MR1 is a highly conserved microbial immune-detection system in mammals. It captures vitamin B–related metabolite antigens from diverse microbes and presents them at the cell surface to stimulate MR1-restricted lymphocytes including mucosal-associated invariant T (MAIT) cells. MR1 presentation and MAIT cell recognition mediate homeostasis through host defense and tissue repair. The cellular mechanisms regulating MR1 cell surface expression are critical to its function and MAIT cell recognition, yet they are poorly defined. Here, we report that human MR1 is equipped with a tyrosine-based motif in its cytoplasmic domain that mediates low affinity binding with the endocytic adaptor protein 2 (AP2) complex. This interaction controls the kinetics of MR1 internalization from the cell surface and minimizes recycling. We propose MR1 uses AP2 endocytosis to define the duration of antigen presentation to MAIT cells and the detection of a microbial metabolic signature by the immune system.
Introduction
Major histocompatibility complex (MHC) molecules display antigens (Ag) for presentation to T lymphocytes and are crucial for their development, activation, and expansion. A range of related MHC molecules are tailored to bind and present chemically diverse Ag. Classical MHC class I and II molecules (MHC-I and -II) present peptides to conventional T cells (Rock et al., 2016), whereas the non-classical MHC-related proteins of the CD1 family and MR1 present lipids and small metabolite Ag, respectively, to unconventional T cells (Corbett et al., 2014; Kjer-Nielsen et al., 2012; Mayassi et al., 2021). Binding of the Ag presented by each MHC molecule occurs at specific locations within the cell, e.g., the ER, recycling endosomes, or late endosomes. MHC molecules follow precise intracellular routes to reach these locations, governed by intrinsic amino acid–encoded motifs and the accessory proteins that recognize them (Barral and Brenner, 2007; Rock et al., 2016; van Endert, 2016). Although MR1 is one of the most conserved mammalian MHC molecules and has a unique Ag presenting function, the molecular mechanisms that regulate MR1 presentation remain unclear.
MR1 presents metabolites to MR1-restricted T cells, such as mucosal-associated invariant T (MAIT) cells (Awad et al., 2020; Gherardin et al., 2016; Harriff et al., 2018; Keller et al., 2017; Le Nours et al., 2019; McWilliam et al., 2020; Salio et al., 2020). The best characterized metabolites presented by MR1 stimulate MAIT cells and are produced by a diverse range of bacteria and fungi through the biosynthesis of vitamin B2 (riboflavin), termed vitamin B-related Ag (VitBAg). The most potent Ag described to date is 5-(2-oxopropylideneamino)-6-D-ribitylaminouracil (5-OP-RU), formed from the reaction of the riboflavin intermediate 5-amino-6-d-ribitylaminouracil (5-A-RU) with methyl glyoxal (Corbett et al., 2014). VitBAg recognition by MAIT cells is involved in tissue repair, immunity against pathogens, cancer, and possibly other functions (Constantinides et al., 2019; Crowther et al., 2020; Leng et al., 2019; Lepore et al., 2017; Meierovics and Cowley, 2016; Meierovics et al., 2013; Wang et al., 2018; Zhao et al., 2021). Understanding the intracellular mechanisms that regulate MR1-Ag presentation is critical to characterize the role of the MR1–MAIT cell axis and devise strategies to manipulate it.
MR1 can acquire 5-OP-RU produced by commensal or infecting extracellular bacteria (Constantinides et al., 2019; Legoux et al., 2019), or derived from microorganisms located intracellularly such as phagocytosed or cytosolic bacteria (Harriff et al., 2016; Le Bourhis et al., 2013; Le Bourhis et al., 2010; McWilliam et al., 2016; Ussher et al., 2016). While the subject of ongoing work, it is emerging that MR1 uses a pathway distinct from other MHC molecules (Harriff et al., 2016; Karamooz et al., 2019; Kulicke et al., 2020; McWilliam et al., 2016; McWilliam et al., 2020; Ussher et al., 2016). In steady-state conditions, little MR1 is displayed at the surface of the antigen-presenting cell, with the majority located within the ER (McWilliam et al., 2016; McWilliam et al., 2020; Salio et al., 2020). Extracellular metabolites are captured by ligand-receptive MR1 residing within the ER, forming a complex which then traffics through the secretory pathway to the cell surface for presentation (Karamooz et al., 2019; Kulicke et al., 2020; McWilliam et al., 2016; McWilliam et al., 2020; Salio et al., 2020). Key to this phenomenon is the formation of a covalent Schiff base bond between the VitBAg and a crucial lysine residue (K43) in MR1 (Corbett et al., 2014; Kjer-Nielsen et al., 2012). The charged K43 sidechain acts as a molecular switch for ER exit; when neutralized by VitBAg binding, or removed by mutation to alanine (K43A), MR1 refolds and progresses out of the ER (Howson et al., 2020; McWilliam et al., 2016; McWilliam and Villadangos, 2017).
MR1 is displayed at the cell surface for several hours, and then these complexes are internalized and degraded (Chua et al., 2011; McWilliam et al., 2016; Ussher et al., 2016). Unlike MHC-I molecules, where the half-life of surface display is related to the affinity of the MHC–peptide interaction, the duration of MR1 surface display is unrelated to the presence or nature of VitBAg loaded on MR1 (Howarth et al., 2004; McWilliam et al., 2016). A small fraction of internalized MR1 can recycle back to the cell surface, and during its transit MR1 can sample endosomal metabolites in a process distinct from the ER-loading mode (Harriff et al., 2016; Karamooz et al., 2019; Lange et al., 2020; McWilliam et al., 2016). However, the relative contributions of these two loading pathways for metabolite presentation are not known (McWilliam and Villadangos, 2020).
We hypothesized that a specific molecular mechanism serves to internalize cell surface MR1 complexes, for degradation or further Ag loading in the endosomes. Here, we employed a CRISPR/Cas9 genome-wide loss-of-function screen to reveal that the adaptor protein 2 (AP2) complex is required for MR1 internalization. The tetrameric AP2 is one of the main complexes that recruits plasma membrane proteins for clathrin-mediated endocytosis (Kaksonen and Roux, 2018). Protein cargo that contain specific endocytic signals in their cytoplasmic domains are recognized by AP2 (Traub and Bonifacino, 2013), and directed into early/sorting endosomes where they can further progress in the endolysosomal route for degradation, or be recycled back to the cell surface (Naslavsky and Caplan, 2018). We identify an atypical AP2-binding motif in the cytoplasmic tail of MR1 that is conserved and fine-tuned for a defined rate of surface display and MR1–metabolite complex decay. This study illuminates a key molecular interaction that regulates microbial Ag presentation by controlling the duration of Ag exposure on the cell surface for presentation to MR1-restricted T cells.
Results
Kinetics of MR1 endocytosis in diverse cell types
First, we asked whether MR1 was endocytosed at different rates in various cell types. Previous observations on MR1 internalization and recycling were made using overexpressed MR1 in the lymphoblastoma cell line C1R, where it took around 2–3 h for 50% of surface MR1 to be internalized and only ∼5% of surface MR1 was recycled (Howson et al., 2020; McWilliam et al., 2016). To determine if endogenously expressed MR1 was similarly regulated, we incubated primary human cells and unmodified cell lines with 5-OP-RU to recruit MR1 to the cell surface and investigated the kinetics of its internalization and recycling (Fig. 1, A and B). To measure internalization, we employed a method developed for the quantitation of internalized proteins by flow cytometry (Liu and Johnston, 2013). Used by us and others (Dumont et al., 2017; Liu et al., 2022; McWilliam et al., 2016), it employs a cell impermeable quenching dye to ablate fluorescence of non-endocytosed material remaining at the cell surface; hence, the fluorescence level after quenching defines the amount internalized. For recycling, a cleavable biotin tag was added to the anti-MR1 antibody. After internalization, remaining surface anti-MR1 antibody was cleaved by a cell impermeable agent. Cells were returned to culture and the reemergence of internalized MR1 (i.e., recycled) was detected by fluorescently conjugated streptavidin.
Discovery of the molecular machinery that internalizes cell surface MR1. (A) The internalization rate of surface MR1 was measured in primary human PBMCs from healthy donors gating on monocytes (Mono.; blue), B cells (black), and T cells (red), or the C1R (black) and THP-1 (blue) cell lines. Cells were cultured overnight with 10 µM Ac-6-FP, then surface MR1 was labeled with anti-MR1 mAb 8F2.F9 conjugated to a nucleic acid-conjugated FIP. Cells were allowed to internalize for up to 4 h, and any remaining surface fluorescence was quenched with a complementary nucleic acid probe conjugated to a quenching dye. MR1 internalization was calculated as a percentage of the initial un-quenched signal. (B) The recycling rate of internalized MR1 of cells treated with Ac-6-FP overnight as in A. Surface MR1 was labeled with 8F2.F9-SS-biotin and allowing to internalize for 1 h. Remaining biotin label at the surface was removed by a cell-impermeable reducing agent, and cells were then incubated for up to 30 min to allow labeled MR1 to recycle back to the cell surface. Recycled MR1 was detected by incubating the cells with streptavidin conjugated to AF647 and measured by flow cytometry. The amount of MR1 recycled was calculated as the percentage of the initial surface MR1 signal prior to biotin removal. (C) The effect of endocytosis inhibitors on MR1 internalization was measured in C1R (left) or THP-1 (right) cells as in A. Cells were incubated with 10 µM 5-OP-RU for 4 h, treated with each inhibitor for 1 h, then internalization after 2 h measured with the FIP in the presence or absence of inhibitors. (D) C1R cells were pulsed with 5-OP-RU for 4 h then washed and chased without ligand for 8 h. The fold change of geometric mean fluorescent intensity (gMFI; left) of MR1 surface expression is shown. C1R cells transduced with lentivirus containing a GeCKO v2 human CRISPR-Cas9 KO library were similarly pulsed with 5-OP-RU for 4 h then chased without ligand for 8 h and stained for surface MR1 (solid black histogram). MR1HIGH cells (5% of population) were sorted and expanded and then sorted again for a second time. MR1 levels in cells cultured without ligand (black line) compared to after 5-OP-RU pulse (dotted line), and unstained cells (gray) are shown for comparison, a representative of three replicates. (E) The viral integration site containing the sgRNA cassette was amplified and sequenced from each sample, and the twice-sorted compared to the unsorted pool samples. Shown is the plot of significance (−log10[P value]) and fold change (Log2[Fold change]) for each gene based on a MAGeCK-RRA analysis. The only significantly enriched hit with a false discovery rate >0.05 was AP2A1 shown in blue. (F) The number of reads for each sgRNA targeting AP2A1 for the unsorted library (gray) is shown compared to the MR1HIGH-sorted replicates (blue). Each dot represents one replicate where the sgRNA was detected. Data are shown as the mean ± SD of replicates from two separate experiments for cell lines (A–D; n = 4), or individual data points for healthy donors (A and B; n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Discovery of the molecular machinery that internalizes cell surface MR1. (A) The internalization rate of surface MR1 was measured in primary human PBMCs from healthy donors gating on monocytes (Mono.; blue), B cells (black), and T cells (red), or the C1R (black) and THP-1 (blue) cell lines. Cells were cultured overnight with 10 µM Ac-6-FP, then surface MR1 was labeled with anti-MR1 mAb 8F2.F9 conjugated to a nucleic acid-conjugated FIP. Cells were allowed to internalize for up to 4 h, and any remaining surface fluorescence was quenched with a complementary nucleic acid probe conjugated to a quenching dye. MR1 internalization was calculated as a percentage of the initial un-quenched signal. (B) The recycling rate of internalized MR1 of cells treated with Ac-6-FP overnight as in A. Surface MR1 was labeled with 8F2.F9-SS-biotin and allowing to internalize for 1 h. Remaining biotin label at the surface was removed by a cell-impermeable reducing agent, and cells were then incubated for up to 30 min to allow labeled MR1 to recycle back to the cell surface. Recycled MR1 was detected by incubating the cells with streptavidin conjugated to AF647 and measured by flow cytometry. The amount of MR1 recycled was calculated as the percentage of the initial surface MR1 signal prior to biotin removal. (C) The effect of endocytosis inhibitors on MR1 internalization was measured in C1R (left) or THP-1 (right) cells as in A. Cells were incubated with 10 µM 5-OP-RU for 4 h, treated with each inhibitor for 1 h, then internalization after 2 h measured with the FIP in the presence or absence of inhibitors. (D) C1R cells were pulsed with 5-OP-RU for 4 h then washed and chased without ligand for 8 h. The fold change of geometric mean fluorescent intensity (gMFI; left) of MR1 surface expression is shown. C1R cells transduced with lentivirus containing a GeCKO v2 human CRISPR-Cas9 KO library were similarly pulsed with 5-OP-RU for 4 h then chased without ligand for 8 h and stained for surface MR1 (solid black histogram). MR1HIGH cells (5% of population) were sorted and expanded and then sorted again for a second time. MR1 levels in cells cultured without ligand (black line) compared to after 5-OP-RU pulse (dotted line), and unstained cells (gray) are shown for comparison, a representative of three replicates. (E) The viral integration site containing the sgRNA cassette was amplified and sequenced from each sample, and the twice-sorted compared to the unsorted pool samples. Shown is the plot of significance (−log10[P value]) and fold change (Log2[Fold change]) for each gene based on a MAGeCK-RRA analysis. The only significantly enriched hit with a false discovery rate >0.05 was AP2A1 shown in blue. (F) The number of reads for each sgRNA targeting AP2A1 for the unsorted library (gray) is shown compared to the MR1HIGH-sorted replicates (blue). Each dot represents one replicate where the sgRNA was detected. Data are shown as the mean ± SD of replicates from two separate experiments for cell lines (A–D; n = 4), or individual data points for healthy donors (A and B; n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Monocytes had the fastest rate of MR1–5-OP-RU complex internalization, with 50% of complexes removed from the cell surface in <1 h, followed by T cells at ∼2 h and B cells at >4 h (Fig. 1 A). We compared C1R cells (a B cell lymphoblastoid cell line) to THP-1 cells (a monocytic line used as MR1-presenting cells [Salio et al., 2020; Ussher et al., 2016]). Both had similar internalization rates with 50% endocytosed by 2 h. Recycling varied between cell types, with the highest degree observed in monocytes (14%), while T and B cells both only recycled ∼2% (Fig. 1 B). C1R cells recycled slightly more than THP-1 (∼7 vs. 4%; Fig. 1 B). Thus, endogenous MR1 is constitutively internalized from primary cells with a small fraction recycled, and these rates vary in a cell type-dependent manner. The rates of endocytosis and recycling of endogenous (or overexpressed [McWilliam et al., 2016]) MR1 in cell lines are within the diverse range observed in primary cells, so the cell lines are valid models for investigating MR1 trafficking.
To investigate the mechanism of MR1 internalization, C1R and THP-1 cells were treated with endocytosis inhibitors, and the rate of MR1 internalization was measured. The inhibitor of clathrin-mediated endocytosis, Pitstop2, blocked MR1 internalization in both cell lines (Fig. 1 C), as well as the uptake of fluorescently labeled transferrin, that binds to the transferrin receptor (TfR). TfR is a well-defined cargo for clathrin-mediated endocytosis (Bellve et al., 2006; Motley et al., 2003; Fig. S1 A). The actin polymerization inhibitor cytochalasin D reduced MR1 internalization in C1R cells only (Fig. 1 C) and transferrin in both cell types (Fig. S1 A). The inhibitor of macropinocytosis, 5-[N-ethyl-N-isopropyl]amiloride, did not affect MR1 internalization or transferrin uptake, although it blocked pinocytosis of labeled dextran (Fig. S1 A). This suggests that MR1 internalization is mediated through clathrin-mediated endocytosis rather than macropinocytosis.
The effect of endocytosis inhibitors on dextran and transferrin uptake. (A) C1R or THP-1 cells were treated with each inhibitor for 1 h, then incubated with TAMRA-conjugated dextran (70 kD) or AF647-conjugated transferrin for 1 h. Internalization after 2 h measured with the FIP in the presence or absence of inhibitors. Dextran and transferrin uptake inhibition by endocytosis inhibitor in C1R and THP-1 cells as indicated. Data are shown as mean ± SD of replicates from two independent experiments (n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test where *, P < 0.05 and **, P < 0.01. (B) Schematic of the AP2 complex recognizing its cargo, illustrating its constituent subunits and the genes encoding each in italics. (C) Relative expression levels of the transcripts for each AP2 subunit among PBMCs. Data obtained from the Immunological Genome Project (upper) and the Human Protein Atlas (lower).
The effect of endocytosis inhibitors on dextran and transferrin uptake. (A) C1R or THP-1 cells were treated with each inhibitor for 1 h, then incubated with TAMRA-conjugated dextran (70 kD) or AF647-conjugated transferrin for 1 h. Internalization after 2 h measured with the FIP in the presence or absence of inhibitors. Dextran and transferrin uptake inhibition by endocytosis inhibitor in C1R and THP-1 cells as indicated. Data are shown as mean ± SD of replicates from two independent experiments (n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test where *, P < 0.05 and **, P < 0.01. (B) Schematic of the AP2 complex recognizing its cargo, illustrating its constituent subunits and the genes encoding each in italics. (C) Relative expression levels of the transcripts for each AP2 subunit among PBMCs. Data obtained from the Immunological Genome Project (upper) and the Human Protein Atlas (lower).
Discovery of the cellular machinery required for MR1 internalization
To discover gene products essential for regulating MR1 internalization, a genome-wide CRISPR-Cas9 loss-of-function library screen was employed, using the GeCKO v2 library which has six single-guide RNAs (sgRNAs) targeting each gene. Recently, we successfully performed similar screens in the C1R cell line (Gherardin et al., 2021; McWilliam et al., 2020); hence, we used the same cell line with a modified screening approach. C1R cells were pulsed with 5-OP-RU for 4 h, causing an approximately sevenfold increase in MR1 surface expression, and then chased in media lacking 5-OP-RU for 8 h, at which point the MR1 surface expression had returned to near baseline levels (Fig. 1 D). We hypothesized that cells where a gene encoding a protein essential for MR1 internalization was deleted would retain high levels of surface MR1 after the ligand-free chase period. We used this 5-OP-RU pulse-chase method on three replicate libraries and sorted the top 5% of MR1HIGH cells sequentially twice. The sgRNAs within this sorted population were amplified and identified by sequencing and compared to the unsorted library. The only significantly enriched gene targeted by sgRNAs in the MR1HIGH sorted cells was AP2 complex subunit α1 (Ap2a1; Fig. 1 E). Five of six sgRNA targeting Ap2a1 were enriched in the MR1HIGH cell population in at least two replicates compared to the unsorted cells (Fig. 1 F). This protein is an integral member of the AP2 heterotetrameric complex, which is involved in sorting cargo for clathrin-mediated endocytosis (Kadlecova et al., 2017). AP2 is composed of four subunits: α (encoded either by two genes, Ap2a1 or Ap2a2); β2 (Ap2b1); µ2 (Ap2m1); and σ2 (Ap2s1; Kirchhausen, 1999; Fig. S1 B). This finding implicates the AP2 complex in MR1 internalization. Considering we saw a higher internalization rate in human blood monocytes compared to T and B cells (Fig. 1 A), we reasoned that differences in AP2 expression might be responsible. Indeed, two transcriptomic datasets revealed that monocytes had the highest expression of three of the four genes encoding AP2 (Fig. S1 C). Further, C1R and THP1 cells, which internalized MR1 similarly, expressed comparable levels of AP2A1 and AP2M1 protein (Fig. S2 A).
CRISPR-Cas9 deletion of AP2 subunits in C1R and THP-1 cells. (A) C1R and THP-1 cells transduced with Ctrl-1 or -2 or single-cell clones deleted for AP2A1 (∆AP2A1-1 or -2) were lysed and immunoblotted for AP2A1, AP2M1, and actin. Probable non-specific band indicated (*). (B) The internalization of MR1 (i) and TfR (ii) in THP-1 cells as measured as in Fig. 1 A. (C) AP2M1 KO was attempted in C1R cells with CRISPR-Cas9 using two different sgRNAs (AP2M1-1 and -2). Cells were lysed and immunoblotted for AP2M1 and actin (i). The AP2M1 gene target site was sequenced, and INDEL analysis was performed with ICE-TIDE. The frequency of INDELs in each cell line is reported in the pie graph (ii): where no INDEL (white) and those not resulting in frame shift (blue) generally do not result in KO, and those INDELs resulting in a frame shift (red) that cause KO. (D) The internalization of MR1 (i) and TfR (ii) in C1R cells transduced with Ctrl-1 or -2 or for AP2M1 (∆AP2M1-1 or -2) as measured in Fig. 1 A. (E) AP2A2 KO was attempted using two different sgRNAs (AP2A2-1 and -2) in WT C1R cells (Ctrl) or cells that were previously KO for AP2A1 (∆AP2A1). The AP2A2 gene target site was sequenced and INDEL analysis performed as in C. The gene sequence of Ap2a2 in single-cell clones of ∆AP2A2 single KOs compared to control cells is aligned, highlighting the INDELs (red) and sgRNA sequences (blue). (F) The internalization of MR1 (i) and TfR (ii) in C1R cells transduced with Ctrl-1 or -2 or for AP2A2 KO clones (combined data for ∆AP2A2-1 and –2) as measured as in Fig. 1 A. Data shown in B, D, and F are the mean ± SD of the two clones from at least two separate experiments (n = 4–6). Molecular weight standards (kD) are shown (A and C). Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData FS2.
CRISPR-Cas9 deletion of AP2 subunits in C1R and THP-1 cells. (A) C1R and THP-1 cells transduced with Ctrl-1 or -2 or single-cell clones deleted for AP2A1 (∆AP2A1-1 or -2) were lysed and immunoblotted for AP2A1, AP2M1, and actin. Probable non-specific band indicated (*). (B) The internalization of MR1 (i) and TfR (ii) in THP-1 cells as measured as in Fig. 1 A. (C) AP2M1 KO was attempted in C1R cells with CRISPR-Cas9 using two different sgRNAs (AP2M1-1 and -2). Cells were lysed and immunoblotted for AP2M1 and actin (i). The AP2M1 gene target site was sequenced, and INDEL analysis was performed with ICE-TIDE. The frequency of INDELs in each cell line is reported in the pie graph (ii): where no INDEL (white) and those not resulting in frame shift (blue) generally do not result in KO, and those INDELs resulting in a frame shift (red) that cause KO. (D) The internalization of MR1 (i) and TfR (ii) in C1R cells transduced with Ctrl-1 or -2 or for AP2M1 (∆AP2M1-1 or -2) as measured in Fig. 1 A. (E) AP2A2 KO was attempted using two different sgRNAs (AP2A2-1 and -2) in WT C1R cells (Ctrl) or cells that were previously KO for AP2A1 (∆AP2A1). The AP2A2 gene target site was sequenced and INDEL analysis performed as in C. The gene sequence of Ap2a2 in single-cell clones of ∆AP2A2 single KOs compared to control cells is aligned, highlighting the INDELs (red) and sgRNA sequences (blue). (F) The internalization of MR1 (i) and TfR (ii) in C1R cells transduced with Ctrl-1 or -2 or for AP2A2 KO clones (combined data for ∆AP2A2-1 and –2) as measured as in Fig. 1 A. Data shown in B, D, and F are the mean ± SD of the two clones from at least two separate experiments (n = 4–6). Molecular weight standards (kD) are shown (A and C). Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData FS2.
AP2A1 is required for MR1 internalization
To confirm that AP2A1 mediates MR1 internalization, we deleted AP2A1 in C1R cells using two distinct sgRNAs and generated clonal populations from single cells (Fig. 2 A). Compared to the WT cells expressing non-targeting control sgRNAs (Ctrl-1 or -2), AP2A1 knockout (KO) cell clones (∆AP2A1-1 and -2) had higher expression of TfR on the cell surface (Fig. 2 B i), indicating a reduction in AP2-mediated internalization, consistent with the reported effect of AP2A1 depletion (Borner et al., 2006; Dugast et al., 2005). Similarly, surface MR1 was also expressed at higher levels on the mutant cells before or after incubation with 5-OP-RU (Fig. 2 B ⅱ). Pulsing with 5-OP-RU, and then chasing without ligand to follow the internalization of surface molecules, revealed a pronounced difference between AP2A1-deleted and unmodified cells (Fig. 2 C i); note that for clarity, the data from the two ∆AP2A1 or Ctrl cell lines are combined in this and subsequent analyses. The proportional rate of MR1 surface decline during the chase without ligand was slightly lower in ∆AP2A1 cells but not significantly different (Fig. 2 C ⅱ). However, the absolute number of MR1 molecules internalized was reduced in AP2A1-deleted cells, with ∼70% lower internalization at 4 h (Fig. 2 D), explaining the higher surface expression (Fig. 2, B and C). The rate of MR1 recycling was also reduced in cells lacking AP2A1 (∼70%; Fig. 2 E). We transfected C1R∆AP2A1 cells with the AP2A1 coding sequence and achieved partial restoration of protein expression (Fig. 2 F). These cells showed decreased surface expression of TfR and MR1, although the decrease of MR1 was small and not statistically significant (Fig. 2 G). However, AP2A1 re-expression increased the internalization rate of MR1 (Fig. 2 H). To confirm the role of the AP2 complex, we deleted AP2A1 in THP-1 cells (Fig. S2 A). We found a reduction in the rate of MR1 internalization, although there was no change in the rate of TfR internalization (Fig. S2 B).
AP2A1 is essential for MR1 internalization and recycling. (A) C1R cells transduced with Ctrl-1 or -2 or ∆AP2A1-1 or -2 were lysed and immunoblotted (IB) for AP2A1 and actin. (B) Surface expression of TfR (i) and MR1 (ii) in Ctrl cells (gray and black) or ∆AP2A1 cells (sky blue and navy) treated without or with 5-OP-RU for 4 h measured by flow cytometry. (C) Surface expression (i) of MR1 in Ctrl or ∆AP2A1 cells (shown is the combined data from both Ctrl-1 and -2, or ∆AP2A1-1 and -2 clones) following 5-OP-RU pulse and then chase in the absence of ligand for the indicated times. The decline in expression (ii) at each time point is calculated as the percent of surface MR1 at the beginning of the chase (t = 4 h). (D and E) The internalization (D) or recycling (E) rates of MR1 and TfR in Ctrl or ∆AP2A1 cells measured as described in Fig. 1, A and B. (F) The levels of AP2A1 and actin in Ctrl, ∆AP2A1 cells, and the latter with AP2A1 reexpressed (∆AP2A1.AP2A1) detected by immunoblotting. (G) Surface expression of TfR (above) and MR1 (below) after 4 h culture with 10 µM Ac-6-FP on the cells from F detected by flow cytometry. (H) Internalization rate of MR1 in the cells from F as described in Fig. 1 A. Data shown are the mean ± SD of replicates from one experiment representing at least two independent experiments (A, B, F, and G; n = 2); and the mean ± SD of two Ctrl and ∆AP2A1 clones from replicates of two separate experiments (B–E and H; n = 4). Molecular weight standards (kD) are shown (A and F). Statistical significance was calculated with one-way (B and G) or two-way (C–E and H) ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData F2.
AP2A1 is essential for MR1 internalization and recycling. (A) C1R cells transduced with Ctrl-1 or -2 or ∆AP2A1-1 or -2 were lysed and immunoblotted (IB) for AP2A1 and actin. (B) Surface expression of TfR (i) and MR1 (ii) in Ctrl cells (gray and black) or ∆AP2A1 cells (sky blue and navy) treated without or with 5-OP-RU for 4 h measured by flow cytometry. (C) Surface expression (i) of MR1 in Ctrl or ∆AP2A1 cells (shown is the combined data from both Ctrl-1 and -2, or ∆AP2A1-1 and -2 clones) following 5-OP-RU pulse and then chase in the absence of ligand for the indicated times. The decline in expression (ii) at each time point is calculated as the percent of surface MR1 at the beginning of the chase (t = 4 h). (D and E) The internalization (D) or recycling (E) rates of MR1 and TfR in Ctrl or ∆AP2A1 cells measured as described in Fig. 1, A and B. (F) The levels of AP2A1 and actin in Ctrl, ∆AP2A1 cells, and the latter with AP2A1 reexpressed (∆AP2A1.AP2A1) detected by immunoblotting. (G) Surface expression of TfR (above) and MR1 (below) after 4 h culture with 10 µM Ac-6-FP on the cells from F detected by flow cytometry. (H) Internalization rate of MR1 in the cells from F as described in Fig. 1 A. Data shown are the mean ± SD of replicates from one experiment representing at least two independent experiments (A, B, F, and G; n = 2); and the mean ± SD of two Ctrl and ∆AP2A1 clones from replicates of two separate experiments (B–E and H; n = 4). Molecular weight standards (kD) are shown (A and F). Statistical significance was calculated with one-way (B and G) or two-way (C–E and H) ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData F2.
The µ2 subunit (AP2M1) of AP2 is responsible for binding cargo proteins bearing tyrosine-based motifs (Fig. S1 B). Hence if AP2 mediates MR1 internalization, AP2M1 is likely to be critical for MR1 internalization. We noted that deletion of AP2A1 reduced the abundance of AP2M1 in C1R and to a lesser extent in THP-1 cells (Fig. S2 A). The reduced AP2M1 protein would result in lower numbers of AP2 complexes (particularly in C1R cells), even if AP2A2 can substitute for AP2A1 as the α subunit, which explains the loss in TfR and MR1 internalization in ∆AP2A1 cells.
We attempted to delete AP2M1 in C1R cells; however, this only yielded partial KOs for this protein (Fig. S2 C). We sequenced the frequency of insertion-deletion mutations (INDEL) and found that there was only ∼50% frame-shifting INDELs in the single-cell clones. Since frame-shifting INDELs are most likely to result in KO, this suggests that we could only achieve heterozygous AP2M1 deletion and that the expression of AP2M1 is essential for cell survival. Nevertheless, AP2M1-depleted cells had a slight reduction in both MR1 and TfR internalizations (Fig. S2 D).
We also explored if AP2A2 was important in MR1 internalization, as we suspected it could compensate for loss of AP2A1. We first attempted to delete AP2A2 in WT (Ctrl) cells, where we could induce 42 and 43% of frame-shifting mutations in the unsorted cell populations. From these, we could generate single-cell ∆AP2A2 KO clones that had marginally reduced MR1 internalization rate, but a slower TfR internalization (Fig. S2, E and F). We next attempted to delete AP2A2 in ∆AP2A1 cells to address if the double KO was viable. However, we could induce very few frame-shift INDELs in the ∆AP2A1 cell lines (Fig. S2 E), and no KO clones could be generated, despite double KOs for other genes being possible in this system (McWilliam et al., 2020). Since cell viability is maintained in the absence of each α subunit, but not both, it confirms that the α subunits are redundant and each can maintain some AP2 internalization in the absence of the other.
Together this supports that the AP2 complex is important for MR1 internalization in distinct cell types. Additionally, MR1 and TfR have different requirements for the α subunits; MR1 requires AP2A1, but not AP2A2, whereas both gene products are required for normal TfR internalization.
The cytoplasmic tail of MR1 encodes a tyrosine-based internalization motif
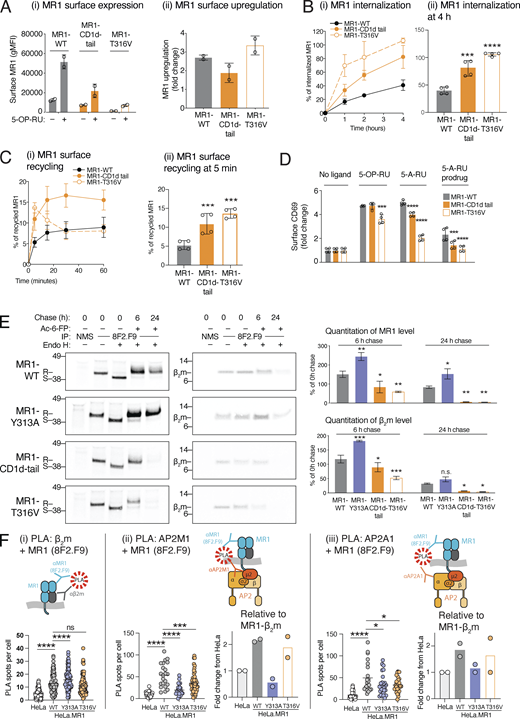
We next sought to determine whether MR1 contains a motif that may mediate AP2 recognition. AP2 binds its plasma membrane cargo through linear sorting motifs, either tyrosine-based or dileucine-based (Boll et al., 1996). The canonical tyrosine-based motif consists of a tyrosine followed by any two amino acids and a bulky hydrophobic amino acid (YXXΦ; Traub and Bonifacino, 2013) although there are some variations from this motif (Royle et al., 2005). We aligned MR1 cytoplasmic domains from 60 diverse mammalian species with available sequences and noted the high conservation of a tyrosine residue (Y313 in human MR1; Fig. 3 A and Table S1). Generally, this was followed by a leucine with less conservation but then a highly conserved proline (P315; commonly found in this Y+2 position in these motifs (Ohno et al., 1998)) followed by a threonine (T316). This suggested a possible tyrosine-based AP2 motif, albeit non-classical as the T316 is not a hydrophobic Φ residue. Notably, the tyrosine, proline, and threonine residues are conserved in 80, 65, and 70% of mammalian sequences analyzed, respectively, including primates, rodents, marsupials, and cetaceans. We also noted three conserved Arg residues (RRR) at the beginning of the cytoplasmic tail (Fig. 3 A).
A conserved tyrosine residue in the MR1 cytoplasmic tail regulates internalization. (A) A consensus logo for the cytoplasmic tail of 60 aligned mammalian MR1 sequences. The height of the residue indicates its conservation, and the width indicates the frequency of a residue at that location. Amino acid properties are indicated by color; neutral (green), hydrophilic (blue), or hydrophobic (black). The human sequence is listed below including numbering for reference. (B) Schematic of the MR1 cytoplasmic tail mutants used in this study. (C) C1R cells transduced with MR1 WT or the indicated mutants were incubated with or without 10 µM Ac-6-FP for 3 h, and surface MR1 was measured by flow cytometry. Shown is expression relative to MR1-WT without ligand (fold change): cells without ligand (i) or after Ac-6-FP (ii). (D) Data from C are shown as a fold change after Ac-6-FP exposure from baseline. (E) The internalization of MR1 WT or mutants after 4 h was measured as in Fig. 1 A. (F and G) The internalization (F) and recycling (G) of surface MR1-WT (black) or -Y313A mutant (red) at the indicated times measured as in Fig. 1, A and B. (H) HeLa cells expressing MR1-WT or -Y313A were cultured with Ac-6-FP overnight and surface MR1 labeled with AF647-conjugated anti-MR1 (8F2.F9; green). Cells were washed and allowed to internalize for 4 h, and then were fixed, permeabilized, and co-stained with markers for endosomes: early (EEA1, red) or late/lysosomes (LAMP1, white) and nuclei (blue). The percentage of MR1+ endosomes overlapping with EEA1+ or LAMP1+ (yellow arrows) were enumerated (right hand side) from n = 20–50 cells from each of two independent experiments. Images are shown as maximum projections of Z-stacks, and the white scale bars represent 10 µM. Column graphs represent the mean ± SD of replicates from two independent experiments (C–H; n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test in C–E, two-way ANOVA with multiple comparison test in F and G, unpaired t-test in H. Significance is shown by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
A conserved tyrosine residue in the MR1 cytoplasmic tail regulates internalization. (A) A consensus logo for the cytoplasmic tail of 60 aligned mammalian MR1 sequences. The height of the residue indicates its conservation, and the width indicates the frequency of a residue at that location. Amino acid properties are indicated by color; neutral (green), hydrophilic (blue), or hydrophobic (black). The human sequence is listed below including numbering for reference. (B) Schematic of the MR1 cytoplasmic tail mutants used in this study. (C) C1R cells transduced with MR1 WT or the indicated mutants were incubated with or without 10 µM Ac-6-FP for 3 h, and surface MR1 was measured by flow cytometry. Shown is expression relative to MR1-WT without ligand (fold change): cells without ligand (i) or after Ac-6-FP (ii). (D) Data from C are shown as a fold change after Ac-6-FP exposure from baseline. (E) The internalization of MR1 WT or mutants after 4 h was measured as in Fig. 1 A. (F and G) The internalization (F) and recycling (G) of surface MR1-WT (black) or -Y313A mutant (red) at the indicated times measured as in Fig. 1, A and B. (H) HeLa cells expressing MR1-WT or -Y313A were cultured with Ac-6-FP overnight and surface MR1 labeled with AF647-conjugated anti-MR1 (8F2.F9; green). Cells were washed and allowed to internalize for 4 h, and then were fixed, permeabilized, and co-stained with markers for endosomes: early (EEA1, red) or late/lysosomes (LAMP1, white) and nuclei (blue). The percentage of MR1+ endosomes overlapping with EEA1+ or LAMP1+ (yellow arrows) were enumerated (right hand side) from n = 20–50 cells from each of two independent experiments. Images are shown as maximum projections of Z-stacks, and the white scale bars represent 10 µM. Column graphs represent the mean ± SD of replicates from two independent experiments (C–H; n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test in C–E, two-way ANOVA with multiple comparison test in F and G, unpaired t-test in H. Significance is shown by *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
We expressed MR1 with point mutations or deletions in the cytoplasmic tail to determine if these conserved features were required for internalization (Fig. 3 B). We mutated conserved residues in the putative AP2 region which was expected to ablate interaction with AP2 and reduce internalization (Ohno et al., 1998): the Y313 residue to Ala (Y313A) or Phe (Y313F), and T316 and P315 to Ala (T316A and P315A). To address the possibility that phosphorylation of T316 might regulate internalization, we mutated it to aspartic acid (T316D) as this residue is often used as a phosphomimetic of threonine. We truncated the tail by placing a stop codon after the RRR arginine triplet (∆P305) or prior (∆R302), mutated each RRR residue to Ala in separate mutants (R302A, R303A, R304A) and tested the MR1-GFP construct where GFP is fused to the C-terminus of the cytoplasmic tail directly following R319 (McWilliam et al., 2016).
WT and mutant MR1 molecules were similarly upregulated at the cell surface following a pulse with ligand (Fig. 3, C and D), indicating the cytoplasmic tail motifs have no role in ER retention or trafficking of MR1 to the surface. All mutant molecules were expressed at similar levels as MR1-WT in the absence of ligands, except MR1-∆R302 and MR1-P315A, which had lower expression (Fig. 3 C). In contrast, MR1-GFP had significantly higher surface expression. Among the residues in the putative AP2 motif, only mutating the Y313 to alanine (Y313A) reduced the rate of MR1 internalization (Fig. 3, E and F). Surprisingly, the ∆P305 tail deletion did not affect internalization, yet the complete loss of the tail including the triplet (∆R302) induced rapid internalization. Individual R to A mutations in this triplet did not affect surface stability. This suggests that the complete arginine triplet is important for surface expression (Fig. 3 C). These positively charged motifs are a trait common in mammalian transmembrane proteins (Sharpe et al., 2010), thus could be required for general membrane stability.
We also found the MR1-GFP construct showed a reduced internalization rate, suggesting GFP interferes with the internalization mechanism. This construct has been used by us and others in previous MR1 localization studies (Harriff et al., 2016; Karamooz et al., 2019; McWilliam et al., 2016). While the steps prior to surface expression do not seem affected by the fusion, the reduced internalization suggests data generated from this and similar constructs should be interpreted cautiously.
Interestingly, the Y313A point mutation phenocopied the loss of AP2A1 with a similar level of reduced internalization rate (Fig. 3 F and Fig. 2 D). This corroborates that MR1 internalization is consequent to its interaction with the AP2 complex mediated by the Y313 residue. However, MR1 recycling rates differed; with the rate of MR1-Y313A molecules only slightly reduced compared to the MR1-WT rate, whereas the loss of AP2A1 reduced recycling by half (Fig. 2 E). We speculate this may be due to an overexpression artifact. It was noted that the level of recycling in overexpressed MR1 (3%) is half that of endogenously expressed MR1 in WT C1R cells (∼6%; compare Fig. 3 G to Fig. 2 E). Therefore, overexpressed MR1 may saturate the recycling mechanism; hence, it cannot be reduced further with the loss of AP2 interaction (the Y313A mutant).
We next investigated the subcellular location of internalized MR1 molecules. We labeled surface MR1 with anti-MR1 (8F2.F9) in live HeLa cells expressing either MR1-WT or -Y313A, and then allowed labeled MR1 to internalize for 4 h. The WT and mutant molecules were expressed at similar levels (Fig. S3). After fixing and labeling of endosomal compartments, we found MR1 both at the plasma membrane and in punctate endosomal-like compartments (Fig. 3 H). We quantitated the amount of MR1+ endosomes coinciding with early endosomes (EEA1+) or late endosomes/lysosome (LAMP1+). We found that MR1-Y313A was less likely to be within both types of endosomes than MR1-WT, with the difference being more pronounced for early rather than late endosomes (Fig. 3 H). Collectively, these data suggest that MR1 contains a conserved tyrosine residue in the cytoplasmic tail that is recognized by the AP2 complex for efficient internalization from the cell surface into early endosomes.
Expression of mutant MR1 molecules in HeLa cells. HeLa cells transduced with MR1 WT or the indicated mutants were incubated with 10 µM Ac-6-FP for 3 h, and surface MR1 measured by flow cytometry. Data shown are the individual means of two independent experiments (n = 2).
Expression of mutant MR1 molecules in HeLa cells. HeLa cells transduced with MR1 WT or the indicated mutants were incubated with 10 µM Ac-6-FP for 3 h, and surface MR1 measured by flow cytometry. Data shown are the individual means of two independent experiments (n = 2).
AP2 interacts with MR1 at the plasma membrane
Our attempts to demonstrate an interaction between the AP2 complex with MR1, or the well-characterized AP2 cargo TfR, by immunoprecipitation were unsuccessful, likely due to the transient nature of AP2-cargo interactions. Instead, we employed the proximity ligation assay (PLA), which can measure protein interactions within fixed cells. We used three combinations of antibodies (Fig. 4) against MR1 and two subunits of AP2 to determine if these two protein complexes interact (within 40 nm), as revealed by distinct fluorescent PLA puncta (Gullberg and Andersson, 2010): (i) antisera against the MR1 cytoplasmic tail (αMR1-CT [McWilliam et al., 2020]) and AP2A1; (ii) the antibody recognizing MR1 luminal domain (8F2.F9) and AP2A1; and (iii) 8F2.F9 and AP2M1. We compared the number of PLA puncta in HeLa cells that express low endogenous levels of MR1 to HeLa overexpressing MR1 (HeLa.MR1) without ligand or with 5-OP-RU, which recruits MR1–β2m complexes to the plasma membrane. For all three PLA experiments, the highest number of PLA puncta were in HeLa.MR1 cells pulsed with 5-OP-RU, compared to HeLa cells pulsed with 5-OP-RU or HeLa.MR1 cells without 5-OP-RU (Fig. 4). This suggests that MR1 does not interact with AP2A1 or AP2M1 when most MR1 is located within the ER, but rather upon recruitment of MR1–VitBAg complexes to the cell surface.
MR1 interacts with AP2 when displayed at the cell surface. HeLa cells transduced with or without MR1 were incubated with or without 5-OP-RU for 4 h and then fixed. After permeabilization, cells were stained for the nucleus, total membranes, and then with antibodies directed to MR1 and AP2 proteins in the following combinations: (i) rabbit anti-MR1-cytosolic tail (αMR1 [CT]) and mouse anti-AP2A1; (ii) rabbit anti-AP2A1 and mouse anti-MR1 luminal domain (clone 8F2.F9); (iii) rabbit anti-AP2M1 and mouse anti-MR1 luminal domain (clone 8F2.F9). The slides were then subjected to PLA. Each PLA spot represents a molecular interaction between MR1 and AP2A1, and these were enumerated in each cell. Images are shown as maximum projections of Z-stacks, and the white scale bars represent 10 µM. Data are shown as the mean ± SD of n = 20–50 cells from each of two independent experiments. Statistical significance was calculated with one-way ANOVA with multiple comparison test where ****, P < 0.0001.
MR1 interacts with AP2 when displayed at the cell surface. HeLa cells transduced with or without MR1 were incubated with or without 5-OP-RU for 4 h and then fixed. After permeabilization, cells were stained for the nucleus, total membranes, and then with antibodies directed to MR1 and AP2 proteins in the following combinations: (i) rabbit anti-MR1-cytosolic tail (αMR1 [CT]) and mouse anti-AP2A1; (ii) rabbit anti-AP2A1 and mouse anti-MR1 luminal domain (clone 8F2.F9); (iii) rabbit anti-AP2M1 and mouse anti-MR1 luminal domain (clone 8F2.F9). The slides were then subjected to PLA. Each PLA spot represents a molecular interaction between MR1 and AP2A1, and these were enumerated in each cell. Images are shown as maximum projections of Z-stacks, and the white scale bars represent 10 µM. Data are shown as the mean ± SD of n = 20–50 cells from each of two independent experiments. Statistical significance was calculated with one-way ANOVA with multiple comparison test where ****, P < 0.0001.
The AP2 complex regulates the duration of surface MR1–VitBAg presentation
We next sought to establish if AP2 is required for loading and presentation of VitBAg, similar to its role promoting MHC-II and CD1 access to endosomes for Ag loading (Dugast et al., 2005; McCormick et al., 2005; Van Kaer et al., 2016). We first employed an MR1 Ag analog tethered to the tetramethylrhodamine fluorophore (MAgA-TAMRA; McWilliam et al., 2020), which allows the detection of MR1 metabolite presentation at the cell surface using TAMRA-recognizing antibodies (Fig. 5 A i). We pulsed C1R.Ctrl, C1R∆AP2A1, or C1R∆MR1 cells with MAgA-TAMRA and then chased without ligand. We could detect MAgA-TAMRA at the surface of WT (Ctrl) cells above the ∆MR1 cells, which gradually returned to near baseline levels at 8 h of chase; in contrast, ∆AP2A1 cells had a prolonged presentation with higher surface MAgA-TAMRA than WT cells at 8 h chase (Fig. 5 A ⅱ). We tested if the same was true for the presentation of 5-OP-RU by measuring the activation of Jurkat cells expressing a canonical MAIT TCR (Jurkat.MAIT), in a similar pulse-chase assay with 5-OP-RU (Fig. 5 B). There was an elevated Jurkat.MAIT cell activation for AP2A1 deleted cells over a 24-h chase period, implying these mutant cells exhibited prolonged display of surface MR1–5-OP-RU complexes. We also compared MAgA-TAMRA presentation in C1R cells overexpressing MR1-WT or -Y313A. As expected, the MR1-Y313A molecules had an extended presentation of MAgA-TAMRA with more at 4 and 8 h chase (Fig. 5 C). We also measured Förster resonance energy transfer (FRET) between the TAMRA moiety and the fluorescent antibody-labeled MR1 to measure MR1–MAgA-TAMRA complexes (McWilliam et al., 2020). This FRET signal only occurs when MAgA-TAMRA is bound to MR1 and thus is a more accurate measure of MR1–metabolite complexes. Similarly, this showed that MR1-Y313A complexes were slower to decay than MR1-WT (Fig. 5 D). Together this shows that MR1–metabolite complex internalization controls presentation, and loss of AP2A1 or preventing the MR1–AP2 interaction with the Y313A mutation results in unregulated and extended antigen presentation.
AP2 internalizes MR1–metabolite surface complexes. (A) (i) The detection of surface presented MAgA-TAMRA is measured using anti-TAMRA antibodies. (ii) Ctrl or ∆AP2A1 or ∆MR1 C1R cells were pulsed with 1 µM of MAgA-TAMRA for 4 h, then chased in ligand-free media for up to 8 h. Surface TAMRA was detected anti-TAMRA antibody by flow cytometry. Representative histogram of surface TAMRA presentation (left) and fold change of surface TAMRA presentation over the time (right). (B) Ctrl or ∆AP2A1 C1R cells were incubated with 10 nM of 5-OP-RU for 4 h, then washed and incubated in ligand-free media for up to 24 h prior overnight co-culture with Jurkat.MAIT cells. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. (C) C1R cells expressing MR1-WT or -Y313A were pulsed with MAgA-TAMRA for 4 h and then chased in ligand-free media for up to 18 h, and TAMRA detected at the surface as in A. (D) Cells were treated as in C and then stained with anti–MR1-AF647. The interaction of MR1-AF647 and MAgA-TAMRA was detected by measuring the FRET signal between AF647 and TAMRA in close proximity. Data shown are mean ± SD of two Ctrl and ∆AP2A1 clones (A and B; n = 4 replicates) or each construct (C and D; n = 4 replicates) from two individual experiments. Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; ***, P < 0.001; and ****, P < 0.0001.
AP2 internalizes MR1–metabolite surface complexes. (A) (i) The detection of surface presented MAgA-TAMRA is measured using anti-TAMRA antibodies. (ii) Ctrl or ∆AP2A1 or ∆MR1 C1R cells were pulsed with 1 µM of MAgA-TAMRA for 4 h, then chased in ligand-free media for up to 8 h. Surface TAMRA was detected anti-TAMRA antibody by flow cytometry. Representative histogram of surface TAMRA presentation (left) and fold change of surface TAMRA presentation over the time (right). (B) Ctrl or ∆AP2A1 C1R cells were incubated with 10 nM of 5-OP-RU for 4 h, then washed and incubated in ligand-free media for up to 24 h prior overnight co-culture with Jurkat.MAIT cells. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. (C) C1R cells expressing MR1-WT or -Y313A were pulsed with MAgA-TAMRA for 4 h and then chased in ligand-free media for up to 18 h, and TAMRA detected at the surface as in A. (D) Cells were treated as in C and then stained with anti–MR1-AF647. The interaction of MR1-AF647 and MAgA-TAMRA was detected by measuring the FRET signal between AF647 and TAMRA in close proximity. Data shown are mean ± SD of two Ctrl and ∆AP2A1 clones (A and B; n = 4 replicates) or each construct (C and D; n = 4 replicates) from two individual experiments. Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; ***, P < 0.001; and ****, P < 0.0001.
Impaired MR1 recycling reduces presentation of a 5-A-RU prodrug
We and others have suggested that recycling of surface MR1 molecules may be a mechanism of Ag loading, whereby during MR1’s access to the endosomal compartment, it may exchange its cargo for new VitBAg such as those released from intracellular pathogens residing in phagosomes (Harriff et al., 2016; Karamooz et al., 2019; McWilliam et al., 2016). Our findings here indicate that deletion of AP2A1 in C1R cells impairs the ability of surface MR1 molecules to be recycled by around 70% (Fig. 2 E). Thus if recycling is critical for MR1–VitBAg presentation, cells lacking AP2A1 would have impaired presentation of endosomal VitBAg. To test this, we employed a recycling-dependent MR1 ligand reported recently (Lange et al., 2020). This synthetic “5-A-RU prodrug” contains a cleavable motif requiring enzymatic digestion in the endosomal compartment, and MR1 recycling is required for efficient presentation (Lange et al., 2020). We compared WT or ∆AP2A1 cells for their ability to present either native 5-A-RU or the 5-A-RU prodrug to Jurkat.MAIT cells (Fig. 6 A). 5-A-RU–treated ∆AP2A1 cells could activate Jurkat.MAIT cells better than WT cells at the highest dose, owing to extended presentation (Fig. 5 A ⅱ). However, the opposite was true for the 5-A-RU prodrug; ∆AP2A1 cells had a reduced ability to present Ag from the prodrug (Fig. 6 A). In contrast, the MR1-Y313A molecules, which did not have impaired recycling, had no reduction in the presentation of Ag from the 5-A-RU prodrug compared to MR1-WT molecules (Fig. 6 B). This confirms that the loss of AP2A1 in C1R cells reduces MR1 recycling.
Significance of MR1 recycling in antigen presentation. (A and B) Ctrl or ∆AP2A1 C1R cells (A) or cells expressing MR1-WT or MR1-Y313A (B) were co-cultured with Jurkat.MAIT cells with the indicated concentrations of 5-A-RU or the 5-A-RU prodrug overnight. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. Representative histogram at 1 µM of the indicated ligand (left) and fold change of CD69 surface expression (right) are shown. (C) Ctrl, ∆AP2A1, or ∆MR1 C1R cells uninfected (UI) or infected with STM, S. flexneri (Sf), or B. thailandensis (Bt). Cells were then co-cultured overnight with Jurkat.MAIT cells in the presence of gentamycin. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. Representative histogram of CD69 expression (left) and the percentage of CD69+ Jurkat.MAIT cells are shown (right). (D) Ctrl or ∆AP2A1 or ∆MR1 C1R cells were pretreated with or without 10 µM 6-FP for 4 h, followed by BFA and 1 µM of MAgA-TAMRA. Surface TAMRA was detected with anti-TAMRA antibody. (E) C1R cells overexpressing MR1 were cultured with MAgA-TAMRA, and MR1–MAgA-TAMRA complexes were immunoprecipitated with anti-TAMRA antibody. The precipitate was then divided into new tubes and incubated with buffers of varying pH with or without 10 µM Ac-6-FP and then washed. The remaining MR1 HC or β2m was detected by immunoblotting (IB). Protein abundance was quantitated with densitometry and shown as a percentage of that at pH 7 for each treatment (below). Molecular weight standards (kD) are shown. Data shown are the mean ± SD of replicates from two independent experiments (A–C and E, n = 4), or representative of two separate experiments (E). Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; and ****, P < 0.0001; or not significant (n.s.). Source data are available for this figure: SourceData F6.
Significance of MR1 recycling in antigen presentation. (A and B) Ctrl or ∆AP2A1 C1R cells (A) or cells expressing MR1-WT or MR1-Y313A (B) were co-cultured with Jurkat.MAIT cells with the indicated concentrations of 5-A-RU or the 5-A-RU prodrug overnight. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. Representative histogram at 1 µM of the indicated ligand (left) and fold change of CD69 surface expression (right) are shown. (C) Ctrl, ∆AP2A1, or ∆MR1 C1R cells uninfected (UI) or infected with STM, S. flexneri (Sf), or B. thailandensis (Bt). Cells were then co-cultured overnight with Jurkat.MAIT cells in the presence of gentamycin. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry. Representative histogram of CD69 expression (left) and the percentage of CD69+ Jurkat.MAIT cells are shown (right). (D) Ctrl or ∆AP2A1 or ∆MR1 C1R cells were pretreated with or without 10 µM 6-FP for 4 h, followed by BFA and 1 µM of MAgA-TAMRA. Surface TAMRA was detected with anti-TAMRA antibody. (E) C1R cells overexpressing MR1 were cultured with MAgA-TAMRA, and MR1–MAgA-TAMRA complexes were immunoprecipitated with anti-TAMRA antibody. The precipitate was then divided into new tubes and incubated with buffers of varying pH with or without 10 µM Ac-6-FP and then washed. The remaining MR1 HC or β2m was detected by immunoblotting (IB). Protein abundance was quantitated with densitometry and shown as a percentage of that at pH 7 for each treatment (below). Molecular weight standards (kD) are shown. Data shown are the mean ± SD of replicates from two independent experiments (A–C and E, n = 4), or representative of two separate experiments (E). Statistical significance was calculated with two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; and ****, P < 0.0001; or not significant (n.s.). Source data are available for this figure: SourceData F6.
MR1 recycling is not required to sample intracellular bacterial Ag
Next, we tested the ability of WT or AP2A1-deleted cells infected with various intracellular bacteria to activate Jurkat.MAIT cells. We chose three bacteria that have riboflavin biosynthesis pathways and distinct intracellular niches: Salmonella enterica serovar Typhimurium (STM), which predominantly resides in a vacuolar space (García-Del Portillo, 2001); Shigella flexneri, which escapes from the phagosome to the cytoplasm within minutes (Schnupf et al., 2019); and Burkholderia thailandensis that occupies the phagosome for several hours before breaching this compartment (French et al., 2011). These infections activated Jurkat.MAIT cells in an MR1-dependent manner since there was increased CD69 expression when these cells were co-cultured with WT (Ctrl) compared to ∆MR1 C1R cells (Fig. 6 C). Despite the loss of recycling in the ∆AP2A1 mutant cells, there was no loss in Jurkat.MAIT activation; indeed, there was a slight increase in all three infections, probably due to prolonged display of MR1–Ag complexes (Fig. 5).
Thirdly, we tested if the exchange of exogenous ligand was dependent on recycling MR1 molecules as previously proposed (Harriff et al., 2016; Karamooz et al., 2019; McWilliam et al., 2016). We monitored the loading of MR1 already present at the cell surface or in recycling endosomes by blocking the traffic of new MR1 molecules from the ER with brefeldin A (BFA), employing MAgA-TAMRA to detect MR1–metabolite complexes (Fig. 6 D). In both control and ∆AP2A1 cells, MAgA-TAMRA presentation was largely reduced with BFA treatment (Fig. 6 D), confirming that MR1–Ag complexes are predominantly formed in the ER (McWilliam et al., 2016; McWilliam et al., 2020). However, after pretreatment with non-stimulatory MR1 ligand 6-FP, which is known to upregulate MR1, there was an increase in the MAgA-TAMRA presentation, and this was MR1 dependent since it did not occur in ∆MR1 cells (Fig. 6 D). This suggests that there is an exchange of 6-FP complexes for MAgA-TAMRA. However, this was similar in WT and ∆AP2A1 cells. Taken together, these data indicate that MR1 recycling is not required for metabolite Ag presentation in this system.
Considering that MR1–ligand exchange does occur (Fig. 6 D and shown in previous studies [Karamooz et al., 2019; Lange et al., 2020; McWilliam et al., 2016]), we sought to understand the physicochemical properties that define MR1–ligand dissociation. For this, we employed MAgA-TAMRA immunoprecipitation, where MR1–MAgA-TAMRA complexes are isolated from cells using antisera against TAMRA as previously shown (McWilliam et al., 2020). We treated these complexes with buffers of decreasing pH (7.0–2.5) in the presence or absence of a second ligand, Ac-6-FP, and washed the immunoprecipitates by repeated resuspension and centrifugation (Fig. 6 E). The final precipitates were then probed by immunoblotting for the presence of MR1 heavy chain (HC) or β2m. We found that in the absence of a second ligand (Fig. 6 E, upper blots), there was no loss of MR1 HC between pH 7 and 5, suggesting very little dissociation of MAgA-TAMRA–MR1 complexes in these conditions. However, with Ac-6-FP present (lower blots), low pH caused reduced MR1 recovery. In addition, we saw a striking loss of β2m at pH < 6.0, regardless of the presence or absence of Ac-6-FP (Fig. 6 E). This suggests that while the MR1–ligand Schiff base bond is relatively stable, a low pH of 5.0 corresponding to that within the lysosome allows the replacement of one ligand by another. However, since the MR1 HC-β2m interaction is highly unstable at pH < 6.0, it is likely that the MR1 heterodimer would dissociate before the ligand exchange can occur. As β2m is required for MR1 surface expression (Abós et al., 2011), its dissociation is expected to prevent MAIT cell recognition.
The cytoplasmic tail of MR1 defines the duration of metabolite Ag surface display
Next, we probed the molecular basis for MR1’s unusual internalization rate, which is slower than for other cargoes that possess canonical AP2 motifs such as the TfR, which is largely internalized within minutes (Briken, 2002; Dumont et al., 2017) or CD1 molecules such as CD1d and CD1b where ∼30–50% is internalized in 1 h (Briken, 2002; Jayawardena-Wolf et al., 2001). Mutation of T316 to alanine had no effect on MR1 internalization, suggesting it is not essential for recognition by AP2. Instead, we hypothesized that the unusual T316 residue in the place of the canonical Φ serves to reduce the affinity of MR1 for AP2, thus preventing its rapid internalization and prolonging the surface display of MR1–metabolite complexes. Given that MR1–β2m heterodimer is unstable in endosomal pH (< 6.0), this may serve to allow display and prevent premature transit through the late endosomes where the heterodimer may be denatured.
To test this hypothesis, we mutated the MR1 tail to generate a canonical YXXΦ motif: (i) we swapped the entire MR1 tail with that of CD1d (MR1-CD1d-tail); and (ii) mutated only the T316 with the Φ residue from human CD1d (Rodionov et al., 1999), valine (MR1-T316V). Both mutants had reduced surface expression, particularly MR1-T316V, but all could upregulate MR1 in response to VitBAg with a similar fold change (Fig. 7 A). Each mutant showed an increased rate of internalization, with more internalized at 4 h for the MR1-CD1d-tail and MR1-T316V mutants (Fig. 7 B). MR1-CD1d-tail and MR1-T316V had increased recycling rates, with the latter showing higher recycling at 5 min, which then decreased after this early time point (Fig. 7 C). We assessed the functional MR1–VitBAg presentation of each mutant by measuring CD69 expression on Jurkat.MAIT cells (Fig. 7 D); both mutants had impaired ability to present 5-A-RU and the 5-A-RU prodrug, and MR1-T316V could also not present 5-OP-RU to the same extent as MR1-WT. Thus, conferral of a canonical YXXΦ motif increased MR1 internalization and recycling, but decreased MR1–VitBAg presentation.
The non-canonical tyrosine-based AP2 motif in MR1 defines the duration of Ag presentation. (A) C1R cells expressing MR1-WT or MR1-CD1d-tail or the mutation MR1-T316V were treated with 10uM 5-OP-RU for 4 h. Surface MR1 expression was analyzed with anti-MR1 (clone 8F2.F9) by flow cytometry and shown as gMFI (i) and the fold change in expression from no ligand (ii). (B) The internalization of MR1-WT, the MR1-CD1d-tail, or MR1-T316V mutants was measured as in Fig. 1 A over 4 h (i) or at 4 h (ii). (C) The recycling rate of MR1-WT, the MR1-CD1d-tail, or MR1-T316V mutants was measured as in Fig. 1 A over 4 h (i) or at 5 min (ii). (D) The activation of Jurkat.MAIT cells co-cultured with C1R cells expressing MR1-WT, the MR1-CD1d-tail, or MR1-T316V. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry, represented as the fold change of gMFI of CD69 surface expression. (E) C1R cells expressing MR1-WT, the MR1-CD1d-tail, or MR1-T316V were metabolically radiolabeled with 35S-methionine/cysteine and then chased for the indicated times in the presence of Ac-6-FP, then MR1–β2m complexes were immunoprecipitated with mAb 8F2.F9. The protein bands were quantitated with densitometry. Molecular weight standards (kD) are shown. (F) HeLa cells transduced without or with MR1-WT or the mutants were incubated with or without 5-OP-RU for 4 h and then fixed. PLA performed as for Fig. 1 A, with antibodies in the following combinations: (i) rabbit anti-β2m and mouse anti-MR1 luminal domain (clone 8F2.F9); (ii) rabbit anti-AP2A1 and mouse anti-MR1 luminal domain (clone 8F2.F9); (iii) rabbit anti-AP2M1 and mouse anti-MR1 luminal domain (clone 8F2.F9). The PLA spots were enumerated, and each dot represents one cell. The average number of PLA spots for ii and iii for each of two experiments were also calculated relative to the number of MR1–β2m spots in i and expressed as a fold change of HeLa cells. Data are shown as mean ± SD of replicates from one experiment representative of two (A; n = 2) or the mean ± SD of replicates from two independent experiments (B–F; n = 4). Statistical significance was calculated with one-way or two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData F7.
The non-canonical tyrosine-based AP2 motif in MR1 defines the duration of Ag presentation. (A) C1R cells expressing MR1-WT or MR1-CD1d-tail or the mutation MR1-T316V were treated with 10uM 5-OP-RU for 4 h. Surface MR1 expression was analyzed with anti-MR1 (clone 8F2.F9) by flow cytometry and shown as gMFI (i) and the fold change in expression from no ligand (ii). (B) The internalization of MR1-WT, the MR1-CD1d-tail, or MR1-T316V mutants was measured as in Fig. 1 A over 4 h (i) or at 4 h (ii). (C) The recycling rate of MR1-WT, the MR1-CD1d-tail, or MR1-T316V mutants was measured as in Fig. 1 A over 4 h (i) or at 5 min (ii). (D) The activation of Jurkat.MAIT cells co-cultured with C1R cells expressing MR1-WT, the MR1-CD1d-tail, or MR1-T316V. Jurkat.MAIT cell activation was measured by CD69 surface expression by flow cytometry, represented as the fold change of gMFI of CD69 surface expression. (E) C1R cells expressing MR1-WT, the MR1-CD1d-tail, or MR1-T316V were metabolically radiolabeled with 35S-methionine/cysteine and then chased for the indicated times in the presence of Ac-6-FP, then MR1–β2m complexes were immunoprecipitated with mAb 8F2.F9. The protein bands were quantitated with densitometry. Molecular weight standards (kD) are shown. (F) HeLa cells transduced without or with MR1-WT or the mutants were incubated with or without 5-OP-RU for 4 h and then fixed. PLA performed as for Fig. 1 A, with antibodies in the following combinations: (i) rabbit anti-β2m and mouse anti-MR1 luminal domain (clone 8F2.F9); (ii) rabbit anti-AP2A1 and mouse anti-MR1 luminal domain (clone 8F2.F9); (iii) rabbit anti-AP2M1 and mouse anti-MR1 luminal domain (clone 8F2.F9). The PLA spots were enumerated, and each dot represents one cell. The average number of PLA spots for ii and iii for each of two experiments were also calculated relative to the number of MR1–β2m spots in i and expressed as a fold change of HeLa cells. Data are shown as mean ± SD of replicates from one experiment representative of two (A; n = 2) or the mean ± SD of replicates from two independent experiments (B–F; n = 4). Statistical significance was calculated with one-way or two-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Source data are available for this figure: SourceData F7.
To determine if the tail mutants affect the half-life of MR1, we radiolabeled cells expressing WT or mutant MR1, and chased these in the presence of the stable ligand Ac-6-FP. We immunoprecipitated MR1–β2m complexes using conformational mAb 8F2.F9 and treated these with endoglycosidase H to distinguish those within the ER (endo H sensitive) and those processed through the Golgi apparatus (endo H resistant). As expected, immediately after the pulse and without Ac-6-FP, virtually all the radiolabeled WT and mutant MR1 molecules were was endo H sensitive (Fig. 7 E, lanes 2 vs. 3). Following 6 h chase in the presence of Ac-6-FP, the majority of MR1 became endo H resistant for all MR1 constructs as the molecules trafficked out of the ER/Golgi apparatus (Fig. 7 E, lane 4). For MR1-WT, there was more MR1 precipitated at 6 h compared to 0 h (∼150%) because the MR1 ligands promote continuous folding and acquisition of the 8F2.F9 epitope. By 24 h, some degradation was evident with only ∼80% of the initial MR1 remaining (Fig. 7 E). For the MR1-Y313A mutant, there was ∼70% more MR1–β2m complexes recovered at 6 h compared to WT. This implies that a proportion of WT molecules undergo degradation by 6 h. At 24 h, a large pool of Y313A still remained. For both the MR1-CD1d-tail and MR1-T316V mutants, the opposite occurred; there was less MR1 recovered at 6 h chase and almost none remaining by 24 h (Fig. 7 E). This confirms that the tail motif of MR1 controls the half-life of mature MR1 molecules.
Finally, we used PLA to test if MR1 tail mutants had altered interaction with AP2. We examined parental HeLa cells to those overexpressing MR1-WT, -Y313A, or -T316V, cultured with 5-OP-RU to recruit MR1 to the cell surface. MR1-WT and -Y313A had similar surface expression, but T316V mutant had very low expression as seen in C1R cells (Fig S3 and Fig. 7 A). We first measured PLA interactions between MR1–β2m to control for differences in the numbers of these complexes. Each cell line with overexpressed MR1 had higher number of PLA spots compared to HeLa cells with low MR1 expression (Fig. 7 F i). Compared to MR1-WT, there were more complexes for MR1-Y313A, supporting the idea that this mutant has a slower rate of degradation, but only slightly less for MR1-T316V. Next, we compared the PLA spots for MR1-AP2M1 and MR1-AP2A1 (Fig. 7, F ⅱ and ⅲ). Both mutant molecules had fewer PLA spots and hence reduced interactions with AP2M1 and AP2A1 than MR1-WT. However, since there were different amounts of MR1–β2m complexes for each cell line, we calculated the number of spots relative to the number of MR1–β2m PLA. This allowed us to determine the relative interaction of each MR1 with AP2 (Fig. 7, F ⅱ and ⅲ). This confirmed that the Y313A mutation caused strikingly lower MR1–AP2 interactions; in contrast the T316V mutation had comparable AP2 interactions to MR1-WT. Altogether, this indicates that the non-canonical tyrosine-based motif of MR1 interacts suboptimally with AP2, resulting in reduced internalization, enhanced survival of folded MR1 and prolonged metabolite presentation.
Discussion
In this study, we show that the display of MR1–metabolite complexes is controlled by their cytoplasmic tail, likely recognized by the AP2 complex, which controls a defined rate of surface expression and minimizes recycling.
AP2 functions to recruit proteins to clathrin-coated pits that then fuse with early/sorting endosomes (Maxfield and McGraw, 2004). As the clathrin coat and AP2 are shed rapidly after internalization, AP2 operates in the initial stages of endocytosis (Maxfield and McGraw, 2004). The fate of endocytosed AP2 cargo is either transit to the endolysosomal route for degradation, or recycling back to the cell surface (Naslavsky and Caplan, 2018). Recycling can occur directly from the early endosomes (fast recycling), or after trafficking to the recycling endosome (slow recycling; Maxfield and McGraw, 2004; Sheff et al., 2002), but does not directly require AP2. While the mechanisms of cargo recycling are not fully resolved (Naslavsky and Caplan, 2018), sorting factors are involved for some proteins such as TfR (Dai et al., 2004).
To understand the role of AP2 in MR1 internalization, we examined firstly AP2A1 KO cells, and secondly cells expressing the MR1-Y313A mutant to prevent AP2 interaction. Importantly, both approaches resulted in a similar phenotype: reduced MR1 internalization leading to the accumulation of MR1–VitBAg complexes on the cell surface and increased MAIT cell activation. This implies that internalization limits MR1 display.
A low level of MR1 recycling is seen in several cell types. This was dependent on AP2 endocytosis since cells lacking AP2A1 had dramatically impaired MR1 recycling. However, it is likely that AP2 plays an indirect role in recycling since it operates at the plasma membrane and not within the early/sorting or recycling endosomal compartment (Maxfield and McGraw, 2004)—perhaps by delivering a fraction of MR1 complexes to the appropriate endosomes that allows recycling.
The discovery that the deletion of AP2A1 impairs most of the recycling in C1R cells allowed us to interrogate for the first time the importance of MR1 recycling on Ag presentation. We found that AP2A1 KO cells were still proficient at presenting Ag from the extracellular medium, or from three intracellular bacteria. Additionally, while we could detect ligand exchange in endogenous levels of MR1 with MAgA-TAMRA, this was also not dependent on recycling and likely occurs at the cell surface. Thus, we conclude that recycling is dispensable for MR1–metabolite Ag presentation.
The inefficient rate of MR1 recycling appears to be governed by its unique cytoplasmic tail motif, since converting the motif to the canonical sequence for AP2 binding dramatically increased recycling. However, this impaired MAIT cell stimulation, suggesting recycling is detrimental for MR1 function. Recycling may bring MR1 into acidic compartments, as seen for CD1 molecules, which will likely denature MR1–β2m complexes as they are unstable at pH < 6.0. This acid instability intriguingly coincides with the acid-labile nature of VitBAg (Mak et al., 2017). Together, these observations strongly suggest that MR1 does not survey acidic compartments through a recycling mechanism. MHC-II and CD1b-d molecules rely on AP2 for their internalization to enable transit to endosomal compartments where Ag loading takes place (Dugast et al., 2005; McCormick et al., 2005; Van Kaer et al., 2016). However, our data suggest AP2 serves to internalize preformed MR1–VitBAg complexes from the cell surface for degradation and is not required for Ag acquisition (Fig. S4).
A schematic for the dominant pathway of metabolite presentation by MR1. MR1 (pink) resides in the ER in a ligand-receptive form. Extracellular or intracellular VitBAg (orange) can load onto MR1 within ER (1). MR1–VitBAg complexes traffic through the Golgi apparatus to the plasma membrane (PM) for presentation (2). Complexes are recognized by the AP2 complex (dark blue) at the cytoplasmic tail and internalized to the early endosomes (EE; 3). The majority are degraded in the late endosomes/lysosomes (LEL; 4) with some recycling back to the cell surface (5). Surface MR1 can exchange its VitBAg for another type (green) at the cell surface (6).
A schematic for the dominant pathway of metabolite presentation by MR1. MR1 (pink) resides in the ER in a ligand-receptive form. Extracellular or intracellular VitBAg (orange) can load onto MR1 within ER (1). MR1–VitBAg complexes traffic through the Golgi apparatus to the plasma membrane (PM) for presentation (2). Complexes are recognized by the AP2 complex (dark blue) at the cytoplasmic tail and internalized to the early endosomes (EE; 3). The majority are degraded in the late endosomes/lysosomes (LEL; 4) with some recycling back to the cell surface (5). Surface MR1 can exchange its VitBAg for another type (green) at the cell surface (6).
MR1 VitBAg presentation and MAIT cell stimulation are conserved among diverse cell types—such as epithelial and professional Ag presenting cells (Jeffery et al., 2016; Le Bourhis et al., 2010; McWilliam et al., 2016; McWilliam et al., 2020). The findings presented here that MR1 surface expression is regulated by its tail and interaction with endocytic machinery offers a mechanism for cell type–specific regulation of MR1 trafficking. Indeed, we noted differences in MR1 internalization and recycling in the cell types examined; primary monocytes had high levels of internalization and recycling compared to lymphocytes, and this correlated with higher expression of most AP2 subunit genes. C1R and THP1 cells also had relatively high internalization. As clearance of plasma membrane MR1–VitBAg complexes limits MAIT cell activation, we speculate that cell types that have greater endocytosis of MR1—such as myeloid or professional presenting cells—may use this to cease metabolite presentation for a more rapid return to homeostasis. Further work that directly compares cell-type MR1 presentation and trafficking are needed to confirm this.
Our data presented here does not prove a direct interaction of MR1 and AP2, but multiple lines of evidence strongly support MR1 as a bona fide AP2 cargo: (i) MR1 internalization is sensitive to an inhibitor of clathrin-mediated endocytosis, the path directed by AP2; (ii) Ap2a1 was the only gene found in our genome-wide screen regulating MR1 internalization; (iii) MR1 endocytosis requires an intact AP2 complex in different cell types; (iv) MR1 requires its conserved tyrosine residue for internalization, a residue recognized by AP2; (v) MR1 is in close proximity to AP2 components in cells, dependent on its tyrosine residue which indicates interaction; and (vi) when MR1’s tail is modified to have a canonical AP2 motif, its internalization is faster, pointing to increased AP2 affinity.
Despite numerous attempts, we could not use immunoprecipitation followed by immunoblotting or mass spectrometry to determine whether MR1 and AP2 interact or are part of a larger complex. Therefore, we used PLA and showed that MR1 and AP2 were in proximity indicative of a direct interaction in fixed cells. While PLA does not demonstrate a direct interaction of two proteins, it reveals if they are within 40 nm, the size of a large protein complex; indeed AP2 itself is ∼10 nm (Kovtun et al., 2020), and MR1 at least 4 nm (Kjer-Nielsen et al., 2012). This might be explained in two ways: (i) MR1-AP2 directly interacts, or (ii) MR1 is simply very close to AP2 in the plasma membrane. However, subsequent PLA analysis showed that the MR1-Y313A tail mutant had no detectable interaction with AP2. Importantly, this mutant is present in the plasma membrane to a greater extent than its WT counterpart, implying that simply being in this location does not cause MR1 to come into proximity with AP2. Therefore, these data strongly support the former explanation that WT MR1 molecules directly interact with AP2. It is important to note that we did not find the T316V mutation increased MR1’s association with AP2. A possible explanation for this is that the T316V mutant does not have an increased association rate with AP2 than MR1-WT, but rather the affinity is higher, and each interaction is more likely to result in an endocytosis event.
In summary, we show that the appropriate display of metabolite Ag by MR1 molecules is governed by its cytoplasmic tail motif, and we pose that this is recognized by the AP2 complex. A balance exists between two residues in the motif for reduced affinity with AP2 that defines MR1’s rate of endocytosis. Y313 increases internalization, whereas T316 was not required; but converting the latter to a Φ residue increased internalization; this suggests that it may function as an “anti-Φ” residue to limit AP2 internalization. We hypothesize therefore that MR1’s motif is tuned for an internalization rate of hours, rather than minutes, as observed for cargo bearing a canonical YXXΦ motif. Further, the lack of a Φ residue reduced recycling—which is detrimental for MR1 Ag presentation. Rather than needing to traffic to endosomal compartments to acquire metabolites, MR1 can efficiently capture ligands prior to translocating to the cell surface, and from there most molecules are eventually endocytosed. Thus, we postulate that AP2 serves as the “off” signal for MR1–VitBAg cell-surface display. We speculate that the distinct AP2 motif of MR1 has arisen to simultaneously enable sufficient exposure time on the surface of the antigen presenting cell to enable presentation to MAIT cells, while limiting exposure to avoid inappropriate responses once the microbial threat has been eliminated.
Materials and methods
Expression constructs and individual CRISPR-Cas9 gene KO
MR1 constructs were expressed in C1R cells using the pMSCV-IRES-GFP II vector (pMIG II, a gift from Dario Vignali, University of Pittsburgh, Pittsburgh, PA; plasmid #52107; Addgene; http://n2t.net/addgene:52107; RRID:Addgene_52107), and MR1-GFP expressed in HeLa cells as previously generated (McWilliam et al., 2020). MR1 mutants were generated through site-directed mutagenesis of WT MR1 sequence using the QuikChange II Site-Directed Mutagenesis Kit (Agilent) and then cloned into the pMIG vector. Transduced cells were sorted based on GFP expression. For overexpression in HeLa cells, MR1 and constructs were cloned into the pLenti-puro vector (Guan et al., 2011). The GFP-TM construct (pHR_EGFPligand; plasmid #79129; Addgene) was previously reported (Roybal et al., 2016).
CRISPR-Cas9 gene KOs were generated using the FUCas9Cherry and FgH1tUTG vectors (Aubrey et al., 2015; plasmid #70182; Addgene; http://n2t.net/addgene:70182; RRID:Addgene_70182; and plasmid # 70183; http://n2t.net/addgene:70183; RRID:Addgene_70183). The sgRNA expression was induced with 1 µg/ml doxycycline, then cells were cloned, expanded, and the KO confirmed by immunoblotting. To minimize off-target effects from one sgRNA, two distinct sgRNAs were designed for each gene using the online Broad Institute sgRNA Designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and the non-targeting control sgRNA were obtained from the GeCKO v2 library. The sgRNA sequences are: Control non-targeting sgRNA 1 (Ctrl-1), 5′-ACGGAGGCTAAGCGTCGCAA-3′; Control non-targeting sgRNA 2 (Ctrl-2), 5′-CGCTTCCGCGGCCCGTTCAA-3′; AP2A1 sgRNA 1 (AP2A1-1), 5′-CGATGGGATGCGGGGGCTCG-3′; AP2A1 sgRNA 2 (AP2A1-2), 5′-TGTGCCCAAAGTCAATGTCA-3′; AP2M1 sgRNA 1 (AP2M1-1) 5′-AAATGTTGGACCGCTTAACG-3′; AP2M1 sgRNA 2 (AP2M1-2), 5′-ATAGAGGAATTCGAAGACCA-3′; AP2A2 sgRNA 1 (AP2A2-1), 5′-TGATGGCGTTGTTGATCAGG-3′; AP2A2 sgRNA 2 (AP2A2-2), 5′-AGGGCCAGGCCCATGAAGGT-3′.
MR1 ligands
MR1 ligands were added directly to the cells in the culture media at the indicated concentration. Stock solutions of Ac-6-FP (Schircks Laboratories), 5-OP-RU (synthesized in DMSO [Mak et al., 2017; Mak et al., 2021]), and MAgA-TAMRA (McWilliam et al., 2020) were stored in DMSO. 5-A-RU was kept in aqueous solution and the 5-A-RU prodrug (compound 10 from Lange et al., 2020) was dissolved in DMSO at 10 mg/ml (Lange et al., 2020).
Human samples and ethics
Buffy coats from healthy human donors were obtained from the Australia Red Cross Blood Service with written and informed consent from the donors with ethics approval from the University of Melbourne Human Research and Ethics Committee (#1035100). Peripheral blood mononuclear cells (PBMCs) were purified by standard density gradient (Ficoll-Paque Plus; GE Healthcare) and preserved in 90% FCS and 10% DMSO in liquid nitrogen.
CRISPR-Cas9 genome-wide screening
Genome-wide KO screening was performed with the human GeCKO v2 CRISPR KO library (a gift from Feng Zhang, Broad Institute, Cambridge, MA [#1000000048; Addgene]). For each of three replicates, the library with ∼500× coverage of each sgRNA was generated by transducing 200 million C1R cells at MOI = 0.3 (Joung et al., 2017). After selection with puromycin (0.5 mg/ml) for 1 wk, each library was expanded then divided into two: (1) for the unsorted reference library that remained in culture throughout the experiment and (2) to enrich the MR1HIGH cells. For the latter, the cells were incubated with 0.5 μM 5-OP-RU for 4 h, after two washes with PBS, cell was incubated without 5-OP-RU in complete DMEM for 8 h. After this, cells were stained for MR1 and MR1HIGH cells were sorted based on top 5% of the histogram. Sorted cells were expanded in culture for another 5 d and then the sorting process was repeated. Finally, 3 d after the second sorting, the unsorted reference library and sorted samples were frozen. DNA was extracted from the cells based on salt precipitation (Chen et al., 2015). The sgRNAs in each replicate was amplified and labeled with designated primers with two-step PCR as described previously using Herculase II fusion DNA polymerase (Agilent). Labeled amplicons were pulled down and purified with Nucleo Mag NGS beads (Macherey-Nagel), then sequenced using NextSeq500 (Illumina). CRISPR/Cas9 library data processing and differential representation analysis was performed using MAGeCK-VISPR (Li et al., 2015).
Internalization and recycling assays
For measure internalization, fluorescence internalization probe (FIP) assays were employed (Liu and Johnston, 2013). FIP-azide (5′-Cy5-TCAGTTCAGGACCCTCGGCT-N3-3′) and the complementary nucleic acid-conjugated quencher (5′-AGCCGAGGGTCCTGAACTGA-BHQ2-3′) were obtained from Integrated DNA Technologies. To determine receptor internalization, cells were stained on ice with FIP-Cy5–conjugated mAbs against MR1 (clone 8F2.F9) or TfR (clone OKT9). FIP-labeled cells were incubated in complete DMEM at 37°C and 10% CO2 for the indicated times. Cells were washed and resuspended in media with or without 1 μM quencher, and fluorescence measured by flow cytometry. The percentage of internalization was calculated as a percentage of the initial un-quenched signal (McWilliam et al., 2016). To measure the recycling of MR1, mAb 8F2.F9 was conjugated to NHS-SS-Biotin (Thermo Fisher Scientific; McWilliam et al., 2016). Surface MR1 was labeled with 8F2.F9-SS-biotin and labeled MR1 allowed to internalize for 1 h at 37°C. Remaining surface biotin label was removed by a cell-impermeable reducing agent, and cells were then incubated for up to 30 min to allow labeled MR1 to recycle back to the cell surface. Recycled MR1 was detected by incubating the cells with streptavidin conjugated to Alexa Fluor 647 and measured by flow cytometry. The amount of MR1 recycled was calculated as the percentage of the initial surface MR1 signal prior to biotin removal. For both assays, cells were incubated with 10uM Ac-6-FP for 16–18 h to promote the trafficking of MR1 to the cell surface (McWilliam et al., 2016).
Human PBMC expression data
The relative expression of the transcripts for each AP2 subunit was obtained from (i) The Immunological Genome Project Ultra Low Input RNAseq dataset (https://www.immgen.org/Databrowser19/HumanExpressionData.html; Heng et al., 2008); and (ii) The Human Protein Atlas version 21.0 “RNA HPA blood cell gene data” (https://www.proteinatlas.org/about/download; Uhlen et al., 2019).
Bacterial infections and Ag presentation assays
Bacterial infections were carried out with STM strain SL1344, S. flexneri, and B. thailandensis. C1R cells were infected with each bacterial species for 2 h at MOI of 10. Cells were then washed with media and incubated with either 50 μg/ml gentamicin for STM and Shigella, or 250 μg/ml for Burkholderia, for 1 h at 37°C to kill extracellular bacteria. Then, cells were co-cultured with Jurkat.MAIT in the presence of 50 μg/ml gentamicin for overnight incubation. For Ag presentation assays with soluble VitBAg (5-OP-RU, 5-A-RU, or the 5-A-RU prodrug), cells were incubated with Jurkat.MAIT cells in the presence or absence of the indicated VitBAg overnight.
Confocal microscopy and PLA
Confocal microscopy images were acquired on a Zeiss LSM780 with a 40× oil lens (numerical aperture 1.4) at room temperature (22°C) and captured with a photo multiplier tube detector by Zen 2012 software. Slides were imaged in 90% glycerol, 20 mM Tris-HCl, pH 8, 0.2 M DABCO (Sigma-Aldrich). Imaging was performed on 10–25 cells from each of two random fields of view, for each of two experiments. Internalized MR1 was followed in HeLa cells overexpressing MR1-WT or Y313A were treated with 10 µM 5-OP-RU for 4 h and then placed on ice to stop protein traffic. Surface MR1 were labeled with mouse anti-MR1 (clone 8F2.F9), then washed and incubated at 37°C in folate-free RPMI (Gibco) with 10% FCS for 4 h to allow internalization. Cells were washed, fixed, and permeabilized in 2% PFA and 0.05% saponin in PBS, after which permeabilized and blocked in 5% normal donkey serum in 0.05% saponin in PBS. Next cells were stained with goat anti-EEA1 and rabbit anti-LAMP1 followed by secondary antibodies (anti-goat Alexa Fluor [AF] 647, anti-rabbit AF568, and anti-mouse AF488). Finally, nuclei were labeled with DAPI (1 mg/ml). Images were analyzed using Imaris software (Bitplane, version 9.6.1). Each cell was segregated using the “Surfaces” function on the MR1 surface stain. The MR1+, EEA1+, or LAMP1+ endosomes were identified using the “Spots” function and overlapping spots were enumerated.
For PLA, HeLa cells overexpressing MR1 or not were seeded onto the coverslips then treated with 10 µM 5-OP-RU as indicated. After washing with PBS, the plasma membrane was stained with the Cell Plasma Membrane Staining kit (ab219941; Abcam; ex. 540 nm, em. 590 nm) as instructed by manufacturer. Fixation, permeabilization, and DAPI staining were performed as above. For each PLA assay, a pair of mouse and rabbit primary antibodies were used to label the indicated targets. PLA was carried out with Far Red Duolink PLA reagents (DUO92008; Sigma-Aldrich; ex. 594, em. 624) with anti-mouse PLUS (DUO92001) and anti-rabbit MINUS (DUO92005) probes according to the manufacturer’s instructions. Images were analyzed using Imaris software (Bitplane, version 9.6.1). Each cell was segregated using the “Surfaces” function on the plasma membrane stain, then PLA spots were enumerated within each cell using the “Spots” function.
Antibodies
For flow cytometry staining of MR1, 8F2.F9 mAb was used conjugated to biotin and detected with streptavidin conjugated to phycoerythrin. TfR was stained with biotin conjugated anti-CD71 (clone OKT9). Analysis of human PBMCs used: anti-CD19-FITC (clone HIB19; BD); anti-CD3-PerCP (clone SK7; BD); and anti-CD14-BUV805 (clone M5E2; BD). Jurkat cell activation was measured with Pacific Blue conjugated anti-CD69 (FN50; Biolegend). The presentation of MAgA-TAMRA on the cell surface was detected with rabbit anti-TAMRA (A6397; Thermo Fisher Scientific) and AF647 conjugated donkey anti-rabbit IgG. MAgA-TAMRA was measured on LSR Fortessa (BD Biosciences) by excitation with 561 nm laser and 585/15 nm emission filter and FRET between MAgA-TAMRA and AF647 by excitation with 561 nm laser and 670/30 nm emission filter (McWilliam et al., 2020). For immunoblotting and immunofluorescence/PLA: mouse anti-MR1 mAb (clone 8G3 [McWilliam et al., 2020]) and rabbit anti-β2m (product ab75853; Abcam). For detection of AP2 proteins: mouse anti-AP2A1 (clone 8/Adaptin a; BD), rabbit anti-AP2A1 (product AHP2432; Bio-rad), mouse anti-AP2M1 (clone 31/AP50; BD), and rabbit anti-AP2M1 (product AHP2434; Bio-rad). For markers of subcellular locations, the early endosome marker EEA1 (clone N-19; Santa Cruz) and late endosome and lysosome marker, LAMP1 (ab24170; Abcam) were used, followed by secondary antibodies: AF647 conjugated anti-goat, AF568 conjugated anti-rabbit, and AF488 conjugated anti-mouse (Life Technologies).
Immunoblotting, immunoprecipitation, and radiolabeling
For immunoblotting, cells were lysed in 0.5% IGEPAL CA-630 (Sigma-Aldrich), 50 mM Tris-Cl, pH 7.4, 5 mM MgCl2 with Complete EDTA-free Protease Inhibitor cocktail (Roche), and nuclei were separated by centrifugation at 13,000 × g for 10 min. Proteins were denatured with LDS sample buffer (Life Technologies) and 100 mM DTT, separated on NuPAGE 4–12% Bis-Tris precast gels (Life Technologies) and immunoblotted on to nitrocellulose membranes using the iBlot system (Life Technologies). Blots were probed with the indicated antibodies and then detected with anti-mouse HRP (NA931; GE Healthcare) or anti-rabbit HRP (NA934; GE Healthcare), developed using Amersham ECL Select Detection Reagent (GE Healthcare) and images captured on an Amersham Imager 600 (GE Healthcare). To test the pH sensitivity of MR1–β2m–MAgA-TAMRA complexes, C1R.MR1 cells were incubated with 20 μM MAgA-TAMRA for 4 h, then lysates prepared as above. The lysate was precleared once with protein G sepharose and then complexes were immunoprecipitated with anti-TAMRA and protein G sepharose. Precipitates were washed three times and then divided equally among six tubes. These were then incubated with phosphate-citrate buffers of varying pH for 5 min at RT, in the presence or absence of 10 μM Ac-6-FP. Precipitates were washed twice with PBS and then subjected to SDS-PAGE and immunoblotting as above.
For radiolabeling, cells were starved for 30 min in methionine and cysteine-free DMEM and then pulsed for 30 min in this media supplemented with 35S-labeled methionine/cysteine (Express Protein Labeling Mix, Perkin Elmer) at 200 μCi/ml. After washing cells were chased in complete DMEM containing 10 µM Ac-6-FP for the indicated times, then washed in PBS and lysed. Lysate was precleared twice with protein G sepharose and normal mouse serum and then twice without serum. MR1 was immunoprecipitated with anti-MR1 clone 8F2.F9 and protein G Sepharose and then precipitates were washed three times and treated with or without endoglycosidase Hf (New England Biolabs) as per the manufacturer’s instructions. Proteins were separated as above and transferred to nitrocellulose membranes, which were then dried and exposed to a storage phosphor screen (GE Healthcare) and imaged on a Typhoon imager (GE Healthcare).
Cytoplasmic tail consensus logo
Mammalian MR1 sequences were collected from NCBI and aligned. The cytoplasmic tail of human MR1 was identified using the transmembrane hidden Markov model online software (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0; Krogh et al., 2001; Sonnhammer et al., 1998), and after alignment of all mammalian MR1 sequences, the other species’ cytoplasmic sequences were inferred from this. Finally, the consensus logo of all sequences was generated using WebLogo (http://weblogo.threeplusone.com; Crooks et al., 2004).
Statistical analysis
Statistical analysis was performed using GraphPad Prism (version 9.1.2). Comparisons between two groups were made using unpaired t tests and paired t test with data from multiple experiments. Comparisons of more than two groups were made using one-way ANOVA for one parameter, or two-way for multiparameter tests, with repeated measures for data from multiple experiments. Each was followed by a multiple comparison test. All groups were analyzed with the assumption of normal distribution, but this was not formally tested. The following P values were considered statistically significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows the effect of endocytosis inhibitors on dextran and transferrin uptake. Fig. S2 shows CRISPR-Cas9 deletion of AP2 subunits in C1R and THP-1 cells. Fig. S3 shows expression of mutant MR1 molecules in HeLa cells. Fig. S4 shows a schematic for the dominant pathway of metabolite presentation by MR1. Table S1 shows mammalian MR1 cytoplasmic tail sequences.
Acknowledgments
We thank the Melbourne Cytometry Platform from the Doherty Institute node and the Biological Optical Microscopy Platform (University of Melbourne) for expert assistance. Finally, we thank Prof. Jennifer Stow for critical reading of this manuscript and expert advice.
E.G. McWilliam is supported by a National Health and Medical Research Council (NHMRC) Ideas Grant (2003192), and J.A. Villadangos is supported by research grants from the Australian Research Council (ARC; DP170102471), NHMRC (1113293), and an NHMRC Senior Research Fellowship (1058193). D.P. Fairlie is an NHMRC Senior Principal Research Fellow (1117017). A. Hachani acknowledges support from a Horizon 2020_MSCA Global Fellowship grant (657766). D.P. Fairlie and J. Rossjohn acknowledge support from the ARC Centre of Excellence in Advanced Molecular Imaging (CE140100011). J.A. Villadangos, J. Rossjohn, and D.P. Fairlie are supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI148407). J. Rossjohn is supported by an ARC Australian Laureate Fellowship, and G.F. Painter is supported by the New Zealand Ministry of Business Innovation and Employment (RTVU 1603).
J. Rossjohn and D.P. Fairlie are named inventors on a patent application (PCT/AU2013/000742, WO2014005194) and J.Y.W. Mak, J. Rossjohn, and D.P. Fairlie are named inventors on another patent application (PCT/AU2015/050148, WO2015149130) involving MR1 ligands for MR1-restricted MAIT cells. The authors declare no competing financial interests.
Author contributions: H.J. Lim led the experiments, wrote the manuscript, and provided intellectual input. J.M. Wubben, C.P. Garcia, S. Cruz-Gomez, J. Deng, J.Y.W. Mak, A. Hachani, R.J. Anderson, G.F. Painter, J. Goyette, S.L. Amarasinghe, M.E. Ritchie, A. Roquilly, D.P. Fairlie, K. Gaus, and J. Rossjohn performed experiments, provided key reagents, analyzed data, and/or provided intellectual input. H.E.G. McWilliam and J.A. Villadangos wrote the manuscript and led the study.
References
Author notes
J.A. Villadangos and H.E.G. McWilliam contributed equally to this paper.
Katharina Gaus died on March 3, 2021.


![Discovery of the molecular machinery that internalizes cell surface MR1. (A) The internalization rate of surface MR1 was measured in primary human PBMCs from healthy donors gating on monocytes (Mono.; blue), B cells (black), and T cells (red), or the C1R (black) and THP-1 (blue) cell lines. Cells were cultured overnight with 10 µM Ac-6-FP, then surface MR1 was labeled with anti-MR1 mAb 8F2.F9 conjugated to a nucleic acid-conjugated FIP. Cells were allowed to internalize for up to 4 h, and any remaining surface fluorescence was quenched with a complementary nucleic acid probe conjugated to a quenching dye. MR1 internalization was calculated as a percentage of the initial un-quenched signal. (B) The recycling rate of internalized MR1 of cells treated with Ac-6-FP overnight as in A. Surface MR1 was labeled with 8F2.F9-SS-biotin and allowing to internalize for 1 h. Remaining biotin label at the surface was removed by a cell-impermeable reducing agent, and cells were then incubated for up to 30 min to allow labeled MR1 to recycle back to the cell surface. Recycled MR1 was detected by incubating the cells with streptavidin conjugated to AF647 and measured by flow cytometry. The amount of MR1 recycled was calculated as the percentage of the initial surface MR1 signal prior to biotin removal. (C) The effect of endocytosis inhibitors on MR1 internalization was measured in C1R (left) or THP-1 (right) cells as in A. Cells were incubated with 10 µM 5-OP-RU for 4 h, treated with each inhibitor for 1 h, then internalization after 2 h measured with the FIP in the presence or absence of inhibitors. (D) C1R cells were pulsed with 5-OP-RU for 4 h then washed and chased without ligand for 8 h. The fold change of geometric mean fluorescent intensity (gMFI; left) of MR1 surface expression is shown. C1R cells transduced with lentivirus containing a GeCKO v2 human CRISPR-Cas9 KO library were similarly pulsed with 5-OP-RU for 4 h then chased without ligand for 8 h and stained for surface MR1 (solid black histogram). MR1HIGH cells (5% of population) were sorted and expanded and then sorted again for a second time. MR1 levels in cells cultured without ligand (black line) compared to after 5-OP-RU pulse (dotted line), and unstained cells (gray) are shown for comparison, a representative of three replicates. (E) The viral integration site containing the sgRNA cassette was amplified and sequenced from each sample, and the twice-sorted compared to the unsorted pool samples. Shown is the plot of significance (−log10[P value]) and fold change (Log2[Fold change]) for each gene based on a MAGeCK-RRA analysis. The only significantly enriched hit with a false discovery rate >0.05 was AP2A1 shown in blue. (F) The number of reads for each sgRNA targeting AP2A1 for the unsorted library (gray) is shown compared to the MR1HIGH-sorted replicates (blue). Each dot represents one replicate where the sgRNA was detected. Data are shown as the mean ± SD of replicates from two separate experiments for cell lines (A–D; n = 4), or individual data points for healthy donors (A and B; n = 4). Statistical significance was calculated with one-way ANOVA with multiple comparison test where *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/12/10.1083_jcb.202110125/1/m_jcb_202110125_fig1.png?Expires=1773830563&Signature=noNj~84dHAU0AJp0IDLxdVikeHTaxG0OIPouzwBLB~EM7lj3rpdwtfviNdnAI7AwLvOb1qwGpvWCVbBhxtXDqXoftkQTc0r2vDWyDvSzH8Yza1SqRTfdvf72IZwvcnEW-zliDUk8Axq2H4Ob8g8bc1EaQGVz1zG0ho6SILqDMUcuus1XyXygKTNh803sKn35D6TSF-BGcCNbHpnRWUmjHdWvbyPfzEYG8ZZcHFN5PQ1Dfdl2KSy3jxpvDGbOzFk4SwwdY8XI4p1FHp57czmX7jnxWpgVMEuIMU~EXKdidfMqZ5l3Ec3P4zxD3~N2UAYGQKm5DiySDrb8A~tvxUhAjA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

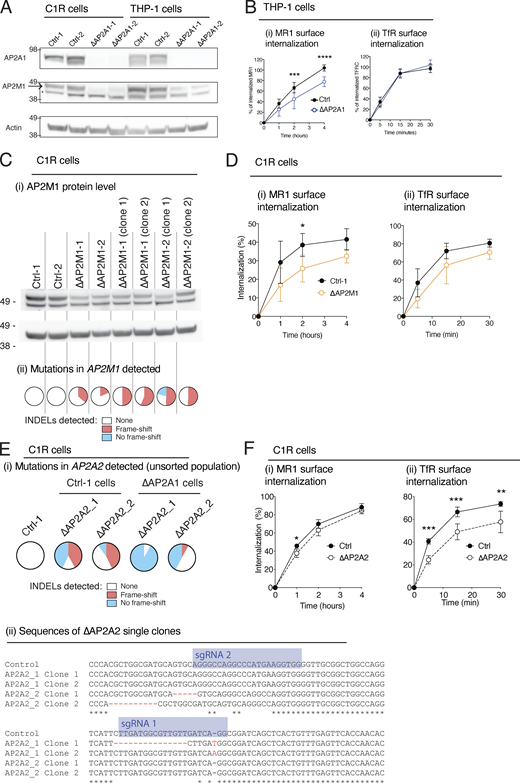
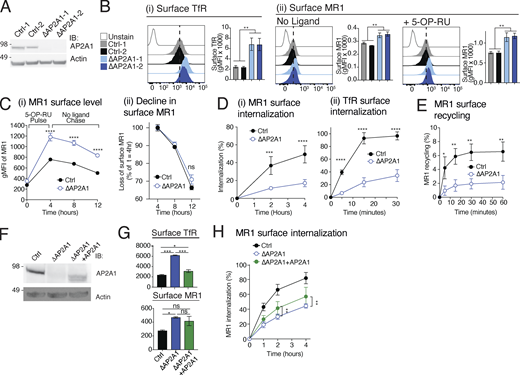


![MR1 interacts with AP2 when displayed at the cell surface. HeLa cells transduced with or without MR1 were incubated with or without 5-OP-RU for 4 h and then fixed. After permeabilization, cells were stained for the nucleus, total membranes, and then with antibodies directed to MR1 and AP2 proteins in the following combinations: (i) rabbit anti-MR1-cytosolic tail (αMR1 [CT]) and mouse anti-AP2A1; (ii) rabbit anti-AP2A1 and mouse anti-MR1 luminal domain (clone 8F2.F9); (iii) rabbit anti-AP2M1 and mouse anti-MR1 luminal domain (clone 8F2.F9). The slides were then subjected to PLA. Each PLA spot represents a molecular interaction between MR1 and AP2A1, and these were enumerated in each cell. Images are shown as maximum projections of Z-stacks, and the white scale bars represent 10 µM. Data are shown as the mean ± SD of n = 20–50 cells from each of two independent experiments. Statistical significance was calculated with one-way ANOVA with multiple comparison test where ****, P < 0.0001.](https://cdn.rupress.org/rup/content_public/journal/jcb/221/12/10.1083_jcb.202110125/1/m_jcb_202110125_fig4.png?Expires=1773830563&Signature=Lk6xZ-PlCuPFc5PKFS2nY5GuO-0nAsYFS4RITgx2hjZbFAKolT7qHQDoPxeUvark3le8M0DHB4xHa0NspQaSHDO8geatKWdexx~IzYJi0layT-BBh47YVD0EwBzV8JUuZP157TpX2moliAYOnGzE-n8OPHtnaj1VLISzitdpHxte0bvYbnKfVNN2bzF2eC~Cbuja2DRLaMHRU4WY3JZ6bNO5ZTG18YgY2Sr-Yp1w3vY48kCP1iyZ1ZO6P-uxiFndoCfHGBiTwzVHFpEMDmSGJfz08P4nUUxh8Cam2GfZpkTDogekwZ4UXSlvvwIyAAluQ3~on8u~m858V~~24b8o5w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)