Chromosome congression requires the stable attachment of microtubules to chromosomes mediated by the kinetochore, a large proteinaceous structure whose mechanism of assembly is unknown. In this paper, we present the finding that a protein called TRAMM (formerly known as TrappC12) plays a role in mitosis. Depletion of TRAMM resulted in noncongressed chromosomes and arrested cells in mitosis. Small amounts of TRAMM associated with chromosomes, and its depletion affected the localization of some kinetochore proteins, the strongest effect being seen for CENP-E. TRAMM interacts with CENP-E, and depletion of TRAMM prevented the recruitment of CENP-E to the kinetochore. TRAMM is phosphorylated early in mitosis and dephosphorylated at the onset of anaphase. Interestingly, this phosphorylation/dephosphorylation cycle correlates with its association/disassociation with CENP-E. Finally, we demonstrate that a phosphomimetic form of TRAMM recruited CENP-E to kinetochores more efficiently than did the nonphosphorylatable mutant. Our study identifies a moonlighting function for TRAMM during mitosis and adds a new component that regulates kinetochore stability and CENP-E recruitment.
Introduction
Replication and proper segregation of the genome of an organism is critical for its survival. Defects in segregation result in aneuploid cells that are hallmarks of genetic disorders and some types of cancer. In humans, a key step in genome segregation is the ability of microtubules to properly attach to replicated chromosomes, a process that is mediated by a macromolecular structure known as the kinetochore (Cheeseman, 2014) that may contain as many as 200 associated proteins (Ohta et al., 2010; Tipton et al., 2012).
In addition to mediating microtubule attachment, the kinetochore also acts as a scaffold upon which proteins involved in the spindle assembly checkpoint (SAC) are recruited (Cleveland et al., 2003). The SAC prevents the onset of anaphase until all chromosomes are properly attached to microtubules (Malmanche et al., 2006). Key SAC proteins such as Mad1, Mad2, BubR1, Bub1, and Cdc20 repress the activity of the anaphase-promoting complex/cyclosome until biorientation of the chromosomes and generation of tension by attached microtubules.
Microtubule attachment to the kinetochore is mediated in large part by a network of proteins referred to as the KMN network, composed of Knl1, and the Mis12 and Ndc80 complexes (Maiato et al., 2004; Cheeseman et al., 2006; Varma and Salmon, 2012). Another key component involved in stabilizing microtubule–kinetochore interactions is CENP-E, a plus end–directed kinesin that binds to microtubules (Putkey et al., 2002). CENP-E also functions in SAC amplification (Abrieu et al., 2000; Weaver et al., 2003), and thus, it is a key regulator of early stages of mitosis. However, depletion of CENP-E prevents some, but not all, chromosomes from binding to microtubules (Schaar et al., 1997; Putkey et al., 2002; Tanudji et al., 2004), suggesting that the protein may be part of a secondary mechanism for chromosome capture (McEwen et al., 2001; Kapoor et al., 2006).
The complex nature of the kinetochore suggests that there must be an ordered recruitment to ensure its proper assembly, and several possibilities for the temporal assembly of the kinetochore have been postulated (Gascoigne and Cheeseman, 2011; Basilico et al., 2014). Although organized into outer and inner layers, there are nevertheless contacts between these layers (Liu et al., 2003; Cheeseman and Desai, 2008; Gascoigne and Cheeseman, 2013; Hornung et al., 2014). Given the large number of kinetochore proteins, it is not unexpected that localization of some proteins is dependent on the function of other proteins (Johnson et al., 2004; Santaguida and Musacchio, 2009).
Our laboratory has been studying the mechanism of membrane trafficking, a process that ensures the proper organization of subcellular compartments. In particular, we have focused on a large protein complex called transport protein particle (TRAPP) that has been shown to regulate several membrane trafficking pathways (Sacher et al., 1998, 2001, 2008; Cai et al., 2005; Lynch-Day et al., 2010). Here, we report the unexpected finding that one TRAPP subunit that we now call TRAMM (previously known as TrappC12 or TTC15), functions in mitosis. Our study leads us to propose a model whereby TRAMM cycles between its role in TRAPP in interphase cells, and its newly identified roles during mitosis where it regulates stability of the kinetochore and participates in CENP-E recruitment.
Results and discussion
Depletion of TRAMM/TrappC12 results in a chromosome congression failure
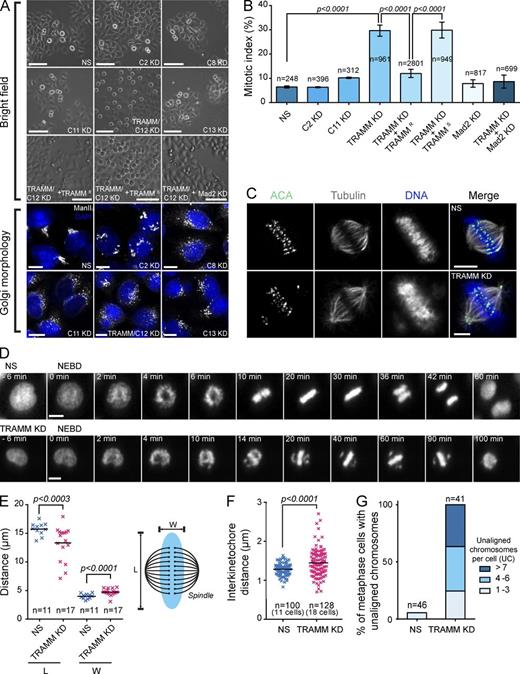
Two forms of the TRAPP complex have been described in mammalian cells called TRAPP II and TRAPP III (Bassik et al., 2013). To study the function of TRAPP III, we depleted HeLa cells of the four subunits specific to TRAPP III (TrappC8, C11, C12, and C13) as well as a core subunit of the complex (TrappC2) using siRNA. Unexpectedly, depletion of the TrappC12 subunit arrested the cells in mitosis (Fig. 1 A). Quantitation of this effect indicated that the mitotic index increased from 6.4 ± 0.43% for a nonspecific siRNA (NS) to 29.7 ± 2.3% after depletion of TrappC12 (Fig. 1 B). A similar result was seen using a second technique called knocksideways (unpublished data; Robinson et al., 2010). Given the additional role of this protein in mitosis, as elaborated upon in this paper, we have renamed the protein TRAMM, reflecting its dual role in the trafficking of membranes and mitosis.
Depletion of TRAMM results in a mitotic arrest as a result of a chromosome congression failure. (A) HeLa cells were photographed by brightfield microscopy at 20× magnification 24 h after treatment with siRNA against the TRAPP subunit indicated or Mad2. In some cases, the cells were cotransfected with siRNA-resistant or -sensitive TRAMM. For simplicity “Trapp” is omitted for each subunit. Depletion was assessed by Golgi morphology using anti–mannosidase II and Western analysis for subunits to which antibodies were available (Fig. S1 A). Bars: (brightfield images) 200 µm; (fluorescence images) 5 µm. (B) Cells from A were quantitated by counting the number of mitotic cells in multiple fields over multiple experiments. Error bars indicate SD. Significance was assessed by an unpaired t test. (C) HeLa cells were fixed 24 h after treatment with nonspecific (NS; top row) or TRAMM/TrappC12-specific (bottom row) siRNA. Staining was performed with antitubulin, ACA, and 7-aminoactinomycin D. Bars, 5 µm. (D) HeLa cells expressing fluorescently labeled histone H2B were treated with NS (top row) or TRAMM/TrappC12-specific (bottom row) siRNA. The cells shown were recorded 20 h after treatment. Frames from Videos 1 and 2 were captured, and times indicated are relative to nuclear envelope breakdown (NEBD). Bars, 10 µm. (E–G) Measurements of chromosome boundary dimensions (E), interkinetochore distances (F), and unaligned chromosomes (G) were made after treatment of HeLa cells as in A. Chromosome boundary length (L) and width (W) were measured between the spindle poles and perpendicular to the poles, respectively. Results are shown ± SD, and significance was assessed by an unpaired t test. KD, knockdown.
Depletion of TRAMM results in a mitotic arrest as a result of a chromosome congression failure. (A) HeLa cells were photographed by brightfield microscopy at 20× magnification 24 h after treatment with siRNA against the TRAPP subunit indicated or Mad2. In some cases, the cells were cotransfected with siRNA-resistant or -sensitive TRAMM. For simplicity “Trapp” is omitted for each subunit. Depletion was assessed by Golgi morphology using anti–mannosidase II and Western analysis for subunits to which antibodies were available (Fig. S1 A). Bars: (brightfield images) 200 µm; (fluorescence images) 5 µm. (B) Cells from A were quantitated by counting the number of mitotic cells in multiple fields over multiple experiments. Error bars indicate SD. Significance was assessed by an unpaired t test. (C) HeLa cells were fixed 24 h after treatment with nonspecific (NS; top row) or TRAMM/TrappC12-specific (bottom row) siRNA. Staining was performed with antitubulin, ACA, and 7-aminoactinomycin D. Bars, 5 µm. (D) HeLa cells expressing fluorescently labeled histone H2B were treated with NS (top row) or TRAMM/TrappC12-specific (bottom row) siRNA. The cells shown were recorded 20 h after treatment. Frames from Videos 1 and 2 were captured, and times indicated are relative to nuclear envelope breakdown (NEBD). Bars, 10 µm. (E–G) Measurements of chromosome boundary dimensions (E), interkinetochore distances (F), and unaligned chromosomes (G) were made after treatment of HeLa cells as in A. Chromosome boundary length (L) and width (W) were measured between the spindle poles and perpendicular to the poles, respectively. Results are shown ± SD, and significance was assessed by an unpaired t test. KD, knockdown.
Careful examination of the TRAMM knockdown-induced mitotic phenotype revealed several chromosomes at the spindle poles (Fig. 1 C). To better understand how the polar chromosome phenotype is established, live-cell imaging was performed using HeLa cells expressing fluorescently tagged histone H2B. We observed that the polar chromosomes never migrated to the metaphase plate, indicating a chromosome congression defect (Fig. 1 D and Videos 1 and 2). These results demonstrate that TRAMM, to the exclusion of other tested TRAPP III subunits, has an unexpected role in mitosis.
HeLa cells expressing mCherry–histone H2B and GalT-GFP were treated with NS. Images were acquired by time-lapse microscopy using a swept-field confocal microscope (Live Scan Swept Field confocal microscope; Nikon) between 8 and 24 h after transfection with the siRNA. Frames were imaged every 2 min. Normal mitosis is seen in these cells.
HeLa cells expressing mCherry–histone H2B and GalT-GFP were treated with NS. Images were acquired by time-lapse microscopy using a swept-field confocal microscope (Live Scan Swept Field confocal microscope; Nikon) between 8 and 24 h after transfection with the siRNA. Frames were imaged every 2 min. Normal mitosis is seen in these cells.
HeLa cells expressing mCherry–histone H2B and GalT-GFP were treated with siRNA targeting TRAMM. Images were acquired by time-lapse microscopy using a swept-field confocal microscope (Live Scan Swept Field confocal microscope; Nikon) between 8 and 24 h after transfection with the siRNA. Frames were imaged every 2 min. Note that the cells arrested in mitosis as a result of the chromosome congression failure remain in this state for the duration of the video.
HeLa cells expressing mCherry–histone H2B and GalT-GFP were treated with siRNA targeting TRAMM. Images were acquired by time-lapse microscopy using a swept-field confocal microscope (Live Scan Swept Field confocal microscope; Nikon) between 8 and 24 h after transfection with the siRNA. Frames were imaged every 2 min. Note that the cells arrested in mitosis as a result of the chromosome congression failure remain in this state for the duration of the video.
TRAMM depletion activates the SAC
To further characterize the TRAMM depletion phenotype, we measured various parameters including length and width of the chromosomal boundary (ChB) during metaphase, interkinetochore distance, and the number of noncongressed chromosomes. After TRAMM depletion, mean ChB width increased from 3.91 to 4.55 µm, whereas mean ChB length decreased from 15.61 to 13.04 µm (Fig. 1 E). A small but significant increase in mean interkinetochore distance on aligned chromosomes was also seen after TRAMM depletion (from 1.28 to 1.45 µm; Fig. 1 F). The number of chromosomes that failed to congress varied from 1 to >10, with the majority of the arrested cells displaying four or more (Fig. 1 G).
The presence of chromosomes at the spindle poles suggests a defect in their biorientation, which would lead to activation of the SAC. Indeed, codepletion of TRAMM and Mad2, a key SAC component, reversed the TRAMM depletion-induced increase in the mitotic index to that of control (Fig. 1, A and B). This suggests that TRAMM depletion leads to activation of the SAC.
A small portion of TRAMM associates with metaphase chromosomes
Given the aforementioned results, we reasoned that a portion of TRAMM may be found in the nucleus. Indeed, cellular fractionation indicated that a small but reproducible amount of TRAMM cofractionated with a nuclear marker (Fig. 2 A). Other TRAPP subunits (TrappC11 and TrappC3) were not found in the nucleus. Although a small amount of TrappC2 was found in the nucleus, this was not unexpected because this protein functions in transcription (Ghosh et al., 2001, 2003). Purification of chromosomes from metaphase-arrested cells showed a small amount of TRAMM associating with the chromosomes, though the overwhelming majority was unassociated (Fig. 2 B). This was consistent with chromosome spreads that showed a cellular staining not typical of stably bound kinetochore proteins (Fig. S1 B).
TRAMM affects kinetochore stability. (A) HeLa cells were fractionated into cytosolic (Cyt) and nuclear (Nucl) fractions and analyzed by Western analysis. A portion of the total lysate (TL) was also included. (B) Chromosomes were purified from HeLa cells that were treated with colcemid. The fraction used for final centrifugation (total lysate) generated supernatant (Spnt) and chromosome (Chr) fractions. (C) Purified mitotic chromosomes were stained for endogenous TRAMM and ACA (+) or with secondary antibodies only (−). Although the majority of chromosomes were not positive for TRAMM staining, some chromosomes showed a TRAMM signal at one or both kinetochores (representative images are shown). Ab, antibody. Bars, 2 µm. (D) HeLa cells were treated with NS or TRAMM-specific siRNA for 24 h. A portion of the cells were fixed and stained with ACA and an antibody against the indicated protein. A second portion was lysed and subjected to Western analysis using an antibody against the indicated protein. Tubulin was used as a loading control. The signal intensity in the table indicates fluorescence intensity of the TRAMM-depleted sample as a percentage of the NS siRNA control ± SD.
TRAMM affects kinetochore stability. (A) HeLa cells were fractionated into cytosolic (Cyt) and nuclear (Nucl) fractions and analyzed by Western analysis. A portion of the total lysate (TL) was also included. (B) Chromosomes were purified from HeLa cells that were treated with colcemid. The fraction used for final centrifugation (total lysate) generated supernatant (Spnt) and chromosome (Chr) fractions. (C) Purified mitotic chromosomes were stained for endogenous TRAMM and ACA (+) or with secondary antibodies only (−). Although the majority of chromosomes were not positive for TRAMM staining, some chromosomes showed a TRAMM signal at one or both kinetochores (representative images are shown). Ab, antibody. Bars, 2 µm. (D) HeLa cells were treated with NS or TRAMM-specific siRNA for 24 h. A portion of the cells were fixed and stained with ACA and an antibody against the indicated protein. A second portion was lysed and subjected to Western analysis using an antibody against the indicated protein. Tubulin was used as a loading control. The signal intensity in the table indicates fluorescence intensity of the TRAMM-depleted sample as a percentage of the NS siRNA control ± SD.
Because small amounts of TRAMM associate with chromosomes, and because the chromosome spreads could not be used to assess its localization on the chromosomes, we purified mitotic chromosomes and stained them for anticentromere antibodies (ACA) and TRAMM. This procedure removes the large amount of nonchromosomal protein, making it possible to determine the localization of the small amounts of TRAMM associating with the chromosomes. Although the majority of chromosomes did not contain TRAMM, some chromosomes had TRAMM associated with either one or both ACA-positive structures (Fig. 2 C). Collectively, these results suggest a weak chromosomal association for TRAMM and that the small amounts of TRAMM on chromosomes associate with ACA-positive structures representing the kinetochore.
Kinetochore structure is affected upon TRAMM depletion
We next addressed what effect TRAMM depletion had on kinetochore structure. To do this, we quantitated the fluorescence intensity of kinetochore components on aligned chromosomes in the presence or absence of TRAMM. The intensity of the ACA signal was used as a reference for the quantitation to which all fluorescence intensities were normalized (Meraldi and Sorger, 2005; Liu et al., 2007). Depletion of TRAMM did not significantly alter the cellular levels of any of the proteins measured (Fig. 2 D). However, dramatic effects on kinetochore localization were seen for several outer layer components including CENP-E, CENP-F, the MIS12 complex and BubR1, but not for Knl1, the NDC80 complex, and INCENP (an inner layer protein; Fig. 2 D and Fig. S2).
The decrease in fluorescence intensity at the kinetochore for CENP-E after TRAMM depletion was the most dramatic and resulted in levels only 6% of those seen using an NS. Although depletion of CENP-E did not affect the overall levels of TRAMM, it similarly resulted in a decrease in the number of kinetochores that were positive for TRAMM (unpublished data). In summary, TRAMM affects the localization of some components of the outer layer of the kinetochore.
CENP-E recruitment to kinetochores is dependent on TRAMM
Our results thus far suggest that TRAMM may cooperate with CENP-E to mediate chromosome congression. Although the TRAMM depletion phenotype is very similar to that of CENP-E depletion (Fig. 3 A), codepletion of TRAMM and CENP-E did not display an exacerbated phenotype (Fig. 3, A and B), suggesting that TRAMM and CENP-E may act together in chromosome congression with depletion of either protein resulting in a similar effect.
Recruitment of CENP-E to the kinetochore is dependent on TRAMM. (A) HeLa cells were transfected with NS or siRNA against TRAMM, CENP-E, or both TRAMM and CENP-E as indicated. After 24 h, the cells were fixed and stained with antitubulin, ACA, and anti–histone H2B. Shown next to each image is a brightfield image of the cells. The mitotic index is indicated in the brightfield images as a percentage ± SD. The intermediate value obtained for the double depletion was seen in four independent experiments. Bars: (fluorescent images) 5 µm; (brightfield images) 100 µm. A Western analysis of each condition is shown to the right of A. (B) After staining the cells, ACA-positive pairs that were not aligned in the metaphase plate were counted and expressed as 1–5 or >5 pairs. (C) TRAPP subunits in a Gal4-DNA binding domain vector (BD; pGBKT7) were transformed into yeast cells and mated with a yeast strain containing CENP-E (amino acids 2,131–2,701) in a Gal4 activation domain vector (AD; pGADT7). To assess autoactivation, each TRAPP subunit was also transformed into a yeast strain with an empty Gal4 activation domain vector (Ø). Growth on DDO (−Trp/−Leu) plates indicates mating between the two strains, whereas growth on TDO (−Trp/−Leu/−His) plates containing 4 mM 3-aminotriazol (3-AT) indicates a protein–protein interaction. (D) HeLa cells were treated with NS or TRAMM-specific siRNA. After 24 h, the cells were left untreated or treated with nocodazole (Noc) as indicated. The cells were then fixed and stained for ACA and anti–CENP-E. Bars, 5 µm. (E) Fluorescence intensity was measured for CENP-E signals in D that colocalized with ACA. The intensity of NS cells not treated with nocodazole was adjusted to 100%. Values from two independent experiments ± SD are indicated. KD, knockdown.
Recruitment of CENP-E to the kinetochore is dependent on TRAMM. (A) HeLa cells were transfected with NS or siRNA against TRAMM, CENP-E, or both TRAMM and CENP-E as indicated. After 24 h, the cells were fixed and stained with antitubulin, ACA, and anti–histone H2B. Shown next to each image is a brightfield image of the cells. The mitotic index is indicated in the brightfield images as a percentage ± SD. The intermediate value obtained for the double depletion was seen in four independent experiments. Bars: (fluorescent images) 5 µm; (brightfield images) 100 µm. A Western analysis of each condition is shown to the right of A. (B) After staining the cells, ACA-positive pairs that were not aligned in the metaphase plate were counted and expressed as 1–5 or >5 pairs. (C) TRAPP subunits in a Gal4-DNA binding domain vector (BD; pGBKT7) were transformed into yeast cells and mated with a yeast strain containing CENP-E (amino acids 2,131–2,701) in a Gal4 activation domain vector (AD; pGADT7). To assess autoactivation, each TRAPP subunit was also transformed into a yeast strain with an empty Gal4 activation domain vector (Ø). Growth on DDO (−Trp/−Leu) plates indicates mating between the two strains, whereas growth on TDO (−Trp/−Leu/−His) plates containing 4 mM 3-aminotriazol (3-AT) indicates a protein–protein interaction. (D) HeLa cells were treated with NS or TRAMM-specific siRNA. After 24 h, the cells were left untreated or treated with nocodazole (Noc) as indicated. The cells were then fixed and stained for ACA and anti–CENP-E. Bars, 5 µm. (E) Fluorescence intensity was measured for CENP-E signals in D that colocalized with ACA. The intensity of NS cells not treated with nocodazole was adjusted to 100%. Values from two independent experiments ± SD are indicated. KD, knockdown.
Given the dramatic effect seen on CENP-E localization after a TRAMM knockdown, we asked whether the two proteins physically interact. Although a stable interaction could not be demonstrated using lysates from cultured cells, a yeast two-hybrid interaction could be demonstrated between TRAMM and CENP-E (Fig. 3 C). No other TRAPP subunit examined displayed such an interaction.
Quantitation of kinetochore protein localization at metaphase (Fig. 2 D) did not address recruitment to the kinetochore that precedes alignment of the chromosomes. To address this, cells were depleted of TRAMM and then microtubule–chromosome attachments were disrupted by a brief treatment with nocodazole. This has been shown to allow for rerecruitment (i.e., a nocodazole-induced increase in fluorescence intensity) of components including CENP-E to the kinetochore (Johnson et al., 2004). We then quantitated the levels of CENP-E at kinetochores (colocalized with ACA). As shown in Fig. 3 (D and E), treatment of the cells with nocodazole resulted in a slight increase in the colocalization of CENP-E with kinetochores. Importantly, colocalization between the CENP-E and ACA signals was greatly reduced in the absence of TRAMM, and the addition of nocodazole did not result in an increase in fluorescence intensity. In contrast, although largely soluble and challenging to assess (Fig. 2, A and B; and Fig. S1 B), nocodazole treatment did not result in an increase in colocalization between TRAMM and ACA in the presence or absence of CENP-E (unpublished data). Therefore, our results suggest that recruitment of CENP-E to the kinetochore is dependent on TRAMM.
Phosphorylation of TRAMM is important for its mechanism of action during mitosis
Because TRAMM is the only component of TRAPP that also functions during mitosis, we reasoned that it may be released from the TRAPP holocomplex during this stage of the cell cycle. As seen in Fig. 4 A, TRAMM from untreated cells had a broad size distribution on a size-exclusion column (fractions 19–25), a portion of which overlapped with the TRAPP complex-containing fractions (not depicted). However, after colcemid treatment, TRAMM displayed a shift to a smaller molecular size, peaking in fractions 24–25, suggesting that TRAMM is indeed no longer part of the TRAPP holocomplex during mitosis.
Reversible phosphorylation of TRAMM is important for its mitotic function. (A) HeLa cells were untreated (Async, asynchronous) or treated with colcemid (Colc). Lysates were fractionated on a Superose 6 size exclusion column and probed for TRAMM. The migration of molecular size standards is indicated above the top blot. The location of the TRAPP complex was determined by Western analysis of TrappC11 (not depicted) and by a previous study using the same column (Bassik et al., 2013). Black lines indicate that intervening lanes have been spliced out. TL, total lysate. (B) Lysates were prepared from HeLa cells that were either untreated (asynchronous), treated with thymidine (Thym), or treated with colcemid. A portion of each lysate was left untreated (−) or treated with λ-phosphatase (λ-PPase; +) before Western analysis. (C) Treatment of cells in B was repeated using HeLa, A549, and HT1080 cells. p-Hist-H3, phospho–histone H3. (D) HeLa cells were arrested at the G1/S boundary by treatment with thymidine, and then, the cells were transferred to regular growth medium containing nocodazole. Samples were removed at the times indicated (hours) and examined by Western analysis. (E) HeLa cells were untreated or treated with either colcemid, RO-3306, or both colcemid and RO-3306, as indicated. Cells were then lysed, and proteins were detected by Western analysis. (F) HeLa cells were arrested in prometaphase by treatment with nocodazole and released into regular growth medium. Samples were removed at the times indicated (hours) and examined by Western analysis. (G) HeLa cells were treated with NS or TRAMM-specific siRNA. In some cases, the cells were cotransfected with a plasmid expressing an siRNA-resistant form of either wild-type (WT) TRAMM, TRAMM-5A, or TRAMM-5D, as indicated. Results are shown ± SD, and significance was assessed by an unpaired t test. KD, knockdown.
Reversible phosphorylation of TRAMM is important for its mitotic function. (A) HeLa cells were untreated (Async, asynchronous) or treated with colcemid (Colc). Lysates were fractionated on a Superose 6 size exclusion column and probed for TRAMM. The migration of molecular size standards is indicated above the top blot. The location of the TRAPP complex was determined by Western analysis of TrappC11 (not depicted) and by a previous study using the same column (Bassik et al., 2013). Black lines indicate that intervening lanes have been spliced out. TL, total lysate. (B) Lysates were prepared from HeLa cells that were either untreated (asynchronous), treated with thymidine (Thym), or treated with colcemid. A portion of each lysate was left untreated (−) or treated with λ-phosphatase (λ-PPase; +) before Western analysis. (C) Treatment of cells in B was repeated using HeLa, A549, and HT1080 cells. p-Hist-H3, phospho–histone H3. (D) HeLa cells were arrested at the G1/S boundary by treatment with thymidine, and then, the cells were transferred to regular growth medium containing nocodazole. Samples were removed at the times indicated (hours) and examined by Western analysis. (E) HeLa cells were untreated or treated with either colcemid, RO-3306, or both colcemid and RO-3306, as indicated. Cells were then lysed, and proteins were detected by Western analysis. (F) HeLa cells were arrested in prometaphase by treatment with nocodazole and released into regular growth medium. Samples were removed at the times indicated (hours) and examined by Western analysis. (G) HeLa cells were treated with NS or TRAMM-specific siRNA. In some cases, the cells were cotransfected with a plasmid expressing an siRNA-resistant form of either wild-type (WT) TRAMM, TRAMM-5A, or TRAMM-5D, as indicated. Results are shown ± SD, and significance was assessed by an unpaired t test. KD, knockdown.
A band of slightly reduced mobility was seen in fractions 24–25 from asynchronous cells (Fig. 4 A). Furthermore, the mobility of TRAMM in colcemid-treated cells was also reduced to 83 from 79 kD. These results suggest that TRAMM may be mitotically phosphorylated. Indeed, colcemid treatment led to the appearance of slower-migrating forms of TRAMM that increased in mobility after phosphatase treatment (Fig. 4 B). Similar results were seen in A549 and HT1080 cells (Fig. 4 C). These results indicate that TRAMM is mitotically phosphorylated.
We next examined the timing of TRAMM phosphorylation. Cells were synchronized at the G1/S boundary by thymidine treatment and then released into medium containing nocodazole. Samples were probed for TRAMM, cyclin B1, and phospho–histone H3. The levels of cyclin B1 are low during G1 phase and increase steadily through S phase, peaking during early mitosis (Pines and Hunter, 1989), whereas phospho–histone H3 appears in G2 and peaks early in mitosis (Hendzel et al., 1997). The appearance of phosphorylated TRAMM was seen at ∼11 h after release from the thymidine treatment (Fig. 4 D). This coincided with the peak of phospho–histone H3 but was preceded by the appearance of cyclin B1. As a further indication of the timing of TRAMM phosphorylation, cells were treated with RO-3306 (an inhibitor of CDK1 that arrests cells at the G2/M boundary), either in the presence or absence of colcemid. As shown in Fig. 4 E, RO-3306 prevented the colcemid-induced phosphorylation of TRAMM. Collectively, our data suggest that TRAMM phosphorylation occurs as cells enter mitosis.
To examine the dephosphorylation of TRAMM, cells were arrested in prometaphase by treatment with nocodazole and then released into medium without nocodazole. Extensive dephosphorylation of TRAMM was seen between 3 and 4 h after release from nocodazole (Fig. 4 F). This coincided with the degradation of cyclin B1, which occurs immediately before entry into anaphase (Clute and Pines, 1999). Collectively, our analysis suggests that TRAMM is phosphorylated as the cells enter mitosis but is dephosphorylated at or before the onset of anaphase.
To determine which residues of TRAMM are phosphorylated, we used a combination of mass spectrometry, bioinformatic predictions, and previously published phosphoproteomic analyses (Dephoure et al., 2008; Mayya et al., 2009; Kettenbach et al., 2011). Our combined approach led us to examine five potential residues: T107, S109, S127, S182, and S184 (Fig. S2). Mutants that had all of these sites changed to either nonphosphorylatable alanine residues (TRAMM-5A) or phosphomimetic aspartic acid residues (TRAMM-5D) were generated and made siRNA resistant. We then examined the ability of these mutants to rescue the TRAMM depletion-induced increase in the mitotic index. As shown in Fig. 4 G, although wild-type TRAMM as well as TRAMM-5A could suppress the increased mitotic index of a TRAMM depletion nearly equally, TRAMM-5D showed a reduced ability to suppress this phenotype. Our results suggest that, although phosphorylation of TRAMM is not necessary, it needs to be reversible for proper function.
TRAMM and CENP-E partially colocalize when TRAMM is hyperphosphorylated
Because the recruitment of CENP-E to the kinetochore is dependent on TRAMM (Fig. 2 D and Fig. 3 D), we reasoned that the two proteins may localize to common structures during mitosis. Therefore, we examined the localization of both CENP-E and TRAMM during various stages of the cell cycle. As seen in Fig. 5 A, during interphase, TRAMM is largely localized to a perinuclear region representing the Golgi (also see Scrivens et al., 2011). Although the expression of CENP-E is minimal during interphase and increases as cells enter mitosis (Yen et al., 1992), overexposure of the CENP-E signal of interphase cells reveals a residual amount of the protein in the nucleus of some cells but clearly none in the Golgi. In prophase, both proteins are scattered throughout the cell, but based on the colocalization channel, some of these punctae appear to colocalize, persisting through metaphase (Fig. 5 A, colocalization channel). By anaphase, when TRAMM is dephosphorylated, CENP-E localizes to the midzone between the separating chromosomes, but TRAMM remains scattered in the cell. The distinct localization of these two proteins is more apparent during telophase and cytokinesis in which TRAMM concentrates on Golgi membranes, whereas CENP-E clearly localizes to the midzone and midbody, consistent with previous studies (Yen et al., 1991; Yao et al., 1997). In summary, although TRAMM and CENP-E display distinct localization patterns from anaphase onwards, there is some level of colocalization during early mitosis when TRAMM is maximally phosphorylated. It is noteworthy that, although TRAMM interacts with CENP-E (Fig. 3 C), the colocalization seen by fluorescence microscopy does not necessarily indicate a physical interaction.
Partial colocalization between TRAMM and CENP-E occurs during early mitosis. (A) HeLa cells were fixed and stained with anti-TRAMM, anti–CENP-E, and DAPI. During prophase, prometaphase, and metaphase, when the signal for both proteins was punctate, a colocalization channel was used to demonstrate colocalization of the signals. Bars, 5 µm. (B) HeLa cells were transfected with V5-tagged wild-type (WT) TRAMM, TRAMM-5A, or TRAMM-5D. The cells were fixed and stained with mouse anti-V5, ACA, and anti–CENP-E. Fluorescence intensity measurements were calculated for CENP-E signals that colocalized with ACA in V5-positive cells. The results shown are ± SD. Significance was assessed with an unpaired t test, and representative cells for each condition are shown to the right of the bar graph. (C) A model showing the phosphorylation (P) and dephosphorylation of TRAMM during the cell cycle as well as its localization. The localization cycle for CENP-E based on A is also shown. See the text for details. CCAN, constitutive centromere-associated network.
Partial colocalization between TRAMM and CENP-E occurs during early mitosis. (A) HeLa cells were fixed and stained with anti-TRAMM, anti–CENP-E, and DAPI. During prophase, prometaphase, and metaphase, when the signal for both proteins was punctate, a colocalization channel was used to demonstrate colocalization of the signals. Bars, 5 µm. (B) HeLa cells were transfected with V5-tagged wild-type (WT) TRAMM, TRAMM-5A, or TRAMM-5D. The cells were fixed and stained with mouse anti-V5, ACA, and anti–CENP-E. Fluorescence intensity measurements were calculated for CENP-E signals that colocalized with ACA in V5-positive cells. The results shown are ± SD. Significance was assessed with an unpaired t test, and representative cells for each condition are shown to the right of the bar graph. (C) A model showing the phosphorylation (P) and dephosphorylation of TRAMM during the cell cycle as well as its localization. The localization cycle for CENP-E based on A is also shown. See the text for details. CCAN, constitutive centromere-associated network.
One prediction from the previous result is that phosphorylated TRAMM may recruit CENP-E to kinetochores more efficiently than nonphosphorylated TRAMM. To test this, cells were transfected with V5 epitope-tagged forms of either TRAMM-5D or TRAMM-5A. The cells were then treated with nocodazole to allow for rerecruitment of CENP-E to kinetochores (as in Fig. 3 D). We then quantitated the colocalization of CENP-E with kinetochores in V5-positive cells by measuring the fluorescence intensity of CENP-E. As shown in Fig. 5 B, TRAMM-5D was able to recruit more CENP-E to kinetochores than TRAMM-5A was able to recruit after nocodazole treatment. This supports the notion that phosphorylated TRAMM participates in the recruitment of CENP-E to kinetochores and dephosphorylation of TRAMM precedes or coincides with the distinct localization patterns for these proteins after the onset of anaphase.
Collectively, our study implicates TRAMM both as a key molecule involved in CENP-E recruitment to kinetochores and more generally in a regulatory role in kinetochore stability. Our data suggest that, although phosphorylation of TRAMM is not necessary for CENP-E recruitment to kinetochores, it enhances this activity. However, reversal of phosphorylation is necessary to allow mitosis to progress. Our results fit with a model (Fig. 5 C) whereby TRAMM is released from the TRAPP complex before or during early mitosis by an as-yet-undetermined mechanism. It is tempting to speculate that phosphorylation of TRAMM in late G2/early mitosis may contribute to its mechanism of release from TRAPP. This would correspond to the time when premitotic Golgi fragmentation occurs (Corda et al., 2012). The appearance of a naturally occurring, phosphorylated TRAMM from asynchronous cells in the lower molecular size fractions corresponding to the peak of TRAMM in colcemid-treated cells (Fig. 4 A) is consistent with the mitotic form being highly phosphorylated and not associated with TRAPP. TRAMM appears to have a weak or transient association with kinetochores. Whether this precedes the kinetochore association of CENP-E has not been determined, but the small amounts that do appear at the kinetochore are not dependent on CENP-E. During anaphase, when cyclin B1 levels precipitously drop, there is a sudden decrease in the level of TRAMM phosphorylation. This may suggest that TRAMM is phosphorylated by the CDK1–cyclin B1 complex, and indeed, several of the phosphorylated residues examined in this study conform to the CDK1–cyclin B1 consensus sequence (S/T-P), although variation in this sequence is known to occur (Errico et al., 2010). It should be noted, however, that although the CDK1–cyclin B1 inhibitor RO-3306 prevented phosphorylation of TRAMM, this was likely caused by its blocking of the cells from entering mitosis and does not necessarily indicate that TRAMM is a CDK1–cyclin B1 substrate.
Several proteins have been reported to associate with CENP-E or affect its localization. Although depletion of some of these proteins, including Nuf2, BubR1, and Aurora B, result in altered CENP-E localization (Ditchfield et al., 2003; Johnson et al., 2004; Liu et al., 2007), depletion of others, including SKAP, do not (Huang et al., 2012). In addition, the kinetochore localization of CENP-E can also be affected by SUMOylation of the protein (Zhang et al., 2008). These studies highlight the complex nature by which CENP-E recruitment to kinetochores is governed and further highlight the fact that its localization can be affected by proteins that have not been shown to directly interact with CENP-E. Compared with previous studies, TRAMM depletion has the most dramatic effect on CENP-E localization yet reported. An interaction between TRAMM and CENP-E was seen using a yeast two-hybrid system but not in cell lysates. This may indicate that the interaction between these proteins is weak and transient in nature. Given the number of kinetochore proteins affected by TRAMM depletion, its role at the kinetochore will likely be complex, and we suggest that TRAMM may interact with other kinetochore proteins to facilitate its interaction with CENP-E (Fig. 5 C, indicated by a question mark). A weak association of TRAMM at the kinetochore is consistent with a recent study demonstrating that chicken TRAMM (TTC15) associates with mitotic chromosomes (Ohta et al., 2010). How TRAMM influences kinetochore stability and recruitment of other kinetochore proteins will rely on identification of its full complement of interacting partners.
Although other proteins functioning in membrane traffic have been reported to have mitotic-specific functions (Royle, 2011), TRAMM is unique in that it associates with one large complex during interphase (TRAPP) and influences another large complex (kinetochore) through a transient association during mitosis. The identification of TRAMM as an important factor involved in kinetochore stability and chromosome congression adds to the complexity of this macromolecular structure.
Materials and methods
Cell culture, drug treatments, and cell synchronization
HeLa, A549, HT1080 cells, and a stable HeLa line expressing the chromatin marker mCherry–histone H2B and GalT-GFP (a gift from D.W. Gerlich [Institute of Molecular Biotechnology of the Austrian Academy of Sciences, Vienna, Austria] constructed from the plasmids pH2B-mCherry-IRES-neo3 and pGalT-GFP-IRES-puro2b) were cultured in DMEM (Wisent) supplemented with 10% (vol/vol) FBS (Wisent) at 37°C in a humidified incubator with 5% CO2. Cells were plated on 18-mm glass coverslips precoated with poly-l-lysine (Thermo Fisher Scientific) for immunostaining and on 35-mm glass-bottom microwell dishes (MatTek Corporation) for time-lapse microscopy.
For drug treatments, cells were treated with colcemid (10 µg/ml for 16 h; Life Technologies), nocodazole (100 ng/ml for the times indicated; Sigma-Aldrich), or RO-3306 (9 µM for 16 h; SelleckChem) in DMEM. To synchronize the cells at the G1/S boundary, HeLa cells in 6- or 10-cm-diameter dishes were treated with 2 mM thymidine for ∼18 h. The cells were washed with PBS and grown in DMEM for 6 h. The cells were then treated with a second dose of thymidine for an additional ∼18 h. The cells were washed with PBS, a sample was removed (0 h), and then the cells were placed in DMEM. After 3 h, nocodazole was added to 100 ng/ml. Samples were harvested at various time intervals as noted in Fig. 4 D. For synchronization in prometaphase, the cells were treated with 100 ng/ml nocodazole for ∼18 h. The cells were washed, a sample was removed (0 h), and the cells were placed in DMEM. Samples were harvested at various time intervals as noted in Fig. 4 F. In both cases, the cells were harvested with 600 µl of lysis buffer (50 mM Tris, pH 7.2, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 1% Triton X-100 [vol/vol], one tablet of protease inhibitor cocktail [Roche], and two tablets of PhosStop [Roche] per 10 ml).
DNA transfection and siRNA treatment
Transfection of plasmids and siRNAs (Table 1) was performed using JetPrime (Polyplus) as per the manufacturer’s protocol. The amounts of DNA used were 0.4, 1, and 5 µg for a single well of a 12- or 6-well plate and a 10-cm dish, respectively. All siRNAs were used at a concentration of 10 nM except for TrappC2, which was used at 20 nM. 16 h before transfection, cells were trypsinized and replated at a density of ∼30–40% confluency. Transfection or siRNA treatment was performed when the cells had reached ∼60% confluency. Cells were analyzed by Western blot analysis or fixed for microscopy 24–48 h after transfection or siRNA treatment.
List of siRNAs used in this study
| Targeted gene | Sequence (5′ → 3′) | Source |
| TRAMM | CGGACAAGCUGAACGAACATT | Life Technologies |
| Mad2 | GGGAAGAGUCGGGACCACAGUUUAU | Life Technologies |
| CENP-E | GGUUGACUCAGAUACUACATT | Life Technologies |
| TrappC2 | UCCAUUUUAUGAACCCAAUTT | Life Technologies |
| TrappC2L | AGCCCUUCGAGACAACGAATT | Life Technologies |
| TrappC4 | CGAAAGAUUUAUGAGAUUUTT | Life Technologies |
| TrappC8 | CAGCUCUCCUAAUACGGUUTT | Life Technologies |
| TrappC9 | GGAGAAAGUCAGCAACUAATT | Life Technologies |
| TrappC10 | GUGCCAACUGGCUGACTTTTT | Abnova |
| TrappC11 | GGAUUUAUAAACUACAAGGATT | Life Technologies |
| TrappC13 | GCAAUCAAGUUGUAAAAGATT | Life Technologies |
| Nonspecific | UAACGACGCGACGACGUAATT | Life Technologies |
| Targeted gene | Sequence (5′ → 3′) | Source |
| TRAMM | CGGACAAGCUGAACGAACATT | Life Technologies |
| Mad2 | GGGAAGAGUCGGGACCACAGUUUAU | Life Technologies |
| CENP-E | GGUUGACUCAGAUACUACATT | Life Technologies |
| TrappC2 | UCCAUUUUAUGAACCCAAUTT | Life Technologies |
| TrappC2L | AGCCCUUCGAGACAACGAATT | Life Technologies |
| TrappC4 | CGAAAGAUUUAUGAGAUUUTT | Life Technologies |
| TrappC8 | CAGCUCUCCUAAUACGGUUTT | Life Technologies |
| TrappC9 | GGAGAAAGUCAGCAACUAATT | Life Technologies |
| TrappC10 | GUGCCAACUGGCUGACTTTTT | Abnova |
| TrappC11 | GGAUUUAUAAACUACAAGGATT | Life Technologies |
| TrappC13 | GCAAUCAAGUUGUAAAAGATT | Life Technologies |
| Nonspecific | UAACGACGCGACGACGUAATT | Life Technologies |
Time-lapse microscopy
For live-cell imaging, stable HeLa cells expressing mCherry–histone H2B and GalT-GFP were plated in 35-mm glass-bottom dishes (glass diameter of 14 mm, glass thickness of 1.5; MatTek Corporation). Time-lapse microscopy was performed beginning at 8 or at 24 h after transfection using a 40× oil objective (NA 1.3), no binning, on an inverted confocal microscope (LiveScan Swept Field; Nikon), Piezo Z stage (Nano-Z100N; Mad City Labs, Inc.), and an electron-multiplying charge-coupled device camera (512 × 512; iXon X3; Andor Technology). The microscope was equipped with an environmental chamber heated to 37°C with 5% CO2. Images were acquired with NIS-Elements Version 4.0 acquisition software every 2 min using a 0.2-s exposure at 0.5-µm increment sizes with a slit size of 50 µm for 15–20 h. Images were viewed and analyzed on Imaris version 7.6 (Bitplane) and ImageJ (National Institutes of Health). Images from the videos with corresponding time points were plotted in Illustrator CS6 (Adobe).
Immunofluorescence microscopy
At 24 h after transfection or at the time points indicated, the medium was carefully removed, and coverslips were gently washed twice with PBS. The cells were then fixed and permeabilized in chilled methanol for 4 min at −20°C. Cells were rehydrated in PBS for 20 min and blocked in 5% normal goat serum (Cell Signaling Technology) in PBS for 40 min at room temperature. Primary antibodies (Table 2) were diluted in 5% normal goat serum and were added to coverslips and incubated for 16 h at 4°C. Cells were then washed two times with PBS for 10 min. Cross-adsorbed secondary antibodies (Life Technologies; Table 2) were applied for 1 h at room temperature. Coverslips were washed two times with PBS for 20 min, mounted with Prolong Gold AntiFade reagent (Life Technologies), and sealed with nail polish. 12-bit images with 1,024 × 1,024–pixel resolution were recorded on a confocal microscope (TCS SP2; Leica) with a 100×, NA 1.3 objective (Olympus) running Leica Confocal Software v.2.0. Z stacks were acquired with a 0.2-µm increment. The stacks were then deconvoluted using AutoQuant X3 software (Media Cybernetics).
List of antibodies used in this study
| Antigen | Type | Host | IF dilution | WB dilution | Size | Catalog number | Source |
| TRAMM | P | m | 1:50 | 1:2,000 | 78 | H00051112-B01P | Abnova |
| TRAMM | P | r | 1:200 | 1:2,500 | 78 | N/A | Sacher laboratory |
| Tubulin | M | m | 1:500 | 1:5,000 | 50 | T6199 | Sigma-Aldrich |
| Knl1 | P | r | 1:100 | 1:2,000 | 265 | NBP1-89223 | Novus Biologicals |
| Dsn1 | P | r | 1:1,000 | 1:2,000 | 41 | GTX120402 | GeneTex |
| Dsn1 | P | r | 1:200 | N/A | 41 | N/A | Desai laboratory |
| Nsl1 | P | r | 1:200 | 1:2,000 | 32 | N/A | Desai laboratory |
| Nnf1 | P | r | 1:200 | 1:2,000 | 23 | N/A | Desai laboratory |
| Hec1 | M | m | 1:200 | 1:1,000 | 74 | ab3613 | Abcam |
| Nuf2 | M | r | 1:200 | 1:1,000 | 54 | ab180945 | Abcam |
| Spc24 | P | r | 1:500 | 1:12,500 | 24 | N/A | Stukenberg laboratory |
| Spc25 | P | r | 1:500 | 1:5,000 | 25 | N/A | Stukenberg laboratory |
| Bub1 | M | m | 1:400 | 1:500 | 122 | ab54893 | Abcam |
| BubR1 | P | r | 1:100 | 1:5,000 | 120 | ab70544 | Abcam |
| INCENP | M | r | 1:100 | 1:1,000 | 105 | ab134112 | Abcam |
| CENP-E | P | r | 1:100 | 1:500 | 316 | ab4163 | Abcam |
| CENP-F | P | r | 1:500 | 1:1,500 | 330 | ab5 | Abcam |
| ZW10 | P | r | 1:200 | 1:1,000 | 89 | N/A | Tagaya laboratory |
| Ska1 | P | r | 1:200 | 1:2,000 | 29 | N/A | Cheeseman laboratory |
| Mis12 | P | r | 1:200 | 1:1,500 | 24 | ab70843 | Abcam |
| Cyclin B1 | P | r | 1:200 | 1:1,000 | 58 | 4138 | Cell Signaling |
| Histone H2B | P | r | 1:200 | 1:1,000 | 15 | ab1790 | Abcam |
| Phospho–histone H3 | M | m | N/A | 1:1,000 | 17 | 3377 | Cell Signaling |
| Mad2 | P | r | 1:200 | 1:20,000 | 25 | A300-301A | Bethyl Laboratories, Inc. |
| TrappC2 | P | r | N/A | 1:1,000 | 16 | N/A | Sacher laboratory |
| TrappC3 | P | r | N/A | 1:1,000 | 20 | N/A | Sacher laboratory |
| TrappC11 | P | r | N/A | 1:1,000 | 129 | N/A | Sacher laboratory |
| ACA | P | h | 1:500 | N/A | N/A | 15-234-0001 | Antibodies, Inc. |
| V5 | M | m | 1:300 | 1:1,000 | N/A | ab27671 | Abcam |
| V5 | P | r | 1:200 | N/A | N/A | ab9116 | Abcam |
| GFP | M | m | N/A | 1:1,000 | 27 | 11814460001 | Roche |
| Secondary IgGs | |||||||
| Alexa Fluor 488 anti–human | CA | g | 1:500 | N/A | N/A | A-11013 | Life Technologies |
| Alexa Fluor 568 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-11031 | Life Technologies |
| Alexa Fluor 594 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-11032 | Life Technologies |
| Alexa Fluor 647 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-21236 | Life Technologies |
| Alexa Fluor 647 anti–rabbit | HCA | g | 1:500 | N/A | N/A | A-21245 | Life Technologies |
| HRP-labeledanti–mouse | P | g | N/A | 1:5,000 | N/A | KP-474-1806 | KPL |
| HRP-labeledanti–rabbit | P | g | N/A | 1:5,000 | N/A | KP-474-1506 | KPL |
| Antigen | Type | Host | IF dilution | WB dilution | Size | Catalog number | Source |
| TRAMM | P | m | 1:50 | 1:2,000 | 78 | H00051112-B01P | Abnova |
| TRAMM | P | r | 1:200 | 1:2,500 | 78 | N/A | Sacher laboratory |
| Tubulin | M | m | 1:500 | 1:5,000 | 50 | T6199 | Sigma-Aldrich |
| Knl1 | P | r | 1:100 | 1:2,000 | 265 | NBP1-89223 | Novus Biologicals |
| Dsn1 | P | r | 1:1,000 | 1:2,000 | 41 | GTX120402 | GeneTex |
| Dsn1 | P | r | 1:200 | N/A | 41 | N/A | Desai laboratory |
| Nsl1 | P | r | 1:200 | 1:2,000 | 32 | N/A | Desai laboratory |
| Nnf1 | P | r | 1:200 | 1:2,000 | 23 | N/A | Desai laboratory |
| Hec1 | M | m | 1:200 | 1:1,000 | 74 | ab3613 | Abcam |
| Nuf2 | M | r | 1:200 | 1:1,000 | 54 | ab180945 | Abcam |
| Spc24 | P | r | 1:500 | 1:12,500 | 24 | N/A | Stukenberg laboratory |
| Spc25 | P | r | 1:500 | 1:5,000 | 25 | N/A | Stukenberg laboratory |
| Bub1 | M | m | 1:400 | 1:500 | 122 | ab54893 | Abcam |
| BubR1 | P | r | 1:100 | 1:5,000 | 120 | ab70544 | Abcam |
| INCENP | M | r | 1:100 | 1:1,000 | 105 | ab134112 | Abcam |
| CENP-E | P | r | 1:100 | 1:500 | 316 | ab4163 | Abcam |
| CENP-F | P | r | 1:500 | 1:1,500 | 330 | ab5 | Abcam |
| ZW10 | P | r | 1:200 | 1:1,000 | 89 | N/A | Tagaya laboratory |
| Ska1 | P | r | 1:200 | 1:2,000 | 29 | N/A | Cheeseman laboratory |
| Mis12 | P | r | 1:200 | 1:1,500 | 24 | ab70843 | Abcam |
| Cyclin B1 | P | r | 1:200 | 1:1,000 | 58 | 4138 | Cell Signaling |
| Histone H2B | P | r | 1:200 | 1:1,000 | 15 | ab1790 | Abcam |
| Phospho–histone H3 | M | m | N/A | 1:1,000 | 17 | 3377 | Cell Signaling |
| Mad2 | P | r | 1:200 | 1:20,000 | 25 | A300-301A | Bethyl Laboratories, Inc. |
| TrappC2 | P | r | N/A | 1:1,000 | 16 | N/A | Sacher laboratory |
| TrappC3 | P | r | N/A | 1:1,000 | 20 | N/A | Sacher laboratory |
| TrappC11 | P | r | N/A | 1:1,000 | 129 | N/A | Sacher laboratory |
| ACA | P | h | 1:500 | N/A | N/A | 15-234-0001 | Antibodies, Inc. |
| V5 | M | m | 1:300 | 1:1,000 | N/A | ab27671 | Abcam |
| V5 | P | r | 1:200 | N/A | N/A | ab9116 | Abcam |
| GFP | M | m | N/A | 1:1,000 | 27 | 11814460001 | Roche |
| Secondary IgGs | |||||||
| Alexa Fluor 488 anti–human | CA | g | 1:500 | N/A | N/A | A-11013 | Life Technologies |
| Alexa Fluor 568 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-11031 | Life Technologies |
| Alexa Fluor 594 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-11032 | Life Technologies |
| Alexa Fluor 647 anti–mouse | HCA | g | 1:500 | N/A | N/A | A-21236 | Life Technologies |
| Alexa Fluor 647 anti–rabbit | HCA | g | 1:500 | N/A | N/A | A-21245 | Life Technologies |
| HRP-labeledanti–mouse | P | g | N/A | 1:5,000 | N/A | KP-474-1806 | KPL |
| HRP-labeledanti–rabbit | P | g | N/A | 1:5,000 | N/A | KP-474-1506 | KPL |
All protein sizes are in kilodaltons. N/A, not applicable; M, monoclonal; P, polyclonal; CA, cross-adsorbed; HCA, highly cross-adsorbed; r, rabbit; m, mouse; g, goat; h, human; IF, immunofluorescence; WB, Western blotting.
Data analysis
Interkinetochore distances were manually measured between the centers of the paired ACA spots from a deconvoluted 3D image using the MeasurementPro module of Imaris software. The analysis was performed in ≥11 cells (total n ≥ 100 kinetochore pairs). To estimate significant differences between means, unpaired two-tailed t tests with Welch’s correction, assuming unequal standard deviation, were performed using Prism (GraphPad Software).
Fluorescence intensities of the kinetochore proteins were determined by measuring the integrated fluorescence intensity within a 9 × 9–pixel square positioned over a single kinetochore and subtracting the background intensity of a 9 × 9–pixel square positioned adjacent to the kinetochore region. For the NS condition, the background-corrected kinetochore intensity was determined relative to a background-corrected ACA reference. For the TRAMM knockdown condition, the kinetochore intensity was divided by the intensity from the NS condition. Metaphase cells from the NS condition and cells arrested in mitosis from the TRAMM knockdown condition were chosen for the analyses. At least five cells (>80 kinetochores) per case from two independent experiments were used.
Cell fractionation
Cell fractionation was performed as previously described (Asai et al., 2003). In brief, HeLa cells were grown in four 15-cm dishes. Cells were washed twice with PBS and then collected by scraping with residual PBS into a 15-ml conical tube. The cells were pelleted by centrifugation at 1,700 rpm for 5 min in a table top centrifuge. The pellet was resuspended by vortexing in 9 ml of buffer A (10 mM Hepes. pH 7.6, 10 mM NaCl, 3 mM CaCl2, and 0.5% NP-40, protease inhibitor cocktail tablets; Roche). A portion of the lysate was removed as the total lysate. The remainder of the lysate was centrifuged at 1,700 rpm in a table top centrifuge to obtain the nuclear fraction. The supernatant was aliquoted into microfuge tubes and centrifuged at 13,000 rpm for 5 min, and the supernatant was kept as the cytoplasmic fraction. The nuclear fraction (∼200–300 µl in volume) was washed three times by resuspending in ∼3 ml of buffer A and pelleting as previously in this paragraph until the pellet was white. After the final wash, the pellet was resuspended in 2 ml of buffer B (buffer A with 10 mM EDTA). The nuclei were lysed by sonication (8× for 10 s on and 10 s off at 25% intensity). The lysed nuclear fraction was centrifuged at 13,000 rpm for 5 min, and the supernatant was saved as the nuclear fraction. For Western analysis, samples were loaded after normalizing the volumes.
Chromosome purification protocol
Purification of mitotic chromosomes was performed essentially as previously described (Kulukian et al., 2009). In brief, mitotic HeLa cells from 28 15-cm dishes (arrested with 50 ng/ml colcemid for 16 h) were collected by washing the mitotic cells off the surface in PBS with a pipette (PIPETMAN; Gilson) and collecting into 50-ml conical tubes. Cells were pelleted by centrifugation at 1,200 rpm for 2 min. The pellets were combined and resuspended in 25 ml of hypotonic buffer MPME (5 mM Pipes, pH 7.2, 10 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, and 2 mM EDTA) and incubated at 37°C for 5 min. The swollen cells (∼1 ml in total volume) were collected by centrifugation at 1,200 rpm for 5 min and then resuspended in 5 ml of ice cold lysis buffer (MPME buffer supplemented with protease inhibitors, 0.5 mM spermine, 1 mM spermidine, 1 mM PMSF, 0.1% digitonin, and phosphatase inhibitors [PhosSTOP]). The cells were disrupted by 10 strokes using a glass dounce homogenizer to produce an initial total lysate. The total lysate was transferred to a 50-ml conical tube and centrifuged at 900 rpm in a table top centrifuge for 1 min to pellet intact nuclei and cell debris. The nuclear and cell debris pellet was rewashed in 3 ml of lysis buffer and combined with the initial supernatant. The NaCl concentration of the combine supernatants was raised to 100 mM, the lysate was split in half, and each portion was placed over a sucrose step gradient (2 ml each of 30%, 40%, 50%, and 60%) prepared with lysis buffer supplemented with 100 mM NaCl. The sucrose gradient was centrifuged at 6,300 rpm in a rotor (SW41Ti; Beckman Coulter) for 30 min at 4°C. The flocculent white layer containing chromosomes was collected at the 40–50% and 50–60% interphase and pooled. The chromosomes were diluted with 15 ml of MPME supplemented with 100 mM NaCl (hsMPME) and homogenized by five strokes with a loose-fitting dounce homogenizer. The homogenate was transferred to a 50-ml conical tube and centrifuged at 4°C in table top centrifuge for 15 min at 3,700 rpm. The supernatant was carefully removed, and the loose chromosomal pellet was resuspended in 2 ml hsMPME with 50% sucrose and dounce homogenized with 10 strokes.
Chromosome spreads
Mitotic HeLa cells from two 10-cm dishes (arrested with 1 µg/ml colcemid for 3 h) were collected by washing the mitotic cells off the surface in growth medium with a PIPETMAN and collecting into a 50-ml conical tube. After centrifugation at 200 g in a table top centrifuge, the supernatant was poured off, and with the remaining medium (∼100–200 µl), the cells were resuspended by vigorously tapping the tube. To the resuspended cells, 5 ml of hypotonic buffer (75 mM KCl) was slowly added on the side of the tube while gently tapping the tube. The cells were allowed to swell for 15 min at room temperature, at which time the cells were pelleted at 1,100 rpm in a table top centrifuge for 5 min. The cells were then washed in PBS and centrifuged as before. The cells were resuspended in ∼100–200 µl PBS and then fixed by adding 5 ml of freshly prepared fixative solution (3:1 methanol/glacial acetic acid) and gently inverting the tube. The cells were pelleted and resuspended in 10 ml of fresh fixative solution. This washing in fixative was then repeated. The final cell pellet was resuspended in 1 ml of fixative solution. Several drops were applied to coverslips by allowing them to drop from a height of 50–100 cm to burst the cells. The slides were dried by first placing them on a tray in a 37°C water bath for 1 h to maintain humidity while the methanol and acetic acid evaporate. At this time, the slides were placed on a bench top to completely dry and then stained with antibodies as described in the section Immunofluorescence microscopy.
Western blotting of fractions
Samples of 30 µg were analyzed on 8%, 10%, or 15% polyacrylamide gels (depending on the protein analyzed). The proteins were transferred to nitrocellulose membranes for 1 h at 100 V or overnight at 30 V. Membranes were blocked with 5% skim milk powder or 5% BSA in PBS-T (PBS with 0.1% Tween 20 [vol/vol]). The primary and secondary antibodies used, and their dilutions, are listed in Table 2. Antibodies were incubated in PBS-T for ∼1 h each. Samples were detected using ECL reagent (GE Healthcare) and exposed to film for various times.
Purification of TRAMM and mass spectrometry
HeLa cells were treated with colcemid for 16 h as described in the section Cell culture, drug treatments, and cell synchronization. Cells from 15-cm dishes were collected and lysed in 1 ml lysis buffer (see Cell culture, drug treatments, and cell synchronization) per plate. From the lysate, 40 mg protein was treated with or without 25 µg of mouse anti-TRAMM overnight at 4°C, and the immune complexes were collected onto protein A–Sepharose beads (20 µl) for 2 h in the cold. Immunoprecipitated material was washed in lysis buffer and eluted off the beads in 50 µl of 0.2-M glycine, pH 2.3, or with Laemmli’s buffer. The glycine eluate was neutralized with ∼8 µl of 1-M Tris, pH 9. The extracted material was concentrated into a stacking gel by SDS-PAGE. The gel was then fixed and stained with Coomassie brilliant blue G according to standard procedure. Excised stained proteins from the stacking zone were in-gel digested according to the procedure by Shevchenko et al. (2006). In brief, the gel slices were destained with ammonium bicarbonate/acetonitrile and dehydrated with acetonitrile. The slices were then rehydrated with trypsin to produce tryptic peptides.
Phosphopeptides were enriched on titanium dioxide (TiO2) beads and eluted with ammonium hydroxide, according to the procedure by Thingholm et al. (2006). In brief, the tryptic peptides were diluted fivefold with 2,5-dihydrobenzoic acid in 80% acetonitrile/2% TFA and loaded onto a TiO2 microcolumn. After washing the column with 2,5-dihydrobenzoic acid, phosphopeptides were eluted in a small volume of 25% ammonium hydroxide. The eluted enriched phosphopeptides were subjected to C18 ultra HPLC reverse phase separation, followed by tandem mass spectrometry analysis on a mass spectrometer (Orbitrap Velos; Thermo Fisher Scientific). Data files were formatted and searched with a Mascot Search engine (Matrix Sciences), with acetamidated cysteines set as fixed modification and phospho-S, T, and Y and oxidized methionine set as variable modifications. The data were validated through the Trans-Proteomic Pipeline of Scaffold software (Proteome Software).
Yeast two hybrid
The ORFs encoding all known TRAPP subunits were cloned into pDONR201 using the Gateway cloning method. The ORFs were then transferred to pGBKT7 (ADH1 promoter, multicopy plasmid) that was made Gateway compatible (Scrivens et al., 2011) and transformed into the yeast strain Y187. A region of CENP-E encoding residues 2,131–2,701 was cloned into pGADT7 (ADH1 promoter, multicopy plasmid) and transformed into AH109 yeast cells. The cells were allowed to mate on rich (YPD [yeast, peptone, dextrose]) medium for ∼24 h and then replicated onto double drop out medium (DDO) lacking tryptophan and leucine, as well as onto triple drop out medium (TDO) lacking tryptophan, leucine, and histidine and containing 4 mM 3-aminotriazol.
Knocksideways and mitotic index determination
A construct consisting of an RNAi-resistant form of TRAMM fused to FK506 binding protein (FKBP) was transfected into HeLa cells containing a mitochondrially localized FKBP-rapamycin binding domain derived from mammalian target of rapamycin (Robinson et al., 2010). Cells were simultaneously treated with siRNA targeting the endogenous TRAMM message. After 16 h, the cells were either left untreated or treated with 200 nM rapamycin for 8 h to allow for sequestration of TRAMM-FKBP at the mitochondria. Brightfield images were then acquired on a microscope (Eclipse TS100; Nikon) with a Plan Fluor 10×/0.3 NA objective (Nikon), and the percentage of cells arrested in mitosis was calculated from a minimum of five different fields over two independent experiments.
Online supplemental material
Fig. S1 shows the level of knockdown of several of the TRAPP subunits examined in Fig. 1 and an image from a chromosome spread made from HeLa cells expressing V5-TRAMM, indicating that some of the ectopically expressed protein associates with ACA-positive structures. Fig. S2 shows sample images of kinetochore proteins after treatment of HeLa cells with NS or siRNA targeting TRAMM used to generate the information in Fig. 2 D. Fig. S3 shows identification and conservation of the phosphorylation sites examined in this study. Video 1 shows HeLa cells expressing mCherry-H2B after treatment with an NS. Video 2 shows HeLa cells expressing mCherry-H2B after treatment with siRNA targeting TRAMM.
Acknowledgments
We are grateful to Rodney Joyette for his technical assistance and for the artwork for Fig. 5 C. We thank Drs. Iain Cheeseman (Whitehead Institute for Biomedical Research, Cambridge, MA), Arshad Desai (University of California, San Diego, San Diego, CA), Paul Maddox (University of North Carolina at Chapel Hill, Chapel Hill, NC), Don Cleveland (University of California, San Diego, San Diego, CA), Daniel Gehrlich, Mitsuo Tagaya (Tokyo University of Pharmacy and Life Sciences, Tokyo, Japan), and Todd Stukenberg (University of Virginia, Charlottesville, VA) for reagents and Dr. Alisa Piekny for constructive conversations throughout this work. All microscopy in this study was performed in the Centre for Microscopy and Cellular Imaging at Concordia University.
This study was supported by grants to M. Sacher from the Canadian Institutes of Health Research, the Natural Sciences and Engineering Research Council of Canada, and the Canada Foundation for Innovation. M. Sacher is a member of the Groupe de Recherche Axé sur la Structure des Protéines network.
The authors declare no competing financial interests.