The cortical endoplasmic reticulum (cER) of yeast underlies the plasma membrane (PM) at specific contact sites to enable a direct transfer of information and material between both organelles. During budding, directed movement of cER to the young bud followed by subsequent anchorage at its tip ensures the faithful inheritance of this organelle. The ER membrane protein Scs2p tethers the cER to the PM and to the bud tip through so far unknown receptors. We characterize Epo1p as a novel member of the polarisome that interacts with Scs2p exclusively at the cell tip during bud growth and show that Epo1p binds simultaneously to the Cdc42p guanosine triphosphatase–activating protein Bem3p. Deletion of EPO1 or deletion of BEM3 in a polarisome-deficient strain reduces the amount of cER at the tip. This analysis therefore identifies Epo1p as a novel and important component of the polarisome that promotes cER tethering at sites of polarized growth.
Introduction
As a result of its asymmetric growth and the polar delivery of its organelles, the yeast Saccharomyces cerevisiae is a preferred model system to study the mechanisms and molecules that are responsible for faithful organelle inheritance in eukaryotic cells (Pruyne et al., 2004). The ER of the yeast harbors enzymes for lipid and sugar synthesis, contributes to the structural organization of the nucleus, and is the site of protein synthesis, membrane translocation, and protein complex maturation (Schuldiner and Schwappach, 2013). Although the ER is a single copy organelle, it is structurally not uniform but can be classified into three clearly distinct domains: the membrane of the nuclear envelope, the cortical ER (cER) located as sheets and tubules underneath the plasma membrane (PM), and ER tubules that connect both ER domains and are also occasionally found in close apposition to mitochondria, peroxisomes, and the endosome/vacuole (Estrada de Martin et al., 2005; Shibata et al., 2010; West et al., 2011; Chen et al., 2013). Distinct and not fully characterized protein complexes organize the contact sites between the membrane of the cER and the other organelles (Prinz, 2014). Particularly, the architectures and compositions of the contact sites between cER and PM are far from understood. The cER is tethered to the PM through at least six different proteins: Ist2p, a multispanning membrane protein of the ER, the three tricalbins (Tcb1–3p), peripheral membrane proteins with a synaptotagmin-like domain structure, and Scs2p and Scs22p, the yeast homologues of the human VAMP (vesicle associated membrane protein)–associated protein (Loewen et al., 2007; Manford et al., 2012; Wolf et al., 2012). The simultaneous deletion of all six proteins removes the close apposition between cER and PM almost completely and causes the accumulation of phosphatidylinositol 4-phosphate (PI4P) at the PM (Manford et al., 2012). This effect very probably reflects the spatial separation of the ER-located phosphatase Sac1p from its PM-based substrate PI4P in these cells. Cells lacking cER–PM tethers also display an up-regulated unfolded protein response (Manford et al., 2012). cER–PM contact sites might thus function as hubs for integrating stress signaling pathways and for transmitting information from the cellular outside to the ER (Babour et al., 2010; Stefan et al., 2013). So far, the PM-located receptor for none of the six ER tethers is known.
Scs2p is unique among the cER tethers in that its single deletion already leads to a severe reduction in the number of cER–PM contact sites (Loewen et al., 2007). Besides serving as a tether, the cytosolic domain of Scs2p binds short FFAT motifs within Osh proteins, the yeast members of a family of oxysterol binding proteins (Loewen et al., 2003; Loewen and Levine, 2005). Osh proteins accumulate at ER–PM contact sites through their lipid-binding pleckstrin homology (PH) domains and the interactions of their FFAT motifs with Scs2p. Once formed, the Osh–Scs2p complexes exchange sterol lipids between both organelles and stimulate the activity of the phosphoinositide phosphatase Sac1p, thereby regulating the levels of PI4P at the PM (Stefan et al., 2011).
Scs2p also contributes to the tethering of the ER to the septins and to the robust inheritance of the cER (Loewen et al., 2007; Chao et al., 2014). As the ER cannot arise de novo, yeast cells have to use a dedicated pathway to guarantee its equal partitioning between mother and daughter during mitosis. This ER inheritance pathway can be divided into three consecutive steps. Initially, ER tubules travel on actin cables into the small growing bud of the cell (Estrada et al., 2003). The tubules are then attached to the bud tip and passively pulled along during the growth of the bud. The cER finally spreads out from these tubules to form the characteristic sheets below the PM. Scs2p, but not its paralogue Scs22p, were shown to be directly involved in attaching the cER to the tip of the growing cell (Loewen et al., 2007). Indirect evidence pointed to the polarisome as part of the tip-specific receptor of Scs2p (Loewen et al., 2007). The polarisome is a multiprotein complex of at least five different proteins (Snyder, 1989; Sheu et al., 1998). During the G1 and S phase of the cell cycle, the polarisome displays a very focused localization below the bud tip before spreading across the entire bud during the S/G2 transition. The constituents of the polarisome are the structural members Spa2p and Pea2p, Bud6p, and the yeast formin Bni1p, which catalyze actin filament formation, and the proteins Msb3p/Msb4p acting as GTPase-activating proteins for the small GTPase Sec4p (Valtz and Herskowitz, 1996; Amberg et al., 1997; Evangelista et al., 1997; Fujiwara et al., 1998; Sheu et al., 1998; Shih et al., 2005; Tcheperegine et al., 2005; Graziano et al., 2011). Sec4p is involved in the tethering and fusion of secretory vesicles to the PM (Salminen and Novick, 1987; Gao et al., 2003). Although all known members of the polarisome were tested at that time for their influence on the localization of Scs2p, the identity of the bud tip–specific receptor for Scs2p remained undisclosed (Loewen et al., 2007).
We characterize the protein encoded by YMR124w as a new member of the polarisome that binds directly to Scs2p and anchors the cER to the bud tip. After the discovery of the ER–mitochondria encounter structure complex as a link between ER tubules and mitochondria, our findings now provide a second detailed molecular characterization of an ER contact site in yeast that assists in the faithful inheritance and correct localization of this organelle (Kornmann et al., 2009).
Results
Epo1p is a new member of the polarisome
Our Split-Ubiquitin (Ub)–based systematic protein interaction analysis of the polarisome revealed the protein encoded by YMR124w (from here on EPO1) as a novel interaction partner of Spa2p and a potentially new member of the polarisome (Moreno et al., 2013; Chao et al., 2014). To substantiate this finding, we fused Epo1p to the Split-Ub module consisting of the C-terminal half of Ub (Cub) and a yeast enzyme involved in uracil synthesis (Ura3p). We screened this fusion (Epo1CRU) against our array of yeast strains expressing 389 different fusions to the N-terminal half of Ub (Nub; Dünkler et al., 2012). This array is enriched in Nub fusions to proteins that are known or suspected to be involved in polar growth, cytoskeleton, cellular architecture and organization, stress responses, and the cell cycle (Hruby et al., 2011). Table 1 lists the hits of this interaction screen, and Fig. 1 A shows the results of a smaller array including many of the newly found interaction partners. Beside Spa2p and Pea2p, two members of the polarisome, we discovered further proteins of the polar cortical domain, indicating a close association of Epo1p to this structure. To unravel the molecular nature of this association, we probed the interaction between Epo1CRU and Nub-Spa2p in a strain lacking PEA2 and the interaction between Epo1CRU and Nub-Pea2p in a strain lacking SPA2 (Fig. 1 B). The two assays suggested that Epo1p is connected to Spa2p and the polarisome through Pea2p. The same strategy revealed that Kel1p, a further member of the polar cortical domain, and Bem3p, a GTPase-activating protein of Cdc42p, bind to Epo1p independently of Pea2p and Spa2p (Fig. 1 B; Zheng et al., 1993; Philips and Herskowitz, 1998; Bidlingmaier and Snyder, 2004; Park and Bi, 2007). The deletion of PEA2 had also no measurable influence on the interaction between Epo1p and Bud6p or Scs2p, indicating that their association to Epo1 does not occur through Pea2p (Fig. 1 B). A screen of Pea2CRU against the Nub array confirmed its interaction with Epo1p and additionally highlighted Kel1p, Kel2p, Scs2p, and Bem3p as four further proteins that interact with both Pea2p and Epo1p (Table 1). The loss of the interaction signal in a strain lacking EPO1 revealed that Epo1p mediates the interactions of Bem3p and Scs2p with Pea2p (Fig. 1 C). The same analysis proved that Kel1p and Spa2p bind to Pea2p independently of Epo1p (Fig. 1 C).
Summary of large-scale Split-Ub protein interaction screens
| Cub fusion | Nub fusion |
| Epo1p | Bem3p, Boi1p, Boi2p, Bud6p, Epo1p, Fks2p, Kel1p, Kel2p, Pea2p, Scs2p, Sec4p, Skt5p, Spa2p |
| Pea2p | Bem3p, Bud14p, Epo1p, Kel1p, Kel2p, Mlc1p, Pea2p, Scs2p, Spa2p |
| Kel1p | Bud14p, Kel1p, Kel2p |
| Cub fusion | Nub fusion |
| Epo1p | Bem3p, Boi1p, Boi2p, Bud6p, Epo1p, Fks2p, Kel1p, Kel2p, Pea2p, Scs2p, Sec4p, Skt5p, Spa2p |
| Pea2p | Bem3p, Bud14p, Epo1p, Kel1p, Kel2p, Mlc1p, Pea2p, Scs2p, Spa2p |
| Kel1p | Bud14p, Kel1p, Kel2p |
All arrayed Nub fusion proteins are listed in Table S1.
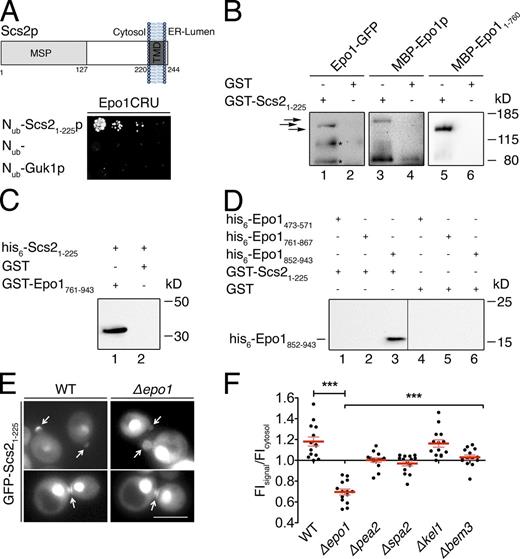
Epo1p interacts with members of the polarisome and Scs2p. (A) Split-Ub interaction assay of 48 yeast strains each coexpressing Epo1CRU with a different Nub fusion. Shown are quadruplets of each strain after 3 d of growth on medium containing 5-FOA. White boxes indicate the fusions that induce the growth of the strain reflecting the interaction between Nub and Cub fusion. The identities of all Nub fusions are revealed in Table S2. (B) Split-Ub interaction assay between Epo1CRU and selected Nub fusion proteins in WT, Δpea2, Δspa2, Δkel1, and Δbem3 cells. Cells were grown to OD600 of 1 and 4 µl of this, and 10-fold serial dilutions were spotted on 5-FOA plates. Nub without a C-terminally attached ORF (Nub−) serves as a control for the specificity of the Split-Ub assays. (C) As in B, but selected interactions of Pea2CRU were compared between WT and Δepo1 cells. (D) Domain structure of Epo1p. Shown as blue rectangles are the three predicted coiled-coil (CC) regions. Numbers indicate amino acid positions of the putative start and end points of each domain. (E) As in A, but with fragments of Epo1p as CRU fusions and 16 independently generated diploids for each experiment shown after 4 d of growth. (F) Protein extracts of bacterial cells expressing his6-Pea2 (lanes 1–4) or yeast cells expressing MYC-tagged Kel1p (lanes 5–8) were incubated with glutathione-coupled Sepharose beads exposing bacterially expressed GST (lanes 1 and 5), GST-Epo1761–943 (lanes 2 and 6), GST-Epo1761–867 (lanes 3 and 7), or GST-Epo1852–943 (lanes 4 and 8). Glutathione eluates were separated by SDS-PAGE and probed with anti-His (lanes 1–4) or anti-MYC (lanes 5–8) antibodies after Western blotting. (G) As in F, except bacterially expressed MBP-Epo1 (lanes 1 and 2) or MBP-Epo11–760 (lanes 3 and 4) were precipitated with bacterially expressed and Sepharose bead immobilized GST (lanes 2 and 4) or GST-Pea2p (lanes 1 and 3). The inputs for the experiments in F and G are shown in Fig. S1. (H) Pea2p mediates the interaction between Epo1p and Spa2p. As in F, except bacterially expressed his6-Spa21–535-SNAP (lanes 1, 2, 7, and 8) or his6-Spa21–488-SNAP (lanes 4, 5, 9, and 10) were first incubated with his6-Pea2 (lanes 1 and 4) or left untreated (lanes 7 and 9) before being incubated with bacterially expressed and immobilized GST-Epo1761–943. The glutathione eluates are shown in lanes 1, 4, 7, and 9. The inputs for the experiment in lane 1 are shown in lanes 2 and 3. The inputs for the experiment in lane 4 are shown in lanes 5 and 6. The inputs for the experiment in lanes 7 and 9 are shown in lanes 8 and 10, respectively. The asterisk indicates a degradation product. Lanes 1–6 show cutouts of the same gel with the vertical line indicating the removal of an empty lane. (I) Architecture of the ER–cell tip tethering complex. Edges connecting nodes indicate direct (black) or potentially indirect (green) interactions. Blue rectangles indicate coiled-coil regions shown in D.
Epo1p interacts with members of the polarisome and Scs2p. (A) Split-Ub interaction assay of 48 yeast strains each coexpressing Epo1CRU with a different Nub fusion. Shown are quadruplets of each strain after 3 d of growth on medium containing 5-FOA. White boxes indicate the fusions that induce the growth of the strain reflecting the interaction between Nub and Cub fusion. The identities of all Nub fusions are revealed in Table S2. (B) Split-Ub interaction assay between Epo1CRU and selected Nub fusion proteins in WT, Δpea2, Δspa2, Δkel1, and Δbem3 cells. Cells were grown to OD600 of 1 and 4 µl of this, and 10-fold serial dilutions were spotted on 5-FOA plates. Nub without a C-terminally attached ORF (Nub−) serves as a control for the specificity of the Split-Ub assays. (C) As in B, but selected interactions of Pea2CRU were compared between WT and Δepo1 cells. (D) Domain structure of Epo1p. Shown as blue rectangles are the three predicted coiled-coil (CC) regions. Numbers indicate amino acid positions of the putative start and end points of each domain. (E) As in A, but with fragments of Epo1p as CRU fusions and 16 independently generated diploids for each experiment shown after 4 d of growth. (F) Protein extracts of bacterial cells expressing his6-Pea2 (lanes 1–4) or yeast cells expressing MYC-tagged Kel1p (lanes 5–8) were incubated with glutathione-coupled Sepharose beads exposing bacterially expressed GST (lanes 1 and 5), GST-Epo1761–943 (lanes 2 and 6), GST-Epo1761–867 (lanes 3 and 7), or GST-Epo1852–943 (lanes 4 and 8). Glutathione eluates were separated by SDS-PAGE and probed with anti-His (lanes 1–4) or anti-MYC (lanes 5–8) antibodies after Western blotting. (G) As in F, except bacterially expressed MBP-Epo1 (lanes 1 and 2) or MBP-Epo11–760 (lanes 3 and 4) were precipitated with bacterially expressed and Sepharose bead immobilized GST (lanes 2 and 4) or GST-Pea2p (lanes 1 and 3). The inputs for the experiments in F and G are shown in Fig. S1. (H) Pea2p mediates the interaction between Epo1p and Spa2p. As in F, except bacterially expressed his6-Spa21–535-SNAP (lanes 1, 2, 7, and 8) or his6-Spa21–488-SNAP (lanes 4, 5, 9, and 10) were first incubated with his6-Pea2 (lanes 1 and 4) or left untreated (lanes 7 and 9) before being incubated with bacterially expressed and immobilized GST-Epo1761–943. The glutathione eluates are shown in lanes 1, 4, 7, and 9. The inputs for the experiment in lane 1 are shown in lanes 2 and 3. The inputs for the experiment in lane 4 are shown in lanes 5 and 6. The inputs for the experiment in lanes 7 and 9 are shown in lanes 8 and 10, respectively. The asterisk indicates a degradation product. Lanes 1–6 show cutouts of the same gel with the vertical line indicating the removal of an empty lane. (I) Architecture of the ER–cell tip tethering complex. Edges connecting nodes indicate direct (black) or potentially indirect (green) interactions. Blue rectangles indicate coiled-coil regions shown in D.
To locate the binding sites for the different ligands on the primary structure of Epo1p, we created CRU fusions of N- and C-terminal fragments of Epo1p and tested them against a subset of its Nub-labeled binding partners (Fig. 1, D and E). We initially divided Epo1p into an N-terminal (Epo11–760) and a C-terminal fragment (Epo1761–943; Fig. 1, D and E). Epo11–760CRU interacted exclusively with Nub-Scs2p, and the C-terminal Epo1761–943CRU retained interactions with all other tested ligands including Scs2p (Fig. 1 E). Deleting the most C-terminal coiled-coil region (CC3) created Epo11–876CRU, which maintained most of the interactions of the full-length Epo1p, except its interaction with Bem3p, which was strongly reduced (Fig. 1 E).
To confirm the Split-Ub–measured interactions in vitro, we precipitated Escherichia coli–expressed, his6-tagged Pea2p (his6-Pea2p) with GST fusions to Epo1761–943, Epo1761–867, and Epo1852–943 (Fig. 1 F). The analysis located the Pea2p binding site onto the CC2 region of Epo1p (Fig. 1 F, lanes 2 and 3). The same GST-labeled fragments of Epo1p could precipitate a MYC epitope-labeled Kel1p from yeast extracts (Fig. 1 F, lanes 6 and 7). These findings indicate that Pea2p and Kel1p might share CC2 as binding site to Epo1p. The maltose-binding protein (MBP) fusion to the full-length Epo1p could be precipitated by glutathione-immobilized GST-Pea2p beads (Fig. 1 G, lane 1). In contrast, the MBP fusion to the N-terminal fragment of Epo1p failed to bind to GST-Pea2p, confirming the Split-Ub–deduced localization of the Pea2p binding site onto the C-terminal CC2 region of Epo1p (Fig. 1 G, lane 3).
To reconstitute a minimal polarisome, we expressed a his6-tagged, N-terminal fragment of Spa2p containing its binding site to Pea2p (his6-Spa21–535) and a second N-terminal fragment of Spa2p lacking this binding site (his6-Spa21–488; Sheu et al., 1998). As predicted by our Split-Ub analysis, both Spa2p fragments did not interact with immobilized GST-Epo1761–943 (Fig. 1 H, lanes 7 and 9; lanes 2, 3, 5, and 6 show the inputs). However, preincubating the GST-Epo1761–943 beads with his6-Pea2p before adding one of the two Spa2p fragments allowed precipitating his6-Spa21–535 but not his6-Spa21–488 (Fig. 1 H, lanes 1 and 3). This experiment showed that a stable ternary complex of Spa2p, Pea2p, and Epo1p can be formed in vitro. Pea2p acts as a bridge linking Epo1p to Spa2p, the core element of the polarisome.
Members of the polarisome are characterized by a very focused localization at the tip of young buds and an accumulation at the bud neck during mitosis (Fig. 2 A). Time-lapse observations of cells coexpressing Epo1p-GFP together with Spa2-Cherry revealed that Epo1p and Spa2p arrive together at the incipient bud site to colocalize at the bud cortex till late into mitosis. Spa2p arrives shortly before Epo1p at the bud neck during mitosis (Fig. 2 B and Video 1). To more accurately determine the time of bud neck appearance, we coexpressed Spa2-GFP or Epo1-GFP together with the Cherry-labeled septin subunit Shs1p. The splitting of the septin rings occurred 6 min after Spa2-GFP and 1 min after Epo1-GFP were first detected at the bud neck (Fig. 2 C and Videos 2 and 3).
Time-lapse analysis of yeast cells coexpressing Spa2-Cherry with Epo1-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every 3 min for 120 min using a DeltaVision life cell imaging unit (Applied Precision). Selected stills are shown in Fig. 2 B.
Time-lapse analysis of yeast cells coexpressing Spa2-Cherry with Epo1-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every 3 min for 120 min using a DeltaVision life cell imaging unit (Applied Precision). Selected stills are shown in Fig. 2 B.
Time-lapse analysis of yeast cells coexpressing Shs1-Cherry with Epo1-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every minute for 85 min using a DeltaVision life cell imaging unit. Selected stills are shown in Fig. 2 C
Time-lapse analysis of yeast cells coexpressing Shs1-Cherry with Epo1-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every minute for 85 min using a DeltaVision life cell imaging unit. Selected stills are shown in Fig. 2 C
Time-lapse analysis of yeast cells coexpressing Shs1-Cherry with Spa2-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every minute for 65 min using a DeltaVision life cell imaging unit. Selected stills are shown in Fig. 2 C.
Time-lapse analysis of yeast cells coexpressing Shs1-Cherry with Spa2-GFP. Cells were incubated at 30°C on SD complete. Frames were taken every minute for 65 min using a DeltaVision life cell imaging unit. Selected stills are shown in Fig. 2 C.
Epo1p is a component of the polarisome. (A) Two-channel microscopy of yeast cells coexpressing Spa2-GFP and Kel1-Cherry (left), Epo1-GFP and Kel1-Cherry (middle), or Epo1-GFP and Spa2-Cherry (right). Each image shows polar-growing cells (left) and cells during mitosis (right). (B) Stills of a time-lapse analysis of cells coexpressing Epo1-GFP and Spa2-Cherry. Pictures start in late G2 (t = 0 min) and end with the outgrowth of a new bud (t = 48 min). (C) Stills of a time-lapse analysis of cells in late mitosis and coexpressing Spa2-GFP and Shs1-Cherry (left) or Epo1-GFP and Shs1-Cherry (middle). t = 0 min indicates the splitting of the septin rings. (right) Calculated time between the first appearance of Spa2-GFP (n = 31) or Epo1-GFP (n = 33) at the bud neck and the splitting of the septin rings (error bars show SEM). Bars: (A and B) 5 µm; (C) 1 µm.
Epo1p is a component of the polarisome. (A) Two-channel microscopy of yeast cells coexpressing Spa2-GFP and Kel1-Cherry (left), Epo1-GFP and Kel1-Cherry (middle), or Epo1-GFP and Spa2-Cherry (right). Each image shows polar-growing cells (left) and cells during mitosis (right). (B) Stills of a time-lapse analysis of cells coexpressing Epo1-GFP and Spa2-Cherry. Pictures start in late G2 (t = 0 min) and end with the outgrowth of a new bud (t = 48 min). (C) Stills of a time-lapse analysis of cells in late mitosis and coexpressing Spa2-GFP and Shs1-Cherry (left) or Epo1-GFP and Shs1-Cherry (middle). t = 0 min indicates the splitting of the septin rings. (right) Calculated time between the first appearance of Spa2-GFP (n = 31) or Epo1-GFP (n = 33) at the bud neck and the splitting of the septin rings (error bars show SEM). Bars: (A and B) 5 µm; (C) 1 µm.
Epo1p is not a core component of the polarisome
Deletion of any one of the polarisome proteins diminishes or abolishes the tip localization of the other polarisome members (Valtz and Herskowitz, 1996; Fujiwara et al., 1998; Segal et al., 2000; Ozaki-Kuroda et al., 2001; Sagot et al., 2002a; Tcheperegine et al., 2005). The polarisome-like distribution during bud growth and cytokinesis of Epo1-GFP was either completely lost or transformed into a more homogenous staining of the entire cortex of the bud when Epo1-GFP–expressing cells carried deletions of either PEA2 or BNI1 (Fig. 3, A and B; and Fig. S2). The bud neck localization of Epo1-GFP was also impaired by the deletion of PEA2 (Figs. 3 B and S2). Conversely, a deletion of EPO1 hardly affected the tiplike distribution of its binding partner Pea2-GFP (Fig. 3 A). To closer characterize the relation between Epo1p and the polarisome, we determined the relative stoichiometries of Epo1p, Spa2p, and Pea2p at the bud tip. Cells coexpressing Spa2-GFP and a histone-Cherry fusion (Hhf2-Cherry) were mixed with cells expressing Pea2-GFP, Epo1-GFP, or Spa2-GFP. The fluorescence intensities (FIs) at the tip of the cells in this mixture were measured, and the Spa2-GFP–expressing reference cells were identified by the Cherry staining of their nuclei. Comparison of the GFP intensities revealed a sixfold higher concentration of Spa2p at the bud tip than Pea2p or Epo1p (Fig. 3 C).
Epo1p is not a core component of the polarisome. (A) Comparison between the bud staining of Epo1-GFP in WT and Δpea2 cells (top) and between the Pea2-GFP staining in WT and Δepo1 cells (bottom). Insets show a reduction of the DIC image of the same cell. Bars, 3 µm. Shown on the right are the means of the intensity profiles (radial sums) of the GFP signals at the bud circumference derived from four cells each. (B, top) Quantification of FI of Epo1-GFP at the tip of small budded and the neck of large budded cells. The mean of the signal intensity at the bud tip or neck were divided by the mean cytosolic intensity measured in the mother cell. (101 < n < 108; error bars show SEM). (bottom) Relative distribution of cells showing either no, a spread, or a focused bud tip signal of Epo1-GFP. Analyzed were the same small-budded cells as in the top quantification. Shown are the means of 100 cells each. (C) Quantification of signal intensity of Pea2-GFP and Epo1-GFP relative to Spa2-GFP at the bud tip of small budded cells (42 < n < 55; error bars show SEM). (D, top) Length/width ratio of daughter cells of WT and selected single-deletion strains (200 < n < 246; error bars show SEM). (bottom) DIC pictures of representative cells of the corresponding genotypes. Bar, 5 µm. ***, P < 0.001.
Epo1p is not a core component of the polarisome. (A) Comparison between the bud staining of Epo1-GFP in WT and Δpea2 cells (top) and between the Pea2-GFP staining in WT and Δepo1 cells (bottom). Insets show a reduction of the DIC image of the same cell. Bars, 3 µm. Shown on the right are the means of the intensity profiles (radial sums) of the GFP signals at the bud circumference derived from four cells each. (B, top) Quantification of FI of Epo1-GFP at the tip of small budded and the neck of large budded cells. The mean of the signal intensity at the bud tip or neck were divided by the mean cytosolic intensity measured in the mother cell. (101 < n < 108; error bars show SEM). (bottom) Relative distribution of cells showing either no, a spread, or a focused bud tip signal of Epo1-GFP. Analyzed were the same small-budded cells as in the top quantification. Shown are the means of 100 cells each. (C) Quantification of signal intensity of Pea2-GFP and Epo1-GFP relative to Spa2-GFP at the bud tip of small budded cells (42 < n < 55; error bars show SEM). (D, top) Length/width ratio of daughter cells of WT and selected single-deletion strains (200 < n < 246; error bars show SEM). (bottom) DIC pictures of representative cells of the corresponding genotypes. Bar, 5 µm. ***, P < 0.001.
Deletions of members of the polarisome generally impair the polarized growth during bud expansion. In contrast to the slightly elongated, ellipsoid wild-type (WT) cells, these mutants display a round curvature (Fig. 3 D). The lack of Epo1p did not alter the bud morphology of the cell nor did it visibly affect the architecture of the actin cytoskeleton (Figs. 3 D and S2). Both observations do not exclude a role of Epo1p in polarized growth but clearly distinguish it from the core components of the polarisome.
The N-terminal coiled-coil domain of Bem3p interacts directly with the C-terminal coiled-coil domains of Epo1p
The C-terminal Rho GTPase-activating protein domain of Bem3p is preceded by a lipid-binding PH, a PX (phox) domain, and an N-terminal region that harbors a predicted coiled-coil sequence (Fig. 4 B; Smith et al., 2002; Mukherjee et al., 2013). We created Nub fusions to different regions of the protein and tested them for their interaction with Epo1CRU. The interaction assay localized the binding site for Epo1p within the N-terminal region of Bem3p (Fig. 4 A). A similar Split-Ub analysis already identified CC3 and potentially CC2 as regions of Epo1p contributing to the binding to Bem3p (Fig. 1 E). We expressed the respective coiled-coil domains of Bem3p and Epo1p separately in E. coli and could show that Bem31–100 binds to CC3 and CC2 (Fig. 4 C). The latter interaction appeared much weaker but was reproducibly detected.
The N-terminal 100 residues of Bem3p interact with the C-terminal coiled-coil regions of Epo1p. (A) Split-Ub assay as in Fig. 1 B but with cells coexpressing Epo1CRU and Nub fusions to Bem3p and its fragments. The Nub fusion to Guk1p should not interact and served as a control for the specificities of the observed interactions. (B) Cartoon indicating the positions of the Nub fragments and the domains of Bem3p. GAP, GTPase-activating domain. (C) As in Fig. 1 F, but with protein extracts of bacterial cells expressing his6-Epo1852–943 (lanes 1, 3, and 4) or his6-Epo1761–867 (lanes 2, 5, and 6) that were incubated with glutathione-coupled Sepharose beads exposing bacterially expressed GST (lanes 3 and 5) or GST-Bem31–100 (lanes 4 and 6). The vertical line indicates the removal of a lane loaded with a molecular mass marker. The inputs of GST and GST-Bem31–100 are shown as lanes 7 and 8 of a Coomassie-stained gel. Arrows indicate the running positions of GST-Bem31–100 and GST.
The N-terminal 100 residues of Bem3p interact with the C-terminal coiled-coil regions of Epo1p. (A) Split-Ub assay as in Fig. 1 B but with cells coexpressing Epo1CRU and Nub fusions to Bem3p and its fragments. The Nub fusion to Guk1p should not interact and served as a control for the specificities of the observed interactions. (B) Cartoon indicating the positions of the Nub fragments and the domains of Bem3p. GAP, GTPase-activating domain. (C) As in Fig. 1 F, but with protein extracts of bacterial cells expressing his6-Epo1852–943 (lanes 1, 3, and 4) or his6-Epo1761–867 (lanes 2, 5, and 6) that were incubated with glutathione-coupled Sepharose beads exposing bacterially expressed GST (lanes 3 and 5) or GST-Bem31–100 (lanes 4 and 6). The vertical line indicates the removal of a lane loaded with a molecular mass marker. The inputs of GST and GST-Bem31–100 are shown as lanes 7 and 8 of a Coomassie-stained gel. Arrows indicate the running positions of GST-Bem31–100 and GST.
Epo1p binds directly to Scs2p, the major VAMP-associated protein homologue in yeast
The newly detected interaction between Epo1p and Scs2p pointed to Epo1p as the missing member of the polarisome that mediates the connection between cER and bud tip (Fig. 1, A–E; Chao et al., 2014). Scs2p consists of an N-terminal cytosolic domain of 225 residues and a C-terminal membrane-spanning segment (Fig. 5 A; Loewen et al., 2007). The Split-Ub analysis of Nub-Scs21–225 and Epo1CRU confined the interaction domain of Scs2p to its cytosolic 225 residues (Fig. 5 A). The precipitations of Epo1-GFP from yeast cell extracts and the precipitation of a fusion between the MBP and Epo1p (MBP-Epo1p) from E. coli extracts by the bacterially expressed GST-Scs21–225 confirmed the direct nature of the Scs2p–Epo1p interaction (Fig. 5 B, lanes 1 and 3). Split-Ub analysis of fragments of Epo1p revealed at least two binding sites for Scs2p. One located within the first N-terminal 760 residues and the other within a region spanning residues 760–943 (Fig. 1 E). The MBP fusion corresponding to the N-terminal fragment of Epo1p (MBP-Epo11–760) could be specifically pulled down with immobilized GST-Scs21–225 (Fig. 5 B, lane 5). The GST fusion to the C-terminal fragment of Epo1p (GST-Epo1761–943) precipitated his6-tagged Scs21–225 from E. coli extracts (Fig. 5 C, lane 1). We could further restrict the C-terminal Scs2-binding site of Epo1p to its CC3 region, as GST-Scs21–225 was able to specifically pull down E. coli–expressed his6-Epo1852–943 (Fig. 5 D, lane 3).
Epo1p binds to two different sites in Scs2p. (A, top) Domain structure of Scs2p. The major sperm domain (MSP) is separated from the C-terminal transmembrane segment (TMD) by a stretch of 93 residues. (bottom) Split-Ub interaction assay as in Fig. 1 B but between Epo1CRU and Nub-Scs21–225 and two Nub fusions that should not interact with Epo1CRU. (B) Eluates of Sepharose bead-coupled GST-Scs21–225 (lanes 1, 3, and 5) or GST (lanes 2, 4, and 6) incubated with yeast extracts containing GFP-tagged Epo1p (lanes 1 and 2) or bacterial extracts containing MBP-Epo1p (lanes 3 and 4) or MBP-Epo11–760 (lanes 5 and 6) were separated by SDS-PAGE. Western blots were probed with anti-GFP (lanes 1 and 2) or anti-MBP antibodies (lanes 3–6). Arrows indicate from top to bottom: MBP-Epo1p, Epo1-GFP, and MBP-Epo11–760. Fragments of Epo1-GFP and MBP-Epo1p running <80 kD and interacting nonspecifically are not depicted. Asterisks indicate proteolytic fragments of Epo1-GFP. (C) As in B but with eluates of Sepharose bead-coupled GST-Epo1761–943 (lane 1) or GST (lane 2) incubated with bacterial extracts containing his6-Scs21–225. The Western blot was probed with anti-His antibodies. (D) As in B, but with eluates of Sepharose bead-coupled GST-Scs21–225 (lanes 1–3) or GST (lanes 4–6) incubated with bacterial extracts containing his6-tagged Epo1473–571 (lanes 1 and 4), Epo1761–867 (lanes 2 and 5), or Epo1852–943 (lanes 3 and 6). The Western blot was probed with anti-His antibodies. The vertical line indicates the removal of a lane loaded with a molecular mass marker. (E) Fluorescence microscopy of WT or Δepo1 cells expressing GFP-Scs21–225 from the PMET17 promoter. Cells were grown in medium containing 70 µM methionine to moderately express the GFP fusion. Top and bottom images show representative yeast cells during polar growth and during cell separation, respectively. Arrows indicate the GFP-Scs21–225 staining at bud tip and bud neck. Bar, 5 µm. (F) Quantification of relative FIs of GFP-Scs21–225 at the bud tips of WT cells and cells lacking the indicated genes (n = 14; error bars show SEM). ***, P < 0.001. The inputs for the experiments in C and D are shown in Fig. S1.
Epo1p binds to two different sites in Scs2p. (A, top) Domain structure of Scs2p. The major sperm domain (MSP) is separated from the C-terminal transmembrane segment (TMD) by a stretch of 93 residues. (bottom) Split-Ub interaction assay as in Fig. 1 B but between Epo1CRU and Nub-Scs21–225 and two Nub fusions that should not interact with Epo1CRU. (B) Eluates of Sepharose bead-coupled GST-Scs21–225 (lanes 1, 3, and 5) or GST (lanes 2, 4, and 6) incubated with yeast extracts containing GFP-tagged Epo1p (lanes 1 and 2) or bacterial extracts containing MBP-Epo1p (lanes 3 and 4) or MBP-Epo11–760 (lanes 5 and 6) were separated by SDS-PAGE. Western blots were probed with anti-GFP (lanes 1 and 2) or anti-MBP antibodies (lanes 3–6). Arrows indicate from top to bottom: MBP-Epo1p, Epo1-GFP, and MBP-Epo11–760. Fragments of Epo1-GFP and MBP-Epo1p running <80 kD and interacting nonspecifically are not depicted. Asterisks indicate proteolytic fragments of Epo1-GFP. (C) As in B but with eluates of Sepharose bead-coupled GST-Epo1761–943 (lane 1) or GST (lane 2) incubated with bacterial extracts containing his6-Scs21–225. The Western blot was probed with anti-His antibodies. (D) As in B, but with eluates of Sepharose bead-coupled GST-Scs21–225 (lanes 1–3) or GST (lanes 4–6) incubated with bacterial extracts containing his6-tagged Epo1473–571 (lanes 1 and 4), Epo1761–867 (lanes 2 and 5), or Epo1852–943 (lanes 3 and 6). The Western blot was probed with anti-His antibodies. The vertical line indicates the removal of a lane loaded with a molecular mass marker. (E) Fluorescence microscopy of WT or Δepo1 cells expressing GFP-Scs21–225 from the PMET17 promoter. Cells were grown in medium containing 70 µM methionine to moderately express the GFP fusion. Top and bottom images show representative yeast cells during polar growth and during cell separation, respectively. Arrows indicate the GFP-Scs21–225 staining at bud tip and bud neck. Bar, 5 µm. (F) Quantification of relative FIs of GFP-Scs21–225 at the bud tips of WT cells and cells lacking the indicated genes (n = 14; error bars show SEM). ***, P < 0.001. The inputs for the experiments in C and D are shown in Fig. S1.
A GFP-fusion to the N-terminal cytosolic domain of Scs2p (GFP-Scs21–225) localizes at the bud tip, neck, and in the nucleus of yeast cells (Loewen et al., 2007; Fig. 5 E). This finding already suggested the existence of a distinct receptor for Scs2p at the bud (Loewen et al., 2007). Former attempts to identify this receptor by following the localization of GFP-Scs21–225 in a panel of 57 deletion strains detected an impaired but not abolished bud tip staining of GFP-Scs21–225 in 10 of the tested strains (Loewen et al., 2007). Interestingly, three of the herein characterized binding partners of Epo1p were found among those hits: Bem3p, Pea2p, and Spa2p. As predicted for the postulated role of Epo1p as the cortical receptor of Scs2p, the bud tip staining of GFP-Scs21–225 was completely lost in Δepo1 cells, whereas residual localized staining was observed in Δpea2, Δspa2, and Δbem3 cells (Fig. 5, E and F). Notably, the appearance of GFP-Scs21–225 at the bud neck of mitotic cells was not influenced by the absence of any of the tested genes (Fig. 5 E).
The membrane-associated proteins Tcb1p, Tcb2p, Tcb3p, and Ist2p act as alternative receptors for tethering the cER to the PM (Manford et al., 2012; Wolf et al., 2012). None of the corresponding Nub fusions revealed interactions with Epo1CRU (Fig. S3).
The tempospatial interaction characteristics identify Epo1p as a tip-specific Scs2p receptor during bud growth
Our experiments argue for a distinct ER–cell tip complex consisting of Scs2p, Epo1p, Bem3p, and the polarisome yet leaving open where and when this complex exists. We therefore performed a SPLIFF analysis of Epo1p to follow its interaction with the other members of this complex in a time and spatially resolved manner (Moreno et al., 2013). SPLIFF is a modification of the Split-Ub technique. Here, the Cub is sandwiched between the autofluorescent Cherry and GFP (CCG; Moreno et al., 2013). Upon the interaction-induced reassociation with a Nub, the native-like Ub is reconstituted, and GFP is cleaved off and rapidly degraded. The subsequent local increase in the ratio of red to green fluorescence indicates where and when the interaction between both proteins took place. The CCG module was attached to the C terminus of Epo1p to create Epo1CCG. To improve the fluorescent signals, we integrated the PMET17 promoter upstream of EPO1CCG. Fluorescence microscopy of the fusion showed that the cortical distribution of the overexpressed Epo1CCG is similar to the natively expressed Epo1-GFP (Fig. 6, A and C). Two yeast cells of opposing mating types, one expressing the Epo1CCG fusion and the other the Nub-labeled interaction partner, were mated, and the interaction between both proteins was monitored online by two channel fluorescence microscopy (Fig. 6 A). This trick allowed us to follow the interaction during the first cell cycle of the newly formed diploid cell. The SPLIFF analysis revealed a complex succession of associations and dissociations of the different binding partners of Epo1p (Fig. 6, A and B). Nub-Kel1p and Nub-Bem3p interacted with Epo1CCG at the polar cortical domain close to the sites of cell fusion, below the tip of growing buds and finally below the membrane at the bud neck of cells shortly before entry into mitosis and cell separation (Fig. 6, A and B). Interaction between Nub-Pea2p and Epo1CCG was first observed at the tip of the growing bud and later during mitosis at the bud neck, whereas Nub-Scs2p interacted with Epo1CCG only at the tip of growing buds (Fig. 6, A and B). This interaction pattern is consistent with a role of Epo1p as a bud tip–specific receptor for Scs2p. Our analysis is not compatible with an Epo1p-dependent anchoring of the cER to the site of cell fusion during mating and to the site of cell separation during cytokinesis (Fig. 6 B).
Epo1p interacts with Scs2p only during polar cell growth. (A) SPLIFF analysis of Epo1CCG. a cells expressing Epo1CCG were mated with α cells expressing (from top to bottom) Nub-Bem3, Nub-Kel1p, Nub-Pea2p, or Nub-Scs2p. Shown are the GFP and Cherry images of cells shortly after cell fusion (cell fusion, left column), during polar growth (bud tip, middle column), and during cell separation (bud neck, right column). Blue frames indicate the areas used for the quantification of the Cherry and GFP fluorescence at the respective phases of the cell cycle. (B) The calculated fractions of converted Epo1CC in cells expressing the indicated Nub fusions were plotted against time. Shown are the means of 7 < n < 13 independent matings (error bars show SEM). Note that Nub-Bni1p and the Nub without an attached yeast protein do not interact with Epo1CCG. The cartoons above the graphs indicate the respective cell cycle stages. The blue rectangles mark the areas used for signal quantification, and the red lines indicate the positions of Epo1CCG and Epo1CC appearance. (C) Microscopy of cells coexpressing (from the left to the right) Spa2-GFP/Kel1-Cherry, Epo1-GFP/Kel1-Cherry, Epo1-GFP/Spa2-Cherry, and Bem3-GFP/Spa2-Cherry. Shown are images of the diploid cells shortly before and after cell fusion. The cartoons above the images were intended to help positioning of the GFP and Cherry fluorescence with respect to the cell. Bars, 5 µm.
Epo1p interacts with Scs2p only during polar cell growth. (A) SPLIFF analysis of Epo1CCG. a cells expressing Epo1CCG were mated with α cells expressing (from top to bottom) Nub-Bem3, Nub-Kel1p, Nub-Pea2p, or Nub-Scs2p. Shown are the GFP and Cherry images of cells shortly after cell fusion (cell fusion, left column), during polar growth (bud tip, middle column), and during cell separation (bud neck, right column). Blue frames indicate the areas used for the quantification of the Cherry and GFP fluorescence at the respective phases of the cell cycle. (B) The calculated fractions of converted Epo1CC in cells expressing the indicated Nub fusions were plotted against time. Shown are the means of 7 < n < 13 independent matings (error bars show SEM). Note that Nub-Bni1p and the Nub without an attached yeast protein do not interact with Epo1CCG. The cartoons above the graphs indicate the respective cell cycle stages. The blue rectangles mark the areas used for signal quantification, and the red lines indicate the positions of Epo1CCG and Epo1CC appearance. (C) Microscopy of cells coexpressing (from the left to the right) Spa2-GFP/Kel1-Cherry, Epo1-GFP/Kel1-Cherry, Epo1-GFP/Spa2-Cherry, and Bem3-GFP/Spa2-Cherry. Shown are images of the diploid cells shortly before and after cell fusion. The cartoons above the images were intended to help positioning of the GFP and Cherry fluorescence with respect to the cell. Bars, 5 µm.
We confirmed this analysis by colocalization studies of the corresponding GFP and Cherry fusions (Fig. 6 C). Spa2p displayed a focused localization at the site of cell fusion that overlapped with the distribution of Epo1p and Bem3p (Fig. 6 C). Kel1p occupied a broader region during this period (Fig. 6 C). After cell fusion, Spa2p stayed focused at this region, whereas Epo1p, Kel1p, and partially Bem3p showed a wider distribution that included the membrane surrounding the opening between both cells (Fig. 6 C). Colocalization between Epo1p and Spa2p was also observed at the cell tip during bud growth and the bud neck during mitosis in haploid cells (Fig. 2 and Video 1).
Deletion of EPO1 disrupts the contact between cER and the bud tip
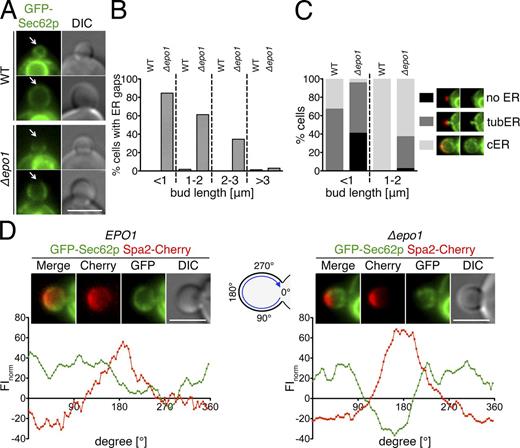
Our analysis predicted that a deletion of EPO1 might impair the inheritance and bud tip association of the cER. We used a fusion of GFP to Sec62p as a marker to compare the distribution of the ER between WT and Δepo1 cells. Sec62p is a membrane-localized member of the ER protein translocation machinery (Fig. 7, A–C).
Epo1p links the cER to the bud tip of the cell. (A) Cells containing (top) or lacking (bottom) EPO1 and expressing GFP-Sec62p as the ER marker were analyzed by fluorescence microscopy. Arrows indicate the cortex at the tip of the cells. (B) Regions of absent cER at the bud tip were counted in GFP-Sec62p–expressing WT and Δepo1 cells. Dashed lines separate the results of cells according to the lengths of their buds (22 < n < 144). (C) Effect of EPO1 deletion on cER inheritance. (left) WT and Δepo1 cells expressing GFP-Sec62p were scored for three stages of cER inheritance (no, tubular, or cER) and classified according to the length of their buds (71 < n < 104). (right) Images of Δepo1 cells (top and middle images) and WT cells (bottom image) expressing GFP-Sec62p and Spa2-Cherry exemplifying the three stages of cER inheritance. The same representative images are shown again in Fig. S3 D. (D) The FI profiles at the circumference of the bud were quantified for WT (left, n = 6) and Δepo1 cells (right, n = 15) coexpressing GFP-Sec62p and Spa2-Cherry. (top) Representative image of the inspected WT and inspected Δepo1 cells. (bottom) FIs plotted against the position at the surface of the bud of the two cells shown in the top images. Intensities are given as relative values to the FIs in the cytosol of the mother cell. Bars, 3 µm.
Epo1p links the cER to the bud tip of the cell. (A) Cells containing (top) or lacking (bottom) EPO1 and expressing GFP-Sec62p as the ER marker were analyzed by fluorescence microscopy. Arrows indicate the cortex at the tip of the cells. (B) Regions of absent cER at the bud tip were counted in GFP-Sec62p–expressing WT and Δepo1 cells. Dashed lines separate the results of cells according to the lengths of their buds (22 < n < 144). (C) Effect of EPO1 deletion on cER inheritance. (left) WT and Δepo1 cells expressing GFP-Sec62p were scored for three stages of cER inheritance (no, tubular, or cER) and classified according to the length of their buds (71 < n < 104). (right) Images of Δepo1 cells (top and middle images) and WT cells (bottom image) expressing GFP-Sec62p and Spa2-Cherry exemplifying the three stages of cER inheritance. The same representative images are shown again in Fig. S3 D. (D) The FI profiles at the circumference of the bud were quantified for WT (left, n = 6) and Δepo1 cells (right, n = 15) coexpressing GFP-Sec62p and Spa2-Cherry. (top) Representative image of the inspected WT and inspected Δepo1 cells. (bottom) FIs plotted against the position at the surface of the bud of the two cells shown in the top images. Intensities are given as relative values to the FIs in the cytosol of the mother cell. Bars, 3 µm.
The deletion of EPO1 delayed the appearance of the cER in very young buds and also slightly increased the persistence of the tubular cER structures in medium-sized buds (Fig. 6 C). In accordance with the cell cycle–specific and tip-restricted interaction between Epo1p and Scs2p, the temporal emergence of large gaps lacking GFP-Sec62p below the tip of small and to a lesser extent of large budded cells figured as the most prominent and striking phenotype of Δepo1 cells (Fig. 7, A and B). These ER gaps strongly coincided with the presence of the polarisome at this site (Fig. 7 D). A very similar phenotype is shared by the deletion of SCS2, the respective ligand of Epo1p at the cER (see Fig. 9, B and C; Loewen et al., 2007). Both observations point to Epo1p and Scs2p as key mediators of the molecular contact between the cER and the bud tip of the cell.
We defined the C-terminal part of Epo1p as a region interacting with the polarisome, Bem3p, and Scs2p, whereas the N-terminal region of Epo1p harbors a second binding site exclusively for Scs2p (Figs. 1 E and 5, C and D). In accordance with its binding to Pea2p, the GFP fusion to Epo1761–943 showed an Epo1p-like distribution with a clear staining of the bud tip and the neck, whereas Epo11–760-GFP showed a much weaker enrichment at sites of polar growth (Fig. S3 B). Neither of the separately expressed fragments of Epo1p complemented the ER inheritance defects of Δepo1 cells (Fig. S3, C and D). We conclude that Epo1p requires its two Scs2p binding sites for stably anchoring the cER to the cell tip.
Polarized growth reveals defects in cER bud tip anchorage
Our experiments identified the polarisome as the predominant receptor for Epo1p at the bud tip. Unexpectedly, Δspa2 or Δpea2 cells displayed a WT-like staining of the cER in their buds (Fig. 8 B). Revealingly, the additional deletion of EPO1 in Δspa2 or Δpea2 cells did not restore the gaps of ER staining that were observed in Δepo1 cells. We therefore rejected the presence of alternative receptors for Epo1p in Δspa2 or Δpea2 cells as explanation for the missing ER gaps and postulated that the lack of focused tip growth prevents the detection of ER gaps in these cells (Fig. 8 D). The extent of ER gaps should thus depend on both the degree of polarized growth and the strength of the connection between cER and bud tip. According to our model, Δepo1 and Δscs2 cells as well as the double deletions Δepo1 Δscs2, Δepo1 Δpea2, Δepo1 Δspa2, and Δepo1 Δbem3 should completely lack cER–cell tip anchorage (Fig. 8 D). Any quantitative differences in cER staining between these genotypes should thus be caused by the differences in the extents of their tip-directed growth. We estimated the latter value by the quotient of length to width of small buds (Fig. 8 A). For strains without an intact Epo1p–Scs2p connection, we plotted the percentage of cells revealing gaps of cER staining at the tip against their length/width ratios (Fig. 8 C). The linear correlation between both parameters as well as the lack of a cER phenotype in Δbud6, Δbni1, Δepo1 Δbni1, and Δepo1 Δbud6 cells supported our hypothesis (Fig. S5).
Correlation between the extent of polar growth and regions of absent cER. (A) Length/width ratio of daughter cells of WT and selected double-deletion strains (200 < n < 206; error bars show SEM; the value for the WT is taken from Fig. 2 D). ***, P < 0.001. (B) Quantification of gaps of ER staining below the bud tip in 1–2 µm buds of WT and selected single- and double-deletion strains expressing HMG1-GFP as marker for the cER. Shown are the means of 40 < n < 149 cells derived from at least two independent strains with a deviation of <8%. (C) The percentages of cells displaying no ER below the bud tip were plotted against their length/width ratios. Values were taken from A and B, respectively. Strains carrying either a deletion of EPO1, SCS2, or both are indicated by the black symbols. The regression line shows the linear correlation between both values in these strains (R2 = 0.92). Blue dots present the values of the WT strain and strains where the connections between cER and bud tip are impaired to an unknown extent. (D) Model of cER anchorage at the cell tip. The top half depicts WT, and the bottom half shows Δepo1 cells. The blue line indicates a newly incorporated cell wall and PM material.
Correlation between the extent of polar growth and regions of absent cER. (A) Length/width ratio of daughter cells of WT and selected double-deletion strains (200 < n < 206; error bars show SEM; the value for the WT is taken from Fig. 2 D). ***, P < 0.001. (B) Quantification of gaps of ER staining below the bud tip in 1–2 µm buds of WT and selected single- and double-deletion strains expressing HMG1-GFP as marker for the cER. Shown are the means of 40 < n < 149 cells derived from at least two independent strains with a deviation of <8%. (C) The percentages of cells displaying no ER below the bud tip were plotted against their length/width ratios. Values were taken from A and B, respectively. Strains carrying either a deletion of EPO1, SCS2, or both are indicated by the black symbols. The regression line shows the linear correlation between both values in these strains (R2 = 0.92). Blue dots present the values of the WT strain and strains where the connections between cER and bud tip are impaired to an unknown extent. (D) Model of cER anchorage at the cell tip. The top half depicts WT, and the bottom half shows Δepo1 cells. The blue line indicates a newly incorporated cell wall and PM material.
The measured value for Δbem3 Δpea2 cells fell onto the linear slope of the established correlation. This finding confirmed that Epo1p uses both, Bem3p and Pea2p, to anchor Scs2p at the bud tip (Fig. 8, C and D). Conversely, the deviation of Δbem3 cells from this linear correlation allowed us to calculate the contribution of Bem3p to the strength of cER–bud tip connection (Fig. 8 C). According to this estimate Bem3p contributes 20%, and the polarisome contributes the remaining activity of 80% (Fig. 8 D). Neither the sequence homologue of Scs2p, Scs22p, nor the sequence homologue of Epo1p, Ylr031w, contributed significantly to the ER–cell tip contact (Fig. S5)
Genetic analysis assigns Epo1p and Scs2p to a distinct path of cER inheritance
To genetically define the relation between SCS2 and EPO1, we sporulated a heterozygous Δscs2 Δepo1 diploid and compared the sizes of the colonies from the obtained spores. No genetic interaction between the two genes was inferred as the single Δscs2 deletion grew as well as the Δepo1 Δscs2 cells (Fig. 9 A). By introducing the cER marker Hmg1-GFP, we compared the extent of cER inheritance between the cells of different genotypes (Wilhovsky et al., 2000). We classified the bud appearance of the cER into five categories: no, tubular, cortical, random ER, and ER gaps below the tip (Fig. 9 D). The analysis was performed with small (≤1 µm in length), medium (1–2 µm), and large (>2 µm) buds. The analysis showed that the phenotypes of the double deletion closely resembled those of the Δscs2 single deletion (Fig. 9 C). We noticed that the amount of ER gaps was more pronounced in Δscs2 and Δepo1 Δscs2 than in Δepo1 cells (Fig. 9, B and C). This difference probably reflects the dual role of Scs2p in anchoring the cER to the bud tip and in tethering it to the PM. As a result of the larger fraction of cells containing no ER, the number of 1-µm buds scored with ER gaps was smaller in Δscs2 Δepo1 than in Δepo1 cells.
Genetic interactions between members of cER inheritance pathways. (A, top) Tetrad analyses of diploid cells heterozygous for Δepo1 and Δptc1 or Δepo1 and Δscs2. (bottom) Quantifications of the diameters of the colonies formed by single spores after 4 d (left) or 5 d (right) of growth (19 < n < 28). Error bars show SEM. ***, P < 0.001. (B) Fluorescence microscopy of Hmg1-GFP–expressing single- and double-deletion mutants derived from tetrad dissection in A. The classification of cER phenotypes into one of the five different groups is indicated below the images, and idealized drawings of their implementations are given at the bottom. Bar, 5 µm. (C) Classification of cER inheritance defects and quantification of their relative abundance in percentages of inspected WT, Δscs2, Δepo1, and Δepo1 Δscs2 cells. The values for each strain were separated according to bud length of the cells. (D) As in C, but with WT, Δptc1, Δepo1, and Δepo1 Δptc1 cells (43 < n < 220).
Genetic interactions between members of cER inheritance pathways. (A, top) Tetrad analyses of diploid cells heterozygous for Δepo1 and Δptc1 or Δepo1 and Δscs2. (bottom) Quantifications of the diameters of the colonies formed by single spores after 4 d (left) or 5 d (right) of growth (19 < n < 28). Error bars show SEM. ***, P < 0.001. (B) Fluorescence microscopy of Hmg1-GFP–expressing single- and double-deletion mutants derived from tetrad dissection in A. The classification of cER phenotypes into one of the five different groups is indicated below the images, and idealized drawings of their implementations are given at the bottom. Bar, 5 µm. (C) Classification of cER inheritance defects and quantification of their relative abundance in percentages of inspected WT, Δscs2, Δepo1, and Δepo1 Δscs2 cells. The values for each strain were separated according to bud length of the cells. (D) As in C, but with WT, Δptc1, Δepo1, and Δepo1 Δptc1 cells (43 < n < 220).
The polarisome recruits and activates the cell wall integrity kinase Slt2p (van Drogen and Peter, 2002). This step is proposed to strengthen the association between ER tubules and actin filaments during bud invasion (Li et al., 2010, 2013). A protein complex consisting of Nbp2p and the phosphatase Ptc1p reverts this activity of Slt2p (Du et al., 2006; Hruby et al., 2011). Consequently, a loss of PCT1 or NBP2 results in pinlike ER structures that seemed fixed to the bud tip. In accordance with their role as activators of Slt2p, the deletion of either SPA2 or PEA2 suppresses the cER inheritance phenotype of Δptc1 cells (Li et al., 2013). To test whether Epo1p shares the role as activator of Slt2p with the other polarisome components, we conducted tetrad analyses of heterozygous Δptc1 Δepo1 or Δnbp2 Δepo1 cells (Figs. 9 A and S4). Instead of a suppression as seen for the deletion of SPA2 and PEA2, the reduced growth of the spores containing the double deletions documented negative genetic interactions between EPO1 and PTC1 or NBP2 (Figs. 9 A and S4; Li et al., 2013). A similar genetic interaction was observed between PTC1 and SCS2 but not between PTC1 and SCS22 (Fig. S4, B and D). The microscopic analysis of the Hmg1-GFP–expressing haploid spores clearly identified additive effects of both PTC1 and EPO1 on the presence of cER in small buds and on the cortical localization of cER in middle-sized and large buds. Conversely, the increased amount of tubular ER seemed exclusively determined by the lack of PTC1, whereas the lack of ER–cell tip contacts was primarily determined by the deletion of EPO1 (Figs. 8 C and 9, B and D).
Sec3p is a component of the exocyst that contributes to the inheritance of the cER in a genetically distinct pathway from SCS2 (Wiederkehr et al., 2003; Loewen et al., 2007). The tetrad analysis of heterozygous Δepo1 Δsec3 diploids revealed a reduced growth of the haploid spores carrying both deletions compared with those carrying the single deletions (Fig. S4 C). We conclude that EPO1 and SCS2 fall into one class of cER inheritance genes that is distinct from the class defined by the genes PTC1, NBP2, or SEC3.
Discussion
The Split-Ub analysis of Epo1p and Pea2p revealed that Scs2p and Bem3p are connected to Pea2p and the polarisome through the common binding partner Epo1p. Our time-resolved in vivo analysis showed that a simultaneous interaction between Epo1p, Scs2p, Bem3p, and Pea2p occurs only during bud growth. This and the partial reconstitution of these interactions in vitro support the existence of a dynamic protein complex consisting of Scs2p, Epo1p, Bem3p, and the polarisome. The analyses of cells lacking Scs2p or Epo1p proved that this complex attaches the ER to the tip of the growing cell (Fig. 8 D). This finding adds to our knowledge about spatially and functionally distinct populations of the cER and helps to explain how the inheritance of the cER is mechanistically achieved (Fehrenbacher et al., 2002; Du et al., 2004; Estrada de Martin et al., 2005). Tubules of the ER invade the bud by an actin-based mechanism and upon entry become attached to the bud tip through binding to Epo1p. Through the continuous growth at the tip the Epo1p–Scs2p connection might then pull the cER actively into the bud. In larger buds, Ptc1p catalyzes the dephosphorylation of still unknown targets to release the ER tubules from actin filaments and to enable the spreading of the cER below the membrane (Li et al., 2010, 2013). Accordingly, Epo1p does not mediate the connection between Scs2p and the PM in the mother cell or the cortex of the bud distant from its tip. Instead, SPLIFF analysis of Epo1p and the correlation between the extent of polarized growth and lack of ER–cell tip contacts suggest that the Scs2p–Epo1p–Bem3p–polarisome complex might exclusively be required to keep ER tubules or the PM-attached cER close to the tip of the bud during tip growth. The connection between Epo1p and Scs2p is dissolved during the M phase of the cell cycle. Whether and how Epo1p assists Scs2p in its newly discovered roles as a ER–septin tether and in spindle positioning remain open questions for future experiments (Chao et al., 2014).
What are the advantages of using a specialized receptor for connecting the ER to the cell tip? We speculate that Scs2p through binding to Epo1p creates a specific contact that meets the specific demands for rapid membrane and cell wall extension at the cell tip. A direct association between PM and ER is incompatible with the spatial requirements during vesicle fusion and endocytosis (West et al., 2011; Stradalova et al., 2012; Wolf et al., 2012). The Scs2/Epo1p-organized connection between ER and cell tip might reduce the distance between the ER and the PM to efficiently supply the tip with cell wall and PM material but at the same time provide sufficient distance between ER and PM to facilitate vesicle fusion. This model would require a structurally asymmetric organization of the polarisome, with one site facing the PM and the other site oriented toward the cytosol and exposing Pea2p/Epo1p to enable the connection to the ER and perhaps other secretory compartments (Fig. 8 D; Gallego et al., 2010; Bi and Park, 2012).
The ER–cell tip complex might thus act as catalyst of polarized growth by promoting three interrelated activities: (1) Bud6p and Bni1p nucleate actin filaments that are used as tracks for the delivery of secretory vesicles to the bud tip (Evangelista et al., 1997; Sagot et al., 2002b; Graziano et al., 2011, 2013). (2) Msb3p and Msb4p enhance the GTP hydrolysis by Sec4p and support vesicle recruitment (Chapa-y-Lazo et al., 2011) and fusion (Gao et al., 2003; Tcheperegine et al., 2005). (3) Epo1p recruits the ER to stimulate vesicle synthesis at the tip and ensures sufficient space between PM and cER at the bud tip to facilitate the flux of these vesicles to the PM.
Different organisms depend on similar mechanism and molecules to organize the numerous elements involved in cell tip extension (Wedlich-Söldner et al., 2002; Harris et al., 2005; Chapa-y-Lazo et al., 2011; Riquelme and Sánchez-León, 2014). Our herein characterized interaction network might thus serve as template for understanding the organization of polar secretion beyond the budding yeast.
Materials and methods
Construction of fusion genes and other molecular manipulations
Gene fusions to the sequence of the N-terminal 35 residues of Ub (Nub), the sequence of the C-terminal 41 residues of Ub (Cub), the sequence of GFP carrying a S65T exchange, the sequence of CHERRY, the sequence coding for a ninefold repetition of the MYC epitope (MYC), or the sequence of the CHERRY-Cub-GFP module (CCG) were performed as previously described (Hruby et al., 2011; Dünkler et al., 2012; Moreno et al., 2013). Specifically, EPO1CRU, EPO1-GFP, and EPO1CCG were constructed by genomic in-frame integration of the CRU, GFP, or the CCG module behind the coding sequence of EPO1. In brief, a PCR fragment spanning 547 bp of the 3‘ end of the EPO1 ORF and containing an EagI and an SalI site at its 5′ and 3′ end, respectively, but lacking its stop codon was cloned in front of the CRU, GFP, CCG, or MYC module on a pRS303, pRS304, or pRS306 vector (Sikorski and Hieter, 1989). The obtained plasmids were linearized by a unique ClaI site within the EPO1 sequence and transformed into yeast. Subsequent homologous recombination restored the respective fusion to full-length EPO1. The successful recombination was verified by colony PCR using diagnostic primer combinations. The 3′ genomic fusions of the CRU, GFP, CHERRY, or MYC cassettes to PEA2, SPA2, KEL1, and BEM3 were obtained accordingly. Genomic PMET17 promoter integrations were achieved by PCR-based one-step replacements as previously described (Janke et al., 2004). EPO11–776CRU was constructed by genomic in-frame integration of the CRU module behind the codon of 776 of EPO1. EPO1761–943CRU and EPO11–876CRU were obtained by inserting PCR fragments spanning the respective ORFs through EagI–SalI restriction sites between the sequence of the PMET17 promoter and the CRU module on the pRS313 vector (Sikorski and Hieter, 1989). EPO11–760-GFP, EPO1761–943-GFP, and the corresponding Cherry fusions were obtained by inserting the PCR fragments spanning the respective ORFs through EagI–SalI restriction sites in frame between the sequence of the PMET17 promoter and GFP or CHERRY on the centromeric pRS313 or pRS315 vector. Fusions of GFP to SEC62 or SCS21–225 were obtained by inserting PCR fragments of the corresponding genes in frame behind the PMET17-GFP cassette on a pRS313 vector. Nub-Scs21–225 was obtained by inserting the sequence coding for the first 225 residues of SCS2 in frame behind the sequences of the CUP1 promoter, Nub, and the HA epitope (PCUP1-Nub-HA module) on a centromeric plasmid harboring a geneticin-resistance gene on the pRS314 backbone (Sikorski and Hieter, 1989; Dünkler et al., 2012). HMG1-GFP containing plasmid pRH475 was provided by R.-P. Jansen (University of Tübingen, Tübingen, Germany) and linearized with StuI and integrated into the ura3-52 locus of the yeast (Wilhovsky et al., 2000). GST or MBP fusions were obtained by placing the ORF of the respective gene or gene fragment in frame behind the E. coli GST sequence on the pGEX-2T plasmid (GE Healthcare) or behind the E. coli MBP sequence on the pMal-c5x plasmid (New England Biolabs, Inc.). Fusions to the human O6-Alkyl-DNA transferase (SNAP-tag; New England Biolabs, Inc.) were expressed from plasmid pAGT-Xpress, a pET-15b derivative (Schneider et al., 2013). Gene fragments were inserted in frame into a multicloning site located between the upstream 6×HIS-tag coding sequence and the downstream SNAP-tag coding sequence. The 6×HIS-tag fusions were obtained by placing the ORF of the respective gene or gene fragment behind the E. coli 6×HIS-tag sequence on the pAC plasmid. A list of plasmids and yeast strains used in this study can be found in Tables S3 and S4.
Growth conditions, yeast strains, and genetic methods
Culture media and yeast genetic methods were performed following standard protocols (Guthrie and Fink, 1991). Media for the Split-Ub interaction assay contained 1 mg/ml 5-fluoro-orotic acid (5-FOA; Thermo Fisher Scientific). All yeast strains used were derivatives of JD47, a segregant from a cross of the yeast strains YPH500 and BBY45 (Dohmen et al., 1995). One-step gene deletions were performed by PCR-based methods using pFA6a-hphNT1, pFA6a-natNT2, pFA6a-kanMX6, and pFA6a-CmLEU2 as templates (Bähler et al., 1998; Janke et al., 2004; Schaub et al., 2006).
Fluorescence microscopy
Yeast cells were prepared for imaging as previously described (Schneider et al., 2013). In brief, overnight cultures were grown in liquid selective synthetic defined (SD) media, the next day, diluted 1:15 in 3 ml of fresh SD medium, and grown for 4–5 h at 30°C to mid–log phase. Cells for length/width analysis were cultured in YPD (yeast extract, peptone, and dextrose). For standard microscopy, cells were spun down and resuspended in 20–50 µl of fresh medium. 3 µl was transferred to a microscope slide, covered with a glass coverslip, and immediately inspected under the microscope. For live-cell imaging, 3 µl cell culture was immobilized by fixing a coverslip with parafilm strips on a custom-designed glass slide containing solid, SD medium with 1.7% agarose. For SPLIFF analysis, equal amounts of a and α cells were mixed, spun down, resuspended in 50 µl fresh medium, and prepared for microscopy (see previously in this paragraph). Pictures were taken immediately or in case of SPLIFF analysis after 45–60 min of incubation at 30°C under the microscope as soon as the first zygotes appeared.
Cells were examined with either a fluorescence microscopy system (DeltaVision; Applied Precision) including a wide-field microscope (IX71; Olympus) or a spinning disc confocal microscope (Axio Observer Z.1; Carl Zeiss). Yeast cell imaging with the DeltaVision system was performed using a steady-state heating chamber, mercury arc lamp, U Plan S Apochromat 100× 1.4 NA oil ∞/0.17/FN26.5 objective, and a camera (CoolSNAP HQ2-ICX285 or Cascade II 512 electron-multiplying charge-coupled device [CCD]; both by Photometrics). Fluorescent proteins were visualized using softWoRx 5.0 software (Applied Precision) and filter sets for EGFP (λ excitation = 470 nm; λ emission = 525 nm) and Cherry (λ excitation = 572 nm; λ emission = 632 nm), respectively. Images were acquired as adapted z series. The confocal spinning disc system is equipped with two-diode laser channels for excitation of GFP (488 nm) and Cherry (561 nm), an electron-multiplying CCD camera (Evolve 512; Photometrics), and a Plan Apochromat 63×/1.4 NA oil differential interference contrast (DIC) ∞/0.17 objective. Images were acquired under the control of the ZEN2012 software (Carl Zeiss) as adapted z series. For live cell imaging experiments, the CCD capture time was adapted to the intensity of GFP and Cherry signal for every fluorescently labeled construct to reduce bleaching and phototoxicity. Cells were kept at 30°C.
Quantitative analysis of microscopy data
All microscopy files were processed and analyzed with ImageJ 64 1.45s (National Institutes of Health). For FI quantification, z stacks were projected to one layer. The FIs of defined areas of bud tip or bud neck were then calculated by the mean value, and after background subtraction, defined as FIsignal. A randomly selected and background corrected area within the cytosol of the mother cell was used to measure the mean value of the cytosolic fluorescence (FIcytosol). The relative FIs at tip or bud neck were then given as FIsignal/FIcytosol.
To estimate the abundance of selected components of the polarisome at the bud tip, we mixed Spa2-GFP–, Pea2-GFP–, or Epo1-GFP–expressing cells (test strains) with an equal amount of cells expressing Spa2-GFP together with Hhf2-Cherry (reference strain). The normalized intensities of the tip signals of both strains (FItest and FIreference) were quantified by subtracting the intensity of an area in the mother cell (areasignal × mean valuecytosol) from the intensity of an equal area around the tip signal (areasignal × mean valuesignal). The molar proportion between the tested protein and Spa2p at the bud tip was then given as FItest/FIreference.
We used the ImageJ oval profile tool to characterize the distribution of cortical Epo1-GFP and Pea2-GFP in different strains. The tool placed an oval on the cortex of daughter cells and measured the radial sums of the FIs of 100 equally spaced points on the oval. The profile of the same oval, but placed outside the cell, was subtracted for background correction. For each strain and fluorescent fusion, the highest FI value of the oval was set to 100, and the obtained lowest value was then subtracted from all other values to give FInorm. The FInorm’s were finally plotted against their position on the oval. The same tool and a similar procedure were used to compare the distributions of GFP-Sec62p and Spa2-Cherry within one cell by measuring the FIs of both channels of 100 equally spaced points along an oval. The profile of the same oval but placed outside the cell was subtracted for background correction. The highest FIs of each channel were set to 100. The mean FIs of 100 randomly chosen points within the cytosol of the mother was finally subtracted to give the profile of normalized FIs of both channels (FInorm).
SPLIFF interaction analysis
SPLIFF analysis was as previously described (Moreno et al., 2013). In brief, PMET17-Epo1CCG–expressing a cells were cultured in liquid SD media without methionine to obtain sufficient expression for the SPLIFF analysis. Nub fusion proteins were separately expressed in α cells. Interaction measurements were initiated by mixing equal amounts of a and α cells. The cell suspension was immobilized on SD agarose pads and subjected to time-lapse analysis. Formation of diploid zygotes was recorded with a five-section z stack in intervals of 0.65 µm at three channels: brightfield, GFP, and Cherry. Sum projections of the green and red fluorescence z stacks of the different time points were created to construct 2D images. To obtain a spatial fluorescence profile over time, the fluorescence signal of the region of interest and a region within the cytosol were selected to calculate the relative FItest’s (RFIs; see previous paragraph). Each FI(t) value was normalized by relating it to the FI values shortly before cell fusions have occurred (FI(start)) through the following equation:
The conversion (FD) of the CCG fusion to the CC fusion was calculated and normalized by the following equation:
Statistical tests
Data evaluation and statistical analysis were performed using Prism. t tests were used to compare the length/width ratio of measured daughter cells and to compare the relative FIs of measured GFP signals. In all other cases, not passing D’Agostino and Pearson omnibus normality test, Mann–Whitney tests were applied.
Tetrad analysis
Single gene deletions were performed in haploid JD47 and JD53 cells by PCR-based methods using plasmids pFA-hphNT1, pFA-natNT2, pFA-kanMX6, and pFA-CmLEU2 (Table S3; Janke et al., 2004). Strains carrying single gene deletions were mated on solid YPD medium to generate heterozygous diploid zygotes. Single colonies were transferred onto GNA presporulation plates (5% d-glucose, 3% nutrient broth, and 1% yeast extract), incubated for 1 d at 30°C, and replica plated to another fresh GNA plate for one more day at 30°C. A small amount of yeast cells were resuspended in 3 ml liquid sporulation medium (1% potassium acetate and 0.005% zinc acetate) and incubated on a roller wheel for 5 d at 23°C followed by 2 d at 30°C to induce spore formation. For dissection of asci, 1 ml spore suspension was pelleted and resuspended in 100 µl zymolyase solution (100 µg/ml in 1 M sorbitol). After a 5-min incubation at 30°C, the cells were washed twice with water and spread on a YPD plate. The spores of each asci were separated by a dissection microscope (MSM 300; Singer Instruments). The genetic compositions of the spores were determined by spotting cells of each colony onto suitable selection media.
Split-Ub interaction analysis
Large scale Split-Ub assays were performed as described using 389 different Nub fusion–expressing yeast strains or only a subset of those and the robot system (RoToR HDA; Singer Instruments; Hruby et al., 2011; Dünkler et al., 2012). To measure interactions between individual Nub and Cub fusion proteins, the yeast cells coexpressing both fusion proteins grown to an OD600 of 1 and 4 µl of this and of 10-fold serial dilutions were spotted onto media containing or lacking 1 mg/ml 5-FOA and selected for the presence of the fusion constructs. Cells were incubated at 30°C, and growth was recorded daily for ≤5 d (Eckert and Johnsson, 2003). For the interaction analysis in the absence of a third gene, the Nub and Cub fusions were introduced individually in strains of opposing mating types that each carried the deletion of the gene of interest. Split-Ub interaction analysis was then performed with the diploid strain obtained by mating of the two Nub- or Cub-expressing deletion strains.
Preparation of yeast cell extracts
For in vitro binding assays, exponentially grown yeast cell cultures were pelleted and washed once in water. The cell pellet was resuspended in 3 ml extraction buffer (20 mM Tris, 80 mM NaCl, 1 mM DTT, and 0.5 mM PMSF) containing protease inhibitor cocktail (Roche). Glass bead lysis (threefold amount beads to cell wet weight) was performed by vortexing 7–10 times for 30 s, interrupted by short pauses on ice. The obtained yeast cell extracts were clarified by centrifugation at 16,000 g for 20 min at 4°C.
Expression and preparation of fusion proteins in E. coli
All proteins were expressed in E. coli cells (BL21; GE Healthcare) at 37°C in 2YT medium for 4 h or in super broth medium at 18°C overnight. Cells were pelleted, washed once with PBS, resuspended in PBS-containing protease inhibitor cocktail (Roche), lysed by lysozyme treatment (1 mg/ml; 30 min on ice), and subjected to sonication with a Bandelin Sonopuls HD 2070 (Reichmann Industrieservice). Extracts were clarified by centrifugation at 40,000 g for 10 min at 4°C. MBP fusion proteins were purified by immobilization on amylose beads for 1 h at 4°C followed by elution with 0.1 ml buffer containing 20 mM Tris-Cl, 0.2 M NaCl, 1 mM EDTA, and 10 mM maltose.
In vitro binding assay
GST-tagged proteins were immobilized on glutathione-coupled Sepharose beads (GE Healthcare). Beads with bound proteins were incubated with either 1.5 ml yeast or 0.5 ml E. coli cell extract for 1 h at 4°C under rotation and washed three times with yeast or E. coli extraction buffer. The bound material was eluted (50 mM Tris and 20 mM reduced glutathione) and analyzed by SDS-PAGE and immunoblotting as previously described (Hruby et al., 2011). To test the suitability of the MBP fusion proteins for in vitro binding assays, we performed a pull-down assay of MBP with GST and GST-Scs2p (Fig. S1 D).
Online supplemental material
Fig. S1 documents the expressions of the fusion proteins used for pull-down analysis shown in Figs. 1 and 5. Fig. S2 is related to Fig. 3 and shows the localizations of Bem3-GFP in WT cells and of Epo1-, Pea2-, and Spa2-GFP in Δbni1 or Δpea2 cells and the actin staining of WT and of Δepo1 cells. Fig. S3 relates to Figs. 1, 5, and 7 and shows the array-based Split-Ub interaction assay between Epo1CRU and the Nub fusions of all ER–PM tethers and the complementation of Δepo1 cells by the expression of Epo1p fragments. Fig. S4 is related to Fig. 9 and displays the tetrad analyses of the following sporulated heterozygous diploids: Δepo1 Δnbp2, Δptc1 Δssc2, Δptc1 Δssc22, and Δepo1 Δsec3. Fig. S5 summarizes the length/width ratios and ER staining of additional yeast deletion strains and relates to Fig. 8. Table S1 lists all tested Nub fusions. Table S2 reveals the identities of all Nub fusions on the Nub array shown in Fig. 1 A. Table S3 lists the constructs used and created in this study. Table S4 lists the yeast strains used and created in this study. Video 1 shows the time-lapse analysis of Epo1-GFP and Spa2-Cherry coexpressing yeast cells. Video 2 shows the time-lapse analysis of Spa2-GFP and Shs1-Cherry coexpressing yeast cells. Video 3 shows the time-lapse analysis of Epo1-GFP and Shs1-Cherry coexpressing yeast cells. All videos are related to Fig. 2.
Acknowledgments
We thank Judith Müller (Ulm University) for discussions and Ute Nussbaumer (Ulm University) for technical assistance.
We acknowledge the support by the Bundesministerium für Bildung und Forschung (BMBF) Initiative SysTec (0315690B) and the BMBF Initiative GerontoSys2 (SyStaR; 03158 94A).
The authors declare no competing financial interests.