Ubiquitylation and phosphorylation control composition and architecture of the cell separation machinery in yeast and other eukaryotes. The significance of septin sumoylation on cell separation remained an enigma. Septins form an hourglass structure at the bud neck of yeast cells that transforms into a split septin double ring during mitosis. We discovered that sumoylated septins recruit the cytokinesis checkpoint protein Fir1 to the peripheral side of the septin hourglass just before its transformation into the double-ring configuration. As this transition occurs, Fir1 is released from the septins and seamlessly relocates between the split septin rings through synchronized binding to the scaffold Spa2. Fir1 binds and carries the membrane-bound Skt5 on its route to the division plane where the Fir1-Skt5 complex serves as receptor for chitin synthase III.
Introduction
The five mitotic septins of the yeast Saccharomyces cerevisiae, Cdc11, Cdc12, Cdc10, Cdc3, and Shs1, assemble into hetero-octameric rods that polymerize end to end into apolar filaments (Woods and Gladfelter, 2021; Marquardt et al., 2021). At the beginning of each cell cycle, septin filaments form a ring-like structure next to a previous division site through which the directed transport of secretory vesicles creates a new bud that grows and matures into the next daughter cell (Chiou et al., 2017; Pruyne and Bretscher, 2000; Chollet et al., 2020). The septin ring expands later into an hourglass structure in which paired septin filaments are closely aligned at the bud neck along the mother–bud axis (DeMay et al., 2011; Ong et al., 2014). The splitting of the hourglass into two rings prominently marks the beginning of cytokinesis. This transformation involves the loss of the paired filaments with a concomitant formation of new filaments perpendicular to the mother–bud axis (DeMay et al., 2011; Chen et al., 2020; Vrabioiu and Mitchison, 2006). The contracting actin myosin ring (AMR) subsequently pulls the newly exposed membrane between the two rings toward the center of the bud neck. Chitin synthase II (Chs2) simultaneously synthesizes a primary septum between the two sheets of the constricted plasma membrane. The concerted actions of chitin synthase III (Chs3) and the β-1,6 glucan synthase Fks1 layer a secondary septum upon the primary septum. Finally, the daughter cell releases hydrolases that remove the primary septum to separate both cells (Meitinger and Palani, 2016; Bhavsar-Jog and Bi, 2017; Onishi et al., 2013).
The split septin double ring (SSDR) performs two important functions during cytokinesis: first, it seals off the membrane of the cytokinesis compartment to keep the membrane-attached proteins within the SSDR (Dobbelaere and Barral, 2004; Wloka et al., 2011). Second, it serves as a platform from which proteins are sequentially released into the cleavage furrow to take part in the different steps of cell separation (Meitinger et al., 2013; Meitinger et al., 2011; Schneider et al., 2013; Fang et al., 2010).
The yeast septins Cdc11, Cdc3, and Shs1 are known to be sumoylated during mitosis and desumoylated shortly before cell separation (Johnson and Blobel, 1999; Takahashi et al., 1999). Sumoylation involves only a minority of the septins and is restricted to the peripheral region of the septin hourglass facing the mother cell (Johnson and Blobel, 1999). The precise timing of septin sumoylation and desumoylation is achieved by the successive export of the small ubiquitin-like modifier (SUMO, Smt3) ligase Siz1 and the Smt3 protease Ulp1 from the nucleus into the cytosol (Makhnevych et al., 2007; Takahashi et al., 2000, 2001; Johnson and Gupta, 2001). Both enzymes return to the nucleus before a new cell cycle starts (Elmore et al., 2011; Makhnevych et al., 2007; Lewis et al., 2007; Panse et al., 2003). Timing and location imply that septin sumoylation plays a role during cytokinesis. Although suggestive, the question of whether this elaborate modification cycle influences any of the functions of the septins remained open for more than 20 years.
The remarkable resilience of cell separation mechanisms in the face of both internal and external stresses suggests that the consecutive activities of cytokinesis are tightly coordinated. Notably, cell separation should only proceed once AMR contraction and membrane closure have been successfully completed. Fir1, a novel checkpoint protein, was recently proposed to regulate this dependency by modulating the release of septum-degrading hydrolases into the extracellular space (Brace et al., 2019). Fir1 emerges at the division site shortly before cell separation, where it is believed to inhibit the kinase activity of Cbk1, a member of the nuclear Dbf2-related/large tumor suppressor (NDR/LATS) kinase family, known to facilitate the separation of mother and daughter cells (Brace et al., 2019; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002; Weiss 2012). Many aspects of this checkpoint mechanism are still not understood. Outstanding questions pertain to the mechanisms governing Fir1’s localization to the bud neck and its ability to detect and correct errors in septum formation.
In this study, we show that the sumoylation of septins initiates a two-step targeting process that escorts Fir1 and Skt5 into the space between the SSDR where the Fir1-Skt5 complex serves as a receptor for Chs3 and promotes chitin synthesis during secondary septum formation.
Results
Sumoylation recruits Fir1 to the peripheral side of the septin hourglass
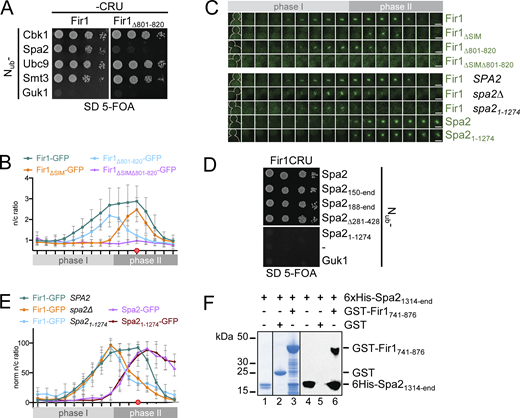
Fir1 is predicted to present multiple linear interaction motifs within a predominantly unfolded structure (Fig. 1 A; Brace et al., 2019; Grinhagens et al., 2020; Jumper et al., 2021). Fir1 binds to Smt3 through its SUMO-interacting motif (SIM) and was recently shown to localize to the bud neck during cell abscission (Hannich et al., 2005; Uzunova et al., 2007; Brace et al., 2019; Grinhagens et al., 2020). To test whether binding to Smt3 is required for Fir1’s localization at the bud neck, we compared the cellular distribution of Fir1-GFP to a mutant of Fir1 that lacked residues 759–767 of the SIM (Fir1∆SIM). Split-Ubiquitin (Split-Ub) analysis proofed that Fir1∆SIM specifically lost its interaction with Smt3 (Fig. 1 B). Fir1-GFP appeared at the bud neck 8–10 min before the septin ring splits and the AMR starts to contract (Fig. 1 C). It remained there for ∼18 min before it rapidly disappeared. Fir1∆SIM-GFP could be detected at the bud neck 10 min later than the wild-type protein, but then showed a similar signal intensity and kinetic of disappearance (Fig. 1 C). Fir1-GFP first appeared as a ring at the mother side of the septin structure (Fig. 1 D). This ring then shifted between the SSDR during the hourglass transition and contracted as cytokinesis continued (Fig. 1 D; Brace et al., 2019). Position and timing of the Fir1 ring within the SSDR coincided with the ring-like structure of Fir1∆SIM-GFP when it first appeared at the bud neck (Fig. 1 C and Fig. S1 A).
Fir1 is recruited to the bud neck by the sumoylated septins. (A) Cartoon of Fir1 indicating the known and herein discovered binding sites to other proteins. Amino acid positions bordering the motifs are indicated. (B) Cutouts of a Split-Ub array of diploid yeast cells expressing genomic Fir1CRU (CRU; C-terminal half of Ubiquitin [Cub]-R-Ura3; left panel), Fir1∆SIMCRU (right panel) together with the indicated Nub fusions of known binding partners of Fir1, or the negative control Nub-Guk1 (Nub, N-terminal half of Ubiquitin). Four independent matings were arrayed as quadruplets on media containing 5-FOA. Colony growth indicates interaction between the fusion proteins. (C) Fluorescence intensities (upper panel) and stills of the bud necks of representative cells (lower panel) coexpressing Fir1-GFP (n = 29) or Fir1∆SIM-GFP (n = 25) and Myo1-Cherry during cytokinesis. The ratio of bud neck fluorescence divided by cytosolic fluorescence (n/c ratio) is shown as mean ± standard deviation (SD). Images were taken every 2 min. The filled red circle (minute 22) indicates the time point of Myo1 disappearance. Phase I indicates association of Fir1 with the septin ring, phase II localization within the SSDR. (D) Deconvoluted images of Fir1-GFP associated with the Cherry-labeled septin ring (Shs1-Cherry) shortly before (left panel), during (middle panel), and shortly after ring splitting (right panel). m and d indicate mother and daughter cells, respectively. (E) As in C but with cells co-expressing Myo1-Cherry with Fir1-GFP in wild type (as reference from C) or siz1∆ cells (n = 22), or with cells coexpressing Shs1-Cherry and Fir1-GFP in wild type (n = 19, Fir1-GFP SEPSUMO) or in sep∆sumo cells (n = 23, Fir1-GFP sep∆SUMO). The orange-filled circle (time point 18) indicates the sudden decrease of the Shs1-Cherry signal. The significance of the differences between Fir1-GFP signal intensities of wild type and the deletion strains are shown in Fig. S1 C. Reintroducing a plasmid-encoded SIZ1 into the siz1∆ cells restored the targeting of Fir1-GFP (Fig. S1 B). Error bars, SD. All scale bars = 3 µm.
Fir1 is recruited to the bud neck by the sumoylated septins. (A) Cartoon of Fir1 indicating the known and herein discovered binding sites to other proteins. Amino acid positions bordering the motifs are indicated. (B) Cutouts of a Split-Ub array of diploid yeast cells expressing genomic Fir1CRU (CRU; C-terminal half of Ubiquitin [Cub]-R-Ura3; left panel), Fir1∆SIMCRU (right panel) together with the indicated Nub fusions of known binding partners of Fir1, or the negative control Nub-Guk1 (Nub, N-terminal half of Ubiquitin). Four independent matings were arrayed as quadruplets on media containing 5-FOA. Colony growth indicates interaction between the fusion proteins. (C) Fluorescence intensities (upper panel) and stills of the bud necks of representative cells (lower panel) coexpressing Fir1-GFP (n = 29) or Fir1∆SIM-GFP (n = 25) and Myo1-Cherry during cytokinesis. The ratio of bud neck fluorescence divided by cytosolic fluorescence (n/c ratio) is shown as mean ± standard deviation (SD). Images were taken every 2 min. The filled red circle (minute 22) indicates the time point of Myo1 disappearance. Phase I indicates association of Fir1 with the septin ring, phase II localization within the SSDR. (D) Deconvoluted images of Fir1-GFP associated with the Cherry-labeled septin ring (Shs1-Cherry) shortly before (left panel), during (middle panel), and shortly after ring splitting (right panel). m and d indicate mother and daughter cells, respectively. (E) As in C but with cells co-expressing Myo1-Cherry with Fir1-GFP in wild type (as reference from C) or siz1∆ cells (n = 22), or with cells coexpressing Shs1-Cherry and Fir1-GFP in wild type (n = 19, Fir1-GFP SEPSUMO) or in sep∆sumo cells (n = 23, Fir1-GFP sep∆SUMO). The orange-filled circle (time point 18) indicates the sudden decrease of the Shs1-Cherry signal. The significance of the differences between Fir1-GFP signal intensities of wild type and the deletion strains are shown in Fig. S1 C. Reintroducing a plasmid-encoded SIZ1 into the siz1∆ cells restored the targeting of Fir1-GFP (Fig. S1 B). Error bars, SD. All scale bars = 3 µm.
Septin sumoylation attaches Fir1 to the septin ring. (A) Deconvoluted images of the bud neck of cells coexpressing Fir1-GFP (left panels) or Fir1∆SIM-GFP (right panels) together with Myo1-Cherry, taken every 2 min, starting 6 min before Myo1 contraction is complete. Overlay, GFP and Cherry channels are shown in the top, middle, and lower panels. Scale bars = 3 µm. (B) As in Fig. 1 C but showing the complementation of siz1∆ cells. Coexpression of plasmid-encoded SIZ1 in siz1∆ cells (pSIZ1; n = 25) rescued Fir1-GFP signal timing and intensity, while the empty vector had no effect (empty plasmid; n = 23). (C) Statistical evaluation of differences in signal intensities of Fir1-GFP in wild type, sep∆sumo, and siz1∆ cells at time point 22 of Fig. 1 E. ****P < 0.0001. Error bars, SD.
Septin sumoylation attaches Fir1 to the septin ring. (A) Deconvoluted images of the bud neck of cells coexpressing Fir1-GFP (left panels) or Fir1∆SIM-GFP (right panels) together with Myo1-Cherry, taken every 2 min, starting 6 min before Myo1 contraction is complete. Overlay, GFP and Cherry channels are shown in the top, middle, and lower panels. Scale bars = 3 µm. (B) As in Fig. 1 C but showing the complementation of siz1∆ cells. Coexpression of plasmid-encoded SIZ1 in siz1∆ cells (pSIZ1; n = 25) rescued Fir1-GFP signal timing and intensity, while the empty vector had no effect (empty plasmid; n = 23). (C) Statistical evaluation of differences in signal intensities of Fir1-GFP in wild type, sep∆sumo, and siz1∆ cells at time point 22 of Fig. 1 E. ****P < 0.0001. Error bars, SD.
To find out whether septin sumoylation is required for the early appearance of Fir1 at the bud neck (phase I targeting), we measured the fluorescence intensity profiles of Fir1-GFP in cells lacking the SUMO ligase Siz1 (siz1∆) or in the congenic strain EJY316, where all sumoylatable lysines were replaced by arginine (sep∆sumo; Table S2; Johnson and Gupta, 2001; Johnson and Blobel, 1999). The Fir1-GFP profiles of both mutants were nearly identical, and in their timing very similar to the profile of Fir1∆SIM-GFP. Both lacked phase I targeting (Fig. 1 E). We conclude that sumoylated septins recruit Fir1 to the periphery of the septin ring shortly before it splits.
During AMR contraction, Fir1-GFP moves from its peripheral position between the SSDR (Fig. 1 D and Fig. S1 A; phase II targeting). Neither deleting the SIM in Fir1 nor preventing septin sumoylation delayed phase II targeting of Fir1-GFP (Fig. 1, C and E). However, the signal intensities of Fir1-GFP in siz1∆ or sep∆sumo cells during phase II targeting were substantially reduced when compared with its signal in wild type cells (Fig. 1 E and Fig. S1 C). These findings argue for a separate receptor that keeps Fir1 at the bud neck after desumoylation of the septins. The partial loss of Fir1 from the SSDR in sumoylation-deficient cells might imply that both targeting steps are somehow coupled.
The polarity scaffold Spa2 attaches Fir1 to the cleavage furrow
We performed a Split-Ub interaction screen to obtain a shortlist of candidate proteins that might anchor Fir1 at the bud neck during phase II targeting (Fig. 2 A and Fig. S2, Table S1; Hruby et al., 2011; Johnsson and Varshavsky, 1994). The polarisome scaffold Spa2 and other subunits of the polarisome were discovered as novel interaction partners of Fir1 (Fig. 2 A; Neller et al., 2015; Tcheperegine et al., 2005; Shih et al., 2005; Dünkler et al., 2021; Valtz and Herskowitz, 1996).
Spa2 recruits Fir1 between the SSDR. (A) Cutouts of a Split-Ub array of 540 diploid yeast cells each expressing genomic Fir1CRU together with a different Nub fusion as in Fig. 1 B. Identities of the Nub fusions are given next to the respective cutouts. Shared colors of the Nub fusions indicate a common process. Red: polarisome; purple: SUMO conjugation; dark blue: exocytosis; light blue: Rho GAPs; yellow: mitotic cyclins; green: phosphatases and their adaptors. The complete analysis is shown in Fig. S2, and the complete list of binding partners in Table S1. (B) As in Fig. 1 C but with cells co-expressing Fir1-GFP with Myo1-Cherry in the presence (as reference from Fig. 1 C) and absence of Spa2 (n = 26), or coexpressing Myo1-Cherry with Spa2-GFP (n = 18). n/c ratios are shown. Reintroducing a plasmid-borne SPA2 into the spa2∆ cells restored the targeting of Fir1-GFP (Fig. S3 B). (C) As in Fig. 1 C but with cells co-expressing Fir1-GFP (as reference from Fig. 1 C) or Fir1∆SIM-GFP with Myo1-Cherry in the presence (as reference from Fig. 1 C) or absence (n = 23) of Spa2. (D) As in Fig. 1 C but with cells coexpressing Fir1750–876-GFP with Myo1-Cherry in the presence (n = 22) and absence (n = 20) of Spa2. Fir1-GFP from Fig. 1 C and Fir1-GFP in spa2∆ from panel B are shown as references. The significance of the differences between Fir1-GFP and Fir1750-876 in wild type and spa2∆ strains is shown in Fig. S3 D. (E) Stills of the bud necks of representative cells corresponding to the intensity profiles shown in B–D. (F) Fluorescence intensity profiles (shown in arbitrary units) along the polarity axes of cells expressing, from left to right, Fir1-GFP, Fir1750–876-GFP in the presence or absence of Spa2, or Spa2-GFP. Numbers and arrows correlate positions on the fluorescence intensity profiles with locations in the cells. Images in B–E were taken every 2 min. Error bars, SD. All scale bars = 3 µm.
Spa2 recruits Fir1 between the SSDR. (A) Cutouts of a Split-Ub array of 540 diploid yeast cells each expressing genomic Fir1CRU together with a different Nub fusion as in Fig. 1 B. Identities of the Nub fusions are given next to the respective cutouts. Shared colors of the Nub fusions indicate a common process. Red: polarisome; purple: SUMO conjugation; dark blue: exocytosis; light blue: Rho GAPs; yellow: mitotic cyclins; green: phosphatases and their adaptors. The complete analysis is shown in Fig. S2, and the complete list of binding partners in Table S1. (B) As in Fig. 1 C but with cells co-expressing Fir1-GFP with Myo1-Cherry in the presence (as reference from Fig. 1 C) and absence of Spa2 (n = 26), or coexpressing Myo1-Cherry with Spa2-GFP (n = 18). n/c ratios are shown. Reintroducing a plasmid-borne SPA2 into the spa2∆ cells restored the targeting of Fir1-GFP (Fig. S3 B). (C) As in Fig. 1 C but with cells co-expressing Fir1-GFP (as reference from Fig. 1 C) or Fir1∆SIM-GFP with Myo1-Cherry in the presence (as reference from Fig. 1 C) or absence (n = 23) of Spa2. (D) As in Fig. 1 C but with cells coexpressing Fir1750–876-GFP with Myo1-Cherry in the presence (n = 22) and absence (n = 20) of Spa2. Fir1-GFP from Fig. 1 C and Fir1-GFP in spa2∆ from panel B are shown as references. The significance of the differences between Fir1-GFP and Fir1750-876 in wild type and spa2∆ strains is shown in Fig. S3 D. (E) Stills of the bud necks of representative cells corresponding to the intensity profiles shown in B–D. (F) Fluorescence intensity profiles (shown in arbitrary units) along the polarity axes of cells expressing, from left to right, Fir1-GFP, Fir1750–876-GFP in the presence or absence of Spa2, or Spa2-GFP. Numbers and arrows correlate positions on the fluorescence intensity profiles with locations in the cells. Images in B–E were taken every 2 min. Error bars, SD. All scale bars = 3 µm.
Split-Ub interaction assay of 540 yeast strains coexpressing P MET17 -Fir1CRU each with a different N ub fusion protein. For every strain, cells of four independent matings were spotted as quadruplets on SD medium containing 5-FOA, 50 µM copper sulfate, and 0 M methionine. Shown is the growth of the diploid yeast cells after 2 d at 30°C. Growth indicates protein–protein interactions.
Split-Ub interaction assay of 540 yeast strains coexpressing P MET17 -Fir1CRU each with a different N ub fusion protein. For every strain, cells of four independent matings were spotted as quadruplets on SD medium containing 5-FOA, 50 µM copper sulfate, and 0 M methionine. Shown is the growth of the diploid yeast cells after 2 d at 30°C. Growth indicates protein–protein interactions.
We considered Spa2 as a candidate receptor as it appeared at the bud neck at about the same time as Fir1∆SIM during phase II targeting (Fig. 2, B, C, and E; Fig. S3 A; Snyder, 1989). Accordingly, the deletion of Spa2 prevented Fir1-GFP from transferring into the SSDR and nearly abolished the bud neck signal of Fir1∆SIM-GFP during both phases (Fig. 2, B, C, and E). Reciprocally, the deletion of Fir1 or the absence of septin sumoylation did not affect the distribution of Spa2 (Fig. S3 C). We conclude that the sumoylated septins recruit Fir1 to the bud neck during phase I targeting, whereas Spa2 acts as a Fir1 receptor during phase II targeting. A C-terminal fragment of Fir1 (Fir1750–876) that starts within the predicted N-terminal β-strand of the SIM lacked phase I targeting and recapitulated the dynamic localization of Spa2-GFP at the bud neck (Fig. 2, D and E; Jumper et al., 2021). Similar to Spa2-Cherry, Fir1750–876-GFP remained at the site of cell division and later transferred to the tip of the growing bud (Fig. 2 F). GFP-fusions of full-length Fir1 and most of its other variants localized to the bud tip at a significantly lower level, almost below the detection limit of our wide-field microscope (Fig. 2 F). The deletion of SPA2 abolished the localization of Fir1750–876-GFP equally and completely at bud neck and tip (Fig. 2, D–F; and Fig. S3 D), thus restricting the potential Spa2-binding site within the C-terminal 126 residues of Fir1.
Spa2 recruits Fir1 into the space between the SSDR. (A) As in Fig. 1 C but with cells co-expressing Fir1-GFP, or Spa2-GFP with Myo1-Cherry. (B) As in Fig. 1 C but showing the complementation of the reduction of the Fir1-GFP bud neck signal in spa2∆ cells. Co-expression of plasmid-encoded SPA2 (pSPA2, n = 28) rescued timing and intensity of the Fir1-GFP signal in spa2∆ cells, while the empty vector had no effect (empty plasmid, n = 26). (C) As in Fig. 1 C. The Spa2-GFP signal at the bud neck remains unaffected in timing and intensity by the deletion of SIZ1 (siz1∆; n = 17) or FIR1 (fir1∆; n = 13). (D) Statistical evaluation of differences in signal intensities at time point 22 of Fig. 2 D between Fir1-GFP in wild type and spa2∆ cells, between Fir1750–876-GFP in wild type and spa2∆ cells, and between Fir1-GFP and Fir1750–876-GFP in wild type and spa2∆ cells. ****P < 0.0001; *P = 0.0174. Error bars, SD. (E) Equal amounts of E. coli extracts containing GST (lanes 1, 3, 5), or GST-Fir1741–876 (2, 4, 6) were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with extracts from E. coli expressing no additional protein (lanes 3, 4) or 6xHis-Spa21,324-end (lanes 1, 2). GST or 6xHis fusions were subsequently detected by anti-GST (lanes 5, 6) or anti-His antibodies (lanes 1–4). Source data are available for this figure: SourceData FS3.
Spa2 recruits Fir1 into the space between the SSDR. (A) As in Fig. 1 C but with cells co-expressing Fir1-GFP, or Spa2-GFP with Myo1-Cherry. (B) As in Fig. 1 C but showing the complementation of the reduction of the Fir1-GFP bud neck signal in spa2∆ cells. Co-expression of plasmid-encoded SPA2 (pSPA2, n = 28) rescued timing and intensity of the Fir1-GFP signal in spa2∆ cells, while the empty vector had no effect (empty plasmid, n = 26). (C) As in Fig. 1 C. The Spa2-GFP signal at the bud neck remains unaffected in timing and intensity by the deletion of SIZ1 (siz1∆; n = 17) or FIR1 (fir1∆; n = 13). (D) Statistical evaluation of differences in signal intensities at time point 22 of Fig. 2 D between Fir1-GFP in wild type and spa2∆ cells, between Fir1750–876-GFP in wild type and spa2∆ cells, and between Fir1-GFP and Fir1750–876-GFP in wild type and spa2∆ cells. ****P < 0.0001; *P = 0.0174. Error bars, SD. (E) Equal amounts of E. coli extracts containing GST (lanes 1, 3, 5), or GST-Fir1741–876 (2, 4, 6) were separated by SDS-PAGE, transferred to nitrocellulose, and incubated with extracts from E. coli expressing no additional protein (lanes 3, 4) or 6xHis-Spa21,324-end (lanes 1, 2). GST or 6xHis fusions were subsequently detected by anti-GST (lanes 5, 6) or anti-His antibodies (lanes 1–4). Source data are available for this figure: SourceData FS3.
Fir1 and Spa2 form a stable protein complex
A stretch within these C-terminal 126 residues of Fir1 is phylogenetically conserved (Fig. 1 A; Fuentes et al., 2022). Deleting this region (Fir1∆801–820) specifically abolished the interaction with Spa2 and caused the premature dissociation of Fir1∆801–820-GFP from the bud neck without affecting its initial phase I targeting (Fig. 3, A–C). The calculated fluorescence intensity profile of Fir1∆801–820-GFP was congruent with the profile of wild-type Fir1-GFP in spa2∆ cells (Fig. 3, B, C, and E) and complementary to the profile of Fir1∆SIM (Fig. 3, B and C). The simultaneous deletion of the potential Spa2-binding site and the SIM motif completely abolished phase I and nearly completely phase II targeting of Fir1∆SIM∆801–820-GFP (Fig. 3, B and C).
Spa2 is the phase II receptor for Fir1. (A) Split-Ub assay of cells coexpressing Fir1CRU (left panel) or Fir1∆801–820CRU (right panel) with Nub fusions to Spa2 and other binding partners of Fir1, or to Guk1 as negative control. Cells were grown to OD600 = 1 and 4 μl, or 4 μl of a 10-fold serial dilution was spotted on media containing 5-FOA and selected for the presence of the Nub and Cub fusions. (B) As in Fig. 1 C but with cells expressing the GFP fusions to the indicated alleles of FIR1 (Fir1∆801–820n = 23; Fir∆SIM∆801–820n = 20; Fir1, Fir1∆SIM as reference from Fig. 1 C). (C) Stills of the bud necks of cells corresponding to the intensity profiles shown in B and E. Scale bars = 3 µm. (D) Split-Ub assay as in A but with cells coexpressing Fir1CRU with the indicated Nub fusions to Spa2 and its mutants, or Nub- and Nub-Guk1 as non-interacting controls. (E) As in Fig. 2 B but with cells coexpressing Myo1-Cherry with Fir1-GFP in spa2∆ (n = 18), or spa21–1,274 (n = 20) cells, and with wild-type cells coexpressing Myo1-Cherry and Spa21–1,274-GFP (n = 20). Fir1-GFP and Spa2-GFP as references from Fig. 1 C and Fig. 2 B. Images in B, C, and E were taken every 2 min. Error bars, SD. (F) Purified 6xHis-Spa1,314-end (lanes 1–6) was incubated with GST-Fir1741–876 (lanes 3, 6) or GST-coupled beads (lanes 2, 5). Glutathione eluates were separated by SDS-PAGE and stained with Coomassie (lanes 1–3), or transferred onto nitrocellulose and stained with anti-His antibody (lanes 4–6). Detection of GST-Fir1741–876 by the anti-His antibody (lane 6) indicates binding of nitrocellulose-detached 6xHis-Spa21,314-end to nitrocellulose fixed GST-Fir1741–876 (Fig. S3 E). Source data are available for this figure: SourceData F3.
Spa2 is the phase II receptor for Fir1. (A) Split-Ub assay of cells coexpressing Fir1CRU (left panel) or Fir1∆801–820CRU (right panel) with Nub fusions to Spa2 and other binding partners of Fir1, or to Guk1 as negative control. Cells were grown to OD600 = 1 and 4 μl, or 4 μl of a 10-fold serial dilution was spotted on media containing 5-FOA and selected for the presence of the Nub and Cub fusions. (B) As in Fig. 1 C but with cells expressing the GFP fusions to the indicated alleles of FIR1 (Fir1∆801–820n = 23; Fir∆SIM∆801–820n = 20; Fir1, Fir1∆SIM as reference from Fig. 1 C). (C) Stills of the bud necks of cells corresponding to the intensity profiles shown in B and E. Scale bars = 3 µm. (D) Split-Ub assay as in A but with cells coexpressing Fir1CRU with the indicated Nub fusions to Spa2 and its mutants, or Nub- and Nub-Guk1 as non-interacting controls. (E) As in Fig. 2 B but with cells coexpressing Myo1-Cherry with Fir1-GFP in spa2∆ (n = 18), or spa21–1,274 (n = 20) cells, and with wild-type cells coexpressing Myo1-Cherry and Spa21–1,274-GFP (n = 20). Fir1-GFP and Spa2-GFP as references from Fig. 1 C and Fig. 2 B. Images in B, C, and E were taken every 2 min. Error bars, SD. (F) Purified 6xHis-Spa1,314-end (lanes 1–6) was incubated with GST-Fir1741–876 (lanes 3, 6) or GST-coupled beads (lanes 2, 5). Glutathione eluates were separated by SDS-PAGE and stained with Coomassie (lanes 1–3), or transferred onto nitrocellulose and stained with anti-His antibody (lanes 4–6). Detection of GST-Fir1741–876 by the anti-His antibody (lane 6) indicates binding of nitrocellulose-detached 6xHis-Spa21,314-end to nitrocellulose fixed GST-Fir1741–876 (Fig. S3 E). Source data are available for this figure: SourceData F3.
Testing different fragments of Spa2 in a Split-Ub interaction assay restricted the Fir1 binding site onto Spa2’s C-terminal 192 residues (Fig. 3 D). A bacterially expressed GST fusion to Fir1741–876 (GST-Fir1741–876) precipitated the purified His-tagged C-terminal domain of Spa2 (His-Spa21,314-end; Fig. 3 F). We conclude that C-terminal domains of Fir1 and Spa2 form a stable complex that anchors Fir1 between the SSDR of dividing cells. Consequently, cells expressing Spa2 without its C-terminal domain (spa21–1,274) lacked phase II targeting of Fir1-GFP, while Spa21–1,274-GFP still binds indistinguishably to the bud neck (Fig. 3, E and C).
Fir1 is a bud neck receptor for Skt5 during cytokinesis
Fir1 might transfer across the septins to carry other proteins piggyback into the space between the SSDR. We measured the fluorescence intensities of GFP fusions to those Fir1 interaction partners that participate in cytokinesis or abscission and compared their profiles between wild type and fir1∆ cells (Fig. 2 A and Fig. 4). Deleting Fir1 did not affect the distributions of Bud14, Hof1, and Msb1 (Fig. S4 A). In contrast, the absence of FIR1 reduced or abolished the phase I targeting of its binding partners Cbk1, Boi1, and Skt5 (Fig. 4, A–C and G; and Fig. S4, B and C; Brace et al., 2019; Grinhagens et al., 2020). While the amounts of Cbk1 and Boi1 recovered to wild type–like levels during phase II targeting, the concentration of Skt5 in the SSDR was significantly reduced (Fig. 4, A–C and Fig. S4 C). Contrary to the other interaction partners, Skt5 seems to require continuous binding to Fir1 for its two-step translocation into the SSDR.
Fir1 carries Skt5 between the SSDR. (A) As in Fig. 1 C but with cells coexpressing Cbk1-GFP together with Myo1-Cherry in the presence (n = 20) or absence (n = 20) of FIR1. (B) As in Fig. 1 C but with cells coexpressing Boi1-GFP with Myo1-Cherry in the presence (n = 20) or absence (n = 25) of FIR1. (C) As in Fig. 1 C but with cells coexpressing Skt5-GFP with Myo1-Cherry in the presence (n = 22) or absence of FIR1 (n = 27), in the absence of SIZ1 (n = 23), or in the absence of SPA2 (n = 21). (D) Images of GFP-Skt5 associated with the Shs1-mCherry–labeled septin ring shortly before (upper left panels from top to bottom: overlay, GFP-, Cherry-channel) and shortly after ring splitting (upper right panels from top to bottom: overlay, GFP, Cherry channel). m and d indicate mother and daughter cell, respectively. Lower panels: Intensity plots in relative fluorescent units of the GFP (green) and Cherry (red) channels along the arrows orthogonal to the septin rings before (left panel) and after (right panel) septin ring splitting. (E) As in Fig. 1 C but with cells coexpressing Chs3-GFP with Myo1-Cherry in wild type (n = 20), in fir1∆ cells (n = 20), or in skt5∆ cells (n = 20). (F) As in Fig. 1 C but with cells co-expressing Chs3-GFP with Shs1-mCherry in wild type (n = 40) or in fir1∆skt5∆ cells (n = 40). The significance of the differences between the profiles in B, C, E, and F are shown in Fig. S4, B–E. (G) Stills of the bud necks of cells corresponding to the intensity profiles shown in C, E, and F. Images were taken every 2 min. Scale bar = 3 µm. (H) Left panel: Mean fluorescence intensities of calcofluor-stained bud necks of wild-type (n = 29), fir1∆ (n = 27), chs2∆ (n = 25), and chs2∆ fir1∆ cells (n = 28). ****P < 0.0001; **P < 0.01; ns = not significant. Right panel: Representative stills of calcofluor-stained cells from each genotype. Error bars, SD. Scale bar = 3 µm.
Fir1 carries Skt5 between the SSDR. (A) As in Fig. 1 C but with cells coexpressing Cbk1-GFP together with Myo1-Cherry in the presence (n = 20) or absence (n = 20) of FIR1. (B) As in Fig. 1 C but with cells coexpressing Boi1-GFP with Myo1-Cherry in the presence (n = 20) or absence (n = 25) of FIR1. (C) As in Fig. 1 C but with cells coexpressing Skt5-GFP with Myo1-Cherry in the presence (n = 22) or absence of FIR1 (n = 27), in the absence of SIZ1 (n = 23), or in the absence of SPA2 (n = 21). (D) Images of GFP-Skt5 associated with the Shs1-mCherry–labeled septin ring shortly before (upper left panels from top to bottom: overlay, GFP-, Cherry-channel) and shortly after ring splitting (upper right panels from top to bottom: overlay, GFP, Cherry channel). m and d indicate mother and daughter cell, respectively. Lower panels: Intensity plots in relative fluorescent units of the GFP (green) and Cherry (red) channels along the arrows orthogonal to the septin rings before (left panel) and after (right panel) septin ring splitting. (E) As in Fig. 1 C but with cells coexpressing Chs3-GFP with Myo1-Cherry in wild type (n = 20), in fir1∆ cells (n = 20), or in skt5∆ cells (n = 20). (F) As in Fig. 1 C but with cells co-expressing Chs3-GFP with Shs1-mCherry in wild type (n = 40) or in fir1∆skt5∆ cells (n = 40). The significance of the differences between the profiles in B, C, E, and F are shown in Fig. S4, B–E. (G) Stills of the bud necks of cells corresponding to the intensity profiles shown in C, E, and F. Images were taken every 2 min. Scale bar = 3 µm. (H) Left panel: Mean fluorescence intensities of calcofluor-stained bud necks of wild-type (n = 29), fir1∆ (n = 27), chs2∆ (n = 25), and chs2∆ fir1∆ cells (n = 28). ****P < 0.0001; **P < 0.01; ns = not significant. Right panel: Representative stills of calcofluor-stained cells from each genotype. Error bars, SD. Scale bar = 3 µm.
Targeting of Bud14, Hof1, and Msb1 is not affected by Fir1. (A) As in Fig. 1 C but with cells co-expressing Myo1-Cherry with Bud14-GFP (upper panel; wild type n = 9; fir1∆ n = 8), Hof1-GFP (middle panel; wild type n = 17; fir1∆ n = 22), or Msb1-GFP (lower panel: wild type n = 13; fir1∆ n = 10) with or without FIR1. (B) Statistical evaluation of differences in signal intensities between Boi1-GFP in the presence and absence of FIR1 at time points 18 and 22 of Fig. 4 B. ****P < 0.0001; ns, not significant. (C) Statistical evaluation of differences in signal intensities between GFP-Skt5 in wild-type cells and strains lacking FIR1, SIZ1, or SPA2 at time points 16 and 24 of Fig. 4 C. ****P < 0.0001; ***P = 0.0005; **P = 0.0066; *P = 0.0166; ns, not significant. (D) Statistical evaluation of differences in signal intensities between Chs3-GFP in the presence and absence of FIR1 or SKT5 at time point 24 of Fig. 4 E. ****P < 0.0001. (E) As in D but for Chs3-GFP in the presence and absence of FIR1 and SKT5 at time point 24 of Fig. 4 F. ****P < 0.0001. Error bars, SD.
Targeting of Bud14, Hof1, and Msb1 is not affected by Fir1. (A) As in Fig. 1 C but with cells co-expressing Myo1-Cherry with Bud14-GFP (upper panel; wild type n = 9; fir1∆ n = 8), Hof1-GFP (middle panel; wild type n = 17; fir1∆ n = 22), or Msb1-GFP (lower panel: wild type n = 13; fir1∆ n = 10) with or without FIR1. (B) Statistical evaluation of differences in signal intensities between Boi1-GFP in the presence and absence of FIR1 at time points 18 and 22 of Fig. 4 B. ****P < 0.0001; ns, not significant. (C) Statistical evaluation of differences in signal intensities between GFP-Skt5 in wild-type cells and strains lacking FIR1, SIZ1, or SPA2 at time points 16 and 24 of Fig. 4 C. ****P < 0.0001; ***P = 0.0005; **P = 0.0066; *P = 0.0166; ns, not significant. (D) Statistical evaluation of differences in signal intensities between Chs3-GFP in the presence and absence of FIR1 or SKT5 at time point 24 of Fig. 4 E. ****P < 0.0001. (E) As in D but for Chs3-GFP in the presence and absence of FIR1 and SKT5 at time point 24 of Fig. 4 F. ****P < 0.0001. Error bars, SD.
Skt5 activates Chs3 and thus determines where and when chitin synthesis occurs (Ono et al., 2000; DeMarini et al., 1997). Recruited by the Bni4-Glc7 complex, Skt5 forms a ring around the incipient bud site and the neck of small budded cells (Larson et al., 2008; DeMarini et al., 1997; Kozubowski et al., 2003). It subsequently spreads out on the plasma membranes of mother and daughter cells before being concentrated independently of Bni1-Glc7 at the bud neck during mitosis (DeMarini et al., 1997; Gohlke et al., 2018; Kozubowski et al., 2003). Microscopy of GFP-Skt5 expressing yeast cells revealed that Skt5, similar to Fir1, contacted the septin ring first at the mother side before entering the space between the SSDR (Fig. 4 D). Inhibiting Smt3 modification of the septins in a siz1∆ strain abolished phase I targeting of Skt5 and decreased the amount of GFP-Skt5 between the SSDR during phase II targeting (Fig. 4, C and G; and Fig. S4 C). The deletion of SPA2 did not influence the targeting of Skt5 to the septin hourglass but substantially reduced its amount between the SSDR (Fig. 4, C and G; and Fig. S4 C). These experiments confirm Fir1 as the mitosis-specific bud neck receptor for Skt5.
Fir1-Skt5 targets Chs3 to the SSDR and stimulates chitin synthesis during cytokinesis
Skt5 activates Chs3 at the bud neck of dividing cells (DeMarini et al., 1997; Kozubowski et al., 2003). The bud neck appearance of Chs3 coincides with the phase II targeting of the Fir1-Skt5 complex (Fig. 4 E; Okada et al., 2020). By comparing the fluorescence intensity profiles between wild type and fir1∆ cells, we could show that fir1∆ cells enriched significantly less Chs3-GFP at the bud neck than wild-type cells (Fig. 4, E and G; and Fig. S4 D). The deletion of Skt5 reduced the Chs3-GFP signal to a similar extent (Fig. 4, E and G; and Fig. S4 D). Chs3-GFP also stayed less focused within the space between the SSDR of fir1∆ and skt5∆ cells (Fig. 4 G). Deleting SKT5 and FIR1 in the same cell (fir1∆ skt5∆) reduced the amount of CHs3-GFP at the bud neck to a similar extent as each single deletion (Fig. 4, F and G; and Fig. S4 E). We conclude that a complex containing Fir1 and Skt5 supports the targeting and/or anchorage of Chs3 during cellular abscission (Reyes et al., 2007).
A deletion of FIR1 should consequently reduce the chitin levels in the secondary septum. Indeed, when we compared wild-type cells with fir1∆ cells, we observed a tendency for fir1∆ cells to produce less chitin in the septum. The effect was, however, not significant (Fig. 4 H). We reasoned that the chitin-rich primary septum might overshadow the influence of Fir1 on the secondary septum. To address this presumption, we compared the calcofluor staining of wild-type cells with cells lacking CHS2 and cells lacking both CHS2 and FIR1. chs2∆ cells displayed an increase in chitin deposition at the bud neck when compared with wild-type cells, whereas the deletion of Fir1 in chs2∆ cells (fir1∆chs2∆) resulted in a threefold reduction in chitin content when compared with chs2∆ cells (Fig. 4 H). We conclude that the Fir1-Skt5 complex stimulates the synthesis of chitin in the secondary septum of dividing cells.
Fir1 contains a separate binding site for Skt5
Skt5 is attached to the plasma membrane by a C-terminally coupled farnesyl moiety (Grabińska et al., 2007). Our findings suggest that Fir1 first links Skt5 to the septin hourglass before the complex relocates between the SSDR. This model implies that the interface of Fir1 for Skt5 should not overlap with those for Smt3 and Spa2. Indeed, Split-Ub interaction analysis of fragments of Fir1 localized the binding site for Skt5 to the supposedly disordered N-terminal 410 residues of the protein (Fig. 5 A and Fig. 1 A; Jumper et al., 2021). A GST fusion to the N-terminal region of Fir1 (GST-Fir11–410) precipitated the bacterial-expressed His-Skt5 and a fragment of Skt5 that harbored its central Sel1-like protein (SEL) domain (His-Skt5146–550; Fig. 5 B and Fig. S5). The Fir1-Skt5 complex could also be reconstituted by incubating nitrocellulose membrane-tethered GST-Fir11–410 with E. coli extracts containing His-Skt5146–550 (Fig. 5 C). We conclude that the interfaces of the Fir1-Skt5 complex are provided by the central SEL domain of Skt5 and the N-terminal half of Fir1.
Skt5 and Fir1 form a protein complex. (A) Split-Ub assay as in Fig. 3 A but with cells coexpressing Fir1CRU (left panel) or Fir11–410CRU (right panel) with the indicated Nub fusions of binding partners of Fir1, or Nub-Guk1 as a negative control. (B) Extracts of E. coli expressing 6xHis-Skt5 (lanes 1, 2, 5, 6), or 6xHis-Skt5146–550 (lanes 3, 4, 7, 8) were incubated with GST-Fir11–410– (lane 1, 3, 5, 7), or GST-coupled beads (lanes 2, 4, 6, 8). Glutathione eluates were separated by SDS-PAGE and stained with Coomassie (lanes 1–4), or transferred onto nitrocellulose and stained with anti-His antibody (lanes 5–8). Input fractions are shown in Fig. S5. (C) Extracts of E. coli cells expressing GST-Fir11–410 (lanes 2, 4, 6, 8), GST (lanes1, 3, 5, 7), or 6xHis-Skt5146–550 (lane 9) were separated by SDS-PAGE, transferred on nitrocellulose and either probed directly with anti-GST antibody (lanes 1–4), or anti-His antibody (lane 9), or incubated first with diluted extracts of E. coli cells expressing no additional protein (lanes 5, 6), or 6xHis-Skt5146–550 (lanes 7, 8), before being incubated with anti-His antibody (lanes 5–8). Source data are available for this figure: SourceData F2.
Skt5 and Fir1 form a protein complex. (A) Split-Ub assay as in Fig. 3 A but with cells coexpressing Fir1CRU (left panel) or Fir11–410CRU (right panel) with the indicated Nub fusions of binding partners of Fir1, or Nub-Guk1 as a negative control. (B) Extracts of E. coli expressing 6xHis-Skt5 (lanes 1, 2, 5, 6), or 6xHis-Skt5146–550 (lanes 3, 4, 7, 8) were incubated with GST-Fir11–410– (lane 1, 3, 5, 7), or GST-coupled beads (lanes 2, 4, 6, 8). Glutathione eluates were separated by SDS-PAGE and stained with Coomassie (lanes 1–4), or transferred onto nitrocellulose and stained with anti-His antibody (lanes 5–8). Input fractions are shown in Fig. S5. (C) Extracts of E. coli cells expressing GST-Fir11–410 (lanes 2, 4, 6, 8), GST (lanes1, 3, 5, 7), or 6xHis-Skt5146–550 (lane 9) were separated by SDS-PAGE, transferred on nitrocellulose and either probed directly with anti-GST antibody (lanes 1–4), or anti-His antibody (lane 9), or incubated first with diluted extracts of E. coli cells expressing no additional protein (lanes 5, 6), or 6xHis-Skt5146–550 (lanes 7, 8), before being incubated with anti-His antibody (lanes 5–8). Source data are available for this figure: SourceData F2.
Inputs forFig. 5 B .E. coli extracts expressing GST-Fir11–410 (lanes 1, 5, 9), GST (lanes 2, 6, 10), 6xHis-Skt5 (lanes 3, 7, 11), or 6xHis-Skt5146–550 (lanes 4, 8, 12) were separated by SDS-PAGE and either stained with Coomassie (lanes 1–4), or transferred to nitrocellulose and stained with anti-GST (lanes 5–8), or anti-His antibodies (lanes 9–12). Source data are available for this figure: SourceData FS5.
Inputs forFig. 5 B .E. coli extracts expressing GST-Fir11–410 (lanes 1, 5, 9), GST (lanes 2, 6, 10), 6xHis-Skt5 (lanes 3, 7, 11), or 6xHis-Skt5146–550 (lanes 4, 8, 12) were separated by SDS-PAGE and either stained with Coomassie (lanes 1–4), or transferred to nitrocellulose and stained with anti-GST (lanes 5–8), or anti-His antibodies (lanes 9–12). Source data are available for this figure: SourceData FS5.
Degradation of Fir1 resets the targeting of Skt5
Fir1 is a substrate of the Cdh1-activated ubiquitin ligase APC and becomes degraded at the end of cytokinesis (Ostapenko et al., 2012). Accordingly, deletion of Cdh1 raised the cytosolic levels of Fir1 and led to prominent tip staining of Fir1-GFP during bud growth (Fig. 6, A–C). Besides a strong cytosolic signal, the fluorescence intensity profile of Fir1-GFP in cdh1∆ cells showed phase I targeting and a stronger and longer enrichment between the SSDR (Fig. 6, A–C; Fig. S6 A). Deviating from Fir1, GFP-Skt5 lacked phase I targeting in cdh1∆ cells and showed a reduced enrichment between the SSDR (Fig. 6, B and D; and Fig. S6 B). We conclude that the degradation of Fir1 at the end of cytokinesis is required to properly reset the mechanism for Skt5 enrichment in the next cell cycle.
Fir1 degradation resets the bud neck targeting of Skt5. (A) As in Fig. 1 C but with cells coexpressing Fir1-GFP together with Shs1-Cherry in the presence (n = 24) or absence of CDH1 (n = 25). The fluorescence intensities are shown as a ratio to extracellular background (n/e; see Materials and methods). (B) Stills of the bud necks of cells corresponding to the intensity profiles shown in A and D. (C) Fir1-GFP fluorescence intensity profiles (arbitrary fluorescent units) along the polarity axis in a wild type (left panels) and a cdh1∆ cell (right panels). Shown from top to bottom: S phase, cytokinesis, G1 phase. Numbers and arrows correlate positions on the fluorescence intensity profiles with locations in the cells. (D) As in A but with cells coexpressing GFP-Skt5 and Shs1-Cherry in the presence (n = 23) or absence (n = 25) of CDH1. The orange-filled circle indicates splitting of the septin ring in A and D. The significance of the differences between the profiles A and D are shown in Fig. S6, A and B. Images in A, B, and D were taken every 2 min. Error bars, SD. Scale bars = 3 µm. (E) A model of how the Fir1-Skt5 complex might transfer into the SSDR. From left to right: Sumoylation at the mother side of the intact septin hourglass concentrates the membrane-bound Fir1-Skt5 complex at the edge of the hourglass. The transformation into the split septin rings reduces the number and length of the septin filaments and switches the direction of the remaining filaments perpendicular to the mother–daughter axis (only shown for the mother side). This process creates temporal holes within the septin grid (only shown for the mother side). Fir1-Skt5 diffuses through the temporal holes of the septin grid along the unoccupied Smt3 sites. The Spa2 trap within the split septin rings and the gradual desumoylation direct the diffusion of the Fir1-Skt5 complex into the space between the SSDR. The holes are filled by repolymerization and Chs3 is captured and activated by the Skt5-Fir1-Spa2 complex.
Fir1 degradation resets the bud neck targeting of Skt5. (A) As in Fig. 1 C but with cells coexpressing Fir1-GFP together with Shs1-Cherry in the presence (n = 24) or absence of CDH1 (n = 25). The fluorescence intensities are shown as a ratio to extracellular background (n/e; see Materials and methods). (B) Stills of the bud necks of cells corresponding to the intensity profiles shown in A and D. (C) Fir1-GFP fluorescence intensity profiles (arbitrary fluorescent units) along the polarity axis in a wild type (left panels) and a cdh1∆ cell (right panels). Shown from top to bottom: S phase, cytokinesis, G1 phase. Numbers and arrows correlate positions on the fluorescence intensity profiles with locations in the cells. (D) As in A but with cells coexpressing GFP-Skt5 and Shs1-Cherry in the presence (n = 23) or absence (n = 25) of CDH1. The orange-filled circle indicates splitting of the septin ring in A and D. The significance of the differences between the profiles A and D are shown in Fig. S6, A and B. Images in A, B, and D were taken every 2 min. Error bars, SD. Scale bars = 3 µm. (E) A model of how the Fir1-Skt5 complex might transfer into the SSDR. From left to right: Sumoylation at the mother side of the intact septin hourglass concentrates the membrane-bound Fir1-Skt5 complex at the edge of the hourglass. The transformation into the split septin rings reduces the number and length of the septin filaments and switches the direction of the remaining filaments perpendicular to the mother–daughter axis (only shown for the mother side). This process creates temporal holes within the septin grid (only shown for the mother side). Fir1-Skt5 diffuses through the temporal holes of the septin grid along the unoccupied Smt3 sites. The Spa2 trap within the split septin rings and the gradual desumoylation direct the diffusion of the Fir1-Skt5 complex into the space between the SSDR. The holes are filled by repolymerization and Chs3 is captured and activated by the Skt5-Fir1-Spa2 complex.
Cdh1 deltetion significantly affects Fir1 and Skt5 localization. (A) Statistical evaluation of differences in signal intensities between Fir1-GFP in the presence and absence of CDH1 at time point 22 of Fig. 6 A. ****P < 0.0001. (B) Statistical evaluation of the differences in signal intensities between GFP-Skt5 in the presence and absence of CDH1 at time point 24 of Fig. 6 D. ****P < 0.0001.
Cdh1 deltetion significantly affects Fir1 and Skt5 localization. (A) Statistical evaluation of differences in signal intensities between Fir1-GFP in the presence and absence of CDH1 at time point 22 of Fig. 6 A. ****P < 0.0001. (B) Statistical evaluation of the differences in signal intensities between GFP-Skt5 in the presence and absence of CDH1 at time point 24 of Fig. 6 D. ****P < 0.0001.
Discussion
Fir1 is the first identified sumoylation-dependent binding partner of the septins. In line with the kinetics of their sumoylation, Fir1 is recruited to the septins shortly before the visible onset of cytokinesis and is released as soon as the transition of the hourglass to the SSDR occurs. The released Fir1 is retained at the bud neck by the perfectly scheduled appearance of its newly discovered binding partner Spa2. The chronology of the interactions moves Fir1 from the periphery of the septin hourglass to the center of the SSDR.
Intuitively, crossing the septin barrier should proceed in two steps: release from the desumoylated septins into the cytosol, followed by binding to the SSDR-located Spa2. However, Fir1’s binding partner Skt5 is attached to the membrane and might prevent the complex from freely entering the cytosol after septin desumoylation (Meissner et al., 2010; Larson et al., 2008; Reyes et al., 2007). Interestingly, GFP-Skt5 and Fir1-GFP have not been observed within endosomal compartments during cytokinesis. This observation, coupled with the finding that septin sumoylation enhances the formation of the Fir1-Skt5 complex within the SSDR, leads us to propose that the Fir1-Skt5 complex traverses the septin barrier directly.
In the initial step of this novel and still hypothetical targeting mechanism, septin sumoylation recruits Skt5 to the periphery of the septin hourglass. As the hourglass transforms into a double ring, a brief opening in the septin gate allows the Fir1-Skt5 complexes to move along their concentration gradient via directed diffusion into the space between the SSDR. Gradual desumoylation, catalyzed by Ulp1 from the maternal side of the ring, prevents backward movement. Meanwhile, Spa2 effectively entraps the incoming Fir1-Skt5 complexes within the double ring (Fig. 6 E).
It is important to note that our hypothesis lacks an independent experimental validation for its central assumption that the septins become more permeable to membrane-associated proteins during the hourglass-SSDR transition (Dobbelaere and Barral, 2004; Wloka et al., 2011). However, it is known from different experimental approaches that the majority of septin filaments disappear during this transition. The remaining septin units exhibit increased mobility and temporarily lose their uniform orientation (DeMay et al., 2011; Dobbelaere et al., 2003; Ong et al., 2014). One of the proposed models to explain this behavior posits the depolymerization of paired filaments, followed by their repolymerization into circumferential filaments (DeMay et al., 2011; Ewers, 2011). In this manner, the transiently dynamic state of the septins might indeed facilitate the movement of Fir1-Skt5 complexes across the forming ring.
Such a permeable septin gate should already allow a significant fraction of Skt5 to passively diffuse into the SSDR without Fir1. Incoming interaction partners like Chs3 or Hof1 might then trap the protein between the SSDR (Oh et al., 2017; DeMarini et al., 1997; Reyes et al., 2007). In agreement with this prediction, we observed that the loss of sumoylated septins, or the deletion of either Fir1 or Spa2, did not completely prevent Skt5 from accumulating between the SSDR (Fig. 4 C). This Fir1-independent enrichment at the bud neck could also explain why Fir1-, Fir1∆SIM-, or Fir1∆801–820-GFP, all retaining their Skt5 binding site, still show some residual accumulation between the SSDR of spa2∆ cells, whereas Fir1750–876-GFP does not (Figs. 2 and 3).
We observed that the absence of sumoylation seems to affect the accumulation of Fir1 between the SSDR more severely than the loss of the Smt3-binding site in Fir1∆SIM (Fig. 1, C and E). This finding might point to an additional role for septin sumoylation on the state of the septins during the hourglass-ring transitions or its properties as a diffusion barrier. These features might facilitate the transfer of Fir1 independently of its SIM and might correspond with findings in mammalian cells, where sumoylation was shown to alter the bundling properties of the septins during cytokinesis (Ribet et al., 2017).
Like other proteins involved in cytokinesis, Fir1 undergoes degradation by the Cdh1-activated APC at the conclusion of the cell cycle (Ostapenko et al., 2012; Tully et al., 2009). Disruption of this degradation results in the accumulation of Fir1 in the cytosol, yet does not affect its targeting during phases I and II. Surprisingly, in cdh1∆ cells, Skt5 was no longer recruited during phase I targeting. This surplus of Fir1 may potentially compete with the Fir1-Skt5 complex for the limited pool of sumoylated septins. Consequently, less Skt5 is recruited to the septins and subsequently enriched between the SSDR. However, alternative explanations remain plausible. This includes the potential presence of an inhibitor that disrupts the formation of the Fir1-Skt5 complex during the final stages of cytokinesis, which subsequently undergoes degradation by Cdh1 in the ensuing G1 phase.
The absence of Fir1 leads to a reduction in chitin content within the secondary septum. This decline in chitin content closely corresponds to the diminished presence of Skt5 and Chs3 at the bud neck of cells lacking Fir1. Consequently, our research has identified Fir1 as the bud neck receptor for Skt5, the pivotal activator of Chs3. Our finding suggests that the unique targeting of Fir1-Skt5 to the bud neck may be a result of the need to prevent premature activation of Chs3 during its conventional transport to the plasma membrane.
Fir1 has recently emerged as a central player in a novel checkpoint mechanism, ensuring that cell separation proceeds only after the successful completion of preceding steps (Brace et al., 2019). This checkpoint can be artificially activated by the deletion of Chs2 or proteins like Cyk3 or Inn1, which promote chitin synthesis in the primary septum (Brace et al., 2019; Foltman et al., 2016; Devrekanli et al., 2012). To compensate for reduced primary septum formation, there is an increased production of Chs3 and Fks1 at the cell division plane (Okada et al., 2020). The deletion of Fir1 exacerbates the growth impairment of these cells and results in a significantly thinner secondary septum (Brace et al., 2019). According to Brace et al., this deletion of Fir1 appears to trigger premature secretion of the Chitinase Cts1, consequently reducing chitin levels in the primary septum.
Our study introduces a novel role for Fir1 by demonstrating that, as part of the Fir1-Skt5 complex, it also stimulates chitin synthesis during cytokinesis. Thus, Fir1 appears to respond to primary septum synthesis defects through two distinct mechanisms: by inhibiting the degradation of the primary septum and, concurrently, promoting chitin synthesis for the secondary septum.
Materials and methods
Growth conditions, cultivation of yeast strains, and genetic methods
All yeast strains were derivatives of JD47, a segregant from a cross of the strains YPH500 and BBY45, and are listed in Table S2 (Dohmen et al., 1995). Yeast strains were cultivated in synthetic defined (SD) or yeast extract peptone dextrose (YPD) media at the indicated temperatures. Media preparation followed standard protocols (Glomb et al., 2020). SD medium for Split-Ub assays contained in addition 1 mg/ml 5-fluoro-orotic acid (5-FOA; Formedium). Gene deletions and promoter replacements by PMET17 were performed by homologous integration of the cassettes derived by PCR from the plasmids pFA6a-hphNT1, pFA6a- natNT2, pFA6a-kanMX6, pFA6a-CmLEU2, or pYM-N35 (Bähler et al., 1998; Janke et al., 2004). E. coli XL1 blue cells were used for plasmid amplification and grown at 37°C in lysogeny broth (LB) medium containing antibiotics. E. coli BL21 cells were used for protein production and were grown in LB or super broth (SB) medium at 37°C.
Generation of plasmids and yeast strains
Detailed lists of all plasmids used in this study are provided in Table S3. Genomic gene fusions were obtained as described (Wittke et al., 1999; Neller et al., 2015; Dünkler et al., 2012). In brief, fusions of GFP, mCHERRY, or CRU to SPA2, MSB1, CBK1, HOF1, BUD14, CHS3, BOI1, SHS1, MYO1, and in some cases to FIR1, were constructed by PCR amplification of the respective C-terminal ORFs without stop codon from genomic DNA. The obtained DNA fragments were cloned via EagI and SalI restriction sites in front of the CRU, GFP, mCherry module on a pRS303, pRS304, or pRS306 vector (Wittke et al., 1999). The plasmids were linearized using a single restriction site within the C-terminal genomic DNA sequence and transformed into yeast. Successful integration was verified by PCR of single yeast colonies with diagnostic primer combinations using a forward primer annealing in the target ORF but upstream of the linearization site, and a reverse primer annealing in the C-terminal module. Most Fir1 fusions were created by introducing a PCR fragment carrying GFP and an TRP1 selection cassette flanked by 45 bp homologous to the 3′-end of the FIR1 ORF and the 5′-end of its 3′ non-translated region, respectively. GFP-SKT5 was generated via CRISPR/Cas9-mediated insertion of a PCR fragment encoding GFP flanked by 45 bp homologous to the region upstream of the start codon of SKT5 and the 5′-end of its ORF. Deletions and base exchanges in the genomic copies of FIR1 and SPA2 were achieved by CRISPR/Cas9 manipulation using plasmid pML104 or pML107 containing specific 20mer guide RNA sequences, and template oligonucleotides for exchanging the information on the genomic DNA (Table S4; Laughery et al., 2015). The correct insertions of PCR fragments and mutations were verified by PCR amplification and sequencing. Gene deletions were obtained by replacing the ORF through single-step homologous recombination with an antibiotic resistance cassette derived by PCR from the plasmids pFA6a-hphNT1, pFA6a- natNT2, pFA6a-kanMX6, pFA6a-CmLEU2, or pYM-N35 (Bähler et al., 1998; Janke et al., 2004).
Fragments of FIR1, SPA2, or SKT5 were expressed as GST or 6xHis fusions in E. coli strains BL21 or BL21 Gold. Fragments for the GST fusions were amplified from yeast genomic DNA using primers containing BamHI/EcoRI restriction sites. The PCR products were fused in-frame behind GST on a pGex2T vector (GE Healthcare). 6xHis-tagged fragments were amplified from yeast genomic DNA using primers containing SfiI restriction sites. The products were inserted into the pES plasmid, downstream and in-frame of a 6xHis-tag.
In vivo Split-Ub interaction analysis
Large-scale Split-Ub assays were performed as described (Dünkler et al., 2012). A library of 540 different α-strains, each expressing a different Nub fusion, was mated with a PMET17FIR1-CRU–expressing a-strain. Diploids were transferred as independent quadruplets on SD media lacking methionine and containing 1 mg/ml 5-FOA, and different concentrations of copper sulfate to adjust the expression of the Nub fusions. For small-scale interaction analysis, a- and α-strains expressing Nub or Cub fusion constructs were mated. The diploid cells were spotted onto SD-FOA medium in four 10-fold serial dilutions starting from OD600 = 1. Growth was recorded at 30°C every day for 2–5 d.
In vitro binding assays
E. coli extracts
An overnight culture of Hisx6-Spa21,314-end– or GST-Fir11–410–expressing E. coli BL21 Gold (GE Healthcare) was diluted into 500 ml of SB medium and incubated at 37°C to an OD600 of 0.8. The cultures were chilled to 18°C and incubated overnight at 18°C in the presence of 1 mM IPTG. Cells were centrifuged at 5,000 × g for 10 min, resuspended in PBS, and centrifuged at 4,000 × g for 10 min. The cell pellet was stored at −80°C. BL21 strains expressing GST-Fir1741–876, 6xHis-SKT5, or 6xHis6-Skt5146–550 were treated as above except that protein expression occurred in SB/1 mM IPTG for 4 h at 37°C.
The cell pellets were washed once in PBS and resuspended in 5 ml PBS containing protease inhibitor cocktail (Roche Diagnostics) and 1 mg/ml lysozyme. After 45 min on ice, cells were lysed by 3 × 2 min sonication with a Bandelin Sonapuls HD 2070 (Reichmann Industrieservice). Extracts were clarified by centrifugation at 40,000 g for 10 min at 4°C. 6xHis-Spa21,314-end–expressing strain BL21 was lysed in IMAC Buffer A (50 mM KH2PO4, pH 8.0, 300 mM NaCl, 20 mM imidazole) by lysozyme treatment and sonication, followed by IMAC purification on an Äkta purifier chromatography system using a 5 ml HisTrap Excel column (GE Healthcare). The column was washed by a linear imidazole gradient (20–70 mM), followed by elution in 200 mM imidazole. Eluted proteins were subsequently transferred on a PD10 column into PBS and concentrated. Enriched 6xHis-Spa21,314-end was finally loaded onto a gel-filtration column (HiLoad 16/600 Superdex 200 pg) equilibrated in 20 mM 2-(N-Morpholino)ethansulfonsäure, 100 mM NaCl, pH 6.7. Peak fractions were pooled and used for the experiments.
Binding assay
All incubation steps were carried out under rotation at 4°C. GST or GST-tagged proteins were immobilized from E. coli extracts on 100 μl Glutathione–Sepharose beads in PBS (GE Healthcare). After incubation for 1 h at 4°C, with either E. coli extracts or purified proteins, the beads were washed three times, the bound material was eluted with GST elution buffer (50 mM Tris, 20 mM reduced glutathione), and subjected to SDS-PAGE followed by Coomassie Blue staining and western blot analysis using anti-His (H1029), or anti-GST (G1160) antibodies (Sigma-Aldrich).
Far-western blot
Enriched GST and GST-Fir11–410 were transferred onto nitrocellulose after SDS-PAGE. The membrane was blocked for 1 h in 20mM Tris, 150 NaCl, 0.1% Tween 20 (TBST) 2% milk powder (wt/vol), washed for 5 min in 10 ml TBST, and incubated for 1 h at 4°C with extracts (20-fold diluted in TBST) from E. coli expressing no additional protein or 6xHis-Skt5146–550. After washing three times for 5 min with 10 ml TBST, the membrane was sequentially incubated at room temperature for 1 h with anti-His (H1029) and conjugated anti-mouse antibodies (A4416; Sigma-Aldrich; Wu et al., 2007).
Fluorescence microscopy
Microscopic observations were performed with two different fluorescence microscopes. The Axio Observer spinning disc confocal microscope (Zeiss) is equipped with an Evolve512 electron-multiplying charge-coupled device camera (Photometrics), a Plan-Apochromat 63×/1.4 oil differential interference contrast (DIC) objective, and a 100×/1.4 oil DIC objective. Fluorescence was excited with 488- and 561-nm diode lasers (Zeiss) and detected with high-efficiency filter sets 38 (GFP) and 45 (Cherry), respectively. Time-lapse experiments were carried out at 30°C in a PeCon Incubator controlled by a PeCon TempModule S1. Operations were performed with the ZEN2.6 (2012) software package (Zeiss).
The DeltaVision system (GE Healthcare) is provided with an IX71 microscope (Olympus), a 100× UPlanSApo 100 × 1.4 oil ∞/0.17/FN26.5 objective (Olympus), a steady-state heating chamber (Weather station by Prescision Control), and a Photofluor LM-75 halogen lamp. The standard live cell filter set (eGFP: excitation 470/40, emission 525/5; dsRed: excitation 580/20, emission 630/60) was used for fluorescence microscopy and images were recorded with either the Cascade II 512 electron-multiplying charge-coupled device camera (Photometrics) or the CoolSNAP HQ2 High-Speed CCD camera (Photometrics). All operations were performed with the SoftWoRx 6.1.3. software (GE Healthcare).
For all microscopic analyses, yeast cultures were grown overnight in SD medium, diluted in 3–4 ml fresh SD medium, and grown for 2–3 h at 30°C to mid-log phase. Exposure times were adapted to the respective GFP- and mCherry-labeled proteins to reduce bleaching and phototoxicity.
Standard time-lapse experiments with the ZEISS microscope were carried out with the 63× objective in a Sarstedt 1-well on cover glass II incubation chamber. 100 μl culture was pipetted onto the glass bottom of the chamber and covered with a slice of standard solid SD medium. Images were taken in intervals of 2 min and obtained with a series of seven z-slices and a distance of 0.5 µm between adjacent z-slices.
Standard time-lapse experiments at the DeltaVision microscope were performed with the 100× objective and the Cascade II 512 camera. 1 ml mid-log phase cell culture was carefully pelleted and resuspended in 50 μl medium. 3 μl of this suspension was transferred on custom-designed glass slides containing solid SD medium with 1.8% agarose and immobilized with a coverslip. Images were taken in intervals of 2 min and obtained with a series of seven z-slices and a distance of 0.5 µm between adjacent z-slices.
Analysis of microscopic data
Image analysis and signal quantifications were carried out with the open source software platforms Image J for the data obtained with the DeltaVision microscope or Fiji for the data obtained with the ZEISS microscope (Schneider et al., 2012; Schindelin et al., 2012).
Quantitative evaluation of time-lapse experiments
For the quantification of bud neck signals in time-lapse series of cells undergoing cytokinesis, we generally employed yeast strains expressing two different fusion proteins. The protein of interest was translationally fused to GFP while mCherry fusions of either Shs1 or Myo1 served as marker proteins for the onset of cytokinesis. Sum projections of all relevant Z planes in both channels served as basis for all measurements. If not noted differently, circular regions of interest (ROI) of exactly 8 pixels in diameter were selected covering the bud neck region (1, bud neck signal), a region without obvious signal within either the daughter or mother cytosol (2, cytosolic signal), and an area outside the cell (3, background). The mean gray value of ROI 1–3 in both channels at 19 time points covering the period of cytokinesis was measured. Data analysis was carried out with Microsoft Excel (Microsoft Excel for Mac, version 16.66.1) and GraphPad Prism (Prism 9 for MacOS, version 9.0.2). The background (3) was subtracted from values (1) and (2) in both channels at every time point and the resultant values (s1) were divided by the cytosolic readings (s2) to obtain the ratio of bud neck to cytosol signal for both channels at each time point (n/c ratio, neck-to-cytosol ratio). The Shs1-Cherry signal drops dramatically when the septin ring splits (defined as minute 18 in this study) while Myo1-Cherry contracts and disappears rapidly at minute 22. To facilitate temporal alignment of the readings originating from different cells, each individual time series was normalized, meaning that the lowest value within each individual series was defined as 0% and the maximum value as 100% (normalized [norm] n/c ratio). The norm n/c ratio values obtained for the Cherry channel were plotted against time and utilized for a visual temporal alignment (facilitated by the loss/drop of the Myo1-/Shs1-Cherry signal) of the individual curves of this experiment. For each experiment, the n/c ratio as well as the norm n/c ratio series were aligned accordingly, and the averages and standard deviations of each time point in the GFP as well as the Cherry channel were determined. Normalization reduces the quantitative deviation between individual curves, facilitates alignment, and is useful for illustrating the relative timing of signals of comparable intensities. In most cases, we preferred to show the n/c ratio as this data set provides additional information on relative signal strength. We are aware that absolute fluorescence signal intensities vary considerably between experiments. The shown quantitative differences could be reproduced repeatedly and independently and represent the visual impression of the pictures very accurately.
Statistical evaluation
For all experiments where the curves of collated protein variants or genotypes displayed reproducibly deviating n/c ratio values, the statistical significance of the difference at selected time points was determined in GraphPad Prism. Kruskal–Wallis ANOVA tests were used for multiple comparisons, whereas Mann–Whitney tests were employed for the comparison of two data sets.
Images time series
For the illustration of bud neck signals, representative cells of the standard time-lapse experiments were chosen and max Z-projections were created in ImageJ or Fiji. Brightness and contrast settings of the GFP and Cherry channels were manually adjusted.
Specifications of selected microscopic experiments
Fig. 1 D and Fig. S1 A: The pictures in Fig. 1 D were obtained with the Deltavision microscope utilizing the 100× objective and the CoolSNAP HQ2 camera. In a short time-lapse experiment, 20 Z-sections at 0.2-µm spacing were taken every 2 min. Images of the GFP and Cherry channels were deconvoluted with the softWoRx 6.1.3 software and subsequently 3D projected in ImageJ. Fig. 2 F: Images shown in Fig. 2 F originated from standard Deltavision time-lapse experiments. A max projection of the GFP channel was superimposed on single Z-sections of the bright field channel. Profiles were measured in single Z-sections of the same cells. A linear ROI with a width of 1 pixel was drawn centrally through budding cells from the mother to the daughter starting and ending outside the cells. The average of the extracellular values of the GFP channel was subtracted from each data point. The value for each data point was divided by the average of the extracellular values of the GFP channel. Fig. 4 D: The images of the bud neck in Fig. 4 D were obtained from a single cell at two different time points with the 100× objective at the Zeiss microscope. Overlays of sum projections of the GFP and Cherry channels are shown. The corresponding profiles were measured in single Z-sections of the same cell. A short linear ROI with a width of 1 pixel was drawn centrally through the bud neck from the mother to the daughter cell. The measurements of both channels were normalized. Fig. 4, E and F: Due to the patchy cytosolic distribution of Chs3-GFP, a circular selection of 13 pixels diameter in the mother cell was used as intracellular reference. Fig. 6 A: Due to the high level of cytosolic Fir1-GFP signal in cdh1∆ cells, the standard procedure for quantitative time-lapse analyses led to an underestimation of the bud neck signal. We thus decided to divide the measured bud neck signal by the reference value obtained outside the cell in wild type and cdh1∆ rather than a cytosolic reference point (n/e ratio, neck to extracellular ratio). Fig. 6 C: Images originate from standard Zeiss time-lapse experiments. A sum projection of the GFP channel was superimposed on a single Z-section of the bright field channel. Profiles were measured in single Z-sections of the same cells. A linear ROI with a width of 1 pixel was drawn centrally from the mother to the daughter starting and ending outside the cells. The value for each data point was divided by the average of the extracellular values of the GFP channel.
Quantitative calcofluor assay
500 μl of a logarithmically grown culture was centrifuged at 8,000 rpm for 3 min, resuspended in 100 μl of medium plus 1 μl of calcofluor White (20 μg/ml), and incubated at room temperature in the dark for 15 min. Cells were washed two times with 1× PBS and incubated for 15 min in the dark with 3.7% formaldehyde. Cells were again washed two times with 1× PBS, resuspended in 100 μl of PBS, and finally used for microscopy. Images were acquired on a Leica TCS SP8 confocal microscope equipped with 40× NA 1.3 oil objective. Fluorescence was excited with a 405-nm diode laser (Leica) and detected spectrally using a HyD detector for a linear signal response. To quantify the amount of chitin at the bud neck of dividing cells, the fluorescence signal was measured along a line (open-source software Fiji, freehand tool, line width 3) across the bud neck. Data analysis was carried out with Microsoft Excel (Microsoft Excel for Mac, version 16.66.1) and GraphPad Prism (Prism 9 for MacOS, version 9.0.2). The statistical significance of the differences between the mean fluorescence intensities of the different strains was determined by one-way ANOVA tests.
Online supplemental material
Fig. S1 shows deconvoluted images of the bud neck of cells expressing Fir1-GFP and Fir1∆SIM-GFP, the complementation of siz1∆ cells, and the statistical evaluation of the fluorescence intensity profiles of Fig. 1 E. Fig. S2 shows the Split-Ub interaction array. Fig. S3 shows the fluorescence intensity profiles of Spa2-GFP in different genetic backgrounds, the complementation of spa2∆ cells, the statistical evaluation of the fluorescence intensity profiles of Fig. 2 D, and the far-western analysis of Fir1750–876 probed with Spa21,324-end. Fig. S4 shows the fluorescence intensity profiles of different bud neck localized proteins in the presence and absence of FIR1 and the statistical evaluation of the fluorescence intensity profiles of Figs. 4, B, C, E, and F. Fig. S5 shows the input fractions of Fig. 5 B. Fig. S6 shows the statistical evaluation of the fluorescence intensity profiles of Fig. 6, A and D. Table S1 lists the Split-Ub discovered interaction partners of Fir1. Tables S2, S3, and S4 list the yeast strains, constructs, and primers used and created in this study.
Data availability
All strains and plasmids are available upon request. Image files of the time-lapse analysis, values of the measured fluorescence intensities, and the calculated ratios are accessible under the DOI 10.5281/zenodo.8405825 through the open-access repository Zenodo.
Acknowledgments
We thank N. Schmid and S. Timmermann for technical assistance. We thank J. Dohmen (University of Cologne, Cologne, Germany) for yeast strains, plasmids, and advice. We thank Dr. Christian Bökel of the Core Facility for Confocal and Multiphoton Microscopy of Ulm University for his support.
The work was funded by grants from the Deutsche Forschungsgemeinschaft to N. Johnsson (Jo 187/8-1).
Author contributions: Conceptualization: N. Johnsson. Investigation: J. Müller, M. Furlan, B. Grupp, and D. Settele. Writing - original draft: N. Johnsson; Writing - review & editing: N. Johnsson, J. Müller, and M. Furlan. Supervision: N. Johnsson and J. Müller. Funding acquisition: N. Johnsson.
References
Author notes
Disclosures: The authors declare no competing interests exist.


![Fir1 is recruited to the bud neck by the sumoylated septins. (A) Cartoon of Fir1 indicating the known and herein discovered binding sites to other proteins. Amino acid positions bordering the motifs are indicated. (B) Cutouts of a Split-Ub array of diploid yeast cells expressing genomic Fir1CRU (CRU; C-terminal half of Ubiquitin [Cub]-R-Ura3; left panel), Fir1∆SIMCRU (right panel) together with the indicated Nub fusions of known binding partners of Fir1, or the negative control Nub-Guk1 (Nub, N-terminal half of Ubiquitin). Four independent matings were arrayed as quadruplets on media containing 5-FOA. Colony growth indicates interaction between the fusion proteins. (C) Fluorescence intensities (upper panel) and stills of the bud necks of representative cells (lower panel) coexpressing Fir1-GFP (n = 29) or Fir1∆SIM-GFP (n = 25) and Myo1-Cherry during cytokinesis. The ratio of bud neck fluorescence divided by cytosolic fluorescence (n/c ratio) is shown as mean ± standard deviation (SD). Images were taken every 2 min. The filled red circle (minute 22) indicates the time point of Myo1 disappearance. Phase I indicates association of Fir1 with the septin ring, phase II localization within the SSDR. (D) Deconvoluted images of Fir1-GFP associated with the Cherry-labeled septin ring (Shs1-Cherry) shortly before (left panel), during (middle panel), and shortly after ring splitting (right panel). m and d indicate mother and daughter cells, respectively. (E) As in C but with cells co-expressing Myo1-Cherry with Fir1-GFP in wild type (as reference from C) or siz1∆ cells (n = 22), or with cells coexpressing Shs1-Cherry and Fir1-GFP in wild type (n = 19, Fir1-GFP SEPSUMO) or in sep∆sumo cells (n = 23, Fir1-GFP sep∆SUMO). The orange-filled circle (time point 18) indicates the sudden decrease of the Shs1-Cherry signal. The significance of the differences between Fir1-GFP signal intensities of wild type and the deletion strains are shown in Fig. S1 C. Reintroducing a plasmid-encoded SIZ1 into the siz1∆ cells restored the targeting of Fir1-GFP (Fig. S1 B). Error bars, SD. All scale bars = 3 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/223/1/10.1083_jcb.202301027/1/m_jcb_202301027_fig1.png?Expires=1773761418&Signature=0uakOgkcuADZwG4a~vkTESkYQBSrhq~CG6EXgjTbb7trzDP8S9kvC0Q4jkszZsp9x-LkO8R5YEcs2~lsME9s5Yk~cfngtpsKwEiNudPsOC2PFoTHgF1dIsVy7XSc~p-h3XzbW1yBNlh6hEmf3jLI5ai0rxqK3pQa6FUCrro80QzjN10l1MUBLAcyvxibMV2zoit5-aUfN~AasGyY7mydFaeG8ggGin86BsFg~2I5FPKOKlgjS5AAMB2Ar7QxqELAcA-zN0ee9~9scKAa3FPMxI261OQTuWSDuqAzDfVbU~XaOWYbYNKmglXKMfdNWCLC6tDe3w~0Y5rcpM~Deiyl1w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)