Although cortical actin plays an important role in cellular mechanics and morphogenesis, there is surprisingly little information on cortex organization at the apical surface of cells. In this paper, we characterize organization and dynamics of microvilli (MV) and a previously unappreciated actomyosin network at the apical surface of Madin–Darby canine kidney cells. In contrast to short and static MV in confluent cells, the apical surfaces of nonconfluent epithelial cells (ECs) form highly dynamic protrusions, which are often oriented along the plane of the membrane. These dynamic MV exhibit complex and spatially correlated reorganization, which is dependent on myosin II activity. Surprisingly, myosin II is organized into an extensive network of filaments spanning the entire apical membrane in nonconfluent ECs. Dynamic MV, myosin filaments, and their associated actin filaments form an interconnected, prestressed network. Interestingly, this network regulates lateral mobility of apical membrane probes such as integrins or epidermal growth factor receptors, suggesting that coordinated actomyosin dynamics contributes to apical cell membrane organization.
Introduction
The cytoskeleton is responsible for detection, transmission, and generation of the mechanical forces that determine individual cell shape and collective cellular behaviors. Whereas microtubules and intermediate filaments mostly pervade the cytosol, actin filaments are concentrated at the cell periphery or cell cortex (Bretscher, 1991), where they facilitate the mechanical stabilization and remodeling of cell shape. Numerous morphogenetic processes are actively driven by actin dynamics, including cell polarization, migration, division, and differentiation (Pantaloni et al., 2001; Pollard and Cooper, 2009). Mechanical forces required for these processes are generated by actin polymerization and depolymerization or via contraction mediated by myosin motors (Martin et al., 2009; Lecuit et al., 2011).
Epithelial cells (ECs) organize into adherent groups, which form the boundary layers that define tissue compartments in multicellular organisms. Such an interface must generate and withstand significant mechanical stresses during tissue remodeling in the course of development, growth, and wound healing. It is therefore of great interest to understand the structural and dynamical aspects of cortical organization and remodeling that enable ECs to exert and sustain such forces.
Individual cells within epithelial monolayers are polarized along an apical–basal axis, a feature that is reflected in aspects of membrane trafficking, ion transport, and cytoskeletal organization (Nelson, 2009). At the basal surface of single-layered epithelia, actin forms stress fibers, made up of antiparallel actomyosin bundles that connect adhesion sites and exert tensile forces on the underlying basement membrane (Katoh et al., 2008). A second prominent actin structure in many polarized ECs is a circumferential ring of actin filaments that are linked to cadherin-based adherens junctions just basal to the apical surface (Danjo and Gipson, 1998). This junctional actin is thought to stabilize cell–cell contacts and to provide the principal structural source of force transmission within epithelia (Lecuit and Wieschaus, 2002).
The apical surface of polarized epithelia is decorated by numerous microvilli (MV). These membrane protrusions are formed around bundles of parallel actin filaments and extend 1–3 µm from the cell surface. In epithelia of the small intestine and kidney, ≤15,000 MV per cell (DeRosier and Tilney, 2000) make up the typical brush border. Brush border MV are interconnected at their bases via a dense meshwork of actin, spectrin, and myosins called the terminal web (Bretscher and Weber, 1978; Hirokawa et al., 1982).
During developmental processes, such as gastrulation (Chuai and Weijer, 2009), as well as wound healing, ECs undergo drastic shape changes to facilitate large-scale tissue morphogenesis. In many cases, ECs undergo an epithelial–mesenchymal transition (EMT) and acquire morphological features that are reminiscent of mesenchymal cells or mesenchymal stem cells (Lee et al., 2006; Mani et al., 2008; Thiery et al., 2009). Specifically, they modify cell–cell and cell–matrix contacts as their typical apical–basal polarity is converted into the front–back polarity characteristic of migratory cells (Nelson, 2009). Actin is then organized in lamellipodia and filopodia at the leading edge of such cells (Burnette et al., 2011). EMTs are common during embryonic development—for example, in neural crest cell delamination and migration (Thiery et al., 2009)—and may underlie the metastatic potential of epithelial tumors (Gupta et al., 2009). Although there has been significant progress in elucidating the molecular basis for regulation of the EMT (Kalluri and Weinberg, 2009), our understanding of the changes in organization of the cortical cytoskeleton that occur during this process remains very limited (Sperry et al., 2010; Xing et al., 2011).
Despite the common perception that MV mainly serve as a device to increase cell surface area, they are likely involved in a variety of additional cellular functions, including regulation of energy metabolism, gating of ion flux, generation and modulation of membrane potential, Ca2+ signaling, and mechanoreception (Lange, 2011). However, the physical and molecular basis for these diverse MV functions is not understood. Apart from the specialized brush borders, ECs often form much less densely packed actin-containing MV with diverse morphologies, including tubelike protrusions (Chinkers et al., 1979; Garbett et al., 2010), ridges (Poole and Müller, 2005), and ruffles (Brunk et al., 1976; Chinkers et al., 1979). Proteins of the ERM (ezrin–radixin–moesin) family (Bretscher et al., 2002) and scaffold proteins, such as EBP50 and PDKZ1 (Garbett et al., 2010; LaLonde et al., 2010), have been shown to play a crucial role in MV assembly and morphogenesis. Although there has been progress in elucidating the molecular composition of individual MV (Garbett et al., 2010; Garbett and Bretscher, 2012; Zwaenepoel et al., 2012), the basis for the morphological diversity of MV has not been addressed. In particular, very little information on MV dynamics and lateral connectivity is available.
Here, we use quantitative live-cell imaging to characterize the spatial organization and dynamics of MV in MDCK cells, a widely used epithelial model cell line. We find that the apical surface of confluent MDCK cells bears a dense array of short and static MV, whereas nonconfluent cells form highly dynamic protrusions, which are more elongated and often oriented parallel to the plane of the membrane. These dynamic MV exhibit complex and spatially correlated patterns of reorganization that depend on myosin II activity. Surprisingly, we found that myosin II is organized into a network of filaments that extend along the entire inner face of the apical membrane of nonconfluent MDCK cells. MV, apical myosin filaments, and their associated actin filaments form an interconnected prestressed network that exhibits highly coordinated motility. Importantly, reorganization of the apical actomyosin network both restricts and facilitates lateral mobility of apical membrane components such as EGF receptors (EGFRs) or integrins and their ligands.
Results
A characteristic MV morphology and organization in nonconfluent ECs
In light of the known morphological diversity of MV or apical membrane protrusions found in mammalian cells, we set out to establish a model system in which different microvillar structures could be clearly differentiated. To monitor organization of MV in living cells, we stably transfected MDCK cells with the actin marker Lifeact-GFP (Riedl et al., 2008). Using live-cell epifluorescence microscopy, we found that the apical surface of confluent MDCK cells was covered with dense arrays of short MV (Fig. 1 A) that protruded from the plasma membrane (Fig. 1 B). In contrast, we found many elongated MV oriented parallel to the apical surface of nonconfluent cells (Fig. 1 C) as well as less protrusive MV (Fig. 1 D). Note that we use the term apical here to refer to the cell surface farthest removed from the glass substrate, irrespective of the polarization state of cells. Formation of elongated apical MV was not a result of Lifeact-GFP expression because similar structures were found in live cells expressing actin-GFP (Fig. S1 A) and in fixed cells stained with Atto 488–phalloidine (Fig. S1 B). Elongated MV were not unique to MDCK cells but were observed on the apical surfaces of various nonconfluent EC lines, including HeLa, MCF-7, and HaCaT cells (Fig. S1 C).
Apical actin organization of ECs. (A–H) Apical actin organization and surface topology of confluent (A, B, E, and F) and nonconfluent (C, D, G, and H) MDCK cells. Images showing filamentous actin structures labeled with Lifeact-GFP in a single focal plane (A and C) or in projections of three planes covering ∼1.5 µm (E and G) are shown. Transmission EM micrographs show varying numbers of protruding MV (B and D), whereas SEM images reveal the topology of apical MV (F and H). (I and J) Quantitative analysis of the topology of MV imaged by SEM, showing length (I) and degree of straightness (J) for confluent and nonconfluent cells (n = 99; error bars: SDs; t test value: ***, P < 10−10). Red lines indicate Gaussian fits. Bars, 2 µm. Cell shapes and the relative positions of the focal planes shown are indicated schematically with color codes corresponding to structures in E and G. au, arbitrary unit.
Apical actin organization of ECs. (A–H) Apical actin organization and surface topology of confluent (A, B, E, and F) and nonconfluent (C, D, G, and H) MDCK cells. Images showing filamentous actin structures labeled with Lifeact-GFP in a single focal plane (A and C) or in projections of three planes covering ∼1.5 µm (E and G) are shown. Transmission EM micrographs show varying numbers of protruding MV (B and D), whereas SEM images reveal the topology of apical MV (F and H). (I and J) Quantitative analysis of the topology of MV imaged by SEM, showing length (I) and degree of straightness (J) for confluent and nonconfluent cells (n = 99; error bars: SDs; t test value: ***, P < 10−10). Red lines indicate Gaussian fits. Bars, 2 µm. Cell shapes and the relative positions of the focal planes shown are indicated schematically with color codes corresponding to structures in E and G. au, arbitrary unit.
To study the density and topology of elongated MV in more detail, we visualized the apical surface of MDCK cells using either projections of successive focal planes (Fig. 1, E and G) or scanning EM (SEM; Fig. 1, F and H). Both approaches confirmed the prevalence of short protruding MV on confluent cells (Fig. 1, E, F, and I), whereas nonconfluent MDCK cells formed slightly longer apical protrusions (Fig. 1, G–I) that were often oriented parallel to the cell surface (Fig. 1 G) and were frequently bent into a bowlike shape (Fig. 1 J). We also observed that some MV responded to an external oscillatory flow (Fig. S1 D), whereas others remained static and membrane associated, indicating that they did not project as far from the apical surface.
We then asked whether the observed differences in morphology and density of MV protrusions were dependent on the time elapsed after seeding or on the position of individual cells within an epithelial sheet. To do so, we systematically evaluated apical actin distribution in MDCK cells by phalloidin staining 1–4 d after seeding. We classified cells according to their position as either single (isolated) cells (I), cells located at the borders of small cell islands (II), or confluent (i.e., surrounded by neighboring cells) cells within small (III) or large (IV) islands (Fig. 2, A and B). We observed that, for confluent cells in categories III and IV, the density of apical protrusions steadily increased with time after seeding (n > 20 cells per data point; Fig. 2 C, green), whereas for nonconfluent cells (categories I and II), this parameter did not increase beyond the 48-h time point (Fig. 2 C, blue). Hence, this difference in MV morphology correlated with cell position, domelike shape, and absence of neighbors.
Factors that influence MV morphology. (A and B) Classification of MDCK cells according to their position within cell layers (schematics and selected regions of representative cells). (C) Relative density of the indicated classes of MV (defined in A and B) in cells cultured for the indicated times. All cells were fixed and stained with Atto 488–phalloidin, and the object density in images was analyzed with an automated computer algorithm (n ≥ 20; error bars: SEMs; t test: *, P = 0.05; ***, P < 0.001). (D) Apical actin organization in a wound-healing experiment. Note the differences between cells at the wound edge (1) and cells within the sheet (2). Arrows indicate basal protrusions. (E) Effects of HGF on apical actin organization (shown here for a single cell). Cells were treated with 100 ng/ml HGF and imaged at different times (given in minutes) after growth factor addition. Bars, 2 µm.
Factors that influence MV morphology. (A and B) Classification of MDCK cells according to their position within cell layers (schematics and selected regions of representative cells). (C) Relative density of the indicated classes of MV (defined in A and B) in cells cultured for the indicated times. All cells were fixed and stained with Atto 488–phalloidin, and the object density in images was analyzed with an automated computer algorithm (n ≥ 20; error bars: SEMs; t test: *, P = 0.05; ***, P < 0.001). (D) Apical actin organization in a wound-healing experiment. Note the differences between cells at the wound edge (1) and cells within the sheet (2). Arrows indicate basal protrusions. (E) Effects of HGF on apical actin organization (shown here for a single cell). Cells were treated with 100 ng/ml HGF and imaged at different times (given in minutes) after growth factor addition. Bars, 2 µm.
Physiological situations in which ECs lose their coherent, sheetlike organization and become motile include wound healing and EMT that occur during gastrulation (Chuai and Weijer, 2009) and neural crest cell delamination (Thiery et al., 2009). During these processes, ECs undergo drastic shape changes that facilitate large-scale tissue morphogenesis. To examine how MV organization is altered during EC morphogenesis, we imaged apical actin organization in MDCK cells during wound healing. We found that cells at the migration front exhibited the elongated MV typical of nonconfluent cells (Fig. 2 D, 1). In contrast, cells further away from the advancing edge retained the denser arrays of short MV typical for confluent cells (Fig. 2 D, 2).
We then tested the effect of hepatocyte growth factor (HGF) on MV morphology. HGF induces dispersion of adherent MDCK cells by promoting dissociation of the cell–cell junctions that hold them together, thus facilitating subsequent migration. This treatment is also used to mimic EMT (Montesano et al., 1991). Before addition of HGF, confluent cells displayed dense arrays of MV on their apical surfaces (Fig. 2 E, 0 min). After several hours exposure to 100 ng/ml HGF, MV progressively increased in length (Fig. 2 E), until the apical surface was completely covered in very long actin-filled protrusions (Fig. 2 E, 300 min). The ERM proteins ezrin, radixin, and moesin are classical components of MV, and ezrin is a known effector of HGF during epithelial morphogenesis (Crepaldi et al., 1997). We therefore tested localization of ERM proteins, as well as their membrane adaptor EBP50, within elongated MV. We found that all four markers colocalized with apical actin in nonconfluent MDCK cells (Fig. S2), indicating that the observed actin structures in nonconfluent cells resemble MV in protein composition, even though they differ in morphology. In summary, we found that bent and elongated MV are a characteristic feature of ECs that are not completely surrounded by other cells and are capable of undergoing morphogenetic changes.
MV in nonconfluent cells exhibit rapid myosin II–dependent dynamics
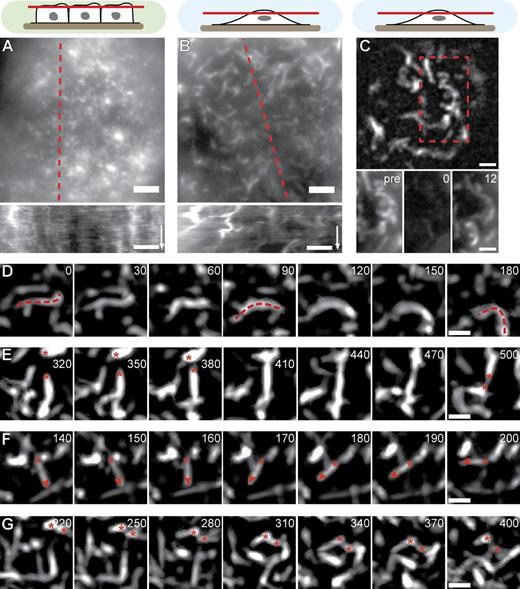
To examine the molecular mechanisms that drive changes in MV morphology and organization in subconfluent cells, we next performed time-lapse fluorescence microscopy on MDCK cells transfected with Lifeact-GFP. In confluent cells, the typical dotted MV structures remained largely static and only exhibited short-range fluctuations (Fig. 3 A and Video 1). In contrast, nonconfluent cells were characterized by extensive lateral rearrangements of their elongated MV (Fig. 3 B and Video 2). This rearrangement was accompanied by rapid turnover of actin, as shown by FRAP experiments with actin-GFP–expressing cells (t1/2 = 4.7 ± 1.0 s [mean ± SD]; n = 21; Fig. 3 C). This recovery did not simply reflect cytosolic diffusion of actin monomers, as recovery of soluble Lifeact-GFP occurred much faster (t1/2 = 2.8 ± 0.7 s [mean ± SD]; n = 18). When we examined apical MV dynamics in greater detail, we frequently observed extensive bending (Fig. 3 D and Video 3) and exchange of connectivity (fusion and fission) between neighboring structures (Fig. 3 E and Video 3). In addition, pivoting of straight MV (Fig. 3 F and Video 3), as well as lateral translation of whole MV (Fig. 3 G and Video 3), was detected.
MV dynamics in confluent cells. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of the apical cortex region of a confluent MDCK cell that has been stably transfected with Lifeact-GFP to label actin. Bar, 2 µm. Frames were taken every 10 s for 10 min. Video to Fig. 3 A.
MV dynamics in confluent cells. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of the apical cortex region of a confluent MDCK cell that has been stably transfected with Lifeact-GFP to label actin. Bar, 2 µm. Frames were taken every 10 s for 10 min. Video to Fig. 3 A.
MV dynamics in nonconfluent cells. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of the apical cortex region of a nonconfluent MDCK cell stably transfected with Lifeact-GFP to label actin. Bars, 2 µm. Frames were taken every 10 s for 10 min. Video to Fig. 3 B.
MV dynamics in nonconfluent cells. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of the apical cortex region of a nonconfluent MDCK cell stably transfected with Lifeact-GFP to label actin. Bars, 2 µm. Frames were taken every 10 s for 10 min. Video to Fig. 3 B.
MV reorganization. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing examples of MV reorganization events in nonconfluent MDCK cells stably transfected with Lifeact-GFP: (1) bending, (2) exchange of connectivity, (3) flickering, and (4) lateral translation. Bar, 2 µm. Frames were taken every 10 s for indicated times. Red asterisks, arrowhead, and circles indicate ends of filaments described in the respective panels. Video to Fig. 3 (D–G).
MV reorganization. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing examples of MV reorganization events in nonconfluent MDCK cells stably transfected with Lifeact-GFP: (1) bending, (2) exchange of connectivity, (3) flickering, and (4) lateral translation. Bar, 2 µm. Frames were taken every 10 s for indicated times. Red asterisks, arrowhead, and circles indicate ends of filaments described in the respective panels. Video to Fig. 3 (D–G).
MV dynamics. (A and B) Qualitative motion analysis of MDCK cells stably transfected with Lifeact-GFP. Kymographs along the dotted red lines reveal lateral actin rearrangement specifically in nonconfluent cells (time arrows, 300 s; Videos 1 and 2). (C) Rapid turnover of actin-GFP shown by FRAP. Red dotted box is magnified on the bottom. Pre, time before FRAP event. (D–G) Image series demonstrating typical MV reorganization through bending (D), exchange of connectivity (E), pivoting (F), and lateral translation (G). Red dotted lines, asterisks, and arrowheads indicate MV contours and ends for the respective behaviors. Also see Video 3. Bars: (A–C) 2 µm; (D–G) 1 µm. Times are given in seconds.
MV dynamics. (A and B) Qualitative motion analysis of MDCK cells stably transfected with Lifeact-GFP. Kymographs along the dotted red lines reveal lateral actin rearrangement specifically in nonconfluent cells (time arrows, 300 s; Videos 1 and 2). (C) Rapid turnover of actin-GFP shown by FRAP. Red dotted box is magnified on the bottom. Pre, time before FRAP event. (D–G) Image series demonstrating typical MV reorganization through bending (D), exchange of connectivity (E), pivoting (F), and lateral translation (G). Red dotted lines, asterisks, and arrowheads indicate MV contours and ends for the respective behaviors. Also see Video 3. Bars: (A–C) 2 µm; (D–G) 1 µm. Times are given in seconds.
More detailed investigation of this lateral translation of whole MV in nonconfluent cells revealed that adjacent MV often moved in a coordinated fashion (Fig. 4, A and B). To determine the spatial extent of correlation in the observed motion, we performed spatiotemporal image correlation spectroscopy (STICS) analysis and found a mean correlation distance of 3 µm (Fig. 4, C and D). As most MV movement was caused by lateral reorganization in the plane of the membrane rather than by polymerization and depolymerization, we hypothesized that myosin II might be the molecule driving the observed dynamics. Indeed, when we inhibited myosin II ATPase activity by treating cells with 50 µM blebbistatin, apical actin reorganization was slowed down significantly (Fig. 4, E–G), and the mean temporal correlation in the overall actin structure increased from 1 to >2.5 min (Fig. 4 F). This effect confirmed that myosin II plays an important role in the lateral motility of apical MV in nonconfluent MDCK cells.
Myosin-dependent motion of apical actin. (A and B) Kymographs along dotted lines in A reveal coordinated motion of MV (B). Arrows indicate correlated motion of neighboring MV (B, 1) and noncorrelated motion of distant MV (B, 2). Time arrows, 500 s. White dotted lines represent viewing guides to better follow trace evolutions. (C) STICS correlation analysis with characteristic spatiotemporal correlation length (inset: 3.3 ± 0.9 µm [mean ± SD]; n = 11). Red lines represent exponential fits. (D) Calculated STICS flow field represented by velocity vectors on a uniform grid. Schematic indicates velocity vectors and correlation length outlined by a radial gradient circle. Arrows indicate direction and magnitude of flow vectors. (E and F) Effect of inhibition of myosin II activity on correlation time in actin reorganization. Sample curves (E) and mean Pearson correlation coefficients (F) are shown for control cells (blue, 62 ± 20 s [mean ± SEM]; n = 8) and cells treated with 50 µM blebbistatin (red, 156 ± 60 s [mean ± SEM]; n = 12). Black lines represent exponential fits. (G) Kymographs from cells analyzed in E and F showing that inhibition of myosin II blocks lateral reorganization. Time arrows, 300 s. Squares, triangles, and circles indicate data points for three representative cells. Bars: (A, B, and G) 2 µm; (D) 1 µm.
Myosin-dependent motion of apical actin. (A and B) Kymographs along dotted lines in A reveal coordinated motion of MV (B). Arrows indicate correlated motion of neighboring MV (B, 1) and noncorrelated motion of distant MV (B, 2). Time arrows, 500 s. White dotted lines represent viewing guides to better follow trace evolutions. (C) STICS correlation analysis with characteristic spatiotemporal correlation length (inset: 3.3 ± 0.9 µm [mean ± SD]; n = 11). Red lines represent exponential fits. (D) Calculated STICS flow field represented by velocity vectors on a uniform grid. Schematic indicates velocity vectors and correlation length outlined by a radial gradient circle. Arrows indicate direction and magnitude of flow vectors. (E and F) Effect of inhibition of myosin II activity on correlation time in actin reorganization. Sample curves (E) and mean Pearson correlation coefficients (F) are shown for control cells (blue, 62 ± 20 s [mean ± SEM]; n = 8) and cells treated with 50 µM blebbistatin (red, 156 ± 60 s [mean ± SEM]; n = 12). Black lines represent exponential fits. (G) Kymographs from cells analyzed in E and F showing that inhibition of myosin II blocks lateral reorganization. Time arrows, 300 s. Squares, triangles, and circles indicate data points for three representative cells. Bars: (A, B, and G) 2 µm; (D) 1 µm.
An isotropic myosin network spanning the apical surface of ECs
As myosin II was essential for MV motility, we determined the localization of myosin at the apical cortex. To this end, we generated cell lines stably expressing GFP fusions to myosin light chain (MLC; MLC-GFP) and myosin IIA heavy chain (MHCA; MHCA-GFP). Ectopic expression levels of MHCA-GFP were comparable to those of endogenous myosin IIA (Fig. S3 A), and we did not observe any changes in cellular morphology upon expression of the fusion proteins. Surprisingly, we found that myosin II formed an extensive network of filaments located immediately below the apical surface of nonconfluent cells (Fig. 5, A and B). Both myosin markers colocalized within filaments (Fig. S3 B), and the network could also be visualized with antibodies against endogenous MHCA (Fig. S3, C and D). In contrast, myosin II in confluent MDCK cells was localized to distinct patches that likely corresponded to the base of MV (Fig. 5, C and D; Hirokawa et al., 1982; Watanabe et al., 2007). The myosin network in nonconfluent cells was branched and exhibited coordinated fluctuations (Fig. 5, E and F; and Video 4) that were sensitive to treatment with 50 µM blebbistatin or the MLC kinase inhibitor ML7 (Fig. S3 E). Their patterns of movement were comparable to those of MV, as confirmed by both spatial and temporal correlation coefficients (Fig. 5, G and H). Finally, the change in myosin organization was also seen in wound-healing experiments, in which cells at the wound edge contained myosin II networks at their apical surfaces (Fig. 5 I, 1), whereas cells deeper within the sheet had a more patchy myosin localization with only a few filamentous connections (Fig. 5 I, 2).
Myosin dynamics. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing isotropic reorganization of the apical myosin network in nonconfluent MDCK cells stably transfected with MHCA-GFP or MLC-GFP. Two focal planes (Δz = 1 µm) were maximum projected and color coded in red (most basal) followed by green and blue (most apical). Bars, 2 µm. Frames were taken every 10 s for 15 min. Video to Fig. 5 (E and F).
Myosin dynamics. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing isotropic reorganization of the apical myosin network in nonconfluent MDCK cells stably transfected with MHCA-GFP or MLC-GFP. Two focal planes (Δz = 1 µm) were maximum projected and color coded in red (most basal) followed by green and blue (most apical). Bars, 2 µm. Frames were taken every 10 s for 15 min. Video to Fig. 5 (E and F).
The apical myosin II network. (A–D) Apical organization of myosin II filaments visualized by labeling of myosin heavy chain II A (MHCA; A and C) or myosin light chain (MLC; B and D) with GFP. Whereas confluent cells exhibit punctate myosin II signals (C and D), nonconfluent MDCK cells form isotropic networks of myosin at their apical cell surface (A and B). (E and F) Dynamic rearrangement of myosin networks labeled with MHCA-GFP (E) or MLC-GFP (F) labeled (time arrow, 400 s; Video 4). Kymographs were taken along the indicated red dotted lines. (G) Representative STICS curve of MLC-GFP dynamics and characteristic correlation length (inset, 2.8 ± 0.8 µm [mean ± SD]; n = 8). Red lines represent exponential fits. (H) Correlation times for myosin network mobility (60 ± 10 s [means ± SEM]; n = 14 [MHCA] and 9 [MLC]). (I) Apical myosin organization in wound-healing experiments. Examples are shown for an isotropic myosin network in a cell close to the wound (1) and more punctate signal of myosin in a cell within the sheet (2). Bars, 2 µm.
The apical myosin II network. (A–D) Apical organization of myosin II filaments visualized by labeling of myosin heavy chain II A (MHCA; A and C) or myosin light chain (MLC; B and D) with GFP. Whereas confluent cells exhibit punctate myosin II signals (C and D), nonconfluent MDCK cells form isotropic networks of myosin at their apical cell surface (A and B). (E and F) Dynamic rearrangement of myosin networks labeled with MHCA-GFP (E) or MLC-GFP (F) labeled (time arrow, 400 s; Video 4). Kymographs were taken along the indicated red dotted lines. (G) Representative STICS curve of MLC-GFP dynamics and characteristic correlation length (inset, 2.8 ± 0.8 µm [mean ± SD]; n = 8). Red lines represent exponential fits. (H) Correlation times for myosin network mobility (60 ± 10 s [means ± SEM]; n = 14 [MHCA] and 9 [MLC]). (I) Apical myosin organization in wound-healing experiments. Examples are shown for an isotropic myosin network in a cell close to the wound (1) and more punctate signal of myosin in a cell within the sheet (2). Bars, 2 µm.
To observe apical actin and myosin structures simultaneously, we generated stably transfected MDCK cells coexpressing Lifeact-mCherry and MHCA-GFP. Remarkably, actin and myosin did not show extensive colocalization, but rather formed a joint network covering the apical surface of cells, made up of alternating stretches of actin and myosin II (Fig. 6, A and B). Although overlap between the two filament systems was low, both actin and myosin exhibited coordinated isotropic motion (Fig. 6 C and Video 5) with similar spatial and temporal correlation coefficients (Fig. 6, D and E). Interconnection between actin and myosin was further confirmed by significant cross-correlation between the respective velocity fields at zero time shift (Fig. 6 F).
Actin–myosin colocalization. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of a nonconfluent MDCK cell double transfected with MHCA-GFP and Lifeact-mCherry. Bar, 2 µm. Frames were taken every 10 s for 15 min. Video to Fig. 6.
Actin–myosin colocalization. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of a nonconfluent MDCK cell double transfected with MHCA-GFP and Lifeact-mCherry. Bar, 2 µm. Frames were taken every 10 s for 15 min. Video to Fig. 6.
The apical actomyosin network. (A–C) Localization and dynamics of actin and myosin in MDCK cells stably transfected with Lifeact-mCherry and MHCA-GFP (Video 5). Both markers label distinct regions (A) that only rarely overlap (yellow in B). A kymograph (C) along the dotted line (overlay) shows correlated motion patterns for both structures. Bars, 2 µm. Time arrow, 400 s. (D and E) Correlation analysis showing similar temporal (D; actin: 53 ± 11 s; myosin: 57 ± 20 s [means ± SD]; n = 10) and spatial (E; actin: 2.1 ± 0.4 µm; myosin: 3.3 ± 0.6 µm [means ± SD]; n = 7) correlation of actin and myosin structures within the same cells. (F) Cross-correlation analysis of STICS velocity. The fact that the mean cross-correlation coefficient (cross-corr. coeff.) peaks at Δt = 0 s indicates interdependence of actin and myosin motion (n = 8; SDs). *, P < 0.05.
The apical actomyosin network. (A–C) Localization and dynamics of actin and myosin in MDCK cells stably transfected with Lifeact-mCherry and MHCA-GFP (Video 5). Both markers label distinct regions (A) that only rarely overlap (yellow in B). A kymograph (C) along the dotted line (overlay) shows correlated motion patterns for both structures. Bars, 2 µm. Time arrow, 400 s. (D and E) Correlation analysis showing similar temporal (D; actin: 53 ± 11 s; myosin: 57 ± 20 s [means ± SD]; n = 10) and spatial (E; actin: 2.1 ± 0.4 µm; myosin: 3.3 ± 0.6 µm [means ± SD]; n = 7) correlation of actin and myosin structures within the same cells. (F) Cross-correlation analysis of STICS velocity. The fact that the mean cross-correlation coefficient (cross-corr. coeff.) peaks at Δt = 0 s indicates interdependence of actin and myosin motion (n = 8; SDs). *, P < 0.05.
A cortical actomyosin network under mechanical tension
Although fluorescent actin markers, such as Lifeact-GFP, can be efficiently used to label bundled actin such as in MV, finer actin structures and individual actin filaments often cannot be distinguished against the high cytosolic background. To indirectly reveal the existence of myosin-associated, load-bearing actin structures within the plane of the apical plasma membrane, we tested whether the apical myosin network in nonconfluent ECs was being subjected to actin-dependent mechanical tension. When we disrupted apical actin structures by exposure to the actin-depolymerizing drug latrunculin A (2 µM), we found that within a few minutes, MV and myosin networks were disrupted at multiple sites and that the remnants of both structures aggregated in randomly dispersed clusters (Fig. 7, A–C; and Video 6), consistent with previous studies using low levels of actin drugs (Verkhovsky et al., 1997; Luo et al., 2013). The clustered MV could also be observed using SEM (Fig. 7 D). This behavior is consistent with the existence of a planar actomyosin network that is under isotropic tension. We confirmed this hypothesis by using laser ablation on MHCA-GFP–expressing cells, in which the myosin network clearly exhibited tension release when locally and rapidly severed (Fig. 7 E). Initial retraction of myosin fibers perpendicular to the cut occurred with 261 ± 51 nm/s (mean ± SD; n = 19), and this experiment also revealed that new myosin filaments were rapidly formed to fill the gap in the network (Fig. 7 E, kymograph).
Disruption of the cell cortex. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing disruption of the apical MV array (left, cells stably transfected with Lifeact-GFP) and myosin network (right, cells stably transfected with MHCA-GFP) at the apical surface of MDCK cells, upon treatment with 2 µM latrunculin A. Maximum projections of three (for actin, Δz = 1.5 µm) or two (for myosin, Δz = 1 µm) focal planes are shown. Bar, 2 µm. Frames were taken every 20 s for 15 min. Video to Fig. 7 (A and B).
Disruption of the cell cortex. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) showing disruption of the apical MV array (left, cells stably transfected with Lifeact-GFP) and myosin network (right, cells stably transfected with MHCA-GFP) at the apical surface of MDCK cells, upon treatment with 2 µM latrunculin A. Maximum projections of three (for actin, Δz = 1.5 µm) or two (for myosin, Δz = 1 µm) focal planes are shown. Bar, 2 µm. Frames were taken every 20 s for 15 min. Video to Fig. 7 (A and B).
Perturbation of network organization. (A–C) Changes in actin (A) and myosin (B and C) organization upon treatment with 2 µM latrunculin A (Video 6). Rupture of networks (C) and formation of clusters (A and B) are shown in selected cells over time. Arrows indicate ends of rupturing myosin filament. (D) Visualization of MV clustering with SEM. (E) Recoil of myosin filaments after scission by laser ablation demonstrates that the apical actomyosin network is under tension. Release of tension appears as the extended gap (arrows) in the kymograph (drawn along dotted line). Bars, 2 µm. Time arrow, 25 s. Times are in minutes (A, B, and D) or seconds (E).
Perturbation of network organization. (A–C) Changes in actin (A) and myosin (B and C) organization upon treatment with 2 µM latrunculin A (Video 6). Rupture of networks (C) and formation of clusters (A and B) are shown in selected cells over time. Arrows indicate ends of rupturing myosin filament. (D) Visualization of MV clustering with SEM. (E) Recoil of myosin filaments after scission by laser ablation demonstrates that the apical actomyosin network is under tension. Release of tension appears as the extended gap (arrows) in the kymograph (drawn along dotted line). Bars, 2 µm. Time arrow, 25 s. Times are in minutes (A, B, and D) or seconds (E).
If connectivity in the apical network relies largely on actomyosin interactions, we reasoned that it should be possible to unbalance the system instead of disrupting it, by only slightly reducing the amount of cortical actin filaments available. Indeed, after treatment with 500 nM latrunculin B for several minutes, apical myosin networks again began to rupture at several positions (Fig. 8 A). However, instead of clustering the remnants, the residual networks started to manifest massive long-distance rearrangements and oscillations (Fig. 8 A and Video 7). Interestingly, stress fibers at the basal cell surface (Fig. 8 A, light blue; and Video 7) were not affected by low latrunculin concentrations and remained static in spite of the fast rearrangements occurring within the apical network. These findings indicate that forces within apical and basal actomyosin assemblies are largely balanced within their respective structures and that local actomyosin assemblies are not necessarily connected into a cell-spanning mesh. In summary, our results are consistent with a joint isotropic 2D network made up of actin and myosin that covers the apical surface of ECs.
Unbalanced dynamics of myosin. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of a MDCK cell stably expressing MHCA-GFP, treated with 400 nM latrunculin B. Green, myosin at apical plane. Cyan, myosin at basal plane. Bar, 5 µm. Frames were taken every 10 s for 15 min starting 6 min after addition of latrunculin B. Video to Fig. 8 A.
Unbalanced dynamics of myosin. Time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) of a MDCK cell stably expressing MHCA-GFP, treated with 400 nM latrunculin B. Green, myosin at apical plane. Cyan, myosin at basal plane. Bar, 5 µm. Frames were taken every 10 s for 15 min starting 6 min after addition of latrunculin B. Video to Fig. 8 A.
Network connectivity and plasma membrane organization. (A) Destabilization of the apical actomyosin network in the presence of low concentrations of 500 nM latrunculin B. Drug treatment induces large-scale reorganization of apical myosin involving the coordinated motion of whole sections of the network (Video 7), whereas basal stress fibers remain unaffected (green: projection of apical 5 µm; cyan: basal focal plane). The kymograph was taken along the indicated red dotted line. (B) Mobility of fluorescent carboxylated beads (red) on the apical cell surface (green: Lifeact-GFP; maximum projection of five subsequent frames at 1 frame/s). (C) Collagen I–coated beads bind to MV and are guided by their motion (kymograph along dotted line). (D) MSD analysis reveals active transport (exponent > 1) of collagen I–coated beads attached to nonconfluent cells (non confl.; 11 cells, n = 426). In contrast, free diffusion (exponent = 1) is seen for carboxylated beads (7 cells, n = 422) and collagen I–coated beads on either nonconfluent cells treated with the integrin β1 inhibitory antibody AIIB2 (5 cells, n = 410) or confluent cells (confl.; 5 cells, n = 668). Beads in B–D had a 100-nm diameter. Results are given as box plots marking 25–75 percentile (boxes), median (lines), and mean value (small boxes). Whiskers indicate range of data points. ***, P < 0.001. (E) α5 Integrin–GFP is concentrated in actin-rich MV but shows rapid exchange, as demonstrated by FRAP analysis (bleached region is shown by the dotted ellipse). Bars: (A and E) 2 µm; (B and C) 1 µm. Time arrows: (A) 300 s; (C) 50 s. Time stamps are in minutes (A) and seconds (B and E).
Network connectivity and plasma membrane organization. (A) Destabilization of the apical actomyosin network in the presence of low concentrations of 500 nM latrunculin B. Drug treatment induces large-scale reorganization of apical myosin involving the coordinated motion of whole sections of the network (Video 7), whereas basal stress fibers remain unaffected (green: projection of apical 5 µm; cyan: basal focal plane). The kymograph was taken along the indicated red dotted line. (B) Mobility of fluorescent carboxylated beads (red) on the apical cell surface (green: Lifeact-GFP; maximum projection of five subsequent frames at 1 frame/s). (C) Collagen I–coated beads bind to MV and are guided by their motion (kymograph along dotted line). (D) MSD analysis reveals active transport (exponent > 1) of collagen I–coated beads attached to nonconfluent cells (non confl.; 11 cells, n = 426). In contrast, free diffusion (exponent = 1) is seen for carboxylated beads (7 cells, n = 422) and collagen I–coated beads on either nonconfluent cells treated with the integrin β1 inhibitory antibody AIIB2 (5 cells, n = 410) or confluent cells (confl.; 5 cells, n = 668). Beads in B–D had a 100-nm diameter. Results are given as box plots marking 25–75 percentile (boxes), median (lines), and mean value (small boxes). Whiskers indicate range of data points. ***, P < 0.001. (E) α5 Integrin–GFP is concentrated in actin-rich MV but shows rapid exchange, as demonstrated by FRAP analysis (bleached region is shown by the dotted ellipse). Bars: (A and E) 2 µm; (B and C) 1 µm. Time arrows: (A) 300 s; (C) 50 s. Time stamps are in minutes (A) and seconds (B and E).
Apical actomyosin dynamics influences lateral membrane organization
Dynamic MV emerge from the cortical actomyosin network as actin-filled membrane protrusions. Such protrusion could directly affect the lateral mobility of membrane-associated factors, either through changes in local topology and membrane composition or via interactions of membrane components with the underlying cytoskeletal elements.
To test this hypothesis, we first monitored lateral mobility of carboxylated and collagen (type I)-coated beads attached to the apical surface of MDCK cells. For robust quantification of bead motility, we automatically tracked individual beads (Fig. S4 A) and calculated their mean squared displacement (MSD; Fig. S4 B). With a mean MSD exponent of 1, carboxylated beads of 100-nm diameter that bound unspecifically to nonconfluent cells exhibited free diffusion, indicating that they were not significantly obstructed by dynamic MV (Fig. S4, A and B, black graph; and Video 8). In contrast, larger beads of 500-nm or 1-µm diameter frequently became stuck between surrounding MV and were therefore actively (MSD exponent > 1) moved along the cell surface via myosin-dependent forces (Fig. S4, A and B, red/blue graphs; and Video 8). Interestingly, we could also observe trapping of 100-nm beads between dynamic MV (Fig. 8 B and Video 9). However, reorganization of MV was rapid enough to not significantly obstruct diffusion of these beads (exponent in MSD analysis did not drop <1 as expected for restricted diffusion). For collagen I–coated beads, we found tight association with MV independent of bead size, reflected in active motion (Fig. 8 D and Fig. S4 C, bottom). As a consequence, beads closely followed the dynamic rearrangement of their MV attachment sites (Fig. 8 C and Video 10). We expected attachment of collagen I beads to MV to be mediated by binding to the main collagen I receptor, integrin α2β1. Indeed, incubation of cells with the β1-blocking antibody AIIB2 (Hall et al., 1990) led to a loss of bead–MV association and to a diffusion-characteristic MSD exponent of 1 (Fig. 8 D). In addition, collagen I beads attached to the apical surface of confluent cells exhibited random mobility (Fig. 8 D), in accordance with the fluctuations seen for short MV in these cells (Fig. 3 A). As β1 integrins are highly expressed in ECs, localization of integrin β1–GFP is difficult to interpret in the presence of endogenous protein (Parsons et al., 2008). To verify whether integrin localization supports our observations of MV-restricted beads, we instead expressed integrin α5–GFP in MDCK cells. We found that integrin α5–GFP was concentrated on dynamic MV (Fig. 8 E). However, in contrast to the immobilized collagen I beads, integrin α5–GFP showed rapid lateral mobility in FRAP experiments (Fig. 8 E).
Size-dependent confinement of beads. Carboxylated red fluorescent polystyrene beads (red) of indicated size were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Bar, 2 µm. Frames were taken every 2 s for 200 s. Video to Fig. 8 (B and C).
Size-dependent confinement of beads. Carboxylated red fluorescent polystyrene beads (red) of indicated size were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Bar, 2 µm. Frames were taken every 2 s for 200 s. Video to Fig. 8 (B and C).
Trapping of carboxylated beads. Carboxylated red fluorescent polystyrene beads (red) were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Frames were taken every 2 s for 200 s. Video to Fig. 8 B.
Trapping of carboxylated beads. Carboxylated red fluorescent polystyrene beads (red) were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Frames were taken every 2 s for 200 s. Video to Fig. 8 B.
Trapping of collagen beads. Collagen I–coated red fluorescent polystyrene beads (red) were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Bar, 2 µm. Frames were taken every 2 s for 200 s. Video to Fig. 8 C.
Trapping of collagen beads. Collagen I–coated red fluorescent polystyrene beads (red) were monitored by time-lapse epifluorescence microscopy on a customized iMIC setup (FEI) at the apical surface of nonconfluent MDCK cells stably transfected with Lifeact-GFP (green). Bar, 2 µm. Frames were taken every 2 s for 200 s. Video to Fig. 8 C.
We found a similar mechanism of kinetic trapping in dynamic MV of nonconfluent MDCK cells for the EGFR (Fig. S5 A). This was also reflected in the guided motion of labeled EGF (Fig. S5 B) and the resulting random distribution within restricted areas (Fig. S5 B, max). Upon binding to nonconfluent cells, both EGF and its receptor were rapidly taken up via endocytosis (Fig. S5 C). In contrast, EGF was hardly bound to the apical surface of confluent cells, where the EGFR was found nearly exclusively at the lateral surface or cell–cell contact regions (Fig. S5 D). Endocytic uptake was consequently minimal in these cells (Fig. S5 D). Collectively, our results strongly suggest that apical actin protrusions, MV motion, and actomyosin network dynamics have a significant effect on the lateral distribution—and function—of a variety of apical surface markers.
Discussion
In this study, we have shown that the apical surface of nonconfluent ECs is covered by elongated and pliable MV that undergo complex, myosin II–driven lateral displacements. Myosin II in these cells is organized into a distinctive, isotropic, and two-dimensional subsurface network of filaments that are interconnected with MV and laterally oriented actin filaments (Fig. 9 A). This apical actomyosin cortex exhibits characteristic coordinated motion, is under mechanical tension, and influences the distribution of various surface markers.
Models for apical actomyosin organization and membrane organization. (A) Hypothetical model for surface topology and actomyosin organization in nonconfluent ECs. Actin filaments (red) form either thick bundles in bent MV or thinner structures (single filaments or thin bundles) that are not resolved as such by fluorescence microscopy (shown here below the plane of the membrane). Myosin (minifilaments) filaments (green) form a two-dimensional network below the apical membrane and are connected to both individual actin filaments and MV (indicated by bent actin filaments at MV roots). The plasma membrane (transparent orange) is tightly connected to MV. Bent MV could consist of a single bundle of actin filaments or of multiple tightly connected bundles oriented orthogonally to the plasma membrane (example shown in the foreground). (B) Schematic depiction of three types of mechanisms by which apical actomyosin dynamics influences the mobility and distribution of plasma membrane components.
Models for apical actomyosin organization and membrane organization. (A) Hypothetical model for surface topology and actomyosin organization in nonconfluent ECs. Actin filaments (red) form either thick bundles in bent MV or thinner structures (single filaments or thin bundles) that are not resolved as such by fluorescence microscopy (shown here below the plane of the membrane). Myosin (minifilaments) filaments (green) form a two-dimensional network below the apical membrane and are connected to both individual actin filaments and MV (indicated by bent actin filaments at MV roots). The plasma membrane (transparent orange) is tightly connected to MV. Bent MV could consist of a single bundle of actin filaments or of multiple tightly connected bundles oriented orthogonally to the plasma membrane (example shown in the foreground). (B) Schematic depiction of three types of mechanisms by which apical actomyosin dynamics influences the mobility and distribution of plasma membrane components.
Bow-shaped MV are found at the apical surface of all of the EC lines that we tested and were easily visualized with several commonly used actin probes. Surprisingly, these structures have not featured prominently in the literature so far. One obvious reason for this is that many studies have focused on polarized and confluent EC, in which MV adopt the prototypical array organization that has traditionally been studied in the highly specialized and biochemically accessible brush border epithelium (Bretscher and Weber, 1978; Hirokawa et al., 1982; Drenckhahn and Dermietzel, 1988). However, scanning electron micrographs from the 1970s show elongated and bent apical protrusions in ECs cultured under specific conditions (Brunk et al., 1976; Bretscher and Weber, 1978; Chinkers et al., 1979). More recently, atomic force microscopy has been used to reveal ridgelike topological structures on the apical surfaces of MDCK and melanoma cells (Poole et al., 2004; Poole and Müller, 2005). In addition, ezrin and its adapter EBP50 have been shown to localize to bent apical protrusions in cultured ECs (Garbett et al., 2010; LaLonde et al., 2010). Importantly, protrusions with the characteristics of bent MV have also been described for epithelia that are undergoing reorganization in developing embryos (Löfberg, 1974; Rauzi et al., 2010).
The pliable form of curved MV in nonconfluent ECs stands in contrast to the stiffer rectilinear organization of actin filaments in classical MV, filopodia, or stress fibers (Chhabra and Higgs, 2007) and also differs markedly from the dendritic organization in lamellipodia (Pollard et al., 2000). Although we currently lack ultrastructural information, the high rate of actin subunit exchange in these MV indicates that individual MV contain bundles of short actin filaments that turn over rapidly. The curved shape of MV likely results from lateral pulling forces exerted by the underlying myosin II filaments. These forces are most probably also responsible for the lateral cortex reorganization implied by the frequent occurrence of fusion and fission events between adjacent MV. Importantly, we found that MV in nonconfluent ECs are mechanically coupled and undergo coordinated lateral motions with characteristic length and time scales. Such coordinated dynamics have so far not been described for individual cultured cells but are typical for networks of flexible polymers and resemble the tissue scale dynamics found in Drosophila melanogaster or Caenorhabditis elegans epithelia undergoing morphogenetic changes (Munro et al., 2004; Martin et al., 2009).
A major finding of our study is that an extensive, isotropic network of myosin filaments lines the inner apical surface of nonconfluent EC. Interestingly, this myosin network is discontinuous, with alternating stretches of filaments and gaps (Fig. 9 A). However, as indicated by the coordinated motions within the network and the collective motion observed in drug perturbation experiments, myosin filaments are likely connected through actin filaments that cannot be visualized against the high background signal of cytosolic actin probe (Fig. 9 A). Myosin II probes are therefore optimally suited to visualize lateral cell cortex organization. In addition, correlation analyses show that the lateral cortical actomyosin network is structurally interlinked with MV. This apical cortex organization constitutes a particularly interesting case of an active biopolymer network in which molecular motors use chemical energy to modify the topology of a semiflexible polymer mesh (Koenderink et al., 2009; Köhler et al., 2011). Such dynamics at the level of whole actin bundles could provide a very efficient mechanism for large-scale reorganization of the cortex, without direct connection to stable reference points, such as those provided by focal adhesions on the basal surface or adherens junctions at cell–cell contacts. In spite of its apparent organizational flexibility, the joint actomyosin network is under considerable tension, as becomes apparent after latrunculin A treatment or focal laser ablation.
Actin bundles in MV are typically connected to the plasma membrane via proteins of the ERM family and their membrane adaptors (Fehon et al., 2010). Hence, the localization of all major ERM proteins, as well as the adaptor EBP50, to bent MV suggests that, during myosin-driven lateral reorganization, actin bundles in MV remain closely associated with the apical plasma membrane. We were therefore not surprised to find that the apical actomyosin network and its dynamics have a direct influence on the mobility and lateral segregation of various plasma membrane markers. The single membrane-spanning receptor α5 integrin exhibits rapid lateral mobility within the apical membrane. However, despite its fast diffusion rate, α5 integrin was highly enriched on bent MV and followed MV dynamics via kinetic association (Fig. 9 B). Our results imply similar scenarios for β1 integrin and EGFR. In contrast, ligands of these kinetically trapped receptors, such as collagen-coated beads and chemically labeled EGF tightly associated with bent MV. This resulted in guided motion following the pattern of actomyosin network motility (Fig. 9 B). Finally, we observed restricted diffusion of beads that were not stably attached to the cell surface but became topologically trapped between adjacent MV (Fig. 9 B). In summary, the apical actomyosin network clearly affects the distribution of membrane markers, either through direct attachment and transport (Brangwynne et al., 2009) or by acting as a diffusion barrier and fence (Ritchie et al., 2003). Restriction of lateral motion through association with actin ridges or cytoskeletal picket fences has been demonstrated previously for several signaling factors (Suzuki et al., 2005; Liu et al., 2010; Jaqaman et al., 2011). Importantly, immobilization of membrane proteins on apical actin structures not only results in reduced lateral mobility of these factors but might at the same time facilitate intermolecular contacts and encounters via the observed active lateral motion of the actomyosin network. Indeed, the constant isotropic rearrangement of bent MV seems well suited to distribute and bring into contact those surface-bound factors, which have been immobilized and concentrated near MV. An exciting task for future studies will be to determine whether the observed actomyosin motion might serve to increase the efficiency of receptor dimerization, receptor–ligand interaction and ultimately signal transduction.
Irrespective of its potential role in membrane organization, the apical actomyosin network is ideally positioned to act as a mechanosensory element. It provides mechanical resistance to external forces, while at the same time being flexible enough to allow for structural adaptations upon mechanical or chemical stimulation during programmed morphogenetic changes. Such a role would be in keeping with the appearance of the actomyosin network in nonconfluent ECs that are still motile and are therefore exposed to a more varied environment. Furthermore, a mechanosensory function might not be limited to the cell cortex, as it has recently been proposed that mechanical forces acting on the cell nucleus directly influence transcriptional activities (Khatau et al., 2010; Mazumder and Shivashankar, 2010; Swift et al., 2013). A flexible, cagelike apical actomyosin network could offer the protection and adaptability required to maintain and control the stress exerted on the nucleus. Thus, elucidating the response of apical actomyosin to local or global changes in the mechanical environment is another exciting challenge for future research.
Materials and methods
Cell culture and transfection
MDCK, HeLa, HaCaT, and MCF-7 cells were grown in 25-cm2 tissue culture flasks (Falcon Company) at 37°C and 5% CO2 in DMEM-GlutaMAX-1 (Gibco) supplemented with 10% FBS (Gibco). For imaging of nonconfluent cells, 200–400 µl cells were seeded in 8-well µ-Slides (Ibidi) at 2 × 104 cells/ml and incubated for 48 h. For observation of confluent monolayers, cells were seeded at 4–5 × 105 cells/ml and incubated for 3–4 d.
All cell transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. To obtain stably transfected lines, cells were selected using 600 µg/ml hygromycin (Roche) and/or 200 µg/ml puromycin (Sigma-Aldrich) for 7–10 d, and selection pressure was maintained during cell culture. Antibiotics were, however, omitted during drug treatments and imaging. Lifeact, MHCA (human; MHC9), MLC (rat; MLC12B), α5 integrin (human; ITGA5), and actin (human β-actin; ACTB) were expressed from the cytomegalovirus promoter using pEGFP-N1 or pEGFP-C1 vectors (Takara Bio Inc.). EGFR-EYFP was expressed from the cytomegalovirus promoter using a pcDNA3.1-derived plasmid obtained from Vibor Laketa (European Molecular Biology Laboratory) and used as previously described (Offterdinger et al., 2004).
Cells were treated with 50 µM blebbistatin (Invitrogen) to inhibit myosin II ATPase activity and with 2 µM latrunculin A (EMD Millipore) or 500 nM latrunculin B (Enzo Life Sciences) to sequester actin monomers. HGF (HumanZyme) was added to a final concentration of 100 ng/ml, and cells were imaged after the indicated incubation times. EGF complexed to Alexa Fluor 555 (Life Technologies) was added to the cells directly before experiments at a final concentration of 2 µg/ml. Uptake of EGF by EGFR-EYFP–expressing MDCK cells was monitored every 20 s for a total length of 30 min.
For wound-healing assays, 6 × 104 cells were seeded on both sides of a 35-mm µ-Dish wound-healing culture insert (Ibidi) and incubated for 24 h. After removal of the insert, cells were again incubated for 16–24 h before imaging. For shear-flow experiments, 5 × 103 cells were seeded in flow chambers (µ-Slide0.2 Luer; Ibidi), incubated for 48 h, and then connected to the pump system (Ibidi), with the perfusion set containing DMEM. Cells were subjected to 20 dyn/cm2 oscillatory shear stress at 0.2 Hz.
Antibodies and beads
Commercial antibodies for immunofluorescence and Western blot analyses used in this study were rabbit anti–nonmuscle myosin IIA (Novus Biologicals), mouse anti–nonmuscle myosin IIA antibody (Abcam), rabbit anti-radixin (Abcam), and mouse anti-moesin (Abcam). Two further antibodies were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development, and maintained by the Department of Biology at the University of Iowa (Iowa city, IA): mouse anti-ezrin (CPTC–Ezrin-1, developed at the National Cancer Institute, Bethesda, MD) and mouse anti–integrin β1 (AIIB2, developed by C.H. Damsky, University of California, San Francisco, San Francisco, CA).
For surface-labeling experiments, carboxyl-coated, red fluorescent polystyrene beads with diameters of 100, 500, or 1,000 nm were used (FluoSpheres; Invitrogen). To induce binding to integrin α2β1, beads were incubated overnight at 4°C in 2 mg/ml type I rat tail collagen solution (PureCol 100 type I collagen; Nutacon). Subsequently, beads were collected by centrifugation for 30 min at 12,000 g. Before use, beads were diluted in PBS, sonicated, and added to the cells for 15 min. Unbound beads were removed by washing with imaging buffer before the experiment.
To inhibit binding of collagen-coated beads, cells were treated with the integrin β1 inhibitory antibody AIIB2 at 1:10 dilution of the supernatant. The antibody was added to the cell culture medium 15 min before addition of collagen-coated beads and was present throughout the experiment.
Immunofluorescence
For immunofluorescence staining, cells were grown on glass coverslips, fixed with 3.2% paraformaldehyde in PBS for 20 min, washed in PBS, permeabilized in 0.3% Triton X-100 in PBS for 5 min, and blocked with 1.25% gelatin and 0.05% saponin in PBS for 20 min before incubation with primary antibody for 1 h. After washing in PBS and incubation in secondary antibodies and phalloidin (Alexa Fluor goat anti–rabbit/mouse [A11008/A11029; Invitrogen] and rhodamine-phalloidin [R415; Invitrogen]) for 1 h in 1.25% gelatin and 0.05% saponin in PBS, cells were washed again in PBS before mounting in Mowiol/DABCO (Roth).
Western analysis
For detection of MHCA in Western blots, equal amounts of cell lysates were separated by SDS-PAGE, transferred to Immobilon-P-membrane (Serva), incubated in primary antibody in the presence of 5% skim milk in TBS-T (TBS with Tween 20) overnight, and labeled with HRP-coupled secondary antibodies.
Transmission EM and SEM
Transmission EM of MDCK cells was performed as described previously (Keil and Steinbrecht, 2010). In brief, cells were fixed for 30 min at 40°C in 0.1 M cacodylate buffer containing 2.5% glutaraldehyde (Sigma-Aldrich) and 5% sucrose at pH 7.2. Cells were then incubated for 30 min at 40°C in cacodylate buffer containing 1% tannic acid. After removal of the fixative, cells were incubated for 1 h at 40°C in 0.1 M cacodylate buffer containing 1% OsO4, washed with double distilled H2O, and stained overnight with 1% aqueous uranyl acetate solution at 60°C. Cells were dehydrated in a graded ethanol series and embedded in Spurr’s medium. Sections (100 nm) were cut using a DiATOME diamond knife on a microtome (Ultracut; Reichert), stained for 30 min in 1% aqueous uranyl acetate solution, and imaged at 120 kV on an electron microscope (CM120; Philips FEI) equipped with a camera (MegaView; Olympus).
For SEM, MDCK cells were incubated for 45 min in 0.1 M phosphate buffer containing 2.5% glutaraldehyde. Subsequently, after three 10-min washing steps in 0.1 M phosphate buffer, cells were subjected to dehydration by successive incubations in 20, 40, 60, 80, and 99% ethanol (EtOH 99%; Sigma-Aldrich) for 10 min each and kept in 99% EtOH overnight. Cells were then dehydrated using a critical point dryer (Polaron E3000; Polaron Instruments) and the standard drying protocol. Cell surfaces were sputter coated with gold (108auto; Cressington Scientific) to a thickness of 6-nm gold and imaged on a scanning electron microscope (SM300; Topcon) at 10–15 kV.
Fluorescence microscopy
Epifluorescence imaging of GFP and RFP fusion proteins was performed on a fully automated iMIC-based microscope from FEI, using an Olympus 100×, 1.4 NA objective and diode-pumped solid-state lasers at 491 nm (75 mW; Calypso; Cobolt) and 561 nm (150 mW; Jive; Cobolt) as light sources. Lasers were selected through an acousto-optical tunable filter and directed through a broadband fiber to the microscope. A galvanometer-driven two-axis scan head was used to adjust laser incidence angles. Images were collected using a camera (Imago-QE SensiCAM; PCO AG). Acquisition was controlled by LiveAcquisition software (TILL Photonics). FRAP of actin-GFP was performed using a third galvanometer-controlled mirror (Polytrope) to switch between wide-field and FRAP modalities.
Confocal microscopy was performed on a spinning-disk system (UltraView Vox; PerkinElmer) based on an inverted microscope (DMI6000 B; Leica) and equipped with 488- and 561-nm lasers, a 100×, 1.47 NA objective (Leica), and a charge-coupled device camera (ImagEM; Hamamatsu Photonics). Image acquisition was controlled by the Volocity 3D software (PerkinElmer).
Ablation of MLC-GFP filaments was performed either on a confocal microscope (FV1000; Olympus) equipped with a 100×, 1.42 NA objective and a pulsed 405-nm picosecond UV laser (Sepia; PicoQuant) or on the aforementioned iMIC-based microscope equipped with a pulsed UV laser (DPSL-355/14; Rapp OptoElectronic). All experiments were performed as previously described (Raabe et al., 2009). In brief, cortical myosin filaments were cut by the pulsed UV laser without damaging the plasma membrane or other intracellular compartments. Initial retraction, as well as the subsequent stabilization, was acquired for the following 30 s at a frame rate of 200 ms. Retraction speeds were calculated from kymographs generated perpendicular to the cut axis. The linear slope of filament traces within the first second after cutting was used to quantify angles and velocities of the retracting filaments. For all measurements, cells were maintained in DMEM at constant temperature (37°C), CO2 level (5%), and humidity.
Image processing and analysis
All images were processed using either Fiji (National Institutes of Health), MATLAB (MathWorks, Inc.), or Volocity 3D (PerkinElmer). Images were contrast adjusted and zoomed for purposes of presentation in the figures only. For image cleanup and denoising, we used either the background subtraction algorithm in Fiji (rolling ball and radius of 50 pixels) or the Block Matching 3D filter (block size of 8 × 8 pixels; Dabov et al., 2007) followed by a local top-hat filter (radius of 5 pixels) in MATLAB (CODE 3). For object density measurements, a MATLAB algorithm was applied to image regions of 150 × 150 pixels. After initial background subtraction and denoising, images were binarized using twice the image mean intensity value as a threshold. Detected objects were thinned to skeletons, single-pixel objects were removed, and the number of objects was counted (CODE 1).
A simple Pearson correlation coefficient was used to calculate similarities between pairs of frames from fluorescence microscopy time-lapse videos to quantify the time scale of structural reorganization. The Pearson correlation coefficient r between two images X and Y is defined as
Here, the subscript denotes the ith pixel of the image, and brackets denote the mean over all pixels in the image. For a given video, we calculated the Pearson correlation coefficient r(X0,Xt) = r(t) of the first frame X0 with each consecutive frame in {Xt} to measure their similarity. Image denoising was applied as described in the previous paragraph, and full images were cut to a region of interest of 250 × 250 pixels before calculating the Pearson correlation coefficient. As r(t) decays over time, corresponding to a structural reorganization, a single exponential decay fit with constant offset was used to determine the characteristic correlation time.
STICS measures the mean velocity of a labeled mobile target in an image template by fitting a Gaussian distribution to the spatiotemporal correlation function (Hebert et al., 2005). Velocity fields were obtained on a uniform grid by performing STICS analyses on neighboring subimage series in space and time. For each grid point (x,y,t) in a given video, STICS analysis was performed on a spatial window {x ± 2dx, y ± 2dy}, and a temporal window of t ± 2dt with the spatial widths of dx = dy = 6 pixels and a temporal width of dt = 8 frames. Nearest-neighbor velocities on the STICS grid have overlapping averaging windows in both space and time, which results in systematically correlated velocities. Thus, nearest-neighbor pairs of velocities were excluded from all correlation analyses to evaluate spatiotemporal correlations in cytoskeletal dynamics using independent values only.
Spatial correlations in STICS velocity fields for a given time-lapse video were quantified using the spatial correlation function (SCF). Following Angelini et al. (2010), the SCF of a vector field v(r,t) at time t is given by
Here, v(r,t) is the velocity vector at the STICS grid position r at time t, and brackets denote averaging over the subscripted variable. ΦR denotes the orientation of R with respect to an arbitrary reference angle. The SCF removes mean drift from the raw STICS velocity vectors. The SCF was fitted with a single exponential decay only considering points out of the STICS window to extract the correlation length scale R0.
Temporal correlations between actin and myosin channels were quantified using the temporal correlation function (TCF):
Here v1 and v2 denote actin and myosin motion fields, respectively, and τ denotes the lag time. TCF1,2(0) equals the mean cross-correlation of the actin and myosin motion fields with no temporal offset (i.e., the mean local flow of the myosin motion field along the direction of the actin field).
MSD analysis was performed using a MATLAB-based algorithm to binarize maximum-projected time-lapse videos of beads on apical surfaces of MDCK cells (three planes; Δz = 1 µm) using adaptable thresholding parameters (CODE 2) and consecutively calculating the center of mass for each binarized object with an x and y diameter <25 pixels (CODE 4). Particles were subsequently tracked using ImageJ Particle Tracker (radius: 3; cutoff: 3; percentile: 0.1%; link range: 2; displacement: 5; National Institutes of Health; Sbalzarini and Koumoutsakos, 2005). All traces longer than 30 frames were analyzed in Origin9.1 (OriginLab) with fitting function y = a · xb (allometric 1). Only MSD exponents b in the range of 0.1 ≤ b ≤ 3 were considered and plotted as box plots marking the 25–75 percentile (box), median (line), and mean value (small box). Reading and writing of TIFF files from MATLAB was performed using customized programs (CODES 5 and 6).
Statistics
Mean values, number of measurements (n), and SD or SEM are provided for all quantified results. Error bars in graphs are explained in the respective legends. Statistical comparison between conditions was performed using the unpaired t test with Welch correction.
Online supplemental material
Fig. S1 shows apical actomyosin organization in different EC lines and with different markers. Fig. S2 shows colocalization of actin with ERM proteins. Fig. S3 shows expression levels and localization of myosin heavy chain and MLC in transfected and untransfected MDCK cells. Fig. S4 shows the procedure used to calculate MSD of beads attached to the apical surface of MDCK cells and control results for beads of different sizes. Fig. S5 shows localization and internalization of EGFR-YFP and EGF–Alexa Fluor 555 in unstimulated and EGF-stimulated MDCK cells. Video 1 shows MV dynamics in confluent cells. Video 2 shows MV dynamics in nonconfluent cells. Video 3 shows examples of MV reorganization. Video 4 shows apical myosin dynamics in MDCK cells. Video 5 shows colocalization of actin and myosin at the apical surface of MDCK cells. Video 6 shows the disruption of the apical MV array and myosin network. Video 7 shows unbalanced myosin dynamics upon treatment with low concentrations of latrunculin B. Video 8 shows size-dependent confinement of carboxylated beads at the apical surface of MDCK cells. Video 9 shows dynamic trapping of carboxylated beads at apical surface of MDCK cells. Video 10 shows trapping of collagen I–coated beads on apical MV. A ZIP file is also included that provides six MATLAB source codes. Code 1 provides the MATLAB source code for object density analysis. Code 2 provides the MATLAB source code for MSD analysis. Code 3 provides the MATLAB source code for image filtering. Code 4 provides the MATLAB source code for center of mass determination of beads. Code 5 provides the MATLAB source code for reading LiveAcquisition TIFF files. Code 6 provides the MATLAB source code for writing TIFF files.
Acknowledgments
We thank Paul Hardy, Carsten Grashoff, and Christian Schuberth for comments on the manuscript.
This work for supported by the Max Planck Society and the German Research Foundation via the SFB863 and the Cells-in-Motion Cluster of Excellence (EXC1003–CiM, University of Münster, Münster, Germany).
The authors declare no competing financial interests.
References
Author notes
C. Klingner and A.V. Cherian contributed equally to this paper.




![Figure 4. Myosin-dependent motion of apical actin. (A and B) Kymographs along dotted lines in A reveal coordinated motion of MV (B). Arrows indicate correlated motion of neighboring MV (B, 1) and noncorrelated motion of distant MV (B, 2). Time arrows, 500 s. White dotted lines represent viewing guides to better follow trace evolutions. (C) STICS correlation analysis with characteristic spatiotemporal correlation length (inset: 3.3 ± 0.9 µm [mean ± SD]; n = 11). Red lines represent exponential fits. (D) Calculated STICS flow field represented by velocity vectors on a uniform grid. Schematic indicates velocity vectors and correlation length outlined by a radial gradient circle. Arrows indicate direction and magnitude of flow vectors. (E and F) Effect of inhibition of myosin II activity on correlation time in actin reorganization. Sample curves (E) and mean Pearson correlation coefficients (F) are shown for control cells (blue, 62 ± 20 s [mean ± SEM]; n = 8) and cells treated with 50 µM blebbistatin (red, 156 ± 60 s [mean ± SEM]; n = 12). Black lines represent exponential fits. (G) Kymographs from cells analyzed in E and F showing that inhibition of myosin II blocks lateral reorganization. Time arrows, 300 s. Squares, triangles, and circles indicate data points for three representative cells. Bars: (A, B, and G) 2 µm; (D) 1 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/207/1/10.1083_jcb.201402037/7/m_jcb_201402037_fig4.jpeg?Expires=1771336211&Signature=GWfPTzNnxaTN-HBN7gDPX1G3g7jAVZ1s4Ib6HcKG1I4mGjU0Qva2bEnwuNaI9PyBlHSP2kCLfSUCckgBO87-Hd60F2CCu9d-u9~qBDnmbQLNocNNkFZI5qr8LcyQb8OHaZ5mtzv8Dr2BKHTcdGwHxnSS0h8VCEzfk98N2WGFBG9al5OKq7k6Ry0qOeo13k64aRp~34mmtzoxHcEWmsHMpA065zS7SE6Lumi77a8ga4f4GaqstO950iEMDJNY-eXJ7r3IJ8fJUo8m0YE-Wa7NnkjQlDzKeYocHwYe~VGjwkJiAdPxDu1MPayIDDYeGw4ncaWsjHVZ3xZteb~NNfUseQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. The apical myosin II network. (A–D) Apical organization of myosin II filaments visualized by labeling of myosin heavy chain II A (MHCA; A and C) or myosin light chain (MLC; B and D) with GFP. Whereas confluent cells exhibit punctate myosin II signals (C and D), nonconfluent MDCK cells form isotropic networks of myosin at their apical cell surface (A and B). (E and F) Dynamic rearrangement of myosin networks labeled with MHCA-GFP (E) or MLC-GFP (F) labeled (time arrow, 400 s; Video 4). Kymographs were taken along the indicated red dotted lines. (G) Representative STICS curve of MLC-GFP dynamics and characteristic correlation length (inset, 2.8 ± 0.8 µm [mean ± SD]; n = 8). Red lines represent exponential fits. (H) Correlation times for myosin network mobility (60 ± 10 s [means ± SEM]; n = 14 [MHCA] and 9 [MLC]). (I) Apical myosin organization in wound-healing experiments. Examples are shown for an isotropic myosin network in a cell close to the wound (1) and more punctate signal of myosin in a cell within the sheet (2). Bars, 2 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/207/1/10.1083_jcb.201402037/7/m_jcb_201402037_fig5.jpeg?Expires=1771336211&Signature=zK-to6Fybefup~ibuuKHbQ55G6E8sw6xhuUund39jtZNXntYm0IvQxiXSjdRoLI12k1G5ZjWuWw8SJ~atX0IwEA6pjew-iB--ishDKXFIWFo04wcMRWp2UOY1C81qLrmlm8jdbDf~jSvAcFJ15SXvBX7e62ApaNWqbHY8Hj5u34zWv4-rgRPgdCksKk9IFglx7v7nnPei~vBPG1hRLIIAPr3WQaY8SD7bQnyKJ6AnS3T-~HHt0SXgEI3Wl9LelTXMSa-QvkSQVNtPWD15d59jPaplCVdr1f1Mf8PwT6ugXdJ~G83jDcQhG6xlOmnFH8W9lRoJa8FMnBE8eyF113new__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. The apical actomyosin network. (A–C) Localization and dynamics of actin and myosin in MDCK cells stably transfected with Lifeact-mCherry and MHCA-GFP (Video 5). Both markers label distinct regions (A) that only rarely overlap (yellow in B). A kymograph (C) along the dotted line (overlay) shows correlated motion patterns for both structures. Bars, 2 µm. Time arrow, 400 s. (D and E) Correlation analysis showing similar temporal (D; actin: 53 ± 11 s; myosin: 57 ± 20 s [means ± SD]; n = 10) and spatial (E; actin: 2.1 ± 0.4 µm; myosin: 3.3 ± 0.6 µm [means ± SD]; n = 7) correlation of actin and myosin structures within the same cells. (F) Cross-correlation analysis of STICS velocity. The fact that the mean cross-correlation coefficient (cross-corr. coeff.) peaks at Δt = 0 s indicates interdependence of actin and myosin motion (n = 8; SDs). *, P < 0.05.](https://cdn.rupress.org/rup/content_public/journal/jcb/207/1/10.1083_jcb.201402037/7/m_jcb_201402037_fig6.jpeg?Expires=1771336211&Signature=GgkrCl9qNBDNP6jxUE8cG~04TEWdFCiZKoEt6TBj8QeFRTC~0l2ZhFGC90wFrJGW2OHT81yjYGi8kaUC5HBjBTPjeqzSXPxS9dlZ~UOsQ1aNtHqm-IdcBNf3PIAX-5Izs-CMirREYp2EAVl-3e~D-zm89YBJGZOfFLoawK6BDOf4FVfiJ15vPCFTSBGHDOM0Ll-UfINGmJRL1Vti4FrYzHzDoh63Ysnq1aAUDxe4NnjjWQn6BGx5F1mwd~805bT5kRBbt56bDZOttMW3GhpcHdru3Is3rDfZm7SimVBJ39bgeJCSGuPiKLWV35tVHkVkzttJMKJZiOJpf9IbqGqKsg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





