Faithful DNA repair is essential to maintain genome integrity. Ultraviolet (UV) irradiation elicits both the recruitment of DNA repair factors and the deposition of histone marks such as monoubiquitylation of histone H2A at lesion sites. Here, we report how a ubiquitin E3 ligase complex specific to DNA repair is remodeled at lesion sites in the global genome nucleotide excision repair (GG-NER) pathway. Monoubiquitylation of histone H2A (H2A-ubiquitin) is catalyzed predominantly by a novel E3 ligase complex consisting of DDB2, DDB1, CUL4B, and RING1B (UV–RING1B complex) that acts early during lesion recognition. The H2A-ubiquitin binding protein ZRF1 mediates remodeling of this E3 ligase complex directly at the DNA lesion site, causing the assembly of the UV–DDB–CUL4A E3 ligase complex (DDB1–DDB2–CUL4A-RBX1). ZRF1 is an essential factor in GG-NER, and its function at damaged chromatin sites is linked to damage recognition factor XPC. Overall, the results shed light on the interplay between epigenetic and DNA repair recognition factors at DNA lesion sites.
Introduction
Nucleotide excision repair (NER) constitutes one of the major DNA repair pathways. It handles various helix-distorting DNA lesions such as 6–4 photoproducts and cyclobutane pyrimidine dimers (CPDs), arising after exposure to UV light (de Laat et al., 1999). Impaired NER activity is associated with several genetic disorders such as Xeroderma pigmentosum, which is characterized by hypersensitivity to sunlight and a predisposition for skin cancer (Friedberg, 2001). Mammalian NER comprises two pathways that differ in the nature of recognizing DNA lesions. Transcription-coupled (TC) NER is confined to regions of active transcription, where stalled RNA polymerase II triggers the DNA damage response. In contrast, global genome (GG) NER represents the transcription-independent recognition of lesions. The recognition step is followed by verification of the lesion by the repair factor XPA and by the formation of the preexcision complex involving TFIIH and its helicase subunits XPB and XPD. Subsequently, the DNA lesion is excised by the endonucleases XPF and XPG, and the gap is filled by DNA polymerases. (Fousteri and Mullenders, 2008; Marteijn et al., 2014).
In GG-NER DNA lesions are recognized by two well-described factors: XPC and DDB2. XPC represents a structure specific DNA binding factor, which specifically binds helix-distorting structures (Sugasawa et al., 1998; Riedl et al., 2003). XPC forms a stable complex with the Rad23 homologs RAD23A or RAD23B, respectively, and centrin2 (Masutani et al., 1994; Araki et al., 2001). This trimeric complex binds to a variety of lesions, triggers NER activity, and rapidly dissociates after binding damaged DNA (Sugasawa et al., 2001; Hoogstraten et al., 2008; Bergink et al., 2012). Efficient recognition of CPDs and 6–4 photoproducts also requires the presence of DDB2 (XPE; Tang et al., 2000; Fitch et al., 2003; Moser et al., 2005; Luijsterburg et al., 2007; Nishi et al., 2009). Loss of functional DDB2 causes defective repair of CPDs, reduced repair of 6–4 photoproducts, and hypersensitivity to UV-induced skin cancer (Rapić-Otrin et al., 2003; Alekseev et al., 2005). DDB2 along with DDB1, the RING-domain protein RBX1, and either of the scaffold proteins CUL4A or CUL4B forms E3 ubiquitin ligase complexes (UV–DDB–CUL4A/B) that catalyze the monoubiquitylation of histones H2A, H3, and H4 (Shiyanov et al., 1999; Groisman et al., 2003; Angers et al., 2006; Wang et al., 2006; Guerrero-Santoro et al., 2008). Importantly, the UV–DDB–CUL4A complex catalyzes the polyubiquitylation of XPC, thereby increasing its affinity for DNA in vitro and contributing to recognition and stable binding of photolesions (Sugasawa et al., 2005).
A prominent histone modification present at DNA damage sites is ubiquitylation of histones H2A, H2AX, and H1 (Bergink et al., 2006; Mailand et al., 2007; Pan et al., 2011; Thorslund et al., 2015). At double-strand breaks (DSBs), ubiquitylation of histones is catalyzed by the E3 ligases RNF168, RNF8, and RING1B (Doil et al., 2009; Pan et al., 2011; Mattiroli et al., 2012; Ui et al., 2015). During NER, H2A ubiquitylation is catalyzed by the E3 ligase RNF8 and the UV–DDB–CUL4A/B complexes (Bergink et al., 2006; Kapetanaki et al., 2006; Guerrero-Santoro et al., 2008; Marteijn et al., 2009). Further, it was demonstrated that H2A ubiquitylation after UV irradiation depends on RING1B (Bergink et al., 2006). RING1B constitutes a subunit of the Polycomb group repressive complex 1 (PRC1), which catalyzes the monoubiquitylation of histone H2A at lysine 119 to silence genes during pluripotency (Wang et al., 2004; Morey and Helin, 2010). Interestingly, at DSBs, H2A ubiquitylation is dependent on the PRC1 subunits BMI-1 and RING1B (Ismail et al., 2010; Chagraoui et al., 2011; Ginjala et al., 2011; Pan et al., 2011). More recently, it was reported that PRC1 mediates DSB-induced gene silencing, linking PRC1 strongly to DSB repair (Ui et al., 2015). Still, it remains unclear how the E3 ligases cross talk and in which sequence they act during DNA repair.
We have previously shown that Zuotin-related factor 1 (ZRF1) binds monoubiquitylated histone H2A via its ubiquitin-binding domain and removes PRC1 from chromatin during cellular differentiation (Richly et al., 2010). Given the significance of H2A ubiquitylation in DNA repair, we have set out to study the roles of RING1B and ZRF1 in NER. Our results reveal that RING1B is the catalytic subunit of a novel DDB–cullin–E3 ligase complex, which ubiquitylates histone H2A early during NER. Further, we discovered that ZRF1 is a switch protein that remodels chromatin-bound E3 ligases during lesion recognition. Hence, our study sheds new light on the interplay of epigenetic and DNA repair recognition factors at DNA lesion sites.
Results
RING1B mediates ubiquitylation of histone H2A after UV irradiation
To distinguish the functions of E3 ligases functioning after UV irradiation, we performed knockdown of RING1B (shRING1B), RNF168 (siRNF168), and the scaffold protein CUL4A (siCUL4A), which is a component of the UV–DDB–CUL4A E3 complex, in HEK293T cells. To assess the recruitment of the respective E3 ligases to chromatin, we cross-linked cells at the given time points after UV irradiation and isolated the chromatin fraction. We measured the relative intensities of H2A ubiquitin and H2A after probing Western blots with H2A antibodies. We observed that the reduction of RING1B hampered the increase of H2A ubiquitylation, whereas knockdown of the other E3 ligases did not significantly alter H2A ubiquitin levels (Fig. 1 A; representative Western blots of the analysis: Figs. 1 B and S1, A and B). We also confirmed that RING1B specifically catalyzes monoubiquitylation of lysine 119 at histone H2A after UV irradiation (Figs. 1 B and S1 C). Additionally, we confirmed that knockdown of CUL4A renders the UV–DDB–CUL4A E3 ligase inactive (Fig. S1 A). To further assess whether RING1B is recruited to DNA damage sites, we performed microirradiation experiments with a 405-nm laser in cells expressing DDB2-GFP and RING1B-YFP fusion proteins (Fig. S1, D–F). We observed that both DDB2 and RING1B show a relatively weak, but significant accumulation to sites of DNA damage, consistent with a previous observation demonstrating RING1B-mediated accumulation of H2A-ubiquitylation at DNA damage sites (Bergink et al., 2006). Further, we did not observe any major difference in cellular ubiquitylation levels upon depletion of RING1B (Fig. S1, G and H) as suggested previously (Bergink et al., 2006). To link RING1B to the NER pathway, we investigated its function performing UV irradiation experiments with the nematode Caenorhabditis elegans (Lans and Vermeulen, 2011; Craig et al., 2012). Compared with wild-type animals treated with a control RNAi (N2/control), we observed a reduction of viability after UV irradiation of the RING1B mutant treated with control RNAi (VC31/control) and upon RNAi-mediated depletion of the NER factor XPC in wild-type worms (N2/xpc-1; Fig. 1 C). Knockdown of XPC in RING1B mutant strains (VC31/xpc-1) did not exhibit further reduction of viability, suggesting that RING1B is epistatic to XPC.
Dissection of E3 ligase functions in UV-mediated DNA damage repair. (A) Quantitative analysis of H2A-ubiquitylation levels. Immunoblots (as in B and Fig. S1, A and B) were probed with histone H2A antibody. The intensities of H2A and H2A-ubiquitin bands were quantified by the ImageJ software. The graphs illustrate the relative H2A ubiquitylation calculated as (H2A ubiquitin)/(H2A + H2A ubiquitin), normalized to Ponceau staining intensity after knockdown of the respective proteins (H2A ubiquitin/H2A). Values are normalized to the value from nonirradiated cells and are given as mean ± SEM (n = 4). (B) Monoubiquitylation of histone H2A at lysine 119 after UV irradiation is mainly catalyzed by RING1B. Chromatin association assays of control and RING1B knockdown HEK293T cells after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The specificity of the H2A-ubiquitin antibody was verified (Fig. S1 C). (C) Epistatic relationship of xpc-1 and spat-3. Wild-type nematodes (N2) or spat-3 mutants (VC31) were fed with either control or xpc-1 RNAi–producing bacteria. The relative viability was analyzed after UV irradiation (200 J/m2). Values are given as mean ± SEM (n = 3). (D) Impact of BMI-1 on RING1B-mediated H2A ubiquitylation after UV irradiation. Chromatin association assays of UV-irradiated HEK293T cells treated with siRNAs (control, BMI-1). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. Relative intensities of H2A ubiquitin/H2A and RING1B abundance after BMI-1 depletion were measured. Values are given as mean ± SEM (n = 4). (E) Epistatic relationship of RING1B and BMI-1 in response to UV irradiation. Relative colony formation potential of control or RING1B knockdown cell lines treated with siRNA was analyzed at different UV doses. Control cells were transfected with either control siRNA (control) or BMI-1 siRNA (BMI-1). RING1B knockdown cell lines were transfected with either control siRNA (RING1B) or BMI-1 siRNA (RING1B + BMI-1). Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 9).
Dissection of E3 ligase functions in UV-mediated DNA damage repair. (A) Quantitative analysis of H2A-ubiquitylation levels. Immunoblots (as in B and Fig. S1, A and B) were probed with histone H2A antibody. The intensities of H2A and H2A-ubiquitin bands were quantified by the ImageJ software. The graphs illustrate the relative H2A ubiquitylation calculated as (H2A ubiquitin)/(H2A + H2A ubiquitin), normalized to Ponceau staining intensity after knockdown of the respective proteins (H2A ubiquitin/H2A). Values are normalized to the value from nonirradiated cells and are given as mean ± SEM (n = 4). (B) Monoubiquitylation of histone H2A at lysine 119 after UV irradiation is mainly catalyzed by RING1B. Chromatin association assays of control and RING1B knockdown HEK293T cells after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The specificity of the H2A-ubiquitin antibody was verified (Fig. S1 C). (C) Epistatic relationship of xpc-1 and spat-3. Wild-type nematodes (N2) or spat-3 mutants (VC31) were fed with either control or xpc-1 RNAi–producing bacteria. The relative viability was analyzed after UV irradiation (200 J/m2). Values are given as mean ± SEM (n = 3). (D) Impact of BMI-1 on RING1B-mediated H2A ubiquitylation after UV irradiation. Chromatin association assays of UV-irradiated HEK293T cells treated with siRNAs (control, BMI-1). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. Relative intensities of H2A ubiquitin/H2A and RING1B abundance after BMI-1 depletion were measured. Values are given as mean ± SEM (n = 4). (E) Epistatic relationship of RING1B and BMI-1 in response to UV irradiation. Relative colony formation potential of control or RING1B knockdown cell lines treated with siRNA was analyzed at different UV doses. Control cells were transfected with either control siRNA (control) or BMI-1 siRNA (BMI-1). RING1B knockdown cell lines were transfected with either control siRNA (RING1B) or BMI-1 siRNA (RING1B + BMI-1). Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 9).
Given the function of PRC1 at DSBs, we next determined whether PRC1 plays a role in H2A ubiquitylation after UV irradiation. Knockdown of BMI-1 displayed only a slight effect on the recruitment of RING1B and the deposition of H2A ubiquitin (Fig. 1 D), which is likely a consequence of reduced RING1B and H2A-ubiquitin basal levels. A colony formation assay showed that knockdown of either RING1B or BMI-1 exhibits a mild reduction of the colony formation potential. Interestingly, simultaneous knockdown of both proteins showed additive reduction of the colony formation potential, suggesting that BMI-1 and RING1B likely exert different functions in the repair of UV-mediated DNA lesions (Fig. 1 E). Notably, we observed a similar relationship performing an epistasis analysis with the C. elegans orthologs of BMI-1 (mig-32) and RING1B (spat-3; Karakuzu et al., 2009; Fig. S1 I).
Collectively, these data suggest a critical role for RING1B in H2A-ubiquitylation in the NER pathway. Opposed to its function at DSBs, RING1B seems to catalyze the ubiquitylation reaction without its PRC1 binding partner BMI-1.
RING1B and DDB2 cooperate in the ubiquitylation of histone H2A
Intrigued by the epistatic relationship of XPC and RING1B, we sought to find out whether RING1B is linked to the NER machinery. We expressed FLAGRING1B in HEK293T cells and performed affinity purifications. As expected, RING1B binds the PRC1 subunit BMI-1 (Wang et al., 2004; Fig. 2 A). Interestingly, RING1B interacts robustly with DDB2, but not with other selected factors of the NER pathway (Figs. 2 A and S2 A). Immunoprecipitation of endogenous RING1B further verified the interaction of DDB2 with RING1B (Fig. 2 B). Likewise, purifications performed with FLAGDDB2 displayed strong binding of RING1B and interaction with its well-characterized binding partners DDB1 and CUL4A (Shiyanov et al., 1999; Fig. 2 C).
RING1B and DDB2 cooperate in H2A ubiquitylation. (A) RING1B interacts with DDB2. Control cells and cells expressing FLAGRING1B were irradiated with UV light. After immunoprecipitation with FLAG-M2-Agarose the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (B) Endogenous immunoprecipitations with RING1B antibodies after UV irradiation. Western blots of the precipitated material were incubated with the indicated antibodies. IgG lanes show unspecific staining of the IgG heavy chains. (C) DDB2 associates with RING1B. Control cells and cells expressing FLAGDDB2 were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (D) Epistatic relationship of RING1B and DDB2 in response to UV irradiation. Relative colony formation potential of control or RING1B knockdown cell lines treated with siRNA was analyzed at different UV dosages. Control cells were transfected with either control siRNA (control) or DDB2 siRNA (DDB2). RING1B knockdown cell lines were transfected with either control siRNA (RING1B) or DDB2 siRNA (RING1B + DDB2). Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 6). (E) Knockdown of RING1B does not impair DDB2 recruitment. Chromatin association assays of control and RING1B knockdown HEK293T cells after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative DDB2 and BMI-1 abundance was calculated. Values are given as mean ± SEM (n = 3). (F) Knockdown of DDB2 shows reduced H2A-ubiquitylation but unaltered BMI-1 recruitment. Chromatin association assays of UV-irradiated HEK293T cells treated with siRNAs (control, DDB2). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitylation and RING1B abundance was calculated. Values are given as mean ± SEM (n = 4).
RING1B and DDB2 cooperate in H2A ubiquitylation. (A) RING1B interacts with DDB2. Control cells and cells expressing FLAGRING1B were irradiated with UV light. After immunoprecipitation with FLAG-M2-Agarose the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (B) Endogenous immunoprecipitations with RING1B antibodies after UV irradiation. Western blots of the precipitated material were incubated with the indicated antibodies. IgG lanes show unspecific staining of the IgG heavy chains. (C) DDB2 associates with RING1B. Control cells and cells expressing FLAGDDB2 were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (D) Epistatic relationship of RING1B and DDB2 in response to UV irradiation. Relative colony formation potential of control or RING1B knockdown cell lines treated with siRNA was analyzed at different UV dosages. Control cells were transfected with either control siRNA (control) or DDB2 siRNA (DDB2). RING1B knockdown cell lines were transfected with either control siRNA (RING1B) or DDB2 siRNA (RING1B + DDB2). Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 6). (E) Knockdown of RING1B does not impair DDB2 recruitment. Chromatin association assays of control and RING1B knockdown HEK293T cells after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative DDB2 and BMI-1 abundance was calculated. Values are given as mean ± SEM (n = 3). (F) Knockdown of DDB2 shows reduced H2A-ubiquitylation but unaltered BMI-1 recruitment. Chromatin association assays of UV-irradiated HEK293T cells treated with siRNAs (control, DDB2). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitylation and RING1B abundance was calculated. Values are given as mean ± SEM (n = 4).
Next, we examined whether DDB2 and BMI-1 interact with RING1B in a mutually exclusive manner. Immunoprecipitating BMI-1 we observed binding of RING1B, but not DDB2 (Fig. S2 B). Overexpression of BMI-1 caused a slight increase in the BMI-1–RING1B interaction but a complete loss of DDB2-RING1B binding (Fig. S2 C). Depletion of BMI-1 had only a slight effect on the DDB2–RING1B interaction (Fig. S2 D). These data suggest that the majority of RING1B is associated with BMI-1 rather than DDB2, which is in agreement with the general function of PRC1 in gene silencing.
To investigate a joint function of DDB2 and RING1B in DNA repair, we performed colony formation assays (Fig. 2 D). After depletion of DDB2 we observed reduced colony formation potential, which is in agreement with a previous study showing impaired survival of XPE patient fibroblasts after UV irradiation (Rapić-Otrin et al., 2003). Similarly, depletion of RING1B exhibited reduced colony formation potential. Simultaneous depletion of both proteins showed no further reduction of colony formation potential, suggesting that RING1B and DDB2 likely act in a common DNA repair pathway. To further support this finding, we analyzed skin biopsy specimens after staining with DDB2 and H2A ubiquitin or RING1B antibodies, respectively (Fig. S2, E, G, and I). We observed a clear correlation of DDB2 with both RING1B and H2A-ubiquitin only in UV exposed skin sections as judged by single cell quantification of staining intensities (Fig. S2, F and H). Depletion of RING1B did not hamper the recruitment of DDB2 or BMI-1 to chromatin after UV irradiation (Fig. 2 E), implying divergent roles for RING1B and BMI-1 in UV-triggered DNA repair. Cells depleted of DDB2 as well as XPE patient fibroblasts exhibited reduced H2A ubiquitylation consistent with a previous study (Kapetanaki et al., 2006) and diminished recruitment of RING1B to chromatin (Figs. 2 F, 4 G, and S2 K). Notably, knockdown of DDB2 did not impair BMI-1 recruitment to chromatin, further uncoupling BMI-1 from H2A ubiquitylation in NER (Figs. 2 F and S2 J).
In sum, these data suggest a functional interplay of DDB2 and RING1B in H2A ubiquitylation during NER.
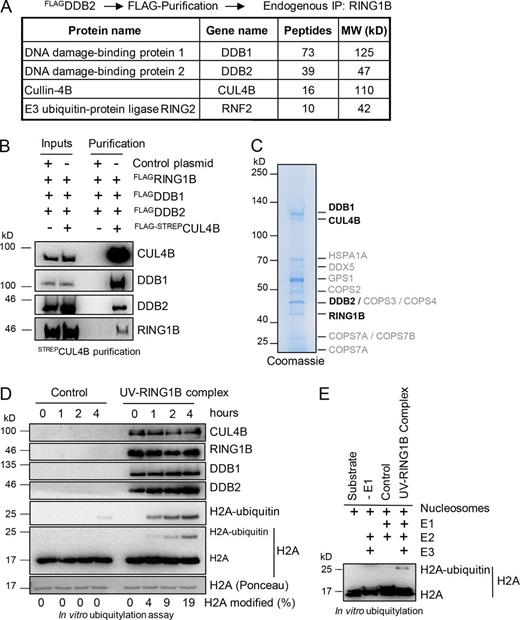
RING1B forms a stable protein complex with CUL4B, DDB1, and DDB2
To reveal the composition of the putative RING1B-DDB2 E3 ligase complex, we expressed FLAGDDB2 in HEK293T cells and performed purifications in UV-irradiated and untreated cells (Fig. 3 A and Table S5). After elution of FLAGDDB2 containing protein complexes with FLAG peptide, we subsequently used the eluate in immunoprecipitations with RING1B antibodies to specifically purify RING1B-DDB2 containing protein complexes. The purified material was subjected to mass spectrometry, identifying DDB1 and CUL4B as the main interactors of RING1B and DDB2 (UV–RING1B complex in Fig. 3 A and Table S5). Furthermore, immunoprecipitations of endogenous DDB1 or RING1B as well as pull-downs with recombinant GST–RING1B and purified DDB1–DDB2 complexes confirmed our findings (Fig. S3, A–D). To verify the assembly of the UV–RING1B E3 ligase complex, we overexpressed FLAGDDB1, FLAGDDB2, and FLAGRING1B with or without FLAG-STREPCUL4B in HEK293T cells (Fig. 3 B). Affinity purifications of CUL4B revealed specific binding of DDB1, DDB2, and RING1B. We further analyzed the interactions of the subunits of the UV–RING1B complex in vitro by pull-down experiments with purified proteins (Fig. S3 E). Collectively, these experiments revealed that RING1B specifically binds to CUL4B and DDB2 but shows no direct interaction with either CUL4A or DDB1 (Fig. S3, F–I). Additionally, to distinguish the UV–RING1B complex from the UV–DDB–CUL4B complex, we performed competition experiments. The E3 ligases RING1B and RBX1 compete for binding to CUL4B as judged by in vitro pull-down experiments with CUL4B (Fig. S3 J). Similarly, in pull-downs with recombinant RBX1 (Fig. S3, K and L) and in immunoprecipitations of endogenous RBX1 after RING1B overexpression (Fig. S3 M), excess RING1B disrupted CUL4B-RBX1 binding.
H2A ubiquitylation after UV irradiation is performed by the UV–RING1B complex. (A) Protein interaction partners of RING1B and DDB2. Mass spectrometry analysis after sequential immunoprecipitations with FLAG and RING1B antibodies revealed DDB1 and CUL4B as main interaction partners of DDB2 and RING1B. A comprehensive list of the identified unique peptides after RING1B and control immunoprecipitations (with or without UV irradiation) is provided in Table S5. (B) Assembly of the UV–RING1B complex. Plasmids expressing FLAGDDB1, FLAGDDB2, and FLAGRING1B were cotransfected in combination with either control plasmid or a plasmid encoding FLAG-STREPCUL4B. After immunoprecipitation with STREP-Tactin beads, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (C) Visualization of the UV–RING1B complex. Purified UV–RING1B complex was subjected to SDS gel electrophoresis and colloidal Coomassie staining. Mass spectrometry analysis revealed the presence of all four subunits (bold). A comprehensive list of unique peptides is provided in Table S6. (D) The UV–RING1B complex catalyzes ubiquitylation of H2A in vitro. Ubiquitylation assays were performed with recombinant H2A, E1 (UBA1), E2 (UBCH5), and either GST (control) or the UV–RING1B complex. Reactions were performed at 37°C, and samples were taken at the indicated time points. Material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. (E) The UV–RING1B complex catalyzes monoubiquitylation of nucleosomal H2A. Ubiquitylation assays were performed with recombinant nucleosomes, E1 (UBA1), E2 (UBCH5), and either GST (control) or UV-RING1B complex. Reactions lacking E1 (−E1) were performed as additional controls. The ubiquitylation assays were performed at 37°C for 5 h, and samples or pure substrate (Substrate) were subjected to Western blotting and probed with H2A antibodies.
H2A ubiquitylation after UV irradiation is performed by the UV–RING1B complex. (A) Protein interaction partners of RING1B and DDB2. Mass spectrometry analysis after sequential immunoprecipitations with FLAG and RING1B antibodies revealed DDB1 and CUL4B as main interaction partners of DDB2 and RING1B. A comprehensive list of the identified unique peptides after RING1B and control immunoprecipitations (with or without UV irradiation) is provided in Table S5. (B) Assembly of the UV–RING1B complex. Plasmids expressing FLAGDDB1, FLAGDDB2, and FLAGRING1B were cotransfected in combination with either control plasmid or a plasmid encoding FLAG-STREPCUL4B. After immunoprecipitation with STREP-Tactin beads, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (C) Visualization of the UV–RING1B complex. Purified UV–RING1B complex was subjected to SDS gel electrophoresis and colloidal Coomassie staining. Mass spectrometry analysis revealed the presence of all four subunits (bold). A comprehensive list of unique peptides is provided in Table S6. (D) The UV–RING1B complex catalyzes ubiquitylation of H2A in vitro. Ubiquitylation assays were performed with recombinant H2A, E1 (UBA1), E2 (UBCH5), and either GST (control) or the UV–RING1B complex. Reactions were performed at 37°C, and samples were taken at the indicated time points. Material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. (E) The UV–RING1B complex catalyzes monoubiquitylation of nucleosomal H2A. Ubiquitylation assays were performed with recombinant nucleosomes, E1 (UBA1), E2 (UBCH5), and either GST (control) or UV-RING1B complex. Reactions lacking E1 (−E1) were performed as additional controls. The ubiquitylation assays were performed at 37°C for 5 h, and samples or pure substrate (Substrate) were subjected to Western blotting and probed with H2A antibodies.
Next, we set out to purify the UV–RING1B complex to test its ubiquitylation capacity in vitro. To this end, we overexpressed FLAGDDB1, FLAGDDB2, FLAGRING1B, and FLAG-STREPCUL4B in HEK293T cells (Fig. S3 N). After enriching for the FLAG-tagged proteins, we selectively purified the UV–RING1B complex. We subjected the purified material to colloidal Coomassie staining (Fig. 3 C) and mass spectrometry (Table S4), which confirmed the specific assembly of the UV–RING1B complex. Importantly, no contamination with chromatin components was found in the purification, ruling out that the assembly of the UV–RING1B complex was generated indirectly through association with chromatin (Tables S5 and S6). Likewise, no other E3 ligases were identified in the affinity purification, excluding unspecific ubiquitylation events when testing the UV–RING1B complex in vitro. To explore whether the purified UV–RING1B complex catalyzes H2A ubiquitylation, we performed in vitro ubiquitylation assays with histone H2A (Fig. 3 D). Compared with control reactions, the UV–RING1B complex strongly increased the specific monoubiquitylation of histone H2A over time. Similarly, the UV–RING1B complex caused monoubiquitylation of nucleosomes at histone H2A in ubiquitylation assays (Fig. 3 E).
In conclusion, we have identified a novel RING1B-containing complex that catalyzes monoubiquitylation of histone H2A.
ZRF1 tethers to the H2A-ubiquitin mark during UV-triggered DNA repair
Monoubiquitylated H2A is bound by ZRF1 during cellular differentiation (Richly et al., 2010). Interestingly, we observed that ZRF1 is recruited to chromatin after UV irradiation and its recruitment is dependent on RING1B (Fig. 4 A). Furthermore, the ubiquitin-binding domain of ZRF1 is required for its association with chromatin after UV irradiation (Fig. 4 B). When inducing local UV damage by irradiation through a micropore membrane, we observed ZRF1 localizing to DNA lesions, which are marked by XPC and XPA (Fig. 4, C and D; and Fig. S4 A), further supporting a role for ZRF1 in UV-mediated DNA repair. We next addressed the association of ZRF1 with DNA lesions in the presence of the RING1B inhibitor PRT4165 (Ismail et al., 2013). Under control conditions, we observed ZRF1 at DNA lesions (Fig. 4 E), whereas administration of the drug abolished H2A ubiquitylation (Fig. S4 B), unscheduled DNA synthesis (UDS) after UV irradiation (Fig. S4 C), and, most importantly, the presence of ZRF1 at the damage site (Figs. 4 E and S4 D). Similarly, ZRF1 recruitment to chromatin was hampered after depletion of the UV–RING1B complex subunit CUL4B or in XPE patient fibroblasts (Fig. 4, F and G). To investigate ZRF1 function in vivo, we analyzed human skin biopsy specimens. ZRF1 and CPD antibody staining signals colocalized only when the skin was exposed to UV light (Fig. S4 E). In addition, single-cell analysis revealed that the relative ZRF1 intensities correlate with the relative intensities of CPDs upon irradiation (Fig. S4, E and F).
Function of ZRF1 in UV-mediated DNA repair. (A) ZRF1 is tethered to chromatin in a RING1B-dependent manner. Chromatin association assays of control and RING1B knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (B) The ubiquitin-binding domain (UBD) is important for tethering ZRF1 to chromatin after UV irradiation. HEK293T cells expressing FLAGZRF1 and FLAGZRF1-ΔUBD were irradiated with UV light, and chromatin was isolated at the indicated time points. De-cross-linked material was subjected to Western blotting and blots were incubated with FLAG-antibody. The relative FLAGZRF1 abundance was calculated. Values are given as mean ± SEM (n = 4). (C and D) ZRF1 localizes to DNA damage sites after UV irradiation. MRC5 fibroblasts expressing mCherry-ZRF1 were UV irradiated (100 J/m2) through a micropore membrane (+ UV) 24 h after transfection. 30 min after irradiation, cells were preextracted and fixed. DNA damage sites were visualized by staining with XPC (C) or XPA (D) antibody. The colocalization of ZRF1 with XPC amounts to 88% ± 1%. The colocalization of ZRF1 with XPA amounts to 73% ± −3%. Nonirradiated control and quantification of the ZRF1 localization at the damage sites are represented in Fig. S4 A. Bar, 10 µm. (E) Inhibition of RING1B affects recruitment of ZRF1 to DNA damage sites. MRC5 fibroblasts expressing mCherry-ZRF1 were treated with PRT4165 or DMSO. Cells were UV-irradiated (100 J/m2) through a micropore membrane. 30 min after irradiation cells were preextracted and fixed. DNA damage sites were visualized by XPC antibody staining. ZRF1 localization to DNA lesions after treatment with DMSO or PRT4165 was quantified (Fig. S4 B). Bar, 10 µm. (F) Depletion of CUL4B impacts H2A ubiquitylation and ZRF1 recruitment. Chromatin association assays of UV irradiated HEK293T cells treated with siRNAs (control, CUL4B). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitin and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (G) Tethering of ZRF1 to chromatin depends on DDB2 during NER. Chromatin association assays in control fibroblasts (GM15876) and XPE (DDB2) fibroblasts (GM01389) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative RING1B and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3).
Function of ZRF1 in UV-mediated DNA repair. (A) ZRF1 is tethered to chromatin in a RING1B-dependent manner. Chromatin association assays of control and RING1B knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (B) The ubiquitin-binding domain (UBD) is important for tethering ZRF1 to chromatin after UV irradiation. HEK293T cells expressing FLAGZRF1 and FLAGZRF1-ΔUBD were irradiated with UV light, and chromatin was isolated at the indicated time points. De-cross-linked material was subjected to Western blotting and blots were incubated with FLAG-antibody. The relative FLAGZRF1 abundance was calculated. Values are given as mean ± SEM (n = 4). (C and D) ZRF1 localizes to DNA damage sites after UV irradiation. MRC5 fibroblasts expressing mCherry-ZRF1 were UV irradiated (100 J/m2) through a micropore membrane (+ UV) 24 h after transfection. 30 min after irradiation, cells were preextracted and fixed. DNA damage sites were visualized by staining with XPC (C) or XPA (D) antibody. The colocalization of ZRF1 with XPC amounts to 88% ± 1%. The colocalization of ZRF1 with XPA amounts to 73% ± −3%. Nonirradiated control and quantification of the ZRF1 localization at the damage sites are represented in Fig. S4 A. Bar, 10 µm. (E) Inhibition of RING1B affects recruitment of ZRF1 to DNA damage sites. MRC5 fibroblasts expressing mCherry-ZRF1 were treated with PRT4165 or DMSO. Cells were UV-irradiated (100 J/m2) through a micropore membrane. 30 min after irradiation cells were preextracted and fixed. DNA damage sites were visualized by XPC antibody staining. ZRF1 localization to DNA lesions after treatment with DMSO or PRT4165 was quantified (Fig. S4 B). Bar, 10 µm. (F) Depletion of CUL4B impacts H2A ubiquitylation and ZRF1 recruitment. Chromatin association assays of UV irradiated HEK293T cells treated with siRNAs (control, CUL4B). De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitin and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (G) Tethering of ZRF1 to chromatin depends on DDB2 during NER. Chromatin association assays in control fibroblasts (GM15876) and XPE (DDB2) fibroblasts (GM01389) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative RING1B and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3).
Collectively, these data suggest that ZRF1 plays a role in UV-triggered DNA repair and that it localizes to the damage site via binding of H2A-ubiquitin.
ZRF1’s function in NER is dependent on XPC
To explore whether ZRF1 interacts with NER factors, we performed affinity purifications after expressing FLAGZRF1 in HEK293T cells (Fig. 5 A). We found the DNA lesion recognition factor XPC interacting with ZRF1, but we did not observe binding of other selected NER factors. Likewise, we found XPC associated with ZRF1 in endogenous immunoprecipitations, confirming the interaction of both proteins (Fig. 5 B). To investigate the interplay between XPC and ZRF1, we analyzed the localization of ZRF1 to lesions sites using DDB2 as a damage marker. Interestingly, we observed reduced colocalization of ZRF1 and DDB2 in XPC patient fibroblasts (Figs. 5 C and S5 A). Next, we analyzed chromatin from XPC patient fibroblasts and control fibroblasts after UV irradiation (Fig. 5 D). We observed reduced levels of ZRF1 despite enhanced RING1B and H2A-ubiquitin levels. Accordingly, siRNA-mediated knockdown of XPC caused a drastic reduction of ZRF1 levels at chromatin after UV irradiation (Fig. S5 B). In contrast, chromatin isolated from XPA patient fibroblasts exhibited no reduction in H2A ubiquitylation, RING1B, and ZRF1 levels as compared with control fibroblasts (Fig. 5 E). These data suggest that H2A ubiquitylation via the UV–RING1B complex and subsequent ZRF1 recruitment predominantly occurs early during DDB2-mediated lesion recognition and likely before the assembly of the DNA incision complex (de Laat et al., 1999; Wakasugi and Sancar, 1999).
ZRF1 interacts with XPC during UV-mediated DNA repair. (A) ZRF1 specifically binds to XPC. Control and FLAGZRF1-expressing cells were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 4%. (B) Endogenous immunopreciptiations with ZRF1 antibodies. Precipitates were subjected to Western blotting, and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (C) ZRF1 localization to DNA damage sites is dependent on XPC. Control fibroblasts and XPC patient fibroblasts expressing both mCherry-ZRF1 and DDB2-GFP were UV irradiated (100 J/m2) through a micropore membrane. Thirty minutes after irradiation, cells were preextracted and fixed. DNA damage sites were visualized by DDB2-GFP. (D) ZRF1 enriches at chromatin after UV irradiation in a XPC-dependent manner. Chromatin association assays with control fibroblasts (GM16248) and XPC patient fibroblasts (GM15983) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitin and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (E) H2A ubiquitylation is not altered in XPA patient fibroblasts. Chromatin association assays with control fibroblasts (GM15876) and XPA fibroblasts (GM04312) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. Relative intensities of H2A-ubiquitin/H2A, ZRF1 and RING1B abundance were measured. Values are given as mean ± SEM (n = 3). (F) Epistasis analysis of ZRF1 and XPC. The relative colony formation potential of control or ZRF1 knockdown cell lines treated with control (Control; ZRF1) or XPC siRNA (XPC; ZRF1+XPC) was analyzed at different UV doses. Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 3).
ZRF1 interacts with XPC during UV-mediated DNA repair. (A) ZRF1 specifically binds to XPC. Control and FLAGZRF1-expressing cells were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 4%. (B) Endogenous immunopreciptiations with ZRF1 antibodies. Precipitates were subjected to Western blotting, and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (C) ZRF1 localization to DNA damage sites is dependent on XPC. Control fibroblasts and XPC patient fibroblasts expressing both mCherry-ZRF1 and DDB2-GFP were UV irradiated (100 J/m2) through a micropore membrane. Thirty minutes after irradiation, cells were preextracted and fixed. DNA damage sites were visualized by DDB2-GFP. (D) ZRF1 enriches at chromatin after UV irradiation in a XPC-dependent manner. Chromatin association assays with control fibroblasts (GM16248) and XPC patient fibroblasts (GM15983) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A-ubiquitin and ZRF1 abundance was calculated. Values are given as mean ± SEM (n = 3). (E) H2A ubiquitylation is not altered in XPA patient fibroblasts. Chromatin association assays with control fibroblasts (GM15876) and XPA fibroblasts (GM04312) after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. Relative intensities of H2A-ubiquitin/H2A, ZRF1 and RING1B abundance were measured. Values are given as mean ± SEM (n = 3). (F) Epistasis analysis of ZRF1 and XPC. The relative colony formation potential of control or ZRF1 knockdown cell lines treated with control (Control; ZRF1) or XPC siRNA (XPC; ZRF1+XPC) was analyzed at different UV doses. Gene knockdown was confirmed by Western blots (not depicted). Values are given as mean ± SEM (n = 3).
Next, we performed an epistasis analysis addressing the common functions of ZRF1 and XPC in NER. We observed a strong reduction in the colony formation potential after irradiating ZRF1 knockdown cells or cells treated with siRNA directed against XPC, respectively (Fig. 5 F), consistent with previous observations in XPC patient fibroblasts (Bohr et al., 1986). Simultaneous knockdown of both factors did not significantly alter the colony formation potential compared with a single knockdown, suggesting that ZRF1 and XPC are likely epistatic in human cells. Additionally, we made similar observations in epistasis experiments using C. elegans (Fig. S5 C). To estimate the contribution of RING1B and ZRF1 in repairing UV-mediated DNA damage, we measured unscheduled DNA synthesis after UV irradiation and removal of CPDs in control fibroblasts, knockdown fibroblasts, and XPA fibroblasts (Fig. 6, A–C). In ZRF1 and RING1B knockdown cells, EdU incorporation was reduced to ∼40% when compared with control cells (Fig. 6 A). Similarly, the removal of CPDs was compromised in ZRF1 and RING1B knockdown fibroblasts (Fig. 6 B).
ZRF1 and RING1B contribute to GG-NER. (A) RING1B and ZRF1 knockdown fibroblasts are defective in UDS after UV irradiation. UDS was measured by EdU incorporation after UV treatment in MRC5 fibroblasts with shRNA-mediated knockdown of the indicated proteins. XPA fibroblasts were used as a positive control. Values are given as mean ± SEM. Data were acquired from three independent experiments (150–300 nuclei per sample). (B) RING1B and ZRF1 knockdown fibroblasts are defective in the removal of CPDs. The CPD removal was analyzed in MRC5 fibroblasts after knockdown of the indicated proteins in MRC5 fibroblasts and in XPA fibroblasts. Cells were irradiated with 10 J/m2 and fixed immediately or 24 or 48 h after irradiation and stained with CPD antibodies. The relative fluorescence intensity was determined. Values are given as mean ± SEM. Data were acquired from three independent experiments (100–200 nuclei per sample). (C) MRC5 fibroblasts were treated with lentiviral particles containing the respective shRNA. Knockdown of the proteins levels was analyzed 48h after infection by Western blotting and incubation with the indicated antibodies. (D) C. elegans knockout mutants for ZRF1 (dnj-11) and RING1B (spat-3) show increased sensitivity toward UV irradiation. Late-L4 larval wild-type worms and the indicated mutants were irradiated with UV light at different doses, and the relative viability was determined by comparing hatched versus dead embryos (unhatched eggs). Values are given as mean ± SEM (n = 3). (E) C. elegans knockout mutants for dnj-11 and for spat-3 show only weak developmental arrest upon somatic UV irradiation. L1 larval worms were irradiated with UV light at different doses. Relative larval-stage stalling was determined after 60 h by using a large particle flow cytometer (BioSorter platform; Union Biometrica), assaying at least 1,000 worms per condition.
ZRF1 and RING1B contribute to GG-NER. (A) RING1B and ZRF1 knockdown fibroblasts are defective in UDS after UV irradiation. UDS was measured by EdU incorporation after UV treatment in MRC5 fibroblasts with shRNA-mediated knockdown of the indicated proteins. XPA fibroblasts were used as a positive control. Values are given as mean ± SEM. Data were acquired from three independent experiments (150–300 nuclei per sample). (B) RING1B and ZRF1 knockdown fibroblasts are defective in the removal of CPDs. The CPD removal was analyzed in MRC5 fibroblasts after knockdown of the indicated proteins in MRC5 fibroblasts and in XPA fibroblasts. Cells were irradiated with 10 J/m2 and fixed immediately or 24 or 48 h after irradiation and stained with CPD antibodies. The relative fluorescence intensity was determined. Values are given as mean ± SEM. Data were acquired from three independent experiments (100–200 nuclei per sample). (C) MRC5 fibroblasts were treated with lentiviral particles containing the respective shRNA. Knockdown of the proteins levels was analyzed 48h after infection by Western blotting and incubation with the indicated antibodies. (D) C. elegans knockout mutants for ZRF1 (dnj-11) and RING1B (spat-3) show increased sensitivity toward UV irradiation. Late-L4 larval wild-type worms and the indicated mutants were irradiated with UV light at different doses, and the relative viability was determined by comparing hatched versus dead embryos (unhatched eggs). Values are given as mean ± SEM (n = 3). (E) C. elegans knockout mutants for dnj-11 and for spat-3 show only weak developmental arrest upon somatic UV irradiation. L1 larval worms were irradiated with UV light at different doses. Relative larval-stage stalling was determined after 60 h by using a large particle flow cytometer (BioSorter platform; Union Biometrica), assaying at least 1,000 worms per condition.
Further analysis of the DNA damage response in the C. elegans germline, which is regarded a measure for GG-NER (Lans and Vermeulen, 2011; Craig et al., 2012), showed that RING1B (spat-3) and XPC (xpc-1) mutants were affected by UV irradiation to a similar extent (Fig. 6 D). ZRF1 mutants (dnj-11) showed a stronger phenotype than XPC mutants (xpc-1), which is only surpassed by XPA mutants (xpa-1). We used a CSB mutant (csb-1) as a control strain, which is defective in TC-NER, but not in GG-NER. This mutant showed UV sensitivity comparable to wild-type animals. We made similar findings using RNAi-mediated knockdown of NER factors RING1B (spat-3) and ZRF1 (dnaj-11; Fig. S5 D). To analyze a potential function of RING1B and ZRF1 in TC-NER, we analyzed the relative larval stage stalling (L1 arrest; Lans and Vermeulen, 2011; Craig et al., 2012). After irradiation with increasing doses of UV light, worms were analyzed microscopically and by sorting on a large-particle sorter (Fig. 6 E; Fig. S5, E and F; and Table S1). Wild-type worms and XPC (xpc-1) and ZRF1 (dnj-11) mutants show larval arrest only at high doses of UV light, whereas CSB (csb-1) and XPA (xpa-1) mutants exhibit very strong phenotypes already at a low UV doses, in line with their defects in the TC-NER pathway (Fig. 6 E).
Collectively, we have identified ZRF1 and RING1B as potential players of GG-NER. ZRF1 recruitment to damaged chromatin is regulated by both its binding partner XPC and H2A ubiquitylation via the UV–RING1B complex.
ZRF1 remodels E3 ligase complexes at the lesion site
To explore the function of ZRF1 at damaged chromatin, we analyzed chromatin from ZRF1 knockdown cells after UV irradiation (Fig. 7 A). Upon depletion of ZRF1, we found enhanced RING1B and H2A-ubiquitylation levels at chromatin consistent with a function of ZRF1 in dislocating RING1B from chromatin (Richly et al., 2010). We next addressed its potential role in dislodging other subunits of the UV–RING1B complex from chromatin. We noticed that depletion of ZRF1 did not alter the recruitment of DDB2 to chromatin (Fig. 7 B). Importantly, however, we observed retention of CUL4B at chromatin, whereas recruitment of CUL4A was impaired. To determine the CUL4A levels at chromatin in control and ZRF1 knockdown cells, we expressed FLAGH2AX and performed affinity purifications (Fig. 7 C). We observed constant levels of DDB2 but reduced levels of CUL4A in the coprecipitate purified from ZRF1 knockdown cells. Similarly, FLAGDDB2 showed diminished association with CUL4A when purified from ZRF1 knockdown cells (Fig. 7 D). These data suggest a potential function for ZRF1 in remodeling the UV–RING1B complex at the DNA damage sites. To follow up on this idea, we analyzed whether the assembly of the UV–DDB–CUL4A complex was compromised in ZRF1 knockdown cells. To that end, we immunoprecipitated HARBX1 in control and ZRF1 knockdown cells (Fig. 7 E). In the coprecipitate, we noticed diminished levels of DDB2 and DDB1 but unaltered CUL4A binding upon ZRF1 knockdown, suggesting that ZRF1 mediates the association of CUL4A–RBX1 with DDB1–DDB2. Next, we tested a function for ZRF1 in remodeling the UV–RING1B complex in vitro. In pull-down experiments with purified proteins, we had noticed that ZRF1, like CUL4B and RING1B, specifically binds DDB2 (Fig. S3, F and G). Hence, we addressed whether ZRF1 competed with CUL4B, DDB1, and RING1B for binding to DDB2 (Fig. 7 F). In pull-downs with GFP-DDB2, we observed that increasing amounts of ZRF1 competes with CUL4B and RING1B binding, whereas the DDB1–DDB2 interaction was unaltered. Experiments using similar amounts of CUL4A, RBX1, and DDB1 showed that ZRF1 did not hamper the interaction of CUL4A and RBX1 with DDB2 (Fig. 7 G).
ZRF1 facilitates the assembly of the UV–DDB–CUL4A E3 ligase complex. (A) ZRF1 displaces RING1B from chromatin during NER. Chromatin association assays of control and ZRF1 knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A ubiquitin and RING1B abundance was calculated. Values are given as mean ± SEM (n = 3). (B) ZRF1 regulates chromatin association of CUL4A and CUL4B. Chromatin association assays of control and ZRF1 knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative CUL4B and CUL4A abundance was calculated. Values are given as mean ± SEM (n = 3). (C) ZRF1 regulates CUL4A association with H2AX containing nucleosomes. Control cells and ZRF1 knockdown cells expressing FLAGH2AX were irradiated with UV. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (D) Knockdown of ZRF1 modulates CUL4A association with DDB2. Control cells and ZRF1 knockdown cells expressing FLAGDDB2 were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (E) Assembly of the UV–DDB–CUL4A E3 ligase is facilitated by ZRF1. Control cells and ZRF1 knockdown HEK293T cells expressing HARBX1 were irradiated with UV light. After immunoprecipitation with HA-specific antibodies the precipitated material was subjected to Western blotting, and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (F) ZRF1 competes with CUL4B and RING1B for DDB2 binding in vitro. GFP and GFP-DDB2 immobilized on beads were incubated with equimolar amounts of purified DDB1, CUL4B, and RING1B and increasing amounts of ZRF1. ZRF1 levels were doubled stepwise reaching an eightfold molar excess of ZRF1 over the other components (relative molarity ZRF1: DDB1–CUL4B–RING1B; lane 3, 1:1; lane 4, 2:1; lane 5, 4:1; lane 6, 8:1). Precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 10%. (G) ZRF1 does not compete with CUL4A and RBX1 for binding to DDB1–DDB2. GFP and GFP-DDB2 immobilized on beads were incubated with equimolar amounts of purified DDB1, CUL4A and RBX1 and increasing amounts of ZRF1. ZRF1 levels were doubled stepwise reaching an eightfold molar excess of ZRF1 (relative molarity ZRF1: DDB1–CUL4A–RBX1; lane 3, 1:1; lane 4, 2:1; lane 5, 4:1; lane 6, 8:1). Precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 10%. (H) ZRF1 mediates the formation of the UV-DDB-CUL4A complex in vitro. GFP and GFP-DDB2 were coupled to beads and incubated with CUL4B, DDB1 and RING1B. After washing, GFP and GFP-DDB2 (UV–RING1B complex) beads were incubated with an estimated fivefold excess of purified CUL4A and RBX1 (lanes 1–3) over the retained UV–RING1B complex. Simultaneously, ZRF1 (lanes 1 and 3) or GST (lane 2) was added to the incubations in equimolar amounts. The precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%.
ZRF1 facilitates the assembly of the UV–DDB–CUL4A E3 ligase complex. (A) ZRF1 displaces RING1B from chromatin during NER. Chromatin association assays of control and ZRF1 knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative H2A ubiquitin and RING1B abundance was calculated. Values are given as mean ± SEM (n = 3). (B) ZRF1 regulates chromatin association of CUL4A and CUL4B. Chromatin association assays of control and ZRF1 knockdown HEK293T cell lines after UV irradiation. De–cross-linked material of the respective time points was subjected to Western blotting and probed with the indicated antibodies. The relative CUL4B and CUL4A abundance was calculated. Values are given as mean ± SEM (n = 3). (C) ZRF1 regulates CUL4A association with H2AX containing nucleosomes. Control cells and ZRF1 knockdown cells expressing FLAGH2AX were irradiated with UV. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (D) Knockdown of ZRF1 modulates CUL4A association with DDB2. Control cells and ZRF1 knockdown cells expressing FLAGDDB2 were irradiated with UV light. After immunoprecipitation with FLAG-M2-agarose, the purified material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 3%. (E) Assembly of the UV–DDB–CUL4A E3 ligase is facilitated by ZRF1. Control cells and ZRF1 knockdown HEK293T cells expressing HARBX1 were irradiated with UV light. After immunoprecipitation with HA-specific antibodies the precipitated material was subjected to Western blotting, and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (F) ZRF1 competes with CUL4B and RING1B for DDB2 binding in vitro. GFP and GFP-DDB2 immobilized on beads were incubated with equimolar amounts of purified DDB1, CUL4B, and RING1B and increasing amounts of ZRF1. ZRF1 levels were doubled stepwise reaching an eightfold molar excess of ZRF1 over the other components (relative molarity ZRF1: DDB1–CUL4B–RING1B; lane 3, 1:1; lane 4, 2:1; lane 5, 4:1; lane 6, 8:1). Precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 10%. (G) ZRF1 does not compete with CUL4A and RBX1 for binding to DDB1–DDB2. GFP and GFP-DDB2 immobilized on beads were incubated with equimolar amounts of purified DDB1, CUL4A and RBX1 and increasing amounts of ZRF1. ZRF1 levels were doubled stepwise reaching an eightfold molar excess of ZRF1 (relative molarity ZRF1: DDB1–CUL4A–RBX1; lane 3, 1:1; lane 4, 2:1; lane 5, 4:1; lane 6, 8:1). Precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 10%. (H) ZRF1 mediates the formation of the UV-DDB-CUL4A complex in vitro. GFP and GFP-DDB2 were coupled to beads and incubated with CUL4B, DDB1 and RING1B. After washing, GFP and GFP-DDB2 (UV–RING1B complex) beads were incubated with an estimated fivefold excess of purified CUL4A and RBX1 (lanes 1–3) over the retained UV–RING1B complex. Simultaneously, ZRF1 (lanes 1 and 3) or GST (lane 2) was added to the incubations in equimolar amounts. The precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%.
Finally, to study ZRF1-mediated remodeling in vitro, we assembled the UV–RING1B complex and analyzed the replacement of CUL4B–RING1B with CUL4A–RBX1 (Fig. 7 H). The addition of purified CUL4A–RBX1 to immobilized UV–RING1B complexes (Fig. 7 H, lane 2) or GFP-loaded beads (lane 1) showed only minimal or no incorporation of CUL4A and RBX1 into the E3 ligase complex. In contrast, in the presence of ZRF1, we noticed a significant replacement of CUL4B–RING1B by CUL4A–RBX1 (lane 3).
In sum, our data suggest that ZRF1 remodels E3 ligase complexes at the lesion site and that it mediates the assembly of the UV–DDB–CUL4A E3 ligase complex.
ZRF1 regulates ubiquitylation of XPC
To confirm that ZRF1 mediates the assembly of the UV–DDB–CUL4A E3 ligase complex, we analyzed the polyubiquitylation of its substrate, XPC (Sugasawa et al., 2005). After UV irradiation of ZRF1 knockdown cells, we observed diminished polyubiquitylation of XPC when compared with control cells (Fig. 8 A). Similarly, immunoprecipitations of ubiquitylated proteins after expressing HAUbiquitin in control, RING1B, and ZRF1 knockdown cells showed a significant reduction of ubiquitylated XPC in knockdowns compared with control (Fig. 8 B). After expression of HAXPC and HISUbiquitin, we immunoprecipitated HAXPC and analyzed its ubiquitylation status (Fig. 8 C). In agreement with our previous data, we observed a significant reduction of XPC ubiquitylation in both knockdown cell lines. Moreover, we expressed HISUbiquitin in control, RING1B, and ZRF1 knockdown cell lines (Fig. 8 D). After UV irradiation of cells, we performed NiNTA pull-down experiments under denaturing conditions to enrich for ubiquitylated proteins. We observed strong ubiquitylation of XPC only in control cells, whereas XPC ubiquitylation levels in ZRF1 and RING1B knockdown cells were reduced. Collectively these experiments suggest that ZRF1 likely regulates XPC ubiquitylation by facilitating the assembly of the UV–DDB–CUL4A complex. RING1B in turn provides a tethering platform for ZRF1, thereby indirectly affecting the remodeling process.
ZRF1 regulates XPC ubiquitylation. (A) ZRF1 facilitates XPC ubiquitylation after UV irradiation. Whole-cell extracts of control and ZRF1 knockdown HEK293T cells from the stated time points were subjected to Western blotting and probed with the indicated antibodies. (B) Role of RING1B and ZRF1 in XPC ubiquitylation. Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HAUbiquitin were irradiated with UV light. After immunoprecipitation with HA-specific antibody, the precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (C) Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HAXPC and HISUbiquitin were irradiated with UV light. After immunoprecipitation with HA-specific antibody, the precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (D) Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HISUbiquitin were irradiated with UV and harvested 1 h after UV exposure. Ubiquitylated proteins were purified by NiNTA agarose under denaturing conditions, and Western blots of the purified material were incubated with the indicated antibodies. (E) The UV–RING1B complex and ZRF1 cooperate during NER. DNA lesions (yellow star) are recognized by the UV-RING1B complex (DDB1–DDB2–CUL4B–RING1B), which catalyzes ubiquitylation of histone H2A (gray sphere). ZRF1 is recruited to the lesion site by XPC and tethers to the H2A-ubiquitin mark. ZRF1 causes the assembly of the UV–DDB–CUL4A complex, which subsequently catalyzes ubiquitylation of XPC.
ZRF1 regulates XPC ubiquitylation. (A) ZRF1 facilitates XPC ubiquitylation after UV irradiation. Whole-cell extracts of control and ZRF1 knockdown HEK293T cells from the stated time points were subjected to Western blotting and probed with the indicated antibodies. (B) Role of RING1B and ZRF1 in XPC ubiquitylation. Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HAUbiquitin were irradiated with UV light. After immunoprecipitation with HA-specific antibody, the precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (C) Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HAXPC and HISUbiquitin were irradiated with UV light. After immunoprecipitation with HA-specific antibody, the precipitated material was subjected to Western blotting and blots were incubated with the indicated antibodies. Inputs correspond to 5%. (D) Control cells and RING1B and ZRF1 knockdown HEK293T cells expressing HISUbiquitin were irradiated with UV and harvested 1 h after UV exposure. Ubiquitylated proteins were purified by NiNTA agarose under denaturing conditions, and Western blots of the purified material were incubated with the indicated antibodies. (E) The UV–RING1B complex and ZRF1 cooperate during NER. DNA lesions (yellow star) are recognized by the UV-RING1B complex (DDB1–DDB2–CUL4B–RING1B), which catalyzes ubiquitylation of histone H2A (gray sphere). ZRF1 is recruited to the lesion site by XPC and tethers to the H2A-ubiquitin mark. ZRF1 causes the assembly of the UV–DDB–CUL4A complex, which subsequently catalyzes ubiquitylation of XPC.
Based on our results, we propose that H2A ubiquitylation by the UV–RING1B complex is catalyzed early during damage recognition (Fig. 8 E). Our data illustrate for the first time how E3 ligase complexes are remodeled at the DNA lesion site. The presented results suggest that ZRF1 acts as a switch protein that remodels E3 ligases at or close to the DNA damage site (Fig. 8 E).
Discussion
Monoubiquitylation of histone H2A is a hallmark of various DNA repair pathways. Nevertheless, it is still a matter of debate how and when different E3 ligases contribute to H2A ubiquitylation during the DNA damage response. Here, we have examined selected E3 ligases involved in UV-induced DNA damage repair. Our data point to RING1B as the main E3 ligase involved in H2A ubiquitylation at lysine 119 early during damage recognition in NER. Depletion of RNF168 or abrogation of UV–DDB–CUL4A E3 ligase function did not cause any significant changes in H2A ubiquitylation after UV irradiation. The UV–DDB–CUL4A E3 complex was previously shown to catalyze ubiquitylation of histone H2A (Kapetanaki et al., 2006). Our data show that the UV–DDB–CUL4A E3 ligase complex functions downstream of ZRF1, suggesting that it might ubiquitylate histone H2A at a later stage in the NER pathway (Fig. 7 D). Hence, we propose that the timing of E3 ligase action is an important feature of NER and other DNA repair pathways. In the same vein, it was demonstrated that RNF8-mediated H2A ubiquitylation is a relatively late event during NER (Marteijn et al., 2009). Our data extend this observation, proposing that E3 ligases operate successively during the DNA damage response. In addition, E3 ligases target different lysines of histone H2A, adding another layer of complexity. For instance, at DSBs, RNF168 catalyzes the ubiquitylation of lysines 13 and 15 (Mailand et al., 2007; Mattiroli et al., 2012), whereas RING1B targets lysine 119 of histone H2A in both DSB repair and NER (Ui et al., 2015). However, understanding the concerted action and the substrate specificity of E3 ligases in DNA repair needs further investigation.
RING1B and H2A ubiquitylation have been implicated in UV-mediated DNA damage repair about a decade ago (Bergink et al., 2006). However, the molecular mechanism of RING1B function still remained unclear. RING1B controls the basal levels of the highly abundant H2A-ubiquitin mark (Matsui et al., 1979; Wang et al., 2004). Thus, it might affect the nuclear pool of free ubiquitin and thereby indirectly ubiquitin signaling during DNA repair (Dantuma et al., 2006). Additionally, it was reported that knockdown of RING1B decreases nuclear ubiquitin levels and thus indirectly reduces histone ubiquitylation at damaged chromatin (Bergink et al., 2006). Our data refute these ideas, as we observe no global changes in the levels of ubiquitylated proteins in RING1B knockdown cells (Fig. S1, G and H). Thus, we rule out an indirect effect of RING1B knockdown, implying a DNA damage–specific role of RING1B in H2A ubiquitylation. In particular, we provide evidence that RING1B constitutes a DNA damage–specific E3 ligase, as it is specifically recruited to DNA lesion sites induced by irradiation with a 405-nm laser (Fig. S1, D–F). This observation is also in agreement with a recent study demonstrating that RING1B is recruited to DSBs to promote local gene silencing (Ui et al., 2015). In light of these findings, we addressed how RING1B interacts with the NER pathway, which is an essential DNA repair pathway implicated in repair of UV-mediated DNA damage. Previously, RING1B had been shown to mediate ubiquitylation of histones H2A and H2AX at DSBs together with its PRC1 binding partner, BMI-1 (Pan et al., 2011; Ui et al., 2015). After UV irradiation, RING1B seems to catalyze H2A ubiquitylation at lysine 119 independent of BMI-1, contrasting its function in DSB repair and during gene silencing. Our data indicate that RING1B binds to the DNA damage recognition factor DDB2. Importantly, DDB2 determines whether RING1B is recruited to chromatin after UV irradiation, suggesting that DDB2 tethers RING1B to the damage site. DDB2 and RING1B represent subunits of a novel E3 ligase complex (UV–RING1B). In this complex, RING1B directly interacts with CUL4B (Fig. S3, E–I), which is in agreement with the common modular composition of cullin-RING E3 ligases (Petroski and Deshaies, 2005). The UV–RING1B complex is reminiscent of the well-described UV–DDB–CUL4A complex consisting of DDB1, DDB2, CUL4A, and RBX1 (Groisman et al., 2003). Our study suggests that DDB1–DDB2 might act as a platform that can either accommodate CUL4B–RING1B or CUL4A–RBX1 modules, respectively. We have demonstrated that the UV–RING1B complex dramatically enhances ubiquitylation of histone H2A in vitro and in vivo. Hence, RING1B mediated monoubiquitylation at lysine 119 in DNA repair is performed by either the PRC1 complex or the UV–RING1B complex.
Because ZRF1 is one of the few known readers of H2A ubiquitin, we hypothesized that it would play a similar role in UV-mediated DNA repair as in cellular differentiation (Richly et al., 2010). In accordance, we observed that binding of ZRF1 to chromatin after UV irradiation depends both on presence of RING1B and its ability to bind H2A ubiquitin. More importantly, ZRF1 localizes to XPA and XPC foci after local irradiation and knockdown of ZRF1 compromises DNA repair as seen by UDS and removal of CPD, describing ZRF1 as a new player in UV-mediated DNA repair. Drug-mediated inhibition of the RING1B activity significantly reduced ZRF1 colocalization with XPC, supporting a role for H2A ubiquitin in tethering ZRF1 to the damage site. On the other hand, UV irradiation–triggered recruitment of ZRF1 to chromatin depends on XPC. This close interplay between ZRF1 and XPC is further reflected by the interaction of both proteins and the epistasis analysis performed with either human cells or C. elegans, supporting a role for ZRF1 in GG-NER. In light of these findings, we speculate that XPC is probably involved in ZRF1’s recruitment to the DNA damage site, whereas the H2A-ubiquitin mark is potentially needed to stably tether ZRF1 to chromatin. Most importantly, ZRF1 mediates the remodeling of E3 ligase complexes at DNA damage sites (Fig. 7 D). Upon recruitment to chromatin, ZRF1 causes the exchange of the cullin-E3 ligase module, whereas DDB1 and DDB2 most probably remain bound to the lesion site. This observation does not exclude that UV–CUL4A complexes are generated independent of ZRF1. Still, our data reflect one plausible succession of events that take place at damaged chromatin. This function of ZRF1 is reminiscent of the Cand1 protein, which promotes the exchange of subunits from cullin–RING complexes (Pierce et al., 2013). We propose that ZRF1 acts in concert with other remodeling complexes or chaperones at chromatin. In fact, ZRF1 was shown to cooperate with the HSP70 chaperone network during protein quality control (Qiu et al., 2006; Jaiswal et al., 2011). It remains to be tested whether ZRF1 cooperates with the HSP70 system, Cand1, or chromatin remodeling complexes during NER.
Materials and methods
Cell lines and transfections
HEK293T and HEK293 cells were cultured in DMEM supplemented with 10% FBS at 37°C and 5% CO2. HeLa Kyoto cells stably expressing cherry-PCNA were cultured in DMEM supplemented with 10% FCS and 1 µM/ml gentamycin and 2.5 µg/ml blasticidin. MRC5 (AG05965), normal skin fibroblasts (GM15876), XPE (GM01389), XPE (GM02415), XPC-complemented (GM16248), XPC (GM15983), XPA-complemented (GM15876), and XPA (GM04312 and GM00710) fibroblasts were purchased from Coriell Cell Repositories and cultured in DMEM, supplemented with 15% FBS. To generate cells stably expressing FLAGRING1B, HEK293 cells were transfected with a pCMV-2b-RING1B-FLAG plasmid and selected with G148 for 14 d. The expression of FLAGRING1B was verified by Western blot.
Transfection of HEK293T cells was either performed by the calcium phosphate coprecipitation method as described previously (Richly et al., 2010) or by Lipofectamine (Invitrogen) transfection. Information on the plasmids used is provided in Table S2.
UV irradiation and drug treatment
Cells were irradiated with 10 J/m2 UV-C using a CL-1000 UV cross-linker (UVP) unless stated otherwise. PRT4165 (Abcam) was used at a concentration of 50 µM as described in Ismail et al. (2013).
Gene inactivation by shRNA/siRNA
HEK293T-shControl, HEK293T-shZRF1, and HEK293T-shRING1B were described previously and generated by transduction of HEK293T cells with retrovirus vector, containing shRNA against ZRF1 or RING1B (Richly et al., 2010). Gene knockdown in MRC5 fibroblasts was performed by introduction of MISSION pLKO.1-shRNA plasmids (Sigma-Aldrich) targeting the respective gene using third generation lentivirus system. Plasmids contained the following sequences (Sigma-Aldrich): control (TRC1/1.5), ZRF1 (TRCN0000254058), RING1B (TRCN0000033697), DDB2 (TRCN0000083995), and XPC (TRCN0000307193).
The siRNA transfections were performed using Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen). The following siRNAs were used in this study: control (SIC001; Sigma-Aldrich), CUL4A (esiRNA EHU011891; Sigma-Aldrich), RNF168 (SMARTpool D-011-22-(01–04); GE Healthcare), DDB2 (SASI_Hs01_00101645, SASI_Hs01_00101647; Sigma-Aldrich), BMI-1 (esiRNA EHU004421; Sigma-Aldrich), CUL4B (esiRNA EHU064911; Sigma-Aldrich), XPC (SASI_Hs01_00086530, SASI_Hs01_00086531; Sigma-Aldrich). Information on shRNA and siRNA sequences used in this study is provided in Table S3.
Chromatin association assays
HEK293T cells (unless stated otherwise) were irradiated with UV and cross-linked by formaldehyde at the indicated time points after UV irradiation. Assays were essentially performed as published (Richly et al., 2010). In brief, cell pellets were resuspended in buffer A (100 mM Tris, pH 7.5, 5 mM MgCl2, 60 mM KCl, 125 mM NaCl, 300 mM sucrose, 1% NP-40, and 0.5 mM DTT) and kept on ice for 10 min. After centrifuging nuclei pellet was lysed in a hypotonic solution (3mM EDTA, 0.2 mM EGTA, and 1 mM DTT) twice. The chromatin-containing pellet was solubilized in 2× Laemmli buffer, sonicated, and boiled to reverse cross-linking. Information on antibodies used for Western blots is provided in Table S4. All experiments were repeated at least three times. Band intensities from Western blots were measured as stated in the figure legends using ImageJ or ImageLab (Bio-Rad) software.
Immunoprecipitations and affinity purifications
Cells were treated with UV and harvested 1 h after exposure unless stated otherwise. Cells were resuspended in buffer A (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl and 0.5 mM DTT, 1 mM PMSF, and protease inhibitors; Roche) and homogenized by 10 strokes in a Dounce homogenizer with a B-type pestle. After centrifugation, nuclei were resuspended in lysis buffer (20 mM Hepes, 150 mM NaCl, 2.5 mM EGTA, 2 mM EDTA, 0.1% Triton X-100, 0.5 mM DTT, 1 mM PMSF, and protease inhibitors; Roche) and sonified using a Diagenode Bioruptor for 20 min on the high setting. To verify sonification efficiency, DNA from the extracts analyzed by agarose gel electrophoresis. Only samples containing DNA of 300 bp or smaller were used in the experiments. Protein extracts were then subjected to centrifugation (21,000 g, 4°C, 15 min), and the supernatant was incubated with antibodies overnight at 4°C. After incubation with protein A agarose beads for 2 h at 4°C, the immune complexes were washed extensively in lysis buffer and material retained on the beads was subjected to Western blotting. Information on antibodies used for immunoprecipitations and Western blots is provided in Table S4.
Affinity purifications using FLAG-M2 agarose beads (Sigma-Aldrich) and Anti-HA Agarose beads (Sigma-Aldrich) were performed using the protocol stated for immunoprecipitations. Purifications involving the STREP tag were performed with STREP-Tactin beads (Iba LifeSciences) and Desthiobiotin (Sigma-Aldrich) according to the manufacturer’s instructions. Purifications involving the GFP tag were performed with GFP-Trap agarose beads (ChromoTek) according to the manufacturer’s instructions. For purification of the proteins used in the in vitro experiments (Fig. S3 E: FLAG-STREPCUL4B, FLAGDDB1, FLAGRING1B, FLAGZRF1, FLAGZRF1, HARBX1, and HACUL4A), the proteins were washed extensively on the beads with lysis buffer containing 1 M NaCl before elution with FLAG or HA peptide (Sigma-Aldrich).
In vitro ubiquitylation assays
In vitro ubiquitylation reactions were performed with 3 µg purified histone H2A (New England Biolabs, Inc.) or 5 µg recombinant nucleosomes (Active Motif), 200 ng purified HIS-UBA1 (E1), 20 ng purified GST-UBC5H (E2), 150 ng purified UV-RING1B (E3), or 150 ng GST (control) in UBAB buffer (25 mM Tris/HCl, pH 7.5, 50 mM NaCl, and 10 mM MgCl2) supplemented with 20 mM ATP, 1.5 mg/ml ubiquitin, 10 mM DTT, and 1 U creatine phosphokinase. Reactions were kept at 37°C for the indicated times and subsequently subjected to Western blotting.
Purification of recombinant proteins
Proteins were purified as suggested by GE Healthcare (GST-tagged proteins) or QIAGEN (His-tagged proteins) after inducing BL21 bacterial strains transformed with the respective plasmids at an OD = 0.5 with 0.2 mM isopropyl-β-D-thiogalactoside for 4 h at 37°C or at 20°C for 14 h. The following recombinant proteins were purchased: H2A (New England Biolabs), Ubiquitin (Boston Biochem), nucleosomes (Active Motif), GST-RBX1 (Novus Biologicals), and RAD23A (Abcam).
GST pull-downs
Purified GST–proteins were bound in equimolar amounts to glutathione beads (Amersham) in binding buffer (20 mM Tris, pH 7.4, 150 mM NaCl, and 0.1% Triton X-100). Loaded beads were washed in the same buffer and used for incubation with purified proteins for 2 h at 4°C. After extensive washing in binding buffer, the retained material was subjected to Western blotting.
Purification of ubiquitin conjugates from cells
Cells expressing HIS-tagged ubiquitin were lysed in lysis buffer (8 M urea, 100 mM NaH2PO4, and 10 mM Tris, pH 8.0) 1 h after UV irradiation. Ubiquitylated proteins were retained on NiNTA agarose after washing with wash buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris, pH 6.3, 300 mM NaCl, and 0.1% Triton X-100) and detected by Western blotting using the indicated antibodies.
Fractionation of cell extracts
HEK293T cells were harvested by trypsinization and the cell pellet was divided in two equal parts. One part was resuspended in Laemmli buffer and sonicated (whole-cell extract), and the other was washed twice with PBS and resuspended in buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease inhibitors, and 0.1% Triton X-100) and cells were incubated for 8 min on ice. Subsequently, cells were spun down (4°C, 1,300 g, 5 min). The supernatant (cytoplasmic fraction) was collected, precipitated with TCA, and resuspended in Laemmli buffer. Nuclei were washed twice with buffer A, resuspended in Laemmli buffer, and sonicated. Whole-cell extract, cytoplasmic, and nuclear fractions were subjected to Western blotting as indicated.
Mass spectrometry analysis
Mass spectrometry sample preparation, measurement and database search were performed as described previously (Bluhm et al., 2016). Gradient lengths of 45 or 105 min were chosen depending on the immunoprecipitated material obtained. Raw files were processed with MaxQuant (version 1.5.2.8) and searched against the Homo sapiens UniProt database (February 25, 2012) using the Andromeda search engine integrated into MaxQuant and default settings were applied. Proteins with at least two peptides, one of them unique, count as identified.
Fluorescence microscopy
Experiments were performed with MRC5 fibroblasts and patient-derived fibroblasts. Cells were transfected with mCherry-ZRF1 and GFP-DDB2 expressing plasmids. Cells were exposed to localized UV damage (100 J/m2) using a micropore membrane with 5-µm pore size as described previously (Katsumi et al., 2001). Preextraction was performed with CSK supplemented with 0.2% Triton X-100 at 30 min after UV and then fixed in 4% PFA. Cells were stained with XPA (Novus Biologicals) or XPC (Cell Signaling Technology) antibodies overnight at 4°C. After washing, coverslips were incubated with Alexa Fluor 488 fluorophore–conjugated secondary antibodies (Thermo Fisher Scientific) and mounted in Vectashield with DAPI. Images were acquired with the LAS AF software (Leica Biosystems) using a TCS SP5 confocal microscope (Leica Biosystems) with a 63×/1.4 oil-immersion objective. For colocalization studies, ∼100 lesions were counted per condition.
Imaging and microirradiation experiments
For microirradiation, HeLa-Kyoto Cherry-PCNA cells were grown on cover slide dishes and transfected with the indicated constructs using polyethylenimine. Imaging and microirradiation experiments were performed using an UltraVIEW VoX spinning-disc confocal system (PerkinElmer) in a closed live-cell microscopy chamber (ACU; PerkinElmer) at 37°C with 5% CO2 and 60% humidity, mounted on a Nikon TI microscope (Nikon). Images were taken with a CFI Apochromat 60×/1.45 NA oil immersion objective. GFP and Cherry or mRFP were imaged with 488 and 561 nm laser excitation and 527 ± 55 and 612 ± 70 nm (full width at half maximum) emission filters, respectively. For microirradiation, a preselected spot (1 µm diameter) within the nucleus was microirradiated for 1,200 ms with the 405-nm laser resulting in 1 mJ. Before and after microirradiation, confocal image series of one midnucleus z section were recorded in 2-s intervals. For evaluation of the accumulation kinetics between 4 and 12 cells were analyzed. Images were first corrected for cell movement (ImageJ plugin StackReg and transformation mode Rigid body), and mean intensity of the irradiated region was divided by mean intensity of the whole nucleus (both corrected for background) using ImageJ software. Maximal accumulation represents the highest ratio from each experiment.
Microscopy on skin biopsy specimens
Human skin sections were taken from material biopsied from patients who had given their written consent and were provided by R. Greinert and B. Volkmer (Dermatology Center Buxtehude, Buxtehude, Germany). Biopsy specimens were taken from either the cheek (UV exposed) or groin (not exposed), and 7-µm cryosections were prepared after freezing in liquid nitrogen. The sections were mounted on glass slides and fixed in 100% MeOH and 100% acetone for 10 min, each at −20°C. For immunostaining, the sections were rehydrated in PBS, and antigen retrieval was performed at 80°C in sodium citrate buffer (10 mM sodium citrate, pH 6.0) overnight. Then the sections were blocked in 4% BSA in PBS for 30 min before the first antibody was applied in 1% BSA, 0.1% Triton X-100 in PBS (ZRF1; self-made), H2A ubiquitin (Cell Signaling Technology), RING1B (self-made; all diluted 1:100), and mouse DDB2 (1:20; Abcam). For CPD detection, DNA was additionally denatured for 3 min in 0.1 N NaOH/70% ethanol after the antigen retrieval followed by dehydration in 70%, 90%, and 100% ethanol. The CPD antibody (Kamiya) was used at a dilution of 1:100. Primary antibodies were incubated for 3 h at room temperature, followed by three washes in PBS. Secondary antibodies (anti–mouse IgG Alexa Fluor 488; Invitrogen; and anti–rabbit IgG-Cy3 and anti–rabbit IgG TexasRed; Jackson ImmunoResearch Laboratories, Inc.) were added at 1:500 for 1 h at room temperature. Sections were then washed three times in PBS and stained with 10 µM DAPI for 10 min before being mounted in Vectashield. Skin sections were imaged using an Axiovert 200 (ZEISS) equipped with a 40× Planneofluar 1.3 NA objective lens and single channels were recorded with a black and white Axicam mRM (ZEISS). Quantification of signals on the single-cell level was performed using ImageJ. After selecting random nuclei in the DAPI channel, the mean and integrated intensities of the red and green channels were measured. All intensities are normalized to the DNA content of the corresponding nucleus. At least 200 nuclei were analyzed in at least three sections.
Colony formation assay
HEK293T control and knockdown cell lines were transfected with the respective siRNAs with Lipofectamine (Invitrogen) according to the manufacturer’s protocol. Details on the respective transfections are given in the figure legends. Cells were plated on tissue culture plates at a density of 1,000 cells per plate 24 h after transfection. Cells were irradiated with the indicated UV dose 48 h after transfection. Colonies were counted 7 d after irradiation. Numbers of colonies formed after UV irradiation were normalized against the non-UV–treated control.
UDS
UDS experiments were performed as described previously (Jia et al., 2015). In brief, MRC5 fibroblasts were transduced with lentiviral particles expressing the respective shRNAs. XPA fibroblasts were used as a positive control. After viral transduction, the cells were serum starved for 24 h, irradiated with UV light (20 J/m2), and incubated with 10 µM EdU (Thermo Fisher Scientific) for 2 h. Alexa Fluor 555 azide (Thermo Fisher Scientific) was conjugated to EdU using the Click-reaction. The coverslips were mounted in Vectashield with DAPI. Images were acquired with the LAS AF software (Leica Biosystems) using a AF-7000 widefield microscope (Leica Biosystems) with a 63×/1.4 oil immersion objective and an ORCA CCD camera (Hamamatsu). Images were analyzed using ImageJ. DAPI was used to define nuclei, and EdU intensity within nuclei was measured after background subtraction. A total of 150–300 nuclei were analyzed per sample. Mean intensities of +UV and −UV conditions for all cells were calculated and used to estimate the DNA repair occurring in the particular sample.
Removal of CPDs
MRC5 fibroblasts were transduced with lentiviral particles expressing the respective shRNAs. XPA fibroblasts were used as a positive control. 24 h after viral transduction, cells were replated on coverslips, exposed to UV light, and fixed at the indicated time points. Cells were stained with CPD antibody (Cosmo Bio) using the manufacturer’s protocol, followed by incubation with Alexa Fluor 488 fluorophore–conjugated secondary antibodies (Thermo Fisher Scientific). The cells were mounted in Vectashield with DAPI, and images were acquired with the LAS AF software (Leica Biosystems) using an AF-7000 widefield microscope (Leica Biosystems) with a 63×/1.4 oil-immersion objective and an ORCA CCD camera (Hamamatsu Photonics). Images were analyzed using ImageJ. DAPI was used to define nuclei, and CPD intensity within nuclei was measured after background subtraction. 100–200 nuclei were analyzed per sample. Mean intensities of +UV and −UV conditions for all cells were calculated and used to estimate the DNA repair occurring in the particular sample.
C. elegans culture
Nematodes were cultured on agar plates at 20°C according to standard procedures. Strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). The following strains were used: wild type (N2 Bristol), VC31/spat-3 (gk22; WBGene00020496), DL74/mig-32 (n4275; WBGene00008684), VC1642/dnj-11 (gk1025; WBGene00001029), RB885/xpc-1(ok734; WBGene00022296), RB1801/csb-1(ok2335; WBGene00000803), and RB864/xpa-1 (ok698; WBGene00006963). Mutant strains were outcrossed at least three times to the wild-type strain (N2).
Measuring DNA damage response in the C. elegans germline
The L4 survival assay was performed as described previously (Craig et al., 2012). In brief, late-L4 larval hermaphrodites were irradiated with different doses of UV light. The damage sensitivity of the meiotic pachytene cells of the germline was measured by determining the survival of embryos produced between 24 and 30 h after L4-stage irradiation.
Measuring DNA damage response in the C. elegans soma via developmental arrest
The L1 development arrest assay was performed as described previously (Craig et al., 2012). In brief, L1-stage worms were synchronized via starvation and irradiated with different doses of UV light. Relative larval-stage stalling was determined after 60 h, when control worms were fully fertile. Larval-stage scoring was done using a large-particle flow cytometer (BioSorter platform; Union Biometrica).
RNAi via feeding
Worms were fed at L1 larval stage with Escherichia coli feeding clones (HT115), which express dsRNAi targeted against a gene of interest. In brief a single colony of a clone was grown overnight in LB containing 100 µg/ml ampicillin (37°C, 200 rpm). Subsequently the clone was induced for 1 h by adding 4 mM IPTG to the LB media. The induced bacteria then was spun down at room temperature and resuspended in nematode growth medium with 4 mM IPTG. L1 larval worms were directly grown in this medium at 20°C until they reached late L4 stage or early adulthood (50–60 h).
Online supplemental material
Fig. S1 shows the function of RING1B in H2A ubiquitylation during UV-triggered DNA repair and recruitment of RING1B to UV-mediated DNA damage sites. Fig. S2 shows the BMI-1 independent interaction of RING1B–DDB2. RING1B, H2A ubiquitin, and DDB2 staining in human skin sections and H2A ubiquitin accumulation after UV irradiation in GM02415 fibroblasts. Fig. S3 shows interactions of UV–RING1B subunits and competition of RING1B and RBX-1 for binding to CUL4B. Fig. S4 shows a quantification of ZRF1 localization to DNA damage sites and its dependency on H2A ubiquitin. Fig. S5 shows the ZRF1 and XPC interplay and effect on UV sensitivity assays in C. elegans. Table S1 shows a data summary of developmental arrest assay in mutant strains. Table S2 lists plasmids used in this study. Table S3 lists the shRNA and siRNA sequences used for this study. Table S4 lists antibodies used in this study. Table S5 provides peptide numbers and protein names for all proteins identified in the mass spectrometry analysis after sequential immunoprecipitations with FLAG and RING1B antibodies. Table S6 provides peptide numbers and protein names for all proteins identified in the mass spectrometry analysis of purified UV–RING1B complex.
Acknowledgments
We thank the members of the Richly laboratory for comments on the manuscript. We thank M. Hanulova for help with processing data for UDS experiments. We thank S. Papaefstathiou, S. Schaffer, and S. Meyer for excellent technical work.
This work was supported by grants from the Boehringer Ingelheim Foundation and the Deutsche Forschungsgemeinschaft (RI-2413/1-1). M.C. Cardoso was supported by a grant from the Bundesministerium für Bildung und Forschung (02NUK017D). We thank the Institute of Molecular Biology Proteomics and Microscopy core facilities for their expert assistance. C. elegans strains were kindly provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
The authors declare no competing financial interests.
References
Author notes
E. Gracheva and S. Chitale contributed equally to this paper.