Linker histone H1 is required for mitotic chromosome architecture in Xenopus laevis egg extracts and, unlike core histones, exhibits rapid turnover on chromatin. Mechanisms regulating the recruitment, deposition, and dynamics of linker histones in mitosis are largely unknown. We found that the cytoplasmic histone chaperone nucleosome assembly protein 1 (Nap1) associates with the embryonic isoform of linker histone H1 (H1M) in egg extracts. Immunodepletion of Nap1 decreased H1M binding to mitotic chromosomes by nearly 50%, reduced H1M dynamics as measured by fluorescence recovery after photobleaching and caused chromosome decondensation similar to the effects of H1M depletion. Defects in H1M dynamics and chromosome condensation were rescued by adding back wild-type Nap1 but not a mutant lacking sites subject to posttranslational modification by glutamylation. Nap1 glutamylation increased the deposition of H1M on sperm nuclei and chromatin-coated beads, indicating that charge-shifting posttranslational modification of Nap1 contributes to H1M dynamics that are essential for higher order chromosome architecture.
Introduction
During mitosis, the duplicated genome undergoes a dramatic structural reorganization, resulting in condensed, resolved chromosomes that can be segregated by the spindle during anaphase. Core histones H2A, H2B, H3, and H4 provide the first level of genome compaction, assembling into stable octameric units around which DNA is encircled to form nucleosomes. Linker histones bind nucleosomes and the linker DNA between them, stabilizing folded or oligomeric conformations of chromatin fibers, regulating transcription, and generating higher order structures required for mitotic chromosome formation (Thoma and Koller, 1977; Thoma et al., 1979; Ausió, 2006; Harshman et al., 2013). However, the precise functions of linker histones have been difficult to define, as their sequences are more divergent than those of core histones, and many isoforms are present in eukaryotic genomes, with multiple, functionally redundant variants present in most cell types (Fan et al., 2003; Happel and Doenecke, 2009).
Xenopus laevis egg extracts have provided a unique opportunity to study the contribution of linker histone H1 to chromosome structure, as the cytoplasm contains a single embryonic isoform called H1M, also known as B4 (Dworkin-Rastl et al., 1994; Saeki et al., 2005). Importantly, H1M is necessary for mitotic chromosome architecture and condensation in egg extracts (Maresca et al., 2005). In contrast to core histones, all H1 isoforms including H1M are highly dynamic components of chromatin, with residence half-times in vivo on the order of seconds, a surprising characteristic for a structural protein (Misteli et al., 2000; Lever et al., 2000; Freedman et al., 2010). Factors regulating this dynamic behavior have not previously been described.
A key feature of both core and linker histones is their positive charge, which neutralizes negatively charged DNA but also causes free histones to be insoluble at physiological salt concentrations in vitro. Naked DNA and free histones bind tightly and nonspecifically to one another under these conditions, forming disordered nonnucleosomal aggregates rather than chromatin (Wilhelm et al., 1978). In vivo, free histones also bind promiscuously to negatively charged cellular components other than DNA, such as RNA complexes and tubulin (Hondele and Ladurner, 2011). To avoid nonspecific interactions and maintain histone solubility in the cytoplasm, cells use a variety of chaperones, which have been shown to be particularly important in the Xenopus egg where excess histone–chaperone complexes are stored in preparation for rapid rounds of chromatin assembly after fertilization (Loyola and Almouzni, 2004; De Koning et al., 2007; Elsässer and D’Arcy, 2012). One such chaperone is nucleosome assembly protein 1 (Nap1), a highly conserved and enigmatic protein that has been found genetically and biochemically associated with factors functioning in a wide variety of processes including cell cycle regulation, meiosis, nuclear import, polarity, and protein translation (Zlatanova et al., 2007). Best characterized as a histone-binding protein, Nap1 promotes assembly of core nucleosomes in vitro and behaves as a chaperone for histones H2A-H2B in vivo (Ishimi et al., 1984; Ishimi and Kikuchi, 1991; Ito et al., 1996; Chang et al., 1997). Interestingly, Nap1 cofractionated with H1M in Xenopus egg extracts subjected to gel filtration and was shown to be required for sperm chromatin remodeling as well as for proper deposition of H1M onto a synthetic dinucleosome template (Shintomi et al., 2005). Furthermore, treatment of isolated HeLa chromatin fibers with recombinant yeast Nap1 removed H1 and induced a more extended chromatin conformation (Kepert et al., 2005). Thus, Nap1 could potentially affect histone deposition and turnover on chromosomes, but its role during the cell cycle in a physiological setting is unclear.
Charge-shifting posttranslational modifications such as acetylation and phosphorylation of both histones and their chaperones have been strongly implicated in regulating deposition of histones onto DNA, causing changes in nucleosomal arrangement and chromatin structure (Korolev et al., 2007; Eitoku et al., 2008; Avvakumov et al., 2011). Nap1 in HeLa cells was identified as a substrate for a rare posttranslational modification called glutamylation (Regnard et al., 2000), which adds glutamate residues to the γ-carboxyl group of an existing glutamate residue within a peptide sequence. This modification was first discovered on α- and β-tubulin (Eddé et al., 1990) and is enriched specifically on mitotic spindle microtubules (Lacroix et al., 2010), but a functional role for glutamylation of Nap1 has not been defined.
In this study, we show that Nap1 is required for linker histone H1M-mediated mitotic chromosome condensation in Xenopus egg extracts and that glutamylation of Nap1 is required for proper deposition and turnover of H1M on chromatin in both interphase and mitosis. We propose that Nap1 chaperone activity drives H1M deposition and dynamics on core nucleosomal arrays, thereby generating the fundamental chromatin template required for complete mitotic chromosome condensation.
Results and discussion
Nap1 is the major HIM chaperone and is required for mitotic chromosome condensation
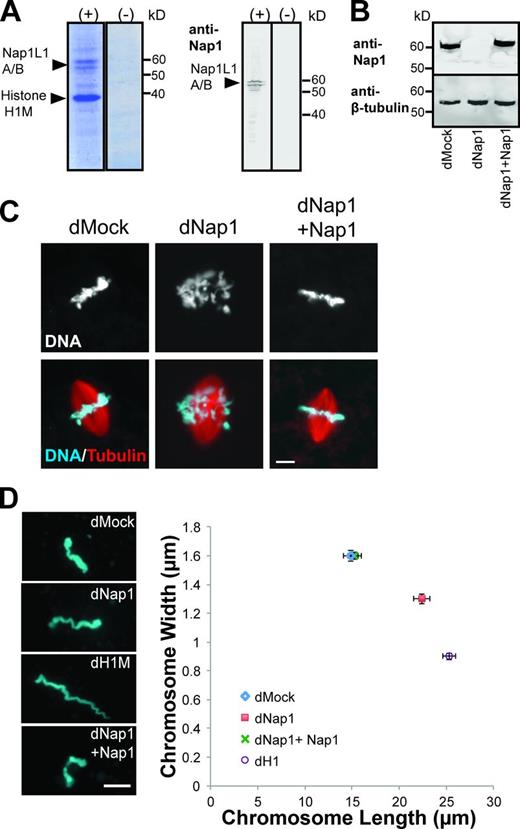
We showed previously that the dynamic behavior of histone H1M depends on the presence of cytoplasm, as H1M-GFP incubated with sperm chromatin in buffer does not exhibit turnover as measured by FRAP (Freedman et al., 2010). To identify cytoplasmic factors contributing to H1M dynamics, we retrieved recombinant H1M that had been added to metaphase-arrested Xenopus egg extracts in a pull-down experiment and observed two major H1M-associated proteins by SDS-PAGE at 56 and 60 kD, which were revealed by mass spectrometry analysis to be two isoforms of the histone chaperone Xenopus Nap1-like 1-A and 1-B, respectively (Fig. 1 A). This interaction has been demonstrated previously (Shintomi et al., 2005), and vertebrate linker histones have also been reported to associate with the chaperones NASP (nuclear autoantigenic sperm protein) and TAF-1/SET (Kato et al., 2011; Wang et al., 2012). Our pull-down experiment indicates that Nap1 is the major histone chaperone binding to H1M in Xenopus egg cytoplasm.
Nap1 binds linker histone H1M in Xenopus egg extracts and is required for normal chromosome condensation. (A) Coomassie-stained gel and Western blot of pull-down with recombinant Strep/His-tagged H1M protein (+) or Strep/His tag alone (−) from Xenopus egg extract. The two bands at 56 and 60 kD were recognized by a Nap1 antibody and identified by 1D liquid chromatography–tandem mass spectrometry as Nap1L1A and Nap1L1B (NCBI reference sequences NP_001082010 and NP_001080547). (B) Western blot of mock-depleted extract (dMock), Nap1-depleted extract (dNap1), and Nap1-depleted extract supplemented with 1 µM recombinant Nap1 (dNap1+ Nap1). β-Tubulin is shown as a loading control. (C) Fluorescence micrographs of spindles and mitotic chromosomes in mock-depleted extract, Nap1-depleted extract, and Nap1-depleted extract supplemented with 1 µM recombinant Nap1. (D) Individual mitotic chromosome dimensions in mock-depleted (dMock), Nap1-depleted (dNap1), H1M-depleted (dH1M) and Nap1-depleted extracts supplemented with 1 µM recombinant Nap1. Photographs on the left show representative images. Images were thresholded, and mean fiber lengths and widths were plotted. n ≥ 80 chromosomes from nine extracts in each condition; error bars are ± SEM, P < 0.001 from unpaired t test for length and breadth between dMock and dNap1, also between dNap1 and dNap1 + Nap1. SDs of chromosome lengths in each condition (µm): dMock = 7.37, dNap1 = 7.98, dNap1 + Nap1 = 6.52, and dH1 = 6.48. SDs of chromosome width in each condition (µm): dMock = 0.35, dNap1 = 0.31, dNap1 + Nap1 = 0.36, and dH1 = 0.23. The distribution of chromosome dimensions is also shown in histograms (Fig. S1, A and B). Bars, 10 µm.
Nap1 binds linker histone H1M in Xenopus egg extracts and is required for normal chromosome condensation. (A) Coomassie-stained gel and Western blot of pull-down with recombinant Strep/His-tagged H1M protein (+) or Strep/His tag alone (−) from Xenopus egg extract. The two bands at 56 and 60 kD were recognized by a Nap1 antibody and identified by 1D liquid chromatography–tandem mass spectrometry as Nap1L1A and Nap1L1B (NCBI reference sequences NP_001082010 and NP_001080547). (B) Western blot of mock-depleted extract (dMock), Nap1-depleted extract (dNap1), and Nap1-depleted extract supplemented with 1 µM recombinant Nap1 (dNap1+ Nap1). β-Tubulin is shown as a loading control. (C) Fluorescence micrographs of spindles and mitotic chromosomes in mock-depleted extract, Nap1-depleted extract, and Nap1-depleted extract supplemented with 1 µM recombinant Nap1. (D) Individual mitotic chromosome dimensions in mock-depleted (dMock), Nap1-depleted (dNap1), H1M-depleted (dH1M) and Nap1-depleted extracts supplemented with 1 µM recombinant Nap1. Photographs on the left show representative images. Images were thresholded, and mean fiber lengths and widths were plotted. n ≥ 80 chromosomes from nine extracts in each condition; error bars are ± SEM, P < 0.001 from unpaired t test for length and breadth between dMock and dNap1, also between dNap1 and dNap1 + Nap1. SDs of chromosome lengths in each condition (µm): dMock = 7.37, dNap1 = 7.98, dNap1 + Nap1 = 6.52, and dH1 = 6.48. SDs of chromosome width in each condition (µm): dMock = 0.35, dNap1 = 0.31, dNap1 + Nap1 = 0.36, and dH1 = 0.23. The distribution of chromosome dimensions is also shown in histograms (Fig. S1, A and B). Bars, 10 µm.
To probe Nap1’s function in chromosome condensation in Xenopus, we generated an antibody against recombinant Nap1-L1B, which recognized both isoforms of Nap1 by Western blotting (Fig. 1 A). Using this antibody, we could efficiently immunodeplete >95% of both Nap1 isoforms from Xenopus egg extract (Fig. 1 B) and examine the effects of Nap1 depletion on mitotic spindle assembly and chromosome structure. Interestingly, chromosomes appeared misaligned at the metaphase plate and were abnormally long and thin (Fig. 1, C and D). These defects were reminiscent of the effects of H1M depletion, characterized by aberrantly elongated chromosomes that extend outside the boundaries of the spindle and segregate poorly at anaphase (Maresca et al., 2005). Addback of 1 µM Nap1-L1B matched endogenous concentrations by Western blotting (Fig. 1 B) and was sufficient to rescue the phenotype (Fig. 1 C). To further analyze and quantify chromosome condensation defects, individual replicated chromosomes were isolated from Xenopus egg extracts, and their physical dimensions were measured. Chromosomes from Nap1-depleted egg extracts were on average 34.8% longer and 19% thinner than those from mock-depleted extracts. By comparison, chromosomes from H1M-depleted egg extracts were 41.9% longer and 43.8% thinner (Fig. 1 D and Fig. S1, A and B). Thus, Nap1 is required for complete mitotic chromosome condensation in egg extracts, but the effects of its depletion are not as pronounced as for H1M.
Nap1 was previously found to associate specifically with B-type cyclins in Xenopus egg and budding yeast extracts and to be a substrate for Cdk1/cyclin B (Kellogg et al., 1995). In addition to incomplete chromosome condensation, aberrant spindle assembly was also sometimes observed upon Nap1 depletion, a defect that was not seen upon H1M depletion (e.g., Fig. S2 B; Maresca et al., 2005). Therefore, although promoting proper chromosome condensation appears to be the major function of Nap1 in egg extracts, it may also have other targets important for mitotic events.
Nap 1 modulates H1M binding to mitotic chromosomes
As chromosome dimensions are known to vary with the amount of H1M in the extract (Freedman and Heald, 2010), we examined the levels of H1M bound to pelleted chromosomes. Chromatin isolated from Nap1-depleted egg extracts was significantly depleted of H1M by a mean of 48.6% by Western blotting, whereas levels of chromosome-bound core histone H2A remained unchanged (Fig. 2 A and Fig. S1 C). By immunofluorescence, mitotic chromosomes showed a reduction in H1M staining intensity by 37.5% (Fig. 2, B and C). The observed decrease in H1M association with chromosomes was not simply caused by codepletion of H1M with Nap1 because cytoplasmic H1M levels were not significantly different between mock- and Nap1-depleted egg extracts (Fig. 2 A and Fig. S1 C). These results indicate that H1M fails to bind chromatin normally in the absence of Nap1, which may then result in incomplete mitotic chromosome condensation. The effects of Nap1 depletion on chromatin are likely indirect and a result of its modulation of H1M activity, since by immunofluorescence and Western blotting, Nap1 was not enriched on mitotic chromosomes or the spindle (unpublished data).
Nap1 is required for H1M-mediated chromosome condensation. (A) Representative Western blot of pelleted chromosomes (left) and cytoplasm (right) from mock- or Nap1-depleted egg extracts probed with antibodies to H1M and histone H2A. Levels of chromatin-bound H2A decreased by a mean of 0.8 ± 0.4% (mean ± SD) in Nap1-depleted extracts, whereas chromatin-bound H1M decreased by a mean of 48.6 ± 4.8%. Levels of H1M in Nap1-depleted cytoplasm decreased by 3.5 ± 1.2%, and levels of H2A decreased by 1.6 ± 0.2% on average. Quantification is shown in Fig. S1 C. (B) Immunofluorescence of mitotic chromosomes sedimented onto coverslips and stained for H1M. (C) Quantification of H1M/DNA fluorescence intensity on structures in B. Means ± SEM. Fluorescence intensity in each condition was background subtracted and normalized to H1-depleted extracts (see Materials and methods). n ≥ 50 structures were evaluated per condition in three separate extracts. *, P < 0.001 from unpaired t test comparing mock-depleted and Nap1-depleted extracts as well as comparing mock-depleted and Nap1-depleted/supplemented with Nap1N6C9. (D) Representative fluorescence micrographs of spindle assembly reactions in mock- or Nap1-depleted extracts supplemented with different concentrations of recombinant H1M. In the dNap1 + H1 condition, >99% of Hoechst-stained structures were hypercompacted chromatin masses with little or no microtubule polymerization around them. Bars, 10 µm.
Nap1 is required for H1M-mediated chromosome condensation. (A) Representative Western blot of pelleted chromosomes (left) and cytoplasm (right) from mock- or Nap1-depleted egg extracts probed with antibodies to H1M and histone H2A. Levels of chromatin-bound H2A decreased by a mean of 0.8 ± 0.4% (mean ± SD) in Nap1-depleted extracts, whereas chromatin-bound H1M decreased by a mean of 48.6 ± 4.8%. Levels of H1M in Nap1-depleted cytoplasm decreased by 3.5 ± 1.2%, and levels of H2A decreased by 1.6 ± 0.2% on average. Quantification is shown in Fig. S1 C. (B) Immunofluorescence of mitotic chromosomes sedimented onto coverslips and stained for H1M. (C) Quantification of H1M/DNA fluorescence intensity on structures in B. Means ± SEM. Fluorescence intensity in each condition was background subtracted and normalized to H1-depleted extracts (see Materials and methods). n ≥ 50 structures were evaluated per condition in three separate extracts. *, P < 0.001 from unpaired t test comparing mock-depleted and Nap1-depleted extracts as well as comparing mock-depleted and Nap1-depleted/supplemented with Nap1N6C9. (D) Representative fluorescence micrographs of spindle assembly reactions in mock- or Nap1-depleted extracts supplemented with different concentrations of recombinant H1M. In the dNap1 + H1 condition, >99% of Hoechst-stained structures were hypercompacted chromatin masses with little or no microtubule polymerization around them. Bars, 10 µm.
As the decondensed chromosomes observed upon Nap1 depletion were also partially depleted of H1M, we reasoned that proper chromosome condensation might be rescued by supplementing additional recombinant H1M to the extract, which is known to condense H1M depleted-chromosomes in a dose-dependent manner (Freedman et al., 2010). H1M is an abundant protein at 1.5–2 µM in Xenopus egg extracts (Maresca et al., 2005). However, addition of 0.4 µM H1M to Nap1-depleted extracts caused chromatin hypercompaction and densely clustered sperm chromosomes that completely failed to support microtubule polymerization and spindle assembly (Fig. 2 D). In contrast, addition of 0.4 µM H1M to mock-depleted extracts did not have deleterious effects, and similar chromosome and spindle aberrations were observed only when ≥3.5 µM of recombinant H1M was added (Fig. 2 D; Freedman et al., 2010). Thus, chromosomes in Nap1-depleted egg extracts were much more sensitive to addition of recombinant H1M, suggesting that its chaperone activity is essential for normal H1M binding to chromatin and condensation activity.
Glutamylation of Nap1 is required for mitotic chromosome condensation and turnover of H1M
Posttranslational modification of histones and their associated chaperones has been shown to regulate deposition and exchange of histones on chromatin (Loyola and Almouzni, 2004; Campos et al., 2010). We therefore explored whether any modifications influenced Nap1’s deposition of H1M onto mitotic chromosomes and if this in turn affected the dynamics of H1M. Recombinant Nap1 incubated in egg extract was retrieved on resin in a pull-down experiment and subjected to mass spectrometry analysis. A total of 12 potential glutamylated residues were detected, with three high-confidence sites found at amino acid residues E30, E31, and E357 (Fig. 3 A). Most of the modified residues were located at the far N and C termini of the protein in disordered regions (Park and Luger, 2006). Glutamylation was also detected on retrieved Nap1 by Western blotting using antibodies raised to glutamylated tubulin peptides (Fig. 3 B; Spano and Frankfurter, 2010). To assay the functional role of Nap1 glutamylation, we generated mutant recombinant Nap1 proteins with glutamic acid residues within these disordered regions changed to aspartic acid (Fig. 3 A). Mutation of six potential glutamylation sites in either the N-terminal (Nap1N6) or C-terminal regions (Nap1C9) alone did not abolish glutamylation of Nap1 as assayed by Western blotting (Fig. 3 B and Fig. S2 A), and these proteins rescued chromosome condensation defects when added back to Nap1-depleted extracts (Fig. S2 B), suggesting that mutation of these residues does not interfere with Nap1 function or stability. Furthermore, mutation of N- and C-terminal sites in combination (Nap1N6C9) did not impair Nap1-induced decondensation of sperm nuclei when added at high levels, similar to wild-type Nap1 (Fig. S2 C). However, significantly reduced glutamylation of Nap1N6C9 was observed, and this mutant was unable to rescue chromosome condensation defects (Fig. 3, C and D; and Fig. S2 B). Depleted extract supplemented with Nap1N6C9 produced chromosomes comparable in dimensions to those in a Nap1-depleted reaction and displayed a similar 43% depletion of H1M from chromosomes by immunofluorescence (Fig. 2, B and C; Fig. 3, C and D; and Fig. S3, A and B). Altogether, these results suggest that glutamylation of Nap1 is required for mitotic chromosome condensation and that preventing this modification alters the deposition of H1M on mitotic chromosomes.
Glutamylation of Nap1 is required for mitotic chromosome condensation. (A) Schematic of the 393–amino acid Xenopus Nap1 protein, including three high-confidence glutamylation sites identified by 1D liquid chromatography–tandem mass spectrometry, marked with asterisks. Sites identified were N-terminal glutamate residues 30 and 31 as well as C-terminal residue 357. Three Nap1 mutants were created to reduce glutamylation by mutating indicated glutamic acid residues in the disordered N and C termini to aspartic acids. Nap1N6 = six glutamic acid residues mutated in the N terminus only. Nap1C9 = nine residues mutated in the C terminus only. Nap1 N6C9 = six N-terminal and nine C-terminal residues mutated together. Mutated sites are indicated in green. (B) Western blot of recombinant wild-type or mutant Nap1 proteins isolated from egg extracts and probed with antibodies that recognize polyglutamylated tubulin (anti-Glu). Nap1N6C9 displayed markedly reduced glutamylation in egg extracts with a 85 ± 3.8% decrease in band intensity (mean percentage ± SD, P = 0.005 from unpaired t test), whereas Nap1N6 and Nap1C9 were decreased by 19 ± 4.4% and 32 ± 5.1%, respectively. Recombinant wild-type Nap1 was pulled down in buffer as a negative control. (C) The Nap1 N6C9 mutant fails to rescue chromosome defects in Nap1-depleted egg extracts. Bar, 10 µm. (D) Chromosome length and width measurements were collected as in Fig. 2 A and averaged for each condition. n ≥ 60 chromosomes from five separate extracts. Error bars are ± SEM. P < 0.005 from unpaired t test comparing mock- and Nap1-depleted extracts as well as mock- and Nap1-depleted/supplemented with Nap1N6C9. SDs of chromosome lengths in each condition (µm): dMock = 6.0, dNap1 = 8.2, dNap1 + Nap1 = 6.9, and dNap1 + Nap1N6C9 = 6.9. SDs of chromosome width in each condition (µm): dMock = 0.34, dNap1 = 0.36, dNap1 + Nap1 = 0.34, and dNap1 + Nap1N6C9 = 0.38. The distribution of chromosome dimensions is also shown in histograms (Fig. S3, A and B).
Glutamylation of Nap1 is required for mitotic chromosome condensation. (A) Schematic of the 393–amino acid Xenopus Nap1 protein, including three high-confidence glutamylation sites identified by 1D liquid chromatography–tandem mass spectrometry, marked with asterisks. Sites identified were N-terminal glutamate residues 30 and 31 as well as C-terminal residue 357. Three Nap1 mutants were created to reduce glutamylation by mutating indicated glutamic acid residues in the disordered N and C termini to aspartic acids. Nap1N6 = six glutamic acid residues mutated in the N terminus only. Nap1C9 = nine residues mutated in the C terminus only. Nap1 N6C9 = six N-terminal and nine C-terminal residues mutated together. Mutated sites are indicated in green. (B) Western blot of recombinant wild-type or mutant Nap1 proteins isolated from egg extracts and probed with antibodies that recognize polyglutamylated tubulin (anti-Glu). Nap1N6C9 displayed markedly reduced glutamylation in egg extracts with a 85 ± 3.8% decrease in band intensity (mean percentage ± SD, P = 0.005 from unpaired t test), whereas Nap1N6 and Nap1C9 were decreased by 19 ± 4.4% and 32 ± 5.1%, respectively. Recombinant wild-type Nap1 was pulled down in buffer as a negative control. (C) The Nap1 N6C9 mutant fails to rescue chromosome defects in Nap1-depleted egg extracts. Bar, 10 µm. (D) Chromosome length and width measurements were collected as in Fig. 2 A and averaged for each condition. n ≥ 60 chromosomes from five separate extracts. Error bars are ± SEM. P < 0.005 from unpaired t test comparing mock- and Nap1-depleted extracts as well as mock- and Nap1-depleted/supplemented with Nap1N6C9. SDs of chromosome lengths in each condition (µm): dMock = 6.0, dNap1 = 8.2, dNap1 + Nap1 = 6.9, and dNap1 + Nap1N6C9 = 6.9. SDs of chromosome width in each condition (µm): dMock = 0.34, dNap1 = 0.36, dNap1 + Nap1 = 0.34, and dNap1 + Nap1N6C9 = 0.38. The distribution of chromosome dimensions is also shown in histograms (Fig. S3, A and B).
Upon observing the changes in H1M binding and the corresponding chromosome condensation defects when Nap1 function was perturbed, we wondered whether such changes could be linked to abnormal H1M turnover. To answer this question, we used a FRAP assay in which chromosomes were assembled in egg extracts double depleted of both Nap1 and H1M. Extracts were then supplemented with endogenous concentrations of recombinant H1M-GFP, which was assayed for turnover on mitotic chromosomes in the absence or presence of recombinant Nap1 proteins (Fig. 4 A). Interestingly, depletion of Nap1 caused a fivefold reduction in the rate of H1M recovery to the photobleached area (half-time of recovery, or t1/2 = 8.6 s, compared with t1/2 = 1.7 s in mock-depleted extracts). Recovery time was rescued by the addition of wild-type Nap1 (t1/2 = 1.8 s), but not Nap1N6C9 (t1/2 = 9.5 s), indicating that Nap1 glutamylation is required for proper H1M turnover. Reducing Nap1 glutamylation also caused a similar reduction in H1M turnover on interphase chromatin, as t1/2 decreased from 2.1 s in mock-depleted interphase extracts to 8.9 s in Nap1-depleted extracts supplemented with Nap1N6C9 (Fig. 4 B). This result indicates that Nap1 glutamylation is required for H1 dynamics throughout the cell cycle. To test whether glutamylation of Nap1 alters its affinity for H1M, recombinant wild-type Nap1 and mutant Nap1N6C9 were retrieved from interphase and metaphase egg extracts in pull-down experiments and probed for H1M. Similar amounts of H1M were found associated with Nap1 under all conditions, suggesting that steady-state H1M binding is not affected by glutamylation (Fig. S2 D).
Glutamylation of Nap1 is required for proper H1 dynamics. (A) FRAP curves of H1M-GFP on mitotic chromosomes in metaphase-arrested egg extracts depleted of Nap1 (dNap1) and supplemented with either wild-type or the nonglutamylated mutant (Nap1N6C9). Curves represent means of n ≥ 10 chromosomes from three separate extracts. Half-lives for fluorescence recovery (t1/2 ± SD; s): dMock = 1.7 ± 0.68, dNap1 = 8.6 ± 0.51, dNap1+Nap1 = 1.7 ± 0.60, and dNap1±Nap1N6C9 = 9.5 ± 0.69. (B) FRAP curves of GFP-H1 on chromatin in metaphase- or interphase-arrested egg extracts depleted of Nap1 and supplemented with Nap1N6C9. Regardless of cell cycle state, Nap1-depleted extract supplemented with Nap1 N6C9 caused a similar significant reduction in H1M recovery time. Curves are means of n ≥ 10 chromosomes from five separate extracts. Half-times for fluorescence recovery (t1/2 ± SD; s): dMock-Mitosis = 1.9 ± 0.57, Nap1N6C9-Mitosis = 9.7 ± 0.67, dMock-Interphase = 2.1 ± 0.59, and Nap1N6C9-Interphase = 8.9 ± 0.70. For both A and B, the photobleach is plotted at time = 0.5 s. All curves were normalized to a baseline fluorescence of 1 at time = 0 s and 0 at time = 0.5 s. Error bars show ±SEM.
Glutamylation of Nap1 is required for proper H1 dynamics. (A) FRAP curves of H1M-GFP on mitotic chromosomes in metaphase-arrested egg extracts depleted of Nap1 (dNap1) and supplemented with either wild-type or the nonglutamylated mutant (Nap1N6C9). Curves represent means of n ≥ 10 chromosomes from three separate extracts. Half-lives for fluorescence recovery (t1/2 ± SD; s): dMock = 1.7 ± 0.68, dNap1 = 8.6 ± 0.51, dNap1+Nap1 = 1.7 ± 0.60, and dNap1±Nap1N6C9 = 9.5 ± 0.69. (B) FRAP curves of GFP-H1 on chromatin in metaphase- or interphase-arrested egg extracts depleted of Nap1 and supplemented with Nap1N6C9. Regardless of cell cycle state, Nap1-depleted extract supplemented with Nap1 N6C9 caused a similar significant reduction in H1M recovery time. Curves are means of n ≥ 10 chromosomes from five separate extracts. Half-times for fluorescence recovery (t1/2 ± SD; s): dMock-Mitosis = 1.9 ± 0.57, Nap1N6C9-Mitosis = 9.7 ± 0.67, dMock-Interphase = 2.1 ± 0.59, and Nap1N6C9-Interphase = 8.9 ± 0.70. For both A and B, the photobleach is plotted at time = 0.5 s. All curves were normalized to a baseline fluorescence of 1 at time = 0 s and 0 at time = 0.5 s. Error bars show ±SEM.
To investigate more mechanistically how Nap1 glutamylation was altering H1M dynamics, we added H1M-GFP to demembranated Xenopus sperm nuclei in buffer in the presence of Nap1 and TTLL4 (tyrosine tubulin ligase–like 4), an enzyme previously shown to glutamylate Nap1 in vitro (van Dijk et al., 2007) that induced Nap1 glutamylation as monitored by Western blotting (Fig. 5 A). Nap1 increased the deposition of H1M on to sperm chromatin, and this was further stimulated in the presence of TTLL4 and wild-type Nap1 but not the glutamylation site mutant Nap1N6C9 (Fig. 5, B and C). The composition of chromatin proteins in sperm nuclei is very different from that of somatic nuclei. Thus, H1M binding to sperm nuclei and mitotic chromosomes may be regulated differently. However, qualitatively similar results with H1M deposition were obtained using chromatin-coated beads in both buffer and in egg extracts, under conditions in which binding of both endogenous H1M and H1M-GFP could be examined (Fig. 5, D–G). Thus, Nap1 polyglutamylation appears to promote H1M deposition onto chromatin.
Nap1 glutamylation promotes H1M deposition on sperm or on chromatin beads in extract and in buffer. (A) Representative Western blot of recombinant wild-type or mutant Nap1 proteins incubated with TTLL4 and probed with antibodies that recognize either Nap1 or glutamylated tubulin (anti-Glu). (B) Fluorescence micrographs of Xenopus sperm nuclei combined with 0.2 µM H1M-GFP alone or with equal concentrations of either Nap1 or Nap1N6C9, in the presence or absence of 1 µM TTLL4 enzyme. Bar, 10 µm. (C) Quantification of H1M/DNA fluorescence intensity on structures in B; means ± SD, n ≥ 50 structures were evaluated per condition in three separate experiments. *, P < 0.001 in unpaired t test. (D) Western blot of isolated chromatin beads probed with antibodies against H1M. Left column shows chromatin beads assembled in H1M-depleted egg extracts showing that H1M is initially absent from the chromatin. H1M-depleted chromatin beads were then incubated in Nap1-depleted extracts supplemented with either recombinant Nap1 or the Nap1N6C9 mutant and 2 µM H1M-GFP. Greater amounts of both endogenous (E) and recombinant GFP-tagged H1M (R) were recruited to beads in the presence of wild-type Nap1 compared with the glutamylation site mutant. Histone H2A is shown as a loading control. (E) Western blot of isolated chromatin beads probed with antibodies against H1M in a similar experiment performed in buffer. H1M-depleted chromatin beads were incubated in Hepes buffer supplemented with either recombinant Nap1 or the Nap1N6C9 mutant and 2 µM H1M-GFP and the TTLL4 enzyme. A small amount of degraded H1M-GFP (D) is seen on the blot (arrow). Histone H2A is shown as a loading control. (F) Quantification of H1M-GFP band intensities in three experiments from three different extracts as in D; means ± SD. Band intensity for the (−)H1M condition was normalized to 0. *, P = 0.005 by unpaired t test comparing wild-type and mutant conditions. (G) Quantification of mean H1M-GFP band intensities in three experiments as in E; means ± SD. *, P = 0.002 by unpaired t test comparing wild-type and mutant conditions.
Nap1 glutamylation promotes H1M deposition on sperm or on chromatin beads in extract and in buffer. (A) Representative Western blot of recombinant wild-type or mutant Nap1 proteins incubated with TTLL4 and probed with antibodies that recognize either Nap1 or glutamylated tubulin (anti-Glu). (B) Fluorescence micrographs of Xenopus sperm nuclei combined with 0.2 µM H1M-GFP alone or with equal concentrations of either Nap1 or Nap1N6C9, in the presence or absence of 1 µM TTLL4 enzyme. Bar, 10 µm. (C) Quantification of H1M/DNA fluorescence intensity on structures in B; means ± SD, n ≥ 50 structures were evaluated per condition in three separate experiments. *, P < 0.001 in unpaired t test. (D) Western blot of isolated chromatin beads probed with antibodies against H1M. Left column shows chromatin beads assembled in H1M-depleted egg extracts showing that H1M is initially absent from the chromatin. H1M-depleted chromatin beads were then incubated in Nap1-depleted extracts supplemented with either recombinant Nap1 or the Nap1N6C9 mutant and 2 µM H1M-GFP. Greater amounts of both endogenous (E) and recombinant GFP-tagged H1M (R) were recruited to beads in the presence of wild-type Nap1 compared with the glutamylation site mutant. Histone H2A is shown as a loading control. (E) Western blot of isolated chromatin beads probed with antibodies against H1M in a similar experiment performed in buffer. H1M-depleted chromatin beads were incubated in Hepes buffer supplemented with either recombinant Nap1 or the Nap1N6C9 mutant and 2 µM H1M-GFP and the TTLL4 enzyme. A small amount of degraded H1M-GFP (D) is seen on the blot (arrow). Histone H2A is shown as a loading control. (F) Quantification of H1M-GFP band intensities in three experiments from three different extracts as in D; means ± SD. Band intensity for the (−)H1M condition was normalized to 0. *, P = 0.005 by unpaired t test comparing wild-type and mutant conditions. (G) Quantification of mean H1M-GFP band intensities in three experiments as in E; means ± SD. *, P = 0.002 by unpaired t test comparing wild-type and mutant conditions.
In conclusion, posttranslational glutamylation of Nap1 is vital to its function, as reducing Nap1 glutamylation yields mitotic chromosomes that are longer, thinner, and H1M depleted. Although some fraction of H1M is still able to bind chromatin, it does so aberrantly, and concomitant abnormalities in higher order chromatin structure occur (Fig. 3 C). Precisely how Nap1 glutamylation affects H1M function remains unclear, but our results indicate that the additional acidic residues allow Nap1 to deposit H1M more efficiently. This may explain effects on H1M turnover as measured by FRAP, but it is also possible that Nap1 and its glutamylation contribute to H1M dissociation from chromatin as well. Because the affinity of Nap1 for H1M is not obviously affected by glutamylation, regulation may be modulated by other factors or occur through the chromatin template. Because the activity of Nap1 appears to be independent of cell cycle state, our results indicate that it is required to generate the basal chromatin structure in both interphase and mitosis, but physical changes on the diffuse state of interphase chromatin are not detectable by light microscopy. Interestingly, recent work showed that Nap1-dependent removal of H1 in somatic cells was required for efficient homologous recombination to allow DNA double-strand break repair (Machida et al., 2014). This suggests that Nap1 also acts as a linker histone chaperone in somatic cells. In summary, our findings shed new light on a physiological pathway regulating linker histone H1 dynamics through posttranslational modification of a known histone chaperone, and future experiments promise to reveal how histone chaperone regulation modulates chromosome structure and function in a variety of contexts.
Materials and methods
Xenopus egg extracts and spindle assembly reactions
Cystostatic factor (CSF)–arrested Xenopus egg extracts were prepared as previously described (Hannak and Heald, 2006; Maresca and Heald, 2006). In brief, metaphase-arrested Xenopus eggs were dejellied and fractionated via centrifugation at 10,200 rpm in a rotor (Sorvall HB-6; Thermo Fisher Scientific). The cytoplasmic fraction was removed and supplemented with energy mix (to a final concentration of 4 mM creatine phosphate, 0.5 mM ATP, 0.5 mM MgCl2, and 0.05 mM EGTA, pH 7.7). For spindle assembly reactions, demembranated sperm nuclei were added to egg extracts, and reactions were cycled through interphase by addition of 0.5 mM CaCl2 in 10 mM Hepes as previously described (Hannak and Heald, 2006; Maresca and Heald, 2006) and then driven into mitosis by addition of fresh CSF-arrested egg extract to generate mitotic spindles and chromosomes. Spindles were visualized by supplementing the extract with X-rhodamine–labeled tubulin (50 µg/ml). Recombinant proteins were added before spindle assembly. To process spindles for immunofluorescence, 25 µl reactions were fixed by addition to 1 ml of dilution buffer (80 mM Pipes, 1 mM MgCl2, 1 mM EGTA, 30% glycerol, 0.5% Triton X-100, and 2.5% formaldehyde). After incubating for 10 min at 23°C, samples were spun onto coverslips through a 5-ml cushion (BRB80 + 40% glycerol) at 10,200 rpm for 15 min using a rotor (Sorvall HB-6). Reactions were also squash fixed for imaging: in brief, 2 µl samples were spotted onto slides, overlaid with 5 µl of spindle fix (48% glycerol, 11% formaldehyde in Marc’s modified Ringer’s solution [5 mM Hepes, pH 7.8, 2 mM KCl, 1 mM MgSO4, 100 mM NaCl, 2 mM CaCl2, and 0.1 mM EGTA], and 5 µg/ml Hoechst 33258) and squashed with a coverslip (Maresca and Heald, 2006; Hannak and Heald, 2006). Images were acquired at 23°C with a fluorescence microscope (BX51; Olympus) controlled by MetaMorph software (Molecular Devices) equipped with TRITC, DAPI, and FITC filters (Chroma Technology, Corp.), a 40× air objective (U Plan Fluor N, NA 0.75; Olympus), a 60× oil objective (Plan Apochromat, NA 1.4), a 100× oil objective (U Plan Fluor, NA 1.30), and a cooled charge-coupled device camera (Orca II; Hamamatsu Photonics).
Cloning and protein purification
Xenopus H1M (B4) cDNA was originally provided by M. Dasso (National Institutes of Health, Bethesda, MD) in a pGEX-KG vector (GE Healthcare). Wild-type Xenopus p60 Nap1/Nap1L1-B in a pTrcHis vector (Invitrogen) used in Shintomi et al. (2005) was a provided by K. Ohsumi (Nagoya University, Nagoya, Japan). Both cDNAs were cloned into a pET51b(+) expression vector (EMD Millipore) with a T7 promoter to introduce an N-terminal Strep II and a C-terminal His tag. H1M-GFP was created by cloning the H1M (B4) sequence described in this paragraph into a pET30 vector with a T7 promoter including an N-terminal GFP tag and a C-terminal His tag. Recombinant H1M and H1M-GFP were both expressed in BL21 (DE3) Escherichia coli (Agilent Technologies) as described in Freedman and Heald (2010), using an overnight induction with 0.5 mM IPTG at 16°C. Bacteria were lysed using BugBuster Protein Extraction reagent (EMD Millipore) in PBS + 500 mM NaCl. H1M-GFP and wild-type H1M were purified from bacterial lysates using NiNTA agarose (QIAGEN) followed by elution with PBS + 500 mM NaCl + 500 mM imidazole. Eluates were then incubated with Strep-Tactin Superflow Plus resin (QIAGEN) for 2 h at 4°C with rotation. The resin was washed several times with PBS + 500 mM NaCl, and the H1M protein was eluted with PBS + 500 mM NaCl + 2.5 mM desthiobiotin, according to the manufacturer’s instructions. Mutant Nap1 proteins (Nap1N6, Nap1C9, and Nap1N6C9) were generated using a site-directed mutagenesis kit (QuikChange II; Agilent Technologies) and cloned into the pET51b(+) vector. Wild-type and mutant Nap1 constructs were expressed in BL21(DE3) E. coli (Agilent Technologies) using an overnight induction with 0.5 mM IPTG at 16°C and two-step affinity purified as for H1M, except 50 mM Hepes + 100 mM NaCl was used in place of PBS + 500 mM NaCl.
Immunodepletion and rescue experiments
Rabbit polyclonal antibodies against full-length Nap1 were raised by Covance and affinity purified from total serum on a HiTrap N-hydroxysuccinimide–activated HP column (GE Healthcare) coupled with recombinant Nap1. Antibodies were eluted with Gentle Ag/Ab Elution Buffer (Thermo Fisher Scientific) and dialyzed into 50 mM Hepes.
H1M was immunodepleted from egg extracts as previously described (Maresca et al., 2005). In brief, 55 µl CSF extract was subjected to two successive 45-min incubations with 40 µg anti-H1M antibody coupled to 200 µl protein A Dynabeads (Invitrogen). To deplete Nap1, 11 µg of affinity-purified Nap1 antibody coupled to 40 µl protein A Dynabeads (Invitrogen) was used to deplete 11 µl of egg extract over two rounds of 45-min incubations. For control (mock depleted) reactions, an equal amount of total rabbit IgG antibody was coupled to beads. Recombinant Nap1-L1B or mutants (Nap1N6, Nap1C9, and Nap1N6C9) were added back to depleted extracts at a final concentration of 1 µM to match endogenous Nap1 levels.
Pull-down experiments
For pull-downs, 10 µM H1M or 1.2 µM Nap1 was incubated in egg extract for 10 min, and then, the mixture was added to equilibrated Strep-Tactin resin. Reactions were incubated at 4°C for 30 min with gentle shaking. Resin was pelleted at 500 g, and the extract supernatant was removed. Resin was then washed 3× with Xenopus buffer (XB; 10 mM Hepes, 50 mM sucrose, 1 mM MgCl2, 0.1 mM CaCl2, and 100 mM KCl), 3× with XB buffer + 0.5% Triton X-100, and 3× with XB buffer, and then, proteins were eluted with SDS sample buffer and subjected to 12% SDS-PAGE.
Immunofluorescence and measurement of individual chromosomes
Egg extract reactions were cycled through interphase to replicate chromatids. Once metaphase structures formed, reactions were diluted 40-fold in XBE2 buffer (10 mM Hepes, 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA, and 50 mM sucrose, pH 7.7) supplemented with 2% formaldehyde and 0.25% Triton X-100. Reactions were fixed at room temperature for 20 min and spun onto coverslips over a 30% glycerol cushion in XBE2 buffer. Coverslips were postfixed in 100% methanol, stained with DAPI, and mounted on microscope slides using Vectashield (Vector Laboratories). Chromosome dimensions were measured as described in Freedman et al. (2010): For quantification of chromosome lengths, individual chromosomes were selected at random, and the structures were thresholded manually in ImageJ (National Institutes of Health). A line was drawn down the long axis of the midline of the chromosome arm, and the distance in pixels was converted to micrometers. For quantification of chromosome width, a perpendicular line was drawn across both chromosome arms at the top and bottom of each chromosome, avoiding centromeric regions. These two values were averaged, and the distance in pixels was converted to micrometers.
For immunofluorescence, coverslips were postfixed in 100% methanol, blocked with PBS–1% BSA, and incubated with primary antibody against H1M (rabbit, generated using full-length H1M protein as described in Maresca and Heald, 2006), diluted 1:1,200 in PBS–1% BSA, washed with PBS–0.1% NP-40, and incubated with a 1:1,000 dilution of secondary antibody (Alexa Fluor 488–labeled anti–rabbit; Invitrogen) before staining with DAPI and mounting on microscope slides using Vectashield. Structures were visualized with a 40× or 60× objective using FITC and DAPI filters and imaged using identical exposure times. Image files were then subjected to automated structure identification and morphometric/intensity colocalization analysis using CellProfiler software (Broad Institute). The FITC/DAPI fluorescence intensity ratio was used for quantification.
Background subtraction of fluorescence intensity in images
To background subtract nonspecific H1M staining from specific staining on structures, extracts were immunodepleted of H1M, stained, and imaged as described in the previous paragraph. Fluorescence intensity in the FITC channel for 30 structures per five separate coverslips were measured and averaged. This quantity was then subtracted from the measured FITC fluorescence intensity for each image analyzed in the CellProfiler software to generate the total fluorescence intensity in the FITC channel.
Chromosome pelleting
To quantify histone levels bound to chromosomes in Xenopus egg extracts, reactions containing at least 4,000 sperm nuclei/µl were cycled through interphase and then driven into metaphase, diluted 1:5 in egg lysis buffer–chromatin isolation buffer (ELB-CIB; 10 mM Hepes, 2.5 mM MgCl2, 50 mM KCl, 250 mM sucrose, 1 mM DTT, 1 mM EDTA, 1 mM spermidine, 1 mM spermine, and 0.1% Triton X-100), and mitotic chromosomes were pelleted through a 3-ml, 0.5-M sucrose cushion in ELB-CIB as described in Banaszynski et al. (2010). Supernatants were removed, and pellets were rinsed twice with ELB-CIB buffer. Pellets were then diluted in SDS sample buffer + Benzonase nuclease (Sigma-Aldrich) to degrade the DNA and subjected to 12% SDS-PAGE and Western blotting.
Western blotting
Antibodies used for Western blotting were diluted in PBS + 2.5% milk. Primary rabbit antibody to full-length Xenopus H1M (Maresca et al., 2005) was used 1:1,500. Primary antibody to histone H2A (rabbit, ab13923; Abcam) was used 1:1,000. Nap1 antisera were used at 1:4,000. Primary antibody to β-tubulin (mouse anti–tubulin E7; Developmental Studies Hybridoma Bank) was used at 1:5,000. Primary antibodies for γ-glutamylation, called TTβIII-glu and TTSG1 (rabbit, created against short synthetic glutamylated peptides derived from sequences from rat brain tubulin) were used at 1:10,000 as described in Spano and Frankfurter (2010). TTβIII-glu and TTSG1 antibodies were provided by D. Shechter (Albert Einstein University, Bronx, NY). Secondary antibodies from Rockland Immunochemicals (goat anti–rabbit IRDye 800 or goat anti–mouse IRDye 700) were used at 1:10,000. Blots were scanned with an Odyssey Infrared Imaging System (LI-COR Biosciences), and band intensity was quantified with ImageJ.
Mass spectrometry
To examine after translational modification of Nap1, recombinant Strep-tagged wild-type Nap1 was added to Nap1-depleted egg extracts and incubated for 30 min at 4°C with gentle shaking. The protein was then retrieved on Strep-Tactin resin, eluted with SDS sample buffer, and subjected to 12% SDS-PAGE. Nap1 bands were excised, extracted from the gel, and adjusted to 8 M urea. After carboxyamidomethylation of cysteines, samples were digested with trypsin and Asp-N proteases and submitted for analysis by either by the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory at University of California, Berkeley or with the assistance of S. Danielson at Thermo Fisher Scientific (San Jose, CA).
Protein identification and quantification were performed with Integrated Proteomics Pipeline (IP2; Integrated Proteomics Applications, Inc.) using ProLuCID/Sequest, DTASelect2, and Census (Tabb et al., 2002; Peng et al., 2003; McDonald et al., 2004; Xu et al., 2006; Cociorva et al., 2007; Park et al., 2008). Under the filtering conditions used, the estimated false discovery rate was <0.1% at both the peptide and protein levels for all samples.
Nap1 glutamylation and H1 deposition assays
Recombinant TTLL4 was provided by C. Janke (Institut Curie Research Centre, Paris, France) and tested for Nap1 glutamylation in vitro as described in Van Dijk et al. (2007). In brief, 0.2 µM recombinant or mutant Nap1 and 1 µM TTLL4 enzyme were combined in buffer (20 µl; 50 mM Hepes, pH 8.0, 400 µM ATP, 2.4 mM MgCl2, 500 µM DTT, and 8 µM glutamate), mixed gently, and incubated at 23°C for 2 h. Reactions were used to assay H1M deposition on chromatin beads or stopped by adding SDS sample buffer and analyzed by Western blotting using Nap1 and γ-glutamylation antibodies.
Nap1 glutamylation reactions were added to demembranated Xenopus sperm nuclei diluted to 20 nuclei/µl in 50 mM Hepes buffer, pH 8.0, so that the final concentration of Nap1 in each 20 µl sperm reaction was 0.2 µM. A final concentration of 0.2 µM of H1M-GFP was then added, and reactions were incubated for 20 min at 23°C, fixed with formaldehyde, stained with Hoechst, and immediately imaged by fluorescence microscopy.
Chromatin beads were prepared as described in Hannak and Heald (2006). In brief, a ratio of 1 µg biotinylated MCP DNA (Bluescript plasmid containing a 5-kb insert of noncoding Drosophila DNA) was coupled to 5 µl of M280 Streptavidin Dynabeads (Invitrogen). Beads were then added to Xenopus egg extracts depleted of H1M to assemble chromatin and cycled through interphase to allow DNA replication. Chromatin beads from 30-µl reactions were then retrieved on a magnetic stand and added to either Nap1-depleted egg extracts or buffer (50 mM Hepes, pH 8.0, 400 µM ATP, 2.4 mM MgCl2, 8 µM glutamate, and 500 µM DTT, containing 3 µM TTLL4) supplemented with 2 µM recombinant wild-type or mutant Nap1 and 2 µM H1M-GFP. After a 1-h incubation at 23°C, chromatin beads were retrieved on a magnetic stand, washed twice with 50 mM Hepes, added to 20 µl SDS sample buffer, and analyzed by Western blotting using H1M antibodies.
FRAP
Reactions supplemented with 1.5 µM H1M-GFP and 500 nuclei/µl extract were cycled through interphase and either arrested by the addition of 100 µg/ml cycloheximide or driven back into mitosis by the addition of fresh metaphase-arrested egg extract. 2 µl of reaction was spotted onto a polyethylene glycol–coated slide, overlaid with a 12-mm circular coverglass, and imaged every 300 ns with a 60× oil objective (Plan Apochromat, NA 1.40) on a confocal microscope (Axiovert 200M; Carl Zeiss). A 3-s photobleach at 100% power for the Argon/488-nm laser was applied to a 1–2-µm-diameter circle on individual metaphase plates or interphase nuclei. Image stacks were aligned and curve fitted using the FRAP Profiler plugin for ImageJ.
Online supplemental material
Fig. S1 shows the distribution of lengths and widths of individual chromosomes in Nap1-depleted egg extracts as well as the quantification of chromatin-bound and cytoplasmic H1M. Fig. S2 shows that reducing N- or C-terminal glutamylation alone does not affect spindle assembly or chromosome condensation and that Nap1N6C9 is competent to decondense Xenopus sperm nuclei. Fig. S3 shows the distribution of lengths and widths of individual chromosomes in Nap1-depleted egg extracts supplemented with Nap1N6C9.
Acknowledgments
We thank Dr. Keita Ohsumi for the Nap1 expression clone and antisera used in preliminary experiments and Dr. David Shechter for anti–γ-glutamyl antibodies and interesting discussions. We also thank Dr. Carsten Janke for the TTLL4 expression clone. We extend special thanks to Lori Kohlstaedt, Sharleen Zhou, and staff at the Vincent J. Coates Proteomics Facility (University of California, Berkeley) for quality assistance with mass spectrometry. We also thank Beno Freedman for advice throughout the project, M. Strzelecka, M. Ellefson-Crowder, and A. Lane for comments on the manuscript, and the Heald laboratory for continued support and helpful discussions.
This work was supported by National Institutes of Health grant R01 GM057839 (R. Heald) and the Flora Lamson Hewlett Fund (R. Heald and K.E. Miller).
The authors declare no competing financial interests.