Despite recent advances in mass spectrometry, proteomic characterization of transport vesicles remains challenging. Here, we describe a multivariate proteomics approach to analyzing clathrin-coated vesicles (CCVs) from HeLa cells. siRNA knockdown of coat components and different fractionation protocols were used to obtain modified coated vesicle-enriched fractions, which were compared by stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative mass spectrometry. 10 datasets were combined through principal component analysis into a “profiling” cluster analysis. Overall, 136 CCV-associated proteins were predicted, including 36 new proteins. The method identified >93% of established CCV coat proteins and assigned >91% correctly to intracellular or endocytic CCVs. Furthermore, the profiling analysis extends to less well characterized types of coated vesicles, and we identify and characterize the first AP-4 accessory protein, which we have named tepsin. Finally, our data explain how sequestration of TACC3 in cytosolic clathrin cages causes the severe mitotic defects observed in auxilin-depleted cells. The profiling approach can be adapted to address related cell and systems biological questions.
Introduction
Vesicle trafficking is a fundamentally important process required for the exchange of proteins and lipids between organelles. Clathrin-coated vesicles (CCVs) are among the most abundant and versatile transport intermediates, which function in trafficking between the trans-Golgi network and endosomes, as well as in endocytosis (Robinson, 2004). Knowledge of the complete protein complement of different types of coated vesicles would significantly enhance our understanding of membrane traffic. In recent years, proteomics has emerged as a powerful tool to determine the composition of subcellular fractions, but the analysis of transport vesicles has remained challenging. Because of the transient nature and low abundance of vesicles, it is difficult to prepare highly enriched fractions with sufficient yields, so only the most prominent vesicle types have yielded to proteomic analysis (Bergeron et al., 2010). Many other vesicle coats, such as retromer (McGough and Cullen, 2011), AP-3, and AP-4 (Robinson, 2004), still await detailed characterization.
A general problem of fractionation-based proteomics is the inevitable detection of contaminants. Modern mass spectrometry is exquisitely sensitive and allows the identification of thousands of proteins from complex mixtures. However, because no subcellular fraction is ever completely pure, one cannot objectively distinguish between proteins genuinely associated with the organelle of interest and copurifying contaminants. Uncharacterized proteins are particularly problematic in this regard, and this was a limitation of early proteomic investigations of CCVs (Blondeau et al., 2004; Girard et al., 2005; for review see McPherson, 2010). Five years ago, we developed a comparative approach to address the issue of contaminants (Borner et al., 2006). Using quantitative mass spectrometry, we compared CCV fractions from tissue culture cells with “mock” CCV fractions obtained from clathrin-depleted cells. This approach allowed us to identify genuine CCV proteins because these proteins were depleted from mock CCVs. Nevertheless, owing to the limited dynamic range of the quantification technique (iTRAQ), the separation of CCV proteins from contaminants was suboptimal, and the list of predicted CCV proteins not comprehensive. We could also not exclude the possibility that some of the proteins depleted from mock CCVs were non-CCV proteins whose fractionation properties were altered by the clathrin knockdown. Finally, our method did not discriminate between endocytic and intracellular CCVs.
Here, we describe a multivariate comparative proteomics approach that overcomes the shortcomings of previous proteomic investigations of CCVs, and also allows us to begin to characterize the functions of the identified proteins. The method is highly flexible, and can be adapted to investigate the composition of low-abundance vesicle coats and protein complexes. Although the focus of this study is the dissection of clathrin-dependent pathways, our data also shed light on the role of clathrin in mitosis, and include the first proteomic analysis of the retromer and AP-4 coats.
Results
The profiling concept
Like all subcellular fractions, our CCV-enriched fraction from HeLa cells is not pure. It is contaminated with abundant protein complexes such as ribosomes and proteasomes, as well other types of coated and noncoated vesicles. As we have previously shown, comparative proteomics of modified CCV fractions can be used to distinguish CCV proteins from copurifying contaminants (Borner et al., 2006). Building on this approach, we performed multiple binary comparisons of CCV fractions prepared under different experimental conditions. Each comparison helps to identify CCV proteins, and also reveals differences between proteins associated with endocytic and intracellular CCVs. We then combined the individual datasets into a systematic “profiling analysis,” which classifies CCV proteins with high specificity, and in addition begins to group both CCV and non-CCV proteins according to function.
Proteomic analysis of modified CCV fractions by SILAC
We used three different conditions to generate modified CCV fractions, each of which was compared with a control CCV fraction. The first condition was to deplete clathrin heavy chain (CHC) by siRNA-mediated knockdown, thereby abolishing the formation of CCVs. When compared with control CCVs, genuine CCV proteins are depleted from this mock CCV fraction, contaminants are unaffected, and intracellular CCV proteins are more strongly depleted than endocytic ones (Fig. 1 A; Borner et al., 2006). The second condition was to deplete auxilin, a protein involved in CCV uncoating (Eisenberg and Greene, 2007). This causes the cell to form membraneless cytosolic clathrin cages (Hirst et al., 2008), which are packed with clathrin-binding proteins and depleted of cargo, and which are selectively enriched in proteins involved in clathrin-mediated endocytosis relative to proteins involved in intracellular transport (Fig. 1 B). Finally, we have modified our original protocol to obtain a CCV fraction with higher purity (Fig. S1), so the third condition was to use the older protocol. All CCV proteins are enriched around twofold in the improved relative to the original CCV fraction (Fig. 1 C).
Three unbiased criteria to identify CCV proteins. (A–C) Western blots of CCV fractions. (D–F) Corresponding proteomic analyses by SILAC and quantitative mass spectrometry. The ratio of depletion or enrichment (fold change) was calculated for each protein, and proteins were ranked from the highest to the lowest ratio. (A and D) Control CCV fractions compared with mock CCV fractions prepared from clathrin-depleted cells. Genuine CCV proteins (AP-1, AP-2, and CIMPR) are depleted from the mock fraction; contaminants (EF-2) are unchanged. The apparent depletion is more pronounced for AP-1 than for AP-2. (B and E) CCV/cage fractions from auxilin-depleted cells, which accumulate membraneless clathrin cages, compared with control CCV fractions. Endocytic proteins (AP-2 and SNX9) are enriched in the cages, whereas transmembrane cargo proteins (CIMPR) are depleted. (C and F) Improved CCV preparation compared with original preparation. CCV proteins are enriched approximately twofold in the improved CCV fraction (see also Fig. S1). To exclude the possibility that any of the changes were caused by altered transcriptional regulation, we performed a comprehensive microarray gene expression analysis (Fig. S2). Positions of protein molecular mass markers are indicated in kilodaltons in A–C. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
Three unbiased criteria to identify CCV proteins. (A–C) Western blots of CCV fractions. (D–F) Corresponding proteomic analyses by SILAC and quantitative mass spectrometry. The ratio of depletion or enrichment (fold change) was calculated for each protein, and proteins were ranked from the highest to the lowest ratio. (A and D) Control CCV fractions compared with mock CCV fractions prepared from clathrin-depleted cells. Genuine CCV proteins (AP-1, AP-2, and CIMPR) are depleted from the mock fraction; contaminants (EF-2) are unchanged. The apparent depletion is more pronounced for AP-1 than for AP-2. (B and E) CCV/cage fractions from auxilin-depleted cells, which accumulate membraneless clathrin cages, compared with control CCV fractions. Endocytic proteins (AP-2 and SNX9) are enriched in the cages, whereas transmembrane cargo proteins (CIMPR) are depleted. (C and F) Improved CCV preparation compared with original preparation. CCV proteins are enriched approximately twofold in the improved CCV fraction (see also Fig. S1). To exclude the possibility that any of the changes were caused by altered transcriptional regulation, we performed a comprehensive microarray gene expression analysis (Fig. S2). Positions of protein molecular mass markers are indicated in kilodaltons in A–C. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
For binary comparisons of CCV fractions, we used stable isotope labeling of amino acids in cell culture (SILAC; Ong et al., 2002) in conjunction with quantitative mass spectrometry. For each identified protein, a ratio of enrichment or depletion in the control relative to a modified CCV fraction was obtained. In a typical experiment, we quantified 1,000–2,000 proteins. We performed triplicate biological repeats of each of our three conditions compared with control, as well as a further comparison based on fractionation properties (Fig. S1). In total, 10 datasets covering 2,527 proteins were acquired (Table S1).
For each dataset, proteins were sorted from highest to lowest ratio and plotted. CCV proteins were depleted up to 25-fold from mock CCVs (Fig. 1 D), and enriched up to 35-fold in clathrin cages (Fig. 1 E), whereas most contaminants had ratios near one. The fractionation analysis (Fig. 1 F) shows clustering of CCV proteins in a narrow bracket of the plot. Hence, under all three conditions, known CCV proteins behaved distinctly from contaminants.
As a first step toward combining the three conditions, we plotted the fold changes of representative CCV proteins and contaminants for all 10 datasets (Fig. 2). For each protein, a characteristic profile of enrichment and depletion was obtained. Intracellular (AP-1) and endocytic (AP-2) CCV proteins had different profiles, and both were distinct from proteasomal proteins. Although there was some variability between repeat experiments, different subunits of the same protein complex always showed closely correlated behavior. This suggested that functionally linked proteins might be identified by their profiles.
CCV proteins and contaminants have distinct profiles. The fold changes of selected CCV proteins (AP-1 and AP-2 subunits) and copurifying proteasomal contaminants (PSMA1 and PSMB1) are shown for 10 SILAC datasets. Proteins that are subunits of the same protein complex have closely correlated profiles. Con/Cla kd 1–3: Triplicate repeats of control versus clathrin-depleted. Aux kd/Con 1–3: Triplicate repeats of auxilin-depleted versus control. Fractionation 1–3: Triplicate repeats of improved versus original CCV preparation. Fractionation 4*: A further comparison based on fractionation properties of CCVs (see Fig. S1, D and E).
CCV proteins and contaminants have distinct profiles. The fold changes of selected CCV proteins (AP-1 and AP-2 subunits) and copurifying proteasomal contaminants (PSMA1 and PSMB1) are shown for 10 SILAC datasets. Proteins that are subunits of the same protein complex have closely correlated profiles. Con/Cla kd 1–3: Triplicate repeats of control versus clathrin-depleted. Aux kd/Con 1–3: Triplicate repeats of auxilin-depleted versus control. Fractionation 1–3: Triplicate repeats of improved versus original CCV preparation. Fractionation 4*: A further comparison based on fractionation properties of CCVs (see Fig. S1, D and E).
Merging the SILAC datasets: Profiling through principal component analysis (PCA)
We first analyzed the 688 proteins that were represented in all 10 SILAC experiments. Each protein’s profile consisted of 10 ratios of enrichment or depletion. To identify groups of proteins with similar profiles from this unwieldy 10-dimensional dataset, we used PCA, a standard tool for the transformation of multivariate data (see “PCA” in the Materials and methods section). By removing redundant linear correlations, PCA can decrease the dimensionality of data to a few derived super-axes termed “principal components.” In turn, this allows the simple visual representation of the information in two- or three-dimensional space. PCA is routinely used in systems biology to harness large datasets and perform cluster analysis (Janes and Yaffe, 2006), but has so far seen limited use in cell biological and proteomics applications (Dunkley et al., 2004; Hall et al., 2009). We performed PCA of our SILAC data with a commercially available program (SIMCA-P+, Umterics; see Table S4 for a step-by-step guide). Through the projection of the 10-dimensional SILAC data on the first two principal components, the dispersion of the data can be represented as a single scatter in only two dimensions (Figs. 3 and S3). Each protein is symbolized by a scatter point. Proximity of proteins indicates similar behavior under the experimental conditions; therefore, proteins with similar profiles are expected to cluster.
PCA reveals clustering of CCV and non-CCV proteins into functional groups. The SILAC data were decorrelated by PCA. The figure shows the projections of the data on the first (x axis) and second (y axis) principal components (scores plot). Each scatter point represents a protein. Distance from the origin of the plot indicates how strongly a protein is affected under the experimental conditions. Close proximity of proteins indicates similar profiles. Only the 688 proteins common to all 10 SILAC sets were analyzed. Subunits of a known protein complex are shown in the same color. Clathrin heavy and light chains, AP-1, and AP-2 form distinct clusters that and are clearly separated from the bulk of the other proteins. Known CCV coat proteins are annotated as AF (accessory factors). Mannose 6-phopshate receptors (M6PR) and lysosomal enzymes (Lys) are established cargo molecules of intracellular CCVs. A fully annotated version of this plot is shown in Fig. S3. Retro, retromer; RPS and RPL, small and large ribosomal subunits; COPI, COPI coat; CCT, CCT chaperone; Sig, signalosome; BLOC, BLOC-1; EIF3, translation initiation factor 3; PHK, phosphorylase kinase. P19, P20, PA: 19S, 20S, and 11S subcomplexes of the proteasome.
PCA reveals clustering of CCV and non-CCV proteins into functional groups. The SILAC data were decorrelated by PCA. The figure shows the projections of the data on the first (x axis) and second (y axis) principal components (scores plot). Each scatter point represents a protein. Distance from the origin of the plot indicates how strongly a protein is affected under the experimental conditions. Close proximity of proteins indicates similar profiles. Only the 688 proteins common to all 10 SILAC sets were analyzed. Subunits of a known protein complex are shown in the same color. Clathrin heavy and light chains, AP-1, and AP-2 form distinct clusters that and are clearly separated from the bulk of the other proteins. Known CCV coat proteins are annotated as AF (accessory factors). Mannose 6-phopshate receptors (M6PR) and lysosomal enzymes (Lys) are established cargo molecules of intracellular CCVs. A fully annotated version of this plot is shown in Fig. S3. Retro, retromer; RPS and RPL, small and large ribosomal subunits; COPI, COPI coat; CCT, CCT chaperone; Sig, signalosome; BLOC, BLOC-1; EIF3, translation initiation factor 3; PHK, phosphorylase kinase. P19, P20, PA: 19S, 20S, and 11S subcomplexes of the proteasome.
The analysis achieved a striking separation of proteins into functional groups. Subunits of known protein complexes formed tight clusters, including adaptor protein (AP) complexes 1–4, COPI, BLOC-1, retromer, ribosomes, and proteasomes. The resolution of complexes was excellent; for example, the ribosomal cluster was resolved into large and small subunits (RPL and RPS), and the proteasomal cluster into 20S core and 19S regulatory subunits (P20 and P19). Importantly, clathrin and the clathrin adaptors AP-1 and AP-2 were well separated from the bulk of the proteins, and clustered in the periphery of the plot. Known CCV cargo proteins, such as mannose 6-phosphate receptors and lysosomal hydrolases, surrounded the AP-1 cluster. Moreover, established AP-1 and AP-2 binding proteins clustered closely with their respective AP complex (accessory factors). Based on these observations, uncharacterized proteins in the vicinity of AP-1 or AP-2 are candidate new CCV proteins.
Automated “reference” clustering allows extensive profiling of proteins not represented in all SILAC datasets
Fig. 3 demonstrates the power of the profiling approach; however, the analysis is restricted to the 688 proteins identified in all 10 SILAC experiments, as PCA requires all proteins to be represented in every dataset. To analyze our data more comprehensively, we developed a novel method for processing SILAC ratios by PCA, which allows the inclusion of proteins with disparate numbers of data points. First, we selected proteins that were identified at least once under each of the three experimental conditions, and in at least four of the 10 experiments. 1,523 proteins matched these specifications (Table S1), more than doubling the number of analyzable proteins. Custom software written in MATLAB 7.0 (MathWorks) was then used to profile individual proteins in the context of a reference set. Reference proteins were represented in all 10 datasets, and belonged to six protein complexes: AP-1, AP-2, AP-3, AP-4, retromer, and ribosomes. For each of the 1,523 candidate proteins, the algorithm first identified the SILAC sets in which the protein was represented. Next, it performed PCA on the reference proteins using only those SILAC sets. The candidate protein was then projected onto its reference plot, and assigned to the nearest cluster. Proteins assigned to either AP-1 or AP-2 were considered candidate CCV proteins (Fig. 4).
Automated PCA of sample CCV proteins. Automated PCA was performed in the context of a reference set (85 proteins belonging to six protein complexes: AP-1, AP-2, AP-3, AP-4, retromer, and ribosomes; Table S1). For each candidate protein, PCA of the reference set was performed using only those datasets in which the candidate protein was represented. The candidate protein was then projected onto the corresponding PCA plot, and the nearest reference cluster was determined. The figure shows the projections on the first two principal components for six sample proteins. The relative positions of reference clusters change with different combinations of available data, but in each case, all reference proteins are correctly grouped into the expected complexes. Candidate proteins are shown in red and reference proteins in black.
Automated PCA of sample CCV proteins. Automated PCA was performed in the context of a reference set (85 proteins belonging to six protein complexes: AP-1, AP-2, AP-3, AP-4, retromer, and ribosomes; Table S1). For each candidate protein, PCA of the reference set was performed using only those datasets in which the candidate protein was represented. The candidate protein was then projected onto the corresponding PCA plot, and the nearest reference cluster was determined. The figure shows the projections on the first two principal components for six sample proteins. The relative positions of reference clusters change with different combinations of available data, but in each case, all reference proteins are correctly grouped into the expected complexes. Candidate proteins are shown in red and reference proteins in black.
In total, the automated cluster analysis identified 136 candidate CCV proteins. These included >93% of the known coat proteins detected in the CCV fraction. The assignment of coat proteins to either AP-1– or AP-2–positive CCVs was equally successful; >91% of proteins were predicted as reported in the literature. Furthermore, our analysis predicted 36 new CCV proteins (Tables 1 and S2).
Vesicle proteins identified through multivariate profiling
| Number | CCVs | Retromer | AP-4 | ||||
| Coat | Cargo | Coat | Coat | ||||
| Intracellular | Shared | Endocytic | Name | Type | Name | Name | |
| 1 | AP1B1 | CLTA | AP2A1 | ARSA | Lys | VPS35 | AP4B1 |
| 2 | AP1G1 | CLTB | AP2A2 | ASAH1 | Lys | VPS29 | AP4E1 |
| 3 | AP1M1 | CLTC | AP2B1 | CTSB | Lys | VPS26A | AP4M1 |
| 4 | AP1S1 | CLTCL1 | AP2M1 | CTSC | Lys | VPS26B | AP4S1 |
| 5 | AP1S2 | BCL9L | AP2S1 | CTSD | Lys | DENND4C* | C17orf56/ |
| 6 | AP1S3 | C10orf88* | AAK1 | CTSL1 | Lys | FNBP1L | Tepsin* |
| 7 | AFTPH | DERPC | AAK1A | CTSZ | Lys | NSF | |
| 8 | AP1GBP1 | DNAJC6/Aux1 | AGFG1 | DNASE2 | Lys | PACSIN2 | |
| 9 | ARF1 | DNM2 | ARRB2 | EPDR1 | Lys | PI4K2B | |
| 10 | C4orf16 | DNMBP | BIN3 | GGH | Lys | SNX2 | |
| 11 | CLINT1 | EIF2C2 | BMP2K* | GLB1 | Lys | SNX3 | |
| 12 | FAM84B* | FAM109A | CLLD6 | GNS | Lys | SNX5 | |
| 13 | GGA2 | FAM109B | DAB2 | HEXB | Lys | SNX6 | |
| 14 | HEATR5B | GAK/Aux2 | DENND1A | LIPA | Lys | TBC1D4 | |
| 15 | MLK4 | HIP1R | EPN1 | MAN2B1 | Lys | TBC1D5 | |
| 16 | RAB4A | NAPA | EPN2 | NEU1 | Lys | ||
| 17 | RAB4B | NAPG | EPS15 | NPC2 | Lys | ||
| 18 | RABGEF1 | NCOA3 | EPS15L1 | PLBD2 | Lys | ||
| 19 | SCYL2 | OCRL | FAM21A | PLOD1 | Lys | ||
| 20 | SNX24 | PIK3C2A | FCHO2 | PLOD2 | Lys | ||
| 21 | TRIM47 | PLEKHJ1 | GAPVD1 | PPGB | Lys | ||
| 22 | VPS45 | PUM1 | HIP1 | PPT2 | Lys | ||
| 23 | STAMBPL1 | ITSN1 | PSAP | Lys | |||
| 24 | TNRC6A | KIAA0196 | TPP1 | Lys | |||
| 25 | TNRC6B | LDLRAP1 | STX6 | SNARE | |||
| 26 | MAP3K7IP1 | STX7 | SNARE | ||||
| 27 | MEA1 | STX8 | SNARE | ||||
| 28 | NECAP1 | STX10 | SNARE | ||||
| 29 | NECAP2 | STX16 | SNARE | ||||
| 30 | NUMB | VTI1A | SNARE | ||||
| 31 | NUMBL | VTI1B | SNARE | ||||
| 32 | PICALM | VAMP4 | SNARE | ||||
| 33 | PTPN23 | ABCA2 | TMD | ||||
| 34 | PUM2 | ANKH | TMD | ||||
| 35 | RALBP1 | ATP2C1 | TMD | ||||
| 36 | REPS1* | ATP7A | TMD | ||||
| 37 | SMAP1 | FAM18B | TMD | ||||
| 38 | SNX9 | HFE | TMD | ||||
| 39 | STAMBP | IGF2R | TMD | ||||
| 40 | STON2 | M6PR | TMD | ||||
| 41 | SYNJ1 | SCAMP2 | TMD | ||||
| 42 | TNK2 | SORT1 | TMD | ||||
| 43 | TSPAN13 | TMD | |||||
| 44 | VLDLR | TMD | |||||
| 45 | YIPF6 | TMD | |||||
| 46 | GRN | Lumenal | |||||
| 47 | HSPA13 | Lumenal | |||||
| Number | CCVs | Retromer | AP-4 | ||||
| Coat | Cargo | Coat | Coat | ||||
| Intracellular | Shared | Endocytic | Name | Type | Name | Name | |
| 1 | AP1B1 | CLTA | AP2A1 | ARSA | Lys | VPS35 | AP4B1 |
| 2 | AP1G1 | CLTB | AP2A2 | ASAH1 | Lys | VPS29 | AP4E1 |
| 3 | AP1M1 | CLTC | AP2B1 | CTSB | Lys | VPS26A | AP4M1 |
| 4 | AP1S1 | CLTCL1 | AP2M1 | CTSC | Lys | VPS26B | AP4S1 |
| 5 | AP1S2 | BCL9L | AP2S1 | CTSD | Lys | DENND4C* | C17orf56/ |
| 6 | AP1S3 | C10orf88* | AAK1 | CTSL1 | Lys | FNBP1L | Tepsin* |
| 7 | AFTPH | DERPC | AAK1A | CTSZ | Lys | NSF | |
| 8 | AP1GBP1 | DNAJC6/Aux1 | AGFG1 | DNASE2 | Lys | PACSIN2 | |
| 9 | ARF1 | DNM2 | ARRB2 | EPDR1 | Lys | PI4K2B | |
| 10 | C4orf16 | DNMBP | BIN3 | GGH | Lys | SNX2 | |
| 11 | CLINT1 | EIF2C2 | BMP2K* | GLB1 | Lys | SNX3 | |
| 12 | FAM84B* | FAM109A | CLLD6 | GNS | Lys | SNX5 | |
| 13 | GGA2 | FAM109B | DAB2 | HEXB | Lys | SNX6 | |
| 14 | HEATR5B | GAK/Aux2 | DENND1A | LIPA | Lys | TBC1D4 | |
| 15 | MLK4 | HIP1R | EPN1 | MAN2B1 | Lys | TBC1D5 | |
| 16 | RAB4A | NAPA | EPN2 | NEU1 | Lys | ||
| 17 | RAB4B | NAPG | EPS15 | NPC2 | Lys | ||
| 18 | RABGEF1 | NCOA3 | EPS15L1 | PLBD2 | Lys | ||
| 19 | SCYL2 | OCRL | FAM21A | PLOD1 | Lys | ||
| 20 | SNX24 | PIK3C2A | FCHO2 | PLOD2 | Lys | ||
| 21 | TRIM47 | PLEKHJ1 | GAPVD1 | PPGB | Lys | ||
| 22 | VPS45 | PUM1 | HIP1 | PPT2 | Lys | ||
| 23 | STAMBPL1 | ITSN1 | PSAP | Lys | |||
| 24 | TNRC6A | KIAA0196 | TPP1 | Lys | |||
| 25 | TNRC6B | LDLRAP1 | STX6 | SNARE | |||
| 26 | MAP3K7IP1 | STX7 | SNARE | ||||
| 27 | MEA1 | STX8 | SNARE | ||||
| 28 | NECAP1 | STX10 | SNARE | ||||
| 29 | NECAP2 | STX16 | SNARE | ||||
| 30 | NUMB | VTI1A | SNARE | ||||
| 31 | NUMBL | VTI1B | SNARE | ||||
| 32 | PICALM | VAMP4 | SNARE | ||||
| 33 | PTPN23 | ABCA2 | TMD | ||||
| 34 | PUM2 | ANKH | TMD | ||||
| 35 | RALBP1 | ATP2C1 | TMD | ||||
| 36 | REPS1* | ATP7A | TMD | ||||
| 37 | SMAP1 | FAM18B | TMD | ||||
| 38 | SNX9 | HFE | TMD | ||||
| 39 | STAMBP | IGF2R | TMD | ||||
| 40 | STON2 | M6PR | TMD | ||||
| 41 | SYNJ1 | SCAMP2 | TMD | ||||
| 42 | TNK2 | SORT1 | TMD | ||||
| 43 | TSPAN13 | TMD | |||||
| 44 | VLDLR | TMD | |||||
| 45 | YIPF6 | TMD | |||||
| 46 | GRN | Lumenal | |||||
| 47 | HSPA13 | Lumenal | |||||
New predicted CCV-, retromer-, or AP-4-associated proteins are shown in bold. Known clathrin coat components were classified as localizing to intracellular, endocytic, or both types of CCVs (“shared”), based on published evidence (Table S2). New clathrin coat proteins were categorized as predicted in this study. Lysosomal enzymes (Lys) are cargo proteins of intracellular CCVs. Most transmembrane cargo proteins are trafficked by both intracellular and endocytic CCVs, and hence do not have an unambiguous known affiliation. For retromer- and AP-4–associated proteins, the analysis was restricted to coat proteins, and is likely to be less comprehensive than for CCV proteins. Asterisks indicate subcellular localization confirmed by microscopy (Fig. 5). TMD, transmembrane domain.
Although the profiling approach was designed to identify CCV proteins, the tight clustering of protein complexes in Fig. 3 suggested that it may also be suitable to predict associations among non-CCV proteins. From the results of the automated profiling analysis, we predicted 15 proteins as part of the retromer cluster, including 10 known and five new associations, and an unknown protein (C17orf56) as a potential accessory factor for the AP-4 complex (Table 1).
Validation of candidate CCV proteins
To test the validity of our predictions, we localized four of the new predicted CCV proteins (BMP2K, REPS1, FAM84B, and C10orf88) by immunofluorescence microscopy (Fig. 5). All four proteins colocalized with the clathrin adaptors AP-1 or AP-2 as predicted. The protein C10orf88 colocalized with AP-1 and AP-2 (the prediction was AP-1); this dual affiliation is also reflected in the PCA plot, which places the protein between AP-1 and AP-2, but closer to AP-1 (Fig. 4). We also localized a predicted new retromer protein (DENND4C), as well as the candidate AP-4 binding partner. DENND4C showed extensive colocalization with the retromer subunit VPS26, and C17orf56-GFP colocalized very closely with AP-4 ε. Collectively, these findings strongly support the accuracy of our predictions of CCV and non-CCV associated proteins.
Subcellular localization of predicted new CCV, retromer, and AP-4–associated proteins. The proteins shown in Fig. 4 were localized by immunofluorescence microscopy. In the case of REPS1, the endogenous protein was detected with a specific antibody. Other proteins were myc- or GFP-tagged as indicated, and transiently expressed. Colocalization with intracellular or plasma membrane clathrin-coated structures was investigated by double labeling with antibodies against AP-1 γ or AP-2 α, respectively. C10orf88-myc colocalizes with AP-1 and AP-2, which necessitated imaging in two different focal plains. For reasons of antibody compatibility, CALM was used in this case to define clathrin-coated structures at the plasma membrane. Overlays show new proteins in red and reference proteins in green. Bars: (large panels) 10 µm; (insets) 2 µm.
Subcellular localization of predicted new CCV, retromer, and AP-4–associated proteins. The proteins shown in Fig. 4 were localized by immunofluorescence microscopy. In the case of REPS1, the endogenous protein was detected with a specific antibody. Other proteins were myc- or GFP-tagged as indicated, and transiently expressed. Colocalization with intracellular or plasma membrane clathrin-coated structures was investigated by double labeling with antibodies against AP-1 γ or AP-2 α, respectively. C10orf88-myc colocalizes with AP-1 and AP-2, which necessitated imaging in two different focal plains. For reasons of antibody compatibility, CALM was used in this case to define clathrin-coated structures at the plasma membrane. Overlays show new proteins in red and reference proteins in green. Bars: (large panels) 10 µm; (insets) 2 µm.
Stoichiometry of the CCV coat
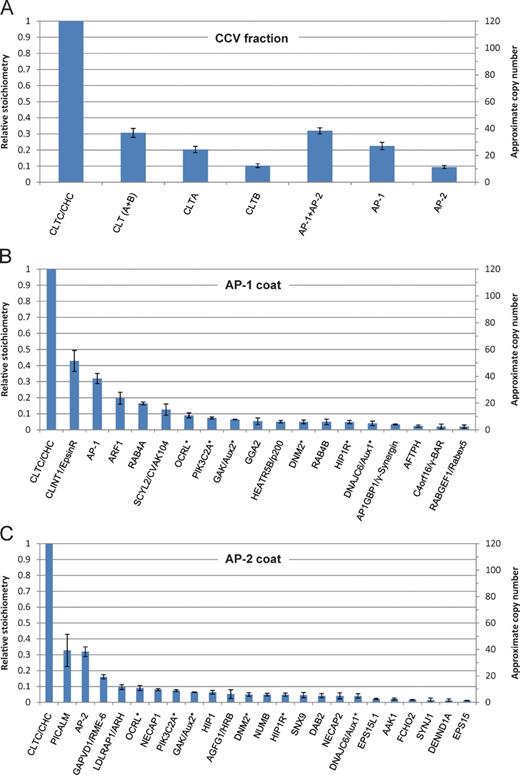
SILAC data can be used to estimate the relative abundance of proteins through modified spectral counting (Ohta et al., 2010), allowing us to perform the first detailed characterization of the CCV coat stoichiometry (Fig. 6). Our analysis suggests that the vesicle inside the clathrin coat is densely covered with AP complexes, with approximately one AP complex per clathrin triskelion. The ratio of clathrin light to heavy chains is ∼1:3, intermediate to what has been reported for rat liver and brain CCVs (Girard et al., 2005). Surprisingly, epsinR appears to be as abundant as AP-1 on intracellular CCVs, and CALM as abundant as AP-2 on endocytic CCVs. EpsinR is a cargo adaptor for the endosomal Q-SNARE vti1b (Hirst et al., 2004), and CALM is required for the sorting of multiple R-SNAREs (Miller et al., 2011). These findings indicate that transport of post-Golgi SNAREs is a major function of CCVs.
Estimated stoichiometry of clathrin coats. To estimate the abundance of major CCV proteins in the CCV fraction, emPAIs were calculated from the SILAC data (Table S1) and normalized to CHC (relative emPAI = 1). The approximate number of copies of each protein in a “typical” CCV with 40 triskelia (Cheng et al., 2007) is also indicated. Error bars indicate SD (n = 3). (A) Relative abundance of APs and clathrin. For every CHC molecule, there are ∼0.3 light chains (CLT A + B) and ∼0.3 AP complexes. AP-1 is approximately twice as abundant as AP-2, which suggests that ∼2/3 of the CCVs in the preparation are intracellular, and ∼1/3 are endocytic. (B) Estimated composition of the intracellular CCV coat, assuming a CHC/AP-1 ratio of ∼3:1. There may be more than one type of intracellular CCV; the figure shows the estimates for a hypothetical average CCV. (C) Estimated composition of the endocytic CCV coat, assuming a CHC/AP-2 ratio of ∼3:1. Asterisks indicate shared proteins.
Estimated stoichiometry of clathrin coats. To estimate the abundance of major CCV proteins in the CCV fraction, emPAIs were calculated from the SILAC data (Table S1) and normalized to CHC (relative emPAI = 1). The approximate number of copies of each protein in a “typical” CCV with 40 triskelia (Cheng et al., 2007) is also indicated. Error bars indicate SD (n = 3). (A) Relative abundance of APs and clathrin. For every CHC molecule, there are ∼0.3 light chains (CLT A + B) and ∼0.3 AP complexes. AP-1 is approximately twice as abundant as AP-2, which suggests that ∼2/3 of the CCVs in the preparation are intracellular, and ∼1/3 are endocytic. (B) Estimated composition of the intracellular CCV coat, assuming a CHC/AP-1 ratio of ∼3:1. There may be more than one type of intracellular CCV; the figure shows the estimates for a hypothetical average CCV. (C) Estimated composition of the endocytic CCV coat, assuming a CHC/AP-2 ratio of ∼3:1. Asterisks indicate shared proteins.
Auxilin depletion causes sequestration of proteins required for cell division
Auxilin depletion not only leads to the accumulation of membraneless clathrin cages, it also causes mitotic arrest (Royle, 2011). To begin to understand the basis of the auxilin knockdown phenotype, we devised a simple profiling approach to determine the protein composition of the clathrin cages. Merging the data from all three auxilin depletion SILAC sets, we identified a group of 104 proteins that were consistently enriched (4–30-fold) in the clathrin cage fraction (Table S3). Many of these proteins had not been included in the CCV profiling analysis because they were detectable only in auxilin-depleted cells, due to their increased abundance. One such protein is TACC3, which functions in cell division and which normally forms a complex with clathrin and two other proteins, GTSE1 and CKAP5/chTOG, on the mitotic spindle (Hubner et al., 2010; Booth et al., 2011). GTSE1 and CKAP5 are also highly enriched in the clathrin cages (Fig. 7 A), which suggests that cage formation and mitotic arrest may be causally linked.
Auxilin depletion causes sequestration of mitotic spindle proteins in clathrin cages. (A) Representative SILAC analysis of CCV fractions from control and auxilin-depleted cells, as performed in Fig. 1 E. The ranks of TACC3, GTSE1, and CKAP5 are indicated. (B) Western blot of whole cell lysates and CCV fractions corresponding to A. TACC3 and CKAP5 are barely detectable in control CCV fractions but highly enriched in CCV fractions from auxilin-depleted cells. In contrast, typical CCV proteins such as SNX9 are readily detectable in control CCVs. MHC-I is shown as a loading control. (C) Fractionation behavior of CKAP5, TACC3, and GTSE1 in control (left) and auxilin-depleted cells (right). In both cases, fractions were prepared using the original or the improved protocol, and compared by SILAC as in Fig. 1 F. CKAP5, GTSE1, and TACC3 (which was undetectable under control conditions) fractionate like clathrin-associated proteins only in auxilin-depleted cells, which indicates a specific association with clathrin cages. (D) Auxilin depletion causes loss of TACC3 from the mitotic spindle in HeLa cells. Bars, 10 µm. (E) HeLa cells were depleted of auxilin, clathrin, or both, or treated with a nontargeting siRNA (control), then stained with Hoechst to visualize DNA. Micronuclei (arrowheads) were detected by automated microscopy. Representative images are shown. Bar, 20 µm. (F) Relative changes in the proportion of micronucleated cells, normalized to nontargeting siRNA-treated cells (control). Error bars show the SEM of four independent repeats (three repeats for untreated cells). (G) Binary comparisons of the treatment groups shown in F, using ANOVA and the post-hoc Tukey-Kramer significance test. The difference between auxilin depletion and combined clathrin/auxilin depletion is highly significant (P < 0.001). The total number of cells scored for every condition is indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Corresponding Western blot analysis of cell lysates. Clathrin and auxilin (isoforms 1 and 2) were effectively depleted in the individual and the combined knockdowns. Positions of protein molecular mass markers are indicated in kilodaltons in B and H. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
Auxilin depletion causes sequestration of mitotic spindle proteins in clathrin cages. (A) Representative SILAC analysis of CCV fractions from control and auxilin-depleted cells, as performed in Fig. 1 E. The ranks of TACC3, GTSE1, and CKAP5 are indicated. (B) Western blot of whole cell lysates and CCV fractions corresponding to A. TACC3 and CKAP5 are barely detectable in control CCV fractions but highly enriched in CCV fractions from auxilin-depleted cells. In contrast, typical CCV proteins such as SNX9 are readily detectable in control CCVs. MHC-I is shown as a loading control. (C) Fractionation behavior of CKAP5, TACC3, and GTSE1 in control (left) and auxilin-depleted cells (right). In both cases, fractions were prepared using the original or the improved protocol, and compared by SILAC as in Fig. 1 F. CKAP5, GTSE1, and TACC3 (which was undetectable under control conditions) fractionate like clathrin-associated proteins only in auxilin-depleted cells, which indicates a specific association with clathrin cages. (D) Auxilin depletion causes loss of TACC3 from the mitotic spindle in HeLa cells. Bars, 10 µm. (E) HeLa cells were depleted of auxilin, clathrin, or both, or treated with a nontargeting siRNA (control), then stained with Hoechst to visualize DNA. Micronuclei (arrowheads) were detected by automated microscopy. Representative images are shown. Bar, 20 µm. (F) Relative changes in the proportion of micronucleated cells, normalized to nontargeting siRNA-treated cells (control). Error bars show the SEM of four independent repeats (three repeats for untreated cells). (G) Binary comparisons of the treatment groups shown in F, using ANOVA and the post-hoc Tukey-Kramer significance test. The difference between auxilin depletion and combined clathrin/auxilin depletion is highly significant (P < 0.001). The total number of cells scored for every condition is indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (H) Corresponding Western blot analysis of cell lysates. Clathrin and auxilin (isoforms 1 and 2) were effectively depleted in the individual and the combined knockdowns. Positions of protein molecular mass markers are indicated in kilodaltons in B and H. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
We first tested by Western blotting whether hyperenrichment of these proteins in the cage fraction was a consequence of altered protein expression or stability (Fig. 7 B). Overall expression levels of TACC3 and CKAP5 were unaffected by auxilin knockdown. Both proteins are barely detectable in control CCVs, but are highly enriched in the clathrin cage fraction. To ensure that the proteins were actually components of the clathrin cages, we next compared cage fractions prepared using the original and the improved preparation protocol by SILAC (as in Fig. 1, C and F). We found that TACC3, GTSE1, and CKAP5 all had enrichment profiles similar to clathrin and other coat components in the auxilin-depleted cells, but behaved like non-CCV proteins in control cells (Fig. 7 C). These data indicate that TACC3, GTSE1, and CKAP5 are not constituents of normal CCVs. However, in auxilin-depleted cells, they become sequestered in clathrin cages, and are hence not available during mitosis. Consistent with this hypothesis, auxilin depletion caused a strong reduction in the levels of TACC3 associated with mitotic spindles (Fig. 7 D).
Two recent reports show that auxilin depletion results in cell cycle arrest and chromosomal missorting (Shimizu et al., 2009; Tanenbaum et al., 2010). Because clathrin itself is also important for mitosis, the authors hypothesized that in the absence of auxilin, a lack of clathrin recycling caused the mitotic defects. However, this hypothesis does not explain why the phenotype of auxilin depletion is more severe than that of clathrin depletion. Our finding that TACC3, GTSE1, and CKAP5 are sequestered into clathrin cages suggests the alternative hypothesis that auxilin depletion deprives cells not only of free cytosolic clathrin, but also of other proteins required for mitosis. If the first hypothesis is correct, then a combined depletion of clathrin and auxilin should be additive, because the trace amounts of clathrin remaining after a clathrin knockdown would be prevented from accessing the spindle. However, if our hypothesis is correct, then the combined depletion should result in a less severe phenotype than auxilin depletion alone, as the lack of clathrin would prevent the formation of cages, and hence liberate TACC3, GTSE1, and CKAP5.
To test these hypotheses, we analyzed mitotic defects in clathrin- and auxilin-depleted cells by automated fluorescence microscopy (Fig. 7 E). We determined the proportion of micronucleated cells, which is a standard measure of chromosome misalignment and mitotic slippage (Schneider et al., 2007). Clathrin depletion caused a moderate increase (2.4-fold), and auxilin depletion caused a strong increase (6.5-fold) in the number of micronucleated cells (Fig. 7, F and G). The joint depletion of clathrin and auxilin significantly improved mitotic fidelity relative to auxilin depletion alone (4.1-fold increase in micronucleated cells). Thus, clathrin depletion can substantially override the effects of auxilin depletion, which is consistent with our hypothesis that auxilin depletion causes sequestration of TACC3 and associated proteins in clathrin cages.
Tepsin: The first AP-4 accessory protein
Although our experiments were designed to identify CCV proteins, the PCA profiling also provides insights into other transport intermediates, including AP-4–coated vesicles. AP-4 was first described 13 years ago and has recently gained prominence because of its links with Alzheimer’s disease and other neurological disorders (Verkerk et al., 2009; Burgos et al., 2010; Abou Jamra et al., 2011). However, to date no cytosolic AP-4–associated proteins have been identified. The finding that an uncharacterized protein, C17orf56, coclusters with AP-4 subunits (Fig. S4) prompted us to investigate this protein further.
Bioinformatic analysis of C17orf56 revealed that it has an epsin N-terminal homology (ENTH) domain, most closely related to that of epsinR (Hirst et al., 2004), so we have named the protein “tepsin” (tetra-epsin). Like AP-4, tepsin is evolutionarily ancient and is found in all five eukaryotic supergroups (Fig. 8 A). Both AP-4 and tepsin have been concomitantly lost from several organisms including yeast, worms, and flies. This suggests that AP-4 and tepsin depend upon each other for function. To explore the relationship between AP-4 and tepsin further, we used fibroblasts from patients with mutations in either the β or the µ subunit of AP-4 (AP4B1*, Abou Jamra et al., 2011; AP4M1*, Verkerk et al., 2009), both of which result in premature stop codons (see Materials and methods). The very similar phenotypes of the patients indicate that both mutations inactivate the AP-4 complex. Western blots of cell homogenates showed reduced levels of other AP-4 subunits in cells from the AP-4–deficient patients, whereas tepsin levels were unchanged. However, both AP-4 and tepsin were undetectable in coated vesicle fractions from the patients’ cells (Fig. 8 B). As shown by immunofluorescence, tepsin has a punctate juxtanuclear pattern in control fibroblasts as well as in primary neurons (Fig. 8, C and D), which colocalizes extensively with AP-4. This pattern is completely absent in fibroblasts from AP-4–deficient patients (Fig. 8 D), which suggests that tepsin requires AP-4 for membrane recruitment. Furthermore, tepsin was coimmunoprecipitated with AP-4 β in extracts from control cells, but not in extracts from AP4M1* patient cells, even though the residual large AP-4 subunits, ε and β4, were still coimmunoprecipitated (Fig. 8 E). This indicates that tepsin interacts only with functional AP-4.
Characterization of tepsin, a new AP-4–associated protein. (A) Schematic diagrams of tepsin from organisms belonging to all five eukaryotic supergroups. All tepsins have an N-terminal ENTH domain and a central VHS/ENTH-like domain. (B) Western blots of whole cell lysates and coated vesicle (CV) fractions from primary human fibroblasts. Tepsin has two isoforms. Although the total amount of tepsin is unchanged in cells with mutations in the AP-4 β and μ subunits (AP4B1* and AP4M1*, respectively), the tepsin no longer co-fractionates with coated vesicles. (C) Immunofluorescence double labeling for tepsin and the AP-4 ε subunit in a primary cortical rat neuron. The two proteins show extensive colocalization. Bar, 10 µm. (D) Immunofluorescence double labeling for tepsin and AP-4 ε in primary human fibroblasts. Although the two proteins colocalize in a punctate perinuclear pattern in control cells, this pattern is absent in AP-4–deficient cells. Bar, 10 µm. (E) Western blots of immunoprecipitates of the AP-4 β subunit from extracts of control and AP-4–deficient fibroblasts, probed for AP-4 subunits and tepsin. Tepsin coimmunoprecipitates with AP-4 only in the control extracts. (F) PNS and coated vesicle fraction from control, AP-4–depleted, and tepsin-depleted HeLa cells. Although tepsin requires AP-4 for incorporation into coated vesicles, AP-4 can still be incorporated in the absence of tepsin. (G) GST pull-downs from tepsin-GFP–expressing HeLa cells. Tagged tepsin specifically interacts with the AP-4 β “ear” domain. (H) GST pull-downs from HeLa cells were analyzed by mass spectrometry. Peptides, unique matched peptides; count, total number of matched peptides. Tepsin is only pulled down with the AP-4 β ear. Positions of protein molecular mass markers are indicated in kilodaltons in B and E–G. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
Characterization of tepsin, a new AP-4–associated protein. (A) Schematic diagrams of tepsin from organisms belonging to all five eukaryotic supergroups. All tepsins have an N-terminal ENTH domain and a central VHS/ENTH-like domain. (B) Western blots of whole cell lysates and coated vesicle (CV) fractions from primary human fibroblasts. Tepsin has two isoforms. Although the total amount of tepsin is unchanged in cells with mutations in the AP-4 β and μ subunits (AP4B1* and AP4M1*, respectively), the tepsin no longer co-fractionates with coated vesicles. (C) Immunofluorescence double labeling for tepsin and the AP-4 ε subunit in a primary cortical rat neuron. The two proteins show extensive colocalization. Bar, 10 µm. (D) Immunofluorescence double labeling for tepsin and AP-4 ε in primary human fibroblasts. Although the two proteins colocalize in a punctate perinuclear pattern in control cells, this pattern is absent in AP-4–deficient cells. Bar, 10 µm. (E) Western blots of immunoprecipitates of the AP-4 β subunit from extracts of control and AP-4–deficient fibroblasts, probed for AP-4 subunits and tepsin. Tepsin coimmunoprecipitates with AP-4 only in the control extracts. (F) PNS and coated vesicle fraction from control, AP-4–depleted, and tepsin-depleted HeLa cells. Although tepsin requires AP-4 for incorporation into coated vesicles, AP-4 can still be incorporated in the absence of tepsin. (G) GST pull-downs from tepsin-GFP–expressing HeLa cells. Tagged tepsin specifically interacts with the AP-4 β “ear” domain. (H) GST pull-downs from HeLa cells were analyzed by mass spectrometry. Peptides, unique matched peptides; count, total number of matched peptides. Tepsin is only pulled down with the AP-4 β ear. Positions of protein molecular mass markers are indicated in kilodaltons in B and E–G. In some cases, the apparent protein molecular weight is shown instead (indicated by ∼); this was estimated from the average migration of the protein relative to molecular weight markers on at least three gels.
To further investigate this association, we prepared coated vesicle fractions from HeLa cells depleted of either AP-4 or tepsin (Fig. 8 F). AP-4 depletion resulted in loss of both AP-4 and tepsin from the vesicle fraction. Conversely, depleting tepsin did not affect the yield of AP-4 in the vesicle fraction, which indicates that AP-4 recruits tepsin rather than vice versa. To identify the binding domain on AP-4 for tepsin, we performed GST pull-downs using the C-terminal “ear” domains of the two large subunits as bait, chosen because most of the accessory proteins that associate with AP-1 or AP-2 do so by interacting with one or both of the ears. We found that GFP-tagged tepsin specifically interacts with the β4 ear (Fig. 8 G). We independently confirmed this result by mass spectrometric analysis of AP-4–ear pull-downs performed in nontransfected HeLa cells (Fig. 8 H). Thus, tepsin is the first reported ear binding partner for any of the non–clathrin-associated AP complexes.
Finally, we explored the dynamics of the AP-4 coat for the first time by carrying out live cell imaging on cells expressing tepsin-GFP (Video 1). Tepsin-GFP puncta were highly mobile, and some appeared to move from the perinuclear area to the cell periphery. This indicates that the AP-4 vesicles and/or the compartments from which they bud are capable of vectorial long-distance transport.
Tepsin-GFP puncta are highly mobile. HeLa cells stably expressing tepsin-GFP were analyzed by time-lapse confocal microscopy using a spinning disk microscope (Cell Observer; Carl Zeiss). A z stack of five optical sections (thickness = ∼1 µm/slice) was imaged every 2.6 s over a period of 175 s. Z stacks were collapsed into maximum intensity projections, and rendered at 15 frames per second (accelerated ∼40×).
Tepsin-GFP puncta are highly mobile. HeLa cells stably expressing tepsin-GFP were analyzed by time-lapse confocal microscopy using a spinning disk microscope (Cell Observer; Carl Zeiss). A z stack of five optical sections (thickness = ∼1 µm/slice) was imaged every 2.6 s over a period of 175 s. Z stacks were collapsed into maximum intensity projections, and rendered at 15 frames per second (accelerated ∼40×).
Discussion
Multivariate profiling maps CCV-dependent pathways with unprecedented accuracy
In this study, we developed a multivariate comparative proteomics approach to characterize clathrin-dependent pathways in HeLa cells. We established several conditions for generating modified CCV fractions, and compared these by quantitative mass spectrometry. Combination of our data through automated PCA profiling allowed the unbiased high-confidence identification and classification of both CCV and non-CCV proteins.
Our analysis identified 136 CCV proteins (89 coat and 47 cargo proteins), including 55 of the 59 known coat proteins present in the CCV fraction (Table S2), which suggests that the profiling approach provided near-complete coverage of CCV proteins expressed in HeLa cells. The four known CCV proteins that were missed are Rab5a–c and β-arrestin 1. Rab5 localizes mostly to endosomes (Stenmark, 2009), whereas arrestins become incorporated into CCVs only upon stimulus (Traub, 2009). Hence, these proteins are not predominantly associated with CCVs, and this was reflected in our analysis. In addition, our list of predicted CCV proteins is completely free of typical contaminants, such as ribosomal proteins. The four predicted CCV proteins chosen for validation showed significant or complete colocalization with clathrin adaptors, which suggests that the number of false positive predictions is likely to be small. Collectively, these data show that the profiling approach provides a comprehensive analysis of CCV proteins in HeLa cells.
A further novel feature of the profiling approach is the unbiased assignment of proteins to endocytic or intracellular CCVs. Our analysis correctly predicted the association of 42 out of 46 known CCV coat proteins (only proteins with an established unambiguous endocytic or intracellular association were scored; Table S2). The 25 predicted new CCV coat proteins can therefore be classified with high confidence.
Among the predicted new CCV proteins are three components of the RNA-induced silencing complex (RISC): TNRC6A/GW182, TNRC6B, and Argonaute-2/EIF2C2. Their profiles indicate that these proteins may associate with both intracellular and endocytic CCVs. Because RISC is required for RNA processing, this was certainly an unexpected finding. Nevertheless, preliminary GST pull-down experiments show that all three proteins can interact with AP-1 (unpublished data), and a recent report shows that the closely related protein TNRC6C can coprecipitate clathrin, AP-1, and AP-2 (Chekulaeva et al., 2011). Our data therefore suggest that CCVs are involved in regulating RISC function or localization.
Finally, our cluster analysis also allowed us to investigate the behavior of proteins whose association with CCVs is controversial. For instance, the AP-3 complex has been proposed to interact with clathrin (Dell’Angelica, 2009), but our data indicate that this is not the case in HeLa cells. The relationship between clathrin and retromer is also unclear (McGough and Cullen, 2011), and in our previous study, we found a partial depletion of retromer from the CCV fraction when clathrin was depleted. Here we find that retromer and CCVs do not cofractionate (Fig. S1), and in the profiling analysis (Fig. 3) the retromer complex forms a distinct cluster relatively close to the AP-1 cluster. Thus, although we cannot rule out the possibility that there might be two pools of retromer, one associated with CCVs and one not, our data indicate that for the most part, clathrin and retromer are not part of the same transport intermediate.
Limitations of the profiling approach
The profiling approach is likely to miss proteins whose association with CCVs is very transient, or which are mostly associated with another organelle. This is particularly relevant for cargo proteins, such as the transferrin receptor, which occupy CCVs only in transit, and which therefore have different profiles from proteins that are mainly associated with CCVs at steady-state. As a result, the list of CCV cargo proteins identified here is likely to be incomplete. Furthermore, our analysis discriminates only between endocytic and intracellular CCVs. There are probably multiple types of intracellular CCVs, which may rely on different APs, such as GGAs and AP-1 (Robinson, 2004). Although we have attempted to interfere with the formation of AP-1–positive CCVs, knockdown of AP-1 had surprisingly little effect on CCV composition, possibly because of up-regulation of compensatory pathways (unpublished data). A more informative approach may be to use acute depletion of AP-1 or GGAs by drug-induced “knocksideways” (Robinson et al., 2010), and we are currently exploring this possibility.
Auxilin knockdown causes sequestration of proteins required for cell division
Auxilin depletion causes cell cycle arrest in a pro-mitotic state, whereas clathrin depletion results in a milder phenotype. Here, we provide a simple explanation for this puzzling observation. Auxilin depletion causes the accumulation of cytosolic clathrin cages, which are packed with clathrin-binding proteins. Our profiling analysis shows that these binding partners include the mitotic spindle protein TACC3, which stabilizes microtubules. Under control conditions, TACC3 recruits free clathrin to the mitotic spindle (Booth et al., 2011); however, in auxilin-depleted cells, clathrin is aggregated into cages, which provide a relatively immobile, high-affinity binding surface. Therefore, TACC3 and its binding partners CKAP5 and GTSE1 become trapped in the clathrin cages, and are no longer available to stabilize spindle microtubules, resulting in severe mitotic defects. We tested this model by quantifying micronucleation in cells depleted of auxilin and clathrin. As predicted, clathrin depletion substantially reversed the effects of auxilin depletion. The incomplete rescue may be explained by timing effects during the 96-h knockdown. Because clathrin is a very stable and abundant protein, auxilin depletion is likely to precede clathrin depletion. Hence, clathrin cages will form initially and cause some micronucleation before clathrin depletion overrides the auxilin phenotype. In addition, it appears that the knockdown efficiency for clathrin may be slightly reduced in the combined clathrin/auxilin depletion (Fig. 7 H).
Finally, it is worth noting that the increase in the proportion of micronucleated cells in auxilin-depleted cells described here (∼6.5-fold) is very similar to that observed in TACC3-depleted cells (∼6-fold; Schneider et al., 2007). This further suggests that sequestration of TACC3 and associated proteins accounts for the auxilin mitosis phenotype.
Profiling of non-CCV proteins
Although our experimental conditions were optimized for CCV proteins, a byproduct of the profiling is a comprehensive cluster analysis of non-CCV proteins, which constitute the vast majority of the proteins present in the CCV fraction. In particular, stable protein complexes such as ribosomes and proteasomes, as well as non-clathrin vesicle coats, such as COPI, retromer, AP-3, and AP-4, are identified as tight clusters in Fig. 3. The discriminating power is highest for peripheral clusters, and central clusters are likely to overlap with functionally unrelated proteins. Nevertheless, the plot provides candidate interacting proteins for all present complexes. As a proof of principle, we used the profiling data to predict retromer and AP-4–associated proteins. For retromer, 10 known and five new components were predicted, and we substantiated the predictions by colocalizing one of the new proteins, DENND4C, with retromer. DENND4C is a guanine nucleotide exchange factor for the endosomal GTPase Rab10 (Yoshimura et al., 2010), and thus is potentially an important new regulator of retromer-mediated trafficking.
We also identified the first cytosolic AP-4–associated protein reported to date, an ENTH domain–containing protein that we have named tepsin. The importance of AP-4, in particular in neurons, is highlighted by several recent studies. First, the Alzheimer’s disease protein, APP, has been shown to interact with AP-4, and there is evidence that its trafficking is AP-4 dependent (Burgos et al., 2010). In addition, several patients have been identified with mutations in each of the four AP-4 subunits who present with severe intellectual disability and progressive spastic paraplegia (Verkerk et al., 2009; Abou Jamra et al., 2011; Moreno-De-Luca et al., 2011). There are many cases of familial intellectual disability and spastic paraplegia where the responsible gene has not yet been identified, and it will be important to determine whether any of these patients have mutations in the tepsin gene.
Why was tepsin not discovered earlier? The most likely explanation is that only small amounts of tepsin are present in AP-4 immunoprecipitations and GST pull-downs, together with a high background of nonspecific proteins. In addition, we suspect that tepsin only associates with AP-4 when it is productively involved in vesicle formation. Two observations support this hypothesis: first, tepsin does not appear to coimmunoprecipitate with defective AP-4 complexes, even though these complexes contain the β4 appendage to which it binds (Fig. 8 E). Second, Western blots of tepsin and AP-4 immunoprecipitates indicate that at steady-state, most of the AP-4 and tepsin are in separate pools, yet spectral counting indicates that they have similar stoichiometries in our coated vesicle fraction (unpublished data). Thus, tepsin is similar in this regard to epsinR and CALM, which are major components of AP-1– and AP-2–positive CCVs, respectively, but which are not tightly associated with cytosolic AP complexes. Because our method is designed to isolate transport intermediates, we are able to detect physiologically relevant interactions that may be missed using more conventional approaches.
Perspective: PCA-based vesicle profiling
This study represents the most comprehensive and stringent analysis of the CCV proteome to date. In addition, it provides a conceptually novel and advantageous approach to subcellular proteomics. Multivariate profiling renders organelle purity much less critical than in primarily fractionation-based proteomics. Here, CCVs were characterized with high sensitivity and specificity in the presence of >90% background proteins, allowing the first detailed characterization of CCVs from human cells. Compared with other comparative proteomics approaches (e.g., Ohta et al., 2010), profiling through PCA is computationally straightforward. Construction of basic PCA projections, such as the scatter plot in Fig. 3, involves minimal processing of the SILAC data, does not require specialist knowledge, and is readily performed with commercially or freely available software. We encourage readers to recreate Fig. 3 with the step-by-step guide provided in the supplemental data (Table S4) to convince themselves of the simplicity of the procedure.
A further advantage of the present approach is that the PCA is blind to the candidate dataset and hence requires no prior training with the proteins of interest, circumventing the need for a large training dataset. Moreover, in introducing automated “reference” clustering, we have shown how PCA-based profiling can accommodate proteins with disparate numbers of data points, a problem that has so far restricted the usefulness of PCA in proteomic applications. Finally, the study also highlights how the sensitivity of the approach extends to other coated vesicles that were not the primary target of the investigation. As a byproduct of CCV profiling, we describe here the first compositional analysis of the AP-4 coat, which has so far defied biochemical purification because of its low abundance. The method may be refocused, for example, by replacing clathrin knockdowns with AP-4 knockdowns, by using different fractionation protocols, or by adding small molecules to perturb a particular pathway. In principle, by choosing suitable conditions, the profiling approach can be adapted to characterize any subcellular fraction, from any cell type, and thus provides a universal tool for mapping protein–protein interactions.
Materials and methods
Cell culture, metabolic labeling, and siRNA-mediated knockdowns
For two-way comparison SILAC experiments, HeLaM cells (Tiwari et al., 1987) were cultured in DME (Sigma-Aldrich) supplemented with 10% (vol/vol) fetal calf serum (Sigma-Aldrich), or in SILAC medium (Thermo-Fisher Scientific) supplemented with 10% (vol/vol) dialyzed fetal calf serum (10,000 MW cut-off; Invitrogen) and “heavy” amino acids (l-arginine-13C615N4:HCl [50 mg/liter] and l-lysine-13C615N2:2HCl [100 mg/liter]; Cambridge Isotope Laboratories) for 7 d to achieve metabolic labeling. The mean incorporation efficiency was ∼97%, as determined by liquid chromatography-tandem mass spectrometry (LC-MSMS). For the three-way comparison SILAC experiment, cells were grown in DME as above, or in SILAC medium supplemented with “medium heavy” l-arginine-13C6 and l-lysine-4,4,5,5-D4 (Cambridge Isotope Laboratories), or in SILAC medium supplemented with “heavy” l-arginine-13C615N4 and l-lysine-13C615N2, for 7 d. SILAC medium was always supplemented with 200 mg/liter l-proline (Sigma-Aldrich). DME and SILAC medium were supplemented with antibiotics (penicillin/streptomycin; no. P0781; Sigma-Aldrich) according to the manufacturer’s instructions. In all SILAC experiments, cells used for control CCV preparations were grown in “heavy” labeled SILAC medium.
All siRNA oligos were purchased from Thermo Fisher Scientific. Knockdown of auxilin 1/DNAJC6 and auxilin 2/GAK was performed with oligo-2 from the siGENOME SMARTpool (DNAJC6, no. D-009885-02) and an siGENOME SMARTpool (GAK, no. M-005005-02). Knockdown of CHC was performed with an ON-TARGETplus SMARTpool (CLTC; no. L-004001-01). Transfection of siRNA was achieved with Oligofectamine (Invitrogen), as described previously (Borner et al., 2006; Hirst et al., 2008). For single-hit knockdowns (72-h protocol), the final concentration of siRNA was 20 nM for clathrin depletion, and 30 nM (i.e., 15 nM + 15 nM) for auxilin 1 and auxilin 2 depletion. For double-hit knockdowns (96 h protocol), an additional second transfection was performed 48 h after the first hit. Control knockdowns were performed with a nontargeting siRNA (no. D-001810-10). The double-hit protocol was used for all immunofluorescence and microarray experiments. For the preparation of CCV fractions, both double- and single-hit protocols were used, as specified in Table S1.
Depletion of AP-4 was achieved with a combined AP-4 µ and AP-4 ε knockdown, using ON-TARGETplus SMARTpools (AP4M1, no. L-011918-01; AP4E1, no. L-021474-00). Knockdown of C17orf56/tepsin was also performed with an ON-TARGETplus SMARTpool (C17orf56, no. L-015821-02). For depletion of AP-4 and C17orf56/tepsin, a double-hit 96-h protocol was used. For the first hit, the final concentration of siRNA was 40 nM for AP-4 (20 nM AP4M1 + 20 nM AP4E1), and 30 nM for C17orf56/tepsin. 48 h after the first hit, a second transfection at half the final concentration of siRNA was performed.
Antibodies and reagents
Western blot analysis and immunofluorescence microscopy were performed with the following antibodies: AP-1 γ (mAb100/3; Sigma-Aldrich), AP-2 α (AP.6, a gift from F. Brodsky, University of California, San Francisco, San Francisco, CA), AP-2 μ and AP-1 μ (anti-AP50; no. 611351; BD; this antibody recognizes both AP-2 μ and AP-1 μ, as verified by siRNA knockdown), AP-3 µ (Simpson et al., 1996), AP-4 ε, β (for Western blotting; Hirst et al., 1999), AP-4 ε (for immunofluorescence microscopy; no. 612019; BD), auxilin 1 and auxilin 2/GAK (this antibody recognizes both auxilin 1 and 2, and was a gift from S. Sever, Harvard Medical School, Boston, MA; Newmyer et al., 2003), CALM (C18; Santa Cruz Biotechnology, Inc.), CKAP5/chTOG (no. 86073; Abcam), CIMPR (1001; this antibody was a gift from P. Luzio, Cambridge Institute for Medical Research, Cambridge, England, UK), CHC/CHC17 (Simpson et al., 1996), EF-2 (C-14; Santa Cruz Biotechnology, Inc.), GFP (3E6; MP Biomedicals), MHC-I (HC10; a gift from P. Lehner, Cambridge Institute for Medical Research), α-myc epitope (9E10 and A14; Santa Cruz Biotechnology, Inc.), nonmuscle myosin 2 (Drenckhahn et al., 1983; the antibody was a gift from J. Kendrick-Jones, Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK), SNX9 (Hirst et al., 2003), TACC3 (no. 56595; Abcam), VPS26 (Seaman, 2004), α-tubulin (no. 18251; Abcam), REPS1 (rabbit pAb; this study), and tepsin (rabbit pAb; this study). The REPS1 antibody was raised against an N-terminal GST fusion protein corresponding to a central part of REPS1 (isoform 2): LEDSADVGDQPGEVGYSGSPAEAPPSKSPSMPSLNQTWPELNQSSEDTAIVHPVPIRMTPSKIHMQEMELKRTGSDHTNPTSPLLVKPSDLLEENKINSSVKFASGNTVDG. The cDNA used for cloning was Integrated Molecular Analysis of Genomes and their Expression (IMAGE) clone no. 5754186. The specificity of the Reps1 antibody was verified by siRNA knockdown. The tepsin antibody was raised against an N-terminal GST fusion protein corresponding to a C-terminal fragment of the long (525 aa) tepsin isoform: DLSRVSDSGSHSGSDSHSGASREPGDLAERVEVVALSDCQQELSLVRTVTRGPRAFLSREEAQHFIKACGLLNCEAVLQLLTCHLRGTSECTQLRALCAIASLGSSDLLPQEHILLRTRPWLQELSMGSPGPVTNKATKILRHFEASCGQLSPARGTSAEPGPTAALPGPSDLLTDAVPLPGSQVFLQPLSSTPVSSRSPAPSSGMPSSPVPTPPPDASPIPAPGDPSEAEARLAESRRWRPERIPGGTDSPKRGPSSCAWSRDSLFAGMELVACPRLVGAGAAAGESCPDAPRAPQTSSQRTAAKE. The cDNA used for cloning was IMAGE clone no. 5430837. The tepsin antibody recognizes both isoforms (presumably splice variants) of the protein (apparent molecular masses are ∼65 and 72 kD). Both bands are sensitive to siRNA knockdown of tepsin, demonstrating the specificity of the antibody (Fig. 8 F). HRP-linked secondary antibodies were purchased from Sigma-Aldrich, and fluorescently labeled secondary antibodies were from Invitrogen. Hoechst 33342 (Invitrogen) was used to stain DNA.
Preparation of CCV-enriched fractions
Control CCV-enriched fractions were prepared as described previously (Borner et al., 2006), with several minor modifications. The most important improvement is the reduced speed of the final pelleting step, which increases the purity of CCVs approximately twofold relative to our previously published protocol. The following is a brief summary of the “improved” protocol:
Four confluent dishes (500 cm2 each) of HeLa cells were scraped into ∼10 ml of buffer A (0.1 M MES, pH 6.5 [adjusted with NaOH], 0.2 mM EGTA, and 0.5 mM MgCl2). Cells were homogenized with a motorized Potter-Elvehjem homogenizer (20 strokes), and centrifuged at ∼4,100 g for 32 min. Supernatants were treated with ribonuclease A at 50 µg/ml for 60 min. Partially digested ribosomes were pelleted by centrifugation (∼4,100 g for 3 min), and discarded. Membranes were pelleted by centrifugation at 55,000 rpm (209,900 g RCFmax) for 40 min in an MLA-80 rotor (Beckman Coulter). Membranes were resuspended in ∼800 µl buffer A using a 1 ml Dounce homogenizer, and mixed with an equal volume of FS buffer (12.5% [wt/vol] Ficoll and 12.5% [wt/vol] sucrose, in buffer A). All subsequent centrifugation steps were performed with a TLA-110 rotor (Beckman Coulter). Samples were spun at 20,000 rpm (21,700 g RCFmax) for 34 min to pellet the bulk of the non-CCV membranes (pellet discarded). Supernatants were diluted with four volumes of buffer A, and centrifuged at 20,000 rpm (21,700 g RCFmax) for 15 min to pellet contaminating glycogen particles (pellet discarded). Supernatants were centrifuged at 35,000 rpm (66,500 g RCFmax) for 30 min to obtain the CCV-enriched fraction (pellet). All preparations were performed at 4°C.
Mock CCV fractions and clathrin-cage enriched fractions were prepared likewise, but from 8–16 confluent dishes (177 cm2 each) of HeLa cells that had been treated with siRNA against CHC, or auxilin 1 and auxilin 2, respectively. “Original” CCV fractions were also prepared as above, but the glycogen pelleting step was omitted, and the final pelleting step was performed at 60,000 rpm (195,500 g RCFmax) for 30 min.
Comparison of CCV fractions by quantitative mass spectrometry
CCV-enriched fractions were resuspended in small volumes (∼100 µl) of nonreducing SDS buffer (4% [wt/vol] SDS and 10 mM Tris-HCl, pH 8.0), heated to 65°C for 3 min, and centrifuged at 16,000 g for 1 min to pellet insoluble material. Samples were stored at −70°C until further use. Protein concentrations were estimated with a BCA assay (Thermo Fisher Scientific).
For binary comparisons, CCV fractions prepared from metabolically labeled and unlabeled cells were pooled (∼20–50 µg protein per fraction; equal amounts pooled). Samples were adjusted with 1:3 volumes of 4× sample buffer (10% [wt/vol] SDS, 40% [vol/vol] glycerol, 8% [vol/vol] 2-mercaptoethanol, and 200 mM Tris-HCl, pH 6.8), reduced at 90°C for 3 min, and separated in a single lane by SDS-PAGE. Gels were stained with Coomassie blue, and each lane was cut into 20 slices. Proteins were reduced, alkylated with iodoacetamide (no. A3221; Sigma-Aldrich), and in-gel digested with trypsin. A detailed protocol can be found in Antrobus and Borner (2011).
LC-MSMS
Tryptic peptides were dried almost to completion in a centrifugal vacuum concentrator (Eppendorf) and resuspended in 10 µl MS solvent (0.1% [vol/vol] TFA and 3% [vol/vol] MeCN). Peptides were analyzed using an LTQ-OrbiTrap XL (Thermo-Fisher Scientific) coupled to a nanoAcquity uPLC (Waters). The uPLC was equipped with a trap column (Symmetry, 180 µm × 20 mm, 5 µm; Waters) and an analytical column (BEH 130, 75 µm × 250 mm, 1.7 µm; Waters) with mobile phase solvent A (0.1% vol/vol formic acid) and solvent B (100% MeCN).
3 µl of tryptic peptides was injected onto the trap column and washed for 3 min at 10 µl/min 99.9% solvent A and 0.1% solvent B. Peptides were eluted to the analytical column and resolved using a variety of gradient methods with the optimum method rising from 3 to 25% solvent B over 90 min, to 40% by 110 min, and to 85% by 115 min. Eluting peptides were sprayed using a 10 µm PicoTip emitter (New Objective) at a spray voltage of 2.3 kV. MS spectra were acquired between m/z 400 and 1,700 at 60,000 resolution (fwhm at m/z 400). Where lock mass was enabled, the polydimethylcyclosiloxane ions (m/z 445.120025) were used. MS to MSMS switching was controlled in an automatic data-dependent acquisition (DDA) fashion with MSMS spectra acquired in the LTQ at a normalized collision energy of 35%. Ions selected for MSMS were excluded from further fragmentation for up to 120 s. All samples were run in duplicate.
Raw files were processed using MaxQuant version 1.0.13.13 (Cox and Mann, 2008) and Mascot v.2.3.0 (Matrix Science). Data were searched against the International Protein Index (IPI) human database (v.3.68) with a concatenated reversed decoy database. Cam (C) was selected as a fixed modification, and oxidized (M), acetyl (Protein N-term), and deamidation (NQ) were selected as variable modifications. Peptide and protein false discovery rate (FDR) were both set to 0.01 and a minimum peptide length of six was required. Ratio calculations were performed on razor and unique peptides with requantify on and a minimum ratio count of 1.
To determine the incorporation efficiency of heavy amino acids, lysates from metabolically labeled cells were prepared, trypsin digested, and analyzed by LC-MSMS. Raw files were processed using MaxQuant version 1.0.13.13 with the requantify function turned off. Output Peptides.txt files were processed using the R statistical package (R Foundation for Statistical Computing) to generate density plots illustrating incorporation rate. Mean incorporation efficiency was ∼97%.
For the mass spectrometric analysis in Fig. 8 H, proteins were digested in-gel with trypsin, and peptides were eluted to an LTQ-OrbiTrap XL using a 50-min gradient. DDA collision-induced dissociation (CID) data were acquired and raw files were exported to Proteome Discoverer 1.2, processed, and searched against the IPI human database v.3.68 using Sequest. Deamidation (N,Q) and oxidation (M) were allowed as potential variable modifications and carbamidomethylation (C) as a fixed modification. A decoy search was performed and an FDR threshold of 1% imposed with reported proteins requiring a minimum of two peptides.
Processing of quantitative mass spectrometry data
The primary MaxQuant output for each SILAC comparison of CCV fractions was a list of identified proteins, a ratio of relative abundance of each protein in the two compared samples (heavy over light label = ratio H/L), and the number of quantification events (observed peptides) that the ratio calculation was based on. Each MaxQuant output file was formatted in an identical manner: Proteins with no gene name were removed. Proteins with (non-normalized) ratios >50 or <0.02 were removed, as these were outside the theoretical range that could be observed with a SILAC labeling efficiency of <98%. In the case of duplicate entries for the same protein, the entry with the smaller number of quantification events was deleted. Ratios were linearly normalized assuming equal protein quantities in both heavy and light labeled samples (Borner et al., 2006). The number of quantification events was used to weight the contribution of individual proteins. In brief, the ratio H/L of each protein was converted into two fractions, FH and FL, which add up to 1 (formula: FH = [H/L]/[1 + H/L]; FL = 1/[1+H/L]). Each fraction was multiplied with the number of quantification events for the corresponding protein as an estimated measure of its relative contribution to protein loading. All multiplied FH and FL values were summed up to obtain total FH and total FL. The ratio of total FH over total FL is an estimate of the protein quantity present in the heavy sample relative to the light sample. If both samples were evenly loaded, the ratio was one. Otherwise, the ratio of total FH over total FL was used to normalize the original H/L ratios by division. Normalization adjustments were generally small (<20% for most datasets, 17.6% on average for all datasets). Table S1 lists the complete processed SILAC data used in this study.
Bioinformatic profiling analysis
For analysis of proteins represented in all 10 SILAC sets, the ratio data were logarithmically transformed, centered, and decorrelated by PCA. Data transformation and PCA were performed with SIMCA-P+ 11.5 (Umetrics), which was also used to generate the scatter plots in Figs. 3, S3, and S4. A Windows-compatible trial version of this program is freely available on the manufacturer’s website (http://www.umetrics.com/simca). Table S4 contains the SILAC dataset and a simple step-by-step guide to recreating Fig. 3 with SIMCA-P+.
For automated cluster analysis, a reference set of 85 proteins was defined (Table S1). This set included subunits of six known protein complexes (AP-1, AP-2, AP-3, AP-4, retromer, and the ribosome). Group assignment of candidate proteins was based on PCA to decorrelate the SILAC ratio data for the reference set. The algorithm was performed using custom software (“RefClus”) written in MATLAB 7 (MathWorks). RefClus is available online (Robinson, 2012). All SILAC ratio data were logarithmically transformed and then centered to the mean of the reference log data. For each candidate protein, the algorithm performed PCA of the reference set using only those SILAC sets in which the candidate protein was represented. The resulting reference PCA plots therefore varied for each combination of available data. In this way, each candidate protein was always represented in every SILAC set used for generating its corresponding reference plot. After confirmation of perfectly successful clustering of the reference data using the K-means iterative algorithm, the candidate data were projected on the principal components of the reference data (the number of principal components was variable because it depended on the available data). For each candidate protein, the squared Euclidean distance from each centroid of every reference cluster was calculated to assign each candidate protein to its nearest reference group. Proteins assigned to the AP-1 or AP-2 reference cluster were considered candidate CCV proteins. Only proteins that had at least four SILAC ratios, and were represented at least once under each experimental condition (clathrin kd, auxilin kd, or fractionation), were included in the automated cluster analysis. 1,523 proteins matched these requirements (Table S1).
In addition to a cluster assignment, the automated analysis also provided the Euclidean distance (ED) of a candidate protein to the nearest (ED1) and second nearest (ED2) reference cluster, as well as the ratio ED2/ED1. A high ratio ED2/ED1 indicates that a protein is very clearly associated with only one reference cluster. An ED2/ED1 near 1 suggests that a protein is contested between the two closest reference clusters. This parameter was used to refine the primary output of the automated analysis:
First, there was a group of proteins that mapped to the space between AP-1 and AP-2. These could be recognized by their assignment to AP-1 as the closest and AP-2 as the second closest cluster, or vice versa. To identify which of these proteins were contested between AP-1 and AP-2 (i.e., not clearly associated with either one of them), we defined an ED2/ED1 <1.6 as a useful empirical cut-off. These proteins are annotated as AP1/AP2 in Table 1 and Table S2; they include several proteins known to be shared between AP-1– and AP-2–positive CCVs. Furthermore, Figs. 3 and S3 suggest that the cluster of known AP-1–associated proteins has an elongated shape that abuts the retromer cluster at the more central end. To define a boundary between the two clusters, we determined which known retromer component was most distal to the retromer reference cluster, and most proximal to the AP-1 reference cluster. This was the protein SNX6, with an EDAP1/EDRetromer of 1.92 (i.e., approximately twice as far away from AP-1 as from retromer). Any retromer-assigned proteins with an EDAP1/EDRetromer <1.92 were reassigned to AP-1. Similarly, the six proteins contested between retromer and AP-2 were reassigned to AP-2.
As a final quality control check, the profiles of all predicted CCV proteins were manually evaluated. Five proteins whose profiles were deemed inconclusive or only loosely similar to known CCV-associated proteins were highlighted as “weak predictions” in Table S2.
The automated clustering was designed primarily to identify CCV proteins. To make predictions for the more central AP-4 and retromer clusters, the stringency of the analysis had to be increased to yield a conservative (and therefore possibly less comprehensive) list of candidate retromer and AP-4–associated proteins. Only proteins with at least eight SILAC ratios were considered, and proteins with predicted transmembrane domains were excluded from the analysis.
All AP-4–assigned proteins were sorted by ED2/EDAP4; candidate AP-4 proteins were expected to have high ratios. The lowest-ranking known AP-4 subunit was AP-4 σ, which was used to define the cut-off. Only proteins with a higher ED2/EDAP4 ratio than AP-4 σ were kept, leaving the four known AP-4 subunits and one new candidate AP-4–associated protein (Table 1).
Proteins assigned to the retromer cluster were sorted by ED2/EDRetromer, and the known retromer subunit with the lowest ratio was used as a cut-off (SNX6). Only proteins with higher ratios were considered candidate retromer-associated proteins. As a further stringency filter, proteins represented in all 10 SILAC sets were sorted by absolute distance to the retromer reference cluster; again, SNX6 was the known retromer subunit with the greatest distance. Proteins that were further away from the retromer reference cluster than SNX6 were discarded. The result was a list of 10 known and five new predicted retromer proteins (Table 1).
It is important to note that absolute distance measurements are only comparable between proteins when their corresponding reference PCA plots are identical (which is the case for proteins present in all 10 SILAC sets). For CCV predictions, proteins with 4–10 SILAC ratios were analyzed, and PCA reference plots were accordingly divergent. Therefore, absolute distances to reference clusters were not compared between candidate CCV proteins. In contrast, the ED2/ED1 ratio is independent of reference plot shape, and was hence used as a tool to refine CCV predictions.
Estimation of relative protein abundance
The SILAC data were used to calculate exponentially modified protein abundance indices (emPAIs) for proteins present in the CCV enriched fraction. emPAI is an approximate measure of protein abundance (Ishihama et al., 2005). Here, a modified version of emPAI was used, as recently described by Ohta et al. (2010). This modified emPAI is based on spectral counts rather than the number of unique peptides, and is hence particularly suitable for large SILAC datasets. For each protein, emPAI was calculated as follows: emPAI = (10[spectral count/MW]) −1.
MW is the molecular weight of the protein. The spectral count is derived from the number of peptides observed for the protein, and the number of times that each peptide was observed (Table S1).
Because clathrin and auxilin depletion affect the abundance of CCV proteins, only SILAC datasets from fractionation experiments (i.e., no knockdown) were used to calculate emPAIs (datasets F1, F2, and F4*; dataset F3 was excluded because it was part of a triple-labeling experiment that included an auxilin knockdown). emPAI values were calculated only for proteins that were identified in all three of these experiments to obtain robust means. Hence, Fig. 6 shows the relative abundance of major CCV coat components; further coat proteins are present in trace amounts. In addition, only well-established CCV proteins were included in Fig. 6.
To calculate emPAIs of AP complexes, the means of the AP-1 γ and AP-1 μ subunits were used for AP-1, and the means of the AP-2 α (isoforms 1 + 2) and AP-2 μ subunits were used for AP-2. The β-subunits were not included, as they are partially shared between AP-1 and AP-2. Peptide data were insufficient to include the AP-2 σ subunit.
The usefulness of emPAI as a tool to estimate protein abundance has been demonstrated convincingly (e.g., Ohta et al., 2010). Nevertheless, the accuracy of emPAI-based quantification is thought to be within ∼70% of the actual value (Ishihama et al., 2005). Because of this relatively large margin of error, the analysis shown in Fig. 6 should be regarded as a rough estimate of the HeLa CCV coat stoichiometry.
Constructs for transient expression
FAM84B and C10orf88 were amplified by PCR from IMAGE clones 5262890 and 5519534, respectively. The 3′ primers included a sequence encoding a single myc tag. The amplified and tagged cDNAs were cloned into the expression vector pIRESneo2 (Takara Bio Inc.). C17orf56/tepsin was amplified from an IMAGE clone (5430837) by PCR, and cloned in-frame into pEGFP-N3 (Takara Bio Inc.). The sequence of all constructs reported here was verified by PCR. Expression-ready C-terminally myc-tagged BMP2K was purchased from OriGene (no. RC215795). The murine DENND4C-GFP construct was as a gift from F. Barr (University of Oxford, Oxford, England, UK; see Yoshimura et al., 2010).
Fluorescence microscopy and analysis of micronuclei frequency
For immunofluorescence microscopy, cells where either fixed in methanol at −20°C or fixed in 3% formaldehyde in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.76 mM KH2PO4, pH 7.4) at room temperature and permeabilized with 0.1% (vol/vol) Triton X-100 (in PBS). For some experiments, cells were pre-permeabilized with 0.05% (wt/vol) saponin (in PBS) for 10–15 s before fixation. Proteins were detected with primary and secondary antibodies listed under “Antibodies and reagents.” Fluorescence microscopy was performed with an Axiovert 200M microscope (Carl Zeiss), equipped with a 63× 1.4 NA objective lens and a charge-coupled device camera (Orca AG; Hamamatsu Photonics), and controlled by OpenLab software version 5.5 (Cellular Imaging and Analysis; Perkin Elmer). Images were analyzed and adjusted for brightness and contrast in Photoshop CS4 (Adobe).
To localize epitope- or GFP-tagged proteins, cells were transfected with the appropriate expression vectors 24–48 h before imaging, using HeLa Monster transfection reagent (Mirus Bio LLC).
For the analysis of micronuclei frequency, control and siRNA-treated Hela cells were seeded into 96-well plates at a range of densities on day four of the five-day (96 h) transfection protocol. On day five, the cells were fixed with 3.3% (vol/vol) formaldehyde in PBS, permeabilized in 0.1% (vol/vol) Triton X-100, and stained with 0.4 ng/ml Hoechst 33342 (Invitrogen) for 40 min at room temperature. The nuclei were imaged with an ArrayScan VTI microscope (Cellomics/Thermo-Fisher Scientific) equipped with a camera (ORCA-ER; Hamamatsu Photonics), using a 20× 0.4 NA objective lens (Carl Zeiss) and the XF100 Hoechst filter. (The fields shown in Fig. 7 E were imaged with a Carl Zeiss 40× 0.5 NA objective lens.) 3–10 wells per condition were imaged in each experiment. The nuclei and micronuclei identification was performed using the proprietary Micronucleus Bioapplication software (Cellomics/Thermo-Fisher Scientific); quantitative analysis was performed within the R statistical package (R Foundation for Statistical Computing). Sparse (<70 cells) or over-confluent (>120 cells) fields were excluded from the analysis, and a fold increase in percentage of cells with at least one micronucleus relative to cells treated with nontargeting siRNA was calculated for each condition. Statistical analysis was performed using one-way analysis of variance (ANOVA) with Tukey-Kramer’s multiple comparison post-test, within Prism 5.0 (GraphPad Software).
Live cell imaging
Movies were captured using a spinning disk microscope (Cell Observer SD; Carl Zeiss) equipped with a 63× 1.4 NA objective lens, a charge-coupled device camera (AxioCam MR), and AxioVision software v4.8 (Carl Zeiss). All imaging was performed at 37°C. Image sequences were processed with AxioVision software, and QuickTime Pro (Apple). For Video 1, a z stack of five optical sections (thickness ∼1 µm/slice) was imaged every 2.6 s over a period of 175 s (67 stacks in total). Z stacks were collapsed into maximum intensity projections, and rendered at 15 frames per second (∼40× accelerated). Live cell imaging was performed on a HeLa cell line that stably expresses the tepsin-GFP construct described in “Constructs for transient expression.”
Western blot analysis
SDS-PAGE and Western blot analysis were performed according to standard protocols. The positions of protein molecular weight markers (PageRuler Plus, Fermentas/Thermo-Fisher; BenchMark Protein Ladder, Invitrogen) are indicated in Figs. 1, 7, 8, S1, and S2. In some panels, the approximate apparent molecular weights of proteins are shown instead (indicated by ∼); they were estimated by comparison with the migration of molecular weight markers. Please note that these values represent reproducible averages from multiple experiments (n ≥ 3), and in some cases deviate considerably from the predicted molecular weights of the proteins. In Fig. 7 B, the loading was 13 µg/lane (lysates), 2 µg/lane (CCV fractions); i.e., 6.5:1. In Fig. 8 F, the loading was 8 µg/lane (postnuclear supernatant [PNS]), 1.6 µg/lane (coated vesicle fractions); i.e., 5:1.
Bioinformatic analysis of tepsin
To look for tepsin homologues, Basic Local Alignment Search Tool (BLAST) searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were performed using the C17orf56 sequence (GenBank/EMBL/DDBJ accession no. NP_653280.1). Organisms belonging to all five eukaryotic supergroups were searched: Homo sapiens (Opisthokonta), Dictyostelium discoideum (Amoebozoa), Arabidopsis thaliana (Archaeplastida), Trypanosoma brucei (Excavata), and Plasmodium falciparum (Stramenopila, Alveolata, and Rhizaria [SAR]/Cryptophyta, Centrohelida, Telonemia, and Haptophyta [CCTH]). Previous studies had shown that all five of these organisms have AP-4 genes (Field et al., 2007). Tepsins were found in all five; the GenBank/EMBL/DDBJ accession nos. are NP_653280, XP_641657, NP_566540, XP_822540, and XP_001348743, respectively. When each of the nonhuman sequences was used in BLAST searches of the National Center for Biotechnology Information human protein database, the top hit in every case was tepsin. HHpred (http://hhpred.tuebingen.mpg.de/hhpred) was used to look for structural similarities with other proteins. All five proteins were predicted to contain an ENTH domain at the N terminus, as well as a second α-helical domain, related to both VHS and ENTH domains, in the middle of the sequence. The sequences were also analyzed using Jpred (Cole et al., 2008; http://www.compbio.dundee.ac.uk/www-jpred/) and predicted to be mainly unstructured outside of the two α-helical domains.
Cell lines for AP-4 experiments
Human fibroblasts from control and AP-4–deficient patients were obtained from G. Mancini (Erasmus Medical Center, Rotterdam, Netherlands; AP4M1*), L. Colleaux (Fondation IMAGINE, Université Paris Descartes, Paris, France; AP4B1*), and A. Raas-Rothschild (Hadassah Hebrew University Medical Center, Jerusalem, Israel; AP4B1*). Both mutations cause very similar phenotypes, including severe intellectual disability, as has been described previously (Verkerk et al., 2009; Abou Jamra et al., 2011). The similarity of the patient phenotypes suggests that both mutations equally disrupt the AP-4 pathway.
The mutation in the AP4B1 gene is an insertion in exon 5, resulting in an early premature stop codon (amino acids 163–739 missing), which causes mRNA instability (Abou Jamra et al., 2011). Any residual truncated AP4B1* protein (162 amino acids) is likely to be completely nonfunctional, and unstable.
The mutation in the AP4M1 gene (Verkerk et al., 2009) is a transversion in intron 14, which results in the skipping of exon 14, causing a late premature stop codon. The resulting truncated AP4M1* protein misses the C-terminal 112 (out of 453) amino acids. This truncated mutant AP4M1* is undetectable by Western blotting (Verkerk et al., 2009), which suggests that it is nonfunctional and unstable. Our immunoprecipitation data (Fig. 8 E) suggest however that some mutant AP4M1* protein can still inefficiently assemble into an AP-4 complex. This mutant AP-4 appears to be nonfunctional, as indicated by the patient phenotype, and our immunofluorescence data (Fig. 8 D).
Fibroblasts were cultured in DME (Sigma-Aldrich) supplemented with 10% (vol/vol) fetal calf serum (Sigma-Aldrich) and antibiotics, as described for HeLa cells (see “Cell culture, metabolic labeling, and siRNA-mediated knockdowns”). Primary cortical neurons from embryonic rats were prepared and provided by C. Freeman (Cambridge Institute for Medical Research, Cambridge, England, UK).
Coated vesicle preparations for AP-4 experiments
Coated vesicle enriched fractions were prepared exactly as CCV fractions, but the final pelleting step was performed at 86,900 g RCFmax (40,000 RPM, TLA-110) for 30 min, which results in slightly higher enrichment of non-CCVs.
Immunoprecipitations and GST pull-downs
For immunoprecipitations of AP-4 β, cell were lysed in PBS-N (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.76 mM KH2PO4, and 1% [vol/vol] IGEPAL CA-630 [Sigma-Aldrich], pH 7.4). Lysates were cleared by centrifugation. Immunoprecipitated complexes were recovered with protein A–Sepharose (no. 17-0780-01; GE Healthcare). See Hirst et al. (1999) for a more detailed protocol. For AP-4 pull-downs, DNA fragments encoding the “ear” domains of AP-4 β and AP-4 ε were amplified from cDNA (from IMAGE clone 2906087, and RZPD clone DKFZp686L12167Q, respectively), and cloned into vector pGEX 4T1 (GE Healthcare). N-terminal GST fusions of the β and ε ear domains were expressed in E. coli. The ear domains had the following sequences: AP-4 β, aa 598–739, GPLIPEENKERVQELPDSGALMLVPNRQLTADYFEKTWLSLKVAHQQVLPWRGEFHPDTLQMALQVVNIQTIAMSRAGSRPWKAYLSAQDDTGCLFLTELLLEPGNSEMQISVKQNEARTETLNSFISVLETVIGTIEEIKS; AP-4 ε, aa 881–1,135, MEIFHPPQSTAASVAKESSLASSFLEETTEYIHSNAMEVCNNETISVSSYKIWKDDCLLMVWSVTNKSGLELKSADLEIFPAENFKVTEQPGCCLPVMEAESTKSFQYSVQIEKPFTEGNLTGFISYHMMDTHSAQLEFSVNLSLLDFIRPLKISSDDFGKLWLSFANDVKQNVKMSESQAALPSALKTLQQKLRLHIIEIIGNEGLLACQLLPSIPCLLHCRVHADVLALWFRSSCSTLPDYLLYQCQKVMEGS. For pull-downs, cells were lysed in PBS-T (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.76 mM KH2PO4, pH 7.4, adjusted to 0.5% Triton X-100 from a 10% stock), and cleared of debris by centrifugation and filtration (0.2 µm). Lysates were adjusted to a protein concentration of ∼2.5 mg/ml, and 50 µg of fusion protein were added as bait for every 4 ml of lysate. The baits and associated proteins were recovered with glutathione Sepharose 4B (no. 17-0756-01; GE Healthcare), and eluted with 2.5% (wt/vol) SDS/50 mM Tris, pH 8.0, at 60°C.
Protein identification by matrix assisted laser desorption/ionization time-of-flight (MALDI-TOF)
Individual protein bands excised from the one-dimensional gel shown in Fig. S1 were analyzed by in-gel trypsin digestion and mass fingerprinting through MALDI-TOF. See Borner et al. (2006) for a more detailed description.
Electron microscopy
For ultrastructural analysis, a control CCV pellet was prepared as described under “Preparation of CCV-enriched fractions” and immediately fixed in 2% paraformaldehyde/2.5% glutaraldehyde (0.1 M sodium cacodylate buffer, pH 6.5) for 1 h at room temperature. The pellet was postfixed with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer, pH 7.3, en bloc stained with 0.5% uranyl acetate in 0.05 M sodium maleate buffer pH 5.2 for 1 h, dehydrated in ethanol, and embedded in Araldite CY212 epoxy resin (Agar Scientific). Ultrathin sections (60–70 nm) were stained with uranyl acetate and Reynolds lead citrate, and viewed in a transmission electron microscope (model CM 100; Philips).
Microarray gene expression analysis
Microarray analysis was performed by the Cambridge Genomic Services (Cambridge, England UK) using an Illumina HumanHT-12 v4 Expression BeadChip.
PCA: A brief introduction
Contemporary high-throughput techniques such as gene array expression analyses or large-scale quantitative proteomic screens produce datasets of considerable complexity. In most cases, the data can be tabulated as a matrix consisting of rows of different observations, with each variable heading a column. An example of this is Table S1, which contains 2,527 proteins (observations) whose change in abundance was measured across 10 SILAC experiments (variables). Such data matrices are difficult to interpret without further processing. Each variable can be depicted as a single axis, but plotting individual variables against each other is practically limited to two or three dimensional representations, which reflect only a small proportion of multidimensional data. However, several computational methods are available to transform multivariate datasets into a more manageable format. The purpose of the transformation is to represent the information contained in the dataset in a way that allows the simple identification of underlying trends (“cluster analysis”; see Janes and Yaffe [2006] for an accessible review on the subject).
PCA can decrease the dimensionality of data by removing redundant linear correlations that exist between variables. If the behaviour of two or more variables is closely linked, their combined contribution to the distribution of the data may be represented using a single new composite variable that weights the optimal contributions from each original variable. A simple example of this is the repeat experiments of the same experimental condition, which are likely to show a high degree of correlation (covariance). They may therefore be “collapsed” into a derived variable that more or less reflects the underlying trend of this condition. But PCA also detects weak or partial correlations between seemingly unrelated experimental conditions.
To derive new composite variables and thus reduce the dimensionality of the data, PCA determines the orthogonal directions of covariances, in order of statistical relevance. These directions are vectors called principal components (PCs); they define the orientation of the new super-axes that describe the reduced dataspace. Two or three PCs are often sufficient to account for most of the variability in the data. Each “observation” (e.g., protein) is associated with a row of measurements, which constitutes its data vector. By multiplying each data vector with a principal component vector, a new composite variable is obtained, which is called a z score. The z score indicates the position of an observation projected along the corresponding principal component. Each z score is thus a super-variable that combines weighted levels of input from the original variables. Because all principal component vectors are orthogonal, the z scores themselves are free of linear correlations.
Each principal component explains only a proportion of the covariance in the data, with the greatest contribution from PC1, and decreasing contributions from each higher-order principal component. Therefore, the first two or three z scores are usually combined into a “scores plot” for data evaluation and cluster analysis. Fig. 3 is an example of this: for each protein, the z scores of PC1 versus PC2 were plotted. Proteins that are strongly affected by the experimental conditions have large (positive or negative) scores, placing them in the periphery of the plot. Importantly, proteins that show similar behaviour across the 10 SILAC experiments have similar sets of scores, and therefore cluster. Three-dimensional scores plots, such as Fig. S4, may confer higher discriminating power, but are often impractical to illustrate in print for large datasets.
Online supplemental material
Fig. S1 shows how analysis of the CCV-enriched fraction by electron microscopy allows the design of an improved preparation protocol. Fig. S2 shows a microarray gene expression analysis of clathrin- and auxilin-depleted cells. Fig. S3 shows how PCA reveals clustering of CCV and non-CCV proteins into functional groups (a fully annotated version of Fig. 3). Fig. S4 shows a three-dimensional PCA scores plot that reveals a novel candidate AP-4–associated protein. Table S1 shows the complete proteomic profiling data. All 2,527 proteins identified in the CCV fraction are listed here, including their corresponding ratio and peptide count data. Table S2 shows predicted CCV proteins. A detailed summary of the results of the profiling analysis is given. Table S3 shows predicted clathrin cage–associated proteins. Table S4 shows the dataset and a step-by-step guide to recreating Fig. 3 (PCA). Video 1 shows that tepsin-GFP puncta are highly mobile.
Acknowledgments
Mass spectrometric identifications in Fig. S1 B were performed by M. Harbour. Reagents were generously provided by G. Mancini (AP4M1* cell line), L. Colleaux and A. Raas-Rothschild (AP4B1* cell line), F. Barr (DENND4C construct), and C. Freeman (rat neurons). We thank P. Luzio, J. Kilmartin, and S. Schuck for their critical feedback, and K. Lilley and S. Hester for technical support during the initial phase of the project.
This work was funded by the Wellcome Trust (RG52996 and RG53217).
References
- AP
adaptor protein
- CCV
clathrin-coated vesicle
- CHC
clathrin heavy chain
- ED
Euclidean distance
- emPAI
exponentially modified protein abundance index
- ENTH
epsin N-terminal homology
- LC-MSMS
liquid chromatography-tandem mass spectrometry
- PCA
principal component analysis
- RISC
RNA-induced silencing complex
- SILAC
stable isotope labeling of amino acids in cell culture
Author notes
R. Antrobus and J. Hirst contributed equally to this paper.