Lipid droplets (LDs) in the nucleus of hepatocyte-derived cell lines were found to be associated with premyelocytic leukemia (PML) nuclear bodies (NBs) and type I nucleoplasmic reticulum (NR) or the extension of the inner nuclear membrane. Knockdown of PML isoform II (PML-II) caused a significant decrease in both nuclear LDs and type I NR, whereas overexpression of PML-II increased both. Notably, these effects were evident only in limited types of cells, in which a moderate number of nuclear LDs exist intrinsically, and PML-II was targeted not only at PML NBs, but also at the nuclear envelope, excluding lamins and SUN proteins. Knockdown of SUN proteins induced a significant increase in the type I NR and nuclear LDs, but these effects were cancelled by simultaneous knockdown of PML-II. Nuclear LDs harbored diacylglycerol O-acyltransferase 2 and CTP:phosphocholine cytidylyltransferase α and incorporated newly synthesized lipid esters. These results corroborated that PML-II plays a critical role in generating nuclear LDs in specific cell types.
Introduction
Lipid droplets (LDs) are recognized as independent organelles that exist widely in prokaryotic and eukaryotic cells (Walther and Farese, 2012; Ohsaki et al., 2014). Lipid esters composing the LD core are used for many pathways, such as β-oxidation and membrane biogenesis. In addition to these functions directly connected with lipid metabolism, LDs are likely to be engaged in other seemingly unrelated activities, such as protein storage and degradation (Fujimoto and Parton, 2011).
LDs are generally thought to form in the ER and distribute in the cytoplasm (Walther and Farese, 2012), but some LDs are observed in the nucleus (Layerenza et al., 2013; Uzbekov and Roingeard, 2013; Wang et al., 2013). It remains uncertain, however, whether the nuclear LDs are generated by some specific mechanism or whether they merely represent cytoplasmic LDs that are erratically entrapped in the nucleus. This study was designed to address this question.
To understand how nuclear LDs form, we examined where in the nucleus LDs are located and found that they are associated with two structures: the type I nucleoplasmic reticulum (NR; Malhas et al., 2011) and the premyelocytic leukemia (PML) nuclear body (NB; Bernardi and Pandolfi, 2007; Lallemand-Breitenbach and de Thé, 2010). In contrast with type II NR, which is the invagination of the entire nuclear envelope and therefore essentially refers to indentation of the nucleus, type I NR is the extension of the perinuclear cistern bound with the inner nuclear membrane (INM) alone (Fig. 1 A). Type I NR was defined only morphologically, and no particular function has been assigned to the structure.
Nuclear LDs in Huh7. Cells used in this and subsequent figures were cultured with 0.4 mM OA for 12 h if not otherwise described. (A) Scheme of type I and type II NR. (B) Serial ultrathin sections (60 nm thick) of cells cultured with 0.2 mM DHA for 2 h to enhance the contrast of membranes. The LD indicated by the arrowhead was verified to be in the nucleoplasm. (C) Immunolabeling of perilipin2 (ADRP), perilipin3 (TIP47), and Rab18. Cytoplasmic LDs were labeled for all three, whereas nuclear LDs were labeled for perilipin3 and Rab18 (arrowheads), but not for perilipin2. Proteins (red), LDs (green), nucleus (blue). The bar graph shows the ratio of labeled LDs in the nucleus and the cytoplasm in one representative experiment out of three (mean ± SEM, n ≥ 99 cells/experiment). (D) Cytoplasmic LD counts were significantly lower in clones deleted with the perilipin2 gene than in the control, but nuclear LD counts were at similar levels. Two independent clones (1 and 2) were examined. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (E) Cells expressing HRP-KDEL were processed by DAB histochemistry to delineate the ER and the nuclear envelope. Nuclear LDs showed a close association with DAB precipitates, either in linear shapes (arrowheads) or in puncta (arrows), which were thought to represent the type I NR sectioned longitudinally or vertically, respectively. See Fig. S2 A for more examples. (F) Type I NR in association with nuclear LDs (arrowhead) was observed in cells without HRP-KDEL expression by conventional EM.
Nuclear LDs in Huh7. Cells used in this and subsequent figures were cultured with 0.4 mM OA for 12 h if not otherwise described. (A) Scheme of type I and type II NR. (B) Serial ultrathin sections (60 nm thick) of cells cultured with 0.2 mM DHA for 2 h to enhance the contrast of membranes. The LD indicated by the arrowhead was verified to be in the nucleoplasm. (C) Immunolabeling of perilipin2 (ADRP), perilipin3 (TIP47), and Rab18. Cytoplasmic LDs were labeled for all three, whereas nuclear LDs were labeled for perilipin3 and Rab18 (arrowheads), but not for perilipin2. Proteins (red), LDs (green), nucleus (blue). The bar graph shows the ratio of labeled LDs in the nucleus and the cytoplasm in one representative experiment out of three (mean ± SEM, n ≥ 99 cells/experiment). (D) Cytoplasmic LD counts were significantly lower in clones deleted with the perilipin2 gene than in the control, but nuclear LD counts were at similar levels. Two independent clones (1 and 2) were examined. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (E) Cells expressing HRP-KDEL were processed by DAB histochemistry to delineate the ER and the nuclear envelope. Nuclear LDs showed a close association with DAB precipitates, either in linear shapes (arrowheads) or in puncta (arrows), which were thought to represent the type I NR sectioned longitudinally or vertically, respectively. See Fig. S2 A for more examples. (F) Type I NR in association with nuclear LDs (arrowhead) was observed in cells without HRP-KDEL expression by conventional EM.
On the other hand, the PML NB is a macromolecular structure in the nuclear matrix to which diverse functions have been ascribed, including transcriptional regulation and cellular senescence. The PML NB core is made of PML protein isoforms, which diverge in the C terminus because of alternative splicing (Bernardi and Pandolfi, 2007), and we found that PML isoform II (PML-II) plays a critical role in nuclear LD formation. The involvement of PML-II, however, was evident only in limited types of cells in which PML-II was targeted to the INM. In those cells, nuclear LDs were found frequently, and the expression of PML-II was correlated with the amount of nuclear LDs and intranuclear membranes, including type I NR. This result indicated that the nuclear LD formation is linked with both the PML NB and type I NR.
We also found that newly synthesized lipid esters are incorporated into nuclear LDs. Consistent with this, diacylglycerol O-acyltransferase 2 (DGAT2) and CTP:phosphocholine cytidylyltransferase α (CCTα), which exist in cytoplasmic LDs and catalyze critical steps of triglyceride and phosphatidylcholine synthesis, respectively (Krahmer et al., 2011; Wilfling et al., 2013), were distributed in nuclear LDs.
The results corroborated that the formation of nuclear LDs involves a specific mechanism and that they exist in specific locations within the nucleus. To our knowledge, this is the first study to show the molecular basis of nuclear LD formation and should therefore provide a solid ground on which to explore the functional roles of nuclear LDs.
Results and discussion
Nuclear LDs have properties that differ from those of cytoplasmic LDs
LDs stained by BODIPY493/503 were frequently observed in the nuclear region of Huh7, a human hepatocarcinoma cell line, but fluorescence microscopy could not determine whether those LDs actually existed in the nucleoplasm or were instead located in the cytoplasmic extension within the type II NR (Fig. 1 A). EM showed that most of those LDs were in the nucleoplasm and not in the type II NR, a result that was verified by observing serial ultrathin sections (Fig. 1 B and see Fig. S2 A for an additional EM result).
Besides Huh7, nuclear LDs were observed frequently in HepG2 and McA-RH7777 (Fig. S1 A), human and rat hepatocarcinoma cell lines, and also in mouse hepatocytes in vivo after high-fat diet feeding (Fig. S1B). In contrast, nuclear LDs were scarce in both HeLa cells and human fibroblasts, even when the cells were cultured with oleic acid (OA) to increase cytoplasmic LDs (Fig. S1 C). Even in cells with abundant cytoplasmic LDs, such as Y1 (mouse steroidogenic cell line; Buonassisi et al., 1962) and OP9 (mouse stromal cell line) differentiated to adipocytes (Wolins et al., 2006), nuclear LDs were hardly observed (Fig. S1 D).
Notably, perilipin2 (ADRP), a constitutive LD protein (Bickel et al., 2009), was virtually absent in nuclear LDs (Fig. 1 C). This result was not caused by poor antibody penetration into the nucleus, because perilipin3 and Rab18 were labeled in a substantial number of nuclear LDs (Fig. 1 C). Consistent with this result, targeted deletion of the perilipin2 gene significantly decreased cytoplasmic LDs but spared nuclear LDs (Fig. 1 D). These results suggested that nuclear LDs might form via a unique mechanism and have different properties than cytoplasmic LDs.
Nuclear LDs are associated with type I NR
To gain insight into nuclear LDs, we examined whether they are correlated with any other structures in the nucleus. First, to exhaustively observe their relationship with nuclear membranes, we used cells expressing HRP-KDEL, which is targeted at the perinuclear space and the ER lumen and can be visualized by DAB histochemistry (Connolly et al., 1994). This method clearly distinguished nuclear LDs from LDs in type II NR, showing that >90% of LDs in the nuclear region were in the nucleoplasm and not in type II NR (Fig. S2 A). More importantly, the method revealed that type I NR, which is the extension of the INM alone (Malhas et al., 2011), ran deep in the nucleoplasm and was closely associated with nuclear LDs (Figs. 1 E and S2 A). The close association of nuclear LDs and type I NR could be observed by conventional EM of Huh7 without HRP-KDEL expression (Fig. 1 F).
Nuclear LDs are associated with PML NBs
Next, we examined the correlation between nuclear LDs and nuclear bodies. Cajal bodies, speckles, and paraspeckles, which were labeled by antibodies to coilin, SC35, and PSF, respectively, did not exhibit a relationship with nuclear LDs (Fig. S2 B). In contrast, PML NBs labeled with anti-PML antibody colocalized with nuclear LDs (Fig. 2 A). Pre-embedding immuno-EM confirmed that PML is located on the surface of nuclear LDs (Fig. 2 B). Furthermore, correlative light and EM demonstrated that nuclear LDs bearing EGFP-tagged PML-I had radiating bristle-like structures, a typical morphologic feature of PML NBs, on their surface (Fig. 2 C; Lallemand-Breitenbach and de Thé, 2010). These results revealed the intimate relationship between nuclear LDs and PML NBs.
Nuclear LDs were associated with PML NBs in Huh7. (A) Colocalization of endogenous PML (red) and nuclear LDs (green; arrowheads). More than 70% of PML and >30% of nuclear LDs were associated with the other in cells cultured with OA for 12 h; the colocalization ratio per LDs increased when the number of nuclear LDs decreased in a lipoprotein-deficient medium. The bar graph shows mean ± SEM of one representative experiment out of three. n = 88 cells. (B) Pre-embedding immuno-EM. PML labeling was observed adjacent to nuclear LDs. (C) Correlative light and EM. Nuclear LDs that harbor EGFP-tagged PML-I showed a radiating bristle-like structure on the surface (arrowheads). (D) Distribution of PML isoforms (red). siRNA-resistant mCherry-PML isoform cDNAs were transfected to cells depleted of endogenous PML. Only PML-II showed prominent colocalization with nuclear LDs (green; arrowheads). The bar graphs (C–E) show mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (E) Overexpression of PML-II, but not of other isoforms, induced a significant increase in nuclear LDs compared with the empty vector control. (F) Knockdown of PML-II decreased nuclear LDs. Two siRNAs targeted at different sequences of PML-II (1 and 2) were used.
Nuclear LDs were associated with PML NBs in Huh7. (A) Colocalization of endogenous PML (red) and nuclear LDs (green; arrowheads). More than 70% of PML and >30% of nuclear LDs were associated with the other in cells cultured with OA for 12 h; the colocalization ratio per LDs increased when the number of nuclear LDs decreased in a lipoprotein-deficient medium. The bar graph shows mean ± SEM of one representative experiment out of three. n = 88 cells. (B) Pre-embedding immuno-EM. PML labeling was observed adjacent to nuclear LDs. (C) Correlative light and EM. Nuclear LDs that harbor EGFP-tagged PML-I showed a radiating bristle-like structure on the surface (arrowheads). (D) Distribution of PML isoforms (red). siRNA-resistant mCherry-PML isoform cDNAs were transfected to cells depleted of endogenous PML. Only PML-II showed prominent colocalization with nuclear LDs (green; arrowheads). The bar graphs (C–E) show mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (E) Overexpression of PML-II, but not of other isoforms, induced a significant increase in nuclear LDs compared with the empty vector control. (F) Knockdown of PML-II decreased nuclear LDs. Two siRNAs targeted at different sequences of PML-II (1 and 2) were used.
PML-II plays a critical role in nuclear LD formation
The association of nuclear LDs and PML NBs prompted us to examine whether PML proteins play any role in nuclear LD formation. Human PML proteins have seven isoforms, i.e., PML-I to PML-VII, which hetero-oligomerize to make PML NBs (Bernardi and Pandolfi, 2007). When EGFP-tagged PML isoforms were expressed in Huh7, all of the isoforms exhibited localization around nuclear LDs (Fig. S3 A), but the result was likely to be affected by hetero-oligomerization with endogenous PML proteins. Therefore, to clearly examine isoform specificity, endogenous PML was knocked down by RNAi, and individual mCherry-tagged PML isoforms were expressed by transfection of siRNA-resistant cDNA. Under these conditions, PML-II showed by far the most frequent colocalization with nuclear LDs (Fig. 2 D), and it was the only isoform that induced a significant increase in nuclear LDs (Fig. 2 E). In contrast, knockdown of PML-II caused a significant decrease in nuclear LDs (Fig. 2 F). These results indicated that PML-II plays a critical role in generating nuclear LDs.
Formation of nuclear LDs is correlated with the PML-II patch formation
The function of PML-II in the nuclear LD formation was thought to be related to its unique C-terminal domain, which contains the nuclear periphery-binding motif (653–681) and the extra nuclear localization motif (717–767; Fig. 3 A; Jensen et al., 2001; Jul-Larsen et al., 2010). The nuclear periphery-binding motif was shown to mediate distribution of PML-II along the nuclear envelope, which will be referred to as the PML-II patch hereafter (Fig. 3 A, arrows; Jul-Larsen et al., 2010). By transfecting siRNA-resistant cDNA after PML-II knockdown, both PML-II (1–716) and the full-length PML-II formed the PML-II patch and induced an increase in nuclear LDs, whereas PML-II (1–652) did not (Fig. 3 B). These results suggested that the ability of PML-II to increase nuclear LDs is correlated with its binding to the nuclear envelope.
Correlation of the PML-II patch and nuclear LDs in Huh7. (A) Distribution of PML-II truncation mutants in cells depleted of endogenous PML-II. PML-II (FL, full length) and PML-II (1–716) showed colocalization with nuclear LDs (green; arrowheads) and linear distribution in the nuclear periphery (arrows), whereas PML-II (1–652) did not show either. (B) PML-II (FL) and PML-II (1–716), but not PML-II (1–652), induced a significant increase in nuclear LDs. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (C) The PML-II patch and nuclear LDs formed concomitantly in a cell type–specific manner. mCherry-PML-II was targeted at both LD-associated PML NBs (arrowheads) and the nuclear periphery (arrows) in Huh7 and U2OS, but mCherry-PML-II distributed only to PML NBs in Y1 and HeLa. (D) Pre-embedding immuno-EM of GFP–PML-II. The labeling was observed on the nucleoplasmic side of the INM. ONM, outer nuclear membrane. (E) LBR and SUN1 were excluded from the PML-II patch region of the nuclear envelope (arrowheads).
Correlation of the PML-II patch and nuclear LDs in Huh7. (A) Distribution of PML-II truncation mutants in cells depleted of endogenous PML-II. PML-II (FL, full length) and PML-II (1–716) showed colocalization with nuclear LDs (green; arrowheads) and linear distribution in the nuclear periphery (arrows), whereas PML-II (1–652) did not show either. (B) PML-II (FL) and PML-II (1–716), but not PML-II (1–652), induced a significant increase in nuclear LDs. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (C) The PML-II patch and nuclear LDs formed concomitantly in a cell type–specific manner. mCherry-PML-II was targeted at both LD-associated PML NBs (arrowheads) and the nuclear periphery (arrows) in Huh7 and U2OS, but mCherry-PML-II distributed only to PML NBs in Y1 and HeLa. (D) Pre-embedding immuno-EM of GFP–PML-II. The labeling was observed on the nucleoplasmic side of the INM. ONM, outer nuclear membrane. (E) LBR and SUN1 were excluded from the PML-II patch region of the nuclear envelope (arrowheads).
Interestingly, the PML-II patch formation occurred only in limited types of cells, i.e., U2OS (human osteosarcoma; Jul-Larsen et al., 2010) and several cells of hepatic origin (this study). Those cells showed frequent nuclear LDs, whereas other cells with few nuclear LDs, e.g., HeLa, did not form the PML-II patch (Fig. 3 C). Importantly, in the latter group of cells, even overexpression of PML-II did not increase nuclear LDs, and GFP–PML-II was not associated with the few existing nuclear LDs. These results reinforce the idea that formation of the PML-II patch and nuclear LDs is correlated. The molecular basis of the cell-type specificity of the PML-II patch formation is not known, but it may not be surprising considering the highly variable proteome of the nuclear envelope (de Las Heras et al., 2013).
PML-II patch and intranuclear membranes
The aforementioned results showed that nuclear LDs are associated with type I NR and PML NBs and that PML-II is involved in nuclear LD formation. To understand the mechanistic connection between these observations, we focused on the PML-II patch. Using immuno-EM, PML-II was observed along the nucleoplasmic surface of the INM (Fig. 3 D), which could explain the defect in the nuclear lamina in the PML-II patch (Fig. S3 B; Jul-Larsen et al., 2010). In addition, we found that lamin B receptor (LBR) and SUN1, transmembrane proteins of INM that are linked with lamin B and lamin A, respectively (Olins et al., 2010; Chang et al., 2015), were also excluded from the PML-II patch (Fig. 3 E).
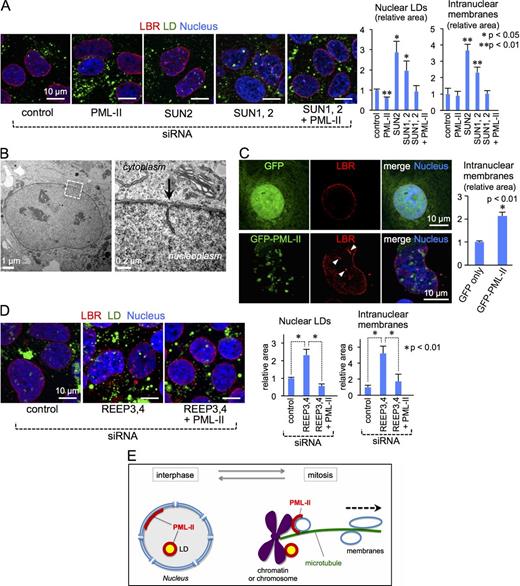
We suspected that depletion of lamins and/or the INM proteins may be conducive to the increase in nuclear LDs, but the knockdown of lamin A/C, lamin B1, and lamin B2, either singly or in combination, did not cause such an increase. On the other hand, the knockdown of SUN proteins significantly increased nuclear LDs (Fig. 4 A). Furthermore, SUN knockdown also increased LBR-positive linear labels within the nucleus (Fig. 4 A), and EM indicated that many of them are type I NR (Figs. 4 B and S3 C). (Based on these results, the LBR labeling inside the nucleus will be hereafter referred to as intranuclear membranes.) The increase in intranuclear membranes was thought to occur because membrane removal from chromatin in early prometaphase was down-regulated by SUN knockdown (Turgay et al., 2014). Notably, the increase in both nuclear LDs and intranuclear membranes by SUN knockdown was cancelled when PML-II was knocked down simultaneously (Fig. 4 A), indicating that PML-II functions upstream of SUN proteins.
Correlation of PML-II, nuclear LDs, and intranuclear membranes. (A) Knockdown of SUN2 alone or both SUN1 and SUN2 in Huh7 caused increases in nuclear LDs (green) and intranuclear membranes labeled for LBR (red), but the increases were cancelled by additional knockdown of PML-II. In this experiment, OA was not added to the culture medium. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (B) Type I NR observed in Huh7 after SUN2 knockdown. See Fig. S3 C for more examples. (C) Overexpression of PML-II in Huh7 significantly increased intranuclear membranes labeled for LBR (arrowheads). The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (D) Knockdown of REEP3/4 in Huh7 significantly increased nuclear LDs (green) and intranuclear membranes (red), but the increases were not observed when both REEP3/4 and PML-II were knocked down. Cells were cultured without OA. (E) Correlation between PML-II and nuclear LDs. In most cell types, LDs and membranes are removed from chromatin or chromosome during mitosis by the mechanism involving SUN and REEP proteins. In the limited types of cells forming the PML-II patch (e.g., Huh7), LDs and membranes are potently tethered to chromatin by PML-II, leading to their frequent presence in the interphase nucleus. As shown in Fig. 5, lipid esters are thought to be synthesized in these intranuclear LDs and membranes.
Correlation of PML-II, nuclear LDs, and intranuclear membranes. (A) Knockdown of SUN2 alone or both SUN1 and SUN2 in Huh7 caused increases in nuclear LDs (green) and intranuclear membranes labeled for LBR (red), but the increases were cancelled by additional knockdown of PML-II. In this experiment, OA was not added to the culture medium. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (B) Type I NR observed in Huh7 after SUN2 knockdown. See Fig. S3 C for more examples. (C) Overexpression of PML-II in Huh7 significantly increased intranuclear membranes labeled for LBR (arrowheads). The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (D) Knockdown of REEP3/4 in Huh7 significantly increased nuclear LDs (green) and intranuclear membranes (red), but the increases were not observed when both REEP3/4 and PML-II were knocked down. Cells were cultured without OA. (E) Correlation between PML-II and nuclear LDs. In most cell types, LDs and membranes are removed from chromatin or chromosome during mitosis by the mechanism involving SUN and REEP proteins. In the limited types of cells forming the PML-II patch (e.g., Huh7), LDs and membranes are potently tethered to chromatin by PML-II, leading to their frequent presence in the interphase nucleus. As shown in Fig. 5, lipid esters are thought to be synthesized in these intranuclear LDs and membranes.
Based on these results and the observation that PML NBs are linked to chromatin (Wang et al., 2004; Chen et al., 2008), we hypothesized that PML-II tethers LDs and membranes to chromatin and that this activity is enhanced in cell types where PML-II binds to the nuclear envelope to form the PML-II patch (Fig. 4 E). This can explain why nuclear LDs and intranuclear membranes occur intrinsically in cells such as Huh7 despite the presence of SUN proteins.
This hypothesis was supported by the following observations. First, overexpression of PML-II induced an increase in both intranuclear membranes and nuclear LDs (Fig. 4 C), and this effect was evident only in cell types forming the PML-II patch (i.e., Huh7 and U2OS) but not in others (e.g., HeLa; Fig. 3 C). Second, an increase in nuclear LDs and intranuclear membranes that was induced by REEP3/4 was also cancelled by simultaneous PML-II knockdown (Fig. 4 D). REEP3/4 functions to clear membranes from the metaphase chromosome (Schlaitz et al., 2013), i.e., in a different phase and therefore most likely by a different mechanism from that of SUN proteins; therefore, the result reinforced the idea that PML-II works as a tether for LDs and membranes. We therefore inferred that enhancement of this PML-II functionality in cells such as Huh7 and U2OS counteracts the removal of LDs and membranes from chromatin/chromosome during mitosis, leading to their presence in the interphase nucleus (Fig. 4 E).
LDs grow inside the nucleus
The aforementioned results suggested that the formation of nuclear LDs proceeds independently from that of cytoplasmic LDs. To test whether lipid esters are directly incorporated into nuclear LDs, Huh7 cells were given a fluorescent fatty acid analog, BODIPY558/568-C12, to metabolically label newly synthesized lipid esters (Kasurinen, 1992). The result showed that nuclear LDs were labeled with an intensity similar to that of cytoplasmic LDs, even when mitosis was inhibited by hydroxyurea, indicating that lipid esters were incorporated into LDs in the interphase nucleus (Fig. 5 A).
Nuclear LDs in Huh7 grow by incorporating newly synthesized lipid esters. (A) Fluorescence microscopic assay. Cells were incubated for 1 h with a fluorescence fatty acid analog BODIPY558/568-C12 (red) and chased for another hour, all in the presence of 1.5 mM hydroxyurea to inhibit mitosis. After fixation, LDs were labeled with BODIPY493/503 (green; top). BODIPY558/568-positive LDs did not appear when treated with 5 µM triacsin C (bottom), which confirmed that the analog was incorporated into LDs as lipid esters. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (B) EM assay. Cells were cultured with either 0.4 mM OA for 17 h (a) or 0.4 mM OA for 9 h and then with 0.2 mM DHA for 8 h (b and c), without (b) or with (c) the presence of 1.5 mM hydroxyurea. The electron density of nuclear LDs was increased by DHA to the same degree as that of cytoplasmic LDs, irrespective of the hydroxyurea treatment. The bar graph shows mean ± SD of pooled data from four independent experiments. n = 156 (cytoplasmic LD) and n = 216 (nuclear LD). (C) DGAT2 in the nucleus. Venus-DGAT2 (green) showed (a) colocalization with RFP-LBR (red) and (b) distribution around nuclear LDs (red; arrowheads). Some of those LDs were positively labeled for PML (blue), which confirmed their presence in the nucleoplasm. (D) CCTα in the nucleus. Endogenous CCTα (red) was labeled around nuclear LDs (green; arrowheads), some of which were also labeled positively for PML (blue). CCTα-EGFP showed a similar distribution (Fig. S3 D).
Nuclear LDs in Huh7 grow by incorporating newly synthesized lipid esters. (A) Fluorescence microscopic assay. Cells were incubated for 1 h with a fluorescence fatty acid analog BODIPY558/568-C12 (red) and chased for another hour, all in the presence of 1.5 mM hydroxyurea to inhibit mitosis. After fixation, LDs were labeled with BODIPY493/503 (green; top). BODIPY558/568-positive LDs did not appear when treated with 5 µM triacsin C (bottom), which confirmed that the analog was incorporated into LDs as lipid esters. The bar graph shows mean ± SD of pooled data from three independent experiments. n ≥ 100 cells/experiment. (B) EM assay. Cells were cultured with either 0.4 mM OA for 17 h (a) or 0.4 mM OA for 9 h and then with 0.2 mM DHA for 8 h (b and c), without (b) or with (c) the presence of 1.5 mM hydroxyurea. The electron density of nuclear LDs was increased by DHA to the same degree as that of cytoplasmic LDs, irrespective of the hydroxyurea treatment. The bar graph shows mean ± SD of pooled data from four independent experiments. n = 156 (cytoplasmic LD) and n = 216 (nuclear LD). (C) DGAT2 in the nucleus. Venus-DGAT2 (green) showed (a) colocalization with RFP-LBR (red) and (b) distribution around nuclear LDs (red; arrowheads). Some of those LDs were positively labeled for PML (blue), which confirmed their presence in the nucleoplasm. (D) CCTα in the nucleus. Endogenous CCTα (red) was labeled around nuclear LDs (green; arrowheads), some of which were also labeled positively for PML (blue). CCTα-EGFP showed a similar distribution (Fig. S3 D).
The result was further tested by EM using docosahexaenoic acid (DHA) to impart a higher electron density to newly synthesized lipid esters (Cheng et al., 2009). With or without hydroxyurea, cytoplasmic and nuclear LDs showed an equivalent increase in electron density (Fig. 5 B), verifying that newly synthesized lipid esters were added to nuclear LDs directly.
The aforementioned observation was further substantiated by the presence of DGAT2 in intranuclear membranes and nuclear LDs (Fig. 5 C), which seemingly corresponded to its distribution in the ER and cytoplasmic LDs (Wilfling et al., 2013). Together with the nuclear presence of diacylglycerol and acyl-CoA (Elholm et al., 2000; Irvine, 2003), which are substrates of DGAT2, this result indicated that triglycerides could be synthesized in the nucleus. Furthermore, CCTα, the penultimate enzyme of the phosphatidylcholine synthetic Kennedy pathway that was reported to exist in cytoplasmic LDs (Krahmer et al., 2011), was also found around nuclear LDs (Figs. 5 D and S3 D). These results corroborated the hypothesis that nuclear LDs can grow by incorporating lipids synthesized in the nucleus.
Significance of nuclear LDs
LDs in the nucleus must have been observed by many researchers, but how nuclear LDs are generated and why their frequency differs drastically among different cell types has not been clarified. This study identified PML-II as a critical factor in nuclear LD formation and found that the diversity among cell types is related to the PML-II patch-forming property. Understanding the molecular mechanism by which PML-II binds to the INM only in limited types of cells should be important in fully understanding PML-II functionality in nuclear LD formation.
The functional significance of nuclear LDs being associated with PML NBs also remains to be clarified, but it is tempting to speculate that nuclear LDs may fine-tune the fatty acid concentration around PML NBs and regulate nuclear receptors, such as peroxisome proliferator-activated receptors (Georgiadi and Kersten, 2012). Considering that PML is a potent activator of fatty acid oxidation (Carracedo et al., 2012), the effects of nuclear LDs on the PML NB function in lipid metabolism warrant further studies. From a pathological viewpoint, it is noteworthy that adenovirus 5 E4 Orf3 binds to the nuclear periphery-binding motif of PML-II, causing disruption of PML NBs (Leppard et al., 2009) and activation of viral gene expression (Berscheminski et al., 2013). The significance of nuclear LDs also needs to be studied from these perspectives.
Materials and methods
Antibodies
Rabbit anti–human Rab18 (Ozeki et al., 2005) and rabbit anti–human TIP47 antibodies (Ohsaki et al., 2006) were prepared as described. Rabbit anti–human PML (Xu et al., 2005) and rabbit anti-CCTα (Gehrig et al., 2008) were donated by K.-S. Chang (The University of Texas MD Anderson Cancer Center, Houston, TX) and N. Ridgway (Dalhousie University, Halifax, Nova Scotia), respectively. Mouse anti–human lamin A/C (4C11; Cell Signaling), rabbit anti-coilin (H-300; Santa Cruz Biotechnology, Inc.), mouse anti-SC35 (S4045), mouse anti-FLAG (M2; Sigma-Aldrich), and mouse anti–human perilipin2 (AP125; Progen) antibodies were obtained from the respective suppliers. Cy3-conjugated donkey anti–rabbit IgG, Cy3-conjugated donkey anti–mouse IgG, Alexa Fluor 647–conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories), and Alexa Fluor 594 FluoroNanogold-conjugated anti–rabbit IgG (Nanoprobes) antibodies were also purchased.
Cell lines
Huh7, HepG2, McA-RH7777, Y1, and HeLa were obtained from the Japanese Collection of Research Bioresources Cell Bank and cultured in DMEM supplemented with 10% FBS and antibiotics at 37°C in a humidified atmosphere of 95% air/5% CO2. Human fibroblasts (H34) and U2OS were donated by H. Ninomiya (Tottori University, Tottori, Japan) and H. Kiyoi (Nagoya University, Nagoya, Japan), respectively. In some experiments, cells were cultured with the indicated concentration of OA or DHA (Sigma-Aldrich) in complex with fatty acid–free BSA (Wako) at a molar ratio of 6:1. Differentiation of Y1 was induced by culturing in DMEM/Ham’s F-12 with 15% horse serum, 2.5% FBS, and 0.4 mM OA (Brasaemle et al., 1997). OP9, provided by K. Kitajima (Tokyo Metropolitan Institute for Medical Science, Tokyo, Japan), was maintained in MEM supplemented with 20% FBS and induced to differentiate by culturing in MEM with 0.2% FBS, 175 nM insulin, and 0.9 mM OA (Wolins et al., 2006).
Mouse hepatocyte
Female ddY mice were fed a high-fat diet (14.4% fat; Quick Fat; CLEA Japan) for 15 wk, and liver tissues were processed for EM observation as described (Yamamura et al., 2014). The experiment conformed to the Guidelines for Proper Conduct of Animal Experiments of the Science Council of Japan and was approved by the Animal Experimentation Committee of the Nagoya University Graduate School of Medicine (approval ID 23432).
Plasmids
pEGFP-C1 and pmCherry-C1 were obtained from Clontech. Plasmid vectors for EGFP-C3-PML (I, II, III, IV, V, and VI; Weidtkamp-Peters et al., 2008), HRP-KDEL (Connolly et al., 1994), mRFP-LBR (Haraguchi et al., 2000), siRNA-resistant HA-REEP4 (Schlaitz et al., 2013), Venus-mouse DGAT2 (Xu et al., 2012), and CCTα-EGFP (Gehrig et al., 2008) were donated by P. Hemmerich (Fritz-Lipmann Institute, Jena, Germany), C. Hopkins (University College London, London, England, UK), T. Haraguchi (Advanced ICT Research Institute, Tokyo, Japan), A.-L. Schlaitz (University of Heidelberg, Heidelberg, Germany), H.Y. Mak (Hong Kong University of Science and Technology, Hong Kong, China), and N. Ridgway, respectively. Plasmids carrying siRNA-resistant cDNA for mCherry-tagged PML isoforms and PML-II deletion mutants were prepared by PCR. siRNA-resistant cDNAs were prepared by site-directed mutagenesis using Pfu Turbo polymerase (Agilent Technologies).
siRNA
siRNAs were synthesized by Japan Bio Service. Target nucleotide sequences of human genes for siRNAs are as follows: scramble control, 5′-AUCCGCGCGAUAGUACGUATT-3′; PML (PML-I 1,181–1,201), 5′-GAUGCAGCUGUAUCCAAGATT-3′ (Nojima et al., 2009); PML-II 1 (PML-II 2,131–2,149), 5′-CAUCCUGCCCAGCUGCAAATT-3′ (Xu et al., 2005); PML-II 2 (PML-II 2,232–2,250), 5′-GUGGCUCAACAACUUUUUUTT-3′; REEP3/4 (REEP4 or REEP3 123–141), 5′-CTGGATTGTTTTTGCACTCTT-3′ (Schlaitz et al., 2013); SUN1 (SUN1 845–863), 5′-UUACCAGGUGCCUUCGAAATT-3′; and SUN2 (SUN2 1,550–1,568), 5′-UGGCAGAGAUGCAGGGCAATT-3′ (Turgay et al., 2014).
Transfection
Cells were transfected with cDNA and siRNA using Lipofectamine 2000 and Lipofectamine RNAiMAX (Invitrogen), respectively. They were analyzed for 2 (cDNA) and 3 d (siRNA) after transfection. For studies using siRNA and siRNA-resistant cDNA, cells were transfected with siRNA on day 0, transfected with siRNA-resistant cDNA on day 2, and analyzed on day 3.
Gene knockout by CRISPR-Cas9 system
Plin2 in Huh7 cells was selectively disrupted according to the standard protocol (Ran et al., 2013). Guide RNA sequences in exon 4 (mRNA 527–549, accession no. NM_001122) that are commonly used in two transcriptional variants were selected using web-based prediction software (CRISPRdirect) and cloned in a Cas9-guide RNA packaged vector, pSpCas9(BB)-2A-GFP (Addgene). The lack of perlipin2 protein expression in two independent cell clones was verified by Western blotting.
Immunofluorescence microscopy and data analysis
Cells were fixed with 3% formaldehyde alone or 3% formaldehyde and 0.015% glutaraldehyde in 0.1 M phosphate buffer for 15 min and permeabilized with 0.01% digitonin in PBS for 30 min (for Rab18), 90% methanol for 10 min at −20°C (for perilipin2 and perilipin3), or 0.5% Triton X-100 in PBS for 10 min (for other proteins). Cells were then incubated in 3% BSA in PBS (for labeling of perilipin2, perilipin3, and Rab18) or 5% normal goat serum in PBS (for other proteins) for 30 min before incubating with primary antibodies. After incubation with secondary antibodies, nuclei were labeled with Hoechst 33342 (Sigma-Aldrich), and LDs were stained with BODIPY493/503, LipidTox Red, or LipidTox DeepRed (Invitrogen) according to the manufacturer’s instructions. Samples were mounted in Mowiol 4–88 to which 2.5% DABCO had been added.
Images of random areas were captured by a confocal laser scanning microscope A1 (Nikon) equipped with a GaASP multidetector using an PlanApo 100× lens with a 1.45 NA, an Axiovert 200 M microscope equipped with an AxioCam MRm CCD camera, and Apotome (Carl Zeiss) using an Apochromat 63× lens with a 1.40 NA or an Axio Imager 2 equipped with an AxioCam506 mono camera and Apotome2 (Carl Zeiss) using a Plan-Neofluar 100× lens with a 1.30 NA. Image acquisition software included NIS-Elements, AxioVision (rel. 4.8), and Zen 2 pro (Carl Zeiss), respectively. The color, brightness, and contrast of presented images were adjusted to the entire images using Adobe Photoshop, version CS3 (Adobe).
For quantification, results from three independent experiments were averaged unless stated otherwise. The number of LDs was counted manually, and the area of interest was measured using ImageJ (National Institutes of Health). The relative area of nuclear LDs was calculated by dividing the sum of all nuclear LD areas by the nuclear area, whereas that of intranuclear membranes was obtained by dividing the sum of the LBR-positive area within the nucleus by the nuclear area. The statistical difference between different experimental groups was examined by t test, unpaired and two-tailed. P-values <0.05 were considered significant.
Incorporation of newly synthesized lipid esters into the existing LDs
For fluorescence microscopic analysis, Huh7 cells precultured with 0.4 mM OA for 12 h were treated with BODIPY 558/568-C12 (Invitrogen) for 1 h to metabolically label newly synthesized lipid esters, rinsed, and cultured for another hour. For the last 2 h of incubation, 1.5 mM hydroxyurea (Sigma-Aldrich) was added to the culture medium with or without 5 µM triacsin C (Santa Cruz Biotechnology, Inc.). After fixation, cells were incubated with BODIPY 493/503 to stain all LDs. For the EM method to visualize newly synthesized lipid esters, cells pretreated with OA for 12 h were cultured with 0.2 mM DHA (Sigma-Aldrich) for 8 h before fixation (Cheng et al., 2009).
EM
For genuine morphologic observation, cells cultured on coverslips were fixed with a mixture of 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for >2 h, postfixed with a mixture of 1% osmium tetroxide and 0.1% potassium ferrocyanide in 0.1 M sodium cacodylate buffer (White et al., 1979), dehydrated, and embedded in epoxy resin. Ultrathin sections were observed using a JEM1011 electron microscope (JEOL) operated at 100 kV. Images were collected using an Orius CCD camera SC200 (Gatan) and DigitalMicrograph software, version 1.83.842 (Gatan). Brightness and contrast were adjusted to the entire images using Adobe Photoshop, version CS3.
The electron density of LDs was quantified by establishing a scale with two reference points in the same micrograph (Cheng et al., 2009). That is, the specimen grid bar for the maximum density (or no transmission) point and the extracellular space for the minimum density (or maximum transmission) point were set at 1 and 0, respectively, in the scale. The measurements were done using ImageJ.
For pre-embedding immuno-EM, cells were fixed with a mixture of 3% formaldehyde and 0.05% glutaraldehyde in 0.1 M sodium phosphate buffer, pH 7.4, for 30 min, permeabilized with 0.01% digitonin in PBS for 30 min, labeled sequentially by a primary antibody and a FluoroNanogold-conjugated secondary antibody (Nanoprobes), and then treated with GoldEnhance (Nanoprobes) to visualize nanogold particles.
Huh7 cells expressing HRP-KDEL (Connolly et al., 1994) were fixed with a mixture of 2% formaldehyde and 1.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 20 min and incubated in the enzyme histochemical reaction solution to form DAB precipitates (Brown and Farquhar, 1989; Jokitalo et al., 2001). Samples were postfixed with osmium tetroxide and processed for EM.
Online supplemental material
Fig. S1 shows that the occurrence of nuclear LDs in different cell types was not correlated with the abundance of cytoplasmic LDs. Fig. S2 presents more EM figures that exhibit an association between nuclear LDs and type I NR, and it also shows that nuclear LDs do not have a correlation with nuclear bodies other than PML NB. Fig. S3 shows the distribution of EGFP-tagged PML isoforms in Huh7 cells expressing endogenous PML, exclusion of lamins from the PML-II patch, additional EM pictures of Huh7 after SUN knockdown, and distribution of CCTα-EGFP in nuclear LDs. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201507122.dv.
Acknowledgments
We are grateful to many researchers for donating materials essential to our study; Drs. Tokuko Haraguchi, Naoko Imamoto, and Elina Ikonen for critical reading of the manuscript; and Ms. Tsuyako Tatematsu for technical assistance.
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (to Y. Ohsaki and T. Fujimoto) and by the Hori Sciences and Arts Foundation, Toyoaki Scholarship Foundation, Ono Medical Research Foundation, and MSD (to Y. Ohsaki).
The authors declare no competing financial interests.