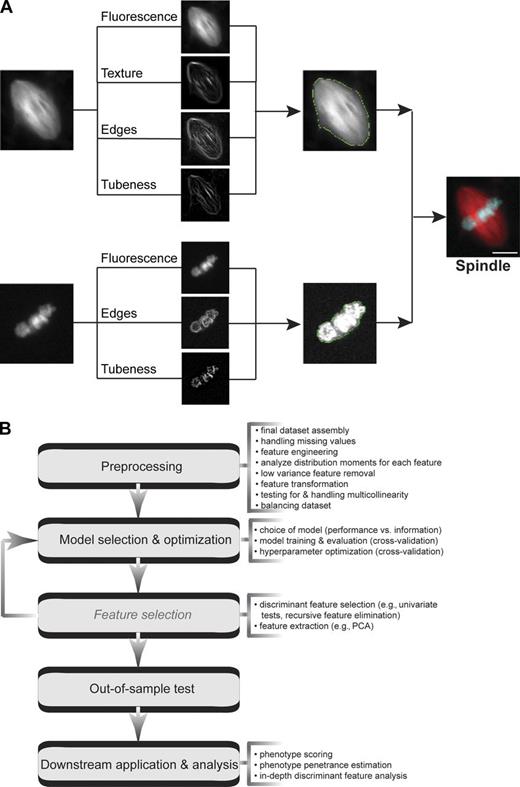
Imaging datasets are rich in quantitative information. However, few cell biologists possess the tools necessary to analyze them. Here, we present a large dataset of Xenopus extract spindle images together with an analysis pipeline designed to assess spindle morphology across a range of experimental conditions. Our analysis of different spindle types illustrates how kinetochore microtubules amplify spindle microtubule density. Extract mixing experiments reveal that some spindle features titrate, while others undergo switch-like transitions, and multivariate analysis shows the pleiotropic morphological effects of modulating the levels of TPX2, a key spindle assembly factor. We also apply our pipeline to analyze nuclear morphology in human cell culture, showing the general utility of the segmentation approach. Our analyses provide new insight into the diversity of spindle types and suggest areas for future study. The approaches outlined can be applied by other researchers studying spindle morphology and adapted with minimal modification to other experimental systems.
Introduction
The spindle is a complex and dynamic macromolecular machine that functions to accurately distribute chromosomes during eukaryotic cell division. Spindle morphology varies across a wide range of cellular environments (Helmke and Heald, 2014; Mitchison et al., 2015). Proper spindle function is essential to maintain ploidy during cell division, and factors that influence spindle assembly and morphology are frequently altered in cancer (Gordon et al., 2012). Despite the clear biological and biomedical significance of the spindle, quantitative descriptions of spindle morphology are limited to a small number of studies in model organisms including yeast (Vizeacoumar et al., 2010), Caenorhabditis elegans (Farhadifar et al., 2015), and humans (Neumann et al., 2010; Sironi et al., 2011; Young et al., 2014). Although these studies are information-rich resources, gaining access to raw data may be problematic, making meta-analysis and data reuse difficult. Furthermore, secondary analyses of datasets that contain only a few measurements per spindle may not be very informative. Our goal was to assemble a large dataset of Xenopus spindle images and develop a pipeline for efficient measurement and analysis that can be broadly applied by cell biologists without specialized skills in automated image analysis.
Although the number of large image datasets is limited, mechanisms that control specific spindle features such as length have been extensively investigated (Goshima and Scholey, 2010). Using in vitro spindle assembly systems, we previously described molecular mechanisms that scale spindle length to cell size between the closely related frog species Xenopus laevis and Xenopus tropicalis (Brown et al., 2007; Loughlin et al., 2011; Helmke and Heald, 2014) and during X. laevis embryonic development (Wilbur and Heald, 2013). Levels of one of the identified spindle scaling factors, the cancer-linked microtubule-associated protein TPX2 (Neumayer et al., 2012), also modulated spindle microtubule organization, suggesting that spindle length and overall architecture could be linked (Helmke and Heald, 2014). This is consistent with work in C. elegans and human cells (Hara and Kimura, 2013; Young et al., 2014). However, with the exception of spindle length, little is known about how spindle features are affected by changes in spindle environment or the relationships among spindle features.
To generate a resource in which input images, raw data, and analysis tools are made available to the community, we created an image analysis pipeline using CellProfiler (Carpenter et al., 2006), an easy to use open source software package. Our analysis generated a dataset rich in quantitative information describing the morphology of more than 36,000 spindles from over 80 independent experiments over several years performed by multiple investigators. We used these data to investigate how spindle architecture, namely size, shape, and microtubule content are altered by different cytoplasmic and DNA components. These observations generate testable hypotheses for how distinct spindle morphologies are established and maintained. We also adapted our pipeline to analyze Lamin B1–stained nuclei in human cell culture, showing the utility of our image analysis approach across experimental systems and subcellular structures.
Results
Xenopus extract is a powerful system to reconstitute spindle assembly in vitro and evaluate the effects of a wide range of perturbations. However, this system presents an automated image analysis challenge because background fluorescence intensity and image texture vary widely within individual images and across experiments. This complicates image segmentation, the process of distinguishing objects of interest from background, because background structures and spindles share many of the same features. To overcome this problem, we developed a segmentation strategy in CellProfiler (Carpenter et al., 2006) in which primary segmentation is carried out on a composite image formed by summing processed versions of the input image in which consistent features of spindle microtubules and mitotic chromatin have been enhanced. For microtubules, these features were tubulin fluorescence, texture (a measure of subspindle coarseness), edges (indicative of a boundary between the spindle and background), and tubeness (a measure of curvilinear structures), and for chromatin, these features were fluorescence, edges, and tubeness. Spindles were then defined as segmented microtubule structures that overlapped with segmented chromatin structures (Fig. 1 A). Once segmented, over 100 features of microtubule and chromatin structures were determined by CellProfiler modules, which resulted in an extensive dataset describing object dimensions, shape, and intensity. We then performed pairwise comparisons of individual spindle features among different experimental conditions. In addition, some datasets were subject to multivariate analysis to generate classification models and identify discriminant image features in an unbiased approach (Fig. 1 B).
Highly accurate spindle segmentation allows quantitative analysis of the Xenopus extract spindle. (A) Segmentation scheme for Xenopus extract spindles relies on fluorescence, texture, edges, and tubeness for microtubule structures and fluorescence, edges, and tubeness for chromatin. Bar, 10 µm. All of the raw images analyzed are available at ftp://xenbaseturbofrog.org/. (B) Multivariate data analysis workflow for the data generated by CellProfiler pipeline. Most of the steps listed were applied to the analysis corresponding to Figs. 5, 6, S2, and S3, and the code is available at https://github.com/MultivariateSpindleResource/Super-Spindle-DataSet.
Highly accurate spindle segmentation allows quantitative analysis of the Xenopus extract spindle. (A) Segmentation scheme for Xenopus extract spindles relies on fluorescence, texture, edges, and tubeness for microtubule structures and fluorescence, edges, and tubeness for chromatin. Bar, 10 µm. All of the raw images analyzed are available at ftp://xenbaseturbofrog.org/. (B) Multivariate data analysis workflow for the data generated by CellProfiler pipeline. Most of the steps listed were applied to the analysis corresponding to Figs. 5, 6, S2, and S3, and the code is available at https://github.com/MultivariateSpindleResource/Super-Spindle-DataSet.
We first applied our pipeline to a dataset of 18,261 spindles formed around duplicated sperm nuclei in X. laevis egg extract from 50 independent spindle reactions. Xenopus extract spindle morphology can be highly variable, and spindle lengths in these reactions ranged from 9 to 106 µm (Fig. 2, A and B). Despite the broad distribution, median spindle lengths for individual experimenters were within 5% of the pooled sample median of 28.8 µm (Fig. 2C). Interestingly, spindle length variation showed a roughly cyclical pattern with respect to the time of year (Fig. S1). Collectively, these data suggest that despite the observed variation in Xenopus extract spindle morphology, metrics such as length are consistent.
Xenopus egg extract spindles vary in morphology but converge to a consistent median length. (A) Examples showing variability in spindle morphology within a single egg extract reaction. Bar, 10 µm. (B) Boxplots for spindle lengths across 50 independent control experiments. Horizontal lines correspond to median spindle lengths. Bottom and top of the boxes are first and third quartiles, respectively; whiskers show highest and lowest values within 1.5 times the interquartile range and outliers are plotted as single points. (C) Distributions of spindle lengths from 50 independent experiments are shown in Fig. 1 B, grouped by three different experimenters.
Xenopus egg extract spindles vary in morphology but converge to a consistent median length. (A) Examples showing variability in spindle morphology within a single egg extract reaction. Bar, 10 µm. (B) Boxplots for spindle lengths across 50 independent control experiments. Horizontal lines correspond to median spindle lengths. Bottom and top of the boxes are first and third quartiles, respectively; whiskers show highest and lowest values within 1.5 times the interquartile range and outliers are plotted as single points. (C) Distributions of spindle lengths from 50 independent experiments are shown in Fig. 1 B, grouped by three different experimenters.
Cytoplasmic and chromosome components both contribute to spindle architecture
To test the general utility of our segmentation approach in the Xenopus extract system, we analyzed different spindle types as defined by the source of DNA and cytoplasm (Fig. 3 A). These included cycled spindles, assembled in X. laevis or X. tropicalis egg cytoplasm around sperm nuclei that had undergone DNA replication; cytostatic factor–arrested (CSF) spindles, assembled in X. laevis egg cytoplasm around unreplicated sperm nuclei; chromatin bead spindles, assembled in X. laevis egg cytoplasm around chromatin-coated beads; and stage 8 spindles, assembled with cytoplasm and nuclei from stage 8 X. laevis embryos.
Cytoplasmic and chromatin composition affect spindle morphology. (A) Example images of the different spindle types. Table shows differences in source of DNA and cytoplasm used to form each spindle type as well as differences in DNA content among spindle types. Image inset indicates the number of structures analyzed for each spindle type. Xla, X. laevis; Xtr, X. tropicalis. Bar, 10 µm. (B) Violin plots showing distributions for spindle length, width, eccentricity, and microtubule density, measured as median rhodamine tubulin fluorescence intensity. ***, P < 10−13 for a two-sample Kolmogorov–Smirnov test with Bonferroni correction for multiple testing compared to X. laevis cycled spindles.
Cytoplasmic and chromatin composition affect spindle morphology. (A) Example images of the different spindle types. Table shows differences in source of DNA and cytoplasm used to form each spindle type as well as differences in DNA content among spindle types. Image inset indicates the number of structures analyzed for each spindle type. Xla, X. laevis; Xtr, X. tropicalis. Bar, 10 µm. (B) Violin plots showing distributions for spindle length, width, eccentricity, and microtubule density, measured as median rhodamine tubulin fluorescence intensity. ***, P < 10−13 for a two-sample Kolmogorov–Smirnov test with Bonferroni correction for multiple testing compared to X. laevis cycled spindles.
Our analysis detected previously reported differences in spindle length between X. laevis and X. tropicalis cycled spindles, as well as between X. laevis cycled and stage 8 spindles (Fig. 3 B). Interestingly, we also found that spindle width was inversely related to the number of chromosomes on the metaphase plate (Fig. 3, A and B). Stage 8 spindles were narrower than spindles formed in egg extract despite accommodating twice as many chromosomes, while X. tropicalis cycled spindles were wider than X. laevis cycled spindles despite containing fewer chromosomes. Bead spindles were revealed to be 10% longer and 27% wider than spindles formed in the same cytoplasm around sperm nuclei with similar DNA content, consistent with previous work showing that chromatin configuration influences spindle structure (Dinarina et al., 2009) and that DNA content does not dramatically affect spindle size (Brown et al., 2007; Wühr et al., 2008). In addition to size differences, we identified previously unreported differences in features describing spindle shape (Fig. 3 B). In addition to aspect ratio, which has been used previously to approximate spindle shape (Young et al., 2014; Crowder et al., 2015), our data contain several measures of shape that use either an elliptical approximation of the spindle or the outline of the spindle generated during segmentation. Interestingly, we found that chromatin bead spindles and X. tropicalis cycled spindles were both more circular (possess lower eccentricity values) than X. laevis cycled and CSF spindles (Fig. 3 B).
We also investigated the relationship between cytoplasm and chromatin source and spindle microtubule content. Over nearly the entire range of spindle sizes observed, spindles formed around duplicated chromosomes in egg cytoplasm possessed higher rhodamine tubulin fluorescence intensity, indicating a higher microtubule density than in the other spindle types examined (Figs. 3 B, 4 A, and S2). To investigate this difference, we compared X. laevis cycled and CSF spindles and reasoned that the decreased microtubule density observed in CSF spindles could be caused by the absence of juxtaposed sister kinetochores and associated kinetochore microtubules (Sawin and Mitchison, 1991; Desai et al., 1997). In support of this hypothesis, addition of inhibitory antibodies against the outer kinetochore protein Nuf2 to cycled spindle reactions, which prevents kinetochore microtubule formation (McCleland et al., 2003), resulted in a significant decrease in spindle microtubule density (Fig. 4 B). Assuming ∼20 microtubules per kinetochore fiber (McEwen et al., 2001) and 100,000 microtubules per spindle (Brugués and Needleman, 2014), kinetochore fibers themselves should constitute only ∼1% of the total microtubule mass of the X. laevis spindle. Therefore, the >20% decrease in microtubule density observed after Nuf2 inhibition suggests that kinetochore microtubules stimulate microtubule formation within the spindle.
Kinetochore fibers increase spindle microtubule density. (A) Relationship between spindle area and total tubulin fluorescence intensity for spindle types showing differences in microtubule density among spindle types. Large plot shows data averaged across 21 equally sized cross-sectional area bins, and small plots show raw data for X. laevis cycled and CSF spindles. (B) Example images and microtubule density violin plots for IgG control (n = 117) and Nuf2 antibody inhibition (n = 230) showing decreased microtubule density after interfering with kinetochore microtubule formation. ***, P < 10−13 for a two-sample Kolmogorov–Smirnov test.
Kinetochore fibers increase spindle microtubule density. (A) Relationship between spindle area and total tubulin fluorescence intensity for spindle types showing differences in microtubule density among spindle types. Large plot shows data averaged across 21 equally sized cross-sectional area bins, and small plots show raw data for X. laevis cycled and CSF spindles. (B) Example images and microtubule density violin plots for IgG control (n = 117) and Nuf2 antibody inhibition (n = 230) showing decreased microtubule density after interfering with kinetochore microtubule formation. ***, P < 10−13 for a two-sample Kolmogorov–Smirnov test.
Mixing of meiotic and mitotic cytoplasm reveals titratable spindle features
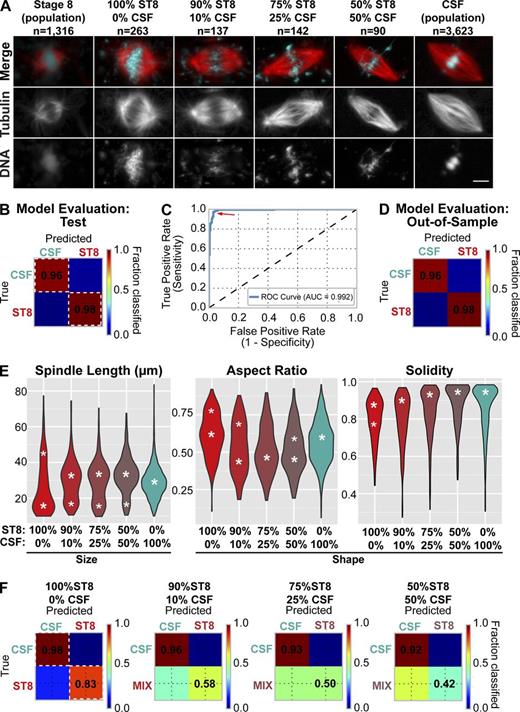
In previous work, we documented the abrupt change in spindle length and morphology that occurs across many animal clades between the female meiotic divisions and the first mitotic division of the zygote, which are separated by a single cell cycle (Crowder et al., 2015). To test the ability of our pipeline to capture the transition in spindle morphology between meiosis and mitosis and probe its biochemical basis, we utilized our existing dataset of spindles formed in cytoplasm from X. laevis meiosis II–arrested eggs (CSF extract) and mitotic embryos (stage 8 extract) and compared spindles formed in mixed reactions that contained different proportions of the two extracts (Fig. 5 A).
Meiotic cytoplasm has a dominant effect on spindle morphology. (A) Example images of X. laevis spindles formed in stage 8, CSF, and mixed extracts. Bar, 10 µm. (B) Cross-validation based evaluation of classification model (generalized linear model, LASSO-penalized logistic regression) on a CSF/stage 8 (ST8) spindle quantification test set (not used for training). Color scale ranges from blue (0% of objects classified to the group) to red (100% of objects classified). White dashed line indicates correctly classified objects with classification accuracy marked. (C) Receiver-operating characteristic (ROC) curve, an alternative assessment of the classification model performance for the model in B with area under the curve (AUC) marked. Optimal probability threshold for the logistic regression model (red arrow indicates maximum sensitivity and specificity) is 0.6. AUC is 0.99 (perfect model, with 100% specificity and 100% sensitivity would have AUC = 1.0). (D) Evaluation of classification model from Fig. 3 A on out-of-sample data. Accuracy for correctly classified spindles is marked (main diagonal, dark red). (E) Violin plots showing distributions of spindle length, aspect ratio, and solidity for structures assembled in stage 8, CSF, or mixed extracts (see also Fig. S3). White asterisks correspond to inferred maxima of distributions (Materials and methods). Major maxima are in bold. (F) The classification model (B–D) accurately distinguished CSF and stage 8 spindles but failed to accurately classify stage 8 spindles in mixed extracts.
Meiotic cytoplasm has a dominant effect on spindle morphology. (A) Example images of X. laevis spindles formed in stage 8, CSF, and mixed extracts. Bar, 10 µm. (B) Cross-validation based evaluation of classification model (generalized linear model, LASSO-penalized logistic regression) on a CSF/stage 8 (ST8) spindle quantification test set (not used for training). Color scale ranges from blue (0% of objects classified to the group) to red (100% of objects classified). White dashed line indicates correctly classified objects with classification accuracy marked. (C) Receiver-operating characteristic (ROC) curve, an alternative assessment of the classification model performance for the model in B with area under the curve (AUC) marked. Optimal probability threshold for the logistic regression model (red arrow indicates maximum sensitivity and specificity) is 0.6. AUC is 0.99 (perfect model, with 100% specificity and 100% sensitivity would have AUC = 1.0). (D) Evaluation of classification model from Fig. 3 A on out-of-sample data. Accuracy for correctly classified spindles is marked (main diagonal, dark red). (E) Violin plots showing distributions of spindle length, aspect ratio, and solidity for structures assembled in stage 8, CSF, or mixed extracts (see also Fig. S3). White asterisks correspond to inferred maxima of distributions (Materials and methods). Major maxima are in bold. (F) The classification model (B–D) accurately distinguished CSF and stage 8 spindles but failed to accurately classify stage 8 spindles in mixed extracts.
First, to determine whether pipeline measurements were sufficient to differentiate between CSF and stage 8 spindles, we used our measurement output to train a generalized linear classification model that used a total of 166 features describing the size, shape, and fluorescence intensity of microtubule and chromatin components of the spindle (Fig. 5, B and C). Importantly, the model accurately classified 96% of CSF and 98% of stage 8 spindles not contained in the training set (Fig. 5 D). We took advantage of this model to identify the primary chromatin and microtubule features that best distinguished the two spindle types (Fig. S3) and to determine whether intermediate morphologies were observed when the two extracts were mixed. We found that some features, such as solidity, which measures spindle edge regularity, exhibited a gradual transition between stage 8-like and CSF-like states (Figs. 5 E and S4). In contrast, spindle length, width, and radius distributions were bimodal, with two peaks corresponding to the stage 8 and CSF peaks discernable even when only 10% (vol/vol) CSF extract was added to embryonic stage 8 cytoplasm (Figs. 5 E and S3, compare red and turquoise distributions). Furthermore, at equal volumes of CSF cytoplasm and stage 8 cytoplasm, the stage 8 spindle length peak was nearly undetectable. The classification model also revealed the potent effect of CSF extract on spindle morphology. Spindles formed in mixed extracts were frequently misclassified, even when only 10% (vol/vol) CSF extract was added, indicating loss of stage 8 spindle characteristics (Fig. 5 F). These experiments reveal that spindle features can possess titratable or switch-like properties and demonstrate that meiotic cytoplasm has a dominant effect on parameters such as spindle length.
TPX2 modulates multiple aspects of spindle architecture
After finding that our pipeline could reliably detect differences among spindle types, we sought to assess its efficacy in characterizing the effects of a single protein on spindle morphology. As a test case, we used an image dataset from our examination of the spindle assembly factor TPX2, which showed that differences in TPX2 concentration between X. tropicalis and X. laevis contribute to differences in spindle size and architecture between the two species (Helmke and Heald, 2014).
We found that addition of 200 nM TPX2 to X. laevis egg extract, which brings its concentration up to that found in X. tropicalis egg extract, also led to previously uncharacterized changes in spindle morphology (Fig. 6, A and B). To determine whether TPX2-supplemented spindles constituted a morphologically distinct class, we trained a classification model to categorize the output data. Because of the highly correlated nature of the dataset (Fig. S4 A), we applied dimensionality reduction strategies to identify the most discriminant features (Fig. S4, B and C). Ultimately, the model was able to accurately assign ∼90% of both control and TPX2-supplemented spindles to their respective classes (Fig. S4 B). Among the most discriminant features of the TPX2 addition phenotype, we found descriptors of spindle size, shape, and spindle microtubule fluorescence intensity (Fig. 6 B).
TPX2 influences spindle size, shape, and microtubule density. (A) Example images of spindles assembled in the absence (n = 1,935) or presence of 200 nM X. laevis TPX2 (n = 2,621), 50 µM Monastrol (n = 278), or 200 nM TPX2Δ7 (n = 1,417; Helmke and Heald, 2014). Bar, 10 µm. (B) Violin plots showing the distribution of spindle length, width, aspect ratio, eccentricity, and microtubule density for spindles formed in control (n = 1,935) or 200 nM TPX2-supplemented extracts (n = 2,621). ***, P < 10−15 for a two-sample Kolmogorov–Smirnov test with Bonferroni correction. (C) Violin plots showing the distributions of spindle length, width, aspect ratio, and microtubule density for spindles formed in control (n = 221), 200 nM TPX2–supplemented (n = 378), and 50 µM Monastrol (Mon)-treated (n = 278) extracts. ***, P < 10−15 for a two-sample Kolmogorov–Smirnov test. (D) Violin plots showing spindle width distributions for structures formed in control (n = 1,006), 200 nM wild-type TPX2–supplemented (n = 914), and 200 nM TPX2Δ7 (Δ7)-supplemented (n = 1,417) extracts. ***, P < × 10−15 for a two-sample Kolmogorov–Smirnov test.
TPX2 influences spindle size, shape, and microtubule density. (A) Example images of spindles assembled in the absence (n = 1,935) or presence of 200 nM X. laevis TPX2 (n = 2,621), 50 µM Monastrol (n = 278), or 200 nM TPX2Δ7 (n = 1,417; Helmke and Heald, 2014). Bar, 10 µm. (B) Violin plots showing the distribution of spindle length, width, aspect ratio, eccentricity, and microtubule density for spindles formed in control (n = 1,935) or 200 nM TPX2-supplemented extracts (n = 2,621). ***, P < 10−15 for a two-sample Kolmogorov–Smirnov test with Bonferroni correction. (C) Violin plots showing the distributions of spindle length, width, aspect ratio, and microtubule density for spindles formed in control (n = 221), 200 nM TPX2–supplemented (n = 378), and 50 µM Monastrol (Mon)-treated (n = 278) extracts. ***, P < 10−15 for a two-sample Kolmogorov–Smirnov test. (D) Violin plots showing spindle width distributions for structures formed in control (n = 1,006), 200 nM wild-type TPX2–supplemented (n = 914), and 200 nM TPX2Δ7 (Δ7)-supplemented (n = 1,417) extracts. ***, P < × 10−15 for a two-sample Kolmogorov–Smirnov test.
Our analysis indicates that in addition to decreasing spindle length, TPX2 addition results in spindles that are wider, more circular, and more microtubule dense than control spindles (Fig. 6, A and B). TPX2 is involved in multiple processes in the spindle, including microtubule nucleation (Gruss et al., 2001) and inhibition of the microtubule motor Eg5 (Ma et al., 2011; Gable et al., 2012; Balchand et al., 2015). Our previous work showed that TPX2-dependent Eg5 inhibition was important for controlling spindle length (Helmke and Heald, 2014). To examine whether the other aspects of the TPX2 phenotype were mediated through Eg5, we added low doses of the Eg5 inhibitor Monastrol (Mayer et al., 1999; Kapoor et al., 2000) to spindle assembly reactions. Monastrol caused a phenotype more pronounced than TPX2 addition for spindle length, shape, and microtubule density, but it did not cause a change in spindle width (Fig. 6, A, C, and D). Instead, we found that addition of the TPX2Δ7 mutant, which has enhanced nucleation capacity (Helmke and Heald, 2014), resulted in a more pronounced spindle width phenotype than addition of wild-type TPX2 (Fig. 6, A and D). These analyses suggest that TPX2 regulates spindle morphology primarily by modulating Eg5-dependent antiparallel microtubule sliding and spindle pole separation, while regulation of spindle width by TPX2 is likely mediated by its stimulation of microtubule nucleation.
Adaptation of the image analysis pipeline to nuclei detects REEP3/4 RNAi phenotypes
Because the use of multiple image features as the basis for segmentation was applicable across a wide range of experimental conditions in the Xenopus extract system, we asked whether this approach could be adapted to other systems and subcellular structures. As a test case, we analyzed Lamin B1–stained nuclei in HeLa cells following RNAi depletion of REEP3 and REEP4, two ER proteins important for membrane clearance from chromatin during cell division (Schlaitz et al., 2013). Similar to the application of our pipeline in Xenopus, using a composite of multiple image processing steps for segmentation allowed us to differentiate between background and the objects of interest (nuclei).
With an effective segmentation strategy, we evaluated the effects of REEP3/4 depletion on nuclear morphology (Fig. 7 A). The previously reported invaginations of the nuclear envelope caused by membrane trapping were detected as an increase in texture (Fig. 7 B) and change in solidity (Fig. 7 C), because the surface of the nuclear envelope became irregular upon REEP3/4 depletion. No significant effect on nuclear size was observed (Fig. 7 D), but REEP3/4-depleted nuclei were more oblong than controls as measured by an increase in eccentricity (Fig. 7 E), an effect that was not previously noted. Similar results were obtained using an independent approach (Fig. S5). Thus, the segmentation approach and image analysis pipeline described here can be easily modified to provide in-depth quantitative analysis of nuclear phenotypes.
Image analysis pipeline can be adapted to other subcellular structures. (A) Example images of control and REEP3/4-depleted nuclei (Schlaitz et al., 2013). Bar, 10 µm. (B) Violin plot of Lamin B1 texture for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (C) Violin plot of nuclear solidity for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (D) Violin plot of nuclear area for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (E) Violin plot of nuclear eccentricity for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763).
Image analysis pipeline can be adapted to other subcellular structures. (A) Example images of control and REEP3/4-depleted nuclei (Schlaitz et al., 2013). Bar, 10 µm. (B) Violin plot of Lamin B1 texture for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (C) Violin plot of nuclear solidity for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (D) Violin plot of nuclear area for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763). (E) Violin plot of nuclear eccentricity for control cells (n = 2,123) and REEP3/4-depleted cells (n = 1,763).
Discussion
Our CellProfiler-based image analysis approach provides a tool for cell biologists interested in evaluating or repurposing large image datasets. Here, we have focused primarily on Xenopus extract spindle images. Although it is possible to generate thousands of spindles in a single experiment, variation in this system has previously limited the quantitative description of spindle morphology to several easily measured features, such as length. In addition, highly heterogeneous background signals complicated automated analysis. To circumvent these difficulties, we developed a strategy that relies on multiple object features as the basis for segmentation. This allowed us to analyze a large dataset of Xenopus spindles and extract morphological parameters. Minor modifications of the pipeline enable analysis of cultured cell nuclei as well as other structures such as microtubule asters. The complete spindle image dataset is available on Xenbase (ftp://xenbaseturbofrog.org/), and step-by-step instructions for implementing the pipeline or adapting the segmentation strategy are provided in the supplemental text.
Our in-depth analysis of thousands of images provides several new insights into the regulation of spindle morphology and illustrates the utility of our approach to repurpose preexisting data and guide future experiments. Analyzing spindles that differed with respect to source of cytoplasm and DNA, we found evidence that kinetochore microtubules contribute a disproportionate amount of microtubule mass to the spindle. We hypothesize that because of their relatively long half-life (Kabeche and Compton, 2012), kinetochore microtubules act as a platform for microtubule branching nucleation (Petry et al., 2013). In addition, we found through cytoplasm-mixing experiments that some spindle features, such as length and width, do not smoothly titrate when meiotic and mitotic extracts are mixed. Instead, their distributions appear to be bimodal and dominated by even a low percentage of meiotic extract. Investigating the biochemical basis of this phenomenon could help elucidate mechanisms underlying the abrupt change from meiotic to mitotic spindle morphology characteristic of many animal species (Crowder et al., 2015). Finally, we applied our pipeline to further define the morphological features that depend on the spindle protein TPX2, showing the complex morphological effects of increasing TPX2 levels during spindle assembly. Detailed analysis of other key spindle microtubule regulators such as XMAP215, whose levels also affect spindle length and mass (Reber et al., 2013), will provide more comprehensive insight into the roles of individual factors that define spindle morphology.
In addition to assessing spindle morphology, this work also highlights the diverse data analysis approaches that can be applied to the pipeline outputs, from traditional pairwise analysis of individual spindle parameters to more complex multidimensional analysis of correlated features to multidimensional analysis using more advanced machine learning techniques. These new tools streamline the process of analyzing spindle morphology, giving holistic, quantitative information, enabling data repurposing, and facilitating the generation of new hypotheses.
Materials and methods
Xenopus extract preparation and use
Xenopus egg extract was prepared and used as previously described (Maresca and Heald, 2006). TPX2 purification and experiments were described previously (Helmke and Heald, 2014); the TPX2 experiments presented here were all carried out with 200 nM of X. laevis TPX2. Xenopus embryo extract was prepared as previously described (Wilbur and Heald, 2013), and extract mixing was performed before the onset of spindle assembly. Nuf2 inhibitory antibody (McCleland et al., 2003) was added before the onset of spindle assembly and was a generous gift from Todd Stukenberg (University of Virginia). Images were taken on an Olympus BX51 epifluorescence microscope controlled by Micromanager software under 40× magnification (0.75 NA, UPlanFl N; Olympus) with TRITC (for rhodamine-labeled tubulin) and DAPI (for DNA) filters (Chroma Technology Corp.) with an Orca-ER cooled CCD camera (Hamamatsu Photonics) at 25°C. Exposure times were 50 ms for TRITC and 20 ms for DAPI, and focus was manually assessed. Vectashield was used as mounting medium.
Growth, transfection, immunostaining, and microscopy of mammalian tissue culture cells
HeLa cells (sourced from ATCC) were cultured in DMEM (containing 4.5 g/l glucose and GlutaMax; Invitrogen) supplemented with 10% fetal bovine serum at 37° C in a humidified 5% CO2 incubator.
siRNA transfections were performed using Lipofectamine RNAiMax (Invitrogen) and an siRNA concentration of 10 nM. For REEP3/4 depletion, Ambion siRNA s37271 was used (Schlaitz et al., 2013). Control cells were transfected with Silencer Negative Control No. 1 siRNA (AM4611).
For immunofluorescence, cells grown on coverslips were fixed 48 h after transfection with siRNAs using 4% formaldehyde in PBS, quenched with 15 mM glycine in PBS, permeabilized with 0.1% Triton X-100 in PBS, and stained with anti-LaminB1 antibody ab16048 (Abcam) at 1:500. Imaging was performed on a microscope (LSM 710; ZEISS).
Image analysis pipeline
Most image analysis was performed on a custom built computer with an Intel 4770K processor (at 3.69 GHz) and 32-Gb 1,600-MHz DDR3 RAM. Human nuclei (149 8-bit images) were also analyzed on a MacBook Air, 1.3-GHz IntelCore i5 (4CPUs), 8 GB RAM. The analysis took 2.5 h. Illumination correction was carried out during image preprocessing using the Background method with spline smoothing of the resulting illumination function. Next, objects were identified using the multiple image feature segmentation scheme outlined in Fig. 1 A. Because the microtubule structures identified using this strategy were significantly larger than the true microtubule structures, we used them as masks for a second round of segmentation based on fluorescence alone. For most of the analyses performed, equal weighting was given to each image feature used for segmentation. However, weighting is end-user tunable in the ImageMath modules and minor weighting modification allows segmentation of spindle images from a wide range of imaging setups. Object size ranges should also be modified in the segmentation modules to accommodate differences in microscope calibration. All the images in this work were taken on an imaging setup with a calibration of 6.18 pixels/µm. Segmentation efficiency was determined manually by evaluating 171 spindles that were randomly chosen and analyzed by the pipeline. Of these, 96.5% were correctly classified by the pipeline, 0% were oversegmented (one object split into multiple), 1.2% were undersegmented (two or more objects joined), 1.8% were false positive (objects mistakenly identified), and 0.5% were false negative (objects not detected).
Raw images are available through Xenbase, the Xenopus genome database (Karpinka et al., 2015), at ftp://xenbaseturbofrog.org/ (choose guest login when prompted), and additional instructions for using and adapting the pipeline are available in the supplemental text.
Data analysis
The CellProfiler pipelines, output data, and code generated in the course of our analysis are available at https://github.com/MultivariateSpindleResource. Quantification datasets for the image processing and analysis pipeline of the spindle contain 203 variables, 12 of which contain metadata (experimental details), and 24 correspond to x and y coordinates of segmented objects within the image matrix. The remaining 167 were used in spindle image analysis. For each of the datasets (corresponding to individual figures), we considered, tested, and performed various steps depicted in Fig. 1 B. Quantification datasets for human nuclei differed slightly depending on the approach used.
For spindle-related datasets, we first analyzed parameters describing the distribution of every feature (e.g., variance, skewness, and kurtosis). Variables were then subjected to Box–Cox transformation (Box and Cox, 1964), scaled, and centered. For classification, we typically tested at least one type of linear model (logistic regression–based generalized linear model) and one type of non-linear model (support vector machines with radial kernel) and used complete, principle component, projected (for dimensionality reduction) and significant features-only versions of the datasets. In the case of principal component-based dimensionality reduction, we projected a 166-feature input dataset on 64 principal components, so that the transformed dataset retained 95% of the original variance (Fig. S4 C). For feature selection, we tested and applied univariate statistics-based filter methods (ANOVA, Kolmogorov–Smirnov, both with Bonferroni correction and Kruskal–Wallis test; Kuhn and Johnson, 2013). If the classification model trained on prescreened features showed good performance (by accuracy, sensitivity, and specificity measurements), we analyzed the most discriminant features more closely. Distributions of individual features were plotted as violin plots and statistical significance between conditions evaluated with Kolmogorov–Smirnov test (with Bonferroni correction for multiple testing). To detect peak locations, we first computed Gaussian kernel density estimate based on selected feature distributions for smoothing, followed by determination of the first derivative of the obtained feature density estimate and determination of the first derivative of the sign of the obtained values (+1 for positive value, 0 for 0, −1 for negative value). Derivative estimates corresponding to −2 or 2 were used as local maxima and minima locations, respectively. Plots were generated using ggplot2 in R (Wickham, 2009), matplotlib (Python), and Microsoft Excel.
Online supplemental material
Fig. S1 shows the cyclical pattern of spindle length with respect to time of year. Fig. S2 shows the data from Fig. 4 A separated by spindle types with best-fit power functions to assess scaling relationships and standard deviation bars to show data spread. Fig. S3 shows individual features of spindles and chromatin formed in mixed meiotic (CSF) and stage 8 extract. Fig. S4 contains information regarding the design and implementation of a model to classify TPX2 supplemented spindles. Fig. S5 contains data generated by a pipeline that uses a different segmentation scheme than that presented for spindles and nuclei in the main text to assess nuclear morphology after REEP3/4 knockdown. Table S1 contains the location of peaks marked in Fig. 3 B. Supplemental methods explains in detail how to use and modify the segmentation scheme of our image analysis pipeline, and the changes we made to our pipeline to segment Lamin B1–stained HeLa cell nuclei are also included.
Acknowledgments
We thank present and past members of the Heald lab for their spindle images. We would also like to thank C. Brownlee, G. Barnes, K. Collins, D. Fletcher, and all other members of the Heald lab for helpful discussions; A. Ettinger for help with FIJI macro language; and A. Lane, R. Gibeaux, and A. Ettinger for comments on the manuscript.
This work was supported by National Institutes of Health grants GM098766 (R. Heald) and GM057839 (R. Heald), the National Science Foundation Graduate Research Fellowship Program (A.W. Grenfell), and National Institutes of Health training grant 2T32GM007232-36 (A.W. Grenfell).
The authors declare no competing financial interests.
Author contributions: A.W. Grenfell, K.J. Helmke, M.E. Crowder, A.-L. Schlaitz, and M. Strzelecka carried out experiments. A.W. Grenfell and M. Strzelecka constructed the pipelines, analyzed images, and performed data analysis. A.W. Grenfell, M. Strzelecka, and R. Heald wrote the paper. A.W. Grenfell and M. Strzelecka prepared the figures.
References
Author notes
A.W. Grenfell and M. Strzelecka contributed equally to this paper.
M. Strzelecka’s present address is QIAGEN, 40724 Hilden, Germany.
M.E. Crowder’s present address is Dept. of Molecular and Cell Biology, University of California, Davis, Davis, CA 95616.
K.J. Helmke’s present address is Bioengineering Department, Stanford University, Stanford, CA 94305.
A.-L. Schlaitz’s present address is Zentrum für Molekulare Biologie der Unversitlät Heidelberg, 69120 Heidelberg, Germany.