Podosomes are dynamic cell adhesions that are also sites of extracellular matrix degradation, through recruitment of matrix-lytic enzymes, particularly of matrix metalloproteinases. Using total internal reflection fluorescence microscopy, we show that the membrane-bound metalloproteinase MT1-MMP is enriched not only at podosomes but also at distinct “islets” embedded in the plasma membrane of primary human macrophages. MT1-MMP islets become apparent upon podosome dissolution and persist beyond podosome lifetime. Importantly, the majority of MT1-MMP islets are reused as sites of podosome reemergence. siRNA-mediated knockdown and recomplementation analyses show that islet formation is based on the cytoplasmic tail of MT1-MMP and its ability to bind the subcortical actin cytoskeleton. Collectively, our data reveal a previously unrecognized phase in the podosome life cycle and identify a structural function of MT1-MMP that is independent of its proteolytic activity. MT1-MMP islets thus act as cellular memory devices that enable efficient and localized reformation of podosomes, ensuring coordinated matrix degradation and invasion.
Introduction
Podosomes are dynamic, actin-rich adhesion structures in a variety of cell types, including macrophages (Linder et al., 1999), dendritic cells (Burns et al., 2001), osteoclasts (Destaing et al., 2003), endothelial cells (Osiak et al., 2005; Moreau et al., 2006), smooth muscle cells (Burgstaller and Gimona, 2005), and neural crest cells (Murphy et al., 2011). Together with the related invadopodia, they comprise the invadosome type of cell–matrix contacts (Linder et al., 2011; Murphy and Courtneidge, 2011).
Podosome-enabled cell invasion is thought to be involved in such diverse functions as immune cell surveillance (Wiesner et al., 2014), endothelial tubulogeneration (Obika et al., 2014), angiogenic sprouting (Rottiers et al., 2009; Seano et al., 2014), and cellular patterning during embryogenesis (Murphy et al., 2011). Accordingly, absence or impaired formation of podosomes has been implicated in a variety of diseases based on defects in cell migration and invasion such as Wiskott–Aldrich syndrome (Linder et al., 1999; Thrasher et al., 2000; Buschman et al., 2009), Frank-ter Haar syndrome (Iqbal et al., 2010), and PAPA (pyogenic arthritis, pyoderma gangrenosum and acne) syndrome (Cortesio et al., 2010; Starnes et al., 2014).
Podosomes display a bipartite architecture, with a core structure consisting of Arp2/3 complex–nucleated F-actin (Linder et al., 2000a) and actin-associated proteins (Linder and Aepfelbacher, 2003) and a ring structure containing adhesion plaque proteins such as vinculin, talin, and paxillin (Linder and Aepfelbacher, 2003). Podosomes are anchored to the substratum by cell–matrix adhesion proteins such as integrins (Zambonin-Zallone et al., 1989; Chellaiah, 2006; Luxenburg et al., 2012) and CD44 (Chabadel et al., 2007). Moreover, individual podosomes are connected by contractile actomyosin cables (Bhuwania et al., 2012; van den Dries et al., 2013a), reflecting the fact that podosomes are organized into higher-ordered groups. Podosomes are highly dynamic organelles with a lifetime of 2–12 min (Destaing et al., 2003). They can be formed de novo, through Arp2/3-dependent actin nucleation (Linder et al., 2000a), or by fission of preexisting podosomes (Evans et al., 2003; Kopp et al., 2006). Moreover, even in steady state, podosomal actin is being turned over approximately three times (Destaing et al., 2003), and the whole structure undergoes cycles of internal stiffness, based on actin turnover and actomyosin contractility (Labernadie et al., 2010).
Degradation of the ECM is a key function of podosomes. Accordingly, podosomes have been shown to recruit matrix-degrading enzymes such as matrix metalloproteinases and ADAMs (a disintegrin and metalloproteinase; Linder et al., 2011; Murphy and Courtneidge, 2011). In particular, the membrane-bound metalloproteinase MT1-MMP has emerged as a critical regulator of matrix degradation of both podosomes and invadopodia (Poincloux et al., 2009). Transport of MT1-MMP–positive vesicles along microtubules to podosomes has been demonstrated, and regulators of this transport, such as the motor proteins kinesin-1 and -2 (Wiesner et al., 2010) or the RabGTPases Rab5a, Rab8a, and Rab14 (Wiesner et al., 2013), have been identified. However, in contrast to invadopodia, actual enrichment of MT1-MMP at bona fide podosomes, including its exposure on the ventral cell surface, has not been demonstrated yet.
Using total internal reflection fluorescence (TIRF) live-cell imaging of primary human macrophages, we now detect surface-exposed MT1-MMP at podosomes and also at dot-like “islets” that are embedded in the ventral plasma membrane. MT1-MMP islets become apparent upon podosome dissolution and are also preferred sites for podosome reemergence. Islet formation is based on the C-terminal cytoplasmic tail of MT1-MMP and its binding to the subcortical actin cytoskeleton. We propose that MT1-MMP islets constitute cellular memory devices that facilitate formation of new podosomes that are well integrated into the regular pattern of podosome groups, ensuring efficient and localized podosome formation and matrix degradation. These findings constitute a further extension of the functional repertoire of podosomes and their components. At the same time, the demonstration of a nonproteolytic function of MT1-MMP in the turnover of podosomes should also provide a new aspect for the study of other adhesion and invasion structures, most notably invadopodia, and their contribution to cell invasion and cancer progression.
Results
Cell surface–exposed MT1-MMP is present at podosomes and at podosome-free islets
To localize cell surface-exposed MT1-MMP in primary human macrophages, we used a pH-sensitive construct (MT1-MMP-pHluorin; Monteiro et al., 2013), which is fluorescent only at an extracellular pH of 7.4 (Miesenböck, 2012). (pHluorin was inserted N terminally of the transmembrane domain and is thus extracellular on the surface-exposed protease.) For visualization of total cellular MT1-MMP, cells were cotransfected with MT1-MMP-mCherry. Confocal imaging of the ventral cell side showed MT1-MMP-mCherry at a central accumulation, corresponding to the Golgi, and in vesicles (Fig. 1 B), consistent with previous results (Wiesner et al., 2010). MT1-MMP-pHluorin was detected at the ventral plasma membrane and at the cell periphery (Fig. 1 A). However, both proteins showed no localization reminiscent of podosome core or ring structures (Fig. 1, A–C). Strikingly, TIRF analysis of the same cells showed a dot-like localization of MT1-MMP-pHluorin at the substrate-attached cell side, reminiscent of podosome cores (Fig. 1, D–F; compare to Fig. 1, A–C). Indeed, MT1-MMP signals colocalized with F-actin–rich podosome cores (Fig. 1, D–F, insets). Moreover, visualization of endogenous MT1-MMP in TIRF revealed a similar dot-like staining, which mostly colocalized with F-actin–rich podosome cores (Fig. 1, G–I and M). Further analyses showed that surface-associated MT1-MMP-pHluroin is present beneath the podosome core structure and surrounded by podosome ring components such as talin (Fig. 1, J–L and N). A colocalization analysis of MT1-MMP-pHluorin and F-actin–rich podosomes (Fig. S1, A–E; n = 1,100 podosomes) showed a colocalization index of 0.57 (Fig. S1 F), with a negative correlation (r = −0.389) between MT1-MMP-pHluorin enrichment at podosomes and podosome core size (Fig. S1 G). This indicates that larger podosomes, such as the peripherally located subpopulation of precursor podosomes that also shows high turnover (Bhuwania et al., 2012), are less likely to accumulate the proteinase than smaller, and generally longer-lived, podosomes.
TIRF microscopy reveals podosome-associated MT1-MMP. (A and B) Micrographs of macrophage expressing MT1-MMP-pHluorin (A and D, green) and MT1-MMP-mCherry (B and E, red), with merges (C and F), with both sets of images showing the same cell. Note that in confocal mode (A–C), surface-exposed MT1-MMP, as indicated by pHluorin-based fluorescence, is not visible at distinct structures, whereas only in TIRF mode (D–F), dot-like accumulations of both MT1-MMP-pHluorin and MT1-MMP-mCherry become visible. (G–I) TIRF micrographs of endogenous MT1-MMP, stained with specific primary antibody and Alexa Fluor 488–labeled secondary antibody (G, green), and F-actin, stained with Alexa Fluor 568–labeled phalloidin (H, red), with merge (I). (J–L) TIRF micrographs of macrophage expressing MT1-MMP-pHluorin (J, red), and mCherry-Talin-1C to visualize podosome ring structures (K, white), with merge (L). White boxes (D–L) indicate detail regions shown as insets. Dotted line indicates cell circumference. Bars: 10 µm; (insets) 1 µm. (M and N) Fluorescence intensity diagrams of single podosomes. Distances are indicated by yellow lines in insets of (G and H) and (J and K). Note that peak of MT1-MMP–based fluorescence matches with that of F-actin (M) and is surrounded by peaks of talin-based fluorescence (N). Measurements are representative for the respective setup, and similar curves have been reproduced multiple times.
TIRF microscopy reveals podosome-associated MT1-MMP. (A and B) Micrographs of macrophage expressing MT1-MMP-pHluorin (A and D, green) and MT1-MMP-mCherry (B and E, red), with merges (C and F), with both sets of images showing the same cell. Note that in confocal mode (A–C), surface-exposed MT1-MMP, as indicated by pHluorin-based fluorescence, is not visible at distinct structures, whereas only in TIRF mode (D–F), dot-like accumulations of both MT1-MMP-pHluorin and MT1-MMP-mCherry become visible. (G–I) TIRF micrographs of endogenous MT1-MMP, stained with specific primary antibody and Alexa Fluor 488–labeled secondary antibody (G, green), and F-actin, stained with Alexa Fluor 568–labeled phalloidin (H, red), with merge (I). (J–L) TIRF micrographs of macrophage expressing MT1-MMP-pHluorin (J, red), and mCherry-Talin-1C to visualize podosome ring structures (K, white), with merge (L). White boxes (D–L) indicate detail regions shown as insets. Dotted line indicates cell circumference. Bars: 10 µm; (insets) 1 µm. (M and N) Fluorescence intensity diagrams of single podosomes. Distances are indicated by yellow lines in insets of (G and H) and (J and K). Note that peak of MT1-MMP–based fluorescence matches with that of F-actin (M) and is surrounded by peaks of talin-based fluorescence (N). Measurements are representative for the respective setup, and similar curves have been reproduced multiple times.
TIRF live-cell imaging of cells coexpressing MT1-MMP-pHluorin and mCherry-Talin-1C confirmed that podosome-localized MT1-MMP-pHluorin often shows dynamic codistribution with podosomes (Fig. S1, H–J; and Video 1). However, in many cells (∼35%), we also observed MT1-MMP–positive spots at the ventral surface that did not colocalize with podosome core or ring components such as F-actin or talin (Fig. 2, A–C; and Video 2). This second group of structures, termed “MT1-MMP islets,” was especially evident in motile or polarized cells with pronounced trailing edges, with 60–70% of polarized cells showing islets. In these cells, podosomes were recruited to protruding areas of the cell (Burns et al., 2001; Linder et al., 2011), whereas MT1-MMP islets were mostly localized toward the trailing edge (Fig. 2, A–C).
Dynamics of MT1-MMP-pHluorin and mCherry-talin-1C (from Fig. S1). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (red; exposure time at 488 nm, 10 ms) and mCherry-Talin-1C (gray; exposure time at 568 nm, 50 ms). Acquisition rate: 20 time points/min; frame rate: 10 fps; sequence: 5 min and 21 s. Bars, 10 µm. Note that MT1-MMP-pHluorin signals are mostly surrounded by mCherry-Talin-1C, indicative of their podosomal localization.
Dynamics of MT1-MMP-pHluorin and mCherry-talin-1C (from Fig. S1). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (red; exposure time at 488 nm, 10 ms) and mCherry-Talin-1C (gray; exposure time at 568 nm, 50 ms). Acquisition rate: 20 time points/min; frame rate: 10 fps; sequence: 5 min and 21 s. Bars, 10 µm. Note that MT1-MMP-pHluorin signals are mostly surrounded by mCherry-Talin-1C, indicative of their podosomal localization.
MT1-MMP is present at podosomes and at podosome-free islets (from Fig. 2, A–C). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (red; exposure time at 488 nm, 200 ms) and mCherry-Talin-1C (gray; exposure time at 568 nm, 200 ms). Acquisition rate: 2 time points/min; frame rate: 30 fps; sequence: 2 h. Bar, 10 µm. Note the presence of MT1-MMP at podosomes and at podosome-free regions termed “islets.”
MT1-MMP is present at podosomes and at podosome-free islets (from Fig. 2, A–C). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (red; exposure time at 488 nm, 200 ms) and mCherry-Talin-1C (gray; exposure time at 568 nm, 200 ms). Acquisition rate: 2 time points/min; frame rate: 30 fps; sequence: 2 h. Bar, 10 µm. Note the presence of MT1-MMP at podosomes and at podosome-free regions termed “islets.”
Surface-exposed MT1-MMP is present at podosomes and at podosome-free islets. (A–C) TIRF micrographs of macrophage expressing MT1-MMP-pHluorin (A, red) and mCherry-Talin-1C (B, white), with merge (C). White boxes indicate two detail regions, in which MT1-MMP is associated with podosomes (left box) and present at podosome-free islets (right box). Leading and trailing edge (e.) are indicated. (D and E). Color-coded analysis of cell shown in A–C for visualization of motile versus static structures. For both fluorescence channels (MT1-MMP-pHluorin [D] and mCherry-Talin-1C [E]), each frame was colored along the spectrum, with subsequent merge into a single image. White boxes indicate detail regions also shown enlarged below. mCherry-Talin-1C presents in varying colors, indicating regular podosome ring dynamics (E′), whereas podosome-free areas show only diffuse background (E″). Podosome-associated MT1-MMP-pHluorin also presents in varying colors, indicative of podosome core dynamics (D′). However, MT1-MMP islets present mostly in white (D″), indicating restricted lateral mobility. Dotted line indicates cell circumference. Bars: 10 µm; (insets) 1 µm.
Surface-exposed MT1-MMP is present at podosomes and at podosome-free islets. (A–C) TIRF micrographs of macrophage expressing MT1-MMP-pHluorin (A, red) and mCherry-Talin-1C (B, white), with merge (C). White boxes indicate two detail regions, in which MT1-MMP is associated with podosomes (left box) and present at podosome-free islets (right box). Leading and trailing edge (e.) are indicated. (D and E). Color-coded analysis of cell shown in A–C for visualization of motile versus static structures. For both fluorescence channels (MT1-MMP-pHluorin [D] and mCherry-Talin-1C [E]), each frame was colored along the spectrum, with subsequent merge into a single image. White boxes indicate detail regions also shown enlarged below. mCherry-Talin-1C presents in varying colors, indicating regular podosome ring dynamics (E′), whereas podosome-free areas show only diffuse background (E″). Podosome-associated MT1-MMP-pHluorin also presents in varying colors, indicative of podosome core dynamics (D′). However, MT1-MMP islets present mostly in white (D″), indicating restricted lateral mobility. Dotted line indicates cell circumference. Bars: 10 µm; (insets) 1 µm.
To analyze the dynamics of podosome-associated MT1-MMP and MT1-MMP islets, TIRF live-cell videos of cells coexpressing MT1-MMP-pHluorin and mCherry-Talin-1C were acquired and color-coded using ImageJ. For both channels, each frame was colored progressively along the spectrum, with subsequent merge into a single image (Fig. 2, D and E). Dynamic structures are thus depicted in varying colors, whereas static structures are shown in white. The mCherry-Talin-1C merge presented in varying colors, indicating regular podosome ring dynamics (Fig. 2 E′), whereas podosome-free areas showed diffuse background (Fig. 2 E″). Podosome-associated MT1-MMP-pHluorin was also depicted in varying colors, indicative of podosome core dynamics (Fig. 2 D′). However, MT1-MMP islets were mostly white (Fig. 2 D″). This indicates that MT1-MMP at podosomes shows a dynamic codistribution with these structures, whereas MT1-MMP at islets displays reduced lateral dynamics.
We next performed a color-coded analysis of TIRF live-cell videos from macrophages coexpressing Lifeact-RFP and MT1-MMP-pHluorin. This analysis showed that podosome cores display variations in length and orientation of respective tracks (Fig. 3, A–A″). In contrast, podosome-associated MT1-MMP-pHluorin showed less variation (Fig. 3, B–B″), with lower lateral mobility (2.9 ± 0.6 µm during 60 min), compared with that of podosomal F-actin (4.2 ± 0.8 µm; Fig. 3 C). Also, duration of continuous tracks of Lifeact-RFP and MT1-MMP-pHluorin from podosome cores was calculated for all podosomes of five cells (n = 1,411; Fig. 4, A and B). Track duration for Lifeact-RFP signals, indicative of podosome core lifetime, was 668.4 ± 25.7 s, in agreement with earlier results (Destaing et al., 2003). In contrast, track duration for MT1-MMP-pHluorin was significantly higher at 809.6 ± 39.3 s (Fig. 4 C). Interestingly, subgroups of both signals persisted throughout the whole recording period (Fig. 4 C), with 18.4 ± 10.5% of podosome cores and 49.9 ± 6.5% of MT1-MMP-pHluorin patches persisting for >50 min (Fig. 4 D).
Podosome-associated MT1-MMP shows reduced lateral mobility compared with podosomal F-actin. (A and B) TIRF micrographs of macrophage expressing Lifeact-RFP to detect F-actin–rich podosome cores (A) and MT1-MMP-pHluorin (B). Images show color-coded merges of time-lapse videos with successive coloration of individual frames along the spectrum, as indicated. White boxes indicate areas of detail images shown below each panel (A′ and B′). (A″ and B″) Shown are tracks of the center of mass in yellow. Note high variability of track orientation and length of podosomes cores, whereas tracks of MT1-MMP-pHluorin are more uniform and short. Dotted line indicates cell circumference. Bar, 10 µm. Drift of the microscope stage was corrected by use of fluorescent beads, indicated by arrows in B. (C) Statistical analysis of lateral displacement of podosomal F-actin and podosomal MT1-MMP-pHluorin. Each dot represents lateral displacement of the respective center of mass of a single podosome-associated signal. Collectively, 1,411 podosomes were evaluated. Red bar indicates mean ± SEM. ****, P < 0.0001.
Podosome-associated MT1-MMP shows reduced lateral mobility compared with podosomal F-actin. (A and B) TIRF micrographs of macrophage expressing Lifeact-RFP to detect F-actin–rich podosome cores (A) and MT1-MMP-pHluorin (B). Images show color-coded merges of time-lapse videos with successive coloration of individual frames along the spectrum, as indicated. White boxes indicate areas of detail images shown below each panel (A′ and B′). (A″ and B″) Shown are tracks of the center of mass in yellow. Note high variability of track orientation and length of podosomes cores, whereas tracks of MT1-MMP-pHluorin are more uniform and short. Dotted line indicates cell circumference. Bar, 10 µm. Drift of the microscope stage was corrected by use of fluorescent beads, indicated by arrows in B. (C) Statistical analysis of lateral displacement of podosomal F-actin and podosomal MT1-MMP-pHluorin. Each dot represents lateral displacement of the respective center of mass of a single podosome-associated signal. Collectively, 1,411 podosomes were evaluated. Red bar indicates mean ± SEM. ****, P < 0.0001.
MT-MMP signals at podosomes persist longer than podosomal F-actin. (A and B) TIRF micrographs of macrophage expressing Lifeact-RFP to detect F-actin–rich podosome cores (A) and MT1-MMP-pHluorin (B). The center of mass of each podosome-associated signal was tracked, and tracks were colored according to their duration, as indicated. (C and D) Statistical analyses of track duration of podosomal F-actin and podosome-associated MT1-MMP. Each dot in C represents track duration of a single podosomal signal. Red bar indicates mean ± SEM. Note higher mean of MT1-MMP-pHluorin signals and also higher amounts of respective signals persisting for >50 min, which is plotted in D as the mean ± SEM percentage of total tracks. **, P < 0.01.
MT-MMP signals at podosomes persist longer than podosomal F-actin. (A and B) TIRF micrographs of macrophage expressing Lifeact-RFP to detect F-actin–rich podosome cores (A) and MT1-MMP-pHluorin (B). The center of mass of each podosome-associated signal was tracked, and tracks were colored according to their duration, as indicated. (C and D) Statistical analyses of track duration of podosomal F-actin and podosome-associated MT1-MMP. Each dot in C represents track duration of a single podosomal signal. Red bar indicates mean ± SEM. Note higher mean of MT1-MMP-pHluorin signals and also higher amounts of respective signals persisting for >50 min, which is plotted in D as the mean ± SEM percentage of total tracks. **, P < 0.01.
MT-MMP islets are formed by podosome dissolution
Several lines of evidence indicated that MT1-MMP islets and podosomes could be related: (1) The mean diameter of MT1-MMP-pHluorin islets was determined as 0.83 ± 0.06 µm, and their density as 23.98 ± 1.54/100 µm2. These values are similar to size (0.86 ± 0.06 µm) and density (24.52 ± 1.90/100 µm2) of podosome cores (Fig. S1, K and L). (2) MT1-MMP at podosomes persists longer than the podosome structure itself (Fig. 4). (3) MT1-MMP islets are localized toward the trailing edge (Fig. 2, A–C), a preferential site of podosome dissolution (Bhuwania et al., 2012). Therefore, we next explored the potential relationship between podosomes and MT1-MMP islets and, specifically, whether MT1-MMP islets appear as a result of podosome dissolution.
To induce synchronized dissolution of a large number of podosomes, cells coexpressing MT1-MMP-pHluorin and Lifeact-RFP were treated with 100 µM of the Arp2/3 complex inhibitor CK-666 and analyzed by TIRF live-cell imaging. Podosome formation and upkeep is critically based on Arp2/3 complex–dependent actin nucleation (Linder et al., 2000a). Consequently, addition of CK-666 led to disruption of podosomes, indicated by the disappearance of the Lifeact-RFP signal from its podosomal localization. This was sometimes also accompanied by unspecific accumulation of F-actin in the cell center (Fig. 5, A–F; and Video 3). Complete disruption of the podosome structure was confirmed in parallel by dislocalization of other key components of the core, such as Arp2 and α-actinin, or of the ring structure, such as talin, vinculin, or paxillin (Table S1). In contrast, most of the dot-like, previously podosome-associated MT1-MMP-pHluorin signals persisted at their location (Fig. 5, A and D), which was especially visible in kymographs of CK-666–treated cells (Fig. 5, G–I). FRAP analysis of CK-666–induced islets (Fig. 5 J) further showed that MT1-MMP-pHluorin shows only moderate turnover in these structures, with a mobile fraction of 21.8% and a half-time of recovery of 5.2 s (Fig. 5 K).
Podosomal MT1-MMP persists beyond disruption of the podosome structure itself (from Fig. 5, A–F). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 600 ms) and Lifeact-RFP (red; exposure time at 568 nm, 50 ms). Acquisition rate: two time points/min; frame rate: 10 fps; sequence: 25 min and 6 s. Bar, 10 µm. Note the persistence of MT1-MMP signals even after CK-666–induced disruption of podosomes, indicated by absence of the F-actin reporter Lifeact-RFP.
Podosomal MT1-MMP persists beyond disruption of the podosome structure itself (from Fig. 5, A–F). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 600 ms) and Lifeact-RFP (red; exposure time at 568 nm, 50 ms). Acquisition rate: two time points/min; frame rate: 10 fps; sequence: 25 min and 6 s. Bar, 10 µm. Note the persistence of MT1-MMP signals even after CK-666–induced disruption of podosomes, indicated by absence of the F-actin reporter Lifeact-RFP.
MT1-MMP accumulations persist beyond disruption of podosomes. TIRF still images of a cell expressing MT1-MMP-pHluorin (A and D, white) and Lifeact-RFP (B and E, red), with merges (C and F), before (A–C) and 30 min after (D–F) addition of Arp2/3 inhibitor CK-666. White boxes in D–F indicate detail regions shown as insets. Note that regions devoid of podosomes (E) still contain dot-like accumulations of MT1-MMP-pHluorin. Bar, 10 µm. (G–I) Kymographs of the cell shown in A–F. Time since start of the experiments is indicated in minutes. CK-666 was added at time point 0. Note that the localized MT1-MMP-pHluorin signal persists also in the absence of podosomes. Bars, 1 µm. (J) FRAP analysis of MT1-MMP-pHluorin turnover at CK-666 induced islets. Gallery shows TIRF micrographs taken from time-lapse movie of MT1-MMP-pHluorin expressing macrophage. Dashed line indicates single bleached islet. Time before and after bleaching is given above each panel. (K) Quantification of FRAP showing MT1-MMP-pHluorin-based, normalized fluorescence intensity over time, with fitted curve in black. Values are given as mean ± SD. For each value, 10 islets from three cells from three different donors were evaluated.
MT1-MMP accumulations persist beyond disruption of podosomes. TIRF still images of a cell expressing MT1-MMP-pHluorin (A and D, white) and Lifeact-RFP (B and E, red), with merges (C and F), before (A–C) and 30 min after (D–F) addition of Arp2/3 inhibitor CK-666. White boxes in D–F indicate detail regions shown as insets. Note that regions devoid of podosomes (E) still contain dot-like accumulations of MT1-MMP-pHluorin. Bar, 10 µm. (G–I) Kymographs of the cell shown in A–F. Time since start of the experiments is indicated in minutes. CK-666 was added at time point 0. Note that the localized MT1-MMP-pHluorin signal persists also in the absence of podosomes. Bars, 1 µm. (J) FRAP analysis of MT1-MMP-pHluorin turnover at CK-666 induced islets. Gallery shows TIRF micrographs taken from time-lapse movie of MT1-MMP-pHluorin expressing macrophage. Dashed line indicates single bleached islet. Time before and after bleaching is given above each panel. (K) Quantification of FRAP showing MT1-MMP-pHluorin-based, normalized fluorescence intensity over time, with fitted curve in black. Values are given as mean ± SD. For each value, 10 islets from three cells from three different donors were evaluated.
These data indicated that MT1-MMP islets are derived from podosome-localized MT1-MMP upon podosome disruption, either induced by CK-666 addition to adherent, podosome-containing cells or during regular podosome turnover. To test whether MT1-MMP islets can also develop independently of podosomes, nonadherent macrophages were seeded under conditions that inhibit podosome formation. These included addition of the integrin-binding peptide RGD, inhibiting integrin-based adhesion (Ruoslahti, 1996), or addition of CK-666 inhibiting Arp2/3-dependent actin nucleation (Nolen et al., 2009) and thus formation of (integrin-based) podosomes, before seeding of cells. Macrophages coexpressing Lifeact-RFP and MT1-MMP-pHluorin seeded in the presence of 10 µM RGD did not attach firmly to the substratum. They also did not develop podosomes, and only a few irregularly sized accumulations of Lifeact-RFP were discernible (Fig. S2, A–C). MT1-MMP-pHluorin was mostly localized to irregularly sized patches (diameter: 2.0–3.5 µm) on the ventral cell side and also to the edges of polarized cells. Colocalization of MT1-MMP-pHluorin with Lifeact-RFP patches was only observed on few occasions. Similar results were obtained using cells treated with CK-666 before seeding (unpublished data). Collectively, these data indicate that podosome formation is a prerequisite for the genesis of MT1-MMP islets and that the appearance of islets is a result of podosome dissolution.
Molecular characterization of MT1-MMP islets
To characterize MT1-MMP islets on the molecular level, a variety of potential components were tested. Macrophages expressing respective overexpression constructs were treated with CK-666 to induce islets, fixed, stained with labeled phalloidin to ensure podosome disruption, and visualized by confocal TIRF microscopy. For detection of endogenous proteins, respective antibodies were used. Tested proteins included components of podosome core or ring structures such as Arp2 (Linder et al., 2000a), α-actinin (Gimona et al., 2003), Tks5 (Burger et al., 2011), or vinculin (Zambonin-Zallone et al., 1989), talin (Zambonin-Zallone et al., 1989), paxillin (Pfaff and Jurdic, 2001), and transmembrane proteins that link podosomes to the underlying matrix, such as β1 (Marchisio et al., 1988), β2 (Marchisio et al., 1988), and β3 (Zambonin-Zallone et al., 1989) integrins, CD44 (Chabadel et al., 2007), and integrin-associated proteins such as integrin-linked kinase (ILK; Griera et al., 2014) or kindlin-3 (Ussar et al., 2006; Table S1). All of these components localized to podosomes but were absent from islets. In case of integrins and CD44, unspecific accumulations of irregular size were observed at the ventral plasma membrane, which did not colocalize with islets. We next tested proteins associated with membrane curvature at invadosomes, such as CIP4 (Linder et al., 2000b) and FBP17 (Tsuboi et al., 2009), and vesicle regulatory proteins such as flotillin-1 and 2 that influence podosomal matrix degradation (Cornfine et al., 2011). Interestingly, CIP4 and FBP17 were absent from both podosomes and islets, whereas flotillin-2 was present at podosomes, but not at islets. Moreover, c-Src also showed no distinct localization at podosomes or islets, consistent with previous results (Linder and Aepfelbacher, 2003), and inhibition of Src activity by addition of 10 µm PP2 did not lead to discernible alterations in islet appearance.
As invadosomes are privileged membrane sites (Oikawa et al., 2008; Yu et al., 2013), we next tested a variety of membrane lipids. Indeed, comparable to invadopodia (Caldieri et al., 2009), we could show that cholesterol, stained by filipin (Gimpl and Gehrig-Burger, 2011), is also present at macrophage podosomes (Fig. S2, D–F). However, cholesterol was not detectable at MT1-MMP islets (Fig. S2, G–I). Apolipoprotein E, a regulator of cholesterol transport (Vance et al., 2006), localized to intracellular vesicles (Fig. S2, J–L) but was absent from both podosomes and islets. Interestingly, from all other probes used for the detection of lipids (Table S1), the phosphatidylinositol 4-phosphate (PI(4)P) sensor OSH2-2xPH-GFP (Balla et al., 2008) gave the clearest signal at podosomes (Fig. S2, M–O). However, we could not detect enrichment of PI(4)P, or of any other tested lipid, at islets (Fig. S2, P–R). Collectively, these experiments pointed to MT1-MMP as the major component of islets.
The presence of MT1-MMP at islets raised the question whether these structures are able to degrade matrix. We thus performed matrix-degradation experiments in the presence of CK-666. MT1-MMP-pHluorin–expressing cells were seeded on rhodamine-labeled gelatin for 4.5 h, to allow adhesion and podosome formation, but before the onset of widespread matrix degradation, as determined by a time-course analysis (for presence of islets in untreated cells on gelatin, see Fig. S2, S–V). CK-666 was added at 4.5 h (Fig. S2 W), and cells remained on the matrix for an additional 2 h before fixation and quantification of matrix degradation (Fig. S2 X), with absence of podosomes in CK-666–treated cells being checked by staining of F-actin (Fig. S2 Y). Importantly, matrix degradation did not increase during the 2-h time period in CK-666–treated cells, when only islets were present, in contrast to control cells that contained podosomes (Fig. S2 X). We conclude from these data that islets are not degradative on gelatin matrix.
MT1-MMP islets are sites of podosome reemergence
Podosomes are often formed at sites of previous podosome localization. We therefore hypothesized that MT1-MMP islets could be sites of renewed podosome emergence. To test this, macrophages expressing MT1-MMP-pHluorin and Lifeact-RFP were imaged using TIRF live-cell microscopy and treated with CK-666 to disrupt podosomes, and thus also induce islets, with subsequent washout to allow podosome reformation. Strikingly, many of the MT1-MMP islets acquired Lifeact-RFP, indicative of podosome reformation (Fig. 6, A–C; and Video 4). Reformation of complete podosomes was confirmed in parallel by staining of markers for podosome ring (vinculin: Fig. S3, A–C; paxillin: Fig. S3, D–F) and core structures (Arp2: Fig. S2, G–I; F-actin: Fig. S2, J–L). To determine the extent of MT1-MMP islet reuse for podosome formation, we also acquired time-lapse videos during CK-666 treatment and washout. Starting with the time point of washout, all frames of the Lifeact-RFP channel were merged into one image, which was submitted to colocalization analysis with the first frame of the MT1-MMP-pHluorin channel, thus reporting renewed podosomal F-actin accumulation at islets. Quantification showed that 67.3 ± 1.1% of MT1-MMP islets were sites of renewed podosome formation within 10 min after CK-666 washout (n = 15 cells, from three donors). Interestingly, islet formation under inhibition of MT-MMP activity by addition of 100 µM of the inhibitor NSC405020 (Remacle et al., 2012) led to similar values of islet reuse (62.1 ± 3.5%), indicating that the proteolytic activity of the protease is not necessary for podosome reemergence (see also catalytically inactive MT1-MMP-E240A in Fig. 8, D and E).
MT1-MMP islets are sites of podosome reemergence (from Fig. 6, A′–C′). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 300 ms) and Lifeact-RFP (red; exposure time at 568 nm, 300 ms). Acquisition rate: six time points/min; frame rate: 10 fps; sequence: 46 min and 40 s. Bar, 10 µm. Note the successive appearance of new podosomes, indicated by Lifeact-RFP, at MT1-MMP islets.
MT1-MMP islets are sites of podosome reemergence (from Fig. 6, A′–C′). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 300 ms) and Lifeact-RFP (red; exposure time at 568 nm, 300 ms). Acquisition rate: six time points/min; frame rate: 10 fps; sequence: 46 min and 40 s. Bar, 10 µm. Note the successive appearance of new podosomes, indicated by Lifeact-RFP, at MT1-MMP islets.
MT1-MMP islets are sites of podosome reemergence. (A–C) MT1-MMP islets are sites of de novo actin nucleation. TIRF still images of a macrophage expressing MT1-MMP-pHluorin (A, green) and Lifeact-RFP (B, red), with merge (C), treated with CK-666, with subsequent washout of the drug to allow reformation of podosomes. Cell circumference indicated by dashed line. Bars: 10 µm; (insets) 1 µm. White boxes indicate areas shown enlarged in A′–C′. Note that in A′ and B′, respective fluorescence signals have been changed to inverted grayscale. Red arrows in A′ and B′ indicate MT1-MMP islets that are also positive for Lifeact-RFP. Note absence of podosomal Lifeact-RFP signals at 7- and 15-min time points, with successive accumulation of Lifeact-RFP signals at MT1-MMP islets. Time after washout of CK-666 is indicated below (C′). (D–I) MT-MMP islets can recruit material generated by podosome fission. TIRF micrograph of macrophage expressing MT1-MMP-pHluorin (D, green) and Lifeact-RFP (E, red), with merge (F). Still images taken from Video 5. White boxes indicate detail regions shown in D′–F′. Note MT1-MMP islet (arrow) recruiting F-actin accumulation generated by fission of a podosome (white arrowheads). Time since start of the experiment is indicated in minutes and second. Bars: 10 µm; (insets) 1 µm. (G–I) Fluorescence intensity diagrams of MT1-MMP-pHluorin and Lifeact-RFP, taken along the dotted line indicated in detail images of G′–I′ at the indicated time points. Note rise of RFP-fluorescence intensity, indicative of increased actin polymerization, in the core before podosome fission, with subsequent redistribution of the daughter podosome to an MT1-MMP islet. Measurements are representative for the respective set-up, and similar curves have been reproduced multiple times.
MT1-MMP islets are sites of podosome reemergence. (A–C) MT1-MMP islets are sites of de novo actin nucleation. TIRF still images of a macrophage expressing MT1-MMP-pHluorin (A, green) and Lifeact-RFP (B, red), with merge (C), treated with CK-666, with subsequent washout of the drug to allow reformation of podosomes. Cell circumference indicated by dashed line. Bars: 10 µm; (insets) 1 µm. White boxes indicate areas shown enlarged in A′–C′. Note that in A′ and B′, respective fluorescence signals have been changed to inverted grayscale. Red arrows in A′ and B′ indicate MT1-MMP islets that are also positive for Lifeact-RFP. Note absence of podosomal Lifeact-RFP signals at 7- and 15-min time points, with successive accumulation of Lifeact-RFP signals at MT1-MMP islets. Time after washout of CK-666 is indicated below (C′). (D–I) MT-MMP islets can recruit material generated by podosome fission. TIRF micrograph of macrophage expressing MT1-MMP-pHluorin (D, green) and Lifeact-RFP (E, red), with merge (F). Still images taken from Video 5. White boxes indicate detail regions shown in D′–F′. Note MT1-MMP islet (arrow) recruiting F-actin accumulation generated by fission of a podosome (white arrowheads). Time since start of the experiment is indicated in minutes and second. Bars: 10 µm; (insets) 1 µm. (G–I) Fluorescence intensity diagrams of MT1-MMP-pHluorin and Lifeact-RFP, taken along the dotted line indicated in detail images of G′–I′ at the indicated time points. Note rise of RFP-fluorescence intensity, indicative of increased actin polymerization, in the core before podosome fission, with subsequent redistribution of the daughter podosome to an MT1-MMP islet. Measurements are representative for the respective set-up, and similar curves have been reproduced multiple times.
Treatment of cells with CK-666 was used to generate sufficient numbers of islets for statistical analysis but is an artificial process. We thus asked whether reemergence of podosomes at islets could also be detected during regular podosome turnover. For this, cells expressing Lifeact-RFP and MT1-MMP-pHluorin were analyzed by TIRF live-cell imaging. Indeed, we found that 25–30% of MT1-MMP islets coincide with sites of renewed podosome reformation. Comparable to podosomes formed upon CK-666 treatment, reformation was based on actin nucleation, as indicated by increasing accumulation of Lifeact-RFP over time. In addition to de novo formation, new podosomes can also be formed by fission from preexisting podosomes and in particular from the subpopulation of precursor podosomes (Evans et al., 2003; Kopp et al., 2006). Strikingly, we observed that ∼30% of the podosomes generated by fission are recruited to nearby MT1-MMP islets (Fig. 6, D–I; and Video 5), indicating that islets can also function as anchoring points for preformed podosome cores.
MT1-MMP islets can recruit material generated by podosome fission (from Fig. S4, A′ – C′). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 300 ms) and Lifeact-RFP (red; exposure time at 568 nm, 300 ms). Acquisition rate: six time points/min; frame rate: 10 fps; sequence: 1 h. Bar, 10 µm. Note fission of podosome, resulting in the generation of a new podosome core, which is subsequently recruited to a MT-MMP islet.
MT1-MMP islets can recruit material generated by podosome fission (from Fig. S4, A′ – C′). TIRF time-lapse video of a primary human macrophage expressing MT1-MMP-pHluorin (green; exposure time at 488 nm, 300 ms) and Lifeact-RFP (red; exposure time at 568 nm, 300 ms). Acquisition rate: six time points/min; frame rate: 10 fps; sequence: 1 h. Bar, 10 µm. Note fission of podosome, resulting in the generation of a new podosome core, which is subsequently recruited to a MT-MMP islet.
The impact of other cytoskeletal components such as microtubules or dynamin on podosome reformation was determined by use of respective inhibitors, such as nocodazole and dynasore, and analysis by TIRF live imaging (Fig. S3, M–O and V–X). Addition of 1 µM nocodazole led to disruption of podosomes, as reported previously (Linder et al., 2000a), but not of islets (Fig. S3, P–R). Similar observations were made upon addition of dynasore (10 µM; Fig. S3, Y, Z, and A1). Strikingly, upon washout of nocodazole, podosomes reformed mostly between islets (Fig. S3, S–U), in contrast to washout of dynasore, with podosomes reforming mostly at islets (Fig. S3, B1, C1, and D1), indicating a differential impact of these cytoskeletal elements on the memory effect of islets.
Collectively, these data show that MT1-MMP islets can serve as sites of podosome reformation. This can be achieved by de novo Arp2/3-dependent actin nucleation or by recruiting material generated through podosome fission. This memory effect is preserved upon previous inhibition of dynamin, but not under previous disruption of microtubules.
MT1-MMP facilitates podosome reformation
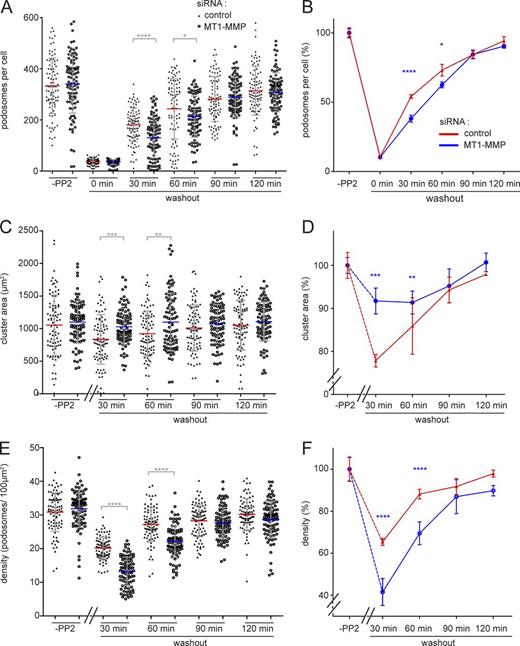
As MT1-MMP islets form sites for podosome reemergence, we next investigated the consequences of MT1-MMP depletion for podosome formation. Cells treated with MT1-MMP–specific siRNA (Wiesner et al., 2013) or control siRNA were processed in a podosome reformation assay. This assay is based on disruption of podosomes by addition of the Src tyrosine kinase inhibitor PP2, with subsequent reformation of podosomes upon washout of the drug (Linder et al., 2000b), followed by semiautomated software analysis (Cervero et al., 2013). (PP2 was used for podosome dissolution as the semiautomated podosome reformation assay has been established for this inhibitor.) As expected, most (>90%) podosomes were disrupted upon PP2 treatment, in both MT1-MMP knockdown and control cells (Fig. 7, A and B). 90 min after washout of the drug, both cell populations had also recovered regular podosome numbers (Fig. 7, A and B). However, MT1-MMP–depleted cells had formed significantly fewer podosomes at time points 30 and 60 min, corresponding to relative decreases of 24% and 13%. Moreover, in control cells, the area of podosome formation was significantly decreased at these 30- and 60-min time points (Fig. 7, C and D). This probably reflects the fact that MT1-MMP islets, which are preferentially used for podosome reformation, show a restricted localization in cells (see Fig. 2). In contrast, MT1-MMP–depleted cells formed new podosomes over a larger area (Fig. 7, C and D). Podosome density, the ratio of podosome number and podosome-covered area, thus showed highly significant decreases of 34% at 30 min and of 19% at 60 min for MT1-MMP–depleted cells (Fig. 7, E and F). These results indicate that the presence of MT1-MMP is not strictly required for podosome reformation. However, MT1-MMP facilitates the generation of new podosomes, leading to faster recovery of regular podosome numbers and densities.
Podosome reformation proceeds less efficiently in cells depleted for MT1-MMP. Statistical evaluations of podosome numbers (A and B), area covered by podosome groups (“clusters”; C and D), or podosome density (podosomes/100 µm2; E and F) during a podosome reformation assay. Parameters were evaluated in cells before disruption of podosomes (“-PP2”) or after treatment with podosome-disrupting PP2 and washout of the drug for the indicated periods, in cells treated with control siRNA (black triangles) or cells treated with MT1-MMP–specific siRNA (open circles). Each dot in A–C represents a single cell, with n = 3 × 30, for cells from three different donors. Diagrams in B, D, and F show data as respective line diagrams. Podosomes are mostly absent at 0 min of the washout (A); podosome-covered area (B) and podosome density (C) were not evaluated for this time point. Note that podosome density of MT1-MMP–depleted cells is significantly different from controls at 30- and 60-min time points (C). Values are given as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For specific values, see Table S2.
Podosome reformation proceeds less efficiently in cells depleted for MT1-MMP. Statistical evaluations of podosome numbers (A and B), area covered by podosome groups (“clusters”; C and D), or podosome density (podosomes/100 µm2; E and F) during a podosome reformation assay. Parameters were evaluated in cells before disruption of podosomes (“-PP2”) or after treatment with podosome-disrupting PP2 and washout of the drug for the indicated periods, in cells treated with control siRNA (black triangles) or cells treated with MT1-MMP–specific siRNA (open circles). Each dot in A–C represents a single cell, with n = 3 × 30, for cells from three different donors. Diagrams in B, D, and F show data as respective line diagrams. Podosomes are mostly absent at 0 min of the washout (A); podosome-covered area (B) and podosome density (C) were not evaluated for this time point. Note that podosome density of MT1-MMP–depleted cells is significantly different from controls at 30- and 60-min time points (C). Values are given as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. For specific values, see Table S2.
Islet formation depends on the LLY motif within the MT1-MMP cytoplasmic tail
We next asked which features of MT1-MMP might enable its localization to podosomes or islets and generated a series of pHluorin-tagged mutants that are (a) deficient in either proteolytic activity (E240A; Rozanov et al., 2001) or (b) oligomerization (DTY/KAF; D385K, T412A, and Y436F; Tochowicz et al., 2011), (c) lack all extracellular domains and are thus defective in ECM binding (ΔNterm), or (d) lack the cytoplasmic tail (ΔCterm). In all cases, the transmembrane domain was included to ensure membrane embedding. (Fig. 8 A). Constructs were coexpressed with Lifeact-RFP in macrophages to assess their subcellular localization by TIRF. The E240A, DTY/KAF and ΔΝterm constructs showed localization to podosomes (Fig. 8, D–I) and islets (see below; Fig. S4), comparable to the wild type (WT; Fig. 8, B and C), whereas the ΔCterm construct mostly did not localize to podosomes or islets but showed a diffuse distribution over the ventral cell surface (Fig. 8, J and K). Importantly, in all cases, the number of podosomes per cell (∼350) was not significantly altered (Fig. 8 L). Collectively, these experiments indicated that the C-terminal cytoplasmic tail of MT1-MMP is necessary and sufficient to ensure localization of the protease at podosomes and islets. Indeed, podosome reformation experiments using CK-666 in cells expressing an siRNA-insensitive ΔNterm construct showed podosome reformation at respective islets comparable to WT MT1-MMP (Fig. S3, E1, F1, G1, H1, I1, J1, K1, L1, and M1), whereas an siRNA-insensitive ΔCterm construct failed to form islets (Fig. S3, N1, O1, and P1).
The cytoplasmic tail of MT1-MMP is necessary for localization of the protease to podosomes. (A) Domain structure of MT1-MMP, containing a propeptide domain absent in the activated protease, a catalytic domain harboring protease activity, a hemopexin domain involved in oligomerization, a transmembrane domain, and a C-terminal cytoplasmic domain. pHluorin was inserted as an intramolecular tag N terminally of the transmembrane domain and is thus extracellular on the surface-exposed protease. First and last amino acid residues are indicated. Yellow asterisks indicate respective point mutants (E240A; DTY/KAF, D385K, T412A, Y436F; LLY/AAA, L571A, L572A, L573A). (B–K) TIRF micrographs of macrophages expressing indicated MT1-MMP-pHluorin constructs (green), together with Lifeact-RFP (red). White boxes indicate areas of detail images shown in black and white insets, with colored merges on the right. Note absence of ΔCterm construct from podosomes (J and K). Bars: 10 µm; (insets) 1 µm. (L and M) Statistical evaluation of podosome numbers in cells expressing indicated MT1-MMP-pHluorin constructs (L) or of MT1-MMP-pHluorin siRNA-insensitive constructs localizing to islets in cells depleted for endogenous MT1-MMP (M). For each value, 3 × 5 cells from three different donors were evaluated. Bars represent mean ± SD. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. For specific values, see Table S2.
The cytoplasmic tail of MT1-MMP is necessary for localization of the protease to podosomes. (A) Domain structure of MT1-MMP, containing a propeptide domain absent in the activated protease, a catalytic domain harboring protease activity, a hemopexin domain involved in oligomerization, a transmembrane domain, and a C-terminal cytoplasmic domain. pHluorin was inserted as an intramolecular tag N terminally of the transmembrane domain and is thus extracellular on the surface-exposed protease. First and last amino acid residues are indicated. Yellow asterisks indicate respective point mutants (E240A; DTY/KAF, D385K, T412A, Y436F; LLY/AAA, L571A, L572A, L573A). (B–K) TIRF micrographs of macrophages expressing indicated MT1-MMP-pHluorin constructs (green), together with Lifeact-RFP (red). White boxes indicate areas of detail images shown in black and white insets, with colored merges on the right. Note absence of ΔCterm construct from podosomes (J and K). Bars: 10 µm; (insets) 1 µm. (L and M) Statistical evaluation of podosome numbers in cells expressing indicated MT1-MMP-pHluorin constructs (L) or of MT1-MMP-pHluorin siRNA-insensitive constructs localizing to islets in cells depleted for endogenous MT1-MMP (M). For each value, 3 × 5 cells from three different donors were evaluated. Bars represent mean ± SD. *, P < 0.05; **, P < 0.001; ***, P < 0.0001. For specific values, see Table S2.
Several scenarios could be envisioned for the mode of action of the C-terminal domain: it could be involved in (a) localizing MT1-MMP to vesicles, (b) regulating the surface exposure of MT1-MMP, or (c) restricting the mobility of the surface-exposed protease. To distinguish between these possibilities, we generated mCherry-fused versions of the ΔCterm construct, and also of the E240A construct, for comparison. (1) Localization at vesicles was tested by coexpression of GFP-fused Rab14 Q70L (Junutula et al., 2004) or Rab22a (Weigert et al., 2004). Comparable to MT1-MMP WT (Wiesner et al., 2013), both ΔCterm and E240A constructs showed prominent localization to Rab14 vesicles (Fig. S4, A–C) or Rab22a vesicles (Fig. S4, D–F). (2) The amount of surface-exposed material was determined by staining of unpermeabilized macrophages using an anti-mCherry antibody and a secondary, Alexa Fluor 647–labeled antibody. The respective surface-associated fluorescence intensity was measured and analyzed (Fig. S4, G–J). This value was not reduced in case of both the ΔCterm and E240A constructs, compared with WT, and was even slightly elevated in case of ΔCterm (Fig. S4 J). These experiments show that absence of the C-terminal domain of MT1-MMP does not influence the ability of the protease to localize to vesicles or to be exposed at the surface of macrophages.
We therefore reasoned that the localization-relevant function of the MT1-MMP C terminus could be to restrict the lateral mobility of the surface-exposed protease, possibly by anchoring it to the cortical cytoskeleton. Treatment of cells with the Arp2/3 inhibitor CK-666 leads to disruption of branched actin networks, including podosome cores, but leaving unbranched actin filaments of the cortex unaffected. We thus disrupted F-actin globally by treatment with both cytochalasin D, to inhibit actin filament elongation and latrunculin A, to sequester actin monomers. First, islets were induced by addition of CK-666 for 30 min to MT1-MMP-pHluorin–expressing cells. Next, 50 µM cytochalasin D and 10 µm latrunculin A were added, and cells were incubated for another 30 min. Strikingly, this led to complete dissolution of MT1-MMP islets, with MT1-MMP-pHluorin being distributed over the plasma membrane (Fig. 9, A–C), indicating that anchoring to the cortical unbranched actin cytoskeleton is important for MT1-MMP localization in islets.
The F-actin anchoring LLY motif in the MT1-MMP C terminus is critical for localization of the protease to podosomes and islets. Confocal micrographs of macrophages expressing Lifeact-RFP (B, E, H, K, and N, red) and MT1-MMP-pHluorin constructs (green), of either WT (A) or an MT1-MMP-LLY/AAA mutant (D, G, J, and M). (A–C) Cells were treated with cytochalasin D and latrunculin A to inhibit formation of both branched and unbranched actin filaments. Note the absence of MT1-MMP islets. (D–I) Cells expressing MT1-MMP-LLY/AAA-pHluorin in a background of endogenous MT1-MMP (not labeled), without (D–F) or with (G–I) the addition of CK-666 to inhibit Arp2/3-generated branched actin filaments. Note localization of the MT1-MMP mutant construct to both podosomes (D–F) and islets (G–I). (J–O) Macrophages treated with control siRNA (J–L) or depleted of endogenous MT1-MMP (M–O) and expressing an siRNA-insensitive MT1-MMP-LLY/AAA-pHluorin construct. Note dispersed localization of the expressed protein, in contrast to enrichment at podosomes. Bars, 10 µm.
The F-actin anchoring LLY motif in the MT1-MMP C terminus is critical for localization of the protease to podosomes and islets. Confocal micrographs of macrophages expressing Lifeact-RFP (B, E, H, K, and N, red) and MT1-MMP-pHluorin constructs (green), of either WT (A) or an MT1-MMP-LLY/AAA mutant (D, G, J, and M). (A–C) Cells were treated with cytochalasin D and latrunculin A to inhibit formation of both branched and unbranched actin filaments. Note the absence of MT1-MMP islets. (D–I) Cells expressing MT1-MMP-LLY/AAA-pHluorin in a background of endogenous MT1-MMP (not labeled), without (D–F) or with (G–I) the addition of CK-666 to inhibit Arp2/3-generated branched actin filaments. Note localization of the MT1-MMP mutant construct to both podosomes (D–F) and islets (G–I). (J–O) Macrophages treated with control siRNA (J–L) or depleted of endogenous MT1-MMP (M–O) and expressing an siRNA-insensitive MT1-MMP-LLY/AAA-pHluorin construct. Note dispersed localization of the expressed protein, in contrast to enrichment at podosomes. Bars, 10 µm.
Indeed, MT1-MMP has been reported to bind to F-actin by its C-terminal region and to thus prevent its diffusion out of invadopodia (Uekita et al., 2001; Yu et al., 2012). To test this, we generated a pHluorin-based, full-length construct of MT1-MMP, in which the three residues critical for F-actin binding were mutated to alanine (LLY/AAA; L571A, L572A, and L573A; Uekita et al., 2001; Yu et al., 2012). MT1-MMP-LLY/AAA-pHluorin was localized to podosomes (Fig. 9, D–F), and its expression did not change the number of podosomes per cell (Fig. 8 L). Subsequent to CK-666 treatment, MT1-MMP-LLY/AAA-pHluorin was also present at islets (Fig. 9, G–I). However, these experiments were performed in cells containing endogenous MT1-MMP, which might provide stabilization through oligomerization. We therefore generated a siRNA-insensitive MT1-MMP-LLY/AAA mutant, expressed it in macrophages depleted for endogenous MT1-MMP, and examined cells by TIRF microscopy. Strikingly, the LLY/AAA mutant construct failed to localize to podosomes in most cells and was diffusely localized over the ventral surface (Fig. 9, M–O), recapitulating the phenotype of the cytochalasin D/latrunculin A treatment.
Next, siRNA-insensitive MT1-MMP-pHluorin mutants were expressed in cells depleted for MT1-MMP and their localization to islets was quantified. This analysis showed that the E240A (Fig. S4, N–P) and DTY/KAF mutants (Fig. S4, Q–S) exhibit a capacity to localize to islets comparable to WT (Fig. 8 M and Fig. S4, K–M), whereas ΔCterm and LLY/AAA mutants showed strongly reduced numbers of cells with islets (Fig. 8 M). The ΔNterm mutant (Fig. S4, T–V) showed intermediate levels of islet formation (Fig. 8 M), indicating a potential influence of the MT1-MMP N terminus.
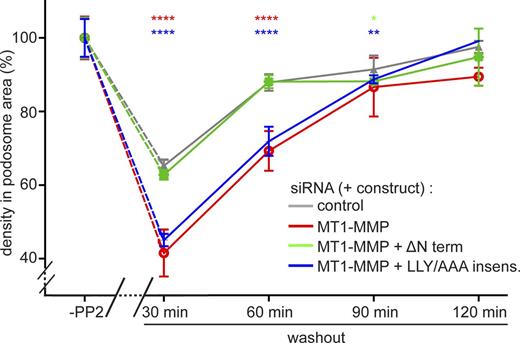
Finally, we tested the capacity of the MT1-MMP C terminus and its LLY motif to restore regular podosome reformation. For this, cells treated with control siRNA or MT1-MMP–specific siRNA were treated with PP2 to disrupt podosomes. In addition, two subsets of MT1-MMP siRNA-treated cells also expressed a pHluorin-fused construct of the MT1-MMP C terminus, including the transmembrane domain (not targeted by the siRNA; “MT1-MMP-ΔNterm”; Fig. 8 A), or of full-length MT1-MMP mutated in the LLY motif (“MT1-MMP-LLY/AAA insens”; Fig. 8 A) and rendered siRNA insensitive. Remarkably, expression of MT1-MMP-ΔNterm was sufficient to restore regular podosome numbers, podosome-covered areas (Fig. S5), and podosome densities (Fig. 10) during reformation,. In contrast, full-length MT1-MMP-LLY/AAA insens was not able to restore these parameters to control values. Instead, the recovery curves closely followed those of cells depleted for MT1-MMP (Figs. 10 and S5). Collectively, these results indicate that the cytoplasmic region of MT1-MMP is crucial for its localization to podosomes. Moreover, the LLY motif within this region is able to stabilize MT1-MMP at podosome-free islets by anchoring the protease to the unbranched cortical actin network. Through this motif, MT1-MMP is able to exert its memory function and to facilitate efficient reformation of podosomes.
The LLY motif in the MT1-MMP C terminus is crucial for the recovery of regular podosome densities during podosome reformation. Statistical evaluation of podosome density (podosomes/100 µm2) during a podosome reformation assay. Parameters were evaluated in cells before disruption of podosomes (“-PP2”) or after treatment with podosome-disrupting PP2 and washout of the drug for the indicated periods, in cells treated with control siRNA (gray) or with MT1-MMP-specific siRNA (red). Subsets of MT1-MMP–depleted cells were also expressing a construct of the MT1-MMP C terminus that is not targeted by the siRNA (ΔNterm; green) or of a full-length MT1-MMP construct mutated in the LLY motif, which was rendered siRNA insensitive (LLY/AAA insens.; blue). n = 3 × 30, for cells from three different donors. Note that podosomes are mostly absent at 0 min of the washout; therefore, podosome density was not evaluated for this time point. Note also that expression of the ΔNterm constructs rescues the recovery of regular podosome density, whereas expression of the LLY/AAA mutant construct does not. Values are given as means ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. For dot diagrams of podosome numbers, podosome area, and podosome density, see Fig. S6. For specific values, see Table S2.
The LLY motif in the MT1-MMP C terminus is crucial for the recovery of regular podosome densities during podosome reformation. Statistical evaluation of podosome density (podosomes/100 µm2) during a podosome reformation assay. Parameters were evaluated in cells before disruption of podosomes (“-PP2”) or after treatment with podosome-disrupting PP2 and washout of the drug for the indicated periods, in cells treated with control siRNA (gray) or with MT1-MMP-specific siRNA (red). Subsets of MT1-MMP–depleted cells were also expressing a construct of the MT1-MMP C terminus that is not targeted by the siRNA (ΔNterm; green) or of a full-length MT1-MMP construct mutated in the LLY motif, which was rendered siRNA insensitive (LLY/AAA insens.; blue). n = 3 × 30, for cells from three different donors. Note that podosomes are mostly absent at 0 min of the washout; therefore, podosome density was not evaluated for this time point. Note also that expression of the ΔNterm constructs rescues the recovery of regular podosome density, whereas expression of the LLY/AAA mutant construct does not. Values are given as means ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. For dot diagrams of podosome numbers, podosome area, and podosome density, see Fig. S6. For specific values, see Table S2.
Discussion
The matrix metalloproteinase MT1-MMP is crucial for the ability of podosomes and invadopodia to degrade ECM (Steffen et al., 2008; Poincloux et al., 2009; Linder et al., 2011; Wiesner et al., 2014) and enables invadosome-forming cells to invade into surrounding tissues. In contrast to invadopodia, which are highly enriched in MT1-MMP (Chen and Wang, 1999; Poincloux et al., 2009), detection of MT1-MMP at bona fide podosomes has proven to be challenging. Previous work demonstrated contact of MT1-MMP–positive vesicles with podosomes (Wiesner et al., 2010), whereas siRNA-mediated knockdown of MT1-MMP in human macrophages (Wiesner et al., 2010) and endothelial cells (Varon et al., 2006) inhibited podosomal matrix degradation. However, in contrast to general assumption, detection of MT1-MMP at the ventral surface of podosomes, as well as its exact localization at podosome core or ring structures, has not been shown. Using a pH-sensitive construct of MT1-MMP in combination with TIRF microscopy, we now demonstrate enrichment of MT1-MMP at the ventral surface of primary macrophages, underneath the core structure of podosomes.
Importantly, we also describe a previously unrecognized localization of MT1-MMP at podosome-free islets that are embedded in the ventral plasma membrane of macrophages. These islets become apparent upon podosome dissolution, as shown by live-cell imaging of podosome turnover, and also by drug-induced disruption of podosomes. Podosomes can thus be viewed as organelles that imprint MT1-MMP islets in the plasma membrane, which persist beyond the lifetime of the actual podosome structure. Indeed, several lines of evidence pointed at a potential link between podosomes and islets: (a) both structures show similar sizes and spacing patterns; (b) MT1-MMP islets tend to be localized toward the trailing edge, a preferential site of podosome dissolution; (c) inhibition of podosome formation during adhesion of cells also led to absence of islets; and (d) podosome-associated MT1-MMP-pHluorin has a longer lifetime than the podosome itself.
During the course of regular podosome turnover, 49.98% of podosome-associated MT1-MMP persisted for >50 min, whereas only 18.49% of podosomes showed a comparable lifetime. Similar values were gained by measuring MT1-MMP-pHluorin fluorescence at islets formed by podosome disruption. Here, MT1-MMP islets persisted for at least 60 min. It is also noteworthy that MT1-MMP at invadopodia of breast cancer cells has previously been shown to persist for >40 min (Monteiro et al., 2013). Considering that invadopodia have a lifetime of more than 1 h, whereas podosomes are turned over within minutes (Linder, 2007), these similar values probably reflect the general ability of MT1-MMP to persist for an extended time at the plasma membrane, rather than being podosome or invadopodia specific.
Lateral movement of podosome core structures is highly variable in the micrometer range. In contrast, podosome-localized MT1-MMP-pHluorin patches showed a more restricted mobility, which probably reflects embedding of the protease in the plasma membrane. This restricted lateral mobility of MT1-MMP was even more evident for MT-MMP islets. In this case, only the net movement of the membrane might provide the driving force for MT1-MMP islets. This could be compared with the well-known sliding of focal adhesions (Ballestrem et al., 2001), the major difference being that focal adhesions are coupled to the ECM via integrins (Wehrle-Haller and Imhof, 2003), whereas this is not a prerequisite for MT1-MMP islets, as discussed below.
An obvious question concerns the molecular composition of MT1-MMP islets. Testing both overexpressed and endogenous proteins, also at high laser power, we could not detect any of the typical podosome core or ring components at MT1-MMP islets, which argues for a complete disruption of the podosome structure. However, recruitment of MT1-MMP to invadosomes has been proposed to involve binding of MT1-MMP to other transmembrane proteins such as integrins and CD44, both of which have been shown to bind MT1-MMP in breast cancer or carcinoma cells (Deryugina et al., 2001; Marrero-Diaz et al., 2009). Therefore, we focused especially on transmembrane cell-matrix adaptors such as CD44, β1, β2, and β3 integrins, as well as integrin interactors such as talin-1 and kindlin-3. Importantly, none of the tested proteins localized to islets. This finding also indicates that the persistence of MT1-MMP at islets after podosome dissolution is not a result of its nature as a transmembrane protein, but must be based on another specific property. We also explored the possibility that islets could be sites of altered membrane curvature, as dendritic cell podosomes can act as sensors of membrane topology (van den Dries et al., 2013b), and podosomes have been shown to contain membrane curvature–associated proteins such as CIP4 (Linder et al., 2000b) and FBP17 (Tsuboi et al., 2009). However, neither protein was detectable at islets. In conclusion, MT1-MMP islets present as domains at the ventral plasma membrane that are devoid of typical podosome proteins and contain MT1-MMP as a major proteinaceous component. Their exact composition should be analyzed in future experiments.
We next explored the possibility that MT1-MMP islets could be enriched in specific lipids. Indeed, the podosome-related invadopodia have been identified as specialized membrane domains enriched in cholesterol (Caldieri and Buccione, 2010; Albrechtsen et al., 2011). Moreover, phosphatidylinositols, including PI(3,4,5) (Yu et al., 2013) and PI(3,4)P (Oikawa et al., 2008) have been implicated in invadosome formation in untransformed (Yu et al., 2013) or Src-transformed (Oikawa et al., 2008) fibroblasts. Strikingly, of all lipids tested (Table S1), only cholesterol and PI(4)P showed a clear and previously unreported enrichment at podosomes. Still, no respective accumulations were found in MT1-MMP islets, reinforcing the notion that the protease itself constitutes a major component of islets.
Surprisingly, a matrix degradation assay showed that islets are not degradative on gelatin matrix. There are several possible explanations for this. (a) MT1-MMP at islets does not cleave gelatin directly and might serve only as an activator of the gelatinase MMP-2, which needs intact podosomes as docking sites for respective vesicles. However, this is unlikely, as MT1-MMP has been identified as a bona fide gelatinase. In fact, gelatin zymography is routinely used as an assay to determine MT1-MMP activity (Evans and Itoh, 2007). (b) MT1-MMP is not active at islets. This is also unlikely, as degradative podosomes are acutely disrupted by CK-666. Islets should thus contain active MT1-MMP. Also, as shown by FRAP, there is no major exchange of MT1-MMP at islets, so MT1-MMP that is active at the time of islet appearance is expected to reside for a certain time at islets. (c) MT1-MMP at islets is potentially active but does not come into contact with matrix material. Considering that islets do not contain integrins or CD44 that establish firm and close contact with the ECM, surface-exposed MT1-MMP could be covered by the cellular glycocalyx, preventing contact with the substratum, analogous to the situation in cancer cells (Paszek et al., 2014). Support in establishing matrix contact might come from the oscillatory protrusion of podosomes (van den Dries et al., 2013a), but in the absence of podosomes, islets probably represent flat, nonprotrusive parts of the plasma membrane that are not in close contact with the matrix. Combined, these effects could explain the observed absence of degradative activity at MT1-MMP islets.
Live-cell imaging of podosome reformation, both during regular podosome turnover and disruption induced by Arp2/3 complex inhibition, showed that podosomes are often formed at MT1-MMP islets. Indeed, >60% of all disruption-induced MT1-MMP islets are reused for podosome formation within 10 min after drug washout. Moreover, analysis of regular podosome turnover also allowed the identification of a second mechanism for islet reuse in podosome formation, through recruitment of material generated by fission of adjacent podosomes. Podosome fission is a process that is typically observed in macrophages, with material being split off from the mother podosome, whereas the newly formed daughter podosome is positioned at a distance that integrates it into the podosome group (Evans et al., 2003; Kopp et al., 2006). How fission-generated podosomes are integrated into the regular podosome pattern has been unclear so far. Podosomes are mostly parts of a higher-ordered group, with actin cables forming the connections between individual podosomes (Luxenburg et al., 2007; Bhuwania et al., 2012). The typical equidistant pattern of podosomes is thus thought to be based on actomyosin contractility generated by these cables (Linder et al., 2011). We now show that also MT1-MMP islets play a role in the regulation of podosome spacing, by acting as sites of podosome reemergence or as anchoring points for fission-generated material that integrate well into the existing pattern of podosome groups.
Mutation and deletion analyses showed that the localization of MT1-MMP to podosomes critically depends on its C-terminal cytoplasmic region. Constructs deficient in either catalytic ability (E240A; Rozanov et al., 2001) or oligomerization (DTY/KAF; Tochowicz et al., 2011) localized to podosomes and islets. The former result is also in line with the presence of MT1-MMP-pHluorin at islets under pharmacological inhibition of its proteolytic activity, using the inhibitor NSC405020. It is noteworthy that NSC405020 targets the hemopexin domain interaction surfaces, thus inhibiting collagenolytic activity, but not the overall catalytic activity of MT1-MMP (Remacle et al., 2012). Moreover, deletion of the MT1-MMP N terminus had no effect on its localization to podosomes or islets. Conversely, removal of the cytoplasmic region resulted in dispersal of the respective construct along the plasma membrane. At the same time, control experiments showed that absence of the MT1-MMP C terminus had no discernible effect on localization of the protease to vesicles and its exposure on the cell surface, whereas the number of podosomes per cell was also unaffected.
Importantly, the MT1-MMP C terminus has been shown to contain F-actin binding ability (Uekita et al., 2001), which also prevents diffusion of MT1-MMP out of invadopodia (Yu et al., 2012). The ability of MT1-MMP and actin to interact directly in biochemical experiments (Yu et al., 2012), however, does not rule out a more indirect mode of interaction in cells. It is also noteworthy that podosome cores contain mostly Arp2/3 complex–generated and thus branched actin networks (Linder et al., 2011), and both podosome dissolution during regular turnover or through CK666-induced inhibition of Arp2/3 complex are expected to leave the unbranched actin cytoskeleton intact. We therefore explored the possibility that the C-terminal region of MT1-MMP could anchor the protease to the unbranched subcortical actin cytoskeleton. This idea was substantiated by global inhibition of branched and unbranched F-actin using both cytochalasin D and latrunculin A, which resulted in dissolution of islets. Moreover, upon depletion of endogenous MT1-MMP, a construct of full-length MT1-MMP deficient in F-actin binding (LLY/AAA) failed to form islets. Collectively, these experiments showed that the F-actin binding ability of MT1-MMP is critical for localization of the protease to both podosomes and islets. Additional interactions with podosome components such as cortactin (Artym et al., 2006) or p130Cas (Pan et al., 2011) may support MT1-MMP localization at podosomes. However, upon podosome dissolution, MT1-MMP islets are strictly dependent for stabilization on binding of the MT1-MMP C terminus to the cortical actin cytoskeleton. In addition, oligomerization of MT1-MMP, both by N- (Tochowicz et al., 2011) and C-terminal parts (Rozanov et al., 2001) of the molecule may support the coherence of islets.
F-actin binding could also explain the observed role of MT1-MMP islets in recruiting core material generated through podosome fission. F-actin binding of individual MT1-MMP C termini within islets is most likely dynamic, and a subset of binding sites are thus expected to be unoccupied at a given time point. These free binding sites could act as anchoring points for F-actin cores generated by fission. Fission-independent reformation of podosomes may be based on a similar mechanism, as MT1-MMP C termini in islets could bind short cytoplasmic actin filaments, which could act as local concentration points for Arp2/3 complex and actin-associated adaptor proteins such as Tks5 (Seals et al., 2005; Oikawa et al., 2008) or cortactin (Artym et al., 2006), thus facilitating local nucleation of branched actin networks and recruitment of further components. In addition, the MT1-MMP C terminus is also a substrate for Src kinase (Nyalendo et al., 2007) and could thus directly concentrate one of the most important upstream regulators of podosomes (Linder and Aepfelbacher, 2003; Oikawa et al., 2008). Ultimately, all of these potential mechanisms could contribute to MT1-MMP–based reformation of podosomes.
Our data thus reveal a novel function for MT1-MMP at podosomes that is independent of its proteolytic activity. MT1-MMP therefore plays crucial roles not only at the endpoint of podosome assembly, by enabling degradation of ECM material, but also by functioning as a subcellular signpost that facilitates formation or anchoring of nascent podosomes at sites of previously disassembled structures.
In conclusion, we report a previously unrecognized localization of MT1-MMP at podosome-free islets at the ventral plasma membrane of primary human macrophages. MT1-MMP islets become apparent upon dissolution of podosomes, and the existence of podosomes is a prerequisite for islet formation. Importantly, MT1-MMP islets serve as sites for podosome reemergence, either by de novo actin nucleation or by recruiting material generated through podosome fission. MT1-MMP islets enable efficient reformation of podosomes as well as upkeep of the typical equidistant pattern within podosome groups. MT1-MMP islets thus function as cellular memory devices that facilitate the continuous and coordinated ability of macrophages to locally degrade and invade the ECM.
Materials and methods
Cell isolation, cell culture, and transfection
Human peripheral blood monocytes were isolated from buffy coats (provided by F. Bentzien, University Medical Center Hamburg-Eppendorf, Hamburg, Germany) and differentiated into macrophages as described previously (Linder et al., 1999). Cells were cultured in RPMI containing 20% autologous serum at 37°C, 5% CO2, and 90% humidity. Monocytes were differentiated for at least 7 d, under addition of 20% human autologous serum. At days 7–10 of culture, macrophages were transiently transfected using the Neon Transfection System (Thermo Fisher Scientific) with standard settings (1,000 V, 40 ms, two pulses).
Antibodies, constructs and reagents
Antibodies were purchased from the following companies: mouse monoclonal anti-MT1-MMP and anti–β-actin from EMD Millipore, rabbit polyclonal anti-Tks5, mouse anti-β3 integrin and mouse anti–α-actinin from Santa Cruz Biotechnology, Inc., mouse anti–β1 integrin from Transduction Labs, mouse anti-vinculin from Sigma-Aldrich, mouse anti-CD44 from Cell Signaling Technology, and mouse anti-Arp2 from Abcam. Rabbit polyclonal anti-Src antibody was purchased from Proteintech. Rabbit polyclonal anti-CIP-4 antibody and pEGFP-CIP4 were gifts from P. Aspenström (Karolinska Institute, Sweden). pLifeact-TagGFP2 and pLifeact-TagRFP2 were purchased from Ibidi. mCherry-flotillin-1 and -2 constructs were gifts from C. Gauthier-Rouvière (Centre de Recherche de Biochimie Macromoléculaire, Montpellier, France); Akt-PH-GFP, PLCδ1-PH-GFP, and TAPP1-PH-GFP were provided by T. Balla (National Institutes of Health, Bethesda, MD); mCherry-Talin1C was provided by W. Ziegler (Hannover Medical School, Hannover, Germany); mTagRFP-T-C-Src-7 was purchased from Addgene (plasmid #58006; deposited by M. Davidson, Florida State University, Tallahassee, FL); Tks5-GFP was a gift from S. Courtneidge (Oregon Health and Science University, Portland, OR); GFP-Rab22aQ64L was provided by J. Donaldson (National Institutes of Health, Bethesda, MD); ILK-GFP, β3 integrin-RFP, and GFP-kindlin-3 were gifts from R. Fässler (Max Planck Institute Martinsried, Munich, Germany); GFP-Rab14 Q70L was a gift from R.H. Scheller (Genentech, San Francisco, CA); ApoE-GFP was a gift from M. Kockx (University of Sydney, Sydney, Australia); and GFP–α-actinin was a gift from M. Gimona (Paracelsus University, Salzburg, Austria). MT1-MMP-mCherry and MT1-MMP-pHluorin, both based on the human MT1-MMP sequence, were provided by P. Chavrier (Institute Curie, Paris, France). Respective mutants were created by introducing point mutations (QuikChange Site-Directed Mutagenesis kit; Stratagene) with the following primers for MT1-MMP-pHluorin E240A: (forward [F]: 5′-TTCCTGGTGGCTGTGCACGCCCTGGGCCATGCCCTGGGGC-3′, reverse [R]: 5′-GCCCCAGGGCATGGCCCAGGGCGTGCACAGCCACCAGGAA-3′). MT1-MMP-pHluorin DTY/KAF was generated by introducing three successive point mutations using the following primers: F: 5′-ATTCGTCTTCTTCAAAGGAAAGAAGCATTGGGTGTTTGAT-3′, R: 5′-ATCAAACACCCAATGCTTCTTTCCTTTGAAGAAGACGAAT-3′; F: 5′-TGGGCCGAGGGCTGCCTGCCGACAAGATTGATGCTGCTCT-3′, R: 5′-AGAGCAGCATCAATCTTGTCGGCAGGCAGCCCTCGGCCCA-3′; and F: 5′-CTTCCGTGGAAACAAGTACTTCCGTTTCAACGAAGAGCTC-3′, R: 5′-GAGCTCTTCGTTGAAACGGTAGTACTTGTTTCCACGGAAG-3′. MT1-MMP ΔCterm-pHluorin was generated by the addition of a stop codon after the pHluorin tag using F: 5′-CCTTGCAGTCTTCTTCTTCTGACGCCATGGGACCCCCAGG-3′, R: 5′-CCTGGGGGTCCCATGGCGTCAGAAGAAGAAGACTGCAAGG-3′; and MT1-MMP LLY/AAA-pHluorin was generated using F: 5′-TGGGACCCCCAGGCGAGCGGCCGCCTGCCAGCGTTCCCTG-3′, R: 5′-CAGGGAACGCTGGCAGGCGGCCGCTCGCCTGGGGGTCCCA-3′. MT1-MMP ΔNterm-pHluorin was generated by the insertion of a HindIII restriction site before the pHluorin tag using F: 5′-ATATATTAAATAAGCTTCATGAGTAAAGGAGAA-3′, R: 5′-TAGATAGGGCCCTCTAGATCAGACCTTGTCC-3′; the amplified fragment was digested with HindIII and XbaI and inserted into pcDNA 3.1. NCS 405020 was purchased from APExBIO, RGD peptide was purchased from Sigma-Aldrich, and cytochalasin D and latrunculin A were purchased from EMD Millipore.
siRNA, siRNA-insensitive constructs, and podosome reformation assay
MT1-MMP-specific siRNA was 5′-AACAGGCAAGCGTGATGCAGA-3′, with control siRNA 5′-AGGTAGTGTAACCGCCTTGTT-3′ targeting firefly luciferase (Wiesner et al., 2010, 2013). siRNA-insensitive MT1-MMP ΔCterm-pHluorin and MT1-MMP LLY/AAA-pHluorin were generated by introducing two successive mutations, first with F: 5′-CTTGCAAGTAACTGGCAAAGCTG-3′, R: 5′-CAGCTTTGCCAGTTACTTGCAAG-3′; followed by F: 5′-GGCAAAGCAGATGCAGACACCAT-3′, R: 5′-ATGGTGTCTGCATCTGCTTTGCC-3′.
Podosome reformation assay
Podosome reformation was performed 72 h after siRNA transfection (Cervero et al., 2013). For live-cell imaging, siRNA transfected cells were retransfected after 48 h with both siRNA and Lifeact-GFP and seeded on glass-bottom live-cell dishes for 24 h before imaging. Podosome reformation was analyzed in the live-cell videos. Numbers were processed in Excel 2013 (Microsoft) and GraphPad Prism 6.
Immunostaining
Macrophages were seeded at a density of 105 cells per glass coverslip (12-mm diameter) and fixed for 10 min in 3.7% formaldehyde, washed three times in PBS, and permeabilized for 10 min in 0.5% Triton X-100. After three washes with PBS, the cells were incubated for 30 min in blocking solution (1% NHS and NGS in PBS), washed briefly in PBS, and incubated for 60 min in the primary antibody solution. Cells were washed three times in PBS and then incubated for 30 min in secondary antibody solution, supplemented with fluorescence-labeled phalloidin when indicated. After three washes in PBS, the coverslips were kept in PBS for imaging.
Microscopy
Images of fixed samples were acquired with a confocal laser-scanning microscope (DMI 6000 with a TCS SP5 AOBS confocal point scanner; Leica) equipped with a oil-immersion HCX PL APO 63× NA 1.4–0.6 objective.
Live-cell imaging
Cells were imaged in RPMI 1640 medium at 37°C. Images were acquired with a spinning disk confocal system (spinning disc CSU22) fitted on an eclipse Ti microscope (Nikon) with oil immersion Plan-Apo 63× NA 1.4 objective and a charge coupled device camera (EM-CCD C-9100-2). Acquisition and processing of images was performed with Volocity Software (Improvision). Cells were seeded on glass-bottomed dishes (35 mm; Ibidi) at a density of 2 × 105 and incubated for 20 h before the start of the experiment. TIRF imaging was performed with an iLAS TIRF unit from Visitron Systems fitted on an eclipse Ti microscope (Nikon) with oil immersion Plan-Apo 63× NA 1.45 and Plan-Apo 100× NA 1.49. To correct for drift of the microscope stage, TetraSpec Microspheres (0.1 µm, fluorescent blue/green/orange/dark red; Thermo Fisher Scientific) were used.
FRAP
Cells were transfected with MT1-MMP-pHluorin and Lifeact-RFP to visualize islets, and 20 h after transfection, single images of cells were taken every 1 s. FRAP was preceded by acquisition of 10 prebleach images; 10 circular regions of interest with individual podosomes were bleached using 20% of a 405-nm laser. Recovery images were taken for additional 120 s. FRAP analysis was performed with Volocity Software (Improvision). Data were processed with Excel 2013 and GraphPad Prism 6. Fluorescence recovery was measured for 10 individual podosomes at each time point for three cells from three donors.
Quantification of MT1-MMP surface exposure
Macrophages were seeded on glass coverslips (12-mm diameter) at a density of 105 and fixed with 3.7% formaldehyde solution (10 min), but not permeabilized. Surface MT1-MMP-mCherry was stained with anti-mCherry antibody followed by an Alexa Fluor 647–conjugated secondary antibody (Wiesner et al., 2010). Quantification of Alexa Fluor 647–based fluorescence intensity was performed using ImageJ. Values of control cells were set to 100%. For comparability, laser intensity was not changed between measurements. For each value, 3 × 30 cells were evaluated. Statistical analysis was performed with Microsoft Excel and GraphPad Prism. Differences between mean values were analyzed using the Student’s t test.
Quantification of F-actin/MT1-MMP-pHluorin colocalization
Macrophages overexpressing MT1-MMP-pHluorin were fixed and stained with phalloidin/Alexa Fluor 647. Overlap images (actin/actin or actin/MT1-MMP-pHluorin) were generated using ImageJ (colocalization plugin), with a threshold ratio of 50%. Generated images (colocalized points 8-bit) were quantified by measuring the number of pixels per area of single podosomes with ImageJ. The colocalization index was defined as the ratio of pixels per area (actin/MT1-MMP-pHluorin) and pixels per area (actin/actin) for each podosome. In total, 1,110 podosomes from four independent cells were analyzed. Statistical analysis was performed with Microsoft Excel and GraphPad Prism software. For a correlation analysis, each podosome was measured by size (diameter of core, labeled by phalloidin/Alexa Fluor 647) using ImageJ. For each podosome, correlation between size and the respective colocalization index was calculated as described in the Statistical analysis and podosome and islet tracking section.
Matrix degradation assay
Analysis of gelatin matrix degradation was performed as described previously (Wiesner et al., 2010).
Statistical analysis and podosome and islet tracking
Podosome and islet measurements and tracking were performed using Fiji (Trackmate macro) and ImageJ version 1.49. Student’s t test was performed using Microsoft Excel and GraphPad Prism software. Statistically significant differences are indicated by single/multiple asterisks (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; and *, P < 0.05) Correlation analysis was calculated and plotted using a correlation plot and linear regression line to visualize the degree of correlation. Pearson correlation coefficient (r) and its square (r2), slope, and significance (P) are shown on the plots.
Online supplemental material
Fig. S1 shows that acquisition of MT1-MMP-pHluorin correlates negatively with podosome size, that surface-exposed MT1-MMP codistributes dynamically with podosomes, and that podosomes and MT1-MMP islets show similar sizes and densities. Fig. S2 shows that the membrane lipids cholesterol and PI(4)P localize to podosomes, but not to MT1-MMP islets, that ApoE-GFP does not localize to podosomes or islets, and that MT1-MMP islets are not degradative on gelatin matrix. Fig. S3 shows the composition of reformed podosomes, the impact of microtubule disruption or dynamin inhibition on islets, and the localization of MT1-MMP mutants to islets. Fig. S4 shows that absence of the cytoplasmic region or catalytic inactivity does not influence MT1-MMP localization to vesicles or cell-surface exposure, and it shows the ability of MT1-MMP mutants to form islets. Fig. S5 shows that the LLY motif in the MT1-MMP C terminus is crucial for the recovery of regular podosome number, area, and density during podosome reformation. Video 1 shows dynamics of MT1-MMP-pHluorin and mCherry-talin-1C. Video 2 shows that MT1-MMP is present at podosomes and at podosome-free islets. Video 3 shows that podosomal MT1-MMP persists beyond disruption of the podosome structure itself. Video 4 shows that MT1-MMP islets are sites of podosome reemergence. Video 5 shows MT1-MMP islets can recruit material generated by podosome fission. Table S1 lists tested potential components of islets. Table S2 provides values for podosome reformation assays and MT1-MMP constructs. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201510043.dv.
Acknowledgments
We thank P. Aspenström for pEGFP-C1-CIP4 and anti-CIP4 antibody; T. Balla for Akt-PH-GFP, PLCδ1-PH-GFP, TAPP1-PH-GFP, and OSH2-2xPH-GFP; F. Bentzien for buffy coats; P. Chavrier for MT1-MMP-pHluorin; S. Courtneidge for Tks5-mCherry; J. Donaldson for GFP-Rab22-Q64L; R. Fässler for ILK, β3 integrin, and kindlin-3 constructs; C. Gauthier-Rouvière for mCherry-flotillin-1 and -2 constructs; M. Gimona for GFP–α-actinin; M. Kockx for ApoE-GFP; R.H. Scheller for GFP-Rab14-Q70L; W. Ziegler for mCherry-Talin-1C; A. Mordhorst for expert technical assistance; the Universitätsklinikum Eppendorf microscope facility for help with microscopy; and M. Aepfelbacher for continuous support. This work is part of the doctoral thesis of K. El Azzouzi.
This work has been supported by Deutsche Forschungsgemeinschaft (LI925/2-2), the Wilhelm Sander-Stiftung (2007.020.2 and 2014.135.1), and the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement FP7-237946 (T3Net).
The authors declare no competing financial interests.



![Figure 2. Surface-exposed MT1-MMP is present at podosomes and at podosome-free islets. (A–C) TIRF micrographs of macrophage expressing MT1-MMP-pHluorin (A, red) and mCherry-Talin-1C (B, white), with merge (C). White boxes indicate two detail regions, in which MT1-MMP is associated with podosomes (left box) and present at podosome-free islets (right box). Leading and trailing edge (e.) are indicated. (D and E). Color-coded analysis of cell shown in A–C for visualization of motile versus static structures. For both fluorescence channels (MT1-MMP-pHluorin [D] and mCherry-Talin-1C [E]), each frame was colored along the spectrum, with subsequent merge into a single image. White boxes indicate detail regions also shown enlarged below. mCherry-Talin-1C presents in varying colors, indicating regular podosome ring dynamics (E′), whereas podosome-free areas show only diffuse background (E″). Podosome-associated MT1-MMP-pHluorin also presents in varying colors, indicative of podosome core dynamics (D′). However, MT1-MMP islets present mostly in white (D″), indicating restricted lateral mobility. Dotted line indicates cell circumference. Bars: 10 µm; (insets) 1 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/213/1/10.1083_jcb.201510043/7/m_jcb_201510043_fig2.jpeg?Expires=1773777627&Signature=EXq1oR1mifxCvGfRJBYwuCnIaFby~aYRoTuJTRaUeRYTkwzrrL74ejt8CyN2-w4a9G6kJQEJA7xLahANL3z4J6DfVTVcEbCkFJV5UGOQ9OJLLqDpzLZ7EhAUX99xOUr9gMEFtlEtFZbyHpXBLvfbe5N~cHgNB7ajkLdBB5NErjZPk8xXQ5uIeJjV2Q2CtYLWp2tgn76v3WMbeb8BV-4oGkfZAN~CKkfnZpfmt83QXhP6r9RCC0JdkMmUQ4wV6HfXUcbFyO2GWz372qdDBoZT6QpbgDvZf6flT-36J7V6UPHca8dzp2oe7nwVbErID-mDVtIDu0vlnU8-1kBgqzwupg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)