The mechanisms by which tumor cells metastasize and the role of endocytic proteins in this process are not well understood. We report that overexpression of the GTPase RAB5A, a master regulator of endocytosis, is predictive of aggressive behavior and metastatic ability in human breast cancers. RAB5A is necessary and sufficient to promote local invasion and distant dissemination of various mammary and nonmammary tumor cell lines, and this prometastatic behavior is associated with increased intratumoral cell motility. Specifically, RAB5A is necessary for the formation of invadosomes, membrane protrusions specialized in extracellular matrix (ECM) degradation. RAB5A promotes RAB4- and RABENOSYN-5–dependent endo/exocytic cycles (EECs) of critical cargos (membrane-type 1 matrix metalloprotease [MT1-MMP] and β3 integrin) required for invadosome formation in response to motogenic stimuli. This trafficking circuitry is necessary for spatially localized hepatocyte growth factor (HGF)/MET signaling that drives invasive, proteolysis-dependent chemotaxis in vitro and for conversion of ductal carcinoma in situ to invasive ductal carcinoma in vivo. Thus, RAB5A/RAB4 EECs promote tumor dissemination by controlling a proteolytic, mesenchymal invasive program.
Introduction
In the initial invasive phase, cancer cells migrate through the basement membrane and through different types of stromal ECM. These 3D structures display diverse physicochemical properties that, while providing a substrate for adhesion and traction, also impose different degrees of mechanical resistance (Friedl and Alexander, 2011). Cancer cells confront these diverse migratory environments by adopting flexible invasive strategies (Friedl and Wolf, 2010). In one such strategy, referred to as “mesenchymal motility,” invasion is achieved by coupling polarized actin-based protrusions with spatially restricted pericellular proteolytic activity in both migrating cells and reactive stromal cells (Egeblad et al., 2010; Kessenbrock et al., 2010). Invasion can also be achieved, however, in a protease-independent fashion (amoeboid motility; Madsen and Sahai, 2010). Although the physiological relevance of amoeboid motility has been questioned (Sabeh et al., 2009), it is likely that both proteolytic- and nonproteolytic modes of invasion exist and cooperate during migration of cancer cells (Wolf et al., 2003). The resulting migration “plasticity” is thought to contribute to the diverse array of cancer invasion routes and programs, tumor heterogeneity, and, ultimately, metastatic dissemination.
Podosomes and invadopodia, collectively defined as invadosomes (Linder et al., 2011), are actin-rich, adhesive protrusions that degrade the ECM via the directed release of proteases (Tarone et al., 1985; Linder et al., 2011). The delivery of the membrane-type 1 matrix metalloprotease (MT1-MMP) to invadosomes is critical for their formation and functionality (Hotary et al., 2003, 2006; Itoh and Seiki, 2006). MT1-MMP delivery to invadosomes can be achieved by its polarized secretion in response to the activation of cell-adhesion receptors (Poincloux et al., 2009), through recycling from late endosomal compartments (Steffen et al., 2008; Yu et al., 2012; Monteiro et al., 2013), and by exosome release (Hoshino et al., 2013).
Some motogenic growth factors, such as EGF and hepatocyte growth factor (HGF), induce invadosomes in a transient and polarized fashion within minutes of stimulation (Yamaguchi et al., 2005; DesMarais et al., 2009; Frittoli et al., 2011). Under these conditions, the cell must interpret the signal in a limited time frame and simultaneously enact several spatially restricted programs leading to actin polymerization, extension of migratory protrusions, and delivery of adhesion molecules and proteases, first and foremost MT1-MMP. It seems reasonable to postulate the existence of a master regulator that orchestrates this sequence of events.
RAB5, a GTPase pivotal in endocytosis (Zerial and McBride, 2001; Zeigerer et al., 2012), is a fitting candidate for this role. We previously demonstrated that RAB5-dependent endocytic/exocytic cycles (EECs) of the small GTPase RAC1 are sufficient to promote: (1) the spatial restriction of RAC1 signaling, leading to the formation of polarized migratory protrusions; (2) elongated cell migration and increased cell velocity; (3) an amoeboid-to-mesenchymal (AMT) switch in the mode of migration; and (4) the acquisition of invasive potential by different tumor cell types (Palamidessi et al., 2008). Here, we report that elevated expression of RAB5A, one of three functionally redundant RAB5 genes, is predictive of increased local and distant relapse in early stage estrogen receptor–positive (ER+), lymph node–negative (N0) breast cancer patients. RAB5A expression is significantly elevated in lymph node metastases with respect to matched human primary breast tumors. At the molecular level, RAB5A promotes RAB4-dependent fast recycling of β3 integrin and MT1-MMP, leading to invadosome formation, degradation, and remodeling of the ECM. These processes are, in turn, crucial for local tumor invasion and dissemination to distant organs. We propose that a RAB5A/RAB4 recycling route is central in promoting proteolytic/mesenchymal invasive programs in human breast cancer.
Results
RAB5A expression is predictive of clinical outcome in breast cancer patients
RAB5 expression is sufficient to promote a mesenchymal mode of cell invasion (Palamidessi et al., 2008). Individual ablation of the three human RAB5 genes (RAB5A/B/C) has also been reported to impair invasion and dissemination of different types of cancer cells (Yu et al., 1999; Torres et al., 2010; Torres and Stupack, 2011; Liu et al., 2011; Onodera et al., 2012; Mendoza et al., 2013; Diaz et al., 2014). However, a recent report indicates that in normal macrophages RAB5A is dispensable for matrix degradation (Wiesner et al., 2013). We thus investigated whether RAB5 could be directly linked to naturally occurring human tumors and to metastasis.
We initially meta-analyzed five published human breast tumor gene expression datasets containing 980 primary breast cancers. This analysis revealed that overexpression of RAB5A, but not RAB5B or RAB5C (not depicted), correlates with poor prognosis (P = 0.031; Fig. 1 A). The prognostic power of RAB5A became progressively more significant in the following subgroups of patients: lymph node negative (N0; P = 0.01); N0 and ER+ (P = 0.003); and grade 2 (G2), N0, ER+ (P = 0.0004) breast tumor patients (Fig. 1, B–D). RAB5A prognostic significance was independent from the proliferation marker Ki67 as assessed in multivariate COX analysis (Fig. 1 E). Additionally, RAB5A levels do not co-vary with the signature associated with proliferation as determined by gene set enrichment analysis (GSEA; Fig. 1 E).
RAB5A predicts poor clinical outcome in breast cancer patients and its expression is elevated in breast cancer lymph node metastases. (A–D) Kaplan–Meier plots for breast cancer patients stratified by RAB5A expression for the following groups of patients: (A) all 980 patients; (B) 695 patients with lymph node negative (N0) breast tumors; (C) 528 patients with lymph node–negative and ER-positive (N0 ER+) breast tumors; (D) 166 patients with lymph node–negative, ER-positive, and Grade 2 (N0 ER+ G2) breast tumors. High RAB5A, >75th percentile; low RAB5A, ≤75th percentile. Y axes = the probability of local and distant relapse free survival (DFS). X axes = years of follow up. P-values are from a log-rank test. (E) RAB5A prognostic significance is independent from Ki67, and its expression is not associated with a proliferation signature. (E, left) Multivariate analysis with a Cox proportional hazard model on prognosis of patients with breast cancer. The RAB5A and Ki67 genes were used as covariates. High RAB5A or Ki67 expression levels, >75th percentile of the normalize expression distribution. Low RAB5A or Ki67 expression levels, ≤75th percentile. HR, hazard ratio by Cox model. 95% CI, 95% confidence intervals of HR. P-value, likelihood ratio χ2 test on the null hypothesis that the parameter estimate for the RAB5A high/low, or Ki67 high/low, covariate is zero. (E, right) Gene set enrichment analysis (GSEA) analysis of a known proliferation signature (Ben-Porath et al., 2008) among genes ranked by signal-to-noise metric based on their correlation with Ki67 high/low or RAB5A high/low. FDR, false discovery rate based on 1,000 random permutations of class labels. (F) IHC staining of RAB5 on FFPE samples of a human primary infiltrating breast tumor (PT) and matched nodal metastasis (NM). N, normal duct; L, normal negative lymphocytes. Boxed regions are enlarged below. Bar, 500 µm. (G) Percentage of low and high RAB5A-expressing primary tumors and metastases. Normal breast tissue scores always range between 0 and 1. **, P < 0.000271 (χ2 test).
RAB5A predicts poor clinical outcome in breast cancer patients and its expression is elevated in breast cancer lymph node metastases. (A–D) Kaplan–Meier plots for breast cancer patients stratified by RAB5A expression for the following groups of patients: (A) all 980 patients; (B) 695 patients with lymph node negative (N0) breast tumors; (C) 528 patients with lymph node–negative and ER-positive (N0 ER+) breast tumors; (D) 166 patients with lymph node–negative, ER-positive, and Grade 2 (N0 ER+ G2) breast tumors. High RAB5A, >75th percentile; low RAB5A, ≤75th percentile. Y axes = the probability of local and distant relapse free survival (DFS). X axes = years of follow up. P-values are from a log-rank test. (E) RAB5A prognostic significance is independent from Ki67, and its expression is not associated with a proliferation signature. (E, left) Multivariate analysis with a Cox proportional hazard model on prognosis of patients with breast cancer. The RAB5A and Ki67 genes were used as covariates. High RAB5A or Ki67 expression levels, >75th percentile of the normalize expression distribution. Low RAB5A or Ki67 expression levels, ≤75th percentile. HR, hazard ratio by Cox model. 95% CI, 95% confidence intervals of HR. P-value, likelihood ratio χ2 test on the null hypothesis that the parameter estimate for the RAB5A high/low, or Ki67 high/low, covariate is zero. (E, right) Gene set enrichment analysis (GSEA) analysis of a known proliferation signature (Ben-Porath et al., 2008) among genes ranked by signal-to-noise metric based on their correlation with Ki67 high/low or RAB5A high/low. FDR, false discovery rate based on 1,000 random permutations of class labels. (F) IHC staining of RAB5 on FFPE samples of a human primary infiltrating breast tumor (PT) and matched nodal metastasis (NM). N, normal duct; L, normal negative lymphocytes. Boxed regions are enlarged below. Bar, 500 µm. (G) Percentage of low and high RAB5A-expressing primary tumors and metastases. Normal breast tissue scores always range between 0 and 1. **, P < 0.000271 (χ2 test).
Next, we examined the expression of RAB5A in a panel of normal human mammary gland tissues (N), primary human breast cancers (T), and their matched lymph node metastases (NM; n = 5, T = 35, M = 35). Immunohistochemical (IHC) analysis revealed that RAB5A was expressed at low levels in normal mammary epithelial, whereas in tumor and lymph node metastasis, its expression varied from barely detectable to highly expressed (Fig. 1 F). Importantly, RAB5A expression was significantly higher in matched lymph node metastases, with respect to their primary tumors (Fig. 1 G, P < 0.000271 χ2 test.). The up-regulation of RAB5A levels between each patient’s primary tumor and node metastasis is also highly significant when calculated with the Wilcoxon signed-rank test (P = 0.0055). Thus, RAB5 overexpression is an event that is selected for during breast cancer progression, possibly because it confers a migratory advantage to tumor cells.
RAB5A is required for local and distant dissemination of breast cancer in vivo
We next established relevant cell model systems to study the involvement of RAB5A in local invasiveness and distant metastatic dissemination. We chose the triple-negative breast cancer MDA-MB-231 and MCF10.DCIS.com cell lines. The former cells are invasive, disseminate to distant organs (Kang et al., 2003), and display relatively elevated levels of RAB5 expression when compared with a panel of breast cancer cell lines (Fig. S1 A). In contrast, MCF10.DCIS.com cells recapitulate features of comedo-type ductal carcinoma in situ (DCIS), upon injection into immunodeficient mice (Miller et al., 2000). Because the three RAB5 genes are functionally redundant (Zeigerer et al., 2012) and essential for mitotic progression (Capalbo et al., 2011; Serio et al., 2011), we generated stable MDA-MB-231 and MCF10.DCIS.com cell lines that express a dominant-negative RAB5A (RAB5AS34N) isoform in a doxycycline-inducible fashion.
We xenografted RAB5AS34N and control MDA-MB-231 cells into mammary fat pads of immunocompromised mice. After 3 wk, mice were fed with a doxycycline to induce the expression of the transgene, which was detected by immunoblotting (Fig. S1 B). We then analyzed local ipsilateral metastasis (Fig. 2 A) and lung metastases after removal of the primary tumor (Fig. 2 B). The sizes of control and RAB5AS34N-expressing primary tumors were indistinguishable (Fig. S1 B). Conversely, there was a significant decrease in both the number and size of local and distant metastatic outgrowths in mice injected with RAB5AS34N-MDA-MB-231 cells compared with mice injected with control cells (Fig. 2, A and B).
RAB5A is required for tumor dissemination and for the conversion from DCIS to infiltrating mammary carcinoma. (A and B) Doxycycline-inducible RAB5AS34N- and control MDA-MB-231 cells were injected into the mammary fat pads of NSG mice. After 3 wk, mice were fed with doxycycline. Metastases were analyzed 4 wk after removal of primary tumor. (A) Ipsilateral metastasis (arrowhead) of control or RAB5AS34N-MDA-MB-231 tumors. (A, right) Quantification (mean ± SEM [error bars]; n = 10 mice/group) of the number and size of disseminated tumors. (B) H&E of control and RAB5AS34N-MDA-MB-231 lung tissue sections (left). The size of metastatic nodules in lungs is the mean tumor area/lung area ± SEM (error bars; n = 9 mice/group repeated in three independent experiments). (C) Mixtures of CFSE-labeled (green) scramble-transfected control (Ctr) and eFluor 670–labeled (red) RAB5A,B,C-depleted MDA-MB-231 cells (0.5 × 106 each) were coinjected into the tail vein of NSG mice. (C, left) Images of lung tissue. (C, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A,B,C-depleted cells/field of lung tissue at the indicated times. **, P < 0.005. (C, right) Efficacy of RAB5 gene silencing by QRT-PCR. **, P < 0.005. (D) Doxycycline-inducible RAB5AS34N- and control-MCF10.DCIS.com cells were injected subcutaneously into NSG mice. After 4 d, mice were fed with doxycycline. (D, left) IHC analyses of control (Ctr) and RAB5AS34N-MCF10.DCIS.com xenografts performed at 1 and 3 wk after doxycycline treatment. Representative images from three independent experiments are shown (n = 5 mice/experimental condition). (D, right) mRNA level of RAB5AS34N expression in control (Ctr) and RAB5AS34N tumors. Error bars indicate SEM. **, P < 0.005. (E) Control (Ctr) and RAB5A-MCF10.DCIS.com cells (5 × 105 each) were labeled with eFluor 670 and injected into NSG mice tail vein. (E, left) Images of lung tissue. (E, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A-MCF10.DCIS.com cells per field at the indicated times. **, P < 0.005. (E, right) RAB5A expression by immunoblotting. Bars: (B) 2 µm; (C) 80 µm; (D) 100 µm; (E) 80 µm.
RAB5A is required for tumor dissemination and for the conversion from DCIS to infiltrating mammary carcinoma. (A and B) Doxycycline-inducible RAB5AS34N- and control MDA-MB-231 cells were injected into the mammary fat pads of NSG mice. After 3 wk, mice were fed with doxycycline. Metastases were analyzed 4 wk after removal of primary tumor. (A) Ipsilateral metastasis (arrowhead) of control or RAB5AS34N-MDA-MB-231 tumors. (A, right) Quantification (mean ± SEM [error bars]; n = 10 mice/group) of the number and size of disseminated tumors. (B) H&E of control and RAB5AS34N-MDA-MB-231 lung tissue sections (left). The size of metastatic nodules in lungs is the mean tumor area/lung area ± SEM (error bars; n = 9 mice/group repeated in three independent experiments). (C) Mixtures of CFSE-labeled (green) scramble-transfected control (Ctr) and eFluor 670–labeled (red) RAB5A,B,C-depleted MDA-MB-231 cells (0.5 × 106 each) were coinjected into the tail vein of NSG mice. (C, left) Images of lung tissue. (C, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A,B,C-depleted cells/field of lung tissue at the indicated times. **, P < 0.005. (C, right) Efficacy of RAB5 gene silencing by QRT-PCR. **, P < 0.005. (D) Doxycycline-inducible RAB5AS34N- and control-MCF10.DCIS.com cells were injected subcutaneously into NSG mice. After 4 d, mice were fed with doxycycline. (D, left) IHC analyses of control (Ctr) and RAB5AS34N-MCF10.DCIS.com xenografts performed at 1 and 3 wk after doxycycline treatment. Representative images from three independent experiments are shown (n = 5 mice/experimental condition). (D, right) mRNA level of RAB5AS34N expression in control (Ctr) and RAB5AS34N tumors. Error bars indicate SEM. **, P < 0.005. (E) Control (Ctr) and RAB5A-MCF10.DCIS.com cells (5 × 105 each) were labeled with eFluor 670 and injected into NSG mice tail vein. (E, left) Images of lung tissue. (E, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A-MCF10.DCIS.com cells per field at the indicated times. **, P < 0.005. (E, right) RAB5A expression by immunoblotting. Bars: (B) 2 µm; (C) 80 µm; (D) 100 µm; (E) 80 µm.
We also investigated the effects of simultaneously silencing the three RAB5 isoforms using siRNAs on the ability of MDA-MB-231 cells to disseminate in vivo. RAB5-depleted and control MDA-MB-231 cells were labeled with different fluorescent vital dyes, co-injected in equal numbers into the tail vein of immunocompromised mice, and then monitored for their ability to colonize the host lung. After 2 h, control and RAB5-depleted cells were present in the lung at a ratio of 1:1. However, at 24 and 96 h, the ratio of control to RAB5-depleted cells had significantly increased (Fig. 2 C). RAB5 knockdown did not significantly alter the cell cycle profile or survival of MDA-MB-231 cells (unpublished data); thus, it presumably affected the ability of these cells to extravasate, spread within, or adhere to lung tissue.
Next, we turned to MCF10.DCIS.com cells, which reproducibly form comedo-type DCIS lesions that spontaneously progress to invasive tumors (Miller et al., 2000; Hu et al., 2008). We subcutaneously injected control and doxycycline-inducible RAB5AS34N-MCF10.DCIS.com cells. After 4 d, we fed mice with doxycycline and monitored tumor growth and progression to invasive disease over time. After 1 wk, control and RAB5AS34N tumors were comparable in size (Fig. S1 C) and presented the typical DCIS histology, with a core of cancer cells delimited by a smooth muscle actin (SMA)-positive myoepithelial layer (Fig. 2 D). However, although MCF10.DCIS.com control lesions lost the myoepithelial layer and progressed to become invasive tumors after 3 wk, RAB5AS34N-expressing cells maintained the typical DCIS histology and their invasiveness was significantly impaired (Fig. 2 D).
To assess whether the elevation of RAB5A expression was sufficient to promote lung dissemination, we generated RAB5A-expressing MCF10.DCIS.com (Fig. 2 E). Control and RAB5A-MCF10.DCIS.com cells were labeled with a fluorescent vital dye, injected into mice tail vein, and monitored for their ability to colonize the host lung. A similar number of control and RAB5A-expressing cells were present in the lung 2 h after injection (Fig. 2 E). However, after 96 h the number of RAB5A-MCF10.DCIS.com cells was significantly higher than that of control cells (Fig. 2 E). RAB5A expression did not significantly alter the cell cycle profile or survival of MCF10.DCIS.com cells (unpublished data); thus, it was sufficient to enhance the ability of these cells to extravasate and adhere to lung tissue.
Collectively, these data indicate that RAB5A function is required, at least in the used model systems, for breast cancer cells to acquire the hallmarks of aggressiveness, both at the level of local invasiveness and of metastatic ability.
RAB5A promotes dissemination in vivo and invasion in vitro of poorly invasive tumor cells
To assess whether elevation of RAB5A levels is also sufficient to promote metastatic behavior in other types of cancer, we turned to HeLa cells, which are poorly invasive and express low levels of RAB5A (Palamidessi et al., 2008; Liu et al., 2011). We engineered control and RAB5A-expressing HeLa cells to also stably and homogenously express GFP-LifeAct; an F-actin binding peptide (Riedl et al., 2008; Fig. S1 D). Next, we xenografted these cells into the mammary fat pads of NSG mice to establish tumors, and then monitored ipsilateral and distant metastasis after removal of the primary tumor. In this assay, HeLa cells behaved similarly to MDA-MB-231 cells: the size, proliferation rate, and extent of apoptosis in control and RAB5A-HeLa primary tumors were indistinguishable (Fig. S1 E). In contrast, there was a significant increase in both the number and size of ipsilateral and distant metastatic outgrowths originating from RAB5A-HeLa tumors, compared with control tumors (Fig. 3, A and B). We then performed intravital microscopy on the primary tumors. Control HeLa tumors were compact masses with defined tumor margins (Fig. 3, C and D). In contrast, RAB5A-HeLa tumors were composed of loosely adherent, scattered cells, with nonhomogeneous, ragged margins (Fig. 3, C and D). Importantly, whereas control HeLa cells displayed little motility within tumors, RAB5A-HeLa cells were elongated, with frequently extended pseudopodia protrusions (Videos 1 and 2), rare circular structures that resembled circular ruffles observed on the dorsal surface of the cell in 2D (CDR), and blebs (Video 2). RAB5A-HeLa cells could also be seen migrating within the tumors (Fig. 3 E and Video 2).
A single plane of EGFP-LifeAct–positive RAB5A-HeLa tumors, where cells are seen either extending an actin-dense circular ruffles (arrowhead) and blebs (arrows). None of these events could be observed in control EGFP-HeLa tumors. The clip was obtained by intravital imaging (TCS SP5 upright DM600 CFS; Leica) as described in the Materials and methods section. Images were taken every 2 min for 54 min.
A single plane of EGFP-LifeAct–positive RAB5A-HeLa tumors, where cells are seen either extending an actin-dense circular ruffles (arrowhead) and blebs (arrows). None of these events could be observed in control EGFP-HeLa tumors. The clip was obtained by intravital imaging (TCS SP5 upright DM600 CFS; Leica) as described in the Materials and methods section. Images were taken every 2 min for 54 min.
Areas of EGFP-LifeAct–positive control and RAB5A-HeLa tumors. Each clip obtained by intravital imaging with multiphoton microscopy (TCS SP5 upright DM600 CFS; Leica) lasts 22 min, with images taken every 2 min. The control video contains nonmotile cells. The RAB5A show rapidly moving cells from the center (arrowheads) to the right bottom side (related to Fig. 3 E).
Areas of EGFP-LifeAct–positive control and RAB5A-HeLa tumors. Each clip obtained by intravital imaging with multiphoton microscopy (TCS SP5 upright DM600 CFS; Leica) lasts 22 min, with images taken every 2 min. The control video contains nonmotile cells. The RAB5A show rapidly moving cells from the center (arrowheads) to the right bottom side (related to Fig. 3 E).
RAB5A is sufficient to promote intratumoral cell motility and distant dissemination. (A) GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A) HeLa cells were injected into the mammary fat pads of NSG mice. Metastases were analyzed 4 wk after removal of primary tumor. (A, left) Ipsilateral metastasis (arrowheads) of control or RAB5A-HeLa tumors. (A, right) Quantitation of number and volume (mean ± SEM [error bars]; n = 10 mice/group) of disseminated tumors nodules. **, P < 0.005. (B, left) H&E and anti-GFP staining of FFPE lung tissue sections. Magnified boxed regions show metastasis. (B, right) The size of metastatic nodules is the mean tumor area/total lung area ± SEM (error bars; n = 9 mice/group repeated in two independent experiments). **, P < 0.005. (C) Tumors from GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A)-HeLa cells injected into the mammary fat pads of NSG mice were analyzed by two-photon microscopy. Green, GFP-LifeAct; gray, collagen structure (SHG). (D) Tumor invasive front visualized: in the top panels by projecting ∼40 serial z sections (green, GFP-LifeAct; gray, collagen structure [SHG]; or, in bottom panels, by IHC. (E) Intratumoral motion analysis of control and RAB5A-HeLa cells was obtained by overlaying 10 differentially colored, consecutive frames of time-lapse recording (Videos 1 and 2; left). Coloring indicates motile cells. The percentage of motility events/field of view/tumor is the mean ± SEM (error bars; n = 45). Bars: (B, left) 2 mm; (B, right) 0.2 mm; (C) 400 µm; (D, top) 200 µm; (D, bottom) 10 µm; (E) 80 µm.
RAB5A is sufficient to promote intratumoral cell motility and distant dissemination. (A) GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A) HeLa cells were injected into the mammary fat pads of NSG mice. Metastases were analyzed 4 wk after removal of primary tumor. (A, left) Ipsilateral metastasis (arrowheads) of control or RAB5A-HeLa tumors. (A, right) Quantitation of number and volume (mean ± SEM [error bars]; n = 10 mice/group) of disseminated tumors nodules. **, P < 0.005. (B, left) H&E and anti-GFP staining of FFPE lung tissue sections. Magnified boxed regions show metastasis. (B, right) The size of metastatic nodules is the mean tumor area/total lung area ± SEM (error bars; n = 9 mice/group repeated in two independent experiments). **, P < 0.005. (C) Tumors from GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A)-HeLa cells injected into the mammary fat pads of NSG mice were analyzed by two-photon microscopy. Green, GFP-LifeAct; gray, collagen structure (SHG). (D) Tumor invasive front visualized: in the top panels by projecting ∼40 serial z sections (green, GFP-LifeAct; gray, collagen structure [SHG]; or, in bottom panels, by IHC. (E) Intratumoral motion analysis of control and RAB5A-HeLa cells was obtained by overlaying 10 differentially colored, consecutive frames of time-lapse recording (Videos 1 and 2; left). Coloring indicates motile cells. The percentage of motility events/field of view/tumor is the mean ± SEM (error bars; n = 45). Bars: (B, left) 2 mm; (B, right) 0.2 mm; (C) 400 µm; (D, top) 200 µm; (D, bottom) 10 µm; (E) 80 µm.
To gain insights into how RAB5A controls motility programs, we monitored invasion into 3D matrices in vitro. We initially set up organotypic gels formed of acid-extracted rat-tail collagen preconditioned with human macrophages to recapitulate the tumor stroma (Nyström et al., 2005). Under these conditions, control HeLa cells survived, but barely entered into organotypic matrices over a 14-d period. In contrast, RAB5A-HeLa cells effectively invaded the matrix in a metalloprotease-dependent manner (Fig. 4, A and B). Inspection by second harmonic generation (SHG) of two-photon microscopy of the 3D organization of the collagen fibrillar meshwork revealed that RAB5A-HeLa cells extensively remodeled the network, generating spaces and gaps, and altering the parallel, orderly orientation of type I collagen fibers (Fig. S1 F). We observed a similar remodeling of skin interstitial tissues around the xenografted tumors resulting from subcutaneous injection of RAB5A-HeLa, but not of control, cells (Fig. S1, G and H). Thus, ectopic RAB5A expression is sufficient to transform a poorly invasive HeLa-originated tumor into a collagen remodeling, invasive tumor.
RAB5A is necessary and sufficient to promote matrix-metalloprotease–dependent invasion into a 3D matrix. (A) RAB5A promotes organotypic cell invasion and remodels stromal collagen. (A, left) H&E-stained cross-sections of GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A) HeLa cells grown for 14 d on organotypic collagen matrix preconditioned with U937 macrophages, in the absence or presence of GM6001. Bar, 400 µm. (B) Quantification of the number of invading cells/condition, with respect to the number of invading RAB5A-HeLa cells, is the mean invasion index ± SEM (error bars) of four independent experiments (n = 45). *, P < 0.01. (C and D) Doxycycline-induced control- and RAB5AS34N-MDA-MB-231 (C) or RAB5AS34N-MCF10.DCIS.com (D) cells were assessed for their invasiveness by placing them on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel and 100 ng/ml HGF were added. Examples of cell migration tracks are shown on the left (Videos 5 and 6). (C and D, right) Cell invasion is expressed as the mean forward invasion index ± SEM (error bars; n = 75 single cells/experiment repeated in four independent assays). *, P < 0.01; **, P < 0.005. (E) Doxycycline-inducible control (empty) or RAB5AS34N-MF10.DCIS.com cells were grown on a thick 1:1 Matrigel/type I collagen mixture and overlaid with diluted Matrigel (2 mg/ml). After 1 wk, cells were treated with doxycycline and/or HGF (20 ng/ml), or were mock treated. Bars, 400 µm. (E, right) The percentage of structures with invasive outgrowths was expressed as mean ± SEM (error bars) of four independent experiments; n = 35. **, P < 0.005.
RAB5A is necessary and sufficient to promote matrix-metalloprotease–dependent invasion into a 3D matrix. (A) RAB5A promotes organotypic cell invasion and remodels stromal collagen. (A, left) H&E-stained cross-sections of GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A) HeLa cells grown for 14 d on organotypic collagen matrix preconditioned with U937 macrophages, in the absence or presence of GM6001. Bar, 400 µm. (B) Quantification of the number of invading cells/condition, with respect to the number of invading RAB5A-HeLa cells, is the mean invasion index ± SEM (error bars) of four independent experiments (n = 45). *, P < 0.01. (C and D) Doxycycline-induced control- and RAB5AS34N-MDA-MB-231 (C) or RAB5AS34N-MCF10.DCIS.com (D) cells were assessed for their invasiveness by placing them on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel and 100 ng/ml HGF were added. Examples of cell migration tracks are shown on the left (Videos 5 and 6). (C and D, right) Cell invasion is expressed as the mean forward invasion index ± SEM (error bars; n = 75 single cells/experiment repeated in four independent assays). *, P < 0.01; **, P < 0.005. (E) Doxycycline-inducible control (empty) or RAB5AS34N-MF10.DCIS.com cells were grown on a thick 1:1 Matrigel/type I collagen mixture and overlaid with diluted Matrigel (2 mg/ml). After 1 wk, cells were treated with doxycycline and/or HGF (20 ng/ml), or were mock treated. Bars, 400 µm. (E, right) The percentage of structures with invasive outgrowths was expressed as mean ± SEM (error bars) of four independent experiments; n = 35. **, P < 0.005.
To further corroborate this notion, we monitored in real time the invasive migration of tumor cells into 3D gels of native type I collagen (Hotary et al., 2003; Sabeh et al., 2004; Li et al., 2008). Native, acid-extracted, type I collagen retains its telopeptide and, unlike pepsin-extracted collagens, polymerizes into a dense fibrillar network, which provides a formidable barrier to invasion, unless cells acquire collagenolytic activity (Sabeh et al., 2009; see also Fig. S2 A and Video 3 [top] for assay standardization). Control HeLa cells could not enter into the native type I collagen matrix. However, RAB5A-HeLa readily invaded and their invasiveness was abrogated by the broad spectrum metalloprotease inhibitor GM6001 (Fig. S2 B and Videos 3 and 4). Conversely, expression of RAB5AS34N in MDA-MB-231 (Fig. 4 C and Video 6) or MCF10.DCIS.com (Fig. 4 D and Video 6) cells almost completely arrested their invasive potential. A similar inhibition of invasion was achieved by siRNA-mediated silencing of the three RAB5 isoforms in MDA-MB-231 cells (Fig. S2 C and Video 6).
High-magnification time lapse of cells moving into 3D collagen in metalloprotease-dependent manner. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of HT1080 cells, treated with control vehicle or broad-spectrum metalloprotease inhibitor GM6001, invading into native type I collagen chamber slides (see also Fig. S2 A). (Bottom) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of HeLa cells transfected with pEGFP alone or in combination with RAB5A, treated with control vehicle or GM6001 and tested for invasion into native type I collagen gel chamber slides (see also Fig. S2 B). Images were taken every 5 min for 13 h. Representative videos of at least six independent experiments are shown.
High-magnification time lapse of cells moving into 3D collagen in metalloprotease-dependent manner. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of HT1080 cells, treated with control vehicle or broad-spectrum metalloprotease inhibitor GM6001, invading into native type I collagen chamber slides (see also Fig. S2 A). (Bottom) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of HeLa cells transfected with pEGFP alone or in combination with RAB5A, treated with control vehicle or GM6001 and tested for invasion into native type I collagen gel chamber slides (see also Fig. S2 B). Images were taken every 5 min for 13 h. Representative videos of at least six independent experiments are shown.
High-magnification time lapse (IX81 microscope; Olympus) of control (left) and RAB5A-expressing HeLa (right) cells moving into 3D collagen. HeLA cells embedded into 2.3 mg/ml acid extracted–only polymerized type I collagen gel. The gel was overlaid with medium containing 100 ng/ml HGF. Images were taken with a 63× objective lens every 20 s for 20 min. Control cells are round and immobile, whereas RAB5-expressing HeLA adopt a typical mesenchymal mode of motility. A representative video of at least four independent experiments is shown. Please note the morphological and migratory changes that occur upon expression of RAB5A. This type of analysis, however, does not allow a precise quantitative evaluation of invasion parameters.
High-magnification time lapse (IX81 microscope; Olympus) of control (left) and RAB5A-expressing HeLa (right) cells moving into 3D collagen. HeLA cells embedded into 2.3 mg/ml acid extracted–only polymerized type I collagen gel. The gel was overlaid with medium containing 100 ng/ml HGF. Images were taken with a 63× objective lens every 20 s for 20 min. Control cells are round and immobile, whereas RAB5-expressing HeLA adopt a typical mesenchymal mode of motility. A representative video of at least four independent experiments is shown. Please note the morphological and migratory changes that occur upon expression of RAB5A. This type of analysis, however, does not allow a precise quantitative evaluation of invasion parameters.
TIRF time-lapse microscopy (IX81; Olympus) of serum-free (SF) HeLa cells transfected with RFP-LifeAct and Fluorescein-MT1-MMP in the presence (bottom) or absence of RAB5A. RAB5A-expressing cells were stimulated with HGF. Videos are representative of at least three independent experiments. Images were taken every 2 s for 5 min. Only representative videos of control and RAB5A-expressing cells stimulated with HGF are shown. Similar results, however, were obtained with either HGF stimulation or RAB5 expression alone, although the concomitant treatment increases the number of cell forming invadosomes as well as degradation activities (not depicted).
TIRF time-lapse microscopy (IX81; Olympus) of serum-free (SF) HeLa cells transfected with RFP-LifeAct and Fluorescein-MT1-MMP in the presence (bottom) or absence of RAB5A. RAB5A-expressing cells were stimulated with HGF. Videos are representative of at least three independent experiments. Images were taken every 2 s for 5 min. Only representative videos of control and RAB5A-expressing cells stimulated with HGF are shown. Similar results, however, were obtained with either HGF stimulation or RAB5 expression alone, although the concomitant treatment increases the number of cell forming invadosomes as well as degradation activities (not depicted).
We next exploited the ability of MCF10.DCIS.com cells to generate invasive outgrowths in 3D basement membrane overlay assays (Lee et al., 2007). We plated control and doxycycline-inducible RAB5AS34N MCF10.DCIS.com cells as single cells onto a gel composed of Matrigel and type I collagen. As expected, control cells generated compact, acini-like structures, which upon addition of HGF formed heterogeneous structures with extended invasive outgrowths (Fig. 4 E; Jedeszko et al., 2009). The invasive outgrowths were abrogated by the expression of RAB5AS34N, which was induced by the addition of doxycycline at the same time as HGF (Fig. 4 E).
Thus, RAB5A is necessary and sufficient to promote a proteolytic mesenchymal program of cell invasion in vitro and tumor dissemination in vivo.
RAB5A is necessary and sufficient for invadosome formation
Invadosomes are actin-rich adhesive platforms that degrade ECM in invasive tumor cells (Linder et al., 2011). Indeed, both MDA-MB-231 and MCF10.DCIS.com cells readily formed functional invadosomes upon HGF stimulation in vitro. Inhibition of RAB5 by RAB5AS34N expression (Fig. 5, A and B), or siRNAs targeting the three RAB5 isoforms (Fig. S2 D), significantly reduced HGF-induced invadosome formation and matrix degradation in MDA-MB-231 and MCF10.DCIS.com cells as well as in other breast cancer cells (Fig. S2, E and F). Conversely, the elevation of RAB5A levels was sufficient to increase invadosomes formation (Fig. S2 G).
RAB5A is essential for HGF-induced invadosome formation. (A and B) Doxycycline-induced control and RAB5AS34N-MDA-MB-231 (A) or RAB5AS34N-MCF10.DCIS.com (B) cells plated onto fluorescently conjugated gelatin were stimulated with HGF (100 ng/ml) for 3 h. (B, left) Images of cells stained with phalloidin (left), fluorescently conjugated gelatin (middle), and merged channels (right). (B, right) Quantification of gelatin degradation was expressed as a degradation index (calculated as described in Materials and methods). Data are the mean ± SEM (error bars; n = 70 cells/experiment in five independent ones). **, P < 0.005. (C, left) TIRF microscopy of RAB5A-HeLa cells. F-actin and RAB5A were detected with phalloidin and anti–RAB5 Ab (RAB5), respectively. (C, right) xz sections of control and RAB5A-HeLa cells plated onto fluorescently conjugated gelatin (red) and stained with phalloidin (green). (D) Control and RAB5A-HeLa cells were plated onto fluorescently conjugated gelatin (middle) overnight under serum-starved conditions and stimulated with suboptimal doses of HGF (1 ng/ml). F-actin and RAB5A were detected with phalloidin and anti-RAB5A Ab (RAB5A), respectively. Insets show magnifications of the boxed regions. Arrows indicate invadosomes. (D, right) Gelatin degradation was expressed as a degradation index (see Materials and methods). Data are the mean ± SEM (n = 50 cells/experiment in four independent ones). **, P < 0.005. Bars: (A–C) 20 µm; (D) 10 µm.
RAB5A is essential for HGF-induced invadosome formation. (A and B) Doxycycline-induced control and RAB5AS34N-MDA-MB-231 (A) or RAB5AS34N-MCF10.DCIS.com (B) cells plated onto fluorescently conjugated gelatin were stimulated with HGF (100 ng/ml) for 3 h. (B, left) Images of cells stained with phalloidin (left), fluorescently conjugated gelatin (middle), and merged channels (right). (B, right) Quantification of gelatin degradation was expressed as a degradation index (calculated as described in Materials and methods). Data are the mean ± SEM (error bars; n = 70 cells/experiment in five independent ones). **, P < 0.005. (C, left) TIRF microscopy of RAB5A-HeLa cells. F-actin and RAB5A were detected with phalloidin and anti–RAB5 Ab (RAB5), respectively. (C, right) xz sections of control and RAB5A-HeLa cells plated onto fluorescently conjugated gelatin (red) and stained with phalloidin (green). (D) Control and RAB5A-HeLa cells were plated onto fluorescently conjugated gelatin (middle) overnight under serum-starved conditions and stimulated with suboptimal doses of HGF (1 ng/ml). F-actin and RAB5A were detected with phalloidin and anti-RAB5A Ab (RAB5A), respectively. Insets show magnifications of the boxed regions. Arrows indicate invadosomes. (D, right) Gelatin degradation was expressed as a degradation index (see Materials and methods). Data are the mean ± SEM (n = 50 cells/experiment in four independent ones). **, P < 0.005. Bars: (A–C) 20 µm; (D) 10 µm.
Interestingly, HeLa cells, which are notoriously poor in degrading the ECM under growing conditions, also acquired this ability in response to HGF stimulation (Fig. S3, A and B). Notably, HGF addition acutely increased the levels of GTP-bound RAB5A (Fig. S3, A and B). Additionally, genetic or functional interference with RAB5 severely inhibited HGF-stimulated matrix proteolysis (Fig. S4 A). Conversely, elevation of RAB5A was sufficient to induce actin-rich clusters on the ventral plasma membrane (PM) in unstimulated cells (Fig. 5, C and D). These structures represented bona fide invadosomes based on: (1) the presence of several prototypical invadosomal markers (Fig. S3 C), (2) their dependency on the nucleation promoting factor, N-WASP (Yamaguchi et al., 2005; Fig. S4, D and E), and (3) their ability to focally degrade the ECM (Fig. 5, C and D).
Thus, RAB5A is necessary and sufficient to promote invadosome formation and focal matrix degradation activity in response to stimulation with motogenic factors.
The RAB4-dependent recycling route is essential for invadosome formation
We used a molecular genetics approach to dissect the circuitry involved in the formation of HGF (or RAB5A)-induced invadosomes. We performed this screening in HeLa cells, which can be efficiently transfected and induced to form CDRs and invadosomes by either HGF stimulation or the expression of RAB5A (Palamidessi et al., 2008). Interference with the major internalization routes by inhibition of clathrin or dynamin, or with recycling pathways by reducing the temperature to 16°C, reduced both RAB5A-induced invadosomes and CDRs (Table S1). Similar inhibition of both these structures was achieved by silencing of ARF6 and RAB35, which are involved in membrane recycling (Palamidessi et al., 2008; Svensson et al., 2008; Zhang et al., 2009; Table S1). In contrast, the inhibition of the fast recycling route by interference with RAB4 or its effector RABENOSYN-5 prevented the formation of invadosomes, but not of CDRs (Table S1). Finally, impairment of the RAB11-dependent slow recycling, RAB7-mediated degradative, or RAB8-biosynthetic secretory exocytic pathways had no effect on the formation of invadosomes or CDRs (Table S1).
We next determined whether RAB4 and RABENOSYN-5 were also necessary for HGF-induced matrix degradation. Silencing of the two RAB4 isoforms (A and B), or expression of a dominant-negative RAB4A mutant reduced (>80%) the number of invadosomes and matrix degradation (Fig. S4, A and B). Similar effects were obtained by silencing RABENOSYN-5, whereas suppression of RAB11 activity had no effect (Fig. S4, A and B).
We also validated these findings in breast cancer cell models. Like in HeLa cells, silencing of RAB4 (or expression of RAB4AS22N) or of RABENOSYN-5, but not of RABAPTIN-5, impaired matrix proteolysis of MCF10.DCIS.com (Fig. 6, A and B). A similar impairment in matrix degradation was also observed in MDA-MB-231 upon interference with RAB4A and -B (Fig. S4, C and D). Thus, growth factor–induced RAB5-dependent invadosomes require a functional RAB4–RABENOSYN-5 recycling pathway. The critical role of RABENOSYN-5 should depend on its ability to physically and simultaneously bind to activated RAB5 and RAB4. Accordingly, although the expression of a siRNA-resistant RABENOSYN-5 wild-type protein restored matrix degradation of RABENOSYN-5 knockdown cells (Fig. 6, C and D), siRNA-resistant RABENOSYN-5 mutants impaired either in the binding to RAB5 or to RAB4 (Eathiraj et al., 2005) failed to do so (Fig. 6, C and D).
The RAB4–RABENOSYN-5 recycling pathway is necessary for HGF-induced invadosome formation. (A) Serum-starved MCF10.DCIS.com cells, transfected with scrambled siRNA (Ctr), siRNAs against RABENOSYN-5 (siRNA RBNS-5) or RAB4A and -B (siRNA RAB4), or RABAPTIN-5. GFP-RAB4AS22N-MCF10.DCIS.com cells were induced or not induced with doxycycline and serum starved. Cells were plated onto fluorescently conjugated gelatin (green), stimulated with HGF for 3 h, or left in serum free conditions (SF), and stained with phalloidin (red). Bar, 15 µm. (A, left) GFP-RAB4AS22N (GFP-RAB4A) expression was verified by immunoblotting. (B) Gelatin degradation was expressed as a degradation index. Data are the mean ± SEM (error bars; n = 75 cells/experiment in four independent ones). *, P < 0.01. Silencing of RAB4A, RAB4B, and RABENOSYN-5 genes was performed using two independent siRNA oligos, which gave identical results (not depicted), and was verified by QRT-PCR (right). **, P < 0.005. (C) MCF10.DCIS.com cells were cotransfected with scrambled siRNA (Ctr) or siRNAs against RABENOSYN-5 (siRNA RBNS-5) together with siRNA-resistant RABENOSYN-5 wild type (resWT-RBNS-5) or mutants impaired in binding to RAB4 (resRAB4-Δ-RBNS-5) or to RAB5 (resRAB5-Δ-RBNS-5), or to both GTPases (resRAB4/5-Δ-RBNS-5) fused to GFP. Cells were plated onto fluorescently conjugated gelatin (red), stimulated with HGF for 3 h, or left in serum free conditions, and stained with DAPI (blue). Arrows, matrix degradation areas. Bar, 20 µm. (D) Gelatin degradation is expressed as a degradation index, as indicated in B. Data are the mean ± SEM (error bars; n = 65 cells/experiment in three independent ones). *, P < 0.01. Silencing of RABENOSYN-5 was verified by QRT-PCR (right). **, P < 0.005.
The RAB4–RABENOSYN-5 recycling pathway is necessary for HGF-induced invadosome formation. (A) Serum-starved MCF10.DCIS.com cells, transfected with scrambled siRNA (Ctr), siRNAs against RABENOSYN-5 (siRNA RBNS-5) or RAB4A and -B (siRNA RAB4), or RABAPTIN-5. GFP-RAB4AS22N-MCF10.DCIS.com cells were induced or not induced with doxycycline and serum starved. Cells were plated onto fluorescently conjugated gelatin (green), stimulated with HGF for 3 h, or left in serum free conditions (SF), and stained with phalloidin (red). Bar, 15 µm. (A, left) GFP-RAB4AS22N (GFP-RAB4A) expression was verified by immunoblotting. (B) Gelatin degradation was expressed as a degradation index. Data are the mean ± SEM (error bars; n = 75 cells/experiment in four independent ones). *, P < 0.01. Silencing of RAB4A, RAB4B, and RABENOSYN-5 genes was performed using two independent siRNA oligos, which gave identical results (not depicted), and was verified by QRT-PCR (right). **, P < 0.005. (C) MCF10.DCIS.com cells were cotransfected with scrambled siRNA (Ctr) or siRNAs against RABENOSYN-5 (siRNA RBNS-5) together with siRNA-resistant RABENOSYN-5 wild type (resWT-RBNS-5) or mutants impaired in binding to RAB4 (resRAB4-Δ-RBNS-5) or to RAB5 (resRAB5-Δ-RBNS-5), or to both GTPases (resRAB4/5-Δ-RBNS-5) fused to GFP. Cells were plated onto fluorescently conjugated gelatin (red), stimulated with HGF for 3 h, or left in serum free conditions, and stained with DAPI (blue). Arrows, matrix degradation areas. Bar, 20 µm. (D) Gelatin degradation is expressed as a degradation index, as indicated in B. Data are the mean ± SEM (error bars; n = 65 cells/experiment in three independent ones). *, P < 0.01. Silencing of RABENOSYN-5 was verified by QRT-PCR (right). **, P < 0.005.
RAB5A- and HGF-mediated matrix degradation requires RAB4-dependent fast recycling of MT1-MMP
The requirement of RAB5- and RAB4-dependent fast EECs for invadosome formation suggests that a membrane-associated matrix degradative cargo, most likely MT1-MMP, should be delivered to nascent invadosomes. Consistently, silencing of MT1-MMP abrogated HGF- and RAB5A-induced matrix degradation and reduced the number of invadosomes (Fig. S5, A and B). As HeLa cells express low levels of MT1-MMP (Zhai et al., 2005), we used fluorescently tagged MT1-MMP to facilitate its visualization. In unstimulated cells, MT1-MMP could be detected on the ventral PM by TIRF microscopy (Video 6) and inside vesicles (Figs. S5 C and S6 D). As previously reported (Remacle et al., 2003), a sizable fraction of these vesicles represented early EEA1-positive endosomes, which were also positive for RAB5, RAB4, and RABENOSYN-5 (Fig. S5 C). These vesicles could also be observed in cells embedded in 3D collagen (Fig. S5 D). Importantly, we confirmed the localization of endogenous MT1-MMP into EEA1-, RABENOSYN-5–, and RAB4-positive endosomes in MCF10.DCIS.com cells (Fig. S5 E). Notably, in these cells, only a fraction of MT1-MMP localizes in LAMP-1–positive late endosomes (Fig. S5 E). Additionally, after treatment with HGF, or ectopic RAB5A expression, MT1-MMP relocalized to ventrally restricted, F-actin–rich structures (Video 6), which were also transiently targeted by RAB5A- and RAB4A-positive early endosomes (Fig. S5 F and Video 7).
DeltaVision time-lapse microscopy (IX71; Olympus) of HGF-stimulated Hela cells cotransfected with RFP-LifeAct and either GFP-RAB5A or GFP-RAB4A. The boxed region was magnified to visualize the motility of endosomal vesicles (see Fig. S5 F). Images were taken every 3 s for 1 min. Videos are representative of >50 cells from of at least three independent experiments.
DeltaVision time-lapse microscopy (IX71; Olympus) of HGF-stimulated Hela cells cotransfected with RFP-LifeAct and either GFP-RAB5A or GFP-RAB4A. The boxed region was magnified to visualize the motility of endosomal vesicles (see Fig. S5 F). Images were taken every 3 s for 1 min. Videos are representative of >50 cells from of at least three independent experiments.
These results suggest that fast recycling of MT1-MMP is essential for HGF- and RAB5A-induced invadosome formation. To provide direct evidence for this possibility, we used an antibody that recognizes the extracellular domain of MT1-MMP to follow recycling of preinternalized metalloprotease back to the PM. HGF stimulation significantly increased the mobilization rate of MT1-MMP, with >60% recycling occurring after 2 min of stimulation (Fig. 7 A). HGF-induced recycling of MT1-MMP was reduced by silencing of RAB5, RAB4, and RABENOSYN-5, but not of integrin β3 expression (see the following paragraph; Fig. 7 A), under conditions in which the steady-state amounts of cell surface MT1-MMP were not altered (Fig. 7 B). Only RAB5 loss impaired MT1-MMP internalization (Fig. 7 C), which suggests that the primary effect of RAB5 is to impede entry into the early endosome from which MT1-MMP is subsequently recycled through a RAB4–RABENOSYN-5 pathway.
HGF-induced fast recycling of MT1-MMP depends onto RAB5, RAB4, and RABENOSYN-5. (A) Cherry-MT1-MMP–expressing HeLa cells were transfected with scrambled siRNA or siRNAs against RAB5A,B,C (siRNA-RAB5) or RAB4A and -B (siRNA RAB4 A/B), RABENOSYN-5 (siRNA RBNS-5), or integrin β3 (siRNA β3). Serum-starved cells were incubated with anti–MT1-MMP antibody at 16°C for 2 h. After a mild acid wash to remove surface antibody, cells were switched to 37°C and stimulated with HGF (100 ng/ml), or left in serum free (SF) conditions. At the indicated time points, cells were fixed, and stained in the absence of permeabilization with FITC-conjugated secondary antibody (green). Cherry-MT1-MMP (red) was detected by epifluorescence. The relative cell surface levels of MT1-MMP were quantified using ImageJ software on nonsaturated images, and expressed as relative cell surface MT1-MMP levels with respect to HGF-stimulated control cells after 15 min (set at 100%). Data are the mean ± SEM (error bars; n = 25 cells repeated in three independent experiments). (B) Steady-state cell surface levels of MT1-MMP. Cherry-MT1-MMP-HeLa cells were transfected with scrambled siRNA or siRNAs against RAB5A,B,C (siRNA-RAB5) or RAB4A and -B (siRNA RAB4 A/B), RABENOSYN-5 (siRNA RBNS-5), or integrin β3 (siRNA β3). Cells were incubated with anti–MT1-MMP antibody at 4°C for 2 h, washed, fixed, and stained in the absence of permeabilization with FITC-conjugated secondary antibody (green). (C) Silencing of RAB5 impairs MT1-MMP internalization. Cherry-MT1-MMP-HeLa cells were transfected with scrambled (Ctr) or anti-RAB5A,B,C siRNAs. Serum-starved cells were incubated with anti–MT1-MMP antibody at 16°C for 2 h. Cells were washed, fixed, permeabilized with 0.1% Triton X-100, and stained with FITC-conjugated secondary antibody (green). Bottom right, efficacy of gene silencing by QRT-PCR. Data are the mean ± SEM (error bars). **, P < 0.005. Bars, 10 µm.
HGF-induced fast recycling of MT1-MMP depends onto RAB5, RAB4, and RABENOSYN-5. (A) Cherry-MT1-MMP–expressing HeLa cells were transfected with scrambled siRNA or siRNAs against RAB5A,B,C (siRNA-RAB5) or RAB4A and -B (siRNA RAB4 A/B), RABENOSYN-5 (siRNA RBNS-5), or integrin β3 (siRNA β3). Serum-starved cells were incubated with anti–MT1-MMP antibody at 16°C for 2 h. After a mild acid wash to remove surface antibody, cells were switched to 37°C and stimulated with HGF (100 ng/ml), or left in serum free (SF) conditions. At the indicated time points, cells were fixed, and stained in the absence of permeabilization with FITC-conjugated secondary antibody (green). Cherry-MT1-MMP (red) was detected by epifluorescence. The relative cell surface levels of MT1-MMP were quantified using ImageJ software on nonsaturated images, and expressed as relative cell surface MT1-MMP levels with respect to HGF-stimulated control cells after 15 min (set at 100%). Data are the mean ± SEM (error bars; n = 25 cells repeated in three independent experiments). (B) Steady-state cell surface levels of MT1-MMP. Cherry-MT1-MMP-HeLa cells were transfected with scrambled siRNA or siRNAs against RAB5A,B,C (siRNA-RAB5) or RAB4A and -B (siRNA RAB4 A/B), RABENOSYN-5 (siRNA RBNS-5), or integrin β3 (siRNA β3). Cells were incubated with anti–MT1-MMP antibody at 4°C for 2 h, washed, fixed, and stained in the absence of permeabilization with FITC-conjugated secondary antibody (green). (C) Silencing of RAB5 impairs MT1-MMP internalization. Cherry-MT1-MMP-HeLa cells were transfected with scrambled (Ctr) or anti-RAB5A,B,C siRNAs. Serum-starved cells were incubated with anti–MT1-MMP antibody at 16°C for 2 h. Cells were washed, fixed, permeabilized with 0.1% Triton X-100, and stained with FITC-conjugated secondary antibody (green). Bottom right, efficacy of gene silencing by QRT-PCR. Data are the mean ± SEM (error bars). **, P < 0.005. Bars, 10 µm.
Thus, after HGF stimulation, MT1-MMP is redirected to defined areas of the PM, namely to invadosomes, primarily via a RAB4 fast recycling route.
Genetic and functional interference with αVβ3-, but not β1-containing integrins inhibits matrix degradation
Invadosomes are integrin-based adhesive, mechanosensory modules. Although different integrins localize to invadosomes, a major role has been attributed to β1 and β3 integrins in invadosome formation (for review see Destaing et al., 2011). To dissect the role of β1 and β3 integrins in RAB5A-induced invadosomes, we initially studied their localization in RAB5A-expressing HeLa cells. Both integrins localized on the ventral PM in serum-starved cells (Video 8). However, only αVβ3, but not β1, integrin colocalized with RAB5A-induced invadosomes both by confocal (Fig. 8 A) and TIRF analysis (Fig. 8 B and Video 8).
TIRF time-lapse microscopy (IX81, Olympus) of serum-free (SF) HeLa cells transfected with RFP-LifeAct and GFP–β3 integrin in the presence (bottom) or absence of RAB5A. RAB5A-expressing cells were stimulated with HGF. Images were taken every 2 s for 3 min. Videos are representative of at least three independent experiments (see also Fig. 8 B). Only representative videos of control and RAB5A-expressing cells stimulated with HGF are shown. Similar results, however, were obtained with either HGF stimulation or RAB5 expression alone, although the concomitant treatment increases the number of cell forming invadosomes as well as degradation activities (not depicted).
TIRF time-lapse microscopy (IX81, Olympus) of serum-free (SF) HeLa cells transfected with RFP-LifeAct and GFP–β3 integrin in the presence (bottom) or absence of RAB5A. RAB5A-expressing cells were stimulated with HGF. Images were taken every 2 s for 3 min. Videos are representative of at least three independent experiments (see also Fig. 8 B). Only representative videos of control and RAB5A-expressing cells stimulated with HGF are shown. Similar results, however, were obtained with either HGF stimulation or RAB5 expression alone, although the concomitant treatment increases the number of cell forming invadosomes as well as degradation activities (not depicted).
Genetic and functional interference with αVβ3, but not α5β1, integrin inhibits RAB5A- and HGF-induced invadosome formation and matrix degradation. (A) Confocal analysis of RAB5A-HeLa cells stained with phalloidin (red) or the indicated Ab (green). (B) Still images from TIRF microscopy time-lapse of HeLa cells transfected with RAB5, GFP-β3, and RFP-LifeAct (see also Video 8). (C) HeLa cells cotransfected with RAB5A and MT1-MMP were subjected to PLA (red dots) with abs against MT1-MMP and integrin β3 or stained with phalloidin (green). Negative controls (Ctr) were cells stained with oligonucleotide-labeled PLA probes alone. Confocal images in A, B, and C are representative of >100 analyzed cells. (D) HeLa cells were transfected with scrambled (Ctr), anti-β1, or anti-β3 siRNA, or incubated with the inhibitory antibodies, 4B4 and LM609, against β1 and αVβ3, respectively. Serum-starved cells were plated onto fluorescently conjugated gelatin (red), stimulated with HGF (100 ng/ml) for 3 h, or left in serum free conditions (SF), and stained with phalloidin (green). Insets are magnifications of boxed regions. Quantification of gelatin degradation was expressed as a degradation index (relative to the area of degradation of control, HGF-stimulated cells normalized for cell number). Data are the mean ± SEM (error bars; n = 60 cells/experiment in five independent experiments). Two independent siRNAs were used for each integrin with similar results. Silencing of β1 and β3 genes was verified by QRT-PCR (right). **, P < 0.005. Arrows in A–D indicate examples of invadosomes. (E) MDA-MB-231 and MCF10.DCIS.com cells were transfected with scrambled (Ctr) or anti-β3 siRNA. Serum-starved cells were then plated onto fluorescently conjugated gelatin (red), stimulated with HGF (100 ng/ml) for 3 h, or left in serum free conditions (SF), and stained with phalloidin (green). Gelatin degradation is expressed as a degradation index. Data are the mean ± SEM (error bars; n = 40 cells/experiment in three independent ones). Two independent siRNAs were used for each integrin with similar results. Silencing β3 genes was verified by QRT-PCR (right). **, P < 0.005. Bars: (A) 15 µm; (B–E) 20 µm.
Genetic and functional interference with αVβ3, but not α5β1, integrin inhibits RAB5A- and HGF-induced invadosome formation and matrix degradation. (A) Confocal analysis of RAB5A-HeLa cells stained with phalloidin (red) or the indicated Ab (green). (B) Still images from TIRF microscopy time-lapse of HeLa cells transfected with RAB5, GFP-β3, and RFP-LifeAct (see also Video 8). (C) HeLa cells cotransfected with RAB5A and MT1-MMP were subjected to PLA (red dots) with abs against MT1-MMP and integrin β3 or stained with phalloidin (green). Negative controls (Ctr) were cells stained with oligonucleotide-labeled PLA probes alone. Confocal images in A, B, and C are representative of >100 analyzed cells. (D) HeLa cells were transfected with scrambled (Ctr), anti-β1, or anti-β3 siRNA, or incubated with the inhibitory antibodies, 4B4 and LM609, against β1 and αVβ3, respectively. Serum-starved cells were plated onto fluorescently conjugated gelatin (red), stimulated with HGF (100 ng/ml) for 3 h, or left in serum free conditions (SF), and stained with phalloidin (green). Insets are magnifications of boxed regions. Quantification of gelatin degradation was expressed as a degradation index (relative to the area of degradation of control, HGF-stimulated cells normalized for cell number). Data are the mean ± SEM (error bars; n = 60 cells/experiment in five independent experiments). Two independent siRNAs were used for each integrin with similar results. Silencing of β1 and β3 genes was verified by QRT-PCR (right). **, P < 0.005. Arrows in A–D indicate examples of invadosomes. (E) MDA-MB-231 and MCF10.DCIS.com cells were transfected with scrambled (Ctr) or anti-β3 siRNA. Serum-starved cells were then plated onto fluorescently conjugated gelatin (red), stimulated with HGF (100 ng/ml) for 3 h, or left in serum free conditions (SF), and stained with phalloidin (green). Gelatin degradation is expressed as a degradation index. Data are the mean ± SEM (error bars; n = 40 cells/experiment in three independent ones). Two independent siRNAs were used for each integrin with similar results. Silencing β3 genes was verified by QRT-PCR (right). **, P < 0.005. Bars: (A) 15 µm; (B–E) 20 µm.
Notably, αVβ3 is primarily recycled through the RAB4 pathway (Roberts et al., 2001), similar to what we report here for MT1-MMP. This suggests that the two proteins may be cotrafficked to invadosomes in response to HGF. In support of this possibility, we showed by proximity ligation in situ that MT1-MMP and αVβ3 partially colocalized in punctate structures in the cytoplasm, as well as on ventral actin clusters (invadosomes) upon HGF stimulation (Fig. 8 C). Finally, we interfered with the activity αVβ3 and β1 integrin using specific inhibitory antibodies or siRNAs. In the case of αVβ3, but not of β1 integrins, this led to the impairment of HGF-induced, RAB5A-dependent invadosomes formation and matrix degradation in HeLa (Fig. 8 D), as well as in MDA-MB-231 and MCF10.DCIS.com (Fig. 8 E). Of note, the extent of MT1-MMP recycling to the PM was independent of integrin β3 (Fig. 7 A), which indicates that this integrin is likely required for the localized PM targeting, but not for the trafficking of MT1-MMP.
RAB5A and RAB4A promote MT1-MMP– and β3-dependent cell invasion into 3D matrices
Our current and previously published data (Palamidessi et al., 2008) support a model in which RAB5 couples elongated protrusions with pericellular proteolysis by controlling RAB4-dependent fast recycling of MT1-MMP and β3 integrin cargos to enable efficient invasion. We verified this model by monitoring in real time the chemotactic invasion of RAB5A-HeLa cells, in which MT1-MMP, RAB4A/B, or β1 or β3 integrin expression had been inhibited, into 3D native type I collagen gels containing HGF. RAB5A-dependent invasion was impaired by silencing of MT1-MMP, RAB4A/B, and β3 integrin, but only marginally affected by silencing of β1 integrin (Fig. 9 A and Video 9). Silencing of RAB4A/B and β3 integrin also impaired MDA-MB-231 invasion (Fig. 9 B and Video 10).
Time-lapse wide field microscopy (Eclipse TE2000-E; Nikon) of RAB5A-expressing HeLa cells were treated with the indicated siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 A). Images were taken every 5 min for 33 h. Videos are representative of at least three independent experiments.
Time-lapse wide field microscopy (Eclipse TE2000-E; Nikon) of RAB5A-expressing HeLa cells were treated with the indicated siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 A). Images were taken every 5 min for 33 h. Videos are representative of at least three independent experiments.
RAB4A and β3 integrin are required for invasion into 3D collagen of breast cancer cells. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of MDA-MB-231 cells transfected with scrambled (Ctr) or anti-RAB4A/B siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 B). (Bottom) Time-lapse wide-field microscopy of doxycycline-inducible RAB4AS22N-expressing and control MCF10.DCIS.com. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 C). Images were taken every 5 min for 30 h. Representative videos of at least four independent experiments are shown.
RAB4A and β3 integrin are required for invasion into 3D collagen of breast cancer cells. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of MDA-MB-231 cells transfected with scrambled (Ctr) or anti-RAB4A/B siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 B). (Bottom) Time-lapse wide-field microscopy of doxycycline-inducible RAB4AS22N-expressing and control MCF10.DCIS.com. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. 9 C). Images were taken every 5 min for 30 h. Representative videos of at least four independent experiments are shown.
The RAB5A/RAB4A circuitry promotes 3D matrix invasion in an MT1-MMP– and β3 integrin–dependent manner, and is dysregulated in invasive breast cancer. (A) RAB5-HeLa cells were transfected with the indicated siRNAs or scrambled control (Ctr), and tested for invasion as described in Fig. 4 (C and D). (A, top) Examples of cell migration tracks (see Video 9). (A, bottom) Quantification of cell invasion is the mean forward invasion index ± SEM (error bars; n = 60 cells/experiment/siRNA repeated in four independent assays). *, P < 0.05; **, P < 0.005. Gene silencing was verified by QRT-PCR (bottom right). **, P < 0.001. (B) MDA-MB-231 cells were transfected with the indicated siRNAs or scrambled control (Ctr) and tested for invasion as described in Fig. 4 (C and D). (B, left) Examples of cell migration tracks (see Video 10). Quantification of cell invasion is expressed (right) as the mean forward invasion index ± SEM (error bars; n = 40 cells/experiment/siRNA repeated in three independent invasion assays). *, P < 0.05; **, P < 0.005. Silencing of the different genes was verified by QRT-PCR (right). **, P < 0.001. (C) Control and RAB4AS22N-MCF10.DCIS.com cells were induced with doxycycline and tested for invasion as described in Fig. 4 (C and D). (C, left) Examples of cell migration tracks (Video 10). (C, right) Quantification of cell invasion is the mean forward invasion index ± SEM (error bars; n = 45 cells/experiments in two independent experiments). (D) Doxycycline-inducible RAB4AS22N- and control-MCF10.DCIS.com cells were injected subcutaneously into NSG mice. After 2 d, mice were fed with doxycycline. Histological (H&E) and IHC analyses with anti–α smooth muscle actin (SMA) of control (Ctr) and RAB4AS22-MCF10.DCIS.com xenografts were performed at 1 and 3 wk after doxycycline treatment. Images are from three independent experiments (n = 6 mice/experimental condition). Bars, 100 µm.
The RAB5A/RAB4A circuitry promotes 3D matrix invasion in an MT1-MMP– and β3 integrin–dependent manner, and is dysregulated in invasive breast cancer. (A) RAB5-HeLa cells were transfected with the indicated siRNAs or scrambled control (Ctr), and tested for invasion as described in Fig. 4 (C and D). (A, top) Examples of cell migration tracks (see Video 9). (A, bottom) Quantification of cell invasion is the mean forward invasion index ± SEM (error bars; n = 60 cells/experiment/siRNA repeated in four independent assays). *, P < 0.05; **, P < 0.005. Gene silencing was verified by QRT-PCR (bottom right). **, P < 0.001. (B) MDA-MB-231 cells were transfected with the indicated siRNAs or scrambled control (Ctr) and tested for invasion as described in Fig. 4 (C and D). (B, left) Examples of cell migration tracks (see Video 10). Quantification of cell invasion is expressed (right) as the mean forward invasion index ± SEM (error bars; n = 40 cells/experiment/siRNA repeated in three independent invasion assays). *, P < 0.05; **, P < 0.005. Silencing of the different genes was verified by QRT-PCR (right). **, P < 0.001. (C) Control and RAB4AS22N-MCF10.DCIS.com cells were induced with doxycycline and tested for invasion as described in Fig. 4 (C and D). (C, left) Examples of cell migration tracks (Video 10). (C, right) Quantification of cell invasion is the mean forward invasion index ± SEM (error bars; n = 45 cells/experiments in two independent experiments). (D) Doxycycline-inducible RAB4AS22N- and control-MCF10.DCIS.com cells were injected subcutaneously into NSG mice. After 2 d, mice were fed with doxycycline. Histological (H&E) and IHC analyses with anti–α smooth muscle actin (SMA) of control (Ctr) and RAB4AS22-MCF10.DCIS.com xenografts were performed at 1 and 3 wk after doxycycline treatment. Images are from three independent experiments (n = 6 mice/experimental condition). Bars, 100 µm.
To assess whether interference with RAB4 impacted the invasive behavior of breast cancer cells in vivo, we generated doxycycline-inducible RAB4AS22N-expressing MCF10.DCIS.com cells. First, we verified that RAB4AS22N impaired invasiveness of these cells into collagen type I matrix in vitro (Fig. 9 C and Video 10). Next, we injected control and RAB4AS22N-MCF10.DCIS.com cells into immunodeficient mice and monitored tumor progression to invasive disease over time. After 1 wk, control and RAB4AS22N-tumors were comparable in size and presented the typical DCIS histology (Fig. 9 D). However, while MCF10.DCIS.com control lesions lost the myoepithelial layer and progressed to become invasive tumors after 3 wk, RAB4AS22N-expressing cells maintained the typical DCIS histology and their invasiveness was impaired (Fig. 9 D).
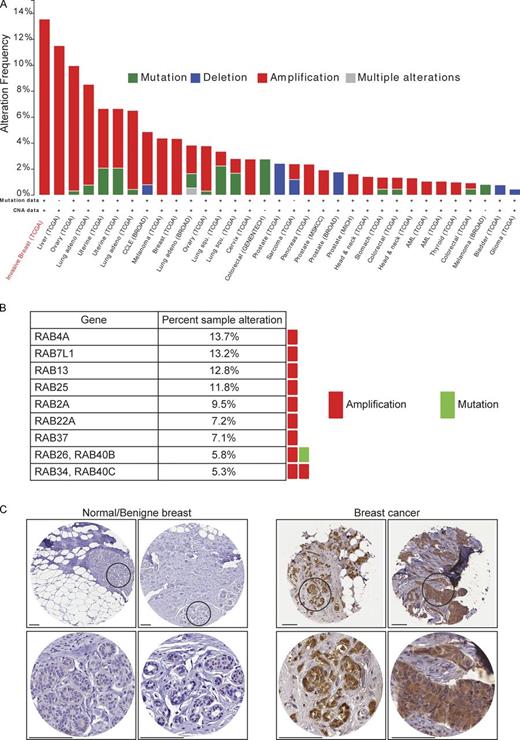
Prompted by this set of findings, we investigated the matter by manually querying the Cancer Genome Atlas (TCGA) database. This analysis revealed that RAB4A is amplified in various tumors (Fig. 10 A). In invasive breast carcinoma, RAB4A is the most frequently amplified RAB GTPase (13.7%; Fig. 10 B). Consistently, analysis of RAB4A expression by IHC in a general tumor tissue microarray confirmed that RAB4A levels are significantly more elevated in various tumors, and most notably in breast cancers, as compared with their normal epithelial tissue (Fig. 10 C and Table S2). Although an analysis in a larger cohort of primary and metastasis samples would be necessary to establish the relevance of RAB4A elevation on breast cancer progression and whether it may act as a driver of tumorigenesis, our results suggest that this event is selected in human tumors and may contribute to their invasive phenotype.
RAB4A is amplified in invasive breast cancer. (A) RAB4A is amplified in various tumors. The cBio Cancer Genomics Portal (http://www.cbioportal.org/public-portal/) was queried for RAB4A across various tumor datasets. The alteration frequency, type of alterations of RAB4, availability of mutation analysis, and Copy Number Alteration (CNA) in the various tumor cohorts are shown. (B) RAB4A is, among the RAB family members, the most frequently amplified gene in invasive breast carcinoma. The percentage of each RAB gene amplification in invasive breast carcinoma reported in the Cancer Genome Atlas (TCGA) is shown. Only the genes with a frequency of amplification >5% are shown. (C) RAB4A is overexpressed in breast cancer. Shown are examples of the data of Table S2. Circled areas are enlarged below. Bars, 100 µm.
RAB4A is amplified in invasive breast cancer. (A) RAB4A is amplified in various tumors. The cBio Cancer Genomics Portal (http://www.cbioportal.org/public-portal/) was queried for RAB4A across various tumor datasets. The alteration frequency, type of alterations of RAB4, availability of mutation analysis, and Copy Number Alteration (CNA) in the various tumor cohorts are shown. (B) RAB4A is, among the RAB family members, the most frequently amplified gene in invasive breast carcinoma. The percentage of each RAB gene amplification in invasive breast carcinoma reported in the Cancer Genome Atlas (TCGA) is shown. Only the genes with a frequency of amplification >5% are shown. (C) RAB4A is overexpressed in breast cancer. Shown are examples of the data of Table S2. Circled areas are enlarged below. Bars, 100 µm.
Discussion
This study demonstrates that RAB5A/RAB4-dependent EECs control the execution of metalloprotease-mediated invasive programs. RAB5A, acting through the RAB4 fast recycling pathway, is necessary and sufficient to redirect adhesive (β3 integrin) and proteolytic molecules (MT1-MMP) to invadosomes, thus allowing their maturation into fully competent ECM degrading structures. Previously, we demonstrated that RAB5-dependent EECs are also indispensable for the spatial restriction of signals leading to the formation of PM protrusions, and the acquisition of mesenchymal motility (Palamidessi et al., 2008). Thus, RAB5 orchestrates the execution of the key events in the mesenchymal program of invasion: invadosome formation and maturation, actin remodeling, and mesenchymal cell motility. The inducibility of this program by growth factors, such as HFG or EGF, argues in favor of its physiological significance. However, the most relevant impact of this pathway appears to be on the acquisition of an invasive phenotype by metastatic cancer cells, as demonstrated by our analysis in invasive-metastatic models, and supported by findings in naturally occurring tumors. We propose, therefore, that RAB5 is a master regulator of tumor invasion programs.
RAB5A controls recycling of MT1-MMP to the PM and its activation
Invadosomes can be induced as a result of mechanosensing or upon stimulation with motogenic soluble factors. In the former case, invadosome formation can be considered as a “default response” that is triggered when tumor cells adhere to the ECM and sense matrix rigidity (Alexander et al., 2008). MT1-MMP is the key effector in this circuitry, as matrix rigidity is sufficient to induce its mobilization from intracellular compartments toward the PM (Alexander et al., 2008). This event has been shown to require RAB8-dependent polarized exocytosis (Bravo-Cordero et al., 2007), though it is less clear whether the originating compartment is represented by the classic biosynthetic/secretory pathway or by the recycling endosomal station (Poincloux et al., 2009). Late endosomal recycling (Steffen et al., 2008; Monteiro et al., 2013) of MT1-MMP as well as exosome secretion (Hoshino et al., 2013) can also contribute to the local delivery of this protease (Steffen et al., 2008; Williams and Coppolino, 2011; Monteiro et al., 2013). Similarly, we found that interference with late endosomal routes by silencing RAB7 reduced, but did not abrogate, RAB5-dependent invasion into dense collagen matrix (unpublished data). This implies that a fraction of MT1-MMP is sorted from early to late endosomes before PM delivery and is consistent with the notion that a cargo can use different trafficking routes depending on cell types, extracellular stimuli, and micro environmental conditions (Yamaguchi et al., 2005; DesMarais et al., 2009; Frittoli et al., 2011).
We demonstrated here that HGF is a potent inducer of invadosome formation, and this process is exquisitely dependent on RAB5A/RAB4 EECs. Thus, the default and inducible modes of invadosome formation might represent two sides of the same “signaling” coin: adhesion- and RTK-stimulated, respectively. Based on our data, these pathways appear to converge on RAB5A and RAB5A-controlled delivery of MT1-MMP to spatially defined regions of the PM. This possibility is corroborated by evidence that fast and slow EEC-mediated turnover of MT1-MMP at the PM is critical to sustain matrix degradation (Hoshino et al., 2012) and further reinforce the notion that RAB5A is a pivotal regulator of the invasive program.
RAB5A couples migratory and invasive protrusions
Cell migration is a four-step process involving actin-propelled leading edge protrusions, transient formation of integrin-mediated focal attachment to the substrates, actomyosin-mediated cell contraction for rear-end retraction, and forward sliding of the cell body (Lauffenburger and Horwitz, 1996). This model is sufficient to describe the events necessary for 2D migration. However, in vivo, cells need to deal with the physical constraints of 3D stromal tissues, which are particularly challenging under situations where the stromal ECM is organized as a dense meshwork. Under these conditions, individual tumor cells must activate pericellular proteolytic activity for ECM degradation and remodeling, and integrate this activity within established steps of cell migration.
How this integration is achieved has so far remained elusive. Based on the present data and our previously published work, we can conceive a model for integration, in which RAB5 early endosomes represent the signaling platform where proteolytic and migratory cues converge to then be transmitted to the cell in an interpretable, spatially restricted fashion. Indeed, we provide evidence that RAB5-dependent EECs are critical to the delivery of MT1-MMP and β3 integrin to spatially restricted regions of the PM where invadosomes form. Previously, we demonstrated that RAB5-dependent recycling is required and sufficient for the delivery of active RAC1 to specific regions of the PM, which imparts spatial restrictions on HGF signaling and leads to the polarized generation of actin-based cellular protrusions (Palamidessi et al., 2008). Thus, RAB5-EECs coordinate all the known steps required for the execution of a successful migratory/invasive program.
RAB5A/RAB4-EECs and MT1-MMP in tumor dissemination
Cancer progression is accompanied and promoted by MMP-dependent ECM remodeling and desmoplasia (Sternlicht et al., 1999). Genetic or pharmaceutical inhibition impairment of MMPs, and specifically of MT1-MMPs, reduces breast metastasis (Hotary et al., 2003, 2006; Martin et al., 2008). Nevertheless, clinical trials targeting MMPs have failed, which suggests that the role of MMPs in cancer is more complicated (Coussens et al., 2002). Indeed, there is a dynamic and reciprocal relationship between ECM deposition, processing, degradation, and malignant progression. Cancer is best viewed as a dynamic, phenotypically plastic, and highly coordinated tissue remodeling process that is tightly regulated by biochemical and mechanical cues (Friedl and Alexander, 2011). RAB5 and RAB4 endosomes may serve as intracellular hubs that integrate diverse cell-autonomous responses to both soluble and insoluble microenvironmental cues. This would enable fast adaptation of the migratory strategy and ultimately promote tumor cell dissemination. It is thus not surprising that elevation of RAB5A and/or amplification of RAB4A may be selected events in a subset of breast cancers to increase their metastatic potential.
Materials and methods
Plasmids, antibodies, and reagents
The plasmids pCDNA3–RAB5AS34N-Myc, pCMV–RABA and pCDNA3–RAB5AQ79L-Myc were gifts from M. Zerial (Max Planck Institute, Dresden, Germany). pCFP-RAB5A was gift from A. Sorkin (University of Pittsburgh School of Medicine, Pittsburgh, PA). pCDNA-RAB4AS22N was a gift from P. van der Sluijs, University Medical Center Utrecht, Utrecht, The Netherlands). pEGFP-DYNAMIN2-K44A was a gift from P. De Camilli (Yale School of Medicine, New Haven, CT). pSR2ARF6T44N and pCDNA3.1-cherry MT1-MMP were gifts from P. Chavrier (Institute Curie, Paris, France). pCDNA3-MT1-MMP was a gift from R. Béliveau (Université du Québec à Montréal, Montréal, Québec, Canada). pEGFP-RAB11S22N, pEGFP-RAB4, pEGFP-RABENOSYN5, and pCDNA3-RAB35S22N were gifts from B. Goud (Institute Curie, Paris, France). pEGFP vector was from Takara Bio Inc.
For lentiviral infection, RAB5AWT, RAB5AS34N, and GFP-RAB4AS22N were cloned in the TET ON pSLIK vector.
pEGFP-res WT-RABENOSYN5 containing silent mismatches in the target sequence of RABENOSYN5 oligo 2 (GGTCTATaATGTGCAAaAAGTGTAT), pEGFP-res RAB4-Δ-RABENOSYN5 (W443A), pEGFP-res RAB5-Δ-RABENOSYN5 (I734E), and pEGFP-res RAB4/5-Δ-RABENOSYN5 (W443A and I734E) were provided by Mutagenex Inc.
Antibodies used were: rabbit anti-RAC 1/2/3 and anti-Caspase3 (Cell Signaling Technology); mouse anti-HA1 (BabCo); rabbit anti-RAB5A (S-19), anti–Integrin β3 (H96), anti-GFP (FL), mouse monoclonal anti-ARP3 (4), and goat anti-EEA1 (N-19; all from Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-clathrin (X22; Thermo Fisher Scientific); mouse monoclonal anti-FAK and anti-VASP (BD); anti-Actin (AC-40); anti-LAMP1; anti-SMA (α smooth muscle actin); mouse monoclonal anti–α Tubulin and anti-RAB4 (1C10; both from Sigma-Aldrich); rabbit anti-GFP (Takara Bio Inc.); anti-Cortactin (4F11; EMD Millipore); mouse monoclonal anti–MT1-MMP (LEM-2/15.8; EMD Millipore); mouse monoclonal anti-αv/β3 (LM609) and anti-α5/β1 (P1D6; Immunological Sciences); mouse monoclonal anti-β1 (4B4; Beckman Coulter); rabbit anti-Rabenosyn5 (No. 155247; Abcam); and rabbit anti–human Vimentin (Vector Laboratories). Mouse monoclonal anti-Myc (Hybridoma No. 9E10) was produced in-house in our departmental antibody facility and was raised against a AEEQKLISEEDLLRKRREQLKHKLEQLRNSCA synthetic peptide of Myc. Rabbit polyclonal anti–N-WASP antibody raised against a full-length, Sf9-expressed rat N-WASP was provided by M. Kirschner (Harvard Medical School, Boston, MA).
The following materials were used: recombinant human HGF (R&D Systems); GM6001 (EMD Millipore); rat-tail type I collagen High Concentration (BD); purified bovine atelo-collagen (PureCol; Advanced BioMatrix); Matrigel Basement Membrane Matrix High Concentration (BD); TRITC/FITC-phalloidin (Sigma-Aldrich); Alexa Fluor 594 protein labeling kit, carboxyfluorescein succinimidyl ester (CFSE), and eFluor 670 (Molecular Probes); Avertin and Trypan blue solution (Sigma-Aldrich); and Picrosirius Red (Polysciences, Inc.).
Cell lines
HeLa, HT1080, BT-20, BT-549, SUM-149, HCC-1428, HS-578T, 4T1, U937, and MDA-MB-231 cells were obtained from ATCC and grown according to ATCC specifications. MCF10.DCIS.com were described in Miller et al. (2000).
CDR and invadosome assays
To assess the formation of CDRs and invadosomes, two observers evaluated duplicate slides independently, and at least 200 cells per experimental condition were analyzed. Data are reported as a ruffling or invadosomes index, normalized to an internal control assumed to be 1 (indicated in the figure legends), and are the mean of at least three experiments performed in duplicate. In absolute terms, in RAB5A-expressing HeLa cells, ∼20% and ∼40% of the cells formed CDRs and invadosomes, respectively. In HGF-treated HeLa cells, ∼20% and ∼30% of the cells formed CDRs and invadosomes, respectively. In some experiments, the formation of CDRs and invadosomes was monitored by real-time microscopy. CDRs generally formed (on average 1–3/cell) within 5–8 min of HGF stimulation. Invadosomes generally formed within 15–60 min of HGF stimulation.
In situ zymography
Labeling of gelatin with the Alexa Fluor 594 dye was performed using the Alexa Fluor 594 protein labeling kit (Invitrogen) according to the manufacturer’s instructions. Glass coverslips (20 mm in diameter) were coated with 2 mg/ml Alexa Fluor 594–gelatin and cross-linked with 0.5% glutaraldehyde. Cells were plated onto matrix-coated coverslips and incubated at 37°C to allow degradation, then fixed with 4% paraformaldehyde, washed in PBS, and stained with specific antibodies and FITC-phalloidin.
The extent of matrix degradation was calculated by determining a degradation index, which represents the total relative area of degradation with respect to total area of degradation in control samples normalized for the number of cells.
MT1-MMP recycling assay
HeLa cells transiently transfected with Cherry-MT1-MMP were plated on coverslips, washed twice with PBS, and incubated at 16°C for 2 h in a serum-free, CO2-independent, L-15 medium Leibovitz (Gibco) containing 10 µg/ml of mouse MT1-MMP antibody that recognizes the extracellular portion of the protease. Under these conditions, recycling is blocked, whereas internalization proceeds, albeit at a reduced rate (Punnonen et al., 1998). The cells were washed with ice-cold PBS and incubated for 5 min at 4°C with mild acid wash buffer (150 mM NaCl, 1 mM CaCl2, and 20 mM acetic acid, pH 4.6), followed by successive rinses with PBS to remove surface antibody. The cells were then transferred to 37°C to allow recycling of internalized MT1-MMP. At various time points, cells were fixed with 4% paraformaldehyde without permeabilization, then blocked in 1% BSA for 30 min followed by incubation for 1 h with the anti–mouse Alexa Fluor 488–conjugated antibody. MT1-MMP surface expression was examined using the 63× (NA 1.4) lens of an inverted microscope (Leica) with constant gain and pinhole parameters. Relative cell surface expression was quantified for at least 30 cells per experimental condition using ImageJ software after having defined manually the cell contour on confocal z sections. The total anti–MT1-MMP ab signal is the sum of cytoplasmic and cell surface signal. Recycled MT1-MMP is calculated as the fraction on the cell surface/total MT1-MMP at each time point.
In situ proximity ligation assay (PLA)
We detected the association in situ between MT1-MMP/Integrin β3 with a Duolink II detection kit (Olink Bioscience), according to the manufacturer’s instructions. Primary antibodies against overexpressed MT1-MMP and endogenous Integrin β3 were incubated in the presence of blocking solution (PBS, 0.05 Tween 20, 1% BSA). This was followed by incubation with secondary antibodies conjugated to oligonucleotides that are ligated to form a closed circle in the presence of Duolink Ligation Solution (Söderberg et al., 2006). In the final step, we added DNA polymerase in order to amplify ligated oligos, which were detected using complementary, fluorescently labeled oligonucleotides.
In vitro 3D assays
3D cell migration.
For 3D collagen gel invasion assays we used type I collagen diluted in 1× DMEM, Hepes (50 mM), NaOH (5 mM), and NaHCO3 (0.12%), at a final concentration of 2.3 mg/ml in the presence of 100 ng/ml HGF. We then casted unpolymerized collagen in home-made migration chambers (0.4 mm thick) at 37°C for 1 h. Cells were added on top of collagen gel. Chambers were sealed with dental glue and incubated at 37°C, 5% CO2. Time-lapse recording was performed using an inverted microscope (Eclipse TE2000-E; Nikon).
3D morphological analysis of cells embedded in Matrigel.
HeLa cells were embedded in Matrigel. To this end, 2 × 105 cells/ml were suspended with Matrigel solution (21 mg/ml) to achieve a final concentration of ∼7 mg/ml at 4°C. 300 µl of this cell suspension was seeded in an 8-well chamber slide (Lab-Tek) and allowed to solidify at 37°C for 4 h. Cells were fixed in 4% paraformaldehyde and stained with anti-RAB5 Ab and phalloidin to detect F-actin. Sequential z sections (at least 30) of embedded cells were obtained by indirect immunofluorescence microscopy using a confocal microscope (TCS SP2 AOBS; Leica).
Organotypic matrix invasion assay
U937 cells (2.5 × 104) were embedded in rat-tail type I collagen (final concentration of 4 mg/ml). After 3 d of incubation at 37°C, gels were treated with antibiotics to kill macrophages and rinsed. We used macrophages instead of fibroblasts, since these cells are known to associate with growing tumors, and they can modify the ECM and release significant amounts of motogenic factors, including EGF and HGF (Qian and Pollard, 2010; Mantovani et al., 2011). GFP-LifeAct-HeLa cells (2 × 104), expressing RAB5 or empty vector as control, were plated on the top of the matrix in complete medium. The gel was then mounted on a plastic bridge and fed from underneath with complete medium (changed daily). After 14 d, the cultures were fixed with 4% PFA, paraffin embedded, and processed by standard methods for hematoxylin and eosin (H&E) staining. The invasion index was calculated by measuring the number of H&E-stained control and RAB5A-HeLa cells that invaded a fixed area of the organotypic gel using the gel upper border as the upper limit. At least 10 areas along the organotypic gel were examined in at least two independent experiments. The error bars represent the SEM.
Organotypic outgrowths in 3D Matrigel
For the analysis of invasive outgrowth of MCF10.DCIS.com cells, an overlay basement membrane assay was carried in the presence of HGF, as described previously (Jedeszko et al., 2009). In brief, 6-well plates were coated with 12 mg/ml native Matrigel (BD) and allowed to solidify for 20 min at 37°C. MCF10.DCIS.com cells (3.0 × 105) were seeded as single cells onto the solidified basement membrane. Cultures were grown in M171 Mammary Epithelial Medium (Invitrogen) supplemented with Mammary Epithelial Growth Supplement (Invitrogen) and 2% Matrigel. After 7 d, cells were incubated with doxycycline and 20 ng/ml of HGF. Medium was replenished every 2 d. MCF10.DCIS.com were imaged in triplicate for development of invasive outgrowths by differential interference contrast (DIC) imaging using a 20× objective lens. Invasive outgrowths were defined as consisting of two or more cells migrating away from their structure of origin. A minimum of 20 images were analyzed for each experimental condition.
Quantitative RT-PCR analysis
Total RNA was isolated with the RNeasy kit method (QIAGEN). 2 µg of RNA were used, with 100 ng of random hexamers, in a reverse-transcription reaction (Superscript Vilo; Invitrogen). 0.1 ng cDNA was amplified, in triplicate, in a reaction volume of 25 µl with 10 pMol of each gene-specific primer and the SYBR-green PCR MasterMix (Applied Biosystems). Real-time PCR was performed on the 14 ABI/Prism 7700 Sequence Detector System (PerkinElmer/Applied Biosystems), using a pre-PCR step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Specificity of the amplified products was confirmed by melting curve analysis (Dissociation Curve TM; PerkinElmer/Applied Biosystems) and by 6% PAGE. Preparations with RNA template without reverse transcription were used as negative controls. Samples were amplified with primers for each gene (for details see the Q-PCR primer list below) and ribosomal RNA 18S as a housekeeping gene. The Ct values were normalized to the 18S curve. The 18S gene was used as a control gene for normalization. Results were quantified using the 2−ΔΔCT method (Livak and Schmittgen, 2001). PCR experiments were performed in triplicate, and standard deviations calculated and displayed as error bars. Primer assay IDs were: MT1-MMP, Hs00237119_m1; β1 Integrin, Hs00559595_m1; β3 Integrin, Hs01001469_m1; RAB5A, Hs00991290_m1; RAB5B, Hs00161184_m1; RAB5C, Hs00428044_m1; RAB4A, Hs00190157_m1; RAB4B, Hs00535053_m1; RAB7A, Hs01115139_m1; RAB7B, Hs00332830_m1; RAB8A, Hs00180479_m1; RAB8B, Hs00213008_m1; RABENOSIN-5 (ZFYVE20), Hs00223482_m1; and RABAPTIN-5, Hs01091595_m1.
siRNA experiments
The following siRNAs were used for knocking down specific genes. Similar results were obtained regardless of the siRNA used. All sequences are 5′ to 3′. CLATHRIN, (1) TCCAATTCGAAGACCAATT; CLATHRIN, (2) CCTGCGGTCTGGAGTCAAC; ARF6 (1), AGCTGCACCGCATTATCAAT; ARF6 (2), and CACCGCATTATCAATGACCGTT were from GE Healthcare. RAB5A (1), GCCAGAGGAAGAGGAGTAGACCTTA; RAB5A (2), GGAGAGTCCGCTGTTGGCAAATCAA; RAB5B (1), GCAGATGACAACAGCTTATTGTTCA; RAB5B (2), GCTATGAACGTGAATGATCTCTTCC; RAB5C (1), TCCGCTTTGTCAAGGGACAGTTTCA; RAB5C (2), CAATGAACGTGAACGAAATCTTCAT; Integrin β1 (1), CCTAAGTCAGCAGTAGGAACATTAT; Integrin β1 (2), GGGAGCCACAGACATTTACATTAAA; Integrin β3 (1), CCTCCAGCTCATTGTTGATGCTTAT; Integrin β3 (2), GAGGCCACGTCTACCTTCACCAATA; RABENOSYN-5 (1), CCACTATGTTGTGGAAGTCAATAAA; RABENOSYN-5 (2), GGTCTATTATGTGCAAGAAGTGTAT; RABAPTIN-5 (1), GTGTTCCAATTACGAAAAA; RABAPTIN-5 (2), CCAAAGCTTTAGGCTATAA; N-WASP(1), TCAAATTAGAGAGGGTGCTCAGCTA; N-WASP (2), CCCTCTTCACTTTCCTCGGCAAGAA; MT1-MMP (1), CCTACGAGAGGAAGGATGGCAAATT; MT1-MMP (2), CCGACAAGATTGATGCTGCTCTCTT; RAB7A (1), CAGGAAACGGAGGTGGAGCTGTACA; RAB7A (2), CCCTAGATAGCTGGAGAGATGAGTT; RAB7B (1), TGACATCAATGTGGTGCAAGCGTTT; RAB7B (2), GCCAGCATCCTCTCCAAGATTATCA; RAB8A (1), GCAAGAGAATTAAACTGCAGATATG; RAB8A (2), GCGAAGACCTACGATTACCTGTTCA; RAB8B (1), GAGAAGCTAGCAATTGACTATGGGA; RAB8B (2), CCTGGGTAACAAATGTGATATGAAT; RAB4A (1), GGAGTGGAATTTGGTTCAAAGATAA; RAB4A (2), GAGAAACCTACAATGCGCTTACTAA; RAB4B (1), CCCAGGAGAATGAGCTGATGTTCCT; and RAB4B (2), AGAGGATGGGCTCTGGCATTCAGTA were from Life Technologies.
For all siRNA experiments, the appropriate scrambled oligos were used as control siRNAs (ctr-siRNA).
Orthotopic xenograft in vivo studies
All animal experiments were performed in accordance with national and international laws and policies. Mice were bred and housed under pathogen-free conditions in our animal facilities at Cogentech Consortium at the FIRC Institute of Molecular Oncology Foundation and at the European Institute of Oncology in Milan.
Subcutaneous tumor development in (NSG) NU/NU nude mice Crl:NU-Foxn1nu.
Before injection, control or RAB5A-expressing HeLa cells were trypsin detached, washed twice, and resuspended in PBS at a final concentration of 1 × 106 cells/13 µl. The cell suspension was then mixed with 5 µl of growth factor–reduced Matrigel and 2 µl Trypan blue solution, and maintained on ice until injection. To establish tumors in mice, cells were injected subcutaneously in the mid-dorsal region of female nude mice, 6–9 wk old. Tumors were allowed to grow for 4 wk. Tumor growth was monitored weekly using digital calipers, and tumor volume was calculated according to the formula: L × W2/2 = mm3. The tumors were excised, fixed in 4% phosphate-buffered formalin, and embedded in paraffin. 5-µm sections of the entire tumor samples were prepared and slides were counterstained with H&E and with anti-RAB5 antibody for the detection of metastases. The Scan Scope XT device and the Aperio Digital pathology system software (Aperio; Leica) were used to detect metastases.
Subcutaneous tumor development in NOD SCID gamma mice.
Control and RAB5AS34N-MCF10.DCIS.com (100,000 cells) were injected subcutaneously into 6–9-wk-old female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (commonly known as the NOD SCID gamma; NSG) mice in 50% Matrigel (BD) as described previously (Hu et al., 2008). Tumors were allowed to grow for 4 d before feeding the animals with doxycycline. 1–3 wk later, xenografts were measured and then snap frozen on dry ice and stored at −80°C for protein or mRNA analysis, or formalin fixed and paraffin embedded for IHC staining.
Mammary fat pad tumor development in NSG mice.
Before injection, control HeLa, RAB5A-HeLa, control-MDA-MB-231, and RAB5AS34N-MDA-MB-231 cells were trypsin detached, washed twice, and resuspended in PBS to a final concentration of 5 × 106 cells/13 µl. The cell suspension was then mixed with 5 µl growth factor–reduced Matrigel and 2 µl Trypan blue solution and maintained on ice until injection. Aseptic conditions under a laminar flow hood were used throughout the surgical procedure. Female NSG mice, 6–9 wk old, were anesthetized with 375 mg/Kg Avertin, laid on their backs, and injected with a 20-µl cell suspension directly in the fourth mammary fad pad. In the case of control and RAB5AS34N-MDA-MB-231, mice were fed with doxycycline after 3 wk. Tumor growth was monitored weekly using digital calipers, and tumor volume was calculated according to the formula: L × W2/2 = mm3. After 4–6 wk, mice were anesthetized with 375 mg/Kg Avertin to remove primary tumors (mastectomy). The tumor mass was carefully removed and the incision closed with wound clips. Tumor metastasis was analyzed after an additional 4 wk. To this end, mice were sacrificed, and ipsilaterally disseminated tumors and lung tissue were removed. For histological evaluation, primary tumors and metastases in lung tissues were fixed in 4% phosphate-buffered formalin and embedded in paraffin. 3-µm sections of the entire lungs were made, and slides were counterstained with H&E and with anti-GFP antibody for the detection of metastases. The Scan Scope XT device and the Aperio Digital pathology system software (Aperio) were used to detect metastases.
Short-term lung colonization assay.
5 × 105 control MDA-MB231 cells (labeled with E-Fluor 670) and 5 × 105 siRNA RAB5A/B/C MDA-MB231 cells (labeled with CFSE) were mixed in 200 µl PBS and injected intravenously. Mice were then sacrificed after 2, 24, and 96 h. The lungs were removed and micrometastases were visualized using a fluorescence microscope and counted.
Intravital microscopy.
We induced anesthesia with a high dose of isoflurane 4%, verified nonresponsiveness, and then performed the surgery under a lower dose of anesthetic (2.5%). Imaging was performed with a 2% dose of anesthetic. Anesthesia at this level maintains the mouse in a nonresponsive state, but permits a nonforced respiratory pattern. A trained operator was present during imaging to enable prompt intervention if the animal were to become responsive during this time.
Image acquisition
Confocal microscopy was performed on a Leica TCS SP2 AOBS confocal microscope controlled by Leica Confocal Software and equipped with violet (405 nm laser diode), blue (argon, 488 nm), yellow (561 nm Solid State Laser), and red (633 nm HeNe Laser) excitation laser lines. A 63× oil-immersion objective lens (HCX Plan-Apochromat 63× NA 1.4 Lbd Bl; Leica) was used for analysis. Image acquisition conditions were set to remove channel crosstalk, optimizing spectral detection bands and scanning modalities. ImageJ software was used for data analysis.
Time-lapse imaging of cell migration was performed on an inverted microscope (Eclipse TE2000-E; Nikon) equipped with an incubation chamber (OKOLab) maintained at 37°C in an atmosphere of 5% CO2. Movies were acquired with a Cascade II 512 (Photometrics) charge-coupled device (CCD) camera controlled by MetaMorph Software (Universal Imaging) using a 10× magnification objective lens (Plan Fluor 10×, NA 0.30). Tracking of cells was performed using the “Manual Tracking” and “Chemotaxis and migration tool” plug-in distributed by ImageJ software.
Total internal reflection fluorescence (TIRF) imaging of cells was performed with a TIRF workstation (Biosystem; Olympus) based on MetaMorph Software (Universal Imaging) and equipped with an incubation chamber (OKOLab) maintained at 37°C in an atmosphere of 5% CO2. For excitation of Alexa Fluor 405, Alexa Fluor 488, and Texas red, a 405-nm laser diode, a 491-nm Ar laser, and a 561-nm Kr laser were used, respectively, coupled to an inverted epifluorescence motorized microscope (IX81; Olympus). Cells plated on glass coverslips were viewed through a high-aperture 60× objective lens (UIS2 60× TIRFM Plan-Apochromat N, NA 1.45; Olympus) with an additional 1.6× magnification lens. Images (12-bit depth) were acquired using an Orca-ER Cooled CCD digital camera (Hamamatsu Photonics).
Intravital microscopy was performed with a confocal microscope (Leica; TCS SP5) on an upright microscope (DM6000 CFS) equipped with blue (argon, 488 nm), yellow (561 nm solid state laser), and red (633 nm solid state laser) excitation laser lines with an 20× water immersion objective lens (HCX Apochromat L 20× NA 1; Leica) and controlled by Leica LAS AF software (Leica). We used a two-photon excitation (2PE) technique with a pulsed infrared laser (Chameleon Ultra II; Coherent) at 980 nm. 3D reconstructions of primary tumors were done using Volocity software (PerkinElmer).
Live cell DeltaVision imaging was performed using a DeltaVision Elite imaging system (Applied Precision) controlled by softWoRx Explorer 2.0 (Applied Precision) equipped with a CCD camera (CoolSNAP HQ2; Photometrics) and an inverted microscope (IX71; Olympus) using a 100× oil-immersion objective lens (UPLS Apochromat 100× NA 1.4; Olympus). The system was equipped with an environmental chamber (Applied Precision) maintained at 37°C in an atmosphere of 5% CO2. Images were acquired for 1 min every 3 s as a single z plane.
Image analysis
For time-lapse imaging of cell migration, the tracking of cells was performed using the “Manual tracking” and “Chemotaxis and migration tool” plug-in distributed by ImageJ software. Notably, the invasion index represents the efficiency of invasion of at least 20 cells along the axis parallel to the direction of invasion into the type I collagen gel. The invasion index was calculated by dividing the net distance covered by a cell over the chemotactic invasion axis (Yc) by the total accumulated distance (Dc). Pearson’s coefficient was calculated using the JACoP plug-in with ImageJ software.
Quantification of motility events by intravital microscopy
Each video generated by intravital two-photon microscopy was analyzed for the following criteria, as described in Wyckoff et al. (2000): (1) cell protrusions (both blebbing and pseudopodia extension were included among cell protrusions, although blebbing was a rare event under the conditions tested); and (2) cell locomotion (defined as a net cell displacement of a cell centroid over time). We monitored the appearance of the above migration features in a field of view of defined size (2.4 × 105 µm2) during 1 h of time lapse. The percentage of time-lapse sequences during which we recorded any of the events specified above was reported. Typically, we monitored at least three distinct fields of view per tumor. Between 10 and 15 tumors/condition were analyzed. Data are expressed as mean ± SEM.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) human or mouse tumor tissue sections were stained with anti-RAB5A or ant-RAB4A antibody by IHC. Samples were rehydrated through xylene and graded alcohols. Antigen retrieval was accomplished using 1 mM EDTA, 0.05% Tween. Samples were incubated with 3% H2O2 for 5 min and then blocked with 2% goat serum in PBS for 1 h. Samples were incubated with primary antibodies for 2 h at RT in 2% goat serum. HRP-conjugated secondary antibodies were used. Samples were developed with DAB and were counterstained with hematoxylin. The IHC stains were scored on a semiquantitative scale (the best that one could do in this type of assays) on a four-level scale: 0 = negative, 1 = weakly positive, 2 = moderate positive, and 3 = strongly positive. For the analysis, samples were considered as Rab5-low if the score was ≤1, and Rab5-high if the score was >1. Normal breast tissue scores always range between 0 and 1. For the matched analysis the ratio of RAB5A expression in node metastasis versus primary tumor was calculated and labeled as not changed if the ratio = 1, up-regulated (in lymph node metastases) if the ratio was >0, and down-regulated (in lymph node metastases) if the ratio was <0.
Human tumor samples used for the analysis of RAB5A expression.
3-µm-thick sections were obtained from paraffin blocks of primary tumors and matched nodal metastases from 35 patients operated at the European Institute of Oncology (IEO) and retrieved from the archival material of the pathology division. The project was approved by the ethical committee of the IEO.
Microarray analysis
Affymetrix gene expression data of five independent cohorts of breast cancer patients were downloaded from Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) using the following accession nos.: GSE4922, GSE6532, GSE2034, GSE7390, and GSE1456. Raw data (CEL files) were normalized using Robust Multi-array Average (RMA) in the R software environment (Irizarry et al., 2003). Follow-up of recurrence (local, regional, or distant) was truncated at 10 yr to standardize the different cohorts of patients analyzed. Patients without evidence of relapse and with a follow up <5 yr were not considered in our analyses. In the original GSE4922 dataset, authors used data on any type of recurrence (local, regional, or distant) or death from breast cancer to determine “disease free survival (DFS) events.” Therefore, it was not possible to distinguish the two parameters in this dataset. To identify RAB5A-overexpressing tumors, we analyzed the log2 normalized gene expression level distribution of RAB5A (probe set ID: 209889_at; HG-U133A chip) across all tumor samples in each independent dataset. We defined RAB5A “high-expressing” tumors as those having RAB5A expression levels greater than the 75th percentile of the expression distribution. Tumors with RAB5A expression levels lower than or equal to the 75th percentile were considered to be RAB5A “low-expressing” tumors.
Online supplemental material
Fig. S1 shows the effects of RAB5A expression in human breast cancer. Fig. S2 shows that RAB5A is necessary and sufficient to induce invasion and invadosomes formation. Fig. S3 shows that RAB5A promotes N-WASP–dependent invadosome formation. Fig. S4 demonstrate that the RAB4–RABENOSYN-5 recycling pathway is necessary for HGF-induced invadosome formation. Fig. S5 shows that MT1-MMP colocalizes with RAB5A and RAB4A endosomes. Table 1 shows membrane trafficking molecules involved in the formation of invasive protrusions. Table 2 shows the number of normal and tumor samples analyzed and the number and percentage of RAB4A high-expressing samples for each screened tissue. Videos 1–3 are intravital real-time analyses of intratumoral cell motility and protrusion extension. Video 4 is a high-magnification time lapse of control and RAB5A-HeLa cells moving into 3D collagen. Video 5 shows time-lapse microscopy of invading breast tumor cells upon deregulation of RAB5A. Video 6 is a TIRF time lapse of RFP-Lifeact and Fluorescein-MT1-MMP in the presence or absence of RAB5A in HeLa. Video 7 shows time-lapse microscopy of RFP-LifeAct and either GFP-RAB5A or GFP-RAB4A in HGF-stimulated HeLa. Video 8 is a TIRF time lapse of RFP-LifeAct and GFP–β3 integrin in the presence or absence of RAB5A in HeLa cells. Video 9 shows that time-lapse microscopy of invading RAB5A-HeLa cells treated with a variety of siRNAs. Video 10 shows time-lapse microscopy of invading breast cancer cells with reduced RAB4A activity or levels. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201403127.dv.
RAB5A is required for invasion into 3D collagen of various cancer cells. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of control and doxycycline-inducible RAB5AS34N-MDA-MB-231 cells tested for invasion into native type I collagen gel chamber slides (see also Fig. 4 C). (Middle) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of control and doxycycline-inducible RAB5AS34N-MCF10.DCIS.com cells tested for invasion into native type I collagen gel chamber slides (see also Fig. 4 D). (Bottom) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of MDA-MB-231 cells transfected with scrambled (Ctr) or anti-RAB5A/B/C siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. S2 C). Images were taken every 5 min for 48 h. Representative videos of at least four independent experiments are shown.
RAB5A is required for invasion into 3D collagen of various cancer cells. (Top) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of control and doxycycline-inducible RAB5AS34N-MDA-MB-231 cells tested for invasion into native type I collagen gel chamber slides (see also Fig. 4 C). (Middle) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of control and doxycycline-inducible RAB5AS34N-MCF10.DCIS.com cells tested for invasion into native type I collagen gel chamber slides (see also Fig. 4 D). (Bottom) Time-lapse wide-field microscopy (Eclipse TE2000-E; Nikon) of MDA-MB-231 cells transfected with scrambled (Ctr) or anti-RAB5A/B/C siRNAs. Cells were placed on one side of a chamber slide in which 2.3 mg/ml acid extracted–only polymerized type I collagen gel containing 100 ng/ml HGF was added (see also Fig. S2 C). Images were taken every 5 min for 48 h. Representative videos of at least four independent experiments are shown.
Acknowledgments
We are indebted to Pascale Romano and Rosalind Gunby for editing the manuscript. We thank Chiara Luise and Federica Pisati for technical assistance with IHC analysis, and Elisa Dama and Fabio Dezi for statistical and bioinformatics support.
Work in the authors’ laboratory is supported by grants from the Associazione Italiana per la Ricerca sul Cancro (to P.P. Di Fiore, IG-14404; and G. Scita, IG 14104), the European Research Council (to P.P. Di Fiore, #233033; and G. Scita, #268836), the Italian Ministries of Education University Research (MIUR) and of Health (to P.P. Di Fiore and G. Scita), the Association for International Cancer Research (to G. Scita, #14-0335), the Ferrari Foundation (to P.P. Di Fiore), and the CARIPLO Foundation (P.P. Di Fiore and G. Scita). C. Malinverno and S. Cortellino are supported by a fellowship form Fondazione Umberto Veronesi (FUV).
The authors declare no competing financial interests.
References
- CFSE
carboxyfluorescein succinimidyl ester
- DCIS
ductal carcinoma in situ
- EEC
endocytic/exocytic cycle
- ER
estrogen receptor
- FFPE
Formalin-fixed, paraffin-embedded
- H&E
hematoxylin and eosin
- HGF
hepatocyte growth factor
- IHC
immunohistochemical
- MT1-MMP
membrane-type 1 matrix metalloprotease
- PLA
proximity ligation assay
- SHG
second harmonic generation
Author notes
E. Frittoli and A. Palamidessi contributed equally to this paper.
P.P. Di Fiore and G. Scita contributed equally to this paper.



![Figure 2. RAB5A is required for tumor dissemination and for the conversion from DCIS to infiltrating mammary carcinoma. (A and B) Doxycycline-inducible RAB5AS34N- and control MDA-MB-231 cells were injected into the mammary fat pads of NSG mice. After 3 wk, mice were fed with doxycycline. Metastases were analyzed 4 wk after removal of primary tumor. (A) Ipsilateral metastasis (arrowhead) of control or RAB5AS34N-MDA-MB-231 tumors. (A, right) Quantification (mean ± SEM [error bars]; n = 10 mice/group) of the number and size of disseminated tumors. (B) H&E of control and RAB5AS34N-MDA-MB-231 lung tissue sections (left). The size of metastatic nodules in lungs is the mean tumor area/lung area ± SEM (error bars; n = 9 mice/group repeated in three independent experiments). (C) Mixtures of CFSE-labeled (green) scramble-transfected control (Ctr) and eFluor 670–labeled (red) RAB5A,B,C-depleted MDA-MB-231 cells (0.5 × 106 each) were coinjected into the tail vein of NSG mice. (C, left) Images of lung tissue. (C, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A,B,C-depleted cells/field of lung tissue at the indicated times. **, P < 0.005. (C, right) Efficacy of RAB5 gene silencing by QRT-PCR. **, P < 0.005. (D) Doxycycline-inducible RAB5AS34N- and control-MCF10.DCIS.com cells were injected subcutaneously into NSG mice. After 4 d, mice were fed with doxycycline. (D, left) IHC analyses of control (Ctr) and RAB5AS34N-MCF10.DCIS.com xenografts performed at 1 and 3 wk after doxycycline treatment. Representative images from three independent experiments are shown (n = 5 mice/experimental condition). (D, right) mRNA level of RAB5AS34N expression in control (Ctr) and RAB5AS34N tumors. Error bars indicate SEM. **, P < 0.005. (E) Control (Ctr) and RAB5A-MCF10.DCIS.com cells (5 × 105 each) were labeled with eFluor 670 and injected into NSG mice tail vein. (E, left) Images of lung tissue. (E, middle) The number (mean ± SEM [error bars]; n = 10) of labeled control and RAB5A-MCF10.DCIS.com cells per field at the indicated times. **, P < 0.005. (E, right) RAB5A expression by immunoblotting. Bars: (B) 2 µm; (C) 80 µm; (D) 100 µm; (E) 80 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/206/2/10.1083_jcb.201403127/6/m_jcb_201403127_fig2.jpeg?Expires=1773759890&Signature=cOxR7p0mq2XYdhPPF1rSzlLge1yoUCwVeTbzwaw50WvRxfKoju7IZXione9JK7MhKto0gqFWtPlTpzUfq6oIa9TzJXI~6cB6xz1I3OBX-WgLBv5Q5LUV8cHBQKHjgCy12fk8XMtcTA6v4ghRoE4Ux5sJi0tIjmdy5Zy0FbXEzH85kCSqrc2bVDOUgwYtQxspzHmhz5mx2D31kfH~JLlJ3oebU0T50qSwmr0cXXc5cNhn7lk5DPJQ6KZ5DyOdBBbZXK9ZU3tpnyqGN0zl-bmoHoFHHC5R4ouuBZpFhjUEqy65ZAWtaevavbg9N7FgIM87uQQMTZau-1ULj5IK-ERS~w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. RAB5A is sufficient to promote intratumoral cell motility and distant dissemination. (A) GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A) HeLa cells were injected into the mammary fat pads of NSG mice. Metastases were analyzed 4 wk after removal of primary tumor. (A, left) Ipsilateral metastasis (arrowheads) of control or RAB5A-HeLa tumors. (A, right) Quantitation of number and volume (mean ± SEM [error bars]; n = 10 mice/group) of disseminated tumors nodules. **, P < 0.005. (B, left) H&E and anti-GFP staining of FFPE lung tissue sections. Magnified boxed regions show metastasis. (B, right) The size of metastatic nodules is the mean tumor area/total lung area ± SEM (error bars; n = 9 mice/group repeated in two independent experiments). **, P < 0.005. (C) Tumors from GFP-LifeAct control (Ctr) or GFP-LifeAct-RAB5A-(RAB5A)-HeLa cells injected into the mammary fat pads of NSG mice were analyzed by two-photon microscopy. Green, GFP-LifeAct; gray, collagen structure (SHG). (D) Tumor invasive front visualized: in the top panels by projecting ∼40 serial z sections (green, GFP-LifeAct; gray, collagen structure [SHG]; or, in bottom panels, by IHC. (E) Intratumoral motion analysis of control and RAB5A-HeLa cells was obtained by overlaying 10 differentially colored, consecutive frames of time-lapse recording (Videos 1 and 2; left). Coloring indicates motile cells. The percentage of motility events/field of view/tumor is the mean ± SEM (error bars; n = 45). Bars: (B, left) 2 mm; (B, right) 0.2 mm; (C) 400 µm; (D, top) 200 µm; (D, bottom) 10 µm; (E) 80 µm.](https://cdn.rupress.org/rup/content_public/journal/jcb/206/2/10.1083_jcb.201403127/6/m_jcb_201403127_fig3.jpeg?Expires=1773759890&Signature=hgU2twJbU~yIjW-FRMGP1MkhJGSnbNLMP8C9bDlEG8WJ22CNh31cAmE9h~~Jmel~VLe3AjUJr-Vcrk2pn0820hY1UPcdnKeyUzUcu4qs6GUoIzePtPP6TT1-gZU36ohmMDcVbiUObYbMQgFIqlU5IpNsZqyq5ByQdcYt03oJjnGwU1kpyG~Fu1ym-rvHWqfXE6vrxhy75ZVD9eX1lSaV7TFZ3GplfBxflRw7w4JxAzKTgkIIwTeK5MORbE89ZWdqtGPbmHOQUY09ER~jWUB~9E1pZDU~iZNnTTV8rxn4q4rGyDCzjiohk83pMSvxhNC93sA4xLH8aqQzp4EowP5zqw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)