Cilia are microtubule-based organelles that assemble via intraflagellar transport (IFT) and function as signaling hubs on eukaryotic cells. IFT relies on molecular motors and IFT complexes that mediate the contacts with ciliary cargo. To elucidate the architecture of the IFT-B complex, we reconstituted and purified the nonameric IFT-B core from Chlamydomonas reinhardtii and determined the crystal structures of C. reinhardtii IFT70/52 and Tetrahymena IFT52/46 subcomplexes. The 2.5-Å resolution IFT70/52 structure shows that IFT52330–370 is buried deeply within the IFT70 tetratricopeptide repeat superhelix. Furthermore, the polycystic kidney disease protein IFT88 binds IFT52281–329 in a complex that interacts directly with IFT70/IFT52330–381 in trans. The structure of IFT52C/IFT46C was solved at 2.3 Å resolution, and we show that it is essential for IFT-B core integrity by mediating interaction between IFT88/70/52/46 and IFT81/74/27/25/22 subcomplexes. Consistent with this, overexpression of mammalian IFT52C in MDCK cells is dominant-negative and causes IFT protein mislocalization and disrupted ciliogenesis. These data further rationalize several ciliogenesis phenotypes of IFT mutant strains.
Introduction
Cilia are hair-like projections found on the surface of most eukaryotic cells, where they function in cell motility, signaling, and sensory reception (Ishikawa and Marshall, 2011). The architecture of cilia is that of a microtubule-based axoneme surrounded by the ciliary membrane, which is continuous with the plasma membrane (Mizuno et al., 2012). Cilium formation relies on intraflagellar transport (IFT) in almost all ciliated organisms (Rosenbaum and Witman, 2002). IFT is mediated by the molecular motors kinesin 2 (Cole et al., 1992, 1993; Walther et al., 1994) and dynein 2/1b (Pazour et al., 1998; Porter et al., 1999; Perrone et al., 2003; Hou et al., 2004), and by the IFT particle (Piperno and Mead, 1997; Cole et al., 1998). The IFT particle is composed of ∼22 subunits that divide into a 6-subunit IFT-A complex and an ∼16-subunit IFT-B complex, and likely mediates the contacts to ciliary motor and cargo proteins (Taschner et al., 2012; Bhogaraju et al., 2013a). Sandwiched between the axoneme and the ciliary membrane, so-called “trains” of IFT particles move bidirectionally between the base and the tip of cilia to deliver building blocks required for growth and also, in some organisms, maintenance of this organelle (Kozminski et al., 1993, 1995; Pigino et al., 2009).
The IFT-B complex was demonstrated to consist of a 9-subunit salt-stable core (IFT88, -81, -74, -70, -52, -46, -27, -25, and -22) and several peripheral subunits (Lucker et al., 2005; Ou et al., 2005; Wang et al., 2009b; Fan et al., 2010; Taschner et al., 2011). IFT22 and IFT27 are small GTPases (Schafer et al., 2006; Adhiambo et al., 2009; Bhogaraju et al., 2011), and the other IFT-B core subunits are predicted to have several protein–protein interaction domains (Taschner et al., 2012). Although IFT22 and IFT27 are prime candidates for regulators of IFT, no GTPase-activating proteins have so far been identified for these small GTPases (Bhogaraju and Lorentzen, 2014). Genetic studies have demonstrated that IFT-B core components are required for ciliogenesis in a wide range of organisms spanning green alga (Chlamydomonas reinhardtii) to vertebrates (Taschner et al., 2012). IFT88 is of particular interest, as the Tg737 mouse model carrying a hypomorphic mutation in this gene is an established model system for autosomal recessive polycystic kidney disease (ARPKD; Moyer et al., 1994; Pazour et al., 2000), and loss-of-function mutations in IFT88 lead to early embryonic lethality caused by hedgehog signaling defects in mice (Huangfu and Anderson, 2005). Currently, knowledge about how proteins of the IFT-B core complex specifically select ciliary cargoes is limited to outer dynein arm transport by IFT46 (Hou et al., 2007; Ahmed et al., 2008) and tubulin transport by IFT81/74 (Bhogaraju et al., 2013b), as well as evidence for the transport of various motility-related factors by the IFT56/TTC26/DYF13 protein (Ishikawa et al., 2014).
Published structural studies of IFT complexes are limited to the electron tomographic reconstructions of IFT trains (Pigino et al., 2009), high-resolution crystal structures of the IFT27/25 subcomplex (Bhogaraju et al., 2011), and the N-terminal calponin homology domain of IFT81 (Bhogaraju et al., 2013b). In this study, we report the novel crystal structures of subcomplexes between IFT70/52 and IFT52/46, and map a number of previously uncharacterized interactions within the complex to arrive at a complete IFT-B core interaction map where the C-terminal domain of IFT52 is key to complex stability. Finally, we test the functional importance of this finding by overexpressing IFT52C in MDCK cells and demonstrate a very strong reduction in ciliogenesis and a loss of IFT88 localization to basal bodies. Taken together, our data provide novel biochemical and structural insights into IFT-B core architecture and assembly and allow for specific manipulation of IFT protein domains to affect in vivo function.
Results
IFT70 binds a conserved proline-rich sequence motif of IFT52
9 of the 22 IFT proteins identified to date form the IFT-B core complex (Lucker et al., 2005; Wang et al., 2009b; Fan et al., 2010). Previously published data showed that the middle region of IFT52 (residues 281–381), but not the N- or C-terminal domains, directly binds IFT70 (Taschner et al., 2011). IFT52281–381 does not contain any domains recognizable by bioinformatics (Taschner et al., 2012) but has a conserved proline-rich sequence motif (residues 330–360; see Fig. 1 A). To define the minimal IFT70-binding region on IFT52, GST-tagged IFT52281–329, IFT52330–360, or IFT52360–381 were used to solubilize and pull down full-length His-tagged IFT70 (Fig. 1 B). The results showed that IFT52330–360 containing the proline-rich region was sufficient to bind and solubilize IFT70. Most of the prolines in IFT52330–360 are conserved from C. reinhardtii to human, which suggests that the IFT70–IFT52 interaction is evolutionarily conserved. The IFT70/52330–360 complex was stable during size-exclusion chromatography (SEC), and we conclude that IFT52330–360 constitutes a minimal IFT70-binding region.
Crystal structure of IFT70/52. (A) Domain composition of the IFT52 protein showing an N-terminal “GIFT” domain (52N), a central region (52M), and a small C-terminal domain (52C). A sequence alignment of residues 330–370 from Chlamydomonas reinhardtii (Cr), Homo sapiens (Hs), Caenorhabditis elegans (Ce), and Drosophila melanogaster (Dm) reveals numerous conserved prolines in the central region. Asterisks, completely conserved; colons, highly conserved; periods, moderately conserved. (B) Pull-down of His-tagged IFT70 with six different GST-tagged IFT52 constructs coexpressed in E. coli demonstrates that residues 330–360 of IFT52 are sufficient to solubilize and pull down IFT70. (C) Two perpendicular views of the 2.5-Å resolution crystal structure of the IFT70/IFT52330–381 complex with α-helices shown as cylinders. IFT70 is shown in blue and IFT52 in red. The 15 TPRs of IFT70 are numbered. (D) Unbiased Fo-Fc electron density map in green at 2.5 σ visualizing the IFT52330–370 peptide (magenta). IFT70 helices are shown as blue cylinders and TPRs blocking the view of the IFT52 peptide have been omitted for better visualization of the IFT52 peptide.
Crystal structure of IFT70/52. (A) Domain composition of the IFT52 protein showing an N-terminal “GIFT” domain (52N), a central region (52M), and a small C-terminal domain (52C). A sequence alignment of residues 330–370 from Chlamydomonas reinhardtii (Cr), Homo sapiens (Hs), Caenorhabditis elegans (Ce), and Drosophila melanogaster (Dm) reveals numerous conserved prolines in the central region. Asterisks, completely conserved; colons, highly conserved; periods, moderately conserved. (B) Pull-down of His-tagged IFT70 with six different GST-tagged IFT52 constructs coexpressed in E. coli demonstrates that residues 330–360 of IFT52 are sufficient to solubilize and pull down IFT70. (C) Two perpendicular views of the 2.5-Å resolution crystal structure of the IFT70/IFT52330–381 complex with α-helices shown as cylinders. IFT70 is shown in blue and IFT52 in red. The 15 TPRs of IFT70 are numbered. (D) Unbiased Fo-Fc electron density map in green at 2.5 σ visualizing the IFT52330–370 peptide (magenta). IFT70 helices are shown as blue cylinders and TPRs blocking the view of the IFT52 peptide have been omitted for better visualization of the IFT52 peptide.
IFT70 adopts a tetratricopeptide repeat (TPR) superhelical structure that wraps around IFT52
The complex between IFT70 and the minimal IFT52330–360 binding region identified above did not yield diffracting crystals. However, an IFT52 construct with an additional 21 residues at the C terminus (IFT52330–381) in complex with IFT70 yielded well-diffracting crystals, and the 2.5-Å structure was determined using single-wavelength anomalous dispersion on a gold derivative (Table 1). The crystal structure reveals that IFT70 consists of 15 consecutive TPRs (named TPR 1–15; Fig. S1 A), which form 720° of superhelical structure (Fig. 1 C). Of the 15 TPRs, only 9 were predicted with high confidence from the primary structure (Taschner et al., 2012), as TPRs 4, 7–10, and 15 all have noncanonical amino acid substitutions in the TPR helices (Fig. S1 B). Several TPRs are connected by α-helices instead of short loops, and TPR15 is capped by an additional C-terminal α36 helix (Fig. 1 C). TPR15 connects to α36 via a β-hairpin loop that folds back on TPRs 9 and 10 to fix the conformation of the IFT70 superhelix. Structures of proteins with a wide variety of cellular functions display significant structural similarity to IFT70 including importin α (Conti et al., 1998), the O-linked GlcNAc sugar transferase (Jínek et al., 2004), and the anaphase promoting complex subunit APC6 (Wang et al., 2009a).
Data collection and refinement statistics
| Data | CrIFT70FL/52330–381 | CrIFT70FL/52330–381 | TtIFT52540–603/46236–347 | TtIFT52540–603/46236–347 |
| Native data | KAu(CN)2 soak | Native data | KAu(CN)2 soak | |
| Data collection | ||||
| Space group | P63 | P63 | P3221 | P3221 |
| Unit cell/Å | a = b = 143.2, c = 88.7 | a = b = 143.2, c = 88.7 | a = b = 84.3, c = 95.5 | a = b = 84.9, c = 95.2 |
| Resolution/Å | 50–2.5 (2.65–2.50) | 50–2.5 (2.65–2.50) | 40–2.3 (2.4–2.3) | 50–2.8 (2.9–2.8) |
| I/sigma (I) | 17.7 (1.3) | 8.0 (1.1) | 17.0 (0.6) | 20.2 (2.0) |
| R-meas | 0.095 (1.83) | 0.152 (1.16) | 0.072 (3.21) | 0.09 (0.95) |
| CC (1/2) | 0.999 (0.481) | 0.995 (0.385) | 1.0 (0.54) | 1.0 (0.90) |
| Completeness | 0.995 (0.972) | 0.993 (0.981) | 0.988 (0.929) | 0.982 (0.891) |
| Anomalous signal | None | 4.5 Å | None | 3.7 Å |
| Data refinement | ||||
| Resolution/Å | 50–2.5 (2.53–2.50) | 40–2.32 (2.46–2.3) | ||
| No of reflections (free test set) | 35,954 (1758) | 17,160 (878) | ||
| Rwork | 0.172 (0.273) | 0.218 (0.391) | ||
| Rfree | 0.236 (0.317) | 0.252 (0.453) | ||
| RMSD bonds | 0.008Å | 0.009Å | ||
| RMSD angles | 1.1° | 1.24° | ||
| Ramachandran plot | ||||
| Favored | 94.0% | 95.3% | ||
| Allowed | 5.4% | 2.9% | ||
| Outliers | 0.6% | 0.6% |
| Data | CrIFT70FL/52330–381 | CrIFT70FL/52330–381 | TtIFT52540–603/46236–347 | TtIFT52540–603/46236–347 |
| Native data | KAu(CN)2 soak | Native data | KAu(CN)2 soak | |
| Data collection | ||||
| Space group | P63 | P63 | P3221 | P3221 |
| Unit cell/Å | a = b = 143.2, c = 88.7 | a = b = 143.2, c = 88.7 | a = b = 84.3, c = 95.5 | a = b = 84.9, c = 95.2 |
| Resolution/Å | 50–2.5 (2.65–2.50) | 50–2.5 (2.65–2.50) | 40–2.3 (2.4–2.3) | 50–2.8 (2.9–2.8) |
| I/sigma (I) | 17.7 (1.3) | 8.0 (1.1) | 17.0 (0.6) | 20.2 (2.0) |
| R-meas | 0.095 (1.83) | 0.152 (1.16) | 0.072 (3.21) | 0.09 (0.95) |
| CC (1/2) | 0.999 (0.481) | 0.995 (0.385) | 1.0 (0.54) | 1.0 (0.90) |
| Completeness | 0.995 (0.972) | 0.993 (0.981) | 0.988 (0.929) | 0.982 (0.891) |
| Anomalous signal | None | 4.5 Å | None | 3.7 Å |
| Data refinement | ||||
| Resolution/Å | 50–2.5 (2.53–2.50) | 40–2.32 (2.46–2.3) | ||
| No of reflections (free test set) | 35,954 (1758) | 17,160 (878) | ||
| Rwork | 0.172 (0.273) | 0.218 (0.391) | ||
| Rfree | 0.236 (0.317) | 0.252 (0.453) | ||
| RMSD bonds | 0.008Å | 0.009Å | ||
| RMSD angles | 1.1° | 1.24° | ||
| Ramachandran plot | ||||
| Favored | 94.0% | 95.3% | ||
| Allowed | 5.4% | 2.9% | ||
| Outliers | 0.6% | 0.6% |
Numbers in parentheses are for the highest resolution shell.
In the IFT70/52330–381 crystal structure, residues 330–370 of IFT52 are visible in the electron density (Fig. 1 D), and adopt an extended conformation, with the exception of residues 364–369, which fold into a short helix. The structure thus reveals that the complete IFT70-binding region of IFT52 comprises residues 330–370. The interaction mode of the IFT70/52 complex is antiparallel such that the N-terminal part of IFT70 binds the C-terminal part of the IFT52330–370 construct. The IFT52 peptide is largely buried in the interior of the IFT70 superhelix (3,174 Å2 of buried surface in the interface) with the exception of residues 356–370, which are partially exposed to solvent (Fig. 2 A). The IFT70–IFT52 interaction is largely hydrophobic, and given the buried position of IFT52 within IFT70, the IFT52330–370 peptide can be considered to form part of the hydrophobic core of IFT70 required for proper folding. This notion is supported by the facts that CrIFT70, although expressed to a high level in Escherichia coli, is completely insoluble in the absence of IFT52 and that the purified IFT70/52 complex is stable in high-salt conditions (up to 2 M NaCl tested; unpublished data).
Interaction mode within the IFT70/52 complex. (A) A semitransparent surface representation of IFT70 (gray) shows that the IFT52 peptide (yellow sticks, blue surface rendering) is mostly occluded within the IFT70 superhelix. The boxed inset shows the contacts between IFT52354–363 and IFT70 with the interacting residues displayed as sticks. (B) Schematics showing the >100 individual interactions between IFT52330–370 and IFT70.
Interaction mode within the IFT70/52 complex. (A) A semitransparent surface representation of IFT70 (gray) shows that the IFT52 peptide (yellow sticks, blue surface rendering) is mostly occluded within the IFT70 superhelix. The boxed inset shows the contacts between IFT52354–363 and IFT70 with the interacting residues displayed as sticks. (B) Schematics showing the >100 individual interactions between IFT52330–370 and IFT70.
The IFT70 superhelix wraps two full turns around the IFT52 peptide, which makes extended contacts along the 15 TPRs of IFT70 (Figs. 1 C and 2 A). Of the >100 contacts between IFT70 and IFT52330–370, most are hydrophobic, with only three salt bridges observed (E361IFT52-R47IFT70, E354IFT52-K93IFT70, and R353IFT52-E144IFT70/E178IFT70; see Fig. 2 B). The numerous conserved proline residues in IFT52330–370 fit snugly into binding pockets often structured by aromatic residues of IFT70. This is exemplified by the tetra-proline sequence (354-PPPP-358) of IFT52 that binds at an aromatic pocket lined by six conserved tyrosine residues of IFT70 (Fig. 2 A, inset). The many conserved prolines of IFT52 furthermore serve important structural roles, as they induce kinks allowing the IFT52330–370 peptide to snake its way through the IFT70 protein (Fig. 2 A). Given the high degree of amino acid conservation of IFT52330–370 (Fig. 1 A) and the IFT52-interacting residues of IFT70 (Fig. S1 A), it is reasonable to assume that the mode of IFT70/52 complex formation described here is conserved across ciliated species.
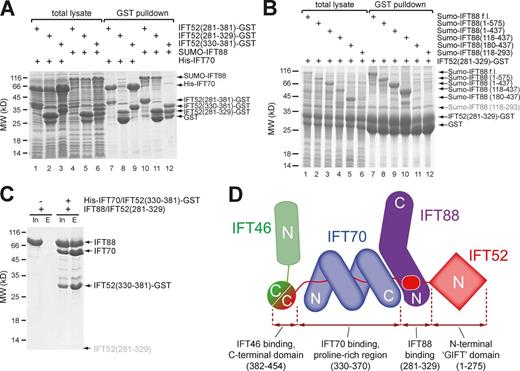
IFT88 binds IFT52281–329 in a complex that interacts directly with IFT70/IFT52330–381
IFT70 and IFT52 are found in a stable tetrameric subcomplex that also includes IFT88 and IFT46 (Taschner et al., 2011). IFT88 was previously shown to interact with the middle part of IFT52 (residues 281–381) and contains 11 TPRs predicted with high confidence (Taschner et al., 2012). Given that residues 330–370 of IFT52 are buried within the IFT70 structure, this region is unlikely to also interact with IFT88. Pull-downs confirm this notion, as GST-tagged IFT52281–329 but not IFT52330–381 interacts with IFT88 (Fig. 3 A). The reciprocal mapping of the IFT52-interacting region on IFT88 using six different constructs revealed that IFT88118–437 is sufficient for IFT52 binding (Fig. 3 B). These data suggest that the IFT88-binding site on IFT52 is located N-terminal to the IFT70 binding site. IFT88 would thus be located at the C-terminal side of the IFT70 superhelix (Fig. 3 D). Interestingly, a highly conserved surface patch is indeed found on the C-terminal part of the IFT70 structure (Fig. S2). To test this notion experimentally, GST-tagged IFT70/52330–381 was used to pull down untagged IFT88/52281–329, demonstrating an interaction in trans (Fig. 3 C) and suggesting the architecture depicted in Fig. 3 D. The question of whether IFT88, like IFT70, adopts the structure of a TPR superhelix that wraps around IFT52 awaits structural elucidation.
IFT88 binds at the C-terminal end of the IFT70 superhelix. (A) GST pull-down demonstrating that GST-IFT52330–381 specifically pulls down IFT70, whereas GST-IFT52281–329 specifically binds to IFT88. Both total bacterial lysate and elution after purification using GSH beads are shown. (B) The reciprocal mapping shows that IFT88118–437 is sufficient for binding to IFT52281–329. (C) The purified GST-tagged IFT70/IFT52330–381 complex pulls down the purified untagged IFT88/52281–329 complex, demonstrating an interaction in trans. The small IFT52281–329 peptide is not visible on the gel but was positively identified by mass spectrometry. (D) Interaction map summarizing the findings from A–C. Despite previously reported interaction between IFT46-IFT88 (Lucker et al., 2010) and IFT46-FT70 (Fan et al., 2010), we do not detect these (see Fig. S3). The IFT88/46 interaction is especially hard to reconcile with our data because the binding sites for these two proteins on IFT52 are separated by the IFT70 superhelix and would thus be ∼100 Å apart. In agreement with the Lucker et al. (2010) study, we do detect interaction of IFT46 and IFT88 in pull-down experiments, but further analysis of the obtained material reveals an aggregate rather than a soluble complex (Fig. S3 C).
IFT88 binds at the C-terminal end of the IFT70 superhelix. (A) GST pull-down demonstrating that GST-IFT52330–381 specifically pulls down IFT70, whereas GST-IFT52281–329 specifically binds to IFT88. Both total bacterial lysate and elution after purification using GSH beads are shown. (B) The reciprocal mapping shows that IFT88118–437 is sufficient for binding to IFT52281–329. (C) The purified GST-tagged IFT70/IFT52330–381 complex pulls down the purified untagged IFT88/52281–329 complex, demonstrating an interaction in trans. The small IFT52281–329 peptide is not visible on the gel but was positively identified by mass spectrometry. (D) Interaction map summarizing the findings from A–C. Despite previously reported interaction between IFT46-IFT88 (Lucker et al., 2010) and IFT46-FT70 (Fan et al., 2010), we do not detect these (see Fig. S3). The IFT88/46 interaction is especially hard to reconcile with our data because the binding sites for these two proteins on IFT52 are separated by the IFT70 superhelix and would thus be ∼100 Å apart. In agreement with the Lucker et al. (2010) study, we do detect interaction of IFT46 and IFT88 in pull-down experiments, but further analysis of the obtained material reveals an aggregate rather than a soluble complex (Fig. S3 C).
The C-terminal domains of IFT52 and IFT46 form a tightly packed heterodimer that constitutes the central bridge between IFT-B core subcomplexes
How do IFT81/74/27/25 and IFT88/70/52/46 subcomplexes associate to form a stable IFT-B core complex? To address this issue, we performed pull-downs and co-purifications with complexes where specific domains were deleted (Fig. 4, A and B; and Fig. S3). The results show that a heterodimeric complex of the C-terminal domains of C. reinhardtii IFT52 (IFT52366–454; IFT52C) and IFT46 (IFT46165–319; IFT46C) binds the IFT81ΔN/74ΔN/27/25ΔC tetramer to form a stable hexameric complex purified by SEC (Fig. 4 A). Consistently, we could reconstitute and purify a stable pentameric complex of Tetrahymena thermophila (Tt) IFT81ΔN/74ΔN/52C/46C/27 (Fig. S4; IFT25 is absent in Tt). As it has been previously shown that IFT52/46 does not bind directly to IFT27/25 (Taschner et al., 2011), we conclude that IFT52C/46C binds to the IFT81/74 subcomplex. Pull-downs with various complexes lacking the C terminus of IFT52 showed that this domain is essential for the interaction between IFT-B core subcomplexes (Fig. 4 C), but, interestingly, it is not sufficient, as GST-IFT52C only pulls down IFT81ΔN/74ΔN/27/25ΔC in the presence of IFT46 (Fig. 4 B). Similarly, GST-IFT46 alone is unable to pull down IFT81ΔN/74ΔN/27/25ΔC (Fig. 4 C, lanes 7–8). From these results, we conclude that both IFT52C and IFT46C are necessary for efficient binding to the IFT81/74/27/25 complex, which is in agreement with several reports using C. reinhardtii mutants, as outlined in the Discussion.
Structural and biochemical analysis of the IFT52C/46C complex. (A) SEC analysis showing that a dimeric IFT52C/46C complex can form a stable complex with IFT81ΔN/74ΔN/27/25ΔC. The IFT81ΔN/74ΔN/27/25ΔC tetramer was incubated with an excess of IFT52382–454/46165–319 (IFT52C/46C), and the resulting products were analyzed by SEC using a Superose 6 column. The chromatogram and the SDS-PAGE analysis of the peak fractions show that stoichiometric levels of the IFT52C/46C are shifted from the low-molecular weight peak 2 to the high molecular weight peak 1. (B) GST pull-down experiments showing that IFT52C is not sufficient for stable binding to IFT81ΔN/74ΔN/27/25ΔC, but that it requires IFT46. IFT81ΔN/74ΔN/27/25ΔC did not bind to GSH beads nonspecifically (lanes 1 and 2), and was not detectably pulled down by GST-IFT52382–454 (GST-IFT52C, lanes 3 and 4). To rule out that IFT52C was not able to bind at all to established interaction partners when GST-tagged, we tested binding to IFT46, which did not bind nonspecifically to the beads (lanes 5 and 6), but was efficiently pulled down by GST-IFT52C (lanes 7 and 8). A mixture of IFT81ΔN/74ΔN/27/25ΔC and IFT46 did not bind nonspecifically to the beads (lanes 9 and 10), but was efficiently pulled down by GST-IFT52C (lanes 11 and 12), showing that IFT52C requires IFT46 for stable binding to IFT81ΔN/74ΔN/27/25ΔC. (C) GST pull-down experiments showing that IFT52C is necessary for linking the IFT88/70/52/46 and IFT81ΔN/74ΔN/27/25ΔC subcomplexes. Whereas untagged IFT81ΔN/74ΔN/27/25ΔC complex did not bind nonspecifically to GSH beads (lanes 1 and 2), it was efficiently pulled down by GST-tagged full-length IFT88/70/52/46 (lanes 3 and 4), but not by GST-tagged IFT88/70/521–381; IFT88/70/52ΔC, lanes 5 and 6). Additionally, IFT46 alone was tested, but showed binding to IFT81ΔN/74ΔN/27/25ΔC (lanes 7 and 8). Binding was restored when a GST-IFT46/His-IFT52382–454 complex was used (GST-IFT46/His-IFT52C, lanes 9 and 10). (D) Model of the 2.3-Å crystal structure of the TtIFT52540–603/46236–347 complex, showing that these two fragments form a stable heterodimeric complex. The N and C termini as well as important secondary structure elements are labeled. The boxed region is shown in detail in E. (E) Detailed view on three hydrophobic TtIFT52 residues together with their respective binding residues in TtIFT46. (F) Pull-down experiments showing that a triple mutant of CrIFT52C (L414E/F418E/I421E, corresponding to L588, L592, and L595 in TtIFT152) is no longer able to bind to CrIFT46 and thus disrupts the hydrophobic interface formed between these two protein domains.
Structural and biochemical analysis of the IFT52C/46C complex. (A) SEC analysis showing that a dimeric IFT52C/46C complex can form a stable complex with IFT81ΔN/74ΔN/27/25ΔC. The IFT81ΔN/74ΔN/27/25ΔC tetramer was incubated with an excess of IFT52382–454/46165–319 (IFT52C/46C), and the resulting products were analyzed by SEC using a Superose 6 column. The chromatogram and the SDS-PAGE analysis of the peak fractions show that stoichiometric levels of the IFT52C/46C are shifted from the low-molecular weight peak 2 to the high molecular weight peak 1. (B) GST pull-down experiments showing that IFT52C is not sufficient for stable binding to IFT81ΔN/74ΔN/27/25ΔC, but that it requires IFT46. IFT81ΔN/74ΔN/27/25ΔC did not bind to GSH beads nonspecifically (lanes 1 and 2), and was not detectably pulled down by GST-IFT52382–454 (GST-IFT52C, lanes 3 and 4). To rule out that IFT52C was not able to bind at all to established interaction partners when GST-tagged, we tested binding to IFT46, which did not bind nonspecifically to the beads (lanes 5 and 6), but was efficiently pulled down by GST-IFT52C (lanes 7 and 8). A mixture of IFT81ΔN/74ΔN/27/25ΔC and IFT46 did not bind nonspecifically to the beads (lanes 9 and 10), but was efficiently pulled down by GST-IFT52C (lanes 11 and 12), showing that IFT52C requires IFT46 for stable binding to IFT81ΔN/74ΔN/27/25ΔC. (C) GST pull-down experiments showing that IFT52C is necessary for linking the IFT88/70/52/46 and IFT81ΔN/74ΔN/27/25ΔC subcomplexes. Whereas untagged IFT81ΔN/74ΔN/27/25ΔC complex did not bind nonspecifically to GSH beads (lanes 1 and 2), it was efficiently pulled down by GST-tagged full-length IFT88/70/52/46 (lanes 3 and 4), but not by GST-tagged IFT88/70/521–381; IFT88/70/52ΔC, lanes 5 and 6). Additionally, IFT46 alone was tested, but showed binding to IFT81ΔN/74ΔN/27/25ΔC (lanes 7 and 8). Binding was restored when a GST-IFT46/His-IFT52382–454 complex was used (GST-IFT46/His-IFT52C, lanes 9 and 10). (D) Model of the 2.3-Å crystal structure of the TtIFT52540–603/46236–347 complex, showing that these two fragments form a stable heterodimeric complex. The N and C termini as well as important secondary structure elements are labeled. The boxed region is shown in detail in E. (E) Detailed view on three hydrophobic TtIFT52 residues together with their respective binding residues in TtIFT46. (F) Pull-down experiments showing that a triple mutant of CrIFT52C (L414E/F418E/I421E, corresponding to L588, L592, and L595 in TtIFT152) is no longer able to bind to CrIFT46 and thus disrupts the hydrophobic interface formed between these two protein domains.
To obtain structural insights into the interaction of the C-terminal domains of IFT52 and IFT46, we expressed such complexes from various species and performed crystallization trials. Whereas the complexes from C. reinhardtii and mouse could not be crystallized due to very high solubility, a complex of the T. thermophila proteins TtIFT52540–603/TtIFT46236–347 yielded well-diffracting crystals. The structure was solved at 2.3 Å resolution and revealed a tightly packed α-helical hetero-dimer (Fig. 4 D) with a total interaction surface area of 1,852 Å2. The IFT52C fragment consists of three helices that bind via mainly hydrophobic interactions to helices of IFT46C (Fig. 4 D). The three helices of IFT52C are unlikely to constitute a domain with a stable conformation on its own, as the relative position of the helices appear to be defined through binding to IFT46 (Fig. 4 D). Helix α3 from IFT52C packs against helices α4 and α5 of IFT46C, completely burying L588, L592, and L595 of IFT52C (Fig. 4 E). Mutation of the three corresponding hydrophobic amino acids in CrIFT52C to glutamates disrupts the interaction with CrIFT46C, as demonstrated by pull-downs (Fig. 4 F). We conclude that a prerequisite for assembly of the IFT-B core is the formation of a stable IFT52/46 subcomplex.
Overexpression of the C-terminal IFT52 fragment severely inhibits ciliogenesis in mammalian cells
Based on the data presented above, we predicted that overexpression of the C-terminal IFT52 fragment would have a dominant-negative effect on ciliogenesis by preventing normal IFT-B complex formation. Such an IFT52C fragment would compete with the endogenous IFT52 protein for binding to IFT46, and the resulting IFT52C–IFT46 complex would bind to the IFT81/74/27/25 subcomplex, effectively leading to a smaller IFT-B core complex lacking IFT88 and IFT70. To test this notion experimentally, we overexpressed GFP-tagged versions of either full-length Mus musculus (Mm) IFT52 or the C-terminal fragment MmIFT52351-C inducibly in MDCK cells, where ciliogenesis can be monitored. Overexpression of full-length IFT52 did not have any measurable impact on ciliogenesis (judged by immuno-stainings for acetylated tubulin; ac-tub), demonstrating that the GFP tag does not significantly interfere with IFT52 function (Fig. 5 A [top] and Fig. 5 B), and that mouse IFT52 can substitute the endogenous canine version in the MDCK cell line, as the mouse GFP-IFT52 was visible in cilia (not depicted). However, when GFP-tagged IFT52C was overexpressed, the percentage of ciliated cells was reduced from 61% to 11%, demonstrating a highly significant defect in cilium formation (Fig. 5 A [bottom] and Fig. 5 B).
Ciliogenesis defect by overexpression of an IFT52 C-terminal fragment. (A) Representative fluorescence microscopy images of inducible overexpression of full-length MmIFT52 or MmIFT52351-C. Cells treated with tetracycline express the fusion protein (EGFP-MmIFT52fl or EGFP-MmIFT52351-C, green in merged image). Cilia are visualized by staining with anti-acetylated tubulin (ac-tub). Hoechst staining indicates nuclei. (B) The mean fraction of ciliated cells upon overexpression of MmIFT52 full length (n = 3; −tet, 1,693 cells; +tet, 1,559 cells) or MmIFT52351-C (n = 3; −tet, 1,696 cells; +tet, 1,670 cells; **, P < 0.001). (C) Representative images of cells overexpressing full-length MmIFT52 or MmIFT52351-C. Centrioles are visualized by immunostaining with anti–γ-tubulin. (D) Quantification of IFT88-positive centrioles. The number of IFT88-positive centrioles is indicated as a fraction of total centriole number. Note that each cell contains two centrioles. Because IFT88 is only present at the mother centriole, the maximum value is 0.5 (n = 3; MmIFT52 full-length −tet, 859 cells; +tet, 713 cells; MmIFT52351-C −tet, 766 cells; +tet, 822 cells; **, P = 0.002). All images are maximum intensity projections from z stacks. Error bars indicate mean ± SEM.
Ciliogenesis defect by overexpression of an IFT52 C-terminal fragment. (A) Representative fluorescence microscopy images of inducible overexpression of full-length MmIFT52 or MmIFT52351-C. Cells treated with tetracycline express the fusion protein (EGFP-MmIFT52fl or EGFP-MmIFT52351-C, green in merged image). Cilia are visualized by staining with anti-acetylated tubulin (ac-tub). Hoechst staining indicates nuclei. (B) The mean fraction of ciliated cells upon overexpression of MmIFT52 full length (n = 3; −tet, 1,693 cells; +tet, 1,559 cells) or MmIFT52351-C (n = 3; −tet, 1,696 cells; +tet, 1,670 cells; **, P < 0.001). (C) Representative images of cells overexpressing full-length MmIFT52 or MmIFT52351-C. Centrioles are visualized by immunostaining with anti–γ-tubulin. (D) Quantification of IFT88-positive centrioles. The number of IFT88-positive centrioles is indicated as a fraction of total centriole number. Note that each cell contains two centrioles. Because IFT88 is only present at the mother centriole, the maximum value is 0.5 (n = 3; MmIFT52 full-length −tet, 859 cells; +tet, 713 cells; MmIFT52351-C −tet, 766 cells; +tet, 822 cells; **, P = 0.002). All images are maximum intensity projections from z stacks. Error bars indicate mean ± SEM.
Our IFT-B core complex architectural map predicts that IFT88 and IFT70 would be excluded from the complex and therefore lost from the ciliary base after overexpression of IFT52C. We therefore set out to determine colocalization of IFT88 with the centriole marker γ-tubulin in cells overexpressing either full-length IFT52 or only the C-terminal fragment. IFT88 colocalization with γ-tubulin at the mother centriole was unperturbed in cells overexpressing full-length IFT52 (Fig. 5 C [top] and Fig. 5 D), but the amount of IFT88 was significantly reduced at the mother centriole after expression of the dominant-negative truncation (Fig. 5 C [bottom] and Fig. 5 D). To the best of our knowledge, this is the first reported example of an IFT protein domain where overexpression has a strong dominant-negative effect on ciliogenesis.
A complete interaction map of the IFT-B core complex
To obtain a complete interaction map for the IFT-B core, several interactions remained to be investigated in detail. First, the interaction of the small GTPase IFT22 to the rest of the complex has not been mapped so far. Second, although we know that IFT27/25 and IFT52C/46C bind to the IFT81/74 subcomplex, the exact binding site on this large coiled-coil complex remained unknown. To address this, we purified recombinant C. reinhardtii IFT22 and tested binding to IFT88/70/52/46 or IFT81/74/27/25. We only detected the formation of a stable pentameric complex when IFT22 was mixed with IFT81/74/27/25 (Fig. S5 A). Because no complex was formed between IFT22 and IFT27/25 (Fig. S5 B), we concluded that IFT22 binds directly to the IFT81/74 subcomplex. IFT81 and IFT74 contain predicted coiled-coil domains that mediate the interactions between the two proteins as well as central linker regions that likely divide the proteins into N-terminal and C-terminal coiled-coil domains (Lucker et al., 2005; Taschner et al., 2012). Whereas full-length IFT81/74 or the C-terminal coiled-coil domains were not soluble when recombinantly expressed, the N-terminal coiled-coil domains with or without the linker regions as well as the short linker regions alone were soluble and could be assayed for interaction with IFT22. Interestingly, a combination of the short linker regions in IFT81 and IFT74 (IFT81390–442 and IFT74398–459, respectively), but not the individual regions, were able to pull down untagged IFT22 (Fig. 6 A). When IFT27/25 or IFT52/46 were mixed with the soluble IFT81/74 complex containing the N-terminal coiled-coil region as well as the central linker, we could not detect any complex formation (Fig. S5, C and D), and we therefore conclude that the C-terminal coiled-coil region of IFT81/74 serves as the attachment point of these subcomplexes. With detailed knowledge about the interactions occurring between IFT-B proteins, we determined a purification strategy for assembling a stable nine-subunit IFT-B core complex from purified pentameric and tetrameric subcomplexes using a combination of recombinant expression in insect cells and E. coli (Fig. 6 B). All nine proteins including IFT74 and IFT81 were present in stoichiometric amounts in the IFT-B nonamer, as judged from the band intensities on the resulting SDS-PAGE (Fig. 6 B). The reconstituted IFT-B core complex on its own does not display any tendency to oligomerize in vitro, and the propensity to form IFT trains likely relies on additional factors such as peripheral IFT-B and IFT-A polypeptides. The data presented here allow us to construct a detailed IFT-B core interaction map (Fig. 6 C), pave the way for structural studies of larger assemblies, and may serve to interpret and design functional cell biological experiments.
Interaction map for the nonameric IFT-B core complex. (A) Untagged IFT22 is pulled down by a His-tagged IFT81390–442/74398–459 linker–region complex but not by His-IFT81390–442 or His-IFT74398–459 alone. (B) SEC profiles for the nonameric IFT-B core complex as well as smaller tetramer and pentameric subcomplexes. SDS-PAGE of the peak fractions from the nonamer purification is shown. (C) A schematic model of the IFT-B core complex summarizing the data presented in this work, with all currently structures shown in a representation with a transparent surface. Note that the exact positions of IFT27/25 and IFT52C/46C on the C-terminal coiled-coil region of IFT81/74 are not known.
Interaction map for the nonameric IFT-B core complex. (A) Untagged IFT22 is pulled down by a His-tagged IFT81390–442/74398–459 linker–region complex but not by His-IFT81390–442 or His-IFT74398–459 alone. (B) SEC profiles for the nonameric IFT-B core complex as well as smaller tetramer and pentameric subcomplexes. SDS-PAGE of the peak fractions from the nonamer purification is shown. (C) A schematic model of the IFT-B core complex summarizing the data presented in this work, with all currently structures shown in a representation with a transparent surface. Note that the exact positions of IFT27/25 and IFT52C/46C on the C-terminal coiled-coil region of IFT81/74 are not known.
Discussion
In this study, we further our understanding of the molecular architecture of the IFT-B core complex by reconstituting this complex in vitro, by mapping previously uncharacterized direct protein–protein interactions, and by presenting two novel high-resolution crystal structures. The C. reinhardtii IFT70/IFT52330–381 structure reveals that IFT70 consists of 15 TPRs, which wrap tightly around the proline-rich and mainly hydrophobic fragment of IFT52. The T. thermophila IFT52540–603/IFT46236–347 structure is purely α-helical and shows that the interaction between these two proteins is mediated through a largely hydrophobic surface. The pronounced hydrophobicity of these interaction interfaces provides a structural explanation for the reported salt stability of the IFT-B core (Lucker et al., 2005).
The interactions between IFT proteins and the ∼600 flagellar proteins in C. reinhardtii (Pazour et al., 2005) still remain largely uncharacterized apart from the reported direct interactions between IFT46 and ODA16 (Ahmed et al., 2008) as well as IFT81/74 and tubulin (Bhogaraju et al., 2013b), in addition to the indication for the transport of motility-related axonemal proteins by IFT56/TTC26/DYF13 (Ishikawa et al., 2014). The structure of the IFT70/52 complex presented in this report shows several surface patches, which are highly conserved between ciliated species (Fig. S2) and are good candidates for ciliary motor or cargo binding sites. Although the identity of such interactors is currently unknown, several potential candidates can be envisaged from recent work in various organisms. In zebrafish, the IFT70 orthologue Fleer has been shown to be a regulator of ciliary tubulin polyglutamylation (Pathak et al., 2007, 2011), a function that is also observed in C. elegans (Pathak et al., 2007) but, intriguingly, not in T. thermophila (Dave et al., 2009). Consequently, IFT70 might be the adapter for tubulin-modifying enzymes, at least in certain organisms. In C. elegans, the IFT70 orthologue DYF-1 has been shown to be important for activation of the homo-dimeric OSM-3 motor (Ou et al., 2005), but a direct interaction between these proteins has not yet been detected. Elucidating the cargo specificities of individual IFT proteins, including IFT70, will certainly be an interesting area of future research.
The complete interaction map of the IFT-B core complex derived here allows us to rationalize a number of previous in vivo observations. Since the discovery of the multisubunit IFT complex (Piperno and Mead, 1997; Cole et al., 1998), several mutant C. reinhardtii strains lacking individual components were analyzed for specific defects in ciliogenesis. Interestingly, despite being part of the same complex, the phenotypes of ift46, ift52, and ift88 mutants are quite different. Whereas ift52 (Brazelton et al., 2001) and ift88 (Pazour et al., 2000) are unable to assemble flagella, ift46 can assemble stunted flagella lacking outer dynein arms (Hou et al., 2007). A partial suppressor strain for ift46 was also identified, which was able to assemble nearly normal flagella but still lacked outer dynein arms (Hou et al., 2007). A more recent study gave the first insights into the effect of the absence of individual IFT-B core proteins on the assembly of the IFT-B complex, showing that certain factors have much more pronounced effects on IFT-B complex stability (Richey and Qin, 2012). The interaction map shown in Fig. 6 C provides a molecular rationale for the various ciliary phenotypes observed in specific IFT-B core complex protein mutants (Fig. 7).
Impact of IFT mutants on IFT-B core complex assembly and stability. (A) Schematics of the wild-type IFT-B core complex. (B) The ift88 mutant displays a stable IFT-B core, but loses peripheral IFT-B subunits and has a strong ciliogenesis defect. (C) The ift52 mutant strain lacks flagella due to a complete disruption of the IFT-B core complex. The schematics show that IFT46, IFT70, and IFT88 (and consequently all the proteins that are linked to the IFT-B core by these factors) are lost from the complex. (D) The ift46 mutant has a partially intact, but unstable, IFT-B core complex and can only assemble short flagella lacking outer dynein arms. In agreement with our data, the IFT52-(81/74/27/25/22) interaction is highly unstable in the absence of the IFT46 C-terminal domain. (E) The Supift461 suppressor strain expresses the C terminus of IFT46, leading to stabilization of the IFT52-(81/74/27/25/22) interface and to an overall stabilization of the IFT-B complex. The cells can assemble much longer flagella than the ift46 strain, but these flagella still lack outer dynein arms.
Impact of IFT mutants on IFT-B core complex assembly and stability. (A) Schematics of the wild-type IFT-B core complex. (B) The ift88 mutant displays a stable IFT-B core, but loses peripheral IFT-B subunits and has a strong ciliogenesis defect. (C) The ift52 mutant strain lacks flagella due to a complete disruption of the IFT-B core complex. The schematics show that IFT46, IFT70, and IFT88 (and consequently all the proteins that are linked to the IFT-B core by these factors) are lost from the complex. (D) The ift46 mutant has a partially intact, but unstable, IFT-B core complex and can only assemble short flagella lacking outer dynein arms. In agreement with our data, the IFT52-(81/74/27/25/22) interaction is highly unstable in the absence of the IFT46 C-terminal domain. (E) The Supift461 suppressor strain expresses the C terminus of IFT46, leading to stabilization of the IFT52-(81/74/27/25/22) interface and to an overall stabilization of the IFT-B complex. The cells can assemble much longer flagella than the ift46 strain, but these flagella still lack outer dynein arms.
Mutations in ift88 lead to a complete loss of flagella (Pazour et al., 2000), but nevertheless the IFT-B complex appears stable in these cells apart from apparently losing the attachment of one or more peripheral IFT-B proteins (Richey and Qin, 2012). The stability of the IFT-B core complex is consistent with our interaction map (see Fig. 7 B), as IFT88 is not required for the attachment of other IFT-B core proteins to the complex. The severe effect on ciliogenesis in the ift88 mutant suggests that IFT88 binds non–IFT-B core proteins with essential roles in ciliogenesis. Mutations in ift52 (in the bld1 strain) lead to a similar phenotype as ift88 mutations with completely “bald” cells (Brazelton et al., 2001). Analysis of IFT complex assembly in this mutant showed that the IFT-B complex is essentially absent (Richey and Qin, 2012). This is in agreement with our interaction map, as IFT52 has an important role in linking IFT88, IFT70, and IFT46 to the rest of the IFT-B core complex (see Fig. 7 C). The fact that IFT88, which on its own is essential for ciliogenesis (Pazour et al., 2000), is no longer able to attach to the rest of the IFT-B core in the ift52 mutant explains the lack of flagella in the bld1 mutant strain, although other defects might contribute to the “bald” phenotype. The ift46 mutant strain differs from ift88 and ift52 because it is able to assemble short axonemes (Hou et al., 2007), and sucrose-density gradient analysis of the IFT-B core complex from this mutant showed that the complex can form to a certain extent, but is unstable and prone to degradation (Richey and Qin, 2012). This is consistent with the finding that the levels of IFT-B proteins are decreased in this strain (Hou et al., 2007). We have previously shown that coexpression of full-length IFT52 with IFT81/74/27/25 in insect cells yields a pentameric complex, although IFT52 is substoichiometric, which suggests only weak association (Taschner et al., 2011). This is in agreement with the data presented in Fig. 4, because IFT52 requires IFT46 to assemble a stable IFT-B core complex. Based on these results, we speculate that in the ift46 strain, IFT52 can partially link the IFT88/70/52 complex with the IFT81/74/27/25/22 complex (Fig. 7 D), but due to the inherent instability of the IFT52–(81/74/27/25/22) interaction, this is not sufficient to support the growth of full-length flagella. Another intriguing phenotype of the ift46 mutant strain is the lack of axonemal outer dynein arms (Hou et al., 2007), showing that IFT46 acts as the binding site for an outer dynein arm adapter, which is consistent with the finding that IFT46 can directly bind to the ODA16 protein (Ahmed et al., 2008). A partial ift46 suppressor strain was identified that was able to assemble much longer flagella than in the ift46 strain but still lacked outer dynein arms (Hou et al., 2007). Interestingly, the authors detected expression of the 3′ end of the IFT46 gene, indicating that the C-terminal domain of IFT46 was present in this strain, and they speculated that this fragment was incorporated into the IFT-B core complex, leading to the observed stabilization of IFT-B protein levels (Fig. 7 E).
With a detailed interaction model of the IFT-B core complex at hand, we wanted to test if we could use this knowledge to modify this complex in vivo in mammalian cell culture. We show that, as predicted by our model, overexpression of the C-terminal fragment of IFT52 has a very strong dominant-negative effect on ciliogenesis and also leads to the absence of IFT88 from basal bodies. The negative effect of this truncation on cilia formation is comparable to siRNA-mediated knockdown of core IFT-B proteins such as IFT88, and thus this finding will allow researchers in the future to abolish ciliogenesis without depletion of IFT proteins. To our knowledge, this is the first report of a dominant-negative IFT protein fragment identified using biochemistry/structural biology. Similar experiments to investigate the functions and effects of other IFT-B core protein domains will be possible based on the interaction map presented in Fig. 6 C. The structural, biochemical, and functional data so far available support the notion of a common architecture of the IFT-B core complex conserved from green algae to humans.
Materials and methods
Cloning, expression, and purification of protein complexes from bacterial cultures
Full-length C. reinhardtii IFT genes and corresponding fragments were cloned into pEC vectors and expressed in E. coli BL21(DE3), as reported previously (Taschner et al., 2011). Plasmids containing the coding sequences of M. musculus IFT-B proteins were a generous gift from G. Pazour (University of Massachusetts Medical School, Worcester, MA). IFT protein coding sequences from T. thermophila were synthesized (GenScript) and subsequently cloned into pEC vectors. To achieve high-level expression, the bacterial cultures were grown at 37°C until the OD600 reached a value of 2, and the temperature was then reduced to 18°C. After the cultures had cooled down, 0.5 mM IPTG was added and the culture was kept shaking overnight. The IFT88/70/52/46 complex was purified as reported previously (Taschner et al., 2011). Briefly, the bacterial pellet from a 6-liter culture of E.coli cells transformed with a combination of four plasmids (pEC-K-GST-IFT88, pEC-S-His-IFT70, pEC-A-His-IFT52, and pETMCN-IFT46) was resuspended in an equal volume of lysis buffer (50 mM phosphate buffer, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM β-mercapto ethanol, 1 mM PMSF, and 10 µg/ml DNaseI), and the cells were lysed by sonication (total energy ∼25 kJ) on ice. The lysate was cleared by centrifugation at 25,000 rpm for 1 h and subsequently passed over a Ni2+-NTA column. The column was washed extensively with high salt buffer (lysis buffer containing 1 M NaCl) and the bound proteins were eluted with an imidazole gradient (0–500 mM in lysis buffer). The eluted proteins were passed through a Glutathione-Sepharose column to enrich for the tetrameric complex containing GST-IFT88. The bound complex was eluted in lysis buffer containing 30 mM reduced glutathione. The obtained material was then dialyzed overnight against buffer A (20 mM Tris, pH 7.5, 50 mM NaCl, and 10% glycerol) in the presence of TEV protease to remove the tag. Both the cleaved tags and the protease were removed using a Ni column, and the flow-through from this column was applied to a Heparin column and eluted using a gradient from buffer A to buffer B (20 mM Tris, pH 7.5, 1 M NaCl, and 10% glycerol). The fractions of interest were diluted 1:4 in buffer A and applied to Mono-Q ion-exchange column from which the bound material was eluted again with a gradient from buffer A to buffer B. Finally, the fractions containing the target complex were concentrated and applied to a HiLoad-Superdex200 column in SEC buffer (10 mM Hepes, pH 7.5, 150 mM NaCl, and 1 mM DTT).
For all other proteins and complexes expressed in bacterial cells, the same basic purification protocol was used. E. coli cells transformed with the various expression plasmids were grown in TB medium supplemented with the appropriate antibiotics, the resuspended pellet was lysed by sonication in lysis buffer, and His-tagged proteins were purified with a Ni2+-NTA column as described for the IFT88/70/52/46 complex. Eluted material was dialyzed overnight against buffer A in the presence of TEV-protease, followed by removal of tags and protease with Ni-column. The flow-through was loaded on a Q-Sepharose column and bound proteins eluted with a gradient from buffer A to buffer B (50 mM to 1 M NaCl). Fractions containing the clean complex were concentrated and applied to a SEC column (HiLoad200 or HiLoad75, depending on the size of the protein/complex) in SEC buffer (10 mM Hepes, pH 7.5, 150 mM NaCl, and 1 mM DTT).
Cloning, expression, and purification of protein complexes from insect cells
For expression in the insect cell line HighFive (Invitrogen), N-terminal truncations of C. reinhardtii IFT81 and IFT74, a C-terminal truncation of IFT25, and full-length IFT27 were cloned in the pFL vector (Fitzgerald et al., 2006; Bieniossek et al., 2008) in order to produce a single virus expressing the IFT81ΔN/74ΔN/27/25ΔX tetramer. PCR-products of untagged IFT81117–655 (IFT81ΔN) and His-tagged IFT27 were cloned into MCS1 using the enzymes SmaI and SphI, and the resulting plasmids were opened with EcoRI and XbaI. PCR products of untagged IFT74128–641 (IFT74ΔN) and IFT251–136 (IFT25ΔC) were also digested with EcoRI and XbaI and then inserted into MCS2 (IFT74ΔN together with IFT81ΔN, IFT25ΔC together with His-IFT27). In the last step, the His-IFT27/IFT25ΔC module was excised with PmeI and AvrII, and inserted into the IFT81ΔN/IFT74ΔN-containing vector, which had been opened with BstZ17I and SpeI. The resulting plasmid was transformed into DH10Bac E. coli cells (Invitrogen), according to the instructions of the manufacturer. The obtained viral genome was used to transfect Sf21 insect cells in 6-well plates (0.8 × 106 cells/well) using CellFectin (Life Technologies) according to the manufacturer’s instructions. 4 d after transfection, the 1 ml of the supernatant was added to a 10-ml Sf21 suspension culture (106 cells/ml). After another 4 d, the culture was centrifuged and the supernatant was used to infect a 500-ml Sf21 suspension culture (0.4 × 106 cells/ml). After 72 h of incubation, the final baculovirus-containing supernatant was harvested and stored at 4°C. The amount of viral supernatant required to produce recombinant protein in the HighFive cell line was determined empirically for each new batch of virus (typically the best results were obtained with 5–10 ml of viral supernatant per liter of HighFive culture).
For expression of the IFT81ΔN/74Δ/27/25ΔC complex, we usually infected a 3-liter HighFive culture and harvested the cells by centrifugation (15 min at 2,000 rpm). After resuspension in lysis buffer (50 mM phosphate buffer, pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM β-mercapto ethanol, 1 mM PMSF, and 25 µg/ml DNaseI), the cells were lysed by sonication on ice (total energy typically 8 kJ), and the lysate was subsequently cleared by centrifugation at 25,000 rpm for 1 h at 4°C. Purification of IFT81ΔN/74Δ/27/25ΔC followed the same protocols as for proteins and complexes expressed in E. coli, except that dialysis and TEV-protease cleavage were omitted. Instead, the eluate from the Ni2+-NTA column was directly applied to the MONO-Q column. For purification of a pentameric IFT81ΔN/74Δ/27/25ΔC/22 complex, the crude insect cell lysate from a 3-liter HighFive culture expressing the IFT81ΔN/74Δ/27/25ΔC tetramer was mixed was crude bacterial lysate from a 2-liter E.coli culture transformed with pEC-A-His-IFT22. The resulting mixed lysate was cleared by centrifugation at 4°C (25,000 rpm, 1 h), and the complex was purified as for the IFT81ΔN/74Δ/27/25ΔC tetramer.
Reconstitution of large IFT-B core complexes
To obtain complexes with more than five subunits, two smaller complexes were mixed either in equimolar amounts (for complexes with a similar size) or with an excess of the smaller complex (for complexes with a clear size difference), and incubated at 4°C for at least 2 h followed by SEC (Superdex 200 or Superose 6, depending on the size of the resulting complex).
GST pull-down experiments
In the case of purified proteins, 0.5 nmol of individual proteins/complexes (either GST-tagged or untagged) were mixed in a total volume of 200 µl of binding buffer (10 mM Hepes, pH 7.5, 150 mM NaCl, and 1 mM DTT) with 20 µl of GSH-resin. After removal of the “input” sample, the proteins were incubated overnight at 4°C. The beads were then washed three times with 1 ml of binding buffer, and the bound material was eluted by incubation of the beads with 40 µl of binding buffer containing 30 mM of reduced glutathione (“elution” sample). In the case of GST pull downs from lysate, the proteins were coexpressed in E. coli strain BL21(DE3). Pellets were resuspended in an equal volume of lysis buffer (50 mM phosphate-buffer, pH 7.5, 150 mM NaCl, 10% glycerol, and 1 mM DTT), and cells were lysed by sonication on ice (20 pulses of 1 s each). The lysate was cleared by centrifugation at 4°C (15,000 rpm for 30 min; Eppendorf centrifuge) and subsequently incubated with 25 µl of GSH-resin equilibrated in lysis buffer. After 2 h of incubation at 4°C, the beads were harvested by centrifugation (4 min at 500 g, 4°C) and washed three times with 1 ml of lysis buffer. Finally, the resin was incubated with 50 µl of elution buffer (lysis buffer supplemented with 30 mM of reduced glutathione), the beads were collected by centrifugation, and the supernatant was removed for analysis of eluted material by SDS-PAGE.
Crystallization, x-ray diffraction data collection, and structure solution
Purified IFT70/52330–381 complex was mixed in equal volume with precipitant solution containing 20% Tacsimate, pH 7.0, and incubated against a reservoir of precipitant solution at 22°C for 4 d until crystals appeared. Crystals were cryo protected with mother liquor supplemented with 25% glycerol and flashed-cooled in liquid nitrogen before x-ray data collection at the Swiss Light Source synchrotron beamline PXII. For the gold derivative, 0.25 µl of mother liquor containing 2 mM of KAu(CN)2 was added to 1 µl drops containing crystals, and after soaking for 3 h, the crystals were back-soaked in mother liquor containing 25% glycerol and flash-cooled in liquid nitrogen, and single-wavelength anomalous dispersion data were collected. Native and derivative x-ray diffraction data were integrated with XDS (Kabsch, 2010) and scaled by the AIMLESS program as part of the CCP4 suite of crystallographic programs (Winn et al., 2011). Gold sites were located and SAD phases calculated in PHENIX autosolve (Adams et al., 2010) to produce an initial experimental map into which most of the amino acid sequences were traced in Coot (Emsley et al., 2010). The model was then refined against the native data, which were isomorphous to the derivative data, using PHENIX (Adams et al., 2010). Comparison of the IFT70/52 complex structure against the entire set of protein structures in the Protein Data Bank (PDB) was done using the DALI server (Holm and Rosenström, 2010), and structure figures were prepared using PYMOL (http://www.pymol.org/). Purified TtIFT52540–603/46236–347 complex was mixed in equal volume with precipitant solution containing 50 mM Tris, pH 8.2, and 0.6 M tri-sodium-citrate, and incubated against a reservoir of precipitant solution at 22°C for 2 d until crystals appeared. Crystals were cryo-protected with mother liquor supplemented with 25% glycerol and flashed-cooled in liquid nitrogen before x-ray data collection. For the gold derivative, 2 µl of mother liquor containing 4 mM of KAu(CN)2 was added to 1-µl drops containing the crystals. After soaking for 30 min, crystals were cryo-protected in mother liquor supplemented with 25% glycerol and 4 mM KAu(CN)2, and flash-cooled in liquid nitrogen. Single-wavelength anomalous dispersion data were then collected. The structure was determined as described for the IFT70/52 complex.
Transgenic cell lines
For inducible overexpression, full-length MmIFT52 or MmIFT52351-C (amino acids 351–426) were cloned into Mlu and Not1 sites of a modified pLVTH, resulting in an N-terminal GFP fusion construct (Wiznerowicz and Trono, 2003). MDCK cells expressing the tTR-KRAB repressor were transfected with lentivirus encoding either the full-length MmIFT52 or MmIFT52351-C. For the induction of protein expression, tetracycline (5 µg/ml, T-3383; Sigma-Aldrich) was added to the cells for 7–10 d before experiments.
Immunofluorescence microscopy measurements
For immunofluorescence staining, the following antibodies were used: monoclonal anti–γ-tubulin (1:500 dilution; mouse-monoclonal; T6557; Sigma-Aldrich), anti-acetylated tubulin (1:3,000 dilution; mouse-monoclonal; T7451; Sigma-Aldrich), and anti-Ift88 (1:200 dilution; rabbit-polyclonal; 13967-1 AP; ProteinTech Group). Antibodies were visualized using Cy5-, Cy3-, or Alexa Fluor 488–labeled secondary antibodies (1:1,000; Jackson ImmunoResearch Laboratories, Inc.). Hoechst 33342 (10–6 mol/liter; H-1399; Molecular Probes) was used for staining nuclei. For staining of cilia, cells were grown on glass coverslips for 7–10 d. The cells were fixed for 10 min at room temperature for ciliogenesis analysis with 4% paraformaldehyde (15170; ElectroMicroscopyS) and for 5 min for IFT88 staining with 50:50 methanol/acetone at −20°C (Kotsis et al., 2008). The presence of cilia was observed with an inverted laser-scanning microscope (LSM 510 Meta NLO; Plan-Apochromat 63×/1.4 NA, 7 z planes, 1 µm z distance, pixel size 279 nm) and IFT88 staining at the basal body with an LSM 510 Meta Duo Live microscope (Plan-Apochromat 100×/1.4 NA, 11 z planes, 0.65 µm z distance, pixel size 88 nm; both from Carl Zeiss). For each independent experiment (n), five random fields of view were selected and z stacks were recorded under equal conditions for scanning. Imaging, measurements, and statistical analysis were performed in a blinded manner. 3D reconstruction and measurements were performed with Imaris software (Bitplane). Ciliated cells were counted based on staining for acetylated tubulin. IFT88/γ-tubulin ratios were determined by software-based detection of γ-tubulin signal using Imaris spot detection. Local intensity measurements of IFT88 and γ-tubulin were performed with background subtraction. Centrioles were considered IFT88 positive if the IFT88/γ-tubulin ratio was >0.5.
Statistical analyses
Statistical analysis was performed in Excel (Microsoft). P-values were calculated with an unpaired t test from the mean data of three independent experiments (n), each represented by five fields of view. All values are given as a mean of n ± SEM. All statistical analyses were considered significant for P-values of <0.05.
Online supplemental material
Fig. S1 shows a multiple sequence alignment for IFT highlighting the position of individual TPRs and indicating residues important for IFT52 binding. Fig. S2 shows conserved surface patches on the IFT70 protein. Fig. S3 shows the results of pull-down experiments between IFT70/88 and IFT46. Fig. S4 shows the purification of IFT-B core subcomplexes from T. thermophila. Fig. S5 shows pull-down experiments to elucidate the binding sites of IFT27/25, IFT22, and IFT52/46 on the IFT81/74 subcomplex. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201408002.dv.
Acknowledgments
We thank the staff at the Swiss Light Source for help with x-ray diffraction data collection, the crystallization facility of the Max Planck Institute of Biochemistry (Munich) for access to crystallization screening, Dr. C. Basquin for static light scattering experiments, and Lissy Weyher at the MPI of Biochemistry core facility for mass spectrometric analyses. We acknowledge M. Morawetz and M. Stiegler for technical assistance with molecular biology, and Michaela Rode for help with insect cell culture. André Mourão is acknowledged for nuclear magnetic resonance analysis of IFT52/46 complexes. We are also grateful to Gregory Pazour for providing plasmids containing the coding sequences for mouse IFT proteins.
This work was funded by an Emmy Noether grant (Deutsche Forschungsgemeinschaft [DFG]; LO1627/1-1), the European Research Council (ERC grant 310343), and by the EMBO Young Investigator program. M. Taschner was the recipient of an Erwin-Schroedinger stipend granted by the Austrian Science Fund (J3148-B12). E.W. Kuehn was funded through DFG KU 1504/4-1. Structural coordinates have been deposited at the Protein Data Bank, accession no. 4UZY for the Cr70/52 structure and 4UZZ for the Tt52/46 structure.
The authors declare no competing financial interests.
Author contributions: M. Taschner carried out the purification and interaction studies of C. reinhardtii and M. musculus IFT proteins and complexes, and determined the crystal structure of CrIFT70/52. P. Braeuer carried out the purification of T. thermophila complexes and determined the crystal structure of TtIFT52/46. E. Lorentzen supervised the biochemical and structural experiments. The cell-based experiments were carried out by F. Kotsis under the supervision of E.W. Kuehn. M. Taschner and E. Lorentzen wrote the paper.