-time biacore experiments. (A) Immunofluorescence analysis of parental and SCRIB KO cells using anti-ZO1 (lanes 1 and 4), anti-E-cadherin antibodies (lanes 2 and 5), and DAPI staining (lanes 3 and 6) at 2, 5, 9, 12, 15, 18, and 21 d of differentiation on filters. (B) SCRIB on SHROOM. The GFP-tagged SHROOM ASD2 domain was captured on a Biacore sensorchip surface using anti-GFP nanobodies (steps 1 and 2). After PBS wash, 50 nM of the SCRIB C-terminal fragment (1,478–1,630) was injected over the functionalized surface in a constant buffer flow (steps 3 and 4). (C) SCRIB on SHROOM/ROCK. The GFP-tagged SHROOM ASD2 domain was captured on a Biacore sensorchip surface using anti-GFP nanobodies (steps 1 and 2). After PBS wash, 5 µM of the ROCK SBD domain (834–913) was injected over the functionalized surface in a constant buffer flow (steps 3 and 4). After PBS wash, 50 nM of SCRIB C-terminal fragment (1,478–1,630) was injected over the SHROOM/ROCK surface in a constant buffer flow (steps 5 and 6).
SCRIB depletion disrupts apicobasal myosin polarization and alters junctional proteins in Caco-2 cells
The contractile actomyosin network controls apical constriction across the apical cell surface and plays an important role in tissue folding during development (Martin and Goldstein, 2014). Thus, we examined the myosin expression pattern in Caco-2 cells stably expressing GFP-tagged non-sarcomeric myosin regulatory light chain MRLC (also known as myosin regulatory light chain 2A). After differentiation on the filter, the parental Caco-2 showed an enrichment of myosin light chain at the apical cell surface and along cell–cell contacts (Fig. 3 A), as previously described (Wayt et al., 2021). In SCRIB KO cells, MRLC apical enrichment was reduced, and instead, strong MRLC accumulation could be detected at the basal cell side (Fig. 3 B). We then assessed myosin activity using anti-phospho-myosin S19 light chain antibody (Fig. 3 C). Parental Caco-2 cells displayed strong enrichment of activated MRLC at the apical side of the cells (Fig. 3, D and F). In contrast, SCRIB KO cells displayed similar p-MRLC staining at the apical and basal poles of the cell (Fig. 3, C, E, and F). Altogether, these results show that SCRIB depletion in Caco-2 cells perturbs the apicobasal polarization of myosin light chain and points toward the role of SCRIB in the polarized control of actomyosin contractility. Given that previous studies have demonstrated SCRIB’s role in stabilizing the interaction between E-cadherin and the catenins (Qin et al., 2005; Awadia et al., 2019) and the essential requirement of E-cadherin for the appropriate localization of myosin2 (Shewan et al., 2005), it is reasonable to speculate that the loss of SCRIB disrupts myosin recruitment throughout epithelial cell contacts.
Myosin light chain localization and activation in SCRIB KO cells. (A) Parental Caco-2 cells stably expressing myosin light chain MYL12A tagged with GFP and filter differentiated for 21 d were analyzed by confocal microscopy. Apical plane (left), basal plane (right), and Z-transversal sections (bottom) are represented. (B) SCRIB KO Caco-2 cells stably expressing myosin light chain MYL12A tagged with GFP and filter-differentiated for 21 d were analyzed by live confocal microscopy. Apical plane (left), basal plane (right), and transversal sections (bottom) are represented. The white arrowhead indicates basal myosin accumulation. (C) Filter-differentiated parental Caco-2 cells (top) and SCRIB KO (bottom) stained with ZO1 (left) and anti-Phospho S19 activated myosin light chain (apical plan, center panel, and basal plan, right panel). (D) Fluorescence intensity quantification of Myosin S19 (red) and Dapi (blue) along the Z axis of filter-differentiated parental Caco-2 cells. Error bars correspond to standard deviations. The graph is the average of 10 measurements. Apical and basal myosin S19 fluorescence intensity maxima are indicated. (E) Fluorescence intensity quantification of Myosin S19 (red) and Dapi (blue) along the Z axis of filter-differentiated SCRIB KO cells. Error bars correspond to standard deviations. The graph is the average of 10 measurements. (F) Basal/apical relative fluorescence myosin S19 ratio of parental and SCRIB KO cells. The P value represents the result of an unpaired two-tailed t test done on the mean of four independent experiments (red, pink, yellow, and red), each of them being the result of 10 measurements (Parental Caco-2 means: 0.9644, 0.6647, 0.5191, and 0.3406; SCRIB KO means: 1.1540, 1.037, 1.007, and 0.8906). (G) Immunostaining with anti-ZO1 (left) and anti-SHROOM2 of parental Caco-2 (top) and SCRIB KO cells (bottom). (H) Quantification of SHROOM2 intensity picks at cell–cell junctions. The results have been obtained on 3 × 15 cells. Bar is 20 µm.
Myosin light chain localization and activation in SCRIB KO cells. (A) Parental Caco-2 cells stably expressing myosin light chain MYL12A tagged with GFP and filter differentiated for 21 d were analyzed by confocal microscopy. Apical plane (left), basal plane (right), and Z-transversal sections (bottom) are represented. (B) SCRIB KO Caco-2 cells stably expressing myosin light chain MYL12A tagged with GFP and filter-differentiated for 21 d were analyzed by live confocal microscopy. Apical plane (left), basal plane (right), and transversal sections (bottom) are represented. The white arrowhead indicates basal myosin accumulation. (C) Filter-differentiated parental Caco-2 cells (top) and SCRIB KO (bottom) stained with ZO1 (left) and anti-Phospho S19 activated myosin light chain (apical plan, center panel, and basal plan, right panel). (D) Fluorescence intensity quantification of Myosin S19 (red) and Dapi (blue) along the Z axis of filter-differentiated parental Caco-2 cells. Error bars correspond to standard deviations. The graph is the average of 10 measurements. Apical and basal myosin S19 fluorescence intensity maxima are indicated. (E) Fluorescence intensity quantification of Myosin S19 (red) and Dapi (blue) along the Z axis of filter-differentiated SCRIB KO cells. Error bars correspond to standard deviations. The graph is the average of 10 measurements. (F) Basal/apical relative fluorescence myosin S19 ratio of parental and SCRIB KO cells. The P value represents the result of an unpaired two-tailed t test done on the mean of four independent experiments (red, pink, yellow, and red), each of them being the result of 10 measurements (Parental Caco-2 means: 0.9644, 0.6647, 0.5191, and 0.3406; SCRIB KO means: 1.1540, 1.037, 1.007, and 0.8906). (G) Immunostaining with anti-ZO1 (left) and anti-SHROOM2 of parental Caco-2 (top) and SCRIB KO cells (bottom). (H) Quantification of SHROOM2 intensity picks at cell–cell junctions. The results have been obtained on 3 × 15 cells. Bar is 20 µm.
In addition to its function at the zonula adherens, SCRIB also interacts with proteins associated with tight junctions. SCRIB directly binds to the tight junction protein ZO-2 that forms a heterodimer with ZO-1 (Métais et al., 2005). In MDCK cells, ZO1 overexpression induces “zigzag” junctions that phenocopy the ones observed in the present study with SCRIB loss of function (Fanning et al., 2012; Tokuda et al., 2014). Additionally, SCRIB was shown to interact directly with SHROOM2 (Amano et al., 2015), an adaptor protein involved in actomyosin regulation and epithelial morphogenesis (Hildebrand, 2005; Hildebrand and Soriano, 1999). Interestingly, SHROOM2 has been shown to bind directly to ZO-1 (Etournay et al., 2007). To test if SHROOM2 localization was impacted by SCRIB KO, we performed immunostaining of endogenous SHROOM2 in parental and SCRIB KO cells. While parental cells show SHROOM2 localization at cell–cell contacts, this recruitment was reduced in the KO cells as shown by the quantification of intensity picks at the cell–cell junction (Fig. 3, G and H).
Characterization of the evolutionarily conserved WRAAR box in SCRIB interaction with SHROOM2
SCRIB C-terminal fragment (aa 1,178–1,488) has been shown to interact with SHROOM2 ASD2 domain (Amano et al., 2015). In turn, the ASD2 domain of SHROOM2 directly binds to Rho-associated coiled-coil kinase (ROCK1) SBD domain and stimulates its kinase activity in vitro (Mohan et al., 2013; Zalewski et al., 2016; Fig. 4 A). After confirming the direct binding of the SCRIB C-terminal part with the SHROOM2 ASD2 domain (aa 1,178–1,488), we further characterized this complex using a Biacore assay. GFP-tagged SHROOM ASD2 domain was produced in HEK/293T cells and subsequently captured on a Biacore sensorchip surface using anti-GFP nanobodies. Analytes (purified GST-tagged SCRIB and ROCK protein domains, Fig. 4 B) were injected over the functionalized surface in a constant buffer flow, and the interaction between domains was monitored in real-time by measuring surface plasmon resonance variations. The binding affinity of SHROOM2 for ROCK1 was of the same order of magnitude as that of SHROOM2 for SCRIB (1 ± 0.15 and 0.96 ± 0.37 µM, respectively; Fig. 4, C and D). No binding could be detected between SCRIB and ROCK1 in the absence of SHROOM2 (Fig. 4 D). We then used the Biacore system to compare the binding behavior of the SCRIB C-terminal domain on the SHROOM2 domain alone or on the preformed SHROOM2:ROCK1 complex (Fig. S3, B and C). These experiments showed that the affinity of SCRIB for the preformed SHROOM2:ROCK1 complex was nearly five times higher than that for ROCK1 alone (Fig. 4, E and F), suggesting the existence of a positive binding cooperativity between the three protein domains. To identify the SCRIB sequence necessary for its interaction with SHROOM, we looked for an evolutionary conserved sequence in the carboxy-terminal region of SCRIB. The protein sequence homology between humans and Drosophila SCRIB/scribble C-terminal regions is poor (Boëda and Etienne-Manneville, 2015). However, a more detailed comparison revealed a small island of conservation surrounding human SCRIB serine 1508 in the corresponding Drosophila sequence (93% similarity over 29 aa; Fig. 4 G). Further phylogenetic analysis showed that this 29 aa amino-acid peptide sequence, that we named the WRAAR box, is an evolutionary conserved motif present in all Metazoan SCRIB genes (Fig. 4 G). Search in the human protein database (Swiss prot) for the WRAAR motif sequence shows that SCRIB is the only protein displaying this short linear motif in the human genome. Interestingly Alpha fold structure database predicts that the WRAAR motif adopts a small alpha helix structure (Fig. 4 G). Of note, the Drosophila WRAAR motif in scribble is located on the alternative exon 24, which is present in only 8 out of the 18 alternative scribble transcripts (according to http://flybase.org). In contrast, the WRAAR motif in human SCRIB is encoded by the ubiquitous exons 32 and 33 and therefore present in all SCRIB transcripts. Deletion of the WRAAR box totally abolished the binding of the SCRIB C-terminal fragment to the SHROOM2 ASD2 domain both in GST pull-down and in Biacore assays (Fig. 4, H and I). Moreover, the WRAAR Box alone fused to GST was able to bind to the SHROOM2 ASD2 domain (Fig. 4 H). To test if SCRIB WRAAR box was able to recruit SHROOM2 ASD2 domain, we artificially targeted SCRIB WT and delta WRAAR C-terminal fragment to the outer mitochondrial membrane of Hela cells using the TOM5 anchor (Postigo et al., 2006). In these cells, we observed that the WT SCRIB sequence triggered the relocation of the ASD2 domain, whereas the mutant delta WRAAR box did not (Fig. 4 J). In a complementary approach, SCRIB KO cells were either rescued by WT full-length SCRIB or by a delta WRAAR mutant. While cells rescued with WT sequence display SHROOM2 immunostaining at the cell junctions, KO cells transfected with the delta WRAAR mutant did not (Fig. 4 K). Altogether these results suggest that the WRAAR box encodes an evolutionary conserved SHROOM2 binding domain. Interestingly, the WRAAR box includes the serine 1508 which has been shown to be phosphorylated by ROCK1 (Amano et al., 2015). Since in our experiments the SCRIB C-terminal fragment containing the WRAAR motif was produced in bacteria, serine 1508 phosphorylation is unlikely a prerequisite for SHROOM binding. Nevertheless, the phosphorylation of SCRIB may influence the formation of the complex in cells. While the similarity of the SCRIB WRAAR sequence between humans, anemones, and corals is high (86% and 93%, respectively), phylogenetic analysis shows that the serine 1508 is not conserved in these cnidarian sequences (Fig. 4 G), suggesting that the acquisition of the SHROOM binding motif evolutionary predated a potential control by ROCK phosphorylation. One hypothesis is that the phosphorylation of SCRIB by ROCK is part of a feedback loop controlling the stability of the SCRIB-SHROOM-ROCK tripartite complex, acquired secondarily during evolution.
SHROOM2/ROCK complex and SCRIB WRAAR box. (A) Schematic representation of the SCRIB, SHROOM2, and ROCK1 proteins with their respective interacting binding domains. (B) Coomassie staining of the indicated protein domains produced as GST fusion proteins in bacteria and used as analytes in Biacore studies. (C) Different binding affinity values obtained for the ROCK1:SHROOM2 (mean 1 ± 0.15 µm) and SCRIB:SHROOM2 (mean 0.96 ± 0.37 µm) protein domain interactions in the Biacore SPR analysis. (D) Schematic representation of the average binding affinity values between each domain measured in this study. (E) Real-time SPR response variations upon injection of three different concentrations of the SCRIB protein fragment (12.5, 25, and 50 nM) over a chip functionalized with the SHROOM2 fragment. A round of binding, wash, and dissociation was recorded for each condition. (F) Real-time SPR response variations upon injection of three different concentrations of the SCRIB protein fragment (12.5, 25, and 50 nM) over a chip functionalized with the SHROOM2: ROCK1 protein complex. A round of binding, wash, and dissociation was recorded for each condition. Each sensorgram is shown after subtraction of the response measured for the same injections on a control surface functionalized with GFP alone. (G) Top: Graphical representation of the protein identity comparison between human SCRIB and Drosophila Scribble protein sequences. (bottom) Sequence alignment of various SCRIB sequences showing WRAAR box sequence conservation during animal evolution. Red residues correspond to amino acids conserved in the human sequence. The S1508 ROCK1 phosphorylation site is indicated. (H) Top: Graphical representation of the different SCRIB C-terminal sequences used in the GST pull-down to map the SHROOM binding site. Bottom: Pull down assay on the HEK 293 cell lysate overexpressing GFP ASD2 domain of SHROOM2 with the indicated GST fusion protein used as affinity resins. Samples were analyzed by Ponceau staining and immunoblotting using anti-GFP. (I) Biacore SPR profile showing a round of binding, wash, and dissociation upon injection of SCRIB WT (solid line) and WRAAR box (dashed line) fragments over the SHROOM ASD2 domain tethered on the chip surface. (J) Hela cells were transfected with the indicated GFP-SCRIB and Myc-SHROOM2 construct. (K) SCRIB KO Caco-2 cells infected with WT SCRIB expressing lentiviruses (top) or Delta WRAAR mutant (bottom) and stained with anti-SCRIB (left) or anti-SHROOM2. Source data are available for this figure: SourceData F4.
SHROOM2/ROCK complex and SCRIB WRAAR box. (A) Schematic representation of the SCRIB, SHROOM2, and ROCK1 proteins with their respective interacting binding domains. (B) Coomassie staining of the indicated protein domains produced as GST fusion proteins in bacteria and used as analytes in Biacore studies. (C) Different binding affinity values obtained for the ROCK1:SHROOM2 (mean 1 ± 0.15 µm) and SCRIB:SHROOM2 (mean 0.96 ± 0.37 µm) protein domain interactions in the Biacore SPR analysis. (D) Schematic representation of the average binding affinity values between each domain measured in this study. (E) Real-time SPR response variations upon injection of three different concentrations of the SCRIB protein fragment (12.5, 25, and 50 nM) over a chip functionalized with the SHROOM2 fragment. A round of binding, wash, and dissociation was recorded for each condition. (F) Real-time SPR response variations upon injection of three different concentrations of the SCRIB protein fragment (12.5, 25, and 50 nM) over a chip functionalized with the SHROOM2: ROCK1 protein complex. A round of binding, wash, and dissociation was recorded for each condition. Each sensorgram is shown after subtraction of the response measured for the same injections on a control surface functionalized with GFP alone. (G) Top: Graphical representation of the protein identity comparison between human SCRIB and Drosophila Scribble protein sequences. (bottom) Sequence alignment of various SCRIB sequences showing WRAAR box sequence conservation during animal evolution. Red residues correspond to amino acids conserved in the human sequence. The S1508 ROCK1 phosphorylation site is indicated. (H) Top: Graphical representation of the different SCRIB C-terminal sequences used in the GST pull-down to map the SHROOM binding site. Bottom: Pull down assay on the HEK 293 cell lysate overexpressing GFP ASD2 domain of SHROOM2 with the indicated GST fusion protein used as affinity resins. Samples were analyzed by Ponceau staining and immunoblotting using anti-GFP. (I) Biacore SPR profile showing a round of binding, wash, and dissociation upon injection of SCRIB WT (solid line) and WRAAR box (dashed line) fragments over the SHROOM ASD2 domain tethered on the chip surface. (J) Hela cells were transfected with the indicated GFP-SCRIB and Myc-SHROOM2 construct. (K) SCRIB KO Caco-2 cells infected with WT SCRIB expressing lentiviruses (top) or Delta WRAAR mutant (bottom) and stained with anti-SCRIB (left) or anti-SHROOM2. Source data are available for this figure: SourceData F4.
Role of the WRAAR box in SCRIB’s selective binding to SHROOM2 and SHROOM4 in controlling apical cell shape
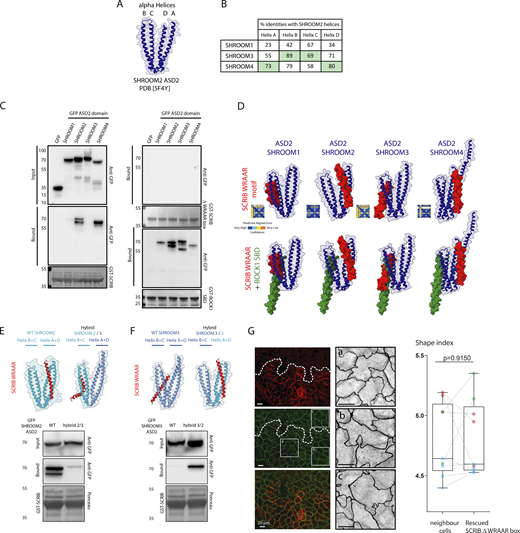
The SHROOM protein family consists of four members, each encoding ASD2 domains (Fig. 5 A; Hagens et al., 2006). The ASD2 domain is comprised of four helices (A–D) that come together to form two coiled-coil segments. The first coiled-coil segment contains helices A and D, while the second segment consists of helices B and C (Fig. 5 B; Zalewski et al., 2016). By comparing the sequences of ASD2 domains, we found that helices A and D of the SHROOM2 ASD2 domain share a higher percentage of identity with SHROOM4, whereas helices B and C show a closer resemblance to SHROOM3. When the C-terminal fragment of SCRIB (amino acids 1,178–1,488) is fused with GST, it can successfully capture the GFP-tagged ASD2 domain from SHROOM2 and SHROOM4, but not from SHROOM1 and SHROOM3 (Fig. 5 C). While no binding was observed with GST-SCRIB delta WRAAR box, all four ASD2 domains were pulled down using the ROCK1 SBD domain fused to GST. To understand the molecular basis for SCRIB’s binding selectivity toward SHROOM2 and SHROOM4 but not SHROOM1 and SHROOM3, we used an alphafold-based structural simulation (Mirdita et al., 2022). The simulation predicts with high confidence that the conserved alpha helices A and D of the ASD2 domain of SHROOM2 and 4 directly interact with the WRAAR alpha helix of SCRIB (Fig. 5 D). Interestingly, this binding is compatible with the ROCK1 SBD domain, which has been shown to occur on the opposing alpha helices B and C. On the other hand, the binding prediction between SHROOM1 and 3 and SCRIB’s WRAAR helix is of low confidence and conflicts with ROCK1 binding (Fig. 5 D). To validate the predicted model, we swapped the helices A and D of SHROOM2 with those of SHROOM3. The introduction of SHROOM3 helices into the SHROOM2 ASD2 domain suppressed its ability to bind to GST-SCRIB Cter resin (Fig. 5 E). Conversely, swapping helices A and D of SHROOM3 with those of SHROOM2 was sufficient to induce SCRIB binding (Fig. 5 F). Altogether, these results show that the WRAAR box sequence binds both SHROOM2 and SHROOM4 ASD2 domains at the level of the coil–coil segment formed by A and D helices. Finally, we assessed the functional role of the WRAAR box by performing rescue experiments infecting KO SCRIB cells with either SCRIB WT or SCRIB delta WRAAR box encoding lentiviruses. After 3 wk of growth on transwell filters, we measured the shape index of rescued cell patches and compared it to the shape index of non-rescued neighboring cells. In contrast to WT-SCRIB, the SCRIB delta WRAAR construct was unable to rescue significantly the shape index of SCRIB KO cells (Fig. 5 G). Altogether, these results show that the SHROOM2/4 binding site encoded by the WRAAR box sequence is an essential module participating in SCRIB control of apical cell shape in Caco-2 cells.
SCRIB binds to SHROOM4. (A) Graphical representation of SHROOM2 ASD2 domain with its four alpha helices A–D. (B) Table displaying the sequence identity between helices A–D in the ASD2 domain of SHROOM2 and the sequences from the ASD2 domains of SHROOM1, SHROOM3, and SHROOM4. (C) GST pull-down assay using GST SCRIB Cter WT, GST SCRIB Cter delta WRAAR, and GST ROCK1 SBD domain from Hek 293 expressing GFP-tagged ASD2 domain from SHROOM1, 2, 3, and 4. Samples were analyzed by Ponceau staining and immunoblotting using anti-GFP. (D) Top: Colabfold protein complex prediction of SCRIB WRAAR alpha helix (red) with ASD2 domain (blue) from SHROOM1, 2, 3, and 4, respectively. Bottom: Colabfold protein prediction of tripartite SCRIB/SHROOM/ROCK1 complex (green: ROCK1 SBD domain). (E and F) GST pull-down assay using GST SCRIB Cter WT from Hek 293 expressing GFP SHROOM2 and SHROOM3 WT and hybrids indicated ASD2 domains. Samples were analyzed as indicated. (G) Right: SCRIB KO Caco-2 cells were infected with lentiviruses expressing WRAAR box SCRIB mutant and differentiated over filters for 21 d. Cells were further stained with anti-ZO1 (green and gray) and anti-SCRIB (red) to identify rescued patches of cells. Left: Shape-index quantification and comparison of neighbor SCRIB KO cells (left) bordering rescued SCRIB WRAAR box clonal cells patches. The P value from a paired two-tailed t test statistic calculated from nine points over three repeats (red, green, and blue) is indicated. Paired experiments are connected with dashed lines. Source data are available for this figure: SourceData F5.
SCRIB binds to SHROOM4. (A) Graphical representation of SHROOM2 ASD2 domain with its four alpha helices A–D. (B) Table displaying the sequence identity between helices A–D in the ASD2 domain of SHROOM2 and the sequences from the ASD2 domains of SHROOM1, SHROOM3, and SHROOM4. (C) GST pull-down assay using GST SCRIB Cter WT, GST SCRIB Cter delta WRAAR, and GST ROCK1 SBD domain from Hek 293 expressing GFP-tagged ASD2 domain from SHROOM1, 2, 3, and 4. Samples were analyzed by Ponceau staining and immunoblotting using anti-GFP. (D) Top: Colabfold protein complex prediction of SCRIB WRAAR alpha helix (red) with ASD2 domain (blue) from SHROOM1, 2, 3, and 4, respectively. Bottom: Colabfold protein prediction of tripartite SCRIB/SHROOM/ROCK1 complex (green: ROCK1 SBD domain). (E and F) GST pull-down assay using GST SCRIB Cter WT from Hek 293 expressing GFP SHROOM2 and SHROOM3 WT and hybrids indicated ASD2 domains. Samples were analyzed as indicated. (G) Right: SCRIB KO Caco-2 cells were infected with lentiviruses expressing WRAAR box SCRIB mutant and differentiated over filters for 21 d. Cells were further stained with anti-ZO1 (green and gray) and anti-SCRIB (red) to identify rescued patches of cells. Left: Shape-index quantification and comparison of neighbor SCRIB KO cells (left) bordering rescued SCRIB WRAAR box clonal cells patches. The P value from a paired two-tailed t test statistic calculated from nine points over three repeats (red, green, and blue) is indicated. Paired experiments are connected with dashed lines. Source data are available for this figure: SourceData F5.
In conclusion, we show that SCRIB plays a crucial role in regulating cell and tissue morphology during Caco-2 intestinal epithelial cell differentiation. SCRIB deficiency disrupts the apicobasal myosin polarization and affects key junctional proteins. Our study also uncovers the importance of the WRAAR box, a conserved motif mediating SCRIB’s interaction with SHROOM2 and SHROOM4, which proved essential for maintaining apical cell shape. We previously showed that the C-terminal part of SCRIB contained, additionally, to the WRAAR box, three spectrin binding motifs, named SADH motifs (Spectrin-Ablim-DMTN Homology), which are crucial for SCRIB cortical dynamics and polarity function (Boëda and Etienne-Manneville, 2015). This study highlights that the C-terminal region of SCRIB is specialized in hosting multiple compact peptide motifs interacting with and influencing the actomyosin cytoskeleton.
Material and methods
Cell lines
Caco-2 cells were obtained from ATCC. For the generation of Caco-2 knock-out cells and to work with a homogenous cell population, a parental clone that displayed a characteristic “Chicken wire”-like ZO-1 pattern when grown on filters for 21 d was preselected as the founder of the parental population. This parental cell line was then used to perform the CRISPR knock-down experiments. Caco-2 cells and Hek/293T (ATCC) were grown in Dulbecco’s Modified Eagle Medium (DMEM + GlutaMAX, 4.5 g/liter D-Glucose; Gibco) supplemented with 10% FBS (fetal bovine serum; Eurobio) and penicillin/streptomycin (Gibco) in 5% CO2 at 37°C.
CRISPR cell lines
SgRNA targeting exon1 of the SCRIB human gene (SgRNA 1: 5′-CAGCGGGATGCACTTGAGCA-3′ and SgRNA 2: 5′-CTGCAACCGGCACGTGGAGT-3′) were designed using the CRISPOR software (Concordet and Haeussler, 2018) to perform the double-nicking strategy. Double-stranded oligos with overhangs were cloned into pMLM3636 vector using BsmBI sites. Lonza transfection kit SE (program B-024) was used to transfect 5 × 105 Caco-2 cells with 2 µg of each pMLM3636 encoding SgRNA 1 and 2 combined with 2 µg of Cas9 D10A encoding plasmid. Single cells were individually seeded in 96-well plates 48 h after transfection, amplified, and further assessed by Western blot for SCRIB depletion using SANTA CRUZ sc-11049 (C-20) antibody and anti-GAPDH (6C5; cat#MAB374; Sigma-Aldrich) for loading control. Genomic DNA from five negative SCRIB clones was extracted (E.Z.N.A Tissue DNA Kit; VWR) and SCRIB exon1 targeted region was amplified by PCR using primers 5′-GAGGAATCACGGGCTGGG-3′ and 5′-CGCGCTTCCTAAGAGTCTGA-3′ (Q5 High-Fidelity DNA Polymerase; NEB). PCR products were subcloned into pCR2.1 using Zero Blunt TOPO PCR cloning kit (Invitrogen). For each CRISPR clone, a minimum of 10 cloned PCR products were sequenced to determine the genomic modification of the different alleles of the targeted region.
Antibodies, plasmids, and protein production
The following antibodies were used in this study: mouse anti-ZO1 (cat#610967; BD Transduction), goat anti-SCRIB (sc-11049 [C-20]; Santa Cruz), anti-mouse phospho-myosin light chain 2 (Ser19; cat#3675; Cell Signalling), and anti-SHROOM2 (cat# HPA05 1646; Sigma-Aldrich). Mus musculus myosin regulatory non-sarcomeric light chain 12A (Myl12a) cDNA was cloned into pLL3.7 lentivirus in fusion with the GFP gene using Gibson Assembly Cloning Kit (NEB). Tagless full-length human SCRIB was cloned into pLL3.7 lentivirus under the control of the CMV promoter after removing the GFP gene. Δ WRAAR motif deletion on pLL3.7 SCRIB vector was performed using the Q5 site-directed mutagenesis kit (NEB). Human cDNA fragments encoding the SCRIB C-terminal domain (1,478–1,630 aa), the SHROOM ASD2 domain 1 (473–852), 2 (mouse protein 1,178–1,488 aa), 3 (1,648–1,997), 4 (1,203–1,493), and the ROCK1 SBD domain (834–913 aa) were amplified by PCR or synthesized by eurofins and cloned into the pMW172-GST vector for bacterial expression and into CB6-GFP for eukaryotic expression. ASD2 hybrids were designed by swapping helices A (1,301–1,351 aa) and D (1,440–1,481 aa) of SHROOM2 with helices A (1,772–1,826 aa) and D (1,915–1,956 aa) of SHROOM3. The sequence from TOM5 protein (KQAAYVAAFLWVSPMIWHLVKKQWK) was fused to the C-terminal part of SCRIB to trigger mitochondria targeting. All GST proteins were expressed in E. coli BL21(DE3) Rosetta strain. Bacterial cell pellets were lysed for 1 h at RT in 150 mM NaCl, 50 mM Tris pH 8, and 25% sucrose supplemented with 5,000 U of lysozyme (Sigma-Aldrich). Cleared supernatants were mixed at 4°C for 90 min with glutathione-sepharose 4B beads (GE Healthcare). The resulting resins were washed three times with PBS containing 200 mM NaCl and 0.1% Triton. Proteins were eluted in 50 mM Tris-Base pH 8.0 buffer supplemented with 100 mM reduced glutathione and then dialyzed overnight against PBS using Slide-A-Lyzer cassettes (Pierce).
IOC
Intestine chips were produced by using the complete human emulation system from Emulate. First, S1 chips were activated and ECM-coated as per manufacturer instructions. Briefly, the chips were activated using ER-1 solution dissolved in ER-2 at a concentration of 0.5 mg/ml (Emulate) and exposed to UV light (36 W, 365 nm) for 20 min. Finally, the chips were rinsed with ER-2 solution and then PBS (Gibco). ECM solution (100 µg/ml of Matrigel [Corning] and 30 μg/ml rat tail Collagen type 1 [Gibco] diluted in DPBS [Gibco]) was then loaded into the activated S1 chips overnight at 4°C and 1 h at 37°C 5% CO2. After a DMEM rinse, a 1.5 million/ml cell suspension was loaded into the upper channel. Cells were left to adhere for 2 h at 37°C 5% CO2. After cell attachment, gentle washing with a warm cell culture medium containing 100 U/ml penicillin and 100 μg/ml streptomycin was performed on the seeded S1 chips. Cells in the chips were cultured statically overnight at 37°C 5% CO2. Finally, the chips were connected to the Pod-1 system (Emulate) and loaded into the Zoë machine (Emulate). After a regulated cycle, the chips were cultured for 3, 7, or 10 d at 37°C, 5% CO2. Medium flowrate was maintained at 30 μl/h. Stretching (10%, 0.15 Hz) was applied from day 1. Cell culture media was equilibrated and refreshed every 48 h.
Permeability assay
Intestine chip was prepared as described above. Each chip was visually inspected for gaps before the assay. FITC-dextran 70 kD (Sigma-Aldrich) at 0.1 mg/ml was loaded into the top channel reservoir feeding the mature gut epithelium. Flowthrough was collected at different time points (2, 4, 6, and 12 h), samples from the top and bottom were collected, and fluorescence was measured in a spectrofluorometer (Infinite M200Pro; Tecan) with an excitation wavelength of 488 nm and an emission wavelength of 520 nm. EDTA at 0, 1, and 50 mM (Sigma-Aldrich) was added to destabilize the endothelial barrier. Permeability assay was performed in triplicate.
Cell culture on transwell and immunofluorescence
Caco-2 cells were grown on Transwell Permeable Supports 0.4 µm polycarbonate membranes (Costar). Briefly, Caco-2 cells were seeded at a density of 2.5 × 105 onto 12-mm diameter transwells (cat#3412) or 4 × 105 onto 24-mm diameter transwells (cat#3401) and cultured with media change every 3 d for 21 d before analysis. Caco-2 cells were then fixed in 4% paraformaldehyde (Electron Microscopy Sciences), diluted in PBS for 10 min, and permeabilized in 0.2% Triton (Merck) for 10 min before being processed for immunofluorescence. After staining, the polycarbonate membranes were cut up with a scalpel and placed on superfrost plus slides (Thermo Fisher Scientific) with cells facing up. Mounting was done by putting a drop of ProLong Diamond Antifade Mountant (Invitrogen) onto the cells and covering it with a coverslip. Lentivirus production and infection were performed according to Rubinson et al. (2003).
Image acquisition
For organoid depth, map analysis Z stack images were acquired on a confocal LSM 700 upright with a 10× air objective (Zeiss Plan-Apochromat 10×/0.45) using 0.5 µm steps. For filter-differentiated Caco-2 cells, Z stacks images were acquired on the same microscope using a 40× oil objective (Plan-Apochromat X40/1.3) using 0.5 µm steps and assembled into a single square panel using 20% overlap between tiles for further segmentation analysis. For GFP-MYL12A live observation, images were acquired using a Perkin-Elmer spinning disk confocal microscope equipped with an EMCCD camera and a 63× 1.4 NA objective with 5% CO2 and at 37°C.
Scanning electron microscopy
To access differentiated Caco-2 cells in the shape of organoids or cell monolayers, PDMS chips and polycarbonate membrane filters were cut in pieces using a scalpel before being fixed with glutaraldehyde (2.5% in 0.1 M cacodylate buffer) for 1 h and washed 3× in distilled H2O. Cells were then treated with three consecutive cycles of osmic acid (1% in 0.1 M cacodylate buffer) 1 h and thiocarbohydrazide (20 min; OTOTO technique) and washed 6× in distilled H2O. Cells were then dehydrated through a graded ethanol series (35%, 70%, 85%, 95%, and 100%) before being treated with hexamethyldisilazane for 10 min for a chemical critical point. Specimens were then sputter-coated with up to 10 Å gold/palladium conductive layer using a gun ionic evaporator PEC 682. Images were acquired on a JEOL JSM 6700F field emission scanning electron microscope operated at 7 kV.
Cell segmentation
For cell segmentation, 3D images were first subjected to a series of image processing steps performed in Fiji (Schindelin et al., 2012). Briefly, 3D images were denoised using a band-pass filter (filter_large = 100, filter_small = 1.5) and then subjected to a background subtraction using a rolling ball algorithm (rolling = 50). For multi-tile acquisitions, images were stitched in 3D using Fiji. The ZO-1 signal was projected in 2D using a maximum intensity projection. Cell–cell junction segmentation was performed using the machine learning WEKA tool (Hornik et al., 2009), and cell segmentation was manually curated in TissueAnalyzer (Aigouy et al., 2010). TissueMiner was applied on each sample to generate the corresponding database containing cell morphology parameters including cell perimeter and cell area. The databases were queried using the R language for further analysis in R or in GraphPad (Prism). The cell shape index was calculated for each cell from the ratio of the cell perimeter to the square root of the cell area.
Depth map calculation
The depth map was computed using an epithelium projection algorithm (Stephane, 2020) which consists of finding the local maxima intensity of the DAPI signal along the depth axis of the chips’ volume. The depth map was then converted into a two-dimensional manifold surface by mapping a triangulation over the image pixels coordinate and using the depth value for height information. Finally, we subdivided the surface into a local regular regions of ∼160 × 160 μm using an Euclidean grid. For each local region, we computed the surface values of the tissue which we compared with the surface value of a theoretical flat surface to obtain the local surface ratio.
Surface plasmon resonance (SPR)
Real-time biosensing experiments were performed on a Biacore T200 instrument (GE Healthcare) equilibrated at 25°C in binding buffer (PBS). Anti-GFP VHH (10 µg/ml; Chromotek) was covalently immobilized on a carboxymethylated dextran-coated sensorchip (GE Healthcare) using NHS/EDC coupling. HEK293T cell lysate transfected with a GFP-tagged SHROOM ASD2 construct was injected over the functionalized surface and stably captured by the anti-GFP VHH. After washing, serially diluted purified GST-tagged SCRIB or ROCK protein domains were injected over the functionalized surface in a constant buffer flow and their affinity for the GFP-ASD2 SHROOM domain was determined by analyzing the real-time SPR response variations using the Biacore T200 evaluation software.
Statistical studies
Statistical significance was determined using two-tailed paired or unpaired Student’s t test with Welsh correction, as referred to in the figure legends. Statistics were completed using Prism 8 (GraphPad). For all analysis, significance was considered P < 0.05.
Structure prediction
Colabfold program (Mirdita et al., 2022) was used to generate interaction predictions between SCRIB WRAAR protein sequence (residues 1,488–1,526 aa) or ROCK1 SBD domain (834–913 aa) and ASD2 domain protein sequence from SHROOM1 (473–852 aa), SHROOM2 (1,178–1,487 aa), SHROOM3 (1,648–1,997 aa), and SHROOM4 (1,203–1,493 aa), respectively. Chimera X program was used to analyze the predicted structure (Pettersen et al., 2021).
Online supplemental material
Data availability
The data are available from the corresponding author upon reasonable request.
Acknowledgments
We are grateful to the Photonic BioImaging—C2RT/C2RA facility of Institut Pasteur for technical support.
This work was supported by the Centre National de la Recherche Scientifique (CNRS; S. Etienne-Manneville), Institut Pasteur (S. Etienne-Manneville), and by the Emergence Grant Canceropole Ile de France (B. Boëda). B Boëda is supported by Institut National de la Santé et de la Recherche Médicale (INSERM).
References
Author notes
Disclosures: The authors declare no competing interests exist.


![Parental and SCRIB KO Caco-2 cells growth on an IOC system. (A) IOC is made of a flexible silicone polymer (polydimethylsiloxane [PDMS]; left panel) in which cells are grown in the upper central channel under continuous perfusion (right panel). (B) Scanning electronic microscopy image of parental (left) and SCRIB KO (right) Caco-2 cells grown 7 d on the IOC system under continuous perfusion. (C) Higher magnifications of SEM images of parental (left) and mutant (right) intestine-like villi. (D) Intestine-like tissues were grown on IOC for 7 d, dissected from the PDMS chips, stained with DAPI, and observed with confocal microscopy. Quantification of folds was performed on central portions of the intestine strips as indicated by the white square. (E) Maximum intensity projection of confocal acquisition of parental (left) and mutant (right) intestine-like villi grown 7 d on the IOC system and stained with phalloidin and DAPI used for depth map quantification. (F) Z-section confocal images corresponding to the double arrow dotted lines shown in E. (G) Depth map of images presented in E. (H) Quantification of the local surface ratio (surface of the folded gut divided by the corresponding plane surface) intestine-like tissues derived from parental and SCRIB KO Caco-2 cells. The P value represents the result of an unpaired two-tailed t test done on the mean of three independent experiments (means WT: 2.364, 2.043, and 2.297 versus means KO: 1.814, 1.839, and 1.859). (I) Maximum intensity projection of confocal acquisition of parental (top) and mutant (bottom) intestine-like villi stained with phalloidin/ZO1. Zooms indicated by the white squares (left) are shown on the right (ZO1 only).](https://cdn.rupress.org/rup/content_public/journal/jcb/222/12/10.1083_jcb.202211113/1/m_jcb_202211113_fig1.png?Expires=1773574351&Signature=FZ-OrXRCSBdU8teyLjf3H7s9b3DFr5XcBWaPc6tQt2-rjxqPuboUnBjg7lR4xMPsSlYOIWsuh2Bq7OZJpUcwK6xxC9pf5ThLTJixdY31VHi0TDcaIVl~Bsc0g-33cDsdZaPfK~SBApXWRiIhlda0TYo1Vf7VMo2lEhWSVXoKQdMVScYGJrqvZV6EgHacVsTet4Q5abWFA2VLnuEdr-6gmulFhrKi0JLMzFKVmpWPYm66yUaUWthD-okH-mu7tTs0-j25zi6LKuKj3x1bynZVYC3wEbzxZZ31BM62PVTXE06p5z2Rvy21GHjfDYH-UsXa39Tulf5S0XHXz6ItgV1ZdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)